Abstract
The regulation of dendritic branching is critical for sensory reception, cell−cell communication within the nervous system, learning, memory, and behavior. Defects in dendrite morphology are associated with several neurologic disorders; thus, an understanding of the molecular mechanisms that govern dendrite morphogenesis is important. Recent investigations of dendrite morphogenesis have highlighted the importance of gene regulation at the posttranscriptional level. Because RNA-binding proteins mediate many posttranscriptional mechanisms, we decided to investigate the extent to which conserved RNA-binding proteins contribute to dendrite morphogenesis across phyla. Here we identify a core set of RNA-binding proteins that are important for dendrite morphogenesis in the PVD multidendritic sensory neuron in Caenorhabditis elegans. Homologs of each of these genes were previously identified as important in the Drosophila melanogaster dendritic arborization sensory neurons. Our results suggest that RNA processing, mRNA localization, mRNA stability, and translational control are all important mechanisms that contribute to dendrite morphogenesis, and we present a conserved set of RNA-binding proteins that regulate these processes in diverse animal species. Furthermore, homologs of these genes are expressed in the human brain, suggesting that these RNA-binding proteins are candidate regulators of dendrite development in humans.
Keywords: Caenorhabditis elegans, dendrite morphogenesis, RNA-binding proteins, posttranscriptional regulation, PVD neurons
Dendrites are neuronal structures that receive sensory and synaptic information and are often elaborately branched to cast a wide receptive field or receive synaptic input from many other cells. Dendritic morphogenesis is a critical step in nervous system development, learning, and memory such that dendritic defects are associated with myriad neurologic disorders such as autism, Alzheimer disease, and schizophrenia (Jan and Jan 2010; Kulkarni and Firestein 2012). Therefore, it is important to understand the molecular genetic mechanisms that underlie dendrite morphogenesis. Although several insights into the molecular controls of dendrite morphogenesis have come from studies that have focused on transcriptional control (Parrish et al. 2006; Ou et al. 2008; Jan and Jan 2010; Iyer et al. 2013), there is increasing evidence that posttranscriptional mechanisms such as mRNA localization and localized translational control are important as well (reviewed by Holt and Schuman 2013). For example, the translational repressor Nanos (Nos) regulates dendrite morphogenesis and branching complexity of Drosophila class IV da neurons. Importantly, dendrite morphogenesis depends on proper localization of nos mRNA to dendrites as well as translational regulation of nos mRNA, both of which are mediated by cis-elements in the nos 3′ untranslated region (UTR; Brechbiel and Gavis 2008). Therefore, elucidating posttranscriptional mechanisms that regulate dendrite morphogenesis is an important research goal.
RNA-binding proteins (RBPs) are important posttranscriptional regulators of gene expression that are involved in mRNA splicing, transport, localization, stability, and translational control. Animal genomes encode a diverse suite of several hundred RBPs (Gamberi et al. 2006; Lee and Schedl 2006; Hogan et al. 2008; Kerner et al. 2011). Although there are some examples of mutations in RBP-encoding genes that are associated with neurologic disorders (Zhou et al. 2014), it is not clear how many RBPs regulate dendrite morphogenesis. Thus far, only one study has reported a systematic screen of a majority of RBP-encoding genes for function in dendrite morphogenesis. Olesnicky et al. (2014) reported that 63 RBP-encoding genes in the Drosophila genome are important for dendrite development in the larval dendritic arborization (da) neurons. However, it is not clear the extent to which RBPs involved in dendrite development are conserved across animal species. To gain insight into this question, we identified C. elegans homologs of the RBPs identified in the Olesnicky et al. (2014) study and tested each for a role in dendrite development in the worm using the multidendritic PVD neuron as a model.
The C. elegans PVD neuron is an excellent model for the molecular genetic investigation of dendrite morphogenesis. The bilateral PVDs, which are located between the epidermis and the body wall muscles, have extensively branched dendritic trees that function as mechanoreceptors, nociceptors, proprioceptors, and cold temperature receptors (Way and Chalfie 1989; Halevi et al. 2002; Tsalik and Hobert 2003; Oren-Suissa et al. 2010; Smith et al. 2010; Albeg et al. 2011; Chatzigeorgiou and Schafer 2011). PVD dendritic trees are stereotypic with primary (1°) branches that project anteriorly and posteriorly from the cell body and menorah- or candelabra-shaped structures extending from the primary branches, which include an orthogonal series of secondary (2°), tertiary (3°), and quaternary (4°) branches (Oren-Suissa et al. 2010; Smith et al. 2010; Figure 1A). Thus, PVD function and morphology are similar to Drosophila da neurons and mammalian polymodal nociceptors (Albeg et al. 2011).
Figure 1.
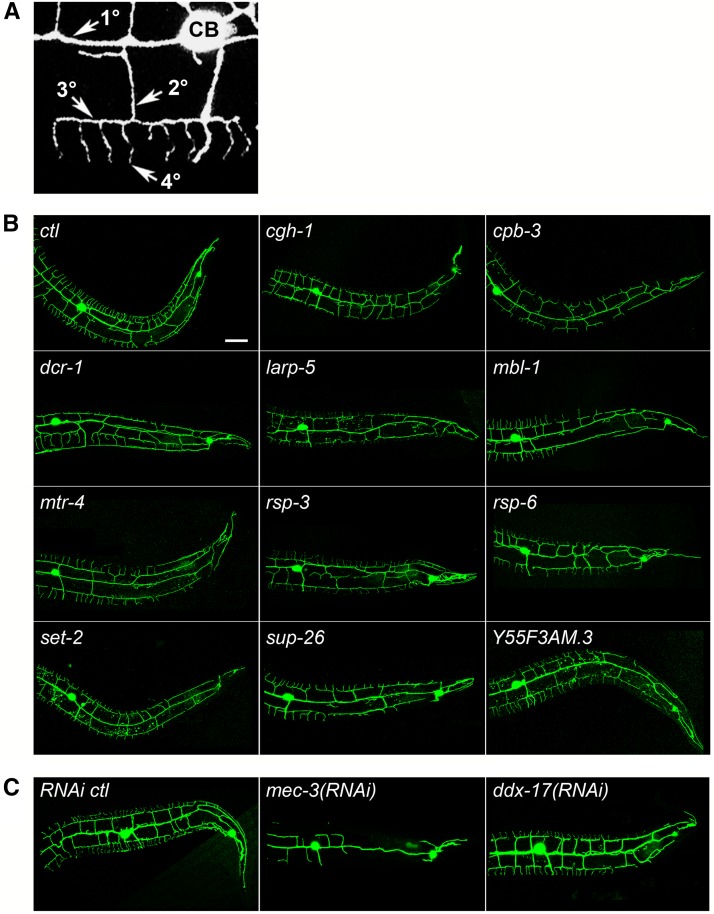
Loss or reduction of RNA-binding protein (RBP) genes results in a decrease in dendritic termini in PVD neurons. (A) PVD dendritic tree morphology includes primary (1°) branches extending from the cell body (CB) and a series of perpendicular secondary (2°), tertiary (3°), and quaternary (4°) branches. (B) Animals carrying a green fluorescent protein (GFP) marker for PVD neurons and a mutation in the RBP gene indicated have reduced dendritic termini compared with the control (ctl). (C) Animals carrying a GFP marker for PVD neurons and treated with RNA interference (RNAi) for the genes indicated have reduced dendritic termini compared with the control. mec-3(RNAi) is a positive control for RNAi and reduced dendrite phenotypes. Posterior is to the right in all images. Bar = 25 μm.
Although little is known about PVD dendrite morphogenesis, we do know that a tripartite ligand-receptor complex, which includes epidermal ligands MNR-1 and SAX-7 and the PVD receptor DMA-1, promotes PVD dendrite branching (Liu and Shen 2012; Salzberg et al. 2013; Dong et al. 2013). In addition, nascent secondary branches are stabilized by HPO-30, a PVD-expressed claudin-like transmembrane protein that likely anchors dendrites to the epidermis (Smith et al. 2013). We also know that the UNC-6/Netrin guidance molecule, the UNC-40/DCC receptor, and the UNC-5/Netrin receptor mediate dendrite self-avoidance (Smith et al. 2012). However, it is still unclear how PVD shape changes are mediated downstream of DMA-1 or Netrin signaling. Mechanical changes to dendrite morphology are, at a most basic level, the result of cytoskeleton dynamics. Interestingly, an RNA interference (RNAi) screen for PVD dendrite defects identified several genes that encode cytoskeleton-associated proteins, such as UNC-116/Kinesin-1 heavy chain, DLI-1/Dynein light intermediate chain, and BICD-1/Bicaudal-D (Aguirre-Chen et al. 2011). An open question is whether the DMA-1, HPO-30, Netrin, or other signaling pathways affect posttranscriptional gene regulation of genes involved in cytoskeleton dynamics, or other genes, to mediate PVD dendrite morphogenesis. Because RBPs are important posttranscriptional regulators, an investigation of the role of RBPs in dendrite morphogenesis may provide insights to this question. Thus far, no RBPs have been implicated in dendrite development in PVDs or other neurons in C. elegans, although a PVD-specific expression profile suggests that 47 RBP-encoding genes are enriched in expression in the PVD over other cells (Smith et al. 2010).
Here we identify the C. elegans homologs of Drosophila RBP genes described as important for da neuron dendrite morphogenesis (Olesnicky et al. 2014) and test each of them for a role in dendrite morphogenesis in PVDs. Using a combination of genetic mutations and RNAi assays, we show that reduction or elimination of function of 12 of the candidate RBP-encoding genes reveals a reduction in the number of dendritic termini in PVD neurons. Some of these genes regulate the number of secondary and tertiary dendrite branches as well. We show that each of these genes is expressed within the PVD neuron, and we use time-course analyses to show that although most of these RBPs affect terminal dendrite branch formation, two of the RBPs are required for dendrite maintenance, and one RBP is required for the timing of PVD dendrite morphogenesis. We examine the subcellular localization of each RBP within PVD neurons and discuss potential molecular roles of each RBP in this context and in the context of previous research. Because each of these RBPs functions in dendrite development in fly and worm and has at least one strong mammalian homolog, it suggests that these RBPs may be important in the development of dendrites in mammals as well. This claim is bolstered by expression data in humans showing that most of the human homologs of RBPs identified in this screen are expressed in the human brain.
Materials and Methods
C. elegans strains
Strains were derived from the Bristol strain N2, grown at 20°, and constructed using standard procedures (Brenner 1974). Mutants screened for dendrite defects are listed in Supporting Information, Table S1. Mutant strains obtained from the Mitani Lab through the C. elegans National Bioresource Project of Japan, the C. elegans Reverse Genetics Core Facility at the University of British Columbia, the C. elegans Reverse Genetics Core Facility at the Oklahoma Medical Research Foundation, and the Million Mutation Project (Thompson et al. 2013) were outcrossed at least four times. PVD dendrites were marked by wdIs52[PF49H12.4::GFP] or wdIs51[PF49H12.4::GFP] (Watson et al. 2008) for screening, rwIs1[Pmec-7::RFP] (Smith et al. 2010) for gene expression studies, and wyIs587[ser-2prom3::myr-mCherry] (Liu and Shen 2012; Dong et al. 2013) for subcellular localization and rescue studies. The sup-26 expression pattern was determined using smIs259[Psup-26::sup-26::GFP] (Mapes et al. 2010). All other transgenes used in this study were constructed as described herein. RNAi was conducted with strain wdIs52; sid-1(pk3321); uIs69[Pmyo-2::mCherry + Punc-119::sid-1] (Calixto et al. 2010).
Imaging and quantification of PVD dendrite morphology
Worms were picked at the life stages indicated, mounted on slides with 2% agarose pads, and immobilized with 600 μM levamisole. Initial screening, time-course studies, and rescue experiments were conducted using a 40× or 63× objective on a Zeiss Axioskop or Leica DM5000B epifluorescence microscope. Dendrites, gene expression patterns, and subcellular localization were imaged with a Leica SP5 spectral confocal microscope at 63× with 0.5 μm per step and Leica LAS software. Secondary, tertiary, and terminal (quaternary and senary) dendrites were counted from the PVD cell body to the posterior end separately on the dorsal side, the ventral side, or both. Although numerous researchers contributed to the primary screen, positive hits were independently verified by confocal microscopy by a single researcher who did not participate in the primary screen. Statistical tests were performed and graphs created with Prism 6.0f software (GraphPad Software, Inc.).
Construction of transgenes and DNA microinjection
All primers used in construction of transgenes described here are given in Table S2. For cgh-1, dcr-1, and mtr-4, presumptive promoter regions were amplified with polymerase chain reaction (PCR) and subcloned into the SphI/KpnI sites of the Fire Lab vector pPD117.01, which carries a multiple cloning site upstream of green fluorescent protein (GFP) with a let-858 3′ UTR. For all other genes, presumptive promoter regions, which include at least 1000 bp upstream of the start codon, or the entire upstream intragenic region, or previously published sequences, were amplified with PCR and subcloned into the pDONR221 vector using Gateway BP Clonase II (Invitrogen). Promoters were then subcloned into pDJK237, a promoterless plasmid with a Gateway cassette upstream of GFP with a 3′ UTR from let-858 derived from pPD117.01, using Gateway LR Clonase II Plus (Invitrogen).
cDNAs were gifts of Y. Kohara or Ding Xue and James Mapes (sources given in Table S2), or were amplified from a cDNA library derived from him-5(e1490) created by Trizol/chloroform and first-strand synthesis by Superscript Reverse Transcriptase III (Invitrogen) and oligo dT primers. In all cases in which multiple isoforms exist, the longest isoform was selected. cDNAs were amplified with PCR without stop codons and were cloned in pDONR221 using Gateway BP Clonase II (Invitrogen). The ser-2 promoter 3 fragment (Tsalik and Hobert 2003) was amplified with PCR and cloned into pDONR P4-P1r and the GFP coding sequence with the let-858 3′ UTR from pPD117.01 was amplified with PCR and cloned into pDONR P2r-P3 using Gateway BP Clonase II (Invitrogen). The cDNAs, ser-2 promoter 3, and GFP were all cloned into pDEST R4-R3 using Gateway LR Clonase II Plus (Invitrogen).
DNA microinjection was performed using standard practices (Mello and Fire 1995). For gene expression pattern studies, unc-76(e911); rwIs1 hermaphrodites were injected with 20 ng/μL GFP plasmid and 60 ng/μL unc-76(+) plasmid. For subcellular localization and rescue studies, wyIs587; unc-76(e911) worms or wyIs587; unc-76(e911); RBP mutation-bearing worms were injected with 10-20 ng/μL GFP plasmid and 60 ng/μL unc-76(+) plasmid.
RNAi
RNAi feeding was performed essentially as described (Kamath and Ahringer 2003). wdIs52; sid-1(pk3321); uIs69[Pmyo-2::mCherry + Punc-119::sid-1] L4 hermaphrodites were placed on Petri plates with nematode growth medium seeded with dsRNA-expressing Escherichia coli. The P0 worms were transferred to a new plate after 24 hr and the progeny from that second plate scored at the young adult stage. dsRNA-expressing E. coli were obtained from the Ahringer lab library (Fraser et al. 2000; Kamath et al. 2003) or were constructed by PCR amplification of a region of the target gene and Gateway Clonase−mediated insertion of that PCR amplicon into a double-T7 pPD129.36 plasmid modified with a Gateway cassette (Invitrogen). In cases in which target genes produce multiple isoforms, RNAi strategies targeted mRNA sequences common to all isoforms. See Table S2 for specific information on the RNAi clones used in this study.
Blast searches
Blast searches were performed using the command line NCBI-BLAST package (version 2.2.25; Camacho et al. 2009). C. elegans protein sequences were aligned to custom BLAST databases using the BLASTp algorithm with default parameters. BLAST databases were assembled from annotated Drosophila melanogaster proteins (genome release 6.01, downloaded from flybase.org on 8/4/2014) and the human protein Refseq database (downloaded from NCBI on 8/4/2014) for alignments between C. elegans and Drosophila and human sequences respectively. Alignment Expect (E) values for all comparisons are reported.
Orthology mapping and expression of human orthologs
To identify human orthologs, the C. elegans genes were mapped on to the Database of Orthologous Groups (OrthoDB; Waterhouse et al. 2013) to determine the OrthoDB identifier for each gene. All human genes assigned to the same OrthoDB identifier were taken as orthologs of the C. elegans gene and used for additional analysis. Data from the Tissue-specific Gene Expression and Regulation (TiGER) database (Liu et al. 2008) was used to assay whether the identified human orthologs are expressed in human brain tissue. Raw data files were downloaded from the TiGER website (http://bioinfo.wilmer.jhu.edu/tiger/, accessed on June 27, 2014) and analyzed using a custom Perl script.
Results
A screen for conserved RBPs that function in C. elegans PVD sensory neuron dendrite morphogenesis
Although RBPs have been shown to be important for the regulation of dendrite development (Ye et al. 2004; Vessey et al. 2008; Bestman and Cline 2008; Brechbiel and Gavis 2008; Olesnicky et al. 2012; Olesnicky et al. 2014), it is not clear the extent to which RBPs are conserved in this process across species. Thus far, only one study has aimed to identify all predicted RBP-encoding genes in a genome that contribute to dendrite morphogenesis. Olesnicky et al. (2014) screened the vast majority of the predicted RBP-encoding genes in the Drosophila genome for a role in dendrite morphogenesis of the larval class IV da sensory neurons and found that 63 of these genes were important. To determine whether these conserved RBPs function in dendrite morphogenesis in other species, we tested the homologs of these 63 RBPs for function in dendrite development using the C. elegans multidendritic PVD sensory neuron.
Using BLASTp, we identified 54 C. elegans homologs of the 63 RBPs reported to function in dendrite morphogenesis in Drosophila (Olesnicky et al. 2014). The relative smaller number of candidate RBPs in C. elegans is caused by several cases in which two Drosophila RBPs are homologous to a single C. elegans RBP and one case in which a Drosophila RBP, specifically Oskar, does not have a clear homolog in C. elegans. We next identified mutations and RNAi treatments to eliminate or knock down the function of each candidate RBP gene in C. elegans. To assay each RBP gene for a role in PVD dendrite morphogenesis, young adult mutant or RNAi-treated animals carrying a GFP marker for PVD neurons were imaged and the number of dendritic termini counted on the dorsal and ventral sides from the cell body to the posterior end of the animal (see Materials and Methods). Thirty of the 54 RBP genes were tested using genetic mutations while we used RNAi to test the remaining 24. Twenty-eight of the mutations we used are presumptive null alleles characterized by small deletions or nonsense substitutions, whereas the other two mutations were previously reported to cause a reduction of function. RNAi was used to test genes for which mutations were unavailable or to bypass pleiotropy and possibly reveal dendrite phenotypes in cases where genetic mutations resulted in embryonic or larval lethality. To increase the efficacy of RNAi in neurons, we used a neuron-sensitized knockdown strategy, which expresses the dsRNA transporter gene sid-1 in the neurons of an otherwise sid-1−mutant animal (Calixto et al. 2010; see the section Materials and Methods). mec-3(RNAi) was used as a positive control for RNAi treatments and produced a reduced branching phenotype in PVDs similar to previously published phenotypes (Aguirre-Chen et al. 2011). The complete list of RBPs tested, BLASTp E values, alleles used, and RNAi treatments is given in Table S1.
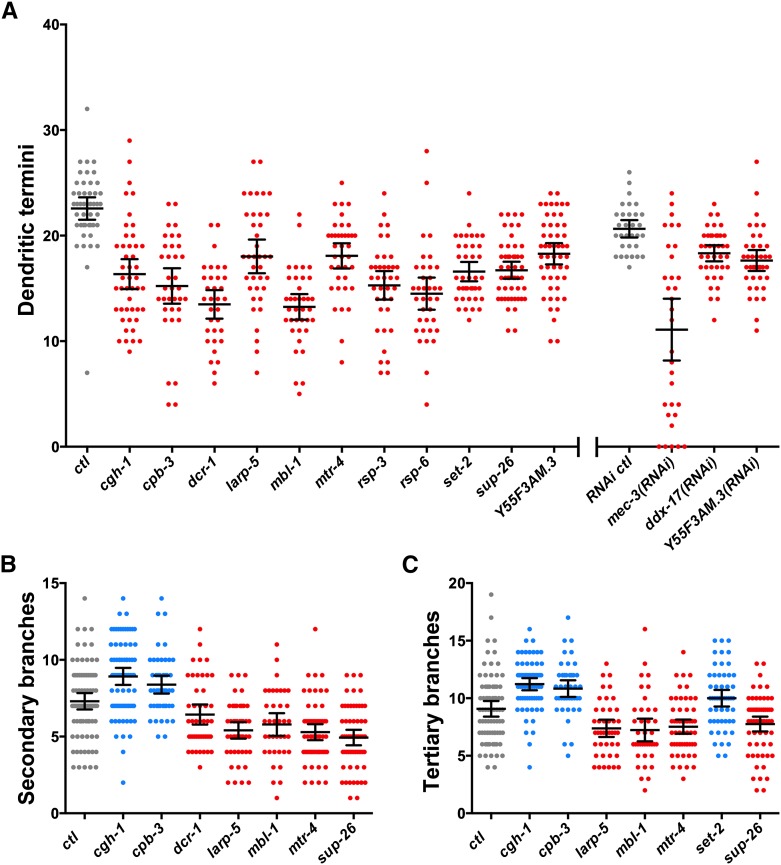
We found that the loss or reduction of function of 12 RBP genes, individually, resulted in a statistically significant reduction in PVD dendritic termini compared with control animals (Table 1, Figure 1, and Figure 2A). Whereas control animals at the young adult stage have an average of 23 dendritic termini in the region scored, mutations in cgh-1, cpb-3, dcr-1, larp-5, mbl-1, mtr-4, rsp-3, rsp-6, set-2, and sup-26 all result in at least a 20% reduction in dendrite termini (Figure 1B and Figure 2A). Although RNAi screening was less effective at identifying RBP genes important for dendrite morphogenesis (see the section Discussion), we did find that ddx-17 and Y55F3AM.3 RNAi-treated animals showed a 14% and 11% respective reduction in the number of dendritic termini compared to untreated control animals (Figure 1C and Figure 2A). After completing the RNAi screen, we tested several point mutations in Y55F3AM.3 generated by the Million Mutation Project (Thompson et al. 2013) to identify a reduction-of-function allele. One allele, gk454899, affects a conserved residue across Drosophila, zebrafish, and mammals and resulted in a 19% reduction of dendritic termini compared to control animals (Table S1, Figure 1B, and Figure 2A).
Table 1. Genes identified as positive hits in a genetic screen for PVD dendrite defects.
| C. elegans RBP | Drosophila RBP | E Value | Predicted Protein |
|---|---|---|---|
| CGH-1 | Gem3 | 6.00E-61 | ATP-dependent DEAD-box RNA helicase |
| CPB-3 | Orb | 7.00E-64 | Cytoplasmic polyadenylation element binding protein |
| DCR-1 | Dcr-1 | 3.00E-120 | RNase III family member; ortholog of Dicer |
| DDX-17 | CG10777 | 5.00E-160 | ATP-dependent DEAD-box RNA helicase |
| LARP-5 | CG11505 | 8.00E-33 | La-related protein with a LARP5 domain |
| MBL-1 | Mbl | 5.00E-45 | CCCH zinc-finger RNA-binding domain regulator of splicing |
| MTR-4 | L(2)35Df | 0 | RNA helicase; homolog of yeast Mtr4p, which is part of the TRAMP complex that is involved in various RNA processing events |
| RSP-3 | SF2 | 4.00E-59 | SR splicing factor required for constitutive splicing and for influencing alternative splicing |
| RSP-6 | X16 | 3.00E-22 | SR splicing factor required for constitutive splicing and for influencing alternative splicing; implicated in transcriptional termination |
| SET-2 | Set1 | 4.00E-68 | RRM domain-containing histone H3K4 methyltransferase |
| SUP-26 | Shep | 6.00E-52 | RRM domain-containing protein; translational repressor |
| Y55F3AM.3 | CG11266 | 1.00E-97 | RRM domain-containing protein with splicing factor RBM39 linker; colocalizes with spliceosomal proteins |
The C. elegans RBPs identified as important for dendrite morphogenesis in PVD neurons are given with an E value from a BLASTp search on D. melanogaster unique protein isoform database. Predicted protein function is given based on Wormbase (WS244). DEAD, Asp-Glu-Ala-Asp; CCCH, motif with three cysteine and one histidine residue; TRAMP, Trf4/Air2/Mtr4p polyadenylation complex; SR, serine/arginine rich; RRM, RNA recognition motif; H3K4, histone 3 lysine 4.
Figure 2.
Quantification of PVD dendrite phenotypes in RNA-binding protein (RBP) mutants and RNA interference (RNAi) knockdowns. Points within each scatter column represent counts of (A) dendritic termini, (B) secondary branches, or (C) tertiary branches from the PVD cell body to the tail on the dorsal or ventral side of the worm. Lines within each column represent the means and the 95% confidence interval of the mean. Results in red and blue are significantly lower or higher respectively than controls (ctl) in gray based on a one-way analysis of variance test with a Fisher's Least Significant Difference multiple comparisons test with a 95% confidence interval.
The Y55F3AM.3 RNAi and mutant studies provide independent verification of the role of this RBP gene in PVD dendrite morphogenesis. To obtain independent verification of the role of other RBP genes in dendrite development, we obtained additional alleles where available. One additional allele of mbl-1 and set-2 and three additional alleles of sup-26 confirmed a reduction of dendritic termini (data not shown). We found that set-2(ok952) animals had a weak reduction in the number of dendrites compared to the n4589 allele (data not shown), which is consistent with a previous report that the ok952 deletion is a hypomorphic allele (Xiao et al. 2011).
We next tested to see whether the loss or reduction of function of these 12 RBP genes also resulted in defects in lower order dendritic branching in PVDs by counting the number of secondary and tertiary branches. Loss or reduction of function of five of the 12 genes, dcr-1, larp-5, mbl-1, mtr-4, and sup-26, resulted in a reduction in the number of secondary branches. Interestingly, loss of function of two of the 12 genes, cgh-1 and cpb-3, resulted in an increase in the number of secondary branches (Figure 2B). Loss of cgh-1, cpb-3, and set-2 resulted in an increase of tertiary branches whereas loss of larp-5, mbl-1, mtr-4, and sup-26 resulted in a reduction of tertiary branches (Figure 2C). Thus, we find that loss of cgh-1 or cpb-3 results in an increase of secondary and tertiary dendrite branches but also causes a roughly 30% reduction of terminal branches (Figure 2). This similarity in the cgh-1 and cpb-3 mutant phenotypes is interesting, given their predicted molecular functions (see the section Discussion). Loss of larp-5, mbl-1, mtr-4, or sup-26 results in a decrease of all orders of dendrite branches (Figure 2). Notably, we did not find any mutations or RNAi treatments that resulted in supernumerary dendritic termini at the young adult stage. The pleiotropic defects in PVD neurons resulting from loss of RBP gene function are summarized in Table 2.
Table 2. Summary of pleiotropic defects in C. elegans PVD architecture resulting from loss of RBP function.
| RBP Gene | Reduction of Terminal Branches | 3° Branch Defect | 2° Branch Defect |
|---|---|---|---|
| cgh-1 | 28% | 24% increase | 21% increase |
| cpb-3 | 33% | 19% increase | 12% increase |
| dcr-1 | 40% | − | 14% decrease |
| ddx-17(RNAi) | 11% | − | − |
| larp-5 | 20% | 19% decrease | 28% decrease |
| mbl-1 | 41% | 20% decrease | 23% decrease |
| mtr-4 | 20% | 17% decrease | 29% decrease |
| rsp-3 | 32% | − | − |
| rsp-6 | 36% | − | − |
| set-2 | 27% | 10% increase | − |
| sup-26 | 26% | 15% decrease | 34% decrease |
| Y55F3AM.3 | 19% | − | − |
RBP, RNA-binding protein.
Finally, we examined all 12 RBP mutants or RNAi treatments for qualitative patterning phenotypes in the PVD. We note that mbl-1 null mutants display a particularly striking and reproducible terminal dendrite branching defect. Although terminal branches near the cell body are similar to controls in terms of length and distribution, mbl-1 mutants have progressively fewer and shorter terminal branches toward the posterior end of the PVD neuron (Figure 1). This finding suggests that mbl-1 is not explicitly required for dendrite branching in general but is specifically important for the patterning of branching posterior to the cell body.
RBPs are required for formation, timing, or maintenance of dendrite branches
A reduction in the number of terminal dendritic branches could be attributable to a failure to form the branches, a delay in branch formation relative to other hallmarks of animal development, or a failure to maintain branches. To distinguish between these possibilities, we conducted a time course analysis of dendrite development by counting dendritic termini of the PVD neuron at the mid-L4 stage, the young adult stage, and 18 hr past the young adult stage (adult).
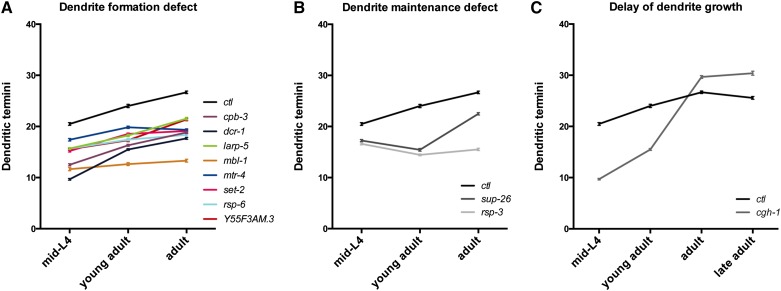
We found a statistically significant reduction of dendritic termini in cpb-3, dcr-1, larp-5, mbl-1, mtr-4, set-2, rsp-6, and Y55F3AM.3 mutants relative to the control at all stages (Figure 3A). This finding suggests that these mutants fail to form the appropriate number of terminal branches and that there is neither a delay nor a maintenance defect.
Figure 3.
A time course analysis of dendrite development reveals defects in dendrite formation, dendrite maintenance, and timing. Control (ctl) or mutant animals were scored for the number of PVD dendritic termini, as before, at the mid-L4, young adult, adult (18 hr after the young adult), and sometimes late adult (48 hr after the young adult) stages. All data points for mutants are significantly different from controls based on a one-way analysis of variance (ANOVA) test with a Fisher’s Least Significant Difference multiple comparisons test with a 95% confidence interval. Data points are the mean values where n = 80 for each genotype. Error bars show the standard error of the mean. (A) Mutants indicated have a dendrite formation defect. (B) Mutants indicated have a dendrite maintenance defect in the L4 stage. Mutant values at each time point are significantly different from each other time point based on a one-way ANOVA test with a Fisher's Least Significant Difference multiple comparisons test with a 95% confidence interval. (C) cgh-1 mutants have a delay in dendritic termini formation.
The time course analyses suggest that sup-26 and rsp-3 are required for dendrite maintenance during the L4 stage. We found that young adult sup-26 mutant animals have a statistically significant, roughly 10% reduction in dendritic termini compared to mid-L4 stage mutant animals (Figure 3B). The number of dendrite termini in sup-26 mutants then increases significantly during the adult stage but does not reach the same level as controls (Figure 3B). Similar to sup-26 mutants, rsp-3 mutants exhibit a statistically significant loss of roughly 9% of dendritic termini in PVD neurons from the mid-L4 to the young adult stage (Figure 3B). Adult rsp-3 mutant animals exhibit a mild but statistically significant increase in the number of dendrites from the young adult stage but never recover beyond the number of dendritic termini observed in mid-L4 animals (Figure 3B). Together, this finding suggests that sup-26 and rsp-3 mutants lose more terminal dendrite branches than they form during the late-L4 stage. However, branch loss ceases during the adult and some growth ensues, albeit weakly in rsp-3 mutants. We also find that sup-26 and rsp-3 mutants have significantly fewer dendrites than controls at the mid-L4 stage (Figure 3B) shortly after fourth order dendrite branch outgrowth is initiated in the early L4 stage (Smith et al. 2010). This finding suggests that sup-26 and rsp-3 mutants are defective in maintenance of terminal branches as soon as they begin to develop or that these mutants fail to form the appropriate number of dendritic termini initially.
Interestingly, we find that cgh-1 mutants are delayed in dendrite development with a 51% reduction at the mid-L4 stage but only a 35% reduction at the young adult stage. As adults and late adults (48 hr past the young adult stage), there is no longer a reduction in dendritic termini in cgh-1 mutants relative to controls. In fact, cgh-1 mutants have statistically significantly more terminal branches than controls at these later stages (Figure 3C). Collectively, the time course analyses demonstrate at least three different etiologies for dendrite defects; hence, the RBPs identified in this study likely employ different molecular mechanisms for dendrite morphogenesis.
RBP genes required for dendrite development are expressed in the PVD neuron
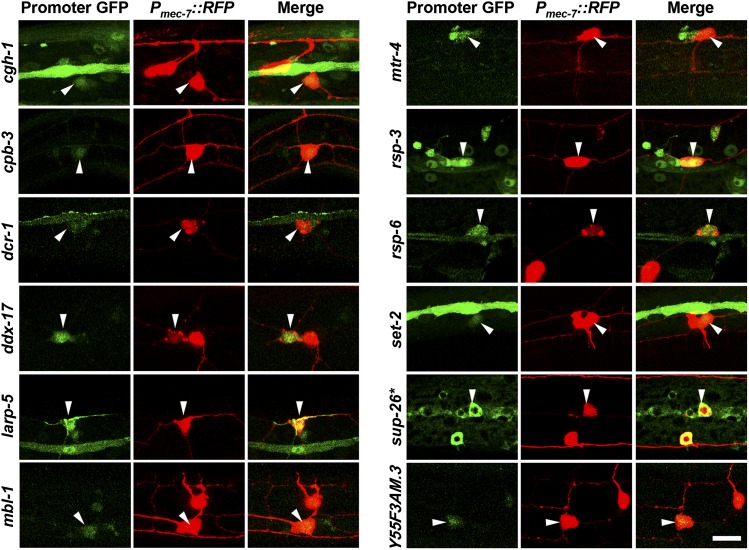
To determine the expression patterns for each of the RBP genes identified in our screen, we expressed the coding sequence of GFP under the control of presumptive promoters for each RBP gene (see the section Materials and Methods). Each of the 12 RBP gene regulatory regions expressed GFP in the PVD neuron (Figure 4). Most of the genes (cgh-1, cpb-3, dcr-1, mtr-4, rsp-3, rsp-6, set-2, and sup-26) are expressed broadly throughout development (data not shown) excluding the germ line, which often silences repetitive DNA such as the extrachromosomal arrays generated by DNA microinjection in worms (Mello et al. 1991; Kelly and Fire 1998). In contrast, ddx-17, larp-5, mbl-1, and Y55F3AM.3 are expressed mostly or exclusively in neurons including the PVD (data not shown). These data demonstrate that RBP genes required for PVD dendrite development are expressed in the PVD, and our findings are consistent with a cell-autonomous function of the RBPs within the PVD neuron. However, RBP gene expression is not specifically restricted to the PVD neuron, suggesting that these RBP genes may play additional roles in other neurons and, in some cases, non-neural tissues.
Figure 4.
RNA-binding protein (RBP) genes that are important for PVD dendrite morphogenesis are expressed in the PVD neuron. Presumptive promoter regions for each RBP gene indicated were fused to green fluorescent protein (GFP) to determine whether they are expressed in PVD neurons, which were marked with Pmec-7::RFP. Arrowheads mark the PVD cell body. *sup-26 expression was determined from a Psup-26::sup-26 cDNA::GFP construct. Bar = 10μm.
Subcellular localization of RBPs suggests how they influence dendrite development
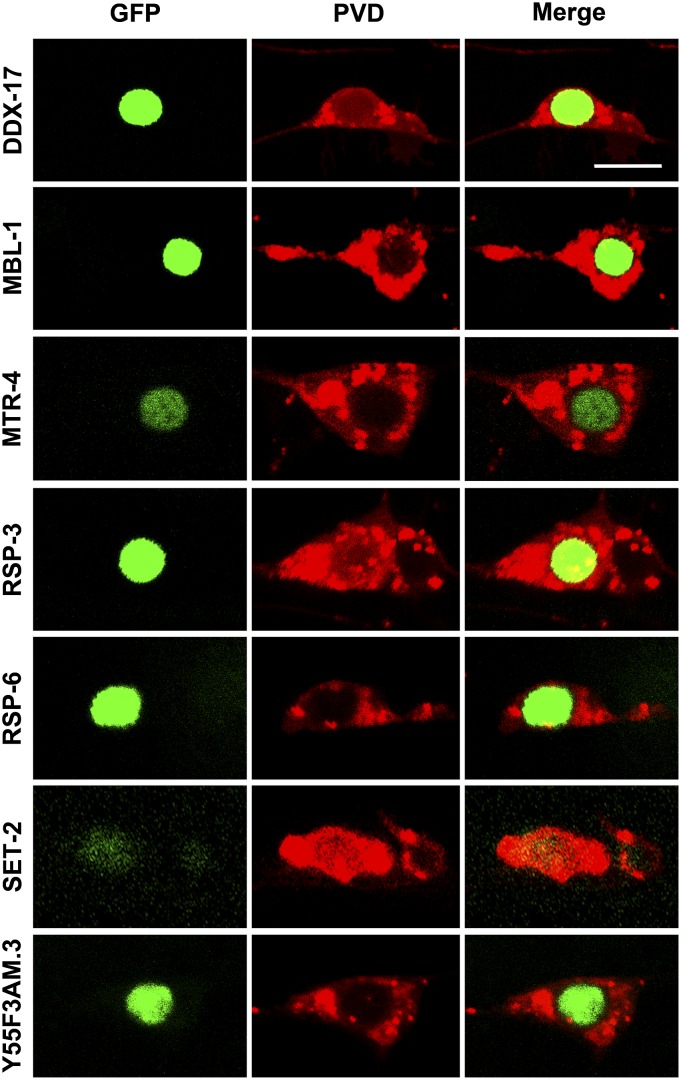
Because RBPs play diverse molecular roles related to posttranscriptional gene regulation, the subcellular localization of any given RBP may offer some insight as to which role(s) it does or does not play. To determine where within the PVD each RBP is localized, we expressed each RBP as a fusion to GFP in PVD neurons using the ser-2prom3 regulatory element (Tsalik and Hobert 2003; see the section Materials and Methods).
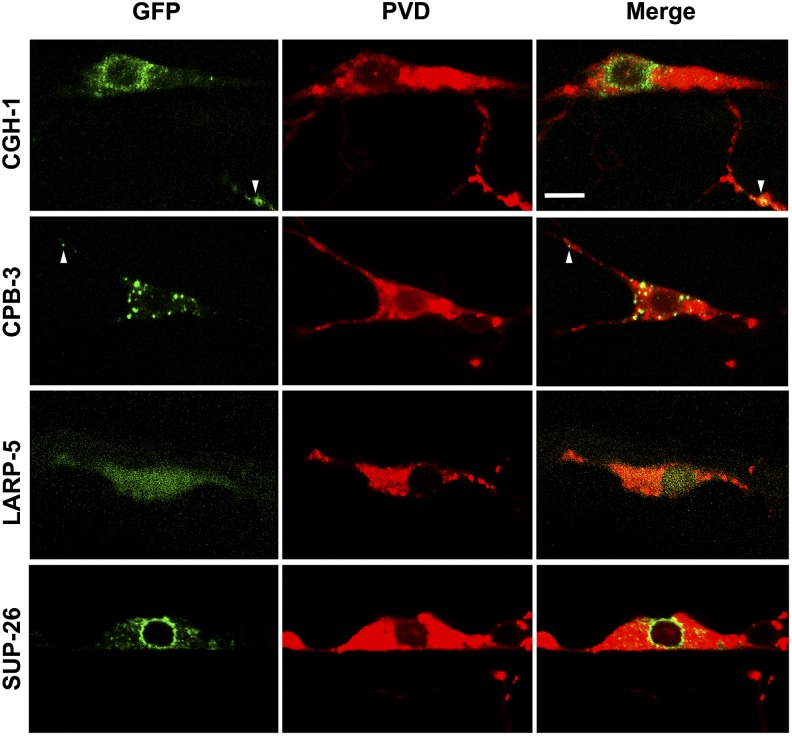
DDX-17, MBL-1, MTR-4, RSP-3, RSP-6, SET-2, and Y55F3AM.3 translation fusions to GFP show clear nuclear expression (Figure 5). This finding is consistent with these RBPs playing roles in RNA processing within the nucleus (see the section Discussion). Conversely, we find that CGH-1, CPB-3, LARP-5, and SUP-26 translational fusions to GFP are localized to the cytoplasm (Figure 6). More specifically, CGH-1::GFP is enriched in the perinuclear region and small puncta are present throughout the cytoplasm, including particles in dendrites (Figure 6). Similarly, CPB-3::GFP also is found in puncta in the cytoplasm of the cell body and dendrites. However, CPB-3 puncta are substantially larger and lack perinuclear enrichment (Figure 6). Unlike CGH-1 and CPB-3, SUP-26::GFP is restricted to the cytoplasm of the cell body and is not found in dendrites. SUP-26 has a striking, perinuclear enrichment with intermediate-sized puncta, relative to CGH-1 and CPB-3, located further from the nucleus but still within the cell body (Figure 6). Finally, LARP-5::GFP is diffuse throughout the cytoplasm, including the dendrites, and the nucleus and does not localize to discrete puncta (Figure 6).
Figure 5.
Nuclear localization of several RNA-binding proteins (RBPs) in PVD neurons. cDNAs for RBP genes indicated were fused to green fluorescent protein (GFP) under the control of a PVD-specific promoter and reveal a nuclear localization. PVDs were marked by ser-2prom3::myr-mCherry. Bar = 5 μm.
Figure 6.
Cytoplasmic localization of CGH-1, CPB-3, LARP-5, and SUP-26 in PVD neurons. cDNAs for RNA-binding protein (RBP) genes indicated were fused to green fluorescent protein (GFP) under the control of a PVD-specific promoter and reveal cytoplasmic localization. PVDs were marked by ser-2prom3::myr-mCherry. Arrowheads indicate GFP-positive particles within dendrites. Bar = 5 μm.
We were not able to generate a visible DCR-1::GFP fusion protein expressed in the PVD neuron. However, anti-DCR-1 immunohistochemistry reveals that DCR-1 is in puncta in the cytoplasm and nucleus of C. elegans germ cells (Beshore et al. 2011). This finding is consistent with known roles of DCR-1/Dicer in the microRNA and RNAi pathways as well as restricting the accumulation of dsRNA from bidirectional transcription (Bernstein et al. 2001; Grishok et al. 2001; White et al. 2014).
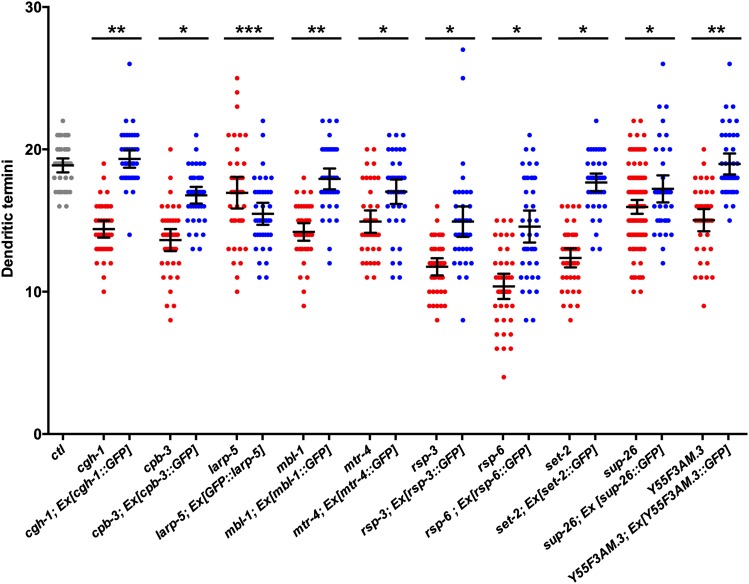
To test whether these RBPs function cell-autonomously in the PVD neuron and to determine if RBP::GFP fusion protein subcellular localization is biologically relevant, we expressed each RBP::GFP transgene specifically in the PVD neuron and tested for its ability to rescue PVD dendrite defects in RBP mutant animals (see the section Materials and Methods). We found that PVD-specific expression of CGH-1, MBL-1, and Y55F3AM.3 fusion proteins to GFP confer a statistically significant complete rescue of the PVD dendritic termini defects in the respective mutants (Figure 7). This strongly suggests that CGH-1, MBL-1, and Y55F3AM.3 each function cell-autonomously in the PVD to control dendrite development. Furthermore, the subcellular localizations inferred from these RBP::GFP fusions proteins are likely accurate because these proteins are functional.
Figure 7.
PVD-specific expression of RNA-binding protein::green fluorescent protein (RBP::GFP) fusion proteins rescues dendrite defects in most of the RBP mutants. Control (ctl) animals (gray), mutant animals (red), and mutant animals bearing an extrachromosomal array (Ex) that expresses the corresponding RBP::GFP fusion protein in the PVD neuron (blue) were scored for the number of dendritic termini using the ser-2prom3::myr-mCherry PVD marker. Points within each scatter column represent counts of dendritic termini from the PVD cell body to the tail on the dorsal or ventral side of the worm. Lines represent the means and the 95% confidence interval of the mean. All of the mutants are significantly lower than the control. *Mutants expressing RBP::GFP are significantly different from mutants and significantly different from controls indicating a partial rescue. **Mutants with RBP::GFP are significantly different from mutants and not significantly different from controls indicating a complete rescue. ***GFP::LARP-5 causes a significant reduction in dendritic termini compared to the larp-5 mutant alone. All statistics are based on a one-way ANOVA test with a Fisher’s Least Significant Difference multiple comparisons test with a 95% confidence interval.
We also found that cpb-3, mtr-4, rsp-3, rsp-6, set-2, and sup-26 dendrite defects are rescued partially by PVD-specific expression of the corresponding RBP::GFP fusion proteins. The reduction of PVD dendritic termini associated with each mutant is significantly ameliorated by expression of the corresponding RBP::GFP fusion protein, but does not return to control levels (Figure 7). This finding suggests that these RBPs function within the PVD neuron to control dendrite development. However, the partial rescue does not allow us to exclude the possibility that RBP expression in other cells is also important for PVD dendrite development. Alternatively, partial rescue may be the result of GFP slightly reducing the activity of the RBPs or non-native expression levels of the RBP::GFP fusion proteins.
We were unable to recover transgenic lines expressing LARP-5::GFP, suggesting that this transgene is toxic. However, we were able to generate transgenic animals expressing GFP::LARP-5. Interestingly, we found that larp-5 mutants expressing a GFP::LARP-5 fusion protein in PVD neurons were not rescued; rather, these animals had significantly lower numbers of dendritic termini than larp-5 mutants alone (Figure 7). It is possible that a GFP tag on LARP-5 disrupts its function. Alternatively, the results may indicate that LARP-5 levels must be precisely controlled for proper dendritic development. This would not be unusual given that the expression levels of multiple RBPs are critical for Drosophila da sensory neuron development. For example, overexpression of nanos, pumilio, and brat all result in more severe dendrite defects than the mutants alone (Ye et al. 2004; Olesnicky et al. 2012). We were unable to test DDX-17::GFP for cell-autonomous function and relevance of its subcellular localization since this RBP was identified from RNAi, and ddx-17 mutants die early in development.
Discussion
Conservation of the role of RBPs in dendrite morphogenesis
Sixty-three RBP genes were reported to function in Drosophila da neuron morphogenesis, suggesting that many aspects of posttranscriptional regulation are important to this process (Olesnicky et al. 2014). Here, we test 54 homologs of the Drosophila suite of dendrite RBPs and show that 12 of them, or roughly 22%, are important for PVD dendrite morphogenesis. This relatively low rate of functional conservation may not be unexpected. Only 21% of C. elegans genes with an ortholog in another eukaryote give obvious RNAi phenotypes (Kamath et al. 2003). In addition, the plasticity of genetic networks between C. elegans and C. briggsae, which share similar body plans and ecological niches despite roughly 20 million years of evolution, is significant; over 25% of orthologs in these worm species have different functions (Verster et al. 2014).
There are few reasons why such a small percentage of the RBP genes required for Drosophila da neuron morphogenesis have homologs with conserved function in C. elegans PVD neurons. Although da neurons and PVD neurons are both complex multidendritic sensory neurons, the PVD has a simpler and more stereotyped shape, which may not require as many posttranscriptional regulatory factors. Another significant difference between da neurons and PVDs is that da neurons undergo extensive pruning and shape changes during development while the PVD neuron does not undergo drastic morphological changes (Williams and Truman 2005a,b; Albeg et al. 2011). Perhaps the reduced need for shape changes in PVDs also reduces the need for posttranscriptional gene regulation mechanisms mediated by RBPs.
Although in this study we specifically tested 54 RBP genes using predominantly predicted null alleles and a neuron-sensitized RNAi strategy, the 12 genes reported are likely an underestimate. Although 10 of 30 genetic mutations tested positive for dendrite defects, our primary RNAi screen identified only two (ddx-17 and Y55F3AM.3) of 24 positive hits. On the basis of the rate of positive hits using genetic mutations, we would have expected a greater percentage of the genes tested by RNAi to also be positive. Moreover, after completing the RNAi screen, we identified some mutations from the Million Mutation Project (Thompson et al. 2013) that affect genes for which no deletion allele was available. Using these alleles, we confirmed Y55F3AM.3 and added larp-5, which was not a hit in the initial RNAi screen. Thus, we conclude that false negatives by RNAi are likely and difficult to avoid. Some of the genes tested by genetic null mutation also may be reported as false negatives in our screen due to maternal rescue in cases where homozygous sterile mutants are scored from heterozygous mothers (see below for discussion of drsh-1/Drosha). Finally, although most alleles tested are predicted nulls, at least two have been reported to be hypomorphic alleles. Homologs of stau-1 are involved in dendrite development in various species, yet we did not find statistically significant defects in PVD neurons using the hypomorphic allele tm2266 (Tang et al. 2001; Barbee et al. 2006; Goetze et al. 2006; Legendre et al. 2013). The sym-2(mn617) allele is also likely to be a hypomorph (Yochem et al. 2004) and did not produce PVD defects in our screen. Although the sym-2 homolog in Drosophila, glo, is required for dendrite development (Brechbiel and Gavis 2008; Olesnicky et al. 2014), this has not been demonstrated for the human homolog, heterogeneous nuclear ribonucleoprotein F.
Although the dendritic roles of several RBP genes identified in the Olesnicky et al. (2014) RNAi screen were confirmed by genetic mutation or second RNAi line, it is possible that some false positives were reported because of a previously unknown problem with some RNAi fly stocks used. A recent study showed that many randomly selected Vienna Drosophila RNAi Center “KK” series RNAi stocks produce nonspecific phenotypes when crossed to certain Gal4 drivers due to an additional and previously uncharacterized RNAi hairpin vector insertion site (Green et al. 2014). We therefore acknowledge this caveat to our assessment of the number of conserved RBPs that function in dendrite morphogenesis in both Drosophila da neurons and C. elegans PVDs. Considering the results of the Drosophila da neuron dendrite screen and this study, the percent of RBPs that play a conserved role in dendrite morphogenesis is likely to exceed the 22% reported here.
mRNA localization and translational regulation contribute to dendrite morphogenesis
It is well documented that mRNAs are transported within RNA−protein complexes (RNPs) into dendrites, and there is increasing evidence that translational control of these mRNAs contributes to dendrite morphology (reviewed by Bramham and Wells 2007; Pimentel and Boccaccio 2014; Tom Dieck et al. 2014; Di Liegro et al. 2014). Therefore, we were not surprised that this study identified RBP genes, such as cgh-1, cpb-3, larp-5, and sup-26, which are implicated in mRNA transport, localization, and/or translation.
cgh-1 encodes an RNA helicase similar to mammalian DDX6/RCK/p54. Both are associated with various RNP granules and are required for mRNA translational repression, possibly by stalling translation by polysomes associated with RNPs (reviewed by Rajyaguru and Parker 2009; Pimentel and Boccaccio 2014). We found that functional CGH-1::GFP protein is enriched in the cell body and in puncta within PVD dendrites (Figure 6 and Figure 7) suggesting that CGH-1 may act as a translational repressor of target mRNAs during their transport into dendritic compartments.
cpb-3 encodes a homolog of mammalian cytoplasmic polyadenylation element binding protein (CPEB), which acts as a translational repressor of target mRNAs but also promotes translation when phosphorylated (reviewed by Villalba et al. 2011). Although invertebrate CPEBs, such as CPB-3, lack the site of phosphorylation (Hasegawa et al. 2006), it is possible that their dual-function as a repressor and activator may be regulated by an alternative mechanism. CPEB has known roles within the nervous system where it is required for learning and memory by regulating synaptic plasticity (Si et al. 2003; Miniaci et al. 2008). In addition, CPEB regulates the transport of mRNAs in dendrites and disruption of CPEB function in frogs led to stunted development of dendritic arbors (Huang et al. 2003; Bestman and Cline 2008). Because CPEB-containing granules are found within dendrites in frogs and a functional CPB-3::GFP protein is found within dendrites in C. elegans (Bestman and Cline 2009; Figure 6 and Figure 7), it suggests that CPEB-dependent translation is important for providing localized sources of proteins within the dendritic compartment, which in turn regulates local dendrite development.
sup-26 encodes an RNA recognition motif-containing protein similar to Drosophila Shep and mammalian RBMS1/2/3. C. elegans SUP-26 has been shown to directly bind to the 3′ UTR of a target mRNA and repress its translation, possibly through a physical interaction with poly(A)-binding protein 1 (Mapes et al. 2010). Drosophila shep was shown recently to be important for neuronal remodeling (Chen et al. 2014), suggesting a broader role in nervous system development. A functional SUP-26::GFP fusion protein is largely perinuclear and absent from dendrites (Figure 6 and Figure 7) and thus may play a role in repressing the translation of target mRNAs upon nuclear export and before they associate with RNPs that take over the role of mRNA localization and translational control.
LARP-5 is homologous to mammalian LARP4 and LARP4B, which encode proteins that bind to the poly(A)-binding protein, associate with polysomes, and promote mRNA stability and translation (Schäffler et al. 2010; Yang et al. 2011). siRNA-mediated knockdown of LARP4 leads to decreased mRNA levels, whereas overexpression of LARP4 increases mRNA levels, strongly suggesting that LARP4 can prolong mRNA half-life (Yang et al. 2011). In C. elegans, LARP-5::GFP is localized to the cytoplasm including the dendrites (Figure 6), which is consistent with association with poly(A)-binding protein and polysomes and suggests that LARP-5 may promote the stability of target mRNAs that are required for dendrite development. However, the subcellular localization of LARP-5 must be considered in context of the caveat that GFP::LARP-5 does not rescue dendrite defects of larp-5 mutants and actually leads to a significant further reduction of dendritic termini. Thus the subcellular localization we observe may not reflect endogenous LARP-5 protein localization (Figure 7).
Taken together, we favor a model whereby SUP-26 translationally represses mRNAs upon nuclear export and before loading with additional RBPs, possibly including CGH-1, CPB-3, and/or LARP-5, which regulate mRNA transport, stability, and translation. In the absence of SUP-26, target mRNAs may be prematurely translated in the cell body where they cannot influence dendrite development. In support of this idea, sup-26 mutants have fewer branches at all orders (Figure 2). In the absence of CGH-1 and CPB-3, target mRNAs may be translated prematurely within dendrites, which could plausibly explain the excess secondary and tertiary branches observed in cgh-1 and cpb-3 mutant PVDs (Figure 2). Terminal branches eventually do form in cgh-1 mutants (Figure 3C), suggesting that over time target mRNAs or their protein products do eventually reach the correct location or exhibit the correct activity. Finally, we suggest that a loss of LARP-5 activity results in decreased mRNA stability and thus a reduction of branching at all orders (Figure 2). These findings highlight the importance of mRNA localization, stability, translational repression, and localized protein synthesis within the developing dendritic arbor.
The role of mRNA splicing in dendrite morphogenesis
One striking finding of this screen is that six (MBL-1, RSP-3, RSP-6, SET-2, Y55F3AM.3, and DDX-17) of the 12 RBPs that we identified as important for PVD dendrite morphogenesis are known or thought to be involved in mRNA splicing, and more specifically alternative splicing.
mbl-1 encodes a homolog of Drosophila Muscleblind (Mbl) and mammalian Muscleblind-like proteins (Mbnl), which are Zinc finger containing proteins that regulate alternative splicing (Begemann et al. 1997; Kanadia et al. 2003; Ho et al. 2004; Pascual et al. 2006; Wang et al. 2008; Sasagawa et al. 2009; Spilker et al. 2012; Wang et al. 2012). Although C. elegans MBL-1 has not been shown definitively to participate in alternative splicing, strong homology to fly Mbl and mammalian Mbnl proteins strongly suggests that this molecular function is conserved (Pascual et al. 2006; Spilker et al. 2012).
rsp-3 and rsp-6 provide another strong link to alternative splicing. These genes encode members of the serine/arginine-rich (SR) family of splicing factors that regulate constitutive and alternative splicing (reviewed by Risso et al. 2012). rsp-3 encodes a protein similar to Drosophila SF2 and human SRSF1/9, and rsp-6 encodes a homolog of Drosophila X16 and mammalian SRSF3/7 splicing factors (Krainer et al. 1991; Kawano et al. 2000; Longman et al. 2000; Vorbrüggen et al. 2000; De La Mata and Kornblihtt 2006).
Although little is known about Y55F3AM.3 and ddx-17, the paucity of information does suggest that these genes may be involved in alternative splicing. Y55F3AM.3 is an RRM-containing protein with a human homolog (RBM39) that co-localizes with spliceosomal proteins and affects alternative splicing for some target genes (Imai et al. 1993; Dowhan et al. 2005; McCracken et al. 2005; Ellis et al. 2008; Huang et al. 2012). ddx-17 encodes a DEAD-box RNA helicase protein similar to human DDX17/p72 (and DDX5/p68), which is a regulator of alternative splicing and co-purifies with the U1snRNP and SR protein SRrp86 (Hönig et al. 2002; Lee 2002; Li et al. 2003). Furthermore, one study suggests that DDX5/p68 facilitates the binding of Mbnl proteins to splicing targets (Laurent et al. 2012), adding another link to alternative splicing.
Finally, set-2 encodes an RRM-containing histone 3 lysine 4 (H3K4) methyltransferase similar to yeast and fly Set1 and human SETD1A and SETD1B (Xu and Strome 2001; Briggs et al. 2001; Miller et al. 2001; Lee and Skalnik 2005; Xiao et al. 2011; Table 1). Set1 is recruited by RNA polymerase II to mediate H3K4 trimethylation (me3) on nearby nucleosomes. Thus, H3K4me3 is a landmark of actively transcribed genes or genes that experience transcription initiation (Miller et al. 2001; Krogan et al. 2003; Tenney and Shilatifard 2005; Guenther et al. 2007). In addition, H3K4me3 enhances splicing efficiency and affects alternative splicing, possibly through its association with the U2 snRNP (Sims et al. 2007; Luco et al. 2010). Furthermore, H3K4me3 is enriched at alternative transcriptional start sites suggesting it may regulate alternative promoter use. This is interesting because the use of alternative transcriptional start sites contributes more to transcriptome diversity than alternative splicing does in the mouse cerebellum (Pal et al. 2011). Because loss of set-2 activity greatly reduces H3K4me3 in worms (Greer et al. 2010), it is attractive to suggest that set-2 may play a role in regulating alternative splicing and/or alternative transcription start sites.
There are several possibilities for how alternative splicing and use of alternative promoters may affect dendrite development. The simplest possibility is that one or more specific alternatively spliced products are important for dendrite development and that defects in alternative splicing reveal defects in dendrite morphology (Olesnicky et al. 2014). Support for this comes from the well-documented prevalence of transcriptome diversity in the nervous system, which is thought to be important for contributing to cellular complexity (Yeo et al. 2004; Lipscombe 2005; Li et al. 2007; Norris and Calarco 2012). For example, alternative splicing of the mammalian Bcl11A transcriptional factor-encoding gene produces a long form that negatively regulates dendrite outgrowth and a short form that antagonizes the long form and thus promotes dendrite outgrowth (Kuo et al. 2009).
An alternative possibility is that some splicing occurs in the dendrites rather than in the nucleus. Several mammalian splicing components have been shown to exit the nucleus and retain function in the dendroplasm, perhaps to provide localized mature mRNAs for localized translation of specific transcripts (Glanzer et al. 2005; Bell et al. 2008). However, all of the putative splicing regulators in this study show a tight nuclear localization as observed with RBP::GFP fusion proteins (Figure 5). Because each of these fusion proteins rescues the mutant dendrite defects (except DDX-17, which was not amenable to testing), it suggests that the strong nuclear localization reflects normal activity (Figure 7). Thus, if any of these factors are involved in activities within dendrites, they must be present in concentrations that are below the level of detection by confocal microscopy. Furthermore, cytoplasmic splicing has thus far not been described in C. elegans.
Proteins associated with splicing also may be important for mRNA localization and/or translational regulation. For example, mammalian Mbnl proteins, which are homologs of MBL-1, are important for targeting hundreds of mRNAs to membranes where they are translated (Adereth et al. 2005; Osborne et al. 2009; Du et al. 2010; Masuda et al. 2012; Wang et al. 2012). Again, we do not favor this model for how the splice factors identified in this study affect dendrite development because their localization is restricted to the nucleus (Figure 5). However, the localization of mRNAs can be influenced by RBPs that never leave the nucleus. For example, localization of ASH1 mRNA in yeast requires the function of Loc1p, an RBP that is exclusively nuclear (Long et al. 2001). Loc1p is required for other RBPs to assemble onto ASH1 mRNA, which then remove Loc1p, and guide its localization (Niedner et al. 2013). Similarly, ZBP2 is a predominantly nuclear RBP that is required for beta-actin mRNA localization in fibroblasts and neurons (Gu et al. 2002). ZBP2 facilitates the binding of ZBP1 to beta-actin mRNAs, which then escorts the mRNA and represses translation (Pan et al. 2007). Thus, there is precedent that nuclear RBPs, including at least one implicated in mRNA splicing, can affect mRNA localization without leaving the nucleus.
Knockdown of several RBP genes encoding splicing factors in Drosophila led to excess dendrite branches in da neurons (Olesnicky et al. 2014). Among these were mbl and x16, both of which are implicated in alternative splicing (Vorbrüggen et al. 2000; Wang et al. 2012). However, mutations in the worm homologs of these genes, mbl-1 and rsp-6, result in a decrease in the number of dendritic termini in PVDs. Although splicing is important in both species, it is possible that the target genes of alternative splicing are not conserved and that these targets regulate different aspects of dendrite morphogenesis.
RNA processing and dendrite morphogenesis
mtr-4 encodes a homolog of yeast Mtr4p, an RNA helicase that modulates the polyadenylation activity of the Trf4/Air2/Mtr4 polyadenylation complex. Trf4/Air2/Mtr4 polyadenylation adds short poly(A) tails to mRNAs and small noncoding RNAs (such as small nucleolar RNAs and transfer RNAs) to mark them for degradation or processing by the nuclear exosome (Liang et al. 1996; Jia et al. 2011; Schmidt and Butler 2013). Although it is difficult to speculate on how a general process such as RNA processing and degradation may lead to a specific phenotype such as dendrite morphology, at least one study has shown that exosome activity is important for nervous system development. Loss of zebrafish EXOSC3 activity, which is required for exosome function, resulted in spinal motor neuron developmental defects and degeneration (Wan et al. 2012).
Is the microRNA/RNAi pathway important for dendrite development?
We found that dcr-1/Dicer mutants have a reduction in the number of terminal dendrite branches. Although this might suggest that the process of dendrite morphogenesis is partly regulated by microRNAs, this is inconsistent with our finding that a null mutation in drsh-1/Drosha does not cause dendrite defects. One possibility is that drsh-1 mutants, which are recovered from heterozygous mothers, are rescued from defects by maternal drsh-1(+). Maternal rescue of adult stage developmental events in drsh-1 mutants has been suggested previously (Denli et al. 2004). There is some evidence that microRNAs are important for dendrite morphogenesis (Bicker et al. 2014). For example, rat miR-134 is localized to dendrites in the hippocampus and negatively regulates the size of dendritic spines. Brain-derived neurotrophic factor positively regulates the size of dendritic spines by alleviating miR-134 repression (Schratt et al. 2006). The microRNA bantam regulates the scaling of da neuron arbors in Drosophila. However, bantam functions within the epidermis, not the da neuron (Parrish et al. 2009). It is unclear from our work if dcr-1 functions in the neuron or non-cell-autonomously. Alternatively, dendrite development may be partly regulated by endogenous RNAi mechanisms, which are dcr-1-dependent but drsh-1-independent, or dcr-1 may have additional roles that do not involve small noncoding RNAs.
RBPs are candidates for dendrite development in humans
By comparing results from genetic screens for dendrite morphogenesis in two similar but distinct neuronal types in distantly related species, we have identified 12 RBPs that we suggest are likely to be involved in dendrite development in many other species as well. Furthermore, these 12 proteins may constitute a minimal set of evolutionarily conserved RBPs required for dendrite development across animal phyla. This is bolstered by the fact that human orthologs of these RBP genes are expressed in the human brain based on the TiGER database (Table 3; Liu et al. 2008). It will be interesting to see whether future functional studies in vertebrates demonstrate a role for these RBPs in dendrite development or whether studies in humans identify any of these RBP genes as mutated in neurological disorders in humans.
Table 3. Human orthologs of C. elegans RBPs implicated in PVD dendrite development.
| C. elegans RBP | OrthoDB ID | Human Ortholog | E Value | Brain Expression |
|---|---|---|---|---|
| CGH-1 | EOG7D85W7 | DDX6 | 1.00E-179 | Expressed |
| CPB-3 | EOG751NG9 | CPEB1 | 5.00E-71 | Expressed |
| DCR-1 | EOG78PV82 | DICER1 | 2.00E-167 | Expressed |
| DDX-17 | EOG7HTHGB | DDX17 | 2.00E-163 | Expressed |
| EOG7HTHGB | DDX5 | 5.00E-163 | Expressed | |
| LARP-5 | EOG74J971 | LARP4B | 2.00E-32 | Expressed |
| EOG74J971 | LARP4 | 1.00E-30 | Expressed | |
| MBL-1 | EOG77M8PC | MBNL2 | 1.00E-32 | Not assayed |
| EOG77M8PC | MBNL3 | 5.00E-31 | Expressed | |
| EOG77M8PC | MBNL1 | 5.00E-30 | Not assayed | |
| MTR-4 | EOG7XSTCX | SKIV2L2 | 0.0 | Expressed |
| RSP-3 | EOG76X620 | SRSF1 | 5.00E-56 | Expressed |
| EOG76X620 | SRSF9 | 3.00E-53 | Not assayed | |
| RSP-6 | EOG73NG5V | SRSF3 | 9.00E-23 | Expressed |
| EOG73NG5V | SRSF7 | 3.00E-22 | Expressed | |
| SET-2 | EOG72JWHB | SETD1A | 6.00E-65 | Expressed |
| EOG72JWHB | SETD1B | 7.00E-60 | Expressed | |
| SUP-26 | EOG71G9V3 | RBMS1 | 4.00E-53 | Expressed |
| EOG71G9V3 | RBMS2 | 8.00E-52 | Expressed | |
| EOG71G9V3 | RBMS3 | 9.00E-52 | Expressed | |
| Y55F3AM.3 | EOG71K63D | RBM39 | 3.00E-95 | Expressed |
| EOG71K63D | RBM23 | 3.00E-53 | Expressed |
Human orthologs were identified using the Database of Orthologous Groups and are displayed with the corresponding OrthoDB IDs. C. elegans proteins were aligned to the human protein RefSeq database using BLASTp and E values are given. Gene expression in the brain was determined using the TiGER database (Liu et al. 2008). RBP, RNA-binding protein; TiGER, Tissue-specific Gene Expression and Regulation.
Supplementary Material
Acknowledgments
We thank Delaine Winkelblech and Brent Wallace for technical support; Tracy Kress for insightful discussions; the Caenorhabditis Genetics Center; the Mitani Lab as part of the National Bio-Resource Project of Japan; James Mapes, Ding Xue, Cody Smith, David Miller, Caitlin Taylor, and Kang Shen for strains; and Yuji Kohara, James Mapes, and Ding Xue for cDNA clones. This work was supported by the National Science Foundation (IOS proposal number 1257703 to D.J.K. and E.C.O.), a CC Mrachek Fellowship to D.J.K., a CC Figge-Bourquin Summer Research Grant to L.K., CC Venture Grants to M.A.S. and G.K., and the CC and UCCS Dean’s offices.
Footnotes
Supporting information is available online at http://www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.017327/-/DC1
Communicating editor: H. D. Lipshitz
Literature Cited
- Adereth Y., Dammai V., Kose N., Li R., Hsu T., 2005. RNA-dependent integrin alpha3 protein localization regulated by the Muscleblind-like protein MLP1. Nat. Cell Biol. 7: 1240–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre-Chen C., Bülow H. E., Kaprielian Z., 2011. C. elegans bicd-1, homolog of the Drosophila dynein accessory factor Bicaudal D, regulates the branching of PVD sensory neuron dendrites. Development 138: 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albeg A., Smith C. J., Chatzigeorgiou M., Feitelson D. G., Hall D. H., et al. , 2011. C. elegans multi-dendritic sensory neurons: morphology and function. Mol. Cell. Neurosci. 46: 308–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbee S. A., Estes P. S., Cziko A. M., Hillebrand J., Luedeman R. A., et al. , 2006. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron 52: 997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begemann G., Paricio N., Artero R., Kiss I., Pérez-Alonso M., et al. , 1997. muscleblind, a gene required for photoreceptor differentiation in Drosophila, encodes novel nuclear Cys3His-type zinc-finger-containing proteins. Development 124: 4321–4331. [DOI] [PubMed] [Google Scholar]
- Bell T. J., Miyashiro K. Y., Sul J. Y., McCullough R., Buckley P. T., et al. , 2008. Cytoplasmic BK(Ca) channel intron-containing mRNAs contribute to the intrinsic excitability of hippocampal neurons. Proc. Natl. Acad. Sci. USA 105: 1901–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E., Caudy A. A., Hammond S. M., Hannon G. J., 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. [DOI] [PubMed] [Google Scholar]
- Beshore E. L., McEwen T. J., Jud M. C., Marshall J. K., Schisa J. A., et al. , 2011. C. elegans Dicer interacts with the P-granule component GLH-1 and both regulate germline RNPs. Dev. Biol. 350: 370–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestman J. E., Cline H. T., 2008. The RNA binding protein CPEB regulates dendrite morphogenesis and neuronal circuit assembly in vivo. Proc. Natl. Acad. Sci. USA 105: 20494–20499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestman J. E., Cline H. T., 2009. The relationship between dendritic branch dynamics and CPEB-Labeled RNP granules captured in vivo. Front Neural Circuits 3: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicker S., Lackinger M., Weiß K., Schratt G., 2014. MicroRNA-132, -134, and -138: a microRNA troika rules in neuronal dendrites. Cell Mol Life Sci 71: 3987–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramham C. R., Wells D. G., 2007. Dendritic mRNA: transport, translation and function. Nat. Rev. Neurosci. 8: 776–789. [DOI] [PubMed] [Google Scholar]
- Brechbiel J. L., Gavis E. R., 2008. Spatial regulation of nanos is required for its function in dendrite morphogenesis. Curr. Biol. 18: 745–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs S. D., Bryk M., Strahl B. D., Cheung W. L., Davie J. K., et al. , 2001. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 15: 3286–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calixto A., Chelur D., Topalidou I., Chen X., Chalfie M., 2010. Enhanced neuronal RNAi in C. elegans using SID-1. Nat. Methods 7: 554–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., et al. , 2009. BLAST+: architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzigeorgiou M., Schafer W. R., 2011. Lateral facilitation between primary mechanosensory neurons controls nose touch perception in C. elegans. Neuron 70: 299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Qu C., Hewes R. S., 2014. Neuronal remodeling during metamorphosis is regulated by the alan shepard (shep) gene in Drosophila melanogaster. Genetics 197: 1267–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli A. M., Tops B. B., Plasterk R. H., Ketting R. F., Hannon G. J., 2004. Processing of primary microRNAs by the microprocessor complex. Nature 432: 231–235. [DOI] [PubMed] [Google Scholar]
- Di Liegro C. M., Schiera G., Di Liegro I., 2014. Regulation of mRNA transport, localization and translation in the nervous system of mammals. Int. J. Mol. Med. 33: 747–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Liu O. W., Howell A. S., Shen K., 2013. An extracellular adhesion molecule complex patterns dendritic branching and morphogenesis. Cell 155: 296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowhan D. H., Hong E. P., Auboeuf D., Dennis A. P., Wilson M. M., et al. , 2005. Steroid hormone receptor coactivation and alternative RNA splicing by U2AF65-related proteins CAPERalpha and CAPERbeta. Mol. Cell 17: 429–439. [DOI] [PubMed] [Google Scholar]
- Du H., Cline M. S., Osborne R. J., Tuttle D. L., Clark T. A., et al. , 2010. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat. Struct. Mol. Biol. 17: 187–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J. D., Llères D., Denegri M., Lamond A. I., Cáceres J. F., 2008. Spatial mapping of splicing factor complexes involved in exon and intron definition. J. Cell Biol. 181: 921–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser A. G., Kamath R. S., Zipperlen P., Martinez-Campos M., Sohrmann M., et al. , 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408: 325–330. [DOI] [PubMed] [Google Scholar]
- Gamberi C., Johnstone O., Lasko P., 2006. Drosophila RNA binding proteins. Int. Rev. Cytol. 248: 43–139. [DOI] [PubMed] [Google Scholar]
- Glanzer J., Miyashiro K. Y., Sul J. Y., Barrett L., Belt B., et al. , 2005. RNA splicing capability of live neuronal dendrites. Proc. Natl. Acad. Sci. USA 102: 16859–16864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetze B., Tuebing F., Xie Y., Dorostkar M. M., Thomas S., et al. , 2006. The brain-specific double-stranded RNA-binding protein Staufen2 is required for dendritic spine morphogenesis. J. Cell Biol. 172: 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, E. WFedele G., Giorgini F., Kyriacou C. P., 2014. A Drosophila RNAi collection is subject to dominant phenotypic effects. Nat. Methods 11: 222–223. [DOI] [PubMed]
- Greer E. L., Maures T. J., Hauswirth A. G., Green E. M., Leeman D. S., et al. , 2010. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature 466: 383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A., Pasquinelli A. E., Conte D., Li N., Parrish S., et al. , 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34. [DOI] [PubMed] [Google Scholar]
- Gu W., Pan F., Zhang H., Bassell G. J., Singer R. H., 2002. A predominantly nuclear protein affecting cytoplasmic localization of beta-actin mRNA in fibroblasts and neurons. J. Cell Biol. 156: 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther M. G., Levine S. S., Boyer L. A., Jaenisch R., Young R. A., 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130: 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halevi S., McKay J., Palfreyman M., Yassin L., Eshel M., et al. , 2002. The C. elegans ric-3 gene is required for maturation of nicotinic acetylcholine receptors. EMBO J. 21: 1012–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa E., Karashima T., Sumiyoshi E., Yamamoto M., 2006. C. elegans CPB-3 interacts with DAZ-1 and functions in multiple steps of germline development. Dev. Biol. 295: 689–699. [DOI] [PubMed] [Google Scholar]
- Ho T. H., Charlet-B N., Poulos M. G., Singh G., Swanson M. S., et al. , 2004. Muscleblind proteins regulate alternative splicing. EMBO J. 23: 3103–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan D. J., Riordan D. P., Gerber A. P., Herschlag D., Brown P. O., 2008. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 6: e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C. E., Schuman E. M., 2013. The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron 80: 648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hönig A., Auboeuf D., Parker M. M., O’Malley B. W., Berget S. M., 2002. Regulation of alternative splicing by the ATP-dependent DEAD-box RNA helicase p72. Mol. Cell. Biol. 22: 5698–5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G., Zhou Z., Wang H., Kleinerman E. S., 2012. CAPER-α alternative splicing regulates the expression of vascular endothelial growth factor165 in Ewing sarcoma cells. Cancer 118: 2106–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. S., Carson J. H., Barbarese E., Richter J. D., 2003. Facilitation of dendritic mRNA transport by CPEB. Genes Dev. 17: 638–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai H., Chan E. K., Kiyosawa K., Fu X. D., Tan E. M., 1993. Novel nuclear autoantigen with splicing factor motifs identified with antibody from hepatocellular carcinoma. J. Clin. Invest. 92: 2419–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer E. P., Iyer S. C., Sullivan L., Wang D., Meduri R., et al. , 2013. Functional genomic analyses of two morphologically distinct classes of Drosophila sensory neurons: post-mitotic roles of transcription factors in dendritic patterning. PLoS ONE 8: e72434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan Y. N., Jan L. Y., 2010. Branching out: mechanisms of dendritic arborization. Nat. Rev. Neurosci. 11: 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., Wang X., Liu F., Guenther U. P., Srinivasan S., et al. , 2011. The RNA helicase Mtr4p modulates polyadenylation in the TRAMP complex. Cell 145: 890–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S., Ahringer J., 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30: 313–321. [DOI] [PubMed] [Google Scholar]
- Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., et al. , 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. [DOI] [PubMed] [Google Scholar]
- Kanadia R. N., Johnstone K. A., Mankodi A., Lungu C., Thornton C. A., et al. , 2003. A muscleblind knockout model for myotonic dystrophy. Science 302: 1978–1980. [DOI] [PubMed] [Google Scholar]
- Kawano T., Fujita M., Sakamoto H., 2000. Unique and redundant functions of SR proteins, a conserved family of splicing factors, in Caenorhabditis elegans development. Mech. Dev. 95: 67–76. [DOI] [PubMed] [Google Scholar]
- Kelly W. G., Fire A., 1998. Chromatin silencing and the maintenance of a functional germline in Caenorhabditis elegans. Development 125: 2451–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerner P., Degnan S. M., Marchand L., Degnan B. M., Vervoort M., 2011. Evolution of RNA-binding proteins in animals: insights from genome-wide analysis in the sponge Amphimedon queenslandica. Mol. Biol. Evol. 28: 2289–2303. [DOI] [PubMed] [Google Scholar]
- Krainer A. R., Mayeda A., Kozak D., Binns G., 1991. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell 66: 383–394. [DOI] [PubMed] [Google Scholar]
- Krogan N. J., Dover J., Wood A., Schneider J., Heidt J., et al. , 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11: 721–729. [DOI] [PubMed] [Google Scholar]
- Kulkarni V. A., Firestein B. L., 2012. The dendritic tree and brain disorders. Mol. Cell. Neurosci. 50: 10–20. [DOI] [PubMed] [Google Scholar]
- Kuo T. Y., Hong C. J., Hsueh Y. P., 2009. Bcl11A/CTIP1 regulates expression of DCC and MAP1b in control of axon branching and dendrite outgrowth. Mol. Cell. Neurosci. 42: 195–207. [DOI] [PubMed] [Google Scholar]
- de la Mata M., Kornblihtt A. R., 2006. RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nat. Struct. Mol. Biol. 13: 973–980. [DOI] [PubMed] [Google Scholar]
- Laurent F. X., Sureau A., Klein A. F., Trouslard F., Gasnier E., et al. , 2012. New function for the RNA helicase p68/DDX5 as a modifier of MBNL1 activity on expanded CUG repeats. Nucleic Acids Res. 40: 3159–3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. G., 2002. RH70, a bidirectional RNA helicase, co-purifies with U1snRNP. J. Biol. Chem. 277: 39679–39683. [DOI] [PubMed] [Google Scholar]
- Lee J. H., Skalnik D. G., 2005. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J. Biol. Chem. 280: 41725–41731. [DOI] [PubMed] [Google Scholar]
- Lee M. H., Schedl T., 2006. RNA-binding proteins (April 18, 2006).WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.79.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeGendre J. B., Campbell Z. T., Kroll-Conner P., Anderson P., Kimble J., et al. , 2013. RNA targets and specificity of Staufen, a double-stranded RNA-binding protein in Caenorhabditis elegans. J. Biol. Chem. 288: 2532–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Hawkins I. C., Harvey C. D., Jennings J. L., Link A. J., et al. , 2003. Regulation of alternative splicing by SRrp86 and its interacting proteins. Mol. Cell. Biol. 23: 7437–7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Lee J. A., Black D. L., 2007. Neuronal regulation of alternative pre-mRNA splicing. Nat. Rev. Neurosci. 8: 819–831. [DOI] [PubMed] [Google Scholar]
- Liang S., Hitomi M., Hu Y. H., Liu Y., Tartakoff A. M., 1996. A DEAD-box-family protein is required for nucleocytoplasmic transport of yeast mRNA. Mol. Cell. Biol. 16: 5139–5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscombe D., 2005. Neuronal proteins custom designed by alternative splicing. Curr. Opin. Neurobiol. 15: 358–363. [DOI] [PubMed] [Google Scholar]
- Liu O. W., Shen K., 2012. The transmembrane LRR protein DMA-1 promotes dendrite branching and growth in C. elegans. Nat. Neurosci. 15: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Yu X., Zack D. J., Zhu H., Qian J., 2008. TiGER: a database for tissue-specific gene expression and regulation. BMC Bioinformatics 9: 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long R. M., Gu W., Meng X., Gonsalvez G., Singer R. H., et al. , 2001. An exclusively nuclear RNA-binding protein affects asymmetric localization of ASH1 mRNA and Ash1p in yeast. J. Cell Biol. 153: 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman D., Johnstone I. L., Cáceres J. F., 2000. Functional characterization of SR and SR-related genes in Caenorhabditis elegans. EMBO J. 19: 1625–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco R. F., Pan Q., Tominaga K., Blencowe B. J., Pereira-Smith O. M., et al. , 2010. Regulation of alternative splicing by histone modifications. Science 327: 996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapes J., Chen J. T., Yu J. S., Xue D., 2010. Somatic sex determination in Caenorhabditis elegans is modulated by SUP-26 repression of tra-2 translation. Proc. Natl. Acad. Sci. USA 107: 18022–18027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda A., Andersen H. S., Doktor T. K., Okamoto T., Ito M., et al. , 2012. CUGBP1 and MBNL1 preferentially bind to 3′ UTRs and facilitate mRNA decay. Sci Rep 2: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S., Longman D., Marcon E., Moens P., Downey M., et al. , 2005. Proteomic analysis of SRm160-containing complexes reveals a conserved association with cohesin. J. Biol. Chem. 280: 42227–42236. [DOI] [PubMed] [Google Scholar]
- Mello C., Fire A., 1995. DNA transformation. Methods Cell Biol. 48: 451–482. [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V., 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T., Krogan N. J., Dover J., Erdjument-Bromage H., Tempst P., et al. , 2001. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. USA 98: 12902–12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miniaci M. C., Kim J. H., Puthanveettil S. V., Si K., Zhu H., et al. , 2008. Sustained CPEB-dependent local protein synthesis is required to stabilize synaptic growth for persistence of long-term facilitation in Aplysia. Neuron 59: 1024–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedner A., Müller M., Moorthy B. T., Jansen R. P., Niessing D., 2013. Role of Loc1p in assembly and reorganization of nuclear ASH1 messenger ribonucleoprotein particles in yeast. Proc. Natl. Acad. Sci. USA 110: E5049–E5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris A. D., Calarco J. A., 2012. Emerging roles of alternative pre-mRNA splicing regulation in neuronal development and function. Front Neurosci 6: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesnicky E. C., Bhogal B., Gavis E. R., 2012. Combinatorial use of translational co-factors for cell type-specific regulation during neuronal morphogenesis in Drosophila. Dev. Biol. 365: 208–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesnicky E. C., Killian D. J., Garcia E., Morton M. C., Rathjen A. R., et al. , 2014. Extensive use of RNA-binding proteins in Drosophila sensory neuron dendrite morphogenesis. G3 (Bethesda) 4: 297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren-Suissa M., Hall D. H., Treinin M., Shemer G., Podbilewicz B., 2010. The fusogen EFF-1 controls sculpting of mechanosensory dendrites. Science 328: 1285–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne R. J., Lin X., Welle S., Sobczak K., O’Rourke J. R., et al. , 2009. Transcriptional and post-transcriptional impact of toxic RNA in myotonic dystrophy. Hum. Mol. Genet. 18: 1471–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Y., Chwalla B., Landgraf M., van Meyel D. J., 2008. Identification of genes influencing dendrite morphogenesis in developing peripheral sensory and central motor neurons. Neural Dev. 3: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S., Gupta R., Kim H., Wickramasinghe P., Baubet V., et al. , 2011. Alternative transcription exceeds alternative splicing in generating the transcriptome diversity of cerebellar development. Genome Res. 21: 1260–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F., Hüttelmaier S., Singer R. H., Gu W., 2007. ZBP2 facilitates binding of ZBP1 to beta-actin mRNA during transcription. Mol. Cell. Biol. 27: 8340–8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish J. Z., Kim M. D., Jan L. Y., Jan Y. N., 2006. Genome-wide analyses identify transcription factors required for proper morphogenesis of Drosophila sensory neuron dendrites. Genes Dev. 20: 820–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish J. Z., Xu P., Kim C. C., Jan L. Y., Jan Y. N., 2009. The microRNA bantam functions in epithelial cells to regulate scaling growth of dendrite arbors in drosophila sensory neurons. Neuron 63: 788–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual M., Vicente M., Monferrer L., Artero R., 2006. The Muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing. Differentiation 74: 65–80. [DOI] [PubMed] [Google Scholar]
- Pimentel J., Boccaccio G. L., 2014. Translation and silencing in RNA granules: a tale of sand grains. Front Mol Neurosci 7: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajyaguru P., Parker R., 2009. CGH-1 and the control of maternal mRNAs. Trends Cell Biol. 19: 24–28. [DOI] [PubMed] [Google Scholar]
- Risso G., Pelisch F., Quaglino A., Pozzi B., Srebrow A., 2012. Regulating the regulators: serine/arginine-rich proteins under scrutiny. IUBMB Life 64: 809–816. [DOI] [PubMed] [Google Scholar]
- Salzberg Y., Díaz-Balzac C. A., Ramirez-Suarez N. J., Attreed M., Tecle E., et al. , 2013. Skin-derived cues control arborization of sensory dendrites in Caenorhabditis elegans. Cell 155: 308–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasagawa N., Ohno E., Kino Y., Watanabe Y., Ishiura S., 2009. Identification of Caenorhabditis elegans K02H8.1 (CeMBL), a functional ortholog of mammalian MBNL proteins. J. Neurosci. Res. 87: 1090–1097. [DOI] [PubMed] [Google Scholar]
- Schäffler K., Schulz K., Hirmer A., Wiesner J., Grimm M., et al. , 2010. A stimulatory role for the La-related protein 4B in translation. RNA 16: 1488–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K., Butler J. S., 2013. Nuclear RNA surveillance: role of TRAMP in controlling exosome specificity. Wiley Interdiscip Rev RNA 4: 217–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt G. M., Tuebing F., Nigh E. A., Kane C. G., Sabatini M. E., et al. , 2006. A brain-specific microRNA regulates dendritic spine development. Nature 439: 283–289. [DOI] [PubMed] [Google Scholar]
- Si K., Giustetto M., Etkin A., Hsu R., Janisiewicz A. M., et al. , 2003. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in aplysia. Cell 115: 893–904. [DOI] [PubMed] [Google Scholar]
- Sims R. J., Millhouse S., Chen C. F., Lewis B. A., Erdjument-Bromage H., et al. , 2007. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol. Cell 28: 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. J., Watson J. D., Spencer W. C., O’Brien T., Cha B., et al. , 2010. Time-lapse imaging and cell-specific expression profiling reveal dynamic branching and molecular determinants of a multi-dendritic nociceptor in C. elegans. Dev. Biol. 345: 18–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. J., Watson J. D., VanHoven M. K., Colón-Ramos D. A., Miller D. M., 2012. Netrin (UNC-6) mediates dendritic self-avoidance. Nat. Neurosci. 15: 731–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. J., O’Brien T., Chatzigeorgiou M., Spencer W. C., Feingold-Link E., et al. , 2013. Sensory neuron fates are distinguished by a transcriptional switch that regulates dendrite branch stabilization. Neuron 79: 266–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilker K. A., Wang G. J., Tugizova M. S., Shen K., 2012. Caenorhabditis elegans Muscleblind homolog mbl-1 functions in neurons to regulate synapse formation. Neural Dev. 7: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S. J., Meulemans D., Vazquez L., Colaco N., Schuman E., 2001. A role for a rat homolog of staufen in the transport of RNA to neuronal dendrites. Neuron 32: 463–475. [DOI] [PubMed] [Google Scholar]
- Tenney K., Shilatifard A., 2005. A COMPASS in the voyage of defining the role of trithorax/MLL-containing complexes: linking leukemogensis to covalent modifications of chromatin. J. Cell. Biochem. 95: 429–436. [DOI] [PubMed] [Google Scholar]
- Thompson O., Edgley M., Strasbourger P., Flibotte S., Ewing B., et al. , 2013. The million mutation project: a new approach to genetics in Caenorhabditis elegans. Genome Res. 23: 1749–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tom Dieck S., Hanus C., Schuman E. M., 2014. SnapShot: local protein translation in dendrites. Neuron 81: 958–958.e1. [DOI] [PubMed] [Google Scholar]
- Tsalik E. L., Hobert O., 2003. Functional mapping of neurons that control locomotory behavior in Caenorhabditis elegans. J. Neurobiol. 56: 178–197. [DOI] [PubMed] [Google Scholar]
- Verster A. J., Ramani A. K., McKay S. J., Fraser A. G., 2014. Comparative RNAi screens in C. elegans and C. briggsae reveal the impact of developmental system drift on gene function. PLoS Genet. 10: e1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vessey J. P., Macchi P., Stein J. M., Mikl M., Hawker K. N., et al. , 2008. A loss of function allele for murine Staufen1 leads to impairment of dendritic Staufen1-RNP delivery and dendritic spine morphogenesis. Proc. Natl. Acad. Sci. USA 105: 16374–16379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalba A., Coll O., Gebauer F., 2011. Cytoplasmic polyadenylation and translational control. Curr. Opin. Genet. Dev. 21: 452–457. [DOI] [PubMed] [Google Scholar]
- Vorbrüggen G., Onel S., Jäckle H., 2000. Localized expression of the Drosophila gene Dxl6, a novel member of the serine/arginine rich (SR) family of splicing factors. Mech. Dev. 90: 309–312. [DOI] [PubMed] [Google Scholar]
- Wan J., Yourshaw M., Mamsa H., Rudnik-Schöneborn S., Menezes M. P., et al. , 2012. Mutations in the RNA exosome component gene EXOSC3 cause pontocerebellar hypoplasia and spinal motor neuron degeneration. Nat. Genet. 44: 704–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E. T., Cody N. A., Jog S., Biancolella M., Wang T. T., et al. , 2012. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell 150: 710–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. C., Hung W. T., Pan H., Chen K. Y., Wu Y. C., et al. , 2008. Growth-dependent effect of muscleblind knockdown on Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 366: 705–709. [DOI] [PubMed] [Google Scholar]
- Waterhouse R. M., Tegenfeldt F., Li J., Zdobnov E. M., Kriventseva E. V., 2013. OrthoDB: a hierarchical catalog of animal, fungal and bacterial orthologs. Nucleic Acids Res. 41: D358–D365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J. D., Wang S., Von Stetina S. E., Spencer W. C., Levy S., et al. , 2008. Complementary RNA amplification methods enhance microarray identification of transcripts expressed in the C. elegans nervous system. BMC Genomics 9: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way C., Chalfie M., 1989. The mec-3 gene of Caenorhabditis elegans requires its own product for maintained expression and is expressed in three neuronal cell types. Genes Dev. 3: 1823–1833. [DOI] [PubMed] [Google Scholar]
- White E., Schlackow M., Kamieniarz-Gdula K., Proudfoot N. J., Gullerova M., 2014. Human nuclear Dicer restricts the deleterious accumulation of endogenous double-stranded RNA. Nat. Struct. Mol. Biol. 21: 552–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D. W., Truman J. W., 2005a Cellular mechanisms of dendrite pruning in Drosophila: insights from in vivo time-lapse of remodeling dendritic arborizing sensory neurons. Development 132: 3631–3642. [DOI] [PubMed] [Google Scholar]
- Williams D. W., Truman J. W., 2005b Remodeling dendrites during insect metamorphosis. J. Neurobiol. 64: 24–33. [DOI] [PubMed] [Google Scholar]
- Xiao Y., Bedet C., Robert V. J., Simonet T., Dunkelbarger S., et al. , 2011. Caenorhabditis elegans chromatin-associated proteins SET-2 and ASH-2 are differentially required for histone H3 Lys 4 methylation in embryos and adult germ cells. Proc. Natl. Acad. Sci. USA 108: 8305–8310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Strome S., 2001. Depletion of a novel SET-domain protein enhances the sterility of mes-3 and mes-4 mutants of Caenorhabditis elegans. Genetics 159: 1019–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Gaidamakov S. A., Xie J., Lee J., Martino L., et al. , 2011. La-related protein 4 binds poly(a), interacts with the poly(a)-binding protein MLLE domain via a variant PAM2w Motif, and can promote mRNA sability. Mol. Cell. Biol. 31: 542–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B., Petritsch C., Clark I. E., Gavis E. R., Jan L. Y., et al. , 2004. Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr. Biol. 14: 314–321. [DOI] [PubMed] [Google Scholar]
- Yeo G., Holste D., Kreiman G., Burge C. B., 2004. Variation in alternative splicing across human tissues. Genome Biol. 5: R74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochem J., Bell L. R., Herman R. K., 2004. The identities of sym-2, sym-3 and sym-4, three genes that are synthetically lethal with mec-8 in Caenorhabditis elegans. Genetics 168: 1293–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Mangelsdorf M., Liu J., Zhu L., Wu J. Y., 2014. RNA-binding proteins in neurological diseases. Sci China Life Sci 57: 432–444. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.