Abstract
Transgenesis in numerous eukaryotes has been facilitated by the use of site-specific integrases to stably insert transgenes at predefined genomic positions (landing sites). However, the utility of integrase-mediated transgenesis in any system is constrained by the limited number and variable expression properties of available landing sites. By exploiting the nonstandard recombination activity exhibited by a phiC31 integrase mutant, we developed a rapid and inexpensive method for isolating landing sites that exhibit desired expression properties. Additionally, we devised a simple technique for constructing arrays of transgenes at a single landing site, thereby extending the utility of previously characterized landing sites. Using the fruit fly Drosophila melanogaster, we demonstrate the feasibility of these approaches by isolating new landing sites optimized to express transgenes in the nervous system and by building fluorescent reporter arrays at several landing sites. Because these strategies require the activity of only a single exogenous protein, we anticipate that they will be portable to species such as nonmodel organisms, in which genetic manipulation is more challenging, expediting the development of genetic resources in these systems.
Keywords: integrase, landing site, phiC31, shuffle, transgene array
THE ability to introduce exogenous DNA sequences (transgenes) into the genome of an organism or cell line (transgenesis) underpins much of modern molecular genetics and experimental biology. Transgenes are used to label cells, to manipulate physiology and behavior, and to test hypotheses about gene function. Hence, it is essential that the DNA sequences carried within a transgene be expressed at functional levels and with high spatial and temporal specificity. Yet endogenous regulatory elements frequently exert a strong influence over the expression of a locally inserted transgene, and it is common for two insertions of the same transgene to behave differently due to their distinct local chromatin environments (Lewis 1950; Spradling and Rubin 1983; Levis et al. 1985; Akhtar et al. 2013). Such position effects (PE) may lead to spatial and temporal mis-regulation, overexpression, or silencing of transgenes, which can seriously confound the interpretation of experimental results.
Transgenesis strategies that allow researchers to control the genomic position of transgenes present an opportunity to standardize PE across transgenes and experiments. Among these, site-specific transgene integration, mediated by a bacteriophage-derived integrase, is an especially popular technique due to its high rate of transgenesis and ability to function in a broad range of species (Smith et al. 2010; Geisinger and Calos 2013). The integrase from phiC31 has been deployed in multiple eukaryotes, including fruit flies (Groth et al. 2004), zebrafish (Mosimann et al. 2013), tobacco (Lutz et al. 2004), and cultured human cells (Groth et al. 2000). Three components are required to integrate a transgene. The first component, the integrase protein (Int), mediates recombination between two specific DNA sequences called attP and attB (Kuhstoss and Rao 1991). The second component is the transgene to be integrated, which carries the attB sequence. The third component consists of a stable, molecularly mapped transposon insertion that carries the attP sequence; this genomic “landing site” serves as the platform into which the transgene is integrated. This organization gives Int-mediated transgenesis incredible flexibility: any transgene with attB can be integrated, and the genomic position of a transgene can be controlled based on landing site selection. Yet despite its considerable advantages, Int-mediated transgenesis is constrained in many systems by a lack of well-characterized landing sites.
The primary measure of landing site quality is whether integrated transgenes express faithfully and at experimentally relevant levels. In the fruit fly Drosophila melanogaster, a number of groups have constructed collections of landing sites (Groth et al. 2004; Venken et al. 2006; Bischof et al. 2007; Markstein et al. 2008; Ni et al. 2009), and two studies have explored the prevalence of PE by comparing the expression of reporter transgenes integrated at multiple landing sites across the genome (Markstein et al. 2008; Pfeiffer et al. 2010). Using a Gal4-inducible luciferase transgene integrated at 20 different landing sites, Markstein et al. (2008) observed both variable Gal4-induced expression and “leaky” expression of luciferase in the absence of Gal4. Furthermore, for a given landing site, the authors observed that the degree of luciferase induction in one tissue or cell type was not predictive of expression in other tissues or cell types (Markstein et al. 2008). Given that many landing sites (perhaps most) are subject to undesirable PE (Markstein et al. 2008; Pfeiffer et al. 2010), it is clear that landing sites should be tested for deleterious PE in the cell types or developmental stages of interest before embarking on an experiment. However, assessing large numbers of landing sites is time- and resource-intensive (Markstein et al. 2008; Pfeiffer et al. 2010) and imposes a substantial burden on the research community. Thus, a strategy that streamlines the screening of landing site candidates would expedite the generation of transgenic resources tailored to specific experimental needs.
Our efforts to devise such a strategy led us to consider alternate enzymatic activities that might complement the normal integration function supplied by Int. Several integrase mutants that exhibit atypical recombination properties have been reported previously; these mutants can catalyze the excision of an integrated sequence and promiscuous recombination in vitro between nonstandard att site pairs (Rowley et al. 2008). We reasoned that the unique properties of these mutants might support the development of novel Int-mediated transgenesis techniques. For example, a mutant Int capable of regenerating a functional landing site by excising a previously integrated reporter would obviate several of the steps required to characterize new landing sites. Likewise, a hyperactive mutant Int, capable of catalyzing promiscuous recombination between attB and one of the hybrid att sequences that flank an integrated transgene, might be useful for inserting an additional transgene at an occupied landing site. Constructing transgene arrays in this fashion would mitigate the need for a large number of landing sites, while simultaneously simplifying complex genetic schemes. These considerations prompted us to investigate the capabilities of several integrase mutants in vivo using D. melanogaster.
From a panel of eight integrase mutants (collectively referred to as “Int*”), we identified several that exhibited high levels of excision and promiscuous recombination. Int* allowed us to develop new techniques and genetic resources in the fly: Using a novel approach that leverages the excisionase activity of Int*, we generated a set of seven new landing sites that are optimized to express transgenes in the central nervous system of adult Drosophila. Notably, we were able to characterize the expression properties of potential landing sites in half as many generations and at a fraction of the cost compared to using traditional methodology. Moreover, this method can be used to isolate landing sites optimized to any desired criteria. We also devised a protocol that employs Int* (as excisionase) and wild-type Int (as integrase) to relocate a previously integrated transgene to a different landing site on another chromosome. Finally, we exploited the relaxed recombination specificity of the Int* integrase activity to construct two-component transgene arrays at several landing sites. These techniques have the potential to significantly extend the integrase-mediated transgenesis platform.
Materials and Methods
Fly husbandry and Drosophila stocks
Flies were reared at 21°–25° on standard cornmeal/molasses food. Crosses were heat-shocked in vials at 37° for 1 hr in a circulating water bath. Injections were performed by Rainbow Transgenic Flies, Inc. (Camarillo, CA) using standard phiC31 transgenesis protocols, unless otherwise indicated.
The following insertions were generated for this study: the genetic sources of Int* ZH-51C[hs-Int*], ZH-86Fb[hs-Int*], P{CaryP}attP2[R57C10-Gal4(w)], P{CaryP}attP2[pJFRC-13xLexAop2-nls-LacZ], and P{CaryP}attP2[pJFRC-10xUAS-tdTomato-nls]. To make ZH-51C[hs-Int*] and ZH-86Fb[hs-Int*], embryos carrying M{vas-int.Dm}ZH-2A; M{3xP3-RFP.attP}ZH-51C or M{vas-int.Dm}ZH-2A; M{3xP3-RFP.attP}ZH-86Fb were injected with the Int* expression construct. G0 adults were crossed to w1118, and integrants were detected among the F1 progeny by mini-white expression. To stabilize the hs-Int* transgene, males with the integrated transgene were crossed to virgins carrying hs-Cre on the homologous chromosome. Crosses were raised at 25° without heat shock, and hs-Int*/hs-Cre progeny males were crossed to balancer virgins. In the F2 generation, flies carrying stabilized hs-Int* were identified by the absence of mini-white.
In addition, we used genotypes including the following insertions, described elsewhere: from Bischof et al. (2007)—M{3xP3-RFP.attP}ZH-51C and M{3xP3-RFP.attP}ZH-86Fb and P{nos-phiC31\int.NLS}X; from Groth et al. (2004)—P{CaryP}attP2; from Markstein et al. (2008)—P{CaryP}attP40; from Pfeiffer et al. (2010)—pJFRC19 integrated into P{CaryP}attP40 and P{CaryP}attP2; pJFRC12 integrated into P{CaryP}attP2; pJFRC2 integrated into the landing sites P{CaryP}attP18, P{CaryP}attP40, P{CaryP}attP2, P{CaryIP}su(Hw)attP8; P{CaryIP}su(Hw)attP5; P{CaryIP}su(Hw)attP1; P{CaryIP}su(Hw)attP2; and PBAC{y+-attP-3B}VK00005; from Jenett et al. (2012)—R11C05-Gal4, R32A11-Gal4, and R42E06-Gal4 integrated into P{CaryP}attP2. Recombinant chromosome dsxGAL4.∆2, fruP1.LexA was a gift of Y. Pan; details on the construction of the two knock-ins can be found in Pan et al. (2011) and Mellert et al. (2010), respectively. Finally, we used the following unpublished reagents: R11C05-LexA integrated into P{CaryP}attP18, P{CaryP}attP40, and P{CaryP}attP2 (a gift of A. Jenett and G. Rubin). Flies that were y1 w1118; CyO, PBac{Delta2-3.Exel}2/amosTft (Bloomington Drosophila Stock Center #8201) were used to mobilize P-element-based landing sites to generate new candidates. Flies that were y1 w67c23; MKRS, P{hsFLP}86E/TM6B, P{Crew}DH2, Tb1 (Bloomington Drosophila Stock Center #1501), and y1 w67c23; snaSco/CyO, P{Crew}DH1 (Bloomington Drosophila Stock Center #1092) were used to stabilize hs-Int* integrants.
Cloning and molecular biology
Int* expression constructs:
Int* variants were synthesized by megaprimer PCR (Ke and Madison 1997) with slight modifications, using pET11-phiC31 (gift of Michele Calos) as the template. First, the Int-coding sequence (CDS) was amplified in separate reactions as incomplete 5′ and 3′ fragments using primers that hybridized to the ends of the Int CDS paired with internal primers designed to introduce the desired point mutation(s). A 1:1 mixture of these two fragments, which overlap in the region to be mutated, served as the template for a second round of amplification using the 5′ and 3′ terminal primers to extend the full-length Int* sequence. The CDS for each Int* variant was subcloned, sequence verified, and then cloned into pMGX-loxP-hsP or pMGXw+yR-hsP. These vectors were derived by replacing the UAS sites and minimal promoter of JFRC2 with the hsp70Bb promoter (Ingolia et al. 1980) and the mCD8-GFP sequence with the Int* CDS. Additionally, pMGXw+yR-hsP contains a DsRed transgene marker between the loxP site and the hsp70Bb promoter.
New reporter constructs:
The CDS for β-galactosidase fused C-terminally to the SV40 nuclear localization signal (nls) was amplified from pNLZ (gift of Y.-L. Huang), sequence verified, and cloned into pJFRC19 (Pfeiffer et al. 2010) in place of myr-GFP, yielding pJFRC-13xLexAop3-nls-LacZ. To clone pJFRC-13xLexAop2-GFP-nls, the CDS for Stinger GFP was amplified from pStinger (Barolo et al. 2000), sequence verified, and then cloned into pJFRC19 (Pfeiffer et al. 2010) in place of myr-GFP. To construct pJFRC-10xUAS-tdTomato-nls, the CDS for tdTomato was amplified minus the stop codon from genomic DNA of flies carrying a codon-optimized tdTomato reporter (gift of B. Pfeiffer). Separately, the tra nls (Barolo et al. 2000) was amplified from w1118 genomic DNA, adding 5′ homology to tdTomato, which placed the NLS in-frame. These fragments were mixed at a 1:1 ratio and used as a template for overlap PCR. The resulting full-length tdTomato-nls CDS was sequence-verified and then cloned into pJFRC2 (Pfeiffer et al. 2010) in place of mCD8-GFP. Removing the mini-white marker from this construct produced pJFRC-10xUAS-tdTomato-nls(w). The plasmid pJFRC-10xUAS-mCD8-tdTomato was a gift of S. Hampel; removing the mini-white marker yielded pJFRC-10xUAS-mCD8-tdTomato(w).
New driver construct:
The plasmid pBPGw-R57C10-Gal4(w) was derived from pBPGw-R57C10-Gal4 (gift of G. Rubin) by removing the mini-white marker.
Genomic DNA prep:
For molecular characterization of new candidate landing sites and Int*-mediated recombination events, genomic DNA was prepared from flies according to a previously published protocol (Schuldiner et al. 2008).
Excisionase assays
Males bearing hs-Int* were crossed to virgins homozygous for P{CaryP}attP18[pJFRC2]. Crosses were heat-shocked to induce Int* when larvae reached the wandering stage, and then F1 males were crossed to white (w1118) virgins. All female progeny in the F2 generation were scored for mini-white (absence indicates loss of the transgene) to calculate the excision frequency for each Int* variant. Excisionase assays using other landing sites were conducted similarly. For excisionase assays using a nongenetic source of Int*, embryos with the genotypes w; JK22C[R11C05-LexA] or w; attP2[13xLexAop2-nls-LacZ] were injected with 3 μg of a helper plasmid encoding Int*-KEE. Adults were crossed to w; amosTft/CyO or w; TM3/TM6B, respectively, and the F1 progeny were scored for mini-white (absence indicates loss of transgene). Int* helper plasmids were derived from the expression constructs (see above) by deleting the attB site and mini-white marker and replacing the SV40 terminator with the 3′ UTR of the germline-expressed bag of marbles gene.
Re-integration assays at reconstituted landing sites
Stocks were established from single males bearing reconstituted P{CaryP}attP40 and P{CaryP}attP2. To compare the integration efficiency of reconstituted and native landing sites, four injection replicates were performed to integrate additional transgenes. For each replicate, all four genotypes (native attP2 and attP40 and reconstituted attP2 and attP40) were injected with the same transgene on the same day. The transgenes used included pJFRC2-10xUAS-mCD8-GFP (twice, on separate days; Pfeiffer et al. 2010), pJFRC-10xUAS-tdTomato-nls(w), and pJFRC-10xUAS-mCD8-tdTomato(w). G0 adults were crossed to w1118 or w; attP2[R57C10-Gal4], respectively, and F1 progeny were screened for integrants by eye color or fluorescence. Separately, standard transgenesis procedures were used to integrate each of the following plasmids into several additional reconstituted landing sites: pBPGUw-R9C11-Gal4 (Pfeiffer et al. 2010), pBPGw-R57C10-Gal4 (Jenett et al. 2012), pBPLexA::p65uw-R32A11-LexA, and pBPLexA::p65uw-R42E06-LexA. The latter two plasmids were a gift of H. Dionne and G. Rubin.
Landing site remobilization and mapping of new insertions
The X-linked insertion P{CaryP}attP18[R11C05-LexA] was remobilized by crossing homozygous virgins to males that express transposase from PBac{Delta2-3.Exel}2, located on the CyO balancer (Bloomington Drosophila Stock Center #8201). Dysgenic male progeny (F1 generation) were crossed to w1118 virgins. In the F2 generation, male progeny that express mini-white (the R11C05-LexA transgene marker) and lack CyO carry novel, stable insertions, which were mapped by standard genetic methods and tested for homozygous viability and fertility. For selected insertions, flanking genomic DNA was amplified by splinkerette PCR (Potter and Luo 2010) at both the 5′ and 3′ ends, sequenced, and aligned to the Drosophila genome by BLAST.
Transgene shuffling
pJFRC19-LexAop2-myr-GFP was genetically shuffled between the landing sites attP40→ZH-86Fb or attP2→ZH-51C using hs-Int* in ZH-86Fb or ZH-51C, respectively. For some experiments, the X-linked insertion P{nos-phiC31\int.NLS}X (Bischof et al. 2007) provided a source of wild-type Int. Virgins carrying the donor and receiver landing sites were crossed to males carrying hs-Int* and marked chromosomes on the other major autosome (e.g., males with hs-Int* at ZH-51C also had MKRS/TM6B). After 3 days, crosses were transferred to fresh vials to start the experiment. Following transfer, crosses were flipped daily for 5 days, resulting in a set of vials for each cross with progeny at the embryonic, L1, L2, or L3 stages. These were heat-shocked and permitted to complete development. Adults were separated at eclosion and crossed within 3 days to w1118. Progeny carrying candidate shuffle events were identified by mini-white expression and the presence of CyO (attP40-donor experiment) or TM6B (attP2-donor experiment). Shuffled transgenes were tested for GFP expression by crossing to a pan-neural LexA. The re-integration site was molecularly verified by PCR with a pair of primers that hybridized within the landing site and the shuffled transgene, respectively.
Two-component array construction
Embryos homozygous for the primary transgene were co-injected with the plasmid bearing the secondary transgene and a helper plasmid encoding Int* in a 3:1 ratio, for a total of 16 μg DNA per injection. G0 adults were crossed to a screening stock (specified below); F1 progeny were screened for the presence of both the primary and the secondary transgenes. For injections presented in Figure 5B, the primary and secondary transgenes and screening stock were as follows (primary | secondary | screening): pair 1—attP2[13xLexAop-nls-lacZ] | pJFRC-10xUAS-tdTnls(w) | attP2[R57C10-gal4(w)]; pair 2—attP2[13xLexAop-nls-lacZ] | pJFRC-Brev-10xUAS-tdTnls(w) | attP2[R57C10-gal4(w)]; pair 3—attP2[13xLexAop-nls-lacZ] | pJFRC-13xLexAop2-GFP-nls | w1118; pair 4—attP2[R11C05-LexA] | pBDP-R57C10-Gal4 | attP2[10xUAS-tdTomato-nls(w)]; and pair 5—attP2[R11C05-LexA] | pBDP-R9C11-Gal4 | attP2[10xUAS-tdTomato-nls(w)]. The reporter array characterized in Figure 5C was obtained from the pair 1 injections, and these flies had the genotype yw; P{CaryP}attP2[pJFRC-13xLexAop2-nlsLacZ+pJFRC-10xUAS-tdTomato::nls(w)]. The reporter array presented in Figure 5D was constructed in the su(Hw)attP8 landing site (Ni et al. 2009), using pJFRC15 as the primary transgene (Pfeiffer et al. 2010) and pJFRC-10xUAS-mCD8::tdTomato as the secondary transgene. The genotype of these flies was yw P{CaryIP}su(Hw)attP8[pJFRC15 + pJFRC-10xUAS-mCD8::tdTomato].
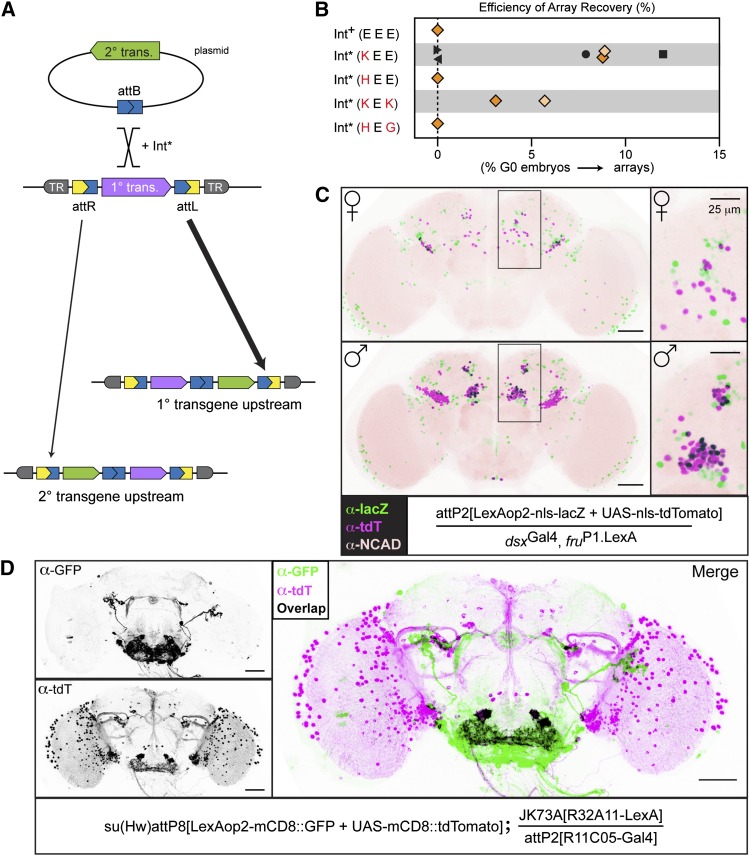
Figure 5.
Int* can be used to assemble arrays of transgenes in a single landing site. (A) Int*-mediated transgene array construction. The hyperactive integrase activity of Int* catalyzes recombination between one of the att sequences flanking the primary transgene (purple) and the attB sequence associated with the secondary transgene (green). In the resulting array, the primary and secondary transgenes are separated by a reconstituted attB sequence, and the entire array is flanked by attR and attL sequences. The site where the secondary transgene integrates determines the order of transgenes in the array; the frequency at which we observed recombination at attL or attR is represented by the width of the arrow under each site (also see Figure S7B). (B) Int*-KEE is superior to other Int* mutants for constructing transgene arrays. The indicated variants were tested for the ability to produce transgene arrays using several primary/secondary transgene pairs. (For an overview of the genetics, see Figure S7A.) The efficiency of array recovery was calculated by dividing the number of G0 crosses yielding an array to the total number screened. With one exception, each data point represents a separate injection with at least 20 germlines screened; Int*-HEE represents only 10 germlines. The first and second rounds of the pilot experiment are indicated by dark and light orange symbols, respectively. Symbols with the same shape indicate experiments with primary and secondary transgenes in common: pair 1, diamonds; pair 2, circle; pair 3, square; pair 4, right-facing arrowhead; pair 5, left-facing arrowhead. See Materials and Methods for transgene pair details. (C) The components of a LexAop2-nls-lacZ/UAS-tdTomato-nls reporter array retain transcriptional independence. Colabeling with nuclear-localized nls-LacZ (green) and tdTomato-nls (magenta) in the adult posterior brain (boxed regions and insets at right) was observed in males but not females, consistent with the published characterization of dsxGAL4.∆2 and fruP1.LexA (Zhou et al. 2014). Cells expressing both reporters appear black. Bars, 100 μm (left); 25 μm (right). (D) The components of a LexAop2-mCD8-GFP/UAS-mCD8-tdTomato reporter array retain transcriptional independence. In combination with R11C05-Gal4 and R32A11-LexA, the transgene array expresses both GFP and tdTomato in spatially distinct patterns, as shown in the single-channel gray-scale immunostaining images on the left. The merged image on the right reveals four cells in the sub-esophageal zone that co-express R11C05-Gal4 and R32A11-LexA. Bar, 100 μm.
Immunohistochemistry
Adult brain and ventral nerve cords were dissected from 3- to 7-day-old females (unless otherwise indicated) in 1× phosphate buffered saline (PBS). Tissue was fixed with 2% paraformaldehyde (Sigma, P1213) overnight (∼16 hr) at 4° with gentle rocking. Following fixation, samples were washed 3× for 20 min each with PAT [1× PBS, 1% BSA (Sigma A-6793), and 0.5% Triton (Sigma X-100)] and then blocked with 3% normal goat serum (NGS) (BioSource, PCN5000) in PAT for 1 hr at room temperature. Samples were incubated overnight with primary antibodies diluted in PAT+NGS at 4° with gentle nutation. Next, samples were washed 3× for 20 min each with PAT and then incubated overnight with secondary antibody diluted in PAT+NGS. Finally, samples were washed again 3× for 20 min each with PAT, rinsed with 1× PBS, and then mounted in VectaShield (Vector Laboratories, Inc., H-1000). Prior to imaging, mounted samples were kept at room temperature for 1 hr or stored at −20°. Primary antibodies used included rat anti-DN-Cadherin [1:50, Developmental Studies Hybridoma Bank (DSHB), DN-Ex #8], mouse anti-β-galactosidase (1:50, DSHB, 40-1a), rabbit anti-DsRed (1:500, Clontech #632496), rabbit anti-GFP (1:500, Invitrogen #A11122), and mouse anti-GFP (1:200, Sigma #G6539). The following goat secondary antibodies from Invitrogen/Life Technologies were used at 1:500 dilution: Alexa Fluor 488 anti-rabbit (A11034), Alexa Fluor 568 anti-rat (A11077), Alexa Fluor 633 anti-rat (A21094), Alexa Fluor 568 anti-rabbit (A11036), and Alexa Fluor 488 anti-mouse (A11029).
Quantitative imaging
For a given experiment, flies were age-matched and dissected on the same day, and then samples were processed identically for immunostaining and imaging. Samples were imaged as 12-bit stacks using either a Zeiss LSM 510 Pascal or a Zeiss LSM 700 laser-scanning confocal microscope. To prepare images for publication, FIJI was used to convert stacks to 8-bit, prepare maximum projections, apply Look-Up Tables (LUTs), and merge channels when applicable. Processed images were saved as TIFFs and then cropped in Adobe Photoshop.
Visual screen of new landing site candidates
Males carrying new insertions of P{CaryP}attP[R11C05-LexA] were crossed to virgins homozygous for P{CaryP}attP2[pJFRC19], which expresses LexA-inducible, myristoylated GFP reporter (Pfeiffer et al. 2010). Progeny were dissected in PBS, and brains were immediately inspected for GFP expression using a fluorescence dissection scope (Olympus, model BX51W1). For each candidate insertion, two to four brains were scored for GFP expression in the ellipsoid body, sub-esophageal zone, optic lobes, and projections from the optic lobes using a qualitative scale. Candidates were discarded for weak or aberrant expression.
Brain alignment and quantification of GFP expression
Brains were dissected from 4- to 7-day-old R11C05-LexA > LexAop2-myrGFP females, fixed, stained with anti-DN-Cad to reveal the neuropil, and then quantitatively imaged for native GFP and neuropil using a confocal microscope. Stacks were aligned to a standard reference brain (Jenett et al. 2012) by neuropil signal, using the JBA algorithm (Peng et al. 2011). Maximum projections were prepared from aligned stacks; a custom FIJI script was used to collect mean fluorescence intensity from the GFP channel for five regions of the image. These included two blank regions to permit normalization of background fluorescence and three regions overlapping the ellipsoid body, the left sub-esophageal zone, and the left optic lobe.
Results
Site-specific transgenesis permits position effects to be standardized across transgenes and obviates the need to map new insertions. One popular method uses the integrase from phiC31 bacteriophage to integrate transgenes at defined genomic positions that contain a phage-derived DNA sequence called attP (Figure 1A) (Groth et al. 2004), which typically has been inserted into the genome randomly via a transposon. Such insertions, called landing sites, serve as platforms into which any transgene bearing the compatible attB sequence can be integrated. To integrate a transgene, Int recombines attP (in the landing site) with the corresponding attB sequence on the plasmid (Figure 1A). Following integration, the transgene is flanked by the hybrid sequences attL and attR, which are each composed of a half-site from both attP and attB (Figure 1A).
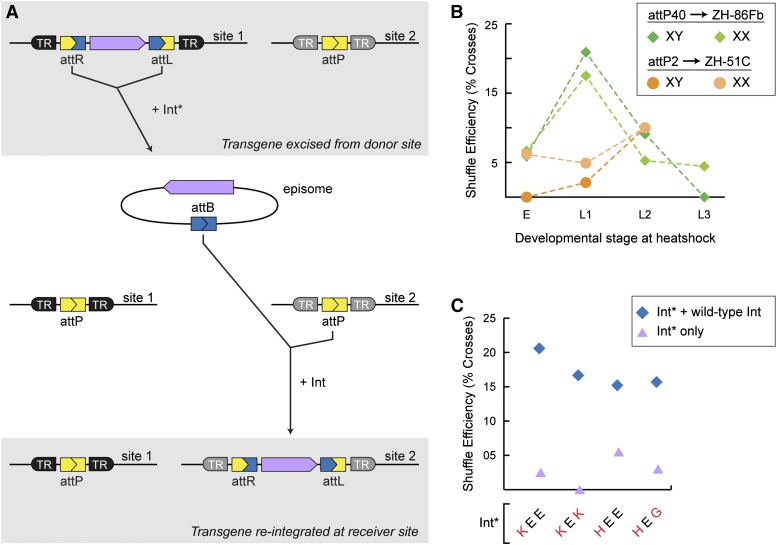
Figure 1.
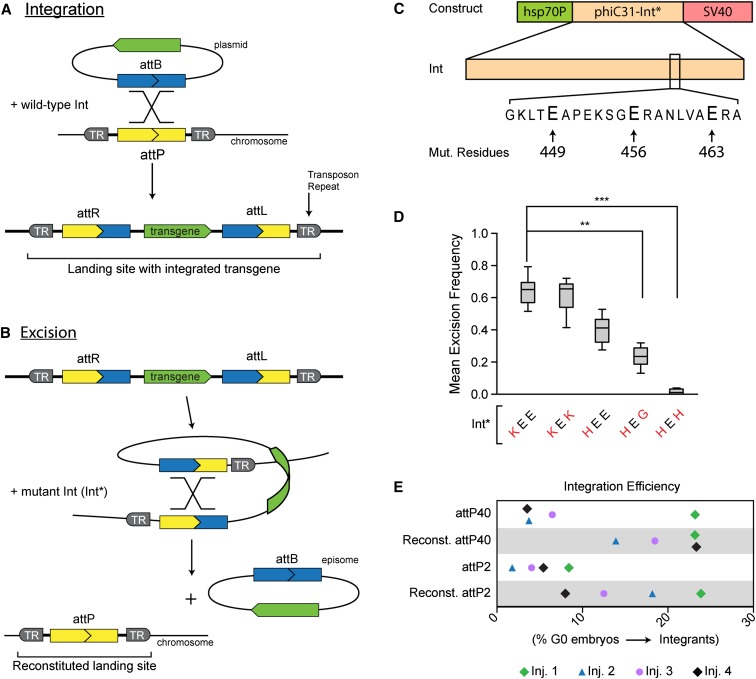
Mutant phiC31-Int (Int*) regenerates functional landing sites in vivo. (A) Wild-type integrase (Int) mediates recombination between the attP and attB sequences. The transgene becomes stably integrated into the chromosome by recombining the attP of a landing site (introduced into the genome via a transposon) with the attB on the plasmid. (B) Mutant integrase (Int*) mediates recombination between the attL and attR sequences flanking the integrated transgene, leading to reconstitution of attP at the landing site and concomitant release of an episome that corresponds to the originally integrated plasmid. (C) The generic expression construct for Int* variants. The hsp70Bb promoter drives Int* expression, and the SV40 poly(A) signal terminates the message. (Bottom) The glutamate residues that were mutated to construct Int* variants. All three residues lie on one face of a coiled-coil motif that is thought to regulate interactions between integrase dimers (Rowley et al. 2008). (D) Excisionase activity of Int* variants in vivo. Int* variants were tested in the male germline for their ability to excise a mini-white-marked transgene (see Figure S1B for crossing scheme). Excision frequency was calculated as the number of female progeny lacking mini-white (the transgene marker) divided by the total number of female progeny. Mean excision frequency was computed by averaging results from 10 crosses. For a comparison of all mutants tested, see Table S1. (E) Transgene integration at reconstituted landing sites. Integration efficiency between native and reconstituted landing sites was compared by targeting each landing site in four separate transgenesis trials (each trial consisted of injections into native and reconstituted attP40 and attP2). Color denotes injections performed on the same day. pJFRC2 (diamonds); pJFRC-10xUAS-tdTomato-nls(w) (circles); pJFRC-10xUAS-mCD8-GFP(w) (triangles). Integration efficiency reflects the number of G0 germlines that gave rise to integrants divided by the number of germlines screened. Each data point typically represents >25 germlines scored, although some injections resulted in fewer fertile, surviving G0 flies. The lower integration efficiencies of the native landing sites may be due to differences in genetic background between these strains and those bearing reconstituted sites.
Since Int cannot recombine attL and attR without an additional viral cofactor, the integration reaction is unidirectional (Kuhstoss and Rao 1991; Khaleel et al. 2011; Farruggio et al. 2012). This feature is beneficial when integrating transgenes, but the ability to excise a previously integrated transgene and reconstitute a functional landing site could be advantageous in other circumstances. Previously, several Int mutants that can catalyze the excision reaction in vitro in the absence of the viral cofactor have been reported (Rowley et al. 2008). We characterized these and additional mutants in vivo for use in three novel techniques that expand the genetic toolkit available in Drosophila.
Mutant integrase reconstitutes functional landing sites in vivo
No DNA sequence is gained or lost during the integration reaction (Kuhstoss and Rao 1991). Thus, reversing the reaction should reconstitute an intact attP sequence (i.e., a functional landing site) by excising an episome that corresponds to the originally integrated transgene (Figure 1B). To establish such an excisionase activity in vivo, we constructed a series of transgenic constructs that express different phiC31 integrase variants (collectively referred to as “Int*”) under the control of a heat-shock promoter (Figure 1C). The Int* variants featured combinations of mutated glutamate residues at positions 449, 456, and 463 (Figure 1C), including three previously reported single mutants (Rowley et al. 2008) and five additional double- or triple-point mutants (Supporting Information, Table S1A). Expression constructs were integrated into the same landing site so Int* variants could be compared without confounding PE. To prevent the Int* transgene from self-excising, we used a Cre-dependent strategy to delete the attR site from one end of the integrated transgene, rendering it immune to the activity of Int* (Figure S1A).
We determined the frequency with which Int* expression induced germline loss of a mini-white-marked transgene integrated into the attP18 landing site. Briefly, males bearing an Int* expression construct and the X-linked transgene were subjected to larval heat shock to induce Int* and then crossed to white virgins upon eclosion (Figure S1B). We inferred the excision frequency for each Int* variant by counting the number of female progeny lacking mini-white. While some Int* variants had virtually undetectable excision, most exceeded 10% (Figure 1D and Table S1A). Two variants excised the tester transgene from more than half of chromosomes. We observed comparable rates of excision in the female germline (data not shown). For the mutants comprising our panel, the major determinant of excisionase activity seems to be the basic character of the residue at position 449. Hence, Int* variants with lysine at position 449 were generally more active than those with histidine, which were generally more active than those with glycine (Table S1A). These results mirror the in vitro activity trend that was obtained when E449 was systematically mutated (Rowley et al. 2008). We also asked whether a nongenetic source of Int* could drive transgene excision from two autosomal landing sites. Embryos were injected with a helper plasmid encoding the most active variant, Int*-KEE. Surviving adults were crossed to white flies, and progeny were assessed for mini-white. We observed appreciable excision, although at reduced rates relative to genetic sources of Int* (data not shown). Finally, to gauge the accessibility of landing sites in the genome generally, we assayed transgene excision frequency from six additional landing sites, including one on each major chromosome arm. Int* successfully excised the tester at each landing site (Table S1B), from which we infer that most sites in the genome are accessible to Int*.
Given our goal of reconstituting functional landing sites in vivo, it is essential that Int*-mediated excision leaves an intact attP site. To verify the integrity of reconstituted attPs following Int*-mediated tester excision, we analyzed DNA sequences from individual progeny males in the immediate vicinity of the anticipated recombination site. Sequencing across this region revealed that each chromosome harbored a perfect attP (18/18 independent events; data not shown). These results predict that the integration efficiency of a reconstituted landing site should be equivalent to the corresponding native site that had never hosted a transgene. We tested this by integrating transgenes into reconstituted attP40 and reconstituted attP2, as well as into native attP40 and attP2. Three different transgenes were separately injected into embryos harboring each attP site. For each injection, integration efficiency was calculated as the ratio of recovered integrants to G0 germlines scored (Figure 1E). While there is significant spread in the data owing to the inherent variability of injections, transgenes were integrated into reconstituted landing sites as efficiently as native sites, consistent with our expectation that reconstituted landing sites are fully functional.
Int* facilitates the identification of landing sites with desirable expression properties
The degree to which a given landing site can support robust and spatiotemporally appropriate expression of resident transgenes is a major consideration in experimental design. Indeed, there is clear evidence that expression properties are variable between landing sites and even vary for a single landing site examined in multiple tissues (Markstein et al. 2008; Pfeiffer et al. 2010). Currently, identifying landing sites with desirable expression properties can be accomplished only through empirical characterization of each landing site in a collection. This requires that a reporter transgene be separately integrated into each landing site, an investment of labor and time that is likely a disincentive to many researchers. A strategy that bypasses this requirement would facilitate screening large numbers of landing sites and enable researchers to isolate purpose-built landing sites.
We employed Int*-mediated transgene excision to facilitate the identification of landing sites with desirable expression properties. First, we mobilized a transposon-born landing site containing an integrated reporter transgene (referred to as attP[tester]) to new genomic positions to generate a panel of landing site candidates. Since these insertions are pre-loaded with a reporter transgene, they can be directly assayed for expression in any cell type of interest. Subsequently, we used Int* to excise the tester and reconstitute the attP sequence at desirable landing sites (Figure 2A). This strategy eliminates the need for separate tester integrations into each landing site and concomitantly reduces the number of fly generations between the isolation of new landing sites and their characterization. As such, this approach enjoys considerable advantages over the traditional method (compared in Figure S2, A and B).
Figure 2.
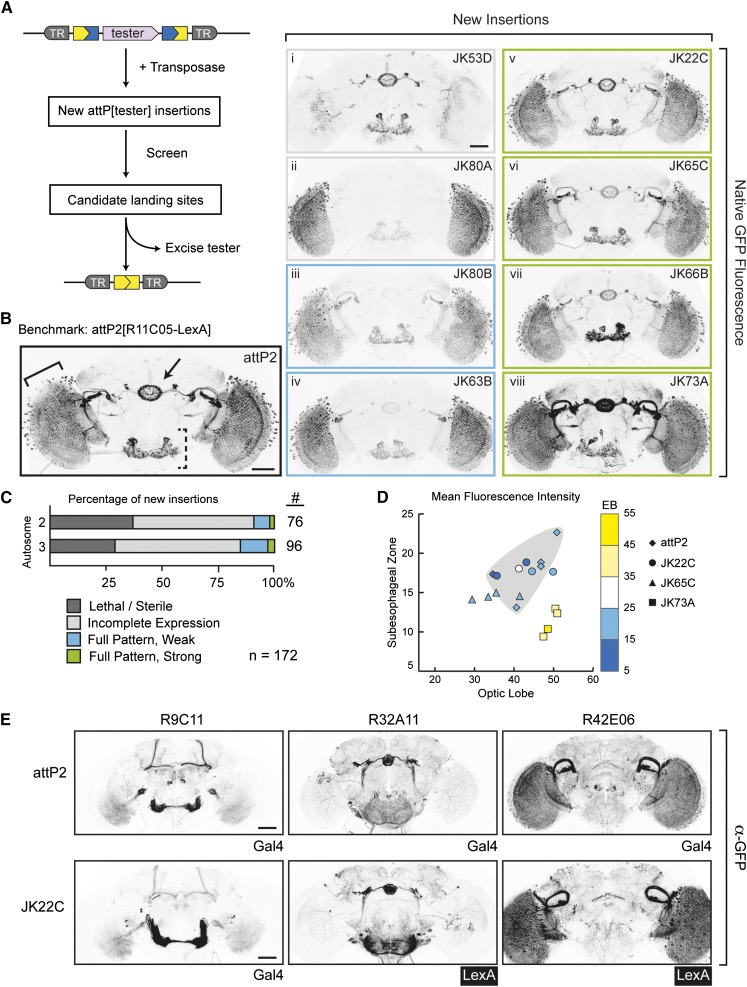
Int*-facilitated identification of landing site candidates that support transgene expression in the nervous system. (A) Diagram of Int*-facilitated site generation. A transposon-born landing site carrying an integrated “tester” transgene is mobilized to create new landing site candidates by random transposition. New insertions are screened to identify candidates that exhibit desirable characteristics, such as homozygous viability and strong expression of the tester. Int*-mediated excision of the tester transgene reconstitutes attP to regenerate a functional landing site from selected candidates. (B) Landing site candidates that recapitulate the expression properties of attP2. New insertions of P{attP[R11C05-LexA]} were visually screened for GFP expression to identify candidates that recapitulate the spatial pattern and expression levels of R11C05-LexA in attP2, which served as the benchmark (shown at left). Examples of new candidate lines (shown at right) were quantitatively imaged for native GFP fluorescence; images show maximum projections of representative confocal stacks. The optic lobe (solid bracket), sub-esophageal zone (dashed bracket), and ellipsoid body (arrow) are indicated for attP2[R11C05-LexA]. Candidate landing site designations (upper right corners) reflect the cytological band of the insertion. Colored outlines surrounding panels i-viii correspond to the expression properties described in C. Bar, 50 μm. (C) Statistics of P{attP18[R11C05-LexA]} mobilization. From 200 mobilization crosses, we recovered 172 new insertions on the large autosomes; no insertions on 4 or Y were recovered. Approximately 80% of new insertions exhibited a phenotype when homozygous or showed an incomplete pattern of GFP expression. Four candidate sites (green, examples in B) expressed LexA comparably to the attP2[R11C05-LexA] benchmark. (D) Comparison of R11C05-LexA expression in the optic lobe, sub-esophageal zone, and ellipsoid body (EB) for attP2 and several candidate landing sites. Brains were quantitatively imaged for native GFP fluorescence; stacks were aligned to a standard brain and maximum-projected; and then mean fluorescence intensity was collected for each region using a custom FIJI script (see Materials and Methods and Figure S3, B and C). Each line is represented by a symbol, with each point denoting one brain. Shading indicates the region bounded by the attP2 samples. (E) Behavior of three additional driver transgenes in JK22C. To test whether JK22C is generally permissive for expressing driver transgenes, additional Gal4 or LexA constructs driven by different “enhancers” (listed at the top) were integrated. For comparison, the expression of each enhancer in attP2 is shown. Brains were immunostained for GFP; images show maximum projections of representative confocal stacks. Bar, 50 μm.
We undertook to isolate new landing sites that support robust expression in the Drosophila adult central nervous system and are homozygous-viable without causing notable phenotypes. Specifically, in light of the large number of transgenes that have been characterized following integration into attP2 (e.g., Jenett et al. 2012; Kvon et al. 2014), we deemed that it would be useful to identify new sites that recapitulate the expression properties of this landing site. We selected a tester transgene with a sparse expression pattern to facilitate the primary screen, a simple visual screen using a fluorescence microscope. R11C05-LexA expresses in several regions of the Drosophila brain, including the ellipsoid body, the sub-esophageal zone, and the optic lobes (Figure 2B), but the limited number of cells expressing this driver ensures that deviations in expression can be easily spotted. Next, we mobilized the X-linked insertion P{CaryP}attP18[R11C05-LexA] using P transposase (Figure S2C), generating 172 new autosomal insertions. Fifty-five of these were homozygous-lethal or sterile (Figure 2C), in line with P mutagenesis screens (Cooley et al. 1988); these insertions were discarded. The remaining landing site candidates (hereafter, “candidates”) were crossed to LexAop2-GFP to characterize the expression properties of R11C05-LexA.
Among the candidates that were visually screened, a large fraction (62%) expressed the R11C05-LexA tester incompletely, as they lacked expression in one or more of the scored brain regions (Figure 2B, i and ii). However, we judged 10 candidates to support expression in the complete R11C05-LexA pattern, with 4 expressing GFP at levels similar to or higher than attP2[R11C05-LexA], which serves as a benchmark for comparison (Figure 2B, v–viii; Figure 2C). These results were corroborated when brains were stained for GFP and quantitatively imaged by confocal microscopy, although minor differences between several candidates and the benchmark were detectable following this higher-resolution analysis (Figure S3A).
To refine the comparison between these 10 candidates and the attP2 benchmark, we developed a proxy measure for GFP expression in different brain regions. Briefly, we quantitatively imaged native GFP fluorescence for each candidate and then computationally aligned image stacks to a reference brain using the neuropil marker N-Cadherin (Peng et al. 2011; Jenett et al. 2012). Samples that failed to achieve a minimum alignment score were excluded from subsequent consideration, as were candidates represented by fewer than four aligned samples (Figure S3B). Next, we measured the mean fluorescence intensity for three regions of the R11C05 pattern: the ellipsoid body, the left optic lobe (primarily the lobula), and the sub-esophageal zone (Figure 2D, brain regions depicted in Figure S3C). Graphing these data in three dimensions gives a sense for how comparable different candidates are for expression levels (Figure 2D); candidates with similar values for all three regions cluster near each other in space. As shown, samples from candidates JK22C and JK65C largely overlap the region defined by the samples from attP2[R11C05-LexA]. In contrast, ellipsoid body expression in samples from candidate JK73A exceeds all other candidates as well as attP2[R11C05-LexA], but expression in the sub-esophageal zone is reduced. These data support the qualitative assessment of the candidates from the visual screen.
The four strongly expressing candidates isolated above are promising landing sites. Yet, it may be that they are narrowly optimized to express R11C05-LexA, rather than being generally permissive for expression of various transgenes. To address this possibility, we reconstituted the landing sites in these candidates via Int*-mediated excision and integrated four additional transgenes in which the expression of Gal4 or LexA was under the control of different enhancer fragments. We assessed the ability of each of these transgenes to drive the appropriate GFP reporter and compared their expression to that observed when the corresponding Gal4 transgene (i.e., Gal4 driven by the corresponding enhancer fragment) was integrated into attP2 (Figure 2E and Figure S4). The results of these comparisons differentiated the JK22C landing site from the other three candidates: transgenes integrated into JK22C recapitulated attP2-like expression in each case, while each of the remaining three landing sites exhibited aberrant expression for at least one transgene (Figure S4; data not shown for the pan-neural driver R57C10-Gal4).
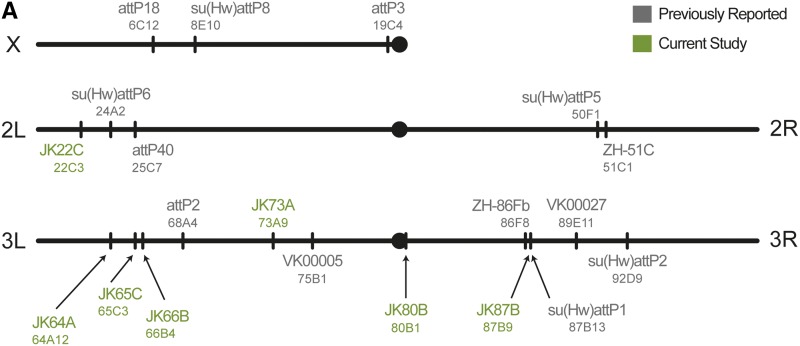
Finally, we investigated the chromosomal distribution of the new landing sites by mapping their genomic positions. We used splinkerette PCR to amplify the genomic DNA flanking each P insertion (Potter and Luo 2010), then aligned this sequence to the reference genome. Thus, we determined the positions of the four strongly expressing sites and the three weakly expressing lines that reproduce the complete R11C05-LexA pattern. These new landing sites were unevenly distributed on 2L and along both arms of the third chromosome, but no candidates mapped to 2R (Figure 3 and Figure S5). The locations of the new landing sites reflect the known bias of P elements to insert at promoters (Spradling et al. 1999; Liao et al. 2000; Bellen et al. 2004), as four of the new landing sites inserted in the 5′ UTR of a gene and the remaining three are within 500 bp of the nearest gene (Figure S5). Conveniently, the genetic map predicts that most of the new landing sites fall at least 5 cM distant from the nearest commonly used landing site, which should facilitate recombination between these sites.
Figure 3.
Cytological location of new landing sites. Cytological location of new landing sites. Sites are named with a JK prefix, according to cytological band. Commonly used sites: attP2 was isolated by Groth et al. (2004). VK sites are from Venken et al. (2006). ZH sites were constructed by Bischof et al. (2007). attP3, attP18, and attP40 were isolated by Markstein et al. (2008). su(Hw) sites are described by Ni et al. (2009).
Int* enables transgenes to be shuffled between landing sites genetically
A transgene excised by Int* is released as an episome that bears a reconstituted attB site. This circular DNA element resembles the plasmid transgenic constructs that are injected into embryos and the intermediates of several genetic techniques for translocating DNA sequences from one position in the genome to another (Golic et al. 1997; Gohl et al. 2011) . Therefore, we tested whether a transgene contained in an Int*-excised episome can be translocated from one landing site to another.
Translocating a transgene (hereafter referred to as “shuffling”) would require two competing enzymatic activities: excisionase to release the transgene from the donor site, and an integrase to re-integrate it at the receiver site (Figure 4A). Several Int* variants exhibit these opposing activities in vitro with distinct ratios of excisionase and integrase activities (Rowley et al. 2008). Therefore, we tested several variants for their ability to shuffle a transgene between landing sites on different chromosomes.
Figure 4.
The coordinated action of Int* and Int can shuffle transgenes between landing sites. (A) Schematic depicting transgene shuffling. (Top) Int* excises the transgene to be relocated, resulting in a circular episome that can diffuse away from the donor landing site (site 1). (Bottom) Int integrates the episome into the receiver landing site (site 2). Int* alone is sufficient for the reaction, but the combined action of wild-type Int and Int* significantly enhances the efficiency and fidelity with which transgenes are shuffled (see C). (B) Shuffling a transgene between landing sites. Int* (combined with wild-type Int) was used to shuffle pJFRC19 to a receiver landing site on another autosome; the crossing scheme for the attP40/ZH-86Fb site pair is presented in Figure S6A. Heat shocks delivered within a broad developmental window were sufficient to shuffle transgenes in either the male or female germlines. Shuffle efficiency was calculated by dividing the number of crosses that yielded a shuffle event by the total number of crosses screened. For the attP40/ZH-86Fb experiment, data from all variants were pooled; only Int*-KEE was used for the ZH-51C/attP2 experiment. Each point represents at least 11 germlines screened. (C) The presence of wild-type Int influences the efficiency of transgene shuffling more than the identity of Int*. For the indicated genotypes, wild-type integrase was expressed in the germline under the control of the nanos promoter (P{nos-phiC31\int.NLS}) (Bischof et al. 2007). Efficiency was calculated as in B. To derive a shuffling efficiency for each Int* variant, results were pooled for males and females of each genotype and for all heat shock time points. Each data point represents at least 24 crosses screened.
We assembled the donor and receiver landing sites in a stock, which we then crossed to flies carrying Int*. Crosses were heat-shocked during the embryonic or larval stages to induce Int*, and the F1 progeny were crossed to white flies (Figure S6A). We screened the F2 progeny to identify individuals in which the transgene marker (mini-white) segregated from the donor site chromosome and cosegregated with the receiver-site chromosome (distinguished by DsRed expression). Flies carrying candidate shuffle events were individually stocked to allow further characterization of the receiver locus. To calculate the efficiency of transgene shuffling for each Int* variant, we divided the number of crosses yielding a candidate shuffle event by the total screened.
For the landing site pair attP40 and ZH-86Fb, candidate shuffle events were recovered following heat shock at each of the developmental stages tested (Figure 4B), although the frequency declined sharply between heat shocks performed during the first and third larval instars. We confirmed proper re-integration for a subset of candidate shuffle events by using PCR to amplify the junction between the receiver landing site and the transgene. In all cases, the product matched the predicted size (data not shown). In addition to correctly targeted shuffle events, we recovered a number of chromosomes that bore the transgene marker (mini-white) but lacked the landing site marker (DsRed). Molecular characterization revealed that a majority of these lines result from re-integration into the Int*-bearing chromosome via recombination between attB and the attL that lies downstream of the hs-Int* transgene (Figure S6B). Similar results were obtained from a second experiment in which the same transgene was shuffled between the landing sites attP2 and ZH-51C (Figure 4B).
Contrary to our expectation, the efficiency of transgene shuffling was uniformly low for each of the Int* variants tested (Figure 4C), possibly due to a failure of Int* to re-integrate the episome efficiently or because Int* vigorously re-excises the transgene once it has been integrated at the receiver landing site. Regardless, we speculated that the presence of wild-type Int during transgene shuffling would shift the balance of both reactions toward integration, favoring the recovery of shuffled transgenes. To test this hypothesis, we incorporated a germline source of wild-type Int (Figure S6A). With this regime (Int plus Int*), shuffle events were recovered at least threefold more frequently for each Int* variant (Figure 4C). The presence of Int during transgene shuffling also yielded an unanticipated benefit. Wild-type Int improves the fidelity of transgene shuffling by curbing the hyperactive integrase activity of Int* (see below), possibly by forming heterodimers with Int*. This restricts re-integration events to those that result from recombination between attP and attB (Figure S6C).
Int* can be used to assemble tandem arrays of transgenes via atypical recombination
The hyperactive integrase activity of Int* can mediate recombination between nonstandard att site pairs in vitro (Rowley et al. 2008) and in vivo (above, transgene shuffling). We wondered whether this activity might be harnessed to sequentially integrate multiple transgenes into a single landing site.
Linking transgenes through Int*-mediated atypical integration is predicted to result in a transgene array (hereafter, “array”) in which the constituents lie in a tandem tail-to-head orientation (Figure 5A). The integration site of each additional component (attL or attR) determines the order of the constituents. Integration into attL results in the primary transgene being upstream; attR yields the reverse order. To avoid ambiguity, we shall name constituents based on the temporal order in which they are integrated. Hence, the first transgene to be integrated is referred to as the primary transgene (Figure 5A, purple bar).
As a pilot experiment, we sought to assemble a two-component array in attP2 that consisted of LexAop2-nls-lacZ as the primary transgene and UAS-tdTomato-nls as the secondary transgene. We tested four Int* variants for the ability to integrate a secondary transgene. Embryos homozygous for the primary transgene were co-injected with a mixture of the secondary transgene and an Int*-expressing plasmid construct. G0 adults were crossed to the appropriate screening stock, and F1 progeny were screened for the presence of both transgenes (Figure S7A). To calculate the efficiency of array recovery for each Int* variant, we divided the number of G0 crosses that produced progeny-carrying array candidates by the total number of G0 germlines screened. Two of the four Int* variants tested gave rise to array candidates from between 2% and 10% of fertile G0 crosses (Figure 5B). Repeating the experiment using only these two variants yielded similar results, with the Int*-KEE showing higher integration frequency.
To characterize array candidates, we confirmed the presence of each constituent transgene by crossing to Gal4 and LexA drivers. The order of primary and secondary transgenes in individual candidate arrays was determined by PCR (Figure S7B). Although Int*-KEE (Int-E449K) has been reported to catalyze recombination equally well between attB and attL or attR in vitro (Rowley et al. 2008), the arrays that we characterized molecularly suggest that attL may be the preferred target in vivo (Figure S7B). Next, we sought to construct four additional arrays at attP2 using Int*-KEE. Two of these experiments failed to produce any candidates; the efficiency of the remaining two was comparable to what we observed during the pilot (Figure 5B).
The ability to construct arrays has a natural application in linking transgenes that must cosegregate, such as pairs of reporter transgenes for colabeling experiments. However, it is essential that the transcriptional units within an array not interfere with one another. To investigate the prevalence of transcriptional crosstalk, we characterized the expression properties of the constituent transgenes within an array composed of LexAop2-nls-lacZ and UAS-tdTomato-nls (Figure S7B). Specifically, we assessed whether expressing one constituent would drive inappropriate expression or suppression of the other constituent.
To establish the baseline expression of the constituents, brains from heterozygous flies were immunostained for nls-LacZ and tdTomato-nls. Neither reporter was detectable in the absence of Gal4 and LexA, indicating that the mere presence of multiple transgenes at a landing site does not induce leaky expression (Figure S7C, top row). Next, we expressed either nls-lacZ or tdTomato-nls under the control of a strong, pan-neural driver to determine whether this would cause expression of the other reporter. In both cases, a small number of cells ectopically expressed the reporter that was not actively driven (Figure S7C, middle and bottom rows). Thus, the constituents of an array retain transcriptional independence in the vast majority of cells, even under extreme conditions.
Finally, we asked whether the expression of one constituent of an array would suppress expression of the other. We selected Gal4- and LexA-expressing lines that were previously shown to have limited overlap in their expression and used these to drive the array constituents. Specifically, fruP1.LexA (Mellert et al. 2010) and dsxGAL4.∆2 (Pan et al. 2011) each express in a limited number of cells in the posterior brain, with a subset of cells in males, but not females, co-expressing (Zhou et al. 2014). Consistent with our expectation, females possessed numerous cells in the posterior brain that expressed one or the other reporter, but no cells were labeled with both nls-LacZ and tdTomato-nls. In contrast, males displayed numerous colabeled cells (Figure 5C), demonstrating that the constituents of the array do not suppress the expression of one another. We corroborated this result using a different pair of Gal4 and LexA drivers to probe the expression of a separate reporter array integrated into a different landing site (Figure 5D). Taken together, these results strongly imply that the constituents of a transgene array have minimal impact on the expression of one another.
Discussion
We reasoned that the unusual recombination activities reported in vitro for several phiC31 integrase mutants could be applied in vivo to expand the available transgenesis toolkit. Using D. melanogaster, we tested integrase variants (Int*) for their ability to excise, integrate, and re-integrate transgenes. Using these activities, we developed methods that simplify the generation of landing sites with specific expression properties, enable the mobilization of an integrated transgene to a different landing site, and allow iterative insertion of multiple transgenes into a single landing site. After demonstrating that several Int* variants could excise transgenes from landing sites across the genome of D. melanogaster, we isolated a set of landing sites with expression properties optimized for the adult central nervous system. We also exploited this activity to genetically mobilize transgenes from one landing site to another. Fortuitously, we also observed that the most active integrase variants catalyze recombination between nonstandard att site pairs (e.g., attB and attL), which we exploited to build transgene arrays at several landing sites. Using Int* to perform any of these applications has distinct advantages over previous strategies.
Int* facilitates isolation of landing sites with specific expression properties
For any experimental system, the quality of available landing sites dictates the utility of site-specific integrases for transgenesis in that system. The Drosophila community benefits from having several landing sites that have been extensively characterized (Venken et al. 2006; Bischof et al. 2007; Markstein et al. 2008; Pfeiffer et al. 2010), allowing researchers to select sites to maximize transgene expression and minimize the impact of PE that might confound the interpretation of results. Nonetheless, there is a limited number of landing sites that are viewed as having desirable expression properties, and several considerations suggest that it will be advantageous, if not necessary, to develop additional landing sites in the future. For example, the expansion of available transactivation systems (Lai and Lee 2006; Luan et al. 2006; Potter et al. 2010) and recombinases (Nern et al. 2011) has spurred the development of increasingly sophisticated genetic techniques that require many transgenes, straining the current set of commonly used landing sites (e.g., Awasaki et al. 2014). Furthermore, in Drosophila it has recently been appreciated that the potential for transvection, a phenomenon where transcriptional elements on one chromosome influence transcription from promoters located at the same position on the homologous chromosome, is widespread across the genome (Bateman et al. 2012; Mellert and Truman 2012). Thus, care must be taken when different transgenes occupy the same landing site on homologous chromosomes if their independent expression is desired. The simple solution to transvection would be to locate transgenes at different landing sites, but this is complicated by the fact that the same transgene in different landing sites is frequently expressed at different levels due to PE (Pfeiffer et al. 2010). For sensitive phenotypes, this variation may not be acceptable.
The variability and unpredictability of PE make it unlikely that the current suite of commonly used landing sites will be sufficient to support robust transgene expression in all tissues or at every developmental stage. Ideally, for experiments involving any cell type at a given developmental stage, there would be the option to integrate transgenes at several landing sites that exhibit minimal PE. Developing a comprehensive landing site toolkit will require identifying additional landing sites specifically selected for these characteristics. Yet isolating landing sites customized to user-defined criteria has been impractical previously, owing to the onerous and expensive process of characterizing the expression properties of landing site candidates. By alleviating these problems, Int*-facilitated landing site generation should empower researchers to isolate purpose-built landing sites, thereby expediting the development of additional genetic resources.
Using the method that we describe, landing sites that are optimized to express transgenes in a particular tissue, cell-type, or during a specific developmental stage are regenerated from attP[tester] insertions. We demonstrated the efficacy of this method by identifying landing sites that are optimized to express driver transgenes in the adult nervous system. Notably, we isolated and characterized landing site candidates in half as many generations and at significantly reduced fly work compared to the traditional method. These savings, coupled with the fact that new candidates can be screened without additional injections, make Int*-facilitated landing site identification orders of magnitude less expensive than the traditional, two-step methodology. Moreover, Int*-mediated landing site isolation has an advantage over other methods for introducing attP sites (e.g., by CRISPR/Cas9-induced recombination) because it allows an unbiased survey of sites in the genome.
The fraction of candidates that are ultimately rejected from a landing site screen underscores the advantage of having a minimally intensive screening strategy. Among the 172 new landing site candidates that we tested (which exceeds the combined total of landing sites previously reported in the literature), a scant seven expressed the full pattern of the tester in our visual screen. Of these, only half expressed at levels comparable to the tester transgene in attP2, which stood as the benchmark. The low frequency at which our screen identified landing sites with the desired expression properties (∼4%) is largely due to the prevalence of PE, although it is also a consequence of our stringent requirement that newly isolated candidates recapitulate the behavior of attP2.
The success of any landing site screen depends on the expression properties of the tester and the scale at which the screen is conducted. The sensitivity of the tester transgene to PE will dictate the number of candidates recovered from a landing site screen, as well as whether the expression properties of candidates extend to additional transgenes. Considering the extreme cases illustrates the point: A tester transgene that is highly sensitive to position effects may yield few or no landing site candidates. Conversely, screening with an unusually robust tester transgene may identify a larger number of candidates, but ultimately few of these may be suitable for expressing more typical transgenes. R11C05-LexA exhibited moderate sensitivity to position effects following integration at several commonly used landing sites (A. Jenett and C. McKellar, personal communication), making it ideal for our purposes. When such data are unavailable prior to initiating a screen for new landing sites, it may be prudent to generate candidates using several different tester transgenes. Another consideration when initiating a landing site screen is the mobility of the progenitor element used to generate new landing site candidates. Similar to other transposon insertions (Bellen et al. 2004, 2011), landing sites can exhibit vastly different transposition frequencies. For example, attempts to mobilize su(Hw)attP8 and attP2 resulted in a lower frequency of new insertions than did attP18 (J.-M. Knapp, unpublished observations). Because lower mobility landing sites yield fewer new candidates for the same amount of fly handling, they should be avoided as progenitor elements for landing site screens.
Finally, we propose that Int*-facilitated landing site identification should be practical in any species or experimental system where transposition and site-specific integration have been demonstrated. Indeed, by reducing the number of separate transgenesis steps required to characterize landing sites, we anticipate that this method can speed the development of genetic resources in these systems.
Int* enables transgene shuffling between landing sites genetically
Int*-mediated transgene shuffling offers an economical alternative to performing repeated injections. We envision that it has several immediate applications, including placing one transgene at multiple landing sites. Likewise, transgene shuffling can facilitate moving an existing transgene collection into a new landing site, such as one that is better suited to planned experiments than the site in which the transgenes were initially integrated. Finally, for situations in which the plasmid construct containing a transgene of interest is unavailable, transgene shuffling offers the only viable strategy that does not require recloning the construct.
Our results suggest several points to bear in mind when shuffling transgenes. First, while Int* mutants possess both integrase and excisionase activities, excision (a unimolecular reaction) is favored over integration (a bimolecular reaction). Because shuffled transgenes are themselves flanked by attR and attL, they remain substrates for the Int* excisionase activity and can be excised from the receiver landing site following re-integration. Thus, including a source of wild-type Int is recommended to favor the recovery of shuffle events. In addition to increasing the recovery of shuffled transgenes, we observed that wild-type Int reduces the occurrence of undesired products that result from nonstandard recombination reactions. Finally, although heat shock during early development was sufficient to shuffle transgenes, the efficiency between a particular pair of sites may vary considerably between developmental stages. Thus, we recommend that users perform heat shocks at multiple time points when attempting to shuffle a transgene between two landing sites for the first time.
While the efficiency of recovering transgene shuffling events is within a practicable range, it might be improved further by reducing the likelihood of undesired integration events. In our experiments, the presence of the attL sequence downstream of hs-Int* appeared to reduce transgene shuffling by competing with the receiver landing site for the episome. Therefore, generating a source of Int* without this sequence (e.g., Int* inserted into the genome via a transposon) should result in increased shuffling efficiency.
Int* enables the construction of transgene arrays in single landing sites
The hyperactive integrase activity of Int* can sequentially integrate multiple transgenes into a single landing site to create an array of transgenes linked in tandem. Int* incorporates successive units by recombining the attB sequence associated with the transgene to be added with attL or attR in the occupied landing site. Thus, Int* can be used to assemble transgene arrays at any landing site, and this technique is compatible with extant transgenes without requiring additional cloning or modification. To demonstrate the feasibility of constructing arrays with Int*, we built two-component arrays at landing sites on the X and third chromosomes.
Transgene arrays have several useful applications. Integrating multiple transgenes in an array can compensate for a shortage of landing sites in systems or species where landing sites are scarce and isolating new sites is impractical. Additionally, for experiments that rely upon cosegregation of transgenes, linking them in an array offers an ideal solution that simultaneously simplifies the experiment by reducing the number of loci that must be tracked. Furthermore, since arrays remain flanked by attL and attR sites following successive integrations, it is theoretically possible to link an arbitrary number of transgenes in an array. Although we have not attempted arrays with more than two components, there is no mechanistic reason to anticipate that integrating a third component will be more difficult than the previous step. Alternatively, wild-type Int could be used to integrate an attP-bearing transgene at the reconstituted attB site to yield a three-component array.
The competing integrase and excisionase activities of Int* result in a diverse mix of products for each iteration of array construction. Therefore, a robust strategy for identifying candidates possessing all components is imperative. Screening for arrays is straightforward when transgene components are differentially marked (e.g., by fluorescence and mini-white expression). Absent these, detecting transgene arrays can be challenging. Indeed, we speculate that our failed array construction attempts were unsuccessful because we could not detect candidates, rather than because no transgene arrays were produced. The development of additional transgene markers should facilitate the construction of transgene arrays using Int* and may be essential when more than two transgenes are to be linked.
Concluding remarks
The experiments presented here offer a proof-of-concept demonstration of three new techniques that extend the phiC31 transgenesis platform. The efficiency of each technique likely can be further improved by optimizing the starting fly stocks, the timing or levels of Int* expression, and the availability of additional genetic sources of Int*. We hope that these techniques will stimulate the development of genetic resources in D. melanogaster and all the diverse species in which phiC31 integrase has been shown to function.
Supplementary Material
Acknowledgments
We thank members of the Simpson lab, Carmen Robinett and Aljoscha Nern, for helpful scientific discussions; Gerald Rubin, Barret Pfeiffer, and Heather Dionne for transgenic reagents; Michele Calos for integrase reagents; the Developmental Studies Hybridoma Bank for antibodies; Janelia Research Campus FlyCore and Scientific Computing members for technical assistance; Emily Nielson for excellent administrative support; the Bloomington Drosophila Stock Center and FlyBase for providing fly stocks and genomic resources, respectively; and Andrew Seeds, Jessica Cande, and Carmen Robinett for comments on the manuscript.
Author contributions: J.M.K. conceived, designed, and executed the experiments. J.M.K. and P.M.C. collected the data. J.M.K. analyzed data and prepared the figures. J.M.K. wrote the manuscript. J.H.S. and J.M.K. edited the manuscript.
Footnotes
Available freely online through the author-supported open access option.
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.173187/-/DC1.
Communicating editor: N. Perrimon
Literature Cited
- Akhtar W., de Jong J., Pindyurin A. V., Pagie L., Meuleman W., et al. , 2013. Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell 154(4): 914–927. [DOI] [PubMed] [Google Scholar]
- Awasaki T., Kao C.-F., Lee Y.-J., Yang C.-P., Huang Y., et al. , 2014. Making Drosophila lineage-restricted drivers via patterned recombination in neuroblasts. Nat. Neurosci. 17: 631–637. [DOI] [PubMed] [Google Scholar]
- Barolo S., Carver L. A., Posakony J. W., 2000. GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. BioTechniques 29(4): 726–732. [DOI] [PubMed] [Google Scholar]
- Bateman J. R., Johnson J. E., Locke M. N., 2012. Comparing enhancer action in cis and in trans. Genetics 191(4): 1143–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen H. J., Levis R. W., Liao G., He Y., Carlson J. W., et al. , 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167(2): 761–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen H. J., Levis R. W., He Y., Carlson J. W., Evans-Holm M., et al. , 2011. The Drosophila gene disruption project: progress using transposons with distinctive site specificities. Genetics 188(3): 731–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F., Basler K., 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104(9): 3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley L., Kelley R., Spradling A., 1988. Insertional mutagenesis of the Drosophila genome with single P elements. Science 239(4844): 1121–1128. [DOI] [PubMed] [Google Scholar]
- Farruggio A. P., Chavez C. L., Mikell C. L., Calos M. P., 2012. Efficient reversal of phiC31 integrase recombination in mammalian cells. Biotechnol. J. 7(11): 1332–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisinger J. M., Calos M. P., 2013. Site-directed insertion of transgenes, pp. 211–239 in Topics in Current Genetics, Vol. 23, edited by Renault S., Duchateau P.. Springer Dordrecht Heidelberg, NY. [Google Scholar]
- Gohl D. M., Silies M. A., Gao X. J., Bhalerao S., Luongo F. J., et al. , 2011. A versatile in vivo system for directed dissection of gene expression patterns. Nat. Methods 8(3): 231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic M. M., Rong Y. S., Petersen R. B., Lindquist S. L., Golic K. G., 1997. FLP-mediated DNA mobilization to specific target sites in Drosophila chromosomes. Nucleic Acids Research 25(18): 3665–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A. C., Olivares E. C., Thyagarajan B., Calos M. P., 2000. A phage integrase directs efficient site-specific integration in human cells. Proc. Natl. Acad. Sci. USA 97(11): 5995–6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A. C., Fish M., Nusse R., Calos M. P., 2004. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166(4): 1775–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia T. D., Craig E. A., McCarthy B. J., 1980. Sequence of three copies of the gene for the major Drosophila heat shock induced protein and their flanking regions. Cell 21(3): 669–679. [DOI] [PubMed] [Google Scholar]
- Jenett A., Rubin G. M., Ngo T.-T. B., Shepherd D., Murphy C., et al. , 2012. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2(4): 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S. H., Madison E. L., 1997. Rapid and efficient site-directed mutagenesis by single-tube “megaprimer” PCR method. Nucleic Acids Res. 25(16): 3371–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaleel T., Younger E., McEwan A. R., Varghese A. S., Smith M. C. M., 2011. A phage protein that binds ϕC31 integrase to switch its directionality. Mol. Microbiol. 80(6): 1450–1463. [DOI] [PubMed] [Google Scholar]
- Kuhstoss S., Rao R. N., 1991. Analysis of the integration function of the streptomycete bacteriophage phi C31. J. Mol. Biol. 222(4): 897–908. [DOI] [PubMed] [Google Scholar]
- Kvon E. Z., Kazmar T., Stampfel G., Yáñez-Cuna J. O., Pagani M., et al. , 2014. Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature 512(7512): 91–95. [DOI] [PubMed] [Google Scholar]
- Lai S.-L., Lee T., 2006. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat. Neurosci. 9(5): 703–709. [DOI] [PubMed] [Google Scholar]
- Levis R., Hazelrigg T., Rubin G. M., 1985. Effects of genomic position on the expression of transduced copies of the white gene of Drosophila. Science 229(4713): 558–561. [DOI] [PubMed] [Google Scholar]
- Lewis E. B., 1950. The phenomenon of position effect, pp. 73–115 in Advances in Genetics, Vol. 3, edited by M. Demerec. Elsevier, Amsterdam; New York. [DOI] [PubMed] [Google Scholar]
- Liao G. C., Rehm E. J., Rubin G. M., 2000. Insertion site preferences of the P transposable element in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 97(7): 3347–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan H., Peabody N. C., Vinson C. R., White B. H., 2006. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron 52(3): 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz K. A., Corneille S., Azhagiri A. K., Svab Z., Maliga P., 2004. A novel approach to plastid transformation utilizes the phiC31 phage integrase. Plant J. 37(6): 906–913. [DOI] [PubMed] [Google Scholar]
- Markstein M., Pitsouli C., Villalta C., Celniker S. E., Perrimon N., 2008. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 40(4): 476–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellert D. J., Truman J. W., 2012. Transvection is common throughout the Drosophila genome. Genetics 191(4): 1129–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellert D. J., Knapp J.-M., Manoli D. S., Meissner G. W., Baker B. S., 2010. Midline crossing by gustatory receptor neuron axons is regulated by fruitless, doublesex and the Roundabout receptors. Development 137(2): 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C., Puller A.-C., Lawson K. L., Tschopp P., Amsterdam A., et al. , 2013. Site-directed zebrafish transgenesis into single landing sites with the phiC31 integrase system. Dev. Dyn. 242(8): 949–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A., Pfeiffer B. D., Svoboda K., Rubin G. M., 2011. Multiple new site-specific recombinases for use in manipulating animal genomes. Proc. Natl. Acad. Sci. USA 108(34): 14198–14203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J.-Q., Liu L.-P., Binari R., Hardy R., Shim H.-S., et al. , 2009. A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics 182(4): 1089–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Robinett C. C., Baker B. S., 2011. Turning males on: activation of male courtship behavior in Drosophila melanogaster. PLoS ONE 6(6): e21144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H., Chung P., Long F., Qu L., A. Jenett et al, 2011. BrainAligner: 3D registration atlases of Drosophila brains. Nat. Methods 8(6): 493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer B. D., Ngo T.-T. B., Hibbard K. L., Murphy C., Jenett A., et al. , 2010. Refinement of tools for targeted gene expression in Drosophila. Genetics 186(2): 735–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter C. J., Luo L., 2010. Splinkerette PCR for mapping transposable elements in Drosophila. PLoS ONE 5(4): e10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter C. J., Tasic B., Russler E. V., Liang L., Luo L., 2010. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell 141(3): 536–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley P. A., Smith M. C. A., Younger E., Smith M. C. M., 2008. A motif in the C-terminal domain of phiC31 integrase controls the directionality of recombination. Nucleic Acids Res. 36(12): 3879–3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner O., Berdnik D., Levy J. M., Wu J. S., Luginbuhl D., et al. , 2008. piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev. Cell 14(2): 227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. C. M., Brown W. R. A., McEwan A. R., Rowley P. A., 2010. Site-specific recombination by phiC31 integrase and other large serine recombinases. Biochem. Soc. Trans. 38(2): 388–394. [DOI] [PubMed] [Google Scholar]
- Spradling A. C., Rubin G. M., 1983. The effect of chromosomal position on the expression of the Drosophila xanthine dehydrogenase gene. Cell 34(1): 47–57. [DOI] [PubMed] [Google Scholar]
- Spradling A. C., Stern D., Beaton A., Rhem E. J., Laverty T., et al. , 1999. The Berkeley Drosophila Genome Project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics 153(1): 135–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K. J. T., He Y., Hoskins R. A., Bellen H. J., 2006. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314(5806): 1747–1751. [DOI] [PubMed] [Google Scholar]
- Zhou C., Pan Y., Robinett C. C., Meissner G. W., Baker B. S., 2014. Central brain neurons expressing doublesex regulate female receptivity in Drosophila. Neuron 83(1): 149–163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.