Abstract
Red soils, which are widely distributed in tropical and subtropical regions of southern China, are characterized by low organic carbon, high content of iron oxides, and acidity and, hence, are likely to be ideal habitats for acidophilic actinomycetes. However, the diversity and biosynthetic potential of actinomycetes in such habitats are underexplored. Here, a total of 600 actinomycete strains were isolated from red soils collected in Jiangxi Province in southeast China. 16S rRNA gene sequence analysis revealed a high diversity of the isolates, which were distributed into 26 genera, 10 families, and 7 orders within the class Actinobacteria; these taxa contained at least 49 phylotypes that are likely to represent new species within 15 genera. The isolates showed good physiological potentials for biosynthesis and biocontrol. Chemical screening of 107 semirandomly selected isolates spanning 20 genera revealed the presence of at least 193 secondary metabolites from 52 isolates, of which 125 compounds from 39 isolates of 12 genera were putatively novel. Macrolides, polyethers, diketopiperazines, and siderophores accounted for most of the known compounds. The structures of six novel compounds were elucidated, two of which had a unique skeleton and represented characteristic secondary metabolites of a putative novel Streptomyces phylotype. These results demonstrate that red soils are rich reservoirs for diverse culturable actinomycetes, notably members of the families Streptomycetaceae, Pseudonocardiaceae, and Streptosporangiaceae, with the capacity to synthesize novel bioactive compounds.
INTRODUCTION
While high-throughput sequencing methods expand our understanding of species diversity in microbial communities and reveal the presence of many new groups that were previously undetected in cultivation studies, the availability of pure cultures is still essential in order to discover new or useful bioactive compounds from microorganisms. As the most prolific secondary metabolite producers, actinomycetes account for approximately 41% of the bioactive microbial metabolites that have been discovered (1). In recent years, with the emergence of antibiotic-resistant microbial pathogens and rediscovery of known bioactive compounds, special habitats in the ocean (2, 3), sponges (4), plants (5, 6), and insects (7) have been found to be potentially important in the search and discovery of bioactive compound-producing strains. Nevertheless, soil is still the predominant source of secondary metabolite-producing actinomycetes. Unexplored and underexplored soil habitats have been found to be a rich source of actinomycetes that produce novel bioactive metabolites (8–10). Acidic soils in China, mostly red soils, are one such habitat.
Red soils are generated by abundant rainfall and high temperature and are widespread in tropical and subtropical regions of southern China, covering about 1.13 million km2 (11). These soils have been subjected to intensive weathering, and mineral nutrients (including Ca, Mg, P, and K) in the soil are mostly eluted, forming the major soil constraints of low organic carbon, high content of iron oxides and exchangeable Al3+, and acidity (12). Chinese red soils thus provide a potential source for the search and discovery of acidophilic and acidotolerant microorganisms. However, despite reports on ammonia-oxidizing and sulfate-reducing prokaryotes in such soils (13, 14), the diversity and biosynthetic potential of actinomycetes in red soils has barely been investigated.
Since the isolation of acidophilic streptomycetes from acidic soil samples by Williams and colleagues (15, 16), studies of acidophilic actinomycetes have been focused mainly on the description of novel taxa and on their phylogenetic diversity (17–22). Acidophilic actinomycetes can be assigned to two groups, namely, a group of moderately acidophilic actinomycetes with an optimal pH of around 6.0 and a group of obligately acidophilic actinomycetes with an optimal pH of no more than 4.0 (23). Obligate acidophiles classified in the class Actinobacteria include members of the genera Actinospica and Streptacidiphilus, while moderate acidophiles described to date are mainly affiliated with the genera Catenulispora and Rugosimonospora and with some groups of Streptomyces (18, 19, 23). Even so, acidophilic actinomycetes are still largely unexplored, and only limited attention has been focused on their natural products. There is evidence that the basis for the traditional use of Jordan's red soils for the treatment of skin infections is the proliferation of antibiotic-producing bacteria inhabiting the soil, notably actinomycetes, and their concomitant production of antimicrobial compounds (24). Furthermore, actinomycetes isolated from acidic soils seem to be a better source of polyether ionophores than their counterparts from other habitats (25). Consequently, it makes good sense to systematically screen actinomycete diversity in acidic red soils in the search for new bioactive compounds.
Jiangxi Province (24°7′ to 29°9′N, 114°02′ to 118°28′E) is one of the typical red soil regions located in southeastern China. The red soils of this region are Acrisols and Ferralsols, which are predominantly derived from Quaternary red earth (6.48%), Tertiary red sandstone (20.38%), and granite (21.92%) (26). The clay minerals are dominated by kaolinite and hydrous mica and also contain a small amount of vermiculite (11). The aim of the present study was to evaluate the biodiversity of culturable actinomycetes in acidic red soils from Jiangxi Province and to highlight their potential to produce novel secondary metabolites. Using combinations of four pretreatments and nine selective media, hundreds of actinomycetes belonging to 26 genera were recovered in pure cultures. Antimicrobial and chitinolytic activity assays and chemical analyses were applied to evaluate the biosynthetic potential and ecological role of the isolates. The results demonstrate that actinomycetes isolated from acidic red soil represent a promising source of new secondary metabolites, some of which have great potential as candidates for the biocontrol of fungal pathogens.
MATERIALS AND METHODS
Collection of soil samples.
A total of 32 acidic soil samples (pH between 2.6 and 6.6) were collected in August 2007 and September 2009 from six red soil locations in Jiangxi Province (see Table S1 in the supplemental material), which has a typical subtropical monsoon climate with a mean annual temperature of 17.8°C and a mean annual rainfall of 1,341 to 1,940 mm. At each sampling site, surface soil from the top 5 to 20 cm was collected and sealed in polyethylene bags. All samples were transported intact at ambient temperature to the laboratory and examined immediately. At any one time, three to five samples from different sites of each location were mixed to form composite samples referred to as sampling locations AU, LJZ, PYH, WS, XS, and YL. Actinomycetes were selectively isolated from these samples.
Selective isolation.
The soil samples were subjected to four treatments, ultrasonication/dilution (27), dispersion and differential centrifugation (DDC) (28), dry heating (120°C, 1 h) (29), and hydration and centrifugation (30), prior to the selective isolation of actinomycetes using nine culture media (M1 to M9); the compositions of these media are given in Table S2 in the supplemental material. All of the media contained cycloheximide, nystatin, and nalidixic acid (each at 50 μg/ml); one set of plates were supplemented with novobiocin (25 μg/ml), but not a second set. The initial pH of each medium was between 5.0 and 5.5. The isolation was done in three batches. In each batch, 200 μl of each of the pretreated 10−2 and 10−3 dilutions were spread, in triplicate, over the surface of four to six of the media, and the inoculated plates were incubated at 28°C for 4 to 6 weeks. Colonies were purified on the same media, and purified isolates preserved at −80°C in 20% (vol/vol) glycerol.
16S rRNA gene sequencing and taxonomic diversity analysis.
Genomic DNA was extracted from representative isolates using the method of Chun and Goodfellow (31). Almost-complete 16S rRNA genes were amplified using universal primers 27f and 1492r (32), and the PCR products were purified and forward and reverse sequenced using an ABI 3730XL sequencer. The chromatograms for the resultant 16S rRNA gene sequences for each strain were checked manually and edited and assembled using the MEGA5.0 package (33). The resultant sequences (>1,350 nucleotides) were aligned using RDP online aligner (34) and grouped into operational taxonomic units (OTUs; equal to phylotypes) using the web-based tool Clusterer (35) and a sequence identity of 99% as recommended by Stach et al. (36). Each singleton and cluster was defined as an OTU. Phylogenetic trees based on the 16S rRNA gene sequences of the representative OTUs and reference sequences were constructed by using the neighbor-joining (37) and maximum-likelihood methods (38) from the MEGA 5.0 package, with 1,000 bootstrap replicates. Bayesian analyses were run in MRBAYES version 3.1.2 (39) for 10,000,000 generations, using the general time-reversible (GTR) model of DNA substitution with gamma-distributed rate variation across invariant sites. Trees were sampled every 100 generations. The first 25,000 trees were discarded, and the remaining 75,000 were used for calculating posterior probabilities (PP) in the majority-rule consensus tree. The calculation of pairwise 16S rRNA gene sequence similarities was achieved using EzTaxon-e (40).
Determination of pH profiles.
Cells (mycelium and/or spores) for the pH tests were harvested by scraping them from glucose-yeast extract-malt extract (GYM; see Table S2 in the supplemental material) plates that had been incubated for 14 days at 28°C. Twenty microliters of the resultant cell suspensions were streaked on GYM plates adjusted to pH 3.5 to 8.0 at intervals of 0.5 pH, using KH2PO4-HCl and KH2PO4-K2HPO4 buffer systems. These preparations were made in duplicate, and the plates were incubated at 28°C for 3 weeks, although plates were checked for growth every 5 days.
Chitin degradation.
Chitinolytic activity was evaluated by growing isolates on synthetic medium M10 (see Table S2 in the supplemental material) containing 0.2% colloidal chitin (41) as the only organic carbon and nitrogen sources; the pH of the medium was 5.5. Plates were incubated at 28°C for 14 days, after which colonies with transparent zones were considered chitin-degrading isolates.
Fermentation, extraction, and chemical analysis.
Strains were fermented on solid agar in petri dishes at 28°C for 7 to 12 days using three media: GYM (M11), mannitol-oatmeal-soy peptone-yeast extract (MOSY; M12), and potato starch-glucose-glycerol (SGG; M13) (see Table S2 in the supplemental material). After fermentation, large agar blocks (100 ml in volume) were cut into small pieces with a sterilized stainless knife and extracted three times with 100 ml ethanol. All organic phases for each sample were pooled and concentrated to dryness in a vacuum, and the residue redissolved in 2 ml dimethyl sulfoxide (DMSO). Twenty microliters of each extract was subjected to high-performance liquid chromatography-UV spectrometry analysis (HPLC-UV/VIS; Shimadzu SPD-M20A) with a linear gradient of 50 to 100% aqueous methanol for 25 min (4.6- by 150-mm column; flow, 1.0 ml/min; photodiode array detector, 190 to 800 nm; Xbridge ODS [octadecylsilyl]). Extracts that did not show UV absorbance were submitted to thin-layer chromatography (TLC)-mass spectrometry (MS) analysis using silica gel 60 F254 (Merck). Fractions were then subjected to mass spectrometric analysis (Thermo-Finnigan LCQ DECA XP, Agilent 1260/6460 triple quadrupole LC-MS, or Waters Xevo G2 quadrupole time of flight-ultraperformance liquid chromatography [QTOF UPLC]-MS), and mass spectra (scanning 100 to 2,000 atomic mass units) were collected in both positive and negative modes. Compounds were identified by the comparison of molecular weights, UV spectra, and retention times with published chemical data from standard databases (e.g., Dictionary of Natural Products [DNP] on DVD, version 22.2, and SciFinder 2007) and references. Some compounds were subjected to nuclear magnetic resonance (NMR) spectroscopic analysis (Bruker AV600 NMR spectrometer), circular dichroism (CD) analysis (Applied Photophysics Chirascan), and optical rotation analyses (Antonpaar MCP-200 automatic polarimeter) for further identification.
Large-scale solid-state fermentation was carried out for strains FXJ1.172 (MOSY, 12 liters), FXJ1.532 (GYM, 10 liters), FXJ1.076 (GYM, 12 liters), and FXJ1.264 (GYM, 25 liters). Compounds were extracted and purified from the fermentation samples and then subjected to structural elucidation, using a procedure described previously (25).
Antimicrobial and antitumor assays.
Isolates were cultivated on GYM (M11) as appropriate to obtain lawns of colonies and then screened for activity toward fungi and bacteria using the agar block method (42). Four pathogenic fungal strains, Aspergillus fumigatus CGMCC 3.772, Fusarium oxysporum CGMCC 3.2830, Candida albicans CGMCC 2.538, and Candida pseudorugosa CGMCC 2.3107, were obtained from the China General Microbiological Culture Collection Center. Five drug-resistant bacterial indicator strains, extended-spectrum β-lactamase (ESBL)-producing Escherichia coli 4-1, Klebsiella pneumoniae 5-1, methicillin-resistant Staphylococcus aureus (MRSA) 1-1, Pseudomonas aeruginosa stain PAO1, and Vibrio cholerae O1 were obtained from the Weifang Medical University, Shandong Province, China. Antitumor activities were tested against human cervical carcinoma cell line HeLa, human colonic carcinoma cell line SW480, and human lung adenocarcinoma cell line A549, using the MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) method (43) and the procedure described by Wang et al. (44). Briefly, cells were incubated in 96-well plates (104 cells per well) in 100 μl Dulbecco's modified Eagle's medium (DMEM) overnight and then in fresh medium containing different concentrations of test compounds (0.2% DMSO and cisplatin were used as negative and positive controls, respectively) for 48 h. The medium was removed, phosphate-buffered saline (PBS) containing 0.5 mg/ml MTT (Sigma) was added, and the plates were incubated in the dark at 37°C for 3 h. Upon removal of MTT, 100 μl of DMSO was added to each well and the plates agitated at 60 rpm for 5 min to dissolve the purple precipitate. The absorbance at 540 nm was determined using a microplate reader (BioTek Synerge H4), and the 50% inhibitory concentrations (IC50s) were read directly from the instrument. An IC50 of <10 μg/ml was recorded as strong antitumor activity, and an IC50 of >100 μg/ml as absence of antitumor activity.
Nucleotide sequence accession numbers.
The 16S rRNA nucleotide sequence data reported in the present study have been deposited in the DDBJ/EMBL/GenBank databases under accession numbers FJ418903 to FJ418907, FJ418910, FJ938343 to FJ938345, FJ938347, FJ938349, FJ938351, FJ938352, FJ938354, HQ537063 to HQ537065, HQ537067, HQ537069, KC137299 to KC137302, KF830662 to KF830690, KF879929 to KF879937, KJ152024 to KJ152065, KM598592 to KM598633, KP126244 to KP126422, KP126424 to KP126428, and KP126430 to KP126459.
RESULTS
Growth pH profiles.
Four treatments and nine media were used to isolate diverse actinomycetes from the red soil samples. Six hundred actinomycete isolates were purified. More than half of them (308) were purified from M6 medium (GTV, pH 5.0 to 5.5; see Table S2 in the supplemental material). The isolates could be assigned to three categories according to their growth on GYM plates at different pHs. Most of the isolates (63%) could grow at pH 4.0 to 7.0, with an optimum at pH 5.5 to 6.0, and failed to grow at pH 3.5 or ≥7.5 and, thus, could be assigned to the moderately acidophilic group. Only 8% of the isolates could be considered obligately acidophilic given that their optimal growth occurred at pH 4.0, with a growth range of pH 3.5 to 7.0. The remaining 29% of the isolates grew well at pH 5.0 to 8.0, poorly at pH 4.5, and optimally at pH 7.5, and hence, were assigned to the acidotolerant group.
Taxonomic diversity.
According to sample sources, cultural characteristics (surface and reverse colony color and diffusible pigments), and morphological features (formation and organization of substrate and aerial mycelium and formation of spores and spore chains), 358 representative isolates were selected for 16S rRNA gene sequencing and OTU calculation. Using 99% sequence identity, the isolates were grouped into 147 OTUs (70 clusters and 77 singletons) spanning 26 genera, 10 families, and 7 orders of the class Actinobacteria (Table 1). The acidophilic isolates were assigned to 23 genera (with the only exceptions being Actinocorallia, Dactylosporangium, and Microtetraspora) and the acidotolerant isolates to 9 genera (Table 1). Members of the family Streptomycetaceae, containing the genera Streptomyces and Streptacidiphilus, accounted for the majority (57.8%) of OTUs, followed by members of the families Pseudonocardiaceae (10.2%) and Streptosporangiaceae (9.5%), each containing six genera. Streptomyces strains predominated and were obtained from each of the sampling locations. Seven genera, Actinomadura, Amycolatopsis, Microbispora, Micromonospora, Nocardia, Nonomuraea, and Streptosporangium, were also frequently isolated from different locations, while the Actinoallomurus strain was isolated only from LJZ (see Table S3 in the supplemental material). The other genera were represented by no more than five isolates. LJZ hosted the most diverse culturable actinomycetes, which belonged to 24 genera. In contrast, XS harbored the isolates of only three genera (see Table S3).
TABLE 1.
Classification of isolates and OTUs delineated by 99% 16S rRNA gene sequence identity
| Ordera | Familya | Genus | No. of: |
Total no. of OTUs | No. of OTUs containing putative new lineages | ||
|---|---|---|---|---|---|---|---|
| Isolates | OTU singletons | OTU clusters | |||||
| Catenulisporales | Catenulisporaceae | 5 | 0 | 2 | 2 | 0 | |
| Catenulispora | 5 | 0 | 2 | 2b | 0 | ||
| Corynebacteriales | Mycobacteriaceae | 2 | 2 | 0 | 2 | 1 | |
| Mycobacterium | 2 | 2 | 0 | 2b | 1b | ||
| Nocardiaceae | 20 | 3 | 6 | 9 | 4 | ||
| Nocardia | 19 | 2 | 6 | 8b | 4b | ||
| Williamsia | 1 | 1 | 0 | 1b | 0 | ||
| Tsukamurellaceae | 1 | 1 | 0 | 1 | 0 | ||
| Tsukamurella | 1 | 1 | 0 | 1b | 0 | ||
| Micromonosporales | Micromonosporaceae | 27 | 3 | 6 | 9 | 3 | |
| Dactylosporangium | 2 | 0 | 1 | 1d | 1d | ||
| Micromonospora | 25 | 3 | 5 | 8c | 2b | ||
| Propionibacteriales | Nocardioidaceae | 4 | 1 | 1 | 2 | 0 | |
| Kribbella | 3 | 0 | 1 | 1b | 0 | ||
| Marmoricola | 1 | 1 | 0 | 1b | 0 | ||
| Pseudonocardiales | Pseudonocardiaceae | 20 | 11 | 4 | 15 | 6 | |
| Actinomycetospora | 3 | 3 | 0 | 3b | 1b | ||
| Actinosynnema | 1 | 1 | 0 | 1b | 1b | ||
| Amycolatopsis | 8 | 3 | 2 | 5b | 2b | ||
| Pseudonocardia | 2 | 2 | 0 | 2b | 1b | ||
| Lentzea | 3 | 1 | 1 | 2c | 1c | ||
| Saccharothrix | 3 | 1 | 1 | 2b | 0 | ||
| Streptomycetales | Streptomycetaceae | 213 | 49 | 36 | 85 | 28 | |
| Streptacidiphilus | 5 | 2 | 1 | 3b | 1b | ||
| Streptomyces | 208 | 47 | 35 | 82c | 27c | ||
| Streptosporangiales | Streptosporangiaceae | 42 | 5 | 9 | 14 | 6 | |
| Microbispora | 10 | 0 | 2 | 2c | 2c | ||
| Microtetraspora | 2 | 0 | 1 | 1d | 0 | ||
| Nonomuraea | 15 | 2 | 4 | 6c | 3b | ||
| Planotetraspora | 1 | 1 | 0 | 1b | 0 | ||
| Sphaerisporangium | 1 | 1 | 0 | 1b | 0 | ||
| Streptosporangium | 13 | 1 | 2 | 3b | 1b | ||
| Thermomonosporaceae | 24 | 2 | 6 | 8 | 1 | ||
| Actinoallomurus | 13 | 0 | 3 | 3b | 0 | ||
| Actinocorallia | 1 | 1 | 0 | 1d | 0 | ||
| Actinomadura | 10 | 1 | 3 | 4c | 1b | ||
| Total | 358 | 77 | 70 | 147 | 49 | ||
The assignment of strains to orders and families is based on the hierarchical classification in the second edition of Bergey's Manual of Systematic Bacteriology (45).
OTUs contain only acidophilic strains.
OTUs contain both acidophilic and acidotolerant strains.
OTUs contain only acidotolerant strains.
At least 49 OTUs assigned to 15 genera showed <99% 16S rRNA gene sequence similarity with type strains of validly published species, of which 27 (55%) OTUs were assigned to the genus Streptomyces (see Table S4), 6 to the family Pseudonocardiaceae, and 6 to the family Streptosporangiaceae. It is worth mentioning that most strains within these OTUs belonged to the acidophilic group (40/49 OTUs, 84/101 strains). Four strains from four of these OTUs were assigned to the obligately acidophilic group, among which three were affiliated with the genus Streptomyces (singleton strains FXJ1.124, FXJ1.257, and FXJ1.300) and the fourth with the genus Nocardia (OTU25, strain FXJ1.333). The remaining acidotolerant strains within these OTUs were mainly associated with the genus Streptomyces (seven singletons and OTU50) as well, and occasionally with the genera Dactylosporangium (OTU53, FXJ1.974 and FXJ1.262), Lentzea (OTU57, FXJ1.357), and Microbispora (OTU37, FXJ1.1062).
Phylogenetic analysis of the families Pseudonocardiaceae, Streptosporangiaceae, and Streptomycetaceae.
The diversity of putative new phylotypes within these families and their relationships with the most closely related species are shown in Fig. 1, 2, and 3.
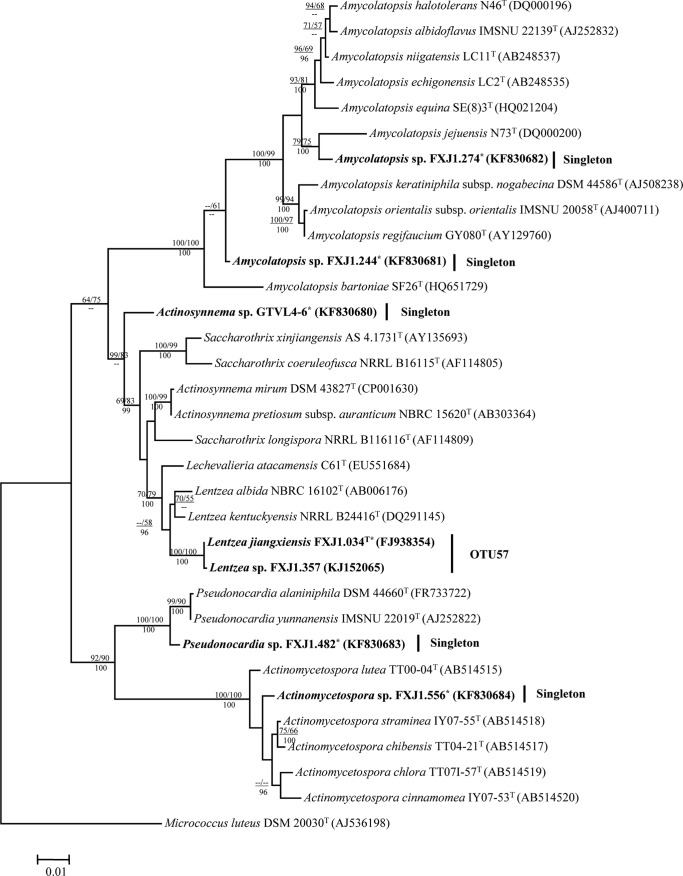
FIG 1.
Maximum-likelihood phylogenetic tree of the family Pseudonocardiaceae based on 16S rRNA gene sequences showing relationships between isolates representing putative novel phylotypes (in boldface font) and related members of the family Pseudonocardiaceae. DDBJ/EMBL/GenBank accession numbers are shown in parentheses. Bootstrap values of the neighbor-joining (NJ) and maximum-likelihood (ML) analyses are shown above the internodes before and after the backslash, respectively, and posterior probabilities of the Bayesian analyses (≥0.95) are shown below the internodes; only values above 50% are given. Bar, 0.01 substitutions per site. The tree was rooted with Micrococcus luteus DSM 20030T. *, strain is moderately acidophilic.
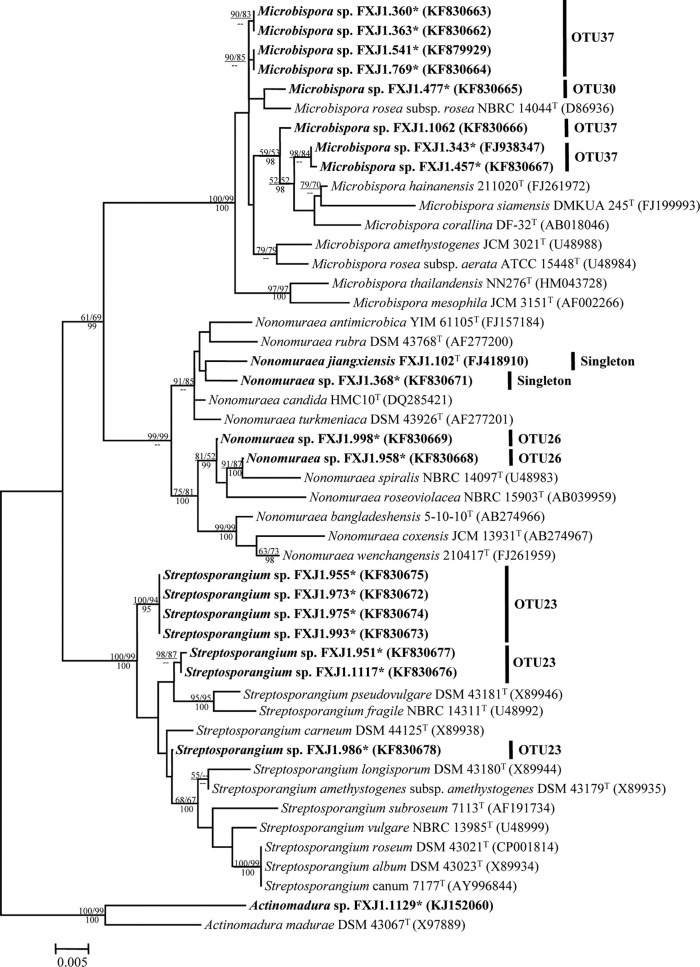
FIG 2.
Maximum-likelihood phylogenetic tree of the family Streptosporangiaceae based on 16S rRNA gene sequences, showing relationships between isolates representing putative novel phylotypes (in boldface font) and related members of the Streptosporangiaceae. DDBJ/EMBL/GenBank accession numbers are shown in parentheses. Bootstrap values of the NJ and ML analyses are shown above the internodes before and after the backslash, respectively, and posterior probabilities of the Bayesian analyses (≥0.95) are shown below the internodes; only values above 50% are given. The tree was rooted by novel isolate Actinomadura sp. strain FXJ1.1129 and Actinomadura madurae DSM 43067T. Bar, 0.005 substitutions per site. *, strain is moderately acidophilic.
FIG 3.
Maximum-likelihood trees based on 16S rRNA gene sequences, showing the phylogenetic positions of Streptomyces isolates representing putative novel phylotypes (in boldface font). DDBJ/EMBL/GenBank accession numbers are shown in parentheses. Bootstrap values of the NJ and ML analyses are shown above the internodes before and after the backslash, respectively, and posterior probabilities of the Bayesian analyses (≥0.95) are shown below the internodes; only values above 50% are given. Bar, 0.01 substitutions per site. *, strain is moderately acidophilic; **, strain is obligately acidophilic.
Seven isolates (FXJ1.274, FXJ1.244, GTVL4-6, FXJ1.034T, FXJ1.357, FXJ1.482, and FXJ1.556) of six OTUs within the family Pseudonocardiaceae were only loosely related to their nearest relatives (16S rRNA gene sequence similarities of 97.1 to 98.7%) and formed six coherent phylogenetic lineages in five of the genera classified in this family (Fig. 1; see also Table S4 in the supplemental material). It is noteworthy that isolate GTVL4-6, which showed the highest sequence similarity (97.5%) to both Actinosynnema pretiosum subsp. auranticum and Lechevalieria atacamensis, formed a stable monophyletic line that was sharply separated from all of the genera in the Pseudonocardiaceae 16S rRNA gene phylogeny. Lentzea isolates FXJ1.034T and FXJ1.357, from AU and PYH, respectively, formed a cluster defined at the 99.9% 16S rRNA similarity level and, hence, are most likely members of the same lineage. The results of a comprehensive polyphasic taxonomic study showed that isolate FXJ1.034T belonged to a new Lentzea species, for which the name Lentzea jiangxiensis has been proposed (46).
Nineteen isolates belonging to six OTUs of the family Streptosporangiaceae were assigned to at least 11 phylogenetic lineages within three genera (Fig. 2; see also Table S4 in the supplemental material). OTU37, which contained seven Microbispora isolates (FXJ1.360, FXJ1.363, FXJ1.541, FXJ1.769, FXJ1.1062, FXJ1.343, and FXJ1.457), was split into three distinct lineages. Strain FXJ1.477 within OTU30 formed a distinct branch sharing its highest 16S rRNA gene sequence similarity (98.8%) with Microbispora rosea subsp. rosea NBRC 14044T. Within the genus Nonomuraea, four isolates (FXJ1.102T, FXJ1.368, FXJ1.998, and FXJ1.958) of three OTUs were located separately in the tree and shared 98.7 to 98.9% similarities with related type strains, and hence, they may represent four lineages. A detailed polyphasic study on strain FXJ1.102T showed that it represented a new Nonomuraea species, Nonomuraea jiangxiensis (47). OTU23, which contained seven isolates (FXJ1.955, FXJ1.973, FXJ1.975, FXJ1.993, FJX1.951, FXJ1.1117, and FXJ1.986) of the genus Streptosporangium, may represent three lineages that shared less than 98.9% similarity with the related species.
Within the genus Streptomyces, the 27 OTUs sharing <99% 16S rRNA gene sequence similarity with their nearest type strains correspondingly formed 27 distinct lineages in the phylogenetic tree (Fig. 3; see also Table S4 in the supplemental material). Among these OTUs, a moderately acidophilic OTU (OTU6) encompassing 11 isolates was recovered only from LJZ; this taxon formed a putative novel phylotype that was most closely related to Streptomyces ferralitis CGMCC 4.1985T (48). Besides these OTUs, another 13 OTUs (six clusters and seven singletons) of this genus formed 15 distinct lineages that shared 99.0 to 99.2% 16S rRNA gene sequence similarities with their nearest species (see Fig. S1).
Antimicrobial and chitinolytic activities.
Nearly one-third of the isolates (193 of 600) showed inhibitory activity against pathogenic bacteria or/and fungi, most frequently against methicillin-resistant S. aureus (MRSA) (13.7%). High levels of activity were also shown against C. albicans (13.2%), F. oxysporum (10.2%), A. fumigatus (7.3%), and C. pseudorugosa (6.8%), but lower levels were shown against the Gram-negative bacteria, namely, ESBL-producing E. coli (5.7%), K. pneumonia (5.0%), P. aeruginosa (4.2%), and V. cholerae (3.8%). Interestingly, the ability to inhibit pathogenic fungi was observed more often among the acidophilic isolates, while in contrast, a higher proportion of acidotolerant isolates were active against Gram-negative bacteria, apart from K. pneumonia (Fig. 4).
FIG 4.
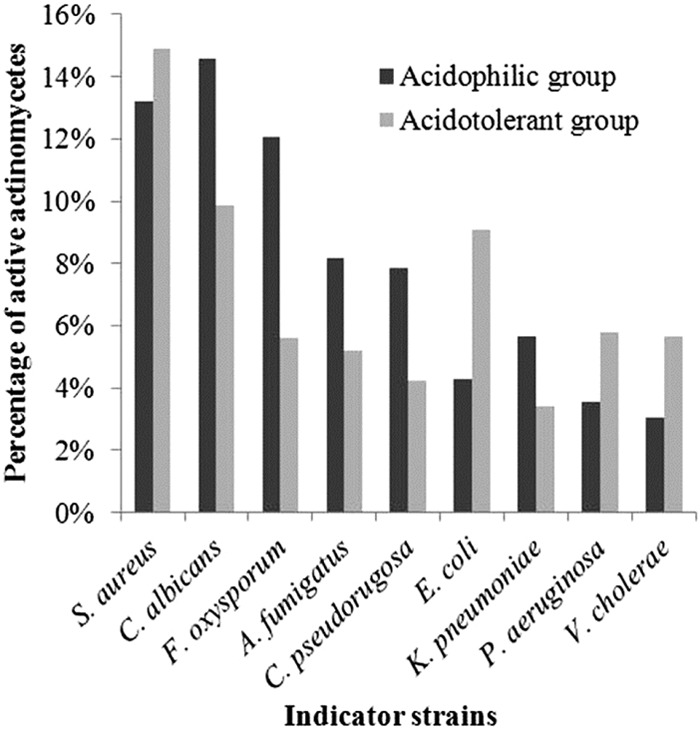
Bioactivity spectra of acidophilic and acidotolerant actinomycetes against nine indicator strains.
Different levels of activity were shown by representatives of the various genera and families (see Table S5 in the supplemental material). Isolates classified within the genus Streptomyces of the family Streptomycetaceae showed the highest rate (52.6%) and broadest spectrum of antagonistic activities. Indeed, isolates of OTU6 displayed remarkable activity against the pathogenic fungi, and some of these strains inhibited the growth of MRSA and the P. aeruginosa strains. Within nonstreptomycete isolates, the highest level of activity, 28.5%, was shown by isolates assigned to the family Streptosporangiaceae, followed by those classified in the families Pseudonocardiaceae (20%) and Catenulisporaceae (20%). Considerable levels of antimicrobial activity were detected in strains assigned to the families Micromonosporaceae (18.5%), Thermomonosporaceae (16.7%), and Nocardiaceae (10%). Isolates classified in the families Mycobacteriaceae, Nocardioidaceae, and Tsukamurellaceae did not display any activity.
A high proportion (78%) of the isolates showed chitinolytic activity, contributed mostly by strains from the family Streptomycetaceae (48.5%), followed by strains of the Streptosporangiaceae (12.4%), Micromonosporaceae (6.7%), Pseudonocardiaceae (4.5%), and Thermomonosporaceae (3.9%). Indeed, the chitinolytic strains within these families represented 97.1% of all of the active strains. It was also interesting that the occurrence of antifungal activity detected among the chitinolytic actinomycetes (39%) exceeded that of the nonchitinolytic isolates (13%). The most pronounced zones of clearing on colloidal chitin plates were shown by isolates from OTU6.
Chemical analysis of fermentation extracts of the isolates.
Chemical analysis was focused on isolates classified in the genera of the above-mentioned seven bioactive families. Following 16S rRNA gene sequence and isolation origin-based dereplication (6), i.e., strains with almost identical 16S rRNA gene sequences and the same isolation origins were dereplicated, isolates from each of the genera (except those within the Streptacidiphilus and Planotetraspora, which grew very poorly on the fermentation media) were selected randomly regardless of the antimicrobial activity they displayed. A total of 107 isolates (see Table S6 in the supplemental material) covering 20 genera were selected for further fermentation studies using three different media (GYM, MOSY, and SGG; see Table S2). Chemical analyses of the extracts revealed that 52 (48.6%) isolates from 13 genera produced detectable amounts of secondary metabolites; 33 of these isolates belonged to the genus Streptomyces (Table 2), and 36 were from the acidophilic group (Table 3; see also Table S7). At least 193 different compounds were found in the fermentation samples, 68 of which were identified as known compounds by using LC-UV/VIS–MS and, partially, by CD and NMR techniques (Table 3; see also Table S7). The compounds identified were assigned to 14 structural classes, within which macrolides (27 compounds, produced by 11 isolates), polyethers (12 compounds, produced by 7 isolates), diketopiperazines (9 compounds, produced by 3 isolates), and siderophores (7 compounds, produced by 5 isolates) accounted for 80.9% of the total (see Table S7). The remaining 125 compounds, which were obtained from 39 isolates classified in 12 genera, mainly Streptomyces (67.2%), were considered to be putatively novel because their molecular weights, UV spectra, or both did not match any published compounds (Table 3). Of the isolates producing putative novel compounds, 27 were from the acidophilic group and 12 from the acidotolerant group (Table 3).
TABLE 2.
Numbers of secondary metabolites produced by isolates subjected to chemical analysis
| Family | Genus | No. ofa: |
|||
|---|---|---|---|---|---|
| Isolates | CPSb | IDCc | PNCc | ||
| Catenulisporaceae | Catenulispora | 2 | 0 | 0 | 0 |
| Micromonosporaceae | Dactylosporangium | 1 | 0 | 0 | 0 |
| Micromonospora | 5 | 0 | 0 | 0 | |
| Nocardiaceae | Nocardia | 4 | 1 (0) | 2 | 0 |
| Williamsia | 1 | 0 | 0 | 0 | |
| Pseudonocardiaceae | Actinomycetospora | 2 | 0 | 0 | 0 |
| Actinosynnema | 1 | 0 | 0 | 0 | |
| Amycolatopsis | 6 | 5 (4) | 10 | 6 | |
| Pseudonocardia | 1 | 1 (1) | 0 | 1 | |
| Lentzea | 1 | 1 (1) | 0 | 4 | |
| Saccharothrix | 2 | 1 (1) | 0 | 6 | |
| Streptomycetaceae | Streptomyces | 65 | 33 (22) | 55 | 84 |
| Streptosporangiaceae | Microbispora | 3 | 2 (2) | 0 | 7 |
| Microtetraspora | 1 | 0 | 0 | 0 | |
| Nonomuraea | 2 | 1 (1) | 0 | 4 | |
| Sphaerisporangium | 1 | 1 (1) | 0 | 2 | |
| Streptosporangium | 3 | 2 (2) | 0 | 4 (5)d | |
| Thermomonosporaceae | Actinoallomurus | 1 | 1 (1) | 0 | 1 |
| Actinocorallia | 1 | 1 (1) | 0 | 1 | |
| Actinomadura | 4 | 2 (2) | 1 | 5 | |
| Total | 107 | 52 (39) | 68 | 125 | |
CPS, compound-producing strain(s); IDC, identified known compound(s); PNC, putative novel compound(s).
Numbers in parentheses indicate the number of strains that produce putative new compounds.
The number of compounds has been dereplicated, i.e., the same compound from different isolates was only counted once.
Streptosporangium sp. strain FXJ1.481 produced a putative new compound identical with that from Actinocorallia sp. strain FXJ1.544, so its PNC number was dereplicated. The nondereplicated number is shown in parentheses.
TABLE 3.
Novel and putatively novel secondary metabolites from the selected isolates
| Genus and strain | Fermentation medium | Compound(s) produced | Structural class | Chemical data |
|
|---|---|---|---|---|---|
| UV (MeOH) | MS (m/z)c | ||||
| Streptomycetes | |||||
| Streptomyces sp. FXJ1.030 | GYM | 1 putative novel compound | —g | 265, 492 | [M-H]− = 1,034.7 |
| Streptomyces sp. FXJ1.050a | SGG | 2 putative novel compounds, related to niphimycins | Macrolide | 225, 231 (sh) | [M-H]− = 1,140.7159; [M+H]+ = 1,142.7312 |
| Streptomyces sp. FXJ1.066 | SGG | 1 putative novel compound | Peptide-type compound | 221, 280 | [M-H]− = 1,022.7 |
| SGG | 5 new products related to spoxazomicins | Siderophore | 243, 302 | [M+H]+ = 775.2, 757.2, 818.3, 789.2, 803.3 | |
| Streptomyces sp. FXJ1.068 | GYM | 1 new analogue of nanchangmycind | Polyether | 234 | [M+Na]+ = 903.7 |
| Streptomyces sp. FXJ1.069a,b | GYM | 5 putative novel compounds | — | 249, 289/295 | [M+Na]+ = 609.6, 609.6 |
| — | 249, 366 | [M+H]+ = 595.5 | |||
| — | 240, 319, 721/731 | [M+H]+ = 333.1, 333.2 | |||
| SGG | 2 putative novel compounds | — | 249 | [M-H]− = 275.1647 | |
| — | 238, 262 | MS no ion current | |||
| Streptomyces sp. FXJ1.075a | MOSY | 3 putative novel compounds | — | 257, 369 | [M+H]+ = 371.4 |
| — | 228 | [M+H]+ = 761.9 | |||
| — | 220, 271, 290 | [M+H]+ = 1,058.9 | |||
| Streptomyces sp. FXJ1.076a | GYM/MOSY | 2 elaiophylin-type new compoundse | Macrolide | 250 | [M+HCOO]− = 759.4320 |
| 249 | [M+HCOO]− = 921.5212 | ||||
| Streptomyces sp. FXJ1.172a | MOSY | 1 new analogue of thienodolind | — | 239, 292, 334 | [M-H]− = 264.9839 |
| GYM | 6 putative novel compounds | — | End absorbance | [M-H]− = 545.5 | |
| — | 230, 297 | [M+H]+ = 526.3427, 540.3597, 554.3679, 554.3745, 568.3896 | |||
| SGG | 6 putative novel compounds | Peptide-type compound | 277/279/277/281/276/277 | [M+H]+ = 742.3423, 798.4035, 812.4167, 826.4343, 840.4512, 920.4258 | |
| Streptomyces sp. FXJ1.234 | SGG | 4 putative novel compounds | — | 253, 291 | [M+Na]+ = 919.4, 1065.6, 1,079.6, 1,051.5 |
| Streptomyces sp. FXJ1.235b | MOSY/SGG | 6 putative novel compounds | Chlorinated compounds | 244, 256, 265, 294,307, 340 | [M+H]+ = 304.0403, 445.0760 |
| — | 243, 256, 265, 305, 332 | [M+H]+ = 270.0809 | |||
| — | 244, 256, 265, 294,307, 338 | [M+H]+ = 437.2 | |||
| — | 244, 256, 265, 294,307, 340 | 2 MS no ion current | |||
| Streptomyces sp. FXJ1.250b | GYM | 1 putative novel compounds | — | 295 | [M-H]− = 407.5 |
| Streptomyces sp. FXJ1.253a | MOSY | 3 putative novel compounds | — | 249/249/269, 253 | MS no ion current |
| Streptomyces sp. FXJ1.264 | GYM | 1 new analogue of etheromycine | Polyether | None | [M-H]− = 927.5659 |
| GYM | 8 putative novel compounds related to oxazolomycins | PKS-NRPS hybridf | 276, 334/334/336/326/327/326/331/365 | [M+H]+ = 568.4, 568.3, 568.3, 452.3, 452.2, 480.2, 480.3, 665.3 | |
| GYM/MOSY | 4 putative novel compounds | Related to siderophore | 270 | [M+H]+ = 670.2627, 670.2666, 684.2830, 684.2815 | |
| Streptomyces sp. FXJ1.275a,b | MOSY | 1 putative novel quinomycin-type compound | Peptide-type compound | 246, 330 | [M+H]+ = 1,151.6 |
| SGG | 2 putative novel compounds | — | 236, 353/236, 354 | MS no ion current | |
| Streptomyces sp. FXJ1.310 | GYM | 1 putative novel compound | — | 264, 493 | [M-H]− = 559.5 |
| Streptomyces sp. FXJ1.408a,b | GYM/MOSY/SGG | 4 brunefungin- or flavopentin-type new compounds | Polyene macrolide | 264, 363 | [M+H]+ = 735.2, the other 3 MS not measured |
| Streptomyces sp. FXJ1.409 | MOSY | 2 putative novel compounds | Related to peptide | 218 (sh), 280, 316 (sh)/325 (sh) | [M+H]+ = 842.2603, 617.1464 |
| Streptomyces sp. FXJ1.414a,b | GYM/MOSY/SGG | 4 brunefungin- or flavopentin-type new compounds | Polyene macrolide | 264, 363 | [M+H]+ = 735.2, the other 3 MS not measured |
| Streptomyces sp. FXJ1.532a,b | GYM | 2 new macrolide compoundse | Macrolide | 229 | [M+Na]+ = 789.4402, 789.4402 |
| Streptomyces sp. FXJ1.535a,b | SGG | 1 putative novel compounds | Angucycline-type compound | 259, 398 | [M-H]− = 413.2692 |
| Streptomyces sp. GTVL2G15b | MOSY | 7 putative novel compounds | — | 250, 282, 364 | [M+H]+ = 295.9, 294.0 |
| — | 252, 270 (sh), 474 | [M-H]− = 459.2 | |||
| — | 291 | MS no ion current | |||
| Actinomycin-like compound | 231, 420, 441 | [M/2+H]+ = 628.3 | |||
| — | 229, 272, 344, 409 | MS no ion current | |||
| — | 228, 275, 338, 432 | [M-H]− = 339.2 | |||
| SGG | 1 Oxachelin analogue | Siderophore | 242, 250, 260 (sh), 303 | Peak degraded before MS | |
| Streptomyces sp. FXJ23ya,b | GYM/SGG | 3 putative novel compounds | — | 243, 366 | MS no ion current |
| — | 211 | [M-H]− = 649.8, 649.8 | |||
| Nonstreptomycete actinomycetes | |||||
| Saccharothrix sp. FXJ1.021a | MOSY | 6 putative novel compounds | Related to anthracycline | 253, 362 | [M+H]+ = 323.0798 |
| 262, 377 | [M+H]+ = 505.2234 | ||||
| 254, 400 | [M+H]+ = 489.2273 | ||||
| 254, 370 | [M+H]+ = 505.2212 | ||||
| 258, 373 | [M+H]+ = 505.2193 | ||||
| 276, 370 | [M+H]+ = 489.2285 | ||||
| Lentzea jiangxiensis FXJ1.034Ta,b | SGG | 4 putative novel compounds | 3 related to siderophore | 285 | [M+Na]+ = 669.2 |
| 205, 245, 312 | [M+H]+ = 654.4 | ||||
| 206, 243, 311 | 2 MS no ion current | ||||
| Nonomuraea jiangxiensis FXJ1.102Ta,b | SGG | 1 putative novel compound related to antibiotic BE 14106 | Macrolactam | 260, 270, 310 (sh) | [M+H]+ = 474.5 |
| SGG | 3 putative novel compounds | — | 236, 289, 318 | [M+H]+ = 972.9 | |
| — | 236, 289, 318 | 2 MS no ion current | |||
| Amycolatopsis sp. FXJ1.244a,b | GYM/MOSY/SGG | 5 new peptides | Peptide | 212 | [M+H]+ = 1,195.6, 1,181.7, 1,181.6, 935.7, 100.2 |
| Actinomadura sp. FXJ1.344a | SGG | 3 homoprejadomycin-type new derivatives | Angucycline | 266, 385/397/401 | [M+H]+ = 659.6, 323.3, 759.1 |
| 1 putative novel compound | — | 225, 314, 429 | [M+H]+ = 637.6 | ||
| Amycolatopsis sp. FXJ1.406a | GYM/MOSY/SGG | 5 new peptides | Peptide | 212 | Similar to those of FXJ1.244 |
| Amycolatopsis sp. FXJ1.428a | SGG | 1 putative novel compound | — | 214, 285 | [M+H]+ = 637.4 |
| Amycolatopsis sp. FXJ1.444a | SGG | 1 putative novel compound | — | 214, 285 | [M+H]+ = 637.4 |
| Sphaerisporangium sp. FXJ1.452a | SGG | 2 putative novel compound | — | 225, 236, 260 (sh), 335 | [M+H]+ = 897.4116, 883.3928 |
| Pseudonocardia sp. FXJ1.453a | SGG | 1 putative novel compound | — | 250, 310 | [M+H]+ = 406.1264 |
| Microbispora sp. FXJ1.457a,b | MOSY | 5 putative novel compounds | — | 262–263, 362–363 | 5 MS no ion current |
| Streptosporangium sp. FXJ1.481a | SGG | 1 putative novel compound identical with that from FXJ1.544 | — | 214, 247 | [M+H]+ = 375.1448 |
| Actinoallomurus sp. FXJ1.503a | SGG | 1 putative novel compound | — | 257 | [M+NH4]+ = 507.3441 |
| Actinocorallia sp. FXJ1.544 | SGG | 1 putative novel compound, identical with that from FXJ1.481 | — | 214, 247 | [M+H]+ = 375.1442 |
| Actinomadura sp. FXJ1.848a | MOSY | 1 putative novel compound | — | 234, 256 (sh), 280 (sh), 308, 320 (sh) | [M+H]+ = 488.2 |
| Microbispora sp. FXJ1.1062b | SGG | 2 putative novel compounds | — | 246 | [M+H]+ = 466.3160, 403.2486 |
| Streptosporangium sp. FXJ1.1111a | SGG | 4 putative novel compounds | Siderophore | 247, 314 | [M+H]+ = 467.2036, 425.1935, 466.2185, 449.1921 |
Strain is assigned to the acidophilic group.
Strain represents a putative new species; FXJ1.034T and FXJ1.102T have been proposed as type strains of Lentzea jiangxiensis sp. nov. and Nonomuraea jiangxiensis sp. nov., respectively (46, 47).
Values accurate to 1 decimal place were generated by regular MS, and values accurate to 4 decimal places were generated by HR-MS.
Compound was subjected to tandem MS analysis.
Compound was subjected to NMR analysis.
PKS-NRPS, polyketide synthase-nonribosomal peptide synthetase.
—, compounds not classified.
The putative new metabolites exhibited a fascinating chemical variety; some of them could be assigned to known structural classes based on their spectroscopic data, including some additional tandem MS and NMR data, and physicochemical properties (Table 3). Compounds with maximum UV absorption at bands 235 to 250 and 300 to 320 nm and which exhibited a red color in Fe3+ solution could be phenolated siderophores; these metabolites were produced by Lentzea sp. strain FXJ1.034T, Streptomyces sp. strain FXJ1.066, Streptomyces sp. strain GTVL2G15, and Streptosporangium sp. strain FXJ1.1111. Compounds with maximum UV absorption near 400 nm and which exhibited a yellow color could be angu-/anthar-cyclines; these metabolites were produced by Actinomadura sp. strain FXJ1.344, Saccharothrix sp. strain FXJ1.021, and Streptomyces sp. strain FXJ1.535. Furthermore, compounds exhibiting characteristic or end UV absorbance with large molecular masses (>1,000 Da) and which showed sequential fragment ion ladders with 100- to 130-Da differences in the MSn spectra could contain peptide components; these metabolites were produced by Amycolatopsis sp. strain FXJ1.244, Amycolatopsis sp. strain FXJ1.406, Streptomyces sp. FXJ1.066, Streptomyces sp. strain FXJ1.172, and Streptomyces sp. strain FXJ1.275. Similarly, compounds showing similar UV absorption with known macrolides of different molecular masses might be new macrolides; these compounds were produced by Amycolatopsis sp. strain FXJ1.274, Nonomuraea jiangxiensis FXJ1.102T (macrolactam), Streptomyces sp. strain FXJ1.050, Streptomyces sp. strain FXJ1.076, Streptomyces sp. strain FXJ1.253, Streptomyces sp. strain FXJ1.408, and Streptomyces sp. strain FXJ1.414. Two new polyether analogues were produced by Streptomyces sp. strain FXJ1.068 and Streptomyces sp. strain FXJ1.264, respectively (25). It was not possible to analyze 19 substances, mainly because ion currents could not be detected in their total ion current (TIC) spectra using at least two different MS machines with both positive and negative modes; these metabolites were derived from six isolates, namely, Lentzea jiangxiensis FXJ1.034T, Microbispora sp. strain FXJ1.457, Streptomyces sp. strain FXJ1.069, Streptomyces sp. FXJ1.253, Streptomyces sp. FXJ1.275, and Streptomyces sp. strain FXJ23y (Table 3).
Identification of novel secondary metabolites and their bioactivities.
Large-scale fermentation, isolation, and structural analysis of the putative novel compounds produced by four selected isolates were performed, and six novel compounds were identified (Fig. 5). Related chemical data (e.g., NMR and high-resolution mass spectrometry [HRMS] data) and detailed structural elucidation are provided in Table 4 and Appendix S1 in the supplemental material.
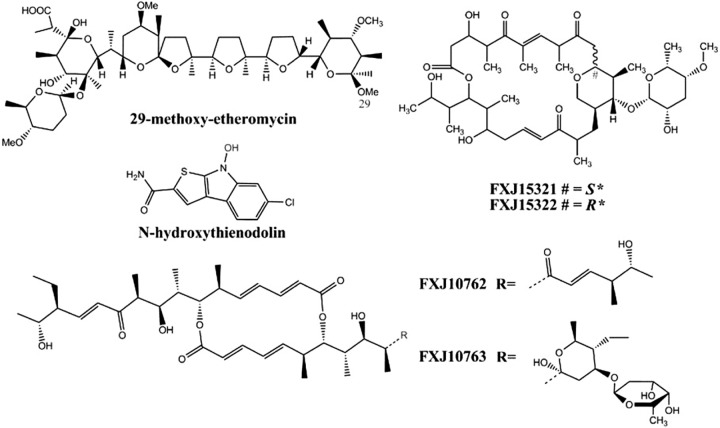
FIG 5.
Novel compounds produced by the red soil isolates. The relative configurations of compounds FXJ15321 and FXJ153221 were tentatively assigned, and their absolute configurations are arbitrary.
TABLE 4.
| Position | FXJ15321 |
FXJ15322 |
||
|---|---|---|---|---|
| δC | δH | δC | δH | |
| 1 | 172.8, C | 172.7, C | ||
| 2 | 39.6, CH2 | 2.43 (dd; 10.8, 15.0) | 39.8, CH2 | 2.42 (dd; 11.4, 15.0) |
| 2.67 (dd; 2.4, 15.0) | 2.70 (dd; 2.4, 15.0) | |||
| 3 | 70.7, CH | 4.27 (m) | 70.9, CH | 4.28 (ddd; 2.4, 9.9, 9.9) |
| 4 | 44.8, CH | 3.49 (m) | 44.8, CH | 3.47 (m) |
| 5 | 204.9, C | 205, C | ||
| 6 | 138.8, C | 136.6, C | ||
| 7 | 140.9, CH | 6.47 (d; 9.6) | 142.6, CH | 6.66 (d; 9.6) |
| 8 | 48.2, CH | 3.85 (m) | 44.5, CH | 4.01 (dq; 6.6, 9.6) |
| 9 | 208.8, C | 210.9, C | ||
| 10 | 40.8, CH2 | 2.35 (dd; 2.4, 16.2) a | 47.2, CH2 | 2.33 (dd; 9.6, 13.8) a |
| 2.92 (dd; 10.8, 16.2) b | 3.03 (dd; 3.6, 13.8) b | |||
| 11 | 73.8, CH | 4.34 (ddd; 2.4, 3.6–4.2, 10.8) | 78.2, CH | 3.52 (ddd; 3.6, 9.6, 9.6) |
| 12 | 38.4, CH | 1.92 (m) | 43.9, CH | 1.53 (ddq; 9.6, 9.6, 6.6,) |
| 13 | 80.1, CH | 3.43 (m) | 83.9, CH | 3.07 (dd; 10.2, 10.2) |
| 14 | 38.1, CH | 1.58 (m) | 39.8, CH | 1.39 (m) |
| 15 | 66.1, CH2 | 3.26 (m) a | 70.1, CH | 2.82 (dd; 12.0, 12.0) a |
| 3.71 (dd; 3.6–4.2; 12.0) b | 3.75 (dd; 4.8; 12.0) b | |||
| 16 | 34.1, CH2 | 1.26 (m) | 32.3, CH2 | 0.88 (m) |
| 2.15 (m) | 2.54 (m) | |||
| 17 | 38, CH | 3.16 (m) | 37.79, CH | 3.08 (m) |
| 18 | 205.9, C | 206, C | ||
| 19 | 131.6, CH | 6.16 (d; 16.2) | 131.4, CH | 6.12 (d; 16.2) |
| 20 | 146.3, CH | 6.97 (ddd; 16.2, 6.6, 6.6) | 146.8, CH | 6.95 (ddd; 16.2, 6.6, 6.6) |
| 21 | 37.9, CH2 | 2.29 (m) | 37.85, CH2 | 2.27 (m) |
| 2.50 (m) | 2.49 (m) | |||
| 22 | 68.6, CH | 3.89 (m) | 68.4, CH | 3.85 (m) |
| 23 | 39.5, CH | 1.90 (m) | 39.6, CH | 1.92 (m) |
| 24 | 78.6, CH | 5.06 (dd; 4.2, 8.4) | 78.5, CH | 5.07 (dd; 3.6, 9.0) |
| 25 | 40, CH | 1.85 (m) | 39.9, CH | 1.86 (m) |
| 26 | 65.6, CH | 4.03 (dq; 2.4, 6.0) | 65.6, CH | 4.07 (dq; 6.6; 2.4–3.0) |
| 27 | 20, CH3 | 1.23 (d; 6.6) | 19.9, CH3 | 1.22 (d; 6.6) |
| 28 | 13.6, CH3 | 1.03 (d; 7.2) | 14, CH3 | 1.01 (d; 7.2) |
| 29 | 10.9, CH3 | 1.96 (s) | 10.6, CH3 | 1.87 (s) |
| 30 | 14.2, CH3 | 1.22 (d; 6.6) | 16, CH3 | 1.21 (d; 6.6) |
| 31 | 12.5, CH3 | 1.03 (d; 7.2) | 12.1, CH3 | 1.07 (d; 6.6) |
| 32 | 16.6, CH3 | 1.05 (d; 7.2) | 18.6, CH3 | 1.01 (d; 7.2) |
| 33 | 8.9, CH3 | 0.93 (d; 7.2) | 8.8, CH3 | 0.92 (d; 7.2) |
| 34 | 9.8, CH3 | 0.97 (d; 7.2) | 9.9, CH3 | 0.98 (d; 7.2) |
| a | 102.9, CH | 4.26 (d; 7.8) | 102.9, CH | 4.33 (d; 7.2) |
| b | 74.3, CH | 3.17 (ddd; 0, 7.2, 7.2) | 74.7, CH | 3.20 (ddd; 0, 7.2, 7.2) |
| c | 37.2, CH2 | 1.17 (m) ca | 37.2, CH2 | 1.18 (m) ca |
| 2.12 (m) cb | 2.13 (ddd; 1.8, 5.4, 12.6) cb | |||
| d | 80.2, CH | 3.24 (m) | 80.3, CH | 3.27 (m) |
| e | 67.4, CH | 3.56 (dq; 6.6, 1.2) | 67.3, CH | 3.55 (dq; 6.0, 1.8) |
| 20.7, CH3 | 1.21 (d; 6.6) | 20.7, CH3 | 1.23 (d; 6.6) | |
| g | 56, CH3 | 3.43 (s) | 55.9, CH3 | 3.44 (s) |
1H-NMR, 600 MHz; 13C-NMR, 150 MHz.
δ in ppm, J in Hz.
As isolates assigned to OTU6 showed broad activity against pathogenic fungi, a representative strain from this phylotype, Streptomyces sp. strain FXJ1.532, was the subject of intensive chemical study. Two new macrolides (FXJ15321 and FXJ15322) (Fig. 5; Table 4; see also Appendix S1) with similar maximum UV absorption at 230 nm and bearing the same molecular mass of 766 Da (stereoisomers) were obtained from the fermentation extract of this isolate. Interestingly, a unique tetrahydropyran ring with its 1,4 position linking the polyketide chains was detected in both structures, a finding that is quite different from the typical 1,5 linkage in many macrolides (e.g., amphotericin and niphimycin) and has not previously been reported. Further HPLC-UV/VIS analyses revealed that these two compounds were produced by other isolates from this phylotype.
Three Streptomyces strains (FXJ1.264, FXJ1.076, and FXJ1.172) with strong anti-MRSA and antifungal bioactivity were also studied. Following bioactivity tracking, three interesting compounds were obtained from Streptomyces sp. FXJ1.264, anthrabenzoxocinone, etheromycin, and a new etheromycin-type derivative, developments which led to the discovery of a new ionophore, 29-methoxy-etheromycin (Fig. 5) (25). Streptomyces sp. FXJ1.076 was shown to produce the well-known bioactive polyether ionophore nigericin (25) and some elaiophylin-type macrolides, two of which, FXJ10762 and FXJ10763 (Fig. 5; see also Appendix S1 in the supplemental material), were found to be novel. Finally, Streptomyces sp. FXJ1.172 synthesized an antimicrobial ionophore identified as lasalocid A (25). When grown on MOSY medium (M12; see Table S2), the fermentation extracts did not form any lasalocid but still showed strong anti-MRSA bioactivity, which led to the discovery of a new bioactive indole-type antibiotic, N-hydroxythienodolin (Fig. 5; see Appendix S1).
The results from the antimicrobial and antitumor assays showed that compounds FXJ15321 and FXJ15322 had weak antitumor activities, with IC50s of 39.5 and 38.2 μg/ml, respectively, against HeLa cell lines and 44.6 and 48.2 μg/ml, respectively, against SW480 cell lines. Compounds FXJ10762 and FXJ10763 exhibited weak anti-MRSA activities, with MICs of 100 to 200 μg/ml, while preliminary experiments revealed that N-hydroxythienodolin showed strong anti-MRSA activity. However, this compound was unstable after purification and rapidly lost its N-hydroxy group (see Appendix S1 in the supplemental material), and it ceased to show any bioactivity against MRSA; hence, it was not possible to accurately measure its MIC. It is not possible at present to determine why the N-hydroxy group is so important for its bioactivity. The properties of all of these compounds are as follows.
FXJ15321.
Pale yellow solid; UV λmax (MeOH) 230; 1H-NMR and 13C-NMR, Table 4; electrospray ionization (ESI)-MS m/z 784 [M+NH4]+, 789 [M+Na]+, 765 [M-H]−, 801 [M+Cl]−; HR-ESI-MS m/z 789.4402 [M+Na]+ (calculated for C41H66O13, 789.4401).
FXJ15322.
Pale yellow solid; UV λmax (MeOH) 230; 1H-NMR and 13C-NMR, Table 4; ESI-MS m/z 784 [M+NH4]+, 789 [M+Na]+, 765 [M-H]−, 801 [M+Cl]−; HR-ESI-MS m/z 789.4402 [M+Na]+ (calculated for C41H66O13, 789.4401).
FXJ10762.
Grayish-yellow solid; [α]D25 = −4° (c = 0.4, MeOH); UV λmax (MeOH) 249; 1H-NMR and 13C-NMR, see Table A2 in Appendix S1 in the supplemental material; ESI-MS m/z 715 [M+H]+, 737 [M+Na]+, 1,451 [2M+Na]+, 713 [M-H]−; HR-ESI-MS m/z 759.4320 [M+HCOO]− (calculated for C41H62O10, 759.4320).
FXJ10763.
Grayish-yellow solid; [α]D25 = +10° (c = 0.21, MeOH); UV λmax (MeOH) 249; 1H-NMR and 13C-NMR, see Table A3 in Appendix S1 in the supplemental material; ESI-MS m/z 894 [M+NH4]+; HR-ESI-MS m/z 921.5228 [M+HCOO]− (calculated for C48H76O14, 921.5212).
N-Hydroxythienodolin.
UV λmax (MeOH) 239, 274, 292, 334; UV λmax [20% MeOH + 80% water (with 0.5‰ NH3OH)] 252, 296, 379; ESI-MS m/z 264 [M-H]−; HR-ESI-MS m/z 264.9878 [M-H]− (calculated for C11H7ClN2O2S, 264.9839).
DISCUSSION
Our study assessed the taxonomic diversity, phylogenetic novelty, and biosynthetic potential of culturable actinomycetes from red soils.
Diversity and novelty of culturable actinomycetes from the red soils.
Six hundred actinomycetes isolated from red soils collected at six sampling locations were assigned to 26 genera that belonged to 10 families based on 16S rRNA gene sequence analysis. Such high diversity of culturable actinomycetes from acidic environments reflected the use of several pretreatment methods and a range of selective media. In comparison, Muramatsu et al. (17) isolated 122 actinomycetes from Malaysian acid soils (pH 4.2 to 5.9) and assigned them to six genera, while Suela et al. (49) examined 2,152 actinobacterial strains from native Cerrado soils (pH 4.1 to 5.5) and classified them into nine genera. In this and the earlier studies, Streptomyces strains were the most frequently isolated and widely distributed among all samples investigated, although Actinoallomurus, Amycolatopsis, Nocardia, and Streptacidiphilus strains were also isolated. However, in the present study, 21 non-Streptomyces genera were isolated, none of which were observed in the previous studies. In contrast, Actinospica, Frankia, Nakamurella, and Kitasatospora were not recovered from the Jiangxi red soil samples, possibly resulting from a low abundance of these genera in the samples. Some bacterialike genera, such as Arthrobacter, Microbacterium, Leifsonia, and Rhodococcus, were not obtained in the present study either, probably due to our predilection for isolating filamentous actinomycetes.
It was also interesting that members of some genera seemed to be discontinuously distributed; for example, Actinoallomurus strains were isolated only from LJZ and Actinomadura and Streptosporangium strains only from the LJZ and WS samples (see Table S3 in the supplemental material). This observation might be attributed to the possibility that the isolates were not a representative sample of the actinobacterial communities present in the soil samples, because actinomycete spores could have come from anywhere as a result of aerial dispersal and might be distributed across all sampling locations. However, other possibilities are that the geographic distance among the sampling locations was great enough (up to 212 km) for dispersal limitation to occur and/or red soils at the different sampling locations possessed different edaphic characteristics and environmental conditions (e.g., pH, moisture, and anthropogenic activity) that influenced the colonization of certain actinomycetes. Culture-independent studies (e.g., pyrosequencing) in combination with measurements of geographic and environmental factors are needed to check such apparent trends.
Although the 97% 16S rRNA gene sequence similarity threshold has been widely used as a boundary for bacterial species delineation (50), this value has been raised to 98.2 to 99.0% for most prokaryotes (51–53) and to 99% for actinobacteria (36) based on comparative studies between 16S rRNA gene sequence similarities and DNA-DNA hybridization (DDH) and average nucleotide identity (ANI) values of whole genomes. Consequently, in this study, we chose a threshold of 99% to cluster strains into OTUs. Using this metric, the lineages within the 49 OTUs that exhibited <99% sequence similarities with related species were considered to represent putatively new species, a finding that was underpinned by the fact that representatives of these lineages formed distinct phyletic lines (Fig. 1 to 3), indicating that many novel taxa in the red soils can be readily cultivated using fairly traditional approaches to selective isolation and characterization. Indeed, approximately 40% of the Streptomyces isolates (83 of the 208 sequenced) (Fig. 3; see also Fig. S1 in the supplemental material) could not be assigned to known species. Similarly, isolates assigned to the families Pseudonocardiaceae and Streptosporangiaceae showed a high degree of diversity and novelty (Fig. 1 and 2), though this was not as apparent with strains classified in the families Micromonosporaceae, Nocardiaceae, and Thermomonosporaceae, which, despite being frequently isolated, showed much lower diversity at both the genus and OTU level, with the majority of isolates being identified as members of known species.
Biosynthetic potential of actinomycetes from the red soils.
Despite the fact that the frequent rediscovery of known compounds weakened the importance and sometimes caused abandonment of discovery of soil actinomycetes in the past decade (10), in this study, 65% of the 193 different secondary metabolites isolated from 107 semirandomly selected red soil actinomycetes were found to be putatively novel. It is also remarkable that 39 of the 52 producer strains (75%) synthesized putative novel compounds. Given these interesting results, we believe that poorly studied soil ecosystems, such as red soils, are a potentially rich source of promising actinomycetes with the capacity to produce new metabolites as drug leads.
As is apparent from a host of studies, including this one, Streptomyces is a prolific source of natural products. In the present study, over 50% of the Streptomyces isolates showed antagonistic activity and had the potential to produce secondary metabolites. In our screening system, 33 of the 52 producers (64%) were Streptomyces, which accounted for 72% (139 of 193) of the secondary metabolites and 67% (84 of 125) of the putative new ones (Table 2). Furthermore, 10 of the 14 putative new Streptomyces producers were responsible for 38 of the putative new compounds (Table 3). In our earlier investigation, we reported that acidic soil-derived streptomycetes were a promising source of polyether ionophores (25), but it is now apparent that these organisms also produce a broad range of antibiotics that include anthracyclines, diketopiperazines, macrolides, and peptides (Table 3; see also Table S7 in the supplemental material). Some of the Streptomyces producers, particularly those representing putative novel Streptomyces species, such as strains FXJ1.069 and FXJ1.235 (Fig. 3; see also Fig. S1), were an especially rich source of new chemical entities (Table 3).
It has also been shown that nonstreptomycetes from previously neglected or unexplored habitats are a useful source of novel bioactive compounds (54, 55). The AntiBiotic Literature (ABL) database demonstrates that members of the family Micromonosporaceae account for the highest proportion (38%) of producing strains among nonstreptomycete families, but this was not the case in our chemical analyses with respect to the six members of the family Micromonosporaceae. Conversely, 4 of the 6 Thermomonosporaceae isolates (67%), 8 of the 13 Pseudonocardiaceae isolates (62%), and 6 of the 10 Streptosporangiaceae isolates (60%) synthesized secondary metabolites under laboratory fermentation conditions; these were mostly acidophilic (16 out of 18 strains) (Table 3; see also Table S7 in the supplemental material) and accounted for almost all (18 of 19) of the producing capacity shown by nonstreptomycetes from the red soils (Table 2). Among these producers, Amycolatopsis, Microbispora, and Streptosporangium strains showed the greatest capacity to produce secondary metabolites. Furthermore, the production of putative novel compounds is a remarkable feature of the nonstreptomycetes from the red soils (Table 3). Metabolites from 16 of the 19 non-Streptomyces producers distributed in 11 genera did not match any published compounds and, thus, are probably novel.
It should be noted, however, that as our chemical screening system was mainly designed to detect fat-soluble compounds, water-soluble compounds with strong polarity (e.g., aminoglycosides and nucleosides) could not be discovered, and this may have led to the omission of potential isolates with broad activity (e.g., Streptomyces sp. strains FXJ1.086 and FXJ1.088). Furthermore, the three fermentation media used in this study may not have been suitable to induce the production of an array of secondary metabolites.
Antifungal characteristics of actinomycetes from the red soils.
The low pH and physicochemical properties characteristic of red soils also tend to be suitable for fungal growth (56). In the present investigation, a higher ratio of antifungal activity was detected among the acidophilic group than among the acidotolerant group (Fig. 4), a result that is in line with previous studies (15, 42). It has been indicated that the biocontrol of fungi can be attributed mainly to the production of antifungal compounds, nutrient competition between antagonists and pathogens, and secretion of extracellular enzymes that degrade fungal cell walls (57). The results of the present study also suggest that red soil actinomycetes may impose antagonistic effects on pathogenic fungi through the production of polyether ionophores, siderophores, and chitinases.
The ability of polyether ionophores to control drug-resistant bacteria and parasitic infections is well known, but these compounds also show a broad spectrum of bioactivities, including antifungal, antiviral, antitumor, herbicidal, and anti-inflammatory activities (58, 59). In this study, a relatively high number of polyether-producing actinomycetes were isolated, notably the acidophilic streptomycetes (Streptomyces sp. strains FXJ1.050, FXJ1.068, FXJ1.076, FXJ1.172, FXJ1.253, FXJ1.264, and FXJ1.907), which showed antifungal activity. Moreover, the purified polyethers (e.g., nigericin and nanchangmycin) without exception exhibit strong antifungal activity, while other, coproduced metabolites (e.g., elaiophylins and milbemycins) showed very weak, if any, activity against fungi in our tests. These facts indicate that polyether ionophores account for most of the antifungal activity of these isolates, for polyethers are able to transport cations across plasma membranes and lead to depolarization and succedent cell death (25, 57). Although polyene macrolide antibiotics are well known as antifungal agents (60), in our study, only two isolates were found to produce this type of compound. We therefore infer that polyene macrolides are probably not the major agents produced by actinomycetes to control fungi in the red soils.
The ability to synthesize siderophores or siderophore-type compounds is another notable characteristic of actinomycetes isolated from the red soils (Table 3; see also Table S7 in the supplemental material). Iron, while abundant in soil, is not readily bioavailable in aerobic environments due to the low solubility of Fe(III); hence, the need of many terrestrial microorganisms to sequester iron from the environment. Indeed, nearly all fungi have the ability to synthesize siderophore molecules (61), which have a high affinity for ferric ions (62). It is possible that filamentous actinomycetes secrete siderophores that might inhibit the growth of phytopathogens in soil by competition for iron (63–65). It was interesting that all of the siderophores detected except desferrioxamine E were produced following cultivation on SGG medium, on which most isolates neither formed visible aerial hyphae nor sporulated. In contrast, very small amounts of siderophores were produced on GYM and MOSY media, on which cultures formed abundant aerial hyphae and spores (Table 3; see also Table S7). It is possible that the nutrients in SGG medium mimic the competitive environment in red soil, because siderophores are produced in response to intracellular iron deficiency and are secreted into the environment to scavenge iron (66); the bacterial developmental step is acutely sensitive to iron limitation, and the competitive acquisition of iron drives developmental arrest (67).
Antagonistic activity against fungi can also be attributed to chitinolysis (68). Among the red soil actinomycetes, we detected a high rate of chitinolytic strains that correlated with a higher occurrence (39%) of activity against fungi than in nonchitinolytic strains (13%). Similar results were observed for endophytic Streptomyces spp. with potential for biocontrol (69). In addition to synergistic effects with secondary metabolites, the complex chitinolytic system within streptomycetes may allow some strains to effectively degrade fungal cell walls solely by secretion of chitinases (70), as may be the case with isolates from OTU6. Members of this phylotype were shown to be outstanding antagonists of pathogenic fungi, probably due to the production of chitinases, as these strains were not found to produce antifungal secondary metabolites.
Future prospects.
The present study confirms the view that red soils constitute an ideal environment for bioprospecting of actinomycetes. To recover the representative diversity of actinomycetes from red soils, it is important to establish effective isolation approaches. Although this study was not designed to evaluate the effectiveness of different pretreatment/medium regimes and we did not count the actinomycete colony numbers on isolation plates, based on the purified isolates, GTV medium (M6; see Table S2 in the supplemental material) appeared to be the most useful for our objectives by yielding the highest diversity (18 genera) of isolates and number of bioactive strains (15) that produced putative novel compounds, suggesting that the addition of soil extract to medium is helpful for the isolation of actinomycetes from red soils. Nevertheless, given the vast diversity of microbes in soil, further efforts need to be made in comprehensive sampling and high-throughput culture-independent analyses to obtain a panorama of the actinobacterial community in red soils and, then, in the development of improved selective isolation strategies and procedures to isolate members of taxa that were not seen in this culture-dependent study; such efforts are ongoing in our laboratory. In addition, given the wealth of interesting strains and compounds found in this study, efforts to elucidate the structures of the putative new compounds and their bioactivities and to identify their biosynthetic gene clusters are also under way. Besides, in light of the large number of acidophilic isolates and their high diversity, putative novelty, and biosynthetic potential compared to those of the acidotolerant group obtained in this study, it would be meaningful to investigate the adaptive strategies of acidophilic actinomycetes in red soil and their ecological interactions with other microorganisms.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yuanming Luo, Qian Wang, and Guomin Ai of the Institute of Microbiology, Chinese Academy of Sciences (IMCAS), for their help with the MS analyses, Xuejun Jiang at the IMCAS for undertaking the antitumor assays, and colleagues from the Red Soil Ecology Experimental Station of CAS at Liujiazhan for assistance in the collection of the soil samples. We are also grateful to Michael Goodfellow at Newcastle University and other anonymous reviewers for their constructive comments on the manuscript and help in editing it.
This research was supported by the National Natural Science Foundation of China (grants 31170010, 31300010, and 31470142) and by the Specialized Research Fund for the State Key Laboratories of China.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03859-14.
REFERENCES
- 1.Berdy J. 2012. Thoughts and facts about antibiotics: where we are now and where we are heading. J Antibiot (Tokyo) 65:441. doi: 10.1038/ja.2012.54. [DOI] [PubMed] [Google Scholar]
- 2.Wagner M, Abdel-Mageed WM, Ebel R, Bull AT, Goodfellow M, Fiedler HP, Jaspars M. 2014. Dermacozines H-J isolated from a deep-sea strain of Dermacoccus abyssi from Mariana Trench sediments. J Nat Prod 77:416–420. doi: 10.1021/np400952d. [DOI] [PubMed] [Google Scholar]
- 3.Gontang EA, Gaudencio SP, Fenical W, Jensen PR. 2010. Sequence-based analysis of secondary-metabolite biosynthesis in marine actinobacteria. Appl Environ Microbiol 76:2487–2499. doi: 10.1128/AEM.02852-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khan ST, Komaki H, Motohashi K, Kozone I, Mukai A, Takagi M, Shin-ya K. 2011. Streptomyces associated with a marine sponge Haliclona sp.; biosynthetic genes for secondary metabolites and products. Environ Microbiol 13:391–403. doi: 10.1111/j.1462-2920.2010.02337.x. [DOI] [PubMed] [Google Scholar]
- 5.Qin S, Xing K, Jiang JH, Xu LH, Li WJ. 2011. Biodiversity, bioactive natural products and biotechnological potential of plant-associated endophytic actinobacteria. Appl Microbiol Biotechnol 89:457–473. doi: 10.1007/s00253-010-2923-6. [DOI] [PubMed] [Google Scholar]
- 6.Janso JE, Carter GT. 2010. Biosynthetic potential of phylogenetically unique endophytic actinomycetes from tropical plants. Appl Environ Microbiol 76:4377–4386. doi: 10.1128/AEM.02959-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim KH, Ramadhar TR, Beemelmanns C, Cao S, Poulsen M, Currieb CR, Clardy J. 2014. Natalamycin A, an ansamycin from a termite-associated Streptomyces sp. Chem Sci 5:4333–4338. doi: 10.1039/C4SC01136H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meklat A, Sabaou N, Zitouni A, Mathieu F, Lebrihi A. 2011. Isolation, taxonomy, and antagonistic properties of halophilic actinomycetes in Saharan soils of Algeria. Appl Environ Microbiol 77:6710–6714. doi: 10.1128/AEM.00326-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nachtigall J, Kulik A, Helaly S, Bull AT, Goodfellow M, Asenjo JA, Maier A, Wiese J, Imhoff JF, Sussmuth RD, Fiedler HP. 2011. Atacamycins A-C, 22-membered antitumor macrolactones produced by Streptomyces sp. C38. J Antibiot (Tokyo) 64:775–780. doi: 10.1038/ja.2011.96. [DOI] [PubMed] [Google Scholar]
- 10.Baltz RH. 2008. Renaissance in antibacterial discovery from actinomycetes. Curr Opin Pharmacol 8:557–563. doi: 10.1016/j.coph.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 11.Xu RK, Zhao AZ, Li QM, Kong XL, Ji GL. 2003. Acidity regime of the red soils in a subtropical region of southern China under field conditions. Geoderma 115:75–84. doi: 10.1016/S0016-7061(03)00077-6. [DOI] [Google Scholar]
- 12.Zhang MK, He ZL. 2004. Long-term changes in organic carbon and nutrients of an Ultisol under rice cropping in southeast China. Geoderma 118:167–179. doi: 10.1016/S0016-7061(03)00191-5. [DOI] [Google Scholar]
- 13.Liu XZ, Zhang LM, Prosser JI, He JZ. 2009. Abundance and community structure of sulfate reducing prokaryotes in a paddy soil of southern China under different fertilization regimes. Soil Biol Biochem 41:687–694. doi: 10.1016/j.soilbio.2009.01.001. [DOI] [Google Scholar]
- 14.He J, Shen J, Zhang L, Zhu Y, Zheng Y, Xu M, Di HJ. 2007. Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ Microbiol 9:2364–2374. doi: 10.1111/j.1462-2920.2007.01358.x. [DOI] [PubMed] [Google Scholar]
- 15.Khan MR, Williams ST. 1975. Studies on the ecology of actinomycetes in soil. VIII. distribution and characteristics of acidophilic actinomycetes. Soil Biol Biochem 7:345–348. [Google Scholar]
- 16.Williams ST, Davies FL, Mayfield CI, Khan MR. 1971. Studies on the ecology of actinomycetes in soil. II. The pH requirements of streptomycetes from two acid soils. Soil Biol Biochem 3:187–199. [Google Scholar]
- 17.Muramatsu H, Murakami R, Ibrahim ZH, Murakami K, Shahab N, Nagai K. 2011. Phylogenetic diversity of acidophilic actinomycetes from Malaysia. J Antibiot (Tokyo) 64:621–624. doi: 10.1038/ja.2011.57. [DOI] [PubMed] [Google Scholar]
- 18.Monciardini P, Cavaletti L, Ranghetti A, Schumann P, Rohde M, Bamonte R, Sosio M, Mezzelani A, Donadio S. 2009. Novel members of the family Micromonosporaceae, Rugosimonospora acidiphila gen. nov., sp. nov., and Rugosimonospora africana sp. nov. Int J Syst Evol Microbiol 59:2752–2758. doi: 10.1099/ijs.0.010231-0. [DOI] [PubMed] [Google Scholar]
- 19.Kim SB, Seong CN, Jeon SJ, Bae KS, Goodfellow M. 2004. Taxonomic study of neutrotolerant acidophilic actinomycetes isolated from soil and description of Streptomyces yeochonensis sp. nov. Int J Syst Evol Microbiol 54:211–214. doi: 10.1099/ijs.0.02519-0. [DOI] [PubMed] [Google Scholar]
- 20.Busti E, Cavaletti L, Monciardini P, Schumann P, Rohde M, Sosio M, Donadio S. 2006. Catenulispora acidiphila gen. nov., sp. nov., a novel, mycelium-forming actinomycete, and proposal of Catenulisporaceae fam. nov. Int J Syst Evol Microbiol 56:1741–1746. doi: 10.1099/ijs.0.63858-0. [DOI] [PubMed] [Google Scholar]
- 21.Cavaletti L, Monciardini P, Schumann P, Rohde M, Bamonte R, Busti E, Sosio M, Donadio S. 2006. Actinospica robiniae gen. nov., sp. nov., and Actinospica acidiphila sp. nov.: proposal for Actinospicaceae fam. nov. and Catenulisporinae subord. nov. in the order Actinomycetales. Int J Syst Evol Microbiol 56:1747–1753. doi: 10.1099/ijs.0.63859-0. [DOI] [PubMed] [Google Scholar]
- 22.Kim SB, Lonsdale J, Seong CN, Goodfellow M. 2003. Streptacidiphilus gen. nov., acidophilic actinomycetes with wall chemotype I and emendation of the family Streptomycetaceae (Waksman and Henrici (1943)AL) emend. Rainey et al. 1997 Antonie Van Leeuwenhoek 83:107–116. doi: 10.1023/A:1023397724023. [DOI] [PubMed] [Google Scholar]
- 23.Bull AT. 2011. Actinobacteria of the extremobiosphere, p 1203–1240. In Horikoshi K, Antranikian G, Bull AT, Robb FT, Stetter KO (ed), Extremophiles handbook. Springer Verlag, Tokyo, Japan. [Google Scholar]
- 24.Falkinham JO, Wall TE, Tanner JR, Tawaha K, Alali FQ, Li C, Oberlies NH. 2009. Proliferation of antibiotic-producing bacteria and concomitant antibiotic production as the basis for the antibiotic activity of Jordan's red soils. Appl Environ Microbiol 75:2735–2741. doi: 10.1128/AEM.00104-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H, Liu N, Xi L, Rong X, Ruan J, Huang Y. 2011. Genetic screening strategy for rapid access to polyether ionophore producers and products in actinomycetes. Appl Environ Microbiol 77:3433–3442. doi: 10.1128/AEM.02915-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu YB, Cai ZC, Xu ZH. 2012. Production and consumption of N2O during denitrification in subtropical soils of China. J Soil Sediments 12:1339–1349. doi: 10.1007/s11368-012-0548-3. [DOI] [Google Scholar]
- 27.Qiu D, Ruan J, Huang Y. 2008. Selective isolation and rapid identification of members of the genus Micromonospora. Appl Environ Microbiol 74:5593–5597. doi: 10.1128/AEM.00303-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hopkins DW, O'Donnell AG, Macnaughton SJ. 1991. Evaluation of a dispersion and elutriation technique for sampling microorganisms from soil. Soil Biol Biochem 23:227–232. doi: 10.1016/0038-0717(91)90056-P. [DOI] [Google Scholar]
- 29.Hayakawa M, Sadakata T, Kajiura T, Nonomura H. 1991. New methods for the highly selective isolation of Micromonospora and Microbispora from soil. J Biosci Bioeng 72:320–326. [Google Scholar]
- 30.Hayakawa M, Otoguro M, Takeuchi T, Yamazaki T, Iimura Y. 2000. Application of a method incorporating differential centrifugation for selective isolation of motile actinomycetes in soil and plant litter. Antonie Van Leeuwenhoek 78:171–185. doi: 10.1023/A:1026579426265. [DOI] [PubMed] [Google Scholar]
- 31.Chun J, Goodfellow M. 1995. A phylogenetic analysis of the genus Nocardia with 16S rRNA gene sequences. Int J Syst Bacteriol 45:240–245. doi: 10.1099/00207713-45-2-240. [DOI] [PubMed] [Google Scholar]
- 32.Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175. In Stackebrandt E, Goodfellow M (ed), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom. [Google Scholar]
- 33.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klepac-Ceraj V, Ceraj I, Polz MF. 2006. Clusterer: extendable Java application for sequence grouping and cluster analyses. Online J Bioinform 7:15–21. [Google Scholar]
- 36.Stach JE, Maldonado LA, Masson DG, Ward AC, Goodfellow M, Bull AT. 2003. Statistical approaches for estimating actinobacterial diversity in marine sediments. Appl Environ Microbiol 69:6189–6200. doi: 10.1128/AEM.69.10.6189-6200.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 38.Felsenstein J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 39.Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 40.Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J. 2012. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721. doi: 10.1099/ijs.0.038075-0. [DOI] [PubMed] [Google Scholar]
- 41.Hsu SC, Lockwood JL. 1975. Powdered chitin agar as a selective medium for enumeration of actinomycetes in water and soil. Appl Microbiol 29:422–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zakaliukina I, Zenova GM. 2007. Antagonistic activity of soil acidophilic actinomycetes. Biol Bull 34:329–332. doi: 10.1134/S1062359007040036. [DOI] [PubMed] [Google Scholar]
- 43.Mosmann T. 1983. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y, Zheng Z, Liu S, Zhang H, Li E, Guo L, Che Y. 2010. Oxepinochromenones, furochromenone, and their putative precursors from the endolichenic fungus Coniochaeta sp. J Nat Prod 73:920–924. doi: 10.1021/np100071z. [DOI] [PubMed] [Google Scholar]
- 45.Ludwig W, Euzéby J, Schumann P, Busse HJ, Trujillo ME, Kämpfer P, Whitman WB. 2012. Road map of the phylum Actinobacteria, p 1–28. In Goodfellow M, Kämpfer P, Busse HJ, Trujillo ME, Suzuki K, Ludwig W, Whitman WB (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 5 Springer, New York, NY. [Google Scholar]
- 46.Li X, Zhang L, Ding Y, Gao Y, Ruan J, Huang Y. 2012. Lentzea jiangxiensis sp. nov., isolated from acidic soil. Int J Syst Evol Microbiol 62:2342–2346. doi: 10.1099/ijs.0.033795-0. [DOI] [PubMed] [Google Scholar]
- 47.Li X, Zhang L, Ding Y, Gao Y, Ruan J, Huang Y. 2012. Nonomuraea jiangxiensis sp. nov., isolated from acidic soil. Int J Syst Evol Microbiol 62:1409–1413. doi: 10.1099/ijs.0.034520-0. [DOI] [PubMed] [Google Scholar]
- 48.Saintpierre-Bonaccio D, Amir H, Pineau R, Lemriss S, Goodfellow M. 2004. Streptomyces ferralitis sp. nov., a novel streptomycete isolated from a New-Caledonian ultramafic soil. Int J Syst Evol Microbiol 54:2061–2065. doi: 10.1099/ijs.0.02891-0. [DOI] [PubMed] [Google Scholar]
- 49.Suela Silva M, Naves SA, Teixeira MK, Ribeiro DD, Schwan RF. 2013. Brazilian Cerrado soil Actinobacteria ecology. Biomed Res Int 2013:503805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stackebrandt E, Goebel BM. 1994. A place for DNA-DNA reassociation and 16S rRNA sequence-analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849. doi: 10.1099/00207713-44-4-846. [DOI] [Google Scholar]
- 51.Kim M, Oh HS, Park SC, Chun J. 2014. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 52.Meier-Kolthoff JP, Göker M, Spröer C, Klenk HP. 2013. When should a DDH experiment be mandatory in microbial taxonomy? Arch Microbiol 195:413–418. doi: 10.1007/s00203-013-0888-4. [DOI] [PubMed] [Google Scholar]
- 53.Stackebrandt E, Ebers J. 2006. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today 33:152–155. [Google Scholar]
- 54.Kurtboke DI. 2012. Biodiscovery from rare actinomycetes: an eco-taxonomical perspective. Appl Microbiol Biotechnol 93:1843–1852. doi: 10.1007/s00253-012-3898-2. [DOI] [PubMed] [Google Scholar]
- 55.Tiwari K, Gupta RK. 2012. Rare actinomycetes: a potential storehouse for novel antibiotics. Crit Rev Biotechnol 32:108–132. doi: 10.3109/07388551.2011.562482. [DOI] [PubMed] [Google Scholar]
- 56.Rousk J, Brookes PC, Bååth E. 2009. Contrasting soil pH effects on fungal and bacterial growth suggest functional redundancy in carbon mineralization. Appl Environ Microbiol 75:1589–1596. doi: 10.1128/AEM.02775-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chet I, Ordentlich A, Shapira R, Oppenheim A. 1990. Mechanisms of biocontrol of soil-borne plant pathogens by rhizobacteria. Plant Soil 129:85–92. doi: 10.1007/BF00011694. [DOI] [Google Scholar]
- 58.Rutkowski J, Brzezinski B. 2013. Structures and properties of naturally occurring polyether antibiotics. Biomed Res Int 2013:162513. doi: 10.1155/2013/162513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kevin DA, Meujo D, Hamann MT. 2009. Polyether ionophores: broad-spectrum and promising biologically active molecules for the control of drug-resistant bacteria and parasites. Expert Opin Drug Dis 4:109–146. doi: 10.1517/17460440802661443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lewis RE. 2010. Polyene antifungal agents, p 281–305. In Pasqualotto AC. (ed), Aspergillosis: from diagnosis to prevention. Springer Science and Business Media, New York, NY. [Google Scholar]
- 61.Haas H. 2003. Molecular genetics of fungal siderophore biosynthesis and uptake: the role of siderophores in iron uptake and storage. Appl Microbiol Biotechnol 62:316–330. doi: 10.1007/s00253-003-1335-2. [DOI] [PubMed] [Google Scholar]
- 62.Neilands JB. 1995. Siderophores: structure and function of microbial iron transport compounds. J Biol Chem 270:26723–26726. doi: 10.1074/jbc.270.45.26723. [DOI] [PubMed] [Google Scholar]
- 63.Tokala RK, Strap JL, Jung CM, Crawford DL, Salove MH, Deobald LA, Bailey JF, Morra MJ. 2002. Novel plant-microbe rhizosphere interaction involving Streptomyces lydicus WYEC108 and the pea plant (Pisum sativum). Appl Environ Microbiol 68:2161–2171. doi: 10.1128/AEM.68.5.2161-2171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Müller G, Raymond KN. 1984. Specificity and mechanism of ferrioxamine-mediated iron transport in Streptomyces pilosus. J Bacteriol 160:304–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Müller G, Matzanke BF, Raymond KN. 1984. Iron transport in Streptomyces pilosus mediated by ferrichrome siderophores, rhodotorulic acid, and enantio-rhodotorulic acid. J Bacteriol 160:313–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Andrews SC, Robinson AK, Rodriguez-Quinones F. 2003. Bacterial iron homeostasis. FEMS Microbiol Rev 27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 67.Traxler MF, Seyedsayamdost MR, Clardy J, Kolter R. 2012. Interspecies modulation of bacterial development through iron competition and siderophore piracy. Mol Microbiol 86:628–644. doi: 10.1111/mmi.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Neeraja C, Anil K, Purushotham P, Suma K, Sarma P, Moerschbacher BM, Podile AR. 2010. Biotechnological approaches to develop bacterial chitinases as a bioshield against fungal diseases of plants. Crit Rev Biotechnol 30:231–241. doi: 10.3109/07388551.2010.487258. [DOI] [PubMed] [Google Scholar]
- 69.Quecine MC, Araujo WL, Marcon J, Gai CS, Azevedo JL, Pizzirani-Kleiner AA. 2008. Chitinolytic activity of endophytic Streptomyces and potential for biocontrol. Lett Appl Microbiol 47:486–491. doi: 10.1111/j.1472-765X.2008.02428.x. [DOI] [PubMed] [Google Scholar]
- 70.Hoster F, Schmitz JE, Daniel R. 2005. Enrichment of chitinolytic microorganisms: isolation and characterization of a chitinase exhibiting antifungal activity against phytopathogenic fungi from a novel Streptomyces strain. Appl Microbiol Biotechnol 66:434–442. doi: 10.1007/s00253-004-1664-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.