ABSTRACT
The Gram-negative curved bacillus Vibrio cholerae causes the severe diarrheal illness cholera. During host infection, a complex regulatory cascade results in production of ToxT, a DNA-binding protein that activates the transcription of major virulence genes that encode cholera toxin (CT) and toxin-coregulated pilus (TCP). Previous studies have shown that bile and its unsaturated fatty acid (UFA) components reduce virulence gene expression and therefore are likely important signals upon entering the host. However, the mechanism for the bile-mediated reduction of TCP and CT expression has not been clearly defined. There are two likely hypotheses to explain this reduction: (i) UFAs decrease DNA binding by ToxT, or (ii) UFAs decrease dimerization of ToxT. The work presented here elucidates that bile or UFAs directly affect DNA binding by ToxT. UFAs, specifically linoleic acid, can enter V. cholerae when added exogenously and are present in the cytoplasm, where they can then interact with ToxT. Electrophoretic mobility shift assays (EMSAs) with ToxT and various virulence promoters in the presence or absence of UFAs showed a direct reduction in ToxT binding to DNA, even in promoters with only one ToxT binding site. Virstatin, a synthetic ToxT inhibitor, was previously shown to reduce ToxT dimerization. Here we show that virstatin affects DNA binding only at ToxT promoters with two binding sites, unlike linoleic acid, which affects ToxT binding promoters having either one or two ToxT binding sites. This suggests a mechanism in which UFAs, unlike virstatin, do not affect dimerization but affect monomeric ToxT binding to DNA.
IMPORTANCE Vibrio cholerae must produce the major virulence factors cholera toxin (CT) and toxin-coregulated pilus (TCP) to cause cholera. CT and TCP production depends on ToxT, the major virulence transcription activator. ToxT activity is negatively regulated by unsaturated fatty acids (UFAs) present in the lumen of the upper small intestine. This study investigated the mechanism for inhibition of ToxT activity by UFAs and found that UFAs directly reduce specific ToxT binding to DNA at virulence promoters and subsequently reduce virulence gene expression. UFAs inhibit ToxT monomers from binding DNA. This differs from the inhibitory mechanism of a synthetic ToxT inhibitor, virstatin, which inhibits ToxT dimerization. Understanding the mechanisms for inhibition of virulence could lead to better cholera therapeutics.
INTRODUCTION
Vibrio cholerae, a Gram-negative curved bacillus possessing a single polar flagellum, is the causative agent of cholera. Cholera is a diarrheal disease contracted by consuming contaminated food or water; it is characterized by severe diarrhea that leads to dehydration and can ultimately cause death if left untreated. Each year, there are an estimated 1.4 million to 4.3 million cholera cases and 20,000 to 142,000 deaths from the disease (1, 2). To cause disease, V. cholerae must colonize the upper small intestine, where it expresses virulence genes, including those that encode the two most important virulence factors: toxin-coregulated pilus (TCP) and cholerae toxin (CT). TCP is required for intestinal colonization, while CT is responsible for the massive secretion of electrolytes and water into the lumen, causing diarrhea. Transcription of these and other virulence genes is regulated by the major virulence transcription activator ToxT.
Virulence gene expression is regulated by what is historically known as the ToxR regulon (3, 4). toxT transcription is activated by two pairs of inner membrane proteins, ToxR/ToxS and TcpP/TcpH, that bind upstream of toxT, as well as by a ToxT positive-feedback loop (5–9). ToxT is a 32-kDa member of the AraC/XylS transcriptional regulator family having a conserved 100-amino-acid DNA binding domain, consisting of two helix-turn-helix motifs, in its C-terminal domain (CTD). Located upstream of all ToxT-activated genes are 13-bp degenerate DNA sequences called toxboxes that vary in configuration at individual genes (10–14). ToxT directly controls transcription not only of genes encoding TCP and CT but also of other accessory virulence genes, including acfA, acfD, aldA, tagA, tarA, tarB, and tcpI (11–13, 15, 16). Except for the aldA promoter, in which there is a single toxbox, there are two toxboxes present at ToxT-activated promoters (11–14, 16). ToxT can bind to single toxboxes as a monomer, but it is thought that full activation only occurs upon ToxT dimerization on the DNA, at least at some genes (12, 13). The ToxT N-terminal domain (NTD) does not share significant sequence homology with other proteins but has some structural similarity to the AraC NTD, which is the domain necessary for AraC dimerization and arabinose binding (17). NTDs of ToxT have been shown to interact when separated from the CTD, although the true role of dimerization in ToxT function is not fully understood (16–22).
There are a number of host and environmental factors that affect ToxT activity, including temperature, pH, bile, and bicarbonate (23–27). Bile is produced by the liver and then subsequently stored in the gallbladder. Upon eating, bile is released into the duodenum, where it acts to solubilize lipids. Bile itself is a complex, heterogeneous mixture that includes bile salts, cholesterol, bilirubin, saturated fatty acids, and unsaturated fatty acids (UFAs). V. cholerae encounters bile early during infection, and it is proposed to be a natural effector of ToxT, as it is found in the places that V. cholerae colonizes (24, 25, 27). V. cholerae has reduced virulence gene expression in the presence of bile and/or UFAs, with increased motility gene expression, biofilm formation, the induction of efflux pumps, and increased amounts of outer membrane proteins OmpU, OmpT, and TolC (24, 25, 28–31). Bile and its UFA components have been previously shown to decrease CT and TCP production, but the mechanism of this effect on virulence gene expression levels is not fully understood (24, 25). Bile also has another role in that it is able to act as a bactericide for the benefit of the host, but enteric bacteria, such as V. cholerae, not only are adapted to live in the presence of bile but potentially can recognize bile as a signal to ensure survival in the host (24, 27). Another ToxT inhibitor is a synthetic molecule called virstatin (21, 22). The mechanism of action for virstatin has been proposed to be through inhibition of ToxT dimerization (21). Whether UFAs and virstatin inhibit ToxT activity via the same mechanism is unknown.
In this study, we showed that UFAs, specifically the unsaturated fatty acid linoleic acid, are able to enter the bacterial cytoplasm, where ToxT is also present. We report that linoleic acid causes decreased transcription of the ToxT-controlled virulence genes assessed. The mechanism by which this occurs is decreased binding affinity at ToxT-activated promoters in the presence of linoleic acid, regardless of the number or configuration of the toxboxes. Our results indicating that UFAs decrease ToxT binding to DNA even at promoters having one ToxT binding site suggest a mechanism in which UFAs affect the ability of monomeric ToxT to bind to DNA, in contrast to the results we obtained using virstatin. Thus, these two ToxT inhibitors exhibit somewhat different modes of action. These results give us a clearer understanding of the regulatory networks controlling virulence gene expression during human infection.
MATERIALS AND METHODS
V. cholerae strains and growth conditions.
All V. cholerae strains used in this study were derived from classical biotype O395. Strains were maintained in Luria broth (LB) containing 20% glycerol and stored at −70°C. All promoter::lacZ fusions used for β-galactosidase assays were made in plasmid pTL61T as described for previous studies (26, 32). Overnight cultures were grown for ∼16 h at 37°C in LB and then diluted 1:40 into LB (pH 6.5) at 30°C for virulence-inducing conditions in the presence or absence of freshly prepared 0.05% bile (sodium choleate), 160 μM linoleic acid, or 100 μM virstatin. V. cholerae strains were grown with a concentration of 100 μg/ml of streptomycin, and strains with plasmid pTL61T were grown with 100 μg/ml of ampicillin.
[14C]linoleic acid uptake.
V. cholerae classical biotype strain O395 was grown overnight in LB at 37°C, subcultured 1:40 in LB (pH 6.5), and grown for 2 h in the absence of linoleic acid. At 2 h, 0.1 μCi of 14C-radiolabeled linoleic acid (58.2 mCi/mmol; Perkin-Elmer) was added for each milliliter of the subculture. Upon addition of the radiolabeled linoleic acid, 1 ml of the culture was immediately centrifuged, and the supernatant was saved to compare to cell pellet counts per minute. The cell pellet was washed 3 times with 1 ml of phosphate-buffered saline (PBS) and centrifuged each time. Cell pellets were resuspended in 100 μl of PBS and added to 5 ml of scintillation cocktail (Fisher Scientific). The same procedure was followed for other aliquots of the subculture at times 5, 15, and 30 min. After uptake, counts per minute were measured for each time point using an LS6000IC liquid scintillation counting system (Beckman).
[14C]linoleic acid fractionation assay.
After overnight growth, V. cholerae O395 classical biotype was subcultured 1:40 in the absence of linoleic acid. After 2 h, 0.1 μCi of 14C-radiolabeled linoleic acid (58.2 mCi/mmol; Perkin-Elmer) was added to 1 ml of the subculture and incubated at room temperature. After 1 h, the bacteria were harvested by centrifugation and washed three times in PBS. Bacteria were then resuspended in 20 mM Tris-HCl (pH 8.5) and 500 mM NaCl, freeze-thawed in an ethanol and dry ice bath, and then thawed at 37°C. This process was repeated for a total of three freeze-thaw cycles. The bacteria were then fractionated by centrifugation for 10 min at 15,000 × g to separate the membrane and cytoplasm. The cytoplasm was taken as the supernatant and the pellet was resuspended in PBS. Each fraction was then measured for counts per minute of each fraction using a LS6000IC liquid scintillation counting system (Beckman).
Western blot analysis.
Immunodetection of ToxT was performed using the same procedure as previously described (26). Briefly, bacteria were harvested after being grown under inducing conditions with dimethyl sulfoxide (DMSO) only, 160 μM linoleic acid, or 100 μM virstatin. Cells were normalized by optical density at 600 nm (OD600) and resuspended in 2× protein buffer. Samples were then boiled, separated by 14% SDS-PAGE, blotted, and blocked for 30 min in Tris-buffered saline (TBS)-Tween buffer. After blocking, the blots were incubated overnight in a 1:2,500 dilution of rabbit polyclonal anti-ToxT serum and washed three times in TBS buffer, and secondary goat anti-rabbit IgG conjugated to alkaline phosphatase (AP) was used at a dilution of 1:5,000 (Southern Biotech). Blots were washed again and then developed using 5-bromo-4-chloro-3-indolylphosphate (BCIP; MP Biomedicals).
To ensure complete fractionation, immunodetection of cytoplasmic ToxT and the outer membrane protein OmpU was performed. After bacteria were separated into cytoplasmic and membrane fractions, with whole-cell lysate used as a control, a 14% SDS-PAGE gel was run, blotted, and blocked as described above. The membrane was then incubated overnight in anti-ToxT serum to detect the cytoplasmic fraction and in a 1:500 dilution of goat polyclonal anti-OmpU serum to detect the membrane fraction. Blots were washed three times in TBS-Tween buffer, and secondary rabbit anti-goat IgG–AP (to detect OmpU) followed by secondary goat anti-rabbit IgG–AP (to detect ToxT) was used. Blots were washed again and then developed using immune BCIP.
β-Galactosidase assays.
Strains to be analyzed were grown overnight in LB at 37°C and then subsequently subcultured 1:40 into LB (pH 6.5) in the presence or absence of 160 μM linoleic acid dissolved in DMSO. Cultures were grown under virulence-inducing conditions (aeration at 30°C) for 3 h and then analyzed. The β-galactosidase assay was then performed by following the same established protocol (33).
qPCR.
As in previous assays, after overnight growth, V. cholerae classical biotype O395 was subcultured 1:40, either with or without 160 μM linoleic acid. RNA from three biological samples under each condition was extracted using the RNeasy Bacteria Protect minikit (Qiagen), and the manufacturer's protocols were followed. DNA contamination was removed using an on-column RNase-free DNase kit (Qiagen) and confirmed to be free of DNA by the absence of bands using logarithmic PCR. To measure the relative mRNA levels of tcpA and aldA, the following primers were used: forward tcpA (5′-ACGCAAATGCTGCTACACAG-3′), reverse tcpA (5′-CCCCTACGCTTGTAACCAAA-3′), forward aldA (5′-TTGGTGGGCATCCTAACAAT-3′), and reverse aldA (5′-ACACCGGCACCTAAACCATA-3′). Quantitative real-time PCR (qPCR) was performed using one-step SYBR green master mix (Invitrogen) and the following program: cDNA synthesis for 10 min at 55°C and a denaturing step for 5 min at 95°C, followed by 35 cycles of 95°C for 10 s and then 55°C for 30 s. The level of each mRNA was normalized to the level of rpoB using primers forward rpoB (5′-ACCTGAAGGTCCAAACATCG-3′) and reverse rpoB (5′-CAAAACCGCCTTCTTCTGTC-3′). The relative levels of transcript with the addition of linoleic were calculated using the threshold cycle (2−ΔΔCT) method and analyzed comparing the ΔΔCT values as previously described (34).
Protein purification.
Maltose binding protein-ToxT fusion (MBP-ToxT) and MBP-cyclic AMP (cAMP) receptor protein (MBP-CRP) purification was performed as previously described using Escherichia coli strain BL21(DE3) with plasmid pMAL-c2E carrying either MBP-ToxT or MBP-CRP (32, 35). Briefly, after MBP-ToxT or MBP-CRP induction by isopropyl-β-d-thiogalactopyranoside (IPTG), cells were lysed by French press and run over an amylose column, and fractions containing MBP-ToxT or MBP-CRP were saved and dialyzed into 50 mM Na2HPO4 (pH 8.0) and 100 mM NaCl and then dialyzed into the same solution containing 20% glycerol to save as a freezer stock at −80°C.
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were performed as previously described (35). Purified MBP-ToxT, or MBP-CRP with cAMP (New England BioLabs), was incubated with DNA probes made by PCR from the promoter sequence of interest using one primer radiolabeled with γ-32P (Perkin-Elmer) by T4 polynucleotide kinase (New England BioLabs). Binding reaction mixtures contained various amounts of MBP-ToxT with a constant 10 μg/ml of salmon sperm DNA as a nonspecific competitor, 10 mM Tris-acetate (pH 7.4), 1 mM potassium EDTA (pH 7.0), 100 mM KCl, 1 mM dithiothreitol (DTT), 0.3 mg/ml of bovine serum albumin (BSA), and 10% glycerol in a volume of 30 μl. To each reaction mixture, a constant concentration of the labeled DNA probe was added. In reaction mixtures containing linoleic acid, the final concentration was 32 μM for each, and for reaction mixtures containing virstatin, the final concentration was 50 μM, except at the aldA promoter, where a higher concentration, 100 μM, was used. All other reaction mixtures contained 3.33% (1 μl in 30 μl) DMSO as a solvent control. Binding reaction mixtures were incubated for 30 min at 37°C and then loaded into a 6% polyacrylamide gel at 4°C. Gels were dried for 1 h and then analyzed by autoradiography.
Binding-curve analysis.
Autoradiographs were analyzed using ImageJ software (NIH) as previously described, with nonspecific binding omitted from further analysis (32). Briefly, to determine the KD (equilibrium dissociation constant) for samples containing either linoleic acid or virstatin and compared to protein bound to DNA without inhibitor, the percentage of protein bound with labeled DNA was determined for each lane. This was then fit to the following equation: percent bound = Bmax × [protein]h/(KDh + [protein]h), where h is the Hill coefficient and Bmax is the amount of bound DNA at which the curve plateaus, which was set to a constraint of 100% using GraphPad Prism 5 software. The KD values for each condition were compared to each other using the extra sum of squares F test to determine if the two values were statistically different.
RESULTS
ToxT protein is produced in the presence of linoleic acid.
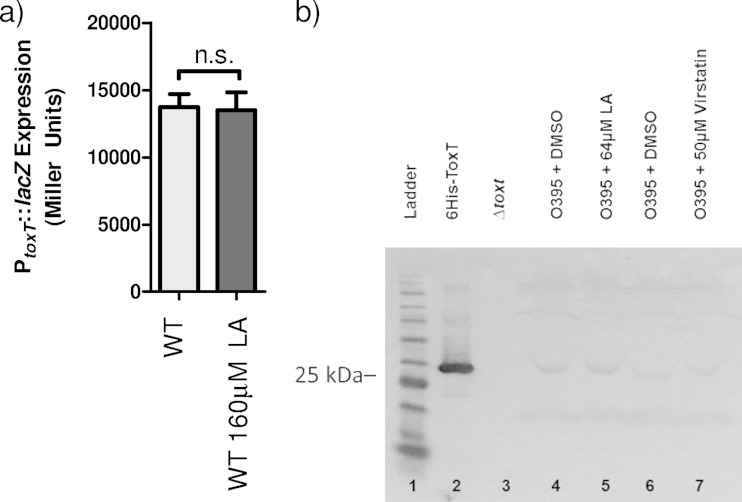
Previous work has demonstrated that UFAs inhibit virulence gene expression by acting through ToxT (25). Oleic acid has been previously shown to negatively regulate TCP and CT expression levels (17). In preliminary experiments, we determined that linoleic acid had the strongest negative effect on ToxT activity of several different UFAs tested (data not shown), and previous studies have shown that linoleic acid makes up the largest proportion of unsaturated fatty acid content in human bile (36). To date, direct interaction of linoleic acid and ToxT has not been observed, although a palmitoleic acid molecule was visualized within the ToxT NTD in the solved crystal structure (17). To begin our study, we investigated whether ToxT protein production was affected by the addition of linoleic acid in the media. It had been previously determined that there are comparable levels of toxT expression in cultures grown with and without linoleic acid (25). To confirm this, we analyzed the β-galactosidase production from a toxT::lacZ reporter plasmid in V. cholerae grown in the presence or absence of linoleic acid (Fig. 1a). When V. cholerae was grown under virulence-inducing conditions (LB pH = 6.5, aeration) in the presence of linoleic acid, the amount of β-galactosidase activity indicated that there was no statistically significant difference (P = 0.799) in toxT expression whether the culture had linoleic acid or lacked it. This is consistent with previous data confirming that linoleic acid does not affect toxT transcription.
FIG 1.
Effect of linoleic acid on PtoxT::lacZ and ToxT expression. (a) Wild-type (WT) V. cholerae was grown without or with 160 μM linoleic acid (LA). Statistical significance was determined by Student's t test. n.s., not significant. (b) Effects of linoleic acid and virstatin on ToxT protein levels. V. cholerae was grown under virulence-inducing conditions (LB [pH 6.5], 30°C, aeration) with DMSO, linoleic acid, or virstatin (lanes 4, 5, 6, and 7). Purified full-length ToxT-6His migrating to ∼32 kDa (lane 2) and a ΔtoxT strain showing no band (lane 3) were used as controls. Samples were normalized by optical density at 600 nm (OD600). The Western blot was probed with anti-ToxT antibody. This is representative of three separate experiments.
To further examine the effect of linoleic acid on ToxT, and confirm that there were no effects of linoleic acid on translation, we carried out Western blot analysis to assess the ToxT protein levels (Fig. 1b). V. cholerae was grown under virulence-inducing conditions in the presence or absence of linoleic acid, and cell extracts were harvested. ToxT-specific polyclonal antibodies were used to detect protein levels. No differences in ToxT levels were observed regardless of the presence or absence of linoleic acid, as a ToxT-specific band was visible in Western blots at the same intensity under both conditions (Fig. 1b, lanes 4 and 5). This band was not detected in the ΔtoxT control strain (Fig. 1b, lane 3). Thus, ToxT was stably produced regardless of the presence of linoleic acid while bacteria were grown under virulence-inducing conditions.
Linoleic acid enters the V. cholerae cytoplasm.
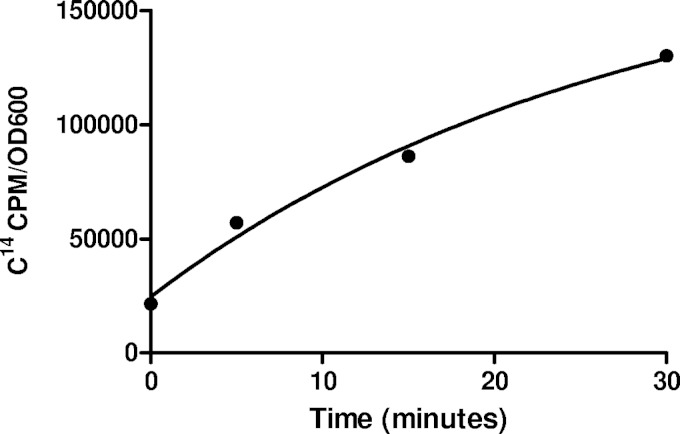
As ToxT protein levels were roughly equivalent with and without linoleic acid, we next assessed whether a direct interaction between linoleic acid and ToxT would be possible. Such an interaction would require that linoleic acid be imported into the bacterial cytosol. To determine whether this occurs, we performed an uptake assay using radiolabeled linoleic acid (Fig. 2). V. cholerae was subcultured under virulence-inducing conditions for 2 h, and then 14C-labeled linoleic acid was added to the culture. Aliquots of the culture were taken at 0, 5, 15, and 30 min after addition of radiolabeled linoleic acid, and radioactivity was quantified by a scintillation counter. The amount of radioactivity found in the cell pellet was compared to that in the supernatant. Results from this assay show that linoleic acid enters the bacteria with a best-fit equation for one-phase association of y = 24,622 + 156,786 × (1 − e−0.036x) and an R2 value of 0.989.
FIG 2.
14C-radiolabeled linoleic acid is taken up by V. cholerae. Classical strain O395 cells were grown under virulence-inducing conditions for 2 h, and then the radiolabeled linoleic acid was added at time zero and analyzed at 5, 15, and 30 min postaddition, measured as counts per minute per optical density unit at 600 nm. The equation of best fit is y = 24,622 + 156,786 × (1 − e−0.036x) and an R2 value of 0.989.
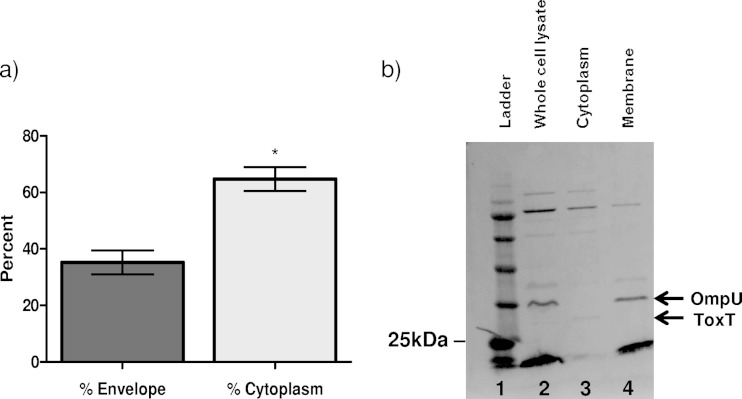
These data, however, do not indicate whether linoleic acid is present inside the bacterium or simply associated with the cell surface or outer membrane. To determine whether linoleic acid enters the cytosol, 14C-labeled linoleic acid was added to V. cholerae cultures grown under virulence-inducing conditions and then the bacteria were fractionated into membrane and cytoplasmic portions. This allowed us to see where in the bacterium linoleic acid was localized: the membrane, the cytoplasm, or both. Cultures were incubated for 1 h with the radiolabeled linoleic acid. These cultures were then pelleted and washed, and the bacteria were separated into cytoplasmic and envelope fractions. The [14C]linoleic acid in each fraction was then quantified using a scintillation counter. Results of this experiment show that most of the linoleic acid was able to enter the cell (Fig. 3a). A control Western blot was performed to ensure complete fractionation of the bacteria and showed no ToxT in the membrane fraction and no OmpU in the cytoplasmic fraction (Fig. 3b). Directly comparing the amounts of linoleic acid in the two fractions showed that about 65% of the total [14C]linoleic acid was present in the cytoplasmic fraction, while 35% was present in the membrane fraction. These data suggest that a significant fraction of extracellular linoleic acid can enter the cytosol, where it could then interact with ToxT.
FIG 3.
14C-radiolabeled linoleic acid is able to enter the cell and enter the cytoplasm. (a) After a 2-h subculture under virulence-inducing conditions, linoleic acid was added to the culture and the culture was allowed to sit for 1 h. After 1 h, V. cholerae was fractionated into envelope and cytoplasmic portions by multiple freeze-thaw cycles. There was significantly more linoleic acid in the cytoplasmic portion than in the envelope portion. Error bars represent standard errors of the means, and statistical significance was determined by Student's t test (*, P < 0.05). (b) Western blot showing fractionation of V. cholerae. Whole-cell lysate (lane 2) shows that both ToxT and OmpU are present, while the cytoplasm (lane 3) only shows ToxT and the membrane (lane 4) only shows OmpU. The Western blot was probed with anti-ToxT and anti-OmpU antibodies. This is representative of three separate experiments.
Linoleic acid decreases ToxT activation of virulence gene transcription.
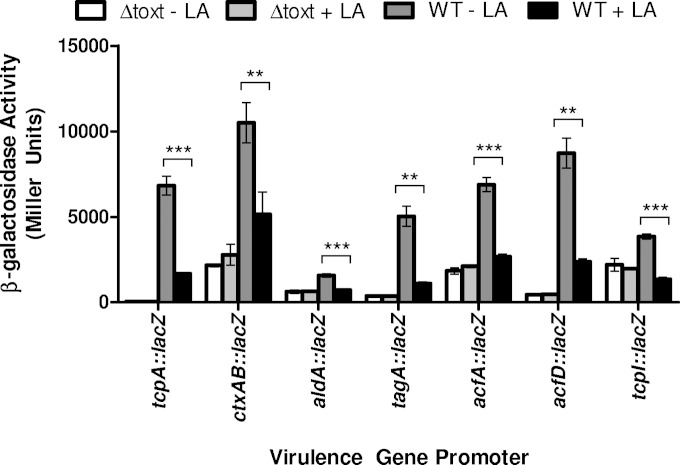
tcpA and ctxAB expression has been previously shown to be inhibited by the addition of oleic acid to the media, but the effects of linoleic acid were not assessed in those studies (17). We assessed whether the inhibitory effect of linoleic acid that we observed extended to a variety of ToxT-activated promoters with various ToxT binding site (toxbox) configurations, including tcpA, ctxAB, acfA, aldA, acfD, tagA, and tcpI. All of these promoters contain two toxboxes in either direct (tcpA and ctxAB) (13) or inverted (acfA, acfD, tagA, and tcpI) (12–14) repeat configurations except for aldA, which contains only one toxbox (16). Plasmid-borne promoter-lacZ fusions were added to the wild-type V. cholerae classical strain as well as an otherwise isogenic ΔtoxT strain to measure transcriptional activity at these promoters with and without 160 μM linoleic acid (Fig. 4). At each of these promoters, linoleic acid had a statistically significant negative effect that prevented ToxT from fully activating transcription at the promoters in wild-type O395. No effect of linoleic acid was observed in the ΔtoxT strain, confirming that linoleic acid acts through ToxT (Fig. 4).
FIG 4.
The negative effect of linoleic acid on various ToxT-activated promoters. Cultures were grown under virulence-inducing conditions with or without 160 μM linoleic acid. β-Galactosidase activity produced from plasmid-borne virulence gene promoter fusion constructs in either wild-type or isogenic ΔtoxT. Statistical significance determined using Student's t test (*, P < 0.005; **, P < 0.0005). Error bars represent standard errors of the means.
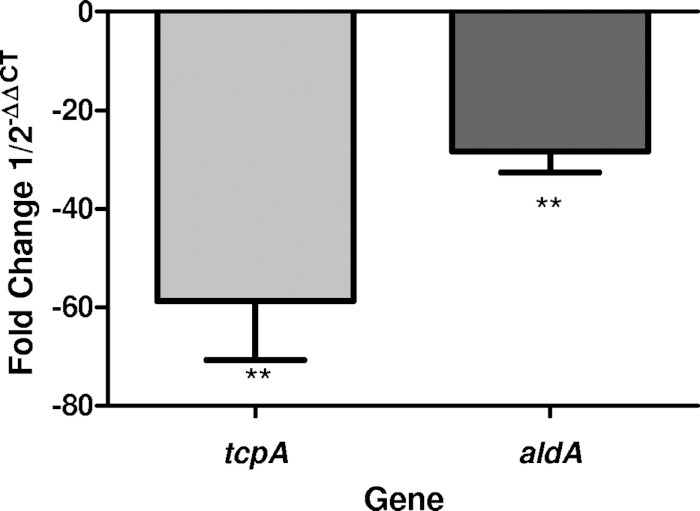
As the aldA promoter activity even in the absence of linoleic acid was much lower than at the other promoters, we performed quantitative real-time PCR (qPCR) to compare expression levels of this promoter to expression levels of the tcpA promoter upon the addition of linoleic acid. Bacteria were cultured using the same methods as for the β-galactosidase assay, and then the RNA was extracted. qPCR was then performed using primers for tcpA and aldA, while using rpoB as the housekeeping gene control. The qPCR results showed that both tcpA and aldA were downregulated upon the addition of linoleic acid to the media (Fig. 5).
FIG 5.
qPCR data of tcpA and aldA comparing cultures grown with and without linoleic acid. Both transcripts were expressed less with the addition of linoleic acid. Data are shown as negative fold change. Statistical significance was determined using Student's t test (**, P < 0.005).
Linoleic acid decreases ToxT-DNA binding affinity.
EMSA was used to determine if linoleic acid is able to directly affect ToxT binding to DNA at the ToxT-activated promoters. This method allows the binding pattern of ToxT to be characterized and gives a rough estimation of the equilibrium dissociation constant (KD), which represents the concentration of ToxT required for binding 50% of the DNA at equilibrium. As the concentration of active ToxT varies from preparation to preparation, the KD values can be compared only per experimental condition and are only estimates. The KD values with and without linoleic acid give a direct comparison of the effect of the fatty acid on the given promoter, as a higher KD value indicates a lower binding affinity of ToxT for the DNA and a lower KD value indicates higher binding affinity.
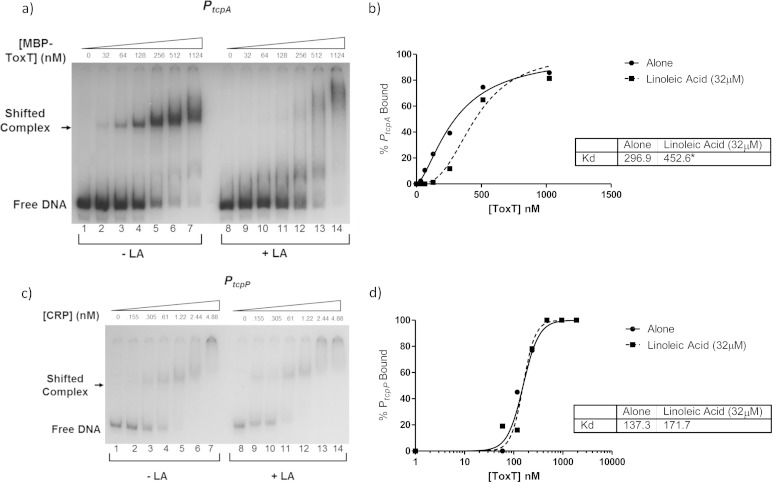
To begin, we used the primary promoter in the tcp operon, PtcpA, as our 32P-labeled DNA probe. Using purified MBP-ToxT, we added increasing amounts of protein to binding reaction mixtures containing a constant concentration of labeled DNA probe. Additionally, either DMSO alone (Fig. 6a, lanes 1 to 7) or 32 μM linoleic acid dissolved in DMSO was added to the reaction mixtures. A representative EMSA is shown in Fig. 6a. Densitometry analysis was performed to calculate the percentage of bound DNA at each concentration of MBP-ToxT, and a binding curve was drawn for each set of reactions to determine the percentage of bound DNA at each concentration of MBP-ToxT (Fig. 6b). In the case of MBP-ToxT binding to PtcpA DNA, the KD without linoleic acid was 296.9 nM and the KD with linoleic acid was 452.6 nM. These values are significantly different and indicate a lower ToxT binding affinity for PtcpA in the presence of linoleic acid. These data strongly suggest that there is a direct interaction between linoleic acid and ToxT.
FIG 6.
Linoleic acid decreases binding affinity of MBP-ToxT to PtcpA. MBP-ToxT binding to PtcpA was analyzed using EMSA. (a) Binding reactions between MBP-ToxT and PtcpA in lanes 1 to 7 were conducted in the absence of linoleic acid. Lanes 8 to 14 were incubated in the presence of 32 μM linoleic acid. Lanes 1 and 8 contained PtcpA DNA in the absence of MBP-ToxT. Subsequent lanes contained a titration of MBP-ToxT with concentrations labeled. (b) Binding curve for the autoradiograph shown in panel a. Densitometry of the autoradiograph was performed with ImageJ software. Circles represent percent PtcpA bound by MBP-ToxT in the absence of linoleic acid. The solid line corresponds to the binding curve for MBP-ToxT. Squares and the dashed line represent percent bound and binding curve, respectively, in the presence of linoleic acid. (c) Autoradiograph of EMSA showing titration of MBP-CRP bound to PtcpP with DMSO (lanes 1 to 7) and linoleic acid (lanes 8 to 14). (d) Binding curves with and without linoleic acid with KD. Autoradiographs of EMSAs presented are representative of three or more independent experiments. The KD for each condition is shown in the inset, and significant difference between the best-fit values of each data set is indicated by an asterisk (P < 0.0001).
To confirm that linoleic acid does not generally inhibit protein-DNA interactions, we used cyclic AMP receptor protein (CRP) as a control, together with the region upstream of tcpP to which CRP binds, as described above. We performed an EMSA comparing CRP binding data with and without the addition of linoleic acid (Fig. 6c) and performed densitometric analysis to determine the binding curves and KD values. The binding curves and KD values were statistically the same for CRP binding to PtcpP with and without linoleic acid (Fig. 6d).
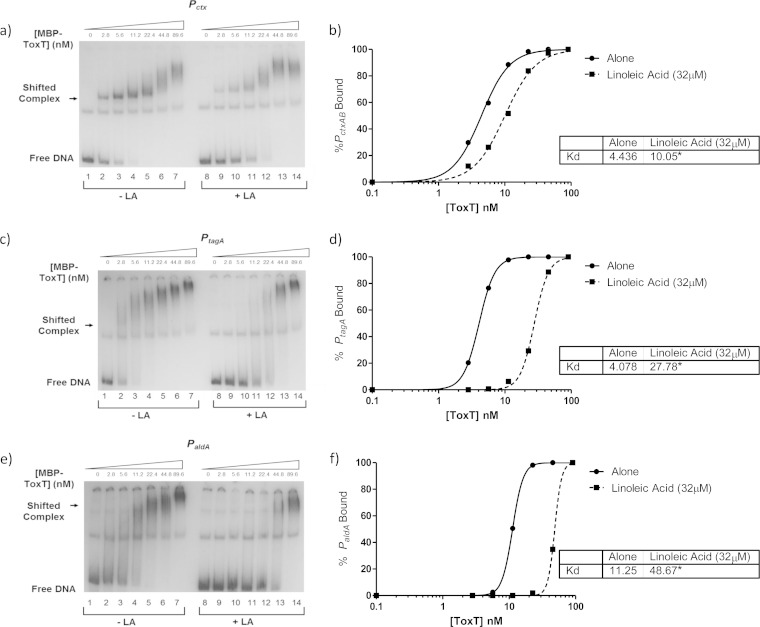
We next determined the binding curves and KD values at other ToxT-activated promoters to determine whether different toxbox configurations may impact the effect of linoleic acid on ToxT binding to DNA (Fig. 7). Equilibrium binding experiments on PtcpA, which has two direct repeat toxboxes, showed a decrease in DNA binding upon addition of linoleic acid. Other promoters have direct and inverted repeat toxbox configurations, and PaldA has only one toxbox (Fig. 7). If linoleic acid could decrease the binding affinity of ToxT for PaldA, it could indicate that the mechanism for ToxT inhibition did not involve effects on dimerization, which had been previously proposed as the mechanism for decreasing virulence gene expression (18). PctxAB contains two directly repeated toxboxes like PtcpA, albeit in the opposite orientation and having different spacing (35). The rest of the ToxT-activated promoters previously mentioned, PacfA, PacfD, PtagA, and PtcpI, each contain toxboxes oriented in inverted repeat configuration as well as variable spacing between the toxboxes (12–14).
FIG 7.
Linoleic acid decreases binding affinity of MBP-ToxT at other virulence promoters. In all radiographs, binding reactions in lanes 1 to 7 are with DMSO only, while binding reactions in lanes 8 to 14 are in the presence of 32 μM linoleic acid. Panels a, c, and e show autoradiographs of MBP-ToxT binding at Pctx, PtagA, and PaldA, respectively. Panels b, d, and f show binding curves corresponding to the autographs for Pctx, PtagA, and PaldA, respectively. All autoradiographs and binding curves are representative of at least three separate experiments. The KD for each condition is shown in the inset, and significant difference between the best-fit values of each data set is indicated by an asterisk (P < 0.0001).
As linoleic acid was shown as described above to decrease promoter activity for all of these ToxT-activated promoters, we determined whether linoleic acid also affected the DNA binding affinity using EMSA with each promoter region. For PctxAB, containing direct repeats, the KD without linoleic acid is 4.436 nM and that with linoleic acid is 10.05 nM, indicating decreased binding affinity of ToxT (Fig. 7a and b). We chose to look at PtagA as a representative of the inverted repeat toxbox promoters. Densitometry and binding-curve analysis again revealed decreased binding affinity of ToxT for the promoter upon the addition of linoleic acid, with KDs of 4.078 nM without linoleic acid and 27.78 nM with linoleic acid (Fig. 7c and d). Lastly, we examined ToxT binding to the single toxbox promoter, PaldA. Figure 7e shows the autoradiograph of ToxT binding to PaldA with and without linoleic acid. The KD once again was increased, going from 11.25 nM without linoleic acid to 48.67 nM with linoleic acid (Fig. 7f). These data, along with the previously described decrease in promoter activity with the addition of linoleic acid as indicated by the β-galactosidase assays, suggest that linoleic acid decreases binding at each promoter regardless of toxbox configuration and spacing.
Another negative ToxT effector, virstatin, acts differently than linoleic acid.
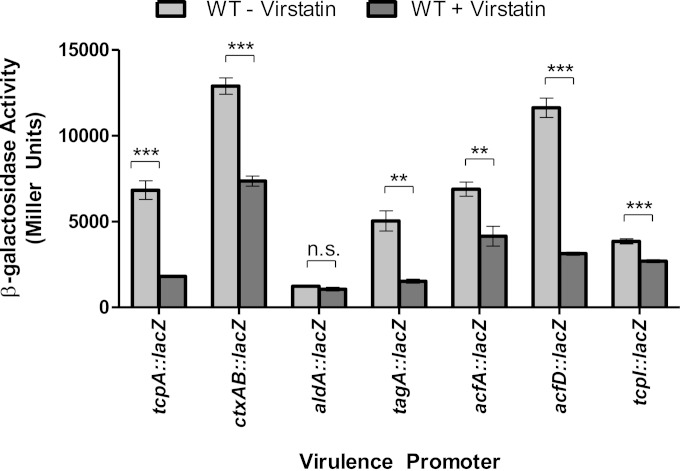
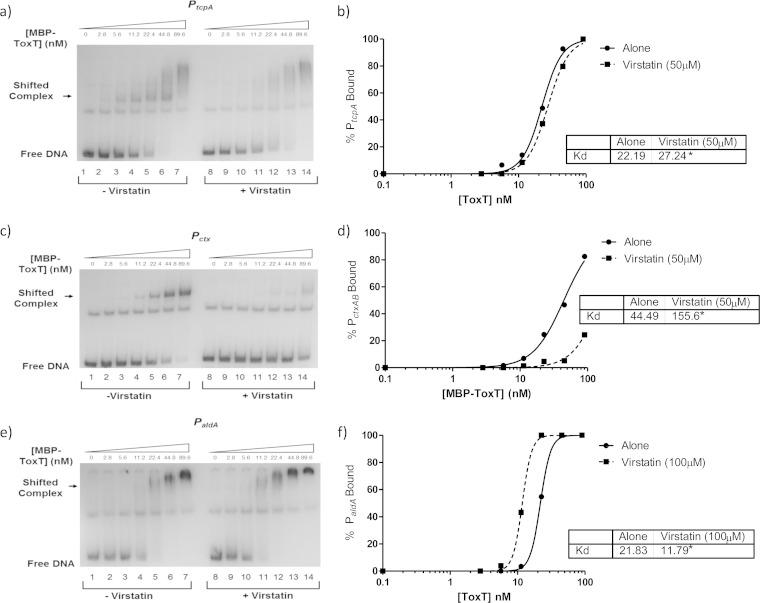
The results described above suggest a direct negative effect of linoleic acid on ToxT binding to DNA. We next examined whether this was also true for another negative ToxT effector, virstatin. Previous studies suggested that virstatin negatively affects ToxT dimerization (21). To begin, we looked at ToxT levels using Western blot analysis to compare V. cholerae grown with and without virstatin (Fig. 1b, lanes 6 and 7). The ToxT levels were unchanged, confirming that ToxT production is not affected by virstatin. We then performed β-galactosidase assays on the previously described ToxT-regulated genes to assess the effects of virstatin (Fig. 8). These data show decreased ToxT activity at all promoters except for aldA, where virstatin had no significant effect. A previous study suggested that the aldA promoter was affected by virstatin (21). We then performed EMSAs to determine whether virstatin affected the DNA binding ability of ToxT at tcpA, ctxAB, and aldA, as representatives of the different toxbox configurations (Fig. 9). Virstatin decreased binding at tcpA and ctxAB promoters, as indicated by analysis of the binding curve and KD (Fig. 9a to d), but did not decrease ToxT binding at the aldA promoter even at a higher concentration of virstatin (Fig. 9e). In fact, the KD significantly decreased, from 21.83 nM to 11.79 nM, with the addition of virstatin, indicating a higher binding affinity of ToxT for the aldA promoter when virstatin is present (Fig. 9f).
FIG 8.
Effect of virstatin on ToxT-regulated virulence gene promoters. Cultures were grown under virulence-inducing conditions with and without virstatin. Promoter activities with and without 100 μM were compared, and at all promoters except for aldA, virstatin caused a significant decrease in promoter activity. Statistical significance was determined using Student's t test (**, P < 0.005; ***, P < 0.0005).
FIG 9.
Virstatin decreases binding affinity of MBP-ToxT at PtcpA and Pctx but not at PaldA. In all radiographs, binding reactions in lanes 1 to 7 are with DMSO only, while binding reactions in lanes 8 to 14 are in the presence of virstatin. A 50 μM concentration of virstatin was used for panels a and c, and 100 μM was used for panel e. Panels a, c, and e show autoradiographs of MBP-ToxT binding at PtcpA, Pctx, and PaldA, respectively. Panels b, d, and f show the binding curves corresponding to the autographs for PtcpA, Pctx, and PaldA, respectively. All autoradiographs and binding curves are representative of at least three separate experiments. The KD for each condition is shown in the inset, and significant difference between the best-fit values of each data set is indicated by an asterisk (P < 0.0005).
DISCUSSION
The activation of virulence gene expression relies on the major virulence transcription activator, ToxT. Previous studies have looked at the involvement of bile, UFAs, and virstatin on V. cholerae virulence gene expression and ToxT dimerization (17, 21, 22, 24, 25, 27). Most of this work with negative effectors has been done only on tcpA and ctxAB; in this study, in addition to these genes, we also examined the effects of negative effectors on ToxT activity at aldA, acfA, acfD, tagA, and tcpI. All ToxT-controlled promoters showed a decrease in expression in the presence of linoleic acid. Prior to entry into the mucous layer of epithelial cells and colonization, V. cholerae must pass through the lumen of the small intestine, where high concentrations of bile and UFAs are present. Linoleic acid must be taken up by V. cholerae and enter the cytoplasm in order to directly interact with ToxT. Here, we show that linoleic acid is able to enter the cell and that the majority of it enters the cytoplasm. One caveat is that there may be periplasmic components present in the cytoplasmic portion in our fractionation studies. However, the observation that a majority of linoleic acid was in this fraction, combined with our other observations that linoleic acid directly impacts DNA binding by ToxT in vitro and ToxT activity in vivo, strongly suggest that at least some linoleic acid is present in the bacterial cytosol, where it directly interacts with ToxT.
Previous work has described the effect of negative ToxT effectors on virulence gene expression, but their direct effect on ToxT DNA binding affinity had not been previously assessed. It has been proposed that the mechanism of action for virstatin and UFAs is inhibition of ToxT dimerization (18, 21). The binding sites for ToxT, toxboxes, and their configurations have been extensively studied (12, 13, 16, 35), but whether toxbox configuration factors into the inhibition of ToxT activity by linoleic acid had not been examined. Here we show that the decrease in binding affinity in response to linoleic acid can be seen at each ToxT-activated promoter that we examined, regardless of toxbox configuration. This suggests that linoleic acid has a general effect on DNA binding by ToxT monomers. Previous work strongly suggested that ToxT binds to DNA as a monomer (12, 13, 16) and that ToxT does not behave as a typical obligatorily dimeric activator, such as AraC.
We also looked at virstatin and its method of action on ToxT. A previous study examined virulence gene promoter activity in the presence of virstatin; however, unlike in that earlier study, the effect of virstatin in our hands was not as pronounced. We did observe that virstatin significantly decreased promoter activity, but to a lesser extent than previously described (Fig. 8) (21). While Shakhnovich et al. saw, at most, 20% promoter activity in the presence of virstatin at the aldA, acfA, and tcpI promoters, we observed a decrease, but not to that degree. Interestingly, we did not see any effect of virstatin on ToxT activation at the single toxbox, aldA promoter with the β-galactosidase assay (Fig. 8). In the same vein, virstatin also did not negatively affect the binding affinity of ToxT for the aldA promoter (Fig. 9e and f). In contrast, we did see a negative effect of linoleic acid on both ToxT-dependent virulence gene promoter activity (Fig. 4) and ToxT binding affinity for PaldA, where the KD increased from 11.25 nM to 48.67 nM (Fig. 7e and f).
These data suggest that virstatin and linoleic acid, although both negative effectors of ToxT activity, do not have the same mechanism for downregulating gene expression. In another study, bacterial two-hybrid assays were performed to look at the effects of various UFAs on dimerization, and the data suggested that another UFA, oleic acid, does affect ToxT dimerization (25). However, linoleic acid was not tested, and that study was done using only the NTD of ToxT instead of intact, full-length ToxT, whose dimerization has never been experimentally observed, at least according to published reports. More testing using full-length ToxT in V. cholerae with this two-hybrid system in the presence of linoleic acid would confirm whether linoleic acid is involved in dimerization. As there was reduced binding of ToxT at the aldA promoter, it is likely that linoleic acid changes the structural conformation of ToxT, leading to inhibited DNA binding.
Understanding the effect of host signals on bacterial pathogenesis is useful for complete understanding of the V. cholerae virulence cascade, as many host signals are used by the bacteria to sense the appropriate location in which virulence gene transcription should be initiated. When passing through the small intestine, V. cholerae encounters many different chemical signals, such as bile and bicarbonate, which either inhibit or activate ToxT-dependent gene transcription. Bile and its UFA components are at high concentrations in the duodenal lumen, in which ToxT is inactive (18, 20, 25, 27). Once the mucous layer is penetrated, the UFA concentration decreases, while the bicarbonate concentration increases. At this point, along the epithelial surface, ToxT becomes active, virulence genes are expressed, and V. cholerae can colonize. UFAs such as linoleic acid likely keep ToxT in a form unable to bind tightly to DNA in the lumen, because production of TCP would lead to bacterial aggregation prior to penetration of the mucous layer and colonization of the epithelial surface would become impossible. According to this model, V. cholerae is able to use both negative and positive effectors of virulence in order to identify and colonize its ideal niche. Our findings here show a direct effect of linoleic acid on ToxT, giving insight into the mechanisms of ToxT gene regulation.
ACKNOWLEDGMENTS
We thank the members of the Withey laboratory for helpful discussions.
This work was supported by PHS grants K22AI071011 and R56AI093622 (to J.H.W.), Bill and Melinda Gates Foundation grant OPP1068124, and Wayne State University funds.
REFERENCES
- 1.World Health Organization (WHO). 2013. Cholera, 2012. Wkly Epidemiol Rec 88:321–334. [PubMed] [Google Scholar]
- 2.Ali M, Lopez AL, You YA, Kim YE, Sah B, Maskery B, Clemens J. 2012. The global burden of cholera. Bull World Health Organ 90:209–218A. doi: 10.2471/BLT.11.093427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peterson KM, Mekalanos JJ. 1988. Characterization of the Vibrio cholerae ToxR regulon: identification of novel genes involved in intestinal colonization. Infect Immun 56:2822–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skorupski K, Taylor RK. 1997. Control of the ToxR virulence regulon in Vibrio cholerae by environmental stimuli. Mol Microbiol 25:1003–1009. doi: 10.1046/j.1365-2958.1997.5481909.x. [DOI] [PubMed] [Google Scholar]
- 5.Häse CC, Mekalanos JJ. 1998. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci U S A 95:730–734. doi: 10.1073/pnas.95.2.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Higgins DE, DiRita VJ. 1994. Transcriptional control of toxT, a regulatory gene in the ToxR regulon of Vibrio cholerae. Mol Microbiol 14:17–29. doi: 10.1111/j.1365-2958.1994.tb01263.x. [DOI] [PubMed] [Google Scholar]
- 7.Krukonis ES, Yu RR, Dirita VJ. 2000. The Vibrio cholerae ToxR/TcpP/ToxT virulence cascade: distinct roles for two membrane-localized transcriptional activators on a single promoter. Mol Microbiol 38:67–84. doi: 10.1046/j.1365-2958.2000.02111.x. [DOI] [PubMed] [Google Scholar]
- 8.Yu RR, DiRita VJ. 1999. Analysis of an autoregulatory loop controlling ToxT, cholera toxin, and toxin-coregulated pilus production in Vibrio cholerae. J Bacteriol 181:2584–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hulbert RR, Taylor RK. 2002. Mechanism of ToxT-dependent transcriptional activation at the Vibrio cholerae tcpA promoter. J Bacteriol 184:5533–5544. doi: 10.1128/JB.184.20.5533-5544.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Busby S, Ebright RH. 1994. Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell 79:743–746. doi: 10.1016/0092-8674(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 11.Richard AL, Withey JH, Beyhan S, Yildiz F, DiRita VJ. 2010. The Vibrio cholerae virulence regulatory cascade controls glucose uptake through activation of TarA, a small regulatory RNA. Mol Microbiol 78:1171–1181. doi: 10.1111/j.1365-2958.2010.07397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Withey JH, DiRita VJ. 2005. Activation of both acfA and acfD transcription by Vibrio cholerae ToxT requires binding to two centrally located DNA sites in an inverted repeat conformation. Mol Microbiol 56:1062–1077. doi: 10.1111/j.1365-2958.2005.04589.x. [DOI] [PubMed] [Google Scholar]
- 13.Withey JH, DiRita VJ. 2006. The toxbox: specific DNA sequence requirements for activation of Vibrio cholerae virulence genes by ToxT. Mol Microbiol 59:1779–1789. doi: 10.1111/j.1365-2958.2006.05053.x. [DOI] [PubMed] [Google Scholar]
- 14.Wood TI, Griffith KL, Fawcett WP, Jair KW, Schneider TD, Wolf RE Jr. 1999. Interdependence of the position and orientation of SoxS binding sites in the transcriptional activation of the class I subset of Escherichia coli superoxide-inducible promoters. Mol Microbiol 34:414–430. doi: 10.1046/j.1365-2958.1999.01598.x. [DOI] [PubMed] [Google Scholar]
- 15.Bradley ES, Bodi K, Ismail AM, Camilli A. 2011. A genome-wide approach to discovery of small RNAs involved in regulation of virulence in Vibrio cholerae. PLoS Pathog 7:e1002126. doi: 10.1371/journal.ppat.1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Withey JH, Dirita VJ. 2005. Vibrio cholerae ToxT independently activates the divergently transcribed aldA and tagA genes. J Bacteriol 187:7890–7900. doi: 10.1128/JB.187.23.7890-7900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowden MJ, Skorupski K, Pellegrini M, Chiorazzo MG, Taylor RK, Kull FJ. 2010. Structure of Vibrio cholerae ToxT reveals a mechanism for fatty acid regulation of virulence genes. Proc Natl Acad Sci U S A 107:2860–2865. doi: 10.1073/pnas.0915021107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Childers BM, Cao X, Weber GG, Demeler B, Hart PJ, Klose KE. 2011. N-terminal residues of the Vibrio cholerae virulence regulatory protein ToxT involved in dimerization and modulation by fatty acids. J Biol Chem 286:28644–28655. doi: 10.1074/jbc.M111.258780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Childers BM, Weber GG, Prouty MG, Castaneda MM, Peng F, Klose KE. 2007. Identification of residues critical for the function of the Vibrio cholerae virulence regulator ToxT by scanning alanine mutagenesis. J Mol Biol 367:1413–1430. doi: 10.1016/j.jmb.2007.01.061. [DOI] [PubMed] [Google Scholar]
- 20.Prouty MG, Osorio CR, Klose KE. 2005. Characterization of functional domains of the Vibrio cholerae virulence regulator ToxT. Mol Microbiol 58:1143–1156. doi: 10.1111/j.1365-2958.2005.04897.x. [DOI] [PubMed] [Google Scholar]
- 21.Shakhnovich EA, Hung DT, Pierson E, Lee K, Mekalanos JJ. 2007. Virstatin inhibits dimerization of the transcriptional activator ToxT. Proc Natl Acad Sci U S A 104:2372–2377. doi: 10.1073/pnas.0611643104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hung DT, Shakhnovich EA, Pierson E, Mekalanos JJ. 2005. Small-molecule inhibitor of Vibrio cholerae virulence and intestinal colonization. Science 310:670–674. doi: 10.1126/science.1116739. [DOI] [PubMed] [Google Scholar]
- 23.Kovacikova G, Lin W, Skorupski K. 2010. The LysR-type virulence activator AphB regulates the expression of genes in Vibrio cholerae in response to low pH and anaerobiosis. J Bacteriol 192:4181–4191. doi: 10.1128/JB.00193-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta S, Chowdhury R. 1997. Bile affects production of virulence factors and motility of Vibrio cholerae. Infect Immun 65:1131–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chatterjee A, Dutta PK, Chowdhury R. 2007. Effect of fatty acids and cholesterol present in bile on expression of virulence factors and motility of Vibrio cholerae. Infect Immun 75:1946–1953. doi: 10.1128/IAI.01435-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abuaita BH, Withey JH. 2009. Bicarbonate induces Vibrio cholerae virulence gene expression by enhancing ToxT activity. Infect Immun 77:4111–4120. doi: 10.1128/IAI.00409-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuhmacher DA, Klose KE. 1999. Environmental signals modulate ToxT-dependent virulence factor expression in Vibrio cholerae. J Bacteriol 181:1508–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bina JE, Mekalanos JJ. 2001. Vibrio cholerae tolC is required for bile resistance and colonization. Infect Immun 69:4681–4685. doi: 10.1128/IAI.69.7.4681-4685.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Provenzano D, Klose KE. 2000. Altered expression of the ToxR-regulated porins OmpU and OmpT diminishes Vibrio cholerae bile resistance, virulence factor expression, and intestinal colonization. Proc Natl Acad Sci U S A 97:10220–10224. doi: 10.1073/pnas.170219997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hung DT, Mekalanos JJ. 2005. Bile acids induce cholera toxin expression in Vibrio cholerae in a ToxT-independent manner. Proc Natl Acad Sci U S A 102:3028–3033. doi: 10.1073/pnas.0409559102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chatterjee A, Chaudhuri S, Saha G, Gupta S, Chowdhury R. 2004. Effect of bile on the cell surface permeability barrier and efflux system of Vibrio cholerae. J Bacteriol 186:6809–6814. doi: 10.1128/JB.186.20.6809-6814.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomson JJ, Withey JH. 2014. Bicarbonate increases binding affinity of Vibrio cholerae ToxT to virulence gene promoters. J Bacteriol 196:3872–3880. doi: 10.1128/JB.01824-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 34.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Dittmer JB, Withey JH. 2012. Identification and characterization of the functional toxboxes in the Vibrio cholerae cholera toxin promoter. J Bacteriol 194:5255–5263. doi: 10.1128/JB.00952-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Berge Henegouwen GP, van der Werf SD, Ruben AT. 1987. Fatty acid composition of phospholipids in bile in man: promoting effect of deoxycholate on arachidonate. Clin Chim Acta 165:27–37. doi: 10.1016/0009-8981(87)90215-4. [DOI] [PubMed] [Google Scholar]