SUMMARY
The global emergence of multidrug-resistant Gram-negative bacteria is a growing threat to antibiotic therapy. The chromosomally encoded drug efflux mechanisms that are ubiquitous in these bacteria greatly contribute to antibiotic resistance and present a major challenge for antibiotic development. Multidrug pumps, particularly those represented by the clinically relevant AcrAB-TolC and Mex pumps of the resistance-nodulation-division (RND) superfamily, not only mediate intrinsic and acquired multidrug resistance (MDR) but also are involved in other functions, including the bacterial stress response and pathogenicity. Additionally, efflux pumps interact synergistically with other resistance mechanisms (e.g., with the outer membrane permeability barrier) to increase resistance levels. Since the discovery of RND pumps in the early 1990s, remarkable scientific and technological advances have allowed for an in-depth understanding of the structural and biochemical basis, substrate profiles, molecular regulation, and inhibition of MDR pumps. However, the development of clinically useful efflux pump inhibitors and/or new antibiotics that can bypass pump effects continues to be a challenge. Plasmid-borne efflux pump genes (including those for RND pumps) have increasingly been identified. This article highlights the recent progress obtained for organisms of clinical significance, together with methodological considerations for the characterization of MDR pumps.
INTRODUCTION
Antibiotic resistance has emerged as a major threat to public health in this century, as evident from global surveillance data (1). Indeed, with the ancient origin and widespread presence of diverse resistance genes (2, 3), the modern evolution of resistance has led to the global emergence and spread of a large number of resistant bacteria that possess sophisticated genotypes and phenotypes against antibiotics. This phenomenon is a consequence of the natural selection process in microorganisms and promotion by human activities over the past 70 years of the antibiotic era (4, 5). In 2013, the U.S. Centers for Disease Control and Prevention (6) listed current resistance threats, of which multidrug-resistant Gram-negative bacteria constitute a large proportion (e.g., Enterobacteriaceae, Acinetobacter, and Pseudomonas). Of the various molecular and biochemical mechanisms of resistance to antibiotics, active efflux of antibiotics in bacteria plays an important role in both intrinsic and acquired multidrug resistance (MDR) of clinical relevance. It also interplays with other resistance mechanisms, such as the membrane permeability barrier, enzymatic inactivation/modification of drugs, and/or antibiotic target changes/protection, in significantly increasing the levels and profiles of resistance.
Energy-dependent drug efflux was discovered in the 1970s, initially with P-glycoprotein in mammalian cells (7) and later with Tet proteins in Escherichia coli isolates resistant to the specific antibiotic class tetracyclines (8). The subsequent discovery in the early 1990s of MDR pumps in E. coli and Pseudomonas aeruginosa, represented by the resistance-nodulation-division (RND) superfamily exporters (9–13), has made an important contribution to our understanding of resistance mechanisms (14). Since then, with rapid technological advances in biochemistry and molecular biology, there have been ever-growing identification and characterization of MDR pumps in numerous bacterial species of public health concern (e.g., in the ESKAPE [Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, P. aeruginosa, and Enterobacter species] pathogens), which compellingly demonstrate their predominant role in clinical settings (15, 16). Meanwhile, efforts of scientists led to the understanding of not only the structural and functional basis of these drug transporters but also their regulation and inhibition. In this review, we aim to provide a comprehensive and up-to-date description of efflux-mediated antibiotic resistance in Gram-negative bacteria.
BIOCHEMISTRY AND GENETICS OF MULTIDRUG EFFLUX PUMPS
Classes of Efflux Pumps
Because there are so many different efflux transporters, the only feasible way for their classification is to use phylogenetic grouping, based on protein sequences. Such a classification for all transporter proteins has been established by Milton Saier's group (17–19) and is available in the Transporter Classification Database (http://www.tcdb.org/). Transporter genes in hundreds of sequenced bacterial genomes are classified in Ian Paulsen's database (20) for each of these genomes (http://www.membranetransport.org/). Among many families of transporters, several contain prominent members of efflux transporters: especially important in bacteria are the RND, MFS (major facilitator superfamily), MATE (multidrug and toxic compound extrusion), SMR (small multidrug resistance), and ABC (ATP-binding cassette) superfamilies or families. ABC transporters utilize ATP hydrolysis as the energy source, but all others are dependent on proton motive force and are thus secondary transporters or proton/drug antiporters.
The transporters also differ in their subcellular organization. The RND pumps, which are all exporters of drugs and toxic cations, are located in the inner membrane (IM) (cytoplasmic membrane) but must interact with the periplasmic adaptor protein (also called membrane fusion protein) and the outer membrane (OM) channel, thus producing a tripartite complex spanning the IM, the periplasm, and the OM (represented by E. coli AcrAB-TolC and P. aeruginosa MexAB-OprM) (see the multicomponent pump depicted in Fig. 1). Some members of the ABC superfamily (e.g., MacB), the MATE family (e.g., MdtK), and even the MFS (e.g., EmrB) (all from E. coli) also are organized in this manner. The tripartite transporters excrete drugs directly into the external medium so that the reentry of drugs requires the slow traversal of the OM, an effective permeability barrier (21, 22). For this reason, these pumps are far more efficient in creating detectable resistance to antibiotics (especially AcrB, a constitutive RND transporter of E. coli [9]) (see Gammaproteobacteria: Enterobacteriaceae, below). In contrast, the pumps that are not organized in this manner and exist as single-component or “singlet” pumps in the IM (Fig. 1), including the vast majority of MFS and SMR pumps, are less effective in producing a detectable decrease in susceptibility, because the drug molecules are excreted only into the periplasm and can spontaneously diffuse back into the cytosol, since most antibiotics are relatively lipophilic molecules that can cross the phospholipid bilayer region of the IM. However, RND pumps, which are thought to capture antibiotics mostly from the periplasm (23, 24), can collaborate with the singlet pumps and thus increase their efficacy (25, 26).
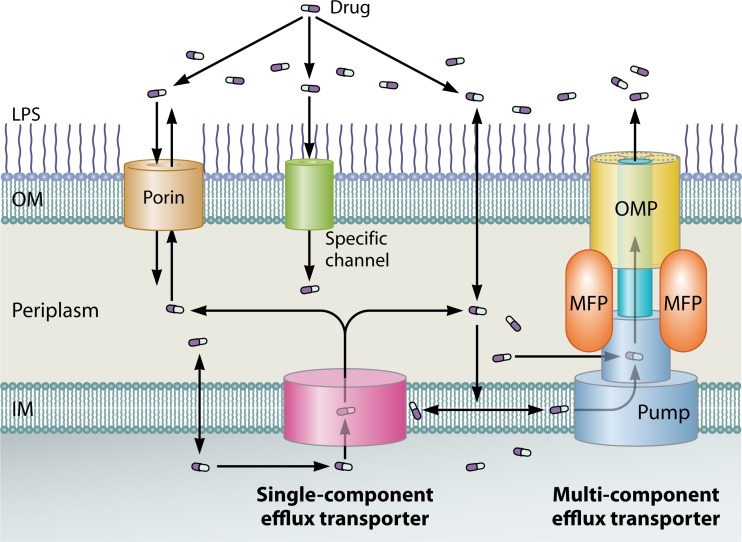
FIG 1.
Location of drug efflux pumps and pathways of drug influx and efflux across the OM and IM in Gram-negative bacteria. The influx of drugs (shown as pills) through the OM occurs in one or more of the following three pathways: porin channels (e.g., OmpF of E. coli and OprF of P. aeruginosa), specific protein channels (e.g., CarO of A. baumannii and OprD of P. aeruginosa for carbapenems), and the LPS-containing asymmetric lipid bilayer region. After their entry into the periplasmic space, the drug molecules can further penetrate the IM via diffusion. However, these drugs can be extruded out of the cell by efflux transporters, which exist as either single-component pumps (“singlet”; e.g., Tet pumps) or multicomponent pumps (e.g., AcrAB-TolC and MexAB-OprM tripartite efflux systems that each typically contain a pump, an OM channel protein [OMP], and an accessory membrane fusion protein [MFP]). While the singlet pumps may take up the drug from the cytosol and the periplasm and function with porins or other types of protein channels to make the efflux process effective, the multicomponent exporters capture their substrates from the periplasm and the IM and directly pump them into the medium. The competition between the influx and efflux processes ultimately determines the steady state of drug molecules in bacterial cells. With the lipophilic drug molecules that cross the OM slowly or the hydrophilic drugs that penetrate the A. baumannii/P. aeruginosa low-permeability porins (i.e., “slow porins”), the efflux mechanism become very effective, thus being able to yield MDR. In contrast, with the less hydrophobic and smaller drug molecules that can rapidly penetrate, for example, E. coli porins, efflux is not effective to counteract drug influx, thus hardly decreasing the concentrations of the drug in the cell.
The most detailed information on the contribution of various pumps to drug susceptibility is available for E. coli K-12, and Table 1 lists data on known and predicted multidrug pumps identified in the Transporter Classification Database mentioned above. An obvious way to detect the contribution of individual pumps is to measure the MICs of drugs in defective mutants. This was done in 2001 by Sulavik and coworkers (27) and showed that the RND transporter AcrB (in cooperation with its periplasmic and OM partners AcrA and TolC) plays a truly predominant role in raising the MIC levels in a wild-type strain. This also creates a problem because deletion of other pumps rarely produces detectable changes in MICs in the presence of the active AcrB-AcrA-TolC system. A similar problem was reported in a study (28) examining the MIC values of nearly 4,000 deletion mutants of all nonessential E. coli genes (the “Keio collection” [29]). Thus, although that study showed that the functions of many metabolic genes have an unsuspected influence on drug sensitivity, in terms of transporter genes, it essentially identified the effect of only the acrAB-tolC complex and nothing else. One possible exception is the deletion of the ycdZ gene, which produced hypersusceptibility to tetracycline and may code for an exporter. However, this conclusion is not supported by a study from the Carol Gross group, who quantitated the growth phenotype of the same set of mutants in the presence of sub-MICs of various drugs (30). This approach is more sensitive than the determination of MIC values and, indeed, as presented in Table 1, showed that the deletion of practically all known and suspected pumps produces hypersusceptibility to at least one agent tested. These results, however, must be interpreted with care, since this approach is very sensitive and could produce false-positive results in spite of efforts to avoid them (Table 1).
TABLE 1.
Phenotypes of proven and putative efflux pump mutants of E. colia
| Family and efflux gene | Deletion phenotypeb (agent[s], fold decrease in MIC) | Overexpression phenotypec (agent[s], fold increase in MIC) | Agent(s) that inhibited growthd | Other substrate(s) or information (reference) |
|---|---|---|---|---|
| RND | ||||
| acrB | ACR, 128; AMP, 4; BAC, 32; CHL, 8; CIP, 4; CLO, >2; DAU, >128; DEO, >2; EB, 256; ERY, 32; FUA, 128; MTX, >8; NAL, 2; NOV, 64; PUR, 64; R6G, 512; SDS, >128; TET, 8; TPP, 256 | ACR, >16; BAC, 32; CHL, 8; CV, 8; DEO, >32; DOR, >64; ERY, 32; NAL, 4; NOR, 8; NOV, 64; R6G, 64; SDS, >8; TMP, >32;TPP, 16 | ACR, AMP, AZM, AZT, BAC, BLE, BS, CER, CHIR, CHL, CHO, CIP, CLR, CPZ, DEO, DOX, EB, ERY, FUA, MEC, MIN, NAL, NIT, NOR, NOV, OXA, PUR, SDS, SPT, SPR, STG, TCH, TCS, TET, TMP, TRX, VER | |
| acrD | No change | DEO, >32; KAN, 2; NOV, 4; SDS, >8 | PAR | |
| acrF | No change | ACR, 8; DEO, 4; DOX, 2; SDS, 4 | PHL, VER | |
| yhiV (mdtF) | No change | CV, 2; DEO, 4; DOR, 8; EB, 4; ERY, 8; R6G, 16; SDS, 4; TPP, 4 | FUA | |
| mdtB | No change | DEO, >32; FOF, 2; NAL, 2; NOR, 2; NOV, 16; SDS, 4 | CIP, SDS | |
| mdtC | No change | DEO, >32; FOF, 2; NAL, 2; NOR, 2; NOV, 16; SDS, 4 | BAT, CSD, NAL, PHL | |
| MFS | ||||
| bcr | ACR, 2; FOF, 4; KAN, 2; TET 4 | CHO, ERY, SMZ | ||
| cmr (cmlA, mdfA) | BC, 4; EB, 4 | ACR, 8; CHL, 16; DOX, 4; EB, 4; NOR, 8; TET, 2; TMP, 4; TPP, 4 | ACR, BAC, CHL, EB | Pentoses (1010) |
| emrB | No change | DEO, 32; PAR, 2; R6G, 2; SDS, 2 | EB, SXT | CCCP, NAL, TSA (139); CER, TLM (15) |
| emrD | No change | BAC, 2; SDS, 2 | AZM, GEN, NIT, OXA, SMT, SPR | Uncouplers (15); pentoses (1010) |
| emrY | No change | CEC, SMZ, TET | H2O2, MIT, NAL, UV irradiation (1011) | |
| yajR | EB, NIT, OXA, SPR, TMP | |||
| yceB | AMX, BAC, MEC, NAL, PAR | |||
| yceE (mdtG) | FOF, 4 | BAC, CAZ, MEC, MTX, PUR, SDS, TLM | ||
| yceL (mdtH) | ATM, EB | |||
| ydeB (marC) | ACT | |||
| ydhC | FUA, SPT | Pentoses (1010) | ||
| yebQ | AMK, CHL, CHO, CSD, DEO, ERY, FOX, NOV, PER, PMB, TET, VER | |||
| yegB (mdtD) | None with agents tested | Iron citrate (in Salmonella) (1012) | ||
| yidY (mdtL) | AMX | |||
| yieO (hsrA) | ACR, BAC, CAR, CHIR, CSD, DOX, EB, FOX, GFF, MIT, SDS, STR | |||
| yjiO (mdtM) | AZT, BLE, CAL, CHIR, CHL, CIP, CPZ, INH, OXA, SMZ, SPR, TET | Binds CHL (1013) | ||
| ynfM | AMP, DEO | Pentoses (1010) | ||
| MATE (MOP) | ||||
| mdtK (ydhE, norM) | BAC, 2; CHL, 2; DEO, 32; DOR, 8; EB, 2; FOF, 2; NOR, 8; PAR, 4; TMP, 4; TPP, 32 | CPZ, FUA, NOR, PUR, STR | ||
| yeeO | ACR, BAC, BIC, CAR, DOR, PUR, VAN, VER | |||
| SMR (DMT) | ||||
| emrE | EB, 4; PAR, 8 | ACR, 16; BAC, 2; EB, 8; PAR, 2 | ACR, EB, FOF, PAR | EB (1014); PAR (1015); ERY (155); cationic osmoprotectants (159) |
| ydgE (mdtI) | DEO, 4; SDS, 2 | ACR, BAC, CHL, DEO, EB, FUA, MTX, PMB, PUR, TRX | Spermidine (1016) | |
| ydgF (mdtJ) | DEO, 4; SDS, 2 | CHO, CSD, ERY, NAL, VER | Spermidine (1016) | |
| ABC | ||||
| macB | No change | ERY, 8 | BAT, BIC, BS, CEC, EB, FOX, NIT, SDS, TCH | |
| LysE | ||||
| argO | AMP, BS, GFF, INH, NAL, PHL, TMP | Arginine (1017) | ||
| Unknown | ||||
| ycdZ | NOR, SDS (not TET) | TET, 1.7- to 2.5-fold decrease in MIC compared to the wild type (28) |
Abbreviations: ACR, acriflavine; ACT, actinomycin D; AMK, amikacin; AMP, ampicillin; AMX, amoxicillin; ATM, aztreonam; AZM, azithromycin; AZT, azidothymidine; BAC, benzalkonium chloride; BAT, bacitracin; BIC, bicyclomycin; BLE, bleomycin; BS, bile salts; CAL, calcofluor; CAR, carbenicillin; CAZ, ceftazidime; CCCP, carbonyl cyanide m-chlorophenylhydrazone; CEC, cefaclor; CER, cerulenin; CHIR, CHIR-900 (an LpxC inhibitor); CHL, chloramphenicol; CHO, cholate; CIP, ciprofloxacin; CLO, cloxacillin; CLR, clarithromycin; CPZ, chlorpromazine; CSD, cefsulodin; CV, crystal violet; DAU, daunomycin; DEO, deoxycholate; DOR, doxorubicin; DOX, doxycycline; EB, ethidium bromide; ERY, erythromycin; FOF, fosfomycin; FOX, cefoxitin; FUA, fusidic acid; GEN, gentamicin; GFF, glufosfomycin (fosfomycin plus glucose-6-P); INH, isoniazid; KAN, kanamycin; MEC, amdinocillin; MIN, minocycline; MIT, mitomycin; MTX, methotrexate; NAL, nalidixic acid; NIT, nitrofurantoin; NOR, norfloxacin; NOV, novobiocin; OXA, oxacillin; PAR, paraquat (methyl viologen); PER, peroxide; PHL, phleomycin; PMB, polymyxin B; PUR, puromycin; R6G, rhodamine 6G; SDS, sodium dodecyl sulfate; SMT, sulfamonomethoxine; SMZ, sulfamethizole; SPR, spiramycin; SPT, spectinomycin; STG, streptonigrin; STR, streptomycin; SXT, trimethoprim-sulfamethoxazole; TCH, taurocholate; TCS, triclosan; TDC, taurodeoxycholate; TET, tetracycline; TLM, thiolactomycin; TMP, trimethoprim; TPP, tetraphenyl phosphonium; TRX, Triton X-100; TSA, tetrachlorosalicylanilide; VAN, vancomycin; VER, verapamil.
See reference 27. Numbers after the abbreviation for the agent name indicate the fold decrease in MIC in comparison with the wild-type strain.
See reference 31. Numbers after the abbreviation for the agent name indicates the fold increase in MIC in comparison with the ΔacrAB parent strain.
Only the antimicrobial agents, dyes, and detergents that significantly inhibited the growth of mutants more than that of the wild type, usually leading to growth scores of <−2 (30), are listed. Furthermore, we confirmed that the inhibition was reproducible by examining the data obtained with different concentrations of the same agent.
A completely different approach is the plasmid-based overexpression of putative efflux genes. This analysis by Nishino and Yamaguchi (31) indeed detected efflux activity in those genes whose activity was difficult to detect by deletion-based approaches. However, these data do not tell us whether the pumps are functioning in the wild-type or even mutant cells, although the level of expression of most pumps can be increased by regulatory signals (see Regulation of Multidrug Efflux Pumps, below).
Reviews describing various types of efflux pumps include those written by Poole (32, 33), Piddock (34–36), Paulsen et al. (37), Saier and others (38), Van Bambeke and others (39), and Higgins (40). A review by Alekshun and Levy (41) is useful, as it also emphasizes the contribution of nonefflux mechanisms of resistance.
RND Transporters
AcrB of Escherichia coli.
The constitutively expressed pump AcrB of Escherichia coli plays a major role in raising the MICs of most antibiotics, due mostly to the fact that it exists as the AcrB-AcrA-TolC tripartite complex so that the exported drug molecules end up in the external medium, not in the periplasm, and thus cannot easily reenter the cells except by crossing the effective OM permeability barrier (24). Therefore, the effectiveness of RND pumps is intimately tied to the strength of the OM barrier; permeabilizing the OM destroys the effect of RND pump-mediated efflux almost as effectively as the inactivation of the pump itself (42, 43). Available reviews on RND pumps include those emphasizing the structure and mechanism (24, 44–49), computational approaches (50), roles in solvent tolerance (51), and functions other than drug resistance (34, 52–54).
AcrB has been studied most intensively as the prototype of RND pumps. It has an extremely wide specificity, including practically all types of antibacterial agents (except aminoglycosides), detergents, microbicides, dyes (Table 1), free fatty acids (55), and even simple solvents (56). In a reconstitution assay (57), AcrB was shown to also extrude modified phospholipids. A common property of these AcrB substrates is the presence of a hydrophobic domain (24, 58). Such a wide specificity appeared surprising at first. However, as pointed out by Neyfakh (59), this may be expected. Thus, when a typical soluble enzyme captures its hydrophilic substrate from the aqueous medium, the process requires the removal of the substrate molecule, already stabilized by its numerous hydrogen-bonding interactions with the surrounding water. Stable binding in the binding site of the enzyme therefore must involve precise, strong interactions with the residues in the site, in order to overcome this energy barrier, and it requires that the site is small and carefully designed to bind stringently only one substrate species. However, with the multidrug pumps that capture drugs with sizeable hydrophobic domains, the drugs are not strongly stabilized in the aqueous environment, as their presence involves a large entropic cost accompanying the ordering of the surrounding water molecules. Hence, the drug binding to the transporter does not require the binding site to be small, tight, and stringent. It can be very large and can thus accommodate a large range of substrates with small decreases in the binding free energy.
For biochemical studies of any transporters, transport studies of membrane vesicles are usually the preferred approach. Nevertheless, with AcrB, this approach was not fruitful, presumably because most of the ligands transported are lipophilic and cannot be accumulated in the intravesicular space due to their readiness in crossing the phospholipid bilayer domain of the membrane. It is also possible that the absolute rate of transport does not need to be high because of the presence of the OM barrier, and this impedes the detection of transport in vesicles. Thus, the first major advance had to rely on the reconstitution of purified AcrB into proteoliposomes, accomplished by Zgurskaya and Nikaido in 1999 (57). In order to circumvent the problem of the spontaneous diffusion of most ligands across the lipid bilayer, this study used an innovative approach relying on the efflux of fluorescently labeled phospholipids into empty “acceptor” vesicles and detected the efflux of conventional ligands through competition with phospholipid efflux. In this way, drugs such as cloxacillin, erythromycin, novobiocin, and fusidic acid (but curiously not chloramphenicol) as well as various bile acids were shown to compete against phospholipid efflux. Furthermore, the half-maximal concentration for inhibition was lowest for bile acid taurocholate (∼15 μM), suggesting that the properties of AcrB were optimized for the exclusion of bile salts, major toxic components in the mammalian intestine, the normal habitat for E. coli. Interestingly, the addition of AcrA to the aqueous phase (in the presence of Mg2+) strongly stimulated phospholipid transport: because lipids had to be transported from one vesicle to another, we hypothesized that AcrA may act by bringing the two vesicles together. This approach also established that AcrB was a proton/drug antiporter, as the transmembrane pH gradient was dissipated accompanying the flux of ligands.
A similar reconstitution assay was successfully used for E. coli AcrD (23), an AcrB homolog that also works with AcrA and TolC as a tripartite transporter. This study is important, as AcrD transports aminoglycosides, which are very hydrophilic and not expected to diffuse spontaneously across the lipid bilayer. Thus, a conventional accumulation assay using radiolabeled aminoglycosides indeed proved their accumulation in proteoliposomes. When streptomycin was added as the substrate to either the more acidic, intravesicular space corresponding to the periplasm or the more alkaline external space corresponding to the cytosol, pumping activity (as detected by the flux of protons) was observed only in the former case, showing clearly that the pump captures its substrate only in the periplasm. Although other aminoglycosides appeared to stimulate the pump activity even when added to the external space, the activities were rather weak, and it seems likely that AcrD (and possibly also other RND pumps) at least prefers to capture its substrates from the periplasm. Interestingly, the addition of AcrA was necessary for the function of AcrD. Because the assay does not require the juxtaposition of vesicles, AcrA is likely to stimulate directly the function of AcrD (and AcrB) by simply binding to the transporter.
Since reconstitution assays are quite cumbersome, methods for quantitative, real-time determination of pumping activity in intact cells were needed. Fluorescent probes [e.g., N-phenyl-1-naphthylamine, ethidium bromide, and 2-(4-dimethylamino)styryl-N-ethylpyridinium iodide] were preloaded into bacterial cells deenergized by uncouplers, and efflux was monitored by fluorescence after reenergization by adding an energy source, such as glucose (60, 61). It is difficult to perform assays of this type in a reproducible manner because some uncouplers remain after reenergization. An optimized, semiquantitative method for E. coli using the fluorescent dye Nile red was reported in 2010 (62). A major step in the intact-cell assay of AcrB was the real-time assay of cephalosporin efflux in E. coli achieved by Nagano and Nikaido in 2009 (63). Those authors measured spectrophotometrically cephalosporin hydrolysis in intact cells by a periplasmic β-lactamase. By comparing the hydrolysis rate with the Vmax and Km of the enzyme, those authors calculated the periplasmic concentrations of the cephalosporins, overcoming the most serious problems in intact-cell assays of efflux. They then obtained the expected influx rate (Vin) of the drug across the OM from the permeability coefficient obtained from uncoupler-poisoned cells and from the difference in the external and periplasmic concentrations of the drug. The difference between Vin and the observed hydrolysis rate then corresponds to the rate of efflux. When the efflux rate of nitrocefin was plotted against the periplasmic concentrations, a Michaelis-Menten-type saturation curve was obtained, showing the Vmax (0.024 nmol/mg/s) and Km (5 μM) of the AcrB-catalyzed efflux process for the first time. Since the expression of AcrB was increased severalfold in the strain used, we estimate that the Vmax is ∼6 pmol/mg/s in wild-type E. coli K-12. This assay was used with conventional cephalosporins and penicillins (63–65); one surprising finding was that a sigmoidal kinetics was often observed in the plots of velocity versus periplasmic drug concentration, suggesting positive cooperativity. Recently, a sigmoidal kinetics was observed for the AcrB-catalyzed efflux of a compound of a very different nature, l-arginine-β-naphthylamide (A. Kinana, A. V. Vargiu, and H. Nikaido, unpublished data), indicating that this is a common feature of the AcrB-catalyzed transport process.
A major advance in the study of AcrB was made when Murakami et al. (66) solved the crystal structure of trimeric AcrB in 2002. This symmetric structure showed that each protomer of AcrB contained a large periplasmic domain, as predicted from the primary sequence. Furthermore, the periplasmic domain was seen to have a large cleft facing the surrounding periplasm. Although binding of ligands to the area close to the cleft was shown by cocrystallization (67–69), this observation did not immediately suggest the mechanisms of drug extrusion (see below). The next big advance in our understanding of AcrB structure and function came with crystallographic analysis of the asymmetric trimer structure, where each protomer takes a unique conformation slightly different from that of its neighbor, elucidated in three laboratories (Fig. 2) (70–72). The work by Murakami and coworkers (70) was especially important, because they succeeded in cocrystallizing AcrB with the substrate minocycline or doxorubicin. In both cases, the substrates were seen in a predominantly hydrophobic pocket within the periplasmic domain, now called the distal binding pocket, close to the center of the trimer and located in one particular protomer, called the binding protomer. The presence of three conformationally different protomers, the access, binding, and extrusion protomers (Fig. 2) (44), suggested a functionally rotating mechanism, in which each protomer goes through a succession of conformational alterations. The distal binding pocket becomes collapsed in the extrusion protomer, consistent with the movement of the drug to the exit gate close to the end of the TolC channel. This concept of conformational cycling or functional rotation was then substantiated by the finding that disulfide cross-linking of nearby residues, although apparently occurring in only one or two protomers, nearly completely inactivated the trimeric complex (73). (The AcrB homolog MexB of P. aeruginosa has been crystallized without [74] and with [75] an added inhibitor, a pyridopyrimidine derivative.) In a similar vein, when the AcrB trimer was produced as a covalently linked single protein, and only one protomeric unit was inactivated in the proton translocation pathway, the entire trimeric complex became inactive (76). Furthermore, when the Cys residues were introduced into only one of the protomeric units, their cross-linking immediately inactivated the function of AcrB trimers, showing that inactivation was not due to a failure of the trimeric assembly.
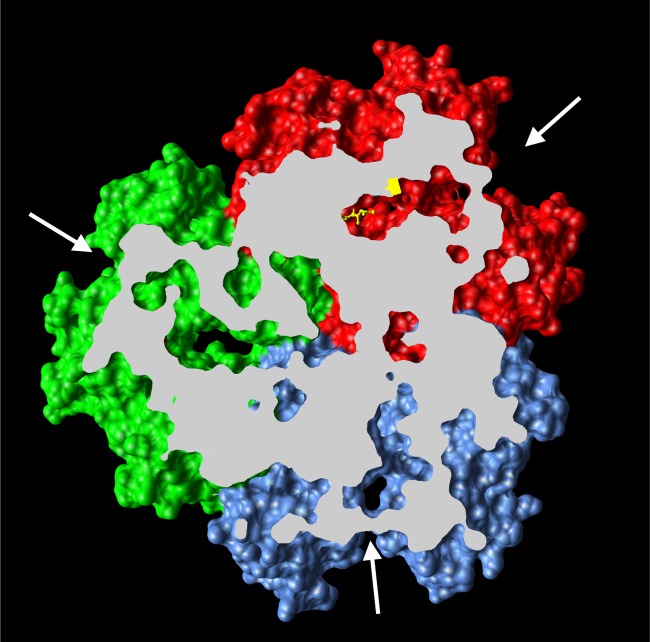
FIG 2.
Drug transport mechanism of AcrB. Shown is the asymmetric crystal structure of AcrB (Protein Data Bank accession number 2DRD), viewed from outside the cell, with the top portion cut off for clarity. Conformational cycling of 3 AcrB protomers, in access (blue), binding (red), and extrusion (green), is seen by cocrystallization of AcrB with its substrate minocycline, shown in a yellow stick model.
More recently, two laboratories (77, 78) showed that large substrates such as macrolides, rifampin, and a dimer of doxorubicin bind to a more proximal binding site in the access protomer, presumably before their eventual movement to the distal pocket concomitant with the conformational change of the protein into the binding protomer. Interestingly, this proximal binding site surrounded by residues Asp566, Phe664, Phe666, Glu673, Arg717, and Asn719 overlaps mostly the periplasmic binding site identified by Yu and collaborators several years earlier (67) in a symmetric AcrB crystal structure. Yu and associates further showed the importance of this binding site by site-directed mutagenesis, finding that a Phe666Ala mutation, for example, results in a drastic decrease of resistance to a wide range of substrates (67). The involvement of residues surrounding this site was also shown by the fact that their Cys-substituted mutants were strongly labeled by an AcrB substrate, boron-dipyrromethene (BODIPY)-maleimide (79). Another symmetrical crystal structure containing a substrate at this position has a deoxycholate molecule (80). The importance of this proximal binding pocket was further emphasized by recent studies (81, 82). The preference of AcrD for β-lactams containing multiple anionic groups, such as carbenicillin, sulbenicillin, and aztreonam, is essentially due to the residues within the proximal pocket (81), while a region in the MexY aminoglycoside pump that corresponds to a proximal binding pocket of AcrB plays a role in aminoglycoside recognition and export (82).
Yet another binding site for drugs was identified within the central cavity of the trimer by cocrystallization (67, 83). Possibly, the initial binding here is followed by the eventual translocation of the substrates to the distal binding pocket, perhaps through the “vestibules” between the protomers (84). Although the functional significance of this binding site could not be ascertained by site-directed mutagenesis, symmetric cocrystals of AcrB with drugs in the central cavity have been reported for ampicillin (68) and linezolid (85).
Dastidar et al. carried out the first effort to use substrate competition for covalent labeling of selected residues for the elucidation of the path of the drug molecules within the large periplasmic domain of AcrB (86). Some residues of Haemophilus influenzae AcrB were converted to Cys and were labeled with fluorescein maleimide. The labeling of Ala288Cys (corresponding to Gly290 of E. coli AcrB and close to the distal binding site) was decreased by the presence of all substrates tested, except ethidium. We followed up on this work by selecting 48 residues that lie on the presumed path(s) of the drugs, converting each residue to Cys, and labeling the Cys residue in intact cells with a hydrophobic, covalent-labeling probe, BODIPY-maleimide (79). Residues outside the predicted path were not labeled at all, even when they were located in the middle of hydrophobic patches. In contrast, most of the tested residues in the distal binding pocket were strongly labeled, as were the residues lining the proximal pocket as well as the entrance and the bottom of the large external cleft between two subdomains (PC1 and PC2 [66]). Finally, by using bulky covalent-labeling reagents, with some residues, we have been able to “clog” the substrate path so that the efflux of a substrate, Nile red, could be blocked.
This study reinforced the importance of the drug binding to the distal binding pocket as a major step in efflux. Site-directed mutagenesis of Phe residues in this pocket (87) indicated that these residues are important for efflux, with the Phe610Ala mutation showing the most widespread effect on many substrates. A molecular dynamics (MD) simulation study of this mutant protein (88) revealed that a substrate, doxorubicin, still bound to the pocket with a strong affinity; the interpretation of these results is described below. Site-directed mutagenesis based on the sequence difference between AcrB and MexB, which show different proficiencies in macrolide efflux, led to the discovery of the importance of Gly616 in AcrB for this function (89); interestingly, this residue is a part of the Gly-rich loop (also called the switch loop), which separates the distal pocket from the proximal pocket (77) and is thought to be critical for translocation of the substrates, especially large molecules such as macrolides. More recently, Eicher et al. showed the coupling of remote alternating-access transport mechanisms for protons and AcrB substrates through a mechanism involving two remote alternating-access conformational cycles within each promoter (90).
Although these mutagenesis studies are valuable, they do not tell us how various substrates bind to the AcrB transporter. As stated above, only a few crystal structures of drug-AcrB complexes are currently available. Thus, computational analysis of drug-AcrB interactions was first initiated with the docking software Autodock Vina (91). Various known substrates of AcrB, including minocycline, docked to the upper part (closer to the exit gate) of the distal binding site, which contains a characteristic crevice (Fig. 3). This is where minocycline and doxorubicin bound in the crystal structures (70). Cefazolin, a nonsubstrate (63), gratifyingly did not bind to the binding pocket. However, other substrates failed to bind to this upper portion of the pocket; as an example, chloramphenicol and solvents such as cyclohexane were predicted to “bind” with a significantly lower binding energy to the lower part of the pocket, which we called a “cave” (Fig. 3) (91).
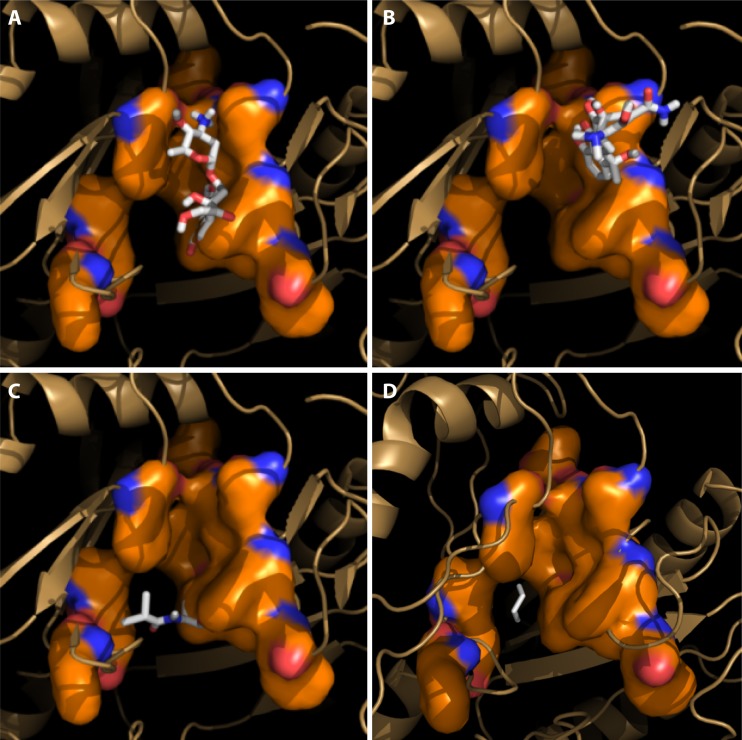
FIG 3.
Interaction of drug substrates and the AcrB-binding protomer analyzed with Autodock Vina docking software. Substrates are shown to bind to either the upper part (groove binder) (doxorubicin [A] and tetracycline [B]) or the lower part (cave binder) (chloramphenicol [C] and cyclohexane [D]) of the distal binding site. (Modified from reference 91.)
To get further insights into the binding-and-efflux process, we examined potential competition between substrates. It had been nearly impossible to show competition among substrates of AcrB by using an MIC assay (92). Still, with a real-time efflux assay with the dye Nile red, we showed that doxorubicin, minocycline, and other tetracyclines as well as tetraphenylphosphonium, but not chloramphenicol, macrolides, deoxycholate, nafcillin, or novobiocin, inhibited dye efflux (62). Also, when a real-time efflux assay of nitrocefin (63) was used, we showed strong inhibition by minocycline, predicted to bind to the upper part of the pocket, like nitrocefin (91). In contrast, a substrate that is not predicted to bind to this part of the pocket, i.e., chloramphenicol, did not inhibit nitrocefin efflux, and actually, there was some hint of stimulation instead (91). The latter phenomenon is discussed in more detail below.
Docking programs, however, have been optimized by using the binding of small, hydrophilic substrates mostly to the binding sites within enzymes. Binding of hydrophobic or amphiphilic ligands to the large binding pockets of transporters is predicted to occur in significantly different ways (59). Thus, we examined in detail the binding of 9 substrates, 2 inhibitors, and 2 nonsubstrates to the distal binding pocket of AcrB by extensive MD simulations (93). This introduced two major improvements over the docking approach. First, water molecules now became a part of the system so that the interaction of amphiphilic and more hydrophilic ligands could be predicted in a much more realistic manner. Second, the distal binding pocket is composed of residues that are on several loop segments in a relatively loosely constructed area of the protein, so movement and rotations of the chains were expected. Indeed, with many ligands, there was an extensive alteration in the shape of the binding site to better accommodate diverse substrates. Interestingly, some of those “cave binders” in the docking approach left the lower area and were found to favor the upper area of the pocket, although the binding appeared to be weak.
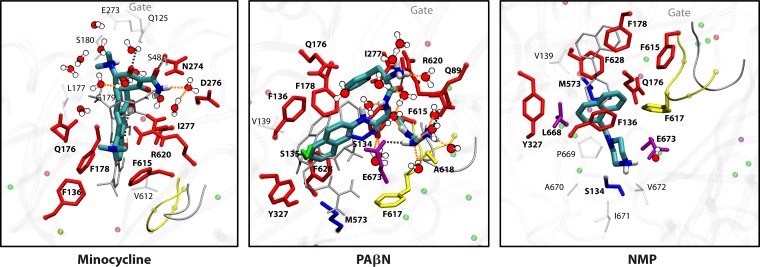
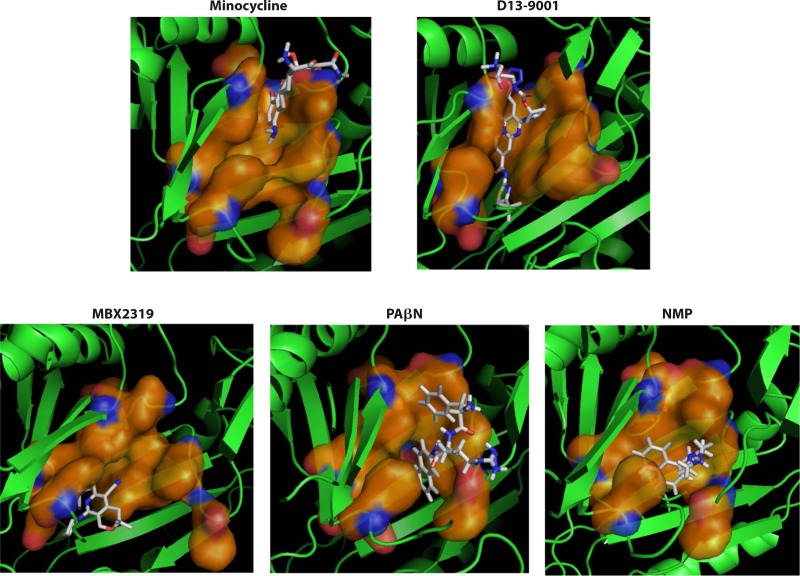
Chloramphenicol was found to slightly accelerate the efflux of nitrocefin (91). This was confirmed by a subsequent careful study, and it was found that solvents such as benzene or cyclohexane produced much more pronounced stimulation of nitrocefin efflux (94). MD simulations suggested that benzene interacts primarily with the Phe-rich hydrophobic domain that comprises the lower portion of the binding pocket and not with the upper subpocket that binds minocycline or nitrocefin (94). Interestingly, the lower portion of the binding pocket is where the hydrophobic part of the inhibitor D13-9001 binds tightly and was named a “hydrophobic trap” by Nakashima et al. (75) (see Efflux Pump Inhibitors, below). Furthermore, recent MD simulations showed that other efflux pump inhibitors (EPIs), such as phenylalanine-arginine-β-naphthylamide (PAβN), 1-(1-naphthylmethyl)-piperazine (NMP), and the new, potent inhibitor MBX2319 (95), all bind tightly to the hydrophobic trap and thereby distort the shape of the rest of the pocket, closing the crevice where minocycline or nitrocefin becomes bound (96). These observations suggest the following. (i) The distal binding pocket is very large, and different ligands prefer different areas of the pocket for binding. Thus, a “typical” substrate, like minocycline or nitrocefin, which has a number of hydrophilic groups, tends to bind to the upper “crevice” area, which is rich in hydrophilic and charged residues and was indeed shown to be involved in substrate binding (93). In contrast, for hydrophobic ligands such as cyclohexane or chloramphenicol, binding to this area is difficult. (ii) A tight interaction with the hydrophobic trap distorts the structure of the crevice, inhibiting the efflux of typical substrates. (iii) A loose interaction with the hydrophobic trap, on the other hand, may enhance the efflux of typical substrates, by either facilitating the interaction of such substrates with the pocket, speeding up the sequence of conformational changes needed for the export of substrates, or both. In any case, the interaction between the AcrB transporter and its substrates/inhibitors/enhancers appears quite complex. In assessing the binding of drugs to the pocket of transporters such as AcrB, we now realize that binding follows the same principles elucidated by the pioneering early crystallographic studies by Brennan and coworkers (for example, see references 97 and 98), carried out by using soluble regulators of MDR pumps, such as QacR, rather than the pumps themselves.
Computer simulation has now become an important approach for studying the mechanism of AcrB function. The movement of a substrate, doxorubicin, from the distal binding pocket to a position close to the exit gate, accompanying the closure of the pocket, was shown by a targeted MD simulation (99). Large substrates, found in the proximal binding pocket in AcrB cocrystals (77), moved substantially in the direction of the distal binding pocket during MD simulation (100). Movement of water molecules was analyzed by simulation (101, 102), and coarse-grained models were used to analyze the conformational transitions (103) as well as the drug pathways (104) within AcrB.
After the binding of the substrate to the distal binding pocket in the binding protomer, the proton(s) must come in from the periplasm to bind to the Asp residue(s) in the transmembrane domain, causing the conformation of the protein to change into the extrusion protomer, thereby squeezing out the substrate to the exit gate by the collapse of the pocket. Asp407 and Asp408 appear to be essential for the energy transduction of AcrB, together with Lys940 and Arg971 (105) as well as Thr978 (106). In the binding protomer, both Asp407 and Asp408 appear to be deprotonated, as the presumably protonated Lys940 side chain is situated between the two Asp side chains. In the extrusion protomer, Lys940 is moved away from the Asp residues and now faces the Thr978 side chain (107). The carboxyl group of Asp408 was indeed shown to have an unusually high pKa of 7.4, which would help in the facile binding and release of the proton under physiological conditions (107). A recent MD simulation study (108) suggests that in the extrusion protomer, Asp408 becomes protonated, but Asp407 remains deprotonated. In this scheme, the translocation of one proton across the IM would be sufficient to cause the conformational changes in AcrB, resulting in the extrusion of the drug molecule(s). The folding and assembly of the AcrB trimer were studied mainly by Wei and associates, who showed that the folding of the monomeric unit precedes trimerization (109) and analyzed the function of a protruding loop that inserts deeply into the neighboring subunit (110).
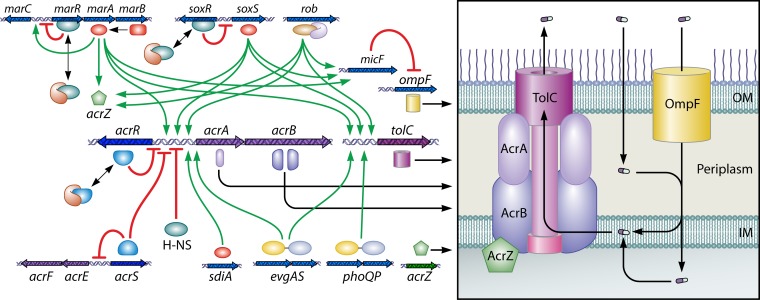
In 2007, when AcrB was crystallized without amplification from E. coli, the symmetric crystals were found to contain a small (110-residue) protein, YajC, associated with the transmembrane domain (68). The yajC gene occurs in the secDF operon (111), but the significance of this association is unclear. Deletion of yajC appeared to make E. coli marginally more susceptible to penicillins (68). Five years later, Hobbs et al. (112) found that a 49-residue protein, renamed AcrZ, associates with the transmembrane domain of AcrB. Deletion of acrZ renders E. coli moderately more susceptible to chloramphenicol and tetracycline. Importantly, AcrZ expression is regulated in the same manner as that of AcrB, by the global regulators MarA, SoxS, and Rob (see below).
AcrB functions as a member of the tripartite machinery including the periplasmic adaptor protein AcrA and the OM channel TolC. The structure of TolC, solved by the Koronakis group (113), shows a trimer containing an OM-spanning β-barrel and a contiguous, long, 12-stranded α-barrel. Among the adaptor proteins, the MexA structure was elucidated first (114). The AcrA structure is similar (115), but only three domains (the long α-hairpin, a lipoyl domain, and a short β-barrel) were elucidated; the membrane-proximal domain had to wait for work by Symmons and others in 2009 (116). Those authors showed by chemical cross-linking that the three domains other than the long α-hairpin interact closely with the periplasmic domain of AcrB. This suggested that the remaining α-hairpin domain was connected to the lower part of the α-barrel of TolC, completing the tripartite assembly. The top of the periplasmic domain of AcrB was cross-linked to the tip of the TolC α-barrel (117), and a model of the tripartite complex was proposed (116). This complex was shown to be stable enough to withstand cell disruption (118). The interaction of AcrA and other adaptor proteins with TolC has been studied by surface plasmon resonance (119), and a similar approach with immobilized AcrB showed that AcrB interacted with TolC, in the absence of AcrA, with a relatively high affinity (KD [equilibrium dissociation constant] of 90 nM) (120).
The number of AcrA molecules per assembly was uncertain in early models. With MacA, another adaptor protein that functions with the ABC transporter MacB (see below), a hexameric crystal in which MacA forms a closed barrel was found (121). Because the diameter of the end of this MacA barrel (composed of α-hairpins) was similar to the diameter of the α-barrel of TolC, and because the tip of the hairpin had amino acids that are conserved among various adaptor proteins, it was proposed that MacB does not directly touch the end of TolC and that the MacA tunnel acts as a bridge between these two proteins (121). A cocrystal of the Cu efflux RND pump CusA with its cognate adaptor protein CusB (122) also shows that the top of the transporter trimer interacts with the lower end of the CusB hexamer to form a channel that is likely to allow the partial insertion of the α-barrel domain of the OM channel CusC. In this model, again, the end of the α-barrel of the OM channel is not in contact with the top of CusA. Finally, in the recently reported electron micrographic structure of the AcrB-AcrA-TolC complex elucidated by the use of an AcrB-AcrA fusion protein, again, the top of AcrB is not in contact with the end of the TolC channel (123). However, there is evidence that these two domains are likely to come into contact in intact cells, as mentioned above, and it seems possible that the tripartite assembly is a dynamic one that could become shorter during the efflux cycle, as was suggested by Su and coworkers (122).
A rather close homolog of AcrB appears to exist in all members of the Enterobacteriaceae and also in many other species (e.g., MexB in P. aeruginosa). The acrB gene forms an operon with the acrA gene coding for the adaptor protein, and a similar arrangement is common with other RND pumps and in other species, especially in Enterobacteriaceae, where the OM component TolC, also serving other transporters, is encoded elsewhere. In contrast, in P. aeruginosa and A. baumannii, where each RND pump tends to operate with its specific adaptor and OM channel, a three-gene operon coding for all three components is more common (see sections on Pseudomonas, Acinetobacter, and Stenotrophomonas, below).
Other RND transporters in E. coli.
AcrF appears to have a wide substrate specificity, similarly to AcrB (124, 125). AcrD is an aminoglycoside efflux pump that works with AcrA and TolC (23). MdtF (YhiV) is likely involved in the extrusion of toxic metabolites during nitrosative stress, such as the nitrosyl derivative of indole, produced during anaerobic growth of E. coli (126). MdtBC is unusual because it contains two different transporter proteins, MdtB and MdtC, and appears to function only as a B2C heterotrimer (127). When overexpressed, it pumps out norfloxacin, novobiocin, cloxacillin, and deoxycholate (15, 31, 128). Site-directed fluorescein maleimide modification studies suggest that MdtC binds the substrate, but MdtB probably functions in other ways, such as initiating the conformational alteration for drug efflux (129).
MFS Transporters
MFS transporters can be classified into at least 74 families on the basis of sequence homology (130). E. coli K-12 contains 70 MFS transporters, 15 of which may be considered drug exporters, as they belong to families 2 and 3 (http://www.membranetransport.org/), which are composed of 12-TMS (transmembrane segment) and 14-TMS members, respectively, of drug/H+ antiporters (37, 130). Most of them, however, are free-standing transporters located in the IM and transport drugs from the cytosol to only the periplasm. Because most antimicrobial agents reach the cytosol usually by diffusion across the membrane bilayer, the pumped-out drug molecules have a good chance of reentering the cytosol through this free-diffusion process, and the transporters of this class are not expected to create high-level resistance. However, constitutive RND pumps, such as AcrAB-TolC and MexAB-OprM, may capture such pumped-out drug molecules in the periplasm and thus synergistically enhance the activity of singlet pumps in producing resistance (Fig. 1). This was first shown in P. aeruginosa (25) and was rediscovered in E. coli nearly a decade later (26). The latter study (26) also made an important point that the contribution of some singlet pumps may have escaped detection because of overlapping specificities; thus, the double deletion of an MFS pump, MdfA, and an SMR pump, EmrE, made E. coli as susceptible as or even more susceptible than the AcrB deletion mutant to cationic agents with intracellular targets, like acriflavine or ethidium. This finding suggests that RND pumps are usually rather inefficient in capturing drugs from the cytosol (although some contrary views have been presented [131]) and that the singlet pumps often play an important role in resistance to agents with intracellular targets. The fact that the plasmid-encoded TetA pump creates significant tetracycline resistance (8) suggests that this synergistic mechanism can sometimes be quite effective.
Although the MFS-type MDR pumps usually do not play a predominant role in resistance, as described below, the singlet drug pumps of the Tet group, usually plasmid encoded, are clinically important in creating tetracycline-specific resistance in many bacterial species. In the current nomenclature, TetA refers to the MFS exporter, and the phylogenetic group to which it belongs is specified by the group within parentheses, as in TetA(B). Currently, the 12-TMS TetA pumps, present in Gram-negative bacteria, contain 13 phylogenetic groups, whereas the 14-TMS Tet pumps, present in Gram-positive bacteria, contain at least 3 groups (132). The plasmid-encoded TetA pumps were the first bacterial drug efflux pumps identified (8, 133). Biochemical studies by the Yamaguchi group showed that their substrate was the magnesium salt of tetracycline (134), and cysteine-scanning mutagenesis followed by labeling studies identified the residues important for substrate binding and proton translocation (135). Interestingly, the Gram-positive pumps Tet(K) and Tet(L) were found to transport monovalent cations, such as Na+, and cation transport was hypothesized to be the original function of such pumps (136). Finally, glycylcyclines such as tigecycline were developed by selecting for derivatives that withstand the presence of Tet pumps and are indeed poor substrates for these tetracycline-specific transporters (137). However, tigecycline is a substrate for the RND pumps of many species, including E. coli, such as AcrAB or AcrEF (138).
A few MFS pumps, however, occur with their own periplasmic adaptor proteins and with OM channels, such as TolC, and presumably produce an efficient tripartite efflux system. In E. coli, these pumps include EmrB (occurring with the cognate adaptor EmrA) and EmrY (with EmrK), which indeed appear to be involved in the efflux of uncouplers and other substrates (Table 1) (15, 31, 139, 140). Importantly, the crystallographic structure of EmrD was determined (141). It is similar in general to those of the other MFS transporters but has a larger central cavity surrounded by hydrophobic and aromatic side chains. It was also noted that the loops connecting H4 and H5, and H10 and H11, protrude into the cytoplasm much more than in the inward transporters of the MFS, such as LacY and GlpT, and that these loops may play a role in substrate recognition and capture (141). A pH-dependent conformational change was also established for EmrD (142). EmrB that occurs with the periplasmic adaptor EmrA appears to assemble in vitro into a dimer of EmrAB dimers (143). If EmrA forms an intermediary channel similar to the AcrA and CusB channels (see above), perhaps the discrepancy between the trimeric TolC and dimeric EmrB may not matter. Alternatively, the dimeric arrangement could be an artifact of the in vitro assembly of the proteins. The EmrA protein was shown to form dimers and trimers in vitro, and interestingly, it bound an efflux substrate, carbonyl cyanide m-chlorophenylhydrazone (CCCP) (a proton uncoupler), with a reasonable affinity (KD of ∼1 μM) (144).
Among the singlet MFS transporters, MdfA, which confers MDR when overproduced (145), has been studied extensively in terms of biochemistry (146). However, its clinical relevance remains unknown, although it plays a major role with the SMR-type transporter EmrE in the efflux of cationic dyes (and presumably other cationic agents, such as quaternary ammonium compounds) (26).
ABC Transporters
In fungi and animal cells, most of the transporters involved in drug efflux belong to the ABC family (7, 147). In Gram-negative bacteria, there are only few examples of ABC family drug efflux pumps, although MsbA, the exporter of biosynthetic intermediates of lipopolysaccharide (LPS), was shown to pump out drugs, including erythromycin, when overexpressed in Lactococcus lactis (148).
The best-studied bacterial ABC drug exporter is MacB of E. coli, which functions together with the periplasmic adaptor MacA and the OM channel TolC (149). MacAB-TolC raises macrolide MIC values when overproduced (149). The isolated MacB shows only trace ATPase activity, which is not stimulated by substrates. However, ATP hydrolysis is very strongly stimulated by the simultaneous presence of MacA (150); these results were confirmed by another laboratory (151), which showed that MacB is a dimer, as expected for an ABC transporter. Finally, an analysis using surface plasmon resonance led to the conclusion that MacA binds to MacB with a nanomolar affinity, and the complex remains stable during the ATP hydrolysis cycle (152); the authors of this study assume that the MacA channel connects a TolC trimer and a MacB dimer, with no direct connection between the latter two. The expression of the MacAB system is stimulated by the heat shock sigma factor σ32 (153). Its physiological function might be related to the export of LPS or its biosynthetic intermediate (154).
SMR Transporters
The proton-motive force-driven SMR transporters belong to the drug/metabolite transporter (DMT) superfamily and are very small, each containing only four TMSs. Thus, unlike MFS transporters, which presumably function as monomers, SMR transporters, which typically exchange incoming H+ with the pumping out of either monocationic (ethidium and tetraphenylphosphonium, etc.) or dicationic (e.g., paraquat) compounds (155), must function as a dimer. They also appear to decrease susceptibility to aminoglycosides when the proteins are overproduced from plasmids (156). However, there was controversy on the issue of whether this was a parallel dimer in which each component monomer was embedded in the same direction within the bilayer or an antiparallel dimer. A crystallographic study clearly shows the antiparallel arrangement within an EmrE dimer (157), but chemical cross-linking favors a parallel arrangement, and it appears that the direction of insertion of the monomeric unit really does not matter for the efflux function (158).
EmrE is one of just a few transporters that produce a drug-hypersusceptible phenotype when the gene is deleted in wild-type E. coli still containing AcrAB (Table 1). One of the characteristic substrates of EmrE is a quaternary ammonium compound, including the endogenous osmoprotectant of E. coli, betaine (159), and thus, EmrE overproduction makes cells more susceptible to hyperosmolarity conditions as well as alkaline-pH media. Using these phenotypes, one study found that OmpW, an OM protein, apparently helps in the removal of such compounds pumped into the periplasm by EmrE (160). This is rather unexpected, as OmpW forms an 8-stranded β-barrel, which usually contains a channel too narrow for solute diffusion. Indeed, its structure shows that its central channel is truncated, although it may open up sideways into the interior of the OM (161). If (and how) quaternary ammonium compounds could diffuse through OmpW is thus an open question; however, we note that AcrAB-TolC is not needed for full resistance to paraquat (26).
There are a few reports suggesting the possible involvement of SMR transporters in resistance in clinical isolates of organisms other than E. coli. The deletion of abeS resulted in significant decreases in MICs of chloramphenicol, ciprofloxacin, and erythromycin in A. baumannii (162), and the deletion of a pair of genes, kpnEF, in K. pneumoniae makes cells hypersusceptible to a wide range of antimicrobials (163). An EmrE homolog contributes to MDR in P. aeruginosa (164). The contribution of SMR transporters to resistance seems to be an important future area of study for clinical microbiologists.
MATE Transporters
The MATE transporters have now become a part of a new superfamily, the multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) flippase superfamily (19), because of their connection to transporters like the LPS flippase RfbX. These transporters are widespread in bacteria and are also found in higher animals and plants.
The first of these transporters identified was the Na+/cationic agent antiporter NorM from Vibrio parahaemolyticus (165), with 12 TMSs. Its homologs pump out cationic dyes, fluoroquinolones, and aminoglycosides into the periplasmic space. Many of these transporters use a gradient of Na+, H+, or both as the energy source. The crystal structure of NorM from Vibrio cholerae indicates an outward-facing conformation with two portals open to the outer leaflet of the IM (166). The structures of NorM from Neisseria gonorrhoeae (167) and its distant, proton-driven Pyrococcus furiosus homolog (168) bound to several substrates show a wide, central substrate-binding cavity. With the latter, protonation of Asp41 bends TMS1 so that the part of the cavity located in the N-terminal half of the protein becomes collapsed, suggesting that this will produce the extrusion of substrates. Furthermore, cyclic peptides representing prototype inhibitors were also cocrystallized, and the best inhibitor appears to bind tightly to the cavity, preventing the bending of TMS1 (168). Finally, the mechanism of the NorM pump was examined with MD simulation (169).
Although genes for the MATE pumps have been cloned from many pathogens, their contribution to resistance in the organisms of origin has not been studied in most cases. One exception is a transporter from Enterobacter cloacae, EmmdR, and its gene disruption contributed to susceptibility to fluoroquinolones and cationic dyes (170).
SYNERGY WITH THE OUTER MEMBRANE BARRIER
Pathways of Drug Influx across the OM
In Gram-negative bacteria, antimicrobial agents must first traverse the OM barrier in order to exert their action (Fig. 1). The OM usually functions as a very effective permeability barrier, because the porin channels are narrow, being only 7 by 11 Å in the E. coli OmpF porin at its constriction point (171), and the bilayer domain of the OM is asymmetric, with its outer leaflet composed only of LPS (172), producing an unusually impermeable bilayer. Furthermore, because several basic amino acid residues are on one side and acidic amino acids are on the opposite side at the constriction zone of the porin channel, the water molecules inside are thought to be strongly oriented in one direction, and this presumably hinders the diffusion of lipophilic drugs, as they must disorganize this assembly of water molecules for penetration (173). Indeed, measurement of influx rates of cephalosporins showed that lipophilicity strongly hinders diffusion through porin channels (174). These considerations suggest that only those drugs that are relatively small, and preferably not too lipophilic, pass through the porin channels relatively rapidly. This group includes β-lactams, fluoroquinolones, tetracycline, chloramphenicol, cycloserine, and aminoglycosides. Aminoglycosides can be fairly large, but as polycations they are likely to become “sucked into” the periplasm by the presence of the interior-negative Donnan potential (175). Aminoglycosides and β-lactams are also essentially prohibited from diffusion through the bilayer region because of the presence of multiple cationic groups and a strongly acidic group, respectively. β-Lactams allow us to measure their OM permeation process precisely by coupling their influx with their subsequent hydrolysis by the periplasmic β-lactamase (174), and the importance of the porin pathway can be ascertained by using porin-deficient mutants in this assay. For other classes of drugs, quantitative assays of the OM penetration process are difficult. However, one can artificially increase the permeability of the OM bilayer region, either by using an agent that disorganizes the LPS leaflet (polymyxin B nonapeptide) (176) or with mutants with partial defects in LPS synthesis (177). The MIC values of the agents mentioned above showed little change under these conditions, suggesting that they predominantly permeate through the nonbilayer pathway, i.e., through the porin channels, at least in E. coli.
The preference for the OM permeation pathway, however, is not absolute. We note in particular that among β-lactams, those that are more hydrophobic (e.g., oxacillin and cloxacillin) or larger (such as some third- or fourth-generation cephalosporins) tend to be hindered in their penetration through porin channels, and for them, permeation through the bilayer region, although slow, may become significant (176, 177).
It should be mentioned here that the situation is very different for organisms such as P. aeruginosa or Acinetobacter species, which do not produce classical E. coli-type trimeric porins that provide a fast influx of small drugs. The major nonspecific porin in these organisms is a homolog of E. coli OmpA, and its major function is structural, that is, to connect the OM to the underlying peptidoglycan (178). The porin function is produced by the alternative folding of only a small fraction of the protein (perhaps ∼2% of the population) to produce ∼16 transmembrane β-strands, and we proposed to call this class of porins “slow porins,” in order to distinguish them from the classical trimeric porins, in which every molecule produces an open channel (179, 180). Because the number of open channels is small, OM permeability is very low, and β-lactams cross the OM of P. aeruginosa at a rate ∼100 times lower than that for the E. coli OM (181, 182). Because of this slow permeation through slow porins, the endogenous RND system MexAB-OprM can compete well with the influx of hydrophilic β-lactams as well as other antibiotics (Fig. 1). Thus, the deletion of a component of this pump complex decreases the MICs of many antibiotics drastically (Table 2). The situation is similar for A. baumannii (183). Hence, with these organisms, even efflux at moderate rates is expected to produce significant increases in β-lactam MICs, and indeed, the genetic deletion of major efflux pumps decreases β-lactam MICs substantially (Table 2) (13, 184). If we increase the OM permeability of P. aeruginosa by adding polymyxin B nonapeptide, we see impressive decreases in antibiotic MICs, comparable to those obtained by the genetic deletion of MexAB-OprM (42).
TABLE 2.
Effect of efflux pump gene inactivation on antibiotic MICsa
| Agent | MIC (μg/ml) |
|||
|---|---|---|---|---|
|
P. aeruginosa PAO1 |
E. coli K-12 |
|||
| Wild type | mexA::tet | Wild type | ΔacrAB | |
| Norfloxacin | >8 | 1 | 0.004 | 0.004 |
| Ciprofloxacin | 2 | 0.1 | 0.01 | 0.0025 |
| Levofloxacin | 0.25b | 0.015b | 0.063c | 0.016c |
| Besifloxacin | 1d | 0.06d | 0.25d | 0.015d |
| Moxifloxacin | 0.8e | 0.05e | ||
| Tetracycline | 8–16 | 0.5f | 1.25 | 0.156 |
| Tigecycline | 8g | 0.25g | 0.5h | 0.125h |
| Chloramphenicol | 16 | 4 | 6.25 | 0.78 |
| Novobiocin | 128 | 16 | 100 | 1.56 |
| Erythromycin | 512i | 64i | 50 | 1.56 |
| Azithromycin | 100 | 6.25 | 8j | 0.5j |
| Benzylpenicillin | >1,024 | 512 | 16k | 8k |
| Cloxacillin | 5,120l | 2.56l | 256k | 2k |
| Ampicillin | 12.5 | 3.12 | ||
| Carbenicillin | 32 | 0.25 | 4k | 1k |
| Azlocillin | 4 | 0.5 | 16k | 4k |
| Piperacillin | 3m | 0.4m | 4c | 0.25c |
| Cefoperazone | 4 | 0.5 | 0.03k | 0.015k |
| Ceftriaxone | 64 | 8 | 0.0015k | 0.0015k |
| Cefepime | 2 | 1 | 0.0075k | 0.0075k |
| Cefpirome | 4 | 2 | 0.015k | 0.015k |
| Imipenem | 2 | 1–2 | 0.12k | 0.12k |
The MIC data for P. aeruginosa are from reference 13, and those for E. coli are from reference 27, unless otherwise indicated.
Data from reference 411, where a mexAB-oprM mutant was used.
Data from reference 95.
Data from reference 1009, where an oprM mutant was used for P. aeruginosa.
Data from reference 392.
Because the mutant strain contains the tet marker, the value for an oprM mutant is used (13).
Data from reference 393.
Data from reference 138.
Data from reference 187.
X.-Z. Li, unpublished data.
Data from reference 188.
Data from reference 390, where a mexAB-oprM mutant was used.
Data from reference 391, where an oprM mutant was used.
Because LPS contains about six, usually all saturated, fatty acid chains in a single molecule, it is expected to produce a strong permeability barrier when organized into an LPS-only leaflet, as in the OM (22). Indeed, when the permeability of the OM bilayer to steroid probes (which are too large and too hydrophobic for passage through porin channels) was examined, it was found to be ∼2 orders of magnitude lower than that of the conventional phospholipid bilayer membranes (185). At the time when this study was carried out, the existence of multidrug efflux transporters was not known. More recently, however, such derivatives of steroid hormones were shown to be the substrates of AcrB (186). Thus, the permeability difference between the OM bilayer and the phospholipid bilayer might not quite reach 100-fold, yet it seems clear that the OM bilayer is an unusually impermeant barrier. Nevertheless, for large, lipophilic agents, the bilayer is the only possible pathway for OM permeation. These agents include macrolides, rifamycins, novobiocin, and fusidic acid (Table 3). Glycopeptides such as vancomycin and teicoplanin are large but not lipophilic. Nevertheless, because of their size, their only possible path to cross the OM is through the bilayer. Since these agents have so much difficulty in crossing the OM, they are usually considered agents effective against only Gram-positive bacteria. These agents can be active against Gram-negative bacteria if the LPS leaflet is breached (176, 177) or if the RND pump is inactivated (13, 187).
TABLE 3.
Molecular sizes and lipophilicities of antimicrobial agents and their relation with the effect of acrAB deletion on MICs in E. coli K-12a
| Agent | Molecular weight | Lipophilicity (XlogP3)c | MIC (μg/ml) for strain |
|
|---|---|---|---|---|
| acrAB+ | ΔacrAB | |||
| Lipophilic and/or large agents | ||||
| Clotrimazole | 345 | 5.0 | >32 | 16 |
| Cloxacillinb | 436 | 2.4 | 256 | 2 |
| Erythromycin | 734 | 2.7 | 50 | 1.56 |
| Fusidic acid | 517 | 5.5 | 400 | 3.125 |
| Methotrexate | 454 | −1.8 | >640 | 80 |
| Novobiocin | 613 | 3.3 | 100 | 1.56 |
| Puromycin | 472 | 0.0 | 100 | 1.56 |
| Rifampin | 823 | 4.0 | 5 | 2.5 |
| SDS | 288 | 4.7 | >12,800 | 100 |
| Small and hydrophilic agents | ||||
| Ampicillin | 349 | −1.1 | 12.5 | 3.12 |
| Cephalothinb | 396 | −0.4 | 4 | 4 |
| Cefoxitinb | 427 | 0.0 | 4 | 1 |
| Ceftazidimeb | 547 | 0.4 | 0.12 | 0.12 |
| Imipenemb | 317 | −0.7 | 0.12 | 0.12 |
| Nalidixic acid | 232 | 1.4 | 3.13 | 1.56 |
| Norfloxacin | 319 | −1.0 | 0.004 | 0.004 |
| Ciprofloxacin | 331 | −1.1 | 0.01 | 0.0025 |
| Chloramphenicol | 323 | 1.1 | 6.25 | 0.78 |
| Tetracycline | 444 | −2.0 | 1.25 | 0.156 |
The MIC data are mostly from reference 27.
MIC data are from reference 188.
The values of logP (the logarithm of the partition coefficient between n-octanol and water) are from PubChem (http://pubchem.ncbi.nlm.nih.gov/) based on the calculation by the XlogP3 method.
Drugs Traversing the OM Mainly through Porin Channels
The RND transporters, which play a predominant role in raising the MIC values of most antibiotics, pump out drugs mostly from the periplasmic space, as mentioned above (see Biochemistry and Genetics of Multidrug Efflux Pumps). They cannot create resistance if the drugs flow into the periplasm across the OM rapidly enough to counteract the rate of efflux. This is especially so because the RND pumps appear to have a relatively low velocity, for example, with AcrB having a velocity of ∼0.3 nmol/s/mg cells (dry weight) for penicillins (65) (or turnover rates of the order of 100/s [see reference 63]). (In E. coli, the chromosomally encoded class C β-lactamase is expressed at a low, constitutive level, and thus, β-lactam MIC values are essentially determined by the balance between influx and active efflux.) On the other hand, ampicillin was measured to cross the OM with a permeability coefficient (P) of 2.8 × 10−4 cm/s (64) or at a rate of P × A × Δc, where A is the area of cell surface (∼128 cm2/mg) and Δc is the concentration difference of the drug across the OM. If the external concentration of ampicillin is 10 μg/ml (3 × 10−5 M), the influx rate is expected to be ∼10 nmol/s/mg, which is much higher than the Vmax of efflux. Thus, efflux has only a barely visible effect on the MIC of this drug in E. coli, as has been ascertained by the use of ΔacrAB mutants (Table 2) (188). Similarly, for relatively small antibiotics such as fluoroquinolones and tetracycline, which are expected to diffuse through the trimeric porin channels rapidly, AcrAB inactivation decreases their MIC values only minimally (Table 2).
Even among the β-lactams, however, more hydrophobic compounds, such as oxacillin or cloxacillin, diffuse through the E. coli porin channels presumably rather slowly. Consequently, active efflux by AcrB strongly affects the MIC values, and the cloxacillin MIC decreases 128-fold, from 256 to 2 μg/ml, upon the deletion of acrB (Tables 2 and 3) (188). Because the acrB deletion hardly affects the ampicillin MIC, we thought previously that cloxacillin must be an exceptionally good substrate of AcrB and that ampicillin was a very poor substrate. Nonetheless, quantitative determination of efflux kinetic parameters (65) showed that these two drugs have similar affinities for AcrB and that the Vmax is higher for ampicillin by only ∼2-fold.
In contrast, in P. aeruginosa, where even small antibiotics must diffuse across the OM slowly via its slow porin, active efflux becomes very effective in increasing MICs, as seen from the fact that MICs of practically any antibiotic are drastically decreased upon the deletion of its major RND pump MexAB-OprM (Table 2). Here the situation with β-lactams becomes somewhat more complex, because with early compounds, hydrolysis by the powerful, inducible chromosomal β-lactamase plays a significant role. The relative lack of an effect of pump deletion on the fourth-generation cephems cefepime and cefpirome (Table 2) may suggest that they are poor substrates of the pump; it may also reflect the extreme stability of these compounds with the chromosomal class C enzyme (189). The imipenem MIC is hardly affected by the pump deletion, but this is because imipenem permeates across the OM much more rapidly than other compounds, by utilizing a specific channel, OprD (190). Thus, the efflux pump, even if it were capable of pumping out imipenem, would be overmatched by the rapid influx of the substrate (Fig. 1).
Drugs Traversing the OM through the Lipid Bilayer Region
Large molecules that cannot diffuse through the porin channels must penetrate the OM by slowly diffusing through the asymmetric bilayer domains, which have similarly low permeability in E. coli and P. aeruginosa (185, 191). Because of their slow influx, active efflux can become extremely effective, particularly when these molecules are preferred substrates for the efflux pumps, as can be seen in the huge decreases of MICs upon genetic inactivation of the main RND pumps (novobiocin and erythromycin) (Tables 2 and 3).
To recapitulate, the multidrug pumps work in synergy with the OM barrier. The pumps can make Gram-negative bacteria resistant only when the influx of the drug across the OM is relatively slow, and thus, efflux should always be considered in relation to the OM penetration process (Fig. 1). This also underscores the problems presented by organisms that produce slow porins, such as P. aeruginosa and Acinetobacter, because the efflux processes there become extremely efficient in increasing the resistance level.
GAMMAPROTEOBACTERIA: ENTEROBACTERIACEAE
Drug efflux pumps are widely distributed in bacterial species. The contribution of representative pumps to resistance and their synergistic interplay with other resistance mechanisms in clinical settings are further described below. The members of the Enterobacteriaceae family discussed in this section all produce high-permeability trimeric porins in their OM, although there are likely differences in the sizes of the channels.
E. coli
E. coli is a commensal resident of human and animal intestinal tracts but also includes various intestinal pathogenic types (enterotoxigenic, enterohemorrhagic, enteroinvasive, enteropathogenic, enteroaggregative, and diffusely adherent E. coli) as well as extraintestinal pathogenic E. coli (192). The major, constitutively expressed RND-type multidrug transporter is AcrB, although E. coli possesses a number of drug pumps of various families (Table 1) (15, 16). AcrB is described above as the prototype example for the structural and biochemical elucidation of RND pump transport mechanisms. The effect of acrAB genetic deletion on antimicrobial susceptibility is shown in Tables 1 to 3. Lipophilic (or large) compounds (e.g., erythromycin, novobiocin, fusidic acid, and cloxacillin) cannot diffuse easily through porins, and consequently, AcrB-catalyzed efflux becomes very effective in raising their MIC values to a range outside clinical utility (Table 3). Thus, efflux is responsible, in synergy with the OM barrier, for making E. coli intrinsically resistant to such compounds. In contrast, compounds that are smaller and usually hydrophilic (with the exception of nalidixic acid and chloramphenicol) (such as ampicillin, cephalothin, imipenem, and fluoroquinolones) can penetrate the OM barrier rapidly through porin channels. Therefore, in wild-type cells, AcrB cannot raise MIC values to a significant extent, although many of these drugs are likely to be good substrates of AcrB (for the case of ampicillin, see reference 65).
These small agents are of course useful in the treatment of E. coli infections. Regarding treatment options, a major advance in the 1960s was the introduction of semisynthetic penicillins and cephalosporins active against Gram-negative bacteria, such as ampicillin and amoxicillin, and the first-generation cephalosporins (e.g., cephalothin). Their efficacy decreased drastically with the spread of plasmids coding for class A β-lactamases (usually of the TEM or SHV type), which could hydrolyze these drugs rapidly. To counter this problem, in the 1980s, extended-spectrum cephems (third-generation cephalosporins) were introduced. They could withstand the assault of class A β-lactamases but were hydrolyzed at sufficient rates by overproduced, chromosomal, class C AmpC β-lactamases in Enterobacter and Proteus, etc. (but not in E. coli). In E. coli, these extended-spectrum cephems eventually became less useful because of the spread of plasmids producing TEM or SHV derivatives as extended-spectrum β-lactamases (ESBLs) that acquired a broadened substrate specificity. More recently, however, plasmids encoding class A CTX-M-type enzymes, which apparently originated from a chromosomal gene in an obscure genus called Kluyvera, have become so prevalent as to replace the older ESBLs (193). One common type, CTX-M-15, hydrolyzes a third-generation agent, cefotaxime, much faster than it does a first-generation compound, cephalothin. Other agents also entered the market around the time of the introduction of the extended-spectrum cephems or somewhat later. These agents include fluoroquinolones, semisynthetic aminoglycosides, and carbapenems.
It should be noted that there has been a steady increase in the resistance of E. coli isolates to the agents mentioned above. In a survey covering 30 years of isolates in Sweden (194), the prevalence of isolates showing “non-wild-type” MICs of ciprofloxacin increased from 0% to 40% in 2009. Drugs that have become essentially useless in recent years include ampicillin (70% showing non-wild-type MIC values), tetracyclines, and trimethoprim (up to 60%). It is even more alarming that these statistics are from Sweden, a country with one of the lowest frequencies of drug-resistant bacteria. The prevalence of E. coli isolates resistant to extended-spectrum cephalosporins was 4.4% in Sweden in 2012 but was much higher in some other European countries, e.g., 31% in Slovenia (195). Efflux mechanisms likely contribute to such a rapid emergence of resistance in the presence of antimicrobial selection pressure, as discussed below.
Fluoroquinolones.
Because fluoroquinolones are not hydrolyzed in bacteria, resistance to these agents involves the OM permeation barrier, active efflux, and mutational alterations to their targets, DNA topoisomerases. The target mutations are well known and are indeed present in practically all resistant strains of clinical origin. However, the first two factors also make frequent contributions, especially in strains exhibiting very high MIC values. Already in 1996, a pioneering study by Everett et al. (196) showed that among 36 E. coli strains with ciprofloxacin MICs of >2 μg/ml, 22 accumulated smaller amounts of ciprofloxacin than the wild-type strains in an energy-dependent manner, suggesting active efflux. That study also found alterations in porin expression, although their identities were not conclusively established. The involvement of efflux was also determined by the finding that fluoroquinolone-resistant isolates tended to be resistant to solvents (197). Since both fluoroquinolones and solvents are substrates of AcrB, this suggests strongly the involvement of this pump, an idea that was later confirmed by the discovery that these isolates had mutations in the marR repressor gene, which resulted in the overproduction of the MarA activator of acrAB transcription (198). The strong overexpression of MarA was also found in most levofloxacin-resistant clinical strains from Japan (199), and the overexpression of AcrA/AcrB was correlated with high levels of norfloxacin resistance among clinical isolates from the United States (200). Since MarA represses the expression of the larger channel porin OmpF (201), the downregulation of OmpF in some strains might have been caused in this manner. Among isolates resistant to amoxicillin, co-trimoxazole, and quaternary ammonium disinfectants, TolC overproduction was common, and this appeared to be caused by MarA or SoxS overproduction (202). High-level fluoroquinolone-resistant strains from China contained mutations in the acrR gene (203). In any case, numerous studies have now confirmed the important role of AcrAB overproduction in high-level fluoroquinolone resistance (188, 200, 204). Although the levels of porins were not examined frequently, a study showed that 10 out of 11 highly fluoroquinolone-resistant isolates had a strongly decreased expression level of OmpF (205). Additionally, a plasmid-encoded fluoroquinolone-specific MFS pump, QepA, was reported in 2007 (206).
β-Lactams.
Most cephalosporins and penicillins are substrates of AcrB. With sophisticated methodologies using intact cells, the kinetic constants for efflux have been determined for about a dozen β-lactams (63–65). Older compounds (e.g., ampicillin, cephalothin, and cephaloridine) were effective against E. coli partly because they were relatively small and hydrophilic and thus penetrated rapidly through the OmpF and OmpC porins (174, 207, 208). As described above (see Synergy with the Outer Membrane Barrier), active efflux cannot increase the MICs of these rapidly penetrating compounds, and indeed, the deletion of acrAB has no or very minor effects on their MICs (188). However, if porin permeability is decreased, efflux would produce a more visible effect. In a study reported in 1981, selection with carbenicillin easily enriched for mutants lacking the wider-channel OmpF and producing only the more restrictive OmpC porin (209). Since carbenicillin is not easily hydrolyzed by the chromosomal AmpC β-lactamase (210), from the vantage point of 2014, we can conclude that the mutant lacking OmpF became resistant to carbenicillin because the balance between influx and active efflux was perturbed. (The reader should be reminded here that even a very slowly penetrating drug would reach a half-equilibrium concentration in the periplasm within 1 min [21]. Thus, counteracting mechanisms such as efflux or enzymatic hydrolysis are absolutely necessary to raise the β-lactam MIC beyond the range needed for inhibiting their targets.) Indeed, one study (209) showed that the mutant became much more resistant to good substrates of AcrB, such as benzylpenicillin (65) or cephaloram, but not to poor substrates of AcrB, such as cefazolin and cephaloridine (63). A similar in vitro selection of porin-deficient strains was achieved by using cefoxitin (211), which is also only slowly hydrolyzed by the chromosomal β-lactamase. Laboratory selection using ceftazidime or ceftibuten, starting from an E. coli strain containing a TEM-1-producing plasmid, also resulted in the loss of OmpF or OmpF and OmpC (212). A clinical example involving the loss of a porin during therapy with a first-generation cephalosporin is described below in the section on Salmonella spp.
The loss or downregulation of porins thus seems an important mechanism for increased resistance to β-lactams in E. coli (as with Enterobacter and Klebsiella, described below), but unfortunately, this has not been examined in most studies of clinical isolates of this species. One would predict (see also above) that such a mechanism may be even more important with the extended-spectrum cephems because they tend to be larger and tend to penetrate more slowly through wild-type porins (208). In rare studies where the porin pattern was examined, its alteration was found. In 1991, seven out of the eight amoxicillin-clavulanate-resistant strains examined were found to produce reduced levels of or no OmpF protein (213). In the era dominated by extended-spectrum cephems, a study of clonally related cefoxitin- and ceftazidime-resistant strains from a Spanish hospital in 2000 showed that all strains were deficient in OmpF and produced AmpC β-lactamase at a high level (214). In a 2003 study of E. coli strains resistant to extended-spectrum cephems, most strains not producing ESBLs had altered patterns usually involving the apparent loss of OmpF (215). The absence of OmpF was also seen in a ceftazidime-resistant strain (216). In these porin-deficient strains, efflux is likely to contribute to the increased MIC values, although an even stronger contribution would probably come from enzymatic hydrolysis.
More recently, the widespread use of carbapenems appears to have selected porin-deficient mutants in E. coli (217, 218), as is also seen in other species of Enterobacteriaceae (see below). This could be because the widely disseminated enzymes cannot yet hydrolyze carbapenems very efficiently. The prevalence of porin-deficient strains is also consistent with the fact that imipenem, a frequently used agent, is most probably a poor substrate of AcrB; thus, porin loss, but not pump overproduction, raises the MIC values. However, other carbapenems, containing larger and often hydrophobic side chains, behave as the substrates of RND pumps (see reference 219, for example).
A priori, a strong overproduction of AcrAB-TolC should be able to increase the MIC values of β-lactams that are the substrates of this pump. However, AcrB is already constitutively expressed, and mutations in the global regulatory systems Mar, Sox, and Rob (see Regulation of Multidrug Efflux Pumps, below) can increase its expression only severalfold and may not produce a strong effect on the MIC, especially when enzymatic hydrolysis also plays a role in resistance. Perhaps this might be why pump overproducers were often not noted among clinical resistant isolates.
Other drugs.
Given its extremely wide specificity, the AcrAB-TolC system obviously makes a major contribution to the increased resistance to other drugs (Tables 1 to 3). Even for rifamycins, which are large lipophilic molecules and expected to cross the OM via its lipid bilayer, the acrAB status has shown a modest impact on rifampin susceptibility. A relatively low level (20 μg/ml) of PAβN, an EPI of the AcrB pump, also decreased the MIC (>128-fold) of rifaximin, a rifamycin derivative, for resistant strains with no target mutations (21, 220), suggesting that efflux can contribute to rifamycin resistance under certain conditions (e.g., strain or species specific) (see also the section on Neisseria for rifampin as an MtrCDE pump substrate, below). However, as expected, the susceptibility of E. coli to rifampin has also been known to be impacted by the OM status (177).
Salmonella spp.
A clinical Salmonella enterica serovar Typhimurium isolate that lost OmpC and became resistant to the earlier-generation cephems was reported in 1987 (221). A patient who received a kidney transplant developed septicemia involving this organism, which developed cephalexin resistance only 3 days after this drug was added to the regimen. The pretherapy strain produced OmpF and OmpC in the laboratory, but the posttherapy isolate produced only OmpF. Presumably, the synthesis of the OmpF porin was strongly repressed by the relatively high salt content within the human body and was thus irrelevant during therapy, and hence, the posttherapy strain allowed only a minimal influx of drugs, which made it more resistant to cephalexin, which was hydrolyzed readily by the plasmid-encoded TEM β-lactamase in both isolates.
In Salmonella, OmpF and OmpC are the major porins, and AcrB is also the major, constitutive multidrug pump, although there are a total of 5 chromosomally encoded RND systems identified (i.e., AcrAB-TolC, AcrA-AcrD-TolC, AcrEF-TolC, MdtABC-TolC, and MsdABC or MsdAB-TolC) (58, 222–224). Importantly, RamA, a positive regulator of the acrAB transcription that does not exist in E. coli, seems to play a major role in AcrAB overproduction (see Regulation of Multidrug Efflux Pumps, below). Thus, for fluoroquinolones, we would expect situations similar to those that have been found for E. coli. In 1993, Piddock and associates found that posttherapy, ciprofloxacin-resistant isolates accumulated much less drug inside the cells (225), a phenomenon that is now interpreted to be a result of AcrB-driven efflux. Although mutations in target DNA topoisomerases are important, very high-level resistance to fluoroquinolones seems to require the additional contribution of increased efflux (188, 226), and this was the case in the DT204 clone (227). Among nontyphoid Salmonella isolates from Spain, ∼40% were nalidixic acid resistant, although their ciprofloxacin MICs were below the clinical resistance breakpoint; in all of these strains, MICs of nalidixic acid (and ciprofloxacin) decreased strongly in the presence of the AcrB inhibitor PAβN at 20 μg/ml (228) (see Efflux Pump Inhibitors, below). In laboratory-selected ciprofloxacin-resistant S. enterica serovar Enteritidis mutants, AcrB overproduction was present, and in one mutant, a deficiency of the OmpF porin was also present (229). About 5% of S. enterica serovar Typhi strains isolated in South Africa were nalidixic acid resistant, and their nalidixic acid and ciprofloxacin MICs decreased strongly in the presence of PAβN, although this inhibitor was used at a high concentration of 40 μg/ml (230). With β-lactams, one might also expect a situation similar to that found in E. coli, i.e., decreased porin expression and efflux contributing to resistance together with hydrolysis by β-lactamases encoded by R plasmids. In fact, in a plasmid-free strain, the contribution of efflux is more pronounced because of the total absence of enzymatic hydrolysis, caused by the absence of the chromosomal ampC gene in this species (231). Thus, in S. enterica serovar Typhimurium, inactivation of acrAB decreases MIC values of penicillins and cephalosporins quite strongly (58).
Efflux is a significant factor for most β-lactams, with the exception of cefazolin, which is too hydrophilic to be a substrate for AcrB (58, 63). These data agree with the actual measurements of AcrB-catalyzed efflux of β-lactams (63–65). However, changes in porins and efflux have been rarely reported for β-lactam-resistant Salmonella isolates. These results may be related to the ecology of these organisms. Salmonella, as a pathogen of farm animals, tends to exist in a strongly clonal manner (232). Therefore, once a clone acquires a resistance plasmid, it will remain and can become the major mechanism of resistance. During the recent period in which extended-spectrum cephems were among the most important agents for therapy, strains containing plasmid-encoded AmpC β-lactamases (e.g., CMY enzymes) and ESBL β-lactamase (e.g., CTX-M) were prevalent among resistant strains (233). They have an advantage over porin-depressed or efflux-enhanced strains, which may have to pay heavily for these unfavorable metabolic changes. In addition, with E. coli (bloodstream) infections, resistant mutants can be selected during the course of therapy, thus favoring porin or efflux mutants. Infections with nontyphoid Salmonella may be caused frequently by strains already containing R plasmids (232, 234). In this regard, there are increasing numbers of reports of multidrug-resistant or extensively drug-resistant isolates of Salmonella that carry the plasmid-encoded OqxAB RND pump with other resistance determinants such as aac(6′)-Ib-cr and/or CTX-M genes (228, 235, 236).
Regarding porin mutants, there are a few reports implicating porin downregulation in β-lactam resistance. Armand-Lefèvre et al. (237) showed that OmpF was completely deficient in an imipenem-resistant, CMY-4 β-lactamase-producing isolate of S. enterica serovar Wien and that the resistance can be reproduced in an OmpF-deleted E. coli strain. Su et al. (234) reported that treatment of a patient infected by S. enterica serovar Typhimurium with ertapenem led to the development of carbapenem resistance concomitant with the mutational inactivation of OmpC, which was the only strongly expressed porin in the parent strain; this confirms the above-mentioned prediction that porin loss would be one mechanism of resistance development during the course of therapy. In a report with results more difficult to interpret, the downregulation of OmpD was correlated with an increased MIC of ceftriaxone (238). Unfortunately, nothing is known about the permeability of the channels of OmpD, although its E. coli homolog, NmpC, showed less single-channel conductance in NaCl than with OmpF and OmpC (239).
Citrobacter spp.
The genus Citrobacter is closely related to Salmonella, and one would expect rather similar situations. Thus, some fluoroquinolone-resistant isolates of Citrobacter freundii were found to accumulate less drug (240). In vitro selection of fluoroquinolone-resistant mutants showed the importance of increased efflux in practically all cases (241). Intrinsic resistance to linezolid is mediated by AcrB in this species as well as others (242). A C. freundii isolate containing the plasmid-encoded RND pump was reported recently (243) (see Drug Efflux Genes on Plasmids, below).
Enterobacter aerogenes and Enterobacter cloacae
In contrast to E. coli, the chromosomally encoded ampC β-lactamase in Enterobacter aerogenes and Enterobacter cloacae is strongly inducible, and thus, penicillins and the first-generation cephalosporins were ineffective for them. The third-generation cephalosporins were initially thought to be resistant to AmpC-catalyzed hydrolysis. However, strongly overproduced AmpC turned out to have a low Vmax but a very high affinity for these drugs (244) and to be capable of producing significant resistance. The fourth-generation cephems and carbapenems are quite effective against non-plasmid-containing strains. Enterobacter spp. produce the constitutive AcrB pump as in E. coli. (Another RND pump gene, eefB, was cloned from E. aerogenes, but its inactivation had no effect on the MICs of antibiotics [245].) They also possess the E. coli-like porins OmpF and OmpC, and hence, one would expect situations in these organisms similar to those seen in E. coli regarding their mechanisms of resistance to β-lactams and fluoroquinolones.
When a large number of β-lactam-resistant strains of Enterobacteriaceae from French hospitals was examined for the production of porins by Pagès and associates in 1996 (246), 6% of resistant strains of E. cloacae and 44% of resistant strains of E. aerogenes lacked detectable porins. The very high fraction of porin deficiency in E. aerogenes is striking. As these bacteria are commensal organisms, they presumably had less chance to acquire R plasmids than did obligatory pathogens like Salmonella, at least up to 1996 (although the plasmids producing ESBLs have become increasingly common in recent years). Furthermore, the resistant strains were probably selected mostly during therapy, favoring mutational mechanisms. Still, the difference in the frequency of porin-deficient strains between E. aerogenes (20 out of 44 resistant strains) and E. cloacae (1 out of 17 resistant isolates) begs for an explanation. The porins of at least one strain of E. cloacae had a five-times-lower permeability than did those of E. coli (244), so perhaps, in this organism, the influx of drugs across the OM is already limited, even without any mutation. Cephaloridine permeation of E. cloacae was found to be 25-fold lower than that of E. coli by another laboratory (247). (The OM permeability of E. cloacae to β-lactams, measured by a novel method [248], was reported to be 20 to 1,000 times lower than that of E. coli. However, the E. coli values were not obtained in the same way, and it is uncertain if a quantitative comparison is warranted.) In contrast, E. aerogenes was found to have a permeability very similar to that of E. coli (249), and thus, the creation of significant levels of resistance may need the downregulation of porins in addition to drug hydrolysis and/or efflux. (OmpF and OmpC from E. aerogenes were studied in detail, and the repression of OmpF biosynthesis by high osmotic pressure resulted in MIC increases of 8- to 16-fold [250] for slowly penetrating compounds such as cefotaxime, ceftazidime, and ceftriaxone [208].) An imipenem-resistant isolate of E. cloacae was deficient in both OmpF and OmpC (247). A carbapenem-resistant mutant of E. cloacae, selected during imipenem therapy, lacked both OmpF and OmpD porins (251); furthermore, ertapenem and meropenem MIC values were decreased strongly in the presence of 40 μg/ml PAβN, although overexpression of AcrB was not evident. (Imipenem does not appear to be a substrate of the P. aeruginosa MexAB-OprM system [13], but meropenem, a derivative with a lipophilic side chain, is a substrate [252]. Ertapenem, another carbapenem with a lipophilic side chain, is also expected to be a substrate for MexAB-OprM and homologous AcrAB.) In vitro selection for E. cloacae mutants with increased MICs of ceftazidime, cefepime, or cefpirome often resulted in strains defective in OmpF (253, 254). A study of ertapenem-resistant isolates from Taiwan (255) found that 43% were altered in the expression of porins (and 96% expressed a multidrug pump). In another study of carbapenem-resistant isolates (256), both OmpF and OmpC porins were totally lacking in two E. aerogenes isolates, whereas variable decreases in porin levels were found in E. cloacae isolates. Similarly, an imipenem-resistant E. aerogenes isolate lacked both OmpF and OmpC (257). A more recent study demonstrated that porin expression often becomes altered when E. aerogenes infection is treated with imipenem (258).
The Pagès group continued their work on E. aerogenes clinical isolates, which resulted in several studies. In one cephalosporin-resistant isolate, there was a missense mutation leading to an altered porin with decreased permeability (259). They also identified an increased efflux of fluoroquinolones and chloramphenicol in some multidrug-resistant strains (260), which is likely to be due to AcrB (261). Finally, frequencies of chloramphenicol-resistant isolates susceptible to the AcrB inhibitor PAβN appeared to have increased between 1995 and 2003, suggesting a larger role for the broad-specificity efflux mechanism in recent years (262).
As would be expected from results with E. coli and Salmonella, a large fraction of fluoroquinolone-resistant isolates of E. cloacae appeared to overproduce a multidrug pump, presumably AcrB (263). Deletion of acrA predictably decreased the MICs of oxacillin, erythromycin, clindamycin, linezolid, ciprofloxacin, chloramphenicol, tetracycline, and tigecycline (264). All multidrug-resistant E. aerogenes strains tested (also resistant to β-lactams) were found to overproduce AcrAB (265), but the effect of an EPI on β-lactam MICs was not examined. Among the large, lipophilic agents that have little activity against these species, macrolides are AcrAB substrates, but a ketolide, telithromycin, appeared to also be pumped out by an additional pump that is susceptible to PAβN inhibition (266). Enterobacter spp. develop resistance to tigecycline through AcrAB overproduction (during ciprofloxacin treatment in one study [267]), and this was caused by the overproduction of the RamA regulator (268), although one study concluded that the overexpression of RamA, an not necessarily that of AcrAB, correlated with the resistance, suggesting the involvement of other efflux pumps (269).
Klebsiella pneumoniae
Klebsiella pneumoniae produces the E. coli-like trimeric porins OmpK36 (an OmpC homolog) (270) and OmpK35 (an OmpF homolog) (271). Their permeability has not been measured in a way that could be compared with that of E. coli porins. However, our recent measurement of ampicillin and benzylpenicillin permeation in strain ATCC 11296 showed that the OM of this species is vastly more permeable to benzylpenicillin than E. coli OmpF, yet the permeability to ampicillin is actually similar (S. Kojima, E. Sugawara, and H. Nikaido, unpublished data), an observation that explains why porin deficiency is needed for resistance in K. pneumoniae. It also produces the ubiquitous AcrAB-TolC pump, whose deletion predictably produces hypersusceptibility to erythromycin, chloramphenicol, nalidixic acid, fluoroquinolones, cefoxitin, and cefotaxime and surprisingly produces hypersusceptibility to gentamicin (272). Thus, some aspects of the resistance mechanisms are expected to be similar to what was found for the species discussed above. Indeed, the loss of porins was reported frequently to be the contributing mechanism of resistance, beginning with a 1985 report by Gutmann and associates (273) that one-step selection with nalidixic acid, trimethoprim, or chloramphenicol yielded mutants showing cross-resistance to all three agents and with detectable changes in OM protein patterns. Later, the selection of a porin-deficient mutant in a patient treated with cefoxitin was reported by the same group (274). During the 1990s, in vitro selection of porin mutants by cefoperazone-sulbactam (275) or cefoxitin (276) was confirmed, and clinical porin-deficient isolates resistant to cefoxitin and extended-spectrum cephems (277, 278) were reported. Porin loss is often caused by the insertion of an insertion sequence (IS) element (279). In the present century, reports on porin loss are so numerous that it is impossible to give an exhaustive list of the literature; we give only a few representative examples (280, 281). We note, however, that clinical isolates resistant to carbapenems often lack porins (257, 282, 283). The deletion of OmpC alone has little effect on antibiotic MICs, but the loss of OmpF has a large effect, as predicted (284). When OmpC or OmpF was expressed from plasmids in a strain expressing an ESBL but lacking both porins, MIC values for most agents decreased significantly, but the extent of the decrease was greater with the expression of OmpF (OmpK35) (271). The difference was larger with more bulky agents: for cefpirome (515 Da), the MIC of 512 μg/ml in the porin-deficient strain was reduced to 4 μg/ml by the expression of OmpC, whereas OmpF expression reduced it to 0.5 μg/ml. The MIC of a large agent, ceftazidime (547 Da), which was >512 μg/ml in the porin-deficient parent strain, was reduced only to 256 μg/ml by the expression of OmpC, whereas OmpF expression decreased it to 2 μg/ml. On the other hand, for a smaller agent, meropenem (383 Da), the expression of either porin decreased the MIC of 4 μg/ml in the parent strain to the same value, 0.03 μg/ml. These results suggest that OmpF produces a more permissive, possibly larger, diffusion channel than does OmpC. (This concept may also explain why there have been some confusing data on the role of OmpF versus OmpC for carbapenem susceptibility [see reference 285].) A similar explanation was invoked to explain the higher level of resistance to a more bulky ceftazidime than a smaller cefotaxime in a strain lacking only OmpF (286); this seems to be a valid hypothesis since the strain produced an ESBL enzyme that hydrolyzed cefotaxime much better, on the basis of Vmax/Km values.
In recent years, K. pneumoniae has become a major nosocomial pathogen causing outbreaks of multidrug-resistant clones. Such clones typically express ESBLs (more recently of the CTX-M type) and/or AmpC-type enzymes. These enzymes obviously make predominant contributions to general β-lactam resistance, thus making carbapenems some of the few remaining effective drugs. Recently, however, porin deficiency combined with enzymes with marginal carbapenemase activity has attracted attention. One of the often encountered types combines a frameshift null mutation of OmpF with a mutational change in OmpC, which inserts a few residues into loop 3 that create the narrow constriction of the channel (for example, see references 283, 285, 287, and 288). The role of porin deficiency is often underestimated. For example, Zhang et al. (285) conclude that porin alterations play only a minor role because the removal of the KPC-2 plasmid reduces the carbapenem MICs drastically. Still, unlike the presence of β-lactamase, the porin deficiency by itself cannot raise the carbapenem MICs substantially. (The proper experiment should have been the addition of the same plasmid to the wild-type and porin-deficient strains.) Considering that active efflux contributes to resistance to agents other than imipenem in organisms with a low-permeability OM, such as P. aeruginosa (252), efflux is likely to also make a contribution in porin-deficient K. pneumoniae, but this remains a topic for future study. Finally, LamB appeared to allow some permeation of carbapenems in a strain deficient in both OmpF and OmpC (289).
The genome of K. pneumoniae shows the presence of a large number of drug efflux pumps similar to those observed in E. coli (290). The overexpression of efflux pumps, most probably that of AcrAB, is important in resistance to certain agents, such as chloramphenicol and tetracycline but especially fluoroquinolones (see references 291 and 292). In an isolate lacking porins and showing high-level fluoroquinolone resistance, an efflux pump was apparently expressed strongly, as judged from the low level of norfloxacin accumulation in the absence of energy poison (293). In an important study of ESBL-producing strains from outbreaks of nosocomial infections (294), ciprofloxacin resistance (caused in part by AcrB overproduction) was strongly correlated with cefoxitin resistance, suggesting that for β-lactam resistance, not only the β-lactamases and porin depletion but also efflux may make a significant contribution. Similarly, ESBL-producing strains often lack porins and show a stronger efflux of fluoroquinolones (295). The significance of efflux in β-lactam resistance was pursued later by the Pagès group (296), who examined cefoxitin-resistant but ceftazidime-susceptible clinical isolates. Although β-lactam resistance is most frequently caused by the plasmid-encoded ESBLs, the ceftazidime susceptibility of these strains did not fit with this idea. Indeed, all the strains examined were devoid of plasmid-encoded β-lactamases and produced only the chromosomal class A enzyme. However, in the presence of PAβN (50 μg/ml [unfortunately an excessive concentration]), the MICs of cefoxitin (and cloxacillin), erythromycin, chloramphenicol, nalidixic acid, and ofloxacin decreased strongly, implicating active efflux as a major factor in resistance. There was little decrease in the ceftazidime MIC; as we mention above, ceftazidime is too hydrophilic to be a good substrate of AcrB. Similarly, studies by Källman et al. (297, 298) are important because they show that AcrAB-mediated efflux contributes to cefuroxime resistance in both K. pneumoniae and E. coli, together with the porin deficiency and β-lactamase-catalyzed hydrolysis. Additionally, in vitro selection with cefoxitin and fluoroquinolones, starting from the drug-susceptible revertants of the clinical strains described above, resulted in AcrB overproduction caused mainly by mutations in ramR and in one case in soxR (299). A recent study linked ramR mutation-driven overexpression of AcrAB to tigecycline resistance in KPC-producing strains (300), although one study suggested that RamA-AcrB overexpression occurred only in about one-half of the tigecycline-resistant isolates (301).
Finally, efflux pumps other than AcrB also contribute to resistance. In addition to MDR mediated by the chromosomally encoded OqxAB pump (302–304), overexpression of the newly identified RND-type KpgABC pump was involved in tigecycline nonsusceptibility due to an IS element insertion in the promoter region of kpgABC (305). The KexD RND pump expressed by cloning provides MDR with the requirement of AcrA accessory and KocC OM proteins (306). KpnEF, an SMR pump, contributes to MDR and may be important in the infection process, because its expression is regulated by the Cpx regulatory system, which also affects capsule production (163). Inactivation of the MFS-type KpnGH pump results in 4- to 10-fold MIC reductions of the third- and fourth-generation cephalosporins, spectinomycin, streptomycin, and tetracycline (307).
Proteus, Providencia, and Morganella spp.
In an extensive study in 1987 (308), it was shown that two major OM proteins (presumably porins) are produced in Proteus mirabilis, Proteus vulgaris, Morganella morganii, Providencia rettgeri, and Providencia alcalifaciens and that the major porin in every case had channel properties similar to those of E. coli OmpF. Furthermore, in vitro selection with cefoxitin led to the isolation of mutants deficient in the major porin in each species. Since the chromosomal AmpC β-lactamases of these species can hydrolyze cefoxitin only extremely slowly and cefoxitin passes through the porin channel only slowly, this selection is similar to what has been achieved with carbenicillin and E. coli K-12, resulting in the loss of the porin with a larger channel. Interestingly, in 4 out of 5 strains, the MIC of tetracycline also increased upon selection with cefoxitin, suggesting that strains derepressed for a multidrug pump, presumably AcrB, were also selected. In a fluoroquinolone-resistant M. morganii isolate selected in the laboratory, there was a downregulation of what appeared to be OmpF and increased norfloxacin uptake in the presence of CCCP, indicating active efflux (309).
The substrate range of AcrB from P. mirabilis includes, as expected, a number of antibiotics, dyes, and detergents (310, 311). A similar range was found in a tigecycline-resistant clinical isolate of M. morganii overexpressing AcrAB, whose inactivation has led to hypersusceptibility to multiple agents, including a 130-fold reduction of the tigecycline MIC value (312). In a 1992 study (313), a cefoxitin-resistant laboratory isolate of P. vulgaris not only lacked OmpF but also accumulated smaller amounts of most fluoroquinolones; although multidrug pumps were not known at that time, it seems clear that resistance was the result of synergy between the decreased OM permeability and AcrB-catalyzed efflux. This study found that porin loss did not diminish the intracellular accumulation of two agents, sparfloxacin and tosufloxacin. Since it is difficult to imagine that these compounds cross the OM by a unique, unconventional route, this result, if confirmed, may indicate that the major pump discriminates between these substrates. Isolates of P. rettgeri from larvae of the oil fly have shown a correlation between natural resistance to a variety of antimicrobials and organic solvent tolerance, a phenomenon indicating strong efflux involvement (314).
Serratia marcescens
Porins of Serratia marcescens are similar to OmpF/OmpC of E. coli in terms of cephalosporin permeation (315). S. marcescens isolates selected in increasingly higher concentrations of cephalosporins, often those compounds that are not easily hydrolyzed by the endogenous β-lactamase, were found to lack one or more porins (316, 317). Similar porin-deficient S. marcescens isolates were also selected by using chloramphenicol, nalidixic acid, or trimethoprim and showed cross-resistance to β-lactams (273); from the vantage point of today, it seems quite likely that at least some mutants were overexpressing efflux pumps. Selection with moxalactam yielded a mutant strain lacking a “42-kDa” porin, or OmpF, and its OM showed a permeability to cephaloridine >100-fold lower than that of the parent strain, in spite of the presence of the “40-kDa” or OmpC porin (318). In contrast, OmpC deletion had little influence on antibiotic susceptibility. Salicylate induction of fluoroquinolone resistance was ascribed to the decreased expression level of OmpF (319); however, increased production of AcrB is likely to have played a more important role.
As expected, fluoroquinolone-resistant strains produced higher levels of an RND pump in Serratia (15). Cosmid cloning of the genes responsible for this phenotype identified sdeA and sdeB, which confer resistance to fluoroquinolones, chloramphenicol, sodium dodecyl sulfate (SDS), and ethidium (320). SdeAB's function is dependent on the TolC-like OM protein HasF, and sdeB inactivation increases the susceptibility to fluoroquinolones and other drugs listed above (321). Exposure to a biocide (cetylpyridinium chloride) led to a mutational upregulation of SdeAB and thus antibiotic resistance (322). SdeAB expression is controlled by a BadM-type repressor, SdeS (323), and a putative MarA-like regulator, SdeR (321). Another RND system (SdeCDE) requires two paired pump genes, sdeDE, but their deletion did not seem to affect the MICs of common antibiotics (320).The RND pump SdeXY produces resistance to fluoroquinolones as well as many substrates of AcrB (16). Its overexpression (with HasF participation) is responsible for tigecycline and fluoroquinolone resistance (324). SdeY, however, has a sequence 84% identical to that of E. coli AcrB and should more properly be called AcrB. Thus, at present, the relative importance of AcrB and SdeB in fluoroquinolone efflux remains unclear, and the expression levels of sdeB in clinical isolates had no clear correlation with their fluoroquinolone resistance levels (325). An MFS pump (SmfY), an SMR pump (SsmE), and an ABC pump (SmdAB) increased the MICs of several drugs when expressed from a plasmid in E. coli (16).
Shigella flexneri
For Shigella flexneri, the involvement of efflux, including AcrAB overproduction, leading to high-level resistance to fluoroquinolones, was reported (326, 327). Inactivation of the MFS pump MdfA-encoding gene led to an 8-fold decease in the norfloxacin MIC (328).
Yersinia enterocolitica and Yersinia pestis
The genus Yersinia, although relatively distantly related to E. coli, produces the classical trimeric porins OmpF and OmpC (329). It also contains genes for RND pumps. In one study, all 41 nalidixic acid-resistant isolates of Y. enterocolitica showed significant decreases in nalidixic acid MICs in the presence of 20 μg/ml PAβN, suggesting a strong contribution of efflux (330). A Mar homolog was identified in Y. pestis, and its overexpression increased, albeit to a small degree, the MICs of several antibiotics, including tetracycline and rifampin (331), presumably by the increased expression of the AcrB-type pump. Moreover, a homolog of a well-known activator of acrAB transcription, Rob, increased ofloxacin MICs >10-fold (332). Indeed, the acrAB deletion resulted in large decreases in MICs of many antimicrobials, including aminoglycosides (333). Lastly, an MFS pump, together with a KefC-like exporter, pumped out cationic antimicrobial peptides in Y. enterocolitica, and their deletion made cells more susceptible to novobiocin and tetracycline (334).
OTHER GAMMAPROTEOBACTERIA: VIBRIO, AEROMONAS, LEGIONELLA, AND PASTEURELLACEAE
This section examines genera and species that are known or suspected to produce high-permeability trimeric porins and thus are similar to Enterobacteriaceae in this respect.
Vibrio spp.
Vibrio spp. are outside the Enterobacteriaceae but not very far in terms of phylogeny. V. cholerae has attracted attention as the causative agent of cholera. Its OM appears to be significantly different from that of Enterobacteriaceae. First, its outer leaflet appears to contain phospholipids in addition to LPS (335), which should allow an unusually rapid permeation of large, lipophilic antibiotics. Indeed, V. cholerae is far more susceptible to such agents, e.g., erythromycin, novobiocin, and rifampin, than are members of the Enterobacteriaceae (336). Second, its major porins, OmpU and OmpV, albeit belonging to the OmpF/OmpC family, are quite different from their enterobacterial representatives in sequences (22) and in permeability. An early study on OmpU (337) found that its single-channel conductivity was ∼3 times higher than that of E. coli OmpF, consistent with the results of a liposome swelling assay revealing a much larger channel than that of OmpF (338). Finally, some of the bile salts apparently bind to the channel interior and block permeation (339), suggesting that these large, planar, lipophilic molecules diffuse through the channel, at least up to the constriction zone. These high-permeability characteristics of the V. cholerae OM are surprising for a gastrointestinal pathogen but may be related to the ecology of this organism as a water dweller.
Because of their genetic relatedness to E. coli, cloning of Vibrio genes in E. coli was not difficult, and a number of efflux systems have been identified in this manner. The V. cholerae genome contains 6 RND transporters (VexAB, VexCD/BreAB, VexEF, VexGH, VexIJK, and VexLM), and most of them require TolC for drug efflux (340–345). VexAB seems to play a predominant role in the efflux of antibiotics (benzylpenicillin, erythromycin, and polymyxin B) and detergents (cholate, SDS, and Triton X-100) (341), although among the 6 RND systems cloned from a non-O1 strain, VexEF conferred the strongest resistance to an E. coli host (343). VexCD (confusingly “renamed” BreAB) is involved in bile efflux (340) and is induced by bile (342). A drug-susceptible phenotype could also be created by adding EPI PAβN or NMP to the parent strain (346). VexGH shows an overlapping substrate specificity and also contributes to the production of cholera toxin (344). In an interesting study (345), it was shown that not only was the expression of VexAB and VexGH enhanced by the CpxAR system, but the expression of this system also was stimulated by the deletion of efflux pumps, presumably due to the accumulation of intracellular metabolites. In V. parahaemolyticus, 12 RND pumps were identified, and when expressed in hypersusceptible E. coli, two-thirds of them produced an MDR phenotype (347–349), although the deletion of one of them, VmeAB, produced little increase in drug susceptibility (347). The isolation of laboratory mutants resistant to deoxycholate showed that VmeTUV is important for the efflux of bile salts (349).
Among the MFS transporters, VceB was the earliest discovered drug transporter in V. cholerae (350); it occurs together with a periplasmic accessory protein, VceA, and a TolC-like OM protein, VceC (16), and contributes to the intrinsic levels of resistance to deoxycholate, CCCP, pentachlorophenol, and nalidixic acid (350). Another MFS transporter, called EmrD-3, was found by cloning in a drug-hypersusceptible E. coli host and produced resistance to various lipophilic agents, such as linezolid (351). Five MFS transporters (each under the control of a Lys-type MfsR regulator) whose deletion causes tetracycline and bile salt hypersusceptibility are known (352). V. parahaemolyticus was the organism in which the first MATE family transporter, NorM, was discovered by cloning in E. coli (353), and in V. cholerae, 5 members of this family were described (16), including the NorM homolog VcmA (15), which increases MICs of fluoroquinolones, ethidium bromide, acriflavine, and doxorubicin when overproduced in E. coli. Two MATE transporters from a multidrug-resistant V. fluvialis isolate provided 2-fold increases in MIC values of ciprofloxacin and norfloxacin when expressed in E. coli (354).
Although many potential efflux transporters are known, there is not much knowledge on their role in clinically relevant situations, possibly because for V. cholerae, antimicrobial chemotherapy plays only a subsidiary role to fluid repletion therapy. The incidence of drug-resistant V. cholerae appears to be increasing, but the mechanisms presumably involve plasmids and integrons in many cases. In a study of fluoroquinolone-resistant V. cholerae clinical isolates, decreased accumulation of norfloxacin was found together with target gene mutations, suggesting the involvement of efflux (15).
Aeromonas spp.
Aeromonas belongs to another order, Aeromonadales. Although the pore-forming activity of an OmpA-like, monomeric protein has been reported (355), this protein presumably contributes only a minor activity to OM permeability, because OmpF/OmpC-like trimeric porins appear to exist in the genome sequences of Aeromonas salmonicida and Aeromonas hydrophila. The genome of A. hydrophila contains 10 RND pump genes as well as genes for MFS, MATE, SMR, and ABC efflux transporters (16). Apparently, an RND system, AheABC, plays a major role in the maintenance of the basal level of intrinsic resistance to most antimicrobials, which is rather similar to that of E. coli, except that A. hydrophila is more susceptible to macrolides like erythromycin and pristinamycin (16). An SMR pump, SugE, produces resistance to tributyltin, a compound that was used in the past as a biocide to prevent biofouling of ships; when introduced into E. coli, this gene increased resistance to chloramphenicol, tetracycline, as well as ethidium bromide (356). Fluoroquinolone-resistant isolates were often seen to have enhanced efflux activity (16). In contrast, another study found little evidence of the contribution of efflux to fluoroquinolone resistance (357), perhaps not surprisingly, because most strains had relatively low (<0.4 μg/ml) MICs of ciprofloxacin.
Legionella spp.
Legionella pneumophila contains the major oligomeric OM protein of 28 kDa (358). Although its single-channel conductance was low (0.1 nS) compared with that of E. coli OmpF (0.7 nS), this may have been caused by the low salt concentration (0.1 M rather than 1 M), and the channel size is difficult to ascertain. L. pneumophila strains show high levels of in vitro susceptibility to a variety of antibiotics, including macrolides, rifamycins, fluoroquinolones, aminoglycosides, and β-lactams (359), probably suggesting a limited role of either OM permeability or an efflux mechanism in intrinsic resistance. However, the fact that in vitro exposure of Legionella to erythromycin or ciprofloxacin selects mostly low-level resistance may be an indication of efflux participation (360), while target mutation-derived moxifloxacin resistance causes high-level MIC increases (8- to 512-fold) (361). The genomes of L. pneumophila strains (3.4 to 3.5 Mb) are smaller than that of E. coli K-12 (4.64 Mb) but still possess a large number of genes encoding efflux pumps, membrane fusion proteins, and OM channel proteins (362, 363). Inactivation of the TolC efflux channel (36% identity to E. coli TolC) increases the susceptibility to erythromycin (16-fold reduction in the MIC) and to benzalkonium chloride, deoxycholate, ethidium bromide, norfloxacin, novobiocin, and rhodamine 6G (2- to 8-fold decreases in MIC values) (362, 363). Moreover, TolC is also involved in the secretion of a lipid-containing surfactant that promotes Legionella motility and displays activity against Legionella (363). Given that intracellular killing of L. pneumophila by antibiotics is required for legionellosis therapy, drug efflux could be an important factor affecting the efficacy of in vivo antibiotic regimens. Particularly, the multiplication of L. pneumophila within macrophages has already limited the choice of antibiotics to those that can penetrate phagocytic cells, such as macrolides, rifamycins, and fluoroquinolones, which are generally good substrates of typical drug pumps.
Pasteurellaceae
Pasteurella multocida.
Pasteurella multocida produces a trimeric porin, OmpH, that is remotely related to the enterobacterial porins in sequence (364). OmpH is similar to E. coil porins in terms of permeability on the basis of the reported single-channel conductivity (364, 365). The genome of this organism contains one AcrB-like transporter, and the inactivation of tolC makes the organism susceptible to a wide range of agents (366). An Msr(E) ABC transporter, together with the phosphotransferase Mph(E), mediates clinically relevant resistance to macrolides, including new veterinary ones, gamithromycin and tildipirosin (367). Efflux-based tetracycline resistance genes from Pasteurella are also usually found on plasmids (16).
Haemophilus influenzae.
Because of its clinical significance and its natural ability to be transformed by naked DNA, many early studies were carried out by using Haemophilus influenzae. H. influenzae produces one trimeric porin, OmpP2, which allows the passage of larger oligosaccharides (up to 1,400 Da) than the E. coli porins (typically excluding solutes of >600 Da) (368). The large size of the porin channel is probably responsible for making this organism susceptible to large, hydrophobic agents such as macrolides. Deficiency in this porin accounts for, at least in large part, resistance to chloramphenicol (369) and some other agents. H. influenzae contains a homolog of E. coli acrB, and its disruption made the organism more susceptible to erythromycin, rifampin, novobiocin, and other toxic agents (370). A later study suggested that AcrAB acted together with TolC and showed that the system also pumps out additional compounds such as ampicillin, fusidic acid, linezolid, puromycin, trimethoprim, cholate, Triton X-100, rhodamine 6G, and hexadecyltrimethylammonium bromide (371). A MATE family pump was cloned and produced resistance when expressed in an efflux-deficient E. coli host (16); however, its inactivation in H. influenzae produced little effect on antimicrobial susceptibility (371).
When a number of clinical isolates were studied for the accumulation of azithromycin with and without the proton conductor CCCP, azithromycin-resistant strains tended to show a higher level of accumulation with CCCP, suggesting a large contribution of efflux (372). Similar results were also obtained for the efflux of telithromycin (16). There has been a rapidly increasing prevalence of ampicillin-resistant clinical isolates that do not produce β-lactamase. Although most of them have mutations in the target penicillin-binding proteins, in a good portion of strains, frameshift mutations in the negative regulator AcrR were discovered, thus suggesting the increased production of AcrAB (16, 373). Isolates showing reduced susceptibility to the formylase inhibitor LBM415 often had mutations in acrR, and such mutations were selected by this agent in vitro (374).
GAMMAPROTEOBACTERIA: PSEUDOMONAS, ACINETOBACTER, AND STENOTROPHOMONAS
The gammaproteobacteria Pseudomonas, Acinetobacter, and Stenotrophomonas are very different from those discussed above, as they do not produce the high-permeability trimeric porins, and the effective OM permeability barrier makes their RND efflux systems highly efficient.
Pseudomonas aeruginosa
In its natural habitat as well as in the hospital, the notorious opportunistic pathogen Pseudomonas aeruginosa is often exposed to fluctuating and hostile external conditions. To maintain cell homeostasis and thrive in challenging environments, it has evolved a strong and selective OM permeability barrier whose effectiveness is reinforced by broad-spectrum exporters. No fewer than 12 different RND-type drug efflux systems have been recognized and characterized in strain PAO1. Although most of these multispecific pumps accommodate antibiotic molecules and confer some degree of resistance when overexpressed from recombinant plasmids, only a minority of them appear to be therapeutically relevant. Indeed, significant constitutive or drug-induced expression levels are required for efflux pumps to contribute to intrinsic and/or acquired resistance to antimicrobial agents. Involvement of other families of transporters in resistance to antibiotics appears to be restricted to a few examples (MATE-type transporter PmpM and SMR-type transporter EmrE) (16, 164).
OM permeability.
P. aeruginosa is characterized by its very low OM permeability due to the presence of the slow porin OprF instead of the classical OmpF/OmpC trimeric porins, as mentioned above (179, 181, 182) and as discussed elsewhere (22). However, this low-OM-permeability property alone does not sufficiently explain the intrinsic MDR of this organism (11, 12, 375) and may also explain (together with the envelope-stabilizing function of OprF) the lack of or rare detection of OprF deficiency in clinical isolates (376–378). On the other hand, P. aeruginosa possesses specific channels, such as OprB, specific for glucose uptake, and OprD, specific for the diffusion of basic amino acids and peptides (22). The latter channel is the primary channel for the entry of carbapenems across the OM, and the reduced expression or loss of OprD has been frequently observed in carbapenem-resistant clinical isolates (379–384), which may also display upregulated drug efflux systems, such as MexEF-OprN, due to the shared regulation of the expression of OprD and MexEF-OprN (carbapenem susceptibility is unlikely affected by MexEF-OprN) (see Regulation of Multidrug Efflux Pumps, below) (385–387).
RND efflux pumps.
(i) MexAB-OprM.
Constitutively expressed, the MexAB-OprM pump was the first RND system characterized in P. aeruginosa (10, 13, 388), although a link between the development of MDR and increased amounts of an OM protein, OprM, had previously been established in in vitro-selected mutants (389) and was also analyzed together with other overproduced 50-kDa OM proteins (11). Reminiscent of E. coli AcrAB-TolC, MexAB-OprM displays an incredibly wide substrate specificity that encompasses structurally very different antibiotics (e.g., β-lactams, including β-lactamase inhibitors and certain carbapenems [except imipenem], aminoglycosides [with low-ionic-strength medium], [fluoro]quinolones, tetracyclines, tigecycline, macrolides, amphenicols, novobiocin, sulfonamides, trimethoprim, cerulenin, and thiolactomycin) (10–13, 16, 164, 389–394) as well as a series of amphiphilic molecules, disinfectants, dyes, solvents, detergents, and C8 to C14 3-oxo-acyl-homoserine lactones involved in quorum sensing (395–400). (A recent study also showed the quorum sensing inhibitors of nonnative N-acylated l-homoserine lactones and derivatives as the substrates of MexAB-OprM [401].) This system thus provides P. aeruginosa with protection against multiple inhibitors that can be encountered in different environments.
MexAB-OprM-overproducing mutants can be readily generated by in vitro selection in the presence of an antibiotic(s) (11, 389, 402), and in vitro studies on reference strains have shown that any mutational event inactivating the mexR (also called nalB), nalC, or nalD gene or impairing the activity of their respective products (MexR, ArmR, and NalD [see Regulation of Multidrug Efflux Pumps, below]) results in the overexpression (≥3-fold) of mexAB-oprM with a concomitant increase in resistance (2- to 16-fold MIC increases) to all the pump substrates compared to baseline levels, with nalC mutants being in general 2-fold more susceptible than the nalB and nalD mutants (393, 403–409). Similar findings were reported for phenotypically but non-genetically characterized multidrug-resistant mutants selected in vitro with various antibiotics (11, 12, 392, 402, 410, 411).
Because clinical strains often accumulate multiple resistance mechanisms, MexAB-OprM overproducers may have quite atypical drug susceptibility profiles and therefore may be underrecognized by medical microbiologists. Methods aimed at measuring the intracellular accumulation of pump substrates are not amenable to routine laboratory practice and not specific because of the large overlaps between RND pump substrate specificities. In several studies, systematic quantification of mexA and/or mexB transcripts by reverse transcription-quantitative PCR (RT-qPCR) demonstrated that these mutants are very prevalent among multiresistant non-cystic fibrosis strains (16, 412–419), including those producing reduced susceptibility to carbapenems (380, 381, 383, 420, 421) and those producing ESBLs or metallo-β-lactamases (385, 422). Rates of MexAB-OprM overproducers of near 50% were recorded even in subpopulations of isolates exhibiting a reduced susceptibility to ticarcillin (≥32 μg/ml) (423). These results agree with those of other experimental approaches based on the use of EPIs (e.g., PAβN and MC-04,124) (60, 424). Interestingly, a detailed analysis of 12 multidrug-resistant MexAB-OprM-overproducing strains revealed an equivalent distribution of nalB, nalC, and nalD mutants among them, highlighting the fact that all these mutant types may emerge in patients (413). Similar data were reported by different investigators (380, 425).
In the absence of other known resistance mechanisms (e.g., enzymatic drug inactivation and drug target alterations), MexAB-OprM dysregulation decreases the susceptibility of clinical isolates to substrate antibiotics from 2- to 8-fold compared to baseline levels (394, 415, 426). According to the susceptibility breakpoints from the Clinical and Laboratory Standards Institute (CLSI) (1029), changes in strain categorization resulting from a maximal effect of the efflux mechanism (8-fold) are limited to a small number of antipseudomonal agents, such as ticarcillin (from S [drug susceptible] to I [intermediate] or R [resistant]), aztreonam (from S to I or R), meropenem (from S to I), ciprofloxacin (from S to I), and levofloxacin (from S to I). Whether this modest impact of MexAB-OprM on drug MICs is therapeutically relevant still awaits to be clarified by clinical investigations (427). Nevertheless, it can reasonably be assumed that even low-level-resistant mutants will survive chemotherapy better than wild-type bacteria if inappropriate agents are used to treat the infection or if only suboptimal drug concentrations reach the infection site because of limited diffusion in vivo or an insufficient antibiotic dosage. As demonstrated in an animal model of infective endocarditis, management of difficult-to-treat infections requires higher doses of a β-lactam if MexAB-OprM-upregulated mutants develop (428).
Another important therapeutic issue relates to whether overexpressed MexAB-OprM can potentiate other resistance mechanisms and thus can enable P. aeruginosa to become recalcitrant to more antibiotic treatments. This does not seem to be the case, as interplays between the pump (active drug efflux) and the β-lactamase AmpC (drug inactivation) or mutations in type II topoisomerases (reduced drug target affinity) result mostly in cooperative rather than synergistic effects on drug resistance levels (383, 412, 415, 417, 429). As shown in in vitro mutants, the overproduction of MexAB-OprM causes only a slight increase in the MIC of carbenicillin when the enzyme AmpC is derepressed (430) (however, since either MexAB-OprM or derepressed AmpC production alone has resulted in high-level carbenicillin resistance, it is apparently difficult for these combined mechanisms to further increase the carbenicillin resistance level). Similarly, the coexpression of two RND pumps simultaneously (e.g., MexAB-OprM plus MexXY or MexEF-OprN) at the most tends to produce additive effects on the MICs of shared substrates, as exemplified with fluoroquinolones (25, 377, 413, 431). Synergistic interactions are expected to occur when multicomponent efflux pumps (e.g., RND systems) and singlet pumps (e.g., TetA/C) operate coordinately to extrude substrates from both the cytoplasm and the periplasmic space up to the external medium (25). Since MexAB-OprM is able to accommodate some carbapenems such as meropenem (252, 402, 430, 432), its contribution to the acquired resistance of clinical strains to these agents was investigated and found to be modest with respect to other mechanisms such as the loss of OprD and carbapenemase production (380, 381, 383, 384). Thus, the role of the pump appears to be obscured by more efficient drug-specific resistance mechanisms in terms of the resistance phenotype of the mutants. However, their role with respect to resistance emergence should not be underestimated, for example, in the development of resistance to pump substrate antimicrobials such as fluoroquinolones (433).
(ii) MexXY-OprM(OprA).
The MexXY proteins, which are encoded by a two-gene locus, need to interact with an OM component in order to form a functional tripartite pump (434–436). Despite the fact that MexXY may accommodate various OM channels (e.g., OpmB) to actively export substrates, OprM appears to be the primary partner in most strains (434, 435, 437). In the phylogenetically distinct clade PA7, the mexXY operon contains a third gene encoding an OprM-like protein, named OprA, which somewhat surprisingly shows a higher level of sequence similarity with OM channels of Burkholderia efflux pumps than with those of P. aeruginosa (438). In the PA7-related strains, the MexXY proteins seem to cooperate with both OprM and OprA, as each of the latter can compensate for the genetically engineered suppression of the other. While the MexXY-OprM/OprA system is able to transport aminoglycosides, fluoroquinolones, macrolides, tetracyclines, tigecycline, and zwitterionic cephalosporins (cefepime and ceftobiprole) (393, 434–436, 439), its contribution to natural resistance is restricted to those agents able to induce mexXY(oprA) expression (219, 434). This occurs when bacteria are exposed to subinhibitory concentrations of ribosome-targeting antibiotics, including chloramphenicol, a poor substrate, if at all, of the pump (440, 441).
Stable overexpression of the MexXY proteins provides P. aeruginosa a 2- to 16-fold-higher level of resistance to all the pump substrates, with OprM being present in amounts apparently sufficient to stoichiometrically cooperate with both the MexXY and MexAB proteins in non-PA7-related strains. The weak promoter identified ahead of oprM within the mexAB-oprM operon (442) could possibly account for an excess of OprM molecules relative to MexAB, thus allowing the interaction of OprM with other extrusion systems without impacting the activity of MexAB-OprM by titration. MexXY(OprA)-overproducing mutants can be easily selected in vitro and in vivo by substrate antibiotics (393, 439, 443, 444) or protein synthesis inhibitors (445), which is consistent with the increased prevalence, sometimes >80%, of such mutants in cystic fibrosis (436, 446–451) and non-cystic fibrosis (384, 385, 414, 416, 417, 420–423, 429, 431, 452) patients worldwide. The abundance of reactive oxygen species in the cystic fibrosis lung environment might explain the high rates of resistant mutants with this pathology (453). Supporting this notion, it was observed that prolonged exposure of P. aeruginosa to H2O2 promoted the emergence of MexXY overproducers in vitro (454). Differential resistance (MIC ratio, ≥4) to cefepime and ceftazidime, which are good and poor substrates of MexXY/OprM(OprA), respectively, has been attributed to pump derepression in some non-cystic fibrosis isolates (425, 455, 456).
Three types of MexXY-overproducing mutants have been characterized so far. In so-called agrZ mutants, the mexZ gene and its product are compromised by a number of nonspecific genetic events (e.g., indels and point mutations) (425, 439, 443, 446–448, 450, 457). In addition to mutations causing mexZ disruption, some generate single amino acid substitutions in the DNA-binding domain, the dimerization domain, or the structure of MexZ, abrogating its repressor activity (416, 452, 458, 459). A second group of mutants, dubbed agrW1, was defined in line with various defects in ribosomal proteins such as L1 (436), L25 (460), L21 and L27 (461), or components of the Fmt bypass (methionyl-tRNAfMet formyltransferase FolD) (445) that ultimately affect protein synthesis. Reminiscent of the effects of ribosome-targeting inhibitors, any mutation impairing the translation process seems to be able to induce PA5471 (ArmZ) expression and subsequently the mexXY(oprA) operon via the MexZ-ArmZ interaction. Finally, in the third group of mutants, named agrW2, mutational activation of the sensor ParS or the response regulator ParR of the two-component system ParRS leads to constitutive expression of the efflux operon (384, 444). A detailed analysis of a collection of non-cystic fibrosis isolates showing moderate, nonenzymatic resistance to aminoglycosides demonstrated the occurrence of the three types of mutants (agrZ, agrW1, and agrW2) among clinical strains (452). In cystic fibrosis patients, the agrZ type seems to predominate over the other two (446–448).
With providing a 2- to 16-fold increase in resistance, the MexXY-OprM(OprA) pump is not expected to change the classification of most clinical strains from S to I or R regarding the pump substrates, unless additional mechanisms are expressed. Therefore, the clinical impact of this low to moderate level of resistance remains uncertain, and as for MexAB-OprM, it is likely to depend upon individual patient conditions and treatment options. Very few studies have examined the potential role of MexXY/OprM(OprA) in clinical outcomes. In a rabbit experimental model of pneumonia treated with intravenous administration of tobramycin, the pump was considered to have a modest influence on animal survival and posttreatment bacterial loads (462). However, tobramycin itself appeared to have poor bacteriological efficacy in this model, contrasting with the quite high survival rates. The increased prevalence of MexXY(OprA) overproducers in the clinical setting, as reported above, can be interpreted as resulting from a positive, adverse effect of the efflux system on either the resilience of P. aeruginosa to chemotherapy (i.e., drug resistance) or its adaptation to the host (e.g., resistance to the immune system or improved fitness). Reinforcement of efflux activity due to mexXY(oprA) derepression may be just one of the multiple means by which the pathogen is able to combine to gradually increase its resistance to potent antimicrobials (460). For instance, the simultaneous overexpression of multiple efflux pumps (e.g., MexAB-OprM, MexXY, and MexEF-OprN) in conjunction with other resistance mechanisms is common in hospital strains (11, 413, 431, 464).
(iii) MexCD-OprJ.
The MexCD-OprJ system is generally not expressed in wild-type strains (465) but is inducible by membrane-damaging agents (see Regulation of Multidrug Efflux Pumps and “Bacterial Stress Responses,” below) (466, 467). Overproduction of the MexCD-OprJ pump in nfxB mutants causes increased resistance to fluoroquinolones, zwitterionic cephalosporins (cefepime and cefpirome), macrolides, chloramphenicol, and tetracyclines concomitant with hypersusceptibility to aminoglycosides and other β-lactams (15, 440, 465, 468). This overexpression results in an apparently deficient production of MexAB-OprM and/or impaired drug inducibility of the intrinsic β-lactamase AmpC, the effects of which were each proposed to account for the higher level of susceptibility of the nfxB mutants to β-lactams (469, 470), although this issue seems to be controversial (471). In dense bacterial communities such as those occurring in cystic fibrosis patients, the nfxB mutations could be less detrimental to P. aeruginosa than in planktonic cells. Some data indeed suggest that in nfxB mutants, the enzyme AmpC leaks out of the cells and concentrates in the surrounding milieu rather than in the periplasm, thus generating whole protection for cells of the biofilm (472). This may be relevant in vivo, as important extracellular AmpC activities have been measured in the sputa of cystic fibrosis patients (473). Whereas the mexXY genes (and their products) are expressed at similar levels in nfxB mutants and wild-type strains, MexCD-OprJ appears to compromise the MexXY/OprM(OprA) drug transport activity and associated resistance to aminoglycosides by downmodulating the production of the protein OprM (472). Analysis of fluoroquinolone-resistant clinical strains identified only a minority of nfxB mutants by comparison with mutants harboring quinolone target alterations (GyrA/B and ParC/E) or overexpressing other efflux pumps [e.g., MexAB-OprM, MexXY/OprM(OprA), and MexEF-OprN] (15, 431, 474). Likewise, MexCD-OprJ overproduction was infrequent in β-lactam-resistant strains (380, 421, 423), including those exhibiting increased MICs of the pump substrate cefepime relative to those of ceftazidime (425, 455). Microbiological follow-up of one mechanically ventilated patient treated with two substrates of MexCD-OprJ, namely, ciprofloxacin (14 days) and cefepime (19 days), revealed the emergence of nfxB mutants over the time, which accounted for a change of bacteria from S to I or R with regard to their susceptibility to fluoroquinolones (CLSI breakpoints) (475). Interestingly, the overexpression of MexCD-OprJ, as with that of MexAB-OprM or MexXY, occurred in 60% of carbapenem-resistant clinical isolates (476). Moreover, although MexCD-OprJ overproduction can be part of the complex resistance mechanisms (464, 476, 477), the genotypic alterations may not correlate with the phenotype (478), likely attributed at least partly to global changes in the physiology and metabolism caused by nfxB mutations (479).
(iv) MexEF-OprN.
In vitro selection of P. aeruginosa mutants cross-resistant to (fluoro)quinolones, chloramphenicol, trimethoprim, and carbapenems (imipenem) while being hypersusceptible to other β-lactams and to aminoglycosides was reported in the early 1990s (480, 481). This type of mutant, named nfxC mutants, is readily selected by fluoroquinolones and chloramphenicol but not by carbapenems (433, 481, 482). The observed resistance phenotype partly relies upon the overproduction of MexEF-OprN, which exports only (fluoro)quinolones, chloramphenicol, trimethoprim, and tetracycline (483, 484). nfxC mutants have rarely been reported in clinical settings (421, 431, 485) and even failed to be detected in several studies systematically investigating clinical resistance mechanisms (385, 414). Such a low prevalence can be attributed to β-lactam hypersusceptibility and/or the impaired virulence of these bacteria, although additional resistance mechanisms may mask the loss of β-lactam resistance in some strains (431). However, these mutants have been found in cystic fibrosis and other patients (478, 486).
(v) Other RND pumps.
There are also additional RND systems in P. aeruginosa, and they are known to often require OprM for efflux activity (16). However, their clinical relevance remains essentially unknown in spite of their involvement in resistance or virulence. MexJK functions with OprM or another OM protein, OpmH, for pumping out erythromycin and triclosan, respectively (16). Several cloned RND pumps were able to confer resistance to a P. aeruginosa or E. coli host deficient in major RND pumps: MexMN-OprM for resistance to macrolides and fluoroquinolones (487); MexPQ-OpmE for resistance to amphenicols (487); MexVW-OprM for resistance to macrolides, chloramphenicol, fluoroquinolones, and tetracycline (488); and MuxABC-OpmB (with two RND components, MuxBC) for resistance to aztreonam, macrolides, novobiocin, and tetracyclines (489). The inactivation of MuxABC-OpmB increases resistance to carbenicillin (490). TriABC-OpmH, with an unusual property of requiring two periplasmic adaptor proteins, TriA and TriB, pumps out triclosan (491), while the CzcCBA (CzrCBA) system is involved in resistance to cadmium, cobalt, and zinc salts (492, 493).
Acinetobacter spp.
Acinetobacter spp. and particularly those belonging to the A. baumannii-A. calcoaceticus complex have emerged globally as common nosocomial and community pathogens with high levels of MDR or pandrug resistance (494, 495). The modest genome size of A. baumannii of ca. 4 Mb has shown the acquired genetic diversity (including at least two dozen genomic resistance islands [A. baumannii resistance islands {AbaR}] of 22 to 121 kb) that provides the molecular basis of almost all types of resistance mechanisms and renders the organism significantly resistant to a large number of antibiotics, biocides, and heavy metals (496–502). Multidrug-resistant isolates or their epidemic clones are frequently isolated from patients after treatment with ciprofloxacin, co-trimoxazole, colistin, imipenem, and/or tigecycline (503–506), highlighting the rapid in vivo evolution of MDR in Acinetobacter.
OM permeability.
Acinetobacter has an OM of exceptionally low permeability that is similar to that of P. aeruginosa and ∼100-fold lower than that of E. coli (to cephalosporins) (183, 507). This is attributed to the lack of a high-permeability trimeric porin found in Enterobacteriaceae (183, 507). Being a close homolog of E. coli OmpA and P. aeruginosa OprF, the major OM protein of A. baumannii (508), monomeric OmpA, was experimentally shown to be the principal nonspecific slow porin (183). However, as with other slow porins such as OprF (180), the majority conformer of OmpA folds as a two-domain protein that is needed for the stabilization of the cell envelope. Thus, the porin function of OmpA is difficult to decipher by using OmpA deletion mutants, which paradoxically show reduced MICs of most antibiotics (183, 509) due to the destabilization of the envelope. It required the precise determination of the OM permeation rates of zwitterionic cephaloridine, which is unlikely to permeate across the bilayer domain of the OM, to show that ompA deletion results in a decrease of OM permeability (183). Plant extracts from Holarrhena antidysenterica permeabilize the OM of extensively drug-resistant isolates and thus restore certain activities of antibiotics (510), similar to those observed with other plant extracts, including coriander oil, geraniol, and ginger compounds (511, 512).
A. baumannii also possesses channel proteins specific to some substrates. CarO (for carbapenem resistance-associated OM protein) functions as an influx channel for carbapenems with a demonstrated imipenem binding site (513–516), similar to P. aeruginosa OprD, which is specific for the uptake of basic amino acids and carbapenems, although OprD and CarO share no recognizable homology (190, 514). CarO is also involved in the influx of l-ornithine and basic amino acids (517). Indeed, the absence of CarO expression, due to gene disruption by an ISAba10 or ISAba825 insertion, correlates well with carbapenem resistance in clinical isolates (502, 514, 518). The observation of extensive genetic diversity of carO within clinical populations suggests horizontal gene transfer as well as assortative gene recombination, likely providing a strategy for A. baumannii survival under different environmental conditions (519). The synergistic interplay between the loss of CarO and pump overproduction contributes to MDR phenotypes (502, 506). Moreover, the loss of additional OM proteins of 31 to 36 kDa is also reported to be associated with carbapenem resistance (520–522). In response to a physiological level of 200 mM NaCl, there is an upregulation of 14 distinct transporter genes, while the expression of carO and 31- to 36-kDa OM protein genes is downregulated. NaCl can induce significant tolerance to aminoglycosides, carbapenems, quinolones, and colistin (523).
RND efflux pumps.
There are a large number of studies that have investigated the role of Acinetobacter efflux pumps in resistance to clinically relevant antibiotics and also to biocides, dyes, and detergents (16, 184). Three RND systems, AdeABC, AdeFGH, and AdeIJK, have been well characterized. AdeABC is apparently not well expressed in wild-type strains (524) but contributes significantly to acquired MDR (including biocide resistance) in clinical isolates obtained worldwide (506, 525–534). These contributions include resistance to tigecycline, a major alternative drug for treating Acinetobacter infections, in isolates covering epidemic clones (502, 526, 531, 533, 535–538). In vitro exposure of susceptible isolates to tigecycline resulted in a >10-fold tigecycline MIC increase that was accompanied by AdeABC hyperproduction (536). There was a difference of tigecycline MICs of 16-fold between isogenic parental and AdeABC-hyperproducing strains (184). A nearly 30-fold overexpression of adeB was observed for tigecycline-nonsusceptible isolates (533). Thus, although tigecycline displays activity against isolates possessing tetracycline-specific resistance mechanisms (ribosomal protection and efflux) (539), broad-substrate-specificity RND pumps play a key role in the emergence of tigecycline resistance in Acinetobacter spp., similar to observations for many other Gram-negative bacteria (16, 138, 268, 310, 312, 324, 393, 540). (Additionally, non-pump-mediated tigecycline insusceptibility occurred due to a deletion mutation in the trm gene that encodes S-adenosyl-l-methionine-dependent methyltransferase [541], showing the complexity of tigecycline resistance mechanisms.) AdeABC overproduction is also seen in carbapenem-resistant A. baumannii isolates (504, 538).
AdeIJK contributes to both intrinsic and acquired MDR (502, 528, 530, 533, 542, 543). Its inactivation in a wild-type strain caused mostly 4- to 16-fold reductions of MICs of β-lactams (aztreonam, ceftazidime, cefepime, and ticarcillin), chloramphenicol, clindamycin, erythromycin, fluoroquinolones (norfloxacin and ciprofloxacin), and tetracyclines (tetracycline, minocycline, and tigecycline) (184, 542). AdeIJK also mediates resistance to biocides, including the frequently used biocides chlorhexidine and triclosan and other disinfectants in hospitals (530, 544). In vitro selection of AdeIJK-overproducing mutants by triclosan was recently demonstrated (544). A synergistic interplay between AdeIJK and AdeABC was observed with resistance to chloramphenicol, fluoroquinolones, and tetracyclines, including tigecycline (184). Recently, the efflux properties of the AdeABC and AdeIJK systems were compared with those of the E. coli AcrAB-TolC pump, and important activity and substrate differences were identified (545). Expressed in a heterologous E. coli host, the AdeABC and AdeIJK pumps were both able to pump out β-lactams, and this activity is masked in Acinetobacter due to endogenous β-lactamases. AdeABC is more effective than AcrAB-TolC in the extrusion of tetracycline but weaker in the efflux of lipophilic β-lactams, novobiocin, and ethidium bromide. AdeIJK is remarkably more active in pumping out multiple agents (except erythromycin) (545).
AdeFGH was initially identified through a microarray assessment of in vitro-selected multidrug-resistant mutants deficient in AdeABC and AdeIJK (546). This pump mediates acquired MDR and shows a broad substrate profile, including fluoroquinolones and tigecycline (184, 547). Although its clinical significance remains largely unknown, AdeFGH was the most overexpressed RND pump in isolates from Canadian hospitals (532).
AdeA-AdeA2-AdeB (containing a pair of AdeAs) is a newly identified AdeAB homolog involved in tigecycline resistance (548). Another putative RND pump, AdeT, possibly involved in aminoglycoside resistance (16), was further confirmed to be present in the genomes of two multidrug-resistant isolates (500). Many other non-A. baumannii species of Acinetobacter also often possess homologs of RND pumps (16, 528, 543, 549).
The critical role of RND pumps in MDR in Acinetobacter emphasizes the need to identify drug candidates that bypass or inhibit efflux mechanisms. Several studies have investigated the potentiation of anti-Acinetobacter activity by EPIs, which often included PAβN and NMP, which themselves have MIC values of ≥400 and 200 to ≥400 μg/ml, respectively (527, 550, 551). At a lower concentration of 25 μg/ml, their effect on reversing resistance is quite limited, while at a higher concentration of 100 μg/ml, NMP has a stronger effect on restoring drug susceptibility than that of PAβN (550); this difference is likely to be due to the poor permeation across the OM by NMP, in contrast to PAβN, which permeates this membrane by perturbing its structure (see Efflux Pump Inhibitors, below). In other studies, PAβN at 10 μg/ml decreased the MIC values of chloramphenicol, clindamycin, and trimethoprim against clinical isolates regardless of the adeFGH expression status mostly 2- to 4-fold (552), while PAβN at 20 μg/ml reduced the nalidixic acid MIC up to 16-fold but displayed little effect on ciprofloxacin susceptibility (553). PAβN and NMP, each at 100 μg/ml, restored susceptibility to fluoroquinolone (2- to 16-fold reduction of MICs) and tigecycline (mostly by a 2-fold MIC decrease) (554). Apparently, these EPIs have a stronger effect on resistance reversal with agents that have relatively high MIC values, such as chloramphenicol, clarithromycin, clindamycin, linezolid, rifampin, and trimethoprim (552, 553, 555), agents whose OM penetration is likely to be slow. Intriguingly, one study revealed that NMP at 64 μg/ml increased the tigecycline MIC 2-fold, although no effect on susceptibility to doxycycline, minocycline, and tetracycline was observed (551). While PAβN at 25 μg/ml decreased rifampin MIC values against rifampin-resistant isolates containing no mutations in the rpoB gene by 16- to 32-fold, NMP at 100 μg/ml showed little effect on rifampin susceptibility (555).
By using a multidrug-resistant isolate, one study tested the effects of a large number of EPIs (at 0.5× MIC), including not only CCCP (not an EPI per se), PAβN, and NMP but also omeprazole, phenothiazines (chlorpromazine, prochlorperazine, and promazine), reserpine, and verapamil (556), on antibiotic susceptibility. Although all agents tested showed decreased ethidium bromide accumulation in intact cells, PAβN, NMP, and phenothiazines were the only agents that restored susceptibility to certain antibiotics by an ≥8-fold decrease in the MIC values. Omeprazole and verapamil showed either no effect on antibiotic susceptibility or an antagonistic impact on tigecycline susceptibility (i.e., an increase of up to 128-fold MIC) (556). Similarly, three mammalian proton pump inhibitors, omeprazole, lansoprazole, and pantoprazole, increased tigecycline MIC values (up to >128-fold) for all six ESKAPE pathogens, including A. baumannii, in a concentration-dependent manner (557). Although the mechanism of this increase in tigecycline MICs is not clear, it should be noted that some compounds can actually enhance the efflux of other drugs through AcrB (94).
Non-RND efflux pumps.
An MFS pump, AmvA, shows a broad substrate specificity, including both antibiotics and disinfectants, and its expression is detectable in clinical isolates (558). AbeM and AbeS pumps that belong to the MATE- and SMR-type exporters, respectively, also accommodate a number of drug substrates (16, 162). AbeM hyperexpression (with moderately increased expression levels of AdeABC and AdeIJK) was observed in imipenem-resistant isolates (559). Three additional AbeM homologs, AbeM2, -3, and -4, were also identified, with no demonstrated role of AbeM4 in MDR (560). Although the clinical importance of AbeM pumps remains unknown, the abeM gene was used with other resistance genes (adeB, adeR, ampC, and ompA) to assess the genetic linkage of multidrug-resistant endemic isolates (529).
Besides these MDR pumps, the MFS pump CraA specifically provides resistance to chloramphenicol (>128-fold MIC increase) (561). Its elevated expression occurs in response to NaCl induction (523). Tetracycline-specific pumps such as Tet(A), Tet(B), and Tet39 were also found (16) and were frequently encoded both in chromosomal genomic resistance islands and by plasmids (496, 500, 562). Plasmid-borne tet(B)-tetR genes were associated with the ISCR2 mobile element in multidrug-resistant isolates (562), suggesting possible rapid horizontal resistance spread. The copresence of the Tet(A) pump and Tet(M) for ribosomal protection was also noted (16). Lastly, a new type of efflux pump, AceI, was found to mediate resistance to chlorhexidine (563). Chlorhexidine itself also induces the expression of AceI and AdeABC. AceI belongs to the bacterial transmembrane pair family and is grouped as a prototype member of the proteobacterial chlorhexidine efflux family (563). Similarly, the exposure of Acinetobacter baylyi to chlorhexidine also induced resistance to chlorhexidine and oxidants (564), which could potentially be attributed to efflux pumps.
Stenotrophomonas maltophilia
Stenotrophomonas maltophilia, found in various environments, including hospital patients and animal sources, is a key emerging opportunistic pathogen in humans that is highly versatile and adaptable. Its genome contains genes encoding numerous major mechanisms of resistance, including drug efflux pumps (565, 566). The MDR phenotypes are attributed to the interplay between low OM permeability (567) and efflux mechanisms (15). The reduction of LPS synthesis is associated with modestly increased susceptibility to several antibiotics (568). The efflux mechanism was initially suggested by the selection of multidrug-resistant isolates by any of several structurally unrelated agents (565, 569). Eight RND-type Sme systems and several other types of drug exporters have been identified (570).
The first RND pump, SmeABC, identified in this species via the construction of a cosmid-based genomic library (571), is attributable to acquired MDR (572, 573). The subsequently characterized SmeDEF pump plays a major role in both intrinsic and acquired MDR in clinical isolates from various sources (392, 572, 574–576), and its overexpression can be readily selected in vitro by conventional antibiotics and also by biocides such as triclosan (565, 577, 578). Inactivation of SmeDEF usually leads to a 2- to 8-fold reduction of the MIC values of fluoroquinolones (including ciprofloxacin, clinafloxacin, and moxifloxacin), tetracyclines (including minocycline and tigecycline), macrolides (erythromycin and azithromycin), chloramphenicol, novobiocin, dyes, and SDS against wild-type and multidrug-resistant isolates (574). In mutants carrying a genetic inactivation of the class B L1 metallo-β-lactamases and class A L2 β-lactamases, the role of RND pumps in resistance to aztreonam, piperacillin, cefepime, and cefpirome (4-fold MIC increases) was demonstrated (565, 571, 574). SmeC overexpression was also associated with L2 β-lactamase production (571). Intriguingly, SmeDEF disruption has little or only a minimal impact on susceptibility to penicillins, cephalosporins, carbapenems, and monobactams (i.e., no or merely a 2-fold MIC decrease in mutants that are also deficient in L1 and L2 β-lactamases) and does not alter rifampin and trimethoprim susceptibility (574).
There are 4 additional RND Sme pumps that all lack a genetically linked OM component (i.e., SmeGH, SmeIJK [paired SmeJK pump], SmeMN, and SmeYZ) and 2 RND pumps containing an OM protein (SmeOP-TolC and SmeVWX) (570, 579). Hypersusceptibility data with the selective inactivation of the SmeC or SmeF OM protein suggest that these OM proteins may function in multiple drug exporters (571, 574), similar to the situations observed for OprM of P. aeruginosa or TolC of E. coli. Although a TolC homolog was also identified in S. maltophilia (580), it is phylogenetically distinct from the SmeC, SmeF, and SmeX OM channels, and hence, its role in any Sme system (except SmeOP [579]) remains unknown. The tolC gene and an upstream pcm gene (encoding protein-l-isoaspartate O-methyltransferase) likely form the pcm-tolC operon, but only TolC inactivation renders the organism susceptible to aminoglycosides and macrolides (580). In fact, the Smlt3926 gene, encoding a TetR repressor, SmeRo, of SmeOP is located immediately upstream of the pcm-tolC operon, and thus, there is a 5-gene cluster comprised of tolC-pcm-smeRo-smeO-smeP. SmeOP-TolC was recently demonstrated to provide resistance to several antibiotics, CCCP, dyes, and detergents (579). The simultaneous hyperexpression of SmeJK (forming one exporter) and SmeZ pumps increases the substrate profiles of a clinical isolate (581). Inactivation of SmeJK significantly increases the susceptibility to aminoglycosides (amikacin, gentamicin, kanamycin, and tobramycin) and the macrolide leucomycin (8- to 16-fold reduction in MIC values), yet the disruption of either SmeJ or SmeK produces merely a 2-fold decrease in the MICs of the tested aminoglycoside agents (582), likely suggesting that SmeJ or SmeK alone may still be functional.
S. maltophilia also possesses ABC and MFS transporters (570). The ABC-type tripartite FuaABC system mediates fusaric acid-inducible resistance to fusaric acid, and this resistance is dependent on the FhuR regulator, which functions as a repressor in the absence of fusaric acid but as an activator in its presence (583). ABC-type MacABC causes intrinsic resistance to aminoglycosides, macrolides, and polymyxins, as its inactivation resulted in 2- to 8-fold reduction of the MIC values of these agents (570, 584). Another SmrA ABC pump conferred resistance to fluoroquinolones, tetracycline, doxorubicin, and dyes when expressed in E. coli (585), and an antibody developed against this pump enhanced antibiotic susceptibility in S. maltophilia (586). An MFS pump, EmrCAB, is involved in the extrusion of hydrophobic toxic agents but is not well expressed intrinsically (587).
ALPHAPROTEOBACTERIA: BRUCELLA, BARTONELLA, AND RICKETTSIA
The alphaproteobacteria contain several species that are major human and animal pathogens, such as Brucella, Bartonella, and Rickettsia.
Brucella spp.
Brucella spp. are facultative intracellular coccobacilli with six recognized species (Brucella abortus, B. canis, B. melitensis, B. neotomae, B. ovis, and B. suis) that display distinct host-pathogen associations. These species are of high clinical significance given their role as causative agents of zoonotic brucellosis, which is reemerging, can be transmitted to humans, and is commonly associated with laboratory-acquired infections (particularly with B. melitensis) (588). Brucella spp. show similar genomes with two circular chromosomes and contain a number of putative drug efflux transporters (589, 590). These species contain trimeric porins homologous to E. coli porins with comparable permeability (591, 592).
Two RND systems, BepDE and BepFG, are involved in resistance to antibiotics and other toxic agents (including doxycycline, a major choice for the treatment of brucellosis) in collaboration with the OM channel BepC (593, 594). Repressed by the BepR regulator, the expression of BepDE was induced by deoxycholate. Inactivation of BepFG produced an increased susceptibility to toxic agents such as dyes but not to conventional antibiotics (594). The inclusion of PAβN resulted in variable reductions in erythromycin and moxifloxacin MIC values (595, 596), consistent with the RND pump's contribution to resistance. A putative BicA macrolide pump is present in B. abortus and B. suis (590). Two MATE pumps, NorMI and NorMII, were identified in B. melitensis, with NorMI being confirmed to yield MDR when expressed in E. coli (16). However, they were not involved in clinical isolates resistant to fluoroquinolones and rifampin (597).
Bartonella spp.
Bartonella spp. exist in mammalian host reservoirs and are linked to several human infections, such as cat scratch disease. While Bartonella species are susceptible to most conventional antibiotic classes, the major resistance mechanism characterized to date is related to target modifications for resistance to fluoroquinolones, macrolides, and rifamycins (598). A VceA MDR pump was identified in Bartonella australis (GenBank accession number YP_007461659). OM porins and efflux components, including a TolC homolog as well as RND systems, were identified (599–601); nevertheless, none of the efflux pumps have been characterized for their role in resistance.
Rickettsia spp.
Rickettsia spp. are obligately intracellular bacteria, including those causing spotted fevers. Rickettsia spp. are often resistant to β-lactams, aminoglycoside, and sulfonamide-trimethoprim but show variable susceptibilities to macrolides (602). Genome comparison of Rickettsia species shows the presence of a number of drug efflux pump genes, such as 6 genes encoding RND pumps and 20 genes encoding ABC transporters in Rickettsia conorii as well as an SMR pump in Rickettsia bellii (602, 603).
BETAPROTEOBACTERIA: ACHROMOBACTER, BURKHOLDERIA, AND NEISSERIA
Often found in natural environmental samples, the betaproteobacteria Achromobacter, Burkholderia, and Neisseria consist of many species (aerobic or facultative) that interact intimately with plants or animals. Nonpathogenic betaproteobacteria were also found to contribute to the evolution of class 1 integrons and resistance in the pathogens of other species (604).
Achromobacter spp.
The nonfermentative Achromobacter bacilli include an emerging opportunistic pathogen in cystic fibrosis patients, Achromobacter xylosoxidans, whose genome contains a range of resistance genes encoding drug-modifying enzymes and efflux pumps (605). The latter include 4 RND systems, additional tripartite efflux systems, and an ABC-type macrolide-specific MacAB transporter (605). Two RND pumps, AxyABM and AxyXY-OprZ, were identified to mediate MDR (606). Inactivation of AxyB produced up to a 20-fold reduction of the MIC values of several third-generation cephalosporins, with no changes in susceptibility to the aminoglycosides amikacin and tobramycin (606). However, the disruption of AxyY rendered a wild-type strain highly susceptible to aminoglycosides (16- to 128-fold MIC decreases for amikacin, gentamicin, netilmicin, and tobramycin), doripenem, erythromycin, and tetracycline (all with 4-fold MIC reductions). In an aminoglycoside-resistant mutant, AxyY inactivation restored its aminoglycoside susceptibility (20- to 192-fold MIC changes), thus revealing that AxyXY pumps out antibiotics often used for treatment of pulmonary infection of cystic fibrosis patients (607).
Burkholderia spp.
The genus Burkholderia contains >40 species that are present in a variety of ecological niches, including soil, water, plants, and animals. MDR is an emerging feature of many isolates such as those of Burkholderia cepacia. A comparison of the OM proteomes of Burkholderia mallei and Burkholderia pseudomallei suggests many similarities, which include a number of porins and efflux pumps (608). The major trimeric porin (Omp38) is a distant relative of E. coli OmpF, but its permeability is likely lower, by 1 or 2 orders of magnitude, than that of OmpF (609). However, another study showed that the single-channel conductance of Omp38 was similar to that of OmpF (610). The genomes of Burkholderia contain multiple replicons such as those in the B. cenocepacia genome that has three chromosomes in each strain with RND pumps encoded in all chromosomes (611). A recent study assessed all 8 RND families with 471 putative RND sequences in 26 completely sequenced Burkholderia genomes (including the virulent, epidemic cystic fibrosis strain J2315) (612). The drug-related HAE-1 RND pumps in the Burkholderia genus vary very much in number, ranging from only 4 in 3 strains of B. mallei to 15 in Burkholderia sp. strain CCGE1002 and 11 to 16 in B. cenocepacia (611–613).
The first RND pump characterized in Burkholderia, CeoAB-OpcM of B. cenocepacia (i.e., RND-10 of the 14 RND systems) (611), mediates resistance to multiple antibiotics, including tigecycline, and is inducible by salicylate, iron starvation, and chloramphenicol (16). While ceoB expression appears weak, the expression of 4 RND pump genes (rnd-3, -9, -11, and -13) is readily detectable. Expression of the rnd-2 gene is also inducible by chloramphenicol (611). RNDs 6 and 7 are twin pumps encoded by the same operon, similar to MdtBC of E. coli (127, 611). Inactivation of the rnd-4 gene led to a 4- to 8-fold increase in susceptibility to aztreonam, ciprofloxacin, levofloxacin, gentamicin, tobramycin, and chloramphenicol (614, 615). With 16 RND pumps present in B. cenocepacia strain J2315, their differential roles in drug resistance of planktonic and biofilm cells were investigated (613). Another study showed the high prevalence of RND pump-overproducing clinical isolates (particularly RND-3 overproducers) (616). However, the data regarding the effect of PAβN on antibiotic susceptibility appeared contradictory. While PAβN at 40 μg/ml did not alter the antibiotic susceptibility of several strains, as also noted with erythromycin in B. pseudomallei (614, 617), a newer study showed that PAβN at 64 μg/ml increased the susceptibility to tigecycline (mostly 32- to 64-fold) in the B. cepacia complex (618). The MICs of PAβN against several strains are 30 to 640 μg/ml (614), suggesting various susceptibilities of the individual strains to PAβN. The RND system encoded by the operon containing the BCAM0925 to BCAM0927 genes is upregulated by chlorpromazine and mediates resistance to azithromycin and chlorhexidine (619).
In B. pseudomallei, containing a dozen RND systems, the widespread expression of 7 RND pumps in clinical strains was evident (612, 620). Three pumps, AmrAB-OprA, BpeAB-OprB, and BpeEF-OprC, were characterized (617, 621–623). While both AmrAB-OprA and BpeAB-OprB mediate intrinsic resistance to aminoglycosides and macrolides (617, 621), expression of these pumps is commonly seen in clinical isolates (620). BpeEF-OprC is responsible for the widespread trimethoprim resistance in both clinical and environmental isolates (624). Interestingly, aminoglycoside- and macrolide-hypersusceptible strains (with gentamicin and azithromycin MICs of ca. 0.5 to 2 μg/ml) were isolated from both clinical and environmental samples and contained a point mutation in amrB that inactivated AmrAB-OprA. The reversion of this mutation via in vitro exposure of the susceptible strains to gentamicin and kanamycin (40 and 30 μg/ml, respectively) was able to restore aminoglycoside and macrolide resistance (625), highlighting the clinical concerns on current gentamicin use (e.g., 4 μg/ml) in medium to isolate B. pseudomallei (626). In B. thailandensis, multidrug-resistant mutants selected by doxycycline overproduced AmrAB-OprA and BpeEF-OprC. When either of these pumps was inactivated, a third pump, BpeAB-OprB, was hyperexpressed, indicating the interplay among the three RND pumps (627), similar to those observed in P. aeruginosa (628) and Salmonella (629). Interestingly, an antagonistic effect between PAβN (at 50 and 200 μg/ml) and aminoglycosides/β-lactams was observed, and the speculation was that PAβN might have induced the overproduction of the AmrAB-OprA and BpeAB-OprB pumps (627). It remains to be determined whether an induction process exists for explaining the above-mentioned observations that PAβN at 40 μg/ml did not alter antibiotic susceptibility in B. pseudomallei (614, 617) and that PAβN at 20 μg/ml did not affect the efflux activity of SmeDEF toward ciprofloxacin and tetracycline in S. maltophilia (630).
Neisseria spp.
Neisseria spp. are commensal bacteria in many animals, with two significant human pathogens, Neisseria gonorrhoeae and Neisseria meningitidis, which have shown resistance emergence (for a recent review, see reference 631). These species possess trimeric porins that display high permeability with anion selectivity (632), and they may render the species more susceptible to anionic penicillins. Low-level MDR is mediated by mutations in genes encoding porins or efflux pumps (15, 633). In fact, simultaneous porin mutation and RND pump overproduction are needed for gonococcal penicillin or ceftriaxone resistance, again supporting the significance of the synergistic interplay between the OM and efflux pumps (634–636). Mutations in the “multiple transferable resistance” gene (mtrR) had long been associated with MDR in N. gonorrhoeae, with an assumed alteration in OM permeability (15), but the gene was then found to control the expression of MtrCDE, the most characterized RND pump in Neisseria that contributes to resistance to antibiotics (including β-lactams, macrolides, and rifampin), detergents, bile salts, gonadal steroidal hormones, as well as host cationic peptides (15, 633, 635–639). Either the deletion or overexpression of MtrCDE produces a 4-fold rifampin MIC change (639). MtrCDE deficiency renders gonococci more susceptible to progesterone (640).
Overexpression of MtrCDE due to mtrR or other regulatory mutations (see Regulation of Multidrug Efflux Pumps, below) has widely been observed in multidrug-resistant clinical isolates (16, 641). This includes interpatient transmission of high-level ceftriaxone-resistant/multidrug-resistant N. gonorrhoeae with MtrCDE overproduction (636). A new study also showed the global gene transcriptional changes as a consequence of MtrCDE overproduction, and these changes included an upregulation of the gene encoding cytochrome c peroxidase that is associated with tolerance to peroxides (642). The newly available crystal structures of MtrD and MtrE (643, 644) also suggest that the transport mechanisms of AcrAB-TolC discussed above (see Biochemistry and Genetics of Multidrug Efflux Pumps) are applicable to the MtrCDE pump. Another RND pump, FarAB-MtrE, of both N. gonorrhoeae and N. meningitidis mediates resistance to antibacterial fatty acids and cationic peptides that are likely present on mucosal surfaces of the host (16, 645). Since many substrates of MtrCDE and FarAB-MtrE are present in the host, these efflux pumps are also involved in bacterial pathogenesis such that in vivo fitness or survival is enhanced by these pumps (640, 641). The fatty acids and bile salts in the gut may likely facilitate Mtr-mediated resistance, as observed for isolates from rectal infections often possessing the MDR phenotype (646). Also, MATE and ABC pumps were also identified with additional tetracycline-specific exporters (16). The clinical importance of three MDR pumps (i.e., MtrCDE, MacAB, and NorM) was recently demonstrated by their impact on extensive or multiple drug resistance of clinical isolates in comparison with clinical susceptibility breakpoints (647). For example, in an extensively resistant isolate, the inactivation of MtrCDE changed the susceptibility status for azithromycin, penicillin, and tetracycline from R to S or I. MacAB inactivation alone also rendered the resistant isolate S, providing an important clinical example of the role of ABC transporters in bacterial drug resistance. The NorM deficiency was also able to make an R-to-I status change for tetracycline (8-fold MIC decrease) and to yield a >32-fold MIC reduction for solithromycin, a fluoroketolide for which the susceptibility breakpoints are as yet unavailable (647). Lastly, the gene encoding an MFS transporter, the Mef macrolide pump, on conjugative plasmids is present mainly in Gram-positive bacteria (15) but was also found in N. gonorrhoeae and Acinetobacter junii. These plasmids were readily transferred in vitro to several Gram-positive and Gram-negative bacteria, including Neisseria (648).
EPSILONPROTEOBACTERIA: CAMPYLOBACTER AND HELICOBACTER
Many of the epsilonproteobacterial species are host associated, such as Campylobacter and Helicobacter, and inhabit a wide variety of ecological niches ranging from gastrointestinal tracts of animals to water/marine reservoirs (649).
Campylobacter spp.
The Campylobacter group contains the two very important human enteropathogens Campylobacter jejuni and Campylobacter coli, which are among the top pathogens responsible for foodborne illnesses worldwide. There is an increasing emergence of resistance to clinically relevant antibiotics such as fluoroquinolones and macrolides, where drug efflux makes a major contribution (16).
Campylobacters possess a major porin, major outer membrane protein (MOMP), which appears to exist in a monomeric form as well as in a weakly associated trimeric form (650, 651) and likely yields OM permeability channels smaller than those of E. coli porins (652, 653). The marked susceptibility of wild-type campylobacter strains to large, hydrophobic antibiotics such as macrolides may suggest the presence of an unusually permeable bilayer domain in the OM.
The genome of C. jejuni indicates the presence of a large number of drug exporters (16). The CmeABC system is the most characterized RND pump in campylobacters and is responsible for both intrinsic and acquired MDR, including resistance to bile salts (654, 655). It is distributed in diverse isolates of animals and humans that are resistant to macrolides, fluoroquinolones, and tetracyclines (16, 656–661). Inactivation of CmeABC reduces significantly the in vitro emergence of fluoroquinolone-resistant mutants, while its overproduction facilitates such selection (659). Although the contribution of another RND pump, CmeDEF, to intrinsic resistance is likely masked by CmeABC, the two pumps interplay synergistically in providing resistance (662). CmeABC also interplays with 23S rRNA target alterations or ribosomal protein modifications in increasing macrolide resistance levels (658). The contribution of CmeABC to MDR was further observed with the synergistic interplay between anti-CmeA and anti-CmeB peptide nucleic acids to sensitize C. jejuni to multiple antimicrobials, thus also suggesting a potential novel approach to combat efflux-mediated MDR by using the pump-component-specific antisense peptide nucleic acids (663). The combinational use of anti-CmeA and anti-CmeB peptide nucleic acids, each at 1 to 4 μM, decreased the MIC values of ciprofloxacin and erythromycin against a wild-type strain 4- to >32-fold, while their separate use alone yielded merely a 2-fold MIC reduction. Although these peptide nucleic acids were effective in reducing the MIC of ciprofloxacin against a gyrA mutant 16-fold, they were able to produce only a 4-fold reduction of the erythromycin MIC for a high-level erythromycin-resistant mutant with a 23S rRNA mutation. An optimization study of these antisense peptide nucleic acids showed the importance of the ribosome-binding sites as the target (664). However, the entry of antibacterial peptides, including peptide nucleic acids, may require importers such as the SbmA/BacA IM proteins that were identified in several Gram-negative bacteria but remain unknown in campylobacters (665).
Tolerance of campylobacters to trisodium phosphate, a highly alkaline agent used to reduce pathogen prevalence on meat, is mediated by the NhaA1/NhaA2 cation/proton transporter and can be hindered by PAβN. This phenomenon was proposed to be attributable to the contribution of RND pumps (666). However, the effect of PAβN on OM permeability could also provide an alternative explanation, since the PAβN concentration used, 64 μg/ml, was quite high.
Campylobacters are widely distributed in food-producing animals, particularly in poultry. Thus, resistance in campylobacters of poultry origin has drawn much attention. Macrolides and fluoroquinolones are the drugs of choice to treat human campylobacteriosis. The exposure of chickens to either a fluoroquinolone (enrofloxacin) in drinking water or a macrolide (tylosin) in animal feed readily resulted in the isolation of CmeABC-overproducing mutants with an MDR phenotype (656, 667). In vitro stepwise exposures of C. jejuni to escalating levels of erythromycin or tylosin have generated a variety of macrolide-resistant mutants (668). The mutations in ribosome proteins L4 and L22 occurred early during this selection (low resistance level, with an erythromycin MIC of 8 to 16 μg/ml), followed by CmeABC overproduction (intermediate level, with an erythromycin MIC of 32 to 256 μg/ml), and finally accompanied by mutations in the 23S rRNA genes (high level, with an erythromycin MIC of ≥256 μg/ml), highlighting that efflux mechanisms likely facilitate the emergence of high-level macrolide resistance, similar to that of high-level fluoroquinolone resistance (659). CmeABC overexpression was partially a result of single-residue changes in the C terminus of the repressor CmeR (668). It should be noted that the multiple-stepwise-selection approach could yield pleiotropic mutations, thus underestimating the importance of the CmeABC-overproducing mutants that also possessed ∼200 up- or downregulated genes (including the upregulation of genes encoding an SMR pump [Cj1173] and two MFS pumps [CmeG and Cj0035c]), and some of these changes may affect physiology and metabolism (668). The latter offers an explanation regarding the growth burdens and fitness cost of macrolide-resistant campylobacters reported by the groups of Zhang and Yuan (669–671).
Additional pumps that are independent of CmeABC and CmeEFG also likely mediate MDR (16) but remain to be further studied. Similar to the above-mentioned gene expression changes in erythromycin-resistant mutants (668), the transcriptional response of C. jejuni to an inhibitory concentration of erythromycin (4 μg/ml; 16× MIC) involved more than a hundred up- or downregulated genes, including two upregulated putative drug efflux operons (Cj0309c-Cj0310c and Cj1173-Cj1174), each encoding a paired SMR transporter. Inactivation of these operons impaired cell growth under conditions of high oxygen levels and colonization in chickens but did not alter the drug susceptibility toward 14 agents tested (including erythromycin) (672). The only MFS drug pump characterized in this species to date is CmeG, and its inactivation renders C. jejuni more susceptible to erythromycin, ciprofloxacin, and H2O2 (673). Finally, a novel ABC transporter, ArsP, was recently identified to mediate resistance to nitarsone and roxarsone, the organic arsenic agents used in poultry production (674).
Helicobacter spp.
Helicobacter species, including Helicobacter pylori, are associated with several important human and animal illnesses. As a dominant species of the human gastric microbiota, H. pylori causes a persistent inflammatory response and is linked to the development of ulcers and gastric cancers (675). While H. pylori infections need to be treated with antibiotics (676), acquired resistance to fluoroquinolones, macrolides, and metronidazole, agents used for the treatment of H. pylori infections, has emerged and is associated with antibiotic consumption (677, 678).
H. pylori displays intrinsic resistance to multiple antimicrobials, including glycopeptides, nalidixic acid, polymyxins, sulfonamides, and trimethoprim (676), suggesting that limited access to drug targets may constitute part of the mechanisms of resistance. Despite its relatively small genome size (1.7 Mb), H. pylori has genes encoding a large number of OM proteins (679), with several porins, including one that forms a large nonspecific channel (680). Nevertheless, the exquisite susceptibility of H. pylori to macrolides (MIC90 of <0.1 μg/ml [676]) suggests, as with campylobacter, that many drugs may permeate the bilayer regions of the OM. Of several RND systems (681), HefABC contributes to MDR (682, 683), and HefDEF (CznBAC) is a metal exporter for cadmium, nickel, and zinc resistance and gastric colonization (684). Four TolC homologs (HefA, HefD, HefG, and HP1489) were identified, and only HefA inactivation rendered the mutants more susceptible to deoxycholate and novobiocin. The simultaneous disruption of HefA and HefD increased metronidazole susceptibility (685), and laboratory-selected resistant mutants overexpressed HefA (686). The expression of HefA was increased following exposure to metronidazole in clinical isolates (687). In an interesting study, H. pylori cultured in the presence of cholesterol became resistant to bile salts and their analogs, and this phenotype required the HefC pump (688). Intriguingly, hefC missense mutations were among the several gene mutations in in vitro-selected isolates with high-level resistance to amoxicillin, and the introduction of a mutated allele to the wild type increased resistance (689). Through examining the whole-genome sequences of clarithromycin-resistant isolates, a new study revealed the gene clusters of TolC homologs involved in clarithromycin susceptibility profiles in individual isolates (690). Given the extremely acidic environments where H. pylori colonizes, the effect of this environment on secondary transporters in in vivo antimicrobial resistance remains to be determined. In this regard, there are a number of putative ABC transporters in H. pylori (679). Inactivation of the ABC-type transporter MsbA rendered the strain more susceptible to erythromycin and glutaraldehyde. MsbA also interplays synergistically with the glutaraldehyde-resistant protein Ost/Imp to enhance hydrophobic drug resistance and LPS biogenesis (691). Various types of putative drug transporters (e.g., 2 RND pumps, 1 MFS pump, 2 MATE pumps, 4 SMR pumps, and 1 ABC pump) were also identified in Helicobacter cinaedi, a pathogen increasingly known for infections in immunocompromised patients (692).
BACTEROIDACEAE AND PREVOTELLACEAE
The anaerobic families Bacteroidaceae and Prevotellaceae constitute a dominant part of the mammalian gut microbiota and play an important role in maintaining human health. However, they may cause anaerobic infections (e.g., blood and intra-abdominal infections such as those associated with Bacteroides fragilis) and exhibit intrinsic resistance to a number of antimicrobials (for a review, see reference 693). Increasing MDR in Bacteroides spp. has been observed in recent years. The OM of B. fragilis contains porins that produce a much lower rate of diffusion of hydrophilic saccharides than those of E. coli porins (16), and this would make an efflux process efficient. However, the OM was shown to allow rapid passage of hydrophobic agents such as rifamycin and clindamycin, likely through its bilayer domain, which was shown to be more lipophilic than that of the enteric bacteria (694). Considering the environments that Bacteroides inhabits, it is not surprising to see large genome sizes (e.g., 5.2 Mb with B. fragilis and 6.3 Mb with Bacteroides thetaiotaomicron) (695, 696), which also include a large number of drug efflux genes, e.g., those encoding 16 putative RND pumps in B. fragilis (697) and 60 predicated drug efflux components in B. thetaiotaomicron (696).
Many RND pump genes of B. fragilis were expressed under laboratory conditions, and some were demonstrated to play a role in intrinsic resistance (463, 697). Inactivation of BmeABC5 yielded a 4-fold reduction in the metronidazole MIC and also increased susceptibility to cefoperazone and SDS (698). Intrinsic resistance to fluoroquinolones is mediated by an MFS NorA pump in B. fragilis and a MATE BexA exporter in B. thetaiotaomicron (15). A macrolide resistance gene, msr(SA) (coding for an ABC exporter), previously found in Gram-positive bacteria, was also detected in B. fragilis (699). Thus, drug efflux is likely a major contributor to resistance in Bacteroides. Indeed, one study confirmed a wide presence of resistance efflux genes in a number of clinical isolates (700). Furthermore, a recent study of a multidrug-resistant B. fragilis isolate led to the identification of a unique conjugative transposon containing a hybrid mosaic of elements, including genes for 3 efflux systems (MefA and ABC exporters of Gram-positive bacteria and RND pump conserved in Bacteroides) (701), again showing the emergence of complex resistance gene assembly.
Prevotella spp. also constitute part of the oral and vaginal microflora and can also cause anaerobic infections. A recent study from the United Kingdom (702) suggested the presence of multiple RND pumps in 5 Prevotella spp. and their contribution to tetracycline resistance in clinical isolates from patients with cystic fibrosis and invasive infections. However, a relatively high PAβN concentration (80 μg/ml) was used.
DRUG EFFLUX GENES ON PLASMIDS
Plasmids have played a critical role in both the emergence and spread of resistance to most classes of antibiotics in bacteria (703–705). Their mobile nature (often linked to transposons/integrons) within or across bacterial species makes it extremely difficult to contain resistance. Moreover, plasmid-mediated mechanisms often confer high-level resistance. Genes encoding various classes of drug pumps have also been identified on numerous plasmids. The typical examples are the well-known single-drug-class efflux genes, such as various tet efflux genes usually found on plasmids and encoding MFS pumps (8, 15, 133). The floR gene, found first in an R plasmid in the florfenicol-resistant fish pathogen Pasteurella piscicida and then in Salmonella and E. coli, also codes for a singlet MFS pump responsible for amphenicol resistance (15). Another efflux gene, mef(B), coexists with the aadA and sul3 resistance genes in plasmids of porcine E. coli isolates and mediates macrolide resistance (706). The plasmid-borne qepA and qepA2 genes encode MFS pumps providing resistance specific to fluoroquinolones (206, 707). These drug-specific exporter genes are limited to encoding the singlet MFS pumps.
In contrast to many MFS pumps, RND family efflux genes generally require the contribution of three gene products in order to produce effective efflux. Most Gram-negative bacteria contain at least one set in their chromosome and in many cases contain several sets of genes for such systems. Hence, the RND-type transporter genes appear to be infrequently present on plasmids. However, the fact that several RND pump-encoding plasmids have been reported to date (Table 4) forces us to consider that such plasmids may exist more widely than previously suspected. Interestingly, most of these plasmids were already isolated decades ago and derived from environments with possibly strong antimicrobial selection pressures (Table 4). Of these plasmids, the oqxAB genes that code for such a pump and a periplasmic adaptor protein were found in a plasmid in an E. coli strain resistant to olaquindox (708), a quinoxaline-di-N-oxide agent once widely used in pigs in Europe. The OqxB transporter shows only 40% identity with AcrB but is more closely related to MexF of P. aeruginosa. Dependent on the host TolC protein, OqxAB has a wide substrate specificity, making E. coli also more resistant to fluoroquinolones, nalidixic acid, chloramphenicol, trimethoprim, benzalkonium chloride, and SDS (709). The oqxAB genes have since increasingly been found on R plasmids from E. coli (710–714) and several other species of Enterobacteriaceae, such as Salmonella and K. pneumoniae (235, 236, 303, 715). The coexistence of the oqxAB and floR or CTX-M gene on the same plasmids was also observed (713, 715). Chromosomal oqxAB was found in clinical isolates of K. pneumoniae and considered a possible source of plasmid-borne oqxAB (302).
TABLE 4.
Conjugative plasmids encoding RND or ABC multicomponent efflux systems of Gram-negative bacteria
| Plasmid(s) (GenBank accession no.) | Isolation yr | Source, country | Host | Inc group | Size (kb) | Efflux component(s) | Pump regulator | Additional resistance gene(s) and transposon(s) | Resistance phenotype of initial host strain | References |
|---|---|---|---|---|---|---|---|---|---|---|
| pMG101 (AF067954) | 1973 | Burn ward, USA | Salmonella enterica serovar Typhimurium | IncHI | 180 | SilABC RND and SilP ABC transporters | SilRS | Unknown | Ampicillin, chloramphenicol, streptomycin, tetracycline, silver | 716, 717 |
| pMCBF1 (AY950444), pMCBF6 (EF107516) | 1993 | Marine, Sweden | Uncultured bacterial biofilm (Pseudomonas putida as recipient) | IncP-1 | 63, 67 | MexEF-OprN RND homolog and MerF/MerT transporters | MerR | merA, merB, merD, merP; Tn5053 or Tn5058 | Mercury | 721, 722 |
| pOLA52 (EU370913) | 1999 | Swine manure, Denmark | Escherichia coli | IncX1 | 52 | OqxAB RND pump | ORF68 (GntR family) | blaTEM; Tn6011 | Ampicillin, carbadox, chloramphenicol, kanamycin, nitrofurantoin, olaquindox, sulfamethoxazole, trimethoprim | 708, 892, 1018, 1019 |
| pB4 (AJ431260) | 1999 | Activated sludge, Germany | Uncultured bacterium (Pseudomonas knackmussii as recipient) | IncP-1β | 79 | MexCD-OprJ RND homolog | NfxB | blaNPS-1, chrABC, strA, strB; Tn5719, Tn5720, Tn5393c | β-Lactams, spectinomycin, streptomycin, tetracycline, chromate | 1020, 1021 |
| pRSB101 (AJ698325) | 2001 | Activated sludge, Germany | Uncultured bacterium (Pseudomonas knackmussii as recipient) | IncQ | 48 | AcrA-ABC permease/ATPase, OprM and TetA MSF pump | AcrR and TetR | aadA2, blaTLA-2, chrA, dhfr1, mph(A), mphR(A); qacEΔ, sulI, Tn402, and intI | Cephalosporins, erythromycin, nalidixic acid, norfloxacin, roxithromycin, spectinomycin, streptomycin, sulfonamides, tetracycline, trimethoprim | 719, 1021 |
| pNDM-CIT (JX182975) | 2010 | Patient, India/France | Citrobacter freundii | IncHI1 | 289 | MexAB-CusC RND homolog | AcrR, HN-S-like protein | blaNDM-1, armA, mel, mph; Tn1548 | β-Lactams, aminoglycoside, fluoroquinolones, macrolides, nitrofurantoin, tetracycline, tigecycline | 243, 724 |
A silver resistance megaplasmid, pMG101, the earliest isolated plasmid containing genes for an RND transporter, was obtained from multidrug-resistant Salmonella in a hospital burn ward where silver sulfadiazine was widely used in the 1970s (Table 4) (716, 717). (Chromosomally encoded efflux and porin loss have also been reported for E. coli [718].) Another tripartite exporter (AcrA-ABC pump-OprM) was encoded by a large plasmid (pRSB101) isolated from a sewage treatment plant (719). In a study of mixed plasmid DNAs from a similar wastewater treatment plant (720), homologs of E. coli acrB and acrD as well as those of P. aeruginosa mexB, mexD, and mexY were found. If these genes are truly derived from plasmids, this result may show us that putting RND pumps on plasmids is not so difficult and may serve as a cautionary note for the future development of MDR in Gram-negative bacteria. Indeed, this worrisome speculation is supported by a recent study that showed the inclusion of the complete P. aeruginosa mexEF-oprN-like genes (ca. 23%, 35%, and 17% identities with MexE, MexF, and OprN, respectively, at the protein level [X.-Z. Li, unpublished data]) into two conjugative broad-host-range plasmids from a marine microbial biofilm (721). Initially isolated as mercury resistance plasmids from marine samples collected in 1993 (722) and maintained in Pseudomonas putida, these two plasmids from an unknown host are largely identical and evolved through homologous recombination, and each plasmid contains transposons and various gene clusters (trb for mating pair function, tra for conjugative gene transfer, mer for mercury resistance, and mexEF-oprN-like genes for the RND pump) (Table 4) (721, 723). When tested for susceptibility to chloramphenicol, nalidixic acid, and trimethoprim, an E. coli isolate carrying one of these two plasmids did not show altered drug susceptibility. However, a Pseudomonas host with a deficiency in endogenous RND pumps and a greater variety of agents could have been used for defining the function of the plasmid-borne RND pump. Lastly, a plasmid from an extremely drug-resistant C. freundii isolate from a patient returning from India (724) contains genes encoding the carbapenemase NDM-1, the ArmA 16S RNA transferase, and an RND system (homologous to AcrR-MexAB-CusC) (243). The RND gene region belongs to part of the CP4-like prophage sequence, and whether this RND pump is functional in providing MDR remains unknown. Nevertheless, these RND pump-encoding plasmids (Table 4) have no doubt provided evidence for sophisticated resistance evolution with possible adverse clinical implications.
REGULATION OF MUTLIDRUG EFFLUX PUMPS
The regulation of drug efflux pump expression involves a variety of complex pathways that typically require the participation of numerous local and global transcriptional regulators or modulators as well as the two-component regulatory systems. Mutational changes of these regulators can lead to altered expression levels of the pumps. Certain pumps are also inducible by various compounds, including antibiotics. The versatile nature of pump regulation is consistent with the widespread presence of numerous differentially expressed drug pumps and their role in resistance and other physiological functions in order to adapt to diverse environments. Below are examples that demonstrate the multilevel regulation of drug exporters. An early review on the regulation of drug transporters was available (725), but since then, major advances have been made.
E. coli Efflux Pumps
AcrAB-TolC.
AcrAB is constitutively expressed at a significant level in E. coli, and a similar situation seems to prevail for AcrAB homologs in other organisms. Although the acrAB and tolC genes are not genetically clustered, their expressions are often regulated by common regulators at multiple levels (Fig. 4). First, the local repressor AcrR, encoded by the acrR gene that is divergently transcribed from acrAB, represses directly both acrAB expression and its own expression (726). A member of the TetR repressor family (727), AcrR functions as a dimeric two-domain molecule, and each monomer contains a smaller N-terminal domain and a larger C-terminal domain. The latter constitutes the ligand-binding domain with a multientrance pocket to accommodate a number of ligands or inducers (e.g., rhodamine 6G), followed by the conformational change of AcrR that cooperatively affects the N-terminal DNA-binding domain (728–730). However, AcrR does not tightly inhibit AcrAB expression, and this allows the constitutive production of AcrAB, conferring intrinsic MDR. Second, other regulators, the AcrS repressor of the AcrEF pump (731), the histone-like nucleoid structuring protein (H-NS) (732), and the quorum-sensing receptor SdiA (733, 734), also participate in the regulation of acrAB. SdiA likely acts as a minor activator since its genetic deletion results in a small (2- to 3-fold) decrease in the MICs of fluoroquinolones (733). Third, as described below, three global regulators, MarA, SoxS, and Rob, play major roles by positively controlling the expression of acrAB, tolC, and micF. The micF transcript inhibits the translation of OmpF porin mRNA.
FIG 4.
Regulation of expression of the AcrAB-TolC efflux system of E. coli. Transcription of the acrAB and tolC genes is not genetically clustered but is often regulated by common regulators at multiple levels. The local repressor AcrR represses acrAB expression directly. Other regulators include the AcrS repressor of the AcrEF system, histone-like nucleoid structuring protein (H-NS), and the SdiA global regulator. Three global regulators, MarA, SoxS, and Rob, positively control the expression of acrAB, tolC, and micF. The micF transcript inhibits the translation of OmpF porin mRNA. The two-component regulatory systems EvgAS and/or PhoQP can enhance acrAB and tolC expression. The red lines show the repression of the transcription of the relevant gene by the repressors AcrR, AcrS, H-NS, MarR, and SoxR. The green arrowed lines reveal the activation of relevant gene expression by the activators MarA, SoxS, SdiA, Rob, EvgS, and PhoP (SoxS and Rob can also stimulate MarA expression). MarB modulates MarA expression. Several regulators can bind with certain ligands (such as antimicrobial agents and metabolites) or be induced by oxidative stress and thus become inactivated (in the case of AcrR, MarR, and SoxR) or activated (in the case of Rob when binding with bile salts or fatty acids). Mutational changes can lead to the inactivation of AcrR, AcrS, MarR, and SoxR. The crystal structures of AcrR, MarA, MarR, SoxR, and Rob are available with identified ligand-binding domains and conformational changes for regulation. Regulation of AcrAB by noncoding RNAs has also been identified (see the text). Overall, under various conditions, these multiple regulation mechanisms can together produce MDR by allowing simultaneously decreased influx (via OmpF porin) and increased efflux (via AcrAB-TolC) of antimicrobial agents, which can be captured by the pump complex from the outer and/or inner leaflets of the IM and the periplasm (but not directly from the cytosol). Expression of acrZ (whose product can be copurified with AcrB) is coregulated with that of acrAB via MarA, SoxS, and Rob.
The multiple antibiotic resistance (mar) locus is a hot spot for mutation (735) and includes two divergent transcriptional units, marRAB and marC, which are transcribed from a common operator/promoter region (marO) (201). (However, marC, with unknown function, is not involved in resistance [736].) The prototypical member of the MarR family regulators, the MarR repressor, is a dimer, with each monomer containing a winged-helix DNA-binding motif. It negatively regulates the expression of marRAB and is critical in determining the expression of the MarA activator (737). MarA is an AraC family transcriptional activator (738) and possesses two similar helix-turn-helix DNA-binding subdomains (739). MarA not only positively controls the expression of marRAB (by binding to marO in a region that differs from where MarR binds) (201) but also is involved in the regulation of >60 genes (740). Promoter discrimination of MarA regulon promoters is mediated by Glu89 of MarA (741). AcrZ, a recently identified small accessory protein of AcrB (112, 123), is also positively regulated transcriptionally by MarA, SoxS, and Rob (112). The level of MarA (and SoxS) is also controlled by proteolytic degradation by the Lon protease (742). The function of MarB remained unknown until 2011 with a phenotypic examination of the E. coli mutant library (Table 1) (30), which revealed that MarB is a fine-tuning player inhibiting MarA and that this inhibition likely occurs via a posttranscriptional process. Two additional global activators, SoxS (the effector of the oxidative stress soxRS regulon) and Rob (743), also belong to the AraC family and can bind to marO and stimulate the gene expressions of the mar regulon, including acrAB and tolC (201). Because MarA and SoxS are very small proteins, they work solely by changing the level of their expression. Their upregulation may occur through the mutational inactivation of their cognate repressors MarR and SoxR, respectively; in clinical isolates showing high levels of fluoroquinolone resistance, mutations inactivating MarR are common (198). In other studies often involving other species of Enterobacteriaceae, fluoroquinolone-resistant mutants also had mutations in soxRS or acrR, as described above in the section dealing with Enterobacteriaceae. Alternatively, these repressors may become inactive through interactions with small molecules, e.g., salicylate, 2,4-dinitrophenol, or plumbagin (15, 744) binding to MarR, or oxidative inactivation of FeS-containing SoxR by superoxide (745). In contrast, Rob is much larger, and the binding of ligands such as dipyridyl (15) or fatty acids and bile salts (746) increases its binding affinity for the promoter region of acrAB, resulting in the upregulation of AcrB. Also, unlike with MarA, only one of Rob's two helix-turn-helix DNA motifs engages the binding site (747).
A two-component system, EvgAS, is also involved in the transcriptional regulation of not only acrAB and tolC but also emrKY, mdtEF, and mdfA (748, 749). Still, tolC expression is dependent on both EvgAS and another two-component system, PhoPQ. These systems constitute a signal transduction cascade and are connected via a small IM protein named B1500 (65 amino acids), whose expression is directly regulated by EvgSA (750). The common regulation of acrAB-tolC by many regulators is further elucidated by the transcriptional activation of two tolC promoters using one binding site (mar box) by MarA, SoxS, and Rob, whereas a different promoter of tolC is activated by EvgAS and PhoPQ (751). Interestingly, a small RNA, RyeB, expressed during the stationary phase, can inhibit TolC expression (752). This type of regulation occurs at the posttranscriptional level through binding of the small RNA to the 5′ untranslated region of mRNA targets, and it requires a small chaperone protein, Hfq. An Hfq mutant is susceptible to several antibiotics and other toxic agents, and this phenotype is dependent on a functional AcrAB, suggesting the involvement of Hfq in AcrAB production (753). Hfq mutants have reduced abilities in fitness, virulence, and biofilm formation, likely due to the effect of Hfq on the regulation of the stationary-phase sigma factor RpoS and the envelope stress response sigma factor RpoE (754). However, the clinical significance of small RNA regulation of MDR efflux systems requires further investigation.
Overall, under diverse conditions, these multiple regulatory mechanisms can together produce MDR by allowing simultaneously decreased influx (via decreased OmpF porin) and increased efflux (via AcrAB-TolC) of various agents (Fig. 4). Indeed, one in vitro-selected high-level ceftazidime-resistant mutant (128-fold MIC increase) had OmpF loss and increased expression levels of acrB, acrD, and acrF with multiple mutations in acrR, marR, and the gene for penicillin-binding protein 3 and overexpressed sdiA, while another mutant with low-level ceftazidime resistance (4-fold MIC increase) showed only increased acrB expression levels due to an acrR mutation (755).
Regulation of other pumps.
The MdtABC and MdtEF RND pumps are regulated by the two-component systems BaeSR and EvgSA, respectively (128, 756). Expressed from the genes downstream of mdtABCD, BaeSR not only activates the expression of mdtABCD (128) but also controls the expression of >60 genes (including acrD) that are part of the BaeSR regulon and involved in signal transduction, the chemotactic response, flagellum biosynthesis, metal homeostasis, and sugar or drug transport (757), providing a major pathway related to the bacterial cell envelope response (758). (BaeSR is in fact required for envelope stress-induced CRISPR [clustered regularly interspaced short palindromic repeat] RNA-mediated DNA silencing that constitutes part of a defense mechanism in E. coli [759].) MdtBC production is increased strongly with subinhibitory concentrations of ciprofloxacin (760), but clinically relevant conditions for its overexpression are not obvious. Functioning through the BaeSR and CpxAR pathways, indole induces the expression of MdtABC and AcrD (756). Tannins, secondary metabolites of plants, can also induce the expression of MdtABC via BaeSR (761). Of the BaeSR regulon, there is a spy gene encoding a periplasmic chaperone (Spy), which shows increased levels in TolC or pump mutants and requires BaeSR and CpxAR systems for full activation (762). The expression of MdtEF is positively regulated by two AraC family activators, YdeO and GadX (763–765), and is also stimulated by N-acetyl-d-glucosamine through catabolite activation (763) and overexpression of a small (85-nucleotide) noncoding DsrA RNA (766). The latter RNA functions as an antisilencer of the H-NS-silenced genes (767). H-NS represses the expression of the efflux operons mdtEF, acrEF, and emrKY (732). Under anaerobic conditions, mdtEF expression involves regulation by the ArcBA system, which antagonizes the effect of the H-NS (768). One study also suggested that the expression of acrAB, emrAB, emrD, emrE, emrKY, mdfA, and ydgFE is relatively stable during the various phases of growth, but mdtEF has the highest expression level at the late stationary phase, and this is mediated by RpoS (769).
Salmonella Efflux Pumps
Similar to the regulation of its E. coli homolog, the Salmonella AcrAB-TolC system is controlled through several regulatory pathways, such as AcrR, MarA, and SoxS (16, 770–773). Paraquat can induce AcrAB production, and this induction is dependent on SoxS (774). A recent study also showed that the expression level of the acrB, acrD, and/or acrF gene was increased when one or multiple acr genes were deleted (629), and this observation was similar to the situation found among the Mex pump genes of P. aeruginosa (628). However, the compensatory acr expression level changes appeared to have only a minimal impact on the drug susceptibility phenotype (except some aminoglycosides) (629).
Another gene locus of ramRA that is widespread in Enterobacteriaceae except E. coli also significantly influences the expression of not only AcrAB but also AcrEF and MdtABC in Salmonella (16, 229, 773, 775, 776). ramRA are transcribed divergently, with RamR repressing ramA expression (775–779). Induced by a variety of environmental signals, including bile salts, indole, and phenothiazines, RamA serves as a small activator protein to contribute to increased AcrAB production (776, 780–782). Bile binds to RamR (778) and in this way is thought to increase the expression of AcrAB. Although an earlier study (780) claimed that cholate and indole bound directly to RamA, the binding of an effector ligand to such a small regulator (only 129 amino acids) is highly unusual. Indole induces AcrAB production by also increasing RamA expression (780). RamA expression can be also increased by the inactivation of AcrAB and is regulated by the ATP-dependent Lon protease (782, 783). (The ramA sequence was present, but escaped notice, within the E. cloacae genomic fragment that was found to downregulate E. coli OmpF in 1990 [784] and was correctly identified in K. pneumoniae later [785].) The TetR-type RamR repressor negatively controls the expression of ramA and ramR itself by binding to the intergenic ramA-ramR promoter region (778), and mutations in ramR lead to increased production of RamA (777). RamR forms complexes with multiple agents (e.g., berberine, crystal violet, dequalinium, ethidium bromide, and rhodamine 6G), and this interaction occurs in a common residue of Phe115 and reduces the DNA-binding affinity of RamR (779). A recent review also highlighted the role of RamRA in the regulation of AcrAB (36). Additionally, the expression of the ABC-type MacAB pump is negatively controlled by PhoPQ (222).
K. pneumoniae Efflux Pumps
Similar to that in E. coli and Salmonella, AcrAB expression in K. pneumoniae, as shown in various clinical isolates or laboratory mutants, involves both negative control by AcrR and positive control by RamA and SoxS (292, 299, 501, 786). RamA and SoxS expressions are affected by RamR and SoxR regulators, respectively (299, 787). RamA can also autoregulate its own expression through binding to two ramA promoter sites (788). ramR is located upstream of the romA and ramA genes and is transcribed divergently (787), and RamR binds to two sites of the romA-ram locus (788). Mutations in ramR can yield hyperproduction of RamA and AcrAB-TolC (300, 787, 788).
Another AraC regulator, RarA, which is encoded by a gene upstream of the chromosomal oqxAB pump genes (chromosomal oqxAB genes are found only in K. pneumoniae to date), also positively controls the expression of AcrAB and OqxAB. RarA is present not only in K. pneumoniae but also in Enterobacter and Serratia (789, 790), and it functions in a regulon that affects the expression of 66 genes (791). The expression of OqxAB is additionally downregulated by a GntR-type regulator, OqxR, encoded by a gene located downstream of oqxAB (790). A point mutation in oqxR was linked to the hyperexpression of rarA and oqxB (792). Intriguingly, the genetic arrangement of rarA-oqxAB-oqxR is observed both in chromosomes and on a plasmid (790). Nearly all tested clinical tigecycline-nonsusceptible isolates (25 out of 26) from China contained mutations in ramR and/or acrR, with about one-third of the isolates carrying the simultaneous overexpression of ramA-acrB and rarA-oqxB (301). The expression of eefABC appears to be induced by an acidic or hyperosmolar environment but not by bile salts (793). The CpxAR system and the LysR-type OxyR regulator are also involved in the positive control of the expression of the acrB, acrD, and/or eefB efflux gene (794, 795).
P. aeruginosa Efflux Pumps
MexAB-OprM.
Despite rather stable expression under standard laboratory conditions, the mexAB-oprM operon is subject to complex and finely tuned regulation. Multiple gene products have been identified to influence mexAB-oprM activity, and these include the regulators MexR, NalD, and ArmR (due to NalC alteration) and a two-component system, RocS2-RocA2 (Fig. 5). MexR, a MarR family repressor, is determined by a self-regulated gene (mexR [also called nalB]) adjacent to mexAB-oprM on the chromosome (403). The binding of MexR as a dimer to the intergenic DNA region carrying the divergent overlapping promoters of mexR and mexAB-oprM (PI) results in the balanced transcription of both mexR and mexAB-oprM, which provides P. aeruginosa a protective baseline level of wide-spectrum efflux activity (405, 796). Despite the presence of another more proximal promoter (PII), PI drives most of the expression of the operon in wild-type cells (796, 797). Interestingly, some in vitro data support the notion that MexR dimerization through the formation of intermonomer disulfide bonds between redox-active cysteines prevents MexR from interacting with its cognate DNA-binding sites and from exerting its repressor activity (798, 799). This redox modulation of MexR was proposed to occur in vivo when bacteria are stressed by oxidative agents (cumene hydroperoxide) or antibiotics (meropenem and nalidixic acid) (798). However, this assumption still needs to be substantiated, as H2O2 (800, 801) and antibiotics such as colistin (802) and tobramycin (803, 804) apparently do not induce mexAB-oprM transcription significantly in planktonic and/or sessile cells.
FIG 5.
Regulation of expression of RND multidrug efflux systems of P. aeruginosa. Of 12 RND pump operons identified in this organism, half of them (presented in green, with the arrows showing their transcriptional directions) are regulated under a local regulator (mostly by a repressor [MexR, NfxB, EsrC, MexZ, or MexL] or by an activator [MexT] encoded by a gene adjacent to the efflux operons) or a two-component system of CzcRS for the czcCBA operon. (RND pump operons with no identified local regulatory genes are not included.) The red lines show the repression of the transcription of the relevant gene by repressors, while the green arrows reveal positive regulation by the regulators. Local repressors are controlled by antirepressor proteins (ArmR and ArmZ) and can also bind to ligands (e.g., antimicrobial agents or metabolites, including quorum-sensing molecules) or be induced under various conditions (oxidative or nitrosative stress or the presence of different agents). The ribosomal proteins L21 and L27, encoded by rplU-rpmA, indirectly upregulate ArmZ and negatively control mexXY expression. Activation of mexXY expression also occurs through positive control by the AmgRS two-component system via the HtpX and PA5528 proteins. Additional regulators encoded by the genes not genetically clustered with the efflux operons also participate in regulation. Mutational changes can also lead to an inactivation of regulators (ArmR, ArmZ, MexR, NalC, NalD, MexS, and MexZ). The crystal structures of MexR and MexZ are available. AlgU is a sigma factor required for the oxidative stress response, and its activity is controlled by Muc, the inner membrane-associated proteins involved in the production of alginate exopolysaccharide and with their encoding genes clustered with algU. Some regulators, including AmpR, BrlR, MvaT, ParRS, and RocS2-RocA2, are involved in the regulation of expression of other genes, e.g., downregulation of OprD by MexT and CzcR (in carbapenem resistance) or LPS modification by BrlR and ParR (affecting polymyxin susceptibility). Certain gene products (e.g., BrlR) are involved in gene regulation in biofilm cells. See the text for detail.
The expression of mexAB-oprM can be modulated positively although indirectly by ArmR, a 53-residue peptide encoded by the second gene of a two-gene operon, PA3720-PA3719 (406). Isothermal titration calorimetry studies demonstrated that ArmR can sequester MexR via an allosteric polypeptide-protein interaction of high affinity, thus alleviating the repressor activity of MexR toward mexAB-oprM (805). Indeed, the conformation of MexR in complex with ArmR is incompatible with DNA binding (806); thus, ArmR may be considered an antirepressor. Unless mutations disrupt the nalC (PA3721) gene, which encodes a TetR family regulator (727) that strongly represses the adjacent PA3720-PA3719 operon, baseline amounts of ArmR are not expected to influence MexAB-OprM production in wild-type cells (406). However, chemostat experiments with strain PAO1 showed that various chlorinated phenols, including the environmental contaminant pentachlorophenol, can induce the expression of the PA3720-PA3719 and mexAB-oprM operons through reversible, noncovalent binding to the NalC protein (807, 808). Recent data, however, showed that pentachlorophenol stimulates MexAB-OprM production, surprisingly, in an ArmR-independent (although MexR-dependent) manner (809). Possibly, the regulatory pathway here involves in vivo-generated catabolite effectors mimicking more specific antimicrobial phenolic compounds than pentachlorophenol that P. aeruginosa may encounter in its natural environment (809). The physiological conditions under which ArmR ultimately activates mexAB-oprM thus remain unclear and require further studies.
The third known regulator of mexAB-oprM is NalD, another TetR-type repressor encoded by the PA3574 gene, which binds to a proximal promoter, PII, upstream of the efflux operon (408, 810). Its DNA binding abolishes PII activity in wild-type bacteria, resulting in mexAB-oprM being expressed essentially from the distal promoter PI (810). In contrast to its homolog TtgR from P. putida, which negatively controls the expression of the TtgABC pump, no ligand that is able to relieve NalD from its operator site has been reported so far (811). Whether NalD-dependent induced expression of mexAB-oprM occurs when P. aeruginosa is challenged with some natural or semisynthetic antimicrobials remains to be elucidated. Moreover, experimentally adding polyethylene glycol to NalD in vitro during a structural study resulted in the contraction of NalD intraprotein chains (812), yet its physiological significance is unknown. Note that MexAB-OprM overproduction with combinational mutations in mexR, nalC, and nalD has been observed in clinical isolates, including epidemic strains (380, 382, 413, 425).
In addition to MexR, ArmR, and NalD, somewhat more complex regulatory circuits control MexAB-OprM expression. This is not really surprising per se in view of the major protective function of the transporter. Thus, mexAB-oprM expression has been reported to be growth phase regulated and to reach a maximum level at the onset of the stationary phase, independently of MexR and of LasR, a transcriptional regulator controlling the production of the quorum-sensing cell-to-cell signal N-3-oxo-dodecanoyl-l-homoserine lactone (3-oxo-C12-HSL) (16, 797, 813). Despite its structural relationship with 3-oxo-C12-HSL, the Rhl quorum-sensing signal N-butanoyl-l-homoserine lactone (C4-HSL) is not a substrate for MexAB-OprM (398, 399). The length of the acyl side chain seems to be an important factor in the binding affinity of acyl-HSL for MexB, with C8 to C14 compounds being better substrates for the pump than C4 to C7 molecules (399, 400). While 3-oxo-C12-HSL does not significantly impact mexAB-oprM transcript levels when added exogenously to P. aeruginosa (400, 814), C4-HSL can induce operon expression quite strongly. Consequently, it was inferred that the C4-HSL molecule might play a role in the growth-phase dependent regulation of MexAB-OprM (814), with MexR not being required for this control (815). In this regard, the local activator MexT of the MexEF-OprN system also displays an inhibitory effect on MexAB-OprM expression in MexEF-OprN-overexpressed nfxC mutants through as-yet-uncharacterized mechanisms (814). A macrolide (azithromycin) can negatively affect MexAB-OprM expression through its impact on the quorum-sensing system (16). However, the interplay between the pump and quorum sensing in P. aeruginosa is far from clear if one considers that the deletion of the mexAB-oprM operon does not result in increased production of elastase in strain PAO1 (398). This result is surprising since 3-oxo-C12-HSL, which signals elastase production, would be expected to reach higher intracellular levels if it is no longer effluxed out of the cells. Moreover, a global transcriptional regulator of the LysR family, AmpR, which also controls the expression of AmpC β-lactamase, is reported to repress mexR expression and thus to increase the level of MexAB-OprM production (816).
Another still open issue relates to the role that MexAB-OprM potentially plays in the increased intrinsic resistance of P. aeruginosa biofilms to antibiotics. In some experiments, the expression levels of mexAB-oprM appeared to have no or a limited influence on the resilience (log reduction of CFU) of in vitro-developed biofilms exposed to ofloxacin, ciprofloxacin, or tetracycline (817). Supporting the notion of a weak impact of the export system on biofilm resistance, mexAB-oprM expression was even found to be repressed by RocA2, a response regulator activated by two distinct sensor kinases (RocS1 and RocS2) that are themselves known to positively control the pilus assembly machinery cluster cupC involved in adherence/microcolony formation (814, 818). Interestingly, BrlR, a biofilm-specific MerR-type regulator, was required to sustain mexAB-oprM expression during an early stage of biofilm development through its binding to the promoter region of the operon (819). Compared to wild-type PAO1 biofilms, PAO1ΔbrlR biofilms were susceptible (as evaluated by the log reduction in the number of viable cells after drug exposure) to norfloxacin, tetracycline, trimethoprim, and aminoglycosides, and they also expressed 4-fold-lower levels of the MexA protein (819, 820), supporting that MexAB-OprM indeed has a role in biofilm antibiotic resistance. (Still, plasmid-borne brlR overexpression produces 4- to 6-fold MIC increases for chloramphenicol, norfloxacin, tetracycline, tobramycin, and trimethoprim in planktonic cells [820]. This is attributed to the regulatory activation of mexAB-oprM and mexEF-oprN by BrlR's binding to the promoter regions of these efflux operons [819].) Both BrlR production and BrlR-DNA binding are stimulated by the secondary messenger cyclic di-GMP (c-di-GMP) (821). Since MexAB-OprM does not accommodate aminoglycosides unless bacteria are cultured in low-ionic-strength medium (164), the reported hypersusceptibility of PAO1ΔbrlR biofilms to tobramycin and kanamycin might be interpreted as the result of OprM being in limiting amounts to form a functional MexXY/OprM system able to extrude these molecules (434). Levels of c-di-GMP are increased by the histidine kinase SagS, a two-component hybrid that is expressed from the early developmental stage of biofilms and that also affects BrlR production (822). Considering that SagS indirectly activates brlR expression at the irreversible attachment step of biofilm development (823), mexAB-oprM expression thus appears to be under the control of at least two distinct and opposite signal-transducing systems (i.e., SagS-BrlR and RocS1/RocS2-RocA2). An additional level of complexity in the biofilm-associated regulation of mexAB-oprM was highlighted by Pamp et al. (824). By using a pmexA-gfp reporter fusion and a dead-cell fluorescence probe, those authors noted an increased expression level of mexA in the metabolically active parts of mature biofilms challenged with colistin. More puzzling were their findings suggesting that MexAB-OprM might contribute to the tolerance of flow chamber-grown biofilms to colistin, as polymyxins (colistin and polymyxin B) do not appear to be transported by the pump (440). Similar conclusions were reached regarding the implication of two other RND pumps, MexCD-OprJ and MuxABC-OpmB, in biofilm recalcitrance to colistin (477). That MexAB-OprM might play a role in tolerance to colistin independent of its drug export activity implies that the pump has broader physiological functions than xenobiotic transport.
MexXY.
As with MexAB-OprM, the MexXY/OprM(OprA) system is also subject to complex, multilevel regulation implying both local and more general regulators (Fig. 5). The very low basal expression level of mexXY(oprA) in wild-type bacteria results from the direct binding of the dimerized, strong repressor MexZ to a 20-bp palindromic sequence encompassing the overlapping promoters of mexXY(oprA) and the adjacent, divergently transcribed gene mexZ (458, 825, 826). Unlike other TetR-type regulators (727), MexZ's DNA binding is not relieved by antibiotics through a direct ligand-regulator interaction but seemingly via indirect protein-protein sequestration, a process relying on the product of the PA5471 gene, named ArmZ, for the antirepressor of mexZ (825–827). It was demonstrated that the induction of mexXY expression in response to protein synthesis inhibitors is totally dependent upon ArmZ and that the expression of armZ itself is induced by ribosome-targeting agents through a sophisticated mechanism of transcriptional attenuation involving a short 13-amino-acid leader peptide, PA5471.1 (828). When the bacteria grow in drug-free medium, the transcribed PA5471.1 sequence is predicted to form a stem-loop structure with adjacent regions of the leader mRNA ahead of PA5471; downstream, another terminator-like stem-loop is allowed to form, which attenuates transcription of the PA5471-PA5470 operon by RNA polymerase (828). Antibiotic interference with the ribosomal machinery and translation of the PA5471.1 gene would thus prevent the formation of these secondary mRNA structures and result in increased expression levels of PA5471 with subsequent mexXY(oprA) activation through ArmZ (828). Additionally, the rplU-rpmA operon encodes the ribosomal proteins L21 and L27. Mutations in its promoter region, as observed in pan-aminoglycoside-resistant mutants, led to the reduced expression of this operon, which was linked to ArmZ-dependent increased MexXY production. Thus, the ribosome-perturbing mutations act in a way reminiscent of mexXY induction mediated by ribosome-targeting antibiotics (461). However, effectors other than ArmZ seem to be required for full MexZ-dependent drug induction of the efflux operon, as the latter still remains inducible in mexZ and mexZ-PA5471 knockout mutants (441, 827). Furthermore, it was demonstrated that exposure of P. aeruginosa to reactive oxygen species such as H2O2 also results in ArmZ-dependent mexXY(oprA) derepression (454). The physiological implications of this induction remain unclear, as the pump itself does not contribute to resistance to reactive oxygen species. Lastly, activation of mexXY expression also occurs through a newly described pathway where the AmgRS two-component system regulates mexXY expression likely via its positive effect on the expression of the htpX and PA5528 genes (which encode an IM-associated protease and a modulator of the FtsH protease, respectively) (829–831).
In addition to ArmZ-MexZ, which functionally links the activation of the pump to ribosome dysfunction, at least two signal transduction regulatory systems interconnect MexXY/OprM(OprA) with other cellular processes. Disruption of the PA2572 gene (which encodes a noncanonical response regulator) or the PA2573 gene (the determinant of a probable methyl-accepting chemotaxis protein) strongly increased mexXY(oprA) expression (832). This led to increased resistance to aminoglycosides but, intriguingly, not to ciprofloxacin, yet another pump substrate (832). As demonstrated by gene inactivation, the putative histidine kinase sensor (PA2571) of both regulators does not seem to initiate the cascade that eventually controls pump expression, suggesting that PA2572 and PA2573 respond to so-far-unknown signals, perhaps through other chemoreceptors. More is known about the regulation of MexXY by the response regulator ParR and the membrane sensor ParS. When activated by mutations or bacterial exposure to subinhibitory concentrations of polycationic agents such as polymyxins (833), this two-component system downregulates oprD expression with a concomitant upregulation of the transcript levels of mexXY(oprA) and the LPS modification operon arnBCADTEF-ugd (444). This coordinated response results in an MDR phenotype due to the complementary effects of drug efflux and reduced OM permeability (LPS modification and OprD loss) (444, 452).
Other Mex pumps.
Other Mex pumps, including MexCD-OprJ and MexEF-OprN, are not typically expressed in wild-type strains. Some of these pumps are regulated by local regulators (Fig. 5), such as the NfxB and EsrC repressors of MexCD-OprJ (465, 834, 835), the MexT activator of MexEF-OprN (836), and the MexL repressor of MexJK (837). With the MexCD-OprJ system, NfxB functions as a multimer with C termini required for multimerization and N termini implicated in DNA binding (834). EsrC is a newly identified second regulator of MexCD-OprJ, and its encoding gene (PA4596) is located downstream of mexCD-oprJ. EsrC exerts its inhibitory impact on mexCD-oprJ expression only when cells are under envelope stress and is dependent on NfxB (835). nfxB mutations in wild-type or mutator strains or as a result of the use of mutagens can occur over the entire nfxB gene (838). Elevated frequencies of nfxB mutations are also evident with the inactivation of the DNA oxidative repair system (839). Expression of mexCD-oprJ can be induced by a number of biocides (e.g., chlorhexidine and benzalkonium chloride), dyes (ethidium bromide), and other membrane-damaging agents (detergents, solvents, polymyxin B, and antimicrobial peptides) (466, 467). Indeed, mexCD-oprJ shows the greatest transcriptomic response among the genes after exposure of P. aeruginosa to chlorhexidine (840). These membrane-damaging agents apparently act to disrupt the cell membranes and result in the production of membrane lipid derivatives that stimulate the membrane-associated Muc proteins and eventually activate the stress response sigma factor AlgU for enhancing MexCD-OprJ production. nfxB mutation-related mexCD-oprJ hyperexpression is also dependent on AlgU (467). However, AlgU is negatively regulated by the global regulator AmpR (816, 818).
The expression of MexEF-OprN is controlled by several regulators (Fig. 5). The LysR-type regulator MexT is involved in controlling the expression of multiple genes in nfxC mutants, including mexEF-oprN, oprD, and genes for virulence factors (836, 841–844). mexT variations have been observed in wild-type strains and MexEF-OprN-hyperproducing nfxC mutants, which possess inactive and active forms of MexT, respectively (845). One gene of the MexT regulon, mexS (encoding an oxidoreductase of unknown function [836]), is of particular importance, as its alteration in nfxC mutants induces mexEF-oprN expression with concomitant development of MDR (846). MexT may interact with MexS, whose mutations enhance MexT-dependent mexEF-oprN expression through the intracellular accumulation of possibly toxic metabolites recognized by MexT as coinducers (846). The consequent production of MexEF-OprN would allow the extrusion of these noxious intermediates. Recent data support this hypothesis, since the exposure of P. aeruginosa to nitrosative stressors such as S-nitrosoglutathione activates mexEF-oprN transcription through MexT (847). The possible MexT-MexS interaction also affects the type III secretion system (848) and the response to disulfide stress [elicited by diazenedicarboxylic acid bis(N,N′-dimethylamide)] that perturbs the thiol-disulfide balance in the cytoplasm (849), thus further confirming the linked regulation among the drug efflux pump, virulence factor production, and the redox stress response. MexEF-OprN contributes to intrinsic resistance to this disulfide stress elicitor only in the presence of MexT (849). However, the functional link between MexS and MexEF-OprN might be more complex than anticipated, as data suggest that overexpressed mexS would also lead to MexT-dependent mexEF-oprN overexpression, at least in certain genetic backgrounds (850). Moreover, mutations in the ParSR system, which is involved in the regulation of MexXY, downregulate the expression of both mexS and mexEF-oprN (387). Interestingly, concomitant upregulation of MexS and MexEF-OprN was seen when P. aeruginosa was exposed to airway epithelial cells releasing unknown efflux-inducing signals (851). An H-NS family protein, MvaT, is a global regulator affecting the expression of hundreds of genes, including mexEF-oprN and others involved in biofilm formation, quorum sensing, and virulence (16, 852–854). The inactivation or mutation of mvaT is linked to MexEF-OprN hyperexpression and a reduction of the OprD level, and this impact on MexEF-OprN is independent of mexT or mexS (855). An mvaT mutant also shows decreased expression of the two-component regulator gene (PA2570) located immediately downstream of the czcABC efflux operon (853). Hyperproduction of MexEF-OprN with mutations in mexT, mexS, and mvaT was confirmed in clinical isolates (431). Genetic inactivation of AmpR yields an increased production of MexEF-OprN with an MDR phenotype (816), and preexposure of P. aeruginosa to subinhibitory concentrations of antibiotics (e.g., imipenem at a concentration as low as 3 ng/ml) induces AmpR production (856). The above-mentioned regulator BrlR is also an activator for MexEF-OprN expression (819).
The CzcCBA metal exporter is upregulated by at least 2 two-component systems, CzcRS (CzrRS) (492) and CopRS (857). Subinhibitory concentrations of zinc or copper salts can induce the expression of czcCBA, czcRS, and copRS. CzcRS and CopRS are also involved in the downregulation of OprD expression with concomitant resistance to carbapenems (16, 493). CzcR further affects various genes involved in virulence, including gene expression of quorum-sensing 3-oxo-C12-HSL and C4-HSL autoinducers (386).
A. baumannii Efflux Pumps
The AdeABC, AdeFGH, and AdeIJK RND pumps are regulated by the AdeRS system (858), AdeL repressor (547), and AdeN repressor (543), respectively. While adeRS and adeL are located upstream of the relevant operons, adeN is found ca. 800 kbp upstream of adeIJK (543). Mutations in AdeS and AdeR (including AdeS truncated by an ISAba1 insertion) are related to AdeABC overproduction (538, 858–860). Functional mutations were found in the conserved domains of AdeRS in AdeABC-hyperexpressing isolates with two mutational hot spots, one in AdeS near His149 and another in the DNA-binding domain of AdeR (861). Another study also identified diverse mutations in AdeRS in extensively drug-resistant isolates with the shared residue substitutions of Gly186Val in AdeS and Ala136Val in AdeR (860).
Inactivation of adeN led to 5-fold adeJ overexpression with resistance to aztreonam, ertapenem, meropenem, minocycline, and tigecycline, and mutants carrying adeN mutations in the region required for dimerization of TetR-type proteins displayed elevated levels of AdeIJK production (543), suggesting that remotely encoded AdeN negatively controls AdeIJK expression. The BaeSR regulatory system was found to positively influence the expression of the AdeA-AdeA2-AdeB system (548). Transcriptional upregulation of the MFS-type transporters MdfA and Tet(A) was observed among >200 up- or downregulated genes in a multidrug-resistant mutant after its exposure to a subinhibitory concentration of tigecycline (862).
S. maltophilia Efflux Pumps
Of 8 RND Sme pumps, two two-component system regulators are linked to SmeABC and SmeYZ, while three TetR family repressors are found for SmeDEF, SmeGH, and SmeOP. SmeABC is positively controlled by SmeSR, as is evident with the reduced MDR due to the smeR deletion (571). Located upstream of the smeDEF operon, smeT encodes the most studied pump repressor SmeT in S. maltophilia (863, 864). The promoter regions of smeDEF and smeT are overlapping, and SmeT provides negative control to SmeDEF and its own expression. A Leu166Gln substitution in SmeT results in higher expression levels of both smeDEF and smeT (863). SmeT functions as a dimer, like other TetR proteins (727), but has two extensions at the N and C termini, with a small binding pocket observed in the TetR family repressors (864). SmeDEF overproduction was also found in an isolate with an IS1246-like element in the smeDEF promoter where SmeT likely binds, suggesting that an IS element may also influence smeDEF expression (576). Triclosan not only selects SmeDEF-overproducing multidrug-resistant isolates (577) but also induces SmeDEF production via its binding to SmeT and subsequent release from the smeT promoter (865). Similarly, natural flavonoids bind to SmeT and thus influence SmeDEF expression (866). While fusaric acid-inducible FuaABC is positively controlled by its local regulator, FuaR (583), another ABC-type MacABC pump was not experimentally proven to be affected by the MacRS two-component system encoded by genes divergently transcribed from macABC (584). The MFS EmrCAB pump is downregulated by the EmrR repressor (587).
Neisseria Efflux Pumps
Expression of the MtrCDE pump is regulated both positively and negatively by several cis- and trans-acting elements (631). The MtrR repressor binds to the mtrR-mtrC intergenic region and downregulates mtrCDE expression (15, 867). MtrR also inhibits the production of FarR, a regulator of the FarAB pump (16). (A genome-wide microarray analysis revealed ∼70 genes whose expression can be repressed or activated by MtrR. One of the repressed genes is rpoH, encoding a stress response sigma factor, and as such, inducible MtrR production also increases gonococcal susceptibility to H2O2 [868].) Two AraC family regulators, the MtrA activator and MpeR repressor, also influence MtrCDE expression either directly or indirectly (637, 869). Several membrane-damaging agents can induce MtrCDE expression through their interaction with MtrA (637). This induction is also dependent on an envelope protein, MtrF, whose expression is negatively controlled by MtrR and MpeR (16, 869). A single-base-pair change located 120 bp upstream of the mtrC start codon generates a second promoter for mtrCDE expression and is sufficient not only to increase mtrCDE expression but also to render such expression independent of the control by MtrR or MtrA (870).
In N. meningitidis, mtrCDE expression appears to be independent of either MtrR or MtrA and is modulated by the Correia repeat enclosed element (CREE) that is inserted in the regulatory region for mtrCDE (871). This modulation involves the posttranscriptional regulation of the mtrCDE transcript by cleavage in the inverted repeat of the CREE (871). The CREEs are repetitive sequences identified in Neisseria spp. (commonly 153 to 157 bp or 104 to 108 bp) with an inverted repeat and a characteristic core (such as the integration host factor) that are involved in gene regulation (872). Intriguingly, CREE deletions do not show an effect on susceptibility to ciprofloxacin, erythromycin, and rifampin (873). However, whether these deletions could affect mtrCDE expression remains unknown.
C. jejuni Efflux Pumps
Located upstream of the cmeABC efflux operon, cmeR encodes the TetR-type repressor CmeR that binds directly to the promoter region of cmeABC (874). Bile salts, one type of the CmeABC substrate (655), can induce cmeABC expression through their interaction with CmeR (875). CmeR is a pleiotropic regulator, and its dimeric structure is unlike that of other TetR-type regulators since it has a large center-to-center distance between two N termini of the dimer and a large flexible multiligand-binding pocket in the C-terminal domain (876). The crystal structure of CmeR with bound bile acids taurocholate and cholate was obtained (877). cmeR mutations yield elevated expression levels of CmeABC with an MDR phenotype (878). One study also identified the negative regulation of CmeABC by another regulator, CosR, which is involved in the regulation of ∼90 genes, including an element of oxidative stress defense, the catalase-encoding katA gene (879). Induction of cmeABC expression by salicylate via its binding to CmeR was demonstrated. Salicylate at 100 μg/ml enabled better growth and survival of C. jejuni in the presence of inhibitory levels of ciprofloxacin, erythromycin, novobiocin, or tetracycline and also enhanced the emergence of fluoroquinolone resistance under antibiotic selection pressure (880). This observation is consistent with the effect of salicylate on the induction of MDR phenotypes in a number of bacteria, including B. cenocepacia, B. fragilis, E. cloacae, E. coli, K. pneumoniae, S. enterica serovar Typhimurium, S. marcescens, S. maltophilia, S. aureus, and Mycobacterium tuberculosis, often through an induction of efflux pump expression (16).
ROLE OF EFFLUX PUMPS IN BIOFILM FORMATION AND RESISTANCE
Bacterial cells can adhere to each other or to an animate or inanimate surface (including that of medical devices); this function is involved in various processes in infections and poses a major challenge to antimicrobial therapy (881). This is because biofilm cells display significantly higher levels of resistance to antimicrobials than do planktonic cells. Mechanisms of biofilm resistance may involve, for example, a low growth rate, altered metabolism and physiology, persister cells, an extracellular biofilm matrix, and an upregulated stress response (881, 882). Since antibiotic efflux pumps are involved in resistance and other functions, their roles in relation to biofilm formation and resistance have been investigated.
Biofilm formation occurs in response to numerous environmental signals and requires specific genes and regulatory circuits. This process involves three stages: initial attachment, maturation, and detachment (883). The impact of drug exporters on biofilm formation varies in different species, so P. aeruginosa mutants deficient in or overproducing RND pumps can still form biofilms (16, 884), while the loss or inhibition of any of 9 MDR pumps (AcrAB, AcrD, AcrEF, MdtABC, MdsABC, EmrAB, MdfA, MdtK, and MacAB) or the TolC OM protein in Salmonella impairs biofilm formation with reduced production of curli (885, 886). Interestingly, the biocide triclosan induces acrAB and marA expression in Salmonella biofilm cells (887). Similarly, E. coli mutants with a genetic deletion of one of the RND (acrAB, acrD, acrEF, mdtABC, and mdtEF), MFS (emrAB, emrD, and emrKY), SMR (emrE), and ABC (macAB) pump genes resulted in reduced biofilm formation (888). Inactivation of macABC in S. maltophilia led to a 50% reduction in biofilm formation (584). During biofilm growth, two uropathogenic E. coli strains showed upregulated expression of 20 transport genes, and the inclusion of an EPI, PAβN or thioridazine, in the medium reduced biofilm formation in P. putida and S. aureus (889). PAβN in combination with an iron chelator showed synergistic activity against P. aeruginosa biofilm development (890). Similar to the role of TolC, a plasmid encoding a TolC homolog, AatA, promoted aggregation and biofilm formation of an enteroaggregative E. coli isolate (891). A large conjugative plasmid encoding the OqxAB pump and type 3 fimbriae enabled E. coli to form a biofilm (892). In P. putida, inactivation of an extracytoplasmic function sigma factor, ECF-10, led to enhanced biofilm formation and the upregulation of the TtgABC pump with increased MDR (893). Upregulation of multiple proteins, including an uncharacterized RND pump and the OmpA and CarO OM proteins, also occurs in A. baumannii biofilms (894). However, despite the overall positive impact of drug exporters on biofilm formation, the inactivation of two RND pumps (i.e., RND-4, RND-9, or both) in a B. cenocepacia strain enhanced biofilm formation (615). These observations of the efflux pump's impact on biofilm formation may partially be attributable to the altered levels of certain signaling molecules in the mutant strains.
Efflux pumps, as one of the major means of conventional antibiotic resistance in planktonic cells, certainly contribute to the survival of biofilm cells in the presence of antibiotics. For instance, in P. aeruginosa, although still having relatively high levels of biofilm-derived resistance, MexAB-OprM-deficient biofilm cells display lower MDR than do cells of the wild-type strain (884). Biofilm resistance to ofloxacin (but not to ciprofloxacin) is dependent on MexAB-OprM expression at a low ofloxacin concentration range, and MexCD-OprJ provides a biofilm-specific mechanism for azithromycin resistance (16). Biofilm cells include distinct subpopulations, and resistance to colistin in metabolically active cells is attributable to the presence of MexAB-OprM and the pmr-mediated LPS modification (824). The survival of these active subpopulations after exposure to membrane-targeting agents (colistin and chlorhexidine) is also linked to not only MexAB-OprM but also MexCD-OprJ and MuxABC-OpmB pumps (477). In E. coli, the plasmid-borne TetA pump and TEM-1 β-lactamase interplay in the presence of subinhibitory levels of antibiotics to induce biofilm resistance to multiple antibiotics, and this is partly due to the induction of the chromosomal pump EmrKY and its regulator EvgAS (140). However, several early studies were unable to establish an additional contribution of AcrAB or MexAB-OprM to biofilm resistance to ciprofloxacin (16).
Recent studies with P. aeruginosa have shown the requirements of SagS and BrlR regulatory proteins for antibiotic resistance in biofilm cells (820, 822) and the linkage of these proteins to the level of the second messenger c-di-GMP (823), which was identified previously as being involved in aminoglycoside-induced biofilm formation and resistance (895). SagS is needed with at least 3 other two-component regulatory systems, BfiRS (biofilm initiation), BfmRS (biofilm maturation), and MifRS (microcolony formation), for the coordination of biofilm formation (822). BrlR, which is specifically expressed in biofilm cells (820), acts as a transcriptional activator for the expression of mexAB-oprM and mexEF-oprN (819). Unlike the MerR-type regulators (e.g., BltR and BmrR) that function in the regulation of MDR pumps of Gram-positive bacteria (896), brlR expression is not induced by the pump substrates. The inactivation of SagS correlates with a reduced level of c-di-GMP, and thus, c-di-GMP is positively regulated by SagS (823). c-di-GMP further positively affects the production of BrlR and also enhances the binding of BrlR to the promoters of BrlR target genes (821, 823). Together, SagS, BrlR, and c-di-GMP form an important signaling network related to the susceptibility-resistance switch and are all required for elevated expression in biofilm cells of the MexAB-OprM and MexEF-OprN pumps, which contribute to resistance in both biofilm and planktonic cells. It should be noted that similar to those in biofilm cells, increased levels of c-di-GMP in planktonic cells also confer elevated resistance to antibiotics to the cells, and this phenotype correlates well with the hyperexpression of brlR, mexA, and mexE (823). Additionally, a PA1875-PA1877-encoded efflux system in P. aeruginosa is composed of an OM protein (OpmL), an ABC exporter, and a membrane fusion protein (HlyD homolog) and contributes to biofilm-specific resistance to ciprofloxacin, gentamicin, and tobramycin (897). In a uropathogenic E. coli strain, inactivation of the rapA gene, encoding a helicase-like protein, does not alter biofilm formation but increases susceptibility to penicillin G in biofilm cells. In biofilm cells of the rapA mutant, the expression levels of 22 genes are reduced, including those encoding a putative MDR pump (YhcQ), a putative carbohydrate transport and metabolism protein (YeeZ), and a transcriptional regulator (SdiA) (898). B. fragilis cells show elevated RND pump expression upon induction by bile salts and also increased possibility of biofilm formation (899).
INVOLVEMENT OF MUTLIDRUG EFFLUX PUMPS IN OTHER FUNCTIONS
The wide distribution and overlapping functions of MDR efflux pumps in bacteria suggest a physiological role of these pumps beyond drug resistance (16, 34, 52, 54, 900). Indeed, there is an increasing understanding of the roles of multidrug pumps in bacterial cell communication, the stress response, fitness, colonization, intracellular survival, and virulence, as discussed below. However, interpretation of these results, based mostly on the properties of deletion mutants of acrB and its homologs, needs some caution because such deletions are known to result in changes in the expressions of the other remaining pumps. For instance, the deletion of acrB in S. enterica serovar Typhimurium results in the overproduction of AcrD and AcrF (629, 771). The mechanism of this regulatory response is still unclear, but it may be related to the function of AcrB (and possibly other constitutively expressed RND pumps in other species) in removing toxic intermediates of metabolism, first proposed by Helling et al. (901) and more recently developed by Rosner and Martin (762, 902).
Bacterial Stress Responses
Increasing evidence has shown that antibiotic exporters also make a great contribution to bacterial stress responses, where these pumps can function as either a preexisting mechanism or an activated resource in response to numerous cellular stresses, and this can be regarded as part of the physiological functions of drug pumps (16, 53, 54). Antibiotics and other chemical substances themselves can cause stress to the microbes and frequently induce the expression of MDR pumps, which in turn increase the capacity to resist the stress. These inducers are also often the substrates of the relevant pumps. Thus, the response to bacterial stress is complex, as demonstrated by phenomic profiling of the E. coli mutants from the Keio collection that showed thousands of phenotypes in response to >100 antibiotic or other drug challenges with the involvement of a large number of transporter genes (Table 1) (30).
The expression of the predominant AcrAB pump can be stimulated in response to the stress posed by bile salts/fatty acids, ethanol, and high salt concentrations, and this elevated level of AcrAB production enables the survival of enteric bacterial cells against bile salt stress in the intestinal tract (16, 55, 903, 904). The MdtEF and EmrAB pumps are also involved in the efflux of free fatty acids (55). A singlet MFS pump, MdtM, protects E. coli from bile salt stress and functions with AcrAB-TolC in a synergistic manner (905). The MATE NorM provides protection against H2O2 killing by possibly extruding compounds that oxidize the guanine of DNA and nucleotides as well as susceptible proteins (906). The macrolide-specific ABC exporter MacAB in Salmonella is induced upon exposure to H2O2 and is essential for survival of S. enterica serovar Typhimurium against oxidative stress. Reactive oxygen species-mediated killing in macrophages is alleviated by MacAB (907). Both SmeIJK and MacABC of S. maltophilia also contribute to tolerance to oxidative and envelope stresses (582, 584). In addition to the role of efflux pumps themselves in the stress response, the levels of several pump or OM protein regulators (e.g., MarA, SoxS, Rob, OmpR, and EnvZ) were elevated in the presence of antibiotic or chemical stress (908). E. coli cells with stress caused by iron starvation showed increased expression levels of MdtF and decreased expression levels of AcrD (16). Reduced resistance to bile, hyperosmotic, oxidative, or nitrosative stress is evident in a K. pneumoniae mutant with an inactivated OxyR regulator and reduced AcrB expression (795). An OM lipoprotein, NlpE, is involved in the envelope stress response mediated by the CpxRA signal transduction pathway, such as in response to misfolded cell envelope proteins (909), and also positively impacts the expression of the AcrD and MdtABC pumps for providing increased resistance (910). Similarly, a reciprocal regulation of RND pumps and the Cpx system occurs in V. cholerae, where Cpx system activation induces the expression of VexAB-TolC and VexGH-TolC, while the inactivation of these pumps stimulates the activation of the Cpx response (345). The KpnEF pump in K. pneumoniae also belongs to the Cpx cell envelope stress regulon involved in the response to bile salt and osmotic stresses (163). BaeSR responds to cell envelope stress (758) and regulates the expression of several RND pumps, as discussed above (see Regulation of Multidrug Efflux Pumps).
Several Mex pumps of P. aeruginosa (MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY) clearly function in stress responses, as described above (see Regulation of Multidrug Efflux Pumps) (Fig. 5). Their regulators (e.g., MexR, EsrC, ArmZ, and MexT) can respond to various stresses caused by, for example, membrane-damaging or ribosome-disrupting agents, reactive oxygen species, and/or nitrosative stress and subsequently cause increased activities of these pumps against the stresses (441, 454, 461, 467, 798, 799, 827, 835, 911). Consistently, P. putida RND pump mutants are categorized into 4 functionally distinct subgroups, and one of them is more susceptible to oxidation-inducing agents (912). In a situation mimicking chronic cystic fibrosis infections with antibiotic therapy, oxidative stress and antibiotic exposure result in the hyperexpression of AmrAB-OprM of Burkholderia vietnamiensis (913).
In Neisseria, the expression of the general stress response sigma factor RpoH is inhibited by the repressor MtrR, and this subsequently impacts the expression of at least 69 RpoH-regulated genes and the levels of gonococcal susceptibility to H2O2 (868). In C. jejuni, an OmpR-type oxidative stress regulator, CosR, provides overall negative regulation of oxidative stress defense proteins, and its expression is significantly decreased by a superoxide generator, paraquat (914). Reduced production of CosR is linked to CmeABC hyperexpression, consistent with the role of CmeABC in responding to oxidative stress (879). The CmeG exporter from C. jejuni also mediates oxidative defense (673).
Fitness, Colonization, and Virulence
Bacterial fitness, colonization, and virulence can be affected by changes in the status of drug pumps and OM proteins. While some resistant mutations may come with a substantial cost in these measurements, others have enhanced abilities. Despite certain exceptions, it is frequently seen that either inactivation or overproduction of an antibiotic efflux pump can add a fitness cost or reduce virulence, suggesting that the native expression status of the pumps may have been optimized for fitness and virulence. However, this observation with efflux-deficient strains may be due to the absent role of efflux pumps in responding to antibiotic killing or stress and may not be directly linked to fitness or virulence per se.
Enteric bacteria.
AcrB or TolC mutants of S. enterica serovar Typhimurium colonize and persist poorly in the intestine of chickens (although this is likely caused by direct inhibition by bile salts, the primary substrates of the pump) but also fail to invade macrophages in vitro (915). MacAB-TolC of E. coli is involved in exporting an extracellular peptide toxin (enterotoxin II) produced by enterotoxigenic E. coli (916). A Salmonella isolate with an inactivated MacAB pump is less virulent, and a mutant deficient in all drug efflux pumps is avirulent in mice (222). In E. cloacae, inactivation of either acrA or tolC led to reduced fitness (both in vitro and in vivo) as well as reduced virulence in a mouse model of systemic infection (264). A deficiency in AcrAB and OqxAB in K. pneumoniae also caused reduced virulence (792). Similarly, a mutant of V. cholerae deficient in RND pumps produced significantly less cholera toxin and fewer toxin-coregulated pili (341, 344), and TolC mutants were deficient in intestinal colonization in mice (917) (but see the comment above on S. enterica serovar Typhimurium). Genetic deletion of one of the five MFS pump genes also produced a colonization defect of V. cholerae in mice (352). Inactivation of RND pumps in V. parahaemolyticus resulted in reduced virulence in a rabbit model (348). In H. pylori, the CznABC metal pump is also essential for urease modulation and gastric colonization (684). Administration of PAβN also decreased the colonization of C. jejuni in chickens (918). Of 9 Salmonella drug transporters assessed for their role in virulence, the MdtABC, MdsABC, and MacAB pumps were required for virulence (222). acrAB and acrEF mutants had an impaired ability to cause mortality of mice by the oral route of infection (again, see the comment above on the primary substrates of AcrAB), and a strain deleted for all 9 pump genes did not cause mortality in mice (222). In contrast, exposure of B. fragilis to bile salts increased, in addition to efflux, bacterial coaggregation and adhesion to intestinal epithelial cells (899). Together, these results suggest the significance of the functional presence of efflux pumps for bacterial fitness and virulence. Nevertheless, the establishment of a link between efflux pumps and virulence needs to be made prudently, since the above-described results can be due to the fact that the major function of RND pumps in enteric bacteria is protection of bacteria against bile salts.
In contrast, it is likely easier to see the relation between fitness or virulence changes and pump overproduction. Tigecycline-resistant E. coli strains with likely AcrAB pump overproduction or LPS alterations have reduced fitness (919). In vitro-selected ceftriaxone-resistant mutants of S. enterica serovar Typhimurium exhibited overexpression of acrAB-tolC and reduced expression of the invA virulence gene with decreased invasion of cultured epithelial cells (920). Quinolone-resistant strains of E. coli (and P. aeruginosa) that accumulated multiple mutations (including gyrase mutations and RND pump overexpression) had reduced fitness, and subsequently, fitness-compensatory mutations affecting DNA supercoiling were acquired for bacterial survival (16).
P. aeruginosa.
The high prevalence of MexAB-OprM overproducers in the clinic tends to indicate that the upregulated system does not strongly impair the pathogenicity of P. aeruginosa. Consistent with the notion that such mutants still retain some degree of virulence, mexAB-oprM-overexpressing strains were isolated from patients with severe infections (385, 422, 429). A cystic fibrosis epidemic strain overproduced MexAB-OprM (and MexXY) and displayed enhanced virulence (921). Strains coisolated from chronically colonized airways of cystic fibrosis patients contained frequent mutations in mexR that may have resulted in MexAB-OprM overproduction (450). Decreased virulence is associated with MuxABC-OpmB inactivation (490). However, the outcome of P. aeruginosa infections is known to depend primarily upon the patient's conditions, and the contribution of bacterial virulence is sometimes not very clear. Results of several in vitro and in vivo experiments support the idea that nalB mutants might have lower fitness and lower virulence than wild-type bacteria (398, 922, 923). In agreement with this notion, it was noticed that some epidemic nalB isolates apparently had reverted to a basal mexAB-oprM expression level via mutations in the PI promoter or the putative ribosome-binding site ahead of the operon (see Regulation of Multidrug Efflux Pumps, above) (412). On the other hand, two observations tend to suggest that MexAB-OprM could play an essential role in baseline virulence under certain conditions through the export of still unidentified factors. First, the inactivation of mexA impaired the fast killing of the worm Caenorhabditis elegans and reduced the mortality rates of infected mice, compared with wild-type bacteria (924). Second, deletion of the whole mexAB-oprM operon appeared to compromise the invasiveness of P. aeruginosa, as judged by its capacity to transmigrate across Madin-Darby canine kidney epithelial cell monolayers and to cause lethal septicemia in mice (925). Concordant results were obtained in this cellular monolayer model with the MexAB-OprM-specific EPI D13-9001 (926). In line with this, it was reported that during the course of chronic lung colonization, cystic fibrosis strains tend to accumulate mutations in mexA and mexB resulting in a functionally deficient MexAB-OprM pump (447, 927). This evolution is parallel to the loss of multiple virulence factors and their regulators (e.g., LasR) over time by cystic fibrosis strains (928). Such MexAB-OprM-defective mutants that also occur in patients with chronic obstructive pulmonary disease are absent from acute infections, suggesting that the pump may indeed contribute to pathogenicity by as-yet-uncharacterized mechanisms.
While a rat model of acute pneumonia failed to demonstrate lower mortality rates in animals infected with MexCD-OprJ-upregulated mutants than in those challenged with a wild-type or nalB strain (929), other results suggest a negative impact of nfxB loss-of-function mutations on bacterial fitness, all forms of motility (swarming, swimming, and twitching), and the production of virulence factors (pyocyanin, caseinase, elastase, and type III secretion system-dependent cytotoxicity) (479, 922, 930). However, the effects of nfxB mutations on virulence factor production are highly variable and clearly strain dependent in clinical isolates (474). From very few clinical observations, it appears that the emergence of nfxB isolates in vivo occurs mainly under long-term treatment with fluoroquinolones (474), which correlates with the ability of these agents to readily select such mutants in vitro at concentrations around the MIC (433, 931). In the in vitro biofilm mode of growth, nfxB mutants arise very easily under ciprofloxacin exposure (932). The notion that the moderate resistance to fluoroquinolones conferred by the pump may lead to therapeutic failures is not always supported by animal models of infection (sepsis and neutropenic mouse thigh models) and pharmacokinetic/pharmacodynamic drug parameters (933). To the best of our knowledge, no fatal cases of infection attributable to MexCD-OprJ-overproducing mutants were reported in the literature. Furthermore, these mutants seem to be unable to cause bloodstream infections (385, 429, 474), likely because of their high susceptibility to serum complement (934), slow growth (474), and probably more general defects (479). The specific conditions that exist in the cystic fibrosis lung environment might be more favorable to the persistence of nfxB subpopulations, as the production of virulence factors and serum resistance are no longer required at the stage of chronic colonization (928). Therefore, the debate about the pathogenicity of these bacteria and why their prevalence in the clinical setting is so low remains open.
Overproduction of MexEF-OprN may also have a fitness cost and contribute to decreased survival and virulence, including impaired type III or VI secretion systems, which deliver toxins to the cytoplasm of the host cells (935). In vitro-selected MexEF-OprN-overproducing nfxC mutants are strongly deficient in the production of major quorum sensing-dependent virulence factors such as pyocyanin, elastase, and rhamnolipids and thereby appear to be avirulent in various infection models (483, 929). This correlates with the decreased production and secretion of the Rhl signaling molecule C4-homoserine lactone and the suppression of rhamnolipid-dependent swarming mobility (936, 937). Köhler et al. demonstrated that the decrease in virulence factor production was due solely to MexEF-OprM overproduction in an nfxC-type mutant and not to pleiotropic effects induced by MexT (936). Moreover, MexT can also downregulate type III secretion, pyocyanin production, and early attachment to a solid surface (polystyrene) independently of MexEF-OprN (842). Transcriptomic analysis revealed 17 genes positively controlled by MexT apart from mexEF-oprN (843). Lastly, c-di-GMP levels negatively affect the expression of the OprD channel, and this explains the improved survival of high-c-di-GMP strains in the presence of imipenem (938).
Other bacteria.
The insertional disruption of a 4-gene operon encoding a putative toluene exporter reduces the lung persistence of A. baumannii (939). A deficiency in the BpeAB-OprB pump of B. pseudomallei is linked to attenuated invasiveness and cytotoxicity toward human lung epithelial and macrophage cells (622). BesABC of Borrelia burgdorferi is essential for virulence in mouse infection (16). The MtrCDE pump enhances gonococcal genital tract infection in female mice (640). Although acrAB deletion did not significantly impact the tissue colonization of Yersinia pestis in mouse pneumonic and septicemic plague models (333), the inactivation of either of the two MarA regulatory homologs decreased lung colonization by 10-fold in a mouse model (940). A heterodimeric ABC transporter, MrtAB, provides resistance to ethidium bromide and is also required for Yersinia pseudotuberculosis to colonize the mesenteric lymph nodes (941). However, overexpressed pumps can also have a biological cost such that S. maltophilia with SmeDEF overproduction displays reduced virulence (16). Additionally, TolC proteins constitute a major component of multicomponent pumps and are required for virulence of a large number of bacteria, including Aggregatibacter actinomycetemcomitans (an oral commensal associated with periodontitis), Brucella suis, and Francisella tularensis (16). Finally, cell polarity in the model species Caulobacter crescentus was linked to efflux-based resistance, where the TipN polarity landmark protein (for directing flagellum placement to the new cell pole) is also essential for induced AcrAB expression (942).
MULTIDRUG EFFLUX PUMPS AS A CHALLENGE IN DRUG DEVELOPMENT
The broad substrate profile of the major multidrug exporters, the need for the drug to traverse the IM, and the OM permeability barrier clearly indicate key challenges in antibiotic development for Gram-negative bacteria (15, 16, 943–945). The second requirement usually means that the drug must be made somewhat lipophilic, and this makes the drug susceptible to multidrug efflux. On top of this, OM penetration requires that neither the size nor the lipophilicity of the drug can be excessive. Nevertheless, the kinetics of AcrB (see “RND Transporters,” above) show that the affinity and the maximal rate of transport by multidrug transporters cover a very wide range, so there is hope for producing drugs that are only poorly pumped out by prevalent RND pumps under clinically relevant conditions. In principle, small, hydrophilic drugs would act as nonsubstrates for multidrug efflux pumps. However, such molecules cannot easily cross the bilayers of the IM and usually must depend on specific (inward) transporters for access to cytosolic targets; in this case, mutant bacterial populations lacking such transporters are likely to develop as resistant clones. These considerations likely explain the lack of new antibiotic classes against Gram-negative bacilli for several decades (946, 947). New antibiotic pipelines are often available only for Gram-positive species and those Gram-negative species lacking significant drug efflux activity and OM barriers (944, 948, 949), and there is a particular need for drugs against multidrug-resistant A. baumannii and P. aeruginosa.
Ribosome-targeting omadacyclines (e.g., amadacycline), as new broad-spectrum aminomethylcyclines, possess activity against tetracycline-specific efflux and ribosome protection mechanisms (950, 951) but are still rendered inactive by the AcrAB-TolC and MexAB-OprM pumps (944). Similarly, the broad-spectrum agent eravacycline (a new fluorocycline) also lacks activity against A. baumannii, B. cenocepacia, and P. aeruginosa (949, 952). Plazomicin is a new aminoglycoside derivative of sisomicin with significant activity against a range of Gram-positive and Gram-negative bacteria (including multidrug-resistant isolates) (953, 954). However, its activity is adversely affected by increased efflux in A. baumannii and P. aeruginosa (955). Moreover, several new cephalosporin–β-lactamase–inhibitor combinational products in clinical trials (e.g., ceftazidime-avibactam, ceftaroline-avibactam, and ceftolozane-tazobactam) (944) are still likely to be the substrates of RND pumps, as are other β-lactams and β-lactamase inhibitors (13, 390, 439, 545), because avibactam cannot reverse efflux-mediated ceftazidime resistance (956), and both ceftaroline and ceftolozane (a new antipseudomonal cephalosporin) are still affected by efflux pump- and/or porin-related resistance mechanisms (although ceftolozane, containing multiple charged groups, appears less impacted by Mex pumps than many other β-lactams and did not select in vitro for pump overproducers in P. aeruginosa, unlike other agents) (417, 957–959). These observations could also illustrate their reduced or lack of synergistic activity against multidrug-resistant A. baumannii and P. aeruginosa (960–962). Additionally, a target-based inhibitor of LpxC (a metalloamidase involved in LPS biosynthesis), CHIR-090, is also a substrate for MexAB-OprM, MexCD-OprJ, and MexEF-OprN (963).
In spite of their synthetic nature, (fluoro)quinolones are often the typical substrates of MDR pumps. The activity spectrum profiles of several newer quinolones in clinical trials (e.g., the fluoroquinolones delafloxacin, finafloxacin, and JNJ-Q2; the nonfluorinated quinolone nemonoxacin; and an isothiazoloquinolone, ACH-702) (944, 964–968) suggest that these agents are substrates of RND pumps. Nevertheless, although still a substrate of the A. baumannii AdeABC and AdeM pumps, a new broad-spectrum fluoroquinolone, DS-8587, was found to have better activity against AdeABC- or AdeM-overproducing mutants than ciprofloxacin and levofloxacin (969), further supported by its efficacy in an animal model (970).
Newer ketolides (e.g., cethromycin and solithromycin) and oxazolidinones (posizolid, radezolid, and tedizolid) are still the agents mainly against Gram-positive bacteria (944), and the lack of potency for Gram-negative bacilli is likely attributable to drug efflux and the OM permeability barrier, as shown with earlier members of their classes (85, 242, 266, 971). For example, solithromycin is clearly a strong substrate for three pumps, MtrCDE, MacAB, and NorM, of different transporter families (647). Again, these characteristics continue to show the need to specifically develop small-molecule inhibitors of RND pumps. Indeed, RND pump inhibition rendered an FtsZ-directed antistaphylococcal prodrug (TXY436) active in vitro against E. coli, K. pneumoniae, and A. baumannii (972).
Additionally, it should be emphasized that the increasing understanding of multidrug transporters certainly facilitates antibiotic discovery and development (16, 36). Hypersusceptible mutants deficient in major RND pumps and/or LPS (9, 22, 27, 164, 177, 390) can often be best used as model organisms for screening potential agents against Gram-positive and Gram-negative organisms.
EFFLUX PUMP INHIBITORS
Active drug efflux, especially that caused by the RND pumps, plays a major role in intrinsic resistance and elevated resistance (in pump overproducers) in Gram-negative bacteria. Thus, if such pumps can now be antagonized by an inhibitor, this may open up a wide possibility of adjuvant therapy. Practically all antibiotics are susceptible to active efflux, and for many, their utility has become limited because of the overproduction of pumps in pathogens. Such inhibitors can make these “old” antibiotics effective again. Furthermore, when we consider that a wide range of antibacterial agents (including lipophilic penicillins, many macrolides, glycopeptides, oxazolidinones, lipoglycopeptides, and the lipopeptide daptomycin) are useful only for treating Gram-positive pathogens and that their poor activity against Gram-negative organisms is caused at least partially by efflux, EPIs could broaden dramatically the spectrum of these agents. Finally, it is important to note that in Salmonella and in other organisms, EPIs were shown to inhibit biofilm formation, which is integral in pathogenesis and requires the presence of RND pumps (886, 889); this gives even more incentive for the development of clinically useful EPIs.
EPIs have been used widely in order to determine if drug resistance in clinical isolates is caused by efflux. However, in some cases, the inhibitors were used under inappropriate conditions (most often too-high concentrations), so interpretation of the results becomes difficult (see also Methodological Considerations, below).
PAβN
Reserpine and verapamil, known inhibitors of P-glycoprotein, were examined in 1991 as potential inhibitors of the Gram-positive pump NorA of S. aureus and were indeed found to inhibit this bacterial transporter (973). Thus, there was a precedent for an EPI. Nevertheless, as detailed by Neyfakh for the NorA homolog Bmr of Bacillus subtilis (59), these Gram-positive MFS transporters pump out mostly fluoroquinolones (which contain a positively charged piperazine moiety) and other cationic compounds, such as doxorubicin, puromycin, cationic dyes, and even the polyamine spermidine. Therefore, it was possible to imagine that the inhibitors, also containing cationic groups, would bind to more or less similar binding sites as the substrates. In contrast, Gram-negative RND pumps, such as MexB and AcrB, have a much wider substrate specificity (Table 2) and transport not only the substrates mentioned above but also neutral compounds (chloramphenicol and solvents) and acidic ones (β-lactams). Many researchers thought that it would not be possible to find inhibitors of such pumps, and in this context, the discovery of PAβN (MC-207,110; 446 Da; reported in 2001 by scientists at Microcide [60]) (Fig. 6) was quite remarkable. This compound was found to be a broad-spectrum inhibitor of three major RND pumps in P. aeruginosa, MexAB-OprM, MexCD-OprJ, and MexEF-OprN, for the efflux of fluoroquinolones and was also found to inhibit the E. coli AcrAB-TolC pump. At 20 μg/ml, it strongly decreased the MICs of a wide range of antibiotics in a MexAB-OprM-overproducing P. aeruginosa strain: those for chloramphenicol, sparfloxacin, erythromycin, and levofloxacin decreased 128-, 128-, 32-, and 32-fold, respectively. On the other hand, MICs of carbenicillin, tetracycline, and ethidium bromide showed only a marginal decrease (<8-fold), although the genetic inactivation of MexAB-OprM caused a huge decrease of the MIC for them (between 32- and 512-fold). Thus, the effectiveness of this inhibitor depends strongly on the nature of the antibiotic, an observation that suggests, for example, that the drugs and the inhibitor may or may not share the same subsite within the binding pocket of the transporter (60). Recently, some substrates of AcrB, such as chloramphenicol, benzene, and cyclohexane, were shown to stimulate the efflux of nitrocefin and cefamandole (94), rather than the expected competitive inhibition, and this observation may also be related to the presence of subsites and to the complexity of the drug extrusion process in the giant AcrB transporter.
FIG 6.
Chemical structures of RND pump inhibitors.
Lomovskaya and colleagues (60) also showed that PAβN is a substrate of these pumps and therefore that their inhibitory action is likely the result of competition in the transport process. PAβN is not a proton conductor. Additionally, as expected for lipophilic peptides containing more than one cationic site, PAβN does permeate the P. aeruginosa OM at high concentrations, especially in strains with defective efflux. However, half-maximal permeation required a concentration of 70 μg/ml even in these cases, and this activity was completely abolished in the presence of 1 mM Mg2+ in the medium (60). The latter points are worth emphasizing because there are recent publications suggesting that the OM-permeating function explains most or even all of the antibiotic-sensitizing activity of PAβN. Matsumoto et al. (974) found that OM permeability, assayed by nitrocefin hydrolysis in ΔtolC cells of E. coli, became increased by PAβN, with the half-maximal increase occurring at ∼16 μg/ml. The assay was conducted in the absence of Mg2+, and the cells may already have been damaged because they contained a high-copy-number plasmid and were grown in the presence of 100 μg/ml ampicillin for the maintenance of the plasmid. Those authors also found that the cells became stained more by Sytox green dye at high concentrations of PAβN and took this as evidence of the permeabilization of the OM. In any case, there seems to be a huge leap from these results to the conclusion that the effect of PAβN is caused mainly by membrane permeabilization. More recently, Lamers et al. (975) showed that a P. aeruginosa mutant derepressed for the production of endogenous β-lactamase leaked out more enzyme into medium when relatively high concentrations (25 to 50 μg/ml) of PAβN were added. As shown by Lomovskaya et al. (60), Mg2+ prevented the permeabilization of the OM; however, this was observed at low concentrations but not at high concentrations of PAβN. Lamers et al. examined the entry of 8-anilino-1-naphthalenesulfonic acid as a marker of OM permeability and found that it was increased in the efflux-deficient mutant only with 50 μg/ml PAβN. Finally, those authors showed that the fluorescence of a cyanine dye was increased in the presence of 25 and 50 μg/ml PAβN and concluded that the IM became depolarized (975). However, because there was no calibration, it is impossible to tell if the “depolarization” was of a magnitude significant for bacterial physiology. These authors (975) found significant effects usually only at high concentrations, and these observations are quite consistent with the initial characterization of PAβN (60). Thus, it is important to interpret the permeabilizing effect of PAβN on the OM in the context of both PAβN concentrations and the specific bacterial strains or species (as noted above, the MIC values of PAβN for different strains of the same species and different species may vary). The molecular basis of pump inhibition by PAβN was examined by exploring its mode of binding to the binding protomer of AcrB via MD simulation (93) and is discussed together with the mode of action of NMP below. (Although ethidium influx rates at different concentrations of PAβN have been used to calculate the Km and Vmax [976], these values have little credibility because such simplistic calculations cannot be applied to a complex system, where spontaneous influx, decreased by the activity of the pump, is measured rather than the pump activity itself.)
One important effect of inhibitors, established with PAβN, was that their presence dramatically decreases the frequency of emergence of resistant mutants, presumably because the concentration of the drug is so far above the MIC all the time (60). Thus, when a wild-type P. aeruginosa strain was plated onto agar with 1 μg/ml levofloxacin (8-fold MIC), resistant mutants appeared at a frequency of 10−7; if 20 μg/ml PAβN was added to the drug, the frequency of appearance of resistant mutants was decreased to only 10−11.
Although the PAβN structure has been modified to decrease acute toxicity to a tolerable level, the presence of two cationic groups led to prolonged accumulation in tissues (presumably in acidic vesicles) and prevented repeated dosing (977). This structural feature was also likely to cause renal toxicity, and thus, the development of this series was abandoned (978). Again, the issue of toxicity is a complex one, and it is unfortunate that researchers now trying to develop new classes of inhibitors are often focused on acute toxicity data alone. In this regard, one may also consider the inhibitory spectrum (broad or narrow) of an EPI as well as the clinical situations where the EPI would be administered as an antibiotic adjuvant.
NMP
The second compound that has been used widely in the laboratory is NMP (226 Da) (Fig. 6), discovered by the examination of a series of aryl piperazines by Bohnert and Kern (979). Its activity is somewhat weaker than that of PAβN, requiring 50 μg/ml to decrease the levofloxacin MIC for an AcrB-overproducing E. coli strain. Interestingly, the activities of NMP and PAβN were quite different in their specificity (980). Thus, NMP was very effective in making E. coli clinical isolates more susceptible to ethidium bromide, for which PAβN had no effect. In contrast, NMP had no effect on susceptibility to rifampin and clarithromycin, for which PAβN showed a strong effect at 25 μg/ml (980). Because NMP is likely to act as a serotonin agonist, it is said to be unlikely to be developed into a clinically useful drug (981).
The molecular basis of inhibition by NMP and PAβN was investigated by MD simulation using the binding protomer of AcrB (93). We first noticed that neither PAβN nor NMP bound exceptionally tightly to the AcrB-binding site; the calculated affinity was in the range of, or even weaker than, that of the typical substrates. This is an observation consistent with our knowledge that these inhibitors can behave as substrates and are pumped out by AcrB (60, 979). As shown in Fig. 7, the typical substrate minocycline bound to the upper subsite within the binding pocket, which is full of hydrophilic residues such as S48, Q151, S180, N274, and D276, and only F178 and F615 were close to the substrate among the six Phe residues lining the pocket. In contrast, both PAβN and NMP bound to the lower part of the pocket that is rich in Phe residues. Thus, PAβN interacted with F136, F178, F615, and F628, and the only hydrophilic residues found nearby were Q176 and E673. NMP interacts with F617, F664, and F666, and no hydrophilic residue is found nearby (except perhaps G675). It may be important that although the docking predicted that both inhibitors bound to the binding site just like the substrate, MD simulation showed that they both moved out of the pocket in the direction of the proximal pocket and ended up straddling the G-loop (also called the F617 loop [78] or switch loop [77]). Since the G-loop is thought to play a critical role in allowing the movement of substrates from the proximal to the distal pocket, one possibility was thought to be that the straddling of the G-loop and the interference in substrate movement might explain the action of metabolites. (However, an alternative interpretation now appears more likely [see below].) A recent study using random mutagenesis of E. coli AcrB identified certain residues near the outer face of the distal substrate-binding pocket (such as G141, N282, A279, and G288) for efflux inhibition by NMP (982).
FIG 7.
MD simulation-predicted binding of the inhibitors PAβN and NMP, with the substrate minocycline shown as a reference. The positions of ligands initially predicted by docking (Autodock Vina) are shown as thin gray sticks, and those in the final phase of MD simulation are shown as thick blue sticks. AcrB residues within 3.5 Å of the ligand are shown in stick models (red, green, or yellow, if they belong to the distal pocket, proximal pocket, or G-loop, respectively). For NMP, two somewhat different equilibrium positions were obtained, and only one is shown here. (Modified from reference 93.)
D13-9001
The Daiichi-Microcide collaboration that produced PAβN continued to produce MexB-specific inhibitors, and one of the final products was D13-9001, a relatively large (669-Da) (Fig. 6) pyridopyrimidine compound (983), which decreased the levofloxacin MIC 8-fold at 2 μg/ml in P. aeruginosa. This compound is also important because it was cocrystallized with MexB and AcrB (75). In that work, the authors showed that D13-9001 is neither a substrate of AcrB nor pumped out by the transporter. Moreover, by using purified AcrB and MexB for isothermal titration calorimetry, those authors showed that the inhibitor was bound tightly by the transporter, with a dissociation constant of ∼1 μM. When the binding energy was calculated from the crystal structure, it was −11.2 kcal/mol for the inhibitor, which can be compared with the much weaker binding (−5.6 kcal/mol) for a typical substrate, minocycline. The crystal structure revealed that the lipophilic portion of this large inhibitor (tert-butylthiazolyl aminocarboxyl pyridopyrimidine) is bound to the bottom of the distal binding pocket and is surrounded by F136, F178, F610, F615, and F628, called a “hydrophobic trap” by those authors. In contrast, the hydrophilic parts of the inhibitor (the piperidine acetoaminoethylene ammonioacetate moiety as well as the tetrazole group) are bound to the upper, groove-like portion of the binding pocket and interact with many less-lipophilic side chains (Fig. 8). This structure then explains the molecular basis of pump inhibition by this agent. First, the bound agent prevents the binding of other substrates to the upper part of the binding pocket. Indeed, purified AcrB bound much less minocycline in the presence of D13-9001 (75). Second, even if the drugs bind to a subsite that does not overlap the inhibitor, the tight binding of the inhibitor will prevent the conformational changes needed for drug efflux through the functional rotation mechanism. Lastly, we note the fact that D13-9001 was effective in inhibiting efflux in intact cells of P. aeruginosa (983), although it is essentially a neutral compound and is unlikely to perturb the OM. However, the two P. aeruginosa-active EPIs, PAβN and D13-9001, contain multiple charged groups and may thus traverse the OM through one of the specific channels, allowing the influx of basic amino acids, peptides, and even some acidic compounds (984).
FIG 8.
Binding of various inhibitors determined by MD simulation. Although the binding of PAβN and NMP was examined previously (93), the simulation process was extended to >300 ns. The orange surface shows the distal binding pocket (defined previously [91]), and the inhibitor molecules are shown in sticks with CPK colors. AcrB is shown in green cartoon models, and the part closer to the viewer was removed for clarity. This figure was drawn by using the program Pymol, on the basis of data reported previously (96).
MBX2391
The pyranopyridine derivative MBX2391, recently developed by Microbiotix, is a strong inhibitor of the AcrB pump in Enterobacteriaceae (95). It is a relatively small (410-Da) (Fig. 6) neutral molecule and shows activity at very low concentrations. In a killing assay with E. coli with a minimally bactericidal concentration of ciprofloxacin, even 0.19 μM (0.08 μg/ml) the inhibitor increased killing significantly, and in the presence of 3 μM (1.2 μg/ml), 99.99% killing was achieved in 4 h, whereas ciprofloxacin alone caused no killing at all. In a nitrocefin efflux assay using intact E. coli cells, this inhibitor at 0.2 μM produced a clear inhibition with a strong increase of the Km. In contrast, much higher concentrations of PAβN could not produce clear signs of inhibition.
Some of us recently examined the binding of MBX2319 to AcrB by MD simulation (96). In this work, we also examined the binding of other inhibitors by the same method. First, we noted that D13-9001 and MBX2319 bound more tightly (−18.2 and −12.5 kcal/mol) than the typical substrate minocycline (−7.2 kcal/mol). D13-9001 bound to AcrB in exactly the same position as that shown previously by X-ray crystallography (75), confirming the reliability of our approach. MBX2319 interestingly also bound to the bottom of the distal binding pocket, or the hydrophobic trap, just like the hydrophobic portion of D13-9001. However, because MBX2319 does not have the hydrophilic side chains of D13-9001, the upper portion of the pocket, where exported drugs usually bind, is not occupied (Fig. 8).
Examination of Fig. 8 suggests furthermore that the upper “groove” portion of the binding pocket becomes closed upon the binding of MBX2319, PAβN, and NMP and occluded by the binding of the hydrophilic parts of D13-9001. Indeed, the docking of minocycline to the inhibitor-AcrB complexes showed that it is impossible for minocycline to bind to any subsite within the binding pocket (96). This observation then suggests the hypothesis most likely at present: the binding of inhibitors distorts the binding pocket so that the binding of substrates becomes difficult. Although our earlier hypothesis of interference with the G-loop cannot be totally discarded, this interpretation clearly does not apply to MBX2319, as it binds away from the G-loop, unlike PAβN or NMP (Fig. 8). Although MBX2319 does not inhibit efflux in P. aeruginosa, this is most likely due to its poor penetration across the OM, as it lacks any charged groups and cannot utilize common specific channels. Indeed, efflux inhibition was seen once P. aeruginosa OM permeability was increased by the simultaneous application of polymyxin B nonapeptide (T. J. Opperman, personal communication).
Other Compounds That Inhibit RND Pumps
Many other types of compounds have been investigated as potential EPIs and reviewed (16, 981, 985–987). Quinoline and pyridoquinoline derivatives were investigated as inhibitors of E. aerogenes AcrAB by the Pagès group (988). The compound showing the highest activity, 2,8-dimethyl-4-(2′-pyrrolidinoethyloxy)quinoline (Fig. 6), reversed chloramphenicol and norfloxacin resistance of clinical isolates substantially; however, it had to be used at a high concentration of 1 mM (989). The same group reported later that the derivatives of 7-chloroquinoline were more potent (1030).
Pharmacia scientists reported that arylpiperidines, such as 2-fluoro-3-(2-chloro-5-bromo-phenylethyl)piperidine (Fig. 6), were capable of inhibiting AcrAB and decreasing linezolid MICs in E. coli at a concentration of 100 μM (990). The Bohnert-Kern group, who discovered NMP as an inhibitor, examined other compounds with an arylpiperidine structure and found that pimozide (Fig. 6) inhibited AcrAB function strongly when Nile red efflux or ethidium influx assays were used but had little effect on the MICs of conventional antibiotics, presumably because its inhibitory action was substrate specific (991). Those researchers found that a selective serotonin reuptake inhibitor, sertraline (Fig. 6), decreased the tetracycline, clarithromycin, and linezolid MICs at 100 μM but had a much smaller effect on oxacillin MICs (992).
Another group of compounds is phenothiazines (Fig. 6), including chlorpromazine. These compounds were reported in 1997 to decrease drug resistance in E. coli (993), but it was unclear if the mechanism involved inhibition of the pump, because those authors showed that phenothiazine derivatives eliminated R plasmids. However, in 2003, Kaatz and others (994) demonstrated that these compounds decrease the MICs of several agents in S. aureus, presumably by inhibiting MFS pumps, although there was a small decrease in membrane potential. A phenothiazine, thioridazine, was found to inhibit the presumably AcrB-catalyzed efflux of ethidium in E. coli at a concentration of 15 μg/ml (995), and similar activity was seen with chlorpromazine (996). Recently, 40 new phenothiazine derivatives were tested with E. coli, and some appeared to inhibit AcrB significantly on the basis of ethidium accumulation assays (997). Still, strangely, the compounds were not effective against S. enterica serovar Enteritidis, and it is unclear which one of the three listed Salmonella strains was used for the assay. Moreover, none of the identified compounds could potentiate susceptibility of AcrAB-overproducing E. coli or S. enterica serovar Enteritidis. Additionally, certain polyamino geranic derivatives at a concentration range of 0.03 to 0.25 mM were able to decrease, by possible efflux inhibition, MIC values of chloramphenicol and nalidixic acid against E. aerogenes and Salmonella 2- to 64-fold (998). When the structures of these compounds are examined (Fig. 6), it is curious that most of the inhibitors contain a hydrophobic polycyclic core, which is likely to be bound in the hydrophobic pocket of the AcrB-binding site. Thus, all inhibitors might share a mode of binding to the transporter protein.
Martins et al. also reported a study in which verapamil (40 to 80 μg/ml) was seen to inhibit the efflux of ethidium at pH 8 in E. coli (999). The effect was seen to decrease in the presence of glucose, a result that led those authors to the very unlikely conclusion that ABC transporters, suggested to be MsbA without any evidence, were responsible for drug efflux at this pH. It should be noted that glucose also generates proton motive force and not only ATP.
Plant extracts often contain EPIs (mostly for pumps of Gram-positive bacteria), and the literature up to 2007 was reviewed previously (1000). Extracts of some plants were found to inhibit the AcrAB pump (1000), but this observation does not appear to have been followed up. Extracts from the genus Berberis are especially known to contain inhibitors of staphylococcal NorA (15). Extracts of such plants decreased ciprofloxacin MICs drastically, especially in highly ciprofloxacin-resistant strains of E. coli and P. aeruginosa, where efflux is likely to play a major role (16). Li et al. found that artesunate, a derivative of artemisinin (used against malaria), potentiated β-lactam activities against E. coli and increased cell accumulation of daunorubicin, possibly through its inhibition of acrAB-tolC expression (1001). However, artesunate concentrations required for such inhibition were quite high, in the range of 32 to 512 μg/ml. Similar to early observations (16), plant extracts containing alkaloids, flavonoids, phenols, triterpenes, and sterols potentiated antibiotic activity against multidrug-resistant E. coli, E. aerogenes, K. pneumoniae, and P. aeruginosa, with 2- to 8-fold decreases in MICs of certain antibiotics tested (but antagonistic effects were also observed with some combinations) (1002). Scientists at Microcide reported that the fermentation product of Streptomyces spp. contained two polycyclic compounds that showed strong potentiation of levofloxacin activity against the P. aeruginosa MexAB-OprM system (reviewed in reference 15). Recently, a gallotannin, 1,2,6-tri-O-galloyl-β-d-glucopyranose from Terminalia chebula fruit, was revealed to potentiate antibiotic activity against multidrug-resistant uropathogenic E. coli, at least partly attributed to its inhibitory effect on efflux pumps (1003). Two compounds derived from human serum increased the accumulation of ethidium bromide and minocycline in A. baumannii cells and potentiated activity of several antibiotics against A. baumannii and P. aeruginosa (1004).
Finally, it should be mentioned that proton conductors such as CCCP are sometimes loosely called inhibitors in the literature. However, they simply abolish the energy source, the proton motive force, for the pump function and do nothing to the pump per se. Furthermore, they are so extremely toxic that there is no chance that they could become drugs. Thus, such compounds have no place in the discussion of pump inhibitors.
METHODOLOGICAL CONSIDERATIONS
The availability of a large amount of bacterial genome data and the rapid development of technology in biochemistry and molecular biology have facilitated research for understanding the significance of drug transporters in antibiotic resistance. To date, various molecular and biochemical approaches have been used to detect and characterize the contribution of efflux pumps to resistance. Table 5 provides a summary of the methodological considerations for these methods. Some approaches have been discussed in different sections above. Here we discuss the application of antimicrobial susceptibility testing (AST) for studying efflux-mediated resistance.
TABLE 5.
Methodological considerations for detection and characterization of drug efflux pumps
| Method | Consideration(s) | References |
|---|---|---|
| Microbiological | ||
| Antimicrobial susceptibility testing ± EPI | Can be a routine assay | 60, 346, 550, 553, 630, 989 |
| Used for development of novel antibacterials | ||
| Requires appropriate EPI and needs to rule out nonefflux inhibitory effects of the EPI | ||
| Unable to provide identity of the pumps | ||
| Genetic and molecular | ||
| PCR | Readily carried out and widely used | 528, 1022, 1023 |
| Can largely screen the distribution of efflux genes | ||
| Multiplex PCR can be used for identifying multiple resistance determinants | ||
| Requires sequences of the pump genes | ||
| RT-PCR | Readily carried out (qualitative and quantitative) and widely used | 199, 200, 380, 381, 383, 385, 413, 417, 420–422, 451, 502, 531, 572, 573, 628, 668, 1024–1027 |
| Can link gene expression with resistance phenotype (without or with an EPI) | ||
| Can assess the impact of factors (e.g., induction) on pump expression | ||
| Requires purification of RNA and that there is no DNA contamination | ||
| Requires sequences of the pump genes | ||
| Requires appropriate controls (e.g., a housekeeping gene) for comparison | ||
| Cloning and expression in native and/or exogenous host and mutational analysis of efflux components | Can be used for determining the function and substrate specificity (including identification of important residues of pump components) | 82, 306, 489, 560, 623 |
| Drug efflux pump-deficient hypersusceptible E. coli can often be used as a host | ||
| Requires appropriate expression vector and host | ||
| Overexpression of a pump may be toxic to the host | ||
| Genetic inactivation | Can be used to assess the role of a specific pump in intrinsic and acquired resistance when combined with susceptibility testing | 10, 13, 27, 28, 30, 164, 560, 574, 623 |
| Can be used to assess the role of pumps beyond drug resistance (e.g., biofilm formation, stress response, fitness, and virulence) | ||
| Can be used to study pump regulation | ||
| Requires appropriate methods to construct mutants | ||
| Genomic/proteomic analysis including a microarray assay | Used to determine the distribution of various classes of pumps, including putative drug pumps and other resistance determinants | 496, 500, 534, 570, 615 |
| Microarray assay may compare a large no. of efflux pump genes and nonefflux genes | ||
| May not reveal a function and needs experimental approaches for confirmation | ||
| Requires certain instrument facilities | ||
| Biochemical | ||
| Cell-based drug accumulation or uptake assay | Can be readily carried out | 9, 11–13, 60–63, 465, 563 |
| Can be developed for high-throughput screening methods for searching for novel antimicrobials and EPIs | ||
| May be used to measure steady-state drug levels | ||
| May be used for transport kinetic studies | ||
| Requires the substrates to be traceable, such as radiolabeled or fluorescent substrates | ||
| An ionophore proton conductor, CCCP, has often been used | ||
| Membrane vesicles | Can be used to demonstrate the efflux process | 133, 1028 |
| Require delicate experimental conditions (e.g., French cell press and radiolabeled substrates) | ||
| Not widely used and mostly demonstrated in E. coli with certain pumps | ||
| Liposome reconstitution transport | Can be used to demonstrate the efflux process | 23, 57, 142, 1028 |
| Requires expression and purification of efflux protein components | ||
| Immunoblot assay | Confirms the presence of pumps | 483, 628, 1027 |
| Quantifies pump expression | ||
| Used to study pump component interactions | ||
| Requires pump component-specific antibodies | ||
| Structural studies | Determines molecular and biochemical basis of efflux pumps and drug-pump interactions | 66–68, 70, 74, 75, 91, 93, 114, 166 |
| Used to search for novel antimicrobials and EPIs | ||
| Requires delicate biochemical experimental conditions for studying crystal structures | ||
| Computer simulations can also be used |
From a clinical microbiology perspective, routine AST conducted with intact cells in the presence and absence of a known EPI may provide a good indication regarding the likely involvement of efflux pumps in resistance. Indeed, given the simplicity of AST, numerous such studies have been undertaken with clinical isolates (Table 5). However, it is of critical importance to predetermine the appropriate concentrations of an EPI and other testing conditions in order to rule out the contribution of nonefflux processes, since even a subinhibitory concentration of an EPI may exert its impact on bacterial growth.
An example for carefully assessing EPIs and establishing criteria for EPIs is the characterization of PAβN (60). This should bring much attention because PAβN has frequently been used as a classical EPI of RND pumps, yet the OM-permeabilizing effect of this agent at high levels may have often been ignored. Moreover, regardless of its mode of action, the potential antibacterial activity of an EPI itself should not be underestimated in combinational AST studies. Either membrane-permeabilizing agents or EPIs could increase antibiotic susceptibility. The MIC values of PAβN against E. coli and P. aeruginosa of different RND pump backgrounds vary, with a range of 32 to 512 μg/ml (60, 974). Thus, studies with carelessly chosen concentrations may inappropriately estimate the role of efflux mechanisms in clinical isolates. Consistent with its nature as a dipeptide amide with two positive charges at physiological pH, PAβN has been noted since its discovery for its membrane-permeabilizing effect (particularly in the absence of a functional MexAB-OprM pump) at a level of 16 μg/ml, especially in Mg2+-poor media (60, 1005). In combinational studies with antibiotics, PAβN has often been used at concentrations of >20 μg/ml (Table 5) (60, 550, 1005). Several more recent studies have also highlighted such OM-permeabilizing effects (974, 975, 1006, 1007), although their conclusions need to be examined carefully, as discussed above (see Efflux Pump Inhibitors). In Burkholderia spp., PAβN at a concentration of up to 200 μg/ml was considered most effective as an EPI (627, 1005). With this high level, its impact on OM permeability should be investigated. Moreover, PAβN at 200 μg/ml induced the expression of AmrAB-OprA and BpeAB-OprB of B. thailandensis (627), and this requires further study regarding the interplay between RND pumps and PAβN.
The literature is unfortunately full of examples of indiscriminate uses of EPIs for assessing the efflux status of both Gram-positive and Gram-negative bacteria. To date, there are more potential EPIs identified for Gram-positive bacteria than for Gram-negative bacteria (16). However, the results obtained with a typical EPI for Gram-positive bacteria such as reserpine for inhibiting RND pumps of Gram-negative bacteria remain to be further assessed (1008, 1009). Hence, caution should be taken in the selection of appropriate EPIs. In this regard, guidance documents for conducting AST or analyzing resistance mechanisms in clinical laboratory settings are available, for example, for β-lactamase identification. However, despite numerous studies using AST in the presence and absence of EPIs with clinical isolates, there are currently no guidelines regarding the choice of appropriate EPIs, the selection of their proper concentrations, and other testing conditions (e.g., standard isolates with known efflux status, media, and quality control). Such a standardized method can be applied together with other approaches (Table 5), particularly the RT-qPCR technique, to characterize efflux-mediated drug resistance in clinical isolates.
CONCLUSIONS
Over the past 2 decades, impressive advances in science and technology have revolutionized our understanding of the significant role that multidrug efflux pumps play in multidrug-resistant Gram-negative bacteria. These pumps have been characterized in a large number of human and animal pathogens, as described in this review. Through numerous studies targeting RND-type AcrAB-TolC and Mex pumps, we have obtained an in-depth understanding of the structural and biochemical basis of both transport mechanisms and substrate profiles of MDR pumps as well as their role in and beyond antibiotic resistance. A better understanding of pump regulation as well as synergistic interactions between these pumps and other resistance mechanisms could provide promising targets for drug discovery. However, even with our appreciation of efflux pumps in MDR, we are still facing challenges in developing novel antibiotics that can bypass the effects of MDR pumps and clinically useful EPIs. Meanwhile, for clinical microbiologists, standardized methods that can readily identify both genotypic and phenotypic contributions of these pumps to MDR in clinical isolates should be established and validated. Furthermore, the broad substrate specificity of these pumps and the rapid selection of pump-overproducing isolates during clinical therapy illustrate the importance of antibiotic stewardship by optimizing antibiotic use and reducing antibiotic overuse.
ACKNOWLEDGMENTS
Research in the laboratory of H.N. has been supported by a grant from the U.S. Public Health Service (AI-09644).
The views in this review do not necessarily reflect those of Health Canada.
We have no disclosable conflicts of interest.
Biographies

Xian-Zhi Li received his medical degree and an M.Sc. in Pharmacology from Luzhou Medical College and West China University of Medical Sciences, respectively, in China in the 1980s. He later obtained another M.Sc. and a Ph.D. in Microbiology from the University of Saskatchewan and Queen's University (Kingston) in Canada, respectively. He is currently a team leader at Health Canada and is involved in microbiological and toxicological assessments of premarket drugs as well as antimicrobial drug policy development. He is also an advisor to the Veterinary Antimicrobial Susceptibility Testing Subcommittee of the Clinical and Laboratory Standards Institute. He has published over 70 peer-reviewed articles on antimicrobial resistance with notable contributions to the seminal discovery and characterization of RND pumps of Pseudomonas and Stenotrophomonas at the University of California, Berkeley, and Queen's University. His interests include antimicrobial resistance evolution, mechanisms, and risk assessment.

Patrick Plésiat received his Pharm.D. and Ph.D. from Franche-Comté University, France. After a postdoctoral year at the University of California, Berkeley, in 1990 to 1991 with Hiroshi Nikaido, he was appointed head of the Bacteriology Department at University Hospital Jean Minjoz, Besançon, France, in 1995 and became professor at the Faculty of Medicine in the same city in 1997. In 2012, he was appointed by the French Ministry of Health as coordinator of the National Reference Center for Antibiotic Resistance. Since the early 1990s, his main research interests have been the porin- and efflux-based mechanisms leading to multidrug resistance in clinical strains of Pseudomonas aeruginosa, with special interest for the MexXY/OprM (OprA) pump. He has published more than 120 peer-reviewed papers. His current research is on the regulation of efflux systems and lipopolysaccharide modifications.

Hiroshi Nikaido received his M.D. and Ph.D. from Keio University in Tokyo, Japan. After a postdoctoral year with Herman Kalckar, he became a faculty member in the Bacteriology Department of Harvard Medical School in 1963. He moved to the University of California, Berkeley, in 1969, first as an Associate Professor of Bacteriology, and is now an emeritus Professor in the Department of Molecular and Cell Biology. At Harvard Medical School, his interest was in the biosynthesis of lipopolysaccharides. In the 1980s, he became interested in the property of the bacterial outer membrane as a permeability barrier, a topic that led to the discovery of bacterial porins. Since the mid-1990s, his main interest has been the multidrug efflux pumps of bacteria, especially AcrB of Escherichia coli. He has published more than 300 peer-reviewed papers, and he is a Fellow of the American Academy of Arts and Science and a member of the National Academy of Sciences.
ADDENDUM IN PROOF
A new family of drug efflux transporters was recently reported (K. A. Hassan, Q. Liu, P. J. F. Henderson, and I. T. Paulsen, mBio 6:e01982-14, 2015, http://dx.doi.org/10.1128/mBio.01982-14). Also, a recent report describes the synthesis of derivatives of MBX2319, some of which are 30 times more potent than the original inhibitor, based on the potentiation of levofloxacin and piperacillin (S. T. Nguyen et al., Bioorg Med Chem, in press). Finally, Blair et al. discovered that AcrB protein of Salmonella Typhimurium from a patient treated with ciprofloxacin had a single amino acid substitution at the binding site, which made the pump more effective for fluoroquinolones and less effective for other antibiotics (J. M. A. Blair et al., Proc Natl Acad Sci U S A, in press, http://dx.doi.org/10.1073/pnas.1419939112).
REFERENCES
- 1.World Health Organization. 2014. Antimicrobial resistance: global report on surveillance. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.D'Costa VM, King CE, Kalan L, Morar M, Sung WW, Schwarz C, Froese D, Zazula G, Calmels F, Debruyne R, Golding GB, Poinar HN, Wright GD. 2011. Antibiotic resistance is ancient. Nature 477:457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 3.Forsberg KJ, Patel S, Gibson MK, Lauber CL, Knight R, Fierer N, Dantas G. 2014. Bacterial phylogeny structures soil resistomes across habitats. Nature 509:612–616. doi: 10.1038/nature13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies J, Davies D. 2010. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finley RL, Collignon P, Larsson DG, McEwen SA, Li X-Z, Gaze WH, Reid-Smith R, Timinouni M, Graham DW, Topp E. 2013. The scourge of antibiotic resistance: the important role of the environment. Clin Infect Dis 57:704–710. doi: 10.1093/cid/cit355. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 7.Gottesman MM, Ling V. 2006. The molecular basis of multidrug resistance in cancer: the early years of P-glycoprotein research. FEBS Lett 580:998–1009. doi: 10.1016/j.febslet.2005.12.060. [DOI] [PubMed] [Google Scholar]
- 8.Levy SB. 1992. Active efflux mechanisms for antimicrobial resistance. Antimicrob Agents Chemother 36:695–703. doi: 10.1128/AAC.36.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma D, Cook DN, Alberti M, Pon NG, Nikaido H, Hearst JE. 1993. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J Bacteriol 175:6299–6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poole K, Krebes K, McNally C, Neshat S. 1993. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol 175:7363–7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X-Z, Livermore DM, Nikaido H. 1994. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: resistance to tetracycline, chloramphenicol, and norfloxacin. Antimicrob Agents Chemother 38:1732–1741. doi: 10.1128/AAC.38.8.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X-Z, Ma D, Livermore DM, Nikaido H. 1994. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: active efflux as a contributing factor to β-lactam resistance. Antimicrob Agents Chemother 38:1742–1752. doi: 10.1128/AAC.38.8.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X-Z, Nikaido H, Poole K. 1995. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother 39:1948–1953. doi: 10.1128/AAC.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nikaido H. 1994. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science 264:382–388. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- 15.Li X-Z, Nikaido H. 2004. Efflux-mediated drug resistance in bacteria. Drugs 64:159–204. doi: 10.2165/00003495-200464020-00004. [DOI] [PubMed] [Google Scholar]
- 16.Li X-Z, Nikaido H. 2009. Efflux-mediated drug resistance in bacteria: an update. Drugs 69:1555–1623. doi: 10.2165/11317030-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saier MH Jr, Tran CV, Barabote RD. 2006. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res 34:D181–D186. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saier MH Jr, Yen MR, Noto K, Tamang DG, Elkan C. 2009. The Transporter Classification Database: recent advances. Nucleic Acids Res 37:D274–D278. doi: 10.1093/nar/gkn862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saier MH Jr, Reddy VS, Tamang DG, Vastermark A. 2014. The Transporter Classification Database. Nucleic Acids Res 42:D251–D258. doi: 10.1093/nar/gkt1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren Q, Kang KH, Paulsen IT. 2004. TransportDB: a relational database of cellular membrane transport systems. Nucleic Acids Res 32:D284–D288. doi: 10.1093/nar/gkh016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nikaido H. 1989. Outer membrane barrier as a mechanism of antimicrobial resistance. Antimicrob Agents Chemother 33:1831–1836. doi: 10.1128/AAC.33.11.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aires JR, Nikaido H. 2005. Aminoglycosides are captured from both periplasm and cytoplasm by the AcrD multidrug efflux transporter of Escherichia coli. J Bacteriol 187:1923–1929. doi: 10.1128/JB.187.6.1923-1929.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikaido H. 1996. Multidrug efflux pumps of Gram-negative bacteria. J Bacteriol 178:5853–5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee A, Mao W, Warren MS, Mistry A, Hoshino K, Okumura R, Ishida H, Lomovskaya O. 2000. Interplay between efflux pumps may provide either additive or multiplicative effects on drug resistance. J Bacteriol 182:3142–3150. doi: 10.1128/JB.182.11.3142-3150.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tal N, Schuldiner S. 2009. A coordinated network of transporters with overlapping specificities provides a robust survival strategy. Proc Natl Acad Sci U S A 106:9051–9056. doi: 10.1073/pnas.0902400106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sulavik MC, Houseweart C, Cramer C, Jiwani N, Murgolo N, Greene J, DiDomenico B, Shaw KJ, Miller GH, Hare R, Shimer G. 2001. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob Agents Chemother 45:1126–1136. doi: 10.1128/AAC.45.4.1126-1136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu A, Tran L, Becket E, Lee K, Chinn L, Park E, Tran K, Miller JH. 2010. Antibiotic sensitivity profiles determined with an Escherichia coli gene knockout collection: generating an antibiotic bar code. Antimicrob Agents Chemother 54:1393–1403. doi: 10.1128/AAC.00906-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nichols RJ, Sen S, Choo YJ, Beltrao P, Zietek M, Chaba R, Lee S, Kazmierczak KM, Lee KJ, Wong A, Shales M, Lovett S, Winkler ME, Krogan NJ, Typas A, Gross CA. 2011. Phenotypic landscape of a bacterial cell. Cell 144:143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishino K, Yamaguchi A. 2001. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J Bacteriol 183:5803–5812. doi: 10.1128/JB.183.20.5803-5812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poole K. 2005. Efflux-mediated antimicrobial resistance. J Antimicrob Chemother 56:20–51. doi: 10.1093/jac/dki171. [DOI] [PubMed] [Google Scholar]
- 33.Poole K. 2007. Efflux pumps as antimicrobial resistance mechanisms. Ann Med 39:162–176. doi: 10.1080/07853890701195262. [DOI] [PubMed] [Google Scholar]
- 34.Piddock LJ. 2006. Multidrug-resistance efflux pumps—not just for resistance. Nat Rev Microbiol 4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- 35.Piddock LJ. 2006. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev 19:382–402. doi: 10.1128/CMR.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piddock LJ. 2014. Understanding the basis of antibiotic resistance: a platform for drug discovery. Microbiology 160:2366–2373. doi: 10.1099/mic.0.082412-0. [DOI] [PubMed] [Google Scholar]
- 37.Paulsen IT, Brown MH, Skurray RA. 1996. Proton-dependent multidrug efflux systems. Microbiol Rev 60:575–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saier MH Jr, Paulsen IT, Sliwinski MK, Pao SS, Skurray RA, Nikaido H. 1998. Evolutionary origins of multidrug and drug-specific efflux pumps in bacteria. FASEB J 12:265–274. [DOI] [PubMed] [Google Scholar]
- 39.Van Bambeke F, Glupczynski Y, Plésiat P, Pechère JC, Tulkens PM. 2003. Antibiotic efflux pumps in prokaryotic cells: occurrence, impact on resistance and strategies for the future of antimicrobial therapy. J Antimicrob Chemother 51:1055–1065. doi: 10.1093/jac/dkg224. [DOI] [PubMed] [Google Scholar]
- 40.Higgins CF. 2007. Multiple molecular mechanisms for multidrug resistance transporters. Nature 446:749–757. doi: 10.1038/nature05630. [DOI] [PubMed] [Google Scholar]
- 41.Alekshun MN, Levy SB. 2007. Molecular mechanisms of antibacterial multidrug resistance. Cell 128:1037–1050. doi: 10.1016/j.cell.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 42.Nikaido H. 1998. The role of outer membrane and efflux pumps in the resistance of Gram-negative bacteria. Can we improve drug access? Drug Resist Updat 1:93–98. [DOI] [PubMed] [Google Scholar]
- 43.Nikaido H. 2001. Preventing drug access to targets: cell surface permeability barriers and active efflux in bacteria. Semin Cell Dev Biol 12:215–223. doi: 10.1006/scdb.2000.0247. [DOI] [PubMed] [Google Scholar]
- 44.Nikaido H, Takatsuka Y. 2009. Mechanisms of RND multidrug efflux pumps. Biochim Biophys Acta 1794:769–781. doi: 10.1016/j.bbapap.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nikaido H. 2011. Structure and mechanism of RND-type multidrug efflux pumps. Adv Enzymol Relat Areas Mol Biol 77:1–60. doi: 10.1002/9780470920541.ch1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murakami S. 2008. Multidrug efflux transporter, AcrB—the pumping mechanism. Curr Opin Struct Biol 18:459–465. doi: 10.1016/j.sbi.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 47.Ruggerone P, Murakami S, Pos KM, Vargiu AV. 2013. RND efflux pumps: structural information translated into function and inhibition mechanisms. Curr Top Med Chem 13:3079–3100. doi: 10.2174/15680266113136660220. [DOI] [PubMed] [Google Scholar]
- 48.Blair JM, Piddock LJ. 2009. Structure, function and inhibition of RND efflux pumps in Gram-negative bacteria: an update. Curr Opin Microbiol 12:512–519. doi: 10.1016/j.mib.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 49.Pos KM. 2009. Drug transport mechanism of the AcrB efflux pump. Biochim Biophys Acta 1794:782–793. doi: 10.1016/j.bbapap.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 50.Ruggerone P, Vargiu AV, Collu F, Fischer N, Kandt C. 2013. Molecular dynamics computer simulations of multidrug RND efflux pumps. Comput Struct Biotechnol J 5:e201302008. doi: 10.5936/csbj.201302008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramos JL, Duque E, Gallegos MT, Godoy P, Ramos-Gonzalez MI, Rojas A, Teran W, Segura A. 2002. Mechanisms of solvent tolerance in Gram-negative bacteria. Annu Rev Microbiol 56:743–768. doi: 10.1146/annurev.micro.56.012302.161038. [DOI] [PubMed] [Google Scholar]
- 52.Poole K. 2008. Bacterial multidrug efflux pumps serve other functions. Microbe Mag 3:179–185. [Google Scholar]
- 53.Poole K. 2012. Stress responses as determinants of antimicrobial resistance in Gram-negative bacteria. Trends Microbiol 20:227–234. doi: 10.1016/j.tim.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 54.Poole K. 2014. Stress responses as determinants of antimicrobial resistance in Pseudomonas aeruginosa: multidrug efflux and more. Can J Microbiol 60:783–791. doi: 10.1139/cjm-2014-0666. [DOI] [PubMed] [Google Scholar]
- 55.Lennen RM, Politz MG, Kruziki MA, Pfleger BF. 2013. Identification of transport proteins involved in free fatty acid efflux in Escherichia coli. J Bacteriol 195:135–144. doi: 10.1128/JB.01477-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsukagoshi N, Aono R. 2000. Entry into and release of solvents by Escherichia coli in an organic-aqueous two-liquid-phase system and substrate specificity of the AcrAB-TolC solvent-extruding pump. J Bacteriol 182:4803–4810. doi: 10.1128/JB.182.17.4803-4810.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zgurskaya HI, Nikaido H. 1999. Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc Natl Acad Sci U S A 96:7190–7195. doi: 10.1073/pnas.96.13.7190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nikaido H, Basina M, Nguyen V, Rosenberg EY. 1998. Multidrug efflux pump AcrAB of Salmonella typhimurium excretes only those β-lactam antibiotics containing lipophilic side chains. J Bacteriol 180:4686–4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neyfakh AA. 2002. Mystery of multidrug transporters: the answer can be simple. Mol Microbiol 44:1123–1130. doi: 10.1046/j.1365-2958.2002.02965.x. [DOI] [PubMed] [Google Scholar]
- 60.Lomovskaya O, Warren MS, Lee A, Galazzo J, Fronko R, Lee M, Blais J, Cho D, Chamberland S, Renau T, Leger R, Hecker S, Watkins W, Hoshino K, Ishida H, Lee VJ. 2001. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob Agents Chemother 45:105–116. doi: 10.1128/AAC.45.1.105-116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murakami S, Tamura N, Saito A, Hirata T, Yamaguchi A. 2004. Extramembrane central pore of multidrug exporter AcrB in Escherichia coli plays an important role in drug transport. J Biol Chem 279:3743–3748. doi: 10.1074/jbc.M308893200. [DOI] [PubMed] [Google Scholar]
- 62.Bohnert JA, Karamian B, Nikaido H. 2010. Optimized Nile red efflux assay of AcrAB-TolC multidrug efflux system shows competition between substrates. Antimicrob Agents Chemother 54:3770–3775. doi: 10.1128/AAC.00620-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nagano K, Nikaido H. 2009. Kinetic behavior of the major multidrug efflux pump AcrB of Escherichia coli. Proc Natl Acad Sci U S A 106:5854–5858. doi: 10.1073/pnas.0901695106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kojima S, Nikaido H. 2013. Permeation rates of penicillins indicate that Escherichia coli porins function principally as nonspecific channels. Proc Natl Acad Sci U S A 110:E2629–E2634. doi: 10.1073/pnas.1310333110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lim SP, Nikaido H. 2010. Kinetic parameters of efflux of penicillins by the multidrug efflux transporter AcrAB-TolC of Escherichia coli. Antimicrob Agents Chemother 54:1800–1806. doi: 10.1128/AAC.01714-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Murakami S, Nakashima R, Yamashita E, Yamaguchi A. 2002. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419:587–593. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- 67.Yu EW, Aires JR, McDermott G, Nikaido H. 2005. A periplasmic drug-binding site of the AcrB multidrug efflux pump: a crystallographic and site-directed mutagenesis study. J Bacteriol 187:6804–6815. doi: 10.1128/JB.187.19.6804-6815.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tornroth-Horsefield S, Gourdon P, Horsefield R, Brive L, Yamamoto N, Mori H, Snijder A, Neutze R. 2007. Crystal structure of AcrB in complex with a single transmembrane subunit reveals another twist. Structure 15:1663–1673. doi: 10.1016/j.str.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 69.Yu EW, McDermott G, Zgurskaya HI, Nikaido H, Koshland DE Jr. 2003. Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science 300:976–980. doi: 10.1126/science.1083137. [DOI] [PubMed] [Google Scholar]
- 70.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. 2006. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 443:173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 71.Seeger MA, Schiefner A, Eicher T, Verrey F, Diederichs K, Pos KM. 2006. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science 313:1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- 72.Sennhauser G, Amstutz P, Briand C, Storchenegger O, Grutter MG. 2007. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol 5:e7. doi: 10.1371/journal.pbio.0050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Seeger MA, von Ballmoos C, Eicher T, Brandstatter L, Verrey F, Diederichs K, Pos KM. 2008. Engineered disulfide bonds support the functional rotation mechanism of multidrug efflux pump AcrB. Nat Struct Mol Biol 15:199–205. doi: 10.1038/nsmb.1379. [DOI] [PubMed] [Google Scholar]
- 74.Sennhauser G, Bukowska MA, Briand C, Grutter MG. 2009. Crystal structure of the multidrug exporter MexB from Pseudomonas aeruginosa. J Mol Biol 389:134–145. doi: 10.1016/j.jmb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 75.Nakashima R, Sakurai K, Yamasaki S, Hayashi K, Nagata C, Hoshino K, Onodera Y, Nishino K, Yamaguchi A. 2013. Structural basis for the inhibition of bacterial multidrug exporters. Nature 500:102–106. doi: 10.1038/nature12300. [DOI] [PubMed] [Google Scholar]
- 76.Takatsuka Y, Nikaido H. 2009. Covalently linked trimer of the AcrB multidrug efflux pump provides support for the functional rotating mechanism. J Bacteriol 191:1729–1737. doi: 10.1128/JB.01441-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Eicher T, Cha HJ, Seeger MA, Brandstatter L, El-Delik J, Bohnert JA, Kern WV, Verrey F, Grutter MG, Diederichs K, Pos KM. 2012. Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc Natl Acad Sci U S A 109:5687–5692. doi: 10.1073/pnas.1114944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nakashima R, Sakurai K, Yamasaki S, Nishino K, Yamaguchi A. 2011. Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket. Nature 480:565–569. doi: 10.1038/nature10641. [DOI] [PubMed] [Google Scholar]
- 79.Husain F, Nikaido H. 2010. Substrate path in the AcrB multidrug efflux pump of Escherichia coli. Mol Microbiol 78:320–330. doi: 10.1111/j.1365-2958.2010.07330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Drew D, Klepsch MM, Newstead S, Flaig R, De Gier JW, Iwata S, Beis K. 2008. The structure of the efflux pump AcrB in complex with bile acid. Mol Membr Biol 25:677–682. doi: 10.1080/09687680802552257. [DOI] [PubMed] [Google Scholar]
- 81.Kobayashi N, Tamura N, van Veen HW, Yamaguchi A, Murakami S. 2014. β-Lactam selectivity of multidrug transporters AcrB and AcrD resides in the proximal binding pocket. J Biol Chem 289:10680–10690. doi: 10.1074/jbc.M114.547794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lau CH, Hughes D, Poole K. 2014. MexY-promoted aminoglycoside resistance in Pseudomonas aeruginosa: involvement of a putative proximal binding pocket in aminoglycoside recognition. mBio 5(2):e01068-14. doi: 10.1128/mBio.01068-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yu EW, Aires JR, Nikaido H. 2003. AcrB multidrug efflux pump of Escherichia coli: composite substrate-binding cavity of exceptional flexibility generates its extremely wide substrate specificity. J Bacteriol 185:5657–5664. doi: 10.1128/JB.185.19.5657-5664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Husain F, Bikhchandani M, Nikaido H. 2011. Vestibules are part of the substrate path in the multidrug efflux transporter AcrB of Escherichia coli. J Bacteriol 193:5847–5849. doi: 10.1128/JB.05759-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hung LW, Kim HB, Murakami S, Gupta G, Kim CY, Terwilliger TC. 2013. Crystal structure of AcrB complexed with linezolid at 3.5 A resolution. J Struct Funct Genomics 14:71–75. doi: 10.1007/s10969-013-9154-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dastidar V, Mao W, Lomovskaya O, Zgurskaya HI. 2007. Drug-induced conformational changes in multidrug efflux transporter AcrB from Haemophilus influenzae. J Bacteriol 189:5550–5558. doi: 10.1128/JB.00471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bohnert JA, Schuster S, Seeger MA, Fahnrich E, Pos KM, Kern WV. 2008. Site-directed mutagenesis reveals putative substrate binding residues in the Escherichia coli RND efflux pump AcrB. J Bacteriol 190:8225–8229. doi: 10.1128/JB.00912-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vargiu AV, Collu F, Schulz R, Pos KM, Zacharias M, Kleinekathofer U, Ruggerone P. 2011. Effect of the F610A mutation on substrate extrusion in the AcrB transporter: explanation and rationale by molecular dynamics simulations. J Am Chem Soc 133:10704–10707. doi: 10.1021/ja202666x. [DOI] [PubMed] [Google Scholar]
- 89.Wehmeier C, Schuster S, Fahnrich E, Kern WV, Bohnert JA. 2009. Site-directed mutagenesis reveals amino acid residues in the Escherichia coli RND efflux pump AcrB that confer macrolide resistance. Antimicrob Agents Chemother 53:329–330. doi: 10.1128/AAC.00921-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Eicher T, Seeger MA, Anselmi C, Zhou W, Brandstatter L, Verrey F, Diederichs K, Faraldo-Gomez JD, Pos KM. 2014. Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB. eLife 3:e03145. doi: 10.7554/eLife.03145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Takatsuka Y, Chen C, Nikaido H. 2010. Mechanism of recognition of compounds of diverse structures by the multidrug efflux pump AcrB of Escherichia coli. Proc Natl Acad Sci U S A 107:6559–6565. doi: 10.1073/pnas.1001460107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Elkins CA, Mullis LB. 2007. Substrate competition studies using whole-cell accumulation assays with the major tripartite multidrug efflux pumps of Escherichia coli. Antimicrob Agents Chemother 51:923–929. doi: 10.1128/AAC.01048-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vargiu AV, Nikaido H. 2012. Multidrug binding properties of the AcrB efflux pump characterized by molecular dynamics simulations. Proc Natl Acad Sci U S A 109:20637–20642. doi: 10.1073/pnas.1218348109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kinana AD, Vargiu AV, Nikaido H. 2013. Some ligands enhance the efflux of other ligands by the Escherichia coli multidrug pump AcrB. Biochemistry 52:8342–8351. doi: 10.1021/bi401303v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Opperman TJ, Kwasny SM, Kim HS, Nguyen ST, Houseweart C, D'Souza S, Walker GC, Peet NP, Nikaido H, Bowlin TL. 2014. Characterization of a novel pyranopyridine inhibitor of the AcrAB efflux pump of Escherichia coli. Antimicrob Agents Chemother 58:722–733. doi: 10.1128/AAC.01866-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vargiu AV, Ruggerone P, Opperman TJ, Nguyen ST, Nikaido H. 2014. Inhibition of E. coli AcrB multidrug efflux pump by MBX2319: molecular mechanism and comparison with other inhibitors. Antimicrob Agents Chemother 58:6224–6234. doi: 10.1128/AAC.03283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schumacher MA, Brennan RG. 2003. Deciphering the molecular basis of multidrug recognition: crystal structures of the Staphylococcus aureus multidrug binding transcription regulator QacR. Res Microbiol 154:69–77. doi: 10.1016/S0923-2508(02)00013-X. [DOI] [PubMed] [Google Scholar]
- 98.Schumacher MA, Miller MC, Brennan RG. 2004. Structural mechanism of the simultaneous binding of two drugs to a multidrug-binding protein. EMBO J 23:2923–2930. doi: 10.1038/sj.emboj.7600288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schulz R, Vargiu AV, Collu F, Kleinekathofer U, Ruggerone P. 2010. Functional rotation of the transporter AcrB: insights into drug extrusion from simulations. PLoS Comput Biol 6:e1000806. doi: 10.1371/journal.pcbi.1000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Feng Z, Hou T, Li Y. 2012. Unidirectional peristaltic movement in multisite drug binding pockets of AcrB from molecular dynamics simulations. Mol Biosyst 8:2699–2709. doi: 10.1039/c2mb25184a. [DOI] [PubMed] [Google Scholar]
- 101.Fischer N, Kandt C. 2011. Three ways in, one way out: water dynamics in the trans-membrane domains of the inner membrane translocase AcrB. Proteins 79:2871–2885. doi: 10.1002/prot.23122. [DOI] [PubMed] [Google Scholar]
- 102.Schulz R, Vargiu AV, Ruggerone P, Kleinekathofer U. 2011. Role of water during the extrusion of substrates by the efflux transporter AcrB. J Phys Chem B 115:8278–8287. doi: 10.1021/jp200996x. [DOI] [PubMed] [Google Scholar]
- 103.Yao XQ, Kenzaki H, Murakami S, Takada S. 2010. Drug export and allosteric coupling in a multidrug transporter revealed by molecular simulations. Nat Commun 1:117. doi: 10.1038/ncomms1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yao XQ, Kimura N, Murakami S, Takada S. 2013. Drug uptake pathways of multidrug transporter AcrB studied by molecular simulations and site-directed mutagenesis experiments. J Am Chem Soc 135:7474–7485. doi: 10.1021/ja310548h. [DOI] [PubMed] [Google Scholar]
- 105.Murakami S, Yamaguchi A. 2003. Multidrug-exporting secondary transporters. Curr Opin Struct Biol 13:443–452. doi: 10.1016/S0959-440X(03)00109-X. [DOI] [PubMed] [Google Scholar]
- 106.Takatsuka Y, Nikaido H. 2006. Threonine-978 in the transmembrane segment of the multidrug efflux pump AcrB of Escherichia coli is crucial for drug transport as a probable component of the proton relay network. J Bacteriol 188:7284–7289. doi: 10.1128/JB.00683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Seeger MA, von Ballmoos C, Verrey F, Pos KM. 2009. Crucial role of Asp408 in the proton translocation pathway of multidrug transporter AcrB: evidence from site-directed mutagenesis and carbodiimide labeling. Biochemistry 48:5801–5812. doi: 10.1021/bi900446j. [DOI] [PubMed] [Google Scholar]
- 108.Yamane T, Murakami S, Ikeguchi M. 2013. Functional rotation induced by alternating protonation states in the multidrug transporter AcrB: all-atom molecular dynamics simulations. Biochemistry 52:7648–7658. doi: 10.1021/bi400119v. [DOI] [PubMed] [Google Scholar]
- 109.Lu W, Zhong M, Wei Y. 2011. Folding of AcrB subunit precedes trimerization. J Mol Biol 411:264–274. doi: 10.1016/j.jmb.2011.05.042. [DOI] [PubMed] [Google Scholar]
- 110.Fang J, Yu L, Wu M, Wei Y. 2013. Dissecting the function of a protruding loop in AcrB trimerization. J Biomol Struct Dyn 31:385–392. doi: 10.1080/07391102.2012.703065. [DOI] [PubMed] [Google Scholar]
- 111.Pogliano KJ, Beckwith J. 1994. Genetic and molecular characterization of the Escherichia coli secD operon and its products. J Bacteriol 176:804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hobbs EC, Yin X, Paul BJ, Astarita JL, Storz G. 2012. Conserved small protein associates with the multidrug efflux pump AcrB and differentially affects antibiotic resistance. Proc Natl Acad Sci U S A 109:16696–16701. doi: 10.1073/pnas.1210093109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- 114.Akama H, Matsuura T, Kashiwagi S, Yoneyama H, Narita S, Tsukihara T, Nakagawa A, Nakae T. 2004. Crystal structure of the membrane fusion protein, MexA, of the multidrug transporter in Pseudomonas aeruginosa. J Biol Chem 279:25939–25942. doi: 10.1074/jbc.C400164200. [DOI] [PubMed] [Google Scholar]
- 115.Mikolosko J, Bobyk K, Zgurskaya HI, Ghosh P. 2006. Conformational flexibility in the multidrug efflux system protein AcrA. Structure 14:577–587. doi: 10.1016/j.str.2005.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Symmons MF, Bokma E, Koronakis E, Hughes C, Koronakis V. 2009. The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc Natl Acad Sci U S A 106:7173–7178. doi: 10.1073/pnas.0900693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tamura N, Murakami S, Oyama Y, Ishiguro M, Yamaguchi A. 2005. Direct interaction of multidrug efflux transporter AcrB and outer membrane channel TolC detected via site-directed disulfide cross-linking. Biochemistry 44:11115–11121. doi: 10.1021/bi050452u. [DOI] [PubMed] [Google Scholar]
- 118.Tikhonova EB, Zgurskaya HI. 2004. AcrA, AcrB, and TolC of Escherichia coli form a stable intermembrane multidrug efflux complex. J Biol Chem 279:32116–32124. doi: 10.1074/jbc.M402230200. [DOI] [PubMed] [Google Scholar]
- 119.Tikhonova EB, Dastidar V, Rybenkov VV, Zgurskaya HI. 2009. Kinetic control of TolC recruitment by multidrug efflux complexes. Proc Natl Acad Sci U S A 106:16416–16421. doi: 10.1073/pnas.0906601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tikhonova EB, Yamada Y, Zgurskaya HI. 2011. Sequential mechanism of assembly of multidrug efflux pump AcrAB-TolC. Chem Biol 18:454–463. doi: 10.1016/j.chembiol.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yum S, Xu Y, Piao S, Sim SH, Kim HM, Jo WS, Kim KJ, Kweon HS, Jeong MH, Jeon H, Lee K, Ha NC. 2009. Crystal structure of the periplasmic component of a tripartite macrolide-specific efflux pump. J Mol Biol 387:1286–1297. doi: 10.1016/j.jmb.2009.02.048. [DOI] [PubMed] [Google Scholar]
- 122.Su CC, Long F, Zimmermann MT, Rajashankar KR, Jernigan RL, Yu EW. 2011. Crystal structure of the CusBA heavy-metal efflux complex of Escherichia coli. Nature 470:558–562. doi: 10.1038/nature09743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Du D, Wang Z, James NR, Voss JE, Klimont E, Ohene-Agyei T, Venter H, Chiu W, Luisi BF. 2014. Structure of the AcrAB-TolC multidrug efflux pump. Nature 509:512–515. doi: 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kobayashi K, Tsukagoshi N, Aono R. 2001. Suppression of hypersensitivity of Escherichia coli acrB mutant to organic solvents by integrational activation of the acrEF operon with the IS1 or IS2 element. J Bacteriol 183:2646–2653. doi: 10.1128/JB.183.8.2646-2653.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Olliver A, Valle M, Chaslus-Dancla E, Cloeckaert A. 2005. Overexpression of the multidrug efflux operon acrEF by insertional activation with IS1 or IS10 elements in Salmonella enterica serovar Typhimurium DT204 acrB mutants selected with fluoroquinolones. Antimicrob Agents Chemother 49:289–301. doi: 10.1128/AAC.49.1.289-301.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang Y, Xiao M, Horiyama T, Zhang Y, Li X, Nishino K, Yan A. 2011. The multidrug efflux pump MdtEF protects against nitrosative damage during the anaerobic respiration in Escherichia coli. J Biol Chem 286:26576–26584. doi: 10.1074/jbc.M111.243261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kim HS, Nagore D, Nikaido H. 2010. Multidrug efflux pump MdtBC of Escherichia coli is active only as a B2C heterotrimer. J Bacteriol 192:1377–1386. doi: 10.1128/JB.01448-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Baranova N, Nikaido H. 2002. The baeSR two-component regulatory system activates transcription of the yegMNOB (mdtABCD) transporter gene cluster in Escherichia coli and increases its resistance to novobiocin and deoxycholate. J Bacteriol 184:4168–4176. doi: 10.1128/JB.184.15.4168-4176.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kim HS, Nikaido H. 2012. Different functions of MdtB and MdtC subunits in the heterotrimeric efflux transporter MdtB2C complex of Escherichia coli. Biochemistry 51:4188–4197. doi: 10.1021/bi300379y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Reddy VS, Shlykov MA, Castillo R, Sun EI, Saier MH Jr. 2012. The major facilitator superfamily (MFS) revisited. FEBS J 279:2022–2035. doi: 10.1111/j.1742-4658.2012.08588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ohene-Agyei T, Lea JD, Venter H. 2012. Mutations in MexB that affect the efflux of antibiotics with cytoplasmic targets. FEMS Microbiol Lett 333:20–27. doi: 10.1111/j.1574-6968.2012.02594.x. [DOI] [PubMed] [Google Scholar]
- 132.Sapunaric FM, Aldema-Ramos M, McMurry LM. 2005. Tetracycline resistance: efflux, mutation, and other mechanisms, p 3–18. In White DG, Alekshun MN, McDermot PF (ed), Frontiers in antimicrobial resistance. A tribute to Stuart B. Levy. ASM Press, Washington, DC. [Google Scholar]
- 133.McMurry L, Petrucci RE Jr, Levy SB. 1980. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc Natl Acad Sci U S A 77:3974–3977. doi: 10.1073/pnas.77.7.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yamaguchi A, Udagawa T, Sawai T. 1990. Transport of divalent cations with tetracycline as mediated by the transposon Tn10-encoded tetracycline resistance protein. J Biol Chem 265:4809–4813. [PubMed] [Google Scholar]
- 135.Tamura N, Konishi S, Yamaguchi A. 2003. Mechanisms of drug/H+ antiport: complete cysteine-scanning mutagenesis and the protein engineering approach. Curr Opin Chem Biol 7:570–579. doi: 10.1016/j.cbpa.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 136.Krulwich TA, Jin J, Guffanti AA, Bechhofer H. 2001. Functions of tetracycline efflux proteins that do not involve tetracycline. J Mol Microbiol Biotechnol 3:237–246. [PubMed] [Google Scholar]
- 137.Petersen PJ, Jacobus NV, Weiss WJ, Sum PE, Testa RT. 1999. In vitro and in vivo antibacterial activities of a novel glycylcycline, the 9-t-butylglycylamido derivative of minocycline (GAR-936). Antimicrob Agents Chemother 43:738–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hirata T, Saito A, Nishino K, Tamura N, Yamaguchi A. 2004. Effects of efflux transporter genes on susceptibility of Escherichia coli to tigecycline (GAR-936). Antimicrob Agents Chemother 48:2179–2184. doi: 10.1128/AAC.48.6.2179-2184.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lomovskaya O, Lewis K. 1992. Emr, an Escherichia coli locus for multidrug resistance. Proc Natl Acad Sci U S A 89:8938–8942. doi: 10.1073/pnas.89.19.8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.May T, Ito A, Okabe S. 2009. Induction of multidrug resistance mechanism in Escherichia coli biofilms by interplay between tetracycline and ampicillin resistance genes. Antimicrob Agents Chemother 53:4628–4639. doi: 10.1128/AAC.00454-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yin Y, He X, Szewczyk P, Nguyen T, Chang G. 2006. Structure of the multidrug transporter EmrD from Escherichia coli. Science 312:741–744. doi: 10.1126/science.1125629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Steed PR, Zou P, Trone KE, McHaourab HS. 2013. Structure and pH-induced structural rearrangements of the putative multidrug efflux pump EmrD in liposomes probed by site-directed spin labeling. Biochemistry 52:7964–7974. doi: 10.1021/bi4012385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tanabe M, Szakonyi G, Brown KA, Henderson PJ, Nield J, Byrne B. 2009. The multidrug resistance efflux complex, EmrAB from Escherichia coli forms a dimer in vitro. Biochem Biophys Res Commun 380:338–342. doi: 10.1016/j.bbrc.2009.01.081. [DOI] [PubMed] [Google Scholar]
- 144.Borges-Walmsley MI, Beauchamp J, Kelly SM, Jumel K, Candlish D, Harding SE, Price NC, Walmsley AR. 2003. Identification of oligomerization and drug-binding domains of the membrane fusion protein EmrA. J Biol Chem 278:12903–12912. doi: 10.1074/jbc.M209457200. [DOI] [PubMed] [Google Scholar]
- 145.Edgar R, Bibi E. 1997. MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition. J Bacteriol 179:2274–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Fluman N, Bibi E. 2009. Bacterial multidrug transport through the lens of the major facilitator superfamily. Biochim Biophys Acta 1794:738–747. doi: 10.1016/j.bbapap.2008.11.020. [DOI] [PubMed] [Google Scholar]
- 147.Cannon RD, Lamping E, Holmes AR, Niimi K, Baret PV, Keniya MV, Tanabe K, Niimi M, Goffeau A, Monk BC. 2009. Efflux-mediated antifungal drug resistance. Clin Microbiol Rev 22:291–321. doi: 10.1128/CMR.00051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Woebking B, Reuter G, Shilling RA, Velamakanni S, Shahi S, Venter H, Balakrishnan L, van Veen HW. 2005. Drug-lipid A interactions on the Escherichia coli ABC transporter MsbA. J Bacteriol 187:6363–6369. doi: 10.1128/JB.187.18.6363-6369.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kobayashi N, Nishino K, Yamaguchi A. 2001. Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J Bacteriol 183:5639–5644. doi: 10.1128/JB.183.19.5639-5644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tikhonova EB, Devroy VK, Lau SY, Zgurskaya HI. 2007. Reconstitution of the Escherichia coli macrolide transporter: the periplasmic membrane fusion protein MacA stimulates the ATPase activity of MacB. Mol Microbiol 63:895–910. doi: 10.1111/j.1365-2958.2006.05549.x. [DOI] [PubMed] [Google Scholar]
- 151.Lin HT, Bavro VN, Barrera NP, Frankish HM, Velamakanni S, van Veen HW, Robinson CV, Borges-Walmsley MI, Walmsley AR. 2009. MacB ABC transporter is a dimer whose ATPase activity and macrolide-binding capacity are regulated by the membrane fusion protein MacA. J Biol Chem 284:1145–1154. doi: 10.1074/jbc.M806964200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lu S, Zgurskaya HI. 2012. Role of ATP binding and hydrolysis in assembly of MacAB-TolC macrolide transporter. Mol Microbiol 86:1132–1143. doi: 10.1111/mmi.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Guisbert E, Yura T, Rhodius VA, Gross CA. 2008. Convergence of molecular, modeling, and systems approaches for an understanding of the Escherichia coli heat shock response. Microbiol Mol Biol Rev 72:545–554. doi: 10.1128/MMBR.00007-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lu S, Zgurskaya HI. 2013. MacA, a periplasmic membrane fusion protein of the macrolide transporter MacAB-TolC, binds lipopolysaccharide core specifically and with high affinity. J Bacteriol 195:4865–4872. doi: 10.1128/JB.00756-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yerushalmi H, Lebendiker M, Schuldiner S. 1995. EmrE, an Escherichia coli 12-kDa multidrug transporter, exchanges toxic cations and H+ and is soluble in organic solvents. J Biol Chem 270:6856–6863. doi: 10.1074/jbc.270.12.6856. [DOI] [PubMed] [Google Scholar]
- 156.Nasie I, Steiner-Mordoch S, Schuldiner S. 2012. New substrates on the block: clinically relevant resistances for EmrE and homologues. J Bacteriol 194:6766–6770. doi: 10.1128/JB.01318-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chen YJ, Pornillos O, Lieu S, Ma C, Chen AP, Chang G. 2007. X-ray structure of EmrE supports dual topology model. Proc Natl Acad Sci U S A 104:18999–19004. doi: 10.1073/pnas.0709387104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Schuldiner S. 2012. Undecided membrane proteins insert in random topologies. Up, down and sideways: it does not really matter. Trends Biochem Sci 37:215–219. doi: 10.1016/j.tibs.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bay DC, Turner RJ. 2012. Small multidrug resistance protein EmrE reduces host pH and osmotic tolerance to metabolic quaternary cation osmoprotectants. J Bacteriol 194:5941–5948. doi: 10.1128/JB.00666-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Beketskaia MS, Bay DC, Turner RJ. 2014. Outer membrane protein OmpW participates with small multidrug resistance protein member EmrE in quaternary cationic compound efflux. J Bacteriol 196:1908–1914. doi: 10.1128/JB.01483-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hong H, Patel DR, Tamm LK, van den Berg B. 2006. The outer membrane protein OmpW forms an eight-stranded β-barrel with a hydrophobic channel. J Biol Chem 281:7568–7577. doi: 10.1074/jbc.M512365200. [DOI] [PubMed] [Google Scholar]
- 162.Srinivasan VB, Rajamohan G, Gebreyes WA. 2009. Role of AbeS, a novel efflux pump of the SMR family of transporters, in resistance to antimicrobial agents in Acinetobacter baumannii. Antimicrob Agents Chemother 53:5312–5316. doi: 10.1128/AAC.00748-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Srinivasan VB, Rajamohan G. 2013. KpnEF, a new member of the Klebsiella pneumoniae cell envelope stress response regulon, is an SMR-type efflux pump involved in broad-spectrum antimicrobial resistance. Antimicrob Agents Chemother 57:4449–4462. doi: 10.1128/AAC.02284-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Li X-Z, Poole K, Nikaido H. 2003. Contributions of MexAB-OprM and an EmrE homolog to intrinsic resistance of Pseudomonas aeruginosa to aminoglycosides and dyes. Antimicrob Agents Chemother 47:27–33. doi: 10.1128/AAC.47.1.27-33.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kuroda T, Tsuchiya T. 2009. Multidrug efflux transporters in the MATE family. Biochim Biophys Acta 1794:763–768. doi: 10.1016/j.bbapap.2008.11.012. [DOI] [PubMed] [Google Scholar]
- 166.He X, Szewczyk P, Karyakin A, Evin M, Hong WX, Zhang Q, Chang G. 2010. Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature 467:991–994. doi: 10.1038/nature09408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lu M, Symersky J, Radchenko M, Koide A, Guo Y, Nie R, Koide S. 2013. Structures of a Na+-coupled, substrate-bound MATE multidrug transporter. Proc Natl Acad Sci U S A 110:2099–2104. doi: 10.1073/pnas.1219901110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Tanaka Y, Hipolito CJ, Maturana AD, Ito K, Kuroda T, Higuchi T, Katoh T, Kato HE, Hattori M, Kumazaki K, Tsukazaki T, Ishitani R, Suga H, Nureki O. 2013. Structural basis for the drug extrusion mechanism by a MATE multidrug transporter. Nature 496:247–251. doi: 10.1038/nature12014. [DOI] [PubMed] [Google Scholar]
- 169.Song J, Ji C, Zhang JZ. 2014. Insights on Na+ binding and conformational dynamics in multidrug and toxic compound extrusion transporter NorM. Proteins 82:240–249. doi: 10.1002/prot.24368. [DOI] [PubMed] [Google Scholar]
- 170.He GX, Thorpe C, Walsh D, Crow R, Chen H, Kumar S, Varela MF. 2011. EmmdR, a new member of the MATE family of multidrug transporters, extrudes quinolones from Enterobacter cloacae. Arch Microbiol 193:759–765. doi: 10.1007/s00203-011-0738-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Cowan SW, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit RA, Jansonius JN, Rosenbusch JP. 1992. Crystal structures explain functional properties of two E. coli porins. Nature 358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 172.Kamio Y, Nikaido H. 1976. Outer membrane of Salmonella typhimurium: accessibility of phospholipid head groups to phospholipase c and cyanogen bromide activated dextran in the external medium. Biochemistry 15:2561–2570. doi: 10.1021/bi00657a012. [DOI] [PubMed] [Google Scholar]
- 173.Schulz GE. 1993. Bacterial porins: structure and function. Curr Opin Cell Biol 5:701–707. doi: 10.1016/0955-0674(93)90143-E. [DOI] [PubMed] [Google Scholar]
- 174.Nikaido H, Rosenberg EY. 1983. Porin channels in Escherichia coli: studies with liposomes reconstituted from purified proteins. J Bacteriol 153:241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Stock JB, Rauch B, Roseman S. 1977. Periplasmic space in Salmonella typhimurium and Escherichia coli. J Biol Chem 252:7850–7861. [PubMed] [Google Scholar]
- 176.Vaara M. 1992. Agents that increase the permeability of the outer membrane. Microbiol Rev 56:395–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Vaara M. 1993. Antibiotic-supersusceptible mutants of Escherichia coli and Salmonella typhimurium. Antimicrob Agents Chemother 37:2255–2260. doi: 10.1128/AAC.37.11.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Koebnik R. 1995. Proposal for a peptidoglycan-associating alpha-helical motif in the C-terminal regions of some bacterial cell-surface proteins. Mol Microbiol 16:1269–1270. doi: 10.1111/j.1365-2958.1995.tb02348.x. [DOI] [PubMed] [Google Scholar]
- 179.Sugawara E, Nestorovich EM, Bezrukov SM, Nikaido H. 2006. Pseudomonas aeruginosa porin OprF exists in two different conformations. J Biol Chem 281:16220–16229. doi: 10.1074/jbc.M600680200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Sugawara E, Nagano K, Nikaido H. 2010. Factors affecting the folding of Pseudomonas aeruginosa OprF porin into the one-domain open conformer. mBio 1(4):e00228-10. doi: 10.1128/mBio.00228-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yoshimura F, Nikaido H. 1982. Permeability of Pseudomonas aeruginosa outer membrane to hydrophilic solutes. J Bacteriol 152:636–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Angus BL, Carey AM, Caron DA, Kropinski AM, Hancock RE. 1982. Outer membrane permeability in Pseudomonas aeruginosa: comparison of a wild-type with an antibiotic-supersusceptible mutant. Antimicrob Agents Chemother 21:299–309. doi: 10.1128/AAC.21.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Sugawara E, Nikaido H. 2012. OmpA is the principal nonspecific slow porin of Acinetobacter baumannii. J Bacteriol 194:4089–4096. doi: 10.1128/JB.00435-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Coyne S, Courvalin P, Perichon B. 2011. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob Agents Chemother 55:947–953. doi: 10.1128/AAC.01388-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Plésiat P, Nikaido H. 1992. Outer membranes of Gram-negative bacteria are permeable to steroid probes. Mol Microbiol 6:1323–1333. doi: 10.1111/j.1365-2958.1992.tb00853.x. [DOI] [PubMed] [Google Scholar]
- 186.Elkins CA, Mullis LB. 2006. Mammalian steroid hormones are substrates for the major RND- and MFS-type tripartite multidrug efflux pumps of Escherichia coli. J Bacteriol 188:1191–1195. doi: 10.1128/JB.188.3.1191-1195.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Li X-Z, Zhang L, Poole K. 2000. Interplay between the MexA-MexB-OprM multidrug efflux system and the outer membrane barrier in the multiple antibiotic resistance of Pseudomonas aeruginosa. J Antimicrob Chemother 45:433–436. doi: 10.1093/jac/45.4.433. [DOI] [PubMed] [Google Scholar]
- 188.Mazzariol A, Cornaglia G, Nikaido H. 2000. Contributions of the AmpC β-lactamase and the AcrAB multidrug efflux system in intrinsic resistance of Escherichia coli K-12 to β-lactams. Antimicrob Agents Chemother 44:1387–1390. doi: 10.1128/AAC.44.5.1387-1390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Nikaido H, Liu W, Rosenberg EY. 1990. Outer membrane permeability and β-lactamase stability of dipolar ionic cephalosporins containing methoxyimino substituents. Antimicrob Agents Chemother 34:337–342. doi: 10.1128/AAC.34.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Trias J, Nikaido H. 1990. Protein D2 channel of the Pseudomonas aeruginosa outer membrane has a binding site for basic amino acids and peptides. J Biol Chem 265:15680–15684. [PubMed] [Google Scholar]
- 191.Plésiat P, Aires JR, Godard C, Kohler T. 1997. Use of steroids to monitor alterations in the outer membrane of Pseudomonas aeruginosa. J Bacteriol 179:7004–7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. 2013. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev 26:822–880. doi: 10.1128/CMR.00022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Bush K. 2013. Proliferation and significance of clinically relevant β-lactamases. Ann N Y Acad Sci 1277:84–90. doi: 10.1111/nyas.12023. [DOI] [PubMed] [Google Scholar]
- 194.Kronvall G. 2010. Antimicrobial resistance 1979-2009 at Karolinska hospital, Sweden: normalized resistance interpretation during a 30-year follow-up on Staphylococcus aureus and Escherichia coli resistance development. APMIS 118:621–639. doi: 10.1111/j.1600-0463.2010.02660.x. [DOI] [PubMed] [Google Scholar]
- 195.European Center for Disease Prevention and Control. 2013. Antimicrobial resistance surveillance in Europe 2012. European Center for Disease Prevention and Control, Stockholm, Sweden. [Google Scholar]
- 196.Everett MJ, Jin YF, Ricci V, Piddock LJ. 1996. Contributions of individual mechanisms to fluoroquinolone resistance in 36 Escherichia coli strains isolated from humans and animals. Antimicrob Agents Chemother 40:2380–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Oethinger M, Kern WV, Goldman JD, Levy SB. 1998. Association of organic solvent tolerance and fluoroquinolone resistance in clinical isolates of Escherichia coli. J Antimicrob Chemother 41:111–114. doi: 10.1093/jac/41.1.111. [DOI] [PubMed] [Google Scholar]
- 198.Oethinger M, Podglajen I, Kern WV, Levy SB. 1998. Overexpression of the marA or soxS regulatory gene in clinical topoisomerase mutants of Escherichia coli. Antimicrob Agents Chemother 42:2089–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Yasufuku T, Shigemura K, Shirakawa T, Matsumoto M, Nakano Y, Tanaka K, Arakawa S, Kinoshita S, Kawabata M, Fujisawa M. 2011. Correlation of overexpression of efflux pump genes with antibiotic resistance in Escherichia coli strains clinically isolated from urinary tract infection patients. J Clin Microbiol 49:189–194. doi: 10.1128/JCM.00827-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Swick MC, Morgan-Linnell SK, Carlson KM, Zechiedrich L. 2011. Expression of multidrug efflux pump genes acrAB-tolC, mdfA, and norE in Escherichia coli clinical isolates as a function of fluoroquinolone and multidrug resistance. Antimicrob Agents Chemother 55:921–924. doi: 10.1128/AAC.00996-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Alekshun MN, Levy SB. 1997. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob Agents Chemother 41:2067–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Buffet-Bataillon S, Le Jeune A, Le Gall-David S, Bonnaure-Mallet M, Jolivet-Gougeon A. 2012. Molecular mechanisms of higher MICs of antibiotics and quaternary ammonium compounds for Escherichia coli isolated from bacteraemia. J Antimicrob Chemother 67:2837–2842. doi: 10.1093/jac/dks321. [DOI] [PubMed] [Google Scholar]
- 203.Wang H, Dzink-Fox JL, Chen M, Levy SB. 2001. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob Agents Chemother 45:1515–1521. doi: 10.1128/AAC.45.5.1515-1521.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Lautenbach E, Metlay JP, Mao X, Han X, Fishman NO, Bilker WB, Tolomeo P, Wheeler M, Nachamkin I. 2010. The prevalence of fluoroquinolone resistance mechanisms in colonizing Escherichia coli isolates recovered from hospitalized patients. Clin Infect Dis 51:280–285. doi: 10.1086/653931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Karczmarczyk M, Martins M, Quinn T, Leonard N, Fanning S. 2011. Mechanisms of fluoroquinolone resistance in Escherichia coli isolates from food-producing animals. Appl Environ Microbiol 77:7113–7120. doi: 10.1128/AEM.00600-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Yamane K, Wachino J, Suzuki S, Kimura K, Shibata N, Kato H, Shibayama K, Konda T, Arakawa Y. 2007. New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob Agents Chemother 51:3354–3360. doi: 10.1128/AAC.00339-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Nikaido H, Rosenberg EY, Foulds J. 1983. Porin channels in Escherichia coli: studies with β-lactams in intact cells. J Bacteriol 153:232–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Yoshimura F, Nikaido H. 1985. Diffusion of β-lactam antibiotics through the porin channels of Escherichia coli K-12. Antimicrob Agents Chemother 27:84–92. doi: 10.1128/AAC.27.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Harder KJ, Nikaido H, Matsuhashi M. 1981. Mutants of Escherichia coli that are resistant to certain β-lactam compounds lack the OmpF porin. Antimicrob Agents Chemother 20:549–552. doi: 10.1128/AAC.20.4.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Galleni M, Amicosante G, Frère JM. 1988. A survey of the kinetic parameters of class C β-lactamases. Cephalosporins and other β-lactam compounds. Biochem J 255:123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Jaffe A, Chabbert YA, Semonin O. 1982. Role of porin proteins OmpF and OmpC in the permeation of β-lactams. Antimicrob Agents Chemother 22:942–948. doi: 10.1128/AAC.22.6.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Perilli M, Segatore B, Tavio M, Setacci D, Celenza G, De Santis F, Pellegrini C, Rossolini GM, Amicosante G. 2007. In vitro selection and characterization of mutants in TEM-1-producing Escherichia coli by ceftazidime and ceftibuten. J Chemother 19:123–126. doi: 10.1179/joc.2007.19.2.123. [DOI] [PubMed] [Google Scholar]
- 213.Reguera JA, Baquero F, Pérez-Diaz JC, Martínez JL. 1991. Factors determining resistance to β-lactam combined with β-lactamase inhibitors in Escherichia coli. J Antimicrob Chemother 27:569–575. doi: 10.1093/jac/27.5.569. [DOI] [PubMed] [Google Scholar]
- 214.Martínez-Martínez L, Conejo MC, Pascual A, Hernández-Allés S, Ballesta S, Ramirez De Arellano-Ramos E, Benedi VJ, Perea EJ. 2000. Activities of imipenem and cephalosporins against clonally related strains of Escherichia coli hyperproducing chromosomal β-lactamase and showing altered porin profiles. Antimicrob Agents Chemother 44:2534–2536. doi: 10.1128/AAC.44.9.2534-2536.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Tenover FC, Raney PM, Williams PP, Rasheed JK, Biddle JW, Oliver A, Fridkin SK, Jevitt L, McGowan JE Jr, Project ICARE. 2003. Evaluation of the NCCLS extended-spectrum β-lactamase confirmation methods for Escherichia coli with isolates collected during Project ICARE. J Clin Microbiol 41:3142–3146. doi: 10.1128/JCM.41.7.3142-3146.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Palasubramaniam S, Subramaniam G, Muniandy S, Parasakthi N. 2007. Extended-spectrum β-lactam resistance due to AmpC hyperproduction and CMY-2 coupled with the loss of OmpK35 in Malaysian strains of Escherichia coli and Klebsiella pneumoniae. Microb Drug Resist 13:186–190. doi: 10.1089/mdr.2007.726. [DOI] [PubMed] [Google Scholar]
- 217.Stapleton PD, Shannon KP, French GL. 1999. Carbapenem resistance in Escherichia coli associated with plasmid-determined CMY-4 β-lactamase production and loss of an outer membrane protein. Antimicrob Agents Chemother 43:1206–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Tangden T, Adler M, Cars O, Sandegren L, Lowdin E. 2013. Frequent emergence of porin-deficient subpopulations with reduced carbapenem susceptibility in ESBL-producing Escherichia coli during exposure to ertapenem in an in vitro pharmacokinetic model. J Antimicrob Chemother 68:1319–1326. doi: 10.1093/jac/dkt044. [DOI] [PubMed] [Google Scholar]
- 219.Masuda N, Sakagawa E, Ohya S, Gotoh N, Tsujimoto H, Nishino T. 2000. Contribution of the MexX-MexY-OprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 44:2242–2246. doi: 10.1128/AAC.44.9.2242-2246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Kothary V, Scherl EJ, Bosworth B, Jiang ZD, Dupont HL, Harel J, Simpson KW, Dogan B. 2013. Rifaximin resistance in Escherichia coli associated with inflammatory bowel disease correlates with prior rifaximin use, mutations in rpoB, and activity of Phe-Arg-β-naphthylamide-inhibitable efflux pumps. Antimicrob Agents Chemother 57:811–817. doi: 10.1128/AAC.02163-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Medeiros AA, O'Brien TF, Rosenberg EY, Nikaido H. 1987. Loss of OmpC porin in a strain of Salmonella typhimurium causes increased resistance to cephalosporins during therapy. J Infect Dis 156:751–757. doi: 10.1093/infdis/156.5.751. [DOI] [PubMed] [Google Scholar]
- 222.Nishino K, Latifi T, Groisman EA. 2006. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol Microbiol 59:126–141. doi: 10.1111/j.1365-2958.2005.04940.x. [DOI] [PubMed] [Google Scholar]
- 223.Nishino K, Nikaido E, Yamaguchi A. 2007. Regulation of multidrug efflux systems involved in multidrug and metal resistance of Salmonella enterica serovar Typhimurium. J Bacteriol 189:9066–9075. doi: 10.1128/JB.01045-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Nishino K, Hayashi-Nishino M, Yamaguchi A. 2009. H-NS modulates multidrug resistance of Salmonella enterica serovar Typhimurium by repressing multidrug efflux genes acrEF. Antimicrob Agents Chemother 53:3541–3543. doi: 10.1128/AAC.00371-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Piddock LJ, Griggs DJ, Hall MC, Jin YF. 1993. Ciprofloxacin resistance in clinical isolates of Salmonella typhimurium obtained from two patients. Antimicrob Agents Chemother 37:662–666. doi: 10.1128/AAC.37.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Chen S, Cui S, McDermott PF, Zhao S, White DG, Paulsen I, Meng J. 2007. Contribution of target gene mutations and efflux to decreased susceptibility of Salmonella enterica serovar Typhimurium to fluoroquinolones and other antimicrobials. Antimicrob Agents Chemother 51:535–542. doi: 10.1128/AAC.00600-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Baucheron S, Imberechts H, Chaslus-Dancla E, Cloeckaert A. 2002. The AcrB multidrug transporter plays a major role in high-level fluoroquinolone resistance in Salmonella enterica serovar Typhimurium phage type DT204. Microb Drug Resist 8:281–289. doi: 10.1089/10766290260469543. [DOI] [PubMed] [Google Scholar]
- 228.Lunn AD, Fabrega A, Sánchez-Cespedes J, Vila J. 2010. Prevalence of mechanisms decreasing quinolone-susceptibility among Salmonella spp. clinical isolates. Int Microbiol 13:15–20. doi: 10.2436/20.1501.01.107. [DOI] [PubMed] [Google Scholar]
- 229.O'Regan E, Quinn T, Pagès JM, McCusker M, Piddock L, Fanning S. 2009. Multiple regulatory pathways associated with high-level ciprofloxacin and multidrug resistance in Salmonella enterica serovar Enteritidis: involvement of ramA and other global regulators. Antimicrob Agents Chemother 53:1080–1087. doi: 10.1128/AAC.01005-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Smith AM, Govender N, Keddy KH, Group for Enteric, Respiratory and Meningeal Disease Surveillance in South Africa. 2010. Quinolone-resistant Salmonella Typhi in South Africa, 2003-2007. Epidemiol Infect 138:86–90. doi: 10.1017/S0950268809990331. [DOI] [PubMed] [Google Scholar]
- 231.Bergstrom S, Lindberg FP, Olsson O, Normark S. 1983. Comparison of the overlapping frd and ampC operons of Escherichia coli with the corresponding DNA sequences in other Gram-negative bacteria. J Bacteriol 155:1297–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Butaye P, Michael GB, Schwarz S, Barrett TJ, Brisabois A, White DG. 2006. The clonal spread of multidrug-resistant non-Typhi Salmonella serotypes. Microbes Infect 8:1891–1897. doi: 10.1016/j.micinf.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 233.Li X-Z, Mehrotra M, Ghimire S, Adewoye L. 2007. β-Lactam resistance and β-lactamases in bacteria of animal origin. Vet Microbiol 121:197–214. doi: 10.1016/j.vetmic.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 234.Su LH, Wu TL, Chiu CH. 2012. Development of carbapenem resistance during therapy for non-typhoid Salmonella infection. Clin Microbiol Infect 18:E91–E94. doi: 10.1111/j.1469-0691.2012.03767.x. [DOI] [PubMed] [Google Scholar]
- 235.Li L, Liao X, Yang Y, Sun J, Li L, Liu B, Yang S, Ma J, Li X, Zhang Q, Liu Y. 2013. Spread of oqxAB in Salmonella enterica serotype Typhimurium predominantly by IncHI2 plasmids. J Antimicrob Chemother 68:2263–2268. doi: 10.1093/jac/dkt209. [DOI] [PubMed] [Google Scholar]
- 236.Wong MH, Yan M, Chan EW, Biao K, Chen S. 2014. Emergence of clinical Salmonella enterica serovar Typhimurium isolates with concurrent resistance to ciprofloxacin, ceftriaxone, and azithromycin. Antimicrob Agents Chemother 58:3752–3756. doi: 10.1128/AAC.02770-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Armand-Lefevre L, Leflon-Guibout V, Bredin J, Barguellil F, Amor A, Pagès JM, Nicolas-Chanoine MH. 2003. Imipenem resistance in Salmonella enterica serovar Wien related to porin loss and CMY-4 β-lactamase production. Antimicrob Agents Chemother 47:1165–1168. doi: 10.1128/AAC.47.3.1165-1168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Hu WS, Chen HW, Zhang RY, Huang CY, Shen CF. 2011. The expression levels of outer membrane proteins STM1530 and OmpD, which are influenced by the CpxAR and BaeSR two-component systems, play important roles in the ceftriaxone resistance of Salmonella enterica serovar Typhimurium. Antimicrob Agents Chemother 55:3829–3837. doi: 10.1128/AAC.00216-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Lakey JH, Watts JP, Lea EJ. 1985. Characterisation of channels induced in planar bilayer membranes by detergent solubilised Escherichia coli porins. Biochim Biophys Acta 817:208–216. doi: 10.1016/0005-2736(85)90022-7. [DOI] [PubMed] [Google Scholar]
- 240.Navia MM, Ruiz J, Ribera A, de Anta MT, Vila J. 1999. Analysis of the mechanisms of quinolone resistance in clinical isolates of Citrobacter freundii. J Antimicrob Chemother 44:743–748. doi: 10.1093/jac/44.6.743. [DOI] [PubMed] [Google Scholar]
- 241.Tavio M, Vila J, Ruiz J, Amicosante G, Franceschini N, Martin-Sánchez AM, de Anta MT. 2000. In vitro selected fluoroquinolone-resistant mutants of Citrobacter freundii: analysis of the quinolone resistance acquisition. J Antimicrob Chemother 45:521–524. doi: 10.1093/jac/45.4.521. [DOI] [PubMed] [Google Scholar]
- 242.Schumacher A, Trittler R, Bohnert JA, Kummerer K, Pagès J-M, Kern WV. 2007. Intracellular accumulation of linezolid in Escherichia coli, Citrobacter freundii and Enterobacter aerogenes: role of enhanced efflux pump activity and inactivation. J Antimicrob Chemother 59:1261–1264. doi: 10.1093/jac/dkl380. [DOI] [PubMed] [Google Scholar]
- 243.Dolejska M, Villa L, Poirel L, Nordmann P, Carattoli A. 2013. Complete sequencing of an IncHI1 plasmid encoding the carbapenemase NDM-1, the ArmA 16S RNA methylase and a resistance-nodulation-cell division/multidrug efflux pump. J Antimicrob Chemother 68:34–39. doi: 10.1093/jac/dks357. [DOI] [PubMed] [Google Scholar]
- 244.Vu H, Nikaido H. 1985. Role of β-lactam hydrolysis in the mechanism of resistance of a β-lactamase-constitutive Enterobacter cloacae strain to expanded-spectrum β-lactams. Antimicrob Agents Chemother 27:393–398. doi: 10.1128/AAC.27.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Masi M, Pagès JM, Villard C, Pradel E. 2005. The eefABC multidrug efflux pump operon is repressed by H-NS in Enterobacter aerogenes. J Bacteriol 187:3894–3897. doi: 10.1128/JB.187.11.3894-3897.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Charrel RN, Pagès JM, De Micco P, Mallea M. 1996. Prevalence of outer membrane porin alteration in β-lactam-antibiotic-resistant Enterobacter aerogenes. Antimicrob Agents Chemother 40:2854–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Lee EH, Nicolas MH, Kitzis MD, Pialoux G, Collatz E, Gutmann L. 1991. Association of two resistance mechanisms in a clinical isolate of Enterobacter cloacae with high-level resistance to imipenem. Antimicrob Agents Chemother 35:1093–1098. doi: 10.1128/AAC.35.6.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Lakaye B, Dubus A, Joris B, Frère JM. 2002. Method for estimation of low outer membrane permeability to β-lactam antibiotics. Antimicrob Agents Chemother 46:2901–2907. doi: 10.1128/AAC.46.9.2901-2907.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Nikaido H, Pagès JM. 2012. Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol Rev 36:340–363. doi: 10.1111/j.1574-6976.2011.00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Bornet C, Saint N, Fetnaci L, Dupont M, Davin-Regli A, Bollet C, Pagès JM. 2004. Omp35, a new Enterobacter aerogenes porin involved in selective susceptibility to cephalosporins. Antimicrob Agents Chemother 48:2153–2158. doi: 10.1128/AAC.48.6.2153-2158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Szabo D, Silveira F, Hujer AM, Bonomo RA, Hujer KM, Marsh JW, Bethel CR, Doi Y, Deeley K, Paterson DL. 2006. Outer membrane protein changes and efflux pump expression together may confer resistance to ertapenem in Enterobacter cloacae. Antimicrob Agents Chemother 50:2833–2835. doi: 10.1128/AAC.01591-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Köhler T, Michea-Hamzehpour M, Epp SF, Pechère JC. 1999. Carbapenem activities against Pseudomonas aeruginosa: respective contributions of OprD and efflux systems. Antimicrob Agents Chemother 43:424–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Fung-Tomc JC, Gradelski E, Huczko E, Dougherty TJ, Kessler RE, Bonner DP. 1996. Differences in the resistant variants of Enterobacter cloacae selected by extended-spectrum cephalosporins. Antimicrob Agents Chemother 40:1289–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Piddock LJ, Traynor EA. 1991. β-Lactamase expression and outer membrane protein changes in cefpirome-resistant and ceftazidime-resistant Gram-negative bacteria. J Antimicrob Chemother 28:209–219. doi: 10.1093/jac/28.2.209. [DOI] [PubMed] [Google Scholar]
- 255.Yang FC, Yan JJ, Hung KH, Wu JJ. 2012. Characterization of ertapenem-resistant Enterobacter cloacae in a Taiwanese university hospital. J Clin Microbiol 50:223–226. doi: 10.1128/JCM.01263-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Doumith M, Ellington MJ, Livermore DM, Woodford N. 2009. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J Antimicrob Chemother 63:659–667. doi: 10.1093/jac/dkp029. [DOI] [PubMed] [Google Scholar]
- 257.Yigit H, Anderson GJ, Biddle JW, Steward CD, Rasheed JK, Valera LL, McGowan JE Jr, Tenover FC. 2002. Carbapenem resistance in a clinical isolate of Enterobacter aerogenes is associated with decreased expression of OmpF and OmpC porin analogs. Antimicrob Agents Chemother 46:3817–3822. doi: 10.1128/AAC.46.12.3817-3822.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Lavigne JP, Sotto A, Nicolas-Chanoine MH, Bouziges N, Pagès JM, Davin-Regli A. 2013. An adaptive response of Enterobacter aerogenes to imipenem: regulation of porin balance in clinical isolates. Int J Antimicrob Agents 41:130–136. doi: 10.1016/j.ijantimicag.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 259.De E, Basle A, Jaquinod M, Saint N, Mallea M, Molle G, Pagès JM. 2001. A new mechanism of antibiotic resistance in Enterobacteriaceae induced by a structural modification of the major porin. Mol Microbiol 41:189–198. doi: 10.1046/j.1365-2958.2001.02501.x. [DOI] [PubMed] [Google Scholar]
- 260.Gayet S, Chollet R, Molle G, Pagès JM, Chevalier J. 2003. Modification of outer membrane protein profile and evidence suggesting an active drug pump in Enterobacter aerogenes clinical strains. Antimicrob Agents Chemother 47:1555–1559. doi: 10.1128/AAC.47.5.1555-1559.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Pradel E, Pagès JM. 2002. The AcrAB-TolC efflux pump contributes to multidrug resistance in the nosocomial pathogen Enterobacter aerogenes. Antimicrob Agents Chemother 46:2640–2643. doi: 10.1128/AAC.46.8.2640-2643.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Chevalier J, Mulfinger C, Garnotel E, Nicolas P, Davin-Regli A, Pagès JM. 2008. Identification and evolution of drug efflux pump in clinical Enterobacter aerogenes strains isolated in 1995 and 2003. PLoS One 3:e3203. doi: 10.1371/journal.pone.0003203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Glatz K, Toth A, Paszti J. 2011. The cyclohexane tolerance and Phe-Arg-β-naphtylamide susceptibility of multidrug-resistant Enterobacter cloacae clinical isolates, and the predominance of one PFGE clone in Hungary. Clin Microbiol Infect 17:1254–1261. doi: 10.1111/j.1469-0691.2010.03408.x. [DOI] [PubMed] [Google Scholar]
- 264.Pérez A, Poza M, Fernández A, del Carmen Fernández M, Mallo S, Merino M, Rumbo-Feal S, Cabral MP, Bou G. 2012. Involvement of the AcrAB-TolC efflux pump in the resistance, fitness, and virulence of Enterobacter cloacae. Antimicrob Agents Chemother 56:2084–2090. doi: 10.1128/AAC.05509-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Tran QT, Dupont M, Lavigne JP, Chevalier J, Pagès JM, Sotto A, Davin-Regli A. 2009. Occurrence of efflux mechanism and cephalosporinase variant in a population of Enterobacter aerogenes and Klebsiella pneumoniae isolates producing extended-spectrum β-lactamases. Antimicrob Agents Chemother 53:1652–1656. doi: 10.1128/AAC.00822-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Chollet R, Chevalier J, Bryskier A, Pagès JM. 2004. The AcrAB-TolC pump is involved in macrolide resistance but not in telithromycin efflux in Enterobacter aerogenes and Escherichia coli. Antimicrob Agents Chemother 48:3621–3624. doi: 10.1128/AAC.48.9.3621-3624.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Hornsey M, Ellington MJ, Doumith M, Scott G, Livermore DM, Woodford N. 2010. Emergence of AcrAB-mediated tigecycline resistance in a clinical isolate of Enterobacter cloacae during ciprofloxacin treatment. Int J Antimicrob Agents 35:478–481. doi: 10.1016/j.ijantimicag.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 268.Keeney D, Ruzin A, Bradford PA. 2007. RamA, a transcriptional regulator, and AcrAB, an RND-type efflux pump, are associated with decreased susceptibility to tigecycline in Enterobacter cloacae. Microb Drug Resist 13:1–6. doi: 10.1089/mdr.2006.9990. [DOI] [PubMed] [Google Scholar]
- 269.Veleba M, De Majumdar S, Hornsey M, Woodford N, Schneiders T. 2013. Genetic characterization of tigecycline resistance in clinical isolates of Enterobacter cloacae and Enterobacter aerogenes. J Antimicrob Chemother 68:1011–1018. doi: 10.1093/jac/dks530. [DOI] [PubMed] [Google Scholar]
- 270.Dutzler R, Rummel G, Alberti S, Hernández-Allés S, Phale P, Rosenbusch J, Benedi V, Schirmer T. 1999. Crystal structure and functional characterization of OmpK36, the osmoporin of Klebsiella pneumoniae. Structure 7:425–434. doi: 10.1016/S0969-2126(99)80055-0. [DOI] [PubMed] [Google Scholar]
- 271.Doménech-Sánchez A, Martínez-Martínez L, Hernández-Allés S, del Carmen Conejo M, Pascual A, Tomás JM, Alberti S, Benedi VJ. 2003. Role of Klebsiella pneumoniae OmpK35 porin in antimicrobial resistance. Antimicrob Agents Chemother 47:3332–3335. doi: 10.1128/AAC.47.10.3332-3335.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Padilla E, Llobet E, Doménech-Sánchez A, Martínez-Martínez L, Bengoechea JA, Alberti S. 2010. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob Agents Chemother 54:177–183. doi: 10.1128/AAC.00715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Gutmann L, Williamson R, Moreau N, Kitzis MD, Collatz E, Acar JF, Goldstein FW. 1985. Cross-resistance to nalidixic acid, trimethoprim, and chloramphenicol associated with alterations in outer membrane proteins of Klebsiella, Enterobacter, and Serratia. J Infect Dis 151:501–507. doi: 10.1093/infdis/151.3.501. [DOI] [PubMed] [Google Scholar]
- 274.Pangon B, Bizet C, Bure A, Pichon F, Philippon A, Regnier B, Gutmann L. 1989. In vivo selection of a cephamycin-resistant, porin-deficient mutant of Klebsiella pneumoniae producing a TEM-3 β-lactamase. J Infect Dis 159:1005–1006. doi: 10.1093/infdis/159.5.1005. [DOI] [PubMed] [Google Scholar]
- 275.Rice LB, Carias LL, Etter L, Shlaes DM. 1993. Resistance to cefoperazone-sulbactam in Klebsiella pneumoniae: evidence for enhanced resistance resulting from the coexistence of two different resistance mechanisms. Antimicrob Agents Chemother 37:1061–1064. doi: 10.1128/AAC.37.5.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Chen HY, Livermore DM. 1993. Activity of cefepime and other β-lactam antibiotics against permeability mutants of Escherichia coli and Klebsiella pneumoniae. J Antimicrob Chemother 32(Suppl B):63–74. [DOI] [PubMed] [Google Scholar]
- 277.Martínez-Martínez L, Hernández-Allés S, Alberti S, Tomás JM, Benedi VJ, Jacoby GA. 1996. In vivo selection of porin-deficient mutants of Klebsiella pneumoniae with increased resistance to cefoxitin and expanded-spectrum cephalosporins. Antimicrob Agents Chemother 40:342–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Ardanuy C, Linares J, Dominguez MA, Hernández-Allés S, Benedi VJ, Martínez-Martínez L. 1998. Outer membrane profiles of clonally related Klebsiella pneumoniae isolates from clinical samples and activities of cephalosporins and carbapenems. Antimicrob Agents Chemother 42:1636–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Hernández-Allés S, Benedi VJ, Martínez-Martínez L, Pascual A, Aguilar A, Tomás JM, Alberti S. 1999. Development of resistance during antimicrobial therapy caused by insertion sequence interruption of porin genes. Antimicrob Agents Chemother 43:937–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Nelson EC, Segal H, Elisha BG. 2003. Outer membrane protein alterations and blaTEM-1 variants: their role in β-lactam resistance in Klebsiella pneumoniae. J Antimicrob Chemother 52:899–903. doi: 10.1093/jac/dkg486. [DOI] [PubMed] [Google Scholar]
- 281.Doménech-Sánchez A, Pascual A, Suarez AI, Alvarez D, Benedi VJ, Martínez-Martínez L. 2000. Activity of nine antimicrobial agents against clinical isolates of Klebsiella pneumoniae producing extended-spectrum β-lactamases and deficient or not in porins. J Antimicrob Chemother 46:858–859. doi: 10.1093/jac/46.5.858. [DOI] [PubMed] [Google Scholar]
- 282.Jacoby GA, Mills DM, Chow N. 2004. Role of β-lactamases and porins in resistance to ertapenem and other β-lactams in Klebsiella pneumoniae. Antimicrob Agents Chemother 48:3203–3206. doi: 10.1128/AAC.48.8.3203-3206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Poulou A, Voulgari E, Vrioni G, Koumaki V, Xidopoulos G, Chatzipantazi V, Markou F, Tsakris A. 2013. Outbreak caused by an ertapenem-resistant, CTX-M-15-producing Klebsiella pneumoniae sequence type 101 clone carrying an OmpK36 porin variant. J Clin Microbiol 51:3176–3182. doi: 10.1128/JCM.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Hernández-Allés S, Conejo M, Pascual A, Tomás JM, Benedi VJ, Martínez-Martínez L. 2000. Relationship between outer membrane alterations and susceptibility to antimicrobial agents in isogenic strains of Klebsiella pneumoniae. J Antimicrob Chemother 46:273–277. doi: 10.1093/jac/46.2.273. [DOI] [PubMed] [Google Scholar]
- 285.Zhang Y, Jiang X, Wang Y, Li G, Tian Y, Liu H, Ai F, Ma Y, Wang B, Ruan F, Rajakumar K. 2014. Contribution of β-lactamases and porin proteins OmpK35 and OmpK36 to carbapenem resistance in clinical isolates of KPC-2-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 58:1214–1217. doi: 10.1128/AAC.02045-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Rasheed JK, Anderson GJ, Yigit H, Queenan AM, Doménech-Sánchez A, Swenson JM, Biddle JW, Ferraro MJ, Jacoby GA, Tenover FC. 2000. Characterization of the extended-spectrum β-lactamase reference strain, Klebsiella pneumoniae K6 (ATCC 700603), which produces the novel enzyme SHV-18. Antimicrob Agents Chemother 44:2382–2388. doi: 10.1128/AAC.44.9.2382-2388.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Garcia-Fernández A, Miriagou V, Papagiannitsis CC, Giordano A, Venditti M, Mancini C, Carattoli A. 2010. An ertapenem-resistant extended-spectrum-β-lactamase-producing Klebsiella pneumoniae clone carries a novel OmpK36 porin variant. Antimicrob Agents Chemother 54:4178–4184. doi: 10.1128/AAC.01301-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Wozniak A, Villagra NA, Undabarrena A, Gallardo N, Keller N, Moraga M, Roman JC, Mora GC, Garcia P. 2012. Porin alterations present in non-carbapenemase-producing Enterobacteriaceae with high and intermediate levels of carbapenem resistance in Chile. J Med Microbiol 61:1270–1279. doi: 10.1099/jmm.0.045799-0. [DOI] [PubMed] [Google Scholar]
- 289.Garcia-Sureda L, Juan C, Doménech-Sánchez A, Alberti S. 2011. Role of Klebsiella pneumoniae LamB porin in antimicrobial resistance. Antimicrob Agents Chemother 55:1803–1805. doi: 10.1128/AAC.01441-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Ramos PI, Picao RC, Almeida LG, Lima NC, Girardello R, Vivan AC, Xavier DE, Barcellos FG, Pelisson M, Vespero EC, Medigue C, Vasconcelos AT, Gales AC, Nicolas MF. 2014. Comparative analysis of the complete genome of KPC-2-producing Klebsiella pneumoniae Kp13 reveals remarkable genome plasticity and a wide repertoire of virulence and resistance mechanisms. BMC Genomics 15:54. doi: 10.1186/1471-2164-15-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Mazzariol A, Zuliani J, Cornaglia G, Rossolini GM, Fontana R. 2002. AcrAB efflux system: expression and contribution to fluoroquinolone resistance in Klebsiella spp. Antimicrob Agents Chemother 46:3984–3986. doi: 10.1128/AAC.46.12.3984-3986.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Schneiders T, Amyes SG, Levy SB. 2003. Role of AcrR and ramA in fluoroquinolone resistance in clinical Klebsiella pneumoniae isolates from Singapore. Antimicrob Agents Chemother 47:2831–2837. doi: 10.1128/AAC.47.9.2831-2837.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Chevalier J, Pagès JM, Eyraud A, Mallea M. 2000. Membrane permeability modifications are involved in antibiotic resistance in Klebsiella pneumoniae. Biochem Biophys Res Commun 274:496–499. doi: 10.1006/bbrc.2000.3159. [DOI] [PubMed] [Google Scholar]
- 294.Gruteke P, Goessens W, Van Gils J, Peerbooms P, Lemmens-Den Toom N, Van Santen-Verheuvel M, Van Belkum A, Verbrugh H. 2003. Patterns of resistance associated with integrons, the extended-spectrum β-lactamase SHV-5 gene, and a multidrug efflux pump of Klebsiella pneumoniae causing a nosocomial outbreak. J Clin Microbiol 41:1161–1166. doi: 10.1128/JCM.41.3.1161-1166.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Martínez-Martínez L, Pascual A, del Carmen Conejo M, Garcia I, Joyanes P, Doménech-Sánchez A, Benedi VJ. 2002. Energy-dependent accumulation of norfloxacin and porin expression in clinical isolates of Klebsiella pneumoniae and relationship to extended-spectrum β-lactamase production. Antimicrob Agents Chemother 46:3926–3932. doi: 10.1128/AAC.46.12.3926-3932.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Pagès JM, Lavigne JP, Leflon-Guibout V, Marcon E, Bert F, Noussair L, Nicolas-Chanoine MH. 2009. Efflux pump, the masked side of β-lactam resistance in Klebsiella pneumoniae clinical isolates. PLoS One 4:e4817. doi: 10.1371/journal.pone.0004817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Källman O, Motakefi A, Wretlind B, Kalin M, Olsson-Liljequist B, Giske CG. 2008. Cefuroxime non-susceptibility in multidrug-resistant Klebsiella pneumoniae overexpressing ramA and acrA and expressing ompK35 at reduced levels. J Antimicrob Chemother 62:986–990. doi: 10.1093/jac/dkn296. [DOI] [PubMed] [Google Scholar]
- 298.Källman O, Giske CG, Samuelsen O, Wretlind B, Kalin M, Olsson-Liljequist B. 2009. Interplay of efflux, impermeability, and AmpC activity contributes to cefuroxime resistance in clinical, non-ESBL-producing isolates of Escherichia coli. Microb Drug Resist 15:91–95. doi: 10.1089/mdr.2009.0883. [DOI] [PubMed] [Google Scholar]
- 299.Bialek-Davenet S, Marcon E, Leflon-Guibout V, Lavigne JP, Bert F, Moreau R, Nicolas-Chanoine MH. 2011. In vitro selection of ramR and soxR mutants overexpressing efflux systems by fluoroquinolones as well as cefoxitin in Klebsiella pneumoniae. Antimicrob Agents Chemother 55:2795–2802. doi: 10.1128/AAC.00156-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Villa L, Feudi C, Fortini D, Garcia-Fernández A, Carattoli A. 2014. Genomics of KPC-producing Klebsiella pneumoniae sequence type 512 clone highlights the role of RamR and ribosomal S10 protein mutations in conferring tigecycline resistance. Antimicrob Agents Chemother 58:1707–1712. doi: 10.1128/AAC.01803-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Sheng ZK, Hu F, Wang W, Guo Q, Chen Z, Xu X, Zhu D, Wang M. 2014. Mechanisms of tigecycline resistance among Klebsiella pneumoniae clinical isolates. Antimicrob Agents Chemother 58:6982–6985. doi: 10.1128/AAC.03808-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Kim HB, Wang M, Park CH, Kim EC, Jacoby GA, Hooper DC. 2009. oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae. Antimicrob Agents Chemother 53:3582–3584. doi: 10.1128/AAC.01574-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Pérez F, Rudin SD, Marshall SH, Coakley P, Chen L, Kreiswirth BN, Rather PN, Hujer AM, Toltzis P, van Duin D, Paterson DL, Bonomo RA. 2013. OqxAB, a quinolone and olaquindox efflux pump, is widely distributed among multidrug-resistant Klebsiella pneumoniae isolates of human origin. Antimicrob Agents Chemother 57:4602–4603. doi: 10.1128/AAC.00725-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Kumar V, Sun P, Vamathevan J, Li Y, Ingraham K, Palmer L, Huang J, Brown JR. 2011. Comparative genomics of Klebsiella pneumoniae strains with different antibiotic resistance profiles. Antimicrob Agents Chemother 55:4267–4276. doi: 10.1128/AAC.00052-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Nielsen LE, Snesrud EC, Onmus-Leone F, Kwak YI, Aviles R, Steele ED, Sutter DE, Waterman PE, Lesho EP. 2014. IS5 element integration, a novel mechanism for rapid in vivo emergence of tigecycline nonsusceptibility in Klebsiella pneumoniae. Antimicrob Agents Chemother 58:6151–6156. doi: 10.1128/AAC.03053-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Ogawa W, Onishi M, Ni R, Tsuchiya T, Kuroda T. 2012. Functional study of the novel multidrug efflux pump KexD from Klebsiella pneumoniae. Gene 498:177–182. doi: 10.1016/j.gene.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 307.Srinivasan VB, Singh BB, Priyadarshi N, Chauhan NK, Rajamohan G. 2014. Role of novel multidrug efflux pump involved in drug resistance in Klebsiella pneumoniae. PLoS One 9:e96288. doi: 10.1371/journal.pone.0096288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Mitsuyama J, Hiruma R, Yamaguchi A, Sawai T. 1987. Identification of porins in outer membrane of Proteus, Morganella, and Providencia spp. and their role in outer membrane permeation of β-lactams. Antimicrob Agents Chemother 31:379–384. doi: 10.1128/AAC.31.3.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Tavio MM, Vila J, Ruiz J, Martin Sánchez AM, Jimenez de Anta MT. 2000. Decreased permeability and enhanced proton-dependent active efflux in the development of resistance to quinolones in Morganella morganii. Int J Antimicrob Agents 14:157–160. doi: 10.1016/S0924-8579(00)00117-5. [DOI] [PubMed] [Google Scholar]
- 310.Visalli MA, Murphy E, Projan SJ, Bradford PA. 2003. AcrAB multidrug efflux pump is associated with reduced levels of susceptibility to tigecycline (GAR-936) in Proteus mirabilis. Antimicrob Agents Chemother 47:665–669. doi: 10.1128/AAC.47.2.665-669.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Saito R, Sato K, Kumita W, Inami N, Nishiyama H, Okamura N, Moriya K, Koike K. 2006. Role of type II topoisomerase mutations and AcrAB efflux pump in fluoroquinolone-resistant clinical isolates of Proteus mirabilis. J Antimicrob Chemother 58:673–677. doi: 10.1093/jac/dkl297. [DOI] [PubMed] [Google Scholar]
- 312.Ruzin A, Keeney D, Bradford PA. 2005. AcrAB efflux pump plays a role in decreased susceptibility to tigecycline in Morganella morganii. Antimicrob Agents Chemother 49:791–793. doi: 10.1128/AAC.49.2.791-793.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Mitsuyama J, Itoh Y, Takahata M, Okamoto S, Yasuda T. 1992. In vitro antibacterial activities of tosufloxacin against and uptake of tosufloxacin by outer membrane mutants of Escherichia coli, Proteus mirabilis, and Salmonella typhimurium. Antimicrob Agents Chemother 36:2030–2036. doi: 10.1128/AAC.36.9.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Kadavy DR, Hornby JM, Haverkost T, Nickerson KW. 2000. Natural antibiotic resistance of bacteria isolated from larvae of the oil fly, Helaeomyia petrolei. Appl Environ Microbiol 66:4615–4619. doi: 10.1128/AEM.66.11.4615-4619.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Hutsul JA, Worobec E. 1997. Molecular characterization of the Serratia marcescens OmpF porin, and analysis of S. marcescens OmpF and OmpC osmoregulation. Microbiology 143:2797–2806. [DOI] [PubMed] [Google Scholar]
- 316.Gutmann L, Chabbert YA. 1984. Different mechanisms of resistance to latamoxef (moxalactam) in Serratia marcescens. J Antimicrob Chemother 13:15–22. doi: 10.1093/jac/13.1.15. [DOI] [PubMed] [Google Scholar]
- 317.Weindorf H, Schmidt H, Martin HH. 1998. Contribution of overproduced chromosomal β-lactamase and defective outer membrane porins to resistance to extended-spectrum β-lactam antibiotics in Serratia marcescens. J Antimicrob Chemother 41:189–195. doi: 10.1093/jac/41.2.189. [DOI] [PubMed] [Google Scholar]
- 318.Ruiz N, Montero T, Hernandez-Borrell J, Vinas M. 2003. The role of Serratia marcescens porins in antibiotic resistance. Microb Drug Resist 9:257–264. doi: 10.1089/107662903322286463. [DOI] [PubMed] [Google Scholar]
- 319.Berlanga M, Vinas M. 2000. Salicylate induction of phenotypic resistance to quinolones in Serratia marcescens. J Antimicrob Chemother 46:279–282. doi: 10.1093/jac/46.2.279. [DOI] [PubMed] [Google Scholar]
- 320.Kumar A, Worobec EA. 2005. Cloning, sequencing, and characterization of the SdeAB multidrug efflux pump of Serratia marcescens. Antimicrob Agents Chemother 49:1495–1501. doi: 10.1128/AAC.49.4.1495-1501.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Begic S, Worobec EA. 2008. The role of the Serratia marcescens SdeAB multidrug efflux pump and TolC homologue in fluoroquinolone resistance studied via gene-knockout mutagenesis. Microbiology 154:454–461. doi: 10.1099/mic.0.2007/012427-0. [DOI] [PubMed] [Google Scholar]
- 322.Maseda H, Hashida Y, Konaka R, Shirai A, Kourai H. 2009. Mutational upregulation of a resistance-nodulation-cell division-type multidrug efflux pump, SdeAB, upon exposure to a biocide, cetylpyridinium chloride, and antibiotic resistance in Serratia marcescens. Antimicrob Agents Chemother 53:5230–5235. doi: 10.1128/AAC.00631-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Maseda H, Hashida Y, Shirai A, Omasa T, Nakae T. 2011. Mutation in the sdeS gene promotes expression of the sdeAB efflux pump genes and multidrug resistance in Serratia marcescens. Antimicrob Agents Chemother 55:2922–2926. doi: 10.1128/AAC.01755-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Hornsey M, Ellington MJ, Doumith M, Hudson S, Livermore DM, Woodford N. 2010. Tigecycline resistance in Serratia marcescens associated with up-regulation of the SdeXY-HasF efflux system also active against ciprofloxacin and cefpirome. J Antimicrob Chemother 65:479–482. doi: 10.1093/jac/dkp475. [DOI] [PubMed] [Google Scholar]
- 325.Dalvi SD, Worobec EA. 2012. Gene expression analysis of the SdeAB multidrug efflux pump in antibiotic-resistant clinical isolates of Serratia marcescens. Indian J Med Microbiol 30:302–307. doi: 10.4103/0255-0857.99491. [DOI] [PubMed] [Google Scholar]
- 326.Yang H, Duan G, Zhu J, Lv R, Xi Y, Zhang W, Fan Q, Zhang M. 2008. The AcrAB-TolC pump is involved in multidrug resistance in clinical Shigella flexneri isolates. Microb Drug Resist 14:245–249. doi: 10.1089/mdr.2008.0847. [DOI] [PubMed] [Google Scholar]
- 327.Azmi IJ, Khajanchi BK, Akter F, Hasan TN, Shahnaij M, Akter M, Banik A, Sultana H, Hossain MA, Ahmed MK, Faruque SM, Talukder KA. 2014. Fluoroquinolone resistance mechanisms of Shigella flexneri isolated in Bangladesh. PLoS One 9:e102533. doi: 10.1371/journal.pone.0102533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Kim JY, Jeon SM, Kim H, Park MS, Kim SH. 2011. A contribution of MdfA to resistance to fluoroquinolones in Shigella flexneri. Osong Public Health Res Perspect 2:216–217. doi: 10.1016/j.phrp.2011.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Brzostek K, Nichols WW. 1990. Outer membrane permeability and porin proteins of Yersinia enterocolitica. FEMS Microbiol Lett 58:275–277. [DOI] [PubMed] [Google Scholar]
- 330.Capilla S, Ruiz J, Goni P, Castillo J, Rubio MC, Jimenez de Anta MT, Gomez-Lus R, Vila J. 2004. Characterization of the molecular mechanisms of quinolone resistance in Yersinia enterocolitica O:3 clinical isolates. J Antimicrob Chemother 53:1068–1071. doi: 10.1093/jac/dkh225. [DOI] [PubMed] [Google Scholar]
- 331.Udani RA, Levy SB. 2006. MarA-like regulator of multidrug resistance in Yersinia pestis. Antimicrob Agents Chemother 50:2971–2975. doi: 10.1128/AAC.00015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Stirrett KL, Ferreras JA, Rossi SM, Moy RL, Fonseca FV, Quadri LE. 2008. A multicopy suppressor screening approach as a means to identify antibiotic resistance determinant candidates in Yersinia pestis. BMC Microbiol 8:122. doi: 10.1186/1471-2180-8-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Lister IM, Raftery C, Mecsas J, Levy SB. 2012. Yersinia pestis AcrAB-TolC in antibiotic resistance and virulence. Antimicrob Agents Chemother 56:1120–1123. doi: 10.1128/AAC.05338-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Bengoechea JA, Skurnik M. 2000. Temperature-regulated efflux pump/potassium antiporter system mediates resistance to cationic antimicrobial peptides in Yersinia. Mol Microbiol 37:67–80. doi: 10.1046/j.1365-2958.2000.01956.x. [DOI] [PubMed] [Google Scholar]
- 335.Paul S, Chaudhuri K, Chatterjee AN, Das J. 1992. Presence of exposed phospholipids in the outer membrane of Vibrio cholerae. J Gen Microbiol 138:755–761. doi: 10.1099/00221287-138-4-755. [DOI] [PubMed] [Google Scholar]
- 336.Dalsgaard A, Serichantalergs O, Pitarangsi C, Echeverria P. 1995. Molecular characterization and antibiotic susceptibility of Vibrio cholerae non-O1. Epidemiol Infect 114:51–63. doi: 10.1017/S0950268800051906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Benz R, Maier E, Chakraborty T. 1997. Purification of OmpU from Vibrio cholerae classical strain 569B: evidence for the formation of large cation-selective ion-permeable channels by OmpU. Microbiologia 13:321–330. [PubMed] [Google Scholar]
- 338.Chakrabarti SR, Chaudhuri K, Sen K, Das J. 1996. Porins of Vibrio cholerae: purification and characterization of OmpU. J Bacteriol 178:524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Pagel M, Delcour AH. 2011. Effects of conjugated and unconjugated bile acids on the activity of the Vibrio cholerae porin OmpT. Mol Membr Biol 28:69–78. doi: 10.3109/09687688.2010.519727. [DOI] [PubMed] [Google Scholar]
- 340.Bina JE, Provenzano D, Wang C, Bina XR, Mekalanos JJ. 2006. Characterization of the Vibrio cholerae vexAB and vexCD efflux systems. Arch Microbiol 186:171–181. doi: 10.1007/s00203-006-0133-5. [DOI] [PubMed] [Google Scholar]
- 341.Bina XR, Provenzano D, Nguyen N, Bina JE. 2008. Vibrio cholerae RND family efflux systems are required for antimicrobial resistance, optimal virulence factor production, and colonization of the infant mouse small intestine. Infect Immun 76:3595–3605. doi: 10.1128/IAI.01620-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Cerda-Maira FA, Ringelberg CS, Taylor RK. 2008. The bile response repressor, BreR, regulates expression of the Vibrio cholerae breAB efflux system operon. J Bacteriol 190:7441–7452. doi: 10.1128/JB.00584-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Rahman MM, Matsuo T, Ogawa W, Koterasawa M, Kuroda T, Tsuchiya T. 2007. Molecular cloning and characterization of all RND-type efflux transporters in Vibrio cholerae non-O1. Microbiol Immunol 51:1061–1070. doi: 10.1111/j.1348-0421.2007.tb04001.x. [DOI] [PubMed] [Google Scholar]
- 344.Taylor DL, Bina XR, Bina JE. 2012. Vibrio cholerae VexH encodes a multiple drug efflux pump that contributes to the production of cholera toxin and the toxin co-regulated pilus. PLoS One 7:e38208. doi: 10.1371/journal.pone.0038208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Taylor DL, Bina XR, Slamti L, Waldor MK, Bina JE. 2014. Reciprocal regulation of RND efflux systems and the Cpx two-component system in Vibrio cholerae. Infect Immun 82:2980–2991. doi: 10.1128/IAI.00025-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Bina XR, Philippart JA, Bina JE. 2009. Effect of the efflux inhibitors 1-(1-naphthylmethyl)-piperazine and phenyl-arginine-β-naphthylamide on antimicrobial susceptibility and virulence factor production in Vibrio cholerae. J Antimicrob Chemother 63:103–108. doi: 10.1093/jac/dkn466. [DOI] [PubMed] [Google Scholar]
- 347.Matsuo T, Hayashi K, Morita Y, Koterasawa M, Ogawa W, Mizushima T, Tsuchiya T, Kuroda T. 2007. VmeAB, an RND-type multidrug efflux transporter in Vibrio parahaemolyticus. Microbiology 153:4129–4137. doi: 10.1099/mic.0.2007/009597-0. [DOI] [PubMed] [Google Scholar]
- 348.Matsuo T, Nakamura K, Kodama T, Mikami T, Hiyoshi H, Tsuchiya T, Ogawa W, Kuroda T. 2013. Characterization of all RND-type multidrug efflux transporters in Vibrio parahaemolyticus. Microbiologyopen 2:725–742. doi: 10.1002/mbo3.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Matsuo T, Ogawa W, Tsuchiya T, Kuroda T. 2014. Overexpression of vmeTUV encoding a multidrug efflux transporter of Vibrio parahaemolyticus causes bile acid resistance. Gene 541:19–25. doi: 10.1016/j.gene.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 350.Colmer JA, Fralick JA, Hamood AN. 1998. Isolation and characterization of a putative multidrug resistance pump from Vibrio cholerae. Mol Microbiol 27:63–72. doi: 10.1046/j.1365-2958.1998.00657.x. [DOI] [PubMed] [Google Scholar]
- 351.Smith KP, Kumar S, Varela MF. 2009. Identification, cloning, and functional characterization of EmrD-3, a putative multidrug efflux pump of the major facilitator superfamily from Vibrio cholerae O395. Arch Microbiol 191:903–911. doi: 10.1007/s00203-009-0521-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Chen S, Wang H, Katzianer DS, Zhong Z, Zhu J. 2013. LysR family activator-regulated major facilitator superfamily transporters are involved in Vibrio cholerae antimicrobial compound resistance and intestinal colonisation. Int J Antimicrob Agents 41:188–192. doi: 10.1016/j.ijantimicag.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 353.Morita Y, Kodama K, Shiota S, Mine T, Kataoka A, Mizushima T, Tsuchiya T. 1998. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob Agents Chemother 42:1778–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Mohanty P, Patel A, Kushwaha Bhardwaj A. 2012. Role of H- and D-MATE-type transporters from multidrug resistant clinical isolates of Vibrio fluvialis in conferring fluoroquinolone resistance. PLoS One 7:e35752. doi: 10.1371/journal.pone.0035752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Lutwyche P, Exner MM, Hancock RE, Trust TJ. 1995. A conserved Aeromonas salmonicida porin provides protective immunity to rainbow trout. Infect Immun 63:3137–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Cruz A, Micaelo N, Felix V, Song JY, Kitamura S, Suzuki S, Mendo S. 2013. sugE: a gene involved in tributyltin (TBT) resistance of Aeromonas molluscorum Av27. J Gen Appl Microbiol 59:39–47. doi: 10.2323/jgam.59.47. [DOI] [PubMed] [Google Scholar]
- 357.Alcaide E, Blasco MD, Esteve C. 2010. Mechanisms of quinolone resistance in Aeromonas species isolated from humans, water and eels. Res Microbiol 161:40–45. doi: 10.1016/j.resmic.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 358.Gabay JE, Blake M, Niles WD, Horwitz MA. 1985. Purification of Legionella pneumophila major outer membrane protein and demonstration that it is a porin. J Bacteriol 162:85–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Bruin JP, Ijzerman EP, den Boer JW, Mouton JW, Diederen BM. 2012. Wild-type MIC distribution and epidemiological cut-off values in clinical Legionella pneumophila serogroup 1 isolates. Diagn Microbiol Infect Dis 72:103–108. doi: 10.1016/j.diagmicrobio.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 360.Nielsen K, Bangsborg JM, Hoiby N. 2000. Susceptibility of Legionella species to five antibiotics and development of resistance by exposure to erythromycin, ciprofloxacin, and rifampicin. Diagn Microbiol Infect Dis 36:43–48. doi: 10.1016/S0732-8893(99)00095-4. [DOI] [PubMed] [Google Scholar]
- 361.Almahmoud I, Kay E, Schneider D, Maurin M. 2009. Mutational paths towards increased fluoroquinolone resistance in Legionella pneumophila. J Antimicrob Chemother 64:284–293. doi: 10.1093/jac/dkp173. [DOI] [PubMed] [Google Scholar]
- 362.Chien M, Morozova I, Shi S, Sheng H, Chen J, Gomez SM, Asamani G, Hill K, Nuara J, Feder M, Rineer J, Greenberg JJ, Steshenko V, Park SH, Zhao B, Teplitskaya E, Edwards JR, Pampou S, Georghiou A, Chou IC, Iannuccilli W, Ulz ME, Kim DH, Geringer-Sameth A, Goldsberry C, Morozov P, Fischer SG, Segal G, Qu X, Rzhetsky A, Zhang P, Cayanis E, De Jong PJ, Ju J, Kalachikov S, Shuman HA, Russo JJ. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305:1966–1968. doi: 10.1126/science.1099776. [DOI] [PubMed] [Google Scholar]
- 363.Ferhat M, Atlan D, Vianney A, Lazzaroni JC, Doublet P, Gilbert C. 2009. The TolC protein of Legionella pneumophila plays a major role in multi-drug resistance and the early steps of host invasion. PLoS One 4:e7732. doi: 10.1371/journal.pone.0007732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Luo Y, Glisson JR, Jackwood MW, Hancock RE, Bains M, Cheng IH, Wang C. 1997. Cloning and characterization of the major outer membrane protein gene (ompH) of Pasteurella multocida X-73. J Bacteriol 179:7856–7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Chevalier G, Duclohier H, Thomas D, Shechter E, Wroblewski H. 1993. Purification and characterization of protein H, the major porin of Pasteurella multocida. J Bacteriol 175:266–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Hatfaludi T, Al-Hasani K, Dunstone M, Boyce J, Adler B. 2008. Characterization of TolC efflux pump proteins from Pasteurella multocida. Antimicrob Agents Chemother 52:4166–4171. doi: 10.1128/AAC.00245-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Michael GB, Eidam C, Kadlec K, Meyer K, Sweeney MT, Murray RW, Watts JL, Schwarz S. 2012. Increased MICs of gamithromycin and tildipirosin in the presence of the genes erm(42) and msr(E)-mph(E) for bovine Pasteurella multocida and Mannheimia haemolytica. J Antimicrob Chemother 67:1555–1557. doi: 10.1093/jac/dks076. [DOI] [PubMed] [Google Scholar]
- 368.Vachon V, Lyew DJ, Coulton JW. 1985. Transmembrane permeability channels across the outer membrane of Haemophilus influenzae type b. J Bacteriol 162:918–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Burns JL, Smith AL. 1987. A major outer-membrane protein functions as a porin in Haemophilus influenzae. J Gen Microbiol 133:1273–1277. [DOI] [PubMed] [Google Scholar]
- 370.Sánchez L, Pan W, Vinas M, Nikaido H. 1997. The acrAB homolog of Haemophilus influenzae codes for a functional multidrug efflux pump. J Bacteriol 179:6855–6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Trepod CM, Mott JE. 2004. Identification of the Haemophilus influenzae tolC gene by susceptibility profiles of insertionally inactivated efflux pump mutants. Antimicrob Agents Chemother 48:1416–1418. doi: 10.1128/AAC.48.4.1416-1418.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Peric M, Bozdogan B, Jacobs MR, Appelbaum PC. 2003. Effects of an efflux mechanism and ribosomal mutations on macrolide susceptibility of Haemophilus influenzae clinical isolates. Antimicrob Agents Chemother 47:1017–1022. doi: 10.1128/AAC.47.3.1017-1022.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Garcia-Cobos S, Campos J, Lazaro E, Roman F, Cercenado E, Garcia-Rey C, Pérez-Vazquez M, Oteo J, de Abajo F. 2007. Ampicillin-resistant non-β-lactamase-producing Haemophilus influenzae in Spain: recent emergence of clonal isolates with increased resistance to cefotaxime and cefixime. Antimicrob Agents Chemother 51:2564–2573. doi: 10.1128/AAC.00354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Dean CR, Narayan S, Daigle DM, Dzink-Fox JL, Puyang X, Bracken KR, Dean KE, Weidmann B, Yuan Z, Jain R, Ryder NS. 2005. Role of the AcrAB-TolC efflux pump in determining susceptibility of Haemophilus influenzae to the novel peptide deformylase inhibitor LBM415. Antimicrob Agents Chemother 49:3129–3135. doi: 10.1128/AAC.49.8.3129-3135.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Livermore DM, Davy KW. 1991. Invalidity for Pseudomonas aeruginosa of an accepted model of bacterial permeability to β-lactam antibiotics. Antimicrob Agents Chemother 35:916–921. doi: 10.1128/AAC.35.5.916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Pumbwe L, Everett MJ, Hancock RE, Piddock LJ. 1996. Role of gyrA mutation and loss of OprF in the multiple antibiotic resistance phenotype of Pseudomonas aeruginosa G49. FEMS Microbiol Lett 143:25–28. doi: 10.1111/j.1574-6968.1996.tb08456.x. [DOI] [PubMed] [Google Scholar]
- 377.Pumbwe L, Piddock LJ. 2000. Two efflux systems expressed simultaneously in multidrug-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother 44:2861–2864. doi: 10.1128/AAC.44.10.2861-2864.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Bratu S, Landman D, Gupta J, Quale J. 2007. Role of AmpD, OprF and penicillin-binding proteins in β-lactam resistance in clinical isolates of Pseudomonas aeruginosa. J Med Microbiol 56:809–814. doi: 10.1099/jmm.0.47019-0. [DOI] [PubMed] [Google Scholar]
- 379.Trias J, Nikaido H. 1990. Outer membrane protein D2 catalyzes facilitated diffusion of carbapenems and penems through the outer membrane of Pseudomonas aeruginosa. Antimicrob Agents Chemother 34:52–57. doi: 10.1128/AAC.34.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Quale J, Bratu S, Gupta J, Landman D. 2006. Interplay of efflux system, ampC, and oprD expression in carbapenem resistance of Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother 50:1633–1641. doi: 10.1128/AAC.50.5.1633-1641.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Gutierrez O, Juan C, Cercenado E, Navarro F, Bouza E, Coll P, Pérez JL, Oliver A. 2007. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa isolates from Spanish hospitals. Antimicrob Agents Chemother 51:4329–4335. doi: 10.1128/AAC.00810-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Tomás M, Doumith M, Warner M, Turton JF, Beceiro A, Bou G, Livermore DM, Woodford N. 2010. Efflux pumps, OprD porin, AmpC β-lactamase, and multiresistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob Agents Chemother 54:2219–2224. doi: 10.1128/AAC.00816-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Riera E, Cabot G, Mulet X, Garcia-Castillo M, del Campo R, Juan C, Canton R, Oliver A. 2011. Pseudomonas aeruginosa carbapenem resistance mechanisms in Spain: impact on the activity of imipenem, meropenem and doripenem. J Antimicrob Chemother 66:2022–2027. doi: 10.1093/jac/dkr232. [DOI] [PubMed] [Google Scholar]
- 384.Fournier D, Richardot C, Muller E, Robert-Nicoud M, Llanes C, Plésiat P, Jeannot K. 2013. Complexity of resistance mechanisms to imipenem in intensive care unit strains of Pseudomonas aeruginosa. J Antimicrob Chemother 68:1772–1780. doi: 10.1093/jac/dkt098. [DOI] [PubMed] [Google Scholar]
- 385.Xavier DE, Picao RC, Girardello R, Fehlberg LC, Gales AC. 2010. Efflux pumps expression and its association with porin down-regulation and β-lactamase production among Pseudomonas aeruginosa causing bloodstream infections in Brazil. BMC Microbiol 10:217. doi: 10.1186/1471-2180-10-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Dieppois G, Ducret V, Caille O, Perron K. 2012. The transcriptional regulator CzcR modulates antibiotic resistance and quorum sensing in Pseudomonas aeruginosa. PLoS One 7:e38148. doi: 10.1371/journal.pone.0038148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Wang D, Seeve C, Pierson LS III, Pierson EA. 2013. Transcriptome profiling reveals links between ParS/ParR, MexEF-OprN, and quorum sensing in the regulation of adaptation and virulence in Pseudomonas aeruginosa. BMC Genomics 14:618. doi: 10.1186/1471-2164-14-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Poole K, Heinrichs DE, Neshat S. 1993. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa: regulation by iron and possible involvement in the secretion of the siderophore pyoverdine. Mol Microbiol 10:529–544. doi: 10.1111/j.1365-2958.1993.tb00925.x. [DOI] [PubMed] [Google Scholar]
- 389.Masuda N, Ohya S. 1992. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob Agents Chemother 36:1847–1851. doi: 10.1128/AAC.36.9.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Li X-Z, Zhang L, Srikumar R, Poole K. 1998. β-Lactamase inhibitors are substrates for the multidrug efflux pumps of Pseudomonas aeruginosa. Antimicrob Agents Chemother 42:399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Nakae T, Saito K, Nakajima A. 2000. Effect of sulbactam on anti-pseudomonal activity of β-lactam antibiotics in cells producing various levels of the MexAB-OprM efflux pump and β-lactamase. Microbiol Immunol 44:997–1001. doi: 10.1111/j.1348-0421.2000.tb02595.x. [DOI] [PubMed] [Google Scholar]
- 392.Zhang L, Li XZ, Poole K. 2001. Fluoroquinolone susceptibilities of efflux-mediated multidrug-resistant Pseudomonas aeruginosa, Stenotrophomonas maltophilia and Burkholderia cepacia. J Antimicrob Chemother 48:549–552. doi: 10.1093/jac/48.4.549. [DOI] [PubMed] [Google Scholar]
- 393.Dean CR, Visalli MA, Projan SJ, Sum PE, Bradford PA. 2003. Efflux-mediated resistance to tigecycline (GAR-936) in Pseudomonas aeruginosa PAO1. Antimicrob Agents Chemother 47:972–978. doi: 10.1128/AAC.47.3.972-978.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Dupont P, Hocquet D, Jeannot K, Chavanet P, Plésiat P. 2005. Bacteriostatic and bactericidal activities of eight fluoroquinolones against MexAB-OprM-overproducing clinical strains of Pseudomonas aeruginosa. J Antimicrob Chemother 55:518–522. doi: 10.1093/jac/dki030. [DOI] [PubMed] [Google Scholar]
- 395.Papadopoulos CJ, Carson CF, Chang BJ, Riley TV. 2008. Role of the MexAB-OprM efflux pump of Pseudomonas aeruginosa in tolerance to tea tree (Melaleuca alternifolia) oil and its monoterpene components terpinen-4-ol, 1,8-cineole, and alpha-terpineol. Appl Environ Microbiol 74:1932–1935. doi: 10.1128/AEM.02334-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Robertson GT, Doyle TB, Du Q, Duncan L, Mdluli KE, Lynch AS. 2007. A novel indole compound that inhibits Pseudomonas aeruginosa growth by targeting MreB is a substrate for MexAB-OprM. J Bacteriol 189:6870–6881. doi: 10.1128/JB.00805-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Li X-Z, Zhang L, Poole K. 1998. Role of the multidrug efflux systems of Pseudomonas aeruginosa in organic solvent tolerance. J Bacteriol 180:2987–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Evans K, Passador L, Srikumar R, Tsang E, Nezezon J, Poole K. 1998. Influence of the MexAB-OprM multidrug efflux system on quorum sensing in Pseudomonas aeruginosa. J Bacteriol 180:5443–5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Pearson JP, Van Delden C, Iglewski BH. 1999. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J Bacteriol 181:1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Minagawa S, Inami H, Kato T, Sawada S, Yasuki T, Miyairi S, Horikawa M, Okuda J, Gotoh N. 2012. RND type efflux pump system MexAB-OprM of Pseudomonas aeruginosa selects bacterial languages, 3-oxo-acyl-homoserine lactones, for cell-to-cell communication. BMC Microbiol 12:70. doi: 10.1186/1471-2180-12-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Moore JD, Gerdt JP, Eibergen NR, Blackwell HE. 2014. Active efflux influences the potency of quorum sensing inhibitors in Pseudomonas aeruginosa. Chembiochem 15:435–442. doi: 10.1002/cbic.201300701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Masuda N, Sakagawa E, Ohya S. 1995. Outer membrane proteins responsible for multiple drug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 39:645–649. doi: 10.1128/AAC.39.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Poole K, Tetro K, Zhao Q, Neshat S, Heinrichs DE, Bianco N. 1996. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob Agents Chemother 40:2021–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Srikumar R, Paul CJ, Poole K. 2000. Influence of mutations in the mexR repressor gene on expression of the MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa. J Bacteriol 182:1410–1414. doi: 10.1128/JB.182.5.1410-1414.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Adewoye L, Sutherland A, Srikumar R, Poole K. 2002. The mexR repressor of the mexAB-oprM multidrug efflux operon in Pseudomonas aeruginosa: characterization of mutations compromising activity. J Bacteriol 184:4308–4312. doi: 10.1128/JB.184.15.4308-4312.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Cao L, Srikumar R, Poole K. 2004. MexAB-OprM hyperexpression in NalC-type multidrug-resistant Pseudomonas aeruginosa: identification and characterization of the nalC gene encoding a repressor of PA3720-PA3719. Mol Microbiol 53:1423–1436. doi: 10.1111/j.1365-2958.2004.04210.x. [DOI] [PubMed] [Google Scholar]
- 407.Saito K, Akama H, Yoshihara E, Nakae T. 2003. Mutations affecting DNA-binding activity of the MexR repressor of mexR-mexA-mexB-oprM operon expression. J Bacteriol 185:6195–6198. doi: 10.1128/JB.185.20.6195-6198.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Sobel ML, Hocquet D, Cao L, Plésiat P, Poole K. 2005. Mutations in PA3574 (nalD) lead to increased MexAB-OprM expression and multidrug resistance in laboratory and clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother 49:1782–1786. doi: 10.1128/AAC.49.5.1782-1786.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Andrésen C, Jalal S, Aili D, Wang Y, Islam S, Jarl A, Liedberg B, Wretlind B, Martensson LG, Sunnerhagen M. 2010. Critical biophysical properties in the Pseudomonas aeruginosa efflux gene regulator MexR are targeted by mutations conferring multidrug resistance. Protein Sci 19:680–692. doi: 10.1002/pro.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Hamzehpour MM, Pechère JC, Plésiat P, Köhler T. 1995. OprK and OprM define two genetically distinct multidrug efflux systems in Pseudomonas aeruginosa. Antimicrob Agents Chemother 39:2392–2396. doi: 10.1128/AAC.39.11.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Lomovskaya O, Lee A, Hoshino K, Ishida H, Mistry A, Warren MS, Boyer E, Chamberland S, Lee VJ. 1999. Use of a genetic approach to evaluate the consequences of inhibition of efflux pumps in Pseudomonas aeruginosa. Antimicrob Agents Chemother 43:1340–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Hocquet D, Bertrand X, Köhler T, Talon D, Plésiat P. 2003. Genetic and phenotypic variations of a resistant Pseudomonas aeruginosa epidemic clone. Antimicrob Agents Chemother 47:1887–1894. doi: 10.1128/AAC.47.6.1887-1894.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Llanes C, Hocquet D, Vogne C, Benali-Baitich D, Neuwirth C, Plésiat P. 2004. Clinical strains of Pseudomonas aeruginosa overproducing MexAB-OprM and MexXY efflux pumps simultaneously. Antimicrob Agents Chemother 48:1797–1802. doi: 10.1128/AAC.48.5.1797-1802.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Henrichfreise B, Wiegand I, Pfister W, Wiedemann B. 2007. Resistance mechanisms of multiresistant Pseudomonas aeruginosa strains from Germany and correlation with hypermutation. Antimicrob Agents Chemother 51:4062–4070. doi: 10.1128/AAC.00148-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Cavallo JD, Hocquet D, Plésiat P, Fabre R, Roussel-Delvallez M, GERPA. 2007. Susceptibility of Pseudomonas aeruginosa to antimicrobials: a 2004 French multicentre hospital study. J Antimicrob Chemother 59:1021–1024. doi: 10.1093/jac/dkm076. [DOI] [PubMed] [Google Scholar]
- 416.Cabot G, Ocampo-Sosa AA, Dominguez MA, Gago JF, Juan C, Tubau F, Rodríguez C, Moya B, Pena C, Martínez-Martínez L, Oliver A, Spanish Network for Research in Infectious Diseases. 2012. Genetic markers of widespread extensively drug-resistant Pseudomonas aeruginosa high-risk clones. Antimicrob Agents Chemother 56:6349–6357. doi: 10.1128/AAC.01388-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Moya B, Beceiro A, Cabot G, Juan C, Zamorano L, Alberti S, Oliver A. 2012. Pan-β-lactam resistance development in Pseudomonas aeruginosa clinical strains: molecular mechanisms, penicillin-binding protein profiles, and binding affinities. Antimicrob Agents Chemother 56:4771–4778. doi: 10.1128/AAC.00680-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Li X-Z. 2012. Multidrug resistance efflux pumps of Pseudomonas aeruginosa: a 10-year update. Chin J Antibiot 37:481–500. [Google Scholar]
- 419.Sacha P, Wieczorek P, Ojdana D, Hauschild T, Milewski R, Czaban S, Poniatowski B, Tryniszewska E. 2014. Expression of MexAB-OprM efflux pump system and susceptibility to antibiotics of different Pseudomonas aeruginosa clones isolated from patients hospitalized in two intensive care units at University Hospital in Bialystok (northeastern Poland) between January 2002 and December 2009. APMIS 122:931–940. doi: 10.1111/apm.12236.. [DOI] [PubMed] [Google Scholar]
- 420.Pournaras S, Maniati M, Spanakis N, Ikonomidis A, Tassios PT, Tsakris A, Legakis NJ, Maniatis AN. 2005. Spread of efflux pump-overexpressing, non-metallo-β-lactamase-producing, meropenem-resistant but ceftazidime-susceptible Pseudomonas aeruginosa in a region with blaVIM endemicity. J Antimicrob Chemother 56:761–764. doi: 10.1093/jac/dki296. [DOI] [PubMed] [Google Scholar]
- 421.Castanheira M, Deshpande LM, Costello A, Davies TA, Jones RN. 2014. Epidemiology and carbapenem resistance mechanisms of carbapenem-non-susceptible Pseudomonas aeruginosa collected during 2009-11 in 14 European and Mediterranean countries. J Antimicrob Chemother 69:1804–1814. doi: 10.1093/jac/dku048. [DOI] [PubMed] [Google Scholar]
- 422.Fehlberg LC, Xavier DE, Peraro PP, Marra AR, Edmond MB, Gales AC. 2012. β-Lactam resistance mechanisms in Pseudomonas aeruginosa strains causing bloodstream infections: comparative results between Brazilian and American isolates. Microb Drug Resist 18:402–407. doi: 10.1089/mdr.2011.0174. [DOI] [PubMed] [Google Scholar]
- 423.Hocquet D, Roussel-Delvallez M, Cavallo JD, Plésiat P. 2007. MexAB-OprM- and MexXY-overproducing mutants are very prevalent among clinical strains of Pseudomonas aeruginosa with reduced susceptibility to ticarcillin. Antimicrob Agents Chemother 51:1582–1583. doi: 10.1128/AAC.01334-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Kriengkauykiat J, Porter E, Lomovskaya O, Wong-Beringer A. 2005. Use of an efflux pump inhibitor to determine the prevalence of efflux pump-mediated fluoroquinolone resistance and multidrug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 49:565–570. doi: 10.1128/AAC.49.2.565-570.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Campo Esquisabel AB, Rodríguez MC, Campo-Sosa AO, Rodríguez C, Martínez-Martínez L. 2011. Mechanisms of resistance in clinical isolates of Pseudomonas aeruginosa less susceptible to cefepime than to ceftazidime. Clin Microbiol Infect 17:1817–1822. doi: 10.1111/j.1469-0691.2011.03530.x. [DOI] [PubMed] [Google Scholar]
- 426.Ziha-Zarifi I, Llanes C, Köhler T, Pechère JC, Plésiat P. 1999. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA-MexB-OprM. Antimicrob Agents Chemother 43:287–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Ong CT, Tessier PR, Li C, Nightingale CH, Nicolau DP. 2007. Comparative in vivo efficacy of meropenem, imipenem, and cefepime against Pseudomonas aeruginosa expressing MexA-MexB-OprM efflux pumps. Diagn Microbiol Infect Dis 57:153–161. doi: 10.1016/j.diagmicrobio.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 428.Boutoille D, Jacqueline C, Le Mabecque V, Potel G, Caillon J. 2009. In vivo impact of the MexAB-OprM efflux system on β-lactam efficacy in an experimental model of Pseudomonas aeruginosa infection. Int J Antimicrob Agents 33:417–420. doi: 10.1016/j.ijantimicag.2008.10.029. [DOI] [PubMed] [Google Scholar]
- 429.Hocquet D, Berthelot P, Roussel-Delvallez M, Favre R, Jeannot K, Bajolet O, Marty N, Grattard F, Mariani-Kurkdjian P, Bingen E, Husson MO, Couetdic G, Plésiat P. 2007. Pseudomonas aeruginosa may accumulate drug resistance mechanisms without losing its ability to cause bloodstream infections. Antimicrob Agents Chemother 51:3531–3536. doi: 10.1128/AAC.00503-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Nakae T, Nakajima A, Ono T, Saito K, Yoneyama H. 1999. Resistance to β-lactam antibiotics in Pseudomonas aeruginosa due to interplay between the MexAB-OprM efflux pump and β-lactamase. Antimicrob Agents Chemother 43:1301–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Llanes C, Köhler T, Patry I, Dehecq B, van Delden C, Plésiat P. 2011. Role of the MexEF-OprN efflux system in low-level resistance of Pseudomonas aeruginosa to ciprofloxacin. Antimicrob Agents Chemother 55:5676–5684. doi: 10.1128/AAC.00101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Okamoto K, Gotoh N, Nishino T. 2002. Alterations of susceptibility of Pseudomonas aeruginosa by overproduction of multidrug efflux systems, MexAB-OprM, MexCD-OprJ, and MexXY/OprM to carbapenems: substrate specificities of the efflux systems. J Infect Chemother 8:371–373. doi: 10.1007/s10156-002-0193-7. [DOI] [PubMed] [Google Scholar]
- 433.Köhler T, Michea-Hamzehpour M, Plésiat P, Kahr AL, Pechère JC. 1997. Differential selection of multidrug efflux systems by quinolones in Pseudomonas aeruginosa. Antimicrob Agents Chemother 41:2540–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Aires JR, Köhler T, Nikaido H, Plésiat P. 1999. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob Agents Chemother 43:2624–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Mine T, Morita Y, Kataoka A, Mizushima T, Tsuchiya T. 1999. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob Agents Chemother 43:415–417. doi: 10.1093/jac/43.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Westbrock-Wadman S, Sherman DR, Hickey MJ, Coulter SN, Zhu YQ, Warrener P, Nguyen LY, Shawar RM, Folger KR, Stover CK. 1999. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob Agents Chemother 43:2975–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Murata T, Gotoh N, Nishino T. 2002. Characterization of outer membrane efflux proteins OpmE, OpmD and OpmB of Pseudomonas aeruginosa: molecular cloning and development of specific antisera. FEMS Microbiol Lett 217:57–63. doi: 10.1111/j.1574-6968.2002.tb11456.x. [DOI] [PubMed] [Google Scholar]
- 438.Roy PH, Tetu SG, Larouche A, Elbourne L, Tremblay S, Ren Q, Dodson R, Harkins D, Shay R, Watkins K, Mahamoud Y, Paulsen IT. 2010. Complete genome sequence of the multiresistant taxonomic outlier Pseudomonas aeruginosa PA7. PLoS One 5:e8842. doi: 10.1371/journal.pone.0008842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Baum EZ, Crespo-Carbone SM, Morrow BJ, Davies TA, Foleno BD, He W, Queenan AM, Bush K. 2009. Effect of MexXY overexpression on ceftobiprole susceptibility in Pseudomonas aeruginosa. Antimicrob Agents Chemother 53:2785–2790. doi: 10.1128/AAC.00018-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Masuda N, Sakagawa E, Ohya S, Gotoh N, Tsujimoto H, Nishino T. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob Agents Chemother 44:3322–3327. doi: 10.1128/AAC.44.12.3322-3327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Jeannot K, Sobel ML, El Garch F, Poole K, Plésiat P. 2005. Induction of the MexXY efflux pump in Pseudomonas aeruginosa is dependent on drug-ribosome interaction. J Bacteriol 187:5341–5346. doi: 10.1128/JB.187.15.5341-5346.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Zhao Q, Li X-Z, Srikumar R, Poole K. 1998. Contribution of outer membrane efflux protein OprM to antibiotic resistance in Pseudomonas aeruginosa independent of MexAB. Antimicrob Agents Chemother 42:1682–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Hocquet D, Muller A, Blanc K, Plésiat P, Talon D, Monnet DL, Bertrand X. 2008. Relationship between antibiotic use and incidence of MexXY-OprM overproducers among clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother 52:1173–1175. doi: 10.1128/AAC.01212-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Muller C, Plésiat P, Jeannot K. 2011. A two-component regulatory system interconnects resistance to polymyxins, aminoglycosides, fluoroquinolones, and β-lactams in Pseudomonas aeruginosa. Antimicrob Agents Chemother 55:1211–1221. doi: 10.1128/AAC.01252-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Caughlan RE, Sriram S, Daigle DM, Woods AL, Buco J, Peterson RL, Dzink-Fox J, Walker S, Dean CR. 2009. Fmt bypass in Pseudomonas aeruginosa causes induction of MexXY efflux pump expression. Antimicrob Agents Chemother 53:5015–5021. doi: 10.1128/AAC.00253-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Vogne C, Aires JR, Bailly C, Hocquet D, Plésiat P. 2004. Role of the multidrug efflux system MexXY in the emergence of moderate resistance to aminoglycosides among Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Antimicrob Agents Chemother 48:1676–1680. doi: 10.1128/AAC.48.5.1676-1680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D'Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, Burns JL, Kaul R, Olson MV. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A 103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Feliziani S, Lujan AM, Moyano AJ, Sola C, Bocco JL, Montanaro P, Canigia LF, Argarana CE, Smania AM. 2010. Mucoidy, quorum sensing, mismatch repair and antibiotic resistance in Pseudomonas aeruginosa from cystic fibrosis chronic airways infections. PLoS One 5:e12669. doi: 10.1371/journal.pone.0012669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Mulcahy LR, Burns JL, Lory S, Lewis K. 2010. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J Bacteriol 192:6191–6199. doi: 10.1128/JB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Qin X, Zerr DM, McNutt MA, Berry JE, Burns JL, Kapur RP. 2012. Pseudomonas aeruginosa syntrophy in chronically colonized airways of cystic fibrosis patients. Antimicrob Agents Chemother 56:5971–5981. doi: 10.1128/AAC.01371-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Llanes C, Pourcel C, Richardot C, Plésiat P, Fichant G, Cavallo JD, Merens A, GERPA Study Group. 2013. Diversity of β-lactam resistance mechanisms in cystic fibrosis isolates of Pseudomonas aeruginosa: a French multicentre study. J Antimicrob Chemother 68:1763–1771. doi: 10.1093/jac/dkt115. [DOI] [PubMed] [Google Scholar]
- 452.Guénard S, Muller C, Monlezun L, Benas P, Broutin I, Jeannot K, Plésiat P. 2014. Multiple mutations lead to MexXY-OprM-dependent aminoglycoside resistance in clinical strains of Pseudomonas aeruginosa. Antimicrob Agents Chemother 58:221–228. doi: 10.1128/AAC.01252-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Galli F, Battistoni A, Gambari R, Pompella A, Bragonzi A, Pilolli F, Iuliano L, Piroddi M, Dechecchi MC, Cabrini G, Working Group on Inflammation in Cystic Fibrosis. 2012. Oxidative stress and antioxidant therapy in cystic fibrosis. Biochim Biophys Acta 1822:690–713. doi: 10.1016/j.bbadis.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 454.Fraud S, Poole K. 2011. Oxidative stress induction of the MexXY multidrug efflux genes and promotion of aminoglycoside resistance development in Pseudomonas aeruginosa. Antimicrob Agents Chemother 55:1068–1074. doi: 10.1128/AAC.01495-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Hocquet D, Nordmann P, El Garch F, Cabanne L, Plésiat P. 2006. Involvement of the MexXY-OprM efflux system in emergence of cefepime resistance in clinical strains of Pseudomonas aeruginosa. Antimicrob Agents Chemother 50:1347–1351. doi: 10.1128/AAC.50.4.1347-1351.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Peña C, Suarez C, Tubau F, Juan C, Moya B, Dominguez MA, Oliver A, Pujol M, Ariza J. 2009. Nosocomial outbreak of a non-cefepime-susceptible ceftazidime-susceptible Pseudomonas aeruginosa strain overexpressing MexXY-OprM and producing an integron-borne PSE-1 β-lactamase. J Clin Microbiol 47:2381–2387. doi: 10.1128/JCM.00094-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Morita Y, Tomida J, Kawamura Y. 2012. Primary mechanisms mediating aminoglycoside resistance in the multidrug-resistant Pseudomonas aeruginosa clinical isolate PA7. Microbiology 158:1071–1083. doi: 10.1099/mic.0.054320-0. [DOI] [PubMed] [Google Scholar]
- 458.Alguel Y, Lu D, Quade N, Sauter S, Zhang X. 2010. Crystal structure of MexZ, a key repressor responsible for antibiotic resistance in Pseudomonas aeruginosa. J Struct Biol 172:305–310. doi: 10.1016/j.jsb.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 459.Jahandideh S. 2013. Diversity in structural consequences of MexZ mutations in Pseudomonas aeruginosa. Chem Biol Drug Des 81:600–606. doi: 10.1111/cbdd.12104. [DOI] [PubMed] [Google Scholar]
- 460.El'Garch F, Jeannot K, Hocquet D, Llanes-Barakat C, Plésiat P. 2007. Cumulative effects of several nonenzymatic mechanisms on the resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob Agents Chemother 51:1016–1021. doi: 10.1128/AAC.00704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Lau CH, Fraud S, Jones M, Peterson SN, Poole K. 2012. Reduced expression of the rplU-rpmA ribosomal protein operon in mexXY-expressing pan-aminoglycoside-resistant mutants of Pseudomonas aeruginosa. Antimicrob Agents Chemother 56:5171–5179. doi: 10.1128/AAC.00846-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Martha B, Croisier D, Durand D, Hocquet D, Plésiat P, Piroth L, Portier H, Chavanet P. 2006. In-vivo impact of the MexXY efflux system on aminoglycoside efficacy in an experimental model of Pseudomonas aeruginosa pneumonia treated with tobramycin. Clin Microbiol Infect 12:426–432. doi: 10.1111/j.1469-0691.2006.01371.x. [DOI] [PubMed] [Google Scholar]
- 463.Pumbwe L, Glass D, Wexler HM. 2006. Efflux pump overexpression in multiple-antibiotic-resistant mutants of Bacteroides fragilis. Antimicrob Agents Chemother 50:3150–3153. doi: 10.1128/AAC.00141-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Bruchmann S, Dotsch A, Nouri B, Chaberny IF, Haussler S. 2013. Quantitative contributions of target alteration and decreased drug accumulation to Pseudomonas aeruginosa fluoroquinolone resistance. Antimicrob Agents Chemother 57:1361–1368. doi: 10.1128/AAC.01581-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, Yamasaki T, Neshat S, Yamagishi J, Li X-Z, Nishino T. 1996. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol Microbiol 21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 466.Morita Y, Murata T, Mima T, Shiota S, Kuroda T, Mizushima T, Gotoh N, Nishino T, Tsuchiya T. 2003. Induction of mexCD-oprJ operon for a multidrug efflux pump by disinfectants in wild-type Pseudomonas aeruginosa PAO1. J Antimicrob Chemother 51:991–994. doi: 10.1093/jac/dkg173. [DOI] [PubMed] [Google Scholar]
- 467.Fraud S, Campigotto AJ, Chen Z, Poole K. 2008. MexCD-OprJ multidrug efflux system of Pseudomonas aeruginosa: involvement in chlorhexidine resistance and induction by membrane-damaging agents dependent upon the AlgU stress response sigma factor. Antimicrob Agents Chemother 52:4478–4482. doi: 10.1128/AAC.01072-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Masuda N, Gotoh N, Ohya S, Nishino T. 1996. Quantitative correlation between susceptibility and OprJ production in NfxB mutants of Pseudomonas aeruginosa. Antimicrob Agents Chemother 40:909–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Gotoh N, Tsujimoto H, Tsuda M, Okamoto K, Nomura A, Wada T, Nakahashi M, Nishino T. 1998. Characterization of the MexC-MexD-OprJ multidrug efflux system in ΔmexA-mexB-oprM mutants of Pseudomonas aeruginosa. Antimicrob Agents Chemother 42:1938–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Masuda N, Sakagawa E, Ohya S, Gotoh N, Nishino T. 2001. Hypersusceptibility of the Pseudomonas aeruginosa nfxB mutant to β-lactams due to reduced expression of the AmpC β-lactamase. Antimicrob Agents Chemother 45:1284–1286. doi: 10.1128/AAC.45.4.1284-1286.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Wolter DJ, Hanson ND, Lister PD. 2005. AmpC and OprD are not involved in the mechanism of imipenem hypersusceptibility among Pseudomonas aeruginosa isolates overexpressing the MexCD-OprJ efflux pump. Antimicrob Agents Chemother 49:4763–4766. doi: 10.1128/AAC.49.11.4763-4766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Mulet X, Moya B, Juan C, Macia MD, Pérez JL, Blazquez J, Oliver A. 2011. Antagonistic interactions of Pseudomonas aeruginosa antibiotic resistance mechanisms in planktonic but not biofilm growth. Antimicrob Agents Chemother 55:4560–4568. doi: 10.1128/AAC.00519-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Giwercman B, Meyer C, Lambert PA, Reinert C, Hoiby N. 1992. High-level β-lactamase activity in sputum samples from cystic fibrosis patients during antipseudomonal treatment. Antimicrob Agents Chemother 36:71–76. doi: 10.1128/AAC.36.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Jeannot K, Elsen S, Köhler T, Attree I, van Delden C, Plésiat P. 2008. Resistance and virulence of Pseudomonas aeruginosa clinical strains overproducing the MexCD-OprJ efflux pump. Antimicrob Agents Chemother 52:2455–2462. doi: 10.1128/AAC.01107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Reinhardt A, Köhler T, Wood P, Rohner P, Dumas JL, Ricou B, van Delden C. 2007. Development and persistence of antimicrobial resistance in Pseudomonas aeruginosa: a longitudinal observation in mechanically ventilated patients. Antimicrob Agents Chemother 51:1341–1350. doi: 10.1128/AAC.01278-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Rodríguez-Martínez JM, Poirel L, Nordmann P. 2009. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 53:4783–4788. doi: 10.1128/AAC.00574-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Chiang WC, Pamp SJ, Nilsson M, Givskov M, Tolker-Nielsen T. 2012. The metabolically active subpopulation in Pseudomonas aeruginosa biofilms survives exposure to membrane-targeting antimicrobials via distinct molecular mechanisms. FEMS Immunol Med Microbiol 65:245–256. doi: 10.1111/j.1574-695X.2012.00929.x. [DOI] [PubMed] [Google Scholar]
- 478.Wolter DJ, Black JA, Lister PD, Hanson ND. 2009. Multiple genotypic changes in hypersusceptible strains of Pseudomonas aeruginosa isolated from cystic fibrosis patients do not always correlate with the phenotype. J Antimicrob Chemother 64:294–300. doi: 10.1093/jac/dkp185. [DOI] [PubMed] [Google Scholar]
- 479.Stickland HG, Davenport PW, Lilley KS, Griffin JL, Welch M. 2010. Mutation of nfxB causes global changes in the physiology and metabolism of Pseudomonas aeruginosa. J Proteome Res 9:2957–2967. doi: 10.1021/pr9011415. [DOI] [PubMed] [Google Scholar]
- 480.Fukuda H, Hosaka M, Hirai K, Iyobe S. 1990. New norfloxacin resistance gene in Pseudomonas aeruginosa PAO. Antimicrob Agents Chemother 34:1757–1761. doi: 10.1128/AAC.34.9.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Rådberg G, Nilsson LE, Svensson S. 1990. Development of quinolone-imipenem cross resistance in Pseudomonas aeruginosa during exposure to ciprofloxacin. Antimicrob Agents Chemother 34:2142–2147. doi: 10.1128/AAC.34.11.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Aubert G, Pozzetto B, Dorche G. 1992. Emergence of quinolone-imipenem cross-resistance in Pseudomonas aeruginosa after fluoroquinolone therapy. J Antimicrob Chemother 29:307–312. doi: 10.1093/jac/29.3.307. [DOI] [PubMed] [Google Scholar]
- 483.Köhler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty LK, Pechère JC. 1997. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol 23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 484.Maseda H, Yoneyama H, Nakae T. 2000. Assignment of the substrate-selective subunits of the MexEF-OprN multidrug efflux pump of Pseudomonas aeruginosa. Antimicrob Agents Chemother 44:658–664. doi: 10.1128/AAC.44.3.658-664.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Fukuda H, Hosaka M, Iyobe S, Gotoh N, Nishino T, Hirai K. 1995. nfxC-type quinolone resistance in a clinical isolate of Pseudomonas aeruginosa. Antimicrob Agents Chemother 39:790–792. doi: 10.1128/AAC.39.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Henrichfreise B, Wiegand I, Luhmer-Becker I, Wiedemann B. 2007. Development of resistance in wild-type and hypermutable Pseudomonas aeruginosa strains exposed to clinical pharmacokinetic profiles of meropenem and ceftazidime simulated in vitro. Antimicrob Agents Chemother 51:3642–3649. doi: 10.1128/AAC.00160-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Mima T, Sekiya H, Mizushima T, Kuroda T, Tsuchiya T. 2005. Gene cloning and properties of the RND-type multidrug efflux pumps MexPQ-OpmE and MexMN-OprM from Pseudomonas aeruginosa. Microbiol Immunol 49:999–1002. doi: 10.1111/j.1348-0421.2005.tb03696.x. [DOI] [PubMed] [Google Scholar]
- 488.Li Y, Mima T, Komori Y, Morita Y, Kuroda T, Mizushima T, Tsuchiya T. 2003. A new member of the tripartite multidrug efflux pumps, MexVW-OprM, in Pseudomonas aeruginosa. J Antimicrob Chemother 52:572–575. doi: 10.1093/jac/dkg390. [DOI] [PubMed] [Google Scholar]
- 489.Mima T, Kohira N, Li Y, Sekiya H, Ogawa W, Kuroda T, Tsuchiya T. 2009. Gene cloning and characteristics of the RND-type multidrug efflux pump MuxABC-OpmB possessing two RND components in Pseudomonas aeruginosa. Microbiology 155:3509–3517. doi: 10.1099/mic.0.031260-0. [DOI] [PubMed] [Google Scholar]
- 490.Yang L, Chen L, Shen L, Surette M, Duan K. 2011. Inactivation of MuxABC-OpmB transporter system in Pseudomonas aeruginosa leads to increased ampicillin and carbenicillin resistance and decreased virulence. J Microbiol 49:107–114. doi: 10.1007/s12275-011-0186-2. [DOI] [PubMed] [Google Scholar]
- 491.Mima T, Joshi S, Gomez-Escalada M, Schweizer HP. 2007. Identification and characterization of TriABC-OpmH, a triclosan efflux pump of Pseudomonas aeruginosa requiring two membrane fusion proteins. J Bacteriol 189:7600–7609. doi: 10.1128/JB.00850-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Hassan MT, van der Lelie D, Springael D, Romling U, Ahmed N, Mergeay M. 1999. Identification of a gene cluster, czr, involved in cadmium and zinc resistance in Pseudomonas aeruginosa. Gene 238:417–425. doi: 10.1016/S0378-1119(99)00349-2. [DOI] [PubMed] [Google Scholar]
- 493.Perron K, Caille O, Rossier C, Van Delden C, Dumas JL, Köhler T. 2004. CzcR-CzcS, a two-component system involved in heavy metal and carbapenem resistance in Pseudomonas aeruginosa. J Biol Chem 279:8761–8768. doi: 10.1074/jbc.M312080200. [DOI] [PubMed] [Google Scholar]
- 494.Dijkshoorn L, Nemec A, Seifert H. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol 5:939–951. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 495.Garcia-Quintanilla M, Pulido MR, Lopez-Rojas R, Pachon J, McConnell MJ. 2013. Emerging therapies for multidrug resistant Acinetobacter baumannii. Trends Microbiol 21:157–163. doi: 10.1016/j.tim.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 496.Fournier PE, Vallenet D, Barbe V, Audic S, Ogata H, Poirel L, Richet H, Robert C, Mangenot S, Abergel C, Nordmann P, Weissenbach J, Raoult D, Claverie JM. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet 2:e7. doi: 10.1371/journal.pgen.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497.Adams MD, Goglin K, Molyneaux N, Hujer KM, Lavender H, Jamison JJ, MacDonald IJ, Martin KM, Russo T, Campagnari AA, Hujer AM, Bonomo RA, Gill SR. 2008. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J Bacteriol 190:8053–8064. doi: 10.1128/JB.00834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Krizova L, Dijkshoorn L, Nemec A. 2011. Diversity and evolution of AbaR genomic resistance islands in Acinetobacter baumannii strains of European clone I. Antimicrob Agents Chemother 55:3201–3206. doi: 10.1128/AAC.00221-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.Zhou H, Zhang T, Yu D, Pi B, Yang Q, Zhou J, Hu S, Yu Y. 2011. Genomic analysis of the multidrug-resistant Acinetobacter baumannii strain MDR-ZJ06 widely spread in China. Antimicrob Agents Chemother 55:4506–4512. doi: 10.1128/AAC.01134-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Zhu L, Yan Z, Zhang Z, Zhou Q, Zhou J, Wakeland EK, Fang X, Xuan Z, Shen D, Li QZ. 2013. Complete genome analysis of three Acinetobacter baumannii clinical isolates in China for insight into the diversification of drug resistance elements. PLoS One 8:e66584. doi: 10.1371/journal.pone.0066584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Bratu S, Landman D, George A, Salvani J, Quale J. 2009. Correlation of the expression of acrB and the regulatory genes marA, soxS and ramA with antimicrobial resistance in clinical isolates of Klebsiella pneumoniae endemic to New York City. J Antimicrob Chemother 64:278–283. doi: 10.1093/jac/dkp186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Rumbo C, Gato E, Lopez M, Ruiz de Alegria C, Fernández-Cuenca F, Martínez-Martínez L, Vila J, Pachon J, Cisneros JM, Rodríguez-Bano J, Pascual A, Bou G, Tomás M, Spanish Group of Nosocomial Infections and Mechanisms of Action and Resistance to Antimicrobials, Spanish Society of Clinical Microbiology and Infectious Diseases, Spanish Network for Research in Infectious Diseases. 2013. Contribution of efflux pumps, porins, and β-lactamases to multidrug resistance in clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother 57:5247–5257. doi: 10.1128/AAC.00730-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Figueiredo S, Poirel L, Croize J, Recule C, Nordmann P. 2009. In vivo selection of reduced susceptibility to carbapenems in Acinetobacter baumannii related to ISAba1-mediated overexpression of the natural blaOXA-66 oxacillinase gene. Antimicrob Agents Chemother 53:2657–2659. doi: 10.1128/AAC.01663-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Higgins PG, Schneiders T, Hamprecht A, Seifert H. 2010. In vivo selection of a missense mutation in adeR and conversion of the novel blaOXA-164 gene into blaOXA-58 in carbapenem-resistant Acinetobacter baumannii isolates from a hospitalized patient. Antimicrob Agents Chemother 54:5021–5027. doi: 10.1128/AAC.00598-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Hua X, Zhou H, Jiang Y, Feng Y, Chen Q, Ruan Z, Yu Y. 2012. Genome sequences of two multidrug-resistant Acinetobacter baumannii strains isolated from a patient before and after treatment with tigecycline. J Bacteriol 194:6979–6980. doi: 10.1128/JB.01887-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Asai S, Umezawa K, Iwashita H, Ohshima T, Ohashi M, Sasaki M, Hayashi H, Matsui M, Shibayama K, Inokuchi S, Miyachi H. 2014. An outbreak of blaOXA-51-like- and blaOXA-66-positive Acinetobacter baumannii ST208 in the emergency intensive care unit. J Med Microbiol 63:1517–1523. doi: 10.1099/jmm.0.077503-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Sato K, Nakae T. 1991. Outer membrane permeability of Acinetobacter calcoaceticus and its implication in antibiotic resistance. J Antimicrob Chemother 28:35–45. doi: 10.1093/jac/28.1.35. [DOI] [PubMed] [Google Scholar]
- 508.Jyothisri K, Deepak V, Rajeswari MR. 1999. Purification and characterization of a major 40 kDa outer membrane protein of Acinetobacter baumannii. FEBS Lett 443:57–60. doi: 10.1016/S0014-5793(98)01679-2. [DOI] [PubMed] [Google Scholar]
- 509.Smani Y, Fabrega A, Roca I, Sánchez-Encinales V, Vila J, Pachon J. 2014. Role of OmpA in the multidrug resistance phenotype of Acinetobacter baumannii. Antimicrob Agents Chemother 58:1806–1808. doi: 10.1128/AAC.02101-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Chusri S, Na-Phatthalung P, Siriyong T, Paosen S, Voravuthikunchai SP. 2014. Holarrhena antidysenterica as a resistance modifying agent against Acinetobacter baumannii: its effects on bacterial outer membrane permeability and efflux pumps. Microbiol Res 169:417–424. doi: 10.1016/j.micres.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 511.Lorenzi V, Muselli A, Bernardini AF, Berti L, Pagès JM, Amaral L, Bolla JM. 2009. Geraniol restores antibiotic activities against multidrug-resistant isolates from Gram-negative species. Antimicrob Agents Chemother 53:2209–2211. doi: 10.1128/AAC.00919-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Wang HM, Chen CY, Chen HA, Huang WC, Lin WR, Chen TC, Lin CY, Chien HJ, Lu PL, Lin CM, Chen YH. 2010. Zingiber officinale (ginger) compounds have tetracycline-resistance modifying effects against clinical extensively drug-resistant Acinetobacter baumannii. Phytother Res 24:1825–1830. doi: 10.1002/ptr.3201. [DOI] [PubMed] [Google Scholar]
- 513.Limansky AS, Mussi MA, Viale AM. 2002. Loss of a 29-kilodalton outer membrane protein in Acinetobacter baumannii is associated with imipenem resistance. J Clin Microbiol 40:4776–4778. doi: 10.1128/JCM.40.12.4776-4778.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Mussi MA, Limansky AS, Viale AM. 2005. Acquisition of resistance to carbapenems in multidrug-resistant clinical strains of Acinetobacter baumannii: natural insertional inactivation of a gene encoding a member of a novel family of b-barrel outer membrane proteins. Antimicrob Agents Chemother 49:1432–1440. doi: 10.1128/AAC.49.4.1432-1440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Siroy A, Molle V, Lemaitre-Guillier C, Vallenet D, Pestel-Caron M, Cozzone AJ, Jouenne T, De E. 2005. Channel formation by CarO, the carbapenem resistance-associated outer membrane protein of Acinetobacter baumannii. Antimicrob Agents Chemother 49:4876–4883. doi: 10.1128/AAC.49.12.4876-4883.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Catel-Ferreira M, Coadou G, Molle V, Mugnier P, Nordmann P, Siroy A, Jouenne T, De E. 2011. Structure-function relationships of CarO, the carbapenem resistance-associated outer membrane protein of Acinetobacter baumannii. J Antimicrob Chemother 66:2053–2056. doi: 10.1093/jac/dkr267. [DOI] [PubMed] [Google Scholar]
- 517.Mussi MA, Relling VM, Limansky AS, Viale AM. 2007. CarO, an Acinetobacter baumannii outer membrane protein involved in carbapenem resistance, is essential for L-ornithine uptake. FEBS Lett 581:5573–5578. doi: 10.1016/j.febslet.2007.10.063. [DOI] [PubMed] [Google Scholar]
- 518.Lee Y, Kim CK, Lee H, Jeong SH, Yong D, Lee K. 2011. A novel insertion sequence, ISAba10, inserted into ISAba1 adjacent to the blaOXA-23 gene and disrupting the outer membrane protein gene carO in Acinetobacter baumannii. Antimicrob Agents Chemother 55:361–363. doi: 10.1128/AAC.01672-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519.Mussi MA, Limansky AS, Relling V, Ravasi P, Arakaki A, Actis LA, Viale AM. 2011. Horizontal gene transfer and assortative recombination within the Acinetobacter baumannii clinical population provide genetic diversity at the single carO gene, encoding a major outer membrane protein channel. J Bacteriol 193:4736–4748. doi: 10.1128/JB.01533-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Clark RB. 1996. Imipenem resistance among Acinetobacter baumannii: association with reduced expression of a 33-36 kDa outer membrane protein. J Antimicrob Chemother 38:245–251. doi: 10.1093/jac/38.2.245. [DOI] [PubMed] [Google Scholar]
- 521.Bou G, Cervero G, Dominguez MA, Quereda C, Martínez-Beltran J. 2000. Characterization of a nosocomial outbreak caused by a multiresistant Acinetobacter baumannii strain with a carbapenem-hydrolyzing enzyme: high-level carbapenem resistance in A. baumannii is not due solely to the presence of β-lactamases. J Clin Microbiol 38:3299–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.del Mar Tomás M, Beceiro A, Pérez A, Velasco D, Moure R, Villanueva R, Martínez-Beltran J, Bou G. 2005. Cloning and functional analysis of the gene encoding the 33- to 36-kilodalton outer membrane protein associated with carbapenem resistance in Acinetobacter baumannii. Antimicrob Agents Chemother 49:5172–5175. doi: 10.1128/AAC.49.12.5172-5175.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Hood MI, Jacobs AC, Sayood K, Dunman PM, Skaar EP. 2010. Acinetobacter baumannii increases tolerance to antibiotics in response to monovalent cations. Antimicrob Agents Chemother 54:1029–1041. doi: 10.1128/AAC.00963-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524.Magnet S, Courvalin P, Lambert T. 2001. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob Agents Chemother 45:3375–3380. doi: 10.1128/AAC.45.12.3375-3380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525.Nemec A, Maixnerova M, van der Reijden TJ, van den Broek PJ, Dijkshoorn L. 2007. Relationship between the AdeABC efflux system gene content, netilmicin susceptibility and multidrug resistance in a genotypically diverse collection of Acinetobacter baumannii strains. J Antimicrob Chemother 60:483–489. doi: 10.1093/jac/dkm231. [DOI] [PubMed] [Google Scholar]
- 526.Ruzin A, Keeney D, Bradford PA. 2007. AdeABC multidrug efflux pump is associated with decreased susceptibility to tigecycline in Acinetobacter calcoaceticus-Acinetobacter baumannii complex. J Antimicrob Chemother 59:1001–1004. doi: 10.1093/jac/dkm058. [DOI] [PubMed] [Google Scholar]
- 527.Valentine SC, Contreras D, Tan S, Real LJ, Chu S, Xu HH. 2008. Phenotypic and molecular characterization of Acinetobacter baumannii clinical isolates from nosocomial outbreaks in Los Angeles County, California. J Clin Microbiol 46:2499–2507. doi: 10.1128/JCM.00367-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528.Lin L, Ling BD, Li X-Z. 2009. Distribution of the multidrug efflux pump genes, adeABC, adeDE and adeIJK, and class 1 integron genes in multiple-antimicrobial-resistant clinical isolates of Acinetobacter baumannii-Acinetobacter calcoaceticus complex. Int J Antimicrob Agents 33:27–32. doi: 10.1016/j.ijantimicag.2008.06.027. [DOI] [PubMed] [Google Scholar]
- 529.Landman D, Butnariu M, Bratu S, Quale J. 2009. Genetic relatedness of multidrug-resistant Acinetobacter baumannii endemic to New York City. Epidemiol Infect 137:174–180. doi: 10.1017/S0950268808000824. [DOI] [PubMed] [Google Scholar]
- 530.Rajamohan G, Srinivasan VB, Gebreyes WA. 2010. Novel role of Acinetobacter baumannii RND efflux transporters in mediating decreased susceptibility to biocides. J Antimicrob Chemother 65:228–232. doi: 10.1093/jac/dkp427. [DOI] [PubMed] [Google Scholar]
- 531.Hornsey M, Ellington MJ, Doumith M, Thomas CP, Gordon NC, Wareham DW, Quinn J, Lolans K, Livermore DM, Woodford N. 2010. AdeABC-mediated efflux and tigecycline MICs for epidemic clones of Acinetobacter baumannii. J Antimicrob Chemother 65:1589–1593. doi: 10.1093/jac/dkq218. [DOI] [PubMed] [Google Scholar]
- 532.Fernando D, Zhanel G, Kumar A. 2013. Antibiotic resistance and expression of resistance-nodulation-division pump- and outer membrane porin-encoding genes in Acinetobacter species isolated from Canadian hospitals. Can J Infect Dis Med Microbiol 24:17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Deng M, Zhu MH, Li JJ, Bi S, Sheng ZK, Hu FS, Zhang JJ, Chen W, Xue XW, Sheng JF, Li LJ. 2014. Molecular epidemiology and mechanisms of tigecycline resistance in clinical isolates of Acinetobacter baumannii from a Chinese university hospital. Antimicrob Agents Chemother 58:297–303. doi: 10.1128/AAC.01727-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Lee SY, Yun SH, Lee YG, Choi CW, Leem SH, Park EC, Kim GH, Lee JC, Kim SI. 2014. Proteogenomic characterization of antimicrobial resistance in extensively drug-resistant Acinetobacter baumannii DU202. J Antimicrob Chemother 69:1483–1491. doi: 10.1093/jac/dku008. [DOI] [PubMed] [Google Scholar]
- 535.Peleg AY, Potoski BA, Rea R, Adams J, Sethi J, Capitano B, Husain S, Kwak EJ, Bhat SV, Paterson DL. 2007. Acinetobacter baumannii bloodstream infection while receiving tigecycline: a cautionary report. J Antimicrob Chemother 59:128–131. doi: 10.1093/jac/dkl441. [DOI] [PubMed] [Google Scholar]
- 536.Peleg AY, Adams J, Paterson DL. 2007. Tigecycline efflux as a mechanism for nonsusceptibility in Acinetobacter baumannii. Antimicrob Agents Chemother 51:2065–2069. doi: 10.1128/AAC.01198-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Bratu S, Landman D, Martin DA, Georgescu C, Quale J. 2008. Correlation of antimicrobial resistance with β-lactamases, the OmpA-like porin, and efflux pumps in clinical isolates of Acinetobacter baumannii endemic to New York City. Antimicrob Agents Chemother 52:2999–3005. doi: 10.1128/AAC.01684-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538.Sun JR, Perng CL, Chan MC, Morita Y, Lin JC, Su CM, Wang WY, Chang TY, Chiueh TS. 2012. A truncated AdeS kinase protein generated by ISAba1 insertion correlates with tigecycline resistance in Acinetobacter baumannii. PLoS One 7:e49534. doi: 10.1371/journal.pone.0049534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Fluit AC, Florijn A, Verhoef J, Milatovic D. 2005. Presence of tetracycline resistance determinants and susceptibility to tigecycline and minocycline. Antimicrob Agents Chemother 49:1636–1638. doi: 10.1128/AAC.49.4.1636-1638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Horiyama T, Nikaido E, Yamaguchi A, Nishino K. 2011. Roles of Salmonella multidrug efflux pumps in tigecycline resistance. J Antimicrob Chemother 66:105–110. doi: 10.1093/jac/dkq421. [DOI] [PubMed] [Google Scholar]
- 541.Chen Q, Li X, Zhou H, Jiang Y, Chen Y, Hua X, Yu Y. 2014. Decreased susceptibility to tigecycline in Acinetobacter baumannii mediated by a mutation in trm encoding SAM-dependent methyltransferase. J Antimicrob Chemother 69:72–76. doi: 10.1093/jac/dkt319. [DOI] [PubMed] [Google Scholar]
- 542.Damier-Piolle L, Magnet S, Bremont S, Lambert T, Courvalin P. 2008. AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob Agents Chemother 52:557–562. doi: 10.1128/AAC.00732-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Rosenfeld N, Bouchier C, Courvalin P, Perichon B. 2012. Expression of the resistance-nodulation-cell division pump AdeIJK in Acinetobacter baumannii is regulated by AdeN, a TetR-type regulator. Antimicrob Agents Chemother 56:2504–2510. doi: 10.1128/AAC.06422-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Fernando DM, Xu W, Loewen PC, Zhanel GG, Kumar A. 2014. Triclosan can select for an AdeIJK overexpressing mutant of Acinetobacter baumannii ATCC 17978 that displays reduced susceptibility to multiple antibiotics. Antimicrob Agents Chemother 58:6424–6431. doi: 10.1128/AAC.03074-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545.Sugawara E, Nikaido H. 2014. Properties of AdeABC and AdeIJK efflux systems of Acinetobacter baumannii compared with those of the AcrAB-TolC system of Escherichia coli. Antimicrob Agents Chemother 58:7250–7257. doi: 10.1128/AAC.03728-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546.Coyne S, Guigon G, Courvalin P, Perichon B. 2010. Screening and quantification of the expression of antibiotic resistance genes in Acinetobacter baumannii with a microarray. Antimicrob Agents Chemother 54:333–340. doi: 10.1128/AAC.01037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 547.Coyne S, Rosenfeld N, Lambert T, Courvalin P, Perichon B. 2010. Overexpression of resistance-nodulation-cell division pump AdeFGH confers multidrug resistance in Acinetobacter baumannii. Antimicrob Agents Chemother 54:4389–4393. doi: 10.1128/AAC.00155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 548.Lin MF, Lin YY, Yeh HW, Lan CY. 2014. Role of the BaeSR two-component system in the regulation of Acinetobacter baumannii adeAB genes and its correlation with tigecycline susceptibility. BMC Microbiol 14:119. doi: 10.1186/1471-2180-14-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 549.Roca I, Espinal P, Marti S, Vila J. 2011. First identification and characterization of an AdeABC-like efflux pump in Acinetobacter genomospecies 13TU. Antimicrob Agents Chemother 55:1285–1286. doi: 10.1128/AAC.01142-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 550.Pannek S, Higgins PG, Steinke P, Jonas D, Akova M, Bohnert JA, Seifert H, Kern WV. 2006. Multidrug efflux inhibition in Acinetobacter baumannii: comparison between 1-(1-naphthylmethyl)-piperazine and phenyl-arginine-β-naphthylamide. J Antimicrob Chemother 57:970–974. doi: 10.1093/jac/dkl081. [DOI] [PubMed] [Google Scholar]
- 551.Bean DC, Wareham DW. 2009. Paradoxical effect of 1-(1-naphthylmethyl)-piperazine on resistance to tetracyclines in multidrug-resistant Acinetobacter baumannii. J Antimicrob Chemother 63:349–352. doi: 10.1093/jac/dkn493. [DOI] [PubMed] [Google Scholar]
- 552.Cortez-Cordova J, Kumar A. 2011. Activity of the efflux pump inhibitor phenylalanine-arginine β-naphthylamide against the AdeFGH pump of Acinetobacter baumannii. Int J Antimicrob Agents 37:420–424. doi: 10.1016/j.ijantimicag.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 553.Ribera A, Ruiz J, Jiminez de Anta MT, Vila J. 2002. Effect of an efflux pump inhibitor on the MIC of nalidixic acid for Acinetobacter baumannii and Stenotrophomonas maltophilia clinical isolates. J Antimicrob Chemother 49:697–698. doi: 10.1093/jac/49.4.697. [DOI] [PubMed] [Google Scholar]
- 554.Golanbar GD, Lam CK, Chu YM, Cueva C, Tan SW, Silva I, Xu HH. 2011. Phenotypic and molecular characterization of Acinetobacter clinical isolates obtained from inmates of California correctional facilities. J Clin Microbiol 49:2121–2131. doi: 10.1128/JCM.02373-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 555.Giannouli M, Di Popolo A, Durante-Mangoni E, Bernardo M, Cuccurullo S, Amato G, Tripodi MF, Triassi M, Utili R, Zarrilli R. 2012. Molecular epidemiology and mechanisms of rifampicin resistance in Acinetobacter baumannii isolates from Italy. Int J Antimicrob Agents 39:58–63. doi: 10.1016/j.ijantimicag.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 556.Yang Y, Chua KL. 2013. Assessment of the effect of efflux pump inhibitors on in vitro antimicrobial susceptibility of multidrug-resistant Acinetobacter baumannii. Int J Antimicrob Agents 42:283–284. doi: 10.1016/j.ijantimicag.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 557.Ni W, Cai X, Liang B, Cai Y, Cui J, Wang R. 2014. Effect of proton pump inhibitors on in vitro activity of tigecycline against several common clinical pathogens. PLoS One 9:e86715. doi: 10.1371/journal.pone.0086715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 558.Rajamohan G, Srinivasan VB, Gebreyes WA. 2010. Molecular and functional characterization of a novel efflux pump, AmvA, mediating antimicrobial and disinfectant resistance in Acinetobacter baumannii. J Antimicrob Chemother 65:1919–1925. doi: 10.1093/jac/dkq195. [DOI] [PubMed] [Google Scholar]
- 559.Hou PF, Chen XY, Yan GF, Wang YP, Ying CM. 2012. Study of the correlation of imipenem resistance with efflux pumps AdeABC, AdeIJK, AdeDE and AbeM in clinical isolates of Acinetobacter baumannii. Chemotherapy 58:152–158. doi: 10.1159/000335599. [DOI] [PubMed] [Google Scholar]
- 560.Eijkelkamp BA, Hassan KA, Paulsen IT, Brown MH. 2011. Development of a high-throughput cloning strategy for characterization of Acinetobacter baumannii drug transporter proteins. J Mol Microbiol Biotechnol 20:211–219. doi: 10.1159/000329836. [DOI] [PubMed] [Google Scholar]
- 561.Roca I, Marti S, Espinal P, Martínez P, Gibert I, Vila J. 2009. CraA, a major facilitator superfamily efflux pump associated with chloramphenicol resistance in Acinetobacter baumannii. Antimicrob Agents Chemother 53:4013–4014. doi: 10.1128/AAC.00584-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Vilacoba E, Almuzara M, Gulone L, Traglia GM, Figueroa SA, Sly G, Fernández A, Centron D, Ramirez MS. 2013. Emergence and spread of plasmid-borne tet(B)::ISCR2 in minocycline-resistant Acinetobacter baumannii isolates. Antimicrob Agents Chemother 57:651–654. doi: 10.1128/AAC.01751-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 563.Hassan KA, Jackson SM, Penesyan A, Patching SG, Tetu SG, Eijkelkamp BA, Brown MH, Henderson PJ, Paulsen IT. 2013. Transcriptomic and biochemical analyses identify a family of chlorhexidine efflux proteins. Proc Natl Acad Sci U S A 110:20254–20259. doi: 10.1073/pnas.1317052110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 564.Fuangthong M, Julotok M, Chintana W, Kuhn K, Rittiroongrad S, Vattanaviboon P, Mongkolsuk S. 2011. Exposure of Acinetobacter baylyi ADP1 to the biocide chlorhexidine leads to acquired resistance to the biocide itself and to oxidants. J Antimicrob Chemother 66:319–322. doi: 10.1093/jac/dkq435. [DOI] [PubMed] [Google Scholar]
- 565.Zhang L, Li XZ, Poole K. 2000. Multiple antibiotic resistance in Stenotrophomonas maltophilia: involvement of a multidrug efflux system. Antimicrob Agents Chemother 44:287–293. doi: 10.1128/AAC.44.2.287-293.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 566.Brooke JS. 2012. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin Microbiol Rev 25:2–41. doi: 10.1128/CMR.00019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 567.Mett H, Rosta S, Schacher B, Frei R. 1988. Outer membrane permeability and β-lactamase content in Pseudomonas maltophilia clinical isolates and laboratory mutants. Rev Infect Dis 10:765–769. doi: 10.1093/clinids/10.4.765. [DOI] [PubMed] [Google Scholar]
- 568.McKay GA, Woods DE, MacDonald KL, Poole K. 2003. Role of phosphoglucomutase of Stenotrophomonas maltophilia in lipopolysaccharide biosynthesis, virulence, and antibiotic resistance. Infect Immun 71:3068–3075. doi: 10.1128/IAI.71.6.3068-3075.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 569.Alonso A, Martínez JL. 1997. Multiple antibiotic resistance in Stenotrophomonas maltophilia. Antimicrob Agents Chemother 41:1140–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 570.Crossman LC, Gould VC, Dow JM, Vernikos GS, Okazaki A, Sebaihia M, Saunders D, Arrowsmith C, Carver T, Peters N, Adlem E, Kerhornou A, Lord A, Murphy L, Seeger K, Squares R, Rutter S, Quail MA, Rajandream MA, Harris D, Churcher C, Bentley SD, Parkhill J, Thomson NR, Avison MB. 2008. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol 9:R74. doi: 10.1186/gb-2008-9-4-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 571.Li X-Z, Zhang L, Poole K. 2002. SmeC, an outer membrane multidrug efflux protein of Stenotrophomonas maltophilia. Antimicrob Agents Chemother 46:333–343. doi: 10.1128/AAC.46.2.333-343.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 572.Chang LL, Chen HF, Chang CY, Lee TM, Wu WJ. 2004. Contribution of integrons, and SmeABC and SmeDEF efflux pumps to multidrug resistance in clinical isolates of Stenotrophomonas maltophilia. J Antimicrob Chemother 53:518–521. doi: 10.1093/jac/dkh094. [DOI] [PubMed] [Google Scholar]
- 573.Cho HH, Sung JY, Kwon KC, Koo SH. 2012. Expression of Sme efflux pumps and multilocus sequence typing in clinical isolates of Stenotrophomonas maltophilia. Ann Lab Med 32:38–43. doi: 10.3343/alm.2012.32.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 574.Zhang L, Li XZ, Poole K. 2001. SmeDEF multidrug efflux pump contributes to intrinsic multidrug resistance in Stenotrophomonas maltophilia. Antimicrob Agents Chemother 45:3497–3503. doi: 10.1128/AAC.45.12.3497-3503.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 575.Alonso A, Martínez JL. 2001. Expression of multidrug efflux pump SmeDEF by clinical isolates of Stenotrophomonas maltophilia. Antimicrob Agents Chemother 45:1879–1881. doi: 10.1128/AAC.45.6.1879-1881.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 576.Gould VC, Avison MB. 2006. SmeDEF-mediated antimicrobial drug resistance in Stenotrophomonas maltophilia clinical isolates having defined phylogenetic relationships. J Antimicrob Chemother 57:1070–1076. doi: 10.1093/jac/dkl106. [DOI] [PubMed] [Google Scholar]
- 577.Sánchez P, Moreno E, Martínez JL. 2005. The biocide triclosan selects Stenotrophomonas maltophilia mutants that overproduce the SmeDEF multidrug efflux pump. Antimicrob Agents Chemother 49:781–782. doi: 10.1128/AAC.49.2.781-782.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 578.García-León G, Salgado F, Oliveros JC, Sánchez MB, Martínez JL. 2014. Interplay between intrinsic and acquired resistance to quinolones in Stenotrophomonas maltophilia. Environ Microbiol 16:1282–1296. doi: 10.1111/1462-2920.12408. [DOI] [PubMed] [Google Scholar]
- 579.Lin CW, Huang YW, Hu RM, Yang TC. 2014. SmeOP-TolCsm efflux pump contributes to the multidrug resistance of Stenotrophomonas maltophilia. Antimicrob Agents Chemother 58:2405–2408. doi: 10.1128/AAC.01974-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 580.Huang YW, Hu RM, Yang TC. 2013. Role of the pcm-tolCsm operon in the multidrug resistance of Stenotrophomonas maltophilia. J Antimicrob Chemother 68:1987–1993. doi: 10.1093/jac/dkt148. [DOI] [PubMed] [Google Scholar]
- 581.Chen CH, Huang CC, Chung TC, Hu RM, Huang YW, Yang TC. 2011. Contribution of resistance-nodulation-division efflux pump operon smeU1-V-W-U2-X to multidrug resistance of Stenotrophomonas maltophilia. Antimicrob Agents Chemother 55:5826–5833. doi: 10.1128/AAC.00317-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 582.Huang YW, Liou RS, Lin YT, Huang HH, Yang TC. 2014. A linkage between SmeIJK efflux pump, cell envelope integrity, and σE-mediated envelope stress response in Stenotrophomonas maltophilia. PLoS One 9:e111784. doi: 10.1371/journal.pone.0111784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 583.Hu RM, Liao ST, Huang CC, Huang YW, Yang TC. 2012. An inducible fusaric acid tripartite efflux pump contributes to the fusaric acid resistance in Stenotrophomonas maltophilia. PLoS One 7:e51053. doi: 10.1371/journal.pone.0051053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 584.Lin YT, Huang YW, Liou RS, Chang YC, Yang TC. 2014. MacABCsm, an ABC-type tripartite efflux pump of Stenotrophomonas maltophilia involved in drug resistance, oxidative and envelope stress tolerances and biofilm formation. J Antimicrob Chemother 69:3221–3226. doi: 10.1093/jac/dku317. [DOI] [PubMed] [Google Scholar]
- 585.Al-Hamad A, Upton M, Burnie J. 2009. Molecular cloning and characterization of SmrA, a novel ABC multidrug efflux pump from Stenotrophomonas maltophilia. J Antimicrob Chemother 64:731–734. doi: 10.1093/jac/dkp271. [DOI] [PubMed] [Google Scholar]
- 586.Al-Hamad A, Burnie J, Upton M. 2011. Enhancement of antibiotic susceptibility of Stenotrophomonas maltophilia using a polyclonal antibody developed against an ABC multidrug efflux pump. Can J Microbiol 57:820–828. doi: 10.1139/w11-076. [DOI] [PubMed] [Google Scholar]
- 587.Huang YW, Hu RM, Chu FY, Lin HR, Yang TC. 2013. Characterization of a major facilitator superfamily (MFS) tripartite efflux pump EmrCABsm from Stenotrophomonas maltophilia. J Antimicrob Chemother 68:2498–2505. doi: 10.1093/jac/dkt250. [DOI] [PubMed] [Google Scholar]
- 588.Traxler RM, Lehman MW, Bosserman EA, Guerra MA, Smith TL. 2013. A literature review of laboratory-acquired brucellosis. J Clin Microbiol 51:3055–3062. doi: 10.1128/JCM.00135-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 589.Paulsen IT, Seshadri R, Nelson KE, Eisen JA, Heidelberg JF, Read TD, Dodson RJ, Umayam L, Brinkac LM, Beanan MJ, Daugherty SC, Deboy RT, Durkin AS, Kolonay JF, Madupu R, Nelson WC, Ayodeji B, Kraul M, Shetty J, Malek J, Van Aken SE, Riedmuller S, Tettelin H, Gill SR, White O, Salzberg SL, Hoover DL, Lindler LE, Halling SM, Boyle SM, Fraser CM. 2002. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc Natl Acad Sci U S A 99:13148–13153. doi: 10.1073/pnas.192319099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 590.Halling SM, Peterson-Burch BD, Bricker BJ, Zuerner RL, Qing Z, Li LL, Kapur V, Alt DP, Olsen SC. 2005. Completion of the genome sequence of Brucella abortus and comparison to the highly similar genomes of Brucella melitensis and Brucella suis. J Bacteriol 187:2715–2726. doi: 10.1128/JB.187.8.2715-2726.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 591.Douglas JT, Rosenberg EY, Nikaido H, Verstreate DR, Winter AJ. 1984. Porins of Brucella species. Infect Immun 44:16–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 592.Roussel G, Matagne A, De Bolle X, Perpete EA, Michaux C. 2012. Purification, refolding and characterization of the trimeric Omp2a outer membrane porin from Brucella melitensis. Protein Expr Purif 83:198–204. doi: 10.1016/j.pep.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 593.Posadas DM, Martin FA, Sabio y Garcia JV, Spera JM, Delpino MV, Baldi P, Campos E, Cravero SL, Zorreguieta A. 2007. The TolC homologue of Brucella suis is involved in resistance to antimicrobial compounds and virulence. Infect Immun 75:379–389. doi: 10.1128/IAI.01349-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 594.Martin FA, Posadas DM, Carrica MC, Cravero SL, O'Callaghan D, Zorreguieta A. 2009. Interplay between two RND systems mediating antimicrobial resistance in Brucella suis. J Bacteriol 191:2530–2540. doi: 10.1128/JB.01198-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 595.Halling SM, Jensen AE. 2006. Intrinsic and selected resistance to antibiotics binding the ribosome: analyses of Brucella 23S rrn, L4, L22, EF-Tu1, EF-Tu2, efflux and phylogenetic implications. BMC Microbiol 6:84. doi: 10.1186/1471-2180-6-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 596.Ravanel N, Gestin B, Maurin M. 2009. In vitro selection of fluoroquinolone resistance in Brucella melitensis. Int J Antimicrob Agents 34:76–81. doi: 10.1016/j.ijantimicag.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 597.Valdezate S, Navarro A, Medina-Pascual MJ, Carrasco G, Saez-Nieto JA. 2010. Molecular screening for rifampicin and fluoroquinolone resistance in a clinical population of Brucella melitensis. J Antimicrob Chemother 65:51–53. doi: 10.1093/jac/dkp389. [DOI] [PubMed] [Google Scholar]
- 598.Biswas S, Rolain JM. 2010. Bartonella infection: treatment and drug resistance. Future Microbiol 5:1719–1731. doi: 10.2217/fmb.10.133. [DOI] [PubMed] [Google Scholar]
- 599.Biswas S, Raoult D, Rolain JM. 2008. A bioinformatic approach to understanding antibiotic resistance in intracellular bacteria through whole genome analysis. Int J Antimicrob Agents 32:207–220. doi: 10.1016/j.ijantimicag.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 600.Li DM, Liu QY, Zhao F, Hu Y, Xiao D, Gu YX, Song XP, Zhang JZ. 2011. Proteomic and bioinformatic analysis of outer membrane proteins of the protobacterium Bartonella henselae (Bartonellaceae). Genet Mol Res 10:1789–1818. doi: 10.4238/vol10-3gmr1153. [DOI] [PubMed] [Google Scholar]
- 601.Rolain JM, Vayssier-Taussat M, Saisongkorh W, Merhej V, Gimenez G, Robert C, Le Rhun D, Dehio C, Raoult D. 2013. Partial disruption of translational and posttranslational machinery reshapes growth rates of Bartonella birtlesii. mBio 4(2):e00115-13. doi: 10.1128/mBio.00115-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 602.Rolain JM, Raoult D. 2005. Genome comparison analysis of molecular mechanisms of resistance to antibiotics in the Rickettsia genus. Ann N Y Acad Sci 1063:222–230. doi: 10.1196/annals.1355.035. [DOI] [PubMed] [Google Scholar]
- 603.Felsheim RF, Kurtti TJ, Munderloh UG. 2009. Genome sequence of the endosymbiont Rickettsia peacockii and comparison with virulent Rickettsia rickettsii: identification of virulence factors. PLoS One 4:e8361. doi: 10.1371/journal.pone.0008361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 604.Gillings M, Boucher Y, Labbate M, Holmes A, Krishnan S, Holley M, Stokes HW. 2008. The evolution of class 1 integrons and the rise of antibiotic resistance. J Bacteriol 190:5095–5100. doi: 10.1128/JB.00152-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 605.Jakobsen TH, Hansen MA, Jensen PO, Hansen L, Riber L, Cockburn A, Kolpen M, Ronne Hansen C, Ridderberg W, Eickhardt S, Hansen M, Kerpedjiev P, Alhede M, Qvortrup K, Burmolle M, Moser C, Kuhl M, Ciofu O, Givskov M, Sorensen SJ, Hoiby N, Bjarnsholt T. 2013. Complete genome sequence of the cystic fibrosis pathogen Achromobacter xylosoxidans NH44784-1996 complies with important pathogenic phenotypes. PLoS One 8:e68484. doi: 10.1371/journal.pone.0068484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 606.Bador J, Amoureux L, Duez JM, Drabowicz A, Siebor E, Llanes C, Neuwirth C. 2011. First description of an RND-type multidrug efflux pump in Achromobacter xylosoxidans, AxyABM. Antimicrob Agents Chemother 55:4912–4914. doi: 10.1128/AAC.00341-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 607.Bador J, Amoureux L, Blanc E, Neuwirth C. 2013. Innate aminoglycoside resistance of Achromobacter xylosoxidans is due to AxyXY-OprZ, an RND-type multidrug efflux pump. Antimicrob Agents Chemother 57:603–605. doi: 10.1128/AAC.01243-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 608.Schell MA, Zhao P, Wells L. 2011. Outer membrane proteome of Burkholderia pseudomallei and Burkholderia mallei from diverse growth conditions. J Proteome Res 10:2417–2424. doi: 10.1021/pr1012398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 609.Siritapetawee J, Prinz H, Krittanai C, Suginta W. 2004. Expression and refolding of Omp38 from Burkholderia pseudomallei and Burkholderia thailandensis, and its function as a diffusion porin. Biochem J 384:609–617. doi: 10.1042/BJ20041102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 610.Suginta W, Mahendran KR, Chumjan W, Hajjar E, Schulte A, Winterhalter M, Weingart H. 2011. Molecular analysis of antimicrobial agent translocation through the membrane porin BpsOmp38 from an ultraresistant Burkholderia pseudomallei strain. Biochim Biophys Acta 1808:1552–1559. doi: 10.1016/j.bbamem.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 611.Guglierame P, Pasca MR, De Rossi E, Buroni S, Arrigo P, Manina G, Riccardi G. 2006. Efflux pump genes of the resistance-nodulation-division family in Burkholderia cenocepacia genome. BMC Microbiol 6:66. doi: 10.1186/1471-2180-6-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 612.Perrin E, Fondi M, Papaleo MC, Maida I, Emiliani G, Buroni S, Pasca MR, Riccardi G, Fani R. 2013. A census of RND superfamily proteins in the Burkholderia genus. Future Microbiol 8:923–937. doi: 10.2217/fmb.13.50. [DOI] [PubMed] [Google Scholar]
- 613.Buroni S, Matthijs N, Spadaro F, Van Acker H, Scoffone VC, Pasca MR, Riccardi G, Coenye T. 2014. Differential roles of RND efflux pumps in antimicrobial drug resistance of sessile and planktonic Burkholderia cenocepacia cells. Antimicrob Agents Chemother 58:7424–7429. doi: 10.1128/AAC.03800-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 614.Buroni S, Pasca MR, Flannagan RS, Bazzini S, Milano A, Bertani I, Venturi V, Valvano MA, Riccardi G. 2009. Assessment of three resistance-nodulation-cell division drug efflux transporters of Burkholderia cenocepacia in intrinsic antibiotic resistance. BMC Microbiol 9:200. doi: 10.1186/1471-2180-9-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 615.Bazzini S, Udine C, Sass A, Pasca MR, Longo F, Emiliani G, Fondi M, Perrin E, Decorosi F, Viti C, Giovannetti L, Leoni L, Fani R, Riccardi G, Mahenthiralingam E, Buroni S. 2011. Deciphering the role of RND efflux transporters in Burkholderia cenocepacia. PLoS One 6:e18902. doi: 10.1371/journal.pone.0018902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 616.Tseng SP, Tsai WC, Liang CY, Lin YS, Huang JW, Chang CY, Tyan YC, Lu PL. 2014. The contribution of antibiotic resistance mechanisms in clinical Burkholderia cepacia complex isolates: an emphasis on efflux pump activity. PLoS One 9:e104986. doi: 10.1371/journal.pone.0104986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 617.Chan YY, Tan TM, Ong YM, Chua KL. 2004. BpeAB-OprB, a multidrug efflux pump in Burkholderia pseudomallei. Antimicrob Agents Chemother 48:1128–1135. doi: 10.1128/AAC.48.4.1128-1135.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 618.Rajendran R, Quinn RF, Murray C, McCulloch E, Williams C, Ramage G. 2010. Efflux pumps may play a role in tigecycline resistance in Burkholderia species. Int J Antimicrob Agents 36:151–154. doi: 10.1016/j.ijantimicag.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 619.Sass A, Marchbank A, Tullis E, Lipuma JJ, Mahenthiralingam E. 2011. Spontaneous and evolutionary changes in the antibiotic resistance of Burkholderia cenocepacia observed by global gene expression analysis. BMC Genomics 12:373. doi: 10.1186/1471-2164-12-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 620.Kumar A, Mayo M, Trunck LA, Cheng AC, Currie BJ, Schweizer HP. 2008. Expression of resistance-nodulation-cell-division efflux pumps in commonly used Burkholderia pseudomallei strains and clinical isolates from northern Australia. Trans R Soc Trop Med Hyg 102(Suppl 1):S145–S151. doi: 10.1016/S0035-9203(08)70032-4. [DOI] [PubMed] [Google Scholar]
- 621.Moore RA, DeShazer D, Reckseidler S, Weissman A, Woods DE. 1999. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob Agents Chemother 43:465–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 622.Chan YY, Chua KL. 2005. The Burkholderia pseudomallei BpeAB-OprB efflux pump: expression and impact on quorum sensing and virulence. J Bacteriol 187:4707–4719. doi: 10.1128/JB.187.14.4707-4719.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 623.Kumar A, Chua KL, Schweizer HP. 2006. Method for regulated expression of single-copy efflux pump genes in a surrogate Pseudomonas aeruginosa strain: identification of the BpeEF-OprC chloramphenicol and trimethoprim efflux pump of Burkholderia pseudomallei 1026b. Antimicrob Agents Chemother 50:3460–3463. doi: 10.1128/AAC.00440-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 624.Podnecky NL, Wuthiekanun V, Peacock SJ, Schweizer HP. 2013. The BpeEF-OprC efflux pump is responsible for widespread trimethoprim resistance in clinical and environmental Burkholderia pseudomallei isolates. Antimicrob Agents Chemother 57:4381–4386. doi: 10.1128/AAC.00660-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 625.Podin Y, Sarovich DS, Price EP, Kaestli M, Mayo M, Hii K, Ngian H, Wong S, Wong I, Wong J, Mohan A, Ooi M, Fam T, Wong J, Tuanyok A, Keim P, Giffard PM, Currie BJ. 2014. Burkholderia pseudomallei isolates from Sarawak, Malaysian Borneo, are predominantly susceptible to aminoglycosides and macrolides. Antimicrob Agents Chemother 58:162–166. doi: 10.1128/AAC.01842-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 626.Howard K, Inglis TJ. 2003. Novel selective medium for isolation of Burkholderia pseudomallei. J Clin Microbiol 41:3312–3316. doi: 10.1128/JCM.41.7.3312-3316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 627.Biot FV, Lopez MM, Poyot T, Neulat-Ripoll F, Lignon S, Caclard A, Thibault FM, Peinnequin A, Pagès JM, Valade E. 2013. Interplay between three RND efflux pumps in doxycycline-selected strains of Burkholderia thailandensis. PLoS One 8:e84068. doi: 10.1371/journal.pone.0084068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 628.Li X-Z, Barré N, Poole K. 2000. Influence of the MexA-MexB-OprM multidrug efflux system on expression of the MexC-MexD-OprJ and MexE-MexF-OprN multidrug efflux systems in Pseudomonas aeruginosa. J Antimicrob Chemother 46:885–893. doi: 10.1093/jac/46.6.885. [DOI] [PubMed] [Google Scholar]
- 629.Blair JM, Smith HE, Ricci V, Lawler AJ, Thompson LJ, Piddock LJ. 2015. Expression of homologous RND efflux pump genes is dependent upon AcrB expression: implications for efflux and virulence inhibitor design. J Antimicrob Chemother 70:424–431. doi: 10.1093/jac/dku380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 630.Sánchez P, Le U, Martínez JL. 2003. The efflux pump inhibitor Phe-Arg-β-naphthylamide does not abolish the activity of the Stenotrophomonas maltophilia SmeDEF multidrug efflux pump. J Antimicrob Chemother 51:1042–1045. doi: 10.1093/jac/dkg181. [DOI] [PubMed] [Google Scholar]
- 631.Unemo M, Shafer WM. 2014. Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin Microbiol Rev 27:587–613. doi: 10.1128/CMR.00010-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 632.Young JD, Blake M, Mauro A, Cohn ZA. 1983. Properties of the major outer membrane protein from Neisseria gonorrhoeae incorporated into model lipid membranes. Proc Natl Acad Sci U S A 80:3831–3835. doi: 10.1073/pnas.80.12.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 633.Pan W, Spratt BG. 1994. Regulation of the permeability of the gonococcal cell envelope by the mtr system. Mol Microbiol 11:769–775. doi: 10.1111/j.1365-2958.1994.tb00354.x. [DOI] [PubMed] [Google Scholar]
- 634.Olesky M, Zhao S, Rosenberg RL, Nicholas RA. 2006. Porin-mediated antibiotic resistance in Neisseria gonorrhoeae: ion, solute, and antibiotic permeation through PIB proteins with penB mutations. J Bacteriol 188:2300–2308. doi: 10.1128/JB.188.7.2300-2308.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 635.Shafer WM, Folster JP. 2006. Towards an understanding of chromosomally mediated penicillin resistance in Neisseria gonorrhoeae: evidence for a porin-efflux pump collaboration. J Bacteriol 188:2297–2299. doi: 10.1128/JB.188.7.2297-2299.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 636.Camara J, Serra J, Ayats J, Bastida T, Carnicer-Pont D, Andreu A, Ardanuy C. 2012. Molecular characterization of two high-level ceftriaxone-resistant Neisseria gonorrhoeae isolates detected in Catalonia, Spain. J Antimicrob Chemother 67:1858–1860. doi: 10.1093/jac/dks162. [DOI] [PubMed] [Google Scholar]
- 637.Rouquette C, Harmon JB, Shafer WM. 1999. Induction of the mtrCDE-encoded efflux pump system of Neisseria gonorrhoeae requires MtrA, an AraC-like protein. Mol Microbiol 33:651–658. doi: 10.1046/j.1365-2958.1999.01517.x. [DOI] [PubMed] [Google Scholar]
- 638.Shafer WM, Qu X, Waring AJ, Lehrer RI. 1998. Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc Natl Acad Sci U S A 95:1829–1833. doi: 10.1073/pnas.95.4.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 639.Hagman KE, Pan W, Spratt BG, Balthazar JT, Judd RC, Shafer WM. 1995. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology 141:611–622. doi: 10.1099/13500872-141-3-611. [DOI] [PubMed] [Google Scholar]
- 640.Jerse AE, Sharma ND, Simms AN, Crow ET, Snyder LA, Shafer WM. 2003. A gonococcal efflux pump system enhances bacterial survival in a female mouse model of genital tract infection. Infect Immun 71:5576–5582. doi: 10.1128/IAI.71.10.5576-5582.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 641.Warner DM, Shafer WM, Jerse AE. 2008. Clinically relevant mutations that cause derepression of the Neisseria gonorrhoeae MtrC-MtrD-MtrE efflux pump system confer different levels of antimicrobial resistance and in vivo fitness. Mol Microbiol 70:462–478. doi: 10.1111/j.1365-2958.2008.06424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 642.Ohneck EA, Goytia M, Rouquette-Loughlin CE, Joseph SJ, Read TD, Jerse AE, Shafer WM. 2015. Overproduction of the MtrCDE efflux pump in Neisseria gonorrhoeae produces unexpected changes in cellular transcription patterns. Antimicrob Agents Chemother 59:724–726. doi: 10.1128/AAC.04148-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 643.Bolla JR, Su CC, Do SV, Radhakrishnan A, Kumar N, Long F, Chou TH, Delmar JA, Lei HT, Rajashankar KR, Shafer WM, Yu EW. 2014. Crystal structure of the Neisseria gonorrhoeae MtrD inner membrane multidrug efflux pump. PLoS One 9:e97903. doi: 10.1371/journal.pone.0097903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 644.Lei HT, Chou TH, Su CC, Bolla JR, Kumar N, Radhakrishnan A, Long F, Delmar JA, Do SV, Rajashankar KR, Shafer WM, Yu EW. 2014. Crystal structure of the open state of the Neisseria gonorrhoeae MtrE outer membrane channel. PLoS One 9:e97475. doi: 10.1371/journal.pone.0097475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 645.Lee EH, Hill SA, Napier R, Shafer WM. 2006. Integration host factor is required for FarR repression of the farAB-encoded efflux pump of Neisseria gonorrhoeae. Mol Microbiol 60:1381–1400. doi: 10.1111/j.1365-2958.2006.05185.x. [DOI] [PubMed] [Google Scholar]
- 646.Morse SA, Lysko PG, McFarland L, Knapp JS, Sandstrom E, Critchlow C, Holmes KK. 1982. Gonococcal strains from homosexual men have outer membranes with reduced permeability to hydrophobic molecules. Infect Immun 37:432–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 647.Golparian D, Shafer WM, Ohnishi M, Unemo M. 2014. Importance of multidrug efflux pumps in the antimicrobial resistance property of clinical multidrug-resistant isolates of Neisseria gonorrhoeae. Antimicrob Agents Chemother 58:3556–3559. doi: 10.1128/AAC.00038-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 648.Luna VA, Cousin S Jr, Whittington WL, Roberts MC. 2000. Identification of the conjugative mef gene in clinical Acinetobacter junii and Neisseria gonorrhoeae isolates. Antimicrob Agents Chemother 44:2503–2506. doi: 10.1128/AAC.44.9.2503-2506.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 649.Campbell BJ, Engel AS, Porter ML, Takai K. 2006. The versatile epsilon-proteobacteria: key players in sulphidic habitats. Nat Rev Microbiol 4:458–468. doi: 10.1038/nrmicro1414. [DOI] [PubMed] [Google Scholar]
- 650.Zhuang J, Engel A, Pagès JM, Bolla JM. 1997. The Campylobacter jejuni porin trimers pack into different lattice types when reconstituted in the presence of lipid. Eur J Biochem 244:575–579. doi: 10.1111/j.1432-1033.1997.t01-1-00575.x. [DOI] [PubMed] [Google Scholar]
- 651.Labesse G, Garnotel E, Bonnel S, Dumas C, Pagès JM, Bolla JM. 2001. MOMP, a divergent porin from Campylobacter: cloning and primary structural characterization. Biochem Biophys Res Commun 280:380–387. doi: 10.1006/bbrc.2000.4129. [DOI] [PubMed] [Google Scholar]
- 652.Page WJ, Huyer G, Huyer M, Worobec EA. 1989. Characterization of the porins of Campylobacter jejuni and Campylobacter coli and implications for antibiotic susceptibility. Antimicrob Agents Chemother 33:297–303. doi: 10.1128/AAC.33.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 653.De E, Jullien M, Labesse G, Pagès JM, Molle G, Bolla JM. 2000. MOMP (major outer membrane protein) of Campylobacter jejuni; a versatile pore-forming protein. FEBS Lett 469:93–97. doi: 10.1016/S0014-5793(00)01244-8. [DOI] [PubMed] [Google Scholar]
- 654.Lin J, Michel LO, Zhang Q. 2002. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob Agents Chemother 46:2124–2131. doi: 10.1128/AAC.46.7.2124-2131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 655.Lin J, Sahin O, Michel LO, Zhang Q. 2003. Critical role of multidrug efflux pump CmeABC in bile resistance and in vivo colonization of Campylobacter jejuni. Infect Immun 71:4250–4259. doi: 10.1128/IAI.71.8.4250-4259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 656.Luo N, Sahin O, Lin J, Michel LO, Zhang Q. 2003. In vivo selection of Campylobacter isolates with high levels of fluoroquinolone resistance associated with gyrA mutations and the function of the CmeABC efflux pump. Antimicrob Agents Chemother 47:390–394. doi: 10.1128/AAC.47.1.390-394.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 657.Randall LP, Ridley AM, Cooles SW, Sharma M, Sayers AR, Pumbwe L, Newell DG, Piddock LJ, Woodward MJ. 2003. Prevalence of multiple antibiotic resistance in 443 Campylobacter spp. isolated from humans and animals. J Antimicrob Chemother 52:507–510. doi: 10.1093/jac/dkg379. [DOI] [PubMed] [Google Scholar]
- 658.Cagliero C, Mouline C, Cloeckaert A, Payot S. 2006. Synergy between efflux pump CmeABC and modifications in ribosomal proteins L4 and L22 in conferring macrolide resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob Agents Chemother 50:3893–3896. doi: 10.1128/AAC.00616-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 659.Yan M, Sahin O, Lin J, Zhang Q. 2006. Role of the CmeABC efflux pump in the emergence of fluoroquinolone-resistant Campylobacter under selection pressure. J Antimicrob Chemother 58:1154–1159. doi: 10.1093/jac/dkl412. [DOI] [PubMed] [Google Scholar]
- 660.Piddock LJ, Griggs D, Johnson MM, Ricci V, Elviss NC, Williams LK, Jorgensen F, Chisholm SA, Lawson AJ, Swift C, Humphrey TJ, Owen RJ. 2008. Persistence of Campylobacter species, strain types, antibiotic resistance and mechanisms of tetracycline resistance in poultry flocks treated with chlortetracycline. J Antimicrob Chemother 62:303–315. doi: 10.1093/jac/dkn190. [DOI] [PubMed] [Google Scholar]
- 661.Rozynek E, Mackiw E, Kaminska W, Tomczuk K, Antos-Bielska M, Dzierzanowska-Fangrat K, Korsak D. 2013. Emergence of macrolide-resistant Campylobacter strains in chicken meat in Poland and the resistance mechanisms involved. Foodborne Pathog Dis 10:655–660. doi: 10.1089/fpd.2012.1333. [DOI] [PubMed] [Google Scholar]
- 662.Akiba M, Lin J, Barton YW, Zhang Q. 2006. Interaction of CmeABC and CmeDEF in conferring antimicrobial resistance and maintaining cell viability in Campylobacter jejuni. J Antimicrob Chemother 57:52–60. doi: 10.1093/jac/dki419. [DOI] [PubMed] [Google Scholar]
- 663.Mu Y, Shen Z, Jeon B, Dai L, Zhang Q. 2013. Synergistic effects of anti-CmeA and anti-CmeB peptide nucleic acids on sensitizing Campylobacter jejuni to antibiotics. Antimicrob Agents Chemother 57:4575–4577. doi: 10.1128/AAC.00605-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 664.Oh E, Zhang Q, Jeon B. 2014. Target optimization for peptide nucleic acid (PNA)-mediated antisense inhibition of the CmeABC multidrug efflux pump in Campylobacter jejuni. J Antimicrob Chemother 69:375–380. doi: 10.1093/jac/dkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 665.Runti G, del Carmen Lopez Ruiz M, Stoilova T, Hussain R, Jennions M, Choudhury HG, Benincasa M, Gennaro R, Beis K, Scocchi M. 2013. Functional characterization of SbmA, a bacterial inner membrane transporter required for importing the antimicrobial peptide Bac7(1-35). J Bacteriol 195:5343–5351. doi: 10.1128/JB.00818-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 666.Riedel CT, Cohn MT, Stabler RA, Wren B, Brondsted L. 2012. Cellular response of Campylobacter jejuni to trisodium phosphate. Appl Environ Microbiol 78:1411–1415. doi: 10.1128/AEM.06556-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 667.Lin J, Yan M, Sahin O, Pereira S, Chang YJ, Zhang Q. 2007. Effect of macrolide usage on emergence of erythromycin-resistant Campylobacter isolates in chickens. Antimicrob Agents Chemother 51:1678–1686. doi: 10.1128/AAC.01411-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 668.Hao H, Yuan Z, Shen Z, Han J, Sahin O, Liu P, Zhang Q. 2013. Mutational and transcriptomic changes involved in the development of macrolide resistance in Campylobacter jejuni. Antimicrob Agents Chemother 57:1369–1378. doi: 10.1128/AAC.01927-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 669.Hao H, Dai M, Wang Y, Peng D, Liu Z, Yuan Z. 2009. 23S rRNA mutation A2074C conferring high-level macrolide resistance and fitness cost in Campylobacter jejuni. Microb Drug Resist 15:239–244. doi: 10.1089/mdr.2009.0008. [DOI] [PubMed] [Google Scholar]
- 670.Almofti YA, Dai M, Sun Y, Haihong H, Yuan Z. 2011. Impact of erythromycin resistance on the virulence properties and fitness of Campylobacter jejuni. Microb Pathog 50:336–342. doi: 10.1016/j.micpath.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 671.Luangtongkum T, Shen Z, Seng VW, Sahin O, Jeon B, Liu P, Zhang Q. 2012. Impaired fitness and transmission of macrolide-resistant Campylobacter jejuni in its natural host. Antimicrob Agents Chemother 56:1300–1308. doi: 10.1128/AAC.05516-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 672.Xia Q, Muraoka WT, Shen Z, Sahin O, Wang H, Wu Z, Liu P, Zhang Q. 2013. Adaptive mechanisms of Campylobacter jejuni to erythromycin treatment. BMC Microbiol 13:133. doi: 10.1186/1471-2180-13-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 673.Jeon B, Wang Y, Hao H, Barton YW, Zhang Q. 2011. Contribution of CmeG to antibiotic and oxidative stress resistance in Campylobacter jejuni. J Antimicrob Chemother 66:79–85. doi: 10.1093/jac/dkq418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 674.Shen Z, Luangtongkum T, Qiang Z, Jeon B, Wang L, Zhang Q. 2014. Identification of a novel membrane transporter mediating resistance to organic arsenic in Campylobacter jejuni. Antimicrob Agents Chemother 58:2021–2029. doi: 10.1128/AAC.02137-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 675.Polk DB, Peek RM Jr. 2010. Helicobacter pylori: gastric cancer and beyond. Nat Rev Cancer 10:403–414. doi: 10.1038/nrc2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 676.Megraud F, Lehours P. 2007. Helicobacter pylori detection and antimicrobial susceptibility testing. Clin Microbiol Rev 20:280–322. doi: 10.1128/CMR.00033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 677.Graham DY, Fischbach L. 2010. Helicobacter pylori treatment in the era of increasing antibiotic resistance. Gut 59:1143–1153. doi: 10.1136/gut.2009.192757. [DOI] [PubMed] [Google Scholar]
- 678.Megraud F, Coenen S, Versporten A, Kist M, Lopez-Brea M, Hirschl AM, Andersen LP, Goossens H, Glupczynski Y. 2013. Helicobacter pylori resistance to antibiotics in Europe and its relationship to antibiotic consumption. Gut 62:34–42. doi: 10.1136/gutjnl-2012-302254. [DOI] [PubMed] [Google Scholar]
- 679.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, Venter JC. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 680.Doig P, Exner MM, Hancock RE, Trust TJ. 1995. Isolation and characterization of a conserved porin protein from Helicobacter pylori. J Bacteriol 177:5447–5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 681.Bina JE, Alm RA, Uria-Nickelsen M, Thomas SR, Trust TJ, Hancock RE. 2000. Helicobacter pylori uptake and efflux: basis for intrinsic susceptibility to antibiotics in vitro. Antimicrob Agents Chemother 44:248–254. doi: 10.1128/AAC.44.2.248-254.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 682.Liu ZQ, Zheng PY, Yang PC. 2008. Efflux pump gene hefA of Helicobacter pylori plays an important role in multidrug resistance. World J Gastroenterol 14:5217–5222. doi: 10.3748/wjg.14.5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 683.Kutschke A, de Jonge BL. 2005. Compound efflux in Helicobacter pylori. Antimicrob Agents Chemother 49:3009–3010. doi: 10.1128/AAC.49.7.3009-3010.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 684.Stahler FN, Odenbreit S, Haas R, Wilrich J, Van Vliet AH, Kusters JG, Kist M, Bereswill S. 2006. The novel Helicobacter pylori CznABC metal efflux pump is required for cadmium, zinc, and nickel resistance, urease modulation, and gastric colonization. Infect Immun 74:3845–3852. doi: 10.1128/IAI.02025-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 685.van Amsterdam K, Bart A, van der Ende A. 2005. A Helicobacter pylori TolC efflux pump confers resistance to metronidazole. Antimicrob Agents Chemother 49:1477–1482. doi: 10.1128/AAC.49.4.1477-1482.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 686.Tsugawa H, Suzuki H, Muraoka H, Ikeda F, Hirata K, Matsuzaki J, Saito Y, Hibi T. 2011. Enhanced bacterial efflux system is the first step to the development of metronidazole resistance in Helicobacter pylori. Biochem Biophys Res Commun 404:656–660. doi: 10.1016/j.bbrc.2010.12.034. [DOI] [PubMed] [Google Scholar]
- 687.Mehrabadi JF, Sirous M, Daryani NE, Eshraghi S, Akbari B, Shirazi MH. 2011. Assessing the role of the RND efflux pump in metronidazole resistance of Helicobacter pylori by RT-PCR assay. J Infect Dev Ctries 5:88–93. doi: 10.3855/jidc.1187. [DOI] [PubMed] [Google Scholar]
- 688.Trainor EA, Horton KE, Savage PB, Testerman TL, McGee DJ. 2011. Role of the HefC efflux pump in Helicobacter pylori cholesterol-dependent resistance to ceragenins and bile salts. Infect Immun 79:88–97. doi: 10.1128/IAI.00974-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 689.Qureshi NN, Gallaher B, Schiller NL. 2014. Evolution of amoxicillin resistance of Helicobacter pylori in vitro: characterization of resistance mechanisms. Microb Drug Resist 20:509–516. doi: 10.1089/mdr.2014.0019. [DOI] [PubMed] [Google Scholar]
- 690.Iwamoto A, Tanahashi T, Okada R, Yoshida Y, Kikuchi K, Keida Y, Murakami Y, Yang L, Yamamoto K, Nishiumi S, Yoshida M, Azuma T. 2014. Whole-genome sequencing of clarithromycin resistant Helicobacter pylori characterizes unidentified variants of multidrug resistant efflux pump genes. Gut Pathog 6:27. doi: 10.1186/1757-4749-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 691.Chiu HC, Lin TL, Yang JC, Wang JT. 2009. Synergistic effect of imp/ostA and msbA in hydrophobic drug resistance of Helicobacter pylori. BMC Microbiol 9:136. doi: 10.1186/1471-2180-9-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 692.Morita Y, Tomida J, Kawamura Y. 2012. Multidrug efflux systems in Helicobacter cinaedi. Antibiotics 1:29–43. doi: 10.3390/antibiotics1010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 693.Wexler HM. 2007. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev 20:593–621. doi: 10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 694.Kobayashi Y, Kanazawa K, Nishino T. 1991. Transmembrane diffusion of hydrophobic antimicrobial agents and cell surface hydrophobicity in Bacteroides fragilis. FEMS Microbiol Lett 65:141–144. [DOI] [PubMed] [Google Scholar]
- 695.Cerdeno-Tarraga AM, Patrick S, Crossman LC, Blakely G, Abratt V, Lennard N, Poxton I, Duerden B, Harris B, Quail MA, Barron A, Clark L, Corton C, Doggett J, Holden MT, Larke N, Line A, Lord A, Norbertczak H, Ormond D, Price C, Rabbinowitsch E, Woodward J, Barrell B, Parkhill J. 2005. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science 307:1463–1465. doi: 10.1126/science.1107008. [DOI] [PubMed] [Google Scholar]
- 696.Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 697.Ueda O, Wexler HM, Hirai K, Shibata Y, Yoshimura F, Fujimura S. 2005. Sixteen homologs of the mex-type multidrug resistance efflux pump in Bacteroides fragilis. Antimicrob Agents Chemother 49:2807–2815. doi: 10.1128/AAC.49.7.2807-2815.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 698.Pumbwe L, Chang A, Smith RL, Wexler HM. 2007. BmeRABC5 is a multidrug efflux system that can confer metronidazole resistance in Bacteroides fragilis. Microb Drug Resist 13:96–101. doi: 10.1089/mdr.2007.719. [DOI] [PubMed] [Google Scholar]
- 699.Cassone M, D'Andrea MM, Iannelli F, Oggioni MR, Rossolini GM, Pozzi G. 2006. DNA microarray for detection of macrolide resistance genes. Antimicrob Agents Chemother 50:2038–2041. doi: 10.1128/AAC.01574-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 700.Eitel Z, Soki J, Urban E, Nagy E, ESCMID Study Group on Anaerobic Infection. 2013. The prevalence of antibiotic resistance genes in Bacteroides fragilis group strains isolated in different European countries. Anaerobe 21:43–49. doi: 10.1016/j.anaerobe.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 701.Husain F, Veeranagouda Y, Boente R, Tang K, Mulato G, Wexler HM. 2014. The Ellis Island effect: a novel mobile element in a multi-drug resistant clinical isolate includes a mosaic of resistance genes from Gram-positive bacteria. Mob Genet Elements 4:e29801. doi: 10.4161/mge.29801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 702.Sherrard LJ, Schaible B, Graham KA, McGrath SJ, McIlreavey L, Hatch J, Wolfgang MC, Muhlebach MS, Gilpin DF, Schneiders T, Elborn JS, Tunney MM. 2014. Mechanisms of reduced susceptibility and genotypic prediction of antibiotic resistance in Prevotella isolated from cystic fibrosis (CF) and non-CF patients. J Antimicrob Chemother 69:2690–2698. doi: 10.1093/jac/dku192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 703.Davies J, Smith DI. 1978. Plasmid-determined resistance to antimicrobial agents. Annu Rev Microbiol 32:469–518. doi: 10.1146/annurev.mi.32.100178.002345. [DOI] [PubMed] [Google Scholar]
- 704.Li X-Z. 2005. Quinolone resistance in bacteria: emphasis on plasmid-mediated mechanisms. Int J Antimicrob Agents 25:453–463. doi: 10.1016/j.ijantimicag.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 705.Carattoli A. 2013. Plasmids and the spread of resistance. Int J Med Microbiol 303:298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 706.Liu J, Keelan P, Bennett PM, Enne VI. 2009. Characterization of a novel macrolide efflux gene, mef(B), found linked to sul3 in porcine Escherichia coli. J Antimicrob Chemother 63:423–426. doi: 10.1093/jac/dkn523. [DOI] [PubMed] [Google Scholar]
- 707.Cattoir V, Poirel L, Nordmann P. 2008. Plasmid-mediated quinolone resistance pump QepA2 in an Escherichia coli isolate from France. Antimicrob Agents Chemother 52:3801–3804. doi: 10.1128/AAC.00638-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 708.Hansen LH, Johannesen E, Burmolle M, Sorensen AH, Sorensen SJ. 2004. Plasmid-encoded multidrug efflux pump conferring resistance to olaquindox in Escherichia coli. Antimicrob Agents Chemother 48:3332–3337. doi: 10.1128/AAC.48.9.3332-3337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 709.Hansen LH, Jensen LB, Sorensen HI, Sorensen SJ. 2007. Substrate specificity of the OqxAB multidrug resistance pump in Escherichia coli and selected enteric bacteria. J Antimicrob Chemother 60:145–147. doi: 10.1093/jac/dkm167. [DOI] [PubMed] [Google Scholar]
- 710.Chen X, Zhang W, Pan W, Yin J, Pan Z, Gao S, Jiao X. 2012. Prevalence of qnr, aac(6′)-Ib-cr, qepA, and oqxAB in Escherichia coli isolates from humans, animals, and the environment. Antimicrob Agents Chemother 56:3423–3427. doi: 10.1128/AAC.06191-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 711.Xu X, Cui S, Zhang F, Luo Y, Gu Y, Yang B, Li F, Chen Q, Zhou G, Wang Y, Pang L, Lin L. 2014. Prevalence and characterization of cefotaxime and ciprofloxacin co-resistant Escherichia coli isolates in retail chicken carcasses and ground pork, China. Microb Drug Resist 20:73–81. doi: 10.1089/mdr.2012.0224. [DOI] [PubMed] [Google Scholar]
- 712.Dotto G, Giacomelli M, Grilli G, Ferrazzi V, Carattoli A, Fortini D, Piccirillo A. 2014. High prevalence of oqxAB in Escherichia coli isolates from domestic and wild lagomorphs in Italy. Microb Drug Resist 20:118–123. doi: 10.1089/mdr.2013.0141. [DOI] [PubMed] [Google Scholar]
- 713.Liu BT, Li L, Fang LX, Sun J, Liao XP, Yang QE, Huang T, Liu YH. 2014. Characterization of plasmids carrying oqxAB in blaCTX-M-negative Escherichia coli isolates from food-producing animals. Microb Drug Resist 20:641–650. doi: 10.1089/mdr.2014.0022. [DOI] [PubMed] [Google Scholar]
- 714.Chandran SP, Diwan V, Tamhankar AJ, Joseph BV, Rosales-Klintz S, Mundayoor S, Lundborg CS, Macaden R. 2014. Detection of carbapenem resistance genes and cephalosporin, and quinolone resistance genes along with oqxAB gene in Escherichia coli in hospital wastewater: a matter of concern. J Appl Microbiol 117:984–995. doi: 10.1111/jam.12591. [DOI] [PubMed] [Google Scholar]
- 715.Li L, Liao XP, Liu ZZ, Huang T, Li X, Sun J, Liu BT, Zhang Q, Liu YH. 2014. Co-spread of oqxAB and blaCTX-M-9G in non-Typhi Salmonella enterica isolates mediated by ST2-IncHI2 plasmids. Int J Antimicrob Agents 44:263–268. doi: 10.1016/j.ijantimicag.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 716.McHugh GL, Moellering RC, Hopkins CC, Swartz MN. 1975. Salmonella typhimurium resistant to silver nitrate, chloramphenicol, and ampicillin. Lancet i:235–240. [DOI] [PubMed] [Google Scholar]
- 717.Gupta A, Matsui K, Lo JF, Silver S. 1999. Molecular basis for resistance to silver cations in Salmonella. Nat Med 5:183–188. doi: 10.1038/5545. [DOI] [PubMed] [Google Scholar]
- 718.Li X-Z, Nikaido H, Williams KE. 1997. Silver-resistant mutants of Escherichia coli display active efflux of Ag+ and are deficient in porins. J Bacteriol 179:6127–6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 719.Szczepanowski R, Krahn I, Linke B, Goesmann A, Puhler A, Schluter A. 2004. Antibiotic multiresistance plasmid pRSB101 isolated from a wastewater treatment plant is related to plasmids residing in phytopathogenic bacteria and carries eight different resistance determinants including a multidrug transport system. Microbiology 150:3613–3630. doi: 10.1099/mic.0.27317-0. [DOI] [PubMed] [Google Scholar]
- 720.Szczepanowski R, Linke B, Krahn I, Gartemann KH, Gutzkow T, Eichler W, Puhler A, Schluter A. 2009. Detection of 140 clinically relevant antibiotic-resistance genes in the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to selected antibiotics. Microbiology 155:2306–2319. doi: 10.1099/mic.0.028233-0. [DOI] [PubMed] [Google Scholar]
- 721.Norberg P, Bergstrom M, Hermansson M. 2014. Complete nucleotide sequence and analysis of two conjugative broad host range plasmids from a marine microbial biofilm. PLoS One 9:e92321. doi: 10.1371/journal.pone.0092321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 722.Dahlberg C, Linberg C, Torsvik VL, Hermansson M. 1997. Conjugative plasmids isolated from bacteria in marine environments show various degrees of homology to each other and are not closely related to well-characterized plasmids. Appl Environ Microbiol 63:4692–4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 723.Norberg P, Bergstrom M, Jethava V, Dubhashi D, Hermansson M. 2011. The IncP-1 plasmid backbone adapts to different host bacterial species and evolves through homologous recombination. Nat Commun 2:268. doi: 10.1038/ncomms1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 724.Poirel L, Ros A, Carricajo A, Berthelot P, Pozzetto B, Bernabeu S, Nordmann P. 2011. Extremely drug-resistant Citrobacter freundii isolate producing NDM-1 and other carbapenemases identified in a patient returning from India. Antimicrob Agents Chemother 55:447–448. doi: 10.1128/AAC.01305-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 725.Grkovic S, Brown MH, Skurray RA. 2002. Regulation of bacterial drug export systems. Microbiol Mol Biol Rev 66:671–701. doi: 10.1128/MMBR.66.4.671-701.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 726.Ma D, Alberti M, Lynch C, Nikaido H, Hearst JE. 1996. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol Microbiol 19:101–112. doi: 10.1046/j.1365-2958.1996.357881.x. [DOI] [PubMed] [Google Scholar]
- 727.Cuthbertson L, Nodwell JR. 2013. The TetR family of regulators. Microbiol Mol Biol Rev 77:440–475. doi: 10.1128/MMBR.00018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 728.Li M, Gu R, Su CC, Routh MD, Harris KC, Jewell ES, McDermott G, Yu EW. 2007. Crystal structure of the transcriptional regulator AcrR from Escherichia coli. J Mol Biol 374:591–603. doi: 10.1016/j.jmb.2007.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 729.Su CC, Yu EW. 2007. Ligand-transporter interaction in the AcrB multidrug efflux pump determined by fluorescence polarization assay. FEBS Lett 581:4972–4976. doi: 10.1016/j.febslet.2007.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 730.Gu R, Li M, Su CC, Long F, Routh MD, Yang F, McDermott G, Yu EW. 2008. Conformational change of the AcrR regulator reveals a possible mechanism of induction. Acta Crystallogr Sect F Struct Biol Cryst Commun 64:584–588. doi: 10.1107/S1744309108016035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 731.Hirakawa H, Takumi-Kobayashi A, Theisen U, Hirata T, Nishino K, Yamaguchi A. 2008. AcrS/EnvR represses expression of the acrAB multidrug efflux genes in Escherichia coli. J Bacteriol 190:6276–6279. doi: 10.1128/JB.00190-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 732.Nishino K, Yamaguchi A. 2004. Role of histone-like protein H-NS in multidrug resistance of Escherichia coli. J Bacteriol 186:1423–1429. doi: 10.1128/JB.186.5.1423-1429.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 733.Rahmati S, Yang S, Davidson AL, Zechiedrich EL. 2002. Control of the AcrAB multidrug efflux pump by quorum-sensing regulator SdiA. Mol Microbiol 43:677–685. doi: 10.1046/j.1365-2958.2002.02773.x. [DOI] [PubMed] [Google Scholar]
- 734.Kim T, Duong T, Wu CA, Choi J, Lan N, Kang SW, Lokanath NK, Shin D, Hwang HY, Kim KK. 2014. Structural insights into the molecular mechanism of Escherichia coli SdiA, a quorum-sensing receptor. Acta Crystallogr D Biol Crystallogr 70:694–707. doi: 10.1107/S1399004713032355. [DOI] [PubMed] [Google Scholar]
- 735.Nicoloff H, Perreten V, Levy SB. 2007. Increased genome instability in Escherichia coli lon mutants: relation to emergence of multiple-antibiotic-resistant (Mar) mutants caused by insertion sequence elements and large tandem genomic amplifications. Antimicrob Agents Chemother 51:1293–1303. doi: 10.1128/AAC.01128-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 736.McDermott PF, McMurry LM, Podglajen I, Dzink-Fox JL, Schneiders T, Draper MP, Levy SB. 2008. The marC gene of Escherichia coli is not involved in multiple antibiotic resistance. Antimicrob Agents Chemother 52:382–383. doi: 10.1128/AAC.00930-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 737.Alekshun MN, Levy SB, Mealy TR, Seaton BA, Head JF. 2001. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 A resolution. Nat Struct Biol 8:710–714. doi: 10.1038/90429. [DOI] [PubMed] [Google Scholar]
- 738.Gallegos MT, Schleif R, Bairoch A, Hofmann K, Ramos JL. 1997. Arac/XylS family of transcriptional regulators. Microbiol Mol Biol Rev 61:393–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 739.Rhee S, Martin RG, Rosner JL, Davies DR. 1998. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc Natl Acad Sci U S A 95:10413–10418. doi: 10.1073/pnas.95.18.10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 740.Barbosa TM, Levy SB. 2000. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J Bacteriol 182:3467–3474. doi: 10.1128/JB.182.12.3467-3474.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 741.Martin RG, Rosner JL. 2011. Promoter discrimination at class I MarA regulon promoters mediated by glutamic acid 89 of the MarA transcriptional activator of Escherichia coli. J Bacteriol 193:506–515. doi: 10.1128/JB.00360-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 742.Griffith KL, Shah IM, Wolf RE Jr. 2004. Proteolytic degradation of Escherichia coli transcription activators SoxS and MarA as the mechanism for reversing the induction of the superoxide (SoxRS) and multiple antibiotic resistance (Mar) regulons. Mol Microbiol 51:1801–1816. doi: 10.1046/j.1365-2958.2003.03952.x. [DOI] [PubMed] [Google Scholar]
- 743.McMurry LM, Oethinger M, Levy SB. 1998. Overexpression of marA, soxS, or acrAB produces resistance to triclosan in laboratory and clinical strains of Escherichia coli. FEMS Microbiol Lett 166:305–309. doi: 10.1111/j.1574-6968.1998.tb13905.x. [DOI] [PubMed] [Google Scholar]
- 744.McMurry LM, Levy SB. 2013. Amino acid residues involved in inactivation of the Escherichia coli multidrug resistance repressor MarR by salicylate, 2,4-dinitrophenol, and plumbagin. FEMS Microbiol Lett 349:16–24. doi: 10.1111/1574-6968.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 745.Hidalgo E, Ding H, Demple B. 1997. Redox signal transduction via iron-sulfur clusters in the SoxR transcription activator. Trends Biochem Sci 22:207–210. doi: 10.1016/S0968-0004(97)01068-2. [DOI] [PubMed] [Google Scholar]
- 746.Rosenberg EY, Bertenthal D, Nilles ML, Bertrand KP, Nikaido H. 2003. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol Microbiol 48:1609–1619. doi: 10.1046/j.1365-2958.2003.03531.x. [DOI] [PubMed] [Google Scholar]
- 747.Kwon HJ, Bennik MH, Demple B, Ellenberger T. 2000. Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nat Struct Biol 7:424–430. doi: 10.1038/75213. [DOI] [PubMed] [Google Scholar]
- 748.Eguchi Y, Oshima T, Mori H, Aono R, Yamamoto K, Ishihama A, Utsumi R. 2003. Transcriptional regulation of drug efflux genes by EvgAS, a two-component system in Escherichia coli. Microbiology 149:2819–2828. doi: 10.1099/mic.0.26460-0. [DOI] [PubMed] [Google Scholar]
- 749.Nishino K, Yamaguchi A. 2002. EvgA of the two-component signal transduction system modulates production of the yhiUV multidrug transporter in Escherichia coli. J Bacteriol 184:2319–2323. doi: 10.1128/JB.184.8.2319-2323.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 750.Eguchi Y, Itou J, Yamane M, Demizu R, Yamato F, Okada A, Mori H, Kato A, Utsumi R. 2007. B1500, a small membrane protein, connects the two-component systems EvgS/EvgA and PhoQ/PhoP in Escherichia coli. Proc Natl Acad Sci U S A 104:18712–18717. doi: 10.1073/pnas.0705768104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 751.Zhang A, Rosner JL, Martin RG. 2008. Transcriptional activation by MarA, SoxS and Rob of two tolC promoters using one binding site: a complex promoter configuration for tolC in Escherichia coli. Mol Microbiol 69:1450–1455. doi: 10.1111/j.1365-2958.2008.06371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 752.Parker A, Gottesman S. 2014. Small RNA regulation of a multidrug efflux pump. FASEB J 28(Suppl):750.1. [Google Scholar]
- 753.Yamada J, Yamasaki S, Hirakawa H, Hayashi-Nishino M, Yamaguchi A, Nishino K. 2010. Impact of the RNA chaperone Hfq on multidrug resistance in Escherichia coli. J Antimicrob Chemother 65:853–858. doi: 10.1093/jac/dkq067. [DOI] [PubMed] [Google Scholar]
- 754.Kulesus RR, Diaz-Pérez K, Slechta ES, Eto DS, Mulvey MA. 2008. Impact of the RNA chaperone Hfq on the fitness and virulence potential of uropathogenic Escherichia coli. Infect Immun 76:3019–3026. doi: 10.1128/IAI.00022-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 755.Tavio MM, Aquili VD, Vila J, Poveda JB. 2014. Resistance to ceftazidime in Escherichia coli associated with AcrR, MarR and PBP3 mutations and overexpression of sdiA. J Med Microbiol 63:56–65. doi: 10.1099/jmm.0.063727-0. [DOI] [PubMed] [Google Scholar]
- 756.Hirakawa H, Inazumi Y, Masaki T, Hirata T, Yamaguchi A. 2005. Indole induces the expression of multidrug exporter genes in Escherichia coli. Mol Microbiol 55:1113–1126. doi: 10.1111/j.1365-2958.2004.04449.x. [DOI] [PubMed] [Google Scholar]
- 757.Nishino K, Honda T, Yamaguchi A. 2005. Genome-wide analyses of Escherichia coli gene expression responsive to the BaeSR two-component regulatory system. J Bacteriol 187:1763–1772. doi: 10.1128/JB.187.5.1763-1772.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 758.Leblanc SK, Oates CW, Raivio TL. 2011. Characterization of the induction and cellular role of the BaeSR two-component envelope stress response of Escherichia coli. J Bacteriol 193:3367–3375. doi: 10.1128/JB.01534-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 759.Pérez-Rodríguez R, Haitjema C, Huang Q, Nam KH, Bernardis S, Ke A, DeLisa MP. 2011. Envelope stress is a trigger of CRISPR RNA-mediated DNA silencing in Escherichia coli. Mol Microbiol 79:584–599. doi: 10.1111/j.1365-2958.2010.07482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 760.Guerrero P, Collao B, Morales EH, Calderon IL, Ipinza F, Parra S, Saavedra CP, Gil F. 2012. Characterization of the BaeSR two-component system from Salmonella Typhimurium and its role in ciprofloxacin-induced mdtA expression. Arch Microbiol 194:453–460. doi: 10.1007/s00203-011-0779-5. [DOI] [PubMed] [Google Scholar]
- 761.Zoetendal EG, Smith AH, Sundset MA, Mackie RI. 2008. The BaeSR two-component regulatory system mediates resistance to condensed tannins in Escherichia coli. Appl Environ Microbiol 74:535–539. doi: 10.1128/AEM.02271-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 762.Rosner JL, Martin RG. 2013. Reduction of cellular stress by TolC-dependent efflux pumps in Escherichia coli indicated by BaeSR and CpxARP activation of spy in efflux mutants. J Bacteriol 195:1042–1050. doi: 10.1128/JB.01996-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 763.Hirakawa H, Inazumi Y, Senda Y, Kobayashi A, Hirata T, Nishino K, Yamaguchi A. 2006. N-Acetyl-D-glucosamine induces the expression of multidrug exporter genes, mdtEF, via catabolite activation in Escherichia coli. J Bacteriol 188:5851–5858. doi: 10.1128/JB.00301-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 764.Nishino K, Senda Y, Yamaguchi A. 2008. The AraC-family regulator GadX enhances multidrug resistance in Escherichia coli by activating expression of mdtEF multidrug efflux genes. J Infect Chemother 14:23–29. doi: 10.1007/s10156-007-0575-Y. [DOI] [PubMed] [Google Scholar]
- 765.Nishino K, Senda Y, Hayashi-Nishino M, Yamaguchi A. 2009. Role of the AraC-XylS family regulator YdeO in multi-drug resistance of Escherichia coli. J Antibiot (Tokyo) 62:251–257. doi: 10.1038/ja.2009.23. [DOI] [PubMed] [Google Scholar]
- 766.Nishino K, Yamasaki S, Hayashi-Nishino M, Yamaguchi A. 2011. Effect of overexpression of small non-coding DsrA RNA on multidrug efflux in Escherichia coli. J Antimicrob Chemother 66:291–296. doi: 10.1093/jac/dkq420. [DOI] [PubMed] [Google Scholar]
- 767.Sledjeski D, Gottesman S. 1995. A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. Proc Natl Acad Sci U S A 92:2003–2007. doi: 10.1073/pnas.92.6.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 768.Deng Z, Shan Y, Pan Q, Gao X, Yan A. 2013. Anaerobic expression of the gadE-mdtEF multidrug efflux operon is primarily regulated by the two-component system ArcBA through antagonizing the H-NS mediated repression. Front Microbiol 4:194. doi: 10.3389/fmicb.2013.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 769.Kobayashi A, Hirakawa H, Hirata T, Nishino K, Yamaguchi A. 2006. Growth phase-dependent expression of drug exporters in Escherichia coli and its contribution to drug tolerance. J Bacteriol 188:5693–5703. doi: 10.1128/JB.00217-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 770.Randall LP, Woodward MJ. 2001. Multiple antibiotic resistance (mar) locus in Salmonella enterica serovar Typhimurium DT104. Appl Environ Microbiol 67:1190–1197. doi: 10.1128/AEM.67.3.1190-1197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 771.Eaves DJ, Ricci V, Piddock LJ. 2004. Expression of acrB, acrF, acrD, marA, and soxS in Salmonella enterica serovar Typhimurium: role in multiple antibiotic resistance. Antimicrob Agents Chemother 48:1145–1150. doi: 10.1128/AAC.48.4.1145-1150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 772.Koutsolioutsou A, Martins EA, White DG, Levy SB, Demple B. 2001. A soxRS-constitutive mutation contributing to antibiotic resistance in a clinical isolate of Salmonella enterica (serovar Typhimurium). Antimicrob Agents Chemother 45:38–43. doi: 10.1128/AAC.45.1.38-43.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 773.Zheng J, Cui S, Meng J. 2009. Effect of transcriptional activators RamA and SoxS on expression of multidrug efflux pumps AcrAB and AcrEF in fluoroquinolone-resistant Salmonella Typhimurium. J Antimicrob Chemother 63:95–102. doi: 10.1093/jac/dkn448. [DOI] [PubMed] [Google Scholar]
- 774.Nikaido E, Shirosaka I, Yamaguchi A, Nishino K. 2011. Regulation of the AcrAB multidrug efflux pump in Salmonella enterica serovar Typhimurium in response to indole and paraquat. Microbiology 157:648–655. doi: 10.1099/mic.0.045757-0. [DOI] [PubMed] [Google Scholar]
- 775.Bailey AM, Ivens A, Kingsley R, Cottell JL, Wain J, Piddock LJ. 2010. RamA, a member of the AraC/XylS family, influences both virulence and efflux in Salmonella enterica serovar Typhimurium. J Bacteriol 192:1607–1616. doi: 10.1128/JB.01517-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 776.Baucheron S, Nishino K, Monchaux I, Canepa S, Maurel MC, Coste F, Roussel A, Cloeckaert A, Giraud E. 2014. Bile-mediated activation of the acrAB and tolC multidrug efflux genes occurs mainly through transcriptional derepression of ramA in Salmonella enterica serovar Typhimurium. J Antimicrob Chemother 69:2400–2406. doi: 10.1093/jac/dku140. [DOI] [PubMed] [Google Scholar]
- 777.Abouzeed YM, Baucheron S, Cloeckaert A. 2008. ramR mutations involved in efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium. Antimicrob Agents Chemother 52:2428–2434. doi: 10.1128/AAC.00084-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 778.Baucheron S, Coste F, Canepa S, Maurel MC, Giraud E, Culard F, Castaing B, Roussel A, Cloeckaert A. 2012. Binding of the RamR repressor to wild-type and mutated promoters of the ramA gene involved in efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium. Antimicrob Agents Chemother 56:942–948. doi: 10.1128/AAC.05444-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 779.Yamasaki S, Nikaido E, Nakashima R, Sakurai K, Fujiwara D, Fujii I, Nishino K. 2013. The crystal structure of multidrug-resistance regulator RamR with multiple drugs. Nat Commun 4:2078. doi: 10.1038/ncomms3078. [DOI] [PubMed] [Google Scholar]
- 780.Nikaido E, Yamaguchi A, Nishino K. 2008. AcrAB multidrug efflux pump regulation in Salmonella enterica serovar Typhimurium by RamA in response to environmental signals. J Biol Chem 283:24245–24253. doi: 10.1074/jbc.M804544200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 781.Sun Y, Dai M, Hao H, Wang Y, Huang L, Almofti YA, Liu Z, Yuan Z. 2011. The role of RamA on the development of ciprofloxacin resistance in Salmonella enterica serovar Typhimurium. PLoS One 6:e23471. doi: 10.1371/journal.pone.0023471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 782.Lawler AJ, Ricci V, Busby SJ, Piddock LJ. 2013. Genetic inactivation of acrAB or inhibition of efflux induces expression of ramA. J Antimicrob Chemother 68:1551–1557. doi: 10.1093/jac/dkt069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 783.Ricci V, Blair JM, Piddock LJ. 2014. RamA, which controls expression of the MDR efflux pump AcrAB-TolC, is regulated by the Lon protease. J Antimicrob Chemother 69:643–650. doi: 10.1093/jac/dkt432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 784.Komatsu T, Ohta M, Kido N, Arakawa Y, Ito H, Mizuno T, Kato N. 1990. Molecular characterization of an Enterobacter cloacae gene (romA) which pleiotropically inhibits the expression of Escherichia coli outer membrane proteins. J Bacteriol 172:4082–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 785.George AM, Hall RM, Stokes HW. 1995. Multidrug resistance in Klebsiella pneumoniae: a novel gene, ramA, confers a multidrug resistance phenotype in Escherichia coli. Microbiology 141:1909–1920. [DOI] [PubMed] [Google Scholar]
- 786.Ruzin A, Immermann FW, Bradford PA. 2008. Real-time PCR and statistical analyses of acrAB and ramA expression in clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother 52:3430–3432. doi: 10.1128/AAC.00591-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 787.Hentschke M, Wolters M, Sobottka I, Rohde H, Aepfelbacher M. 2010. ramR mutations in clinical isolates of Klebsiella pneumoniae with reduced susceptibility to tigecycline. Antimicrob Agents Chemother 54:2720–2723. doi: 10.1128/AAC.00085-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 788.Rosenblum R, Khan E, Gonzalez G, Hasan R, Schneiders T. 2011. Genetic regulation of the ramA locus and its expression in clinical isolates of Klebsiella pneumoniae. Int J Antimicrob Agents 38:39–45. doi: 10.1016/j.ijantimicag.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 789.Veleba M, Schneiders T. 2012. Tigecycline resistance can occur independently of the ramA gene in Klebsiella pneumoniae. Antimicrob Agents Chemother 56:4466–4467. doi: 10.1128/AAC.06224-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 790.Veleba M, Higgins PG, Gonzalez G, Seifert H, Schneiders T. 2012. Characterization of RarA, a novel AraC family multidrug resistance regulator in Klebsiella pneumoniae. Antimicrob Agents Chemother 56:4450–4458. doi: 10.1128/AAC.00456-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 791.De Majumdar S, Veleba M, Finn S, Fanning S, Schneiders T. 2013. Elucidating the regulon of multidrug resistance regulator RarA in Klebsiella pneumoniae. Antimicrob Agents Chemother 57:1603–1609. doi: 10.1128/AAC.01998-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 792.Bialek-Davenet S, Lavigne JP, Guyot K, Mayer N, Tournebize R, Brisse S, Leflon-Guibout V, Nicolas-Chanoine MH. 2015. Differential contribution of AcrAB and OqxAB efflux pumps to multidrug resistance and virulence in Klebsiella pneumoniae. J Antimicrob Chemother 70:81–88. doi: 10.1093/jac/dku340. [DOI] [PubMed] [Google Scholar]
- 793.Coudeyras S, Nakusi L, Charbonnel N, Forestier C. 2008. A tripartite efflux pump involved in gastrointestinal colonization by Klebsiella pneumoniae confers a tolerance response to inorganic acid. Infect Immun 76:4633–4641. doi: 10.1128/IAI.00356-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 794.Srinivasan VB, Vaidyanathan V, Mondal A, Rajamohan G. 2012. Role of the two component signal transduction system CpxAR in conferring cefepime and chloramphenicol resistance in Klebsiella pneumoniae NTUH-K2044. PLoS One 7:e33777. doi: 10.1371/journal.pone.0033777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 795.Srinivasan VB, Mondal A, Venkataramaiah M, Chauhan NK, Rajamohan G. 2013. Role of OxyRKP, a novel LysR-family transcriptional regulator, in antimicrobial resistance and virulence in Klebsiella pneumoniae. Microbiology 159:1301–1314. doi: 10.1099/mic.0.065052-0. [DOI] [PubMed] [Google Scholar]
- 796.Evans K, Adewoye L, Poole K. 2001. MexR repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa: identification of MexR binding sites in the mexA-mexR intergenic region. J Bacteriol 183:807–812. doi: 10.1128/JB.183.3.807-812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 797.Sánchez P, Rojo F, Martínez JL. 2002. Transcriptional regulation of mexR, the repressor of Pseudomonas aeruginosa mexAB-oprM multidrug efflux pump. FEMS Microbiol Lett 207:63–68. doi: 10.1111/j.1574-6968.2002.tb11029.x. [DOI] [PubMed] [Google Scholar]
- 798.Chen H, Hu J, Chen PR, Lan L, Li Z, Hicks LM, Dinner AR, He C. 2008. The Pseudomonas aeruginosa multidrug efflux regulator MexR uses an oxidation-sensing mechanism. Proc Natl Acad Sci U S A 105:13586–13591. doi: 10.1073/pnas.0803391105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 799.Chen H, Yi C, Zhang J, Zhang W, Ge Z, Yang CG, He C. 2010. Structural insight into the oxidation-sensing mechanism of the antibiotic resistance of regulator MexR. EMBO Rep 11:685–690. doi: 10.1038/embor.2010.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 800.Chang W, Small DA, Toghrol F, Bentley WE. 2005. Microarray analysis of Pseudomonas aeruginosa reveals induction of pyocin genes in response to hydrogen peroxide. BMC Genomics 6:115. doi: 10.1186/1471-2164-6-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 801.Salunkhe P, Topfer T, Buer J, Tummler B. 2005. Genome-wide transcriptional profiling of the steady-state response of Pseudomonas aeruginosa to hydrogen peroxide. J Bacteriol 187:2565–2572. doi: 10.1128/JB.187.8.2565-2572.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 802.Cummins J, Reen FJ, Baysse C, Mooij MJ, O'Gara F. 2009. Subinhibitory concentrations of the cationic antimicrobial peptide colistin induce the pseudomonas quinolone signal in Pseudomonas aeruginosa. Microbiology 155:2826–2837. doi: 10.1099/mic.0.025643-0. [DOI] [PubMed] [Google Scholar]
- 803.Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, Greenberg EP. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- 804.Anderson GG, Moreau-Marquis S, Stanton BA, O'Toole GA. 2008. In vitro analysis of tobramycin-treated Pseudomonas aeruginosa biofilms on cystic fibrosis-derived airway epithelial cells. Infect Immun 76:1423–1433. doi: 10.1128/IAI.01373-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 805.Daigle DM, Cao L, Fraud S, Wilke MS, Pacey A, Klinoski R, Strynadka NC, Dean CR, Poole K. 2007. Protein modulator of multidrug efflux gene expression in Pseudomonas aeruginosa. J Bacteriol 189:5441–5451. doi: 10.1128/JB.00543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 806.Wilke MS, Heller M, Creagh AL, Haynes CA, McIntosh LP, Poole K, Strynadka NC. 2008. The crystal structure of MexR from Pseudomonas aeruginosa in complex with its antirepressor ArmR. Proc Natl Acad Sci U S A 105:14832–14837. doi: 10.1073/pnas.0805489105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 807.Muller JF, Stevens AM, Craig J, Love NG. 2007. Transcriptome analysis reveals that multidrug efflux genes are upregulated to protect Pseudomonas aeruginosa from pentachlorophenol stress. Appl Environ Microbiol 73:4550–4558. doi: 10.1128/AEM.00169-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 808.Ghosh S, Cremers CM, Jakob U, Love NG. 2011. Chlorinated phenols control the expression of the multidrug resistance efflux pump MexAB-OprM in Pseudomonas aeruginosa by interacting with NalC. Mol Microbiol 79:1547–1556. doi: 10.1111/j.1365-2958.2011.07544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 809.Starr LM, Fruci M, Poole K. 2012. Pentachlorophenol induction of the Pseudomonas aeruginosa mexAB-oprM efflux operon: involvement of repressors NalC and MexR and the antirepressor ArmR. PLoS One 7:e32684. doi: 10.1371/journal.pone.0032684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 810.Morita Y, Cao L, Gould G, Avison MB, Poole K. 2006. nalD encodes a second repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa. J Bacteriol 188:8649–8654. doi: 10.1128/JB.01342-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 811.Terán W, Krell T, Ramos JL, Gallegos MT. 2006. Effector-repressor interactions, binding of a single effector molecule to the operator-bound TtgR homodimer mediates derepression. J Biol Chem 281:7102–7109. doi: 10.1074/jbc.M511095200. [DOI] [PubMed] [Google Scholar]
- 812.Yu J, Chen W, Wu C, Chen H. 2014. PEG-protein interaction induced contraction of NalD chains. PLoS One 9:e96616. doi: 10.1371/journal.pone.0096616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 813.Jimenez PN, Koch G, Thompson JA, Xavier KB, Cool RH, Quax WJ. 2012. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol Mol Biol Rev 76:46–65. doi: 10.1128/MMBR.05007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 814.Maseda H, Sawada I, Saito K, Uchiyama H, Nakae T, Nomura N. 2004. Enhancement of the mexAB-oprM efflux pump expression by a quorum-sensing autoinducer and its cancellation by a regulator, MexT, of the mexEF-oprN efflux pump operon in Pseudomonas aeruginosa. Antimicrob Agents Chemother 48:1320–1328. doi: 10.1128/AAC.48.4.1320-1328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 815.Sawada I, Maseda H, Nakae T, Uchiyama H, Nomura N. 2004. A quorum-sensing autoinducer enhances the mexAB-oprM efflux-pump expression without the MexR-mediated regulation in Pseudomonas aeruginosa. Microbiol Immunol 48:435–439. doi: 10.1111/j.1348-0421.2004.tb03533.x. [DOI] [PubMed] [Google Scholar]
- 816.Balasubramanian D, Schneper L, Merighi M, Smith R, Narasimhan G, Lory S, Mathee K. 2012. The regulatory repertoire of Pseudomonas aeruginosa AmpC β-lactamase regulator AmpR includes virulence genes. PLoS One 7:e34067. doi: 10.1371/journal.pone.0034067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 817.Brooun A, Liu S, Lewis K. 2000. A dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 44:640–646. doi: 10.1128/AAC.44.3.640-646.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 818.Sivaneson M, Mikkelsen H, Ventre I, Bordi C, Filloux A. 2011. Two-component regulatory systems in Pseudomonas aeruginosa: an intricate network mediating fimbrial and efflux pump gene expression. Mol Microbiol 79:1353–1366. doi: 10.1111/j.1365-2958.2010.07527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 819.Liao J, Schurr MJ, Sauer K. 2013. The MerR-like regulator BrlR confers biofilm tolerance by activating multidrug efflux pumps in Pseudomonas aeruginosa biofilms. J Bacteriol 195:3352–3363. doi: 10.1128/JB.00318-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 820.Liao J, Sauer K. 2012. The MerR-like transcriptional regulator BrlR contributes to Pseudomonas aeruginosa biofilm tolerance. J Bacteriol 194:4823–4836. doi: 10.1128/JB.00765-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 821.Chambers JR, Liao J, Schurr MJ, Sauer K. 2014. BrlR from Pseudomonas aeruginosa is a c-di-GMP-responsive transcription factor. Mol Microbiol 92:471–487. doi: 10.1111/mmi.12562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 822.Gupta K, Marques CN, Petrova OE, Sauer K. 2013. Antimicrobial tolerance of Pseudomonas aeruginosa biofilms is activated during an early developmental stage and requires the two-component hybrid SagS. J Bacteriol 195:4975–4987. doi: 10.1128/JB.00732-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 823.Gupta K, Liao J, Petrova OE, Cherny KE, Sauer K. 2014. Elevated levels of the second messenger c-di-GMP contribute to antimicrobial resistance of Pseudomonas aeruginosa. Mol Microbiol 92:488–506. doi: 10.1111/mmi.12587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 824.Pamp SJ, Gjermansen M, Johansen HK, Tolker-Nielsen T. 2008. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol Microbiol 68:223–240. doi: 10.1111/j.1365-2958.2008.06152.x. [DOI] [PubMed] [Google Scholar]
- 825.Matsuo Y, Eda S, Gotoh N, Yoshihara E, Nakae T. 2004. MexZ-mediated regulation of mexXY multidrug efflux pump expression in Pseudomonas aeruginosa by binding on the mexZ-mexX intergenic DNA. FEMS Microbiol Lett 238:23–28. doi: 10.1111/j.1574-6968.2004.tb09732.x. [DOI] [PubMed] [Google Scholar]
- 826.Yamamoto M, Ueda A, Kudo M, Matsuo Y, Fukushima J, Nakae T, Kaneko T, Ishigatsubo Y. 2009. Role of MexZ and PA5471 in transcriptional regulation of mexXY in Pseudomonas aeruginosa. Microbiology 155:3312–3321. doi: 10.1099/mic.0.028993-0. [DOI] [PubMed] [Google Scholar]
- 827.Hay T, Fraud S, Lau CH, Gilmour C, Poole K. 2013. Antibiotic inducibility of the mexXY multidrug efflux operon of Pseudomonas aeruginosa: involvement of the MexZ anti-repressor ArmZ. PLoS One 8:e56858. doi: 10.1371/journal.pone.0056858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 828.Morita Y, Gilmour C, Metcalf D, Poole K. 2009. Translational control of the antibiotic inducibility of the PA5471 gene required for mexXY multidrug efflux gene expression in Pseudomonas aeruginosa. J Bacteriol 191:4966–4975. doi: 10.1128/JB.00073-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 829.Lee S, Hinz A, Bauerle E, Angermeyer A, Juhaszova K, Kaneko Y, Singh PK, Manoil C. 2009. Targeting a bacterial stress response to enhance antibiotic action. Proc Natl Acad Sci U S A 106:14570–14575. doi: 10.1073/pnas.0903619106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 830.Lau CH, Fraud S, Jones M, Peterson SN, Poole K. 2013. Mutational activation of the AmgRS two-component system in aminoglycoside-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother 57:2243–2251. doi: 10.1128/AAC.00170-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 831.Lau CH, Krahn T, Gilmour C, Mullen E, Poole K. 2015. AmgRS-mediated envelope stress-inducible expression of the mexXY multidrug efflux operon of Pseudomonas aeruginosa. Microbiologyopen 4:121–135. doi: 10.1002/mbo3.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 832.McLaughlin HP, Caly DL, McCarthy Y, Ryan RP, Dow JM. 2012. An orphan chemotaxis sensor regulates virulence and antibiotic tolerance in the human pathogen Pseudomonas aeruginosa. PLoS One 7:e42205. doi: 10.1371/journal.pone.0042205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 833.Fernández L, Gooderham WJ, Bains M, McPhee JB, Wiegand I, Hancock RE. 2010. Adaptive resistance to the “last hope” antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob Agents Chemother 54:3372–3382. doi: 10.1128/AAC.00242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 834.Purssell A, Poole K. 2013. Functional characterization of the NfxB repressor of the mexCD-oprJ multidrug efflux operon of Pseudomonas aeruginosa. Microbiology 159:2058–2073. doi: 10.1099/mic.0.069286-0. [DOI] [PubMed] [Google Scholar]
- 835.Purssell A, Fruci M, Mikalauskas A, Gilmour C, Poole K. 2015. EsrC, an envelope stress-regulated repressor of the mexCD-oprJ multidrug efflux operon in Pseudomonas aeruginosa. Environ Microbiol 17:186–198. doi: 10.1111/1462-2920.12602. [DOI] [PubMed] [Google Scholar]
- 836.Köhler T, Epp SF, Curty LK, Pechère JC. 1999. Characterization of MexT, the regulator of the MexE-MexF-OprN multidrug efflux system of Pseudomonas aeruginosa. J Bacteriol 181:6300–6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 837.Chuanchuen R, Gaynor JB, Karkhoff-Schweizer R, Schweizer HP. 2005. Molecular characterization of MexL, the transcriptional repressor of the mexJK multidrug efflux operon in Pseudomonas aeruginosa. Antimicrob Agents Chemother 49:1844–1851. doi: 10.1128/AAC.49.5.1844-1851.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 838.Monti MR, Morero NR, Miguel V, Argarana CE. 2013. nfxB as a novel target for analysis of mutation spectra in Pseudomonas aeruginosa. PLoS One 8:e66236. doi: 10.1371/journal.pone.0066236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 839.Mandsberg LF, Ciofu O, Kirkby N, Christiansen LE, Poulsen HE, Hoiby N. 2009. Antibiotic resistance in Pseudomonas aeruginosa strains with increased mutation frequency due to inactivation of the DNA oxidative repair system. Antimicrob Agents Chemother 53:2483–2491. doi: 10.1128/AAC.00428-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 840.Nde CW, Jang HJ, Toghrol F, Bentley WE. 2009. Global transcriptomic response of Pseudomonas aeruginosa to chlorhexidine diacetate. Environ Sci Technol 43:8406–8415. doi: 10.1021/es9015475. [DOI] [PubMed] [Google Scholar]
- 841.Ochs MM, McCusker MP, Bains M, Hancock RE. 1999. Negative regulation of the Pseudomonas aeruginosa outer membrane porin OprD selective for imipenem and basic amino acids. Antimicrob Agents Chemother 43:1085–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 842.Tian ZX, Mac Aogain M, O'Connor HF, Fargier E, Mooij MJ, Adams C, Wang YP, O'Gara F. 2009. MexT modulates virulence determinants in Pseudomonas aeruginosa independent of the MexEF-OprN efflux pump. Microb Pathog 47:237–241. doi: 10.1016/j.micpath.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 843.Tian ZX, Fargier E, Mac Aogain M, Adams C, Wang YP, O'Gara F. 2009. Transcriptome profiling defines a novel regulon modulated by the LysR-type transcriptional regulator MexT in Pseudomonas aeruginosa. Nucleic Acids Res 37:7546–7559. doi: 10.1093/nar/gkp828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 844.Lister PD, Wolter DJ, Hanson ND. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 845.Maseda H, Saito K, Nakajima A, Nakae T. 2000. Variation of the mexT gene, a regulator of the MexEF-oprN efflux pump expression in wild-type strains of Pseudomonas aeruginosa. FEMS Microbiol Lett 192:107–112. doi: 10.1111/j.1574-6968.2000.tb09367.x. [DOI] [PubMed] [Google Scholar]
- 846.Sobel ML, Neshat S, Poole K. 2005. Mutations in PA2491 (mexS) promote MexT-dependent mexEF-oprN expression and multidrug resistance in a clinical strain of Pseudomonas aeruginosa. J Bacteriol 187:1246–1253. doi: 10.1128/JB.187.4.1246-1253.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 847.Fetar H, Gilmour C, Klinoski R, Daigle DM, Dean CR, Poole K. 2011. mexEF-oprN multidrug efflux operon of Pseudomonas aeruginosa: regulation by the MexT activator in response to nitrosative stress and chloramphenicol. Antimicrob Agents Chemother 55:508–514. doi: 10.1128/AAC.00830-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 848.Jin Y, Yang H, Qiao M, Jin S. 2011. MexT regulates the type III secretion system through MexS and PtrC in Pseudomonas aeruginosa. J Bacteriol 193:399–410. doi: 10.1128/JB.01079-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 849.Fargier E, Mac Aogain M, Mooij MJ, Woods DF, Morrissey JP, Dobson AD, Adams C, O'Gara F. 2012. MexT functions as a redox-responsive regulator modulating disulfide stress resistance in Pseudomonas aeruginosa. J Bacteriol 194:3502–3511. doi: 10.1128/JB.06632-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 850.Uwate M, Ichise YK, Shirai A, Omasa T, Nakae T, Maseda H. 2013. Two routes of MexS-MexT-mediated regulation of MexEF-OprN and MexAB-OprM efflux pump expression in Pseudomonas aeruginosa. Microbiol Immunol 57:263–272. doi: 10.1111/1348-0421.12032. [DOI] [PubMed] [Google Scholar]
- 851.Frisk A, Schurr JR, Wang G, Bertucci DC, Marrero L, Hwang SH, Hassett DJ, Schurr MJ. 2004. Transcriptome analysis of Pseudomonas aeruginosa after interaction with human airway epithelial cells. Infect Immun 72:5433–5438. doi: 10.1128/IAI.72.9.5433-5438.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 852.Vallet I, Diggle SP, Stacey RE, Camara M, Ventre I, Lory S, Lazdunski A, Williams P, Filloux A. 2004. Biofilm formation in Pseudomonas aeruginosa: fimbrial cup gene clusters are controlled by the transcriptional regulator MvaT. J Bacteriol 186:2880–2890. doi: 10.1128/JB.186.9.2880-2890.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 853.Castang S, McManus HR, Turner KH, Dove SL. 2008. H-NS family members function coordinately in an opportunistic pathogen. Proc Natl Acad Sci U S A 105:18947–18952. doi: 10.1073/pnas.0808215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 854.Castang S, Dove SL. 2012. Basis for the essentiality of H-NS family members in Pseudomonas aeruginosa. J Bacteriol 194:5101–5109. doi: 10.1128/JB.00932-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 855.Westfall LW, Carty NL, Layland N, Kuan P, Colmer-Hamood JA, Hamood AN. 2006. mvaT mutation modifies the expression of the Pseudomonas aeruginosa multidrug efflux operon mexEF-oprN. FEMS Microbiol Lett 255:247–254. doi: 10.1111/j.1574-6968.2005.00075.x. [DOI] [PubMed] [Google Scholar]
- 856.Kumari H, Balasubramanian D, Zincke D, Mathee K. 2014. Role of Pseudomonas aeruginosa AmpR on β-lactam and non-β-lactam transient cross resistance upon pre-exposure to sub-inhibitory concentrations of antibiotics. J Med Microbiol 63:544–555. doi: 10.1099/jmm.0.070185-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 857.Caille O, Rossier C, Perron K. 2007. A copper-activated two-component system interacts with zinc and imipenem resistance in Pseudomonas aeruginosa. J Bacteriol 189:4561–4568. doi: 10.1128/JB.00095-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 858.Marchand I, Damier-Piolle L, Courvalin P, Lambert T. 2004. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob Agents Chemother 48:3298–3304. doi: 10.1128/AAC.48.9.3298-3304.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 859.Lopes BS, Amyes SG. 2013. Insertion sequence disruption of adeR and ciprofloxacin resistance caused by efflux pumps and gyrA and parC mutations in Acinetobacter baumannii. Int J Antimicrob Agents 41:117–121. doi: 10.1016/j.ijantimicag.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 860.Sun JR, Perng CL, Lin JC, Yang YS, Chan MC, Chang TY, Lin FM, Chiueh TS. 2014. AdeRS combination codes differentiate the response to efflux pump inhibitors in tigecycline-resistant isolates of extensively drug-resistant Acinetobacter baumannii. Eur J Clin Microbiol Infect Dis 33:2141–2147. doi: 10.1007/s10096-014-2179-7. [DOI] [PubMed] [Google Scholar]
- 861.Yoon EJ, Courvalin P, Grillot-Courvalin C. 2013. RND-type efflux pumps in multidrug-resistant clinical isolates of Acinetobacter baumannii: major role for AdeABC overexpression and AdeRS mutations. Antimicrob Agents Chemother 57:2989–2995. doi: 10.1128/AAC.02556-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 862.Hua X, Chen Q, Li X, Yu Y. 2014. Global transcriptional response of Acinetobacter baumannii to a subinhibitory concentration of tigecycline. Int J Antimicrob Agents 44:337–344. doi: 10.1016/j.ijantimicag.2014.06.015. [DOI] [PubMed] [Google Scholar]
- 863.Sánchez P, Alonso A, Martínez JL. 2002. Cloning and characterization of SmeT, a repressor of the Stenotrophomonas maltophilia multidrug efflux pump SmeDEF. Antimicrob Agents Chemother 46:3386–3393. doi: 10.1128/AAC.46.11.3386-3393.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 864.Hernández A, Maté MJ, Sánchez-Diaz PC, Romero A, Rojo F, Martínez JL. 2009. Structural and functional analysis of SmeT, the repressor of the Stenotrophomonas maltophilia multidrug efflux pump SmeDEF. J Biol Chem 284:14428–14438. doi: 10.1074/jbc.M809221200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 865.Hernández A, Ruiz FM, Romero A, Martínez JL. 2011. The binding of triclosan to SmeT, the repressor of the multidrug efflux pump SmeDEF, induces antibiotic resistance in Stenotrophomonas maltophilia. PLoS Pathog 7:e1002103. doi: 10.1371/journal.ppat.1002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 866.García-León G, Hernández A, Hernando-Amado S, Alavi P, Berg G, Martínez JL. 2014. A function of the major quinolone resistance determinant of Stenotrophomonas maltophilia SmeDEF is the colonization of the roots of the plants. Appl Environ Microbiol 80:4559–4565. doi: 10.1128/AEM.01058-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 867.Hoffmann KM, Williams D, Shafer WM, Brennan RG. 2005. Characterization of the multiple transferable resistance repressor, MtrR, from Neisseria gonorrhoeae. J Bacteriol 187:5008–5012. doi: 10.1128/JB.187.14.5008-5012.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 868.Folster JP, Johnson PJ, Jackson L, Dhulipali V, Dyer DW, Shafer WM. 2009. MtrR modulates rpoH expression and levels of antimicrobial resistance in Neisseria gonorrhoeae. J Bacteriol 191:287–297. doi: 10.1128/JB.01165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 869.Folster JP, Shafer WM. 2005. Regulation of mtrF expression in Neisseria gonorrhoeae and its role in high-level antimicrobial resistance. J Bacteriol 187:3713–3720. doi: 10.1128/JB.187.11.3713-3720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 870.Ohneck EA, Zalucki YM, Johnson PJ, Dhulipala V, Golparian D, Unemo M, Jerse AE, Shafer WM. 2011. A novel mechanism of high-level, broad-spectrum antibiotic resistance caused by a single base pair change in Neisseria gonorrhoeae. mBio 2(5):e00187-11. doi: 10.1128/mBio.00187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 871.Rouquette-Loughlin CE, Balthazar JT, Hill SA, Shafer WM. 2004. Modulation of the mtrCDE-encoded efflux pump gene complex of Neisseria meningitidis due to a Correia element insertion sequence. Mol Microbiol 54:731–741. doi: 10.1111/j.1365-2958.2004.04299.x. [DOI] [PubMed] [Google Scholar]
- 872.Snyder LA, Cole JA, Pallen MJ. 2009. Comparative analysis of two Neisseria gonorrhoeae genome sequences reveals evidence of mobilization of Correia repeat enclosed elements and their role in regulation. BMC Genomics 10:70. doi: 10.1186/1471-2164-10-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 873.Enriquez R, Abad R, Chanto G, Corso A, Cruces R, Gabastou JM, Gorla MC, Maldonado A, Moreno J, Muros-Le Rouzic E, Sorhouet C, Vazquez JA. 2010. Deletion of the Correia element in the mtr gene complex of Neisseria meningitidis. J Med Microbiol 59:1055–1060. doi: 10.1099/jmm.0.021220-0. [DOI] [PubMed] [Google Scholar]
- 874.Lin J, Akiba M, Sahin O, Zhang Q. 2005. CmeR functions as a transcriptional repressor for the multidrug efflux pump CmeABC in Campylobacter jejuni. Antimicrob Agents Chemother 49:1067–1075. doi: 10.1128/AAC.49.3.1067-1075.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 875.Lin J, Cagliero C, Guo B, Barton YW, Maurel MC, Payot S, Zhang Q. 2005. Bile salts modulate expression of the CmeABC multidrug efflux pump in Campylobacter jejuni. J Bacteriol 187:7417–7424. doi: 10.1128/JB.187.21.7417-7424.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 876.Gu R, Su CC, Shi F, Li M, McDermott G, Zhang Q, Yu EW. 2007. Crystal structure of the transcriptional regulator CmeR from Campylobacter jejuni. J Mol Biol 372:583–593. doi: 10.1016/j.jmb.2007.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 877.Lei HT, Shen Z, Surana P, Routh MD, Su CC, Zhang Q, Yu EW. 2011. Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni. Protein Sci 20:712–723. doi: 10.1002/pro.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 878.Cagliero C, Maurel MC, Cloeckaert A, Payot S. 2007. Regulation of the expression of the CmeABC efflux pump in Campylobacter jejuni: identification of a point mutation abolishing the binding of the CmeR repressor in an in vitro-selected multidrug-resistant mutant. FEMS Microbiol Lett 267:89–94. doi: 10.1111/j.1574-6968.2006.00558.x. [DOI] [PubMed] [Google Scholar]
- 879.Hwang S, Zhang Q, Ryu S, Jeon B. 2012. Transcriptional regulation of the CmeABC multidrug efflux pump and the KatA catalase by CosR in Campylobacter jejuni. J Bacteriol 194:6883–6891. doi: 10.1128/JB.01636-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 880.Shen Z, Pu XY, Zhang Q. 2011. Salicylate functions as an efflux pump inducer and promotes the emergence of fluoroquinolone-resistant Campylobacter jejuni mutants. Appl Environ Microbiol 77:7128–7133. doi: 10.1128/AEM.00763-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 881.Romling U, Kjelleberg S, Normark S, Nyman L, Uhlin BE, Akerlund B. 2014. Microbial biofilm formation: a need to act. J Intern Med 276:98–110. doi: 10.1111/joim.12242. [DOI] [PubMed] [Google Scholar]
- 882.Mah TF. 2012. Biofilm-specific antibiotic resistance. Future Microbiol 7:1061–1072. doi: 10.2217/fmb.12.76. [DOI] [PubMed] [Google Scholar]
- 883.O'Toole G, Kaplan HB, Kolter R. 2000. Biofilm formation as microbial development. Annu Rev Microbiol 54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 884.De Kievit TR, Parkins MD, Gillis RJ, Srikumar R, Ceri H, Poole K, Iglewski BH, Storey DG. 2001. Multidrug efflux pumps: expression patterns and contribution to antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 45:1761–1770. doi: 10.1128/AAC.45.6.1761-1770.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 885.Baugh S, Ekanayaka AS, Piddock LJ, Webber MA. 2012. Loss of or inhibition of all multidrug resistance efflux pumps of Salmonella enterica serovar Typhimurium results in impaired ability to form a biofilm. J Antimicrob Chemother 67:2409–2417. doi: 10.1093/jac/dks228. [DOI] [PubMed] [Google Scholar]
- 886.Baugh S, Phillips CR, Ekanayaka AS, Piddock LJ, Webber MA. 2014. Inhibition of multidrug efflux as a strategy to prevent biofilm formation. J Antimicrob Chemother 69:673–681. doi: 10.1093/jac/dkt420. [DOI] [PubMed] [Google Scholar]
- 887.Tabak M, Scher K, Hartog E, Romling U, Matthews KR, Chikindas ML, Yaron S. 2007. Effect of triclosan on Salmonella typhimurium at different growth stages and in biofilms. FEMS Microbiol Lett 267:200–206. doi: 10.1111/j.1574-6968.2006.00547.x. [DOI] [PubMed] [Google Scholar]
- 888.Matsumura K, Furukawa S, Ogihara H, Morinaga Y. 2011. Roles of multidrug efflux pumps on the biofilm formation of Escherichia coli K-12. Biocontrol Sci 16:69–72. doi: 10.4265/bio.16.69. [DOI] [PubMed] [Google Scholar]
- 889.Kvist M, Hancock V, Klemm P. 2008. Inactivation of efflux pumps abolishes bacterial biofilm formation. Appl Environ Microbiol 74:7376–7382. doi: 10.1128/AEM.01310-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 890.Liu Y, Yang L, Molin S. 2010. Synergistic activities of an efflux pump inhibitor and iron chelators against Pseudomonas aeruginosa growth and biofilm formation. Antimicrob Agents Chemother 54:3960–3963. doi: 10.1128/AAC.00463-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 891.Imuta N, Nishi J, Tokuda K, Fujiyama R, Manago K, Iwashita M, Sarantuya J, Kawano Y. 2008. The Escherichia coli efflux pump TolC promotes aggregation of enteroaggregative E. coli 042. Infect Immun 76:1247–1256. doi: 10.1128/IAI.00758-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 892.Norman A, Hansen LH, She Q, Sorensen SJ. 2008. Nucleotide sequence of pOLA52: a conjugative IncX1 plasmid from Escherichia coli which enables biofilm formation and multidrug efflux. Plasmid 60:59–74. doi: 10.1016/j.plasmid.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 893.Tettmann B, Dotsch A, Armant O, Fjell CD, Overhage J. 2014. Knock-out of extracytoplasmic function sigma factor ECF-10 affects stress resistance and biofilm formation in Pseudomonas putida KT2440. Appl Environ Microbiol 80:4911–4919. doi: 10.1128/AEM.01291-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 894.Cabral MP, Soares NC, Aranda J, Parreira JR, Rumbo C, Poza M, Valle J, Calamia V, Lasa I, Bou G. 2011. Proteomic and functional analyses reveal a unique lifestyle for Acinetobacter baumannii biofilms and a key role for histidine metabolism. J Proteome Res 10:3399–3417. doi: 10.1021/pr101299j. [DOI] [PubMed] [Google Scholar]
- 895.Hoffman LR, D'Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. 2005. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436:1171–1175. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- 896.Heldwein EE, Brennan RG. 2001. Crystal structure of the transcription activator BmrR bound to DNA and a drug. Nature 409:378–382. doi: 10.1038/35053138. [DOI] [PubMed] [Google Scholar]
- 897.Zhang L, Mah TF. 2008. Involvement of a novel efflux system in biofilm-specific resistance to antibiotics. J Bacteriol 190:4447–4452. doi: 10.1128/JB.01655-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 898.Lynch SV, Dixon L, Benoit MR, Brodie EL, Keyhan M, Hu P, Ackerley DF, Andersen GL, Matin A. 2007. Role of the rapA gene in controlling antibiotic resistance of Escherichia coli biofilms. Antimicrob Agents Chemother 51:3650–3658. doi: 10.1128/AAC.00601-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 899.Pumbwe L, Skilbeck CA, Nakano V, Avila-Campos MJ, Piazza RM, Wexler HM. 2007. Bile salts enhance bacterial co-aggregation, bacterial-intestinal epithelial cell adhesion, biofilm formation and antimicrobial resistance of Bacteroides fragilis. Microb Pathog 43:78–87. doi: 10.1016/j.micpath.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 900.Martínez JL, Sánchez MB, Martínez-Solano L, Hernández A, Garmendia L, Fajardo A, Alvarez-Ortega C. 2009. Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol Rev 33:430–449. doi: 10.1111/j.1574-6976.2008.00157.x. [DOI] [PubMed] [Google Scholar]
- 901.Helling RB, Janes BK, Kimball H, Tran T, Bundesmann M, Check P, Phelan D, Miller C. 2002. Toxic waste disposal in Escherichia coli. J Bacteriol 184:3699–3703. doi: 10.1128/JB.184.13.3699-3703.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 902.Rosner JL, Martin RG. 2009. An excretory function for the Escherichia coli outer membrane pore TolC: upregulation of marA and soxS transcription and Rob activity due to metabolites accumulated in tolC mutants. J Bacteriol 191:5283–5292. doi: 10.1128/JB.00507-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 903.Ma D, Cook DN, Alberti M, Pon NG, Nikaido H, Hearst JE. 1995. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol 16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 904.Prouty AM, Brodsky IE, Falkow S, Gunn JS. 2004. Bile-salt-mediated induction of antimicrobial and bile resistance in Salmonella Typhimurium. Microbiology 150:775–783. doi: 10.1099/mic.0.26769-0. [DOI] [PubMed] [Google Scholar]
- 905.Paul S, Alegre KO, Holdsworth SR, Rice M, Brown JA, McVeigh P, Kelly SM, Law CJ. 2014. A single-component multidrug transporter of the major facilitator superfamily is part of a network that protects Escherichia coli from bile salt stress. Mol Microbiol 92:872–884. doi: 10.1111/mmi.12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 906.Guelfo JR, Rodríguez-Rojas A, Matic I, Blazquez J. 2010. A MATE-family efflux pump rescues the Escherichia coli 8-oxoguanine-repair-deficient mutator phenotype and protects against H2O2 killing. PLoS Genet 6:e1000931. doi: 10.1371/journal.pgen.1000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 907.Bogomolnaya LM, Andrews KD, Talamantes M, Maple A, Ragoza Y, Vazquez-Torres A, Andrews-Polymenis H. 2013. The ABC-type efflux pump MacAB protects Salmonella enterica serovar Typhimurium from oxidative stress. mBio 4(6):e00630-13. doi: 10.1128/mBio.00630-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 908.Viveiros M, Dupont M, Rodrigues L, Couto I, Davin-Regli A, Martins M, Pagès JM, Amaral L. 2007. Antibiotic stress, genetic response and altered permeability of E. coli. PLoS One 2:e365. doi: 10.1371/journal.pone.0000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 909.Otto K, Silhavy TJ. 2002. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc Natl Acad Sci U S A 99:2287–2292. doi: 10.1073/pnas.042521699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 910.Nishino K, Yamasaki S, Hayashi-Nishino M, Yamaguchi A. 2010. Effect of NlpE overproduction on multidrug resistance in Escherichia coli. Antimicrob Agents Chemother 54:2239–2243. doi: 10.1128/AAC.01677-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 911.Morita Y, Sobel ML, Poole K. 2006. Antibiotic inducibility of the MexXY multidrug efflux system of Pseudomonas aeruginosa: involvement of the antibiotic-inducible PA5471 gene product. J Bacteriol 188:1847–1855. doi: 10.1128/JB.188.5.1847-1855.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 912.Godoy P, Molina-Henares AJ, de la Torre J, Duque E, Ramos JL. 2010. Characterization of the RND family of multidrug efflux pumps: in silico to in vivo confirmation of four functionally distinct subgroups. Microb Biotechnol 3:691–700. doi: 10.1111/j.1751-7915.2010.00189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 913.Jassem AN, Forbes CM, Speert DP. 2014. Investigation of aminoglycoside resistance inducing conditions and a putative AmrAB-OprM efflux system in Burkholderia vietnamiensis. Ann Clin Microbiol Antimicrob 13:2. doi: 10.1186/1476-0711-13-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 914.Hwang S, Kim M, Ryu S, Jeon B. 2011. Regulation of oxidative stress response by CosR, an essential response regulator in Campylobacter jejuni. PLoS One 6:e22300. doi: 10.1371/journal.pone.0022300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 915.Buckley AM, Webber MA, Cooles S, Randall LP, La Ragione RM, Woodward MJ, Piddock LJ. 2006. The AcrAB-TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell Microbiol 8:847–856. doi: 10.1111/j.1462-5822.2005.00671.x. [DOI] [PubMed] [Google Scholar]
- 916.Yamanaka H, Kobayashi H, Takahashi E, Okamoto K. 2008. MacAB is involved in the secretion of Escherichia coli heat-stable enterotoxin II. J Bacteriol 190:7693–7698. doi: 10.1128/JB.00853-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 917.Bina JE, Mekalanos JJ. 2001. Vibrio cholerae tolC is required for bile resistance and colonization. Infect Immun 69:4681–4685. doi: 10.1128/IAI.69.7.4681-4685.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 918.Lin J, Martínez AL. 2006. Effect of efflux pump inhibitors on bile resistance and in vivo colonization of Campylobacter jejuni. J Antimicrob Chemother 58:966–972. doi: 10.1093/jac/dkl374. [DOI] [PubMed] [Google Scholar]
- 919.Linkevicius M, Sandegren L, Andersson DI. 2013. Mechanisms and fitness costs of tigecycline resistance in Escherichia coli. J Antimicrob Chemother 68:2809–2819. doi: 10.1093/jac/dkt263. [DOI] [PubMed] [Google Scholar]
- 920.Li L, Yang YR, Liao XP, Lei CY, Sun J, Li LL, Liu BT, Yang SS, Liu YH. 2013. Development of ceftriaxone resistance affects the virulence properties of Salmonella enterica serotype Typhimurium strains. Foodborne Pathog Dis 10:28–34. doi: 10.1089/fpd.2012.1216. [DOI] [PubMed] [Google Scholar]
- 921.Salunkhe P, Smart CH, Morgan JA, Panagea S, Walshaw MJ, Hart CA, Geffers R, Tummler B, Winstanley C. 2005. A cystic fibrosis epidemic strain of Pseudomonas aeruginosa displays enhanced virulence and antimicrobial resistance. J Bacteriol 187:4908–4920. doi: 10.1128/JB.187.14.4908-4920.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 922.Sánchez P, Linares JF, Ruiz-Diez B, Campanario E, Navas A, Baquero F, Martínez JL. 2002. Fitness of in vitro selected Pseudomonas aeruginosa nalB and nfxB multidrug resistant mutants. J Antimicrob Chemother 50:657–664. doi: 10.1093/jac/dkf185. [DOI] [PubMed] [Google Scholar]
- 923.Abdelraouf K, Kabbara S, Ledesma KR, Poole K, Tam VH. 2011. Effect of multidrug resistance-conferring mutations on the fitness and virulence of Pseudomonas aeruginosa. J Antimicrob Chemother 66:1311–1317. doi: 10.1093/jac/dkr105. [DOI] [PubMed] [Google Scholar]
- 924.Mahajan-Miklos S, Rahme LG, Ausubel FM. 2000. Elucidating the molecular mechanisms of bacterial virulence using non-mammalian hosts. Mol Microbiol 37:981–988. doi: 10.1046/j.1365-2958.2000.02056.x. [DOI] [PubMed] [Google Scholar]
- 925.Hirakata Y, Srikumar R, Poole K, Gotoh N, Suematsu T, Kohno S, Kamihira S, Hancock RE, Speert DP. 2002. Multidrug efflux systems play an important role in the invasiveness of Pseudomonas aeruginosa. J Exp Med 196:109–118. doi: 10.1084/jem.20020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 926.Hirakata Y, Kondo A, Hoshino K, Yano H, Arai K, Hirotani A, Kunishima H, Yamamoto N, Hatta M, Kitagawa M, Kohno S, Kaku M. 2009. Efflux pump inhibitors reduce the invasiveness of Pseudomonas aeruginosa. Int J Antimicrob Agents 34:343–346. doi: 10.1016/j.ijantimicag.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 927.Vettoretti L, Plésiat P, Muller C, El Garch F, Phan G, Attree I, Ducruix A, Llanes C. 2009. Efflux unbalance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob Agents Chemother 53:1987–1997. doi: 10.1128/AAC.01024-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 928.Hauser AR, Jain M, Bar-Meir M, McColley SA. 2011. Clinical significance of microbial infection and adaptation in cystic fibrosis. Clin Microbiol Rev 24:29–70. doi: 10.1128/CMR.00036-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 929.Cosson P, Zulianello L, Join-Lambert O, Faurisson F, Gebbie L, Benghezal M, Van Delden C, Curty LK, Köhler T. 2002. Pseudomonas aeruginosa virulence analyzed in a Dictyostelium discoideum host system. J Bacteriol 184:3027–3033. doi: 10.1128/JB.184.11.3027-3033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 930.Linares JF, Lopez JA, Camafeita E, Albar JP, Rojo F, Martínez JL. 2005. Overexpression of the multidrug efflux pumps MexCD-OprJ and MexEF-OprN is associated with a reduction of type III secretion in Pseudomonas aeruginosa. J Bacteriol 187:1384–1391. doi: 10.1128/JB.187.4.1384-1391.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 931.Morero NR, Monti MR, Argarana CE. 2011. Effect of ciprofloxacin concentration on the frequency and nature of resistant mutants selected from Pseudomonas aeruginosa mutS and mutT hypermutators. Antimicrob Agents Chemother 55:3668–3676. doi: 10.1128/AAC.01826-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 932.Macia MD, Pérez JL, Molin S, Oliver A. 2011. Dynamics of mutator and antibiotic-resistant populations in a pharmacokinetic/pharmacodynamic model of Pseudomonas aeruginosa biofilm treatment. Antimicrob Agents Chemother 55:5230–5237. doi: 10.1128/AAC.00617-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 933.Griffith DC, Corcoran E, Lofland D, Lee A, Cho D, Lomovskaya O, Dudley MN. 2006. Pharmacodynamics of levofloxacin against Pseudomonas aeruginosa with reduced susceptibility due to different efflux pumps: do elevated MICs always predict reduced in vivo efficacy? Antimicrob Agents Chemother 50:1628–1632. doi: 10.1128/AAC.50.5.1628-1632.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 934.Martínez-Ramos I, Mulet X, Moya B, Barbier M, Oliver A, Alberti S. 2014. Overexpression of MexCD-OprJ reduces Pseudomonas aeruginosa virulence by increasing its susceptibility to complement-mediated killing. Antimicrob Agents Chemother 58:2426–2429. doi: 10.1128/AAC.02012-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 935.Olivares J, Alvarez-Ortega C, Linares JF, Rojo F, Köhler T, Martínez JL. 2012. Overproduction of the multidrug efflux pump MexEF-OprN does not impair Pseudomonas aeruginosa fitness in competition tests, but produces specific changes in bacterial regulatory networks. Environ Microbiol 14:1968–1981. doi: 10.1111/j.1462-2920.2012.02727.x. [DOI] [PubMed] [Google Scholar]
- 936.Köhler T, van Delden C, Curty LK, Hamzehpour MM, Pechère JC. 2001. Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J Bacteriol 183:5213–5222. doi: 10.1128/JB.183.18.5213-5222.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 937.Lamarche MG, Deziel E. 2011. MexEF-OprN efflux pump exports the Pseudomonas quinolone signal (PQS) precursor HHQ (4-hydroxy-2-heptylquinoline). PLoS One 6:e24310. doi: 10.1371/journal.pone.0024310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 938.Nicastro GG, Kaihami GH, Pereira TO, Meireles DA, Groleau MC, Deziel E, Baldini RL. 2014. Cyclic-di-GMP levels affect Pseudomonas aeruginosa fitness in the presence of imipenem. Environ Microbiol 16:1321–1333. doi: 10.1111/1462-2920.12422. [DOI] [PubMed] [Google Scholar]
- 939.Wang N, Ozer EA, Mandel MJ, Hauser AR. 2014. Genome-wide identification of Acinetobacter baumannii genes necessary for persistence in the lung. mBio 5(3):e01163-14. doi: 10.1128/mBio.01163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 940.Lister IM, Mecsas J, Levy SB. 2010. Effect of MarA-like proteins on antibiotic resistance and virulence in Yersinia pestis. Infect Immun 78:364–371. doi: 10.1128/IAI.00904-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 941.Crimmins GT, Mohammadi S, Green ER, Bergman MA, Isberg RR, Mecsas J. 2012. Identification of MrtAB, an ABC transporter specifically required for Yersinia pseudotuberculosis to colonize the mesenteric lymph nodes. PLoS Pathog 8:e1002828. doi: 10.1371/journal.ppat.1002828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 942.Kirkpatrick CL, Viollier PH. 2014. Synthetic interaction between the TipN polarity factor and an AcrAB-family efflux pump implicates cell polarity in bacterial drug resistance. Chem Biol 21:657–665. doi: 10.1016/j.chembiol.2014.02.018. [DOI] [PubMed] [Google Scholar]
- 943.Silver LL. 2011. Challenges of antibacterial discovery. Clin Microbiol Rev 24:71–109. doi: 10.1128/CMR.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 944.Pucci MJ, Bush K. 2013. Investigational antimicrobial agents of 2013. Clin Microbiol Rev 26:792–821. doi: 10.1128/CMR.00033-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 945.Cohen BE. 2014. Functional linkage between genes that regulate osmotic stress responses and multidrug resistance transporters: challenges and opportunities for antibiotic discovery. Antimicrob Agents Chemother 58:640–646. doi: 10.1128/AAC.02095-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 946.Boucher HW, Talbot GH, Benjamin DK Jr, Bradley J, Guidos RJ, Jones RN, Murray BE, Bonomo RA, Gilbert D, Infectious Diseases Society of America. 2013. 10 × ′20 progress—development of new drugs active against Gram-negative bacilli: an update from the Infectious Diseases Society of America. Clin Infect Dis 56:1685–1694. doi: 10.1093/cid/cit152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 947.Tamma PD, Cosgrove SE, Maragakis LL. 2012. Combination therapy for treatment of infections with Gram-negative bacteria. Clin Microbiol Rev 25:450–470. doi: 10.1128/CMR.05041-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 948.Piddock LJ. 2012. The crisis of no new antibiotics—what is the way forward? Lancet Infect Dis 12:249–253. doi: 10.1016/S1473-3099(11)70316-4. [DOI] [PubMed] [Google Scholar]
- 949.Butler MS, Blaskovich MA, Cooper MA. 2013. Antibiotics in the clinical pipeline in 2013. J Antibiot (Tokyo) 66:571–591. doi: 10.1038/ja.2013.86. [DOI] [PubMed] [Google Scholar]
- 950.Macone AB, Caruso BK, Leahy RG, Donatelli J, Weir S, Draper MP, Tanaka SK, Levy SB. 2014. In vitro and in vivo antibacterial activities of omadacycline, a novel aminomethylcycline. Antimicrob Agents Chemother 58:1127–1135. doi: 10.1128/AAC.01242-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 951.Draper MP, Weir S, Macone A, Donatelli J, Trieber CA, Tanaka SK, Levy SB. 2014. Mechanism of action of the novel aminomethylcycline antibiotic omadacycline. Antimicrob Agents Chemother 58:1279–1283. doi: 10.1128/AAC.01066-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 952.Sutcliffe JA, O'Brien W, Fyfe C, Grossman TH. 2013. Antibacterial activity of eravacycline (TP-434), a novel fluorocycline, against hospital and community pathogens. Antimicrob Agents Chemother 57:5548–5558. doi: 10.1128/AAC.01288-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 953.Landman D, Kelly P, Backer M, Babu E, Shah N, Bratu S, Quale J. 2011. Antimicrobial activity of a novel aminoglycoside, ACHN-490, against Acinetobacter baumannii and Pseudomonas aeruginosa from New York City. J Antimicrob Chemother 66:332–334. doi: 10.1093/jac/dkq459. [DOI] [PubMed] [Google Scholar]
- 954.Walkty A, Adam H, Baxter M, Denisuik A, Lagace-Wiens P, Karlowsky JA, Hoban DJ, Zhanel GG. 2014. In vitro activity of plazomicin against 5,015 Gram-negative and Gram-positive clinical isolates obtained from patients in Canadian hospitals as part of the CANWARD study, 2011-2012. Antimicrob Agents Chemother 58:2554–2563. doi: 10.1128/AAC.02744-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 955.Aggen JB, Armstrong ES, Goldblum AA, Dozzo P, Linsell MS, Gliedt MJ, Hildebrandt DJ, Feeney LA, Kubo A, Matias RD, Lopez S, Gomez M, Wlasichuk KB, Diokno R, Miller GH, Moser HE. 2010. Synthesis and spectrum of the neoglycoside ACHN-490. Antimicrob Agents Chemother 54:4636–4642. doi: 10.1128/AAC.00572-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 956.Mushtaq S, Warner M, Livermore DM. 2010. In vitro activity of ceftazidime+NXL104 against Pseudomonas aeruginosa and other non-fermenters. J Antimicrob Chemother 65:2376–2381. doi: 10.1093/jac/dkq306. [DOI] [PubMed] [Google Scholar]
- 957.Clark C, McGhee P, Appelbaum PC, Kosowska-Shick K. 2011. Multistep resistance development studies of ceftaroline in Gram-positive and -negative bacteria. Antimicrob Agents Chemother 55:2344–2351. doi: 10.1128/AAC.01602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 958.Bulik CC, Christensen H, Nicolau DP. 2010. In vitro potency of CXA-101, a novel cephalosporin, against Pseudomonas aeruginosa displaying various resistance phenotypes, including multidrug resistance. Antimicrob Agents Chemother 54:557–559. doi: 10.1128/AAC.00912-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 959.Cabot G, Bruchmann S, Mulet X, Zamorano L, Moya B, Juan C, Haussler S, Oliver A. 2014. Pseudomonas aeruginosa ceftolozane-tazobactam resistance development requires multiple mutations leading to overexpression and structural modification of AmpC. Antimicrob Agents Chemother 58:3091–3099. doi: 10.1128/AAC.02462-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 960.Sader HS, Castanheira M, Flamm RK, Farrell DJ, Jones RN. 2014. Antimicrobial activity of ceftazidime-avibactam against Gram-negative organisms collected from U.S. medical centers in 2012. Antimicrob Agents Chemother 58:1684–1692. doi: 10.1128/AAC.02429-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 961.Karlowsky JA, Adam HJ, Baxter MR, Lagace-Wiens PR, Walkty AJ, Hoban DJ, Zhanel GG. 2013. In vitro activity of ceftaroline-avibactam against Gram-negative and Gram-positive pathogens isolated from patients in Canadian hospitals from 2010 to 2012: results from the CANWARD surveillance study. Antimicrob Agents Chemother 57:5600–5611. doi: 10.1128/AAC.01485-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 962.Sader HS, Farrell DJ, Castanheira M, Flamm RK, Jones RN. 2014. Antimicrobial activity of ceftolozane/tazobactam tested against Pseudomonas aeruginosa and Enterobacteriaceae with various resistance patterns isolated in European hospitals (2011-12). J Antimicrob Chemother 69:2713–2722. doi: 10.1093/jac/dku184. [DOI] [PubMed] [Google Scholar]
- 963.Caughlan RE, Jones AK, Delucia AM, Woods AL, Xie L, Ma B, Barnes SW, Walker JR, Sprague ER, Yang X, Dean CR. 2012. Mechanisms decreasing in vitro susceptibility to the LpxC inhibitor CHIR-090 in the Gram-negative pathogen Pseudomonas aeruginosa. Antimicrob Agents Chemother 56:17–27. doi: 10.1128/AAC.05417-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 964.Almer LS, Hoffrage JB, Keller EL, Flamm RK, Shortridge VD. 2004. In vitro and bactericidal activities of ABT-492, a novel fluoroquinolone, against Gram-positive and Gram-negative organisms. Antimicrob Agents Chemother 48:2771–2777. doi: 10.1128/AAC.48.7.2771-2777.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 965.Adam HJ, Laing NM, King CR, Lulashnyk B, Hoban DJ, Zhanel GG. 2009. In vitro activity of nemonoxacin, a novel nonfluorinated quinolone, against 2,440 clinical isolates. Antimicrob Agents Chemother 53:4915–4920. doi: 10.1128/AAC.00078-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 966.Higgins PG, Stubbings W, Wisplinghoff H, Seifert H. 2010. Activity of the investigational fluoroquinolone finafloxacin against ciprofloxacin-sensitive and -resistant Acinetobacter baumannii isolates. Antimicrob Agents Chemother 54:1613–1615. doi: 10.1128/AAC.01637-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 967.Morrow BJ, He W, Amsler KM, Foleno BD, Macielag MJ, Lynch AS, Bush K. 2010. In vitro antibacterial activities of JNJ-Q2, a new broad-spectrum fluoroquinolone. Antimicrob Agents Chemother 54:1955–1964. doi: 10.1128/AAC.01374-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 968.Pucci MJ, Podos SD, Thanassi JA, Leggio MJ, Bradbury BJ, Deshpande M. 2011. In vitro and in vivo profiles of ACH-702, an isothiazoloquinolone, against bacterial pathogens. Antimicrob Agents Chemother 55:2860–2871. doi: 10.1128/AAC.01666-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 969.Higuchi S, Onodera Y, Chiba M, Hoshino K, Gotoh N. 2013. Potent in vitro antibacterial activity of DS-8587, a novel broad-spectrum quinolone, against Acinetobacter baumannii. Antimicrob Agents Chemother 57:1978–1981. doi: 10.1128/AAC.02374-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 970.Higuchi S, Kurosaka Y, Uoyama S, Yoshida K, Chiba M, Ishii C, Fujikawa K, Karibe Y, Hoshino K. 2014. Anti-multidrug-resistant Acinetobacter baumannii activity of DS-8587: in vitro activity and in vivo efficacy in a murine calf muscle infection model. J Infect Chemother 20:312–316. doi: 10.1016/j.jiac.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 971.Bohnert JA, Schuster S, Fahnrich E, Trittler R, Kern WV. 2007. Altered spectrum of multidrug resistance associated with a single point mutation in the Escherichia coli RND-type MDR efflux pump YhiV (MdtF). J Antimicrob Chemother 59:1216–1222. doi: 10.1093/jac/dkl426. [DOI] [PubMed] [Google Scholar]
- 972.Kaul M, Zhang Y, Parhi AK, Lavoie EJ, Pilch DS. 2014. Inhibition of RND-type efflux pumps confers the FtsZ-directed prodrug TXY436 with activity against Gram-negative bacteria. Biochem Pharmacol 89:321–328. doi: 10.1016/j.bcp.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 973.Neyfakh AA, Bidnenko VE, Chen LB. 1991. Efflux-mediated multidrug resistance in Bacillus subtilis: similarities and dissimilarities with the mammalian system. Proc Natl Acad Sci U S A 88:4781–4785. doi: 10.1073/pnas.88.11.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 974.Matsumoto Y, Hayama K, Sakakihara S, Nishino K, Noji H, Iino R, Yamaguchi A. 2011. Evaluation of multidrug efflux pump inhibitors by a new method using microfluidic channels. PLoS One 6:e18547. doi: 10.1371/journal.pone.0018547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 975.Lamers RP, Cavallari JF, Burrows LL. 2013. The efflux inhibitor phenylalanine-arginine β-naphthylamide (PAβN) permeabilizes the outer membrane of Gram-negative bacteria. PLoS One 8:e60666. doi: 10.1371/journal.pone.0060666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 976.Paixao L, Rodrigues L, Couto I, Martins M, Fernandes P, de Carvalho CC, Monteiro GA, Sansonetty F, Amaral L, Viveiros M. 2009. Fluorometric determination of ethidium bromide efflux kinetics in Escherichia coli. J Biol Eng 3:18. doi: 10.1186/1754-1611-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 977.Watkins WJ, Landaverry Y, Leger R, Litman R, Renau TE, Williams N, Yen R, Zhang JZ, Chamberland S, Madsen D, Griffith D, Tembe V, Huie K, Dudley MN. 2003. The relationship between physicochemical properties, in vitro activity and pharmacokinetic profiles of analogues of diamine-containing efflux pump inhibitors. Bioorg Med Chem Lett 13:4241–4244. doi: 10.1016/j.bmcl.2003.07.030. [DOI] [PubMed] [Google Scholar]
- 978.Lomovskaya O, Bostian KA. 2006. Practical applications and feasibility of efflux pump inhibitors in the clinic—a vision for applied use. Biochem Pharmacol 71:910–918. doi: 10.1016/j.bcp.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 979.Bohnert JA, Kern WV. 2005. Selected arylpiperazines are capable of reversing multidrug resistance in Escherichia coli overexpressing RND efflux pumps. Antimicrob Agents Chemother 49:849–852. doi: 10.1128/AAC.49.2.849-852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 980.Kern WV, Steinke P, Schumacher A, Schuster S, von Baum H, Bohnert JA. 2006. Effect of 1-(1-naphthylmethyl)-piperazine, a novel putative efflux pump inhibitor, on antimicrobial drug susceptibility in clinical isolates of Escherichia coli. J Antimicrob Chemother 57:339–343. doi: 10.1093/jac/dki445. [DOI] [PubMed] [Google Scholar]
- 981.Zechini B, Versace I. 2009. Inhibitors of multidrug resistant efflux systems in bacteria. Recent Pat Antiinfect Drug Discov 4:37–50. doi: 10.2174/157489109787236256. [DOI] [PubMed] [Google Scholar]
- 982.Schuster S, Köhler S, Buck A, Dambacher C, Konig A, Bohnert JA, Kern WV. 2014. Random mutagenesis of the multidrug transporter AcrB from Escherichia coli for identification of putative target residues of efflux pump inhibitors. Antimicrob Agents Chemother 58:6870–6878. doi: 10.1128/AAC.03775-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 983.Yoshida K, Nakayama K, Ohtsuka M, Kuru N, Yokomizo Y, Sakamoto A, Takemura M, Hoshino K, Kanda H, Nitanai H, Namba K, Yoshida K, Imamura Y, Zhang JZ, Lee VJ, Watkins WJ. 2007. MexAB-OprM specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 7: highly soluble and in vivo active quaternary ammonium analogue D13-9001, a potential preclinical candidate. Bioorg Med Chem 15:7087–7097. doi: 10.1016/j.bmc.2007.07.039. [DOI] [PubMed] [Google Scholar]
- 984.Eren E, Vijayaraghavan J, Liu J, Cheneke BR, Touw DS, Lepore BW, Indic M, Movileanu L, van den Berg B. 2012. Substrate specificity within a family of outer membrane carboxylate channels. PLoS Biol 10:e1001242. doi: 10.1371/journal.pbio.1001242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 985.Mahamoud A, Chevalier J, Alibert-Franco S, Kern WV, Pagès J-M. 2007. Antibiotic efflux pumps in Gram-negative bacteria: the inhibitor response strategy. J Antimicrob Chemother 59:1223–1229. doi: 10.1093/jac/dkl493. [DOI] [PubMed] [Google Scholar]
- 986.Pagès JM, Amaral L, Fanning S. 2011. An original deal for new molecule: reversal of efflux pump activity, a rational strategy to combat Gram-negative resistant bacteria. Curr Med Chem 18:2969–2980. doi: 10.2174/092986711796150469. [DOI] [PubMed] [Google Scholar]
- 987.Bhardwaj AK, Mohanty P. 2012. Bacterial efflux pumps involved in multidrug resistance and their inhibitors: rejuvinating the antimicrobial chemotherapy. Recent Pat Antiinfect Drug Discov 7:73–89. doi: 10.2174/157489112799829710. [DOI] [PubMed] [Google Scholar]
- 988.Pagès JM, Masi M, Barbe J. 2005. Inhibitors of efflux pumps in Gram-negative bacteria. Trends Mol Med 11:382–389. doi: 10.1016/j.molmed.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 989.Chevalier J, Bredin J, Mahamoud A, Mallea M, Barbe J, Pagès JM. 2004. Inhibitors of antibiotic efflux in resistant Enterobacter aerogenes and Klebsiella pneumoniae strains. Antimicrob Agents Chemother 48:1043–1046. doi: 10.1128/AAC.48.3.1043-1046.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 990.Thorarensen A, Presley-Bodnar AL, Marotti KR, Boyle TP, Heckaman CL, Bohanon MJ, Tomich PK, Zurenko GE, Sweeney MT, Yagi BH. 2001. 3-Arylpiperidines as potentiators of existing antibacterial agents. Bioorg Med Chem Lett 11:1903–1906. doi: 10.1016/S0960-894X(01)00330-4. [DOI] [PubMed] [Google Scholar]
- 991.Bohnert JA, Schuster S, Kern WV. 2013. Pimozide inhibits the AcrAB-TolC efflux pump in Escherichia coli. Open Microbiol J 7:83–86. doi: 10.2174/1874285801307010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 992.Bohnert JA, Szymaniak-Vits M, Schuster S, Kern WV. 2011. Efflux inhibition by selective serotonin reuptake inhibitors in Escherichia coli. J Antimicrob Chemother 66:2057–2060. doi: 10.1093/jac/dkr258. [DOI] [PubMed] [Google Scholar]
- 993.Molnar J, Hever A, Fakla I, Fischer J, Ocsovski I, Aszalos A. 1997. Inhibition of the transport function of membrane proteins by some substituted phenothiazines in E. coli and multidrug resistant tumor cells. Anticancer Res 17:481–486. [PubMed] [Google Scholar]
- 994.Kaatz GW, Moudgal VV, Seo SM, Kristiansen JE. 2003. Phenothiazines and thioxanthenes inhibit multidrug efflux pump activity in Staphylococcus aureus. Antimicrob Agents Chemother 47:719–726. doi: 10.1128/AAC.47.2.719-726.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 995.Viveiros M, Martins A, Paixao L, Rodrigues L, Martins M, Couto I, Fahnrich E, Kern WV, Amaral L. 2008. Demonstration of intrinsic efflux activity of Escherichia coli K-12 AG100 by an automated ethidium bromide method. Int J Antimicrob Agents 31:458–462. doi: 10.1016/j.ijantimicag.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 996.Martins A, Machado L, Costa S, Cerca P, Spengler G, Viveiros M, Amaral L. 2011. Role of calcium in the efflux system of Escherichia coli. Int J Antimicrob Agents 37:410–414. doi: 10.1016/j.ijantimicag.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 997.Takacs D, Cerca P, Martins A, Riedl Z, Hajos G, Molnar J, Viveiros M, Couto I, Amaral L. 2011. Evaluation of forty new phenothiazine derivatives for activity against intrinsic efflux pump systems of reference Escherichia coli, Salmonella Enteritidis, Enterococcus faecalis and Staphylococcus aureus strains. In Vivo 25:719–724. [PubMed] [Google Scholar]
- 998.Brunel JM, Lieutaud A, Lome V, Pagès JM, Bolla JM. 2013. Polyamino geranic derivatives as new chemosensitizers to combat antibiotic resistant Gram-negative bacteria. Bioorg Med Chem 21:1174–1179. doi: 10.1016/j.bmc.2012.12.030. [DOI] [PubMed] [Google Scholar]
- 999.Martins A, Spengler G, Rodrigues L, Viveiros M, Ramos J, Martins M, Couto I, Fanning S, Pagès JM, Bolla JM, Molnar J, Amaral L. 2009. pH modulation of efflux pump activity of multi-drug resistant Escherichia coli: protection during its passage and eventual colonization of the colon. PLoS One 4:e6656. doi: 10.1371/journal.pone.0006656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000.Stavri M, Piddock LJV, Gibbons S. 2007. Bacterial efflux pump inhibitors from natural sources. J Antimicrob Chemother 59:1247–1260. doi: 10.1093/jac/dkl460. [DOI] [PubMed] [Google Scholar]
- 1001.Li B, Yao Q, Pan XC, Wang N, Zhang R, Li J, Ding G, Liu X, Wu C, Ran D, Zheng J, Zhou H. 2011. Artesunate enhances the antibacterial effect of β-lactam antibiotics against Escherichia coli by increasing antibiotic accumulation via inhibition of the multidrug efflux pump system AcrAB-TolC. J Antimicrob Chemother 66:769–777. doi: 10.1093/jac/dkr017. [DOI] [PubMed] [Google Scholar]
- 1002.Touani FK, Seukep AJ, Djeussi DE, Fankam AG, Noumedem JA, Kuete V. 2014. Antibiotic-potentiation activities of four Cameroonian dietary plants against multidrug-resistant Gram-negative bacteria expressing efflux pumps. BMC Complement Altern Med 14:258. doi: 10.1186/1472-6882-14-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1003.Bag A, Chattopadhyay RR. 2014. Efflux-pump inhibitory activity of a gallotannin from Terminalia chebula fruit against multidrug-resistant uropathogenic Escherichia coli. Nat Prod Res 28:1280–1283. doi: 10.1080/14786419.2014.895729. [DOI] [PubMed] [Google Scholar]
- 1004.Blanchard C, Barnett P, Perlmutter J, Dunman PM. 2014. Identification of Acinetobacter baumannii serum-associated antibiotic efflux pump inhibitors. Antimicrob Agents Chemother 58:6360–6370. doi: 10.1128/AAC.03535-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1005.Biot FV, Valade E, Garnotel E, Chevalier J, Villard C, Thibault FM, Vidal DR, Pagès JM. 2011. Involvement of the efflux pumps in chloramphenicol selected strains of Burkholderia thailandensis: proteomic and mechanistic evidence. PLoS One 6:e16892. doi: 10.1371/journal.pone.0016892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1006.Vidal-Aroca F, Meng A, Minz T, Page MG, Dreier J. 2009. Use of resazurin to detect mefloquine as an efflux-pump inhibitor in Pseudomonas aeruginosa and Escherichia coli. J Microbiol Methods 79:232–237. doi: 10.1016/j.mimet.2009.09.021. [DOI] [PubMed] [Google Scholar]
- 1007.Iino R, Nishino K, Noji H, Yamaguchi A, Matsumoto Y. 2012. A microfluidic device for simple and rapid evaluation of multidrug efflux pump inhibitors. Front Microbiol 3:40. doi: 10.3389/fmicb.2012.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1008.Ribera A, Jurado A, Ruiz J, Marco F, Del Valle O, Mensa J, Chaves J, Hernandez G, Jimenez de Anta MT, Vila J. 2002. In vitro activity of clinafloxacin in comparison with other quinolones against Stenotrophomonas maltophilia clinical isolates in the presence and absence of reserpine. Diagn Microbiol Infect Dis 42:123–128. doi: 10.1016/S0732-8893(01)00335-2. [DOI] [PubMed] [Google Scholar]
- 1009.Shinabarger DL, Zurenko GE, Hesje CK, Sanfilippo CM, Morris TW, Haas W. 2011. Evaluation of the effect of bacterial efflux pumps on the antibacterial activity of the novel fluoroquinolone besifloxacin. J Chemother 23:80–86. doi: 10.1179/joc.2011.23.2.80. [DOI] [PubMed] [Google Scholar]
- 1010.Koita K, Rao CV. 2012. Identification and analysis of the putative pentose sugar efflux transporters in Escherichia coli. PLoS One 7:e43700. doi: 10.1371/journal.pone.0043700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1011.Han X, Dorsey-Oresto A, Malik M, Wang JY, Drlica K, Zhao X, Lu T. 2010. Escherichia coli genes that reduce the lethal effects of stress. BMC Microbiol 10:35. doi: 10.1186/1471-2180-10-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1012.Frawley ER, Crouch ML, Bingham-Ramos LK, Robbins HF, Wang W, Wright GD, Fang FC. 2013. Iron and citrate export by a major facilitator superfamily pump regulates metabolism and stress resistance in Salmonella Typhimurium. Proc Natl Acad Sci U S A 110:12054–12059. doi: 10.1073/pnas.1218274110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1013.Holdsworth SR, Law CJ. 2012. Functional and biochemical characterisation of the Escherichia coli major facilitator superfamily multidrug transporter MdtM. Biochimie 94:1334–1346. doi: 10.1016/j.biochi.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 1014.Purewal AS, Jones IG, Midgley M. 1990. Cloning of the ethidium efflux gene from Escherichia coli. FEMS Microbiol Lett 56:73–76. [DOI] [PubMed] [Google Scholar]
- 1015.Morimyo M, Hongo E, Hama-Inaba H, Machida I. 1992. Cloning and characterization of the mvrC gene of Escherichia coli K-12 which confers resistance against methyl viologen toxicity. Nucleic Acids Res 20:3159–3165. doi: 10.1093/nar/20.12.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1016.Higashi K, Ishigure H, Demizu R, Uemura T, Nishino K, Yamaguchi A, Kashiwagi K, Igarashi K. 2008. Identification of a spermidine excretion protein complex (MdtJI) in Escherichia coli. J Bacteriol 190:872–878. doi: 10.1128/JB.01505-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1017.Nandineni MR, Gowrishankar J. 2004. Evidence for an arginine exporter encoded by yggA (argO) that is regulated by the LysR-type transcriptional regulator ArgP in Escherichia coli. J Bacteriol 186:3539–3546. doi: 10.1128/JB.186.11.3539-3546.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1018.Hansen LH, Sorensen SJ, Jorgensen HS, Jensen LB. 2005. The prevalence of the OqxAB multidrug efflux pump amongst olaquindox-resistant Escherichia coli in pigs. Microb Drug Resist 11:378–382. doi: 10.1089/mdr.2005.11.378. [DOI] [PubMed] [Google Scholar]
- 1019.Sorensen AH, Hansen LH, Johannesen E, Sorensen SJ. 2003. Conjugative plasmid conferring resistance to olaquindox. Antimicrob Agents Chemother 47:798–799. doi: 10.1128/AAC.47.2.798-799.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1020.Droge M, Puhler A, Selbitschka W. 2000. Phenotypic and molecular characterization of conjugative antibiotic resistance plasmids isolated from bacterial communities of activated sludge. Mol Gen Genet 263:471–482. doi: 10.1007/s004380051191. [DOI] [PubMed] [Google Scholar]
- 1021.Tauch A, Schluter A, Bischoff N, Goesmann A, Meyer F, Puhler A. 2003. The 79,370-bp conjugative plasmid pB4 consists of an IncP-1β backbone loaded with a chromate resistance transposon, the strA-strB streptomycin resistance gene pair, the oxacillinase gene blaNPS-1, and a tripartite antibiotic efflux system of the resistance-nodulation-division family. Mol Genet Genomics 268:570–584. doi: 10.1007/s00438-002-0785-z. [DOI] [PubMed] [Google Scholar]
- 1022.Rose S, Desmolaize B, Jaju P, Wilhelm C, Warrass R, Douthwaite S. 2012. Multiplex PCR to identify macrolide resistance determinants in Mannheimia haemolytica and Pasteurella multocida. Antimicrob Agents Chemother 56:3664–3669. doi: 10.1128/AAC.00266-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1023.Yuan J, Xu X, Guo Q, Zhao X, Ye X, Guo Y, Wang M. 2012. Prevalence of the oqxAB gene complex in Klebsiella pneumoniae and Escherichia coli clinical isolates. J Antimicrob Chemother 67:1655–1659. doi: 10.1093/jac/dks086. [DOI] [PubMed] [Google Scholar]
- 1024.Mesaros N, Glupczynski Y, Avrain L, Caceres NE, Tulkens PM, Van Bambeke F. 2007. A combined phenotypic and genotypic method for the detection of Mex efflux pumps in Pseudomonas aeruginosa. J Antimicrob Chemother 59:378–386. doi: 10.1093/jac/dkl504. [DOI] [PubMed] [Google Scholar]
- 1025.Cabot G, Ocampo-Sosa AA, Tubau F, Macia MD, Rodríguez C, Moya B, Zamorano L, Suarez C, Pena C, Martínez-Martínez L, Oliver A. 2011. Overexpression of AmpC and efflux pumps in Pseudomonas aeruginosa isolates from bloodstream infections: prevalence and impact on resistance in a Spanish multicenter study. Antimicrob Agents Chemother 55:1906–1911. doi: 10.1128/AAC.01645-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1026.Pérez A, Poza M, Aranda J, Latasa C, Medrano FJ, Tomás M, Romero A, Lasa I, Bou G. 2012. Effect of transcriptional activators SoxS, RobA, and RamA on expression of multidrug efflux pump AcrAB-TolC in Enterobacter cloacae. Antimicrob Agents Chemother 56:6256–6266. doi: 10.1128/AAC.01085-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1027.Roy S, Datta S, Viswanathan R, Singh AK, Basu S. 2013. Tigecycline susceptibility in Klebsiella pneumoniae and Escherichia coli causing neonatal septicaemia (2007-10) and role of an efflux pump in tigecycline non-susceptibility. J Antimicrob Chemother 68:1036–1042. doi: 10.1093/jac/dks535. [DOI] [PubMed] [Google Scholar]
- 1028.Lewinson O, Adler J, Poelarends GJ, Mazurkiewicz P, Driessen AJ, Bibi E. 2003. The Escherichia coli multidrug transporter MdfA catalyzes both electrogenic and electroneutral transport reactions. Proc Natl Acad Sci U S A 100:1667–1672. doi: 10.1073/pnas.0435544100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1029.CLSI. 2014. Performance standards for antimicrobial susceptibility testing. Twenty-fourth informational supplement. M100-S24. CLSI, Wayne, PA. [Google Scholar]
- 1030.Ghisalberti D, Mahamoud A, Chevalier J, Baitiche M, Martino M, Pagès JM, Barbe J. 2006. Chloroquinolines block antibiotic efflux pumps in antibiotic-resistant Enterobacter aerogenes isolates. Int J Antimicrob Agents 27:565–569. doi: 10.1016/j.ijantimicag.2006.03.010. [DOI] [PubMed] [Google Scholar]