SUMMARY
Escherichia coli sequence type 131 (ST131) and Klebsiella pneumoniae ST258 emerged in the 2000s as important human pathogens, have spread extensively throughout the world, and are responsible for the rapid increase in antimicrobial resistance among E. coli and K. pneumoniae strains, respectively. E. coli ST131 causes extraintestinal infections and is often fluoroquinolone resistant and associated with extended-spectrum β-lactamase production, especially CTX-M-15. K. pneumoniae ST258 causes urinary and respiratory tract infections and is associated with carbapenemases, most often KPC-2 and KPC-3. The most prevalent lineage within ST131 is named fimH30 because it contains the H30 variant of the type 1 fimbrial adhesin gene, and recent molecular studies have demonstrated that this lineage emerged in the early 2000s and was then followed by the rapid expansion of its sublineages H30-R and H30-Rx. K. pneumoniae ST258 comprises 2 distinct lineages, namely clade I and clade II. Moreover, it seems that ST258 is a hybrid clone that was created by a large recombination event between ST11 and ST442. Epidemic plasmids with blaCTX-M and blaKPC belonging to incompatibility group F have contributed significantly to the success of these clones. E. coli ST131 and K. pneumoniae ST258 are the quintessential examples of international multidrug-resistant high-risk clones.
INTRODUCTION
The Enterobacteriaceae, especially Escherichia coli and Klebsiella pneumoniae, are common causes of nosocomial and community infections among humans (1). One of the most urgent areas of antimicrobial drug resistance is the rapid evolution of fluoroquinolone, cephalosporin, and carbapenem resistance among Enterobacteriaceae, which has spread globally during the last decade. Recently, the World Health Organization (WHO) released a report entitled Antimicrobial Resistance: Global Report on Surveillance 2014 (2). This report focused on antibiotic resistance among bacteria responsible for common, serious infections, including bloodstream-associated infections (BSIs), urinary tract infections (UTIs), and intra-abdominal infections (IAIs). It states that resistance to certain antibiotics among frequently isolated bacteria is rife in certain parts of the globe.
For E. coli, this report states the following: “Resistance to one of the most widely used antibacterial medicines (i.e., the fluoroquinolones) for the treatment of UTIs caused by E. coli is very widespread. In the 1980s, when these drugs were first introduced, resistance was virtually zero. Today, there are countries in many parts of the world where this treatment is now ineffective in more than half of patients” (2). Specifically, for K. pneumoniae, the WHO report states the following: “Resistance to the last resort agents (i.e., carbapenem antibiotics) for the treatment of life-threatening infections caused by a common intestinal bacteria, K. pneumoniae has spread to all regions of the world. K. pneumoniae is a major cause of hospital-acquired infections such as pneumonia, BSIs, infections in newborns and intensive care unit patients. In some countries, because of resistance, carbapenem antibiotics would not work in more than half of people treated for K. pneumoniae infections” (2).
The global dissemination of drug-resistant organisms is troublesome for medical practitioners because it decreases the available options for appropriate treatment. This contributes to increased patient mortality and morbidity (3). Therefore, it is not surprising that the global spread of drug resistance was recently recognized as a major threat to human health (4). Antimicrobial resistance endangers some of the accomplishments of modern medicine; e.g., in a world without antibiotics, it would be difficult to provide effective chemotherapy to cancer patients, since most will die due to opportunistic multidrug-resistant (MDR) infections.
This pandemic comes at a time when Moore's law has enabled the analysis of large amounts of sequencing data, enabling molecular epidemiologists to study antimicrobial resistance (5). The analysis of such data will enable bioinformaticians to determine the global dissemination of clones and plasmids responsible for antimicrobial resistance in ways that were not feasible 10 years ago. Hopefully, such new insights can yield urgently needed clues on how to prioritize tactics for limiting the global spread of multidrug resistant Enterobacteriaceae.
“Eminent or successful” bacterial clones are a powerful source for the propagation of antimicrobial-resistant genetic components (i.e., genes, integrons, transposons, and plasmids) (6). They are able to provide stable platforms for the maintenance and propagation of genes responsible for antimicrobial resistance and have played an essential role in the recent global emergence of multidrug resistance among Gram-negative organisms, especially the Enterobacteriaceae.
Plasmids are extrachromosomal elements of circular DNA present in bacteria, which replicate independently of the host genome. The horizontal transfer of plasmids containing resistance genes is an essential mechanism for the dispersion of antimicrobial resistance (7). This free movement of plasmid-borne genes responsible for drug resistance has also been central to the recent and rapid global increase in antimicrobial resistance (8). Antimicrobial resistance plasmids can broadly be divided into 2 main groups, namely, the narrow-host-range group, which most often belongs to incompatibility group F (IncF), and the broad-host-range group, which belongs to the IncA/C, IncL/M, and IncN. Broad-host-range plasmids can easily be transferred between different species, while narrow-host-range plasmids tend to be restricted to species (9). Epidemic resistance plasmids belonging to IncF with divergent replicon types (e.g., FIA, FIB, and FII) have the ability to acquire resistance genes and then rapidly disseminate among Enterobacteriaceae, especially among certain clones within species (9).
The focus of molecular epidemiologists when investigating antimicrobial-resistant bacteria has been on analyzing chromosomal DNA, because the initial spread of successful clones did not necessarily require an in-depth investigation of the role that plasmids played in the dissemination of drug resistance genes. Molecular and bioinformatics tools for analyzing large amounts of plasmid DNA combined with refined techniques for large-scale chromosomal analysis will likely provide a better understanding of the ceaseless movement of drug-resistant genes throughout the microbial world (5).
The pandemics caused by multidrug-resistant E. coli and K. pneumoniae (including fluoroquinolone-, cephalosporin-, and carbapenem-resistant isolates) are due mostly to the global dissemination of certain high-risk clones, namely, E. coli sequence type 131 (ST131) and K. pneumoniae ST258. There is a strong relationship of E. coli ST131 and K. pneumoniae ST258 with IncF epidemic plasmids containing FIA and FII replicon types. The reasons for the particular success of these high-risk clones and their association with certain epidemic resistance plasmids are uncertain. However, their ability to spread swiftly is beyond dispute.
This review provides insights into how antibiotic resistance evolves and spreads in bacterial populations and attempts to highlight recent information about the continuous interplay between bacterial clones and antimicrobial resistance plasmids. This article expands the knowledge regarding the role, the importance, and the interdependence of high-risk clones and epidemic plasmids in global spread among multidrug-resistant Enterobacteriaceae. It also provides an overview on rapid laboratory methods that will aid clinicians and infection control measures in combating the spread of high-risk clones.
EXPANDED-SPECTRUM β-LACTAMASES
Overview
Antibiotic therapy with cephalosporins that contain the oxyimino side chain (i.e., cefotaxime, ceftazidime, ceftriaxone, and cefepime) is considered one of the choice treatments for serious infections due to Klebsiella spp. and E. coli. Moreover, carbapenem-resistant E. coli and Klebsiella spp. are troublesome for medical physicians, since these drugs are often the last efficient treatment left for serious infections (1).
β-Lactamases are the most common cause of resistance to various β-lactam agents. Several different schemes have traditionally been used for the classification of β-lactamases. These enzyme can be divided into either classes A, B, C, and D (referred to as the molecular or Ambler classification based on amino acid sequences) or groups 1, 2, and 3 (referred to as the functional or Bush-Jacoby classification based on substrate and inhibitor profiles). Class A, C, and D β-lactamases are serine enzymes, while class B β-lactamases require divalent zinc ions for their activity (10). Group 1 is referred to as serine cephalosporinases, and group 2 is referred to as serine β-lactamases, which includes penicillinases and broad-spectrum enzymes, while group 3 is also called metallo-β-lactamases (MBLs) (10).
For the sake of simplicity, we use the term “expanded-spectrum” β-lactamases in this review article to stipulate those enzymes with activity against the cephalosporins with oxyimino side chains and/or the carbapenems. These enzymes consist of the class C plasmid-mediated or imported AmpC β-lactamases (e.g., CMY types), class A extended-spectrum β-lactamases (ESBLs) (e.g., CTX-M, SHV, and TEM types), and carbapenemases (e.g., KPC types [class A]; MBLs, e.g., VIM, IPM, and NDM types [class B]; and the oxacillinases, e.g., OXA-48-like enzymes [class D]). The production of expanded-spectrum β-lactamases causes nonsusceptibility to various β-lactam agents, and Enterobacteriaceae with these enzymes are often coresistant to a variety of other classes of antibiotics. The most common global type of ESBL is the CTX-M β-lactamases, while the NDM, OXA-48, and KPC β-lactamases are the most frequent carbapenemases among nosocomial and community isolates of Enterobacteriaceae (11). Infections with ESBL- and carbapenemase-producing Enterobacteriaceae in the developed world are sometimes associated with travel to a region of endemicity (12). The features of the expanded-spectrum β-lactamases are outlined in Table 1.
TABLE 1.
Characteristics of Enterobacteriaceae that produce expanded-spectrum β-lactamases
| Enzyme | Enterobacteriaceae member | Class | Example(s) | Spectrum of activity | Agent inhibited | Area(s) of endemicity | Molecular epidemiology |
|---|---|---|---|---|---|---|---|
| Extended-spectrum β-lactamases | E. coli | A | CTX-M-14, -15, others | Penicillins | Clavulanic acid | Worldwide | ST131 |
| K. pneumoniae | TEM, SHV | Cephalosporins | Tazobactam | IncF plasmids | |||
| Others (rare) | Monobactams | Sulbactam | ISEcp1 | ||||
| Plasmid-mediated or imported AmpC β-lactamases | K. pneumoniae | C | CMY, FOX, ACT, MOX, DHA | Penicillins | Cloxacillin | Worldwide | IncA/C, other plasmids |
| E. coli | Cephalosporins (not cefepime) | Boronic acid | |||||
| Salmonella spp. | Cephamycins | Avibactam | |||||
| Others (rare) | Monobactams | ||||||
| Metallo-β-lactamases | K. pneumoniae | B | NDM-1 | Penicillins | Metal chelators, e.g., EDTA, dipicolinic acid | Japan (IMP), Taiwan (IMP), Indian subcontinent (NDM), Balkan states (NDM) | IncA/C, IncN, other plasmids |
| E. coli | IMP, VIM | Cephalosporins | Greece (VIM) | Class I integrons | |||
| Others (rare) | Cephamycins | ||||||
| Carbapenems | |||||||
| KPC carbapenemases | K. pneumoniae | A | KPC-2, -3 | Penicillins | Clavulanic acid (weak) | USA, Greece, Italy, Israel, China, Brazil, Colombia, Argentina | Tn4401 |
| Enterobacter spp. | Others | Cephalosporins | Tazobactam (weak) | IncFII plasmids | |||
| E. coli | Cephamycins | Boronic acid | CC258 | ||||
| Others | Carbapenems | Avibactam | |||||
| OXA β-lactamases | K. pneumoniae | D | OXA-48, OXA-163 | Penicillins | NaCl | Turkey | Tn1999 |
| E. coli | OXA-181 | Temocillin | North Africa (Morocco, Tunisia) | IncL/M plasmids | |||
| Others (rare) | OXA-204 | β-Lactamase inhibitor combinations | |||||
| OXA-232 | Carbapenems |
CTX-M β-Lactamases
ESBLs provide resistance to most of the β-lactam drugs and are inhibited by certain β-lactamase inhibitors (13) (see Table 1 for a summary on which Enterobacteriaceae produce these enzymes and the classification, spectrum of activity, inhibition properties, types, regions of endemicity, and molecular epidemiology of ESBLs). The SHV or TEM types of ESBLs were common during the 1980s and 1990s (13), while the CTX-M types became prominent since 2000 (14). Currently, the CTX-M enzymes are present in various types of bacteria from all continents, being especially frequent in E. coli (15).
The CTX-M types belong to the molecular class A or functional group 2be β-lactamases and include at least six lineages (i.e., CTX-M-1-like, CTX-M-2-like, CTX-M-8-like, CTX-M-9-like, CTX-M-25-like, and KLUC-like) that differ from each other by ≥10% amino acid homology (14). The association of plasmids carrying blaCTX-M that belong to incompatibility groups IncF, IncN, and IncK with certain insertion sequences (ISs) (e.g., ISEcp1 or ISCR1) is able to capture and mobilize blaCTX-M genes effectively among members of the Enterobacteriaceae. IS elements can also act as strong promoters for the high-level expression of blaCTX-M (16) (Table 1). CTX-M-15 is the most universal type of ESBL among E. coli isolates and has been associated with the presence of a clone named ST131 (17). It seems that this clone or sequence type accounted for the global distribution and increase in the prevalence of E. coli with blaCTX-M-15 during the early to mid-2000s (more details are provided in the high-risk clone section, below).
E. coli strains with CTX-M enzymes are responsible for nosocomial and community UTIs, BSIs, and IAIs (14), and risk factors include the preceding use of antibiotics and visiting certain regions of endemicity (18). The worldwide dissemination of E. coli with CTX-M enzymes has been very efficient and involved health care settings, community, livestock, companion animals, wildlife, and the environment (19). Studies have also shown high transmission rates of E. coli with blaCTX-M genes within households (20).
The CTX-M pandemic significantly contributed to the rapid global increase in the rate of cephalosporin resistance among Enterobacteriaceae with subsequent increased usage of the carbapenems for the medication of infections due to these MDR bacteria. Unfortunately, CTX-M-producing E. coli isolates are often coresistant to various antibiotic classes, which include co-trimoxazole, the aminoglycosides, and the fluoroquinolones (15). This has important clinical implications because some of these drugs (e.g., co-trimoxazole) are popular oral treatment options for community-acquired uncomplicated lower UTIs. Fortunately, fosfomycin, amdinocillin, and nitrofurantoin retain sufficient activity against a high percentage of E. coli isolates with blaCTX-M genes (16).
CTX-M-producing E. coli is an important component among global multidrug-resistant bacteria and should be regarded as a major target for surveillance, infection control, and fundamental investigations in the field of antimicrobial drug resistance (21).
AmpC β-Lactamases or Cephalosporinases
Enterobacteriaceae with AmpC β-lactamases are important causes of cephalosporin and cephamycin resistance (see Table 1 for a summary on which Enterobacteriaceae produce imported or plasmid-mediated class C cephalosporinases and the classification, spectrum of activity, inhibition properties, types, regions of endemicity, and molecular epidemiology of plasmid-mediated AmpC β-lactamases). Since high-risk clones and epidemic plasmids do not play important roles in the global dissemination of AmpC enzymes, we refer the reader to some excellent review articles on this topic (22–24).
KPC β-Lactamases
The most clinically significant class A carbapenemases are the KPC (i.e., Klebsiella pneumoniae carbapenemase) types (25) (see Table 1 for a summary on which Enterobacteriaceae produce these enzymes and the classification, spectrum of activity, inhibition properties, types, regions of endemicity, and molecular epidemiology of KPCs). KPC-2 and -3 are the most prevalent isoenzymes among KPCs, and bacteria with these β-lactamases are nonsusceptible to a variety of β-lactam drugs, including the majority of β-lactamase inhibitor combinations (26). KPC enzymes are especially prevalent in Klebsiella spp. and are found to a lesser extent in Enterobacter spp. (25).
Several hospital outbreaks, most often due to K. pneumoniae with blaKPC-2 and blaKPC-3, have been reported in North America (especially the United States), South America (Colombia and Argentina), Europe (Greece, Italy, and Poland), Asia (China), and the Middle East (Israel) (26–28). Regions where KPC-producing bacteria are endemic are shown in Table 1 (28). K. pneumoniae ST258 isolates with blaKPC-2 and blaKPC-3 have significantly contributed to the worldwide distribution of this resistance trait (more details are provided in the section on high-risk clones, below) (28).
Plasmids with blaKPC in association with the mobile element Tn4401 are responsible for the effective spread of these genes among different types of Enterobacteriaceae (29) and, with other antibiotic resistance determinants on the same plasmid, provide an easy mechanism for carbapenemase genes to effectively spread as hitchhiker genes, even in the absence of carbapenem selection (30). Enterobacteriaceae with blaKPC are often multidrug resistant to classes such as the aminoglycosides, fluoroquinolones, and co-trimoxazole (31).
NDM β-Lactamases
The MBL designated NDM was described for K. pneumoniae and E. coli from Sweden during the late 2000s. This patient was previously admitted to a New Delhi hospital in India (32) (see Table 1 for a summary of which Enterobacteriaceae produce these enzymes and the classification, spectrum of activity, inhibition properties, types, regions of endemicity, and molecular epidemiology of NDMs). Patients visiting certain high-risk regions and then returning to their respective home countries with NDM-producing bacteria have been described, and these enzymes are some of the most common carbapenemases identified in countries such as Canada, the United Kingdom, and France (11). Since high-risk clones and epidemic plasmids do not seem to play important roles in the global dissemination of NDMs, we refer the reader to some excellent review articles and recent reports on this topic (11, 33–36).
OXA-48-Like β-Lactamases
The molecular class D β-lactamases are commonly referred to as OXAs and comprise >400 enzymes, with some variants that possess carbapenemase activity (also referred to as carbapenem-hydrolyzing class D β-lactamases [CHDLs]) (37) (see Table 1 for a summary of which Enterobacteriaceae produce these enzymes and the classification, spectrum of activity, inhibition properties, types, regions of endemicity, and molecular epidemiology of CHDLs in Enterobacteriaceae). CHDLs are common in Acinetobacter spp., but the OXA-48-like types of CHDLs are most often encountered among the Enterobacteriaceae (37, 38). OXA-163 is different from other OXA-48-like enzymes in that this enzyme has the ability to hydrolyze the oxyimino cephalosporins. Enterobacteriaceae with OXA-163 are common in certain South American countries, especially Argentina (39). Since high-risk clones and epidemic plasmids do not seem to play an important role in the global dissemination of OXA-48 derivatives, we refer the reader to some excellent review articles on this topic (37, 38).
INTERNATIONAL MULTIDRUG-RESISTANT HIGH-RISK CLONAL LINEAGES
Various definitions have been used to describe the criteria for resistance among MDR Enterobacteriaceae. We use the recent definition of MDR Enterobacteriaceae that was adopted by the European Center for Disease Prevention and Control (40). This definition states that a MDR Enterobacteriaceae isolate is nonsusceptible to at least 1 drug in >3 antimicrobial categories, including aminoglycosides, cephalosporins (divided into 3 groups), cephamycins, antipseudomonal penicillins with β-lactamase inhibitors, penicillins, penicillins with β-lactamase inhibitors, monobactams, carbapenems, folate pathway inhibitors, glycylcyclines, fluoroquinolones, phenicols, phosphonic acids, polymyxins, and tetracyclines.
A bacterial clone refers to the progeny of one bacterial cell through asexual reproduction, implying that the same clonal lineage consists of very closely related isolates that have recently diverged from a common ancestor (41). However, bacterial genomes are plastic and are subjected to genome rearrangements (i.e., deletions and insertion sequences, etc.) and, to various extents, to localized recombinational events. Thus, bacterial isolates assigned to the same clone may not be identical, as recent descendants of the same common ancestor may differ somewhat in genotype (42). Therefore, the strict definition of a clone tends to be loosened slightly in bacteriology, and clones are defined as isolates that are indistinguishable or highly similar to each other, as identified by using a particular molecular typing procedure.
It is important to take into account that the identification of clones depends very much on the molecular typing technique used. The most common molecular techniques currently used by molecular epidemiologists to determine if isolates are clonally related are multilocus sequence typing (MLST), pulsed-field gel electrophoresis (PFGE), PCR typing (such as enterobacterial repetitive intergenic consensus [ERIC] or randomly amplified polymorphic DNA [RAPD] methods), and multilocus variable-number tandem-repeat analysis (MLVA) (43). PFGE is a technique with some of the highest levels of discrimination for typing of bacteria, and in the mid-1990s, Tenover and colleagues provided guidelines to define clones or clusters (i.e., possibly related with 4 to 6 band differences) (44). MLST and MLVA are less discriminatory than PFGE, and clones identified by these techniques often have different pulsotypes; e.g., an international collection of K. pneumoniae ST258 strains (identified by using MLST) consisted of 4 different pulsotypes that showed a distinctive geographical distribution (45).
In this article, the term clone refers to any bacterium propagated from a single colony isolated at a specific time and place showing common phylogenetic origins. This implies that such isolates have similar traits, as determined by methods (e.g., biochemical and molecular typing methods) indicating that they belong to the same group or lineage and possess a common ancestor. This implies that whole-genome sequencing will provide all of the information necessary to identify clones (43). The term clone has also become useful in molecular epidemiology, particularly in the study of possible relationships between isolates from different geographical areas. It has become well recognized that not all isolates of pathogenic species are necessarily equal and that in a typical pathogenic species, a small number of clones, clusters, or lineages are greatly overrepresented among those isolates recovered from particular types of infection (46).
Short of whole-genome sequencing, MLST has often been used to type several members of the Enterobacteriaceae, especially E. coli and K. pneumoniae. This genotyping method assigns all isolates that are related to each other to a certain sequence type or strain type using a numerical system (e.g., ST1 and ST2, etc.). Two standardized MLST schemes are widely used to type E. coli (e.g., Achtman and Pasteur schemes), while the Pasteur MLST scheme is often utilized for K. pneumoniae (for more details on MLST, including the Achtman and Pasteur MLST schemes, see the section on laboratory methods, below).
As described previously, an eminent or successful bacterial clone is a powerful source for the propagation of genetic antimicrobial-resistant components (i.e., genes, integrons, transposons, and plasmids) (6). Drug-resistant determinants are provided to the offspring in a vertical fashion, and such eminent or high-risk clones increase the prevalence of antibiotic resistance by their enhanced ability to survive and reproduce efficiently. Moreover, due to the ability of high-risk clones to survive for long periods of time, they also play import roles in the horizontal transfer of drug resistance determinants to other bacteria, acting as efficient donors and recipients.
International multidrug-resistant high-risk clones have a global distribution and can remain viable for prolonged time periods in diverse areas (47). High-risk clones have acquired certain adaptive traits that increase their pathogenicity and survival skills, which is accompanied by the acquisition of antibiotic resistance determinants. These clones have the tenacity and flexibility to accumulate and then provide resistance and virulence genes to other isolates. High-risk clones have contributed to the spread of global multidrug resistance through the transmission of different types of genetic platforms, including plasmids, and resistance genes among Gram-negative bacteria (6). Examples of some multidrug-resistant high-risk clones among Enterobacteriaceae are E. coli ST38, ST69, ST131, ST155, ST393, ST405, and ST648 and K. pneumoniae ST14, ST37, ST147, and ST258 (47).
High-risk clones most likely possess some types of biological factors that lead to increased “fitness,” providing these strains with a Darwinian edge over other isolates of the same species (46). Such advantages will provide them with the abilities to outcontest other bacteria and become the principal part of the bacterial populace in that area. This will provide these clones with increased opportunities to spread as well as time to acquire antimicrobial drug resistance determinants from other bacteria. The spread of multidrug-resistant high-risk clones is especially facilitated by the selective pressures of antimicrobial drugs present in health care settings and used during food animal husbandry.
To qualify as an international multidrug-resistant high-risk clone, clones must possess the following characteristics (47): (i) a global distribution, (ii) an association with various antimicrobial resistance determinants, (iii) the ability to colonize and persist in hosts for long time intervals (>6 months), (iv) the ability for effective transmission among hosts, (v) enhanced pathogenicity and fitness, and (vi) the ability to cause severe and/or recurrent infections.
ESCHERICHIA COLI SEQUENCE TYPE 131
Initial Studies Pertaining to E. coli ST131
Extraintestinal pathogenic E. coli (ExPEC) is an important cause of nosocomial and community infections in humans (especially UTIs, BSIs, and IAIs) (48). Resistance to certain antimicrobial classes (especially the fluoroquinolones and cephalosporins) among ExPEC isolates was rare before 2000 but has increased exponentially since the mid- to late 2000s (49).
During the mid-2000s, molecular typing of E. coli with blaCTX-M-15 using PFGE in the United Kingdom and Canada identified different pulsotypes among these isolates. These pulsotypes were related but showed <80% similarity and did not fulfill the Tenover criteria for relatedness (44). The pulsotypes from the United Kingdom were named clones A to E, while the Canadian pulsotypes were named clones 15A and 15AR (related to A) (50, 51). In 2008, investigators from Spain and France collected E. coli with blaCTX-M-15 from different countries, including Spain, France, Canada (including isolates from PFGE clusters 15A and 15AR), Portugal, Switzerland, Lebanon, India, Kuwait, and South Korea (52, 53). Both groups of investigators performed MLST on this collection, and ST131 was present in all the countries that provided isolates for the studies (52, 53). Clusters A to E from the United Kingdom were later identified as also belonging to ST131 (54). All isolates of multidrug-resistant E. coli ST131 typed as belonging to serotype O25b:H4 belonged to phylogenetic group B2 and harbored IncF types of plasmids containing blaCTX-M-15 (52, 53). The results of these two initial studies suggested that ST131 had appeared simultaneously in the community setting and seemingly unrelatedly in separate areas of the globe without any obvious link between the patients. The results suggested that the appearance of ST131 was due to a contaminated common source (e.g., water or food) and/or was being introduced into different regions via travelers returning to their respective home countries.
Subsequently, ST131 with CTX-M-15 was described in the United Kingdom (54), the rest of Canada (55), Italy (56), Turkey (57), Croatia (58), Japan (59), the United States (60), South Africa (61), Brazil (62), and Norway (63). Healthy Parisians in France were also found to be rectally colonized with E. coli ST131 without CTX-M β-lactamases (64). Interestingly, ST131 showed a high prevalence among fluoroquinolone-resistant ESBL-negative E. coli urinary isolates obtained from Canada (65). It quickly became apparent that E. coli ST131 strains with blaCTX-M-15 were present among global isolates recovered from the community setting (66); hospitals (67); long-term-care settings, including nursing homes (68); and, interestingly, companion animals (69).
Molecular epidemiologists were intrigued with this unexpected global appearance of E. coli ST131 mostly in the community setting, apparently in the same time frame but without any obvious connections between these patients. The first studies to provide insight into this fascinating issue occurred in Calgary, Canada, and Auckland, New Zealand. The Auckland study reported a series of patients that presented to a local health care facility with community-onset UTIs due to CTX-M-15-producing E. coli. This was the first encounter of those authors with E. coli with blaCTX-M-15, and of special interest was that all the patients had recently visited India as tourists or emigrated from India (70). The population-based surveillance study from Calgary showed that recent travel (i.e., within the previous 3 months) to the Indian subcontinent, the African continent, and the Middle East was linked with notable high risks for developing community-onset UTIs (including upper UTIs) with CTX-M-producing E. coli (71). A follow-up study by the same investigators in Calgary showed that these UTIs in returning travelers were due mostly to ST131 strains with blaCTX-M-15 (72).
A retrospective molecular epidemiology investigation from Calgary, Canada, during 2000 to 2007 highlighted that E. coli ST131 with blaCTX-M-15 had appeared during the mid-2000s as an important etiology of community-acquired health care-associated BSIs, especially during the closing period of that study (66). Community-onset cases were categorized into community-acquired or health care-associated infections (73). Health care-associated infections included patients attending community clinics, patients from long-term-care facilities, or patients with infections that occurred within the first 48 h of admittance to an acute-care facility. E. coli ST131 was more likely to be multidrug resistant, to possess the aac(6′)-Ib-cr gene, and to trigger community-onset BSIs, especially those secondary to upper UTIs.
It seems that the unexpected and sudden global appearance of E. coli with blaCTX-M-15 in the community was related to the emergence of ST131, which was then followed by the subsequent expansion of this multidrug-resistant clone. Global surveillance studies have shown that >50% of E. coli isolates from community specimens obtained on the Indian subcontinent contain ESBLs (74). Therefore, it is possible that the rectal colonization of travelers returning from certain regions of endemicity could possibly have been critical for the early global spread of ST131 (12). However, it seems that international travel is not essential for the current E. coli ST131 global pandemic: a follow-up study during 2011 from the Calgary region of Canada showed that international travelers to regions of endemicity such as India had rectal colonization rates for E. coli ST131 similar to those of nontravelers (75).
The sudden worldwide appearance of E. coli with CTX-M-15 was most likely due to the acquisition of certain IncF epidemic plasmids harboring blaCTX-M-15 by a high-risk clone such as ST131. The combination of drug-resistant epidemic plasmids harboring multiple antibiotic resistance determinants with the increased fitness of the high-risk clone due to several virulence factors enabled ST131 to move effortlessly between the community, different hospitals, and long-term-care facilities. The horizontal transfer of plasmids with blaCTX-M-15 between E. coli ST131 and non-ST131 isolates was also described in certain areas such as France, Spain, and Portugal (76).
Investigators from the United States and Canada characterized ∼200 ExPEC isolates with various levels of co-trimoxazole and fluoroquinolone resistance. These urine isolates originated from Manitoba (Canada) and were acquired during the early 2000s (65). The overall prevalence rate of ST131 was 23%, and of special interest was that 99% of isolates of this clone were fluoroquinolone resistant, but only 2% were resistant to the cephalosporins (65). Results from this study provided some evidence that E. coli ST131 was initially a fluoroquinolone-resistant clone and later acquired plasmids harboring blaCTX-M genes.
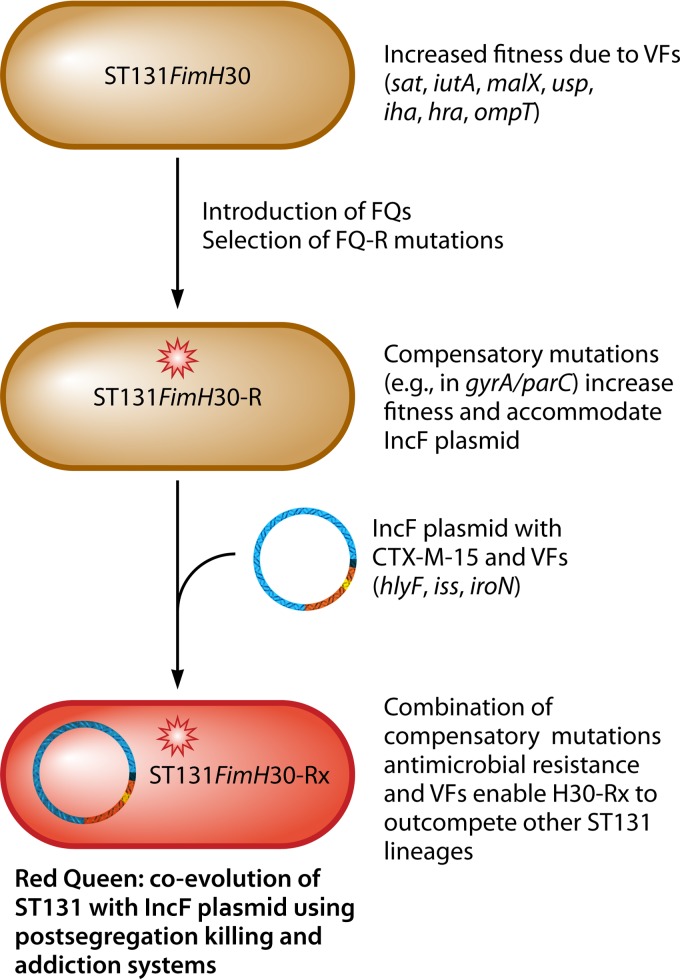
One of the first in vitro studies that investigated E. coli ST131 for the presence of different virulence factors related to ExPEC was performed by Johnson and colleagues (77). Those researchers investigated 127 ExPEC isolates obtained in the United States during 2007 from the SENTRY and MYSTIC surveillance programs. The overall prevalence rate of ST131 was 17%, and this clone was specifically associated with multidrug resistance and had a significantly higher overall virulence score than did non-ST131 E. coli isolates. It is interesting to note that the presence of some virulence factors, such as the uropathogenic specific protein (usp), outer membrane protein (ompT), secreted autotransporter toxin (sat), aerobactin receptor (iutA), and pathogenicity island marker (malX), corresponded specifically to ST131. The results of that study demonstrated that ST131 showed a typical virulence profile (compared to those of other ExPEC isolates), and it seemed that multidrug resistance in association with some virulence factors may be essential for the ecological triumph of this clone. Subsequent studies from different investigators have shown similar virulence profiles among ST131 isolates (19). Of special interest is the low prevalence of classical ExPEC-associated virulence factors (i.e., pap [P fimbriae], cnf1 [cytotoxic necrotizing factor], and hlyD [alpha-hemolysin]) among isolates that belong to ST131. The precise roles of these virulence factor genes remain to be elucidated; however, it is possible that certain recognized virulence factors possibly play a part in the fitness and adaption of ST131 (78). Virulence factor genes such as sat, iutA, malX, usp, iha, hra, and ompT might increase the ability of E. coli ST131 to efficiently colonize human tissues rather than being responsible for virulence and causing infection (79).
As described above, PFGE analysis showed that E. coli ST131 is not a single entity and can be separated into various pulsotypes. Molecular surveillance has demonstrated that ST131 isolates with similar PFGE clusters are present in different regions or countries (53, 76, 80). A study that investigated a global collection of ST131 isolates (isolates were obtained from 9 countries in North America, South America, Europe, the Middle East, Asia, and Oceania) identified 15 distinct pulsotypes; nearly half of the ST131 isolates belonged to four distinctive clusters showing a global distribution (81). However, ST131 isolates with less related pulsotypes can be present within the same location (80, 81). Johnson and colleagues genotyped >500 global ST131 ExPEC isolates obtained from the late 1960s until 2010 from multiple materials, including materials from humans, animals, and the environment, by PFGE (82). This study proved that ST131 is highly divergent when PFGE is used to analyze this clone. Those researchers also showed that a certain number of closely related pulsotypes preside internationally, and these so-called “high-frequency” clusters appeared only during the last 10 years or so (82). A retrospective molecular surveillance study from Calgary, Canada, from 2000 until 2010 identified a prominent closely related pulsotype of ST131 causing a significant increase in the number of ESBL-producing E. coli strains isolated from blood cultures (83). This cluster became especially dominant during 2009 and 2010.
Although ST131 first came to the attention of the medical community because of its particular association with ESBL-producing E. coli, especially ExPEC with blaCTX-M-15, it became apparent during the late 2000s that most isolates of this clone were initially ESBL negative and resistant to the fluoroquinolones (19). Global surveillance studies have shown that ST131 was a fluoroquinolone-resistant clone during the early 2000s and became strongly associated with blaCTX-M-15 toward the end of the 2000s (84).
Recent Developments Pertaining to E. coli ST131
The presence of ST131 among human clinical E. coli isolates varies by geographic region and host population. Recent surveillance studies have shown that the overall prevalence ranges from ∼10% to nearly 30% of all E. coli clinical isolates (84).
Epidemiology and clinical issues.
Like other ExPEC isolates, ST131 causes a variety of extraintestinal infections, including BSIs, pneumonia, UTIs, IAIs, and wound infections. ST131 is strongly associated with community-onset infections, especially in patients with regular contact with health care settings. This was initially illustrated by infections due to ESBL-producing isolates (66, 83) and more recently by a population-based cohort study from the United States (85). Interestingly, E. coli ST131 has not been responsible for large nosocomial outbreaks in intensive care or high-care units.
Infections with ST131 are most common among the elderly, and this sequence type has a high prevalence among residents of nursing homes and long-term-care facilities (85). ST131 isolates have also been detected in nonhuman sources such as companion animals, other animals, food sources, and the environment (19). However, the prevalence of ST131 is substantially higher among humans than in animals, produce, or the environment (86). It seems that the ST131 pandemic is primarily a human-based phenomenon and that this clone has somehow successfully adapted to human hosts. However, global studies regarding the prevalence of ST131 among nonhuman hosts are lacking.
There is a clear association between previous antibiotic consumption and colonization followed by infection due to ESBL-producing E. coli, including ST131 isolates. Antimicrobial agents such as the fluoroquinolones and cephalosporins have most often been implicated in selection for colonization and subsequent infections due to E. coli ST131 (85). The high prevalence of ST131 in community environments is associated with the extensive use of antimicrobial agents (i.e., the outpatient setting of health care settings, nursing homes, and long-term-care centers). This indirectly supports the notion that antimicrobial usage played a role in the selection of ST131. However, the underlying basis for why E. coli ST131 has not been implicated in nosocomial outbreaks remains an enigma.
Population structure.
The most prevalent lineage within ST131 is named fimH30 because it contains the H30 variant of the type 1 fimbrial adhesin gene fimH (87). The fimH30 lineage was first identified among ST131 isolates obtained from different geographical regions and sources by means of subtyping of >1,000 historical and recent ExPEC isolates (both ST131 and non-ST131) using a combination of typing strategies, including sequencing of the fimH, gyrA, and parC genes; MLST; and PFGE (87). Johnson and colleagues observed that the fimH30 ST131 lineage consisted of nearly 70% of recent fluoroquinolone-resistant E. coli isolates, while it remained infrequent (i.e., <1%) among fluoroquinolone-susceptible ST131 isolates. The ST131 fimH30 lineage first appeared in the early 2000s, became prominent in the mid-2000s, and then expanded rapidly during the late 2000s. The majority of strains of the fimH30 lineage have very close genetic similarity, suggesting that they originated from a single fimH30-carrying ancestor (88). It seems that the dramatic global emergence of fluoroquinolone-resistant ExPEC ST131has been driven by the clonal expansion and dissemination of the fimH30 lineage. Additional support for a single ancestor was provided by the linkage of the ST131 fimH30 lineage with a specific gyrA and parC allele combination, despite proof for the widespread generation of different gyrA and parC allele combinations among non-H30 lineages (87). A PCR-based assay that detects fimH30-specific single-nucleotide polymorphisms (SNPs) is available to rapidly and cost-effectively detect the H30 lineage (89).
Price and colleagues, using whole-genome sequencing combined with phylogenetic SNP analysis, recently identified two important sublineages within the ST131 fimH30 lineage, called H30-R and H30-Rx, because of their extensive antimicrobial resistance profiles (88). Those authors initially analyzed just over 500 ST131 isolates collected between 1967 and 2011 from different geographic regions by using PFGE and found that the fimH30 lineage showed significantly different pulsotypes irrespective of whether the isolates were susceptible or resistant to a fluoroquinolone or contained blaCTX-M. These findings suggested that fluoroquinolone-resistant mutations and CTX-M genes were horizontally acquired over a long period of time. However, when 105 of the isolates with significantly different pulsotypes underwent next-generation sequencing (NGS) combined with SNP analysis to reconstruct the phylogeny of ST131, the fluoroquinolone-resistant fimH30 lineage and strains carrying fimH30 with blaCTX-M-15 formed a tight cluster irrespective of whether their pulsotypes were different. Moreover, when those authors performed additional sequencing with increased coverage, they found that within the fluoroquinolone-resistant fimH30 lineage, isolates with blaCTX-M-15 formed a distinct cluster, named the H30-Rx sublineage, that was separated from ESBL-negative fluoroquinolone-resistant fimH30 isolates by 3 core genome SNPs (88).
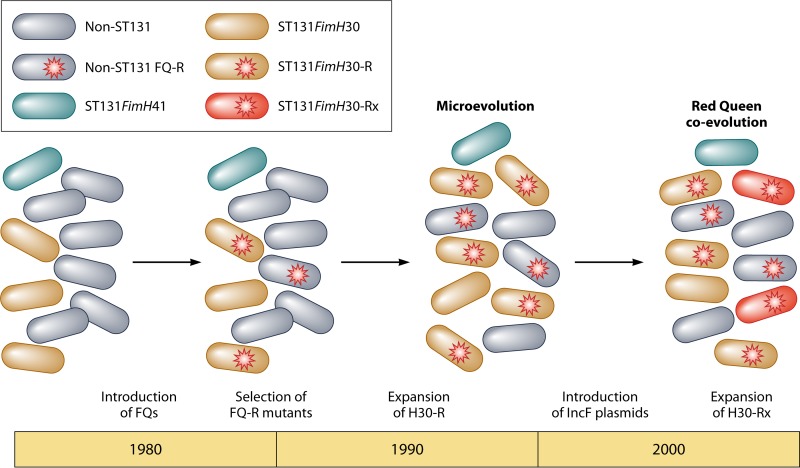
The phylogenetic analysis undertaken by Price and colleagues indicated that the ST131 fimH30 lineage comprised a series of nested sublineages that were most likely derived from a single common fluoroquinolone-susceptible fimH30 ancestor (84). These investigators named these sublineages as follows: the H30-R sublineage or subclone, which shows fluoroquinolone resistance without blaCTX-M-15, and the H30-Rx sublineage or subclone, which shows fluoroquinolone resistance with blaCTX-M-15 (88). This clonal structure results in a succession of antimicrobial resistance among the fimH30-associated lineages, from the most susceptible H30 lineage (fluoroquinolone susceptible and CTX-M negative), to the more resistant H30-R sublineage (fluoroquinolone resistant and CTX-M negative), to the most extensively resistant H30-Rx sublineage (fluoroquinolone resistant and CTX-M positive). Whole-genome sequencing combined with SNP analysis indicated that the clonal expansion of the ST131 fimH30 lineage is the most dominant and important vehicle for the increasing global prevalence of fluoroquinolone resistance and blaCTX-M-15 among ExPEC strains. These results were later independently confirmed by Petty and colleagues (90). The population structure of the E. coli ST131 fimH30 lineage, H30 sublineages, and other ST131-associated lineages is illustrated in Fig. 1.
FIG 1.
Population structure of the Escherichia coli ST131 fimH30 lineage, H30 sublineages, and other ST131-associated lineages. FQ-R, fluoroquinolone resistant; FQ-S, fluoroquinolone sensitive.
The prevalences and distributions of the fimH30 lineage and the H30-R and H30-Rx ST131 sublineages were initially described for populations within the United States. A study of E. coli clinical isolates from various U.S. veterans centers obtained during 2011 showed that the prevalence of ST131 was only 7% among fluoroquinolone-susceptible ExPEC isolates, 78% among fluoroquinolone-resistant ExPEC isolates, and 64% among ESBL-producing E. coli isolates; the ST131 fimH30 lineage accounted for 12.5% of fluoroquinolone-susceptible ST131 isolates, which increased to 95% of fluoroquinolone-resistant and 98% of ESBL-producing ST131 isolates (89). Similarly, in a case-control study conducted in the region of Chicago, IL, >50% of ESBL-producing ExPEC isolates were identified as ST131; 98% were further characterized as fimH30, while 92% were characterized as H30-Rx (91). These studies supported the notion that there is a strong alliance linking the H30-Rx sublineage with blaCTX-M-15. However, not all ST131 isolates with CTX-M-15 are members of the H30-Rx sublineage.
In a population-based study of consecutively collected ExPEC isolates in Minnesota, the prevalence of the ST131 fimH30 lineage was high among elderly patients (i.e., >60 years of age), while infections due to non-H30 ST131 isolates were more common among younger patients (i.e., <2 years of age) (92). In a different multicenter U.S. study that analyzed >1,600 ExPEC isolates, clonotype CH40-30 (which corresponded to the fimH30 lineage of ST131) was the most prevalent clonotype among all ExPEC isolates and was statistically associated with recurrent or persistent UTIs and the presence of sepsis (93). The next-generation sequencing study by Price and colleagues described above also observed that sepsis was significantly associated with infections due to the H30-Rx sublineage (88).
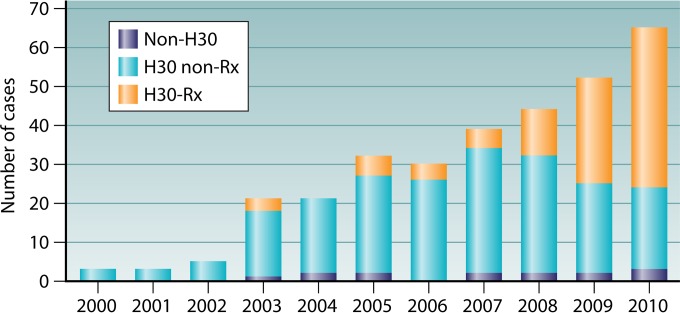
The first study outside the United States to determine the prevalence and distribution of the ST131 fimH30 lineage and its respective sublineages was a retrospective population-based surveillance study investigating BSIs due to fluoroquinolone-resistant ExPEC in Calgary, Canada (2000 to 2010) (94). This study demonstrated that the deluge of the H30-Rx sublineage toward the latter part of the study period was responsible for the significant increase in the prevalence of ST131 and fluoroquinolone-resistant ExPEC (94) (Fig. 2). This study recognized the relationship of H30-Rx with primary sepsis and BSIs due to prostate biopsies. This sublineage was also more likely to contain aac(6′)-lb-cr.
FIG 2.
Numbers of isolates of Escherichia coli ST131 and the H30-R and H30-Rx sublineages isolated from blood in the Calgary Region from 2000 to 2010. (Adapted from reference 94.)
O16:H5 H41 and other ST131 lineages.
As described above, several investigators noticed during the late 2000s that E. coli ST131 strains with ESBLs exhibited different pulsotypes (52, 55, 60, 61, 66). Interestingly, some ST131 isolates belonged to a well-defined cluster that was separated from the more prominent O25b:H4 clade and tested negative by Clermont ST131 PCR (170) (for more details on Clermont PCR, see the section on laboratory methods, below). This cluster contained blaCTX-M-14 and was susceptible to the fluoroquinolones, and with additional serotype and fimH allele characterization, it tested positive for O16:H5 and belonged to the fimH41 lineage (81). Of interest, the O16 and O25b serotypes within ST131 were identified as representing different STs by using the Pasteur Institute MLST scheme, but isolates with these different serotypes were recognized as the same ST by the Achtman MLST scheme (84).
The O16:H5 ST131 fimH41 lineage had distinct combinations of gyrA and parC alleles, and surveillance data showed that this lineage comprised 1 to 5% of E. coli ST131 isolates (95). The fimH41 lineage is associated with resistance to trimethoprim-sulfamethoxazole and gentamicin (compared to the fimH30 ST131 lineage), while ESBLs and fluoroquinolone resistance are rarely detected with this lineage. O16:H5 ST131 fimH41 isolates with blaOXA-48 in the United Arab Emirates and Morocco have recently been described (96). A rapid PCR screening test was developed to detect the fimH41 lineage and combined the O16 rfb gene with the ST131-specific mdh and gyrB alleles (95) (for more details on this PCR, see the section on laboratory methods, below).
Two additional ST131 lineages with the O25b:H4 serotype but showing significant different pulsotypes from the main fimH30 cluster were recently reported (81). These isolates belonged to the fimH22 and fimH35 lineages and were also associated with blaCTX-M-15, fluoroquinolone resistance, and the presence of aac(6′)-lb-cr. O25b:H4 ST131 fimH22 isolates with blaKPC from Argentina and Colombia were recently described (96). The population structure of the E. coli ST131 fimH30, fimH41, fimH22, and fimH35 lineages as well as the H30-R and H30-Rx sublineages is illustrated in Fig. 1.
Virulence.
A Spanish study proposed a new classification system for ST131, where it is classified into four virulence-associated groups (i.e., groups A, B, C, and D) (also named virotypes) (97). Those investigators performed in vitro virulence characterization on a large global collection of ST131 isolates and identified four virulence factors that showed a distinctive distribution among the different virotypes. These virulence factors include afa (GenBank accession number FM955459) (Afa/Dr adhesion), iroN (catecholate siderophore receptor), ibeA (invasion of brain endothelium), and sat (secreted autotransporter toxin) (97). Virotypes A and B were related to antibiotic nonsusceptibility, blaCTX-M-15, and aac(6′)-lb-cr. Virotype C was present in several countries and was linked to invasive infections compared to other virotypes such as virotypes A and B (97).
A U.S. study described the ExPEC-associated virulence profiles of ST131 and its sublineages (H30-R and H30-Rx) among a large E. coli collection from the Midwest near the Great Lakes region (91). Those researchers identified distinctive virulence profiles among these sublineages, with the H30-Rx sublineage having the highest aggregative virulence score (91). Colpan and colleagues showed that the fluoroquinolone-susceptible ST131 fimH30 lineage had a significantly different virulence profile from that of non-ST131 fluoroquinolone-susceptible isolates. Moreover, as this lineage gained resistance to the fluoroquinolones and acquired ESBLs, isolates tended to progressively accumulate additional virulence factors, especially among the ST131 fimH30 lineage that acquired CTX-M-15 (89).
E. coli ST131 and carbapenemases.
The development of resistance to the carbapenems in community pathogens such as E. coli is worrisome to the medical fraternity (49). NDM and OXA-48 are the most prevalent carbapenemases present in nosocomial and community isolates of E. coli, while the VIM, IPM, and KPC β-lactamases are not yet commonly encountered in this species (11). Infections with carbapenemase-producing E. coli often occur in patients who had recently visited certain regions of endemicity, such as the Indian subcontinent for NDMs and North Africa or Turkey for OXA-48 (12).
Due to the revolutionary worldwide triumph of ST131, carbapenemases among isolates of this clone have been carefully monitored by molecular epidemiologists. The carbapenemase gene blaNDM was first identified in ST131 isolates during 2010 from patients in Chicago and Paris (98, 99). Both patients had previously visited the Indian subcontinent. This was followed by case reports of ST131 with blaVIM from Italy (100); ST131 with blaKPC from Ireland (101), France (102), the United States (103), Italy (104), Taiwan (105), and China (106); ST131 with blaOXA-48 from the United Kingdom (107), Ireland (108), Algeria (109), and Spain (110); and ST131 with blaIMP from Taiwan (111). The largest cluster of ST131 with carbapenemases was recently described in Pittsburgh, PA (112). The authors of that study characterized 20 isolates of KPC-producing E. coli: 60% belonged to the ST131 fimH30 lineage, while the blaKPC plasmid belonged to the IncF type with FIIk replicons.
A recent study by the SMART and AstraZeneca global surveillance programs showed that 35% of 116 carbapenemase-producing E. coli isolates belonged to ST131, which was associated with fluoroquinolone resistance, the presence of blaKPC, the fimH30 lineage, and virotype C (96). Genes for ESBLs such as blaCTX-M-15 and blaSHV-12 were also present in some of the ST131 isolates with carbapenemases. ST131 was also isolated from Argentina, China, Colombia, Ecuador, India, Italy, Jordan, Morocco, Panama, Philippines, Puerto Rico, Thailand, Turkey, the United Arab Emirates, the United States, and Vietnam. E. coli ST131 with carbapenemases poses a significant new public health risk due to its worldwide distribution and relationship with the dominant fimH30 lineage.
Does ST131 Qualify as an International Multidrug-Resistant High-Risk Clone?
The pandemic emergence of E. coli ST131, and specifically its H30-R and H30-Rx sublineages, occurred over <10 years. It is a remarkable antimicrobial resistance success story rivaling the global pandemics caused by clones within methicillin-resistant Staphylococcus aureus (i.e., ST5, ST8, and ST36) and Streptococcus pneumoniae (i.e., ST236 and ST320).
Global distribution and prevalence.
E. coli ST131 was initially described among isolates with blaCTX-M-15 from the following countries: Canada, France, Switzerland, Portugal, Spain, Kuwait, Lebanon, India, and South Korea (52, 53). Subsequently, ST131 was identified among ESBL, non-ESBL, fluoroquinolone-resistant, and fluoroquinolone-susceptible E. coli isolates from all corners of the world (19). If investigators decide to determine the presence of ST131 among E. coli isolates collected from human sources, they will most likely detect ST131 among their collection. ST131 is still overrepresented among antimicrobial-resistant ExPEC isolates; recent global surveillance showed that ST131 consistently accounts for ∼60% to 80% of fluoroquinolone-resistant isolates and 50 to 60% of ESBL-producing isolates but only 0% to 7% of fluoroquinolone-susceptible isolates (84). E. coli ST131 has a true global distribution and is present among ExPEC isolates on all continents, except possibly Antarctica.
Association with antimicrobial resistance mechanisms.
E. coli ST131 was initially described in ExPEC isolates with CTX-M-15 and today is known to also be associated with fluoroquinolone resistance (83, 89, 91). Population genetics indicated that fluoroquinolone resistance in the ST131 fimH30 lineage is due mostly to gyrA1AB and parC1aAB mutations in gyrase and topoisomerase IV, respectively (87). The gyrA1AB and parC1aAB mutations are present in 71% of fluoroquinolone-resistant and in 62% of ESBL-positive E. coli isolates from various Veterans Administration hospitals in the United States (89). ST131 is also associated with the presence of the plasmid-mediated quinolone resistance determinant aac(6′)-lb-cr (17). This resistance determinant causes decreased susceptibility to the fluoroquinolones ciprofloxacin and norfloxacin as well as resistance to the aminoglycosides tobramycin and amikacin.
Various expanded-spectrum β-lactamases (e.g., CTX-Ms, CMYs, SHV ESBLs, and TEM ESBLs) have been detected in ST131 isolates (19), with CTX-M-15 in combination with TEM-1 and OXA-30 being by far the most common. The presence of carbapenemases is still rare among ST131 isolates, although some enzymes have been reported in different parts of the world (19).
Other resistance determinants that have been characterized in ST131 isolates include mph(A) (responsible for resistance to the macrolides), catB4 (responsible for resistance to chloramphenicol), tetA (responsible for tetracycline resistance), dfrA7 (responsible for trimethoprim resistance), aadA5 (responsible for streptomycin resistance), and sul1 (responsible for sulfonamide resistance). Therefore, isolates belonging to ST131 are most often nonsusceptible to the cephalosporins, monobactams, fluoroquinolones, trimethoprim-sulfamethoxazole, and the aminoglycosides (19).
Ability to colonize human hosts.
Because intestinal colonization with ExPEC is believed to be a prerequisite for extraintestinal infection, it is possible that the enhanced ability to colonize the intestinal tract is partly responsible for the widespread dissemination of ST131. Several studies have shown that asymptomatic individuals can be rectally colonized with E. coli ST131 (19). In a mouse model of intestinal colonization, an ST131 isolate surpassed other commensal E. coli isolates mixed in a 1:1 ratio and administered enterally into streptomycin-pretreated mice. Although that study was limited by its evaluation of only a single ST131 isolate, it provided evidence that some ST131 isolates have the ability to efficiently colonize the intestine, bladder, and kidney of mice (113). It is unclear whether the prevalence or duration of intestinal colonization in humans is different for ST131 compared to other E. coli isolates. It is also unclear if the dominant fimH30 ST131 lineage, including the sublineages (i.e., H30-R and H30-Rx), has intestinal colonization abilities different from those of other ST131 lineages (e.g., fimH41, fimH35, and fimH22).
The prevalence of E. coli ST131 among rectal isolates of E. coli varied considerably with the population studied, geographical regions, host characteristics, the presence of resistance mechanisms, as well as the time frame when the study was conducted: rectal colonization rates of ST131 that varied from 0 to 44% have been reported (19).
Effective transmission among hosts.
The transmission of ST131 between different household family members (father to daughter, daughter to mother, and sister to sister) and companion animals (dogs and cats in particular) has previously been documented (19, 84). A study from Switzerland showed that ST131 isolates were more likely to be transmitted between members of the same family than within patients in the hospital environment (20). A French day care center recently reported that 7 children were rectally colonized with ST131 that produced CTX-M-15, suggesting that this clone was effectively transmitted within the day care setting (114).
It seems that E. coli ST131 can effectively be transmitted between members of the same family, and this might have played an important role in the emergence and dissemination of this sequence type within the community setting. However, whether ST131 is more efficiently transmitted than other E. coli isolates is uncertain and deserves additional studies.
Enhanced pathogenicity and fitness.
In several molecular epidemiological studies, ST131 isolates, compared to non-ST131 ExPEC isolates, consistently had a large number of ExPEC-associated virulence factor genes and had significantly high aggregate virulence scores (77, 115). Moreover, within ST131, the fimH30 lineage and specifically the H30-Rx sublineage had characteristic virulence profiles with higher virulence scores than those of non-H30 ST131 E. coli isolates, most likely playing an important role in the overall virulence and fitness of this sublineage (91).
In vitro studies that investigated the maximal growth rate and the ability of ST131 to produce biofilms showed that this clone had a high metabolic potential, most likely by increasing the fitness and ability of ST131 to establish intestinal colonization for long periods of time (116). In vivo studies that investigated the virulence potential of ST131 in animal models indicated that this sequence type kills mice (117) but is less virulent than non-ST131 E. coli in the Caenorhabditis elegans and zebra fish embryo models (118).
Cause of severe and/or recurrent infections.
It is unclear if ST131 isolates cause more severe infections than other ExPEC E. coli isolates, but clinical epidemiological data suggest that ST131 isolates are more likely to cause upper UTIs than lower UTIs (84). In a series of studies from Australia, ST131 accounted for 30% of pyelonephritis isolates among ExPEC isolates from women, versus only 13% of cystitis isolates and 4% of fecal isolates. Similar prevalence trends were seen among men and children (116). A study from the United Kingdom showed similar results, where the prevalence of ST131 was 21% among bacteremia isolates, compared to only 7% among urinary isolates (119).
Whether ST131 is associated with worse clinical outcomes than other ExPEC isolates is unclear. Some studies suggest that ST131 is more likely to cause persistent or recurrent UTIs (85), while other investigators have found no significant difference in outcomes of infections with ST131 versus other ExPEC isolates (120). When investigators adjusted for host factors, no differences in cure or mortality were found between patients with infections due to ST131 and those with infections due to other ExPEC isolates (93). Of special interest, isolates of the H30-Rx sublineage of ST131 have demonstrated statistical and epidemiological associations with sepsis (88).
In summary, E. coli ST131 isolates clearly have all of the essential characteristics that define a high-risk clone (Table 2). In fact, this sequence type might be the quintessential example of an international multidrug-resistant high-risk clone.
TABLE 2.
Characteristics of Escherichia coli ST131 and Klebsiella pneumoniae ST258 that define them as high-risk clones
| Characteristic | Description |
|
|---|---|---|
| Escherichia coli ST131 | Klebsiella pneumoniae ST258 | |
| Global distribution | Endemic to all continents except Antarctica | ST258 is endemic to the USA, Israel, Greece, Italy, Poland, and Colombia; ST11 has been reported in China and Brazil; ST512 has been reported in Israel, Italy, and Colombia; ST340 has been reported in Brazil and Greece |
| Association with various antimicrobial resistance determinants | Various but associated with fluoroquinolone resistance and CTX-M-15 (CTX-M-14 to a lesser extent) | Various but associated with KPC-2 and KPC-3 |
| Ability to colonize and persist in hosts for long periods of time | Rectal colonization for up to 6 mo | Rectal colonization for up to 12 mo |
| Effective transmission among hosts | Transmission among family members | Successful nosocomial transmission for months after introduction |
| Enhanced pathogenicity and fitness | Higher aggregate ExPEC-associated virulence scores; high metabolic potential and biofilm production | Unclear |
| Causes severe and/or recurrent infections | More likely to cause upper UTIs and recurrent UTIs, and the H30-Rx sublineage is associated with sepsis | Mortality rates are higher than with non-ST258 K. pneumoniae (most likely due to the patient's underlying conditions) |
KLEBSIELLA PNEUMONIAE SEQUENCE TYPE 258
Initial Studies Pertaining to K. pneumoniae ST258
The rate at which carbapenem resistance in K. pneumoniae has disseminated globally and the consequences thereof have raised cause for alarm among the medical community at large. To date, blaKPC has been found in >100 different STs, but this pandemic is driven primarily by the spread of KPC-producing K. pneumoniae isolates that are members of clonal complex 258 (CC258) (121). CC258 (the founder member is ST292) consists of one predominant ST, namely, ST258, and, to a lesser extent, ST11, ST340, and ST512, which are single-locus variants (SLVs) of ST258 (30, 121). K. pneumoniae ST258 is a prototype of a high-risk clone, and recent information about the epidemiology, genetic rearrangement, and evolution of this successful clone has provided insights into the global spread of antimicrobial drug resistance.
K. pneumoniae with blaKPC was first identified in a non-ST258 isolate during 1996 in the Southern United States (122). During the late 1990s to early 2000s, there were sporadic reports of K. pneumoniae with blaKPC from the Northeastern United States; however, large outbreaks due to related isolates were not described (123). In 2009, the U.S. Centers for Disease and Prevention in collaboration with investigators from Israel performed MLST on K. pneumoniae with blaKPC, and they identified ST258 among isolates from the New York area collected during 2005 (124). As time progressed, ST258 was detected in geographically diverse regions of the United States, and in 2009, it became apparent that ST258 was the predominant clone in this country, being responsible for 70% of K. pneumoniae isolates with blaKPC obtained from different parts of the country (125). During the mid-2000s, Israel experienced several nosocomial outbreaks of infections due to K. pneumoniae with blaKPC that were caused by a clone (identified by PFGE) named clone Q (124). Interestingly, clone Q has a pulsotype similar to that of ST258, present in the United States. This was followed by global reports of ST258 among K. pneumoniae isolates with blaKPC from countries such as Greece (126), Norway, Sweden (127), Italy (128), Poland (129), Canada (130), Brazil (131), and South Korea (132), suggesting that this ST had characteristics of international multidrug-resistant high-risk clones. Recent reports from Israel (133) and Italy (134) demonstrated the endemicity and persistence of CC258 over time while remaining the predominant clone among K. pneumoniae isolates with blaKPC. Interestingly, Israel has seen an overall dramatic decrease in the incidence of KPCs among K. pneumoniae isolates, but ST258 still remains the most predominant clone (133).
The other SLVs from CC258 have the following global distribution: ST11 is the major ST among K. pneumoniae isolates with blaKPC from Asia (especially China) (135) and has also been described in Latin America (especially Brazil) (121). Other STs that belong to CC258 have been reported in Colombia (i.e., ST512), Italy (i.e., ST512), Israel (i.e., ST512), Brazil (i.e., ST340), and Greece (i.e., ST340) (121).
Recent Developments Pertaining to K. pneumoniae ST258
Population structure.
The diversity in K. pneumoniae genomes is due primarily to mobile genes that move frequently by horizontal transfer between bacteria, including plasmids, phages, integrative conjugative elements (ICEs), and insertion elements (IEs) (30). DeLeo and colleagues recently performed whole-genome sequencing on two K. pneumoniae ST258 urinary isolates from New Jersey and then did supplementary sequencing on a different global collection of just over 80 CC258 clinical isolates (136). A phylogenetic single-nucleotide polymorphism (SNP) analysis of the core genomes of these isolates showed that K. pneumoniae ST258 isolates belonged to two well-defined lineages, named clade I and clade II. Clade I was associated with KPC-2, and clade II was associated with KPC-3. The genetic divergence between these two clades occurred in a 215-kb area that included the genetic material used for capsule polysaccharide biosynthesis (cps), an important virulence factor for K. pneumoniae.
That same group then compared the genetic structures of the cps regions and dispersal of SNPs in the core genomes of ST258 clades I and II with other K. pneumoniae sequence types (i.e., ST11, ST442, and ST42) (137). Kreiswirth and colleagues found a 1.1-Mbp area in ST258 clade II that is identical to that of ST442, while the remaining part of the ST258 genome was homologous to that of ST11. This indicated that ST258 clade II is a hybrid or crossbreed clone that was created by a large recombination event between ST11 and ST442. Those investigators then identified the same cps regions in ST42 and ST258 clade I. The similarity of the areas surrounding the cps regions from ST42, ST258 clade I, and ST258 clade II indicated that ST258 clade I evolved from ST258 clade II due to the replacement of the cps region from ST42. Recent developments regarding the population structure of K. pneumoniae ST258 are illustrated in Fig. 3.
FIG 3.
Population structure of Klebsiella pneumoniae ST258. ICE, integrative conjugative element; cps, capsule polysaccharide biosynthesis gene region.
The integrative conjugative element ICEKp258.2 contains gene clusters for a type IV pilus (i.e., pilV) and a type III restriction-modification system. A type IV pilus increases the exchange of plasmid DNA between bacteria and facilitates the attachment of bacteria to objects. The type IV pilus may therefore facilitate the movement of resistance genes (137). A type III restriction-modification system could also be responsible for “host specificity,” i.e., preventing the exchange of certain mobile elements, including plasmids, among bacteria (137). A study from Israel investigated the specific association of ICEKp258.2 with K. pneumoniae ST258 and other sequence types by searching for the presence of pilV and showed that this gene cluster was present only among ST258 isolates but was absent in non-ST258 K. pneumoniae isolates (including SLVs of ST258 such as ST11) (138). Their results shed some light on the different behaviors of ST258 and its close relative ST11: ST258 (which contains ICEKp258.2) is specifically associated with KPC and narrow-host-range IncF plasmids (30), while ST11 (which lacks ICEKp258.2) is associated with various carbapenemases (KPC, VIM, IMP, NDM, and OXA-48) on a broad range of plasmids (e.g., IncF, IncA/C, InL/M, IncN, and nontypeable plasmids) (139–141). Therefore, the restriction of plasmids and specific mobile elements by a type III restriction-modification system on ICEKp258.2 may explain some of the differences observed between these SLVs (i.e., ST258 and ST11). Moreover, pilV may also in part explain the high transmissibility and survival abilities of ST258 on living and nonliving surfaces. It is possible that ICEKp258.2 contributes significantly to the ecological success of K. pneumoniae ST258 (137).
Association with antimicrobial resistance mechanisms.
High-risk clones act as stable hosts that harbor resistance genes, transposons, integrons, and plasmids, which allow such elements to spread with such clones (6). One of the most consistent hallmarks of K. pneumoniae ST258 is its association with multidrug resistance determinants (30). The majority of antimicrobial-resistant determinants present in ST258 are plasmid mediated, and this clone often contained more than one plasmid, each with multiple individual resistant determinants (136). A recent study that used next-generation sequencing with long reads examined the contents on plasmids in a single ST258 isolate. Those investigators identified four plasmids containing 24 different resistance genes (142). Several investigators have shown that K. pneumoniae ST258 isolates have multiple antimicrobial resistance determinants responsible for aminoglycoside resistance [i.e., aac(6′)-Ib, aadA2, and aph(3′)-Ia], β-lactam resistance (i.e., blaKPCs, blaOXA-9, blaSHV-11, and blaTEM-1), fluoroquinolone resistance (i.e., oqxA and oqxB), macrolide-lincosamide-streptogramin B resistance (i.e., mphA), chloramphenicol resistance (i.e., catA1), trimethoprim resistance (i.e., dfrA12), and sulfonamide resistance (i.e., sul1) (142, 143). ST258 isolates can also decrease the number of porin channels, which leads to nonsusceptibility to additional classes of antimicrobial drugs, leaving clinicians with very limited treatment options (144).
The emergence of colistin resistance among ST258 isolates is a worrisome finding since this antimicrobial agent has remained one of the last-line salvation therapies for infections due to K. pneumoniae isolates with blaKPC (144). Colistin resistance among ST258 isolates has been documented in several diverse geographical regions, which likely indicates that colistin-resistant ST258 mutants can be selected over time after the introduction of this agent for the treatment of infections due to K. pneumoniae ST258 (144–146).
Ability to colonize human hosts.
A recent study from Germany investigated the rectal colonization of patients following an outbreak due to K. pneumoniae ST258 and showed that 25% of patients remained colonized for up to 1 year after the outbreak was contained. Some patients were persistently colonized even beyond this time (147). A study from Greece found that 73% of the patients were rectally colonized with K. pneumoniae ST258 with blaKPC on average within 9 days of exposure to this clone (148). Similar data have been reported from Italy, which showed the effective colonization of neonatal patients by K. pneumoniae ST258 (149) as well as long-term acute-care populations in the United States (150). However, in contrast to E. coli populations, ST258 isolates have remained present largely among nosocomial patients with health care associations and are not readily identified among community dwellers.
Transmission within the health care setting.
A detailed analysis of a recent intensive care unit (ICU) outbreak due to K. pneumoniae ST258 in the United States indicated that this clone can be very successfully transmitted between hosts for a long period of time (i.e., months) after the identification of the initial index patient (151). In Italy, the successful spread of ST258 occurred rapidly within the hospital setting (128). The most notable and pronounced outbreak due to ST258 was experienced in Israel after the introduction of this clone during the early to mid-2000s (124). There were sharp increases in the numbers of cases and outbreaks in Israel during 2006 to 2007 in acute-care hospitals, which were driven primarily by ST258. After the nationwide implementation of infection control guidelines during the late 2000s to limit the spread of K. pneumoniae with blaKPC, there has been a steep decline in the incidence of infections due to ST258 throughout Israel (133).
Enhanced pathogenicity and fitness.
Capsular polysaccharide is a recognized virulence factor enabling K. pneumoniae to evade phagocytosis (152). Recent papers have highlighted the likely importance of unique capsular polysaccharides within ST258 and their probable importance in promoting the global success of this clone (136). Characterization of the genome regions of different clades of ST258 (i.e., clade I and clade II) showed that that capsule polysaccharide biosynthesis regions cps-1 and cps-2 (also referred to as cps207-2 and cpsBO-4) are significantly different between clades I and II and are likely involved in the global distribution of these clades (137, 153). This cps region of diversification could be advantageous for K. pneumoniae ST258 clades I and II, which manages to change the polysaccharide structure as a mechanism to evade host defenses. Capsule switching is used by certain bacteria to escape the host immune response (154). It is possible that the DNA interchange that occurs up- and downstream of the cps regions may be an important method used by K. pneumoniae to quickly expand and change over time (154). It would be interesting to determine what selection process is responsible for capsule switching among K. pneumoniae ST258 clades I and II.
As described above, pilV is present on ICEKp258.2 and may in part be responsible for the high transmissibility and survival abilities of K. pneumoniae ST258. This element is present in both clades of ST258, but it seems to have been acquired after the large recombination event between ST11 and ST442 to create ST258 clade II. It seems that the genetic material for KPC and the ICEKp258.2 element were independently acquired at two distinct points in time (136). A type IV secretion system might have been beneficial to K. pneumoniae ST258 by increasing the fitness and survival abilities of this clone (137).
It is unclear if ST258 isolates are more virulent than other K. pneumoniae isolates. ST258 lacks well-characterized K. pneumoniae virulence factors, including the K1, K2, and K5 capsular antigen genes; the aerobactin genes; and the regulator of mucoid phenotype gene, rmpA (156). A recent study demonstrated that ST258 is nonvirulent in animal models, is highly susceptible to serum killing, and can rapidly undergo phagocytosis (156). A different group showed that not all ST258 isolates produce the same results in a mouse lethality model, but similar results were obtained in a moth (Galleria mellonella) virulence model (157). There has been evidence of virulence in a nematode model with Caenorhabditis elegans killing, and this effect was not diminished when the isolates were cured of blaKPC plasmids (158). This study suggested that the blaKPC plasmids contained additional factors that could result in persistence but not necessarily enhanced virulence. There was also some strain-to-strain variability in the nematode model.
In summary, K. pneumoniae ST258 clearly has all of the essential characteristics that define a high-risk clone (Table 2).
LABORATORY METHODS FOR DETECTION OF E. COLI ST131 AND K. PNEUMONIAE ST258
Due to the unprecedented success of E. coli ST131 and K. pneumoniae ST258, several investigators have designed rapid methods for the detection of these sequence types to aid molecular epidemiologists and surveillance studies (Table 3). Such techniques have facilitated the enhanced detection of these sequence types as part of surveillance programs. These techniques include PCR, PFGE, DiversiLab repetitive PCR typing, abbreviated MLVA, and, more recently, MALDI-TOF mass spectrometry.
TABLE 3.
Laboratory methods for detection of Escherichia coli ST131 and Klebsiella pneumoniae ST258
| Method | Characteristic(s) of detection of: |
|
|---|---|---|
| Escherichia coli ST131 | Klebsiella pneumoniae ST258 | |
| NGS | High-resolution, accurate, and reproducible; not yet routine | High-resolution, accurate, and reproducible; not yet routine |
| MLST | Gold standard; expensive and time-consuming; 2 schemes (Achtman and Pasteur) | Gold standard; expensive and time-consuming; Pasteur scheme |
| PFGE | Used during the late 2000s; poor method since ST131 consists of different pulsotypes | Used during the late 2000s; poor method since ST258 consists of different pulsotypes |
| Repetitive-sequence-based PCR typing | Standardized fingerprinting kit; rapid and expensive | Standardized fingerprinting kit; rapid and expensive |
| MLVA | Rapid, cost-effective, and comparable to MLST | Not yet described |
| PCR | Several techniques; rapid and inexpensive for screening a large no. of isolates | Several techniques; rapid and inexpensive for screening a large no. of isolates; multiplex for clades I and II |
| MALDI-TOF MS | Rapid and inexpensive; not yet routine | Not yet described |
Next-generation sequencing (NGS) is a promising new technology with substantial potential for clinical microbiology (159). This technique uses PCR to amplify individual DNA molecules that are immobilized on a solid surface, enabling molecules to be sequenced in parallel, leading to decreased costs and rapid turnaround (160). NGS provides high-resolution DNA data in a rapid fashion, which can be used for accurate typing of medically important organisms. Several E. coli ST131 and K. pneumoniae ST258 isolates have undergone NGS (88), and it is likely that this technique, combined with more user-friendly and rapid bioinformatics, will most likely become the gold standard for the identification of these sequence types in the near future.
Multilocus Sequence Typing
MLST is a sequence-based molecular typing method that determines the nucleotide sequences of some (i.e., 6 to 10) conserved housekeeping genes (161). This makes it suitable for continuous surveillance and is excellent for collating data obtained from several separate sources via Web-based databases. MLST is often used to examine the evolutionary relationships among bacteria, which implies some form of common ancestry among isolates with the same ST. Relatedness between isolates determined by using MLST is based on polymorphisms within conserved housekeeping genes. MLST determines the population structures of different species and has led to the identification of certain global antimicrobial-resistant STs or clones (161). MLST does not have the necessary discrimination to detect genetic changes among isolates involved in outbreak situations (43).
MLST is perfect for tracing multidrug-resistant clones or STs across the globe (161). Unfortunately, MLST is expensive, since it often uses Sanger sequencing, but this technique remains the benchmark for the identification of E. coli ST131 and K. pneumoniae ST258.
As described above, there are 2 MLST schemes that are often used to identify E. coli ST131. The Achtman scheme (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli) uses the gene sequences of seven housekeeping genes: adk (adenylate kinase), fumC (fumarate hydratase), gyrB (DNA gyrase), icd (isocitrate/isopropylmalate dehydrogenase), mdh (malate dehydrogenase), purA (adenylosuccinate dehydrogenase), and recA (ATP/GTP binding motif). The Pasteur Institute E. coli scheme (http://www.pasteur.fr/recherche/genopole/PF8/mlst/EColi.html) uses eight housekeeping genes: dinB (DNA polymerase), icdA (isocitrate dehydrogenase), pabB (p-aminobenzoate synthase), polB (polymerase II [Pol II]), putP (proline permease), trpA (tryptophan synthase subunit A), trpB (tryptophan synthase subunit B), and uidA (beta-glucuronidase). The scheme from the Pasteur institute tends to identify different sequence types among ST131 isolates (as identified by the Achtman scheme). Most investigators use the Achtman MLST scheme to routinely identify ST131.
The Pasteur Institute is the most popular MLST scheme used to identify K. pneumoniae ST258 (162). (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html). This scheme uses the following seven housekeeping genes: rpoB (beta-subunit of RNA polymerase), gapA (glyceraldehyde-3-phosphate dehydrogenase), mdh (malate dehydrogenase), pgi (phosphoglucose isomerase), phoE (phosphorine E), infB (translation initiation factor 2), and tonB (periplasmic energy transducer).
Pulsed-Field Gel Electrophoresis
PFGE is the benchmark for molecular typing of medically important bacteria during outbreak investigations (43). PFGE involves the cutting of bacterial DNA into fragments of various sizes, followed by separation into fingerprints by gel electrophoresis, and this technique is often used to track clones or clusters during outbreak investigations (163). Unfortunately, PFGE is arduous, and it takes days to obtain results, while personnel need to be technically skilled in this method to obtain reliable and reproducible results. Additionally, comparison of various pulsotypes created in different locations remains a problem.
PFGE was extensively used during the late 2000s to recognize isolates that belonged to E. coli ST131 (66) and K. pneumoniae ST258 (125). However, PFGE is not a very good method to identify both sequence types because they consist of different pulsotypes that display high levels of intralineage genetic variations (45, 81). Both high-risk clones often display <80% similarity and often do not satisfy the criteria for relatedness outlined previously by Tenover et al. (44).
Repetitive-Sequence-Based PCR Typing
Three separate studies, from Canada, the United Kingdom, and the United States, evaluated the ability of the DiversiLab fingerprinting kit, a type of repetitive-element PCR, to identify E. coli ST131 (164, 165) and K. pneumoniae ST258 (166). The DiversiLab system allowed the discrimination of ST131 isolates from isolates of other E. coli sequence types and of ST258 isolates from isolates of other K. pneumoniae sequence types. The DiversiLab system offers kit-based standardized typing results in a rapid fashion but is unfortunately rather expensive, and this prohibited its widespread utilization and the implementation of this technique for the recognition of E. coli ST131 and K. pneumoniae ST258.
Multilocus Variable-Number Tandem-Repeat Analysis
An abbreviated version of MLVA for the detection of E. coli ST131 that uses a benchtop capillary electrophoresis instrument was developed by Nielsen et al. (167). The exclusion of two loci affected the discriminatory power of the abbreviated MLVA, making it less discriminatory than the original assay. However, this approach can be used as a high-throughput assay and provided rapid typing results for E. coli ST131 that were comparable to those provided by MLST and PFGE.
PCR
PCR-based techniques offer rapid and inexpensive methodologies (compared to MLST and PFGE) to identify E. coli ST131 and K. pneumoniae ST258. These techniques are the most popular approaches used to screen for ST131 and ST258 among large numbers of E. coli and K. pneumoniae isolates. The PCR screening methods are based on the recognition of different SNPs within the O serogroup and housekeeping genes specific for E. coli ST131 (19) and K. pneumoniae ST258 (168).
Several PCR screening methods are widely available to determine whether an E. coli isolate belongs to ST131. These techniques include a PCR for SNPs within the 5′ portion of the rfb locus specific for O25b (169) and SNPs within the pabB gene (170). Johnson and colleagues designed a sequencing method based on SNPs within the mdh and gyrB genes (65). A novel and rapid technique based on sequencing of the fumC and fimH loci (called CH typing or clonotyping) identified the fimH30 ST131 lineage as clonotype CH40-30 (171). Blanco and colleagues designed a triplex PCR that was based on the detection of the O25b rfb allele, blaCTX-M-15, and the afa/draBC virulence factor (172). A recent PCR screening test combined the O16 rfb variant with the ST131-specific alleles of mdh and gyrB that can distinguish between the two serogroups of ST131, namely, O25b (fimH30) and O16 (fimH41) (95). PCR methods are also available for the identification of the fimH30 lineage and the H30-Rx sublineage of ST131 (87, 91, 95). The identification of the H30-Rx sublineage requires an additional sequencing step to be performed. Note that PCR-based screening of E. coli ST131 may infrequently identify isolates that belong to clonal complex 131 (which varies from ST131 by one or two alleles) as belonging to ST131 and very rarely can misidentify non-ST131 E. coli isolates as belonging to ST131.
The multiplex real-time PCR for the detection of K. pneumoniae ST258 described by Chen and colleagues targets 2 SNPs in the tonB allele that are unique to ST258 and related STs (168). This PCR also included primers against the gapA allele (present in all K. pneumoniae isolates), which serves as an internal control. The regular multiplex PCR method described by Adler and colleagues detects the presence or absence of 2 genes, namely pilv-l and prp (138). Whole-genome sequencing has shown that the pilv-l allele is specific for ST258 and related STs. Both studies included ST258 and ST512 as positive controls in their evaluation. Chen and colleagues also developed a multiplex PCR for the identification of different lineages within ST258, i.e., clade I and clade II (173).
Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry
MALDI-TOF MS is a high-throughput methodology based on the identification of the mass-to-charge ratios of peptides and small proteins, most of which are ribosomal (174). It has been increasingly adopted by clinical microbiology laboratories all over the world for quick and reliable microbial identification to the species level based on the comparison of mass spectral fingerprints (obtained from single colonies or crude extracts) with those in previously established reference databases. The use of MALDI-TOF MS for bacterial typing is based on the principle that sequence variations within a given taxon or subspecies will be translated into the corresponding protein or peptide sequences and supported by a similar clustering obtained from either mass spectral data or genomic sequences, highlighting the usefulness of peak patterns as taxonomic markers (175). The advantages of simplicity, speed, and low cost associated with MALDI-TOF MS and its potential application on a large-scale basis might be useful to enable timely and appropriate diagnosis, infection control, and individual patient management decisions.
Japanese and Portuguese investigators evaluated MALDI-TOF MS for the identification of E. coli ST131. The study from Japan used Flexanalysis software to successfully distinguish ST131 (O25b and O16 serotypes) from other sequence types with a sensitivity of 97.0% and a specificity of 91.5% (176). The study from Portugal showed that MALDI-TOF MS fingerprinting analysis was able to discriminate between ST131, ST69, ST405, and ST39 isolates and between phylogenetic group B2 ST131 isolates and other phylogenetic group B2 isolates that did not belong to ST131 (177). MALDI-TOF MS is a promising tool for the detection of E. coli ST131 that might be available for routine use in the near future.
To our knowledge, studies that have evaluated MALDI-TOF MS for the detection of K. pneumoniae ST258 have not yet been reported.
ROLE OF PLASMIDS IN HIGH-RISK E. COLI ST131 AND K. PNEUMONIAE ST258 CLONES
Overview
Since plasmids are often maintained and exchanged independently of the bacterial host genome, they undergo evolutionary changes that are unrelated to the host. To understand the interdependence of the acquisition of plasmids and their subsequent dissemination in high-risk clones, a brief description of the classification of and similarities and differences among plasmids is required.
Plasmids consist of extrachromosomal DNA and require independent mechanisms for maintenance and coinheritance into daughter cells (9). Large portions of plasmid DNA show high degrees of plasticity, consisting of several types of mobile elements, such as insertion elements and transposons. This plasticity has challenged studies investigating the evolutionary history and relatedness of different plasmids, even when investigators used the latest sequencing technologies. However, DNAs that are required for the replication of plasmids are conserved and can therefore be used as regions for the comparison and classification of different plasmids obtained over different time periods, from different geographical areas, and from different bacteria. This is referred to as the “incompatibility group typing” scheme, which is based on the characterization of unique or conserved replication areas on different plasmids and is used primarily to demonstrate the relatedness and behavior of different plasmid groups (178, 179). It is important to remember that not all plasmids can be typed with this technique; plasmids that do not show results with incompatibility typing are often referred to as nontypeable. Plasmids that belong to different incompatibility groups exhibit variable behavior characteristics (e.g., narrow-bacterial-host-range plasmids versus broad-host-range plasmids and high-copy-number plasmids versus low-copy-number plasmids).
The classification scheme of broad-host-range versus narrow-host-range Proteobacteria plasmids is based on the diversity of the bacterial hosts into which plasmids can be transferred and then successfully maintained in a sustainable manner (180). The narrow-host-range group of plasmids most often belongs to incompatibility group F (IncF), which contains different replicons (e.g., FIA, FIB, and FII), while the broad-host-range group includes the IncA/C and IncN types (among others). Broad-host-range plasmids can easily be transferred between different species, while narrow-host-range plasmids tend to be restricted to certain species or even clones within species. This concept is important when evaluating plasmids in high-risk clones, as there is a predominance of narrow-range plasmids (specifically IncF with certain β-lactamases [e.g., blaCTX-M-15, blaKPC-2, and blaKPC-3]) in these clones. These plasmids have recently been termed “epidemic resistance plasmids” due to their propensity to acquire resistance genes and rapid dissemination among the Enterobacteriaceae (9). IncF plasmids use postsegregational killing and addiction systems to ensure their propagation among high-risk clones. IncF plasmids are often present in E. coli, and up to 70% of plasmids characterized in human and avian E. coli isolates belonged to IncF with different replicons (181).
Plasmids have coevolved with bacteria, providing them with rapid ways to evolve and adapt through the accumulation of point mutations, deletions, and insertions. Plasmids can provide intact functional genes to various bacteria that will assist them in adapting to harsh conditions and unsuitable environments (182). Plasmids often contain various combinations of virulence, fitness, and antimicrobial resistance genes and have contributed significantly to the success of antimicrobial-resistant bacteria (183).
Once a resistance plasmid has been transferred to and has replicated in a new bacterial host, the presence of antimicrobial agents will create artificial selective pressure that will select for a bacterial population with such resistance plasmids. However, in the absence of antimicrobial agents, the resistance plasmid may be retained only transiently by the bacterium if it is unstable within the host (184). The long-term stability and persistence of resistance plasmids in various bacterial populations during the absence of antibiotic selection pressures has received surprisingly little attention. Very little is known about the impact of plasmid stability on the long-term survival and host range of resistance plasmids, especially in the absence of selection pressure created by antimicrobial agents.
As described above, there is a predominance of narrow-range plasmids (i.e., IncF) among high-risk clones such as E. coli ST131 and K. pneumoniae ST258, and it seems that such clones with IncF plasmids provide a stable environment for antimicrobial resistance genes (185). Antimicrobial resistance determinants on epidemic plasmids provide a selective advantage to high-risk clones and are likely central to their success (186, 187). IncF plasmids are diverse, are complex, tend to have a narrow host range, and are present in low copy numbers (188). The concept of incompatibility for this group is somewhat of a misnomer, with several descriptions of multiple replicons being present in the same plasmid with one replicon remaining silent while another replicon is acting as the area of replication (188).
Plasmids Associated with E. coli ST131
The initial description of E. coli ST131 in 2008 by Coque and colleagues included the characterization of eight plasmids harboring blaCTX-M-15 in this ST (52). The sizes of the plasmids varied between 100 kb and 150 kb, with 3 different restriction fragment length polymorphism patterns, and they all belonged to IncF with FII and FIA replicons.
The first plasmids from ST131 to be entirely sequenced were obtained in the United Kingdom and included IncF plasmids (i.e., pEK499 and pEK516) and IncI1 plasmid pEK204 (189). Plasmid pEK499 turned out to be a fusion of FII and FIA replicons and contained the following genes for antimicrobial determinants: blaTEM-1, blaCTX-M-15, blaOXA-1, aac(6′)-Ib-cr, mph(A), catB4, tetA, dfrA7, aadA5, and sul1. pEK499 also harbored the following postsegregational killing systems that are responsible for stable plasmid inheritance during vertical spread: the hok-mok killing protein and modulator system, the pemI-pemK toxin-antitoxin system, the vagC-vagD virulence-associated genes, and the ccdA-ccdB toxin-antitoxin system. These postsegregational killing systems could be responsible for the persistence of pEK499-like plasmids among ST131 isolates, which can occur even in the absence of antibiotic selection pressure. IncF plasmid pEK516 contains the FII replicon with the following antimicrobial resistance genes: blaTEM-1, blaCTX-M-15, blaOXA-1, aac(6′)-Ib-cr, aac(3)-II, catB4, and tetA. Plasmids pEK516 and pEK499 display 75% DNA sequence similarity, despite pEK516 being substantially smaller than pEK499. Plasmid pEK516 also carries the pemI-pemK and hok-mok postsegregational killing systems as well as the type I partitioning locus parM and stbB. Plasmid pEK204 belongs to a different incompatibility group, IncI1 (i.e., a broad-host-range type of plasmid) and contains blaCTX-M-3 and blaTEM-1 accompanied by an ISEcp1-like mobile element upstream of blaCTX-M-3. These initial plasmid studies were then followed by several global molecular epidemiology studies that characterized plasmids associated with CTX-M enzymes among ST131 isolates. The majority of the plasmids associated with blaCTX-M-15 belonged to IncF and contained the fused FIA-FII or FII replicons (19). IncF plasmids present in E. coli ST131 can also encode β-lactamases other than CTX-M-15, such as CTX-M-14, SHV-2, and SHV-12, while blaCTX-M-15 was also identified on plasmids that belong to other incompatibility groups, such as IncI1, IncN, and IncA/C, as well as being on pir-type plasmids (19).
The narrow-host-range IncF and broad-host-range plasmids (i.e., IncN, IncN2, IncI1, IncHI2, IncL/M, IncA/C, IncK, IncX4, and IncU) as well as the rolling circle replication (RCR) plasmid families have been associated with CTX-M enzymes, but the IncF plasmids are by far the most common Inc type detected in E. coli ST131 isolates harboring blaCTX-M (190). blaCTX-M-15 has been found mainly on IncF plasmids with fused FIA-FII or FII replicons in ST131, whereas Inc plasmids with different replicons (i.e., FIB) have been identified in non-ST131 ExPEC (191). Some IncF plasmids with blaCTX-M-15 also carry multiple virulence determinants (e.g., iutA ompT, hlyF, iss, and iroN) (192). IncF plasmids provide various antimicrobial resistance determinants and virulence factors that confer additional selective and significant advantages for the ST131 host. IncF plasmids also use postsegregational killing and addiction systems to ensure their propagation and maintenance within E. coli ST131 and have played an essential role in the dissemination of blaCTX-M-15 among isolates of this high-risk clone, even in the absence of artificial selection pressure created by antimicrobial agents. The introduction of such IncF plasmids created the H30-Rx sublineage, and in some areas, this sublineage is more prevalent than the H30-R sublineage (Fig. 2) (94). Banerjee and colleagues have shown that isolates of the H30-Rx sublineage with blaCTX-M-15-containing plasmids had significantly higher virulence scores than did isolates of the H30-R sublineage and non-H30 ST131 isolates, implying greater virulence potential created by the presence of such IncF plasmids and possibly providing an explanation for the high prevalence of this sublineage (91). A different study showed that as ST131 gained resistance to the fluoroquinolones and acquired ESBLs, isolates (especially those that had acquired blaCTX-M-15) tended to accumulate additional virulence factors, thus implying that the virulence factors on plasmids contribute to the success of the H30-Rx sublineage (89).
Plasmids Associated with K. pneumoniae ST258
Several different KPC-containing plasmids have been identified in ST258. They belong to IncF (with FIIk1, FIIk2, and FIA replicons), IncI2, IncX, IncA/C, IncR, and ColE1, and these plasmids often contain various genes encoding nonsusceptibility to different antimicrobial drugs (30). However, the most predominant blaKPC-carrying plasmid type associated with K. pneumoniae ST258 is IncF with FIIk replicons (185). The first blaKPC-carrying plasmid identified in ST258 (named clone Q at that time) was obtained in 2006 from Israel and was named pKpQIL (193). This plasmid is a 113-kb IncF plasmid with an FIIk2 replicon containing Tn4401a, with a backbone very similar to that of plasmid pKPN4, first characterized in 1994 in a non-KPC antimicrobial-resistant K. pneumoniae isolate obtained in Massachusetts (193).
A retrospective plasmid analysis of K. pneumoniae strains with blaKPC from the New York and New Jersey areas isolated during the early 2000s showed that ST258 contained blaKPC-2 and blaKPC-3 on pKpQIL-like plasmids, which were nearly identical to the Israeli plasmid pKpQIL described in 2006 (194). pKpQIL-like plasmids from the New York and New Jersey isolates were associated mostly with blaKPC-2 and to a lesser extent with blaKPC-3, whereas the pKpQIL plasmids from Israel were associated mainly with blaKPC-3 (194, 195). This suggests that ST258 isolates with blaKPC-3 on pKpQIL plasmids were introduced into Israel from the United States during the mid-2000s (a founder effect), followed by clonal expansion in Israel. The IncFIIk plasmids are also the most common plasmids identified among ST258 isolates with blaKPC from several different geographically diverse areas, including Canada, Poland, the United States, Israel, Brazil, Italy, and Norway (131, 185, 196). There also appears to be an association between different plasmid Inc groups and ST258 clades I and II. The blaKPC-3-associated IncI2 plasmids and blaKPC-3-associated IncFIA plasmids were found exclusively in clade II, while the pKpQIL-associated IncFIIk2 plasmids with blaKPC-2 were detected in both clades I and II (136). pKpQIL plasmids were not restricted to only ST258 and were present in 33% of non-ST258 K. pneumoniae isolates in the New York area (194).
The complete sequencing of plasmids associated with ST258 from large collections reveals that they are evolving over time through large genetic rearrangements (185, 197, 198). This process is creating hybrid plasmids, as was previously described in Italy, with ST258 isolates containing two different IncF plasmids, namely, pKpQIL-IT and pKPN-IT, as well as a ColE-like plasmid with blaKPC-2 (199). Both pKpQIL-IT and pKPN-IT have a very high degree of homology to the historic plasmids pKPN4 and pKPN3 from a non-KPC-producing K. pneumoniae strain isolated in 1994 (199). This suggests that certain ancestral plasmids are particularly well suited to Enterobacteriaceae such as K. pneumoniae and are good candidates for sustaining the presence of blaKPC through multiple independent insertion and transposition events. This is further supported by a recent study from South Korea that demonstrated that ancestral plasmids were present among ST258 isolates from various geographical regions and were obtained as early as 2002 (143).
Logic might dictate that when a single blaKPC plasmid enters ST258 isolates at a single time point, it will develop diversity over time. However, it appears that K. pneumoniae ST258 isolates acquired various types of blaKPC plasmids at multiple time points and in different geographical locations around the globe, which was then followed by the notable local spread of ST258 isolates with the specific KPC plasmid (30).
It seems that the presence of plasmids with blaKPC is central to the success of ST258. The loss of pKpQIL by ST258 isolates has limited the ability of these isolates to successfully disseminate compared to other K. pneumoniae isolates without blaKPC and ST258 isolates with pKpQIL (187). This suggests that blaKPC in combination with other virulence or persistence factors on pKpQiL-like plasmids promoted the fitness and survival of ST258 isolates. This is further supported by the epidemiological observation that non-ST258 K. pneumoniae isolates with blaKPC did not demonstrate the same global success as did ST258 isolates with blaKPC. It appears that the successful global dissemination and survival of K. pneumoniae ST258 are dependent in part on the combination of blaKPC on IncF plasmids with factors inherently present on the chromosome of this high-risk clone (186).
IncI2 with blaKPC-3 can also successfully pair with ST258 and was recently detected in 23% of ST258 isolates from the New York and New Jersey areas (200). Interestingly, this IncI2 plasmid also contained type IV pili, which may contribute to the successful dissemination of K. pneumoniae ST258.
Broad-Host-Range Plasmids
Broad-host-range plasmids (i.e., IncA/C, IncL/M, and IncN) are rare in E. coli ST131 and K. pneumoniae ST258 isolates (19, 30). This is a curious finding, and it seems that at this time, broad-range plasmids do not play a meaningful role in the success and global dominance of K. pneumoniae ST258 and E. coli ST131.
CONCLUSION
There is no doubt that the high-risk E. coli ST131 and K. pneumoniae ST258 clones are important human pathogens, have spread extensively throughout the world, and are responsible for the sudden increase in multidrug resistance among E. coli (17) and K. pneumoniae (121) isolates, respectively. ST131 is responsible for extraintestinal infections (especially UTIs), is most often fluoroquinolone resistant, and can also be linked with the production of CTX-M-15 (19), while ST258 is known to cause UTIs, respiratory tract infections, and BSIs and is associated with carbapenemase production, most often KPC-2 and KPC-3 (201) (Table 4).
TABLE 4.
Summary of Escherichia coli ST131 and Klebsiella pneumoniae ST258
| Characteristic | Descriptiona |
|
|---|---|---|
| Escherichia coli ST131 | Klebsiella pneumoniae ST258 | |
| Clonal complex | CC131 (with ST131 being dominant) | CC258 (with ST258 being dominant; also includes ST11, ST340, ST437, and ST512) |
| Multidrug-resistant high-risk clone | Yes | Yes |
| Region(s) of endemicity | Global | USA, Greece, Italy, Israel, Columbia |
| Resistance determinants | Various but associated with fluoroquinolone resistance and CTX-M-15 (CTX-M-14 to a lesser extent) | Various but associated with KPC-2 and KPC-3 |
| Mobile element | ISEcp1 | Tn4401 |
| Plasmids | Various incompatibility groups, but IncF with virulence factors is the most common | Various incompatibility groups, but IncFIIk is the most common |
| Clades/lineages | O25 fimH30 lineage that consists of the H30-R and H30-Rx sublineages; O16 fimH41 lineage | Clade I associated with KPC-2; clade II associated with KPC-3 |
| Population biology | H30-R sublineage is associated with fluoroquinolone resistance, and H30-Rx is associated with CTX-M-15 | Clade II is a hybrid of ST11 and ST442; clade I is a hybrid of clade II and ST42 |
| Methods of acquisition | Community-onset health care associated, LTCFs | Nosocomial, LTCFs |
| Types of infection | UTIs, IAIs, BSIs | UTIs, RTIs, BSIs |
| Mortality | Unclear if the rate is higher than that with non-ST131 E. coli | Higher rate than that with non-ST258 K. pneumoniae (most likely due to the patient's underlying conditions) |
UTIs, urinary tract infections; IAIs, intra-abdominals infections; BSIs, bloodstream-associated infections; RTIs, respiratory tract infections; LTCFs long-term-care facilities.
Biological evolution is defined as “descent with modification within a defined population,” with the sole purpose of adaptation to environmental changes (202). This definition encompasses microevolution (i.e., changes in gene frequency within a population from one generation to the next) and macroevolution (i.e., the descent of different species from a common ancestor over many generations). Evolution provides the means for members of a population to adapt, survive, and reproduce in an effective manner. For evolution to progress in an orderly and efficient fashion, a selection process for desirable traits is required. Natural selection is due to the interaction of a population with the environment, while artificial selection (also referred to as selective breeding) occurs due to human intervention. Antimicrobial therapy with the subsequent selection of antimicrobial-resistant mutations is considered to be one of the most efficient artificial selection pressures introduced by humans (203).
E. coli ST131 has long been “flying under the antimicrobial resistance radar” and therefore did not generate the same awareness as other multidrug-resistant bacteria. Retrospective molecular epidemiology surveys have demonstrated that ST131 was rare among E. coli isolates from the 1990s and early 2000s (84, 94). However, during the mid- to late 2000s, the numbers of fluoroquinolone-resistant ST131 isolates increased substantially, which was followed by an explosive increase in the number of isolates with blaCTX-M-15 toward the end of the 2000s (19). The reasons for this sudden increase are not clear or well understood. Recent studies utilizing advanced technologies such as whole-genome sequencing combined with phylogenetic analysis have demonstrated that a lineage within ST131, named fimH30, emerged during the early 2000s. This was then followed by the rapid expansion of its sublineages H30-R and H30-Rx, which were responsible for the global emergence of fluoroquinolone resistance and CTX-M-producing ExPEC isolates. The widespread use of antimicrobials possibly selected for ST131, resulting in the emergence of ST131. The strong association between ST131 and fluoroquinolone resistance suggested that the use of fluoroquinolones has contributed to the expansion of the H30-R ST131 sublineage. Likewise, the use of oxyimino cephalosporins may have driven the expansion of the H30-Rx sublineage, which is strongly associated with CTX-M-15. The selection pressures created by the widespread use of the fluoroquinolones and the oxyimino cephalosporins dramatically changed the population structure of E. coli, especially among ExPEC isolates, and created one of the most prevalent and extensive global antimicrobial-resistant E. coli sublineages known to the medical community (84). This is an excellent example of microevolution in action due to the artificial selection pressure created by the use of antimicrobial agents (Fig. 4).
FIG 4.
Change in the population structure of Escherichia coli due to selection pressures created by the fluoroquinolones (FQ) and the oxyimino cephalosporins FQ-R, fluoroquinolone resistant.
The prevalences of certain virulence genes such as sat, iutA, malX, usp, iha, hra, and ompT are consistently high among ST131 isolates (compared to non-ST131 ExPEC isolates), and these genes might increase the versatility and competitiveness of ST131 and the capacity of ST131 to effectively infect the human body (Fig. 5). The exact role of these virulence genes remains to be explained; however, it is possible that a combination of factors contributes to the fitness of ST131 rather than being directly involved in the pathogenesis of infection (79). In vivo studies suggest that fluoroquinolone-resistant E. coli isolates undergo compensatory mutations and become as fit as fluoroquinolone-susceptible isolates (46).
FIG 5.
Possible factors that make the Escherichia coli ST131 fimH30 lineage such a successful high-risk clone. VFs, virulence factors; FQ-R, fluoroquinolone resistant.
This brings up some intriguing issues. Is E. coli ST131 inherently more fit than other ExPEC clones and therefore better able to survive in certain environments? Is this “fitness” due to certain virulence factors (e.g., sat, iutA, malX, usp, iha, hra, and ompT) that provided ST131 with opportunities to be exposed to and acquire certain narrow-host-range IncF plasmids? Did the gyrA-parC compensatory mutations (i.e., gyrA1AB and parC1aAB), selected by fluoroquinolones, reduce the fitness costs associated with IncF plasmids? Is it perhaps possible that the mutations accommodating IncF plasmids increased the fitness of ST131 isolates, enabling them, with the aid of certain virulence factors and antimicrobial resistance determinants on the CTX-M-15 plasmids, to outcompete other ExPEC clones? Colpan and colleagues have shown that fluoroquinolone-susceptible E. coli ST131 isolates have different virulence factor profiles than those of fluoroquinolone-susceptible non-ST131 isolates, and as ST131 isolates gained resistance to the fluoroquinolones and acquired ESBLs (especially CTX-M-15), they tended to accumulate additional virulence factors (89). Furthermore, in vitro studies have shown that ST131 has high metabolic potential and an enhanced ability to produce biofilms (19). These studies suggest that the inherent combination of different virulence factors, biofilm production, and metabolic potential provides ST131 isolates with the means to be “more fit” than other ExPEC clones. Moreover, it seems that ST131 (especially the fimH30 lineage) has the ability to counteract the fitness cost associated with antimicrobial resistance and also tends to accumulate virulence factors, as this clone/lineage is becoming progressively more antimicrobial resistant (89). With its combination of multidrug resistance and ecological success, the ST131 fimH30 lineage counters the hypothesis that increased antimicrobial resistance entails a significant fitness cost. E. coli ST131 is most likely the best example of an international multiresistant high-risk clone (Fig. 5).
The role of IncF plasmids with blaCTX-M-15 in the success of ST131 is another intriguing issue. IncF plasmids provide various antimicrobial resistance determinants, but some plasmids also encode multiple virulence factors that confer additional selective and significant advantages for the ST131 host (192). IncF plasmids use postsegregational killing and addiction systems to ensure their propagation among high-risk clones and have played an essential role in the dispersion of blaCTX-M-15 among E. coli ST131 isolates by creating the H30-Rx sublineage. A population-based molecular surveillance study (2000 to 2010) from Calgary showed that the H30-Rx sublineage with blaCTX-M-15 became the most dominant sublineage toward the later part of the study period (94) (Fig. 2). It is therefore possible that the coevolution of certain sublineages within high-risk clones (i.e., H30-Rx) and epidemic plasmids (e.g., IncF) provided rapid and continual adaptation for certain sublineages, giving them the additional ability to outcompete other sublineages without such epidemic plasmids (Fig. 5). This is consistent with both the macro- and microevolutionary versions of the Red Queen hypothesis, as demonstrated recently by using the microscopic roundworm Caenorhabditis elegans as a host and the bacterium Serratia marcescens as a parasite to create a host-parasite coevolutionary system (204) (Fig. 4 and 5).
Recent molecular studies have shown that K. pneumoniae ST258 consists of 2 distinct lineages, namely, clade I and clade II (136). Clade I was specifically associated with blaKPC-2, and clade II was associated with blaKPC-3 (Table 4). The genetic differentiation of the two clades occurred in a 215-kb region that included genes responsible for capsule polysaccharide biosynthesis, indicating that these 2 clades had distinct evolutionary pathways. Additional investigation showed that ST258 clade II is a cross-hybrid clone that was created by a large recombination event between ST11 and ST442 (137). Moreover, it seems that ST258 clade I was created by the cps replacement in ST258 clade II that originated from ST42 (Fig. 3). The integrative conjugative element ICEKp258.2 contains gene clusters for a type IV pilus (i.e., pilV) and a type III restriction-modification system. pilV on ICEKp258.2 may in part be responsible for the high transmissibility and durability of ST258 on foreign surfaces, and it seems that this integrative conjugative element contributes significantly to the epidemiological success of K. pneumoniae ST258 (137).
The use of antimicrobial agents will continue to create selection pressure that gives high-risk clones the opportunity to become effective intestinal colonizers and provides opportunities for them to cause infections. To prescribe more effective empirical antimicrobial therapy, clinicians will need to have an increased awareness of E. coli ST131 and K. pneumoniae ST258, more specifically the prevalence and extensive antimicrobial resistance capabilities of these high-risk clones, including which patients are at risk for infection. E. coli ST131 and K. pneumoniae ST258 cause infections similar to those caused by non-ST131 E. coli and non-ST258 K. pneumoniae isolates, respectively, and clinical risk factors most likely will be insufficient to reliably identify patients with an increased risk of infections by these clones. Therefore, the medical community needs cost-effective diagnostic tests that have the ability to rapidly detect E. coli ST131 and K. pneumoniae ST258. PCR- and sequence-based approaches have been developed and reliably detect both clones in a matter of hours (Table 3), but unfortunately, due to cost and personnel issues, these methods are difficult to implement routinely in a diagnostic laboratory. Such approaches might provide timely and appropriate antimicrobial therapy that can help to improve clinical outcomes with infections due to E. coli ST131 and K. pneumoniae ST258.
E. coli ST131, K. pneumoniae ST258, and their lineages/clades are now major threats to public health due to their worldwide distribution and association with multidrug resistance. The recent emergence of E. coli ST131 with carbapenemases poses a significant additional public health menace due to its relationship with the very dominant fimH30 lineage.
Future studies should be undertaken to determine if antibiotic stewardship and which infection control bundles will decrease colonization and infections by E. coli ST131 and K. pneumoniae ST258. Additionally, vaccines that target E. coli ST131 serotypes O25b and O16 and those that target the cps region of K. pneumoniae ST258 clades I and II should also be developed. This might reduce the reservoir of asymptomatic individuals colonized with these high-risk clones and will reduce the spread of and infection due to E. coli ST131 and K. pneumoniae ST258.
ACKNOWLEDGMENTS
This work was supported in part by a research grant from Calgary Laboratory Services (grant number 10006465).
J.D.D.P. previously received research funds from Merck and AstraZeneca. A.J.M. has received research funds from Rempex Pharmaceuticals. G.P. has nothing to declare.
Biographies

Amy J. Mathers, M.D., is an Assistant Professor of Medicine and Pathology at the University of Virginia in the Division of Infectious Diseases and International Health, where she serves as Clinical Director of Antimicrobial Stewardship and Assistant Director of Clinical Microbiology. She received her bachelor's of science in Molecular Biology at Humboldt State University in California. She completed her medical degree at Loyola Stritch School of Medicine in Chicago, IL. During Internal Medicine Residency at the Maine Medical Center in Portland, ME, she investigated the strain diversity of Borrelia burgdorferi. After completing her Infectious Diseases fellowship at the University of Virginia, she transitioned to faculty in 2009. Focusing on the urgent clinical problem of carbapenem resistance in Enterobacteriaceae, she has been evaluating detection methods and molecular transmission of carbapenemase genes for the last 8 years. Using next-generation sequencing, she seeks to better understand the origin, transmission, and sustainability of genes of antimicrobial resistance.

Gisele Peirano, Ph.D., is a Research Associate in the Microbiology section of Calgary Laboratory Services in Calgary, Alberta, Canada. She graduated from Rio de Janeiro State University (1997) and received her M.Sc. (2000) and Ph.D. (2005) in Bacteriology from the Oswaldo Cruz Institute, Rio de Janeiro, Brazil. She was awarded a Foreign Affairs, Trade and Development Canada postdoctoral fellowship at the Department of Pathology and Laboratory Medicine, University of Calgary, Calgary, AB, Canada. Her main research interest is resistance to antimicrobial agents among Gram-negative bacteria, especially the molecular epidemiology of newer β-lactamases such as extended-spectrum β-lactamases and carbapenemases. She has published over 40 papers in this field.
Johann D. D. Pitout, M.D., is a Medical Microbiologist and Professor at the University of Calgary and Calgary Laboratory Services. His main research interests are resistance to antimicrobial agents among Gram-negative bacteria, especially the laboratory detection, characterization, and molecular epidemiology of bacteria with newer β-lactamases such as AmpC, extended-spectrum β-lactamases, and carbapenemases. He has also been involved in population-based surveillance studies and studies of the role of high-risk clones among bacteria producing these newer types of β-lactamases. He has published over 120 papers in this field.
REFERENCES
- 1.Paterson DL. 2006. Resistance in gram-negative bacteria: Enterobacteriaceae. Am J Infect Control 34:S20–S28. doi: 10.1016/j.ajic.2006.05.238. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. 2014. Antimicrobial resistance: global report on surveillance 2014. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 3.Schwaber MJ, Navon-Venezia S, Kaye KS, Ben-Ami R, Schwartz D, Carmeli Y. 2006. Clinical and economic impact of bacteremia with extended-spectrum-β-lactamase-producing Enterobacteriaceae. Antimicrob Agents Chemother 50:1257–1262. doi: 10.1128/AAC.50.4.1257-1262.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Infectious Diseases Society of America. 2010. The 10 × ‘20 initiative: pursuing a global commitment to develop 10 new antibacterial drugs by 2020. Clin Infect Dis 50:1081–1083. doi: 10.1086/652237. [DOI] [PubMed] [Google Scholar]
- 5.Reuter S, Ellington MJ, Cartwright EJ, Koser CU, Torok ME, Gouliouris T, Harris SR, Brown NM, Holden MT, Quail M, Parkhill J, Smith GP, Bentley SD, Peacock SJ. 2013. Rapid bacterial whole-genome sequencing to enhance diagnostic and public health microbiology. JAMA Intern Med 173:1397–1404. doi: 10.1001/jamainternmed.2013.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woodford N, Turton JF, Livermore DM. 2011. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev 35:736–755. doi: 10.1111/j.1574-6976.2011.00268.x. [DOI] [PubMed] [Google Scholar]
- 7.Carattoli A. 2013. Plasmids and the spread of resistance. Int J Med Microbiol 303:298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother 53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bush K, Jacoby GA. 2010. Updated functional classification of β-lactamases. Antimicrob Agents Chemother 54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nordmann P, Poirel L. 2014. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect 20:821–830. doi: 10.1111/1469-0691.12719. [DOI] [PubMed] [Google Scholar]
- 12.van der Bij AK, Pitout JD. 2012. The role of international travel in the worldwide spread of multiresistant Enterobacteriaceae. J Antimicrob Chemother 67:2090–2100. doi: 10.1093/jac/dks214. [DOI] [PubMed] [Google Scholar]
- 13.Paterson DL, Bonomo RA. 2005. Extended-spectrum β-lactamases: a clinical update. Clin Microbiol Rev 18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canton R, Coque TM. 2006. The CTX-M β-lactamase pandemic. Curr Opin Microbiol 9:466–475. doi: 10.1016/j.mib.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 15.Pitout JD, Laupland KB. 2008. Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis 8:159–166. doi: 10.1016/S1473-3099(08)70041-0. [DOI] [PubMed] [Google Scholar]
- 16.D'Andrea MM, Arena F, Pallecchi L, Rossolini GM. 2013. CTX-M-type β-lactamases: a successful story of antibiotic resistance. Int J Med Microbiol 303:305–317. doi: 10.1016/j.ijmm.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Peirano G, Pitout JD. 2010. Molecular epidemiology of Escherichia coli producing CTX-M β-lactamases: the worldwide emergence of clone ST131 O25:H4. Int J Antimicrob Agents 35:316–321. doi: 10.1016/j.ijantimicag.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez-Bano J, Pascual A. 2008. Clinical significance of extended-spectrum β-lactamases. Expert Rev Anti Infect Ther 6:671–683. doi: 10.1586/14787210.6.5.671. [DOI] [PubMed] [Google Scholar]
- 19.Nicolas-Chanoine MH, Bertrand X, Madec JY. 2014. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev 27:543–574. doi: 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hilty M, Betsch BY, Bogli-Stuber K, Heiniger N, Stadler M, Kuffer M, Kronenberg A, Rohrer C, Aebi S, Endimiani A, Droz S, Muhlemann K. 2012. Transmission dynamics of extended-spectrum β-lactamase-producing Enterobacteriaceae in the tertiary care hospital and the household setting. Clin Infect Dis 55:967–975. doi: 10.1093/cid/cis581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Talbot GH, Bradley J, Edwards JE Jr, Gilbert D, Scheld M, Bartlett JG. 2006. Bad bugs need drugs: an update on the development pipeline from the Antimicrobial Availability Task Force of the Infectious Diseases Society of America. Clin Infect Dis 42:657–668. doi: 10.1086/499819. [DOI] [PubMed] [Google Scholar]
- 22.Jacoby GA. 2009. AmpC β-lactamases. Clin Microbiol Rev 22:161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanson ND. 2003. AmpC β-lactamases: what do we need to know for the future? J Antimicrob Chemother 52:2–4. doi: 10.1093/jac/dkg284. [DOI] [PubMed] [Google Scholar]
- 24.Philippon A, Arlet G, Jacoby GA. 2002. Plasmid-determined AmpC-type β-lactamases. Antimicrob Agents Chemother 46:1–11. doi: 10.1128/AAC.46.1.1-11.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Queenan AM, Bush K. 2007. Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev 20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walther-Rasmussen J, Hoiby N. 2007. Class A carbapenemases. J Antimicrob Chemother 60:470–482. doi: 10.1093/jac/dkm226. [DOI] [PubMed] [Google Scholar]
- 27.Deshpande LM, Rhomberg PR, Sader HS, Jones RN. 2006. Emergence of serine carbapenemases (KPC and SME) among clinical strains of Enterobacteriaceae isolated in the United States Medical Centers: report from the MYSTIC Program (1999-2005). Diagn Microbiol Infect Dis 56:367–372. doi: 10.1016/j.diagmicrobio.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 28.Nordmann P, Cuzon G, Naas T. 2009. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis 9:228–236. doi: 10.1016/S1473-3099(09)70054-4. [DOI] [PubMed] [Google Scholar]
- 29.Naas T, Cuzon G, Villegas MV, Lartigue MF, Quinn JP, Nordmann P. 2008. Genetic structures at the origin of acquisition of the β-lactamase blaKPC gene. Antimicrob Agents Chemother 52:1257–1263. doi: 10.1128/AAC.01451-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. 2014. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol 22:686–696. doi: 10.1016/j.tim.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poirel L, Pitout JD, Nordmann P. 2007. Carbapenemases: molecular diversity and clinical consequences. Future Microbiol 2:501–512. doi: 10.2217/17460913.2.5.501. [DOI] [PubMed] [Google Scholar]
- 32.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nordmann P, Poirel L, Walsh TR, Livermore DM. 2011. The emerging NDM carbapenemases. Trends Microbiol 19:588–595. doi: 10.1016/j.tim.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 35.Woodford N, Wareham DW, Guerra B, Teale C. 2014. Carbapenemase-producing Enterobacteriaceae and non-Enterobacteriaceae from animals and the environment: an emerging public health risk of our own making? J Antimicrob Chemother 69:287–291. doi: 10.1093/jac/dkt392. [DOI] [PubMed] [Google Scholar]
- 36.Johnson AP, Woodford N. 2013. Global spread of antibiotic resistance: the example of New Delhi metallo-β-lactamase (NDM)-mediated carbapenem resistance. J Med Microbiol 62:499–513. doi: 10.1099/jmm.0.052555-0. [DOI] [PubMed] [Google Scholar]
- 37.Poirel L, Potron A, Nordmann P. 2012. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother 67:1597–1606. doi: 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]
- 38.Poirel L, Bonnin RA, Nordmann P. 2012. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob Agents Chemother 56:559–562. doi: 10.1128/AAC.05289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poirel L, Castanheira M, Carrer A, Rodriguez CP, Jones RN, Smayevsky J, Nordmann P. 2011. OXA-163, an OXA-48-related class D β-lactamase with extended activity toward expanded-spectrum cephalosporins. Antimicrob Agents Chemother 55:2546–2551. doi: 10.1128/AAC.00022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 41.Dijkshoorn L, Ursing BM, Ursing JB. 2000. Strain, clone and species: comments on three basic concepts of bacteriology. J Med Microbiol 49:397–401. [DOI] [PubMed] [Google Scholar]
- 42.Spratt BG. 2004. Exploring the concept of clonality in bacteria. Methods Mol Biol 266:323–352. doi: 10.1385/1-59259-763-7:323. [DOI] [PubMed] [Google Scholar]
- 43.Sabat AJ, Budimir A, Nashev D, Sa-Leao R, van Dijl J, Laurent F, Grundmann H, Friedrich AW, ESCMID Study Group of Epidemiological Markers . 2013. Overview of molecular typing methods for outbreak detection and epidemiological surveillance. Euro Surveill 18(4):pii=20380 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20380. [DOI] [PubMed] [Google Scholar]
- 44.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33:2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lascols C, Peirano G, Hackel M, Laupland KB, Pitout JD. 2013. Surveillance and molecular epidemiology of Klebsiella pneumoniae isolates that produce carbapenemases: first report of OXA-48-like enzymes in North America. Antimicrob Agents Chemother 57:130–136. doi: 10.1128/AAC.01686-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riley LW. 2014. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin Microbiol Infect 20:380–390. doi: 10.1111/1469-0691.12646. [DOI] [PubMed] [Google Scholar]
- 47.Baquero F, Tedim AP, Coque TM. 2013. Antibiotic resistance shaping multi-level population biology of bacteria. Front Microbiol 4:15. doi: 10.3389/fmicb.2013.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 49.Pitout JD. 2012. Extraintestinal pathogenic Escherichia coli: an update on antimicrobial resistance, laboratory diagnosis and treatment. Expert Rev Anti Infect Ther 10:1165–1176. doi: 10.1586/eri.12.110. [DOI] [PubMed] [Google Scholar]
- 50.Pitout JD, Church DL, Gregson DB, Chow BL, McCracken M, Mulvey MR, Laupland KB. 2007. Molecular epidemiology of CTX-M-producing Escherichia coli in the Calgary Health Region: emergence of CTX-M-15-producing isolates. Antimicrob Agents Chemother 51:1281–1286. doi: 10.1128/AAC.01377-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woodford N, Ward ME, Kaufmann ME, Turton J, Fagan EJ, James D, Johnson AP, Pike R, Warner M, Cheasty T, Pearson A, Harry S, Leach JB, Loughrey A, Lowes JA, Warren RE, Livermore DM. 2004. Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum beta-lactamases in the UK. J Antimicrob Chemother 54:735–743. doi: 10.1093/jac/dkh424. [DOI] [PubMed] [Google Scholar]
- 52.Coque TM, Novais A, Carattoli A, Poirel L, Pitout J, Peixe L, Baquero F, Canton R, Nordmann P. 2008. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerg Infect Dis 14:195–200. doi: 10.3201/eid1402.070350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nicolas-Chanoine MH, Blanco J, Leflon-Guibout V, Demarty R, Alonso MP, Canica MM, Park YJ, Lavigne JP, Pitout J, Johnson JR. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J Antimicrob Chemother 61:273–281. doi: 10.1093/jac/dkm464. [DOI] [PubMed] [Google Scholar]
- 54.Lau SH, Kaufmann ME, Livermore DM, Woodford N, Willshaw GA, Cheasty T, Stamper K, Reddy S, Cheesbrough J, Bolton FJ, Fox AJ, Upton M. 2008. UK epidemic Escherichia coli strains A-E, with CTX-M-15 β-lactamase, all belong to the international O25:H4-ST131 clone. J Antimicrob Chemother 62:1241–1244. doi: 10.1093/jac/dkn380. [DOI] [PubMed] [Google Scholar]
- 55.Peirano G, Richardson D, Nigrin J, McGeer A, Loo V, Toye B, Alfa M, Pienaar C, Kibsey P, Pitout JD. 2010. High prevalence of ST131 isolates producing CTX-M-15 and CTX-M-14 among extended-spectrum-β-lactamase-producing Escherichia coli isolates from Canada. Antimicrob Agents Chemother 54:1327–1330. doi: 10.1128/AAC.01338-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cagnacci S, Gualco L, Debbia E, Schito GC, Marchese A. 2008. European emergence of ciprofloxacin-resistant Escherichia coli clonal groups O25:H4-ST 131 and O15:K52:H1 causing community-acquired uncomplicated cystitis. J Clin Microbiol 46:2605–2612. doi: 10.1128/JCM.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yumuk Z, Afacan G, Nicolas-Chanoine MH, Sotto A, Lavigne JP. 2008. Turkey: a further country concerned by community-acquired Escherichia coli clone O25-ST131 producing CTX-M-15. J Antimicrob Chemother 62:284–288. doi: 10.1093/jac/dkn181. [DOI] [PubMed] [Google Scholar]
- 58.Literacka E, Bedenic B, Baraniak A, Fiett J, Tonkic M, Jajic-Bencic I, Gniadkowski M. 2009. blaCTX-M genes in Escherichia coli strains from Croatian hospitals are located in new (blaCTX-M-3a) and widely spread (blaCTX-M-3a and blaCTX-M-15) genetic structures. Antimicrob Agents Chemother 53:1630–1635. doi: 10.1128/AAC.01431-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suzuki S, Shibata N, Yamane K, Wachino J, Ito K, Arakawa Y. 2009. Change in the prevalence of extended-spectrum-β-lactamase-producing Escherichia coli in Japan by clonal spread. J Antimicrob Chemother 63:72–79. doi: 10.1093/jac/dkn463. [DOI] [PubMed] [Google Scholar]
- 60.Peirano G, Costello M, Pitout JD. 2010. Molecular characteristics of extended-spectrum β-lactamase-producing Escherichia coli from the Chicago area: high prevalence of ST131 producing CTX-M-15 in community hospitals. Int J Antimicrob Agents 36:19–23. doi: 10.1016/j.ijantimicag.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 61.Peirano G, van Greune CH, Pitout JD. 2011. Characteristics of infections caused by extended-spectrum β-lactamase-producing Escherichia coli from community hospitals in South Africa. Diagn Microbiol Infect Dis 69:449–453. doi: 10.1016/j.diagmicrobio.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 62.Peirano G, Asensi MD, Pitondo-Silva A, Pitout JD. 2011. Molecular characteristics of extended-spectrum β-lactamase-producing Escherichia coli from Rio de Janeiro, Brazil. Clin Microbiol Infect 17:1039–1043. doi: 10.1111/j.1469-0691.2010.03440.x. [DOI] [PubMed] [Google Scholar]
- 63.Naseer U, Haldorsen B, Tofteland S, Hegstad K, Scheutz F, Simonsen GS, Sundsfjord A. 2009. Molecular characterization of CTX-M-15-producing clinical isolates of Escherichia coli reveals the spread of multidrug-resistant ST131 (O25:H4) and ST964 (O102:H6) strains in Norway. APMIS 117:526–536. doi: 10.1111/j.1600-0463.2009.02465.x. [DOI] [PubMed] [Google Scholar]
- 64.Leflon-Guibout V, Blanco J, Amaqdouf K, Mora A, Guize L, Nicolas-Chanoine MH. 2008. Absence of CTX-M enzymes but high prevalence of clones, including clone ST131, among fecal Escherichia coli isolates from healthy subjects living in the area of Paris, France. J Clin Microbiol 46:3900–3905. doi: 10.1128/JCM.00734-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnson JR, Menard M, Johnston B, Kuskowski MA, Nichol K, Zhanel GG. 2009. Epidemic clonal groups of Escherichia coli as a cause of antimicrobial-resistant urinary tract infections in Canada, 2002 to 2004. Antimicrob Agents Chemother 53:2733–2739. doi: 10.1128/AAC.00297-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pitout JD, Gregson DB, Campbell L, Laupland KB. 2009. Molecular characteristics of extended-spectrum-β-lactamase-producing Escherichia coli isolates causing bacteremia in the Calgary Health Region from 2000 to 2007: emergence of clone ST131 as a cause of community-acquired infections. Antimicrob Agents Chemother 53:2846–2851. doi: 10.1128/AAC.00247-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oteo J, Diestra K, Juan C, Bautista V, Novais A, Perez-Vazquez M, Moya B, Miro E, Coque TM, Oliver A, Canton R, Navarro F, Campos J. 2009. Extended-spectrum β-lactamase-producing Escherichia coli in Spain belong to a large variety of multilocus sequence typing types, including ST10 complex/A, ST23 complex/A and ST131/B2. Int J Antimicrob Agents 34:173–176. doi: 10.1016/j.ijantimicag.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 68.Rooney PJ, O'Leary MC, Loughrey AC, McCalmont M, Smyth B, Donaghy P, Badri M, Woodford N, Karisik E, Livermore DM. 2009. Nursing homes as a reservoir of extended-spectrum β-lactamase (ESBL)-producing ciprofloxacin-resistant Escherichia coli. J Antimicrob Chemother 64:635–641. doi: 10.1093/jac/dkp220. [DOI] [PubMed] [Google Scholar]
- 69.Pomba C, da Fonseca JD, Baptista BC, Correia JD, Martinez-Martinez L. 2009. Detection of the pandemic O25-ST131 human virulent Escherichia coli CTX-M-15-producing clone harboring the qnrB2 and aac(6′)-Ib-cr genes in a dog. Antimicrob Agents Chemother 53:327–328. doi: 10.1128/AAC.00896-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Freeman JT, McBride SJ, Heffernan H, Bathgate T, Pope C, Ellis-Pegler RB. 2008. Community-onset genitourinary tract infection due to CTX-M-15-producing Escherichia coli among travelers to the Indian subcontinent in New Zealand. Clin Infect Dis 47:689–692. doi: 10.1086/590941. [DOI] [PubMed] [Google Scholar]
- 71.Laupland KB, Church DL, Vidakovich J, Mucenski M, Pitout JD. 2008. Community-onset extended-spectrum β-lactamase (ESBL) producing Escherichia coli: importance of international travel. J Infect 57:441–448. doi: 10.1016/j.jinf.2008.09.034. [DOI] [PubMed] [Google Scholar]
- 72.Pitout JD, Campbell L, Church DL, Gregson DB, Laupland KB. 2009. Molecular characteristics of travel-related extended-spectrum-β-lactamase-producing Escherichia coli isolates from the Calgary Health Region. Antimicrob Agents Chemother 53:2539–2543. doi: 10.1128/AAC.00061-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Friedman ND, Kaye KS, Stout JE, McGarry SA, Trivette SL, Briggs JP, Lamm W, Clark C, MacFarquhar J, Walton AL, Reller LB, Sexton DJ. 2002. Health care-associated bloodstream infections in adults: a reason to change the accepted definition of community-acquired infections. Ann Intern Med 137:791–797. doi: 10.7326/0003-4819-137-10-200211190-00007. [DOI] [PubMed] [Google Scholar]
- 74.Hawser SP, Bouchillon SK, Hoban DJ, Badal RE, Hsueh PR, Paterson DL. 2009. Emergence of high levels of extended-spectrum-β-lactamase-producing Gram-negative bacilli in the Asia-Pacific region: data from the Study for Monitoring Antimicrobial Resistance Trends (SMART) program, 2007. Antimicrob Agents Chemother 53:3280–3284. doi: 10.1128/AAC.00426-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Peirano G, Laupland KB, Gregson DB, Pitout JD. 2011. Colonization of returning travelers with CTX-M-producing Escherichia coli. J Travel Med 18:299–303. doi: 10.1111/j.1708-8305.2011.00548.x. [DOI] [PubMed] [Google Scholar]
- 76.Novais A, Pires J, Ferreira H, Costa L, Montenegro C, Vuotto C, Donelli G, Coque TM, Peixe L. 2012. Characterization of globally spread Escherichia coli ST131 isolates (1991 to 2010). Antimicrob Agents Chemother 56:3973–3976. doi: 10.1128/AAC.00475-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. 2010. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin Infect Dis 51:286–294. doi: 10.1086/653932. [DOI] [PubMed] [Google Scholar]
- 78.Pitout JD. 2012. Extraintestinal pathogenic Escherichia coli: a combination of virulence with antibiotic resistance. Front Microbiol 3:9. doi: 10.3389/fmicb.2012.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dobrindt U. 2005. (Patho-)genomics of Escherichia coli. Int J Med Microbiol 295:357–371. doi: 10.1016/j.ijmm.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 80.Gibreel TM, Dodgson AR, Cheesbrough J, Fox AJ, Bolton FJ, Upton M. 2012. Population structure, virulence potential and antibiotic susceptibility of uropathogenic Escherichia coli from Northwest England. J Antimicrob Chemother 67:346–356. doi: 10.1093/jac/dkr451. [DOI] [PubMed] [Google Scholar]
- 81.Peirano G, van der Bij AK, Freeman JL, Poirel L, Nordmann P, Costello M, Tchesnokova VL, Pitout JD. 2014. Characteristics of Escherichia coli sequence type 131 isolates that produce extended-spectrum β-lactamases: global distribution of the H30-Rx sublineage. Antimicrob Agents Chemother 58:3762–3767. doi: 10.1128/AAC.02428-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Johnson JR, Nicolas-Chanoine MH, Debroy C, Castanheira M, Robicsek A, Hansen G, Weissman S, Urban C, Platell J, Trott D, Zhanel G, Clabots C, Johnston BD, Kuskowski MA. 2012. Comparison of Escherichia coli ST131 pulsotypes, by epidemiologic traits, 1967-2009. Emerg Infect Dis 18:598–607. doi: 10.3201/eid1804.111627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Peirano G, van der Bij AK, Gregson DB, Pitout JD. 2012. Molecular epidemiology over an 11-year period (2000 to 2010) of extended-spectrum β-lactamase-producing Escherichia coli causing bacteremia in a centralized Canadian region. J Clin Microbiol 50:294–299. doi: 10.1128/JCM.06025-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Banerjee R, Johnson JR. 2014. A new clone sweeps clean: the enigmatic emergence of Escherichia coli sequence type 131. Antimicrob Agents Chemother 58:4997–5004. doi: 10.1128/AAC.02824-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Banerjee R, Johnston B, Lohse C, Porter SB, Clabots C, Johnson JR. 2013. Escherichia coli sequence type 131 is a dominant, antimicrobial-resistant clonal group associated with healthcare and elderly hosts. Infect Control Hosp Epidemiol 34:361–369. doi: 10.1086/669865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Platell JL, Johnson JR, Cobbold RN, Trott DJ. 2011. Multidrug-resistant extraintestinal pathogenic Escherichia coli of sequence type ST131 in animals and foods. Vet Microbiol 153:99–108. doi: 10.1016/j.vetmic.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 87.Johnson JR, Tchesnokova V, Johnston B, Clabots C, Roberts PL, Billig M, Riddell K, Rogers P, Qin X, Butler-Wu S, Price LB, Aziz M, Nicolas-Chanoine MH, Debroy C, Robicsek A, Hansen G, Urban C, Platell J, Trott DJ, Zhanel G, Weissman SJ, Cookson BT, Fang FC, Limaye AP, Scholes D, Chattopadhyay S, Hooper DC, Sokurenko EV. 2013. Abrupt emergence of a single dominant multidrug-resistant strain of Escherichia coli. J Infect Dis 207:919–928. doi: 10.1093/infdis/jis933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Price LB, Johnson JR, Aziz M, Clabots C, Johnston B, Tchesnokova V, Nordstrom L, Billig M, Chattopadhyay S, Stegger M, Andersen PS, Pearson T, Riddell K, Rogers P, Scholes D, Kahl B, Keim P, Sokurenko EV. 2013. The epidemic of extended-spectrum-β-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. mBio 4(6):e00377-13. doi: 10.1128/mBio.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Colpan A, Johnston B, Porter S, Clabots C, Anway R, Thao L, Kuskowski MA, Tchesnokova V, Sokurenko EV, Johnson JR, VICTORY Investigators . 2013. Escherichia coli sequence type 131 (ST131) subclone H30 as an emergent multidrug-resistant pathogen among US veterans. Clin Infect Dis 57:1256–1265. doi: 10.1093/cid/cit503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Petty NK, Ben Zakour NL, Stanton-Cook M, Skippington E, Totsika M, Forde BM, Phan MD, Gomes Moriel D, Peters KM, Davies M, Rogers BA, Dougan G, Rodriguez-Bano J, Pascual A, Pitout JD, Upton M, Paterson DL, Walsh TR, Schembri MA, Beatson SA. 2014. Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci U S A 111:5694–5699. doi: 10.1073/pnas.1322678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Banerjee R, Robicsek A, Kuskowski MA, Porter S, Johnston BD, Sokurenko E, Tchesnokova V, Price LB, Johnson JR. 2013. Molecular epidemiology of Escherichia coli sequence type 131 and its H30 and H30-Rx subclones among extended-spectrum-β-lactamase-positive and -negative E. coli clinical isolates from the Chicago region, 2007 to 2010. Antimicrob Agents Chemother 57:6385–6388. doi: 10.1128/AAC.01604-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Banerjee R, Johnston B, Lohse C, Chattopadhyay S, Tchesnokova V, Sokurenko EV, Johnson JR. 2013. The clonal distribution and diversity of extraintestinal Escherichia coli isolates vary according to patient characteristics. Antimicrob Agents Chemother 57:5912–5917. doi: 10.1128/AAC.01065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tchesnokova V, Billig M, Chattopadhyay S, Linardopoulou E, Aprikian P, Roberts PL, Skrivankova V, Johnston B, Gileva A, Igusheva I, Toland A, Riddell K, Rogers P, Qin X, Butler-Wu S, Cookson BT, Fang FC, Kahl B, Price LB, Weissman SJ, Limaye A, Scholes D, Johnson JR, Sokurenko EV. 2013. Predictive diagnostics for Escherichia coli infections based on the clonal association of antimicrobial resistance and clinical outcome. J Clin Microbiol 51:2991–2999. doi: 10.1128/JCM.00984-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Peirano G, Pitout JD. 2014. Fluoroquinolone-resistant Escherichia coli sequence type 131 isolates causing bloodstream infections in a Canadian region with a centralized laboratory system: rapid emergence of the H30-Rx sublineage. Antimicrob Agents Chemother 58:2699–2703. doi: 10.1128/AAC.00119-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Johnson JR, Clermont O, Johnston B, Clabots C, Tchesnokova V, Sokurenko E, Junka AF, Maczynska B, Denamur E. 2014. Rapid and specific detection, molecular epidemiology, and experimental virulence of the O16 subgroup within Escherichia coli sequence type 131. J Clin Microbiol 52:1358–1365. doi: 10.1128/JCM.03502-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Peirano G, Bradford PA, Kazmierczak KM, Badal RE, Hackel M, Hoban DJ, Pitout JD. 2014. Global incidence of carbapenemase-producing Escherichia coli ST131. Emerg Infect Dis 20:1928–1931. doi: 10.3201/eid2011.141388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Blanco J, Mora A, Mamani R, Lopez C, Blanco M, Dahbi G, Herrera A, Marzoa J, Fernandez V, de la Cruz F, Martinez-Martinez L, Alonso MP, Nicolas-Chanoine MH, Johnson JR, Johnston B, Lopez-Cerero L, Pascual A, Rodriguez-Bano J, Spanish Group for Nosocomial Infections. 2013. Four main virotypes among extended-spectrum-β-lactamase-producing isolates of Escherichia coli O25b:H4-B2-ST131: bacterial, epidemiological, and clinical characteristics. J Clin Microbiol 51:3358–3367. doi: 10.1128/JCM.01555-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bonnin RA, Poirel L, Carattoli A, Nordmann P. 2012. Characterization of an IncFII plasmid encoding NDM-1 from Escherichia coli ST131. PLoS One 7:e34752. doi: 10.1371/journal.pone.0034752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Peirano G, Schreckenberger PC, Pitout JD. 2011. Characteristics of NDM-1-producing Escherichia coli isolates that belong to the successful and virulent clone ST131. Antimicrob Agents Chemother 55:2986–2988. doi: 10.1128/AAC.01763-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mantengoli E, Luzzaro F, Pecile P, Cecconi D, Cavallo A, Attala L, Bartoloni A, Rossolini GM. 2011. Escherichia coli ST131 producing extended-spectrum β-lactamases plus VIM-1 carbapenemase: further narrowing of treatment options. Clin Infect Dis 52:690–691. doi: 10.1093/cid/ciq194. [DOI] [PubMed] [Google Scholar]
- 101.Morris D, Boyle F, Ludden C, Condon I, Hale J, O'Connell N, Power L, Boo TW, Dhanji H, Lavallee C, Woodford N, Cormican M. 2011. Production of KPC-2 carbapenemase by an Escherichia coli clinical isolate belonging to the international ST131 clone. Antimicrob Agents Chemother 55:4935–4936. doi: 10.1128/AAC.05127-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Naas T, Cuzon G, Gaillot O, Courcol R, Nordmann P. 2011. When carbapenem-hydrolyzing β-lactamase KPC meets Escherichia coli ST131 in France. Antimicrob Agents Chemother 55:4933–4934. doi: 10.1128/AAC.00719-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kim YA, Qureshi ZA, Adams-Haduch JM, Park YS, Shutt KA, Doi Y. 2012. Features of infections due to Klebsiella pneumoniae carbapenemase-producing Escherichia coli: emergence of sequence type 131. Clin Infect Dis 55:224–231. doi: 10.1093/cid/cis387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Accogli M, Giani T, Monaco M, Giufre M, Garcia-Fernandez A, Conte V, D'Ancona F, Pantosti A, Rossolini GM, Cerquetti M. 2014. Emergence of Escherichia coli ST131 sub-clone H30 producing VIM-1 and KPC-3 carbapenemases, Italy. J Antimicrob Chemother 69:2293–2296. doi: 10.1093/jac/dku132. [DOI] [PubMed] [Google Scholar]
- 105.Ma L, Siu LK, Lin JC, Wu TL, Fung CP, Wang JT, Lu PL, Chuang YC. 2013. Updated molecular epidemiology of carbapenem-non-susceptible Escherichia coli in Taiwan: first identification of KPC-2 or NDM-1-producing E. coli in Taiwan. BMC Infect Dis 13:599. doi: 10.1186/1471-2334-13-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cai JC, Zhang R, Hu YY, Zhou HW, Chen GX. 2014. Emergence of Escherichia coli sequence type 131 isolates producing KPC-2 carbapenemase in China. Antimicrob Agents Chemother 58:1146–1152. doi: 10.1128/AAC.00912-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dimou V, Dhanji H, Pike R, Livermore DM, Woodford N. 2012. Characterization of Enterobacteriaceae producing OXA-48-like carbapenemases in the UK. J Antimicrob Chemother 67:1660–1665. doi: 10.1093/jac/dks124. [DOI] [PubMed] [Google Scholar]
- 108.Morris D, McGarry E, Cotter M, Passet V, Lynch M, Ludden C, Hannan MM, Brisse S, Cormican M. 2012. Detection of OXA-48 carbapenemase in the pandemic clone Escherichia coli O25b:H4-ST131 in the course of investigation of an outbreak of OXA-48-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 56:4030–4031. doi: 10.1128/AAC.00638-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Agabou A, Pantel A, Ouchenane Z, Lezzar N, Khemissi S, Satta D, Sotto A, Lavigne JP. 2014. First description of OXA-48-producing Escherichia coli and the pandemic clone ST131 from patients hospitalised at a military hospital in Algeria. Eur J Clin Microbiol Infect Dis 33:1641–1646. doi: 10.1007/s10096-014-2122-y. [DOI] [PubMed] [Google Scholar]
- 110.Fernandez J, Montero I, Fleites A, Rodicio MR. 2014. Cluster of Escherichia coli isolates producing a plasmid-mediated OXA-48 β-lactamase in a Spanish hospital in 2012. J Clin Microbiol 52:3414–3417. doi: 10.1128/JCM.01271-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yan JJ, Tsai LH, Wu JJ. 2012. Emergence of the IMP-8 metallo-β-lactamase in the Escherichia coli ST131 clone in Taiwan. Int J Antimicrob Agents 40:281–282. doi: 10.1016/j.ijantimicag.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 112.O'Hara JA, Hu F, Ahn C, Nelson J, Rivera JI, Pasculle AW, Doi Y. 2014. Molecular epidemiology of KPC-producing Escherichia coli: occurrence of ST131-fimH30 subclone harboring pKpQIL-like IncFIIk plasmid. Antimicrob Agents Chemother 58:4234–4237. doi: 10.1128/AAC.02182-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vimont S, Boyd A, Bleibtreu A, Bens M, Goujon JM, Garry L, Clermont O, Denamur E, Arlet G, Vandewalle A. 2012. The CTX-M-15-producing Escherichia coli clone O25b:H4-ST131 has high intestine colonization and urinary tract infection abilities. PLoS One 7:e46547. doi: 10.1371/journal.pone.0046547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Blanc V, Leflon-Guibout V, Blanco J, Haenni M, Madec JY, Rafignon G, Bruno P, Mora A, Lopez C, Dahbi G, Dunais B, Anastay M, Branger C, Moreau R, Pradier C, Nicolas-Chanoine MH. 2014. Prevalence of day-care centre children (France) with faecal CTX-M-producing Escherichia coli comprising O25b:H4 and O16:H5 ST131 strains. J Antimicrob Chemother 69:1231–1237. doi: 10.1093/jac/dkt519. [DOI] [PubMed] [Google Scholar]
- 115.Van der Bij AK, Peirano G, Pitondo-Silva A, Pitout JD. 2012. The presence of genes encoding for different virulence factors in clonally related Escherichia coli that produce CTX-Ms. Diagn Microbiol Infect Dis 72:297–302. doi: 10.1016/j.diagmicrobio.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 116.Kudinha T, Johnson JR, Andrew SD, Kong F, Anderson P, Gilbert GL. 2013. Escherichia coli sequence type 131 as a prominent cause of antibiotic resistance among urinary Escherichia coli isolates from reproductive-age women. J Clin Microbiol 51:3270–3276. doi: 10.1128/JCM.01315-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Johnson JR, Porter SB, Zhanel G, Kuskowski MA, Denamur E. 2012. Virulence of Escherichia coli clinical isolates in a murine sepsis model in relation to sequence type ST131 status, fluoroquinolone resistance, and virulence genotype. Infect Immun 80:1554–1562. doi: 10.1128/IAI.06388-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lavigne JP, Vergunst AC, Goret L, Sotto A, Combescure C, Blanco J, O'Callaghan D, Nicolas-Chanoine MH. 2012. Virulence potential and genomic mapping of the worldwide clone Escherichia coli ST131. PLoS One 7:e34294. doi: 10.1371/journal.pone.0034294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Alhashash F, Weston V, Diggle M, McNally A. 2013. Multidrug-resistant Escherichia coli bacteremia. Emerg Infect Dis 19:1699–1701. doi: 10.3201/eid1910.130309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chung HC, Lai CH, Lin JN, Huang CK, Liang SH, Chen WF, Shih YC, Lin HH, Wang JL. 2012. Bacteremia caused by extended-spectrum-β-lactamase-producing Escherichia coli sequence type ST131 and non-ST131 clones: comparison of demographic data, clinical features, and mortality. Antimicrob Agents Chemother 56:618–622. doi: 10.1128/AAC.05753-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC. 2001. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Moland ES, Black JA, Ourada J, Reisbig MD, Hanson ND, Thomson KS. 2002. Occurrence of newer β-lactamases in Klebsiella pneumoniae isolates from 24 U.S. hospitals. Antimicrob Agents Chemother 46:3837–3842. doi: 10.1128/AAC.46.12.3837-3842.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Navon-Venezia S, Leavitt A, Schwaber MJ, Rasheed JK, Srinivasan A, Patel JB, Carmeli Y, Israeli KPC Kpn Study Group . 2009. First report on a hyperepidemic clone of KPC-3-producing Klebsiella pneumoniae in Israel genetically related to a strain causing outbreaks in the United States. Antimicrob Agents Chemother 53:818–820. doi: 10.1128/AAC.00987-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-Venezia S, Carmeli Y, Brolund A, Giske CG. 2009. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother 53:3365–3370. doi: 10.1128/AAC.00126-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Giakkoupi P, Papagiannitsis CC, Miriagou V, Pappa O, Polemis M, Tryfinopoulou K, Tzouvelekis LS, Vatopoulos AC. 2011. An update of the evolving epidemic of blaKPC-2-carrying Klebsiella pneumoniae in Greece (2009-10). J Antimicrob Chemother 66:1510–1513. doi: 10.1093/jac/dkr166. [DOI] [PubMed] [Google Scholar]
- 127.Samuelsen O, Naseer U, Tofteland S, Skutlaberg DH, Onken A, Hjetland R, Sundsfjord A, Giske CG. 2009. Emergence of clonally related Klebsiella pneumoniae isolates of sequence type 258 producing plasmid-mediated KPC carbapenemase in Norway and Sweden. J Antimicrob Chemother 63:654–658. doi: 10.1093/jac/dkp018. [DOI] [PubMed] [Google Scholar]
- 128.Richter SN, Frasson I, Franchin E, Bergo C, Lavezzo E, Barzon L, Cavallaro A, Palu G. 2012. KPC-mediated resistance in Klebsiella pneumoniae in two hospitals in Padua, Italy, June 2009-December 2011: massive spreading of a KPC-3-encoding plasmid and involvement of non-intensive care units. Gut Pathog 4:7. doi: 10.1186/1757-4749-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Baraniak A, Izdebski R, Herda M, Fiett J, Hryniewicz W, Gniadkowski M, Kern-Zdanowicz I, Filczak K, Lopaciuk U. 2009. Emergence of Klebsiella pneumoniae ST258 with KPC-2 in Poland. Antimicrob Agents Chemother 53:4565–4567. doi: 10.1128/AAC.00436-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chan WW, Peirano G, Smyth DJ, Pitout JD. 2013. The characteristics of Klebsiella pneumoniae that produce KPC-2 imported from Greece. Diagn Microbiol Infect Dis 75:317–319. doi: 10.1016/j.diagmicrobio.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 131.Andrade LN, Curiao T, Ferreira JC, Longo JM, Climaco EC, Martinez R, Bellissimo-Rodrigues F, Basile-Filho A, Evaristo MA, Del Peloso PF, Ribeiro VB, Barth AL, Paula MC, Baquero F, Canton R, Darini AL, Coque TM. 2011. Dissemination of blaKPC-2 by the spread of Klebsiella pneumoniae clonal complex 258 clones (ST258, ST11, ST437) and plasmids (IncFII, IncN, IncL/M) among Enterobacteriaceae species in Brazil. Antimicrob Agents Chemother 55:3579–3583. doi: 10.1128/AAC.01783-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yoo JS, Kim HM, Yoo JI, Yang JW, Kim HS, Chung GT, Lee YS. 2013. Detection of clonal KPC-2-producing Klebsiella pneumoniae ST258 in Korea during nationwide surveillance in 2011. J Med Microbiol 62:1338–1342. doi: 10.1099/jmm.0.059428-0. [DOI] [PubMed] [Google Scholar]
- 133.Adler A, Hussein O, Ben-David D, Masarwa S, Navon-Venezia S, Schwaber MJ, Carmeli Y, Post-Acute-Care Hospital Carbapenem-Resistant Enterobacteriaceae Working Group. 2015. Persistence of Klebsiella pneumoniae ST258 as the predominant clone of carbapenemase-producing Enterobacteriaceae in post-acute-care hospitals in Israel, 2008-13. J Antimicrob Chemother 70:89–92. doi: 10.1093/jac/dku333. [DOI] [PubMed] [Google Scholar]
- 134.Gaiarsa S, Comandatore F, Gaibani P, Corbella M, Dalla Valle C, Epis S, Scaltriti E, Carretto E, Farina C, Labonia M, Landini MP, Pongolini S, Sambri V, Bandi C, Marone P, Sassera D. 2015. Genomic epidemiology of Klebsiella pneumoniae in Italy and novel insights into the origin and global evolution of its resistance to carbapenem antibiotics. Antimicrob Agents Chemother 59:389–396. doi: 10.1128/AAC.04224-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yang J, Ye L, Guo L, Zhao Q, Chen R, Luo Y, Chen Y, Tian S, Zhao J, Shen D, Han L. 2013. A nosocomial outbreak of KPC-2-producing Klebsiella pneumoniae in a Chinese hospital: dissemination of ST11 and emergence of ST37, ST392 and ST395. Clin Microbiol Infect 19:E509–E515. doi: 10.1111/1469-0691.12275. [DOI] [PubMed] [Google Scholar]
- 136.DeLeo FR, Chen L, Porcella SF, Martens CA, Kobayashi SD, Porter AR, Chavda KD, Jacobs MR, Mathema B, Olsen RJ, Bonomo RA, Musser JM, Kreiswirth BN. 2014. Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 Klebsiella pneumoniae. Proc Natl Acad Sci U S A 111:4988–4993. doi: 10.1073/pnas.1321364111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chen L, Mathema B, Pitout JD, DeLeo FR, Kreiswirth BN. 2014. Epidemic Klebsiella pneumoniae ST258 is a hybrid strain. mBio 5(3):e01355-14. doi: 10.1128/mBio.01355-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Adler A, Khabra E, Chmelnitsky I, Giakkoupi P, Vatopoulos A, Mathers AJ, Yeh AJ, Sifri CD, De Angelis G, Tacconelli E, Villegas MV, Quinn J, Carmeli Y. 2014. Development and validation of a multiplex PCR assay for identification of the epidemic ST-258/512 KPC-producing Klebsiella pneumoniae clone. Diagn Microbiol Infect Dis 78:12–15. doi: 10.1016/j.diagmicrobio.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 139.Liu Y, Wan LG, Deng Q, Cao XW, Yu Y, Xu QF. 2015. First description of NDM-1-, KPC-2-, VIM-2- and IMP-4-producing Klebsiella pneumoniae strains in a single Chinese teaching hospital. Epidemiol Infect 143:376–384. doi: 10.1017/S0950268814000995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Voulgari E, Gartzonika C, Vrioni G, Politi L, Priavali E, Levidiotou-Stefanou S, Tsakris A. 2014. The Balkan region: NDM-1-producing Klebsiella pneumoniae ST11 clonal strain causing outbreaks in Greece. J Antimicrob Chemother 69:2091–2097. doi: 10.1093/jac/dku105. [DOI] [PubMed] [Google Scholar]
- 141.Villa J, Viedma E, Brañas P, Mingorance J, Chaves F. 2014. Draft whole-genome sequence of OXA-48-producing multidrug-resistant Klebsiella pneumoniae KP_ST11_OXA-48. Genome Announc 2(4):e00737-14. doi: 10.1128/genomeA.00737-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Villa L, Capone A, Fortini D, Dolejska M, Rodriguez I, Taglietti F, De Paolis P, Petrosillo N, Carattoli A. 2013. Reversion to susceptibility of a carbapenem-resistant clinical isolate of Klebsiella pneumoniae producing KPC-3. J Antimicrob Chemother 68:2482–2486. doi: 10.1093/jac/dkt235. [DOI] [PubMed] [Google Scholar]
- 143.Lee Y, Kim BS, Chun J, Yong JH, Lee YS, Yoo JS, Yong D, Hong SG, D'Souza R, Thomson KS, Lee K, Chong Y. 2014. Clonality and resistome analysis of KPC-producing Klebsiella pneumoniae strain isolated in Korea using whole genome sequencing. Biomed Res Int 2014:352862. doi: 10.1155/2014/352862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Clancy CJ, Chen L, Hong JH, Cheng S, Hao B, Shields RK, Farrell AN, Doi Y, Zhao Y, Perlin DS, Kreiswirth BN, Nguyen MH. 2013. Mutations of the ompK36 porin gene and promoter impact responses of sequence type 258, KPC-2-producing Klebsiella pneumoniae strains to doripenem and doripenem-colistin. Antimicrob Agents Chemother 57:5258–5265. doi: 10.1128/AAC.01069-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bogdanovich T, Adams-Haduch JM, Tian GB, Nguyen MH, Kwak EJ, Muto CA, Doi Y. 2011. Colistin-resistant, Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae belonging to the international epidemic clone ST258. Clin Infect Dis 53:373–376. doi: 10.1093/cid/cir401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jayol A, Poirel L, Brink A, Villegas MV, Yilmaz M, Nordmann P. 2014. Resistance to colistin associated with a single amino acid change in protein PmrB among Klebsiella pneumoniae isolates of worldwide origin. Antimicrob Agents Chemother 58:4762–4766. doi: 10.1128/AAC.00084-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lubbert C, Lippmann N, Busch T, Kaisers UX, Ducomble T, Eckmanns T, Rodloff AC. 2014. Long-term carriage of Klebsiella pneumoniae carbapenemase-2-producing K pneumoniae after a large single-center outbreak in Germany. Am J Infect Control 42:376–380. doi: 10.1016/j.ajic.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 148.Papadimitriou-Olivgeris M, Marangos M, Fligou F, Christofidou M, Sklavou C, Vamvakopoulou S, Anastassiou ED, Filos KS. 2013. KPC-producing Klebsiella pneumoniae enteric colonization acquired during intensive care unit stay: the significance of risk factors for its development and its impact on mortality. Diagn Microbiol Infect Dis 77:169–173. doi: 10.1016/j.diagmicrobio.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 149.Giuffre M, Bonura C, Geraci DM, Saporito L, Catalano R, Di Noto S, Nociforo F, Corsello G, Mammina C. 2013. Successful control of an outbreak of colonization by Klebsiella pneumoniae carbapenemase-producing K. pneumoniae sequence type 258 in a neonatal intensive care unit, Italy. J Hosp Infect 85:233–236. doi: 10.1016/j.jhin.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 150.Endimiani A, Depasquale JM, Forero S, Perez F, Hujer AM, Roberts-Pollack D, Fiorella PD, Pickens N, Kitchel B, Casiano-Colon AE, Tenover FC, Bonomo RA. 2009. Emergence of blaKPC-containing Klebsiella pneumoniae in a long-term acute care hospital: a new challenge to our healthcare system. J Antimicrob Chemother 64:1102–1110. doi: 10.1093/jac/dkp327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Snitkin ES, Zelazny AM, Thomas PJ, Stock F, NISC Comparative Sequencing Program Group, Henderson DK, Palmore TN, Segre JA. 2012. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med 4:148ra116. doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Domenico P, Salo RJ, Cross AS, Cunha BA. 1994. Polysaccharide capsule-mediated resistance to opsonophagocytosis in Klebsiella pneumoniae. Infect Immun 62:4495–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.D'Andrea MM, Amisano F, Giani T, Conte V, Ciacci N, Ambretti S, Santoriello L, Rossolini GM. 2014. Diversity of capsular polysaccharide gene clusters in Kpc-producing Klebsiella pneumoniae clinical isolates of sequence type 258 involved in the Italian epidemic. PLoS One 9:e96827. doi: 10.1371/journal.pone.0096827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Croucher NJ, Klugman KP. 2014. The emergence of bacterial “hopeful monsters.” mBio 5(4):e01550-14. doi: 10.1128/mBio.01550-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Reference deleted.
- 156.Tzouvelekis LS, Miriagou V, Kotsakis SD, Spyridopoulou K, Athanasiou E, Karagouni E, Tzelepi E, Daikos GL. 2013. KPC-producing, multidrug-resistant Klebsiella pneumoniae sequence type 258 as a typical opportunistic pathogen. Antimicrob Agents Chemother 57:5144–5146. doi: 10.1128/AAC.01052-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Diago-Navarro E, Chen L, Passet V, Burack S, Ulacia-Hernando A, Kodiyanplakkal RP, Levi MH, Brisse S, Kreiswirth BN, Fries BC. 2014. Carbapenem-resistant Klebsiella pneumoniae exhibit variability in capsular polysaccharide and capsule associated virulence traits. J Infect Dis 210:803–813. doi: 10.1093/infdis/jiu157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lavigne JP, Cuzon G, Combescure C, Bourg G, Sotto A, Nordmann P. 2013. Virulence of Klebsiella pneumoniae isolates harboring blaKPC-2 carbapenemase gene in a Caenorhabditis elegans model. PLoS One 8:e67847. doi: 10.1371/journal.pone.0067847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pallen MJ, Loman NJ, Penn CW. 2010. High-throughput sequencing and clinical microbiology: progress, opportunities and challenges. Curr Opin Microbiol 13:625–631. doi: 10.1016/j.mib.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 160.Koser CU, Ellington MJ, Cartwright EJ, Gillespie SH, Brown NM, Farrington M, Holden MT, Dougan G, Bentley SD, Parkhill J, Peacock SJ. 2012. Routine use of microbial whole genome sequencing in diagnostic and public health microbiology. PLoS Pathog 8:e1002824. doi: 10.1371/journal.ppat.1002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sullivan CB, Diggle MA, Clarke SC. 2005. Multilocus sequence typing: data analysis in clinical microbiology and public health. Mol Biotechnol 29:245–254. doi: 10.1385/MB:29:3:245. [DOI] [PubMed] [Google Scholar]
- 162.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tenover FC. 2007. Rapid detection and identification of bacterial pathogens using novel molecular technologies: infection control and beyond. Clin Infect Dis 44:418–423. doi: 10.1086/510684. [DOI] [PubMed] [Google Scholar]
- 164.Lau SH, Cheesborough J, Kaufmann ME, Woodford N, Dodgson AR, Dodgson KJ, Bolton EJ, Fox AJ, Upton M. 2010. Rapid identification of uropathogenic Escherichia coli of the O25:H4-ST131 clonal lineage using the DiversiLab repetitive sequence-based PCR system. Clin Microbiol Infect 16:232–237. doi: 10.1111/j.1469-0691.2009.02733.x. [DOI] [PubMed] [Google Scholar]
- 165.Pitout JD, Campbell L, Church DL, Wang PW, Guttman DS, Gregson DB. 2009. Using a commercial DiversiLab semiautomated repetitive sequence-based PCR typing technique for identification of Escherichia coli clone ST131 producing CTX-M-15. J Clin Microbiol 47:1212–1215. doi: 10.1128/JCM.02265-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Endimiani A, Hujer AM, Perez F, Bethel CR, Hujer KM, Kroeger J, Oethinger M, Paterson DL, Adams MD, Jacobs MR, Diekema DJ, Hall GS, Jenkins SG, Rice LB, Tenover FC, Bonomo RA. 2009. Characterization of blaKPC-containing Klebsiella pneumoniae isolates detected in different institutions in the Eastern USA. J Antimicrob Chemother 63:427–437. doi: 10.1093/jac/dkn547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Nielsen JB, Albayati A, Jorgensen RL, Hansen KH, Lundgren B, Schonning K. 2013. An abbreviated MLVA identifies Escherichia coli ST131 as the major extended-spectrum beta-lactamase-producing lineage in the Copenhagen area. Eur J Clin Microbiol Infect Dis 32:431–436. doi: 10.1007/s10096-012-1764-x. [DOI] [PubMed] [Google Scholar]
- 168.Chen L, Chavda KD, Mediavilla JR, Zhao Y, Fraimow HS, Jenkins SG, Levi MH, Hong T, Rojtman AD, Ginocchio CC, Bonomo RA, Kreiswirth BN. 2012. Multiplex real-time PCR for detection of an epidemic KPC-producing Klebsiella pneumoniae ST258 clone. Antimicrob Agents Chemother 56:3444–3447. doi: 10.1128/AAC.00316-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Clermont O, Johnson JR, Menard M, Denamur E. 2007. Determination of Escherichia coli O types by allele-specific polymerase chain reaction: application to the O types involved in human septicemia. Diagn Microbiol Infect Dis 57:129–136. doi: 10.1016/j.diagmicrobio.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 170.Clermont O, Dhanji H, Upton M, Gibreel T, Fox A, Boyd D, Mulvey MR, Nordmann P, Ruppe E, Sarthou JL, Frank T, Vimont S, Arlet G, Branger C, Woodford N, Denamur E. 2009. Rapid detection of the O25b-ST131 clone of Escherichia coli encompassing the CTX-M-15-producing strains. J Antimicrob Chemother 64:274–277. doi: 10.1093/jac/dkp194. [DOI] [PubMed] [Google Scholar]
- 171.Weissman SJ, Johnson JR, Tchesnokova V, Billig M, Dykhuizen D, Riddell K, Rogers P, Qin X, Butler-Wu S, Cookson BT, Fang FC, Scholes D, Chattopadhyay S, Sokurenko E. 2012. High-resolution two-locus clonal typing of extraintestinal pathogenic Escherichia coli. Appl Environ Microbiol 78:1353–1360. doi: 10.1128/AEM.06663-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Blanco M, Alonso MP, Nicolas-Chanoine MH, Dahbi G, Mora A, Blanco JE, Lopez C, Cortes P, Llagostera M, Leflon-Guibout V, Puentes B, Mamani R, Herrera A, Coira MA, Garcia-Garrote F, Pita JM, Blanco J. 2009. Molecular epidemiology of Escherichia coli producing extended-spectrum β-lactamases in Lugo (Spain): dissemination of clone O25b:H4-ST131 producing CTX-M-15. J Antimicrob Chemother 63:1135–1141. doi: 10.1093/jac/dkp122. [DOI] [PubMed] [Google Scholar]
- 173.Chen L, Chavda KD, Findlay J, Peirano G, Hopkins K, Pitout JD, Bonomo RA, Woodford N, DeLeo FR, Kreiswirth BN. 2014. Multiplex PCR for identification of two capsular types in epidemic KPC-producing Klebsiella pneumoniae sequence type 258 strains. Antimicrob Agents Chemother 58:4196–4199. doi: 10.1128/AAC.02673-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Clark AE, Kaleta EJ, Arora A, Wolk DM. 2013. Matrix-assisted laser desorption ionization–time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology. Clin Microbiol Rev 26:547–603. doi: 10.1128/CMR.00072-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lartigue MF. 2013. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry for bacterial strain characterization. Infect Genet Evol 13:230–235. doi: 10.1016/j.meegid.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 176.Matsumura Y, Yamamoto M, Nagao M, Tanaka M, Machida K, Ito Y, Takakura S, Ichiyama S. 2014. Detection of extended-spectrum-β-lactamase-producing Escherichia coli ST131 and ST405 clonal groups by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 52:1034–1040. doi: 10.1128/JCM.03196-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Novais A, Sousa C, de Dios Caballero J, Fernandez-Olmos A, Lopes J, Ramos H, Coque TM, Canton R, Peixe L. 2014. MALDI-TOF mass spectrometry as a tool for the discrimination of high-risk Escherichia coli clones from phylogenetic groups B2 (ST131) and D (ST69, ST405, ST393). Eur J Clin Microbiol Infect Dis 33:1391–1399. doi: 10.1007/s10096-014-2071-5. [DOI] [PubMed] [Google Scholar]
- 178.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 179.Villa L, Garcia-Fernandez A, Fortini D, Carattoli A. 2010. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother 65:2518–2529. doi: 10.1093/jac/dkq347. [DOI] [PubMed] [Google Scholar]
- 180.Suzuki H, Yano H, Brown CJ, Top EM. 2010. Predicting plasmid promiscuity based on genomic signature. J Bacteriol 192:6045–6055. doi: 10.1128/JB.00277-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Johnson TJ, Wannemuehler YM, Johnson SJ, Logue CM, White DG, Doetkott C, Nolan LK. 2007. Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates. Appl Environ Microbiol 73:1976–1983. doi: 10.1128/AEM.02171-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Heuer H, Smalla K. 2012. Plasmids foster diversification and adaptation of bacterial populations in soil. FEMS Microbiol Rev 36:1083–1104. doi: 10.1111/j.1574-6976.2012.00337.x. [DOI] [PubMed] [Google Scholar]
- 183.Rasko DA, Webster DR, Sahl JW, Bashir A, Boisen N, Scheutz F, Paxinos EE, Sebra R, Chin CS, Iliopoulos D, Klammer A, Peluso P, Lee L, Kislyuk AO, Bullard J, Kasarskis A, Wang S, Eid J, Rank D, Redman JC, Steyert SR, Frimodt-Moller J, Struve C, Petersen AM, Krogfelt KA, Nataro JP, Schadt EE, Waldor MK. 2011. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med 365:709–717. doi: 10.1056/NEJMoa1106920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Smets BF, Barkay T. 2005. Horizontal gene transfer: perspectives at a crossroads of scientific disciplines. Nat Rev Microbiol 3:675–678. doi: 10.1038/nrmicro1253. [DOI] [PubMed] [Google Scholar]
- 185.Chen L, Chavda KD, Melano RG, Jacobs MR, Levi MH, Bonomo RA, Kreiswirth BN. 2013. Complete sequence of a blaKPC-2-harboring IncFIIK1 plasmid from a Klebsiella pneumoniae sequence type 258 strain. Antimicrob Agents Chemother 57:1542–1545. doi: 10.1128/AAC.02332-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chmelnitsky I, Shklyar M, Hermesh O, Navon-Venezia S, Edgar R, Carmeli Y. 2013. Unique genes identified in the epidemic extremely drug-resistant KPC-producing Klebsiella pneumoniae sequence type 258. J Antimicrob Chemother 68:74–83. doi: 10.1093/jac/dks370. [DOI] [PubMed] [Google Scholar]
- 187.Adler A, Paikin S, Sterlin Y, Glick J, Edgar R, Aronov R, Schwaber MJ, Carmeli Y. 2012. A swordless knight: epidemiology and molecular characteristics of the blaKPC-negative sequence type 258 Klebsiella pneumoniae clone. J Clin Microbiol 50:3180–3185. doi: 10.1128/JCM.00987-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Osborn AM, da Silva Tatley FM, Steyn LM, Pickup RW, Saunders JR. 2000. Mosaic plasmids and mosaic replicons: evolutionary lessons from the analysis of genetic diversity in IncFII-related replicons. Microbiology 146(Part 9):2267–2275. [DOI] [PubMed] [Google Scholar]
- 189.Woodford N, Carattoli A, Karisik E, Underwood A, Ellington MJ, Livermore DM. 2009. Complete nucleotide sequences of plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrob Agents Chemother 53:4472–4482. doi: 10.1128/AAC.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Zhao WH, Hu ZQ. 2013. Epidemiology and genetics of CTX-M extended-spectrum beta-lactamases in Gram-negative bacteria. Crit Rev Microbiol 39:79–101. doi: 10.3109/1040841X.2012.691460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Shin J, Choi MJ, Ko KS. 2012. Replicon sequence typing of IncF plasmids and the genetic environments of blaCTX-M-15 indicate multiple acquisitions of blaCTX-M-15 in Escherichia coli and Klebsiella pneumoniae isolates from South Korea. J Antimicrob Chemother 67:1853–1857. doi: 10.1093/jac/dks143. [DOI] [PubMed] [Google Scholar]
- 192.Zong Z. 2013. Complete sequence of pJIE186-2, a plasmid carrying multiple virulence factors from a sequence type 131 Escherichia coli O25 strain. Antimicrob Agents Chemother 57:597–600. doi: 10.1128/AAC.01081-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Leavitt A, Chmelnitsky I, Carmeli Y, Navon-Venezia S. 2010. Complete nucleotide sequence of KPC-3-encoding plasmid pKpQIL in the epidemic Klebsiella pneumoniae sequence type 258. Antimicrob Agents Chemother 54:4493–4496. doi: 10.1128/AAC.00175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Chen L, Chavda KD, Melano RG, Jacobs MR, Koll B, Hong T, Rojtman AD, Levi MH, Bonomo RA, Kreiswirth BN. 2014. Comparative genomic analysis of KPC-encoding pKpQIL-like plasmids and their distribution in New Jersey and New York hospitals. Antimicrob Agents Chemother 58:2871–2877. doi: 10.1128/AAC.00120-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Leavitt A, Chmelnitsky I, Ofek I, Carmeli Y, Navon-Venezia S. 2010. Plasmid pKpQIL encoding KPC-3 and TEM-1 confers carbapenem resistance in an extremely drug-resistant epidemic Klebsiella pneumoniae strain. J Antimicrob Chemother 65:243–248. doi: 10.1093/jac/dkp417. [DOI] [PubMed] [Google Scholar]
- 196.Tofteland S, Naseer U, Lislevand JH, Sundsfjord A, Samuelsen O. 2013. A long-term low-frequency hospital outbreak of KPC-producing Klebsiella pneumoniae involving intergenus plasmid diffusion and a persisting environmental reservoir. PLoS One 8:e59015. doi: 10.1371/journal.pone.0059015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Chmelnitsky I, Shklyar M, Leavitt A, Sadovsky E, Navon-Venezia S, Ben Dalak M, Edgar R, Carmeli Y. 2014. Mix and match of KPC-2 encoding plasmids in Enterobacteriaceae-comparative genomics. Diagn Microbiol Infect Dis 79:255–260. doi: 10.1016/j.diagmicrobio.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 198.Chen L, Chavda KD, Melano RG, Hong T, Rojtman AD, Jacobs MR, Bonomo RA, Kreiswirth BN. 2014. Molecular survey of the dissemination of two blaKPC-harboring IncFIA plasmids in New Jersey and New York hospitals. Antimicrob Agents Chemother 58:2289–2294. doi: 10.1128/AAC.02749-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.García-Fernández A, Villa L, Carta C, Venditti C, Giordano A, Venditti M, Mancini C, Carattoli A. 2012. Klebsiella pneumoniae ST258 producing KPC-3 identified in Italy carries novel plasmids and OmpK36/OmpK35 porin variants. Antimicrob Agents Chemother 56:2143–2145. doi: 10.1128/AAC.05308-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Chen L, Chavda KD, Al Laham N, Melano RG, Jacobs MR, Bonomo RA, Kreiswirth BN. 2013. Complete nucleotide sequence of a blaKPC-harboring IncI2 plasmid and its dissemination in New Jersey and New York hospitals. Antimicrob Agents Chemother 57:5019–5025. doi: 10.1128/AAC.01397-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. 2012. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin Microbiol Rev 25:682–707. doi: 10.1128/CMR.05035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Woese CR. 1987. Bacterial evolution. Microbiol Rev 51:221–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.de Visser JA, Krug J. 2014. Empirical fitness landscapes and the predictability of evolution. Nat Rev Genet 15:480–490. doi: 10.1038/nrg3744. [DOI] [PubMed] [Google Scholar]
- 204.Morran LT, Schmidt OG, Gelarden IA, Parrish RC II, Lively CM. 2011. Running with the Red Queen: host-parasite coevolution selects for biparental sex. Science 333:216–218. doi: 10.1126/science.1206360. [DOI] [PMC free article] [PubMed] [Google Scholar]