SUMMARY
Pioneer transcription factors (TFs) access silent chromatin and initiate cell fate changes, using diverse types of DNA binding domains (DBDs). FoxA, the paradigm pioneer TF, has a winged helix DBD that resembles linker histone and thereby binds its target sites on nucleosomes and in compacted chromatin. Herein we compare the nucleosome and chromatin targeting activities of Oct4 (POU DBD), Sox2 (HMG box DBD), Klf4 (zinc finger DBD), and c-Myc (bHLH DBD), which together reprogram somatic cells to pluripotency. Purified Oct4, Sox2, and Klf4 proteins can bind nucleosomes in vitro, and in vivo they preferentially target silent sites enriched for nucleosomes. Pioneer activity relates simply to the ability of a given DBD to target partial motifs displayed on the nucleosome surface. Such partial motif recognition can occur by coordinate binding between factors. Our findings provide insight into how pioneer factors can target naïve chromatin sites.
INTRODUCTION
Silent chromatin is packed with nucleosomes, acting as a barrier to targeting by most transcription factors (TFs) (Adams and Workman, 1995; Mirny, 2010). However, a select group of transcription factors (TFs) known as pioneer factors have the combined ability to access their target sites in silent chromatin and initiate cell fate changes (Iwafuchi-Doi and Zaret, 2014; Zaret and Carroll, 2011). The winged-helix DBD of the pioneer factor FoxA (Clark et al., 1993), which is similar to that of linker histone (Ramakrishnan et al., 1993), allows the protein to bind its DNA motif exposed on a nucleosome and access to silent chromatin (Cirillo and Zaret, 1999; Cirillo et al., 1998, 2002). Such activity is necessary for liver induction (Lee et al., 2005). Other TFs involved in cell reprogramming can target their sites in silent chromatin (Montserrat et al., 2013; Soufi et al., 2012; Takahashi and Yamanaka, 2006; Wapinski et al., 2013), but they possess DBDs that differ from that of FoxA. Whether such reprogramming factors directly bind nucleosomes and how the structures of their respective DBDs relate to nucleosome binding and hence, pioneer activity, has not been assessed.
Transcription factors containing major structural classes of DBDs, including Pit-Oct-Unc (POU), Sry-related High Mobility Group (HMG), Zinc Fingers (ZF), and basic-Helix-Loop-Helix (bHLH), represented by O, S, K, and M, respectively, have been used in the most dramatic example of cellular reprogramming: the conversion of differentiated cells into induced pluripotent stem cells (Takahashi and Yamanaka, 2006). We previously compared genomic chromatin features of human fibroblasts, prior to the ectopic expression of OSKM, to where the factors first bind the genome during their initial expression (Soufi et al., 2012). This allowed us to assess how OSKM target pre-existing states in chromatin, as opposed to assessing chromatin states after the factors are bound. The data showed that Oct4, Sox2, and Klf4, but not c-Myc, could function as pioneers during reprogramming by virtue of their ability to mostly target “closed” chromatin sites that are DNaseI-resistant and “naïve” by virtue of lacking evident active histone modifications (Soufi et al., 2012). Recently, single molecule imaging analysis using fluorescently-tagged proteins monitored in living cells proposed that Sox2 guides Oct4 to its target sites (Chen et al., 2014); the chromatin status of the sites was unknown. However, we previously found that the ectopic Oct4 and Sox2 bind most extensively to separate sites in chromatin (Soufi et al., 2012), leaving open how the bulk of chromatin targeting is achieved. While many of initial binding events were promiscuous and not retained in pluripotent cells, many others occurred at target genes that are required for conversion to pluripotency.
Ascl1, Pax7, and Pu.1, have emerged as pioneer transcription factors based on targeting closed chromatin and their ability to reprogram cells, though assessments of direct interaction with nucleosomes has been lacking (Barozzi et al., 2014; Budry et al., 2012; Wapinski et al., 2013). In light of the bHLH factor c-Myc being unable to bind closed chromatin on its own (Soufi et al., 2012), it was surprising that Ascl1, another bHLH factor, can bind closed chromatin during reprogramming fibroblasts to neuron-like cells (Wapinski et al., 2013). Studies that have examined the correlation between co-existing TF binding and nucleosome occupancy, without characterizing the “pre-bound” chromatin state, could not address questions about initial chromatin access.
Generating induced pluripotent stem (iPS) cells, using the OSKM factors, has proven to be highly valuable for research, with great potential for regenerative medicine (Robinton and Daley, 2012). In an attempt to increase the efficiency of reprogramming, efforts have focused on explaining how somatic cells respond to the ectopic expression of OSKM (Buganim et al., 2013; Papp and Plath, 2013; Soufi, 2014). To gain insights into the molecular mechanisms that impart OSKM access to closed chromatin, we measured the fundamental interaction between the factors and nucleosomes, in vivo and in vitro, by three mutually supportive approaches: biochemical assays, genomics, and structural analysis. We find that the inherent ability of DBDs to recognize one face of DNA on nucleosome, as seen by targeting a part of their canonical motif on nucleosome-enriched sequences in chromatin, is the primary determinant of pioneer factor activity. These findings can explain the pioneer activity of a diverse set of reprogramming factors containing different structural classes of DBDs as well as the synergistic behavior of pioneer and non-pioneer factors.
RESULTS
O, S, K, and M show a range of nucleosome binding in vitro
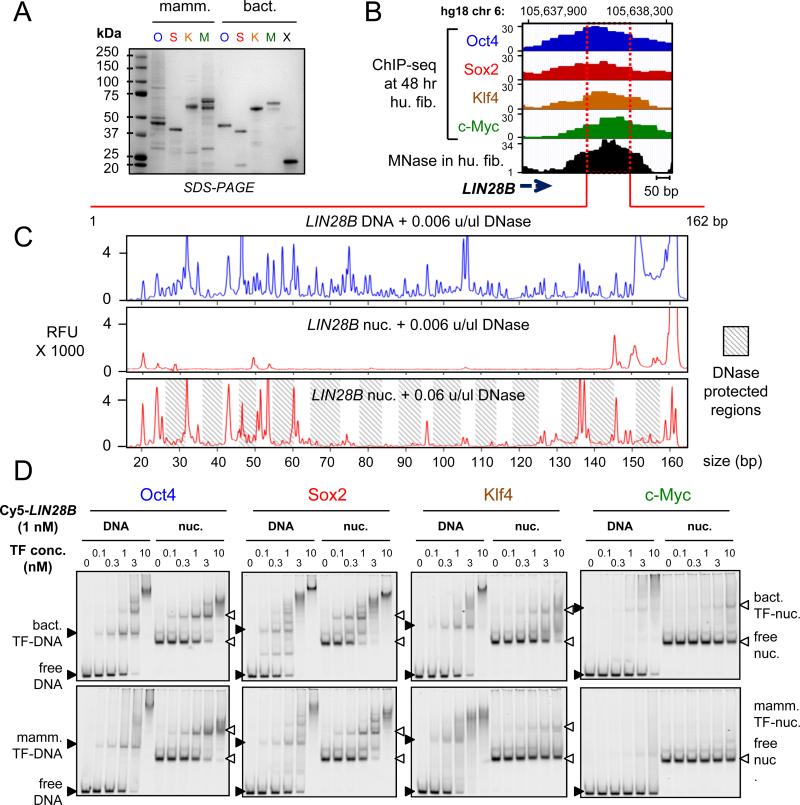
The interaction of full-length O, S, K, and M, as used in reprogramming, with nucleosomes is not known. Therefore, we purified and refolded the full-length O, S, and K factors, along with c-Myc and its obligate heterodimerization partner Max from bacterial cells, representing post-translationally unmodified proteins (Figures 1A, S1A). We also obtained the full-length O, S, K and M expressed in human HEK293 cells and purified under native conditions, representing post-translationally modified versions of the proteins (Figure 1A). To quantify the DNA binding activities of the proteins, the apparent equilibrium dissociation constants (Kd) were determined using two different methods: from the decrement in the amount of free DNA (total Kd) and from the appearance of the first DNA-bound complex (specific Kd), in electrophoretic mobility shift assays (EMSA). As expected, the bacterial (bact.) and the mammalian (mamm.) – expressed, recombinant O, S, K, and M proteins bound to DNA probes containing canonical motifs, as previously reported for the purified DBDs (Farina et al., 2004; Nakatake et al., 2006; Rodda et al., 2005) (Figure S1B, Table 1), with much lower affinity to non-specific DNA sequences of the same length (Figure S1C). The bact. reconstituted Myc:Max heterodimers formed a complex that migrated more slowly than Max homodimers, and no protein-DNA complexes with similar mobility to Max homodimers were observed even at the highest concentrations, confirming that the c-Myc:Max preparation did not contain Max homodimers (Figure S1B). The mamm. c-Myc did not show any specific DNA binding activity in the absence of its partner Max, as seen previously (Wechsler et al., 1994). These data demonstrate that the recombinant full-length OSKM proteins were highly active in specific DNA binding.
Figure 1. O, S, K, and M display differential affinity to nucleosomes in vitro.
(A) Recombinant purified mammalian and bacterial O, S, K, M, and bacterial Max (X) proteins analyzed bySDS-PAGE and Coomassie staining. The respective OSKMX bands run at the expected sizes when compared to the sizes of protein standards. The OSKM DNA binding activity and specificity is shown in Figure S1A-C.
(B) O, S, K, and M ChIP-seq profiles (blue, red, orange, and green, respectively) 48 hr post-induction, and MNase-seq profile (black) in fibroblasts across the LIN28B locus within the displayed genomic location.
(C) DNase-I footprinting showing the protection of LIN28B-DNA before and after nucleosome reconstitution in vitro. Electropherograms of 5’-6FAM end-labeled LIN28B (top strand) oligonucleotides generated by digesting free DNA (blue) and nucleosomal DNA (red) with DNaseI. The amount of DNase-I used are indicated on top of each panel. Shaded boxes represent the DNase-I protected regions within LIN28B-nuc in the expected ~ 10 bp pattern. See Figure S1D for details about nucleosome reconstitution.
(D) Representative EMSA showing the affinity of increasing amounts of recombinant O, S, K, and M proteins (bact. top panels and mamm. bottom panels) to Cy5-labelled LIN28B-DNA (left panels) and LIN28B-nucleosome (right panels). EMSA of O, S K, and M to DNA probes containing specific and non-specific targets are shown in Figure S1B and S1C.
Table 1.
Recombinant O, S, K, and M show a range of affinities to nucleosomes. (related to Figure 1)
| Oct4 | Sox2 | Klf4 | c-Myc | |||||
|---|---|---|---|---|---|---|---|---|
| Apparent Kd (nM) | bact. | mamm. | bact. | mamm. | bact. | mamm. | bact. | mamm. |
| total canonical | 0.61 | 0.64 | 0.37 | 0.98 | 2.49 | 1.46 | 1.88 | ND |
| specific canonical | 0.76 | 1.04 | 0.45 | 1.50 | 3.18 | 1.95 | 0.77 | ND |
| total LIN28B DNA | 0.75 | 0.93 | 0.38 | 1.46 | 1.25 | 0.41 | 8.28 | ND |
| specific LIN28B DNA | 0.92 | 2.05 | 0.68 | 3.83 | 2.26 | 1.12 | 6.25 | ND |
| total LIN28B nuc. | 1.09 | 1.34 | 0.34 | 1.06 | 5.96 | 3.45 | ND | ND |
| specific LIN28B nuc. | 1.17 | 1.84 | 0.39 | 1.43 | 7.21 | 13.97 | ND | ND |
Apparent dissociation constants (Kd) were derived from EMSA to represent the relative affinities of bacterial (bact.) and mammalian (mamm.) O, S, K, and M to their canonical sites, LIN28B free DNA, and LIN28B nucleosomes (nuc.). Apparent Kd were derived from two separate binding curves representing two experimental replicates, fitted to the experimental data within R2 values of ~ 0.97, and expressed in nM units. Apparent Kd were quantified from the fractional decrement of free DNA or nuc, designated as “total” binding, or from the first bound-DNA/nuc complexes, representing “specific” binding.
To measure the direct interactions between OSKM and nucleosomes, we identified a nucleosome-enriched site in the fibroblast genome that is efficiently targeted by OSKM (Soufi et al., 2012), focusing on the LIN28B locus that is important for reprogramming and pluripotency (Shyh-Chang et al., 2013; Yu et al., 2007). RNA-seq data showed that LIN28B is silent in human fibroblasts and remains silent after 48 hr OSKM induction, revealing that OSKM binding precedes LIN28B gene activation (data not shown). We selected a region downstream of the LIN28B poly(A) site that is strongly enriched for a nucleosome in pre-induced human fibroblasts, as measured by MNase-seq (Kelly et al., 2012) and was targeted by all four factors at 48 hr post-induction (Figure 1B). We used PCR on human fibroblast DNA to generate a 162 bp, Cy5-labelled LIN28B-DNA, which was assembled into nucleosomes (LIN28B-nuc) by salt gradient dilution with purified recombinant human histones (Figure S1D). The nucleosomes exhibited protection from low concentrations of DNase-I except at the ends of the LIN28B fragment, compared to free DNA, indicating translational positioning around the center of the 162 bp LIN28B sequence (Figure 1C, top two boxes), similar to the observed position of the center of the MNase-seq peak (Figure 1B and 1C). Ten-fold higher concentrations of DNase generated an approximately 10 bp DNase-cleavage repeat pattern on LIN28B-nuc, reflecting rotational positioning of nucleosomes within the population (Figure 1C, bottom).
It is generally accepted that nucleosomes act as a barrier to DNA binding by TFs (see Introduction), though exceptions have been noted (Perlmann and Wrange, 1988). Interestingly, Oct4, Sox2, and Klf4, but not c-Myc:Max, showed binding to the LIN28B-nuc (Figure 1D). Remarkably, both mamm. and bact. Oct4 and Sox2 showed similar or lower apparent Kd values for LIN28B-nuc compared to LIN28B-DNA, indicating similar or higher affinity to nucleosome than to free DNA (Figure 1D, Table 1). On the other hand, Klf4 was able to bind LIN28B-nuc with a higher apparent Kd value compared to free DNA, indicating substantial nucleosome binding, but at a lower affinity than to free DNA (Figure 1D, Table 1). c-Myc:Max did not yield saturated binding to LIN28B-nuc, even at the highest concentrations of protein used, and thus the apparent Kd must be in the μM range (Figure 1D, Table 1). In conclusion, both mammalian and bacterial expressed O, S, K, and M exhibit the same relative range of affinities to LIN28B-nuc and O, S, and K have an independent nucleosome binding activity.
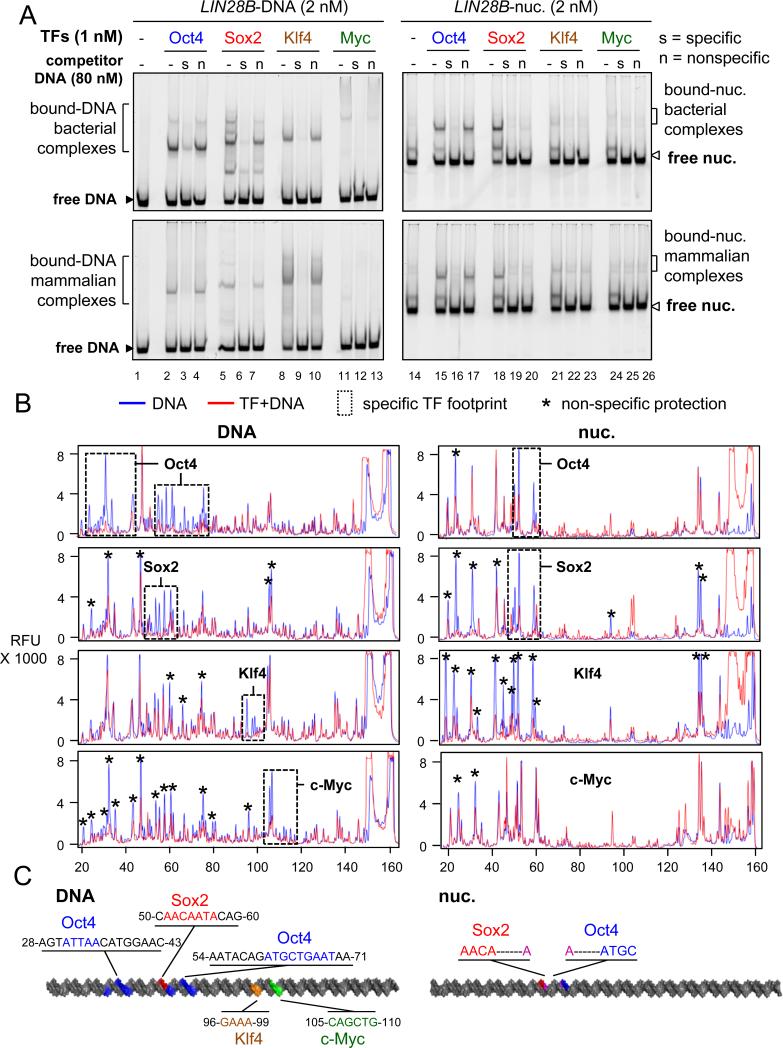
Specific and non-specific DNA interactions contribute to nucleosome binding
It is well recognized that TFs show both sequence-specific and non-specific interactions with their DNA targets (Biggin, 2011). To measure the contribution of specificity on OSK binding to LIN28B nucleosomes, we carried out EMSA in the presence of increasing amounts of specific and non-specific DNA sequences that we had already characterized as competitors (Figure S1B, S1C, Table 1). EMSA competition experiments show that a 40-fold molar excess of non-labelled DNA probes containing specific binding sites, but not probes containing non-specific sequences, can displace LIN28B-DNA complexes with each of the OSKM proteins, indicating specific interaction with LIN28B-DNA (Figure 2A, left panel), similar to OSKM interaction with their canonical sites (Figure S2A). As expected, bact. and mamm. O, S, or K in complexes with LIN28B-nuc were displaced in the presence of a 40X molar excess of unlabeled, specific competitors (Figures 2A, lane 16, 19, and 22). A 40X or lower (range from 5X to 20X) molar excess of non-specific DNA failed to displace bact. and mamm. Oct4 from the LIN28B-nuc (Figures 2A, lane 17; S2B, lanes 14-16), demonstrating specific binding by Oct4 to the nucleosomes in vitro.
Figure 2. The contribution of non-specific binding to nucleosome targeting in vitro.
(A) Representative EMSA showing the affinity of recombinant O, S, K, M proteins (bact. top panels and mamm. bottom panels) to LIN28B-DNA (left panels) and LIN28B-nucleosome (right panels) in the presence of 40 fold molar excess of specific competitor ( “s” lanes) or non-specific competitor (“n” lanes) or absence of competitor (“-“ lanes). Competition assays showing the specificity of O, S, K, and M to their canonical DNA probes and to LIN28B DNA and nucleosome under lower titration of competitor is shown in Figure S2.
(B) DNase-I footprinting showing the protection of LIN28B-DNA (left panels) and LIN28B-nuc (right panels) in the absence (blue lines) or presence (red lines) of O, S, K, and M. Electropherograms of 5’-6FAM end-labeled LIN28B (top strand) oligonucleotides generated by DNase-I digestion of DNA (0.006 U) and nucleosomal DNA (0.06 U). Dashed boxes and stars represent specific and non-specific sites protected by O, S, K, and M, respectively.
(C) A cartoon representation of the 162 bp LIN28B DNA (left) and nucleosome (right) highlighting the binding sites of O, S, K, and M in vitro in blue, red, orange, and green, respectively, as measured by DNase-I footprining. The protected DNA sequences are indicated.
By contrast, a 40X excess of non-specific DNA competed almost all of Sox2 and Klf4 from binding to LIN28B-nuc (Figure 2A lanes 20 and 23).Importantly, lower levels of non-specific competitor, from 5X to 20X, did not compete to the same extent as specific competitor with LIN28B-nuc for binding either Sox2 or Klf4 (Figure S2C and S2D, compare lanes 10, to 11-13 vs 14-16). Thus, both specific and non-specific interactions contribute to Sox2 and Klf4 binding to nucleosomes in vitro.
DNase footprinting showed that each of the O, S, K, and M factors protect sequences on LIN28B free DNA that resemble their canonical motifs (Figure 2B, 2C, dash boxes). In addition, at the concentrations used for footprinting, Sox2, Klf4, and c-Myc also show non-specific protection of the LIN28B free DNA (Figure 2B, peaks labeled by asterisks). DNase footprinting of LIN28B-nuc bound to Oct4 and Sox2 show that the factors protect part of their canonical motifs, agreeing with the specific binding to nucleosomes seen with EMSA competition experiments (Figure 2B, 2C). However, Sox2 and Klf4 protect both specific and non-specific nucleotides on LIN28B-nuc, supporting the non-specific contribution of Sox2 and Klf4 to nucleosomes as seen in EMSA competition experiments (Figure 2B). The Klf4 binding site is close to the predicted nucleosome dyad axis, where DNase cleavage is minimal; thus precluding an accurate assessment of specific footprinting. Expectedly, c-Myc showed minimal protection of LIN28B-nuc, confirming the weak affinity to nucleosomes. Altogether, the O, S, and K reprogramming factors employ specific and nonspecific nucleosome interactions to different extents.
Range of nucleosome binding in vitro is observed in genome targeting in vivo
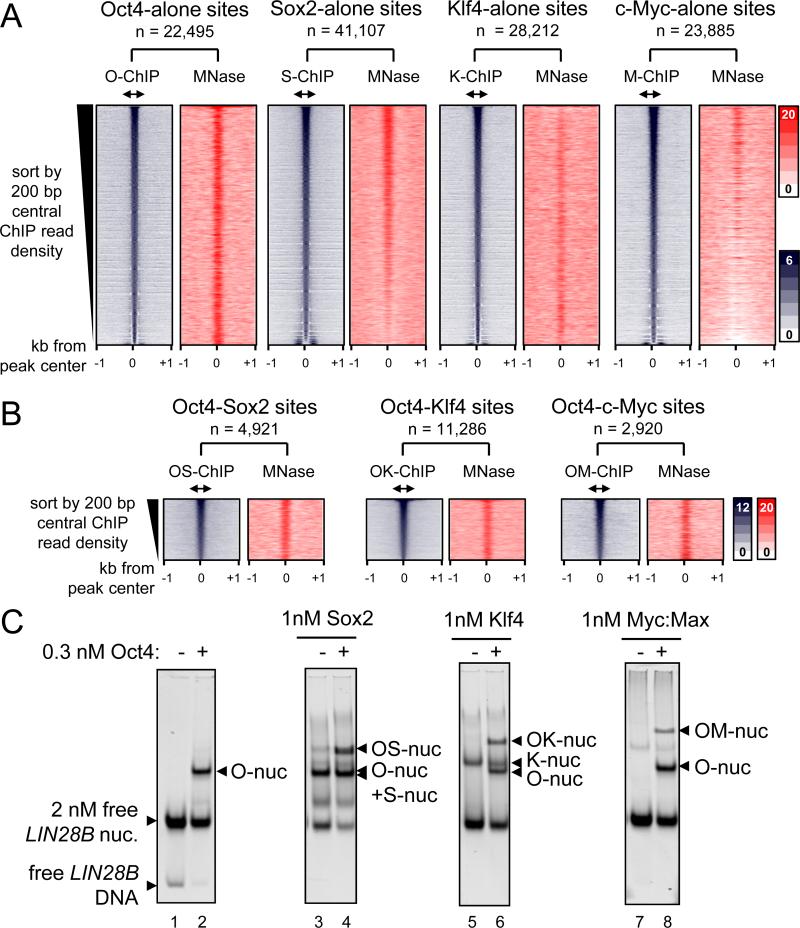
We assessed whether OSKM, 48 hr post-induction, targeted sites with pre-existing nucleosome enrichment in fibroblast chromatin. Pooling seven replicates from the MNase-seq data set (GSM543311) allowed a high-resolution map of nucleosomes with 6.6-fold genome coverage. First, we curated the sites where O, S, K, or M targeted alone, by identifying O, S, K, or M peaks that are 500 bp or more apart from each other. The sites were arranged in rank order by the number of ChIP-seq tags in the central 200 bp, from high to low affinity sites. This analysis confirms that each of the O, S, K, and M factors is highly enriched at the central 200 bp within a 2 kb region (Figure 3A, blue boxes). Interestingly, Sox2 bound most frequently alone (n = 41,107) compared to Oct4 (n = 22,495), Klf4 (n = 28,212), and c-Myc (n = 23,885). Subsequently, MNase tags across the respective 2 kb regions were counted, reflecting local nucleosome enrichment. Read-density heatmaps showed a range of nucleosome enrichment at the central 200 bp regions that were targeted by O, S, K, or M factors alone (Figure 3A, red boxes). Notably, Oct4 targets were the most highly enriched for nucleosomes, followed by Sox2, and then Klf4 throughout the respective TF rank-ordered binding profiles. By contrast, MNase tags in the c-Myc targeted sites were diminished. Also, we did not observe pre-phased arrays of nucleosomes at OSKM target sites, indicating that the initial association with nucleosomes proceeds repositioning, if any. Remarkably, the extent of nucleosome targeting of O, S, K, and M in vivo correlates with the relative abilities of the factors to bind nucleosomes in vitro (Figure 1D, Table 1).
Figure 3. O, S, K, and M display a range of nucleosome targeting in vivo.
(A) Read density heatmaps (in color scales) showing the intensity of O, S, K, and M ChIP-seq signal (blue) and MNase-seq (red) spanning ± 1 kb from the center of the O, S, K, and M peaks where each factor binds alone within 500 bp threshold. The analyzed sequences were organized in rank order, from high to low number ChIP-seq reads within the central 200 bp (double arrows). The number of targeted sites is indicated.
(B) As in (A), but showing where the OS, OK, and OM factors peaks are within 100 bp or less apart from each other. The full possible OSKM combinations are shown in Figure S3.
(C) The binding affinity of S, K, and M (1 nM) in the presence of Oct4 (0.3 nM) to LIN28B nucleosomal DNA (lanes; 4, 6, and 8, respectively) or absence of Oct4 (lanes; 3, 5, and 7). The binding of Oct4 on its own (lane 2) and free LIN28B nucleosomes (lane 1) are indicated. The histone content of the nucleosome bound complexes are shown in Figure S4.
To assess the contribution of non-specific binding in vivo, we counted the number of O, S, K, and M peaks at 48 hr post-induction as function of FDR threshold. Remarkably, while O, K, and M peak numbers begin to stabilize above an FDR of 0.5% (used in our study) (slopes of 1.6, 1.5, and 1.3 respectively), the number of Sox2 peaks continues to increase (slope of 2.1) with higher FDR (Figure S3A). Thus it appears that Sox2 employs a measure of non-specific targeting in vivo, as we observed in vitro.
O, S, K, and/or M synergistic targeting of nucleosomes in vivo and in vitro
It has been previously suggested that transcription factors can access nucleosomal DNA by cooperative binding in order to compete with histones (Polach and Widom, 1996). To investigate the contribution of synergy between O, S, K, and/or M to nucleosome targeting, we studied sites that were co-targeted by multiple factors within a range of 100 bp or less from each other; i.e. within one nucleosome. In general, we observed that all possible O, S, K, and/or M combinations targets were enriched for nucleosomes except for KM targets, and the co-bound sites, on average, were more enriched for nucleosomes than singly bound sites (Figures 3B and S3B). Notably, there were more S, K, and/or M combinations that included Oct4 and showed higher nucleosome enrichment at initially targeted sites, compared to binding combinations lacking Oct4 (Figures 3B and S3; compare panels C-I to J-M). For example, c-Myc showed the most nucleosome targeting when co-bound with Oct4, followed by with Sox2, while c-Myc showed weak targeting to nucleosomes with Klf4 (Figure S3; compare panels E to K and M). Interestingly, the KM combination was the most frequent at nucleosome-depleted promoters, similar to KM targeting DNase hypersensitive regions (Soufi et al., 2012) (Figure S3M; red plot). Nevertheless, KM still targeted nucleosome-enriched sites at TSS-distal regions (Figure S3M; blue plot).
To further investigate synergistic targeting with Oct4, we assessed binding by each of the bact. Sox2, Klf4, and c-Myc:Max (1 nM) to the reconstituted LIN28B-nuc (2 nM) in the presence of low amounts of Oct4 (0.3 nM). EMSA showed that all the three recombinant proteins are able to bind with Oct4 to nucleosomal DNA in vitro, forming higher order complexes (Figures 3C). Notably, c-Myc:Max binding to LIN28B-nuc was enabled in the presence of Oct4 (Figures 3C, right panel). To assess the presence of histones in the LIN28B-nuc in the complexes, we transferred the proteins from an EMSA gel to a PVDF membrane and blotted for H3 and H2B (Figure S4). Though the c-Myc antibody was the weakest, all LIN28B-nuc-bound complexes showed detectable amounts of H3, and to a lesser extent H2B, indicating the factors bind together to nucleosomes. In summary, Oct4, Sox2, and Klf4 enable c-Myc to target nucleosomal sites both in vivo and in vitro.
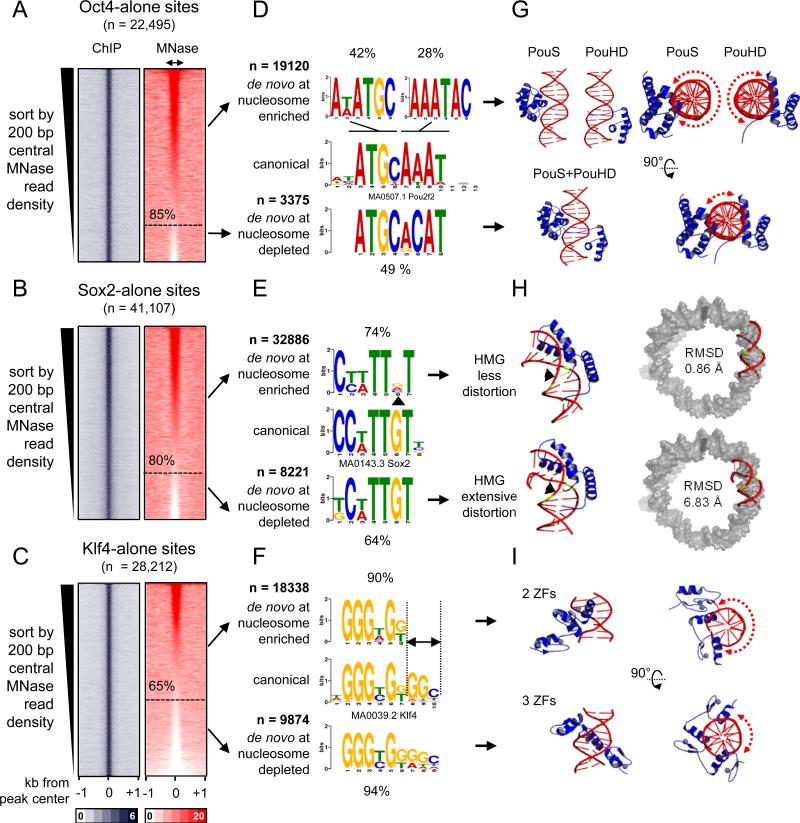
O, S, and K separately recognize partial motifs on nucleosomes
To identify DNA motifs that are associated with O, S, K alone targeting to nucleosomes in vivo, the respective targeted sites were rank ordered according to nucleosome enrichment in the central 200 bp. This allowed us to separate nucleosome-enriched from nucleosome-depleted regions that were individually targeted by O, S, or K. By these criteria, 85%, 80%, and 65% of the genomic sites initially targeted by Oct4, Sox2, and Klf4, respectively, were enriched for nucleosomes (Figure 4A-C, red boxes). We used de novo motif analysis, separately analyzing the targets that were enriched for nucleosomes (Figure 4A-C, red boxes, upper portion) from those that were depleted of nucleosomes; i.e., free DNA targets (Figure 4A-C, red boxes, lower portion). While O, S, and K primarily targeted sequences similar to their canonical motifs at nucleosome-depleted and nucleosome-enriched sites, motifs occurring at nucleosome-enriched sites showed distinctive features (Figure 4D-F).
Figure 4. O, S, and K recognize partial motifs on nucleosomes.
(A-C) Same as in Figure 3A, but the sites were organizing in a descending rank order according to the MNase-seq tags within the central 200 bp. The nucleosome enriched sites were separated from the nucleosome depleted sites (dashed line) for each factor. (D-F) Logo representations of de novo motifs identified in the O, S, and K nucleosome-enriched targets (top) and nucleosome-depleted targets (bottom). The motifs were aligned to canonical motifs (middle). The number of targets analyzed and percentage of motif enrichments are indicated.
(G-I) Cartoon representations of the 3-D structures of O (PDB-3L1P), S (PDB-1GT0), and K (PDBs-2WBS and 2WBU) DBDs in complexes with DNA containing canonical motifs. Side and top views are shown for O and K and dashed curved arrows are shown to represent the extent of exposed DNA surface (G and I). The 3-D structure of the less distorted DNA (top) and extensively distorted DNA (bottom) were superimposed on nucleosomal DNA (PDB-3LZ0, gray) to display the extent Sox2-nucleosome binding compatibility by measuring RMSD of the fit.
Strikingly, while Oct4 targeted its canonical octamer sequence at nucleosome-depleted sites (~49% of n=3375), Oct4 targeted hexameric motifs resembling one or another half of the octamer motif at nucleosome-enriched sites (42% and 28%, respectively, of n=19,120) (Figure 4D). Sox2 targeted its canonical HMG box motif at nucleosome-depleted sites (64% of n=8221), while targeting a more degenerate motif lacking the sixth “G” nucleotide in the nucleosomal motif (~ 74% out of n=32,886) (Figure 4E, arrowhead). Finally, Klf4 alone targeted its nonameric motif at nucleosome-depleted sites (94% of n=9874), whereas Klf4 targeted a hexameric motif that was missing the three terminal nucleotides at nucleosome-enriched sites (90% of n=18,338) (Figure 4F, see dashed lines).
These findings agree with the above DNase footprinting of LIN28B-nuc bound to the factors (Figure 2B, right panels), with Oct4 and Sox2 protecting a part of their canonical motifs on one side of the LIN28B-nuc DNA (Figure 2B, 2C; right). On free DNA, Klf4 protected the first three nucleotides of its motif on the upper strand while protecting the remaining 6 nucleotides of its motif on the bottom strand (Figure S5A). However, Klf4 did not protect the first three nucleotides on the upper strand of LIN28B-nuc, as they were not exposed to DNaseI digestion, indicating that Klf4 may be interacting with part of its motif exposed on the other strand (Figure 2B, 2C).
These data show that the O, S, and K factors can independently target nucleosomes using partial or degenerate motifs, and that each of the factors targets their full canonical motif in the absence of nucleosomes at a target site. Targeting of partial motifs at nucleosomal sites by OS or OK together also reveals partial motifs for each of the factors (data not shown).
The molecular basis for O, S, and K nucleosomal targeting
In order to define the molecular basis that govern O, S, and K interactions with nucleosomal DNA, we interrogated the three dimensional structures of O, S, and K DBDs in complexes with their canonical motifs that were deposited in the RCSB protein data bank. Oct4 contains a bipartite POU domain, composed of an N-terminal POU-specific (POUS) and a C-terminal POU-homeodomain (POUHD), separated by a linker region. The X-ray structure of Oct4-POU-DNA complex confirms that the POUS and POUHD each bind one half of the octameric motif on DNA (Esch et al., 2013) (Figure 4G, lower panels). The truncated POUS and POUHD can bind their respective half motif DNA probes in vitro, independently from each other (Verrijzer et al., 1992). Interestingly, the isolated DNA-bound state of either POUS or POUHD accommodates less than half of the DNA surface across the circumference of the double helix (DNA surface occupied 606 and 718 Å2, respectively), leaving the opposite DNA surface solvent-exposed and potentially free to interact with histones in a nucleosome conformation (Figure 4G, red dashed arrows in upper panels). However, once both POUS and POUHD are bound to the full motif (1321 Å2), less than a quarter of the DNA circumference is solvent-exposed and hence would be incompatible with nucleosome binding, due to steric hindrance (Figure 4G, red dashed arrow in lower panel). Thus, the two POU domains do not target directly adjacent half sites on nucleosomes, as seen in free DNA, but the exposure of the separate half sites on nucleosomes is enough for Oct4 initial targeting.
Sox2 binds DNA through its HMG box, inducing a sharp bend and widening of the minor groove (Reményi et al., 2003) (Figure 4H, lower left panel). Our motif analysis showed that Sox2 targets a degenerate motif within nucleosomes, missing one “G” nucleotide at the sixth position (Figure 4E). This “G” nucleotide is positioned at the angle of the induced bend and makes direct contacts with the N46 residue at the N-terminal tail of Sox2-HMG (Reményi et al., 2003) (Figure 4E, 4H, arrowhead ). Remarkably, mutation of this one amino acid (N46Q) within Sox2-HMG results in a significant decrease in DNA-bending ability without affecting DNA-binding (Scaffidi and Bianchi, 2001). In transient transfection assays, the Sox2-N46Q mutant displays higher transactivation activity from the Fgf4 enhancer compared to Sox2 wild type (Scaffidi and Bianchi, 2001). Furthermore, mutation of the “G” nucleotide in the sixth position of the motif has the unique ability, among all mutations tested, to abolish DNA-bending by wild-type Sox2 (Scaffidi and Bianchi, 2001). Together these data indicate that Sox2 would not induce extensive DNA-distortion when targeting the nucleosomal motif, since that motif lacks the “G” nucleotide. To further support these observations we superimposed the 3D structure of DNA bound by wild-type Sox2 and Sox2-N46Q mutant on nucleosomal DNA and after 1000 cycles refinement we calculated the root-mean-square deviation (RMSD) as a measure of the average distance between the phosphate backbone for the best fit. These analysis reveal that the less distorted DNA is more compatible with nucleosomal DNA (RMSD=0.86 Å) compared to the extensively distorted DNA (RMSD=6.83 Å) (Figure 4H, right panel). In conclusion, our data indicates that Sox2 engages nucleosomes by recognizing a degenerate motif that involves less DNA distortion, better filling the curvature and widened minor groove of DNA around the histone octamer.
Klf4 recognizes the nonameric DNA motif using all three C2H2-type ZFs (three nucleotides per ZF) located at the C-terminus (Schuetz et al., 2011) (Figure 4F). However, we identified a hexameric motif, lacking the last three nucleotides, enriched within nucleosomal targets (Figure 4F, 90%). Mutagenic studies have shown that the hexameric motif represents the minimal essential binding site for Klf4 (Shields, 1998). Recently, X-ray crystallography has revealed the structures of Klf4 bound to the hexameric and nonameric sites (Schuetz et al., 2011) (Figure 4I). Klf4 uses its two most C-terminal ZFs, out of the three, to recognize the hexameric motif, occupying one side of the DNA double helix (595 Å2) and leaving more than half of the opposite surface potentially free to interact with histones in a nucleosome (Figure 4I, red dashed arrow in upper right panel). Klf4 bound to the nonameric motif, with all three ZFs, fills up more than half of the DNA surface (847 Å2) and would hinder binding to nucleosomes (Figure 4I, red dashed arrow in lower right panel). This analysis suggests that Klf4 employs two of its three ZFs to engage nucleosomes.
Interestingly, the observed adaptability of O, S, and K to recognize partial motifs correlates with the apparent flexibility of their respective DBDs that we modeled during their transition from the DNA-free to the DNA-bound states (Figure S5B—G).
c-Myc recognizes a partial motif enriched on nucleosomes through co-binding with other factors
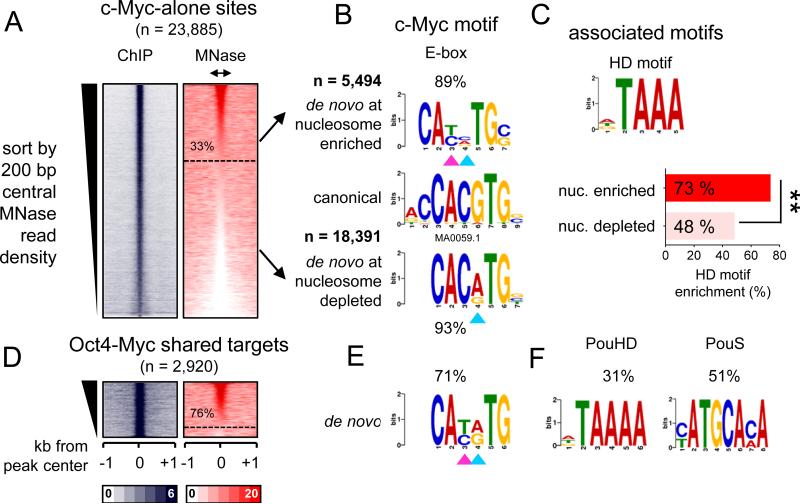
Using the partitioning method in Figure 4A-C, a subset of c-Myc targeted sites (33%, n = 5494) were enriched for nucleosomal DNA, while the majority of sites (77%, n = 18,391) did not exhibit enrichment (Figure 5A). Motif analysis revealed that c-Myc nucleosomal targets were enriched for an E-box motif that is missing the two central nucleotides (CANNTG) compared to the canonical E-box (CACGTG) (Figure 5B, double arrowheads in top panel). However, nucleosome-depleted targets were enriched for a less degenerate E-box motif that we and others have previously reported to be associated with c-Myc binding at enhancers (Lin et al., 2012; Nie et al., 2012; Soufi et al., 2012) (Figure 5B, single arrowhead in bottom panel). Interestingly, c-Myc-alone (i.e., without OSK) nucleosomal targets were additionally enriched for a homeobox (73%) motif that is highly similar to the POUHD motif, compared to nucleosome depleted sites (48%) (Figure 5C). Likewise, the majority of c-Myc sites that co-targeted with Oct4 (76%, n=2219) that are enriched for nucleosomes contain centrally a degenerate E-box motif similar to that identified in nucleosomal c-Myc-alone targets (Figure 5D, E). The separate halves of the POU motif were also enriched at the OM targeted sites, indicating that Oct4 uses one or the other DBD while co-binding with c-Myc (Figure 5F). In conclusion, c-Myc targets nucleosomal sites either with O, S, K or with endogenous homeodomain factors, recognizing a centrally degenerate E-box motif.
Figure 5. c-Myc recognition of degenerate E-box on nucleosome is assisted by binding with co-factors.
(A-F) Same as shown in Figure 4A-F, but for c-Myc alone and OM targets. (C) The enrichment of an associated motif (HD) is measured within c-Myc alone targets containing or depleted from nucleosomes. The data indicate that c-Myc is driven to a degenerate E-box on nucleosomes, in part, by homeodomain factors **p<0.001.
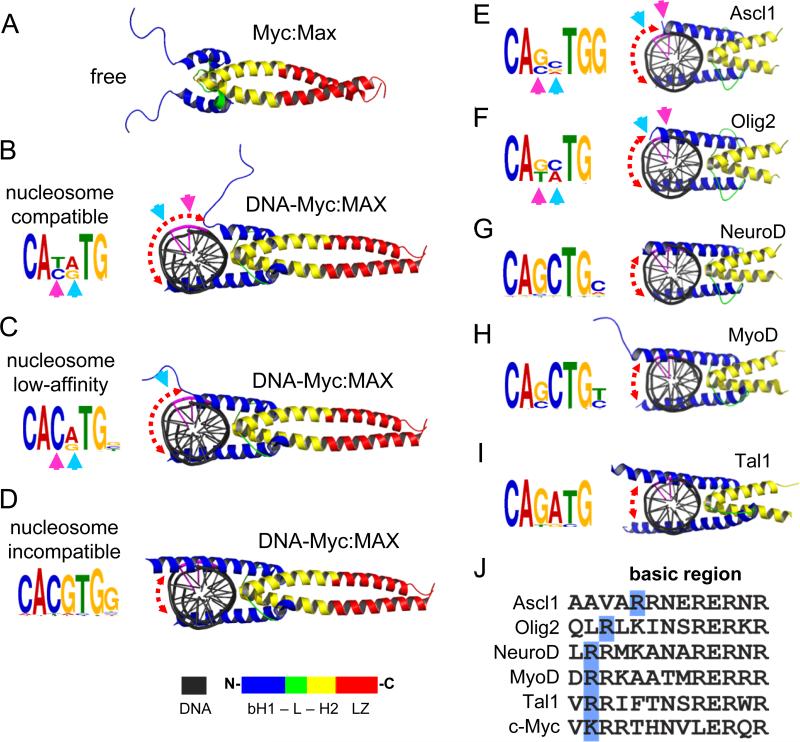
The basic region of bHLH domain, not bound to DNA, appears to be unfolded in solution (Sauvé et al., 2004) (Figure 6A, S6A). Upon DNA binding, the basic region folds as an extension of helix-1 and will be referred to as basic-helix-1 (bH) (Nair and Burley, 2003) (Figure 6D, S6B blue helices). Notably, the most conserved four nucleotides of the E-box (CANNTG) face towards the interaction interface between bHLH and DNA, while the degenerate central two nucleotides (CANNTG) face the exterior part of the DNA helix (Figure 6B, see cyan and magenta arrowheads). The transition between DNA-free and DNA-bound by molecular morphing indicates that the bH follows a gradual folding trajectory across the major groove of DNA (Figure 6A-D; S6B). The interaction between a partially folded bHLH and the CANNTG drives the initial recognition of the E-box without making contacts with the central nucleotides (NN), resulting in the centrally degenerate E-box motif that we observed for c-Myc at the nucleosome-enriched sites (Figure 6B).
Figure 6. The folding extent of bHLH basic helix-1 on DNA anti-correlates with targeting centrally-degenerate E-box motifs on nucleosomes.
(A-D) The folding trajectory of basic helix-1 of c-Myc upon DNA-binding showing the possible conformations of c-Myc:Max heterodimers (B and C) that are compatible with nucleosome binding. See Figure S6A for c-Myc Morph. The initial DNA-free state (A) and the fully folded DNA-bound state (D), which is incompatible with nucleosome-binding, are indicated. The associated motifs for each c-Myc:Max conformation are shown in the left. See Figure S6B for Mitf structure in complexes with E-box with variable central nucleotides.
(E-I) Cartoon representations of various bHLH reprogramming factors in complexes with DNA containing their canonical motifs (right). The de novo motifs identified for each factor from ChIP-seq data are indicated (left). The cyan and pink arrows represent the position of the exposed nucleotides within the central E-box motif not making base-contacts with the relative bHLH conformation. The central two nucleotides (CANNTG) are colored in purple in the DNA cartoon. The color scheme of the bHLH along with leucine zipper (LZ) is shown at the bottom.
(J) Alignment of amino-acid sequences of the basic region of Ascl1, Olig2, NeuroD, MyoD, Tal1 and c-Myc. The last basic residue at the C-terminal end is highlighted in blue. See Figure S6D, E for MNase enrichment and motif analysis of Asl1.
Importantly, the partially folded c-Myc only occupies one half the DNA helix surface, leaving the other half solvent-exposed and potentially nucleosome compatible (Figure 6B, red dashed arrow). Apparently, the partially folded c-Myc-DNA complex requires further assistance from other factors such as Oct4 or other homeodomain-containing proteins to remain associated with DNA. The interaction between a partially folded bHLH and a centrally degenerate E-Box motif has been observed by X-ray crystallography for Mitf, which shares 86% sequence homology across the basic region with c-Myc (Figure S6C) (Pogenberg et al., 2012). Once fully folded, the c-Myc bHLH adopts a rigid structure, stabilizing DNA binding and resulting in less-degenerate E-box motif, which would be incompatible with nucleosomes (Figure 6D). We conclude that partially-unfolded c-Myc targets a centrally-degenerate E-box motif, thereby adapting to a nucleosome template when assisted by other factors.
Predicting pioneer activity among different bHLH factors in reprogramming
To gain insights on how bHLH proteins may differentially target nucleosomes in reprogramming, we examined the 3-D structures of a range of bHLH-DNA complexes that have been used in reprogramming experiments (Longo et al., 2008; Ma et al., 1994; El Omari et al., 2013). Interestingly, the basic helix-1 from the different bHLH domains extends across the DNA helix to variable extents (Figure 6E-I). Motif analysis was also carried out on genomic sites bound by these factors from available ChIP-seq data. Notably, in conjunction with our findings on c-Myc, the length of the bH α-helix negatively correlates with the degeneracy of the central nucleotides (CANNTG) of the de novo motifs that we identified for each factor (Figure 6E-I).
To further test this correlation, we examined the recent findings that the bHLH factor Ascl1 can act as a pioneer factor during reprogramming fibroblasts to neurons (Wapinski et al., 2013). We measured nucleosome enrichment in pre-induced mouse embryonic fibroblasts (MEF) within Ascl1 initial targets in MEFs after 48 hr induction (Teif et al., 2012; Wapinski et al., 2013). Unlike c-Myc, the majority of Ascl1 sites (73%, n=3019) were enriched for nucleosomes (Figure S6D). Importantly, the basic helix-1 of Ascl1 is considerably shorter compared to that of c-Myc, leaving more of the DNA surface solvent exposed (Figure 6E). Similar to c-Myc, Ascl1 target nucleosomes were enriched (99.3%) for an E-box motif with degenerate central two nucleotides (CANNTG) compared to the E-box seen in 98.7% of sites depleted from nucleosomes (Figure S6E). Ascl1 nucleosomal targets contain an extra “G” nucleotide at the 3’-end of the E-box motif, which is missing in the nucleosome-depleted sites, resulting in more specific targeting of nucleosomes despite the centrally degenerate E-box (Figures 6E, S6E).
Ascl1 and Olig2 exhibited the shortest bH regions, by molecular modelling, compared to X-ray crystals of NeuroD, MyoD, and Tal1, with longer bHs. To verify that the observed bH lengths were not due to the methodology, we examined the amino acid composition of the basic regions in all bHLH factors (Figure 6J). The bH-DNA interaction is mainly driven by positively charged residues (and hence the name basic). Interestingly, the Ascl1 bH ends at the last (N-terminal end) basic residue (arginine), which is positioned further upstream (toward the C-terminus) compared to the other factors (Figure 6J, R residues in blue boxes). The last basic residue of Olig2-bH falls in between Ascl1 and the rest of the factors. In conclusion, the basic helix-1 of pioneer bHLH factors such as Ascl1 is intrinsically shorter, allowing the factors to bind nucleosomes more efficiently.
DISCUSSION
The introduction of a defined set of TFs, such as OSKM, into differentiated cells can result in cell fate conversion (Takahashi and Yamanaka, 2006) and yet it has been clear that the different factors have different contributions or “strengths” in cell type conversion. This provided the basis for our effort to tackle the long-standing problem of how TFs initially target their sites in closed chromatin. The pioneer factor theory partly answers this question by suggesting that a select group of TFs, such as FoxA, access closed chromatin by a direct interaction with nucleosomal DNA through a DBD that resembles the structure of a linker histone (Zaret and Carroll, 2011, Iwafuchi-Doi and Zaret, 2014). We previously found that the diverse set of DBDs exhibited by O, S, K and M, which are structurally different from a linker histone, have differential abilities to access closed chromatin (Soufi et al., 2012). Here we revealed that the relative tendencies of O, S, K, and M to initially target nucleosomal sites in reprogramming reflect their inherent ability to bind nucleosomes in vitro and their ability to recognize partial motifs on nucleosomes in vivo. This is different from what was observed for FoxA1, which recognizes the same motif on free DNA and nucleosomes (Cirillo et al., 1998; Li et al., 2011). Factors that cannot bind nucleosomes on their own, such as c-Myc, associate with other factors to target degenerate E-boxes on nucleosomes. Our new approach is in contrast to the previous predictions of pioneer factors by fitting fully-folded DBDs, in their naked DNA-bound state, on nucleosomes through a docking mechanism.
We found that the bipartite POU domain of Oct4 can target partial motifs exposed on nucleosomes using separate PouS or PouHD domains. The single motif targeted by each domain is longer than each half of the octamer motif, thus providing greater binding specificity than a half-motif. In addition, mass-spectroscopy analysis has identified histones as interacting partners of Oct4 in mouse ES cells (Pardo et al., 2010), indicating an additional affinity contribution by protein-histone interactions. The bipartite domain-Pax family of TFs can bind DNA using both domains and still occupy half of the DNA surface, and would therefore be compatible with nucleosome binding (Garvie et al., 2001; Xu et al., 1999) (Figure S7, right, compared to POU TFs). This agrees with the finding that Pax7 is a pioneer factor that uses full motif recognition during initial targeting (Budry et al., 2012). Thus, bipartite TFs have to either employ one DBD or position both DBDs on the same surface of DNA in order to interact with nucleosomes. Notably, the pioneer activity of a Zebrafish homologue of an Oct protein was observed during the maternal-to-zygote transition (Lee et al., 2013; Leichsenring et al., 2013), suggesting that targeting nucleosomal sites may be a general method for de novo programming of the genome.
The high affinity of Sox2 for nucleosomes may be due to the pre-bent conformation of DNA, which widens the DNA minor groove and favors initial minor groove sensing. While bending naked DNA by Sox2 requires minimal work (Privalov et al., 2009), the energy cost would impede Sox2 to further bend DNA on nucleosomes. We find that Sox2 would not further bend nucleosomal DNA because it recognizes a partial motif that diminishes the extreme bending of the full motif. Sox family members share the recognition of the core motif but display diverse preferences outside the core in naked DNA (Badis et al., 2009). Our findings reveal greater flexibility with regard to Sox2 core motif preferences on nucleosomes than was previously recognized. In addition, we showed evidence for both specific and nonspecific binding by Sox2 in vitro and in vivo. The stable, motif-driven targeting by Sox2 on nucleosomes in the ChIP-Seq data shows much lower co-binding with Oct4 (Soufi et al., 2012) than seen in live imaging (Chen et al., 2014), leaving open whether the latter approach depicts nucleosomal or free DNA binding during genome scanning.
Klf4 showed higher affinity to free DNA compared to nucleosomes in vitro and its initial targets in vivo were enriched for nucleosomes, though less so than compared to Oct4 and Sox2. Klf4 targets nucleosomes in vivo using two out of its three zinc fingers, recognizing a hexameric motif. This explains how the affinity of Klf4 to nucleosomes is lower than that to free DNA. The pioneer factor GATA4 binds nucleosomes modestly in vitro (Cirillo and Zaret, 1999) and targets a hexameric motif in vivo (Zheng et al., 2013). Notably, GATA4 only contains two zinc fingers. The Gils zinc finger family 1 (Gli1) greatly enhances reprogramming when co-expressed with OSK (Maekawa et al., 2011). Interestingly, despite containing 5 ZFs, Glis1 only employs two ZFs (number four and five) to recognize its targets (Pavletich and Pabo, 1993). The repressor ZFP57/Kap1, which is known to be associated with closed chromatin, also recognizes a hexameric motif despite containing an array of seven zinc fingers (Quenneville et al., 2011). This suggests that zinc finger proteins in general may use two zinc fingers to initially target hexameric motifs exposed on nucleosomes. Klf4 also showed non-specific interactions with nucleosomes, suggesting a similar genome searching mechanism as Sox2.
Various examples have been reported on the over-expression of bHLH factors in cancer, including c-Myc, Tal1, and Olig2 (Lin et al., 2012; Nie et al., 2012; Palii et al., 2011; Sanda et al., 2012; Suvà et al., 2014). In all of these cases, the bHLH factors have been associated with degenerate E-box motifs and co-binding with other factors. We propose that the extent to which basic helix-1 lays on DNA and co-binds with pioneer factors is reflected in the recognized motif, predicting bHLH ability to bind nucleosomes and access closed chromatin. Interestingly, the mutation of two amino acids within the basic helix1 that interacts with central E-box makes the non-myogenic bHLH factor E12 able to convert fibroblasts to muscle cells (Davis and Weintraub, 1992). The homeodomain factor PBX primes MyoD targets to induce myogenic potential (Maves et al., 2007). Furthermore, the hematopoietic TAL1-E45 heterodimer employs one of the two bHLH domains using LMO2 as an adapter to interact with GATA1 (El Omari et al., 2013). Hence, in addition to their intrinsic structures, bHLH factors co-binding with DNA-binding and non-DNA binding proteins appear to be involved in stabilizing the interaction of the partially folded bHLH factors to nucleosomes. These features are relevant to the multitude of bHLH factors functioning in development, cancer, and reprogramming experiments.
The differential ability of TFs to recognize their target sites on nucleosomes supports a hierarchical model where pioneer factors are the first to gain access to their targets in silent chromatin. We also observe that the initial targeting can occur for non-pioneer proteins when they bind in conjunction with pioneer factors that allow the former to recognize their DBDs to a reduced motif that is compatible with nucleosome binding. Further studies are needed to understand the secondary events that lead to subsequent changes in local chromatin structure and the formation of large complexes at gene regulatory sequences. By understanding the mechanistic basis by which certain transcription factors are especially capable of initiating cell fate changes, we hope to modulate the process and ultimately control cell fates at will.
EXPERIMENTAL PROCEDURES
Protein expression and purification
We made the bacterial expression plasmids pET-28B-huOct4, pET-28B-huSox2, pET-28B-huKlf4, and pET-28B-huMyc encoding the full length human O, S, K, M, respectively, fused to an N-terminal 6X histidine tag. The recombinant proteins were expressed in E. Coli Rosetta (DE3) pLysS (Novagen # 70956-3) and purified using a nickel charged column under denaturing conditions The mammalian expressed human OSKM recombinant proteins were obtained from OriGene (Oct4 #TP311998, Sox2 #TP300757, Klf4 #TP306691, c-Myc #TP301611). See Extended Experimental Procedures for more details of this and following sections.
Nucleosome reconstitution
The 162 bp LIN28B DNA fragment was created by PCR with end-labeled primers. The fluorescent-tagged DNA fragments were gel extracted and further purified using ion-exchange liquid chromatography by MonoQ (GE Healthcare). The nucleosomes were reconstituted by mixing purified human H2A/H2B dimers and H3/H4 tetramers with LIN28B-DNA at 1:1 molar ratio of histone octamer:DNA using a salt-urea gradient.
DNA binding reactions
Cy5 end-labelled DNA containing specific or non-specific sites, LIN28B-DNA, and LIN28B-nucleosomes were incubated with recombinant proteins in 10 mM Tris-HCl (pH7.5), 1 mM MgCl2, 10 uM ZnCl2, 1 mM DTT, 10 mM KCl, 0.5 mg/ml BSA, 5% glycerol at room temperature for 60 min. Free and bound DNA were separated on 4% non-denaturing polyacrylamide gels run in 0.5X Tris–borate–EDTA and visualized using a PhosphorImager. The intensity of Cy5 fluorescence was quantified using Multi-Gauge software (Fujifilm Science lab) to generate binding curves for Kd analysis.
DNase footprinting was carried out by treating free DNA or nucleosomes, 6FAM 5’ end-labelled, with DNase-I (Worthington) in the absence or presence of TFs. The end-labelled digested fragments were separated by capillary electrophoresis in ABI 96-capillary 3730XL Sequencer (Applied Biosystems).
Genomic data analysis
The O, S, K, and M ChIP-seq aligned data along with the called peaks (FDR-controlled at 0.005) were obtained from GEO (GSE36570) (Soufi et al., 2012). The MNase-seq data (GSM543311) (Kelly et al., 2012) were aligned to build version NCBI36/HG18 of the human genome and seven replicates were pooled together generating 145,546,004 unique reads. The MNase-seq reads were extended to 150 bp to cover one nucleosome and thus resulting in 6.6 fold genome coverage.
Motif analysis were carried out using the MEME-ChIP suit v.4.9.1 available at http://meme.nbcr.net (Machanick and Bailey, 2011).
Supplementary Material
ACKNOWLEDGEMENTS
We thank G. Donahue for advice and G. Blobel, R. Marmorstein, M. Iwafuchi-Doi, and D. Nicetto for comments on the manuscript. The work was supported by NIH grant P01GM099134 to K.S.Z.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
A.S and K.Z conceived the study and designed the experiments. A.S and M.F.G. carried out EMSA and nucleosome reconstitution. A.S, A.J, and M.P performed MNase-seq data analysis. A.S and N.O carried out the recombinant protein purification experiments. A.S performed the motif and 3-D structure analysis. A.S and K.Z contributed to supervision of personnel, data interpretation, and writing the manuscript.
REFERENCES
- Adams CC, Workman JL. Binding of disparate transcriptional activators to nucleosomal DNA is inherently cooperative. Mol. Cell. Biol. 1995;15:1405–1421. doi: 10.1128/mcb.15.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barozzi I, Simonatto M, Bonifacio S, Yang L, Rohs R, Ghisletti S, Natoli G. Coregulation of Transcription Factor Binding and Nucleosome Occupancy through DNA Features of Mammalian Enhancers. Mol. Cell. 2014;54:844–857. doi: 10.1016/j.molcel.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin MD. Animal transcription networks as highly connected, quantitative continua. Dev. Cell. 2011;21:611–626. doi: 10.1016/j.devcel.2011.09.008. [DOI] [PubMed] [Google Scholar]
- Budry L, Balsalobre A, Gauthier Y, Khetchoumian K, L'honoré A, Vallette S, Brue T, Figarella-Branger D, Meij B, Drouin J. The selector gene Pax7 dictates alternate pituitary cell fates through its pioneer action on chromatin remodeling. Genes Dev. 2012;26:2299–2310. doi: 10.1101/gad.200436.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Jaenisch R. Mechanisms and models of somatic cell reprogramming. Nat. Rev. Genet. 2013;14:427–439. doi: 10.1038/nrg3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhang Z, Li L, Chen B-C, Revyakin A, Hajj B, Legant W, Dahan M, Lionnet T, Betzig E, et al. Single-Molecule Dynamics of Enhanceosome Assembly in Embryonic Stem Cells. Cell. 2014;156:1274–1285. doi: 10.1016/j.cell.2014.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo LA, Zaret KS. An Early Developmental Transcription Factor Complex that Is More Stable on Nucleosome Core Particles Than on Free DNA. Mol. Cell. 1999;4:961–969. doi: 10.1016/s1097-2765(00)80225-7. [DOI] [PubMed] [Google Scholar]
- Cirillo LA, McPherson CE, Bossard P, Stevens K, Cherian S, Shim EY, Clark KL, Burley SK, Zaret KS. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J. 1998;17:244–254. doi: 10.1093/emboj/17.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- Davis R, Weintraub H. Acquisition of myogenic specificity by replacement of three amino acid residues from MyoD into E12. Science (80-. ) 1992;256:1027–1030. doi: 10.1126/science.1317057. [DOI] [PubMed] [Google Scholar]
- Esch D, Vahokoski J, Groves MR, Pogenberg V, Cojocaru V, Vom Bruch H, Han D, Drexler HCA, Araúzo-Bravo MJ, Ng CKL, et al. A unique Oct4 interface is crucial for reprogramming to pluripotency. Nat. Cell Biol. 2013;15:295–301. doi: 10.1038/ncb2680. [DOI] [PubMed] [Google Scholar]
- Farina A, Faiola F, Martinez E. Reconstitution of an E box-binding Myc:Max complex with recombinant full-length proteins expressed in Escherichia coli. Protein Expr. Purif. 2004;34:215–222. doi: 10.1016/j.pep.2003.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvie CW, Hagman J, Wolberger C. Structural studies of Ets-1/Pax5 complex formation on DNA. Mol. Cell. 2001;8:1267–1276. doi: 10.1016/s1097-2765(01)00410-5. [DOI] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, Zaret KS. Pioneer transcription factors in cell reprogramming. Genes Dev. 2014;28:2679–2692. doi: 10.1101/gad.253443.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly TK, Liu Y, Lay FD, Liang G, Berman BP, Jones PA. Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res. 2012;22:2497–2506. doi: 10.1101/gr.143008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Friedman JR, Fulmer JT, Kaestner KH. The initiation of liver development is dependent on Foxa transcription factors. Nature. 2005;435:944–947. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Takacs CM, Bazzini AA, DiVito KR, Fleming ES, Giraldez AJ. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature. 2013;503:360–364. doi: 10.1038/nature12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichsenring M, Maes J, Mössner R, Driever W, Onichtchouk D. Pou5f1 transcription factor controls zygotic gene activation in vertebrates. Science. 2013;341:1005–1009. doi: 10.1126/science.1242527. [DOI] [PubMed] [Google Scholar]
- Li Z, Schug J, Tuteja G, White P, Kaestner KH. The nucleosome map of the mammalian liver. Nat. Struct. Mol. Biol. 2011;18:742–746. doi: 10.1038/nsmb.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Lovén J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional Amplification in Tumor Cells with Elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo A, Guanga GP, Rose RB. Crystal structure of E47-NeuroD1/beta2 bHLH domain-DNA complex: heterodimer selectivity and DNA recognition. Biochemistry. 2008;47:218–229. doi: 10.1021/bi701527r. [DOI] [PubMed] [Google Scholar]
- Ma PC, Rould MA, Weintraub H, Pabo CO. Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell. 1994;77:451–459. doi: 10.1016/0092-8674(94)90159-7. [DOI] [PubMed] [Google Scholar]
- Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa M, Yamaguchi K, Nakamura T, Shibukawa R, Kodanaka I, Ichisaka T, Kawamura Y, Mochizuki H, Goshima N, Yamanaka S. Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature. 2011;474:225–229. doi: 10.1038/nature10106. [DOI] [PubMed] [Google Scholar]
- Maves L, Waskiewicz AJ, Paul B, Cao Y, Tyler A, Moens CB, Tapscott SJ. Pbx homeodomain proteins direct Myod activity to promote fast-muscle differentiation. Development. 2007;134:3371–3382. doi: 10.1242/dev.003905. [DOI] [PubMed] [Google Scholar]
- Mirny LA. Nucleosome-mediated cooperativity between transcription factors. Proc. Natl. Acad. Sci. U. S. A. 2010;107:22534–22539. doi: 10.1073/pnas.0913805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montserrat N, Nivet E, Sancho-Martinez I, Hishida T, Kumar S, Miquel L, Cortina C, Hishida Y, Xia Y, Esteban CR, et al. Reprogramming of Human Fibroblasts to Pluripotency with Lineage Specifiers. Cell Stem Cell. 2013;13:341–350. doi: 10.1016/j.stem.2013.06.019. [DOI] [PubMed] [Google Scholar]
- Nair SK, Burley SK. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell. 2003;112:193–205. doi: 10.1016/s0092-8674(02)01284-9. [DOI] [PubMed] [Google Scholar]
- Nakatake Y, Fukui N, Iwamatsu Y, Masui S, Takahashi K, Yagi R, Yagi K, Miyazaki J.-i., Matoba R, Ko MSH, et al. Klf4 Cooperates with Oct3/4 and Sox2 To Activate the Lefty1 Core Promoter in Embryonic Stem Cells. Mol. Cell. Biol. 2006;26:7772–7782. doi: 10.1128/MCB.00468-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Omari K, Hoosdally SJ, Tuladhar K, Karia D, Hall-Ponselé E, Platonova O, Vyas P, Patient R, Porcher C, Mancini EJ. Structural basis for LMO2-driven recruitment of the SCL:E47bHLH heterodimer to hematopoietic-specific transcriptional targets. Cell Rep. 2013;4:135–147. doi: 10.1016/j.celrep.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palii CG, Perez-Iratxeta C, Yao Z, Cao Y, Dai F, Davison J, Atkins H, Allan D, Dilworth FJ, Gentleman R, et al. Differential genomic targeting of the transcription factor TAL1 in alternate haematopoietic lineages. EMBO J. 2011;30:494–509. doi: 10.1038/emboj.2010.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Plath K. Epigenetics of Reprogramming to Induced Pluripotency. Cell. 2013;152:1324–1343. doi: 10.1016/j.cell.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo M, Lang B, Yu L, Prosser H, Bradley A, Babu MM, Choudhary J. An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell Stem Cell. 2010;6:382–395. doi: 10.1016/j.stem.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavletich NP, Pabo CO. Crystal structure of a five-finger GLI-DNA complex: new perspectives on zinc fingers. Science. 1993;261:1701–1707. doi: 10.1126/science.8378770. [DOI] [PubMed] [Google Scholar]
- Perlmann T, Wrange O. Specific glucocorticoid receptor binding to DNA reconstituted in a nucleosome. EMBO J. 1988;7:3073–3079. doi: 10.1002/j.1460-2075.1988.tb03172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogenberg V, Ogmundsdóttir MH, Bergsteinsdóttir K, Schepsky A, Phung B, Deineko V, Milewski M, Steingrímsson E, Wilmanns M. Restricted leucine zipper dimerization and specificity of DNA recognition of the melanocyte master regulator MITF. Genes Dev. 2012;26:2647–2658. doi: 10.1101/gad.198192.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polach KJ, Widom J. A Model for the Cooperative Binding of Eukaryotic Regulatory Proteins to Nucleosomal Target Sites. J. Mol. Biol. 1996;258:800–812. doi: 10.1006/jmbi.1996.0288. [DOI] [PubMed] [Google Scholar]
- Privalov PL, Dragan AI, Crane-Robinson C. The cost of DNA bending. Trends Biochem. Sci. 2009;34:464–470. doi: 10.1016/j.tibs.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, Baglivo I, Pedone PV, Grimaldi G, Riccio A, et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol. Cell. 2011;44:361–372. doi: 10.1016/j.molcel.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan V, Finch JT, Graziano V, Lee PL, Sweet RM. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature. 1993;362:219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- Reményi A, Lins K, Nissen LJ, Reinbold R, Schöler HR, Wilmanns M, Remenyi A, Scholer HR. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 2003;17:2048–2059. doi: 10.1101/gad.269303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinton DA, Daley GQ. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda DJ, Chew J-L, Lim L-H, Loh Y-H, Wang B, Ng H-H, Robson P. Transcriptional regulation of nanog by OCT4 and SOX2. J. Biol. Chem. 2005;280:24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- Sanda T, Lawton LN, Barrasa MI, Fan ZP, Kohlhammer H, Gutierrez A, Ma W, Tatarek J, Ahn Y, Kelliher MA, et al. Core transcriptional regulatory circuit controlled by the TAL1 complex in human T cell acute lymphoblastic leukemia. Cancer Cell. 2012;22:209–221. doi: 10.1016/j.ccr.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvé S, Tremblay L, Lavigne P. The NMR solution structure of a mutant of the Max b/HLH/LZ free of DNA: insights into the specific and reversible DNA binding mechanism of dimeric transcription factors. J. Mol. Biol. 2004;342:813–832. doi: 10.1016/j.jmb.2004.07.058. [DOI] [PubMed] [Google Scholar]
- Scaffidi P, Bianchi ME. Spatially precise DNA bending is an essential activity of the sox2 transcription factor. J. Biol. Chem. 2001;276:47296–47302. doi: 10.1074/jbc.M107619200. [DOI] [PubMed] [Google Scholar]
- Schuetz A, Nana D, Rose C, Zocher G, Milanovic M, Koenigsmann J, Blasig R, Heinemann U, Carstanjen D. The structure of the Klf4 DNA-binding domain links to self-renewal and macrophage differentiation. Cell Mol Life Sci. 2011;68:3121–3131. doi: 10.1007/s00018-010-0618-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields J. Identification of the DNA sequence that interacts with the gut-enriched Kruppel-like factor. Nucleic Acids Res. 1998;26:796–802. doi: 10.1093/nar/26.3.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, Zhu H, Yvanka de Soysa T, Shinoda G, Seligson MT, Tsanov KM, Nguyen L, Asara JM, Cantley LC, Daley GQ. Lin28 Enhances Tissue Repair by Reprogramming Cellular Metabolism. Cell. 2013;155:778–792. doi: 10.1016/j.cell.2013.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A. Mechanisms for enhancing cellular reprogramming. Curr. Opin. Genet. Dev. 2014;25:101–109. doi: 10.1016/j.gde.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvà ML, Rheinbay E, Gillespie SM, Patel AP, Wakimoto H, Rabkin SD, Riggi N, Chi AS, Cahill DP, Nahed BV, et al. Reconstructing and Reprogramming the Tumor-Propagating Potential of Glioblastoma Stem-like Cells. Cell. 2014;157:580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Teif VB, Vainshtein Y, Caudron-Herger M, Mallm J-P, Marth C, Höfer T, Rippe K. Genome-wide nucleosome positioning during embryonic stem cell development. Nat. Struct. Mol. Biol. 2012;19:1185–1192. doi: 10.1038/nsmb.2419. [DOI] [PubMed] [Google Scholar]
- Verrijzer CP, Alkema MJ, van Weperen WW, Van Leeuwen HC, Strating MJ, van der Vliet PC. The DNA binding specificity of the bipartite POU domain and its subdomains. EMBO J. 1992;11:4993–5003. doi: 10.1002/j.1460-2075.1992.tb05606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski OL, Vierbuchen T, Qu K, Lee QY, Chanda S, Fuentes DR, Giresi PG, Ng YH, Marro S, Neff NF, et al. Hierarchical Mechanisms for Direct Reprogramming of Fibroblasts to Neurons. Cell. 2013;155:621–635. doi: 10.1016/j.cell.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler DS, Papoulas O, Dang CV, Kingston RE. Differential binding of c-Myc and Max to nucleosomal DNA. Mol Cell Biol. 1994;14:4097–4107. doi: 10.1128/mcb.14.6.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu HE, Rould MA, Xu W, Epstein JA, Maas RL, Pabo CO. Crystal structure of the human Pax6 paired domain-DNA complex reveals specific roles for the linker region and carboxy-terminal subdomain in DNA binding. Genes Dev. 1999;13:1263–1275. doi: 10.1101/gad.13.10.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng R, Rebolledo-Jaramillo B, Zong Y, Wang L, Russo P, Hancock W, Stanger BZ, Hardison RC, Blobel GA. Function of GATA factors in the adult mouse liver. PLoS One. 2013;8:e83723. doi: 10.1371/journal.pone.0083723. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.