Abstract
Motile Escherichia coli cells track gradients of attractant and repellent chemicals in their environment with transmembrane chemoreceptor proteins. These receptors operate in cooperative arrays to produce large changes in the activity of a signaling kinase CheA in response to small changes in chemoeffector concentration. Recent research has provided much deeper understanding of the structure and function of core receptor signaling complexes and the architecture of higher-order receptor arrays, which in turn has led to new insights into the molecular signaling mechanisms of chemoreceptor networks. Current evidence supports a new view of receptor signaling in which stimulus information travels within receptor molecules through shifts in the dynamic properties of adjoining structural elements rather than through a few discrete conformational states.
Keywords: transmembrane signaling, kinase control, cooperative network
Bacterial chemoreceptors: high-sensitivity, high-gain signaling
Motile microbes detect and follow chemical gradients in their environment by means of transmembrane chemoreceptors known as methyl-accepting chemotaxis proteins (MCPs). MCPs mediate chemotactic behaviors in many varieties of Bacteria and Archaea, but are best understood in Escherichia coli, which has long served as the model organism for bacterial chemotaxis research (Box 1). Most studies have focused on the two most abundant chemoreceptors: Tar, the aspartate and maltose receptor, and Tsr, the serine receptor. Chemoreceptors form stable core signaling complexes with two cytoplasmic proteins, CheA, a histidine autokinase, and CheW, which couples CheA to receptor control. Core signaling complexes in turn are organized into supramolecular arrays that enable the receptor ensemble to detect small changes in chemoeffector concentration and, through cooperative signaling interactions, to produce large changes in CheA kinase activity. A sensory adaptation system adjusts detection sensitivity to ambient conditions, allowing chemoreceptors to operate over a wide concentration range.
Box 1. Chemotactic signaling in E. coli.
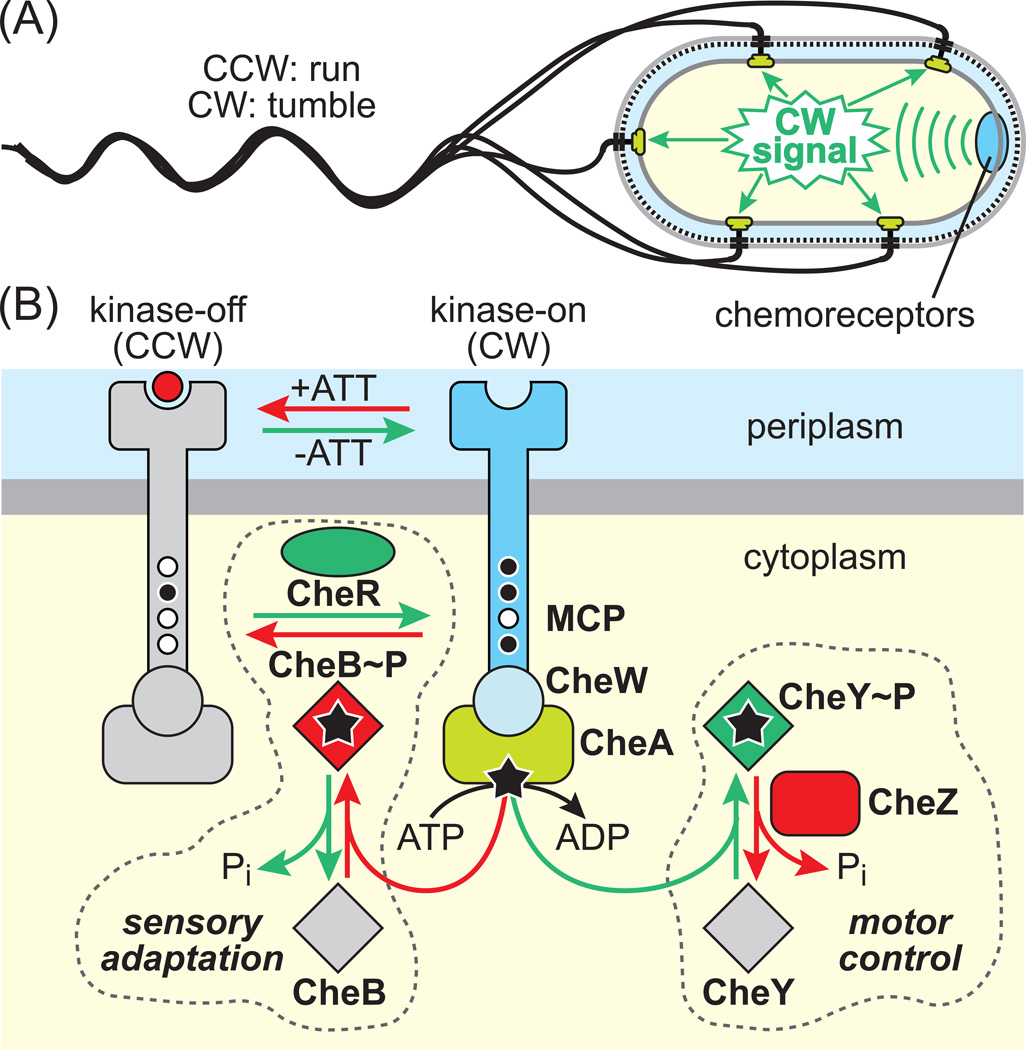
Motile E. coli cells have 4–6 peritrichous flagella, each driven by a rotary motor powered by protonmotive force. The default direction of motor rotation is counter-clockwise (CCW) and produces forward swimming (‘runs’). Transmembrane chemoreceptors form signaling complexes that organize as clusters in the membrane, the largest of which are at the cell pole(s). These receptor arrays control the cell’s swimming behavior by generating clockwise (CW) motor signals that cause random directional changes (‘tumbles’) (Figure IA). In the absence of chemoeffector gradients, the cells swim in a random walk of runs and tumbles. When swimming in chemoeffector gradients, the cells respond to temporal changes in attractant or repellent levels, sensed by their clustered chemoreceptors, by suppressing CW signals during runs that carry them in favorable directions. E. coli swims toward sugars (glucose, galactose, ribose, and maltose), amino acids (aspartic acid and serine), dipeptides, pyrimidines, and electron acceptors (oxygen, nitrate, and fumarate) and away from potentially harmful compounds (fatty acids, alcohols, and some divalent cations). Recent studies have also demonstrated E. coli attractant responses to AI-2, a general quorum-sensing signal, [89] and to a metabolite of norepinephrine [90].
Chemoreceptors form ternary signaling complexes with CheA, a histidine autokinase, and CheW, which couples CheA activity to receptor control (Figure IB). Receptor signaling complexes exhibit kinase-on and kinase-off output states. The cell’s swimming behavior reflects the proportions of receptor signaling complexes in the ON and OFF states. Attractant stimuli shift receptors toward the OFF state, slowing the flux of CheA phosphoryl groups to two response regulators, CheB and CheY. Phospho-CheY interacts with the flagellar basal body to trigger CW rotation, but is short-lived, owing to rapid dephosphorylation by its phosphatase CheZ. The short half-life of phospho-CheY allows the cell to trigger rapid motor responses to chemotactic stimuli.
Phospho-CheB is part of a sensory adaptation feedback circuit that resets the ON-OFF equilibrium to its pre-stimulus poise, enabling cells to monitor temporal changes in chemoeffector concentrations as they move about and to detect such stimuli over a wide concentration range. The sensory adaptation system adjusts receptor output through covalent modification of several glutamyl residues in the cytoplasmic portion of the MCP molecule. CheR, a dedicated methyltransferase, interacts with OFF-state receptors and catalyzes a glutamyl methylation reaction that shifts output toward the ON state. CheB, a dedicated methylesterase, interacts with ON-state MCPs and hydrolyzes glutamyl methyl groups to glutamic acid, shifting receptors toward the OFF state. The OFF-state substrate preference of CheR and the ON-state preference of CheB account for the negative feedback character of the sensory adaptation circuit. Phosphorylation of CheB, which enhances its catalytic activity many-fold, governs the rate of the sensory adaptation process.
Box 1, Figure I Components and signaling logic of the E. coli chemotaxis pathway
(A) Control of cell swimming behavior by chemoreceptor clusters. Flagellar motors turn in the CCW direction by default. Receptors generate signals (green) that elicit episodic CW motor rotation. Cytoplasm, yellow; inner membrane, gray; periplasmic space, light blue; peptidoglycan, black and dashed; outer membrane, light gray.
(B) Two-state model of receptor signaling. Components shown in gray are inactive forms. Green components and reactions enhance phospho-CheY (CW signal) production; red components and reactions reduce phospho-CheY levels. Small circles on receptor molecules (MCP) indicate unmodified (white) and methylated (black) sensory adaptation sites. Stars indicate signaling phosphoryl groups.
How do chemoreceptors process stimulus and sensory adaptation signals? How do they control CheA activity in response to those signals? What is the structure of the core receptor signaling complex? How are those units networked to produce cooperative signaling behavior? Over the past few years of chemoreceptor research, molecular answers to these questions have come into sharper focus. In this brief review we summarize evidence for an emerging dynamics-based view of receptor operation and how it can account for transmission of stimulus and sensory adaptation signals through chemoreceptor molecules. We also summarize recent advances in deciphering the structural organization of receptor signaling complexes and the new mechanistic insights that work has provided. Interested readers may wish to consult other recent reviews for additional coverage of these topics [1–6].
Structural features of chemoreceptor molecules
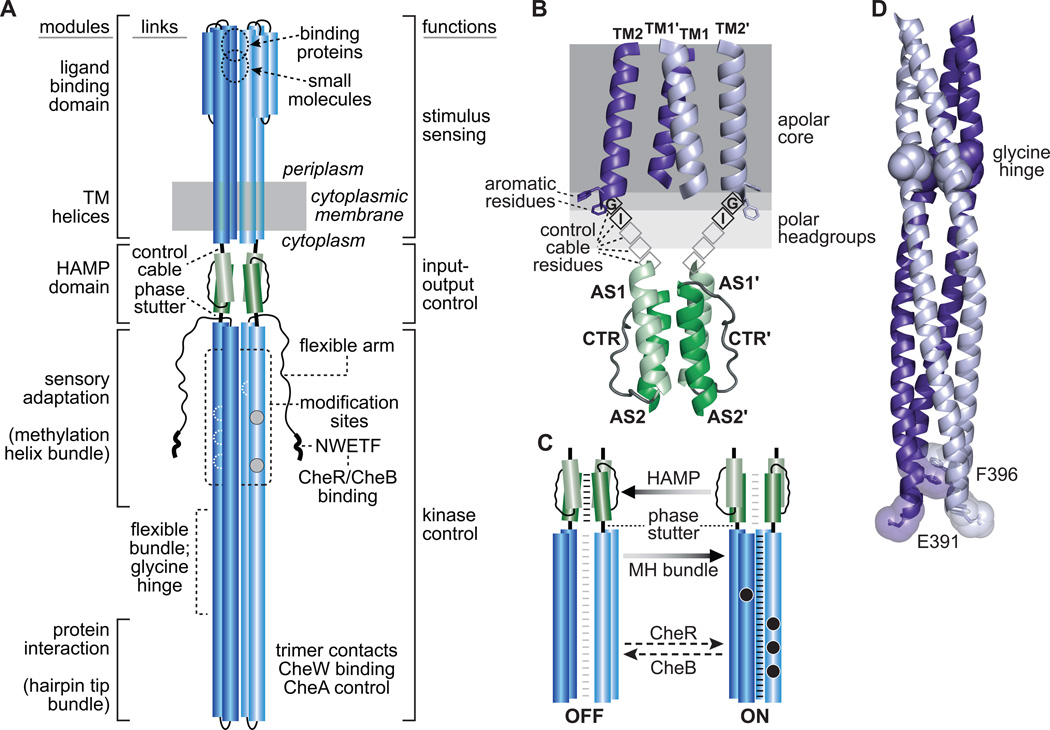
Tar and Tsr molecules are homodimers; their subunits are ~550 amino acids in length and have mainly α-helical secondary structure, defining three functional elements: (i) a sensing module comprising a periplasmic ligand binding domain bounded by four membrane-spanning helices, (ii) a cytoplasmic kinase control domain comprising an antiparallel, 4-helix coiled-coil bundle containing adaptational modification sites and determinants for binding and regulating CheA kinase, and (iii) an intervening HAMP domain that mediates signaling transactions between the sensing and kinase control elements [7] (Figure 1A). Distinctive structural features link adjacent elements and are central to the mechanisms of signal transmission in receptor molecules: A five-residue control cable and a four-residue phase stutter flank each HAMP subunit; a flexible region containing a glycine hinge links the sensory adaptation and protein interaction helix bundles (Figure 1A).
Figure 1. Signal transmission in chemoreceptor dimers.
(A) Architectural features of receptor molecules. Cylindrical segments represent alpha-helical secondary structures, drawn approximately to scale. The two protomers of the homodimer are shown in different shades of blue. Each protomer contains four adaptational modification sites (gray and white circles) common to Tar and Tsr. Gray sites are synthesized as glutaminyl residues and subsequently converted to glutamyl residues by CheB action; white sites are synthesized as glutamyl residues. Sites on the two helices in back are shown as dashed, white outlines. White rectangles in the flexible bundle region represent glycine hinge residues. The 4-helix bundle of the cytoplasmic kinase control domain ends with an unstructured linker segment at the C-terminus of each subunit (thin wavy line). A pentapeptide sequence (NWETF) at the very C-terminus provides a binding site for the CheR and CheB modification enzymes of the sensory adaptation system.
(B) Structure of the TM bundle-control cable-HAMP region of Tar and Tsr. The transmembrane (TM) helices form a 4-helix bundle with interactions between the TM1 and TM1' helices at the dimer interface. Attractant stimuli promote ~2 Å inward piston movement of one of the TM2 helices, which is transmitted through the five control cable residues to modulate the structural stability of the HAMP domain. The first two control cable residues of Tar and Tsr play critical roles in transmembrane signaling, whereas the sidechain character of the other control cable residues has little effect on function.
(C) Dynamic-bundle model of the signaling interplay between the HAMP and MH bundles. The model [41] proposes that the packing stabilities of the HAMP and methylation helix (MH) bundles are coupled in opposition and vary over a range of conformations. Light gray horizontal lines represent weak inter-helix packing forces; black lines represent strong bundle-packing forces. The sensory adaptation system (CheR and CheB enzymes) also modulates MH bundle stability. Unmodified adaptation sites (white circles) destabilize MH packing and promote kinase-off output. Methylated sites (black circles) stabilize MH packing and promote kinase-on output.
(D) The cytoplasmic tip of Tsr showing residues that probably influence tip conformation and dynamics. These structural features are conserved in Tar and most other MCPs. Helices are shown as backbones, with glycine hinge residues in space-fill mode. Side-chain atoms of residues F396 and E391 at the tip are shown as transparent spheres enclosing sticks. Note the stacking interaction of the F396 sidechains in the interior of the 4-helix bundle and the solvent-exposed orientation of E391.
The highly conserved protein interaction hairpin tip – the defining sequence motif of MCPs [8] – directs the assembly and operation of receptor signaling complexes. The tip contains determinants for binding CheA [9–11] and CheW [10, 12, 13] and for forming trimers of receptor dimers [14, 15]. The five E. coli members of the MCP family (Tar, Tsr, Tap, Trg, and Aer) have identical trimer contact residues, enabling low-abundance receptors (Tap, Trg, and Aer) to participate in signaling teams with high-abundance partners (Tar and Tsr) [15, 16].
Signal transmission in chemoreceptor molecules
The signaling properties of Tar and Tsr generally conform to two-state models involving transitions between kinase-on (ON) and kinase-off (OFF) CheA activity states. Chemoeffector concentration changes, sensed as changes in ligand occupancy, promote conformational changes in receptor molecules that shift the ON-OFF equilibrium, thereby triggering flagellar motor responses. The cell’s sensory adaptation system subsequently restores pre-stimulus CheA activity through covalent modifications at specific residues in the receptor cytoplasmic domain (Figure 1C). CheR, an MCP-specific methyltransferase, preferentially interacts with receptor molecules in the OFF state and shifts them toward the ON state by converting glutamyl residues to glutamyl methyl esters. CheB, an MCP-specific methylesterase and deamidase, preferentially interacts with ON-state receptors and shifts them toward the OFF state, either by hydrolyzing methylated glutamates, or by irreversibly deamidating glutaminyl residues to glutamic acid at some adaptation sites in newly synthesized receptors. Although solitary receptors cannot assemble active signaling complexes, they nevertheless undergo modification changes following an attractant stimulus, demonstrating that signal transmission between ligand-binding and kinase control domains is an intrinsic property of chemoreceptor dimers [17].
Despite seemingly binary output behavior, receptors may use dynamics-based control mechanisms for internal signal transmission. An attractive idea, supported by genetic and biochemical observations, is that contiguous receptor signaling elements are linked in structural opposition such that more stable packing in one segment drives weaker packing and increased dynamics in flanking segments, producing an alternating sequence of dynamic shifts that can transmit conformational signals over long distances with a low-energy input, enabling stimulus molecules with small binding free energies to drive transitions [1, 7]. Less defined is the nature of those dynamic interactions. Potential contributors include: gaps in the four-helix bundle or changes in its supercoiling [18, 19], partial helix unwinding [20], helical bending at defined loci [21–24], bulging of one helix in the four-helix bundle [25], axial helix rotation [26], trimer dynamics [27], hairpin tip flexibility [28] and alternative stacking of hydrophobic side chain rings near the hairpin tip [29].
The periplasmic domains of Tar and Tsr contain sites for direct binding of aspartate and serine, respectively, and sites that detect other chemoeffectors through interaction with ligand-occupied periplasmic binding proteins (Figure 1A). Both types of binding sites span the subunit interface. Binding of a single ligand molecule creates conformational asymmetry in the receptor dimer sufficient to initiate a stimulus response. A large body of evidence identifies the key conformational change as a small (~2 Å) movement of one membrane-spanning helix (TM2, the ‘signaling helix’) normal to the plane of the cytoplasmic membrane [30–33]. These stimulus-induced TM2 piston displacements impinge, via a five-residue control cable, on the HAMP domain (Figure 1B).
A wide variety of transmembrane signaling proteins in microbes contain HAMP domains, typically deployed at the cytoplasmic face of the inner membrane, where they relay stimulus information between input sensing and output signaling domains [34, 35]. HAMP protomers comprise two amphiphilic helices (AS1 and AS2) joined by a non-helical connector (CTR). In homodimeric signaling proteins these HAMP elements form parallel, 4-helix coiled-coil bundles, stabilized predominantly by hydrophobic packing forces. The few available high-resolution HAMP structures exhibit various bundle-packing geometries that mainly differ in the precise alignment and register of the helices [36–38]. Although high-resolution structures have not been determined for the HAMP domains of Tar and Tsr, they appear to be 4-helix bundles based on in vitro and in vivo disulfide crosslinking studies [24, 39], extensive mutational analyses [40–43], and structural studies of Aer2, an MCP from Pseudomonas aeruginosa [37, 44].
The nature of kinase-on and kinase-off HAMP signaling states remains controversial [45]. On one hand, some structural and molecular dynamics studies have assigned alternative output states to discrete HAMP conformations [44, 46, 47]. On the other hand, mutational studies of the Tsr HAMP domain and a growing body of structural and molecular dynamics findings, suggest that HAMP domains shift output states through a dynamics-based mechanism, such as changes in bundle-packing stability [34, 45, 48–50]. A dynamic-bundle model best accounts for HAMP action in chemoreceptors [34, 40–43, 51]. For example, Tsr molecules lacking a HAMP domain are locked in kinase-on output, demonstrating that a specific HAMP structure is not required to attain the ON state [43]. Single amino acid replacements that are expected to reduce HAMP packing stability also shift Tsr output toward the kinase-on state but still produce kinase-off responses to high serine concentrations [42, 51]. These behaviors imply that unstably packed HAMP bundles allow kinase-on output, whereas kinase-off output involves an active override of that default state, most likely by a better-organized, less dynamic HAMP bundle (Figure 1C).
The dynamic-bundle signaling model proposes that an inward TM2 piston displacement, acting through the control cable, elicits kinase-off output by enhancing the packing stability of HAMP. Mutational studies of Tar and Tsr suggest that their control cables have helical secondary structures [52, 53]. A helical five-residue connection between TM2 and AS1 would force those helices substantially out-of-register, most likely reducing HAMP bundle stability. TM2 piston motions might relax this destabilizing structural input by creating a kink or swivel in the control cable helix. The first two residues of the control cable (G and I in both Tar and Tsr) are critical for input control, whereas a variety of amino acids at the remaining three control cable positions can support essentially normal signaling function [52–54].
How TM2 piston motions modulate control cable helicity remains a mystery. Aromatic residues at both the periplasmic and cytoplasmic ends of TM2 are thought to constrain TM2 displacements by partitioning at the lipid tail - headgroup interface [55, 56]. Repositioning these aromatic residues shifts the ON-OFF equilibrium and signal output of Tar [55, 56], but mutant receptors lacking aromatic belt residues at one interface are still capable of signaling [54, 57, 58], suggesting that the transmembrane signaling mechanism involves multiple structural interactions. Conceivably, piston displacements of TM2 could influence sidechain interactions of the first two critical control cable residues with the membrane interfacial region. Alternatively, bending of the signaling TM2 helix within the plane of the membrane could alter alignment of the TM2-AS1 helices to regulate HAMP stability [20].
Changes in HAMP packing stability modulate receptor output signals through their structural interplay with the adjoining methylation helix (MH) bundle (Figure 1C). The HAMP domains of signaling proteins connect to output helices through a characteristic ‘phase stutter’ that produces a 4-residue shift between the packing registers of the HAMP AS2 helices and adjoining output helices [34, 41, 59, 60]. The phase stutter linkage probably couples the packing stabilities of the HAMP and MH bundles in opposition [41]: Tight packing of HAMP should weaken MH bundle packing; loose packing of HAMP should permit tight MH bundle packing (Figure 1C). The signaling consequences of adaptational modifications are consistent with this picture. Adaptation sites 1–4 of Tar and Tsr lie on solvent-exposed faces of the methylation helices in regions of high negative charge density where charge repulsion destabilizes helix packing [61]. Unmethylated, anionic sites (glutamyl residues) would be expected to further destabilize MH packing, whereas neutralization of those groups (glutaminyl or glutamyl methyl ester residues) should enhance packing of the MH bundle [61, 62]. In accord with the dynamic-bundle model, receptors in low modification states have OFF-shifted output, whereas receptors in high modification states have on-shifted output (Figure 1C). Tsr has a fifth methylation site (E502) whose signaling properties are also consistent with this mechanistic picture [63].
Changes in the packing stability of the MH bundle must in turn influence the conformation or dynamic motions of kinase control domain at the receptor tip to control CheA activity. Modulation of helix-helix packing stability by engineered inter-helix disulfide bonds and truncations of helix-packing ‘knob’ residues revealed a ‘yin-yang’ relationship between the MH and tip bundles: The ON state is favored by both enhanced packing stability in the MH bundle and by reduced packing stability in the tip bundle [19]. The MH and tip bundles are joined by a potentially flexible region [8] with conserved glycine residues at its midpoint [21] (Figure 1D) that might serve as a structural hinge in receptor function. How the flexible region transmits signaling changes between the sensory adaptation and protein interaction regions of the receptor remains an open question.
Two conserved residues in the hairpin tip (F396 and E391 in Tsr) provide some insights into the structural nature of receptor output states. E391 lies at the solvent-exposed turn in the hairpin tip (Figure 1D). Nonpolar amino acid replacements at this position, which might alter stability of the hairpin turn, cause fast switching of the cell’s flagellar motors, implying that tip dynamics play a role in kinase control [28]. F396 lies at a buried internal position in the tip bundle (Figure 1D). The F396 sidechains from each protomer stack on one another, stabilizing the dimer interface at the tip. Long molecular dynamics simulations revealed flips of the F396 stacking interaction and concomitant conformational changes in the tip that might represent alternative signal states [29]. Cysteine-directed crosslinking studies have also demonstrated state-dependent motions of the receptor tip residues that interact with CheA [9] and CheW [12] that might be central to the kinase control mechanism.
The core chemotaxis signaling complex
The core chemotaxis signaling complex is the smallest assembly of chemotaxis proteins capable of all core chemosensory functions, including kinase activation and control [64]. In vitro reconstitution (detailed in Box 2) rigorously defined the stoichiometry of the core complex as two receptor trimers-of-dimers, one CheA homodimer, and two molecules of the coupling protein CheW (Figure 2A) and demonstrated that this assembly is an independent signaling unit capable of receptor-mediated kinase regulation in response to attractant and adaptation signals [23, 64, 65]. The core complex is not only the fundamental unit of sensory activity, but also the fundamental structural and assembly unit of higher order arrays [66, 67] that are characteristic of chemotaxis systems across bacterial diversity [68].
Box 2. Chemoreceptors in Nanodiscs.
Core signaling complexes can be reconstituted with purified, intact proteins using Nanodiscs to provide a native environment for transmembrane chemoreceptors [23, 64, 65]. Nanodiscs are ~10 nm plugs of lipid bilayer rendered water-soluble by a belt of amphipathic membrane scaffold protein [91]. They provide water-soluble units of native lipid bilayer [92] in which purified transmembrane chemoreceptors, essentially inactive in detergent, can be incorporated and thereby regain activity [93, 94]. Activities restored depend on the number of potentially interacting receptors in the Nanodisc. Isolated dimers bind ligand, are modified by adaptational enzymes, perform transmembrane signaling, and couple protein conformation to ligand occupancy and adaptational modification [17]. Three receptor dimers inserted in parallel in a Nanodisc, and thus capable of making trimers, can form signaling complexes that activate kinase 750-fold, almost as well as receptors in native membrane, and control that activity as a function of ligand occupancy [23, 64, 65, 93]. Thus the central functions of kinase activation and control are properties of individual core signaling complexes.
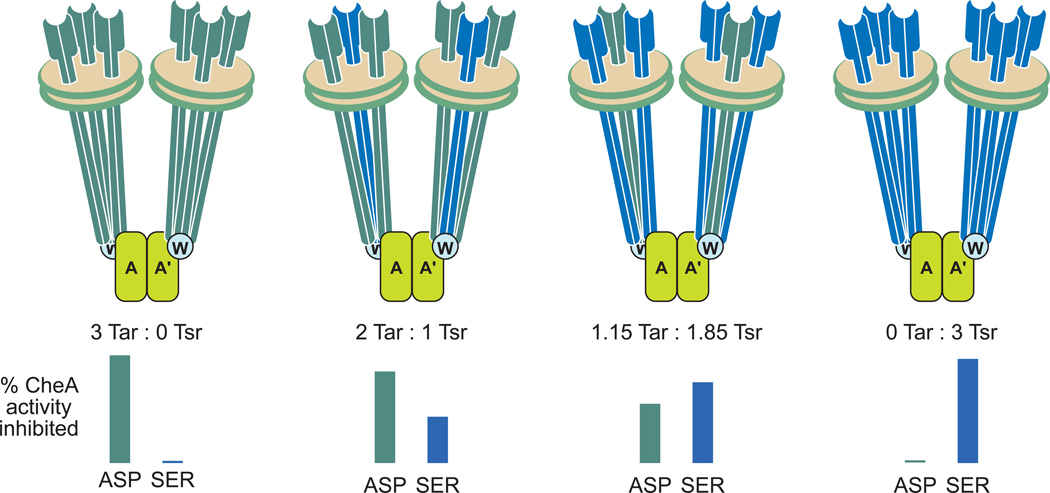
Mixed-receptor core complexes, in which Nanodisc-inserted trimers of receptor dimers contain two receptors with different ligand specificities, can be isolated using receptors carrying different affinity tags [65]. Changing assembly ratios can vary the relative amount of the two. Kinase inhibition in mixed complexes as a function of receptor ligand and trimer composition provided insight into the coupling of receptor occupancy to kinase (Figure I). For isolated signaling complexes with mixed trimers of aspartate receptor Tar and serine receptor Tsr, kinase activity was only partially inhibited by saturation with either ligand, and inhibition was less for signaling complexes containing lower proportions of the ligand-occupied receptor [65]. Several potential modes of receptor-kinase coupling were inconsistent with the data: kinase inhibition generated by ligand occupancy of any dimer in the trimer, by occupancy of a majority of dimers or proportional to the percentage of ligand-occupied dimers. Instead, the data could be explained if the structural asymmetry of receptor-kinase contacts observed in EM-tomography [10, 66, 67] creates functional asymmetry. Specifically, only one dimer in a receptor trimer contacts kinase and only one contacts CheW. The data suggest that kinase inhibition is transmitted through one of these direct interactions but not through receptor dimer-dimer interfaces and that inhibition passes allosterically with ~2/3 efficiency between kinase protomers [65]. Neighboring dimers in a trimer influence each other’s operational ligand affinity and cooperativity but only one of the three dimers mediates kinase inhibition upon ligand occupancy.
Box 2, Figure I. Kinase inhibition in mixed-receptor core signaling complexes
The cartoons represent soluble core signaling complexes assembled using Nanodisc-embedded trimers of receptor dimers with the indicated ratios of Tar to Tsr. The bar graphs show the extent of kinase inhibition at saturation with the Tar ligand aspartate (ASP) and the Tsr ligand serine (SER), respectively. See this box and reference [65] for descriptions of the experiments and results.
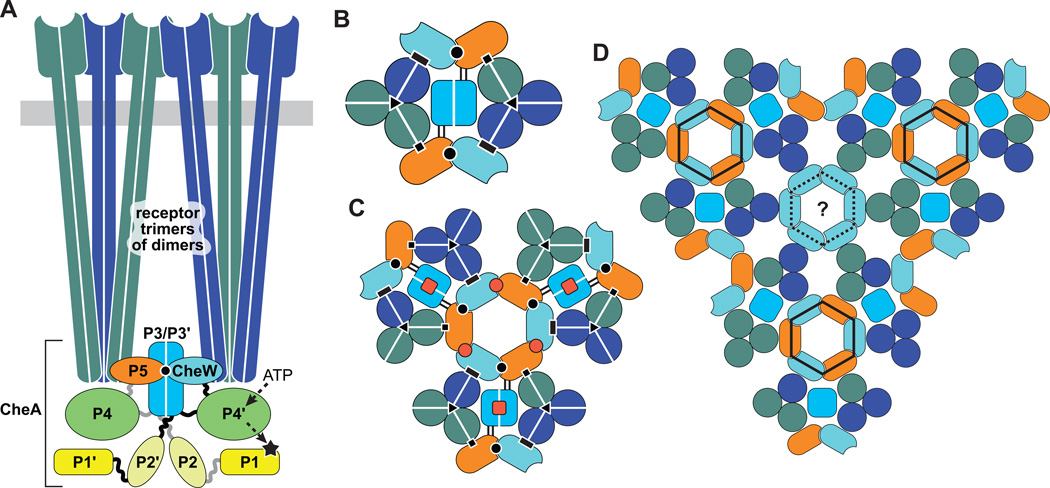
Figure 2. Receptor core complexes and arrays.
The same fill color conventions for the various signaling components and domains are used in all panels. White lines in panels A–C separate the protomers of homodimeric molecules.
(A) Core complex, the minimal unit of receptor signaling. Two receptor trimers of dimers and two CheW molecules are needed to activate and control a CheA dimer. The trimers can contain receptors with different detection specificities (dark green and dark blue). CheA protomers have five domains: P1 (phosphorylation site); P2 (CheB and CheY binding); P3 (dimerization domain); P4 (catalysis, P1 and ATP binding); P5 (receptor/CheW coupling and activity control). A binding interaction between CheW and its CheA-P5 paralog (black circle) is critical for core complex assembly.
(B) Cross-section through the CheA-P5/CheW baseplate of a core complex viewed from the cytoplasmic membrane toward the protein interaction tips of the receptors. Black symbols indicate protein-protein contacts involved in core complex assembly and function: P5-receptor (squares); CheW-receptor (rectangles); P5 [subdomain 1]-CheW [subdomain 2] (circles); trimer contacts between inner subunits of receptor dimers (triangles). Parallel black lines between the P5 and P3 domains of CheA indicate the linkers flanking the P4 domain, a likely route for signaling conformational changes in the core complex.
(C) Signaling connections between core complexes in the receptor array that may confer response amplification. Red squares indicate P3-P3' interactions that could transmit allosteric signals between CheA protomers. Red circles denote interface 2 interactions between P5 (subdomain 2) and CheW (subdomain 1) that could transmit allosteric signals through hexagonal P5-CheW rings.
(D) Proposed organization of core complexes in the receptor array. In addition to hexagonal P5-CheW rings (solid black line), hexagonal rings of CheW might also exist (broken black line).
Structural models of core complexes and their higher order assemblies have been generated by fitting electron densities from cryo-electron tomography of E. coli minicells with X-ray crystallographic structures of protein components and their sub-complexes [66, 67, 69]. In the deduced structure of the core complex (Figure 2A, 2B), the CheA dimer bridges two receptor trimers of dimers through interaction of its P5 domains with one receptor dimer in each of the two trimers of dimers. The two CheW proteins also bind to a receptor dimer in each trimer of dimers (Figure 2B). The four-helix bundle of the CheA P3/P3' dimerization domain is positioned between the two receptor trimers, parallel to their long axes, but does not appear to be in stable physical contact with other core complex components [70] (Figure 2A). Each CheA protomer also binds to one CheW through its P5 domain (Figure 2A and 2B). CheW and the CheA P5 domain are paralogs; each has two structurally similar subdomains. In the core complex, subdomain 1 of CheA P5 binds to subdomain 2 of CheW. This interaction links each CheA protomer to a receptor dimer in the opposite trimer of dimers, again bridging the two trimers. It follows that the two receptor trimers of dimers of the core complex are connected by the CheA dimerization domain and by two P5-CheW heterodimers. Binding of the P5-CheW heterodimer to the two receptor trimers of dimers positions the CheA ATP-binding domain (P4) membrane-distal to P5, and its phosphorylation site domain (P1) and CheY/CheB-binding domain (P2) membrane-distal to P4 [66, 69]. The contacts between receptor, CheA, and CheW proteins make the core complex a stable entity that persists through numerous manipulations in free solution [64, 65] and contribute to the ultra-stability of higher order complex assemblies [71–73].
Signal transmission in core complexes
Different E. coli receptors have identical trimer contact residues at their cytoplasmic tips and readily form mixed trimers of dimers [15]. Incorporation of chemoreceptors with different ligand specificities into the same soluble, Nanodisc-based core complex offers a useful platform with which to characterize coupling of receptors to each other and to the kinase. Such experiments indicate a signaling asymmetry in the trimer of dimers: Only one receptor is capable of inhibiting kinase activity [65]. However, heterologous non-signaling dimers affect the signaling dimer by reducing its apparent ligand affinity [65]. The unexpected signaling asymmetry is consistent with observations that (i) in mixed receptor trimers without CheA and CheW, ligand occupancy of one kind of receptor has differential structural effects on occupied versus unoccupied receptor dimers [74], (ii) in signaling complexes ligand binding and receptor modification have asymmetric effects on receptor conformation and coupling of receptor to kinase [75], and (iii) patterns of kinase inhibition by mixed heterologous receptors in arrays in vivo [76] and in vitro [77] are similar to the patterns for core complexes. Importantly, there is asymmetry in the deduced structure of the core complex: Only one dimer in a trimer of receptor dimers has physical contact with the kinase (via a P5 domain), only one dimer, a different one, has physical contact with a CheW (Figure 2A, 2B) [10, 66, 67], and the third has no interaction partner or may interact with one CheW that is part of an exclusively CheW hexagonal ring (Figure 2D) [10, 66, 67]. Together, these observations support the idea of functional asymmetry in which only one-third of the receptors in a core complex and presumably in higher order assemblies inhibit kinase as a function of ligand occupancy (Box 2).
The mechanism of CheA control in core complexes
CheA autophosphorylation is a trans reaction, involving interaction of the P1 domain in one protomer with the P4 domain of the other protomer (Figure 2A). MCP signaling complexes modulate CheA autophosphorylation activity over more than a 100-fold range, most likely through changes in receptor conformation or dynamic motions that allosterically regulate P1-P4' interactions and/or the kinase active site. A cryo-electron microscopy (cyro-EM) study of mutant receptor arrays with locked outputs indicated that receptors influence the dynamic behavior of CheA domains. Signaling complexes in the ON state had highly mobile CheA P1 and P2 domains, whereas those domains were much less mobile in receptor complexes locked in the OFF state [69]. The enhanced ON-state mobility may also include movements of the P4 domain [1, 69]
The structural features of core complexes are consistent with an allosteric mechanism of control in which receptors regulate P1-P4 interactions or the kinase active site by manipulating other CheA regions. The linker segments at each end of the P4 domain offer mechanistic possibilities because changes in their primary structures are known to influence CheA activity [78, 79]. Alterations in the P4-P5 linker can affect CheA activation; alterations in the P3-P4 linker can affect both basal and activated CheA reaction rates. Thus, stimulus-dependent manipulation of one or both linkers by the receptor, through its binding contacts with CheW and P5, might modulate CheA autophosphorylation activity by imposing conformational changes on the P4 domain. Those changes might shift P4's ATP-binding or catalytic determinants or its binding interactions with the P1 domain. Residues in the A helix of the P1 domain (a 4-helix bundle) promote a productive docking interaction with the P4 domain [80]. Residues in the D helix of P1 may promote a nonproductive docking interaction with P4 [81]. Perhaps manipulation of P4 through its flanking linkers controls CheA activity by presenting or occluding one of these P1 docking sites. Additionally, residue(s) in the receptor tip, acting as catalytic determinants or pseudosubstrate sites, might directly augment or inhibit the CheA autophosphorylation reaction. Available experimental evidence cannot distinguish among these mechanistic possibilities.
Signal amplification and sensory adaptation in receptor arrays
Core signaling complexes organize into higher-order arrays through hexagonal CheA P5-CheW rings, consisting of alternating P5-CheW interactions at interface 1, present in core complexes, and at interface 2, unique to the array (Figure 2C; Box 3). In the chemosensory array, attractant binding to a single receptor can regulate ~35 CheA kinase proteins [82]. This amplification of the attractant signal requires long-range signaling through the array. A likely route for that signal transmission is through P5-CheW Interface 2, which serves as the lone bridge between adjacent core complexes in the array [66, 67] (Figure 2C, 2D). The extended array might be additionally networked by hexagonal CheW rings, which have been proposed to interact with the receptor molecules that do not make contact with either CheW or P5 in a core complex (Figure 2D) [66].
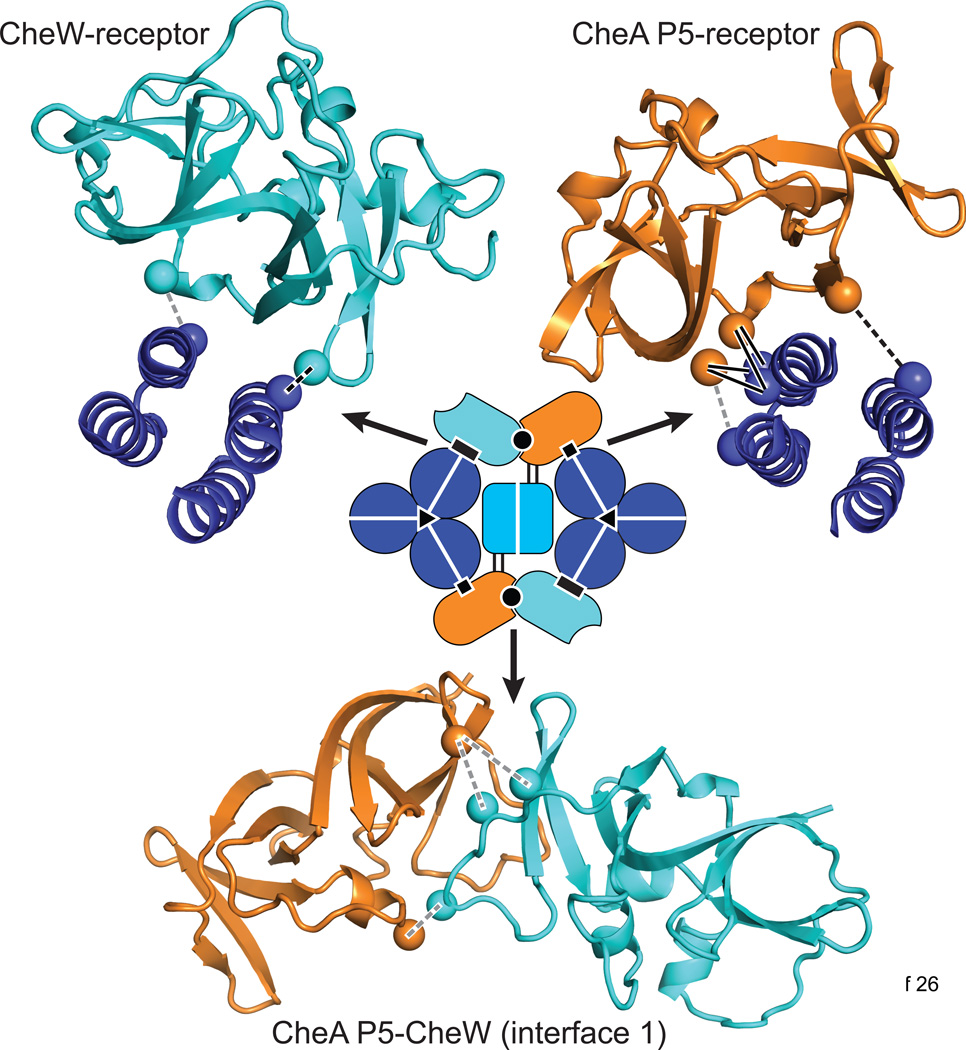
Box 3. Protein-protein contacts in the receptor array.
Repeating protein-protein contacts anchor the three core components (receptors, CheW, and CheA) in the core unit and the extended array. Recent findings from independent cryo-EM, X-ray crystallography, NMR, disulfide mapping, and molecular modeling studies have revealed key structural and dynamical features of three of those interfaces in the core unit, as summarized in Figure I [9–13, 66, 67, 95]. (i) Near the cytoplasmic hairpin tip of one receptor trimer-of dimers, the P5 regulatory domain of CheA contacts the N-terminal helix of one receptor protomer, forming a stable, high-affinity interface revealed by recent disulfide mapping [9] and crystallographic studies [9, 10]. Kinase-off receptor signals likely pass through this P5-receptor interface and appear to involve a rotation of the tight P5-receptor helix complex, as detected by attractant-triggered changes in disulfide formation rates across the interface [9]. (ii) The contact between CheW and the receptor is believed to be homologous to the P5-receptor interface, owing to the homologous structures of P5 and CheW, and to their equivalent placements in the pseudo-symmetric, hexagonal P5-CheW ring [10, 66, 67]. Disulfide mapping studies in cellular arrays provided direct support for this view and revealed an attractant-triggered rearrangement that appears to be similar or identical to that observed at the P5-receptor interface [12]. (iii) Interface 1 between P5 and CheW is critical to core complex assembly and function [96–98]. Disulfide mapping [95] showed that this interface is accurately portrayed by the known crystal structure of a P5-CheW complex [99], and revealed attractant-triggered decreases in disulfide formation rates. The latter findings suggest that attractant signals may reduce the local dynamics of P5-CheW interface 1, perhaps representing a core complex change capable of propagation into the P5-CheW ring [65, 95]. A fourth interface in the core complex, the trimer-of-dimer contacts between receptor molecules, is also critical for core complex assembly and function [27]. The trimer axis might serve as a conformational fulcrum that allows rotational motions of the outer receptor helices during signaling [29].
In the extended array, a second P5-CheW interaction (interface 2) joins core complexes through a hexagonal P5-CheW ring (Figure 2). A crystal structure of this interface exists [10], but has not yet been explored by disulfide mapping or mutational analyses. Because interface 2 bridges adjacent core units in the larger array, it presumably transmits attractant-triggered changes between them. Thus, interface 2 may be largely or exclusively responsible for the dramatic cooperative signaling properties of the native chemosensory array.
Box 3, Figure I. Three key interfaces in the core signaling complex
Colors of the atomic backbone structures are keyed to the core complex schematic: receptor dimer (blue), CheW (cyan), and CheA P5 (orange). Spheres indicate the β-carbon atoms of the cysteine crosslinking reporters that identified interface residues. Solid black lines indicate disulfide bonds whose formation rates are insensitive to signaling state. Dashed black lines indicate crosslinks that form more readily in the presence of attractant; dashed gray lines indicate crosslinks that form less readily in the presence of attractant. In vitro disulfide crosslinking experiments provided data for the P5-receptor [9] and P5-CheW [95] interfaces. Data for the CheW-receptor interface came from in vivo disulfide crosslinking experiments [12].
The molecular basis of long-range signal transmission is not yet known, but could involve changes in the dynamic behaviors of the interconnected array components. Such changes must be relatively subtle ones because arrays in different signaling states exhibit the same overall architecture [69, 70]. Although attractant stimuli do not disrupt receptor clusters, they do elicit detectable changes in receptor packing and signaling cooperativity that are consistent with altered dynamic behaviors of the array elements [83]. A plausible model is that attractant binding switches a core complex from a dynamic ON state to a more static OFF state and that those dynamic shifts propagate to neighboring core complexes through the P5-CheW rings and to neighboring P5-CheW rings through the connecting CheA dimers. Ensuing covalent modifications of the receptor molecules presumably restore the prestimulus dynamic behavior of the core complexes to achieve sensory adaptation.
High-speed observations of the flagellar motors on E. coli cells recently provided dramatic evidence of cooperative behavior in receptor arrays [84, 85]. Two motors on the same cell reversed in synchrony. The motor that was closer to the cell’s receptor cluster reversed first, followed by the second motor with a time lag determined by its distance from the first motor and consistent with the intracellular diffusion rate of phospho-CheY, the CW rotational signal. These observations imply that a cell’s receptor cluster produces phospho-CheY signals in a pulsatile manner, consistent with essentially all-ON or all-OFF array behavior.
Concluding remarks
The advent of detailed structural models of receptor molecules, their core signaling complexes, and their higher-order arrays has opened the door to deeper molecular understanding of these extraordinary signal-processing devices. The idea that bacterial chemoreceptors convey stimulus information through dynamic changes rather than a few discrete structural states is an appealing one, but supporting data will not come easily because protein motions occur over a vast range of timescales that are exceedingly difficult to monitor under native physiological conditions. Which timescales are most relevant to signal transmission? How can those motions be documented and measured in working receptor molecules? Over the next five years of chemoreceptor research, methods capable of revealing protein dynamic behaviors, such as hydrogen/deuterium exchange rates [86, 87] and electron paramagnetic resonance [88], should make valuable contributions to our understanding of receptor signaling. In addition, novel combinations of experimental approaches, for example molecular dynamics simulations on well-characterized mutant receptors, will help to address the technical challenges posed by dynamics-based signaling mechanisms (Box 4).
Box 4. Outstanding questions.
What are the mechanisms of signal transmission between structural modules in the chemoreceptor cytoplasmic domain?
How does the HAMP domain convert an input helix piston motion into an output conformational and/or dynamic change in the cytoplasmic domain?
What is the molecular mechanism of allosteric coupling among chemoreceptors in a trimer of dimers?
What are the mechanisms of kinase control in core complexes?
How are kinase control signals transmitted between core complexes in hexagons and across the extended array?
If signaling involves changes in dynamics, what is the relevant timescale and how can those changes be detected?
If signaling involves changes in both dynamics and average structure, how can we tell which is more important?
What conformational or dynamic differences determine the substrate properties of receptor molecules for the enzymes of sensory adaptation?
Highlights.
Escherichia coli uses transmembrane chemoreceptors to detect environmental chemicals.
Chemoreceptors convey signals through dynamic shifts in coupled structural elements.
Core signaling complexes contain six receptors coupled to one kinase molecule.
Networked core complexes form cooperative, high-amplification signaling arrays.
Acknowledgements
Research in the authors’ laboratories was supported by NIH research grants GM19559 (J.S.P.), GM29963 (G.L.H.) and GM40731 (J.J.F.). We regret that the limited scope of this article precluded a more comprehensive coverage of the literature on bacterial chemoreceptors.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Falke JJ, Piasta KN. Architecture and signal transduction mechanism of the bacterial chemosensory array: Progress, controversies, and challenges. Curr. Opin. Struc. Biol. 2014;29:85–94. doi: 10.1016/j.sbi.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sourjik V, Wingreen NS. Responding to chemical gradients: bacterial chemotaxis. Curr. Opin. Cell Biol. 2012;24:262–268. doi: 10.1016/j.ceb.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Porter SL, et al. Signal processing in complex chemotaxis pathways. Nature reviews. Microbiology. 2011;9:153–165. doi: 10.1038/nrmicro2505. [DOI] [PubMed] [Google Scholar]
- 4.Krell T, et al. Diversity at its best: bacterial taxis. Environ Microbiol. 2011;13:1115–1124. doi: 10.1111/j.1462-2920.2010.02383.x. [DOI] [PubMed] [Google Scholar]
- 5.Sourjik V, Armitage JP. Spatial organization in bacterial chemotaxis. EMBO J. 2010;29:2724–2733. doi: 10.1038/emboj.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hazelbauer GL, Lai WC. Bacterial chemoreceptors: providing enhanced features to two-component signaling. Curr. Opin. Microbiol. 2010;13:124–132. doi: 10.1016/j.mib.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hazelbauer GL, et al. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends. Biochem. Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alexander RP, Zhulin IB. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc. Natl. Acad. Sci. USA. 2007;104:2885–2890. doi: 10.1073/pnas.0609359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piasta KN, et al. Defining a key receptor-CheA kinase contact and elucidating its function in the membrane-bound bacterial chemosensory array: a disulfide mapping and TAM-IDS Study. Biochemistry. 2013;52:3866–3880. doi: 10.1021/bi400385c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X. The 3.2 A resolution structure of a receptor: CheA:CheW signaling complex defines overlapping binding sites and key residue interactions within bacterial chemosensory arrays. Biochemistry. 2013;52:3852–3865. doi: 10.1021/bi400383e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, et al. CheA-receptor interaction sites in bacterial chemotaxis. J. Mol. Biol. 2012;422:282–290. doi: 10.1016/j.jmb.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pedetta A, et al. Signalling-dependent interactions between the kinase-coupling protein CheW and chemoreceptors in living cells. Mol. Microbiol. 2014;93:1144–1155. doi: 10.1111/mmi.12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vu A. The receptor-CheW binding interface in bacterial chemotaxis. J. Mol. Biol. 2012;415:759–767. doi: 10.1016/j.jmb.2011.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim KK, et al. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 15.Studdert CA, Parkinson JS. Crosslinking snapshots of bacterial chemoreceptor squads. Proc. Natl. Acad. Sci. USA. 2004;101:2117–2122. doi: 10.1073/pnas.0308622100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gosink KK, et al. Signaling interactions between the aerotaxis transducer Aer and heterologous chemoreceptors in Escherichia coli. J. Bacteriol. 2006;188:3487–3493. doi: 10.1128/JB.188.10.3487-3493.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amin DN, Hazelbauer GL. The chemoreceptor dimer is the unit of conformational coupling and transmembrane signaling. J. Bacteriol. 2010;192:1193–1200. doi: 10.1128/JB.01391-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu W. A possible degree of motional freedom in bacterial chemoreceptor cytoplasmic domains and its potential role in signal transduction. Int. J. Biochem. Mol. Biol. 2011;2:99–110. [PMC free article] [PubMed] [Google Scholar]
- 19.Swain KE, et al. Engineered socket study of signaling through a four-helix bundle: evidence for a yin-yang mechanism in the kinase control module of the aspartate receptor. Biochemistry. 2009;48:9266–9277. doi: 10.1021/bi901020d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park H, et al. Transmembrane signaling of chemotaxis receptor Tar: insights from molecular dynamics simulation studies. Biophys. J. 2011;100:2955–2963. doi: 10.1016/j.bpj.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coleman MD, et al. Conserved glycine residues in the cytoplasmic domain of the aspartate receptor play essential roles in kinase coupling and on-off switching. Biochemistry. 2005;44:7687–7695. doi: 10.1021/bi0501479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall BA, et al. Mechanism of bacterial signal transduction revealed by molecular dynamics of Tsr dimers and trimers of dimers in lipid vesicles. PLoS Comput Biol. 2012;8:e1002685. doi: 10.1371/journal.pcbi.1002685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li M. Chemotaxis kinase CheA is activated by three neighbouring chemoreceptor dimers as effectively as by receptor clusters. Mol. Microbiol. 2011;79:677–685. doi: 10.1111/j.1365-2958.2010.07478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swain KE, Falke JJ. Structure of the conserved HAMP domain in an intact, membrane-bound chemoreceptor: a disulfide mapping study. Biochemistry. 2007;46:13684–13695. doi: 10.1021/bi701832b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pollard AM, et al. The structure of a soluble chemoreceptor suggests a mechanism for propagating conformational signals. Biochemistry. 2009;48:1936–1944. doi: 10.1021/bi801727m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferris HU, et al. Axial helix rotation as a mechanism for signal regulation inferred from the crystallographic analysis of the E. coli serine chemoreceptor. J. Struct. Biol. 2014;186:349–356. doi: 10.1016/j.jsb.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 27.Gosink KK, et al. Mutational analysis of N381, a key trimer contact residue in Tsr, the Escherichia coli serine chemoreceptor. J. Bacteriol. 2011;193:6452–6460. doi: 10.1128/JB.05887-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mowery P, et al. Different signaling roles of two conserved residues in the cytoplasmic hairpin tip of Tsr, the Escherichia coli serine chemoreceptor. J. Bacteriol. 2008;190:8065–8074. doi: 10.1128/JB.01121-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ortega DR, et al. A phenylalanine rotameric switch for signal-state control in bacterial chemoreceptors. Nature communications. 2013;4:2881. doi: 10.1038/ncomms3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Falke JJ, Hazelbauer GL. Transmembrane signaling in bacterial chemoreceptors. Trends. Biochem. Sci. 2001;26:257–265. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Falke JJ, Erbse AH. The piston rises again. Structure. 2009;17:1149–1151. doi: 10.1016/j.str.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall BA, et al. Transmembrane helix dynamics of bacterial chemoreceptors supports a piston model of signalling. PLoS Comput Biol. 2011;7:e1002204. doi: 10.1371/journal.pcbi.1002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Falke JJ. Piston versus scissors: chemotaxis receptors versus sensor His-kinase receptors in two-component signaling pathways. Structure. 2014;22:1219–1220. doi: 10.1016/j.str.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parkinson JS. Signaling mechanisms of HAMP domains in chemoreceptors and sensor kinases. Annu. Rev. Microbiol. 2010;64:101–122. doi: 10.1146/annurev.micro.112408.134215. [DOI] [PubMed] [Google Scholar]
- 35.Dunin-Horkawicz S, Lupas AN. Comprehensive analysis of HAMP domains: implications for transmembrane signal transduction. J. Mol. Biol. 2010;397:1156–1174. doi: 10.1016/j.jmb.2010.02.031. [DOI] [PubMed] [Google Scholar]
- 36.Hulko M, et al. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell. 2006;126:929–940. doi: 10.1016/j.cell.2006.06.058. [DOI] [PubMed] [Google Scholar]
- 37.Airola MV, et al. Structure of concatenated HAMP domains provides a mechanism for signal transduction. Structure. 2010;18:436–448. doi: 10.1016/j.str.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang C, et al. Mechanistic insights revealed by the crystal structure of a histidine kinase with signal transducer and sensor domains. PLoS Biol. 2013;11:e1001493. doi: 10.1371/journal.pbio.1001493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watts KJ, et al. Structure-function relationships in the HAMP and proximal signaling domains of the aerotaxis receptor Aer. J. Bacteriol. 2008;190:2118–2127. doi: 10.1128/JB.01858-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ames P, et al. Mutational analysis of the connector segment in the HAMP domain of Tsr, the Escherichia coli serine chemoreceptor. J. Bacteriol. 2008;190:6676–6685. doi: 10.1128/JB.00750-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Q, et al. Mutational analyses of HAMP helices suggest a dynamic bundle model of input-output signalling in chemoreceptors. Mol. Microbiol. 2009;73:801–814. doi: 10.1111/j.1365-2958.2009.06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Q, et al. Biphasic control logic of HAMP domain signalling in the Escherichia coli serine chemoreceptor. Mol. Microbiol. 2011;80:596–611. doi: 10.1111/j.1365-2958.2011.07577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ames P, et al. HAMP domain structural determinants for signalling and sensory adaptation in Tsr, the Escherichia coli serine chemoreceptor. Mol. Microbiol. 2014;91:875–886. doi: 10.1111/mmi.12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Airola MV, et al. HAMP domain conformers that propagate opposite signals in bacterial chemoreceptors. PLoS Biol. 2013;11:e1001479. doi: 10.1371/journal.pbio.1001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stewart V. The HAMP signal-conversion domain: static two-state or dynamic three-state? Mol. Microbiol. 2014;91:853–857. doi: 10.1111/mmi.12516. [DOI] [PubMed] [Google Scholar]
- 46.Ferris HU, et al. The mechanisms of HAMP-mediated signaling in transmembrane receptors. Structure. 2011;19:378–385. doi: 10.1016/j.str.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 47.Watts KJ, et al. Different conformations of the kinase-on and kinase-off signaling states in the Aer HAMP domain. J. Bacteriol. 2011;193:4095–4103. doi: 10.1128/JB.01069-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mechaly AE, et al. Segmental helical motions and dynamical asymmetry modulate histidine kinase autophosphorylation. PLoS Biol. 2014;12:e1001776. doi: 10.1371/journal.pbio.1001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klose D, et al. Light-induced switching of HAMP domain conformation and dynamics revealed by time-resolved EPR spectroscopy. FEBS Lett. 2014;588:3970–3976. doi: 10.1016/j.febslet.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 50.Zhu L. The HAMP signal relay domain adopts multiple conformational states through collective piston and tilt motions. PLoS Comput Biol. 2013;9:e1002913. doi: 10.1371/journal.pcbi.1002913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lai RZ, Parkinson JS. Functional suppression of HAMP domain signaling defects in the E. coli serine chemoreceptor. J. Mol. Biol. 2014;426:3642–3655. doi: 10.1016/j.jmb.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kitanovic S, et al. Mutational analysis of the control cable that mediates transmembrane signaling in the E. coli serine chemoreceptor. J. Bacteriol. 2011;193:5062–5072. doi: 10.1128/JB.05683-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wright GA, et al. Mutational analysis of the transmembrane helix 2-HAMP domain connection in the Escherichia coli aspartate chemoreceptor Tar. J. Bacteriol. 2011;193:82–90. doi: 10.1128/JB.00953-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adase CA, et al. Residues at the cytoplasmic end of transmembrane helix 2 determine the signal output of the TarEc chemoreceptor. Biochemistry. 2013;52:2729–2738. doi: 10.1021/bi4002002. [DOI] [PubMed] [Google Scholar]
- 55.Miller AS, Falke JJ. Side chains at the membrane-water interface modulate the signaling state of a transmembrane receptor. Biochemistry. 2004;43:1763–1770. doi: 10.1021/bi0360206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Draheim RR, et al. Tryptophan residues flanking the second transmembrane helix (TM2) set the signaling state of the Tar chemoreceptor. Biochemistry. 2005;44:1268–1277. doi: 10.1021/bi048969d. [DOI] [PubMed] [Google Scholar]
- 57.Adase CA, et al. The residue composition of the aromatic anchor of the second transmembrane helix determines the signaling properties of the aspartate/maltose chemoreceptor Tar of Escherichia coli. Biochemistry. 2012;51:1925–1932. doi: 10.1021/bi201555x. [DOI] [PubMed] [Google Scholar]
- 58.Draheim RR, et al. Tuning a bacterial chemoreceptor with protein-membrane interactions. Biochemistry. 2006;45:14655–14664. doi: 10.1021/bi061259i. [DOI] [PubMed] [Google Scholar]
- 59.Danielson MA, et al. Cysteine and disulfide scanning reveals a regulatory alpha-helix in the cytoplasmic domain of the aspartate receptor. J. Biol. Chem. 1997;272:32878–32888. doi: 10.1074/jbc.272.52.32878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stewart V, Chen LL. The S helix mediates signal transmission as a HAMP domain coiled-coil extension in the NarX nitrate sensor from Escherichia coli K-12. J. Bacteriol. 2010;192:734–745. doi: 10.1128/JB.00172-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Starrett DJ, Falke JJ. Adaptation mechanism of the aspartate receptor: electrostatics of the adaptation subdomain play a key role in modulating kinase activity. Biochemistry. 2005;44:1550–1560. doi: 10.1021/bi048089z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Winston SE, et al. Evidence that the adaptation region of the aspartate receptor is a dynamic four-helix bundle: cysteine and disulfide scanning studies. Biochemistry. 2005;44:12655–12666. doi: 10.1021/bi0507884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Han XS, Parkinson JS. An unorthodox sensory adaptation site in the Escherichia coli serine chemoreceptor. J. Bacteriol. 2014;196:641–649. doi: 10.1128/JB.01164-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li M, Hazelbauer GL. Core unit of chemotaxis signaling complexes. Proc. Natl. Acad. Sci. USA. 2011;108:9390–9395. doi: 10.1073/pnas.1104824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li M, Hazelbauer GL. Selective allosteric coupling in core chemotaxis signaling complexes. Proc. Natl. Acad. Sci. USA. 2014;111:15940–15945. doi: 10.1073/pnas.1415184111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu J. Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells. Proc. Natl. Acad. Sci. USA. 2012;109:E1481–E1488. doi: 10.1073/pnas.1200781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Briegel A, et al. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc. Natl. Acad. Sci. USA. 2012;109:3766–3771. doi: 10.1073/pnas.1115719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Briegel A, et al. Universal architecture of bacterial chemoreceptor arrays. Proc. Natl. Acad. Sci. USA. 2009;106:17181–17186. doi: 10.1073/pnas.0905181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Briegel A, et al. The mobility of two kinase domains in the Escherichia coli chemoreceptor array varies with signalling state. Mol. Microbiol. 2013;89:831–841. doi: 10.1111/mmi.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Briegel A, et al. Activated chemoreceptor arrays remain intact and hexagonally packed. Mol. Microbiol. 2011;82:748–757. doi: 10.1111/j.1365-2958.2011.07854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Erbse AH, Falke JJ. The core signaling proteins of bacterial chemotaxis assemble to form an ultrastable complex. Biochemistry. 2009;48:6975–6987. doi: 10.1021/bi900641c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Slivka PF, Falke JJ. Isolated bacterial chemosensory array possesses quasi- and ultrastable components: functional links between array stability, cooperativity, and order. Biochemistry. 2012;51:10218–10228. doi: 10.1021/bi301287h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Piasta KN, Falke JJ. Increasing and decreasing the ultrastability of bacterial chemotaxis core signaling complexes by modifying protein-protein contacts. Biochemistry. 2014;53:5592–5600. doi: 10.1021/bi500849p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vaknin A, Berg HC. Direct evidence for coupling between bacterial chemoreceptors. J. Mol. Biol. 2008;382:573–577. doi: 10.1016/j.jmb.2008.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Amin DN, Hazelbauer GL. Chemoreceptors in signalling complexes: shifted conformation and asymmetric coupling. Mol. Microbiol. 2010;78:1313–1323. doi: 10.1111/j.1365-2958.2010.07408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sourjik V, Berg HC. Functional interactions between receptors in bacterial chemotaxis. Nature. 2004;428:437–441. doi: 10.1038/nature02406. [DOI] [PubMed] [Google Scholar]
- 77.Lai RZ, et al. Cooperative signaling among bacterial chemoreceptors. Biochemistry. 2005;44:14298–14307. doi: 10.1021/bi050567y. [DOI] [PubMed] [Google Scholar]
- 78.Wang X, et al. Computational and experimental analyses reveal the essential roles of interdomain linkers in the biological function of chemotaxis histidine kinase CheA. J. Am. Chem. Soc. 2012;134:16107–16110. doi: 10.1021/ja3056694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang X, et al. The linker between the dimerization and catalytic domains of the CheA histidine kinase propagates changes in structure and dynamics that are important for enzymatic activity. Biochemistry. 2014;53:855–861. doi: 10.1021/bi4012379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nishiyama S, et al. Mutational analysis of the P1 phosphorylation domain in Escherichia coli CheA, the signaling kinase for chemotaxis. J. Bacteriol. 2014;196:257–264. doi: 10.1128/JB.01167-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hamel DJ, et al. Chemical-shift-perturbation mapping of the phosphotransfer and catalytic domain interaction in the histidine autokinase CheA from Thermotoga maritima. Biochemistry. 2006;45:9509–9517. doi: 10.1021/bi060798k. [DOI] [PubMed] [Google Scholar]
- 82.Sourjik V, Berg HC. Receptor sensitivity in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA. 2002;99:123–127. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Frank V, Vaknin A. Prolonged stimuli alter the bacterial chemosensory clusters. Mol. Microbiol. 2013;88:634–644. doi: 10.1111/mmi.12215. [DOI] [PubMed] [Google Scholar]
- 84.Fukuoka H, et al. Direct imaging of intracellular signaling components that regulate bacterial chemotaxis. Sci Signal. 2014;7:ra32. doi: 10.1126/scisignal.2004963. [DOI] [PubMed] [Google Scholar]
- 85.Terasawa S, et al. Coordinated reversal of flagellar motors on a single Escherichia coli cell. Biophys. J. 2011;100:2193–2200. doi: 10.1016/j.bpj.2011.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Koshy SS, et al. Hydrogen exchange mass spectrometry of functional membrane-bound chemotaxis receptor complexes. Biochemistry. 2013;52:8833–8842. doi: 10.1021/bi401261b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Koshy SS, et al. Hydrogen exchange differences between chemoreceptor signaling complexes localize to functionally important subdomains. Biochemistry. 2014;53:7755–7764. doi: 10.1021/bi500657v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Samanta D, et al. Bacterial chemoreceptor dynamics correlate with activity state and are coupled over long distances. Proc. Natl. Acad. Sci. USA. 2015;112:2455–2460. doi: 10.1073/pnas.1414155112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hegde M, et al. Chemotaxis to the quorum-sensing signal AI-2 requires the Tsr chemoreceptor and the periplasmic LsrB AI-2-binding protein. J. Bacteriol. 2011;193:768–773. doi: 10.1128/JB.01196-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pasupuleti S, et al. Chemotaxis of Escherichia coli to Norepinephrine (NE) Requires Conversion of NE to 3,4-Dihydroxymandelic Acid. J. Bacteriol. 2014;196:3992–4000. doi: 10.1128/JB.02065-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Denisov IG, et al. Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J. Am. Chem. Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 92.Amin DN, Hazelbauer GL. Influence of membrane lipid composition on a transmembrane bacterial chemoreceptor. J. Biol. Chem. 2012;287:41697–41705. doi: 10.1074/jbc.M112.415588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Boldog T, et al. Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc. Natl. Acad. Sci. USA. 2006;103:11509–11514. doi: 10.1073/pnas.0604988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Boldog T, et al. Using Nanodiscs to create water-soluble transmembrane chemoreceptors inserted in lipid bilayers. Methods Enzymol. 2007;423:317–335. doi: 10.1016/S0076-6879(07)23014-9. [DOI] [PubMed] [Google Scholar]
- 95.Natale AM, et al. Structure, function, and on-off switching of a core unit contact between CheA kinase and CheW adaptor protein in the bacterial chemosensory array: A disulfide mapping and mutagenesis study. Biochemistry. 2013;52:7753–7765. doi: 10.1021/bi401159k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhao J, Parkinson JS. Mutational analysis of the chemoreceptor-coupling domain of the Escherichia coli chemotaxis signaling kinase CheA. J. Bacteriol. 2006;188:3299–3307. doi: 10.1128/JB.188.9.3299-3307.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhao J, Parkinson JS. Cysteine-scanning analysis of the chemoreceptor-coupling domain of the Escherichia coli chemotaxis signaling kinase CheA. J. Bacteriol. 2006;188:4321–4330. doi: 10.1128/JB.00274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Miller AS, et al. CheA kinase of bacterial chemotaxis: chemical mapping of four essential docking sites. Biochemistry. 2006;45:8699–8711. doi: 10.1021/bi060580y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Park SY, et al. Reconstruction of the chemotaxis receptor-kinase assembly. Nat Struct Mol Biol. 2006;13:400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]