Abstract
Selecting near-native conformations from the immense number of conformations generated by docking programs remains a major challenge in molecular docking. We introduce DockRank, a novel approach to scoring docked conformations based on the degree to which the interface residues of the docked conformation match a set of predicted interface residues. Dock-Rank uses interface residues predicted by partner-specific sequence homology-based protein–protein interface predictor (PS-HomPPI), which predicts the interface residues of a query protein with a specific interaction partner. We compared the performance of DockRank with several state-of-the-art docking scoring functions using Success Rate (the percentage of cases that have at least one near-native conformation among the top m conformations) and Hit Rate (the percentage of near-native conformations that are included among the top m conformations). In cases where it is possible to obtain partner-specific (PS) interface predictions from PS-HomPPI, DockRank consistently outperforms both (i) ZRank and IRAD, two state-of-the-art energy-based scoring functions (improving Success Rate by up to 4-fold); and (ii) Variants of DockRank that use predicted interface residues obtained from several protein interface predictors that do not take into account the binding partner in making interface predictions (improving success rate by up to 39-fold). The latter result underscores the importance of using partner-specific interface residues in scoring docked conformations. We show that DockRank, when used to re-rank the conformations returned by ClusPro, improves upon the original ClusPro rankings in terms of both Success Rate and Hit Rate. DockRank is available as a server at http://einstein.cs.iastate.edu/DockRank/.
Keywords: protein complex structure prediction, protein–protein docking, partner-specific protein–protein interface residue prediction, docking scoring functions, sequence homologs, homo-interologs
INTRODUCTION
The 3D structures of complexes formed by interacting proteins are valuable sources of information needed to understand the structural basis of interactions and their role in pathways that orchestrate key cellular processes. High-throughput methods such as yeast-2-hybrid (Y2H) assays provide a source of information about possible pairwise interactions between proteins, but not the structures of the corresponding complexes.1 Because of the expense and effort associated with X-ray crystallography or NMR experiments to determine 3D structures of protein complexes, the gap between the number of possible interactions and the number of experimentally determined structures is rapidly expanding. Hence, there is considerable interest in computational methods for determining the structures of complexes formed by proteins. This is especially important in the case of complexes resulting from transient and nonobligate interactions, which tend to be partner-specific2,3 and play important roles in cellular communication and signaling pathways.3,4 When the structures of individual proteins are known or can be predicted with sufficiently high accuracy, docking methods can be used to predict the 3D conformation of complexes formed by two or more interacting proteins, to identify and prioritize drug targets in computational drug design,5 and to potentially validate6,7 or to provide putative structural models8,9 for interactions determined using high-throughput methods such as Y2H assays.
In general, solving the protein–protein docking problem involves three steps10–13: (1) generation of candidate conformations (models or decoys) by sampling the space of possible conformations of complexes formed by the given component proteins. The resulting large number (typically thousands to tens of thousands) of putative conformations are ranked and filtered using some criteria (e.g., geometric complementarity assessed using the Fast Fourier Transform (FFT)); (2) clustering of the top ranked conformations and often ranking the resulting clusters by their size, that is, the number of conformations contained in the clusters; (3) refinement of docked structures and final conformation selection.
Substantial efforts have been dedicated to the design of scoring (ranking) functions for docking programs. Scoring functions in the literature can be broadly classified into four types: 1) geometric complementarity-based scoring functions; 2) energy-based scoring functions; 3) knowledge-based scoring functions; 4) hybrid functions that combine the scoring functions of the first three types.14–17 Geometric complementarity-based scoring functions represent an early generation of scoring functions used in docking programs. Vakser and coworkers18 introduced FFT to calculate the geometric fit between a receptor and ligand. The fast processing speed of FFT made a full conformational space search possible. Scoring functions of this type were successfully applied on bound protein–protein docking but could not perform well for unbound protein–protein docking because of the conformational changes upon binding.11 Energy-based scoring functions are designed to approximate the binding free energy of protein–protein assemblies.19–21 They usually consist of weighted energy terms for van der Waals interactions, electrostatic interactions, and solvation energies. Knowledge-based scoring functions can be grouped into three subtypes. (a) Knowledge-based weighted correlations,22,23 which take into consideration the complementarity of physicochemical properties to overcome the limitations of scoring functions that rely on geometric complementarity alone; (b) Knowledge-based pairwise potentials24–26 derived from observed statistical frequency of amino acid/atom contacts in databases of solved protein structures; (c) Machine-learning-based scoring methods, which can be further divided into three sub-types: (c1) Classifiers trained (on a data set of near-native and non-native protein conformations) to predict whether a query docked conformation is near-native or non-native27,28; (c2) Classifiers that are trained using data extracted from a set of protein complexes to predict the interface residues of proteins, which are then used to rank docked conformations29,30; (c3) Consensus scoring methods31,32 that use a weighted combination of the scores of multiple scoring functions to score conformations.
Despite recent advances in methods for scoring docked conformations, including those used by the state-of-theart docking programs, there is considerable room for improvement in methods for efficiently and reliably identifying near-native conformations from the large number of candidate conformations generated by docking programs.10,33 Hence, there is a need for computationally efficient scoring functions that can reliably distinguish the near-native conformations from non-native ones.
Against this background, we propose DockRank, a novel approach to scoring docked conformations. The intuition behind DockRank is as follows: From among the large number of docked conformations produced by docking a protein A with protein B, a scoring function that preferentially selects conformations that preserve the interface between A and B in the native state of the complex A: B will be able to successfully identify near-native conformations. However, since a goal of docking is to identify near-native conformation, the actual interface residues of the complex A: B in its native state are unknown, and hence cannot be used for scoring conformations. However, if we can reliably predict the residues that constitute the interface between A and B, we should be able to use the degree of agreement between the predicted interface residues and the interface residues of each docked conformation to score the conformations.
While a broad range of computational methods for protein–protein interface prediction have been proposed in the literature (reviewed in Refs. 34–36), barring a few exceptions,37–39 the vast majority of such methods focus on predicting the protein–protein interface residues of a query protein, without taking into account its specific interaction partner(s). Because most transient protein interactions tend to be partner-specific (PS),2 and reliably predicting transient binding sites presents a challenge for nonpartner-specific (NPS) prediction methods (i.e., interface predictors that do not take into consideration, a protein's binding partner in predicting interface residues),4,40,41 DockRank makes use of partner-specific sequence homology-based protein–protein interface predictor (PS-HomPPI),42 a sequence homology-based predictor of interface residues between a given pair of potentially interacting proteins.
PS-HomPPI has been shown to reliably predict the interface residues between a pair of interacting proteins whenever a homo-interolog, that is, a complex structure formed by the respective sequence homologs of the given pair of proteins, is available.42,43 PS-HomPPI has been shown to be effective at predicting interface residues in transient complexes associated with reversible, often highly specific, interactions. Hence, PS-HomPPI offers an especially attractive protein–protein interface prediction method for ranking docked conformations.
Given a docking case, that is, a pair of proteins A and B that are to be docked with each other, DockRank uses PS-HomPPI to predict the interface residues between A and B. It then compares the predicted interface residues with the interface residues in each of the docked conformations of A: B produced by the docking program. The greater the similarity of the interface of a docked conformation with the predicted interface from PS-HomPPI, the higher the rank of the corresponding conformation among all docked conformations. DockRank's reliance on partner-specific interface predictions is what distinguishes it from existing scoring functions that use predicted interfaces to rank docked conformations.29,30
In this study, we first compare the performance of DockRank with several state-of-the-art energy-based scoring functions: ZRank,20,44 IRAD19 and the energy functions built-in ClusPro 2.0.15,17,45 We then evaluate the performance of DockRank variants that use predicted interface residues obtained from several nonpartner-specific protein interface predictors. We also evaluate DockRank on several targets (docking cases) of the Critical Assessment of PRedicted Interactions (CAPRI).12,33 Finally, we illustrate that DockRank complements homology modeling methods for protein complexes such as superimposition and multimeric threading.46–48
An online implementation of DockRank is available at http://einstein.cs.iastate.edu/DockRank/.
MATERIALS AND METHODS
Decoy sets
In this study, for different purposes, we used two benchmark decoy sets: ZDock3-BM3 and ClusPro2-BM3. ZDock3-BM3 is used to compare DockRank with other scoring functions, and ClusPro2-BM3 is used to evaluate the extent to which DockRank can improve upon the ranking of conformations produced by docking programs such as ClusPro.15,17,45 ZDock3-BM3 decoy set faithfully reflects the initial population of conformations generated by ZDock 3.044 before the conformations are clustered or otherwise post-processed. ClusPro2-BM3 corresponds to top 30 conformations that represent the clusters of conformations output by ClusPro 2.0.15,17,45 We also evaluated DockRank on five CAPRI targets and compared with 29 CAPRI scorer groups.
ZDock3-BM3
Docking Benchmark 3.0 (BM3)49 consists of a set of nonredundant nonobligate complexes (3.25Å or better resolution, determined using X-ray crystallography) from three biochemical categories: enzyme-inhibitor, antibody-antigen, and “others”. This data set includes complexes that are categorized into three difficulty groups for benchmarking docking algorithms: Rigid-body (88 complexes), Medium (19), and Difficult (17), based on the conformational change upon binding. Obligate complexes are filtered out manually. BM3 originally had 124 cases. 2VIS (rigid-body), 1K4C (rigid-body), 1FC2 (rigid-body), 1N8O (rigid-body) were deleted because the bound complexes and the corresponding unbound complexes have different number of chains. 1K74 (rigid-body) was deleted because the sequence of chain D in the bound complex is different from the corresponding unbound chain 1ZGY_B. There are finally 119 docking complexes: Rigid-body (83 complexes), Medium (19), and Difficult (17). A set of 54,000 decoys for each case generated using ZDock 3.0 was downloaded from http://zlab.umassmed.edu/zdock/decoys.shtml. Despite the large number of generated decoys, there are only 97 cases that have at least one near-native structure (e.g., a decoy with interface Cα atom Root Mean Square Deviation I–RMSD ≤ 2:5Å). Out of these 97 cases, our homology-based protein–protein interface predictor, PS-HomPPI,42 returned interface predictions for only 67 cases. Therefore, our final decoy set consists of decoys generated for these 67 cases (see Supporting Information Table S1 for the PDB50 IDs for these cases).
ClusPro2-BM3
ClusPro2-BM3 was also generated from the 119 cases in BM3 using ClusPro 2.0 program. For each docking case ClusPro returned 30 conformations. ClusPro2-BM3_31 set of 31 cases was generated using the following selection criteria: i) each case should have at least one hit (i.e., a docked conformation with ligand Root Mean Square Deviation L2RMSD ≤ 10Å); ii) PS-HomPPI interface predictions are available for the proteins in that complex. To evaluate the capability of DockRank to give top ranks to meaningful though incorrect conformations in cases for which ClusPro returns no hits, another set of 45 cases, ClusPro2-BM3_45, was generated by relaxing the definition of a hit to include conformations with L2RMSD ≤ 15Å (see Supporting Information Table S1 and S2 for the corresponding PDB IDs for ClusPro2-BM3_31 set and ClusPro2-BM3_45 set).
CAPRI uploader decoys
CAPRI is a community wide competition of computational protein complex modeling and scoring methods (http://www.ebi.ac.uk/msd-srv/capri/). In the scoring experiments of the CAPRI competition, docking groups uploaded 100 models for each target, which are referred to as “uploader models” and can be downloaded by the participating scorers to rank and submit their selected top 10 models. Through e-mail communications with the CAPRI organizers, we obtained the uploader models and the corresponding CAPRI model classification files for Targets 30 (trimer), 35 (dimer), 36 (dimer), 41 (dimer), 47 (hexamer), and 50 (trimer). We discarded the data for Target 36 because none of the uploader models is near-native. We also discarded models with no chain IDs at all, because DockRank, which is based on the predicted interfaces between chains, requires the information of protein chain boundaries. Specifically, 60 uploader models for Target 30, 99 uploader models for Target 41 and 299 uploader models for Target 50 have no chain IDs at all and were discarded from this study. We were left with a total of 1,283 uploader models for Target 30, of which two models are acceptable models (based on CAPRI criteria13); a total of 499 uploader models for Target 35, of which three models are acceptable models; a total of 1,100 uploader models for Target 41, of which 230, 115 and two models are acceptable, medium, and high-quality models, respectively; a total of 1051 models for Target 47, of which 26, 307, and 278 models are acceptable, medium, and high-quality models, respectively; and a total of 1,152 models for Target 50, of which 84 and 21 models are acceptable and medium quality models, respectively. Because high-quality bound complexes were publicly available for Target 47 at the time of the competition, docking was not needed; instead, the prediction of interface water molecule positions was the real challenge. To evaluate the prediction of interface water molecule positions for Target 47, we used the CAPRI criteria fw (nat), which is the of fraction of actual water-mediated contacts in the target bound complex that are correctly predicted in the docked model. A pair of amino acids, each from the receptor or the ligand, is called a water-mediated contact if both of the amino acids have at least one heavy atom within a 3.5Å distance of the same water molecule. fw(nat) is analogous to an evaluation term in the protein interface prediction literature [73, 74], Sensitivity (see Supporting Information Text S3). Based on fw(nat), each model of Target 47 was classified as bad (0:0 ≤ fw(nat) < 0:1), fair (0:1 ≤ fw(nat) < 0:3), good (0:3 fw(nat) < 0:5), excellent :5 fw≤(nat) < 0:8), and outstanding (0:8 fw (nat) < 1:0).
Partner-specific sequence homology-based protein–protein interface predictor
DockRank uses the predicted interfaces by PS-HomPPI to rank docked models. PS-HomPPI is a sequence homology-based method for partner-specific protein–protein interface residue prediction.42,43 PS-HomPPI uses the experimentally determined interfaces of homo-interologs (homologous interacting proteins) to infer those of a query protein pair. PS-HomPPI is described in detail in Ref. 42, and we briefly summarize it in Supporting Information Text S1 and Figure S1.
To avoid using the target bound complexes as templates due to the redundancy of the PDB (Protein Data Bank),50 highly similar homo-interologs were removed. Specifically, for query A: B and its homologous interacting pair A’ : B’, we also discard the interacting protein pair A’ : B’ if (A and A’) and (B and B’) share ≥ 90% sequence identity. Each case in the decoy sets has bound and unbound proteins. Unbound proteins were used by docking programs to generate docked models and their sequences were used by PS-HomPPI to predict interfaces. The bound complexes were used to evaluate the ranking schemes of docked models. The bound complex of each case (although most bound complexes are probably removed in the first filter of highly similar homologs) was also explicitly deleted from the homo-interolog list, and was not used in later prediction.
Databases used by PS-HomPPI
Three databases are used by PS-HomPPI to make inter-face predictions
ProtInDB51 (version Sep 27th 2012) and S2C DB52 (version Sep 27th, 2012): Used by PS-HomPPI to calculate the interface residues of homo-interologs. ProtInDB is a protein–protein interface residues database (http://einstein.cs.iastate.edu/protInDb/). It contains protein complexes with at least two interacting chains in PDB. S2C DB is used to map the calculated interface residues based on ProtInDB to the whole protein sequences.
BLAST nr_pdbaa_s2c: Used by BLASTP 2.2.27+53 to search for close sequence homologs. It is built based on ProtInDB and S2C DB. Only protein chains existing in ProtInDB are included into nr_pdbaa_s2c. We built a nonredundant database for BLAST queries from the S2C fasta formatted database. To generate the nonredundant BLAST database, we grouped proteins with identical sequences into one entry. As of Sep 27th 2012, nr_pdbaa_s2c contains 38,478 sequences and 9,294,363 total letters.
Interface definition
Interface residues are defined as residues with at least one atom that is within a distance of 5Å from any atoms in the interaction partner chain.
DockRank's scoring function
Given a pair of proteins A and B that are to be docked against each other by a docking program, we use PS-HomPPI to predict the interface residues between A and B. We represent predicted and docked interfaces as binary vectors in which “1” means interface residue and “0” noninterface residue. We then compare the binary vectors of interface residues between A and B predicted by PS-HomPPI with the interface residues in each of the conformations of the complex A: B produced by the docking program. The docked conformation with the greatest interface similarity with the predicted interface residues is assigned the top rank.
Many similarity measures for binary vectors have been proposed (See Ref. 54 for a review). Among these, only Russell-Rao, SoKal-Michener, and Rogers-Tanmoto(-a) measures are defined in the case when both sequences consist of all 0 elements (which is the case when there are no interface residues between the corresponding protein chains and both PS-HomPPI and the docking model correctly predict no interface residues). Because the numbers of interface and noninterface residues are highly unbalanced, we used weighted SoKal-Michener metric to measure the similarity between the interface and noninterface residues in a protein chain A (in complex with chain B) encoded in the form of binary sequences and based on PS-HomPPI predictions and the docked conformation, respectively,
where N is the length of the binary sequence, S11 and S00 are the numbers of positions where the two sequences match with respect to interface residues and noninterface residues, respectively, and β is a weighting factor (0 < β < 1) that is used to balance the number of matching interface residues against the number of matching noninterface residues.
The weighting factor β is defined as a PS-interface residue ratio. In this study, we set β=0:08, which is calculated using a set of transient interaction proteins [71] with experimentally determined interfaces (see Supporting Information Text S2 for details).
Only the interface residues between the receptor and the ligand are used to rank docked models. When the predicted interface vector is a zero vector, it is NOT used in ranking docked models.
For each docked conformation we calculate one score using our scoring function. When a protein complex consists of more than two chains, multiple interface similarities are calculated by pairing each chain of the receptor with each chain of the ligand, and they were weighted (based on the prediction confidence zones of PS-HomPPI) and avaraged to get a final DockRank score. In this study, the weight of the interface similarity is 1 if the predicted interface is from Safe Zone of PSHomPPI, 1 for Twilight Zone, and 0.001 for Dark Zone.
Evaluation of scoring functions
We used Root Mean Square Deviations (RMSDs) to assess the structural difference between each conformation and the corresponding bound complex (target complex). L-RMSD (Ligand-RMSD) is the backbone RMSD between the ligand in the docked conformation and the bound ligand after superimposing the receptor of the docked conformation and that of bound complex. I-RMSD (Interface RMSD) is calculated through two steps: first, map the interface residues of the bound complex to the docked conformation using sequence alignments; second, superimpose the 3D structure of the bound interface of the bound complex onto that of the mapped interface of the docked conformation, and calculate the backbone RMSD as I-RMSD.
We used Success Rate and Hit Rate to evaluate different scoring functions. We define the Success Rate of a scoring function as the percentage of the docking cases in the data set for which at least one near-native structure (hit) is among the m top conformations according to the scoring function. For example, a Success Rate of 25% for top m = 1 predictions means that for 25% of the total test docking cases, the highest ranked conformation is a near-native conformation. Hit Rate is defined as the percentage of hits that are included among the set of m top-ranked conformations. For example, a Hit Rate of 25% for the top m predictions means that 25% of the total hits are found in the top m ranked predictions. Hit Rate measures the enrichment of hits among top ranked conformations.
Upper bound: We supply actual PS-interfaces extracted from bound complexes to DockRank's scoring function to rank the conformations in order to obtain the upper bound of DockRank's scoring function.
RESULTS
DockRank Outperforms Energy-based Scoring Functions
We compared the performance of DockRank with two energy-based scoring functions, ZRank20,44 and IRAD,19 on a subset of 67 docking cases from ZDock3-BM3 benchmark decoy set (see Methods for details). ZRank and IRAD are two energy-based scoring functions developed by the ZDOCK group.44 The ZRank scoring function is a linear combination of atom-based potentials, and it has been shown to be one of top scoring functions in CAPRI.33 IRAD is an improved version of ZRank that augments the ZRank scoring function with residue-based potentials.19 For each of the 67 docking cases, ZDock 3.0 generates 54,000 candidate decoys; however, in 55 of 67 cases the number of total hits (near-native conformations with interface Root Mean Square Deviation I–RMSD 2:5Å19) generated by ZDock 3.0 is fewer than 200 [Supporting Information Fig. S2(A)]. Thus, identifying the hits from a large population of candidate conformations presents a significant challenge.
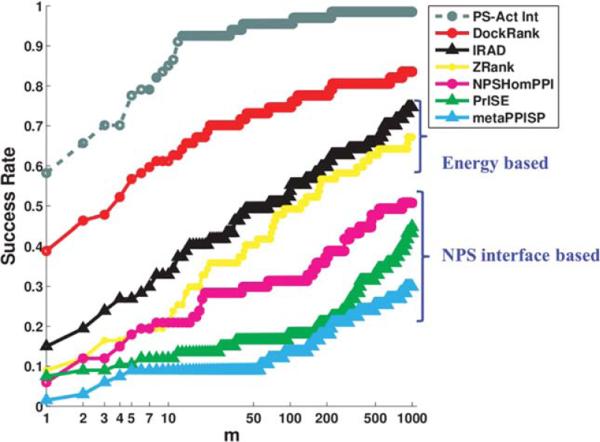
Figure 1 compares the Success Rate of DockRank (red) with that of ZRank (yellow) and IRAD (black) on the ZDock3-BM3 benchmark decoy set. DockRank consistently has a significantly higher Success Rate than both IRAD and ZRank for choices of m ranging from 1 to 1000 (see Materials and Methods section for details). If we limit our comparison to the top ranked conformations (m = 1), DockRank has a Success Rate of 39%, which means that for 39% of the docking cases the highest ranked conformation selected by DockRank is a near-native conformation (a hit), whereas the Success Rates of ZRank and IRAD are 9 and 15%, respectively. When we consider the conformations ranked among the top 10 (m = 10), DockRank achieves a Success Rate of 61%, as compared to 21 and 33% achieved by ZRank and IRAD, respectively. Thus, DockRank improves Success Rate by a factor of 1.8 (61/33 when m = 10) to 4.3 (39/9 when m = 1). Because the 54,000 conformations for each docking case in ZDock3-BM3 are not clustered, each docking case is likely to have a large number of highly similar conformations. In light of this fact, the success of Dock-Rank in ranking near-native conformations among the top 10 conformations is especially encouraging.
Figure 1.
Success rates of DockRank and other scoring schemes on the ZDock3-BM3 decoy set plotted against the top m conformations. The x-axis is plotted on a logarithmic scale to emphasize the region of top ranks. The Success Rate of DockRank (red, using predicted interface residues from a partner-specific predictor, PS-HomPPI) is compared with two energy-based scoring functions, IRAD (black) and ZRank (yellow), and with variants of DockRank using interface residues predicted by three state-of-the-art protein interface residue predictors: NPS-HomPPI (pink), PrISE (green), and meta-PPISP (blue). NPS-HomPPI, PrISE, and meta-PPISP are nonpartner-specific (NPS) interface predictors. The success rate of DockRank's scoring function supplied with partner-specific actual interface residues (labeled as “PS-Act Int,” gray-dashed line) is plotted to define the upper bound of DockRank's scoring function. Studied here are 67 docking cases that have at least one hit (a conformation with I–RMSD ≤ 2:5å) among 54,000 candidate decoys and for which PS-HomPPI is able to return interface predictions.
A comparison of the Hit Rates of DockRank with those of other scoring functions, including IRAD and ZRank, on the ZDock3-BM3 benchmark decoy set is provided in Supporting Information Figure S3. Dock-Rank has a higher Hit Rate than IRAD and ZRank in 53 of 67 cases.
Partner-specific interface predictions can be used to reliably rank docked conformations
Li and Kihara55 had concluded that predicted interfaces cannot be used to reliably identify near-native conformations, based on their study using predicted interfaces from meta-PPISP,56 a nonpartner-specific interface predictor, to rank docked conformations using their scoring function. In light of the fact that most existing protein interface predictors are nonpartner-specific, it is of interest to examine the extent to which partner-specific interface predictions can improve the reliability of identifying near-native docked conformations. To answer this question, we compared the performance of DockRank using predicted interface residues from PS-HomPPI (a partner-specific predictor), with variants of DockRank using interface residues predicted by three state-of-the-art nonpartner-specific protein interface residue predictors: i) NPS-HomPPI,42 a sequence homology-based nonpartner-specific method for predicting the interface residues, which has been shown to outperform several structure-based and sequence-based interface predictors when sequence homologs of the query protein can be reliably identified; ii) PrISE,57 a local structural similarity based method for predicting protein–protein interfaces that has been shown to outperform other structure-based nonpartner-specific interface prediction methods; iii) meta-PPISP,56 a consensus method that takes as input the scores of three other structure-based machine-learning predictors cons-PPISP,58,59 PINUP,60 and Promate.29
Our results on the ZDock3-BM3 decoy set show that DockRank (using PS-HomPPI predicted interface residues) significantly outperforms DockRank variants that use predictions from the nonpartner-specific interface predictors NPS-HomPPI, meta-PPISP, and PrISE based on three criteria. First, in terms of Success Rate, DockRank successfully places at least one near-native conformation within the top ranked conformations (m = 1) for 39% docking cases (i.e., a Success Rate of 39%), while Dock-Rank variants that use predicted nonpartner-specific interfaces can only pick out a near-native conformation to top rank for 1%–7% of docking cases (Fig. 1). This translates to an improvement in Success Rate by a factor of 39 (39%/1%) when m = 1. Second, in terms of Hit Rate, the top 1000 ranked conformations selected by DockRank contain more hits than those selected by DockRank variants that use predicted nonpartner-specific interfaces in 43 of 67 docking cases (Supporting Information Figure S3). Third, in terms of I-RMSD, top ranked conformations selected by DockRank have statistically significantly lower I-RMSDs than those selected by DockRank variants that use predicted nonpartner-specific interfaces (as shown by the boxplot of I-RMSDs and by the nonparametric Nemenyi Test [75] at a significance level of 0.05, see Figure 2 and Supporting Information Figure S4).
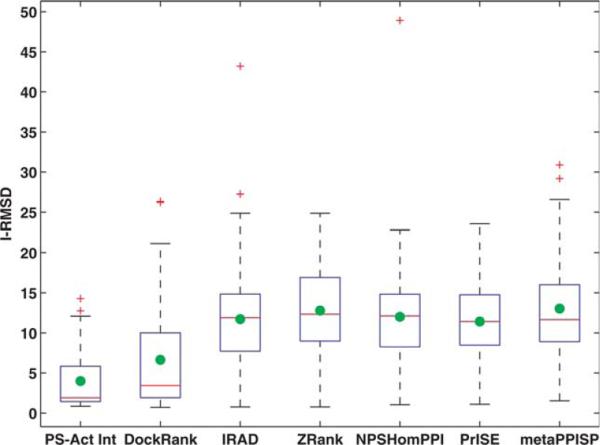
Figure 2.
I-RMSDs of top ranked conformations selected by different docking scoring methods on the ZDock3-BM3 decoy set. The lower (Q1), middle (Q2), and upper (Q3) quartiles of each box are 25th, 50th, and 75th percentile. Interquartile range IQR is Q3–Q1. Any data value that lies more than 1.5 × IQR lower than the first quartile or 1.5 × IQR higher than the third quartile is considered an outlier, which is labeled with a red cross. The whiskers extend to the largest and smallest value that is not an outlier. Averages are marked by green dots. Studied here are 67 cases for which ZDock 3.0 is able to generate at least one hit (I–RMSD 2:5Å) and for which PS-HomPPI is able to return interface predictions. DockRank's scoring function supplied with actual partner-specific interfaces (PS-Act Int) set the lower bound of I-RMSDs of top ranked conformations that DockRank's scoring function can select.
Because DockRank ranks conformations based on a measure of similarity between the predicted interface and the interface defined by a docked conformation, it is natural to ask how well DockRank can be expected to perform if it were provided with the best possible partner-specific interface predictions. In order to estimate an upper-limit for the performance of the DockRank's scoring function, we tested its performance using the actual partner-specific interfaces extracted from the bound complexes. Figure 1 shows that DockRank, when supplied with the actual interface residues for a docking case (grey), ranks at least one hit (a near-native conformation) among the top 10 conformations for 85% of the docking cases. When supplied with interfaces predicted by PS-HomPPI, DockRank ranks at least one hit among the top 10 conformations for 61% of the docking cases. Also, DockRank's scoring function, when supplied with actual partner-specific interface residues, is able to rank ≥50% of all hits among the top 1000 conformations in 60 of 67 docking cases (Supporting Information Fig. S3). These results demonstrate that the scoring function used by DockRank can reliably place near-native conformations among the top ranked conformations when it is provided with reliable partner-specific interface residues. Furthermore, the gap between the performance of Dock-Rank when supplied with interface residues predicted by PS-HomPPI (red line in Figure 1) and the performance of DockRank when supplied with actual partner-specific interface residues (grey dashed line) suggest that there is considerable room for improving the performance of DockRank by improving the reliability of partner-specific protein–protein interface prediction.
In the following text, unless otherwise specified, we refer to our ranking method using partner-specific interface residues predicted by PS-HomPPI as DockRank.
DockRank identifies conformations with lower RMSD at the interface than other scoring functions
We compared the top ranked conformations identified by different scoring functions with respect to I-RMSDs. Figure 2 shows the distributions of I-RMSDs of top ranked conformations (m = 1) selected by different scoring schemes. The top ranked conformations selected by DockRank have two to four times smaller average and median I-RMSDs than those selected by other scoring schemes. The average I-RMSD value of top ranked conformations selected by DockRank is 6.6Å, compared with 11.7, 12.8, 12.0, 11.4, and 13.0Å of those selected by IRAD, ZRank, NPS-HomPPI, PrISE, and meta-PPISP, respectively. The median I-RMSD value of top ranked conformations selected by DockRank is 3.4Å, compared with 11.9, 12.3, 12.11, 11.4, and 11.7Å for IRAD, ZRank, NPS-HomPPI, PrISE, and meta-PPISP, respectively.
Our statistical analysis further shows that when performance is measured in terms of I-RMSD of the top ranked conformations, DockRank significantly outperforms other scoring schemes (see Supporting Information Fig. S4 and Text S4).
DockRank improves upon the success rate, average hit rate, and L-RMSD of ClusPro-ranked conformations
Existing docking programs such as ClusPro typically use built-in scoring functions to rank the conformations they produce. In this context, it is natural to ask whether Dock-Rank, when used to re-rank the docked conformations returned by the docking program using its own built-in scoring schemes, can improve upon the original ranking. For this comparison, we chose ClusPro 2.0,15–17,45 because it has been reported to have superior performance in recent CAPRI competitions.33 Briefly, ClusPro is built on top of a FFT-based rigid docking program PIPER.16 PIPER rotates and translates the ligand with ~109 positions relative to the receptor. PIPER's scoring function, which contains terms of shape complementarity, electro-static, and pairwise potentials, is applied to these candidate conformations, and returns the top 1000 conformations to ClusPro's clustering algorithm. ClusPro ranks the conformations by cluster size. ClusPro also provides two scores from PIPER: Lowest Energy and Center Energy.
We applied ClusPro to the 119 cases in Docking Benchmark 3.0 (See Methods for details about this dataset). In 47 of the 119 cases, ClusPro 2.0 generated at least one hit (a conformation with L–RMSD ≤ 10Å).1 Among these 47 cases, PS-HomPPI is able to return partner-specific interface predictions for 31 cases. Thus, our experiment is limited to only these 31 cases, which we refer to as ClusPro2-BM3_31 benchmark set. In each case, ClusPro returns about 30 ranked conformations. We re-ranked the conformations returned by ClusPro using DockRank and compared the resulting ranking of conformations with the original ranking produced by ClusPro.
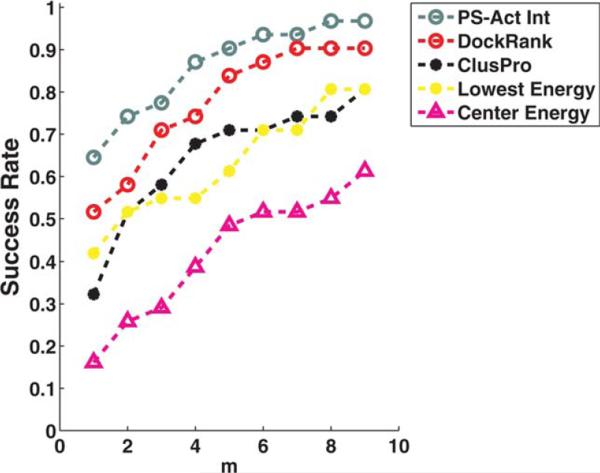
Figure 3 compares the Success Rates of DockRank based on actual partner-specific interfaces (PS-Act Int, grey), DockRank using PS-HomPPI predicted interfaces (Dock-Rank, red) and the three ClusPro scoring schemes: ClusPro (cluster-size based, black), Lowest Energy (yellow), and Center Energy (magenta). DockRank's re-ranking process identifies at least one near-native conformation (hit) for more docking cases than ClusPro scoring schemes (i.e., DockRank improves on the ClusPro scoring schemes in terms of Success Rate). For example, applied to the pre-filtered docked models by ClusPro, DockRank is able to select at least one hit to top rank (m = 1) for 52% cases (i.e., a Success Rate of 52%), whereas the original Success Rates of ClusPro cluster-size based scoring scheme is 32%, ClusPro Lowest Energy based is 42%, and ClusPro Center Energy based is 16% (Fig. 3 at m = 1). In addition, Dock-Rank improves the average Hit Rate of top ranked models (m = 1) selected by ClusPro scoring functions to 0.38 from 0.22, 0.29, and 0.15 produced by ClusPro cluster-size based, Lowest Energy, and Center Energy scoring functions, respectively (Supporting Information Fig. S5).
Figure 3.
Success rates of DockRank and ClusPro scoring functions on the ClusPro2-BM3_31 decoy set plotted against the top m conformations. The x-axis is plotted on a logarithmic scale to emphasize the region of top ranks. ClusPro scoring functions (default cluster-size based, center energy-based, lowest energy-based) were applied on the original docked conformations (109 docked conformations per docking case) generated by ClusPro's underlying docking program, PIPER. DockRank was applied on the docked conformations (30 of the 109 docked conformations per case) output by ClusPro scoring functions. The success rate of DockRank's scoring function supplied with partner-specific interface-based ranking (gray-dashed line) is also plotted to show the upper bound of the success rate of DockRank's scoring function. Studied here are 31 cases for which ClusPro 2.0 is able to return at least one hit (a docked conformation with L–RMSD 10Å) and for which PSHomPPI is able to return interface predictions. We limited m (the number of top ranked conformations) to 9, since ClusPro returned only nine conformations for one of the docking cases, 1PPE.
Our statistical analysis (see Supporting Information Fig. S6 and Text S5 for details) shows that at a significance level of 0.05, the mean L-RMSDs of the top ranked conformations identified by DockRank are significantly lower than those selected by ClusPro Center Energy. However, although DockRank produces top ranked conformations with lower average L-RMSD values than those produced by ClusPro and ClusPro Lowest Energy scoring functions, the difference is not statistically significant. This lack of a statistically significant difference could be due to the limited power of the nonparametric test, which is exacerbated by the small number of cases (31) and the small number of docked conformations (30) used in the comparisons.
It must be noted that this experiment should not be interpreted as a direct comparison between DockRank and ClusPro because ClusPro scoring functions have access to ~109 decoys, generated by PIPER,16 whereas DockRank has access to only a very small set (~30 conformations) of these 109 conformations. Also, DockRank in this experiment is used to re-rank these ~30 conformations output by ClusPro as opposed to the entire set of 109 conformations.
DockRank identifies useful conformations with lower ligand RMSD for further refinement
ClusPro 2.0 generated at least one near-native conformation with L–RMSD 10Å for only a small proportion of cases (47 out of 119). It is interesting to consider whether DockRank can select useful, although incorrect, conformations in cases for which ClusPro returns no hits. The conformations output by ClusPro are the representative conformations selected from clusters of docked conformations that share similar 3D conformations. When no hit (L–RMSD ≤ 10Å) is returned by ClusPro, it is still possible that the cluster, from which the representative conformation is chosen, may contain hits. We examined the docking cases with at least one docked conformation with L–RMSD 15Å in order to study DockRank's ability to indirectly identify clusters that might contain actual hits, and DockRank's ability to identify meaningful conformations for further refinements.
There are 45 cases that have at least one docked conformation with L–RMSD 15Å, and have interface predictions returned by PS-HomPPI. We chose to further analyze these 45 cases, which we refer to as the ClusPro2-BM3_45 decoy set. For each case, we calculate the average L-RMSD of the top five conformations selected by a scoring function.
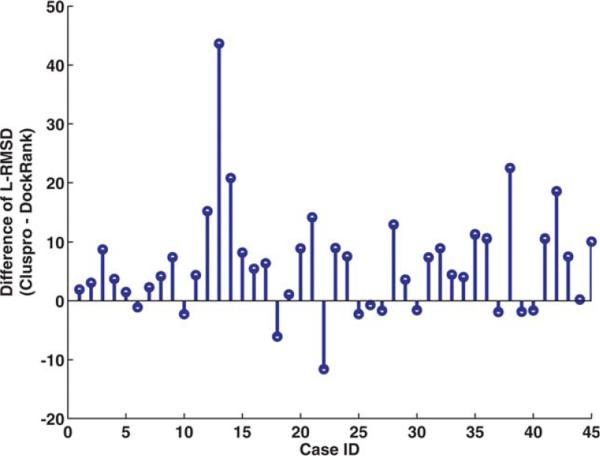
Figure 4 shows the difference between the average of L-RMSDs of top conformations produced by ClusPro and DockRank scoring functions in each case. Positive values correspond to cases where the top ranked Dock-Rank conformations have a lower average L-RMSD than the top ranked ClusPro conformations. In 34 of 45 (75.6%) cases, top conformations selected by DockRank have lower L-RMSD than their ClusPro counterparts. Futhurmore, a pairwise Wilcoxon signed rank test shows that top five conformations selected by DockRank have significantly lower averaged L-RMSD than those selected by ClusPro (p-value < 0.0001).
Figure 4.
The difference between the averages of L-RMSDs of top models between DockRank and ClusPro Rank on each case of the ClusPro2-BM3_45 decoy set. Positive values correspond to cases where the top five ranked DockRank conformations have a lower average L-RMSD than those ranked by ClusPro.
Error analysis
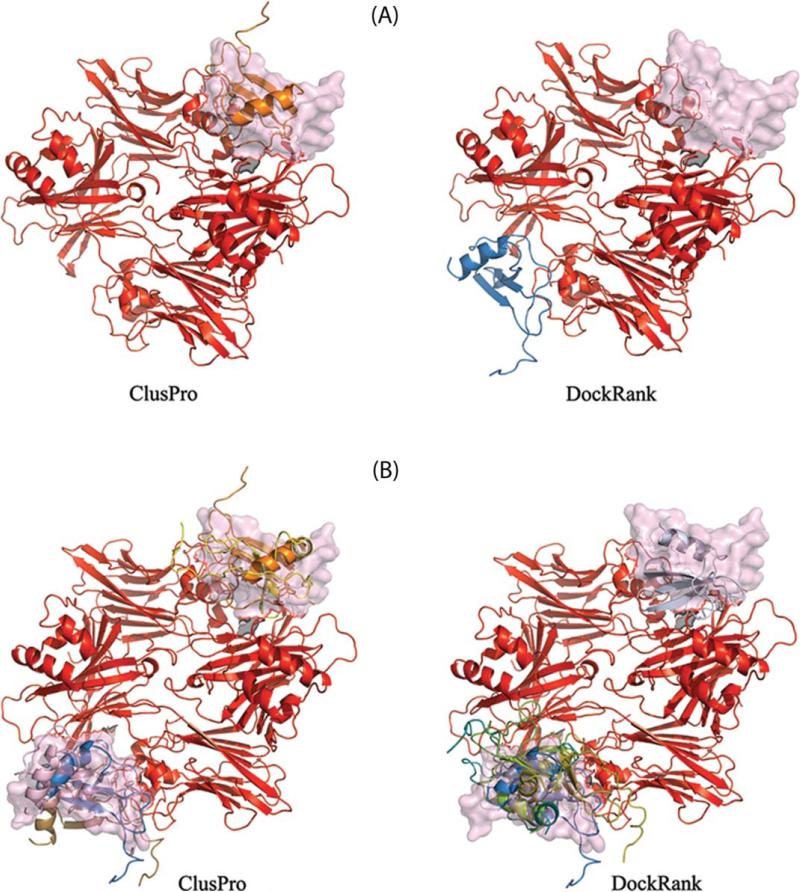
Although in general, DockRank identifies top ranked conformations with lower L-RMSD than those produced by ClusPro, there are some exceptions, for example, docking case 22 (PDB ID 1ML0) in Figure 4. Figure 5(A) illustrates the top ranked ClusPro and DockRank conformations. Note that the docked ligand position in the top DockRank conformation (blue ribbon) is out of place relative to its correct position, that is, the position of the experimentally determined bound ligand (in pink surface representation). However, the interface residues predicted by PS-HomPPI for the 1ML0 case are drawn from the homologs in Safe Zone and hence are expected to be reliable.42 We further note that the structure of the receptor (red ribbon) is symmetric. This raises the possibility that the ligand may bind on both sides of the symmetric receptor as opposed to only one side.
Figure 5.
The top conformations ranked by DockRank and ClusPro for docking case 1ML0. The red ribbon is the receptor. (A) The highest ranked models selected by ClusPro (ligand shown as gold ribbon) and DockRank (ligand shown as blue ribbon), with the bound ligand that is included in the BM3 dataset (pink surface representation). (B) The top five ranked models (ribbons) with two bound ligands from PISA. Figure generated using PYMOL [70].
Examination of the biological assembly structure of 1ML0 downloaded from the PISA web server61 reveals that the 1ML0 bound complex has two identical ligands, which bind on each side of the receptor. The authors of BM3 (Docking Benchmark 3.0) chose to include only one ligand chain in the docking case in Docking Benchmark 3.0 and excluded the other ligand chain. DockRank was able to select docked ligand positions on both sides of the receptor. The ligand of the top ranked conformation selected by DockRank is located close to the bound ligand that was omitted from Docking Benchmark 3.0. The top five conformations selected by ClusPro and DockRank for case 1ML0 are shown in Figure 5(B). Both ClusPro and DockRank have successfully selected conformations with ligands bound to both sides of the receptor.
We recalculated the L-RMSD for all docked conformations for case 1ML0 by considering both of the two identical bound ligands to correspond to native ligand positions. We found that the highest ranking DockRank model is that with the lowest L-RMSD. However, ClusPro identifies four hits (L–RMSD 10Å) among its top five models, whereas DockRank selects only one (albeit, one with the lowest L-RMSD).
We also identified a second case, 1RLB, for which PS-HomPPI returned Safe Zone interface predictions, but DockRank assigned high ranks to conformations with unusually large L-RMSDs. As in the case of 1ML0, it turned out that one of the bound ligands of 1RLB was left out from Docking Benchmark 3.0, and DockRank, in fact, improved the rankings of docked models returned by ClusPro (see Supporting Information Text S6 for details).
DockRank performance on CAPRI uploader models
In the scoring experiments of the CAPRI competition, docking groups were invited to upload a set of 100 predicted models for each target (referred as “uploader models”). The scorer groups ranked the uploader models and submitted their top 10 models.11,33 We evaluated DockRank on the uploader models for Targets 30 (trimer), 35 (dimer), 41(dimer), 47 (hexamer), and T50 (trimer), and compared the quality of the top 10 models selected by DockRank with those selected by the CAPRI scorer groups. 29 CAPRI scorer groups, including both human scorers and web server scorers, evaluated at least one of these targets. We removed the target PDB bound complex from our set of templates. To avoid using complexes that are essentially the same as the target complex due to the redundancy of PDB, we also removed any template A’ : B’ if the sequence homolog A’ and the query protein chain A share ≤90% sequence identity AND B’ and B share ≤90% sequence identity.
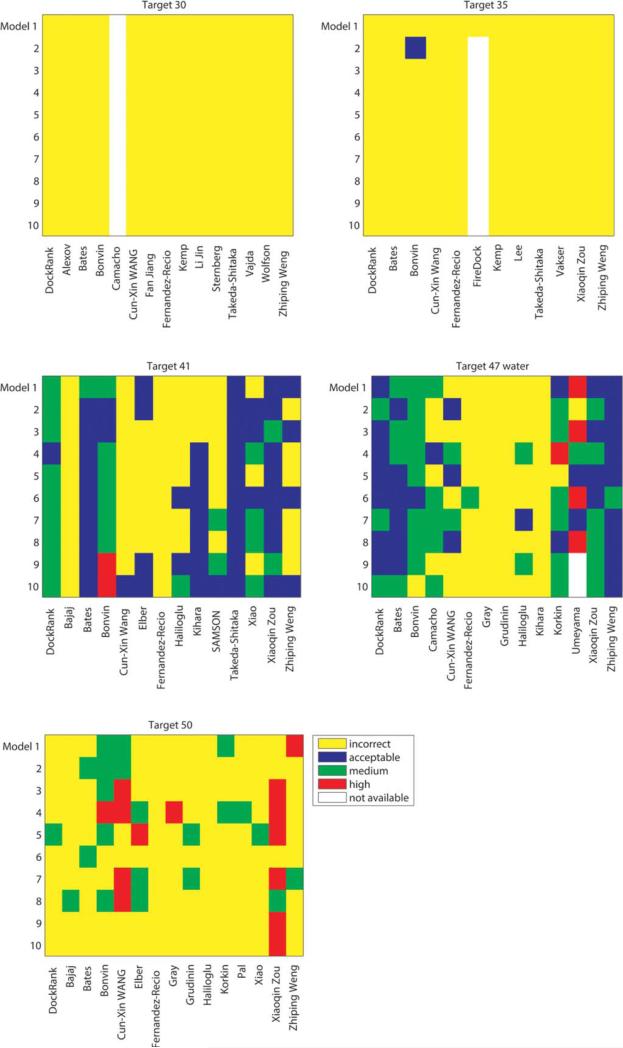
Figure 6 summarizes the performance of DockRank and the CAPRI scorers on the five targets. Target 30 had only two acceptable models and 0 medium/high models out of 1343 uploader models. Neither DockRank nor any of the scorer groups was able to identify either of the acceptable models for Target 30. Target 35 had three acceptable models and 0 medium/high models out of 499 uploader models, and only the Bonvin group was able to rank an acceptable model at rank 2. For Target 41, DockRank selected one acceptable model and nine medium quality models, while the best CAPRI scorer for the same target, the Bonvin group, selected two acceptable models, six medium quality models, and two high-quality models. For Target 47, because high-quality templates are publicly available, no docking was required, and the prediction of the water-mediated interactions was the true challenge for this target. Although DockRank does not use any information regarding water molecules of the template structures, it is able to select seven fair models (0:1 fw(nat) < 0:3) and three good models (0:3 ≤ fw(nat) < 0.5) in terms of interface water positions, which is comparable to the performance of the top CAPRI performers for Target 47. For Target 50, DockRank can predict interfaces for only a subset of query chains but was able to identify one acceptable model (rank 5).
Figure 6.
The rankings of top 10 selected models by DockRank and the CAPRI scorers.
These results suggest that DockRank performed competitively with other top CAPRI scorers on these five targets. To quantitatively evaluate the performance of each scorer group, we assigned a score to each model based on its quality (0 for incorrect models, 1 for an acceptable model, 2 for a medium quality model, and 3 for a high-quality model), and used the sum of the scores of the top 10 models selected by each scorer group to assess the scorer group's performance on one target (see Table I). On the five CAPRI targets tested, the Bonvin group was the best scorer; they selected at least one acceptable or better model for four of the five targets, and their models have the largest sum of model scores, representing the overall highest model quality compared with those selected by other scorers. The other groups: Zou, Dock-Rank, the groups of Bates, Weng, and Wang (ordered according to the sum of their model scores) were able to select at least one acceptable or better model for three of the five targets. The rest of scorers performed well on fewer than three targets. Based on this comparison, DockRank ranks #3 overall (Table I). However, no definitive conclusions can be drawn regarding how the different scoring methods perform relative to one another until more CAPRI targets are made available.
Table I.
The Comparison of DockRank with the 29 CAPRI Scorers
| Scorer groups | Target 30 | Target 35 | Target 41 | Target 47 water | Target 50 |
|---|---|---|---|---|---|
| 1 Bonvin | 0 | 1 | 20 | 17 | 7 |
| 2 Zou | – | 0 | 12 | 16 | 13 |
| 3 DockRank | 0 | 0 | 19 | 13 | 1 |
| 4 Bates | 0 | 0 | 11 | 14 | 2 |
| 5 Weng | 0 | 0 | 4 | 11 | 3 |
| 6 Wang | 0 | 0 | 1 | 7 | 10 |
| 7 Korkin | – | – | – | 17 | 2 |
| 8 Umeyama | – | – | – | 16 | – |
| 9 Xiao | – | – | 11 | – | 1 |
| 10 Camacho | – | – | – | 11 | – |
| Alexov | 0 | – | – | – | – |
| Bajaj | – | – | 0 | – | 1 |
| Elber | – | – | 4 | – | 5 |
| FireDock | – | 0 | – | – | – |
| Fernandez-Recio | 0 | 0 | 0 | 2 | 0 |
| Gray | – | – | – | 0 | 2 |
| Grudinin | – | – | – | 0 | 2 |
| Haliloglu | – | – | 4 | 5 | 0 |
| Jiang | 0 | – | – | – | – |
| Jin | 0 | – | – | – | – |
| Kemp | 0 | 0 | – | – | – |
| Kihara | – | – | 7 | 0 | – |
| Lee | – | 0 | – | – | – |
| Pal | – | – | – | – | 1 |
| SAMSON | – | – | 5 | – | – |
| Sternberg | 0 | – | – | – | – |
| Takeda-Shitaka | 0 | 0 | 10 | – | – |
| Vajda | 0 | – | – | – | – |
| Vakser | – | 0 | – | – | – |
| Wolfson | 0 | – | – | – | – |
Incorrect, acceptable, medium, and high-quality models are assigned with a score of 0, 1, 2, and 3, respectively. Dash denotes that a scorer did not submit any model for that target. The number in each cell is the sum of the scores of the 10 models selected by a scorer group. Ranks of the top 10 scorers are shown. All other scorers are listed alphabetically.
DockRank complements homology modeling
Because DockRank uses homologous complexes to infer interfaces and to rank docked conformations, it could be argued that superimposing or threading component structures onto the homointerologs (templates) used by Dock-Rank would provide a simpler method for modeling protein complexes, thereby obviating the need for docking and hence scoring the docked conformations using Dock-Rank. Although our purpose in developing DockRank was to provide reliable ranking of docked structures, not to generate docked models, we examined the utility of Dock-Rank in relation to homology modeling methods,46,47 such as superimposition or multimeric threading by conducting the following two experiments:
Experiment 1
For all dimer cases, we generated conformations by using TM-align62 to superimpose unbound component structures onto the templates (homo-interologs) used by DockRank to predict interfaces. Among 51 dimers considered, there are 19 cases (37%) for which all superimposed models are near-native (Supporting Information Table S4, section B), 19 cases (37%) for which at least one superimposed model is near-native and at least one model is incorrect (Supporting Information Table S4, section A), and 13 cases (25%) for which no superimposed model is near-native (Supporting Information Table S4, section C).
For the 19 cases with at least one near-native superimposed model and at least one incorrect model, we used DockRank to rank the resulting models. The results summarized in Supporting Information Table S4 section A show that DockRank reliably assigns lower ranks to incorrect models in 18 of 19 cases. DockRank is able to differentiate the correct, that is, near-native, models from the incorrect, that is, non-near-native, models whereas sequence similarity fails to distinguish between them.
Among the 13 cases in which none of the superimposed models is correct (suggesting that the structures of the templates for these cases are not close enough for homology modeling to reliably predict the structure of the protein complexes), PS-HomPPI reliably predicts both sides of the interface in five cases (sensitivities range from 0.51 to 0.96, i.e., 51–96% of the actual interface residues are correctly predicted as interface residues; Specificities range from 0.40 to 0.96, that is, 40–96% of the predicted interface residues are actual interface residues). The three cases are 1H1V (difficult), 2OT3 (difficult), 2CFH (medium), 1ZHI (rigid-body), and 2AJF (rigid-body), where rigid-body, medium, and difficult denote the (increasing) levels of difficulty of docking as measured by the extent of conformational change upon binding (according to Docking Benchmark 3.0).
Experiment 2
We consider proteins that have more than two chains and for which no complete templates are available. We identified seven such cases, for which PS-HomPPI templates can be identified for only a subset of query chains and hence current approaches to superimposition or threading cannot be used (see Table II and Supporting Information Table S5).
Table II.
The Performance of DockRank on Cases That Have More Than Two Chains But Only a Subset of Chains Have Templates
| Case ID (level of conformational changes) | ClusPro2-BM3 |
ZDock3-BM3 |
PS-HomPPI Interface predictiona | ||||
|---|---|---|---|---|---|---|---|
| DockRank | ClusPro | DockRank | IRAD | ZRank | |||
| Group 1 | 1GP2 (medium) | – | – | Acceptable (1) | Acceptable (85) | Acceptable (133) | Very Good for two chains |
| 1K5D (medium) | – | – | Medium (2) Acceptable (1) | Medium (933) Acceptable (12) | Medium (4295) Acceptable (84) | Very Good for two chains | |
| 1JM0 (difficult) | – | – | Acceptable (3168) | NO RANKING | Acceptable (44899) | Very Good for two chains | |
| 1F51 (rigid-body) | Acceptable (1) | Acceptable (15) | High (2) Medium (1) Acceptable (6) | High (10918) Medium (1) Acceptable (13) | High (15370) Medium (3) Acceptable (67) | Good for one chain, ok for another chain. | |
| Group II | 1E6J (rigid-body) | Acceptable (18) | Acceptable (9) | Medium (22169) Acceptable (7037) | Medium (2) Acceptable (17) | Medium (1) Acceptable (8) | Good for one chain. |
| 2FD6 (rigid-body) | – | – | Medium (25373) Acceptable (9277) | Medium (21) Acceptable (1) | Medium (63) Acceptable (15) | Ok for one chain. | |
| Group III | 2HMI (difficult) | – | – | Acceptable (23048) | NO RANKING | Acceptable (272) | Bad |
According to the CARPI criteria, a docked model is classified into four categories: (1) a high-quality model, if its I–RMSD ≤ 1Å; (2) a medium quality model, if its I–RMSD ≤ 2Å; (3) an acceptable model if its I–RMSD ≤ 4Å.
The numbers in the parentheses are the ranks of the first model belonging to the corresponding category.
See Supporting Information Table S5 for details.
We evaluated the interface prediction performance of PS-HomPPI, and compared the scoring performance of DockRank with that of IRAD, ZRank, and ClusPro on two decoy sets of ZDock3-BM3 and ClusPro2-BM3. In order to examine the impact of the reliability of PSHomPPI predictions on the performance of DockRank in ranking conformations, we divided the cases into three groups: (i) Group I: cases in which PS-HomPPI can reliably predict both sides of the interface; (ii) Group II: cases in which PS-HomPPI can reliably predict only one side of the interface; (iii) Group III: cases in which PS-HomPPI cannot reliably predict interacting residues on either side of the interface (see Supporting Information Table S5). Three of the four cases in Group I belong to the “difficult” or “medium” categories (corresponding to large vs. medium conformational change upon binding) of the Docking Benchmark 3.0. The reliability of PS-HomPPI in predicting interfaces for these cases demonstrates that, as expected, our sequence-based interface predictor PS-HomPPI is robust to large conformational changes. For all four cases, DockRank significantly improved the ranking of docked conformations (Table II). For example, for case 1GP2, DockRank ranked one acceptable model as number 1, in contrast to IRAD and ZRank, whose top ranked acceptable models are ranked at position 85 and 133, respectively. This result shows that when both sides of the interface are reliably predicted, even for a subset of the query chains, DockRank is able to improve the rankings of docked conformations. As expected, for the two cases in Group II and the one case in Group III, DockRank cannot rank docked conformations reliably. However, for six of seven multiple-chain cases in this experiment, PS-HomPPI is able to reliably predict the interfaces for at least one chain (Supporting Information Table S5), which can be used to provide important constraint information for docking programs.
From the results of the experiments, we conclude that structure superimposition can be a viable alternative for generating all-atom models of complexes when templates that share high global sequence similarity are available, and DockRank can be used to reliably differentiate the correct superimposed models from incorrect ones. In the case of multimeric complexes, for which PS-HomPPI can identify only partial templates for a subset of query chains, we find that PS-HomPPI reliably predicts interface residues for at least one side of the interface. When both sides of an interface can be reliably predicted by PS-HomPPI, DockRank can significantly improve the rankings of docked models. When only one side of an interface can be reliably predicted, DockRank cannot improve the ranking of docked models, but the interfaces predicted by PS-HomPPI can be used to constrain docking and thus potentially improve the quality of the resulting models. The ability of our ranking method, DockRank, and our interface predictor, PS-HomPPI, to reliably make use of dimeric protein complex templates in cases with more than two chains is of special interest in light of the fact that solving 3D structures using experimental methods is usually more difficult for large protein complexes with multiple chains than for small protein complexes.
DISCUSSION
Selecting near-native conformations from a large number of decoys generated by a docking program remains a challenging problem in computational molecular docking.10 In this study, we presented DockRank—a novel scoring method for protein–protein docking based on predicted interfaces. The proposed scoring function relies on a measure of similarity between interfaces of docked models and predicted interfaces by a partner-specific interface predictor, PS-HomPPI.
The major conclusions of this study are as follows: (1) Comparisons of DockRank with two state-of-the-art energy-based docking scoring functions, ZRank, and IRAD, show that DockRank consistently outperforms both on a decoy set of 67 docking cases for which PSHomPPI is able to predict the interface residues between the receptor and the ligand and for which ZDock 3.0 is able to generate at least one hit among 54,000 candidate decoys per case. These results suggest the viability of DockRank as an alternative to complex energy-based scoring functions in cases where it is possible to obtain reliable partner-specific interface predictions. (2) Comparisons of DockRank variants that use different sources of predicted interfaces underscore the importance of using partner-specific interface predictions to rank docked conformations. (3) DockRank is able to improve upon the Success Rate, average Hit Rate, and L-RMSD of the conformations returned by ClusPro. (4) DockRank performs competitively with the CAPRI top scorers on the five targets we assessed. (5) DockRank complements structural superimposition and threading methods in important ways. In cases where sufficiently good templates can be identified for structural superimposition or threading methods to be applicable, DockRank can be used to reliably differentiate near-native superimposed models from incorrect superimposed models; in cases where homology modeling cannot be directly applied (i.e., cases in which only a subset of chains have templates), PS-HomPPI can reliably identify at least one side, and in some cases both sides, of the interface allowing DockRank to reliably rank docked models. (6) The results of our comparison of DockRank using actual partner-specific interfaces as opposed to PS-HomPPI predicted partner-specific interface residues for scoring docked conformations suggests that there is significant room for improving the performance of DockRank by improving the reliability of partner-specific interface residue predictions.
We hypothesize that the poor performance of Dock-Rank variants that use interface residues predicted by nonpartner-specific interface predictors, in comparison with DockRank using PS-HomPPI predicted interfaces, is at least in part explained by the fact that the docking cases used in this study correspond to nonobligate protein complexes. Most nonobligate complexes are transient interactions,63 which tend to be highly partner-specific.3,4,42 A perfect nonpartner-specific protein interface predictor can reliably predict, at best, the union of all the actual interface residues of a protein with all of its possible binding partners. Therefore, interface residues predicted by even the perfect nonpartner-specific interface predictor on the two proteins (A and B) that make up a docking case will include not only the actual interface between receptor A and ligand B, but also residues that play a role in interactions of A with partners other than B, and B with partners other than A. Thus, the interface residues predicted by nonpartner-specific predictors have a high rate of false positives when used in partner-specific setting. And these “false positive interface residues” may falsely give top ranks to docked conformations that have interfaces near these “false positive” interfaces, resulting in the corresponding deterioration in the quality of ranked conformations. This underscores the critical role of a reliable partner-specific interface predictor in the effectiveness of DockRank, at least for docking cases that correspond to transient interactions. The results of our experiments with DockRank using nonpartner-specific interface predictions are consistent with the observation made by Li and Kihara55 that nonpartner-specific interface predictions cannot reliably rank docked conformations. However, the reliable performance of DockRank using partner-specific interface predictions indicates that predicted partner-specific interfaces can indeed be used to rank docked conformations more reliably than other state-of-the-art scoring functions, when interacting templates for both sides of the query interactions are available.
To facilitate comparisons with new scoring functions that may be developed in the future, the DockRank scores on the decoy sets used in this study are available to the community. Docked conformations generated using different ClusPro energy functions, L-RMSD for each docked conformation, ClusPro scores, and the recalculated L-RMSDs of models of 1ML0 and 1RLB after including both identical bound ligand chains, are also available at http://einstein.cs.iastate.edu/DockRank/supplementaryData_journal.html.
Docking cases with large conformational changes upon binding present challenges for most scoring schemes. It is challenging both for docking programs to generate sufficient numbers of near-native conformations for such cases and for interface residue predictors to make reliable interface predictions. Therefore, it is important to evaluate the performance of scoring functions on complexes with different conformational change levels. Complexes in the BM3 dataset are classified into three groups based on the degree of conformational changes upon binding. We studied the performance of our underlying interface predictor, PS-HomPPI, with respect to different conformational change levels to indirectly study the performance of DockRank in ranking docked models of cases with different conformational changes. For each interface prediction, PS-HomPPI provides an interface prediction confidence zone (Safe/Twilight/Dark Zone) based on the degree of sequence similarity of the homo-interologs used for interface inferences. Because the effects of conformational changes and the prediction confidence zones on the performance of PS-HomPPI may be confounded, we summarized the interface prediction performance of PS-HomPPI into nine subgroups with respect to three levels of conformational changes and three prediction confidence zones (Table III). The results in Table III show that the performance of PS-HomPPI is insensitive to conformational changes upon binding and it is clearly correlated with the prediction confidence zones: the higher confidence, the more reliable the interface predictions. Because both the interface prediction of PSHomPPI and the calculation of DockRank scores are insensitive to the conformational changes, we can expect that the ranking of conformations produced by Dock-Rank is relatively insensitive to conformational changes upon binding.
Table III.
Interface Residue Prediction Performance of PS-HomPPI on the BM3 Dataset with Three Different Prediction Confidence Zones on Three Levels of Conformational Changes upon Binding
| Confidence zones | Conformational change upon binding | Number of predictions (out of 372) | CC | F1 | Specificity | Sensitivity |
|---|---|---|---|---|---|---|
| Safe | Rigid | 138 | 0.63 | 0.64 | 0.71 | 0.65 |
| Medium | 22 | 0.59 | 0.59 | 0.72 | 0.59 | |
| Difficulty | 26 | 0.56 | 0.55 | 0.74 | 0.55 | |
| Twilight | Rigid | 34 | 0.46 | 0.52 | 0.55 | 0.59 |
| Medium | 4 | 0.38 | 0.4 | 0.43 | 0.38 | |
| Difficulty | 4 | 0.54 | 0.6 | 0.58 | 0.63 | |
| Dark | Rigid | 4 | 0.12 | 0.19 | 0.19 | 0.38 |
| Medium | 0 | – | – | – | – | |
| Difficulty | 0 | – | – | – | – | |
| Average | 232/372 = 62% | 0.58 | 0.59 | 0.67 | 0.61 |
Only the interfaces between the receptors and ligands are predicted and used by DockRank in ranking docked models. During the evaluation, we consider each partner-specific-predicted receptor–ligand interface as one prediction. For example, for a complex AB:C with two receptor chains A and B and one ligand chain C, we consider four predictions: A|A:B, A|A:C, B|B:A, C|C:A, where A|A:B means the interface of A that interacts with its binding partner B. Sometimes, part of a protein may not have interface predictions from PS-HomPPI because of the lack of aligned residues from putative sequence homologs. These residues are not considered in the evaluation here, because they are not used by DockRank in ranking docked models. Correlation coefficient (CC), F1, specificity, and sensitivity are performance measurements of interface predictions (see Supporting Information Text S3 for definitions). The higher their values, the more reliable the predictions.
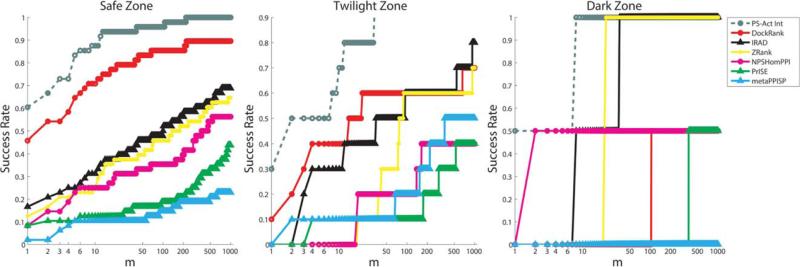
This allows us to investigate the performance of Dock-Rank as a function of prediction confidence, independent of the extent of conformational change upon binding. We studied the Success Rates and Hit Rates of DockRank on cases with only Safe, Twilight, and Dark Zone interface predictions (See Fig. 7).2 As expected, DockRank performs best with Safe Zone interface predictions. Although the performance of DockRank degrades in the Twilight Zone, it still outperforms other scoring functions in terms of the Success Rate. In Dark Zone (2 docking cases), DockRank is able to rank a hit at the rank of 100 out of 54,000 models for one case, but fails to find any hits for the other case among the top 1000 conformations.
Figure 7.
The interface prediction of PS-HomPPI on the ZDock3-BM3 decoy set in different interface prediction confidence zones. A docking case with more than one receptor-ligand chain pair may have predicted interfaces from different confidence zones. Only cases with predictions in a single confidence zone and with at least one hit are studied here. Fifty-eight cases have only Safe Zone interface predictions, out of which ZDock 3.0 is able to generate at least one hit for 48 cases. Fourteen cases have only Twilight Zone interface predictions, out of which 10 cases have at least one hit. Two cases have only Dark Zone interface predictions, of which two cases have at least one hit.
Different weights can be assigned to predicted interfaces with different prediction confidences when calculating the scores for each docked conformation. In this study, we used weights 1, 1, and 0.001 for the Safe, Twilight, and Dark Zone interface predictions, respectively. Our web server allows users to set different weights for interface predictions from PS-HomPPI when ranking docked models: higher weight for Safe Zone predicted interfaces and lower weight for Dark Zone predicted interfaces.
Although superimposition or threading provides a simpler method for modeling protein complexes, they have several important limitations. To the best of our knowledge: (i) superimposition and threading methods cannot adjust the relative orientation of proteins to generate more accurate interface structures, which can be important in many practical applications; (ii) threading can generate only partial models aligned with the templates provided and the resulting models usually contain only C-alpha atoms, necessitating the use of other programs to add side chains and fill in unaligned portions of the structures; and (iii) in the case of large proteins with more than two chains or domains, it could be difficult to find complete templates or even partial templates that can be assembled into a complete template. Even in cases where it is possible to do so, the process relies heavily on the relative orientation of the shared template proteins with other proteins and on the structural similarity between the shared template proteins; inaccuracies in these steps can cumulatively decrease the accuracy of the resulting models. Our results show that DockRank complements structural superimposition and threading approaches in important ways. DockRank's ability to make use of partial templates could be useful in light of the fact that experimentally solving the structure of multiple-chain/domain protein complexes is harder than solving that of proteins with fewer chains/domains.
The applicability of DockRank is limited by the availability of partner-specific interface predictions used by DockRank. At present, PS-HomPPI, the sequence homology-based partner-specific interface predictor42 used by DockRank, can reliably predict interface residues only in settings where reliable homo-interologs of a docking case are available. For example, PS-HomPPI returns interface predictions for 85 out 119 cases comprising Docking Benchmark 3.0 (which translates to a coverage of 71%, data not shown). The limited coverage of PS-HomPPI limits the applicability of DockRank to the subset of docking cases for which homo-interologs are available. The coverage of PS-HomPPI, and hence that of DockRank, can be expected to increase as more and more complexes are deposited in PDB. Expanding the applicability of DockRank to a much broader range of docking cases than currently possible calls for the development of alternatives to PS-HomPPI. To identity templates, PS-HomPPI currently uses the sequence-sequence alignment method BLASTP, which has relatively low sensitivity in detecting remote homologs. This limits the ability of PS-HomPPI to predict interfaces for cases that have templates with conserved interfaces but low sequence similarity. Work in progress is aimed at exploring alternatives to BLASTP, for example, profile–profile alignments, to detect remote sequence homologs, and thereby enable PS-HomPPI to identify more templates with high local sequence similarity and conserved interface residues. In addition, we are also developing machine-learning approaches to partner-specific interface prediction that do not rely on homo-interologs, as well as hybrid methods that combine the complementary strengths of homology-based partner-specific methods, such as PS-HomPPI, and partner-specific machine-learning predictors trained using sequence and structural features of component proteins to be docked against each other. Because the performance of DockRank in ranking conformations is limited ultimately by the reliability of available partner-specific interface information, any advances in methods for reliable partner-specific interface prediction or for that matter, high-throughput interface identification methods, can be leveraged to obtain corresponding improvements in the performance of DockRank.
Because of the high computational cost of exploring the large conformational space of complexes formed by several protein chains, there has been increasing interest in utilizing knowledge of the actual or predicted interface residues between a pair of proteins to constrain the exploration of docked configurations to those that are consistent with the predicted interfaces (thus improving the computational efficiency of docking and the accuracy of docking33,64–69). This raises the possibility of using PS-HomPPI predicted interfaces as a source of constraints to limit or bias the conformational space sampled by docking algorithms.
Supplementary Material
ACKNOWLEDGMENTS
The authors sincerely thank Usha Muppirala for providing ClusPro docked models; the CAPRI organizers, Marc Lensink, and Joel Janin, for sharing the CAPRI uploader models and their helpful email communications; and Ben Lewis for helpful suggestions on the manuscript. The authors also thank Guang Song at Iowa State University for the use of his computer cluster for some of the analyses. Any opinion, finding, and conclusions contained in this article are those of the authors and do not necessarily represent the views of the National Science Foundation.
Grant sponsor: National Institutes of Health; grant number: GM066387 (VH and DD); Grant sponsor: National Science Foundation (VH at National Science Foundation).
Footnotes
Additional Supporting Information may be found in the online version of this article.
In our experiments on the ZDock3-BM3 benchmark decoy set, for direct comparisons with published results on the same decoy set, we adopted the quality measures for evaluating the docked conformations used by ZDock3-BM3 set, that is the IRMSDs, which were precalculated and provided by the authors of ZDock3-BM3 along with the decoys. In our experiments on the ClusPro decoy set that we generated using ClusPro 2.0 webserver, we used L-RMSD as the quality measure for the docked conformations. Using L-RMSD, we measure the ability of DockRank to select docked conformations with meaningful 3D conformations of the ligand; using I-RMSD, we evaluate DockRank's ability to select docked conformations with correct 3D interface area.).
When a docking case has more than two protein chains (hence more than one pair of query proteins), the multiple pairwise interface predictions may have different confidence values. When a case has different confidence zones, it is not further analyzed here.
REFERENCES
- 1.Kuzu G, Keskin O, Gursoy A, Nussinov R. Constructing structural networks of signaling pathways on the proteome scale. Curr Opin Struct Biol. 2012;22:367–377. doi: 10.1016/j.sbi.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 2.Nooren I, Thornton J. Diversity of protein–protein interactions. EMBO J. 2003;22:3486–3492. doi: 10.1093/emboj/cdg359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Acuner Ozbabacan S, Engin H, Gursoy A, Keskin O. Transient protein–protein interactions. Protein Eng Des Sel. 2011;24:635–648. doi: 10.1093/protein/gzr025. [DOI] [PubMed] [Google Scholar]
- 4.Nooren I, Thornton J. Structural characterisation and functional significance of transient protein-protein interactions. J Mol Biol. 2003;325:991–1018. doi: 10.1016/s0022-2836(02)01281-0. [DOI] [PubMed] [Google Scholar]
- 5.Grosdidier S, Fernandez-Recio J. Protein-protein docking and hot-spot prediction for drug discovery. Curr Pharm Des. 2012;18:4607–4618. doi: 10.2174/138161212802651599. [DOI] [PubMed] [Google Scholar]
- 6.Kim PM, Lu LJ, Xia Y, Gerstein MB. Relating three-dimensional structures to protein networks provides evolutionary insights. Sci Signal. 2006;314:1938. doi: 10.1126/science.1136174. [DOI] [PubMed] [Google Scholar]
- 7.Katebi A, Kloczkowski A, Jernigan R. Structural interpretation of protein-protein interaction network. BMC Struct Biol. 2010;10:S4. doi: 10.1186/1472-6807-10-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mosca R, Pons C, Fern andez-Recio J, Aloy P. Pushing structural information into the yeast interactome by high-throughput protein docking experiments. PLoS Comput Biol. 2009;5:e1000490. doi: 10.1371/journal.pcbi.1000490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mosca R, C eol A, Aloy P. Interactome3D: adding structural details to protein networks. Nat Methods. 2012;10:47–53. doi: 10.1038/nmeth.2289. [DOI] [PubMed] [Google Scholar]
- 10.Halperin I, Ma B, Wolfson H, Nussinov R. Principles of docking: an overview of search algorithms and a guide to scoring functions. Proteins. 2002;47:409–443. doi: 10.1002/prot.10115. [DOI] [PubMed] [Google Scholar]
- 11.Lensink M, M endez R, Wodak S. Docking and scoring protein complexes: CAPRI 3rd Edition. Proteins. 2007;69:704–718. doi: 10.1002/prot.21804. [DOI] [PubMed] [Google Scholar]
- 12.Vajda S, Kozakov D. Convergence and combination of methods in protein-protein docking. Curr Opin Struct Biol. 2009;19:164–170. doi: 10.1016/j.sbi.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Méndez R, Leplae R, De Maria L, Wodak S. Assessment of blind predictions of protein–protein interactions: current status of docking methods. Proteins. 2003;52:51–67. doi: 10.1002/prot.10393. [DOI] [PubMed] [Google Scholar]
- 14.Martin O, Schomburg D. Efficient comprehensive scoring of docked protein complexes using probabilistic support vector machines. Proteins. 2008;70:1367–1378. doi: 10.1002/prot.21603. [DOI] [PubMed] [Google Scholar]
- 15.Comeau S, Gatchell D, Vajda S, Camacho C. ClusPro: an automated docking and discrimination method for the prediction of protein complexes. Bioinformatics. 2004;20:45. doi: 10.1093/bioinformatics/btg371. [DOI] [PubMed] [Google Scholar]
- 16.Kozakov D, Brenke R, Comeau S, Vajda S. PIPER: An FFT-based protein docking program with pairwise potentials. Proteins. 2006;65:392–406. doi: 10.1002/prot.21117. [DOI] [PubMed] [Google Scholar]
- 17.Kozakov D, Hall D, Beglov D, Brenke R, Comeau S, et al. Achieving reliability and high accuracy in automated protein docking: ClusPro, PIPER, SDU, and stability analysis in CAPRI rounds 13–19. Proteins. 2010;78:3124–3130. doi: 10.1002/prot.22835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katchalski-Katzir E, Shariv I, Eisenstein M, Friesem A, Aflalo C, et al. Molecular surface recognition: determination of geometric fit between proteins and their ligands by correlation techniques. Proc Natl Acad Sci. 1992;89:2195. doi: 10.1073/pnas.89.6.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vreven T, Hwang H, Weng Z. Integrating atom-based and residue-based scoring functions for protein–protein docking. Protein Sci. 2011;20:1576–1586. doi: 10.1002/pro.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pierce B, Weng Z. ZRANK: reranking protein docking predictions with an optimized energy function. Proteins. 2007;67:1078–1086. doi: 10.1002/prot.21373. [DOI] [PubMed] [Google Scholar]
- 21.Gray J, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, et al. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J Mol Biol. 2003;331:281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 22.Heuser P, Schomburg D. Optimised amino acid specific weighting factors for unbound protein docking. BMC Bioinformatics. 2006;7:344. doi: 10.1186/1471-2105-7-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Philipp H, Dietmar S. Combination of scoring schemes for protein docking. BMC Bioinformatics. 2007;8:279. doi: 10.1186/1471-2105-8-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moont G, Gabb H, Sternberg M. Use of pair potentials across protein interfaces in screening predicted docked complexes. Proteins. 1999;35:364–373. [PubMed] [Google Scholar]
- 25.Liu S, Vakser I. DECK: Distance and environment-dependent, coarse-grained, knowledge-based potentials for protein-protein docking. BMC Bioinformatics. 2011;12:280. doi: 10.1186/1471-2105-12-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Chen R, Weng Z. RDOCK: Refinement of rigid-body protein docking predictions. Proteins. 2003;53:693–707. doi: 10.1002/prot.10460. [DOI] [PubMed] [Google Scholar]
- 27.Bordner A, Gorin A. Protein docking using surface matching and supervised machine learning. Proteins. 2007;68:488–502. doi: 10.1002/prot.21406. [DOI] [PubMed] [Google Scholar]
- 28.London N, Schueler-Furman O. Funnel hunting in a rough terrain: learning and discriminating native energy funnels. Structure. 2008;16:269–279. doi: 10.1016/j.str.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 29.Neuvirth H, Raz R, Schreiber G. ProMate: a structure based prediction program to identify the location of protein-protein binding sites. J Mol Biol. 2004;338:181–199. doi: 10.1016/j.jmb.2004.02.040. [DOI] [PubMed] [Google Scholar]
- 30.Huang B, Schroeder M. Using protein binding site prediction to improve protein docking. Gene. 2008;422:14–21. doi: 10.1016/j.gene.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 31.Bourquard T, Bernauer J, Az e J, Poupon A. A collaborative filtering approach for protein-protein docking scoring functions. PloS One. 2011;6:e18541. doi: 10.1371/journal.pone.0018541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernauer J, Aze J, Janin J, Poupon A. A new protein protein docking scoring function based on interface residue properties. Bioinformatics. 2007;23:555. doi: 10.1093/bioinformatics/btl654. [DOI] [PubMed] [Google Scholar]
- 33.Lensink MF, Wodak SJ. Docking and scoring protein interactions: CAPRI 2009. Proteins. 2010;78:3073–3084. doi: 10.1002/prot.22818. [DOI] [PubMed] [Google Scholar]
- 34.Zhou H, Qin S. Interaction-site prediction for protein complexes: a critical assessment. Bioinformatics. 2007;23:2203–2209. doi: 10.1093/bioinformatics/btm323. [DOI] [PubMed] [Google Scholar]
- 35.Ezkurdia I, Bartoli L, Fariselli P, Casadio R, Valencia A, et al. Progress and challenges in predicting protein–protein interaction sites. Briefings Bioinform. 2009;10:233–246. doi: 10.1093/bib/bbp021. [DOI] [PubMed] [Google Scholar]
- 36.Tuncbag N, Kar G, Keskin O, Gursoy A, Nussinov R. A survey of available tools and web servers for analysis of protein–protein interactions and interfaces. Briefings Bioinform. 2009;10:217–232. doi: 10.1093/bib/bbp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H, Segal E, Ben-Hur A, Li Q, Vidal M, et al. Insite: a computational method for identifying protein-protein interaction binding sites on a proteome-wide scale. Genome Biol. 2007;8:R192. doi: 10.1186/gb-2007-8-9-r192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chung J, Wang W, Bourne P. High-throughput identification of interacting protein-protein binding sites. BMC Bioinformatics. 2007;8:223. doi: 10.1186/1471-2105-8-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahmad S, Mizuguchi K. Partner-aware prediction of interacting residues in protein-protein complexes from sequence data. PloS One. 2011;6:e29104. doi: 10.1371/journal.pone.0029104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panchenko A, Kondrashov F, Bryant S. Prediction of functional sites by analysis of sequence and structure conservation. Protein Sci. 2004;13:884–892. doi: 10.1110/ps.03465504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Vries SJ, Bonvin AM. How proteins get in touch: interface prediction in the study of biomolecular complexes. Curr Protein Pept Sci. 2008;9:394–406. doi: 10.2174/138920308785132712. [DOI] [PubMed] [Google Scholar]
- 42.Xue L, Dobbs D, Honavar V. HomPPI: a class of sequence homology based protein-protein interface prediction methods. BMC Bioinformatics. 2011;12:244. doi: 10.1186/1471-2105-12-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xue L, Jordan R, El-Manzalawy Y, Dobbs D, Honavar V. Ranking docked models of protein-protein complexes using predicted partner-specific protein-protein interfaces: a preliminary study. In: Proceedings of the 2nd ACM Conference on Bioinformatics, Computational Biology and Biomedicine. ACM. 2011:441–445. doi: 10.1145/2147805.2147866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hwang H, Vreven T, Pierce B, Hung J, Weng Z. Performance of ZDOCK and ZRANK in CAPRI rounds 13–19. Proteins. 2010;78:3104–3110. doi: 10.1002/prot.22764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Comeau S, Gatchell D, Vajda S, Camacho C. ClusPro: a fully automated algorithm for protein–protein docking. Nucleic Acids Res. 2004;32:W96–W99. doi: 10.1093/nar/gkh354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mukherjee S, Zhang Y. Protein-protein complex structure predictions by multimeric threading and template recombination. Structure. 2011;19:955–966. doi: 10.1016/j.str.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen H, Skolnick J. M-TASSER: an algorithm for protein quaternary structure prediction. Biophys J. 2008;94:918–928. doi: 10.1529/biophysj.107.114280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu L, Lu H, Skolnick J. MULTIPROSPECTOR: an algorithm for the prediction of protein–protein interactions by multimeric threading. Proteins. 2002;49:350–364. doi: 10.1002/prot.10222. [DOI] [PubMed] [Google Scholar]
- 49.Hwang H, Pierce B, Mintseris J, Janin J, Weng Z. Protein–protein docking benchmark version 3.0. Proteins. 2008;73:705–709. doi: 10.1002/prot.22106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berman H, Westbrook J, Feng Z, Gilliland G, Bhat T, et al. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. [Sep. 27th 2012];ProtinDB-PROTein-protein INterface residues Data Base. Available at: http://protindb.cs.iastate.edu/index.py.
- 52. [Sep. 27th 2012];S2C-A database correlating sequence and atomic coordinate residue numbering in the Protein Data Bank. Available at: http://dunbrack.fccc.edu/Guoli/s2c/index.php.
- 53.Altschul S, Madden T, Schäffer A, Zhang J, Zhang Z, et al. Gapped BLAST and PSIBLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang B, Srihari S. Binary vector dissimilarity measures for handwriting identification. Proc. SPIE. 2003;5010:29. [Google Scholar]
- 55.Li B, Kihara D. Protein docking prediction using predicted protein-protein interface. BMC Bioinformatics. 2012;13:7. doi: 10.1186/1471-2105-13-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qin S, Zhou H. meta-PPISP: a meta web server for protein-protein interaction site prediction. Bioinformatics. 2007;23:3386. doi: 10.1093/bioinformatics/btm434. [DOI] [PubMed] [Google Scholar]
- 57.Jordan R, Yasser E, Dobbs D, Honavar V. Predicting protein-protein interface residues using local surface structural similarity. BMC Bioinformatics. 2012;13:41. doi: 10.1186/1471-2105-13-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou H, Shan Y. Prediction of protein interaction sites from sequence profile and residue neighbor list. Proteins. 2001;44:336–343. doi: 10.1002/prot.1099. [DOI] [PubMed] [Google Scholar]
- 59.Chen H, Zhou H. Prediction of interface residues in protein–protein complexes by a consensus neural network method: test against NMR data. Proteins. 2005;61:21–35. doi: 10.1002/prot.20514. [DOI] [PubMed] [Google Scholar]
- 60.Liang S, Zhang C, Liu S, Zhou Y. Protein binding site prediction using an empirical scoring function. Nucleic Acids Res. 2006;34:3698–3707. doi: 10.1093/nar/gkl454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Janin J, Bahadur RP, Chakrabarti P. Protein-protein interaction and quaternary structure. Q Rev Biophys. 2008;41:133–180. doi: 10.1017/S0033583508004708. [DOI] [PubMed] [Google Scholar]
- 64.de Vries S, van Dijk A, Bonvin A. WHISCY: What information does surface conservation yield? Application to data-driven docking. Proteins. 2006;63:479–489. doi: 10.1002/prot.20842. [DOI] [PubMed] [Google Scholar]
- 65.de Vries S, van Dijk A, Krzeminski M, van Dijk M, Thureau A, et al. HADDOCK versus HADDOCK: new features and performance of HADDOCK2. 0 on the CAPRI targets. Proteins. 2007;69:726–733. doi: 10.1002/prot.21723. [DOI] [PubMed] [Google Scholar]
- 66.Dominguez C, Boelens R, Bonvin A. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 67.de Vries S, Bonvin A. CPORT: a consensus interface predictor and its performance in prediction-driven docking with HADDOCK. PLoS One. 2011;6:e17695. doi: 10.1371/journal.pone.0017695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tovchigrechko A, Vakser I. Development and testing of an automated approach to protein docking. Proteins. 2005;60:296–301. doi: 10.1002/prot.20573. [DOI] [PubMed] [Google Scholar]
- 69.Tovchigrechko A, Vakser I. GRAMM-X public web server for protein–protein docking. Nucleic Acids Res. 2006;34:W310–W314. doi: 10.1093/nar/gkl206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schrödinger LLC. The PyMOL molecular graphics system, version 1.3r1. 2010 [Google Scholar]
- 71.Mintseris J, Weng Z. Structure, function, and evolution of transient and obligate protein–protein interactions. Proc Natl Acad Sci USA. 2005;102:10930. doi: 10.1073/pnas.0502667102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hubbard S, Thornton J. NACCESS, Computer Program, Department of Biochemistry and Molecular Biology. University College London; 1993. [Google Scholar]
- 73.Witten I, Frank E, Hall M. Data Mining: Practical machine learning tools and techniques. Morgan Kaufmann; 2011. [Google Scholar]
- 74.Baldi P, Brunak S, Chauvin Y, Andersen C, Nielsen H. Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics. 2000;16:412–424. doi: 10.1093/bioinformatics/16.5.412. [DOI] [PubMed] [Google Scholar]
- 75.Dem sar J. Statistical comparisons of classifiers over multiple data sets. J Mach Learn Res. 2006;7:1–30. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.