Abstract
Tissue morphogenesis comprises the self-organized creation of various patterns and shapes. Although detailed underlying mechanisms are still elusive in many cases, an increasing amount of experimental data suggests that chemical morphogen and mechanical processes are strongly coupled. Here, we develop and test a minimal model of the axis-defining step (i.e., symmetry breaking) in aggregates of the Hydra polyp. Based on previous findings, we combine osmotically driven shape oscillations with tissue mechanics and morphogen dynamics. We show that the model incorporating a simple feedback loop between morphogen patterning and tissue stretch reproduces a wide range of experimental data. Finally, we compare different hypothetical morphogen patterning mechanisms (Turing, tissue-curvature, and self-organized criticality). Our results suggest the experimental investigation of bigger (i.e., multiple head) aggregates as a key step for a deeper understanding of mechanochemical symmetry breaking in Hydra.
Introduction
During embryogenesis the organism develops from a single cell to a complex entity composed of tissues showing different types of patterns (1). Chemical patterns are patterns of diffusing signaling molecules involved in tissue development, called morphogens (2–4). In contrast, mechanical patterns are related to shape/geometry and (visco-)elastic properties of cells or tissues, which are expressed in terms of geometry, strain, stress, or stiffness (5,6). During many developmental steps, chemical and mechanical patterns appear simultaneously, and it is frequently difficult to distinguish cause from effect.
However, the traditional view of morphogenesis is dominated by a purely chemical approach, where mechanical patterns are regarded as simple consequences of chemical prepatterns (7). According to this theory, morphogenesis is a hierarchical sequence of chemical patterning processes, where each pattern depends critically on the previous one.
Increasing amounts of experimental data show that mechanical or geometric cues can actively influence morphogenetic processes and cell differentiation (for review, see Chanet and Martin (8) and Farge (9)). This suggests that the interplay between chemical and mechanical processes could lead to pattern formation in many cases (10,11). Moreover, purely chemical models propose the existence of morphogens behaving in a certain way (e.g., long-range inhibitors for Turing (4) or the Gierer-Meinhardt (12) models), but these morphogens have not been identified after several decades of research (13). Finally, at least some developmental processes do not critically depend on the existence of chemical prepatterning; from lower animals (14) to organs of mammals (15,16), it has been shown that they may evolve from dissociated and randomly aggregated cells.
For these reasons, the need for fresh modeling approaches, integrating mechanical and chemical processes during development, has been recently stressed (17). Although the number of mechanical models of morphogenesis has strongly increased during the last decade (18), approaches considering mechanochemical processes during tissue development are still in the early stages (10,19,20).
To study the mechanochemistry of development, the Hydra polyp is a suitable organism. Hydra is an established biological model organism for morphogenesis (21,22) and it has been traditionally used to apply purely chemical pattern formation models—such as in the seminal article of Alan Turing (4) and in related approaches (3,12,23,24). Hitherto, several morphogens involved in early pattern formation (i.e., axis formation) have been identified in Hydra: Members of the Wnt signaling pathway are locally expressed during early head formation, with Wnt3 constituting the earliest marker of this signal cascade (25,26). Furthermore, production of the permanently expressed Ks1 morphogen appears to be upregulated in response to early signals of head formation (27,28) whereas its graded expression inversely correlates with the head-forming potential in adult Hydra polyps (29).
However, these molecules are not good candidates for morphogens in Turing-type models; note that the long-range inhibitors are still elusive in Hydra (13,30,31). It has also been recently shown that in this organism, tissue mechanics is indispensable during early pattern formation (19), suggesting that purely chemical models are insufficient.
Hydra is attractive from a modeling point of view. It has a simple body plan and the complexity can be reduced by studying the so-called Hydra aggregates (14). Here, de novo pattern formation and regeneration of polyps can be observed from a bulk of randomly mixed and aggregated cells. Initially, these cells form a hollow tissue sphere, which exhibits osmotically driven shape oscillations: Due to water influx, the tissue sphere swells until tissue rupture takes place, which is followed by a rapid collapse and tissue healing; then the process starts again (32). After several oscillations (phase I), aggregates suddenly change their homogeneous structure to a chemically and mechanically inhomogeneous system (phase II), which defines the first axis-defining step in Hydra called symmetry breaking (19,25,32). In contrast to recent findings in yeast (33), the location of tissue rupture in Hydra aggregates does not seem to determine the final body axis: Before symmetry breaking, the rupture point of each cycle occurs at a different location (19).
In some systems, there is evidence that pattern formation is driven by motile cells (34), which can explain why corresponding molecular species are not found. However, experimental observations exclude such a mechanism in Hydra. Cellular patterning requires a range of cell populations with distinct properties (34). In contrast, pattern formation and symmetry breaking in Hydra can be achieved in aggregates with varying cell composition (14) and axis formation can be induced by purely chemical cues (35).
Thus, in this article, we investigate possible mechanochemical processes underlying symmetry breaking in Hydra aggregates. We propose a model combining osmotically driven shape oscillations (19,36) and morphogen dynamics and test its ability to reproduce recent experimental results. Furthermore, we compare previously proposed models for symmetry breaking, such as diffusion-driven instability (the Turing model (12,19)), self-organized criticality (i.e., SOC (27,37)), and curvature-increasing mechanisms (10) with respect to their predictions of dynamics of this process. Our results are discussed in the context of morphogens associated with symmetry breaking and head formation, such as Ks1 and Wnt (25,26,28).
Materials and Methods
Model
In this section, based on experimental data, we develop an osmomechanochemical (OMC) model for symmetry breaking in early Hydra aggregates. This model combines osmotic processes with tissue mechanics and morphogen dynamics. Because the exact mechanism of morphogen clustering during body axis formation in Hydra is unknown, we focus on the activator-inhibitor model exhibiting Turing instability. However, we consider and compare different alternative mechanisms proposed previously in the literature (such as SOC and curvature-increasing models), which we investigate for simplicity on spheres of fixed radius without tissue mechanics.
The osmomechanochemical model
The OMC model describes dynamics of two interacting and diffusing morphogen species in an elastically deforming tissue surface which undergoes osmosis-driven shape oscillations.
Dynamics of tissue deformations are based on a gradient flow minimizing the free energy, accounting for osmotic pressure, tissue stretching, bending, and local morphogen concentrations. Together with Turing-type morphogen dynamics, we obtain a model in the form of coupled nonlinear partial differential equations of fourth-order.
Most importantly, we propose the following feedback loop between morphogens and tissue mechanics (see Fig. 1): There exists a morphogen that locally lowers the resistance to lateral tissue stretch (MC1), and patterning of this morphogen critically depends on lateral tissue stretch (MC2).
Figure 1.
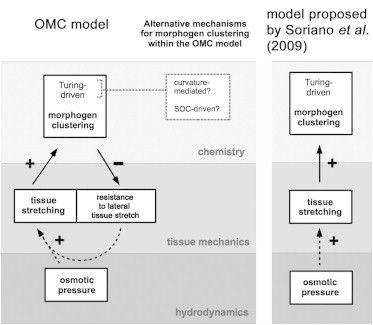
Proposed interactions among morphogens, tissue mechanics, and osmotic processes during symmetry breaking in Hydra aggregates. (Left-hand side) OMC model (presented within this study); (right-hand side) model of Soriano et al. (19). (Continuous arrows) Experimentally motivated model assumptions; (dotted arrows) physical relationships.
These assumptions on the mechanochemical coupling are based on the observation that symmetry breaking in Hydra spheres involves formation of an early head, having a head-organizer in the center (13). The early head can be characterized in different ways: mechanically as a weak spot in the tissue (19,25,27) (i.e., a lower resistance to lateral tissue stretching), or chemically as the local expression of characteristic morphogens, such as multiple Wnt (25,26) or Ks1 (28) molecules. Furthermore, mechanochemical symmetry breaking requires the shape oscillations mentioned above, leading to periodic tissue stretch (19).
The OMC model constitutes an extension of the reaction-diffusion model proposed by Soriano et al. (19) for Hydra symmetry breaking, but differs in the following points (Fig. 1).
-
1.
Soriano et al. (19) consider influence of tissue stretch on morphogen patterning instead of a feedback loop.
-
2.
We explicitly model tissue deformations, whereas Soriano et al. (19) incorporates the shape oscillations more phenomenologically.
-
3.
In contrast to Soriano et al. (19), the OMC model does not require the assumption of viscoelasticity to reproduce experimental results.
Mathematical modeling framework
To describe mathematically the dynamics of a Hydra aggregate, we use a continuous approach, which is justified by the large number of cells (≈104) in the system (19). The cell bilayer forming a hollow tissue sphere is at any time t approximated by a closed two-dimensional surface Γ(t), embedded in three-dimensional space. The evolution of Γ(t) is given by a diffeomorphic time-dependent representation , parameterized over the unit sphere S2 ⊂ ℝ3. Thus, Γ(t) is the image of with for a T ∈ ℝ>0.
Local concentrations of the morphogens A and B at time t are given by continuous functions ΦA, ΦB on the deforming tissue surface Γ(t), defined as morphogen concentrations per cell volume, Φi(t): Γ(t)→ ℝ≥0, i ∈ {A,B}. In order to achieve a consistent formulation with chemical processes being defined on S2 rather than on Γ, we redefine Φi accordingly. This can be done by using the fact that we can identify material points on Γ(t) with , because is smooth and bijective. Thus, for each , t ∈ [0,T], and i ∈ {A,B}, we define the function ϕi:
Energy terms describing elastic behavior
In this study we treat the tissue as purely elastic and not viscoelastic. Although corresponding timescales are within the range of viscoelastic processes, previous studies showed that purely elastic models reproduce Hydra tissue behavior (e.g., Krahe et al. (38), Mombach et al. (39), and Kücken et al. (36)) and can be used as an approximation. Furthermore, we will show that effects that have been previously ascribed to viscoelastic properties of Hydra can be explained by elastic tissue behavior.
Elastic tissue deformations are based on minimization of free energy , composed of energy terms related to tissue bending, stretching, and osmotic pressure. Free energy formulations are expressed in terms of surface integrals, , or volume integrals, , where Ω is the domain inside, i.e., Γ = ∂Ω. We will abbreviate Γ = Γ(t) and Ω = Ω (t). In the parametric formulation, due to the divergence theorem, these time-dependent integrals are given by and , respectively. Here, is the outer unit normal vector and is a surface measure, the latter representing the local area of the tissue and depending on the parametric function .
Osmotic processes and pressure
It has been previously shown that early Hydra aggregates undergo osmotically induced shape oscillations, consisting of repeated tissue swelling, rupture/collapse, and healing cycles (32,36). These oscillations appear to be required for symmetry breaking (19). To integrate this aspect, we follow the approach of Fütterer et al. (32), assuming a Darcy-type law for the temporal development of the total water volume V(t) inside the tissue shell. This is (in the absence of elastic resistance) given by
| (1) |
Here, is the initial (unstressed) total tissue area, α is a constant depending on temperature and water permeability, and (Cin – Cout) is the difference in ion concentrations per unit of volume from the inside to the outside of the Hydra shell (36). Although Cin depends on time, it has been shown that it does not change appreciably after an initial equilibration (36). Hence, for the sake of simplicity, we use a constant for effective ion concentration difference between the inside and the outside of the aggregate, ΔCeff := Cout(t >> 0) – Ctn(t >> 0). With Eq. 1, it follows for the preferred aggregate volume that
| (2) |
We include Eq. 2 in an energy term elastically penalizing a possible overpressure in the interior of the aggregate, the latter quantified by deviations of this volume from the preferred value V(t). This energy is given by
| (3) |
where κ1 balances osmotic pressure with elastic tissue resistance. Furthermore, Eq. 3 is only valid if the tissue sphere is intact. For consideration of tissue rupture and pressure release, we refer to the next subsection.
Tissue stretching, rupture, and healing
Lateral tissue stretching is assumed to play a major role during symmetry breaking in Hydra spheres (19). Hence, unlike in the previous approaches (10,40), we consider a locally compressible surface. We introduce the energy term
| (4) |
which is a generalization of the Hook energy for a spring (41). Deviations of the local tissue area from the initial (relaxed) value are elastically penalized, weighted by the stretch rigidity κ2.
The osmotically induced swelling of the Hydra tissue shell Γ ends due to local tissue rupture, where pressure is released from a small hole until healing takes place, and the swelling process starts again (19,27,32). Because topological changes of Γ are difficult to handle mathematically (and are not the primary aim of this study), we introduce the effect of tissue rupture and healing on a more phenomenological level: If local tissue stretch exceeds a certain threshold β, i.e., , we set the osmotic pressure to zero by setting for a certain time period t ∈ [trupt, theal[, and
afterwards. Hence, we obtain a pressure release in the moment of tissue rupture trupt, and a pressure regeneration starting from the moment of completed tissue healing theal. In the following, we have set β = (1.25)2, which matches the experimental observation that rupture occurs, if, for the relative radius R = (r – r0)/r0, it holds R > 0.25, where r0 is the initial and r this aggregate radius (36).
To account for point (MC1) of the mechanochemical coupling, we set κ2 = κ2(ΦA, ΦB), i.e., we assume that the resistance to stretching depends on local morphogen concentrations. However, a simple linear relationship between tissue stiffness and morphogen concentrations would be unrealistic, because we expect saturation for high morphogen concentrations. We set
| (5) |
where ΦA has been chosen over ΦB for simplicity. The interpretation of Eq. 5 is that the stretch rigidity κ2 decreases with increasing morphogen concentration ΦA and then shows a saturation, i.e., κ2 → (ξ − λ) for ΦA → ∞. Using the parameter values in the Supporting Material, we obtain a maximum effective reduction of κ2 due to ΦA by a factor of ∼0.4, which could be achieved in vivo by locally lowering the cell-cell-contacts.
Bending
To describe the resistance of the tissue surface to bending, we use the Helfrich free energy (42), which is given by
| (6) |
Here, H is the mean curvature and κ3 is the bending rigidity (40). Because tissue curvature effects are assumed to play a minor role within this model, we neglect modifications of bend due to tissue compressibility, which would include changes in bending stiffness κ3 due to local stretching or compression.
Evolution equations for the deforming tissue surface
The total energy = bend + comp + osm of the Hydra tissue shell is given as sum of the energy terms Eqs. 3, 4, and 6. Within this study, we assume that κ1 >> κ2,κ3, because the swelling rate appears to have only a weak dependence on the elasticity of the Hydra shell (36). However, the energy terms for bending and stretching are not negligible even for high values of κ1 (the latter leading to a global volume constraint), because κ2 and κ3 are connected to local curvatures and stretches, which are not uniquely determined by a prescribed volume.
Evolution of the deforming Hydra shell up to time T > 0 is now given by the L2-gradient flow of the total energy
| (7) |
where dt(.) is the total time derivative, L is a kinetic coefficient, is the surface normal vector, and denotes the strong formulation of the variation perpendicular to the deformed surface Γ. Thus, we simplify our system considering only normal surface deformations. This is motivated by the fact that the main deforming force resulting from osmotic pressure acts purely normally. The gradient flow Eq. 7 leads to the minimization of the free energy , hence to the mechanical equilibrium. However, gradient flows may also describe realistic dynamics of deforming surfaces when we consider overdamped systems, e.g., biological surfaces embedded in a fluid (40,43–45). For more details in corresponding definitions, further information on the calculation of gradient flows, detailed calculation of variations, and technical remarks, we refer to the Supporting Material and to Mercker et al. (40).
Morphogen dynamics
Instead of using gradient flows of to derive dynamics of chemicals (10), the evolution of morphogens is assumed to be independent of . This is motivated by the fact that morphogen molecules are not part of the mechanical surface itself, but only may indirectly influence tissue mechanics. Hence, morphogen dynamics do not, per se, depend on .
For the chemical patterning during symmetry breaking in Hydra aggregates, we follow the ideas of Soriano et al. (19) using a reaction-diffusion equation for morphogen patterning. In particular, we modify the minimum model of Koch and Meinhardt (46), considering pattern formation of an activator ΦA and an inhibitor ΦB on a deformed sphere, given by the equations
| (8) |
| (9) |
Here, DA and DB are diffusion rates; is the surface Laplace operator; and CA and CB are reaction rates, comprising production and removal. Equations 8 and 9 are strongly related to those presented by Plaza et al. (47), because we also incorporate tissue curvature and advection, where the latter is in our case naturally given due to the Lagrangian description of Γ (10). However, we additionally consider tissue stretch instead of growth processes.
To include point (MC2) of the mechanochemical coupling, according to Soriano et al. (19), we assume that diffusion rates depend on lateral tissue stretch. We set
| (10) |
i.e., diffusion of the inhibitor ΦB is accelerated if the tissue is laterally stretched, but shows a saturation effect. Alternatively, one could reduce activator diffusion as a response to tissue stretch (19). Using the parameter values as presented within the Supporting Material, local tissue stretch leads to a maximal effective increase of DB by a factor of ∼100. In vivo, underlying mechanisms could be selective channels, which close and open depending on tissue stretch (19).
Possible morphogen patterning mechanisms
The mechanochemical coupling (i.e., the mutual coupling of morphogen dynamics with tissue stretch) is the most important part of the OMC model. We emphasize that the choice of a Turing mechanism for morphogen pattern formation is somewhat arbitrary within this model, because we primarily focus on the mechanochemical aspects of Hydra symmetry breaking, with less attention to the exact chemical mechanism. Thus, Eqs. 8 and 9 could be replaced by alternative mechanisms (Fig. 2).
Figure 2.
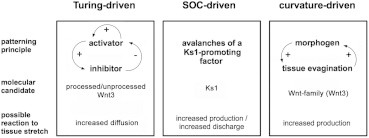
Three alternative mechanisms for morphogen patterning within the context of the OMC model, i.e., coupled to tissue stretch and leading to symmetry breaking in Hydra.
To shed more light on the underlying chemical patterning mechanisms, we compare Turing models with two alternatives (SOC and curvature-increasing models) that have been also proposed to explain symmetry breaking in Hydra-aggregates (4,37,40). In particular, we concentrate on the dependence of the symmetry breaking time tSB on the initial aggregate radius r0. To reduce complexity, at this stage we do not couple those models with tissue deformations.
Turing patterning
Turing models have been used to explain morphogen pattern formation and axis development in Hydra for more than 60 years (4,19,23,48), and have been numerically studied on different tissue geometries including spheres (47,49). We investigated if these models are able to reproduce different experimental data. A Turing model is a nonlinear reaction-diffusion system with diffusion-driven instability (DDI), in which the system admits a spatially homogeneous steady state us that is 1) stable with respect to spatially homogeneous perturbations, but 2) unstable with respect to spatially nonhomogeneous perturbations.
Considering the nonlinear system on spheres of radius r0 for initial conditions being a small perturbation of us, the solution is well approximated by the solution of the linearized system, whose maximum grows like . Here, is the largest eigenvalue of the linearized system and is a coefficient depending on the initial condition. We observed that for the same initial condition, grows linearly with the radius r0.
Defining the symmetry breaking time as the moment where a certain threshold is exceeded, we obtain
Tissue curvature-driven patterning
Recently, an alternative to Turing models has been proposed, where the patterning of only one diffusing morphogen type is driven by a simple positive feedback loop between tissue curvature and morphogen production (40). The morphogen is assumed to drive tissue bending in one direction (e.g., outward curving by apical constriction), and tissue curvatures are assumed to drive local morphogen production in turn. Thus, the interplay between morphogen production and actively generated and passively adapting tissue curvatures leads to pattern formation without reliance on molecular long-range inhibitors (40). Although there is so far no experimental evidence for this mechanism in Hydra, mechanisms using mechanical cues as long-range inhibitors might explain why the identification of appropriate molecular (Turing-) inhibitors still fails. Hence, we investigated if the model of Mercker et al. (40) reproduces the experimental data for the symmetry-breaking time tSB. For details concerning the tested parameter space and the definition of tSB in this context, we refer the reader to Computational Methods.
SOC-driven patterning
The morphogen Ks1 has been shown to be a molecular marker for early head formation in Hydra (28). Further studies have documented that fractal expression patterns of Ks1 in Hydra aggregates show behavior of SOC. Based on these observations, Gamba et al. (37) proposed a certain SOC mechanism for Ks1 expression, originally used in the modeling of earthquakes (50). Here, Ks1 patterning is based on nearest-neighbor interactions of cells. Gamba et al. (37) demonstrated that this model reproduces different experimental observations, such as fractal appearance and size-distribution of Ks1-positive cell clusters and their localization depending on temperature gradients. Thus, we investigated whether the temporal behavior of this model can also reproduce different experimental data. For further details regarding corresponding techniques and the definition of tSB, we refer the reader to the next subsection.
Computational methods
In this subsection, we present computational methods for the investigation of the models. For details concerning the parameterization, we refer to the Supporting Material.
General computation schemes
For simulations of the OMC model and the curvature-increasing model, we used the finite-element library GASCOIGNE 3D (51), approximating the fourth-order partial differential equations in a mixed formulation. For spatial discretization, we used biquadratic elements with ≥8659 grid points; for time discretization, a semi-implicit Euler scheme. For further details of the computation scheme we refer to Mercker et al. (10,40). Computations of the SOC model and of the inverse eigenvalue functions related to the Turing pattern are carried out in C++ and MATLAB 7.11 (The MathWorks, Natick, MA), respectively.
Data analysis of the OMC model
Most of the following definitions are based on the experimental methods as described in Soriano et al. (19). The radius r of (deformed) simulated aggregates Γ = ∂Ω is defined as r = (A/(4π))1/2, with tissue surface area . Changes in aggregate shape symmetry are quantified by the factor b/a with minor and major inner aggregate axis b and a, respectively. Here, a is defined as the radius of the smallest possible sphere Ba with Ba ⊃ Ω, and b as the radius of the largest possible sphere Bb with Bb ⊂ Ω and the same center as Ba. The symmetry breaking time tSB is defined as the moment at which the system geometry initially reaches a certain asymmetry, namely if b/a < 0.95 holds. Swelling rate s has been determined by linear regression of r(t) for t ∈ [0,trupt[, where trupt is the moment of first tissue rupture, averaged over the first three cycles.
Data analysis of the Turing model
We consider a linear reaction-diffusion system ∂tu = DΔu + Ju on a sphere of radius r0. Here, u is a vector of two or three components, D is the diagonal matrix of the diffusion coefficients, and J is the Jacobian of the kinetic system evaluated at the homogeneous steady state. DDI occurs if 1) J has only eigenvalues with negative real parts, and 2) there is a value such that the matrix has an eigenvalue with positive real part. Using the Routh-Hurwitz theorem (Gantmacher (52)), we see that these conditions require a sufficient spread of the diffusion coefficients and that J is not s-stable (compare to Satnoianu et al. (53) for the general definition and the Supporting Material for systems of two and three equations).
We use the method of least-squares under nonlinear constraints to find parameters (entries of D and J) such that there is a minimal deviation between the experimentally given symmetry breaking time and , where λ1(r0) is the largest eigenvalue of J1(r0).
The initial guess for fitting involves randomly generated matrices D0 and J0, where D0 is diagonal with nonnegative ordered entries and J0 is in the Turing space (compare to the Supporting Material).
Data analysis of the curvature increasing model
In Mercker et al. (40), it has been shown that the parameter space of the curvature-increasing model can be reduced to three independent constants , , and . Here, influences only the timescale, controls the size of appearing morphogen patches (and scales with the system size), and influences the distance between these patches and does not depend on system size (40). Within this model, we define the symmetry breaking time tSB as the time at which a certain morphogen concentration Φ is exceeded, namely if . To fit the model-based curve of tSB(r0) to experimental data, for each value , we varied the temporal (i.e., ) and spatial scaling of the curvature-increasing model such that a minimum of the deviation between model and experimental data has been achieved. Because system size itself has been varied, has been always set to a constant.
Data analysis of the SOC model
We used the SOC model and parameters as proposed in Gamba et al. (37). The only difference is a square lattice instead of a hexagonal lattice, as originally proposed by Olami et al. (50). We numerically verified that our square lattice model qualitatively reproduces the behavior of the hexagonal model, such as size distribution of Ks1-positive expression domains at different time-steps, including criticality (results not shown). Knowing that an aggregate of size r0 = 138 μm corresponds to a network of c = 104 cells (19), we deduce the relation c = 104r02/1382 to convert the network size into the initial radius r0 of a Hydra shell.
As proposed by Gamba et al. (37), we assume that the axis of the Hydra aggregate is locked at the moment where a certain Ks1-positive cluster size is exceeded, i.e., the symmetry breaking moment tSB. Alternatively, we define tSB as the moment when Ks1 expression becomes scale free, which appears to experimentally coincide with symmetry breaking (27).
To compare the behavior of tSB(r0) between the SOC model and the experiments, we varied the temporal scaling of the SOC model as to minimize the deviations between numerical and experimental data.
Temperature gradients
In experiments with Hydra aggregates it has been shown that temperature gradients direct the orientation of the body axis and the localization of the head, which has been reproduced by simulations of the corresponding SOC model (27,37). To investigate the influence of temperature on the outcome of the curvature-increasing and Turing models, we introduce linear gradients along the z axis in each reaction or diffusion constant k within the corresponding model. Especially, we consider the values k + dT k at the upper end and k − dT k at the lower end of the aggregate. We performed for each model and each dT ∈ {0.0, 0.2, 0.5} a total of 20 simulations. Head orientation has been subsequently evaluated by calculating the angle θ between the center of the first appearing morphogen patch and 0°, the latter defining the coldest part of the aggregate. We always use projections of the three-dimensional sphere on the z,x plane.
Results and Discussion
Behavior of the OMC model
In the following, we use simulations of the OMC model to investigate osmomechanochemical mechanisms leading to symmetry breaking in Hydra aggregates.
Shape oscillations
To begin, we investigate osmotically induced shape oscillations of an aggregate before symmetry breaking by simulating the OMC model for short times, which minimizes the influence of chemical processes on the system. We show that shape oscillations generated by the OMC model agree with corresponding experimental data. We consider the oscillations of a system with initial radius r0 = 138 μm within different osmotic milieus, the latter given by ΔCeff. Corresponding simulation results fit the experimental data: Evolution of the radius r follows a sawtooth time pattern with a nearly linear swelling and frequency increasing with higher inner ion concentration (Fig. 3, A and D). Furthermore, averaging the swelling rate s over the first three cycles reveals that s depends linearly on the ion concentration difference ΔCeff (Fig. 3, B and E). Finally, changing the initial system size r0 but keeping the ion concentrations constant reveals (similarly to experiments) an almost linear dependence of s on the radius r0 in the range of 100–200 μm (Fig. 3, C and F). However, especially for larger systems, simulation results reveal a saturation effect of s(r0). In summary, numerical results suggest that our system reproduces the experimentally observed shape oscillations qualitatively and quantitatively.
Figure 3.
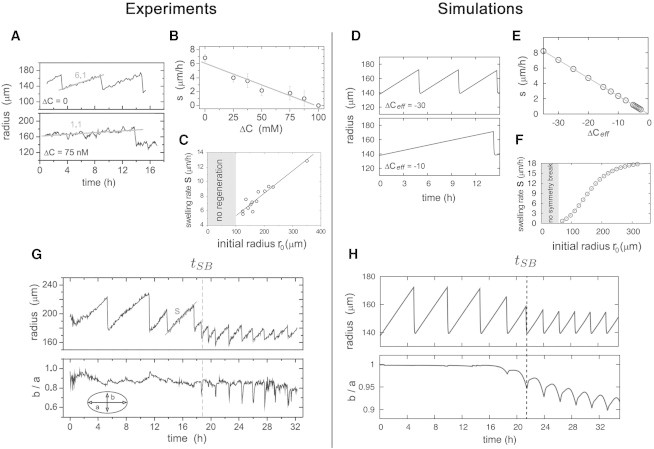
Numerical simulations and comparison with experiments considering osmotically driven shape oscillations. (A–F) Frequency and slope of sawtooth-like shape oscillations depend on both ion concentration and aggregate size. (G and H) Evolution of radius r and ratio between the minor and major inner axes b/a. Symmetry breaking time tSB coincides with low values of b/a. (A, B, and G are taken with permission from Soriano et al. (19) and C from Kücken et al. (36).) For (D) and (E), we used a system of initial size r0 = 138 μm.
Based on this, in the following, we investigate the OMC model for times sufficient for the system to pass from phase I (before symmetry breaking) to phase II (after symmetry breaking) to allow chemical processes play a noticeable role. In agreement with experiments (19,32), we observe in our simulations a relatively abrupt change in the mechanochemical properties of the system: At tSB, the sawteeth are smaller (see Fig. 3, G and H, upper panel) and the previously constant value b/a ≈ 1 starts to peak at lower values (see Fig. 3, G and H, lower panel), with the latter indicating the loss of symmetry. Furthermore, an important observation is that the times at which b/a reaches minima coincide with moments of tissue collapse in both experiments and simulations.
With the change in mechanical symmetry, the sphere also develops a chemical asymmetry at tSB: Whereas morphogens appear equally distributed before symmetry breaking (Fig. 4 A), after tSB, a distinct morphogen patch becomes apparent (Fig. 4 C). Due to the proposed feedback loop, this patch causes a local weakening of the tissue leading to both the mechanical system asymmetry (i.e., low values of b/a) and earlier tissue rupture reducing the sawteeth size. This local tissue weakening probably causes an effect shared by simulations and experimental observations (19): In phase I, tissue rupture occurs at random places whereas in phase II, rupture only occurs at the center of the morphogen patch, representing an early organizer (results not shown). Finally, simulations also show instances of anisotropic inflation-deflation in phase II, as described for aggregates by Soriano et al. (19) (Fig. 4 C, right-hand side). Interestingly, the difference with respect to experimental data is that here, an opposed motion is only restricted to one side of the aggregate. In contrast, the data of Soriano et al. (19) show this behavior at two opposite points.
Figure 4.
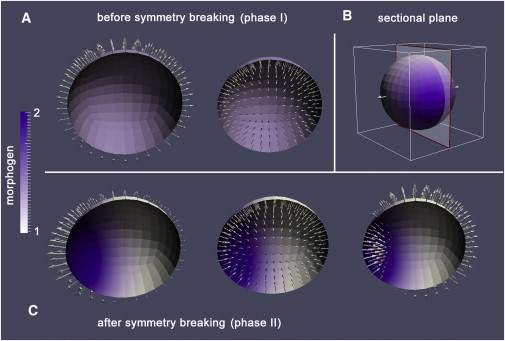
Snapshots of numerical simulations before and after symmetry breaking. (A) Symmetrical behavior before symmetry breaking changes to an asymmetrical shape and chemical patterning after symmetry breaking (C). (B) Sectional plane used for (A) and (C). (Open arrows) Direction of tissue motion; (purple color) high levels of activator morphogen ΦA. Please note that there are moments of anisotropic inflation-deflation after symmetry breaking (lower panel, right-hand side). All simulations consider the OMC model of initial size r0 = 138 μm. To see this figure in color, go online.
Symmetry breaking time
To further investigate our model, we compared simulations to experimental dependence of tSB on swelling rate s, ion concentration difference ΔCeff, and system size r0. Unexpectedly, simulations also reproduce the experimentally observed nonlinear dependence of tSB on swelling rate and ion concentration difference, respectively. This behavior has been previously ascribed to viscoelastic cell properties (19) and was not expected in a purely elastic system. Because the simulation results reproduce experiment strikingly well (Fig. 5, B and C), this behavior must be linked to shape oscillations rather than to rheological cell properties.
Figure 5.
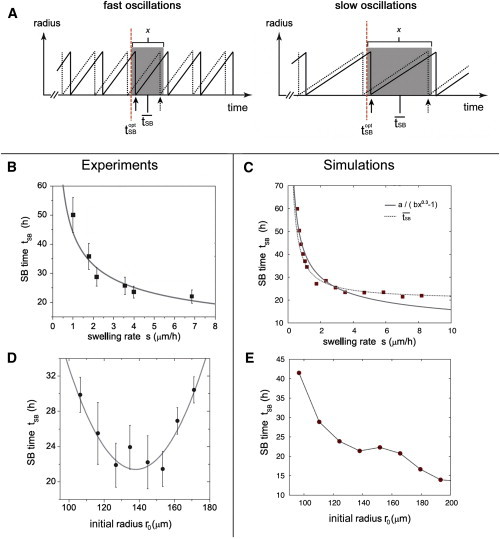
Scheme, simulations, and experiments considering symmetry breaking time tSB. (A) Schematic dependence of tSB on shape oscillations. The molecular mechanism of symmetry breaking allows tSB to occur earliest at . Solid (dashed) lines represent sawteeth with tSB close to (away from) (solid and dashed arrows indicate corresponding moments of tSB, respectively). Delays of tSB are caused by the fact that tSB always coincides with the moment of largest tissue stretch (upper tip of sawteeth). Thus, tSB happens always within a certain interval (shaded in dark gray) with length x and mean value , the latter depending on , swelling rate s, and radius r. (C and D) The tSB value depending on swelling rate s in experiments (B) and our simulations (C). The curve (dashed line in C) fits tSB(s) better than the function a/(bx0.3 – 1) (solid lines in B and C), the latter previously ascribed to viscoelastic cell properties (19). (D and E) Experimentally determined dependence of tSB on system radius r differs distinctly between experiments (D) and numerical data (E), the latter based on Turing-like patterning. (B) and (D) are taken with permission from Soriano et al. (19).) To see this figure in color, go online.
We propose that, instead, a simple delaying impact of shape oscillations on symmetry breaking time tSB is the cause for this behavior (see Fig. 5 A): As we see in Fig. 3, G and H, tSB coincides with moments of maximal tissue stretch, i.e., tissue rupture at upper tip of a sawtooth. This means that even if the system could establish symmetry breaking at time , the actual symmetry breaking time tSB is delayed, depending on the moment when the next tissue rupture occurs for a . Hence, tSB occurs within the interval , where x is the time of an oscillation (see Fig. 5 A). The exact location of may differ for different aggregates due to individual differences; however, the mean value is given by . Trigonometry provides that , where ω = rrupt – r0, rrupt is the radius at time trupt and s is the swelling rate. Based on Fig. 5 C, we roughly estimate considering s → ∞. As we can see in Fig. 5 C, (dashed line) approximates tSB(s) better than the function a/(bx0.3 − 1) previously proposed in Soriano et al. (19) (solid lines in Fig. 5, B and C). Hence, a purely elastic tissue model appears to be sufficient to describe the main aspects of Hydra tissue behavior.
Which morphogen patterning mechanism is involved?
So far, all presented simulation results are partly independent of the chemical aspect of symmetry breaking and pattern formation: Until now only the fact that tissue stretch causes morphogen patterning was required, but not what this dependence and the patterning mechanism looks like in detail.
To understand the role of chemistry in the process of symmetry breaking, we considered the dependence of tSB on the initial aggregate radius r0. Here, the simulation results for the OMC model strongly differ from the experimental results (Fig. 5, D and E). As a next step, we compared experimental data (Fig. 5 D) to the results for the Turing-, SOC-, and curvature-increasing models. These purely chemical models have been investigated in systems without shape oscillations. In contrast, the experimental results depicted in Fig. 5 D are the result of both chemical and mechanical effects influencing tSB. This means that, in a direct comparison, we assume 1) that the delaying impact of shape oscillations is negligible in the considered range of r0; and that 2) parameters depending on periodic tissue stretch can be represented by constant values, i.e., we assume that a tissue constantly stretched at a medium level behaves approximately like the oscillating system.
The first assumption can be easily justified by pointing out that the above-mentioned delaying effect is only visible for swelling rates x ≤ 4 μm/h (Fig. 5, B and C), which corresponds (within the ion concentration regime used for Fig. 5 D) to aggregate sizes <100 μm (see Fig. 3 G), i.e., below the range considered in in Fig. 5 D. Regarding the second assumption, we cannot exclude that the oscillating system produces (beside the delaying impact) other effects we cannot describe using constant parameters. One possibility: finite size synchronization effects between oscillating mechanical and chemical processes (54) that may cause fluctuations in Ks1-expression depend on the sphere size in a nontrivial way. Nevertheless, we assume that the following studies present the main characteristics of the three investigated pattern formation models, and we postpone coupling them to shape oscillations to future research.
Turing patterning
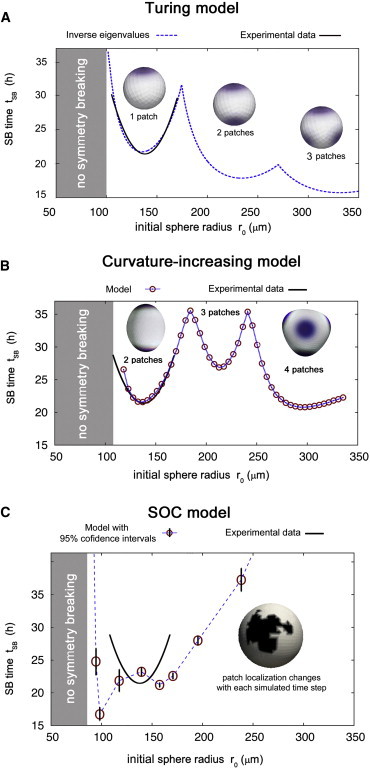
Although we found several different sets of parameters for Turing systems of two and three equations approximating the experimental data provided in Fig. 5 D, they were all indistinguishable to the naked eye. One example is given in Fig. 6 A. As we can see, the Turing mechanism fits experimental data quite well, including the loss of symmetry breaking below sizes of r0 < 100 μm. For larger systems, our numerical data predict an acceleration of symmetry breaking with characteristic upward peaks of tSB at each system size where an additional morphogen patch appears. However, to our knowledge, corresponding experimental data are not available so far. Hence, the experimental investigation of tSB for larger (multiple head) aggregates could serve as a key step to validate an underlying DDI mechanism in Hydra aggregates. In contrast to experimental observations (27), simulations of the Turing model do not show dependence of axis orientation on temperature gradients (Fig. 7).
Figure 6.
Different models considering symmetry breaking time tSB depending on system size. (A) Turing model. (B) Curvature-increasing model. (C) SOC model. (Black solid lines) Approximated experimental data taken from Soriano et al. (19). (Dashed blue lines and red points) Model data. (Gray shaded) Regions without symmetry breaking. To see this figure in color, go online.
Figure 7.
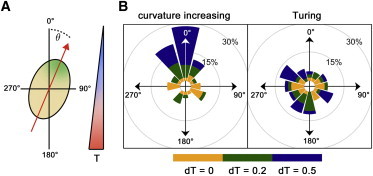
Angular orientation of the Hydra aggregate head orientation within a temperature gradient, simulated for different patterning mechanisms. (A) Schematic view. (B) Rosette-diagram of the curvature-increasing versus the Turing model. 0° corresponds to the cold side. To see this figure in color, go online.
The morphogen Wnt3 fulfills properties of the activator molecule: It appears early during head formation (26); is strongly restricted to the organizer region (25); and its regulation contains elements for an autoregulatory feedback (via the molecules Tcf and β-catenin) and a repressor (31)—as required within Eq. 8. However, a possible molecular candidate for the long-range inhibitor is still missing, and for both Wnt3-activator and -inhibitor molecules, various binding sites have been identified (31). Furthermore, it has been recently proposed that Wnt molecules may act as both activators and, after processing, inhibitors (24).
Tissue curvature-driven patterning
As depicted in Fig. 6 B, the curvature-increasing mechanism fits experimental data, including the loss of symmetry breaking for small aggregates. However, the behavior of tSB for larger aggregates differs distinctly from that of a Turing-driven mechanism: Above r0 ≈ 110 μm, symmetry breaking occurs, starting with two patches for 110 μm ⪅ r0 ⪅ 180 μm, three patches for 180 μm ⪅ r0 ⪅ 240 μm, and four patches for r0 ⪆ 240 μm. Hence, in this model, the smallest possible number of morphogen patches is 2, in contrast to one organizer appearing experimentally in small aggregates. However, two patches still define one body axis. Additional mechanisms are required leading to the selection and dominance of one organizer. In accordance with experimental data (27,37), simulations of the curvature-increasing model reveal that the orientation of the body axis depends critically on temperature gradients: Defining the first of the two (40) appearing morphogen patches as the head, we find that weak temperature gradients (dT = 0.2) result in preferred head formation at the cold (θ ≈ 0°) or the warm (θ ≈ 180°) site, while stronger gradients (dT = 0.5) lead to heads only at the cold side (Fig. 7 B).
Wnt3 molecules appear early during head formation and are restricted to outwardly curved tissue regions, such as head and budding zones (25,26). Thus, one of the various binding sites regulating Wnt3-expression (31) could be influenced by local tissue deformations, finally stimulating Wnt3-expression in evaginated tissue regions. In turn, it has been experimentally shown that Wnt-signaling induces local tissue evaginations in Hydra (35), which could constitute the other part of the curvature-driven feedback loop. In contrast to Turing models, we do not need the existence of a molecular long-range inhibitor, because this role is played by regions where the tissue is not outwardly curved (40).
To obtain a more complete model, such curvature-driven morphogen patterning might need additional coupling to osmomechanical processes, as assumed for Turing-driven patterning within the OMC model (Fig. 1). The morphogen could again lower the resistance to lateral tissue stretch if the production of this morphogen depends on tissue curvature as well as tissue stretch (Fig. 2). Thus, tissue curvature and stress could jointly influence morphogen patterning, leading to symmetry breaking only in case of periodic shape oscillations. Numerical investigation of such a coupling is postponed to future research.
SOC-driven patterning
We tested different possible mechanisms defining symmetry breaking in the context of the SOC model. Assuming that the exceedance of a critical (relative) Ks1-positive cluster size, , locks the head localization, the dependence of tSB on system size shows neither a parabolic shape nor a loss of symmetry breaking for small systems, but changes monotonously. The same holds true if we define the moment of scale-free Ks1 expression as the moment of symmetry breaking (results not shown). However, if we assume that a critical absolute Ks1-positive cluster size defines the axis, the shape of tSB(r0) is qualitatively similar to experimental results: for small aggregates, we do not observe symmetry breaking whereas for larger systems, tSB shows an almost parabolic shape (Fig. 6 C). In contrast to the Turing and the curvature-induced mechanism (Fig. 6, A and B), here, tSB increases monotonously for larger systems with 150 μm ⪅ r0 ⪅ 350 μm. We find the best match to experimental data for , which is of the same order as the critical sizes estimated in Gamba et al. (37). Although the agreement of these simulations with experimental data is not as good as in the case of the Turing or the curvature-increasing model, the exact shape of tSB(r0) could be influenced by other parameters of the SOC model, such as neighbor numbers or production rates, which have not been varied in our studies.
As mentioned above, the SOC mechanism reproduces the dependence of head localization on weak and strong temperature gradients, as previously shown in Soriano et al. (27) and Gamba et al. (37).
Assuming that Ks1-patterning defines the head localization, it might act upstream of morphogen expression patterns related to the organizer, such as Wnt3 (25,26). Although a direct interaction between Ks1 and Wnt3-regulatory elements probably does not take place (31), interestingly, regulatory elements of Ks1 and Wnt3 were both identified to specifically bind the same type of molecules—nuclear proteins from the basal tissue (29,31).
Similarly to the Turing- or the curvature-driven patterning, a complete model of symmetry breaking might involve the coupling of the SOC mechanism to OMC shape oscillations of the Hydra aggregate (Fig. 1). A natural coupling involves Ks1-expressing cells lowering the resistance to lateral tissue stretch, and in turn, tissue stretch accelerating protein production and/or discharge processes (Fig. 2).
Conclusions
In this article we model interplay among osmotic, chemical, and mechanical processes leading to symmetry breaking in Hydra aggregates. Simulations demonstrate that a simple feedback loop between tissue mechanics and morphogen patterning can qualitatively and quantitatively reproduce a wide range of experimental data. In particular, we assume that tissue stretch induces morphogen clustering, and morphogen lowers the resistance to lateral stretching. To elucidate the chemical part of this mechanochemical interplay, we compared previously proposed morphogen patterning models, namely the Turing, SOC, and curvature-increasing models. Comparison to experimental data neither strongly supports nor excludes any of these mechanisms. We showed that the dependence of symmetry breaking time tSB on system size r0 distinctly differs among these three mechanisms for aggregates with r0 ⪆ 180 μm. Thus, we suggest experimental investigations of larger aggregates as a key step to gain further insights into the molecular mechanisms of Hydra symmetry breaking.
Author Contributions
M.M., A.K., and A.M.-C. designed research; M.M. developed mathematical models; M.M. and A.K. performed research, developed numerical tools, and analyzed data; and M.M., A.M.-C., and A.K. wrote the article.
Acknowledgments
M.M. and A.M.-C. have been supported by the European Research Council Starting Grant Biostruct (No. 210680) and the Emmy Noether Program of the Deutsche Forschungsgemeinschaft.
Editor: Rong Li.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supporting Material
Supporting Citations
References (55,56) appear in the Supporting Material.
References
- 1.Gilbert S. Palgrave Macmillan; New York: 2013. Developmental Biology. [Google Scholar]
- 2.Harold F.M. From morphogenes to morphogenesis. Microbiology. 1995;141:2765–2778. doi: 10.1099/13500872-141-11-2765. [DOI] [PubMed] [Google Scholar]
- 3.Meinhardt H. Academic Press; London, UK: 1982. Models of Biological Pattern Formation. [Google Scholar]
- 4.Turing A.M. The chemical basis of morphogenesis. Philos. Trans. R. Soc. Lond. B. 1953;237:37–72. [Google Scholar]
- 5.Gjorevski N., Nelson C.M. The mechanics of development: models and methods for tissue morphogenesis. Birth Defects Res. C Embryo Today. 2010;90:193–202. doi: 10.1002/bdrc.20185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beloussov L.V. Mechanically based generative laws of morphogenesis. Phys. Biol. 2008;5:015009. doi: 10.1088/1478-3975/5/1/015009. [DOI] [PubMed] [Google Scholar]
- 7.Beloussov L.V., Grabovsky V.I. A geometro-mechanical model for pulsatile morphogenesis. Comput. Methods Biomech. Biomed. Eng. 2003;6:53–63. doi: 10.1080/1025584021000047641. [DOI] [PubMed] [Google Scholar]
- 8.Chanet S., Martin A.C. Mechanical force sensing in tissues. Prog. Mol. Biol. Transl. Sci. 2014;126:317–352. doi: 10.1016/B978-0-12-394624-9.00013-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farge E. Mechanotransduction in development. Curr. Top. Dev. Biol. 2011;95:243–265. doi: 10.1016/B978-0-12-385065-2.00008-6. [DOI] [PubMed] [Google Scholar]
- 10.Mercker M., Marciniak-Czochra A., Hartmann D. Modeling and computing of deformation dynamics of inhomogeneous biological surfaces. SIAM J. Appl. Math. 2013;73:1768–1792. [Google Scholar]
- 11.Brouzés E., Farge E. Interplay of mechanical deformation and patterned gene expression in developing embryos. Curr. Opin. Genet. Dev. 2004;14:367–374. doi: 10.1016/j.gde.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 12.Gierer A., Meinhardt H. A theory of biological pattern formation. Kybernetik. 1972;12:30–39. doi: 10.1007/BF00289234. [DOI] [PubMed] [Google Scholar]
- 13.Bode H.R. The head organizer in Hydra. Int. J. Dev. Biol. 2012;56:473–478. doi: 10.1387/ijdb.113448hb. [DOI] [PubMed] [Google Scholar]
- 14.Gierer A., Berking S., Trenkner E. Regeneration of Hydra from reaggregated cells. Nat. New Biol. 1972;239:98–101. doi: 10.1038/newbio239098a0. [DOI] [PubMed] [Google Scholar]
- 15.Lü S.H., Lin Q., Wang C.Y. Self-assembly of renal cells into engineered renal tissues in collagen/Matrigel scaffold in vitro. J. Tissue Eng. Regen. Med. 2012;6:786–792. doi: 10.1002/term.484. [DOI] [PubMed] [Google Scholar]
- 16.Junttila S., Saarela U., Vainio S. Functional genetic targeting of embryonic kidney progenitor cells ex vivo. J. Am. Soc. Nephrol. 2014 doi: 10.1681/ASN.2013060584. Published online September 8, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Urdy S. On the evolution of morphogenetic models: mechano-chemical interactions and an integrated view of cell differentiation, growth, pattern formation and morphogenesis. Biol. Rev. Camb. Philos. Soc. 2012;87:786–803. doi: 10.1111/j.1469-185X.2012.00221.x. [DOI] [PubMed] [Google Scholar]
- 18.Wyczalkowski M.A., Chen Z., Taber L.A. Computational models for mechanics of morphogenesis. Birth Defects Res. C. Embryo Today. 2012;96:132–152. doi: 10.1002/bdrc.21013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soriano J., Rüdiger S., Ott A. Mechanogenetic coupling of Hydra symmetry breaking and driven Turing instability model. Biophys. J. 2009;96:1649–1660. doi: 10.1016/j.bpj.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allena R., Muñoz J.J., Aubry D. Diffusion-reaction model for Drosophila embryo development. Comput. Methods Biomech. Biomed. Eng. 2013;16:235–248. doi: 10.1080/10255842.2011.616944. [DOI] [PubMed] [Google Scholar]
- 21.Grimmelikhuijzen C., Schaller H. Hydra as a model organism for the study of morphogenesis. Trends Biochem. Sci. 1979;4:265–267. [Google Scholar]
- 22.Galliot B. Hydra, a fruitful model system for 270 years. Int. J. Dev. Biol. 2012;56:411–423. doi: 10.1387/ijdb.120086bg. [DOI] [PubMed] [Google Scholar]
- 23.Meinhardt H., Gierer A. Pattern formation by local self-activation and lateral inhibition. BioEssays. 2000;22:753–760. doi: 10.1002/1521-1878(200008)22:8<753::AID-BIES9>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 24.Meinhardt H. Modeling pattern formation in Hydra: a route to understanding essential steps in development. Int. J. Dev. Biol. 2012;56:447–462. doi: 10.1387/ijdb.113483hm. [DOI] [PubMed] [Google Scholar]
- 25.Hobmayer B., Rentzsch F., Holstein T.W. WNT signaling molecules act in axis formation in the diploblastic metazoan Hydra. Nature. 2000;407:186–189. doi: 10.1038/35025063. [DOI] [PubMed] [Google Scholar]
- 26.Lengfeld T., Watanabe H., Holstein T.W. Multiple Wnts are involved in Hydra organizer formation and regeneration. Dev. Biol. 2009;330:186–199. doi: 10.1016/j.ydbio.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 27.Soriano J., Colombo C., Ott A. Hydra molecular network reaches criticality at the symmetry-breaking axis-defining moment. Phys. Rev. Lett. 2006;97:258102. doi: 10.1103/PhysRevLett.97.258102. [DOI] [PubMed] [Google Scholar]
- 28.Weinziger R., Salgado L.M., Bosch T.C. Ks1, an epithelial cell-specific gene, responds to early signals of head formation in Hydra. Development. 1994;120:2511–2517. doi: 10.1242/dev.120.9.2511. [DOI] [PubMed] [Google Scholar]
- 29.Endl I., Lohmann J.U., Bosch T.C.G. Head-specific gene expression in Hydra: complexity of DNA-protein interactions at the promoter of ks1 is inversely correlated to the head activation potential. Proc. Natl. Acad. Sci. USA. 1999;96:1445–1450. doi: 10.1073/pnas.96.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bode H.R. Axial patterning in hydra. Cold Spring Harb. Perspect. Biol. 2009;1:a000463. doi: 10.1101/cshperspect.a000463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamura Y., Tsiairis C.D., Holstein T.W. Autoregulatory and repressive inputs localize Hydra Wnt3 to the head organizer. Proc. Natl. Acad. Sci. USA. 2011;108:9137–9142. doi: 10.1073/pnas.1018109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fütterer C., Colombo C., Ott A. Morphogenetic oscillations during symmetry breaking of regenerating Hydra vulgaris cells. Europhys. Lett. 2003;64:137–143. [Google Scholar]
- 33.Bonazzi D., Julien J.-D., Minc N. Symmetry breaking in spore germination relies on an interplay between polar cap stability and spore wall mechanics. Dev. Cell. 2014;28:534–546. doi: 10.1016/j.devcel.2014.01.023. [DOI] [PubMed] [Google Scholar]
- 34.Hiscock T.W., Megason S.G. Mathematically guided approaches to distinguish models of periodic patterning. Development. 2015;142:409–419. doi: 10.1242/dev.107441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Philipp I., Aufschnaiter R., Hobmayer B. Wnt/β-catenin and noncanonical Wnt signaling interact in tissue evagination in the simple eumetazoan Hydra. Proc. Natl. Acad. Sci. USA. 2009;106:4290–4295. doi: 10.1073/pnas.0812847106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kücken M., Soriano J., Nicola E.M. An osmoregulatory basis for shape oscillations in regenerating Hydra. Biophys. J. 2008;95:978–985. doi: 10.1529/biophysj.107.117655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gamba A., Nicodemi M., Ott A. Critical behavior and axis defining symmetry breaking in Hydra embryonic development. Phys. Rev. Lett. 2012;108:158103. doi: 10.1103/PhysRevLett.108.158103. [DOI] [PubMed] [Google Scholar]
- 38.Krahe M., Wenzel I., Fütterer C. Fluctuations and differential contraction during regeneration of Hydra vulgaris tissue toroids. New J. Phys. 2013;15:035004. [Google Scholar]
- 39.Mombach J., Almeida R., Glazier J. Bursts and cavity formation in Hydra cells aggregates: experiments and simulations. Physica A. 2008;297:495–508. [Google Scholar]
- 40.Mercker M., Hartmann D., Marciniak-Czochra A. A mechanochemical model for embryonic pattern formation: coupling tissue mechanics and morphogen expression. PLoS ONE. 2013;8:e82617. doi: 10.1371/journal.pone.0082617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janssen, M. 2013. Ein kontinuierliches dynamisches Modell der räumlichen entwicklung von Gewebe. Diploma thesis. University of Heidelberg, Heidelberg, Germany.
- 42.Helfrich W. Elastic properties of lipid bilayers: theory and possible experiments. Z. Naturforsch. C. 1973;28:693–703. doi: 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- 43.Taniguchi T. Shape deformation and phase separation dynamics of two-component vesicles. Phys. Rev. Lett. 1996;76:4444–4447. doi: 10.1103/PhysRevLett.76.4444. [DOI] [PubMed] [Google Scholar]
- 44.Li J., Zhang H., Yang Y. A discrete, space variation model for studying the kinetics of shape deformation of vesicles coupled with phase separation. Macromol. Theory Sim. 2006;15:432–439. [Google Scholar]
- 45.Mercker M., Richter T., Hartmann D. Sorting mechanisms and communication in phase-separating coupled monolayers. J. Phys. Chem. B. 2011;115:11739–11745. doi: 10.1021/jp204127g. [DOI] [PubMed] [Google Scholar]
- 46.Koch A., Meinhardt H. Biological pattern formation: from basic mechanisms to complex structures. Rev. Mod. Phys. 1994;66:1481–1507. [Google Scholar]
- 47.Plaza R., Sanchez-Garduno F., Maini P. The effect of growth and curvature on pattern formation. J. Dyn. Differ. Equ. 2004;16:1093–1121. [Google Scholar]
- 48.Meinhardt H. Models of biological pattern formation: from elementary steps to the organization of embryonic axes. Curr. Top. Dev. Biol. 2008;81:1–63. doi: 10.1016/S0070-2153(07)81001-5. [DOI] [PubMed] [Google Scholar]
- 49.Varea C., Aragón J.L., Barrio R.A. Turing patterns on a sphere. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Top. 1999;60(4 Pt B):4588–4592. doi: 10.1103/physreve.60.4588. [DOI] [PubMed] [Google Scholar]
- 50.Olami Z., Feder H.J., Christensen K. Self-organized criticality in a continuous, nonconservative cellular automaton modeling earthquakes. Phys. Rev. Lett. 1992;68:1244–1247. doi: 10.1103/PhysRevLett.68.1244. [DOI] [PubMed] [Google Scholar]
- 51.Becker, R., M. Braack, …, B. Vexler. 2005. GASCOIGNE 3D—a finite element toolbox. http://www.gascoigne.uni-hd.de.
- 52.Gantmacher F.R. Vol. 2. Chelsea; New York: 1959. (The Theory of Matrices). [Google Scholar]
- 53.Satnoianu R.A., Menzinger M., Maini P.K. Turing instabilities in general systems. J. Math. Biol. 2000;41:493–512. doi: 10.1007/s002850000056. [DOI] [PubMed] [Google Scholar]
- 54.Kuramoto Y. Springer; Berlin, Germany: 1984. Chemical Oscillators, Waves and Turbulence. [Google Scholar]
- 55.Jensen H. Cambridge Lecture Notes in Physics. Cambridge University Press; Cambridge, UK: 1998. Self-organized criticality. [Google Scholar]
- 56.Zhong-Can O., Helfrich W. Bending energy of vesicle membranes: general expressions for the first, second, and third variation of the shape energy and applications to spheres and cylinders. Phys. Rev. A. 1989;39:5280–5288. doi: 10.1103/physreva.39.5280. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


