Abstract
Mammalian mitochondrial DNA (mtDNA) is a high-copy maternally inherited genome essential for aerobic energy metabolism. Mutations in mtDNA can lead to heteroplasmy, the co-occurence of two different mtDNA variants in the same cell, which can segregate in a tissue-specific manner affecting the onset and severity of mitochondrial dysfunction. To investigate mechanisms regulating mtDNA segregation we use a heteroplasmic mouse model with two polymorphic neutral mtDNA haplotypes (NZB and BALB) that displays tissue-specific and age-dependent selection for mtDNA haplotypes. In the hematopoietic compartment there is selection for the BALB mtDNA haplotype, a phenotype that can be modified by allelic variants of Gimap3. Gimap3 is a tail-anchored member of the GTPase of the immunity-associated protein (Gimap) family of protein scaffolds important for leukocyte development and survival. Here we show how the expression of two murine Gimap3 alleles from Mus musculus domesticus and M. m. castaneus differentially affect mtDNA segregation. The castaneus allele has incorporated a uORF (upstream open reading frame) in-frame with the Gimap3 mRNA that impairs translation and imparts a negative effect on the steady-state protein abundance. We found that quantitative changes in the expression of Gimap3 and the paralogue Gimap5, which encodes a lysosomal protein, affect mtDNA segregation in the mouse hematopoietic tissues. We also show that Gimap3 localizes to the endoplasmic reticulum and not mitochondria as previously reported. Collectively these data show that the abundance of protein scaffolds on the endoplasmic reticulum and lysosomes are important to the segregation of the mitochondrial genome in the mouse hematopoietic compartment.
Keywords: mitochondria, mitochondrial DNA, mice, segregation, Gimap
MAMMALIAN mitochondrial DNA (mtDNA) is a maternally inherited small circular multicopy genome that encodes 13 proteins that are essential subunits of four of the five complexes required for mitochondrial oxidative phosphorylation. Germline or somatic-cell mtDNA mutations lead to the co-occurrence of two or more sequence variants in a cell, a state known as heteroplasmy. In the absence of selection, the segregation of mtDNA sequence variants is neutral and can be modeled as a random walk (Chinnery and Samuels 1999); however, in some cases there is preferential selection for a mtDNA sequence variant that is dependent upon the nucleotide sequence, tissue, and nuclear background (Battersby and Shoubridge 2001; Battersby et al. 2003, 2005; Jokinen and Battersby 2013; Burgstaller et al. 2014). The majority of pathogenic mtDNA mutations are heteroplasmic and some mutations display skewed segregation patterns in somatic tissues. (Larsson et al. 1990; Boulet et al. 1992; Kawakami et al. 1994; Dunbar et al. 1995; Fu et al. 1996; Chinnery et al. 1997, 1999; Weber et al. 1997), which can affect the onset and severity of mitochondrial dysfunction. Currently, the molecular basis for this regulation of the mitochondrial genome is largely unknown (Jokinen and Battersby 2013).
To study mtDNA segregation in mammals we use a heteroplasmic mouse model with two neutral mtDNA variants derived from two old inbred mouse strains, BALB/c and NZB (Jenuth et al. 1996, 1997). These mtDNA haplotypes, referred to as BALB and NZB, differ at 90 nucleotide positions (Hagstrom et al. 2014) and have been stably transmitted through the female germline of this mouse model for ∼20 years (Jenuth et al. 1996). There is no selection for either mtDNA haplotype during transmission as the heteroplasmy level in the offspring follows a Gaussian distribution (Jenuth et al. 1996; Wai et al. 2008). However, postnatally there is age-dependent selection of one haplotype over the other in three tissue types (liver, kidney, and hematopoietic tissues), while all other tissues in these mice are neutral with respect to mtDNA segregation (Jenuth et al. 1997). In the liver and kidney there is selection for the NZB haplotype to fixation (Jenuth et al. 1997; Battersby and Shoubridge 2001; Battersby et al. 2003, 2005). In contrast, the hematopoietic tissues (bone marrow, blood, thymus, and spleen) select against the NZB mtDNA haplotype, which can be modeled as an exponential decay (Battersby et al. 2005). This tissue-specific mtDNA segregation is best treated as a quantitative genetic phenotype and one that cannot be explained by detectable functional differences in the mitochondrial respiratory chain or by differential rates in mtDNA replication (Battersby and Shoubridge 2001).
To uncover the molecular basis of this mtDNA regulation we have used forward genetic approaches in mice. On the nuclear background of M. m. domesticus in several different mouse strains (BALB/c, C3H, C57BL/6J, DBA, NZB, and 129Sv) there are no differences in these tissue-specific mtDNA segregation phenotypes (Battersby et al. 2003, 2005). In contrast, crosses onto a M. m. castaneus nuclear background have a significant effect on these mtDNA segregation phenotypes (Battersby et al. 2003, 2005). This allowed us to identify three nuclear loci that affect mtDNA segregation in a tissue-specific manner (Battersby et al. 2003). At one of these loci we have successfully cloned Gimap3 (Jokinen et al. 2010), which can modulate the segregation of mtDNA in leukocytes.
GTPase of the immunity-associated proteins (Gimap) are encoded in a conserved cluster of seven to eight genes found only in vertebrates with an ortholog in plants (Krucken et al. 2004). In mammals, Gimap expression appears to be restricted to hematopoietic tissues and is important for leukocyte development and survival, although the molecular basis for these functions is poorly understood (Krucken et al. 2004; Nitta and Takahama 2007; Schulteis et al. 2008; Barnes et al. 2010; Chen et al. 2011). Gimaps can be tail anchored or soluble and are structurally related to Septins, Tocs, and Dynamins (Schwefel et al. 2010). Based on structural studies the membrane anchored Gimaps form GTP-dependent homo-oligomers with low inherent GTP hydrolysis activity that stabilizes these oligomers allowing them to act as scaffolds (Schwefel et al. 2010, 2013).
Gimap3 and Gimap5 are paralogues in this gene cluster and 84% identical at the amino acid level, differing only at the N and C termini (Krucken et al. 2004). Both have a short transmembrane domain at the C terminus for insertion into a lipid bilayer. The intracellular localization of Gimap5 has been controversial with data reporting insertion into many different organelles. However, data using species-specific monoclonal antibodies against Gimap5 in T cells robustly demonstrate that in humans, mice, and rats the protein is anchored into the lysosomal membrane (Wong et al. 2010). So far there is only one report for the localization of Gimap3 suggesting it is mitochondrial (Daheron et al. 2001). In mice, both genes are expressed and appear to be important for T-cell development (Nitta et al. 2006) and possibly in a cooperative way (Yano et al. 2014). In contrast, GIMAP3 in humans appears to be a pseudogene (Krucken et al. 2004). Complete loss of Gimap5 function in mice appears to be catastrophic for the hematopoietic compartment, producing a decrease of lineage-committed hematopoietic progenitors leading to a reduction of T and B lymphocytes, NK and NK T cells, altered erythropoiesis, and early lethality (Schulteis et al. 2008; Barnes et al. 2010; Chen et al. 2011). In rats the loss of Gimap5 function is milder, as a premature termination in Gimap5 of the BioBreeding rat results only in a T-cell lymphopenia, which is a susceptibility factor for autoimmunity in this diabetic rodent model (Hornum et al. 2002; MacMurray et al. 2002). Both Gimap3 and Gimap5 have been shown to interact with Bcl2 family members (Nitta et al. 2006; Chen et al. 2011), although the function of these interactions on membrane surfaces has yet to be elucidated.
Here, we investigated in more detail the basis by which Gimap3 is important for mtDNA segregation in the mouse hematopoietic compartment. We show that the tail-anchor sequence of Gimap3 localizes the protein to the endoplasmic reticulum (ER) and not the mitochondrial outer membrane as previously reported. The steady-state abundance of Gimap3 in leukocytes appears to be a critical factor for the regulation of mtDNA segregation in mouse hematopoietic tissues. Our data also implicate a role for the paralogue Gimap5 in mtDNA segregation. Together our findings demonstrate that the abundance of two membrane-bound protein scaffolds on the endoplasmic reticulum and lysosomes are important modulators to the segregation of the mitochondrial genome in mouse leukocytes.
Materials and Methods
Animals
The Regional State Administrative Agency of Southern Finland (ESAVI) approved all mouse work. The Gimap5tm1Wlr knockout mouse and the heteroplasmic mice were maintained on the BALB/c nuclear background and housed under standard conditions applied by the University of Helsinki Laboratory Animal Center. The animals in experiments were housed and sampled in random groups. Both male and female mice were used in the analyses.
Cell culture
Cells (murine embryonic fibroblasts, COS7, NIH3T3) were cultured under standard conditions (DMEM, 10% fetal bovine serum, glutamax). Splenocytes were freshly isolated from BALB/c mice (Battersby et al. 2005). MG132 (20 μM) (Sigma) was dissolved in DMSO.
cDNAs, plasmids, and retroviral expression
Full-length cDNAs (BALB Gimap3, BALB Gimap5, CAST Gimap3, BALB Gimap3-omp25, YFP-Gimap3, YFP-(263-301) Gimap3, RFP-Gimap3, pGFP-omp25 (Nemoto and De Camilli 1999) (gift from Hans Spelbrink) were cloned into Gateway (Invitrogen) converted retroviral expression vectors (pBABE-puro, pMYS-IRES-Neo, or pMXs-IRES-Blasticidin) or pcDNA3.1 for transient transfections. Retroviral vectors were transfected (Jetprime, Polyplus) into the Phoenix amphotropic packaging line for virus production to infect recipient cells (MEFs and COS-7). Following antibiotic selection, cells were used in experiments. For transient expression, plasmids were transfected (Jetprime, Polyplus) into COS-7 and HEK293 cells. The GFP–Sec61β plasmid was a gift from Benoit Kornmann).
Immunoblotting and antibodies
Cells or tissue samples were solubilized in phosphate buffered saline, 1% dodecyl-maltoside, 1 mM PMSF (phenylmethylsulfonyl fluoride), complete protease inhibitor (Roche). Protein concentrations were measured by the Bradford assay (Bio-Rad). Equal amounts of proteins were separated by Tris-glycine SDS–PAGE and transferred to nitrocellulose by semidry transfer. Primary antibodies were incubated overnight at +4° and detected the following day with secondary HRP conjugates (Jackson ImmunoResearch) using enhanced chemiluminescence (Cell Signaling) with film or a Bio-Rad chemi-doc imaging station. Gimap3 antibodies were generated in rabbits with the following peptides: BALB variant, amino acids 2–16 (ETLQNVVTGGKKGGC) and amino acids 259–272 (C-EGSWVLKVLPIGKK) and CAST variant, amino acids 32–45 (RIPVYTTDHLRCPDS). Sources for commercial antibodies: Chromotek (anti-GFP); Mitosciences (Sdha); Millipore (Porin); Proteintech Group (Atp5B, Calnexin, and RPL18a); Sigma (anti-HA and Atg5), Santa Cruz (Tom40). Representative immunoblotting data from multiple independent biological experiments were cropped in Photoshop with only linear corrections applied. When quantified, Western blots were analyzed with Bio-Rad chemi-doc imaging station and Image Lab software.
In vitro protein synthesis
Proteins were synthesized with TNT T7-coupled reticulocyte lysate system (Promega) according to manufacturer’s instructions using plasmids with Gimap3 or YFP cDNA constructs cloned into pcDNA3.1. Each reaction also included Superase RNAse inhibitor (Ambion) and [35S]Met-Cys (EasyTag, Perkin Elmer) and incubated for 25 min at 30°. The reaction time was chosen based upon linear radiolabel incorporation for YFP (Figure S2). Equal aliquots from each reaction mix were separated by SDS–PAGE with the gel dried and exposed to a Phosphoscreen for scanning with a Typhoon 9400 (GE Healthcare) for quantification.
Histodenz gradients
Mouse spleens were homogenized in buffer (200 mM mannitol, 70 mM sucrose, 10 mM HEPES, 1 mM EGTA pH 7.5) with a Teflon pestle and then centrifuged twice at 600 × g for 10 min. The post-nuclear suspension was mixed with an equal volume of 50% Histodenz (Sigma) (10 mM HEPES, 1 mM EGTA pH 7.5). A discontinuous Histodenz gradient was then formed (10, 15, 20, 25, and 30%) to separate organelles (Okado-Matsumoto and Fridovich 2001). Samples were centrifuged in a SW32.1Ti rotor for 90 min at 52 000 × g at +4°. Fractions were collected from the top, precipitated with TCA, and then resuspended in 1× Laemmli buffer. Samples were then subjected to SDS–PAGE and immunoblotting.
Sucrose gradient centrifugation
HEK293 cells from a 150-mm plate were treated with 100 μg/ml cycloheximide upon harvesting and lysed in 450 μl of lysis buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 100 μg/ml cycloheximide, 1% Triton X-100) with Superase RNAse inhibitor (Ambion) for 20 min and centrifuged for 5 min at 13,000 × g at 4°. The supernatant was loaded on a 16-ml linear 10–50% sucrose gradient (20 mM HEPES pH 7.6, 100 mM KCl, 5 mM MgCl2) and centrifuged for 200 min at 28,000 rpm at 4° (Beckman, SW 32.1 Ti). Eighty-two equal volume (200 μl) fractions were collected from the top. A total of 100 μl of each fraction was used for measuring RNA absorbance at 254 nm. The remaining fractions were pooled, combining five consecutive fractions, for TCA precipitation, heated at 95° for 5 min and separated by 12% SDS–PAGE for immunoblotting.
Mitochondrial DNA analysis
All sampled mouse tissues were immediately frozen in liquid nitrogen and stored at −80°. Total DNA was extracted with phenolchloroform and mtDNA heteroplasmy level determined as described in Battersby and Shoubridge (2001). Modeling selection for mtDNA in the hematopoietic tissues of heteroplasmic mice was as described in Battersby et al. (2005). Data sets were tested first for normality then subject to parametric statistics (two-tailed t-test).
RNA analysis
All tissues sampled were frozen in liquid nitrogen and stored at −80°. Total RNA was extracted with Trizol (Invitrogen) then treated with DNaseI to eliminate potential DNA contamination. Allelic expression of Gimap3 was measured from total RNA amplified by semiquantitative RT–PCR compared to a standard curve with equimolar mixtures of CAST/Ei and BALB/c RNA (r2 = 0.97) and quantified with a Typhoon 9400. Data were analyzed statistically using a Mann–Whitney U-test.
For Northern blotting total RNA was isolated from frozen spleens and thymi homogenized in Trizol (Invitrogen) with Precellys 24 tissue homogenizer (Bertin technologies) with mixed beads and a homogenization cycle of 5500 rpm for 25 sec. Total RNA was isolated by Trizol from stable retroviral cell line controls and treated with DNaseI (New England Biolabs). A total of 15 μg of RNA for tissue samples and 5 µg for the cell line controls (concentration determined with a Nanodrop1000 spectrophotometer) were separated on a 1.2% agarose–formaldehyde gel and transferred by neutral transfer to Hybond-N+ membrane (GE healthcare). The membrane was UV cross-linked and consecutively probed with oligonucleotide probes for Gimap3 and Atp5B. The probes were 5′ radiolabeled with T4 PNK (New England Biolabs) and purified with Illustra ProbeQuant G-50 microcolumn (GE Healthcare). Hybridization (25% formamide, 7% SDS, 1% BSA, 0.25 M sodium phosphate pH 7.2, 1 mM EDTA pH 8.0, 0.25 M NaCl2) was performed for 16–20 hr at 37°. Membranes were washed (2× SSC/0.1% SDS and 0.2× SSC/0.1% SDS) and then dried for exposure on Storage Phosphor screens (GE Healthcare), which were scanned and quantified with a Typhoon 9400 (Amersham).
Microscopy
Cells were grown on glass coverslips and fixed in fresh 4% paraformaldehyde. Coverslips were mounted with DABCO/ MOWIOL on glass slides for imaging. Confocal images were taken on a Zeiss META 510 with the 63× Plan-Apochromat (1.4 NA) objective using the Argon (488 nm) and HeNe1 (543 nm) lasers or a Leica TCS Sp8 CARS with a 63× APO (1.2 NA) water objective using the Argon (488 nm) and DPSS (561 nm) lasers. Fluorescence microscopy was carried out on a Zeiss Axioplan2 using a 63× (1.25 NA) or 100× (1.3 NA) objective. Image processing and line profiles were performed using Image J. Only linear corrections were applied to images.
iTRAQ analysis
Three mouse spleens per genotype were homogenized (200 mM mannitol, 70 mM sucrose, 10 mM HEPES, 1 mM EGTA pH 7.5) with a Teflon pestle. Differential centrifugation was used to enrich for the membrane compartment containing Gimap3 and Gimap5. The postnuclear lysate (two 600 × g for 10 min) was first centrifuged at 5000 × g for 10 min and the resulting pellet was discarded. The supernatant was then centrifuged for 20 000 × g for 40 min. This pellet was solubilized and a total of 120 µg of protein per sample was precipitated using 2-D Clean Up Kit (GE Healthcare). The precipitated proteins were dissolved into 30 µl of iTRAQ dissolution buffer, and 2 µl of each sample was subjected to SDS–PAGE and silver staining to control for protein amount in the samples after precipitation. Protein alkylation, trypsin digestion and labeling of the resulting peptides were performed according to manufacturer’s instructions (Applied Biosystems). After labeling, the samples were pooled and dried, and the peptides were fractionated by strong cation exchange chromatography using an Ettan HPLC system (Amersham Biosciences) connected to a Polysulfoethyl A column. Each selected SCX fraction containing labeled peptides was then processed with C18 Spin Columns (Pierce) according to the manufacturer instructions before being analyzed twice with nano-LC-ESI-MS/MS using Ultimate 3000 nano-LC (Dionex) and QStar Elite mass spectrometer (Applied Biosystems) as previously described (Lietzen et al. 2011). MS data were acquired automatically using Analyst QS 2.0 software. iTRAQ analyses were performed on three biological replicate samples.
Protein identification and relative quantitation were performed with Paragon search algorithm (Shilov et al. 2007) using ProteinPilot 4 interface (Applied Biosystems). Data files from both technical replicates of an iTRAQ sample set were processed together. Database searches were performed against mouse protein sequences in UniProt database (v. 2012-08-13 with 16 540 mouse entries). The search criteria were cysteine alkylation with MMTS, trypsin digestion, biological modifications allowed, thorough search and detected protein threshold of 95% confidence (Unused ProtScore >1.3). Additionally, automatic bias correction was used to correct for uneven protein loading. The false discovery rates were calculated as previously described (Elias and Gygi 2007) and were <1% for all biological replicates.
Relative quantity fold change of 0.66 or 1.5 was used as a threshold for changed proteins, and proteins were included in the data set only if they either exhibited similar change in at least two of the biological replicates, or a statistically significant quantitation change in one of the replicates and no changes in the others. Biological process for the identified proteins was retrieved from UniProtKB.
Results
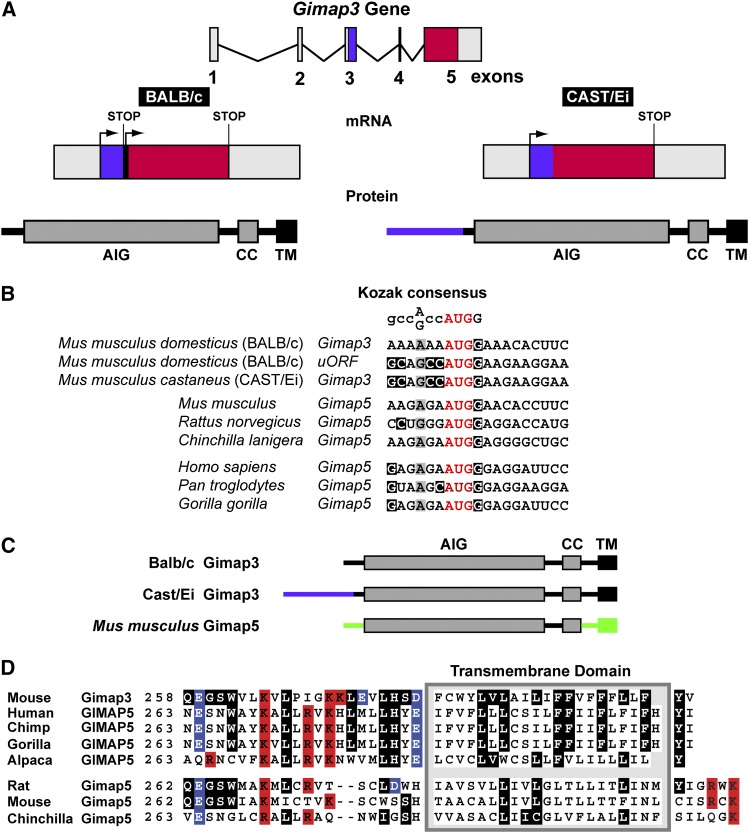
Previously we identified M. musculus allelic variants of Gimap3 that affect mtDNA segregation in hematopoietic tissues (Jokinen et al. 2010). In the common lab mouse strains with a M. m. domesticus background, such as BALB/c, Gimap3 contains five exons and transcription produces an mRNA with two AUG codons as potential start codons for translation initiation (Figure 1A). The first AUG encodes an upstream open reading frame (uORF) of 60 amino acids, while translation initiation on the second AUG generates a 301-amino-acid protein (Figure 1A and Supporting Information, Figure S1) that contains the conserved GTPase domain that defines the Gimap family. Surprisingly, the uORF has a greater Kozak consensus sequence around the AUG than that seen for the downstream reading frame of Gimap3 (Figure 1B). Unless indicated otherwise, hereafter Gimap3 refers to the allele found among the common lab strains with a M. m. domesticus background that encodes the 301-amino-acid protein (Jokinen et al. 2010). In contrast, the subspecies M. m. castaneus has a mutation in the splice acceptor site of exon 4 causing a splicing defect leading to the absence of exon 4 from the CAST Gimap3 mRNA (Figure 1A) (Jokinen et al. 2010). In this CAST allele the AUG of the uORF is used for translation initiation producing a 359 amino acid protein (Figure 1A and Figure S1). These two allelic variants of Gimap3 are identical at the amino-acid-sequence level within all of the known functional domains and differ only at the N terminus, which has an additional 58 amino acids in the CAST/Ei version (Figure 1A) (Jokinen et al. 2010). Linkage mapping and transgenic expression of the CAST Gimap3 cDNA established that this allele significantly decreases the rate of mtDNA segregation in the mouse hematopoietic compartment (Jokinen et al. 2010).
Figure 1.
The organization of the Gimap3 gene in mice. (A) A graphical illustration of the Gimap3 gene, mRNA, and protein derived from BALB/c and CAST/Ei alleles. Red indicates the main coding sequence for Gimap3, while purple indicates the sequence of the uORF. CC, coiled coil; TM, transmembrane; AIG, Gimap family domain. (B) Alignment of the predicted starter methionine from Gimap3 and Gimap5 across selected mammalian taxa against the Kozak consensus sequence. (C) A graphical illustration of the Gimap3 and Gimap5 proteins with color differences indicating sequence divergence between the two proteins. (D) An amino acid alignment of the C terminus of Gimap3 against Gimap5 from selected mammalian taxa. Predicted transmembrane domain with flanking charged amino acids: positive in red and negative in blue. Amino acids in Gimap3 that are conserved with Gimap5 appear in white type on a black background.
In the annotated genomes of most mammals Gimap3 appears to be absent from the Gimap gene cluster. In humans, GIMAP3 appears to be a pseudogene (Krucken et al. 2004) and it is not clear whether this gene function has been completely lost or taken over by GIMAP5. However, the high identity in nucleotide sequence between Gimap3 and Gimap5 can be a complicating factor in the robustness of the genome annotations with respect to the presence of Gimap3 in these taxa. Thus, it is still an open question whether Gimap3 is specific to mice. Aligning the C-terminal amino acid sequence of mouse Gimap3 against that of Gimap5 from a number of mammalian taxa demonstrates that the rodent Gimap5 possesses a difference in the charge balance around the tail-anchor sequence and in the amino acid composition of the transmembrane domain (Figure 1D). These features are critical for intracellular localization of tail-anchor proteins (Rapaport 2003). Based upon this protein alignment of the C terminus it would appear that Gimap5 in Rodentia has diverged in sequence from other taxa of the Mammalia lineage.
Gimap3 is anchored into the endoplasmic reticulum
The intracellular localization of tail-anchored Gimap family members has been controversial (Wong et al. 2010). The original localization study for Gimap3 demonstrated that the tail-anchor sequence of the protein was required for membrane insertion and was proposed to be the outer mitochondrial membrane (Daheron et al. 2001). Unfortunately closer examination of the data suggests that the findings could not properly distinguish between organelles. We decided to revisit the intracellular localization for Gimap3 in more detail.
The first test was to determine with which organelles endogenous Gimap3 would sediment on a Histodenz gradient. A postnuclear spleen lysate was separated in a stepwise discontinuous Histodenz gradient to determine the sedimentation profile of Gimap3. Fractions were collected and used for immunoblotting using polyclonal antisera against Gimap3 (Figure S1). The sedimentation profile for endogenous Gimap3 was similar to the ER chaperone Calnexin but not to mitochondrial proteins in the outer mitochondrial membrane (Porin) and the matrix (Sdha) (Figure 2A).
Figure 2.
Gimap3 localizes to the endoplasmic reticulum. (A) Sedimentation profile of mouse postnuclear spleen lysate through a discontinuous Histodenz gradient. Fractions were separated by SDS–PAGE followed by immunoblotting with the indicated antibodies. Canx is an ER chaperone. Sdha and Porin are mitochondrial proteins localized in the matrix and outer membrane respectively. (B) A graphical illustration of the Gimap3 constructs used in microscopy. (C) Confocal images of COS-7 cells cotransfected with a chimeric RFP–Gimap3 and ER marker GFP–Sec61B. Below, line scans of the red and green channels. (D) Confocal images and inset magnifications of COS-7 cells cotransfected with the chimeric RFP–Gimap3 and the mitochondrial marker GFP–omp25. (E) COS-7 cells transfected with a construct containing only the C-terminal transmembrane domain of Gimap3 fused to YFP. (F) COS-7 cells transfected with a construct containing the full-length Gimap3 fused to the C terminus of YFP. (G) COS-7 cells cotransfected with RFP–Gimap3 and a chimeric YFP–Gimap3 missing the 32 C-terminal amino acids replaced with the omp25 sequence motif for mitochondrial localization. Scale bars, 10 µm.
Unfortunately, our polyclonal antibody against Gimap3 was ineffective for indirect immunofluorescence and thus unable to be used for determining intracellular localization by light microscopy. Therefore, we generated a chimeric Gimap3 with RFP fused at the N terminus of the protein to circumvent this problem (Figure 2B). This RFP construct should not affect the membrane targeting of the protein since tail-anchored proteins require only the C-terminal transmembrane domain and flanking residues for membrane insertion and retention (Rapaport 2003). Stable retroviral transduction or transient expression of this Gimap3 construct consistently localized to the ER and not mitochondria (Figure 2, C and D). Only the 39 amino acids at the C terminus were required for this ER localization (Figure 2, E and F). Furthermore, replacing the last 32 C-terminal amino acids with a tail-anchor sequence from Omp25, a bona fide outer mitochondrial membrane protein (Nemoto and De Camilli 1999) redirects Gimap3 to the outer mitochondrial membrane (Figure 2G). Collectively, these data suggest that Gimap3 localizes to the ER.
The expression level of Gimap3 modulates mtDNA segregation
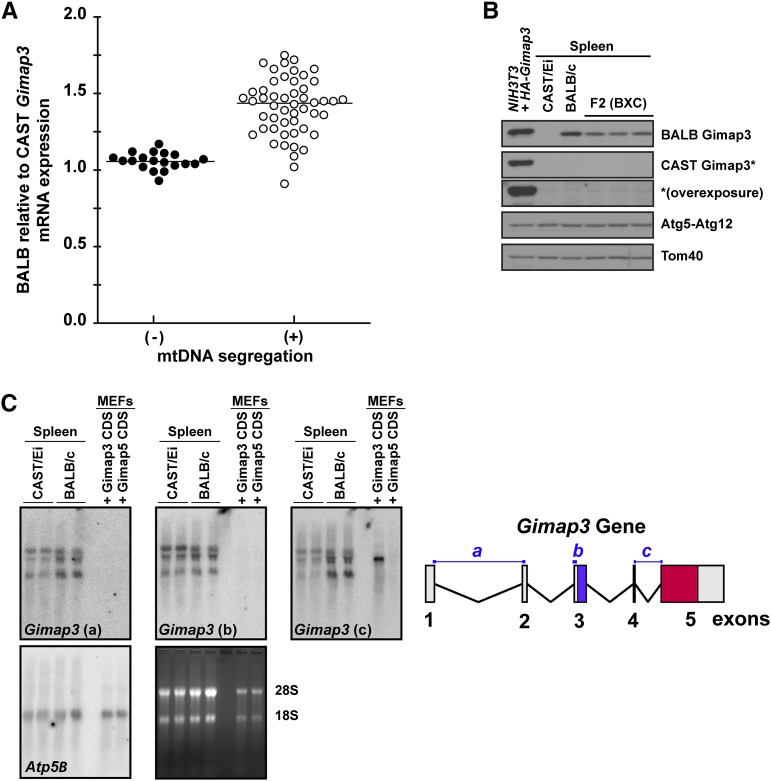
To investigate how allelic expression of Gimap3 modulates mtDNA segregation in the spleen, we first tested whether there were quantitative differences in the expression of these two murine alleles. The mRNA expression was quantified of both the CAST/Ei and BALB/c alleles in 3-month-old mice from an F2 (BALB/c × CAST/Ei) cross, which were heterozygous for these two Gimap3 alleles. At 3 months of age most F2 mice display quantitative differences in the mtDNA segregation rate in hematopoietic tissues and are considered positive for segregation (Jokinen et al. 2010). However, there is also a subset of mice in this F2 cross that at 3 months of age can be identified while mtDNA segregation cannot yet be detected in hematopoietic tissues (Jokinen et al. 2010). These mice were considered negative for segregation. A semiquantitative RT–PCR approach was used to quantify the expression differences of these two alleles in the F2 mice utilizing a primer set that bind in exons 3 and 5 to amplify across exon 4, which is only 35 nucleotides long with a stretch of poly(A)’s and the only unique feature within the Gimap3 mRNA to distinguish between the BALB/c and CAST/Ei alleles. The mRNA expression was quantified against a standard curve generated with equimolar ratios of BALB/c and CAST/Ei RNA. In mice positive for mtDNA selection there was a significantly stronger expression from the BALB Gimap3 allele relative to the CAST Gimap3 allele (P < 0.001) (Figure 3A).
Figure 3.
Gimap3 expression affects mtDNA segregation in mouse spleen. (A) The expression level of the BALB/c Gimap3 mRNA relative to CAST/Ei Gimap3 by semiquantitative RT–PCR in F2 (BALB/c × CAST/Ei) mice heterozygous at the Gimap3 locus. Mice were either positive (+) (n = 51) or negative (−) (n = 19) for mtDNA segregation. Mann–Whitney Test, P < 0.001. (B) Immunoblotting of total cell lysates from the spleen of BALB/c, CAST/Ei or F2 (BALB/c × CAST/Ei) mice. A NIH3T3 cell line was transduced with retroviruses expressing HA-tagged version of both the BALB/c Gimap3 and CAST/Ei Gimap3 was used as positive control. (*) Extended exposure with the CAST Gimap3 antibody. (C) Northern blot of total spleen RNA from CAST/Ei and BALB/c mice with three independent Gimap3 probes (a–c) and a control probe for Atp5B. As positive and negative controls for the Gimap3 probes total RNA was loaded from MEFs ectopically expressing the coding sequence (CDS) of BALB/c Gimap3 and Gimap5. On the right, a graphical illustration of the Gimap3 gene and location of the oligonucleotide probes.
Next, we investigated whether the difference in Gimap3 mRNA expression resulted in a corresponding effect on the steady-state protein levels. Spleen homogenates were prepared from BALB/c, CAST/Ei, and F2 (BALB/c × CAST/Ei) mice heterozygous at the Gimap3 locus for immunoblotting. Both Gimap3 protein variants can be expressed in cultured cells and detected by immunoblotting using our allele-specific polyclonal antibodies (Figure 3B and Figure S1). To our surprise there was no detectable Gimap3 protein (Figure 3B) in spleen lysates from CAST/Ei mice. This is in contrast to the abundant protein expression of the BALB/c Gimap3 (Figure 3B). In F2 (BALB/c × CAST/Ei) mice we also could not detect the CAST Gimap3 protein. Moreover, in these F2 mice the abundance of the BALB Gimap3 protein was reduced ∼50% compared to spleens from BALB/c mice (Figure 3B), suggesting a dosage effect on the steady-state protein abundance of Gimap3.
To confirm the expression of the CAST/Ei Gimap3 mRNA, we analyzed total spleen RNA by Northern blotting. Using three independent oligonucleotide probes that hybridize against sequences from exons 1–2, exon 3, and exons 4–5, we detected three transcripts (Figure 3C). All three probes were selected against sequence unique to Gimap3 and not found in Gimap5 (Figure 3C). As a positive control we used total RNA from murine embryonic fibroblasts ectopically expressing the BALB/c Gimap3 cDNA consisting only of exons 4 and 5, which can be recognized only by probe c (Figure 3C). Probe c bound weakly to the CAST/Ei mRNA, which was not surprising, considering half of the probe recognizes nucleotides in exon 4, a region missing from the CAST/Ei mRNA. Assessing Gimap3 expression using probe b, which hybridizes to exon 3, showed robust expression of both the BALB/c and CAST/Ei alleles in the spleen. These results confirm that although both Gimap3 alleles were expressed at the mRNA level, only the BALB/c variant could be detected in mouse spleens as a mature protein by immunoblotting.
The N terminus of CAST Gimap3 impairs protein synthesis
The inability to detect the CAST Gimap3 protein in vivo may be due to repressed synthesis of the polypeptide chain or in post-translational processing, such as cleavage of the N terminus and differential turnover. We pursued the hypothesis that protein synthesis of the CAST Gimap3 was impaired because this allele has incorporated a uORF into the coding sequence for this protein. The presence of a uORF within a gene can act as a potent in cis post-transcriptional regulator of the main downstream ORF whereby protein synthesis of the uORF stalls ribosomes repressing translation of the downstream ORF (Morris and Geballe 2000; Calvo et al. 2009).
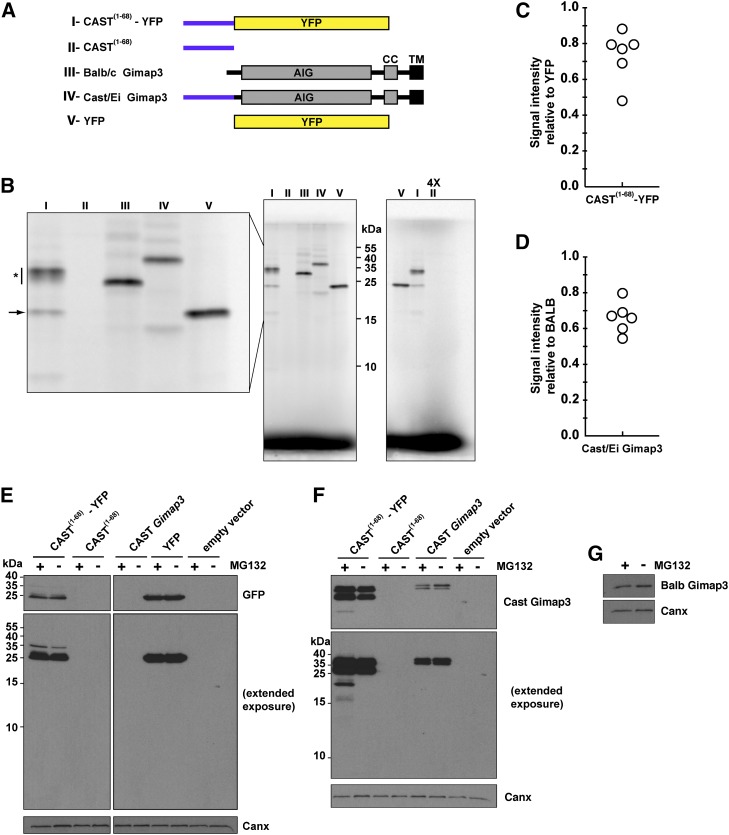
We wanted to first evaluate whether there was any difficulty in the synthesis of the uORF alone or fused to a nonnative protein such as YFP. We generated constructs comprising amino acids 1–68 of CAST Gimap3 alone or fused to YFP at the N terminus (Figure 4A). This stretch of polypeptides represents the only unique amino acids at the N terminus between the BALB and CAST Gimap3 (Jokinen et al. 2010). The objective was to express the constructs both in vitro and in cultured cells to test if there was difficulty in the synthesis of this polypeptide sequence and whether there was any post-translation regulation in the cell to modulate the steady-state protein abundance. We used a T7-promoter-coupled reticulocyte lysate assay with radiolabeled [35S]methionine and cysteine to express these proteins in vitro. The linearity for the 35S-radiolabel incorporation was established using YFP as a template cloned into the same vector backbone (Figure S2). Hereafter, the same conditions were used for expressing all constructs off the T7 promoter in vitro. Strikingly there was no detectable expression of the CAST1–68 fragment alone (Figure 4B). Moreover, fusion of this CAST1–68 fragment to YFP reduced the in vitro expression of the full-length protein compared to YFP (Figure 4, B and C). A prominent feature with the synthesis of CAST1–68–YFP was consistent smearing of the mature protein (Figure 4B), which may be suggestive of stalled synthesis. In addition, there were smaller fragments generated with the CAST1–68–YFP template, one of which comigrated with the full-length YFP (Figure 4B). Next we assessed the in vitro synthesis of full-length CAST vs. BALB Gimap3 (Figure 4, A and B). There was a reduction in the synthesis of the CAST Gimap3 compared to the BALB allele using the same T7-expression-coupled assay (Figure 4D). These data suggest that the N terminus of CAST Gimap3 exerts a negative effect on protein synthesis in vitro.
Figure 4.
The N terminus of CAST Gimap3 impairs protein synthesis. (A) A graphical illustration of the Gimap3 constructs used. AIG, Gimap family domain; CC, coiled coil; TM, transmembrane domain. (B) SDS–PAGE of in vitro synthesis of proteins from a T7 promoter with [35S]methionine and cysteine. Lanes numbered according to the constructs in A. (*) Consistent smearing of labeled protein. Arrow indicates labeled protein from CAST(1–68)–YFP that comigrates with full-length YFP (lane V). Right: Four times excess sample was loaded for construct II. (C) Quantification of CAST1–68–YFP in vitro production relative to YFP from six independent experiments. For the quantification, the smeared signal for CAST(1–68)–YFP (* in B) was included along with the mature protein. (D) Quantification of CAST Gimap3 in vitro production relative to BALB Gimap3 from six independent experiments. (E and F) Immunoblotting of total cell lysates from HEK293 cells transfected with the indicated cDNA constructs and treated with the indicated antibodies. Cells were treated with or without MG132 7 hr prior to collection. (G) Immunoblotting of total cell lysates from HEK293 cells transfected with a Gimap3 cDNA construct and treated with the indicated antibodies. Cells were treated with or without MG132 7 hr prior to collection.
To evaluate the role of the uORF on protein synthesis in cultured cells we transiently transfected the plasmids into HEK293 cells followed by immunoblotting 24 hr post-transfection. For the CAST1–68–YFP construct, most of the immunodetectable protein with an anti-GFP antibody comigrated through SDS–PAGE with full-length GFP and not near the expected molecular size of 35 kD as seen in vitro (Figure 4, B and E). Extended exposure with the anti-GFP antibody could detect a weaker band with the approximate size of the mature CAST1–68–YFP (Figure 4E), showing that this protein can be synthesized in vivo. In contrast, using the CAST-specific polyclonal antibody we detected two prominent bands approximately of equal intensity for CAST1–68–YFP that were more abundant than the steady-state level of full-length CAST Gimap3 (Figure 4F). There also was no detectable protein for the CAST1–68, a finding consistent with the in vitro expression (Figure 4, B and F). Since the abundance of the full-length CAST–YFP differs between in vitro and in vivo, synthesis suggests that addtional factors are important for the steady-state level of the protein.
Stalled protein synthesis on ribosomes generates a stress to the cell, which typically activates mRNA surveillance pathways (Lykke-Andersen and Bennett 2014). As part of this stress response, polypeptide chains on stalled ribosomes are rapidly degraded by the proteasome (Lykke-Andersen and Bennett 2014) and thus stabilized only in the presence of proteasomal inhibitors, such as MG132. To test for evidence of the proteasome in modulating the abundance of the uORF we incubated cells with MG132. In cells expressing the CAST1–68–YFP cDNA we observed smaller-molecular-weight fragments when treating with MG132 and using the CAST-specific antibody, but not the monoclonal anti-GFP antibody that detects only the mature protein (Figure 4, E and F). We also observed a shift with MG132 in the proportion of the two immunodetectable fragments expressed by CAST Gimap3 (Figure 4F). In contrast, MG132 had no effect on the steady-state abundance of Gimap3 (Figure 4G). Since the CAST-specific antibody recognizes a stretch of 15 amino acid residues found only in the N terminus of the protein (Figure S1), the immunodetection of stable smaller-molecular-weight fragments when the proteasomal function is blocked may be attributable to the partial synthesis of the CAST1–68–YFP because of ribosome stalling.
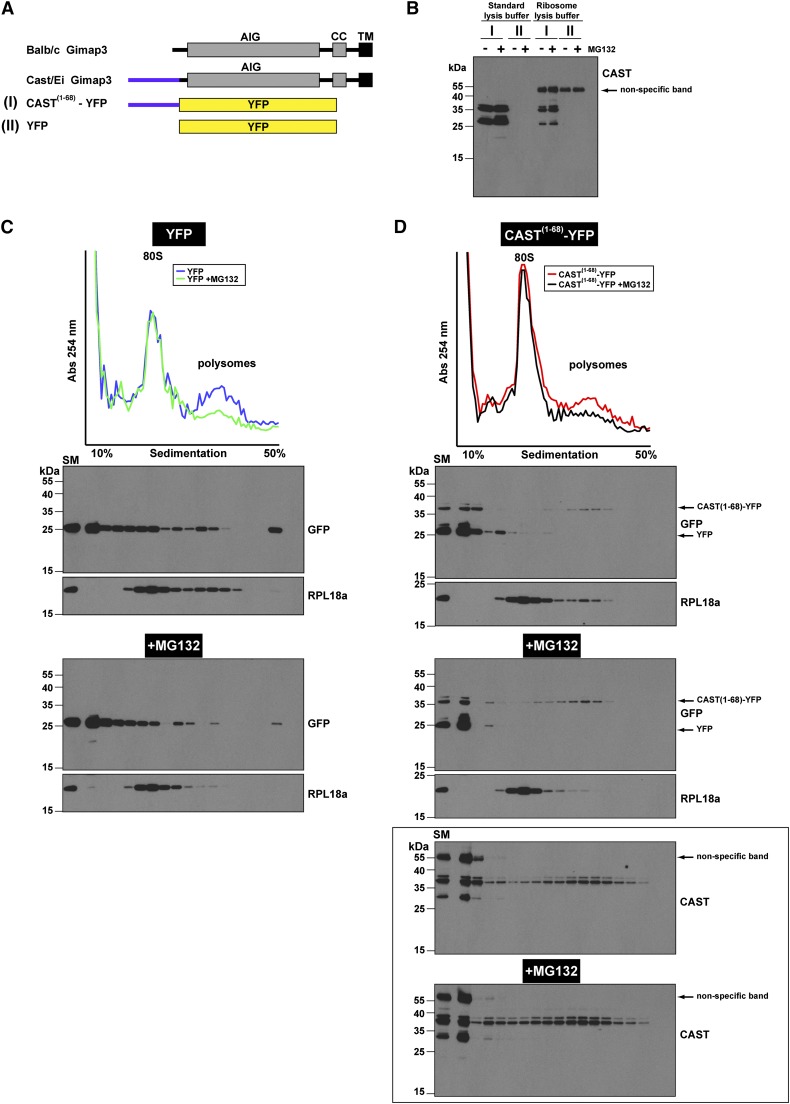
Next, we wanted to confirm this interpretation by testing whether the synthesis of CAST1–68–YFP on polysomes was affected following the treatment of cells with MG132. Isokinetic sucrose density gradients were used to separate ribosomes and polysomes from treated cells (Figure 5). We found an increase in the CAST1–68–YFP on polysomes following MG132 treatment (Figure 5) suggesting that the uORF fused to YFP leads to proteasomal turnover. Moreover, this analysis demonstrates that the predominant protein generated from the CAST1–68–YFP cDNA comigrates with the molecular size of YFP when we used the anti-GFP antibody for immunoblotting (Figure 5, C and D). This is consistent with our previous findings (Figure 4E) and indicates the potential for ribosomes scanning through the uORF sequence to initiate translation on the AUG of YFP. Using the CAST antibody we also detected another truncated protein that was >25 kD (Figure 5D, bottom) and must contain an N terminus with the CAST polypeptide sequence. Collectively, these data suggest that the uORF in Gimap3 mediates a negative effect on protein synthesis, possibly by stalling synthesis on ribosomes, and may account for the decreased abundance of CAST Gimap3 in mouse spleen.
Figure 5.
The CAST(1–68) polypeptide impairs protein synthesis and leads to accumulation on polysomes following proteasome inhibition. (A) A graphical illustration of the Gimap3 and YFP constructs. AIG, Gimap family domain; CC, coiled coil; TM, transmembrane domain. (B) Immunoblotting with the CAST polyclonal antibody comparing immunoreactivity of HEK293 lysates generated with different buffers and detergent combinations. The standard lysis buffer is used in Figure 4, whereas the ribosome lysis buffer is optimized for polysome profiles. (C and D) Polysome profiles of YFP and CAST(1–68)–YFP. Total cell lysates from HEK293 cells transfected with the indicated cDNAs were prepared with the ribosome lysis buffer and separated on 10–50% sucrose gradients. SM, starting material. Top: Absorbance at 254 nm of 82 fractions collected from the top. Below, immunoblotting of TCA precipitated fractions from five pooled aliquots from the top with the indicated antibodies and with equal film exposures ±MG132
Gimap3 and Gimap5 are important for mtDNA segregation
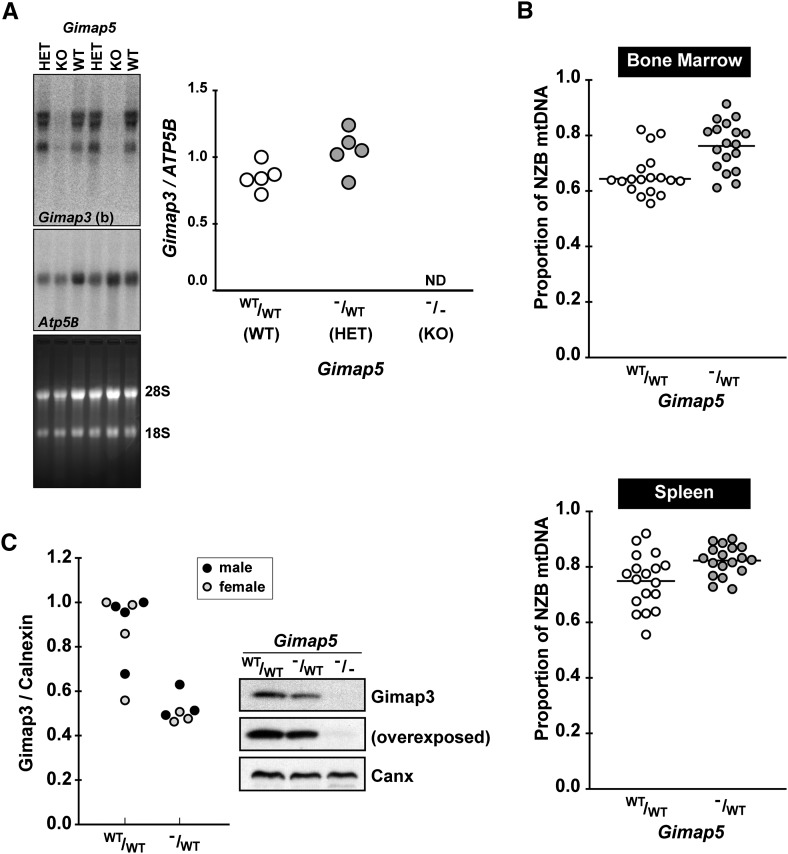
Our original cloning of Gimap3 as a modifier of mtDNA segregation (Jokinen et al. 2010) was unable to determine whether other Gimap family members, in particular the paralogue Gimap5, may also contribute to this mtDNA phenotype. Recent evidence points to a cooperative genetic function between Gimap3 and Gimap5 in lymphocyte development (Yano et al. 2014). To test Gimap5 for a role in mtDNA segregation, we took advantage of a germline knockout mouse Gimap5tm1Wlr (Schulteis et al. 2008). Homozygous Gimap5tm1Wlr mice exhibit a severe hematopoietic stem cell defect, which leads to a reduced number of mature lymphocytes and lack of detectable NK cells (Schulteis et al. 2008). In Gimap5tm1Wlr/WT there are no reported defects in hematopoiesis or on the mRNA expression of other Gimap family members in the thymus; these mice were described only as having a 50% reduction in Gimap5 expression (Schulteis et al. 2008). This dosage effect on Gimap5 expression in heterozygotes has been consistently observed with two other independent murine Gimap5 null mutations at both the mRNA and protein level in Barnes et al. (2010) and Yano et al. (2014), suggesting a tight correlation between Gimap5 gene expression and protein abundance.
To test whether changes in the expression of Gimap5 affect mtDNA segregation, we crossed our heteroplasmic mouse model (NZB and BALB mtDNA haplotypes) to Gimap5tm1Wlr. Since both mouse lines were already established on the BALB/c nuclear background, no further backcrossing was required. In the spleen of homozygous Gimap5tm1Wlr mice there are severe cellular and morphological abnormalities: a ninefold reduction in splenic T cells and a twofold reduction in total splenic cellularity (Schulteis et al. 2008). These pathological features in the spleens of Gimap5tm1Wlr homozygote mice complicate the analysis of mtDNA segregation because the cellular composition of the organ is dramatically altered, which in turn affects Gimap3 expression in this tissue (Figure 6A and Figure S3). Therefore, we focused our mtDNA segregation analysis on Gimap5tm1Wlr/WT because these mice are phenotypically indistinguishable from wild type and display no signs of pathology or cellularity defects in the spleen, but have a 50% decrease in Gimap5 abundance.
Figure 6.
Genetic loss of Gimap5 affects Gimap3 and mtDNA segregation. (A) Northern blot of spleen total RNA from wild-type Gimap5, Gimap5tm1Wlr/WT, and homozygote Gimap5tm1Wlr mice hybridized with Gimap3 (b) probe and control probe Atp5B. Right: Quantification of Gimap3 mRNA compared to Atp5B. (B) Decreased selection against the NZB mtDNA genome in the bone marrow and spleen of Gimap5tm1Wlr/WT compared to wild-type littermate controls. n = 18 for each group; (*) P < 0.01 two-tailed t-test; means are indicated by the line through the scatter plot. Mice were 70 days of postnatal age at the time of sampling. (C) A representative immunoblotting of Gimap3 from whole-cell lysates from isolated splenocytes of wild type, Gimap5tm1Wlr/WT and homozygote Gimap5tm1Wlr mice. Left: Quantification of Gimap3 abundance compared to Calnexin.
In the hematopoietic compartment of wild-type heteroplasmic mice mtDNA segregation can be modeled as an exponential decay, so that by 100 days of post-natal life there is an ∼50% decrease in the proportion of NZB mtDNA in leukocytes of both myeloid and lymphoid lineages (Battersby et al. 2005). We found a significant difference in mtDNA segregation of Gimap5tm1Wlr/WT in both the spleen and bone marrow compared to wild-type littermate controls (Figure 6B). The effect is more modest in the spleen compared to the bone marrow (Figure 6B) because within the spleen the supporting stromal cells are neutral with respect to mtDNA segregation (Battersby et al. 2005).
Since we found that changes in Gimap3 abundance were accompanied with an effect on mtDNA segregation, we wanted to test whether in Gimap5tm1Wlr/WT there was any effect on the abundance of Gimap3. Total cell lysates were prepared from isolated splenocytes for immunoblotting demonstrating a 50% reduction in Gimap3 protein abundance compared to wild type (Figure 6C) even though there was no difference in the expression of the Gimap3 mRNA (Figure 6A). These findings provide further support for a cooperative function between Gimap3 and Gimap5 and indicate that the steady-state abundance of these two proteins is an important factor for the segregation of mtDNA haplotypes in mouse leukocytes.
A 50% reduction of Gimap3 and Gimap5 protein levels was sufficient to significantly modulate mtDNA segregation in leukocytes and suggests that the identification of other factors dependent upon Gimap5 may provide insight into the molecular connection to the segregation of the mitochondrial genome. To identify such factors we used a quantitative proteomic approach with iTRAQ (isobaric tags for relative and absolute quantification) labeling followed by LC-MS/MS analysis of postnuclear membrane enrichments generated from spleens of wild type and Gimap5tm1Wlr heterozygote littermates (File S1 and File S2)(Lietzen et al. 2011). Three independent biological replicates for each genotype were used for the iTRAQ analysis, which led to the identification of >1100 distinct proteins (File S1). We were unable to quantify Gimap3 and Gimap5 using this approach with any confidence because the sequence homology between these two proteins is very high (84% identity) and the number of unique peptide sequences detected by LC-MS/MS was inadequate for confident identification and iTRAQ quantification. In Gimap5tm1Wlr/WT we detected robust changes in the abundance of five proteins that all have a connection to the regulation of the cytoskeleton and cell motility via small GTPase signaling (Table1). The abundance of these proteins was also further affected in Gimap5tm1Wlr homozygotes (File S2). Since Gimap5 is a membrane-anchored lysosomal protein, this points to a quantitative effect of Gimap5 expression on the interaction between organelles and the cytoskeleton during cell motility as an important factor in mtDNA segregation of hematopoietic cell lineages in mice.
Table 1. Proteins whose abundance in mouse splenocytes is dependent upon Gimap5: iTRAQ analysis comparing Gimap5WT/WT to Gimap5tm1Wlr/WT littermates.
| Protein | Fold change het/wt | Biological process |
|---|---|---|
| Cyfip2 | 0.55 | Component of WAVE complex for assembly of F-actin complex |
| Arhgdia | 0.57 | Rho protein signal transduction, regulation of protein localization |
| Vasp | 0.57 | Actin cytoskeleton reorganization |
| Elmo1 | 1.83 | Actin filament-based process, phagocytosis |
| S100A9 | 2.12 | Actin cytoskeleton reorganization, leukocyte chemotaxis, phagocytosis |
Discussion
The mechanisms governing the segregation of mitochondrial genomes are still poorly understood despite considerable efforts since the discovery of pathogenic mtDNA mutations (Jokinen and Battersby 2013). We followed up our cloning of Gimap3 as a modifier of mtDNA segregation in mice to determine how allelic variants of this gene influence the phenotype. The most important finding from this study is that expression differences in Gimap3 and Gimap5 appear to be a critical factor to the segregation of the mitochondrial genome in mouse hematopoietic tissues.
Regulation of Gimap3 expression is complex and possibly involves cell-specific modulators. The incorporation of a uORF into the coding sequence of the M. m. castaneus Gimap3 has a negative effect on protein synthesis potentially through translation repression. Our data are consistent with a variable penetrance of the uORF on the CAST Gimap3, as the expression and stability of the mature protein decreases steadily from in vitro synthesis to ectopic expression in nonhematopoietic cell types to the undetectable levels in vivo in mouse leukocytes. This is in contrast to the BALB Gimap3 that can be robustly detected in the mouse. Since our polyclonal antibodies do not recognize both protein alleles, we cannot exclude the possibility of N-terminal processing of CAST Gimap3 in mice. UORFs have been described as translation regulatory sequences and are estimated to be present in >40% of human and mouse transcripts (Calvo et al. 2009). The mechanism of action for uORFs often involves repressing translation of the downstream (main) open reading frame (Morris and Geballe 2000; Calvo et al. 2009). Such a regulatory mechanism allows for rapid changes in the protein abundance upon stress conditions, when trans factors can release the uORF repression of translation (Barbosa et al. 2013). Our data also indicate that ribosomes may scan through this uORF sequence and initiate translation on a downstream AUG even though there is stronger sequence conservation to the Kozak consensus in the uORF. Further work is required to elucidate the regulatory role of the uORF on the expression of Gimap3 alleles in mice.
We show that Gimap3 localizes to the ER and not mitochondria as previously reported. How then could a resident protein of the ER affect mtDNA segregation? Interactions between the ER and mitochondria play a key role in many aspects of mitochondrial homeostasis, such as morphology of the mitochondrial reticulum (division and tethering of the organelle), calcium handling, lipid transfer, and transmission of the mitochondrial genome among others (de Brito and Scorrano 2008; Kornmann et al. 2009; Friedman et al. 2011; Hoppins et al. 2011; Connerth et al. 2012; Rowland and Voeltz 2012; Korobova et al. 2013; Hajnoczky et al. 2014). Any number of these functions could be important to mtDNA segregation.
Why the need to restrict the expression of these Gimap protein scaffolds to the hematopoietic compartment? Leukocytes have a relatively small cytosolic volume with few organelles by comparison to most cell types and are one of the most dynamic cell types in mammals as they migrate throughout the body by chemotaxis. Chemotaxis and receptor signaling at the immunological synapse both require dynamic and coordinated reorganization of organelle distribution, especially mitochondria (Campello et al. 2006; Quintana et al. 2007). During chemotaxis, leukocytes become polarized with mitochondria relocalized to the uropod. Disrupting this mitochondrial relocalization impairs the ability of a leukocyte to migrate (Campello et al. 2006; Quintana et al. 2007). Recent reports from mammalian systems suggest that interactions with the actin cytoskeleton are needed for mitochondrial division (Korobova et al. 2013) and dynamics during formation of the immunological synapse (Schwindling et al. 2010). The stoichiometric changes in Gimap5tm1Wlr heterozygotes of factors interacting with the actin cytoskeleton could suggest that these Gimap scaffolds are important for the dynamic organization and coordination of organelle movement during chemotaxis. Consistent with this interpretation is the recent work on Gimap4 showing that this soluble member of the Gimap family mediates an interaction between the trans Golgi network and the actin cytoskeleton to sustain for interferon gamma secretion (Heinonen et al. 2014). Since Gimap3 and Gimap5 reside in the membrane of different organelles, it is tempting to hypothesize that these proteins could help mediate organelle distribution, organization, and contact within the cell, which in turn would affect segregation of the mitochondrial genome in leukocyte lineages.
The absence of Gimap3 in most current mammalian genome annotations could indicate that the gene function has been lost or taken over by Gimap5, or could be due to poor resolution of current genome annotations because of the high sequence similarity between Gimap3 and Gimap5. Therefore the overall importance of Gimap3 across mammals, let alone vertebrates, is at present difficult to conclude, as it might be restricted to mice. However, analysis of the C-terminal protein sequence of Gimap5 across mammalian taxa suggests the evolution of Gimap5 in Rodentia differs considerably from other mammalian clades. Clearly, further research is required to resolve this issue.
The regulation of mtDNA segregation in metazoans remains a complex genetic problem. In the majority of cases, selection for specific mtDNA haplotypes in mammals is not based upon detectable differences in mitochondrial oxidative phosphorylation or on the replication rate of the genome (Jokinen and Battersby 2013). Thus, in mammals the identity of regulatory mechanisms that coordinate the segregation of the mitochondrial genome remain intractable despite considerable effort by the field (Jokinen and Battersby 2013). The forward genetic approaches we have taken have provided a degree of insight into the nuclear control of mtDNA segregation. Nonetheless, progress remains slow as the problem can be investigated only in mice because cultured cells cannot faithfully replicate the mtDNA segregation patterns observed in mammals (Jokinen and Battersby 2013). In the end, selection for a particular mtDNA haplotype within a specific tissue can occur only at three levels: the cell, organelle, or genome. Two fundamental components are required for such a mechanism to work: signal generation that distinguishes mtDNA variants or mutations and the molecular machinery to act on or interpret those signals. Such a signal can be generated only at the sequence level, nucleic acid and or polypeptide, or by the functional consequences derived from those sequences. There appears to be tissue-specific differences in either the generation or interpretation of these signals in our mouse model. Based upon our findings, the segregation of mtDNA haplotypes in the mouse hematopoietic compartment may be dependent upon the regulation of an intrinsic property of these cell types to coordinate organelle interactions as they migrate throughout the body.
Supplementary Material
Acknowledgments
We thank Uwe Richter, Anu Wartiovaara, and Howy Jacobs for discussion. We thank the Biomedicum Imaging Unit (BIU) for the confocal facilities. The Academy of Finland Center of Excellence—FinMIT, the University of Helsinki, and the Jane and Aatos Erkko Foundation support research in the BJB laboratory. R.J. was supported by Helsinki Biomedical Graduate Program, Emil Aaltonen Foundation, Biomedicum-Helsinki Foundation, the Waldemar von Frenckell Foundation, and the Finnish Cultural Foundation. H.W. was supported by National Institute of Allergy and Infectious Diseases AI42380. T.A.N.’s laboratory is supported by the Academy of Finland (135628 and 140950) and the University of Helsinki. A.J.K. and J.K.H. were supported by grants from the Academy of Finland and the Sigrid Juselius Foundation. The authors declare no conflict of interest.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.175596/-/DC1.
Communicating editor: J. C. Schimenti
Literature Cited
- Barbosa C., Peixeiro I., Romao L., 2013. Gene expression regulation by upstream open reading frames and human disease. PLoS Genet. 9: e1003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes M. J., Aksoylar H., Krebs P., Bourdeau T., Arnold C. N., et al. , 2010. Loss of T cell and B cell quiescence precedes the onset of microbial flora-dependent wasting disease and intestinal inflammation in Gimap5-deficient mice. J. Immunol. 184: 3743–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battersby B. J., Shoubridge E. A., 2001. Selection of a mtDNA sequence variant in hepatocytes of heteroplasmic mice is not due to differences in respiratory chain function or efficiency of replication. Hum. Mol. Genet. 10: 2469–2479. [DOI] [PubMed] [Google Scholar]
- Battersby B. J., Loredo-Osti J. C., Shoubridge E. A., 2003. Nuclear genetic control of mitochondrial DNA segregation. Nat. Genet. 33: 183–186. [DOI] [PubMed] [Google Scholar]
- Battersby B. J., Redpath M. E., Shoubridge E. A., 2005. Mitochondrial DNA segregation in hematopoietic lineages does not depend on MHC presentation of mitochondrially encoded peptides. Hum. Mol. Genet. 14: 2587–2594. [DOI] [PubMed] [Google Scholar]
- Boulet L., Karpati G., Shoubridge E. A., 1992. Distribution and threshold expression of the tRNA(Lys) mutation in skeletal muscle of patients with myoclonic epilepsy and ragged-red fibers (MERRF). Am. J. Hum. Genet. 51: 1187–1200. [PMC free article] [PubMed] [Google Scholar]
- Burgstaller J. P., Johnston I. G., Jones N. S., Albrechtova J., Kolbe T., et al. , 2014. MtDNA segregation in heteroplasmic tissues is common in vivo and modulated by haplotype differences and developmental stage. Cell Rep. 7: 2031–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo S. E., Pagliarini D. J., Mootha V. K., 2009. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl. Acad. Sci. USA 106: 7507–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campello S., Lacalle R. A., Bettella M., Manes S., Scorrano L., et al. , 2006. Orchestration of lymphocyte chemotaxis by mitochondrial dynamics. J. Exp. Med. 203: 2879–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Yu M., Dai X., Zogg M., Wen R., et al. , 2011. Critical role for Gimap5 in the survival of mouse hematopoietic stem and progenitor cells. J. Exp. Med. 208: 923–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnery P. F., Samuels D. C., 1999. Relaxed replication of mtDNA: a model with implications for the expression of disease. Am. J. Hum. Genet. 64: 1158–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnery P. F., Howell N., Lightowlers R. N., Turnbull D. M., 1997. Molecular pathology of MELAS and MERRF: the relationship between mutation load and clinical phenotypes. Brain 120(10): 1713–1721. [DOI] [PubMed] [Google Scholar]
- Chinnery P. F., Zwijnenburg P. J., Walker M., Howell N., Taylor R. W., et al. , 1999. Nonrandom tissue distribution of mutant mtDNA. Am. J. Med. Genet. 85: 498–501. [PubMed] [Google Scholar]
- Connerth M., Tatsuta T., Haag M., Klecker T., Westermann B., et al. , 2012. Intramitochondrial transport of phosphatidic acid in yeast by a lipid transfer protein. Science 338: 815–818. [DOI] [PubMed] [Google Scholar]
- Daheron L., Zenz T., Siracusa L. D., Brenner C., Calabretta B., 2001. Molecular cloning of Ian4: a BCR/ABL-induced gene that encodes an outer membrane mitochondrial protein with GTP-binding activity. Nucleic Acids Res. 29: 1308–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brito O. M., Scorrano L., 2008. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456: 605–610. [DOI] [PubMed] [Google Scholar]
- Dunbar D. R., Moonie P. A., Jacobs H. T., Holt I. J., 1995. Different cellular backgrounds confer a marked advantage to either mutant or wild-type mitochondrial genomes. Proc. Natl. Acad. Sci. USA 92: 6562–6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias J. E., Gygi S. P., 2007. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4: 207–214. [DOI] [PubMed] [Google Scholar]
- Friedman J. R., Lackner L. L., West M., DiBenedetto J. R., Nunnari J., et al. , 2011. ER tubules mark sites of mitochondrial division. Science 334: 358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu K., Hartlen R., Johns T., Genge A., Karpati G., et al. , 1996. A novel heteroplasmic tRNAleu(CUN) mtDNA point mutation in a sporadic patient with mitochondrial encephalomyopathy segregates rapidly in skeletal muscle and suggests an approach to therapy. Hum. Mol. Genet. 5: 1835–1840. [DOI] [PubMed] [Google Scholar]
- Hagstrom E., Freyer C., Battersby B. J., Stewart J. B., Larsson N. G., 2014. No recombination of mtDNA after heteroplasmy for 50 generations in the mouse maternal germline. Nucleic Acids Res. 42: 1111–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnoczky G., Booth D., Csordas G., Debattisti V., Golenar T., et al. , 2014. Reliance of ER-mitochondrial calcium signaling on mitochondrial EF-hand Ca binding proteins: miros, MICUs, LETM1 and solute carriers. Curr. Opin. Cell Biol. 29C: 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinonen M. T., Kanduri K., Lahdesmaki H. J., Lahesmaa R., Henttinen T. A., 2015. Tubulin- and actin-associating GIMAP4 is required for IFN-gamma secretion during Th cell differentiation. Immunol. Cell Biol. 93: 158–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S., Collins S. R., Cassidy-Stone A., Hummel E., Devay R. M., et al. , 2011. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J. Cell Biol. 195: 323–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornum L., Romer J., Markholst H., 2002. The diabetes-prone BB rat carries a frameshift mutation in Ian4, a positional candidate of Iddm1. Diabetes 51: 1972–1979. [DOI] [PubMed] [Google Scholar]
- Jenuth J. P., Peterson A. C., Fu K., Shoubridge E. A., 1996. Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA. Nat. Genet. 14: 146–151. [DOI] [PubMed] [Google Scholar]
- Jenuth J. P., Peterson A. C., Shoubridge E. A., 1997. Tissue-specific selection for different mtDNA genotypes in heteroplasmic mice. Nat. Genet. 16: 93–95. [DOI] [PubMed] [Google Scholar]
- Jokinen R., Battersby B. J., 2013. Insight into mammalian mitochondrial DNA segregation. Ann. Med. 45: 149–155. [DOI] [PubMed] [Google Scholar]
- Jokinen R., Marttinen P., Sandell H. K., Manninen T., Teerenhovi H., et al. , 2010. Gimap3 regulates tissue-specific mitochondrial DNA segregation. PLoS Genet. 6: e1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami Y., Sakuta R., Hashimoto K., Fujino O., Fujita T., et al. , 1994. Mitochondrial myopathy with progressive decrease in mitochondrial tRNA(Leu)(UUR) mutant genomes. Ann. Neurol. 35: 370–373. [DOI] [PubMed] [Google Scholar]
- Kornmann B., Currie E., Collins S. R., Schuldiner M., Nunnari J., et al. , 2009. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325: 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova F., Ramabhadran V., Higgs H. N., 2013. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science 339: 464–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krucken J., Schroetel R. M., Muller I. U., Saidani N., Marinovski P., et al. , 2004. Comparative analysis of the human gimap gene cluster encoding a novel GTPase family. Gene 341: 291–304. [DOI] [PubMed] [Google Scholar]
- Larsson N. G., Holme E., Kristiansson B., Oldfors A., Tulinius M., 1990. Progressive increase of the mutated mitochondrial DNA fraction in Kearns–Sayre syndrome. Pediatr. Res. 28: 131–136. [DOI] [PubMed] [Google Scholar]
- Lietzen N., Ohman T., Rintahaka J., Julkunen I., Aittokallio T., et al. , 2011. Quantitative subcellular proteome and secretome profiling of influenza A virus-infected human primary macrophages. PLoS Pathog. 7: e1001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J., Bennett E. J., 2014. Protecting the proteome: Eukaryotic cotranslational quality control pathways. J. Cell Biol. 204: 467–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMurray A. J., Moralejo D. H., Kwitek A. E., Rutledge E. A., Van Yserloo B., et al. , 2002. Lymphopenia in the BB rat model of type 1 diabetes is due to a mutation in a novel immune-associated nucleotide (Ian)-related gene. Genome Res. 12: 1029–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris D. R., Geballe A. P., 2000. Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 20: 8635–8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto Y., De Camilli P., 1999. Recruitment of an alternatively spliced form of synaptojanin 2 to mitochondria by the interaction with the PDZ domain of a mitochondrial outer membrane protein. EMBO J. 18: 2991–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta T., Takahama Y., 2007. The lymphocyte guard-IANs: regulation of lymphocyte survival by IAN/GIMAP family proteins. Trends Immunol. 28: 58–65. [DOI] [PubMed] [Google Scholar]
- Nitta T., Nasreen M., Seike T., Goji A., Ohigashi I., et al. , 2006. IAN family critically regulates survival and development of T lymphocytes. PLoS Biol. 4: e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okado-Matsumoto A., Fridovich I., 2001. Subcellular distribution of superoxide dismutases (SOD) in rat liver: Cu,Zn-SOD in mitochondria. J. Biol. Chem. 276: 38388–38393. [DOI] [PubMed] [Google Scholar]
- Quintana A., Schwindling C., Wenning A. S., Becherer U., Rettig J., et al. , 2007. T cell activation requires mitochondrial translocation to the immunological synapse. Proc. Natl. Acad. Sci. USA 104: 14418–14423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D., 2003. Finding the right organelle: targeting signals in mitochondrial outer-membrane proteins. EMBO Rep. 4: 948–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland A. A., Voeltz G. K., 2012. Endoplasmic reticulum-mitochondria contacts: function of the junction. Nat. Rev. Mol. Cell Biol. 13: 607–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulteis R. D., Chu H., Dai X., Chen Y., Edwards B., et al. , 2008. Impaired survival of peripheral T cells, disrupted NK/NKT cell development, and liver failure in mice lacking Gimap5. Blood 112: 4905–4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwefel D., Frohlich C., Eichhorst J., Wiesner B., Behlke J., et al. , 2010. Structural basis of oligomerization in septin-like GTPase of immunity-associated protein 2 (GIMAP2). Proc. Natl. Acad. Sci. USA 107: 20299–20304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwefel D., Arasu B. S., Marino S. F., Lamprecht B., Kochert K., et al. , 2013. Structural insights into the mechanism of GTPase activation in the GIMAP family. Structure 21: 550–559. [DOI] [PubMed] [Google Scholar]
- Schwindling C., Quintana A., Krause E., Hoth M., 2010. Mitochondria positioning controls local calcium influx in T cells. J. Immunol. 184: 184–190. [DOI] [PubMed] [Google Scholar]
- Shilov I. V., Seymour S. L., Patel A. A., Loboda A., Tang W. H., et al. , 2007. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 6: 1638–1655. [DOI] [PubMed] [Google Scholar]
- Wai T., Teoli D., Shoubridge E. A., 2008. The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat. Genet. 40: 1484–1488. [DOI] [PubMed] [Google Scholar]
- Weber K., Wilson J. N., Taylor L., Brierley E., Johnson M. A., et al. , 1997. A new mtDNA mutation showing accumulation with time and restriction to skeletal muscle. Am. J. Hum. Genet. 60: 373–380. [PMC free article] [PubMed] [Google Scholar]
- Wong V.W.Y., A. E. Saunders, A. Hutchings, J. C. Pascall, C. Carter et al, 2010. The autoimmunity-related GIMAP5 GTPase is a lysosome-associated protein. Self Nonself 1: 259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K., Carter C., Yoshida N., Abe T., Yamada A., et al. , 2014. Gimap3 and Gimap5 cooperate to maintain T-cell numbers in the mouse. Eur. J. Immunol. 44: 561–572. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.