Abstract
The distribution of fitness effects (DFE) of new mutations is a key parameter in determining the course of evolution. This fact has motivated extensive efforts to measure the DFE or to predict it from first principles. However, just as the DFE determines the course of evolution, the evolutionary process itself constrains the DFE. Here, we analyze a simple model of genome evolution in a constant environment in which natural selection drives the population toward a dynamic steady state where beneficial and deleterious substitutions balance. The distribution of fitness effects at this steady state is stable under further evolution and provides a natural null expectation for the DFE in a population that has evolved in a constant environment for a long time. We calculate how the shape of the evolutionarily stable DFE depends on the underlying population genetic parameters. We show that, in the absence of epistasis, the ratio of beneficial to deleterious mutations of a given fitness effect obeys a simple relationship independent of population genetic details. Finally, we analyze how the stable DFE changes in the presence of a simple form of diminishing-returns epistasis.
Keywords: distribution of fitness effects, evolutionary equilibrium, mutation–selection–drift balance
MUTATIONS are the ultimate source of evolutionary change. Consequently, the distribution of fitness effects (DFE) is a key parameter determining the course of evolution. The DFE of new mutations controls the rate of adaptation to a new environment (Gerrish and Lenski 1998; Good et al. 2012), the genetic architecture of complex traits (Eyre-Walker 2010), and the expected patterns of genetic diversity and divergence (Sawyer and Hartl 1992). To predict any of these quantities, we must first understand the shape of the DFE.
Many attempts have been made to measure the DFE or predict it from biological principles (Eyre-Walker and Keightley 2007). Some studies have sampled directly from the DFE by measuring the fitnesses of independently evolved lines (Zeyl and Devisser 2001; Burch et al. 2007; Schoustra et al. 2009) or libraries of mutant genotypes (Wloch et al. 2001; Sanjuán et al. 2004; Kassen and Bataillon 2006; McDonald et al. 2011). In other experiments, the fates of tracked lineages provide information about the scale and shape of the DFE (Imhof and Schlötterer 2001; Rozen et al. 2002; Perfeito et al. 2007; Frenkel et al. 2014). In natural populations, the DFE leaves a signature in patterns of molecular diversity and divergence, which may be used for inference (reviewed in Keightley and Eyre-Walker 2010). A separate body of work attempts to derive the DFE from simple biophysical models of RNA (Cowperthwaite et al. 2005) or protein (Wylie and Shakhnovich 2011).
Although these experimental and biophysical approaches can provide some insight into the shape of the DFE, they are necessarily specific to a particular organism in a particular environment. In principle, the effects of mutations depend on many biological details that vary from system to system, and it is not clear whether any general predictions are possible. However, all organisms have one thing in common: they are shaped by the process of evolution. While other phenotypes are under different selective pressures in different organisms and environments, fitness is the common currency of natural selection. It is therefore interesting to ask whether we should expect evolution to produce distributions of fitness effects with a predictable shape.
One well-known attempt to predict the shape of the DFE from evolutionary principles is the extreme value theory argument of Gillespie and Orr (Gillespie 1983, 1984, 1991; Orr 2003). This framework assumes that a well-adapted organism is likely to have one of the fittest available genotypes and that the fitnesses of neighboring genotypes are drawn independently from a common distribution. Gillespie and Orr argued that, under these circumstances, the fitness effects of beneficial mutations will follow an exponential distribution (provided the overall distribution of genotype fitnesses satisfies some technical conditions). This prediction has spawned a large body of theory (reviewed in Orr 2010). However, attempts to validate the theory empirically have had mixed results (e.g., Kassen and Bataillon 2006; Rokyta et al. 2008).
A limitation of extreme value theory is that it neglects the evolutionary process that produced the current genotype. Instead, it assumes that the high-fitness genotype is chosen randomly from among genotypes with similar fitness. However, different high-fitness genotypes may have very different mutational neighborhoods, and evolution does not select among these at random. Rather, it will tend to be biased toward regions of genotype space with particular properties, generating high-fitness genotypes with nonrandom mutational neighborhoods. This bias can lead to DFEs that are not well characterized by extreme value theory.
Here, we use an explicit evolutionary model to study how natural selection shapes the DFE in a constant environment. When a population first encounters a given environment, it will either adapt by accumulating beneficial mutations or decline in fitness in the face of an excess of deleterious mutations (Muller’s ratchet). As a population increases in fitness, opportunities for fitness improvement are converted to chances for deleterious back mutation, and the fraction of mutations that are beneficial declines. Conversely, if a population declines in fitness, the fraction of mutations that are beneficial increases. Eventually, the opposing forces of natural selection and Muller’s ratchet balance, and the population reaches a steady state in which fitness neither increases nor decreases on average (Woodcock and Higgs 1996; McVean and Charlesworth 2000; Comeron and Kreitman 2002; Rouzine et al. 2003, 2008; Seger et al. 2010; Goyal et al. 2012). The approach to this steady-state fitness has been observed in laboratory populations (Silander et al. 2007).
As the rate of adaptation slows, the population will also approach an equilibrium state at the molecular level (Mustonen and Lässig 2009). This equilibrium is characterized by a detailed balance, in which beneficial and deleterious mutations of the same absolute effect have equal substitution rates (Berg and Lässig 2003; Berg et al. 2004; Sella and Hirsh 2005; Mustonen and Lässig 2010; Schiffels et al. 2011; McCandlish et al. 2014). Detailed balance holds for every effect size and therefore defines an equilibrium distribution of fitness effects that is stable under the evolutionary process. This distribution serves as a natural null model for the DFE in a “well-adapted” population.
Below, we describe how the shape of the equilibrium DFE depends on the population genetic parameters and the strength of epistatic interactions across the genome. We find that, in the absence of epistasis, the equilibrium DFE has a particularly simple form and that all of the population genetic details may be summarized by a single parameter. Surprisingly, this result holds across regimes featuring very different mutational dynamics, ranging from the weak-mutation case where the equilibrium state is given by Wright’s single-locus mutation–selection–drift balance (Wright 1931) to situations where linked selection is widespread and Wright’s results do not apply (McVean and Charlesworth 2000; Comeron and Kreitman 2002). We then show how epistasis changes both the shape of the equilibrium DFE and its dependence on the population genetic process.
Model
We model a population of N haploid individuals with an L-site genome, a per-genome per-generation mutation rate U, and a per-genome per-generation recombination rate R. Each site has two alleles, one conferring a fitness benefit relative to the other. The (log) fitness difference, , between the two alleles at each site is drawn independently from an underlying distribution with mean . We assume that the relevant fitness differences are small, so that differences between linear and log fitness can be neglected and the standard diffusion limit applies. We initially assume no epistatic interactions among sites: the relative fitness effect of each site is independent of the allelic state of all other sites. This simplest case functions as a null model against which deviations due to epistasis may be compared. In a later section, we expand the model to include some forms of epistasis (including the differences between additive and multiplicative fitness effects).
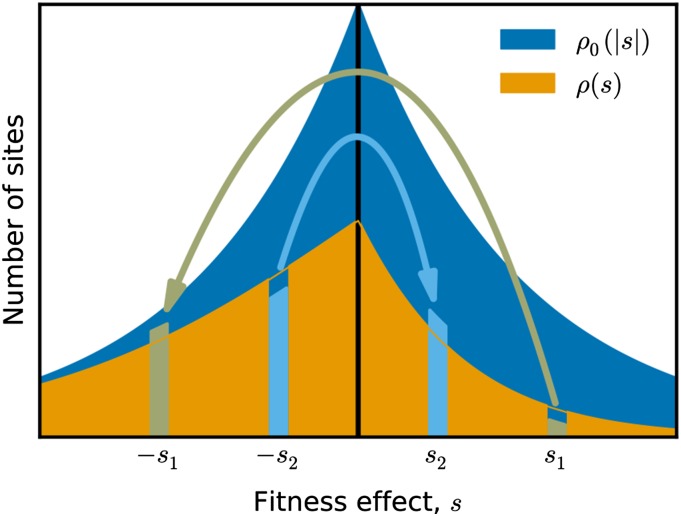
In this model, the distribution of fitness effects, , is determined by the distribution of absolute effects, , and the genotypic state (Figure 1). Sites carrying the deleterious allele have the potential to experience a beneficial mutation and, thus, contribute to the positive side of the DFE. Conversely, sites carrying the beneficial allele contribute to the negative side. We can therefore write the DFE as a sum over the effects of individual sites:
| (1) |
where is the Dirac δ-function, is the absolute effect at site i, and if site i carries the beneficial allele and otherwise. Every mutation modifies the DFE slightly by changing the allelic state at one site, removing the focal mutation from the DFE while creating the opportunity for a back mutation with opposite effect. As the population evolves, the DFE changes until the rate of beneficial substitutions equals the rate of deleterious substitutions at every site. At this steady state, the mean change in fitness is zero and the average distribution of fitness effects is constant.
Figure 1.
The distribution of fitness effects of mutations, , is the product of the distribution of absolute effect sizes, , and the allelic state of each site. A mutation changes the DFE by changing the allelic state at that site. For example, beneficial mutation with effect creates a potential back mutation with effect . Likewise, a deleterious mutation with effect becomes the site of potential beneficial mutation with effect .
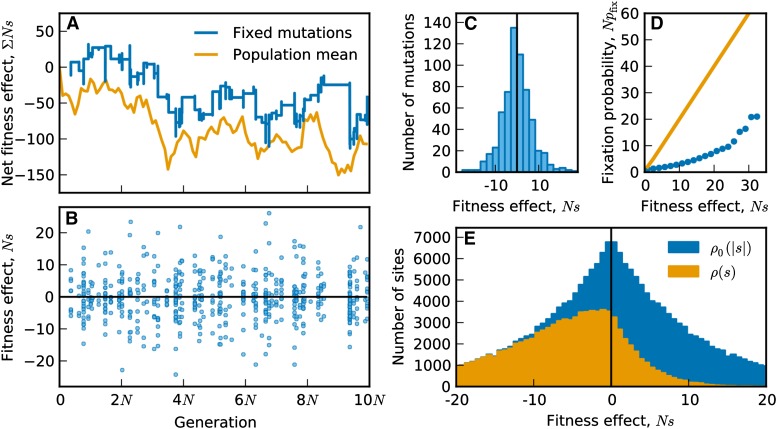
An example of this steady state, generated by a Wright–Fisher simulation of our model, is shown in Figure 2. As observed by Seger et al. (2010), the equilibrium state is not static. Instead, the mean fitness of the population fluctuates over long timescales (Figure 2A, orange line) due to the cumulative fitness effect of multiple beneficial and deleterious substitutions [Figure 2, A (blue line) and B]. Consistent with the steady-state assumption, the fitness effects of fixed mutations are roughly symmetric about zero (Figure 2C), with deviations due to the relatively small size of the sample shown here. The magnitudes of these fixed fitness effects reflect the population dynamics as well as the shape of the equilibrium DFE; in this example, they are at most of order . We discuss the relationship between the fitness effects of fixed mutations and the population parameters below.
Figure 2.
An example of the steady-state dynamics, generated by a Wright–Fisher simulation of our model [, , , , ]. (A) Time course of the mean fitness of the population and the cumulative effect of fixed mutations. (B) The fitness effect of each fixed mutation vs. its fixation time. (C) Histogram of the fitness effects of all fixed mutations. (D) The fixation probability of a beneficial mutation as a function of its fitness effect. Simulation results are shown as circles; the single-locus theory prediction in the absence of linked selection is shown as a solid line. (E) The distribution of absolute effects (blue) and distribution of fitness effects (orange) measured at the end of the simulation.
Our example simulation also illustrates complications that arise due to linked selection. For example, in Figure 2B, we see that mutations mostly fix in clusters. The phenomenon of clustered fixations is a signature of linked selection that has been predicted in theory (Park and Krug 2007) and observed in experimental and natural populations (Nik-Zainal et al. 2012; Strelkowa and Lässig 2012; Lang et al. 2013). In Figure 2D we also see that linked selection reduces the fixation probability of beneficial mutations relative to the standard single-locus prediction (Wright 1931) in a way that cannot be summarized by a simple reduction in the effective population size.
Finally, our simulations reveal the equilibrium shape of the DFE (Figure 2E). In this example, the underlying distribution of absolute effects, , is exponential with mean However, the equilibrium distribution of beneficial effects, , falls off much faster. In fact, almost no beneficial mutations are available with effects greater than
Analysis
The stable distribution of fitness effects
To obtain analytical expressions for the steady-state DFE in our model, we focus on the large L limit, where we can neglect differences in the DFE between genotypes that segregate simultaneously in the population. Furthermore, in the large L limit, the law of large numbers guarantees that fluctuations in the shape of the DFE will be small, even as the mean fitness of the population fluctuates considerably (Figure 2). With this assumption, the average DFE evolves according to the differential equation
| (2) |
where is the fixation probability of a mutation with effect s (Schiffels et al. 2011). The first term on the right-hand side of Equation 2 represents the substitution rate of deleterious mutations with absolute effect , which is the product of the mutation supply rate and the fixation probability. Likewise, the second term gives the substitution rate of beneficial mutations with effect s. Equation 2 captures the fact that each mutation changes the DFE slightly by converting a beneficial mutation to a potential deleterious mutation or vice versa (Figure 1).
At long times, the DFE evolves toward the equilibrium state , in which the substitution rates of beneficial and deleterious mutations exactly balance for every value of s. Setting the time derivative in Equation 2 to zero yields the equilibrium DFE ratio:
| (3) |
We can rewrite Equation 3 in terms of the underlying distribution of absolute effects and the equilibrium state of the genome:
| (4) |
Equation 4 shows that the equilibrium DFE is determined by the relative probabilities of fixation of beneficial and deleterious mutations (Mustonen and Lässig 2007; Schiffels et al. 2011). Unfortunately, there is no general expression for these fixation probabilities because they depend on the effects of linked selection (Hill and Robertson 1966). Moreover, these dynamics of linked selection depend on the shape of the DFE, so the right-hand side of Equation 4 implicitly depends on (Schiffels et al. 2011).
Fortunately, there are two limits of our model where simple expressions for are available. In the limit that mutations are rare (), each mutation fixes or goes extinct independently. Thus, we can use the single-locus probability of fixation (Fisher 1930; Wright 1931),
| (5) |
Substituting Equation 5 into Equation 4 yields
| (6) |
The factor on the right in Equation 6 is characteristic of allelic states in the familiar single-locus mutation–selection–drift equilibrium (Wright 1931).
In the opposite extreme, where the mutation rate is very high, previous work has shown that the probability of fixation depends exponentially on s,
| (7) |
where is the average pairwise coalescence time (Neher et al. 2013; Good et al. 2014). Note that linked selection alters the functional form of and hence cannot be captured by a simple reduction in effective population size. In this strong mutation limit, substituting Equation 7 into Equation 4 shows that the equilibrium DFE has the form
| (8) |
Surprisingly, the shape of the equilibrium DFE has the same dependence on s in both limiting regimes. This is because the ratio falls off exponentially with s when mutation is weak as well as when it is very strong, even though the fixation probabilities have different forms. The fact that the DFE ratio has the same form in two very different limiting regimes suggests that the result may be general. We therefore propose that
| (9) |
where is the scale at which the DFE ratio falls off with s. This single scale encapsulates all of the effects of linked selection and their dependence on the underlying parameters. In the weak mutation limit, , while in the strong mutation limit, . A similar crossover has been noted in models of rapidly adapting populations (Neher and Shraiman 2011; Schiffels et al. 2011; Good et al. 2012; Weissman and Barton 2012; Neher et al. 2013).
To test this conjecture, we calculated the DFE ratio, , across a broad range of parameters for an asexual population with exponential . For each set of parameters, we found the evolutionary equilibrium by varying the initial fraction of sites fixed for the deleterious allele and recording the fitness change in the simulation. We varied the length of the simulations to verify convergence to the steady state (Supporting Information, File S1 and Figure S1). As predicted, we found that the DFE ratio declines exponentially with s for all population parameters (Figure 3A). Similar results are obtained for other choices of and in the presence of recombination (Figure S1). The observed values of varied over three orders of magnitude for the parameters tested (Figure 3A, inset). When mutation is weak, , in accordance with the single-locus intuition. Figure 3A confirms that the limiting analysis above is general: for the purpose of determining the equilibrium DFE, the net result of the complicated mutational dynamics can be summarized by the single parameter .
Figure 3.
The equilibrium DFE and steady-state substitution rate. (A) The equilibrium ratio of the beneficial mutation rate to the deleterious mutation rate for mutations with absolute effect , averaged over 100 replicate simulations. Fitness effects are scaled by a parameter , fitted to the average final DFE for each parameter set. The color of each line indicates the value of for each parameter set (inset). The solid black line shows . (B) The substitution rate of mutations with absolute effect , , declines with the scaled fitness effect . Upper and lower solid curves give the analytical results for the strong- and weak-mutation limits, respectively. All results here are for an asexual population with exponential . The results for other choices of and for are equivalent (Figure S2).
The steady-state substitution rate
While the form of the equilibrium DFE is independent of the mutational dynamics, other features of the steady state depend in detail on the extent of linked selection. For example, as shown in Figure 2, A–C, the steady state is characterized by a constant “churn” of fixations. The distribution of fitness effects of the mutations that fix is symmetrical and its shape is determined by the substitution rate K as a function of . To compare across simulations with different overall mutation rates and underlying DFEs, we define a normalized substitution rate by dividing by the rate at which mutations of effect arise. Using Equations 5 and 7, we can make analytical predictions about the substitution rate in both the weak and strong mutation limits. We find that
| (10) |
Note that has a different functional form in the two limits.
Figure 3B shows the observed substitution rates in our simulations as a function of the scaled fitness effect . Here, the values of the scaling parameter are the values fitted to the equilibrium DFE for each parameter set. The two limiting predictions from Equation 10 are shown as solid curves. These predictions bracket the observed substitution rates. As expected, when , the substitution rates approach the weak mutation limit. On the other hand, when , there is a higher rate of substitution for each value , approaching but not achieving the strong mutation limit.
The relationship between the substitution rate and the effect of the mutation has two notable features. First, the substitution rate declines with the fitness effect, because at equilibrium large-effect sites are almost always fixed for the beneficial allele. Second, unlike the DFE ratio, the substitution rate is not a function of the scaled parameter alone. Instead, populations with large tend to have higher substitution rates of mutations with any given effect than populations with small , due to the effects of linked selection.
The coalescent timescale determines the equilibrium DFE
So far we have treated as a fitting parameter, but we now argue that it can be interpreted in terms of a fundamental timescale of the evolutionary process. Equation 9 shows that is the scale at which a mutation transitions from being effectively neutral to experiencing the effects of selection. This scale is set by the coalescent timescale on which the future common ancestor of the population is determined (Good and Desai 2014). For example, a deleterious mutation with cost s is typically purged from the population in generations. If is much shorter than the time it takes to choose a future common ancestor, the mutant lineage will be eliminated before it has an opportunity to fix. On the other hand, if is much larger than the coalescent timescale, selection will not have enough time to influence the fate of the mutant. We therefore expect to be of order the inverse of the coalescent timescale.
The coalescent timescale depends on the complicated interplay between drift, selection, and interference. Thus, it is difficult to predict from the underlying parameters. Furthermore, the coalescent timescale and the DFE depend on one another and change together as the population evolves. Fortunately, this timescale also determines an independent quantity: the level of neutral diversity within the population. Therefore, we should be able to predict from measurements of diversity in our simulated populations.
To test this expectation, we introduced neutral mutations into our equilibrium simulations and measured the average number of pairwise differences, π, normalized by the expected diversity in a neutrally evolving population of the same size (). As expected, Figure 4 shows that is inversely proportional to . Furthermore, the observed relationship interpolates between the strong-mutation prediction and the weak-mutation prediction . Thus, we can predict the fitted DFE ratio parameter from the neutral pairwise diversity up to an constant.
Figure 4.
Pairwise diversity predicts the equilibrium DFE ratio. For each parameter set, we fitted the scaling parameter to the equilibrium DFE and measured the average number of pairwise differences, π, normalized by the expected diversity in a neutrally evolving population of the same size, .
Diminishing-returns epistasis
In the previous sections, we considered a model without epistasis, where the fitness effect of each site is independent of the state of all other sites. While this provides a useful null model, it is interesting to consider the effect of epistatic interactions on the equilibrium distribution of fitness effects. There are many possible models of epistasis that we could consider. Here, we focus on a simple example suggested by recent empirical work: a general pattern of diminishing-returns epistasis (Chou et al. 2011; Khan et al. 2011; Wiser et al. 2013; Kryazhimskiy et al. 2014).
In the simplest case, this type of epistasis arises when fitness is a nonlinear function of a phenotypic trait. Here, the fitness effect of a mutation is not fixed, but depends on the state of the genome through the current phenotypic value. Specifically, we consider a single fitness-determining phenotypic trait, ξ, controlled by L additive sites,
| (11) |
where is the phenotypic effect of site i and is an indicator variable denoting the allelic state at that site. The fitness of an individual with phenotype ξ is then given by some function . [Note that this form of epistasis includes the case where mutations have additive effects on linear rather than log fitness. In this case, we can take ξ to be linear fitness and .]
With diminishing-returns epistasis, Equation 2 no longer applies because the fitness effect of a mutation depends on the current value of the phenotype. However, because we assume that mutations interact additively at the level of phenotype, we can write an analogous equation for the distribution of phenotypic effects, ,
| (12) |
where is the fitness effect of a mutation of phenotypic effect z that occurs in an individual with phenotype ξ. By analogy to Equation 4, the equilibrium distribution of phenotypic effects, , is then given by
| (13) |
where is the distribution of absolute phenotypic effects and is the equilibrium phenotypic value, which depends on the strength of epistasis. To find the equilibrium distribution of fitness effects, we must change variables from phenotypic effect to fitness effect:
| (14) |
Although Equation 14 is difficult to interpret in general, we can gain qualitative insight by considering the limit of weak diminishing-returns epistasis, where we can expand in the form
| (15) |
Here is a constant that determines the scale at which epistatic effects become important, and since we assume diminishing returns. In this limit, we have
| (16) |
where we have defined
| (17) |
Note that epistasis introduces dependence on the log derivative of the fixation probability: . Unlike the ratio , this quantity does depend on the details of the evolutionary dynamics. For example, in the strong-mutation limit the log derivative is a positive constant, while in the weak-mutation limit it is a positive and increasing function of s. As a result, epistasis has an influence on the equilibrium distribution of fitness effects that cannot be captured by the parameter .
To see the effect of epistasis on the shape of the DFE more concretely, consider the case where the strong-mutation limit applies and . Under these conditions, and the first-order correction in Equation 16 is positive for and negative for . As a result, there are more weakly beneficial mutations and fewer strongly beneficial mutations available in the presence of diminishing-returns epistasis than the nonepistatic analysis would predict.
Discussion
The evolutionary process shapes the distribution of available mutations. Here, we have calculated the equilibrium DFE that evolution produces in a simple null model of a finite genome with no epistasis. Across a wide range of parameters, this equilibrium DFE has the property that falls off exponentially with s. This property holds despite very different population dynamics for different parameters. It is also independent of the shape of the underlying DFE and rate of recombination. The rate of exponential decline depends on the coalescent timescale, which can be predicted from the neutral diversity in the population.
Our results for the equilibrium DFE are strikingly different from earlier attempts to deduce features of the DFE from extreme value theory arguments (Gillespie 1983, 1984, 1991; Orr 2003). According to extreme value theory, the DFE of a well-adapted population depends only on the distribution of genotype fitnesses and not on the particular evolutionary history that brought the population to its well-adapted state. In contrast, we have shown here that the population genetic process (e.g., the historical population size and the mutation rate) can strongly influence the both the shape and the scale of the equilibrium DFE, even when the distribution of genotype fitnesses is held constant. As an example, consider the case where is a half-normal distribution, so that the equilibrium DFE is given by
| (18) |
The equilibrium DFE is thus determined by two scales: , the scale of the underlying DFE, and , the fitness scale at which sites feel the effects of selection strongly. When , selection barely biases the allelic states and is Gaussian. Conversely, when , the equilibrium DFE falls off exponentially for large s. This simple example shows that the shape of the DFE can strongly depend on both the population genetic parameters and the shape of the underlying genotype distribution, and there is no reason to expect it to be exponential in general. In contrast, our analysis predicts that in the absence of epistasis the equilibrium DFE ratio should have a simple exponential form; this can in principle be directly tested experimentally.
Our prediction for the DFE ratio has the same form as standard mutation–selection–drift balance at a single locus, where N is replaced by an effective population size, , which can be estimated from neutral diversity. This drift-barrier intuition forms the basis for many previous empirical studies (Loewe and Charlesworth 2006; Lohmueller et al. 2008; Sung et al. 2012) and theoretical work on the evolution of the mutation rate (Lynch 2011). To some extent, the robustness of this single-locus prediction is surprising, given that it appears to hold even when sites do not evolve independently. Our analysis shows how this simple result emerges more generally and illustrates how it breaks down in the presence of epistasis. In addition, we have shown that the single-locus analysis fails to predict the substitution rate. Thus, while drift-barrier arguments can correctly predict the probability that a given locus is fixed for the beneficial allele, they will often substantially underestimate the rate of fixation of both beneficial and deleterious alleles, even after accounting for the reduction in effective population size.
The continued high rate of fixation illustrates the dynamic nature of the equilibrium that we study here. Rather than approaching a static fitness peak, a population adapting to a constant environment will eventually approach a state of detailed balance. In this state, the rate of substitution of beneficial mutations with a given effect is exactly equal to the rate of substitution of deleterious mutations of the same magnitude. Thus, the mean fitness does not change on average, while the rate of molecular evolution remains high. Depending on the underlying parameters, this population genetic limit to optimization can occur long before any absolute physiological limits become relevant.
In the present work, we have studied only the simplest model of the evolution of the DFE. This null model has several key limitations, which present interesting avenues for future work. Most importantly, we have focused only on evolution in a constant environment. We expect a similar steady state to arise in a fluctuating environment, provided that the statistics of these fluctuations remain constant through time (Gillespie 1991; Mustonen and Lässig 2009). To analyze this more complex situation, we need to understand the distribution of pleiotropic effects of mutations across environmental conditions and how this pleiotropy affects fixation probabilities.
Another important limitation of our model is that we have considered only one specific form of epistasis: a general diminishing-returns model suggested by recent microbial evolution experiments (Chou et al. 2011; Khan et al. 2011; Wiser et al. 2013; Kryazhimskiy et al. 2014). This type of epistasis leads to an excess of weakly beneficial mutations relative to the nonepistatic case, in a way that crucially depends on the population genetic parameters. However, many other types of epistasis may also be common in natural populations. For example, idiosyncratic interactions between specific mutations, including sign epistasis, have been observed in several systems (Weinreich et al. 2006; De Vos et al. 2013). We also often expect to observe modular interactions, in which only the first mutation in each module can confer a fitness effect (Tenaillon et al. 2012). In principle, these and other alternative forms of epistasis can also change the shape of the equilibrium DFE. A quantitative characterization of these changes for more general models of epistasis is an interesting avenue for future research.
Despite these limitations, our analysis provides a useful null model for how the process of evolution shapes the distribution of fitness effects. Our results suggest that experiments should seek to measure the DFE ratio, Equation 3, which in the absence of epistasis is independent of the mutational dynamics or the underlying distribution of effects. Deviations from the null prediction may be informative about the global structure of epistasis or the evolutionary history of the population.
Supplementary Material
Acknowledgments
We thank Daniel Balick, Ivana Cvijović, Elizabeth Jerison, Sergey Kryazhimskiy, David McCandlish, Michael McDonald, Richard Neher, Jeffrey Townsend, and two reviewers for useful discussions and helpful comments on the manuscript. Simulations in this article were run on the Odyssey cluster supported by the FAS Division of Science, Research Computing Group at Harvard University. This work was supported in part by National Science Foundation (NSF) graduate research fellowships (to D.P.R. and B.H.G.), the James S. McDonnell Foundation, the Alfred P. Sloan Foundation, the Harvard Milton Fund, grant PHY 1313638 from the NSF, and grant GM104239 from the National Institutes of Health (to M.M.D.).
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.173815/-/DC1.
Communicating editor: J. Hermisson
Literature Cited
- Berg, J., and M. Lässig, 2003 Stochastic evolution of transcription factor binding sites. Biophysics (Moscow) 48, Suppl. 1: 36–44. [Google Scholar]
- Berg J., Willmann S., Lässig M., 2004. Adaptive evolution of transcription factor binding sites. BMC Evol. Biol. 4: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch C. L., Guyader S., Samarov D., Shen H., 2007. Experimental estimate of the abundance and effects of nearly neutral mutations in the RNA virus ϕ6. Genetics 176: 467–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H.-H., Chiu H.-C., Delaney N. F., Segrè D., Marx C. J., 2011. Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science 332: 1190–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J. M., Kreitman M., 2002. Population, evolutionary, and genomic consequences of interference selection. Genetics 161: 389–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowperthwaite M. C., Bull J., Meyers L. A., 2005. Distributions of beneficial fitness effects in RNA. Genetics 170: 1449–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos M. G. J., Poelwijk F. J., Battich N., Ndika J. D. T., Tans S. J., 2013. Environmental dependence of genetic constraint. PLoS Genet. 9: e1003580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A., 2010. Genetic architecture of a complex trait and its implications for fitness and genome-wide association studies. Proc. Natl. Acad. Sci. USA 107: 1752–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A., Keightley P. D., 2007. The distribution of fitness effects of new mutations. Nat. Rev. Genet. 8: 610–618. [DOI] [PubMed] [Google Scholar]
- Fisher R. A., 1930. The distribution of gene ratios for rare mutations. Proc. R. Soc. Edinb. 50: 204–219. [Google Scholar]
- Frenkel E. M., Good B. H., Desai M. M., 2014. The fates of mutant lineages and the distribution of fitness effects of beneficial mutations in laboratory budding yeast populations. Genetics 196: 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrish P., Lenski R., 1998. The fate of competing beneficial mutations in an asexual population. Genetica 127: 127–144. [PubMed] [Google Scholar]
- Gillespie J., 1983. A simple stochastic gene substitution model. Theor. Popul. Biol. 23: 202–215. [DOI] [PubMed] [Google Scholar]
- Gillespie J., 1984. Molecular evolution over the mutational landscape. Evolution 38: 1116–1129. [DOI] [PubMed] [Google Scholar]
- Gillespie J., 1991. The Causes of Molecular Evolution. Oxford University Press, New York. [Google Scholar]
- Good B. H., Desai M. M., 2014. Deleterious passengers in adapting populations. Genetics 198: 1183–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good B. H., Rouzine I. M., Balick D. J., Hallatschek O., Desai M. M., 2012. Distribution of fixed beneficial mutations and the rate of adaptation in asexual populations. Proc. Natl. Acad. Sci. USA 109: 4950–4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good B. H., Walczak A. M., Neher R. A., Desai M. M., 2014. Genetic diversity in the interference selection limit. PLoS Genet. 10: e1004222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal S., Balick D. J., Jerison E. R., Neher R. A., Shraiman B. I., et al. , 2012. Dynamic mutation-selection balance as an evolutionary attractor. Genetics 191: 1309–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. G., Robertson A., 1966. The effect of linkage on limits to artificial selection. Genet. Res. 8: 269–294. [PubMed] [Google Scholar]
- Imhof M., Schlötterer C., 2001. Fitness effects of advantageous mutations in evolving Escherichia coli populations. Proc. Natl. Acad. Sci. USA 98: 1113–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassen R., Bataillon T., 2006. Distribution of fitness effects among beneficial mutations before selection in experimental populations of bacteria. Nat. Genet. 38: 484–488. [DOI] [PubMed] [Google Scholar]
- Keightley P. D., Eyre-Walker A., 2010. What can we learn about the distribution of fitness effects of new mutations from DNA sequence data? Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 1187–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A. I., Din D. M., Schneider D., Lenski R. E., Cooper T. F., 2011. Negative epistasis between beneficial mutations in an evolving bacterial population. Science 332: 1193–1196. [DOI] [PubMed] [Google Scholar]
- Kryazhimskiy S. K., Rice D. P., Jerison E. R., Desai M. M., 2014. Global epistasis makes adaptation predictable despite sequence-level stochasticity. Science 344: 1519–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G. I., Rice D. P., Hickman M. J., Sodergren E., Weinstock G. M., et al. , 2013. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature 500: 571–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewe L., Charlesworth B., 2006. Inferring the distribution of mutational effects on fitness in Drosophila. Biol. Lett. 2: 426–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller K. E., Indap A. R., Schmidt S., Boyko A. R., Hernandez R. D., et al. , 2008. Proportionally more deleterious genetic variation in European than in African populations. Nature 451: 994–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., 2011. The lower bound to the evolution of mutation rates. Genome Biol. Evol. 3: 1107–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCandlish, D. M., J. Otwinowski, and J. B. Plotkin, 2014 On the role of epistasis in adaptation. arXiv:1401.2508.
- McDonald M. J., Cooper T. F., Beaumont H. J., Rainey P. B., 2011. The distribution of fitness effects of new beneficial mutations in Pseudomonas fluorescens. Biol. Lett. 7: 98–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G. A. T., Charlesworth B., 2000. The effects of Hill–Robertson interference between weakly selected mutations on patterns of molecular evolution and variation. Genetics 155: 929–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustonen V., Lässig M., 2007. Adaptations to fluctuating selection in Drosophila. Proc. Natl. Acad. Sci. USA 104: 2277–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustonen V., Lässig M., 2009. From fitness landscapes to seascapes: non-equilibrium dynamics of selection and adaptation. Trends Genet. 25: 111–119. [DOI] [PubMed] [Google Scholar]
- Mustonen V., Lässig M., 2010. Fitness flux and ubiquity of adaptive evolution. Proc. Natl. Acad. Sci. USA 107: 4248–4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher R., Shraiman B. I., 2011. Genetic draft and quasi-neutrality in large facultatively sexual populations. Genetics 188: 975–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher R. A., Kessinger T. A., Shraiman B. I., 2013. Coalescence and genetic diversity in sexual populations under selection. Proc. Natl. Acad. Sci. USA 110: 15836–15841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S., Loo P. Van, Wedge D. C., Alexandrov L. B., Greenman C. D., et al. , 2012. The life history of 21 breast cancers. Cell 149: 994–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H., 2003. The distribution of fitness effects among beneficial mutations. Genetics 163: 1519–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., 2010. The population genetics of beneficial mutations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Krug J., 2007. Clonal interference in large populations. Proc. Natl. Acad. Sci. USA 104: 18135–18140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfeito L., Fernandes L., Mota C., Gordo I., 2007. Adaptive mutations in bacteria: high rate and small effects. Science 317: 813. [DOI] [PubMed] [Google Scholar]
- Rokyta D. R., Beisel C. J., Joyce P., Ferris M. T., Burch C. L., et al. , 2008. Beneficial fitness effects are not exponential for two viruses. J. Mol. Evol. 67: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzine I., Brunet E., Wilke C., 2008. The traveling-wave approach to asexual evolution: Muller’s ratchet and the speed of adaptation. Theor. Popul. Biol. 73: 24–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzine I. M., Wakeley J., Coffin J. M., 2003. The solitary wave of asexual evolution. Proc. Natl. Acad. Sci. USA 100: 587–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen D. E., de Visser J. A., Gerrish P. J., 2002. Fitness effects of beneficial mutations in microbial populations. Curr. Biol. 12: 1040–1045. [DOI] [PubMed] [Google Scholar]
- Sanjuán R., Moya A., Elena S. F., 2004. The distribution of fitness effects caused by single-nucleotide substitutions in an RNA virus. Proc. Natl. Acad. Sci. USA 101: 8396–8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer S. A., Hartl D. L., 1992. Population genetics of polymorphism and divergence. Genetics 132: 1161–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffels S., Szöllösi G., Mustonen V., Lässig M., 2011. Emergent neutrality in adaptive asexual evolution. Genetics 189: 1361–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoustra S. E., Bataillon T., Gifford D. R., Kassen R., 2009. The properties of adaptive walks in evolving populations of fungus. PLoS Biol. 7: e1000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seger J., Smith W. A., Perry J. J., Hunn J., Kaliszewska Z. A., et al. , 2010. Gene genealogies strongly distorted by weakly interfering mutations in constant environments. Genetics 184: 529–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sella G., Hirsh A. E., 2005. The application of statistical physics to evolutionary biology. Proc. Natl. Acad. Sci. USA 102: 9541–9546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silander O. K., Tenaillon O., Chao L., 2007. Understanding the evolutionary fate of finite populations: the dynamics of mutational effects. PLoS Biol. 5: e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strelkowa N., Lässig M., 2012. Clonal interference in the evolution of influenza. Genetics 192: 671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung W., Ackerman M. S., Miller S. F., Doak T. G., Lynch M., 2012. Drift-barrier hypothesis and mutation-rate evolution. Proc. Natl. Acad. Sci. USA 109: 18488–18492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O., Rodrguez-Verdugo A., Gaut R. L., McDonald P., Bennett A. F., et al. , 2012. The molecular diversity of adaptive convergence. Science 335: 457–461. [DOI] [PubMed] [Google Scholar]
- Weinreich D. M., Delaney N. F., DePristo M. A., Hartl D. L., 2006. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312: 111–114. [DOI] [PubMed] [Google Scholar]
- Weissman D. B., Barton N. H., 2012. Limits to the rate of adaptive substitution in sexual populations. PLoS Genet. 8: e1002740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiser M. J., Ribeck N., Lenski R. E., 2013. Long-term dynamics of adaptation in asexual populations. Science 342: 1364–1367. [DOI] [PubMed] [Google Scholar]
- Wloch D. M., Szafraniec K., Borts R. H., Korona R., 2001. Direct estimate of the mutation rate and the distribution of fitness effects in the yeast Saccharomyces cerevisiae. Genetics 159: 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock G., Higgs P. G., 1996. Population evolution on a multiplicative single-peak fitness landscape. J. Theor. Biol. 179: 61–73. [DOI] [PubMed] [Google Scholar]
- Wright S., 1931. Evolution in Mendelian populations. Genetics 16: 97–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie C. S., Shakhnovich E. I., 2011. A biophysical protein folding model accounts for most mutational fitness effects in viruses. Proc. Natl. Acad. Sci. USA 108: 9916–9921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeyl C., DeVisser J. A. G., 2001. Estimates of the rate and distribution of fitness effects of spontaneous mutation in Saccharomyces cerevisiae. Genetics 157: 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.