Abstract
Allopolyploidization is accompanied by changes in gene expression that are thought to contribute to phenotypic diversification. Here we describe global changes in the single-celled cotton fiber proteome of two natural allopolyploid species (Gossypium hirsutum and G. barbadense) and living models of their diploid parents using two different proteomic approaches. In total, 1323 two-dimensional gel electrophoresis spots and 1652 identified proteins by isobaric tags for relative and absolute quantitation were quantitatively profiled during fiber elongation. Between allopolyploids and their diploid A- and D-genome progenitors, amounts of differential expression ranged from 4.4 to 12.8%. Over 80% of the allopolyploid proteome was additively expressed with respect to progenitor diploids. Interestingly, the fiber proteome of G. hirsutum resembles the parental A-genome more closely, where long, spinable fiber first evolved, than does the fiber proteome of G. barbadense. More protein expression patterns were A-dominant than D-dominant in G. hirsutum, but in G. barbadense, the direction of expression-level dominance switched from the D-genome to the A-genome during fiber development. Comparison of developmental changes between the two allopolyploid species revealed a high level of proteomic differentiation despite their shared ancestry, relatively recent evolutionary divergence, and similar gross morphology. These results suggest that the two allopolyploid species have achieved superficially similar modern fiber phenotypes through different evolutionary routes at the proteome level. We also detected homeolog-specific expression for 1001 proteins and present a novel approach to infer the relationship between homeolog-specific and duplicate expression patterns. Our study provides a proteomic perspective on understanding evolutionary consequences of allopolyploidization, showing how protein expression has been altered by polyploidization and subsequently has diversified among species.
Keywords: additivity, allopolyploid, expression-level dominance, proteomics, Gossypium
POLYPLOIDY is now recognized as a fundamental process in plant evolution, and all flowering plant genomes are known to have experienced several or more rounds of genome doubling in their evolutionary history (Jiao et al. 2011). Compared to the intraspecific genome duplication of autopolyploidy, the formation of allopolyploids entails the merging and doubling of diverged genomes, which has been proposed as an important mechanism of functional and phenotypic evolution driven by structural and regulatory divergence between parental genomes and the attendant duplication of genetic materials (Wendel 2000; Wendel and Doyle 2004; Comai 2005; Doyle et al. 2008; Leitch and Leitch 2008; Soltis and Soltis 2009; Finigan et al. 2012; Madlung 2013; Madlung and Wendel 2013; Ainouche and Wendel 2014).
A large and growing body of literature has demonstrated that allopolyploidy is accompanied by a series of non-Mendelian interactions and processes, including chromosomal rearrangement and variation (Ramsey and Schemske 2002; Szadkowski et al. 2010; Xiong et al. 2011; Chester et al. 2012), DNA sequence elimination (Shaked et al. 2001; Ozkan et al. 2003; Blanc and Wolfe 2004; Han et al. 2005; Skalicka et al. 2005; Anssour et al. 2009; Buggs et al. 2009; Tate et al. 2009; Schnable et al. 2011), recombination between homeologous chromosomes (Gaeta et al. 2007; Szadkowski et al. 2010) and genes (Salmon et al. 2010; Flagel et al. 2012), epigenetic modification (Madlung et al. 2002; Salmon et al. 2005; Bottley et al. 2006; Chen 2007; Gaeta et al. 2007; Kovarik et al. 2008; Ni et al. 2009; Bottley 2014), and differences in small RNA regulation (Kovarik et al. 2008; Ha et al. 2009; Kenan-Eichler et al. 2011; Ng et al. 2011; Woodhouse et al. 2014). With respect to gene expression, a common observation is that the increase of genetic information in allopolyploids leads to different transcriptomic repatterning relative to parental species (Grover et al. 2012a). In particular, nonadditive expression has been studied in allopolyploid species from the perspectives of expression-level dominance, transgressive expression, and homoelog expression bias (reviewed in Yoo et al. 2014). The first two phenomena describe the total expression of a homeolog pair and up-/down-regulation relative to the diploid parents, whereas homeolog expression bias quantifies cases of unequal contribution of two homeologs to total gene expression (Grover et al. 2012a; Yoo et al. 2014).
The advent and subsequent widespread use of microarray and next-generation sequencing technologies have led to a rapid increase in exploration of gene expression at the transcriptional level (Adams et al. 2003; Hegarty et al. 2005; Bottley et al. 2006; Wang et al. 2006; Gaeta et al. 2007; Flagel et al. 2008; Hovav et al. 2008; Chaudhary et al. 2009; Rapp et al. 2009; Buggs et al. 2010a; Buggs et al. 2010b; Chague et al. 2010; Chelaifa et al. 2010; Flagel and Wendel 2010; Koh et al. 2010; Bardil et al. 2011; Chelaifa et al. 2013; Yoo et al. 2013). However, the transcriptome itself is insufficient for understanding the end products of gene expression and phenotypic outcomes (Rose et al. 2004; Thelen and Peck 2007; Karr 2008; Vogel and Marcotte 2012; Ponnala et al. 2014). Because proteins are the major catalysts of cellular activities, the phenotype of an organism arguably may be more directly related to protein abundance and function than to transcriptional abundance (Karr 2008; Diz et al. 2012). Thus, comparative proteomics offers an important perspective on evolutionary processes.
Only a few studies have examined the outcome of gene expression at the protein level in allopolyploids with respect to their diploid progenitors. Using a two-dimensional gel electrophoresis (2-DE) approach or isobaric tags for relative and absolute quantitation (iTRAQ), interspecific comparisons of protein presence and abundance have been performed in wheat (Bahrman and Thiellement 1987; Islam et al. 2003), Brassica napus (Albertin et al. 2006, 2007), Arabidopsis (Ng et al. 2012), Tragopogon (Koh et al. 2012), and cotton (Hu et al. 2011) that demonstrated examples of nonadditive expression at the protein level. In cotton seeds (Hu et al. 2011), a high degree of proteomic variation was found among diploid and allopolyploid proteomes, with a biased accumulation of seed storage proteins from one of the two progenitors. iTRAQ, a gel-free method, was used to investigate proteomic variation with respect to polyploidy in Arabidopsis (Ng et al. 2012) and Tragopogon (Koh et al. 2012) and to study evolutionary changes accompanying two independent cotton domestication events (Hu et al. 2013, 2014). The 2-DE and iTRAQ methods are known to produce complementary results (Rose et al. 2004; Thelen and Peck 2007). For example, only proteins within a narrow range of molecular mass, isoelectric point (pI), and hydrophobicity can be resolved by 2-DE analysis, and identifying proteins is time-consuming and often insufficient to assign functional annotations to the expression patterns of interest. In iTRAQ analysis, protein quantification is based upon MS signals of labeling tags attached to peptides, which allows quantitative comparisons between differentially labeled samples but leaves relative protein abundance within the same sample difficult to measure. It is not clear, for interspecific comparisons in evolutionary studies, how these technical differences between the two approaches would affect proteomic inferences in a manner comparable to those described for transcriptomic studies.
Our goal in this study was to use both 2-DE and iTRAQ proteomic methods to investigate the effects of genome doubling on allopolyploid cotton proteomes. One key advantage of this system is that we could select a relatively simple morphologic structure for evolutionary analysis, namely, the single-celled epidermal trichome colloquially termed cotton “fiber.” A second advantage is that we were able to simultaneously study two natural allopolyploid species, Gossypium hirsutum (AD1) and G. barbadense (AD2), that diversified from the same polyploidy event approximately 1–2 million years ago (Grover et al. 2012b; Wendel et al. 2012) and that were independently domesticated in parallel at least 5000 years ago (Wendel et al. 2012). These allopolyploids were studied in parallel relative to models of their diploid progenitors, the A-genome species G. arboreum (A2) and the D-genome species G. raimondii (D5). This work offered us the opportunity to determine the proteomic consequences of polyploidization and generated a rich database to potentially provide insights into proteomic underpinnings of cotton fiber development.
Materials and Methods
Plant materials and protein extraction
Four Gossypium species were used in this study, including two natural allopolyploids, G. hirsutum (AD1) cultivar Acala Maxxa and G. barbadense (AD2) cultivar Pima S-6, and models of their diploid progenitors, the A-genome species G. arboreum (A2) and the D-genome species G. raimondii (D5) (Figure 1A). Three plants per species, as biological replicates, were grown in the Bessey Hall Greenhouse at Iowa State University. Flowers were tagged at anthesis, and developing bolls were harvested at 10 and 20 days postanthesis (dpa), representing the key developmental stages (Kim and Triplett 2001; Wilkins and Arpat 2005; Haigler et al. 2012) of primary wall synthesis and fast elongation (10 dpa) and the transition to secondary wall synthesis (20 dpa). Collected bolls were dissected immediately after harvest, and ovules were frozen in liquid nitrogen and stored at −80°.
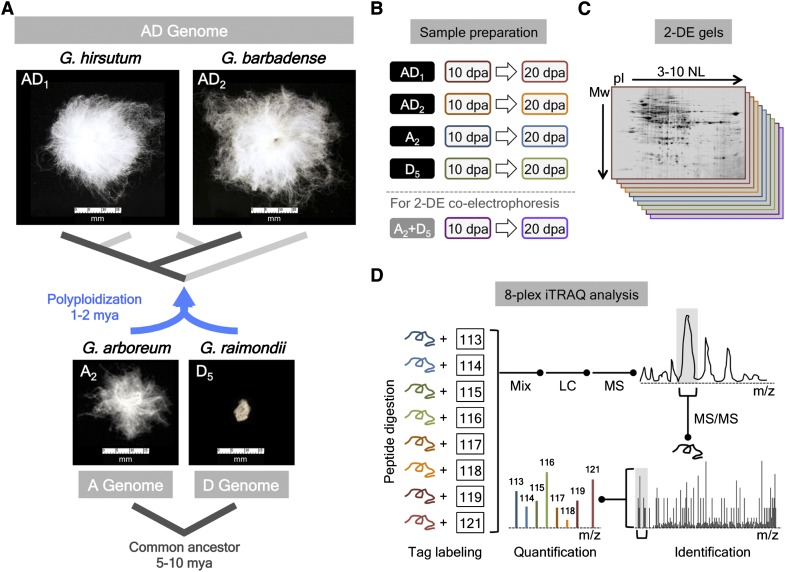
Figure 1.
Comparative proteomics of diploid and allopolyploid cotton fibers. (A) Phylogenetic framework of diploid and allopolyploid Gossypium illustrating the evolutionary history of allopolyploid AD-genome cottons that diversified from a common polyploidization event between the A- and D-genome diploids. A representative image of a single seed with attached fibers is shown for each species. (B) For sample preparation, total proteins were extracted from 10- and 20-dpa fibers in each species for 2-DE and iTRAQ analyses, and in addition, the diploid A- and D-genome protein extracts were mixed to a ratio of 1:1 for coelectrophoresis in 2-DE analysis. (C) The extracted proteins and diploid protein mixtures were separated in 2-DE experiments with a nonlinear IEF range of pH 3–10, and the resulting gels were subjected to image analysis for spot detection and protein quantification based on spot volumes. (D) In iTRAQ analyses, extracted proteins were separately digested and labeled with iTRAQ tags, and the combined peptide mixture was subjected to liquid chromatography (LC) coupled with tandem mass spectrometry (MS) analyses. The iTRAQ reagents allow simultaneous identification and quantitation of proteins in eight different samples.
2-DE and iTRAQ proteomic analyses
Total proteins were isolated from fiber tissues as described in Hu et al. (2013), and proteomic experiments were performed using both the gel-based 2-DE (Figure 1, B and C) and LC-based iTRAQ (Figure 1D) techniques (Hu et al. 2014). The 2-DE experiments were carried out as described in Hu et al. (2011, 2014). Comparative quantification of spot volumes was conducted to assess differential expression for contrasts of interest. Significant changes were identified based on an ANOVA with a P-value of <0.05 and a fold-change cutoff of >1.2 or <0.8.
iTRAQ analysis was conducted using the iTRAQ Reagents 8plex Kit according to the manufacturer’s instructions (AB Sciex, Foster City, CA). The diploid A2 proteins were labeled with iTRAQ tags 113 (10 dpa) and 114 (20 dpa), D5 proteins were labeled with tags 115 (10 dpa) and 116 (20 dpa), AD2 proteins were labeled with tags 117 (10 dpa) and 118 (20 dpa), and AD1 proteins were labeled with tags 119 (10 dpa) and 121 (10 dpa), respectively. After labeling, the peptides were fractionated with strong cation exchange (SCX) using an Agilent HPLC System 1260 with a polysulfoethyl A column (2.1 × 100 mm, 5 µm, 300 Å; PolyLC, Columbia, MD), and 12 final fractions were lyophilized and analyzed using a quadrupole time-of-flight QSTAR Elite MS/MS system (AB Sciex), as described previously (Hu et al. 2014). Protein identification from the MS/MS data was performed by a thorough database search considering biological modifications and amino acid substitutions using the Paragon search algorithm in the ProteinPilot version 4.5 software (AB Sciex), with the cutoff set to a confidence level of 95% and the global false discovery rate (FDR) under 1.0% (Supporting Information, Figure S1, Table S1). Proteins were quantified with at least three spectra based on peak intensities of iTRAQ labeling tags coupled with normalization and bias correction, as described previously (Hu et al. 2014). An expression change was considered to be significant only when the protein fold change was quantified as >1.2 or <0.8 with P < 0.05 in at least two of three biological replicates, along with a Fisher’s combined probability of <0.05 (Fisher 1948).
Protein functional analysis and hierarchical clustering
Identified fiber proteins were annotated with gene ontology categories using Blast2GO (Conesa et al. 2005) and assigned to protein families using PANTHER (Mi et al. 2010). Functional enrichment analyses were performed using the Single Enrichment Analysis (SEA) tool of agriGO (Du et al. 2010). For SEA, a list of proteins of interest was compared to the reference list of identified Gossypium proteins to acquire enriched GO terms using Fisher’s test with P < 0.05 and a minimum of five mapping entries. Hierarchical clustering with regular bootstrap probability (BP) and approximately unbiased bootstrap (AU) P-values was performed using the R software package pvclust (Suzuki and Shimodaira 2006), specifying average linkage and Pearson’s correlation distance metric with 10,000 iterations.
Differentiation of protein orthologs and homeologs
Three protein databases were used for iTRAQ protein identification, including a nonredundant Gossypium protein database and two separate A- and D-genome diploid databases, as described by Hu et al. (2013). To comprehensively identify fiber proteins from multiple cotton species, the MS/MS data were first searched against the nonredundant Gossypium protein database. This step provided identification of protein orthologs in A- and D-genome diploids and their counterparts in allopolyploids representing the combined expression of homeolog pairs (AT and DT, where the subscript indicates the specific genome in the allopolyploid). The derived data were used to analyze the total expression level of homeolog pairs for a given protein in allopolyploids relative to expression levels in the parental diploids in parallel to 2-DE analyses.
To distinguish the expression patterns between homeologs in allopolyploids, i.e., homeolog-specific expression, the MS/MS data were subsequently searched against separate diploid A- and D-genome databases using the same ProteinPilot parameters described earlier. Based on the amino acid difference between protein orthologs in the diploid protein databases, A- or D-specific peptides were identified by comparing the separately generated peptide lists. For an A- or D-specific protein in allopolyploids, i.e., the AT or DT homeolog, expression ratios were calculated based on only the peptides specific to the corresponding diploid genome; to be considered differentially expressed significantly between sample conditions, a protein homeolog needed to be diagnosed with at least three genome-specific peptides from at least two of the replicates and a fold change ≠ 1 with P < 0.05 using Student’s t-test.
Additivity test and categorization of expression patterns in allopolyploid proteomes
The hypothesis of additive expression in the allopolyploids was tested using 2-DE data only, where a spot was considered additive if its expression in the allopolyploid genome was statistically equivalent to the average of expression levels in the parental A- and D-genomes. Coelectrophoresis of a 1:1 mixture of A2 and D5 proteins was conducted to obtain the average expression of the diploid parents (Figure 1B, below dash line). To ensure the reliability of this experiment, coelectrophoretic values were compared to the arithmetic average of parental values by Student’s t-tests (P < 0.05). Any protein expression in allopolyploids that significantly deviated from the coelectrophoretic value was considered to be nonadditive in the form of up- or down-regulation relative to the average parental values.
For both 2-DE and iTRAQ data, protein expression in allopolyploids was categorized into six possible patterns based on their expression levels relative to the diploid parents (illustrated in Figure 5). First, conserved expression was inferred if the expression level in the allopolyploid was statistically equivalent to that in both diploid parents; if it was equivalent to the value of only one parent, expression-level dominance was inferred in the direction of this parent irrespective of whether expression was increased or decreased relative to the other parent, as in Grover et al. (2012a). For protein expression levels that were significantly different from both parental values, median or transgressive expression was inferred depending on whether the expression in the allopolyploid statistically fell between or outside the range of the two diploid parents.
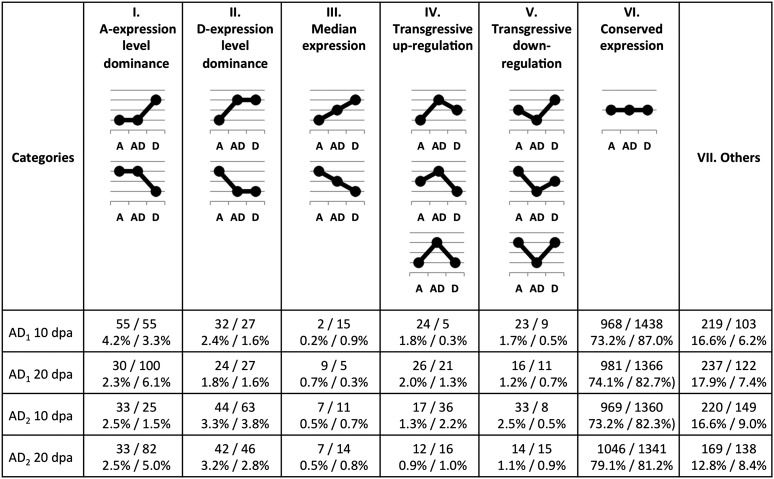
Figure 5.
Categorization of protein expression in allopolyploid proteomes at each developmental stage. Possible patterns of allopolyploid (AD) expression relative to their diploid parents (A and D) were tabulated for each category. Results based on 2-DE and iTRAQ data sets are presented in pairs. For example, for the developmental stage 20 dpa within G. barbadense (AD2), 33 and 82 proteins estimated by 2-DE and iTRAQ analyses, respectively, fall into expression category I, which accounts for 2.5% of 1323 2-DE spots and 5.0% of 1652 iTRAQ proteins profiled.
Results
Comparative proteomics of cotton fibers using 2-DE and iTRAQ methods in parallel
To establish a comparative fiber proteome database, cotton fibers were examined in two natural allopolyploids, G. hirsutum (AD1) and G. barbadense (AD2), and representatives of their diploid progenitors, G. arboreum (A2) and G. raimondii (D5), using two key developmental stages per species—10 and 20 dpa—representing fast fiber elongation and the transition from primary to secondary wall synthesis, respectively. For the twenty-four 2-DE gels using a nonlinear isoelectric focusing range (IEF) of pH 3–10, a total of 1323 protein spots was detected and reproducibly cross-matched (Figure S1 and File S1). The log10-transformed spot-volume distribution was near normal for each sample condition, and its dynamic range represented proteins spanning approximately three orders of magnitude of expression within cotton fiber cells (Figure S2). The quantitative 2-DE spot profiles were used for intra- and interspecific comparisons of global protein features in the subsequent analyses (File S2).
To complement the 2-DE approach, iTRAQ was carried out for protein identification and quantification (Figure 1D). A total of 1652 fiber proteins was identified (95% confidence level) with a less than 1% FDR (Table S1, File S3, and File S4), among which 1001 proteins were diagnosed with ortholog-specific peptides with respect to A- and D-genome diploid species, as well as homeolog-specific (AT, DT, or both) peptides in allopolyploid species, accounting for 60% of their fiber proteome characterized. The identified fiber proteins represent almost all PANTHER protein functional families encoded in the Gossypium genome, with over a third of the proteins classified into oxidoreductase (12.4%), nucleic acid–binding protein (11.5%), and hydrolase (11.0%) categories (Figure S3).
Differential protein expression during fiber development
To study the developmental profiles of cotton fiber proteins, we first performed pairwise comparison between 10- and 20-dpa fibers to assess the proteome dynamics during fiber development within each of the four species studied. Based on 2-DE spot profiles, 4.5–9.6% of the fiber proteomes were differentially expressed during fiber development from 10 to 20 dpa depending on the species, with more developmental changes observed in both diploids (A2: 127 spots; D5: 97 spots) and in G. hirsutum (AD1: 100 spots) than in the other allopolyploid cotton, G. barbadense (AD2: 59 spots; P < 0.05, Fisher’s exact test). Using the iTRAQ approach, a similar level of proteomic dynamics was detected, with 4.1–9.9% of identified proteins being differentially expressed from 10 to 20 dpa; however, contrary to the 2-DE results, AD2 exhibited the highest number of expression changes (163 proteins), with fewer changes observed in the other three species (AD1: 68 proteins; A2: 67 proteins; D5: 116 proteins; P < 0.05, Fisher’s exact test). When comparing the lists of differentially expressed proteins across species, both proteomic approaches revealed only small overlaps of the fiber-developmental variation; expression changes of only about 1% of fiber protein features were shared between two diploid species or between the two allopolyploid species (Figure 2, A and B). This demonstrates that proteome differences during fiber development are mostly unique to each species.
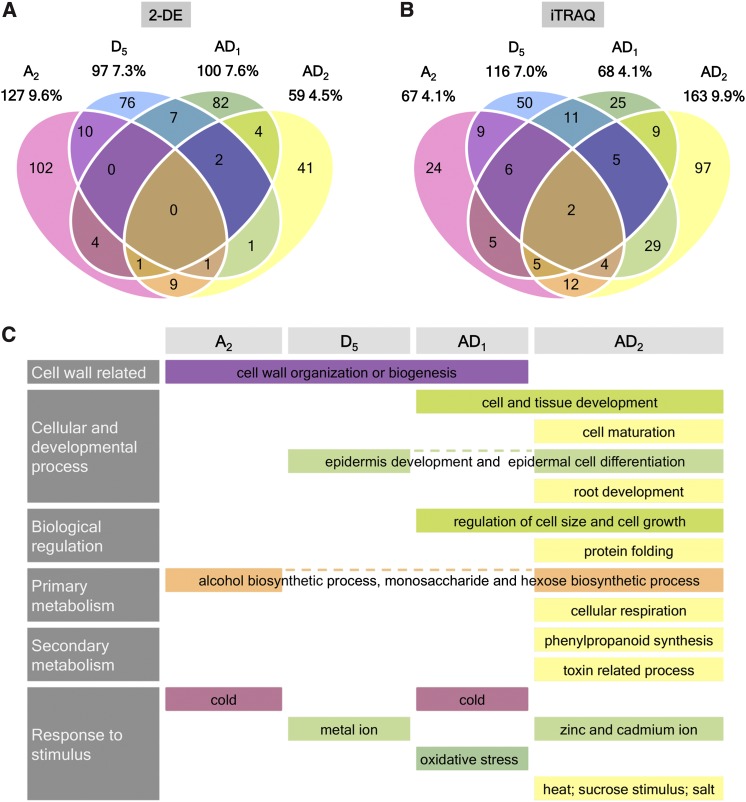
Figure 2.
Differential protein expression and biological processes from 10 to 20 dpa. Venn diagram shows the numbers of differentially expressed proteins between species using (A) 2-DE, and (B) iTRAQ. (C) Functional enrichment analysis revealed important biological processes during fiber elongation within each species. Detailed GO terms and enrichment tests are listed in File S5.
To gain insight into the cellular functions and biological processes associated with fiber development in the different cotton species, functional enrichment analyses were performed for differentially expressed proteins from iTRAQ analysis (File S5). As shown in Figure 2C, comparisons of the significantly enriched GO terms showed that proteins related to cell wall organization or biogenesis were differentially regulated in both the diploid species and AD1, but not in AD2, despite more GO terms being enriched in AD2 from 10 to 20 dpa. Among the cellular and developmental processes, interestingly, epidermis development and epidermal cell differentiation were represented by AD2 and its D-genome progenitor, which has short, nonspinable fiber. Therefore, the rapid fiber growth and elongation from 10 to 20 dpa, as the signature phenotype for both domesticated allopolyploid species, is likely to be related to proteins of cell and tissue development and regulation of cell size and cell growth, which were enriched in both AD1 and AD2. Among metabolic processes, AD2 and its A-genome progenitor exhibited functional enrichment of alcohol biosynthetic process, monosaccharide biosynthetic process, and hexose biosynthetic process, which are known to play essential roles in energy production and to supply building blocks for cell wall synthesis. In the category of cellular response to stimulus, proteins involved in various stress responses were differentially regulated in all four cotton species, including cold (A2 and AD1), metal ion (D5 and AD2), oxidative stress (AD1), and heat, salt, and sucrose stimulus (AD2).
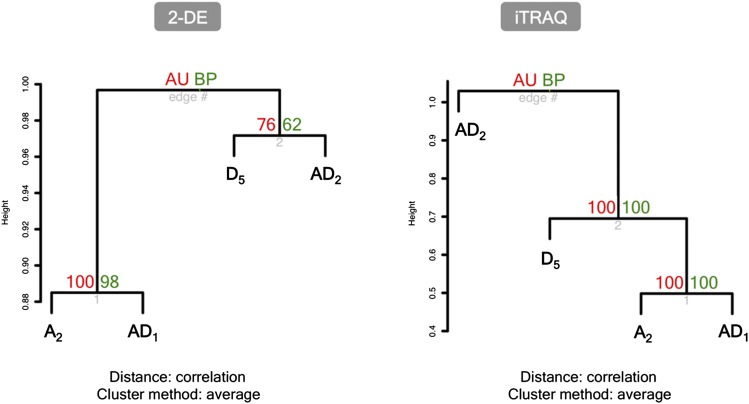
Based on the log2-transformed expression ratios of 20–10 dpa within each species, hierarchical clustering analysis revealed a cluster of A2 and AD1 proteomes (over 95% BP values) using both the 2-DE and iTRAQ data sets (Figure 3). This higher level of developmental similarity between G. hirsutum and its diploid A-genome progenitor G. arboreum also was confirmed by pairwise concordance tests, where the most significant positive correlations were consistently detected between A2 and AD1 (Figure S4). These results suggested that the proteomic changes during fiber development vary substantially among different cotton species, even between the allopolyploid species that trace to the same allopolyploid event 1–2 million years ago. Compared to the allopolyploid AD2 species G. barbadense, the protein-level changes in developing fibers of G. hirsutum are more similar to those of the A-genome diploid progenitor G. arboreum.
Figure 3.
Hierarchical clustering of diploid and allopolyploid cotton according to developmental expression changes. Log2 fold changes of 20 vs. 10 dpa were analyzed for 2-DE and iTRAQ data sets. The percentages of bootstrap probabilities (BP) and approximately unbiased bootstrap probabilities (AU) indicated how strongly each cluster is supported by the data.
Proteomic variation among diploid and allopolyploid cottons
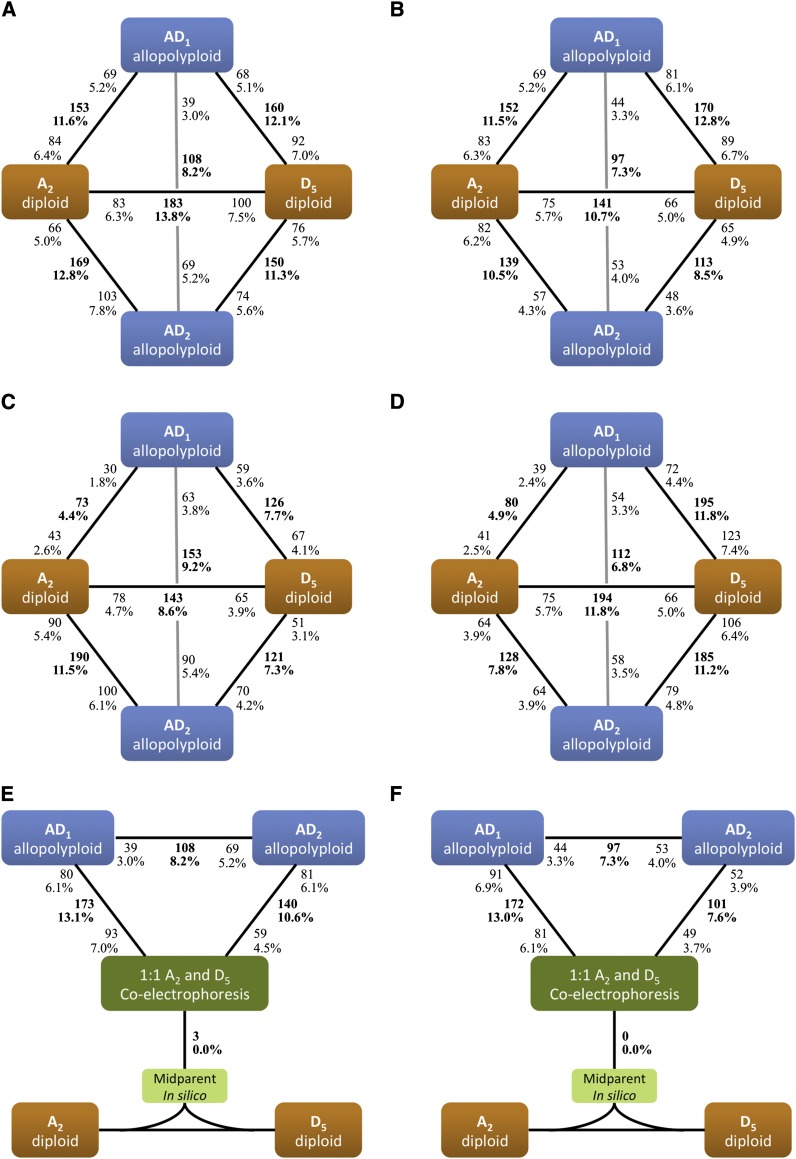
To explore variability of protein expression patterns among diploid and allopolyploid cotton fibers, we directly compared proteomes of different species at the same developmental time point. As shown in Figure 4, A and B, 2-DE profiling revealed 183 (13.8%) and 141 (10.7%) spots that were differentially expressed between the diploid progenitors at 10 and 20 dpa, respectively, a higher level of proteomic divergence than that between the two allopolyploid species, where 108 (8.2%) and 97 (7.3%) proteins were differentially expressed at 10 and 20 dpa, respectively (P < 0.05, Fisher’s exact test). For each allopolyploid, similar amounts of differential expression were found relative to the two diploid progenitors; for example, at 10 dpa (Figure 4A, upper triangle), statistically equivalent amounts of AD1 proteins were differentially expressed compared to A2 and D5 (11.6 vs. 12.1%, respectively; P > 0.05, Fisher’s exact test). As such, according to the 2-DE results, the allopolyploid proteomes were symmetrically different during fiber development from the two parental diploid proteomes.
Figure 4.
Number of proteins differentially expressed among diploid and allopolyploid species. Bold text indicates the total number and percentage of proteins differentially expressed in each comparison. The total number of proteins is partitioned into both directions using bold text; e.g., of the 153 proteins differentially expressed between G. hirsutum and G. arboreum in A, 69 and 84 were up-regulated in the former and latter species, respectively. For 2-DE comparisons at 10 (A) and 20 dpa (B), percentages were calculated based on a total of 1323 spots detected on gels. For iTRAQ comparisons at 10 (C) and 20 dpa (D), percentages were calculated based on 1652 nonredundant proteins. Using coelectrophoresis as control, additivity of protein expression in allopolyploids was tested at 10 (E) and 20 dpa (F) for 2-DE data.
The proteomic variations resulting from iTRAQ analysis exhibited several differences relative to the 2-DE results, as shown in Figure 4, C and D. For example, the percentages of differential expression between parental diploids and between the two allopolyploids were statistically equivalent at 10 dpa (8.6 vs. 9.2%, respectively; P > 0.05, Fisher’s exact test) (Figure 4C), whereas at 20 dpa, more differentially expressed proteins were found between parental diploids than between the two allopolyploids (11.8 vs. 6.8%; P < 0.05, Fisher’s exact test), in agreement with the 2-DE patterns (Figure 4, A and B). For both allopolyploids, the amounts of differential expression relative to diploid parents were unbalanced or asymmetrical (P < 0.05; Fisher’s exact test); i.e., global expression patterns in allopolyploids were biased toward one parental diploid. The proteomic expression of allopolyploid AD1 was closer to that of A2 than to that of D5 at both time points. Interestingly, the directional bias changed for AD2 during fiber development; i.e., overall protein expression more closely mirrored D5 at 10 dpa but switched toward A2 at 20 dpa.
GO enrichment analyses of differentially expressed proteins were performed for the iTRAQ data (File S5). Between the two diploid species, differentially regulated biological processes were related to cell death, cellular homeostasis, phenylpropanoid biosynthesis, and various responses to stimulus at 10 dpa, while biological processes of cell development, epidermal cell differentiation, transport, secondary metabolism, stress, and stimulus response were enriched at 20 dpa. Interestingly, all these biological processes also were overrepresented between the two allopolyploid species AD1 and AD2 at 10 dpa, in addition to primary metabolism of protein, carbohydrate, and nucleotide. At 20 dpa, far fewer GO terms were enriched [17 vs. 93 biological processes (P) in File S5], related to phenylpropanoid biosynthesis, glycolysis, cell wall organization, and response to stress and metal ion. These results suggest a high degree of functional divergence between the two allopolyploid species with respect to fiber development, especially at 10 dpa. When comparing the allopolyploid proteomes to those of their diploid parents, more biological processes in AD1 were different relative to D5 than to A2 (10 dpa: 38 vs. 6; 20 dpa: 99 vs. 5) (File S5), in agreement with the asymmetrical pattern of differential expression (Figure 4, C and D), suggesting that the developing fiber proteome of G. hirsutum is functionally more similar to that of its A-genome diploid progenitor. In contrast to AD1, more GO terms were overrepresented between AD2 and A2 than between AD2 and D5 at 10 dpa, and the numbers of enriched GO terms were similar at 20 dpa (10 dpa: 48 vs. 23; 20 dpa: 16 vs. 14) (File S5). As expected from the high degree of functional divergence between AD1 and AD2, only a few biological processes in allopolyploid cotton were commonly altered relative to each of the diploid species, and they were related to response to chemical stimulus, including reactive oxygen species and inorganic substances.
Additivity test and expression-level dominance in allopolyploid cotton
As a prelude to test the additivity of parental contributions to the allopolyploid proteome, coelectrophoreses of 1:1 A2 and D5 proteins were compared to the average of their independent 2-DE values; only three spots were significantly different using this comparison at 10 dpa, and no differences were found at 20 dpa, much lower than expected by chance at a 1% FDR (13 of 1323 spots). These results justify the use of coelectrophoretic patterns as a control to detect spots in allopolyploids that deviate significantly in abundance from the average of the parental diploids, i.e., to detect nonadditive protein expression. As shown in Figure 4, E and F, approximately 13% of the spots were detected as nonadditive in AD1 at both time points, while the percentage of nonadditive spots in AD2 decreased from 10.6% at 10 dpa to 7.6% at 20 dpa; these nonadditive expressions were equally distributed in the direction of up- and down-regulation relative to the parental average (P > 0.05, Fisher’s exact test).
Regardless of protein expression relative to the average of parental diploids, categorization of expression patterns in allopolyploids was applied to 2-DE and iTRAQ data sets in parallel with respect to each of the diploid parents. Thus, we binned proteins into six possible categories as follows: most of the fiber proteins, accounting for more than 70% of the allopolyploid proteomes, displayed conserved expression with levels in both parents (Figure 5, category VI), while fewer than 1% displayed expression values that were intermediate to but statistically different from those of both parents when the two parents exhibited differential expression (Figure 5, category III). Expression-level dominance of either parental genome (Figure 5, categories I and II) was evident for 4.1–7.8% of the proteins expressed in allopolyploid fibers when statistical equivalence of expression levels was diagnosed between the allopolyploid and only one of its diploid progenitors. According to both the 2-DE and iTRAQ data sets, AD1 exhibited higher amounts of expression-level dominance in the direction of the A-genome than the D-genome (P < 0.05, Fisher’s exact test); following Grover et al. (2012a), expression-level dominance displayed by AD1 was unbalanced toward the A-genome parent. Likewise, unbalanced expression-level dominance of AD2 was suggested by iTRAQ data only, the direction of which, however, was toward the D-genome at 10 dpa but switched to the A-genome at 20 dpa. Novel expression patterns of transgressive up-regulation (Figure 5, category V) or down-regulation (Figure 5, category IV) were identified for 0.8–3.8% of the proteins in allopolyploid fibers. The remaining proteins that were statistically excluded from these categories were grouped together (Figure 5, category VII), accounting for a higher percentage of 2-DE spots than iTRAQ proteins, which may reflect the variable sensitivities of differential expression analysis of these two methods owing to their technical differences.
Homeolog-specific expression in allopolyploid cotton
Among the 1001 proteins identified with genome-diagnostic peptides with respect to at least one of the two diploid parental genomes, 423 proteins in allopolyploids had diagnostic peptides detected for both homeologs (AT and DT) in the fiber proteomes. For each identified peptide, iTRAQ analysis enabled relative quantification of expression levels in fiber proteins from diploid and polyploid species (File S6). For example, a D-genome-specific protein can be detected in the diploid D-genome and allopolyploid fibers, with their quantitative comparison represented by expression level of a DT homeolog in allopolyploid relative to that of the parental protein in the diploid D-genome (i.e., DT/D); likewise, the AT/A ratios can be calculated for an A-genome-specific protein. Because an A- or D-genome-specific protein, in principle, does not exist in the other diploid genome, unexpected false expression of the genome-diagnostic peptides (measured as Afalse/D ≥ 1 or Dfalse/A ≥ 1; P < 0.05, Student’s t-test) can be used to filter out problematic proteins, whose iTRAQ signals were possibly affected by background noise and/or inaccurate A vs. D amino acid differences present in the current diploid protein databases.
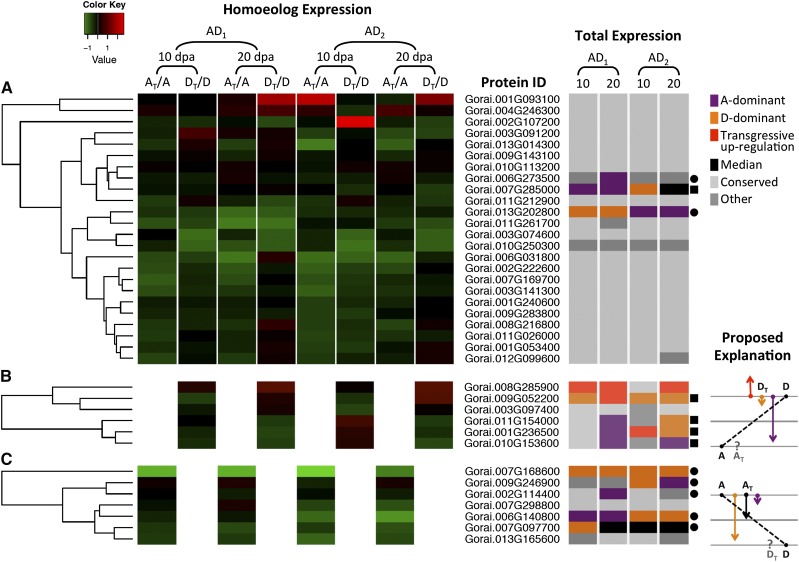
To enable the statistical inference of AT/A and DT/D ratios, an orthologous and homeologous protein was considered only when at least three genome-diagnostic peptides were measured from at least two of the three biological replicates. A total of 37 proteins that met these criteria was examined for their homeolog-specific expression patterns (Figure 6, left heat map). Notably, the AT/A and DT/D ratios were mostly below 1 (green in heat map), suggesting lower expression levels for homeologs relative to their parental proteins in the diploids. This observation was expected given that the total expression of a protein in allopolyploid cotton is the summed value of AT and DT expression and that equal amounts of fiber proteins were studied from diploid and polyploid cotton species. Interestingly, a few cases were found with ratios above 1 (red in heat map), indicating at least a twofold up-regulation of the corresponding homeologs based upon the additivity hypothesis in allopolyploids.
Figure 6.
Coanalysis of homeolog expression and total protein expression categories in allopolyploid cotton. Based on expression ratios of AT/A and DT/D on a log2 scale, heat maps were plotted separately for proteins detected with genome-diagnostic peptides from both diploid genomes (A) and from only the D-genome (B) or the A-genome (C). Up- and down-regulation are shown in red and green, respectively; black corresponds to no significant change. For a given protein represented by a row, its identified category for total expression of both homeologs is shown in the columns on the right. Proteins with unequal expression between A- and D-genome diploids are denoted by a dot if A > D and a square if A < D. In B and C, a proposed explanation for expression-level dominance with respect to homeolog expression is depicted, where total expression of the duplicate pair is mainly explained by expression of the homeolog with higher parental expression levels. For example, for Gorai 001G236500 in B, genome-diagnostic peptides were detected only for the DT homeolog, and a higher expression level was observed in the diploid D-genome than in A-genome (denoted by a square); total expression of both homeologs exhibited transgressive up-regulation (AD2 at 10 dpa) when the DT homeolog was up-regulated relative to that of the parental protein in the diploid D-genome, while A-dominant (AD1 at 20 dpa) and D-dominant (AD1 at 20 dpa) total expression patterns were observed when the DT homeolog was down-regulated to different extents, respectively.
For proteins that had both AT and DT homeologs detected (Figure 6A), even when differential homeolog-specific regulations were apparent with unequal AT/A and DT/D ratios (e.g., Gorai.002G107200, third row from top), their total expression in allopolyploids was mostly “conserved” with respect to equivalent diploid expression (Figure 6A, light gray in right columns). Only three proteins exhibiting unequal parental expressions (marked by black dots and squares) also were detected with expression-level dominance. Interestingly, the total expression of a glycosyl hydrolase protein (Gorai.013G202800) was A-dominant in G. barbadense, while its homeologs appeared to be down-regulated more to achieve a significantly lower expression in G. hirsutum, which was D-dominant.
For proteins detected with genome-diagnostic peptides from only one diploid genome (Figure 6, B and C), nonconservative patterns including median, A-dominant, and D-dominant expressions were categorized for over 70% of proteins, which were associated with unequal A- and D-genome parental expression (marked by black dots and squares). By examining the DT-only proteins with higher expression levels of D- vs. A-genome parents, up-regulation of DT relative to its D-genome parent was coupled with transgressive up-regulation, while down-regulation of DT appeared to lead to A-dominant or D-dominant expression depending on the magnitude of down-regulation (illustrated by the right panel in Figure 6B). In contrast, when the A-genome parent had a higher expression level than the D-genome parent for AT-only proteins, the total expression patterns were associated with down-regulation of AT relative to its A-genome parent at variable magnitudes (right panel in Figure 6C). One possible explanation for the phenomenon observed in both cases is that when expression of the homeolog that is expressed at a lower level in the parental state is negligible, the alternative homeolog is the determinant of total expression. In other words, a causal relationship can be proposed between homeolog-specific and total expression, where the latter is mainly explained by regulation of the homeolog from the diploid parent that exhibits higher expression levels. When genome-diagnostic peptides are detected for both homeologs (Figure 6A), this causal relationship cannot be inferred, and accordingly, patterns of total expression are inferred to reflect regulatory changes of both homeologs.
Discussion
Here we present a comparative approach to investigate proteome modifications in a single-celled structure, the cotton fiber, caused by allopolyploidization. With the enhanced analytical power provided by using both 2-DE and the gel-free iTRAQ methods for proteomic profiling, two natural allopolyploid cotton species were studied in parallel. By demonstrating the protein-level consequences of gene-expression evolution, such as expression-level dominance and homeolog expression bias, our results provide a broad view of proteomic developmental dynamics, evolutionary divergence, and homeolog composition in allopolyploids relative to their progenitor diploid species.
Complementary 2-DE and iTRAQ proteomics of allopolyploidy
Our 2-DE and iTRAQ analyses each present a quantitative and interspecific profile of fiber proteomes that includes more than 1000 protein features. Comparison of the parental G. arboreum and G. raimondii proteomes revealed that approximately 10% of the proteins were differentially expressed between the two diploids, which have diverged for ∼5–10 million years, during which time they have accumulated ∼3.6% synonymous and 0.9% nonsynonymous nucleotide differences (Flagel et al. 2012) and ∼1.8% amino acid differences between orthologous genes (Hu et al. 2013). The proteomic divergence revealed by the profiling experiments reported here is much lower than that previously observed for cotton seeds, where only 50% of the protein spots were qualitatively shared by the two diploids (Hu et al. 2011). In Tragopogon, 3.2% differential protein expression was reported for leaf proteomes between the diploids T. dubius and T. porrifolius, which have about 3.5% synonymous nucleotide divergence (Koh et al. 2012). These results suggest that protein expression differences vary largely across tissue types and are hard to predict from genetic divergence alone, especially when comparisons are made across tissue or organ types that vary in their cellular complexity. Compared to differences between the two diploids, fewer differential expression changes were identified between two allopolyploid cotton species, G. hirsutum and G. barbadense, according to 2-DE data, which is in agreement with their closer evolutionary relationship (1–2 million years of divergence compared to 5–10 million years for the diploids), whereas this pattern was not shown by iTRAQ analysis because of technical differences between the two methods. Although mostly consistent results were derived from 2-DE and iTRAQ data in our analysis, our data demonstrate that the two platforms offer different and hence complementary perspectives on proteomic evolution.
In 2-DE analysis, different proteomes are resolved into two-dimensional spot maps for quantification with the assumption that each spot represents one protein. However, because of protein microheterogeneity caused by post-translational modifications, proteolytic degradation, and other causes, protein products from one gene may exhibit multiple spots, thereby affecting quantitative sensitivity and accuracy (Wu et al. 2006). As reported previously for cotton seeds, although compositions of major seed storage proteins were relatively conserved across species, their numerous isoforms contributed up to 50% of the interspecific variation (Hu et al. 2011). Besides protein isoforms, protein comigration (i.e., multiple proteins locating within one spot) and in silico comigration due to incorrect spot matching across gels pose an even greater challenge in quantification, especially for evolutionary studies when proteomes from different species are compared. Thus the protein expression change inferred from 2-DE analyses reflects confounding effects of both protein abundance and post-translational modifications, which also provides an opportunity to investigate evolutionary processes that modify protein-level-specific properties. In contrast, protein microheterogeneity is less problematic in iTRAQ analysis, where quantification is assessed at the peptide level.
In the iTRAQ platform, labeling and pooling samples have a key advantage over single-stain 2-DE gels by reducing experimental variation and raising confidence in quantitative assessments of protein changes. A second and critical advantage with respect to the study of allopolyploids is that with appropriate data from reference genomes, it is possible to derive information on homeolog-specific protein expression. The first application of iTRAQ for this purpose was described recently for allopolyploid Tragopogon (Koh et al. 2012): by specifying the genome-specific protein sequences between diploid progenitors as well as subgenomes in allopolyploids, genome-specific peptides for a given pair of homeolog proteins were examined for differential expression, which identified two cases of biased expression of a parental homeolog in the natural allopolyploid T. mirus but not in the F1 hybrid or synthetic polyploid. Since that initial report, homeolog expression bias has been discovered in developing fiber proteomes of two allopolyploid cottons, G. barbadense (Hu et al. 2013) and G. hirsutum (Hu et al. 2014). In this study, we demonstrate a quantitative method to examine homeologs relative to parental expression levels (i.e., AT/A and DT/D) and show how homeolog specificity contributes to total expression in allopolyploid proteomes (see below).
A potential pitfall of iTRAQ analysis results from the same factors that make this approach so efficient, i.e., relative protein quantification depending on the ratio of iTRAQ reagents. For a given protein, between-sample expression ratios are extracted and subjected to differential expression and other statistical tests; this is statistically less flexible than comparing direct measurement of expression levels, such as spot volumes in 2-DE analysis and read numbers in RNA-seq analysis. In our analysis, for example, only the 2-DE spot volumes were used to test the additivity of parental contributions to the allopolyploid proteome. Despite the reported application in Arabidopsis (Ng et al. 2012), the use of iTRAQ ratios for additivity tests is statistically problematic, and the in silico average of parental ratios is difficult to justify without validation using values derived from mixed samples of parental proteins. Taken together, 2-DE and iTRAQ methods provide complementary information for comprehensive proteomic profiling (Rose et al. 2004; Thelen and Peck 2007; Diz et al. 2012), with each technology offering different strengths.
Nonadditive expression and unbalanced expression-level dominance in fiber proteomes
Our 2-DE data demonstrated that 7–13% of the fiber proteome in allopolyploid cotton species is differentially expressed relative to the averaged expression of homeologs from its two progenitor diploids. This offers one perspective on proteomic evolution accompanying diploid divergence of 5–10 million years and subsequent genomic merger and doubling 1–2 million years ago. A corollary is that most fiber proteins are additively expressed. The degree of proteomic divergence for cotton fibers is lower than that previously observed in Brassica root and leaf proteomes (25–38%) (Albertin et al. 2006) and for the seed proteome in cotton (34%) (Hu et al. 2011). This might be the result of the difference in the complexity of tissues employed. In contrast to leaf and seed, cotton fiber is a single cell; thus, we can expect less complexity in the fiber proteome and perhaps a more narrowly canalized development program.
The term “expression-level dominance,” modified from the earlier “genome dominance” (Rapp et al. 2009), was suggested by Grover et al. (2012a) to describe the phenomenon in which the total expression level in an allopolyploid resembles that of one of the two parents. As first described in cotton for the leaf transcriptome by Rapp et al. (2009), microarray-profiled gene expression levels in a synthetic allopolyploid genome mimicked those in the parental D-genome more often than those in the A-genome. Using a more sensitive transcriptomic approach (RNA-seq), a similar pattern of unbalanced expression-level dominance was demonstrated in synthetic allopolyploid cotton, while the direction was reversed in an F1 diploid hybrid and two natural allopolyploids that favored the parental A-genome (Yoo et al. 2013). At the protein level, the seed proteome in cotton was characterized for one allopolyploid species using 2-DE, where 16% more 2-DE spots in the G. hirsutum proteome were shared with those in the D-parent than in the A-parent, and more proteins exhibited expression-level dominance favoring the D-genome than the A-genome (Hu et al. 2011).
Here we complement these earlier studies with an expanded experimental design using two different proteomic approaches as well as two different allopolyploid species. A consensus observation of this work is unbalanced expression-level dominance for 10-dpa fibers in the direction of the A-genome, which leads to the suggestion that there is rapid change during fiber development in a genomically biased fashion. Because of the small numbers of fiber proteins categorized (Figure 5), no cellular function or biological processes were enriched with respect to these expression patterns. Nonetheless, the functional significance and underlying regulatory bases of protein expression-level dominance likely are fruitful avenues for future exploration. It is also worth noting that because fiber traits of modern cultivars have been the major target of crop domestication, the patterns of proteomic evolution observed here reflect the superimposed effects of natural polyploidization and human-mediated selection, as discussed further below.
To distinguish the homeologous contributions to duplicated gene expression in allopolyploids, one common practice in RNA-seq analysis is parsing the pairs of homeolog-specific transcripts by single-nucleotide polymorphisms and comparing the homeolog-specific reads of diploids and allopolyploids (A/D, AT/DT, AT/A, and DT/D) to study unequal expression patterns (i.e., homeolog expression bias) (Grover et al. 2012a) and the regulation of homeologs. A recent transcriptomic analysis in allopolyploid cotton showed that homeologs contributed equally for most genes regardless of the diploid parental states, while the total expression-level dominance toward one parental genome is often associated with up- or down-regulation of the homeolog from the “nondominant” genome (Yoo et al. 2013). Here, at the protein level, only homeologous expression relative to parental state (AT/A and DT/D) was examined owing to the limitation of iTRAQ quantification. Despite this limitation, expression-level dominance caused by modifying either or both homeologs was evident at the protein level.
Divergent proteomic development following allopolyploidization
The two natural allopolyploid species G. hirsutum and G. barbadense represent divergent branches from a monophyletic allopolyploidization event that occurred 1–2 million years ago (Wendel and Cronn 2003; Grover et al. 2012b). With both allopolyploids exhibiting the notable morphologic trait of long, spinable cotton fibers, it has long been hypothesized that allopolyploid cotton preferentially displays the physiologic profile and molecular machinery of its A-genome progenitor because it is in this genomic group that long fiber first evolved. Fiber elongation rate is dramatically higher in G. hirsutum and the A-genome diploids compared to fibers from the D-genome diploid G. raimondii, especially during primary cell wall synthesis from 10 to 20 dpa (Applequist et al. 2001). Although the most advanced modern cultivars of G. barbadense produce longer fibers than G. hirsutum cultivars at maturity, their growth curves generally overlap (Chen et al. 2012). The developmental divergence observed here between these two species at the proteome level, represented by 5–10% of fiber proteins, is consistent with our previous work describing developing fiber proteomes in G. hirsutum (Hu et al. 2014) and G. barbadense (Hu et al. 2013). A notable observation, and one that bears future study, is that there is little overlap in protein change during development in the two species (Figure 2, A and B), suggesting that the cellular processes and biological functions specifically enriched during fiber elongation in G. barbadense may lead to new understanding of the elite fiber traits found in the latter species (Figure 2C). Consistent with this, a high degree of divergence was evident between the two allopolyploid proteomes. An additional intriguing observation is that the fiber proteome of G. hirsutum resembles the parental A-genome more closely than does the fiber proteome of G. barbadense, while the direction of G. barbadense expression-level dominance switches from D-genome to A-genome during fiber development (Figure 3 and Figure S4). These results suggest that following allopolyploidization and parallel domestication, the two allopolyploid species have achieved superficially similar modern fiber phenotypes through different evolutionary routes at the proteome level, a result also shown at the transcriptomic level (Zhu et al. 2011; Chen et al. 2012; Lacape et al. 2012). At present, the relationship between these interspecific differences and their individual patterns of homeolog usage, if any, remains obscure.
Conclusions
Gene-expression evolution accompanying allopolyploidization, including expression-level dominance and homeolog expression bias, has been studied at the transcriptomic level but rarely evaluated at the protein level. Only a handful of species (Brassica, Tragopogon, Arabidopsis, and cotton) have been examined to date, each using a single proteomic technique. Here we demonstrate the protein-level consequences of gene regulation using two complementary proteomic strategies, which we apply to a readily harvested single-celled structure. This enabled a more comprehensive documentation of fiber proteomes in response to evolutionary change. Two natural cotton allopolyploids of the same origin were used to study the general consequences of polyploidization, which led to a consensus discovery of A-biased expression-level dominance at the same fiber elongation stage. Interestingly, divergent paths of proteomic modification were exhibited by the two allopolyploids, each domesticated for similar reasons but from different wild progenitors. Future work will reveal whether the proteomic distinctions between the two species reflect the effects of strong directional selection that unknowingly targeted different components of the proteomic network or whether, instead, this happened during evolutionary divergence prior to domestication. Our analyses also provide a database and framework to identify and characterize key protein and metabolic pathways corresponding to species evolution and phenotypic diversity in cotton.
Supplementary Material
Acknowledgments
We thank Kara Grupp and Anna Tuchin for help in tissue collection and Dharminder Pathak for help in the 2-DE analyses. We acknowledge the Protein and Proteomics Facility of Iowa State University for technical assistance in the 2-DE analyses and the Proteomics and Mass Spectrometry Facility of the University of Florida’s Interdisciplinary Center for Biotechnology Research for assistance in LC-MS ⁄MS analysis. The LC-MS/MS system was funded by National Institutes of Health Grant 1S10RR025418-01 to SC. This work was funded by Cotton Incorporated and the NSF Plant Genome Research Program, both to JFW.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.174367/-/DC1.
Communicating editor: J. A. Birchler
Literature Cited
- Adams K. L., Cronn R., Percifield R., Wendel J. F., 2003. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl. Acad. Sci. USA 100: 4649–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainouche M., Wendel J., 2014. Polyploid speciation and genome evolution: Lessons from recent allopolyploids, pp. 87–113 in Evolutionary Biology: Genome Evolution, Speciation, Coevolution and Origin of Life, edited by Pontarotti P. Springer, New York. [Google Scholar]
- Albertin W., Alix K., Balliau T., Brabant P., Davanture M., et al. , 2007. Differential regulation of gene products in newly synthesized Brassica napus allotetraploids is not related to protein function nor subcellular localization. BMC Genomics 8: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertin W., Balliau T., Brabant P., Chevre A. M., Eber F., et al. , 2006. Numerous and rapid nonstochastic modifications of gene products in newly synthesized Brassica napus allotetraploids. Genetics 173: 1101–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anssour S., Krugel T., Sharbel T. F., Saluz H. P., Bonaventure G., et al. , 2009. Phenotypic, genetic and genomic consequences of natural and synthetic polyploidization of Nicotiana attenuata and Nicotiana obtusifolia. Ann. Bot. 103: 1207–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applequist W. L., Cronn R., Wendel J. F., 2001. Comparative development of fiber in wild and cultivated cotton. Evol. Dev. 3: 3–17. [DOI] [PubMed] [Google Scholar]
- Bahrman N., Thiellement H., 1987. Parental genome expression in synthetic wheats (Triticum turgidum sp. × T. tauschii sp.) revealed by two-dimensional electrophoresis of seedling proteins. Theor. Appl. Genet. 74: 218–223. [DOI] [PubMed] [Google Scholar]
- Bardil A., de Almeida J. D., Combes M. C., Lashermes P., Bertrand B., 2011. Genomic expression dominance in the natural allopolyploid Coffea arabica is massively affected by growth temperature. New Phytol. 192: 760–774. [DOI] [PubMed] [Google Scholar]
- Blanc G., Wolfe K. H., 2004. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16: 1679–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottley A., 2014. Epigenetic variation amongst polyploidy crop species, pp. 33–46 in Epigenetics in Plants of Agronomic Importance: Fundamentals and Applications. Springer, New York. [Google Scholar]
- Bottley A., Xia G. M., Koebner R. M., 2006. Homoeologous gene silencing in hexaploid wheat. Plant J. 47: 897–906. [DOI] [PubMed] [Google Scholar]
- Buggs R. J., Chamala S., Wu W., Gao L., May G. D., et al. , 2010a Characterization of duplicate gene evolution in the recent natural allopolyploid Tragopogon miscellus by next-generation sequencing and Sequenom iPLEX MassARRAY genotyping. Mol. Ecol. 19(Suppl 1): 132–146. [DOI] [PubMed] [Google Scholar]
- Buggs R. J., Doust A. N., Tate J. A., Koh J., Soltis K., et al. , 2009. Gene loss and silencing in Tragopogon miscellus (Asteraceae): comparison of natural and synthetic allotetraploids. Heredity 103: 73–81. [DOI] [PubMed] [Google Scholar]
- Buggs R. J., Elliott N. M., Zhang L., Koh J., Viccini L. F., et al. , 2010b Tissue-specific silencing of homoeologs in natural populations of the recent allopolyploid Tragopogon mirus. New Phytol. 186: 175–183. [DOI] [PubMed] [Google Scholar]
- Chague V., Just J., Mestiri I., Balzergue S., Tanguy A. M., et al. , 2010. Genome-wide gene expression changes in genetically stable synthetic and natural wheat allohexaploids. New Phytol. 187: 1181–1194. [DOI] [PubMed] [Google Scholar]
- Chaudhary B., Flagel L., Stupar R. M., Udall J. A., Verma N., et al. , 2009. Reciprocal silencing, transcriptional bias and functional divergence of homeologs in polyploid cotton (Gossypium). Genetics 182: 503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelaifa H., Chague V., Chalabi S., Mestiri I., Arnaud D., et al. , 2013. Prevalence of gene expression additivity in genetically stable wheat allohexaploids. New Phytol. 197: 730–736. [DOI] [PubMed] [Google Scholar]
- Chelaifa H., Monnier A., Ainouche M., 2010. Transcriptomic changes following recent natural hybridization and allopolyploidy in the salt marsh species Spartina × townsendii and Spartina anglica (Poaceae). New Phytol. 186: 161–174. [DOI] [PubMed] [Google Scholar]
- Chen X., Guo W., Liu B., Zhang Y., Song X., et al. , 2012. Molecular mechanisms of fiber differential development between G. barbadense and G. hirsutum revealed by genetical genomics. PLoS ONE 7: e30056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. J., 2007. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu. Rev. Plant Biol. 58: 377–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester M., Gallagher J. P., Symonds V. V., Cruz da Silva A. V., Mavrodiev E. V., et al. , 2012. Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae). Proc. Natl. Acad. Sci. USA 109: 1176–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L., 2005. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6: 836–846. [DOI] [PubMed] [Google Scholar]
- Conesa A., Gotz S., Garcia-Gomez J. M., Terol J., Talon M., et al. , 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- Diz A. P., Martinez-Fernandez M., Rolan-Alvarez E., 2012. Proteomics in evolutionary ecology: linking the genotype with the phenotype. Mol. Ecol. 21: 1060–1080. [DOI] [PubMed] [Google Scholar]
- Doyle J. J., Flagel L. E., Paterson A. H., Rapp R. A., Soltis D. E., et al. , 2008. Evolutionary genetics of genome merger and doubling in plants. Annu. Rev. Genet. 42: 443–461. [DOI] [PubMed] [Google Scholar]
- Du Z., Zhou X., Ling Y., Zhang Z., Su Z., 2010. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38: W64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finigan P., Tanurdzic M., Martienssen R., 2012. Origins of novel phenotypic variation in polyploids, pp. 57–76 in Polyploidy and Genome Evolution, edited by Soltis P. S., Soltis D. E. Springer, Berlin. [Google Scholar]
- Fisher R. A., 1948. Questions and answers #14. Am. Stat. 2: 30–31. [Google Scholar]
- Flagel L., Udall J., Nettleton D., Wendel J., 2008. Duplicate gene expression in allopolyploid Gossypium reveals two temporally distinct phases of expression evolution. BMC Biol. 6: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel L. E., Wendel J. F., 2010. Evolutionary rate variation, genomic dominance and duplicate gene expression evolution during allotetraploid cotton speciation. New Phytol. 186: 184–193. [DOI] [PubMed] [Google Scholar]
- Flagel L. E., Wendel J. F., Udall J. A., 2012. Duplicate gene evolution, homoeologous recombination, and transcriptome characterization in allopolyploid cotton. BMC Genomics 13: 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaeta R. T., Pires J. C., Iniguez-Luy F., Leon E., Osborn T. C., 2007. Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19: 3403–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover C. E., Gallagher J. P., Szadkowski E. P., Yoo M. J., Flagel L. E., et al. , 2012a Homoeolog expression bias and expression level dominance in allopolyploids. New Phytol. 196: 966–971. [DOI] [PubMed] [Google Scholar]
- Grover C. E., Grupp K. K., Wanzek R. J., Wendel J. F., 2012b Assessing the monophyly of polyploid Gossypium species. Plant Syst. Evol. 298: 1177–1183. [Google Scholar]
- Ha M., Lu J., Tian L., Ramachandran V., Kasschau K. D., et al. , 2009. Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc. Natl. Acad. Sci. USA 106: 17835–17840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigler C. H., Betancur L., Stiff M. R., Tuttle J. R., 2012. Cotton fiber: a powerful single-cell model for cell wall and cellulose research. Front. Plant Sci. 3: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F., Fedak G., Guo W., Liu B., 2005. Rapid and repeatable elimination of a parental genome-specific DNA repeat (pGc1R–1a) in newly synthesized wheat allopolyploids. Genetics 170: 1239–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegarty M. J., Jones J. M., Wilson I. D., Barker G. L., Coghill J. A., et al. , 2005. Development of anonymous cDNA microarrays to study changes to the Senecio floral transcriptome during hybrid speciation. Mol. Ecol. 14: 2493–2510. [DOI] [PubMed] [Google Scholar]
- Hovav R., Udall J. A., Chaudhary B., Rapp R., Flagel L., et al. , 2008. Partitioned expression of duplicated genes during development and evolution of a single cell in a polyploid plant. Proc. Natl. Acad. Sci. USA 105: 6191–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G., Houston N. L., Pathak D., Schmidt L., Thelen J. J., et al. , 2011. Genomically biased accumulation of seed storage proteins in allopolyploid cotton. Genetics 189: 1103–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G., Koh J., Yoo M. J., Grupp K., Chen S., et al. , 2013. Proteomic profiling of developing cotton fibers from wild and domesticated Gossypium barbadense. New Phytol. 200: 570–582. [DOI] [PubMed] [Google Scholar]
- Hu G., Koh J., Yoo M. J., Pathak D., Chen S., et al. , 2014. Proteomics profiling of fiber development and domestication in upland cotton (Gossypium hirsutum L.). Planta 240: 1237–1251. [DOI] [PubMed] [Google Scholar]
- Islam N., Tsujimoto H., Hirano H., 2003. Proteome analysis of diploid, tetraploid and hexaploid wheat: towards understanding genome interaction in protein expression. Proteomics 3: 549–557. [DOI] [PubMed] [Google Scholar]
- Jiao Y., Wickett N. J., Ayyampalayam S., Chanderbali A. S., Landherr L., et al. , 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100. [DOI] [PubMed] [Google Scholar]
- Karr T. L., 2008. Application of proteomics to ecology and population biology. Heredity 100: 200–206. [DOI] [PubMed] [Google Scholar]
- Kenan-Eichler M., Leshkowitz D., Tal L., Noor E., Melamed-Bessudo C., et al. , 2011. Wheat hybridization and polyploidization results in deregulation of small RNAs. Genetics 188: 263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J., Triplett B. A., 2001. Cotton fiber growth in planta and in vitro: models for plant cell elongation and cell wall biogenesis. Plant Physiol. 127: 1361–1366. [PMC free article] [PubMed] [Google Scholar]
- Koh J., Chen S., Zhu N., Yu F., Soltis P. S., et al. , 2012. Comparative proteomics of the recently and recurrently formed natural allopolyploid Tragopogon mirus (Asteraceae) and its parents. New Phytol. 196: 292–305. [DOI] [PubMed] [Google Scholar]
- Koh J., Soltis P. S., Soltis D. E., 2010. Homeolog loss and expression changes in natural populations of the recently and repeatedly formed allotetraploid Tragopogon mirus (Asteraceae). BMC Genomics 11: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovarik A., Dadejova M., Lim Y. K., Chase M. W., Clarkson J. J., et al. , 2008. Evolution of rDNA in Nicotiana allopolyploids: a potential link between rDNA homogenization and epigenetics. Ann. Bot. 101: 815–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacape J. M., Claverie M., Vidal R. O., Carazzolle M. F., Guimaraes Pereira G. A., et al. , 2012. Deep sequencing reveals differences in the transcriptional landscapes of fibers from two cultivated species of cotton. PLoS ONE 7: e48855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch A. R., Leitch I. J., 2008. Genomic plasticity and the diversity of polyploid plants. Science 320: 481–483. [DOI] [PubMed] [Google Scholar]
- Madlung A., 2013. Polyploidy and its effect on evolutionary success: old questions revisited with new tools. Heredity 110: 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madlung A., Masuelli R. W., Watson B., Reynolds S. H., Davison J., et al. , 2002. Remodeling of DNA methylation and phenotypic and transcriptional changes in synthetic Arabidopsis allotetraploids. Plant Physiol. 129: 733–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madlung A., Wendel J. F., 2013. Genetic and epigenetic aspects of polyploid evolution in plants. Cytogenet. Genome Res. 140: 270–285. [DOI] [PubMed] [Google Scholar]
- Mi H., Dong Q., Muruganujan A., Gaudet P., Lewis S., et al. , 2010. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 38: D204–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng D. W., Zhang C., Miller M., Palmer G., Whiteley M., et al. , 2011. Cis- and trans-regulation of miR163 and target genes confers natural variation of secondary metabolites in two Arabidopsis species and their allopolyploids. Plant Cell 23: 1729– 1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng D. W., Zhang C., Miller M., Shen Z., Briggs S. P., et al. , 2012. Proteomic divergence in Arabidopsis autopolyploids and allopolyploids and their progenitors. Heredity 108: 419–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z., Kim E. D., Ha M., Lackey E., Liu J., et al. , 2009. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457: 327–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkan H., Tuna M., Arumuganathan K., 2003. Nonadditive changes in genome size during allopolyploidization in the wheat (Aegilops-Triticum) group. J. Hered. 94: 260–264. [DOI] [PubMed] [Google Scholar]
- Ponnala L., Wang Y., Sun Q., van Wijk K. J., 2014. Correlation of mRNA and protein abundance in the developing maize leaf. Plant J. 78: 424–440. [DOI] [PubMed] [Google Scholar]
- Ramsey J., Schemske D. W., 2002. Neopolyploidy in flowering plants. Annu. Rev. Ecol. Syst. 33: 589–639. [Google Scholar]
- Rapp R. A., Udall J. A., Wendel J. F., 2009. Genomic expression dominance in allopolyploids. BMC Biol. 7: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose J. K., Bashir S., Giovannoni J. J., Jahn M. M., Saravanan R. S., 2004. Tackling the plant proteome: practical approaches, hurdles and experimental tools. Plant J. 39: 715–733. [DOI] [PubMed] [Google Scholar]
- Salmon A., Ainouche M. L., Wendel J. F., 2005. Genetic and epigenetic consequences of recent hybridization and polyploidy in Spartina (Poaceae). Mol. Ecol. 14: 1163–1175. [DOI] [PubMed] [Google Scholar]
- Salmon A., Flagel L., Ying B., Udall J. A., Wendel J. F., 2010. Homoeologous nonreciprocal recombination in polyploid cotton. New Phytol. 186: 123–134. [DOI] [PubMed] [Google Scholar]
- Schnable J. C., Springer N. M., Freeling M., 2011. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc. Natl. Acad. Sci. USA 108: 4069–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaked H., Kashkush K., Ozkan H., Feldman M., Levy A. A., 2001. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 13: 1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalicka K., Lim K. Y., Matyasek R., Matzke M., Leitch A. R., et al. , 2005. Preferential elimination of repeated DNA sequences from the paternal, Nicotiana tomentosiformis genome donor of a synthetic, allotetraploid tobacco. New Phytol. 166: 291–303. [DOI] [PubMed] [Google Scholar]
- Soltis P. S., Soltis D. E., 2009. The role of hybridization in plant speciation. Annu. Rev. Plant Biol. 60: 561–588. [DOI] [PubMed] [Google Scholar]
- Suzuki R., Shimodaira H., 2006. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22: 1540–1542. [DOI] [PubMed] [Google Scholar]
- Szadkowski E., Eber F., Huteau V., Lode M., Huneau C., et al. , 2010. The first meiosis of resynthesized Brassica napus, a genome blender. New Phytol. 186: 102–112. [DOI] [PubMed] [Google Scholar]
- Tate J. A., Joshi P., Soltis K. A., Soltis P. S., Soltis D. E., 2009. On the road to diploidization? Homoeolog loss in independently formed populations of the allopolyploid Tragopogon miscellus (Asteraceae). BMC Plant Biol. 9: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen J. J., Peck S. C., 2007. Quantitative proteomics in plants: choices in abundance. Plant Cell 19: 3339–3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C., Marcotte E. M., 2012. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 13: 227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Tian L., Lee H. S., Wei N. E., Jiang H., et al. , 2006. Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 172: 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel J. F., 2000. Genome evolution in polyploids. Plant Mol. Biol. 42: 225–249. [PubMed] [Google Scholar]
- Wendel J. F., Cronn R. C., 2003. Polyploidy and the evolutionary history of cotton, pp. 139–186 in Advances in Agronomy. Academic Press, New York. [Google Scholar]
- Wendel J. F., Doyle J. J., 2004. Polyploidy and evolution in plants, pp. 97–117 in Plant Diversity and Evolution: Genotypic and Phenotypic Variation in Higher Plants, edited by Henry R. J. CABI, Wallingford, UK. [Google Scholar]
- Wendel J. F., Flagel L. E., Adams K. L., 2012. Jeans, genes, and genomes: cotton as a model for studying polyploidy, pp. 181–207 in Polyploidy and Genome Evolution, edited by Soltis P. S., Soltis D. E. Springer, Berlin. [Google Scholar]
- Wilkins T. A., Arpat A. B., 2005. The cotton fiber transcriptome. Physiol. Plant. 124: 295–300. [Google Scholar]
- Woodhouse M. R., Cheng F., Pires J. C., Lisch D., Freeling M., et al. , 2014. Origin, inheritance, and gene regulatory consequences of genome dominance in polyploids. Proc. Natl. Acad. Sci. USA 111: 5283–5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W. W., Wang G., Baek S. J., Shen R. F., 2006. Comparative study of three proteomic quantitative methods, DIGE, cICAT, and iTRAQ, using 2D gel- or LC-MALDI TOF/TOF. J. Proteome Res. 5: 651–658. [DOI] [PubMed] [Google Scholar]
- Xiong Z., Gaeta R. T., Pires J. C., 2011. Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc. Natl. Acad. Sci. USA 108: 7908–7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo M. J., Liu X., Pires J. C., Soltis P. S., Soltis D. E., 2014. Nonadditive gene expression in polyploids. Annu. Rev. Genet. 48: 485–517. [DOI] [PubMed] [Google Scholar]
- Yoo M. J., Szadkowski E., Wendel J. F., 2013. Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity 110: 171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Han X., Lv J., Zhao L., Xu X., et al. , 2011. Structure, expression differentiation and evolution of duplicated fiber developmental genes in Gossypium barbadense and G. hirsutum. BMC Plant Biol. 11: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.