Significance
This paper describes a novel strategy for predicting the function of terpene synthases. Functional assignment of terpene synthases is a daunting task because product selectivity is not high in many terpene synthases, and mutations in and near the active sites of selective enzyme can result in synthesis of different products. Using a homology model of an unknown terpene synthase, we developed an algorithm that predicted the enzyme synthesizes a linear triquinane. We confirmed this prediction; specifically, the enzyme converts farnesyl diphosphate to a linear triquinine sesquiterpene: (5S,7S,10R,11S)-cucumene. The findings highlight the potential for using computational approaches to assist in the discovery and characterization of unknown terpene synthases.
Keywords: sesquiterpene, cyclase, structure-function, annotation
Abstract
Terpenoids are a large structurally diverse group of natural products with an array of functions in their hosts. The large amount of genomic information from recent sequencing efforts provides opportunities and challenges for the functional assignment of terpene synthases that construct the carbon skeletons of these compounds. Inferring function from the sequence and/or structure of these enzymes is not trivial because of the large number of possible reaction channels and products. We tackle this problem by developing an algorithm to enumerate possible carbocations derived from the farnesyl cation, the first reactive intermediate of the substrate, and evaluating their steric and electrostatic compatibility with the active site. The homology model of a putative pentalenene synthase (Uniprot: B5GLM7) from Streptomyces clavuligerus was used in an automated computational workflow for product prediction. Surprisingly, the workflow predicted a linear triquinane scaffold as the top product skeleton for B5GLM7. Biochemical characterization of B5GLM7 reveals the major product as (5S,7S,10R,11S)-cucumene, a sesquiterpene with a linear triquinane scaffold. To our knowledge, this is the first documentation of a terpene synthase involved in the synthesis of a linear triquinane. The success of our prediction for B5GLM7 suggests that this approach can be used to facilitate the functional assignment of novel terpene synthases.
Terpenoid compounds form a large group of natural products synthesized by many species of plants, fungi, and bacteria. To date, more than 70,000 of these compounds have been identified, comprising ∼25% of all of the known natural products (Dictionary of Natural Products database, dnp.chemnetbase.com/intro/index.jsp). Terpenoids and molecules containing terpenoid fragments are produced from two basic five-carbon building blocks: isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) (1, 2). IPP and DMAPP are then converted into a series of terpenoid diphosphate esters of increasing chain lengths by chain length selective polyprenyl diphosphate synthases to give geranyl diphosphate (GPP, C10), farnesyl diphosphate (FPP, C15), geranylgeranyl diphosphate (GGPP, C20), and higher five-carbon homologs (3). These molecules are substrates for terpene synthases, which catalyze hydroxylation, elimination, cyclization, and rearrangement reactions to produce much of the structural diversity of terpenoid molecules found in nature (4, 5).
The two main classes of terpene synthases (class I and class II) are distinguished from one another by the mechanisms they use to initiate carbocationic cyclization and rearrangement reactions and by their respective folds (6, 7). Class I terpene synthases use active site Mg2+ ions to bind and activate the diphosphate moieties of their substrates as leaving groups to generate highly reactive allylic carbocations. In those terpene synthases that catalyze cyclization reactions, the carbocations alkylate distal double bonds in the hydrocarbon chain. The initial cyclization is often followed by additional cyclizations and rearrangements to ultimately produce a myriad of different carbon skeletons. Class II terpene synthases generate an electrophilic tertiary carbocation by protonation of a trisubstituted double bond mediated by an acidic residue (e.g., Asp) in their active sites. These enzymes then mediate cyclization and rearrangement reactions to give a second large group of terpenoid carbon skeletons.
Streptomyces clavuligerus is a gram-positive bacterium that, like most other Streptomyces spp., produces a wide variety of natural products, including penicillin, clavulanic acid, and holomycin (8, 9). The genome of S. clavuligerus ATCC 27604 encodes 20 putative terpene synthases (10). Two of the enzymes are presumed to be geosmin synthases based on their high sequence identity (>60%), with known homologs from other Streptomyces spp. (11, 12). In 2011, Hu et al. reported that two other terpene synthases in this organism are involved in biosynthesis of T-muurolol and δ-cadinene (10). In the same year, Nakano et al. reported the characterization of two additional terpene synthases, a monoterpene cyclase that produced 1,8-cineole and a promiscuous acyclic terpene synthase that produced linalool and nerolidol (13, 14). While this manuscript was in the final stages of preparation, the function of an additional terpene synthase, which is a protein variant of the subject of this investigation, was reported to synthesize a linear triquinane of undefined stereochemistry (15). The functions of the remaining terpene synthases from S. clavuligerus are unknown.
Many terpene synthases are known to be promiscuous. The promiscuity of terpene synthases is perhaps most dramatically illustrated by γ-humulene synthase from Abies grandis, which produces 52 different sesquiterpenes (16). Promiscuity in these enzymes is not surprising given the multiple cyclization and rearrangement steps that the substrates often undergo during catalysis, any one of which can be altered by perturbations to the microenvironment of the active site. Promiscuity is often seen in mutagenesis experiments where a few key mutations are typically sufficient to alter the product selectivity of the enzymes (17, 18). As a result, prediction of function for terpene synthases is still a very challenging task in the field of computational biology.
One of the key objectives of the enzyme function initiative (EFI) is the development of methods that facilitate functional assignments for unknown terpene synthases as was previously demonstrated for the polyprenyl transferases (3, 19). Quantum mechanics-only and quantum mechanics/molecular mechanics (QM/MM) calculations have provided important insights about the mechanisms of reactions catalyzed by terpene synthases (20–24). However, gas phase QM calculations cannot predict functions for specific terpene synthases in the absence of protein structures. QM/MM calculations, which take the enzyme structures into account, are computationally too expensive for making predictions, i.e., for uncharacterized enzymes, because the product chemical space for terpene synthases is huge. We recently described a mechanism-based carbocation docking approach to predict functions of triterpene synthases (25). However, the approach is limited because the carbocation library used for docking contains only known carbocations believed to participate in characterized terpene synthase reactions, implying that novel functions will not be predicted. Two key problems must be solved to predict novel functions for terpene synthases from protein sequences. First, an efficient algorithm must be developed for automatically enumerating all possible carbocations; second, the calculations must be carried out in the presence of crystal structures or homology models for the enzymes, to evaluate the compatibility of the carbocationic intermediates with the active site. We developed a code, iGen, which allowed us to systematically enumerate carbocations in the gas phase. For the second problem, we now report an automated computational workflow for predicting terpene synthase product skeletons from protein sequences. Application of this workflow aided the discovery and characterization of a sesquiterpene synthase (Uniprot: B5GLM7) from S. clavuligerus.
Results
Sequence Similarity Network for the Terpene Synthase-Like 2 Subgroup.
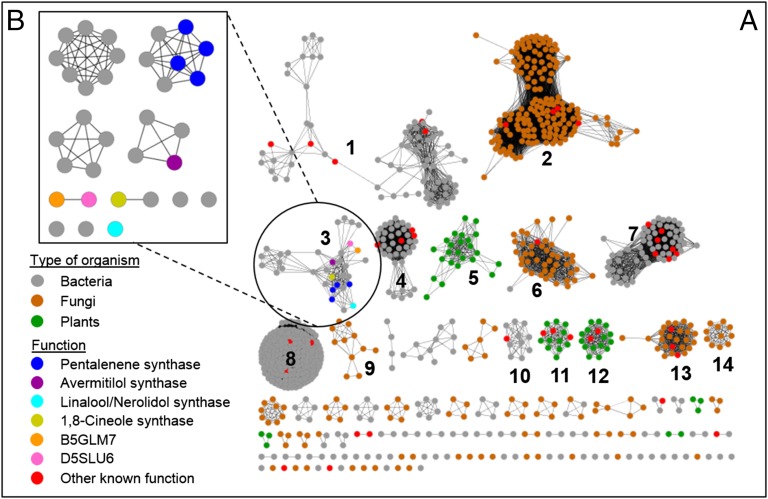
Class I and class II terpene synthases can be distinguished from one another based on sequence homology. The class I terpene synthases, which are related to polyprenyl diphosphate synthases, have a single all α-helix α-domain (PF03936) and are found mostly in bacteria and fungi. The Structure-Function Linkage Database (SFLD) (26) assigns 931 protein sequences to the class I terpene synthases, which are listed in the “terpene synthase-like 2 subgroup” (26). We constructed sequence similarity networks using these sequences (26). At an e-value cutoff of 10−50, these enzymes segregate into 14 main clusters containing 10 or more members (Fig. 1A). Terpene synthases with similar functions sometimes group in the same clusters, as illustrated by epi-isozizaene synthases (cluster 4), 2-methylisoborneol synthases (cluster 7), aristolochene synthases (cluster 13), and geosmin synthases (cluster 8) (27–30). (See Fig. S1 for a list of the products’ structures.)
Fig. 1.
Sequence similarity network of terpene synthase-like 2 subgroup (A) with an e-value of 10−50 and (B) zoom in at cluster 3 with an e-value of 10−75. D5SLU6 is a protein variant of B5GLM7 that overlaps with B5GLM7 and possesses an additional 16 residues at the N-terminal of the protein.
Other clusters contain multiple, seemingly unrelated functions. For example, cluster 3 contains pentalenene synthases, an avermitilol synthase, an acyclic terpene synthase that produces nerolidol and linalool, and a monoterpene cyclase that produces 1,8-cineole (13, 14, 31, 32). The product diversity observed in this cluster suggests that many of the unknown terpene synthases in this cluster that are annotated as pentalenene synthases may be incorrect. Interestingly, when cluster 3 was viewed at a higher resolution (e-value = 10−75), avermitilol synthase, linalool/nerolidol synthase, and 1,8-cineole synthase are separated from the pentalenene synthases (Fig. 1B). Cluster 3 also contains 1 of the 20 putative terpene synthases encoded in the genome of S. clavuligerus (Uniprot: B5GLM7), which is annotated as a putative pentalenene synthase. Although the enzyme is located in the same cluster as pentalenene synthase in the low-resolution map (e-value = 10−50), the enzyme does not cluster with a known terpene synthase when viewed at higher resolution (Fig. 1). This observation suggests that the function of B5GLM7 might differ from those of the known terpene synthases in the cluster. Sequence alignments with B5GLM7 revealed that it has a 46–50% sequence similarity (33–37% sequence identity) with terpene synthases of known function in the cluster (Table S1).
Workflow and Prediction of Product Skeletons.
Our workflow for prediction of product skeletons uses the amino acid sequence for a terpene synthase as the input and predicts the most likely product skeletons. We anticipated that it would be challenging to predict the complete structure of the product, i.e., including all stereochemical assignments, and therefore simplified the predictions to focus instead on the cyclic skeletons of the carbocations without saturated alkyl side chains.
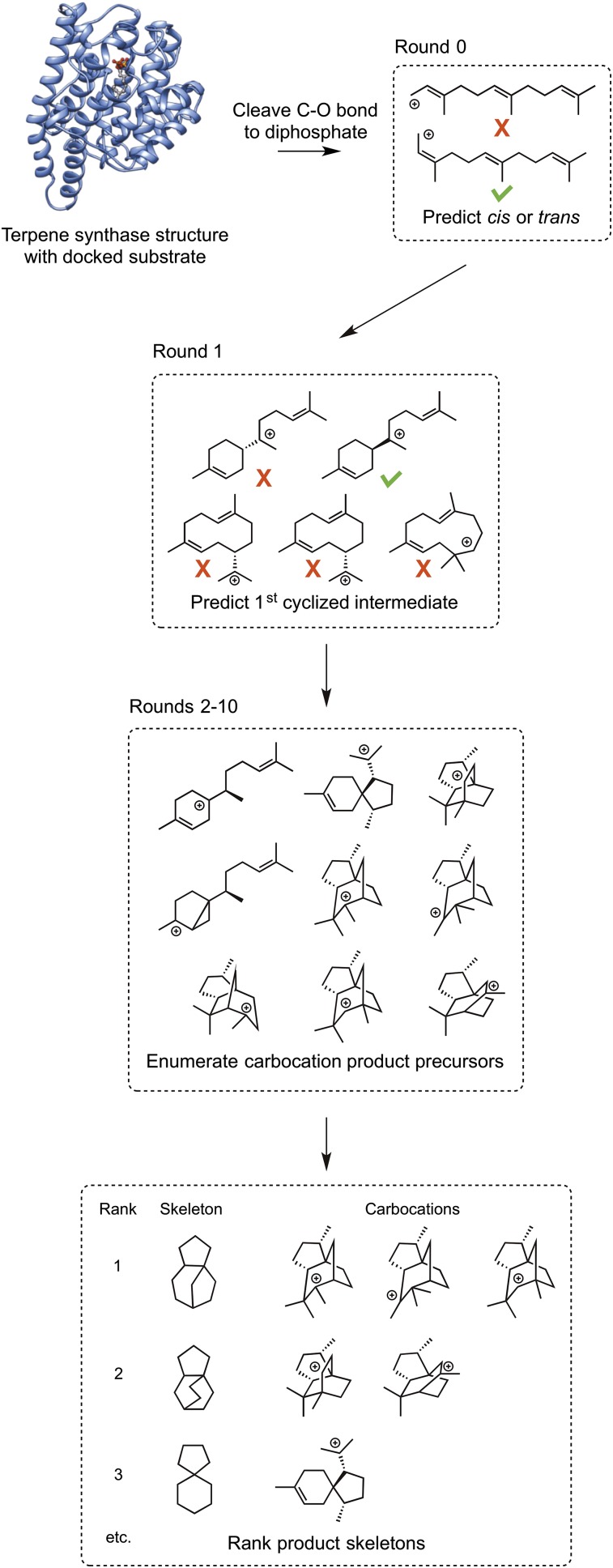
The workflow consists of homology modeling, substrate docking, enumerating carbocationic intermediates in the active site, and ranking product skeletons (Fig. 2). We use iGenPro to automatically enumerate possible carbocationic intermediates in the enzyme active site. Briefly, iGenPro starts with a terpene synthase crystal structure or homology model with substrate docked, and computes carbocationic reactions in the active site (Fig. 2 and Movie S1). The structures are evaluated by QM and docking scores to eliminate those predicted to have high energies or poor binding. Those that pass this filter become reactants for the next round. Finally, carbocations are grouped by skeletons and then are ranked according to a combined reaction energy and heuristic reaction barrier score. We expect that this score reflects a balance of thermodynamics and kinetics of the formation of the carbocations. The best ranking carbocation is used to determine the final rank of the skeleton (Fig. 2).
Fig. 2.
Computational workflow for the prediction of terpene synthase products.
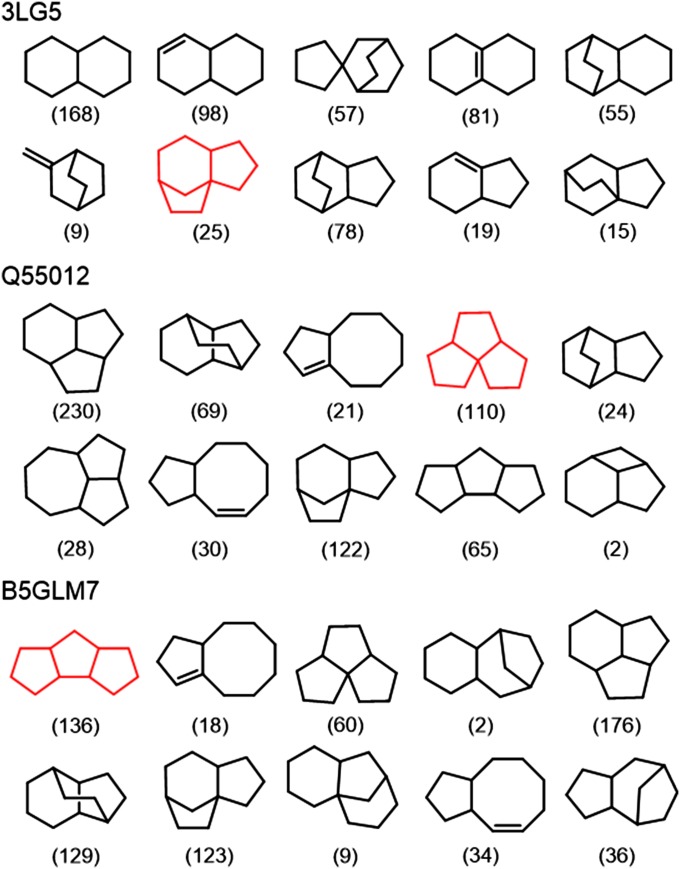
To validate our workflow, we made retrospective predictions for epi-isozizaene synthase [Protein Data Bank (PDB) ID code 3LG5] and pentalenene synthase (UniProt: Q55012) (Fig. 3 and Table S2). Epi-isozizaene synthase was selected because it is the most closely related enzyme to B5GLM7 with a crystal structure in a “closed” active conformation. Q55012 was selected because B5GLM7 was previously annotated as a “putative pentalenene synthase.” Although an apo crystal structure for Q55012 is available (PDB ID code 1PS1), the protein is in the unreactive apo conformation and lacks 20 residues at its C terminus. We constructed a homology model of Q55012 by using 3LG5 as template. FPP was correctly predicted as the substrate for both enzymes (Table S2). During the carbocation enumeration, 1,927 and 2,286 carbocationic intermediates were generated for 3LG5 and Q55012, respectively (Table S2). For 3LG5, the correct product precursor for epi-isozizaene was generated and ranked seventh (Fig. 3). The correct angular triquinane product precursor was generated for the Q55012 homology model and ranked fourth (Fig. 3). These results suggest that our predictions can predict the correct skeletons within the top 10 hits.
Fig. 3.
Top 10 predicted skeletons for 3LG5, Q55012, and B5GLM7. The number in the parentheses indicates the number of enumerated carbocations for each skeleton.
FPP was also the predicted substrate for B5GLM7. The top 10 predicted product skeletons are shown in Fig. 3. Our predictions suggested that the initial cyclization of the farnesyl cation in B5GLM7 gives a trans-humulyl cation, which is similar to the first step during the biosynthesis of pentalenene (33). However, the highest ranked product skeleton predicted for B5GLM7 was a linear triquinane (Fig. 3). In addition, the population of this skeleton was much higher than that for Q55012 (136 vs. 65; Fig. 3). Although linear triquinanes have been isolated from fungi and marine organisms, there were no known terpene synthases that synthesize this carbon skeleton when the predictions were made (34, 35).
Biochemical Characterization of B5GLM7.
Incubation of recombinant S. clavuligerus B5GLM7 with FPP gave five sesquiterpene hydrocarbons with molecular ions at m/z 204 (Fig. S2). GPP and GGPP were not substrates for the enzyme. Kinetic constants for the reaction were calculated from initial velocity measurements at different [FPP]: kcat = (1.2 ± 0.1) × 10−3 s−1, KM = 8 ± 2 μM, and kcat/KM = 1.4 × 102 M−1⋅s−1 (Fig. S2). Although the catalytic efficiency of B5GLM7 is low, it is within the 102- to 106-M−1⋅s−1 range reported for terpene synthases (10, 27, 29, 32, 36).
The mass spectrum of compound 5 matched that reported for β-caryophyllene in the National Institute of Standards and Technology database. Matches were not found for compounds 1–4. Compound 1 gave an exact mass at m/z 204.1878, consistent with a molecular formula of C15H24. NMR spectra were recorded in CDCl3, C6D6, and 3:1 (vol/vol) CDCl3:C6D6 to resolve all of the 1H resonances (Table S3 and Dataset S1). 1H, 1H-13C heteronuclear single quantum correlation (HSQC) and 1H-13C heteronuclear multiple-bond correlation (HMBC) spectra for compound 1 revealed the presence of an olefinic proton (H2, 5.03 ppm), a disubstituted olefinic carbon (C2, 127.6 ppm), and one trisubstituted olefinic carbon (C1, 154.0 ppm), which along with an unsaturation number of 4 suggests that compound 1 is a tricyclic hydrocarbon with one double bond (see Fig. 4 for numbering). Four methyl groups were seen in the HSQC spectrum, two of which are a geminal pair (C12, 30.2 ppm; H12, 1.10 ppm and C13, 28.3 ppm; H13, 1.04 ppm) attached to a quaternary carbon (C3, 50.8 ppm); a methyl (C14, 29.8 ppm; H14, 1.07 ppm) attached to a quaternary carbon (C7, 55.3 ppm); and a methyl (C15, 20.1 ppm; H15, 1.03 ppm) attached to a tertiary carbon (C10, 43.8 ppm; H10, 1.54 ppm). The remaining connectivities for methylene and methine carbons were deduced from 1H-13C HMBC and correlation spectroscopy spectra. Tertiary proton H11 was assigned from the long range 1H-13C couplings seen between H11 and C1, C2, C5, C7, C10, C14, and C15, whereas related couplings were seen between H5 and C1 and C2 and C4, in addition to 1H-1H couplings between H5 and H4 and H6. The connectivity of the remaining diastereotropic protons H4, H6, H8, and H9 were readily determined from the long-range 1H-13C coupling constants between the respective protons and their neighboring carbon atoms. Our NMR assignments agree with those recently reported by Yamada et al., except we assigned the resonances at H6 to 0.85 ppm, 1.77 ppm and at H4 to 1.26 ppm, 1.84 ppm (15).
Fig. 4.
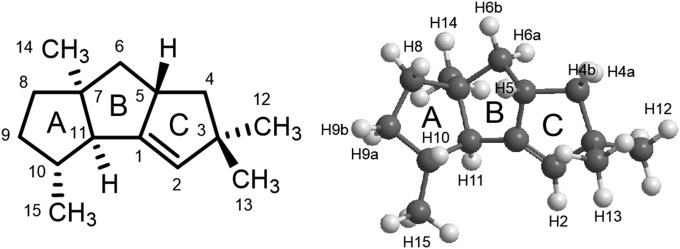
Two- and three-dimensional structure of (5S,7S,10R,11S)-cucumene.
The relative stereochemistry of compound 1 was deduced from the 1D nuclear Overhauser effect (NOE) data listed in Table S4 and Dataset S2. NOE cross-peaks between H11 and the C14 methyl group suggested a cis fusion of rings B and C (Fig. 4). The orientation of the bridgehead proton H5 relative to the BC ring fusion was deduced from strong cross-peaks between H5 and H4b and between H6a and the C14 methyl group. Substantially weaker cross-peaks were seen for H5-H4a, H5-H6a, and H14 methyl-H6b. A cross-peak between H5 and H10 allowed us to determine the configuration of the last stereo center. Distances between hydrogen atoms in the eight possible diastereomers for compound 1 were determined from density functional theory (DFT)-optimized structures (Fig. S3) and are listed in Table S4 with distances between hydrogen atoms being color coded. The NOE data most closely match those calculated for structures P1 and P5. NOEs between H5 and H10 and between H9a and the C14 methyl group indicate that compound 1 is the P1 diastereomer shown in Fig. 4. Although it is not obvious from the 2D structure of compound 1, H5 and H10 are 2.7 Å apart in the P1 diastereomer (Table S4) and ≥4 Å in the other seven. Also, the distance between H9a and the C14 methyl in P1 is 2.5 Å in P1 and 4.2 Å in P5. Finally, 13C chemical shifts were calculated for the eight diastereomers using gauge independent atomic orbital-based quantum mechanical calculations to estimate the nuclear magnetic shielding associated with each molecular configuration (37). The mean absolute deviations (|Δδ|av) of the calculated and experimental 13C chemical shifts for the eight diastereomers was |Δδ|av = 1.6 ppm for the P1 diastereomer vs. |Δδ|av = 2.3–4.8 ppm for P2–P7 (Dataset S3).
The absolute stereochemistry of compound 1 was determined by comparison of its electronic circular dichroism (ECD) spectrum with a spectrum computed for the rigid carbon skeleton of the P1 diastereomer (38). We examined the robustness of this method with four rigid monoterpenes, with excellent matches for (−) and (+)-borneol, camphene, limonene, and α-pinene (Dataset S4). The ECD spectrum of compound 1 compared favorably with the calculated spectra for the (5S,7S,10R,11S) diastereomer (Dataset S4). A 2D structure for compound 1 was published when this manuscript was in the final stage of preparation (15). The authors named the sesquiterpene iso-hirsutene; however, we prefer cucumene, in recognition of the proposal that the methyl substitution patterns of the two carbon skeletons arise from nontrivial differences in the mechanisms for their formation (39).
Discussion
Bacteria, fungi, and plants synthesize a large group of structurally diverse cyclic terpenoids. In bacteria, the genes that encode the biosynthetic enzymes are frequently located in clusters, which are typically identified by the presence of a gene for a putative terpene synthase (28, 40). The explosion of newly sequenced microbial genomes has facilitated genome-mining efforts to discover new terpenoid metabolites. This process typically involves expressing the genes in the biosynthesis clusters and determining the function of each enzyme (40–42). This process is a nontrivial endeavor, and it is desirable to have some indication of the potential novelty of a metabolite when selecting which biosynthesis clusters to study. Because terpene synthases determine the skeletons of the biosynthetic metabolites, knowledge of their functions is an important first step in assessing the potential novelty of metabolites in a pathway.
Terpene synthases catalyze some of the most complex reactions in nature, where the substrates undergo a series of complex chemical transformations. For example, presilphiperfolan-8β-ol synthase catalyzes the highly selective seven-step transformation of FPP to presilphiperfolan-8β-ol (39, 43). Each of these steps is exquisitely sensitive to the steric and electrostatic environment of the active site, and single mutations are often sufficient to substantially alter the structures of the products. Thus, although members of the terpene synthase family can be identified by phylogenetic comparisons, their functions cannot. It is costly and time-consuming to biochemically establish the function of most terpene synthases.
We now demonstrated, as a proof of concept, that computational approaches can be used to facilitate the characterization of terpene synthases at multiple levels. The protocols that we developed for assessing the function of a terpene synthase from its amino acid sequence are not computationally intensive but sufficiently powerful to be useful for identifying potential targets with novel or interesting functions. The steps in the protocol involve (i) construction of a homology model for the unknown terpene synthase, (ii) enumerating carbocations in the active site of the model, and (iii) ranking of the carbocations to provide a ranked list of potential carbon skeletons for the products. Other QM methods such as DFT should in principle provide more accurate predictions. However, due to the high computational cost, such methods are not applied to the enumeration workflow. This strategy was successfully demonstrated for a terpene synthase (B5GLM7) from S. clavuligerus whose function was unknown when this work was performed. The highest-ranked carbon skeleton was a C15 linear triquinane, and on biochemical characterization, we discovered that B5GLM7 converts FPP into (5S,7S,10R,11S)-cucumene, a linear triquinane sesquiterpene. Our calculations predicted three different cucumene stereoisomers (P1, P2, and P5 in Fig. S3), and the correct stereoisomer ranked highest among the three.
Comparisons between pentalenene synthase (Q55012) and cucumene synthase (B5GLM7) suggest that the different predictions for the two enzymes can be attributed to the differences in a few active site residues. An alignment of the homology models for the enzymes revealed a good overlap between the structures except for two loop regions (circled) that are remote from the active site (Fig. S4). Several of the active site residues, Leu53/Leu54, Phe57/Phe58, Met73/Met74, Phe77/Phe78, Trp308/Trp310, and Tyr315/Tyr317, are conserved in Q55012/B5GLM7, respectively. However, others, Phe76/Tyr77, Tyr150/His155, Ile177/Ser182, Asn215/Ile219, and Glu311/His309, are not. Although we are uncertain about the contributions of each of these mutations to the enzyme’s specificity, site-directed mutagenesis should provide information about the importance of these residues.
The putative mechanism for formation of the carbon skeleton for cucumene is closely related to those suggested for linear and angular triquinane sesquiterpenes (39). A key step in all of these pathways is the 1,11 ring closure of the trans-farnesyl cation to give a trans-humulyl cation. This suggestion is supported for cucumene by the coformation of caryophyllene as a minor product. The trans-humulyl cation can then undergo two additional cyclizations to form the protoilludyl cation, which is the putative precursor for sesquiterpene hydrocarbons with protoilludane, hirsutane, pleuroteilane, tremulane, ceratopicane, cerapicane, and sterpurane carbon skeletons. Although a different reaction channel was originally proposed for pentalenene, recent isotopically sensitive branching experiments suggest that the protoilludyl cation is also the precursor for formation of the pentalenane skeleton (33, 36). The protoilludyl cation is a likely intermediate in the formation of cucumene (39).
We anticipate that the ultimate metabolite of the biosynthesis gene cluster containing B5GLM7 is an oxidized derivative of cucumene. A putative cytochrome P450 oxidase (Uniprot: B5GLM9) and a multicopper oxidase (Uniprot: B5GLM3) are located upstream and downstream of B5GLM7, respectively. Cytochrome monooxygenases are often involved in oxidative modification reactions in terpene biosynthesis (44). For example, eight linear triquinane metabolites with cucumane, hirsutane, and ceratopicane carbon skeletons isolated from Macrocystidia cucumis were oxidized at two or more positions (45). Typically, linear triquinane metabolites described in the literature were isolated from fungi in the phylum basidiomycota and often possess antifungal, antibacterial, and/or cytotoxic activity (46). These molecules have been targets for numerous synthetic studies (47, 48).
In summary, we developed a computational approach for predicting the hydrocarbon skeletons of products synthesized by terpene synthases based on homology models constructed from their amino acid sequences. The procedure was applied retrospectively to two known terpene synthases and gave the correct carbon skeleton for the products as the fourth and seventh ranked structures of 120 and 216 candidates, respectively. Finally, the procedure was applied to a terpene synthase of unknown function and the top ranked linear triquinane structure matched that of the product whose structure was determined subsequently. We anticipate this approach will be useful for identifying bacterial gene clusters that direct the biosynthesis of structurally interesting terpenoid metabolites.
Methods
Automatic Enumeration of Carbocations (Algorithm of the iGenPro Code).
Using the predicted model for the substrate-bound enzyme (i.e., the Michaelis complex, in this case with FPP), iGenPro first cleaved the C-O bond of the substrate, keeping the diphosphate moiety in the receptor structure. The obtained receptor structure was used for further molecular mechanics minimizations. The OPLS 2005 force field was used throughout the study (49). In round 0, the trans-carbocation derived from the allylic substrate was copied and transformed into its cis isomer by rotating about the C2-C3 bond (Fig. 2). The estimated relative binding affinities of both isomers of the linear carbocation were then calculated using molecular mechanics energies, and the one with better docking score was moved forward to round 1. iGenPro then generated cyclic carbocationic intermediates (Fig. 2). If the trans isomer was chosen, 1,6-cyclization intermediates were automatically discarded because that mode of cyclization requires the cis isomer of the allylic intermediate. Again, only one cyclized intermediate, with the best docking score, was retained in round 1. Thus, in rounds 0 and 1, the algorithm predicts the first cyclized intermediate, in a manner similar to our previous reaction channel prediction for the triterpenoid synthases (25).
From round 2, iGenPro enumerated a large number of carbocationic intermediates in the active site (more than 15,000 for each enzyme). Briefly, the molecular connectivity of the reactant carbocation was changed according to five predefined reaction types: (i) intramolecular alklation of double bonds; (ii) alkyl shift (excluding 1,2-methyl shift); (iii) hydride shift; (iv) 1,2-methyl shift; and (v) proton transfer. The simplified molecular-input line-entry system string was used for rejecting duplicates. MM minimizations were performed on the product carbocations in the presence of the receptor structure. The receptor was frozen in all MM minimizations. A semiempirical QM method, RM1, was used to remove high-energy intermediates (50, 51). QM calculations were only performed on the ligands. Molecular mechanics energies, using partial charges from the QM calculates, were used to reject carbocations with poor geometric (steric clashes) or electrostatic complementarity to the binding site. We used a threshold value of −40 kcal/mol, an empirical value from our docking studies, to reject carbocations. We were unable to enumerate all possible carbocations in the active site of the terpene synthase because the computational cost would be prohibitive. Instead, we ran full enumerations from rounds 2–4, and from round 5 onward, only the top 300 carbocations with the lowest energies were retained. This process was repeated until round 10, with the assumption that product would be formed within 10 steps.
Ranking Product Skeletons.
The output of iGenPro is a network of carbocationic intermediates, where nodes are carbocations and edges are different reaction types. In principle, it would be possible to estimate barriers between the intermediates using QM/MM methods; however, the computational time required is currently prohibitive. Instead, we estimated the barriers using a heuristic. For each carbocation, we use the graph traversal algorithm to find all of the reaction pathways automatically and then calculate the energetic spans for all these pathways (52). Finally, we select the lowest energetic span as the reaction barrier for this carbocation.
We then ranked all carbocationic intermediates by using a combined Z-score of QM energy and reaction barrier. The Z-score of a raw value x is defined by Eq. 1, where µ is the mean of the population (in this study is the average energy or energy barrier for all enumerated carbocations) and σ is the SD of the population.
The combined Z-score for the QM energies and estimated reaction barriers is a sum of individual Z-scores of the two attributes (Eq. 2).
Each carbocation was converted into a neutral product by adding a hydride on the positively charged carbon. These potential reaction products were then classified into skeletons by removing aliphatic side chains
| [1] |
| [2] |
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants GM-093342 (to S.C.A., M.P.J., and C.D.P.) and GM-25521 (to C.D.P.). J.-Y.C. was supported by the Agency for Science, Technology and Research International Fellowship, Singapore.
Footnotes
Conflict of interest statement: M.P.J. is a consultant to Schrodinger LLC, which distributes software used in this study. The authors declare no competing financial interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1505127112/-/DCSupplemental.
References
- 1.Zhao L, Chang WC, Xiao Y, Liu HW, Liu P. Methylerythritol phosphate pathway of isoprenoid biosynthesis. Annu Rev Biochem. 2013;82:497–530. doi: 10.1146/annurev-biochem-052010-100934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vranová E, Coman D, Gruissem W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu Rev Plant Biol. 2013;64:665–700. doi: 10.1146/annurev-arplant-050312-120116. [DOI] [PubMed] [Google Scholar]
- 3.Wallrapp FH, et al. Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily. Proc Natl Acad Sci USA. 2013;110(13):E1196–E1202. doi: 10.1073/pnas.1300632110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sacchettini JC, Poulter CD. Creating isoprenoid diversity. Science. 1997;277(5333):1788–1789. doi: 10.1126/science.277.5333.1788. [DOI] [PubMed] [Google Scholar]
- 5.Christianson DW. Unearthing the roots of the terpenome. Curr Opin Chem Biol. 2008;12(2):141–150. doi: 10.1016/j.cbpa.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christianson DW. Structural biology and chemistry of the terpenoid cyclases. Chem Rev. 2006;106(8):3412–3442. doi: 10.1021/cr050286w. [DOI] [PubMed] [Google Scholar]
- 7.Gao Y, Honzatko RB, Peters RJ. Terpenoid synthase structures: A so far incomplete view of complex catalysis. Nat Prod Rep. 2012;29(10):1153–1175. doi: 10.1039/c2np20059g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kenig M, Reading C. Holomycin and an antibiotic (MM 19290) related to tunicamycin, metabolites of Streptomyces clavuligerus. J Antibiot (Tokyo) 1979;32(6):549–554. doi: 10.7164/antibiotics.32.549. [DOI] [PubMed] [Google Scholar]
- 9.Tahlan K, Anders C, Jensen SE. The paralogous pairs of genes involved in clavulanic acid and clavam metabolite biosynthesis are differently regulated in Streptomyces clavuligerus. J Bacteriol. 2004;186(18):6286–6297. doi: 10.1128/JB.186.18.6286-6297.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu Y, Chou WK, Hopson R, Cane DE. Genome mining in Streptomyces clavuligerus: Expression and biochemical characterization of two new cryptic sesquiterpene synthases. Chem Biol. 2011;18(1):32–37. doi: 10.1016/j.chembiol.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang J, He X, Cane DE. Geosmin biosynthesis. Streptomyces coelicolor germacradienol/germacrene D synthase converts farnesyl diphosphate to geosmin. J Am Chem Soc. 2006;128(25):8128–8129. doi: 10.1021/ja062669x. [DOI] [PubMed] [Google Scholar]
- 12.Cane DE, He X, Kobayashi S, Omura S, Ikeda H. Geosmin biosynthesis in Streptomyces avermitilis. Molecular cloning, expression, and mechanistic study of the germacradienol/geosmin synthase. J Antibiot (Tokyo) 2006;59(8):471–479. doi: 10.1038/ja.2006.66. [DOI] [PubMed] [Google Scholar]
- 13.Nakano C, Kim HK, Ohnishi Y. Identification and characterization of the linalool/nerolidol synthase from Streptomyces clavuligerus. ChemBioChem. 2011;12(16):2403–2407. doi: 10.1002/cbic.201100501. [DOI] [PubMed] [Google Scholar]
- 14.Nakano C, Kim HK, Ohnishi Y. Identification of the first bacterial monoterpene cyclase, a 1,8-cineole synthase, that catalyzes the direct conversion of geranyl diphosphate. ChemBioChem. 2011;12(13):1988–1991. doi: 10.1002/cbic.201100330. [DOI] [PubMed] [Google Scholar]
- 15.Yamada Y, et al. Novel terpenes generated by heterologous expression of bacterial terpene synthase genes in an engineered Streptomyces host. J Antibiot (Tokyo) 2015 doi: 10.1038/ja.2014.171. , 10.1038/ja.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steele CL, Crock J, Bohlmann J, Croteau R. Sesquiterpene synthases from grand fir (Abies grandis). Comparison of constitutive and wound-induced activities, and cDNA isolation, characterization, and bacterial expression of delta-selinene synthase and gamma-humulene synthase. J Biol Chem. 1998;273(4):2078–2089. doi: 10.1074/jbc.273.4.2078. [DOI] [PubMed] [Google Scholar]
- 17.Yoshikuni Y, Martin VJ, Ferrin TE, Keasling JD. Engineering cotton (+)-delta-cadinene synthase to an altered function: Germacrene D-4-ol synthase. Chem Biol. 2006;13(1):91–98. doi: 10.1016/j.chembiol.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 18.Li R, et al. Reprogramming the chemodiversity of terpenoid cyclization by remolding the active site contour of epi-isozizaene synthase. Biochemistry. 2014;53(7):1155–1168. doi: 10.1021/bi401643u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerlt JA, et al. The Enzyme Function Initiative. Biochemistry. 2011;50(46):9950–9962. doi: 10.1021/bi201312u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian BX, Eriksson LA. Catalytic mechanism and product specificity of oxidosqualene-lanosterol cyclase: A QM/MM study. J Phys Chem B. 2012;116(47):13857–13862. doi: 10.1021/jp3091396. [DOI] [PubMed] [Google Scholar]
- 21.Weitman M, Major DT. Challenges posed to bornyl diphosphate synthase: diverging reaction mechanisms in monoterpenes. J Am Chem Soc. 2010;132(18):6349–6360. doi: 10.1021/ja910134x. [DOI] [PubMed] [Google Scholar]
- 22.Hong YJ, Tantillo DJ. Branching out from the bisabolyl cation. Unifying mechanistic pathways to barbatene, bazzanene, chamigrene, chamipinene, cumacrene, cuprenene, dunniene, isobazzanene, iso-γ-bisabolene, isochamigrene, laurene, microbiotene, sesquithujene, sesquisabinene, thujopsene, trichodiene, and widdradiene sesquiterpenes. J Am Chem Soc. 2014;136(6):2450–2463. doi: 10.1021/ja4106489. [DOI] [PubMed] [Google Scholar]
- 23.Isegawa M, Maeda S, Tantillo DJ, Morokuma K. Predicting pathways for terpene formation from first principles: Routes to known and new sesquiterpenes. Chem Science. 2014;5(4):1555–1560. [Google Scholar]
- 24.Tantillo DJ. Biosynthesis via carbocations: Theoretical studies on terpene formation. Nat Prod Rep. 2011;28(6):1035–1053. doi: 10.1039/c1np00006c. [DOI] [PubMed] [Google Scholar]
- 25.Tian BX, et al. Predicting the functions and specificity of triterpenoid synthases: A mechanism-based multi-intermediate docking approach. PLOS Comput Biol. 2014;10(10):e1003874. doi: 10.1371/journal.pcbi.1003874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akiva E, et al. The structure-function linkage database. Nucleic Acids Res. 2014;42(Database issue):D521–D530. doi: 10.1093/nar/gkt1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin X, Hopson R, Cane DE. Genome mining in Streptomyces coelicolor: Molecular cloning and characterization of a new sesquiterpene synthase. J Am Chem Soc. 2006;128(18):6022–6023. doi: 10.1021/ja061292s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Komatsu M, Tsuda M, Omura S, Oikawa H, Ikeda H. Identification and functional analysis of genes controlling biosynthesis of 2-methylisoborneol. Proc Natl Acad Sci USA. 2008;105(21):7422–7427. doi: 10.1073/pnas.0802312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Felicetti B, Cane DE. Aristolochene synthase: Mechanistic analysis of active site residues by site-directed mutagenesis. J Am Chem Soc. 2004;126(23):7212–7221. doi: 10.1021/ja0499593. [DOI] [PubMed] [Google Scholar]
- 30.Cane DE, Watt RM. Expression and mechanistic analysis of a germacradienol synthase from Streptomyces coelicolor implicated in geosmin biosynthesis. Proc Natl Acad Sci USA. 2003;100(4):1547–1551. doi: 10.1073/pnas.0337625100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lesburg CA, Zhai G, Cane DE, Christianson DW. Crystal structure of pentalenene synthase: Mechanistic insights on terpenoid cyclization reactions in biology. Science. 1997;277(5333):1820–1824. doi: 10.1126/science.277.5333.1820. [DOI] [PubMed] [Google Scholar]
- 32.Chou WK, et al. Genome mining in Streptomyces avermitilis: Cloning and characterization of SAV_76, the synthase for a new sesquiterpene, avermitilol. J Am Chem Soc. 2010;132(26):8850–8851. doi: 10.1021/ja103087w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zu L, et al. Effect of isotopically sensitive branching on product distribution for pentalenene synthase: Support for a mechanism predicted by quantum chemistry. J Am Chem Soc. 2012;134(28):11369–11371. doi: 10.1021/ja3043245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liermann JC, et al. Hirsutane-type sesquiterpenes with uncommon modifications from three basidiomycetes. J Org Chem. 2010;75(9):2955–2961. doi: 10.1021/jo100202b. [DOI] [PubMed] [Google Scholar]
- 35.Chang CH, Wen ZH, Wang SK, Duh CY. Capnellenes from the Formosan soft coral Capnella imbricata. J Nat Prod. 2008;71(4):619–621. doi: 10.1021/np0706116. [DOI] [PubMed] [Google Scholar]
- 36.Seemann M, et al. Pentalenene synthase. Analysis of active site residues by site-directed mutagenesis. J Am Chem Soc. 2002;124(26):7681–7689. doi: 10.1021/ja026058q. [DOI] [PubMed] [Google Scholar]
- 37.Bifulco G, Dambruoso P, Gomez-Paloma L, Riccio R. Determination of relative configuration in organic compounds by NMR spectroscopy and computational methods. Chem Rev. 2007;107(9):3744–3779. doi: 10.1021/cr030733c. [DOI] [PubMed] [Google Scholar]
- 38.Berova N, Di Bari L, Pescitelli G. Application of electronic circular dichroism in configurational and conformational analysis of organic compounds. Chem Soc Rev. 2007;36(6):914–931. doi: 10.1039/b515476f. [DOI] [PubMed] [Google Scholar]
- 39.Quin MB, Flynn CM, Schmidt-Dannert C. Traversing the fungal terpenome. Nat Prod Rep. 2014;31(10):1449–1473. doi: 10.1039/c4np00075g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tetzlaff CN, et al. A gene cluster for biosynthesis of the sesquiterpenoid antibiotic pentalenolactone in Streptomyces avermitilis. Biochemistry. 2006;45(19):6179–6186. doi: 10.1021/bi060419n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeyhle P, et al. Genome-based discovery of a novel membrane-bound 1,6-dihydroxyphenazine prenyltransferase from a marine actinomycete. PLoS ONE. 2014;9(6):e99122. doi: 10.1371/journal.pone.0099122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin HC, et al. Generation of complexity in fungal terpene biosynthesis: Discovery of a multifunctional cytochrome P450 in the fumagillin pathway. J Am Chem Soc. 2014;136(11):4426–4436. doi: 10.1021/ja500881e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang CM, Hopson R, Lin X, Cane DE. Biosynthesis of the sesquiterpene botrydial in Botrytis cinerea. Mechanism and stereochemistry of the enzymatic formation of presilphiperfolan-8beta-ol. J Am Chem Soc. 2009;131(24):8360–8361. doi: 10.1021/ja9021649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cane DE, Ikeda H. Exploration and mining of the bacterial terpenome. Acc Chem Res. 2012;45(3):463–472. doi: 10.1021/ar200198d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hellwig V, et al. New triquinane-type sesquiterpenoids from Macrocystidia cucumis (Basidiomycetes) Eur J Org Chem. 1998;1998(1):73–79. [Google Scholar]
- 46.Abraham WR. Bioactive sesquiterpenes produced by fungi: Are they useful for humans as well? Curr Med Chem. 2001;8(6):583–606. doi: 10.2174/0929867013373147. [DOI] [PubMed] [Google Scholar]
- 47.Takeuchi T, Iinuma H, Iwanaga J, Takahashi S, Takita T. Coriolin, a new Basidiomycetes antibiotic. J Antibiot (Tokyo) 1969;22(5):215–217. doi: 10.7164/antibiotics.22.215. [DOI] [PubMed] [Google Scholar]
- 48.Hu QY, Zhou G, Corey EJ. Application of chiral cationic catalysts to several classical syntheses of racemic natural products transforms them into highly enantioselective pathways. J Am Chem Soc. 2004;126(42):13708–13713. doi: 10.1021/ja046154m. [DOI] [PubMed] [Google Scholar]
- 49.Banks JL, et al. Integrated Modeling Program, Applied Chemical Theory (IMPACT) J Comput Chem. 2005;26(16):1752–1780. doi: 10.1002/jcc.20292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anonymous 2014. Schrödinger Suite. Impact Version 6.5; Prime Version 3.8; MacroModel, Version 10.5 (Schrödinger, Inc., New York)
- 51.Rocha GB, Freire RO, Simas AM, Stewart JJ. RM1: A reparameterization of AM1 for H, C, N, O, P, S, F, Cl, Br, and I. J Comput Chem. 2006;27(10):1101–1111. doi: 10.1002/jcc.20425. [DOI] [PubMed] [Google Scholar]
- 52.Kozuch S, Shaik S. How to conceptualize catalytic cycles? The energetic span model. Acc Chem Res. 2011;44(2):101–110. doi: 10.1021/ar1000956. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.