SUMMARY
N6-methyldeoxyadenosine (6mA or m6A) is a DNA modification preserved in prokaryotes to eukaryotes. It is widespread in bacteria, and functions in DNA mismatch repair, chromosome segregation, and virulence regulation. In contrast, the distribution and function of 6mA in eukaryotes have been unclear. Here we present a comprehensive analysis of the 6mA landscape in the genome of Chlamydomonas using new sequencing approaches. We identified the 6mA modification in 84% of genes in Chlamydomonas. We found that 6mA mainly locates at ApT dinucleotides around transcription start sites (TSS) with a bimodal distribution, and appears to mark active genes. A periodic pattern of 6mA deposition was also observed at base resolution, which is associated with nucleosome distribution near the TSS, suggesting a possible role in nucleosome positioning. The new genome-wide mapping of 6mA and its unique distribution in the Chlamydomonas genome suggest potential regulatory roles of 6mA in gene expression in eukaryotic organisms.
INTRODUCTION
Covalent modifications of individual bases in DNA can encode inheritable genetic information beyond the four canonical DNA bases (Bird, 2007). Methylations of DNA, including 5mC (Sasaki and Matsui, 2008) and 6mA (Wion and Casadesus, 2006), are the most abundant modifications in both prokaryotic and eukaryotic organisms. The well-studied 5mC modification in multicellular eukaryotes regulates diverse cellular and developmental processes (Law and Jacobsen, 2010; Smith and Meissner, 2013); however, the biological function of 6mA in eukaryotes is still unclear.
6mA is known to be present in the genomic DNA of viruses, bacteria, protists, fungi, and algae, and has been detected in plant DNA and mosquito DNA (Ratel et al., 2006). In bacteria, 6mA plays crucial roles in the regulation of DNA mismatch repair (Messer and Noyer-Weidner, 1988), chromosome replication (Lu et al., 1994), cell defense, cell cycle regulation (Collier et al., 2007), transcription and virulence (Low et al., 2001). The maps of 6mA in several bacteria strains have been obtained by using single-molecule real-time (SMRT) sequencing (Fang et al., 2012; Murray et al., 2012).
Besides bacteria, certain unicellular eukaryotes also contain 6mA in their genomes. For instance, the protozoan Tetrahymena (Hattman et al., 1978), Oxytricha fallox (Rae and Spear, 1978), and Paramecium aurelia (Cummings et al., 1974) have relatively abundant 6mA but little 5mC. On the other hand, green algae Chlamydomonas reinhardtii (Hattman et al., 1978) and Volvox carteri (Babinger et al., 2001) possess both 6mA and 5mC. Although common in bacteria, no corresponding restriction endonucleases have been reported in these species. Therefore, 6mA in these unicellular eukaryotic genomes has long been suspected of possessing functions other than exclusion of foreign DNA or viruses (Ehrlich and Zhang, 1990). Additionally, evidences for the existence of 6mA in plants, insects, and mammals have also been reported.
Chlamydomonas reinhardtii (referred to hereafter as Chlamydomonas) is a unicellular green alga that has been widely used as a model organism to study photosynthesis, eukaryotic flagella, and biomass production (Merchant et al., 2007). The high level (~0.3-0.5 mol%) of 6mA in the nuclear DNA of Chlamydomonas (Hattman et al., 1978) prompted us to study its distribution and function, which could help to decipher the long mystery of 6mA in eukaryotes and to develop bioengineering tools that may facilitate biomass and biofuel production (Radakovits et al., 2010).
In this study, we employed/developed several methods for mapping 6mA sites in genomic DNA. We first applied 6mA immunoprecipitation sequencing, or 6mA-IP-seq, which is an antibody-based profiling method to obtain the genome-wide distribution of 6mA. We then developed a 6mA-CLIP-exo strategy of employing photo-crosslinking followed by exonuclease digestion to achieve a much higher resolution. Lastly, we developed a restriction enzyme-based 6mA sequencing, or 6mA-RE-seq, to detect 6mA sites at single nucleotide resolution in genome-wide. Application of these three approaches to the Chlamydomonas genome revealed that 6mA marks more than 14,000 genes, accounting for 84% of all Chlamydomonas genes. This methylation is highly enriched around transcription start sites (TSS) with a bimodal distribution and significant local depletion at TSS. We used RNA-seq to quantify gene expression, and found that the presence of 6mA is correlated with actively expressed genes. This pattern is distinct from that of 5mC, which accumulates mostly in gene bodies in Chlamydomonas. At single nucleotide resolution, we also discovered that 6mA is enriched around TSS but exhibits an unexpected, strongly periodic pattern, suggesting controlled deposition of 6mA in association with nucleosome spacing. Nucleosome profiling revealed that 6mA around TSS occurs primarily within the linker DNA between nucleosomes. Our data show that 6mA is an abundant DNA mark associated with actively expressed genes in Chlamydomonas. These methods and results should stimulate future functional investigations of 6mA in Chlamydomonas and other eukaryotic organisms.
RESULTS
6mA is a Stable Modification in Chlamydomonas Genomic DNA
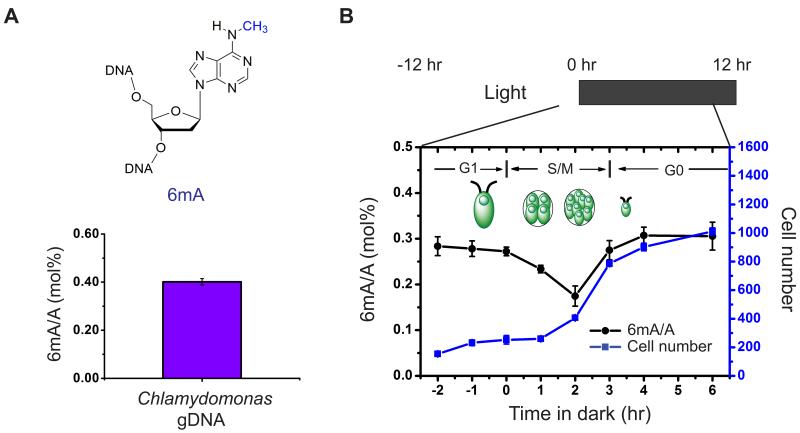
To accurately quantify the level of 6mA in genomic DNA, we applied an LC-MS/MS assay using pure 6mA nucleoside as an external standard (Figures S1A, S1B) (Jia et al., 2011). In agreement with the previous data (Hattman et al., 1978), we detected ~0.4 mol% of 6mA (6mA/A) in the genomic DNA isolated from Chlamydomonas cultured in mixotrophic conditions, i.e. Tris-Acetate-Phosphate (TAP) medium under constant light (Figure 1A).
Figure 1. The Presence and Conservation of 6mA in Chlamydomonas Genomic DNA.
(A) The presence of 6mA in isolated Chlamydomonas genomic DNA is determined by UHPLC-QQQ-MS/MS. Ratios of 6mA/A are shown (n = 6, mean ± S.E.M.).
(B) The level of 6mA decreases at the beginning of S/M phase during DNA replication, and increases back to the original level at the late stage of S/M phase. During a multiple fission cell cycle in naturally synchronized cells, cells begin to replicate its DNA 1 hr after dark and divide two times, leading to ~4-fold increase of the total cell number. Ratios of 6mA/A are shown on the left axis (n = 4, mean ± S.E.M.) with calibrated cell concentration (cells/uL) shown on the right axis (n = 2, mean ± S.E.M.).
See also Figure S1.
To determine if 6mA is stable during cell growth, we monitored the 6mA level during a multiple fission cell cycle in naturally synchronized cells induced by a 12 hr/12 hr light/dark cycle in minimal media cultures (Bisova et al., 2005). Under such growth conditions, cells grow in size during the light phase (G1 phase), and then undergo two to three rapid rounds of alternating DNA replications and cellular divisions (S/M phase) from 1 hr to 5 hr after entering the dark phase. Cells were mostly synchronized and rapidly divided under this light-dark phase transition according to cell counting measured by flow cytometry (Figure 1B). The proportion of 6mA in genomic DNA was measured before and after the switch from light to dark. Our results showed that the overall 6mA level in genomic DNA decreased by ~40% in 2 hr after dark, corresponding to the time period when DNA was replicated. This level then rose quickly back to the original level within 2 hr. This result indicated that 6mA is installed on the newly synthesized DNA within a short time period after DNA replication and is stably maintained during cell proliferation (Figure 1B).
Genome-wide Mapping of 6mA with 6mA-IP-Seq
Although the existence of 6mA in Chlamydomonas has been known, its distribution/localizations are unclear. To generate a de novo map of the genome-wide distribution of 6mA, we applied 6mA-IP-seq. Similar to the methylated DNA immunoprecipitation (MeDIP) (Weber et al., 2005) that has been widely applied to enrich 5mC-containing DNA fragments, we sought to use a 6mA-specific antibody to enrich the 6mA-containing DNA fragments. An antibody that recognizes the N6-methyladenine base has recently been applied to genome-wide profiling of 6mA sites in RNA (Dominissini et al., 2012; Meyer et al., 2012). By performing dot-blot assay on synthesized 6mA-containing DNA oligonucleotide, we confirmed that this anti-6mA antibody can also specifically recognize 6mA in both single-stranded and double-stranded DNA (Figure S2).
We then isolated genomic DNA from Chlamydomonas, and fragmented it into 200-400 base pairs by sonication. The fragmented DNA was ligated to an adapter with specific index sequence (Figure 2), which was then denatured to single-stranded DNA, and immunoprecipitated using the anti-6mA antibody. The captured DNA was eluted through the competition with 6mA single nucleotide, and PCR amplified to construct the DNA library (Figure 2). Simultaneously, an input library was obtained by PCR amplification of the ligated DNA before immunoprecipitation. Both libraries were subjected to high-throughput sequencing. The obtained sequencing reads were mapped to a reference genome of Chlamydomonas (JGI version 9.1), and 6mA sites were identified using a peak-detection algorithm (Zhang et al., 2008). The false detection rate (FDR) was estimated to be below 0.01.
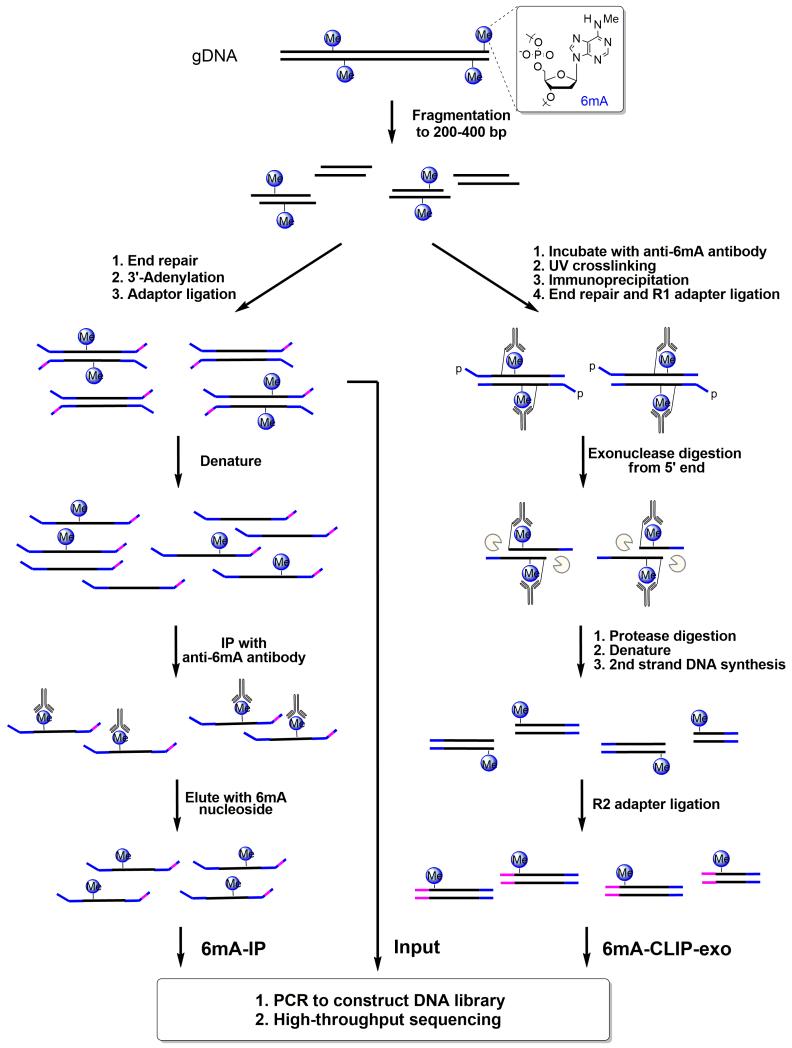
Figure 2. Schematic Diagram of 6mA-IP-seq and 6mA-CLIP-exo.
For 6mA-IP-seq (left), fragmented genomic DNA (gDNA) is ligated to a Y-shaped adapter with specific index sequence, denatured, and immunoprecipitated using anti-6mA antibody. The captured DNA is eluted with 6mA single nucleotide, and PCR amplified to construct the DNA library. Simultaneously, the input library was obtained from the ligated DNA before immunoprecipitation. For 6mA-CLIP-exo (right), fragmented gDNA is incubated with anti-6mA antibody, crosslinked by 254 nm UV irritation, and immunoprecipitated. The crosslinked DNA is ligated to adapter R1 on beads, followed by 5′ to 3′ exonuclease digestion. Antibody-protected DNA is preserved, and a 2nd-strand DNA synthesis is performed after protease digestion of the antibody. A second ligation to adapter R2 provides the template for PCR amplification to construct the library for high-throughput sequencing. Boundaries were determined by the sequencing ends of the 6mA-CLIP-exo-seq to provide a high resolution localization of 6mA.
See also Figure S2.
6mA Bases are Highly Enriched around TSS with a Bimodal Distribution
We performed 6mA-IP-seq on Chlamydomonas cultured under mixotrophic (constant light) or heterotrophic (constant dark) conditions in TAP medium during the pre-stationary phase. For each condition, we performed two biological replicates. After peak calling, we identified 25,803 and 28,982 high confidence 6mA peaks in light samples, and 22,005 and 21,016 peaks in dark samples (FDR<0.01), respectively. Among them, more than 95% of the peaks mutually occur in both replicate samples, indicating the high reproducibility of our approach (Figure S3A). About 88% of the peaks are common under both light and dark conditions, suggesting a faithful installation/maintenance mechanism of 6mA at specific genomic regions. Consistent with the previous measurements that 6mA was only detected in Chlamydomonas nuclear DNA but not chloroplast DNA, all the 6mA peaks were mapped to the nuclear genome but not the chloroplast genome. To our surprise, we observed that 6mA is highly enriched around the transcription start sites (TSS) of 14,868 genes, constituting 84% of all the genes in the Chlamydomonas genome (Figure 3A). A closer examination of the distribution revealed that the 6mA sites enriched around TSS (−500 to +800 bp, ~91% of all 6mA peaks) exhibit a bimodal distribution with a significant local depletion at TSS. The summit of the peak tends to locate within 500 bp downstream of TSS (Figure 3A). The rest of the 6mA peaks (~9%) not associated with TSS do not show specific patterns, and reside in both gene bodies and intergenic regions. The average peak width of the identified peaks is around 320 bp, which is consistent with the fragmentation size of our sequenced DNA (200-400 bp). We cannot quantify the number of methylation sites under each 6mA peak; however, some peaks are noticeably broader with certain peaks containing multiple sub-peak summits, suggesting the presence of multiple methylation sites in these regions (Figure 3B). Thus, our observation revealed a region-specific bimodal methylation pattern of 6mA highly enriched around TSS.
Figure 3. A Bimodal Distribution of 6mA around Transcription Start Sites.
(A) Distribution of 6mA peaks around TSS measured by 6mA-IP-seq. 6mA is enriched around TSS with a bimodal distribution and a local depletion at TSS. 6mA occupancy represents the reads coverage averaged by gene number in 6mA-IP-seq.
(B) Snapshot of 6mA peak determined by both 6mA-IP-seq and 6mA-CLIP-exo in specific gene loci. 6mA peaks can be detected both upstream and downstream of TSS in single direction promoter region and bidirectional promoter region. Some enrichment peaks are located in the first and second introns. Boundaries of 6mA-CLIP-exo-seq on both DNA strands were marked by magenta and blue color. Regions between the two nearest boundaries were determined as a 6mA-containing sequence. Black arrows indicate the transcription direction.
(C) Distribution of 6mA peaks around TSS measured by 6mA-CLIP-exo. The enrichment of 6mA near TSS shows a similar pattern as that obtained using 6mA-IP-seq. In addition, several spikes could be observed from the large peak.
(D) The dinucleotide sequence ApT is enriched in 6mA-CLIP-exo peaks, including CATG.
See also Figure S3.
6mA-CLIP-exo with Immunoprecipitation, Photo-crosslinking, and Exonuclease Digestion
Inspired by chromatin immunoprecipitation followed by exonuclease digestion (ChIP-exo), a method to map the locations at which a protein binds to the genome (Rhee and Pugh, 2012), we introduced photo-crosslinking after the antibody-based 6mA enrichment (Chen et al., 2015) followed by exonuclease digestion in an attempt to identify 6mA peaks with higher resolution. DNA/antibody complexes were covalently crosslinked with UV irradiation before being captured by magnetic Protein A beads. The crosslinked DNA was ligated to adapter R1 before being treated with two 5′-3′ exonucleases, Lambda exonuclease and RecJf exonuclease, to digest the DNA from the 5′ end. The presence of crosslinked antibody stopped the exonuclease digestion before the crosslinking site. Antibody was then removed by proteinase K digestion, and DNA fragments were recovered for primer extension. The dsDNA product was ligated to adapter R2 and sequenced (Figure 2). By mapping the read ends to the Chlamydomonas genome, we determined the boundary sites of antibody-protected regions, which contain one or more 6mA sites. As expected, we successfully improved the resolution to ~33 bp (Figure 3B), and identified 30,899 6mA-containing sequences with 67% overlapping with 6mA peaks identified from 6mA-IP-seq. Meanwhile, 73% of 6mA peaks from 6mA-IP-seq contain at least one 6mA-containing sequence identified from 6mA-CLIP-exo (Figure S3B). These higher resolution 6mA peaks showed the same enrichment around TSS with a bimodal distribution, a local depletion at TSS, and a potential periodic pattern (Figures 3C and S3C). A motif search revealed multiple high frequency sequences (Figure 3D), most of which contain an ApT dinucleotide motif (Figure 3D), reminiscent of the CpG methylation in most eukaryotic organisms and suggesting ApT as the general consensus sequence.
Validation of Individual Methylation Sites
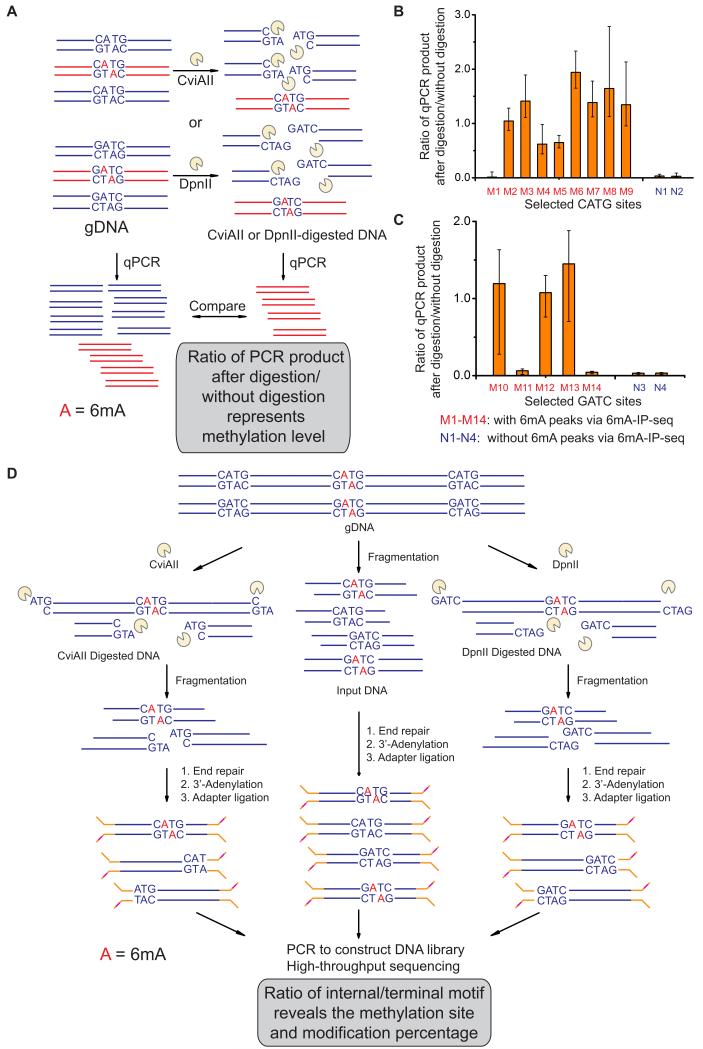
The methylation status of 6mA in specific motif sites can be validated by digestion with restriction enzymes originating from bacteria and viruses that are sensitive to 6mA methylation. For instance, CviAII is sensitive to 6mA and only digests the unmethylated CATG sequence (Zhang et al., 1992), whereas DpnII cuts only the unmethylated GATC sequence (Vovis and Lacks, 1977). We then applied the restriction-enzyme-digestion assay followed by quantitative PCR (6mA-RE-qPCR) to quantitatively evaluate the methylation status on specific motif sequences (Figure 4A). In this assay, we treated the isolated genomic DNA with CviAII or DpnII overnight to fully digest the unmethylated recognition motifs. We then designed PCR primers to specifically amplify the region flanked by the candidate 6mA site. In principle, the percentage of 6mA in the target 6mA site could be determined by quantitative PCR (qPCR) amplification of the restriction enzyme digested genomic DNA using undigested genomic DNA as a control given that 6mA hinders digestion. This strategy was tested by analyzing nine specific CATG sites and five specific GATC sites within identified 6mA peaks from 6mA-IP-seq and 6mA-CLIP-exo, along with two CATG sites and two GATC sites in regions that are not methylated based on 6mA-IP-seq results. The 6mA-RE-qPCR assay identified 8/9 of these CATG sites and 3/5 GATC sites within 6mA peaks to be completely or partially (a lower methylation frequency in a population of DNA molecules) methylated. The control sites not identified by 6mA-IP-seq were not methylated by this assay (Figures 4B and 4C). Therefore, this assay provides locus-specific validation of the 6mA-IP-seq results.
Figure 4. Single Site Detection of 6mA Using Methylation Sensitive Restriction Enzymes.
(A) Schematic diagram of 6mA-RE-qPCR for validation of specific 6mA. Restriction enzymes CviAII or DpnII that are sensitive to 6mA methylation in CATG or GATC were used to digest the unmethylated CATG or GATC sites in genomic DNA, respectively. The undigested CATG or GATC sites represent the methylated fraction, and can be PCR-amplified by using primers that cover these sites.
(B, C) qPCR results of 11 selected CATG sites and 7 GATC sites validated the accuracy of 6mA-IP-seq. After CviAII- or DpnII-mediated digestion, qPCR was performed using specific primers covering these sites. Relative abundances of undigested CATG or GATC sites were calculated from the ΔCt value between digested and undigested DNA samples (n = 3, mean ± S.E.M.).
(D) Schematic diagram of 6mA-RE-seq. gDNA is digested with CviAII or DpnII, sonicated to small fragments around 100 base pair, and constructed into sequencing libraries. The ratio for CATG or GATC internal of sequence reads versus at the end of sequence reads of a specific genomic site represents the relative methylation to unmethylation ratio. An input sample from gDNA without CviAII- or DpnII-based digestion serves as a control.
Genome-Wide Identification of Single 6mA Sites Using 6mA-RE-seq
The Chlamydomonas genome is GC rich (G+C content 64%) and around 120 million base pairs (Merchant et al., 2007). The ~0.4 mol% 6mA/A ratio corresponds to ~85,000 fully methylated 6mA sites. Our 6mA-IP-seq identified roughly 25,000 peaks with each peak potentially covering multiple 6mA sites (most of them are fully methylated at ~100%, see below), consistent with 6mA-IP-seq results showing that most 6mA peaks in the Chlamydomonas genome cluster around TSS sites. The 6mA-CLIP-exo results revealed several high-frequency sequences that include CATG and GATC. After we validated these two sequences as genuine 6mA methylation sites that mark TSS regions in Chlamydomonas, we sought to develop a high-throughput assay to map 6mA methylation in these selected sequences genome-wide at single-base resolution and to quantitatively determine the modification percentage at each site.
Genomic DNA was isolated and treated with CviAII or DpnII, and then sonicated to around 300 base pair fragments, end-repaired by T4 DNA polymerase, 3′-adenylated, and ligated to DNA adapters. The unmethylated CATG or GATC motifs would be digested and should be enriched at the end of the DNA fragments. The methylated motifs should resist restriction enzyme-mediated digestion and be present in the internal locations of DNA fragments. After PCR amplification of the fragments, a DNA library can be prepared for high-throughput sequencing. The ratio of a specific CATG or GATC sites with sequence reads internal versus at the end represents the relative methylation to unmethylation ratio. An input sample from genomic DNA without enzyme digestion serves as a control. Through mapping sequencing reads to the reference genome, we can identify the methylation status for every CATG or GATC motif genome-wide. We named this approach, as diagrammed in Figure 4D, 6mA-RE-seq and applied it to Chlamydomonas genomic DNA. While the specificity of DpnII to non-methylated DNA has been well characterized (Vovis and Lacks, 1977), the specificity of CviAII in cutting only non-methylated but not hemi- or fully-methylated sequences was further confirmed using synthetic DNA probes (Figure S4A).
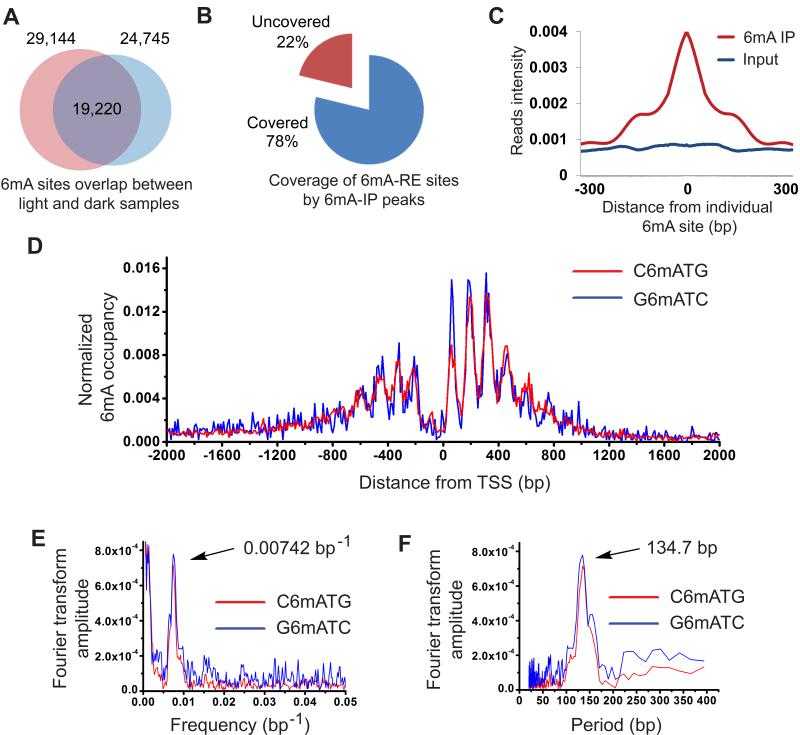
By applying 6mA-RE-seq to two biologically independent samples of Chlamydomonas grown under constant light or dark conditions, we obtained a high-resolution 6mA map of all CATG and GATC motifs in the Chlamydomonas genome. As expected, most of the sequencing reads were initiated with ATG or GATC for samples digested by CviAII or DpnII, which resulted from the digestion of unmethylated CATG or GATC sites, respectively (Figures S4B and S4C). Meanwhile, the intact CATG or GATC motifs that appear internal to the sequencing reads were counted as specific 6mA sites. We developed a bioinformatics algorithm with which to calculate the methylation level of individual 6mA sites within corresponding genomic sequences by calculating the ratio of reads obtained from fragment terminals to total reads of each site. We successfully identified 24,970 and 19,778 C6mATG sites with high confidence (FDR<0.01) in light and dark samples, respectively. In the same samples, 4,967 and 4,174 high confidence G6mATC sites were found in the same samples. Among the methylated sites discovered, 15,883 C6mATG sites and 3,337 G6mATC sites were identified from both light and dark samples, showing consistency of the method and reinforcing 6mA as a persistent DNA modification in Chlamydomonas (Figure 5A). These single 6mA sites include methylation sites that we have also validated using 6mA-RE-qPCR (Figures 4B, 4C, and Table S1). The sites without methylation based on 6mA-IP-seq and 6mA-RE-qPCR results were determined to be unmethylated by 6mA-RE-seq as well (Figures 4B, 4C, and Table S1). Approximately 78% (13,076/15,883 for C6mATG, and 2,069/3,337 for G6mATC) of the total detected sites overlap with 6mA peaks identified by 6mA-IP-seq (Figure 5B). We plotted base-resolution 6mA sites that overlap with corresponding 6mA peaks as identified from 6mA-IP-seq. The 6mA peaks are highly enriched around the identified single 6mA sites, with peak summits right on top of the single 6mA sites (Figure 5C). In addition, most of these methylation sites are close to 100% methylated, as indicated by the ratio of internal versus terminal sequencing reads (Figures S5A and S5B).
Figure 5. Single-Nucleotide Resolution Map of 6mA.
(A) Overlap of two 6mA-RE-seq samples under light and dark growth conditions. The majority of methylation sites were detected in both samples, indicating the consistency of this method.
(B) A majority of the detected single 6mA sites by 6mA-RE-seq are covered by 6mA peaks identified by 6mA-IP-seq.
(C) Overlap of 6mA sites identified by 6mA-RE-seq with the 6mA peak identified by 6mA-IP-seq.
(D) 6mA occupancy around TSS normalized to the CATG and GATC distribution. A periodic pattern of 6mA around TSS could be observed for both C6mATG and G6mATC motifs.
(E) Fourier transformation of 6mA distribution peaks.
(F) Periods of the corresponding frequency in Fourier transformation. The dominant period length is 134.7 bp.
See also Figure S5.
We performed an extended motif search based on the newly identified sites to examine if there is any additional preference of nucleotides flanking the CATG or GATC sequence; however, no additional consensus nucleotides were observed (Figure S5C). Considering the high frequency of CATG and GATC all over the genome (588,209 CATGs and 144,087 GATCs), the methylated sites occupy only 3-4% of all available motifs. However, the identified CATG and GATC methylations represent ~30% (24,970/85,000) and ~6% (4,967/85,000) of all genomic 6mA sites, respectively. On the other hand, there are ~28% of the 6mA-IP-seq peaks that do not contain any CATG or GATC sequences along the entire genomic regions, indicating the presence of other 6mA sites in distinct sequence contexts besides these two motifs. Interestingly, individual 6mA sites located at these two different sequence contexts tend to cluster in short regions (Figure S5D). We also observed multiple CATG and GATC motifs in a single peak identified from 6mA-IP-seq, and the peak length linearly correlates with the number of CATG or GATC motifs present in the region (Figure S5E). Taken together, these results indicate that 6mA methylation occurs mainly to ApT in multiple sequence motifs that tend to cluster together.
Periodic Distribution of 6mA near TSS Sites
To further understand the methylation specificity, we calculated the density of individual fully methylated 6mA sites around TSS (over 90% 6mA sites are close to fully methylated). Strikingly, we observed an apparent periodic pattern of 6mA distribution near the TSS region (Figure 5D). To rule out the possibility that a biased distribution of the CATG or GATC sequences caused the periodic distribution pattern, we normalized the 6mA site frequency according to motif occurrence within each region (Figure S5F). Of particular note is an obvious discontinuity between peaks upstream and downstream of TSS, which corresponds to a local depletion at TSS (Figure 5D). Fourier analysis of the periodic profile showed that the frequency is one per 130-140 bp for both downstream and upstream 6mA peaks (Figure 5E and 5F). The observed periodic pattern is similar to the one observed in the 6mA-CLIP-exo result which is independent of sequence bias (Figure S3C). The pattern is also conserved in both biologically independent samples, and is independent of culture conditions. Both motif sequences show exactly the same pattern (Figures S5G). For comparison with the fully methylated sites, we also analyzed the distribution of partially methylated sites (< 60% methylated measured by 6mA-RE-seq, corresponding to less than 10% of all 6mA sites). These partially methylated sites are evenly distributed without any obvious pattern or periodicity (Figure S5H). It is possible that the occurrence of these sites is governed by different mechanisms than those associated with the periodic, peri-TSS sites in Chlamydomonas.
6mA Preferentially Locates at Linker DNA between Two Adjacent Nucleosomes
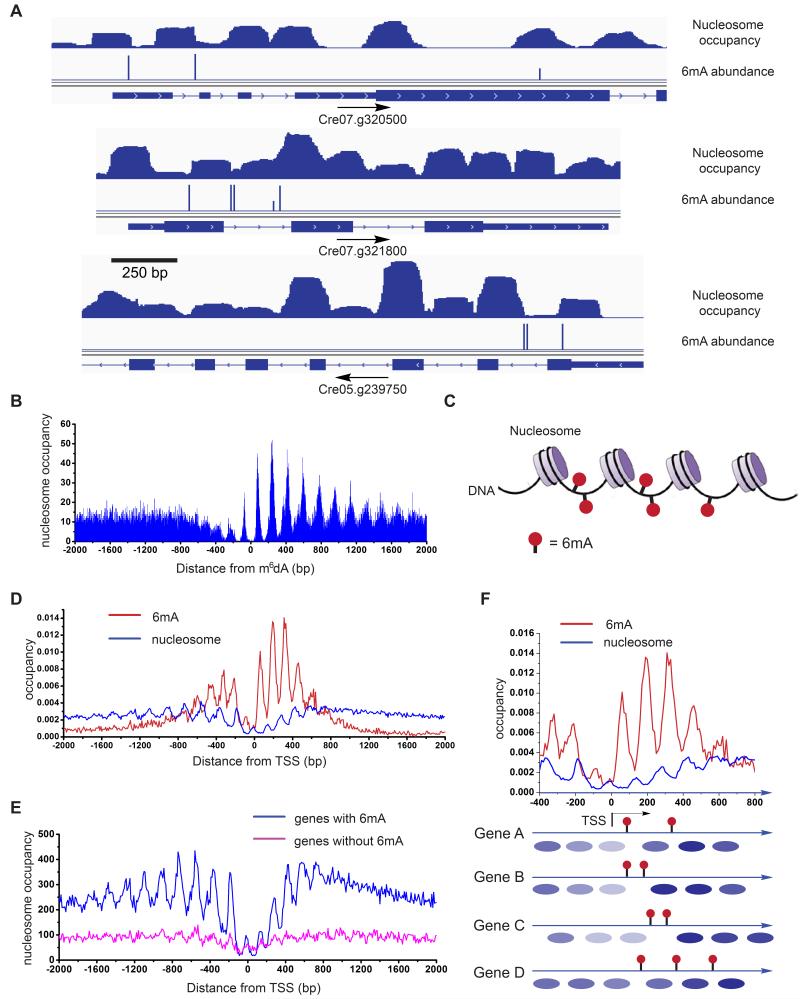
The periodic distribution pattern of 6mA around TSS prompted us to study its correlation with nucleosome positioning. We performed nucleosome foot printing followed by high-throughput sequencing (Chodavarapu et al., 2010) to reveal the exact position of each nucleosome in the Chlamydomonas genome. Briefly, micrococcal nuclease (MNase) was used to digest unprotected DNA between nucleosomes while leaving the nucleosome-occupied DNA intact; the intact DNA was then subjected to library preparation and high-throughput sequencing. After MNase digestion, the purified DNA showed a clear band with ~150 bp length; the DNA is composed of the nucleosome-protected segments (Figure S6A). These DNA segments were fully sequenced by paired-end sequencing. The length distribution is enriched around 147 bp (Figure S6B), which perfectly matches the reported value for Chlamydomonas (Lodha and Schroda, 2005). When we mapped the nucleosomes and 6mA locations to the Chlamydomonas genome, we found that most of the 6mA sites locate between two adjacent nucleosomes (Figure 6A). We then analyzed the statistical distribution of nucleosomes relative to individual 6mA sites, which revealed that the peaks of the closest nucleosomes are enriched ~75 bp upstream and ~78 bp downstream of the 6mA sites (Figure 6B). This pattern further supports that 6mA is mostly present in regions corresponding to the linker DNA between two adjacent nucleosomes (Figure 6C). The analysis of nucleosome-6mA correlation also showed that the downstream nucleosomes possess a progression with a steady phase of 170-180 bp periodicity (Figure S6C), while the upstream nucleosomes are relatively loosely phased, and this tight periodicity disappears around 2 to 3 nucleosomes away from the 6mA site.
Figure 6. 6mA Resides at the DNA Linker Region between Adjacent Nucleosomes.
(A) Distribution of nucleosome and 6mA in selected genes. 6mA mainly lies at the boundary region of nucleosomes. Nucleosome occupancy is shown on the first line, 6mA sites identified from 6mA-RE-seq are shown on the second line. Genome annotations are shown on the bottom line.
(B) Nucleosome occupancy around 6mA sites. 0 defines the 6mA site, with downstream noted as positive. Nucleosomes reside adjacent to but not on the 6mA site. Nucleosomes downstream of 6mA sites show a constant period of ~170-180 bp.
(C) Schematic models of the relationship between nucleosome distribution and 6mA in genomic DNA showing that 6mA mainly distributes in the linker DNA between two adjacent nucleosomes.
(D) Distribution profiles of 6mA and nucleosome around TSS showing they are mostly inversely correlated.
(E) Nucleosomes exhibit a more consistent phase in relation to TSS in genes marked with 6mA than genes without 6mA.
(F) Schematic illustration of the relationship between nucleosome positioning and 6mA location in individual genes. 6mA does not reside on nucleosome-wrapped DNA.
See also Figure S6.
6mA May Contribute to the Positioning of Nucleosomes in Chlamydomonas
To further understand the relationship between nucleosome distribution and 6mA, we plotted their density around TSS. We found that the periodic pattern of average nucleosome occupancy around TSS in Chlamydomonas has distinct features compared to other species (Figure 6D): first, the density of nucleosomes around TSS is much lower than that in gene body regions and upstream promoter regions; second, the periodicity between two nucleosomes is centered at 183 bp upstream of TSS, but has multiple period values downstream of TSS, including 171, 151, and 128 bp (Figures S6D). Previous studies of nucleosome distribution in Chlamydomonas and other organisms revealed that nucleosome-depleted regions (NDRs) are on average ~155-160 bp around TSS, and nucleosomes downstream of the NDRs are strictly phased in a 165-185 bp period, depending on the length of linker region between two adjacent nucleosomes (Huff and Zilberman, 2014; Lodha and Schroda, 2005). The multiple periodic values we observed could be a result of convolution between the regular nucleosome periodicity of ~170 bp and the 6mA-influenced periodicity of 130-140 bp downstream of TSS on DNA. Nonetheless, when we compared the nucleosome distribution with the 6mA distribution around TSS, we found that they correlates with each other with ~180 degree phase shift, consistent with our finding that 6mA preferentially locates at linker regions. To probe the relationship of 6mA distribution and nucleosome positioning in detail, we divided all the genes into two groups: with or without 6mA around TSS. Interestingly, nucleosomes phase well for genes that contain 6mA around TSS, whereas the nucleosome phase pattern was weak for genes without 6mA (Figures 6E and S6E). Taking these results together, we propose a model in which the DNA 6mA modification either restricts or marks the positions of nucleosomes near TSS in Chlamydomonas (Figure 6F). The 130-140 periodic pattern of 6mA leads to out-of-phase distribution and partial occupancy of nucleosomes around TSS. For example, if the distance between two adjacent 6mA sites is larger than the length of a nucleosome, such as 270 bp, one nucleosome may reside between two adjacent 6mA, in place depending on the sequence content. If the distance between two adjacent 6mA sites is shorter than 150 bp, such as 135 bp; nucleosome will be missing, leaving a nucleosome-free region between them (Figures 6A and 6F). The distribution pattern of 6mA may restrict the pattern of nucleosome positioning for each gene, such that the genome-wide pattern of nucleosome is correlated with 6mA distribution pattern.
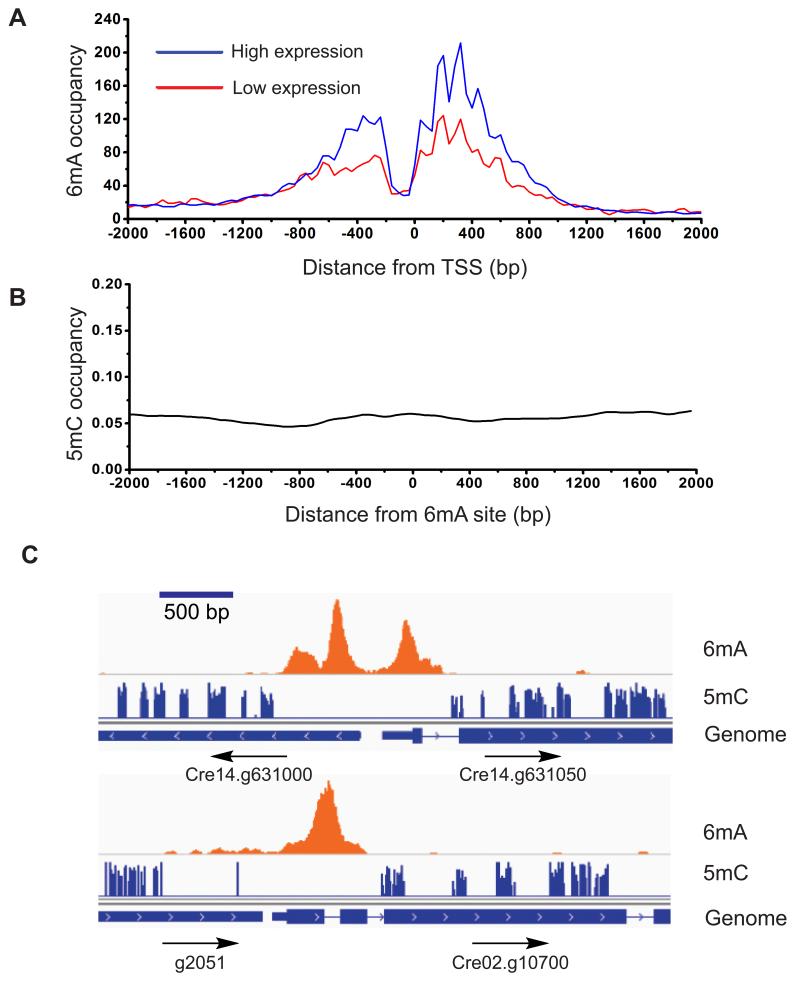
6mA Marks the TSS Regions of Actively Transcribed Genes
The bimodal localization of 6mA around transcription start sites prompted us to investigate its relationship with gene expression. We used RNA-seq to analyze the expression of individual genes. We divided genes into two groups: high expression (80% of all genes) and low expression (20%), and plotted their 6mA peak abundances obtained from 6mA-CLIP-exo experiments (Figure 7A). We found a general trend that genes with lower expression tend to have low occupancies of 6mA around TSS regions. Specifically, among the 16% of genes without 6mA, ~64% are categorized as low expression or non-active genes. Correspondingly, on a genome-wide level, genes with 6mA around TSS express significantly higher than genes without 6mA (Figure S7A). The widely-studied 5mC methylation typically plays repressive roles in the regulation of gene expression. However, our results reveal that 6mA marks the TSS regions of actively transcribed genes in Chlamydomonas. Studies have shown that 6mA can reduce the stability of the DNA duplex due to the requirement of unfavorable trans-configuration for base-pairing. The presence of 6mA may lower the energy required for opening up the DNA duplex (Engel and von Hippel, 1978). Based on the observed periodic distribution pattern, the tightly controlled deposition of 6mA is associated with nucleosome phasing around TSS. These 6mA modifications could affect nucleosome positioning, or recruit protein factors analogous to methyl-CpG-binding proteins as potential ‘readers’ to impact transcription initiation (Sternberg, 1985). Indeed, barley nuclear extract has been shown to contain specific 6mA-binding proteins, and 6mA embedded within GATC at the promoter region can increase the transcription activity of a transfected plasmid (Rogers and Rogers, 1995).
Figure 7. Correlation of 6mA with Active Genes.
(A) The 6mA methylation is correlated with active genes. Two groups of genes with high (FPKM ≥ 1) and low (FPKM <1) expression levels are plotted with its methylation level determined from 6mA-CLIP-exo-seq. 6mA occupancy represents the reads coverage that are normalized to gene counts of each category in 6mA-CLIP-exo. FPKM stands for Fragments Per Kilobase Of Exon Per Million Fragments Mapped.
(B) No correlation was observed between the distributions of 5mC and 6mA. Distance between 5mC and 6mA was plotted, showing no correlation between the two.
(C) Selected examples showing that 5mC mainly appears in the gene body, while 6mA mainly resides near TSS region. 6mA peaks identified from 6mA-IP-seq are shown on the first line, 5mC sites identified from previous results are shown on the second line. Genome annotations are shown on the bottom line.
See also Figure S7.
To study potential effects of 6mA on gene regulation, we profiled the mRNA transcriptome of algae cultured under constant light and dark conditions, and found 4,866 differentially expressed genes. In parallel, we used the restriction enzyme based method to quantify the methylation level of individual 6mA site under light and dark conditions. 6mA levels in most genes were similar under both light and dark conditions (Figure S7B). These results suggest that 6mA is a general mark of TSS regions that could be actively transcribed. Transcription factors and other factors may play more direct roles in determining the exact expression levels of individual genes.
6mA and 5mC Mark Distinct Regions in the Chlamydomonas Genome
As 5mC is also present in high abundance in the Chlamydomonas genome, we wondered if any relationship exists between these two DNA base modifications. Chlamydomonas has an unusual pattern of 5mC methylation: overall it has less CpG methylation compared to multicellular eukaryotes, but possesses all three types of methylation of CpG, CHG, and CHH enriched in exons of genes, and has only CpG methylation enriched in repeats and transposons (Feng et al., 2010). We compared bisulfite sequencing data of 5mC with the 6mA distribution that we generated. There is no specific enrichment pattern of 5mC distribution around TSS regions (Figure S7C), and 5mC generally do not co-localize with 6mA (Figure 7B). 5mC appears mostly in gene bodies with a much broader distribution, and is absent near TSS regions (Figure 7C and S7C). In addition, 5mC has been proposed to be negatively correlated with gene expression in general (Jones, 2012); we did not observe a strong correlation between the gene expression and 5mC occupancy around TSS region (Figure S7D). This analysis indicates that 6mA and 5mC are two distinct marks in Chlamydomonas genome: 6mA may contribute to chromatin structures that enable initiation of gene transcription, while 5mC may contribute to transposon silencing, imprinting, exon definition and affect transcription elongation (Cerutti et al., 1997).
DISCUSSION
The 6mA and 5mC modifications are both abundant in the genome of the green algae Chlamydomonas reinhardtii. We showed that the total 6mA level is robustly maintained during cell proliferation. We applied 6mA-IP-seq and further developed 6mA-CLIP-exo to profile 6mA genome-wide using antibodies that specifically recognize and enrich N6-methylated adenine. We found that 6mA mainly resides around transcription start sites with a bimodal distribution. The results from 6mA-CLIP-exo at higher resolution revealed that 6mA deposition occurs mainly at ApT dinucleotides within multiple sequence contexts. At least two sequence motifs, CATG and GATC, are confirmed by a restriction enzyme digestion assay using CviAII and DpnII that are sensitive to 6mA. We then applied this restriction-enzyme-based 6mA-RE-seq strategy to Chlamydomonas genomic DNA and obtained genome-wide 6mA maps at single-nucleotide resolution. The identified 6mA sites within these two specific sequences account for ~1/3 of the total 6mA in genomic DNA. 6mA sites within other sequence contexts likely show similar distribution patterns (Figures 3D and S3C).
The results from the high-resolution maps of 6mA in two specific sequences not only validate the IP-based profiling data, but also uncover a periodic pattern of 6mA. This periodicity may mark special features of transcription initiation in Chlamydomonas, and could be related to nucleosome positioning around TSS. Indeed, we performed nucleosome footprinting coupled with high-throughput sequencing, and the results revealed a periodic pattern of nucleosome occupancy that correlates with the periodicity of 6mA distribution, but is ~180 degree out of phase around the TSS region. The individual 6mA sites exclusively mark the linker DNA between two adjacent nucleosomes. We propose two possible interpretations for this exclusive behavior. One possibility is that, unlike the nucleosome-wrapped DNA, the linker DNA is exposed and can thus be accessed for methylation. The other possibility is that the locations of 6mA sites contribute to the precise positioning of nucleosomes. Our results favor the latter hypothesis for the following reasons: first, we have shown that nucleosomes around TSS sites exhibit very low densities. Low occupancy nucleosomes unlikely serve as determining factors for 6mA deposition since it occurs at almost 100% at most of these sites. On the other hand, a high density of 6mA might act to reprogram the positioning of nucleosomes around TSS regions. Second, nucleosomes are likely more dynamic than the covalent 6mA mark on DNA in the TSS regions during transcription initiation. Precedence for a role of base methylation in affecting chromatin structure exists: 5mC has been shown to contribute to nucleosome positioning in other eukaryotes (Huff and Zilberman, 2014). Additionally, 6mA may mark the TSS region for more efficient transcription initiation. Although it has been well known that the first intron is always important for transgenic gene expression in Chlamydomonas (Eichler-Stahlberg et al., 2009), the mechanism was unclear. We provide evidence that 6mA can reside in the first intron (Examples shown in Figure 3B). The periodic distribution, its specific location on the linker DNA between two adjacent nucleosomes at TSS, and its marking of gene activation all suggest that this unique DNA mark contributes to nucleosome positioning and transcription initiation.
We have shown that 6mA shares little correlation with 5mC in the Chlamydomonas genome, indicating that they are controlled through different pathways and likely exhibit distinct functions. Our transcriptome analysis found an association of 6mA with gene activation; whereas, 5mC appears to negatively correlate with gene expression. Studies of 5mC have dominated notions of DNA epigenetics in eukaryotes, in particular in vertebrates, because of the critical roles played by 5mC. As shown here, 6mA can also be an important mark that could mark/affect gene activation in eukaryotes. Analogous to 5mC recognition by methyl-CpG-binding proteins, proteins that specifically recognize 6mA at TSS may exist; these proteins could interact with or be part of transcription initiation complexes that contribute to gene activation. It is also possible that 6mA may coordinate with other epigenetic factors such as histone modifications that are also enriched around the TSS region. Highly dense and narrow distributions of modifications such as H3K9 acetylation (H3K9ac) and H3K4 trimethylation (H3K4me3) near transcription start sites have been associated with constitutive expression of genes involved in translation in Arabidopsis (Ha et al., 2011). Cooperative interactions among 6mA, histone modification, and transcriptional factors could serve as a general mechanism for transcription activation in Chlamydomonas and possibly other eukaryotic organisms.
The E. coli Dam DNA methyltransferase methylates the N6 position of adenine at GATC sites. Compared to prokaryotic 6mA modification in genomic DNA, the 6mA methylation in the Chlamydomonas genome exists in a more complex manner with multiple potential sequences mainly centered on ApT, resembling eukaryotic 5mC methylation of CpG. The methyltransferases that are involved in establishing or maintaining the patterns of 6mA sites remain to be determined (Iyer et al., 2011). It should be noted that Greer and Shi et al. (Greer et al., 2015) have recently discovered two enzymes, MAD-1 and DMT-1, which can install or remove 6mA in the genome of Caenorhabditis elegans, respectively.
In summary, our study has demonstrated that 6mA is an abundant DNA modification in the Chlamydomonas genome. It is enriched specifically around transcription start sites and preferentially marks actively transcribed genes. A periodic distribution pattern with depletion at the TSS coupled with an almost exclusive marking of the linker DNA between adjacent nucleosomes indicates a process of controlled deposition as well as functional roles in nucleosome positioning and transcriptional initiation. While 5mC is well known to mark gene repression at promoter and enhancer sites in vertebrates, we show in this work that a different DNA base modification, 6mA, flanks transcription start sites and marks actively transcribed genes. The ribose version of 6mA modification (with 2′-OH) exists as the most abundant internal mRNA modification in almost all eukaryotes. It has recently been shown to be reversible and plays important regulatory functions (Fu et al., 2014). We suspect that 6mA could be widely present in eukaryotic genomes as well; in certain species 6mA may carry important roles in regulating gene expression; in other organisms, 6mA may play complementary roles to 5mC at different stages of development.
EXPERIMENTAL PROCEDURES
6mA-IP-seq
Isolated genomic DNA was diluted to 100-200 ng/uL using TE buffer, and sonicated in 130 uL scale to 200-400 bp using a Covaris Focused-ultrasonicator. End repair, 3′-adenylation, and adapter ligation were performed. The ligated and purified DNA was denatured at 95 °C, and chilled on ice. A portion of 10 uL DNA was saved as input. The rest of the DNA was combined with 3 ug of anti-6mA antibody (Synaptic Systems) in 500 uL of 1× IP buffer, and incubated at 4 °C for 6 hr. At the same time, 40 uL of Protein A magnetic beads was washed twice in 0.5 mL of 1× IP buffer, and pre-blocked in 0.5 mL of 1× IP buffer containing 20 ug/uL of Bovine Serum Album at 4 °C for 6 hr. The Protein A beads were washed twice with 0.5 mL of 1× IP buffer, added to the DNA-6mA antibody mixture, and incubated at 4 °C with gentle rotation overnight. The beads were then washed four times with 0.5 mL of 1× IP buffer. Methylated DNA was eluted twice by 100 uL of elution buffer containing 6mA monophosphate at 4 °C for 1 hr. The two elusion solutions were combined, to which 20 uL of NaOAc (3 M, pH 5.3), 500 uL of EtOH, and 0.5 uL of glycogen (20 ng/uL) were added. The solution was frozen at −80 °C overnight, and centrifuged at 14,000×g for 20 min at 4 °C. The precipitated DNA was dissolved in 7 uL of ddH2O, PCR amplified for 15 to 18 cycles, purified by Ampure beads, and suspended in 16 uL of re-suspension buffer to yield the sequencing library.
6mA-CLIP-exo
Genomic DNA was sonicated to around 200 bp and immunoprecipitated by using anti-6mA antibody. The antibody-DNA complex was then covalently crosslinked using UV 254 nm irradiation, followed by a procedure similar to ChIP-exo (Rhee and Pugh, 2012). The library was constructed with Illumina-compatible adapters and primers and applied to Illumina HiSeq 2000 sequencer with single-end reads. The raw data were aligned by bowtie and the peaks were called by MACE (model-based analysis of ChIP-exo). See Extended Experimental Procedures for detailed procedures.
6mA-RE-seq
Restriction enzyme digestion was performed by treating 1 ug of gDNA with 5 uL of CviAII or DpnII restriction enzyme (5 U/uL) at 25 °C or 37 °C overnight. The digested and non-digested DNA (200 ng each) were fragmented into ~100 bp by sonication, and sequencing libraries were constructed according to Illumina TruSeq DNA sample preparation procedures.
Detection of 6mA peaks from 6mA-IP-seq and 6mA-RE-seq
Reads were mapped to the Chlamydomonas genome (JGI) Version 9.1, with parameters and scripts as described in the Extended Experimental Procedures.
Supplementary Material
Highlights.
Genome-wide profiling reveals a bimodal distribution of 6mA enriched around TSS
6mA marks active genes in Chlamydomonas
A periodic pattern of 6mA at base resolution correlates with nucleosome positioning
6mA exclusively marks DNA linkers between adjacent nucleosomes around TSS
ACKNOWLEDGMENTS
We thank Dr. Q. Jin for help with HPLC-QQQ-MS/MS and Dr. Pieter W. Faber for help with high-throughput sequencing. This work is partially supported by National Institutes of Health (R01 HG006827 to C.H.) and C. H. is a Howard Hughes Medical Institute investigator. The Mass Spectrometry Facility of the University of Chicago is funded by National Science Foundation (CHE-1048528). S. F. Reichard, MA edited the manuscript.
Footnotes
ACCESSION NUMBERS
Sequencing data have been deposited into the Gene Expression Omnibus (GEO) under the accession number GSE62690.
Supplemental Information includes Extended Experimental Procedures, seven figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.cell.
AUTHOR CONTRIBUTIONS
Y.F., G.Z.L., and C.H. conceived the project. Y.F., G.Z.L., and K.C. designed and performed experiments with help from, X.D., M.Y., Z.H., J.L., X.L., X.W., and Q.J. G.Z.L. conducted bioinformatics analysis. D.H. helped on data analysis. L.C.D. helped on high-throughput sequencing, L.M. helped with experiment design. Y.F., G.Z.L., and C.H. wrote the manuscript with input from K.C. and L.M.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Babinger P, Kobl I, Mages W, Schmitt R. A link between DNA methylation and epigenetic silencing in transgenic Volvox carteri. Nucleic Acids Res. 2001;29:1261–1271. doi: 10.1093/nar/29.6.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- Bisova K, Krylov DM, Umen JG. Genome-wide annotation and expression profiling of cell cycle regulatory genes in Chlamydomonas reinhardtii. Plant Physiol. 2005;137:475–491. doi: 10.1104/pp.104.054155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti H, Johnson AM, Gillham NW, Boynton JE. Epigenetic silencing of a foreign gene in nuclear transformants of Chlamydomonas. Plant Cell. 1997;9:925–945. doi: 10.1105/tpc.9.6.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Lu Z, Wang X, Fu Y, Luo G-Z, Liu N, Han D, Dominissini D, Dai Q, Pan T, et al. High-Resolution N6-Methyladenosine (m6A) Map Using Photo-Crosslinking-Assisted m6A Sequencing. Angew. Chem. Int. Ed. 2015;54:1587–1590. doi: 10.1002/anie.201410647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Bernatavichute YV, Chen P-Y, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier J, McAdams HH, Shapiro L. A DNA methylation ratchet governs progression through a bacterial cell cycle. Proc. Natl. Acad. Sci. USA. 2007;104:17111–17116. doi: 10.1073/pnas.0708112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings DJ, Tait A, Goddard JM. Methylated bases in DNA from Paramecium aurelia. Biochim. Biophys. Acta. 1974;374:1–11. doi: 10.1016/0005-2787(74)90194-4. [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Ehrlich M, Zhang X-Y. Chapter 10 Naturally Occurring Modified Nucleosides in DNA. In: Charles WG, Kenneth CTK, editors. Journal of Chromatography Library. Elsevier; 1990. pp. B327–B362. [Google Scholar]
- Eichler-Stahlberg A, Weisheit W, Ruecker O, Heitzer M. Strategies to facilitate transgene expression in Chlamydomonas reinhardtii. Planta. 2009;229:873–883. doi: 10.1007/s00425-008-0879-x. [DOI] [PubMed] [Google Scholar]
- Engel JD, von Hippel PH. Effects of methylation on the stability of nucleic acid conformations. Studies at the polymer level. J Biol Chem. 1978;253:927–934. [PubMed] [Google Scholar]
- Fang G, Munera D, Friedman DI, Mandlik A, Chao MC, Banerjee O, Feng Z, Losic B, Mahajan MC, Jabado OJ, et al. Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat. Biotechnol. 2012;30:1232–1239. doi: 10.1038/nbt.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng SH, Cokus SJ, Zhang XY, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. Conservation and divergence of methylation patterning in plants and animals. Proc. Natl. Acad. Sci. USA. 2010;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat. Rev. Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- Greer EL, Blanco MA, Gu L, Sendinc E, Liu J, Aristizábal-Corrales D, Hsu C-H, Aravind L, He C, Shi Y. Identification of N6-adenine methylation in C. elegans DNA. Cell. 2015 doi: 10.1016/j.cell.2015.04.005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Ng DW, Li WH, Chen ZJ. Coordinated histone modifications are associated with gene expression variation within and between species. Genome Res. 2011;21:590–598. doi: 10.1101/gr.116467.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S, Kenny C, Berger L, Pratt K. Comparative study of DNA methylation in three unicellular eucaryotes. J. Bacteriol. 1978;135:1156–1157. doi: 10.1128/jb.135.3.1156-1157.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff JT, Zilberman D. Dnmt1-independent CG methylation contributes to nucleosome positioning in diverse eukaryotes. Cell. 2014;156:1286–1297. doi: 10.1016/j.cell.2014.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Abhiman S, Aravind L. Natural history of eukaryotic DNA methylation systems. Prog. Mol. Biol. Transl. Sci. 2011;101:25–104. doi: 10.1016/B978-0-12-387685-0.00002-0. [DOI] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodha M, Schroda M. Analysis of chromatin structure in the control regions of the chlamydomonas HSP70A and RBCS2 genes. Plant Mol. Biol. 2005;59:501–513. doi: 10.1007/s11103-005-0450-0. [DOI] [PubMed] [Google Scholar]
- Low DA, Weyand NJ, Mahan MJ. Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect Immun. 2001;69:7197–7204. doi: 10.1128/IAI.69.12.7197-7204.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Campbell JL, Boye E, Kleckner N. SeqA: a negative modulator of replication initiation in E. coli. Cell. 1994;77:413–426. doi: 10.1016/0092-8674(94)90156-2. [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Marechal-Drouard L, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer W, Noyer-Weidner M. Timing and targeting: the biological functions of Dam methylation in E. coli. Cell. 1988;54:735–737. doi: 10.1016/s0092-8674(88)90911-7. [DOI] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray IA, Clark TA, Morgan RD, Boitano M, Anton BP, Luong K, Fomenkov A, Turner SW, Korlach J, Roberts RJ. The methylomes of six bacteria. Nucleic Acids Res. 2012;40:11450–11462. doi: 10.1093/nar/gks891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radakovits R, Jinkerson RE, Darzins A, Posewitz MC. Genetic engineering of algae for enhanced biofuel production. Eukaryot. Cell. 2010;9:486–501. doi: 10.1128/EC.00364-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae PM, Spear BB. Macronuclear DNA of the hypotrichous ciliate Oxytricha fallax. Proc. Natl. Acad. Sci. USA. 1978;75:4992–4996. doi: 10.1073/pnas.75.10.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratel D, Ravanat JL, Berger F, Wion D. N6-methyladenine: the other methylated base of DNA. Bioessays. 2006;28:309–315. doi: 10.1002/bies.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. ChIP-exo method for identifying genomic location of DNA-binding proteins with near-single-nucleotide accuracy. Curr Protoc Mol Biol. 2012 doi: 10.1002/0471142727.mb2124s100. Chapter 21, Unit 21.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers JC, Rogers SW. Comparison of the effects of N6-methyldeoxyadenosine and N5-methyldeoxycytosine on transcription from nuclear gene promoters in barley. Plant J. 1995;7:221–233. doi: 10.1046/j.1365-313x.1995.7020221.x. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat. Rev. Genet. 2008;9:129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- Sternberg N. Evidence that adenine methylation influences DNA-protein interactions in Escherichia coli. J. Bacteriol. 1985;164:490–493. doi: 10.1128/jb.164.1.490-493.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vovis GF, Lacks S. Complementary action of restriction enzymes endo R-DpnI and Endo R-DpnII on bacteriophage f1 DNA. J. Mol. Biol. 1977;115:525–538. doi: 10.1016/0022-2836(77)90169-3. [DOI] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Wion D, Casadesus J. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nature Rev. Microbiol. 2006;4:183–192. doi: 10.1038/nrmicro1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Nelson M, Nietfeldt JW, Burbank DE, Van Etten JL. Characterization of Chlorella virus PBCV-1 CviAII restriction and modification system. Nucleic Acids Res. 1992;20:5351–5356. doi: 10.1093/nar/20.20.5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.