Abstract
Hepatitis C virus (HCV) replicates in membrane associated, highly ordered replication complexes (RCs). These complexes include viral and host proteins necessary for viral RNA genome replication. The interaction network among viral and host proteins underlying the formation of these RCs is yet to be thoroughly characterized. Here, we investigated the association between NS4B and NS5A, two critical RC components. We characterized the interaction between these proteins using fluorescence resonance energy transfer and a mammalian two-hybrid system. Specific tryptophan residues within the C-terminal domain (CTD) of NS4B were shown to mediate this interaction. Domain I of NS5A, was sufficient to mediate its interaction with NS4B. Mutations in the NS4B CTD tryptophan residues abolished viral replication. Moreover, one of these mutations also affected NS5A hyperphosphorylation. These findings provide new insights into the importance of the NS4B–NS5A interaction and serve as a starting point for studying the complex interactions between the replicase subunits.
Keywords: Hepatitis C virus, Protein–protein interaction, Viral replication, Fluorescence resonance energy transfer (FRET), Endoplasmic reticulum (ER)
Introduction
Hepatitis C virus (HCV) infection affects millions worldwide and is one of the leading causes of chronic liver disease. Prolonged HCV infection may lead to liver cirrhosis and liver failure and is associated with the development of liver cancer (El-Serag, 2011). HCV is a positive strand RNA virus, that like other members of this group, replicates in membrane associated replication complexes (RC). These altered membranes are thought to be endoplasmic reticulum (ER) derived and to contain in addition to the viral genome and proteins, a variety of host factors necessary for viral replication (Miller and Krijnse-Locker, 2008).
Once the positive strand RNA genome enters the host cell, its single open reading frame is translated into a polyprotein. This polyprotein is co- and post-translationally processed to yield three structural proteins (core, E1 and E2), an ion-channel (p7), and six non-structural proteins (NS2, NS3, NS4A, NS4B, NS5A, NS5B) (Grakoui et al., 1993b). The minimal set of proteins essential for viral replication consists of NS3, NS4A, NS4B, NS5A and NS5B (Lohmann et al., 1999). Most of these proteins have known activities: NS3 is a helicase and a serine protease that uses NS4A as a cofactor. NS5B is the viral RNA-dependent RNA polymerase (Ago et al., 1999; Bressanelli et al., 1999; Grakoui et al., 1993a; Hijikata et al., 1993; Lesburg et al., 1999). The exact role of NS4B and NS5A, which are essential for viral replication and have a role in assembly, is still not fully understood.
NS4B is a 27 kDa, ER localized, small, hydrophobic transmembrane protein thought to induce the membranous web: modified membrane elements that comprise the viral RC (Egger et al., 2002). NS4B contains three major segments: a cytoplasmic N-terminal part (a.a. 1 to ~69), shown to contain two amphipathic helices (AH1 and AH2) (Elazar et al., 2004; Gouttenoire et al., 2010); a central domain that contains four putative transmembrane segments (a.a. ~70 to ~191) and a cytoplasmic C-terminal part (a.a. ~191 to 261) that contains two conserved helices (H1 and H2) (Gouttenoire et al., 2009; Jones et al., 2009; Lundin et al., 2003). Three main features of NS4B are thought to contribute to its activity in the induction of the membranous web formation: The first is its ability to form oligomers, mediated by several domains of the protein, mainly AH2 and elements at the C-terminus (Gouttenoire et al., 2010; Paul et al., 2011; Yu et al., 2006). The second feature is the ability NS4B’s AH2 domain to promote lipid vesicle aggregation (Cho et al., 2010). The third feature is the ability NS4B’s N-terminal domain to post-translationally translocate from the cytosolic face of the ER into its lumen. NS5A was found to negatively affect this translocation, suggesting that NS5A influences the topology of NS4B (Lundin et al., 2006).
NS5A is a 56–58 kDa phosphorylated protein, occurring in different phosphorylation states, the exact role of these different phosphorylation states in the viral life cycle is still unclear. Adaptive mutations reducing the amount of hyperphosphorylated NS5A increase viral replication (Appel et al., 2005b; Blight et al., 2000; Katze et al., 2000; Tanji et al., 1995b). Furthermore, inhibition of NS5A hyperphosphorylation by kinase inhibitors yields similar consequences (Neddermann et al., 2004). Yet, low levels of hyperphosphorylated NS5A seem to be essential for viral replication since viral replication is inhibited upon its complete loss (Appel et al., 2005a; Fridell et al., 2011). The increase in viral replication caused by reduced levels of the NS5A hyperphosphorylated form inversely correlates with viral production, leading to the notion that the hyperphosphorylated form of NS5A has adverse effects on viral replication but is required for viral assembly (Masaki et al., 2008; Miyanari et al., 2007; Pietschmann et al., 2009; Tellinghuisen et al., 2008a). It was also proposed to regulate the interface between replication and assembly (Tellinghuisen et al., 2008a). Several non-structural proteins including NS4B were found to facilitate NS5A phosphorylation (Koch and Bartenschlager, 1999; Neddermann et al., 1999). The N-terminal domain of NS5A contains an amphipathic helix that mediates its membrane association (Elazar et al., 2003). The rest of the protein is organized in three domains (I, II, and III), which are separated by repetitive low-complexity sequences (Tellinghuisen et al., 2004). Domain I has been attributed to the replicase activity of NS5A (Tellinghuisen et al., 2005). Although a large part of domain II could be deleted without effecting replication or production of the virus (Appel et al., 2008), deletion of a specific region or deletion of the whole domain abolished viral replication (Appel et al., 2008; Tellinghuisen et al., 2008b). In contrast, domain III has been shown to be dispensable for RNA replication (Appel et al., 2008; Tellinghuisen et al., 2008a). Mutations and insertions in domain III showed impact on virus particle assembly (Appel et al., 2008). Domain I was crystallized as a dimer, leading to the notion that NS5A functions as a multimer (Love et al., 2009; Tellinghuisen et al., 2005).
Traditionally, NS4B was thought to be the sole driving force for the induction of the membrane alterations that harbor the RCs (Egger et al., 2002). It was recently shown that double membrane vesicles that appear in infected cells upon the initiation of viral replication are induced by NS5A, highlighting a critical role for both NS4B and NS5A in this initial step in the establishment of the viral RC (Romero-Brey et al., 2012). A genetic interaction between NS4B and NS5A was reported, where a mutation in the C-terminal domain of NS4B that abolished viral replication was rescued by a pseudoreversion in NS4B and an additional mutation in domain I of NS5A (Paul et al., 2011). Similarly, replacement of the NS4B C-terminal domain from a genotype 2a genome with sequences from genotype 1b attenuated virus production. The phenotype was rescued by two adaptive mutations one in the C-terminal of NS4B and the other in NS5A domain III (Han et al., 2013).
NS4B and NS5A were previously shown to physically interact (Aligo et al., 2009; Dimitrova et al., 2003; Gao et al., 2004), however, in spite of their aforementioned functional association, this interaction was never characterized, most likely due to the hydrophobic nature of NS4B that complicates biochemical analysis. Using immunofluorescence, fluorescence resonance energy transfer (FRET) and a mammalian two-hybrid system approach, we confirmed this interaction. The C-terminal domain of NS4B and domain I of NS5A, were found to contain the major determinants that mediate the NS4B–NS5A interaction. Furthermore, mutations in NS4B’s C-terminal domain that affected this interaction abolished viral replication. One of these mutations also inhibited NS5A phosphorylation. These results may provide additional insights into organization of the viral RC, towards an informed model of its functional mechanism.
Results
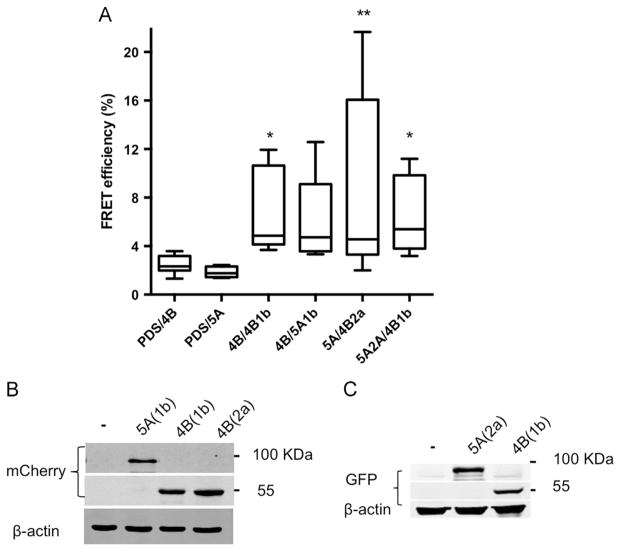
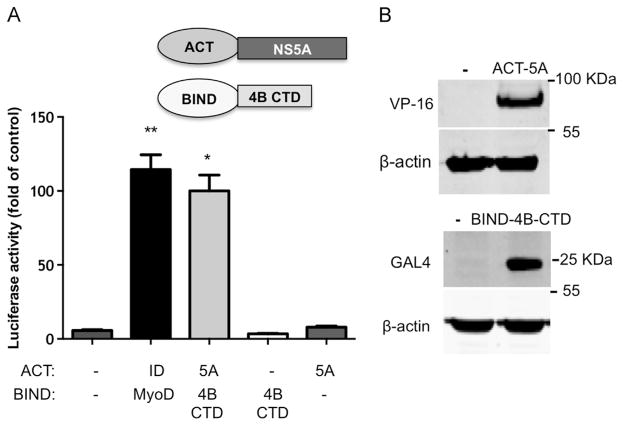
NS4B and NS5A are both known to be essential components of HCVs replicase machinery and as such are expected to colocalize within RCs (Lohmann et al., 1999). As a baseline for our forthcoming experiments, their localization in replicon cells was confirmed. To this end, Bart-HA cells (Huh7 cells harboring a subgenomic 1b replicon with an HA tag insertion in NS5A) were used for immuno-fluorescence analysis with specific antibodies. Both proteins displayed a typical reticular ER and a speckle-like pattern, thought to represent the membrane rearrangements harboring the viral RCs (Egger et al., 2002; Ide et al., 1996; Polyak et al., 1999; Tanji et al., 1995a). NS4B and NS5A colocalized in these structures (Fig. S1). NS4B is a highly hydrophobic transmembrane protein, previous studies reported unsuccessful attempts to study its interaction with NS5A using immunoprecipitation of ectopically expressed proteins or from infected cells (Blight, 2011; Han et al., 2013). Thus, we used an experimental approach that allows analysis of proteins in their native intact membranes namely, Fluorescence Resonance Energy Transfer (FRET). FRET occurs between two fluorophores with partially overlapping emission of one (the donor) and excitation of the other (the acceptor) when the two molecules are very close to each other (less then 10 nm). The efficiency of the FRET is inversely proportional to the distance to the power of 6. Thus, FRET between two fluorescently tagged molecules is a strong indication for interaction (Kenworthy, 2001). In acceptor photobleaching, occurrence of FRET is detected as the increase in the fluorescence emission of the dequenched donor fluorophore following photobleaching of the acceptor. Here FRET was carried out using repetitive partial acceptor photobleaching to demonstrate a correlation between acceptor photobleaching and donor fluorescence increase (Fig. S2). Full length NS4B or NS5A from HCV genotype 1b or 2a were each fused to GFP or mCherry that were then used as a donor-acceptor pair for FRET analysis, following their coexpression in Huh7 cells (Fig. 1A). GFP-tagged Pendrin (PDS) a plasma membrane iodide transporter that upon overexpression is essentially retained and accumulated in the ER (Shepshelovich et al., 2005) was used as a negative control. NS4B (genotype 1b), previously shown by FRET to oligomerize, was used as a positive control (Gouttenoire et al., 2010). As expected, a significant FRET signal (mean EF 6.6±3.5, p<P≤0.03) was observed in cells coexpressing GFP- and mCherry-tagged NS4B, while insignificant FRET was detected in cells expressing PDS together with NS4B or NS5A. A significant FRET signal, comparable to the NS4B positive control, was observed in cells coexpressing NS5A-GFP and NS4B-mCherry from genotype 2a (mean EF 5.5±7.2, p<0.001). A lower, yet noticeable FRET signal was detected in cells coexpressing NS5A-mCherry and NS4B-GFP from genotype 1b (mean EF 6.0±3.8). Chimeras containing NS4B of genotype 1b and NS5A of genotype 2a have been shown to replicate (Han et al., 2013). Thus, we tested if NS4B and NS5A could interact intergenotypically. Indeed FRET was observed between NS5A from genotype 2a and NS4B from genotype 1b (mean EF 6.5±3.2, p<0.005), with FRET efficiency levels comparable to those pre-formed using NS4B and NS5A from the same genotype as a FRET pair. The proper and equivalent co-expression levels of the above-mentioned proteins were assessed using western blot analysis with primary anti-GFP and mCherry antibodies (Fig. 1B). To further confirm the physical interaction between these proteins, a mammalian two-hybrid system was used. In this system one of potential interacting partners is fused to the DNA binding domain of a transcription factor (GAL4) and the second to a transcription factor activation domain (VP16). Plasmids expressing these two fusion proteins are transfected into cells, together with a vector containing GAL4 binding sites upstream of a minimal TATA box that controls the expression of a luciferase reporter gene. Association of the DNA-binding domain and the transcriptional activation domain due to an interaction between the fusion proteins results in transcriptional activation of the firefly luciferase reporter gene. The mammalian two-hybrid interaction happens in the nucleus (fusion proteins contain a nuclear localization signal) and thus was unlikely to occur with the full length transmembrane NS4B. Although a recent study provided some evidence that the N-terminal of NS4B can interact with NS5A (Choi et al., 2013), the N-terminal domain of NS4B is thought to translocate into the ER lumen (Lundin et al., 2006) and thus, might be less accessible for interaction with other proteins. The central segment of NS4B is mostly composed of transmembrane domains, interspaced with relatively short connecting loops (Lundin et al., 2003), thus we decided to focus on the C-terminal domain of the protein. To this end, the C-terminal domain of NS4B (a.a. 192–261) or NS5A from genotype 2a were cloned into both pACT (VP16 fusion) and pBind (GAL4 fusion) mammalian two-hybrid vectors. Luciferase activity was determined 48 h post-transfection of these vectors into Huh7 cells. The positive control consisted of MyoD and Id, two myogenic proteins known to interact (Finkel et al., 1993). Luciferase activity levels comparable to the positive control were obtained when pACT-NS5A and pBIND-NS4B C-terminal domain were coexpressed in Huh7 cells (Fig. 2A). Reverse experiments where NS4B C-terminal domain was expressed from pACT and NS4B from pBIND were unsuccessful (not shown). This phenomenon of specific inserts showing an asymmetric vector preference in such assays was previously described (Finkel et al., 1993). To rule out self-activation of the system by the proteins themselves, pACT-NS5A and pBIND-NS4B C-terminal domain were both co-expressed with the empty reciprocal plasmid. Luciferase activity in these experiments was comparable to background levels, indicating that NS5A and the C-terminal domain of NS4B do not activate luciferase transcription by themselves. The correct and comparable expression of both proteins was assessed using western blot with VP-16 and GAL4 specific antibodies and is shown in Fig. 2B. Taken together these results confirm the physical interaction between NS4B and NS5A.
Fig. 1.
FRET analysis of NS4B–NS5A interaction. A. Huh7 cells were co-transfected with the indicated GFP or mCherry tagged NS4B and NS5A combinations. GFP-tagged pendrin (PDS) was used as a negative control. Acceptor photo-bleaching FRET analysis was performed 24 h post-transfection. Fluorescence of donor and acceptor was measured as a function of time during acceptor photobleaching. Intensities within a region of interest were analyzed and are plotted in the graphs as mean FRET efficiency values. Asterisks denote statistical significance in an unpaired Student t test. **, P≤0.01; *, P≤0.05. B. Comparable expression levels of the constructs shown in panel A as determined by Western blot using GFP and mCherry specific antibodies. Molecular mass markers are indicated on the right. Monoclonal mouse anti-β-actin antibody was used as a loading control.
Fig. 2.
The C terminus of NS4B interacts with NS5A in a mammalian two-hybrid system. A. Huh7 cells were transfected with the indicated plasmid combinations. In this assay the interaction between full length NS5A and the C terminal domain (CTD) of NS4B (a.a. 192–261) was examined (see scheme). The culture media was collected 48 h post-transfection and tested for SEAP activity as a control for transfection efficiency. Followed by a luciferase activity assay performed on cell lysates. Luciferase activity results are presented as a percentage relative to the NS5A and NS4B CTD value. Values are (mean±SD) from triplicate wells. The graph is a representative result from three independent experiments. Asterisks denote statistical significance in an unpaired Student t test *P=0.01; **P=0.008. B. Comparable expression levels of the constructs are shown in panel B as determined by Western blot using VP16 and GAL4 antibodies. Monoclonal mouse anti-β-actin antibody was used as a loading control. Molecular mass markers are indicated on the right.
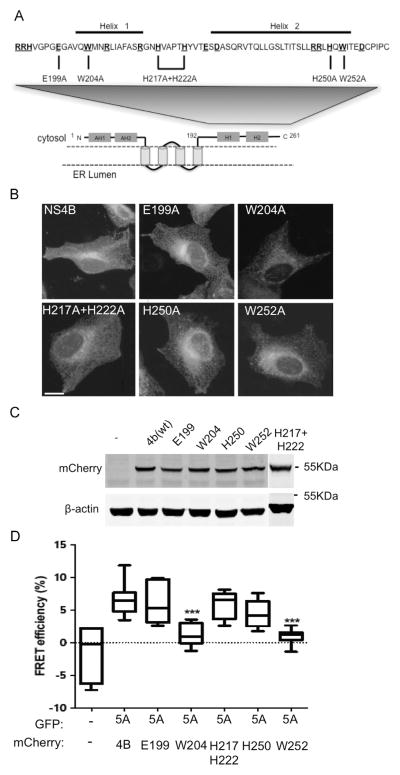
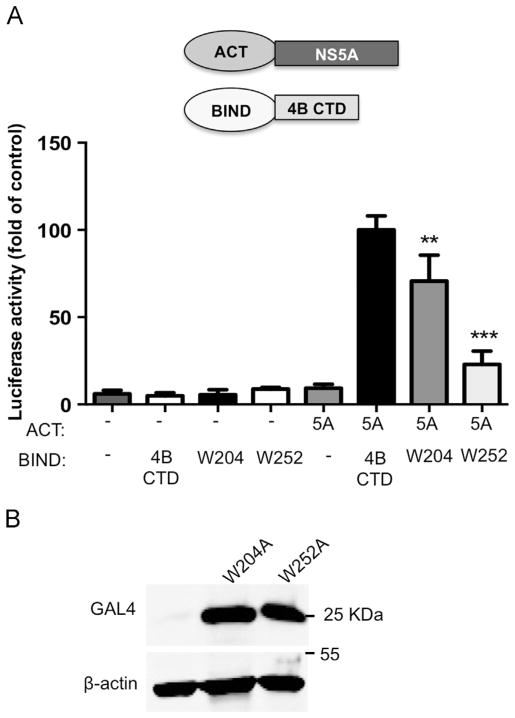
To pinpoint the residues responsible for this interaction, point mutations were inserted into residues in the C-terminal of NS4B. The ability of the NS4B mutants to interact with NS5A was determined using FRET. Our rationale in selecting the residues for mutagenesis was based on the following hypotheses: The interaction between NS4B and NS5A is conserved among the HCV subtypes and thus the residues to be mutated should be well-conserved; charged residues often provide the electrostatic surface required for protein–protein interactions (Sheinerman et al., 2000); thus mutations were planned to replace conserved charged residues. In general, residues belonging to the arginine-rich RNA binding domain of NS4B or conserved residues in their close vicinity were not replaced (Einav et al., 2008). Similarly residues E226 shown to alter NS4B’s protein stability, D256 shown to alter polyprotein processing, and D228 shown to be part of the NS4B NTPase motif, were not substituted as well (Blight, 2011; Einav et al., 2004). In addition to conserved charged residues two tryptophan residues were replaced. These changes were based on the fact that a tryptophan residue in the C-terminal domain of NS4B were previously highlighted as a possible candidate involved in protein–protein interactions (Lindström et al., 2006). Furthermore, tryptophan residues were previously found to be statistically overrepresented in sites of protein–protein interactions (Sillerud and Larson, 2005). Thus, muta-genesis of conserved tryptophan residues was included in our analysis as well. Fig. 3A shows the topology of NS4B, highlighting the cytosolic C-terminal domain. The sequence of this region is shown above and the location of the conserved charged residues is specified. The indicated residues (E199; W204; H217+H222; H250 and W252) were replaced by alanine in the context of NS4B-pmCherry for FRET analysis. To confirm that the indicated mutations did not alter the localization of NS4B, the mutated plasmids were overexpressed in Huh7 cells and their localization was examined using confocal microscopy (Fig. 3B). All of the mutants showed a localization pattern similar to wild type NS4B (top left). Furthermore, all of the mutants showed similar expression levels, comparable to the wild type, indicating that these mutations did not have any significant effect on the expression levels of NS4B (Fig. 3C). The effect of the mutations on the NS4B–NS5A interaction was determined by FRET analysis using acceptor photo-bleaching. These experiments were performed in U2OS cells, a cell line widely used for imaging experiments, due to its well-defined morphology. FRET efficiency values in these cells were similar to those obtained in Huh7 cells (compare the wild types Fig. 1A in Huh7 with U2OS in Fig. 3D). When coexpressed with NS5A, the NS4B mutants E199, H217+H222, and H250 displayed FRET efficiency levels comparable to the wild type control. In contrast, mutations in two tryptophan residues, W204 and W252, significantly reduced FRET efficiency (mean Ef=1.3±0.45 and 1±0.4 respectively compared to 6.5±0.5, p<0.0001). To confirm the role of W204 and W252 in the NS4B–NS5A interaction, the effect of these mutations on their interaction in the two-hybrid system was examined (Fig. 4). In agreement with the FRET results, mutagenesis of W252 had a significant effect on the NS4B–NS5A interaction, as indicated by a 77% reduction in the luciferase activity compared to the wild type control (p<0.0001). While W204 seemed to have a less pronounced effect in this system compared to the FRET experiments (luciferase activity was reduced only by 30% compared to wild type), this effect was, however, still significant (p<0.002). To rule out the possibility of self-activation by the mutants, each mutant was transfected with an empty pACT plasmid. Luciferase activity compared to the background was observed in all of these experiments. To confirm that the mutations did not affect NS4B’s expression in this system, expression was assessed by western blot with GAL4 specific antibodies (Fig. 4B). The expression of the NS4B was not affected by the mutations in this system. Taken together, these data suggest that these two tryptophan residues have a role in NS4B–NS5A interaction.
Fig. 3.
Mutations in the C terminal domain of NS4B affect its interaction with NS5A. A. Schematic representation of the predicted topology of NS4B. The C terminal domain is highlighted. The amino acid sequence of the C terminal domain from genotype 2a is indicated (genbank accession BAB32872.1). Conserved residues are bolded and underlined (for simplicity, labeled residues include both completely conserved residues and residues replaced with a similar amino acid). A black line above the sequence indicates the location of the two C-terminal helices. The inserted mutations are indicated below. B. Subcellular localization of wild type and mutant NS4B constructs was verified using confocal microscopy. Wild type NS4B-mCherry or mutants thereof were transfected into Huh7 cells grown on coverslips. Images were captured 24 h posttransfection using a confocal microscope. Bar=10 μM. C. Comparable expression of the NS4B mutants was confirmed by Western blot. Huh7 cells were transfected with wild type NS4B-mCherry or mutated NS4B constructs, as indicated. The cells were harvested 48 h post-transfection and the expression of the constructs was analyzed using mCherry antibodies. Monoclonal mouse anti-actin antibody was used as a loading control. Molecular mass markers are indicated on the right. D. Acceptor photo-bleaching FRET analysis of the NS4B C-terminal mutants. U2OS cells were co-transfected with the indicated NS4B and NS5A combinations. Acceptor photo-bleaching FRET analysis was performed 24 h post-transfection (as described in Fig. 1). Asterisks denote statistical significance in an unpaired Student t test. ***, P<0.0005.
Fig. 4.
C-terminal mutations within NS4B affect its interaction with NS5A in the two-hybrid system. A. Huh7 cells were transfected with the indicated plasmid combinations. The culture media was collected 48 h post-transfection and tested for SEAP activity as a control for transfection efficiency, followed by a luciferase activity assay performed on cell lysates. Luciferase activity results are presented as a percentage relative to the NS5A and NS4B CTD value. Values are (mean±SD) from triplicate wells. The graph represents three independent experiments. Asterisks denote statistical significance in an unpaired Student t test ***P<0.0005; **P<0.005. B. Expression levels of the mutant NS4B constructs are shown in panel A, as determined by Western blot using GAL4 antibodies. Monoclonal mouse anti-β-actin antibody was used as a loading control. Molecular mass markers are indicated on the right.
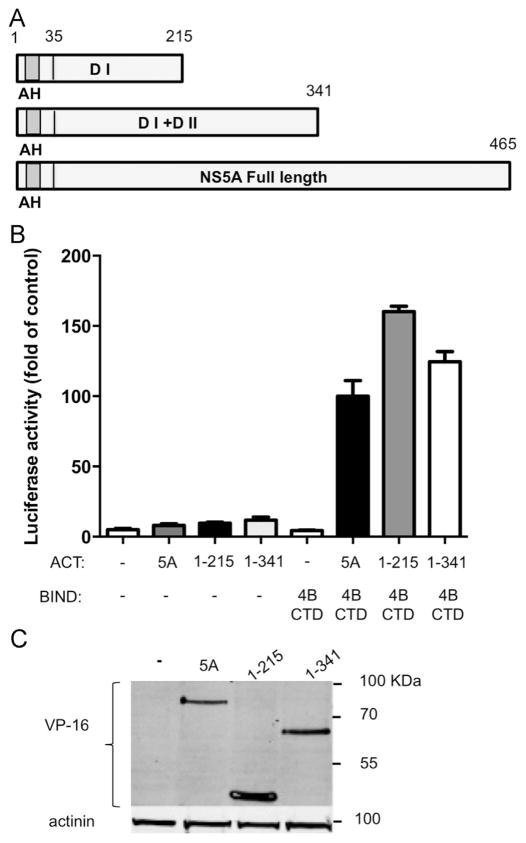
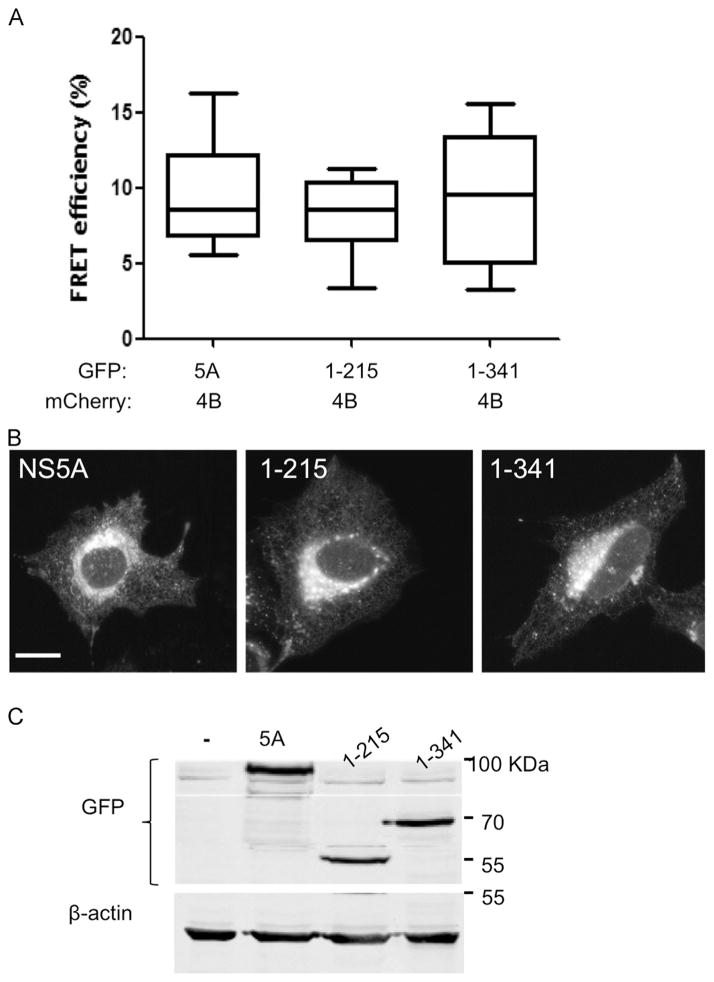
Next, we sought to identify the NS5A domains participating in this interaction. Since the various NS5A domains were linked to different stages in the viral life cycle, identification of a specific domain involved could give us an insight into the functional role of this interaction. The amphipathic helix in domain I is responsible for the correct localization of NS5A. Thus, all of the utilized constructs contained this domain. Two deletions were prepared, a deletion of both DII and DIII (a.a. 1–215), and a DIII deletion (1–341, see scheme in Fig. 5A). The effect of these deletions on the NS4B–NS5A interaction was tested using the two-hybrid system (Fig. 5B). Both deletion mutants were still able to interact with NS4B showing luciferase levels comparable to the full-length control. Interestingly, both deletion mutants showed notably elevated luciferase activities compared to full-length NS5A. This might be explained by the elevated expression levels of the DI deletion mutant (Fig. 5C). To further confirm these results, we performed complementary FRET experiments with similar deletion mutants. In agreement with the two-hybrid results, all the tested combinations displayed similar FRET efficiencies (mean Ef=9.6±0.9 for the wild type, 8±0.8 for DI and 9.3±1.1, Fig. 6A) for DI and DII. The deletions mutants were still able to localize correctly (Fig. 6B) and displayed similar expression levels (Fig. 6C). Together this suggests that a.a 1–215 are sufficient for NS5A’s interaction with NS4B. In attempt to further narrow down the interaction domain two additional deletion mutants were cloned into the mammalian two-hybrid vector. The first deletion contained the most N-terminal part of NS5A that harbors the AH (a.a. 1–34). The second included domain I without the AH (a.a. 35–215). The effect of both of these deletions on the NS5A–NS4B interaction was tested in the mammalian two-hybrid system (Fig. S3A). Transfection of the AH only construct (a.a. 1–35) with the NS4B C-terminal domain of NS4B showed only a minor elevation above the background. While luciferase activity levels of similarly transfected domain I without the AH (a.a. 35–215) were similar to background levels. Several reasons could explain this result; the deletion might have altered the folding of the protein and damaged an important motif. Or the fusion of such a short peptide to VP16 and GAL4 might have masked a region important for the interaction. The correct and comparable expression levels of the plasmids were determined using western blot (Fig. S3B).
Fig. 5.
NS5A a.a 1–215 are sufficient to mediate its interaction with NS4B. A. Schematic representation of the three NS5A constructs used. The N-terminal amphipathic helix (AH) is labeled in gray; Domain I and II are abbreviated as DI and DII respectively. Amino acid numbers are indicated above. B. The interaction between NS5A domains and NS4B CTD was determined using the two-hybrid system. Full length NS5A, NS5A a.a 1–215 or NS5A a.a 1–341 from genotype 2a were cloned into the appropriate two hybrid vectors. NS4B CTD and NS5A constructs were transfected into Huh7 cells as indicated. The culture media was collected 48 h post-transfection and tested for SEAP activity as a control for transfection efficiency. Followed by a luciferase activity assay preformed on the cell lysates. Luciferase activity results are presented as a percentage relative to the full length NS5A and NS4B CTD value. Values are (mean±SD) from triplicate wells. The graph is a representative result from three independent experiments. Values are (mean±SD) from triplicate wells. C. Expression levels of the NS5A deletion mutants shown in panel A and B. Expression levels were determined in Huh7 cells using western blot with VP16 antibodies. Monoclonal mouse anti- β-actin antibody was used as a loading control. Molecular mass markers are indicated on the right.
Fig. 6.
FRET analysis of the NS5A deletion mutants. A. U2OS cells were co-transfected with the indicated NS4B and NS5A combinations. Acceptor photo-bleaching FRET analysis was performed 24 h post-transfection (as described in Fig. 1). B. Subcellular localization of the NS5A deletion mutants. U2OS cells plated on coverslips were transfected with the indicated NS5A expressing constructs. Subcellular localization was determined using confocal microscopy 24 h post-transfection. C. Comparable expression of the NS5A deletion mutants was determined using western blot with anti-GFP antibodies. Monoclonal mouse anti-β-actin antibody was used as a loading control. Molecular mass markers are indicated on the right.
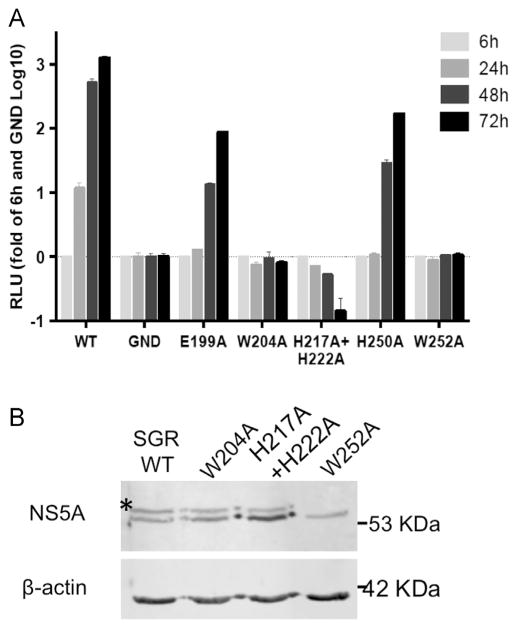
To assess the possible functional consequences of this interaction, we tested the ability of all the aforementioned mutants to replicate in the context of the J6/JFH luciferase reporter virus ((Tscherne et al., 2006), Fig. 7A). While mutations in E199A and H250A effect viral replication only moderately, the mutations W204A, H217A+H222A and W252 completely abolished viral replication. Similar results were obtained for these residues in previous studies (Blight, 2011; Jones et al., 2009; Paul et al., 2011). These data demonstrate the important role of these residues in the formation and function of the viral replication complex. NS4B was found, among other proteins, to be essential for the hyperphosphorylation of NS5A (Koch and Bartenschlager, 1999; Neddermann et al., 1999). Deletions in the C-terminal of NS4B abolished NS5A hyperphosphorylation (Koch and Bartenschlager, 1999). To examine if the point mutations disturbing the NS4B–NS5A interaction, affect NS5A phosphorylation, a vaccinia virus/T7 polymerase expression system was used (Sutter et al., 1995). Huh7 cells were infected with T7 polymerase expressing vaccinia virus, followed by transfection with the replication deficient genomes, expressed from a mutated JFH-1 subgenomic replicon DNA (pSGR-JFH1). The cells were analyzed using western blot with an NS5A specific antibody 20 h post-transfection. Typical bands at 56 and 58 kDa representing basal and hyperphosphorylated forms of NS5A, respectively, were observed for the wild type replicon and for the W204A and the H217A+H222A mutants (Fig. 7B). In contrast, the hyperphosphorylated form of NS5A was completely abolished in the replicon carrying the W252A mutation. These findings are in agreement with results of the two-hybrid interaction assay, where a significant yet moderate effect of W204A on the NS4B–NS5A interaction was observed. On the other hand, W252A had a pronounced effect on the NS4B–NS5A interaction in both FRET and mammalian two-hybrid assays, suggesting that the W204 NS4B mutation might still retain some of its NS5A binding capability while W252A mutation abolishes this interaction. Our results support the notion that the NS4B–NS5A interaction is necessary for viral replication and may be involved in mediating the NS5A hyperphosphorylation.
Fig. 7.
Mutations in the C-terminus of NS4B affect HCV replication and NS5A phosphorylation. A. Huh7.5 cells were transfected with in vitro transcribed RNA from a monocistronic reporter virus (J6/JFH(5′C19Rluc2AUbi)) encoding a full-length infectious J6/JFH genome with the Renilla luciferase reporter. Luciferase activity was determined in cell lysates at 6, 24, 48, and 72 h posttransfection. Luciferase activity in relative light units (RLU) is plotted for each time point. The data was normalized to the 6 h values that reflect transfection efficiency and to background luciferase levels determined using a replicon with a mutation in the viral RNA dependent RNA polymerase. Results represent mean values±SEM from two independent experiments performed in triplicates. B. Expression of pSGR-JFH-1 with the NS4B C terminal mutations using the vaccinia virus infection/transfection system. Huh7 cells were infected with a T7 polymerase expressing modified vaccinia virus, followed by transfection with the indicated pSGR DNA. To monitor the NS5A phosphorylation status, the cells were lysed 20 h post-transfection and analyzed by western blot using the 9E10 anti-NS5A monoclonal antibody. Monoclonal mouse anti-β-actin antibody was used as a loading control. Shown is a representative blot from 2 independent experiments. Asterisk indicates the hyperphosphorylated form of NS5A. Molecular mass markers are indicated on the right.
Discussion
NS4B and NS5A are essential for HCV replication and assembly. Several lines of evidence indicate that these two proteins interact functionally and genetically. However, although previously detected (Aligo et al., 2009; Dimitrova et al., 2003; Gao et al., 2004), their physical interaction was never fully characterized. Here, we confirmed this interaction using two independent methods FRET of ectopically expressed proteins; and the mammalian two-hybrid system.
Efforts to perform FRET experiments with a full-length replicon containing a GFP insertion within NS5A and ectopically expressed NS4B were not successful. This may be attributed to the inaccessibility of the GFP under these conditions. Specific conserved residues in the C-terminal domain of NS4B, W204 and W252 were found to have a central role in mediating this interaction. Mutations in these residues abolished viral replication.
HCV replicates in highly ordered multi-subunit membrane associated RCs (Gao et al., 2004). In addition to the previously characterized complex heterotypic interactions that occur within RCs, a growing list of viral proteins including NS4B, NS5A and most recently NS4A were reported to function as dimers or oligomers (Dimitrova et al., 2003; Gouttenoire et al., 2010; Kohlway et al., 2013; Love et al., 2009; Tellinghuisen et al., 2005; Yu et al., 2006). Interestingly the W204A mutant that affects the NS4B–NS5A interaction was previously shown to impair NS4B heterotypic oligomerization as well (Paul et al., 2011). Similar results regarding W204A were obtained in our hands, however, the effect of W252A was inconclusive due to high variability (FRET efficiency did not significantly differ from the wild type, not shown). Confirmation that these proteins bind the same domain deserves further studies. If proven, this possibility should be taken under consideration when a model for HCV RC assembly is formulated.
NS5A apparently associates with the RC via interaction with other NS proteins. This was shown using fluorescence recovery after photobleaching (FRAP) analysis by demonstrating its increased mobility in the absence of the other NS proteins (Jones et al., 2007). FRAP analysis of NS5A-GFP expressed as a part of the HCV polyprotein was used to determine the possible role of NS4B in mediating the mobility of NS5A. Deletion of NS4B from this construct significantly increased the mobility of NS5A (Jones et al., 2009). NS5A-GFP expressed from a polyprotein in which NS4B was carrying the W252A mutation displayed a similar mobility to the NS4B deletion supporting the notion that NS4B has a role in the association of NS5A with the RC. In agreement with our results Jones et al. showed that W252A abolishes viral replication, prevents NS4B induced foci formation and NS5A hyperphosphorylation (Jones et al., 2009).
Efforts to isolate reversions from replicons carrying either W204A or W252A mutations were unsuccessful, both in our hands and others (Lindström et al., 2006; Paul et al., 2011) demonstrating the importance of these residues. However, replication of a W252A mutant replicon could be partially rescued by a wild type replicon or a replicon defective in NS5A (Jones et al., 2009). Suggesting that NS4B could interact with NS5A supplied in trans. NS4B is a membrane protein, with relatively slow mobility and thus does not readily transfer between foci. This fact, together with the low efficiency of trans-complementation, suggests that complementation probably occurs within a single focus (Gretton et al., 2005; Jones et al., 2009).
The function of NS5A phosphorylation is still not understood, although mutations that lower the amount of the hyperphosphorylated form of NS5A are believed to stimulate viral replication (Appel et al., 2005b; Blight et al., 2000; Katze et al., 2000; Tanji et al., 1995b). Nevertheless, minimal levels of hyperphosphorylated NS5A seem to be essential for viral replication, since viral replication is inhibited upon its complete loss (Appel et al., 2005a; Fridell et al., 2011). In agreement with this notion, Jones et al. found a direct correlation among the ability of NS4B to form foci, the increased mobility of NS5A and its phosphorylation state (Jones et al., 2009). In our study, the hyperphosphorylated state of NS5A expressed from a replicon DNA with a W204A mutation resembled the wild type, while only basal-phosphorylated NS5A was present in a W252A containing replicon.
NS4B was shown to be essential for NS5A phosphorylation (Koch and Bartenschlager, 1999; Neddermann et al., 1999). To produce and maintain its hyperphosphorylated state, NS5A might need to be in an appropriate conformation, recruit host kinases or other assisting factors and to protect its phosphorylated sites from phosphatases (Koch and Bartenschlager, 1999; Neddermann et al., 1999). These roles may be provided by its interaction with NS4B. Although both of our mutations inhibited viral replication and affected the NS4B–NS5A interaction, some prominent differences exist which may explain the ability of the W204A mutation to support hyperphsophorylation of NS5A. While both of the mutations reduced FRET efficiency by comparable levels, significant differences between the mutations were observed in the two-hybrid assay, where W204A had only a moderate effect on the NS4B–NS5A binding. In our hands, viral replication was abolished to the same extent with both mutants, while Paul et al. reported that W204A was able to support low levels of replication (Paul et al., 2011). These data suggest that W204A retains some of its NS5A binding activity and this residual activity is sufficient to modulate NS5A hyperphosphorylation. In contrast, W252A seems to be a major mediator of this interaction; its mutagenesis causes detrimental effects on the ability of the virus to replicate.
Interestingly, a double H217A+H222A mutation did not affect NS5A phosphorylation. This mutation also did not affect the NS4B–NS5A interaction, but it was still unable to support viral RNA replication. The fact that the H222A mutation was previously shown to inhibit the NS4B–NS4B heterotypic interaction (Paul et al., 2011) might account for the inability of this mutant to support viral replication. Nevertheless, this result further highlights the functional differences between these interactions.
Our results point to domain I of NS5A as the area responsible for the interaction. Interestingly, resistance mutants to several of the most advanced direct acting antivirals targeting NS5A map to this region (for a recent review see (Lim and Gallay, 2014)). Our results might suggest that modulation of the NS4B–NS5A interaction could be added to the list of potential NS5A direct acting antivirals targets.
Experimental procedures
Cell culture and transfections
Huh7, Huh7.5 and Huh7 cells harboring a subgenomic 1b replicon with an in frame hemagglutinin (HA) tag insertion in NS5A (Bart-HA, (Sklan et al., 2007)) were propagated in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% (v/v) fetal calf serum, 1% (v/v) penicillin/streptomycin and 1% (v/v) of MEM Non-Essential Amino Acids (Biological Industries, Bet-Haemek, Israel). The Bart-HA cells were grown in the presence of 0.7 μg/ml G418 (Calbiochem, San Diego, CA). U2OS cells were propagated in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. Lipofectamine 2000 (Invitrogen, Carlsbad, CA) or polyethyleneimine (Polysciences, Warrington, PA) were used for plasmid DNA transfections of subconfluent Huh7 and U2OS cells.
Plasmids
The plasmid expressing green fluorescent protein (GFP) or mCherry fusions with NS4B from HCV genotype 2a were created by amplifying the NS4B coding sequence from pFL-J6/JFH DNA and subcloning the obtained insert into pEGFP-N1 and pmCherry-N1 (Clontech, Palo Alto, CA). The pmCherry-N1 plasmid expressing NS4B from genotype 1b fused to mCherry was generated by amplifying NS4B from Bart79I, a genotype 1b subgenomic replicon (Einav et al., 2004), and cloning the obtained insert into pmCherry-N1. The plasmids expressing NS5A(2a)-GFP, NS5A(1b)-mCherry and pEF-NS4B (1b)-GFP were previously described (Einav et al., 2004; Nevo-Yassaf et al., 2012). The plasmids expressing the different NS5A domains fused to GFP were generated by amplifying NS5A a.a. 1–247 or NS5A a.a. 1–352 from the NS5A(2a)-GFP vector to generate the insert that was ligated into the HindIII/AgeII restriction sites of pEGFP-N1. GFP tagged pendrin (GFP-PDS) was previously described (Shepshelovich et al., 2005). To generate the pACT and pBIND plasmids for the mammalian two hybrid system, NS5A of its fragments were amplified from NS5A-GFP(2a) and subcloned into the XbaI/NotI sites of pACT or pBIND. Similarly the C-terminal domain of NS4B (a.a. 192–261) was amplified from NS4B-mCherry (2a), and the PCR products were digested with XbaI and NotI and ligated into pACT or pBIND. pSGR-JFH1 was kindly provided by T. Wakita (National Institutes of Infectious Diseases, Japan) has been previously described (Kato et al., 2003). Mutagenesis was performed using either tail-to-tail PCR (Hemsley et al., 1989) or the QuickChange II site-directed mutagenesis kit (Strategene, La Jolla, CA).
Western blot analysis
Proteins were extracted using cell lysis buffer (300 mM NaCl, 100 mM Tris–HCl pH 8, 0.2 mM EDTA, 10% Glycerol) supplemented with a protease inhibitor cocktail (Sigma). The cell lysates were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane. Membranes were then blocked for 1 h in blocking solution [Phosphate buffer saline (PBS) containing 5% non-fat milk (Sigma, Israel)] followed by incubation with the appropriate primary antibody. Primary antibodies used were mouse monoclonal anti-GAL4 and VP-16 antibodies (Santa Cruz biotechnology, Dallas, TX), rabbit polyclonal anti-RFP antibody (MBL, Woburn, MA) and a rabbit anti-GFP polyclonal antibody (Abcam, Cambridge, MA). Monoclonal mouse antibodies against β actin and Actin were from Sigma and Santa Cruz, respectively. The mouse monoclonal 9E10 antibody against NS5A, was kindly provided by Charles M. Rice (Rockfeller University). The blots were then incubated with the corresponding IR-Dye conjugated secondary antibodies (Li-cor, Lincoln, NE). Proteins were visualized using a LI-COR infrared imager (Odyssey).
In-vitro transcription and electroporation of HCV RNAs
In vitro transcripts of J6/JFH (5′C19Rluc2AUbi) were prepared as described Lindenbach et al., 2005; Tscherne et al., 2006. Briefly, plasmid DNA was linearized using XbaI. The linearized DNA template was purified using phenol:chloroform extraction and ethanol precipitation. The linearized template was transcribed with T7 RNA polymerase using a MEGAscript™ T7 kit (Ambion, Austin, TX). Reaction mixtures were incubated for 3 h at 37 °C and then subjected to DNase treatment for an additional 15 min at 37 °C. The integrity of the RNA was analyzed by non-denaturing agarose gel electrophoresis and the yield was determined spectrophotometrically. For the electroporation, Huh7.5 cells were grown to 60–80% confluency, trypsinized and washed twice in cold RNAse-free PBS (BioWhitaker, Walkersville, MD). Cells were resuspended in cold PBS at a concentration of 1.5 × 107 cells/ml; 0.4 ml of the cell suspension was mixed with 5 μg in vitro-transcribed RNA. The mixture was dispensed into a 2-mm gap width cuvette (BTX, San Diego, CA) and electroporation was performed using a BTX model 830 electroporator (820 V, 5 × 99-μs pulses given at 220-ms intervals). Cells were left to recover for 15 min at room temperature and then mixed with 10 ml of pre-warmed (37 °C) growth medium. Cells were seeded in six-well plates, luciferase levels were determined 6, 24, 48 and 72 h post-electroporation. For the luciferase assay electroporated cells were washed twice with PBS and lysed with reporter lysis buffer (Promega, Madison, WI) followed by 15 min of shaking at room temperature, luciferase activity was determined using Renilla luciferase substrate (Coletrazine, Calbiochem) and a Mithras LB 940 multimode microplate reader (Berthold technologies, Bad Wildbad Germany)
Mammalian two-hybrid experiments
NS4B and NS5A were subcloned into pACT and pBIND plasmids (CheckMate; Promega). The appropriate plasmids (500 ng of each) were transfected together with the reporter plasmid pG5luc into Huh7 cells plated in 24-well plates at a confluence of 60–80%. A plasmid expressing secreted alkaline phosphatase (SEAP, 50 ng) was added to each transfection reaction as a transfection efficiency control. Media from the cells was removed 48 h post-transfection and analyzed for SEAP activity using the Phospha-Light System (Applied biosystems, Foster city, CA). The cells were lysed with luciferase reporter lysis buffer (Promega) and luciferase activity was determined using a Mithras LB 940 multimode microplate reader (Berthold technologies).
Infection and transfection
Huh7 cells were infected with modified vaccinia Ankara virus (MVA) expressing the T7 RNA polymerase (Sutter et al., 1995). Following an incubation of 1 h at 37 °C, the cells were washed twice with Optimem (Invitrogen) and wild type or mutant pSGR-JFH1 DNA was transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. The cells were supplemented with growth media 4 h post-transfection and harvested for Western blot analysis with mouse monoclonal 9E10 antibody against NS5A 20 h post-transfection.
FRET analysis using acceptor photobleaching
Acceptor photobleaching was essentially carried out as described (Yaffe et al., 2012). Briefly, Huh7 or U2OS cells were grown on coverslips and transfected with NS4B and NS5A fused to mCherry (acceptor) or GFP (donor) using polyethyleneimine. GFP-PDS was used as a negative control. The cells were fixed 24 h post-transfection. A region of interest containing the mCherry acceptor was repeatedly photobleached in 4 to 6 cycles using 564 nm HeNe laser. Changes in the fluorescence intensity of the GFP donor and photobleached acceptor were measured using 488 nm Ar and 564 nm HeNe lasers. FRET efficiencies (EF) from at least 10 different cells were calculated using the equation:
where Dpost is the fluorescence intensity of the donor after acceptor photobleaching and Dpre is the fluorescence intensity of the donor before acceptor photobleaching. All calculated FRET efficiency values were normalized to levels of photobleached acceptor calculated as the percent of reduction in the initial fluorescent intensity (prior to photobleaching).
Supplementary Material
Acknowledgments
This work was supported by the Israel Science Foundation Grant, the Marguerite Stolz Research Fund (E.H.S) and NIH RO1AI087917 (J.S.G.). We thank Charles Rice (The Rockefeller University, New York, NY), Takaji Wakita (National Institutes of Infectious Diseases, Tokyo, Japan) and Eran Bacharach (Tel Aviv University) for providing reagents.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.virol.2014.10.021.
Footnotes
Immunofluorescence microscopy
Bart-HA cells were cultured on coverslips, fixed with 2% form-aldyhyde, permeabilized with 0.2 % saponin and blocked with 3 % fetal bovine serum (FBS) followed by immunolabeling with an monoclonal primary antibody against NS4B (Abcam) and an anti-HA rabbit polyclonal antibody (Santa Cruz Biotechnology). The secondary antibodies were Alexa 488 goat anti-mouse antibody (Invitrogen) and Cy3 goat anti-rabbit (Jackson ImmunoResearch, West Grove, PA). Images were acquired using a Zeiss Pascal confocal laser scanning microscope.
References
- Ago H, Adachi T, Yoshida A, Yamamoto M, Habuka N, Yatsunami K, Miyano M. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Structure. 1999;7:1417–1426. doi: 10.1016/s0969-2126(00)80031-3. [DOI] [PubMed] [Google Scholar]
- Aligo J, Jia S, Manna D, Konan KV. Formation and function of hepatitis C virus replication complexes require residues in the carboxy-terminal domain of NS4B protein. Virology. 2009;393:68–83. doi: 10.1016/j.virol.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel N, Herian U, Bartenschlager R. Efficient rescue of hepatitis C virus RNA replication by trans-complementation with nonstructural protein 5A. J Virol. 2005a;79:896–909. doi: 10.1128/JVI.79.2.896-909.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel N, Pietschmann T, Bartenschlager R. Mutational analysis of hepatitis C virus nonstructural protein 5A: potential role of differential phosphorylation in RNA replication and identification of a genetically flexible domain. J Virol. 2005b;79:3187–3194. doi: 10.1128/JVI.79.5.3187-3194.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel N, Zayas M, Miller S, Krijnse-Locker J, Schaller T, Friebe P, Kallis S, Engel U, Bartenschlager R. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 2008;4:e1000035. doi: 10.1371/journal.ppat.1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blight KJ. Charged residues in hepatitis C virus NS4B are critical for multiple NS4B functions in RNA replication. J Virol. 2011;85:8158–8171. doi: 10.1128/JVI.00858-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- Bressanelli S, Tomei L, Roussel A, Incitti I, Vitale RL, Mathieu M, De Francesco R, Rey FA. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc Natl Acad Sci USA. 1999;96:13034–13039. doi: 10.1073/pnas.96.23.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho NJ, Dvory-Sobol H, Lee C, Cho SJ, Bryson P, Masek M, Elazar M, Frank CW, Glenn JS. Identification of a class of HCV inhibitors directed against the nonstructural protein NS4B. Sci Transl Med. 2010;2(15):15ra16. doi: 10.1126/scitranslmed.3000331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M, Lee S, Choi T, Lee C. A hepatitis C virus NS4B inhibitor suppresses viral genome replication by disrupting NS4B’s dimerization/multimerization as well as its interaction with NS5A. Virus Genes. 2013 doi: 10.1007/s11262-013-0956-5. [DOI] [PubMed] [Google Scholar]
- Dimitrova M, Imbert I, Kieny MP, Schuster C. Protein–protein interactions between hepatitis C virus nonstructural proteins. J Virol. 2003;77:5401–5414. doi: 10.1128/JVI.77.9.5401-5414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger D, Wolk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol. 2002;76:5974–5984. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einav S, Elazar M, Danieli T, Glenn JS. A nucleotide binding motif in hepatitis C virus (HCV) NS4B mediates HCV RNA replication. J Virol. 2004;78:11288–11295. doi: 10.1128/JVI.78.20.11288-11295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einav S, Gerber D, Bryson PD, Sklan EH, Elazar M, Maerkl SJ, Glenn JS, Quake SR. Discovery of a hepatitis C target and its pharmacological inhibitors by microfluidic affinity analysis. Nat Biotechnol. 2008;26:1019–1027. doi: 10.1038/nbt.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- Elazar M, Cheong KH, Liu P, Greenberg HB, Rice CM, Glenn JS. Amphipathic helix-dependent localization of NS5A mediates hepatitis C virus RNA replication. J Virol. 2003;77:6055–6061. doi: 10.1128/JVI.77.10.6055-6061.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elazar M, Liu P, Rice CM, Glenn JS. An N-terminal amphipathic helix in hepatitis C virus (HCV) NS4B mediates membrane association, correct localization of replication complex proteins, and HCV RNA replication. J Virol. 2004;78:11393–11400. doi: 10.1128/JVI.78.20.11393-11400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T, Duc J, Fearon ER, Dang CV, Tomaselli GF. Detection and modulation in vivo of helix-loop-helix protein–protein interactions. J Biol Chem. 1993;268:5–8. [PubMed] [Google Scholar]
- Fridell RA, Qiu D, Valera L, Wang C, Rose RE, Gao M. Distinct functions of NS5A in hepatitis C virus RNA replication uncovered by studies with the NS5A inhibitor BMS-790052. J Virol. 2011;85:7312–7320. doi: 10.1128/JVI.00253-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Aizaki H, He JW, Lai MMC. Interactions between viral non-structural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J Virol. 2004;78:3480–3488. doi: 10.1128/JVI.78.7.3480-3488.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouttenoire J, Montserret R, Kennel A, Penin F, Moradpour D. An amphipathic alpha-helix at the C terminus of hepatitis C virus nonstructural protein 4B mediates membrane association. J Virol. 2009;83:11378–11384. doi: 10.1128/JVI.01122-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouttenoire J, Roingeard P, Penin F, Moradpour D. Amphipathic alpha-helix AH2 is a major determinant for the oligomerization of hepatitis C virus nonstructural protein 4B. J Virol. 2010;84:12529–12537. doi: 10.1128/JVI.01798-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A, McCourt DW, Wychowski C, Feinstone SM, Rice CM. A second hepatitis C virus-encoded proteinase. Proc Natl Acad Sci USA. 1993a;90:10583–10587. doi: 10.1073/pnas.90.22.10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A, Wychowski C, Lin C, Feinstone SM, Rice CM. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993b;67:1385–1395. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gretton SN, Taylor AI, McLauchlan J. Mobility of the hepatitis C virus NS4B protein on the endoplasmic reticulum membrane and membrane-associated foci. J Gen Virol. 2005;86:1415–1421. doi: 10.1099/vir.0.80768-0. [DOI] [PubMed] [Google Scholar]
- Han Q, Manna D, Belton K, Cole R, Konan KV. Modulation of hepatitis C virus genome encapsidation by nonstructural protein 4B. J Virol. 2013;87:7409–7422. doi: 10.1128/JVI.03523-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsley A, Arnheim N, Toney MD, Cortopassi G, Galas DJ. A simple method for site-directed mutagenesis using the polymerase chain reaction. Nucl Acids Res. 1989;17:6545–6551. doi: 10.1093/nar/17.16.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata M, Mizushima H, Akagi T, Mori S, Kakiuchi N, Kato N, Tanaka T, Kimura K, Shimotohno K. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J Virol. 1993;67:4665–4675. doi: 10.1128/jvi.67.8.4665-4675.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide Y, Zhang L, Chen M, Inchauspe G, Bahl C, Sasaguri Y, Padmanabhan R. Characterization of the nuclear localization signal and subcellular distribution of hepatitis C virus nonstructural protein NS5A. Gene. 1996;182:203–211. doi: 10.1016/s0378-1119(96)00555-0. [DOI] [PubMed] [Google Scholar]
- Jones DM, Gretton SN, McLauchlan J, Targett-Adams P. Mobility analysis of an NS5A-GFP fusion protein in cells actively replicating hepatitis C virus subgenomic RNA. J Gen Virol. 2007;88:470–475. doi: 10.1099/vir.0.82363-0. [DOI] [PubMed] [Google Scholar]
- Jones DM, Patel AH, Targett-Adams P, McLauchlan J. The hepatitis C virus NS4B protein can trans-complement viral RNA replication and modulates production of infectious virus. J Virol. 2009;83:2163–2177. doi: 10.1128/JVI.01885-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Date T, Miyamoto M, Furusaka A, Tokushige K, Mizokami M, Wakita T. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology. 2003;125:1808–1817. doi: 10.1053/j.gastro.2003.09.023. [DOI] [PubMed] [Google Scholar]
- Katze MG, Kwieciszewski B, Goodlett DR, Blakely CM, Neddermann P, Tan SL, Aebersold R. Ser(2194) is a highly conserved major phosphorylation site of the hepatitis C virus nonstructural protein NS5A. Virology. 2000;278:501–513. doi: 10.1006/viro.2000.0662. [DOI] [PubMed] [Google Scholar]
- Kenworthy AK. Imaging protein–protein interactions using fluorescence resonance energy transfer microscopy. Methods. 2001;24:289–296. doi: 10.1006/meth.2001.1189. [DOI] [PubMed] [Google Scholar]
- Koch JO, Bartenschlager R. Modulation of hepatitis C virus NS5A hyperphosphorylation by nonstructural proteins NS3, NS4A, and NS4B. J Virol. 1999;73:7138–7146. doi: 10.1128/jvi.73.9.7138-7146.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlway A, Pirakitikulr N, Barrera FN, Potapova O, Engelman DM, Pyle AM, Lindenbach BD. Hepatitis C virus RNA replication and virus particle assembly require specific dimerization of the NS4A protein transmembrane domain. J Virol. 2013 doi: 10.1128/JVI.02052-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesburg CA, Cable MB, Ferrari E, Hong Z, Mannarino AF, Weber PC. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat Struct Biol. 1999;6:937–943. doi: 10.1038/13305. [DOI] [PubMed] [Google Scholar]
- Lim PJ, Gallay PA. Hepatitis C NS5A protein: two drug targets within the same protein with different mechanisms of resistance. Curr Opin Virol. 2014;8C:30–37. doi: 10.1016/j.coviro.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- Lindström H, Lundin M, Häggström S, Persson MAA. Mutations of the Hepatitis C virus protein NS4B on either side of the ER membrane affect the efficiency of subgenomic replicons. Virus Res. 2006;121:169–178. doi: 10.1016/j.virusres.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- Love RA, Brodsky O, Hickey MJ, Wells PA, Cronin CN. Crystal structure of a novel dimeric form of NS5A domain I protein from hepatitis C virus. J Virol. 2009;83:4395–4403. doi: 10.1128/JVI.02352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin M, Lindstrom H, Gronwall C, Persson MA. Dual topology of the processed hepatitis C virus protein NS4B is influenced by the NS5A protein. J Gen Virol. 2006;87:3263–3272. doi: 10.1099/vir.0.82211-0. [DOI] [PubMed] [Google Scholar]
- Lundin M, Monne M, Widell A, Von Heijne G, Persson MA. Topology of the membrane-associated hepatitis C virus protein NS4B. J Virol. 2003;77:5428–5438. doi: 10.1128/JVI.77.9.5428-5438.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaki T, Suzuki R, Murakami K, Aizaki H, Ishii K, Murayama A, Date T, Matsuura Y, Miyamura T, Wakita T, Suzuki T. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J Virol. 2008;82:7964–7976. doi: 10.1128/JVI.00826-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S, Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol. 2008;6:363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- Neddermann P, Clementi A, De Francesco R. Hyperphosphorylation of the hepatitis C virus NS5A protein requires an active NS3 protease, NS4A, NS4B, and NS5A encoded on the same polyprotein. J Virol. 1999;73:9984–9991. doi: 10.1128/jvi.73.12.9984-9991.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neddermann P, Quintavalle M, Di Pietro C, Clementi A, Cerretani M, Altamura S, Bartholomew L, De Francesco R. Reduction of hepatitis C virus NS5A hyperphosphorylation by selective inhibition of cellular kinases activates viral RNA replication in cell culture. J Virol. 2004;78:13306–13314. doi: 10.1128/JVI.78.23.13306-13314.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo-Yassaf I, Yaffe Y, Asher M, Ravid O, Eizenberg S, Henis YI, Nahmias Y, Hirschberg K, Sklan EH. Role for TBC1D20 and Rab1 in hepatitis C virus replication via interaction with lipid droplet-bound nonstructural protein 5A. J Virol. 2012;86:6491–6502. doi: 10.1128/JVI.00496-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D, Romero-Brey I, Gouttenoire J, Stoitsova S, Krijnse-Locker J, Moradpour D, Bartenschlager R. NS4B self-interaction through conserved C-terminal elements is required for the establishment of functional hepatitis C virus replication complexes. J Virol. 2011;85:6963–6976. doi: 10.1128/JVI.00502-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietschmann T, Zayas M, Meuleman P, Long G, Appel N, Koutsoudakis G, Kallis S, Leroux-Roels G, Lohmann V, Bartenschlager R. Production of infectious genotype 1b virus particles in cell culture and impairment by replication enhancing mutations. PLoS Pathog. 2009;5:e1000475. doi: 10.1371/journal.ppat.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak SJ, Paschal DM, McArdle S, Gale MJ, Jr, Moradpour D, Gretch DR. Characterization of the effects of hepatitis C virus nonstructural 5A protein expression in human cell lines and on interferon-sensitive virus replication. Hepatology. 1999;29:1262–1271. doi: 10.1002/hep.510290438. [DOI] [PubMed] [Google Scholar]
- Romero-Brey I, Merz A, Chiramel A, Lee JY, Chlanda P, Haselman U, Santarella-Mellwig R, Habermann A, Hoppe S, Kallis S, Walther P, Antony C, Krijnse-Locker J, Bartenschlager R. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 2012;8:e1003056. doi: 10.1371/journal.ppat.1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheinerman FB, Norel R, Honig B. Electrostatic aspects of protein–protein interactions. Curr Opin Struct Biol. 2000;10:153–159. doi: 10.1016/s0959-440x(00)00065-8. [DOI] [PubMed] [Google Scholar]
- Shepshelovich J, Goldstein-Magal L, Globerson A, Yen PM, Rotman-Pikielny P, Hirschberg K. Protein synthesis inhibitors and the chemical chaperone TMAO reverse endoplasmic reticulum perturbation induced by overexpression of the iodide transporter pendrin. J Cell Sci. 2005;118:1577–1586. doi: 10.1242/jcs.02294. [DOI] [PubMed] [Google Scholar]
- Sillerud LO, Larson RS. Design and structure of peptide and peptidomimetic antagonists of protein–protein interaction. Curr Protein Pept Sci. 2005;6:151–169. doi: 10.2174/1389203053545462. [DOI] [PubMed] [Google Scholar]
- Sklan EH, Staschke K, Oakes TM, Elazar M, Winters M, Aroeti B, Danieli T, Glenn JS. A Rab-GAP TBC domain protein binds hepatitis C virus NS5A and mediates viral replication. J Virol. 2007;81:11096–11105. doi: 10.1128/JVI.01249-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter G, Ohlmann M, Erfle V. Non-replicating vaccinia vector efficiently expresses bacteriophage T7 RNA polymerase. FEBS Lett. 1995;371:9–12. doi: 10.1016/0014-5793(95)00843-x. [DOI] [PubMed] [Google Scholar]
- Tanji Y, Hijikata M, Satoh S, Kaneko T, Shimotohno K. Hepatitis C virus-encoded nonstructural protein NS4A has versatile functions in viral protein processing. J Virol. 1995a;69:1575–1581. doi: 10.1128/jvi.69.3.1575-1581.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanji Y, Kaneko T, Satoh S, Shimotohno K. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J Virol. 1995b;69:3980–3986. doi: 10.1128/jvi.69.7.3980-3986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellinghuisen TL, Foss KL, Treadaway J. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 2008a;4:e1000032. doi: 10.1371/journal.ppat.1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellinghuisen TL, Foss KL, Treadaway JC, Rice CM. Identification of residues required for RNA replication in domains II and III of the hepatitis C virus NS5A protein. J Virol. 2008b;82:1073–1083. doi: 10.1128/JVI.00328-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellinghuisen TL, Marcotrigiano J, Gorbalenya AE, Rice CM. The NS5A protein of hepatitis C virus is a zinc metalloprotein. J Biol Chem. 2004;279:48576–48587. doi: 10.1074/jbc.M407787200. [DOI] [PubMed] [Google Scholar]
- Tellinghuisen TL, Marcotrigiano J, Rice CM. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature. 2005;435:374–379. doi: 10.1038/nature03580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tscherne DM, Jones CT, Evans MJ, Lindenbach BD, McKeating JA, Rice CM. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J Virol. 2006;80:1734–1741. doi: 10.1128/JVI.80.4.1734-1741.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe Y, Shepshelovitch J, Nevo-Yassaf I, Yeheskel A, Shmerling H, Kwiatek JM, Gaus K, Pasmanik-Chor M, Hirschberg K. The MARVEL transmembrane motif of occludin mediates oligomerization and targeting to the basolateral surface in epithelia. J Cell Sci. 2012;125:3545–3556. doi: 10.1242/jcs.100289. [DOI] [PubMed] [Google Scholar]
- Yu GY, Lee KJ, Gao L, Lai MM. Palmitoylation and polymerization of hepatitis C virus NS4B protein. J Virol. 2006;80:6013–6023. doi: 10.1128/JVI.00053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.