Abstract
In all living cells, DNA is the storage medium for genetic information. Being quite stable, DNA is well-suited for its role in storage and propagation of information, but RNA is also covalently included in DNA through various mechanisms. Recent studies also demonstrate useful aspects of including ribonucleotides in the genome during repair. Therefore, our understanding of the consequences of RNA inclusion into bacterial genomic DNA is just beginning, but with its high frequency of occurrence the consequences and potential benefits are likely to be numerous and diverse. In this review, we discuss the processes that cause ribonucleotide inclusion in genomic DNA, the pathways important for ribonucleotide removal and the consequences that arise should ribonucleotides remain nested in genomic DNA.
Introduction
DNA is a remarkably stable medium for the storage and replication of genetic information, and it carries out this role in all living cells. RNA is more prone to hydrolysis of the phosphodiester backbone than DNA due to the 2′-OH of the ribose sugar, which has the potential to carry out nucleophilic attack of the 3′-PO4− (Oivanen et al., 1998). It is becoming increasingly evident that the division between RNA and DNA is not absolute, in that ribonucleotides are frequently covalently nested in genomic DNA (Williams and Kunkel, 2014; Caldecott, 2014; Brown and Suo, 2011). Of course this has the potential to make the genome less stable, and ribonucleotides within genomic DNA must be removed and replaced with DNA.
DNA replication can be adversely affected by rNMPs in the template strand, and any nicks present in DNA due to hydrolysis at rNMPs can lead to replication stress with the potential for generating lethal double-stranded breaks (Pizzi et al., 2014; Yao et al., 2013; Reijns et al., 2012). However, there are also some useful aspects of having ribonucleotides in the genome, and in this review we summarize and discuss the mechanisms of insertion, removal and the consequences that result from the incorporation of ribonucleotides into genomic DNA. We briefly discuss the eukaryotic literature, however, we place strong emphasis on what is known in bacterial organisms. For readers more interested in studies emphasizing the removal of ribonucleotides from eukaryotic DNA, we wish to direct you to following recent reviews (Williams and Kunkel, 2014; Caldecott, 2014).
1.0 Primase
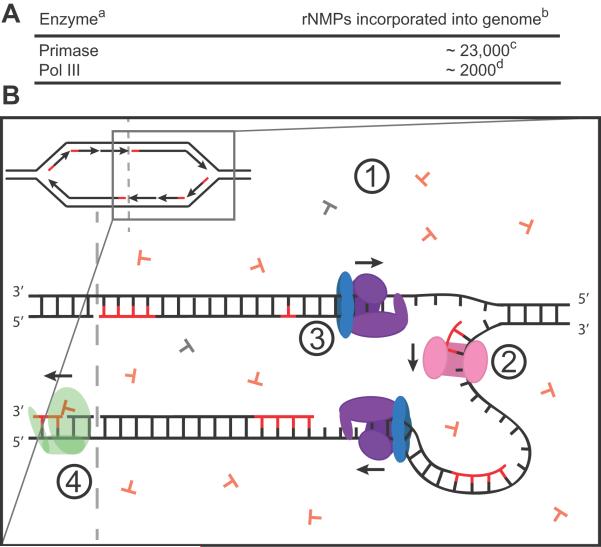
It has long been known that DNA polymerases require a primer for DNA synthesis to occur [for review (Kornberg and Baker, 1992)]. In bacteria, DnaG (primase) provides this function (Bouche et al., 1975; McFadden and Denhardt, 1974). Primase is a DNA-dependent RNA polymerase that is required to catalyze de novo synthesis of small RNA stretches serving to prime leading and lagging strand synthesis during replication (Bouche et al., 1975). The overwhelming majority of RNA in genomic DNA is due to the activity of primase (Figure 1).
Figure 1. Ribonucleotide incorporation into bacterial DNA.
A) A table displaying current information on polymerases that incorporate rNMPs into DNA with the expected number of incorporations per newly replicated strand per round of replication. a Based on E. coli proteins (Yao et al., 2013). b Estimated for the replication of a single strand of the E. coli chromosome (Rowen and Kornberg, 1978). B1) During replication of DNA, concentrations of rNTPs exceed those of dNTPs, increasing the likelihood of rNTP misincorporation by a replicative DNA polymerase (Buckstein et al., 2008a; Ferraro et al., 2010; Nick McElhinny et al., 2010a). B2) Primase (pink), a DNA dependent RNA polymerase, synthesizes RNA, priming replication by pol III (Kuchta and Stengel, 2010). B3) Pol III (purple, β-clamp blue) misincorporates rNTPs during replication of the newly synthesized strand (Yao et al., 2013). B4) Pol I (green) removes RNA primers left by primase after pol III loading (Ide et al., 1993a; Ogawa and Okazaki, 1984).
DnaG synthesizes 10–15 consecutive ribonucleotides, forming an RNA primer approximately every 1.5 kbps when coupled to the replicative helicase (Rowen and Kornberg, 1978; van der Ende et al., 1985; Corn and Berger, 2006). For a bacterial genome size of roughly 4000 kbps, this would result in lagging strand replication of 2000 kbps per new chromosome and approximately 20,000 rNMPs incorporated per chromosome strand per replication event. For comparison, approximately 150 million ribonucleotides are expected to be incorporated during Okazaki fragment synthesis of the human nuclear genome [review (Williams and Kunkel, 2014)]. Although primase is the major contributor of ribonucleotide stretches in genomic DNA, Okazaki fragment maturation results in the efficient removal of these RNA stretches in bacteria through the actions of DNA polymerase I (pol I) and RNase HI (Ogawa and Okazaki, 1984). Thus, although primase is responsible for considerable rNMP incorporation into genomic DNA, RNA primers are rapidly removed from the genome, as Okazaki fragment maturation matches pace with the replication forks in E. coli and B. subtilis, which move at 500 and 750 nt/sec, respectively (Wang et al., 2007; Breier et al., 2005; Sanders et al., 2010).
2.0 Nucleotide pools in vivo
The focus of this review is on the incorporation of ribonucleotides into the genome by DNA polymerases and the pathways responsible for their correction. An important factor affecting proper substrate selection by DNA polymerases is the cellular concentration of dNTPs relative to rNTPs. By all accounts, rNTP pools exceed dNTP pools in organisms ranging from E. coli to humans (Traut, 1994; Buckstein et al., 2008b; Nick McElhinny et al., 2010b; Ferraro et al., 2010). For E. coli, the rNTP:dNTP ratios range from 1.8-fold in the case of rUTP:dTTP to as high as 600-fold for rATP:dATP (Buckstein et al., 2008b). Likewise, in S. cerevisiae the ratios range from 36:1 to 190:1 for rCTP:dCTP and rATP:dATP, respectively (Nick McElhinny et al., 2010b). Therefore, DNA polymerases are initially challenged to select the correct dNTP among the more abundant rNTPs (Figure 1). Such a challenge invariably contributes to rNTP incorporation in place of dNTPs during genome replication. Below, we discuss the tendency of bacterial DNA polymerases to incorporate ribonucleotides in place of deoxyribonucleotides.
3.0 Ribonucleotide incorporation by DNA Polymerases
The ability of DNA polymerases to discriminate between rNTPs and dNTPs has been extensively studied in vitro [reviewed in (Brown and Suo, 2011; Joyce, 1997)]. The general feature that contributes to sugar selection in the DNA polymerase active site is “exclusion” of the 2′-OH on the ribose sugar moiety by a tyrosine, phenylalanine or glutamic acid residue side chain (Figure 2) (Kasiviswanathan and Copeland, 2011; Astatke et al., 1998; Beck et al., 2002; Bonnin et al., 1999; Brown et al., 2010; Cases-Gonzalez et al., 2000; DeLucia et al., 2003; Gao et al., 1997; Gardner and Jack, 1999; Patel and Loeb, 2000; Yang et al., 2002). The use of a steric gate residue side chain to limit rNTP utilization by DNA polymerases is common, although some DNA polymerases from the X-family use a segment of the protein backbone to reduce rNTP incorporation (Garcia-Diaz et al., 2005; Pelletier et al., 1994; Sawaya et al., 1997; Brown et al., 2010). In addition, DNA polymerase domains change from an “open” to “closed” conformation during catalysis and the dynamics of this transition can be altered in the presence of rNTPs providing another mechanism for sugar discrimination (Joyce et al., 2008). Interestingly, depending on the type of DNA polymerase and the specific rNTP:dNTP pair, sugar discrimination can range from just over 2-fold discrimination against rNTPs relative to dNTPs to over a million-fold preference for dNTPs [reviewed in (Brown and Suo, 2011; Joyce, 1997)].
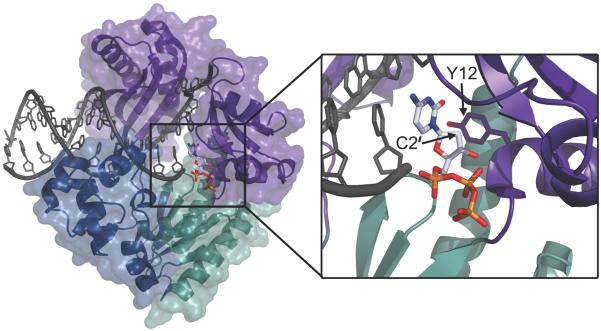
Figure 2. Steric gate residues clash with ribonucleotides.
DNA polymerase IV from Sulfolobus solfataricus (PDB 4QW8) is shown as a ribbon diagram bound to template DNA and a nucleotide (dCTP) represented as sticks (Gaur et al., 2014). The palm (green), thumb (blue) and finger (purple) subdomains have a semitransparent surface. The zoomed view demonstrates the close proximity of the deoxyribose C2′ from dCTP with the tyrosine steric gate side chain (Y12). Oxygen atoms are colored in red, nitrogen in blue, and phosphorous in orange.
Most polymerases are also limited in the number of rNTPs that can be incorporated consecutively, which prevents long stretches from occurring even when rNTPs are plentiful (Astatke et al., 1998; Zhu and Shuman, 2005b; Ruiz et al., 2003; Nick McElhinny and Ramsden, 2003). This is not related to the steric gate, but instead stems from the developing RNA/DNA hybrid adopting an A-form conformation. This demonstrates an additional mechanism by which ribonucleotide incorporation is minimized (Astatke et al., 1998). In this section, we consider and discuss the diverse range of rNTP utilization by replicative, repair and lesion bypass bacterial DNA polymerases.
3.1 Ribonucleotide incorporation by DNA Polymerase I
In bacteria, DNA polymerase I (pol I) has important roles in lagging strand replication, DNA repair and ribonucleotide excision repair (RER) (see below) [for review (Kornberg and Baker, 1992; Vaisman et al., 2014)]. The wild type enzyme can misincorporate rNTPs, particularly rCTP and rGTP, the extent of which depends on nucleotide pool ratios and the catalytic metal ion. The Klenow fragment of E. coli pol I readily incorporates rCTP and rGTP when rNTP:dNTP ratios are elevated to 1000:1 (Astatke et al., 1998; Ide et al., 1993b), while maintaining stringent selection against rUTP and rATP (Astatke et al., 1998). Kinetic analysis demonstrates that Klenow shows about a 3000-fold discrimination for dCTP relative to rCTP and nearly a million fold discrimination against rUTP. Therefore, the range of rNTP:dNTP discrimination is exceedingly broad depending on the rNTP:dNTP pair. Mutation of the steric gate (E710A) imparted substantial rNMP incorporation when rNTP:dNTP ratios were 1:1 and abolished the strong discrimination against rUTP and rATP. In fact, the steric gate variant showed very similar levels of incorporation for all four rNTPs even at rNTP:dNTP ratios of 10:1 (Astatke et al., 1998).
It has long been established that E. coli pol I is more capable of incorporating rNTPs when Mn2+ is used instead of Mg2+ during in vitro synthesis reactions. Mn2+ distorts the active site in a way that allows for increased rNTP utilization, particularly in the case of rCTP and rGTP (Tabor and Richardson, 1989; Van de Sande et al., 1972). In addition, mutation of residues in the O-helix can also elevate rNTP incorporation, though not to the same extent as variants of the steric gate (Astatke et al., 1998). One possibility is that the residue(s) on the O-helix help orient the sugar moiety on the incoming rNTP to clash with the steric gate. These examples illustrate the importance of active site geometry as well as the steric gate in sugar discrimination. The extent to which pol I-dependent ribonucleotide incorporation takes place and impacts genome stability in vivo is not yet understood. Because pol I is critical for Okazaki fragment maturation and resynthesis during DNA repair, there are ample opportunities for pol I-dependent inclusion of rNMPs into genomic DNA.
3.2 Ribonucleotide Incorporation by Replicative DNA Polymerases
In bacteria the main replicative DNA polymerase is a family C-type DnaE or PolC enzyme [for review (McHenry, 2011; Johnson and O'Donnell, 2005)], while in eukaryotes the leading strand polymerase, pol ε, and the lagging strand polymerase, pol δ, are B-family replicases (Braithwaite and Ito, 1993; Pursell et al., 2007). Studies in S. cerevisiae have demonstrated that rNMPs are incorporated into genomic DNA in vivo and in vitro by eukaryotic pols ε, δ and α (Nick McElhinny et al., 2010a; Nick McElhinny et al., 2010b; Sparks et al., 2012; Williams and Kunkel, 2014). These studies have shown that approximately 10,000 rNMP misincorporations are expected during a single round of DNA replication for S. cerevisiae (Nick McElhinny et al., 2010a; Nick McElhinny et al., 2010b). Extrapolating the rNMP incorporation rates for S. cerevisiae and mice to humans predicts approximately ~3 million rNMP errors are made per round of replication for a human cell (Nick McElhinny et al., 2010b; Reijns et al., 2012). Therefore, in eukaryotes, rNMPs embedded into genomic DNA represent the most abundant nucleotide in need of repair.
Using E. coli DNA polymerase III (pol III), as a model for C-family replicases, it was shown that a ribonucleotide incorporation event can be expected every 2,300 bps (Yao et al., 2013). Considering the rate of ribonucleotide incorporation in vitro, Yao et al. expect nearly 2,000 misincorporation events per round of replication for the E. coli genome (Yao et al., 2013). If we consider that E. coli pol III makes a dNTP base pairing error only once every ~7.9 rounds of replication, it becomes clear that sugar errors are by far the most frequent mistake made by bacterial replicative DNA polymerases (Lee et al., 2012). Of course the number of 2,000 per round of replication for E. coli pol III does not take into consideration the possible contributions of DNA pol I, II, IV or V to sugar error during genome replication.
Of the roughly 2,000 errors made by E. coli pol III approximately 1,500 of these are expected to be rAMP followed by over 300 misinsertions of rCMP, a little over 100 misinsertions of rGMP and only 6 rUMP (Yao et al., 2013). Therefore, pol III shows excellent exclusion of rUTP during replication. In vivo studies of the low GC Gram-positive bacterium Bacillus subtilis showed that rNMPs are indeed incorporated into genomic DNA (Yao et al., 2013). B. subtilis uses both DnaE and PolC for chromosomal replication and the contributions of each of these replicases to sugar error are not yet established. Given that S. cerevisiae, E. coli, mouse, B. subtilis and Chlamydophila pneumoniae have all shown a significant amount of rNMP misinsertions either in vitro, in vivo or both, it seems likely that ribonucleotide incorporation by replicative DNA polymerases is a biologically conserved event (Lu et al., 2012b; Nick McElhinny et al., 2010a; Nick McElhinny et al., 2010b; Reijns et al., 2012; Yao et al., 2013).
3.3 Ribonucleotide incorporation by Y-family DNA polymerases
Y-family DNA polymerases are DNA damage inducible and function in lesion bypass on damaged template DNA [for review (Sutton, 2009; Goodman and Woodgate, 2013; Waters et al., 2009; Jarosz et al., 2007; Sale et al., 2012)]. The activity of Y-family DNA polymerases during lesion bypass can result in dNTP mispairing and ribonucleotide incorporation (Goodman and Woodgate, 2013; Sutton, 2009; McDonald et al., 2012; Vaisman et al., 2012; Vaisman et al., 2014). The rate of ribonucleotide incorporation has been studied for a variety of Y-family polymerases and their ability to perform sugar discrimination ranges from 2.5-fold in the case of Mycobacterium smegmatis DinB2 to several thousand-fold in the case of Sulfolobus solfataricus DinB homolog (Dbh) (DeLucia et al., 2003; Ordonez et al., 2014). Therefore, although Y-family polymerases are expected to provide a limited amount of DNA synthesis during chromosomal replication in non-stressed cells, these polymerases do incorporate rNMPs and vary widely in their ability to do so.
The archeon Sulfolobus solfataricus Y-family pol Dbh discriminates 3,400-fold for dGTP relative to rGTP and shows even higher discrimination for dCTP on a frameshift substrate (DeLucia et al., 2003). Study of a steric gate variant of Dbh, DbhF12A, revealed reduced discrimination of dGTP over rGTP from 3,400-fold to only four-fold (DeLucia et al., 2003). Structural information on S. solfataricus polymerase Dpo4 demonstrated that the steric gate residue side chain (Y12) would clash with a 2′-OH group of an incoming rNTP (Figure 2). Importantly, the tyrosine side chain also stacks with the sugar ring while its backbone amide hydrogen bonds with the sugar 3′-OH. These multiple contacts reveal how mutating the steric gate (Y12A) decreases the efficiency of incorporation for both dNTPs and rNTPs while simultaneously reducing sugar discrimination (Kirouac et al., 2011). It also emphasizes the multiple roles played by the steric gate in stabilizing the incoming nucleotide during the polymerization reaction.
E. coli DNA polymerase V (UmuD′2C) in the presence of RecA is proficient at incorporating ribonucleotides while replicating undamaged DNA in vitro [for review of pol V Mut (Patel et al., 2010) and (Vaisman et al., 2012)]. Although mutation of the steric gate in other Y-family polymerases does not convert these to primer-dependent RNA polymerases mainly because extension is inefficient (DeLucia et al., 2003), a UmuCY11A steric gate variant incorporates stretches of RNA from a primer almost as efficiently as the wild type enzyme synthesizes DNA (Vaisman et al., 2012; Kuban et al., 2012). E. coli pol V can be error-prone and contributes nearly 100-fold to UV induced mutagenesis (Goodman and Woodgate, 2013; Sutton, 2009). Interestingly, the UmuCY11A variant caused over a 12-fold decrease in UV-induced mutagenesis (Vaisman et al., 2012; Kuban et al., 2012). Further genetic study of umuCY11A showed that the reduction in mutagenesis was due to its incorporation of rNMPs followed by their subsequent removal and resynthesis by DNA pol I. The propensity of UmuCY11A to incorporate rNMPs was later harnessed to identify gene products important for RER in E. coli (McDonald et al., 2012; Vaisman et al., 2013; Vaisman et al., 2014) (see below Section 4.4).
The work described above shows that pol V can naturally incorporate rNMPs at a substantial rate. Such a finding may have implications for other translesion DNA polymerases, where these pols could use rNTPs in place of dNTPs when dNTP concentrations are low. Mycobacterium smegmatis has three DinB paralogs (Ordonez et al., 2014). Recent work on Mycobacteriumsmegmatis DinB2 (pol IV) has demonstrated that this “translesion” DNA polymerase naturally incorporates rNMPs at a high rate (Ordonez et al., 2014). DinB2 has a leucine residue in place of the canonical tyrosine or phenyalanine at the steric gate position. DinB2 has < 4-fold discrimination for dNTPs relative to rNTPs. Furthermore, DinB2 can synthesize significant stretches of consecutive rNMPs (>10) demonstrating that this enzyme is a primer and template-dependent RNA polymerase. Moreover, DinB2-like enzymes with leucine or a similar amino acid side chain in place of the canonical steric gate side chain are phylogenetically conserved among many bacteria. This work strongly suggests that some Y-family polymerases may incorporate rNMPs in place of dNMPs in vivo. One possible benefit from this activity is that DinB2 could fill in gaps during repair with a stretch of ribonucleotides during stationary phase or other quiescent phases when dNTPs are particularly scarce (Ordonez et al., 2014). The resulting “ribo patch” would allow for temporary repair with rNMPs which could subsequently be removed and replaced with dNMPs through RER (Ordonez et al., 2014). The model of temporary ribo patch repair is very attractive, although it has yet to be demonstrated in vivo.
3.4 Ribonucleotide incorporation by polymerases during non-homologous end joining
Many bacteria can directly rejoin double-strand DNA breaks using Non-Homologous End Joining (NHEJ). This pathway relies on a Ku homodimer that recognizes the broken DNA ends and recruits an ATP-dependent Ligase (LigD). Most LigD homologs have a polymerase (LigD-POL) and phosphoesterase (LigD-PE) domain in addition to their ligase domain (LigD-LIG) [for review (Matthews and Simmons, 2014; Shuman and Glickman, 2007)]. Together, the multiple activities of LigD collaborate to modify and ultimately rejoin broken DNA ends.
NHEJ is critical when bacteria are not replicating and therefore lack a homologous copy of their chromosome for recombination. Consequently, this repair pathway operates during conditions where cellular dNTP levels are at their lowest. It is perhaps not surprising that all three enzymatic domains of LigD have unique properties related to ribonucleotides. The polymerase domain is related to primase and fills in 5′ overhangs at broken DNA ends (Gong et al., 2005). LigD-POL uses rNTPs more efficiently than dNTPs for synthesis. In the presence of rNTPs it also has a lower fidelity, is limited to adding only four nucleotides before further extension is inhibited, and can add a nontemplated base resulting in frameshift mutations (Zhu and Shuman, 2005b; Yakovleva and Shuman, 2006; Gong et al., 2005; Pitcher et al., 2005; Pitcher et al., 2007; Zhu and Shuman, 2006).
LigD-PE is capable of both removing 3′-PO4 groups that can persist after DNA damage as well as stretches of RNA created by LigD-POL until only a single ribonucleotide remains (Zhu and Shuman, 2005a; Zhu et al., 2005; Zhu and Shuman, 2006). The PE domain plays a minor role in NHEJ in vivo, which indicates that this activity is only important for repair at a minority of double-strand breaks (Aniukwu et al., 2008; Bhattarai et al., 2014). Even when active, the PE domain leaves at least one ribonucleotide 3′ to the break, which stimulates the activity of the LigD-LIG domain (Zhu and Shuman, 2008). Thus, the POL, LIG and PE domains work together to mediate repair of DNA breaks using ribonucleotides.
In eukaryotic cells, the error prone pol μ also modifies DNA ends prior to NHEJ. It has a glycine in place of the bulky, aromatic tyrosine normally found at the steric gate and therefore can readily use rNTPs in place of dNTPs during synthesis (Nick McElhinny and Ramsden, 2003; Martin et al., 2013). This also results in a ribonucleotide 3′ to the break, which stimulates Ligase IV to subsequently seal the synapsed DNA ends (Nick McElhinny and Ramsden, 2003). Thus, the use of ribonucleotides to fill in broken DNA ends and stimulate ligation when the dNTP pool is low is a common theme. However, it comes at the cost of sealing in a ribonucleotide at the repair junction, which must subsequently be corrected by RER. Considering LigD-POL may modify all DNA breaks in stationary Mycobacterium smegmatis cells exposed to ionizing radiation (Bhattarai et al., 2014), NHEJ may represent a considerable source of rNMP incorporation in bacteria under adverse conditions.
4.0 Removal of rNMPs from Genomic DNA
RNase H enzymes cleave RNA in an RNA/DNA hybrid. Bacterial RNase H enzymes have established roles in DNA replication, DNA repair and transcription [for review (Kogoma, 1997)]. RNases H from bacteria are categorized into type 1 and type 2 enzymes based on conservation of their primary structure [for review (Tadokoro and Kanaya, 2009)]. RNase HI is a type 1 RNase H, while type 2 enzymes include RNases HII and HIII. Bacteria usually contain either RNases HI and HII or RNases HII and HIII [for review (Ohtani et al., 1999b; Tadokoro and Kanaya, 2009)] (Figure 3A). Here we discuss a few examples of RNase H enzymes. It should be noted that prokaryotic RNase H enzymes show impressive diversity in their activity and substrate recognition. For more information please see the following reviews (Ohtani et al., 1999b; Tadokoro and Kanaya, 2009).
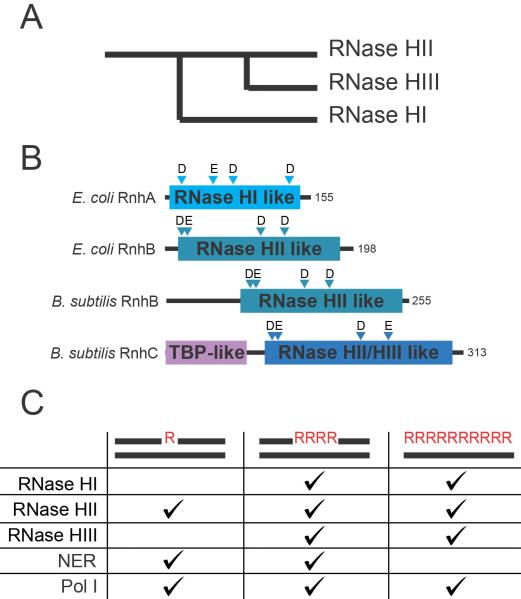
Figure 3. Bacterial recognition and removal of rNMPs.
A) Schematic evolutionary history of the family of RNase H enzymes with respect to sequence similarity (Ohtani et al., 1999b). B) Domain structures of functional E. coli and B. subtilis RNases H. Triangles denote the location and identity of the residues stabilizing the metal ions necessary for catalytic activity (Tadokoro and Kanaya, 2009). C) The ability of the RNases H, NER and pol I to recognize or engage in repair of different RNA/DNA hybrids (Tadokoro and Kanaya, 2009; Vaisman et al., 2013; Vaisman et al., 2014).
In addition to the RNase H system employed for the removal of RNA from genomic DNA, topo I removes single rNMPs in S. cerevisiae and nucleotide excision repair (NER) can remove both single rNMPs and stretches of rNMPs in E. coli (Kim et al., 2011; Vaisman et al., 2013; Cai et al., 2014). In this section, we review the processes impacting removal of rNMPs incorporated into genomic DNA.
4.1 Bacterial RNase HI
The E. coli rnhA gene encodes RNase HI, which cleaves stretches of RNA in an RNA/DNA hybrid, generating a 5′-PO4− (Miller et al., 1973; Berkower et al., 1973). RNase HI enzymes are divided into two groups: (1) the archaeal-type hybrid binding domain (HBD) HBD-RNase HI and (2) the E. coli type RNase HI (Tadokoro and Kanaya, 2009). The HBD recognizes the RNA/DNA hybrid and is important for enzyme activity (Nowotny et al., 2008; Nowotny et al., 2005) (Figure 3B–C). In bacterial systems, E. coli RNase HI is the best-understood enzyme with respect to its biochemical activity and the physiological consequences that ensue in rnhA-deficient cells (Ogawa and Okazaki, 1984; Ogawa et al., 1984; Hong and Kogoma, 1993). E. coli RNase HI cleaves stretches of RNA with a minimum of four consecutive ribonucleotides (Hogrefe et al., 1990). The major in vivo functions of RNase HI in E. coli are to remove R-loops resulting from transcription and to aid pol I in processing Okazaki fragments during lagging strand synthesis (Kogoma et al., 1993; Ogawa and Okazaki, 1984; Hong et al., 1995; Hong et al., 1996; Kogoma and von Meyenburg, 1983). In contrast, ribonucleotide incorporation errors by replicative DNA polymerases are unlikely to generate a substrate for RNase HI, although “ribo patches” incorporated by Y-family polymerases could generate a suitable substrate for RNase HI. It should be noted that several bacteria lack RNase HI and instead have RNase HIII. RNase HIII appears to provide an analogous function to RNase HI (see Section 4.3).
4.2 Bacterial RNase HII
E. coli rnhB (RNase HII) was first identified as a multicopy suppressor of the rnh− temperature-sensitive phenotype (Itaya, 1990). Enzymatically, it cleaves both single ribonucleotides and stretches of consecutive ribonucleotides (Itaya, 1990; Ohtani et al., 1999a; Ohtani et al., 2000). Therefore, RNase HII could serve as a backup or in conjunction with RNase HI in the processing of Okazaki fragments, while also providing the role of removing single rNMPs incorporated during DNA replication. Interestingly, most complete archaeal genomes show a single RNase H enzyme, RNase HII, demonstrating the strong conservation of this enzyme among prokaryotes (Ohtani et al., 2004). Deletion of the rnhB gene (RNase HII) from prokaryotes or inactivation of RNase H2 from eukaryotes shows the accumulation of ribonucleotides in vivo, as judged by alkaline sensitivity of isolated genomic DNA (Lu et al., 2012b; McDonald et al., 2012; Nick McElhinny et al., 2010b; Yao et al., 2013; Reijns et al., 2012). In bacterial systems loss of RNase HII does not impart a growth disadvantage or a strong phenotype. In B. subtilis loss of RNase HII activity increases overall spontaneous mutagenesis about 2-fold, while in E. coli loss of RNase HII does not affect mutation rate (Yao et al., 2013).
The focus of our review is on bacterial enzymes, however it should be noted that an RNase H2 defect is embryonic lethal in mice, and people with the rare autosomal disorder Aicardi-Goutieres syndrome (AGS) have neurological dysfunction linked to mutations in genes important for nucleic acid metabolism including defects in RNase H2 (Reijns and Jackson, 2014; Figiel et al., 2011; Crow et al., 2006a; Crow et al., 2006b). Therefore, the phenotype resulting from RNase H2 inactivation is more striking in mammals, which likely stems from eukaryotes relying more heavily on RNase H2 for RNA removal in vivo than prokaryotes. Furthermore, it is worth mentioning that superimposition of crystal structures of Thermotoga maritima RNase HII and Bacillus stearothermophilus RNase HIII allowed for identification of amino acid residues in the catalytic subunit of Saccharomyces cerevisiae RNase H2 responsible for each of its dual roles in hydrolysis of singly-misinserted rNMPs in DNA and digestion of R-loops. Two amino acid substitutions in its catalytic subunit yielded S. cerevisiae RNase H2 with greatly reduced efficiency for hydrolysis of covalent junctions between rNMPs and DNA, but maintained the ability to efficiently hydrolyze the RNA strand of an RNA/DNA hybrid (Chon et al., 2013). The fact that these substitutions in the S. cerevisiae protein were able to be rationally designed based on structural data from RNases HII and HIII from two distinct bacterial systems highlights the impressive conservation of these enzymes and the importance of continued study of both eukaryotic and prokaryotic ribonucleotide correction.
4.3 Bacterial RNase HIII
While the genomes of most bacteria and eukaryotes encode both RNase HI and HII genes, several bacteria, including Bacillus subtilis, B. stearothermophilus, Streptococcus pneumoniae, Chlamydophila pneumoniae, and Aquifex aeolicus, lack RNase HI and instead contain RNase HIII [for review (Kanaya, 2001)]. RNase HIII is considered a type 2 RNase H and is related to RNase HII by sequence and structural comparison (Ohtani et al., 1999a; Ohtani et al., 1999b) (Figure 3B–C). Even though RNase HIII is considered a type 2 enzyme, biochemical characterization indicates it is functionally analogous to E. coli RNase HI (Lu et al., 2012b; Ohtani et al., 1999a). RNase HIII enzymes differ from other RNases H in that they contain a TATA-Box binding Protein (TBP)-like N-terminal domain and a distinctive active site motif modification (DEDE) where a glutamic acid replaces the aspartic acid found in type 1 and other type 2 RNases H (DEDD) (Chon et al., 2006).
In vitro, RNase HIII enzymes cleave RNA/DNA substrates containing four or more rNMPs supporting the hypothesis that RNase HIII serves as a functional replacement for bacteria that lack RNase HI (Itaya et al., 1999). For a long time the function of the large N-terminal TBP-like domain, unique to RNase HIII, was unknown. Recently the function of this domain has been determined to be important for substrate binding (Jongruja et al., 2012; Miyashita et al., 2011). Truncation of the N-terminal domain decreases enzymatic efficiency in vitro and recent data in B. stearothermophilus have shown that six amino acids in this N-terminal domain when mutated individually to alanine show reduced enzymatic activity and/or reduced substrate binding (Miyashita et al., 2011). Purified Thermovibrio ammonificans RNase HIII TBP domain alone is sufficient to bind an RNA/DNA hybrid, but does not bind DNA/DNA or RNA/RNA duplex molecules (Figiel and Nowotny, 2014; Miyashita et al., 2011). The crystal structure of T. ammonificans RNase HIII bound to an RNA/DNA hybrid substrate reveals two potential “steric gate” residues in the TBP-like domain that interact with the DNA strand of the RNA/DNA substrate and would sterically clash with the 2′-OH of RNA, but this has not been experimentally tested (Figiel and Nowotny, 2014).
RNase HIII enzymes are not known to cleave at single rNMPs, with one interesting exception. Chlamydophila pneumoniae RNase HIII (Cpn-HII) can complement an E. coli rnhB knockout when cells are grown in the presence of high Mn2+ concentrations. Alternatively Cpn-HIII can also complement an E. colirnhA knockout when grown in the presence of Mg2+ (Lu et al., 2012b). Purification and subsequent in vitro characterization demonstrated that Cpn-HIII cleaves at a single rNMP in the presence of Mn2+ but not with Mg2+. Therefore Cpn-HIII is the first RNase HIII that we are aware of to be shown to cleave at stretches of four or more rNMPs in RNA-DNA/DNA hybrids and at a single rNMP nested in duplex DNA. This activity has been attributed to Ser 94, which binds the substrate for Cpn-HIII at the RNA-DNA junction, possibly shifting the conformation and allowing a G(R/K)G motif to form hydrogen bonds with the single rNMP. The G(R/K)G motif is responsible for recognizing single ribonucleotides in RNase HII enzymes (Lu et al., 2012a) and is not conserved amongst all RNases HIII. Nicking activity at single rNMPs in the presence of Mn2+ has yet to be detected in RNase HIII from any other organism (Lu et al., 2012a). Interestingly, high levels of Mn2+ also inhibit RNase HII in Chlamydophila pneumoniae, suggesting a possible adaptation to high levels of manganese (Lu et al., 2012).
4.4 Ribonucleotide Excision Repair (RER)
Ribonucleotide excision repair (RER) refers to the removal of rNMPs and their replacement with dNMPs. Any process that removes rNMPs from DNA can be referred to as RER; however, here we will focus on the RNase HII-dependent pathway because this represents the dominant process for removing rNMPs from genomic DNA. The RER pathway for S. cerevisiae has been reconstituted with purified proteins to understand the discrete steps (Sparks et al., 2012). During this process, RNase H2 nicks a single ribonucleotide generating a 5′-PO4−, the processivity clamp PCNA and Pol δ or ε catalyze strand displacement synthesis followed by removal of the RNA-containing flap by FEN1 or Exo I. The nick is then sealed by DNA ligase to complete RER (Sparks et al., 2012).
In bacteria, RER is less clear and we expect considerable redundancy in this pathway. Woodgate and co-workers have gained insight into RER in E. coli by taking advantage of the propensity of UmuCY11A to readily incorporate rNMPs (Vaisman et al., 2014). In E. coli RER, RNase HII nicks at the rNMP generating a 5′-PO4− rNMP at the nick. DNA pol I, by nick translation, can remove the RNA-containing strand while gap filling with dNMPs (Vaisman et al., 2014). The nick is then sealed by DNA ligase to finish repair. In E. coli, pol I was shown to have a critical role in RER although pol III can function in the absence of pol I (Vaisman et al., 2014). Furthermore, pol I is most-likely responsible for removing the 5′ flap although redundancy with other 5′ flap endo- and 5′ to 3′ single stranded exonucleases capable of providing the same function has been shown (Vaisman et al., 2014).
4.5 Removal of Ribonucleotides by Topo I and NER
In addition to the well-characterized role for the RNases H in removal of ribonucleotides from DNA, other pathways also participate. In yeast, topoisomerase I removes single embedded rNMPs yielding a 2–5 bp deletion, which can be mitigated by helicase Srs2 in collaboration with Exo I (Kim et al., 2011; Sekiguchi and Shuman, 1997; Williams et al., 2013; Potenski et al., 2014). Woodgate and co-workers again used the frequent rNMP incorporation by UmuCY11A variant and found that nucleotide excision repair (NER) serves as a backup pathway for removal of rNMPs in cells deficient for rnhB [(Vaisman et al., 2013) for review (Cai et al., 2014)].
Specifically, this work showed that base excision repair and mismatch repair have minimal to no effect on ribonucleotide excision repair in vivo. Their work did show that UvrABC incises RNA in duplex DNA containing a single, two, or five consecutive rNMPs in a purified system (Vaisman et al., 2013). The involvement of NER in removal of rNMPs from genomic DNA was an important step forward in identifying other pathways capable of removing ribonucleotide misincorporations. How does NER recognize single rNMPs in DNA? A recent model has been proposed where the 2′-OH in the ribose sugar forms electrostatic and hydrogen bonding interactions with tyrosine residues in the β-hairpin of UvrB resulting in “damage” recognition (Cai et al., 2014). Although this model needs to be tested biochemically, it provides a basis for understanding ribose sugar recognition by NER. It is not yet known if eukaryotic NER also recognizes and incises at rNMPs in genomic DNA. If so, it will provide yet another pathway capable of removing rNMPs from genomic DNA in eukaryotic cells.
5.0 Consequences of rNMPs in Genomic DNA
Phosphodiester bonds 3′ to rNMPs are inherently less stable than those 3′ to dNMPs due to the presence of the 2′-OH, which can act as a nucleophile to attack the 3′-PO4−, yielding 5′-OH and a cyclic 2′, 3′-PO4− that is quickly hydrolyzed to 2′- or 3′-PO4− (Oivanen et al., 1998). Furthermore, rNMPs in the template strand for DNA synthesis can stall or slow the DNA polymerase (Yao et al., 2013). Should ribonucleotides go unrepaired in DNA, this could have severe consequences for genome integrity. However, the ubiquity and great frequency of ribonucleotide incorporation by DNA polymerases in systems from bacteria to mammals suggests that organisms may have evolved ways to benefit from rNMPs embedded in DNA. Indeed, a role for ribonucleotides in DNA during NHEJ has already been discussed (Section 3.4) and will not be covered further in this section. A long-standing body of evidence also suggests that DNA replication is able to initiate from transcripts that anneal to template DNA (R-loops) in a process known as constitutive stable DNA replication (cSDR). Here, we discuss potential consequences of ribonucleotide incorporation into and hybridization with DNA, including genome instability, DNA mismatch repair and DNA replication.
5.1 Genome instability
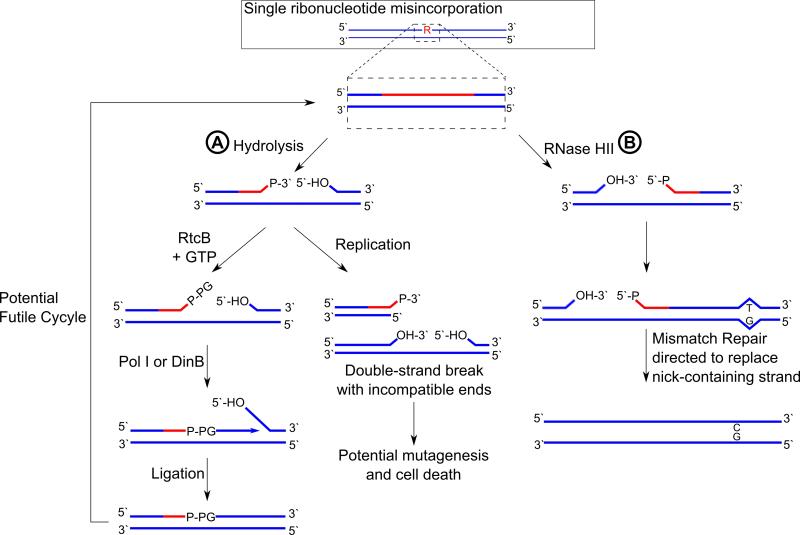
Ribonucleotides that go unrepaired in DNA can hydrolyze to yield 2′- or 3′-PO4−. Interestingly, an RNA ligase found in all three domains of life, RtcB, may play a role in repair of 3′-PO4− in DNA (Englert et al., 2012; Popow et al., 2011; Tanaka et al., 2011; Das et al., 2013). Work performed using E. coli RtcB revealed that as part of its catalytic mechanism, RtcB becomes covalently linked to GMP, which is then transferred to the 3′-PO4− to form a 3′-guanylate (DNA-3′-P-GMP) intermediate (Figure 4A). When acting on single-stranded DNA or RNA, RtcB then catalyzes attack of the 5′-OH on the DNA-3′-P-GMP such that a new phosphodiester bond is formed, with GMP as a leaving group. Completion of ligation requires that RtcB act on a single-stranded substrate, but it is able to guanylate 3′-PO4− in nicked duplex DNA (Das et al., 2013). The 3′-guanylate cap is also able to protect DNA from exonucleolytic digestion and can be used to prime DNA synthesis by Klenow fragment and Mycobacterium smegmatis DinB1, which therefore seals the rGMP into the DNA (Das et al., 2014). The in vivo relevance of this process has not been tested, nor has the effect of RtcB on a 3′-PO4− following an rNMP at a nick in duplexed DNA. Further study of the biochemical pathways processing hydrolyzed DNA 3′ to ribonucleotides will be of great interest in understanding how bacteria avoid death when rNMPs go unrepaired in genomic DNA (Figure 4A).
Figure 4. Consequences of ribonucleotides in genomic DNA.
Shown is a single ribonucleotide embedded in DNA, which can have many consequences to genome integrity. A) If the ribonucleotide remains in the genome it is prone to hydrolysis, producing a 2′- or 3′-PO4− (Oivanen et al., 1998). The resulting nick could cause a double-stranded break during the next round of DNA replication, or it may be guanylated by an RtcB-like enzyme so that it can be used to prime DNA synthesis by pol I or DinB (Das et al., 2014). This would result in the original rNMP as well as the rGMP added by RtcB to remain in the genome. Thus, such a mechanism for tolerance of rNMPs in DNA is likely to represent a futile cycle and may even serve to exacerbate problems caused by rNMPs. B) RNase HII can generate a nick 5′ to the rNMP, which can then direct DNA mismatch repair to target the rNMP-containing strand for replacement (Ghodgaonkar et al., 2013; Lujan et al., 2013).
Action of RNase HII on single rNMPs in DNA results in a 5′-rNMP (see section 4.2). RER in eukaryotic systems and bacteria has been discussed (section 4.4), but there are a number of enzymes that could act aberrantly on the 5′-PO4− at a 5′-rNMP generated by RNase HII enzymes. For instance, it was recently determined that ATP-dependent DNA ligases frequently carry out abortive ligation in vitro when presented with a 5′-rNMP at a nick in DNA (Tumbale et al., 2014). Abortive ligation is the process by which a DNA ligase begins to act on a 5′-PO4− but ultimately fails to complete ligation, resulting in a 5′-adenylated product. In eukaryotes, archaea, and viruses all DNA ligases identified are ATP-dependent (Wilkinson et al., 2001). However, all bacteria encode NAD(+)-dependent DNA ligases (Wilkinson et al., 2001), and the E. coli DNA ligase, LigA is able to complete ligation of DNA to an oligonucleotide with a 5′-rNMP end (Tumbale et al., 2014). This is an interesting finding, given that both ATP- and NAD(+)-dependent DNA ligases employ a reaction mechanism in which 5′-adenylation is an intermediate. It is an intriguing possibility that use of NAD(+)-dependent DNA ligases that are apparently able to carry out complete ligation at 5′-rNMPs in bacteria may circumvent the need for aprataxin homologs responsible for reversing 5′-adenylation in eukaryotic systems (Tumbale et al., 2014). Interestingly, aprataxin is also able to resolve a 3′-guanylate cap to a 3′-PO4− (Das et al., 2014). It is not known whether bacteria which also express ATP-dependent DNA ligases exhibit abortive ligation on 5′ rNMP ends or whether NAD(+)-dependent ligases act on 5′-rNMPs in vivo. If they do, then this action would likely be antagonistic to RER and may represent a backup pathway for dealing with 5′-rNMPs in DNA should RER fail after RNase HII nicking.
We have discussed the implications of rNMPs in DNA with respect to their increased rate of hydrolysis, but what happens during DNA replication when rNMPs are in the template strand? On average, E. coli pol III takes ~3 ms to replicate over a dNMP in template DNA in vitro (Yao et al., 2013). When pol III encounters rUMP in the template the pause is increased to ~14 ms, and with rGMP the pause is ~90 ms (Yao et al., 2013). This represents a 30-fold increase in the time required for pol III to replicate over rGMP. This large effect on DNA polymerase kinetics is not entirely surprising, given that the C3′-endo sugar pucker of an rNMP in DNA will distort the geometry of the local DNA toward the A-form (Rychlik et al., 2010; DeRose et al., 2012). In eukaryotic systems, loss of RNase H2 results in what is likely a replication-dependent DNA damage response (Allen-Soltero et al., 2014; Reijns et al., 2012). The effect of template rNMPs on DNA polymerase accuracy is not clear, but there is a role for rNMPs and RER in the accuracy of the overall process of DNA replication, specifically through their effect on DNA mismatch repair.
5.2 Mismatch Repair
We have already covered a beneficial role for rNMP incorporation into genomic DNA (Zhu and Shuman, 2008) (Section 3.4). Does rNMP incorporation have other important biological roles? In E. coli Dam methylation at d(GATC) sequences provides the basis for strand discrimination during mismatch repair (Iyer et al., 2006; Lahue et al., 1989). In most bacteria and all eukaryotic organisms the mechanism used by the mismatch repair machinery to distinguish the nascent strand from the template strand has remained unclear. Several lines of evidence demonstrate that nicks in the DNA can direct mismatch repair to the error containing strand in Streptococcus pneumoniae and several eukaryotic organisms (Holmes et al., 1990; Thomas et al., 1991; Lacks et al., 1982). A prominent model is that the DNA ends corresponding to Okazaki fragments transiently formed during discontinuous DNA synthesis provide strand discrimination signals for mismatch repair (Pavlov et al., 2003). However, in eukaryotes and most bacteria the strand discontinuities that are used by the mismatch repair pathway to target correction to the nascent leading strand have remained unclear.
Using variants of the leading strand (pol ε) and lagging strand (pol δ) polymerases in S. cerevisiae that increase rNMP incorporation it was shown that loss of RER through inactivation of RNase H2 reduced the efficiency of mismatch repair on the leading strand only (Lujan et al., 2013; Ghodgaonkar et al., 2013). Furthermore, a single rNMP and RNase H2 provide an initiation site for mismatch repair targeted to the rNMP-containing strand in vitro (Ghodgaonkar et al., 2013). In B. subtilis, loss of RNase HII increases mutation rate suggesting a possible role in mismatch repair, although the mechanism underlying the mutagenesis is unknown (Yao et al., 2013). Considering these studies, evidence is emerging that removal of rNMPs contribute to the efficiency of mismatch repair during replication by providing an entry point for mismatch repair to correct dNMP errors in the nascent leading strand.
5.3 cSDR
Constitutive stable DNA replication (cSDR) is the process by which DNA replication is able to proceed in bacteria from sites termed oriK in the absence of the normal DnaA and oriC dependent mode of replication initiation (Kogoma, 1997). Loss of RNase HI activity in E. coli leads to cSDR, which is dependent on transcription and the presence of active RecA [for review (Kogoma, 1997)]. This has led to the model that RecA mediates strand invasion of nascent transcripts such that they anneal to the template DNA strand forming an R-loop. In the absence of RNase HI, the R-loop is not degraded and the 3′-OH of the annealed transcript can serve as a primer for DNA replication. Loss of RecG, a helicase capable of displacing R-loops (Vincent et al., 1996), in E. coli also leads to cSDR, though apparently not to the extent that it will allow replication of the entire chromosome in the absence of DnaA activity (Rudolph et al., 2013; Hong et al., 1995). Loss of recG and rnhA (RNase HI) together is synthetically lethal in E. coli (Hong et al., 1995), and it is therefore hypothesized that unresolved R-loops lead to cell death. Ectopic expression of UvsW, a bacteriophage T4 helicase which is able to unwind R-loops in vitro (Dudas and Kreuzer, 2001), suppresses synthetic lethality of deletion of rnhA and recG together (Carles-Kinch et al., 1997), further supporting the notion that unresolved R-loops in E. coli result in cell death.
Mechanistic evidence that cSDR may be able to initiate from R-loops in vitro comes from the observation that E. coli pol III is able to use codirectionally transcribed mRNA at a halted RNA polymerase elongation complex as a primer for DNA synthesis (Pomerantz and O'Donnell, 2008). Interestingly, the replicative helicase does not dissociate from the DNA upon collision with a halted elongation complex, but rather, RNA polymerase dissociates from the RNA/DNA hybrid so that the mRNA may prime DNA synthesis. Use of the mRNA in this way does require a collision between the DNA polymerase and the halted elongation complex, and until the collision occurs no DNA synthesis is initiated from the mRNA molecule (Pomerantz and O'Donnell, 2008).
Marker frequency analysis originally suggested the existence of four or five discrete oriK sites on the E. coli chromosome in a strain lacking rnhA and oriC (de Massy et al., 1984). Recent work suggests that oriK sites are either more cryptic than previously thought or are randomly distributed. Two studies of genome-wide marker frequency suggest models for the origin of cSDR in E. coli lacking recG or rnhA. Lloyd and co-workers show that in the absence of recG, replication appears to initiate from within the Tus/ter trap. Deletion of tus therefore allows recG-deficient E. coli to survive in the absence of DnaA-initiated DNA replication. It was concluded that replication fork collisions in the Tus/ter trap allow replication fork restart in the absence of recG, thereby leading to cSDR (Rudolph et al., 2013). However, it is unclear by what mechanism these DnaA-independent replication forks come into being so that they may then collide at the normal site of replication termination in the absence of Tus. Kreuzer and co-workers show that E. coli lacking rnhA, tus, and dnaA display the greatest sequencing coverage nearby the site of normal replication termination, with lowest coverage in this strain near oriC. Treatment of cells lacking rnhA and dnaA with hydroxyurea (HU) increases sequencing coverage at particular locations, but it is unclear whether overinitiation of replication upon HU treatment occurs by the same mechanism as initiation at an oriK site. The authors suggest that it is likely that the strongest oriK site(s) exist near terA (Maduike et al., 2014). Another possibility is that replication is able to initiate at R-loops randomly along the chromosome, and that replication forks, which progress codirectionally with transcription and therefore toward the site of replication termination do so with fewer replication/transcription conflicts. This model suggests that in the population of chromosomes sequenced the highest coverage is achieved near terA due to the mass action of replication initiating randomly but progressing more efficiently codirectionally with transcription, culminating in a higher chromosomal copy number in that region.
cSDR has been fairly well studied in E. coli, which, as discussed above, contains RNases HI and HII. cSDR has not been studied in bacteria expressing RNases HII and HIII, and it is not known whether loss of RNase HIII in these bacteria will allow DNA replication in the absence of dnaA or oriC. Thus, further study of cSDR in a diverse range of bacteria has the potential to reveal both similarities and differences between the functions of RNases H in bacteria expressing RNases HI and HII compared to those expressing RNases HII and HIII.
6. Conclusion
Ribonucleotide incorporation into genomic DNA occurs ubiquitously in living organisms. Despite most polymerases having structural deterrents against incorporation of both single and consecutive rNMPs, both are still incorporated. This can serve many biological roles including providing primers for DNA replication, allowing DNA gaps to be filled when dNTP pools are low, and providing a means for the cell to identify the nascent leading strand during mismatch repair. However, regardless of the nature of inclusion, ribonucleotide incorporation creates a high risk for the cell. Consequently, an arsenal of enzymes is employed to continuously cull rNMPs from the genome. By further investigating these systems we can begin to reconcile how cells tolerate the highly dynamic exchange of ribonucleotides with the chemically stable genomic DNA and the far-reaching consequences of their interplay on bacterial physiology.
Acknowledgements
We have tried to focus our review on more recent advances in understanding rNMP incorporation and correction. We wish to thank all of the scientists who have contributed to our understanding of rNMP incorporation and removal from genomic DNA. We also wish to thank two anonymous reviewers for their comments on this work.
This work was funded by a grant from the NIH (GM107312) to L.A.S. We also thank the NIH Genetics Training Grant (T32 GM007544) for supporting J.W.S. and the NIH Biotechnology Training Grant (T32 GM008353) for support of J.R.R.
Footnotes
Declaration of Interest The authors declare no conflict of interest.
References
- Allen-Soltero S, Martinez SL, Putnam CD, Kolodner RD. A saccharomyces cerevisiae RNase H2 interaction network functions to suppress genome instability. Mol Cell Biol. 2014;34:1521–1534. doi: 10.1128/MCB.00960-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aniukwu J, Glickman MS, Shuman S. The pathways and outcomes of mycobacterial NHEJ depend on the structure of the broken DNA ends. Genes Dev. 2008;22:512–527. doi: 10.1101/gad.1631908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astatke M, Ng K, Grindley ND, Joyce CM. A single side chain prevents Escherichia coli DNA polymerase I (Klenow fragment) from incorporating ribonucleotides. Proc Natl Acad Sci U S A. 1998;95:3402–3407. doi: 10.1073/pnas.95.7.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck J, Vogel M, Nassal M. dNTP versus NTP discrimination by phenylalanine 451 in duck hepatitis B virus P protein indicates a common structure of the dNTP-binding pocket with other reverse transcriptases. Nucleic Acids Res. 2002;30:1679–1687. doi: 10.1093/nar/30.7.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkower I, Leis J, Hurwitz J. Isolation and characterization of an endonuclease from Escherichia coli specific for ribonucleic acid in ribonucleic acid-deoxyribonucleic acid hybrid structures. J Biol Chem. 1973;248:5914–5921. [PubMed] [Google Scholar]
- Bhattarai H, Gupta R, Glickman MS. DNA Ligase C1 mediates the LigD independent NHEJ pathway of Mycobacterium smegmatis. J Bacteriol. 2014 doi: 10.1128/JB.01832-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnin A, Lazaro JM, Blanco L, Salas M. A single tyrosine prevents insertion of ribonucleotides in the eukaryotic-type phi29 DNA polymerase. Journal of Molecular Biology. 1999;290:241–251. doi: 10.1006/jmbi.1999.2900. [DOI] [PubMed] [Google Scholar]
- Bouche JP, Zechel K, Kornberg A. dnaG gene product, a rifampicin-resistant RNA polymerase, initiates the conversion of a single-stranded coliphage DNA to its duplex replicative form. J. Biol. Chem. 1975;250:5995–6001. [PubMed] [Google Scholar]
- Braithwaite DK, Ito J. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 1993;21:787–802. doi: 10.1093/nar/21.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breier AM, Weier HU, Cozzarelli NR. Independence of replisomes in Escherichia coli chromosomal replication. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:3942–3947. doi: 10.1073/pnas.0500812102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JA, Fiala KA, Fowler JD, Sherrer SM, Newmister SA, Duym WW, Suo Z. A novel mechanism of sugar selection utilized by a human X-family DNA polymerase. J Mol Biol. 2010;395:282–290. doi: 10.1016/j.jmb.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JA, Suo Z. Unlocking the sugar “steric gate” of DNA polymerases. Biochemistry. 2011;50:1135–1142. doi: 10.1021/bi101915z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckstein MH, He J, Rubin H. Characterization of nucleotide pools as a function of physiological state in Escherichia coli. Journal of Bacteriology. 2008a;190:718–726. doi: 10.1128/JB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckstein MH, He J, Rubin H. Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J Bacteriol. 2008b;190:718–726. doi: 10.1128/JB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Geacintov NE, Broyde S. Ribonucleotides as nucleotide excision repair substrates. DNA Repair (Amst) 2014;13:55–60. doi: 10.1016/j.dnarep.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott KW. Molecular biology. Ribose--an internal threat to DNA. Science. 2014;343:260–261. doi: 10.1126/science.1248234. [DOI] [PubMed] [Google Scholar]
- Carles-Kinch K, George JW, Kreuzer KN. Bacteriophage T4 UvsW protein is a helicase involved in recombination, repair and the regulation of DNA replication origins. Embo J. 1997;16:4142–4151. doi: 10.1093/emboj/16.13.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases-Gonzalez CE, Gutierrez-Rivas M, Menendez-Arias L. Coupling ribose selection to fidelity of DNA synthesis. The role of Tyr-115 of human immunodeficiency virus type 1 reverse transcriptase. The Journal of Biological Chemistry. 2000;275:19759–19767. doi: 10.1074/jbc.M910361199. [DOI] [PubMed] [Google Scholar]
- Chon H, Matsumura H, Koga Y, Takano K, Kanaya S. Crystal structure and structure-based mutational analyses of RNase HIII from Bacillus stearothermophilus: a new type 2 RNase H with TBP-like substrate-binding domain at the N terminus. J Mol Biol. 2006;356:165–178. doi: 10.1016/j.jmb.2005.11.017. [DOI] [PubMed] [Google Scholar]
- Chon H, Sparks JL, Rychlik M, Nowotny M, Burgers PM, Crouch RJ, Cerritelli SM. RNase H2 roles in genome integrity revealed by unlinking its activities. Nucleic AcidsRes. 2013;41:3130–3143. doi: 10.1093/nar/gkt027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corn JE, Berger JM. Regulation of bacterial priming and daughter strand synthesis through helicase-primase interactions. Nucleic Acids Res. 2006;34:4082–4088. doi: 10.1093/nar/gkl363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, Black DN, van Bokhoven H, Brunner HG, Hamel BC, Corry PC, Cowan FM, Frints SG, Klepper J, Livingston JH, Lynch SA, Massey RF, Meritet JF, Michaud JL, Ponsot G, Voit T, Lebon P, Bonthron DT, Jackson AP, Barnes DE, Lindahl T. Mutations in the gene encoding the 3'-5' DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat Genet. 2006a;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C, Aicardi J, Babul-Hirji R, Baumann C, Baxter P, Bertini E, Chandler KE, Chitayat D, Cau D, Dery C, Fazzi E, Goizet C, King MD, Klepper J, Lacombe D, Lanzi G, Lyall H, Martinez-Frias ML, Mathieu M, McKeown C, Monier A, Oade Y, Quarrell OW, Rittey CD, Rogers RC, Sanchis A, Stephenson JB, Tacke U, Till M, Tolmie JL, Tomlin P, Voit T, Weschke B, Woods CG, Lebon P, Bonthron DT, Ponting CP, Jackson AP. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nature genetics. 2006b;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- Das U, Chakravarty AK, Remus BS, Shuman S. Rewriting the rules for end joining via enzymatic splicing of DNA 3'-PO4 and 5'-OH ends. Proc Natl Acad Sci U S A. 2013;110:20437–20442. doi: 10.1073/pnas.1314289110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das U, Chauleau M, Ordonez H, Shuman S. Impact of DNA3'pp5'G capping on repair reactions at DNA 3' ends. Proc Natl Acad Sci U S A. 2014;111:11317–11322. doi: 10.1073/pnas.1409203111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy B, Fayet O, Kogoma T. Multiple origin usage for DNA replication in sdrA(rnh) mutants of Escherichia coli K-12. Initiation in the absence of oriC. J Mol Biol. 1984;178:227–236. doi: 10.1016/0022-2836(84)90141-4. [DOI] [PubMed] [Google Scholar]
- DeLucia AM, Grindley ND, Joyce CM. An error-prone family Y DNA polymerase (DinB homolog from Sulfolobus solfataricus) uses a 'steric gate' residue for discrimination against ribonucleotides. Nucleic Acids Research. 2003;31:4129–4137. doi: 10.1093/nar/gkg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRose EF, Perera L, Murray MS, Kunkel TA, London RE. Solution structure of the Dickerson DNA dodecamer containing a single ribonucleotide. Biochemistry. 2012;51:2407–2416. doi: 10.1021/bi201710q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudas KC, Kreuzer KN. UvsW protein regulates bacteriophage T4 origin-dependent replication by unwinding R-loops. Mol Cell Biol. 2001;21:2706–2715. doi: 10.1128/MCB.21.8.2706-2715.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englert M, Xia S, Okada C, Nakamura A, Tanavde V, Yao M, Eom SH, Konigsberg WH, Soll D, Wang J. Structural and mechanistic insights into guanylylation of RNA-splicing ligase RtcB joining RNA between 3'-terminal phosphate and 5'-OH. Proc Natl Acad Sci U S A. 2012;109:15235–15240. doi: 10.1073/pnas.1213795109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro P, Franzolin E, Pontarin G, Reichard P, Bianchi V. Quantitation of cellular deoxynucleoside triphosphates. Nucleic Acids Res. 2010;38:e85. doi: 10.1093/nar/gkp1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figiel M, Chon H, Cerritelli SM, Cybulska M, Crouch RJ, Nowotny M. The structural and biochemical characterization of human RNase H2 complex reveals the molecular basis for substrate recognition and Aicardi-Goutieres syndrome defects. J Biol Chem. 2011;286:10540–10550. doi: 10.1074/jbc.M110.181974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figiel M, Nowotny M. Crystal structure of RNase H3-substrate complex reveals parallel evolution of RNA/DNA hybrid recognition. Nucleic Acids Res. 2014;42:9285–9294. doi: 10.1093/nar/gku615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Orlova M, Georgiadis MM, Hendrickson WA, Goff SP. Conferring RNA polymerase activity to a DNA polymerase: a single residue in reverse transcriptase controls substrate selection. Proc Natl Acad Sci U S A. 1997;94:407–411. doi: 10.1073/pnas.94.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Diaz M, Bebenek K, Krahn JM, Kunkel TA, Pedersen LC. A closed conformation for the Pol lambda catalytic cycle. Nat Struct Mol Biol. 2005;12:97–98. doi: 10.1038/nsmb876. [DOI] [PubMed] [Google Scholar]
- Gardner AF, Jack WE. Determinants of nucleotide sugar recognition in an archaeon DNA polymerase. Nucleic Acids Res. 1999;27:2545–2553. doi: 10.1093/nar/27.12.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur V, Vyas R, Fowler JD, Efthimiopoulos G, Feng JY, Suo Z. Structural and kinetic insights into binding and incorporation of L-nucleotide analogs by a Y-family DNA polymerase. Nucleic Acids Res. 2014;42:9984–9995. doi: 10.1093/nar/gku709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghodgaonkar MM, Lazzaro F, Olivera-Pimentel M, Artola-Boran M, Cejka P, Reijns MA, Jackson AP, Plevani P, Muzi-Falconi M, Jiricny J. Ribonucleotides misincorporated into DNA act as strand-discrimination signals in eukaryotic mismatch repair. Mol Cell. 2013;50:323–332. doi: 10.1016/j.molcel.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Bongiorno P, Martins A, Stephanou NC, Zhu H, Shuman S, Glickman MS. Mechanism of nonhomologous end-joining in mycobacteria: a low-fidelity repair system driven by Ku, ligase D and ligase C. Nat Struct Mol Biol. 2005;12:304–312. doi: 10.1038/nsmb915. [DOI] [PubMed] [Google Scholar]
- Goodman MF, Woodgate R. Translesion DNA polymerases. Cold Spring Harb Perspect Biol. 2013;5:a010363. doi: 10.1101/cshperspect.a010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogrefe HH, Hogrefe RI, Walder RY, Walder JA. Kinetic analysis of Escherichia coli RNase H using DNA-RNA-DNA/DNA substrates. J Biol Chem. 1990;265:5561–5566. [PubMed] [Google Scholar]
- Holmes J, Jr., Clark S, Modrich P. Strand-specific mismatch correction in nuclear extracts of human and Drosophila melanogaster cell lines. Proc Natl Acad Sci U S A. 1990;87:5837–5841. doi: 10.1073/pnas.87.15.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong X, Cadwell GW, Kogoma T. Escherichia coli RecG and RecA proteins in R-loop formation. Embo J. 1995;14:2385–2392. doi: 10.1002/j.1460-2075.1995.tb07233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong X, Cadwell GW, Kogoma T. Activation of stable DNA replication in rapidly growing Escherichia coli at the time of entry to stationary phase. Mol Microbiol. 1996;21:953–961. doi: 10.1046/j.1365-2958.1996.591419.x. [DOI] [PubMed] [Google Scholar]
- Hong X, Kogoma T. Absence of a direct role for RNase HI in initiation of DNA replication at the oriC site on the Escherichia coli chromosome. J Bacteriol. 1993;175:6731–6734. doi: 10.1128/jb.175.20.6731-6734.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide H, Okagami M, Murayama H, Kimura Y, Makino K. Synthesis and characterization of oligonucleotides containing the alpha-anomer of deoxyadenosine to study its influence on DNA replication. Biochem Mol Biol Int. 1993a;31:485–491. [PubMed] [Google Scholar]
- Ide H, Yagi R, Yamaoka T, Kimura Y. Misincorporation of ribonucleotides by DNA polymerase during in vitro DNA replication. Nucleic Acids Symp Ser. 1993b:133–134. [PubMed] [Google Scholar]
- Itaya M. Isolation and characterization of a second RNase H (RNase H II) encoded by the rnhB gene. Proc. Natl Acad. Sci. U.S.A. 1990;87:8587–8591. doi: 10.1073/pnas.87.21.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itaya M, Omori A, Kanaya S, Crouch RJ, Tanaka T, Kondo K. Isolation of RNase H genes that are essential for growth of Bacillus subtilis 168. Journal of Bacteriology. 1999;181:2118–2123. doi: 10.1128/jb.181.7.2118-2123.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- Jarosz DF, Godoy VG, Walker GC. Proficient and accurate bypass of persistent DNA lesions by DinB DNA polymerases. Cell Cycle. 2007;6:817–822. doi: 10.4161/cc.6.7.4065. [DOI] [PubMed] [Google Scholar]
- Johnson A, O'Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- Jongruja N, You DJ, Angkawidjaja C, Kanaya E, Koga Y, Kanaya S. Structure and characterization of RNase H3 from Aquifex aeolicus. FEBS J. 2012;279:2737–2753. doi: 10.1111/j.1742-4658.2012.08657.x. [DOI] [PubMed] [Google Scholar]
- Joyce CM. Choosing the right sugar: how polymerases select a nucleotide substrate. Proc Natl Acad Sci U S A. 1997;94:1619–1622. doi: 10.1073/pnas.94.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce CM, Potapova O, Delucia AM, Huang X, Basu VP, Grindley ND. Fingers-closing and other rapid conformational changes in DNA polymerase I (Klenow fragment) and their role in nucleotide selectivity. Biochemistry. 2008;47:6103–6116. doi: 10.1021/bi7021848. [DOI] [PubMed] [Google Scholar]
- Kanaya S. Prokaryotic type 2 RNases H. Methods Enzymol. 2001;341:377–394. doi: 10.1016/s0076-6879(01)41165-7. [DOI] [PubMed] [Google Scholar]
- Kasiviswanathan R, Copeland WC. Ribonucleotide discrimination and reverse transcription by the human mitochondrial DNA polymerase. J Biol Chem. 2011;286:31490–31500. doi: 10.1074/jbc.M111.252460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Huang SN, Williams JS, Li YC, Clark AB, Cho JE, Kunkel TA, Pommier Y, Jinks-Robertson S. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332:1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirouac KN, Suo Z, Ling H. Structural mechanism of ribonucleotide discrimination by a Y-family DNA polymerase. J Mol Biol. 2011;407:382–390. doi: 10.1016/j.jmb.2011.01.037. [DOI] [PubMed] [Google Scholar]
- Kogoma T. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol. Mol. Biol. Rev. 1997;61:212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogoma T, Hong X, Cadwell GW, Barnard KG, Asai T. Requirement of homologous recombination functions for viability of the Escherichia coli cell that lacks RNase HI and exonuclease V activities. Biochimie. 1993;75:89–99. doi: 10.1016/0300-9084(93)90029-r. [DOI] [PubMed] [Google Scholar]
- Kogoma T, von Meyenburg K. The origin of replication, oriC, and the dnaA protein are dispensable in stable DNA replication (sdrA) mutants of Escherichia coli K-12. Embo J. 1983;2:463–468. doi: 10.1002/j.1460-2075.1983.tb01445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A, Baker TA. DNA Replication. W. H. Freeman and Company; New York: 1992. [Google Scholar]
- Kuban W, Vaisman A, McDonald JP, Karata K, Yang W, Goodman MF, Woodgate R. Escherichia coli UmuC active site mutants: effects on translesion DNA synthesis, mutagenesis and cell survival. DNA Repair (Amst) 2012;11:726–732. doi: 10.1016/j.dnarep.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchta RD, Stengel G. Mechanism and evolution of DNA primases. Biochim Biophys Acta. 2010;1804:1180–1189. doi: 10.1016/j.bbapap.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacks SA, Dunn JJ, Greenberg B. Identification of base mismatches recognized by the heteroduplex-DNA-repair system of Streptococcus pneumoniae. Cell. 1982;31:327–336. doi: 10.1016/0092-8674(82)90126-x. [DOI] [PubMed] [Google Scholar]
- Lahue RS, Au KG, Modrich P. DNA mismatch correction in a defined system. Science. 1989;245:160–164. doi: 10.1126/science.2665076. [DOI] [PubMed] [Google Scholar]
- Lee H, Popodi E, Tang H, Foster PL. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc Natl Acad Sci U S A. 2012;109:E2774–2783. doi: 10.1073/pnas.1210309109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Hou J, Wang Y, Liu J. Involvement of Ser94 in RNase HIII from Chlamydophila pneumoniae in the recognition of a single ribonucleotide misincorporated into double-stranded DNA. Biochim Biophys Acta. 2012a;1824:859–865. doi: 10.1016/j.bbapap.2012.04.003. [DOI] [PubMed] [Google Scholar]
- Lu Z, Liang R, Liu X, Hou J, Liu J. RNase HIII from Chlamydophila pneumoniae can efficiently cleave double-stranded DNA carrying a chimeric ribonucleotide in the presence of manganese. Mol Microbiol. 2012b;83:1080–1093. doi: 10.1111/j.1365-2958.2012.07990.x. [DOI] [PubMed] [Google Scholar]
- Lujan SA, Williams JS, Clausen AR, Clark AB, Kunkel TA. Ribonucleotides are signals for mismatch repair of leading-strand replication errors. Molecular Cell. 2013;50:437–443. doi: 10.1016/j.molcel.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduike NZ, Tehranchi AK, Wang JD, Kreuzer KN. Replication of the Escherichia coli chromosome in RNase HI-deficient cells: multiple initiation regions and fork dynamics. Mol Microbiol. 2014;91:39–56. doi: 10.1111/mmi.12440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MJ, Garcia-Ortiz MV, Esteban V, Blanco L. Ribonucleotides and manganese ions improve non-homologous end joining by human Polmu. Nucleic Acids Res. 2013;41:2428–2436. doi: 10.1093/nar/gks1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews LA, Simmons LA. Bacterial Non-Homologous End Joining Requires Teamwork. J Bacteriol. 2014 doi: 10.1128/JB.02042-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JP, Vaisman A, Kuban W, Goodman MF, Woodgate R. Mechanisms employed by Escherichia coli to prevent ribonucleotide incorporation into genomic DNA by Pol V. PLoS Genet. 2012;8:e1003030. doi: 10.1371/journal.pgen.1003030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden G, Denhardt DT. Mechanism of replication of phi x174 single-stranded DNA. IX. Requirement for the Escherichia coli dnaG protein. J Virol. 1974;14:1070–1075. doi: 10.1128/jvi.14.5.1070-1075.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHenry CS. Breaking the rules: bacteria that use several DNA polymerase IIIs. EMBO Rep. 2011;12:408–414. doi: 10.1038/embor.2011.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller HI, Riggs AD, Gill GN. Ribonuclease H (hybrid) in Escherichia coli. Identification and characterization. J Biol Chem. 1973;248:2621–2624. [PubMed] [Google Scholar]
- Miyashita S, Tadokoro T, Angkawidjaja C, You DJ, Koga Y, Takano K, Kanaya S. Identification of the substrate binding site in the N-terminal TBP-like domain of RNase H3. FEBS Lett. 2011;585:2313–2317. doi: 10.1016/j.febslet.2011.05.064. [DOI] [PubMed] [Google Scholar]
- Nick McElhinny SA, Kumar D, Clark AB, Watt DL, Watts BE, Lundstrom EB, Johansson E, Chabes A, Kunkel TA. Genome instability due to ribonucleotide incorporation into DNA. Nature chemical biology. 2010a;6:774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Ramsden DA. Polymerase mu is a DNA-directed DNA/RNA polymerase. Mol Cell Biol. 2003;23:2309–2315. doi: 10.1128/MCB.23.7.2309-2315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Watts BE, Kumar D, Watt DL, Lundstrom EB, Burgers PM, Johansson E, Chabes A, Kunkel TA. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proceedings of the National Academy of Sciences of the United States of America. 2010b;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotny M, Cerritelli SM, Ghirlando R, Gaidamakov SA, Crouch RJ, Yang W. Specific recognition of RNA/DNA hybrid and enhancement of human RNase H1 activity by HBD. Embo J. 2008;27:1172–1181. doi: 10.1038/emboj.2008.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Okazaki T. Function of RNase H in DNA replication revealed by RNase H defective mutants of Escherichia coli. Mol Gen Genet. 1984;193:231–237. doi: 10.1007/BF00330673. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Pickett GG, Kogoma T, Kornberg A. RNase H confers specificity in the dnaA-dependent initiation of replication at the unique origin of the Escherichia coli chromosome in vivo and in vitro. Proc Natl Acad Sci U S A. 1984;81:1040–1044. doi: 10.1073/pnas.81.4.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani N, Haruki M, Morikawa M, Crouch RJ, Itaya M, Kanaya S. Identification of the genes encoding Mn2+-dependent RNase HII and Mg2+-dependent RNase HIII from Bacillus subtilis: classification of RNases H into three families. Biochemistry. 1999a;38:605–618. doi: 10.1021/bi982207z. [DOI] [PubMed] [Google Scholar]
- Ohtani N, Haruki M, Morikawa M, Kanaya S. Molecular diversities of RNases H. J Biosci Bioeng. 1999b;88:12–19. doi: 10.1016/s1389-1723(99)80168-6. [DOI] [PubMed] [Google Scholar]
- Ohtani N, Haruki M, Muroya A, Morikawa M, Kanaya S. Characterization of ribonuclease HII from Escherichia coli overproduced in a soluble form. Journal of biochemistry. 2000;127:895–899. doi: 10.1093/oxfordjournals.jbchem.a022684. [DOI] [PubMed] [Google Scholar]
- Ohtani N, Yanagawa H, Tomita M, Itaya M. Identification of the first archaeal Type 1 RNase H gene from Halobacterium sp. NRC-1: archaeal RNase HI can cleave an RNA-DNA junction. Biochem J. 2004;381:795–802. doi: 10.1042/BJ20040153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oivanen M, Kuusela S, Lonnberg H. Kinetics and Mechanisms for the Cleavage and Isomerization of the Phosphodiester Bonds of RNA by Bronsted Acids and Bases. Chem Rev. 1998;98:961–990. doi: 10.1021/cr960425x. [DOI] [PubMed] [Google Scholar]
- Ordonez H, Uson ML, Shuman S. Characterization of three mycobacterial DinB (DNA polymerase IV) paralogs highlights DinB2 as naturally adept at ribonucleotide incorporation. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M, Jiang Q, Woodgate R, Cox MM, Goodman MF. A new model for SOS-induced mutagenesis: how RecA protein activates DNA polymerase V. Crit Rev Biochem Mol Biol. 2010;45:171–184. doi: 10.3109/10409238.2010.480968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel PH, Loeb LA. Multiple amino acid substitutions allow DNA polymerases to synthesize RNA. J Biol Chem. 2000;275:40266–40272. doi: 10.1074/jbc.M005757200. [DOI] [PubMed] [Google Scholar]
- Pavlov YI, Mian IM, Kunkel TA. Evidence for preferential mismatch repair of lagging strand DNA replication errors in yeast. Curr Biol. 2003;13:744–748. doi: 10.1016/s0960-9822(03)00284-7. [DOI] [PubMed] [Google Scholar]
- Pelletier H, Sawaya MR, Kumar A, Wilson SH, Kraut J. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- Pitcher RS, Brissett NC, Picher AJ, Andrade P, Juarez R, Thompson D, Fox GC, Blanco L, Doherty AJ. Structure and function of a mycobacterial NHEJ DNA repair polymerase. J Mol Biol. 2007;366:391–405. doi: 10.1016/j.jmb.2006.10.046. [DOI] [PubMed] [Google Scholar]
- Pitcher RS, Tonkin LM, Green AJ, Doherty AJ. Domain structure of a NHEJ DNA repair ligase from Mycobacterium tuberculosis. J Mol Biol. 2005;351:531–544. doi: 10.1016/j.jmb.2005.06.038. [DOI] [PubMed] [Google Scholar]
- Pizzi S, Sertic S, Orcesi S, Cereda C, Bianchi M, Jackson AP, Lazzaro F, Plevani P, Muzi-Falconi M. Reduction of hRNase H2 activity in Aicardi-Goutieres syndrome cells leads to replication stress and genome instability. Hum Mol Genet. 2014 doi: 10.1093/hmg/ddu485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz RT, O'Donnell M. The replisome uses mRNA as a primer after colliding with RNA polymerase. Nature. 2008;456:762–766. doi: 10.1038/nature07527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popow J, Englert M, Weitzer S, Schleiffer A, Mierzwa B, Mechtler K, Trowitzsch S, Will CL, Luhrmann R, Soll D, Martinez J. HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science. 2011;331:760–764. doi: 10.1126/science.1197847. [DOI] [PubMed] [Google Scholar]
- Potenski CJ, Niu H, Sung P, Klein HL. Avoidance of ribonucleotide-induced mutations by RNase H2 and Srs2-Exo1 mechanisms. Nature. 2014;511:251–254. doi: 10.1038/nature13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns MA, Jackson AP. Ribonuclease H2 in health and disease. Biochem Soc Trans. 2014;42:717–725. doi: 10.1042/BST20140079. [DOI] [PubMed] [Google Scholar]
- Reijns MA, Rabe B, Rigby RE, Mill P, Astell KR, Lettice LA, Boyle S, Leitch A, Keighren M, Kilanowski F, Devenney PS, Sexton D, Grimes G, Holt IJ, Hill RE, Taylor MS, Lawson KA, Dorin JR, Jackson AP. Enzymatic removal of ribonucleotides from DNA is essential for Mammalian genome integrity and development. Cell. 2012;149:1008–1022. doi: 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowen L, Kornberg A. Primase, the dnaG protein of Escherichia coli. An enzyme which starts DNA chains. J Biol Chem. 1978;253:758–764. [PubMed] [Google Scholar]
- Rudolph CJ, Upton AL, Stockum A, Nieduszynski CA, Lloyd RG. Avoiding chromosome pathology when replication forks collide. Nature. 2013;500:608–611. doi: 10.1038/nature12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz JF, Juarez R, Garcia-Diaz M, Terrados G, Picher AJ, Gonzalez-Barrera S, Fernandez de Henestrosa AR, Blanco L. Lack of sugar discrimination by human Pol mu requires a single glycine residue. Nucleic Acids Res. 2003;31:4441–4449. doi: 10.1093/nar/gkg637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rychlik MP, Chon H, Cerritelli SM, Klimek P, Crouch RJ, Nowotny M. Crystal structures of RNase H2 in complex with nucleic acid reveal the mechanism of RNA-DNA junction recognition and cleavage. Mol Cell. 2010;40:658–670. doi: 10.1016/j.molcel.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol. 2012;13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders GM, Dallmann HG, McHenry CS. Reconstitution of the B. subtilis replisome with 13 proteins including two distinct replicases. Mol Cell. 2010;37:273–281. doi: 10.1016/j.molcel.2009.12.025. [DOI] [PubMed] [Google Scholar]
- Sawaya MR, Prasad R, Wilson SH, Kraut J, Pelletier H. Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry. 1997;36:11205–11215. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- Sekiguchi J, Shuman S. Site-specific ribonuclease activity of eukaryotic DNA topoisomerase I. Mol Cell. 1997;1:89–97. doi: 10.1016/s1097-2765(00)80010-6. [DOI] [PubMed] [Google Scholar]
- Shuman S, Glickman MS. Bacterial DNA repair by non-homologous end joining. Nat Rev Microbiol. 2007;5:852–861. doi: 10.1038/nrmicro1768. [DOI] [PubMed] [Google Scholar]
- Sparks JL, Chon H, Cerritelli SM, Kunkel TA, Johansson E, Crouch RJ, Burgers PM. RNase H2-Initiated Ribonucleotide Excision Repair. Molecular Cell. 2012 doi: 10.1016/j.molcel.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MD. Coordinating DNA polymerase traffic during high and low fidelity synthesis. Biochim Biophys Acta. 2009;1804:1167–1179. doi: 10.1016/j.bbapap.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S, Richardson CC. Effect of manganese ions on the incorporation of dideoxynucleotides by bacteriophage T7 DNA polymerase and Escherichia coli DNA polymerase I. Proc Natl Acad Sci U S A. 1989;86:4076–4080. doi: 10.1073/pnas.86.11.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro T, Kanaya S. Ribonuclease H: molecular diversities, substrate binding domains, and catalytic mechanism of the prokaryotic enzymes. FEBS J. 2009;276:1482–1493. doi: 10.1111/j.1742-4658.2009.06907.x. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Meineke B, Shuman S. RtcB, a novel RNA ligase, can catalyze tRNA splicing and HAC1 mRNA splicing in vivo. J Biol Chem. 2011;286:30253–30257. doi: 10.1074/jbc.C111.274597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DC, Roberts JD, Kunkel TA. Heteroduplex repair in extracts of human HeLa cells. J Biol Chem. 1991;266:3744–3751. [PubMed] [Google Scholar]
- Traut TW. Physiological concentrations of purines and pyrimidines. Molecular and cellular biochemistry. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- Tumbale P, Williams JS, Schellenberg MJ, Kunkel TA, Williams RS. Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity. Nature. 2014;506:111–115. doi: 10.1038/nature12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisman A, Kuban W, McDonald JP, Karata K, Yang W, Goodman MF, Woodgate R. Critical amino acids in Escherichia coli UmuC responsible for sugar discrimination and base-substitution fidelity. Nucleic Acids Res. 2012;40:6144–6157. doi: 10.1093/nar/gks233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisman A, McDonald JP, Huston D, Kuban W, Liu L, Van Houten B, Woodgate R. Removal of misincorporated ribonucleotides from prokaryotic genomes: an unexpected role for nucleotide excision repair. PLoS Genet. 2013;9:e1003878. doi: 10.1371/journal.pgen.1003878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisman A, McDonald JP, Noll S, Huston D, Loeb G, Goodman MF, Woodgate R. Investigating the mechanisms of ribonucleotide excision repair in Escherichia coli. Mutat Res Fundam Mol Mech Mutagen. 2014;761:21–33. doi: 10.1016/j.mrfmmm.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Sande JH, Loewen PC, Khorana HG. Studies on polynucleotides. 118. A further study of ribonucleotide incorporation into deoxyribonucleic acid chains by deoxyribonucleic acid polymerase I of Escherichia coli. J Biol Chem. 1972;247:6140–6148. [PubMed] [Google Scholar]
- van der Ende A, Baker TA, Ogawa T, Kornberg A. Initiation of enzymatic replication at the origin of the Escherichia coli chromosome: primase as the sole priming enzyme. Proc. Natl. Acad. Sci. U.S.A. 1985;82:3954–3958. doi: 10.1073/pnas.82.12.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent SD, Mahdi AA, Lloyd RG. The RecG branch migration protein of Escherichia coli dissociates R-loops. J Mol Biol. 1996;264:713–721. doi: 10.1006/jmbi.1996.0671. [DOI] [PubMed] [Google Scholar]
- Wang JD, Sanders GM, Grossman AD. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell. 2007;128:865–875. doi: 10.1016/j.cell.2006.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Minesinger BK, Wiltrout ME, D'Souza S, Woodruff RV, Walker GC. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev. 2009;73:134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson A, Day J, Bowater R. Bacterial DNA ligases. Mol Microbiol. 2001;40:1241–1248. doi: 10.1046/j.1365-2958.2001.02479.x. [DOI] [PubMed] [Google Scholar]
- Williams JS, Kunkel TA. Ribonucleotides in DNA: origins, repair and consequences. DNA Repair (Amst) 2014;19:27–37. doi: 10.1016/j.dnarep.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Smith DJ, Marjavaara L, Lujan SA, Chabes A, Kunkel TA. Topoisomerase 1-mediated removal of ribonucleotides from nascent leading-strand DNA. Molecular Cell. 2013;49:1010–1015. doi: 10.1016/j.molcel.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovleva L, Shuman S. Nucleotide misincorporation, 3'-mismatch extension, and responses to abasic sites and DNA adducts by the polymerase component of bacterial DNA ligase D. J Biol Chem. 2006;281:25026–25040. doi: 10.1074/jbc.M603302200. [DOI] [PubMed] [Google Scholar]
- Yang G, Franklin M, Li J, Lin TC, Konigsberg W. A conserved Tyr residue is required for sugar selectivity in a Pol alpha DNA polymerase. Biochemistry. 2002;41:10256–10261. doi: 10.1021/bi0202171. [DOI] [PubMed] [Google Scholar]
- Yao NY, Schroeder JW, Yurieva O, Simmons LA, O'Donnell ME. Cost of rNTP/dNTP pool imbalance at the replication fork. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:12942–12947. doi: 10.1073/pnas.1309506110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Shuman S. Novel 3'-ribonuclease and 3'-phosphatase activities of the bacterial non-homologous end-joining protein, DNA ligase D. J Biol Chem. 2005a;280:25973–25981. doi: 10.1074/jbc.M504002200. [DOI] [PubMed] [Google Scholar]
- Zhu H, Shuman S. A primer-dependent polymerase function of pseudomonas aeruginosa ATP-dependent DNA ligase (LigD) J Biol Chem. 2005b;280:418–427. doi: 10.1074/jbc.M410110200. [DOI] [PubMed] [Google Scholar]
- Zhu H, Shuman S. Substrate specificity and structure-function analysis of the 3'-phosphoesterase component of the bacterial NHEJ protein, DNA ligase D. J Biol Chem. 2006;281:13873–13881. doi: 10.1074/jbc.M600055200. [DOI] [PubMed] [Google Scholar]
- Zhu H, Shuman S. Bacterial nonhomologous end joining ligases preferentially seal breaks with a 3'-OH monoribonucleotide. J Biol Chem. 2008;283:8331–8339. doi: 10.1074/jbc.M705476200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Wang LK, Shuman S. Essential constituents of the 3'-phosphoesterase domain of bacterial DNA ligase D, a nonhomologous end-joining enzyme. J Biol Chem. 2005;280:33707–33715. doi: 10.1074/jbc.M506838200. [DOI] [PubMed] [Google Scholar]