Abstract
Six cfr-harboring methicillin-resistant Staphylococcus aureus (MRSA) isolates, which belonged to the same clone of sequence type 5 (ST5)-staphylococcal cassette chromosome mec element II (SCCmec II)-spa t311, were investigated in this study. Complete sequencing of a cfr-carrying plasmid, pLRSA417, revealed an 8,487-bp fragment containing a Tn4001-like transposon, cfr, orf1, and ISEnfa4. This segment, first identified in an animal plasmid, pSS-01, was observed in several plasmids from clinical coagulase-negative staphylococci in China, suggesting that the cfr gene, which might originate from livestock, was located in the same mobile element and disseminated among different clinical staphylococcal species.
TEXT
Methicillin-resistant Staphylococcus aureus (MRSA), an important pathogen, is resistant not only to β-lactams but usually also to several other antibiotics. Linezolid is an important alternative for the treatment of infections with MRSA. Alterations in domain V of the 23S rRNA gene, most frequently the G2576T mutation, are the main mechanism contributing to linezolid resistance among staphylococcal isolates (1). In addition, mutations in ribosomal proteins L3 and L4 have been associated with decreased susceptibility to linezolid (2, 3). Staphylococci can also exhibit linezolid resistance by acquisition of the cfr gene, which was originally identified in a bovine Staphylococcus sciuri isolate in 2000 (4) and was subsequently detected in a clinical MRSA isolate in 2005 (5). So far, cfr-carrying staphylococci have spread worldwide, even causing several outbreaks (1).
In China, the cfr gene has been extensively detected, and its genetic environment has been well characterized in both Gram-positive and Gram-negative bacteria of animal origin (6). However, linezolid resistance had not been described in clinical staphylococci in China until the emergence of 17 isolates of methicillin-resistant coagulase-negative staphylococci (MRCoNS) in our hospital in 2011 (7). Soon, linezolid resistance in human clinical CoNS seems to have become an increasing problem in China. Several linezolid-resistant clinical isolates of Staphylococcus capitis (n = 9), Staphylococcus cohnii (n = 6), Staphylococcus haemolyticus (n = 1), Staphylococcus epidermidis (n = 1), and Staphylococcus hominis (n = 1) from Shenyang (n = 3), Beijing (n = 3), Hangzhou (n = 10), Rui'an (n = 1), and Nanjing (n = 1) have been reported (8–11). To date, there has been no report of linezolid-resistant MRSA (LRSA) of human origin in China. In the current study, the molecular epidemiology of six cfr-harboring MRSA isolates and the genetic environment of the cfr gene were investigated.
The 2nd Affiliated Hospital of Zhejiang University is a 2,000-bed comprehensive tertiary care hospital in Hangzhou, China. Six LRSA isolates were obtained from sputum samples from six patients in the neurology intensive care unit (NICU) between April and July 2013. All patients were suffering from cerebral hemorrhage accompanied by pulmonary infection. Other diagnostic samples from six patients with LRSA, including those from blood, cerebrospinal fluid, and feces, were negative for LRSA. No patient received linezolid therapy, except patient 6 was treated for 17 days during the prior month (Table 1). Patients with this organism were placed in isolation rooms under strict contact precautions. No new LRSA isolates have been identified since August 2013.
TABLE 1.
Clinical characteristics of patients with LRSA
| Patient no. | Isolate | Collection date (mo/yr) | Sex | Age (yr) | Days of LRSA isolation/no. of days in ICU | Antibiotic therapy within 2 wk prior to LRSA isolationa | Outcome |
|---|---|---|---|---|---|---|---|
| 1 | LRSA417 | 4/2013 | Male | 66 | 16/50 | TZP | Survived |
| 2 | LRSA422 | 4/2013 | Male | 65 | 3/13 | None | Died |
| 3 | LRSA531 | 5/2013 | Male | 63 | 9/26 | CMN, TZP | Survived |
| 4 | LRSA608 | 6/2013 | Female | 61 | 22/28 | MOX, MEM, TEC | Survived |
| 5 | LRSA621 | 6/2013 | Male | 40 | 15/22 | MEM, TZP, AK | Survived |
| 6 | LRSA726 | 7/2013 | Male | 45 | 33/58 | AK, MEM, PB | Died |
TZP, piperacillin-tazobactam; CMN, cefminox; MOX, moxifloxacin; MEM, meropenem; TEC, teicoplanin; AK, amikacin; PB, polymyxin B.
The MICs were determined by Etest. The cfr gene and 23S rRNA mutations were examined by PCR and sequence analysis (5, 12). Molecular typing was performed by pulsed-field gel electrophoresis (PFGE) (13), multilocus sequence typing (MLST) (14), staphylococcal cassette chromosome mec element (SCCmec) typing (15), spa typing (16), and Panton-Valentine leukocidin (pvl) gene detection (17). The location of the cfr gene in six MRSA isolates was determined by S1-nuclease PFGE and Southern blot hybridization (18). Plasmid DNA was sequenced using the Illumina HiSeq 2000 platform, and reads were assembled using the CLC Workbench program (version 5.5; CLC bio, Aarhus, Denmark). The gaps between contigs were closed by primer walking. The putative open reading frames (ORFs) were identified using the FramePlot 4.0beta program. The Vector NTI program (Invitrogen, CA) was used for annotation of the DNA sequence. Twenty-six pairs of PCR primers (see Table S1 in the supplemental material) were designed based on the whole assembled sequence and were used for analysis of other cfr-carrying plasmids in this study.
Six S. aureus isolates showed similar susceptibility profiles, with a linezolid MIC of 8 μg/ml. All isolates were resistant to oxacillin, cefoxitin, chloramphenicol, clindamycin, ciprofloxacin, gentamicin, erythromycin, and tetracycline but were susceptible to vancomycin, teicoplanin, rifampin, tigecycline, and trimethoprim-sulfamethoxazole. All LRSA isolates were positive for the cfr gene, and no 23S rRNA mutations were detected. All isolates with indistinguishable PFGE band patterns belonged to the same clone of sequence type 5 (ST5)-SCCmec II-spa t311 and were negative for the pvl gene.
The cfr gene of six LRSA isolates was located on a plasmid (see Fig. S1 in the supplemental material). One cfr-carrying plasmid, pLRSA417, was sequenced, and a circular closed sequence of 39,504 bp was obtained (GenBank accession no. KJ922127). pLRSA417 consisted of 40 putative genes for deduced proteins of ≥50 amino acids (Fig. 1). PCR mapping demonstrated that another five LRSA isolates and five representative MRCoNS, which were isolated from our hospital (7), contained the same cfr-carrying plasmid.
FIG 1.
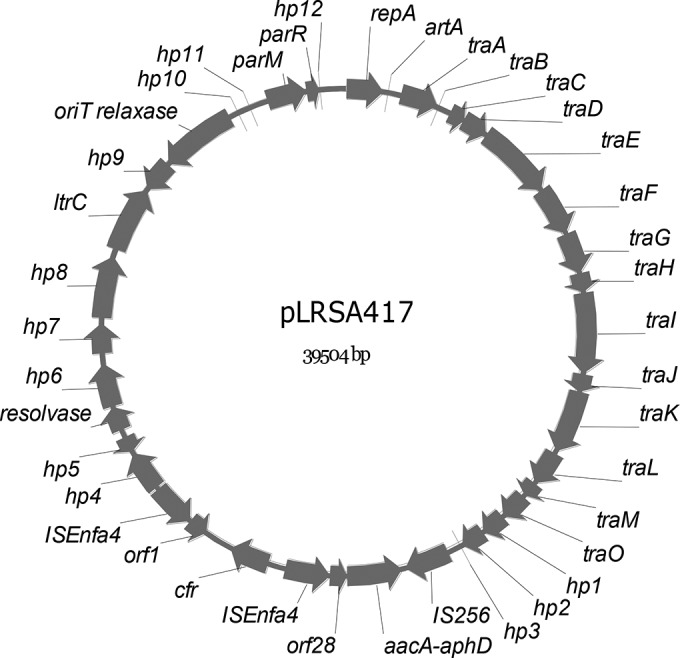
Genetic map of pLRSA417. Coding regions of >50 amino acids are represented by arrows indicating the direction of transcription, and the corresponding genes are annotated. hp1 to hp12 represent putative genes encoding hypothetical proteins.
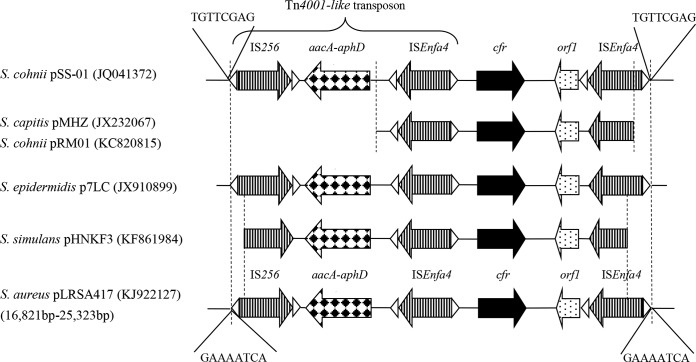
An 8,487-bp fragment containing a Tn4001-like transposon, cfr, orf1, and ISEnfa4 was flanked by two 8-bp target site duplications (TSDs), the signature of a transposition event. This DNA fragment, flanked by different TSDs, showed a sequence identical to that of pSS-01 from S. cohnii of swine origin in China (18) except for three single-nucleotide polymorphisms and two deletions. A similar genetic environment surrounding the cfr gene can be observed in several plasmids from clinical CoNS, including pMHZ in S. capitis from Hangzhou (9), pRM01 and pRA01 in S. cohnii from Beijing and Rui'an (10), p7LC in S. epidermidis from the United States (19), and pHNKF3 from a pig in Guangzhou (Fig. 2). It seemed that the genetic structures of the Tn4001-like transposon, cfr, and ISEnfa4 play an important role in the mobility of cfr in different staphylococcal plasmids of different origins. In addition to ISEnfa4, another insertion sequence, IS21-558, was found in staphylococci from both humans and swine in China (18). These observations further proved that the genetic environment of the cfr gene in clinical staphylococci in China was closely related to that in livestock isolates, and the transmission of cfr-carrying fragments and/or plasmids between staphylococci from animals and humans appears to be likely.
FIG 2.
Schematic representation and comparison of the cfr genetic environment. Linear genetic maps of plasmids are presented, with the accession numbers given in parentheses. Genes and their corresponding transcription orientations are indicated by arrows with various shading patterns. The region within the two dotted lines between plasmid maps illustrates that they share high homology (>95% nucleotide identity). The structure of a Tn4001-like transposon is indicated. The triangles represent the inverted repeats of the respective mobile elements, and target site duplications are presented as 8-bp sequences.
Two outbreaks of cfr-carrying MRSA and S. epidermidis in Madrid, Spain, and Ohio were described in 2010 (20–22). Both outbreaks were associated with nosocomial transmission and prior linezolid exposure. In the current study, however, linezolid therapy seemed not to contribute to the emergence of resistant strains, according to records regarding previous antimicrobial therapy. Notably, 16 MRCoNS with the same cfr-carrying plasmid isolated from ICUs were identified in the previous 3 years. CoNS, which are usually ignored, might become the reservoir of the cfr gene and result in the dissemination of the resistance gene.
In conclusion, this is the first report of clonal spread of cfr-harboring MRSA in China. The same cfr-carrying plasmid disseminated among multiple staphylococcal species in ICUs of our hospital. The cfr gene, carried by a similar mobile genetic organization, appears to easily transfer horizontally among bacteria of both human and animal origins in China.
Supplementary Material
ACKNOWLEDGMENTS
We declare no conflicts of interest.
This project was supported by the Zhejiang Provincial Natural Science Foundation of China (grant LQ13H200001).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.04580-14.
REFERENCES
- 1.Gu B, Kelesidis T, Tsiodras S, Hindler J, Humphries RM. 2013. The emerging problem of linezolid-resistant Staphylococcus. J Antimicrob Chemother 68:4–11. doi: 10.1093/jac/dks354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Locke JB, Hilgers M, Shaw KJ. 2009. Mutations in ribosomal protein L3 are associated with oxazolidinone resistance in staphylococci of clinical origin. Antimicrob Agents Chemother 53:5275–5278. doi: 10.1128/AAC.01032-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mendes RE, Deshpande LM, Farrell DJ, Spanu T, Fadda G, Jones RN. 2010. Assessment of linezolid resistance mechanisms among Staphylococcus epidermidis causing bacteraemia in Rome, Italy. J Antimicrob Chemother 65:2329–2335. doi: 10.1093/jac/dkq331. [DOI] [PubMed] [Google Scholar]
- 4.Schwarz S, Werckenthin C, Kehrenberg C. 2000. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob Agents Chemother 44:2530–2533. doi: 10.1128/AAC.44.9.2530-2533.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toh SM, Xiong L, Arias CA, Villegas MV, Lolans K, Quinn J, Mankin AS. 2007. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol Microbiol 64:1506–1514. doi: 10.1111/j.1365-2958.2007.05744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen J, Wang Y, Schwarz S. 2013. Presence and dissemination of the multiresistance gene cfr in Gram-positive and Gram-negative bacteria. J Antimicrob Chemother 68:1697–1706. doi: 10.1093/jac/dkt092. [DOI] [PubMed] [Google Scholar]
- 7.Cai JC, Hu YY, Zhang R, Zhou HW, Chen GX. 2012. Linezolid-resistant clinical isolates of meticillin-resistant coagulase-negative staphylococci and Enterococcus faecium from China. J Med Microbiol 61:1568–1573. doi: 10.1099/jmm.0.043729-0. [DOI] [PubMed] [Google Scholar]
- 8.Cui L, Wang Y, Li Y, He T, Schwarz S, Ding Y, Shen J, Lv Y. 2013. cfr-mediated linezolid-resistance among methicillin-resistant coagulase-negative staphylococci from infections of humans. PLoS One 8:e57096. doi: 10.1371/journal.pone.0057096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang XJ, Chen Y, Yang Q, Qu TT, Liu LL, Wang HP, Yu YS. 2013. Emergence of cfr-harbouring coagulase-negative staphylococci among patients receiving linezolid therapy in two hospitals in China. J Med Microbiol 62:845–850. doi: 10.1099/jmm.0.051003-0. [DOI] [PubMed] [Google Scholar]
- 10.Chen H, Wu W, Ni M, Liu Y, Zhang J, Xia F, He W, Wang Q, Wang Z, Cao B, Wang H. 2013. Linezolid-resistant clinical isolates of enterococci and Staphylococcus cohnii from a multicentre study in China: molecular epidemiology and resistance mechanisms. Int J Antimicrob Agents 42:317–321. doi: 10.1016/j.ijantimicag.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 11.Huang Y, Xu Y, Liu G, Mei Y, Xia W, Xu T, Gu B, Pan S. 2014. Emergence of linezolid resistance in a clinical Staphylococcus capitis isolate from Jiangsu Province of China in 2012. J Thorac Dis 6:E48–E53. doi: 10.3978/j.issn.2072-1439.2014.02.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kehrenberg C, Schwarz S. 2006. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob Agents Chemother 50:1156–1163. doi: 10.1128/AAC.50.4.1156-1163.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ip M, Lyon DJ, Chio F, Enright MC, Cheng AF. 2003. Characterization of isolates of methicillin-resistant Staphylococcus aureus from Hong Kong by phage typing, pulsed-field gel electrophoresis, and fluorescent amplified-fragment length polymorphism analysis. J Clin Microbiol 41:4980–4985. doi: 10.1128/JCM.41.11.4980-4985.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 38:1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oliveira DC, de Lencastre H. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 46:2155–2161. doi: 10.1128/AAC.46.7.2155-2161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harmsen D, Claus H, Witte W, Rothganger J, Claus H, Turnwald D, Vogel U. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol 41:5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lina G, Piemont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, Vandenesch F, Etienne J. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis 29:1128–1132. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Zhang W, Wang J, Wu C, Shen Z, Fu X, Yan Y, Zhang Q, Schwarz S, Shen J. 2012. Distribution of the multidrug resistance gene cfr in Staphylococcus species isolates from swine farms in China. Antimicrob Agents Chemother 56:1485–1490. doi: 10.1128/AAC.05827-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LaMarre J, Mendes RE, Szal T, Schwarz S, Jones RN, Mankin AS. 2013. The genetic environment of the cfr gene and the presence of other mechanisms account for the very high linezolid resistance of Staphylococcus epidermidis isolate 426-3147L. Antimicrob Agents Chemother 57:1173–1179. doi: 10.1128/AAC.02047-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morales G, Picazo JJ, Baos E, Candel FJ, Arribi A, Pelaez B, Andrade R, de la Torre MA, Fereres J, Sanchez-Garcia M. 2010. Resistance to linezolid is mediated by the cfr gene in the first report of an outbreak of linezolid-resistant Staphylococcus aureus. Clin Infect Dis 50:821–825. doi: 10.1086/650574. [DOI] [PubMed] [Google Scholar]
- 21.Sánchez García M, De la Torre MA, Morales G, Peláez B, Tolón MJ, Domingo S, Candel FJ, Andrade R, Arribi A, García N, Martínez Sagasti F, Fereres J, Picazo J. 2010. Clinical outbreak of linezolid-resistant Staphylococcus aureus in an intensive care unit. JAMA 303:2260–2264. doi: 10.1001/jama.2010.757. [DOI] [PubMed] [Google Scholar]
- 22.Bonilla H, Huband MD, Seidel J, Schmidt H, Lescoe M, McCurdy SP, Lemmon MM, Brennan LA, Tait-Kamradt A, Puzniak L, Quinn JP. 2010. Multicity outbreak of linezolid-resistant Staphylococcus epidermidis associated with clonal spread of a cfr-containing strain. Clin Infect Dis 51:796–800. doi: 10.1086/656281. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.