ABSTRACT
Transposon mutagenesis with next-generation sequencing (TnSeq) is a powerful approach to annotate gene function in bacteria, but existing protocols for TnSeq require laborious preparation of every sample before sequencing. Thus, the existing protocols are not amenable to the throughput necessary to identify phenotypes and functions for the majority of genes in diverse bacteria. Here, we present a method, random bar code transposon-site sequencing (RB-TnSeq), which increases the throughput of mutant fitness profiling by incorporating random DNA bar codes into Tn5 and mariner transposons and by using bar code sequencing (BarSeq) to assay mutant fitness. RB-TnSeq can be used with any transposon, and TnSeq is performed once per organism instead of once per sample. Each BarSeq assay requires only a simple PCR, and 48 to 96 samples can be sequenced on one lane of an Illumina HiSeq system. We demonstrate the reproducibility and biological significance of RB-TnSeq with Escherichia coli, Phaeobacter inhibens, Pseudomonas stutzeri, Shewanella amazonensis, and Shewanella oneidensis. To demonstrate the increased throughput of RB-TnSeq, we performed 387 successful genome-wide mutant fitness assays representing 130 different bacterium-carbon source combinations and identified 5,196 genes with significant phenotypes across the five bacteria. In P. inhibens, we used our mutant fitness data to identify genes important for the utilization of diverse carbon substrates, including a putative d-mannose isomerase that is required for mannitol catabolism. RB-TnSeq will enable the cost-effective functional annotation of diverse bacteria using mutant fitness profiling.
IMPORTANCE
A large challenge in microbiology is the functional assessment of the millions of uncharacterized genes identified by genome sequencing. Transposon mutagenesis coupled to next-generation sequencing (TnSeq) is a powerful approach to assign phenotypes and functions to genes. However, the current strategies for TnSeq are too laborious to be applied to hundreds of experimental conditions across multiple bacteria. Here, we describe an approach, random bar code transposon-site sequencing (RB-TnSeq), which greatly simplifies the measurement of gene fitness by using bar code sequencing (BarSeq) to monitor the abundance of mutants. We performed 387 genome-wide fitness assays across five bacteria and identified phenotypes for over 5,000 genes. RB-TnSeq can be applied to diverse bacteria and is a powerful tool to annotate uncharacterized genes using phenotype data.
INTRODUCTION
Experimental tools to systematically determine gene function are needed to keep up with the pace of microbial genome sequencing. One method that holds promise is the high-throughput analysis of mutant phenotypes, which has been used to assign functions to poorly characterized genes in diverse bacteria (1–4). High-throughput mutant fitness profiling in bacteria is commonly performed by mixing a large number of transposon mutants and monitoring their abundance in a competitive growth assay with next-generation sequencing (5).
A number of approaches have been developed for transposon insertion site sequencing in bacteria, including TnSeq (6), TraDIS (7), HITS (8), INSeq (9), and TnLE-seq (10). While these methods differ in how the sequencing libraries are prepared, they are all conceptually similar; the genomic DNA (gDNA) at the transposon insertion site serves as the “tag” for identifying each strain, and the importance of each gene for fitness is estimated from the number of reads that correspond to insertions within that gene. However, these protocols (referred to collectively as “TnSeq” in this paper) have not been applied to hundreds of experimental conditions, probably because the preparation of sequencing libraries is laborious. For example, the TraDIS or HITS protocols involve DNA shearing, DNA end repair, adapter ligation, and PCR, with multiple purification steps in between (7, 8). Also, the original TnSeq protocol (6) and the INSeq protocol (9) require multiple enzymatic and purification steps. Further, the latter two approaches were originally restricted to the mariner transposon, which inserts only at TA sites, and are therefore not ideal for genomes with high GC content. While the TnSeq protocol has been updated and can be used with any transposon (11), DNA shearing and multiple enzymatic steps are still required.
An alternative approach to measure strain fitness in a pooled, competitive assay is to quantify DNA bar codes, with the requirement that the DNA bar codes are previously associated with the mutations in the mixed library. The best example of this strategy is the Saccharomyces cerevisiae deletion collection (12), where sequence-defined DNA bar codes were incorporated into each deletion strain, enabling the pooling and parallel analysis of mutant fitness using DNA microarrays or, more recently, BarSeq, or DNA bar code sequencing (13). We previously extended the S. cerevisiae DNA bar code strategy to bacteria and Candida albicans by incorporating the same sequence-defined DNA bar codes into transposons (2, 14, 15). While this approach was successfully applied to multiple microorganisms, it was laborious to apply to each new organism because it required archiving individual mutant strains. Also, the number of mutant strains in each pool was limited to about 4,000 by the number of available DNA bar codes. Nevertheless, assaying the abundance of DNA bar codes is much simpler than TnSeq and requires only the PCR amplification of the DNA bar codes from total genomic DNA. To illustrate the scalability of the DNA bar code approach, thousands of genome-wide fitness assays have been performed in S. cerevisiae (16, 17) and hundreds have been performed in the bacteria Shewanella oneidensis MR-1 and Zymomonas mobilis ZM4 (1), far more than have been done with TnSeq approaches to date.
Here, we describe a new method for measuring gene fitness in bacteria, random bar code transposon-site sequencing (RB-TnSeq), which combines the advantages of TnSeq (large numbers of mutant strains with no archiving) and assaying DNA bar codes (easy and scalable quantification). A mutant library needs to be characterized only a single time to link the transposon insertion location to one of millions of random DNA bar codes incorporated in the transposon. All subsequent fitness assays utilize BarSeq. We describe the application of RB-TnSeq to five bacteria and demonstrate that the method is reproducible, compares favorably to other methods, and is scalable to many experimental conditions.
RESULTS
Overview of RB-TnSeq.
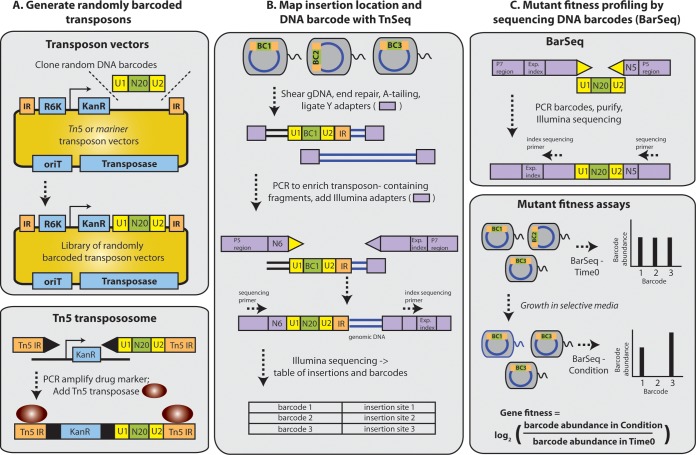
The RB-TnSeq approach is summarized in Fig. 1. In brief, we have decoupled the characterization of a complex transposon mutant library from the determination of strain and gene fitness, which requires only the relative quantification of DNA bar code abundance using BarSeq (13). DNA bar-coded transposons (Fig. 1A) are used to generate large bacterial mutant populations with the aim that each mutant strain in the population carries a single transposon insertion containing a unique DNA bar code. Each mutant library is characterized by a single TnSeq-like approach to link the transposon insertion location to its associated random DNA bar code, but this is done just once instead of for every fitness assay (Fig. 1B).
FIG 1 .
Overview of RB-TnSeq. (A) (Top) We converted both Tn5 and mariner transposon delivery vectors into RB-TnSeq vectors by cloning millions of unique DNA bar codes (N20) flanked by common PCR priming sites (U1 and U2) near the edge of the transposon’s inverted repeat (IR). (Bottom) We generated a randomly bar-coded transpososome by first PCR amplifying the kanamycin resistance gene with oligonucleotides containing Tn5 IRs and the random DNA bar code region and then adding Tn5 transposase. All three systems can be used to mutagenize bacteria by electroporation or (for Tn5 and mariner vectors) conjugation. Regardless of system or delivery method, the goal is to generate a large transposon mutant population such that each strain contains a unique DNA bar code. (B) A randomly bar-coded transposon mutant library is characterized using a protocol similar to HITS (8) or TraDIS (7). Here, we refer to this protocol generically as “TnSeq.” In TnSeq, genomic DNA is sheared, end repaired, and ligated with Illumina Y adapters. Transposon-containing DNA fragments are enriched by PCR with one primer specific to the Y adapter and a second primer specific to the transposon. Both the DNA bar code and the transposon insertion site are identified in a single 150-nucleotide Illumina sequencing read. The TnSeq results are a table of bar codes and associated transposon insertion locations. (C) (Top) In BarSeq, the DNA bar codes are PCR amplified using oligonucleotides that bind the common U1 and U2 regions. Both oligonucleotides contain adapter sequences for Illumina sequencing. One of the oligonucleotides contains an experiment index and enables multiplexing of multiple BarSeq experiments on a single lane of Illumina sequencing. (Bottom) Competitive mutant fitness assays are performed by comparing the abundance of the DNA bar codes with BarSeq before (time zero) and after (condition) selective growth. In this simple example, the gene associated with bar code 2 has reduced fitness; the gene associated with bar code 3 has enhanced fitness.
To identify phenotypes and gain insight into gene function, bar-coded transposon mutant populations are subject to competitive growth assays. In these experiments, the relative abundance of each mutant strain changes depending on the impact of the underlying gene mutation on the fitness of that strain. In traditional TnSeq experiments, changes in mutant strain abundance are assessed by the laborious TnSeq protocol for every condition. This limits the number of conditions that can be reasonably assessed. In RB-TnSeq, mutant fitness assays are replaced by the simple, inexpensive, and scalable BarSeq assay (18) (Fig. 1C).
Generation of complex mutant populations using randomly bar-coded transposons.
To demonstrate the broad utility of RB-TnSeq, we developed a set of reagents that encompass different transposons and different delivery systems. First, we converted Tn5 and mariner transposon delivery vectors into RB-TnSeq vectors by cloning millions of random 20-nucleotide DNA bar codes near the edge of each transposon’s inverted repeat (Fig. 1A). We also generated an RB-TnSeq-compatible transpososome (19) by using a simple PCR to add random bar codes and inverted repeats and then adding Tn5 transposase enzyme (Fig. 1A). Using these diverse reagents, we generated genome-wide transposon mutant libraries in five bacteria: the model bacterium Escherichia coli BW25113 (a K-12 strain; parent strain of the Keio deletion collection [20]), the marine heterotroph Phaeobacter inhibens DSM 17395, the chromium-reducing bacterium Pseudomonas stutzeri RCH2, and two metal-reducing bacteria of the genus Shewanella, Shewanella amazonensis SB2B and S. oneidensis MR-1. To emphasize the flexibility of the RB-TnSeq approach, the five mutant libraries used two different transposon vectors (mariner and Tn5) or a Tn5 transpososome (Table 1).
TABLE 1 .
Summary and coverage of mutant libraries and BarSeq statistics
| Category | Escherichia coli BW25113 | Phaeobacter inhibens DSM 17395 | Pseudomonas stutzeri RCH2 | Shewanella amazonensis SB2B | Shewanella oneidensis MR-1 |
|---|---|---|---|---|---|
| Summary of mutant libraries | |||||
| Mutant library name | KEIO_ML9 | Phaeo_ML1 | psRCH2_ML7 | SB2B_ML5 | MR1_ML3 |
| Transposon | Tn5 transpososome | Tn5 vector | mariner vector | mariner vector | Tn5 vector |
| Method of delivery | Electroporation | Conjugation | Conjugation | Conjugation | Conjugation |
| No. of strains with unique bar codesa | 152,018 | 217,394 | 166,448 | 389,329 | 181,569 |
| % of bar codes with intact vectorb | NAg | <0.1 | 0.6 | 1 | 1 |
| Protein-coding genes | |||||
| Total no. | 4,146 | 3,875 | 4,265 | 3,645 | 4,467 |
| No. with central insertions | 3,728 | 3,453 | 3,548 | 3,278 | 3,778 |
| No. with fitness estimates (% of total) | 3,471 (84) | 3,085 (80) | 3,335 (78) | 3,078 (84) | 3,661 (82) |
| Median no. of strains per genec | 16 | 23 | 20 | 47 | 15 |
| No. with significant phenotyped | 960 | 888 | 1,045 | 1,084 | 1,219 |
| Insertion bias | |||||
| % toward coding strand of genes | 54 | 52 | 51 | 53 | 52 |
| Mean/median no. of reads per genee | 1.6 | 1.8 | 2.2 | 1.6 | 2.2 |
| BarSeq statistics | |||||
| % of reads with sample and bar code | 90 | 93 | 94 | 89 | 87 |
| % of bar codes that map to a straine | 92 | 42 | 57 | 61 | 66 |
| No. of reads per million for the median genee,f | 92 | 43 | 48 | 73 | 46 |
Only strains with insertions in the genome are included.
May represent integration events of entire transposon plasmid into genome.
Includes only genes for which we report fitness estimates and only strains that were used to make those estimates.
Genes with a significant phenotype in at least one BarSeq fitness experiment (|t| > 4).
Computed with time-zero samples.
Includes reads that have no multiplex or bar code in the denominator.
NA, not applicable.
Characterization of bar-coded transposon mutant libraries using TnSeq.
For each mutant library, we performed TnSeq to characterize the mutant library by simultaneously mapping the transposon insertion location and the identity of the linked DNA bar code in a single Illumina read. Our protocol is similar to TraDIS or HITS (7, 8), and involves shearing genomic DNA, end repair, ligating adapters, and PCR that amplifies the transposon junction using primers that are complementary to the adapter and to the transposon, along with several purification steps (Fig. 1B). After filtering out chimeric reads and nonunique DNA bar codes (see Materials and Methods), we identified over 100,000 transposon insertions with unique DNA bar codes in each of the five mutant libraries (Table 1). For each mutant library, the mapped insertions were moderately biased (no regions are underrepresented by more than 2-fold relative to the average) across the genome with little strand or coverage bias (Table 1; see Fig. S1 in the supplemental material). With this coverage, these mutant libraries are ideal for interrogating the mutant fitness of nearly all nonessential protein-coding genes using BarSeq.
Mutant fitness profiling by sequencing pools of random DNA bar codes.
We used BarSeq, or deep sequencing of DNA bar codes, to quantify the abundance of transposon insertion strains and calculate gene fitness in a competitive growth assay. BarSeq was originally developed for sequence-defined DNA bar codes, such as those used in the Saccharomyces cerevisiae deletion collection (13). To our knowledge, this is the first use of BarSeq with randomized bar codes in a microbial fitness experiment. To determine the fitness of each gene under a given condition, we compared the abundance of the bar codes for different strains of a gene (independent transposon insertions) before and after growth selection (Fig. 1C; see Materials and Methods for details). More specifically, the fitness of a strain is the log2 change in abundance during growth (typically 4 to 6 generations); the fitness of a gene is roughly the average of the fitness of the strains that have insertions within that gene. The fitness data are normalized so that the typical gene has a fitness of zero (see Materials and Methods).
Given that genomic regions near the origin of replication can have a higher copy number in dividing cells, we normalized the fitness data by chromosomal position in each experiment. To assess the accuracy of this normalization, we empirically determined the genome abundance bias in different samples of the S. amazonensis mutant library by whole-genome sequencing (see Fig. S2 in the supplemental material). We found that our normalization algorithm, which is based solely on BarSeq data, accurately controls for variation in copy number (see Fig. S2).
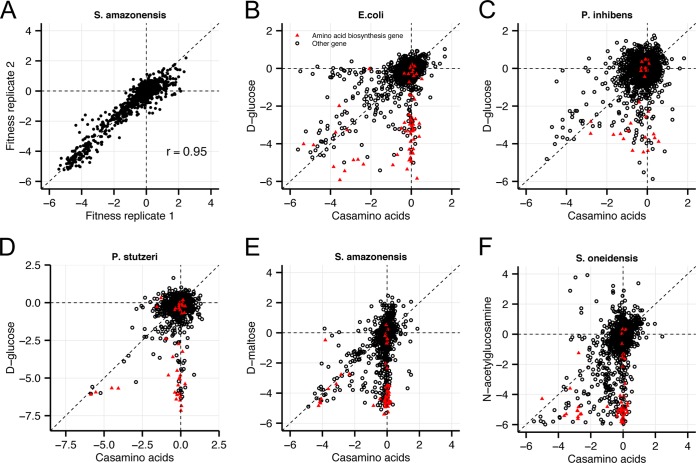
We compared BarSeq fitness data collected from defined minimal medium with either a single carbon substrate or a mixture of amino acids (Casamino Acids) for each of the five bacteria to assess the reproducibility and biological significance of the method. For each BarSeq experiment, we identified genes with significant phenotypes using a t-like statistic that takes into account the consistency of the fitness of all the mutants of that gene (see Materials and Methods). Genes with |t| of >4 have highly significant phenotypes that are largely reproducible in biological replicate experiments (Fig. 2A). We found that mutants of predicted amino acid biosynthetic genes often exhibited reduced fitness in defined medium relative to medium supplemented with Casamino Acids for each of the five bacteria, confirming that BarSeq produces expected biological results in diverse bacteria (Fig. 2B to F).
FIG 2 .
Validation of BarSeq fitness data. (A) Comparison of gene fitness for two biological replicates of S. amazonensis SB2B grown in defined medium with d-maltose as a carbon source. (B to F) Comparison of gene fitness in a defined medium with Casamino Acids (x axis) or a single carbon source (y axis) for E. coli (B), P. inhibens (C), P. stutzeri (D), S. amazonensis (E), and S. oneidensis (F). Genes annotated with the functional role “amino acid biosynthesis” by TIGRFAMs (38) are marked as red triangles.
Comparison of RB-TnSeq to other technologies.
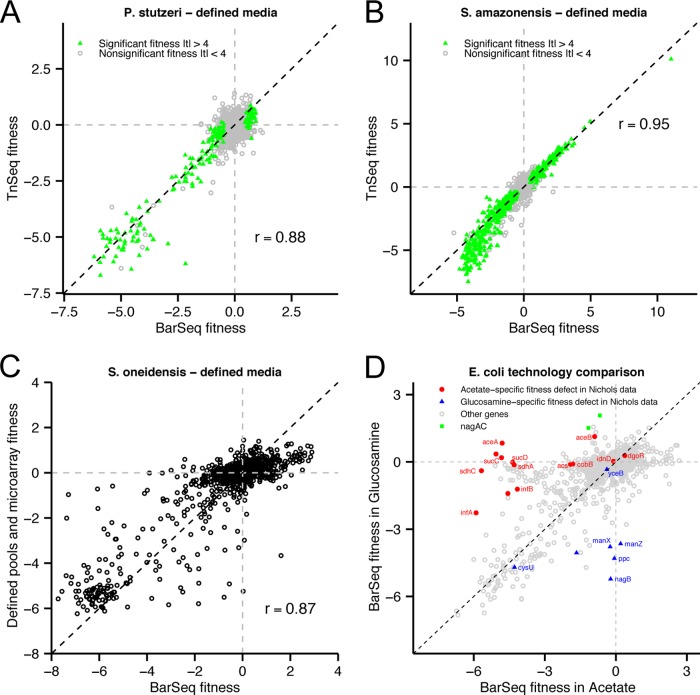
A number of approaches have been developed to assay the fitness of bacterial mutants in high-throughput sequencing, either with pooled competitive growth assays or with individual mutants. Given that these approaches are well established, we compared them to RB-TnSeq to assess the quality of our approach. First, we compared gene fitness results obtained from random BarSeq to results obtained from sequencing the transposon insertion junction (TnSeq). The latter method of using the genomic insertion location as a “tag” is the basic principle of all current transposon-sequencing techniques. For both P. stutzeri and S. amazonensis, we found that gene fitness was highly correlated regardless of whether BarSeq or TnSeq was performed on the same genomic DNA samples (Fig. 3A and B). Therefore, simply assaying the abundance of random DNA bar codes with BarSeq generates data equivalent to those derived from sequencing transposon insertion junctions, which is a far more complicated, expensive, and laborious protocol.
FIG 3 .
Comparison of RB-TnSeq to other technologies. (A) Comparison of gene fitness for P. stutzeri grown in a defined medium with glucose as determined with BarSeq (x axis) or sequencing the transposon-genome insertion junctions (TnSeq; y axis), starting from the same samples of genomic DNA. Genes marked in green have statistically significant phenotypes as determined by BarSeq. The dashed black line marks x = y. (B) Same as panel A for S. amazonensis grown in a defined medium with d,l-lactate. (C) Comparison of S. oneidensis gene fitness in defined medium with l-lactate calculated from BarSeq (x axis) and previously described data that used mutant libraries with defined DNA bar codes and microarrays to assay strain abundance (y axis) (2). The dashed black line marks x = y. (D) BarSeq fitness data for E. coli genes grown in acetate (x axis) or glucosamine (y axis) as the sole source of carbon. Genes marked in red have an acetate-specific fitness defect while those marked in blue have a glucosamine-specific fitness defect in the Nichols et al. data set, with thresholds of S < −5 and S > −2 (4).
Second, we compared S. oneidensis gene fitness data from random BarSeq to fitness data obtained from our previously described method that utilized archived transposon mutants, sequence-defined DNA bar codes (14), and microarrays to assay strain abundance (2). In this previous study, we used the genome-wide fitness data to identify phenotypes for and to annotate the functions of poorly characterized S. oneidensis genes, and we verified many of the putative phenotypes by growing the individual mutant strains (2). Therefore, the comparison of these old data to our new BarSeq-generated fitness data provides an additional measure of the biological significance of the RB-TnSeq method. For growth in a defined medium with l-lactate as the carbon source, gene fitness is strongly correlated (r = 0.87), albeit with a greater dynamic range for BarSeq, as has been previously observed (Fig. 3C) (13).
Lastly, we compared our E. coli BarSeq fitness data to similar data obtained from the high-throughput imaging of individual mutants on plates (4). Both methods successfully identified genes specifically involved in the catabolism of either acetate (aceAB, acs, cobB, infAB, sdhAC, and sucCD) or glucosamine (manXZ, nagB, and ppc) (Fig. 3D). However, in the imaging data, there were a few genes that were identified as specifically sick with acetate (dgoR and idnD) or glucosamine (cysU and yceB) that were not identified by our BarSeq data (Fig. 3D). DgoR, IdnD, and YceB are not expected to be involved in the catabolism of either substrate, whereas mutants in cysU are reported to be auxotrophic in defined medium and are expected to exhibit reduced fitness with either carbon source (21). In addition, only the competitive fitness assay with BarSeq identified nagA and nagC as detrimental to growth with glucosamine (fitness of >1.5 for both genes), as previously reported (22) (Fig. 3D). Given that up to half of bacterial genes are detrimental to fitness under some laboratory growth conditions (1), the ability of the BarSeq method to identify these phenotypes is a key advantage of the RB-TnSeq approach.
Scalability of BarSeq with random bar codes.
The primary benefit of RB-TnSeq is that once a mutant library is characterized by TnSeq, all fitness assays use BarSeq, which greatly simplifies sample processing and increases throughput. To demonstrate the high throughput of RB-TnSeq, we performed 501 BarSeq mutant fitness assays across the five bacteria, with a focus on identifying genes involved in the uptake and catabolism of different carbon sources. Of these 501 experiments, 387 (77%) passed our metric thresholds for successful experiments (see below and Materials and Methods). Most experiments with poorer-quality metrics used our first PCR method for BarSeq (95°C denaturing) and were largely restricted to the higher-GC-content bacteria P. inhibens and P. stutzeri (see Materials and Methods for details on the two PCR conditions used for BarSeq). Among the 387 successful experiments, we studied 163 different bacterium-condition combinations, including 130 different bacterium-carbon source combinations, all but 5 with at least two biological replicates. The median correlation between all replicate experiments was 0.92. Across the 387 experiments, we identified significant phenotypes for 5,196 genes (|t| of >4 and false discovery rate under 2%; see Materials and Methods), ranging from 888 genes in P. inhibens to 1,219 genes in S. oneidensis (Table 1).
The expense of RB-TnSeq is dominated by the cost of sequencing, which depends on how many samples can be multiplexed together. In yeast, using a defined set of roughly 12,000 DNA bar codes, groups have reported multiplexing up to 96 samples (18). Here, we show that 48 to 96 random DNA bar code BarSeq experiments can be multiplexed in one lane of a HiSeq system without reducing the quality of the data. In a lane of the HiSeq system, we usually obtain around 200 million reads (median, 215 million), and typically, over 90% of those have both a multiplexing tag and a 20-nucleotide bar code with flanking sequences, leaving 194 million reads in the median run. However, many of those bar codes are not usable due to bar code reuse (the same bar code mapping to two or more insertions), sequencing error, TnSeq mapping ambiguities, or mapping to the vector that was used to construct the transposon library. Depending on the mutant library, 42 to 92% of those reads are usable (Table 1), which would leave 81 million to 178 million informative reads. The E. coli library had the highest fraction of usable reads, presumably because the PCR product used to generate the transpososome was so diverse and a bar code mapping to 2 or more locations was rare. Ideally, the usable reads would be uniformly distributed across samples. If the multiplexing were perfectly even, then we would obtain around 1.7 million reads per sample with 48-way multiplexing for the worst library (P. inhibens) or 1.9 million reads per sample for E. coli with 96-way multiplexing. However, we have routinely observed an ~2-fold variation in the number of reads per sample, leaving some samples with about 1 million reads or a bit less. Of those 1 million reads, about 10% will map to insertions that are not in genes, and another 20% will map to insertions that are near the edges of genes. Accounting for these losses, ~700,000 reads remain for some samples. If we have 3,000 genes, and the skewness in gene coverage (the mean divided by the median) is around 2, then that reduces to around 110 reads for the typical gene. In theory, 50 reads would suffice to quantify fitness to a standard error of around 0.3 . In practice, our experiments had 183 to 383 reads for the typical gene (25th to 75th percentile of experiments). So in theory, the vast majority of samples have sufficient coverage to accurately estimate the fitness of most genes.
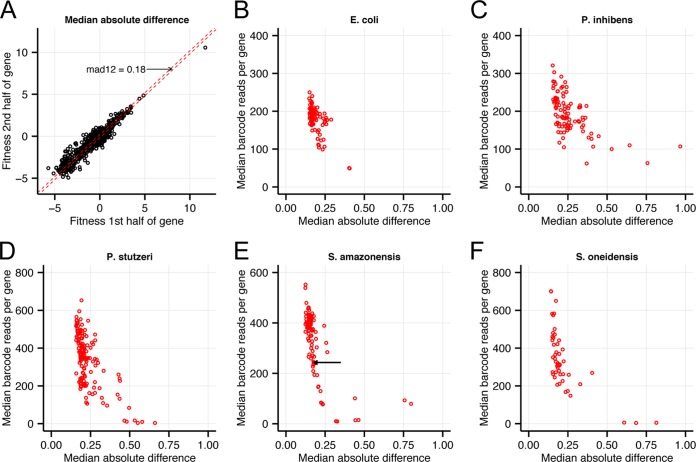
To test how coverage affects the quality of our data in practice, we examined the consistency of the fitness data between the two halves of each gene. As shown for a typical experiment in Fig. 4A, for most genes, the two halves give similar results. To quantify how noisy the fitness values are, we use the median absolute difference between the first- and second-half fitness values (mad12) (Fig. 4A). We consider a mad12 of <0.5 to be an acceptable amount of noise. Across all 501 BarSeq experiments (Fig. 4B to F), there is a trend toward lower mad12 as the number of reads for the median genes increases, but the effect is slight once the typical gene has more than 100 reads. In all organisms, a few experiments are much noisier than expected, given how many reads we have for the typical gene, and these tend to be experiments in which at least one gene is strongly detrimental, with a fitness above 6, implying an over-64-fold increase in the abundance of mutants in that gene(s). It is not obvious why data from these experiments are so much noisier, as for most of these experiments, we still collected adequate amounts of data for the typical gene. These conditions may select for secondary mutations in the detrimental genes, or there may be stochastic effects in exiting the long lag phase that is associated with many of these conditions (23).
FIG 4 .
Consistency of fitness values versus number of reads. (A) Consistency of fitness data between the two halves of each gene (10 to 50% or 50 to 90%), for Shewanella amazonensis SB2B growing in a defined medium with d,l-lactate as the carbon source. To summarize this plot, we computed the median of the absolute difference (mad12) between the two values. Half of the genes are between the dashed lines, which show x = y − mad12 and x = y + mad12. (B to F) Consistency of fitness data as measured by mad12 (x axis) versus number of reads for median gene (y axis), with a separate panel for each organism and a point for each genome-wide fitness experiment. In panel E, the arrow highlights the experiment shown in panel A. Control experiments (time zero) are not included in the plots.
To demonstrate the effective multiplexing of 96 samples, consider the “set1” lane for E. coli, which included 96 samples, including 4 time-zero samples and experiments in 47 different carbon or nitrogen sources. One of the time-zero samples had virtually no reads (1,174 reads with bar codes), but every other sample had at least 1.3 million reads with usable bar codes. Of the 92 experimental samples, 89 passed our quality metrics (see Materials and Methods), with at least 50 reads for the typical gene and a mad12 under 0.5. The three exceptions had strong positive selection, with each experiment containing at least one gene having a fitness value of ~6. Among the successful experiments, the correlation of gene fitness values between biological replicates was high, ranging from 0.79 to 0.97 (median, 0.95). Overall, we confirmed that RB-TnSeq is scalable and that multiplexing 48 to 96 samples per lane yields accurate estimates of gene fitness.
Identification of carbon utilization genes in P. inhibens.
Here, we briefly discuss the P. inhibens results, first by comparing our mutant fitness data to carbohydrate utilization genes recently identified by a metabolomics, proteomics, and comparative genomics study (24). For reference, the fitness data for the P. inhibens genes described below are presented in Fig. 5.
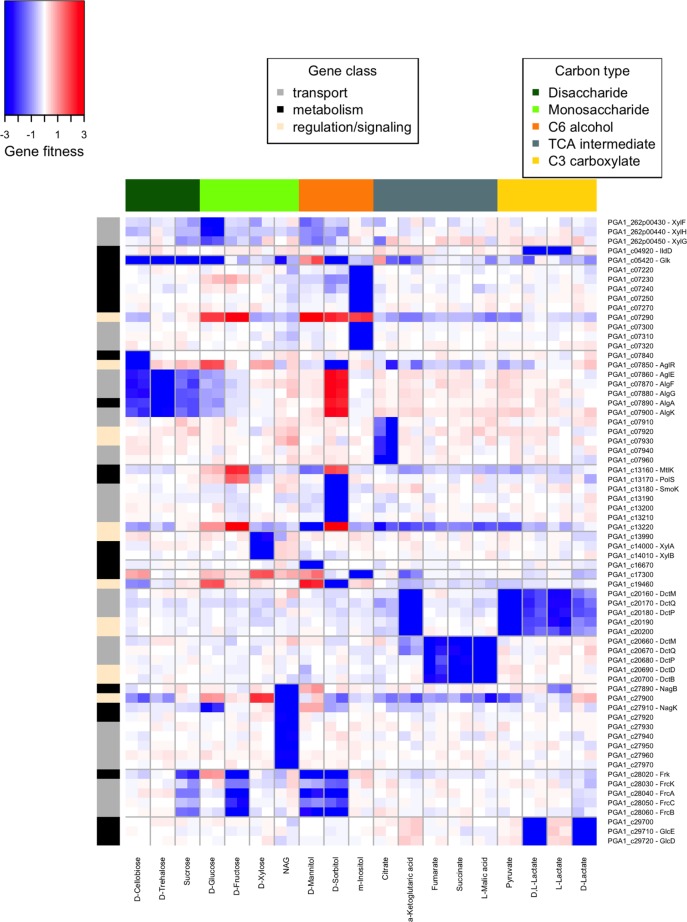
FIG 5 .
Carbon utilization genes of P. inhibens. Heat map of gene fitness values for select P. inhibens genes (y axis) with significant phenotypes during growth in defined medium with one or more carbon sources (x axis). For illustration purposes, genes with fitness values of less than −3 were set to −3. Similarly, genes with fitness values of greater than 3 were set to 3. Operons and other chromosomally clustered genes are split by horizontal gray lines.
In general, our mutant phenotype data are in strong agreement with the gene functions proposed by Wiegmann and colleagues (24). For example, transport genes predicted to be involved in the utilization of N-acetylglucosamine (NAG; c27930:c27970), sucrose (algEFGK; xylFHG), glucose (xylFHG), succinate (DctMQP6; encoded by c20660:c20680), and fructose (frcACB) are required for optimal fitness during growth with these substrates (Fig. 5). Similarly, the fitness data support the putative regulators for the utilization of NAG (c27900), xylose (c13990), disaccharides (aglR), and mannitol (c13220). However, because BarSeq-based mutant fitness profiling can be readily applied to many experimental conditions and provides a fitness measure for the majority of genes, we were able to profile 30 different carbon sources and to use this comprehensive data set to identify new phenotypes. For instance, we found that the putative d-mannose isomerase (c16670) is specifically required for mannitol utilization in P. inhibens (Fig. 5). Similarly, we found that mannitol utilization in S. amazonensis also requires a homologous d-mannose isomerase (Sama_0560; fitness, <−3), as previously predicted (25). We speculate that the mannitol catabolism pathway in P. inhibens proceeds through a mannose intermediate: mannitol is first oxidized to mannose by mannitol dehydrogenase (MtlK, c13160), followed by the conversion of mannose to fructose by mannose isomerase. In addition, we identified multiple genes specifically involved in the utilization of m-inositol (c07220:c07250, c07270, and c07290:c07320), citrate (c07910:c07940 and c07960), and d-lactate (c29700:c29720). Lastly, we found that a second P. inhibens DctMQP6 gene cluster (c20160:c20200) is specifically required for the utilization of lactate, pyruvate, and α-ketoglutaric acid (Fig. 5).
We also identified a number of carbon utilization genes in P. inhibens with complex phenotypic patterns, including genes that are required for growth with some carbon substrates and detrimental in others (Fig. 5). For example, the disaccharide transporter encoded by algEFGK is important for growth with cellobiose, sucrose, and trehalose but is detrimental to growth with sorbitol (average gene fitness, >2). Similarly, mannitol dehydrogenase and the mannitol regulator encoded by c13220 are important for mannitol catabolism but detrimental to growth with either fructose or sorbitol (Fig. 5). The enhanced fitness phenotypes could be due to costly protein activity, as described for the lactose permease in E. coli (26), or due to altered regulation, as described for nagA and nagC in E. coli glucosamine catabolism (22). Indeed, a number of P. inhibens genes involved in either signaling or transcription have complex phenotypes, including the putative m-inositol repressor c07290, the disaccharide regulator aglR, the mannitol regulator c13220, the uncharacterized regulator encoded by c19460, and the NAG regulator c27900, which is detrimental to growth on xylose and d-glucose.
DISCUSSION
RB-TnSeq depends on making a library of mutants that is diverse and reasonably balanced across the entire genome and in which most of the strains have unique bar codes. The first two challenges are not specific to our method. For some organisms, it may be difficult to generate enough mutants, and colony counts can be misleading. In our first E. coli libraries, the insertions were dense near the origin of replication but sparse near the terminus, which indicated that the cells that we used were growing too rapidly. To identify these problems quickly, we recommend making several mutant libraries under different conditions for a given bacterium and sequencing them at low coverage on a platform that gives rapid results (i.e., Illumina’s MiSeq).
For many bacteria, the bar-coded delivery vectors described here, containing mariner or Tn5 transposons marked with kanamycin resistance, should suffice to generate mutant libraries. Alternatively, if the bacterium can be electroporated, then the bar codes can be added to a custom construct that has a different promoter or resistance marker by using PCR, followed by electroporating a DNA-transposase complex, as we report here for E. coli. However, for applications where a modified suicide plasmid is needed, then adding the random bar codes requires an additional cloning step to generate a large and diverse vector library. In order to generate a usable mutant library, it is essential that the majority of mutant strains are marked with a unique DNA bar code. Our method therefore requires the total number of unique DNA bar codes available to be in significant excess over the number of transposon mutants in the final library. This was not a challenge here, but if there are fewer than a million bar codes, then the mutant libraries should have at most a few hundred thousand strains. Conversely, the statistical methods reported here may require modification if there are so many strains that there are very few reads for the typical strain in the typical sample.
In summary, we describe a scalable method for measuring gene fitness in diverse bacteria. The 96 samples of E. coli discussed above required just one person-week of effort: this included recovering the mutant library from the freezer, growing the library under 50 different conditions, extracting genomic DNA from 96 samples, performing 96 BarSeq PCRs, mixing and purifying the PCR products, and sending them to the sequencing center for loading onto a single lane of the Illumina HiSeq system. In contrast, the largest effort with TnSeq-like protocols that we are aware of included just 42 different samples (27). This work describes 387 successful RB-TnSeq fitness experiments, which is probably more than in the entire TnSeq literature. Although all of our experiments assayed growth in pure culture, pooled mutant fitness assays can also be used to assay growth in cocultures (28), growth in mice (29), motility (2), or survival (2) or to identify strains with altered morphology after separation with a cell sorter (30). We also hope to extend our approach to strains that have multiple transposon insertions so that we can assay genetic interactions. Lastly, our random bar code approach should be useful for studying other microorganisms with tools for insertional mutagenesis, including fungi, algae, and archaea.
MATERIALS AND METHODS
Strains and standard growth conditions.
Shewanella amazonensis SB2B was a gift of James Tiedje (Michigan State University). Escherichia coli strain BW25113 was purchased from the Coli Genetic Stock Center. The E. coli conjugation donor strain WM3064 was a gift of William Metcalf (University of Illinois). Shewanella oneidensis MR-1 (ATCC 700550) and Phaeobacter inhibens (DSM 17395) were purchased from the indicated public repositories. Pseudomonas stutzeri RCH2 was a gift of Romy Chakraborty (Lawrence Berkeley National Laboratory). We routinely cultured the E. coli strains WM3064 and BW25113 and S. amazonensis SB2B in Luria-Bertani broth (LB) at 37°C. For culturing WM3064, we supplemented LB with diaminopimelic acid (DAP) to a final concentration of 300 µM. S. oneidensis MR-1 and P. stutzeri RCH2 were routinely cultured in LB at 30°C. We typically cultured P. inhibens in marine broth (Difco 2216) at 25°C. A full list of strains used in this study is contained in Table S1 in the supplemental material.
Construction of bar-coded mariner transposon vector.
The randomly bar-coded mariner transposon delivery vector pKMW3 is derived from pHIMAR-RB1 (31) and the vectors pKMW1 and pKMW2 constructed as part of this study. To generate pKMW2, we replaced a 338-bp region of pHIMAR-RB1 with a linker containing SbfI and FseI restriction sites near the mariner transposon inverted repeat (IR). Specifically, we PCR amplified pHIMAR-RB1 with oligonucleotides pHIMAR_eng_oligo1 and pHIMAR_eng_oligo2. The linker oligonucleotide pHIMAR_eng_oligo4 (ACACTGGCAGAGCATTACGCCCTGCAGGATGCAATGGGCCGGCCAGACCGGGGACTTATCAGCCAACCTGTTATGT; SbfI, FseI, and mariner IR sites in bold) was amplified with pHIMAR_eng_oligo5 and pHIMAR_eng_oligo6. We used Gibson assembly to clone these two PCR products together and generate pKMW2. pKMW1 is a vector library containing ~10 million unique, random 20-nucleotide DNA bar codes. To construct pKMW1, we PCR amplified the randomly bar-coded oligonucleotide pRL27_Eng_oligo19 (GATGTCCACGAGGTCTCTNNNNNNNNNNNNNNNNNNNNCGTACGCTGCAGGTCGAC) with oligonucleotides Amp_barcodes_FOR_Gateway_SbfI and Amp_barcodes_REV_Gateway_FseI and cloned the resulting PCR product into the Gateway donor vector pDONR/Zeo (Invitrogen) using the Gateway BP reaction according to the manufacturer’s protocols (Invitrogen). In pRL27_Eng_oligo19, the N’s represent the random 20-nucleotide DNA bar code sequence and the italic regions are the common priming sites U1 and U2 used to PCR amplify the bar codes (see “BarSeq” below). To construct pKMW3, we cut the DNA bar codes and their common priming sites from pKMW1 with SbfI and FseI, gel purified the insert, and ligated the bar codes into SbfI- and FseI-digested pKMW2. The ligations were electroporated into E. coli pir+ competent cells (Epicentre), and transformants were selected in liquid LB with 50 µg/ml kanamycin. To enable conjugation, we purified pKMW3 plasmid DNA from pir+ cells, electroporated the plasmid library into the E. coli conjugation strain WM3064, and selected transformants in liquid LB supplemented with 50 µg/ml kanamycin and DAP. All primers and vectors used in this study are contained in Tables S2 and S3 in the supplemental material, respectively. Additional details on PCR and cloning conditions used in this study are available on request.
Construction of bar-coded Tn5 transposon vector.
We used a strategy similar to the construction of pKMW3 to generate the randomly bar-coded Tn5 delivery vector pKMW7. To construct the intermediate vector pKMW4, we replaced a 518-bp region of pRL27 (32) with a linker region containing SbfI and FseI sites near a Tn5 IR. Specifically, we amplified the pRL27 backbone with pRL27_Eng_oligo22 and pRL27_Eng_oligo23 and the linker oligonucleotide pRL27_Eng_oligo24 (CCTGCAGGATGCAATGGGCCGGCCGGTTGAGATGTGTATAAGAGACAGTCGAC; SbfI, FseI, and Tn5 IR sites in bold) with oligonucleotides pRL27_Eng_oligo25 and pRL27_Eng_oligo26. We used Gibson assembly to clone the two PCR products together and generate pKMW4. To add the random DNA bar codes to pKMW4, we SbfI and FseI digested the Amp_barcodes_FOR_Gateway_SbfI and Amp_barcodes_REV_Gateway_FseI amplified DNA bar codes (from pRL27_Eng_oligo19) and directly ligated these bar codes into SbfI- and FseI-digested pKMW4 to construct the bar-coded vector library pKMW7. We transformed the ligations into E. coli pir+ cells, isolated plasmid DNA, and transformed the purified pKMW7 vectors into WM3064 as described for pKMW3. The primary difference from our pKMW3 strategy was the source of the DNA bar codes: pKMW3 used the vector library pKMW1 and pKMW7 used a PCR product.
After transfer of pKMW3 or pKMW7 into the conjugation donor, we used BarSeq (see below) to verify that the bar codes were diverse. We performed a separate MiSeq run for each library. To avoid inflating the diversity due to sequencing errors, we required every nucleotide in each bar code to have a minimum quality score of 30 (Q30) and we filtered out bar codes that were just 1 nucleotide different from another bar code. For pKMW3, from 6.9 million usable reads, we observed 3.2 million different bar codes. For pKMW7, from 13.2 million usable reads, we observed 8.9 million different bar codes. For both pKMW3 and pKMW7, many of the reads are for bar codes that were seen just once or twice (44% and 75%, respectively). Given our quality threshold, at most 0.001 × 20 = 2% of the bar codes should be erroneous, so we expect that few of these rare bar codes are sequencing errors. Indeed, when we analyzed a time-zero sample of the E. coli library with BarSeq and the same Q30 cutoff, 95% of the reads mapped to the pool definition, and from 1.7 million reads, we observed just 169,000 different bar codes. A caveat with this comparison is that the E. coli data were collected with Illumina HiSeq, whereas the data for pKMW3 and pKMW7 were collected with Illumina MiSeq. Overall, the true diversity of both pKMW3 and pKMW7 is probably higher than the 3 to 9 million bar codes that we observed.
Generation of bar-coded EZ:Tn5 transpososome.
We prepared a custom EZ:Tn5, randomly bar-coded transpososome (19) by PCR with oligonucleotides containing Tn5-specific inverted repeat (IR) and a random 20-bp DNA bar code flanked by the common BarSeq PCR priming sites U1 and U2. Specifically, we used oligonucleotides EZ_Forward_Kan_pRL27 (CTGTCTCTTATACACATCTTGTGTCTCAAAATCTCTGATGTTAC) and EZ_Reverse_Kan_pRL27 (CTGTCTCTTATACACATCTGTCGACCTGCAGCGTACGNNNNNNNNNNNNNNNNNNNNAGAGACCTCGTGGACATCTTAGAAAAACTCATCGAGCATCAA) to amplify the kanamycin resistance gene from pRL27, where the bold regions represent Tn5 IR, the N’s are the random 20-bp DNA bar code, and the italic regions are the common BarSeq PCR priming sites U1 and U2. We performed PCR in a 100-µl total volume with 25 µmol of each primer, 3 ng of pRL27 template, and Phusion DNA polymerase (New England Biolabs) under the following cycling conditions: 98°C for 30 s, followed by 15 cycles of 10 s at 98°C, 30 s at 58°C, and 1 min at 72°C and by a final extension at 72°C for 5 min. We treated the PCR with DpnI for 1 h at 37°C to digest the template pRL27 vector and purified the final PCR product with AMPure beads (Beckman Coulter). The final, randomly barcoded EZ:Tn5 transpososome was prepared by adding 6 µl of the PCR product (100 ng/µl in Tris-EDTA [TE] buffer), 12 µl EZ:Tn5 transposase (Epicentre), and 6 µl 100% glycerol. The transpososome reaction mixture was mixed, incubated at room temperature for 30 min, and stored as aliquots at −20°C.
Transposon mutant library construction. (i) Escherichia coli BW25113.
We electroporated 1 µl of the DNA bar-coded EZ:Tn5 transpososome into electrocompetent BW25113 cells prepared from mid-log-phase cells grown at 25°C and washed with 10% glycerol. To increase the size of the mutant library, we performed 24 separate electroporation reactions. Postelectroporation, we pooled all cells, performed an outgrowth in the absence of selection in SOC medium for 1 h at 37°C, and plated the cells on LB agar plates supplemented with 50 µg/ml kanamycin. To construct the final E. coli mutant library KEIO_ML9, we scraped together kanamycin-resistant colonies into LB with 50 µg/ml kanamycin, diluted the mutant library to an optical density at 600 nm (OD600) of 0.25 in fresh LB medium with 50 µg/ml kanamycin, and grew the mutant library to a final OD600 of 1.0. We added glycerol to a final concentration of 10%, made multiple 1-ml −80°C freezer stocks of the mutant library, and collected cell pellets for genomic DNA extraction (for TnSeq).
(ii) Phaeobacter inhibens.
We mutagenized P. inhibens by conjugation with a pool of donor WM3064 carrying the pKMW7 Tn5 vector library (strain APA766). Briefly, we combined equal amounts (as determined by OD600) of mid-log-phase recipient P. inhibens and APA766 cells and conjugated the mixture on 0.45-µm nitrocellulose filters (Millipore) overlaid on marine broth agar supplemented with DAP. After 12 h of conjugation at 25°C, the cells on the filters were resuspended in marine broth and plated on marine broth agar containing 300 µg/ml kanamycin. After 3 days of growth, we scraped together kanamycin-resistant colonies into marine broth with 300 µg/ml kanamycin, diluted the mutant library back to a starting OD600 of 0.25 in 100 ml of marine broth with 300 µg/ml kanamycin, and grew the culture at 25°C to a final OD600 of 1.5. We added glycerol to a final volume of 10%, made multiple 1-ml −80°C freezer stocks, and collected cell pellets for genomic DNA extraction (for TnSeq). The final P. inhibens mutant library was designated Phaeo_ML1.
(iii) Pseudomonas stutzeri RCH2.
We created the psRCH2_ML7 transposon mutant library by conjugating P. stutzeri with WM3064 harboring the pKMW3 mariner transposon vector library (APA752). We combined equal cell numbers of mid-log-phase P. stutzeri RCH2 and APA752, conjugated them for 6 h at 30°C on 0.45-µm nitrocellulose filters (Millipore) overlaid on LB agar plates containing DAP, and plated the resuspended cells on LB plates with 50 µg/ml kanamycin to select for mutants. After 2 days of growth at 30°C, we scraped the kanamycin-resistant colonies into LB, determined the OD600 of the mixture, and diluted the mutant library back to a starting OD600 of 0.2 in 250 ml of LB with 50 µg/ml kanamycin. We grew the diluted mutant library at 30°C to a final OD600 of 1.0, added glycerol to a final volume of 10%, made multiple 1-ml −80°C freezer stocks, and collected cells for genomic DNA extraction.
(iv) Shewanella amazonensis SB2B.
To construct mutant library SB2B_ML5, we conjugated saturated cultures of S. amazonensis SB2B (recipient) and the donor strain APA752 at a donor/recipient ratio of 1:4 for 8 h at 37°C on 0.45-µm nitrocellulose filters (Millipore) overlaid on LB agar plates supplemented with DAP. We scraped the conjugation reaction mixtures into LB, plated the cells on LB agar plates supplemented with 100 µg/ml kanamycin, and incubated the plates at 37°C. After 2 days of growth, we scraped and combined kanamycin-resistant colonies into LB with 100 µg/ml kanamycin, determined the OD600 of the mixture, and diluted the mutant library back to a starting OD600 of 0.2 in 250 ml of LB with 100 µg/ml kanamycin. We grew the mutant library at 37°C to a final OD600 of 1.5, added glycerol to a final volume of 15%, made multiple 1-ml −80°C freezer stocks, and pelleted cells for genomic DNA extraction.
(v) Shewanella oneidensis MR-1.
We mutagenized S. oneidensis by conjugation with donor strain APA766. Equal volumes of the mid-log-phase-grown donor APA766 and recipient S. oneidensis were mixed and spotted onto 0.45-µm nitrocellulose filters (Millipore) overlaid on LB agar plates supplemented with DAP. After 5 h of conjugation at 30°C, the filters were resuspended in LB, and the cells were plated on LB agar with 50 µg/ml kanamycin. After 2 days of growth, we scraped together kanamycin-resistant colonies into LB with 50 µg/ml kanamycin, diluted the mutant library back to a starting OD600 of 0.2 in 100 ml of LB with 50 µg/ml kanamycin, and grew the culture at 30°C to a final OD600 of 1.5. We added glycerol to a final volume of 10%, made multiple 1-ml −80°C freezer stocks, and collected cell pellets for genomic DNA extraction (for TnSeq). The final S. oneidensis mutant library was designated MR1_ML3.
TnSeq sequencing library preparation.
To generate Illumina-compatible sequencing libraries to link random DNA bar codes to transposon insertion sites, we first isolated genomic DNA from cell pellets of the mutant libraries with the DNeasy kit (Qiagen). Genomic DNA was quantified with a Qubit double-stranded DNA (dsDNA) HS (high sensitivity) assay kit (Invitrogen), and 1 µg was fragmented by ultrasonication to an average size of 300 bp with a Covaris S220 focused ultrasonicator. To remove DNA fragments of unwanted size, we performed a double size selection using AMPure XP beads (Beckman Coulter) according to the manufacturer’s instructions. The final fragmented and size-selected genomic DNA was quality assessed with a DNA1000 chip on an Agilent Bioanalyzer. Illumina library preparation involves a cascade of enzymatic reactions, each followed by a cleanup step with AMPure XP beads. Fragmentation generates genomic DNA templates with a mixture of blunt ends and 5′ and 3′ overhangs. End repair, A-tailing, and adapter ligation reactions were performed on the fragmented DNA using the NEBNext DNA Library preparation kit for Illumina (New England Biolabs), according to the manufacturer’s recommended protocols. For the adapter ligation, we used 0.5 µl of a 15 µM double-stranded Y adapter, prepared by annealing Mod2_TS_Univ (ACGCTCTTCCGATC*T) and Mod2_TruSeq (Phos-GATCGGAAGAGCACACGTCTGAACTCCAGTCA). In the preceding oligonucleotides, the asterisk and Phos represent phosphorothioate and 5′ phosphate modifications, respectively. To specifically amplify transposon insertion sites by PCR, we used the transposon-specific primer Nspacer_barseq_pHIMAR (ATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNNNCGCCCTGCAGGGATGTCCACGAG), which contains a random hexamer and transposon-specific sequence on the 3′ end and an Illumina TruSeq sequence on the 5′ end, and one of 16 primers (see P7_MOD_TS_index primers in Table S2 in the supplemental material) containing the Illumina P7 end. For the transposon-insertion site enriching PCR, we used JumpStart Taq DNA polymerase (Sigma) in a 100-µl total volume with the following PCR program: 94°C for 2 min and 25 cycles of 94°C 30 s, 65°C for 20 s, and 72°C for 30 s, followed by a final extension at 72°C for 10 min. For the E. coli mutant library (KEIO_ML9), we replaced Nspacer_barseq_pHIMAR with Nspacer_barseq_universal (ATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNNNGATGTCCACGAGGTCT). The final PCR product was purified using AMPure XP beads according to the manufacturer’s instructions, eluted in 25 µl of water, and quantified on an Agilent Bioanalyzer with a DNA1000 chip. For all five mutant libraries, we first sequenced the TnSeq libraries on an Illumina MiSeq to assess the quality of the mutant library. The P. inhibens, P. stutzeri, S. amazonensis, and S. oneidensis TnSeq libraries were paired end sequenced (2 × 150 bp) on an Illumina MiSeq using the MiSeq reagent kit v2 (300 cycles). For KEIO_ML9, we performed single-end sequencing (1 × 150 bp) with the MiSeq reagent kit v3 (150 cycles). Each TnSeq library was also sequenced on either the HiSeq 2000 or HiSeq 2500 system (Illumina) to map a greater fraction of the mutant population.
TnSeq data analysis.
TnSeq reads were analyzed with a custom perl script (MapTnSeq.pl). For each read, the script looks for the flanking sequences U1 and U2 around the bar code and requires an exact match of 5 nucleotides on each side as well as a minimum quality score of 10 for each nucleotide in the bar code. This ensures that the bar code is of the correct length and is likely to be the correct sequence. The script then identifies the end of the transposon terminal repeat and requires an exact match for the last 5 nucleotides. The bar code and the end of the terminal repeat are allowed to be up to 2 nucleotides away from the expected position in the read. If the sequence past the end of the terminal repeat is 15 nucleotides or more, it is compared to the genome sequence and to the intact delivery plasmid (if any) with BLAT (33). Only hits with over 90% identity and a BLAT score (matches minus mismatches) of at least 15 are considered. Also, hits that begin more than 3 nucleotides after the junction are considered unreliable (see below). Similarly, some reads map to more than one location, and these are considered unreliable if the second-best score is up to 5 less than the best score.
A complication of interpreting our TnSeq data arises because the transposon insertion junctions are enriched by PCR. Given that we are mapping two unique regions (the bar code and genome insertion location) split by a transposon region that is identical in all mutants, we expect to see artifacts derived from chimeric PCR, as has been previously observed in 16S rRNA sequencing data (34, 35). One outcome of chimeric PCR is that a unique DNA bar code that is predominantly associated with a single insertion location will also appear linked to secondary insertion sites at a significantly lower frequency.
To empirically demonstrate that our RB-TnSeq approach generates chimeric PCR artifacts, we isolated 96 individual, randomly bar-coded P. stutzeri transposon mutant strains, and for 63 of these strains, we successfully identified the bar code and the insertion location of the transposon by arbitrary PCR and Sanger sequencing. To enable a direct comparison to the individual mutant results, we pooled all 96 strains and performed TnSeq. TnSeq identified matching bar codes for 52 of the 63 strains and predominantly linked each of them to roughly the same site that was identified by Sanger sequencing. For 41 of the strains, the two methods mapped to within 2 nucleotides of each other. On average, 93% of the TnSeq reads mapped to the primary location and the other 7% of the reads mapped to diverse other locations, with the second most common location accounting for just 0.7% of the reads (see Fig. S3 in the supplemental material).
Given the mapped reads, a set of bar codes that consistently map to a unique location in the genome is identified with a custom perl script (DesignRandomPool.pl). Because of chimeric PCR (as described above), a bar code will generally not map to a single location with 100% consistency, but bar codes that legitimately map to more than one location in the genome need to be removed (see Fig. S3 in the supplemental material). A bar code is considered to map uniquely if the bar code matches reliably to its primary location (to the exact nucleotide) at least 10 times, if these primary matches account for 75% of the reads for that bar code, and if the second most frequent location occurs with at most 1/8 of the frequency of the primary location. In practice, we were able to use a large fraction (42 to 92%) of the bar codes (Table 1). Finally, recurring errors in bar codes are removed by looking for cases where two bar codes map to the same location and are identical up to a single nucleotide error. The more common bar code is assumed to be correct, and the other is removed.
For the comparison of TnSeq and BarSeq fitness data (Fig. 3A and B), the TnSeq data were analyzed in the same way as the BarSeq data (see below), once the data were in the form of a count for each insertion location in each sample.
Genome sequencing.
We sequenced genomic DNA from select S. amazonensis mutant library samples using the NEBNext DNA Library Prep kit for Illumina according to the manufacturer’s protocol (New England Biolabs). Briefly, 1 µg of genomic DNA was fragmented by ultrasonication to an average size of 300 bp with a Covaris S220 focused ultrasonicator. After end-repair, A-tailing, and ligation of the same Y adapter used for TnSeq, we size selected 400-bp products with AMPure XP beads. The sequencing libraries were eluted in 25 µl of water and quantified on a Bioanalyzer with a DNA1000 chip (Agilent). We performed paired-end sequencing (2 × 150 bp) on an Illumina MiSeq using MiSeq reagent kit v2 (300 cycles). The genome sequencing data were mapped to the genome using bowtie 0.12.8 (36).
Competitive mutant fitness assays.
A single aliquot of a mutant library was thawed, inoculated into 25 ml of medium supplemented with kanamycin, and grown to mid-log phase. For each bacterium, we used the same medium, kanamycin concentration, and growth temperature that were used for the mutant library construction. After the mutant library recovered and reached mid-log phase, we collected cell pellets as a common reference for BarSeq (termed time-zero samples) and used the remaining cells to set up competitive mutant fitness assays under different experimental conditions at a starting OD600 of 0.02. For carbon source utilization experiments in defined medium, we washed the recovered cells twice in the 2× defined medium without an added carbon source prior to inoculation. The defined medium formulations used for each experiment are contained in Data Set S1 in the supplemental material. The mutant library experiments were grown either in glass tubes with 10-ml volumes or in the wells of a 48-well microplate (700 µl per well). We grew the microplates in Tecan Infinite F200 readers with orbital shaking and OD600 readings every 15 min. In general, all of the mutant library assays reached saturated growth, and we typically collected ~2 × 109 cells (~1 ml of a 1.0-OD600 culture) for genomic DNA extraction. For the microplate experiments, we combined the contents of two replicate wells (1.4-ml total volume) prior to collecting the pellet. Mutant library cell pellets were typically stored at −80°C prior to genomic DNA extraction.
BarSeq.
We isolated genomic DNA from mutant library samples either using the DNeasy Blood and Tissue kit (Qiagen) or in an automated, 96-well format with a QIAxtractor (Qiagen). Genomic DNA was quantified with the Quant-iT dsDNA BR assay kit (Invitrogen). We performed BarSeq PCR in a 50-µl total volume with 20 µmol of each primer and 150 to 200 ng of template genomic DNA. In this study, we used two sets of BarSeq PCR primers and two PCR conditions. Our original design utilized a common forward oligonucleotide (Forward_primer_Nbarseq) and one of 48 reverse primers (HX_R_primer_Nbarseq) with unique 8-bp indexes that were sequenced “in line” with the random DNA bar code from a single Illumina sequencing primer. The second BarSeq design utilized a common reverse primer (BarSeq_P1) and one of 96 forward primers (BarSeq_P2_ITXXX) containing unique 6-bp TruSeq indexes that were sequenced using a separate index primer (see, in the supplemental material, Table S2 for oligonucleotide details and Data Set S1 for details on which BarSeq oligonucleotides were used for each experiment). We found no differences in data quality between the two sets of BarSeq primers (data not shown). The second oligonucleotide design (with TruSeq indexes) is automatically demultiplexed by the Illumina software, and we therefore recommend the use of this set of primers.
We used two PCR conditions for BarSeq, which surprisingly gave results of different quality. Our original BarSeq PCRs were performed with Phusion polymerase (New England Biolabs) in the presence of dimethyl sulfoxide (DMSO): 95°C for 4 min and 25 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, followed by a final extension for 7 min at 72°C. We found that fitness data generated with this PCR method (termed here BarSeq95) were noisy for some experiments, particularly for the higher-GC-content bacteria P. inhibens and P. stutzeri. Specifically, we found that adjacent genes (not in the same operon) often had correlated fitness and that these patterns were influenced by the local GC content of the chromosome (data not shown). We speculate that the genomic DNA (gDNA) was not sufficiently denatured at 95°C and that the efficiency of denaturation is affected by GC content. Therefore, during the course of this work, we transitioned to a new BarSeq method: Q5 DNA polymerase with Q5 GC enhancer and the standard Q5 reaction buffer (New England Biolabs) under the following cycling conditions: 98°C for 4 min followed by 25 cycles of 30 s at 98°C, 30 s at 55°C, and 30 s at 72°C, followed by a final extension at 72°C for 5 min. We term this method BarSeq98, as the vast majority of the GC-associated noise was removed by denaturing at 98°C instead of 95°C. We recommend using the BarSeq98 protocol for all microbes, regardless of the organism’s GC content. Details on which BarSeq method was used for each experiment are contained in Data Set S1 in the supplemental material.
Equal volumes (10 µl) of the individual BarSeq PCRs were pooled, and 200 µl of the pooled PCR product was purified with the DNA Clean and Concentrator kit (Zymo Research). The final BarSeq library was eluted in 30 µl water and quantified using the Qubit dsDNA HS assay kit. The BarSeq libraries were sequenced on either an Illumina HiSeq 2000 or an Illumina HiSeq 2500 instrument. We usually multiplex 48 samples per lane, but for the E. coli library, for which most of the bar codes are informative, we multiplexed 96 samples per lane. For early test runs for S. amazonensis, we multiplexed only 24 samples per lane, and there are a few other lanes that for convenience contained 40 to 45 samples instead of the full 48 samples; note that many of these multiplexed samples are not described in this work.
BarSeq data analysis and calculation of gene fitness.
BarSeq reads were converted to a table of the number of times that each bar code was seen in each sample using a custom perl script (MultiCodes.pl). The script requires an exact match to the 8 nucleotides at the beginning of the read that identify the sample (“inline” indexes), or relies on Illumina software for demultiplexing (TruSeq P7 indexes), depending on the primer design (see “BarSeq” above). The script also requires an exact match for the 9 nucleotides upstream of the bar code. We did not check the quality scores for the bar code or the sequence downstream of the bar code (the -minQuality 0 option). However, bar codes that do not match exactly an expected bar code are ignored in later stages of the analysis.
Given a table of bar codes, where they map in the genome, and their counts in each sample, we estimate strain fitness and gene fitness values and their reliability with a custom R script (FEBA.R). Roughly, strain fitness is the normalized log2 ratio of counts between the treatment sample (i.e., after growth in a certain medium) and the reference “time-zero” sample. Gene fitness is the weighted average of the strain fitness, and a t score is computed based on the consistency of the strain fitness values for each gene. Ideally, the time-zero and treatment samples are sequenced in the same lane. Also, we usually have multiple replicates of any given time zero, with independent extraction of genomic DNA and independent PCR with a different index. We sum the per-strain counts across replicate time-zero samples.
(i) Gene fitness.
The gene fitness is the weighted average of the strain fitness. Strains with more reads have less noisy fitness estimates and are weighted more highly, but not too highly, as rare outlying strains can have many reads (possibly indicating positive selection for a secondary mutation). Also, because the per-strain fitness values are very noisy, information from other strains (other insertions in the same gene) is used to help estimate them.
In more detail, we first select a subset of strains and genes that have adequate coverage in the time-zero samples (3 reads per strain and 30 reads per gene, considering only the adequate strains). Only strains that lie within the central 10 to 90% of a gene are considered. Then, for each experiment,
where ψ is a “pseudocount” (see below) to avoid having a very noisy estimate when the counts are low, nAfter is the bar code count in the treatment, and n0 is the bar code count in the control time zero. If the counts are not too small and the noise in the per-strain counts follows the Poisson distribution (i.e., the variance is equal to the mean), then the variance of the per-strain fitness estimate will be approximately:
This estimate of the strain variance can be derived from normal fluctuations in log2 (nAfter/n0) and Poisson noise in the counts (i.e., the variance of a count equals its true value). In reality, the counts have a bit more than Poisson noise; this will be addressed by the t-like test statistic.
The (unnormalized) gene fitness fu is then the weighted average of the strain fitness, with the weight inversely proportional to the strain variance. This is the optimal estimate if there are no unaccounted sources of noise. However, because there are (rare) outlier strains with high counts, we impose a ceiling on the weight wi, with the maximum weight being that of a strain with 20 reads in each sample:
(ii) Pseudocounts.
A natural value for ψ is sum(nAfter)/sum(n0), where the sum is over all strains in all genes. However, because there is limited information in the reads for each strain, the resulting gene fitness values will be biased toward zero. For example, consider a gene with very low fitness so that nAfter is around 0 for all strains. For simplicity, assume that we have similar numbers of reads for each sample so that ψ = 1. Then, the per-strain fitness will be −log2(1 + n0). If the typical strain has only 7 reads, then strain fitness will be around −3 and so will per-gene fitness, even though the correct value is much lower. Intuitively, we should use the information from other strains to adjust the pseudocount and reduce this bias. Actually, we set:
For genes with just 1 or 2 strains, gene_factor is 1, as we have insufficient information to improve the estimate. Otherwise, we compute a preliminary fitness value for each gene as the median of the preliminary fitness values for the strains, using a pseudocount of 1. The gene fitness values are normalized so that the median is zero, and then gene_factor is 2fu.
A simple simulation illustrates that the more complicated pseudocounts give less biased estimates of gene fitness (see Fig. S4 in the supplemental material). Also, compared to naively summing the count across the gene, the weighted averages are much less sensitive to outlier strains (see Fig. S4). This approach yields better consistency between gene fitness estimates for the first and second halves of genes than naively summing counts across the entire gene, as is typically done when analyzing TnSeq experiments (see Fig. S4).
(iii) Normalization.
The unnormalized gene fitness values fu (from the weighted average of strain fitness values) are normalized separately for each scaffold because of potential bias during genomic DNA extraction. For large scaffolds, we try to remove the bias from the variation of DNA copy number across the chromosome (see Results and also Fig. S2 in the supplemental material). To estimate this bias, we use the smoothed median of the gene fitness values across each large scaffold. The largest functional cluster in bacteria that we know of contains 51 motility genes in Sinorhizobium meliloti. We use the median within a window of 251 genes so that genuine biological effects are unlikely to be removed; also, an odd window size is computationally convenient. We subtract the local median (the estimated bias) from the gene fitness values. Then, for each large scaffold, we subtract the mode (as estimated from the maximum of the kernel density); this reflects an assumption that most genes have subtle or no effects on fitness (37). For smaller scaffolds, we just subtract the median.
(iv) t-like test statistic.
To estimate the reliability of the fitness measurement for each gene f, we use a moderated t statistic:
where σ is a small constant (we use 0.1) that represents uncertainty in the normalization for small fitness values, Ve represents the estimated variance, and Vn represents the naive variance. We estimate the variance in two different ways. First, to prevent underestimates of the variance in some cases, we use an alternate “naive” variance estimate, Vn, based on the best-case Poisson noise:
Second, we look at the observed consistency of the measurements for the gene and take into account data from other genes, especially if there are few strains for this gene:
where n is the number of strains and Vg is a prior estimate of the variance in gene fitness. Note that the left term in the estimated variance is a weighted sum of squared differences of strain fitness for the gene, and its expectation is n − 1 times the variance in fg. (This is a consequence of weighting by 1/variance.) Thus, the expectation of this estimate of the variance is correct.
The intuition behind our prior variance estimate Vg is that the a priori noise in a gene’s fitness measurement depends on the total number of reads. To estimate the overall reliability of the gene measurements, we estimate per-gene fitness using only insertions within 10 to 50% and 50 to 90% of each gene. Genes without at least 15 time-zero reads on each side are excluded. The median absolute difference between the two halves of each gene (mad12) gives a robust estimate of the overall level of noise in the experiment. To convert from mad12 to the variance in the typical gene Vt, we use Vt = mad122/[2 × q(0.75)]2, where q(0.75) = 0.674 is the 75th percentile of the normal distribution. It is easy to show that the expectation of the absolute difference of two random values from the standard normal distribution is . An additional factor of 2 arises because the variance in the fitness estimate for a typical gene should be half the variance of the first-half or second-half estimates. As the variance of a gene’s fitness estimate should decline linearly with additional reads, we then have Vg = Vt × [Vn/median (Vn)], where the median is over genes used to estimate Vt.
To verify that the moderated t statistic has a good fit to the standard normal distribution when there are no genuine fitness differences, we performed control comparisons between replicate time-zero samples (see Fig. S5 in the supplemental material).
For each organism, the number of false positives for genes with phenotypes was estimated as nFalsePositivesInControls × nExperiments/nControlExperiments. Control experiments are comparisons of one time-zero sample versus another from the same day and in the same Illumina sequencing lane. Time-zero samples from old PCR conditions (95°C denaturing) for P. inhibens and P. stutzeri were excluded. The false discovery rate was estimated as EstimatedFalsePositives/nGenesWithPhenotypes and was under 2% for each organism.
(v) Assessment of experiment quality.
We classified the quality of each BarSeq fitness experiment using the following rules: the median gene had at least 50 BarSeq counts (gMed ≥ 50), the median absolute difference in fitness between the first and second halves of the genes was less than or equal to 0.5 (mad12 ≤ 0.5), the Spearman correlation in fitness between the first and second halves of the genes was at least 0.1 (cor12 ≥ 0.1), the correlation between gene GC content and fitness was less than or equal to 0.2 (gccor ≤ 0.2), the Spearman correlation of adjacent genes on different strains was no greater than 0.25 (adjcor ≤ 0.25), and the experiment was not a time-zero sample. A more detailed description of these metrics and their values for each BarSeq experiment are contained in Data Set S1 in the supplemental material. Overall, 387 of the 501 BarSeq assays that we performed met each of these metrics and were deemed successful.
(vi) Polar effects.
We believe that polar effects, in which an insertion in a gene has a phenotype because it disrupts the expression of downstream genes, are not a major factor in our data. To test for polar effects, we looked for cases where the upstream gene in an operon has a strong phenotype but the downstream gene does not, or vice versa. If polar effects are common, then whenever the downstream gene is important for fitness, the upstream gene should be important as well; so, if genes in an operon have different fitness values, it should usually be the upstream gene that has the stronger phenotypes. We considered cases where one gene in an operon had a fitness of <−1 and a t of <−4 and the other gene had a fitness at least 0.75 higher. In each bacterium, cases where only the upstream gene had the phenotype were somewhat more common, with ratios ranging from 1.25:1 (for P. inhibens) to 1.38:1 (for P. stutzeri). Using simpler pools of mutants and microarray to quantify the abundance of each strain, we previously reported a ratio of 1.5 for a similar test in S. oneidensis MR-1 (2).
Another way to examine polar effects is to consider in which orientation the transposon is inserted. Our transposons do not contain transcription terminators (as far as we know), so any termination of transcription should be due to the action of rho on untranslated transcripts. If the antibiotic resistance marker is in the opposite orientation from the mutated gene, then rho might terminate transcription anywhere in the transposon. But, if the antibiotic resistance marker is in the same orientation as the gene, then this should not occur. To test whether the orientation of the insertion has an effect, we analyzed 2,470 genes from S. amazonensis SB2B that had sufficient sequencing coverage of insertions in both orientations (at least 50 reads in the time-zero sample). To look for systematic effects, we took the median across defined-medium experiments. Insertions in either orientation had very similar effects (r = 0.97).
Code availability.
Code for analyzing RB-TnSeq data is available at https://bitbucket.org/berkeleylab/feba. All analyses were performed with release 1.0.0. Raw sequence data and processed fitness values are available from http://genomics.lbl.gov/supplemental/rbarseq/ along with scripts for reproducing all of our results.
SUPPLEMENTAL MATERIAL
Fitness experiments and media. Download
Insertion coverage of transposon mutant libraries. Frequency of unique transposon insertion locations across the main chromosome of E. coli (A), P. inhibens (B), P. stutzeri (C), S. amazonensis (D), and S. oneidensis (E). Download
Normalization of BarSeq mutant fitness data. (A) Genome sequencing counts plotted in 10-kB intervals (y axis) by chromosome position (x axis) for two samples of the S. amazonensis mutant library: growth in defined medium with d-glucose as a carbon source and the time-zero control (grown in LB). The defined-growth sample was collected in stationary phase, while the time-zero control was collected during the exponential phase of growth. Note the biased genome sequencing counts for the time-zero control relative to the defined-medium sample. (B) For the two samples in panel A, the median log2 ratio of the genome sequencing counts plotted in 10-kB intervals (y axis) by chromosome position (x axis). The green line represents the estimated bias of the fitness data, as determined solely from BarSeq data for the same two samples (see Materials and Methods for details). (C) Unnormalized gene fitness data (y axis) plotted by chromosome position (x axis) for S. amazonensis grown in a defined medium with d-maltose as the carbon source. The green line is the BarSeq-derived estimate of chromosome position bias from gDNA copy number variation between the d-maltose-grown sample and the time-zero control. (D) The fitness data in panel C normalized by our estimate of gDNA copy variation (the green line in panel C). Postnormalization, the green line is set at y = 0. Download
Filtering out of chimeric reads from TnSeq. (A) From a set of 96 transposon mutants of P. stutzeri, we identified both the insertion location and bar code for 63 of the strains individually by arbitrary PCR and Sanger sequencing. In addition, we pooled the 96 strains and performed TnSeq. For 52 of the 63 strains mapped by arbitrary PCR, TnSeq identified the same insertion location and bar code. For these 52 strains, the x axis shows the number of TnSeq reads that mapped the bar code to its correct location, and the y axis shows the number of times that the bar code mapped to all other locations. The green line represents the best fit for the data with a linear regression model (roughly 13:1; the 95% confidence region is in gray). The dotted red line shows x = y. (B) Number of TnSeq reads mapped to the primary location (x axis) and to the most frequent secondary location (y axis) for 1,000 randomly selected bar codes from the full P. stutzeri mutant library (psRCH2_ML7). Only bar codes with at least 10 TnSeq reads for their primary location were considered for inclusion. Jitter was added to the secondary count, and for bar codes that mapped to only one location, the secondary count is shown as 0.5. A bar code is considered informative if (i) at least 75% of its reads map to the primary location and (ii) its second most frequent location is at least 8 times less frequent than its primary location. Strains that map primarily to the intact transposon vector were not included. The dotted red line shows x = y. (C) Same as panel B for the full S. amazonensis mutant library SB2B_ML5. Download
Validation of our approach for estimating gene fitness with simulated and genuine data. Our simulation includes 2,000 genes whose fitness follows a normal distribution with a mean of 0 and a standard deviation of 1.5. Each gene has 20 strains, and each strain’s fitness equals the gene fitness except for 100 randomly selected outlier strains, which have a fitness of +6. In the time-zero sample, each strain follows a binomial distribution with an expectation of 20 reads. In the treatment sample, each strain follows a binomial distribution with an expectation of 20 × 2**fitness/sum(2**fitness) reads. (A to C) Estimated gene fitness versus true gene fitness in the simulation, for ‘“naive” gene fitness from the log2 ratios of the total counts for the strains (A), our approach (B), or our approach without the per-gene factor in the pseudocount (C). (D and E) Consistency of fitness data across the two halves of genes for S. amazonensis growing in a defined d,l-lactate medium, computed using the “naive” approach (D) or our approach (E). Download
Distributions of our moderated t-statistic for control comparisons between replicate time-zero samples. Each distribution is shown as a quartile-quartile plot, and each gray line represents a different comparison. A perfect fit to the standard normal distribution would be on the x = y line (in black). Outliers with |t| > 5 are marked. (A) E. coli; (B) P. inhibens; (C) P. stutzeri; (D) S. amazonensis; (E) S. oneidensis. Download
Strains used in this study
Oligonucleotides used in this study
Vectors used in this study
ACKNOWLEDGMENTS
RB-TnSeq development was funded by ENIGMA. The work conducted by ENIGMA was supported by the Office of Science, Office of Biological and Environmental Research of the U.S. Department of Energy, under contract DE-AC02-05CH11231. The P. inhibens, E. coli, and S. amazonensis BarSeq mutant fitness data were supported by Laboratory Directed Research and Development (LDRD) funding from Berkeley Lab, provided by the Director, Office of Science, of the U.S. Department of Energy under contract DE-AC02-05CH11231 and a Community Science Project from the Joint Genome Institute to M.J.B., J.B., A.P.A., and A.D. The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy under contract no. DE-AC02-05CH11231.
K.M.W., M.N.P., M.J.B., J.B., G.B., A.P.A., and A.D. designed the research. K.M.W., R.J.W., J.S.L., J.H., C.A.H., and A.D. performed the experiments. M.N.P. and A.D. analyzed the data. K.M.W., M.N.P., and A.D. wrote the paper.
Footnotes
Citation Wetmore KM, Price MN, Waters RJ, Lamson JS, He J, Hoover CA, Blow MJ, Bristow J, Butland G, Arkin AP, Deutschbauer A. 2015. Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. mBio 6(3):e00306-15. doi:10.1128/mBio.00306-15.
REFERENCES
- 1.Deutschbauer A, Price MN, Wetmore KM, Tarjan DR, Xu Z, Shao W, Leon D, Arkin AP, Skerker JM. 2014. Towards an informative mutant phenotype for every bacterial gene. J Bacteriol 196:3643–3655. doi: 10.1128/JB.01836-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deutschbauer A, Price MN, Wetmore KM, Shao W, Baumohl JK, Xu Z, Nguyen M, Tamse R, Davis RW, Arkin AP. 2011. Evidence-based annotation of gene function in Shewanella oneidensis MR-1 using genome-wide fitness profiling across 121 conditions. PLoS Genet 7:e1002385. doi: 10.1371/journal.pgen.1002385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuehl JV, Price MN, Ray J, Wetmore KM, Esquivel Z, Kazakov AE, Nguyen M, Kuehn R, Davis RW, Hazen TC, Arkin AP, Deutschbauer A. 2014. Functional genomics with a comprehensive library of transposon mutants for the sulfate-reducing bacterium Desulfovibrio alaskensis G20. mBio 5(3):e01041-14. doi: 10.1128/mBio.01041-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nichols RJ, Sen S, Choo YJ, Beltrao P, Zietek M, Chaba R, Lee S, Kazmierczak KM, Lee KJ, Wong A, Shales M, Lovett S, Winkler ME, Krogan NJ, Typas A, Gross CA. 2011. Phenotypic landscape of a bacterial cell. Cell 144:143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Opijnen T, Camilli A. 2013. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat Rev Microbiol 11:435–442. doi: 10.1038/nrmicro3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Opijnen T, Bodi KL, Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods 6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langridge GC, Phan MD, Turner DJ, Perkins TT, Parts L, Haase J, Charles I, Maskell DJ, Peters SE, Dougan G, Wain J, Parkhill J, Turner AK. 2009. Simultaneous assay of every Salmonella typhi gene using one million transposon mutants. Genome Res 19:2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gawronski JD, Wong SM, Giannoukos G, Ward DV, Akerley BJ. 2009. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc Natl Acad Sci U S A 106:16422–16427. doi: 10.1073/pnas.0906627106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodman AL, Wu M, Gordon JI. 2011. Identifying microbial fitness determinants by insertion sequencing using genome-wide transposon mutant libraries. Nat Protoc 6:1969–1980. doi: 10.1038/nprot.2011.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fels SR, Zane GM, Blake SM, Wall JD. 2013. Rapid transposon liquid enrichment sequencing (TnLE-seq) for gene fitness evaluation in underdeveloped bacterial systems. Appl Environ Microbiol 79:7510–7517. doi: 10.1128/AEM.02051-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein BA, Tenorio EL, Lazinski DW, Camilli A, Duncan MJ, Hu LT. 2012. Identification of essential genes of the periodontal pathogen Porphyromonas gingivalis. BMC Genomics 13:578. doi: 10.1186/1471-2164-13-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian K-D, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Güldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kötter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang C-Y, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 13.Smith AM, Heisler LE, Mellor J, Kaper F, Thompson MJ, Chee M, Roth FP, Giaever G, Nislow C. 2009. Quantitative phenotyping via deep bar code sequencing. Genome Res 19:1836–1842. doi: 10.1101/gr.093955.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oh J, Fung E, Price MN, Dehal PS, Davis RW, Giaever G, Nislow C, Arkin AP, Deutschbauer A. 2010. A universal TagModule collection for parallel genetic analysis of microorganisms. Nucleic Acids Res 38:e146–e146. doi: 10.1093/nar/gkq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh J, Fung E, Schlecht U, Davis RW, Giaever G, St Onge RP, Deutschbauer A, Nislow C. 2010. Gene annotation and drug target discovery in Candida albicans with a tagged transposon mutant collection. PLoS Pathog 6:e1001140. doi: 10.1371/journal.ppat.1001140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hillenmeyer ME, Ericson E, Davis RW, Nislow C, Koller D, Giaever G. 2010. Systematic analysis of genome-wide fitness data in yeast reveals novel gene function and drug action. Genome Biol 11:R30. doi: 10.1186/gb-2010-11-3-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee AY, St Onge RP, Proctor MJ, Wallace IM, Nile AH, Spagnuolo PA, Jitkova Y, Gronda M, Wu Y, Kim MK, Cheung-Ong K, Torres NP, Spear ED, Han MK, Schlecht U, Suresh S, Duby G, Heisler LE, Surendra A, Fung E, Urbanus ML, Gebbia M, Lissina E, Miranda M, Chiang JH, Aparicio AM, Zeghouf M, Davis RW, Cherfils J, Boutry M, Kaiser CA, Cummins CL, Trimble WS, Brown GW, Schimmer AD, Bankaitis VA, Nislow C, Bader GD, Giaever G. 2014. Mapping the cellular response to small molecules using chemogenomic fitness signatures. Science 344:208–211. doi: 10.1126/science.1250217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith AM, Heisler LE, St Onge RP, Farias-Hesson E, Wallace IM, Bodeau J, Harris AN, Perry KM, Giaever G, Pourmand N, Nislow C. 2010. Highly-multiplexed bar code sequencing: an efficient method for parallel analysis of pooled samples. Nucleic Acids Res 38:e142. doi: 10.1093/nar/gkq368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goryshin IY, Jendrisak J, Hoffman LM, Meis R, Reznikoff WS. 2000. Insertional transposon mutagenesis by electroporation of released Tn5 transposition complexes. Nat Biotechnol 18:97–100. doi: 10.1038/72017. [DOI] [PubMed] [Google Scholar]
- 20.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joyce AR, Reed JL, White A, Edwards R, Osterman A, Baba T, Mori H, Lesely SA, Palsson BØ, Agarwalla S. 2006. Experimental and computational assessment of conditionally essential genes in Escherichia coli. J Bacteriol 188:8259–8271. doi: 10.1128/JB.00740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alvarez-Añorve LI, Calcagno ML, Plumbridge J. 2005. Why does Escherichia coli grow more slowly on glucosamine than on N-acetylglucosamine? Effects of enzyme levels and allosteric activation of GlcN6P deaminase (NagB) on growth rates. J Bacteriol 187:2974–2982. doi: 10.1128/JB.187.9.2974-2982.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boulineau S, Tostevin F, Kiviet DJ, Wolde ten PR, Nghe P, Tans SJ. 2013. Single-cell dynamics reveals sustained growth during diauxic shifts. PLoS One 8:e61686. doi: 10.1371/journal.pone.0061686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiegmann K, Hensler M, Wöhlbrand L, Ulbrich M, Schomburg D, Rabus R. 2014. Carbohydrate catabolism in Phaeobacter inhibens DSM 17395, a member of the marine Roseobacter clade. Appl Environ Microbiol 80:4725–4737. doi: 10.1128/AEM.00719-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodionov DA, Yang C, Li X, Rodionova IA, Wang Y, Obraztsova AY, Zagnitko OP, Overbeek R, Romine MF, Reed S, Fredrickson JK, Nealson KH, Osterman AL. 2010. Genomic encyclopedia of sugar utilization pathways in the Shewanella genus. BMC Genomics 11:494. doi: 10.1186/1471-2164-11-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eames M, Kortemme T. 2012. Cost-benefit tradeoffs in engineered lac operons. Science 336:911–915. doi: 10.1126/science.1219083. [DOI] [PubMed] [Google Scholar]
- 27.Van Opijnen T, Camilli A. 2012. A fine scale phenotype-genotype virulence map of a bacterial pathogen. Genome Res 22:2541–2551. doi: 10.1101/gr.137430.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer B, Kuehl J, Deutschbauer AM, Price MN, Arkin AP, Stahl DA. 2013. Variation among Desulfovibrio species in electron transfer systems used for syntrophic growth. J Bacteriol 195:990–1004. doi: 10.1128/JB.01959-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rey FE, Gonzalez MD, Cheng J, Wu M, Ahern PP, Gordon JI. 2013. Metabolic niche of a prominent sulfate-reducing human gut bacterium. Proc Natl Acad Sci U S A 110:13582–13587. doi: 10.1073/pnas.1312524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burke C, Liu M, Britton W, Triccas JA, Thomas T, Smith AL, Allen S, Salomon R, Harry E. 2013. Harnessing single cell sorting to identify cell division genes and regulators in bacteria. PLoS One 8:e60964. doi: 10.1371/journal.pone.0060964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bouhenni R, Gehrke A, Saffarini D. 2005. Identification of genes involved in cytochrome c biogenesis in Shewanella oneidensis, using a modified mariner transposon. Appl Environ Microbiol 71:4935–4937. doi: 10.1128/AEM.71.8.4935-4937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larsen RA, Wilson MM, Guss AM, Metcalf WW. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch Microbiol 178:193–201. doi: 10.1007/s00203-002-0442-2. [DOI] [PubMed] [Google Scholar]
- 33.Kent WJ. 2002. BLAT—the blast-like alignment tool. Genome Res 12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methé B, DeSantis TZ, Human Microbiome Consortium, Petrosino JF, Knight R, Birren BW. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol 71:7724–7736. doi: 10.1128/AEM.71.12.7724-7736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price MN, Deutschbauer AM, Skerker JM, Wetmore KM, Ruths T, Mar JS, Kuehl JV, Shao W, Arkin AP. 2013. Indirect and suboptimal control of gene expression is widespread in bacteria. Mol Syst Biol 9:660. doi: 10.1038/msb.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peterson JD, Umayam LA, Dickinson T, Hickey EK, White O. 2001. The comprehensive microbial resource. Nucleic Acids Res 29:123–125. doi: 10.1093/nar/29.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fitness experiments and media. Download
Insertion coverage of transposon mutant libraries. Frequency of unique transposon insertion locations across the main chromosome of E. coli (A), P. inhibens (B), P. stutzeri (C), S. amazonensis (D), and S. oneidensis (E). Download
Normalization of BarSeq mutant fitness data. (A) Genome sequencing counts plotted in 10-kB intervals (y axis) by chromosome position (x axis) for two samples of the S. amazonensis mutant library: growth in defined medium with d-glucose as a carbon source and the time-zero control (grown in LB). The defined-growth sample was collected in stationary phase, while the time-zero control was collected during the exponential phase of growth. Note the biased genome sequencing counts for the time-zero control relative to the defined-medium sample. (B) For the two samples in panel A, the median log2 ratio of the genome sequencing counts plotted in 10-kB intervals (y axis) by chromosome position (x axis). The green line represents the estimated bias of the fitness data, as determined solely from BarSeq data for the same two samples (see Materials and Methods for details). (C) Unnormalized gene fitness data (y axis) plotted by chromosome position (x axis) for S. amazonensis grown in a defined medium with d-maltose as the carbon source. The green line is the BarSeq-derived estimate of chromosome position bias from gDNA copy number variation between the d-maltose-grown sample and the time-zero control. (D) The fitness data in panel C normalized by our estimate of gDNA copy variation (the green line in panel C). Postnormalization, the green line is set at y = 0. Download
Filtering out of chimeric reads from TnSeq. (A) From a set of 96 transposon mutants of P. stutzeri, we identified both the insertion location and bar code for 63 of the strains individually by arbitrary PCR and Sanger sequencing. In addition, we pooled the 96 strains and performed TnSeq. For 52 of the 63 strains mapped by arbitrary PCR, TnSeq identified the same insertion location and bar code. For these 52 strains, the x axis shows the number of TnSeq reads that mapped the bar code to its correct location, and the y axis shows the number of times that the bar code mapped to all other locations. The green line represents the best fit for the data with a linear regression model (roughly 13:1; the 95% confidence region is in gray). The dotted red line shows x = y. (B) Number of TnSeq reads mapped to the primary location (x axis) and to the most frequent secondary location (y axis) for 1,000 randomly selected bar codes from the full P. stutzeri mutant library (psRCH2_ML7). Only bar codes with at least 10 TnSeq reads for their primary location were considered for inclusion. Jitter was added to the secondary count, and for bar codes that mapped to only one location, the secondary count is shown as 0.5. A bar code is considered informative if (i) at least 75% of its reads map to the primary location and (ii) its second most frequent location is at least 8 times less frequent than its primary location. Strains that map primarily to the intact transposon vector were not included. The dotted red line shows x = y. (C) Same as panel B for the full S. amazonensis mutant library SB2B_ML5. Download
Validation of our approach for estimating gene fitness with simulated and genuine data. Our simulation includes 2,000 genes whose fitness follows a normal distribution with a mean of 0 and a standard deviation of 1.5. Each gene has 20 strains, and each strain’s fitness equals the gene fitness except for 100 randomly selected outlier strains, which have a fitness of +6. In the time-zero sample, each strain follows a binomial distribution with an expectation of 20 reads. In the treatment sample, each strain follows a binomial distribution with an expectation of 20 × 2**fitness/sum(2**fitness) reads. (A to C) Estimated gene fitness versus true gene fitness in the simulation, for ‘“naive” gene fitness from the log2 ratios of the total counts for the strains (A), our approach (B), or our approach without the per-gene factor in the pseudocount (C). (D and E) Consistency of fitness data across the two halves of genes for S. amazonensis growing in a defined d,l-lactate medium, computed using the “naive” approach (D) or our approach (E). Download
Distributions of our moderated t-statistic for control comparisons between replicate time-zero samples. Each distribution is shown as a quartile-quartile plot, and each gray line represents a different comparison. A perfect fit to the standard normal distribution would be on the x = y line (in black). Outliers with |t| > 5 are marked. (A) E. coli; (B) P. inhibens; (C) P. stutzeri; (D) S. amazonensis; (E) S. oneidensis. Download
Strains used in this study
Oligonucleotides used in this study
Vectors used in this study