Drosophila immune cells undergo four stereotypical developmental migrations to populate the embryo. Wash is a downstream effector of Rho1 and establishes Rho1>Wash>Arp2/3 as the regulatory pathway controlling the cytoskeleton during one of these developmental hemocyte migrations in a WASH regulatory complex–independent manner.
Abstract
Drosophila immune cells, the hemocytes, undergo four stereotypical developmental migrations to populate the embryo, where they provide immune reconnoitering, as well as a number of non–immune-related functions necessary for proper embryogenesis. Here, we describe a role for Rho1 in one of these developmental migrations in which posteriorly located hemocytes migrate toward the head. This migration requires the interaction of Rho1 with its downstream effector Wash, a Wiskott–Aldrich syndrome family protein. Both Wash knockdown and a Rho1 transgene harboring a mutation that prevents Wash binding exhibit the same developmental migratory defect as Rho1 knockdown. Wash activates the Arp2/3 complex, whose activity is needed for this migration, whereas members of the WASH regulatory complex (SWIP, Strumpellin, and CCDC53) are not. Our results suggest a WASH complex–independent signaling pathway to regulate the cytoskeleton during a subset of hemocyte developmental migrations.
INTRODUCTION
Guided developmental cell migrations require complex signal integration and regulation of actin and microtubule cytoskeletons to provide dynamic cell infrastructure and communication in a three-dimensional setting (Rodriquez, 2003; Yamazaki et al., 2005; Wood and Jacinto, 2007; Yamaguchi and Condeelis, 2007; Insall and Machesky, 2009). Drosophila hemocytes (macrophages) provide an excellent in vivo cell migration model, as they undergo characteristic developmental migrations, as well as chemotactic migrations toward wounds or pathogen invasion (Tepass et al., 1994; Strong et al., 2005; Wood et al., 2006; Wood and Jacinto, 2007; Siekhaus et al., 2010). Hemocytes are highly polarized and exhibit exuberant actin-rich cellular protrusions (Figure 1A). Hemocytes originate in the head mesoderm at stage 10 and undergo four major stereotypical developmental migrations (Figure 1B). 1) A subset of hemocytes leave the head mesoderm in stage 10 embryos and travel to the posterior tail region through an invasive epithelial transmigration. 2) In stage 12–14 embryos, the hemocytes remaining in the head region migrate posteriorly along the ventral midline and dorsal vessel. 3) Also in stage 12–14 embryos, tail hemocytes (the transmigrated hemocytes that are now at the posterior end of embryo after germ-band retraction) move anteriorly along the ventral midline. 4) During stage 14, hemocytes migrating toward each other from the anterior and posterior ends of the embryo along the ventral midline converge and then undergo lateral migrations from the midline to form three parallel lines of circulating cells. Platelet-derived growth factor/vascular endothelial growth factor–receptor-related (PVR) signaling has been proposed to control these hemocyte developmental migrations (Wood et al., 2006). The PVR ligands Pvf2 and Pvf3 are expressed in the embryonic ventral midline beginning at stages 10 and 12, respectively, where they are proposed to drive head and tail hemocyte migration before being reduced before lateral hemocyte migration during stage 14 (Wood et al., 2006). Recent studies, however, show that Pvf2 and Pvf3 only contribute to the first transmigration step, where they coordinate invasion through the head and tail epithelial barriers (Parsons and Foley, 2013). This transmigration step has been shown to involve RhoL, a Rho-family GTPase that is expressed specifically in hemocytes, which regulates invasion, adhesion, and Rap1 localization (Siekhaus et al., 2010).
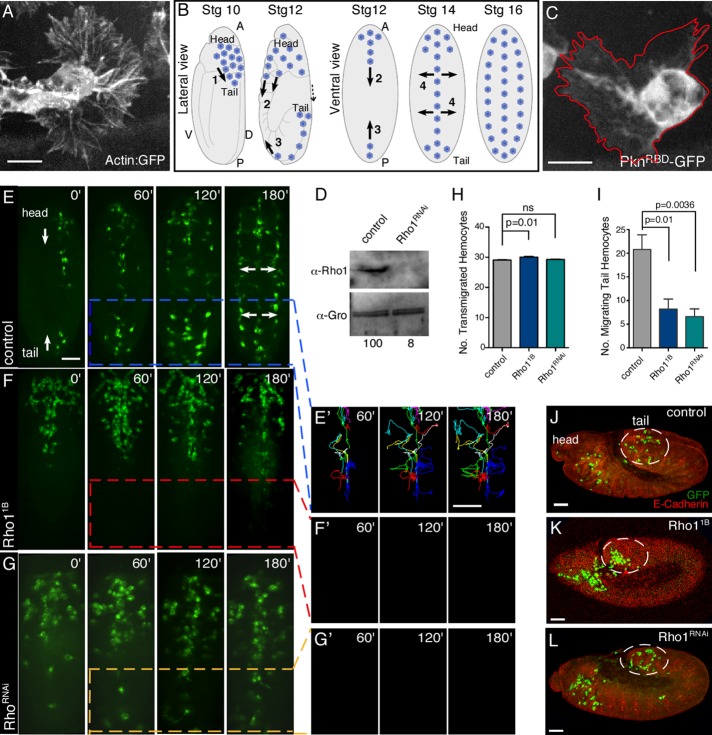
FIGURE 1:
Rho1 participates in a subset of hemocyte developmental migrations. (A) Confocal surface projection micrograph of an otherwise wild-type hemocyte expressing the Actin5C-GFP reporter. (B) Schematic of Drosophila hemocyte developmental migrations in stage 10–16 embryos from lateral or ventral perspectives. The major events indicated are 1) transmigration of hemocytes from the head to the tail, 2) posterior migration of head hemocytes along ventral midline, 3) anterior migration of tail hemocytes along ventral midline, and 4) lateral migration of hemocytes from the ventral midline to form three parallel lines. Head and tail are indicated. A, anterior; P, posterior; D, dorsal; V, ventral. (C) Confocal surface projection micrograph of an otherwise wild-type hemocyte expressing a Rho1 biosensor (UAS-PknRBD-GFP) under the control of the Pxn-Gal4 driver. (D) Western analysis of protein extracts from control (Pxn-Gal4) and Rho1RNAi hemocytes probed with antibodies recognizing Rho1 (α-Rho1(P1D9)) and a loading control (α-Gro). The normalized levels of Rho1 are indicated (with control set to 100). (E–G′) Time-lapse series of surface projections (ventral view) of migrating hemocytes expressing GFP in control (Pxn-Gal4) (E), Rho11B (F), and Rho1RNAi (G) embryos. Head and tail are indicated (E). Hemocytes that began their developmental migrations in the tail were tracked for 180 min (E′, F′, G′). Note that all of the tail hemocytes present in G arrived from the anterior. (H, I) Quantification of the number of transmigrated tail hemocytes (H) and number of these tail hemocytes that migrate anteriorly (I) in control (Pxn-Gal4), Rho11B, and Rho1RNAi embryos (n ≥ 20). (J–L) Confocal surface projection (lateral view) micrographs of stage 10 control (Pxn-Gal4) (J), Rho11B (K), and Rho1RNAi (L) embryos stained for DE-cadherin (red) and GFP (green). Transmigrated hemocytes are circled. All results are given as means ± SEM; p values are indicated. Scale bars, 10 μm (A, C), 40 μm (E–G′, J–L).
Hemocyte migration in response to developmental or chemotactic stimuli is initiated through polarized dynamic membrane protrusions and endocytosis. The driving force for these membrane protrusions is localized polymerization of cortical actin filaments (Rodriquez, 2003; Takenawa and Suetsugu, 2007; Insall and Machesky, 2009). One family of proteins that mediate membrane–cortical cytoskeleton interactions, as well as vesicle trafficking, is the Wiskott–Aldrich syndrome (WAS) family of proteins (Takenawa and Suetsugu, 2007). The founding members of WAS family proteins, WASp and SCAR/WAVE, function downstream of Rho-family GTPases (Cdc42 and Rac, respectively), where they activate the Arp2/3 complex, resulting in the nucleation of branched actin filaments required for a variety of processes, such as cell migration, endocytosis, exocytosis, and vesicle trafficking (Millard et al., 2004; Chhabra and Higgs, 2007; Campellone and Welch, 2010). SCAR was recently shown to affect hemocyte developmental migrations, whereas WASp is not required for hemocyte dispersal (Evans et al., 2013).
WASH, a more recently identified subclass of the WAS family of proteins, is highly conserved (Linardopoulou et al., 2007; Liu et al., 2009; Campellone and Welch, 2010; Rottner et al., 2010). Wash was first examined in Drosophila, where it was shown to function downstream of the Rho1 small GTPase along with the Spire and Cappuccino (formin) actin nucleation factors to control actin and microtubule dynamics during Drosophila oogenesis (Liu et al., 2009). A similar interaction between mammalian WASH and Rho or Cdc42 has not been observed, although a weak interaction with Rac1 was reported (Jia et al., 2010). Similar to other WAS-family proteins, WASH activates actin filament nucleation by the Arp2/3 complex through its conserved C-terminal VCA domain (Millard et al., 2004; Linardopoulou et al., 2007; Liu et al., 2009). Drosophila and mouse WASH are essential genes that are required many times throughout development (Linardopoulou et al., 2007; Liu et al., 2009; Dong et al., 2013; Piotrowski et al., 2013; Xia et al., 2013; Verboon et al., 2015). Conditional WASH knockdown in mouse hematopoietic stem cells results in defective blood production associated with anemia and severe cytopenia (Xia et al., 2014). Studies in mammalian cell lines and Dictyostelium suggest that WASH functions primarily in a multiprotein complex that regulates endosome shape and trafficking in an Arp2/3-dependent manner (Derivery et al., 2009; Gomez and Billadeau, 2009; Duleh and Welch, 2010; Harbour et al., 2012; Park et al., 2013; Piotrowski et al., 2013). This WASH regulatory complex (SHRC; including Strumpellin, SWIP, CCDC53, and FAM21) shares structural features with the SCAR/WAVE regulatory complex (WRC) proteins, as well as some sequence similarity; however the means by which they are activated appears to be different (Jia et al., 2010; Burianek and Soderling, 2013). The SHRC has been shown to regulate endosomal recycling by localizing and polymerizing actin at the surface of mature lysosomes (Gomez et al., 2012; Helfer et al., 2013; King et al., 2013; Park et al., 2013). WASH has also been shown to affect α5β1 integrin recycling, which is necessary for invasive migration (Zech et al., 2011). Consistent with this, WASH is up-regulated in invasive breast cancers and a breast cancer cell line (Leirdal et al., 2004; Nordgard et al., 2008; Moreira et al., 2013).
Here we describe a role for the Rho1 small GTPase and its downstream effector Wash to regulate in an SHRC-independent manner the third stereotypical developmental hemocyte migration, in which posterior hemocytes migrate anteriorly along the ventral midline.
RESULTS
Rho1 is required for the third stereotypical hemocyte developmental migration
Rho1 is expressed in hemocytes (Figure 1D). Expression of a conditional Rho1 biosensor (reporter for active Rho1 expression; PknRBD–green fluorescent protein (GFP); see Materials and Methods) in hemocytes (Pxn-Gal4; UAS-PknRBD-GFP) showed that activated Rho1 protein is not specifically enriched at the leading edge of a migrating hemocyte (Figure 1C).
To examine the role of Rho1 in hemocyte developmental migrations, we performed time-lapse analyses of stage 10–14 embryos expressing UAS-driven GFP under the control of hemocyte-specific Gal4 drivers (Pxn-Gal4, srp.Hemo-Gal4, or Crq-Gal4; Bruckner et al., 2004; Strong et al., 2005; Wood et al., 2006; Evans and Wood, 2011). We examined Rho11B mutant embryos (Rho11B; srp.Hemo-GAL4, UAS:GFP) in which both the hemocytes and tissue through which they migrate are mutant and embryos in which Rho1 was knocked down specifically in the hemocytes using a UAS–RNA interference (RNAi) line for Rho1 (UAS-Rho1v12734; Pxn-GAL4, UAS-GFP, referred to as Rho1RNAi; Figure 1, D–G; Dietzl et al., 2007). In both cases, hemocytes were observed to transmigrate properly from the head to the tail (Figure 1, H and J–L). Hemocytes present in the head migrated posteriorly and subsequently underwent lateral migration from the midline to form the three parallel rows of circulating hemocytes (Figure 1, E–G, and Supplemental Video S1). However, whereas hemocytes undergo the first epithelial transmigration to the tail region in Rho1 mutants, fewer of these tail hemocytes in Rho11B (8.2 ± 2.1; n = 5) and Rho1RNAi (6.6 ± 1.6; n = 5) migrate anteriorly along the ventral midline than in control embryos (w; Pxn-Gal4, UAS-GFP; 20.8 ± 3.1, n = 5; p = 0.0100 and 0.0036, respectively; note that the tail hemocytes present in Figure 1G arrived from the anterior; Figure 1, E–G′ and I). These tail hemocytes in Rho1-mutant embryos do not contribute to the circulating hemocytes but instead remain in the tail region. Our results suggest that these tail hemocytes become distinct from the anterior hemocytes: even though they transmigrate and are correctly positioned in the tail in Rho1 mutants, they appear to have lost their ability to sense and/or respond properly to migratory signals.
Wash is required for the third stereotypical hemocyte developmental migration
WAS-family proteins, through their interactions with Rho-family GTPases, play key roles in the sophisticated regulation of cellular protrusions and membrane trafficking through their tight spatial and temporal coordination of actin dynamics and other cellular machineries (Stradal et al., 2004; Takenawa and Suetsugu, 2007; Campellone et al., 2008). To examine the role of Wash in hemocyte developmental migrations, we performed time-lapse analyses of stage 10–14 embryos in which Wash was depleted specifically in hemocytes using two different UAS-RNAi lines for Wash (w; Pxn-GAL4, UAS-GFP; UAS-washv24642, referred to as washRNAi(2), and w; Pxn-GAL4, UAS-GFP; UAS-washv39769, referred to as washRNAi(3); Figure 2, A–E, and Supplemental Video S2; Dietzl et al., 2007). In both cases, hemocyte transmigration from the head to the tail was similar to control embryos (w; Pxn-Gal4, UAS-GFP; Figure 2, D, F, and G), consistent with Wash interacting physically with Rho1 (Figure 2, H and I; Liu et al., 2009). We performed pull-down experiments using bacterially expressed glutathione S-transferase (GST)–Rho-family GTPases and in vitro Wash (SHRC absent) or Drosophila embryo lysates (SHRC present). Our results show that Wash interacts directly with GTP-bound Rho1 but not RhoL, Cdc42, or Rac (Figure 2I), regardless of the presence of the WASH regulatory complex. This interaction occurs in vivo, as Wash is immunoprecipitated by Rho1 from whole-cell extracts of 0- to 2-h Drosophila embryos (Figure 2H). Similar to Rho1, Wash-depleted hemocytes present in the head migrated posteriorly and underwent the subsequent lateral migration from the midline to form the three parallel lines of circulating hemocytes (Figure 2, A–B′, and Supplemental Video S2). However, Wash knockdown significantly reduces the number of hemocytes that migrate anteriorly from the tail along the ventral midline from 20.8 ± 3.1 (n = 5) hemocytes in control embryos to 6.2 ± 2.1 (n = 5) and 8.4 ± 1.8 (n = 5) hemocytes in washRNAi(2) and washRNAi(3), respectively (p = 0.0043 and 0.0083, respectively; Figure 2, A–B′ and E, and Supplemental Video S2). As a result, most of the hemocytes present in the tail region in washRNAi(2)- and washRNAi(3)-mutant embryos have migrated posteriorly from the head. Similar to Rho1, expression of fluorescently tagged Wash (Wash-mChFP; see Materials and Methods) does not exhibit specific accumulation at the leading edge in migrating hemocytes (Figure 2J). In contrast to our findings, Wash was reported to play no role in hemocyte dispersal based on analysis of wash loss-of-function mutants (Evans et al., 2013). Given that wash mutants survive until the pupal stage (Linardopoulou et al., 2007), this difference may be due to perdurance of Wash activity in the wash loss-of-function mutants compared with its knockdown in RNAi-treated embryos. Thus our results suggest that each of the hemocyte developmental migrations may require distinct Rho-family GTPase proteins (i.e., RhoL for hemocyte transmigration; Rho1 for tail hemocyte migration).
FIGURE 2:
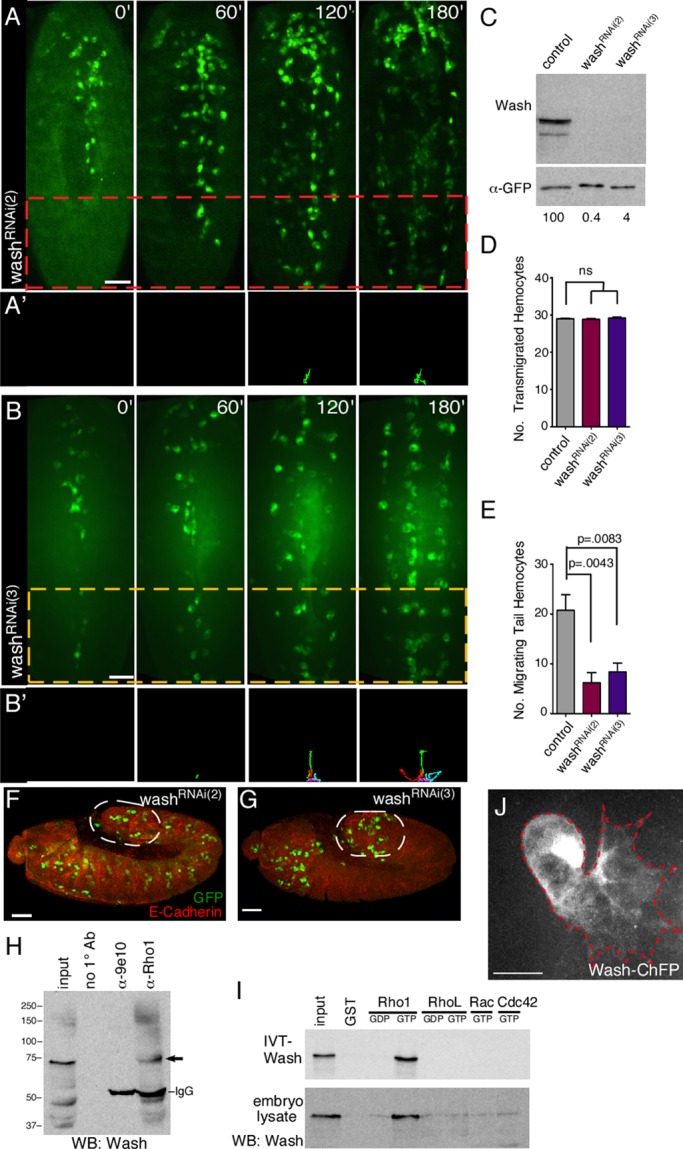
Wash participates in a subset of hemocyte developmental migrations. (A–B′) Time-lapse series of ventral surface projections of washRNAi(2) (A, A′) and washRNAi(3) (B, B′) migrating hemocytes expressing GFP. Hemocytes that began their developmental migrations in the tail were tracked for 180 min (A′, B′). (C) Western analysis of protein extracts from control (Pxn-Gal4), washRNAi(2), and washRNAi(3) hemocytes probed with antibodies recognizing Wash (α-Wash(P3H3)) and a loading control (α-GFP). The normalized levels of Wash are indicated (with control set to 100). (D, E) Quantification of the number of transmigrated tail hemocytes (D) and number of these tail hemocytes that migrate anteriorly (E) in control (Pxn-Gal4), washRNAi(2), and washRNAi(3) embryos (n ≥ 20). (F, G) Confocal surface projection micrographs (lateral view) of stage 10 washRNAi(2) (F) and washRNAi(3) (G) embryos stained for DE-cadherin (red) and GFP (green). Transmigrated hemocytes are circled. (H) Western blot of immunoprecipitations (IPs) from Drosophila embryo extracts performed with no primary antibody, a nonspecific antibody (9e10), and Rho1 antibody (α-Rho1(P1D9)) and probed with antibodies recognizing Wash (α-Wash(P3H3); arrow). (I) GST pull-down assays with 35S-labeled, in vitro–translated, full-length Wash protein (IVT-Wash; top; Wash complex absent) or Drosophila embryo extracts (embryo lysate; bottom; Wash complex present) and GST alone or GST-Rho family GTPases loaded with GDP or GTP as indicated. A 5% input is shown. The resulting Western blot of the embryo lysate pull downs (bottom) was probed with antibodies recognizing Wash (α-Wash(P3H3)). (J) Confocal surface projection micrograph of an otherwise wild-type hemocyte expressing a Wash-ChFP fusion protein. All results are given as means ± SEM; p values are indicated. Scale bars, 40 μm (A, B, F, G), 10 μm (J).
Wash binding to Rho1 maps to the surface-exposed N-terminal KDQ region of Rho1
We mapped the Wash-binding domain on Rho1 using deletions and point mutations described previously (Figure 3; Magie et al., 2002). We find that Wash binding on Rho1 maps to a surface-exposed region between the phosphate-binding loop and the effector domain of Rho1 (Z fragment; Figure 3, A and B). Wash binding to Rho1 can be greatly reduced by substituting three alanines for the KDQ amino acids (aa 27–29) within this Rho1 domain (referred to as KDQ/AAA) but not by the substitution of nearby residues (F39V or KQVE/AAAA; Figure 3, A and D). Reciprocally, we mapped and identified an N-terminal region of Wash (fragment A; aa 1–124) as the region of Wash required for its binding to Rho1 (Figure 3, B–D).
FIGURE 3:
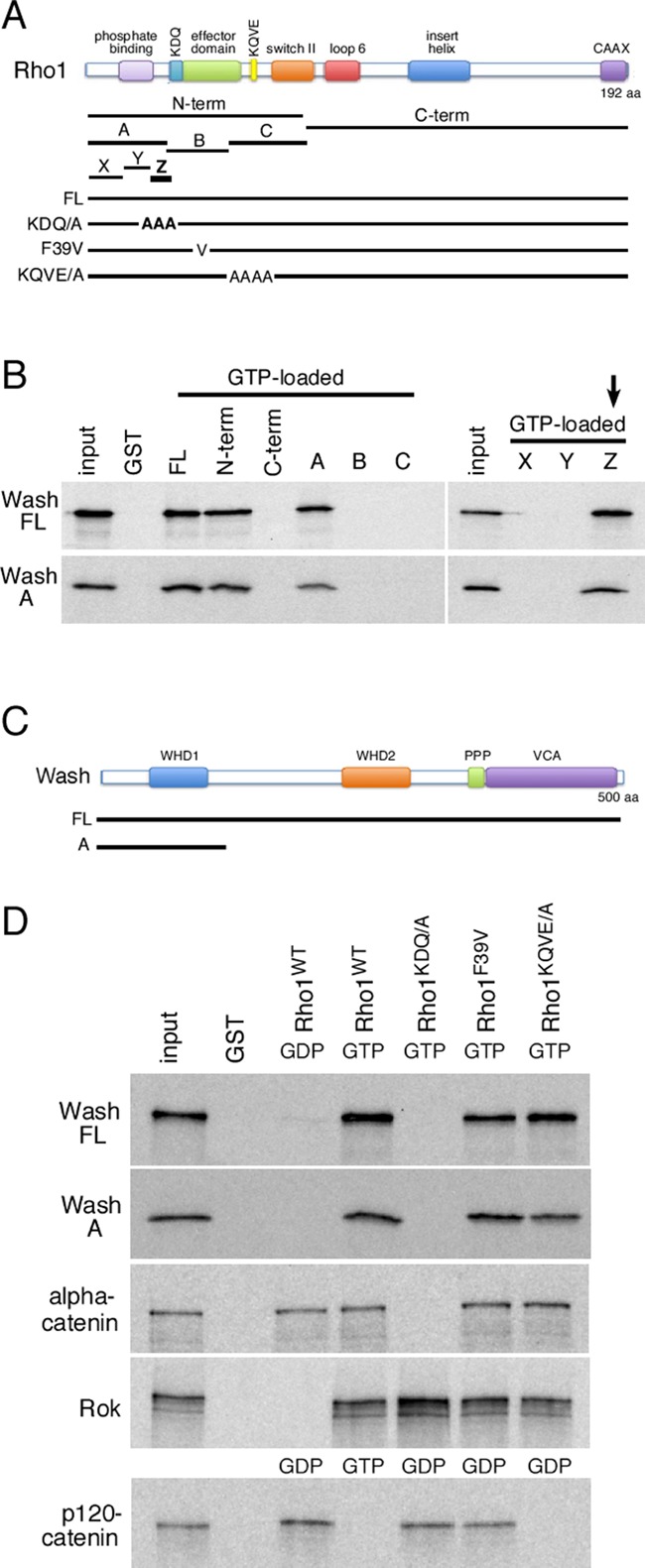
Wash interacts directly with Rho1, and mutations that prevent Wash binding to Rho1 disrupt tail hemocyte migration. (A) Schematic of the Rho1 protein, indicating its domains, the fragments used for mapping, and the location of point/substitution mutations. (B) GST pull-down assays with 35S-labeled, in vitro–translated, full-length Wash or Wash fragment A (aa 1–124) proteins and GST alone or GST fusions of full-length or fragments of Rho1 GTPase loaded with GTP. A 10% input is shown. The smallest fragment of Rho1 (Z) that interacts with Wash is indicated (arrow). (C) Schematic of the Wash protein, indicating its domains and the position of the Wash A fragment. (D) GST pull-down assays with 35S-labeled, in vitro–translated, full-length Wash (Wash FL), Wash fragment A (Wash A), α-catenin, Rok, or p120-catenin proteins and GST alone or GST-Rho1 proteins. The GST-Rho1 proteins were loaded with GDP or GTP as indicated. A 10% input is shown.
Mutations that prevent Wash binding to Rho1 disrupt hemocyte migration
To confirm that Wash is the downstream effector for Rho1 in tail hemocyte migrations, we generated lines expressing a wild-type Rho1 transgene driven by the endogenous Rho1 promoter (Rho11B P{Rho1WT}) or a Rho1 transgene with the KDQ/AAA substitution mutation that disrupts Wash binding (Rho11B P{Rho1KDQ/AAA}) in the background of the Rho11B-null mutant such that the only Rho1 activity comes from the transgene (Figure 4, A–B′, Supplemental Figure S1, A–E′′, and Supplemental Video S3; see Materials and Methods). The wild-type Rho1 transgene fully rescues the Rho11B-mutant phenotype in hemocytes: hemocytes in Rho11B P{Rho1WT} embryos behaved essentially like wild type (21.2 ± 3.0; n = 6 posterior hemocytes compared with 20.8 ± 3.1; n = 5 for control; Figure 4, A, A′, D, and E). However, hemocytes in Rho11B embryos expressing the transgene that disrupts Wash binding (Rho11B P{Rho1KDQ/AAA}) exhibited tail hemocyte migration defects similar to those in wash and Rho1 mutants (Figure 4, B, B′, D, and E). Hemocytes present in the head in Rho11B P{Rho1KDQ/AAA} embryos migrated posteriorly and underwent subsequent lateral migrations from the midline to form the three parallel lines of circulating hemocytes (Figure 4B and Supplemental Video S3). However, similar to that observed with Rho1 and Wash knockdown, there is significant reduction in the number of hemocytes that migrate anteriorly from the tail along the ventral midline (21.2 ± 3.0 [n = 6] hemocytes in control [Rho11B P{Rho1WT}] embryos compared with 3.2 ± 1.8 [n = 5] hemocytes in Rho11B P{Rho1KDQ/AAA}; p = 0.0011; Figure 4, B, B′, D, and E, and Supplemental Video S3).
FIGURE 4:
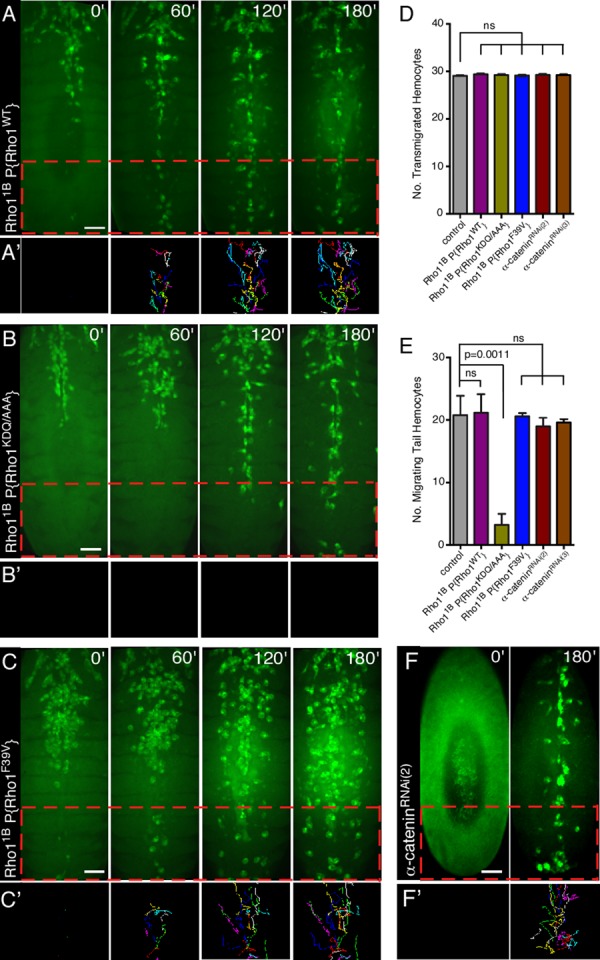
Mutations that prevent Wash binding to Rho1 disrupt hemocyte migration. (A–C′) Time-lapse series of ventral surface projections of migrating hemocytes expressing GFP in Rho11B P{Rho1WT} (A), Rho11B P{Rho1KDQ/A} (B), and Rho11B P{Rho1F39V} (C) embryos. Hemocytes that began their developmental migrations in the tail were tracked for 180 min (A′, B′, C′). (D, E) Quantification of the number of transmigrated tail hemocytes (D) and number of these tail hemocytes that migrate anteriorly (E) in control (Pxn-Gal4), Rho11B P{Rho1WT}, Rho11B P{Rho1KDQ/A}, Rho11B P{Rho1F39V}, α-cateninRNAi(2) and α-cateninRNAi(3) embryos (n ≥ 20). (F, F') Time-lapse series of ventral surface projections of migrating hemocytes expressing GFP in an α-cateninRNAi(2) embryo. Hemocytes that began their developmental migrations in the tail were tracked for 180'. All results are given as means ± SEM; p values are indicated. Scale bars, 40 μm (A–C', F, F').
As a specificity control, we examined another, nearby substitution mutation in Rho1's effector domain (F39V), which does not affect Wash binding (Figure 3, A and D, Supplemental Figure S1, E–E′′, and Supplemental Video S3). We generated a transgenic line expressing a Rho1 transgene with the F39V point mutation in the background of the Rho11B-null mutant (Rho11B P{Rho1F39V}) and found that all hemocyte developmental migrations are indistinguishable from control embryos (Rho11B P{Rho1WT}; Figure 4, C, C′, D, and E). Our results indicate that proper tail hemocyte developmental migration is dependent on a Rho1/Wash interaction.
Of interest, we previously identified the Rho1 KDQ region as the region required for Rho1 binding to α-catenin (Figure 3, A and D; Magie et al., 2002). However, using time-lapse video analyses of stage 10–14 embryos in which α-catenin was depleted specifically in hemocytes using two different UAS-RNAi lines for α-catenin (w; Pxn-GAL4, UAS-GFP; UAS-α-cateninKK107916, referred to as α-cateninRNAi(2), and y1 sc* v1; P{TRiP.HMS00317}attP2, referred to as α-cateninRNAi(3)), we find that α-catenin is not required for the third stereotypical hemocyte developmental migration (19.0 ± 1.4 [n = 6] migrating tail hemocytes in α-cateninRNAi(2) embryos and 19.6 ± 0.2 [n = 5] migrating tail hemocytes in α-cateninRNAi(3) embryos, compared with 20.8 ± 3.1 [n = 5] in control embryos; p = 0.1439; Figure 4, F, F′, D and E, Supplemental Figure S1, F and G, and Supplemental Video S3; Dietzl et al., 2007). Thus, despite the ability of Rho1 to bind two downstream effectors through its KDQ region, it is the binding to Wash that is relevant in the context of hemocyte developmental migrations, indicating that Rho-family GTPases have context-dependent specificity for their effector choices.
Knockdown of WASH regulatory complex subunits does not affect hemocyte developmental migrations
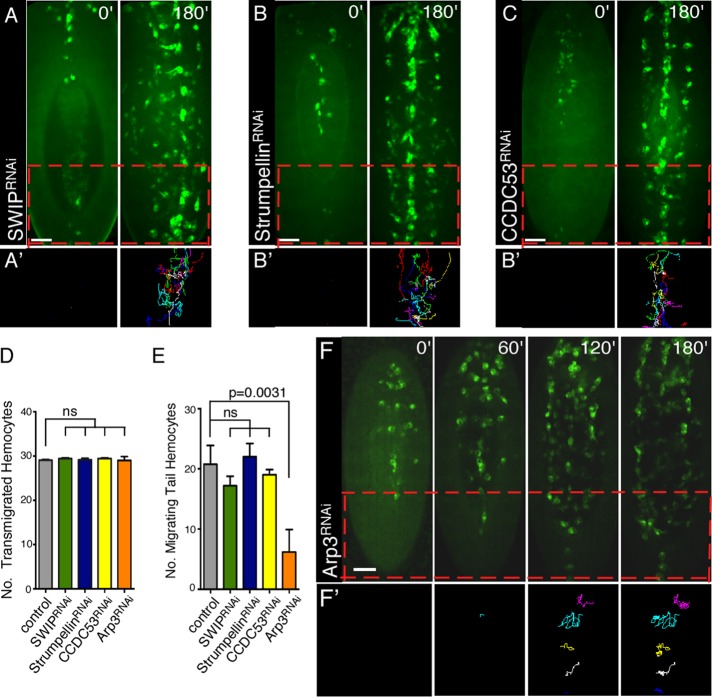
Mammalian WASH has been shown to function as part of a multiprotein complex (SHRC; Jia et al., 2010; Burianek and Soderling, 2013). Although Drosophila has orthologues of the SHRC proteins, whether WASH always functions as part of this SHRC complex is not known. WASH, as part of the SHRC, has been proposed to regulate the recycling of receptors and other proteins via its regulation of intracellular vesicle trafficking (Park et al., 2013). To determine whether endosome recycling of receptors/factors/components was required for the Rho1>Wash–mediated tail hemocyte migrations, we performed time-lapse video analyses of stage 10–14 embryos in which the SHRC members SWIP/CG13957, Strumpellin/CG12272, and CCDC53/CG7429 were depleted specifically in hemocytes using UAS-RNAi lines (w; Pxn-GAL4, UAS-GFP; UAS-CG13957/SWIPv105966, referred to as SWIPRNAi; w; Pxn-GAL4, UAS-GFP; UAS-Strumpellinv107954, referred to as StrumpellinRNAi; and w; Pxn-GAL4, UAS-GFP; UAS-CCDC53v110316, referred to as CCDC53RNAi; Dietzl et al., 2007). Of interest, we find that all hemocyte developmental migrations in SHRC-knockdown hemocytes are indistinguishable from wild-type embryos (Figure 5, A–E, Supplemental Figure S1, H–J, and Supplemental Video S4). In particular, knockdown of SHRC members does not significantly reduce the number of posterior hemocytes that transmigrate to the tail or that migrate anteriorly from the tail along the ventral midline (17.2 ± 1.6 [n = 10 in SWIPRNAi embryos, 22.0 ± 2.2 [n = 5] in StrumpellinRNAi embryos, and 19.0 ± 0.9 [n = 5] in CCDC53RNAi embryos, compared with 20.8 ± 3.1 [n = 5] in control embryos; Figure 5, D and E). Thus our results reveal an SHRC-independent role for Wash in regulating tail hemocyte migration.
FIGURE 5:
Wash functions through the Arp2/3 complex, but not the WASH regulatory complex, for tail hemocyte migration. (A–C′) Time-lapse series of ventral surface projections of migrating hemocytes expressing GFP in SWIPRNAi (A), StrumpellinRNAi (B), or CCDC53RNAi (C) embryos. Hemocytes that began their developmental migrations in the tail were tracked for 180 min (A′, B′, C′). (D, E) Quantification of the number of transmigrated tail hemocytes (D) and number of these tail hemocytes that migrate anteriorly (E) in control (Pxn-Gal4), SWIPRNAi, StrumpellinRNAi, CCDC53RNAi, and Arp3RNAi embryos (n ≥ 20). (F, F′) Time-lapse series of ventral surface projections of migrating hemocytes expressing GFP in Arp3RNAi embryos. Hemocytes that began their developmental migrations in the tail were tracked for 180 min (F′). All results are given as means ± SEM; p values are indicated. Scale bars, 40 μm (A–C′, F, F′).
Knockdown of Arp2/3 function mimics Wash's affects on hemocyte migrations
Polymerization of actin filaments against cellular membranes provides the driving force for cellular processes such as migration and endocytosis (Millard et al., 2004; Takenawa and Suetsugu, 2007; Campellone and Welch, 2010). Mutations specifically disrupting Wash's actin nucleation activity do not exist; however, Wash's actin nucleation activity is dependent on its interaction with the Arp2/3 complex (Linardopoulou et al., 2007; Liu et al., 2009). We performed time-lapse video analyses of stage 10–14 embryos in which Arp3 (Arp2/3 complex subunit) was knocked down specifically in hemocytes (w; UAS-Arp3v36520; Pxn-GAL4, UAS-GFP referred to as Arp3RNAi; Figure 5, D–F′, Supplemental Figure S1K, and Supplemental Video S4; Dietzl et al., 2007). Similar to Wash knockdown, hemocytes transmigrated properly from the head to the tail in Arp3RNAi-knockdown embryos, and fewer of these tail hemocytes migrated anteriorly from the tail (6.2 ± 1.7 [n = 5] compared with 20.8 ± 3.1 [n = 5] in control embryos; p = 0.0031; Figure 5, D and E), suggesting that Wash's actin nucleation activity is required for this migration. Arp3RNAi-knockdown hemocytes are also less organized, resulting in the simultaneous formation of three lines of hemocytes migrating posteriorly from the head rather than the lateral movement of hemocytes from the ventral midline observed in control embryos (Figure 5, F and F′, and Supplemental Video S4), consistent with the Arp2/3 complex functioning downstream of other WAS family proteins as well (Evans et al., 2013).
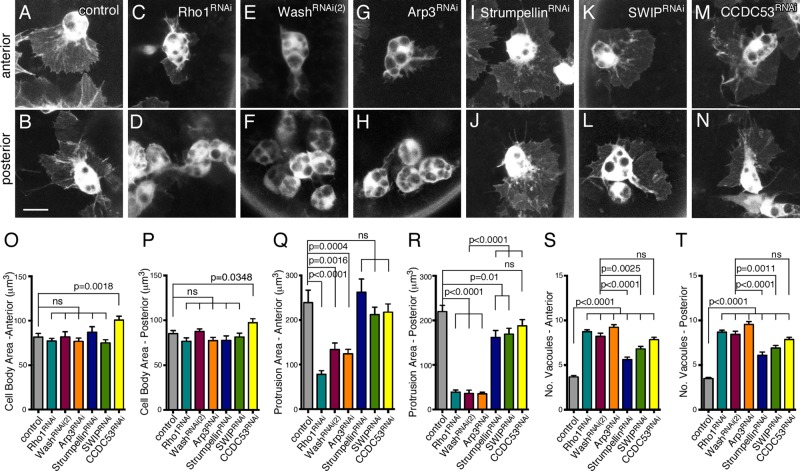
Cellular protrusions are severely reduced in Rho1>Wash>Arp2/3, but not SHRC, posterior hemocytes
Wash, through activation of the Arp2/3 complex, mediates branched actin nucleation required for cellular processes, including lamellipodial formation and endocytosis/exocytosis. Because active cell migration requires dynamic cellular protrusions (lamellipodia and filopodia), we examined cell protrusions in Rho1, Wash, and Arp3 anterior and posterior hemocytes, as well as those in SHRC subunits (Figure 6, A–N). Hemocyte cell body size (both anterior and posterior) was not significantly different in any of the knockdown mutants compared with those in control hemocytes, except for CCDC53, which showed a modest increase in size due to the presence of enlarged vacuoles (Figure 6, O and P). Strikingly, posterior hemocytes in Rho1RNAi-, washRNAi(2)-, and Arp3RNAi-knockdown embryos, but not those knocked down for SHRC subunits, exhibit severe reduction in cellular protrusion area, consistent with their lack of migration (219.8 ± 14.2 μm2 for control [n = 22] compared with 39.1 ± 4.4 μm2 for Rho1RNAi [p < 0.0001, n = 27], 36.0 ± 7.3 μm2 for washRNAi(2) [p < 0.0001, n = 20], 35.5 ± 3.4 μm2 for Arp3RNAi [p < 0.0001, n = 32], 162.1 ± 15.2 μm2 for StrumpellinRNAi [p = 0.0090, n = 27], 169.3 ± 13.2 μm2 for SWIPRNAi [p = 0.0123, n = 23], and 187.8 ± 14.0 μm2 for CCDC53RNAi [p = 0.1181, n = 26]; Figure 6, B, D, F, H, J, L, N, and R). Although StrumpellinRNAi, SWIPRNAi, and CCDC53RNAi posterior hemocytes show a modest decrease in protrusion area, it is unlikely to be a major player, as the area of protrusions in washRNAi(2) compared with the SHRC subunits is also significantly different (p < 0.0001 for each; Figure 6R). Of interest, Rho1RNAi, washRNAi(2), and Arp3RNAi anterior hemocytes, but not those in SHRC subunits, also show a moderate reduction in cellular protrusion area that is not sufficient to prevent their developmental migration (239.2 ± 27.9 μm2 for control [n = 35] compared with 78.3 ± 8.0 μm2 for Rho1RNAi [p < 0.0001, n = 31], 133.8 ± 14.8 μm2 for washRNAi(2) [p = 0.0016, n = 26], 124.3 ± 10.0 μm2 for Arp3RNAi [p = 0.0004, n = 32], 262.2 ± 29.4 μm2 for StrumpellinRNAi [p = 0.5834, n = 24], 212.3 ± 16.6 μm2 for SWIPRNAi [p = 0.4107, n = 27], and 217.6 ± 18.5 μm2 for CCDC53RNAi [p = 0.5349, n = 30]; Figure 6Q).
FIGURE 6:
Cellular protrusions are severely reduced in Rho1>Wash>Arp2/3, but not SHRC, posterior hemocytes. (A–N) Confocal projections of anterior (A, C, E, G, I, K) or posterior (B, D, F, H, J, L) hemocytes in equivalently staged embryos expressing GFP in control (A, B), Rho1RNAi (C, D), washRNAi(2) (E, F), Arp3RNAi (G, H), StrumpellinRNAi (I, J), SWIPRNAi (K, L), and CCDC53RNAi (M, N) backgrounds. Scale bars, 10 μm (A–N). (O–T) Quantification of hemocyte cell body size (O, P), protrusion area (Q, R), and number of vacuoles (S, T) in anterior (O, Q, S) or posterior (P, R, T) hemocytes from control or Rho1RNAi, washRNAi(2), Arp3RNAi, StrumpellinRNAi, SWIPRNAi, and CCDC53RNAi embryos at the onset of posterior to anterior migration. All results are given as means ± SEM; p values are indicated.
In examining Rho1>Wash>Arp2/3 hemocytes, the number of vacuoles present in both the anterior and posterior hemocyte cell bodies was prominent in comparison to control hemocytes (anterior hemocytes: 3.7 ± 0.2 for control [n = 24] compared with 8.7 ± 0.2 for Rho1RNAi [p < 0.0001, n = 51], 8.2 ± 0.4 for washRNAi[2] [p < 0.0001, n = 29], and 9.2 ± 0.3 for Arp3RNAi [p < 0.0001, n = 43]; and posterior hemocytes: 3.5 ± 0.1 for control [n = 30] compared with 8.7 ± 0.2 for Rho1RNAi [p < 0.0001, n = 49], 8.5 ± 0.3 for washRNAi[2] [p < 0.0001, n = 29], and 9.5 ± 0.3 for Arp3RNAi [p < 0.0001, n = 31]; Figure 6, S and T). Hemocytes knocked down for SHRC subunits have a less prominent but significant effect on the number of anterior (3.7 ± 0.2 for control compared with 5.6 ± 0.3 for StrumpellinRNAi [p < 0.0001, n = 31], 6.8 ± 0.3 for SWIPRNAi [p < 0.0001, n = 38], and 7.8 ± 0.3 for CCDC53RNAi [p < 0.0001, n = 38]); and posterior (3.5 ± 0.1 for control compared with 6.1 ± 0.4 for StrumpellinRNAi [p < 0.0001, n = 30], 6.9 ± 0.3 for SWIPRNAi [p < 0.0001, n = 41], and 7.9 ± 0.3 for CCDC53RNAi [p < 0.0001, n = 42]) hemocyte vacuoles (Figure 6, S and T). This increased number of vacuoles in Wash and SHRC subunits likely reflects a separate, SHRC-dependent role for Wash in hemocytes and is consistent with previously proposed roles for WASH in endocytosis and/or exocytosis.
DISCUSSION
Guided cell migrations, essential for normal development and maintenance functions of animal body plans and organ systems, require tight spatial and temporal coordination of cytoskeleton and cytoskeleton–membrane interactions. We find that the Rho1 small GTPase, through its downstream effector Wash, regulates one of the four hemocyte stereotypical developmental migrations in which posterior tail hemocytes migrate anteriorly along the ventral midline. Wash activates the Arp2/3 complex, whose activity is needed for this migration, whereas members of the SHRC (SWIP/CG13957, Strumpellin/CG12272, and CCDC53/CG7429) are not. Mapping of the interaction domain of Wash with Rho1 shows that Rho1 uses the same surface-exposed region to interact with either Wash or α-catenin. Despite this overlap, Rho1 uses Wash, not α-catenin, as its downstream effector for this function, even though α-catenin is present in hemocytes, indicating that Rho-family GTPases have context-dependent specificity for their effector choices.
WASP is known to function downstream of and bind directly to the Cdc42 small GTPase, whereas SCAR/WAVE proteins function as part of a larger multimeric complex (WRC) and associate indirectly with the Rac small GTPase (Takenawa and Suetsugu, 2007). The situation with Wash is not yet clear. Similar to SCAR, mammalian WASH has been shown to function as part of a multiprotein complex (SHRC; Jia et al., 2010; Burianek and Soderling, 2013). No interaction (direct or indirect) has been observed between mammalian WASH and Rho or Cdc42, although a weak interaction with Rac1 was reported (Jia et al., 2010). In contrast, the Drosophila Wash protein has been shown to bind directly to Rho1 but not to Rac, Cdc42, or RhoL (Liu et al., 2009; this study). Although SHRC proteins are present and form a complex in Drosophila, we find that they are not required for posterior hemocyte developmental migrations. Thus our results reveal an SHRC-independent role for Wash in regulating tail hemocyte migration. Wash and the SHRC subunits exhibit an increased number of vacuoles in both anterior and posterior hemocyte cell bodies, consistent with a separate, SHRC-dependent, role for Wash in hemocytes. Taken together, our results suggest that Wash can exist as part of a multiprotein complex like the SCAR/WAVE proteins and function independently of such a complex (similar to WASP) in a context-dependent manner. At least in Drosophila, when Wash functions separately from other SHRC proteins, it does so by binding directly to and functioning downstream of the Rho1 small GTPase. It is also possible that because there are more WAS family members in mammals than in flies, each fly protein might have functions that overlap with multiple mammalian proteins. In the future, it will be interesting to see how Wash chooses its context-dependent means of action.
During migration, Rho's established role is to work at the posterior of a cell to retract the tail through myosin contractility (Spiering and Hodgson, 2011). However, other studies show that active Rho can also function at the leading edge of migrating cells, where it promotes membrane ruffling and facilitates cell motility (Kurokawa and Matsuda, 2005; Machacek et al., 2009). The Rho1>Wash>Arp2/3 pathway in hemocyte migrations could be functioning to induce the formation of branched actin networks in the leading edge of cells necessary for migration or provide a framework for organizing receptors at the cell surface into clusters to facilitate sorting and/or signaling. Consistent with this possibility, Rho1>Wash>Arp2/3 posterior hemocytes exhibit severely reduced protrusion areas and fail to migrate anteriorly. Alternatively, the Rho1>Wash>Arp2/3 pathway could be activated on the surface of endosomes near the leading edge, which is important for receptor recycling and/or signaling. A new actin-dependent mechanism of long-range vesicle transport was reported that relies on the recruitment of the linear nucleation promotion factors formin-2 and Spire1/2 to assemble an extensive actin meshwork on the surface of vesicles (Pfender et al., 2011). Given that Drosophila Wash was shown to interact with Capu (formin) and Spire (Liu et al., 2009), it could be functioning in signal transduction and/or recycling through such a mechanism independent of the SHRC. Indeed, Rickettsia has been shown to use two independent actin polymerization mechanisms for its motility, one requiring activation of the Arp2/3 complex, and one requiring Sca2, a mimic of host formins (Reed et al., 2012). Consistent with these possibilities, tail hemocytes appear to have lost their ability to sense and/or respond to developmental signals or are unable to receive additional signals not required by anterior hemocytes.
A pattern is emerging in which distinct Rho-family GTPases are required to regulate each of the stereotyped Drosophila hemocyte migrations, often through a WAS-family protein. RhoL has been shown to function during the first developmental migration, in which a subset of head hemocytes undergoes an invasive transmigration to populate the posterior tail region of the embryo (Siekhaus et al., 2010). In addition, the WAS family member SCAR is required for hemocyte motility (Evans et al., 2013) and affects the ability of the midline hemocytes, which migrate laterally to generate three rows of circulating hemocytes. SCAR requires its SCAR/WAVE (Hem/Kette/Nap1) complex (Evans et al., 2013), which usually functions downstream of the Rac small GTPase (Takenawa and Suetsugu, 2007). A requirement for Rac in hemocyte developmental migrations has not yet been reported. Our data demonstrate that Rho1, through its effector Wash, regulates the third stereotyped migration in an Arp2/3-dependent manner. Thus hemocyte developmental migrations provide a superb model in which to systematically study Rho GTPases and explore their interactions with WAS-family proteins.
MATERIALS AND METHODS
Fly strains and genetics
Flies were cultured and crossed on yeast/cornmeal/molasses/malt extract medium and maintained at 25°C. The following alleles were used: Rho11B (Magie and Parkhurst, 2005) and Rho11B P{Rho1WT}, Rho11B P{Rho1KDQ/AAA}, and Rho11B P{Rho1F39V} (this study). The following UAS-driven RNAi stocks from the Vienna Drosophila Resource Center Stock Center (Vienna, Austria) were used: w1118; P{GD4726}v12734/CyO (Rho1RNAi); w1118; P{GD932}v24642 (washRNAi(2)); w1118; P{GD7950}v39769 (washRNAi(3)); P{KK111410}VIE-260B (Arp3RNAi); P{KK108277}VIE-260B (Arpc1RNAi); P{KK107916}VIE-260B (α-cateninRNAi(2)); P{KK101329}VIE-260B (SWIPRNAi); P{KK101498}VIE-260B (StrumpellinRNAi); and P{KK100545}VIE-260B (CCDC53RNAi; Dietzl et al., 2007). The following TRiP line was used: y1 sc* v1; P{TRiP.HMS00317}attP2 (α-cateninRNAi(3); Bloomington Stock Center, Bloomington, IN). Hemocyte-specific drivers used were y1 w*; P{crq-GAL4}2 (Bloomington Stock Center); P{Pxn-GAL4}II, P{UAS-GFP}II; P{Pxn-GAL4}III, P{UAS-GFP}III (Strong et al., 2005); and P{srp.Hemo-Gal4}III, P{UAS-GFP}III (Tucker et al., 2011). The following UAS-driven, fluorescently tagged reporter lines were used: P{UAS-PknRBD-GFP}27, P{UAS-mRFP-actin}13, and P{UAS-Wash-ChFP}30 (this study).
Generation of GFP/cherry fluorescent protein fusion constructs and transgenics
UAS-Wash–cherry fluorescent protein (ChFP) was made by fusing ChFP (Shaner et al., 2004) N-terminal to the ATG of the full-length Wash open reading frame and subsequently cloned as a KpnI–NheI fragment into the KpnI–XbaI sites of the pUASp vector (Rorth, 1998). UAS-PknRBD-GFP was made by fusing amino acids 1–339 of the Rho1 downstream effector PKN containing a G58A point mutation (Lu and Settleman, 1999) N-terminal of GFP and subsequently cloned as a KpnI–NotI fragment into the KpnI–NotI sites of the pUASp vector. These constructs were used to make germline transformants as previously described (Spradling, 1986). The resulting transgenic lines (P{w+; UAS-ChFP-Wash}20 and P{w+; UAS-PknRBD-GFP}27) were mapped to a single chromosome and shown to have nonlethal insertions.
Generation of Rho11B P{Rho1WT} and Rho1 point mutation constructs and transgenics
A ∼7-kb HindIII–MluI genomic fragment encompassing the entire Rho1 gene was subcloned into the Casper transformation vector (Pirrotta, 1988). The intronic regions were then removed by substituting wild-type cDNA sequence (Rho1WT) or the cDNA with point mutations generated by PCR (Rho1KDQ/AAA: KDQ to AAA; Rho1F39V: F39 to V; and Rho1KQVE/AAAA: KQVE to AAAA) for the genomic sequence between the ATG and stop codons. These constructs were used to make germline transformants as previously described (Spradling, 1986). Transgenic lines that mapped to chromosome 2 and that had nonlethal insertions were kept. The resulting transgenic lines (P{w+; Rho1WT}, P{w+; Rho1KDQ/AAA}, P{w+; Rho1F39V}, and P{w+; Rho1KQVE/AAAA}) were recombined onto the Rho11B-null chromosome to assess the contribution of the particular Rho1 transgene. The resulting recombinants are essentially gene replacements, as Rho1 activity is only provided by the transgene. These transgenes rely not on overexpression, but on the spatial and temporal expression driven by the endogenous Rho1 promoter itself. We analyzed a minimum of three lines per construct and checked all lines to confirm that the levels and spatial distribution of their expression is indistinguishable from wild type. The wild-type version of this transgene fully rescues the maternal and zygotic phenotypes of the Rho11B loss-of-function mutation.
Confocal image acquisition
Appropriately staged embryos were fixed, stained, and mounted onto glass slides according to standard procedures (Abreu-Blanco et al., 2012). The following primary antibodies were used to stain the embryos: rabbit anti-GFP (1:125; Molecular Probes/Invitrogen, Rockford, IL) and rat anti-DCad2 (1:100; Developmental Studies Hybridoma Bank, Iowa City, IA). The secondary antisera used were donkey anti-rabbit Alexa 488 and donkey anti-rat Alexa 568 (1:1000; Molecular Probes/Invitrogen). Confocal microscopy was performed using a Zeiss LSM-510M or Zeiss LSM-780 microscope (Carl Zeiss, Jena, Germany) with excitation at 488 or 543 nm and emission collection with BP-500-550 or LP560 filter, respectively. A Plan-Apochromat 20×/0.75 dry objective or a Plan-NeoFluor 40×/1.3 oil objective was used for imaging. A 30-μm length of the embryo periphery was imaged in 1.5-μm steps. Images were processed in ImageJ (National Institutes of Health, Bethesda, MD) and assembled with Canvas 8 software (Deneva Systems).
Live image acquisition
For transmigration studies, embryos were collected on grape agar plates at 25°C for 1 h and then aged at 25°C for 7 h. The transmigrated hemocytes were counted halfway through germ-band retraction. For migration studies, embryos were collected on grape agar plates at either 1) 25°C for 1 h and then aged at 18°C for 16 h (developmental studies) or 20 h (reporter studies), or 2) at 29°C for 1 h and then aged at 29°C for 6.5 h (developmental studies). The two regimens yielded indistinguishable results. Embryos were then dechorionated, dried for 5 min, transferred onto strips of glue dried onto No. 1.5 coverslips, and covered with series 700 halocarbon oil (Halocarbon Products, River Edge, NJ). For developmental studies, 24-μm stacks were taken once per minute with a 0.4-μm step size over the course of 3 h. For reporter studies, 15-μm stacks were taken once per minute with a 0.25-μm step size over the course of 1 h.
The following microscopes were used for reporter and some developmental studies: 1) an UltraVIEW VoX Confocal Imaging System (PerkinElmer, Waltham, MA) in a Nikon Eclipse Ti stand (Nikon Instruments, Melville, NY) with 60×/1.4 numerical aperture (NA), 20×/0.75 NA, or 40×/1.3 NA objective lens and controlled by Volocity software, version 5.3.0 (PerkinElmer). Images were acquired at 491 and 561 nm with a Yokogawa CSU-X1 confocal spinning disk head equipped with a Hamamatsu C9100-13 electron-multiplying charge-coupled device (EMCCD) camera (PerkinElmer), and 2) a Nikon LiveScan Swept Field Confocal (for Nikon by Prairie Technologies, Middleton, WI) mounted on a Nikon Eclipse Ti with 20×/0.45 NA objectives lens, using the NIS-Elements AR 3.0 as acquisition software (Nikon Instruments). Images were acquired with a 491-nm laser, and a Photometrics QuantEM 512SC EMCCD camera (Photometrics, Tucson, AZ).
Image analysis
Images were acquired on Nikon Live, Volocity, or Zeiss LSM software and exported as tiffs. Maximum intensity xy-projections of 24 μm (developmental migration) or 1–10 μm (reporter and fixed tissue studies) were generated. Hemoctyes were counted, tracked, and outlined in ImageJ using the base package and Manual Tracking plug-in (http://rsbweb.nih.gov/ij/plugins/).
Western blots
Hemocytes were recovered from larvae as described (Stofanko et al., 2008). The resulting hemocytes were mixed with lysis buffer (50 mM Tris, pH 7.8, 150 mM NaCl, 1% NP-40), flash frozen in liquid nitrogen, and stored at −80°C. Samples were normalized to a loading control (GFP; Gro) and blotted using standard procedures. The following antibodies were used: anti-Rho1 (P1D9; 1:50; Magie et al., 2002); anti-Wash mouse polyclonal (1:200; P3H3; Liu et al., 2009; Rodriguez-Mesa et al., 2012); anti-Arp3 (1:50; Stevenson et al., 2002); anti-GFP (1:1000; Sigma-Aldrich, St. Louis, MO); anti-SWIP/CG13957 (1:400; this study); anti-Strumpellin/CG12272 (1:200; this study); anti-CCDC53/CG7429 (1:200; this study); goat anti-rabbit Alexa 568 (1:15,000; Invitrogen); donkey anti-mouse horseradish peroxidase (1:15,000; Molecular Probes), and, from the Developmental Studies Hybridoma Bank, anti-Groucho (1:300; Gro) and anti–α-catenin (1:10; DCAT-1).
Antibody generation
Polyclonal antiserum against the Drosophila WASH regulatory complex subunits Strumpellin, SWIP, CCDC53, and FAM21 were generated by immunizing Balb/c BYJ Rb(8.12) 5BNR/J mice (strain 1802; Jackson Labs, Bar Harbor, ME) with a mixture of two proteins each that comprised GST fused to their N-terminal or C-terminal halves, except for CCDC53 (CG7429), which was the full-length protein (aa 1–177) fused to GST. Drosophila Strumpellin (CG12272): N-terminal (aa 1–618) or C-terminal (aa 618–1192). Drosophila SWIP (CG13957): N-terminal (aa 1–566) or C-terminal (aa 566–1103). FAM21 (CG16742): N-terminal (aa 1–830) or C-terminal (aa 830–1470).
Immunoprecipitations and pull-down assays
GST-Rho1 wild-type and point mutations were previously described (Magie et al., 2002). Wash constructs were described previously (Liu et al., 2009). Protein expression, in vitro translations, immunoprecipitations, and GST pull-down assays were performed as previously described (Magie et al., 2002; Magie and Parkhurst, 2005; Rosales-Nieves et al., 2006). Drosophila wild-type 0- to h embryo whole-cell extract was a gift from T. Tsukiyama (Fred Hutchinson Cancer Research Center, Seattle, WA).
Statistical analyses
Prism graphing software was used to organize data, generate graphs, and perform statistical analysis. In all cases, the mean was graphed with error bars representing ±SEM. Statistical analysis was performed using an unpaired t test, with Welch's correction in cases in which variance between data sets was significantly different. p < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Bina Sugumar, James Watts, and Parkhurst lab members for their experimental help and comments on the manuscript. We thank Utpal Banerjee, Lynn Cooley, Craig Magie, Bill Theurkauf, Toshi Tsukiyama, the Bloomington, Kyoto, and Vienna Stock Centers, TRiP at Harvard Medical School, the Developmental Studies Hybridoma Bank, and the M.J. Murdoch Charitable Trust for flies, antibodies, spinning disk microscope, and other reagents used in this study. This work was supported by National Institutes of Health Grant GM097083 to S.M.P.
Abbreviations used:
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- mCHFP
mCherry fluorescent protein
- PVR
platelet-derived growth factor/endothelial growth factor-receptor-related
- SHRC
WASH regulatory complex
- WAS
Wiskott Aldrich syndrome
- WRC
WAVE regulatory complex.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-08-1266) on March 4, 2015.
*These authors contributed equally.
REFERENCES
- Abreu-Blanco MT, Verboon JM, Liu R, Watts JJ, Parkhurst SM. Drosophila embryos close epithelial wounds using a combination of cellular protrusions and an actomyosin purse string. J Cell Sci. 2012;125:5984–5997. doi: 10.1242/jcs.109066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckner K, Kockel L, Duchek P, Luque CM, Rorth P, Perrimon N. The PDGF/VEGF receptor controls blood cell survival in Drosophila. Dev Cell. 2004;7:73–84. doi: 10.1016/j.devcel.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Burianek LE, Soderling SH. Under lock and key: spatiotemporal regulation of WASP family proteins coordinates separate dynamic cellular processes. Semin Cell Dev Biol. 2013;24:258–266. doi: 10.1016/j.semcdb.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Cheng HC, Robbins D, Siripala AD, McGhie EJ, Hayward RD, Welch MD, Rosen MK, Koronakis V, Leong JM. Repetitive N-WASP-binding elements of the enterohemorrhagic Escherichia coli effector EspF(U) synergistically activate actin assembly. PLoS Pathog. 2008;4:e1000191. doi: 10.1371/journal.ppat.1000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra ES, Higgs HN. The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol. 2007;9:1110–1121. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]
- Derivery E, Sousa C, Gautier JJ, Lombard B, Loew D, Gautreau A. The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev Cell. 2009;17:712–723. doi: 10.1016/j.devcel.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Dong B, Kakihara K, Otani T, Wada H, Hayashi S. Rab9 and retromer regulate retrograde trafficking of luminal protein required for epithelial tube length control. Nat Commun. 2013;4:1358. doi: 10.1038/ncomms2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duleh SN, Welch MD. WASH and the Arp2/3 complex regulate endosome shape and trafficking. Cytoskeleton (Hoboken) 2010;67:193–206. doi: 10.1002/cm.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans IR, Ghai PA, Urbancic V, Tan KL, Wood W. SCAR/WAVE-mediated processing of engulfed apoptotic corpses is essential for effective macrophage migration in Drosophila. Cell Death Differ. 2013;20:709–720. doi: 10.1038/cdd.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans IR, Wood W. Understanding in vivo blood cell migration—Drosophila hemocytes lead the way. Fly. 2011;5:110–114. doi: 10.4161/fly.5.2.14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez TS, Billadeau DD. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev Cell. 2009;17:699–711. doi: 10.1016/j.devcel.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez TS, Gorman JA, de Narvajas AA, Koenig AO, Billadeau DD. Trafficking defects in WASH-knockout fibroblasts originate from collapsed endosomal and lysosomal networks. Mol Biol Cell. 2012;23:3215–3228. doi: 10.1091/mbc.E12-02-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour ME, Breusegem SY, Seaman MN. Recruitment of the endosomal WASH complex is mediated by the extended “tail” of Fam21 binding to the retromer protein Vps35. Biochem J. 2012;442:209–220. doi: 10.1042/BJ20111761. [DOI] [PubMed] [Google Scholar]
- Helfer E, Harbour ME, Henriot V, Lakisic G, Sousa-Blin C, Volceanov L, Seaman MN, Gautreau A. Endosomal recruitment of the WASH complex: active sequences and mutations impairing interaction with the retromer. Biol Cell. 2013;105:191–207. doi: 10.1111/boc.201200038. [DOI] [PubMed] [Google Scholar]
- Insall RH, Machesky LM. Actin dynamics at the leading edge: from simple machinery to complex networks. Dev Cell. 2009;17:310–322. doi: 10.1016/j.devcel.2009.08.012. [DOI] [PubMed] [Google Scholar]
- Jia D, Gomez TS, Metlagel Z, Umetani J, Otwinowski Z, Rosen MK, Billadeau DD. WASH and WAVE actin regulators of the Wiskott-Aldrich syndrome protein (WASP) family are controlled by analogous structurally related complexes. Proc Natl Acad Sci USA. 2010;107:10442–10447. doi: 10.1073/pnas.0913293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JS, Gueho A, Hagedorn M, Gopaldass N, Leuba F, Soldati T, Insall RH. WASH is required for lysosomal recycling and efficient autophagic and phagocytic digestion. Mol Biol Cell. 2013;24:2714–2726. doi: 10.1091/mbc.E13-02-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K, Matsuda M. Localized RhoA activation as a requirement for the induction of membrane ruffling. Mol Biol Cell. 2005;16:4294–4303. doi: 10.1091/mbc.E04-12-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leirdal M, Shadidy M, Rosok O, Sioud M. Identification of genes differentially expressed in breast cancer cell line SKBR3: potential identification of new prognostic biomarkers. Int J Mol Med. 2004;14:217–222. [PubMed] [Google Scholar]
- Linardopoulou EV, Parghi SS, Friedman C, Osborn GE, Parkhurst SM, Trask BJ. Human subtelomeric WASH genes encode a new subclass of the WASP family. PLoS Genet. 2007;3:e237. doi: 10.1371/journal.pgen.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Abreu-Blanco MT, Barry KC, Linardopoulou EV, Osborn GE, Parkhurst SM. Wash functions downstream of Rho and links linear and branched actin nucleation factors. Development. 2009;136:2849–2860. doi: 10.1242/dev.035246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Settleman J. The role of rho family GTPases in development: lessons from Drosophila melanogaster. Mol Cell Biol Res Commun. 1999;1:87–94. doi: 10.1006/mcbr.1999.0119. [DOI] [PubMed] [Google Scholar]
- Machacek M, Hodgson L, Welch C, Elliott H, Pertz O, Nalbant P, Abell A, Johnson GL, Hahn KM, Danuser G. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magie CR, Parkhurst SM. Rho1 regulates signaling events required for proper Drosophila embryonic development. Dev Biol. 2005;278:144–154. doi: 10.1016/j.ydbio.2004.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magie CR, Pinto-Santini D, Parkhurst SM. Rho1 interacts with p120ctn and alpha-catenin, and regulates cadherin-based adherens junction components in Drosophila. Development. 2002;129:3771–3782. doi: 10.1242/dev.129.16.3771. [DOI] [PubMed] [Google Scholar]
- Millard TH, Sharp SJ, Machesky LM. Signalling to actin assembly via the WASP (Wiskott-Aldrich syndrome protein)-family proteins and the Arp2/3 complex. Biochem J. 2004;380:1–17. doi: 10.1042/BJ20040176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira CG, Jacinto A, Prag S. Drosophila integrin adhesion complexes are essential for hemocyte migration in vivo. Biol Open. 2013;2:795–801. doi: 10.1242/bio.20134564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordgard SH, Johansen FE, Alnaes GI, Bucher E, Syvanen AC, Naume B, Borresen-Dale AL, Kristensen VN. Genome-wide analysis identifies 16q deletion associated with survival, molecular subtypes, mRNA expression, and germline haplotypes in breast cancer patients. Genes Chromosomes Cancer. 2008;47:680–696. doi: 10.1002/gcc.20569. [DOI] [PubMed] [Google Scholar]
- Park L, Thomason PA, Zech T, King JS, Veltman DM, Carnell M, Ura S, Machesky LM, Insall RH. Cyclical action of the WASH complex: FAM21 and capping protein drive WASH recycling, not initial recruitment. Dev Cell. 2013;24:169–181. doi: 10.1016/j.devcel.2012.12.014. [DOI] [PubMed] [Google Scholar]
- Parsons B, Foley E. The Drosophila platelet-derived growth factor and vascular endothelial growth factor-receptor related (Pvr) protein ligands Pvf2 and Pvf3 control hemocyte viability and invasive migration. J Biol Chem. 2013;288:20173–20183. doi: 10.1074/jbc.M113.483818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfender S, Kuznetsov V, Pleiser S, Kerkhoff E, Schuh M. Spire-type actin nucleators cooperate with Formin-2 to drive asymmetric oocyte division. Curr Biol. 2011;21:955–960. doi: 10.1016/j.cub.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski JT, Gomez TS, Schoon RA, Mangalam AK, Billadeau DD. WASH knockout T cells demonstrate defective receptor trafficking, proliferation, and effector function. Mol Cell Biol. 2013;33:958–973. doi: 10.1128/MCB.01288-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V. Vectors for P-mediated transformation in Drosophila. Biotechnology. 1988;10:437–456. doi: 10.1016/b978-0-409-90042-2.50028-3. [DOI] [PubMed] [Google Scholar]
- Reed SC, Serio AW, Welch MD. Rickettsia parkeri invasion of diverse host cells involves an Arp2/3 complex, WAVE complex and Rho-family GTPase-dependent pathway. Cell Microbiol. 2012;14:529–545. doi: 10.1111/j.1462-5822.2011.01739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriquez AA. Botulinum toxin for spasmodic dysphonia. Phys Med Rehabil Clin N Am. 2003;14:767–779. doi: 10.1016/s1047-9651(03)00046-9. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Mesa E, Abreu-Blanco MT, Rosales-Nieves AE, Parkhurst SM. Developmental expression of Drosophila Wiskott-Aldrich Syndrome family proteins. Dev Dyn. 2012;241:608–626. doi: 10.1002/dvdy.23742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth P. Gal4 in the Drosophila female germline. Mech Dev. 1998;78:113–118. doi: 10.1016/s0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
- Rosales-Nieves AE, Johndrow JE, Keller LC, Magie CR, Pinto-Santini DM, Parkhurst SM. Coordination of microtubule and microfilament dynamics by Drosophila Rho1, Spire and Cappuccino. Nat Cell Biol. 2006;8:367–376. doi: 10.1038/ncb1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottner K, Hanisch J, Campellone KG. WASH, WHAMM and JMY: regulation of Arp2/3 complex and beyond. Trends Cell Biol. 2010;20:650–661. doi: 10.1016/j.tcb.2010.08.014. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Siekhaus D, Haesemeyer M, Moffitt O, Lehmann R. RhoL controls invasion and Rap1 localization during immune cell transmigration in Drosophila. Nat Cell Biol. 2010;12:605–610. doi: 10.1038/ncb2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiering D, Hodgson L. Dynamics of the Rho-family small GTPases in actin regulation and motility. Cell Adh Migr. 2011;5:170–180. doi: 10.4161/cam.5.2.14403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC. P Element-Mediated Transformation. Oxford, UK: IRL Press; 1986. [Google Scholar]
- Stevenson V, Hudson A, Cooley L, Theurkauf WE. Arp2/3-dependent pseudocleavage [correction of psuedocleavage] furrow assembly in syncytial Drosophila embryos. Curr Biol. 2002;12:705–711. doi: 10.1016/s0960-9822(02)00807-2. [DOI] [PubMed] [Google Scholar]
- Stofanko M, Kwon SY, Badenhorst P. A misexpression screen to identify regulators of Drosophila larval hemocyte development. Genetics. 2008;180:253–267. doi: 10.1534/genetics.108.089094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stradal TE, Rottner K, Disanza A, Confalonieri S, Innocenti M, Scita G. Regulation of actin dynamics by WASP and WAVE family proteins. Trends Cell Biol. 2004;14:303–311. doi: 10.1016/j.tcb.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Strong DM, Nelson K, Pierce M, Stramer SL. Preventing disease transmission by deceased tissue donors by testing blood for viral nucleic acid. Cell Tissue Bank. 2005;6:255–262. doi: 10.1007/s10561-005-2834-4. [DOI] [PubMed] [Google Scholar]
- Takenawa T, Suetsugu S. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat Rev Mol Cell Biol. 2007;8:37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- Tepass U, Fessler LI, Aziz A, Hartenstein V. Embryonic origin of hemocytes and their relationship to cell death in Drosophila. Development. 1994;120:1829–1837. doi: 10.1242/dev.120.7.1829. [DOI] [PubMed] [Google Scholar]
- Tucker PK, Evans IR, Wood W. Ena drives invasive macrophage migration in Drosophila embryos. Dis Model Mech. 2011;4:126–134. doi: 10.1242/dmm.005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verboon JM, Rincon-Arano H, Werwie TR, Delrow JJ, Scalzo D, Nandakumar V, Groudine M, Parkhurst SM. Wash interacts with lamin and affects global nuclear organization. Curr Biol. 2015;25:804–810. doi: 10.1016/j.cub.2015.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W, Faria C, Jacinto A. Distinct mechanisms regulate hemocyte chemotaxis during development and wound healing in Drosophila melanogaster. J Cell Biol. 2006;173:405–416. doi: 10.1083/jcb.200508161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W, Jacinto A. Drosophila melanogaster embryonic haemocytes: masters of multitasking. Nat Rev Mol Cell Biol. 2007;8:542–551. doi: 10.1038/nrm2202. [DOI] [PubMed] [Google Scholar]
- Xia P, Wang S, Du Y, Zhao Z, Shi L, Sun L, Huang G, Ye B, Li C, Dai Z, et al. WASH inhibits autophagy through suppression of Beclin 1 ubiquitination. EMBO J. 2013;32:2685–2696. doi: 10.1038/emboj.2013.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P, Wang S, Huang G, Zhu P, Li M, Ye B, Du Y, Fan Z. WASH is required for the differentiation commitment of hematopoietic stem cells in a c-Myc-dependent manner. J Exp Med. 2014;211:2119–2134. doi: 10.1084/jem.20140169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Condeelis J. Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim Biophys Acta. 2007;1773:642–652. doi: 10.1016/j.bbamcr.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki D, Kurisu S, Takenawa T. Regulation of cancer cell motility through actin reorganization. Cancer Sci. 2005;96:379–386. doi: 10.1111/j.1349-7006.2005.00062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zech T, Calaminus SD, Caswell P, Spence HJ, Carnell M, Insall RH, Norman J, Machesky LM. The Arp2/3 activator WASH regulates alpha5beta1-integrin-mediated invasive migration. J Cell Sci. 2011;124:3753–3759. doi: 10.1242/jcs.080986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.