Abstract
RNAs play various roles, not only as the genetic codes to synthesize proteins, but also as the direct participants of biological functions determined by their underlying high-order structures. Although many computational methods have been proposed for analyzing RNA structures, their accuracy and efficiency are limited, especially when applied to the large RNAs and the genome-wide data sets. Recently, advances in parallel sequencing and high-throughput chemical probing technologies have prompted the development of numerous new algorithms, which can incorporate the auxiliary structural information obtained from those experiments. Their potential has been revealed by the secondary structure prediction of ribosomal RNAs and the genome-wide ncRNA function annotation. In this review, the existing probing-directed computational methods for RNA secondary and tertiary structure analysis are discussed.
1. Background
RNA molecules, including both coding RNAs and non-coding RNAs (ncRNAs), play much more vital roles in the biological systems than what was suggested in the central dogma [1–3]. Their functions are not only encoded in the primary sequences [4], but also originate from the secondary and the tertiary structures [5–7]. Some well-known instances are the cloverleaf-like structure of tRNAs and the kink-turn structural motifs which server as important sites for protein recognition. Given the fact that most of transcripts (~90%) in typical eukaryotic genomes are ncRNAs, fully understanding RNAs and their functions is impossible without studying the high-order structures. However, the determination of RNA structures is not a trivial task. The traditional high-resolution techniques, such as X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy, are very time consuming and hard to implement. On the other hand, the RNA structure folding algorithms [8–11] and the RNA functional annotation algorithms [12–14] are not accurate and efficient enough for the large RNAs and the genome-wide data sets.
The chemical probing technique, also named “structure probing” or “footprinting”, provides a new way of studying RNA structures. RNAs of interest are treated with the chemical reagents which may modify the specific nucleotides with certain structural features. These modifications can act as stops for the primer extension, and their positions in the sequence can be detected by reverse transcription. Over the last 30 years chemical probing has been adopted for the study of RNA structures [15–17]. Recently more and more new protocols have been proposed to tackle the problems related to RNA structures. One of the most widely used probing experiments is to detect the paired and the unpaired bases. In these experiments, chemical reagents can form stable adducts with the flexible nucleotides in the loop regions, but not the protected bases in the stack regions. Some typical reagent choices are dimethyl sulfate (DMS) [18], kethoxal (KT) [19], diethyl pyrocarbonate (DEPC) [20], and CMCT [21]. None of them can react with all four RNA bases, e.g., DMS can only be applied to N1-adenine and N3-cytidine; KT can only be applied to N1 and N2 of guanine. A new protocol, selective 2’-hydroxyl acylation analyzed by primer extension (SHAPE) [22, 23], can involve reactions with all bases. Moreover, SHAPE is insensible to the solvent accessibility and RNA size, which makes it an excellent choice for characterizing the structure features of large RNAs. RNase enzyme is another important type of reagent for probing RNA secondary structures. Instead of adducting to nucleotides, RNase catalyzes the degradation of the single- or double-stranded regions into smaller segments [24, 25]. As a higher-order conformation which interlinks the packed secondary structure modules with through-space interactions, tertiary structure can also be analyzed with chemical probing experiments. For example, hydroxyl radicals generated by Fe(II)-EDTA catalyst can cleave the specific sites at RNA backbone proximal in space to the location of the bound Fe(II)-EDTA. Hence the long range interactions of the Fe(II) adducted nucleotides can be determined [26, 27]. Cross-linking technique adopts a different strategy to detect juxtaposed nucleotides in three-dimensional space. It bridges the nearby nucleotides in an RNA by using bifunctional reagents [28] or UV-irritation [29]. The products of the reaction can be characterized by mass mapping or sequencing experiments.
The introduction of next generation sequencing (NGS) leads to the development of genome-wide RNA structure probing protocols. Many high-throughput protocols, such as SHAPE-seq [30, 31], PARS [32–34], FragSeq [35], Map-seq [36], dsRNA-seq [37], CIRS-seq [38], and DMS-based high-throughput sequencing [39, 40], have been applied to the transcriptomes of various species. These experiments provide comprehensive insights into the structural features of the coding regions. In addition, the genome-wide sequencing also reveals the structural characterization of substantial ncRNAs, especially the lncRNAs [34, 39]. Recent studies show that the mutations and the dysregulations of lncRNAs are directly linked to many human diseases, ranging from neurodegeneration to cancer [41–45]. On the other hand, single-molecule probing has been combined with massive parallel sequencing to target the RNAs with complex structures. For RNA viruses, the functionally active structures are vital during their life cycle [46]. The global and local chemical probing of various viruses, such as the human immunodeficiency virus (HIV) [47], hepatitis C virus [48], influenza A virus [49] and the dengue virus [50], detected several potential regulatory motifs. Considering the limitations of traditional methods for RNA structure analysis, the rapid explosion of probing data coming from the high-throughput sequencing experiments will certainly enhance our understanding of human diseases.
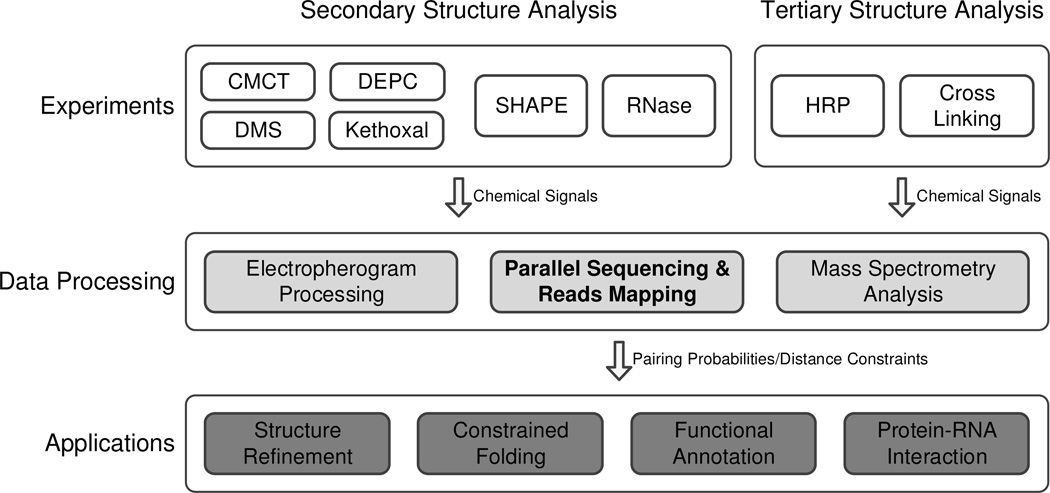
The embedded structural information in the probing data can be quantified, and then incorporated into the computational method. The first breakthrough was in the field of RNA secondary structure folding. By integrating reactivities as extra pseudo energy terms into the nearest neighbor energy model, the secondary structure prediction accuracy of Escherichia coli 16S rRNA can be increased greatly [51]. This successful application suggests the great potential in using reactivities to assist the computational analysis of RNA structure. This review will introduce the existing chemical probing-directed computational methods and their applications (Figure 1). In the discussion section we will also propose the possible directions of future research.
Figure 1.
The hierarchical overview of the RNA high-order structure analysis with probing-based computational methods. The white blocks represent chemical experiments and the shaded blocks represent the computational processing modules. Secondary structure and tertiary structure analysis adopt different protocols with different reagents. The output signals of the top-layer experiments are converted into reactivities, indicating pairing probabilities in secondary structure analysis or distance constraints in tertiary structure analysis, at the mid-layer. Finally the reactivities are incorporated into traditional algorithms at the bottom-layer.
2. The computation of reactivities
In the chemical probing experiments, the modifications on the flexible nucleotides can be located by the 5’-end labeled primer extension. The lengths of the cDNA fragments imply the positions of the modified sites, and the number of the mapped fragments at each site indicates its reactive degree [47]. Traditionally, gel electrophoresis (GE) had been utilized to visualize the results of probing experiments. Analyzing the gel images is a tedious work, so computational methods are required to automate and accelerate the procedure. SAFA [52] (https://simtk.org/home/safa) is a semi-automated analyzing tool for gel quantification. The users only need to edit the intermediate results guided by a graphic user interface. In recent years, most of the single-molecule probing protocols began to make use of capillary electrophoresis (CE) for sequencing. However, the traditional algorithms designed for DNA CE sequencing may not be suitable for quantifying structural probing reactivities [53]. The major issues are signal decay correction, x-axis and y-axis scaling, signal alignment, sequence alignment and peak fitting. CAFA [54] (https://simtk.org/home/cafa) offers a CE analyzing method for the chemical probing experiment which focuses on peak detection and fitting. ShapeFinder [53] (http://giddingslab.org/software) adds a peak and sequence alignment step to refine the fitting. It still requires users to select parameters and adjust the alignment manually. FAST [55] (http://glennlab.stanford.edu/software.html) improves the efficiency of CE analysis by automating the x-axis and y-axis scaling. QuShape [56] (http://www.chem.unc.edu/rna/qushape/) is presented as an updated version of the ShapeFinder by introducing new alignment and scaling algorithms. To align hundreds of capillaries together, two tools are provided by the Das lab: HiTRACE [57] (https://simtk.org/home/hitrace) and HiTRACE-Web [58] (http://hitrace.org/). The output intensity data of HiTRACE can be further processed with a likelihood-based framework [59, 60]. Recently, HiTRACE is also extended to allow CE processing standardization [61].
Compared to the reactivity computation of CE traces, the processing of reads generated by high-throughput sequencing-based probing protocols is more straightforward. First, the mapping of reads to the reference genome infers the sites of modification (normally 1nt upstream of the mapped reads). Second, the number of reads mapped to each site indicates its reactivity. Based on the two features, there are two groups of methods that quantify the read counts to reactivity values. The first group of methods normalizes the read counts directly. For examples, the raw read count of each site can be normalized by that of the most reactive base in a given window [39]; FragSeq computes pseudo counts based on the raw counts in a transcript, and then the pseudo counts are normalized such that they sum up to 1 [35]; PARS normalizes all read counts by sequencing depth, and then the log ratios of normalized counts for V1 RNase (cleaves the double-stranded RNA) and for S1 RNase (cleaves the single-stranded RNA) are computed. Notice that normalization is a general idea and can be applied to almost all the scenarios. On the other hand, the second type relies on the sophisticated statistical methods, which only model the specific protocols. A representative case is SHAPE-Seq, which assumes the number of times an RNA exposing to the chemical reagent follows the Poisson distribution. Based on this hypothesis and two observations (the read counts in (+) and (−) experiments), it employs maximum likelihood estimation to compute three parameters for each site (the rate of the Poisson distribution, adduct probability for each site, and reverse transcription termination probability) [62]. And the probability of modifying one site is defined as its reactivity. A later study suggests that the Poisson distribution assumption is not necessary [63]. The experimental result shows that the simplified model has similar results to the original one. Another interesting approach is proposed in SeqFold (http://ouyanglab.jax.org/seqfold) to process the PARS data [64]. Hypergeometric test is performed for each site to see if it is enriched with V1 or S1 reads. The high false discovery rates (FDR) in the hypothesis testing mark the significant single-stranded and double-stranded sites. To identify the protein-binding sites in RNAs, Probrna [65] (http://yiplab.cse.cuhk.edu.hk/probrna/) also uses the PARS data with statistical methods. In this algorithm, an extended linear Poisson model is proposed to express the relationship between the nucleotide properties and the observed read counts. This model also assumes that the read counts of paired and unpaired nucleotides following Poisson distribution with different parameters. By using the expectation maximization (EM) algorithm to fit the observation, the optimal assignment of the structure features (paired/unpaired) is used as the reactivities.
3. The analysis of RNA secondary structure
The probing data are not strong enough to predict RNA secondary structure directly due to its lack of interacting information. Originally, reactivities were just superimposed on the predicted secondary structures to verify the correctness [66–69]. These structures are determined by either the single-molecule folding algorithms [8–10, 70] or the comparative methods [11, 71–74]. However, both of them have problems: for single-molecule folding, the energy parameters are approximated [9], and the noise from random sequences prevents it from discovering new RNAs [75, 76]; for comparative methods, obtaining a precise structural alignment itself is a challenging problem, and the false positive rates are too high to screen the genome-wide data sets [77–79]. Considering the strengths and weaknesses of chemical probing and computational methods, their integration can generate much superior secondary structure models. All the methods mentioned in this section are summarized in Table 1.
Table 1.
Software packages for RNA secondary structure prediction discussed in this review.
| Software Name | Probing Protocol |
Computational Method |
Feature | Strategy |
|---|---|---|---|---|
| RNAstructure [10, 51] | SHAPE, DMS | MFE model | N/A | Add pseudo energies to the stack regions. |
| RNAsc [81] | SHAPE | MFE model | N/A | Add pseudo energies to all the nucleotides. |
| Optimal pseudo energy term* [82] | SHAPE | Statistical method | N/A | Maximize the posterior probability of the predicted structure given the probing data. |
| SeqFold [64] | PARS, SHAPE, SHAPE-Seq, FragSeq | Statistical method | N/A | Find the cluster of structures in the sampling space whose profile is “nearest” to the probing data. |
| RNApbfold [89] | SHAPE | Least square approximation | Ensemble | Find a structure ensemble minimizing the total error of energy model and probing data. |
| MutualFold [90] | SHAPE | MFE model | Alternative structures | Fold two alternative structures simultaneously by considering their mutual constraints in probing data. |
| ShapeKnots [94] | SHAPE | Heuristic method | Pseudoknot | Construct structures based on the entropic cost of pseudoknots. |
: In this work, the pseudo energy term is proposed without implementation.
One straightforward application of probing data is to use the pairing attributes of bases as constraints in the folding algorithm. Nevertheless, A-U, G-C pairs at helix ends, G-U pairs and nucleotides adjacent to G-U pairs can also be adducted by probing reagents. A pioneering approach using these probing features is implemented in the RNAstructure package [80] (http://rna.urmc.rochester.edu/RNAstructure.html). In this method, reactivities are still treated as hard constraints: the energy of a base pair is set to the positive infinity if they are prohibited to be pairing. The most significant improvement is that the lowly reactive pairs are allowed at the ends of helices. Experimental results show that the secondary structure of the E. coli 5S rRNA is much more accurately predicted when the probing constraints are used than when the energy model is used alone (from 26.3% to 86.8%). In the later version of RNAstructure, a novel soft-constrained algorithm is presented, in which the SHAPE reactivity of i-th site is converted into a pseudo free energy term [51]:
The default values for the intercept b and slope m are −0.8 kcal/mol and 2.6 kcal/mol, respectively. Pseudo energies are added once to the nucleotides at the helix end and twice to the interior bases. This new mechanism improves the sensitivity and the positive predictive value (PPV) of the secondary structure prediction for E. coli 16S rRNA from ~50% (without SHAPE reactivities) to ~97% (with SHAPE reactivities).
The success of rRNA structure prediction inspires people to bring forward many other probing-directed folding algorithms. RNAsc [81] (http://bioinformatics.bc.edu/clotelab/RNAsc/index.spy) argues that RNAstructure has biases because reactivities are only applied to the stack regions. To solve the problem, RNAsc defines the Boltzmann weights for the i-th base in the RNA as follows:
D measures the discrepancy between the predicted structural property x (0 for paired and 1 for unpaired) and the normalized reactivity qi (∈[0,1]). β is a scaling parameter. By integrating it into the computation of partition function, the ensemble distance between the probing data and the predicted structure can be optimized.
Both RNAstructure and RNAsc incorporate reactivities into the minimum free energy (MFE) model as pseudo energies. This strategy has been generalized into a new statistical model based on a joint probability P(α, x, π, θ, ψ) [82]: α is the observed probing data, x is the RNA sequence, π is the inferred secondary structure, θ is the set of energy parameters and ψ is the underlying likelihood model generating α from π. Hence the folding problem is transformed into finding a structure π which maximizes the posterior probability P(π|α, x, θ, ψ). Because P(π|x, θ) is given by the Boltzmann equation, θ can be dropped by integrating the structure ensemble. Thus the optimal secondary structure follows the equation:
It shows that the probing-directed folding algorithm should add the log probability of probing data α given the RNA structure π. Assuming that the reactivity αi is only dependent on πi, ΔG’i=RTlogP(αi|πi) is the appropriate pseudo energy for i-th base, where P(αi|πi) can be inferred from the empirical reactivity distributions.
Besides detecting the structure with the lowest folding energy, searching in suboptimal structures is also a common way of predicting RNA secondary structures [83, 84]. SeqFold [64] adopts this idea to find the optimal solution sampled from the Boltzmann-weighted ensemble [85]. Like RNAsc, the chemical signals of probing experiments are standardized into the Structural Preference Profiles (SPP), whose values range from 0 to 1 (0 for paired and 1 for unpaired). The Manhattan distance between the SPPs of the probing data and of each sampled structure is calculated to evaluate their similarity. The structures that have the shortest distance to the reactivities are recorded as the “nearest neighbors” to the probing data. Then the cluster containing the maximum number of neighbors is selected, and its centroid is identified as the secondary structure of the sequence.
In real biological systems, some RNAs exhibit complex structural dynamics [86–88]. Then the reactivities of those RNAs would come from multiple structures instead of one. RNApbfold (https://github.com/wash/probing) considers the entire Boltzmann ensemble of the sequence. The experimental signals are modeled as position-specific “perturbations” which distort the frequencies of structures in the ensemble [89]. The optimal perturbation vector is computed by minimizing the weighted sum of perturbed energies and the discrepancy between observed and predicted pairing probabilities
where ε is the perturbation vector, pi(εi) and qi are the predicted and observed pairing probability of the i-th site, σ and τ are the parameters estimating the variances of the energies and discrepancy. The term ε can be incorporated into an extended McCaskill’s algorithm to calculate the weighted partition function.
Finding the alternative structures in the ensemble is not an easy task due to the exponentially increasing search space. MutualFold (http://genome.ucf.edu/MutualFold/) proposes to recover two RNA structures within solution (ON/OFF conformations of riboswitch and ribozyme) by folding them simultaneously with the help of reactivities [90]. The predicted results must satisfy: (1) The sum of their free energy is minimized, and (2) The discrepancy between the expected and the observed reactive profiles is minimized. Since one structure can both constrain and be constrained by the folding of the other structure, a direct enumeration of possible structures and the corresponding constraints becomes very time consuming. To solve the problem, the “significant stacks” (> 4 base pairs) are retrieved from the RNA sequence. Then the mutual folding problem can be transformed into detecting the optimal stack configuration of both structures. Because the number of significant stacks is usually much smaller than the length of the sequence, the stack-guided algorithm can be applied to real cases, such as adenine riboswitch and TPP riboswitch, with acceptable time and space complexity.
Pseudoknot is a rare but crucial structural motif in RNA functions [91, 92]. Normally, it is ignored in the secondary structure prediction to avoid the highly time consuming computation [93]. Given the fact that pseudoknot sites can be detected by the chemical probing data, ShapeKnots [94] (http://rna.urmc.rochester.edu/Text/ShapeKnots.html) extends the SHAPE-directed secondary structure model in RNAstructure to consider one pseudoknot. Based on the target sequence and its SHAPE reactivities, the pseudoknot-free MFE structure and 99 suboptimal structures are predicted. A helix in one suboptimal structure is discarded if it shares more than 50% of its nucleotides with a helix in the MFE structure. For each remaining helix, a set of structures is generated by prohibiting its members from pairing. After that, the helix is inserted back as a potential pseudoknot. The energies of the constructed structures containing pseudoknots are adjusted, and the ones with the lowest energies are reported.
No base pairing information can be inferred from the common probing experiments. By incorporating mutagenesis, mutate-and-map converts the “one-dimensional” information obtained from the chemical probing technique into a “two-dimensional” map. Mutating one base in a pair will make its partner accessible to the chemical reagent. As a result, the reactivity of the exposed base is changed. This strategy has been used to verify single interaction [95, 96]. Mutate-and-map extends the idea to change all bases one by one, and then analyze the “map” of reactivities for all mutated sequences. It first was applied to a DNA/RNA double helix, in which the base pair interactions can be inferred by using Z-score [97]. In [98], a more sophisticated pipeline based on Z-score and RNA structure properties was proposed to analyze a “MedLoop hairpin”. It has been shown that this strategy can be applied to real RNAs, such as tRNA, ribozyme and riboswitch, and the prediction accuracy is better than RNAstructure which is based on the one-dimensional SHAPE [99]. (The discussion about the potential defects of the experiments can be found in [60, 100].)
Probing data can also be used to annotate the functions of ncRNAs. Generally, RNA function annotation relies on the structural alignment algorithm [13, 101, 102] whose efficiency is much lower than the sequence alignment. Some methods predict the secondary structures of the target sequences first and then convert them into sequential information to reduce the time complexity. However, the computational intensity is still high (O(n3)), and the prediction accuracy is also impacted [103]. ProbeAlign [104] (http://genome.ucf.edu/ProbeAlign/) makes use of reactivities generated by the high-throughput sequencing-based probing experiments to achieve both efficiency and accuracy. In this algorithm, the query is a profile of a known RNA family and the targets are RNA sequences with reactivities. The pairing partnership in the query is ignored, while the partial structural information of targets embedded in the probing data is retrieved to guide the alignment. Therefore it is similar to a sequence alignment algorithm (O(n2)). The benchmark results show that ProbeAlign outperforms the state-of-the-art tools with higher efficiency.
Recently, various high-throughput sequencing-based probing experiments have been applied to the genome-wide analysis of RNA structural profiles [105]. PARS reveals the structural properties of the yeast and the human transcriptomes [33, 34]. The average reactivities in the coding and UTR regions show that for yeast, coding regions are more structured, while for human, the UTR regions are more structured. In addition, a periodic structural signal (a cycle of 3 nucleotides) is detected in the coding regions with discrete Fourier transform. Last but not least, the correlation between PARS scores and RNA properties, such as the translation efficiency and the biological functions, can be evaluated by statistical hypothesis testing. Structure-seq uses similar scheme to analyze the in vivo regulatory features of Arabidopsis [40]. In [39], the structural difference between the in vivo and in vitro transcripts is studied by using Pearson correlation coefficient and the Gini index. The results show that the mRNAs in the yeast and the human genomes are much less structured in vivo than in vitro. Folding energy landscape can also be studied with high-throughput sequencing-based probing data. PARTE monitors the RNA structures in the whole yeast genome across five different temperatures [106]. The melting temperature (TE) for each nucleotide is computed by detecting the sharp transition with an adaptive regression model [69]. Many interesting features are observed from the TE landscape, e.g., on average the functional ncRNAs melt under a higher temperature than the one affecting mRNAs.
4. The analysis of RNA tertiary structure
Tertiary structures are essential for the functions of many ncRNAs [107, 108]. X-ray crystallography and NMR spectroscopy can provide high-resolution 3D structural conformation, but these methods are hard to implement [109] and NMR is generally limited to small RNAs [110]. On the other hand, many computational methods have been proposed to de novo predict RNA tertiary structures [111–115], but their accuracy is relatively low, especially for large molecules. To yield a better result, the coarse-grained through-space interactions embedded in the chemical probing data can be melded into the computational analysis as constraints. An overview of the probing-based tertiary structure prediction methods discussed in this section can be seen in Table 2.
Table 2.
Methods for RNA tertiary structure prediction discussed in this review.
| Method Name | Probing Protocol | Computational Method |
Strategy |
|---|---|---|---|
| Sequence-encoded cleavage [110] | HRP + SHAPE | DMD | Place Fe(II)-EDTA at specific nucleotides and find their neighbors by using HRP. |
| iFoldRNA-HRP [119] | HRP | DMD | Convert the HRP reactivities into the numbers of neighbors and use them as the constraints. |
| RING-MaP [120] | SHAPE-MaP | DMD | Model the correlation among the reactive sites in one RNA strand as through-space interaction. |
| MOCHA [122] | HRP | FARNA | Generate 2D mapping between interacted nucleotides by performing HRP on RNAs randomly. |
| MS3D [124, 125] | Bifunctional crosslinking | MC-Sym | Detect spatially conjugated nucleotides by using mass mapping technique. |
Discrete molecular dynamics (DMD) is a rapid simulation algorithm for three-dimensional protein structure prediction [116, 117]. By incorporating base pairing and stacking information, DMD has been extended to explore the conformational space of RNA molecules [118]. In this algorithm, a coarse-grained structural model is employed to reduce the computational intensity: each nucleotide is represented by three hard beads (phosphate, sugar and base), and the whole RNA sequence is simplified as a “bead-on-a-string” chain. For inter-atomic interactions, the potential energies are approximated as stepwise functions; for neighboring bonds, beads interact via an infinitely high square well potential. Thus during the search of the minimum energy in the landscape, the simulation procedure involves a series of collision events when the potential step borders are encountered. Distance constraints obtained from probing experiments, including base pairing and long range interaction, can provide additional information to refine the predictions of this model. The three-dimensional structure of yeast tRNAAsp has been analyzed with a sequence-encoded cleavage protocol [110]. The base pairing information of the RNA is determined by SHAPE. To detect the long range constraints, some consecutive base pairs in the native tRNA are mutated to bind methidiumpropyl-EDTA (MPE). After that, Fe(II)-EDTA moieties are placed at the sites marked by MPE. Then the through-space distances from each tethered site to the other sites can be evaluated with the reactive intensity of hydroxyl radical probing (HRP). It can be seen that this protocol has two limitations: (1) the mutations cannot disturb the structure of the native RNA; (2) the cleavage intensity is not at single-nucleotide resolution. In [119], solution HRP is performed on RNAs directly without using the mutated markers. An important observation for the method is that the HRP reactivity of a base is inversely proportional to the number of its through-space neighboring nucleotides, namely “contacts”. Thus for each nucleotide, a threshold contact number derived from its reactivity is assigned as an indirect measurement of the long range interaction distances. During the DMD simulation, repulsion potential is incurred if the contacts of a given nucleotide exceed its threshold. Not only HRP, DMS-based probing can also be adopted to retrieve the through-space constraints [120]. The chemical adduct experiment is optimized to yield multiple modifications in an RNA strand. The modifications are caused by a “breathing” mechanism in which the dynamically interacted nucleotides become transiently accessible to DMS. SHAPE-MaP [121] can provide a mutational profile for the sequence by integrating non-complementary nucleotides to adduct sites during the reverse transcription. The dependence of two mutations is checked by using χ2-test with SHAPE-MaP reactivities, and its strength is evaluated by using Pearson’s phi metric. In the DMD simulation, free energy bonuses are assigned to interacting pairs whose correlation coefficients are greater than the threshold value.
FARNA (Fragment Assembly of RNA) [111] is another de novo RNA tertiary structure prediction algorithm which may integrate constraints from probing data. In this algorithm, the possible local conformations (torsion angles) of an RNA sequence are drawn from a tri-nucleotide fragment library constructed with a crystal structure of 23S rRNA (PDB: 1ffk). The sampling of the global conformation is guided by an energy function, in which the pairing potential is dependent on the relative coordinates and the coplanarity of two bases. The high-throughput contact mapping information obtained from MOCHA (Multiplexed •OH Cleavage Analysis) can be incorporated into FARNA as an additional energy term [122]. Phosphorothioate and Fe(II) are attached to a randomly selected site on the target RNA and the experiment is controlled to yield approximately one modification (for both reagents) per RNA. The sites of HRP cleavage agent can be detected through backbone scission at phosphorothioate, and the corresponding cleaved nucleotides are indicated by the HRP data. Their mapping, which can be read out from a two-dimensional gel electrophoresis, reveals the through-space contacts (~25Å) in the tertiary structure.
Chemical crosslinking is also utilized in the RNA tertiary structure prediction. One major method focusing on using this technique is MS3D [123]. It has been successfully applied to the investigation of the pseudoknot in feline immunodeficiency virus (FIV) [124] and the HIV-1 ψ-RNA [125]. The bifunctional reagents, NM (Nitrogen mustard) and PDG (1,4-phenyl-diglyoxal), are used to bridge the spatially conjugated nucleotides at a relatively high resolution (NM: ~9Å, PDG: ~7.5Å). The sites of contacted nucleotides are determined by mass spectrometric analysis and tandem sequencing. In addition, the secondary structure of the target RNA is probed with CMCT, DMS and KT. Both types of information are used in the molecular modeling of MC-sym [113]. Like the strategy used by MOCHA, MC-sym defines the RNA structures as connected nucleotide cyclic motifs (NCMs), including lone-pair loop NCMs and double-stranded NCMs. All the possible NCM backbone templates are retrieved from the crystal structures in PDB as the references for the sequential construction of RNA tertiary structure. The initial results of MC-sym are refined by CNS (Crystallography and NMR system) [126], which is developed for high-resolution crystallographic or NMR structure determination. Only the predicted structures that obey the binding rules employed by CNS are accepted.
5. Conclusion and future perspectives
In this paper, we have discussed the computational approaches that integrate chemical probing information into the analysis of RNA secondary or tertiary structures. The base-pairing attributes interpreted from the probing data improve the performance of many applications, such as the secondary structure prediction of large RNAs, the detection of switchable structures, and the genome-wide annotation of ncRNAs. In addition, the spatial distance constraints inferred by correlated chemical reactions also support the RNA tertiary structure folding algorithm to be an alternate for X-ray crystallography and NMR spectroscopy.
Despite the rapid progress of the probing-based RNA structure analysis, there are still challenges in how to compute and use the reactivities. The first challenge is to distinguish the useful reactivities from the background. The cleavage and modification on the RNA strands can be affected by diverse structural factors other than base pairing interactions, e.g. the solvent accessibility and the protein-bind activities. Consequently the utilization of the probing data should not be merely restricted to measure the pairing probabilities, and the discrimination of the target reactivities and the noise would be essential to use them properly. Second, the expressed nucleotides from the high-throughput sequencing-based protocols may not be sufficient enough for the analysis of RNA structures. In most of NGS experiments, e.g. RNA-seq, the mapping of a read means all nucleotides in the covered genomic region are expressed. However, in structure probing experiments, one read only shows the pairing attribute of one base. So relatively, the sequencing depth of the structure probing experiment is much lower than that of the RNA-seq experiment. According to our observation, a large amount of nucleotides do not have reactivity values in the output of current high-throughput protocols. Moreover, the high-throughput sequencing technique also raises an issue of computational efficiency. The genome-wide structure probing data set provides substantial new evidences for the large-scale analysis involving the RNA family member searching and the novel RNAs discovery. Naturally, the corresponding algorithms are very time consuming, because the secondary structure folding and alignment need to consider the 2D base pairing interactions. With the additional probing information, we might simplify these interactions, or even the existing energy and statistical models of RNA secondary structure, to reduce the complexity of the existing algorithms. Last, the variation among the existing protocols requires a uniform model to handle the data from different sources. All the current algorithms either focus on one specific type of data, or treat different probing data with different methods. Both of the solutions are not user-friendly, and cannot be scaled to new protocols.
The structure determination of ncRNAs, in particular lncRNAs, and the analysis of genome-wide data sets can be further improved if we overcome these challenges. Although the current soft constrained folding algorithm yields highly accurate results for the rRNAs in E. coli [51], the simulation results show that for some other rRNAs, the performance may not be improved by using probing data directly [127]. With regards to the first challenge, the accuracy of structure prediction depends on the interpretation of the integrated probing data. Naturally the reactivities of the SHAPE protocol are only suggested to be indicators of the paired or unpaired bases. However, from the statistical analysis in the RMDB database [128], it can be seen that the empirical reactivity distributions of the hairpin loop, internal loop, and the junction are different. Thus the secondary structure prediction results should be better if the pseudo energies of different loops are distinguished before they are incorporated into the energy model. What’s more, comparative methods could be extended to recover the reactivities of lowly expressed nucleotides by associating the probing data of similar RNAs together. Several approaches have been proposed to infer the structures of one RNA by differentiating the reactivities of two reagents [129, 130], or by comparing the reactivities of the native RNA with its mutations [99]. Considering the fast pace of high-throughput probing data deposition, the reactivity comparison among different RNAs can be realized in the future. The consensus structure of multiple homologous RNAs generated from both the sequence conservation and the probing data correlation will be much more accurate and significant.
Chemical probing is a powerful technique for RNA structure analysis.
Probing signals can be quantified as reactivities.
Reactivities can be used for computational RNA structure analysis.
Challenges of probing-based computational methods are discussed.
Acknowledgements
This work was supported by the National Institute of General Medical Science of the National Institutes of Health, USA (R01GM102515).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Eddy SR. Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- 2.Huttenhofer A, Schattner P. The principles of guiding by RNA: chimeric RNA-protein enzymes. Nat. Rev. Genet. 2006;7:475–482. doi: 10.1038/nrg1855. [DOI] [PubMed] [Google Scholar]
- 3.Storz G. An expanding universe of noncoding RNAs. Science. 2002;296:1260–1263. doi: 10.1126/science.1072249. [DOI] [PubMed] [Google Scholar]
- 4.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 5.McManus CJ, Graveley BR. RNA structure and the mechanisms of alternative splicing. Curr. Opin. Genet. Dev. 2011;21:373–379. doi: 10.1016/j.gde.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noller HF. RNA structure: reading the ribosome. Science. 2005;309:1508–1514. doi: 10.1126/science.1111771. [DOI] [PubMed] [Google Scholar]
- 7.Wan Y, Kertesz M, Spitale RC, Segal E, Chang HY. Understanding the transcriptome through RNA structure. Nat. Rev. Genet. 2011;12:641–655. doi: 10.1038/nrg3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofacker IL, Fontana W, Stadler PF, Bonhoeffer LS, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structures. Monatsh. Chem. 1994;125:167–188. [Google Scholar]
- 9.Jaeger JA, Turner DH, Zuker M. Improved predictions of secondary structures for RNA. Proc. Natl. Acad. Sci. USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hofacker IL, Fekete M, Stadler PF. Secondary structure prediction for aligned RNA sequences. J. Mol. Biol. 2002;319:1059–1066. doi: 10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
- 12.Bafna V, Zhang S. FastR: fast database search tool for non-coding RNA; Proc. IEEE Comput. Syst. Bioinform. Conf; 2004. pp. 52–61. [DOI] [PubMed] [Google Scholar]
- 13.Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein RJ, Eddy SR. RSEARCH: finding homologs of single structured RNA sequences. BMC Bioinformatics. 2003;4:44. doi: 10.1186/1471-2105-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peattie DA, Gilbert W. Chemical probes for higher-order structure in RNA. Proc. Natl. Acad. Sci. USA. 1980;77:4679–4682. doi: 10.1073/pnas.77.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krol A, Carbon P. A guide for probing native small nuclear RNA and ribonucleoprotein structures. Methods Enzymol. 1989;180:212–227. doi: 10.1016/0076-6879(89)80103-x. [DOI] [PubMed] [Google Scholar]
- 17.Stern S, Moazed D, Noller HF. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- 18.Tijerina P, Mohr S, Russell R. DMS footprinting of structured RNAs and RNA-protein complexes. Nat. Protoc. 2007;2:2608–2623. doi: 10.1038/nprot.2007.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brow DA, Noller HF. Protection of ribosomal RNA from kethoxal in polyribosomes. Implication of specific sites in ribosome function. J. Mol. Biol. 1983;163:27–46. doi: 10.1016/0022-2836(83)90028-1. [DOI] [PubMed] [Google Scholar]
- 20.Singer B. All oxygens in nucleic acids react with carcinogenic ethylating agents. Nature. 1976;264:333–339. doi: 10.1038/264333a0. [DOI] [PubMed] [Google Scholar]
- 21.Fritz JJ, Lewin A, Hauswirth W, Agarwal A, Grant M, Shaw L. Development of hammerhead ribozymes to modulate endogenous gene expression for functional studies. Methods. 2002;28:276–285. doi: 10.1016/s1046-2023(02)00233-5. [DOI] [PubMed] [Google Scholar]
- 22.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2'-hydroxyl acylation and primer extension (SHAPE) J. Am. Chem. Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 23.Wilkinson KA, Merino EJ, Weeks KM. Selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 24.Auron PE, Weber LD, Rich A. Comparison of transfer ribonucleic acid structures using cobra venom and S1 endonucleases. Biochemistry. 1982;21:4700–4706. doi: 10.1021/bi00262a028. [DOI] [PubMed] [Google Scholar]
- 25.Ziehler WA, Engelke DR. Probing RNA structure with chemical reagents and enzymes. Curr. Protoc. Nucleic Acid Chem. 2001 doi: 10.1002/0471142700.nc0601s00. Chapter 6:Unit 6.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tullius TD, Greenbaum JA. Mapping nucleic acid structure by hydroxyl radical cleavage. Curr. Opin. Chem. Biol. 2005;9:127–134. doi: 10.1016/j.cbpa.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 27.Han H, Dervan PB. Visualization of RNA tertiary structure by RNA-EDTA.Fe(II) autocleavage: analysis of tRNA(Phe) with uridine-EDTA.Fe(II) at position 47. Proc. Natl. Acad. Sci. USA. 1994;91:4955–4959. doi: 10.1073/pnas.91.11.4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nygard O, Nika H. Identification by RNA-protein cross-linking of ribosomal proteins located at the interface between the small and the large subunits of mammalian ribosomes. EMBO J. 1982;1:357–362. doi: 10.1002/j.1460-2075.1982.tb01174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Juzumiene D, Shapkina T, Kirillov S, Wollenzien P. Short-range RNA-RNA crosslinking methods to determine rRNA structure and interactions. Methods. 2001;25:333–343. doi: 10.1006/meth.2001.1245. [DOI] [PubMed] [Google Scholar]
- 30.Lucks JB, Mortimer SA, Trapnell C, Luo S, Aviran S, Schroth GP, Pachter L, Doudna JA, Arkin AP. Multiplexed RNA structure characterization with selective 2'-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq) Proc. Natl. Acad. Sci. USA. 2011;108:11063–11068. doi: 10.1073/pnas.1106501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loughrey D, Watters KE, Settle AH, Lucks JB. SHAPE-Seq 2.0: systematic optimization and extension of high-throughput chemical probing of RNA secondary structure with next generation sequencing. Nucleic Acids Res. 2014;42:0. doi: 10.1093/nar/gku909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wan Y, Qu K, Ouyang Z, Chang HY. Genome-wide mapping of RNA structure using nuclease digestion and high-throughput sequencing. Nat. Protoc. 2013;8:849–869. doi: 10.1038/nprot.2013.045. [DOI] [PubMed] [Google Scholar]
- 33.Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, Segal E. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wan Y, Qu K, Zhang QC, Flynn RA, Manor O, Ouyang Z, Zhang J, Spitale RC, Snyder MP, Segal E, Chang HY. Landscape and variation of RNA secondary structure across the human transcriptome. Nature. 2014;505:706–709. doi: 10.1038/nature12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Underwood JG, Uzilov AV, Katzman S, Onodera CS, Mainzer JE, Mathews DH, Lowe TM, Salama SR, Haussler D. FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat. Methods. 2010;7:995–1001. doi: 10.1038/nmeth.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seetin MG, Kladwang W, Bida JP, Das R. Massively parallel RNA chemical mapping with a reduced bias MAP-seq protocol. Methods Mol. Biol. 2014;1086:95–117. doi: 10.1007/978-1-62703-667-2_6. [DOI] [PubMed] [Google Scholar]
- 37.Zheng Q, Ryvkin P, Li F, Dragomir I, Valladares O, Yang J, Cao K, Wang LS, Gregory BD. Genome-wide double-stranded RNA sequencing reveals the functional significance of base-paired RNAs in Arabidopsis. PLoS Genet. 2010;6:e1001141. doi: 10.1371/journal.pgen.1001141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Incarnato D, Neri F, Anselmi F, Oliviero S. Genome-wide profiling of mouse RNA secondary structures reveals key features of the mammalian transcriptome. Genome Biol. 2014;15:491. doi: 10.1186/s13059-014-0491-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- 41.Pasmant E, Sabbagh A, Vidaud M, Bieche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 2011;25:444–448. doi: 10.1096/fj.10-172452. [DOI] [PubMed] [Google Scholar]
- 42.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, 3rd, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tano K, Akimitsu N. Long non-coding RNAs in cancer progression. Front Genet. 2012;3:219. doi: 10.3389/fgene.2012.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hofacker IL, Stadler PF, Stocsits RR. Conserved RNA secondary structures in viral genomes: a survey. Bioinformatics. 2004;20:1495–1499. doi: 10.1093/bioinformatics/bth108. [DOI] [PubMed] [Google Scholar]
- 47.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr, Swanstrom R, Burch CL, Weeks KM. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lukavsky PJ, Otto GA, Lancaster AM, Sarnow P, Puglisi JD. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat. Struct. Biol. 2000;7:1105–1110. doi: 10.1038/81951. [DOI] [PubMed] [Google Scholar]
- 49.Priore SF, Kierzek E, Kierzek R, Baman JR, Moss WN, Dela-Moss LI, Turner DH. Secondary structure of a conserved domain in the intron of influenza A NS1 mRNA. PLoS One. 2013;8:e70615. doi: 10.1371/journal.pone.0070615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chapman EG, Moon SL, Wilusz J, Kieft JS. RNA structures that resist degradation by Xrn1 produce a pathogenic Dengue virus RNA. Elife. 2014;3:e01892. doi: 10.7554/eLife.01892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc. Natl. Acad. Sci. USA. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC. ShapeFinder: a software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA. 2008;14:1979–1990. doi: 10.1261/rna.1166808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mitra S, Shcherbakova IV, Altman RB, Brenowitz M, Laederach A. High-throughput single-nucleotide structural mapping by capillary automated footprinting analysis. Nucleic Acids Res. 2008;36:e63. doi: 10.1093/nar/gkn267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pang PS, Elazar M, Pham EA, Glenn JS. Simplified RNA secondary structure mapping by automation of SHAPE data analysis. Nucleic Acids Res. 2011;39:e151. doi: 10.1093/nar/gkr773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karabiber F, McGinnis JL, Favorov OV, Weeks KM. QuShape: rapid, accurate, and best-practices quantification of nucleic acid probing information, resolved by capillary electrophoresis. RNA. 2013;19:63–73. doi: 10.1261/rna.036327.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoon S, Kim J, Hum J, Kim H, Park S, Kladwang W, Das R. HiTRACE: high-throughput robust analysis for capillary electrophoresis. Bioinformatics. 2011;27:1798–1805. doi: 10.1093/bioinformatics/btr277. [DOI] [PubMed] [Google Scholar]
- 58.Kim H, Cordero P, Das R, Yoon S. HiTRACE-Web: an online tool for robust analysis of high-throughput capillary electrophoresis. Nucleic Acids Res. 2013;41:W492–W498. doi: 10.1093/nar/gkt501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kladwang W, VanLang CC, Cordero P, Das R. Understanding the errors of SHAPE-directed RNA structure modeling. Biochemistry. 2011;50:8049–8056. doi: 10.1021/bi200524n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cordero P, Kladwang W, VanLang CC, Das R. Quantitative dimethyl sulfate mapping for automated RNA secondary structure inference. Biochemistry. 2012;51:7037–7039. doi: 10.1021/bi3008802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kladwang W, Mann TH, Becka A, Tian S, Kim H, Yoon S, Das R. Standardization of RNA chemical mapping experiments. Biochemistry. 2014;53:3063–3065. doi: 10.1021/bi5003426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aviran S, Trapnell C, Lucks JB, Mortimer SA, Luo S, Schroth GP, Doudna JA, Arkin AP, Pachter L. Modeling and automation of sequencing-based characterization of RNA structure. Proc. Natl. Acad. Sci. USA. 2011;108:11069–11074. doi: 10.1073/pnas.1106541108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aviran S, Lucks JB, Pachter L. RNA structure characterization from chemical mapping experiments; 49th Allerton Conference on Communication, Control, and Computing; 2011. pp. 1743–1750. [Google Scholar]
- 64.Ouyang Z, Snyder MP, Chang HY. SeqFold: genome-scale reconstruction of RNA secondary structure integrating high-throughput sequencing data. Genome Res. 2013;23:377–387. doi: 10.1101/gr.138545.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu X, Wong TK, Lu ZJ, Chan TF, Lau TC, Yiu SM, Yip KY. Computational identification of protein binding sites on RNAs using high-throughput RNA structure-probing data. Bioinformatics. 2014;30:1049–1055. doi: 10.1093/bioinformatics/btt757. [DOI] [PubMed] [Google Scholar]
- 66.Inoue T, Cech TR. Secondary structure of the circular form of the Tetrahymena rRNA intervening sequence: a technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc. Natl. Acad. Sci. USA. 1985;82:648–652. doi: 10.1073/pnas.82.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ehresmann C, Baudin F, Mougel M, Romby P, Ebel JP, Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987;15:9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Merryman C, Moazed D, McWhirter J, Noller HF. Nucleotides in 16S rRNA protected by the association of 30S and 50S ribosomal subunits. J. Mol. Biol. 1999;285:97–105. doi: 10.1006/jmbi.1998.2242. [DOI] [PubMed] [Google Scholar]
- 69.Wilkinson KA, Merino EJ, Weeks KM. RNA SHAPE chemistry reveals nonhierarchical interactions dominate equilibrium structural transitions in tRNA(Asp) transcripts. J. Am. Chem. Soc. 2005;127:4659–4667. doi: 10.1021/ja0436749. [DOI] [PubMed] [Google Scholar]
- 70.Markham NR, Zuker M. UNAFold: software for nucleic acid folding and hybridization. Methods Mol. Biol. 2008;453:3–31. doi: 10.1007/978-1-60327-429-6_1. [DOI] [PubMed] [Google Scholar]
- 71.Washietl S, Hofacker IL, Stadler PF. Fast and reliable prediction of noncoding RNAs. Proc. Natl. Acad. Sci. USA. 2005;102:2454–2459. doi: 10.1073/pnas.0409169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ge P, Zhang S. Incorporating phylogenetic-based covarying mutations into RNAalifold for RNA consensus structure prediction. BMC Bioinformatics. 2013;14:142. doi: 10.1186/1471-2105-14-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rivas E, Eddy SR. Noncoding RNA gene detection using comparative sequence analysis. BMC Bioinformatics. 2001;2:8. doi: 10.1186/1471-2105-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Knudsen B, Hein J. RNA secondary structure prediction using stochastic context-free grammars and evolutionary history. Bioinformatics. 1999;15:446–454. doi: 10.1093/bioinformatics/15.6.446. [DOI] [PubMed] [Google Scholar]
- 75.Workman C, Krogh A. No evidence that mRNAs have lower folding free energies than random sequences with the same dinucleotide distribution. Nucleic Acids Res. 1999;27:4816–4822. doi: 10.1093/nar/27.24.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rivas E, Eddy SR. Secondary structure alone is generally not statistically significant for the detection of noncoding RNAs. Bioinformatics. 2000;16:583–605. doi: 10.1093/bioinformatics/16.7.583. [DOI] [PubMed] [Google Scholar]
- 77.Babak T, Blencowe BJ, Hughes TR. Considerations in the identification of functional RNA structural elements in genomic alignments. BMC Bioinformatics. 2007;8:33. doi: 10.1186/1471-2105-8-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stricklin SL. Noncoding RNA Genes in Caenorhabditis Elegans. St. Louis: Washington University; 2006. [Google Scholar]
- 79.Babak T, Blencowe BJ, Hughes TR. A systematic search for new mammalian noncoding RNAs indicates little conserved intergenic transcription. BMC Genomics. 2005;6:104. doi: 10.1186/1471-2164-6-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zarringhalam K, Meyer MM, Dotu I, Chuang JH, Clote P. Integrating chemical footprinting data into RNA secondary structure prediction. PLoS One. 2012;7:e45160. doi: 10.1371/journal.pone.0045160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eddy SR. Computational analysis of conserved RNA secondary structure in transcriptomes and genomes. Annu. Rev. Biophys. 2014;43:433–456. doi: 10.1146/annurev-biophys-051013-022950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kwakman JH, Konings DA, Hogeweg P, Pel HJ, Grivell LA. Structural analysis of a group II intron by chemical modifications and minimal energy calculations. J. Biomol. Struct. Dyn. 1990;8:413–430. doi: 10.1080/07391102.1990.10507813. [DOI] [PubMed] [Google Scholar]
- 84.Banerjee AR, Jaeger JA, Turner DH. Thermal unfolding of a group I ribozyme: the low-temperature transition is primarily disruption of tertiary structure. Biochemistry. 1993;32:153–163. doi: 10.1021/bi00052a021. [DOI] [PubMed] [Google Scholar]
- 85.Ding Y, Lawrence CE. A statistical sampling algorithm for RNA secondary structure prediction. Nucleic Acids Res. 2003;31:7280–7301. doi: 10.1093/nar/gkg938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Garst AD, Edwards AL, Batey RT. Riboswitches: structures and mechanisms. Cold Spring Harb Perspect Biol. 2011;3:a003533. doi: 10.1101/cshperspect.a003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schultes EA, Bartel DP. One sequence, two ribozymes: implications for the emergence of new ribozyme folds. Science. 2000;289:448–452. doi: 10.1126/science.289.5478.448. [DOI] [PubMed] [Google Scholar]
- 88.Zhuang X, Kim H, Pereira MJ, Babcock HP, Walter NG, Chu S. Correlating structural dynamics and function in single ribozyme molecules. Science. 2002;296:1473–1476. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]
- 89.Washietl S, Hofacker IL, Stadler PF, Kellis M. RNA folding with soft constraints: reconciliation of probing data and thermodynamic secondary structure prediction. Nucleic Acids Res. 2012;40:4261–4272. doi: 10.1093/nar/gks009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhong C, Zhang S. Simultaneous folding of alternative RNA structures with mutual constraints: an application to next-generation sequencing-based RNA structure probing. J. Comput. Biol. 2014;21:609–621. doi: 10.1089/cmb.2013.0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pleij CW. Pseudoknots: a new motif in the RNA game. Trends Biochem. Sci. 1990;15:143–147. doi: 10.1016/0968-0004(90)90214-v. [DOI] [PubMed] [Google Scholar]
- 92.Staple DW, Butcher SE. Pseudoknots: RNA structures with diverse functions. PLoS Biol. 2005;3:e213. doi: 10.1371/journal.pbio.0030213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lyngso RB, Pedersen CN. RNA pseudoknot prediction in energy-based models. J. Comput. Biol. 2000;7:409–427. doi: 10.1089/106652700750050862. [DOI] [PubMed] [Google Scholar]
- 94.Hajdin CE, Bellaousov S, Huggins W, Leonard CW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. Proc. Natl. Acad. Sci. USA. 2013;110:5498–5503. doi: 10.1073/pnas.1219988110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Duncan CD, Weeks KM. SHAPE analysis of long-range interactions reveals extensive and thermodynamically preferred misfolding in a fragile group I intron RNA. Biochemistry. 2008;47:8504–8513. doi: 10.1021/bi800207b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pyle AM, Murphy FL, Cech TR. RNA substrate binding site in the catalytic core of the Tetrahymena ribozyme. Nature. 1992;358:123–128. doi: 10.1038/358123a0. [DOI] [PubMed] [Google Scholar]
- 97.Kladwang W, Das R. A mutate-and-map strategy for inferring base pairs in structured nucleic acids: proof of concept on a DNA/RNA helix. Biochemistry. 2010;49:7414–7416. doi: 10.1021/bi101123g. [DOI] [PubMed] [Google Scholar]
- 98.Kladwang W, Cordero P, Das R. A mutate-and-map strategy accurately infers the base pairs of a 35-nucleotide model RNA. RNA. 2011;17:522–534. doi: 10.1261/rna.2516311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kladwang W, VanLang CC, Cordero P, Das R. A two-dimensional mutate-and-map strategy for non-coding RNA structure. Nat. Chem. 2011;3:954–962. doi: 10.1038/nchem.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Leonard CW, Hajdin CE, Karabiber F, Mathews DH, Favorov OV, Dokholyan NV, Weeks KM. Principles for understanding the accuracy of SHAPE-directed RNA structure modeling. Biochemistry. 2013;52:588–595. doi: 10.1021/bi300755u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hochsmann M, Toller T, Giegerich R, Kurtz S. Local similarity in RNA secondary structures. Proc. IEEE Comput. Soc. Bioinform. Conf. 2003;2:159–168. [PubMed] [Google Scholar]
- 102.Will S, Reiche K, Hofacker IL, Stadler PF, Backofen R. Inferring noncoding RNA families and classes by means of genome-scale structure-based clustering. PLoS Comput. Biol. 2007;3:e65. doi: 10.1371/journal.pcbi.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tseng HH, Weinberg Z, Gore J, Breaker RR, Ruzzo WL. Finding non-coding RNAs through genome-scale clustering. J Bioinform. Comput. Biol. 2009;7:373–388. doi: 10.1142/s0219720009004126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ge P, Zhong C, Zhang S. ProbeAlign: incorporating high-throughput sequencing-based structure probing information into ncRNA homology search. BMC Bioinformatics. 2014;15(Suppl 9):S15. doi: 10.1186/1471-2105-15-S9-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mortimer SA, Kidwell MA, Doudna JA. Insights into RNA structure and function from genome-wide studies. Nat. Rev. Genet. 2014;15:469–479. doi: 10.1038/nrg3681. [DOI] [PubMed] [Google Scholar]
- 106.Wan Y, Qu K, Ouyang Z, Kertesz M, Li J, Tibshirani R, Makino DL, Nutter RC, Segal E, Chang HY. Genome-wide measurement of RNA folding energies. Mol. Cell. 2012;48:169–181. doi: 10.1016/j.molcel.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat. Rev. Genet. 2009;10:833–844. doi: 10.1038/nrg2683. [DOI] [PubMed] [Google Scholar]
- 108.Woodson SA. Compact intermediates in RNA folding. Annu. Rev. Biophys. 2010;39:61–77. doi: 10.1146/annurev.biophys.093008.131334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ke A, Doudna JA. Crystallization of RNA and RNA-protein complexes. Methods. 2004;34:408–414. doi: 10.1016/j.ymeth.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 110.Gherghe CM, Leonard CW, Ding F, Dokholyan NV, Weeks KM. Native-like RNA tertiary structures using a sequence-encoded cleavage agent and refinement by discrete molecular dynamics. J. Am. Chem. Soc. 2009;131:2541–2546. doi: 10.1021/ja805460e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Das R, Baker D. Automated de novo prediction of native-like RNA tertiary structures. Proc. Natl. Acad. Sci. USA. 2007;104:14664–14669. doi: 10.1073/pnas.0703836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Martinez HM, Maizel JV, Jr, Shapiro BA. RNA2D3D: a program for generating, viewing, and comparing 3-dimensional models of RNA. J. Biomol. Struct. Dyn. 2008;25:669–683. doi: 10.1080/07391102.2008.10531240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Parisien M, Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452:51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 114.Sharma S, Ding F, Dokholyan NV. iFoldRNA: three-dimensional RNA structure prediction and folding. Bioinformatics. 2008;24:1951–1952. doi: 10.1093/bioinformatics/btn328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhao Y, Huang Y, Gong Z, Wang Y, Man J, Xiao Y. Automated and fast building of three-dimensional RNA structures. Sci. Rep. 2012;2:734. doi: 10.1038/srep00734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dokholyan NV, Buldyrev SV, Stanley HE, Shakhnovich EI. Discrete molecular dynamics studies of the folding of a protein-like model. Fold. Des. 1998;3:577–587. doi: 10.1016/S1359-0278(98)00072-8. [DOI] [PubMed] [Google Scholar]
- 117.Ding F, Dokholyan NV. Simple but predictive protein models. Trends Biotechnol. 2005;23:450–455. doi: 10.1016/j.tibtech.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 118.Ding F, Sharma S, Chalasani P, Demidov VV, Broude NE, Dokholyan NV. Ab initio RNA folding by discrete molecular dynamics: from structure prediction to folding mechanisms. RNA. 2008;14:1164–1173. doi: 10.1261/rna.894608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ding F, Lavender CA, Weeks KM, Dokholyan NV. Three-dimensional RNA structure refinement by hydroxyl radical probing. Nat. Methods. 2012;9:603–608. doi: 10.1038/nmeth.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Homan PJ, Favorov OV, Lavender CA, Kursun O, Ge X, Busan S, Dokholyan NV, Weeks KM. Single-molecule correlated chemical probing of RNA. Proc. Natl. Acad. Sci. USA. 2014;111:13858–13863. doi: 10.1073/pnas.1407306111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Siegfried NA, Busan S, Rice GM, Nelson JA, Weeks KM. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP) Nat. Methods. 2014;11:959–965. doi: 10.1038/nmeth.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Das R, Kudaravalli M, Jonikas M, Laederach A, Fong R, Schwans JP, Baker D, Piccirilli JA, Altman RB, Herschlag D. Structural inference of native and partially folded RNA by high-throughput contact mapping. Proc. Natl. Acad. Sci. USA. 2008;105:4144–4149. doi: 10.1073/pnas.0709032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Young MM, Tang N, Hempel JC, Oshiro CM, Taylor EW, Kuntz ID, Gibson BW, Dollinger G. High throughput protein fold identification by using experimental constraints derived from intramolecular cross-links and mass spectrometry. Proc. Natl. Acad. Sci. USA. 2000;97:5802–5806. doi: 10.1073/pnas.090099097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yu ET, Zhang Q, Fabris D. Untying the FIV frameshifting pseudoknot structure by MS3D. J. Mol. Biol. 2005;345:69–80. doi: 10.1016/j.jmb.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 125.Yu ET, Hawkins A, Eaton J, Fabris D. MS3D structural elucidation of the HIV-1 packaging signal. Proc. Natl. Acad. Sci. USA. 2008;105:12248–12253. doi: 10.1073/pnas.0800509105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta. Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 127.Sukosd Z, Swenson MS, Kjems J, Heitsch CE. Evaluating the accuracy of SHAPE-directed RNA secondary structure predictions. Nucleic Acids Res. 2013;41:2807–2816. doi: 10.1093/nar/gks1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cordero P, Lucks JB, Das R. An RNA Mapping DataBase for curating RNA structure mapping experiments. Bioinformatics. 2012;28:3006–3008. doi: 10.1093/bioinformatics/bts554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Steen KA, Rice GM, Weeks KM. Fingerprinting noncanonical and tertiary RNA structures by differential SHAPE reactivity. J. Am. Chem. Soc. 2012;134:13160–13163. doi: 10.1021/ja304027m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rice GM, Leonard CW, Weeks KM. RNA secondary structure modeling at consistent high accuracy using differential SHAPE. RNA. 2014;20:846–854. doi: 10.1261/rna.043323.113. [DOI] [PMC free article] [PubMed] [Google Scholar]