ABSTRACT
Bovine respiratory disease (BRD) is a common health problem for both dairy and beef cattle, resulting in significant economic loses. In order to identify viruses associated with BRD, we used a metagenomics approach to enrich and sequence viral nucleic acids in the nasal swabs of 50 young dairy cattle with symptoms of BRD. Following deep sequencing, de novo assembly, and translated protein sequence similarity searches, numerous known and previously uncharacterized viruses were identified. Bovine adenovirus 3, bovine adeno-associated virus, bovine influenza D virus, bovine parvovirus 2, bovine herpesvirus 6, bovine rhinitis A virus, and multiple genotypes of bovine rhinitis B virus were identified. The genomes of a previously uncharacterized astrovirus and picobirnaviruses were also partially or fully sequenced. Using real-time PCR, the rates of detection of the eight viruses that generated the most reads were compared for the nasal secretions of 50 animals with BRD versus 50 location-matched healthy control animals. Viruses were detected in 68% of BRD-affected animals versus 16% of healthy control animals. Thirty-eight percent of sick animals versus 8% of controls were infected with multiple respiratory viruses. Significantly associated with BRD were bovine adenovirus 3 (P < 0.0001), bovine rhinitis A virus (P = 0.005), and the recently described bovine influenza D virus (P = 0.006), which were detected either alone or in combination in 62% of animals with BRD. A metagenomics and real-time PCR detection approach in carefully matched cases and controls can provide a rapid means to identify viruses associated with a complex disease, paving the way for further confirmatory tests and ultimately to effective intervention strategies.
IMPORTANCE Bovine respiratory disease is the most economically important disease affecting the cattle industry, whose complex root causes include environmental, genetics, and infectious factors. Using an unbiased metagenomics approach, we characterized the viruses in respiratory secretions from BRD cases and identified known and previously uncharacterized viruses belonging to seven viral families. Using a case-control format with location-matched animals, we compared the rates of viral detection and identified 3 viruses associated with severe BRD signs. Combining a metagenomics and case-control format can provide candidate pathogens associated with complex infectious diseases and inform further studies aimed at reducing their impact.
INTRODUCTION
Bovine respiratory disease (BRD) is the most common and costly problem in the cattle industry, accounting for 70 to 80% of morbidity and 40 to 50% of mortality in U.S. feedlots (1, 2). The cattle industry is one of the largest agricultural sectors of the United States economy, with approximately three-quarters of a million farms raising cattle. The annual costs of BRD have been estimated at over 1 billion dollars per year (3, 4).
In BRD, bacterial infections are thought to be opportunistic infections precipitated by viral infections causing damage to the respiratory epithelium (5, 6). The use of prophylactic antibiotic treatment has been shown to be of limited utility in reducing BRD, and concerns exist about decreasing efficacy and the spread of antibiotic resistance to bovine or human pathogens (7). A review of the efficacy of bacterial vaccination in feedlot cattle for Histophilus somni, Mannheimia haemolytica, and Pasteurella multocida indicated little to no benefit (5, 8). Overall, neither immunization nor antimicrobial therapies have noticeably reduced the prevalence or severity of BRD (4).
Prior studies have implicated several viruses in BRD (2, 6, 9, 10). Studies testing for one or a few viruses or measuring seroconversions have reported the following viruses as associated with BRD: bovine herpesvirus 1 (BHV-1) (9, 11) (Herpesviridae), bovine viral diarrhea virus (BVDV) (in the Flaviviridae family) (12, 13), bovine parainfluenza type 3 virus (PI3V) and bovine respiratory syncytial virus (BRSV) (both in the Paramyxoviridae family) (9, 14, 15), bovine adenovirus 3 (BAdV3) (10, 16), and bovine coronavirus (BoCV) (15, 17, 18).
The use of viral metagenomics has recently allowed the rapid genetic characterization of viral genomes and has found widespread applications in animal viral discovery (19–22). Studies of farm animals have revealed the presence of both known and new viruses occurring at high frequency in healthy and sick animals (20, 23–27). Using deep sequencing, the Schmallenberg bunyavirus was recently identified in the plasma of cows with fever, decreased milk production, and diarrhea and was shown to induce symptoms following inoculations (28). A novel astrovirus was recently identified in brain tissues as the likely cause of a common neurological condition in cattle (29, 30).
In order to characterize the respiratory virome of cattle with BRD, we analyzed nasopharyngeal and pharyngeal recess swabs from 50 young dairy calves with symptoms of BRD and 50 healthy controls collected by the Bovine Respiratory Disease Complex Coordinated Agricultural Project (BRDC-CAP) (31). Using partial and complete viral genomes, we then designed real-time PCR assays to measure the frequency of infection and viral loads in animals with BRD and location-matched healthy controls to identify which respiratory viruses were associated with this disease.
MATERIALS AND METHODS
California dairy calves between the ages of 27 and 60 days of age housed in hutches were closely monitored and considered for enrollment as either BRD cases or controls, based on the assignment of calf health scores using a respiratory screening tool developed by McGuirk (32). Calves were observed and assigned a numeric health score (0 for normal, 1 for slightly abnormal, 2 for abnormal, and 3 for severely abnormal) for rectal temperature, cough, nasal discharge, and the greater of the two scores for eye discharge, and ear tilt. Calves with a cumulative score of ≥5 were enrolled in the BRDC-CAP study as a BRD case. The calves with BRD studied here were selected among those with the most severe signs, with cumulative scores of 8 to 12. Immediately thereafter, a calf adjacent to the BRD case that possessed a cumulative score of ≤3 was enrolled as a matched control for this study.
All case and control calves had nasopharyngeal and pharyngeal recess swabs collected. Samples were collected from the nasopharyngeal region by utilizing a six-inch sterile unguarded polyester swab that was inserted five inches into a clean naris and rotated against the surface for 15 s. Thereafter, the swab was removed and placed into 3 ml of viral transport medium (minimum essential medium, HCO3, HEPES, gentamicin, amphotericin B). The pharyngeal recess was swabbed using a 27-inch sterile guarded swab with a polyester fiber tip (Kalajian Industries, Signal Hill, CA). The swab was rotated against the pharyngeal recess surface for 15 s, retracted back into the guarding sheath, and placed into the same tube of viral transport medium containing the nasopharyngeal swab.
Viral metagenomics.
Ten pools of five randomly selected BRD samples were assembled using 32 μl from each sample. The resulting 160-μl pools were passed through a 200-nm filter, and the resulting filtrates were incubated for 1.5 h in a cocktail of DNase and RNase enzymes (53). Nucleic acids were then extracted using the QIAquick viral RNA column purification system according to the manufacturer's instructions. Reverse transcription was performed using a 28-base oligonucleotide whose 3′ end consisted of eight Ns (all four nucleotides at each of the eight 3′ positions) and whose 5′-end 20 bases consisted of an arbitrarily designed sequence (primer N1, CCTTGAAGGCGGACTGTGAGNNNNNNNN). Following denaturation of the first-strand cDNA product, a complementary strand was synthesized using Klenow fragment DNA polymerase extension (New England BioLabs). The resulting double-stranded cDNA and DNA were then PCR amplified using AmpliTaq Gold DNA polymerase and a 20-base primer (the same as that described above but without the 8 Ns) (33). The randomly amplified nucleic acid was then subjected to the NexteraXT library preparation protocol (Illumina) according to the manufacturer's instructions and sequenced using Illumina's HiSeq platform. A total of 23.7 million reads were generated, fewer than the 100 to 150 million expected due to suboptimal cluster formation.
Bioinformatic analysis.
An in-house analysis pipeline running on a 32-node Linux cluster was used to process the data. One-hundred-base reads from one lane of a HiSeq run were debarcoded using vendor software from Illumina. Human reads and bacterial reads were subtracted by mapping these reads to human reference genome hg19 and bacterial RefSeq genomes release 66 using bowtie2 (34). Reads were considered duplicates if bases 5 to 55 from 5′ end were identical. One random copy of duplicates was kept. Low-sequencing-quality tails were trimmed using a Phred quality score of 10 as the threshold. Adaptor and primer sequences were trimmed using the default parameters of VecScreen (35). The cleaned reads were de novo assembled using an in-house de novo sequence assembler (74) consists of SOAPdenovo2 (36), ABySS (37), meta-Velvet (38), and CAP3 (39). The assembled contigs, along with singlets, were aligned to an in-house viral proteome database using BLASTx.
Sequences belonging to different viral families are reanalyzed manually to identify sequence diversity and viral strains (Geneious version R6). Viral contigs from each pool and across pools were reassembled with high-sequence-identity criteria. Reads representing the same viral genome with high sequence identities (>95%) were reassembled into a larger contig or genome. Reads representing different viral strains (sequence identities of <95%) (e.g., the different BRBV strains) were individually identified and analyzed and then confirmed by specific PCR and Sanger sequencing. Finally, reads were mapped to the final viral sequence to account for the number of reads to that sequence (Geneious version R6).
Genome extension and RACE.
Gaps in the viral genomes were filled using the Superscript III-Platinum Taq one-step reverse transcription-PCR (RT-PCR) kit for RNA sequences shorter than 2,000 bp and the TaKaRa Ex Taq PCR kit for DNA sequences shorter than 2,000 bp. Longer gaps were obtained by bridging PCR amplification with TaKaRa LA Taq. In the case of RNA genomes, random cDNA templates were generated prior to PCR amplification with Prime Script reverse transcriptase (TaKaRa) and a random hexamer. Extremities of RNA genomes were obtained using Clontech SMARTerII rapid amplification of cDNA ends (RACE) cDNA kits followed by amplification with LA Taq (TaKaRa).
Next-generation sequencing of individual samples.
When deemed necessary due to a large number of gaps and/or an incomplete genome, individual samples were prepared for next-generation sequencing in a manner analogous to the one described above. Individual samples were resequenced using an Illumina MiSeq Platform.
Case-control real-time PCR.
Viral transport medium supernatants from nasopharyngeal and pharyngeal recess swabs from asymptomatic control calves (cumulative health scores ranging from 0 to 2) were obtained in a manner analogous to that used for symptomatic animals. Total nucleic acid was extracted from 140 μl using the QIAquick viral RNA extraction kit (Qiagen). Random cDNA was generated using ProtoScript II reverse transcriptase (New England BioLabs). The reaction mix for the reverse transcription consisted of 2 μl of a solution of 1.0 M KCl, 0.10 M Tris base, 0.0025 M MgCl26H2O (pH 8.3) and a solution of 100 mM Tris, 25 mM MgCl26H2O, 10% Tween 20, 10% NP-40 mixed at a 1:1 ratio, 0.2 μl 25 mM deoxynucleoside triphosphate (dNTP), 0.075 μl 100 N random hexamer mix, 0.5 μl RiboLock RNase inhibitor (Thermo Fisher Scientific), and 0.5 μl ProtoScript II reverse transcriptase (New England BioLabs). A 3-μl portion of the above-described premix was mixed with 17 μl of total extracted nucleic acid and incubated at 25°C for 10 min, 42°C for 30 min, and 95°C for 10 min to produce cDNA. For DNA virus screening, the cDNA synthesis step was omitted, and total nucleic acid was used as a template. Real-time PCR amplification was performed in a Roche 480 thermocycler using a Fam-Zen TaqMan probe specific to the virus of interest. The primer-probe set for each virus investigated by real-time PCR and the amplicon lengths are summarized in Table S1 in the supplemental material. The reaction mix for each sample consisted of 15 mM KCl, 40 mM Tris HCl, 25 μg bovine serum albumin (BSA), 5 mM Mg2+, 20 mM dNTP, 1 μM each primer, 0.2 μM TaqMan probe, and 0.75 units Fastart Taq polymerase (Roche). Amplification followed the following cycling profile: a 1-min initial denaturation step at 95°C followed by 45 cycles of 30 s of denaturation at 95°C and 1 min of extension at 60°C. A threshold cycle (CT) value of 45 indicates viral nucleic acid below the level of detection. A lower CT value indicates higher viral loads. Because the real-time PCRs were not calibrated with known concentrations of target nucleic acids, the CT values reflect only relative viral loads for the same virus in different samples and cannot be compared across different viruses.
Sequence and phylogenetic analysis.
The open reading frame (ORF) of the viral nucleotide sequences were analyzed with Geneious version R6. Sequences were aligned using MAFFT with the E-INS-I alignment strategy and previously described parameters (33). Maximum-likelihood (ML) trees were generated from translated protein sequences using RAxML with Dayhoff similarity matrix parameters (40) ML trees were run with 100 bootstrap replications with a gamma distribution for rates over sites. Midpoint rooting was conducted using MEGA (41).
Nucleotide sequence accession numbers.
Complete or partial viral genome sequences determined in this study were submitted to GenBank under the accession numbers given in Table 1.
TABLE 1.
Viruses identified by metagenomics, with GenBank accession numbers
| Virus | Family | GenBank accession no. | Contig length (bp) | No. of sequence reads | Closest relative | Amino acid identity (%) |
|---|---|---|---|---|---|---|
| Bovine adenovirus 3 | Adenoviridae | KP264982 | 2,547 | 4,279 | Bovine adenovirus 3 | 98 (Hexon) |
| Bovine adeno-associated virus | Parvoviridae | KP264981 | 4,279 | 3,481 | Bovine adeno-associated virus | 98 (Rep) |
| Bovine rhinitis A virus BSRI4 | Picornaviridae | KP264974 | 7,207 | 11,109 | Bovine rhinitis A virus | 94 (polyprotein) |
| Bovine rhinitis B virus | ||||||
| BSRI1 | Picornaviridae | KP264980 | 7,359 | 1,224 | Bovine rhinitis B virus | 96 (polyprotein) |
| BSRI2 | Picornaviridae | KP264976 to KP264979 | 1,527 | 985 | Bovine rhinitis B virus | 92 (3D) |
| BSRI3 | Picornaviridae | KP264975 | 6,995 | 430 | Bovine rhinitis B virus | 89 (polyprotein) |
| Bovine influenza D virus BSRI5 | Orthomyxoviridae | 304 | Swine influenza D/OK virus | 92–98 (multiple) | ||
| Bovine astrovirus BSRI1 | Astroviridae | KP264970 | 6,099 | 1,666 | Bovine astrovirus K08/51 | 71 (RdRp) |
| Picobirnavirus | ||||||
| BSRI1 | Picobirnaviridae | KP264972 | 1,286 | 21 | Human picobirnavirus | 33 (RdRp) |
| BSRI2 | Picobirnaviridae | KP264973 | 1,366 | 123 | Human picobirnavirus | 34 (RdRp) |
| Bovine parvovirus 2 BSRI | Parvoviridae | KP264971 | 5,331 | 79 | Bovine parvovirus 2 | 96 (NS1) |
| Bovine herpesvirus 6 | Herpesviridae | 19 | Bovine herpesvirus 6 | 96 to 100 (multiple) |
RESULTS
Viral metagenomics.
Deep nasal swabs collected from 50 animals with extreme symptoms of BRD were randomly distributed in 10 pools of five samples that were then processed for viral metagenomics using the Illumina HiSeq platform. After quality control and removal of duplicates 11.5 million reads were analyzed. Following de novo assembly, both the contigs and singlets were compared to the viral RefSeq database in GenBank for translated protein similarity to the proteins of all eukaryotic viruses using BLASTx. The greatest number of viral hits were as follows: bovine rhinitis A virus > bovine adenovirus 3 > adeno-associated virus > bovine rhinitis B virus > astrovirus BSRI1 > bovine influenza D virus > picobirnaviruses (PBVs) > bovine parvovirus 2 > bovine herpesvirus 6.
We then generated complete or near-complete genome sequences for a subset of these viruses (Table 1).
(i) BAdV3.
Deep sequencing revealed 4,279 sequences related to bovine adenovirus 3 (BAdV3), providing coverage of 82.6% of the total genome length. A complete hexon sequence was obtained, which shared 98% amino acid identity to that of bovine adenovirus 3 (AF030154) (Table 1).
(ii) BAAV.
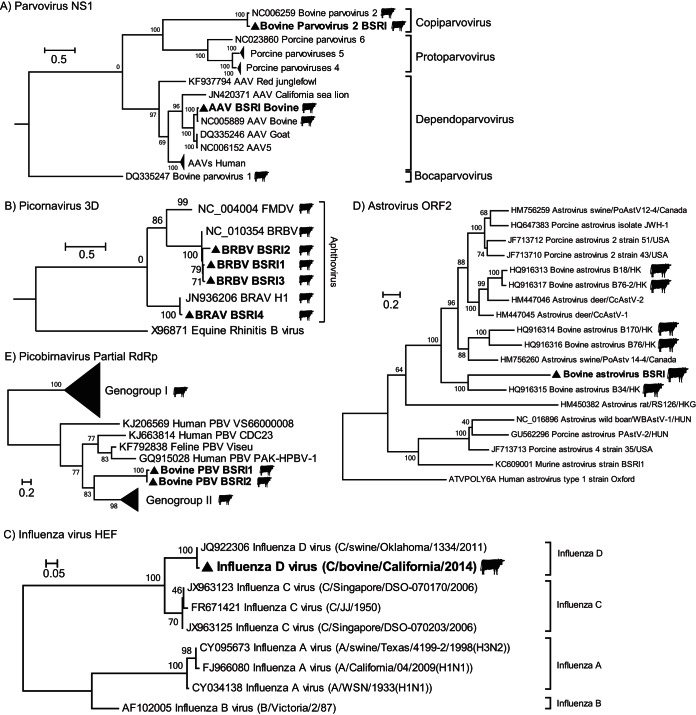
Bovine adeno-associated virus (BAAV) was detected with 3,481 sequences. The assembled genome contains replicase- and capsid-encoding genes and shared 98.5% amino acid identity in the replicase with previously described BAAV in the species Dependoparvovirus B and hexon 99% identity in the capsid protein. Reads assembled into a complete genome contig that shared 97% nucleotide identity to the BAAV reference genome (AY388617) (Table 1). Phylogenetic analysis confirmed it to be closely related to another bovine dependovirus BAAV originally found as a contaminant of two isolates of bovine adenoviruses (42) (Fig. 1A).
FIG 1.
Phylogenetic analysis of different viral protein sequences. Viruses characterized in this study are labeled with triangles. The phylogenetic analysis was performed based on the following: parvovirus and dependoparvovirus nonstructural proteins (A); picornavirus RNA-dependent RNA polymerase proteins of the Aphthovirus genus, including BRAV BSRI and BRBV BSRI1/2/3, other available members of these two species, and ERAV and FMDV species (B); the influenza D virus hemagglutinin-esterase region, including representatives of other genera in the Orthomyxoviridae family (C); astrovirus ORF2 capsid proteins, including representatives from various host species (D); and partial picobirnavirus RdRp proteins from major genogroups, including representative from other species (E).
(iii) BRAV and BRBV.
A total of 11,109 reads with sequences identities to bovine rhinitis A virus (BRAV) were detected (Table 1). A near-complete genome (KP264974) was assembled, resulting in a 7,207-base sequence. The complete polyprotein showed 94% identity to that of the reference BRAV genome and single currently available BRAV genome (JN936206) (Table 1). A total of 512 bases of the 5′ untranslated region (UTR) were generated and were 91% identical to that of the reference genome.
Another 2,639 sequence reads were more closely related to bovine rhinitis B viruses (BRBVs). Further sequence analysis revealed the presence of several BRBV strains. With the use of de novo sequence assembly combined with RT-PCR amplification of gapped regions and 3′ RACE, we obtained two nearly complete (strains BSRI1 and -3) and one partial (strain BSRI2) genome sequences. The near-complete genome of BRBV BSRI1 was comprised of 7,359 bases, including the complete polyprotein, 420-base 5′ UTR, and 68-base 3′ UTR, excluding the poly(A) tail. The polyprotein of BRBV BSRI1 shared 96% identity to that of BRBV reference genome (NC_010354). The entire polyprotein of BRBV BSRI3 shared 89% identity to the BRBV reference. The third strain, BRBV BSRI2, was incomplete, with only partial L-VP4-VP2-VP3-VP1 (with 80% identity to the BRBV reference), partial 2C-3B (91%), and complete 3D (92%).
The phylogenetic analysis therefore revealed multiple, closely related bovine rhinitis viruses from the genus Aphthovirus present in the bovine respiratory disease sample set (Fig. 1B).
(iv) Bovine influenza D virus.
Viral sequences closely related to swine influenza D/OK virus (JQ922308), including segment 4, which encodes the hemagglutinin-esterase fusion (HEF) glycoprotein, were identified (92 to 98%). PCR amplification and Sanger sequencing were used to identify an individual sample within a pool containing influenza virus. Further deep sequencing of that sample generated 304 reads with coverage of 72% of the genome. These sequences shared 92 to 98% protein identity to the seven segments of swine influenza D/OK virus (see Fig. S1 in the supplemental material). Our finding is consistent with the closely related genomes that were recently described in bovine respiratory disease samples (43–45, 75). Maximum-likelihood phylogenetic analysis of the nucleotide sequence of the HEF fragment is depicted in Fig. 1C.
(v) BoAstV BSRI1.
Using 1,666 sequence reads in combination with gap-filling RT-PCR, RACE, and Sanger dideoxy sequencing, we obtained a complete genome of a divergent bovine astrovirus (BoAstV) (BSRI1). The BoAstV BSRI1 genome is 6,099 nucleotides (nt) long, with a GC content of 50.9% (KP264970) (Table 1). Consistent with other members of the Astroviridae family, BoAstV BSRI1 contains two open reading frames (ORF1 and ORF2). ORF1 is segregated into two coding regions, ORF1a and ORF1b, which encode nonstructural and RdRp proteins, respectively. The highly conserved AAAAAAC sequence, which constitutes the ORF1a/b ribosomal frameshift site in astroviruses, was identified at residues 2246 to 2252. Additionally, the conserved promoter sequence UUUGGAGNGGNGGACCNAAN14AUGNC was detected at the start of ORF2. The RdRp ORF1b shared 71% protein identities of a bovine astrovirus recently identified in Hong Kong (Table 1).
BoAstV BSRI1belongs to astrovirus genogroup III, as shown by phylogenetic analysis of the of the ORF2 capsid sequence (Fig. 1D). ORF2 of BoAstV BSRI1 is most closely related to that of bovine astrovirus B34/HK (HQ916315), although with only 43% protein identity.
(vi) BPV.
Two divergent bovine picobirnaviruses (BPVs) were identified. Reverse transcription, PCR amplification, and Sanger sequencing confirmed the presence of these picobirnaviruses in two samples, both of which underwent further individual deep sequencing, and 1,286- and 1,366-bp contigs were obtained. BLASTx analysis of the two partial RdRp sequence revealed that they shared 33 to 34% protein identity to human PBV VS6600008 (KJ206569) (Table 1). Maximum-likelihood phylogenetic analysis of the partial RdRp revealed a clear divergence between both viral sequences and members of genogroups I and II (Fig. 1E).
Unexpectedly, both segments contained numerous UGA stop codons. One picobirnavirus (BPV BSRI1) has six and BPV BSRI2 had three premature RdRp stop codons (KP264972 and KP264973). The presence of these stop codons was detected in both pooled and individual deep-sequencing runs using separate RNA extracts, as well as by RT-PCR and Sanger sequencing. Premature stop codons have been reported in other picobirnavirus RdRp genes (46, 47). Active RdRp may be provided by minority wild-type helper picobirnavirus in the same cell or may be translated despite premature stop codons by readthrough induced by RNA stem-loop structures.
(vii) Bovine parvovirus 2.
Seventy-nine sequences with an average of 82.36% nucleotide identity to bovine parvovirus 2 (AF406966) were identified (Table 1). The sample positive for this genome was identified by PCR and individually deep sequenced. We obtained 4,833 additional sequences that assembled together and allowed for extension of the viral genome of 5,331 bases. Amino acid comparison of the translated NS1-coding sequence revealed 96% identity to that of ungulate copiparvovirus 1 species (AF406966) (48), placing it within the same species group as confirmed by phylogenetic analysis (Fig. 1A).
(viii) BHV-6.
Deep sequencing revealed 19 sequences with an average protein identity of 96 to 100% to multiple proteins of bovine herpesvirus 6 (BHV-6) (Table 1).
Virus detection rates in BRD cases versus controls.
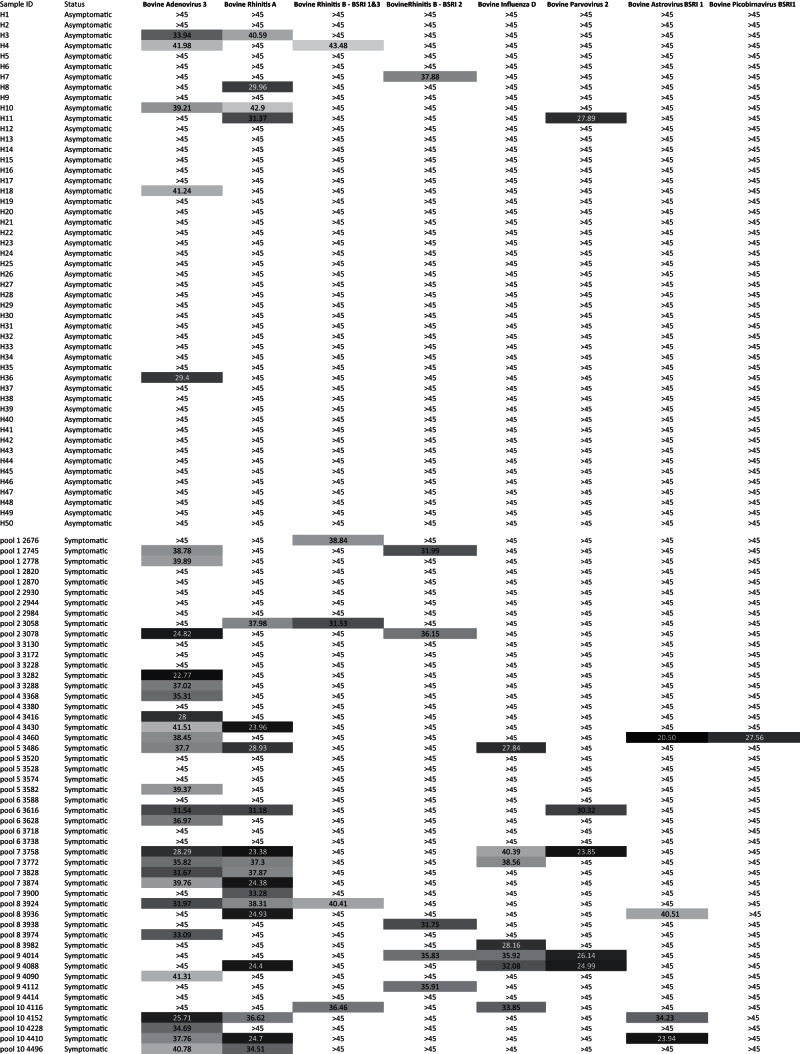
To determine which of these viruses were associated with BRD, real-time PCR assays were designed and CT values were measured for nucleic acids in nasal secretions from 50 BRD cases and 50 healthy controls. The PCR primers and TaqMan probes used are listed in Table S1 in the supplemental material. Fisher's exact test was used to determine whether the rates of detection of tested viruses were significantly higher in BRD cases than in matched controls. The results from the real-time PCR case-control study are shown in Fig. 2, and P values are summarized in Table 2.
FIG 2.
Real-time PCR CT values for eight bovine viruses in respiratory samples from 50 animals with BRD and 50 healthy controls. Bovine rhinitis B virus strains BSRI1 and -3 were screened using the same real-time PCR assay targeting their conserved RdRp regions.
TABLE 2.
Virus detection rates in BRD cases versus controls
| Virus | Viral family | No. positive/total (% positive) |
P valuea | |
|---|---|---|---|---|
| Asymptomatic animals | Animals with BRD | |||
| Bovine adenovirus 3 | Adenoviridae | 5/50 (10) | 24/50 (48) | <0.0001* |
| Bovine rhinitis A virus BSRI4 | Picornaviridae | 4/50 (8) | 12/50 (24) | 0.009* |
| Bovine rhinitis B virus | ||||
| BSRI1 + -3 | Picornaviridae | 1/50 (2) | 4/50 (8) | 0.36 |
| BSRI2 | Picornaviridae | 1/50 (2) | 5/50 (10) | 0.2 |
| Bovine influenza D virus BSRI1 | Orthomyxoviridae | 0/50 (0) | 7/50 (14) | 0.012* |
| Bovine parvovirus 2 BSRI | Parvoviridae | 1/50 (2) | 4/50 (8) | 0.36 |
| Bovine astrovirus BSRI1 | Astroviridae | 0/50 (0) | 4/50 (8) | 0.117 |
| Bovine picobirnavirus BSRI1 | Picobirnaviridae | 0/50 (0) | 1/50 92) | 1 |
*, significant.
(i) Bovine adenovirus 3.
Twenty-four samples (48%) obtained from symptomatic cows were positive for bovine adenovirus 3. In the asymptomatic control group, five cows (10%) were positive for the virus. A significantly different rate of detection was using Fisher's exact test (P < 0.0001).
(ii) Bovine rhinitis A virus BSRI4.
Twelve animals (24%) from the symptomatic group had detectable amounts of bovine rhinitis A virus RNA. In the asymptomatic group, four animals (8%) tested positive for the presence of the BSRI4 strain. A significantly different rate of detection was calculated using Fisher's exact test (P = 0.009).
(iii) Bovine rhinitis B virus strains BSRI1 and -2.
We designed real-time PCR assays that targeted the conserved RdRp sequence of BRBV BSRI1 plus -3 and BSRI2 separately. Among the symptomatic animals, four (8%) were positive for strain BSRI1 plus -3, while strain BSRI2 was detected in five (10%) animals. The asymptomatic group had one animal each positive for BSRI1 plus -3 and BSRI2. The resulting P values were nonsignificant at 0.36 and 0.20 for strains BSRI1 plus -3 and BSRI2, respectively.
(iv) Bovine influenza D virus.
Seven animals (14%) within the symptomatic group were positive for influenza D virus. None of the animals from the symptomatic group was found to be positive for the virus. A significantly different rate of detection was calculated using Fisher's exact test (P = 0.012).
(v) Bovine parvovirus 2 strain BSRI.
Among the symptomatic cows, four (8%) tested positive for parvovirus 2 BSRI, while one animal in the asymptomatic control group also had detectable levels of the virus. The resulting P value was nonsignificant at 0.36.
(vi) Bovine astrovirus BSRI.
Four animals (8%) within the symptomatic group were positive for bovine astrovirus BSRI1. Astrovirus RNA was not detected in any of the 50 asymptomatic animals. The resulting P value were nonsignificant at 0.117.
(vii) Bovine picobirnavirus.
Only a single animal with BRD and none of the healthy animals tested positive for picobirnavirus BSRI1, yielding a nonsignificant P value.
(viii) Other viruses.
Based on the small numbers of reads generated in the Illumina data set, we did not design real-time PCR assays for the bovine picobirnaviruses BSRI2 and herpesvirus 6 genomes. Because adeno-associated viruses have not been associated with disease, this virus was not analyzed further.
DISCUSSION
Our metagenomics analysis revealed a large numbers of different viruses present in the respiratory secretions of animals with BRD in bovine feedlots. Viruses included both close relatives of known viruses and “new” viruses (highly divergent from those currently reported in GenBank). When the distribution of these viruses in nasal secretions was compared between animals with BRD and healthy controls, bovine adenovirus 3 (P < 0.0001), bovine rhinitis A virus (P = 0.005), and bovine influenza D virus (P = 0.006) were each associated with BRD. The rates of detection of these tentative viral pathogens in animals with BRD were 48% for BAdV3, 30% for BRAV, and 14% for bovine influenza D virus. Bovine astrovirus BSRI infection was found only in BRD cases (4/50) which did not reach statistical significance, with a P value of 0.117. The set of severe symptoms defining BRD used here was therefore associated with 3 viruses. A high level of coinfections was also detected, with more than one virus in 38% of BRD cases versus 8% in controls. Whether coinfections or specific combinations of viruses are more likely to result in BRD will require analyzing larger number of cases and controls. A recent study of calves from Ireland with BRD, testing for BVDV, BoCV, BoHV-1, BRSV, and PI3V, reported a virus detection rate of 35%, with 40% of these infected animals coinfected with two or more viruses (49).
In 32% of BRD cases analyzed here, none of the 8 tested viruses were detected. Viruses present below levels of detections, deeper in the respiratory tract, or present only earlier in disease progression may account for such apparently virus-negative cases. Infections with bacterial respiratory pathogens alone may also account for these virus-negative cases. Infections with viruses with no homolog among all the eukaryotic viral genomes in GenBank, and therefore not recognizable using BLASTx, could also account for such cases.
Bovine adenovirus is highly seroprevalent in feedlot calves, with ∼50% of animals reported to seroconvert during the first month in feedlots, and has been associated with increased fever although not reduced weight gain (50). Intratracheal inoculation of 4-months-old colostrum-deprived calves also induced fever and other symptoms (51). Here BAdV3 was the virus most commonly detected in nasal secretions (48%) and was most strongly associated with BRD (P < 0.0001). Because replication-competent BAdV3 strains have been used as vaccine vectors for other infections (52, 53), candidate BAdsV3 vaccines may fortuitously already be available for testing.
In 2013 a highly divergent influenza virus was described in a pig with influenza-like illness; this virus was unable to productively reassort with human influenza C virus and was phylogenetically and antigenically distinct enough to be classified as new genus, named Influenzavirus D in the Orthomyxoviridae family (43–45). Related viruses were reported in Chinese cattle (54). The cellular tropism of this virus was wider than that of a human influenza C virus and was not inhibited at elevated temperatures. In 2014, RT-PCR testing of 208 nasal swab samples from BRD cases revealed that 5% were positive with either of two antigenically distinct strains (D/OK and D/660). No healthy controls were tested (75). Here we report a rate of detection of influenza D virus RNA of 14%, with none of 50 healthy controls positive, using real-time PCR. Of the seven influenza D virus RNA-positive animals, all but one were coinfected, most frequently with adenovirus 3 (n = 4) or BPV2 (n = 3).
Bovine rhinitis A virus (BRAV) is one of four picornavirus species in the Aphthovirus genus, together with foot-and-mouth disease virus (FMDV), equine rhinitis A virus (ERAV), and bovine rhinitis B viruses (BRBV) (http://www.picornaviridae.com/aphthovirus/brv/brv.htm). BRAV consists of up to 3 serotypes (55), but a single BRAV genome is currently available in GenBank (JN936206). A single genome of BRBV is also currently in GenBank (NC_010354) (56); this genome is from a viral isolate derived from a specific-pathogen-free calf that developed respiratory disease (57). Here, one nearly complete genome of bovine rhinitis A virus and two of bovine rhinitis B viruses were sequenced. Real-time PCR for all three viruses showed only BRAV BSRI4 to be significantly associated with BRD.
The bovine astrovirus BSRI1 genetically characterized here was found in 4/50 BRD cases and in none of the 50 healthy controls (P value of 0.117). While these results are suggestive of a possible involvement in a fraction of BRD cases, the study of a larger number of cases and controls will be needed to confirm significant enrichment in BRD. Astroviruses are commonly associated with gastrointestinal symptoms, particularly in very young or immunodeficient humans (58–60), and are commonly found in animal feces (26, 61, 62). Astroviruses have also been found in the brains of humans (63) and animals (64), including cows, with neurological symptoms (29, 30), indicating that their host range can extend beyond enteric tissues. Because all 4 BRD cases with this astrovirus also contained bovine adenovirus 3 and/or bovine rhinitis A virus, it remains possible that its presence aggravates symptoms induced by other viral infections.
Picobirnaviruses are a group of genetically diverse viruses highly prevalent in the feces of both healthy and diarrheic humans and animals (65–68). In humans, PBVs are associated with diarrhea mainly in immunosuppressed individuals (69). Bovine PBVs have been described (70, 71). Here a single infection was detected in a BRD animal coinfected with both bovine adenovirus 3 and astrovirus BSRI1.
A viral metagenomics approach to characterize all viruses present, combined with real-time PCR testing of biological samples from well-matched BRD cases and healthy controls, can provide a simple approach to the study of a complex infectious disease. Multiple factors may influence the outcome of viral infections, including preexisting immunity, stressors, changes in diet, and antibiotic treatment (1–4). Mingling of large number of animals of different ages will also increase the amount and diversity of exposures to viral and bacterial pathogens and affect the outcome of infections. Despite the multifactorial aspect of BRD, the identification of viruses associated with this complex disease paves the way for further studies, including replication of our findings using animals from different herds, animal challenges to test viral pathogenicity, and ultimately vaccination to measure the impact of reducing viral infections. Multivalent vaccination against viruses (BHV-1, PI3V, BVDV, and BRSV) has been shown to reduce respiratory symptoms in subsequently virally challenged animals (72), and such studies could be extended to the potential viral pathogens reported here.
Except for bovine adenovirus 3, the viruses traditionally tested for in BRD, namely, BHV-1, PI3V, BVDV, BoCV, and BRSV (2, 6, 9, 10), were not detected here. Five DNA viruses recently reported in a viral metagenomics study of beef (muscle tissue) (27) were also not detected in these nasal secretions from animals with BRD. Because of the nonspecific nature of the viral metagenomics method used, the nondetection of these viruses reflects either their absence or their presence below the viral loads of those viruses that were successfully detected. The mix of respiratory viruses infecting cattle may also vary widely between herds, in different geographic regions, or when sampled during different years or even seasons (73). Determination of whether the respiratory viruses detected here reflect those infecting other cattle with BRD throughout the United States and other countries will therefore require studies of respiratory samples collected at different times from different locations.
Supplementary Material
ACKNOWLEDGMENTS
We thank Steven Head (Scripps Research Institute) for DNA sequencing, Jana Sachsenröder for help making the HiSeq library, and Lani Montalvo for help and advice with real-time PCR.
Support was provided by BSRI and NIH grant R01 HL105770 to Eric Delwart. The samples used were provided by the Bovine Respiratory Disease Consortium Coordinated Agricultural Project, J. Womack (PD), USDA Agriculture and Food Research Initiative competitive grant no. 2011-68004-30367 from the USDA National Institute of Food and Agriculture.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00064-15.
REFERENCES
- 1.Apley M. 2014. The clinical syndrome of BRD: what it is and what it is not. Anim Health Res Rev 15:135–137. doi: 10.1017/S1466252314000152. [DOI] [PubMed] [Google Scholar]
- 2.Hilton WM. 2014. BRD in 2014: where have we been, where are we now, and where do we want to go? Anim Health Res Rev 15:120–122. doi: 10.1017/S1466252314000115. [DOI] [PubMed] [Google Scholar]
- 3.Griffin D. 1997. Economic impact associated with respiratory disease in beef cattle. Vet Clin North Am Food Anim Pract 13:367–377. [DOI] [PubMed] [Google Scholar]
- 4.McVey DS. 2009. BRD research needs in the next 10-20 years. Anim Health Res Rev 10:165–167. doi: 10.1017/S1466252309990247. [DOI] [PubMed] [Google Scholar]
- 5.Griffin D, Chengappa MM, Kuszak J, McVey DS. 2010. Bacterial pathogens of the bovine respiratory disease complex. Vet Clin North Am Food Anim Pract 26:381–394. doi: 10.1016/j.cvfa.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Mosier D. 2014. Review of BRD pathogenesis: the old and the new. Anim Health Res Rev 15:166–168. doi: 10.1017/S1466252314000176. [DOI] [PubMed] [Google Scholar]
- 7.Portis E, Lindeman C, Johansen L, Stoltman G. 2012. A ten-year (2000-2009) study of antimicrobial susceptibility of bacteria that cause bovine respiratory disease complex—Mannheimia haemolytica, Pasteurella multocida, and Histophilus somni—in the United States and Canada. J Vet Diagn Invest 24:932–944. doi: 10.1177/1040638712457559. [DOI] [PubMed] [Google Scholar]
- 8.Larson RL, Step DL. 2012. Evidence-based effectiveness of vaccination against Mannheimia haemolytica, Pasteurella multocida, and Histophilus somni in feedlot cattle for mitigating the incidence and effect of bovine respiratory disease complex. Vet Clin North Am Food Anim Pract 28:97–106. doi: 10.1016/j.cvfa.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Ellis JA. 2009. Update on viral pathogenesis in BRD. Anim Health Res Rev 10:149–153. doi: 10.1017/S146625230999020X. [DOI] [PubMed] [Google Scholar]
- 10.Härtel H, Nikunen S, Neuvonen E, Tanskanen R, Kivelä SL, Aho R, Soveri T, Saloniemi H. 2004. Viral and bacterial pathogens in bovine respiratory disease in Finland. Acta Vet Scand 45:193–200. doi: 10.1186/1751-0147-45-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muylkens B, Thiry J, Kirten P, Schynts F, Thiry E. 2007. Bovine herpesvirus 1 infection and infectious bovine rhinotracheitis. Vet Res 38:181–209. doi: 10.1051/vetres:2006059. [DOI] [PubMed] [Google Scholar]
- 12.Neibergs H, Zanella R, Casas E, Snowder GD, Wenz J, Neibergs JS, Moore D. 2011. Loci on Bos taurus chromosome 2 and Bos taurus chromosome 26 are linked with bovine respiratory disease and associated with persistent infection of bovine viral diarrhea virus. J Anim Sci 89:907–915. doi: 10.2527/jas.2010-3330. [DOI] [PubMed] [Google Scholar]
- 13.Yeşilbağ K, Förster C, Ozyiğit MO, Alpay G, Tuncer P, Thiel HJ, König M. 2014. Characterisation of bovine viral diarrhoea virus (BVDV) isolates from an outbreak with haemorrhagic enteritis and severe pneumonia. Vet Microbiol 169:42–49. doi: 10.1016/j.vetmic.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 14.Castleman WL, Lay J C, Dubovi EJ, Slauson DO. 1985. Experimental bovine respiratory syncytial virus infection in conventional calves: light microscopic lesions, microbiology, and studies on lavaged lung cells. Am J Vet Res 46:547–553. [PubMed] [Google Scholar]
- 15.Larsen LE, Tjørnehøj K, Viuff B, Jensen NE, Uttenthal A. 1999. Diagnosis of enzootic pneumonia in Danish cattle: reverse transcription-polymerase chain reaction assay for detection of bovine respiratory syncytial virus in naturally and experimentally infected cattle. J Vet Diagn Invest 11:416–422. doi: 10.1177/104063879901100505. [DOI] [PubMed] [Google Scholar]
- 16.Darbyshire JH, Jennings AR, Omar AR, Dawson PS, Lamont PH. 1965. Association of adenoviruses with bovine respiratory diseases. Nature 208:307–308. doi: 10.1038/208307a0. [DOI] [PubMed] [Google Scholar]
- 17.Hick PM, Read AJ, Lugton I, Busfield F, Dawood KE, Gabor L, Hornitzky M, Kirkland PD. 2012. Coronavirus infection in intensively managed cattle with respiratory disease. Aust Vet J 90:381–386. doi: 10.1111/j.1751-0813.2012.00978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saif LJ. 2010. Bovine respiratory coronavirus. Vet Clin North Am Food Anim Pract 26:349–364. doi: 10.1016/j.cvfa.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belák S, Karlsson OE, Blomström AL, Berg M, Granberg F. 2013. New viruses in veterinary medicine, detected by metagenomic approaches. Vet Microbiol 165:95–101. doi: 10.1016/j.vetmic.2013.01.022. [DOI] [PubMed] [Google Scholar]
- 20.Delwart E. 2012. Animal virus discovery: improving animal health, understanding zoonoses, and opportunities for vaccine development. Curr Opin Virol 2:344–352. doi: 10.1016/j.coviro.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delwart E. 2013. A roadmap to the human virome. PLoS Pathog 9:e1003146. doi: 10.1371/journal.ppat.1003146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosario K, Breitbart M. 2011. Exploring the viral world through metagenomics. Curr Opin Virol 1:289–297. doi: 10.1016/j.coviro.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Lager KM, Ng TF, Bayles DO, Alt DP, Delwart EL, Cheung AK. 2012. Diversity of viruses detected by deep sequencing in pigs from a common background. J Vet Diagn Invest 24:1177–1179. doi: 10.1177/1040638712463212. [DOI] [PubMed] [Google Scholar]
- 24.Li L, Kapoor A, Slikas B, Bamidele OS, Wang C, Shaukat S, Masroor MA, Wilson ML, Ndjango JB, Peeters M, Gross-Camp ND, Muller MN, Hahn BH, Wolfe ND, Triki H, Bartkus J, Zaidi SZ, Delwart E. 2010. Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J Virol 84:1674–1682. doi: 10.1128/JVI.02109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L, Shan T, Soji OB, Alam MM, Kunz TH, Zaidi SZ, Delwart E. 2011. Possible cross-species transmission of circoviruses and cycloviruses among farm animals. J Gen Virol 92:768–772. doi: 10.1099/vir.0.028704-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shan T, Li L, Simmonds P, Wang C, Moeser A, Delwart E. 2011. The fecal virome of pigs on a high-density farm. J Virol 85:11697–11708. doi: 10.1128/JVI.05217-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang W, Li L, Deng X, Kapusinszky B, Delwart E. 2014. What is for dinner? Viral metagenomics of US store bought beef, pork, and chicken. Virology 468–470:303–310. doi: 10.1016/j.virol.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoffmann B, Scheuch M, Höper D, Jungblut R, Holsteg M, Schirrmeier H, Eschbaumer M, Goller KV, Wernike K, Fischer M, Breithaupt A, Mettenleiter TC, Beer M. 2012. Novel orthobunyavirus in Cattle, Europe, 2011. Emerg Infect Dis 18:469–472. doi: 10.3201/eid1803.111905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bouzalas IG, Wüthrich D, Walland J, Drögemüller C, Zurbriggen A, Vandevelde M, Oevermann A, Bruggmann R, Seuberlich T. 2014. Neurotropic astrovirus in cattle with nonsuppurative encephalitis in europe. J Clin Microbiol 52:3318–3324. doi: 10.1128/JCM.01195-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li L, Diab S, McGraw S, Barr B, Traslavina R, Higgins R, Talbot T, Blanchard P, Rimoldi G, Fahsbender E, Page B, Phan TG, Wang C, Deng X, Pesavento P, Delwart E. 2013. Divergent astrovirus associated with neurologic disease in cattle. Emerg Infect Dis 19:1385–1392. doi: 10.3201/eid1909.130682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neibergs HL, Seabury CM, Wojtowicz AJ, Wang Z, Scraggs E, Kiser J, Neupane M, Womack JE, Van Eenennaam A, Hagevoort GR, Lehenbauer TW, Aly S, Davis J, Taylor JF. 2014. Susceptibility loci revealed for bovine respiratory disease complex in pre-weaned Holstein calves. BMC Genomics 15:1164. doi: 10.1186/1471-2164-15-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGuirk SM. 2008. Disease management of dairy calves and heifers. Vet Clin North Am Food Anim Pract 24:139–153. doi: 10.1016/j.cvfa.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng TF, Marine R, Wang C, Simmonds P, Kapusinszky B, Bodhidatta L, Oderinde BS, Wommack KE, Delwart E. 2012. High variety of known and new RNA and DNA viruses of diverse origins in untreated sewage. J Virol 86:12161–12175. doi: 10.1128/JVI.00869-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGinnis S, Madden TL. 2004. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res 32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J, Wu G, Zhang H, Shi Y, Liu Y, Yu C, Wang B, Lu Y, Han C, Cheung DW, Yiu SM, Peng S, Xiaoqian Z, Liu G, Liao X, Li Y, Yang H, Wang J, Lam TW, Wang J. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res 19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Namiki T, Hachiya T, Tanaka H, Sakakibara Y. 2012. MetaVelvet: an extension of Velvet assembler to de novo metagenome assembly from short sequence reads. Nucleic Acids Res 40:e155. doi: 10.1093/nar/gks678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang X, Madan A. 1999. CAP3: a DNA sequence assembly program. Genome Res 9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 41.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt M, Katano H, Bossis I, Chiorini JA. 2004. Cloning and characterization of a bovine adeno-associated virus. J Virol 78:6509–6516. doi: 10.1128/JVI.78.12.6509-6516.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hause BM, Collin EA, Liu R, Huang B, Sheng Z, Lu W, Wang D, Nelson EA, Li F. 2014. Characterization of a novel influenza virus in cattle and swine: proposal for a new genus in the Orthomyxoviridae family. mBio 5(2):e00031-14. doi: 10.1128/mBio.00031-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hause BM, Ducatez M, Collin EA, Ran Z, Liu R, Sheng Z, Armien A, Kaplan B, Chakravarty S, Hoppe AD, Webby RJ, Simonson RR, Li F. 2013. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog 9:e1003176. doi: 10.1371/journal.ppat.1003176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheng Z, Ran Z, Wang D, Hoppe AD, Simonson R, Chakravarty S, Hause BM, Li F. 2014. Genomic and evolutionary characterization of a novel influenza-C-like virus from swine. Arch Virol 159:249–255. doi: 10.1007/s00705-013-1815-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Green J, Gallimore CI, Clewley JP, Brown DW. 1999. Genomic characterisation of the large segment of a rabbit picobirnavirus and comparison with the atypical picobirnavirus of Cryptosporidium parvum. Arch Virol 144:2457–2465. doi: 10.1007/s007050050658. [DOI] [PubMed] [Google Scholar]
- 47.Ng TF, Vega E, Kondov NO, Markey C, Deng X, Gregoricus N, Vinjé J, Delwart E. 2014. Divergent picobirnaviruses in human feces. Genome Announc 2(3):e00415-14. doi: 10.1128/genomeA.00415-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allander T, Emerson SU, Engle RE, Purcell RH, Bukh J. 2001. A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Proc Natl Acad Sci U S A 98:11609–11614. doi: 10.1073/pnas.211424698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Neill R, Mooney J, Connaghan E, Furphy C, Graham DA. 2014. Patterns of detection of respiratory viruses in nasal swabs from calves in Ireland: a retrospective study. Vet Rec 175:351. doi: 10.1136/vr.102574. [DOI] [PubMed] [Google Scholar]
- 50.Mattson DE, Norman BB, Dunbar JR. 1988. Bovine adenovirus type-3 infection in feedlot calves. Am J Vet Res 49:67–69. [PubMed] [Google Scholar]
- 51.Lehmkuhl HD, Smith MH, Dierks RE. 1975. A bovine adenovirus type 3: isolation, characterization, and experimental infection in calves. Arch Virol 48:39–46. doi: 10.1007/BF01320564. [DOI] [PubMed] [Google Scholar]
- 52.Baxi MK, Deregt D, Robertson J, Babiuk LA, Schlapp T, Tikoo SK. 2000. Recombinant bovine adenovirus type 3 expressing bovine viral diarrhea virus glycoprotein E2 induces an immune response in cotton rats. Virology 278:234–243. doi: 10.1006/viro.2000.0661. [DOI] [PubMed] [Google Scholar]
- 53.Brownlie R, Kumar P, Babiuk LA, Tikoo SK. 2015. Recombinant bovine adenovirus-3 co-expressing bovine respiratory syncytial virus glycoprotein G and truncated glycoprotein gD of bovine herpesvirus-1 induce immune responses in cotton rats. Mol Biotechnol 57:58–64. doi: 10.1007/s12033-014-9801-x. [DOI] [PubMed] [Google Scholar]
- 54.Jiang WM, Wang SC, Peng C, Yu JM, Zhuang QY, Hou GY, Liu S, Li JP, Chen JM. 2014. Identification of a potential novel type of influenza virus in bovine in China. Virus Genes 49:493–496. doi: 10.1007/s11262-014-1107-3. [DOI] [PubMed] [Google Scholar]
- 55.Yamashita H, Akashi H, Inaba Y. 1985. Isolation of a new serotype of bovine rhinovirus from cattle. Brief report. Arch Virol 83:113–116. doi: 10.1007/BF01310969. [DOI] [PubMed] [Google Scholar]
- 56.Hollister JR, Vagnozzi A, Knowles NJ, Rieder E. 2008. Molecular and phylogenetic analyses of bovine rhinovirus type 2 shows it is closely related to foot-and-mouth disease virus. Virology 373:411–425. doi: 10.1016/j.virol.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 57.Reed SE, Tyrrell DA, Betts AO, Watt RG. 1971. Studies on a rhinovirus (EC11) derived from a calf. I. Isolation in calf tracheal organ cultures and characterization of the virus. J Comp Pathol 81:33–40. [DOI] [PubMed] [Google Scholar]
- 58.Afrad MH, Karmakar PC, Das SK, Matthijnssens J, Ahmed F, Nahar S, Faruque AS, Rahman MZ, Rahman M, Azim T. 2013. Epidemiology and genetic diversity of human astrovirus infection among hospitalized patients with acute diarrhea in Bangladesh from 2010 to 2012. J Clin Virol 58:612–618. doi: 10.1016/j.jcv.2013.09.028. [DOI] [PubMed] [Google Scholar]
- 59.Guix S, Bosch A, Pintó RM. 2005. Human astrovirus diagnosis and typing: current and future prospects. Lett Appl Microbiol 41:103–105. doi: 10.1111/j.1472-765X.2005.01759.x. [DOI] [PubMed] [Google Scholar]
- 60.Wunderli W, Meerbach A, Güngör T, Guengoer T, Berger C, Greiner O, Caduff R, Trkola A, Bossart W, Gerlach D, Schibler M, Cordey S, McKee TA, Van Belle S, Kaiser L, Tapparel C. 2011. Astrovirus infection in hospitalized infants with severe combined immunodeficiency after allogeneic hematopoietic stem cell transplantation. PLoS One 6:e27483. doi: 10.1371/journal.pone.0027483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li L, Shan T, Wang C, Côté C, Kolman J, Onions D, Gulland FM, Delwart E. 2011. The fecal viral flora of California sea lions. J Virol 85:9909–9917. doi: 10.1128/JVI.05026-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang W, Li L, Deng X, Kapusinszky B, Pesavento PA, Delwart E. 2014. The fecal virome of cats in an animal shelter. J Gen Virol 95:2553–2564. doi: 10.1099/vir.0.069674-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quan PL, Wagner TA, Briese T, Torgerson TR, Hornig M, Tashmukhamedova A, Firth C, Palacios G, Baisre-De-Leon A, Paddock CD, Hutchison SK, Egholm M, Zaki SR, Goldman JE, Ochs HD, Lipkin WI. 2010. Astrovirus encephalitis in boy with X-linked agammaglobulinemia. Emerg Infect Dis 16:918–925. doi: 10.3201/eid1606.091536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blomström AL, Widén F, Hammer AS, Belák S, Berg M. 2010. Detection of a novel astrovirus in brain tissue of mink suffering from shaking mink syndrome by use of viral metagenomics. J Clin Microbiol 48:4392–4396. doi: 10.1128/JCM.01040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kapusinszky B, Minor P, Delwart E. 2012. Nearly constant shedding of diverse enteric viruses by two healthy infants. J Clin Microbiol 50:3427–3434. doi: 10.1128/JCM.01589-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Masachessi G, Martínez LC, Giordano MO, Barril PA, Isa BM, Ferreyra L, Villareal D, Carello M, Asis C, Nates SV. 2007. Picobirnavirus (PBV) natural hosts in captivity and virus excretion pattern in infected animals. Arch Virol 152:989–998. doi: 10.1007/s00705-006-0900-2. [DOI] [PubMed] [Google Scholar]
- 67.Phan TGAK, Wang BA, Rose CA, Lipton RKA, Delwart HLA, Eric E. 2011. The fecal viral flora of wild rodents. PLoS Pathog 7:e1002218. doi: 10.1371/journal.ppat.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Leeuwen M, Williams MM, Koraka P, Simon JH, Smits SL, Osterhaus AD. 2010. Human picobirnaviruses identified by molecular screening of diarrhea samples. J Clin Microbiol 48:1787–1794. doi: 10.1128/JCM.02452-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grohmann GS, Glass RI, Pereira HG, Monroe SS, Hightower AW, Weber R, Bryan RT. 1993. Enteric viruses and diarrhea in HIV-infected patients. Enteric Opportunistic Infections Working Group. N Engl J Med 329:14–20. [DOI] [PubMed] [Google Scholar]
- 70.Ghosh S, Kobayashi N, Nagashima S, Naik TN. 2009. Molecular characterization of full-length genomic segment 2 of a bovine picobirnavirus (PBV) strain: evidence for high genetic diversity with genogroup I PBVs. J Gen Virol 90:2519–2524. doi: 10.1099/vir.0.013987-0. [DOI] [PubMed] [Google Scholar]
- 71.Malik YS, Chandrashekar KM, Sharma K, Haq AA, Vaid N, Chakravarti S, Batra M, Singh R, Pandey AB. 2011. Picobirnavirus detection in bovine and buffalo calves from foothills of Himalaya and Central India. Trop Anim Health Prod 43:1475–1478. doi: 10.1007/s11250-011-9834-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Salt JS, Thevasagayam SJ, Wiseman A, Peters AR. 2007. Efficacy of a quadrivalent vaccine against respiratory diseases caused by BHV-1, PI3V, BVDV and BRSV in experimentally infected calves. Vet J 174:616–626. doi: 10.1016/j.tvjl.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 73.Autio T, Pohjanvirta T, Holopainen R, Rikula U, Pentikäinen J, Huovilainen A, Rusanen H, Soveri T, Sihvonen L, Pelkonen S. 2007. Etiology of respiratory disease in non-vaccinated, non-medicated calves in rearing herds. Vet Microbiol 119:256–265. doi: 10.1016/j.vetmic.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Deng X, Naccache SN, Ng T, Federman S, Li L, Chiu CY, Delwart EL. An ensemble strategy that significantly improves de novo assembly of microbial genomes from metagenomic next-generation sequencing data. Nucleic Acids Res, in press. doi: 10.1093/nar/gkv002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Collin EA, Sheng Z, Lang Y, Ma W, Hause BM, Li F. 2015. Cocirculation of two distinct genetic and antigenic lineages of proposed influenza D virus in cattle. J Virol 89:1036–1042. doi: 10.1128/JVI.02718-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.