Abstract
Nonpathogenic-invasive Escherichia coli (InvColi) bacteria are suitable for genetic transfer into mammalian cells and may act as a vehicle for RNA Interference (RNAi) in vivo. Cyclooxygenase-2 (COX-2) is overexpressed in ulcerative colitis (UC) and Crohn's disease (CD), two inflammatory conditions of the colon and small intestine grouped as inflammatory bowel disease (IBD). We engineered InvColi strains for anti-COX-2 RNAi (InvColishCOX2), aiming to investigate the in vivo feasibility of a novel COX-2 silencing strategy in a murine model of colitis induced by dextran sulfate sodium (DSS). Enema administrations of InvColishCOX2 in DSS-treated mice led to COX-2 downregulation, colonic mucosa preservation, reduced colitis disease activity index (DAI) and increased mice survival. Moreover, DSS/InvColishCOX2-treated mice showed lower levels of circulating pro-inflammatory cytokines and a reduced colitis-associated shift of gut microbiota. Considering its effectiveness and safety, we propose our InvColishCOX2 strategy as a promising tool for molecular therapy in intestinal inflammatory diseases.
Introduction
Engineered Escherichia coli bacteria expressing invasin (Inv) and listeriolysin O (HlyA) genes (from Yersinia pseudotuberculosis and Listeria monocytogenes, respectively) are able to invade mammalian cells and promote a functional gene transfer from bacteria to the host.1,2,3 Exploiting the high potential of this phenomenon, Xiang et al. defined a “trans-kingdom RNAi” strategy to silence β-1 catenin gene in mammalian cells using nonpathogenic E. coli expressing Inv/HlyA (InvColi) and specific short hairpin RNA (shRNA).4 It is well known that inflammation and cyclooxygenase-2 (COX-2) gene overexpression contribute to colorectal cancer (CRC) development and progression5,6,7 but effective pharmacological strategies based on COX-2 inhibition are still needed for the prevention and/or treatment of CRC.8,9,10 In view of this, our research group recently developed an InvColi-based approach to efficiently and specifically silence COX-2 in human CRC cells in vitro. This strategy strongly impaired both the proliferative and invasive behavior of cancer cells and had a significant anti-inflammatory effect.11
COX-2 is also overexpressed in the colonic mucosa of patients with inflammatory bowel disease (IBD), mainly Crohn's disease (CD) and ulcerative colitis (UC).12 IBD has a higher incidence in Western countries and is characterized by chronic inflammation of the gastrointestinal tract and recurrent ulceration of the bowel.13 Typical symptoms of IBD are diarrhea, abdominal pain, bleeding, anemia, and weight loss. In long-standing cases, both CD and UC may also be associated with an increased risk of CRC development.14,15 Although the precise etiology of IBD is still unknown, a number of reports have indicated that these disorders are caused by an abnormal immune response to enteric antigens. In genetically susceptible individuals, deregulation of both innate and acquired immunity mechanisms leads to a chronic intestinal inflammation, as shown by a strong imbalance in the cytokine and chemokines network.16,17 Because of functional redundancy, pleiotropy and multiple mechanisms of regulations, cytokine network should be considered as a complex and dynamic system, playing a key role in the initiation and perpetuation of inflammation.18 Several studies support the importance of the microbiota and microbe-mucosa crosstalk in the pathogenesis of IBD, showing that IBD patients have a distorted, low-diverse intestinal microbiota structure.19 In fact, colon inflammation has been associated with a reduction of key immunomodulatory microbiota groups like Lactobacillaceae, Bifidobacteriaceae, Faecalibacterium prausnitzii as well as other components of the Clostridium clusters IV-XIVa and a parallel increase in proinflammatory pathobionts such as Enterobacteriaceae and Enterococcales.20,21 Thus, it is likely that the IBD-associated microbiota possesses a pro-inflammatory structure which establishes inflammation and consolidates the disease.
Aminosalicylates (e.g., 5-ASA), corticosteroids and immunosuppressive agents (e.g., azathioprine and mercaptopurine) represent the first-line therapy for IBD whereas other drugs such as metronidazole and methotrexate, broad-spectrum antibiotics, probiotics or biological drugs such as monoclonal antibodies (e.g., Infliximab and Adalimumab, anti-TNF-α chimeric immunoglobulins) provide alternative therapies.22 Recent advances in biotechnology have led to the development of innovative strategies for IBD treatment and drug delivery in colonic mucosa.23,24 In this regard, molecular therapies based on genetically modified bacteria have proved to be effective and safe, but they raise concerns about survival and propagation in the environment.25,26
There is currently no ideal animal model for IBD, and several methods have been proposed to induce colitis in mice, rats and other animals. Dextran sulfate sodium (DSS)-induced colitis is one of the most commonly used models and reflects many of the clinical features of UC.27 Once it has entered into colonocytes, DSS impairs major cellular functions, causing cell cycle arrest and apoptosis.28 By interfering with intestinal barrier function, DSS is also able to stimulate local and systemic inflammation by locally increasing COX-2 expression and secreting a variety of cytokines and other inflammatory mediators that spread from the colon to the blood.29 Colitis severity depends on the commensal bacterial strains maintained in gnotobiotic animals30 and DSS treatment has been associated with a major pro-inflammatory compositional shift of the intestinal microbiota.31 Our group recently reported a longitudinal analysis of inflammation and microbiota dynamics in a model of mild chronic DSS-induced colitis in mice.32
Aim of this paper was to demonstrate the in vivo feasibility, effectiveness and safety of an RNAi-based/InvColi-driven strategy for COX-2 silencing in the murine model of DSS-induced colitis. Such approach might open promising new perspectives for the prevention/treatment of IBD and CRC.
Results
InvColishCOX2 treatment inhibits DSS-induced colitis
We first designed an RNAi-based system to silence COX-2 in murine cells. By transfecting CT26 mouse colon carcinoma cell line with RNAi vectors (pSUPER, pS), we evaluated the efficiency of anti-COX-2 shRNA targeting two different regions of COX-2 mRNA (Supplementary Figure S1a). After transfection, both pSshCOX2A and pSshCOX2B induced a significant downregulation of COX-2 mRNA compared to the control (pSNC), in the presence or absence of phorbol 12-myristate 13-acetate (PMA) stimulation.33 Importantly, a higher silencing effect was observed after the cotransfection of both vectors in the same sample (Supplementary Figure S1b). To support our RNAi data, we performed a real-time PCR assay based on three different COX-2 amplicons (AMP1-3, Supplementary Figure S1a) and, as expected, we observed the presence of cleaved fractions of COX-2 mRNA on sites “A” and “B” after RNAi induction. In fact, the ratios AMP2/AMP1 and AMP3/AMP1 were significantly lower in pSshCOX2A and pSshCOX2B transfected CT26 cells compared to pSNC, respectively (Supplementary Figure S1c,d).
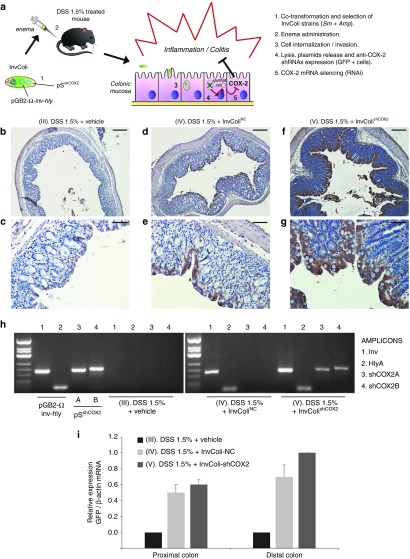
The in vivo efficacy and feasibility of our InvColi/RNAi-based strategy for COX-2 silencing was investigated in a murine model of DSS-induced colitis recently developed by our group.32 For a schematic description of the experimental design and approach, please refer to Figure 1a and Supplementary Figure S2a (see also Materials and Methods section). Briefly, InvColi strains were created cotransforming E. coli (DH5α) with pSNC (empty), pSshCOX2A or pSshCOX2B (expressing the two different shCOX2 under a mammalian promoter) and pGB2-Ω-inv-hly plasmids. Colitis in mice was induced by 9 days DSS 1.5% oral administration whereas InvColi strains treatment (NC or shCOX2A/B mix) was based on repeated enema administrations (×5). COX-2 silencing in colonic mucosa was induced after InvColishCOX2 internalization in epithelial cells followed by bacterial lysis and pSshCOX2A/B plasmids release (Figure 1a). We investigated the ability of InvColi strains to invade and transfect colon cells of DSS-treated mice after enema administration (MOI 1:100). Since pS plasmids carry an expression cassette for the green fluorescent protein (GFP) with a mammalian promoter, GFP protein expression was evaluated by IHC in FFPE colon sections from DSS/InvColi-treated mice and compared to the negative control (DSS alone, Figure 1b,c). IHC analysis was carried out on the distal part of colon specimens taken at time D7 (SC). Basing on our observations, both InvColiNC and InvColishCOX2A/B bacteria were able to efficiently penetrate colon cells and release pS plasmids, leading to GFP expression (Figure 1d–g). The analysis of IHC images at high magnification (400×) showed that epithelial cells in colon mucosa were largely infected/transfected by InvColi bacteria (group IV and V) but it was possible to detect also GFP-positive cells of the immune system, both in colon mucosa and submucosa (in particular group IV, characterized by a significantly higher immune system infiltration). In fact, a number of monocytes, macrophages, neutrophil granulocytes, lymphocytes, and plasma cells were GFP-positive (Supplementary Figure S3). The invasion/transfection of colon cells was further demonstrated assaying the presence of pGB2-Ω-inv-hly and pSshCOX2A/B plasmids in total DNA extracted from colon specimens (Figure 1h) and evaluating GFP mRNA expression by real-time PCR (Figure 1i).
Figure 1.
InvColishCOX2 strategy is suitable for genetic material transfer in vivo. (a) E. coli (DH5α) is cotransformed with pGB2-Ω-inv-hly and pSshCOX2 plasmids to obtain the InvColishCOX2 strain, subsequently selected and administered via enema to colitic mice treated with DSS 1.5%. InvColishCOX2 bacteria penetrate colon epithelial cells and promote after endocytic lysis the expression of two short hairpin RNA (shRNA) targeting COX-2 mRNA. (b–g) GFP protein expression was evaluated by immunohistochemistry in FFPE colon specimens from experimental mice group III (DSS 1.5% + vehicle), IV (DSS 1.5% + InvColiNC), and V (DSS 1.5% + InvColishCOX2). Colon specimens were collected on day 7 (D7, start colitis, SC). (b,d,f) Scale bar = 200 µm; magnification ×50. (c,e,g) Scale bar = 50 µm; magnification ×200. Vehicle = LB medium. (h) PCR analysis of Inv-(1), HlyA-(2), shCOX2A-(3), and shCOX2B-(4) amplicons was performed on pGB2-Ω-inv-hly, pSshCOX2A/B purified plasmids and on total DNA extracted from group III, IV, and V (colon specimens). PCR products were analyzed after electrophoresis on 2.5% agarose gel. (i) GFP mRNA expression was analyzed by real-time PCR and normalized against β-actin mRNA levels. Relative expression was calculated referring to group V (distal colon, time D7) and data represent the mean + SEM of three independent analyses (n = 3 per group).
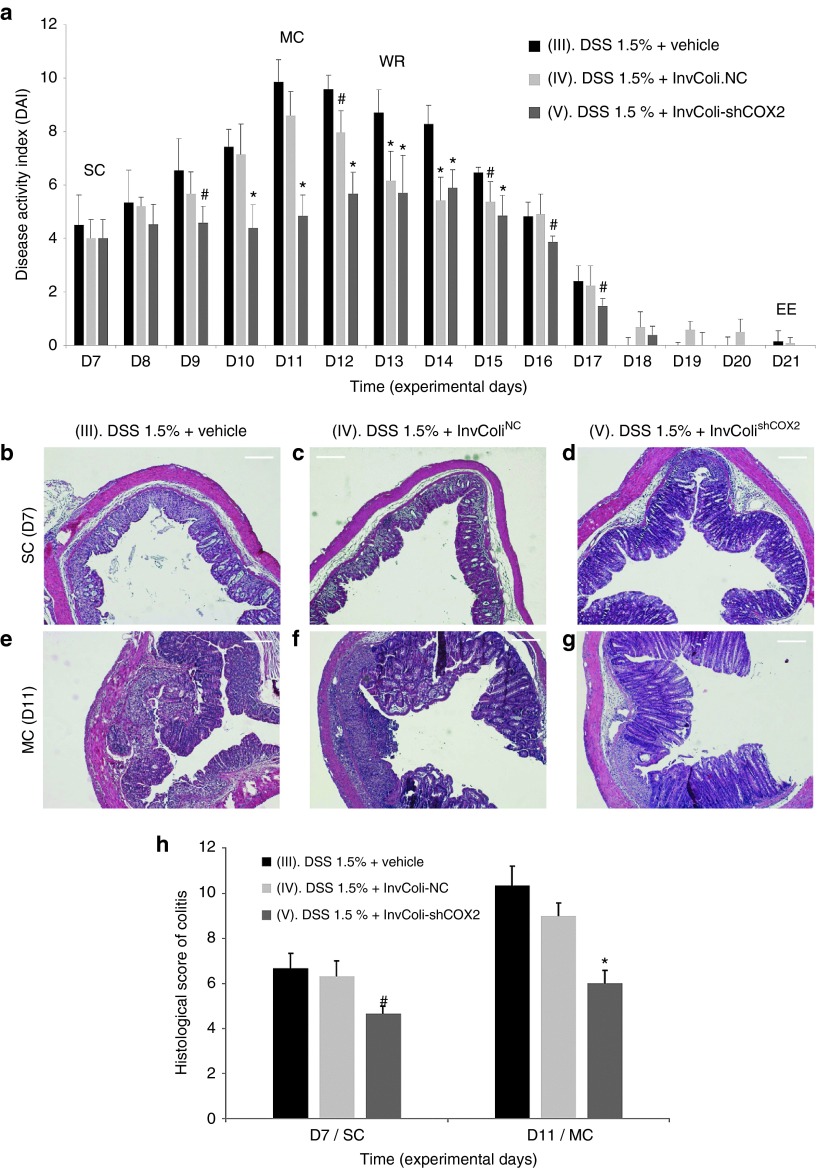
The effect of DSS/InvColishCOX2 treatments was evaluated considering the disease activity index (DAI) from the start of colitis (SC/D7) to the end of the experiment (EE/D21), calculated as the sum of weight loss, stool consistency and bleeding scores (Figure 2a; Supplementary Figures S2b and S4a–c). As expected, mice from colitis-negative control groups I and II (no DSS treatment) did not show any symptom of disease with all parameters scoring zero (data not shown). On the contrary, all DSS-treated mice (groups III, IV, and V) started to display symptoms of colitis after 7 days of oral DSS 1.5% administration (time D7, start colitis, SC). Mice from colitis-positive control groups treated with bacterial vehicle (group III) or InvColiNC (group IV) were characterized by significant weight loss (ranging from 0 to ~20%), decreased stool consistency and increased bleeding, reaching a DAI peak at time D11 (maximum colitis, MC). Moreover, subjects from both experimental groups III and IV (n = 2 and n = 1, respectively) were sacrificed as they had reached the pathological endpoints prematurely. Interestingly, colitic mice treated with InvColishCOX2 (group V) were characterized by lower scores of Δweight (%), stool consistency and bleeding, resulting in a significantly reduced DAI with a shifted peak at time D14. Figure 2a clearly shows that a significant amelioration of colitis-related symptoms can be induced by treating mice with repeated enema administration of InvColishCOX2 bacteria. It is also worth noting that InvColiNC exerted an effect on DSS-treated mice, especially during the weight recovery phase (from D12 to D15), favoring a more rapid decline of colitis DAI.
Figure 2.
Effect of the InvColishCOX2 strategy on DSS-induced colitis. (a) The biological effect of InvColi/RNAi-mediated COX-2 silencing was evaluated considering the disease activity index (DAI) of colitis, calculated by the combined score of weight loss, stool consistency, and bleeding (see also Supplementary Figures S2b and S4). All parameters were scored from experimental day 7 (D7) to experimental day 21 (D21). Experimental mice groups: (III) “DSS 1.5% + vehicle”; (IV) “DSS 1.5% + InvColiNC”; (V) “DSS 1.5% + InvColishCOX2.” Vehicle, LB medium; EE, end of experiment; MC, maximum colitis; SC, start colitis; WR, weight recovery. Data represent the mean + SD of independent measurements; per group: n = 12 (D7), n = 9 (D8–D11), n = 6 (D12–D13), n = 3 (D14–D21). *P < 0.01; #P < 0.05. (b,g) A histological analysis was carried out on FFPE colon specimens from group III, IV, and V. Colon specimens were collected on day 7 (D7, start colitis, SC) and day 11 (D11, maximum colitis, MC). Scale bar = 200 µm; magnification ×50. (h) Quantification of histological differences between experimental mice group III, IV, and V at times D7 and D11, based on the scoring parameters shown in Supplementary Figure S2c. Data represent the mean + SEM of independent measurements (n = 3 per group). *P < 0.01. #P < 0.05.
Clinical scores were also supported by the histological evaluation of colon specimens taken from the colocecal junction to the anus of treated mice. Compared to the colitis-negative controls (groups I and II, data not shown), at time D7 (SC) specimens from groups III, IV, and V were characterized by clear histological changes in colonic mucosa due to DSS-induced inflammation. In particular, we observed immune system infiltration, architectural abnormalities and mucosal erosion, even though significant differences were still evident between the three groups at epithelial level. DSS alone and “DSS + InvColiNC” led to diffuse crypt disappearance and depletion of goblet cells unlike the InvColishCOX2-treated group, in which these histological changes were much less evident (Figure 2b–d). Importantly, these differences between groups III, IV, and V were significantly higher at time D11 (MC) since colon specimens from the “DSS + InvColishCOX2” group showed only moderate loss of epithelium, elongation of colonic mucosa and immune system infiltration while specimens from the “DSS + vehicle” group were characterized by massive immune system infiltration, mucosal erosion and widespread colonic mucosa distortion with a loss of goblet cells and crypts. Compared to group III, group IV was characterized by a lower mucosa distortion and a lower loss of globet cells and cripts (Figure 2e–g). Histological differences between DSS-treated mice groups (III, IV, and V) were scored and quantified (Figure 2h and Supplementary Figure S2c).
InvColishCOX2 treatment inhibits COX-2 expression and inflammation
We investigated the efficiency of COX-2 mRNA silencing mediated by InvColishCOX2 by analyzing colon specimens from colitic mice at different time points (SC/D7 and MC/D11) and performing a real-time PCR assay based on three different COX-2 amplicons (AMP1-3, Figure 3a). Even though COX-2 mRNA (AMP1/β-actin) was slightly detectable in normal colon mucosa (groups I and II, data not shown), its expression was strongly increased after DSS 1.5% treatment (groups III, IV, and V). Nevertheless, enema administration of InvColishCOX2 in DSS-treated mice (group V) promoted a significant decrease in COX-2 mRNA level, compared to groups III and IV (Figure 3b, dark gray bars). Moreover, we confirmed the presence of cleaved fractions of COX-2 mRNA on sites “A” and “B” due to RNAi induction in group V. In fact, both COX-2 mRNA ratios AMP2/AMP1 and AMP3/AMP1 were significantly lower in InvColishCOX2-treated mice, compared to AMP1/β-actin level and to control groups III and IV (Figure 3b, light gray bars). COX-2 protein expression in colon specimens was analyzed by Western blot (Supplementary Figure S5a) and immunohistochemistry. As expected, COX-2 was slightly detectable in IHC colon samples from untreated mice (groups I and II, data not shown). At time D7 (start of colitis), DSS 1.5% treatment induced a high COX-2 expression, particularly in colon epithelium. Nevertheless, enema administration of InvColishCOX2 in DSS-treated mice (group V) promoted a significant decrease in COX-2 protein level in colon epithelial cells, compared to groups III and IV (Figure 3c–e). At time D11 (maximum of colitis), COX-2 protein was strongly overexpressed across the entire mucosa but in the presence of InvColishCOX2 we observed a reduced level (Figure 3f–h). In particular, COX-2 protein resulted particularly induced in those colonic areas with higher colitis-related features, much more evident in specimens from groups III and IV, compared to specimens from InvColishCOX2-treated group V (Figure 3f–h). High-magnification IHC images are shown in Supplementary Figure S5b–g.
Figure 3.
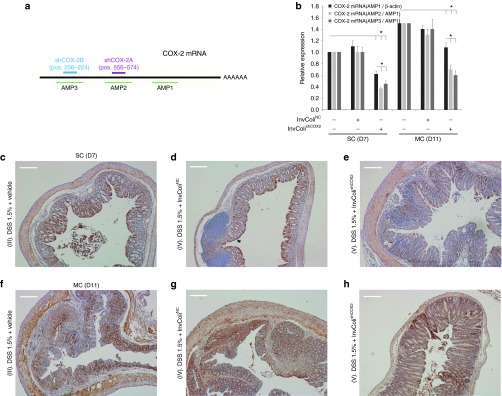
InvColishCOX2 treatment induces COX-2 silencing in murine colon mucosa. (a) Two different sequences of short hairpin RNA (shRNA) were tested for murine COX-2 mRNA silencing and three different amplicons were considered to evaluate COX-2 mRNA levels (b) COX-2 expression was analyzed in fresh colon samples from mice, immediately processed after sacrifice. Total COX-2 mRNA expression (AMP1) was normalized against β-actin mRNA expression whereas AMP2 and AMP3 levels were normalized against AMP1 level. Relative expression was calculated referring to the negative control (DSS-treated/InvColi-untreated mice, time D7) and data represent the mean + SEM of three independent analyses (n = 3 per group). *P < 0.01. (c–h) COX-2 protein expression was evaluated by immunohistochemistry in FFPE colon specimens from experimental mice group III (DSS 1.5% + vehicle), IV (DSS 1.5% + InvColiNC), and V (DSS 1.5% + InvColishCOX2). For IHC analysis, colon specimens were collected on day 7 (D7, start colitis, SC) and day 11 (D11, maximum colitis, MC). Scale bar = 200 µm; magnification ×50.
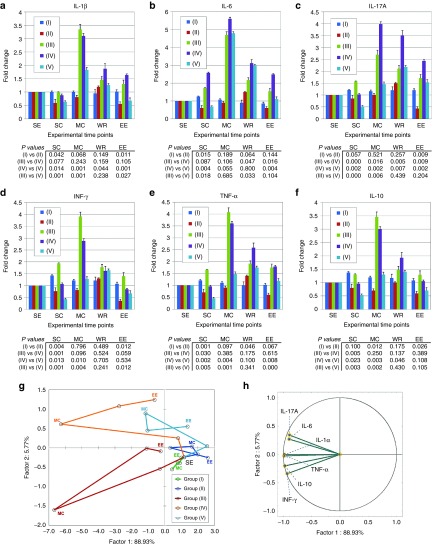
The anti-inflammatory action of InvColishCOX2 treatment was further investigated by analyzing the cytokine profile associated with colitis. In particular, plasma levels of IL-1β, IL-6, IL-17A, INF-γ, TNF-α (pro-inflammatory), and IL-10 (anti-inflammatory) were detected in blood samples from all experimental mice groups (I–V) at five different time points (SE, SC, MC, WR, and EE) (Figure 4a–f). Whereas the cytokine level in blood from untreated mice (group I) did not show any considerable variation from time D7/SC to time D21/EE, DSS 1.5% oral administration in group III induced a strong increase in all cytokines assayed, showing peak levels at time D11/MC (a three to fivefold increase range, compared to the basal level at time D0). Interestingly, InvColishCOX2 treatment (group V) induced in mice a specific change in the cytokine profile associated with colitis. Comparing group V to group III, we recorded a significant decreased level of cytokines at times D7 (all cytokines), D11 (all cytokines, except IL-6) and D21 (IL-1β, TNF-α, and INF-γ) whereas we observed increased level only for IL-6 at time D13. In group V, peak levels of cytokines were recorded at time D11/MC (IL-1β and IL-6) and time D13 (IL-17A, INF-γ, TNF-α, and IL-10). Data collected from experimental mice groups II and IV showed that InvColi treatment might have an effect on inflammatory cytokine profile per se. In the absence of colitis, InvColishCOX2 administration induced in group II a significant decrease in cytokine levels at times D7 (IL-1β, IL-6, INF-γ, and TNF-α), D11 (IL-10) and D21 (IL-1β, IL-17A, INF-γ, and IL-10), compared to group I. Moreover, InvColiNC administration in colitic mice from group IV led to a significant variation in plasmatic cytokines at times D7 (lower IL-17A, INF-γ, TNF-α, and IL-10 levels), D11 (higher IL-17A level), D13 and D21 (higher IL-6 and IL-17A levels), compared to group III.
Figure 4.
InvColishCOX2 treatment exerts anti-inflammatory effects in colitic mice. (a–f) The inflammatory cytokine profile was evaluated in plasma samples from all experimental mice groups (I–V). Blood samples were collected at five different time points: SE (start of experiment, D0), SC (start of colitis, D7), MC (maximum of colitis, D11), WR (weight recovery, D13), and EE (end of experiment, D21). IL-1β, IL-6, IL-17A, IFN-γ, TNF-α, and IL-10 expression analysis was performed using a Luminex-based multiplexed bead immunoassay. Cytokine concentration values (pg/ml, data not shown) were calculated considering the dilution factor of plasma samples and variations in graphs are expressed as fold change, from the basal level of each cytokine at time D0. Data represent the mean + SD of independent measurements; per group: n = 12 (D0 and D7), n = 9 (D11), n = 6 (D13), n = 3 (D21). Statistical significance (P values) for differences between mice groups is included in the figure below each graph (group I versus II, group III versus IV, group IV versus V, and group III versus V). (g,h) Principal component analysis (PCA) was performed on the entire cytokine dataset considering the appropriate variables (cytokines; n = 6) and total cases (five experimental groups at five different time points; n = 25). The relations among variables and cases were highlighted by plotting them in the space generated by the factor axes 1 and 2. Green: “NO DSS + vehicle” (group I); blue: “NO DSS + InvColishCOX2” (group II); red: “DSS 1.5% + vehicle” (group III); orange: “DSS 1.5% + InvColiNC” (group IV); cyan: “DSS 1.5% + InvColishCOX2 (group V). Consecutive time points are connected by the line.
In order to better evaluate the correlation between inflammatory cytokine profiles and animal treatments (DSS and/or InvColi), we performed a principal components analysis (PCA) on the entire dataset (Figure 4g,h). Considering the appropriate variables (six cytokines) and total cases (five experimental groups at five different time points), the PCA plot in Figure 4g clearly shows that colitic groups III and IV are characterized by a large variation on the factor axes (lower-left and upper-left side, respectively), whereas the spatial distribution of colitic group V (treated with InvColishCOX2) is significantly reduced, mapping more similarly to the distribution of healthy groups I and II (lower-right and upper-right side, respectively). The two principal components accounted for 94.7% of the total variation (88.93% Factor 1 and 5.77% Factor 2) and the correlation between the variables and the factor axes in the PCA plot is shown in Figure 4h. Locating nearly across the correlation circle, all cytokines strongly contributed to the spatial distribution of the cases in the PCA plot.
InvColishCOX2 treatment modulates intestinal microbiota modifications induced by colitis
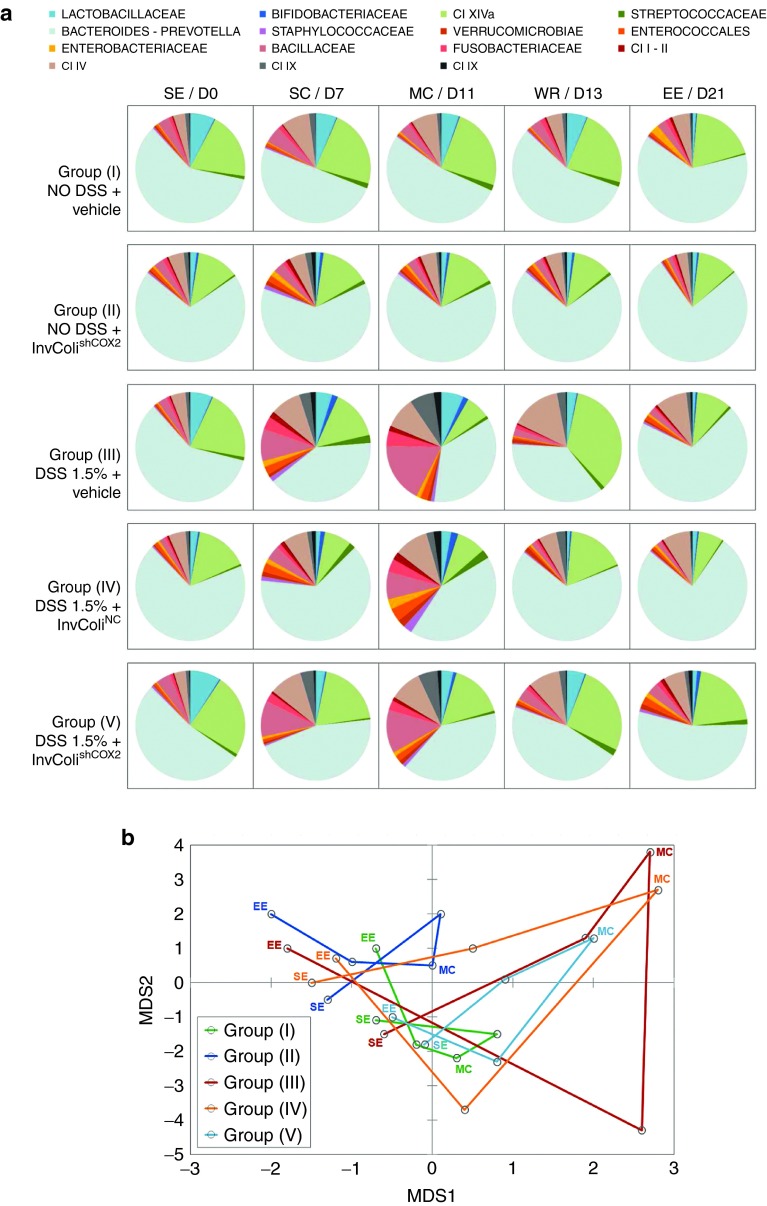
To evaluate the impact of InvColishCOX2 administration on the microbial ecology of DSS-induced colitis, we compared the temporal dynamics of the fecal microbiota structure in all experimental mice groups (I–V) at five different time points (SE, SC, MC, WR, and EE). For each time point, the fecal microbiota structure was determined by using the phylogenetic universal platform HTF-Microbi.Array (Figure 5a). At the compositional level, InvColishCOX2 treatment in group V proved to be effective in lowering the DSS-dependent increase in potential colitogenic pathobionts, such as members of Enterococcales, Enterobacteriaceae, Bacillaceae, Fusobacteriaceae, and Clostridium (I, II, IX, XI), showing a significant difference from groups III and IV (Figure 5a). In colitic mice, the presence of InvColiNC bacteria seemed to have only a partial positive effect due to the reduction of Bacillaceae and Clostridium IX whereas in healthy group II, treated only with InvColishCOX2, we did not observed significant differences on the dynamics of the fecal microbiota structure, compared to group I. Importantly, even after five repeated enema administrations of InvColi (MOI 1:100), no significant/uncontrolled increase in E. coli (a well known member of Enterobacteriaceae) was recorded in mice fecal microbiota from experimental groups II, IV, and V (Figure 5a). The principal coordinate analysis (PCoA) of the Euclidean distance between samples also showed a peculiar temporal dynamics of the intestinal microbiota for the five mice groups (Figure 5b). Samples from mice groups with higher colitis score (in particular, groups III and IV) segregated together in the upper-right side of the PCoA plot, characterized by positive values of both multidimensional scaling 1 and 2 (MDS1 and MDS2, which together account for the largest variation in the dataset). Interestingly, InvColishCOX2 treatment (group V) was effective in modulating and in particular inhibiting the microbiota progression towards a colitis configuration (upper-right side of the PCoA plot), also favoring the recovery of a healthy configuration (lower left side of the PCoA plot) (Figure 5b).
Figure 5.
InvColishCOX2 impairs colitis-associated intestinal microbiota modifications. (a) The relative abundance profile of mice fecal microbiota components is reported for all five experimental mice groups (I–V) at the following time points: SE/D0 (start of experiment), SC/D7 (start of colitis), MC/D11 (maximum of colitis), WR/D13 (weight recovery), and EE/D21 (end of experiment, D21). The microbiota was characterized using the fully validated phylogenetic DNA microarray platform HTF-Microbi.Array. (b) The principal coordinate analysis (PCoA) of the Euclidean distances between the fecal microbiota of each mice groups at five different time points is shown. Green: “NO DSS + vehicle” (group I); blue: “NO DSS + InvColishCOX2” (group II); red: “DSS 1.5% + vehicle” (group III); orange: “DSS 1.5% + InvColiNC” (group IV); cyan: “DSS 1.5% + InvColishCOX2 (group V). Consecutive time points are connected by the line. MDS1 and MDS2 = multidimensional scaling 1 and 2.
Discussion
Engineered E. coli expressing invasin and listeriolysin O genes (InvColi) can be used to transfer genetic material from bacteria to mammalian cells.1,2,3,4 Recently, we developed an InvColi/RNAi-based technique to silence the COX-2 enzyme in human colon cancer cells in vitro.11 Encouraged by the promising results obtained, we aimed to demonstrate the in vivo feasibility of this COX-2 silencing technique in a murine DSS-induced model of colitis.27,32 Prolonged DSS 1.5% oral administration in mice leads to weight loss, decreased stool consistency and increased bleeding, and these typical clinical symptoms of colitis are coupled with marked inflammation and COX-2 overexpression in colon mucosa. This model is characterized by a mild-chronic and self-limiting colitis, with a low mortality rate in mice and less dramatic intestinal injuries compared to 2.5 or 3% DSS-induced colitis.34 Thus, the DSS 1.5%-induced colitis model results more suitable to evaluate the efficacy of therapeutic agents both from a clinical and a histological point of view.
First, data collected in the present work demonstrate that InvColi strains engineered by our group are able to successfully vehiculate genetic material in vivo and to promote the ectopic expression of transgenes of interest in colon tissue (e.g., GFP reporter gene, shCOX2). In particular, it is possible to target and transfect different types of murine colon cells using InvColi strategy, such as epithelial cells and cells of the immune system (both phagocytic and not phagocytic). Second, InvColishCOX2 enema treatment in DSS-colitic mice preserves colon mucosa integrity, delays the onset of colitis and reduces the disease activity index (DAI) of colitis, also leading to increased mice survival. Importantly, these clinical observations are supported by evidence of a significant impairment of COX-2 expression in the colonic mucosa of InvColishCOX2-treated mice, proving the high efficacy of our innovative strategy for COX-2 silencing in vivo.
The therapeutic benefits of InvColishCOX2 administration are likely to stem from a global anti-inflammatory effect. In fact, COX-2 inhibition mediated by InvColishCOX2 is also strongly associated with a marked decrease in plasma levels of colitis-associated inflammatory cytokines (in particular IL-1β, IL-17A, INF-γ, and TNF-α).17,18,32,35 The PCA analysis on the entire cytokine dataset provides an overview on the differences between experimental mice groups, related to their inflammatory profiles. Interestingly, InvColishCOX2 treatment clearly modifies the cytokine variations in colitic mice, shifting their profile to the healthy mice side. Our data show increased levels of circulating IL-10 (an anti-inflammatory cytokine) in DSS-treated mice, significantly lowered by InvColishCOX2 administration. Since we tested the efficacy of our strategy in a model of mild-chronic colitis, this phenomenon might be supported by the evidence that chronic DSS-induced colitis, compared to acute models, is characterized by high levels of IL-10 in murine plasma.32,35 Moreover, variation of IL-10 levels observed in our model might be explained considering that a pro-inflammatory pathway is normally associated with an anti-inflammatory pathway activation. IL-10 can be induced by COX-2 enzyme products (such as PGE2) during inflammation as autoregulatory negative-feedback mechanism.36 Thus, the reduced IL-10 expression in InvColishCOX2-treated mice might be directly dependent on COX-2 silencing. IL-6 is the only pro-inflammatory cytokine that is not lowered by the InvColishCOX2 treatment but it is worth noting that InvColiNC administration leads to a strong increase in IL-6 levels during colitis. Presumably, the lack of IL-6 inhibition in InvColishCOX2-treated mice might be due to the unavoidable administration of bacteria (and LPS) in an inflamed tissue context.
InvColishCOX2 treatment inhibits the colitis-associated intestinal microbiota shift and favors the recruitment of a healthy layout. In particular, InvColishCOX2 administration significantly impairs the DSS-dependent increase in pro-inflammatory pathobionts (such as members of Enterococcales, Enterobacteriaceae, Bacillaceae, Fusobacteriaceae, and Clostridium)21 which consolidate colitis by supporting inflammation in the gut. As a further mechanism of protection, this phenomenon fully supports the anti-inflammatory and therapeutic action of InvColishCOX2 bacteria.
We hypothesize that InvColi strains might have a biological effect per se in the amelioration process of colitis, apart from COX-2 silencing. In fact, the administration of InvColiNC to DSS-treated mice favors a more rapid decline of colitis-associated DAI during the weight recovery phase and modifies the cytokine profile, especially related to INF-γ expression (as demonstrated by the significant variation on the factor 2-axis in PCA plot). Moreover, InvColiNC treatment seems to promote a shift toward a healthy state of microbiota during the recovery phase of colitis. Similarly, InvColishCOX2 strain decreases the plasma level of pro-inflammatory cytokines in DSS-untreated mice. This phenomenon is much more evident during enema administrations and it might be closely related both to the silencing of COX-2 (normally expressed in colonic mucosa) and to the presence in colon of InvColi strain itself. Our hypothesis is supported by previous works on the therapeutic effect of nonpathogenic E. coli strains “Nissle 1917” on ulcerative colitis.37,38
Considering the data collected in this article, we believe that the efficacy of our InvColishCOX2 treatment might rely on a combined effect deriving from both COX-2 RNAi and InvColi nonpathogenic strain usage. COX-2 silencing is a prerequisite for colitis impairment, but this effect is surely strengthened by InvColi as therapeutical vehicle. Very importantly, we did not observe any uncontrolled colonization of engineered InvColi bacteria, local and/or systemic infection or adverse inflammatory/immune response after repeated InvColi enema administrations in mice. Currently, IBD therapy is based on the use of anti-inflammatory molecules, immunomodulatory agents and biological drugs as monoclonal antibodies.22 Anti-inflammatory molecules and immunodulators acts by strongly and a specifically inhibiting inflammatory response, but their long term use may raise concerns due to the onset of severe side effects. Biologic drugs are designed to selectively neutralize a single inflammatory mediator. Despite their selectivity, also these drugs are associated to severe side effects linked to a strong immunosuppression or allergic reactions. Still far from replacing conventional drugs for IBD treatment, InvColi strategy has several advantages and may be surely improved and provide fascinating insights toward novel molecular therapies. In fact, both gene silencing and gene overexpression can be applied to this technology having a high efficiency in the transfer of genetic material in vivo. The expression of anti-COX-2 shRNA driven by a constitutive mammalian promoter surely represents a limitation to InvColishCOX2 strain usage. Loss of integrity of the inflamed colonic epithelium may result in bacterial translocation over the mucosa and a widespread distribution/expression of the transgene2 but, as mentioned above, we did not observe any adverse effect related to bacterial systemic dissemination in InvColi-treated mice (e.g., bacteremia or sepsis). However, the possibility of a spatial/temporal control of therapeutic gene expression may strongly improve our strategy and its selectivity. Fortunately, the use of an inducible regulatory promoter can be easily applied to the InvColi-mediated expression system, as already demonstrated by our group.11
We can conclude that our study provides an example of an innovative and feasible RNAi-based in vivo approach for the treatment of colitis and demonstrates that a gene therapy based on InvColi bacteria may be attainable. The InvColi vehiculating strategy apparently acts as an effective controlled system, self-inactivating and lacking intrinsic toxicity. We do not exclude that such a strategy may be applied for the treatment of inflammatory bowel disease and, more importantly, colorectal cancer.
Material and Methods
RNAi plasmids and InvColi strains. Empty pSUPER.retro vector (Oligoengine, Seattle, WA), based on the murine stem cell virus (MSCV) genome, was considered as the negative control (pSNC). pSUPER.retro vectors expressing two different sequences of shRNA against murine COX-2 mRNA (pSshCOX2A and pSshCOX2B) were created according to procedures described elsewhere.6 Sequences for shCOX2A: FW: 5′-GATCCCCGGGAAATAAGGAGCTTCCTTTCAAGAGAAGGAAGCTCCTTATTTCCCTTTTTTA-3′; REV: 5′-AGCTTAAAAAAGGGAAATAAGGAGCTTCCTTCTCTTGAAAGGAAGCTCCTTATTTC CCGGG-3′. Sequences for shCOX2B: FW: 5′-GATCCCCGGATTTGACCAGTATAAGTTTCAAGAGAACTTATACTGGTCAAATCCTTTTTTA-3′; REV: 5′-AGCTTAAAAAAGGATTTGACCAGTATAAGTTCTCTTGAAACTTATACTGGTCAAATCCGGG-3′. E. coli (DH5α) strains were cotransformed with pGB2-Ω-inv-hly (SmR+, kindly provided by Dr Catherine Grillot-Courvalin, Institut Pasteur, Paris, France) and pSUPER.retro (AmpR+) plasmids to obtain E. coli invasive strains carrying the two different shCOX2 expression vectors (InvColiCOX2A and InvColiCOX2B) and the empty plasmid as negative control (InvColiNC), respectively. InvColi bacteria were grown in LB medium.
Mice and treatments. Sixty male 8-week-old C57BL/6 mice were purchased from Charles River Laboratories (Lecco, Italy). Animals were housed in a controlled environment and in collective cages at 22 ± 2 °C and 50% humidity, under a 12 hours light/dark cycle. Mice were allowed to acclimate to these conditions for 9 days (ninth day = D0) before inclusion in the experiment and had free access to food and water throughout the study. Colitis was induced in mice by oral administration of 1.5% dextran sulfate sodium in tap water (DSS, TdB Consultancy, Uppsala, Sweden). DSS solution was freshly prepared every day and administered to mice for 9 days (D1-9), followed by 12 days of tap water (D10-21). The average amount of DSS taken was recorded daily. Enema administrations were prepared as follow: 100 µl of LB (vehicle); 100 µl of LB + InvColiNC (1 × 105 total cells; approximate MOI 1:100); 100 µl of LB + InvColishCOX2 (0.5 × 105 InvColishCOX2A cells + 0.5 × 105 InvColishCOX2B cells; approximate MOI 1:100). Enemas were administered on days 3, 5, 7, 9, and 11 (D3, D5, D7, D9 and D11). The experimental design is schematized in Supplementary Figure S2a. Mice were divided into five experimental groups (n = 12 mice/group): (I) “NO DSS + vehicle”; (II) “NO DSS + InvColishCOX2”; (III) “DSS 1.5% + vehicle”; (IV) “DSS 1.5% + InvColiNC”; (V) “DSS 1.5% + InvColishCOX2.” All mouse experiments were approved by the Institutional Review Board of the University of Bologna and performed according to Italian and European guidelines.
Colon specimens. Mice were anesthetized and sacrificed by cervical dislocation on days 7, 11, 13, and 21 (D7, D11, D13, and D21). Colon was excised and rinsed with saline solution. Depending on subsequent experiments, colon specimens were fixed in 4% formalin for paraffin embedding (FFPE) or immediately used for DNA, RNA and protein extraction.
Immunohistochemistry. FFPE colon tissue sections (4 µm) were mounted on slides, deparaffinized with xylene and rehydrated through a series of graded alcohols. Sections were then incubated overnight at 4 °C with anti-GFP (Abcam, Cambridge, UK) or anti-COX-2 antibody (Cayman Chemicals, Ann Arbor, MI) at a 1:400 or 1:200 dilution in PBS/BSA-1.5%, respectively. Sections were then incubated with secondary anti-rabbit antibody for 15 minutes at room temperature and then in 3,3-diaminobenzidine tetrahydrochloride (DAKO) for 1 minute. Sections were counterstained with hematoxylin.
Plasmids detection in colon specimens. Fresh colon samples were repeatedly washed in TBS, minced on ice and homogenized. Samples were centrifuged, resuspended in DNA buffer containing proteinase K 100 µg/ml, RNase A 20 µg/ml and SDS 1% and incubated 3 hours at 50 °C. Total DNA was then extracted using standard phenol/chloroform procedure. The presence of Inv, HlyA, shCOX2A and shCOX2B amplicons was evaluated by PCR assay followed by agarose gel electrophoresis analysis. As positive control, PCR was performed also on purified pGB2-Ω-inv-hly, pSshCOX2A and pSshCOX2B plasmids. Inv primer pair: 5′-GCCAATAAGGAGCAGGAGAC-3′ and 5′-CCAAGGAGCCAGCCAATC-3′ (225 bp amplicon); HlyA primer pair: 5′-CATTTCACATCGTCCATCTATTTG-3′ and 5′-TTACCGTTCTCCACCATTCC-3′ (100 bp amplicon); shCOX2 forward primer: 5′-CATCAACCCGCTCCAAGG-3′; shCOX2A reverse primer: 5′-TTGAAAGGAAGCTCCTTATTTCC-3′ (235 bp amplicon); shCOX2B reverse primer: 5′-GTA TAA GTT CTC TTG AAA CTT ATA C-3′ (247 bp amplicon).
Disease activity index. Disease activity index (DAI) was calculated by the combined score of weight loss, stool consistency, and bleeding, as detailed in Supplementary Figure S2b. All parameters were scored from the start of colitis (SC) on day 7 (D7) to the end of the experiment (EE) on day 21 (D21).
Histological evaluation of colitis. FFPE colon tissue sections (4 µm) were mounted on slides, deparaffinized with xylene and rehydrated through a series of graded alcohols. Sections were stained with hematoxylin-eosin and observed for histological assessment of epithelial damage by a pathologist in a blinded manner. The histological degree of colitis was measured according to a scoring system based on the following features: loss of epithelium, crypt elongation, depletion of goblet cells, plasma cell infiltration.39 For further details, please refer to Supplementary Figure S2c.
RNA extraction and real-time PCR. Total RNA from colon samples was extracted using Trizol reagent (Life Technologies, Carlsbad, CA) according to the manufacturer's instructions. Extracted RNA samples were treated with DNase I to remove any genomic DNA contamination using DNA-free kit (Ambion, Austin, TX) and reverse-transcripted using RevertAid First Strand cDNA Synthesis Kit and random hexamer primers (Thermo Scientific, Waltham, MA). GFP, COX-2 and β-actin mRNA levels were analyzed by real-time PCR using SYBR Select Master Mix and StepOnePlus system (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. The melting curve data were collected to check PCR specificity. Each cDNA sample was analyzed in triplicate. GFP and COX-2 mRNA levels were normalized against β-actin mRNA and relative expressions were calculated using the 2−ΔΔCt formula. GFP primer pair: 5′-CAG TGC TTC AGC CGC TAC-3′ and 5′-GAT GTT GCC GTC CTC CTT G-3′ (204 bp amplicon); COX-2 primer pair (AMP1): 5′-TTC TCT ACA ACA ACT CCA TCC TC-3′ and 5′-GCA GCC ATT TCC TTC TCT CC-3′ (247 bp amplicon); COX-2 primer pair (AMP2): 5′-CCC TTC CTC CCG TAG CAG-3′ and 5′-CAA ACA TCA TAT TTG AGC CTT GG-3′ (144 bp amplicon); COX-2 primer pair (AMP3): 5′-TCA GCC AGG CAG CAA ATC-3′ and 5′-TCT TGT CAG AAA TTC AGG TGT AG-3′ (146 bp amplicon); β-actin primer pair: 5′-ACC AAC TGG GAC GAC ATG GAG-3′ and 5′-GTG GTG GTG AAG CTG TAG CC-3′ (380 bp amplicon).
Determination of plasma cytokine levels. Blood samples (~100 µl) were collected from mice tail vein on days 0, 7, 11, 13, and 21 (D0, D7, D11, D13, and D21) and transferred to eppendorf tubes containing sodium citrate. Blood was centrifuged at 1,000g for 10 minutes and plasma was collected and stored at −80 °C. Cytokine profile analysis was performed using a Luminex-based multiplexed mouse bead immunoassay-kit (Bio-Rad, Hercules, CA). The six-plex assays (IL-1β, IL-6, IL-17A, IFN-γ, TNF-α, IL-10) were performed in 96-well filter plates using a Bio-Plex 200 instrument (Bio-Rad) and following the manufacturer's instructions. Sample concentrations were estimated from the standard curve using a fifth-order polynomial equation and expressed as pg/ml after adjusting for the dilution factor (Bio-Plex Manager software 5.0). Samples below the detection limit of the assay were recorded as zero.
Characterization of the intestinal microbiota. The intestinal mice microbiota was characterized using the fully validated phylogenetic DNA microarray platform HTF-Microbi.Array. Targeting 33 phylogenetically related groups, this LDR-based Universal Array covers up to 95% of the mammalian gut microbiota.40 Gut microbiota analysis was performed on days 0, 7, 11, 13, and 21 (D0, D7, D11, D13, and D21). Total DNA from fecal material was extracted using the QIAamp DNA Stool Mini Kit (Qiagen, Limburg, The Netherlands) according to the modified protocol previously reported.41 Final DNA concentration was determined using NanoDrop ND-1000 (NanoDrop Technologies, Wilmington, DE). A nearly full-length portion of 16S rDNA gene was amplified using universal forward primer 27F and reverse primer 1492R, according to the protocol previously described.41 PCR amplifications were performed in a Biometra Thermal Cycler T Gradient (Biometra, Göttingen, Germany). PCR products were purified using the High Pure PCR Cleanup Microkit (Roche, Mannheim, Germany), eluted in 30 µl of sterile water and quantified with NanoDrop ND-1000. Slide chemical treatment, array production, LDR protocol and hybridization conditions were performed as previously reported.42 Briefly, LDR reactions were carried out in a final volume of 20 µl containing 500 fmol of each LDR-UA HTF-Microbi.Array probe, 50 fmol of PCR product and 25 fmol of the synthetic template (5′-AGC CGC GAA CAC CAC GAT CGA CCG GCG CGC GCA GCT GCA GCT TGC TCA TG-3′). LDR products were hybridized on Universal Arrays, setting the probe annealing temperature to 60 °C. All arrays were scanned and processed according to the protocol and parameters already described.41 Fluorescence intensities were normalized on the basis of the synthetic ligation control signal. The relative abundance of each bacterial group was obtained by calculating the relative fluorescence contribution of the corresponding HTF-Microbi.Array probe as a percentage of the total fluorescence.
Statistical analyses. Statistical analysis was carried out using SigmaStat v. 3.5 (Systat Software, San Jose, CA). Data are expressed as mean + SD or mean + SEM of at least three independent determinations. Student's t-test or analysis of variance was used to assess the statistical significance of the differences. Differences were considered statistically significant at P < 0.01 and P < 0.05. The PCA analysis of cytokine expression dataset was performed using Statistica v. 10 (StatSoft, Tulsa, OK). The Euclidean distance of HTF-Microbi.Array relative abundance profiles was used to perform PCoA and analysis was accomplished using the R packages Made4, Vegan, and Stats (www.cran.org).
SUPPLEMENTARY MATERIAL Figure S1. Murine COX-2 mRNA silencing induced by RNAi. Figure S2. Schematic representation of the experimental design. Figure S3. Colon cells express GFP after InvColiNC invasion/transfection. Figure S4. Numeric value and graphical representation of Dweight percentages calculated for all experimental mice groups during the experiment (from time D7 to D21). Figure S5. InvColishCOX2-mediated COX-2 silencing.
Acknowledgments
We gratefully acknowledge Verena Stenico, Alessandro Di Loreto, and Loredana Baffoni for the kind help in the PCA analysis of cytokine dataset. This work was supported by a grant from University of Bologna (RFO 2012 to E.S.) and by a grant from Association for International Cancer Research AICR (Grant No. 11–0738 to E.S.). The authors declare no conflict of interest.
Supplementary Material
Murine COX-2 mRNA silencing induced by RNAi.
Schematic representation of the experimental design.
Colon cells express GFP after InvColiNC invasion/transfection.
Numeric value and graphical representation of Dweight percentages calculated for all experimental mice groups during the experiment (from time D7 to D21).
InvColishCOX2-mediated COX-2 silencing.
References
- Grillot-Courvalin C, Goussard S, Huetz F, Ojcius DM, Courvalin P. Functional gene transfer from intracellular bacteria to mammalian cells. Nat Biotechnol. 1998;16:862–866. doi: 10.1038/nbt0998-862. [DOI] [PubMed] [Google Scholar]
- Castagliuolo I, Beggiao E, Brun P, Barzon L, Goussard S, Manganelli R.et al. (2005Engineered E. coli delivers therapeutic genes to the colonic mucosa Gene Ther 121070–1078. [DOI] [PubMed] [Google Scholar]
- Critchley-Thorne RJ, Stagg AJ, Vassaux G. Recombinant Escherichia coli expressing invasin targets the Peyer's patches: the basis for a bacterial formulation for oral vaccination. Mol Ther. 2006;14:183–191. doi: 10.1016/j.ymthe.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Xiang S, Fruehauf J, Li CJ. Short hairpin RNA-expressing bacteria elicit RNA interference in mammals. Nat Biotechnol. 2006;24:697–702. doi: 10.1038/nbt1211. [DOI] [PubMed] [Google Scholar]
- Spisni E, Tomasi V. Bicknell R, Lewis CE, Ferrara N. Tumour Angiogenesis. Oxford University Press; Oxford; 1997. Involvement of prostanoid in angiogenesis; pp. pp. 291–300. [Google Scholar]
- Strillacci A, Griffoni C, Spisni E, Manara MC, Tomasi V. RNA interference as a key to knockdown overexpressed cyclooxygenase-2 gene in tumour cells. Br J Cancer. 2006;94:1300–1310. doi: 10.1038/sj.bjc.6603094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansone P, Piazzi G, Paterini P, Strillacci A, Ceccarelli C, Minni F.et al. (2009Cyclooxygenase-2/carbonic anhydrase-IX up-regulation promotes invasive potential and hypoxia survival in colorectal cancer cells J Cell Mol Med 139B3876–3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon SD, McMurray JJ, Pfeffer MA, Wittes J, Fowler R, Finn P.et al.; Adenoma Prevention with Celecoxib (APC) Study Investigators 2005Cardiovascular risk associated with celecoxib in a clinical trial for colorectal adenoma prevention N Engl J Med 3521071–1080. [DOI] [PubMed] [Google Scholar]
- Bresalier RS, Sandler RS, Quan H, Bolognese JA, Oxenius B, Horgan K.et al.; Adenomatous Polyp Prevention on Vioxx (APPROVe) Trial Investigators 2005Cardiovascular events associated with rofecoxib in a colorectal adenoma chemoprevention trial N Engl J Med 3521092–1102. [DOI] [PubMed] [Google Scholar]
- Strillacci A, Griffoni C, Valerii MC, Lazzarini G, Tomasi V, Spisni E. RNAi-based strategies for cyclooxygenase-2 inhibition in cancer. J Biomed Biotechnol. 2010;2010:828045. doi: 10.1155/2010/828045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strillacci A, Griffoni C, Lazzarini G, Valerii MC, Di Molfetta S, Rizzello F.et al. (2010Selective cyclooxygenase-2 silencing mediated by engineered E. coli and RNA interference induces anti-tumour effects in human colon cancer cells Br J Cancer 103975–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer II, Kawka DW, Schloemann S, Tessner T, Riehl T, Stenson WF. Cyclooxygenase 2 is induced in colonic epithelial cells in inflammatory bowel disease. Gastroenterology. 1998;115:297–306. doi: 10.1016/s0016-5085(98)70196-9. [DOI] [PubMed] [Google Scholar]
- Fiocchi C. Inflammatory bowel disease: etiology and pathogenesis. Gastroenterology. 1998;115:182–205. doi: 10.1016/s0016-5085(98)70381-6. [DOI] [PubMed] [Google Scholar]
- Bernstein CN, Blanchard JF, Kliewer E, Wajda A. Cancer risk in patients with inflammatory bowel disease: a population-based study. Cancer. 2001;91:854–862. doi: 10.1002/1097-0142(20010215)91:4<854::aid-cncr1073>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Santiago C, Pagán B, Isidro AA, Appleyard CB. Prolonged chronic inflammation progresses to dysplasia in a novel rat model of colitis-associated colon cancer. Cancer Res. 2007;67:10766–10773. doi: 10.1158/0008-5472.CAN-07-1418. [DOI] [PubMed] [Google Scholar]
- Hisamatsu T, Kanai T, Mikami Y, Yoneno K, Matsuoka K, Hibi T. Immune aspects of the pathogenesis of inflammatory bowel disease. Pharmacol Ther. 2013;137:283–297. doi: 10.1016/j.pharmthera.2012.10.008. [DOI] [PubMed] [Google Scholar]
- Geremia A, Biancheri P, Allan P, Corazza GR, Di Sabatino A. Innate and adaptive immunity in inflammatory bowel disease. Autoimmun Rev. 2014;13:3–10. doi: 10.1016/j.autrev.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Neurath MF. Cytokines in inflammatory bowel disease. Nat Rev Immunol. 2014;14:329–342. doi: 10.1038/nri3661. [DOI] [PubMed] [Google Scholar]
- Nell S, Suerbaum S, Josenhans C. The impact of the microbiota on the pathogenesis of IBD: lessons from mouse infection models. Nat Rev Microbiol. 2010;8:564–577. doi: 10.1038/nrmicro2403. [DOI] [PubMed] [Google Scholar]
- Baek SJ, Kim SH, Lee CK, Roh KH, Keum B, Kim CH.et al. (2014Relationship between the severity of diversion colitis and the composition of colonic bacteria: a prospective study Gut Liver 8170–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Seo SU, Chen GY, Núñez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- Pithadia AB, Jain S. Treatment of inflammatory bowel disease (IBD) Pharmacol Rep. 2011;63:629–642. doi: 10.1016/s1734-1140(11)70575-8. [DOI] [PubMed] [Google Scholar]
- Ocampo SM, Romero C, Aviñó A, Burgueño J, Gassull MA, Bermúdez J.et al. (2012Functionally enhanced siRNA targeting TNFa attenuates DSS-induced colitis and TLR-mediated immunostimulation in mice Mol Ther 20382–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takedatsu H, Mitsuyama K, Mochizuki S, Kobayashi T, Sakurai K, Takeda H.et al. (2012A new therapeutic approach using a schizophyllan-based drug delivery system for inflammatory bowel disease Mol Ther 201234–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steidler L, Hans W, Schotte L, Neirynck S, Obermeier F, Falk W.et al. (2000Treatment of murine colitis by Lactococcus lactis secreting interleukin-10 Science 2891352–1355. [DOI] [PubMed] [Google Scholar]
- Steidler L, Neirynck S, Huyghebaert N, Snoeck V, Vermeire A, Goddeeris B.et al. (2003Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10 Nat Biotechnol 21785–789. [DOI] [PubMed] [Google Scholar]
- Wirtz S, Neufert C, Weigmann B, Neurath MF. Chemically induced mouse models of intestinal inflammation. Nat Protoc. 2007;2:541–546. doi: 10.1038/nprot.2007.41. [DOI] [PubMed] [Google Scholar]
- Araki Y, Bamba T, Mukaisho K, Kanauchi O, Ban H, Bamba S.et al. (2012Dextran sulfate sodium administered orally is depolymerized in the stomach and induces cell cycle arrest plus apoptosis in the colon in early mouse colitis Oncol Rep 281597–1605. [DOI] [PubMed] [Google Scholar]
- Bento AF, Leite DF, Marcon R, Claudino RF, Dutra RC, Cola M.et al. (2012Evaluation of chemical mediators and cellular response during acute and chronic gut inflammatory response induced by dextran sodium sulfate in mice Biochem Pharmacol 841459–1469. [DOI] [PubMed] [Google Scholar]
- Samanta AK, Torok VA, Percy NJ, Abimosleh SM, Howarth GS. Microbial fingerprinting detects unique bacterial communities in the faecal microbiota of rats with experimentally-induced colitis. J Microbiol. 2012;50:218–225. doi: 10.1007/s12275-012-1362-8. [DOI] [PubMed] [Google Scholar]
- Berry D, Schwab C, Milinovich G, Reichert J, Ben Mahfoudh K, Decker T.et al. (2012Phylotype-level 16S rRNA analysis reveals new bacterial indicators of health state in acute murine colitis ISME J 62091–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Fazio L, Cavazza E, Spisni E, Strillacci A, Centanni M, Candela M.et al. (2014Longitudinal analysis of inflammation and microbiota dynamics in a model of mild chronic dextran sulfate sodium-induced colitis in mice World J Gastroenterol 202051–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crofford LJ, Wilder RL, Ristimäki AP, Sano H, Remmers EF, Epps HR.et al. (1994Cyclooxygenase-1 and -2 expression in rheumatoid synovial tissues. Effects of interleukin-1 beta, phorbol ester, and corticosteroids J Clin Invest 931095–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose WA, 2nd, Sakamoto K, Leifer CA. Multifunctional role of dextran sulfate sodium for in vivo modeling of intestinal diseases. BMC Immunol. 2012;13:41. doi: 10.1186/1471-2172-13-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alex P, Zachos NC, Nguyen T, Gonzales L, Chen TE, Conklin LS.et al. (2009Distinct cytokine patterns identified from multiplex profiles of murine DSS and TNBS-induced colitis Inflamm Bowel Dis 15341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedi H, Norbert G. Inhibition of IL-6, TNF-a and cyclooxygenase-2 protein expression by prostglandine E2-induced IL-10 in bone marrow-derivade dendritic cells. Cellular Immun. 2004;228:99–109. doi: 10.1016/j.cellimm.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Rembacken BJ, Snelling AM, Hawkey PM, Chalmers DM, Axon AT. Non-pathogenic Escherichia coli versus mesalazine for the treatment of ulcerative colitis: a randomised trial. Lancet. 1999;354:635–639. doi: 10.1016/s0140-6736(98)06343-0. [DOI] [PubMed] [Google Scholar]
- Kruis W, Fric P, Pokrotnieks J, Lukás M, Fixa B, Kascák M.et al. (2004Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine Gut 531617–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iba Y, Sugimoto Y, Kamei C, Masukawa T. Possible role of mucosal mast cells in the recovery process of colitis induced by dextran sulfate sodium in rats. Int Immunopharmacol. 2003;3:485–491. doi: 10.1016/S1567-5769(02)00299-0. [DOI] [PubMed] [Google Scholar]
- Castiglioni B, Rizzi E, Frosini A, Sivonen K, Rajaniemi P, Rantala A.et al. (2004Development of a universal microarray based on the ligation detection reaction and 16S rrna gene polymorphism to target diversity of cyanobacteria Appl Environ Microbiol 707161–7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candela M, Rampelli S, Turroni S, Severgnini M, Consolandi C, De Bellis G.et al. (2012Unbalance of intestinal microbiota in atopic children BMC Microbiol 1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consolandi C, Severgnini M, Castiglioni B, Bordoni R, Frosini A, Battaglia C.et al. (2006A structured chitosan-based platform for biomolecule attachment to solid surfaces: application to DNA microarray preparation Bioconjug Chem 17371–377. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Murine COX-2 mRNA silencing induced by RNAi.
Schematic representation of the experimental design.
Colon cells express GFP after InvColiNC invasion/transfection.
Numeric value and graphical representation of Dweight percentages calculated for all experimental mice groups during the experiment (from time D7 to D21).
InvColishCOX2-mediated COX-2 silencing.