ABSTRACT
HAMP domains are α-helical coiled coils that often transduce signals from extracytoplasmic sensing domains to cytoplasmic domains. Limited structural information has resulted in hypotheses that specific HAMP helix movement changes downstream enzymatic activity. These hypotheses were tested by mutagenesis and cysteine cross-linking analysis of the PhoQ histidine kinase, essential for resistance to antimicrobial peptides in a variety of enteric pathogens. These results support a mechanistic model in which periplasmic signals which induce an activation state generate a rotational movement accompanied by a tilt in α-helix 1 which activates kinase activity. Biochemical data and a high-confidence model of the PhoQ cytoplasmic domain indicate a possible physical interaction of the HAMP domain with the catalytic domain as necessary for kinase repression. These results support a model of PhoQ activation in which changes in the periplasmic domain lead to conformational movements in the HAMP domain helices which disrupt interaction between the HAMP and the catalytic domains, thus promoting increased kinase activity.
IMPORTANCE
Most studies on the HAMP domain signaling states have been performed with chemoreceptors or the HAMP domain of Af1503. Full-length structures of the HAMP-containing histidine kinases VicK and CpxA or a hybrid between the HAMP domain of Af1503 and the EnvZ histidine kinase agree with the parallel four-helix bundle structure identified in Af1503 and provide snapshots of structural conformations experienced by HAMP domains. We took advantage of the fact that we can easily regulate the activation state of PhoQ histidine kinase to study its HAMP domain in the context of the full-length protein in living cells and provide biochemical evidence for different conformational states experienced by Salmonella enterica serovar Typhimurium PhoQ HAMP domain upon signaling.
INTRODUCTION
Bacteria have developed multiple mechanistic strategies to sense and respond to extracellular environments. The ability to correctly sense different environmental stimuli via extracellular protein components and transduce these signals to intracellular domains is crucial for bacterial survival. Two-component regulatory systems (TCS) comprised of sensor histidine kinases (HK) and phosphorylated transcriptional regulators play a central role in the bacterial response to extracellular cues (1, 2). The Salmonella enterica serovar Typhimurium PhoPQ TCS is essential for virulence for mice and humans and survival within acidified macrophage phagosomes (3, 4). PhoQ is a homodimeric HK that detects changes in the environment via its periplasmic domain. Two transmembrane helices that flank the PhoQ periplasmic domain anchor PhoQ within the inner membrane. PhoQ kinase activity is activated by acidic pH and cationic antimicrobial peptides (CAMPs) (5, 6) and repressed by millimolar concentrations of divalent cations (7). A cytosolic HAMP domain connects the second PhoQ transmembrane domain to the other two cytoplasmic domains: a dimerization domain containing the phospho-histidine residue (DHp) and the catalytic/ATP-binding domain (CA) (8). PhoQ cytoplasmic domains interact with the cognate response regulator of PhoQ, PhoP. Upon sensing a stimulus, PhoQ undergoes autophosphorylation in a conserved histidine residue, and subsequently this phosphate group is transferred to a conserved aspartate residue on PhoP. Once phosphorylated, PhoP controls the expression of a large regulon which includes genes involved in invasion, motility, transport of small molecules, acid tolerance, antimicrobial peptide resistance, and specific outer membrane modifications (4, 9–16). Correct timing of PhoQ activation is essential for virulence, as strains with mutations in PhoQ that result in increased net PhoP phosphorylation are attenuated (17). Therefore, extracellular information must travel from the periplasmic domain to the catalytic domain in an accurate and controlled manner.
HAMP domains are frequently found connecting extracellular and transmembrane domains to cytoplasmic catalytic domains and have been shown to be essential for correct signal transduction (18–20). Changes of only one amino acid in this domain are known to bias the enzymatic output of the protein (18, 19). HAMP domains are often present in transmembrane homodimer signaling proteins such as histidine kinases, adenylyl cyclases, methyl-accepting chemoreceptors, and phosphatases (21–23) as well as diguanylate cyclases and phosphodiesterases (24). HAMP domains are ~50 amino acids long and possess only a few conserved residues. HAMP domain structures are characteristic of coiled-coil domains, consisting of two α-helices, amphipathic sequence 1 (AS1) and AS2 (25), joined by an unstructured connector region. In 2006, Hulko et al. provided the first nuclear magnetic resonance (NMR) structure of a HAMP domain from Af1503, a protein of unknown function from the hyperthermophile Archaeoglobus fulgidus (26). Af1503 revealed a symmetric homodimeric parallel coiled-coil structure with unusual complementary x-da helical packing, which prompted the authors to propose a model in which signal transduction occurs by a 26° concerted rotation of the helices, converting the observed complementary x-da packing into more canonical a-d packing (26). Other models of signal transduction via HAMP domains propose vertical helix displacements coupled to helix rotations, tilts, and repacking of the connector (27), or the existence of the HAMP domain as a dynamic bundle, where either too stable or too loose HAMP conformations give rise to kinase-off phenotypes, and kinase-on phenotypes would be present only in bundles of intermediate stability (25, 28, 29).
In this study, we explored the function and structure of the PhoQ HAMP domain for this receptor that can be regulated in vivo in an off and on state, and these studies support a specific mechanistic model of PhoQ activation through its HAMP domain.
RESULTS
Cysteine cross-linking within the PhoQ HAMP domain supports a model with the same general topology as known structures.
Four models of the PhoQ HAMP domain (residues 217 to 266) were generated to investigate conformational changes that contribute to signaling through this domain. I-TASSER (30, 31) was used to construct models using as templates the structures of Af1503 (2L7H and 2L7I) and Aer2 HAMP1 (3LNR and 4I3M) in two different conformations. The model with the highest confidence score (C score) (31) was obtained when Aer2 HAMP1 (3LNR) was used as the template (Table 1). The β-β′ carbon distance of the residues located at the dimer interface was measured for each of the four models (Table 1). This approach revealed differences between the models that can be tested by a disulfide mapping approach. Furthermore, PhoQ has known activation and repression signals, which allowed us to test the relevance of the generated models using in vivo cysteine cross-linking, under conditions of repression and activation.
TABLE 1 .
β-β carbon distance for each residue calculated for each of the models
| Residuea | Distance (Å) |
|||
|---|---|---|---|---|
| Af1053 | Af1053A291F | Aer2 HAMP1 | Aer2 HAMP1L44H | |
| I221 | 8.4 | 11 | 6 | 6.4 |
| L224* | 10.2 | 11.3 | 8 | 10.9 |
| A225* | 13.3 | 15.1 | 13.2 | 11.3 |
| V228 | 8.8 | 9.3 | 8.3 | 8.3 |
| L247 | 11.6 | 10.7 | 12.5 | 15.6 |
| L250 | 7.1 | 6.5 | 7.9 | 10.6 |
| L254* | 8.9 | 10.1 | 10.6 | 11.1 |
| L257* | 4.6 | 7.5 | 6.9 | 5.9 |
| S260* | 9 | 13.3 | 11.6 | 9.9 |
| C score | 0.22 | 0.3 | 0.41 | 0.35 |
Asterisks denote residues for which statistically significant changes were observed by disulfide cross-link assay upon PhoQ activation.
We performed a symmetric disulfide mapping study (32) and used the redox catalyst Cu(II) (1,10-phenanthroline) to initiate the oxidative reaction (33). For this purpose, the two native cysteine residues at positions 392 and 395 were replaced by serines, so that only the engineered single cysteines, introduced into the PhoQ HAMP domain dimer interface, could serve as reporters and result in intermolecular cross-links. This cysteineless PhoQ protein was then engineered with single cysteine substitutions along the predicted HAMP domain dimer interface in residues I221, L224, A225, V228, L231, E232, L247, L250, N253, L254, L257, and S260. The resultant PhoQ homodimers possess two cysteine residues at symmetric positions. The different PhoQ cysteine mutants were expressed in the ΔphoQ::tetR phoN::phoA Salmonella Typhimurium reporter strain (MB101) (5) and tested for the appropriate response to an activating signal to determine which of these strains could be used to assess conformational changes associated with signaling (Fig. 1A). Similar levels of PhoQ protein were observed for all cysteine mutants, as determined by Western blotting (data not shown). In addition, the circular dichroism spectrum of the purified PhoQ HAMP domain is typical of an α-helical fold (see Fig. S1 in the supplemental material), consistent with the likelihood that the structure of the PhoQ HAMP domain is predominantly an α-helical coiled coil and that the models could reflect a conformation state experienced by the PhoQ HAMP domain. While most cysteine substitutions resulted in slightly reduced activity but a normal response to the activating signal, L231C and E232C mutations resulted in constitutive phenotypes and N253C produced an inactive PhoQ protein (Fig. 1A) and therefore could not be analyzed in a state of activation and repression. The antimicrobial peptide C18G was chosen as the signal for PhoQ activation, since other relevant medium conditions which can be used to activate and repress PhoQ, including acidic pH and different concentrations of divalent cations, may interfere with the disulfide cross-link reaction catalyzed by Cu(II) (1,10-phenanthroline). Aliquots from the cultures grown in the presence or absence of the activating signal were assayed in parallel for PhoQ-activated gene alkaline phosphatase reporter activity and for the disulfide cross-link state of PhoQ.
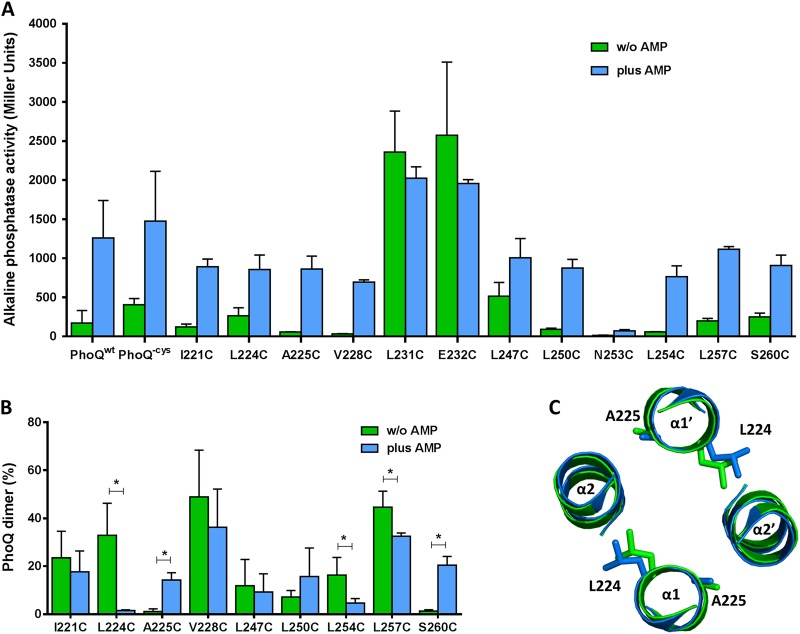
FIG 1 .
Disulfide cross-linking shows PhoQ HAMP domain present in distinct packing conformations in the presence and absence of the activating signal, antimicrobial peptide C18G (AMP). (A) Alkaline phosphatase activity of the PhoQ HAMP cysteine mutants. Cultures were grown in N minimal medium supplemented with 1 mM MgCl2 in the presence (blue) and absence (green) of the antimicrobial peptide C18G. All graphed values are the means and standard deviations and are representative of at least three independent trials. (B) Percent PhoQ dimer was calculated and plotted for each PhoQ cysteine mutant in the absence and in the presence of the activating signal, C18G. Graphed values are the means and standard deviations and are representative of at least three independent trials. A statistical t test was performed (*, P ≤ 0.05). (C) Cross section of the superposition of the PhoQ HAMP models based on Aer2 HAMP1 (green) and Aer2 HAMP1L44H (blue).
As expected from residues located at the dimer interface, and consistent with the models generated, each one of the PhoQ residues tested formed disulfide cross-links under at least one of the conditions assayed (Fig. 1B). Three cysteines were also engineered in residues in which the models estimated β-β′ carbon distances of >20 Å and tested for the ability to form cross-links (R226, D233, and R252). No disulfide formation was observed for these three PhoQ mutant proteins despite normal protein amounts and activity (data not shown). Thus, consistent with the model generated, the PhoQ HAMP domain structure is likely to be similar to that of Af1503 and Aer2 HAMP1.
PhoQ cysteine cross-linking supports a model in which rotation and helical tilt movements are involved in PhoQ signal transduction through the HAMP domain.
Analysis of the disulfide cross-linking experimental data shows that only the residues at positions 224, 225, 254, 257, and 260 present statistically significant changes in cross-linking levels when assayed under conditions of activation (Fig. 1B, blue) or repression (Fig. 1B, green). The comparison of the β-β′ carbon distances for these residues between the two PhoQ HAMP models shows that the model variants based on Aer2 HAMP1 are most consistent with the disulfide cross-linking differences observed (Table 1; Fig. 1B). There is a clear opposite cross-link pattern between the adjacent residues at positions 224 and 225 in α-helix 1 and also between the residues at positions 254 and 260 in α-helix 2 in the presence or absence of an activating signal (Fig. 1B). Although this result is indicative of a rotational movement similar to the one proposed by Hulko et al. (26), the β-β′ carbon distances of the two models based on Af1503 do not support this simple explanation (Table 1). The increase in disulfide formation for PhoQA225C and PhoQS260C in the presence of C18G is suggestive of a transition from an a-d conformation (Af1503A291F model) to a complementary x-da conformation (Af1503 model) where the flanking residues at e (in α1) or g (in α2) positions on an a-d conformation are now part of the four helix bundle packing core at a (in α1) or d (in α2) positions, respectively (Fig. 1B; also, see Fig. S2 in the supplemental material). However, when the β-β′ carbon distances between these two PhoQ models (Af1503 and Af1503A291F) are compared, only the results for residues A225 and S260 are consistent with such a transition (Table 1). Therefore, a purely rotational movement of the PhoQ HAMP is unlikely to explain the cysteine cross-link results. However, the Aer2 HAMP1-based models that present both rotational and tilting motions are consistent with activation-associated changes in cross-linking. A PhoQ HAMP structure similar to the native Aer2 HAMP1 appears to represent the repressed state (Fig. 1C, green), while the Aer2 HAMP1L44H represents the PhoQ HAMP domain activated state (Fig. 1C, blue) (Table 1).
Comparison of the two PhoQ HAMP models based on Aer2 HAMP1 predicts that most regulation-associated changes occur at the top of α-helices 1 and 2. There is a 1.3-Å shift at the top of α1 that is predicted as a result of a 3° tilt in α1. In addition, there is loss of helical structure at the top of α2 in the PhoQ HAMP domain Aer2 HAMP1L44H-based model (see Fig. S3 in the supplemental material). These changes are similar to but much more modest than those observed for the native Aer2 HAMP1 and Aer2 HAMP1L44H (34). Though these models are not supported by an actual in vitro static structure of PhoQ, these in vivo results are most consistent with both helical rotation and tilting movements participating in the transition to the activation state of the PhoQ HAMP domain.
Highly activating mutations are found at the membrane-distal portion of PhoQ HAMP domain.
The remarkably active and constitutive phenotypes of PhoQL231C and PhoQE232C indicated that amino acid substitutions in the PhoQ HAMP domain can be isolated which result in PhoQ mutant proteins that spend more time in the activation state than the wild-type protein. Therefore, we carried out a random mutagenesis screen of the PhoQ HAMP domain (W215 to N266) to identify activated mutants in the presence of a repressing signal to further our understanding of the changes that occur in the HAMP domain associated with PhoQ activation. The ΔphoQ S. Typhimurium reporter strain (MB101) (5) was transformed with the phoQ mutant pool and plated on LB agar containing 10 mM MgCl2 (repressive conditions) and XP (5-bromo-4-chloro-3-indolyl phosphate dipotassium salt; colorimetric indication of PhoA activity). After 3 rounds of independent random mutagenesis experiments, 48 single mutants were identified which showed activity above wild-type levels on reporter LB agar plates and contained a single mutation on the PhoQ HAMP domain (Fig. 2A). Six PhoQ HAMP mutants were considered to have levels of PhoA activity similar to that of the wild type (<2-fold over wild-type activity when grown in N minimal medium supplemented with 10 mM MgCl2) (see Table S1 in the supplemental material). From the remaining 42 single mutants identified, 26 showed more than 5-fold activity and 16 had a >10-fold increase in reporter activity compared to the wild type (Fig. 2A and B; also, see Table S1 in the supplemental material). Alkaline phosphatase activity was measured under PhoQ activating and repressive conditions for all the mutants. Several of these mutants could not be further derepressed by growth in low-metal-containing medium (PhoQE232K, PhoQE232V, PhoQR236H, PhoQR236S, PhoQL254F, PhoQN255T and PhoQN255I) (see Fig. S4 in the supplemental material). PhoQE232K is the most derepressed allele, showing a >35-fold increase in activity over the wild type (see Table S1 in the supplemental material) under repressive conditions. PhoQT48I, the classical PhoP-constitutive allele (35), which is a periplasmic allele that in part simulates signal transduction (4, 36), was included as a reference, indicating by comparison the remarkable activation imparted from a change in residue 232 from glutamate to lysine (Fig. 2B; also, see Fig. S4 in the supplemental material).
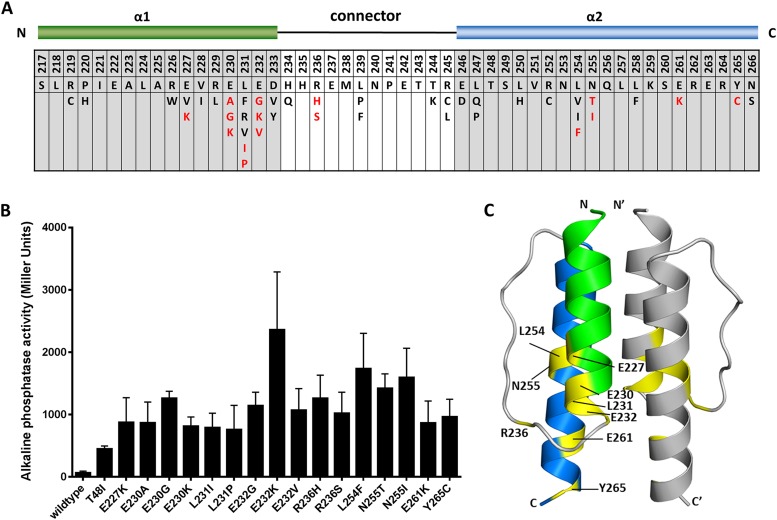
FIG 2 .
Amino acid changes in PhoQ HAMP domain that confer increased activity under repressive conditions. (A) The schematic at the top is a linear representation of PhoQ HAMP domain. α1 (green) and α2 (blue) represent the two α-helices of PhoQ HAMP domain joined by a connector region. Residue numbers relative to full-length PhoQ are indicated in the first row of the chart. The second row shows the PhoQ HAMP domain wild-type sequence. Presented in the bottom row are the amino acid substitutions identified for each position. In red are the amino acid mutations that result in a highly derepressed PhoQ protein. (B) Alkaline phosphatase activity of PhoQ HAMP mutants that show more than 10-fold activity over the wild type under repressive conditions. A reporter fusion between PhoP-dependent acid phosphatase (PhoN) and PhoA was used to measure activation. Cultures were grown in N minimal medium supplemented with 10 mM MgCl2. All graphed values are the means and standard deviations and are representative of at least three independent trials. (C) Model of the PhoQ HAMP domain based on Aer2 HAMP1. One monomer is shown in color with α1 in green and α2 in blue; the other monomer is grey. Residues where highly activating mutations were identified are in yellow, and their residue numbers are shown.
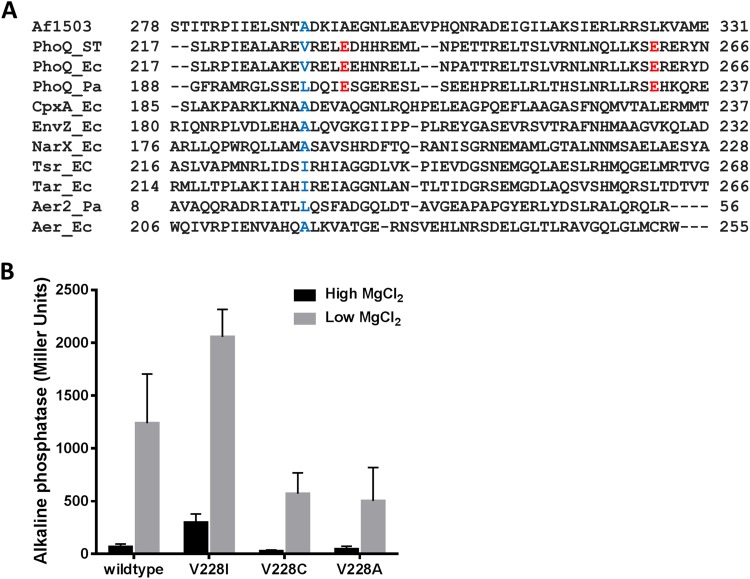
Interestingly, when mapped onto the model of PhoQ HAMP domain, the highly activating mutations (10-fold above wild-type levels) are all found in residues located in the membrane-distal half of the PhoQ HAMP domain (Fig. 2C). Two important residue networks can be distinguished. The first one is around residue V228 (E227, L254, and N255), and the second one comprises residues E230, L231, E232, R236, and E261. The size of the side chain of the residue equivalent to V228 in Af1503 (A291), Tar (I227), Tsr (I229), and EnvZ (A193) has been shown to be an important determinant for bundle conformation and therefore signal outcome (22, 26, 37, 38). In Af1503, amino acid substitutions at alanine 291 that sequentially increase the size of the side chain result in the progressive decrease of activity of chimeric proteins through the proposed transition from a complementary x-da helical packing of the HAMP domain to a conformation closer to the canonical knobs-into-holes packing (26, 39, 40). Residues with small side chains at this position are conserved in some HAMP domains (26) (Fig. 3A). In S. Typhimurium PhoQ, this position is occupied by valine 228, and the substitution of isoleucine for this residue results in increased PhoQ activity (Fig. 2A and 3B), whereas substitution by residues with smaller side chains such as cysteine or alanine results in decreased activity (Fig. 3B). The same is observed for other residues in the same packing layer, such as the L254F mutation, which results in a highly derepressed protein, whereas N253C is locked in an “off” conformation (Fig. 1A and 2B). Therefore, a correlation between the size of the side chain and activity at position 228 is also true for PhoQ, and in the case of PhoQ, a direct relationship is observed between the size of the side chain and PhoQ activity (Fig. 3B). This has also been observed for the HAMP domain of the EnvZ histidine kinase where substitution of the correspondent residue, alanine 193, by valine or leucine in the chimeric protein Tez1A1 resulted in a constitutive phenotype (37).
FIG 3 .
Correlation between the size of the side chain at position 228 and activity in PhoQ. (A) ClustalO primary sequence alignment of the HAMP domains from Af1503 (GenBank no. O28769), PhoQ (P23837, D0ZV89, and Q9I4F8), CpxA (P0AE82), EnvZ (P0AEJ4), NarX (P0AFA2), Tsr (P02942), Tar (P07017), Aer2 (Q9I6V6), and Aer (P50466). ST, S. Typhimurium; Ec, E. coli; Pa, P. aeruginosa. The residues represented in blue are at the equivalent position as A291 in Af1503. In red are the two glutamate residues conserved in PhoQ. (B) Alkaline phosphatase activity of PhoQ mutants with amino acid substitutions at position 228. Cultures were grown in N minimal medium supplemented with 10 mM MgCl2 (high) or 10 µM MgCl2 (low). All values are means and standard deviations and are representative of at least three independent trials.
Of the 16 highly active mutants, 11 have amino acid substitutions in residues E230, L231, E232 (α1), R236 (connector), and E261 (α2) localized at the membrane-distal half of the PhoQ HAMP domain, emphasizing the importance of this region in maintaining PhoQ in a repressed state (Fig. 2A and C). Charge changes or potential helix-breaking mutations at these positions result in higher PhoQ activity levels. R236 occupies a position in the connector known to be important for bundle stability (25, 20). Purification and circular dichroism analysis of the wild-type HAMP domain and two HAMP mutants (HAMPE232K and HAMPR236H) revealed an ellipticity pattern typical of an alpha-helical fold with the characteristic minima of 208 nm and 222 nm (see Fig. S1 in the supplemental material). HAMPR236H exhibits loss of ellipticity, which may be caused by a less helical structure. Therefore, we can assume that, at least for the HAMP mutants tested, the PhoQ HAMP domain retains an alpha-helical fold similar to that of the wild type and that activation in some cases, such as in PhoQR236H, could result through partial loss of helical structure, possibly like the one suggested by the PhoQ HAMP domain Aer2 HAMP1L44H-based model (see Fig. S3 in the supplemental material).
The PhoQ repressed state could be stabilized by an interaction between the membrane-distal half of the HAMP domain and the loop connecting the dimerization domain to the catalytic domain.
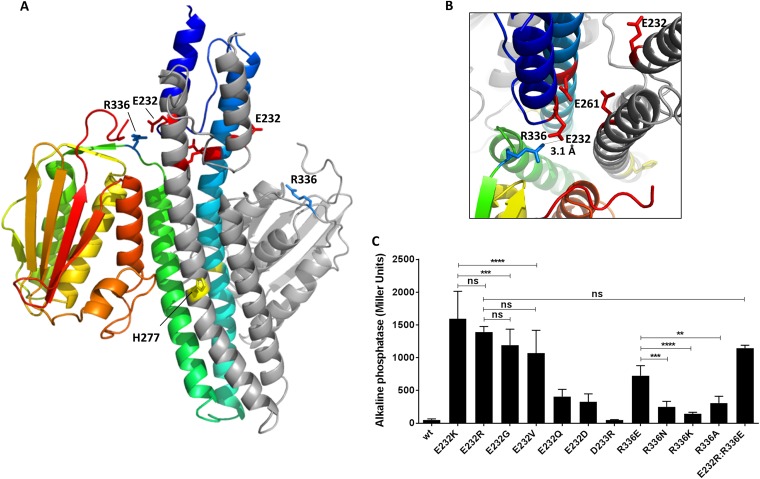
Direct interaction between the HAMP and other protein domains have been proposed to be involved in modulating signal response in other proteins, such as Aer (41), DhNIK1 (42), and CetA (43). The recent structure of the full cytoplasmic domain of the HK CpxA showed its HAMP domain in close proximity to its CA domain in the repressed state (44). Structural evidence has identified several interacting residues between the DHp and the CA domain in the HKs EnvZ and CpxA (39, 44), which are close homologs of PhoQ and share the same domain organization. These interactions are believed to be important in maintaining the HK in a kinase-off state by sequestering the CA away from the phosphorylated histidine (39, 44). Most of the highly activating mutations in PhoQ were found at the membrane-distal half of the PhoQ HAMP domain, suggesting that a possible mechanism of PhoQ regulation could occur through the direct interaction between the HAMP and the CA domains of PhoQ. Therefore, a model of the full cytoplasmic domain of PhoQ (residues 217 to 487) was generated using I-TASSER (30, 31) using the repressed (4BIU:A) and activated (4BIU:B) CpxA chains as templates to identify residues that could be involved in such an interaction (Fig. 4A). The model generated presented high confidence, quality, and similarity scores: C scores of 1.34 (repressed) and 1.04 (activated) and TM scores of 0.90 (repressed) and 0.86 (activated). Interestingly, the PhoQ model obtained indicates that a salt bridge between residues E232 and R336 is possible (Fig. 4B). We hypothesized that this interaction could help stabilize the repressed state and that the loss of such an interaction could represent the mechanism behind the high levels of activity observed for PhoQE232K. A primary sequence alignment revealed that these two residues are very conserved in most PhoQ homologs (see Fig. S5 in the supplemental material), even in the PhoQ protein of Pseudomonas aeruginosa, which shares only 34% identity at the protein level with S. Typhimurium PhoQ and lacks the relevant periplasmic helices that allow it to function as an antimicrobial peptide receptor (5, 45). A series of targeted mutations were generated, and activity was assayed under repressive conditions to provide support for the idea that R336 could interact with E232. First, the charge was reversed for the similarly charged aspartate at residue 233, generating PhoQD233R. The phenotype of PhoQE232K/R versus PhoQD233R confirms the specificity of the phenotype observed, as PhoQD233R behaved like PhoQwt, indicating that maximal PhoQ derepression is specific to changes at position 232. As hypothesized, the R336E mutation results in PhoQ derepression, but not to the same extent as in PhoQE232K/R. Other amino acid changes at position 336 also result in partial derepression but not to the same level as the replacement of the arginine with the oppositely charged glutamate. For both positions (232 and 336), the charge and the size of the side chain appear to be important factors to keep PhoQ in a repressed state, since all the PhoQ mutants tested result in a partially derepressed protein (Fig. 4C). Therefore, the results identified the glutamate and arginine at positions 232 and 336, respectively, as essential for stabilization of the repressed state of PhoQ, possibly through the salt bridge predicted in the PhoQ cytoplasmic domain structural model (Fig. 4B). However, if these two domains do interact as part of the PhoQ repressed state, the interactions between these domains are likely to be more complicated than a simple salt bond between these residues, as reversing the residues at positions 232 and 336 (PhoQE232R:R336E [Fig. 4C] and PhoQE232K:R336E [data not shown]) still resulted in significant PhoQ activity.
FIG 4 .
Model of PhoQ full cytoplasmic domain showing a possible interaction between the bottom of the PhoQ HAMP domain and a loop connecting the dimerization domain to the catalytic domain. (A) I-TASSER model of PhoQ (residues 217 to 487) using as a model the repressed (color) and activated (grey) CpxA chains as the template. Residues E232 (red), E261 (red), H277 (yellow), and R336 (blue) are represented as sticks. (B) Cross section of the model showing a possible salt bridge between residues E232 (red) and R336 (blue), shown as sticks. (C) Alkaline phosphatase activity of PhoQ targeted mutants grown in N minimal medium under repressive conditions (10 mM MgCl2). One-way analysis of variance (ANOVA) was performed. ns, not significant; **, P ≤ 0.005; ***, P ≤ 0.001; ****, P ≤ 0.0001.
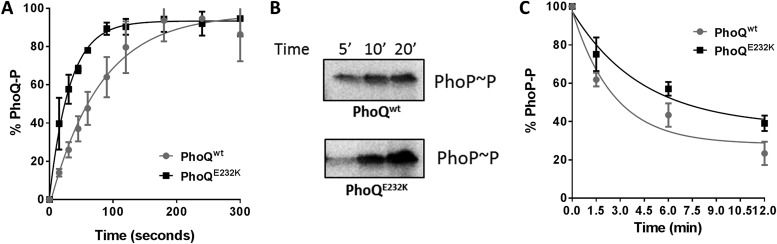
PhoQE232K has faster autophosphorylation activity.
The high levels of PhoQ-activated gene expression under PhoQE232K-repressive conditions suggested that its enzymatic activities were dramatically altered compared to those of the wild-type protein. Higher levels of the PhoA reporter activity can be the result from an increase in either the PhoQ autokinase activity or its phosphotransfer activity or alternatively can be the product of a decrease in the PhoQ phosphatase activity. A shift in the enzymatic activity of PhoQ toward any of these states can potentially create a bigger pool of phosphorylated PhoP in the cell and a consequent increase in PhoA reporter activity. For example, in PhoQT48I (the classical constitutive PhoQ mutant), the constitutive phenotype was shown to be the result of an increase in the net phosphorylation of PhoP (46), which was later attributed solely to a decrease in the phosphatase activity of PhoQT48I (4, 36). If PhoQE232K does indeed result in the release of the catalytic domain and correct positioning of H277 to adopt a kinase-competent conformation, then it should have increased kinase activity. In order to measure the different enzymatic activities of PhoQ, membranes from S. Typhimurium PhoQwt- and PhoQE232K-expressing strains were isolated as described previously (47) and used in autophosphorylation, phosphotransfer, and phosphatase assays. As shown in Fig. 5A, PhoQE232K has faster autophosphorylation activity than PhoQwt, suggesting that indeed PhoQE232K spends more time in a kinase-on state. Phosphotransfer and phosphatase activities were also compared between PhoQwt and PhoQE232K (Fig. 5B and C) using S. Typhimurium PhoQ-enriched membranes and purified PhoP (36). Interestingly, PhoP phosphorylation occurs faster in PhoQwt-containing reaction mixtures than in those containing PhoQE232K, since they exhibit higher levels of PhoP phosphorylated at 5 min (Fig. 5B). However, phosphorylated PhoP accumulates to higher levels in PhoQE232K-containing samples at later time points (Fig. 5B), most probably due to the decrease in the phosphatase activity of this protein (Fig. 5C). Also, if PhoQE232K is more often in a kinase-on state, the interaction site with PhoP may not be as accessible, which could, as observed, result in decreased phosphotransfer at earlier time points and decreased phosphatase activity. Therefore, it seems to be the combined effect of increased autophosphorylation and decreased phosphatase activity in PhoQE232K that leads to a bigger pool of phosphorylated PhoP in the cell. This is most probably due to the destabilization of the PhoQ kinase-off state initiated by conformational changes in the HAMP domain of PhoQE232K propagated to downstream domains. In PhoQ, these changes could result through the disruption of interactions between the CA and DHp and also possibly the HAMP domain, such as the putative interaction between E232 and R336 proposed here.
FIG 5 .
Kinetic activities of PhoQwt and PhoQE232K. (A) PhoQ autophosphorylation. S. Typhimurium PhoQ-enriched membranes were incubated with [γ-33P]ATP. Reactions were stopped at the indicated time points, and the products were separated by SDS-PAGE and exposed overnight to a phosphorimager screen. The fraction of phosphorylated PhoQ was determined for each time point. The data were analyzed using the nonlinear regression model “one phase decay.” (B) Phosphotransfer activity. S. Typhimurium PhoQ-enriched membranes were incubated with purified PhoP in the presence of [γ-33P]ATP. Reactions were stopped at different time points, and the products were separated by SDS-PAGE and exposed overnight to a phosphorimager screen. (C) Dephosphorylation of phosphorylated PhoP (PhoP~P). Purified PhoP~P was incubated with membranes containing overexpressed PhoQwt or PhoQE232K. Reactions were stopped at different time points, and the products were separated by SDS-PAGE and exposed overnight to a phosphorimager screen. The data were analyzed using the nonlinear regression model “one phase decay.” All values are means and standard deviations and are representative of at least three independent trials.
DISCUSSION
In this study, we performed a functional and structural analysis of the S. Typhimurium PhoQ HAMP domain in order to understand the conformational changes associated with PhoQ activation. Our work demonstrates, through cysteine cross-linking experiments, that the HAMP domain of PhoQ can adopt different conformations under repressive or activating conditions. Comparison of the cysteine cross-link data with two different models for HAMP signaling shows that rotation accompanied by a tilting movement of alpha-helix 1 could be involved in PhoQ HAMP domain conformational changes that lead to activation. Random mutagenesis identified activating mutations in the PhoQ HAMP domain. These mutations were integrated with a structural model of the PhoQ HAMP domain which revealed that most of them were located at the membrane-distal half of the PhoQ HAMP domain. These data led to the hypothesis that a charged region in the HAMP domain could directly interact with the PhoQ CA domain to keep it in a state of repression. Specific conformational changes identified in the HAMP domain would then disturb this interaction, thus promoting kinase activity. This hypothesis was tested through directed mutational analysis of these residues and biochemical characterization of the purified mutants. Here, we combine a variety of biochemical and structural results to provide a working model of the events of signal transduction propagated from a periplasmic receptor through the membrane and HAMP domain to the dimerization and kinase domains in the PhoQ HK.
Several mechanisms have been proposed for signal transduction via HAMP domains (26–29, 48). The structures of HAMP domains from different proteins have shown that it can be present in different packing states either in a complementary x-da packing conformation (26, 27, 39) or in a more canonical a-d packing conformation (27, 44, 49), and signaling could result from the interconversion between these two states. The HAMP structure with its multiple equilibrium or dynamic states, as proposed in the dynamic-bundle model (28, 29), could lend itself to the evolution of different conformational changes as a mechanism of signal transduction. Translational movements, helix rotations, tilts, and repacking of the connector have been proposed; some mutational and structural data support these ideas for signal transduction through HAMP domains, and it is plausible that this simple domain could lend itself to many different solutions of signal transduction (27, 40, 50).
Cysteine cross-linking and disulfide mapping studies have been used previously to successfully probe the structure and conformational changes of the aspartate chemoreceptor, Tar (50), and the aerotaxis receptor, Aer, HAMP domains (51). The rate of disulfide formation which typically increases with residue proximity is dependent on the frequency of collision of the sulfhydryls groups as well as on the orientation of the cysteine side chains and their accessibility to the oxidizing reagent (33, 52). Therefore, comparison of the β-β carbon distance between two residues in a model, and the disulfide reactivity of engineered cysteines at those positions can be used to estimate the accuracy of in silico models. We capitalized on the ability to regulate PhoQ activation to document cysteine cross-linking changes, in particular residues in the context of the full-length PhoQ protein assayed in vivo in response to physiological signals. These changes are consistent with a rotational movement accompanied by a tilt in α1 and loss of helical structure at the top of α2 (Fig. 1; also, see Fig. S3 in the supplemental material). Large diagonal displacements in PhoQ transmembrane domains propagated to the HAMP and DHp domains have recently been proposed to be involved in signal transduction in this protein as well as for other HKs (53). Interestingly, similar movements to those identified in this study have been proposed to be involved in signal transduction in Aer (54). Cysteine cross-linking studies suggest that in this protein, upon signaling there is a tilting motion that brings the bottom of the 4-helix bundle closer together and the top further apart. This movement is accompanied by rotation of α2 and a proposed helical discontinuity at the top of α2 (51, 54, 55). The discontinuity in α2 of Aer is thought to be the result of direct lateral interactions with its PAS domain (41, 54). In PhoQ, a discontinuity in α2 is also observed in one of the models and supported by the lower-than-expected disulfide reactivity for the residue at position 250 (Fig. 1B) and the loss of ellipticity observed for HAMPR236H (see Fig. S1 in the supplemental material). Residue 250 is expected to occupy a position similar to those of residues 221, 228, and 257, and therefore, in a continuous α-helix it should exhibit similar disulfide reactivity. Residue V228 in PhoQ occupies a position that in Af1503 serves as a knob in a socket at the dimer interface and has been identified in this as well as in other HAMP domains as important for signal transduction (22, 26, 38, 50, 29). In PhoQ, amino acid substitutions that result in increased side chain volume at this position give rise to derepressed proteins, and the opposite is also true: smaller side chain volume at position 228 results in decreased PhoQ activity. Interestingly, a similar pattern is observed for the other residues localized in the same packing layer as V228 (N253 and L254). The N253C mutant is locked in an “off” conformation, whereas the L254F mutant is a constitutively active PhoQ mutant. It is noteworthy that mutation of the neighboring residues E227 and N255 also resulted in highly derepressed PhoQ.
As for the Aer and Tar HAMP domains (50, 51), most residue substitutions that confer a constitutively active PhoQ phenotype are localized at the membrane-distal half of the PhoQ HAMP domain (Fig. 2C). However, in PhoQ, two negatively charged residues appear to be uniquely conserved at key positions 232 and 261. In the Tar HAMP domain, residue A231, equivalent to position 232 in PhoQ, has also been identified as important for signal transduction by stabilizing the repressed state (50). Position 261 occupies a stutter position in what is defined as the S-helix (Fig. 3A), which is thought to prevent constitutive activation of adjacent output domains (56, 57). Although also important for signaling in other HAMP domains, these positions are usually not charged (Fig. 3A), suggesting that the presence of negatively charged residues at these positions in PhoQ is important for function. Substitution of glutamate 232 by lysine resulted in the highest increase in activity over the wild type under repressive conditions. In vitro assays showed that the increase in activity in PhoQE232K was due in part to faster autokinase activity. This result is in agreement with a structural model of the PhoQ cytoplasmic domain which shows that the negatively charged distal portion of the HAMP may help to stabilize the kinase-off state through a direct interaction with the downstream domains. Therefore, PhoQwt is most likely in a phosphatase-dominant conformation when in the absence of a signal, which is consistent with observations that higher-than-physiologic levels of expression of PhoQ inhibit PhoP-activated gene expression (S. I. Miller, unpublished observations). The glutamate at position 232 and arginine 336 appear to be optimal in keeping PhoQ in the repressed state through a possible salt bridge between these two residues. Only PhoQE232Q and PhoQE232D that can still potentially form an interaction with R336 show lower levels of derepression. However, reversal of the charge at both positions did not result in a wild-type phenotype, suggesting that the conformational changes initiated in the HAMP domain of PhoQ HAMPE232K/R are not favorable for interaction with residue 336 and may be sufficient to optimally position H277 for the phosphorylation reaction. This idea was supported with biochemical data, indicating that increased kinase activity significantly contributed to the PhoQE232K phenotype.
All together, our results captured an important transition between the two helical packing states of the PhoQ HAMP domain. However, both the amplitude of the movement or the frequency of transition from one state to another could differ between signals. It is plausible that the low-pH activating signal could result in a different conformational change of the HAMP domain than the signal tested in this work, since these signals are known to be additive (6), and the change of the cytoplasmic pH could alter the HAMP domain structure. Premature or inappropriate activation of PhoQ has a high fitness cost for the bacteria, since PhoQ is a master regulator of over 200 genes. In addition, constitutive PhoQ mutants are avirulent (17), underscoring the importance for S. Typhimurium to keep PhoQ in a repressed state until its activation is necessary for survival. In PhoQ, the HAMP domain may not only modulate but also tightly control its activity via a possible interaction between the HAMP and downstream domains.
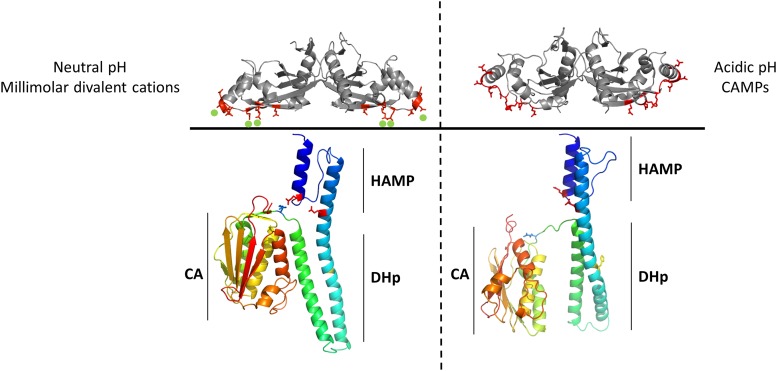
These results lead to an integrated model of PhoQ kinase activation. In the absence of an activating signal, the PhoQ periplasmic domain is tethered to the membrane via divalent cation bridges (58), and the cytoplasmic catalytic domain is in a repressed state with dominant phosphatase activity. The repressed state is stabilized by an interaction between residues in the HAMP domain and in the loop that connects the DHp to the CA domain. Upon sensing of an activating signal, a change in conformation in the PhoQ periplasmic PAS domain occurs which is propagated through the transmembrane region, resulting in α-helical rotation and tilting of α-helix 1 in the PhoQ HAMP domain, disrupting the interaction between residues in the HAMP, DHp, and CA domains and freeing the catalytic domain to adopt an autophosphorylation-competent conformation, shifting the enzymatic activity of PhoQ to predominantly kinase activity and leading to activation of the PhoPQ regulon (Fig. 6).
FIG 6 .
Model of PhoQ activation. In the absence of an activating signal (left), the PhoQ periplasmic domain is tethered to the membrane via divalent cation bridges between the acidic patch of the PhoQ periplasmic domain (red sticks) and the negatively charged phospholipid heads via the presence of metal ions (green spheres), and the cytoplasmic CA domain is in a repressed state with dominant phosphatase activity. Interactions between the CA and the DHp domains as well as between the HAMP domain (present in an a-d conformation) and a loop connecting the CA to the DHp help stabilize the repressed state. Upon sensing of an activating signal (right), a change in conformation in the PhoQ periplasmic domain occurs which is propagated through the transmembrane region, resulting in loosening and rotation of the PhoQ HAMP domain, possibly to a conformation closer to a complementary x-da packing. The interactions between the HAMP, DHp, and CA domains that kept PhoQ in an inactive state are disrupted, freeing the CA to adopt a kinase-competent conformation.
MATERIALS AND METHODS
Bacterial growth conditions.
Strains were grown in LB or N minimal medium supplemented with 100 µg·ml−1 ampicillin or 30 µg·ml−1 chloramphenicol where appropriate and various concentrations of MgCl2.
phoQ HAMP domain random mutagenesis screen and site-directed mutagenesis.
A GeneMorph II random mutagenesis kit (Stratagene) was used to generate a pool of randomly mutagenized pMB106 (pBAD24-phoQ) via PCR mutagenesis with primers that specifically target the PhoQ HAMP domain. S. Typhimurium MB101 was transformed with the pool of mutagenized plasmids and plated on LB containing the chromogenic alkaline phosphatase substrate XP (5-bromo-4-chloro-3-indolyl phosphate dipotassium salt), ampicillin, and 10 mM MgCl2. S. Typhimurium colonies containing plasmids carrying derepressed phoQ alleles appear blue on plates. Dark blue colonies were chosen, and their plasmids were sequenced to identify mutations on the phoQ gene. Site-directed mutagenesis was performed using the QuikChange protocol and primer design guidelines from Stratagene.
Alkaline phosphatase assay.
PhoP activation by the different PhoQ alleles was examined using the alkaline phosphatase reporter phoN::TnphoA as previously described (45). MB101 was freshly transformed before each assay with plasmids carrying the various phoQ alleles; three independent clones were chosen and assayed within 2 days. Activity assays were performed in triplicate in at least three independent trials.
Protein purification.
The phoQ HAMP domain of wild-type, E232K, and R236H sequences was amplified by PCR and cloned into the BamHI and XhoI restriction sites of pGEX-4T-2 (GE Healthcare) as a C-terminal fusion protein to the glutathione S-transferase protein (GST). The expression plasmids containing the three different alleles were transformed into BL21 Rosetta(DE3) competent cells (Novagen). Cells were grown overnight in LB containing the appropriate antibiotics diluted 1:100 in the same medium and cultured at 37°C to an optical density at 600 nm (OD600) of ~0.6 to 0.8, induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and cultured overnight at 22°C. Next day cells were harvested, resuspended in 25 mM NaPO4 (pH 7.2 for HAMPwt and HAMPR236H or pH 6.8 for HAMPE232K) and 150 mM NaCl, and lysed using a homogenizer (EmulsiFlex-C3; Avestin). The lysate was then centrifuged at 10,000 × g for 30 min at 4°C and the supernatant loaded onto a 5-ml GSTrap FF column (GE Healthcare). After extensive washing, the GST tag was cleaved by incubation with thrombin (Sigma) overnight at room temperature. The PhoQ HAMP domain was eluted in the same buffer as above plus antiproteases, concentrated, and loaded onto a Superdex 200 16/60 (GE Healthcare) size exclusion column. Collected fractions were visualized on a Mini-PROTEAN TGX Any kD precast gel (Bio-Rad), and clean fractions containing PhoQ HAMP domain were pooled and subsequently analyzed.
PhoP was purified as previously described (36) with the following modifications. Cultures of BL21(DE3)pLysE pET-PhoPHis were grown at 37°C to an OD600 of ~0.8 and induced for 3 h at 22°C with 0.5 mM IPTG. Cells were harvested, resuspended in 50 mM HEPES (pH 7.0), and disrupted using a homogenizer (EmulsiFlex-C3; Avestin). Following centrifugation at 10,000 × g for 30 min at 4°C, the supernatant was loaded onto a 1-ml HiTrap heparin HP column (GE Healthcare). After extensive washing in the same buffer, PhoP was eluted with a 0 to 2 M NaCl gradient in 50 mM HEPES (pH 7.0) buffer. Pooled PhoP-containing fractions were dialyzed against 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, and 50 mM imidazole and subsequently loaded onto a 5-ml HiTrap chelating HP column (GE Healthcare) previously charged with NiSO4. After extensive wash in the same buffer, PhoP was eluted using a 0 to 1 M imidazole gradient in 50 mM Tris-HCl (pH 7.5) and 50 mM NaCl. Fractions containing PhoP were pooled, dialyzed overnight at 4°C against 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, and 10% glycerol, and aliquots were stored at −80°C.
Circular dichroism.
Circular dichroism (CD) data were collected on an AVIV 62A DS spectrometer. A 20 μM concentration of each protein was used for far-UV CD wavelength scan (200 nm to 260 nm) at 10°C. All measurements were carried out in 25 mM NaPO4 (pH 7.2 for HAMPwt and HAMPR236H and pH 6.8 for HAMPE232K) and 150 mM NaCl and corrected against buffer alone.
PhoQ-enriched S. Typhimurium membrane preparation.
S. Typhimurium MB101 transformed with pBAD24 carrying the various phoQ alleles was grown at 37°C in LB supplemented with ampicillin. Once cultures reached an OD600 of ~0.6 to 0.8, PhoQ expression was induced by addition of 0.2% arabinose and further incubation at 37°C for 4 h. Cells were harvested by centrifugation, and membranes were prepared as previously described (47). The total amount of protein in the membranes was measured using the Bradford reagent (Bio-Rad) in the presence of 0.1% SDS. Western blotting using anti-PhoQ antibodies, raised against PhoQ periplasmic domain, was used to verify that similar amounts of PhoQ were present in the different samples.
Autophosphorylation, phosphotransfer, and phosphatase assay.
PhoQ in vitro autophosphorylation, phosphotransfer, and phosphatase assays were performed as described previously (36), with the exceptions that [γ-33P]ATP was used for all radioactive experiments instead of [γ-32P]ATP. Reactions were stopped by the addition of Laemmli SDS protein sample buffer, and the products were loaded directly onto Mini-PROTEAN TGX Any kD precast gels (Bio-Rad). Similar amounts of PhoQ protein were used in the assays as detected by Western blotting. For the phosphotransfer experiments, samples were taken at 5, 10, and 20 min after the reactions were started.
PhoQ cysteine cross-link.
S. Typhimurium MB101 carrying the different pBAD24 containing phoQ alleles was grown overnight in N minimal medium supplemented with the appropriate antibiotics and 1 mM MgCl2 in the presence or absence of 5 µg/ml C18G. Approximately 2 × 108 CFU were exposed to a solution of 0.6 mM Cu(II) (1,10-phenanthroline) (59), a membrane-permeant reagent that catalyzes disulfide bond formation. Reaction mixtures were incubated for 10 min at room temperature and stopped by addition of protein loading buffer containing 25 mM N-ethyl maleimide (NEM), 25 mM EDTA, and 8 M urea. Samples were heated at 95°C for 5 min and the proteins were separated on a Mini-PROTEAN TGX Any kD precast gel (Bio-Rad). The presence of PhoQ was inspected by Western blotting using anti-PhoQ antibodies directed against the PhoQ periplasmic domain. PhoQ monomer and dimer bands were quantified using ImageJ. The percentage of PhoQ dimer was calculated by dividing the intensity of the disulfide dimer band by the sum of the intensities of the monomer and dimer bands and multiplying the result by 100.
Structural models.
I-TASSER (30, 31) was used to model the PhoQ HAMP domain (residues 217 to 266) using Af1503 (2L7H), Af1503A291F (2L7I), Aer2 HAMP1 (3LNR), and Aer2 HAMP1L44H (4I3M) as templates.
The full cytoplasmic domain of PhoQ model (residues 217 to 487) was generated by using I-TASSER (30, 31). The repressed (4BIU:A) and activated (4BIU:B) chains of CpxA were used as templates.
The C score is a confidence score and is usually in the range of −5 to 2, where higher values signify higher confidence (31). The TM score measures structure similarity and has a value between 0 and 1. A TM score of >0.5 is generally attributed to proteins with the same topology.
SUPPLEMENTAL MATERIAL
Circular dichroism (CD) of purified HAMPwt (●), HAMPE232K (■), and HAMPR236H (▲). Download
Helical wheel representation of a four-helix bundle in an x-da packing conformation (left) and in an a-d packing conformation (right). The corresponding PhoQ residues are indicated. Download
Model of PhoQ HAMP domain based on Aer2 HAMP1L44H. One monomer is shown in color with α1 in green and α2 in blue, and the other monomer is in grey. Download
Comparison of phosphatase activity between activating (10 µM MgCl2) and repressive (10 mM MgCl2) growth conditions. Download
ClustalO primary sequence alignment of several PhoQ homologs. Download
Fold activation of the different PhoQ alleles compared to the wild type grown under PhoQ-repressive conditions.
ACKNOWLEDGMENTS
This work was supported by NIH grant R01AI030479 (to S.I M.) and a postdoctoral fellowship to S.M. from the Fundacao para a Ciencia e a Tecnologia, Portugal (SFRH/BPD/42392/2007) cofunded by Programa Operacional Potencial Humano and Fundo Social Europeu.
We thank Rachel E. Klevit for critically reading the manuscript and Kevin G. Hicks for helpful discussions.
S.M. and S.I.M. designed the experiments, analyzed the data, and wrote the paper; S.M. and K.R.H. performed the experiments.
Footnotes
Citation Matamouros S, Hager KR, Miller SI. 2015. HAMP domain rotation and tilting movements associated with signal transduction in the PhoQ sensor kinase. mBio 6(3):e00616-15. doi:10.1128/mBio.00616-15.
REFERENCES
- 1.Krell T, Lacal J, Busch A, Silva-Jiménez H, Guazzaroni ME, Ramos JL. 2010. Bacterial sensor kinases: diversity in the recognition of environmental signals. Annu Rev Microbiol 64:539–559. doi: 10.1146/annurev.micro.112408.134054. [DOI] [PubMed] [Google Scholar]
- 2.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 3.Aranda A, Swanson JA, Loomis WP, Miller SI. 1992. Salmonella typhimurium activates virulence gene transcription within acidified macrophage phagosomes. Proc Natl Acad Sci U S A 89:10079–10083. doi: 10.1073/pnas.89.21.10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller SI, Kukral AM, Mekalanos JJ. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci U S A 86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 6.Prost LR, Daley ME, Le Sage V, Bader MW, Le Moual H, Klevit RE, Miller SI. 2007. Activation of the bacterial sensor kinase PhoQ by acidic pH. Mol Cell 26:165–174. doi: 10.1016/j.molcel.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 7.García Véscovi E, Soncini FC, Groisman EA. 1996. Mg2+ as an extracellular signal: environmental regulation of salmonella virulence. Cell 84:165–174. doi: 10.1016/S0092-8674(00)81003-X. [DOI] [PubMed] [Google Scholar]
- 8.Dutta R, Qin L, Inouye M. 1999. Histidine kinases: diversity of domain organization. Mol Microbiol 34:633–640. doi: 10.1046/j.1365-2958.1999.01646.x. [DOI] [PubMed] [Google Scholar]
- 9.Adams P, Fowler R, Kinsella N, Howell G, Farris M, Coote P, O’Connor CD. 2001. Proteomic detection of PhoPQ- and acid-mediated repression of salmonella motility. Proteomics 1:597–607. doi:. [DOI] [PubMed] [Google Scholar]
- 10.Bader MW, Navarre WW, Shiau W, Nikaido H, Frye JG, McClelland M, Fang FC, Miller SI. 2003. Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Mol Microbiol 50:219–230. doi: 10.1046/j.1365-2958.2003.03675.x. [DOI] [PubMed] [Google Scholar]
- 11.Bearson BL, Wilson L, Foster JW. 1998. A low pH-inducible, PhoPQ-dependent acid tolerance response protects Salmonella typhimurium against inorganic acid stress. J Bacteriol 180:2409–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Behlau I, Miller SI. 1993. A PhoP-repressed gene promotes Salmonella typhimurium invasion of epithelial cells. J Bacteriol 175:4475–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalebroux ZD, Matamouros S, Whittington D, Bishop RE, Miller SI. 2014. PhoPQ regulates acidic glycerophospholipid content of the Salmonella Typhimurium outer membrane. Proc Natl Acad Sci U S A 111:1963–1968. doi: 10.1073/pnas.1316901111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fields PI, Groisman EA, Heffron F. 1989. A salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science 243:1059–1062. doi: 10.1126/science.2646710. [DOI] [PubMed] [Google Scholar]
- 15.Guo L, Lim KB, Gunn JS, Bainbridge B, Darveau RP, Hackett M, Miller SI. 1997. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science 276:250–253. doi: 10.1126/science.276.5310.250. [DOI] [PubMed] [Google Scholar]
- 16.Soncini FC, García Véscovi E, Solomon F, Groisman EA. 1996. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J Bacteriol 178:5092–5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller SI, Mekalanos JJ. 1990. Constitutive expression of the phoP regulon attenuates salmonella virulence and survival within macrophages. J Bacteriol 172:2485–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Appleman JA, Stewart V. 2003. Mutational analysis of a conserved signal-transducing element: the HAMP linker of the Escherichia coli nitrate sensor NarX. J Bacteriol 185:89–97. doi: 10.1128/JB.185.1.89-97.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park H, Inouye M. 1997. Mutational analysis of the linker region of EnvZ, an osmosensor in Escherichia coli. J Bacteriol 179:4382–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parkinson JS. 2010. Signaling mechanisms of HAMP domains in chemoreceptors and sensor kinases. Annu Rev Microbiol 64:101–122. doi: 10.1146/annurev.micro.112408.134215. [DOI] [PubMed] [Google Scholar]
- 21.Aravind L, Ponting CP. 1999. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol Lett 176:111–116. doi: 10.1111/j.1574-6968.1999.tb13650.x. [DOI] [PubMed] [Google Scholar]
- 22.Butler SL, Falke JJ. 1998. Cysteine and disulfide scanning reveals two amphiphilic helices in the linker region of the aspartate chemoreceptor. Biochemistry 37:10746–10756. doi: 10.1021/bi980607g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams SB, Stewart V. 1999. Functional similarities among two-component sensors and methyl-accepting chemotaxis proteins suggest a role for linker region amphipathic helices in transmembrane signal transduction. Mol Microbiol 33:1093–1102. doi: 10.1046/j.1365-2958.1999.01562.x. [DOI] [PubMed] [Google Scholar]
- 24.Dunin-Horkawicz S, Lupas AN. 2010. Comprehensive analysis of HAMP domains: implications for transmembrane signal transduction. J Mol Biol 397:1156–1174. doi: 10.1016/j.jmb.2010.02.031. [DOI] [PubMed] [Google Scholar]
- 25.Ames P, Zhou Q, Parkinson JS. 2008. Mutational analysis of the connector segment in the HAMP domain of Tsr, the Escherichia coli serine chemoreceptor. J Bacteriol 190:6676–6685. doi: 10.1128/JB.00750-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hulko M, Berndt F, Gruber M, Linder JU, Truffault V, Schultz A, Martin J, Schultz JE, Lupas AN, Coles M. 2006. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell 126:929–940. doi: 10.1016/j.cell.2006.06.058. [DOI] [PubMed] [Google Scholar]
- 27.Airola MV, Watts KJ, Bilwes AM, Crane BR. 2010. Structure of concatenated HAMP domains provides a mechanism for signal transduction. Structure 18:436–448. doi: 10.1016/j.str.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Q, Ames P, Parkinson JS. 2011. Biphasic control logic of HAMP domain signalling in the Escherichia coli serine chemoreceptor. Mol Microbiol 80:596–611. doi: 10.1111/j.1365-2958.2011.07577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Q, Ames P, Parkinson JS. 2009. Mutational analyses of HAMP helices suggest a dynamic bundle model of input-output signalling in chemoreceptors. Mol Microbiol 73:801–814. doi: 10.1111/j.1365-2958.2009.06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bass RB, Butler SL, Chervitz SA, Gloor SL, Falke JJ. 2007. Use of site-directed cysteine and disulfide chemistry to probe protein structure and dynamics: applications to soluble and transmembrane receptors of bacterial chemotaxis. Methods Enzymol 423:25–51. doi: 10.1016/S0076-6879(07)23002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Careaga CL, Falke JJ. 1992. Structure and dynamics of Escherichia coli chemosensory receptors. Engineered sulfhydryl studies. Biophys J 62:209–216, 217–219. doi: 10.1016/S0006-3495(92)81806-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Airola MV, Sukomon N, Samanta D, Borbat PP, Freed JH, Watts KJ, Crane BR. 2013. HAMP domain conformers that propagate opposite signals in bacterial chemoreceptors. PLoS Biol 11:e1001479. doi: 10.1371/journal.pbio.1001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kier LD, Weppelman RM, Ames BN. 1979. Regulation of nonspecific acid phosphatase in salmonella: phoN and phoP genes. J Bacteriol 138:155–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanowar S, Martel A, Moual HL. 2003. Mutational analysis of the residue at position 48 in the Salmonella enterica serovar typhimurium PhoQ sensor kinase. J Bacteriol 185:1935–1941. doi: 10.1128/JB.185.6.1935-1941.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kishii R, Falzon L, Yoshida T, Kobayashi H, Inouye M. 2007. Structural and functional studies of the HAMP domain of EnvZ, an osmosensing transmembrane histidine kinase in Escherichia coli. J Biol Chem 282:26401–26408. doi: 10.1074/jbc.M701342200. [DOI] [PubMed] [Google Scholar]
- 38.Lai RZ, Parkinson JS. 2014. Functional suppression of HAMP domain signaling defects in the E. coli serine chemoreceptor. J Mol Biol 426:3642–3655. doi: 10.1016/j.jmb.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferris HU, Dunin-Horkawicz S, Hornig N, Hulko M, Martin J, Schultz JE, Zeth K, Lupas AN, Coles M. 2012. Mechanism of regulation of receptor histidine kinases. Structure 20:56–66. doi: 10.1016/j.str.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 40.Ferris HU, Dunin-Horkawicz S, Mondéjar LG, Hulko M, Hantke K, Martin J, Schultz JE, Zeth K, Lupas AN, Coles M. 2011. The mechanisms of HAMP-mediated signaling in transmembrane receptors. Structure 19:378–385. doi: 10.1016/j.str.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 41.Campbell AJ, Watts KJ, Johnson MS, Taylor BL. 2010. Gain-of-function mutations cluster in distinct regions associated with the signalling pathway in the PAS domain of the aerotaxis receptor, aer. Mol Microbiol 77:575–586. doi: 10.1111/j.1365-2958.2010.07231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meena N, Kaur H, Mondal AK. 2010. Interactions among HAMP domain repeats act as an osmosensing molecular switch in group III hybrid histidine kinases from fungi. J Biol Chem 285:12121–12132. doi: 10.1074/jbc.M109.075721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elliott KT, Zhulin IB, Stuckey JA, DiRita VJ. 2009. Conserved residues in the HAMP domain define a new family of proposed bipartite energy taxis receptors. J Bacteriol 191:375–387. doi: 10.1128/JB.00578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mechaly AE, Sassoon N, Betton JM, Alzari PM. 2014. Segmental helical motions and dynamical asymmetry modulate histidine kinase autophosphorylation. PLoS Biol 12:e1001776. doi: 10.1371/journal.pbio.1001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prost LR, Daley ME, Bader MW, Klevit RE, Miller SI. 2008. The PhoQ histidine kinases of Salmonella and Pseudomonas spp. are structurally and functionally different: evidence that pH and antimicrobial peptide sensing contribute to mammalian pathogenesis. Mol Microbiol 69:503–519. doi: 10.1111/j.1365-2958.2008.06303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gunn JS, Hohmann EL, Miller SI. 1996. Transcriptional regulation of salmonella virulence: a PhoQ periplasmic domain mutation results in increased net phosphotransfer to PhoP. J Bacteriol 178:6369–6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le Moual H, Quang T, Koshland DE Jr. 1998. Conformational changes in the cytoplasmic domain of the Escherichia coli aspartate receptor upon adaptive methylation. Biochemistry 37:14852–14859. doi: 10.1021/bi980343y. [DOI] [PubMed] [Google Scholar]
- 48.Gushchin I, Gordeliy V, Grudinin S. 2013. Two distinct states of the HAMP domain from sensory rhodopsin transducer observed in unbiased molecular dynamics simulations. PLoS One 8:e66917. doi: 10.1371/journal.pone.0066917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang C, Sang J, Wang J, Su M, Downey JS, Wu Q, Wang S, Cai Y, Xu X, Wu J, Senadheera DB, Cvitkovitch DG, Chen L, Goodman SD, Han A. 2013. Mechanistic insights revealed by the crystal structure of a histidine kinase with signal transducer and sensor domains. PLoS Biol 11:e1001493. doi: 10.1371/journal.pbio.1001493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swain KE, Falke JJ. 2007. Structure of the conserved HAMP domain in an intact, membrane-bound chemoreceptor: a disulfide mapping study. Biochemistry 46:13684–13695. doi: 10.1021/bi701832b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watts KJ, Johnson MS, Taylor BL. 2008. Structure-function relationships in the HAMP and proximal signaling domains of the aerotaxis receptor aer. J Bacteriol 190:2118–2127. doi: 10.1128/JB.01858-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hughson AG, Hazelbauer GL. 1996. Detecting the conformational change of transmembrane signaling in a bacterial chemoreceptor by measuring effects on disulfide cross-linking in vivo. Proc Natl Acad Sci U S A 93:11546–11551. doi: 10.1073/pnas.93.21.11546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Molnar KS, Bonomi M, Pellarin R, Clinthorne GD, Gonzalez G, Goldberg SD, Goulian M, Sali A, DeGrado WF. 2014. Cys-scanning disulfide crosslinking and Bayesian modeling probe the transmembrane signaling mechanism of the histidine kinase, PhoQ. Structure 22:1239–1251. doi: 10.1016/j.str.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watts KJ, Johnson MS, Taylor BL. 2011. Different conformations of the kinase-on and kinase-off signaling states in the aer HAMP domain. J Bacteriol 193:4095–4103. doi: 10.1128/JB.01069-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watts KJ, Ma Q, Johnson MS, Taylor BL. 2004. Interactions between the PAS and HAMP domains of the Escherichia coli aerotaxis receptor aer. J Bacteriol 186:7440–7449. doi: 10.1128/JB.186.21.7440-7449.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anantharaman V, Balaji S, Aravind L. 2006. The signaling helix: a common functional theme in diverse signaling proteins. Biol Direct 1:25. doi: 10.1186/1745-6150-1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stewart V, Chen LL. 2010. The S helix mediates signal transmission as a HAMP domain coiled-coil extension in the NarX nitrate sensor from Escherichia coli K-12. J Bacteriol 192:734–745. doi: 10.1128/JB.00172-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cho US, Bader MW, Amaya MF, Daley ME, Klevit RE, Miller SI, Xu W. 2006. Metal bridges between the PhoQ sensor domain and the membrane regulate transmembrane signaling. J Mol Biol 356:1193–1206. doi: 10.1016/j.jmb.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 59.Chen X, Koshland DE Jr. 1998. Probing the structure of the cytoplasmic domain of the aspartate receptor by targeted disulfide cross-linking. Biochemistry 37:5335. doi: 10.1021/bi9750293. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Circular dichroism (CD) of purified HAMPwt (●), HAMPE232K (■), and HAMPR236H (▲). Download
Helical wheel representation of a four-helix bundle in an x-da packing conformation (left) and in an a-d packing conformation (right). The corresponding PhoQ residues are indicated. Download
Model of PhoQ HAMP domain based on Aer2 HAMP1L44H. One monomer is shown in color with α1 in green and α2 in blue, and the other monomer is in grey. Download
Comparison of phosphatase activity between activating (10 µM MgCl2) and repressive (10 mM MgCl2) growth conditions. Download
ClustalO primary sequence alignment of several PhoQ homologs. Download
Fold activation of the different PhoQ alleles compared to the wild type grown under PhoQ-repressive conditions.