Abstract
Gene regulatory information guides development and shapes the course of evolution. To test conservation of gene regulation within the phylum Nematoda, we compared the functions of putative cis-regulatory sequences of four sets of orthologs (unc-47, unc-25, mec-3 and elt-2) from distantly-related nematode species. These species, Caenorhabditis elegans, its congeneric C. briggsae, and three parasitic species Meloidogyne hapla, Brugia malayi, and Trichinella spiralis, represent four of the five major clades in the phylum Nematoda. Despite the great phylogenetic distances sampled and the extensive sequence divergence of nematode genomes, all but one of the regulatory elements we tested are able to drive at least a subset of the expected gene expression patterns. We show that functionally conserved cis-regulatory elements have no more extended sequence similarity to their C. elegans orthologs than would be expected by chance, but they do harbor motifs that are important for proper expression of the C. elegans genes. These motifs are too short to be distinguished from the background level of sequence similarity, and while identical in sequence they are not conserved in orientation or position. Functional tests reveal that some of these motifs contribute to proper expression. Our results suggest that conserved regulatory circuitry can persist despite considerable turnover within cis elements.
Author Summary
To explore the phylogenetic limits of conservation of cis-regulatory elements, we used transgenesis to test the functions of enhancers of four genes from several species spanning the phylum Nematoda. While we found a striking degree of functional conservation among the examined cis elements, their DNA sequences lacked apparent conservation with the C. elegans orthologs. In fact, sequence similarity between C. elegans and the distantly related nematodes was no greater than would be expected by chance. Short motifs, similar to known regulatory sequences in C. elegans, can be detected in most of the cis elements. When tested, some of these sites appear to mediate regulatory function. However, they seem to have originated through motif turnover, rather than to have been preserved from a common ancestor. Our results suggest that gene regulatory networks are broadly conserved in the phylum Nematoda, but this conservation persists despite substantial reorganization of regulatory elements and could not be detected using naïve comparisons of sequence similarity.
Introduction
Similar expression patterns of orthologous genes imply similarity of developmental programs in different species. Numerous such examples have been uncovered, including hox [1], dlx [2], and dpp/BMP [3] genes, as well as genetic programs regulating photoreceptor [4] and muscle [5] development in distantly related bilaterian animals. Largely based on these and similar findings, a current view of evolution of development emerged that emphasizes the conservation of the genetic “toolkit” within animals and the relative importance of regulatory changes in driving morphological change [6].
The mechanisms responsible for expression pattern conservation are less clear, however. One possibility is that ancestral gene regulatory programs are strictly retained. An alternative is that expression similarity is mediated by divergent regulatory processes [7,8], a phenomenon known as “developmental system drift” [9]. Regulatory rewiring of the latter type is known to occur even when individual components of the diverged networks are highly conserved developmental regulators [10–12]. One way to probe the evolution of regulatory linkages is with enhancer swap experiments, in which cis-regulatory DNA from one species is used to drive expression of a reporter gene in another species (reviewed in [13]). The resulting pattern of gene expression can be compared to the pattern driven by the endogenous regulatory element, with the similarities and differences giving evidence of conservation and divergence in the gene regulatory network.
We wanted to assess the conservation of gene regulatory programs among distantly-related members of the phylum Nematoda, a group of morphologically similar worms with mostly small, vermiform bodies. This body plan is largely conserved, with numbers of certain neuronal subtypes nearly identical in even deeply diverged taxa [14,15], and the intestine arising from a clonal cell lineage [16] in most (but not all, see [17]) nematodes studied. However, instances of developmental divergence have been documented in this clade [18–23]. We therefore performed enhancer swap experiments with regulatory elements of genes expressed in two subsets of neurons and in the developing intestine. By examining the function of cis regulatory sequences from four different nematode species in transgenic C. elegans, we sought to determine the extent of cis-regulatory conservation within this phylum.
Results
Selection of species and genes
The phylum Nematoda is comprised of animals with simple vermiform body plans and diverse life-history strategies. To look for evidence of gene regulatory conservation across this phylum, we carried out a series of enhancer-swap experiments between several distantly-related nematodes and a C. elegans host. Regulatory regions from orthologous C. elegans genes driving the mCherry reporter were co-expressed as controls with the exogenous cis elements driving expression of the GFP gene. This approach allows us to isolate and compare cis-regulatory functions of the two orthologous regulatory elements in a common trans-regulatory background. Any observed differences can then be attributed to the divergence of the cis-regulatory DNA.
We sought broad coverage of the phylum, which is hypothesized to have diversified in the Silurian [24]. Representatives from two basally branching nematode groups have sequenced genomes [25]. These are the Chromodorea (comprised of Clades III-V) and the Dorylaimia (Clade I). No Enoplia (Clade II) genomes have been sequenced to date. For this study we used C. elegans [26] as the transgenic host species, and its congeneric C. briggsae [27] to test divergence of regulatory elements among close relatives (both are from Clade V). The next most closely related nematode species is Meloidogyne hapla (Clade IV, [28,29]), followed by Brugia malayi (Clade III, [30,31]). Finally, as a representative of Clade I, we used Trichinella spiralis [32]. Divergence of Clade I was one of the earliest events in nematode evolution. The relationships among these five species are shown in Fig 1. We leveraged both this phylogeny and the amenability of C. elegans to genetic manipulation to create a series of comparisons of expression of cis-regulatory elements from progressively more distantly-related species in transgenic C. elegans. C. elegans have been used as transgenic hosts of regulatory DNA from a number of different species (reviewed, along with similar studies using Drosophila melanogaster, in [13]), however, to our knowledge, this study is the first explicit test of the relationship between evolutionary relatedness and conservation of cis-regulatory function among a set of genes.
Fig 1. Phylogenetic relationships between the species used in this study.
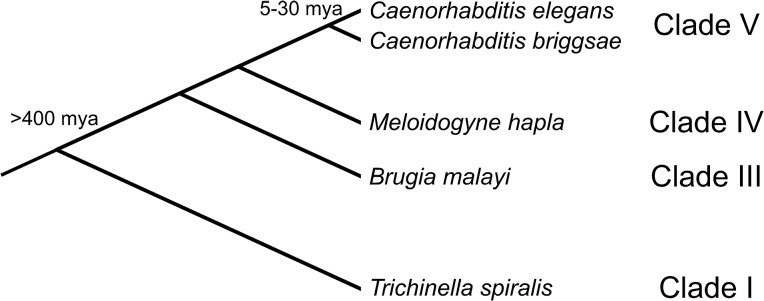
Topology of the tree and clade assignments are after [25].
While our selection of species gave us unprecedented ability to test the phylogenetic limits of regulatory conservation, it also rendered reciprocal transgenesis infeasible due to the complex modes of reproduction of the parasitic species.
We selected genes that have considerable conservation of their coding sequences and are single-copy orthologs among the species. Two genes are expressed in GABAergic neurons in Caenorhabditis nematodes, unc-47 [33,34] and unc-25 [35]. We have previously investigated the evolution of their regulation within this clade [36–39]. The third gene, mec-3, is expressed in another neuronal cell type, the touch-receptor neurons, in C. elegans [40,41]. The regulatory region of the C. briggsae ortholog of mec-3 has previously been shown to drive gene expression in C. elegans [42]. Finally, we chose the gene elt-2, which is expressed in the endoderm [43,44], and shows evidence of regulatory conservation outside the genus Caenorhabditis [45]. These cis-regulatory elements are expressed in different cell types, and drive expression of terminal differentiation genes (unc-47 and unc-25) as well as transcription factors (mec-3 and elt-2). Where possible (see Materials and Methods), the putative regulatory regions we investigated ranged from the start of recognizable protein-coding sequence conservation with C. elegans on the 3’ end to the next upstream coding element on the 5’ end. This choice of putative regulatory sequences in no way depended on non-coding conservation between species.
Regulatory elements from distantly-related nematodes retain some, but not all, functions when swapped into C. elegans
unc-47
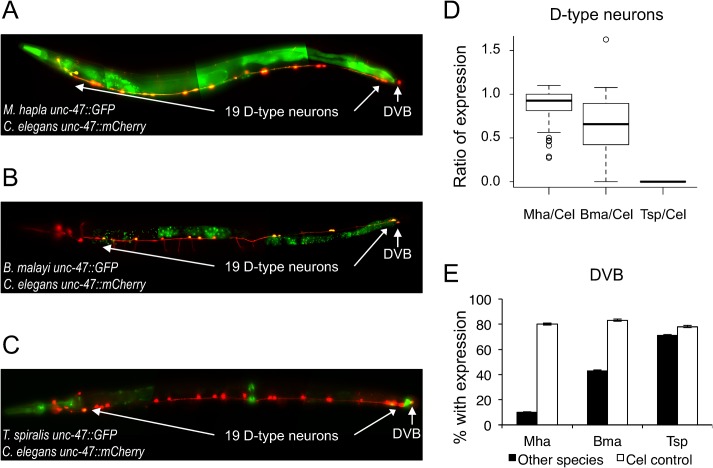
The cis-regulatory elements of the unc-47 genes from all three distant relatives drove gene expression in C. elegans in portions of the endogenous GABAergic neuronal expression pattern (Fig 2). The cells that we examined with particular attention were the D-type neurons in the ventral nerve cord, and the post-anal neuron DVB. The unc-47 cis-regulatory elements from C. briggsae, C. brenneri, and C. remanei, and C. japonica all drove strong and consistent expression in these cells [36–38]. The M. hapla and B. malayi unc-47 cis-regulatory elements drove substantially weaker (S1A and S1B Fig) expression that was less consistent than that of the C. elegans unc-47 cis element (Fig 2A, 2B and 2D). The upstream region of the T. spiralis gene failed to direct expression in the D-type neurons (Figs 2C, 2D and S1C). However, expression in DVB showed the opposite pattern. Both the M. hapla and B. malayi unc-47 regulatory DNA drove expression far less consistently than the C. elegans element (Fig 2E). In contrast, the T. spiralis ortholog directed bright and consistent expression in DVB that was not significantly different from C. elegans expression (Fisher’s Exact test, p = 0.3304), as well as the head neuron RIS (Figs 2C, 2E and S1D). Both of these cells are GABAergic neurons that endogenously express unc-47 in C. elegans. T. spiralis regulatory DNA drove expression in the vulval and rectal epithelia, as well as the neuron PVT, common sites of ectopic expression), and several unidentified head neurons (Fig 2C). Expression patterns were consistent across independently generated transgenic strains (S2 Table).
Fig 2. unc-47 regulatory sequences from distantly-related nematodes drive expression in C. elegans.
(A-C) C. elegans unc-47 regulatory sequence drives expression of mCherry in all transgenic strains; (A) M. hapla, (B) B. malayi, (C) T. spiralis unc-47 regulatory sequences drive expression of GFP. Animals were photographed at 400x magnification. Images are false-colored composites of single animals. Separate GFP and mCherry images are shown in S1 Fig. (D) Ratios of the number of D-type neurons expressing GFP/mCherry in individuals carrying indicated transgene pairs (see Materials and Methods and S1 Table for total counts). T. spiralis cis-element drives no expression in the D-type neurons. M. hapla and B. malayi differ in their fidelity to the C. elegans expression pattern (Kruskal-Wallis test, p = 2.095×10−9). (E) Percentage of individuals with expression in the cell DVB from the heterologous regulatory element (black) and the C. elegans regulatory element (white) for each transgene pair (see Materials and Methods and S1 Table for total counts). Error bars show 95% confidence intervals for the proportion expressing.
unc-25
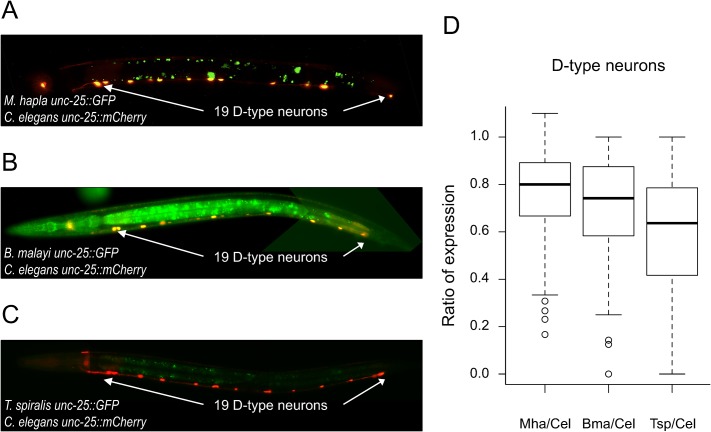
The regulatory elements of unc-25 from C. elegans, C. briggsae, C. brenneri, C. remanei, and C. japonica directed consistent expression in the D-type GABAergic neurons, which constitute the majority of the endogenous expression pattern of this gene [38,39]. The same was the case for their M. hapla, B. malayi, and T. spiralis orthologs, despite the great phylogenetic distances separating these species (Figs 3A–3D and S2). Note that while the T. spiralis unc-47 cis-regulatory DNA did not drive expression in D-type neurons (Fig 2C and 2D), the unc-25 upstream region from this same species did so (Fig 3C and 3D). Expression patterns were consistent across independently generated transgenic strains (S2 Table).
Fig 3. unc-25 regulatory sequences from distantly-related nematodes drive expression in C. elegans.
(A-C) C. elegans unc-25 regulatory sequence drives expression of mCherry in all transgenic strains; (A) M. hapla, (B) B. malayi, (C) T. spiralis unc-25 regulatory sequences drive expression of GFP. Animals were photographed at 400x magnification. Images are false-colored composites of single animals. Separate GFP and mCherry images are shown in S2 Fig. (D) Ratios of the number of D-type neurons expressing GFP/mCherry in individuals carrying each transgene pair (see Materials and Methods and S1 Table for total counts). While each strain drives expression in D-type neurons, the three strains do show differences in their distributions of the ratios of cells expressing GFP relative to mCherry (Kruskal-Wallis test, p = 1.53×10−5).
mec-3
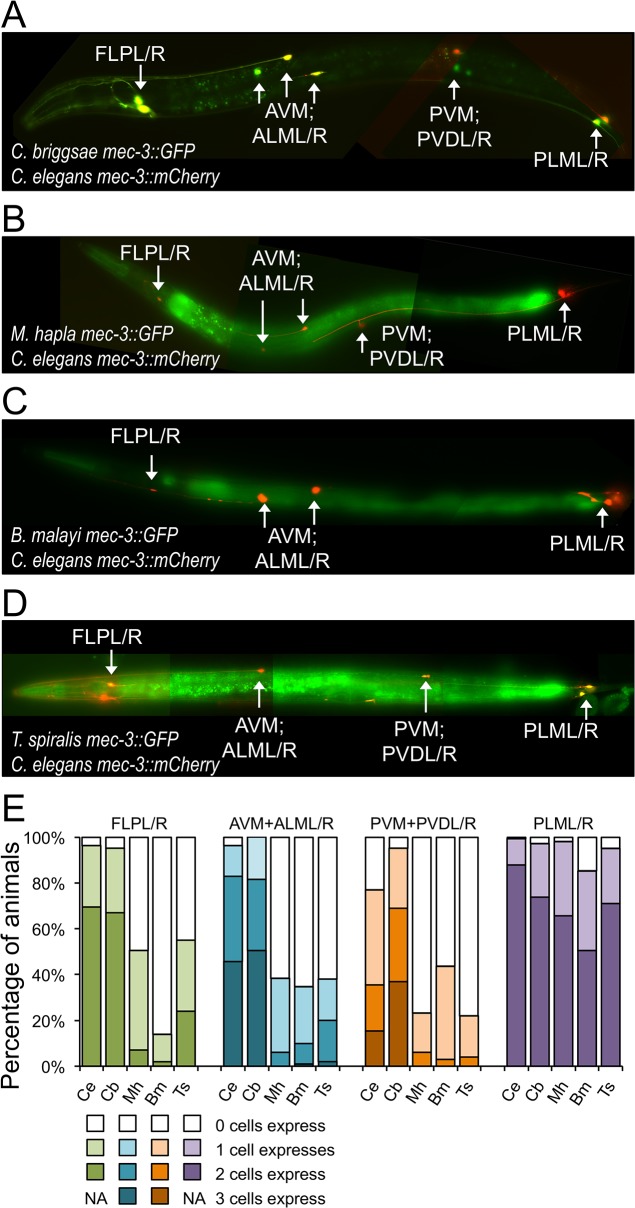
The DNA upstream of orthologs of mec-3 drove expression in the six touch-receptor neurons (ALML/R, AVM, PVM, and PLML/R) and two additional, extensively branched, pairs of mechanosensory neurons, FLPL/R in the head, and PVDL/R in the posterior midbody (Fig 4); these cells constitute the endogenous gene expression pattern in C. elegans [40]. Expression was strong and consistent in all cells from both C. elegans and C. briggsae cis-regulatory elements (Fig 4A, 4E and S3). While none of the regulatory elements completely failed to drive expression in any of these cells, those from the three distantly-related nematodes directed dramatically less consistent expression in all cells but PLML/R in the tail (Fig 4E). In addition to driving expression in the mechanosensory neurons as expected, the B. malayi cis element also drove ectopic expression in several ventral cord neurons (S4 Fig). Expression patterns were consistent across independently generated transgenic strains (S2 Table).
Fig 4. mec-3 regulatory sequences from distantly-related nematodes drive expression in C. elegans.
(A-D) C. elegans mec-3 regulatory sequence drives expression of mCherry in all transgenic strains; (A) C. briggsae, (B) M. hapla, (C) B. malayi, (D) T. spiralis mec-3 regulatory sequences drive expression of GFP. Animals were photographed at 400x magnification. Images are false-colored composites of single animals. Separate GFP and mCherry images are shown in S3 Fig. (E) Percentage of animals with 0 (white), 1 (light), 2 (mid-tone), or in some cases 3 (darkest shade) cells with reporter gene expression of GFP directed by the heterologous elements and mCherry directed by the C. elegans element. The data are based on counting over 100 individuals carrying each transgene pair (see Materials and Methods and S1 Table for total counts). The C. elegans data are derived from averaging the number of animals expressing mCherry across all mec-3 carrying strains shown here (see Materials and Methods and S1 Table). A maximum of two cells for the FLPs and PLMs and three cells for AVM+ALMs and PVM+PVDs could have expression.
elt-2
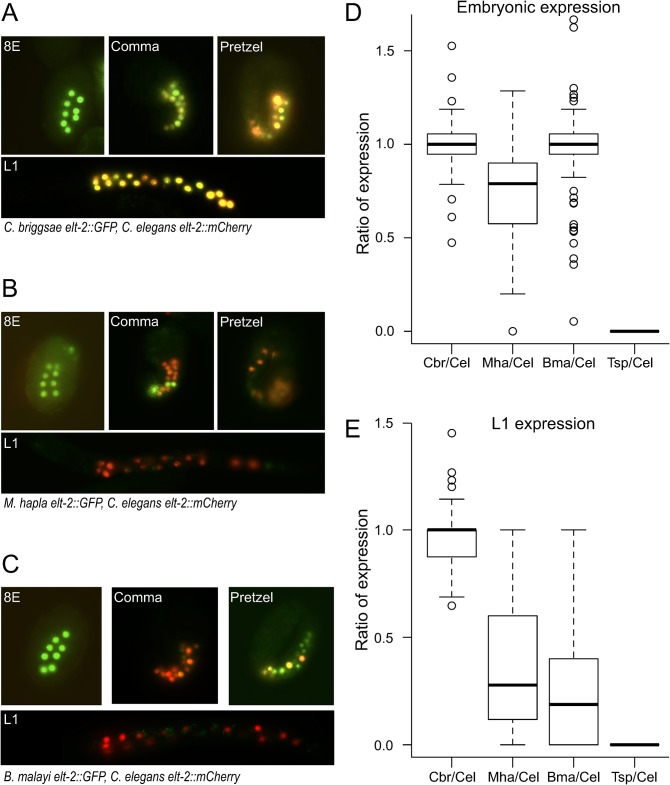
The C. elegans elt-2 gene is endogenously expressed in the endodermal cells throughout development [46], and appears to have the same expression in a Clade V nematode, Haemonchus contortus [45]. The upstream regions of orthologs of elt-2 drove the expected endodermal expression that was first detectable at the 4E stage, and consistently visible at the embryonic 8E stage (Figs 5 and S5). The elt-2 regulatory DNA of C. briggsae, M. hapla, and B. malayi all drove consistent expression at the embryonic stages (Fig 5A–5D). However, only the C. elegans and C. briggsae elt-2 upstream sequences directed consistent expression in the first larval stage (Fig 5E), and beyond. The sequence upstream of T. spiralis elt-2 did not drive any detectable expression in any tissue at any stage (Fig 5E). The M. hapla elt-2 cis-regulatory DNA occasionally drove expression in cells anterior to the developing gut, but this expression was not consistent and diminished in later stages earlier than gut expression (Figs 5B and S5B). Expression patterns were consistent across independently generated transgenic strains (S2 Table).
Fig 5. elt-2 regulatory sequences from distantly-related nematodes drive expression in C. elegans.
(A-C) C. elegans elt-2 regulatory sequence drives expression of mCherry in all transgenic strains; (A) C. briggsae, (B) M. hapla, (C) B. malayi elt-2 regulatory sequences drive expression of GFP. 8E, comma, pretzel and L1 refer to three characteristic embryonic stages and the first larval stage, respectively. Animals were photographed at 400x magnification. Images are false-colored composites of single animals. Separate GFP and mCherry images are shown in S5 Fig. (D) Ratios of the number of E-cell descendants in pretzel stage embryos expressing GFP/mCherry. While the Mha/Cel ratios are significantly different from the other two (Wilcoxon rank sum test, p<10−14), Bma/Cel and Cbr/Cel do not differ (Wilcoxon rank sum test, p = 0.99). (E) Ratios of the number of E-cell descendants in L1 larvae expressing GFP/mCherry. While Cbr/Cel was not significantly different from Bma/Cel at the pretzel stage, at the L1 stage Cbr/Cel is significantly different from both Bma/Cel and Mha/Cel (Wilcoxon rank sum test, p<2.2×10−16). See Materials and Methods and S1 Table for total counts.
Orthologous regulatory sequences from distantly-related nematodes have little similarity
All but one of the 12 regulatory sequences from distantly related species that we tested in C. elegans directed expression in at least a subset of the expected cells, so some degree of functional conservation is preserved even at these great phylogenetic distances. Since the putative regulatory regions from the distant relatives were selected without regard for non-coding conservation, we next examined them for sequence similarity with the C. elegans orthologs. We did not know, a priori, what types of sequence similarity to expect, and did not find any extended sequence conservation. For this reason, we conducted three types of sequence comparison to ascertain the extent of sequence similarity between C. elegans and each of the distantly-related nematodes.
First, we created dotplots, which depict the positions of nucleotide strings of a certain length that are shared by the C. elegans unc-47 sequence and a sequence from another nematode (10 bp examples shown in Fig 6A–6D). Only the C. briggsae cis element displayed substantial evidence of sequence conservation, represented by collinear blocks of sequence with conserved spacing upstream of the translation start site (upper right diagonal, Fig 6A). Not only do the distantly-related nematodes lack any such collinear blocks of sequence (evidence of conservation), they lack much in with way of sequence similarity as well, with only a few scattered motifs found in both the C. elegans unc-47 upstream region and those upstream regions from M. hapla, B. malayi, and T. spiralis (Fig 6B–6D).
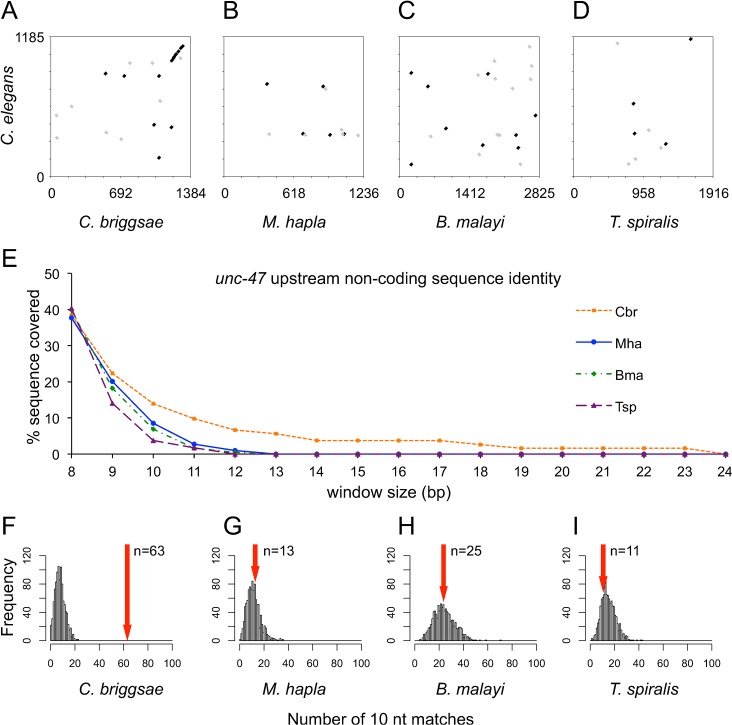
Fig 6. Sequence similarity of orthologous unc-47 cis elements.
(A-D) Dotplots showing perfect sequence identity (in 10 nucleotide windows) of the unc-47 upstream sequences in direct (black) and reverse (gray) orientations between C. elegans and (A) C. briggsae, (B) M. hapla, (C) B. malayi, (D) T. spiralis. (E) Percentage (y-axis) of unc-47 upstream sequences composed of short blocks of sequence identity with C. elegans across various window sizes (x-axis). (F-I) Distributions of the number of perfect 10 nucleotide matches between 1000 replicates of reshuffled C. elegans unc-47 upstream sequence and the upstream unc-47 sequences of (F) C. briggsae, (G) M. hapla, (H) B. malayi, (I) T. spiralis. Trinucleotide frequencies of the original C. elegans unc-47 upstream sequence were retained. Red arrows indicate the actual number of 10 nucleotide matches between C. elegans and each ortholog. Only C. briggsae has more than would be expected by chance (see Results). Note that B. malayi sequence is approximately twice as long as that of C. elegans, C. briggsae, and M. hapla.
We looked more closely at the few 10 bp motifs in each of the divergent sequences that are shared with the C. elegans cis element (Fig 6B–6D). Since the functional units of cis-regulatory elements are thought to be short binding sites, we next hypothesized that the divergent cis elements might be enriched for such short, shared motifs. We tested this in two ways. First, we broke the sequences down into their component k-mers, and asked what percentage of the total sequence length was made up of k-mers shared with the C. elegans sequence. For example, by definition, 100% of the M. hapla unc-47 cis element is made up of 1-mers (A, T, G, or C) that are also found in the C. elegans unc-47 cis element. Approximately 40% of the examined cis elements of C. briggsae and the other 3 nematodes are made up of 8-mers that are also found in the C. elegans sequence (Fig 6E), suggesting that window sizes shorter than 9 nucleotides are not likely to be informative for this comparison. For 9-mers, slight differences in the proportion of shared sequence can be detected among species; at window sizes of 10–12 nucleotides, the difference between C. briggsae and the distantly-related nematodes becomes apparent (Fig 6E). Note that the B. malayi unc-47 cis element, while it functions remarkably better than the T. spiralis ortholog (Fig 2), is not substantially more similar in sequence to the C. elegans regulatory element. None of the three distantly-related nematodes had any identical sequence blocks longer than 12 nucleotides, and blocks longer than 10 nucleotides were primarily low-complexity polynucleotide sequences (S6 Fig), while C. briggsae had identical sequences of up to 23 nucleotides in length (Fig 6E). Alignments showing all of the identical sequence matches in the unc-47 upstream regions that are 9 nucleotides or longer can be found in S6 Fig. These identical blocks are not enriched proximal to the start of the coding sequence. Similar levels of conservation were found for unc-25, mec-3, and elt-2 as well (S7–S9 Figs).
The next method that we used to test whether the orthologous cis elements were enriched for short motifs shared with the C. elegans unc-47 upstream region compared the number of shared motifs detected with the number that might be expected by chance. Here, “chance” refers to a random reordering of the C. elegans sequence that preserves nucleotide, dinucleotide, or trinucleotide frequencies. For each of the four genes, we reshuffled the C. elegans sequence 1000 times. The cis elements from C. briggsae, M. hapla, B. malayi, and T. spiralis were compared to each of the 1000 reshuffled C. elegans sequences, and we calculated the numbers of nucleotide blocks (length 8–12) that were identical between each reshuffled C. elegans cis element and each of the orthologs. This provided empirically derived distributions of sequence identity that could be expected solely as a result of basic nucleotide composition properties. The results for tests of 10 nucleotide blocks are shown in Fig 6F–6I. For the 1000 comparisons between the reshuffled C. elegans sequences and the other nematode’s upstream unc-47 sequence, the number of identical motifs was plotted (Fig 6F–6I). The number of motifs form distributions centered between about 10–20 motifs per reshuffled sequence, depending on the length of the ortholog. Comparing the actual number of conserved blocks of various lengths between C. elegans cis elements and their orthologs revealed that only C. briggsae had more sequence identity than our “chance” rearrangements, with 63 identical 10-mers (Fig 6F). The other three distantly-related nematodes’ sequences had no more similarity than expected by chance, with numbers of shared 10-mers that fell close to the means of the distributions (Fig 6G–6I). The same was true for the upstream noncoding sequences of unc-25, mec-3, and elt-2 (S10 Fig).
Orthologous cis elements share short putative regulatory motifs
Comparisons of noncoding sequence identity did not reveal any substantially conserved regions likely to be responsible for the functional conservation of orthologous cis elements. And yet, 11 of the 12 cis-regulatory elements from deeply diverged nematodes drove gene expression in C. elegans that recapitulated at least some of the expected endogenous expression pattern. We therefore searched the orthologous sequences for motifs known to be functionally important in the C. elegans sequences. Expression of unc-47 is regulated by direct binding of UNC-30 [47] to TAATCC sites. Mutations to this motif abolish expression in the D-type neurons [47]. Perhaps functional conservation of the unc-47 cis elements from distantly-related nematodes is due to the presence of this and other short sequences below the level of detection in our naive sequence comparison.
Searching for the TAATCC site revealed a perfect match, including one flanking base pair on either side in C. briggsae, with similar spacing from the translational start site (Fig 7). The noncoding sequence upstream of M. hapla unc-47 has three instances of this motif, all on the reverse strand, with additional identical nucleotides flanking the core site (Fig 7). The upstream sequences from B. malayi and T. spiralis lack perfect matches to this consensus, but do have 5/6 bp core matches with some additional flanking identity (Fig 7). Either these close matches are divergent cis-regulatory sites, hinting at evolved differences in TF-TFBS recognition, or else there is more to the control of expression in D-type neurons than we have recognized in C. elegans thus far.
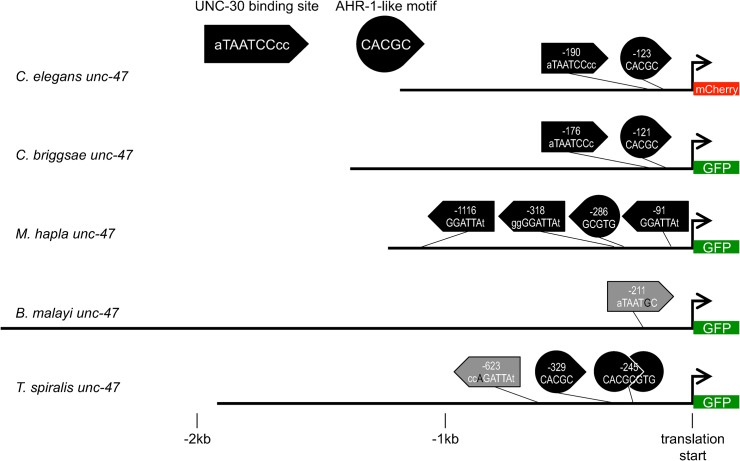
Fig 7. Matches to motifs responsible for the C. elegans unc-47 gene expression pattern can be found in orthologous sequences.
Cartoons depicting sequences upstream of the translation start sites (bent arrows) of unc-47 orthologs. These were fused to mCherry (C. elegans) or GFP (all others). UNC-30 (box arrow) and AHR-1-like (rounded arrow) motifs are shown, in uppercase letters; conserved flanking nucleotides shown in lowercase. Locations of motifs relative to the endogenous translation start sites are indicated. Where the best match has a single mismatch with the consensus sequence, it is shown by a gray shaded arrow with the mismatched nucleotide shown in black.
The C. elegans UNC-30 binding site controls expression in D-type neurons, but not in DVB in the tail or AVL, RIS, or the RMEs in the head [47]. One site that contributes to expression in DVB, RIS, and AVL is the AHR-1-like motif [37]. This motif has the sequence CACGC and is conserved in sequence and position between C. elegans, C. briggsae, C. brenneri, and C. remanei [37]. A match to this motif is found on the reverse strand of the M. hapla unc-47 cis element (Fig 7). A palindromic sequence CACGCGTG, that is, two overlapping AHR-1-like motifs on opposite strands, along with an additional single instance of this motif, are present upstream of the T. spiralis unc-47 gene (Fig 7).
Similar motif-matching analyses were carried out for the other three sets of orthologous cis-regulatory elements. Matches to motifs known to be necessary for function in C. elegans were identified in almost all tested orthologs from distantly-related nematodes (S1 Text; S11–S13 Figs). However, the occurrence of even multiple instances of motifs corresponding to transcription factor binding sites should not be construed as evidence of conservation. First, these motifs are not found any more frequently than in randomly reshuffled C. elegans sequences. We explicitly estimated the probability of finding these motifs in the randomly reshuffled C. elegans sequences. The probability of finding TAATCC (the UNC-30 binding site) in sequences preserving the single-nucleotide composition of the C. elegans cis element was 0.558, conserving dinucleotides it was 0.378, and trinucleotides it was 0.566. The probabilities of finding CACGC (the AHR-1-like motif) in these same sequences were 0.632, 0.606, 0.674, respectively. Second, these motifs were routinely found in the cis elements of the other genes we examined (S3 Table). Third, these motifs are often found on the opposite strand, suggesting that, while individual motifs are born and die, this sequence turnover maintains at least one instance of the motif in each of the orthologous regulatory elements. Therefore, identical motifs are not, strictly speaking, conserved.
Motif similarity identifies functional sites in diverged orthologs
It is suggestive that the unc-47 regulatory sequence from a distantly-related nematode that retains the best function in D-type neurons, that of M. hapla, has the best match to the UNC-30 binding site. Similarly, the regulatory sequence with the best function in DVB—T. spiralis unc-47—has the best matches to the AHR-1-like motif. We therefore tested the contribution of these motifs to functional conservation.
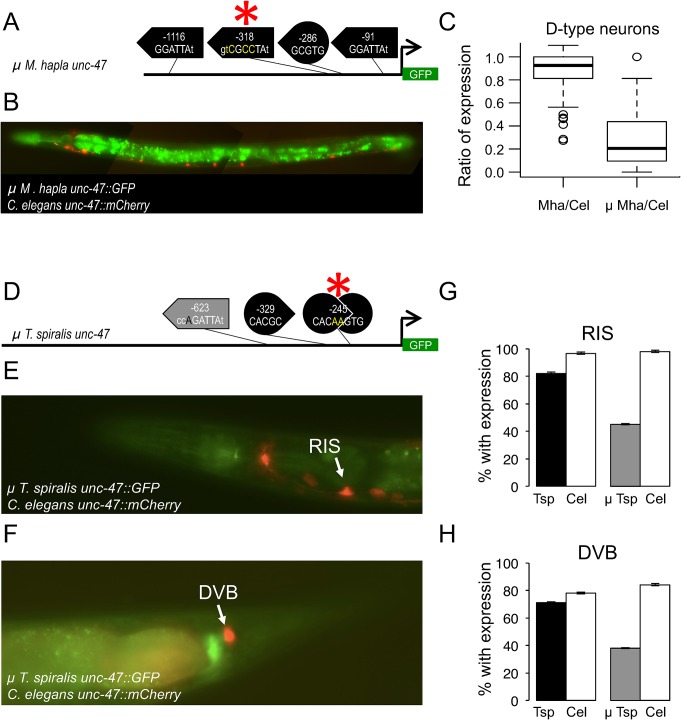
We introduced mutations into an UNC-30 binding motif in the M. hapla unc-47 cis element. This motif was selected because it shares the longest similarity in the flanking sequences with the UNC-30 binding site of the C. elegans unc-47 cis element (Fig 8A). The mutant M. hapla unc-47 sequence directed less consistent expression in the D-type neurons than its wild-type counterpart (Fig 8B, 8C and S14A), suggesting that this UNC-30 motif contributes to control of gene expression in the D-type neurons.
Fig 8. Mutations in putative transcription factor binding sites disrupt cis-regulatory functions.
(A) Cartoon depicting the mutations made to the M. hapla unc-47 cis sequence, with the affected motif designated by the asterisk and the sequence changes shown in yellow. (B) Mutant M. hapla unc-47 cis element drives expression of GFP. C. elegans unc-47 regulatory sequence drives expression of mCherry. Animals were photographed at 400x magnification. Images are false-colored composites of single animals. Separate GFP and mCherry images are shown in S14 Fig. (C) Ratios of the number of D-type neurons expressing GFP/mCherry for wild-type (Mha/Cel) and mutant (μ Mha/Cel) cis-regulatory elements. Each ratio is based on counting over 100 individuals carrying each transgene pair (see Materials and Methods and S1 Table for total counts). The two are significantly different (Kruskal-Wallis test, p < 2.2 ×10−16). (D) Cartoon depicting the mutations made to the T. spiralis unc-47 cis sequence, with the affected motif designated by the asterisk and the sequence changes shown in yellow. Mutant T. spiralis unc-47 cis element drives expression of GFP in RIS (E) and DVB (F). Wild-type C. elegans unc-47 regulatory sequence drives expression of mCherry. Animals were photographed at 400x magnification. Images are false-colored composites of single animals. Separate GFP and mCherry images are shown in S14 Fig. Percentage of individuals with expression in RIS (G) and DVB (H) from the T. spiralis wild-type (black) and mutant (μ Tsp, gray) cis-regulatory elements (see Materials and Methods and S1 Table for total counts). The mutated T. spiralis unc-47 cis element drives substantially less consistent expression than the wild-type, in RIS (Fisher’s Exact Test, p = 4.564×10−6) and DVB (Fisher’s Exact Test, p = 2.536× 10−6). Expression of C. elegans unc-47 regulatory sequence is shown in white; this control is not significantly different for either RIS (p = 0.7056) or DVB (p = 0.3775). Error bars represent 95% confidence interval for the proportion expressing.
Next, we introduced mutations into the palindromic double AHR-1-like motif of the T. spiralis unc-47 element, eliminating the consensus sequence on both strands (Fig 8D). This resulted in a substantial decrease in the fraction of animals expressing the transgene in RIS and DVB (Figs 8E–8H and S14B). This suggests that the palindromic AHR-1-like motif upstream of the T. spiralis unc-47 gene is partially responsible for expression in DVB and RIS, just as the AHR-l-like motif is in C. elegans [37].
Neither mutation eliminated expression in the affected cells entirely, implying that these sites contribute to but are not strictly essential for expression. This could be due to the redundancy of these binding sites in both cis elements (Fig 7). As another possible explanation, consider the case of the B. malayi unc-47 element that lacks good matches for either the UNC-30 or the AHR-1-like motifs, and yet is reasonably well expressed in both the D-type neurons and in DVB. It is possible that some orthologous cis elements retain functional conservation via sequences that can be recognized by C. elegans transcription factors, but that we currently cannot recognize as functional.
Discussion
We investigated cis-regulatory function in an explicitly evolutionary framework. The extent of divergence between the species involved in this study ranged from that of congenerics (C. elegans and C. briggsae) to the deepest in the phylum Nematoda (C. elegans and T. spiralis). This allowed us to test how regulatory information breaks down over time. Transgenic experiments were conducted in the “common garden” of C. elegans to control for the effects of trans-regulatory divergence and to focus comparisons on the cis elements (see Potential Caveats in Materials and Methods). We selected the putative regulatory regions without regard for non-coding sequence similarity, which then permitted us to comment on the relationship between functional and sequence conservation. We used a standard methodology to look at the cis elements of four genes, allowing us to make four generalizations.
First, despite the vast spans of evolutionary time that we sampled—the most distantly-related species diverged perhaps as long as 400 million years ago [24]—the majority of the cis-regulatory elements exhibited appreciably conserved gene regulatory function in C. elegans (Table 1). Two reasons compelled us to focus explicitly on the conserved, rather than divergent, aspects of expression. First, at such great phylogenetic distances, any conservation might be less expected than divergence. Second, for technical reasons, we can only know the endogenous function of the C. elegans regulatory elements, not the patterns driven by the divergent cis elements in their native species (see Potential Caveats in Materials and Methods). Our findings are consistent with previous reports of functional conservation of cis-regulatory elements between distantly-related members of the same phylum, most extensively tested in arthropods and chordates [48,49]. Although there have been a number of reports of functional conservation of cis elements between different phyla [50–56], this is not true for cis elements of all genes tested [39,57]. It is possible that the evolutionary dynamics of regulatory elements may be sufficiently idiosyncratic to preclude general conclusions about the “outer limits” of cis-regulatory conservation.
Table 1. Summary of gene expression driven by parasitic nematode cis elements relative to C. elegans expression.
| M. hapla | B. malayi | T. spiralis | |
|---|---|---|---|
| unc-47 | Weak, missing DVB cell consistently | Weak, patchy | Absent in 24/26 cells consistently, expressed in 2/26 consistently, ectopic expression in non-neuronal cell types |
| unc-25 | Correct but weak | Correct but weak | Correct but weak |
| mec-3 | Inconsistent in anterior cells, but very consistent in PLMs | Inconsistent in anterior cells, but very consistent in PLMs; ectopic in VC neurons | Inconsistent in anterior cells, but very consistent in PLMs |
| elt-2 | Indistinguishable until hatching, reduced in larvae, absent in adults | Indistinguishable until hatching, reduced in larvae, absent in adults | No expression |
Second, in most cases cis-regulatory elements from more distant relatives have retained less function than elements from closer relatives. However, there are notable exceptions and, importantly, the pattern of functional divergence that we observed reflects modular organization of cis-regulatory elements—separable elements control different aspects of expression [58–60]. Due to modularity of cis elements, evolution can “tinker” with some functions while avoiding pleiotropic effects on others [61]. In C. elegans, expression of unc-47 is controlled by different mechanisms in D-type neurons and DVB, RIS, and AVL [37,47]. Accordingly, we see that whereas the T. spiralis unc-47 element is not expressed in D-type neurons, it functions relatively well in DVB and RIS (Figs 2 and S1). In contrast, the M. hapla unc-47 element is expressed well in the D-type neurons, but not in DVB (Fig 2). Similarly, the elt-2 elements from M. hapla and B. malayi are expressed reasonably well during embryogenesis, but not in later stages (Fig 5). We consider this good evidence for separate regulation of pattern, timing, and levels of expression, as well as substantiating evidence that the weak expression of some of these regulatory elements is due to genuine divergence of regulatory information rather than experimental artifacts of weak transgene expression. We conclude that modular organization of cis elements manifests in different rates of divergence for different aspects of expression patterns [62] and may be quite common [13]. Mechanisms controlling spatial, temporal, and levels of expression may be particularly prone to different rates of divergence (e.g. [45,63]).
Third, despite their substantially conserved functions, the regulatory elements of all species but C. briggsae have not retained more sequence similarity than would be expected by chance. This finding is consistent with previous reports that suggested that conservation of cis-regulatory function does not, strictly speaking, require extended sequence conservation [64–70]. Since different types of regulatory elements evolve under different constraints [36], relying on sequence conservation to find cis-regulatory elements might bias discovery to only particular types of elements with highly constrained sequences [71]. Additionally, because sequences of different elements evolve at different rates [38], it is not a priori clear how distant the species to be compared should be to discover cis elements of different types. Even when some short stretches of identical nucleotides are discovered between distantly-related orthologous cis elements, this should not be taken as evidence of conservation. This is because many short matches will always be found by chance, particularly in regions with biased nucleotide composition. For instance, the co-occurrence of the UNC-30 and AHR-1-like motifs upstream of unc-47 orthologs (Fig 7) is more plausibly explained by a birth-and-death process rather than strict conservation, considering that these motifs are found on opposite strands of DNA in different species.
Fourth, despite the lack of extended sequence conservation, for all four genes we could readily identify motifs corresponding to transcription factor binding sites previously identified as functionally important for regulation of C. elegans orthologs. The motifs that we tested contributed to gene regulation of the orthologous cis elements, implying that gene regulatory output can be conserved, even among distantly-related organisms, as long as key gene regulatory connections—“kernels” [10,72] or “input-output devices” [73]—are maintained. This further reinforces the view that when developmental programs evolve, the regulatory “toolkit” controlling major patterning and cell-type specification programs remains relatively static [6]. Of course, the mere presence of these short motifs is not likely to be sufficient to explain regulatory output. For instance, we can find chance matches to GATA motifs important for elt-2 expression in many of the other sequences we tested, which do not drive expression in the intestinal precursor cells. Similarly, we can find matches to the AHR-1-like motif (that regulates unc-47 expression) in the elt-2 cis elements of C. elegans, C. briggsae, and T. spiralis, none of which drive expression in DVB, RIS, or AVL.
In this study we aimed to understand how the patterns of divergence of gene regulatory mechanisms between closely related species scale up over long evolutionary times. Models have predicted [74] that regulatory control can be shifted from one site to another within a cis-regulatory sequence; if these sites arise somewhat stochastically, longer wait times increase the likelihood of new sites originating and being optimized. These new sites could diminish the strength of purifying selection acting on ancestral motifs [75]. On shorter evolutionary time scales, new motifs do not have the time to arise, so function relies on conservation of existing sites [76]. As the same process plays out over different timespans, cis-regulatory conservation remains common among close relatives, but is mostly absent among more distantly-related species.
Naturally, the rates of divergence and motif turnover are different for different genes. An important factor determining the rate of evolution could be the organization of a cis element, whether it is flexible [77,78] or constrained [79], a billboard or an enhanceosome [80,81]. Modeling suggests that some enhancer sequences are inherently more prone to higher rates of turnover than others [74]. Better understanding of the structure of cis-regulatory elements may provide clues to their evolution [70,82,83].
Practically, our results advocate the use of C. elegans as a convenient and reliable experimental system for testing the functions of putative regulatory elements from nematode species, many of them parasites of major economic and medical significance, that are not amenable to transgenic studies [45]. Furthermore, the fact that C. elegans has been a genetic model system for decades means that the wealth of information about gene regulation in this species could be leveraged into hypothesis-driven investigation of non-model organisms.
As discussed above, functionally conserved sequences can retain no more sequence conservation than would be expected by chance. Indeed, motifs that mediate functional conservation, namely transcription factor binding sites, are short enough that they would be likely to be found by chance in sequences of the lengths of these cis elements. By the measures of sequence conservation we applied, including alignment-free methods, M. hapla does not have appreciably greater sequence similarity to C. elegans than does T. spiralis. Nevertheless, M. hapla cis elements of all four tested genes drive more consistent and correct expression in transgenic C. elegans than elements from T. spiralis do. This means that some sequence properties were retained to a greater extent by the more closely related species. Identification of these properties would lead to a better understanding of function and evolution of gene regulatory elements.
Materials and Methods
Cloning of cis-regulatory elements
Orthologous genes from C. briggsae, M. hapla, B. malayi, and T. spiralis were identified as best tblastn/blastx matches with the C. elegans protein sequence. For C. briggsae, B. malayi, and T. spiralis, the genome browser on Wormbase was used. For M. hapla, the genome browser at www.hapla.org was used. Forward primers were designed proximal to the next upstream gene, or failing that the 5’-most part of the contig on which the orthologous coding sequence was found. Reverse primers were selected to make in-frame translational fusions with GFP in the 5’-most part of the gene with protein coding sequence similarity with C. elegans. The only cases in which this was not possible were B. malayi and T. spiralis elt-2, in which protein-coding conservation started deep in the protein-coding sequence, and the fusions were generated in the first exon. A previous study of elt-2 from a parasitic nematode, the less divergent Haemonchus contortus [84], found that despite protein sequence divergence from C. elegans, the H. contortus protein retained function when expressed transgenically in C. elegans by a C. elegans heat shock promoter, so this increases our confidence that these can be elt-2 orthologs despite coding sequence divergence. In all cases, the start codon of the ortholog was included in the fusion. To generate reporter transgenes, upstream non-coding sequences were PCR amplified from genomic DNA and cloned upstream of GFP into the Fire vector pPD95.75, or upstream of mCherry (for C. elegans genes), which was inserted in place of GFP in a modified vector pPD95.75 [85]. elt-2 transgenes carried a nuclear localization signal upstream of GFP or mCherry. Prior to injection, all transgenes were sequenced to ensure accuracy.
Transgenes and strains
We injected a mixture (5 ng/μL (for C. briggsae; 10 ng/μL for the other species) promoter::GFP plasmid, 5 or 10 ng/μL promoter::mCherry plasmid, 5 ng/μL pha-1 rescue transgene, 100 ng/μL salmon sperm DNA) into temperature-sensitive C. elegans pha-1(e2123) strain [86]. Transformants were selected at 25°C. Multiple strains were examined for each transgenic construct. Statistical analyses of consistency of expression patterns between strains and individuals are presented in S2 Table, since extrachromosomal transgenes are known to have more variable expression than integrated transgenes. Our previous reports [36,37] thoroughly addressed the similarity of expression driven by transgenes of different types—extrachromosomal, multicopy integrated, and single-copy integrated. We found that while the strength of the signal increases with multiple copies, and variability increases with extrachromosomal transgenes, the patterns generated by these different methods are consistent.
The structures of extrachromosomal transgene arrays are generally not known. Although there is a possibility of cross-talk between promoters from different species if they land close enough when the DNA is concatenated, we mitigate against this by including an excess of salmon sperm DNA and vector sequence to create distance between the promoters and reduce the repetitiveness of the arrays. We measure expression in multiple independent strains. We also tested several of the highly divergent promoters alone, without a coexpressed C. elegans-DNA-driven reporter (S15 Fig). Without the coexpressed mCherry marker, cells were more difficult to identify, so counts were not attempted for these strains, but expression was observed in the same subsets of cells that it was observed in coexpressing lines.
The coexpressing strains also allowed us to control for the mosaicism inherent in extrachromosomal transgenes. Since the transgenes are concatenated, mCherry and GFP are inherited together by cells, and if array loss or silencing causes the loss of expression of one marker, the other will also disappear. This is why, for most of our quantification, we describe expression as the ratio of mCherry (control) positive cells that also express GFP (see Figs 2–5 and 7).
Mutagenesis of unc-47 cis elements
We tested the functions of motifs corresponding to consensus sequences of binding sites of UNC-30 [47] (TAATCC) and AHR-1-like [37] (CACGC). Motifs were identified using the ConsensusSequence feature on the GeneGrokker web server (https://genegrokker.biology.uiowa.edu). Of the several UNC-30 motifs in the M. hapla unc-47 element, we selected for mutagenesis the longest extended match to the C. elegans sequence: aTAATCCcc (reverse complement, since the motif is found on the (-) strand). This motif was mutagenized to aTAGGCGac (changes highlighted). Of the several matches to the AHR-1-like motif in the T. spiralis unc-47 element, the motif selected for mutagenesis was a palindromic sequence (CACGCGTG), which matches two overlapping instances of the AHR-1-like motif (one on each strand). This sequence was mutagenized to CACAAGTG, changing the CACGC sequence on the (+) strand to CACAA and on the (-) strand to CACTT.
All mutations were introduced by PCR with overlapping, opposite-facing primers carrying the mutant sequence. Primers were used to amplify plasmid DNA carrying the wild-type sequence. Following PCR, the reaction was digested with the methylation sensitive restriction enzyme DpnI to selectively digest the wild-type plasmid template. A second PCR reaction was performed, amplifying the mutagenized cis element and some flanking vector sequence. This PCR product was purified and digested for directional cloning back into the expression vector. Mutations were verified by sequencing before microinjection.
Microscopy
Mixed-stage populations of C. elegans carrying transgenes were grown with abundant food. Worms of appropriate stages were selected. These were immobilized on agar slides with 10 mM sodium azide in M9 buffer. The slides were examined on a Leica DM5000B compound microscope under 400-fold magnification, except in S4 and S15 Figs, which include micrographs taken at 1000-fold magnification (as labeled). Exposure times varied as necessary for each transgene. Each photograph showing worms in figures is composed of several images of the same individual capturing anterior, middle, and posterior sections, as well as shallow and deep focus. False-colored composite images were generated with QCapturePro. Brightness, contrast, and scaling of images were adjusted where necessary in final display items.
The stronger background visible in the GFP images relative to their mCherry counterparts may have several explanations. First, GFP has higher background relative to mCherry, and the autofluorescence of the gut is detectable with GFP filters. Second, longer exposure times were necessary to capture expression of the more weakly expressing exogenous cis-regulatory elements. Finally, GFP fluorescence in the gut is a known site of off-target expression [38]. Worms were also injected with a subset of the GFP transgenes carrying the other nematode’s cis elements alone (without a C. elegans mCherry control), and results were consistent (S1 and S2 Tables, S15 Fig).
Cell counting
Young adult individuals were examined for gene expression, except for elt-2, in which case pretzel stage embryos and L1 larvae were counted. Worms without any visible fluorescence were assumed to have lost the transgene and were ignored. Presence of mCherry was a precondition for the worm to be counted, but without regard for the strength or completeness of the mCherry expression pattern.
Sequence analysis
Motifs matching between C. elegans and each orthologous cis element (identified by the Mirror tool on the GeneGrokker web server https://genegrokker.biology.uiowa.edu) were mapped back to the orthologous sequence, and the total amount of the sequence covered by blocks of conservation of different sizes is plotted in Fig 6A.
Empirical p-values for the sequence similarity of the C. elegans elements to their orthologs were calculated by generating 1000 reshuffled replicates of the C. elegans sequence. Replicates were generated using single, di-, and tri-nucleotide sampling from the C. elegans sequence. Each replicate was compared to each ortholog and scored for similarity in windows of different sizes. The distributions of these similarity scores were plotted (Fig 6F–6I). The actual number of observed motif matches between the C. elegans sequence and its relevant orthologs were indicated on those distributions. The reported p-value is equal to the number of shuffled replicates that had more motif matches than the actual number, divided by 1000. Only C. briggsae had more similar motifs than would be expected by chance.
Potential caveats
We used multicopy extrachromosomal transgenes, which could have made the detected levels of expression higher and less consistent than what would have been produced by single-copy transgenes. In previous work [36,37] we did determine that, at least in the case of unc-47 from C. elegans and C. briggsae, the nature of the transgene (multi- vs. single-copy, extrachromosomal vs. integrated) did not change the pattern, but rather the amount and consistency of expression. If the same principle holds for the genes examined here, the conserved patterns we detected represent the cell types where the foreign cis elements are truly active in C. elegans, but the expression levels could be overestimated. The fact that in most instances only subsets of the overall pattern were conserved suggests that artificially higher expression levels were not solely responsible for the conserved expression patterns we detected.
Any apparent divergence—i.e. incongruence between the pattern driven by the C. elegans cis element and its orthologs—could be due to cis-regulatory changes (in the function of the donor element), trans-regulatory changes (in the function of transcription factor(s) in C. elegans), or due to the experimental combination of the two. In addition, endogenous expression patterns may have diverged between C. elegans and other species. For technical reasons, it is difficult to determine endogenous patterns of gene expression in divergent parasitic nematodes used in this study. It is even more difficult to generate transgenic animals in these species. These technical limitations make it essentially impossible to assess divergence in endogenous expression patterns or to disentangle their causes (that is, cis vs. trans changes). For these reasons, we focused on enumerating similarities, rather than differences in expression. Our tests actually underestimate the extent of regulatory conservation, because a failure of a cis element from a distant nematode when tested in C. elegans may reflect a genuine divergence in cis-regulation in that species that was compensated in trans, therefore maintaining the same overall expression pattern.
Supporting Information
(A-C) C. elegans unc-47 regulatory sequence drives expression of mCherry in all transgenic strains; (A) M. hapla, (B) B. malayi, (C) T. spiralis unc-47 regulatory sequences drive expression of GFP. (D) T. spiralis unc-47 element drives expression of GFP in RIS and DVB. Animals photographed at 400x magnification. Images are mosaics of single animals.
(PDF)
(A-C) C. elegans unc-25 regulatory sequence drives expression of mCherry in all transgenic strains; (A) M. hapla, (B) B. malayi, (C) T. spiralis unc-25 regulatory sequences drive expression of GFP. Animals photographed at 400x magnification. Images are mosaics of single animals.
(PDF)
(A-D) C. elegans mec3 regulatory sequence drives expression of mCherry in all transgenic strains; (A) C. briggsae, (B) M. hapla, (C) B. malayi, (D) T. spiralis mec-3 regulatory sequences drive expression of GFP. Animals photographed at 400x magnification. Images are mosaics of single animals.
(PDF)
(A) B. malayi mec-3 regulatory sequence drives expression of GFP. Animal photographed at 400x magnification, ventral cord at bottom. Image is a mosaic of single animals. (B) 1000x magnification of animal with ventral side up. (C) 1000x magnification of animal with ventral side up, vulva at center.</SI_Caption>
(PDF)
(A-C) C. elegans unc-25 regulatory sequence drives expression of mCherry in all transgenic strains; (A) C. briggsae, (B) M. hapla, (C) B. malayi elt-2 regulatory sequences drive expression of GFP. Stages shown are 8E, comma, and pretzel embryonic stages, with L1 larval stage below. Animals photographed at 400x magnification.</SI_Caption>
(PDF)
All blocks of sequence identity in window sizes shown for each comparison with positions within the upstream non-coding sequence.
(DOCX)
Motifs with identity between C. elegans and orthologous unc-25 upstream sequences. All blocks of sequence identity in window sizes shown for each comparison with positions within the upstream non-coding sequence.
(DOCX)
Motifs with identity between C. elegans and orthologous mec-3 upstream sequences. All blocks of sequence identity in window sizes shown for each comparison with positions within the upstream non-coding sequence.
(DOCX)
Motifs with identity between C. elegans and orthologous elt-2 upstream sequences. All blocks of sequence identity in window sizes shown for each comparison with positions within the upstream non-coding sequence.
(DOCX)
Graphs showing the proportion of sequence similarity over different window sizes for unc-25, mec-3, and elt-2. The number of identical 10 nt blocks between each of the nematode relatives and the C. elegans upstream sequence of the same genes shown in red, on histograms showing the number of predicted 10 nt matches between the relatives’ sequences and 1000 reshuffled C. elegans sequences that preserve tri-nucleotide frequencies.
(PDF)
Cartoons depicting all orthologous upstream unc-25 sequences fused to mCherry (C. elegans) or GFP (all others) near the translation start site (bent arrow), or further downstream. Exons are thick black boxes, introns are gray lines. UNC-30 (box arrow) consensus motifs are shown above, in uppercase letters; conserved flanking nucleotides shown in lowercase. Locations of motifs relative to the endogenous translation start site are indicated. Daggers denote binding sites found to be bound by UNC-30. See S1 Text.
(PDF)
Cartoons depicting the all orthologous upstream mec-3 sequences fused to GFP near the translation start site (bent arrow) or further downstream. Exons are thick black boxes, introns are gray lines. UNC-86 (triangle) and MEC-3 (fletched arrow) consensus motifs are shown above. Locations of motifs relative to the endogenous translation start site are indicated. Several distal motifs are omitted from C. elegans, C. briggsae, and B. malayi. See S1 Text.
(PDF)
Cartoons depicting the all orthologous upstream elt-2 sequences fused to GFP near the translation start site (bent arrow). Gut-enriched extended GATA motif (sideways heart) and generic GATA motif (vertical lines) are shown above. Locations of motifs relative to the endogenous translation start site are indicated. See S1 Text.</SI_Caption>
(PDF)
(A) Mutant M. hapla unc-47 element drives expression of GFP. C. elegans unc-47 regulatory sequence drives expression of mCherry. Images are mosaics of single animals. Mutant T. spiralis unc-47 regulatory element drives expression of GFP, but not consistently in RIS (B) or DVB (C). μ denotes mutated unc-47 cis elements. C. elegans unc-47 regulatory sequence drives expression of mCherry in these cells. All animals photographed at 400x magnification.
(PDF)
(A) M. hapla unc-47::GFP is expressed in several D-type neurons of the ventral cord. (B) T. spiralis unc-47::GFP is expressed in RIS and DVB. (C) M. hapla mec-3::GFP is expressed in the head neuron FLP.
(PDF)
(XLSX)
(XLSX)
(DOCX)
Acknowledgments
We thank the following people for genomic DNA of the parasitic worms: David Bird (North Carolina State University) for M. hapla, Susan Haynes (NIAID/NIH) for B. malayi, Dante Zarlenga (USDA) for T. spiralis. We thank Erin Aprison, Daniel Matute, Chip Ferguson, Marty Kreitman, John Reinitz, and Urs Schmidt-Ott for critical reading of the manuscript, and members of the Ruvinsky laboratory for helpful discussions.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by an NIH pre-doctoral training grant (5 T32 HD055164) and an NSF Graduate Research Fellowship to KLG, an NSF Graduate Research Fellowship to RKA, and grant support from the NSF (IOS-0843504, IOS-1355093) to IR. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Slack JMW, Holland PWH, Graham CF (1993) The Zootype and the Phylotypic Stage. Nature 361: 490–492. [DOI] [PubMed] [Google Scholar]
- 2. Panganiban G, Irvine SM, Lowe C, Roehl H, Corley LS, et al. (1997) The origin and evolution of animal appendages. Proc Natl Acad Sci U S A 94: 5162–5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Holley SA, Jackson PD, Sasai Y, Lu B, De Robertis EM, et al. (1995) A conserved system for dorsal-ventral patterning in insects and vertebrates involving sog and chordin. Nature 376: 249–253. [DOI] [PubMed] [Google Scholar]
- 4. Gehring WJ (2005) New perspectives on eye development and the evolution of eyes and photoreceptors. J Hered 96: 171–184. [DOI] [PubMed] [Google Scholar]
- 5. Fukushige T, Brodigan TM, Schriefer LA, Waterston RH, Krause M (2006) Defining the transcriptional redundancy of early bodywall muscle development in C. elegans: evidence for a unified theory of animal muscle development. Genes Dev 20: 3395–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carroll SB (2008) Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134: 25–36. 10.1016/j.cell.2008.06.030 [DOI] [PubMed] [Google Scholar]
- 7. Takahashi H, Mitani Y, Satoh G, Satoh N (1999) Evolutionary alterations of the minimal promoter for notochord-specific Brachyury expression in ascidian embryos. Development 126: 3725–3734. [DOI] [PubMed] [Google Scholar]
- 8. Oda-Ishii I, Bertrand V, Matsuo I, Lemaire P, Saiga H (2005) Making very similar embryos with divergent genomes: conservation of regulatory mechanisms of Otx between the ascidians Halocynthia roretzi and Ciona intestinalis. Development 132: 1663–1674. [DOI] [PubMed] [Google Scholar]
- 9. True JR, Haag ES (2001) Developmental system drift and flexibility in evolutionary trajectories. Evol Dev 3: 109–119. [DOI] [PubMed] [Google Scholar]
- 10. Hinman VF, Davidson EH (2007) Evolutionary plasticity of developmental gene regulatory network architecture. Proc Natl Acad Sci U S A 104: 19404–19409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rebeiz M, Stone T, Posakony JW (2005) An ancient transcriptional regulatory linkage. Dev Biol 281: 299–308. [DOI] [PubMed] [Google Scholar]
- 12. Goltsev Y, Hsiong W, Lanzaro G, Levine M (2004) Different combinations of gap repressors for common stripes in Anopheles and Drosophila embryos. Dev Biol 275: 435–446. [DOI] [PubMed] [Google Scholar]
- 13. Gordon KL, Ruvinsky I (2012) Tempo and Mode in Evolution of Transcriptional Regulation. Plos Genet 8: e1002432–e1002432. 10.1371/journal.pgen.1002432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guastella J, Johnson CD, Stretton AOW (1991) Gaba-Immunoreactive Neurons in the Nematode Ascaris. J Comp Neurol 307: 584–597. [DOI] [PubMed] [Google Scholar]
- 15. Johnson C, Stretton A (1987) GABA-immunoreactivity in inhibitory motor neurons of the nematode Ascaris. J Neurosci 7: 223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Voronov D, Panchin Y (1998) Cell lineage in marine nematode Enoplus brevis. Development 125: 143–150. [DOI] [PubMed] [Google Scholar]
- 17. Wiegner O, Schierenberg E (1999) Regulative development in a nematode embryo: a hierarchy of cell fate transformations. Dev Biol 215: 1–12. [DOI] [PubMed] [Google Scholar]
- 18. Goldstein B (2001) On the evolution of early development in the Nematoda. Philos Trans R Soc Lond B Biol Sci 356: 1521–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schulze J, Schierenberg E (2011) Evolution of embryonic development in nematodes. Evodevo 2: 18 10.1186/2041-9139-2-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kiontke K, Barrière A, Kolotuev I, Podbilewicz B, Sommer R, et al. (2007) Trends, stasis, and drift in the evolution of nematode vulva development. Curr Biol 17: 1925–1937. [DOI] [PubMed] [Google Scholar]
- 21. Hill RC, de Carvalho CE, Salogiannis J, Schlager B, Pilgrim D, et al. (2006) Genetic flexibility in the convergent evolution of hermaphroditism in Caenorhabditis nematodes. Dev Cell 10: 531–538. [DOI] [PubMed] [Google Scholar]
- 22. Wang X, Chamberlin HM (2004) Evolutionary innovation of the excretory system in Caenorhabditis elegans. Nat Genet 36: 231–232. [DOI] [PubMed] [Google Scholar]
- 23. Verster AJ, Ramani AK, McKay SJ, Fraser AG (2014) Comparative RNAi screens in C. elegans and C. briggsae reveal the impact of developmental system drift on gene function. PLoS Genet 10: e1004077 10.1371/journal.pgen.1004077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rota-Stabelli O, Daley AC, Pisani D (2013) Molecular timetrees reveal a Cambrian colonization of land and a new scenario for ecdysozoan evolution. Curr Biol 23: 392–398. 10.1016/j.cub.2013.01.026 [DOI] [PubMed] [Google Scholar]
- 25. Blaxter M (2011) Nematodes: The Worm and Its Relatives. PLoS Biol 9: e1001050 10.1371/journal.pbio.1001050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. The C. elegans Sequencing Consortium (1998) Genome Sequence of the Nematode C. elegans: A Platform for Investigating Biology. Science 282: 2012–2018. [DOI] [PubMed] [Google Scholar]
- 27. Stein LD, Bao Z, Blasiar D, Blumenthal T, Brent MR, et al. (2003) The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol 1: E45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Abad P, Gouzy J, Aury J-M, Castagnone-Sereno P, Danchin EGJ, et al. (2008) Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat Biotechnol 26: 909–915. 10.1038/nbt.1482 [DOI] [PubMed] [Google Scholar]
- 29. Opperman CH, Bird DM, Williamson VM, Rokhsar DS, Burke M, et al. (2008) Sequence and genetic map of Meloidogyne hapla: A compact nematode genome for plant parasitism. Proc Natl Acad Sci U S A 105: 14802–14807. 10.1073/pnas.0805946105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Blaxter M, Daub J, Guiliano D, Parkinson J, Whitton C (2002) The Brugia malayi genome project: expressed sequence tags and gene discovery. Trans R Soc Trop Med Hyg 96: 7–17. [DOI] [PubMed] [Google Scholar]
- 31. Ghedin E, Wang S, Spiro D, Caler E, Zhao Q, et al. (2007) Draft genome of the filarial nematode parasite Brugia malayi. Science 317: 1756–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mitreva M, Jasmer DP, Zarlenga DS, Wang Z, Abubucker S, et al. (2011) The draft genome of the parasitic nematode Trichinella spiralis. Nat Genet 43: 228–235. 10.1038/ng.769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McIntire SL, Reimer RJ, Schuske K, Edwards RH, Jorgensen EM (1997) Identification and characterization of the vesicular GABA transporter. Nature 389: 870–876. [DOI] [PubMed] [Google Scholar]
- 34. McIntire SL, Jorgensen E, Horvitz HR (1993) Genes required for GABA function in Caenorhabditis elegans. Nature 364: 334–337. [DOI] [PubMed] [Google Scholar]
- 35. Jin Y, Jorgensen E, Hartwieg E, Horvitz HR (1999) The Caenorhabditis elegans Gene unc-25 Encodes Glutamic Acid Decarboxylase and Is Required for Synaptic Transmission But Not Synaptic Development. J Neurosci 19: 539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Barriere A, Gordon KL, Ruvinsky I (2011) Distinct Functional Constraints Partition Sequence Conservation in a cis-Regulatory Element. PLoS Genet 7: e1002095 10.1371/journal.pgen.1002095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barrière A, Gordon KL, Ruvinsky I (2012) Coevolution within and between regulatory loci can preserve promoter function despite evolutionary rate acceleration. PLoS Genet 8: e1002961 10.1371/journal.pgen.1002961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Barrière A, Ruvinsky I (2014) Pervasive Divergence of Transcriptional Gene Regulation in Caenorhabditis Nematodes. PLoS Genet 10: e1004435 10.1371/journal.pgen.1004435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ruvinsky I, Ruvkun G (2003) Functional tests of enhancer conservation between distantly related species. Development 130: 5133–5142. [DOI] [PubMed] [Google Scholar]
- 40. Way JC, Chalfie M (1988) mec-3, a homeobox-containing gene that specifies differentiation of the touch receptor neurons in C. elegans. Cell 54: 5–16. [DOI] [PubMed] [Google Scholar]
- 41. Way JC, Chalfie M (1989) The mec-3 gene of Caenorhabditis elegans requires its own product for maintained expression and is expressed in three neuronal cell types. Genes Dev 3: 1823–1833. [DOI] [PubMed] [Google Scholar]
- 42. Xue D, Finney M, Ruvkun G, Chalfie M (1992) Regulation of the mec-3 gene by the C.elegans homeoproteins UNC-86 and MEC-3. EMBO J 11: 4969–4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fukushige T, Hawkins MG, McGhee JD (1998) The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev Biol 198: 286–302. [PubMed] [Google Scholar]
- 44. Hawkins MG, McGhee JD (1995) elt-2, a Second GATA Factor from the Nematode Caenorhabditis elegans. J Biol Chem 270: 14666–14671. [DOI] [PubMed] [Google Scholar]
- 45. Gilleard JS (2004) The use of Caenorhabditis elegans in parasitic nematode research. Parasitology 128: S49–S70. [DOI] [PubMed] [Google Scholar]
- 46. McGhee JD (2012) The Caenorhabditis elegans intestine. Wiley Interdiscip Rev Dev Biol 2: 347–367. 10.1002/wdev.93 [DOI] [PubMed] [Google Scholar]
- 47. Eastman C, Horvitz HR, Jin YS (1999) Coordinated transcriptional regulation of the unc-25 glutamic acid decarboxylase and the unc-47 GABA vesicular transporter by the Caenorhabditis elegans UNC-90 homeodomain protein. J Neurosci 19: 6225–6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rebeiz M, Castro B, Liu F, Yue F, Posakony JW (2012) Ancestral and conserved cis-regulatory architectures in developmental control genes. Dev Biol 362: 282–294. 10.1016/j.ydbio.2011.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Navratilova P, Fredman D, Hawkins TA, Turner K, Lenhard B, et al. (2009) Systematic human/zebrafish comparative identification of cis-regulatory activity around vertebrate developmental transcription factor genes. Dev Biol 327: 526–540. 10.1016/j.ydbio.2008.10.044 [DOI] [PubMed] [Google Scholar]
- 50. Blanco J, Girard F, Kamachi Y, Kondoh H, Gehring WJ (2005) Functional analysis of the chicken delta1-crystallin enhancer activity in Drosophila reveals remarkable evolutionary conservation between chicken and fly. Development 132: 1895–1905. [DOI] [PubMed] [Google Scholar]
- 51. Brugger SM, Merrill AE, Torres-Vazquez J, Wu N, Ting M-C, et al. (2004) A phylogenetically conserved cis-regulatory module in the Msx2 promoter is sufficient for BMP-dependent transcription in murine and Drosophila embryos. Development 131: 5153–5165. [DOI] [PubMed] [Google Scholar]
- 52. Clarke SL, VanderMeer JE, Wenger AM, Schaar BT, Ahituv N, et al. (2012) Human developmental enhancers conserved between deuterostomes and protostomes. PLoS Genet 8: e1002852 10.1371/journal.pgen.1002852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shimeld SM, Purkiss AG, Dirks RPH, Bateman O a, Slingsby C, et al. (2005) Urochordate betagamma-crystallin and the evolutionary origin of the vertebrate eye lens. Curr Biol 15: 1684–1689. [DOI] [PubMed] [Google Scholar]
- 54. Xu PX, Zhang X, Heaney S, Yoon a, Michelson a M, et al. (1999) Regulation of Pax6 expression is conserved between mice and flies. Development 126: 383–395. [DOI] [PubMed] [Google Scholar]
- 55. Royo JL, Maeso I, Irimia M, Gao F, Peter IS, et al. (2011) Transphyletic conservation of developmental regulatory state in animal evolution. Proc Natl Acad Sci U S A 108: 14186–14191. 10.1073/pnas.1109037108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ayyar S, Negre B, Simpson P, Stollewerk A (2010) An arthropod cis-regulatory element functioning in sensory organ precursor development dates back to the Cambrian. BMC Biol 8: 127 10.1186/1741-7007-8-127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Locascio A, Aniello F, Amoroso A, Manzanares M, Krumlauf R, et al. (1999) Patterning the ascidian nervous system: structure, expression and transgenic analysis of the CiHox3 gene. Development 126: 4737–4748. [DOI] [PubMed] [Google Scholar]
- 58. Arnone M, Davidson E (1997) The hardwiring of development: organization and function of genomic regulatory systems. Development 124: 1851–1864. [DOI] [PubMed] [Google Scholar]
- 59. Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, et al. (2003) The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol 20: 1377–1419. [DOI] [PubMed] [Google Scholar]
- 60. Kimura-Yoshida C, Kitajima K, Oda-Ishii I, Tian E, Suzuki M, et al. (2004) Characterization of the pufferfish Otx2 cis-regulators reveals evolutionarily conserved genetic mechanisms for vertebrate head specification. Development 131: 57–71. [DOI] [PubMed] [Google Scholar]
- 61. Stern DL (2000) Perspective: Evolutionary developmental biology and the problem of variation. Evolution (N Y) 54: 1079–1091. [DOI] [PubMed] [Google Scholar]
- 62. He Z, Eichel K, Ruvinsky I (2011) Functional Conservation of Cis-Regulatory Elements of Heat-Shock Genes over Long Evolutionary Distances. PLoS One 6: e22677 10.1371/journal.pone.0022677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Franks RR, Hough-Evans BR, Britten RJ, Davidson EH (1988) Spatially deranged though temporally correct expression of Strongylocentrotus purpuratus actin gene fusion in transgenic embryos of a different sea urchin family. Genes Dev 2: 1–12. [DOI] [PubMed] [Google Scholar]
- 64. Cooper GM, Brown CD (2008) Qualifying the relationship between sequence conservation and molecular function. Genome Res 18: 201–205. 10.1101/gr.7205808 [DOI] [PubMed] [Google Scholar]
- 65. Dutilh BE, Huynen MA, Snel B (2006) A global definition of expression context is conserved between orthologs, but does not correlate with sequence conservation. BMC Genomics 7: 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Fisher S, Grice EA, Vinton RM, Bessling SL, McCallion AS (2006) Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science 312: 276–279. [DOI] [PubMed] [Google Scholar]
- 67. McGaughey DM, Vinton RM, Huynh J, Al-Saif A, Beer MA, et al. (2008) Metrics of sequence constraint overlook regulatory sequences in an exhaustive analysis at phox2b. Genome Res 18: 252–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nelson AC, Wardle FC (2013) Conserved non-coding elements and cis regulation: actions speak louder than words. Development 140: 1385–1395. 10.1242/dev.084459 [DOI] [PubMed] [Google Scholar]
- 69. Taher L, McGaughey DM, Maragh S, Aneas I, Bessling SL, et al. (2011) Genome-wide identification of conserved regulatory function in diverged sequences. Genome Res 21: 1139–1149. 10.1101/gr.119016.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Erives A, Levine M (2004) Coordinate enhancers share common organizational features in the Drosophila genome. Proc Natl Acad Sci U S A 101: 3851–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Aparicio S, Morrison A, Gould A, Gilthorpe J, Chaudhuri C, et al. (1995) Detecting Conserved Regulatory Elements with the Model Genome of the Japanese Puffer Fish, Fugu Rubripes. Proc Natl Acad Sci U S A 92: 1684–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hinman VF, Nguyen AT, Cameron RA, Davidson EH (2003) Developmental gene regulatory network architecture across 500 million years of echinoderm evolution. Proc Natl Acad Sci U S A 100: 13356–13361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Stern DL, Orgogozo V (2008) The loci of evolution: How predictable is genetic evolution? Evolution (N Y) 62: 2155–2177. 10.1111/j.1558-5646.2008.00450.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bullaughey K (2011) Changes in Selective Effects Over Time Facilitate Turnover of Enhancer Sequences. Genetics 187: 567–U328. 10.1534/genetics.110.121590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. He BZ, Holloway AK, Maerkl SJ, Kreitman M (2011) Does Positive Selection Drive Transcription Factor Binding Site Turnover? A Test with Drosophila Cis-Regulatory Modules. Plos Genet 7: e1002053 10.1371/journal.pgen.1002053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Brown CD, Johnson DS, Sidow A (2007) Functional architecture and evolution of transcriptional elements that drive gene coexpression. Science 317: 1557–1560. [DOI] [PubMed] [Google Scholar]
- 77. Liberman LM, Stathopoulos A (2009) Design flexibility in cis-regulatory control of gene expression: synthetic and comparative evidence. Dev Biol 327: 578–589. 10.1016/j.ydbio.2008.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Smith RP, Taher L, Patwardhan RP, Kim MJ, Inoue F, et al. (2013) Massively parallel decoding of mammalian regulatory sequences supports a flexible organizational model. Nat Genet 45: 1021–1028. 10.1038/ng.2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Papatsenko D, Goltsev Y, Levine M (2009) Organization of developmental enhancers in the Drosophila embryo. Nucleic Acids Res 37: 5665–5677. 10.1093/nar/gkp619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Arnosti DN, Kulkarni MM (2005) Transcriptional enhancers: Intelligent enhanceosomes or flexible billboards? J Cell Biochem 94: 890–898. [DOI] [PubMed] [Google Scholar]
- 81. Papatsenko D, Levine M (2007) A rationale for the enhanceosome and other evolutionarily constrained enhancers. Curr Biol 17: R955–R957. [DOI] [PubMed] [Google Scholar]
- 82. Swanson CI, Schwimmer DB, Barolo S (2011) Rapid Evolutionary Rewiring of a Structurally Constrained Eye Enhancer. Curr Biol 21: 1186–1196. 10.1016/j.cub.2011.05.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Crocker J, Tamori Y, Erives A (2008) Evolution acts on enhancer organization to fine-tune gradient threshold readouts. PLoS Biol 6: e263 10.1371/journal.pbio.0060263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Couthier A, Smith J, McGarr P, Craig B, Gilleard JS (2004) Ectopic expression of a Haemonchus contortus GATA transcription factor in Caenorhabditis elegans reveals conserved function in spite of extensive sequence divergence. Mol Biochem Parasitol 133: 241–253. [DOI] [PubMed] [Google Scholar]
- 85.Xu S, Fire A, Seydoux G, Okkema P (1995) Fire Lab Vector Kit. June 1995.
- 86. Granato M, Schnabel H, Schnabel R (1994) pha-1, a selectable marker for gene-transfer in C. elegans. Nucleic Acids Res 22: 1762–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A-C) C. elegans unc-47 regulatory sequence drives expression of mCherry in all transgenic strains; (A) M. hapla, (B) B. malayi, (C) T. spiralis unc-47 regulatory sequences drive expression of GFP. (D) T. spiralis unc-47 element drives expression of GFP in RIS and DVB. Animals photographed at 400x magnification. Images are mosaics of single animals.
(PDF)
(A-C) C. elegans unc-25 regulatory sequence drives expression of mCherry in all transgenic strains; (A) M. hapla, (B) B. malayi, (C) T. spiralis unc-25 regulatory sequences drive expression of GFP. Animals photographed at 400x magnification. Images are mosaics of single animals.
(PDF)
(A-D) C. elegans mec3 regulatory sequence drives expression of mCherry in all transgenic strains; (A) C. briggsae, (B) M. hapla, (C) B. malayi, (D) T. spiralis mec-3 regulatory sequences drive expression of GFP. Animals photographed at 400x magnification. Images are mosaics of single animals.
(PDF)
(A) B. malayi mec-3 regulatory sequence drives expression of GFP. Animal photographed at 400x magnification, ventral cord at bottom. Image is a mosaic of single animals. (B) 1000x magnification of animal with ventral side up. (C) 1000x magnification of animal with ventral side up, vulva at center.</SI_Caption>
(PDF)
(A-C) C. elegans unc-25 regulatory sequence drives expression of mCherry in all transgenic strains; (A) C. briggsae, (B) M. hapla, (C) B. malayi elt-2 regulatory sequences drive expression of GFP. Stages shown are 8E, comma, and pretzel embryonic stages, with L1 larval stage below. Animals photographed at 400x magnification.</SI_Caption>
(PDF)
All blocks of sequence identity in window sizes shown for each comparison with positions within the upstream non-coding sequence.
(DOCX)
Motifs with identity between C. elegans and orthologous unc-25 upstream sequences. All blocks of sequence identity in window sizes shown for each comparison with positions within the upstream non-coding sequence.
(DOCX)
Motifs with identity between C. elegans and orthologous mec-3 upstream sequences. All blocks of sequence identity in window sizes shown for each comparison with positions within the upstream non-coding sequence.
(DOCX)
Motifs with identity between C. elegans and orthologous elt-2 upstream sequences. All blocks of sequence identity in window sizes shown for each comparison with positions within the upstream non-coding sequence.
(DOCX)
Graphs showing the proportion of sequence similarity over different window sizes for unc-25, mec-3, and elt-2. The number of identical 10 nt blocks between each of the nematode relatives and the C. elegans upstream sequence of the same genes shown in red, on histograms showing the number of predicted 10 nt matches between the relatives’ sequences and 1000 reshuffled C. elegans sequences that preserve tri-nucleotide frequencies.
(PDF)
Cartoons depicting all orthologous upstream unc-25 sequences fused to mCherry (C. elegans) or GFP (all others) near the translation start site (bent arrow), or further downstream. Exons are thick black boxes, introns are gray lines. UNC-30 (box arrow) consensus motifs are shown above, in uppercase letters; conserved flanking nucleotides shown in lowercase. Locations of motifs relative to the endogenous translation start site are indicated. Daggers denote binding sites found to be bound by UNC-30. See S1 Text.
(PDF)
Cartoons depicting the all orthologous upstream mec-3 sequences fused to GFP near the translation start site (bent arrow) or further downstream. Exons are thick black boxes, introns are gray lines. UNC-86 (triangle) and MEC-3 (fletched arrow) consensus motifs are shown above. Locations of motifs relative to the endogenous translation start site are indicated. Several distal motifs are omitted from C. elegans, C. briggsae, and B. malayi. See S1 Text.
(PDF)
Cartoons depicting the all orthologous upstream elt-2 sequences fused to GFP near the translation start site (bent arrow). Gut-enriched extended GATA motif (sideways heart) and generic GATA motif (vertical lines) are shown above. Locations of motifs relative to the endogenous translation start site are indicated. See S1 Text.</SI_Caption>
(PDF)
(A) Mutant M. hapla unc-47 element drives expression of GFP. C. elegans unc-47 regulatory sequence drives expression of mCherry. Images are mosaics of single animals. Mutant T. spiralis unc-47 regulatory element drives expression of GFP, but not consistently in RIS (B) or DVB (C). μ denotes mutated unc-47 cis elements. C. elegans unc-47 regulatory sequence drives expression of mCherry in these cells. All animals photographed at 400x magnification.
(PDF)
(A) M. hapla unc-47::GFP is expressed in several D-type neurons of the ventral cord. (B) T. spiralis unc-47::GFP is expressed in RIS and DVB. (C) M. hapla mec-3::GFP is expressed in the head neuron FLP.
(PDF)
(XLSX)
(XLSX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.