Summary
Leukemic cells disrupt normal patterns of blood cell formation, but little is understood about the mechanism. We investigated if leukemic cells alter functions of normal hematopoietic stem and progenitor cells. Exposure to chronic myelogenous leukemia (CML) caused normal mouse hematopoietic progenitor cells to divide more readily, altered their differentiation, and reduced their reconstitution and self-renewal potential. Interestingly, the normal bystander cells acquired gene expression patterns resembling their malignant counterparts. Therefore, much of the leukemia signature is mediated by extrinsic factors. Indeed, IL-6 was responsible for most of these changes. Compatible results were obtained when human CML were cultured with normal human hematopoietic progenitor cells. Furthermore, neutralization of IL-6 prevented these changes and treated the disease.
Keywords: HSC, CML, IL-6, differentiation, cytokines
Introduction
Most hematopoietic stem cells (HSCs) reside in the bone marrow and self-renew as necessary to maintain their numbers (Mercier et al., 2012). Additionally, a fraction of HSCs develop into progenitor cells that become lineage-restricted, and undergo extensive proliferation and differentiate to produce mature hematopoietic cells (Mayle et al., 2013; Venezia et al., 2004; Wilson et al., 2008). However, these normal processes are severely compromised with leukemia (Colmone et al., 2008; Hartwell et al., 2013; Hu et al., 2009; Krause et al., 2013; Schepers et al., 2013). While this could result from overcrowding by leukemic cells, it has been shown to occur with even low leukemic burden (Colmone et al., 2008). Considerable progress has been made in defining cells within marrow that support normal hematopoiesis (Morrison and Scadden, 2014). Referred to as niches, these environments are thought to include multipotent stromal cells (MSC), osteoblasts, and endothelial cells. Additionally, there is now evidence that leukemia alters their functions (Raaijmakers et al., 2010; Reynaud et al., 2011; Schepers et al., 2013; Zhang et al., 2012). However, consequences of those changes and the direct impact of the leukemic cells on stem and progenitor cells have not been adequately explored.
Myeloproliferative neoplasms are clonal disorders propagated by transformed HSCs. Chronic myelogenous leukemia (CML) is one such disorder, and it is characterized by a reciprocal translocation of the t(9;22)(q34;q11) loci. As a result, transformed cells express the BCR/ABL fusion protein (Ben-Neriah et al., 1986; Hooberman et al., 1989; Levine and Gilliland, 2008; Savona and Talpaz, 2008; Sawyers, 1999; Witte, 1988). This deregulated tyrosine kinase promotes leukemic growth by disrupting signaling pathways involved in cell survival, proliferation, and differentiation. The chronic phase of CML presents with increased numbers of circulating progenitors, anemia and splenomegaly (Petzer et al., 1996). At this time, leukemia-initiating cells (LIC) that can propagate disease are still present (Schemionek et al., 2010; Zhang et al., 2010) and retain the ability to make all blood cells, generating a vast expansion of malignant myeloid cells that displace normal hematopoiesis (Fialkow et al., 1977). In mice, these transformed progenitors are phenotypically similar to normal HSCs and are enriched within the Lin− Sca1+ c-KitHi fraction of bone marrow (KSL) (Holyoake et al., 1999; Hu et al., 2006; Maguer-Satta et al., 1996; Wang et al., 1998). Furthermore, processes that control normal HSC functions are also essential for LICs maintenance (Heidel et al., 2012; Lessard and Sauvageau, 2003; Reynaud et al., 2011; Warr et al., 2011; Zhao et al., 2007).
The chronic phase of CML cannot be efficiently modeled by transplantation of human cells into immunedeficient mice (Dazzi et al., 1998; Zhang et al., 2010). Therefore, our laboratory developed a BCR-ABL inducible mouse model that results in expression of this oncogenic fusion under the control of a tetracycline (Tet)-regulated 3’ enhancer of the murine stem cell leukemia (SCL) gene (Koschmieder et al., 2005; Schemionek et al., 2010). SCL-tTA × BCR-ABL double transgenic mice develop a disease similar to the chronic phase CML observed in patients. BCR-ABL expression following tetracycline withdrawal results in neutrophilic leukocytosis and splenomegaly. The chronic phase is characterized by progressive myeloid expansion, with accumulation of myeloid progenitors and mature granulocytes in the marrow and peripheral blood (PB) (Koschmieder et al., 2005; Schemionek et al., 2010). These CML cells are functionally heterogeneous and capable of maintaining a normal hierarchical differentiation process (Reynaud et al., 2011; Zhang et al., 2012).
This model made it possible to study normal hematopoietic cells while in close proximity to their leukemic counterparts, by transplanting inducible leukemic transgenic marrow cells together with normal marrow cells, and monitoring the effects of exposure of leukemic cells on normal HSPC function. Importantly, the leukemic cells altered the properties of normal HSCs and progenitors, and preventing these environmental changes have important therapeutic implications for the disease.
Results
Leukemic CML cells induce a myeloproliferative effect on normal hematopoietic cells
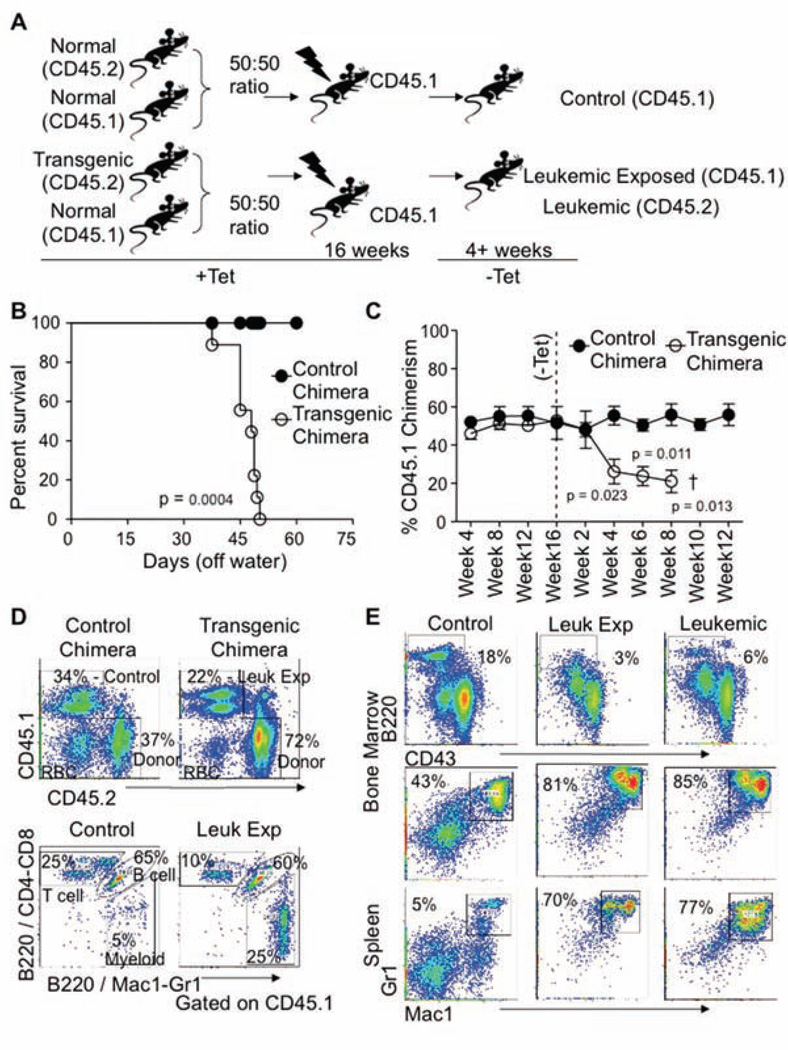
Previous studies from our laboratory and others using this inducible model of CML identified leukemia-initiating activity in the stem/progenitor fraction of the marrow (Reynaud et al., 2011; Schemionek et al., 2010; Zhang et al., 2010). To explore the effects of leukemic cells on normal hematopoietic cells (leukemic-exposed), C57BL/6-CD45.1 (normal) plus SCL-tTA×BCR-ABL-CD45.2 (transgenic) whole marrow cells were transplanted into lethally irradiated C57BL/6-CD45.1 mice. As controls, 2×106 whole marrow cells from C57BL/6-CD45.1 (normal) plus the same number from C57BL/6-CD45.2 (normal) mice were transplanted (Figure 1A). Recipients were then kept on tetracycline (Tet) water for 16 weeks to establish homeostasis within the chimeric mice. Upon removal of Tet, mice bearing transgenic cells developed a disease similar to chronic phase CML with severe myeloid cell expansion, coincident with reduction of lymphoid lineage cells in the marrow, spleen, and peripheral blood. The disease progressed such that ~4 weeks after induction, elevated peripheral blood leukocyte counts were observed (Figure S1A, available online), with increased cellularity in the spleen and decreased cellularity in the marrow. Chimeric mice with transgenic cells succumbed to the disease with the median survival times of 46 days after Tet withdrawal (Figure 1B). All mice transplanted with leukemic cells developed splenomegaly compared to control transplanted mice (Figure S1B). Leukemic cells altered the chimerism of normal cells (CD45.1+, leukemic-exposed) within these same mice from the time of induction as compared to the control transplants. Chimerism is shown as the percentage of CD45.1 cells to the total CD45 positive cells (CD45.1+ and CD45.2+) (Figure 1C).
Figure 1. Experimental design and establishment of a chimera chronic leukemia chimeric model.
A) Overview of the experimental design for generation of CML chimeras, 2×106 whole bone marrow (WBM) cells from either C57BL/6 mice (CD45.2; Control) or Scl-tTa-BCR-ABL (CD45.2; Transgenic) mice mixed with 2×106 C57BL/6 congenic for the CD45 locus (CD45.1; Normal) and transplanted into mice on tetracycline. Chimerism was established for 16 weeks before tetracycline withdrawal and induction of BCR-ABL in those cells carrying the two transgenes. Tissues cells were obtained 4 weeks after induction and CD45.1+ cells from control transplants; leukemic-exposed (CD45.1+ cells from leukemic mice), or leukemic cells (CD45.2+ transgenic cells from leukemic mice) were analyzed.
(B) Survival curves of control and transgenic chimeric mice after Tet withdrawal.
(C) The CD45.1 chimerism results are based on staining of lymphoid (B220/CD3) and myeloid (Mac1/Gr1) cells in peripheral blood versus CD45.2 (control or leukemic) over time, n=4 per time point. Error bars represent ± SEM.
(D) Representative FACS profiles from 4 experiments characterizing the peripheral blood of control or leukemic mice showing the chimerism of CD45.1 versus CD45.2 from control or transgenic chimeric mice 4 weeks after Tet removal [Upper Panel]. FACS profiles of CD45.1 cells in the peripheral blood of control or leukemic mice at 4 weeks post Tet removal. Cells were stained for B220/CD3 and B220/Mac1-Gr1. Cells that stain positive for both fluorochromes are B lineages (B220+), while those that stain only for B220/CD4 and CD8 are T lineage cells (CD4/8), and those cells that are positive for only B220/Mac1-Gr1 are of the myeloid lineage (Mac1-Gr1) [Lower Panel].
(E) Representative FACS profiles from the bone marrow gating for CD45.1+ B lineage cells (B220+ CD43+/−) of either control or transgenic chimeric mice at 4 weeks post Tet withdrawal. Dot plots from leukemic cells (CD45.2) from the transgenic chimeric mice are shown for comparison [right]. Similarly, bone marrow and spleens from control and transgenic chimeric mice were analyzed for the CD45.1+ myeloid lineage cells (Mac1+ and Gr1+) and leukemic cells (CD45.2+) are shown for comparison, n=5. See also Figure S1.
Flow cytometry analysis of the peripheral blood from chimeric mice showed increased numbers of donor (CD45.2+) cells in leukemic transplants compared to that observed in the control mice. Similar to that of the leukemic population, the leukemic-exposed (CD45.1+ normal from the leukemic mice) cells also have increased myeloid lineages with losses in both B and T lineages in the peripheral blood (Figure 1D and Figure S1C). To demonstrate that CD45.1 marked only normal cells lacking the two transgenes, populations of CD45.1+ and CD45.2+ cells were sorted from leukemic mice and only the CD45.2+ sorted cells contained both transgenes by PCR analysis, SCL-tTA and BCR-ABL. (Figure S1D). Increased percentage and absolute numbers of myeloid cells could also be observed seen in the normal cells of the bone marrow and spleens of the leukemic mice as compared to control mice, with a similar phenotypic representation to that of the leukemic cells (CD45.2+) (Figure 1E). Therefore, our chimeric mouse model replicates the disease found in primary transgenic mice. Importantly, accumulation of myeloid lineage cells at the expensive of lymphoid cells was observed in the non-transformed CD45.1+ leukemic-exposed cells similar to that of the leukemic cells expressing the BCR-ABL transgene.
Alterations in normal stem and progenitors during leukemia
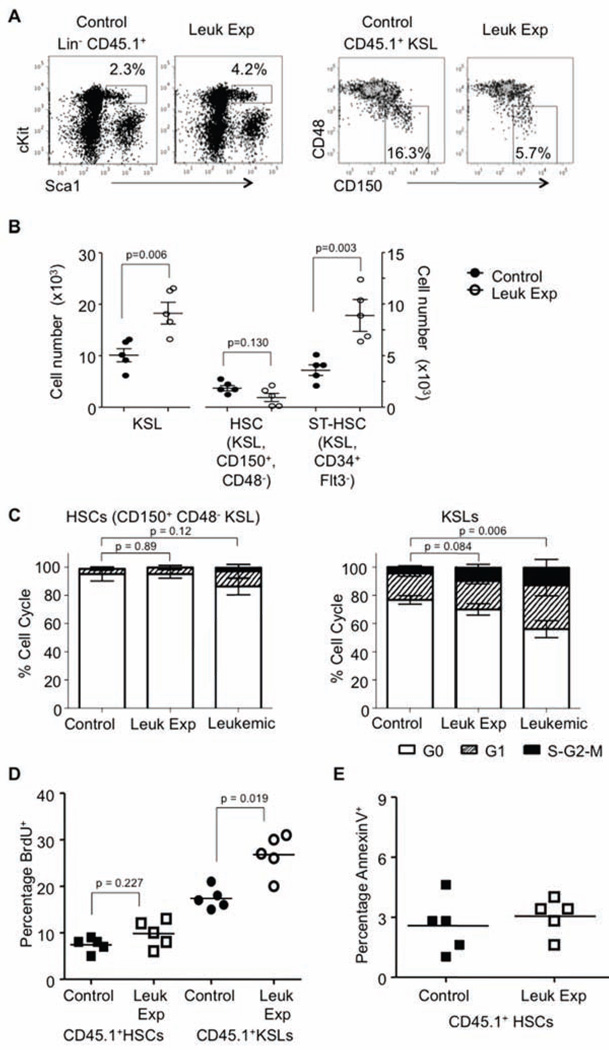
As an initial approach to understand the alterations described above in mature lineages, we performed detailed analyses of stem and progenitor cells in chimeric marrow. Mice were analyzed at 4 weeks after leukemic induction, and subsets of early progenitor populations were evaluated: LinckitHi Sca1+ (KSL, referred to as progenitors), long-term HSCs (HSCs: CD150+ CD48− KSL, or SLAM+ KSL, referred to as stem cells) and short-term HSCs (ST-HSCs: CD34+ Flt3− KSL) (Wilson et al., 2008) (Figure 2A and Figure S2A). Absolute numbers of normal leukemic-exposed HSCs were not significantly altered in the BM of mice with CML. Meanwhile there were significant increases in the leukemic-exposed total KSL and GMP, including specific increases in ST-HSCs and MPP, as compared to controls (Figure 2B and Figure S2A). Therefore, the CML leukemic environment had an altered capacity to support normal progenitors cells, changing the distribution of these populations within 4 weeks of disease.
Figure 2. Normal progenitor profiles within the leukemic environment.
(A) Flow cytometry profiles for control and leukemic-exposed progenitors (CD45.1+) of Lin− ckitHi Sca1+ (KSL) and Lin− ckitHi Sca1+ CD150+ CD48− (HSC), in either control or leukemic marrow at 4 weeks post Tet withdrawal, n=5.
(B) Numbers of control and leukemic-exposed (CD45.1+) progenitors were calculated per femur for Lin− ckitHi Sca1+ (KSL), Lin− ckitHi Sca1+ CD150+ CD48− (HSC), or Lin− ckitHi Sca1+ CD34+, Flt3− (STHSC) in either control or leukemic marrow, n=5. Error bars represent ± SEM.
(C) CD45.1+ HSCs (Lin− ckitHi Sca1+ CD150+ CD48−; HSC) or CD45.1+ KSL (Lin− ckitHi Sca1+; progenitors) were sorted from either control or leukemic environments, and then cell cycle status was determined using pyronin Y and Hoescht, n=5. Error bars represent ± SD.
(D) BrdU incorporation of CD45.1+ HSCs (KSL CD150+ CD48−) from control or leukemic mice was quantified, and lines indicate the mean, n=5.
(E) AnnexinV staining of control or leukemic-exposed HSC to determine the apoptosis within this subset of progenitors, and lines indicate the mean, n=5. See also Figure S2.
We then assessed the proliferation status and cell cycle of these leukemic-exposed stem and progenitor cells. However, there were no observed changes in the HSCs in cycle. Meanwhile, CD45.1+ progenitors from the leukemic environment were induced to proliferate (Figure 2C and Figure S2B). It was previously demonstrated that leukemic BCR-ABL+ HSCs have a hyperproliferative phenotype (Schemionek et al., 2010; Zhang et al., 2010). However, the leukemic-exposed HSCs showed little evidence of proliferation as assessed by BrdU incorporation. Conversely, many leukemic-exposed KSL were dividing (Figure 2D and Figure S2C). The increase of BrdU incorporation could be from DNA damage by the leukemic environment, however, there was no evidence of increased apoptosis in HSCs from the leukemic environment (Figure 2E and Figure S2D). Thus, the leukemic environment promoted proliferation of normal progenitors but not the HSCs from which they derive.
Leukemic cells compromise functions of normal stem and progenitor cells
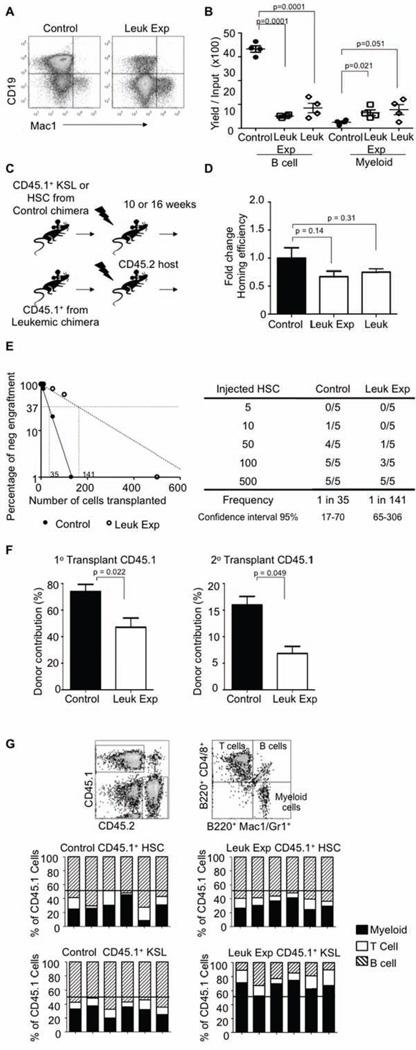
We wondered if co-habitation with leukemic cells caused intrinsic changes in otherwise normal stem and progenitor cells. Normal CD45.1+ KSLs were sorted from either control or leukemic mice and co-cultured on OP9 stromal cells to allow differentiation to the B lineage. It has been previously shown that in addition to B cells, myeloid cells can also be produced inefficiently under these conditions (Mohtashami et al., 2010; Wang et al., 2005; Welner et al., 2008). The yield of B lineage cells produced from normal KSL was severely decreased by leukemic exposure with only modest increases in myeloid lineage cells in this assay, similar to that observed when using leukemic cells (Figure 3A and B).
Figure 3. Stem and progenitor cell memory of leukemic exposure.
(A) CD45.1+ KSLs from either control or leukemic mice were sorted and placed in OP9 co-culture assay. Flow cytometry analysis of CD19 and Mac1 these cultures after 14 days is shown, n=4.
(B) Quantification of B cells (CD19+) and myeloid lineage cells (Mac1+) from OP9 co-cultures were plotted from CD45.1+ KSLs from either control or leukemic-exposed and CD45.2+ leukemic KSL 14 days after initiation, n=4. Error bars represent ± SEM.
(C) Cartoon showing control or leukemic-exposed CD45.1+ KSLs or HSCs (Lin− ckitHi Sca1+ CD150+ CD48−) were sorted from the primary chimeras and transplanted into sublethally irradiated C57BL/6 (CD45.2) hosts. KSLs transplanted mice were analyzed at 10 weeks post transplant, while HSC transplanted mice were analyzed after 16+ weeks post transplant.
(D) CD45.2+ leukemic HSCs and CD45.1+ HSCs were sorted from control and/or leukemic mice and then labeled with CFSE (5(6)-Carboxyfluorescein N-hydroxysuccinimidyl ester). These cells were then transplanted into lethally irradiated mice for 12 hours at which time the femurs were analyzed for the presence of CFSE and CD45.1+ cells. Plots shows homing efficiency relative to the control HSCs, n=5. Error bars represent ± SD.
(E) Limiting dilution CRU (competitive repopulation unit) assay is shown. The indicated numbers of control or leukemic-exposed HSCs were transplanted along with 2×105 BM of a competitor (CD45.2+) into lethally irradiated hosts (CD45.2+). Reconstitution was evaluated in the blood at 16 weeks post-transplantation (p value = 0.0001). Mice with CD45.1+ chimerism < 0.3% were considered as non-responders.
(F) For serial transplantation, 2×106 sorted CD45.1+ bone marrow cells from either control and appropriately adjusted per HSC numbers of CD45.1+ cells from leukemic mice were transplanted into lethally irradiated CD45.2 hosts. Rounds of transplantations with performed at 16-week intervals. The proportion of peripheral blood generated from the CD45.1+ cells in recipient mice was determined for each round of transplantation. Bar graphs indicate chimerism after each round of transplantation, n = 4. Error bars represent ± SD.
(G) Peripheral blood samples from either 10 weeks post-transplant for CD45.1+ KSLs or 16 weeks post-transplant for CD45.1+ HSCs transplants, respectively, were stained with CD45.1, CD45.2, and DAPI for viability. The bars show percentages of the donor type cells (CD45.1) that expressed myeloid (Gr-1 and CD11b, filled), B (B220, striped), or T (CD4/8, open) lineage markers in the blood of individual recipients.
Many methods have been used for HSC purification (Challen et al., 2009), but they are functionally defined through transplantation (Kiel et al., 2005; Spangrude et al., 1995; Szilvassy et al., 1990). To determine how the leukemic cells and their altered environment impact normal HSCs or KSL (progenitors), we transplanted control or leukemic-exposed HSCs into irradiated CD45.2 congenic recipients (Figure 3C). At 12 hours after transplantation, no significant defects in homing were observed from the HSC transplanted mice, similar to results obtained with homing of leukemic cells (Figure 3D). To test whether the observed increase in leukemic-exposed progenitors reflects differences in function, we performed competitive repopulation transplants with limiting numbers of control or leukemic-exposed HSCs. Interestingly, an approximate 4-fold reduction in stem cell activity results from exposure to leukemic cells (Figure 3E). Therefore, a more rigorous test of HSC function was then performed with serial transplantation assays. Since there were modest differences in HSC numbers between control and leukemic-exposed populations, we adjusted for these in the serial transplantations. Residence in a leukemic environment diminished repopulation capability in one round of transplantation, and this was magnified when the cells were re-transplanted to additional recipients (Figure 3F). From similarly transplanted mice, lymphoid (B/T) and myeloid reconstitution in the peripheral blood was determined 16+ weeks post-transplant. Leukemic-exposed HSCs showed no skewing to either myeloid or lymphoid lineages (Figure 3G top). However, when recipient mice were transplanted with KSL/progenitors and analyzed at 8-10 weeks post-transplant, residence in the leukemic environment increased their myeloid cell production at the expense of lymphoid lineages (Figure 3G bottom). Therefore, leukemic exposure caused discrete alterations in hematopoietic progenitors. KSL/progenitors had reduced competency for generation of lymphopoiesis. Whereas, numbers of more rigorously defined HSCs were mostly unaffected and they could produce all blood lineage cells. However, HSCs were not unscathed because their self-renewal and re-populating ability on transplant were compromised.
Normal progenitors resemble transformed CML with respect to molecular features
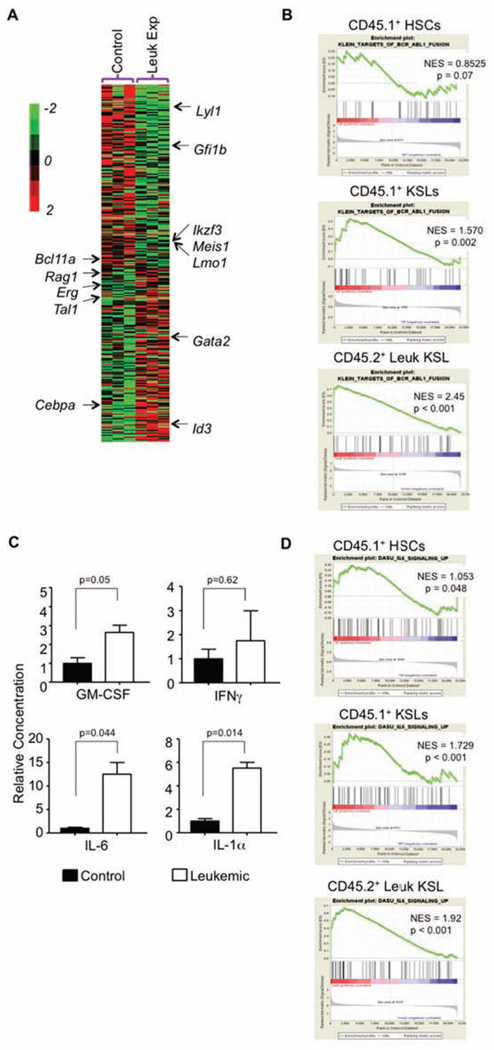
Bystander effects of residence within the leukemic environment were then explored with genome-wide gene profiling of purified control or leukemic-exposed HSCs and KSLs. The data was interrogated with several unbiased approaches, focusing on lineage specific transcription factors that could account for changes in function and differentiation toward the myeloid lineage (Figure 4A). Gene Set Enrichment Analysis (GSEA) was used to seek similarities between these normal progenitors and ones with a CML signature. Surprisingly, leukemic-exposed KSLs acquired a BCRABL+ signature of the leukemic cells with which they reside. In contrast, transcription patterns in leukemic-exposed HSCs were altered but not to the degree in which they resembled the leukemic signature (Figure 4B).
Figure 4. Microarray data points to cytokine changes in the progenitor cells from the leukemic exposure.
(A) Hierarchical clustering with average linkage was used to generate the heatmap of lineage specific transcription factors from the control and leukemic-exposed progenitors.
(B) Enrichment plots of GSEA analysis using control, leukemic-exposed, and leukemic HSCs and KSLs expression data against a list of BCR-ABL related gene changes.
(C) We used an antibody based protein array to measure cytokine changes in the sera from our chimera mice, both control and transgenic. Results are shown based on the normalization of the control sera. Error bars represent ± SD.
(D) Enrichment plots of GSEA using control, leukemic-exposed and leukemic HSCs and KSLs expression data against a list of IL-6-related gene changes. See also Figure S3.
We wanted to further investigate the HSCs microarray data to uncover why we see differences in self-renewal and engraftment upon transplantation. There were 1468 differentially expressed genes in the leukemic-exposed HSCs versus control HSCs (Figure S3A). GSEA was used to determine the correlation of each dataset with known “stem-ness” related genes. We observed bias in control HSCs relative to leukemic-exposed HSC, further supporting their ability to repopulate mice upon transplantation (Figure S3B). Significant to this study were changes observed in signaling pathways potentially altered by the leukemic environment which might account for the changes in lineage specification specific to the KSL/progenitors. Several pathways were shown to be dysregulated in the leukemic-exposed KSLs (Figure S3C) and might account for the changes we see in engraftment; these will be a focus of our future work. Unlike our findings with KSL/progenitors, the leukemic-exposed HSCs showed no consistent lineage bias in myeloid or lymphoid lineage specific transcription factors (Figure S3D).
From our analysis of leukemic-exposed HSCs and KSL/progenitors, we thought that the inflammatory environment might mediate many of the observed changes. From our model, we measured serum cytokine levels (Figure 4C). Consistent with data from others (Reynaud et al., 2011; Zhang et al., 2012), several inflammatory cytokines, including GM-CSF, IL-6 and IL-1α were significantly increased during leukemogenesis. These inflammatory cytokines provide one way that the leukemic environment might impose malignant cell properties on untransformed cells that reside there, as seen with the BCR-ABL GSEA above (Figure 4B). Additionally, GSEA analysis showed there was an inflammatory signature in the leukemic cells that was absent in the leukemic-exposed cells, suggesting the leukemic cells are the source of these cytokines (Figure S3E) (Kleppe et al., 2015). From the current literature and known functions of each of these cytokines, IL-6 was further investigated for this cytokine’s role in altering the normal KSLs functions within the CML environment. We again used GSEA analysis to ask if normal KSLs from the leukemic mice were aligned with an “IL-6 signature”, as this receptor has little to no expression in HSC (Figure S3F) (Maeda et al., 2005). Indeed, this was the case for the leukemic-exposed progenitors but not leukemic-exposed HSCs (Figure 4D). Our own observations were consistent with recent findings that IL-6 was elevated in the leukemic cells of this mouse model (Figure S3G) (Reynaud et al., 2011).
Furthermore, we wondered if current therapy was able to reverse the impact of normal hematopoiesis within CML specifically with regards to IL-6 production. Currently, imatinib (IM) is the first line treatment for CML. Therefore, IM was started two weeks after Tet withdrawal, a time-point when disease was consistently initiated as determined by flow cytometry. Mice were treated by gavage every other day for four weeks. After this treatment, B lymphopoiesis and myeloid differentiation were restored to their normal ratios (Figure S3H). Additionally, we sorted CD45.1+ KSLs from the control, imatinib-treated leukemic, or leukemic mice and determined their ability to make B lineage cells in vitro. Indeed, treated mice had restored B lineage potential (Figure S3I). Moreover, the levels of IL-6 in the serum were reduced to near normal levels, even in the presence of minimal residual disease (2-15% CD45.2+ cells) (Figure S3J), The phenomenon of altered inflammatory environment by IM has been recently observed by others as well (Zhang et al., 2010). Taken together, the leukemic bystander effects are dependent on tyrosine kinase activity, can drive IL-6 production, and are likely reversible.
The leukemic environment changes normal progenitors through IL-6
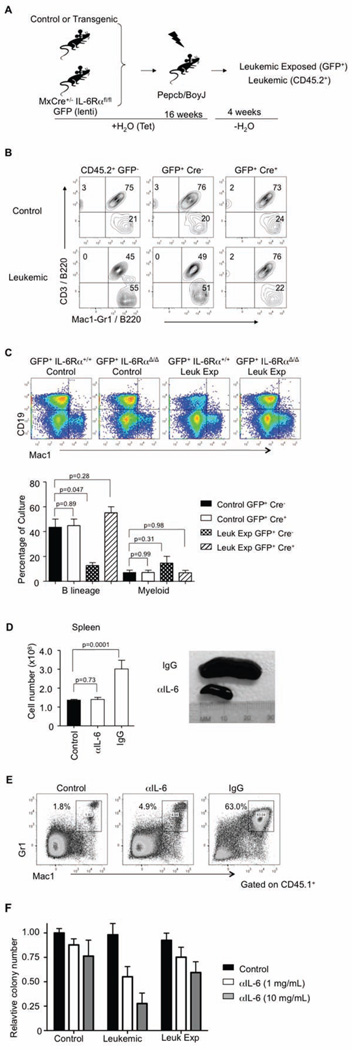
The above findings raised questions related to reversibility or prevention of disease through protection of normal hematopoiesis by manipulation of the CML inflammatory environment. We designed experiments to determine if the observed IL-6 dysregulation contributed to the changes described above in the leukemic-exposed stem and progenitor cells. Therefore, we analyzed chimeric mice lacking essential components of the IL-6 signaling pathway, specifically IL-6Rα. Consequently, MxCre− IL-6Rαflox/flox and MxCre+ IL-6Rαflox/flox mice were treated with polyI:C every other day for 3 total injections to excise IL-6Rα. At this point, the mice had no obvious abnormalities in hematopoiesis. Since these mice were on a CD45.2+ background similar to our BCR-ABL leukemic model, we used lineage depleted marrow cells from the MxCre− IL-6Rαflox/flox and MxCre+ IL-6RαΔ/Δ mice and labeled these two groups of cells with a lentiviral GFP. GFP+ cells were sorted and co-transplanted with control or transgenic (BCR-ABL) bone marrow into lethally irradiated congenic recipients and kept on tetracycline treatment to prevent disease (Figure 5A). At 16 weeks post-transplantation, we induced the expression of BCR-ABL (Tet removal) and the GFP+ cells were then analyzed for their ability to make either lymphoid or myeloid lineage cells (Figure 5B). MxCre+ GFP+ cells maintained their B lymphopoiesis even though they were in a leukemic environment, implicating IL-6 signaling in lymphoid lineage suppression and myeloid expansion. Furthermore, GFP+ MxCre+ IL-6RαΔ/Δ KSLs from the leukemic mice had no intrinsic changes as assayed by our B lineage differentiation in vitro assay (Figure 5C). Even though normal differentiation from the leukemic-exposed cells was restored in the IL-6RαΔ/Δ transplanted cells, this did not hinder the progression of CML disease in the CD45.2+ cells.
Figure 5. Inhibition of IL-6 signaling protects normal progenitor cells.
(A) Cartoon of the method used to determine if leukemic-exposed KSLs skewing to the myeloid lineage was dependent on IL-6Rα signaling. CD45.2+ BCR-ABL leukemic cells were transplanted with CD45.2+ GFP+ MxCre+ IL-6RαΔ/Δ cells or CD45.2+ GFP+ MxCre− IL-6Rαflox/flox cells and kept on tetracycline for 16 weeks before removal of Tet.
(B) Contour plots from staining of peripheral blood gating on GFP+ and GFP− cells for B220/CD3 and B220/Mac1-Gr1. Top panels show the control chimeria while the both show the leukemic. GFP+ MxCre− cells (center panels) contain IL-6Rαflox/flox cells. GFP+ MxCre+ cells are excised for IL-6Rα (right panels), n=4.
(C) GFP+ KSLs cells from control or leukemic mice were sorted and co-cultured on OP9 stromal cells for B lineage differentiation. Flow cytometry plots show CD19 versus Mac1 staining from control and leukemic mice containing either IL-6Rαflox/flox or MxCre+ IL-6RαΔ/Δ cells. Quantification of the percentages of B lineage cells and myeloid cells derived from these cultures are plotted, n=4. Error bars represent ± SD.
(D) Figure of splenic numbers and morphology from IgG and anti-IL-6 treated leukemic mice are shown. Mice were injected i.v. with anti-IL-6 diluted in PBS (0.2 mg/kg). Antibody injected controls were similarly injected with IgG in PBS. Two weeks after removal from Tet, mice were injected ever other day for 6 weeks. Mice were then euthanized and total cell numbers from the spleens of control, IgG leukemic and anti-IL-6 leukemic treated mice were obtained, n=5. Error bars represent ± SD.
(E) Flow cytometry plots of CD45.1+ myeloid cells (Mac1+ Gr-1+) from the spleens of control or leukemic IgG and anti-IL-6-treated mice. Spleens were stained for viable CD45.1, CD45.2, Mac1, Gr1, n=4.
(F) Sorted control, leukemic-exposed and leukemic KSL were cultured in colony assays in the presence or absence of anti-IL-6 at two doses. Colonies were counted and relative colony numbers were determined compared to untreated cells for each population, n=3. Error bars represent ± SD. See also Figure S4.
For therapeutic applications where targeting the inflammatory environment might rescue normal differentiation as well as perturb the leukemic cells, we attempted to block IL-6 using an antibody against the cytokine. Therefore, chimeras were prepared and 16 weeks later, leukemia was induced by withdrawal of Tet. After two weeks off Tet when increased CD45.2+ cells are observed in the peripheral blood, groups of mice received either IgG control antibody or an anti-IL-6 every other day (0.2mg/kg) for 6 weeks. Remarkably, the anti-IL-6 treated mice showed no signs of disease such as splenomegaly or myeloid expansion in hematopoietic tissues (Figure 5D and E). Only residual disease could be detected in the bone marrow of these mice (Figure S4A). It should be noted that this treatment did not eliminate the leukemic initiating cells, as mice taken off anti-IL-6 therapy eventually developed CML like disease (Figure S4B). These treatments were unique to imatinib (IM) and anti-IL-6, as treatment with anti-GM-CSF or anti-IL-1α did not lead to significant cell death of leukemic bone marrow cells in culture (Figure S4C). Moreover, treatment with anti-IL-6 in colony assays with control, leukemic-exposed, or leukemic KSL showed that the leukemic cells were specifically sensitive to this treatment with respect to colony formation (Figure 5F). These data suggest that the BCR-ABL leukemic environment promotes the disease and impacts the neighboring non-transformed cells through IL-6. Furthermore, this environment can be altered through blocking the inflammatory signature.
IL-6 disrupts normal stem and progenitor cell function in human CML
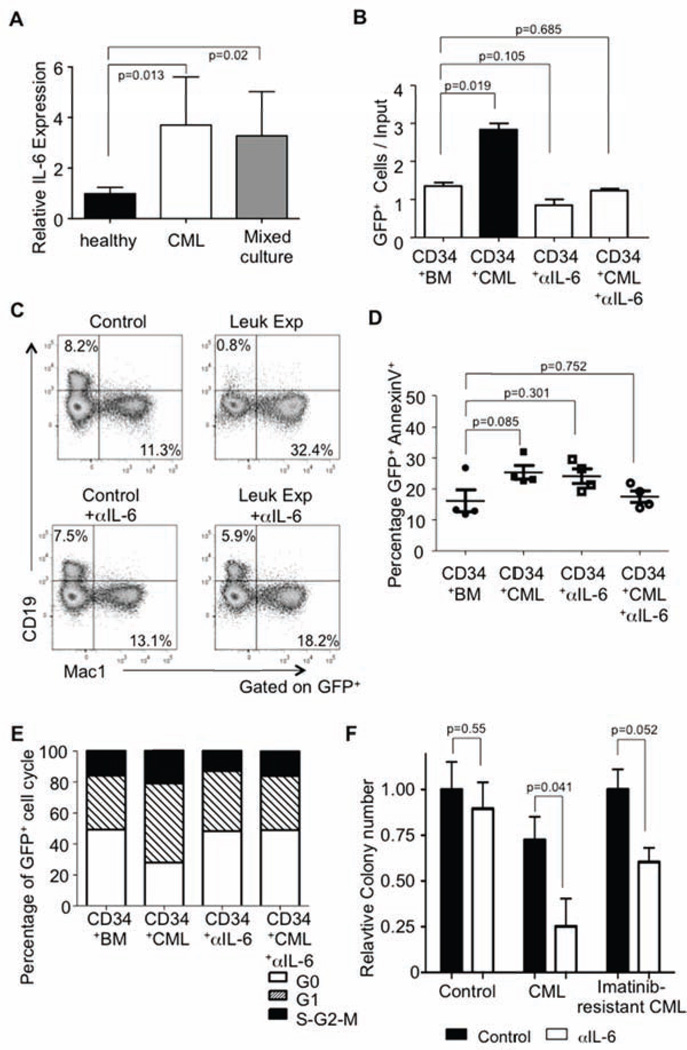
These experimental animal findings beg the question of relevance to human disease, but chronic phase CML has not been satisfactorily reproduced by transplantation into immunodeficient mice (Dazzi et al., 1998)(as well as our own observations, RSW and GA). Therefore, we exploited culture approaches similar to those described to evaluate bystander changes in our mouse model. We cultured lentiviral GFP labeled sorted CD34+ adult bone marrow cells were mixed with bone marrow from either control (healthy) or chronic phase CML patients. As seen with our mouse model of CML, human cultures had increase IL-6 as assessed by RT-PCR (Figure 6A). Furthermore, CML BM exposure caused GFP+ CD34+ progenitors to expand more than twice that observed from co-culture with normal patient BM (Figure 6B). This increase in GFP+ cells was inhibited by addition of an anti- IL-6 antibody. Additionally, CD34+ progenitors co-cultured with CML BM had reduced B lineage potential as assayed during stromal cell co-cultures. Similar to the mouse studies, treatment with anti-IL-6 was able to restore B lineage potential (Figure 6C). From these culture conditions, we were able to determine that CML cells did not significantly increase apoptosis of the GFP marked cells (Figure 6D). Moreover, CML cells not only caused expansion of the GFP+ CD34+ progenitors but also their division, a phenomenon that was also IL-6 dependent (Figure 6E). As seen with our murine studies, anti-IL-6 was able to inhibit colony formation of both chronic phase and imatinib-resistance CML bone marrow patient samples (Figure 6F). We conclude that changes in cytokine levels, specifically those of IL-6, in the CML environment are responsible for altered differentiation and function of normal stem and progenitor cells. Our models underscore the significance of the leukemic cells and environment on conditioning both leukemic and normal stem and progenitor cells. We conclude that changes in cytokine levels, specifically those of IL-6, in the CML are mostly responsible for altered differentiation and function of normal stem and progenitor cells while promoting the disease.
Figure 6. Increased proliferation and skewed differentiation of normal human CD34+ progenitors when mixed with hCML cells.
(A) Cytokine IL-6 mRNA levels from cultured patient bone marrow (healthy, chronic phase CML or mixed) was measured against GADPH and normalized to healthy controls, n = 5. Error bars represent ± SD.
(B) Bar graph showing the yield per input of GFP+ cells when cultured with control or CML bone marrow with or without anti-IL-6. Bead enriched CD34+ cells from human adult bone marrow were labeled with a lentiviral GFP reporter. These cells were then cultured with whole adult bone marrow from healthy patients (control) or those with chronic phase CML (leukemic-exposed) on stromal cells (MS5). Additionally, wells contained either IgG or anti-IL-6 as indicated (5 µg/mL). These cells were cultured for 96 hours with FL, TPO, and IL3 and then counted and analyzed by flow cytometry for total GFP+ cells, n=3. Error bars represent ± SD.
(C) CD34+ GFP marked cells were co-cultured with healthy bone marrow (Control) or chronic phase CML (leukemic-exposed) on stromal cells. These cultures were maintained with IL-7, SCF, and Flt3L for one week and then an additional three weeks with only SCF and Flt3L. As indicated, some wells contained anti-IL-6. After the four week culture, GFP+ human cells were analyzed by flow cytometry for expression of CD19 and Mac1, n=3.
(D) GFP+ CD34+ cells from either control or leukemic-exposed cultures were stained for AnnexinV to determine the relative apoptosis in these conditions, and lines indicate the mean, n=3. Error bars represent ± SEM.
(E) Sorted GFP+ CD34+ cells were analyzed from control and CML cultures (with or without anti-IL-6) after staining with PyroninY and Hoescht to determine cell cycle status, n=3.
F) Bone marrow from healthy, chronic phase CML or imatinib-resistant CML was cultured in Methocult4435 for colony formation with or without human anti-IL-6. Colony numbers are presented as relative to the healthy untreated control, n=2. Error bars represent ± SD.
Discussion
Our models underscore the significance of the leukemic cells on conditioning both leukemic and normal stem and progenitor cells. First, and confirming the recent work of others (Begley and Ellis, 2012; Krause et al., 2013; Reynaud et al., 2011; Schepers et al., 2013), leukemic cells alter neighboring non-transformed hematopoietic cells; however, the direct impact on stem and progenitor cell function was not previously addressed. Second, these leukemic-exposed normal cell counterparts acquire characteristics of the transformed cells, with changes dependent on their stage of differentiation. That is, HSC are altered differently from the changes observed in KSLs/progenitors. Characteristics imprinted on leukemic-exposed HSCs included reconstitution defects but not homing or differentiation potential. Alterations in KSLs were much more striking. Exposure of these progenitors to leukemic cells induced loss of lymphopoietic potential and enhanced myeloid potency. In addition, the normal progenitors entered cell cycle and mimicked leukemic cells with respect to gene expression. Third, an IL-6 signature was prominent in the transcriptome profile, leading us to functional studies involving this cytokine. Furthermore, IL-6 signaling was necessary for acquisition of the characterized bystander properties by normal progenitors. Finally, human leukemic cells perturbed neighboring progenitors through the production of IL-6 similar to that demonstrated by our mouse model. We believe these observations will focus attention on how changes in normal hematopoietic cells that reside near transformed ones in bone marrow can lead to therapeutics targeting the leukemic cells by altering the inflammatory profile from the disease.
Our model was previously shown to closely resemble the chronic phase of human CML (Koschmieder et al., 2005). Moreover, we are encouraged that two recent reports are closely relevant to our study and show the reproducibility of the science (Begley and Ellis, 2012; Schepers et al., 2013; Zhang et al., 2012). In these studies, they lethally irradiated our inducible BCR-ABL transgenic or wild-type controls. The animals were then used as recipients of HSPCs from leukemic or normal marrow from non-irradiated mice. The leukemic host, mainly the stromal niche, was a poor host for normal hematopoiesis. It is important to note however that one group did not record IL-6 overexpression from the environment but rather the changes in the cellular environment (Zhang et al., 2012).
In our work, we closely monitored the normal hematopoietic cells over time following BCR-ABL activation and observed rapid changes in progenitors. After one month of leukemic exposure, KSL numbers were greatly expanded, and they had almost completely lost their ability to generate lymphocytes. This might be explained in terms of selective growth of myeloid precursors and/or redirection away from lymphopoiesis. HSC changes were more subtle and obvious only in transplantation assays, a phenotype that remained even after treating the mice with anti-IL-6. Although we saw little changes in proliferation and differentiation of HCS, these changes may occur at earlier stages of the disease, particularly at onset from perturbation of leukemic stem cells within a similar niche (Zhang et al., 2012). Our data and others suggest that HSC express little to no IL-6Rα and may therefore be less responsive to this specific inflammatory cytokine (Maeda et al., 2005). We are interested to determine other cytokines or environmental changes specifically mediating changes in HSCs. However, we can conclude that residence near leukemic cells provides several disadvantages to cells lacking the BCR-ABL fusion protein. Their inability to self-renew and sustain balanced hematopoiesis would allow them to be out-competed by transformed cells over time.
It has previously been suggested that cytokines create a pro-inflammatory environment during CML development, and that this provides a proliferative advantage to leukemic cells (Holyoake et al., 1999; Zhang et al., 2012; Zhang and Ren, 1998). We observed a conspicuous IL-6 signature in leukemic-exposed progenitors, and there is precedent for the importance of this factor in CML. Two groups observed that serum IL-6 was elevated in sera of leukemic animals (Reynaud et al., 2011; Zhang et al., 2012). Additionally, IL-6 secretion increases with CML development in patients (Anand et al., 1998; Reynaud et al., 2011; Schmidt et al., 2011; Zhang et al., 2012). Furthermore, Reynaud et al. found that disease progression was inhibited when BCR-ABL was induced in hematopoietic cells of IL-6 deficient mice (Reynaud et al., 2011). IL-6 is also capable of blocking a discrete early stage of lymphopoiesis (Maeda et al., 2005). We are not the first to find this inflammatory signature in CML; however, we were able to use antibody therapy to target the inflammatory environment in this model of leukemia.
We found that IL-6 drives normal progenitor changes in our chimeric animal model of CML. Indeed, treatment of the mice with neutralizing anti-IL-6 allowed progenitor composition and differentiation to proceed normally for at least six weeks after BCR-ABL activation. Tumor initiating cells must have been spared, because the disease developed when antibody therapy was discontinued. HSC can propagate CML disease, but are unlikely to be direct targets of IL-6 because they lack critical receptor components (Raaijmakers et al., 2010; Reynaud et al., 2011; Schemionek et al., 2010). Thus, factors other than IL-6 might account for our finding that untransformed HSC had reduced self-renewal ability after residence in leukemic marrow and during therapy. Moreover, Schepers et al. provide evidence that this loss of HSC function could be accomplished by reprogramming stromal cells as a result of the leukemic cells or environment (Schepers et al., 2013). In that context, it is interesting that in human CML, chemokines can inhibit the proliferation of normal hematopoietic cells while sparing transformed ones, and similar interactions may be occurring within the niche (Cashman et al., 1998; Eaves et al., 1993a; Eaves et al., 1993b). The source of IL-6 produced in CML is still in question and might involve multiple types of BCR-ABL transformed cells. The study by Zhang et al., noted above, suggested that radio-resistant stromal cells were not an important source of IL-6 in a transplantation model similar to ours (Zhang et al., 2012). On the other hand, inflammation driven by TLR ligands and T cell produced interferons can elicit IL-6 production in stromal elements (Zhao et al., 2014).
In addition to the results we observed in mice, neutralization of IL-6 protected human CD34+ cells when exposed to CML cells. Given the contribution of IL-6 to a wide range of human inflammatory diseases, clinically effective strategies have already been developed for its neutralization (Tanaka et al., 2011). Our results suggest that this approach might be an effective therapy in drug resistant CML or other leukemias with pro-inflammatory signatures. Specifically, those in which the leukemic cells are also found to have an adverse impact on untransformed bystander hematopoietic cells. From studying the non-transformed cells in other models of leukemia, we hope to find and target additional cell autonomous environmental changes that are necessary for propagation of the leukemic clones.
Experimental Procedures
Mice/Human samples
Inducible, transgenic SCL-tTa/BCR-ABL mice (Koschmieder et al., 2005) were backcrossed into the B6 background and were maintained with administration of tetracycline in the drinking water (0.5 g/L). BCR-ABL expression was induced by tetracycline withdrawal for a minimum of three weeks. Mice were housed in a sterile barrier facility approved by the IUCAC at the Beth Israel Deaconess Medical Center. Human samples were collected by informed consent and IRB approved at the Beth Israel Deaconess Medical Center.
Flow cytometry
Single-cell suspensions from various organs were analysed by flow cytometry using the following monoclonal antibodies conjugated with phycoerythrin (PE), PE–CY7, fluorescein isothiocyanate, allophycocyanin (APC), APC–Cy7 or eFluor 450 obtained from BD Pharmingen (BD), BioLegend or eBioscience: Mac-1/CD11b (M1/70), Gr-1 (8C5), CD3 (KT31.1), CD4 (GK1.5), CD8 (53-6.7), B220 (RA3-6B2), CD19 (1D3), TER119 (TER-119), Sca1 (E13-161-7), c-Kit (2B8), CD16/32 (2.4G3), Thy-1.2 (53-2.1), CD135 (AF2 10.1), CD48 (HM48-1), CD45.1 (A20), CD45.2 (104) and CD150 (TC15-12F12.2). Stained cells were analysed with an LSRII flow cytometer and sorted using a FACSAria II (BD Biosciences). Viable cells were identified by DAPI exclusion. Human stem cells / primitive progenitors were isolated by sorting CD34+CD38− cells. Diva software (BD) and FlowJo (Tree Star) were used for data acquisition and analysis, respectively.
Proliferation and Cell Cycle Analysis
For proliferation analysis BrdU was administered intraperitoneally (1 mg/mouse), BM cells were obtained 16 hr post-injection and BrdU incorporation into stem and progenitor populations were analyzed (Cappella et al., 2008). Cell cycle was also analyzed by PyroninY and Hoerscht labeling as previously described (Staber et al., 2013).
Limiting dilution assay
HSC or total bone marrow cells were collected as CD45.1+ from leukemic or control mice 4 weeks following removal of tetracycline water; then the number of cells indicated to be injected per mouse was sorted into individual wells of a 96-well plate containing 104 wild-type CD45.2+ bone marrow (rescue cells) in PBS. The contents of individual wells were injected into the retro-orbital venous sinus of lethally irradiated CD45.2+ recipients (600 rads twice with a 3 h interval). Peripheral blood was obtained from each mouse each month after transplantation for at least 4 months and analyzed by FACS. Cells were stained with anti-CD45.1 and CD45.2 antibodies to distinguish donor-derived cells from the host cells, as well as lineage-specific antibodies Mac1, Gr1, B220 and CD3 to identify myeloid, B and T lineages. A recipient mouse was considered positive if CD45.1+CD45.2− cells were present in myeloid and B and/or T cells and also comprised more than 0.3% of the cells in the peripheral blood.
Bone Marrow Tranplantation
For initial experimental setup, 2×106 CD45.1+ bone marrow cells and 2×106 CD45.2 bone marrow cells from SCLtTa/BCR-ABL mice were transplanted through retro-orbital injection into lethally irradiated CD45.1 mice. Mice were maintained on tetracycline before (2 weeks) and after transplantation for 16 weeks to establish chimerism before induction of the transgenes.
Microarray Analysis
RMA was run for all CEL files generated in this study to obtain the probe-set expression data. The probe-set expression data were then normalized using the Cross-Correlation method (Lit). Gene set enrichment analysis (GSEA) was carried out with the normalized data by comparing the leukemic-exposed HSCs or KSLs to their corresponding controls, respectively. The gene set database used in GSEA was the msigdb v3. For heatmap presentation of the significantly changed transcription factors in leukemic-exposed versus the control setting, the normalized data were further log2 transformed and the subtraction of the mean of the means was carried out for each gene. Also, hierarchical clustering with average linkage was used to generate the heatmap.
Statistical Analysis
The statistical significances were assessed by Student’s unpaired t-test using the GraphPad software.
Accession Number
GEO accession number GSE59337 for microarray data.
Supplementary Material
Significance.
It has generally been assumed that normal hematopoietic stem and progenitor cells (HSPC) are simply out-competed for space by malignant cells. We have shown that the leukemic cells are able to modify the differentiation potential of non-transformed cells while promoting their own maturation. The leukemic environment imprints functional and transcriptional programs onto the non-transformed progenitors. However, by specifically blocking the IL-6 cytokine, we could inhibit the differentiation bias and eliminate the bulk of the disease. Additionally, these observations were also found in human CML cells and leukemic-exposed human progenitors could be protected by anti-IL-6. These findings implicate a drug targetable mechanism that could account for significant malignancy related abnormalities in CML.
Acknowledgments
This work was supported by grants CA66996 and HL112719 from the National Institutes of Health, by the Singapore Ministry of Health's National Medical Research Council under its Singapore Translational Research (STaR) Investigator Award, and by the National Research Foundation Singapore and the Singapore Ministry of Education under its Research Centres of Excellence initiative. RSW is supported by the José Carreras Leukemia Foundation (FIJC F11/01).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information
Supplemental Information includes five figures and can be found with this article online.
References
- Anand M, Chodda SK, Parikh PM, Nadkarni JS. Abnormal levels of proinflammatory cytokines in serum and monocyte cultures from patients with chronic myeloid leukemia in different stages, and their role in prognosis. Hematological oncology. 1998;16:143–154. doi: 10.1002/(sici)1099-1069(199812)16:4<143::aid-hon628>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Begley CG, Ellis LM. Drug development: Raise standards for preclinical cancer research. Nature. 2012;483:531–533. doi: 10.1038/483531a. [DOI] [PubMed] [Google Scholar]
- Ben-Neriah Y, Daley GQ, Mes-Masson AM, Witte ON, Baltimore D. The chronic myelogenous leukemia-specific P210 protein is the product of the bcr/abl hybrid gene. Science. 1986;233:212–214. doi: 10.1126/science.3460176. [DOI] [PubMed] [Google Scholar]
- Cappella P, Gasparri F, Pulici M, Moll J. A novel method based on click chemistry, which overcomes limitations of cell cycle analysis by classical determination of BrdU incorporation, allowing multiplex antibody staining. Cytometry Part A : the journal of the International Society for Analytical Cytology. 2008;73:626–636. doi: 10.1002/cyto.a.20582. [DOI] [PubMed] [Google Scholar]
- Cashman JD, Eaves CJ, Sarris AH, Eaves AC. MCP-1, not MIP-1alpha, is the endogenous chemokine that cooperates with TGF-beta to inhibit the cycling of primitive normal but not leukemic (CML) progenitors in long-term human marrow cultures. Blood. 1998;92:2338–2344. [PubMed] [Google Scholar]
- Challen GA, Boles N, Lin KK, Goodell MA. Mouse hematopoietic stem cell identification and analysis. Cytometry Part A : the journal of the International Society for Analytical Cytology. 2009;75:14–24. doi: 10.1002/cyto.a.20674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmone A, Amorim M, Pontier AL, Wang S, Jablonski E, Sipkins DA. Leukemic cells create bone marrow niches that disrupt the behavior of normal hematopoietic progenitor cells. Science. 2008;322:1861–1865. doi: 10.1126/science.1164390. [DOI] [PubMed] [Google Scholar]
- Dazzi F, Capelli D, Hasserjian R, Cotter F, Corbo M, Poletti A, Chinswangwatanakul W, Goldman JM, Gordon MY. The kinetics and extent of engraftment of chronic myelogenous leukemia cells in non-obese diabetic/severe combined immunodeficiency mice reflect the phase of the donor's disease: an in vivo model of chronic myelogenous leukemia biology. Blood. 1998;92:1390–1396. [PubMed] [Google Scholar]
- Eaves C, Udomsakdi C, Cashman J, Barnett M, Eaves A. The biology of normal and neoplastic stem cells in CML. Leukemia & lymphoma. 1993a;11(Suppl 1):245–253. doi: 10.3109/10428199309047894. [DOI] [PubMed] [Google Scholar]
- Eaves CJ, Cashman JD, Wolpe SD, Eaves AC. Unresponsiveness of primitive chronic myeloid leukemia cells to macrophage inflammatory protein 1 alpha, an inhibitor of primitive normal hematopoietic cells. Proceedings of the National Academy of Sciences of the United States of America. 1993b;90:12015–12019. doi: 10.1073/pnas.90.24.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fialkow PJ, Jacobson RJ, Papayannopoulou T. Chronic myelocytic leukemia: clonal origin in a stem cell common to the granulocyte, erythrocyte, platelet and monocyte/macrophage. The American journal of medicine. 1977;63:125–130. doi: 10.1016/0002-9343(77)90124-3. [DOI] [PubMed] [Google Scholar]
- Hartwell KA, Miller PG, Mukherjee S, Kahn AR, Stewart AL, Logan DJ, Negri JM, Duvet M, Jaras M, Puram R, et al. Niche-based screening identifies small-molecule inhibitors of leukemia stem cells. Nature chemical biology. 2013;9:840–848. doi: 10.1038/nchembio.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidel FH, Bullinger L, Feng Z, Wang Z, Neff TA, Stein L, Kalaitzidis D, Lane SW, Armstrong SA. Genetic and pharmacologic inhibition of beta-catenin targets imatinib-resistant leukemia stem cells in CML. Cell stem cell. 2012;10:412–424. doi: 10.1016/j.stem.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holyoake T, Jiang X, Eaves C, Eaves A. Isolation of a highly quiescent subpopulation of primitive leukemic cells in chronic myeloid leukemia. Blood. 1999;94:2056–2064. [PubMed] [Google Scholar]
- Hooberman AL, Carrino JJ, Leibowitz D, Rowley JD, Le Beau MM, Arlin ZA, Westbrook CA. Unexpected heterogeneity of BCR-ABL fusion mRNA detected by polymerase chain reaction in Philadelphia chromosome-positive acute lymphoblastic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:4259–4263. doi: 10.1073/pnas.86.11.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Shen H, Tian C, Yu H, Zheng G, XuFeng R, Ju Z, Xu J, Wang J, Cheng T. Kinetics of normal hematopoietic stem and progenitor cells in a Notch1-induced leukemia model. Blood. 2009;114:3783–3792. doi: 10.1182/blood-2009-06-227843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Swerdlow S, Duffy TM, Weinmann R, Lee FY, Li S. Targeting multiple kinase pathways in leukemic progenitors and stem cells is essential for improved treatment of Ph+ leukemia in mice. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:16870–16875. doi: 10.1073/pnas.0606509103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Kleppe M, Kwak M, Koppikar P, Riester M, Keller M, Bastian L, Hricik T, Bhagwat N, McKenney AS, Papalexi E, et al. JAK-STAT Pathway Activation in Malignant and Nonmalignant Cells Contributes to MPN Pathogenesis and Therapeutic Response. Cancer discovery. 2015 doi: 10.1158/2159-8290.CD-14-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koschmieder S, Gottgens B, Zhang P, Iwasaki-Arai J, Akashi K, Kutok JL, Dayaram T, Geary K, Green AR, Tenen DG, et al. Inducible chronic phase of myeloid leukemia with expansion of hematopoietic stem cells in a transgenic model of BCR-ABL leukemogenesis. Blood. 2005;105:324–334. doi: 10.1182/blood-2003-12-4369. [DOI] [PubMed] [Google Scholar]
- Krause DS, Fulzele K, Catic A, Sun CC, Dombkowski D, Hurley MP, Lezeau S, Attar E, Wu JY, Lin HY, et al. Differential regulation of myeloid leukemias by the bone marrow microenvironment. Nature medicine. 2013;19:1513–1517. doi: 10.1038/nm.3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- Levine RL, Gilliland DG. Myeloproliferative disorders. Blood. 2008;112:2190–2198. doi: 10.1182/blood-2008-03-077966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K, Baba Y, Nagai Y, Miyazaki K, Malykhin A, Nakamura K, Kincade PW, Sakaguchi N, Coggeshall KM. IL-6 blocks a discrete early step in lymphopoiesis. Blood. 2005;106:879–885. doi: 10.1182/blood-2005-02-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguer-Satta V, Petzer AL, Eaves AC, Eaves CJ. BCR-ABL expression in different subpopulations of functionally characterized Ph+ CD34+ cells from patients with chronic myeloid leukemia. Blood. 1996;88:1796–1804. [PubMed] [Google Scholar]
- Mayle A, Luo M, Jeong M, Goodell MA. Flow cytometry analysis of murine hematopoietic stem cells. Cytometry Part A : the journal of the International Society for Analytical Cytology. 2013;83:27–37. doi: 10.1002/cyto.a.22093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier FE, Ragu C, Scadden DT. The bone marrow at the crossroads of blood and immunity. Nature reviews Immunology. 2012;12:49–60. doi: 10.1038/nri3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohtashami M, Shah DK, Nakase H, Kianizad K, Petrie HT, Zuniga-Pflucker JC. Direct comparison of Dll1- and Dll4-mediated Notch activation levels shows differential lymphomyeloid lineage commitment outcomes. Journal of immunology. 2010;185:867–876. doi: 10.4049/jimmunol.1000782. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505:327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petzer AL, Eaves CJ, Lansdorp PM, Ponchio L, Barnett MJ, Eaves AC. Characterization of primitive subpopulations of normal and leukemic cells present in the blood of patients with newly diagnosed as well as established chronic myeloid leukemia. Blood. 1996;88:2162–2171. [PubMed] [Google Scholar]
- Raaijmakers MH, Mukherjee S, Guo S, Zhang S, Kobayashi T, Schoonmaker JA, Ebert BL, Al-Shahrour F, Hasserjian RP, Scadden EO, et al. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature. 2010;464:852–857. doi: 10.1038/nature08851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud D, Pietras E, Barry-Holson K, Mir A, Binnewies M, Jeanne M, Sala-Torra O, Radich JP, Passegue E. IL-6 controls leukemic multipotent progenitor cell fate and contributes to chronic myelogenous leukemia development. Cancer cell. 2011;20:661–673. doi: 10.1016/j.ccr.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savona M, Talpaz M. Getting to the stem of chronic myeloid leukaemia. Nature reviews Cancer. 2008;8:341–350. doi: 10.1038/nrc2368. [DOI] [PubMed] [Google Scholar]
- Sawyers CL. Chronic myeloid leukemia. The New England journal of medicine. 1999;340:1330–1340. doi: 10.1056/NEJM199904293401706. [DOI] [PubMed] [Google Scholar]
- Schemionek M, Elling C, Steidl U, Baumer N, Hamilton A, Spieker T, Gothert JR, Stehling M, Wagers A, Huettner CS, et al. BCR-ABL enhances differentiation of long-term repopulating hematopoietic stem cells. Blood. 2010;115:3185–3195. doi: 10.1182/blood-2009-04-215376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers K, Pietras EM, Reynaud D, Flach J, Binnewies M, Garg T, Wagers AJ, Hsiao EC, Passegue E. Myeloproliferative neoplasia remodels the endosteal bone marrow niche into a self-reinforcing leukemic niche. Cell stem cell. 2013;13:285–299. doi: 10.1016/j.stem.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T, Kharabi Masouleh B, Loges S, Cauwenberghs S, Fraisl P, Maes C, Jonckx B, De Keersmaecker K, Kleppe M, Tjwa M, et al. Loss or inhibition of stromal-derived PlGF prolongs survival of mice with imatinib-resistant Bcr-Abl1(+) leukemia. Cancer cell. 2011;19:740–753. doi: 10.1016/j.ccr.2011.05.007. [DOI] [PubMed] [Google Scholar]
- Spangrude GJ, Brooks DM, Tumas DB. Long-term repopulation of irradiated mice with limiting numbers of purified hematopoietic stem cells: in vivo expansion of stem cell phenotype but not function. Blood. 1995;85:1006–1016. [PubMed] [Google Scholar]
- Staber PB, Zhang P, Ye M, Welner RS, Nombela-Arrieta C, Bach C, Kerenyi M, Bartholdy BA, Zhang H, Alberich-Jorda M, et al. Sustained PU.1 levels balance cell-cycle regulators to prevent exhaustion of adult hematopoietic stem cells. Molecular cell. 2013;49:934–946. doi: 10.1016/j.molcel.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szilvassy SJ, Humphries RK, Lansdorp PM, Eaves AC, Eaves CJ. Quantitative assay for totipotent reconstituting hematopoietic stem cells by a competitive repopulation strategy. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:8736–8740. doi: 10.1073/pnas.87.22.8736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Narazaki M, Kishimoto T. Anti-interleukin-6 receptor antibody, tocilizumab, for the treatment of autoimmune diseases. FEBS letters. 2011;585:3699–3709. doi: 10.1016/j.febslet.2011.03.023. [DOI] [PubMed] [Google Scholar]
- Venezia TA, Merchant AA, Ramos CA, Whitehouse NL, Young AS, Shaw CA, Goodell MA. Molecular signatures of proliferation and quiescence in hematopoietic stem cells. PLoS biology. 2004;2:e301. doi: 10.1371/journal.pbio.0020301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Pierce LJ, Spangrude GJ. Lymphoid potential of primitive bone marrow progenitors evaluated in vitro. Annals of the New York Academy of Sciences. 2005;1044:210–219. doi: 10.1196/annals.1349.026. [DOI] [PubMed] [Google Scholar]
- Wang JC, Lapidot T, Cashman JD, Doedens M, Addy L, Sutherland DR, Nayar R, Laraya P, Minden M, Keating A, et al. High level engraftment of NOD/SCID mice by primitive normal and leukemic hematopoietic cells from patients with chronic myeloid leukemia in chronic phase. Blood. 1998;91:2406–2414. [PubMed] [Google Scholar]
- Warr MR, Pietras EM, Passegue E. Mechanisms controlling hematopoietic stem cell functions during normal hematopoiesis and hematological malignancies. Wiley interdisciplinary reviews Systems biology and medicine. 2011;3:681–701. doi: 10.1002/wsbm.145. [DOI] [PubMed] [Google Scholar]
- Welner RS, Pelayo R, Nagai Y, Garrett KP, Wuest TR, Carr DJ, Borghesi LA, Farrar MA, Kincade PW. Lymphoid precursors are directed to produce dendritic cells as a result of TLR9 ligation during herpes infection. Blood. 2008;112:3753–3761. doi: 10.1182/blood-2008-04-151506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W, Jaworski M, Offner S, Dunant CF, Eshkind L, Bockamp E, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- Witte ON. Closely related BCR/ABL oncogenes are associated with the distinctive clinical biologies of Philadelphia chromosome positive chronic myelogenous and acute lymphocytic leukemia. Current topics in microbiology and immunology. 1988;141:42–49. doi: 10.1007/978-3-642-74006-0_7. [DOI] [PubMed] [Google Scholar]
- Zhang B, Ho YW, Huang Q, Maeda T, Lin A, Lee SU, Hair A, Holyoake TL, Huettner C, Bhatia R. Altered microenvironmental regulation of leukemic and normal stem cells in chronic myelogenous leukemia. Cancer cell. 2012;21:577–592. doi: 10.1016/j.ccr.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Strauss AC, Chu S, Li M, Ho Y, Shiang KD, Snyder DS, Huettner CS, Shultz L, Holyoake T, et al. Effective targeting of quiescent chronic myelogenous leukemia stem cells by histone deacetylase inhibitors in combination with imatinib mesylate. Cancer cell. 2010;17:427–442. doi: 10.1016/j.ccr.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ren R. Bcr-Abl efficiently induces a myeloproliferative disease and production of excess interleukin-3 and granulocyte-macrophage colony-stimulating factor in mice: a novel model for chronic myelogenous leukemia. Blood. 1998;92:3829–3840. [PubMed] [Google Scholar]
- Zhao C, Blum J, Chen A, Kwon HY, Jung SH, Cook JM, Lagoo A, Reya T. Loss of beta-catenin impairs the renewal of normal and CML stem cells in vivo. Cancer cell. 2007;12:528–541. doi: 10.1016/j.ccr.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JL, Ma C, O'Connell RM, Mehta A, Diloreto R, Heath JR, Baltimore D. Conversion of danger signals into cytokine signals by hematopoietic stem and progenitor cells for regulation of stress-induced hematopoiesis. Cell stem cell. 2014;14:445–459. doi: 10.1016/j.stem.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.