ABSTRACT
Herpes simplex virus (HSV) is a widespread pathogen that causes epithelial lesions with recurrent disease that manifests over a lifetime. The lifelong aspect of infection results from latent viral infection of neurons, a reservoir from which the virus reactivates periodically. Recent work has demonstrated the breadth of genetic variation in globally distributed HSV strains. However, the amount of variation or capacity for mutation within one strain has not been well studied. Here we developed and applied a streamlined new approach for assembly and comparison of large DNA viral genomes such as HSV-1. This viral genome assembly (VirGA) workflow incorporates a combination of de novo assembly, alignment, and annotation strategies to automate the generation of draft genomes for large viruses. We applied this approach to quantify the amount of variation between clonal derivatives of a common parental virus stock. In addition, we examined the genetic basis for syncytial plaque phenotypes displayed by a subset of these strains. In each of the syncytial strains, we found an identical DNA change, affecting one residue in the gB (UL27) fusion protein. Since these identical mutations could have appeared after extensive in vitro passaging, we applied the VirGA sequencing and comparison approach to two clinical HSV-1 strains isolated from the same patient. One of these strains was syncytial upon first culturing; its sequence revealed the same gB mutation. These data provide insight into the extent and origin of genome-wide intrastrain HSV-1 variation and present useful methods for expansion to in vivo patient infection studies.
IMPORTANCE
Herpes simplex virus (HSV) infects more than 70% of adults worldwide, causing epithelial lesions and recurrent disease that manifests over a lifetime. Prior work has demonstrated that HSV strains vary from country to country and between individuals. However, the amount of variation within one strain has not been well studied. To address this, we developed a new approach for viral genome assembly (VirGA) and analysis. We used this approach to quantify the amount of variation between sister clones of a common parental virus stock and to determine the basis of a unique fusion phenotype displayed by several variants. These data revealed that while sister clones of one HSV stock are more than 98% identical, these variants harbor enough genetic differences to change their observed characteristics. Comparative genomics approaches will allow us to explore the impacts of viral inter- and intrastrain diversity on drug and vaccine efficacy.
INTRODUCTION
Human herpes simplex viruses (HSV) are large DNA viruses of the family Herpesviridae which infect at mucosal surfaces before moving into the peripheral nervous system (1). In neurons, HSV establishes a unique latent form of infection, where periods of quiescence are interspersed with bursts of reactivation that reseed epithelial cells and additional neurons and enable transfer to new hosts (2–4). HSV infections present a continuing public health challenge because the latent viral reservoir in neurons can lead to lifelong disease and viral shedding. Present statistics put the seroprevalence of HSV-1 at over 60% of the adult population of the United States and that of HSV-2 at 20% (5). Although historically, HSV-1 was the main causative agent of oral lesions and HSV-2 of genital lesions, an increasing percentage of genital herpesvirus cases are due to HSV-1 infection (6–8). This shift creates a need to understand the mechanisms of diversity and selection of HSV-1 and -2 during human infection and to understand the selective forces and bottlenecks that influence coinfection and viral adaptation (9–11).
Recently, we used a combination of Sanger sequencing and Illumina high-throughput sequencing to characterize the genomes of a collection of HSV-1 strains from six countries around the world (12). The genetic diversity revealed by this study included coding and noncoding variations, resulting from mutational mechanisms including single nucleotide polymorphisms (SNPs), insertions and deletions, variations in homopolymer tracts and other types of short sequence repeats, and recombination. In contrast to these disparate sources of variation, past estimates of mutation rate in HSV have focused solely on substitution events in conserved regions (13–18). For HSV-1, these estimates range from 1 × 10−8 to 1 × 10−9 substitutions per site per year, which overlooks the variability contributed by the multiple mechanisms listed above. In addition to demonstrating the breadth of genetic diversity, these findings highlighted a range of highly conserved DNA and amino acid regions that may serve as useful vaccine or therapeutic targets as well as diagnostic markers.
One unexpected finding of this study was that almost half of the newly sequenced genomes harbored frameshifts or deletions in proteins thought to play important functional roles in vivo (e.g., UL11, UL13, gH [UL22], UL56) (12, 19–24). In addition, one-quarter of the newly sequenced genomes contained evidence of polymorphic populations; e.g., a majority of sequence reads spanned a deleted UL56 region, and a minority of reads showed an intact UL56 gene. Such findings raised the possibility that viral populations consist of multiple variants, at least when propagated in vitro. In a 2006 study, Sato et al. found that rare subpopulations of the laboratory strain HSV-1 KOS and several clinical isolates had the ability to activate Toll-like receptor 2 (TLR2) in addition to the more standard TLR9 activation (25, 26). In a previous study, we found variants of HSV-1 F with an intact UL13 kinase gene or with a homopolymer-based frameshift (26). The UL13 gene is crucial for in vivo spread but is not required for growth in vitro (20, 27, 28). The origin of this intrastrain homogeneity and its role in human infections are unknown.
At present, whole-genome deep sequencing approaches are the most effective way to detect variations in a viral populations that occur anywhere in the viral genome and with varying penetrance (29, 30). However, the current approaches of rapid, homology-based methods that simply align new sequences to a reference genome are insufficient to detect the many variations that occur, such as rearrangements, large insertions and deletions, and frameshifts. De novo assembly approaches that do not rely on reference sequences are more complex and resource-intensive but allow comprehensive comparison of all classes and degrees of variation. Applying these approaches to HSV genomes has been challenging because of the G+C-rich genome, hundreds of variable-number tandem repeats (VNTRs), and large inverted structural repeats. Here we present a customized approach to overcome these challenges. The methods are used to streamline the assembly of new HSV genomes and to characterize variants of a single strain. We demonstrated these applications by sequencing four HSV-1 variants that share a syncytial, or fusogenic, spread phenotype. We found an identical DNA mutation in the gB fusion protein in all four syncytial variants. The identical gB cytoplasmic tail mutation has been previously described in both spontaneous and engineered HSV syncytial strains, and it is causative of a syncytial phenotype (31–33). These data raise questions about how selective pressure and frequency of mutations interact to form new genetic variants within a given strain and how widespread such variation is outside the laboratory. By facilitating rapid assembly of new HSV-1 genomes, these approaches will inform future studies of both inter- and intrastrain viral variation and evolution.
RESULTS
Syncytial variants found in classical HSV-1 stocks.
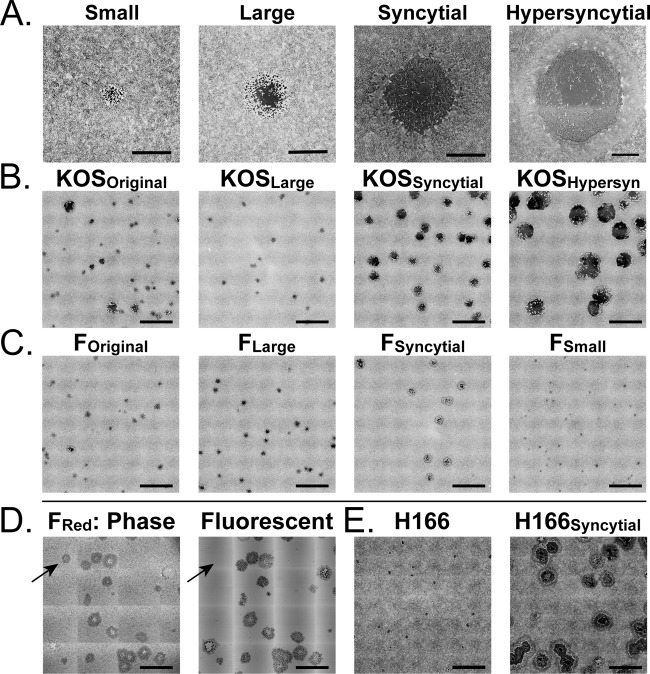
Analysis of HSV-1 stocks often reveals different visual plaque phenotypes when they are propagated in vitro. We have observed this phenomenon in many laboratory-adapted strains as well as in all low-passage-number clinical isolates grown in the lab (34–36). The plaque phenotypes range from small foci of infection to larger zones of cytopathic effect (CPE) and can include fusiform or syncytial plaques of different sizes (Fig. 1A). Most studies reduce this diversity by picking a single plaque, plating it to limiting dilution, picking a second derivative plaque, and repeating the process until homogeneity is achieved. The source of the initial diversity of plaque morphologies remains unexplained. Moreover, such diversity raised the possibility of additional underlying genetic variation that may manifest in phenotypes observed only in vivo (e.g., affecting latency, reactivation, or transmission) or under alternative selective pressures such as drug treatment or neutralizing antibodies. To determine the extent of genetic variation in subcloned plaques of a single strain, we separated and purified several of the most easily identified syncytial variants of two common laboratory strains of HSV-1, F and KOS (Fig. 1B and C), for which intrastrain variability was previously detected (25, 26). At 72 h postinfection (hpi), plaque size varied from 1 to 18 mm2 for HSV-1 KOS and from 1 to 5 mm2 for HSV-1 F (Fig. 2A and B). Although the vast majority of plaques displayed the normal cell rounding CPE, we observed syncytial plaques in both stocks (15% in KOS and 5% in F) (Table 1). We selected plaques representing each phenotype and performed two additional rounds of plaque purification and reselection of the identical plaque phenotype. We termed the starting stock the original (e.g., HSV-1 KOSOriginal) and named the clonal plaque variants by their respective phenotypes (e.g., HSV-1 KOSSyncytial). Reversion or conversion to other plaque phenotypes in these stocks was rare, at less than 1% (Fig. 1B and C; Table 1).
FIG 1 .
Plaque morphologies in HSV-1 stocks, before and after plaque purification. Lab-passaged stocks of classical HSV-1 KOS and F strains contain multiple plaque morphologies. (A) With multiple rounds of limiting dilution, distinct plaque morphologies can be separated into populations that breed true and exhibit greatly reduced diversity (see Table 1 for details). (B) HSV-1 KOS can be separated into large, syncytial, and hypersyncytial variants. (C) HSV-1 F can be separated into large, syncytial, and small variants. (D) We used a previously described BAC-cloned HSV-1 F genome that has been modified to encode an mRFP fusion to the viral capsid (38). Recovery of this cloned genome into mammalian cells regenerates the plaque-morphology diversity observed in HSV-1 F stocks. Phase and fluorescence images reveal that in addition to diversity in plaque morphology, fluorescently tagged viral stocks exhibit diversity in fluorescence (additional images can be found in Fig. S1 in the supplemental material). (E) The low-passage-number clinical isolates HSV-1 H166 and H166Syncytial are distinct strains isolated from the cerebrospinal fluid of the same patient during an episode of viral meningitis. The size and appearance of cytopathic and syncytial plaques mirror those seen in the lab-isolated variants of HSV-1 KOS and F. Viruses were plated at limiting dilutions on monolayers of Vero cells and then fixed and stained with methylene blue at 72 hpi. Images were exported from Nikon NIS-Elements software. Contrast was inverted using Adobe Photoshop to show plaques more clearly. Bars: 1 mm (A), 5 mm (B, C, and E), and 2 mm (D). Further quantifications of plaque size distribution and frequency are found in Fig. 2 and Tables 1 and 2.
FIG 2 .

Distribution of plaque morphologies before and after plaque purification. The graph depicts the number of plaques with a given area for each variant of plaque morphology shown in Fig. 1. (A) Distribution of the original HSV-1 KOS stock and the large, syncytial, and hypersyncytial variants. Bins include the number of plaques with areas up to the value shown on the x axis (bins of 1 mm for HSV-1 KOS subclones). (B) Distribution of the original HSV-1 F stock and the large, small, and syncytial variants. Bins of 0.5 mm were used for HSV-1 F subclones. (C) Size distribution of plaques from clinical HSV-1 strains H166 and H166Syncytial, using bins of 1 mm. Solid black lines indicate parental virus stocks, green lines indicate variants with standard cytopathic effect (CPE), and purple lines indicate syncytial variants. All plaques were quantified at 72 hpi. Note that the x axis scale varies across panels.
TABLE 1 .
Quantification of plaque diversity in original and plaque-purified HSV-1 populations
| HSV-1 strain (no. of plaques) | % plaque type |
Avg size |
|||
|---|---|---|---|---|---|
| Cell rounding | Syncytial | Hypersyncytial | Area (mm2) | Diam (mm) | |
| KOSOriginal (351) | 85.2 | 14.0 | 0.9 | 1.2 | 1.2 |
| KOSLarge (208) | 100 | 0 | 0 | 0.8 | 1.0 |
| KOSSyncytial (239) | 0 | 99.6 | 0.4 | 3.7 | 2.2 |
| KOSHypersyncytial (127) | 0 | 0.8 | 99.2 | 12.5 | 4.0 |
| FOriginal (234) | 95.3 | 4.7 | 0 | 0.7 | 1.0 |
| FLarge (195) | 100 | 0 | 0 | 1.0 | 1.1 |
| FSyncytial (119) | 0 | 100 | 0 | 2.3 | 1.7 |
| FSmall (122) | 100 | 0 | 0 | 0.3 | 0.6 |
| FRed (199) | 94.0 | 6.0 | 0 | 1.6 | 1.4 |
| H166 (65) | 100 | 0 | 0 | 0.7 | 1.0 |
| H166Syncytial (43) | 0 | 100 | 0 | 6.1 | 2.8 |
Variants recur in a BAC-cloned HSV-1 strain.
A single genome of HSV-1 F strain was previously captured in a bacterial artificial chromosome (BAC) (37) and modified by replacing the VP26 capsid protein gene (UL35) with a hybrid gene fusing monomeric red fluorescent protein (mRFP) coding sequences to the 5′ end of the UL35 open reading frame (mRFP1-VP26) (38). We received an aliquot of the virus stock produced from this BAC (HSV-1 F-GS2822; termed HSV-1 FRed here). Upon first plating, we detected a variety of plaque phenotypes, similar to that seen in HSV-1 FOriginal (Fig. 1D). Although syncytial variants are a minority of the overall population (Table 2), the capacity to regenerate this phenotype must lie in the single genome originally cloned into the BAC. Furthermore, the presence of mRFP-VP26 enabled us to examine fluorescence as well as syncytial plaque variants. We observed at least five visible plaque types: cell rounding CPE with capsid-based mRFP (81%), syncytial plaques with capsid-based mRFP (8%), dark nonfluorescent plaques with cell rounding CPE (10%), dark nonfluorescent plaques that were syncytial (1%), and, very rarely, plaques with diffuse red fluorescence that appeared to be non-capsid associated (see Fig. S1 in the supplemental material). In size and proportion, these plaques resemble those in the starting laboratory stock of strain F described above (data not shown). Since the BAC represents a single HSV genome, the capacity to produce the plaque diversity must arise either at the time of BAC DNA transfection into mammalian cells to recover infectious virus or during propagation of the recovered virus. This BAC derived virus has been in culture for a limited number of passages, indicating that genetic diversity arises quickly. As with HSV-1 FOriginal, we can plaque purify HSV-1 FRed to generate a stock that harbors only the majority plaque phenotype of standard CPE with capsid-based fluorescence (data not shown).
TABLE 2 .
Quantification of plaque diversity in BAC-passaged HSV-1 FRed
It seemed likely that these spontaneously occurring variants within each strain (e.g., KOS or F) would be closely related. The variants within each strain had nearly identical restriction fragment length polymorphisms (RFLP) patterns (see Fig. S2 in the supplemental material). We subjected these genomes to Illumina high-throughput sequencing to test their genomic similarity, to determine the number of background mutations, and to pinpoint the causative mutation of the syncytial phenotype.
VirGA: genome sequencing, assembly, and comparison workflow.
We developed a streamlined and facile approach for de novo virus genome assembly and comparison. Previously, we compared diverse alphaherpesvirus genomes using single-end Illumina sequencing data. Technical improvements in sequencing hardware and chemistry now allow longer paired-end reads. These improvements increase the amount of sequence data available and provide spatial separation data that can be used to assemble small sequences into longer blocks. To improve on prior approaches, we wrote computer code for assembly steps that had previously been done by hand, and we standardized approaches to check the validity of assembled genomes. The resulting viral genome assembly (VirGA) approach provides a fully automated procedure to assemble linear draft genome sequences from Illumina high-throughput sequencing (HTSeq) data (see Fig. S3 in the supplemental material for an overview).
VirGA involves initial quality filtering of HTSeq data, followed by de novo assembly of short sequence reads, and then a secondary assembly to join large blocks of continuous sequence (contigs). Next we place these large sequence blocks in order by comparison to a reference sequence of a related strain. An initial comparison of the new draft genome to the reference provides a quick assessment of coding sequences, major differences from the reference genome, and quality of the draft genome (see Fig. S4 in the supplemental material for an overview). Wherever possible, we used previously published tools with proven reliability, and developed custom approaches to join these in series. In several places, such as ordering of blocks of sequence based on the reference genome, we used previously developed methods in a novel way to attain the desired outcome.
We combined up to six strains of HSV-1 for Illumina paired-end sequencing. Using a library of 500-bp fragments, we obtained 100 or 300 bp paired-end reads from these fragments (Table 3). In general, 5 to 15 million sequence reads were obtained for each genome, and after quality filtering, we used 3 to 11 million sequence reads for de novo assembly (described below). Serial assemblies using progressively fewer data indicated that as few as 200,000 sequence reads (300-bp paired-end reads) would allow ~90% of the genome to have a coverage depth of >100 and approximately 90% of the proteins to be intact and gap free (see Fig. S5 in the supplemental material). However, using larger amounts of sequence data reduced the number and size of gaps and improved the assemblies further. Initial filtering steps included removal of sequences matching the genome of the host cells used to grow virus. The quantity of contaminating host DNA varies with each viral nucleocapsid DNA preparation, so, for instance, a much larger percentage of reads (23%, or 3.8 million of the original 16.5 million) (Table 3) were filtered from the HSV-1 FLarge sequencing reads than from the FSyncytial data.
TABLE 3 .
Sequencing statistics for HSV-1 variants
| HSV-1 strain | Paired-end read length (bp) | No. of reads |
Genome length | % with depth of ≥100 | GenBank accession no. | |
|---|---|---|---|---|---|---|
| Raw | Used for assembly | |||||
| KOSLarge | 100 | 14,595,444 | 10,365,343 | 150,147 | 95.5 | KM222721 |
| KOSSyncytial | 100 | 22,860,146 | 17,850,556 | 150,970 | 98.3 | KM222722 |
| KOSHypersyncytial | 100 | 27,307,331 | 21,072,432 | 150,585 | 98.8 | KM222723 |
| FLarge | 100 | 16,543,482 | 9,258,828 | 149,692 | 98.2 | KM222724 |
| FSyncytial | 100 | 14,666,531 | 11,134,103 | 148,997 | 96.6 | KM222725 |
| H166 | 300 | 10,497,790 | 8,244,309 | 152,409 | 99.8 | KM222726 |
| H166Syncytial | 300 | 5,732,563 | 3,267,339 | 152,262 | 96.7 | KM222727 |
For de novo assembly of these large DNA virus genomes, we used the short sequence assembly and K-mer expansion (SSAKE) program (39). Although our first explorations of herpesvirus genome assembly used just one round of de novo assembly, subsequent experience demonstrated the advantages of combining multiple de novo assembly approaches (40, 41). We produced multiple different outputs by varying the software parameters (see Materials and Methods and also see Text S1 in the supplemental material for details). The sequence blocks, or contigs, produced by eight variations of SSAKE de novo assembly were combined using a second round of de novo assembly (40, 41). We used Celera assembly software to combine the SSAKE sequences into even larger blocks of sequence (see Materials and Methods for details) (42). We then aligned the input sequence reads to the new draft genome, to assess coverage depth and check for polymorphisms. This revealed that patterns of coverage depth across the genome supersede HSV-1 strain variation (Fig. 3A and B). It suggests that conserved features of the HSV-1 genome, such as tandem repeats or G+C-rich sequences in the internal repeat regions, present a challenge for sequencing and/or assembly algorithms.
FIG 3 .
Deep sequence coverage of HSV-1 variants from each strain group. Preprocessed sequence reads were aligned to the new draft genomes to assess the coverage depth of each assembly. Coverage depth is plotted on a log10 scale (y axis) across the length of the HSV-1 genome (x axis). Major regions of the HSV-1 genome are diagrammed below the x axis, including the long and short internal repeats (IRL and IRS). Genomes and coverage are shown in a trimmed format (12), where the terminal copies of IRL and IRS are not included. Coverage tracks are overlaid for (A) HSV-1 KOS variants, (B) HSV-1 F variants, and (C) HSV-1 H166 and H166Syncytial. The total number of sequence reads obtained was different for each HSV-1 strain, which affects overall coverage depth (Table 3). However, peaks and valleys of coverage depth fall in similar locations on the HSV-1 genome, with the internal repeats (IRL and IRS) showing the most variability in coverage.
These large contigs were assembled into a draft genome by comparison to a reference sequence (HSV-1 strain 17; GenBank accession no. JN555585). In past work, we used manual comparison to the reference, via BLAST-based homology searches. To automate this process and make it less subjective, we used the Mugsy (43) aligner to determine regions of homology between the new contigs and the reference genome. We then developed a custom algorithm, called maf_net, to join the regions with the best homology to the reference genome. This process created a linear draft version of each genome, whose quality was analyzed further (see Materials and Methods for details; also, see Fig. S3 and S4 in the supplemental material for examples). Each genome was annotated, deposited in GenBank (see below and Table 3 for accession IDs), and compared to the other sequences.
Syncytial variants of HSV-1 KOS and F have identical gB tail mutations.
To determine the extent of variation among subclones of the same parental HSV-1 stock, we compared the percent identity at the DNA level between the genomes of KOSLarge, KOSSyncytial, and KOSHypersyncytial (99.5% identity) and of FLarge and FSyncytial (98.6% identity) (see Fig. S6 in the supplemental material). The metric of percent DNA identity encompasses SNPs, insertions, deletions, variations in short sequence repeats, genes and intergenic regions, and all classes of coding changes (silent, missense, and nonsense). DNA sequence alignment and phylogenetic clustering revealed that intrastrain variants clustered most closely with each other and with previously sequenced isolates of the same strains (see Fig. S6). At the amino acid level, the variants of HSV-1 KOS harbored missense or nonsense mutations in 17 proteins, while missense variations occurred in 15 proteins between the variants FLarge and FSyncytial (see Table S1 for the list). This is approximately half the number of variant proteins (27) (see Table S1 for the list) found between two published isolates of McKrae that were grown and sequenced by separate labs (44, 45).
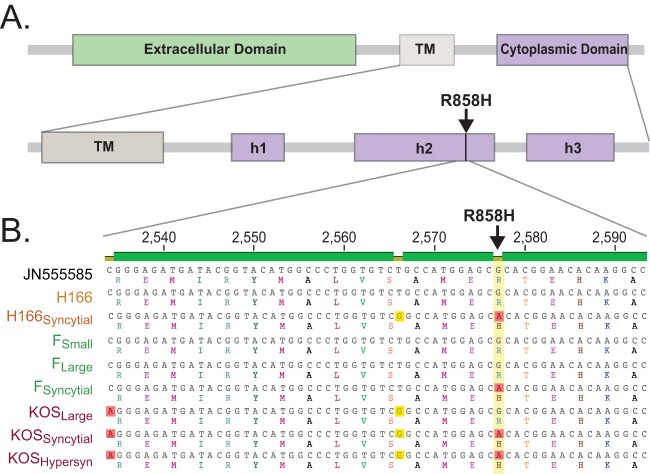
To map the source of the syncytial phenotypes observed in Fig. 1, we searched for amino acid differences present in all syncytial strains but absent from any of the nonsyncytial strains. Accordingly, we first compared all strains back to the reference strain HSV-1 17 and then compared the amino acid differences observed in KOSSyncytial, KOSHypersyncytial, and FSyncytial to those found in KOSLarge and FLarge. The only amino acid difference found only in all three syncytial strains, and absent from both nonsyncytial variants, was residue R858H in the cytoplasmic tail of the core HSV-1 fusion protein glycoprotein B (UL27) (Fig. 4A). The DNA difference underlying this amino acid change was also identical in syncytial strains and absent from nonsyncytial strains. We confirmed this mutation by Sanger sequencing (Fig. 4B). The gB R858H mutation has been previously observed to occur spontaneously in the lab and has been purposely engineered into strains as well (31–33). In all cases, the presence of R858H is sufficient to induce syncytium formation.
FIG 4 .
Syncytial variation R858H in glycoprotein B (gB [UL27]). (A) Diagram of the core fusion glycoprotein B (gB [UL27]) with labeling of domains adapted from reference 65 (TM, transmembrane domain; h, helix). (B) DNA sequence alignment for a segment of the gB (UL27) tail, showing the G-to-A nucleotide mutation that is shared by all syncytial strains and is absent from all nonsyncytial strain (yellow column with red boxes; open reading frame [ORF] position 2,577). Amino acid translations are shown beneath each DNA sequence, demonstrating the resulting R-to-H mutation in the gB protein. Two silent DNA variations are visible in this region as well; one is shared by all KOS variants (red boxes; ORF position 2,534), while the other is coincidentally shared by H166Syncytial as well (yellow boxes; ORF position 2,566).
Clinical syncytial strain HSV-1 H166 contains identical mutations.
Although syncytial cells are a hallmark of HSV infection in many histological preparations, purely syncytial strains are rarely isolated from patients. However, such strains are frequently observed to occur spontaneously during HSV-1 propagation in the lab. This finding raises the question of whether these strains arise solely under lab conditions or represent the expansion of a minority population present in the patient and in the original lesion or tissue sample. Heller et al. described one example of a clinical strain, H166Syncytial, that was syncytial upon first isolation from a patient with viral proctitis and subsequent viral meningitis (46). Strain H166Syncytial was present upon first culture from both cerebrospinal fluid (CSF) and rectal samples from this patient, making it highly unlikely that the virus spontaneously converted at the same time in both samples. In the CSF sample, H166Syncytial co-occurred with a genetically unrelated HSV-1 strain (labeled HSV-1 H166), and in the rectal sample, it co-occurred with a genetically unrelated HSV-2 strain (46). We sequenced strains H166Syncytial and H166 to investigate the basis of the syncytial phenotype in this non-laboratory-based variant of HSV-1.
HSV-1 strains H166Syncytial and H166 were sequenced and assembled as described above, except that the newer MiSeq 300-bp paired-end sequencing technology was used (Fig. 3C; Table 3). Heller et al. previously found by RFLP analysis that H166syn and H166 were unrelated strains of HSV-1 (46). We confirmed these findings by whole-genome comparison. Unlike the clonal variants above, these strains are only 97.2% identical at the DNA level and have coding differences in 63 of the 74 proteins encoded by the viral genome (see Fig. S6 and Table S1 in the supplemental material for details). In the gB (UL27) locus of H166Syncytial, but not of H166, we found the same DNA and amino acid mutations as observed in the syncytial variants of KOS and F (Fig. 4B). This finding suggests that syncytial variants of HSV-1 can and do arise in patients during active states of diseases such as meningitis and proctitis.
DISCUSSION
Due to the availability of high-throughput sequencing data and analysis software (VirGA), our results demonstrate for the first time the degree of genetic relatedness and variation that results from subcloning members of a parental HSV-1 strain population (either by plaque picking or by cloning in BACs). Although the subcloned variants of HSV-1 KOS and F differ in only 1 to 2% of their genomes, they have distinctly different plaque morphologies and harbor missense coding variations in 15 to 17 proteins. These variations will need to be assessed singly, in detail, and in a variety of assays, to determine their biological impact(s). These data suggest that while plaque purification can produce a virus stock with visually homogeneous plaques, there is no guarantee that picking a plaque that resembles the parental population will also retrieve a genome that contains the most common genotype of the parental population. As we discovered previously, when we unintentionally plaque purified a UL13 kinase mutant of strain HSV-1 F, it is possible to purify a visually and phenotypically normal virus in vitro and still have that mutant be nonrepresentative of the majority of HSV-1 F strain plaques (26).
The VirGA sequencing method described here has the capacity to speed de novo viral genome assembly and genotype comparison among strains, as demonstrated here using clonal variants and clinical HSV-1 strains as examples. VirGA can handle multiple types of input DNA: bulk preparations of DNA from cultured virus stocks, minimally passaged clinical isolates, or direct isolates of patient samples containing abundant host DNA. VirGA provides a uniform and automated method to remove contaminating host DNA sequences and to assemble the best possible draft genome from the remaining viral sequence coverage. This approach will facilitate the complete genetic comparison of existing lab-cultured strains, and it will also enable large-scale comparison of hundreds of minimally passaged clinical strains. As methods for DNA isolation and library preparation improve, VirGA will be useful for the high-throughput comparison of uncultured isolates as well.
As with all deep-sequencing endeavors, these approaches raise questions that were not apparent when the project started. We used plaque size and syncytial phenotypes as easily scored measures to quantitate variation in our virus stocks, and we also anticipated that there might be underlying genetic variation unrelated to these phenotypes. Deep sequencing promised to reveal which of the many previously described syncytial mutations might be cause of the observed morphologies; these include a variety of mutations in gB (UL27) (31–33, 47, 48) but also mutations in gH (UL22), UL20, UL24, gK (UL53), and gD (US6) (49–55). We were surprised to find that the disparate HSV-1 F and KOS syncytial variants harbored identical gB tail mutations. What are the mechanisms that lead to this common variation: in vitro selection pressure during multiple rounds of passaging or a DNA-sequence hot spot for mutations?
The clinical isolates H166 and H166Syncytial provide some insight, since the H166Syncytial genome carries an identical gB tail mutation. These isolates were taken from a patient suffering coincident proctitis and meningitis (46). The H166Syncytial isolate was present in the originating patient upon first culturing of both rectal and cerebrospinal fluid (CSF) samples, and the isolates were collected over a week apart. The observation of the syncytial plaque morphology in two distinct isolations, from independent body sites and culture dates, makes it unlikely to be an artifact of culturing or in vitro selection pressure. Nonsyncytial isolates were cultured from the same samples, one of which is the HSV-1 H166 strain sequenced here. The other was a nonsyncytial HSV-2 isolate. Finding an identical gB tail mutation in H166Syncytial, as observed in the two spontaneous in vitro isolates FSyncytial and KOSSyncytial, suggests that these mutations can arise spontaneously in vivo and are not solely the result of in vitro selective pressure.
The authors who first described H166 and H166Syncytial noted that it is unusual for HSV-1 to cause meningitis (46). Although H166 was detected only in the CSF, H166Syncytial was detected in both CSF and rectal samples. This finding raises the possibility that despite its syncytial characteristics, H166Syncytial was able to transit from the rectal lesion to the meninges of the brain. Syncytial isolates tested in animal models generally do not spread well from the periphery, so this transit may well be linked to the coincident presence of syncytial and nonsyncytial HSV strains (34, 56, 57). However, prior studies suggest that syncytial strains are more lethal than nonsyncytial strains when introduced via direct brain injection in animal models (34). This raises the interesting question of whether syncytial strains may enhance viral fitness in vivo, for instance by providing alternative mechanisms of cell-to-cell spread. Surprisingly, at least three prior cases of HSV-1 and -2 coinfection have been described, where a syncytial isolate was isolated at the same time as a nonsyncytial, unrelated strain from the same patient (58–60). One of these isolates was fully characterized and also found to have a gB cytoplasmic-tail mutation, this time in the HSV-2 isolate (58). Direct pairings of syncytial and nonsyncytial viruses in animal models will help to discern whether syncytial variants contribute to fitness in vivo. In addition, genome-wide surveillance of intra- and interstrain diversity in ongoing human infections will contribute to our understanding of the role of viral variation(s) in human disease. The viral genome assembly approach described here will assist in exploring the rate at which HSV evolves and adapts under selective pressures in vivo.
MATERIALS AND METHODS
Virus stocks and DNA preparation.
HSV-1 strain F was received from Ejercito and colleagues (61), HSV-1 strain KOS was received from Ramachandran and colleagues (62), and HSV-1 strains H166 and H166Syncytial were received from Dix and colleagues (34, 46). The BAC-derived virus HSV-1 F-GS2822 (referred to here as HSV-1 FRed for clarity) was received from Antinone and Smith (38) as a virus stock that had already been recovered into cells. Variants of strains F and KOS (including FRed) were grown on monolayers of African green monkey kidney cells (Vero cells; ATCC CCL-81). Stocks were grown Dulbecco’s minimum essential medium (DMEM; HyClone) containing 2% fetal bovine serum (FBS; HyClone) and penicillin-streptomycin (Life Technologies). Virus was plated to limiting dilution from originally received stocks, and plaques of various sizes or phenotypes were chosen for further propagation. Plaques capable of maintaining phenotype through three rounds of plating and plaque selection were expanded for sequencing. Virus strains H166 and H166Syncytial were grown on human lung fibroblast cells (MRC5 cells; ATCC CCL-171) without plaque purification and with minimal expansion before sequencing.
Viral nucleocapsid DNA was isolated as previously described (41, 63, 64). Briefly, host cells were infected at a high multiplicity of infection (MOI = 5); both cells and media were collected at 24 h postinfection (hpi). Pellets were rinsed, resuspended, extracted twice with Freon, and pelleted through a glycerol step gradient. Viral nucleocapsids were lysed using SDS and proteinase K, double extracted using phenol-chloroform, and ethanol precipitated. Viral DNA was resuspended in TE (10 mM Tris, pH 7.6; 1 mM EDTA).
Plaque area and phenotype quantification.
To obtain individual plaques of each strain, serial dilutions of each virus were made; dilutions ranged from 10−5 to 10−7. Six-well plates with confluent monolayers of Vero cells were inoculated with serial dilutions and incubated for 1 h, and then the inoculum was replaced with medium containing 1% methylcellulose. At 72 hpi, the monolayers were washed twice with 1× phosphate-buffered saline (PBS; HyClone) and stained with 0.5% methylene blue in 70% methanol for at least 30 min.
Image capture and analysis were performed on a Nikon Eclipse Ti-E inverted microscope, with Nikon NIS-Elements AR imaging software (version 4.13). Unstained plates were imaged after washing with PBS but before staining with methylene blue; stained plates were imaged after drying. The motorized microscope stage used a map of a 6-well plate with x, y, and z coordinates of the middle of each well, to maintain consistent imaging parameters for all plates. Large 7- by 7-tile images of each well were taken to obtain an overall depiction of the contents of each well. Image stitching was performed by using default settings in NIS-Elements. Plaques were analyzed in NIS-Elements by drawing a region of interest (ROI) around each countable plaque. Countable plaques were defined as plaques that were not distorted or overlapping another plaque. Once ROIs were identified, the area (in square millimeters) of each plaque was measured using the NIS-Elements Perform Measurement (No Binary) function.
The same ROIs were used to score the phenotype of each analyzed plaque, using a numbering system that corresponded to the type of cytopathic effect observed for each plaque. Plaques scored as showing cytopathic effect (CPE) displayed normal cell rounding after infection. Plaques scored as syncytial had a fused center, with the appearance of multiple nuclei within a shared membrane. Syncytial plaques often had a halo of fused cells, with a center that resembled a paint splatter. Syncytial plaques were typically around 1.5 to 2 times larger than normal CPE variants (Table 1). Plaques scored as hypersyncytial plaques had the same morphology as syncytial plaques but were significantly larger, i.e., about 3 times larger than normal CPE plaques (Table 1).
Illumina sequencing.
Barcoded sequencing libraries for each virus were prepared following the manufacturer’s protocol for sequencing of genomic DNA (Illumina TruSeq DNA sample preparation kit). Viral nucleocapsid DNA (1 to 5 µg) was used as the input for library preparation. Five to six libraries were multiplexed per flow cell lane; the DNA fragment size selected for each library was centered at 550 bp. Libraries of HSV-1 KOS and F subclones were sequenced at the Sequencing Core Facility of the Lewis-Sigler Institute for Integrative Genomics (Princeton University), using paired-end, 100-bp sequencing protocols on an Illumina HiSeq2000 with version 2 chemistry. Illumina real-time analysis (RTA 1.12.4.2) software provided image analysis and base calling under default settings. Libraries of HSV-1 H166 and H166Syncytial strains were sequenced on an Illumina MiSeq at Pennsylvania State University, using 300-bp paired-end, version 3 sequencing chemistry. Image analysis and base calling were done under default settings with MiSeq Control software (MCS) version 2.3.0.
VirGA.
Viral genome assembly (VirGA) and the open-source components that it uses are described at https://bitbucket.org/szparalab. VirGA is a collection of shell, perl, and python scripts that combine common bioinformatics software packages to assemble high-confidence draft genomes from raw Illumina sequencing reads. VirGA also utilizes a custom set of scripts called the Virus Assembly Pipeline (VAMP) tools (available in the same repository) for genome linearization and comparison; these scripts are based on our previous work assembling HSV and pseudorabies virus genomes (26, 41). VirGA is designed for both desktop and cluster-computing usage and is packaged with a self-extracting installer. The VirGA workflow is comprised of four steps: (i) raw read preprocessing, (ii) de novo contig assembly, (iii) genome linearization and annotation, and (iv) assembly assessment (see Fig. S3 in the supplemental material). These steps are described in further detail in the supplemental materials and methods (see Text S1 in the supplemental material). VirGA can be initiated at any of the four steps, for instance, to allow reannotation of a finished genome that has had gaps filled in by PCR. The VirGA workflow supports multithreading for faster processing, when parallel processing cores are available. It can be run using a scheduling system (e.g., PBS or Torque) or interactively (bash) and in conjunction with cluster-based module software systems. All scripts and settings used for each run are stored with the output, to allow easy record keeping and preservation of parameters for future replication (see Fig. S4 in the supplemental material). Complete records of all VirGA outputs from this study are archived at https://scholarsphere.psu.edu/collections/sf268c193.
PCR validation of regions of interest.
PCR was used to validate several regions of interest. Primers for these are listed in Table S2 in the supplemental material. Dilute nucleocapsid DNA was boiled for 10 min and snap cooled on ice for 5 min. Amplification was conducted on an Eppendorf Mastercycler, in a 50-µl reaction volume containing 27.5 µl nuclease-free water, 1 µl TaKaRa Ex DNA polymerase (TaKaRa), 5 µl TaKaRa 10× buffer, 1 µl each of 2 µM concentrations of forward and reverse primers, 1.2 M betaine (Sigma), 2% dimethyl sulfoxide (DMSO; Thermo), 0.5 µl of 400 µM concentrations of deoxynucleoside triphosphates, and 1 µl of the template DNA. The reaction conditions were as follows: 95°C for 3 min; then 25 to 35 cycles (depending upon ease of amplification) of 30 s at 95°C, 30 s at 50°C, 1 min at 68°C; and finally 5 min at 68°C. Products were separated by electrophoresis and gel purified before Sanger sequencing.
Nucleotide sequence accession numbers.
The HSV genome sequences determined here have been deposited in GenBank under the following accession numbers: HSV-1 KOSLarge, KM222721; HSV-1 KOSSyncytial, KM222722; HSV-1 KOSHypersyncytial, KM222723; HSV-1 FLarge, KM222724; HSV-1 FSyncytial, KM222725; HSV-1 H166, KM222726; HSV-1 H166Syncytial, KM222727.
SUPPLEMENTAL MATERIAL
Supplemental materials and methods. This document includes a detailed description of the VirGA workflow, along with details for the genetic distance analysis and RFLP methods. Download
HSV-1 FRed includes cell-rounding and syncytial plaques with either punctate, diffuse, or dark fluorescence resulting from its mRFP-capsid label. HSV-1 FRed is a virus stock that was generated from a cloned BAC copy of the HSV-1 F genome. Recovery of HSV-1 FRed into mammalian cells recreates the plaque morphology diversity observed in HSV-1 F stocks. The mRFP-capsid label reveals additional variation that affects the fluorescent label. (A) The majority of plaques showed standard cell-rounding CPE, with punctate fluorescence from the mRFP-capsid fusion protein (see Table 2 for quantitation). (B) The next most frequent variants were syncytial plaques with punctate capsid-based fluorescence. (C) Rarely, plaques revealed diffuse fluorescence. (D and E) Up to 1 in 10 plaques were dark and lacked fluorescence. Cell-rounding (D) and syncytial (E) variants are shown here, next to ordinary fluorescent plaques, to demonstrate that the lack of fluorescence was not due to image acquisition settings. Download
RFLP analysis of HSV-1 KOS and F subclones confirms draft genomes but obscures syncytial differences. Restriction fragment length polymorphism analysis (RFLP) revealed no major differences in digest patterns, for subclones of a given strain. (A) BamHI and (B) HindIII RFLPs were analyzed for subclones of HSV-1 KOS and F described in Fig. 1. Marker lanes (sizes are in kilobases) are shown to the left of each gel. Download
Overview of HSV genome sequencing using the viral genome assembly (VirGA) workflow. The VirGA workflow requires an input of high-throughput Illumina sequence read data from the viral genome of interest. We generated this by expanding a viral stock, isolating viral nucleocapsid DNA, preparing a library of genome fragments, and collecting high-throughput, paired-end sequence reads using an Illumina HiSeq or MiSeq instrument. In VirGA step 1, host sequences and quality-reducing contaminants are removed. In step 2, the viral sequences are assembled into long stretches of continuous sequence (contigs) by the use of two de novo assemblers, SSAKE and Celera. In step 3, these long stretches of sequence are arranged in order by comparison to a reference genome. Gaps can be closed by using the GapFiller program to search for overlapping sequences in the input data. Annotations are transferred from the reference genome to the new draft genome at this stage. In step 4, the original sequence reads are aligned to the draft consensus to check the assembly quality. Best practices in HSV genome assembly involve wet-bench validations of each assembly, such as PCR verification of key differences or RFLP analysis of genome orientation. Download
Example of a VirGA output summary. As part of its output, VirGA generates an interactive HTML file for each draft genome assembly, which can be opened in any web browser. This file summarizes statistics about the assembly and links to additional files. This image includes excerpted sections from a full VirGA output. Complete records of all VirGA outputs from this study are archived at https://scholarsphere.psu.edu/collections/sf268c193. The VirGA report summary includes statistics about the new draft genome, such as length, percent with coverage depth of >100-fold, number of gaps, and number of intact (gap-free) proteins. Links are provided to alignments of each gene and protein versus the reference genome; these are grouped into those without errors (green text) and those needing user attention (red text). Alignments for noncoding features are included as well. Below the summary, the extensive VirGA detailed report includes statistics on the number of sequence reads filtered out during the preprocessing steps, the number of contigs produced during SSAKE and Celera assembly, the gaps closed by GapFiller, and the results of quality assessment when the sequence reads are aligned to the new draft genome. From this extensive report, only a histogram of sequence read quality per base is shown here. Download
Serial assemblies demonstrate how increasing amounts of input data produce improved viral genome assemblies. We used VirGA to generate serial assemblies of both 100-bp paired-end sequence reads (A) and 300-bp paired-end sequence reads (B), doubling the number of input reads each time (x axis). For each assembly, we calculated the percentage of the genome with sequencing coverage depth greater than or equal to 100-fold (solid line) and the percentage of proteins that were assembled intact (gap free; dashed line) and counted the undetermined bases (“N” in the final assembly; blue histograms). (A) For 100-bp, paired-end data, we observed that a minimum of 400,000 reads were needed to produce an HSV-1 assembly with 90% of the proteins intact. (B) For 300-bp, paired-end data, we observed that just 200,000 reads were needed to produce an HSV-1 assembly with 90% of the proteins intact. Results were verified with serial assemblies of data from HSV-1 FSyncytial and H166Syncytial as well (data not shown). Download
Genetic distance diagram, DNA identity, and protein difference statistics summarize relatedness of HSV-1 strains and subclones. We followed the same approach as used previously (Szpara et al., J. Virol, 88:1209–1227, 2014) to generate a genetic distance tree from whole-genome alignments of the strains and subclones shown (see Materials and Methods for details). Branch points show confidence values. We calculated the percent DNA identity and number of variant proteins for selected sets of sequences; these are shown on the right. NCBI accession numbers for strains not sequenced in this paper (marked with asterisks) are as follows: H129, GU734772; 17, JN555585; McKrae, JQ730035 and JX142173; KOS, JQ780693 and JQ673480. Download
List of proteins that vary between subclones of each strain discussed in the text.
Primers for the PCR validations described in Materials and Methods.
ACKNOWLEDGMENTS
This work was supported by K22 AI095384 (M.L.S.) and the Virus Pathogens Resource (ViPR), a Bioinformatics Resource Center (BRC) funded by NIAID. Additional support was provided by a Center Grant (NIH/NIGMS P50 GM071508) and NIH P40 RR 018604 (L.W.E. and M.L.S.).
We appreciate the kind contributions of HSV strains for this study by Richard Dix, Paul Kinchington, Bernard Roizman, and Greg Smith. We thank members of the Enquist and Szpara labs for their contributions and feedback, including Daniel Renner for ongoing VirGA support.
Footnotes
Citation Parsons LR, Tafuri YR, Shreve JT, Bowen CD, Shipley MM, Enquist LW, Szpara ML. 2015. Rapid genome assembly and comparison decode intrastrain variation in human alphaherpesviruses. mBio 6(2):e02213-14. doi:10.1128/mBio.02213-14.
REFERENCES
- 1.Roizman B, Sears E. 1996. Herpes simplex viruses and their replication, p 1043–1107. In Fields BN, Knipe DM, Howley PM (ed), Fields virology, 3rd ed. Lippincott Raven, Philadelphia, PA. [Google Scholar]
- 2.Granstedt AE, Bosse JB, Thiberge SY, Enquist LW. 2013. In vivo imaging of alphaherpesvirus infection reveals synchronized activity dependent on axonal sorting of viral proteins. Proc Natl Acad Sci U. S. A. 110:E3516–E3525. doi: 10.1073/pnas.1311062110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koyuncu OO, Hogue IB, Enquist LW. 2013. Virus infections in the nervous system. Cell Host Microbe 13:379–393. doi: 10.1016/j.chom.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steiner I, Kennedy PG, Pachner AR. 2007. The neurotropic herpes viruses: herpes simplex and varicella-zoster. Lancet Neurol 6:1015–1028. doi: 10.1016/S1474-4422(07)70267-3. [DOI] [PubMed] [Google Scholar]
- 5.Bradley H, Markowitz LE, Gibson T, McQuillan GM. 2014. Seroprevalence of herpes simplex virus types 1 and 2—United States, 1999–2010. J Infect Dis 209:325–333. doi: 10.1093/infdis/jit458. [DOI] [PubMed] [Google Scholar]
- 6.Samra Z, Scherf E, Dan M. 2003. Herpes simplex virus type 1 is the prevailing cause of genital herpes in the Tel Aviv area, Israel. Sex Transm Dis 30:794–796. doi: 10.1097/01.OLQ.0000079517.04451.79. [DOI] [PubMed] [Google Scholar]
- 7.Garceau R, Leblanc D, Thibault L, Girouard G, Mallet M. 2012. Herpes simplex virus type 1 is the leading cause of genital herpes in New Brunswick. Can J Infect Dis Med Microbiol 23:15–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kortekangas-Savolainen O, Orhanen E, Puodinketo T, Vuorinen T. 2014. Epidemiology of genital herpes simplex virus type 1 and 2 infections in southwestern Finland during a 10-year period (2003–2012). Sex Transm Dis 41:268–271. doi: 10.1097/OLQ.0000000000000101. [DOI] [PubMed] [Google Scholar]
- 9.Norberg P. 2010. Divergence and genotyping of human alpha-herpesviruses: an overview. Infect Genet Evol 10:14–25. [DOI] [PubMed] [Google Scholar]
- 10.Nishiyama Y, Kimura H, Daikoku T. 1991. Complementary lethal invasion of the central nervous system by nonneuroinvasive herpes simplex virus types 1 and 2. J Virol 65:4520–4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paludan SR, Bowie AG, Horan KA, Fitzgerald KA. 2011. Recognition of herpesviruses by the innate immune system. Nat Rev Immunol 11:143–154. doi: 10.1038/nri2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szpara ML, Gatherer D, Ochoa A, Greenbaum B, Dolan A, Bowden RJ, Enquist LW, Legendre M, Davison AJ. 2014. Evolution and diversity in human herpes simplex virus genomes. J Virol 88:1209–1227. doi: 10.1128/JVI.01987-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakaoka H, Kurita K, Iida Y, Takada S, Umene K, Kim YT, Ren CS, Nahmias AJ. 1994. Quantitative analysis of genomic polymorphism of herpes simplex virus type 1 strains from six countries: studies of molecular evolution and molecular epidemiology of the virus. J Gen Virol 75:513–527. doi: 10.1099/0022-1317-75-3-513. [DOI] [PubMed] [Google Scholar]
- 14.McGeoch DJ, Cook S. 1994. Molecular phylogeny of the alphaherpesvirinae subfamily and a proposed evolutionary timescale. J Mol Biol 238:9–22. doi: 10.1006/jmbi.1994.1264. [DOI] [PubMed] [Google Scholar]
- 15.McGeoch DJ, Dolan A, Ralph AC. 2000. Toward a comprehensive phylogeny for mammalian and avian herpesviruses. J Virol 74:10401–10406. doi: 10.1128/JVI.74.22.10401-10406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drake JW, Hwang CB. 2005. On the mutation rate of herpes simplex virus type 1. Genetics 170:969–970. doi: 10.1534/genetics.104.040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Norberg P, Tyler S, Severini A, Whitley R, Liljeqvist J, Bergström T. 2011. A genome-wide comparative evolutionary analysis of herpes simplex virus type 1 and varicella zoster virus. PLoS One 6:1–8. doi: 10.1371/journal.pone.0022527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolb AW, Ané C, Brandt CR. 2013. Using HSV-1 genome phylogenetics to track past human migrations. PLoS One 8:e76267. doi: 10.1371/journal.pone.0076267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klopfleisch R, Klupp BG, Fuchs W, Kopp M, Teifke JP, Mettenleiter TC. 2006. Influence of pseudorabies virus proteins on neuroinvasion and neurovirulence in mice. J Virol 80:5571–5576. doi: 10.1128/JVI.02589-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coller KE, Smith GA. 2008. Two viral kinases are required for sustained long distance axon instruments transport of a neuroinvasive herpesvirus. Traffic 9:1458–1470. doi: 10.1111/j.1600-0854.2008.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baines JD, Roizman B. 1992. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J Virol 66:5168–5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim I-J, Chouljenko VN, Walker JD, Kousoulas KG. 2013. Herpes simplex virus 1 glycoprotein M and the membrane-associated protein ul11 are required for virus-induced cell fusion and efficient virus entry. J Virol 87:8029–8037. doi: 10.1128/JVI.01181-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forrester A, Farrell H, Wilkinson G, Kaye J, Davis-Poynter N, Minson T. 1992. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J Virol 66:341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kehm R, Rösen-Wolff A, Darai G. 1996. Restitution of the UL56 gene expression of HSV-1 HFEM led to restoration of virulent phenotype; deletion of the amino acids 217 to 234 of the UL56 protein abrogates the virulent phenotype. Virus Res 40:17–31. doi: 10.1016/0168-1702(96)80248-6. [DOI] [PubMed] [Google Scholar]
- 25.Sato A, Linehan MM, Iwasaki A. 2006. Dual recognition of herpes simplex viruses by TLR2 and TLR9 in dendritic cells. Proc Natl Acad Sci U. S. A. 103:17343–17348. doi: 10.1073/pnas.0605102103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szpara ML, Parsons L, Enquist LW. 2010. Sequence variability in clinical and laboratory isolates of herpes simplex virus 1 reveals new mutations. J Virol 84:5303–5313. doi: 10.1128/JVI.00312-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coulter LJ, Moss HW, Lang J, McGeoch DJ. 1993. A mutant of herpes simplex virus type 1 in which the UL13 protein kinase gene is disrupted. J Gen Virol 74:387–395. doi: 10.1099/0022-1317-74-3-387. [DOI] [PubMed] [Google Scholar]
- 28.Purves FC, Roizman B. 1992. The UL13 gene of herpes simplex virus 1 encodes the functions for posttranslational processing associated with phosphorylation of the regulatory protein alpha 22. Proc Natl Acad Sci U. S. A. 89:7310–7314. doi: 10.1073/pnas.89.16.7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Renzette N, Bhattacharjee B, Jensen JD, Gibson L, Kowalik TF. 2011. Extensive genome-wide variability of human cytomegalovirus in congenitally infected infants. PLoS Pathog 7:e1001344. doi: 10.1371/journal.ppat.1001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Renzette N, Gibson L, Bhattacharjee B, Fisher D, Schleiss MR, Jensen JD, Kowalik TF. 2013. Rapid intrahost evolution of human cytomegalovirus is shaped by demography and positive selection. PLoS Genet 9:e1003735. doi: 10.1371/journal.pgen.1003735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bzik DJ, Fox BA, DeLuca NA, Person S. 1984. Nucleotide sequence of a region of the herpes simplex virus type 1 gB glycoprotein gene: mutations affecting rate of virus entry and cell fusion. Virology 137:185–190. doi: 10.1016/0042-6822(84)90022-9. [DOI] [PubMed] [Google Scholar]
- 32.Silverman JL, Greene NG, King DS, Heldwein EE. 2012. Membrane requirement for folding of the herpes simplex virus 1 gB cytodomain suggests a unique mechanism of fusion regulation. J Virol 86:8171–8184. doi: 10.1128/JVI.00932-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gage PJ, Levine M, Glorioso JC. 1993. Syncytium-inducing mutations localize to two discrete regions within the cytoplasmic domain of herpes simplex virus type 1 glycoprotein B. J. Virol 67:2191–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dix RD, McKendall RR, Baringer JR. 1983. Comparative neurovirulence of herpes simplex virus type 1 strains after peripheral or intracerebral inoculation of BALB/c mice. Infect Immun 40:103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bower JR, Mao H, Durishin C, Rozenbom E, Detwiler M, Rempinski D, Karban TL, Rosenthal KS. 1999. Intrastrain variants of herpes simplex virus type 1 isolated from a neonate with fatal disseminated infection differ in the ICP34.5 gene, glycoprotein processing, and neuroinvasiveness. J Virol 73:3843–3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sacks SL, Wanklin RJ, Reece DE, Hicks KA, Tyler KL, Coen DM. 1989. Progressive esophagitis from acyclovir-resistant herpes simplex. Clinical roles for DNA polymerase mutants and viral heterogeneity? Ann Intern Med 111:893–899. doi: 10.7326/0003-4819-111-11-893. [DOI] [PubMed] [Google Scholar]
- 37.Tanaka M, Kagawa H, Yamanashi Y, Sata T, Kawaguchi Y. 2003. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J Virol 77:1382–1391. doi: 10.1128/JVI.77.2.1382-1391.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Antinone SE, Smith GA. 2010. Retrograde axon transport of herpes simplex virus and pseudorabies virus: a live-cell comparative analysis. J Virol 84:1504–1512. doi: 10.1128/JVI.02029-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warren RL, Sutton GG, Jones SJ, Holt RA. 2007. Assembling millions of short DNA sequences using SSAKE. BioInformatics 23:500–501. doi: 10.1093/bioinformatics/btl629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cunningham C, Gatherer D, Hilfrich B, Baluchova K, Dargan DJ, Thomson M, Griffiths PD, Wilkinson GW, Schulz TF, Dargan DJ, Davison AJ. 2010. Sequences of complete human cytomegalovirus genomes from infected cell cultures and clinical specimens. J Gen Virol 91:605–615. doi: 10.1099/vir.0.015891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szpara ML, Tafuri YR, Parsons L, Shamim SR, Verstrepen KJ, Legendre M, Enquist LW. 2011. A wide extent of inter-strain diversity in virulent and vaccine strains of alphaherpesviruses. PLoS Pathog 7:e1002282. doi: 10.1371/journal.ppat.1002282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, Flanigan MJ, Kravitz SA, Mobarry CM, Reinert KH, Remington KA, Anson EL, Bolanos RA, Chou H-H, Jordan CM, Halpern AL, Lonardi S, Beasley EM, Brandon RC, Chen L, Dunn PJ, Lai Z, Liang Y, Nusskern DR, Zhan M, Zhang Q, Zheng X, Rubin GM, Adams MD, Venter CJ. 2000. A whole-genome assembly of drosophila. Science 287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- 43.Angiuoli SV, Salzberg SL. 2011. Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics 27:334–342. doi: 10.1093/bioinformatics/btq665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watson G, Xu W, Reed A, Babra B, Putman T, Wick E, Wechsler SL, Rohrmann GF, Jin L. 2012. Sequence and comparative analysis of the genome of HSV-1 strain McKrae. Virology 433:528–537. doi: 10.1016/j.virol.2012.08.043. [DOI] [PubMed] [Google Scholar]
- 45.Macdonald SJ, Mostafa HH, Morrison LA, Davido DJ. 2012. Genome sequence of herpes simplex virus 1 strain McKrae. J Virol 86:9540–9541. doi: 10.1128/JVI.01469-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heller M, Dix RD, Baringer JR, Schachter J, Conte JE Jr.. 1982. Herpetic proctitis and meningitis: recovery of two strains of herpes simplex virus type 1 from cerebrospinal fluid. J Infect Dis 146:584–588. doi: 10.1093/infdis/146.5.584. [DOI] [PubMed] [Google Scholar]
- 47.Engel JP, Boyer EP, Goodman JL. 1993. Two novel single amino acid syncytial mutations in the carboxy terminus of glycoprotein B of herpes simplex virus type 1 confer a unique pathogenic phenotype. Virology 192:112–120. doi: 10.1006/viro.1993.1013. [DOI] [PubMed] [Google Scholar]
- 48.Silverman JL, Sharma S, Cairns TM, Heldwein EE. 2010. Fusion-deficient insertion mutants of herpes simplex virus type 1 glycoprotein B adopt the trimeric postfusion conformation. J Virol 84:2001–2012. doi: 10.1128/JVI.01791-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dolter KE, Ramaswamy R, Holland TC. 1994. Syncytial mutations in the herpes simplex virus type 1 gK (UL53) gene occur in two distinct domains. J Virol 68:8277–8281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hutchinson L, Graham FL, Cai W, Debroy C, Person S, Johnson DC. 1993. Herpes simplex virus (HSV) glycoproteins B & K inhibit cell fusion induced by HSV syncytial mutants. Virology 196:514–531. doi: 10.1006/viro.1993.1507. [DOI] [PubMed] [Google Scholar]
- 51.Melancon JM, Foster TP, Kousoulas KG. 2004. Genetic analysis of the herpes simplex virus Type 1 ul20 Protein domains involved in cytoplasmic virion envelopment and virus-induced cell Fusion. J Virol 78:7329–7343. doi: 10.1128/JVI.78.14.7329-7343.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Melancon JM, Luna RE, Foster TP, Kousoulas KG. 2005. Herpes simplex virus type 1 gK is required for gB-mediated virus-induced cell fusion, while neither gB and gK nor gB and ul20p function redundantly in virion de-envelopment. J Virol 79:299–313. doi: 10.1128/JVI.79.1.299-313.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bertrand L, Leiva-Torres GA, Hyjazie H, Pearson A. 2010. Conserved residues in the UL24 protein of herpes simplex virus 1 are important for dispersal of the nucleolar protein nucleolin. J Virol 84:10436. doi: 10.1128/JVI.01538-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pogue-Geile KL, Spear PG. 1987. The single base pair substitution responsible for the syn phenotype of herpes simplex virus type 1, strain MP. Virology 157:67–74. doi: 10.1016/0042-6822(87)90314-X. [DOI] [PubMed] [Google Scholar]
- 55.Rauch DA, Rodriguez N, Roller RJ. 2000. Mutations in herpes simplex virus glycoprotein D distinguish entry of free virus from cell-cell spread. J Virol 74:11437–11446. doi: 10.1128/JVI.74.24.11437-11446.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wheeler CE, Canby CM. 1960. Herpes simplex virus: characteristics of a strain which produces unusually large multinucleated giant cells in tissue culture. Arch Dermatol 82:141–149. doi: 10.1001/archderm.1960.01580030085011. [DOI] [PubMed] [Google Scholar]
- 57.Shimizu F, Satoh J, Tada M, Kumagai K. 1978. Suppression of in vitro growth of virulent and avirulent herpes simplex viruses by cell-mediated immune mechanisms, antibody, and interferon. Infect Immun 22:752–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muggeridge MI, Grantham ML, Johnson FB. 2004. Identification of syncytial mutations in a clinical isolate of herpes simplex virus 2. Virology 328:244–253. doi: 10.1016/j.virol.2004.07.027. [DOI] [PubMed] [Google Scholar]
- 59.Aurelian L, Smerdel S, Kessler II, Kulcar Z. 1974. Three Yugoslav herpes simplex viruses: biologic and antigenic properties and formation of giant cells in vitro by a cervical isolate. J Infect Dis 129:465–469. doi: 10.1093/infdis/129.4.465. [DOI] [PubMed] [Google Scholar]
- 60.Terni M, Roizman B. 1970. Variability of herpes simplex virus: isolation of two variants from simultaneous eruptions at different sites. J Infect Dis 121:212–216. doi: 10.1093/infdis/121.2.212. [DOI] [PubMed] [Google Scholar]
- 61.Ejercito PM, Kieff ED, Roizman B. 1968. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol 2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 62.Ramachandran S, Knickelbein JE, Ferko C, Hendricks RL, Kinchington PR. 2008. Development and pathogenic evaluation of recombinant herpes simplex virus type 1 expressing two fluorescent reporter genes from different lytic promoters. Virology 378:254–264. doi: 10.1016/j.virol.2008.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Szpara ML, Tafuri YR, Enquist LW. 2011. Preparation of viral DNA from nucleocapsids. J Vis Exp JoVE 2011(54):e3151. doi: 10.3791/3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith GA, Enquist LW. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J Virol 73:6405–6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Silverman JL, Heldwein EE. 2013. Mutations in the cytoplasmic tail of herpes simplex virus 1 gH reduce the fusogenicity of gB in transfected cells. J Virol 87:10139–10147. doi: 10.1128/JVI.01760-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental materials and methods. This document includes a detailed description of the VirGA workflow, along with details for the genetic distance analysis and RFLP methods. Download
HSV-1 FRed includes cell-rounding and syncytial plaques with either punctate, diffuse, or dark fluorescence resulting from its mRFP-capsid label. HSV-1 FRed is a virus stock that was generated from a cloned BAC copy of the HSV-1 F genome. Recovery of HSV-1 FRed into mammalian cells recreates the plaque morphology diversity observed in HSV-1 F stocks. The mRFP-capsid label reveals additional variation that affects the fluorescent label. (A) The majority of plaques showed standard cell-rounding CPE, with punctate fluorescence from the mRFP-capsid fusion protein (see Table 2 for quantitation). (B) The next most frequent variants were syncytial plaques with punctate capsid-based fluorescence. (C) Rarely, plaques revealed diffuse fluorescence. (D and E) Up to 1 in 10 plaques were dark and lacked fluorescence. Cell-rounding (D) and syncytial (E) variants are shown here, next to ordinary fluorescent plaques, to demonstrate that the lack of fluorescence was not due to image acquisition settings. Download
RFLP analysis of HSV-1 KOS and F subclones confirms draft genomes but obscures syncytial differences. Restriction fragment length polymorphism analysis (RFLP) revealed no major differences in digest patterns, for subclones of a given strain. (A) BamHI and (B) HindIII RFLPs were analyzed for subclones of HSV-1 KOS and F described in Fig. 1. Marker lanes (sizes are in kilobases) are shown to the left of each gel. Download
Overview of HSV genome sequencing using the viral genome assembly (VirGA) workflow. The VirGA workflow requires an input of high-throughput Illumina sequence read data from the viral genome of interest. We generated this by expanding a viral stock, isolating viral nucleocapsid DNA, preparing a library of genome fragments, and collecting high-throughput, paired-end sequence reads using an Illumina HiSeq or MiSeq instrument. In VirGA step 1, host sequences and quality-reducing contaminants are removed. In step 2, the viral sequences are assembled into long stretches of continuous sequence (contigs) by the use of two de novo assemblers, SSAKE and Celera. In step 3, these long stretches of sequence are arranged in order by comparison to a reference genome. Gaps can be closed by using the GapFiller program to search for overlapping sequences in the input data. Annotations are transferred from the reference genome to the new draft genome at this stage. In step 4, the original sequence reads are aligned to the draft consensus to check the assembly quality. Best practices in HSV genome assembly involve wet-bench validations of each assembly, such as PCR verification of key differences or RFLP analysis of genome orientation. Download
Example of a VirGA output summary. As part of its output, VirGA generates an interactive HTML file for each draft genome assembly, which can be opened in any web browser. This file summarizes statistics about the assembly and links to additional files. This image includes excerpted sections from a full VirGA output. Complete records of all VirGA outputs from this study are archived at https://scholarsphere.psu.edu/collections/sf268c193. The VirGA report summary includes statistics about the new draft genome, such as length, percent with coverage depth of >100-fold, number of gaps, and number of intact (gap-free) proteins. Links are provided to alignments of each gene and protein versus the reference genome; these are grouped into those without errors (green text) and those needing user attention (red text). Alignments for noncoding features are included as well. Below the summary, the extensive VirGA detailed report includes statistics on the number of sequence reads filtered out during the preprocessing steps, the number of contigs produced during SSAKE and Celera assembly, the gaps closed by GapFiller, and the results of quality assessment when the sequence reads are aligned to the new draft genome. From this extensive report, only a histogram of sequence read quality per base is shown here. Download
Serial assemblies demonstrate how increasing amounts of input data produce improved viral genome assemblies. We used VirGA to generate serial assemblies of both 100-bp paired-end sequence reads (A) and 300-bp paired-end sequence reads (B), doubling the number of input reads each time (x axis). For each assembly, we calculated the percentage of the genome with sequencing coverage depth greater than or equal to 100-fold (solid line) and the percentage of proteins that were assembled intact (gap free; dashed line) and counted the undetermined bases (“N” in the final assembly; blue histograms). (A) For 100-bp, paired-end data, we observed that a minimum of 400,000 reads were needed to produce an HSV-1 assembly with 90% of the proteins intact. (B) For 300-bp, paired-end data, we observed that just 200,000 reads were needed to produce an HSV-1 assembly with 90% of the proteins intact. Results were verified with serial assemblies of data from HSV-1 FSyncytial and H166Syncytial as well (data not shown). Download
Genetic distance diagram, DNA identity, and protein difference statistics summarize relatedness of HSV-1 strains and subclones. We followed the same approach as used previously (Szpara et al., J. Virol, 88:1209–1227, 2014) to generate a genetic distance tree from whole-genome alignments of the strains and subclones shown (see Materials and Methods for details). Branch points show confidence values. We calculated the percent DNA identity and number of variant proteins for selected sets of sequences; these are shown on the right. NCBI accession numbers for strains not sequenced in this paper (marked with asterisks) are as follows: H129, GU734772; 17, JN555585; McKrae, JQ730035 and JX142173; KOS, JQ780693 and JQ673480. Download
List of proteins that vary between subclones of each strain discussed in the text.
Primers for the PCR validations described in Materials and Methods.