Abstract
The nuclear pore is a large and complex biological machine, mediating all signal-directed transport between the nucleus and the cytoplasm. The vertebrate pore has a mass of ~120 million daltons or 30 times the size of a ribosome. The large size of the pore, coupled to its tight integration in the nuclear lamina, has hampered the isolation of pore complexes from vertebrate sources. We have now developed a strategy for the purification of nuclear pores from in vitro assembled annulate lamellae (AL), a cytoplasmic mimic of the nuclear envelope that lacks a lamina, nuclear matrix, and chromatin-associated proteins. We find that purified pore complexes from annulate lamellae contain every nuclear pore protein tested. In addition, immunoblotting reveals the presence of soluble transport receptors and factors known to play important roles in the transport of macromolecules through the pore. While transport factors such as Ran and NTF2 show only transient interaction with the pores, a number of soluble transport receptors, including importin β, show a tight association with the purified pores. In summary, we report that we have purified the vertebrate pore by biochemical criteria; silver staining reveals ~40–50 distinct protein bands.
Keywords: Nuclear pore, annulate lamellae, purification, nucleoporin, importin, vertebrate
The regulated transport of macromolecules across the nuclear envelope is critical for the creation and maintenance of the discrete identities of the nucleus and cytosol. Traffic through the nuclear envelope utilizes the nuclear pore complex, a large proteinaceous structure in excess of 120 million daltons in mass (1–3). As such, the nuclear pore represents one of the largest biological machines within the cell. The pore itself consists of three stacked octagonally symmetric rings, with the central ring composed of spoke-like structures supporting a central transporter (Figure 1B). Extending from the cytoplasmic ring are eight short filaments (1,2,4,5). These filaments, composed in part of the nuclear pore proteins (or nucleoporins) Nup214/CAN and Nup358/RanBP2, contain the initial binding sights for nuclear import receptors and their cargo (6–14). Extending from the nuclear ring are eight thin filaments terminating in a smaller ring. This nuclear pore ‘basket’ minimally contains the nucleoporins Nup93, Nup98, and Nup153, and plays roles both in export from the nucleus and in the terminal steps of import (6,11,15–19). Although roughly 18 vertebrate pore proteins have been discovered, a significant fraction remains unknown.
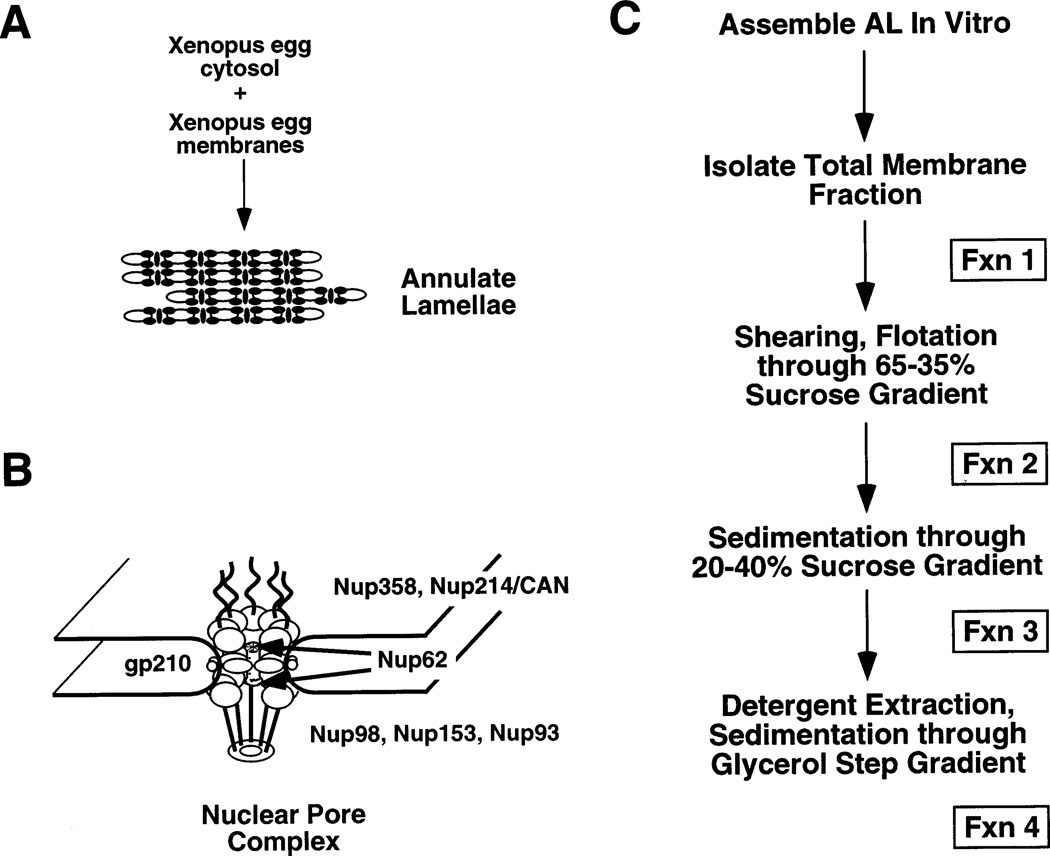
Figure 1. A. A schematic showing the combination of Xenopus laevis egg cytosol and membranes to assemble AL.
B. A structural model derived from electron microscopy and immunoelectron microscopy (19) showing the substructures of the nuclear pore and placement of a fraction of the proteins of the nuclear pore. C. A flow chart of the pore purification strategy developed here.
In higher organisms, the nuclear pore complex is tightly integrated into the nuclear lamina, which lines the inner nuclear membrane. The highly polymerized and insoluble network of lamina filaments has represented one of the greatest obstacles to the purification of nuclear pores from animal sources. Another potential obstacle is the presence of the highly insoluble nuclear matrix (20). Traditionally, high concentrations of detergents and urea were used following nuclease and salt extraction of the nucleus to obtain a nuclear pore complex-lamina-matrix fraction (20). As the name implies, these preparations contain the lamina and its associated polypeptides. More recently, a procedure using the detergent Empigen BB allows one to solubilize nucleoporins from this pore complex-lamina fraction (21,22). It has not yet been reported whether this procedure is specific for nucleoporins, or whether other nuclear envelope proteins are also released. However, that strategy does not yield intact pores. A major goal in the study of nuclear transport is the isolation and definition of the components of the highly complex vertebrate nuclear pore.
In contrast to vertebrate systems, intact nuclear pore complexes from yeast have been successfully purified to homogeneity (23,24). The ability to isolate intact nuclear pores from this organism may in large part be due to the lack of a detectable lamina in yeast (23). We reasoned that annulate lamellae (AL) would represent an attractive starting material to attack the purification of vertebrate nuclear pores, since AL are large cytoplasmic stacks of membranes containing abundant, closely packed pore complexes (25–27). These pores are morphologically identical to the pore complexes in the nuclear envelope and have been shown to bind fluorescently labeled nuclear transport factors when the latter were microinjected into oocytes (28). The observation that they are found in cells with high proliferative capability suggests that AL represent a storage form of nuclear pores. Indeed, 80% of the pore complexes in a mature Xenopus oocyte are present in cytoplasmic stacks of AL (29). Annulate lamellae do not contain a detectable lamina. Moreover, as AL can be readily and efficiently assembled in vitro in Xenopus egg extracts (26,30), they seemed an ideal starting point for the purification of vertebrate nuclear pore complexes. In this study, using in vitro assembled AL, we have been able to purify a preparation that contains all known nucleoporins. In addition, non-pore proteins, whether nuclear, cytoplasmic, or integral membrane proteins, are excluded. The behavior of individual soluble transport factors has been assessed and differs one from another, indicating that each of these factors has a different affinity for the pore complex. The most purified pore fraction contains approximately 40–50 distinct bands in silver stained gels, and individual pore proteins can readily be identified as distinct bands on such gels. We conclude by biochemical criteria that we have purified vertebrate pores.
Results
The strategy for biochemical purification of vertebrate pores
To attempt to purify vertebrate nuclear pores, we took advantage of the ability of extracts prepared from Xenopus eggs to assemble large amounts of pore complexes into annulate lamellae (26,27). Pores assembled into annalate lamellae appear morphologically normal and, in vivo, have been shown to be able to bind microinjected importin β (28). Previous attempts to purify pore complexes from vertebrate nuclear envelopes have been hampered by the tight integration of the pores into the lamina. However, pores have been successfully purified from S. cerevisiae, an organism that is thought to lack a nuclear lamina (23). Since there is no immunologically or morphologically detectable lamina associated with vertebrate annulate lamellae in vivo (26), we reasoned that it should be possible to isolate pores from this organelle. To attempt a biochemical purification of pores, AL were assembled in large amounts in Xenopus egg extracts and different purification steps were tried. At each step, the criteria for purification was retention of a key nuclear pore protein, Nup62 (see Figure 1B for a schematic of localization of a set of known nuclear pore proteins). After extensive trials, the steps summarized in Figure 1C were found to maintain Nup62, while purifying the structures containing this nucleoporin away from non-pore proteins. The final strategy was as follows: annulate lamellae were allowed to assemble for 3 h, then the total membrane fraction that contained assembled AL and all other membranous organelles of the cell was isolated by centrifugation (Figure 1C, Fraction 1). The total cellular membranes were sheared by passage through a small gauge syringe needle to shear annulate lamellae from attached ER. The AL were then separated from the bulk of other membranous organelles, such as ER and Golgi, by flotation through a 65–30% sucrose density gradient. The band corresponding to the annulate lamellae was removed and concentrated by centrifugation to generate Fraction 2. At this point in the purification, the major remaining membrane contaminants were found to consist of mitochondria and some residual ER attached to the AL (data not shown). The mitochondria and ER were removed by centrifugation through a linear sucrose gradient; the denser annulate lamellae pelleted to yield Fraction 3. The purified AL were resuspended in buffer containing 20% DMSO, which we found was essential to stabilize the nuclear pores, as it is in the case of the smaller yeast nuclear pores (23). The membranes surrounding the AL pore complexes were solubilized by the addition of a mixture of the detergents β-octylglucoside and CHAPS. The relative concentration and types of these detergents were optimized to allow retention of Nup62 and gp210, while removing the ER integral membrane protein ribophorin (27). Neither of the detergents alone removed the ER integral membrane protein contaminants, except at very high concentrations which solubilized the nucleoporins themselves. Other detergents, such as Triton-X100, Mega 10, sodium cholate, and sodium deoxycholate either did not remove the ER proteins or, at the other extreme, completely solubilized the nucleoporins (B.R. Miller, data not shown). The putative pore complexes treated with β-octylglucoside/CHAPS were pelleted through a glycerol cushion to isolate the protein-dense nuclear pore structures away from solubilized membranes, generating Fraction 4.
Figure 2 shows the profile of the nucleoporin Nup62 through a typical purification. In the final purification strategy, the bulk of Nup62 (approximately 50% of the amount present in Fraction 1) could be recovered in the final fraction. A typical purification was performed starting with 20 ml of egg extract (~1 g of total protein) and yielded between 1 and 1.5 mg of protein in the final pore fraction (Figure 1, Fraction 4). Based on Nup62, the estimated purification factor for this protocol is roughly 250-fold over the initial cytosol and membrane components. Based on the higher incorporation and maintenance of Nup93 (see below), we estimate that the purification factor for the pores here is ~400–600-fold over the starting AL assembly mixture. (See Table 1 for a typical purification.) To determine the theoretical maximal purification achievable and compare this to our actual purification, we used the value of 2.5 × 108 pores/oocyte that has been observed in vivo (10). This predicts the total mass of nucleoporin protein in an egg to be 5 × 10−2 µg. Given that an average Xenopus egg contains ~15–30 µg of total protein (B.R. Miller, unpublished data), the theoretical maximal purification factor of pores would be on the order of 300–600-fold (15–30 µg/0.05 µg). Thus, through the use of our purification strategy, we have obtained approximately the maximal purification of pores possible.
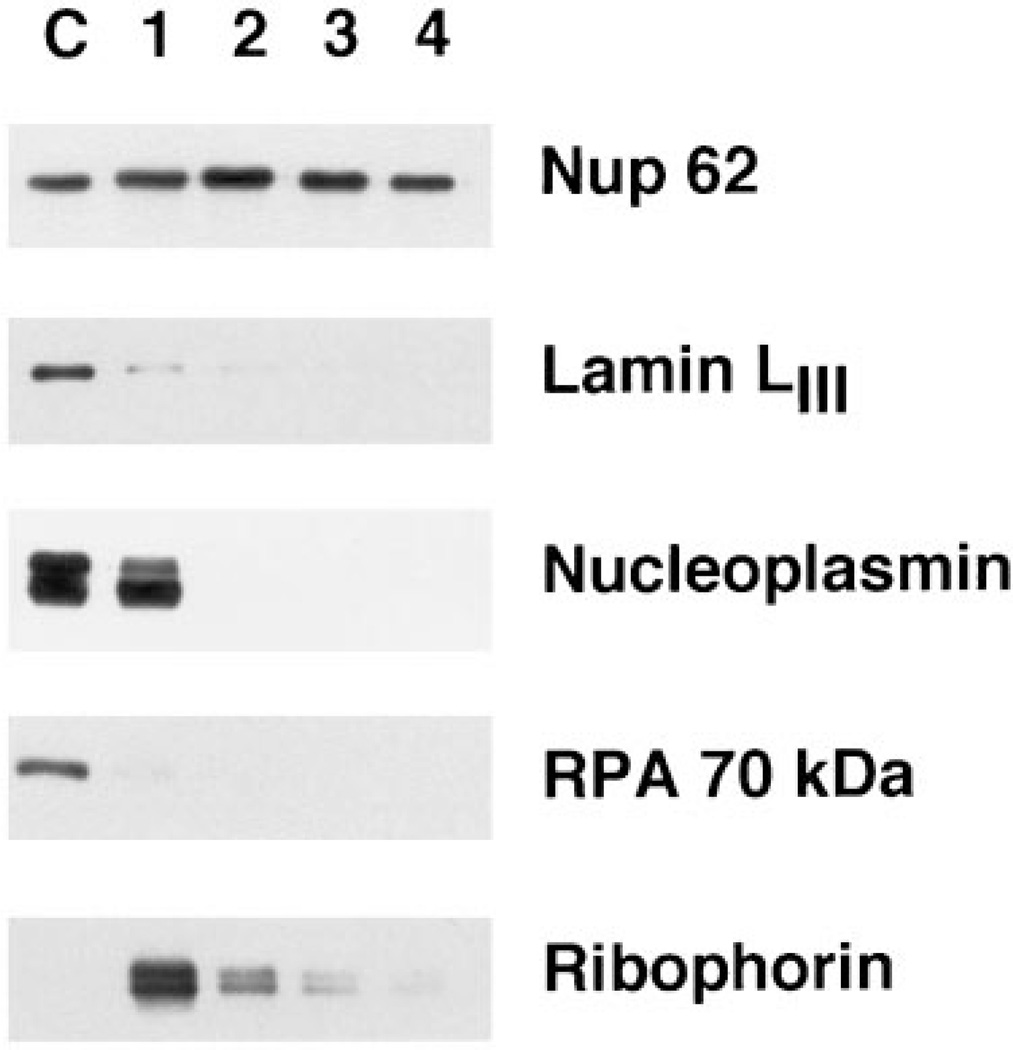
Figure 2. Western blots probing for the presence of a nucleoporin (Nup62), three nuclear antigens (lamin LIII, nucleoplasmin, and the 70-kDa subunit of RPA), and the ER integral membrane protein, ribophorin.
Fractions 1–4 correspond to those in Figure 1C and contain AL or AL fractions derived from the equivalent of 0.25 µl of extract, while lane C contains 0.025 µl of cytosol as a marker.
Table 1.
Purification of AL pores
| Fraction | Protein (mg) | Yield (%) | Approximate fold purification | |
|---|---|---|---|---|
| Total extract | 1 100 | 100 | 1 | |
| I | Crude membrane | 109 | 80 | 8 |
| II | Flotation | 35 | 75 | 24 |
| III | Velocity gradient | 26 | 70 | 29 |
| IV | Detergent extract | 1.4 | 55 | 432 |
Pores were purified as described in Materials and Methods, and the fold-enrichment was determined by densitometric scanning of western blots of the fractions, probing for the nucleoporin Nup93. Fractions I–IV correspond to those in Figure 1C.
Proteins of the inner nuclear envelope, nucleoplasm, and ER do not co-purify with pores
The first steps in the purification removed a number of non-pore proteins, both nuclear and integral membrane proteins (Figure 2, Fractions 1 and 2). Lamin LIII, known to remain soluble in Xenopus egg cytosol in the absence of nuclear formation, was removed in the early steps of purification (Figure 2), confirming both the purification and the previously described lack of a detectable lamina associated with AL (35,26). The highly abundant nuclear protein, nucleoplasmin, showed some association with the initial crude membrane fraction derived from the AL assembly reaction (Fraction 1). However, all nucleoplasmin was removed during the first gradient. The initial association could possibly be due to the presence of multiple nuclear localization sequences in the nucleoplasmin pentamer causing an association with transport receptors bound to the pores (see below). The DNA-binding protein, RPA (Figure 2), which is known to use an import receptor distinct from nucleoplasmin (36), showed little interaction with AL pores and was easily removed during purification.
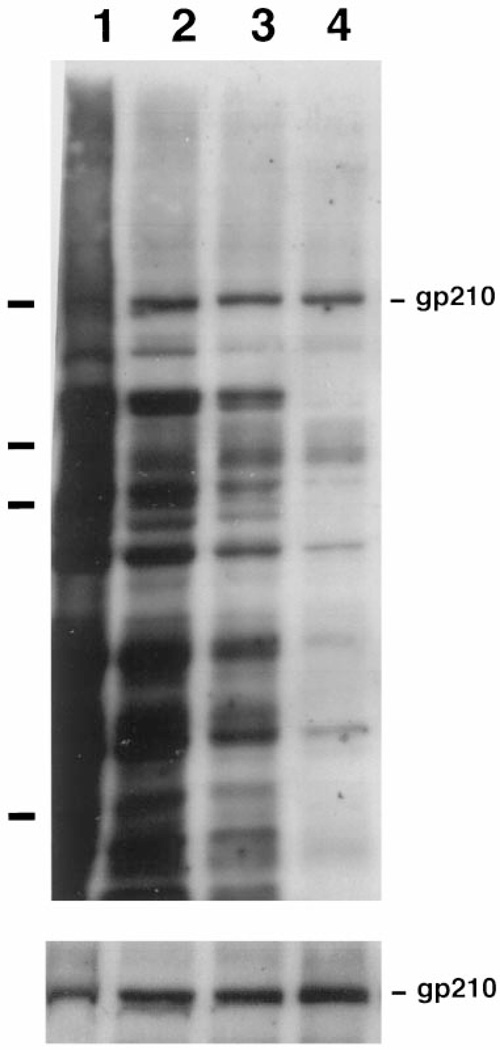
Since AL are assembled into and are physically contiguous with the endoplasmic reticulum both in vivo (37) and in vitro (26,27), it would not be unexpected to find large amounts of ER proteins contaminating AL pore preparations. Indeed, even after the mild shearing of the membrane fraction, a significant amount (~ 10%) of the integral ER membrane protein ribophorin was present in the AL pore preparation after flotation through a sucrose gradient (Figure 2, Fraction 2). However, the subsequent two purification steps removed virtually all of the ribophorin (Figure 2, Fraction 4). This suggests that the vast majority of the ER is removed from the pores by our purification strategy. It remained a formal possibility, however, that detergent extraction specifically removed only ribophorin from the pores, rather than removing all membrane proteins. Another concern was that the detergent extraction would prove too harsh and remove the integral membrane proteins of the nuclear pore itself. To address this, we assessed the presence of glycosylated membrane proteins throughout the purification by probing with HRP-labeled Conconavalin. Con A recognizes mannosyl residues and reacts with a wide variety of glycosylated ER and Golgi integral membrane proteins, as well as with the nuclear pore integral membrane protein gp210 (38). As shown in Figure 3, a large number of Con A-reactive polypeptides are present in the total membrane fraction (Figure 3, upper panel, Fraction 1). Many of these are removed from the pore preparation by the flotation step (Fraction 2), but there remained a significant number of glycosylated proteins present before detergent extraction (Fraction 3). After detergent extraction, the vast majority of glycosylated proteins was removed, leaving as the most prominent band a protein species at ~ 210 kDa (Fraction 4). This protein proved to be the integral pore membrane protein gp210, as determined by immunoblotting the fractions with a polyclonal antibody raised to rat gp210 [Figure 3, lower panel, Fractions 1–4; (39)]. We conclude that our final purification strategy removes the bulk of contaminating integral membrane proteins, as well as soluble proteins, while retaining Nup62 and the pore membrane protein gp210 in a co-fractionating structure.
Figure 3. The integral pore membrane protein gp210 specifically co-purifies with the pores after detergent solubilization.
The upper panel represents a blot of Fractions 1–4 probed with Conconavalin A-HRP in order to detect complex carbohydrate glycoproteins. The position of gp210 is indicated on the left. The bars on the right indicate the position of the molecular weight markers (180, 116, 88, and 45 kDa). The lower panel represents a blot of the same fractions probed with antisera generated to rat gp210.
All tested nucleoporins co-purify with vertebrate pores
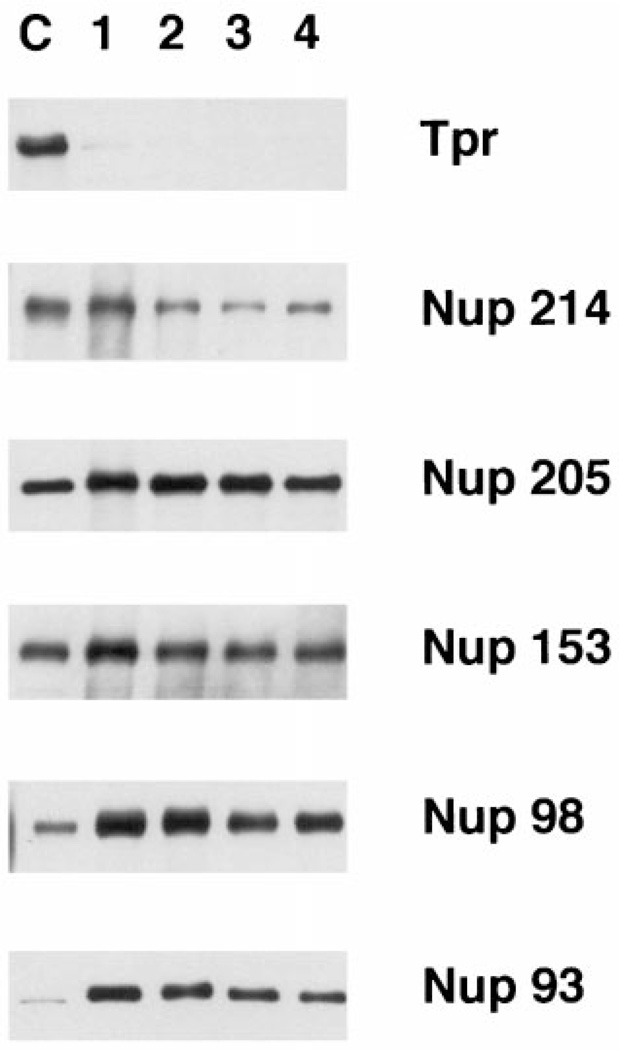
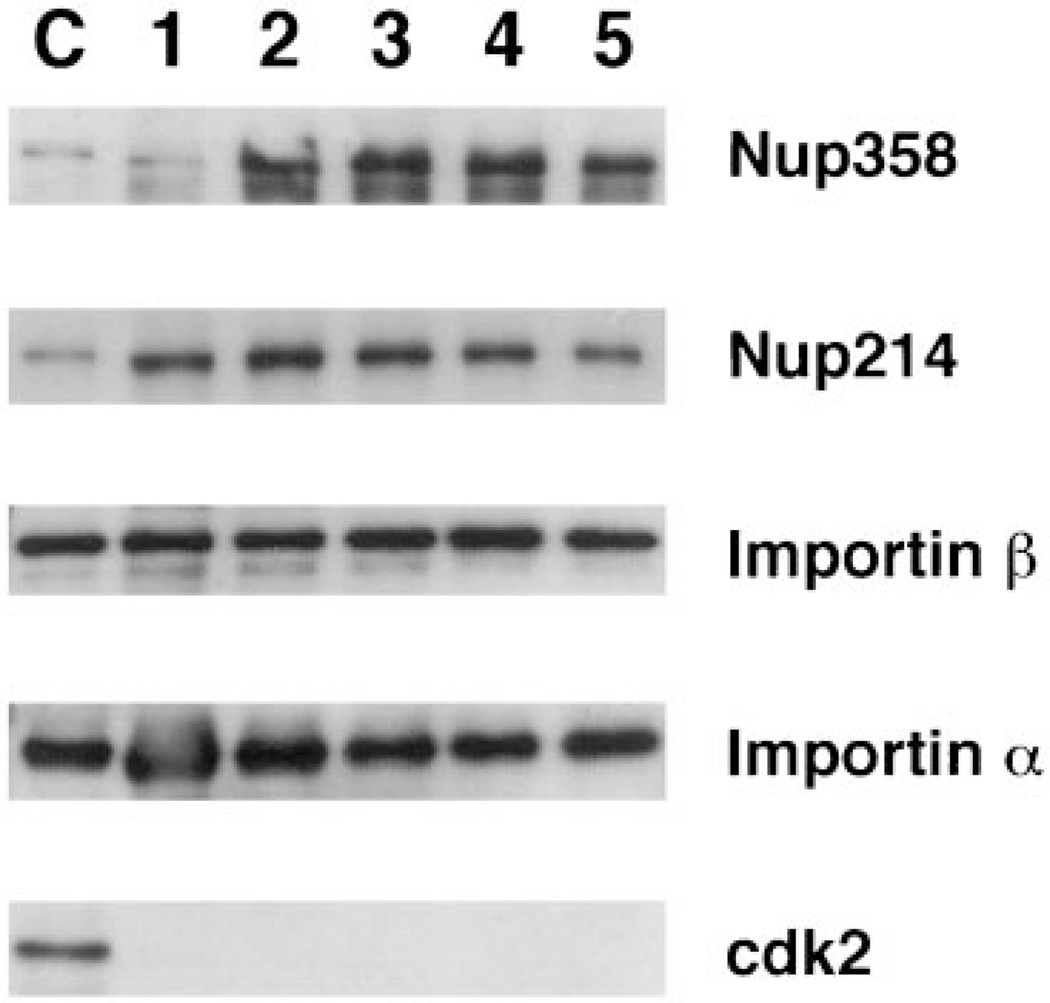
To further characterize the putative purified pores, we next assessed the presence of other known nucleoporins by immunoblotting (Figure 4). The large filamentous protein Tpr resides on long fibers extending from nuclear pores into the nucleus (39,40). Tpr is not associated with annulate lamellae in vivo (39). Tpr has been hypothesized to tie some aspect of pore function to the nuclear interior, rather than itself being a pore protein, and indeed the yeast homolog of Tpr localizes telomeres and silenced genes to the nuclear periphery (41). We found that Tpr did not cofractionate with the purified pores here (Figure 4; top panel). In striking contrast, five known nuclear pore proteins Nup214/CAN, Nup205, Nup153, Nup98, and Nup93 all showed co-purification with Nup62 and gp210 throughout the purification steps (Figure 4) (31,42,43,32,33,16,11). In fact, every pore protein tested to date shows co-purification using this protocol (including Nup155, Gle2, and Nup84 (44–46); data not shown).
Figure 4. Known nucleoporins co-purify as a unit in pore purification.
A panel of blots containing the same purification fractions as in Figure 1C was probed with affinity purified antisera raised against the pore proteins indicated on the right of the figure. As in Figure 2, only the relevant portion of the blots are shown for clarity. The fractions are loaded as in Figure 2. Tpr is not a pore protein in AL pores in vivo (6) or in vitro (shown here).
Co-fractionation of Nup153, Nup98, and Nup93 with the purified pores was extremely high (Figure 4). Nup153, Nup98, and Nup93 have all been localized on or near the nuclear basket of the pore (16,42,47). Nup153 is a pore protein located at the distal ring of the basket on the nuclear side of the pore and might be expected to be the most sensitive of the three to the stresses of purification. However, Nup153 was strongly maintained throughout the purification, arguing that the strategy allows for good preservation of protein–protein interactions. Nup205 was equally well maintained during purification.
When cytoplasmic fibril proteins were examined, Nup358/RanBP2 showed good co-purification throughout (Figure 5). The nucleoporin Nup214/CAN showed good incorporation into AL (Figures 4 and 5), but was sometimes lost during purification (Figure 4). Nup214/CAN is a component of the fibrils extending from the cytoplasmic side of the pore (43,9). Some element of these peripheral fibrils or Nup214/CAN itself may be more delicate. Proteins of the central core of the pore, the nuclear basket, and the other proteins of the cytoplasmic fibrils are well maintained throughout purification.
Figure 5. Association of importin α and β with AL pores.
AL pores were assembled from an independent batch of Xenopus egg extract, and the pores purified as in Figure 1C. The fractions were loaded and are labeled as in Figure 2, with the exception that lane 5 represents AL pores subjected to a second detergent extraction as described in the Materials and Methods.
Nuclear transport receptors co-purify with vertebrate pores, but Ran and NTF2 do not
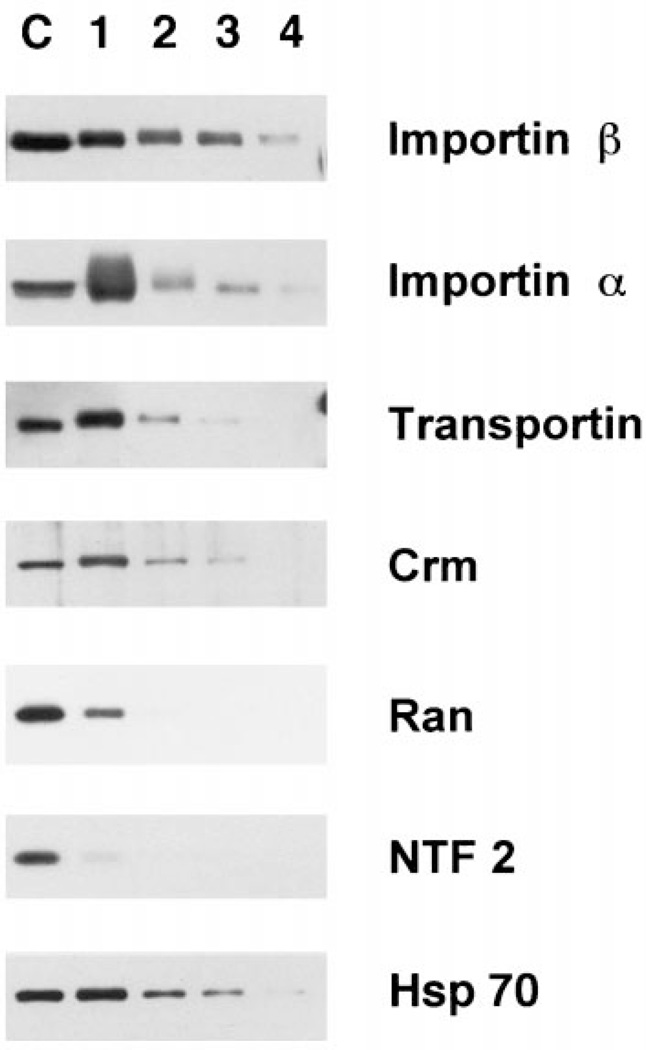
Having good evidence that we had purified the vertebrate pore, we next assayed for the presence of proteins that have been shown to either interact directly with the nuclear pore or play some role in transport through the pore. As demonstrated, nuclear proteins not involved with transport specifically, such as lamin LIII, nucleoplasmin, cdk2, and RPA 70, showed little affinity for the purified pores after the first purification step. In contrast, a number of the nuclear transport receptors showed affinity for the pores through several steps of purification (Figures 5 and 6). The most stable pore association was seen for importin β (Figures 5 and 6), which enriches with the purified pores. This nuclear import receptor has been demonstrated to stably interact with Nup358 in the cytoplasmic fibrils and with Nup153 in the basket of the assembled nuclear pores of somatic cells (11,48,49). Thus, the finding of importin β associated with our purified pore preparation through Fraction 3 (Figure 6, Fractions 1–3) and in some preps throughout purification (Figure 5, Fractions 1–4) is an additional argument for a retention of functional pore interactions. Hsp70 also showed a strong pattern of association with the pore complexes (Figure 6, Fractions 1–3). Hsp70 has been proposed to potentially stabilize some NLS-cargo-importin complexes, and several studies have indicated a role for Hsp70 in NLS-mediated nuclear import (23,50,51). Therefore, the Hsp70 initially present in the AL preparation may be bound to import complexes or, alternately, to some aspect of the pore.
Figure 6. Soluble transport factors show differential association with AL pores during purification.
These blots were loaded as in Figure 2, electrophoresed, transferred to PVDF, and probed with antisera to the transport receptors and transport factors indicated to the right of the figure.
Importin α, which in vivo is bound to pores through its association with importin β (52–55), showed a tremendous degree of association with the first pore fraction (Figure 6, lane 1). In this preparation importin α was removed during the late steps of purification (Figure 6). In other purifications, however, α was maintained through purification to a greater level (Figure 5). The import receptor, transportin, associated with the pore through fewer purification steps (Figure 6). One possible explanation for why this factor interacts with the pores less tightly than importin β may be that importin β has a higher affinity for the NPC relative to the other factors (48,53,56). Both importin β and transportin associate with Nup153 (11,17,48) which is on the nuclear side of the pore. In vivo, importin β can be seen to remain associated with the nucleoplasmic side of the pore after Ran-GTP releases importin α and its cargo into the nuclear interior (53). As such, importin β would be expected to have a relatively high affinity for one or more pore basket nucleoporins, such as Nup153. However, unlike importin β, importin α and transportin are released from the pore and proceed into the nucleoplasm (57,58). It is therefore likely that the interaction of transportin with the nuclear pore is of a more transitory nature than that of importin β and this transitory nature is reflected in Figure 6 as a looser association with the purified pores.
The nuclear export receptor Crm also showed significant interaction with the AL pores (Figure 6). In vivo, a trimeric complex of Crm/RanGTP/export cargo binds strongly to Nup214/CAN on the cytoplasmic fibrils during the terminal step of export. The decrease in Crm binding in our purification may result from the loss of Nup214/CAN from the fibrils as purification proceeds. Alternately, since AL are assembled in cytosol and cytoplasmic Ran is presumably in the GDP-bound form, the Crm in our preparations would lack Ran-GTP and should thus not associate in its typical export complex. On the other hand, Crm1 is re-imported into the nucleus in vivo and thus would be expected to interact with the cytoplasmic face of the nuclear pore under RanGDP conditions (59–62). The nucleoporin(s) with which Crm interacts during its import have yet to be identified. The affinity for Crm that we observe with purified pores (Figure 6) may more likely result from this import-related interaction.
One of the more surprising observations we found was the very low affinity of Ran and NTF2 for the purified pores (Figure 6). These proteins were both completely removed by the flotation gradient, despite the presence of physiological buffer conditions. Even upon long exposure, no NTF2 was visibly associated with the pores past the flotation gradient. NTF2 has been shown to bind in vitro to RanGDP and to FG repeat-containing nucleoporins, such as Nup62, and in fact NTF2 can be isolated by such binding (63–65). It has recently been shown that NTF2 is a carrier protein that imports Ran into the nucleus (66–71). Ran is likely in the GDP-bound form in the cytoplasmic extract; thus, we would expect to find both NTF2 and Ran associated with the AL pores and to find them stably associated. A possible explanation for the very low binding of Ran and NTF2 to purified pores might simply be that the binding of these two factors to the pore is normally transient in nature. Only a small fraction of either Ran or NTF2 may be in contact with the pore at any given time. This would also be true of importin β and Crm, yet these two proteins show a more stable association with AL pores. These differences may simply reflect the relative affinity of the interaction of these various transport factors with the pore. Other options are presented in the Discussion.
SDS-PAGE analysis of purified pores
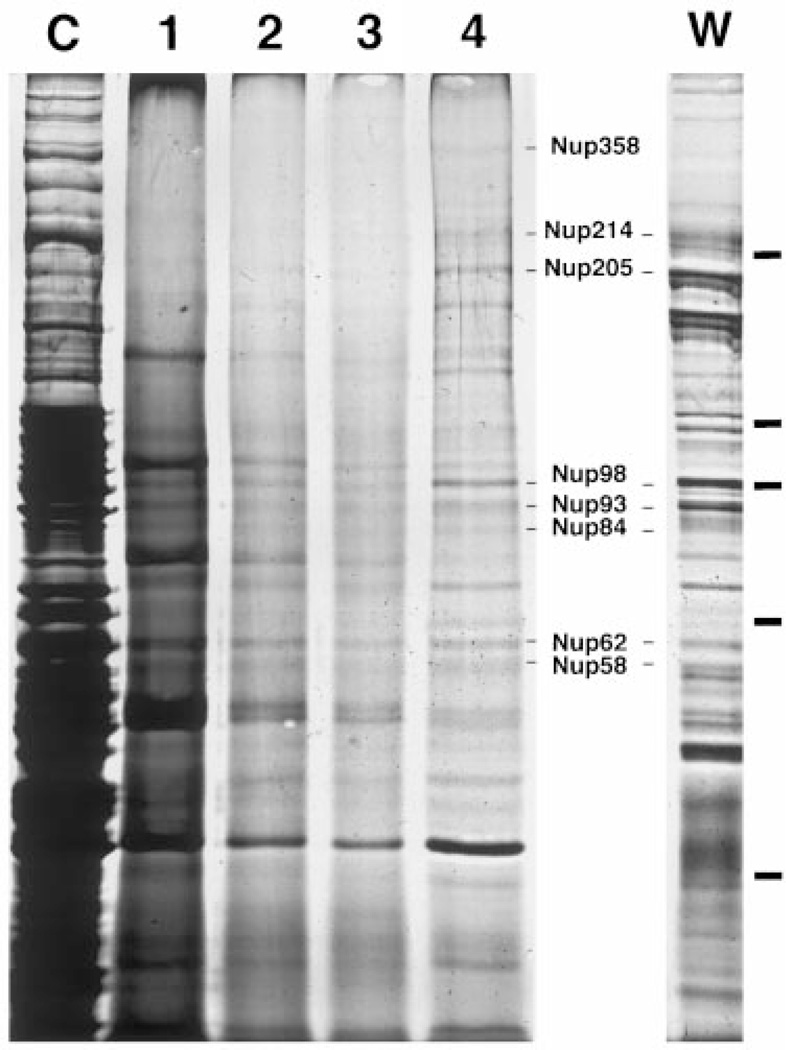
The isolation of a preparation with all the molecular hallmarks of purified pore complexes allows one to analyze its protein composition. When the composition of the purified pores was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and silver staining, a reproducible pattern of roughly ~40–50 different bands was observed in the final fraction (Figure 7, lane 4). The finding that the staining intensities of the known pore polypeptides are comparable to one another, as well as to the other bands, suggests that the preparation is largely composed of nuclear pore proteins. An exception is the hyperabundant band at ~48 kDa molecular weight. This band was subjected to peptide sequencing, and found to be actin. Given in the extract, it is not surprising that it would be found as a contaminant in the pore preparation. However, we can readily detect individual, known pore proteins within the final fraction. As shown in Figure 7, these include the known nucleoporins Nup358/RanBP2, Nup214/CAN, Nup205, Nup98, Nup93, Nup84, Nup62, and Nup58 (Figure 7, compare lanes 4 and W; (16,32,45,72,73)). The identity of each of these known proteins was extensively confirmed by side-by-side comparison with immunoprecipitates (not shown) from the purified pores using individual affinity purified anti-nucleoporin antibodies.
Figure 7. Individual pore proteins can be detected as distinct bands in the most purified fraction by silver staining.
Samples of the pore purification fractions were run on a 7.5% glycerol gradient gel and silver stained. The relative load of each fraction was adjusted for visibility as follows: lanes C and 1, 1×; lanes 2 and 3, 2×; and lane 4, 4×. Also shown is the profile of WGA-binding proteins from Xenopus egg cytosol (bound in low salt) in the lane marked W. The positions of multiple known pore proteins are indicated, verified by immunoprecipitation with specific antisera (data not shown). The bars on the right side of the figure indicate the position of the MW markers (180, 116, 97, 66, and 45 kDa).
Discussion
We have successfully developed a strategy for the purification of vertebrate nuclear pore complexes from in vitro assembled annulate lamellae. We chose this approach over the more traditional, but unsuccessful starting material of isolated rat liver nuclei for several reasons. The main rational for the use of annulate lamellae as a source of pores was to avoid the problems associated with the tight integration of pore complexes into the lamina and nuclear matrix present in somatic nuclei. Release of pore proteins from such nuclear preparations generally requires the use of denaturants and disassembly of the pores in order to liberate the pore proteins from the tightly polymerized lamina/matrix. Since annulate lamellae do not have any detectable lamins associated with them, we are able to release structural units which contain all tested pore proteins from the membranes using mild, non-ionic detergents.
Electron microscopic studies of pore complexes in amphibian oocyte nuclear envelopes have shown that there are pronounced differences in the structures emanating from the nuclear and cytoplasmic faces of the pore. We found that proteins of the nuclear pore basket, the central transporter, nuclear pore membrane, and the cytoplasmic fibrils were maintained in our preparation and co-purified together. The fact that we find every known pore protein tested to be present supports previous in vivo electron microscopic observations arguing that AL pore are equivalent to those assembled in the nuclear envelope (26,37,74). A strong model is that annulate lamellae are a storage depot of nuclear pores in highly proliferative cells. In fact, up to 80% of the pore complexes found in a mature Xenopus oocyte are present in annulate lamellae, while only 20% are found in the oocyte nuclear envelope (29). Because a developing Xenopus embryo must assemble 4000 cells, each with ~3000 pores comprised solely from the components present in the egg, it would therefore stand to reason that each of the pores present in oocyte AL would need to contain the complete complement of nucleoporins.
Examination of the behavior of individual nucleoporins during purification revealed almost complete uniformity in their assembly into AL and their association with the pores. Most of the pore proteins, such as Nup205, Nup155, Nup153, Nup98, Nup93, Nup62, gp210, and Gle2 were almost quantitatively incorporated and maintained in the AL pores throughout purification. Nup214/CAN was the only nucleoporin that at times showed a fragile association with the pores (compare Figures 4 and 5). Since Nup93 and Nup62 have been shown to be very important for the assembly and/or function of the pore complex, these two proteins, for example, would be expected to have a stable association with the pore (72,31,75–77,16). In contrast, Nup214/CAN has been localized to the periphery of the NPC on the cytoplasmic fibrils (43). As peripheral structures, the cytoplasmic fibrils may be somewhat fragile compared to more central structures.
In addition to the retention of nucleoporins throughout the purification, soluble transport receptors also showed copurification. Some, such as importin β and Hsp70, remain associated with the nucleoporins all the way to the detergent extraction step. As discussed above, importin β in vivo strongly associates with basket components of the pore, such as Nup153, at the end of its import cycle (11,17,53). As such, it might be expected to have a high affinity for nucleoporins in the purified pore. Other transport receptors are also known to interact genetically or in immunoprecipitations with distinct nucleoporins (9,11,17,19,54,60,64,70,78–86). Thus, their presence in the purified pores is compelling support for the maintenance of structural integrity of many aspects of the pore. The observation that Hsp70 is also fairly tightly associated with the purified pores is intriguing. One explanation is that Hsp70 is bound to one or more import receptors, which in turn are directly associated with the pore (50). Alternatively, Hsp70 may play a role in mediating pore assembly and be found in the pore preparation by virtue of this role.
It was unexpected to find that neither Ran nor NTF2 showed any appreciable association with the AL pores during purification. This is especially intriguing in light of the fact that Ran/NTF2 has been shown to bind to distinct nucleoporins on columns in both the GDP bound state (e.g. to Nup62), and in the GTP form (e.g. to Nup358/RanBP2) (63,73). As mentioned above, the lack of affinity of Ran and NTF2 to the purified pores may simply reflect a very transient association of Ran with the nuclear pore complex. Another possibility is that AL pores may be modified in some way to limit futile cycling of Ran molecules through these cytoplasmic pore complexes. Interestingly, limitation of such futile interaction might prove especially important for Ran in vivo, as Ran has been shown to play a role in a large number of cellular processes other than nuclear transport, including its newly discovered role in the organization of the mitotic spindle and in nuclear assembly (87–94).
In this study, we have chosen a biochemical approach to purification and characterization of the vertebrate pore, rather than one based on morphological criteria, e.g. electron microscopic monitoring of pores. Although we do not know the degree of structural integrity of the pores purified from AL, we find that all the tested pore proteins co-fractionate as a unit (Figures 2–5). We also know that multiple known nucleoporin sub-complexes are preserved in the purified pores, based on co-immunoprecipitation (B.R. Miller, data not shown). In addition, the transport receptor importin β remains associated with the pores through all but detergent extraction. Given the extremely rapid sedimentation of the pore proteins through high concentrations of glycerol, it is highly unlikely that our preparation yields single pores. The sedimentation is more characteristic of the clusters of pores characteristic of annulate lamellae.
In the purified vertebrate pores, we readily detect individual known nucleoporins by silver staining. We estimate that purification from 20 ml of extract yields a ~400–600-fold purification of the pores (Table 1), which is close to the theoretical maximum possible purification of AL pores in this system. A typical preparation of 1.5 mg total AL pore protein should therefore contain between 5 and 20 µg of each individual pore protein. The fact that known nucleoporins are readily identifiable by silver stained SDS-PAGE of the final fraction indicates that the purified pore preparation can now be used to directly identify novel nucleoporins by peptide microsequencing.
In the pore preparation, we observe ~40–50 distinct bands in the most purified fraction. In immunoblots of the purified preparation, all anti-nucleoporin antibodies gave single bands (with the possible exception of Nup214, which showed an additional 85 kDa band; data not shown.) The purified yeast nuclear pore has been shown to contain 30 distinct nucleoporins (95). A larger number of proteins in the vertebrate nuclear pore could be due to increased complexity due to its larger mass (120 MDa vs. 60 MDa in yeast). Indeed, the vertebrate pore also differs significantly in structure and volume from the yeast pore (3,24,96). It is worth noting that the original estimates of the number of proteins in the vertebrate pore were calculations based entirely on the observed mass of the pore (120 million daltons), divided by 16 to reflect the 8-fold radial and 2-fold apparent mirror symmetry of the pore (97). This calculation resulted in a predicted pore ‘substructure’ molecular mass of 7.5 million Da. At the time, only a few pore proteins had been identified. In consequence, an estimate of an average pore protein size of 100 kDa was used, which then yielded an estimate of ~75 different proteins per pore. A range of 50–100 different proteins was given to reflect the uncertainties inherent in those initial assumptions. The number of bands observed in Figure 7 is near the low end of the range of this estimate. It is, however, possible that the ~40–50 proteins observed in our preparation represent an underestimate of the true number of unique vertebrate pore proteins if certain proteins can not be seen due to poor silver staining properties or are masked behind other more prominent bands. Although this work represents a very large effort, clearly an even greater amount of analysis will be needed to characterize each protein component in the purified vertebrate pores.
The vertebrate pore has been shown to be morphologically, and in the cases of a significant number of proteins, compositionally distinct from the yeast nuclear pore (24,42,43,98–100). This may in part be due to certain fundamental differences in the biology (e.g. closed versus open mitosis) or possibly due to the billion years of evolutionary divergence between yeast and vertebrates (101). Yeast nuclear pores are unsurpassed for providing genetic insights into the structure and function of the pore complex, but they are not readily manipulable by biochemical techniques. In this report, we describe a strategy for the purification of vertebrate pore complexes from a system that allows for the ready manipulation of the pore complex by standard biochemical techniques. To date, the Xenopus system allows for both in vitro manipulation of pore composition and purification of these pores.
Finally, a powerful use for the purified vertebrate pores is that it allows one to determine whether candidate polypeptides, detected in other assays, are bona fide nuclear pore proteins, once antibodies to the candidate proteins have been raised. Previously, the only way to identify a protein as a nucleoporin was by positive immunoelectron microscopy on nuclear envelopes. Thus, the ability to biochemically purify nuclear pore complexes reported here should now allow the elucidation of vertebrate pore complex constituents and structure.
Materials and Methods
Antibodies
Monoclonal antibodies directed against the transport factors importin α and β, transportin, Ran, NTF2, and Hsp70 were obtained from Transduction Laboratories (Lexington, KY) and used according to the manufacturer’s instructions. Affinity purified antibodies directed against Nup62, Nup98, Nup214/CAN, Nup153, Nup93, Tpr, RPA 70 kDa, and gp210 have all been previously described (11,27,31–34). Nup358/RanBP2 was detected with mAb 414 (BABCo, Berkeley, CA). Monoclonal antibodies directed against nucleoplasmin and ribophorin were obtained from Dr. Gregory Leno (University of Mississippi Medical Center, Jackson, MS) and Dr. David Meyer (UCLA), respectively. Rabbit antibodies reacting with Nup205 were generated against an Escherichia coli fusion protein containing amino acids 673–1232 of human Nup205 (16). Fusion proteins were expressed and purified according to the manufacturer’s instructions (Qiagen, Stanford, CA). Rabbit antisera directed against rat gp210 have been previously described (27). Rabbit antisera against human Crm1 were the gift of Dr. Gerard Grosveld (St. Jude’s Childrens Hospital, Tennessee). Antibodies to cdk2 were obtained from Dr. John Newport (UCSD).
Purification of annulate lamellae pores
Egg extracts were prepared essentially as in (27), with the omission of the second clarification spin of the cytosol. Buffer A contained 50 mM KCl, 2.5 mM MgCl2, 10 mM HEPES, pH 7.4. In vitro, AL assembly reactions were set up as in (27), with the exception that the reaction was scaled up to 20 ml. After pores had assembled for 3 h at room temperature, two volumes of cold egg lysis buffer (ELB) (Buffer A plus 250 mM sucrose) containing 15 µg/ml RNAse A were added and the reaction was placed on ice for 30 min, then split into two aliquots. The total membrane fraction from each aliquot was isolated by centrifugation through a 6-ml cushion of ELB500 (Buffer A plus 500 mM sucrose) in an SW28 rotor at 113000 × g for 30 min. The membrane pellets were each resuspended in 5 ml of ELB, combined, then sheared by repeated passage through a 27-g needle. A small aliquot was removed for western blot analysis (Fraction 1). The resuspended sheared membranes (10 ml) were adjusted to 65% sucrose by the addition of 30 ml of 2.5 M sucrose. Aliquots (10 ml) were overlaid with buffered sucrose as follows; 5 ml each of 50% and 40%, 8 ml each of 37.5% and 35%. The flotation gradients were centrifuged at 72000 × g for 20 h in an SW28 rotor. The pellicle at the top of the gradients, which contained the bulk of the cellular membranes (non-AL), was carefully removed by aspiration, discarded, and the sides of the tubes wiped clean with a Kimwipe. The crude AL membranes were then collected from the 50–40% interface, diluted with an equal volume of buffer A, and pelleted at 72000 × g for 30 min in an SW41 rotor. The AL pellets (Fraction 2) were resuspended in 2 ml of ELB, homogenized as before, and 0.5 ml aliquots were layered on top of four 5-ml 20–40% linear sucrose gradients in buffer A. These velocity gradients were centrifuged at 50000 × g for 15 min in an SW55 rotor, and the bottom 0.5 ml and pellet were collected together. The AL were diluted with an equal volume of buffer A, and pelleted at 50000 × g for 10 min in an SW55 rotor. The pore-containing pellets (Fraction 3) were resuspended in 9 ml of buffer B (buffer A made in 20% DMSO) containing 20% glycerol, and 1/20 volume of 20% β-octylglucoside, 10% CHAPS (in buffer B + 20% glycerol) was added. The detergent-extracted pores were briefly vortexed to mix, and incubated on ice for 10 min. Aliquots (1.5 ml) were overlaid onto step gradients composed of 1.5 ml of 40% and 1.5 ml of 60% glycerol in buffer B containing 0.1% β-octylglucoside. The step gradients were spun at 50000 × g for 15 min in a TLA100 rotor, and the pores (Fraction 4) were recovered from the pellet.
In some preparations, the AL pores were subjected to an additional round of detergent extraction. For this, the material from six glycerol gradients was resuspended in 3 ml of buffer B containing 20% glycerol, extracted with detergent, and centrifuged through two glycerol step gradients exactly as in the previous step.
SDS-PAGE and immunoblotting
For immunoblotting, samples were resolved on SDS-PAGE gels, transferred to PVDF and blotted as in (27). For silver-stained gels, the separating gel was cast as a 7.5% acrylamide gel containing a 0–25% glycerol gradient. Staining was performed according to the manufacturer’s instructions (BioRad, Richmond, CA). Molecular weight markers were purchased from Sigma (St. Louis, MO).
Preparation of WGA-binding proteins as markers
Egg cytosol (2 ml), prepared as above, was diluted 3-fold with 10 mM HEPES, 2.5 mM MgCl2, and incubated with 200 µl of wheat germ agglutinin (WGA)-sepharose (EY Labs, San Mateo) at 4°C for 2 h with continuous tumbling. The beads were collected by brief centrifugation, and washed 5 × with the same buffer. The bound WGA-binding proteins were eluted with one bed volume of high sugar buffer (HSB; 0.5 M N-acetylglucosamine [GlcNAc], 16 mM N,N′,N″ triacetylchitotriose [TCT] in ELB), and stored in small aliquots at −80°C until use.
References
- 1.Hinshaw JE, Carragher BO, Milligan RA. Architecture and design of the nuclear pore complex. Cell. 1992;69:1133–1141. doi: 10.1016/0092-8674(92)90635-p. [DOI] [PubMed] [Google Scholar]
- 2.Akey CW, Radermacher M. Architecture of the Xenopus nuclear pore complex revealed by three-dimensional cryo-electron microscopy. J Cell Biol. 1993;122:1–19. doi: 10.1083/jcb.122.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pante N, Aebi U. Towards understanding the three-dimensional structure of the nuclear pore complex at the molecular level. Curr Opin Struct Biol. 1994;4:187–196. [Google Scholar]
- 4.Goldberg MW, Allen TD. The nuclear pore complex: three-dimensional surface structure revealed by field-emission, in-lens, scanning electron microscopy, with underlying structure uncovered by proteolysis. J Cell Sci. 1993;106:261–274. doi: 10.1242/jcs.106.1.261. [DOI] [PubMed] [Google Scholar]
- 5.Wiese C, Goldberg MW, Allen TD, Wilson KL. Nuclear envelope assembly in Xenopus extracts visualized by scanning EM reveals a transport-dependent ‘envelope smoothing’ event. J Cell Sci. 1997;110:1489–1502. doi: 10.1242/jcs.110.13.1489. [DOI] [PubMed] [Google Scholar]
- 6.Moroianu J, Hijikata M, Blobel G, Radu A. Mammalian karyopherin alpha 1 beta and alpha 2 beta heterodimers: alpha 1 or alpha 2 subunit binds nuclear localization signal and beta subunit interacts with peptide repeat-containing nucleoporins. Proc Natl Acad Sci USA. 1995;92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu J, Matunis MJ, Kraemer D, Blobel G, Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J Biol Chem. 1995;270:14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- 8.Saitoh H, Cooke CA, Burgess WH, Earnshaw WC, Dasso M. Direct and indirect association of the small GTPase ran with nuclear pore proteins and soluble transport factors: studies in Xenopus laevis egg extracts. Mol Biol Cell. 1996;7:1319–1334. doi: 10.1091/mbc.7.9.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti KG, Fransen J, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole CN, Hammell CM. Nucleocytoplasmic transport: driving and directing transport. Curr Biol. 1998;8:R368–R372. doi: 10.1016/s0960-9822(98)70239-8. [DOI] [PubMed] [Google Scholar]
- 11.Shah S, Tugendreich S, Forbes D. Major binding sites for the nuclear import receptor are the internal nucleoporin Nup153 and the adjacent nuclear filament protein Tpr. J Cell Biol. 1998;141:31–49. doi: 10.1083/jcb.141.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hood JK, Silver PA. In or out? Regulating nuclear transport. Curr Opin Cell Biol. 1999;11:241–247. doi: 10.1016/s0955-0674(99)80032-5. [DOI] [PubMed] [Google Scholar]
- 13.Kehlenbach RH, Dickmanns A, Kehlenbach A, Guan T, Gerace L. A role for RanBP1 in the release of CRM1 from the nuclear pore complex in a terminal step of nuclear export. J Cell Biol. 1999;145:645–657. doi: 10.1083/jcb.145.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zolotukhin AS, Felber BK. Nucleoporins nup98 and nup214 participate in nuclear export of human immunodeficiency virus type 1 Rev. J Virol. 1999;73:120–127. doi: 10.1128/jvi.73.1.120-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radu A, Moore MS, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- 16.Grandi P, Dang T, Pane N, Shevchenko A, Mann M, Forbes D, Hurt E. Nup93, a vertebrate homologue of yeast Nic96p, forms a complex with a novel 205-kDa protein and is required for correct nuclear pore assembly. Mol Biol Cell. 1997;8:2017–2038. doi: 10.1091/mbc.8.10.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shah S, Forbes DJ. Separate nuclear import pathways converge on the nucleoporin Nup153 and can be dissected with dominant-negative inhibitors. Curr Biol. 1998;8:1376–1386. doi: 10.1016/s0960-9822(98)00018-9. [DOI] [PubMed] [Google Scholar]
- 18.Kasper LH, Brindle PK, Schnabel CA, Pritchard CE, Cleary ML, van Deursen JM. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol Cell Biol. 1999;19:764–776. doi: 10.1128/mcb.19.1.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ullman K, Shah S, Powers M, Forbes D. The nucleoporin Nup153 plays a critical role in multiple types of nuclear export. Mol Biol Cell. 1999;10:649–664. doi: 10.1091/mbc.10.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dwyer N, Blobel G. A modified procedure for the isolation of a pore complex-lamina fraction from rat liver nuclei. J Cell Biol. 1976;70:581–591. doi: 10.1083/jcb.70.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fontoura BM, Blobel G, Matunis MJ. A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J Cell Biol. 1999;144:1097–1112. doi: 10.1083/jcb.144.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rout MP, Blobel G. Isolation of the yeast nuclear pore complex. J Cell Biol. 1993;123:771–783. doi: 10.1083/jcb.123.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Q, Rout MP, Akey CW. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol Cell. 1998;1:223–234. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]
- 25.Kessel RG. The structure and function of annulate lamellae: porous cytoplasmic and intranuclear membranes. Int Rev Cytol. 1983;82:181–303. doi: 10.1016/s0074-7696(08)60826-8. [DOI] [PubMed] [Google Scholar]
- 26.Dabauvalle MC, Loos K, Merkert H, Scheer U. Spontaneous assembly of pore complex-containing membranes (‘annulate lamellae’) in Xenopus egg extract in the absence of chromatin. J Cell Biol. 1991;112:1073–1082. doi: 10.1083/jcb.112.6.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meier E, Miller B, Forbes DJ. Nuclear pore complex assembly studied with a biochemical assay for annulate lamellae formation. J Cell Biol. 1995;129:1459–1472. doi: 10.1083/jcb.129.6.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cordes VC, Rackwitz HR, Reidenbach S. Mediators of nuclear protein import target karyophilic proteins to pore complexes of cytoplasmic annulate lamellae. Exp Cell Res. 1997;237:419–433. doi: 10.1006/excr.1997.3806. [DOI] [PubMed] [Google Scholar]
- 29.Cordes VC, Reidenbach S, Franke WW. High content of a nuclear pore complex protein in cytoplasmic annulate lamellae of Xenopus oocytes. Eur J Cell Biol. 1995;68:240–255. [PubMed] [Google Scholar]
- 30.Meier E, Miller BR, Forbes DJ. Nuclear pore complex assembly studied with a biochemical assay for annulate lamellae formation. J Cell Biol. 1995 doi: 10.1083/jcb.129.6.1459. In Press:0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finlay DR, Meier E, Bradley P, Horecka J, Forbes DJ. A complex of nuclear pore proteins required for pore function. J Cell Biol. 1991;114:169–183. doi: 10.1083/jcb.114.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macaulay C, Meier E, Forbes DJ. Differential mitotic phosphorylation of proteins of the nuclear pore complex. J Biol Chem. 1995;270:254–262. doi: 10.1074/jbc.270.1.254. [DOI] [PubMed] [Google Scholar]
- 33.Powers MA, Macaulay C, Masiarz FR, Forbes DJ. Reconstituted nuclei depleted of a vertebrate GLFG nuclear pore protein, p97, import but are defective in nuclear growth and replication. J Cell Biol. 1995;128:721–736. doi: 10.1083/jcb.128.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walter J, Sun L, Newport J. Regulated chromosomal DNA replication in the absence of a nucleus. Mol Cell. 1998;1:519–529. doi: 10.1016/s1097-2765(00)80052-0. [DOI] [PubMed] [Google Scholar]
- 35.Kessel RG. The annulate lamellae – from obscurity to spotlight. Electron Microsc Rev. 1989;2:257–348. doi: 10.1016/0892-0354(89)90003-8. [DOI] [PubMed] [Google Scholar]
- 36.Jullien D, Gorlich D, Laemmli UK, Adachi Y. Nuclear import of RPA in Xenopus egg extracts requires a novel protein XRIPalpha but not importin alpha. EMBO J. 1999;18:4348–4358. doi: 10.1093/emboj/18.15.4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kessel RG. Origin, differentiation, distribution and possible functional role of annulate lamellae during spermatogenesis in Drosophila melanogaster. J Ultrastruct Res. 1981;75:72–96. doi: 10.1016/s0022-5320(81)80101-3. [DOI] [PubMed] [Google Scholar]
- 38.Gerace L, Blum A, Blobel G. Immunocytochemical localization of the major polypeptides of the nuclear pore complex-lamina fraction. Interphase and mitotic distribution. J Cell Biol. 1978;79:546–566. doi: 10.1083/jcb.79.2.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cordes VC, Reidenbach S, Rackwitz HR, Franke WW. Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J Cell Biol. 1997;136:515–529. doi: 10.1083/jcb.136.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zimowska G, Aris JP, Paddy MR. A Drosophila Tpr protein homolog is localized both in the extrachromosomal channel network and to nuclear pore complexes. J Cell Sci. 1997;110:927–944. doi: 10.1242/jcs.110.8.927. [DOI] [PubMed] [Google Scholar]
- 41.Galy V, Olivo-Marin JC, Scherthan H, Doye V, Rascalou N, Nehrbass U. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature. 2000;403:108–112. doi: 10.1038/47528. [DOI] [PubMed] [Google Scholar]
- 42.Sukegawa J, Blobel G. A nuclear pore complex protein that contains zinc finger motifs, binds DNA, and faces the nucleoplasm. Cell. 1993;72:29–38. doi: 10.1016/0092-8674(93)90047-t. [DOI] [PubMed] [Google Scholar]
- 43.Kraemer D, Wozniak RW, Blobel G, Radu A. The human CAN protein, a putative oncogene product associated with myeloid leukemogenesis, is a nuclear pore complex protein that faces the cytoplasm. Proc Nat Acad Sci. 1994;91:1519–1523. doi: 10.1073/pnas.91.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Radu A, Blobel G, Wozniak RW. Nup155 is a novel nuclear pore complex protein that contains neither repetitive sequence motifs nor reacts with WGA. J Cell Biol. 1993;121:1–9. doi: 10.1083/jcb.121.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bastos R, Ribas de Pouplana L, Enarson M, Bodoor K, Burke B. Nup84, a novel nucleoporin that is associated with CAN/Nup214 on the cytoplasmic face of the nuclear pore complex. J Cell Biol. 1997;137:989–1000. doi: 10.1083/jcb.137.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iovine MK, Wente SR. A nuclear export signal in Kap95p is required for both recycling the import factor and interaction with the nucleoporin GLFG repeat regions of Nup116p and Nup100p. J Cell Biol. 1997;137:797–811. doi: 10.1083/jcb.137.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Powers MA, Forbes DJ, Dahlberg JE, Lund E. The vertebrate GLFG nucleoporin, Nup98, is an essential component of multiple RNA export pathways. J Cell Biol. 1997;136:241–250. doi: 10.1083/jcb.136.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakielny S, Shaikh S, Burke B, Dreyfuss G. Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J. 1999;18:1982–1995. doi: 10.1093/emboj/18.7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yaseen NR, Blobel G. Two distinct classes of Ran-binding sites on the nucleoporin Nup-358. Proc Natl Acad Sci USA. 1999;96:5516–5521. doi: 10.1073/pnas.96.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okuno Y, Imamoto N, Yoneda Y. 70-kDa heat-shock cognate protein colocalizes with karyophilic proteins into the nucleus during their transport in vitro. Exp Cell Res. 1993;206:134–142. doi: 10.1006/excr.1993.1129. [DOI] [PubMed] [Google Scholar]
- 51.Shulga N, Roberts P, Gu Z, Spitz L, Tabb MM, Nomura M, Goldfarb DS. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: a role for heat shock protein 70 during targeting and translocation. J Cell Biol. 1996;135:329–339. doi: 10.1083/jcb.135.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gorlich D, Kostka S, Kraft R, Dingwall C, Laskey RA, Hartmann E, Prehn S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr Biol. 1995;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- 53.Gorlich D, Vogel F, Mills AD, Hartmann E, Laskey RA. Distinct functions for the two importin subunits in nuclear protein import. Nature. 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- 54.Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- 55.Adam SA. Transport pathways of macromolecules between the nucleus and the cytoplasm [In Process Citation] Curr Opin Cell Biol. 1999;11:402–406. doi: 10.1016/S0955-0674(99)80056-8. [DOI] [PubMed] [Google Scholar]
- 56.Ohno M, Fornerod M, Mattaj IW. Nucleocytoplasmic transport: the last 200 nanometers. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- 57.Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pollard VW, Michael WM, Nakielny S, Siomi MC, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 59.Fornerod M, Ohno M, Yoshida M, Mattaj I. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 60.Neville M, Stutz F, Lee L, Davis LI, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 61.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 62.Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 63.Paschal BM, Gerace L. Identification of NTF2, a cytosolic factor for nuclear import that interacts with nuclear pore complex protein p62. J Cell Biol. 1995;129:925–937. doi: 10.1083/jcb.129.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clarkson WD, Kent HM, Stewart M. Separate binding sites on nuclear transport factor 2 (NTF2) for GDP-Ran and the phenylalanine-rich repeat regions of nucleoporins p62 and Nsp1p. J Mol Biol. 1996;263:517–524. doi: 10.1006/jmbi.1996.0594. [DOI] [PubMed] [Google Scholar]
- 65.Hu T, Guan T, Gerace L. Molecular and functional characterization of the p62 complex, an assembly of nuclear pore complex glycoproteins. J Cell Biol. 1996;134:589–601. doi: 10.1083/jcb.134.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Melchior F, Guan T, Yokoyama N, Nishimoto T, Gerace L. GTP hydrolysis by Ran occurs at the nuclear pore complex in an early step of protein import. J Cell Biol. 1995;131:571–581. doi: 10.1083/jcb.131.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wong DH, Corbett AH, Kent HM, Stewart M, Silver PA. Interaction between the small GTPase Ran/Gsp1p and Ntf2p is required for nuclear transport. Mol Cell Biol. 1997;17:3755–3767. doi: 10.1128/mcb.17.7.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Feldherr C, Akin D, Moore MS. The nuclear import factor p10 regulates the functional size of the nuclear pore complex during oogenesis. J Cell Sci. 1998;111:1889–1896. doi: 10.1242/jcs.111.13.1889. [DOI] [PubMed] [Google Scholar]
- 69.Moore MS. Ran and nuclear transport. J Biol Chem. 1998;273:22857–22860. doi: 10.1074/jbc.273.36.22857. [DOI] [PubMed] [Google Scholar]
- 70.Ribbeck K, Lipowsky G, Kent HM, Stewart M, Gorlich D. NTF2 mediates nuclear import of Ran. EMBO J. 1998;17:6587–6598. doi: 10.1093/emboj/17.22.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smith A, Brownawell A, Macara IG. Nuclear import of Ran is mediated by the transport factor NTF2. Curr Biol. 1998;8:1403–1406. doi: 10.1016/s0960-9822(98)00023-2. [DOI] [PubMed] [Google Scholar]
- 72.Finlay DR, Forbes DJ. Reconstitution of biochemically altered nuclear pores: transport can be eliminated and restored. Cell. 1990;60:17–29. doi: 10.1016/0092-8674(90)90712-n. [DOI] [PubMed] [Google Scholar]
- 73.Yokoyama N, Hayashi N, Seki T, Panté N, Ohba T, Nishii K, Kuma K, Hayashida T, Miyata T, Aebi U, et al. A giant nucleopore protein that binds Ran/TC4. Nature. 1995;376:184–188. doi: 10.1038/376184a0. [DOI] [PubMed] [Google Scholar]
- 74.Macaulay C, Forbes DJ. Reconstitution of nuclear pore assembly and function. Cell Dev Biol. 1996;7:475–486. [Google Scholar]
- 75.Grandi P, Doye V, Hurt EC. Purification of NSP1 reveals complex formation with ‘GLFG’ nucleoporins and a novel nuclear pore protein NIC96. EMBO J. 1993;12:3061–3071. doi: 10.1002/j.1460-2075.1993.tb05975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pante N, Bastos R, McMorrow I, Burke B, Aebi U. Interactions and three-dimensional localization of a group of nuclear pore complex proteins. J Cell Biol. 1994;126:603–617. doi: 10.1083/jcb.126.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bailer SM, Berlin WK, Starr CM, Hanover JA. Characterization of nuclear pore protein p62 produced using baculovirus. Prot Express Purif. 1995;6:546–554. doi: 10.1006/prep.1995.1072. [DOI] [PubMed] [Google Scholar]
- 78.Iovine MK, Watkins JL, Wente SR. The GLFG repetitive region of the nucleoporin Nup116p interacts with Kap95p, an essential yeast nuclear import factor. J Cell Biol. 1995;131:1699–1713. doi: 10.1083/jcb.131.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Aitchison JD, Blobel G, Rout MP. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- 80.Marelli M, Aitchison JD, Wozniak RW. Specific binding of the karyopherin Kap121p to a subunit of the nuclear pore complex containing Nup53p, Nup59p, and Nup170p. J Cell Biol. 1998;143:1813–1830. doi: 10.1083/jcb.143.7.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kamei Y, Yuba S, Nakayama T, Yoneda Y. Three distinct classes of the alpha-subunit of the nuclear pore-targeting complex (importin-alpha) are differentially expressed in adult mouse tissues. J Histochem Cytochem. 1999;47:363–372. doi: 10.1177/002215549904700310. [DOI] [PubMed] [Google Scholar]
- 82.Kose S, Imamoto N, Tachibana T, Yoshida M, Yoneda Y. beta-subunit of nuclear pore-targeting complex (importin-beta) can be exported from the nucleus in a Ran-independent manner. J Biol Chem. 1999;274:3946–3952. doi: 10.1074/jbc.274.7.3946. [DOI] [PubMed] [Google Scholar]
- 83.Liu Y, Guo W, Tartakoff PY, Tartakoff AM. A Crm1p-independent nuclear export path for the mRNA-associated protein, Npl3p/Mtr13p. Proc Natl Acad Sci USA. 1999;96:6739–6744. doi: 10.1073/pnas.96.12.6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moy TI, Silver PA. Nuclear export of the small ribosomal subunit requires the ran-GTPase cycle and certain nucleoporins. Genes Dev. 1999;13:2118–2133. doi: 10.1101/gad.13.16.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ribbeck K, Kutay U, Paraskeva E, Gorlich D. The translocation of transportin-cargo complexes through nuclear pores is independent of both Ran and energy. Curr Biol. 1999;9:47–50. doi: 10.1016/s0960-9822(99)80046-3. [DOI] [PubMed] [Google Scholar]
- 86.Seedorf M, Damelin M, Kahana J, Taura T, Silver PA. Interactions between a nuclear transporter and a subset of nuclear pore complex proteins depend on Ran GTPase. Mol Cell Biol. 1999;19:1547–1557. doi: 10.1128/mcb.19.2.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kornbluth S, Dasso M, Newport J. Evidence for a dual role for TC4 protein in regulating nuclear structure and cell cycle progression. J Cell Biol. 1994;125:705–719. doi: 10.1083/jcb.125.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sazer S. The search for the primary function of the Ran GTPase continues. Trends Cell Biol. 1996;6:81–85. doi: 10.1016/0962-8924(96)80992-5. [DOI] [PubMed] [Google Scholar]
- 89.Carazo-Salas RE, Guarguaglini G, Gruss OJ, Segref A, Karsenti E, Mattaj IW. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 1999;400:178–181. doi: 10.1038/22133. [DOI] [PubMed] [Google Scholar]
- 90.Kalab P, Pu RT, Dasso M. The ran GTPase regulates mitotic spindle assembly. Curr Biol. 1999;9:481–484. doi: 10.1016/s0960-9822(99)80213-9. [DOI] [PubMed] [Google Scholar]
- 91.Ohba T, Nakamura M, Nishitani H, Nishimoto T. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran [see comments] Science. 1999;284:1356–1358. doi: 10.1126/science.284.5418.1356. [DOI] [PubMed] [Google Scholar]
- 92.Wilde A, Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 1999;284:1359–1362. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- 93.Hetzer M, Bilbao-Cortes D, Walther TC, Gruss OJ, Mattaj IW. GTP hydrolysis by Ran is required for nuclear envelope assembly. Mol Cell. 2000;5:1013–1024. doi: 10.1016/s1097-2765(00)80266-x. [DOI] [PubMed] [Google Scholar]
- 94.Zhang C, Clarke PR. Chromatin-independent nuclear envelope assembly induced by Ran GTPase in Xenopus egg extracts. Science. 2000;288:1429–1432. doi: 10.1126/science.288.5470.1429. [DOI] [PubMed] [Google Scholar]
- 95.Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex. Composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–652. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Goldberg MW, Wiese C, Allen TD, Wilson KL. Dimples, pores, star-rings, and thin rings on growing nuclear envelopes: evidence for structural intermediates in nuclear pore complex assembly. J Cell Sci. 1997;110:409–420. doi: 10.1242/jcs.110.4.409. [DOI] [PubMed] [Google Scholar]
- 97.Forbes DJ. Structure and function of the nuclear pore complex. Annu Rev Cell Biol. 1992;8:495–527. doi: 10.1146/annurev.cb.08.110192.002431. [DOI] [PubMed] [Google Scholar]
- 98.Greber UF, Senior A, Gerace L. A major glycoprotein of the nuclear pore complex is a membrane-spanning polypeptide with a large lumenal domain and a small cytoplasmic tail. EMBO J. 1990;9:1495–1502. doi: 10.1002/j.1460-2075.1990.tb08267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hallberg E, Wozniak RW, Blobel G. An integral membrane protein of the pore membrane domain of the nuclear envelope contains a nucleoporin-like region. J Cell Biol. 1993;122:513–521. doi: 10.1083/jcb.122.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wozniak RW, Blobel G, Rout MP. POM152 is an integral protein of the pore membrane domain of the yeast nuclear envelope. J Cell Biol. 1994;125:31–42. doi: 10.1083/jcb.125.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gouy M, Li WH. Molecular phylogeny of the kingdoms Animalia, Plantae, and Fungi. Mol Biol Evol. 1989;6:109–122. doi: 10.1093/oxfordjournals.molbev.a040536. [DOI] [PubMed] [Google Scholar]