Abstract
The eukaryotic genome is assembled into distinct types of chromatin. Gene-rich euchromatin has active chromatin marks, while heterochromatin is gene-poor and enriched for silencing marks. In spite of this, genes native to heterochromatic regions are dependent on their normal environment for full expression. Expression of genes in autosomal heterochromatin is reduced in male flies mutated for the noncoding roX RNAs, but not in females. roX mutations also disrupt silencing of reporter genes in male, but not female, heterochromatin, revealing a sex difference in heterochromatin. We adopted a genetic approach to determine how this difference is regulated, and found no evidence that known X chromosome counting elements, or the sex determination pathway that these control, are involved. This suggested that the sex chromosome karyotype regulates autosomal heterochromatin by a different mechanism. To address this, candidate genes that regulate chromosome organization were examined. In XX flies mutation of Topoisomerase II (Top2), a gene involved in chromatin organization and homolog pairing, made heterochromatic silencing dependent on roX, and thus male-like. Interestingly, Top2 also binds to a large block of pericentromeric satellite repeats (359 bp repeats) that are unique to the X chromosome. Deletion of X heterochromatin also makes autosomal heterochromatin in XX flies dependent on roX and enhances the effect of Top2 mutations, suggesting a combinatorial action. We postulate that Top2 and X heterochromatin in Drosophila comprise a novel karyotype-sensing pathway that determines the sensitivity of autosomal heterochromatin to loss of roX RNA.
Introduction
Approximately 30% of the Drosophila genome is heterochromatic [1]. Many cytological and molecular features distinguish gene-poor heterochromatin from gene-rich euchromatin. Heterochromatin forms a compact, relatively inaccessible domain with ordered nucleosome arrays [2]. Heterochromatic loci tend to be near the nuclear periphery during interphase. Heterochromatin is characterized by repetitive DNA sequences, low levels of histone acetylation, hypomethylation at H3K4 and H3K79 and enrichment for Heterochromatin Protein 1 (HP1) [3]. Although relatively gene-poor, Drosophila heterochromatin harbors hundreds of protein coding genes (heterochromatic genes) [1, 4]. The native heterochromatic environment has been shown essential for full expression of some of these genes, and disruption of heterochromatin lowers their expression [5–7].
Euchromatic genes also rely on their native chromatin context, and stochastic silencing is observed when a euchromatic gene is placed in a heterochromatic environment, a phenomenon known as Position Effect Variegation (PEV). PEV represents variable spreading of inactivation over the euchromatic gene, producing irregular silencing [3]. PEV is extraordinarily sensitive to heterochromatin integrity. For example, mutation of a single copy of Su(Var)2–5, encoding HP1, elevates expression of variegating reporters inserted in heterochromatic regions. This effect, called suppression of PEV, enables identification of genes involved in heterochromatin formation and silencing.
Drosophila heterochromatin is typically not thought of as sexually dimorphic. However, recent studies suggest that heterochromatin in male and female flies differs. Reduction in HP1 results in preferential lethality and higher gene misregulation in males [8]. Mutation of the Drosophila roX1 and roX2 RNAs (RNA on the X 1 and -2) is a potent suppressor of PEV for autosomal insertions in male flies, but not in females [9]. A genome-wide reduction in the expression of autosomal heterochromatic genes is also observed in roX1 roX2 males [9]. These findings suggest a general disruption of autosomal heterochromatin in roX1 roX2 mutants that is limited to males. Sexually dimorphic heterochromatin could stem from differential sensitivity to reduced levels of factors necessary in both sexes, or by differences in the establishment or maintenance of heterochromatin in males and females. We refer to heterochromatin as masculine if roX RNA is necessary for normal PEV, and a feminine if roX is unnecessary. This designation does not require knowledge of the mechanism through which roX influences heterochromatin. Interestingly, the roX RNAs are also essential for X chromosome dosage compensation, another male-limited process [10]. roX RNAs assemble with the Male Specific Lethal (MSL) proteins to form a complex that is targeted to X-linked genes. Enzymatic activities within the MSL complex modify chromatin at X-linked genes, leading to increased transcription in male flies. Most of the MSL proteins are also required for full expression of autosomal heterochromatic genes in males [9]. The only member of the MSL complex that is unnecessary for heterochromatic genes is the Male Specific Lethal 2 (MSL2) protein. This is surprising as MSL2, a key regulator of X chromosome dosage compensation, is the sole member of the MSL complex with strictly male-limited expression. This raises intriguing questions about how the sexual dimorphism of heterochromatin is determined. We postulated that heterochromatic sex is under genetic control, and conducted experiments aimed at determining the signal that regulates this process.
Using a PEV reporter assay we demonstrated that feminization of heterochromatin is independent of female-limited components of the Drosophila sex determination pathway. Furthermore, neither MSL2 nor the Y chromosome directs heterochromatin masculinization. We then examined the numerator elements, components of the X chromosome counting mechanism, and saw no effect on heterochromatic sex. This suggests that a novel signal, perhaps direct sensing of karyotype, could be involved. As flies pair homologous chromosomes, the sex chromosome karyotype could be detected by the presence of unpaired chromatin in XY or XO flies. Screening of viable mutations that influence chromosome organization and homologue pairing revealed that Topoisomerase II (Top2) contributes to the feminization of autosomal heterochromatin in XX flies. Top2 promotes homologue pairing, consistent with pairing-dependent detection of sex chromosome karyotype. However, Top2 also binds satellite repeats that make up over 10 Mb of pericentric X heterochromatin [11]. Interestingly, loss of X-heterochromatin partially masculinizes autosomal heterochromatin in XX flies also. We propose that Top2 and pericentromeric X heterochromatin together control the sexual differentiation of heterochromatin in Drosophila melanogaster. The ubiquity of Top2 and repetitive sequences suggests a general mechanism for direct detection of karyotype.
Results
Two metrics of autosomal heterochromatic integrity are disrupted in roX1 roX2 (roX) males, but not females. First, expression of heterochromatic genes on the autosomes decreases in male larvae carrying the severely affected roX1 SMC17A roX2Δ chromosome [9]. Second, adult male escapers with the partial loss of function roX1 ex33 roX2Δ chromosome display a dramatic suppression of PEV at autosomal insertions. However, no suppression of PEV or reduction in heterochromatic gene expression is detected in females with these roX mutations. These observations were surprising because the roX RNAs were not thought to play a role outside of X chromosome dosage compensation. In addition, autosomal heterochromatin is not overtly sexually dimorphic. Variegating insertions typically behave similarly in males and females, and the autosomal heterochromatic genes that are misregulated in roX males rarely display sex-biased expression [9]. The underlying cause of the differences in male and female heterochromatin is completely unknown. In this study, we used a genetic approach to examine this question.
Suppression of PEV increases black abdominal pigmentation from variegating y + reporters (Fig 1A, S1A Fig) and red eye pigmentation from variegating w +mW.hs reporters (S1B Fig). The 3rd chromosome insertion KV24 displays y + PEV in both sexes and the 2nd chromosomal insertion KV20 displays PEV in males, but typically produces less than 1 spot on each female abdomen. Suppression of PEV in roX1 ex33 roX2Δ males was observed for all the autosomal insertions tested, but no effect was observed in roX1 ex33 roX2Δ females, revealing an effect that is not unique to a specific insertion site or reporter (Fig 1A, S1 Fig and [9]).
Fig 1. Heterochromatin masculinization is revealed by position effect variegation (PEV).
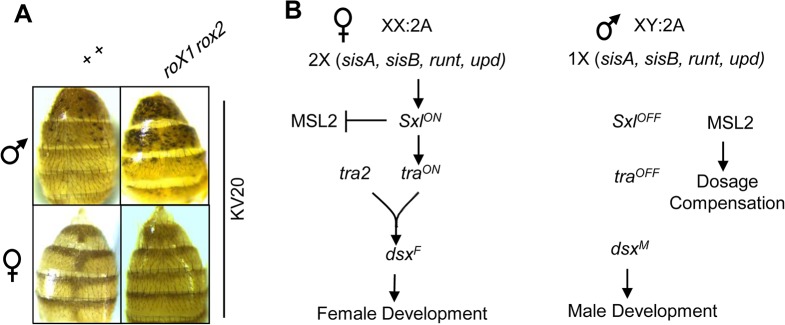
(A) PEV of a y + marker in the KV20 insertion produces black abdominal spots. Suppression of PEV in yw roX1 ex33 roX2Δ /Y; KV20/+ males increases pigmentation (top). Females (bottom) typically produce less than one spot per female, and no suppression of PEV is detected in yw roX1 ex33 roX2Δ; KV20/+ females (right). (B) Somatic sex determination in flies is controlled by the number of X chromosomes. Two copies of X-linked numerator elements (sisA, sisB, runt and upd) turn on Sexlethal (Sxl) expression in XX embryos. Sxl blocks dosage compensation by preventing translation of MSL2 in XX embryos. Sxl ensures productive splicing of transformer (tra) mRNA. tra and transformer2 (tra2) induce the female-specific isoform of doublesex (dsx F). Only dsx M is produced in males. The Dsx transcription factors coordinate visible somatic differentiation. Additional tra and tra2 targets (not shown) regulate differentiation of the nervous system.
To understand how this difference in fly heterochromatin arises, we conducted a screen for the genetic determinants of heterochromatin sexual dimorphism. This screen encompassed the sex determination pathway as well as elements of the sex chromosome karyotype. Matched genotypes differing only at the roX genes were generated to determine if heterochromatin is masculine (roX1 roX2 mutation suppresses PEV) or feminine (no or minor suppression of PEV in roX1 roX2 mutants) in each genetic background. Drosophila sex determination is triggered by the X chromosome dose (X:A, Fig 1B). The Y chromosome is believed to have no role in Drosophila sex determination. The two X chromosomes in female embryos initiate early expression of Sexlethal (Sxl) [12]. Sxl induces productive transformer (tra) splicing [13]. Tra and Transformer 2 (Tra2) direct splicing of the female isoform of the doublesex transcription factor (dsx F). Conversely, in XY embryos Sxl is not expressed [14, 15]. Sxl represses MSL2 translation [16–18]. As MSL2 is a key protein in X chromosome dosage compensation, this limits dosage compensation to males. The absence of Sxl in males also prevents tra expression, resulting in the production of default male isoform of dsx (dsx M). We hypothesized that genes in the sex determination pathway, or the Y chromosome, might control the observed sexual dimorphism of heterochromatin.
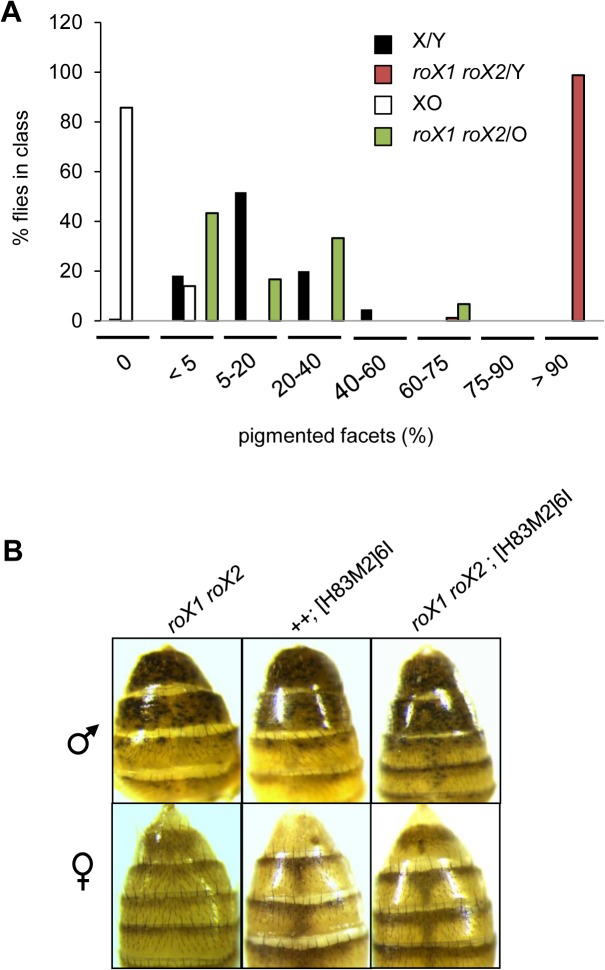
We first considered the possibility that a male-limited factor masculinizes heterochromatin. The Y chromosome is thought to act as a sink for heterochromatin proteins, and thus has epigenetic effects throughout the genome [19, 20]. We generated males with a variegating w +mW.hs marker (insertion 118E-10) that were wild type for the roX genes or carried the partial loss of function roX1 ex33 mutation and a deletion of roX2, a combination that allows over 20% escaper males. Eyes of control males (yw/Y; 118E-10/+) have an average of 20% pigmented facets (black bars, Fig 2A), but yw roX ex33 roX2/Y; 118E-10/+ males display over 90% pigmentation, a dramatic suppression of PEV (red bars, Fig 2A). The absence of a Y chromosome in XO males frees heterochromatin proteins to reinforce silencing and enhance PEV at other loci [20]. As expected, PEV was enhanced in XO males with wild type roX genes, almost 90% of which have no eye pigmentation (yw/O; 118E-10/+; white bars in Fig 2A). We then asked whether PEV in XO males was suppressed by roX mutations, and found that all yw roX ex33 roX2/O; 118E-10/+ males display at least some eye pigmentation (green bars in Fig 2A). Since the loss of roX suppresses PEV in otherwise identical XO males (compare white and green bars in Fig 2A), we conclude that the presence of the Y chromosome is not responsible for masculine heterochromatin in males.
Fig 2. Neither the Y chromosome nor MSL2 direct heterochromatin masculinization.
(A) Eye pigmentation was examined in flies with a variegating marker (w +mW.hs) in the 118E-10 insertion. In XY males (black and red bars) loss of roX (red) dramatically suppresses PEV. XO males display stronger silencing (white and green bars) but loss of roX (green) still suppresses PEV. Full genotypes and number of individuals scored are: black, yw/Y; 118E-10/+, 110, white, yw/O; 118E-10/+, 21, red, yw roX1 ex33 roX2Δ /Y; 118E-10/+, 83 and green, yw roX1 ex33 roX2Δ /O; 118E-10/+, 30. (B) MSL2 does not masculinize XX heterochromatin. Ectopic MSL2 expression from the [H83M2]6I transgene does not lead to suppression of PEV in yw roX1 ex33 roX2Δ females. PEV of y + KV20 is suppressed in yw roX1 ex33 roX2Δ males, and remains unchanged by increased MSL2 expression. At least 50 flies were scored per genotype. Representative male (top) and female (bottom) adults are presented.
The protein Male Specific Lethal-2 (MSL2) binds the roX RNAs and is the only male-limited member of the dosage compensation complex [21–23]. To determine if MSL2 plays a role in heterochromatin masculinization, we expressed MSL2 from the [H83M2]6I transgene in XX females with a variegating y + reporter (insertion KV20), and compared females that were either wild type or mutated for the roX genes [23–25]. This, and following studies utilize roX2Δ a simple deletion that facilitates stock construction [26]. PEV in females expressing MSL2 is not influenced by roX mutations (Fig 2B, bottom). In contrast, roX mutations suppress PEV in males of matched genetic background (Fig 2B, top). This is consistent with a study finding that MSL2 is not required for full expression of autosomal heterochromatic genes in males [9]. As MSL2 appears to have no role in either measure of sexually dimorphic heterochromatin, we conclude that it does not masculinize heterochromatin.
Loss of roX RNAs in males leads to relocalization of MSL proteins to the chromocenter, a structure composed of pericentromeric heterochromatin from all chromosomes. Identical MSL mislocalization is also observed in roX1 roX2 females that ectopically express MSL2 [9]. In spite of the abnormal recruitment of MSL proteins to the chromocenter, no disruption of heterochromatic gene expression or PEV can be detected in roX1 roX2 females that ectopically express MSL2 (Fig 2B and [9]). We conclude that mislocalization of MSL proteins does not produce the disruptions in heterochromatin function that are observed in roX1 roX2 mutants.
We then addressed the possibility that female-limited proteins in the somatic sex determination pathway feminize autosomal heterochromatin. If this is the case, mutations in this pathway will masculinize heterochromatin in XX flies (Fig 3A). We tested Sexlethal (Sxl), tranformer2 (tra2) and doublesex (dsx), representing different levels in the sex determination hierarchy (Fig 1B, left). As these genes direct female somatic differentiation, mutations produce XX intersexes or pseudomales with male-like body pigmentation and altered genital morphology. dsx 1 is amorphic and dsx D produces the male splice form. XX; dsx 1 /dsx D flies are fully masculinized. We generated X/Y; dsx 1 /dsx D and XX; dsx 1 /dsx D flies with KV20 and the yw roX1 ex33 roX2Δ chromosome. Masculinized XX; dsx 1 /dsx D flies were distinguished from XY flies by the absence of a marked Y chromosome (B s Y). Masculinization increased abdominal pigmentation, allowing detection of more y + spots in XX flies. Because of this, comparisons must be between flies with the same dsx status. Although yw roX1 ex33 roX2Δ/ B s Y; KV20/+; dsx 1 /dsx D males displayed strong suppression of PEV in comparison to males with wild type roX, no suppression of PEV was observed in XX; dsx 1 /dsx D pseudomales upon loss of roX (compare yw roX1 ex33 roX2Δ; KV20/+; dsx 1 /dsx D and yw; KV20/+; dsx 1 /dsx D, Fig 3B).
Fig 3. The somatic sex determination pathway and numerator elements do not control heterochromatin feminization.
(A) Scheme for identification of genetic regulators of heterochromatic sex. Heterochromatin is masculine if loss of roX suppresses PEV of an autosomal reporter. If a gene in the sex determination cascade normally feminizes XX heterochromatin, mutation of that gene will masculinize XX heterochromatin, leading to suppression of PEV in roX mutants. (B) tra2 and dsx do not feminize heterochromatin. yw roX1 ex33 roX2Δ / B s Y; KV20/+ males with tra2 B, tra2 ts1 or dsx 1 /dsx D mutations display suppression of PEV, detected by increased abdominal pigmentation (gray bars at right). XX pseudomales and intersexes display a modest increase in spots, consistent with masculinization of pigmentation patterns (hatched bars). However, no suppression of PEV is observed in roX pseudomales. Full genotypes (left to right) are: yw; KV20/+, yw; KV20/+; dsx 1 /dsx D, yw; tra2 B KV20/ tra2 B, yw; tra2 TS1 KV20/ tra2 TS1, yw roX1 ex33 roX2Δ; KV20/+, yw roX1 ex33 roX2Δ; KV20/+; dsx 1 /dsx D, yw roX1 ex33 roX2Δ; tra2 B KV20/ tra2 B, yw roX1 ex33 roX2Δ; tra2 TS1 KV20/ tra2 TS1). Twenty-50 individuals of each genotype were scored. (C) Sxl mutations do not masculinize XX heterochromatin. Representative XY (top) and XX (bottom) flies are shown. XY flies with Sxl mutations suppress PEV upon loss of roX function (right two panels). XX Sxl M1,f3 /Sxl 2593 pseudomales display partial masculinization of genitalia and pigmentation, but no suppression of PEV is observed upon roX mutation. (D) Abdominal pigmentation in Sxl adults. Full genotypes of XY flies (gray bars): yw/Y; KV20/+, 75 flies, yw Sxl 2593 /Y; KV20/+, 75 flies, yw Sxl M1,f3/Y; KV20/+, 64 flies, yw roX1 ex33 Sxl M1,f3 roX2Δ/Y; KV20/+, 17 flies, yw roX1 ex33 Sxl 2593 roX2Δ Y KV20/+, 37 flies. Full genotypes of XX flies (hatched bars): yw Sxl M1,f3 / yw Sxl 2593 KV20/+, 21 flies, yw roX1 ex33 Sxl M1,f3 roX2Δ/ yw roX1 ex33 Sxl 2593 roX2Δ KV20/+, 10 flies. (p-value ***<0.00001, n.s = non-significant). (E) Numerator elements do not feminize XY heterochromatin. Overexpression of SisA and SisB is indicated by ++. Full genotypes: ywSxl f1/Y; 2XP(w +mC,sisA +)+ 2XP(w +mC,sc sisB+)/KV20 and yw roX1 ex33 Sxl f1 roX2Δ/Y; 2XP(w +mC,sisA +) +2XP(w +mC,sc sisB+)/KV20. Data was derived from over 20 individuals per genotype. *** indicates p-value <0.00001.
We next tested the tra2 ts1 and tra2 B mutations. tra2 ts1 is a temperature sensitive hypomorph and tra2 B is a null allele. Loss of tra2 has no visible effect on XY flies but masculinizes XX flies. We generated XX and XY tra2 mutants carrying KV20 and yw roX1 ex33 roX2Δ. Loss of roX suppressed PEV in tra2/ tra2 males (Fig 3B). In contrast, XX; tra2/ tra2 pseudomales mutated for roX displayed no suppression of PEV (Fig 3B).
Although dsx and tra2 do not regulate heterochromatin sexual differentiation, it remained possible that Sxl, the master regulator of sexual determination, did so through a different pathway. Since null Sxl mutations are embryonic lethal in XX zygotes, we tested a heteroallelic combination, Sxl M1,f3 /Sxl 2593, that produces masculinized XX adult escapers. The roX genes and Sxl are X-linked, necessitating generation of two roX1 ex33 Sxl roX2Δ chromosomes. Control masculinized XX adults (yw Sxl M1,f3 / yw Sxl 2593; KV20/+) emerged late and displayed developmental defects and partial sexual transformation (Fig 3C, bottom). Similar to XX flies masculinized by tra2 and dsx, a few abdominal spots were visible. However, mutation of roX had no effect on PEV in XX flies that were masculinized by Sxl mutations (Fig 3D, hatched bars). In contrast, XY males mutated for roX and Sxl displayed strong suppression of PEV (Fig 3C and 3D). This supports the idea that sexual differentiation of heterochromatin is independent of the somatic sex determination pathway. One caveat is that this test requires adult escapers, preventing use of null Sxl alleles. It remains possible that a novel Sxl function is retained in the heteroallelic combination tested. Sxl regulates roX1 expression by repression of MSL2 [27, 28]. One possibility is that Sxl regulates heterochromatic sexual differentiation by modulating roX1 levels in early embryos. For example, high roX1 RNA concentrations could establish male heterochromatin. Repression of MSL2 by Sxl in females reduces roX1 levels. Arguing against this idea is the observation that ectopic expression of MSL2 fails to masculinize heterochromatin. Furthermore, roX1 is abundant in early embryos of both sexes, and pseudomales generated using a similar heteroallelic Sxl combination have elevated roX1 levels[29][30]. However, none of these manipulations activate dosage compensation or roX1 expression to the level observed in normal males. Nevertheless, the stability of heterochromatic sex in genetic backgrounds mutated for tra and dsx suggests genetic regulation at the level of Sxl or above.
A mechanism that detects sex chromosome karyotype could bypass the sex determination cascade altogether. One way this could occur is if the X chromosome counting mechanism that turns on Sxl in XX embryos also controls a second pathway that leads to heterochromatin feminization. Proteins from the X-linked sisterless A and B (sisA and sisB), unpaired (upd) and runt (runt) genes, collectively known as numerator elements, promote early Sxl expression in XX embryos [31–34]. Elevated sisA and sisB expression is benign in XX flies but turns on Sxl expression in XY flies, a lethal situation that can be overcome by mutating Sxl [35, 36]. We examined heterochromatin sexual differentiation in XY flies with multiple sisA and sisB transgenes and the Sxl f1 mutation. We found normal PEV in control males that have wild type roX and overexpress sisA and sisB, but strong suppression of PEV when roX mutations are introduced into this genotype, revealing stable heterochromatin masculinization (Fig 3E). We conclude that sisA and sisB, key components of the X chromosome counting mechanism, do not feminize heterochromatin.
Another possible mechanism for detection of karyotype involves chromosome pairing. Interphase chromosomes of Drosophila are paired throughout development [37–39]. All homologs pair in females, but the structurally dissimilar X and Y chromosomes of males remain unpaired. In theory, unpaired chromatin in XY and XO cells could signal the male karyotype.
To investigate this possibility, we examined several genes that regulate homolog pairing in Drosophila [39, 40]. Three pairing promoters, Topoisomerase II (Top2), Dynein Heavy chain-64c (Dhc64c) and Microcephalin-1 (MCPH1), and three anti-pairers, condensin II subunits Cap-H2 and Cap-D3, and Female sterile (1) homeotic (fs(1)h) were examined. Some of these are essential, requiring the use of partial loss of function mutations, or heteroallelic combinations that produce adult escapers. HP1, an anti-pairing gene, was not selected for the screen, as mutation of HP1 is a potent suppressor of PEV regardless of sex. If fully paired chromosomes signal the XX karyotype, and this in turn regulates heterochromatic sex, mutation of anti-pairers will increase pairing, leading to feminization of autosomal heterochromatin in XY animals. We generated XX and XY flies with KV20 and viable mutations in individual anti-pairers. Each was constructed with wild type or mutated roX genes. Abdominal spots were minimal, but unchanged, in roX mutant females. Males with Cap-H2 Z0019, Cap-D3 c07081 or fs(1)h 1 mutations continued to suppress PEV when mutated for roX (S2 Fig, compare gray and black bars). We conclude that mutation of these anti-pairing factors does not lead to feminization of heterochromatin in males.
We then tested mutations in pairing promoters. These mutations reduce pairing, a condition that could mimic the unpaired chromatin of males. If unpaired chromatin signals the XY karyotype, reduced pairing in XX flies could inappropriately masculinize heterochromatin. We first generated individual XX and XY flies with loss of function mutations in Dhc64c or MCPH1, KV20, and wild type or mutated for the roX genes. XY flies mutated for Dhc64c or MCPH1 continued to show suppression of PEV when mutated for roX (yw roX1 ex33 roX2ΔY; MCPH1 0978 KV20 / MCPH1 0978 and yw roX1 ex33 roX2ΔY; KV20 /+; dhc64c 6-10 / dhc64c 8-1) (S2 Fig, gray bars). However, no masculinization of heterochromatin was apparent in females mutated for Dhc64c or MCPH1 (S2 Fig, hatched bars).
We then tested Top2, a pairing promoter with critical roles in nuclear organization, cell division and DNA repair. Since loss of Top2 is lethal, the complementing heteroallelic Top2 17-1 /Top2 17-3 combination was used [41]. Each mutation is individually lethal, but Top2 17-1 /Top2 17-3 flies display >50% viability. Top2 17-1 (S791F) in the WHD domain reduces protein accumulation, but Top2 17-3 (L471Q) in the TOPRIM domain produces stable, full-length protein (S3A Fig). We generated Top2 17-1 /Top2 17-3 XX and XY flies with variegating y + (KV24 insertion) that were in addition either wild type or mutated for the roX genes. The switch to the 3rd chromosome KV24 was necessitated by our inability to recover a recombinant second chromosome with KV20 and Top2. We observed that Top2 17-1 /Top2 17-3 itself suppressed PEV in males, but not in females, thus identifying an additional difference in the heterochromatin of males and females (Fig 4A and 4B). Surprisingly, Top2 17-1 /Top2 17-3 females displayed highly significant suppression of PEV upon loss of roX, suggesting masculinization of XX heterochromatin by Top2 mutation (Fig 4B). However, mutation of Top2 does not otherwise sexually transform XX flies, which display female morphology.
Fig 4. Mutation of Topoisomerase II (Top2) masculinizes XX heterochromatin.

(A) PEV is suppressed in males mutated for roX or Top2. Ectopic MLE or MSL1 expression does not restore PEV in roX or Top2 mutants. Twenty-50 flies of each genotype were scored. p-values: ** <0.0001; *** <0.00001; n.s non-significant. (B) Suppression of PEV in Top2 females mutated for roX. Pigmentation displays little or no increase in XX flies mutated for roX or Top2 alone (left three bars). Simultaneous mutation of roX and Top2 leads to suppression of PEV. Over expression of MLE, but not MSL1, partially restores PEV in roX and Top2 females (right two bars). (for A, B) Wild type (+) and mutant (-) for indicated genes; (+++) overexpressing transgenes. Full genotypes (left to right) yw; KV24/+, yw roX1 ex33 roX2Δ; KV24/+, yw; Top2 17-1 /Top2 17-3; KV24/+, yw roX1 ex33 roX2Δ; [H83MLE]/+; KV24/+, yw roX1 ex33 roX2Δ; +/+; KV24/[H83M1]Z1, yw roX1 ex33 roX2Δ; Top2 17-1 /Top2 17-3; KV24/+, yw roX1 ex33 roX2Δ; Top2 17-1 /Top2 17-3 [H83MLE]; KV24/+, yw roX1 ex33 roX2Δ; Top2 17-1 /Top2 17-3; KV24/[H83M1]Z1. (C) Overexpression of MLE rescues Top2 lethality in both sexes. yw; Top2 17-1/CyO y + females were mated to yw; Top2 17-3/CyO y + or yw; Top2 17-3[H83 MLE] /CyO y + males. Survival of yw;Top2 17-1/ Top2 17-3 (black) and yw; Top2 17-1/ Top2 17-3[H83 MLE] (gray) was calculated by setting recovery of flies with CyOy + to 100%. Data was compiled from at least 3 replicate matings. (D) Overexpression of MSL1 does not rescue Top2 17-1/ Top2 17-3 survival. yw; Top2 17-1/ In(2LR)GlaBc females were mated to yw/Y; Top2 17-3/In(2LR)GlaBc; [H83M1]Z1/+ males. Survival of yw;Top2 17-1/ Top2 17-3 (black) and yw;Top2 17-1/Top2 17-3; [H83M1]Z1/+ (gray) was calculated by setting recovery of flies with In(2LR)GlaBc to 100%. Survival is derived from 5 replicate matings.
Top2 was the sole pairing promoter that altered the sexual differentiation of heterochromatin, raising questions about the precise molecular function that is disrupted by the mutations used. Top2 17-1 /Top2 17-3 males are fertile, but embryos deposited by Top2 17-1 /Top2 17-3 females fail to hatch (S3B Fig). No evidence of DNA replication could be detected in these embryos by DNA staining (not shown), consistent with meiotic or mitotic failure [42]. We conclude that meiosis, fertilization or embryonic development of Top2 17-1 /Top2 17-3 mutants requires maternal provision of wild type Top2.
We then examined polytene preparations from wild type and Top2 17-1 /Top2 17-3 larvae to determine if there was a visible effect on chromosome organization. Similar heteroallelic Top2 mutants have been shown to disrupt the male X-chromosome [41]. We scored chromosome morphology as abnormal if banding was diffuse and puffy if the chromosome was bloated along its entire length. Chromosomes from Top2 mutants are more susceptible to breaking, suggesting fragility. Seventy percent of male nuclei from Top2 mutants had abnormal or puffy X chromosomes (S3C Fig, black arrows), but only 14% of X chromosomes from wild-type males were scored as abnormal. Top2 mutant females and wild type females display similar levels of X chromosome abnormality (10–15%). Fifty percent of nuclei from Top2 mutants had partially unpaired homologs, in contrast to 15% from wild type larvae (S3C Fig, white arrows, S1 Table). The size, position and extent of unpairing varied between nuclei, and unpaired regions were equally prevalent in males and females. As most of the genome remains paired, this defect appears relatively minor. In summary, examination of chromosomes suggests selective disruption of male X-chromosome polytenization in Top2 mutant larvae and homolog pairing that remains largely intact.
We then examined homolog pairing using a genetic assay. Pairing enables enhancers from one mutant allele to drive the promoter of a different allele, thus restoring expression (transvection). Transvection at yellow (y) is detected by increased pigmentation. While y 82f29 is a deletion of upstream enhancer elements, y 1#8 retains enhancers but lacks a promoter. Transvection in y 82f29 /y 1#8 flies restores body, wing and bristle color (S3D Fig). y 3c3 lacks a bristle enhancer and the y promoter, but retains a wing enhancer. Transvection in y 82f29 /y 3c3 flies restores wing pigmentation (S3 Fig). Flies homozygous for any one of these alleles have light bodies, wings and bristles. Heteroallelic y 82f29 /y 1#8 and y 82f29 /y 3c3 flies in wild type and Top2 17-1 /Top2 17-3 mutant backgrounds displayed equivalent transvection (S3D and S3E Fig). We conclude that Top2 17-1 /Top2 17-3 mutants retain sufficient homolog pairing to support transvection at y. Although no defect in y pairing was observed by this test, it is formally possible that the Top2 mutants we tested are defective for pairing at other loci.
The y 2 allele is produced by a Gypsy insulator that prevents wing and body enhancers from contacting the promoter. Top2 is necessary for Gypsy insulation, and loss of Top2 restores pigmentation in the wing and body of y 2 flies [43]. We examined insulator function by comparing pigmentation in y 2 males that are wild type and Top2 17-1 /Top2 17-3. No increase in body or wing color could be detected in y 2/Y; Top2 17-1 /Top2 17-3 flies (S3F Fig). We conclude that the Top2 17-1 /Top2 17-3 flies retain Gypsy insulator function, consistent with tests of other viable heteroallelic Top2 combinations [44].
Top2 was recently reported to participate in dosage compensation [45]. In support of this idea, a physical interaction between Top2 and Maleless (MLE), an RNA helicase that is a member of the dosage compensation complex, was detected. Based on this, and the disruption of X chromosome morphology in male Top2 17-1 /Top2 17-3 mutants, we asked whether Top2 17-1 /Top2 17-3 affects males more strongly than females. Interestingly, Top2 17-1 /Top2 17-3 flies do not display male-preferential lethality, suggesting that these mutations do not affect the dosage compensation function of Top2 (Fig 4C, black bars). The association between Top2 and MLE prompted us to ask whether overexpression of MLE from a transgene [H83 MLE] could influence the survival of Top2 17-1 /Top2 17-3 flies. MLE overexpression dramatically rescued Top2 17-1 /Top2 17-3 mutants of both sexes (Fig 4C, gray). However, no rescue of Top2 mutants was achieved by overexpression of another member of the dosage compensation complex, male-specific lethal 1 (msl1) (Fig 4D). Our data supports the idea of an interaction between Top2 and MLE, but the lack of sex-specificity of rescue argues against a role that is limited to dosage compensation.
The increased survival of Top2 17-1 /Top2 17-3 mutants upon MLE overexpression prompted us to ask if MLE could restore heterochromatin function in Top2 mutants. To address this we generated Top2 17-1 /Top2 17-3 mutants that overexpress MLE, carry the KV24 reporter and are either wild type or mutant for the roX genes. Increased MLE expression failed to restore PEV in males mutated for roX and Top2 (Fig 4A). In contrast, expression of MLE in roX and Top2 mutant females achieved significant restoration of PEV (Fig 4B). However, overexpression of MSL1 failed to restore PEV in roX and Top2 mutant females (Fig 4B). Taken together, these findings support the idea that a Top2—MLE interaction is necessary for a process other than compensation, but the basis for the sex-specific effect of MLE on restoration of female PEV is speculative at present. However, MLE is part of the MSL complex, making it plausible that recruitment of MLE to the male X chromosome reduces its availability for interaction with Top2 on autosomal heterochromatin, producing the observed differences in response to overexpression.
The involvement of Top2 in a process that may be triggered by sex chromosome karyotype suggested an alternative mechanism. Over 10 Mb of X heterochromatin is composed of satellite repeats (359 bp repeats) that are unique to the X chromosome [39, 46]. Interestingly, the 359 bp repeats bind Top2 in interphase nuclei [11, 47]. This suggested the possibility that an interaction between X heterochromatin and Top2 determines differential heterochromatin sensitivity to loss of roX. If this is the case, deletion of X heterochromatin may act similarly to Top2 mutation. The X;Y translocation Zhr 1 replaces X heterochromatin with part of the Y chromosome [48, 49]. We generated roX mutant females that were heterozygous for Zhr 1 and carry KV20 (yw roX1 ex33 roX2Δ Zhr 1 / yw roX1 ex33 roX2Δ +; KV20/+). Interestingly, weak suppression of PEV was observed in roX females with a single Zhr 1 chromosome, but not in Zhr 1 females wild type for roX (Fig 5A). As removal of one copy of X heterochromatin generates XX females that now depend on roX for normal autosomal PEV, loss of X heterochromatin partially masculinizes autosomal heterochromatin in these flies.
Fig 5. Pericentromeric X heterochromatin contributes to feminization of autosomal heterochromatin in XX flies.

X heterochromatin was deleted by the X;Y translocation Zhr 1. (A) Females with one or two Zhr 1 chromosomes suppress PEV upon loss of roX. The KV20 reporter, which normally produces <1 spot/abdomen, was used. roX and Top2 mutations are indicated by (-). Full genotypes (left to right): yw; KV20/+, yw roX1 ex33 roX2Δ; KV20/+, yw/yw Zhr 1; KV20/+, yw roX1 ex33 roX2Δ + / yw roX1 ex33 roX2Δ Zhr 1; KV20/+, yw Zhr 1 /yw Zhr 1; KV20/+, yw roX1 ex33 roX2Δ Zhr 1 / yw roX1 ex33 roX2Δ Zhr 1; KV20/+. Averages are derived from 20–50 flies of each genotype. *** indicates p-value <0.00001. (B) Loss of Top2 further masculinizes heterochromatin in Zhr 1/+ females. Greater suppression of PEV is observed in roX females mutated for Top2 and with Zhr 1. This study uses the KV24 reporter, producing about 30 spots/female in a wild type background. Full genotypes (left to right): yw; KV24 /+, yw roX1 ex33 roX2Δ; KV24 /+, yw; Top2 17-1/Top2 17-3; KV24 /+, yw roX1 ex33 roX2Δ; Top2 17-1/ Top2 17-3; KV24 /+, yw/yw Zhr 1; KV24 /+, yw roX1 ex33 roX2Δ Zhr 1/ yw roX1 ex33 roX2Δ KV24 /+, yw roX1 ex33 roX2Δ Zhr 1/ yw roX1 ex33 roX2Δ Top2 17-1/ Top2 17-3; KV24 /+. Bars with coarse hatching are reproduced from Fig 4 for comparison. *** p-value <0.00001.
The involvement of Top2 in homolog pairing, and its localization at the 359 bp repeats, suggested the possibility that a large block of unpaired 359 bp repeats itself could signal the XY karyotype. If this is the case, Zhr 1 /Zhr 1 females, which have no unpaired 359 bp repeats, should display feminine heterochromatin. In contrast to this expectation, we found increased suppression of PEV in homozygous Zhr 1 females that lack roX (Fig 5A, right). However, no suppression of PEV was observed in homozygous Zhr 1 females with wild type roX. Suppression of PEV is thus not due solely to the differing chromatin content of Zhr 1 chromosomes. Our findings are consistent with an interaction between Top2 and X heterochromatin determining heterochromatin sensitivity to roX, but do not support the hypothesis that unpaired chromatin in the XY or XO nucleus is a factor.
The suppression of PEV in roX females with one or two Zhr 1 alleles is weak (contrast with suppression of PEV in roX1 roX2 males, Fig 3B). To determine if the effects of Top2 and Zhr 1 mutations are additive, we generated Zhr 1 /+ females mutated for Top2 and compared PEV in the presence and absence of roX. These females displayed greater suppression of PEV upon loss of roX than females mutated for Zhr 1 or Top2 alone, supporting the idea that Top2 and pericentric X heterochromatin act together (Fig 5B).
If the dose of X-heterochromatin acts as a signal for karyotype, duplication of this region in XY flies should feminize their heterochromatin. We attempted to generate XY flies with a duplication of X heterochromatin on the Y chromosome (Zhr + Y) to test this idea [11]. Unfortunately, no roX1 roX2/ Zhr + Y males were recovered, suggesting a genetic incompatibility between chromosomes in this mating.
Discussion
Autosomal heterochromatin is typically not thought of as differing in males and females, but sexually dimorphic PEV has also been observed in mice, where a variegating transgene is more highly expressed in females [50]. This study found that both SRY and sex chromosome karyotype determine silencing. Importantly, this reveals that sexual dimorphism of autosomal heterochromatin is not limited to Drosophila. One attractive possibility is that both male and female flies require roX RNA for heterochromatic silencing, but male heterochromatin is more sensitive to loss of roX. The idea that roX RNAs might in fact also function in females is supported by the modest suppression of PEV sometimes observed in roX1 roX2 females (Figs 4B, 5A and 5B). Although the roX RNAs are typically thought of as male-limited, roX1 is abundantly expressed in early embryos of both sexes, and thus is available in females [51]. While we do not yet understand the rationale for the sex differences in autosomal heterochromatin in flies, the presence of a large, heterochromatic Y chromosome ensures that males have considerably more total heterochromatin than females. It is plausible that the chromatin content of XY cells drove a compensatory adaptation in male flies [8, 9].
The identification of Top2 as a regulator of heterochromatic sexual dimorphism suggests that maintenance of normal chromatin organization plays a role in sex differences based on karyotype. However, the involvement of Top2 in numerous processes complicates analysis. For example, Top2 is itself required to maintain PEV in otherwise wild type males, but not in females. This provides additional evidence for the sexual dimorphism of autosomal heterochromatin, and is in agreement with a role for Top2 in chromatin condensation [52, 53]. However, it also suggests dual roles for Top2 in karyotype detection and heterochromatin maintenance.
Top2 has been reported to participate in the male-limited process of dosage compensation in studies using chemical inhibition or RNAi knockdown [45]. These manipulations produced a 2-fold reduction of expression in a plasmid-based model for dosage compensation. A physical association between Top2 and a single member of the MSL complex, the RNA/DNA helicase MLE, was also detected in these studies. Top2 has also been found with chromatin-bound MSL proteins in S2 cells, but, as Top2 is an abundant component of chromatin, this is unsurprising [54]. Our studies, performed with heteroallelic Top2 mutants, confirm a genetic interaction between MLE and Top2, but this appears equally important in males and females, and thus not limited to dosage compensation. The different methods by which Top2 activity was reduced in these studies may be responsible for this disparity. Interactions between helicases and Top2 are prevalent in other species. Yeast Top2 binds the Sgs1 helicase and mammalian Top2α interacts with BLM, the Bloom Syndrome helicase, and RNA helicase A, orthologous to MLE [55–57]. Disruption of the BLM-Top2α interaction leads to chromosome damage, and Top2 interaction with Sgs1 is required for decatenation in vivo. These interactions are thus important for genomic integrity. The nature of the Top2-MLE interaction remains an interesting question. Drosophila Top2 does associate with RNA, and it is possible that the helicase activity of MLE regulates this association [58]. We speculate that overexpression of MLE stabilizes mutant Top2 or supports its activity, increasing the survival of Top2 mutants of both sexes. An intriguing possibility, suggested by the association of the DEAD/H box RNA helicase P68 with mouse centromeric repeats, is that MLE promotes recruitment of Top2 to the 359 bp repeats [59].
The identification of Top2 as a pairing promoter suggested that X chromosome pairing could signal karyotype, but questions about the functions that are deficient in Top2 mutants complicate interpretation. Some function must be retained in Top2 17-1 /Top2 17-3 mutants because adult escapers are recovered. However, embryos from Top2 17-1 /Top2 17-3 mothers fail to initiate development, revealing a requirement for maternally deposited wild type Top2. It is possible that maternal Top2 is also sufficient to rescue near-normal pairing, transvection and insulation in Top2 17-1 /Top2 17-3 flies. Indeed, studies with a similar heteroallelic Top2 combination found no defect in pairing of the 359 bp repeats [44]. This study, like ours, used larvae that received maternal Top2, potentially obscuring a requirement for Top2 in this process.
Top2 is enriched on the pericentric 359 bp repeats, and deletion of X-heterochromatin additively enhances masculinization of autosomal heterochromatin by Top2 mutations. This prompted the idea that differences in karyotype may be detected by interaction of Top2 and a sequence within X-heterochromatin, possibly the 359 bp repeats. Several scenarios for how this might occur are possible. XX flies have double the X-heterochromatin of XY flies. An absolute difference in the amount of Top2-bound X heterochromatin could distinguish the male and female karyotypes (Fig 6A, left). It is also possible that higher free Top2 in males, with a single copy of the 359 bp repeats, is the source of a karyotype-specific signal (Fig 6A, right). This idea is supported by enhanced masculinization upon deletion of X heterochromatin. Although we obtained no evidence supporting the idea that unpaired chromatin signals the male karyotype, it remains possible that pairing of X heterochromatin, either dependent or independent of Top2, signals the XX karyotype (Fig 6B and 6C). For example, Top2-independent pairing of X-heterochromatin might occur, but association of Top2 with this region could be necessary to detect the paired status (Fig 6C).
Fig 6. Models for detection karyotype detection.

The absolute amount of X heterochromatin (A) or pairing of X heterochromatin (B, C) could generate a signal specifying the XX karyotype. XX flies have two copies of X heterochromatin (thick lines) but XY flies have one. Top2 (red) binds the 359 bp repeats (gray). (A) The absolute amount of Top2-bound 359 bp chromatin (top, left) or free Top2 in males (top, right) could generate a karyotype-specific signal. Mutant Top2 (A, bottom) is deficient in a function necessary for generation of the signal. Non-359 bp X-heterochromatin is shown in white. (B) Top2-dependent pairing of X heterochromatin could signal the XX karyotype (top). Mutant Top2 (bottom) fails to support normal pairing. (C) Top2-independent pairing of X-heterochromatin requires Top2 to generate or transmit a signal. Top2 mutants (bottom) are deficient in this process.
Numerous sex determination strategies have arisen in heterogametic organisms. Each utilizes a primary signal that orchestrates the process of becoming female or male. Recent studies have highlighted the complexity of gene regulation at the bottom of the fly sex determination cascade [60–63]. In contrast, the chromosome counting mechanism at the top of the cascade was long thought to be the exclusive source of differences between the sexes [12, 31, 64]. Our findings suggest that the sex chromosomes of flies have additional ways of modulating phenotype. These findings are in accord with recent studies in multiple organisms documenting regulation by sex chromosome karyotype, rather than the conventional sex determination pathway (reviewed by [30]). Indeed, an analysis in fly heads revealed that most sex-biased gene regulation is not mediated by tra [61]. While some of this likely depends on upstream elements in the sex determination and dosage compensation cascade, the regulatory basis of a significant proportion of the genes identified by this study remains unknown. Our current findings are most easily interpreted as evidence that chromosome-specific repetitive sequences, and proteins that interact with these sequences, produce differences in the nuclear environment that reflect sex chromosome karyotype. We postulate that this leads to the differences in male and female autosomal heterochromatin that we have observed. The universality of repetitive sequences and Top2 in higher eukaryotes suggests a general mechanism that could operate in other heterogametic organisms.
Materials and Methods
Fly strains
Flies were maintained at 25°C on standard cornmeal–agar fly food. Unless otherwise noted, mutations are described in [65]. roX1 mutations have been described [10, 29, 66]. Elimination of roX2 was accomplished by a viable deletion (roX2Δ) or a lethal deletion complemented by a cosmid carrying essential genes but lacking roX2 [10] [26]. Variegating insertions used as reporters in this study are described [67, 68]. A 4th chromosome insertion of P[hsp26-pt, hsp70-w], marked with w +mW.hs (118E-10) and 2nd (KV20) and 3rd (KV24) chromosome insertions of P[SUPor-P], marked with w +mC and yellow (y +) reporters were used. These reporters were selected to facilitate stock construction, but key findings were validated with multiple reporters. Top2 17-1 and Top2 17-3 mutations were generously provided by A. Hohl, C. T. Wu and P. Geyer [41]. Additional mutations are as follows: Cap-D3 c07081 [69], Cap-H2 Z0019 [70], MCPH1 0978 [71], Dhc64c 8-1 [72], [w +-hsp83 MLE] [73], [w +-hsp83 MSL2]6I and [w +-hsp83 MSL1]Z1 [23, 74], 2XP(w +mC,sisA +)+2XP(w +mC,sc sisB+) [36, 75]. Descriptions of Sxl 2593, Sxl M1F3, Tra2 B, Tra2 ts1, Dsx 1, Dsx D, Top2 17-1, Top2 17-3, Cap-D3 c07081, Cap-H2 Z0019, MCPH1 0978, Dhc64c 6-10, Dhc64c 8-1, fs(1)h 1, Zhr + Y and Zhr 1 are available on Flybase (http://www.flybase.org). All other strains used in this study were obtained from the Bloomington Drosophila Stock Center.
Transvection and insulator assays
Restoration of pigmentation by transvection at y is a standard measure of homolog pairing [76–78]. Pigmentation was scored in 1–2 days old flies on a scale of 1–4, where 1 is the no pigmentation and 4 is wild type levels. At least 100 flies of each genotype were scored. The y 2 Gypsy insertion contains an insulator that disrupts communication between the y enhancer and promoter [76]. Flies were aged for 24 h before scoring on the pigmentation scale described above. At least 25 flies from two independent crosses were scored. Significance was determined by a Student’s T-test. Images were obtained using a Zeiss Discovery V8 stereo microscope.
Supporting Information
PEV of y + in KV24 (3rd chromosome) is visible as black abdominal spots in both sexes and is suppressed in roX males (top), but not in roX females (bottom). PEV of w +mW.hs in 118E-10 (4th chromosome) is detected by eye pigmentation. roX males (top), but not females (bottom), suppress 118E-10 PEV. 118E-10 was examined in the yw roX1 ex33 Df(1)52;[4Δ4.3]/+ background, which is mutated for roX1 and roX2 and lacks other w markers, enabling visualization of the w +mW.hs reporter.
(PDF)
Heterochromatic sex was determined in flies mutated for anti-pairers (Cap-H2, Cap-D3 and fs(1)h) and pairing promoters (MCPH1 and Dhc64c). All flies carried the y + KV20 reporter. Flies mutated for each pairing regulator were generated in wild type (++) and yw roX1 ex33 roX2Δ mutant backgrounds. Almost no abdominal pigmentation was observed in XX flies wild type (white) or mutated (hatched) for both roX genes. In contrast, PEV in XY flies (black) is suppressed in roX mutants (dark gray). A slight enhancement of PEV is detected in Cap-D3 mutant flies, consistent with previous reports of condensin mutations as PEV enhancers [79, 80]. Fifteen-50 flies were counted for each genotype.
(PDF)
A) The Top2 mutations disrupt different domains. Missense mutations Top2 17-1 (WHD domain) and Top2 17-3 (TOPRIM domain). B) Top2 17-1 /Top2 17-3 males are fertile but Top2 17-1 /Top2 17-3 females are sterile. Both mutations are homozygous lethal. C) Characteristic abnormalities in a polytene preparation from a Top2 17-1 /Top2 17-3 male larvae. A puffy X chromosome (black arrow) and homolog unpairing (white arrows) are visible. One hundred-250 nuclei from at least 5 larvae were scored for each genotype. D) Transvection restores yellow expression. y 82f29 is a deletion of upstream enhancer elements. y 1#8 retains enhancers but lacks a promoter. y 3c3 lacks a bristle enhancer and the promoter, but retains a wing enhancer. Pairing between y 82f29 and y 1#8 or y 3c3 enables enhancers on the homolog to drive the y 82f29 promoter, restoring expression. Drawing based on [77]. Wing and body pigmentation was ranked from 1 (no pigmentation) to 4 (wild type). Flies homozygous for each allele have light body and wing color (1,1). Transvection in y 82f29 /y 1#8 flies restores wing and body color near wild-type levels (3, 3). Transvection in y 82f29 /y 3c3 flies restores wing pigmentation only (3, 1). Transvection is not disrupted in Top2 17-1 /Top2 17-3 mutants (shaded). Flies were aged 1–2 days before scoring and photography. At least 100 flies were scored for each genotype. E) Representative abdomens showing y transvection. Full genotypes are: y 82f29 /y 1#8; Top2 m / Cyo, y 82f29 /y 1#8; Top2 17-1 /Top2 17-3. F) Top2 mutations do not disrupt Gypsy insulation. Loss of pigmentation in y 2 requires the Top2-dependent Gypsy insulator. Loss of insulation enhances body pigmentation. Full genotypes are: y 2/Y; +/+, y 2/Y; Top2 m /CyO and y 2/Y; Top2 17-1 /Top2 17-3. At least 25 flies of each genotype were aged for 24 h before scoring.
(PDF)
Polytene preparations from control (+/+, reference yw strain) and yw; Top2 17-1/ Top2 17-3 larvae were examined for disrupted morphology and local unpairing. The incidence of abnormality, and total nuclei scored, is in parentheses. Chromosomes with a diffuse banding pattern and those bloated along the entire chromosome length were scored as abnormal. Nuclei with any visible unpairing of homologs was scored as positive for unpairing.
(PDF)
Acknowledgments
We thank Drs. A. Hohl, P. Geyer, C.T. Wu, J. Erickson, P. Ferree, J. Lucchessi, S. Elgin, G. Karpen, G. Bosco, M. Longworth, L.A. Lee, W. Hays and R. Kelley for generously providing fly strains. We are grateful to Dr. A. Hohl for sharing her PhD dissertation with us, and J. Butts for assistance in preparation of this manuscript. The Bloomington Drosophila Stock Center (NIH P40OD018537) provided strains used in this work.
Data Availability
All relevant data are within the paper and supporting information files.
Funding Statement
This work was supported by National Science Foundation 0641121 VHM and National Institutes of Health NIH 093110 VHM.
References
- 1. Smith CD, Shu S, Mungall CJ, Karpen GH. The Release 5.1 annotation of Drosophila melanogaster heterochromatin. Science. 2007;316(5831):1586–91. 10.1126/science.1139815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huisinga KL, Brower-Toland B, Elgin SC. The contradictory definitions of heterochromatin: transcription and silencing. Chromosoma. 2006;115(2):110–22. 10.1007/s00412-006-0052-x . [DOI] [PubMed] [Google Scholar]
- 3. Elgin SC, Reuter G. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harbor perspectives in biology. 2013;5(8):a017780 10.1101/cshperspect.a017780 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gatti M, Pimpinelli S. Functional elements in Drosophila melanogaster heterochromatin. Annual review of genetics. 1992;26:239–75. 10.1146/annurev.ge.26.120192.001323 . [DOI] [PubMed] [Google Scholar]
- 5. Yasuhara JC, Wakimoto BT. Oxymoron no more: the expanding world of heterochromatic genes. Trends in genetics: TIG. 2006;22(6):330–8. 10.1016/j.tig.2006.04.008 . [DOI] [PubMed] [Google Scholar]
- 6. Lu BY, Emtage PC, Duyf BJ, Hilliker AJ, Eissenberg JC. Heterochromatin protein 1 is required for the normal expression of two heterochromatin genes in Drosophila. Genetics. 2000;155(2):699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schulze SR, McAllister BF, Sinclair DA, Fitzpatrick KA, Marchetti M, Pimpinelli S, et al. Heterochromatic genes in Drosophila: a comparative analysis of two genes. Genetics. 2006;173(3):1433–45. 10.1534/genetics.106.056069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu LP, Ni JQ, Shi YD, Oakeley EJ, Sun FL. Sex-specific role of Drosophila melanogaster HP1 in regulating chromatin structure and gene transcription. Nat Genet. 2005;37(12):1361–6. 10.1038/ng1662 . [DOI] [PubMed] [Google Scholar]
- 9. Deng X, Koya SK, Kong Y, Meller VH. Coordinated regulation of heterochromatic genes in Drosophila melanogaster males. Genetics. 2009;182(2):481–91. 10.1534/genetics.109.102087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Meller VH, Rattner BP. The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. The EMBO journal. 2002;21(5):1084–91. 10.1093/emboj/21.5.1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ferree PM, Barbash DA. Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS biology. 2009;7(10):e1000234 10.1371/journal.pbio.1000234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Salz HK, Erickson JW. Sex determination in Drosophila: The view from the top. Fly. 2010;4(1):60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Boggs RT, Gregor P, Idriss S, Belote JM, McKeown M. Regulation of sexual differentiation in D. melanogaster via alternative splicing of RNA from the transformer gene. Cell. 1987;50(5):739–47. . [DOI] [PubMed] [Google Scholar]
- 14. Cline TW. The interaction between daughterless and sex-lethal in triploids: a lethal sex-transforming maternal effect linking sex determination and dosage compensation in Drosophila melanogaster. Developmental biology. 1983;95(2):260–74. . [DOI] [PubMed] [Google Scholar]
- 15. Salz HK, Cline TW, Schedl P. Functional changes associated with structural alterations induced by mobilization of a P element inserted in the Sex-lethal gene of Drosophila. Genetics. 1987;117(2):221–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bashaw GJ, Baker BS. The regulation of the Drosophila msl-2 gene reveals a function for Sex-lethal in translational control. Cell. 1997;89(5):789–98. . [DOI] [PubMed] [Google Scholar]
- 17. Gebauer F, Merendino L, Hentze MW, Valcarcel J. The Drosophila splicing regulator sex-lethal directly inhibits translation of male-specific-lethal 2 mRNA. Rna. 1998;4(2):142–50. [PMC free article] [PubMed] [Google Scholar]
- 18. Kelley RL, Wang J, Bell L, Kuroda MI. Sex lethal controls dosage compensation in Drosophila by a non-splicing mechanism. Nature. 1997;387(6629):195–9. 10.1038/387195a0 . [DOI] [PubMed] [Google Scholar]
- 19. Lemos B, Araripe LO, Hartl DL. Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science. 2008;319(5859):91–3. 10.1126/science.1148861 . [DOI] [PubMed] [Google Scholar]
- 20. Weiler KS, Wakimoto BT. Heterochromatin and gene expression in Drosophila. Annual review of genetics. 1995;29:577–605. 10.1146/annurev.ge.29.120195.003045 . [DOI] [PubMed] [Google Scholar]
- 21. Ilik IA, Quinn JJ, Georgiev P, Tavares-Cadete F, Maticzka D, Toscano S, et al. Tandem stem-loops in roX RNAs act together to mediate X chromosome dosage compensation in Drosophila. Mol Cell. 2013;51(2):156–73. 10.1016/j.molcel.2013.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maenner S, Muller M, Frohlich J, Langer D, Becker PB. ATP-dependent roX RNA remodeling by the helicase maleless enables specific association of MSL proteins. Mol Cell. 2013;51(2):174–84. 10.1016/j.molcel.2013.06.011 . [DOI] [PubMed] [Google Scholar]
- 23. Kelley RL, Solovyeva I, Lyman LM, Richman R, Solovyev V, Kuroda MI. Expression of msl-2 causes assembly of dosage compensation regulators on the X chromosomes and female lethality in Drosophila. Cell. 1995;81(6):867–77. . [DOI] [PubMed] [Google Scholar]
- 24. Bellen HJ, Levis RW, Liao G, He Y, Carlson JW, Tsang G, et al. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167(2):761–81. 10.1534/genetics.104.026427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Konev AY, Yan CM, Acevedo D, Kennedy C, Ward E, Lim A, et al. Genetics of P-element transposition into Drosophila melanogaster centric heterochromatin. Genetics. 2003;165(4):2039–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Menon DU, Meller VH. A role for siRNA in X-chromosome dosage compensation in Drosophila melanogaster. Genetics. 2012;191(3):1023–8. 10.1534/genetics.112.140236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bai X, Alekseyenko AA, Kuroda MI. Sequence-specific targeting of MSL complex regulates transcription of the roX RNA genes. EMBO J. 2004;23(14):2853–61. 10.1038/sj.emboj.7600299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rattner BP, Meller VH. Drosophila male-specific lethal 2 protein controls sex-specific expression of the roX genes. Genetics. 2004;166(4):1825–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meller VH, Wu KH, Roman G, Kuroda MI, Davis RL. roX1 RNA paints the X chromosome of male Drosophila and is regulated by the dosage compensation system. Cell. 1997;88(4):445–57. . [DOI] [PubMed] [Google Scholar]
- 30. Arnold AP. The end of gonad-centric sex determination in mammals. Trends in genetics: TIG. 2012;28(2):55–61. 10.1016/j.tig.2011.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Erickson JW, Cline TW. Key aspects of the primary sex determination mechanism are conserved across the genus Drosophila. Development. 1998;125(16):3259–68. . [DOI] [PubMed] [Google Scholar]
- 32. Erickson JW, Cline TW. A bZIP protein, sisterless-a, collaborates with bHLH transcription factors early in Drosophila development to determine sex. Genes Dev. 1993;7(9):1688–702. . [DOI] [PubMed] [Google Scholar]
- 33. Van Doren M, Ellis HM, Posakony JW. The Drosophila extramacrochaetae protein antagonizes sequence-specific DNA binding by daughterless/achaete-scute protein complexes. Development. 1991;113(1):245–55. . [DOI] [PubMed] [Google Scholar]
- 34. Younger-Shepherd S, Vaessin H, Bier E, Jan LY, Jan YN. deadpan, an essential pan-neural gene encoding an HLH protein, acts as a denominator in Drosophila sex determination. Cell. 1992;70(6):911–22. . [DOI] [PubMed] [Google Scholar]
- 35. Sefton L, Timmer JR, Zhang Y, Beranger F, Cline TW. An extracellular activator of the Drosophila JAK/STAT pathway is a sex-determination signal element. Nature. 2000;405(6789):970–3. 10.1038/35016119 . [DOI] [PubMed] [Google Scholar]
- 36. Cline TW. Evidence that sisterless-a and sisterless-b are two of several discrete "numerator elements" of the X/A sex determination signal in Drosophila that switch Sxl between two alternative stable expression states. Genetics. 1988;119(4):829–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Apte MS, Meller VH. Homologue pairing in flies and mammals: gene regulation when two are involved. Genetics research international. 2012;2012:430587 10.1155/2012/430587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stevens N. A study of the germ cells of certain Diptera with reference to the heterochromosomes and the phenomena of synapsis. J Exp Zool. 1908;5:359–74. [Google Scholar]
- 39. Williams BR, Bateman JR, Novikov ND, Wu CT. Disruption of topoisomerase II perturbs pairing in drosophila cell culture. Genetics. 2007;177(1):31–46. 10.1534/genetics.107.076356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Joyce EF, Williams BR, Xie T, Wu CT. Identification of genes that promote or antagonize somatic homolog pairing using a high-throughput FISH-based screen. PLoS genetics. 2012;8(5):e1002667 10.1371/journal.pgen.1002667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hohl AM, Thompson M, Soshnev AA, Wu J, Morris J, Hsieh TS, et al. Restoration of topoisomerase 2 function by complementation of defective monomers in Drosophila. Genetics. 2012;192(3):843–56. 10.1534/genetics.112.144006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hughes SE, Hawley RS. Topoisomerase II Is Required for the Proper Separation of Heterochromatic Regions during Drosophila melanogaster Female Meiosis. PLoS genetics. 2014;10(10):e1004650 10.1371/journal.pgen.1004650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ramos E, Torre EA, Bushey AM, Gurudatta BV, Corces VG. DNA topoisomerase II modulates insulator function in Drosophila. PloS one. 2011;6(1):e16562 10.1371/journal.pone.0016562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hohl AM. Understanding the role of Topoisomerase 2 in chromosome associations. PhD (Doctor of Philosophy) Thesis. [PhD Thesis]: University of Iowa; 2012.
- 45. Cugusi S, Ramos E, Ling H, Yokoyama R, Luk KM, Lucchesi JC. Topoisomerase II plays a role in dosage compensation in Drosophila. Transcription. 2013;4(5):238–50. . [DOI] [PubMed] [Google Scholar]
- 46. Lohe AR, Hilliker AJ, Roberts PA. Mapping simple repeated DNA sequences in heterochromatin of Drosophila melanogaster. Genetics. 1993;134(4):1149–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kas E, Laemmli UK. In vivo topoisomerase II cleavage of the Drosophila histone and satellite III repeats: DNA sequence and structural characteristics. The EMBO journal. 1992;11(2):705–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sawamura K, Yamamoto MT. Cytogenetical localization of Zygotic hybrid rescue (Zhr), a Drosophila melanogaster gene that rescues interspecific hybrids from embryonic lethality. Molecular & general genetics: MGG. 1993;239(3):441–9. . [DOI] [PubMed] [Google Scholar]
- 49. Sawamura K, Yamamoto MT, Watanabe TK. Hybrid lethal systems in the Drosophila melanogaster species complex. II. The Zygotic hybrid rescue (Zhr) gene of D. melanogaster. Genetics. 1993;133(2):307–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wijchers PJ, Yandim C, Panousopoulou E, Ahmad M, Harker N, Saveliev A, et al. Sexual dimorphism in mammalian autosomal gene regulation is determined not only by Sry but by sex chromosome complement as well. Developmental cell. 2010;19(3):477–84. 10.1016/j.devcel.2010.08.005 . [DOI] [PubMed] [Google Scholar]
- 51. Meller VH. Initiation of dosage compensation in Drosophila embryos depends on expression of the roX RNAs. Mechanisms of development. 2003;120(7):759–67. . [DOI] [PubMed] [Google Scholar]
- 52. Tsai SC, Valkov N, Yang WM, Gump J, Sullivan D, Seto E. Histone deacetylase interacts directly with DNA topoisomerase II. Nat Genet. 2000;26(3):349–53. 10.1038/81671 . [DOI] [PubMed] [Google Scholar]
- 53. Lupo R, Breiling A, Bianchi ME, Orlando V. Drosophila chromosome condensation proteins Topoisomerase II and Barren colocalize with Polycomb and maintain Fab-7 PRE silencing. Mol Cell. 2001;7(1):127–36. . [DOI] [PubMed] [Google Scholar]
- 54. Wang CI, Alekseyenko AA, LeRoy G, Elia AE, Gorchakov AA, Britton LM, et al. Chromatin proteins captured by ChIP-mass spectrometry are linked to dosage compensation in Drosophila. Nature structural & molecular biology. 2013;20(2):202–9. 10.1038/nsmb.2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhou K, Choe KT, Zaidi Z, Wang Q, Mathews MB, Lee CG. RNA helicase A interacts with dsDNA and topoisomerase IIalpha. Nucleic Acids Res. 2003;31(9):2253–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Russell B, Bhattacharyya S, Keirsey J, Sandy A, Grierson P, Perchiniak E, et al. Chromosome breakage is regulated by the interaction of the BLM helicase and topoisomerase IIalpha. Cancer Res. 2011;71(2):561–71. 10.1158/0008-5472.CAN-10-1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Watt PM, Louis EJ, Borts RH, Hickson ID. Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell. 1995;81(2):253–60. . [DOI] [PubMed] [Google Scholar]
- 58. Meller VH, McConnell M, Fisher PA. An RNase-sensitive particle containing Drosophila melanogaster DNA topoisomerase II. The Journal of cell biology. 1994;126(6):1331–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Enukashvily N, Donev R, Sheer D, Podgornaya O. Satellite DNA binding and cellular localisation of RNA helicase P68. Journal of cell science. 2005;118(Pt 3):611–22. 10.1242/jcs.01605 . [DOI] [PubMed] [Google Scholar]
- 60. Ito H, Sato K, Koganezawa M, Ote M, Matsumoto K, Hama C, et al. Fruitless recruits two antagonistic chromatin factors to establish single-neuron sexual dimorphism. Cell. 2012;149(6):1327–38. 10.1016/j.cell.2012.04.025 . [DOI] [PubMed] [Google Scholar]
- 61. Sanders LE, Arbeitman MN. Doublesex establishes sexual dimorphism in the Drosophila central nervous system in an isoform-dependent manner by directing cell number. Developmental biology. 2008;320(2):378–90. 10.1016/j.ydbio.2008.05.543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hoxha V, Lama C, Chang PL, Saurabh S, Patel N, Olate N, et al. Sex-specific signaling in the blood-brain barrier is required for male courtship in Drosophila. PLoS genetics. 2013;9(1):e1003217 10.1371/journal.pgen.1003217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fagegaltier D, Konig A, Gordon A, Lai EC, Gingeras TR, Hannon GJ, et al. A Genome-Wide Survey of Sexually Dimorphic Expression of Drosophila miRNAs Identifies the Steroid Hormone-Induced miRNA let-7 as a Regulator of Sexual Identity. Genetics. 2014. 10.1534/genetics.114.169268 . [DOI] [PMC free article] [PubMed]
- 64. Robinett CC, Vaughan AG, Knapp JM, Baker BS. Sex and the single cell. II. There is a time and place for sex. PLoS biology. 2010;8(5):e1000365 10.1371/journal.pbio.1000365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lindsley DL, Zimm GG. The Genome of Drosophila melanogaster. San Diego, California.: Academic Press,; 1992. [Google Scholar]
- 66. Deng X, Rattner BP, Souter S, Meller VH. The severity of roX1 mutations is predicted by MSL localization on the X chromosome. Mechanisms of development. 2005;122(10):1094–105. 10.1016/j.mod.2005.06.004 . [DOI] [PubMed] [Google Scholar]
- 67. Yan CM, Dobie KW, Le HD, Konev AY, Karpen GH. Efficient recovery of centric heterochromatin P-element insertions in Drosophila melanogaster. Genetics. 2002;161(1):217–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sun FL, Cuaycong MH, Craig CA, Wallrath LL, Locke J, Elgin SC. The fourth chromosome of Drosophila melanogaster: interspersed euchromatic and heterochromatic domains. Proc Natl Acad Sci U S A. 2000;97(10):5340–5. 10.1073/pnas.090530797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Longworth MS, Herr A, Ji JY, Dyson NJ. RBF1 promotes chromatin condensation through a conserved interaction with the Condensin II protein dCAP-D3. Genes Dev. 2008;22(8):1011–24. 10.1101/gad.1631508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hartl TA, Smith HF, Bosco G. Chromosome alignment and transvection are antagonized by condensin II. Science. 2008;322(5906):1384–7. 10.1126/science.1164216 . [DOI] [PubMed] [Google Scholar]
- 71. Rickmyre JL, Dasgupta S, Ooi DL, Keel J, Lee E, Kirschner MW, et al. The Drosophila homolog of MCPH1, a human microcephaly gene, is required for genomic stability in the early embryo. Journal of cell science. 2007;120(Pt 20):3565–77. 10.1242/jcs.016626 . [DOI] [PubMed] [Google Scholar]
- 72. Gepner J, Li M, Ludmann S, Kortas C, Boylan K, Iyadurai SJ, et al. Cytoplasmic dynein function is essential in Drosophila melanogaster. Genetics. 1996;142(3):865–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Morra R, Smith ER, Yokoyama R, Lucchesi JC. The MLE subunit of the Drosophila MSL complex uses its ATPase activity for dosage compensation and its helicase activity for targeting. Molecular and cellular biology. 2008;28(3):958–66. 10.1128/MCB.00995-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chang KA, Kuroda MI. Modulation of MSL1 abundance in female Drosophila contributes to the sex specificity of dosage compensation. Genetics. 1998;150(2):699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gonzalez AN, Lu H, Erickson JW. A shared enhancer controls a temporal switch between promoters during Drosophila primary sex determination. Proc Natl Acad Sci U S A. 2008;105(47):18436–41. 10.1073/pnas.0805993105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Geyer PK, Green MM, Corces VG. Tissue-specific transcriptional enhancers may act in trans on the gene located in the homologous chromosome: the molecular basis of transvection in Drosophila. The EMBO journal. 1990;9(7):2247–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Morris JR, Chen JL, Geyer PK, Wu CT. Two modes of transvection: enhancer action in trans and bypass of a chromatin insulator in cis. Proc Natl Acad Sci U S A. 1998;95(18):10740–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Morris JR, Chen J, Filandrinos ST, Dunn RC, Fisk R, Geyer PK, et al. An analysis of transvection at the yellow locus of Drosophila melanogaster. Genetics. 1999;151(2):633–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Dej KJ, Ahn C, Orr-Weaver TL. Mutations in the Drosophila condensin subunit dCAP-G: defining the role of condensin for chromosome condensation in mitosis and gene expression in interphase. Genetics. 2004;168(2):895–906. 10.1534/genetics.104.030908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cobbe N, Savvidou E, Heck MM. Diverse mitotic and interphase functions of condensins in Drosophila. Genetics. 2006;172(2):991–1008. 10.1534/genetics.105.050567 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PEV of y + in KV24 (3rd chromosome) is visible as black abdominal spots in both sexes and is suppressed in roX males (top), but not in roX females (bottom). PEV of w +mW.hs in 118E-10 (4th chromosome) is detected by eye pigmentation. roX males (top), but not females (bottom), suppress 118E-10 PEV. 118E-10 was examined in the yw roX1 ex33 Df(1)52;[4Δ4.3]/+ background, which is mutated for roX1 and roX2 and lacks other w markers, enabling visualization of the w +mW.hs reporter.
(PDF)
Heterochromatic sex was determined in flies mutated for anti-pairers (Cap-H2, Cap-D3 and fs(1)h) and pairing promoters (MCPH1 and Dhc64c). All flies carried the y + KV20 reporter. Flies mutated for each pairing regulator were generated in wild type (++) and yw roX1 ex33 roX2Δ mutant backgrounds. Almost no abdominal pigmentation was observed in XX flies wild type (white) or mutated (hatched) for both roX genes. In contrast, PEV in XY flies (black) is suppressed in roX mutants (dark gray). A slight enhancement of PEV is detected in Cap-D3 mutant flies, consistent with previous reports of condensin mutations as PEV enhancers [79, 80]. Fifteen-50 flies were counted for each genotype.
(PDF)
A) The Top2 mutations disrupt different domains. Missense mutations Top2 17-1 (WHD domain) and Top2 17-3 (TOPRIM domain). B) Top2 17-1 /Top2 17-3 males are fertile but Top2 17-1 /Top2 17-3 females are sterile. Both mutations are homozygous lethal. C) Characteristic abnormalities in a polytene preparation from a Top2 17-1 /Top2 17-3 male larvae. A puffy X chromosome (black arrow) and homolog unpairing (white arrows) are visible. One hundred-250 nuclei from at least 5 larvae were scored for each genotype. D) Transvection restores yellow expression. y 82f29 is a deletion of upstream enhancer elements. y 1#8 retains enhancers but lacks a promoter. y 3c3 lacks a bristle enhancer and the promoter, but retains a wing enhancer. Pairing between y 82f29 and y 1#8 or y 3c3 enables enhancers on the homolog to drive the y 82f29 promoter, restoring expression. Drawing based on [77]. Wing and body pigmentation was ranked from 1 (no pigmentation) to 4 (wild type). Flies homozygous for each allele have light body and wing color (1,1). Transvection in y 82f29 /y 1#8 flies restores wing and body color near wild-type levels (3, 3). Transvection in y 82f29 /y 3c3 flies restores wing pigmentation only (3, 1). Transvection is not disrupted in Top2 17-1 /Top2 17-3 mutants (shaded). Flies were aged 1–2 days before scoring and photography. At least 100 flies were scored for each genotype. E) Representative abdomens showing y transvection. Full genotypes are: y 82f29 /y 1#8; Top2 m / Cyo, y 82f29 /y 1#8; Top2 17-1 /Top2 17-3. F) Top2 mutations do not disrupt Gypsy insulation. Loss of pigmentation in y 2 requires the Top2-dependent Gypsy insulator. Loss of insulation enhances body pigmentation. Full genotypes are: y 2/Y; +/+, y 2/Y; Top2 m /CyO and y 2/Y; Top2 17-1 /Top2 17-3. At least 25 flies of each genotype were aged for 24 h before scoring.
(PDF)
Polytene preparations from control (+/+, reference yw strain) and yw; Top2 17-1/ Top2 17-3 larvae were examined for disrupted morphology and local unpairing. The incidence of abnormality, and total nuclei scored, is in parentheses. Chromosomes with a diffuse banding pattern and those bloated along the entire chromosome length were scored as abnormal. Nuclei with any visible unpairing of homologs was scored as positive for unpairing.
(PDF)
Data Availability Statement
All relevant data are within the paper and supporting information files.