Abstract
We present a new statistical test of association between a trait and genetic markers, which we theoretically and practically prove to be robust to arbitrarily complex population structure. The statistical test involves a set of parameters that can be directly estimated from large-scale genotyping data, such as that measured in genome-wide association studies (GWAS). We also derive a new set of methodologies, called a genotype-conditional association test (GCAT), shown to provide accurate association tests in populations with complex structures, manifested in both the genetic and environmental contributions to the trait. We demonstrate the proposed method on a large simulation study and on the Northern Finland Birth Cohort study. In the Finland study, we identify several new significant loci that other methods do not detect. Our proposed framework provides a substantially different approach to the problem from existing methods, such as the linear mixed model and principal component approaches.
INTRODUCTION
Performing genome-wide tests of association between a trait and genetic markers is one of the most important research efforts in modern genetics [1–3]. However, a major problem to overcome is how to test for associations in the presence of population structure [4]. Human populations are often structured in the sense that the genotype frequencies at a particular locus are not homogeneous throughout the population. Rather, there is heterogeneity in the genotype frequencies among individuals (correlated with variables such as geography or ancestry). At the same time, there may be other loci and non-genetic factors that also correlate with this genotype frequency heterogeneity, which in turn are correlated with the trait of interest. When this occurs, genetic markers become spuriously statistically associated with the trait of interest despite the fact that there is no biological connection.
The importance of addressing association testing in structured populations is evidenced by the existence of a large literature of methods proposed for this problem [5, 6]. The well-established methods all take a similar strategy in that the trait is modeled in terms of the genetic markers of interest, while attempting to adjust for genetic structure. Two popular approaches are to correct population structure by including principal components of genotypes as adjustment variables [7, 8] or by fitting a linear mixed effects model involving an estimated kinship or covariance matrix from the individuals’ genotypes [9, 10]. Previous work investigating the limitations of these two methods includes Wang, et al. (2013) [11]. These two approaches have been shown to be based on a common model that make differing assumptions about how the kinship or covariance matrices are utilized in the model [5]. This common model does not allow for non-genetic (e.g., environmental) contributions to the trait to be dependent with population structure. The linear mixed effects model requires that the genetic component is composed of small effects that additively are well-approximated by the Normal distribution. The model itself is therefore an approximation, and it is not yet possible to theoretically prove that a test based on this model is robust to structure for the more general class of relevant models that we investigate.
By taking a substantially different approach that essentially reverses the placement of the trait and genotype in the model, we formulate and provide a theoretical solution to the problem of association testing in structured populations for both quantitative and binary traits under general assumptions about the complexity of the population structure and its relationship to the trait through both genetic and non-genetic factors. This theoretical solution directly leads to a method for addressing the problem in practice that differs in key ways from the mixed model and principal component approaches. The method is straightforward: a model of structure is first estimated from the genotypes, and then a logistic regression is performed where the SNP genotypes are logistically regressed on the trait plus an adjustment based on the fitted structure model. The coefficient corresponding to the trait is then tested for statistical significance.
This association-testing framework is robust to general forms of population genetic structure, as well as to non-genetic effects that are dependent or correlated with population genetic structure (for example, lifestyle and environment may be correlated with ancestry) and with heteroskedasticity that is dependent on structure. We introduce an implementation of this test, called “genotype conditional association test” (GCAT). We show the proposed method corrects for structure on simulated data with a quantitative trait and compares favorably to existing methods. We also apply the method to the Northern Finland Birth Cohort data [12] and identify several new associated loci that have not been identified by existing methods. For example, the proposed method is the only one to identify a SNP (rs2814982) associated with height, which we note is linked to another SNP (rs2814993) that has been associated with skeletal frame size [13]. We discuss the advantages and disadvantages of the proposed framework with existing approaches, and we conclude that the proposed framework will be useful in future studies as sample sizes and the complexity of structure increase.
RESULTS
Simulation Studies
We performed an extensive set of simulations to demonstrate that the proposed test is robust to population structure and to assess its power to detect true associations (full technical details in Online Methods and Supplementary Note). We compared the proposed test to its Oracle version (where model (3) from Online Methods and test-statistic (6) from Supplementary Note are used with the true individual-specific allele frequency values imputed). We also included in the simulation studies three important and popular methods: (i) the method of adjusting the trait and genotypes by principal components computed from the full set of genotypes [8] and (ii) two implementations of the linear mixed effects model (LMM) approach [9, 10], specifically EMMAX by Kang et al. (2010) [10] and GEMMA by Zhou and Stephens (2012) [15]. The methods are abbreviated as “PCA,” “LMM-EMMAX,” and “LMM-GEMMA.”
For each of 33 simulation configurations, we simulated and analyzed 100 GWAS data sets from a quantitative trait model (equation (1) from Online Methods), for a grand total of 3300 simulated data sets. Each simulation scenario involved m = 100,000 simulated SNPs on n individuals, where n ranged from 940 to 5000 depending on the scenario. For a given simulated study, we therefore obtained a set of 100,000 p-values, one per SNP. So-called “spurious associations” occur when the p-values corresponding to null (non-associated) SNPs are artificially small. For a given p-value threshold t, we expect there to be m0 × t false positives among the m0 p-values corresponding to null SNPs, where m0 = 100,000–10 in our case. At the same time, we can calculate the observed number of false positive simply by counting how many of the null SNP p-values are less than or equal to t. The excess observed false positives are spurious associations. A method properly accounts for structure when the average difference is zero. The best one can do on a study-by-study basis is captured by the Oracle method, which according to our theory is immune to structure and provides the correct null distribution.
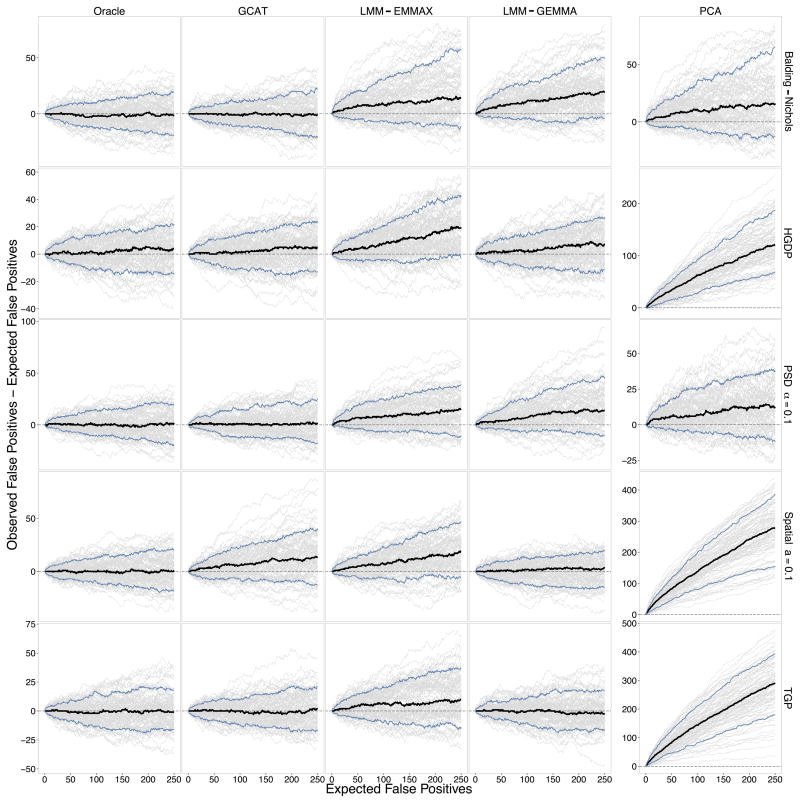
Fig. 2 shows the excess in observed false positives vs. the expected number of false positives for the Oracle, GCAT (proposed), PCA, and both implementations of LMM under five configurations of structure for a quantitative trait variation apportionment corresponding to genetic=5%, non-genetic=5%, and noise=90%. It can be seen that the proposed GCAT method performs similarly to the Oracle test, whereas PCA tends to suffer from an excess of spurious associations.
Figure 2.
Performance of association testing methods. One-hundred quantitative trait GWAS studies were simulated in each of the Balding-Nichols, HGDP, TGP, PSD (α =0.1), and Spatial (a =0.1) simulation scenarios (see Online Methods for definitions of each) to compare the Oracle, GCAT (proposed), LMM-EMMAX, LMM-GEMMA, and PCA testing methods. The variance contributions to the trait are genetic=5%, non-genetic=5%, and noise=90%. The difference between the observed number of false positives and expected number of false positives is plotted against the expected number of false positives under the null hypothesis of no association for each simulated study (grey lines), the average of those differences (black line), and the middle 90% (blue lines). All simulations involved m =100,000 SNPs, so the range of the x-axis corresponds to choosing a significance threshold of up to p-value ≤ 0.0025. The difference on the y-axis is the number of “spurious associations.” PCA is shown on a separate y-axis since it usually has a much larger maximum than the other methods. The Oracle method is where the true population structure parameters are inputted into the proposed test (see Results), which we have theoretically proven always corrects for structure (see Supplementary Note).
We found from using the distributed binary executable EMMAX software and our own implementation that EMMAX required a 10-fold increase in computational time over the proposed method and PCA when analyzing n = 5000 individuals. Therefore, it was not reasonable to apply EMMAX to all 3300 simulated GWAS data sets. We limited comparisons with EMMAX to five representative structure configurations. GEMMA was computationally more efficient, though still significantly slower than GCAT or our implementation of PCA.
Supplementary Figs. 1–8 show results from the remaining set of simulations from all 33 simulation configurations. Due to the computational constraints mentioned above for EMMAX, the additional simulations feature only results from GEMMA for LMM methods.
In comparing the statistical power among the methods (Supplementary Figs. 9–17), we found that the Oracle, GCAT, and PCA performed similarly well, while the two LMM methods sometimes showed a loss or gain in power depending on the scenario. We also carried out analogous simulations on binary traits simulated (from trait model equation (2) in Online Methods) and we found that all methods performed similarly well in terms of producing correct p-values that were robust to structure. This result agrees with the comparisons made between PCA and a linear mixed effects model in Astle and Balding (2009) [5].
Analysis of the Northern Finland Birth Cohort Data
We applied the proposed method to the Northern Finland Birth Cohort (NFBC) genome-wide association study data [12], which includes several metabolic traits and height (Supplementary Figure 18). This study has also been analyzed by the LMM and PCA methods, as well as a standard analysis uncorrected for structure [10]. We carried out association analyses with the proposed method on the 10 traits that were also analyzed using the other methods (Table 1). After processing the data, including filtering for missing data, minor allele frequencies, and departures from Hardy-Weinberg equilibrium, the data were composed of m = 324,160 SNPs and n = 5027 individuals (Supplementary Note). The LFA model of population structure was estimated from a subset of the data where markers were at least 200 kbp apart.
Table 1.
Number of significant loci at genome-wide significance (p-value < 7.2×10−8) for each of the 10 traits from the Northern Finland Birth Cohort data. Each method was performed with a subsequent genomic control inflation factor correction applied (denoted by +GC). The counts for LMM+GC, PCA+GC, and Uncorr+GC were obtained from Table 2 in Kang et al. (2010). In this case LMM is EMMAX-LMM.
| Trait | Abbreviation | GCAT+GC | LMM+GC | PCA+GC | Uncorr+GC |
|---|---|---|---|---|---|
| Body Mass Index | BMI | 0 | 0 | 0 | 0 |
| C-reactive Protein | CRP | 2† | 2 | 2 | 2 |
| Diastolic blood pressure | DBP | 0 | 0 | 0 | 0 |
| Glucose | GLU | 3 | 2 | 2 | 2 |
| HDL Cholesterol | HDL | 4 | 4 | 2 | 4 |
| Height | Height | 1 | 0 | 0 | 0 |
| Insulin | INS | 0 | 0 | 0 | 0 |
| LDL Cholesterol | LDL | 4 | 3 | 3 | 3 |
| Systolic blood pressure | SBP | 0 | 0 | 0 | 0 |
| Triglycerides | TG | 2 | 3 | 2 | 2 |
|
| |||||
| Total | 16 | 14 | 11 | 13 | |
Result when the Box-Cox transformation was not applied to the CRP trait. The result is 1 when the transformation is applied.
Most traits showed only approximate Normal distributions, so we applied a Box-Cox Normal transformation to all traits so that they satisfy the model assumptions. We noted that C-reactive Protein (CRP) and Triglycerides (TG) traits followed an exponential distribution more closely, so it was unnecessary to transform these two traits. The developed theory can be extended to exponential distributed quantitative traits as well.
The 20 most significant SNPs for each of the 10 traits are shown in Supplementary Table 1. Kang et al. (2010) utilized a genome-wide significance threshold of p-value < 7.2×10−8 as proposed in ref. [16], so we also utilized this threshold for comparative purposes. The numbers of loci found to be significant for each method are shown in Table 1. Whereas our proposed method identifies 16 significant loci, the other methods identify 11 to 14 loci.
We identified three new loci that were not identified by the other methods. None of the other methods identified any significant associations for the height trait. However, we identified rs2814982 on chromosome 6 as being statistically associated with height (Supplementary Table 1). This SNP is located ~70kbp from another SNP, rs2814993, which has been associated with skeletal frame size in a previous study [13]. Additionally, rs2814993 was the fifth most significant SNP for height. For the LDL cholesterol trait, we identified a significant association with rs11668477, which was significantly associated with LDL cholesterol in a different study [17]. Finally, there were significant associations between the glucose (GLU) trait and a cluster of SNPs (rs3847554, rs1387153, rs1447352, rs7121092) proximal to the MTNR1B locus; variation at this locus has been associated with glucose in a previous study [18].
As described in Sabatti et al. (2009) [12], the NFBC data show modest levels of inflation due to population structure as measured by the genomic control inflation factor (GCIF) [19] of test statistics from an uncorrected analysis. The population structure present among these individuals may be subtler and manifested on a finer scale than other settings. Noting that the GCAT approach does not attempt to adjust for a polygenic background, the GCIF values calculated for the proposed method (Supplementary Table 2) were found to be in line with what is expected for polygenic traits where no structure is present [20], providing evidence that the proposed method adequately accounts for structure.
DISCUSSION
We considered models of quantitative and binary traits involving genetic effects and non-genetic effects in the presence of arbitrarily complex population structure. We allowed for the non-genetic effects to be confounded with population genetic structure since structure, ancestry, geography, lifestyle, and environment – all factors potentially involved in complex traits – may be highly dependent with one another. A mathematical argument showed that under these models, it is most reasonable to account for this confounding in the genotypes, but it is not tractable to do so in the non-genetic effects. This follows because we have many instances of genotypes that can be jointly modeled to provide reliable estimates of structure, but the non-genetic effects are never directly observed and we do not have repeated instances of them. In general it is not possible to estimate a latent variable that accounts for the confounding between structure and non-genetic effects.
These observations led us to propose an inverse regression approach to testing for associations, where the association is tested by modeling genotype variation in terms of the trait plus model terms accounting for structure. In this model, the terms accounting for structure were based on the logistic factor analysis (LFA) approach that we have proposed [14], although the general form of the association test can incorporate other methods that estimate population structure. We mathematically proved under general assumptions that the trait term in the model is non-zero only when the genetic marker is truly associated with the trait, regardless of the population structure. We demonstrated that the implemented test properly accounts for structure in a large body of simulated studies that included a wide range of population structures. We also applied the method to 10 traits from the Northern Finland Birth Cohort genome-wide association study. The proposed method identified three new loci associated with the traits, including being the only method among those we considered that identifies a locus associated with the height trait. Overall, we showed that the proposed method compares favorably to existing methods and we also noted that it has favorable computational requirements compared to existing methods.
As GWAS increase in sample size and levels of complexity of population structure, it is important to develop methods that properly account for structure and that scale well with sample size. Whereas we found that the popular principal components adjustment does not properly account for structure, we also found that the mixed model approach performs reasonably well. However, the mixed model approach involves estimating a n × n kinship matrix (where n is the number of individuals in the study) and its current implementation does not scale well with sample size. The kinship matrix quickly becomes computationally unwieldy when n grows large, and the possibility of the estimated kinship matrix becoming overwhelmed by noise is a concern [21]. In the Northern Finland Birth Cohort data, the mixed model approach required us to estimate 12 million parameters, whereas the proposed method involved estimating 25-thousand parameters, a ~500-fold decrease. A study involving n = 10,000 individuals with the same complexity of structure requires estimating about 50-million parameters in the mixed model kinship matrix, whereas the proposed method requires estimating 50-thousand parameters, a ~1000-fold decrease. In addition, estimating the structure in the proposed method primarily uses singular value decomposition, for which a rich literature of computational techniques exists. We utilized a Lanczos bidiagonalization algorithm [22], which scales approximately linearly with respect to n for d ≪ n, where is d the number of latent variables used in the LFA model of population structure (Online Methods). The proposed method is well equipped to scale to massive GWAS and can take advantage of future advances for computing singular value decomposition.
The key assumption to verify in utilizing the proposed GCAT approach is that population structure observed in the SNP genotypes is adequately modeled and estimated. One can test for associations among SNPs that show convincing empirical evidence that the model of structure is reasonably well-behaved; this can be directly tested on the genotype data as previously demonstrated in our logistic factor analysis (LFA) model of structure [14]. For example, on the Northern Finland Birth Cohort Study, we empirically verified that utilizing the LFA model with dimension d = 6 accounted for structure reasonably well for the great majority of SNPs. The linear mixed effects model (LMM) approach and principal components (PCA) approach make trait model assumptions that may be difficult to verify in practice (Online Methods and Supplementary Note). In cases where the probabilistic model that we assume is not validated on the data, a different model should be utilized. For example, our probabilistic model does not account for closely related individuals.
We anticipate that the proposed genotype conditional association test (GCAT) will be useful for future studies. The framework we have developed should facilitate its extension to traits modeled according to distributions not considered here while maintaining our theoretical proof that the test accounts for population structure in the presence of non-genetic effects also confounded with structure.
ONLINE METHODS
Population Structure Model
Suppose that there are n individuals, each with m measured SNP genotypes. The genotype for SNP i in individual j is denoted by xij ∈ {0,1,2}, i = 1,2, …, m, j = 1,2, …, n. We collected these SNP genotypes into an m × n matrix X, where the (i, j) entry is xij. We denote the genotypes for individual j by xj = (x1j, x2j, …, xmj)T.
We utilize our recently developed framework that flexibly models complex population structures for diallelic loci [14]. As described in Results, z is an unobserved latent variable that is assumed to capture heterogeneity in allele frequencies among individuals and can be interpreted as capturing the effect of population structure. (Note that, as described in Results, z also captures information on non-genetic contributions to the trait, such as those related to lifestyle and environment.) For a SNP i, the allele frequency πi can be viewed as being a function of z, πi(z). For a random sample of n individuals, we therefore have implicitly sampled unobserved z1, z2, …, zn with resulting allele frequencies πi(z1), πi(z2), …, πi(zn) for SNP i. In Hao et al. (2013) [14], we formulate and estimate a model for m SNPs simultaneously while providing a flexible parameterization of the form of πi(z).
For shorthand, πij ≡ πi(zj) is the allele frequency for SNP i conditioned on the ancestry state of individual j. The πij values may be called “individual-specific allele frequencies” [14]. These allele frequencies can be collected into an m × n matrix F, where the (i,j) entry is πij. Note that E[xij/2|zj] = πij, and when Hardy-Weinberg equilibrium holds, xij|zj ~ Binomial(2, πij). We utilize the framework from Hao et al. (2013) [14], called “logistic factor analysis” (LFA), that allows the simultaneous estimation of all πij from a given genotype data set X. Specifically, it provides estimates of latent variables that form a linear basis of the quantities, which turns out is the most convenient scale on which to estimate a model of structure for the proposed testing framework. It should be noted that other well-behaved estimates of πij may be utilized as well. Further details are provided in Supplementary Note.
Trait Models
We assume a trait (either quantitative or binary) has been measured on each individual, which we denote by yj, j = 1,2, …, n. We consider the following models of quantitative and binary traits. We write the trait models in terms of additive genetic effects, but the framework can be extended to account for dominance models and interactions, and the models can also incorporate adjustment variables that capture known sources of trait variation.
The quantitative trait model is
| (1) |
where βi is the genetic effect of SNP i on the trait, λj is the random non-genetic effect, and εj is the random noise variation. To allow the interdependence of structure, lifestyle, and environment, we assume that xj = (x1j, …, xm,j)T, λj, and may all be functions of zj. We assume that , which allows for heteroskedasticity of the random noise variation. The distribution of λj can remain unspecified, although we assume that λj and zj may be dependent random variables. The population genetic model summarized shows how the distribution of , depends on zj. Without having observed zj, it follows that , λj, and εj are dependent random variables; however, we assume that conditional on zj, these random variables are independent.
The binary trait model is
| (2) |
where again βi is the genetic effect of SNP i on the trait, λj is the non-genetic effect, and we allow for the case that xj and λj may be dependent due to the common confounding latent variable zj as described for the quantitative trait model.
We have shown that the linear mixed effects model and principal components approaches involve more restrictive assumptions about the trait models utilized in testing for associations (Supplementary Note).
Association Test Immune to Population Structure
We have derived a statistical hypothesis test of association that is equivalent to testing whether βi = 0 for each SNP i in the above trait models (1) and (2), and whose null distribution does not depend on structure or the non-genetic effects correlated with structure, making it immune to spurious associations due to structure (Supplementary Note). Specifically, the test allows for general levels of complexity in structure because the test is based on adjusting for structure according to individual-specific allele frequencies.
We have proved a theorem (Supplementary Note) that shows that βi = 0 in models (1) and (2) implies that bi = 0 in the following model:
| (3) |
for all j = 1,2, …, n. This establishes a model that can be used to test for associations in place of models (1) and (2). Note that the non-genetic effects, heteroskedasticity, and polygenic background do not appear in the above model used to test for associations. This is important because under our general assumptions, these terms can be difficult or even impossible to estimate in practice. Furthermore, testing for association under this model means that the test will have a valid null distribution regardless of the form of the non-genetic effects, heteroskedasticity, and polygenic background.
As fully detailed in Supplementary Note, an association statistic whose null distribution is known can be constructed by testing whether bi = 0 in the above model, which we have shown is a valid test if βi = 0 in traits models (1) and (2). Briefly, the testing procedure works as follows:
Formulate and estimate a model of population structure that provides well-behaved estimates of the logit(πij) values. We specifically use the logistic factor analysis (LFA) approach of ref. [14], which has been shown to provide a accurate linear basis of the logit(πij) values.
For each SNP i, perform a logistic regression of the SNP genotypes on the trait values plus the model terms that estimate the values. Also, perform a logistic regression of the SNP genotypes on only the model terms that estimate , where the trait is now excluded from the fit. These two model fits are compared via a likelihood ratio statistic, where the larger the statistic, the more evidence there is that bi ≠ 0.
Calculate a p-value for each SNP, which is done based on our result that when the null hypothesis of no association is true, βi = 0 in models (1) and (2), then the above statistic follows a distribution for large sample sizes.
In our implementation, d estimated logistic factors (from LFA [14]) are included as covariates, which serve as the model terms that estimate the values.
We call our proposed test the “genotype-conditional association test” (GCAT). As a general concept, such an approach is sometimes called an inverse regression model because the trait and genotype are reversed in the regression.
Simulated Data
The complete simulation study on quantitative traits involved population structure constructed in 11 different ways for each of three different apportionments of variance among genetic effects, non-genetic effects, and random variation that all contribute to variation in the trait. Therefore, each configuration involved a constructed allele frequency matrix F and values assigned to variances , and Var(εj) from model (1). For each of these 33 =11×3 configurations, we simulated 100 GWAS data sets, for a grand total of 3300 studies.
We simulated allele frequencies: (i) from the Balding-Nichols model [23] based on allele-frequency and FST estimates calculated on the HapMap data set (Balding-Nichols); (ii) subject to structure estimated from two real data sets: the Human Genome Diversity Project (HGDP) and the 1000 Genomes Project (TGP); (iii) at four different levels of admixture by varying the parameter α (defined in Supplementary Note) in the Pritchard-Stephens-Donnelly (PSD) model [24], which is an extension of the Balding-Nichols model; and (iv) for four different types of spatially defined structure (Spatial) by varying the parameter a (defined in Supplementary Note). We intentionally simulated challenging population structures, having in mind that future GWAS such as the forthcoming “Genotype Tissue Expression” program (GTEx) data may involve particularly challenging forms of structure.
In order to provide an extra challenge to the proposed test, we simulated the allele frequencies from a model that differs from the LFA model (equation 4 in Supplementary Note). We generated allele frequencies parameterized by F = ΓS, where F is the matrix of πij values, G is an m × d matrix and S is the d × n matrix that encapsulates the structure (with d = 3). This model captures as special cases the Balding-Nichols model and the PSD model [14]. It was also intended to provide an advantage to the PCA and LMM methods because the structure is manifested on the observed genotype scale [14], which is the same scale on which both methods estimate structure.
We simulated 10 truly associated SNPs whose effect sizes are distributed according to a Normal distribution. All genotypes were simulated to be in linkage equilibrium so that true and false positives are unambiguous. We set the variances , and Var(εj) to be: (5%, 5%, 90%), (10%, 0%, 90%), and (10%, 20%, 70%). Setting these variances enforced a certain overall level of genetic contribution to the trait; therefore our simulation study results were minimally affected by the choice of 10 truly associated SNPs and the Normal distribution on their effect sizes. In each simulation scenario, we simulated data for m = 100,000 SNPs and n = 5000 individuals, except HGDP necessarily restricted us to n = 940 individuals and TGP to n = 1500 individuals. The dimension of the structure was set to d = 3, although we carried out the same simulations for d = 6 and the results were quantitatively very similar and qualitatively equivalent.
Additional details on the simulations can be found in Supplementary Note.
Supplementary Material
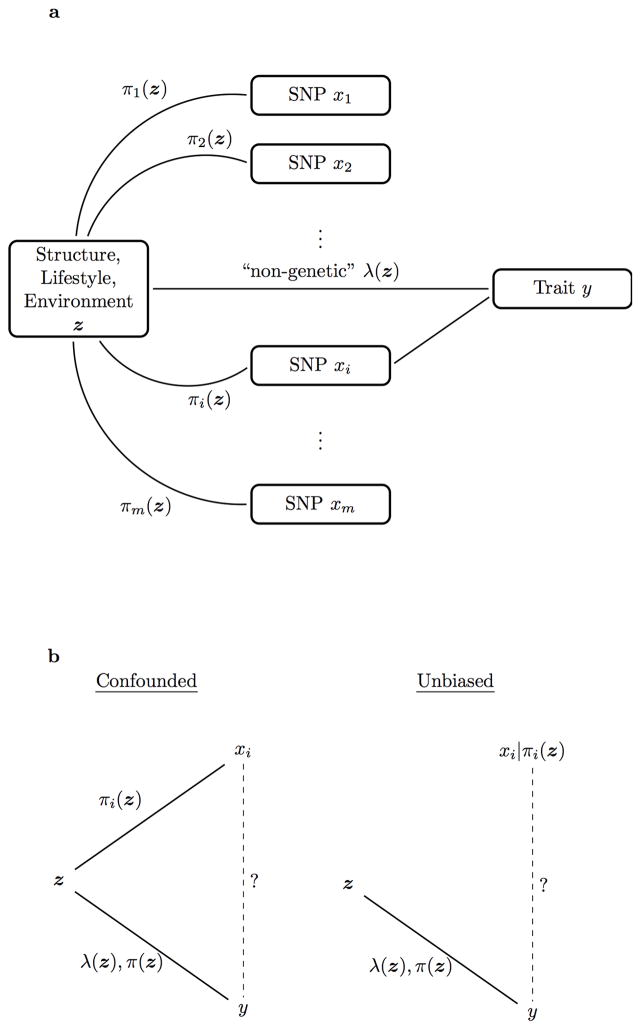
Figure 1.
Rationale for the proposed test of association. (a) A graphical model describing population structure and its effects on a trait of interest. Population structure is captured by a common latent variable z among a set of loci xi (i =1,2, …, m), via the allele frequencies πi(z). When one locus has a causal effect on the trait, this induces spurious associations with other loci affected by population structure. At the same time, population structure may be correlated with lifestyle and environment as these are all possibly related to ancestry and geography. (b) Accounting for confounding due to latent population structure. Left panel: A test for association between the ith SNP xi and trait y without taking into account z will produce a spurious association due to the fact that both xi and y are confounded with z. Right panel: A test for association between xi|πi(z) and y will be an unbiased because conditioning on πi(z) breaks the relationship between z and xi.
Acknowledgments
This research was supported in part by NIH grant R01 HG006448. The Northern Finland Birth Cohort data were collected by the STAMPEED: Cardiovascular Health Study (CHS) GWAS, made available through dbGaP Study Accession phs000226.v2.p1. A full list of contributors to the STAMPEED study can be found at its dbGaP web site.
Footnotes
URLs
The proposed method has been implemented in open source software, available at http://github.com/StoreyLab/gcat/.
Author Contributions
JDS designed the study and wrote the manuscript. MS, WH, and JDS developed theory and methods. MS and WH analyzed data. WH implemented software.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.McCarthy MI, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9(5):356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 2.Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10(4):241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 3.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pritchard JK, Rosenberg NA. Use of unlinked genetic markers to detect population stratification in association studies. Am J Hum Genet. 1999;65(1):220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Astle W, Balding DJ. Population structure and cryptic relatedness in genetic association studies. Stat Sci. 2009;24:451–471. [Google Scholar]
- 6.Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nat Rev Genet. 2010;11(7):459–463. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang S, Zhu X, Zhao H. On a semiparametric test to detect associations between quantitative traits and candidate genes using unrelated individuals. Genet Epi. 2003;24(1):44–56. doi: 10.1002/gepi.10196. [DOI] [PubMed] [Google Scholar]
- 8.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 9.Yu J, et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet. 2006;38(2):203–208. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- 10.Kang HM, et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42(4):348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang K, Hu X, Peng Y. An analytical comparison of the principal component method and the mixed effects model for association studies in the presence of cryptic relatedness and population stratification. Hum Hered. 2013;76(1):1–9. doi: 10.1159/000353345. [DOI] [PubMed] [Google Scholar]
- 12.Sabatti C, et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat Genet. 2009;41(1):35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soranzo N, et al. Meta-analysis of genome-wide scans for human adult stature identifies novel loci and associations with measures of skeletal frame size. PLoS Genet. 2009;5(4):e1000445. doi: 10.1371/journal.pgen.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hao W, Song M, Storey JD. Probabilistic models of genetic variation in structured populations applied to global human studies. 2013 doi: 10.1093/bioinformatics/btv641. arXiv:1312.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44(7):821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet Epi. 2008;32(3):227–234. doi: 10.1002/gepi.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandhu MS, et al. LDL-cholesterol concentrations: a genome-wide association study. Lancet. 2008;371(9611):483–491. doi: 10.1016/S0140-6736(08)60208-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prokopenko I, et al. Variants in MTNR1B influence fasting glucose levels. Nat Genet. 2009;41(1):77–81. doi: 10.1038/ng.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 20.Yang J, et al. Genomic inflation factors under polygenic inheritance. Euro J Hum Genet. 2011;19(7):807–812. doi: 10.1038/ejhg.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Witten DM, Tibshirani R, Hastie T. A penalized matrix decomposition, with applications to sparse principal components and canonical correlation analysis. Biostat. 2009;10(3):515–534. doi: 10.1093/biostatistics/kxp008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baglama J, Reichel L. Restarted block Lanczos bidiagonalization methods. Num Algo. 2006;43:251–272. [Google Scholar]
- 23.Balding DJ, Nichols RA. A method for quantifying differentiation between popu- lations at multi-allelic loci and its implications for investigating identity and paternity. Genetics. 1995;96:3–12. doi: 10.1007/BF01441146. [DOI] [PubMed] [Google Scholar]
- 24.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000 Jun;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.