Summary
Artificial microRNAs (amiRNAs) are used for selective gene silencing in plants. However, current methods to produce amiRNA constructs for silencing transcripts in monocot species are not suitable for simple, cost‐effective and large‐scale synthesis. Here, a series of expression vectors based on Oryza sativa MIR390 (OsMIR390) precursor was developed for high‐throughput cloning and high expression of amiRNAs in monocots. Four different amiRNA sequences designed to target specifically endogenous genes and expressed from OsMIR390‐based vectors were validated in transgenic Brachypodium distachyon plants. Surprisingly, amiRNAs accumulated to higher levels and were processed more accurately when expressed from chimeric OsMIR390‐based precursors that include distal stem–loop sequences from Arabidopsis thaliana MIR390a (AtMIR390a). In all cases, transgenic plants displayed the predicted phenotypes induced by target gene repression, and accumulated high levels of amiRNAs and low levels of the corresponding target transcripts. Genome‐wide transcriptome profiling combined with 5′‐RLM‐RACE analysis in transgenic plants confirmed that amiRNAs were highly specific.
Keywords: RNA silencing, artificial microRNA, MIRNA precursor, Brachypodium distachyon, monocot, Arabidopsis thaliana, technical advance
Significance Statement
A series of amiRNA vectors based on Oryza sativa MIR390 (OsMIR390) precursor were developed for simple, cost‐effective and large‐scale synthesis of amiRNA constructs to silence genes in monocots. Unexpectedly, amiRNAs produced from chimeric OsMIR390‐based precursors including Arabidopsis thaliana MIR390a distal stem‐loop sequences accumulated elevated levels of highly effective and specific amiRNAs in transgenic Brachypodium distachyon plants.
Introduction
MicroRNAs (miRNAs) are a class of ≈21 nt long endogenous small RNAs that posttranscriptionally regulate gene expression in eukaryotes (Bartel, 2004). In plants, DICER‐LIKE1 processes MIRNA precursors with imperfect self‐complementary foldback structures into miRNA/miRNA* duplexes (Bologna and Voinnet, 2014). Typically, one strand of the miRNA duplex is sorted into an ARGONAUTE (AGO) protein according to the identity of the 5′‐terminal nucleotide (nt) of the miRNA (Mi et al., 2008; Montgomery et al., 2008; Takeda et al., 2008) and/or to other sequence or structural properties of the miRNA duplex (Zhu et al., 2011; Endo et al., 2013; Zhang et al., 2014). Plant miRNAs target transcripts with highly complementary sequence through direct AGO‐mediated endonucleolytic cleavage, or through other cleavage‐independent mechanisms (Axtell, 2013).
Artificial miRNAs (amiRNAs) can be produced accurately by modifying the miRNA/miRNA* sequence within a functional MIRNA precursor (Alvarez et al., 2006; Schwab et al., 2006). AmiRNAs have been used in plants to selectively and effectively knockdown reporter and endogenous genes, non‐coding RNAs and viruses (Ossowski et al., 2008; Tiwari et al., 2014). Recently, cost‐ and time‐effective methods to generate large numbers of amiRNA constructs were developed and validated for eudicot species (Carbonell et al., 2014). These included a series of eudicot amiRNA vectors based on Arabidopsis thaliana MIR390a (AtMIR390a) precursor, whose relatively short distal stem–loop allows the cost‐effective synthesis and cloning of the amiRNA inserts into ‘B/c’ expression vectors (Carbonell et al., 2014). In monocots, OsMIR528 precursor has been used successfully to express amiRNAs for silencing endogenous genes in rice (Warthmann et al., 2008; Butardo et al., 2011; Chen et al., 2012a,b). However, OsMIR528‐based cloning methods have not been optimized for efficient generation of monocot amiRNA constructs.
In this report, a series of amiRNA expression vectors for high‐throughput cloning and high‐level expression in monocot species are described and tested. These vectors contain a truncated sequence from Oryza sativa MIR390 (OsMIR390) precursor in a configuration that allows the direct cloning of amiRNAs. OsMIR390‐based amiRNAs were generally more accurately processed and accumulated to higher levels in transgenic Brachypodium distachyon (Brachypodium) when processed from chimeric precursors (OsMIR390‐AtL) containing Arabidopsis thaliana (Arabidopsis) MIR390a (AtMIR390a) distal stem–loop sequences. Functionality of OsMIR390‐AtL‐based amiRNAs was confirmed in Brachypodium transgenic plants that displayed the predicted phenotypes, accumulated high levels of amiRNAs and low levels of the corresponding target transcripts. Moreover, genome‐wide transcriptome profiling in combination with 5′‐RLM‐RACE analysis confirmed that the amiRNAs were highly specific. We also describe a cost‐optimized alternative to generate amiRNA constructs for eudicots, as amiRNAs produced from chimeric AtMIR390a‐based precursors including AtMIR390a basal stem and OsMIR390 short distal stem–loop sequences are highly expressed, accurately processed, and effective in target gene knockdown in A. thaliana.
Results and Discussion
AmiRNA vectors based on the OsMIR390 precursor
Previously, the short AtMIR390a precursor was selected as the backbone for high‐throughput cloning of amiRNAs in a series of vectors for eudicot species (Carbonell et al., 2014). These vectors allow a zero‐background, oligonucleotide cloning strategy that requires no enzymatic modifications, PCR steps, restriction digestions, or DNA fragment isolation (Carbonell et al., 2014). The short distal stem–loop (Figure 1a) of AtMIR390a precursor provides a cost advantage by reducing the length of synthetic oligonucleotides corresponding to the amiRNA precursor sequence. To develop a comparable system for monocot species, a search for conserved, short Oryza sativa (rice) MIRNA (OsMIRNA) precursors that could be adapted for amiRNA vectors was done. Rice MIRNA precursors were analyzed as they have been subjected to extensive prior analysis (Arikit et al., 2013). The distal stem–loop length of 142 OsMIRNA precursor sequences (median length = 54 nt, Figure 1b) from 23 conserved miRNA families (Table S1) revealed that the OsMIR390 precursor was one of the shortest (16 nt). Moreover, OsMIR390 contains the shortest distal stem–loop of all 51 sequenced MIR390 precursors from 36 species (median length = 47 nt; Figure 1b and Table S2), including those from maize (ZmaMIR390a and ZmaMIR390b), sorghum (SbiMIR390a) and B. distachyon (BdiMIR390) with lengths of 137, 148, 134 and 107 nt respectively. The MIR390 family is among the most deeply conserved miRNA families in plants (Axtell et al., 2006; Cuperus et al., 2011).
Figure 1.
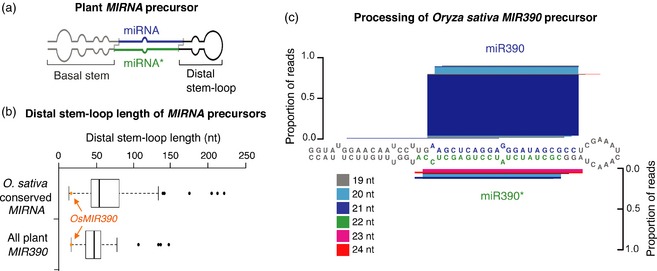
Oryza sativa MIR390 (OsMIR390) is an accurately processed, conserved MIRNA precursor with a particularly short distal stem–loop.
(a) Diagram of a canonical plant MIRNA precursor (adapted from Cuperus et al., 2011). miRNA guide and miRNA* strands are highlighted in blue and green, respectively. Distal stem–loop and basal stem regions are highlighted in black and grey, respectively.
(b) Distal stem–loop length of O. sativa conserved MIRNA precursors and of all plant catalogued MIR390 precursors. Box‐plot showing the distal stem–loop length of O. sativa conserved MIRNA precursors and all catalogued MIR390 precursors. The distal stem–loop length of OsMIR390 is highlighted with an orange dot and indicated with an orange arrow. Outliers are represented with black dots.
(c) OsMIR390 precursor processing diagram. miR390 and miR390* nucleotides are highlighted in blue and green, respectively. Proportion of small RNA reads for the entire OsMIR390 precursor are plotted as stacked bar graphs. Small RNAs are color‐coded by size. Publicly available small RNA data sets from rice grains, roots, shoots, leaves and inflorescences (Heisel et al., 2008; Zhu et al., 2008; Johnson et al., 2009; Zhou et al., 2009; He et al., 2010) were analyzed.
Publicly available small RNA data sets from rice (Heisel et al., 2008; Zhu et al., 2008; Johnson et al., 2009; Zhou et al., 2009; He et al., 2010) were analyzed to assess the OsMIR390 precursor processing accuracy. Approximately 70% of reads mapping to the OsMIR390 foldback correspond to the authentic 21‐nt miR390 guide strand (Figure 1c). Given the short distal stem–loop sequence and relatively accurate precursor processing characteristics, OsMIR390 was selected as the backbone for amiRNA vector development.
A set of amiRNA cloning vectors based on OsMIR390 and named ‘OsMIR390‐B/c’ (from OsMIR390‐B saI/ c cdB) was developed for rapid cloning of amiRNAs (Figure S1 and Table 1). OsMIR390‐B/c vectors include a truncated OsMIR390 precursor sequence whose miRNA/distal stem–loop/amiRNA* region was substituted by a DNA cassette containing the counter‐selectable ccdB gene (Bernard and Couturier, 1992) flanked by two BsaI sites. AmiRNA inserts corresponding to amiRNA/OsMIR390‐distal‐stem–loop/amiRNA* sequences are synthesized using two overlapping and partially complementary 60‐base oligonucleotides (Figure S2). Forward and reverse oligonucleotides must have 5′‐CTTG and 5′‐CATG overhangs, respectively, for direct cloning into OsMIR390‐based vectors (Figure S2).
Table 1.
OsMIR390‐BsaI/ccdB (‘B/c’) vectors for direct cloning of amiRNAs
| Vector | Bacterial antibiotic resistance | Plant antibiotic resistance | GATEWAY use | Backbone | Promoter | Terminator | Plant species tested |
|---|---|---|---|---|---|---|---|
| pENTR‐OsMIR390‐B/c | Kanamycin | – | Donor | pENTR | – | – | – |
| pMDC123SB‐OsMIR390‐B/c | Kanamycin | BASTA | – | pMDC123 | CaMV 2x35S | nos | N. benthamiana |
| pMDC32B‐OsMIR390‐B/c |
Kanamycin Hygromycin |
Hygromycin | – | pMDC32 | CaMV 2x35S | nos |
N. benthamiana
B. distachyon |
| pH7WG2B‐OsMIR390‐B/c | Spectinomycin | Hygromycin | – | pH7WG2 | Os Ubiquitin | CaMV | B. distachyon |
OsMIR390‐B/c vectors include pMDC32B‐OsMIR390‐B/c, pMDC123SB‐OsMIR390‐B/c and pH7WG2B‐OsMIR390‐B/c plant expression vectors, each of which contains an exclusive combination of regulatory sequences and plant and bacterial antibiotic resistance genes (Figure S1 and Table 1). Additionally, a GATEWAY‐compatible entry vector named pENTR‐OsMIR390‐B/c was developed for rapid amiRNA insert cloning and posterior recombination into the GATEWAY expression vector of choice (Figure S1 and Table 1).
High accumulation of amiRNAs derived from chimeric precursors in Brachypodium calli
To test amiRNA expression from OsMIR390 precursors, transformed B. distachyon calli containing amiRNA constructs expressing miR390 or modified versions of several miRNAs from Arabidopsis (amiR173‐21, amiR472‐21 or amiR828‐21) (Cuperus et al., 2010) were analyzed (Figure 2a). In addition, the same amiRNAs were expressed from a chimeric precursor (OsMIR390‐AtL) composed of the OsMIR390 basal stem and AtMIR390a distal stem–loop (Figures 2a and S3). Each amiRNA was also expressed from the reciprocal chimeric precursors (AtMIR390a‐OsL) containing the AtMIR390a basal stem and OsMIR390 distal stem–loop (Figures 2a and S4). A 35S:GUS construct expressing the ß‐glucuronidase transcript was used as negative control.
Figure 2.
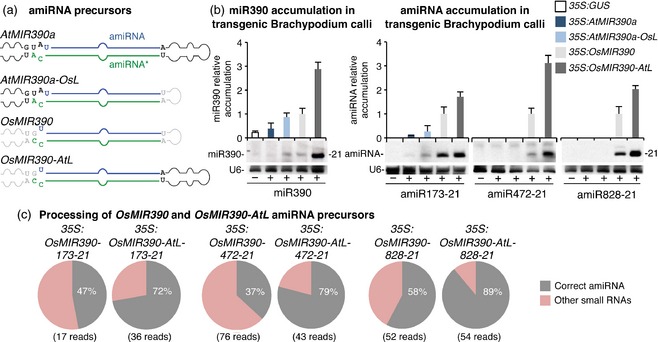
Comparative analysis of accumulation and processing of several amiRNAs produced from AtMIR390a, AtMIR390a‐OsL, OsMIR390 and OsMIR390‐AtL precursors in Brachypodium transgenic calli.
(a) Diagrams of AtMIR390a, AtMIR390a‐OsL, OsMIR390 and OsMIR390‐AtL precursors. Nucleotides corresponding to the miRNA guide strand are in blue, and nucleotides of the miRNA* strand are in green. Other nucleotides from AtMIR390a and OsMIR390 precursors are in black and grey, respectively. Shapes of AtMIR390a and OsMIR390 precursors are in black and grey, respectively.
(b) Accumulation of miR390 (left) and of several 21‐nucleotide amiRNAs (right) expressed from the AtMIR390a, AtMIR390a‐OsL, OsMIR390 or OsMIR390‐AtL precursors in Brachypodium transgenic calli. Mean (n = 3) relative amiRNA levels + standard deviation (SD) when expressed from the OsMIR390 (light grey, amiRNA level = 1.0). Only one blot from three biological replicates is shown. U6 RNA blot is shown as loading control.
(c) Processing analysis of OsMIR390 and OsMIR390‐AtL amiRNA precursors. Pie charts show the percentage of reads corresponding to accurately processed 21‐nt mature amiRNAs (grey sectors) or to other small RNAs (pink sectors).
Surprisingly, miR390 accumulated to highest levels when expressed from the chimeric OsMIR390‐AtL precursor compared with each of the other three precursors (P ≤ 0.001 for all pairwise t‐test comparisons; Figure 2b). Moreover, each amiRNA expressed from OsMIR390‐AtL chimeric precursors also accumulated to significantly higher levels when compared with the other precursors (P < 0.026 for all pairwise t‐test comparisons; Figure 2b). miR390 and each amiRNA derived from authentic AtMIR390a or chimeric AtMIR390a‐OsL precursors accumulated to low or non‐detectable levels, indicating that the AtMIR390a stem is suboptimal for the accumulation and/or processing of amiRNAs in Brachypodium.
To assess the accuracy of precursor processing, small RNA libraries from samples expressing OsMIR390‐AtL‐based amiRNAs were prepared and sequenced (Figure 2c). For comparative purposes, small RNA libraries from samples containing amiRNAs produced from authentic OsMIR390 precursors were also analyzed. In each case, the majority of reads mapping to the chimeric OsMIR390‐AtL precursors corresponded to correctly processed 21 nt amiRNAs (Figure 2c). In contrast, processing of authentic OsMIR390 precursors including amiRNA sequences was less accurate, as revealed in each case by a lower proportion of reads corresponding to correctly processed sequences (Figure 2c).
Gene silencing in Brachypodium and Arabidopsis by amiRNAs derived from chimeric precursors
To assess the functionality of OsMIR390‐AtL‐derived amiRNAs in silencing target transcripts in Brachypodium, BRASSINOSTEROID‐INSENSITIVE 1 (BdBRI1), CINNAMYL ALCOHOL DEHYDROGENASE 1 (BdCAD1), CHLOROPHYLLIDE A OXYGENASE (BdCAO) and SPOTTED LEAF 11 (BdSPL11) gene transcripts were targeted by amiRNAs expressed from the chimeric OsMIR390‐AtL and from authentic OsMIR390 precursors (Figure 3a). The sequences for amiR‐BdBri1, amiR‐BdCad1, amiR‐BdCao and amiR‐BdSpl11 (Figure S5) were designed using the ‘P‐SAMS amiRNA Designer’ tool (http://p-sams.carringtonlab.org). Plants expressing 35S:GUS were used as negative controls. Phenotypes of transgenic plants, amiRNA accumulation, processing of amiRNA precursors, and target transcript accumulation were analyzed in Brachypodium T0 transgenic lines.
Figure 3.

Functionality of amiRNAs produced from authentic OsMIR390‐ or chimeric OsMIR390‐AtL‐based precursors in Brachypodium T0 transgenic plants.
(a) OsMIR390‐ and OsMIR390‐AtL‐based precursors containing Bri1‐, Cad1‐, Cao and Spl11‐amiRNAs. Nucleotides corresponding to the miRNA guide and miRNA* strands are in blue and green, respectively; nucleotides from AtMIR390a or OsMIR390 precursors are in black or grey, respectively, except those that were modified to preserve authentic AtMIR390a or OsMIR390 precursor secondary structures (in red).
(b–e) Representative images of plants expressing amiRNAs from OsMIR390‐AtL or OsMIR390 precursors, or the control construct.
(b) Adult control plant (left), or plants expressing 35S:OsMIR390‐Bri1 (center) or 35S:OsMIR390‐AtL‐Bri1 (right).
(c) Adult control plant (left), or plants expressing 35S:OsMIR390‐Cad (center) or 35S:OsMIR390‐AtL‐Cad1 (bottom).
(d) Adult control plant (left), or plants expressing 35S:OsMIR390‐Cao (center) or 35S:OsMIR390‐AtL‐Cao (right).
(e) Adult control plant (left), or plants expressing 35S:OsMIR390‐Spl11 (center) or 35S:OsMIR390‐AtL‐Spl11 (right).
Sixteen out of 20 and 11 out of 17 transgenic lines containing 35S:OsMIR390‐AtL‐Bri1 or 35S:OsMIR390‐Bri1, respectively, which were predicted to have brassinosteroid signalling defects, had reduced height and altered architecture (Figures 3b and S6 and Table S3). Most organs, particularly leaves, exhibited a contorted phenotype since the earliest stages of development (Figure 3b). Inflorescences had reduced size (Figure 3b), and contained smaller seeds compared to control lines (Figure S6). AmiR‐BdBri1‐induced phenotypes were similar to those described for the Brachypodium bri1 T‐DNA mutants from the BrachyTAG collection (Thole et al., 2012). These phenotypes are consistent with the expectation of plants with brassinosteroid signalling defects (Zhu et al., 2013). All 27 transgenic lines containing 35S:OsMIR390‐AtL‐Cad1, and 52 out of 55 lines including 35S:OsMIR390‐Cad1, exhibited reddish coloration of lignified tissues such as tillers, internodes and nodes (Figure 3c and Table S3), as expected from Cad1 knockdown and loss of function mutant analyses (Bouvier d'Yvoire et al., 2013; Trabucco et al., 2013).
Each of 27 35S:OsMIR390‐AtL‐Cao‐expressing plants, and 12 of 12 of 35S:OsMIR390‐Cao‐expressing plants exhibited light green color compared with control plants (Figure 3d and Table S3), as expected due to reduction in chlorophyllide a to b conversion during chlorophyll b synthesis (Tanaka et al., 1998; Oster et al., 2000; Philippar et al., 2007). Biochemical analysis of chlorophyll content in transgenic lines confirmed that chlorophyll b content in 35S:OsMIR390‐AtL‐Cao and 35S:OsMIR390‐Cao lines was reduced to approximately 57 and 67%, respectively, compared with levels measured in control plants (Figure S7). Carotenoid content was also notably reduced (to almost 50%) in lines expressing amiR‐BdCao from chimeric or authentic precursors (Figure S7), as observed before in Arabidopsis cao mutants (Philippar et al., 2007). Finally, 39 of 43 transgenic lines containing 35S:OsMIR390‐AtL‐Spl11, and 22 of 24 35S:OsMIR390‐Spl11‐expressing plants displayed a spontaneous cell death phenotype characterized by the development of necrotic lesions in leaves (Figure 3e). This was consistent with expectations based on phenotypes of SPL11‐knockdown amiRNA rice lines (Zeng et al., 2004). Phenotypes induced by all four sets of amiRNAs were heritable in self‐pollinated T1 plants expressing OsMIR390‐ or OsMIR390‐AtL‐based amiRNA precursors from pMC32B vectors containing 35S regulatory sequences (Table S4).
Quantitative real‐time RT‐PCR (RT‐qPCR) assays were used to measure the accumulation of amiRNA‐target transcripts in Brachypodium transgenic lines expressing OsMIR390‐AtL‐ or OsMIR390‐based amiRNAs. All target transcripts were expressed to significantly reduced levels compared with control plants (P < 0.005 for all pairwise t‐test comparisons, Figure 4a) in transgenic lines expressing the specific amiRNA. No significant differences were observed in target mRNA levels between lines expressing OsMIR390‐AtL‐ or OsMIR390‐based amiRNAs.
Figure 4.

amiRNA and target mRNA accumulation analysis in Brachypodium T0 transgenic plants.
(a) Mean relative level ± standard error (SE) of B. distachyon BdBRI1, BdCAD1, BdCAO and BdSPL11 mRNAs after normalization to BdSAMDC, BdUBC, BdUBI4 and BdUBI10, as determined by quantitative real‐time RT‐PCR (35S:GUS = 1.0 in all comparisons).
(b) Accumulation of amiRNAs in Brachypodium transgenic plants. In each blot the amiRNA accumulation of a single independent transgenic line per construct is analyzed. U6 RNA blot is shown as a loading control.
AmiR‐BdBri1, amiR‐BdCao and amiR‐BdSpl11 produced from chimeric OsMIR390‐AtL precursors were also expressed using pH7WG2B‐based constructs that contain the rice ubiquitin (UBI) regulatory sequences. Each of the three UBI promoter‐driven amiRNAs induced the expected phenotypes in a relatively high proportion of Brachypodium T0 lines (Table S3), and in the one case tested (amiR‐BdSpl11), phenotypes were heritable in the T1 generation (Table S4).
Finally, we tested if the reciprocal chimeric AtMIR390a‐OsL precursor could be used to express amiRNAs efficiently in eudicots. The synthesis of AtMIR390a‐OsL‐based constructs requires shorter oligonucleotides than the generation of AtMIR390a‐based constructs, and therefore would be a further cost‐optimized alternative. As shown in Nicotiana benthamiana and Arabidopsis assays, AtMIR390‐OsL precursors are accurately processed (Appendix S1 and Figures S8–S10). Indeed, amiRNAs produced from chimeric AtMIR390a‐OsL precursors are highly expressed, accurately processed and highly effective in target gene knockdown in T1 Arabidopsis transgenic plants (Appendix S1, Figures S9–S11 and Table S5). Moreover, amiRNA‐induced phenotypes were still obvious in T2 plants confirming the heritability of the effects (Table S6). Therefore, the use of AtMIR390a‐OsL precursors may be an attractive alternative to express effective amiRNAs in eudicots in a cost‐optimized manner.
Accuracy of processing of OsMIR390 and OsMIR390‐AtL chimeric precursors in Brachypodium
The accumulation of each amiRNA from chimeric and OsMIR390 precursors was analyzed by RNA blot analysis in T0 transgenic lines exhibiting phenotypes induced by amiRNAs (Figure 4b). In most cases, OsMIR390‐AtL‐derived amiRNAs accumulated to higher levels and as more uniform RNA species (Figure 4b). AmiRNAs from the OsMIR390 precursor accumulated to rather low levels (except in transgenic lines containing 35S:OsMIR390‐Cao) and generally as multiple species (Figure 4b).
Small RNA libraries from transgenic lines expressing amiRNAs from chimeric OsMIR390‐AtL or authentic OsMIR390 precursors were prepared to further analyze processing and accumulation of the amiRNA species (Figure 5). Three of the four amiRNAs produced from chimeric OsMIR390‐AtL precursors accumulated predominantly as 20‐nt species (Figure 5a,c,d); only amiR‐BdCad1 accumulated mainly as a 21 nt RNA (Figure 5b). Processing of authentic OsMIR390 precursors generally resulted in a high proportion of small RNAs of diverse sizes, except for OsMIR390‐Cad1 precursors (Figure 5).
Figure 5.
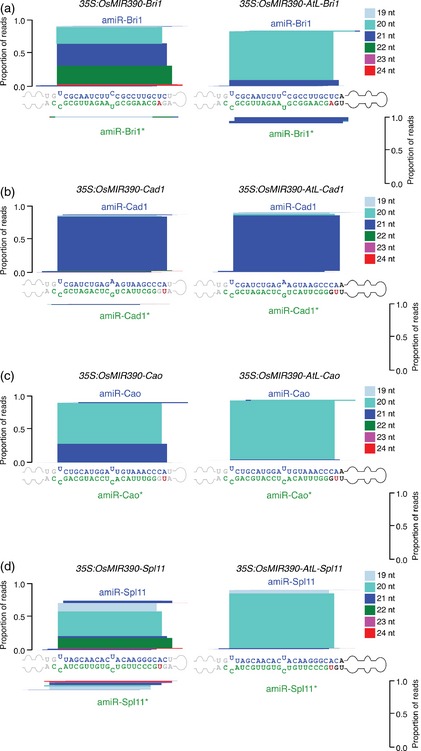
Mapping of amiRNA reads from OsMIR390‐AtL‐ or OsMIR390‐based precursors expressed in Brachypodium T0 transgenic plants.
Analysis of amiRNA and amiRNA* reads in plants expressing (a) amiR‐BdBri1, (b) amiR‐BdCad1, (c) amiR‐BdCao or (d) amiR‐BdSpl11. amiRNA guide and amiRNA* strands are highlighted in blue and green, respectively. Nucleotides from the AtMIR390a or OsMIR390 precursors are in black and grey, respectively, except those that were modified to preserve the corresponding authentic precursor secondary structure (in red). Proportion of small RNA reads are plotted as stacked bar graphs. Small RNAs are color‐coded by size.
The reasons explaining the accumulation of OsMIR390a‐AtL‐based amiRNAs that are 1 nt‐shorter than expected are not clear. AmiRNAs shorter than expected and differing on their 3′ end were also described using AtMIR319a precursors in Arabidopsis (Schwab et al., 2006). Importantly, a recent study has shown that amiRNA efficacy is not affected by the loss of the base‐pairing at the 5′ end of the target site (Liu et al., 2014). Regardless, the inaccurate processing of an amiRNA precursor leading to the accumulation of diverse small RNA populations could conceivably induce undesired off‐target effects. This potential complication argues against using authentic OsMIR390 precursors to express amiRNAs in Brachypodium and possibly other monocot species.
Reads from the amiRNA* strands from each of the OsMIR390 and OsMIR390‐AtL‐derived precursors were under‐represented, relative to the amiRNA strands (Figure 5). The rational P‐SAMS design tool uniformly specifies an amiRNA* strand containing an AGO‐non‐preferred 5′ G residue, which likely promotes amiRNA* degradation.
High specificity of amiRNA derived from chimeric precursors in Brachypodium
To assess amiRNA‐target specificity at a genome‐wide level, transcript libraries from control (35S:GUS) and amiRNA‐expressing lines were generated and analyzed. Only lines expressing amiRNAs from the more accurately processed OsMIR390‐AtL precursors were analyzed. Differential gene expression analyses were done by comparing, in each case, the transcript libraries obtained from four independent control lines with those obtained from four independent amiRNA‐expressing lines exhibiting the expected phenotypes. In total, 494, 1847 and 818 genes were differentially expressed in plants expressing amiR‐BdBri1, amiR‐BdCao and amiR‐BdSpl11, respectively (Figure 6 and Data S1). In contrast, only 21 genes were differentially expressed in plants expressing amiR‐BdCad1 (Figure 6 and Data S1). The high number of differentially expressed genes in amiR‐BdBri1‐, amiR‐BdCao‐ and amiR‐BdSpl11‐expressing lines may reflect the complexity of the corresponding targeted gene pathways involving hormone signalling, photosynthesis and cell death/pathogen resistance respectively. As expected, BdCAD1, BdCAO and BdSPL11 were differentially underexpressed in plants expressing amiR‐BdCad1, amiR‐BdCao and amiR‐BdSpl11, respectively (q < 0.01, Wald test) (Figure 6 and Data S1). However, BdBRI1 was not called as differentially expressed (q = 0.42, Wald test) (Figure 6 and Data S1) despite being notably downregulated in 35S:OsMIR390‐AtL‐Bri1 plants as shown by RT‐qPCR analysis (Figure 4a). Because the power of statistical tests involving count data decreases with lower count numbers (Rapaport et al., 2013), this result could be explained by the low accumulation of BdBRI1 even in control plants (Figure S12 and Data S2). Therefore, the differential expression analysis on RNA‐Seq data approach may not be appropriate to evaluate the differential expression of genes with genuine low expression and/or low coverage, as suggested before (Rapaport et al., 2013).
Figure 6.
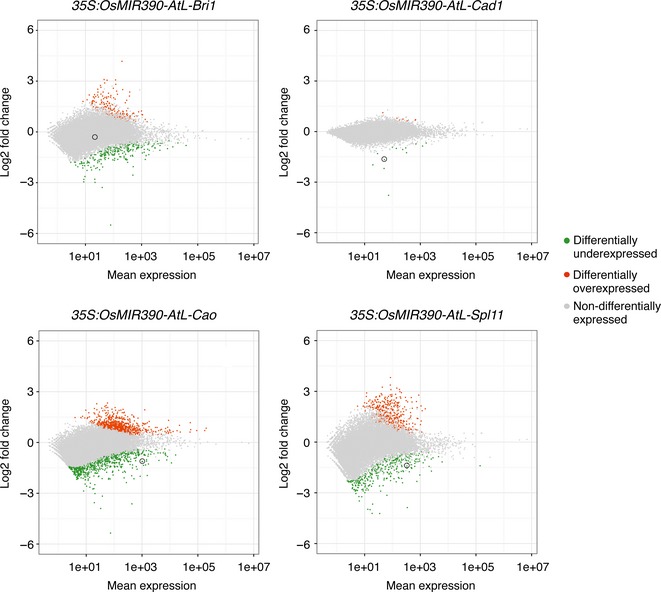
Transcriptome analysis of transgenic Brachypodium plants expressing amiRNAs from chimeric OsMIR390‐AtL precursors. MA plots show log2 fold change versus mean expression of genes for each 35S:OsMIR390‐AtL amiRNA line compared with the control lines (35S:GUS). Green, red and grey dots represent differentially underexpressed, differentially overexpressed or non‐differentially expressed genes, respectively, in each amiRNA versus control comparison. The position of expected amiRNA targets is indicated with a circle.
To assess potential off‐target effects of the amiRNAs, targetfinder (Fahlgren and Carrington, 2010) was used to generate a genome‐wide list of potential candidate targets that share relatively high sequence complementarity with each amiRNA. targetfinder ranks the potential amiRNA targets based on a Target Prediction Score (TPS) assigned to each amiRNA‐target interaction. Scores range from 1 to 11, that is, from highest to lowest levels of sequence complementarity between the small RNA and putative target RNA. Indeed, when designing amiRNAs with the ‘P‐SAMS amiRNA Designer’ tool, ‘optimal’ amiRNAs are selected when: (i) their interaction with the desired target has a TPS = 1; and (ii) no other amiRNA‐target interactions have a TPS < 4. Therefore, direct off‐target effects with amiRNAs described here can only occur through amiRNA‐target RNA interactions with a TPS in the [4, 11] interval. It was hypothesized that off‐target effects, if due to base‐pairing between amiRNAs and the affected transcripts, would be reflected by the presence of differentially underexpressed genes corresponding to target RNAs with lower TPS scores in the [4, 11] interval. Therefore, we next analyzed for all targetfinder‐predicted targets for each amiRNA if their corresponding genes were differentially underexpressed in amiRNA‐expressing lines versus controls.
As expected from P‐SAMS design, BdCad1, BdCao and BdSpl11 were the only genes differentially underexpressed in the [1, 4] TPS interval in plants expressing amiR‐BdCad1, amiR‐BdCao and amiR‐BdSpl11, respectively (Figure 7, Data S3). On the other hand, 2958, 1290, 1528 and 1533 genes corresponded to target RNAs with calculated TPS scores in the [4, 11] interval in targetfinder analyses including amiR‐BdBri1, amiR‐BdCad1, amiR‐BdCao and amiR‐BdSpl11, respectively (Figure 7). In all cases, the number of differentially underexpressed genes corresponding to predicted targets with a TPS in the [4, 11] interval was low (Figure 7, upper panels). Moreover, in each of the four cases the proportion of differentially underexpressed genes among targetfinder‐predicted targets was also low in the [4, 11] TPS interval (Figure 7, bottom panels). Indeed, in this same interval, 0.84, 1.31 and 0.78% of the genes were differentially underexpressed in amiR‐BdBri1‐, amiR‐BdCao‐, and amiR‐BdSpl11‐expressing lines, respectively. In each case, this percentage was lower than the percentage of differentially underexpressed genes from transcripts with a TPS not included in the [4, 11] interval in the same samples (1.12, 3.74 and 1.55% respectively). In amiR‐BdCad‐expressing lines, although the percentage of genes differentially expressed in the [4, 11] interval (0.07%) was higher compared to the percentage of genes differentially underexpressed in the [4, 11] interval (0.04%), this difference was not statistically significant (P = 0.45, Fisher exact test). Together, these results indicate that globally targetfinder‐predicted targets were not preferentially downregulated in the amiRNA‐expressing lines.
Figure 7.
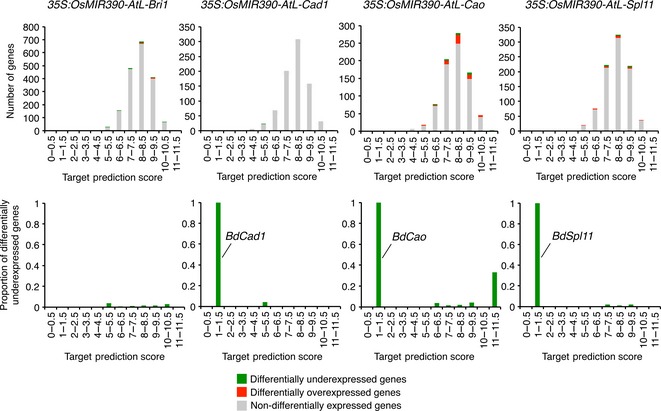
Differential expression analysis of targetfinder‐predicted off‐targets for each amiRNA versus control comparison. Histograms show the total number of genes (top panels) or the proportion of differentially underexpressed genes (bottom panels) in each target prediction score bin. Green, red and grey bars represent differentially underexpressed, differentially overexpressed or non‐differentially expressed genes, respectively. In bottom panels, the name of the expected target gene is indicated when the target gene is the only gene differentially underexpressed in the corresponding bin.
Next, we used 5′‐RLM‐RACE to test for amiRNA‐directed off‐target cleavage of under‐represented transcripts. This analysis detects 3′ cleavage products expected from small RNA‐guided cleavage events. Only targetfinder‐predicted targets with a TPS ≤ 7 were included in the analysis, as targets with higher score are not considered likely to be cleaved, according to previous studies (Addo‐Quaye et al., 2008). For all specific targets, 3′ cleavage products of the expected size were detected in samples expressing the corresponding amiRNA, but not in control samples expressing 35S:GUS (Figure 8). Sequencing analysis confirmed that the majority of sequences comprising these products, in each case, contained a canonical 5′ end position predicted for small RNA‐guided cleavage (Figure 8). In contrast, for all potential off‐target transcripts, no obvious amiRNA‐guided cleavage products were detected in either amiRNA‐expressing or 35S:GUS lines (Figure 8). Additionally, sequencing analysis failed to detect even low‐level amiRNA‐guided cleavage products among potential off‐targets (Figure 8).
Figure 8.
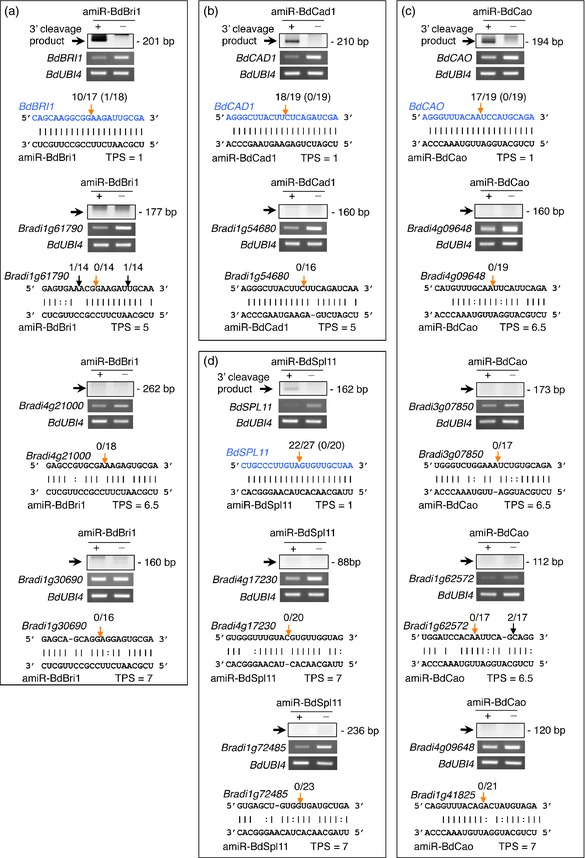
5′ RLM‐RACE mapping of target and potential off‐target cleavage guided by amiRNAs in plants expressing (a) amiR‐BdBri1, (b) amiR‐BdCad1, (c) amiR‐BdCao and (d) amiR‐BdSpl11.
At the top of each panel, ethidium bromide‐stained gels show 5′‐RLM‐RACE products corresponding to the 3′ cleavage product from amiRNA‐guided cleavage (top gel), and RT‐PCR products corresponding to the gene of interest (middle gel) or control BdUBI4 gene (bottom gel). The position and size of the expected amiRNA‐based 5′‐RLM‐RACE products are indicated. At the bottom of each panel, the predicted base‐pairing between amiRNAs and prospective target RNAs is shown. The sequence and the name of authentic target mRNAs are in blue. For each authentic or predicted target mRNA, the expected amiRNA‐based cleavage site is indicated by an orange arrow. Other sites are indicated with a black arrow. The proportion of cloned 5′‐RLM‐RACE products at the different cleavage sites is shown for amiRNA‐expressing lines, with that of control plants expressing 35S:GUS shown in brackets. TPS refers to ‘Target Prediction Score’.
High amiRNA specificity was previously indicated for AtMIR319a‐derived amiRNAs in Arabidopsis based on genome‐wide expression profiling (Schwab et al., 2006). However, a recent and systematic processing analysis of AtMIR319a‐based amiRNA precursors in petunia (Guo et al., 2014) showed that multiple small RNA variants are generated from different regions of the precursor, and that many of these small RNAs meet the required criteria for amiRNA design (Schwab et al., 2006). Here, the fact that chimeric OsMIR390‐AtL precursors produce high levels of accurately processed amiRNAs not only in Brachypodium (Figures 2, 4 and 5) but also in a eudicot species such as N. benthamiana (Figure S8), strongly suggests that these precursors will be functional in a wide range of species.
Conclusions
We have developed and validated a series of expression vectors based on the OsMIR390 precursor for high‐throughput cloning and high expression of amiRNAs in monocots. OsMIR390‐B/c‐based vectors allow the direct cloning of amiRNAs in a zero‐background strategy that does not require oligonucleotide modifications, PCRs, restriction digestions, or isolation of DNA fragments. Thus, OsMIR390‐B/c‐based vectors are particularly attractive for generating large‐scale amiRNA construct libraries for silencing genes in monocots.
‘P‐SAMS amiRNA Designer’ tool was used to design four different amiRNAs, each of which was aimed to target specifically one Brachypodium gene transcript. We show that chimeric OsMIR390‐AtL precursors including OsMIR390 basal stem and AtMIR390a distal stem–loop were processed more accurately, and the resulting amiRNAs generally accumulated to higher levels than amiRNAs derived from authentic OsMIR390 precursors in Brachypodium transgenic plants. Each P‐SAMS‐designed amiRNA induced the expected phenotypes, and specifically decreased expression of the expected target gene. Chimeric OsMIR390‐AtL precursors designed using P‐SAMS, therefore, are likely to be highly effective and specific in silencing genes in monocot species.
Experimental Procedures
Plant materials and growth conditions
Arabidopsis thaliana Col‐0 and N. benthamiana plants were grown as described (Carbonell et al., 2014). Brachypodium distachyon 21‐3 plants were grown in a chamber under long day conditions (16/8 h photoperiod at 200 μmol m−2 s−1) and 24°C/18°C temperature cycle.
Arabidopsis thaliana plants were transformed and grown as described (Carbonell et al., 2014). Embryogenic calli from B. distachyon 21‐3 plants were transformed as described (Vogel and Hill, 2008). Photographs of plants were taken as described (Carbonell et al., 2014).
DNA constructs
pENTR‐OsMIR390‐BsaI construct was generated by ligating into pENTR (Life Technologies; http://www.lifetechnologies.com) the DNA insert resulting from the annealing of oligonucleotides BsaI‐OsMIR390‐F and BsaI‐OsMIR390‐R. Rice ubiquitin 2 promoter and maize ubiquitin promoter‐hygromycin cassettes were transferred into the GATEWAY binary destination vector pH7WG2 (Karimi et al. 2002) to generate pH7WG2‐OsUbi. pH7WG2‐OsMIR390‐BsaI, pMDC123SB‐OsMIR390‐BsaI and pMDC32‐OsMIR390‐BsaI were obtained by LR recombination between pENTR‐OsMIR390‐BsaI and pH7WG2‐OsUbi, pMDC32B (Carbonell et al., 2014) and pMDC123SB (Carbonell et al., 2014), respectively. A modified ccdB cassette (Carbonell et al., 2014) was inserted between the BsaI sites of pENTR‐OsMIR390‐BsaI, pMDC123SB‐OsMIR390‐BsaI, pMDC32B‐OsMIR390‐BsaI and pH7WG2‐OsMIR390‐BsaI to produce pENTR‐OsMIR390‐B/c, pMDC123SB‐OsMIR390‐B/c, pMDC32B‐OsMIR390‐B/c and pH7WG2‐OsMIR390‐B/c, respectively. Finally, an undesired BsaI site was disrupted in pH7WG2‐OsMIR390‐B/c to generate pH7WG2B‐OsMIR390‐B/c. The sequences of the OsMIR390‐B/c‐based amiRNA vectors are in Appendix S2. The following amiRNA vectors for monocots are available from Addgene (http://www.addgene.org/): pENTR‐OsMIR390‐B/c (Addgene plasmid 61468), pMDC32B‐OsMIR390‐B/c (Addgene plasmid 61467) pMDC123SB‐OsMIR390‐B/c (Addgene plasmid 61466) and pH7WG2B‐OsMIR390‐B/c (Addgene plasmid 61465). pMDC32B‐AtMIR390a‐B/c (Addgene plasmid 51776) was described before (Carbonell et al., 2014).
AmiRNA constructs including pMDC32B‐AtMIR390a‐OsL‐173‐21, pMDC32B‐AtMIR390a‐OsL‐472‐21, pMDC32B‐AtMIR390a‐OsL‐828‐21, pMDC32B‐AtMIR390a‐OsL‐Ch42, pMDC32B‐AtMIR390a‐OsL‐Ft, pMDC32B‐AtMIR390a‐OsL‐Trich, pMDC32B‐OsMIR390, pMDC32B‐OsMIR390‐AtL, pMDC32B‐OsMIR390‐173‐21, pMDC32B‐OsMIR390‐173‐21‐AtL, pMDC32B‐OsMIR390‐472‐21, pMDC32B‐OsMIR390‐AtL‐472‐21, pMDC32B‐OsMIR390‐828‐21, pMDC32B‐OsMIR390‐AtL‐828‐21, pMDC32B‐OsMIR390‐Bri1, pMDC32B‐OsMIR390‐AtL‐Bri1, pMDC32B‐OsMIR390‐Cao, pMDC32B‐OsMIR390‐AtL‐Cao, pMDC32B‐OsMIR390‐Cad1, pMDC32B‐OsMIR390‐AtL‐Cad1, pMDC32B‐OsMIR390‐Spl11, pMDC32B‐OsMIR390‐AtL‐Spl11, pH7WG2B‐OsMIR390‐Bri1‐AtL, pH7WG2B‐OsMIR390‐Cao‐AtL, and pH7WG2B‐OsMIR390‐Spl11‐AtL were generated as detailed in the following section. Control construct pH7WG2‐GUS was generated by LR recombination between pENTR‐GUS (Life technologies) and pH7GW2‐OsUbi. pMDC32‐GUS construct was used before (Montgomery et al., 2008). The sequence of all amiRNA precursors used here are in Appendix S3. The sequence of all oligonucleotides used are in Table S7.
amiRNA oligonucleotide design and cloning
Sequences of the amiRNAs expressed in A. thaliana have been described previously (Schwab et al., 2006; Felippes and Weigel, 2009; Liang et al., 2012; Carbonell et al., 2014). Sequences of both the amiRNAs expressed in Brachypodium and the oligonucleotides used for cloning in OsMIR390‐B/c vectors, were designed with the ‘P‐SAMS amiRNA Designer’ tool (http://p-sams.carringtonlab.org). The predicted targets for all the amiRNAs used in this study are shown in Table S8.
The generation of constructs to express amiRNAs from authentic AtMIR390a precursors was described before (Carbonell et al., 2014). Detailed oligonucleotide design for amiRNA cloning in OsMIR390, OsMIR390‐AtL and AtMIR390a‐OsL precursors is given in Figures S2, S3 and S4, respectively. The amiRNA cloning procedure is described in Appendix S4. The name and sequence of the oligonucleotides used for cloning amiRNA sequences are in Table S7.
Transient expression assays
N. benthamiana leaves were agroinfiltrated with A. tumefaciens GV3101 strain as described (Carbonell et al., 2014).
RNA blot assays
Total RNA extraction from Arabidopsis, Brachypodium or N. benthamiana and subsequent RNA blot assays were done as described (Cuperus et al., 2010). Table S7 includes the name and sequences of the oligonucleotides used as probes in small RNA blots.
Quantitative real‐time RT‐qPCR
RT‐qPCR reactions and analyses were done as described (Carbonell et al., 2014). The oligonucleotides used for RT‐qPCR are listed in Table S7 (and are named with the prefix ‘q’). The expression levels of target transcripts were calculated relative to four A. thaliana (AtACT2, AtCPB20, AtSAND and AtUBQ10) or B. distachyon (BdSAMDC, BdUBC18, BdUBI4 and BdUBI10) reference genes as described (Carbonell et al., 2014).
5′‐RLM‐RACE
5′ RNA ligase‐mediated rapid amplification of cDNA ends (5′‐RLM‐RACE) was done using the GeneRacer™ kit (Life Technologies) but omitting the dephosphorylation and decapping steps. Total RNA (2 μg) was ligated to the GeneRacer RNA Oligo Adapter. The GeneRacer Oligo dT primer was then used to prime first strand cDNA synthesis in reverse transcription reaction. An initial PCR was done by using the GeneRacer 5′ and 3′ primers. The 5′ end of cDNA specific to each mRNA was amplified with the GeneRacer 5′ Nested primer and a gene specific reverse primer. For each gene, control PCR reactions were done using gene specific forward and reverse primers. Oligonucleotides used are listed in Table S7. 5′‐RLM‐RACE products were treated as described (Cuperus et al., 2010).
Chlorophyll and carotenoid extraction and analysis
Pigments from Brachypodium leaf tissue (40 mg of fresh weight) were extracted with 5 ml 80% (v/v) acetone in the dark at room temperature for 24 h, and centrifuged at 1000 g for 2 min. One hundred microlitres of supernatant was diluted 1:2 with 80% (v/v) acetone and loaded to flat bottom 96‐well plates. Absorbance was measured from 400 to 750 nm wavelengths in a SpectrMax M2 microplate reader (Molecular Devices, Sunnyvale, CA, USA) using the software softmax pro 5 (Molecular Devices). Content in chlorophyll a, chlorophyll b, and carotenoids was calculated with the following formulas: chlorophyll a (mg/L in extract) = 12.21 * Absorbance663 nm − 2.81 * Absorbance647 nm; chlorophyll b (mg/L in extract) = 20.13 * Absorbance647 nm − 5.03 * Absorbance663 nm; carotenoid (mg/L in extract) = [1000 * Absorbance470 nm − 3.27 * chlorophyll a (mg/L) − 104 * Chlorophyll b (mg/L)]/227.
Preparation of small RNA libraries
Approximately 50–100 μg of Arabidopsis, Brachypodium or Nicotiana total RNA were treated essentially as before (Carbonell et al., 2012; Gilbert et al., 2014), with the difference that small RNA libraries were barcoded at the amplicon PCR reaction step with the standard 5′PCR oligonucleotide (P5) and an indexed 3′ PCR oligonucleotide (i1‐i8, i10 or i11) (Table S7). Library multiplexing was followed by sequencing analysis using a HiSeq 2000 sequencer (Illumina, http://www.illumina.com/).
Small RNA sequencing data analysis
Small RNA sequencing data analysis was done as described (Carbonell et al., 2014). Custom scripts to process small RNA data sets are available at https://github.com/carringtonlab/srtools. Small RNA sequencing libraries from transgenic Arabidopsis inflorescences and Brachypodium calli or leaves, and from N. benthamiana agroinfiltrated leaves, are described in Table S9. O. sativa small RNA data sets used in the processing analysis of authentic OsMIR390 presented in Figure 1(b) were described previously (Cuperus et al., 2010).
Preparation of strand‐specific transcript libraries
Ten microgram of total RNA extracted from four independent lines per construct were treated with TURBO DNase I (Life Technologies). Ribosomal RNAs were depleted from samples by Ribo‐Zero Magnetic Kit ‘Plant Leaf’ (Epicentre, http://www.epibio.com/) treatment. cDNA synthesis followed by strand‐specific transcript library preparation were made as described with minor modifications (Wang et al., 2011; Carbonell et al., 2012). These included the fragmentation with metal ions during 4 min at 95°C of Ribo‐Zero treated RNAs, and the use of 14 cycles in the linear PCR reaction. Y‐shape adaptors were generated by annealing DNA adaptors 1 and 2, and PE‐F oligonucleotide was combined with one indexed oligonucleotide (PE‐R‐N701 to PE‐R‐N710) in the linear PCR (Table S7). DNA amplicon analysis, quantification and sequencing were done as described (Carbonell et al., 2014).
Transcriptome analysis
FASTQ files were de‐multiplexed with the parseFastq.pl perl script (https://github.com/carringtonlab/srtools). Sequencing reads from each de‐multiplexed transcript library were mapped to B. distachyon transcriptome (v2.1; Phytozome 10, http://phytozome.jgi.doe.gov/pz/portal.html) using Butter (Axtell, 2014) and allowing one mismatch. Differential gene expression analysis was done using DESeq2 (Love et al., 2014) with a false discovery rate of 1%. For each 35S:GUS versus 35S:OsMIR390‐AtL pairwise comparison, genes having no expression (0 gene counts) in at least five of the eight samples were removed from the analysis. Differential gene expression analysis results are shown in Data S1.
targetfinder v1.7 (https://github.com/carringtonlab/TargetFinder) (Fahlgren and Carrington, 2010) was used to obtain a ranked list of potential off‐targets for each amiRNA. RNA‐Seq libraries from transgenic Brachypodium leaves are described in Table S10.
Accession Numbers
A. thaliana genes and corresponding locus identifiers are: AtACT2 (AT3G18780), AtCBP20 (AT5G44200), AtCH42 (AT4G18480), AtCPC (AT2G46410), AtETC2 (AT2G30420), AtFT (AT1G65480), AtSAND (AT2G28390), AtTRY (AT5G53200) and AtUBQ10 (AT4G05320). B. distachyon genes and corresponding locus identifiers are: BdBRI1 (Bradi2g48280), BdCAD1 (Bradi3g06480), BdCAO (Bradi2g61500), BdSAMDC (Bradi5g14640), BdSPL11 (Bradi4g04270), BdUBC18 (Bradi4g00660), BdUBI4 (Bradi3g04730) and BdUBI10 (Bradi1g32860). The miRBase (http://mirbase.org) (Kozomara and Griffiths‐Jones, 2014) locus identifiers of the conserved rice MIRNA precursors and plant MIR390 precursors (Figure 1b) are detailed in Tables S1 and S2, respectively. High‐throughput sequencing data were deposited in the Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession number SRP052754.
Supporting information
Figure S1. OsMIR390‐B/c vectors for direct cloning of amiRNAs.
Figure S2. Generation of constructs to express amiRNAs from authentic OsMIR390 precursors.
Figure S3. Generation of constructs to express amiRNAs from chimeric OsMIR390‐AtL precursors.
Figure S4. Generation of constructs to express amiRNAs from chimeric AtMIR390a‐OsL precursors.
Figure S5. Base‐pairing of amiRNAs and Brachypodium target mRNAs.
Figure S6. Plant height and seed length analyses in Brachypodium T0 transgenic plants expressing amiR‐BdBri1 from authentic OsMIR390 or chimeric OsMIR390‐AtL precursors.
Figure S7. Quantification of amiR‐BdCao‐induced phenotype in Brachypodium 35S:OsMIR390‐AtL‐Cao, 35S:OsMIR390‐Cao and 35S:GUS T0 transgenic lines.
Figure S8. Comparative analyses of the accumulation and processing of several amiRNAs derived from AtMIR390a, AtMIR390a‐OsL, OsMIR390 and OsMIR390‐AtL precursors in Nicotiana benthamiana leaves.
Figure S9. Base‐pairing of amiRNAs and Arabidopsis target transcripts.
Figure S10. Functionality in Arabidopsis T1 transgenic plants of amiRNAs derived from AtMIR390a‐based chimeric precursors containing Oryza sativa distal stem‐loop sequences (AtMIR390a‐OsL).
Figure S11. AmiRNA‐induced phenotype quantification in Arabidopsis transgenic plants expressing amiR‐AtFt (left) and amiR‐AtCh42 (right) from AtMIR390a or chimeric AtMIR390a‐OsL precursors.
Figure S12. Target accumulation determined by RNA‐Seq analysis in transgenic Brachypodium plants including 35S:OsMIR390‐AtL‐based or 35S:GUS constructs.
Table S1. MiRbase locus identifiers of Orzya sativa conserved MIRNA precursors.
Table S2. MiRbase locus identifiers of plant MIR390 precursors.
Table S3. AmiRNA phenotypic penetrance in Brachypodium T0 transgenic plants.
Table S4. AmiRNA phenotypic penetrance in Brachypodium T1 transgenic plants.
Table S5. AmiRNA phenotypic penetrance in Arabidopsis T1 transgenic plants.
Table S6. AmiRNA phenotypic penetrance in Arabidopsis T2 transgenic plants.
Table S7. DNA, LNA and RNA oligonucleotides.
Table S8. Sequences and predicted targets for all amiRNAs analyzed.
Table S9. High‐throughput small RNA libraries from Arabidopsis, Brachypodium or Nicotiana benthamiana plants.
Table S10. High‐throughput strand‐specific transcript RNA libraries from independent Brachypodium T0 transgenic lines.
Data S1A. Differential gene expression analysis between 35S:GUS and 35S:OsMIR390‐AtL‐Bri1 Brachypodium samples.
Data S1B. Differential gene expression analysis between 35S:GUS and 35S:OsMIR390‐AtL‐Cad1 Brachypodium samples.
Data S1C. Differential gene expression analysis between 35S:GUS and 35S:OsMIR390‐AtL‐Cao Brachypodium samples.
Data S1D. Differential gene expression analysis between 35S:GUS and 35S:OsMIR390‐AtL‐Spl11 Brachypodium samples.
Data S2. Gene counts in RNA‐Seq libraries from 35S:GUS, 35S:0sMIR390‐AtL‐Bri1, 35S:OsMIR390‐AtL‐Cad1, 35S:0sMIR390‐AtL‐Cao and 35S:OsMIR390‐AtL‐Spl11 transgenic Brachypodium lines.
Data S3A. amiR‐BdBri1 predicted off‐targets differentially underexpressed in 35S:OsMIR390‐AtL‐Bri1 transgenic Brachypodium plants.
Data S3B. amiR‐BdCad1 predicted off‐targets differentially underexpressed in 35S:OsMIR390‐AtL‐Cad1 transgenic Brachypodium plants.
Data S3C. amiR‐BdCao predicted off‐targets differentially underexpressed in 35S:OsMIR390‐AtL‐Cao transgenic Brachypodium plants.
Data S3D. amiR‐BdSpl11 predicted off‐targets differentially underexpressed in 35S:OsMIR390‐AtL‐Spl11 transgenic Brachypodium plants.
Appendix S1. Characterization of AtMIR390a‐OsL‐based amiRNAs in eudicots.
Appendix S2. DNA sequence of B/c vectors used for direct cloning of amiRNAs in zero‐background vectors containing the OsMIR390 sequence.
Appendix S3. FASTA sequences of all amiRNA‐producing MIRNA precursors analyzed.
Appendix S4. Protocol to clone amiRNAs in BsaI/ccdB‐based (‘B/c’) vectors including the OsMIR390 precursor.
Acknowledgements
We thank Goretti Nguyen, Robyn Stevens, Jacob Mreen, Fangfang Ma and Madison Schniers for invaluable technical assistance, and Zacchery R. Smith for his initial contribution to develop the pH7WG2B‐OsMIR390‐B/c vector. Noah Fahlgren was supported by a USDA AFRI NIFA Postdoctoral Fellowship (MOW‐2012‐01361). This work was supported by grants from the National Science Foundation (MCB‐1231726, MCB‐1330562) and National Institutes of Health (AI043288) to James C. Carrington, and from the Department of Energy (DOE DE‐SC0006627) to Todd C. Mockler.
References
- Addo‐Quaye, C. , Eshoo, T.W. , Bartel, D.P. and Axtell, M.J. (2008) Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 18, 758–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez, J.P. , Pekker, I. , Goldshmidt, A. , Blum, E. , Amsellem, Z. and Eshed, Y. (2006) Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell, 18, 1134–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arikit, S. , Zhai, J. and Meyers, B.C. (2013) Biogenesis and function of rice small RNAs from non‐coding RNA precursors. Curr. Opin. Plant Biol. 16, 170–179. [DOI] [PubMed] [Google Scholar]
- Axtell, M.J. (2013) Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 64, 137–159. [DOI] [PubMed] [Google Scholar]
- Axtell, M.J. (2014) Butter: high‐precision genomic alignment of small RNA‐Seq data. bioRxiv, doi.org/10.1101/007427. [Google Scholar]
- Axtell, M.J. , Jan, C. , Rajagopalan, R. and Bartel, D.P. (2006) A two‐hit trigger for siRNA biogenesis in plants. Cell, 127, 565–577. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116, 281–297. [DOI] [PubMed] [Google Scholar]
- Bernard, P. and Couturier, M. (1992) Cell killing by the F plasmid CcdB protein involves poisoning of DNA‐topoisomerase II complexes. J. Mol. Biol. 226, 735–745. [DOI] [PubMed] [Google Scholar]
- Bologna, N.G. and Voinnet, O. (2014) The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu. Rev. Plant Biol. 65, 473–503. [DOI] [PubMed] [Google Scholar]
- Bouvier d'Yvoire, M. , Bouchabke‐Coussa, O. , Voorend, W. et al. (2013) Disrupting the cinnamyl alcohol dehydrogenase 1 gene (BdCAD1) leads to altered lignification and improved saccharification in Brachypodium distachyon . Plant J. 73, 496–508. [DOI] [PubMed] [Google Scholar]
- Butardo, V.M. , Fitzgerald, M.A. , Bird, A.R. et al. (2011) Impact of down‐regulation of starch branching enzyme IIb in rice by artificial microRNA‐ and hairpin RNA‐mediated RNA silencing. J. Exp. Bot. 62, 4927–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell, A. , Fahlgren, N. , Garcia‐Ruiz, H. , Gilbert, K.B. , Montgomery, T.A. , Nguyen, T. , Cuperus, J.T. and Carrington, J.C. (2012) Functional analysis of three Arabidopsis ARGONAUTES using slicer‐defective mutants. Plant Cell, 24, 3613–3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell, A. , Takeda, A. , Fahlgren, N. , Johnson, S.C. , Cuperus, J.T. and Carrington, J.C. (2014) New generation of artificial microRNA and synthetic trans‐acting small interfering RNA vectors for efficient gene silencing in Arabidopsis. Plant Physiol. 165, 15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. , Jiang, S. , Zheng, J. and Lin, Y. (2012a) Improving panicle exsertion of rice cytoplasmic male sterile line by combination of artificial microRNA and artificial target mimic. Plant Biotechnol. J. 11, 336–343. [DOI] [PubMed] [Google Scholar]
- Chen, M. , Wei, X. , Shao, G. , Tang, S. , Luo, J. and Hu, P. (2012b) Fragrance of the rice grain achieved via artificial microRNA‐induced down‐regulation ofOsBADH2. Plant Breed. 131, 584–590. [Google Scholar]
- Cuperus, J.T. , Carbonell, A. , Fahlgren, N. , Garcia‐Ruiz, H. , Burke, R.T. , Takeda, A. , Sullivan, C.M. , Gilbert, S.D. , Montgomery, T.A. and Carrington, J.C. (2010) Unique functionality of 22‐nt miRNAs in triggering RDR6‐dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 17, 997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus, J.T. , Fahlgren, N. and Carrington, J.C. (2011) Evolution and functional diversification of MIRNA genes. Plant Cell, 23, 431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo, Y. , Iwakawa, H.O. and Tomari, Y. (2013) Arabidopsis ARGONAUTE7 selects miR390 through multiple checkpoints during RISC assembly. EMBO Rep. 14, 652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren, N. and Carrington, J.C. (2010) miRNA target prediction in plants. Methods Mol. Biol. 592, 51–57. [DOI] [PubMed] [Google Scholar]
- Felippes, F.F. and Weigel, D. (2009) Triggering the formation of tasiRNAs in Arabidopsis thaliana: the role of microRNA miR173. EMBO Rep. 10, 264–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, K.B. , Fahlgren, N. , Kasschau, K.D. , Chapman, E.J. , Carrington, J.C. and Carbonell, A. (2014) Preparation of multiplexed small RNA libraries from plants. Bio Protoc. 4, e1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Y. , Han, Y. , Ma, J. , Wang, H. , Sang, X. and Li, M. (2014) Undesired small RNAs originate from an artificial microRNA precursor in transgenic petunia (Petunia hybrida). PLoS ONE, 9, e98783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, G. , Zhu, X. , Elling, A.A. et al. (2010) Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell, 22, 17–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisel, S.E. , Zhang, Y. , Allen, E. , Guo, L. , Reynolds, T.L. , Yang, X. , Kovalic, D. and Roberts, J.K. (2008) Characterization of unique small RNA populations from rice grain. PLoS ONE, 3, e2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, C. , Kasprzewska, A. , Tennessen, K. , Fernandes, J. , Nan, G.L. , Walbot, V. , Sundaresan, V. , Vance, V. and Bowman, L.H. (2009) Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res. 19, 1429–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi, M. , Inze, D. and Depicker, A. (2002) GATEWAY vectors for Agrobacterium‐mediated plant transformation. Trends Plant Science, 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Kozomara, A. and Griffiths‐Jones, S. (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, G. , He, H. , Li, Y. and Yu, D. (2012) A new strategy for construction of artificial miRNA vectors in Arabidopsis. Planta, 235, 1421–1429. [DOI] [PubMed] [Google Scholar]
- Liu, Q. , Wang, F. and Axtell, M.J. (2014) Analysis of complementarity requirements for plant microRNA targeting using a Nicotiana benthamiana quantitative transient assay. Plant Cell, 26, 741–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I. , Huber, W. and Anders, S. (2014) Moderated estimation of fold change and dispersion for RNA‐Seq data with DESeq2. bioRxiv, doi.org/10.1101/002832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi, S. , Cai, T. , Hu, Y. et al. (2008) Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell, 133, 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery, T.A. , Howell, M.D. , Cuperus, J.T. , Li, D. , Hansen, J.E. , Alexander, A.L. , Chapman, E.J. , Fahlgren, N. , Allen, E. and Carrington, J.C. (2008) Specificity of ARGONAUTE7‐miR390 interaction and dual functionality in TAS3 trans‐acting siRNA formation. Cell, 133, 128–141. [DOI] [PubMed] [Google Scholar]
- Ossowski, S. , Schwab, R. and Weigel, D. (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 53, 674–690. [DOI] [PubMed] [Google Scholar]
- Oster, U. , Tanaka, R. , Tanaka, A. and Rudiger, W. (2000) Cloning and functional expression of the gene encoding the key enzyme for chlorophyll b biosynthesis (CAO) from Arabidopsis thaliana . Plant J. 21, 305–310. [DOI] [PubMed] [Google Scholar]
- Philippar, K. , Geis, T. , Ilkavets, I. , Oster, U. , Schwenkert, S. , Meurer, J. and Soll, J. (2007) Chloroplast biogenesis: the use of mutants to study the etioplast‐chloroplast transition. Proc. Natl Acad. Sci. USA, 104, 678–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport, F. , Khanin, R. , Liang, Y. , Pirun, M. , Krek, A. , Zumbo, P. , Mason, C.E. , Socci, N.D. and Betel, D. (2013) Comprehensive evaluation of differential gene expression analysis methods for RNA‐seq data. Genome Biol. 14, R95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab, R. , Ossowski, S. , Riester, M. , Warthmann, N. and Weigel, D. (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell, 18, 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, A. , Iwasaki, S. , Watanabe, T. , Utsumi, M. and Watanabe, Y. (2008) The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol. 49, 493–500. [DOI] [PubMed] [Google Scholar]
- Tanaka, A. , Ito, H. , Tanaka, R. , Tanaka, N.K. , Yoshida, K. and Okada, K. (1998) Chlorophyll a oxygenase (CAO) is involved in chlorophyll b formation from chlorophyll a . Proc. Natl Acad. Sci. USA, 95, 12719–12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thole, V. , Peraldi, A. , Worland, B. , Nicholson, P. , Doonan, J.H. and Vain, P. (2012) T‐DNA mutagenesis in Brachypodium distachyon . J. Exp. Bot. 63, 567–576. [DOI] [PubMed] [Google Scholar]
- Tiwari, M. , Sharma, D. and Trivedi, P.K. (2014) Artificial microRNA mediated gene silencing in plants: progress and perspectives. Plant Mol. Biol. 86, 1–18. [DOI] [PubMed] [Google Scholar]
- Trabucco, G.M. , Matos, D.A. , Lee, S.J. , Saathoff, A.J. , Priest, H.D. , Mockler, T.C. , Sarath, G. and Hazen, S.P. (2013) Functional characterization of cinnamyl alcohol dehydrogenase and caffeic acid O‐methyltransferase in Brachypodium distachyon . BMC Biotechnol. 13, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, J. and Hill, T. (2008) High‐efficiency Agrobacterium‐mediated transformation of Brachypodium distachyon inbred line Bd21‐3. Plant Cell Rep. 27, 471–478. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Si, Y. , Dedow, L.K. , Shao, Y. , Liu, P. and Brutnell, T.P. (2011) A low‐cost library construction protocol and data analysis pipeline for Illumina‐based strand‐specific multiplex RNA‐seq. PLoS ONE, 6, e26426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warthmann, N. , Chen, H. , Ossowski, S. , Weigel, D. and Herve, P. (2008) Highly specific gene silencing by artificial miRNAs in rice. PLoS ONE, 3, e1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, L.R. , Qu, S. , Bordeos, A. , Yang, C. , Baraoidan, M. , Yan, H. , Xie, Q. , Nahm, B.H. , Leung, H. and Wang, G.L. (2004) Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U‐box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell, 16, 2795–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Niu, D. , Carbonell, A. , Wang, A. , Lee, A. , Tun, V. , Wang, Z. , Carrington, J.C. , Chang, C.E. and Jin, H. (2014) ARGONAUTE PIWI domain and microRNA duplex structure regulate small RNA sorting in Arabidopsis. Nat. Commun. 5, 5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X. , Sunkar, R. , Jin, H. , Zhu, J.K. and Zhang, W. (2009) Genome‐wide identification and analysis of small RNAs originated from natural antisense transcripts in Oryza sativa . Genome Res. 19, 70–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Q.H. , Spriggs, A. , Matthew, L. , Fan, L. , Kennedy, G. , Gubler, F. and Helliwell, C. (2008) A diverse set of microRNAs and microRNA‐like small RNAs in developing rice grains. Genome Res. 18, 1456–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, H. , Hu, F. , Wang, R. , Zhou, X. , Sze, S.H. , Liou, L.W. , Barefoot, A. , Dickman, M. and Zhang, X. (2011) Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell, 145, 242–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.Y. , Sae‐Seaw, J. and Wang, Z.Y. (2013) Brassinosteroid signalling. Development, 140, 1615–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. OsMIR390‐B/c vectors for direct cloning of amiRNAs.
Figure S2. Generation of constructs to express amiRNAs from authentic OsMIR390 precursors.
Figure S3. Generation of constructs to express amiRNAs from chimeric OsMIR390‐AtL precursors.
Figure S4. Generation of constructs to express amiRNAs from chimeric AtMIR390a‐OsL precursors.
Figure S5. Base‐pairing of amiRNAs and Brachypodium target mRNAs.
Figure S6. Plant height and seed length analyses in Brachypodium T0 transgenic plants expressing amiR‐BdBri1 from authentic OsMIR390 or chimeric OsMIR390‐AtL precursors.
Figure S7. Quantification of amiR‐BdCao‐induced phenotype in Brachypodium 35S:OsMIR390‐AtL‐Cao, 35S:OsMIR390‐Cao and 35S:GUS T0 transgenic lines.
Figure S8. Comparative analyses of the accumulation and processing of several amiRNAs derived from AtMIR390a, AtMIR390a‐OsL, OsMIR390 and OsMIR390‐AtL precursors in Nicotiana benthamiana leaves.
Figure S9. Base‐pairing of amiRNAs and Arabidopsis target transcripts.
Figure S10. Functionality in Arabidopsis T1 transgenic plants of amiRNAs derived from AtMIR390a‐based chimeric precursors containing Oryza sativa distal stem‐loop sequences (AtMIR390a‐OsL).
Figure S11. AmiRNA‐induced phenotype quantification in Arabidopsis transgenic plants expressing amiR‐AtFt (left) and amiR‐AtCh42 (right) from AtMIR390a or chimeric AtMIR390a‐OsL precursors.
Figure S12. Target accumulation determined by RNA‐Seq analysis in transgenic Brachypodium plants including 35S:OsMIR390‐AtL‐based or 35S:GUS constructs.
Table S1. MiRbase locus identifiers of Orzya sativa conserved MIRNA precursors.
Table S2. MiRbase locus identifiers of plant MIR390 precursors.
Table S3. AmiRNA phenotypic penetrance in Brachypodium T0 transgenic plants.
Table S4. AmiRNA phenotypic penetrance in Brachypodium T1 transgenic plants.
Table S5. AmiRNA phenotypic penetrance in Arabidopsis T1 transgenic plants.
Table S6. AmiRNA phenotypic penetrance in Arabidopsis T2 transgenic plants.
Table S7. DNA, LNA and RNA oligonucleotides.
Table S8. Sequences and predicted targets for all amiRNAs analyzed.
Table S9. High‐throughput small RNA libraries from Arabidopsis, Brachypodium or Nicotiana benthamiana plants.
Table S10. High‐throughput strand‐specific transcript RNA libraries from independent Brachypodium T0 transgenic lines.
Data S1A. Differential gene expression analysis between 35S:GUS and 35S:OsMIR390‐AtL‐Bri1 Brachypodium samples.
Data S1B. Differential gene expression analysis between 35S:GUS and 35S:OsMIR390‐AtL‐Cad1 Brachypodium samples.
Data S1C. Differential gene expression analysis between 35S:GUS and 35S:OsMIR390‐AtL‐Cao Brachypodium samples.
Data S1D. Differential gene expression analysis between 35S:GUS and 35S:OsMIR390‐AtL‐Spl11 Brachypodium samples.
Data S2. Gene counts in RNA‐Seq libraries from 35S:GUS, 35S:0sMIR390‐AtL‐Bri1, 35S:OsMIR390‐AtL‐Cad1, 35S:0sMIR390‐AtL‐Cao and 35S:OsMIR390‐AtL‐Spl11 transgenic Brachypodium lines.
Data S3A. amiR‐BdBri1 predicted off‐targets differentially underexpressed in 35S:OsMIR390‐AtL‐Bri1 transgenic Brachypodium plants.
Data S3B. amiR‐BdCad1 predicted off‐targets differentially underexpressed in 35S:OsMIR390‐AtL‐Cad1 transgenic Brachypodium plants.
Data S3C. amiR‐BdCao predicted off‐targets differentially underexpressed in 35S:OsMIR390‐AtL‐Cao transgenic Brachypodium plants.
Data S3D. amiR‐BdSpl11 predicted off‐targets differentially underexpressed in 35S:OsMIR390‐AtL‐Spl11 transgenic Brachypodium plants.
Appendix S1. Characterization of AtMIR390a‐OsL‐based amiRNAs in eudicots.
Appendix S2. DNA sequence of B/c vectors used for direct cloning of amiRNAs in zero‐background vectors containing the OsMIR390 sequence.
Appendix S3. FASTA sequences of all amiRNA‐producing MIRNA precursors analyzed.
Appendix S4. Protocol to clone amiRNAs in BsaI/ccdB‐based (‘B/c’) vectors including the OsMIR390 precursor.


