Abstract
Point-of-care (POC) diagnostic platforms have the potential to enable low-cost, large-scale screening. As no single biomarker is shed by all ovarian cancers, multiplexed biomarker panels promise improved sensitivity and specificity to address the unmet need for early detection of ovarian cancer. We have configured the programmable bio-nano-chip (p-BNC) - a multiplexable, microfluidic, modular platform - to quantify a novel multimarker panel comprising CA125, HE4, MMP-7 and CA72-4. The p-BNC is a bead-based immunoanalyzer system with a credit-card-sized footprint that integrates automated sample metering, bubble and debris removal, reagent storage and waste disposal, permitting POC analysis. Multiplexed p-BNC immunoassays demonstrated high specificity, low cross-reactivity, low limits of detection suitable for early detection, and a short analysis time of 43 minutes. Day-to-day variability, a critical factor for longitudinally monitoring biomarkers, ranged between 5.4–10.5%, well below the biological variation for all four markers. Biomarker concentrations for 31 late-stage sera correlated well (R2 = 0.71 to 0.93 for various biomarkers) with values obtained on the Luminex® platform. In a 31 patient cohort encompassing early- and late-stage ovarian cancers along with benign and healthy controls, the multiplexed p-BNC panel was able to distinguish cases from controls with 68.7% sensitivity at 80% specificity. Utility for longitudinal biomarker monitoring was demonstrated with pre-diagnostic sera from 2 cases and 4 controls. Taken together, the p-BNC shows strong promise as a diagnostic tool for large-scale screening that takes advantage of faster results and lower costs while leveraging possible improvement in sensitivity and specificity from biomarker panels.
Keywords: Ovarian Cancer, Multiplex, Biomarker, Microfluidic, Point-of-Care
Introduction
In 2013, an estimated 22,000 women in the United States were diagnosed with ovarian cancer and 14,000 women died from the disease (1). While ovarian cancer is not one of the most common forms of cancer, it has one of the highest case-to-fatality rates, possibly due to the lack of distinctive symptoms in the early stages of disease (2). Superior surgical management combined with carboplatin and paclitaxel chemotherapy have improved treatment, but there is still less than a 30% cure rate overall (3). Significantly, when the disease is detected in early stage (stage I), survival rates up to 90% can be achieved (4). However, only 20–25% of cases are diagnosed at an early stage (5). Additionally, there is no screening test currently recommended for the general population at average risk. Given the low prevalence of ovarian cancer in post-menopausal women (1 in 2500), any screening test for ovarian cancer must maintain a high specificity (99.6%) with a sensitivity >75% for pre-clinical disease to achieve a positive predictive value (PPV) of 10% (i.e., 10 operations per case of ovarian cancer detected) (4).
Transvaginal sonography (TVS) has been evaluated for early detection of ovarian cancer, but its poor specificity prompts 30 operations for each case of ovarian cancer diagnosed (6,7). The blood biomarker Cancer Antigen 125 (CA125) has been used in clinics for more than three decades to monitor patient response to treatment and to detect ovarian cancer recurrence (8). Despite the clear correlation of elevated CA125 levels to the growth of many ovarian cancers, clinical utility for early detection remains to be established (5,9). A two-stage screening strategy that utilizes rising CA125 to prompt TVS in a small fraction of women is being evaluated by the United Kingdom Collaborative trial of Ovarian Cancer Screening (UKCTOCS) and a Normal Risk Ovarian Cancer Screening Trial in the United States (10–12). Both trials indicate that no more than three operations will be required to detect each case of ovarian cancer (11,12). While the specificity is adequate, it remains to be seen whether adequate sensitivity can be achieved. Early stage disease has been detected in both trials, but the two-stage screening method’s impact on survival and mortality will not be determined until the conclusion of UKCTOCS trial in 2015. One potential limitation of the initial stage of this strategy is that CA125 is only elevated in sera from 50–60% of women with early stage ovarian cancer at the time of conventional diagnosis and only around 80% of all ovarian cancers express CA125 at the tissue level (5). To overcome the limited sensitivity of CA125, several multimarker panels have been proposed that improve sensitivity compared to CA125 alone (13,14). Improved sensitivity, however, needs to be maintained without compromising the high specificity of CA125, critical to achieve the minimum PPV required for early detection. In addition to detecting cancers that fail to express adequate quantities of CA125, additional biomarkers might detect disease earlier than CA125, conferring greater lead time (15). From an assessment of 96 biomarkers utilizing xMAP® bead-based immunoassay technology, several multi-marker panels were identified for early detection of ovarian cancer (16). The most promising 8 markers of the study were tested further using ELISA and after evaluating sensitivity, specificity and biological variability, a combination of CA125, HE4, MMP-7 and CA72-4 was chosen as the most promising panel with the highest sensitivity and specificity (unpublished data). Each of these biomarkers has been linked to abnormal function of ovarian cancer cells. CA125 has been implicated in cell adhesion (17,18), HE4 in ovarian cancer cell migration and adhesion (19,20), MMP-7 in degrading extracellular matrix proteins in invasion and spread of cancer cells (21,22) and CA72-4 in inducing transformation of cells and tumorigenicity (23).
Multiplexing strategies for protein biomarkers have been created and commercialized with success. The most widely used method is currently the Luminex® flow cytometric bead-based system (24). While currently available multiplexing strategies are capable of analyzing multiple biomarkers concurrently and have been used for biomarker discovery (16), they require significant laboratory infrastructure and in the case of ovarian cancer-specific multimarker assays require an overnight incubation. Microfluidic multiplexing systems have been proposed to overcome these challenges by utilizing the inherent advantages of lower sample and reagent volume (25). Microfluidic strategies are also amenable to a point-of-care (POC) format due to their economical use of sample and reagents, rapid assay times and integration of the systems required for immediate analysis (26). Electrochemical immunoassay strategies in the microfluidic regime have been applied to multiplexing ovarian cancer biomarkers and have demonstrated good analytical performance, but long incubation times (10 hours) preclude the use of this method at the POC (27). Another proposed POC platform for ovarian cancer is incapable of multiplexing and suffers from a limited dynamic range despite 1 hour analysis times (28).
Multiplexing biomarkers in a rapid, POC diagnostic test that can quantitate the clinically relevant range of values has the potential to revolutionize the use of biomarkers in clinical practice. In the last decade, our laboratory has continued to evolve the programmable bio-nano-chip (p-BNC) for POC diagnostics (29,30). The programmability of the agarose bead sensors combined with the customizable array allows for a wide-range of multiplexed panels for disease detection or overall health classification (31,32). The agarose bead-based immunoanalyzers are made in-house and incorporated in credit-card sized disposable cards that integrate multiple on-card processes required for analysis, including waste-containment, and are designed to interact with an in-house, in-development portable reader containing all fluid actuation, optics and analysis software to rapidly quantitate protein biomarker levels, permitting analysis at the POC (33).
A POC diagnostic platform such as the p-BNC that can quantify multiplex biomarker panels may aid the first-line of screening for ovarian cancer and permit immediate TVS without the need to schedule a second appointment. Such a strategy may enable rapid and often more cost-effective means of analysis for large-scale screening methodologies, bypassing the need to transfer samples to a remote clinical laboratory (34). By decreasing the lag-time between sample analysis, results, and possible referral to TVS, fewer patient visits will be required, reducing anxiety, travel time and associated costs. These projected reduced costs have the potential to improve screening methods such as Risk of Ovarian Cancer Algorithm (ROCA) by making three month screenings more feasible (35,36).
Similar to ROCA, levels of multiple biomarkers can be monitored over time to account for permanently elevated readings from unrelated conditions rather than using a strict cut-off value to indicate risk of disease. A custom baseline for each biomarker is created for individual patients rather than using an average, effectively creating a personalized cut-off range that when breached indicates elevated risk (37). The potential improvement of using time-point data for all four biomarkers in a single algorithm is still being investigated with UKCTOCS patient samples at MD Anderson Cancer Center, but proteins other than CA125 may be elevated earlier in certain types of ovarian cancers, demonstrating another advantage of a multiplexable screening method. While longitudinally monitoring values has shown promise in helping with early detection, additional attention must be paid to the precision of the diagnostic system. Here, the assay/system variations must be significantly lower than the biomarker variations within and between individuals to permit longitudinal monitoring.
In this study, we program the p-BNC for high sensitivity, low cross-reactivity multiplex analysis of a biomarker panel comprising CA125, HE4, MMP-7 and CA72-4. We explore the analytical performance of the system and demonstrate clinical application of the multiplex p-BNC for ovarian cancer sera of various stages along with healthy controls. We further explore the utility of the multiplexed p-BNC to assess longitudinal pre-clinical sera of women that develop ovarian cancer.
Materials and Methods
Immunoreagents preparation
All reagents (capture and detection antibodies, antigen) utilized in p-BNC immunoassays were prepared with SuperBlock (PBS) Blocking Buffer (Thermo Fisher Scientific Inc., Waltham, MA) to limit nonspecific binding and enhance reagent stability. To develop heterologous double determinant immunoassays in the p-BNC format, murine monoclonal anti-human CA125 (clone M11), anti-human HE4 (clone 2H5) and anti-human CA72-4 (clone CC49) antibodies were utilized to capture CA125, HE4, and CA72-4 respectively. For detection of captured ligands, anti-CA125 (clone OC125), anti-HE4 (clone 3D8) and anti-CA72-4 (clone B72.3) murine monoclonal antibodies were employed. Each of these reagents was generously supplied by Fujirebio Diagnostics, Inc., Malvern, PA. To measure MMP-7, murine monoclonal anti-MMP-7 (clone 111433) was used to capture the ligand and goat anti-MMP-7 polyclonal antibody was used for detection. Anti-MMP-7 antibodies were purchased from R&D Systems, Inc., Minneapolis, MN. Capture antibodies were anchored on glyoxylated agarose microspheres manufactured in our laboratory using previously reported reductive amination protocols at a concentration of 320ng per agarose bead sensor (32). Primary amino groups of detecting antibodies were conjugated to TFP ester moieties of appropriate fluorophores – AlexaFluor® 488 (for HE4, MMP-7 and CA72-4 antibodies) and Oregon Green® 488 (for CA125 antibody) to form stable dye-antibody conjugates (Thermo Fisher Scientific Inc., Waltham, MA). Immunoassay standards for calibration curves were prepared by generating appropriate dilutions using purified protein antigen stocks for CA125, HE4 and CA72-4 (Fujirebio, Diagnostics Inc., Malvern, PA) and EIA antigen standard for MMP-7 (R&D Systems Inc., Minneapolis, MN).
Programmable bio-nano-chip card fabrication
The p-BNC was constructed with alternate layers of 3M™ 9500PC double-sided adhesive (3M Company, St. Paul, MN) and 3M™ AF4300 polyethylene terephthalate (3M Company, St. Paul, MN), which were patterned xurographically with a SummaCut D75 (Summa Inc., Seattle, WA) and a Graphtec FC2250 plotter cutter (Graphtec America, Inc., Irvine, CA), respectively. The resulting 7-layer card featured a network of microfluidic channels for sample delivery and metering, detecting antibody reconstitution and delivery, and washing. Air vents were built into the card using hydrophobic SurePVDF membranes (EMD Millipore, Billerica, MA) to mitigate bubbles and an 8μm Whatman Nuclepore Track-Etch membrane (GE Healthcare, Fairfield, CT) was used for in-line filtration. Glass fiber conjugate pads (EMD Millipore, Billerica, MA) cut into 2×15mm rectangles were used to store detecting antibody in the card and mixers were patterned into the polyethylene terephthalate layer’s channels with the Graphtec plotter cutter to ensure homogenous mixing and consistent delivery of fluids across bead array. The 4×5 array, designed with hexagon-shaped wells to localize individual agarose bead sensors, was cast in an UV-curable photopolymer (Norland Products Inc., Township, NJ) from a custom machined aluminum mold. The array was embedded in the card and was sealed with a cyclo olefin polymer thermoplastic plastic cover (Zeon Corp., Tokyo, Japan) to permit optical access following functionalization with agarose beads on which the sandwich immunoassays were completed. Design rationale and engineering optimization that led to the final design will be reported elsewhere.
Assay execution and image analysis
All assays were performed using the in-house xurography-patterned microfluidic card as described in the previous section, using NE-1000 syringe pumps (New Era Pump Systems Inc., Farmingdale, NY) to variably control fluid flow. A volume of 12μL of detecting antibody cocktail containing 0.15μg of CA125, 0.15μg of HE4, 0.6μg of CA72-4 and 0.015μg of MMP-7 detecting antibodies prepared in SuperBlock (PBS) Blocking Buffer was deposited onto the glass fiber pad. Agarose beads functionalized with capture antibody specific to the analyte of interest (CA125, HE4, MMP-7 or CA72-4) were localized to predetermined wells to permit spatial identification and consequently multiplexing. Four redundant sensor beads per analyte permitted elimination of agarose beads with visual signal obstruction and enabled evaluation of within-assay variance. In addition to the bead sensors for analytes, two negative control beads coupled with IgGs for irrelevant analytes (IL1-β/cTnI antibodies) and two positive control beads (goat anti-mouse immunoglobulin G (IgG) antibodies) were localized to the wells. Following localization of functionalized beads and corresponding controls, 100μL of sample was loaded into each card through the sample entry port, which was then sealed using a double-sided adhesive cover. For clinical samples, 50μL of sample was diluted with 50μL of SuperBlock (PBS) Buffer and the subsequent 100μL was loaded into the card for analysis. Two syringes of SuperBlock (PBS) Buffer were connected to the two main entry ports of the card to supply pressure driven flow inside the card. By actuating the first pump, the 100μL sample was filtered in-card and passed over and through the beads for a period of 30 minutes. After a 1-minute rinse at 100μL/min to wash unbound antigen, the detecting antibody was eluted from the pad and introduced to the bead array at the rate of 10μL/min with the second syringe. After 7 minutes, a final rinse of 100μL/min for 5 minutes was utilized to wash unbound detecting antibody. The total assay time was 43 minutes with a total assay volume of 770μL.
Images were acquired using 10X magnification on a modified Olympus (Tokyo, Japan) BXFM epifluorescent microscope and analyzed using ImageJ (NIH) with previously described custom image analysis macros to determine the mean fluorescence intensity along the perimeter of the bead (38). Optical obstruction of the agarose sensor beads via bubbles or sample/reagent debris resulted in the rejection of the bead sensor for quantitative analysis.
Calibration curves were generated and fitted to a standard four-parameter logistic regression with SigmaPlot 12.0 (Systat Software, Inc., San Jose, CA) software. Unknown concentrations for samples were interpreted from the standard curve utilizing the same software. Microsoft Excel (Microsoft Corp., Redmond, WA) was used to analyze precision study data and MedCalc (MedCalc Software, Ostend Belgium) was used to generate receiver operating characteristics curves (ROC) and box plots for clinical sample concentrations.
Precision study
Human serum-based Liquichek™ Tumor Marker Controls (Bio-Rad Laboratories, Hercules, CA) of three different concentrations (low, medium and high range of each analyte) were assessed on the p-BNC in triplicate over three days to assess within-day and between-day variation of the multiplexed assay on the p-BNC. CA125 is included in the manufacturer’s preparation of the controls and corresponding antigen levels of HE4, CA72-4 and MMP-7 were spiked in each of the three levels (concentrations reported in Supplementary Information Table 1). The intra-assay variation was defined as the variation between redundant bead sensors in a single assay run, whereas the inter-assay precision was defined as the coefficient of variation between different runs over separate days. The coefficient of variation was averaged across the three concentration levels to report the final inter- and intra-assay precision.
Clinical samples
Plasma and serum samples were collected and stored at −80°C at the MD Anderson Cancer Center (Houston, TX) gynecologic oncology tumor bank following standard IRB-approved protocols. Three independent banked sample sets were used in this study, totaling 95 serum samples from 68 patients. For method validation, sera from 31 advanced stage (III–IV) patients were assayed. To evaluate the clinical performance of the p-BNC multiplexed system, sera from healthy individuals (n=7), patients with benign gynecological conditions (n=8), patients with early-stage ovarian cancer (n=7), and patients with late-stage ovarian cancer (n=9) were assayed. To test the potential of the p-BNC to assay sequential samples from the same women, plasma samples were assayed that had been collected at multiple (2–12) time points from women who had remained healthy (n=4) and who had developed ovarian cancer (n=2) during the course of the study. Samples were acquired from the MD Anderson Normal Risk Ovarian Cancer Study (NROS) Study Serum Bank (12). All samples were thawed at 4°C before use and diluted two-fold in SuperBlock (PBS) Buffer prior to analysis on the p-BNC. For method validation, sera were measured on custom multiplex assays for CA125, HE4, MMP-7 and CA72-4 (with CA125 and MMP-7 assays from EMD Millipore (Billerica, MA) multiplexed with CA72-4 and HE4 assays developed in-house) at MD Anderson Cancer Center on the Luminex MAGPIX® magnetic bead-based immunoassay system.
Results
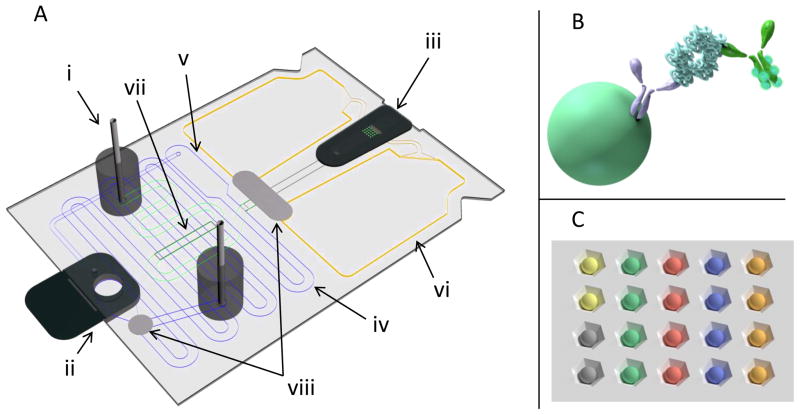
The p-BNC assay card (Fig. 1) is a lab-on-a-chip platform that features a bead-based sensor core and a fully integrated microfluidic network that facilitates on-card sample preparation and metering, reagent storage, mixing, bubble and debris removal, and secure waste containment. By design, the p-BNC assay card minimizes benchtop sample and reagent preparation steps and associated laboratory tools and infrastructure, which is critical for POC analysis. Agarose beads represent the sensor core on which the sandwich immunoassays occur for quantification of biomarker analytes (Fig. 1B). The supporting microfluidic environment, coupled with the 3-D structure of the agarose beads permit optimal analyte capture and detection to achieve low limits of detection and short analysis times. Further, due to the pre-defined spatial arrangement of the beads on the chip (Fig. 1C), the p-BNC permits multiplexing of several biomarkers together. Following sample input, the sample is automatically metered on the card and processed to remove bubbles and debris. The buffer reagents push the sample to the beads, permitting analyte molecules to be captured on the immunosensor (antigen delivery) followed by removal of unbound analytes (wash step). The detecting antibody stored on the glass fiber pad is then eluted by the buffer and delivered to the beads to complete the immunoassay sandwich and is followed by a wash step prior to signal capture. All elements of the card, including the footprint were designed in parallel with a portable optical analyzer to ensure rapid translation of the device to a fully portable POC assay system.
Figure 1.
Illustration of [A] disposable p-BNC card with [i] syringe pump flow adapters, [ii] sample entry port and [iii] bead array holder. The [iv] sample loop contains an [v] overflow chamber, ensuring a 100μL dose of sample for each card. The right pump adapter flows buffer into the sample channel, displacing the sample into the bead-holding chip and through the beads into the [vi] waste. The second pump elutes stored detecting antibody off the [vii] glass fiber pad and into the chip. The flow rate of the second pump is increased to remove the unbound reagents and to perform the final rinse for imaging. All fluid pumped into the card passes through [viii] bubble traps to remove bubbles and an 8μM filter to prevent debris from reaching the bead sensors. A molecular schematic of the agarose bead is shown in [B], demonstrating a completed sandwich immunoassay. [C] shows a zoomed-in illustration of the bead array holder with different colors indicating different bead sensor types permitting multiplexing based on spatial identification.
In this work, we configure a four biomarker panel comprising CA125, HE4, MMP-7 and CA 72-4 to the p-BNC. This biomarker panel has been discovered as an optimal panel in distinguishing early stage ovarian cancer patients from corresponding controls resulting from discovery efforts at MD Anderson Cancer Center. As biomarkers are added or removed the p-BNC system can rapidly be re-configured by replacing, adding or removing bead-types in the array, and therefore reduce the time from biomarker discovery to endpoint analysis.
Assay Optimization
To develop POC-suitable assays for the multiplex panel, the assay ranges were defined based on clinical requirements. An assay suitable for early detection biomarkers in a POC setting should be capable of measuring the relevant range of biomarkers in a short period of time. The dynamic range to encompass the early detection and healthy range for individuals for all four biomarkers were defined based on previous work (16). To function in a POC setting, a total assay time of no longer than one hour was set as the upper limit to include sample incubation, detecting antibody delivery and wash steps to remove unbound antigen and detecting antibody. To measure the four biomarkers on the p-BNC within one hour over a dynamic range with small volumes of plasma, extensive optimization was required. The four biomarker assays were developed individually, beginning with the identification of an optimal matched pair. Based on previous work, an optimal concentration of 320 ng/bead of capturing antibody was coupled to the beads to permit high sensitivity assays (30). A 30 minute sample (antigen) incubation time was chosen for delivery to permit adequate analyte capture while maintaining the short assay times required for a POC assay. A 7 minute incubation with detecting antibody was chosen to permit sufficient time to complete the sandwich immunoassay without increasing background. Flow rates were also optimized to permit proper antigen interaction with the capture antibodies (3.3μL/min), adequate delivery time and consistent dosing of detecting antibody (10μL/min), and to assure sufficient washing without dislodging or destroying bead sensors (100μL/min). Detection antibody optimization involved evaluation of different dilutions from undiluted to a 1:40 dilution. Higher dye loading of the detecting antibody than recommended by manufacturer’s protocols (achieved by overnight incubation rather than 1 hour incubation) resulted in significantly increased signal without a corresponding increase in background noise for detection antibodies. This strategy increased the number of fluorophore moieties per antibody and was found to be suitable for CA125 and CA72-4 assays. Higher fluorescent intensity at a given signal was achieved with AlexaFluor® 488 for HE4, CA72-4 and MMP-7 antibodies, whereas Oregon Green® 488 outperformed AlexaFluor® 488 for CA125. In addition, a 10X magnification significantly improved signal-to-noise ratio (SNR) over the previously used 4X magnification for the p-BNC, due to the advantage derived from the higher numerical aperture. Additionally, a background subtraction method was employed in ImageJ to reduce background noise and further improve the SNR. These optimization steps were necessary to achieve the performance required for the individual immunoassays. A combination of these optimization steps resulted in performance suitable for POC time constraints and early detection assay range.
Analytical performance and validation
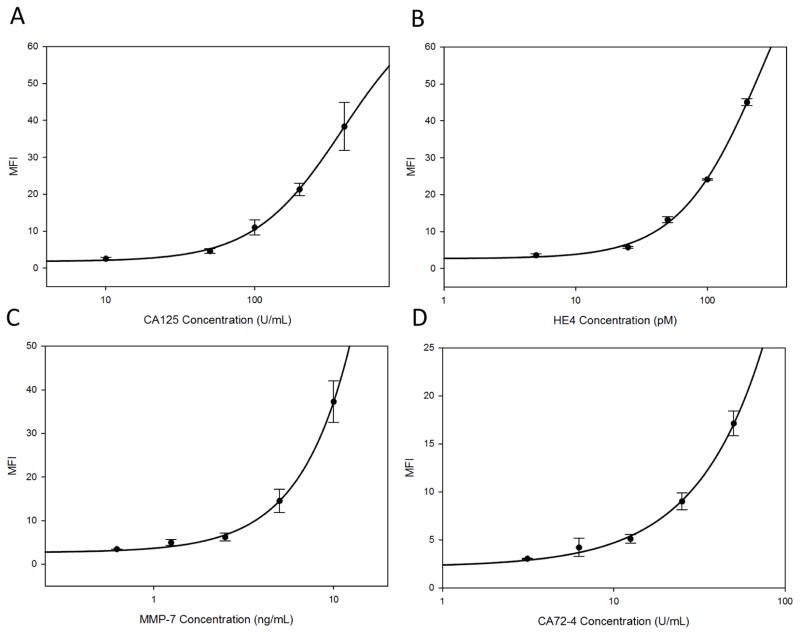
Following optimization as described above, the dose-response curves for the resulting multiplexed immunoassays for each of the four individual markers, fitted to a four-parameter logistic equation are shown in Fig. 2. Standard curves were generated by serial dilution of a standard concentration in a multiplex cocktail of the four antigen standards, encompassing ranges required to cover the biomarker range of interest (healthy women and patients with early stage ovarian cancers): for CA125 0–400U/mL, HE4 0–200pM, CA72-4 0–50U/mL and MMP-7 0–10ng/mL. The multiplexed limits of detection, evaluated as the concentration on the 4-parameter logistical equation corresponding to three standard deviations above zero, for the four markers are shown in Table 1 and displayed in the lower range of each dose response curve in Supplementary Fig. 1. All four assays are capable of reliable measurements below the cut-off values established for healthy individuals and reported in literature: CA125 35U/mL (12), HE4 40–85pM (age-dependent) (39), CA72-4 8.5U/mL (40) and MMP-7 7.4ng/mL (41). Reliable measurements of biomarker concentrations below cut-off levels are essential for establishing baselines for longitudinal biomarker monitoring. The intra-assay precision that measures variation between bead-sensors for a given analyte and the inter-assay precision that measures the variation between independent assays for a given analyte were evaluated for three different concentrations encompassing the dynamic range of the assay and were averaged (Table 1). Intra-assay precision ranged from 5.0% to 9.4%, whereas the inter-assay (conducted on three consecutive days) precision ranged from 5.4% to 10.5% and demonstrate low assay-specific variability. Longitudinal algorithms for biomarkers rely on within-person and between-person biological variation and also the rise in biomarkers levels from the baseline (36). The within- and between-assay precision were both well below the biological variation of the individual markers, determined previously through ELISA (manuscript in preparation).
Figure 2.
Calibration curves showing variation of mean fluorescent intensity (MFI) across concentration ranges tested for [A] CA125, [B] HE4, [C] MMP-7 and [D] CA72-4 obtained on the p-BNC in a multiplexed format and fitted to a four-parameter logistic regression curve. The error bars indicate intra-assay variation measurement between analyte-specific beads. The calibration curves indicate suitability for measurement for low concentrations of biomarkers found in healthy and early disease states with low intra-assay variation across the entire range of measured concentrations. Plots of the lower range of each biomarker dose curve with a linear fit are included in Fig. 1 of Supplementary Information.
Table 1.
Multiplexed limits of detection (LOD) and intra- and inter-assay precision values for CA125, HE4, MMP-7 and CA72-4
| Biomarker | Multiplex LOD | Inter-assay Precision | Intra-assay Precision |
|---|---|---|---|
| CA125 | 1.8 U/mL | 10.5% | 9.4% |
| HE4 | 2.3 pM | 5.4% | 5.0% |
| MMP-7 | 0.2 ng/mL | 8.5% | 8.7% |
| CA72-4 | 1.7 U/mL | 8.3% | 9.1% |
Cross-reactivity
Arguably the most significant challenge with multiplex immunoassays is the presence of cross-reactivity between the detecting antibodies and capturing antibodies for the different analytes. In an ideal multiplexed assay, only the capturing antibody and detecting antibody specific to the analyte of interest will form an immunoassay sandwich, while interactions with non-analyte specific antibodies and antigen will be minimal. However, in reality, several non-ideal interactions are possible between the reagent components of the various multiplex assays (42) that in turn can lead to decrease or increase in specific signal, which may impact the analytical performance of the multiplexed immunoassay in comparison to the singleplex assay. Additionally, cross-reactivity can also occur over the entire dynamic range, precluding multiplexing of the candidates of interest. In order to minimize cross-reactivity, all multiplex reagents were chosen with the other three analytes serving as negative controls. If any signal was observed on the bead-sensors for the non-specific analyte, other matched pair antibody configurations were explored.
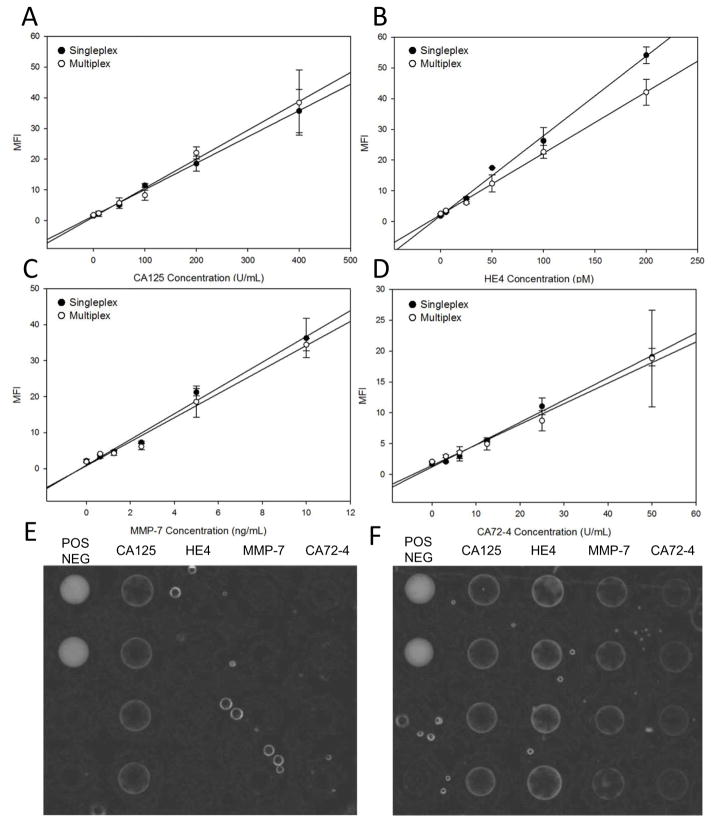
Additionally, while optimizing variables such as flow rates, volumes and detecting antibody dilutions, all four bead-sensors were used to select assay conditions that minimized cross-reactivity. To explore the cross-reactivity across the entire range of analyte concentrations, both the singleplex assays (with the analyte, capturing antibody and detecting antibody of interest) and multiplex assays (with all four analytes and their corresponding capture and detecting antibodies) were completed. The calibration curves for the singleplex and multiplex assays obtained are shown in Fig. 3. The curves obtained from both methods overlap significantly, demonstrating that the signals measured at a given concentration are similar for both singleplex and multiple assays. Less than 10% variation was noted between the singleplex and multiplex dose responses demonstrating negligible cross-reactivity. The four detecting antibodies were combined on a single reagent pad during all multiplexed assays, demonstrating initial compatibility for long-term, in-card storage. Notably, the LODs observed post-multiplexing were comparable to the individual assay LODs, demonstrating that the assay performance was retained upon multiplexing. Thus, reliable measurements may be obtained upon multiplexing without loss of assay performance and with comparable signal intensities and minimal cross-reactivity.
Figure 3.
Calibration curves obtained for [A] CA125, [B] HE4, [C] MMP-7 and [D] CA72-4 in a singleplex (black circles) and multiplex (white circles) format. Akin to ELISA, the singleplex experiments only employed the antigen of interest and the corresponding antibody for the sandwich immunoassay, whereas the multiplex experiments assayed a cocktail of all four antigens with a cocktail of all four detection antibodies. The multiplex assays for each of the four analytes are superimposable with the individual singleplex assays demonstrating negligible cross-reactivity from the introduction of other reagents due to multiplexing. Representative images (contrast enhanced) of a singleplex assay [E] and a multiplex assay [F], where the singleplex demonstrates specific signal for the analyte of interest (CA125) and the multiplex demonstrates specific signals for a cocktail of analytes.
Clinical validation
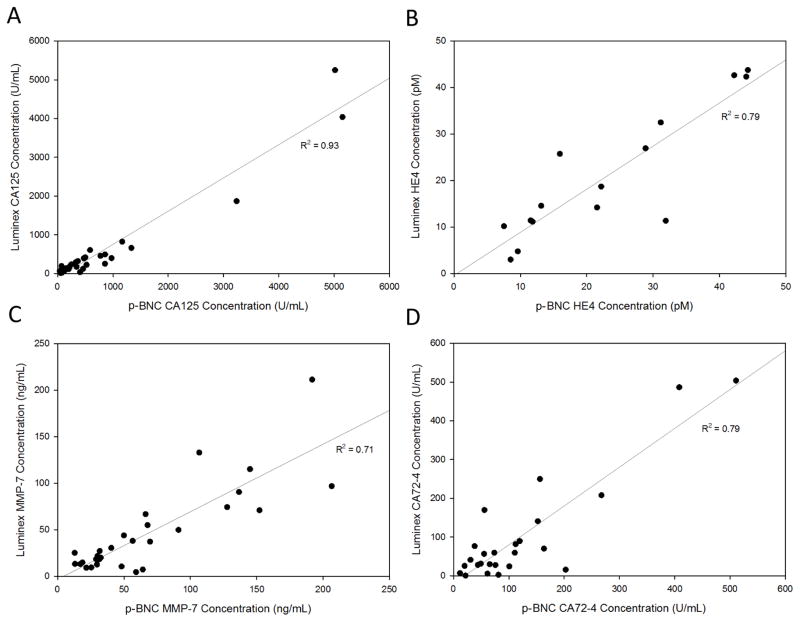
To compare the clinical measurements obtained on the multiplexed p-BNC to laboratory standard multiplex measurements on the Luminex MAGPIX® instrument, we measured sera obtained from 31 late-stage ovarian cancer patients using both methodologies. The method comparison plot (Fig. 4 and Supplementary Fig. 2) shows the concentrations obtained from both methods for a given sample, plotted against each other. The correlation coefficient obtained for all four biomarkers ranged from 0.71 to 0.93, demonstrating good correlation between the two methods.
Figure 4.
Correlation between the concentrations obtained on the p-BNC multiplex assays and Luminex MagPix® multiplex assays for [A] CA125, [B] HE4, [C] MMP-7 and [D] CA72-4 for measurements in the 31 advanced-stage ovarian cancer sera. Plots demonstrate good correlation between the two methods. Plots omitting higher value points and focusing on the lower range of values is included in Fig. 2 of Supplementary Information.
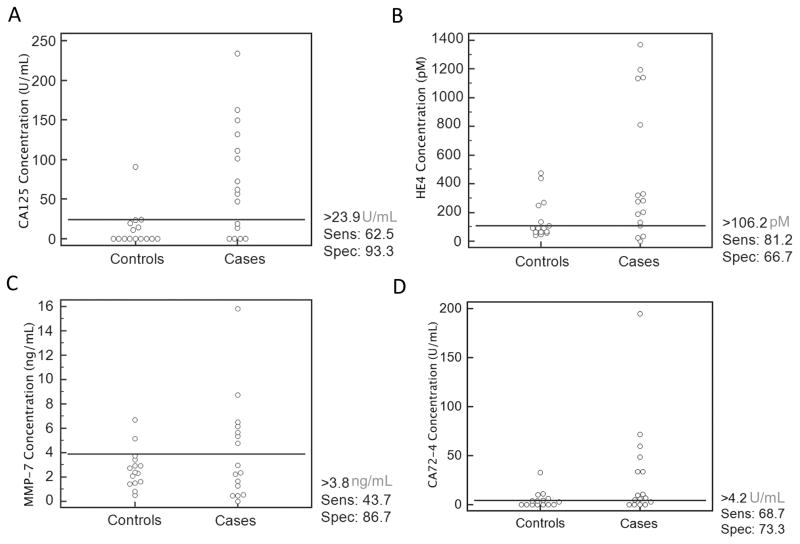
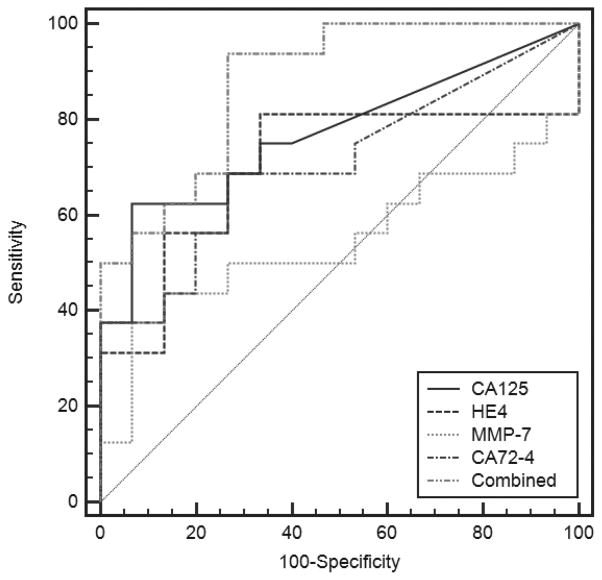
Following method validation, we chose to explore the ability of the multiplexed biomarker panel assessed on the p-BNC to distinguish ovarian cancer patients from patients with non-malignant gynecological conditions and from healthy women. For this purpose, we assayed 31 sera from: 7 healthy women, 8 patients with benign gynecological conditions, 7 patients with early-stage ovarian cancer and 9 patients with late-stage ovarian cancer. The results obtained are shown as dot plots in Fig. 5, displaying the concentrations of individual biomarkers in cases and controls along with suggested cut-off levels for the given cohort of samples. The specificity and sensitivity derived from utilizing these cut-offs are indicated beside the dot plots. The multiplexed concentrations obtained on the p-BNC are capable of distinguishing between cases and controls. Also, the cut-off concentrations for these individual biomarkers obtained on the p-BNC are comparable with previously reported values for these markers (12, 39–41). In addition, the specificities and sensitivities reported for the individual biomarkers for ovarian cancer are concordant with values reported in the literature. The values of all four biomarkers were combined utilizing a logistic regression model utilizing the MedCalc statistical software and the ROC curves for the multi-marker combination and the individual biomarkers are shown in Fig. 6. The multi-marker combination shows improved performance over individual biomarkers alone, performing with a sensitivity of 68.7% at 80.0% specificity, demonstrating the improved sensitivity from using multi-marker panels.
Figure 5.
Dot plots of [A] CA125, [B] HE4, [C] MMP-7 and [D] CA72-4 measured from a set of clinical samples containing healthy and benign patients (controls, n=15) and early-stage and late-stage ovarian cancer patients (cases, n=16). Cut-off levels for each biomarker were determined in MedCalc by achieving highest sensitivity (including as many cases above cut-off as possible) without sacrificing significant specificity (including as many controls below cut-off as possible) and are shown by the horizontal line in each plot demonstrating the clinical performance of the p-BNC to distinguish between various disease states.
Figure 6.
Receiver operating characteristic (ROC) curves for CA125, HE4, MMP-7, CA72-4 and a combination of the four using a logistic regression model. ROC curves were determined from a clinical sample cohort of 31 patients comprising healthy, benign, early-stage and late-stage ovarian cancer patients. Areas under the curve were 0.77 (CA125), 0.70 (HE4), 0.55 (MMP-7), 0.71 (CA72-4) and 0.88 (combined) indicating that the multiplex panel out-performed the individual markers as assessed on the p-BNC.
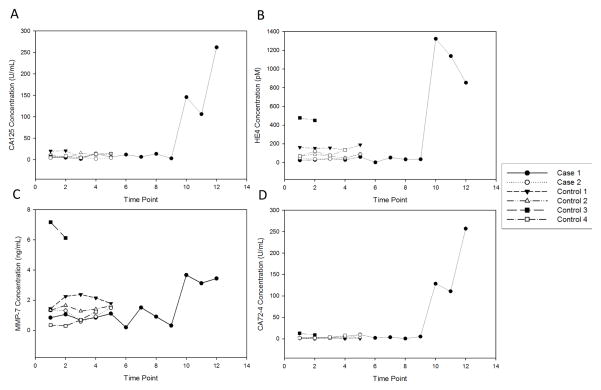
To explore the ability of the p-BNC to perform serial measurements of biomarkers over time, we chose a cohort of six individuals whose sera were collected annually over multiple years. Of these six women, two developed ovarian cancer during the study period while the other four did not develop cancer and were hence deemed healthy. The biomarker profiles of these individuals as obtained on the p-BNC are shown in Fig. 7. For each of these individual biomarkers, the healthy individuals have their own baseline that remained flat during the course of the study. In one of the two cases, the biomarker concentrations rose sharply in comparison to the horizontal plots for the controls. In case 2 (isolated in Fig. 3 of Supplementary Information), CA125 levels did not increase substantially while the other three biomarkers rose prior to diagnosis, demonstrating the importance of additional biomarker information.
Figure 7.
Longitudinal plasma from six women with multiple time-points of sample collection for each patient were measured on the p-BNC and values for [A] CA125, [B] HE4, [C] MMP-7 and [D] CA72-4 are shown over time. Two women (case 1 and 2) developed ovarian cancer and four other women (controls 1-4) did not. The longitudinal profiles show rise in biomarker concentrations for cases in comparison to flat profiles for controls. The time point in years of each measurement is listed in Supplementary Table 2.
While it is premature to determine clinical utility with the small sample cohort assessed, the study nonetheless demonstrates the ability of the p-BNC to reliably measure biomarkers to distinguish disease states based on a multiplex panel and also shows strong promise for serial measurements of longitudinal biomarkers. The four-biomarker panel is being validated with Luminex® in 88 cases and 538 controls from the UKCTOCS trial and will be reported elsewhere.
Discussion
POC diagnostic platforms have the potential to transform clinical diagnostics by significantly reducing the turn-around time between sample analysis and biomarker results that can be interpreted by the clinician. Microfluidic-based POC systems are inherently associated with low sample volumes and analysis times and in the case of a system such as the p-BNC, also permit multiplexing. Unlike traditional ELISAs, where only one biomarker may be assessed, multiplexing offers additional advantages of assessing multiple biomarkers simultaneously, thus reducing required sample volumes and analysis times and allowing incorporation of multimarker signatures. Particularly, in the case of ovarian cancer detection, this additional biomarker information has the potential to improve sensitivity while maintaining specificity. Previously we adapted the p-BNC for measurements of CA125 in sera with analytical performance comparable to ELISAs, but with a shortened analysis time of 43 minutes (30). In this work, we reduced the sample volume 10-fold and incorporated additional biomarkers from a novel ovarian cancer multimarker panel on the p-BNC to permit multiplexing. This multimarker panel, including CA125, HE4, MMP-7 and CA72-4, was chosen based on sensitivity and specificity values, which will be published elsewhere after completion of validation. The p-BNC leverages the microfluidic regime to reduce assay time and cost while enabling multiplexing and providing a path for a POC ovarian cancer screening test.
Analytical performance of the multiplexed assay on the p-BNC demonstrated the ability of the platform to measure a wide range of concentrations of CA125, HE4, MMP-7 and CA72-4 (Fig. 2), including values found in most healthy patient samples. Cross-reactivity between the four assays was found to be minimal, assuring comparable performance between the singleplex and multiplex assays for the individual analytes (Fig. 3). Method validation using a cohort of late-stage ovarian cancer patients showed good correlation for all four markers (R2 > 0.71, Fig. 4) with flow-cytometric bead-based immunoassay system (Luminex®), the current research laboratory standard for multiplex immunoassays. Additionally, a pilot study cohort of healthy, benign, early-stage and late-stage patients assessed on the p-BNC demonstrated the ability of the p-BNC to differentiate cases from controls with acceptable specificity and sensitivity. All patient samples required only 50μL of sample per assay or 12.5μL per analyte.
The longitudinal monitoring of biomarkers could significantly improve diagnostic performance of biomarkers by creating personalized baselines rather than utilizing arbitrary cut-offs that could result in higher false positive rates (43). The cost- and time-reduction of POC tests could benefit a longitudinal monitoring strategy such as ROCA by reducing two patient visits (sample collection and TVS referral) to one and therefore make three month screenings more feasible for patients with rising levels of biomarkers. This strategy can also be applied to high risk patients with familial history of ovarian or breast cancer or with predisposing mutations such as BRCA1/2, demonstrated to need more frequent screenings in the United Kingdom Familial Ovarian Cancer Screening Study (44). The ability to process the patient sample and obtain clinically interpretable results within a single visit will permit TVS on the same visit, avoiding anxiety and additional travel time. A POC test used for longitudinal monitoring requires analytical precision (within- and between-assay) that is much lower than the biological variation of patient samples. Indeed, the p-BNC precision study demonstrated lower analytical %CVs in comparison to the biological variation in healthy individuals for each of these biomarkers. In addition, assessment of longitudinal sera from 2 ovarian cancer patients and 4 healthy controls demonstrated the use of the p-BNC for longitudinal monitoring. Clearly, all four individual biomarkers showed flat baselines in healthy patients and elevated significantly following a period of flat levels in one of the two cases. In the second case with low CA125, additional biomarkers rose indicating the potential of biomarker complementarity permitted by use of a multimarker panel.
The panel used in this manuscript has shown initial promise in early detection of ovarian cancer, but, importantly, the programmability of the p-BNC facilitates replacing, adding or removing biomarkers if better combinations emerge from clinical trials, which is crucial in the diagnosis of such a complex disease as ovarian cancer, where the depth of knowledge of the field is constantly expanding. Other classes of biomarkers such as autoantibodies, nucleic acids or cells may also be assessed on the p-BNC (33), thus permitting the p-BNC to rapidly adapt to the most promising discoveries in the field.
Parallel developments are underway to finalize a portable optical instrument that will fully mobilize the p-BNC assay card and enable analysis at the POC. The portable analyzer will replace the bench-top epifluorescent microscope, syringe pumps, and desktop computer used in this study. Further, progress has been made to translate the p-BNC assay card into a more cost-effective injection-molded format, following best practices in design for manufacturing. The availability of injection-molded disposables in this area has potential to accelerate the translation of the assay system to real world application. Additionally, the consistency resulting from mass manufacturing of injection-molded cards should serve to lower imprecision compared to the in-house made cards used in this study. The assay conditions derived in this work have served in developing the technical specifications for these advancements of the system.
In this study, a POC-amenable microfluidic platform was adapted for rapid analysis of a multiplex biomarker panel with low detection limits and high precision with analysis times shorter than comparable commercial methods. With a short analysis time and inclusion of novel markers for early ovarian cancer detection, this platform shows strong promise as a potential POC screening method for ovarian cancer, where patients could receive results promptly enough to be referred to TVS in the same visit. Reduced costs and easier accessibility to results could also assist in longitudinally monitoring biomarker values over time, which has shown some promise in helping detect early-stage ovarian cancer.
Supplementary Material
Acknowledgments
Financial Support
J. McDevitt acknowledges support from the Cancer Prevention and Research Institute of Texas (CPRIT) (RP101382) and the National Cancer Institute (NCI) Ovarian Cancer SPORE (P50 CA 83639). K. Lu and R. Bast acknowledge support from the MD Anderson CCSG NCI P30 CA16672, the Mossy Foundation, Golfers Against Cancer and Stuart and Gaye Lynn Zarrow.
We would like to thank Fujirebio Diagnostics, Inc. for generously providing antibodies and antigen. We would also like to thank Joseph Celestino for assistance in obtaining clinical samples from the MD Anderson tumor bank.
Footnotes
Disclosure of Potential Conflicts of Interest
R. Bast receives royalties from Fujirebio Diagnostics, Inc. for discovery of CA125 and is a member of the advisory board for Vermillion. J. McDevitt serves as the scientific founder for SensoDX, LLC.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA: A Cancer Journal for Clinicians. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Buys SS, Partridge E, Black A, Johnson CC, Lamerato L, Isaacs C, et al. Effect of Screening on ovarian cancer mortality: The Prostate, Lung, Colorectal and Ovarian (PLCO) cancer screening randomized controlled trial. J Am Med Assoc. 2011;305:2295–303. doi: 10.1001/jama.2011.766. [DOI] [PubMed] [Google Scholar]
- 3.Bast RC, Jr, Skates S, Lokshin A, Moore RG. Differential diagnosis of a pelvic mass. Int J Gynecol Cancer. 2012;22:S5–S8. doi: 10.1097/IGC.0b013e318251c97d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Badgwell D, Bast RC. Early detection of ovarian cancer. Dis Markers. 2007;23:397–410. doi: 10.1155/2007/309382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barakat R, Markman M, Randall ME. Principles and Practice of Gynecologic Oncology. In: Fleming G, Ronnett B, Seidman J, Zaino R, Rubin S, editors. Epithelial Ovarian Cancer. 5. Philadelphia: Lippincott Williams & Wilkins; 2009. pp. 794–795. [Google Scholar]
- 6.Bourne TH, Campbell S, Reynolds KM, Whitehead MI, Hampson J, Royston P, et al. Screening for early familial ovarian cancer with transvaginal ultrasonography and colour blood flow imaging. Brit Med J. 1993;306:1025. doi: 10.1136/bmj.306.6884.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valentin L, Skoog L, Epstein E. Frequency and type of adnexal lesions in autopsy material from postmenopausal women: ultrasound study with histological correlation. Ultrasound Obstet Gynecol. 2003;22(3):284–9. doi: 10.1002/uog.212. [DOI] [PubMed] [Google Scholar]
- 8.Verheijen R, Mensdorff-Pouilly von S. CA 125: fundamental and clinical aspects. Semin Cancer Biol. 1999;9:117–124. doi: 10.1006/scbi.1998.0114. [DOI] [PubMed] [Google Scholar]
- 9.Bast RC, Jr, Brewer M, Zou C, Hernandez MA, Daley M, Ozols R, et al. Prevention and early detection of ovarian cancer: mission impossible? Recent Res Cancer. 2007;147:91–100. doi: 10.1007/978-3-540-37696-5_9. [DOI] [PubMed] [Google Scholar]
- 10.Menon U, Skates S, Lewis S. Prospective study using the risk of ovarian cancer algorithm to screen for ovarian cancer. J Clin Oncol. 2005;23(31):7919–26. doi: 10.1200/JCO.2005.01.6642. [DOI] [PubMed] [Google Scholar]
- 11.Menon U, Gentry-Maharaj A, Hallett R, Ryan A. Sensitivity and specificity of multimodal and ultrasound screening for ovarian cancer, and stage distribution of detected cancers: results of the prevalence screen of the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS) Lancet Oncol. 2009;10(4):327–40. doi: 10.1016/S1470-2045(09)70026-9. [DOI] [PubMed] [Google Scholar]
- 12.Lu KH, Skates S, Hernandez MA, Bedi D, Bevers T, Leeds L, et al. A 2-stage ovarian cancer screening strategy using the Risk of Ovarian Cancer Algorithm (ROCA) identifies early-stage incident cancers and demonstrates high positive predictive value. Cancer. 2013;119(19):3454–61. doi: 10.1002/cncr.28183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gogoi R, Srinivasan S, Fishman DA. Progress in biomarker discovery for diagnostic testing in epithelial ovarian cancer. Expert Rev Mol Diagn. 2006;6:627–37. doi: 10.1586/14737159.6.4.627. [DOI] [PubMed] [Google Scholar]
- 14.Tanyi JL, Scholler N. Oncology biomarkers for gynecologic malignancies. Front Biosci (Elite Ed) 2012;4:1097–110. doi: 10.2741/e444. [DOI] [PubMed] [Google Scholar]
- 15.Anderson GL, McIntosh M, Wu L, Barnett M, Goodman G, Thorpe JD, et al. Assessing lead time of selected ovarian cancer biomarkers: a nested case–control study. J Natl Cancer Inst. 2010;102(1):26–38. doi: 10.1093/jnci/djp438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yurkovetsky Z, Skates S, Lomakin A, Nolen B, Pulsipher T, Modugno F, et al. Development of a multimarker assay for early detection of ovarian cancer. J Clin Oncol. 2010;28:2159–66. doi: 10.1200/JCO.2008.19.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Brien TJ, Beard JB, Underwood LJ, Shigemasa K. The CA 125 Gene: A Newly discovered extension of the glycosylated N-Terminal domain doubles the size of this extracellular superstructure. Tumour Biol. 2002;23:154–69. doi: 10.1159/000064032. [DOI] [PubMed] [Google Scholar]
- 18.O’Brien TJ, Beard JB, Underwood LJ, Dennis RA, Santin AD, York L. The CA 125 Gene: an extracellular superstructure dominated by repeat sequences. Tumour Biol. 2001;22:348–66. doi: 10.1159/000050638. [DOI] [PubMed] [Google Scholar]
- 19.Drapkin R, Horsten von H, Lin Y, Mok S. Human epididymis protein 4 (HE4) is a secreted glycoprotein that is overexpressed by serous and endometrioid ovarian carcinomas. Cancer Res. 2005;65(6):2162–9. doi: 10.1158/0008-5472.CAN-04-3924. [DOI] [PubMed] [Google Scholar]
- 20.Lu R, Sun X, Xiao R, Zhou L, Gao X, Guo L. Human epididymis protein 4 (HE4) plays a key role in ovarian cancer cell adhesion and motility. Biochem Biophys Res Commun. 2012;419:274–80. doi: 10.1016/j.bbrc.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 21.Westermarck J, Kahari VM. Regulation of matrix metalloproteinase expression in tumor invasion. FASEB J. 1999;13(8):781–92. [PubMed] [Google Scholar]
- 22.Sternlicht MD, Werb Z. How matrix metalloproteinases regulate cell behavior. Annu Rev Cell Dev Biol. 2001;17:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kufe DW. Mucins in cancer: function, prognosis and therapy. Nat Rev Cancer. 2009;9:874–85. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vignali D. Multiplexed particle-based flow cytometric assays. J Immunol Methods. 2000;243(1–2):243–55. doi: 10.1016/s0022-1759(00)00238-6. [DOI] [PubMed] [Google Scholar]
- 25.Sorger PK. Microfluidics closes in on point-of-care assays. Nat Biotechnol. 2008;26:1345–6. doi: 10.1038/nbt1208-1345. [DOI] [PubMed] [Google Scholar]
- 26.Soper SA, Brown K, Ellington A, Frazier B, Garcia-Manero G, Gau V, et al. Point-of-care biosensor systems for cancer diagnostics/prognostics. Biosens Bioelectron. 2006;21:1932–42. doi: 10.1016/j.bios.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Wu Y, Xue P, Hui KM, Kang Y. A paper-based microfluidic electrochemical immunodevice integrated with amplification-by-polymerization for the ultrasensitive multiplexed detection of cancer biomarkers. Biosens Bioelectron. 2014;52:180–7. doi: 10.1016/j.bios.2013.08.039. [DOI] [PubMed] [Google Scholar]
- 28.Wang S, Ge L, Yan M, Yu J, Song X, Ge S, et al. 3D microfluidic origami electrochemiluminescence immunodevice for sensitive point-of-care testing of carcinoma antigen 125. Sensor Actuator B-Chem. 2013;176:1–8. [Google Scholar]
- 29.Goodey A, Ali M, Neikirk D, McDevitt J. A microchip-based multianalyte assay system for the assessment of cardiac risk. Anal Chem. 2002;74(13):3030–6. doi: 10.1021/ac011150a. [DOI] [PubMed] [Google Scholar]
- 30.Raamanathan A, Simmons GW, Christodoulides N, Floriano PN, Furmaga WB, Redding SW, et al. Programmable Bio-Nano-Chip systems for serum CA125 quantification: toward ovarian cancer diagnostics at the point-of-care. Cancer Prev Res. 2012;5:706–16. doi: 10.1158/1940-6207.CAPR-11-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weigum S, Floriano P, Christodoulides N, McDevitt J. Cell-based sensor for analysis of EGFR biomarker expression in oral cancer. Lab Chip. 2007;7:995–1003. doi: 10.1039/b703918b. [DOI] [PubMed] [Google Scholar]
- 32.Christodoulides N, Dharshan P, Wong J, Floriano PN, Neikirk D, McDevitt JT. A Microchip-based assay for Interleukin-6. Methods Mol Biol. 2007;385:131–44. doi: 10.1007/978-1-59745-426-1_10. [DOI] [PubMed] [Google Scholar]
- 33.Jokerst J, Jacobson J, Bhagwandin B, Floriano P, Christodoulides N, McDevitt J. Programmable Nano-Bio-Chip sensors: analytical meets clinical. Anal Chem. 2010;82(5):1571–9. doi: 10.1021/ac901743u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rasooly A, Jacobson J. Development of biosensors for cancer clinical testing. Biosens Bioelectron. 2006;21(10):1851–8. doi: 10.1016/j.bios.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 35.Jacobs IJ, Skates S, Davies AP, Woolas RP, Jeyerajah A, Weidemann P, et al. Risk of diagnosis of ovarian cancer after raised serum CA 125 concentration: a prospective cohort study. BMJ. 1996;313:1355–8. doi: 10.1136/bmj.313.7069.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skates SJ, Menon U, MacDonald N, Rosenthal AN, Oram DH, Knapp RC, Jacobs IJ. Calculation of the risk of ovarian cancer from serial CA-125 values for preclinical detection in postmenopausal women. J Clin Oncol. 2003;21:206s–210s. doi: 10.1200/JCO.2003.02.955. [DOI] [PubMed] [Google Scholar]
- 37.Skates S, Jacobs I, Knapp R. Ovarian Cancer. CRC Press; 1998. pp. 187–190. [Google Scholar]
- 38.Jokerst JV, Chou J, Camp JP, Wong J, Lennart A, Pollard AA, et al. Location of biomarkers and reagents within agarose beads of a Programmable Bio-Nano-Chip. Small. 2011;7:613–24. doi: 10.1002/smll.201002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Urban N, Thorpe J, Karlan BY, McIntosh MW, Palomares MR, Daly MB, et al. Interpretation of Single and Serial Measures of HE4 and CA125 in Aysmptomatic Women at High Risk for Ovarian Cancer. Cancer Epidemiol. Biomarkers Prev. 2012;21:2087–94. doi: 10.1158/1055-9965.EPI-12-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yurkovetsky ZR, Linkov FY, Malehorn DE, Lokshin AE. Multiple biomarker panels for early detection of ovarian cancer. Future Oncol. 2006;2:733–41. doi: 10.2217/14796694.2.6.733. [DOI] [PubMed] [Google Scholar]
- 41.Leelawat K, Sakchinabut S, Narong S, Wannaprasert J. Detection of serum MMP-7 and MMP-9 in cholangiocarcinoma patients: evaluation of diagnostic accuracy. BMC Gastroenterol. 2009;9:30. doi: 10.1186/1471-230X-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ellington AA, Kullo IJ, Bailey KR, Klee GG. Antibody-based protein multiplex platforms: technical and operational challenges. Clin Chem. 2010;56:186–93. doi: 10.1373/clinchem.2009.127514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dorigo O, Berek JS. Personalizing CA125 Levels for Ovarian Cancer Screening. Canc Prev Res. 2011;4:1356–9. doi: 10.1158/1940-6207.CAPR-11-0378. [DOI] [PubMed] [Google Scholar]
- 44.Rosenthal AN, Fraser L, Manchanda R, Badman P, Philpott S, Mozersky J, et al. Results of annual screening in phase I of the United Kingdom Familial Ovarian Cancer Screening Study highlight the need for strict adherence to screening schedule. J Clin Oncol. 2013;31:49–57. doi: 10.1200/JCO.2011.39.7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.