Abstract
Candida albicans, a commensal fungus of the oral microbiome, causes oral candidiasis in humans with localized or systemic immune deficiencies. Secreted aspartic proteinases (Saps) are a family of 10 related proteases and are virulence factors due to their proteolytic activity, as well as their roles in adherence and colonization of host tissues. We found that mice infected sublingually with C. albicans cells overexpressing Sap6 (SAP6 OE and a Δsap8 strain) had thicker fungal plaques and more severe oral infection, while infection with the Δsap6 strain was attenuated. These hypervirulent strains had highly aggregative colony structure in vitro and higher secreted proteinase activity; however, the levels of proteinase activity of C. albicans Saps did not uniformly match their abilities to damage cultured oral epithelial cells (SCC-15 cells). Hyphal induction in cells overexpressing Sap6 (SAP6 OE and Δsap8 cells) resulted in formation of large cell-cell aggregates. These aggregates could be produced in germinated wild-type cells by addition of native or heat-inactivated Sap6. Sap6 bound only to germinated cells and increased C. albicans adhesion to oral epithelial cells. The adhesion properties of Sap6 were lost upon deletion of its integrin-binding motif (RGD) and could be inhibited by addition of RGD peptide or anti-integrin antibodies. Thus, Sap6 (but not Sap5) has an alternative novel function in cell-cell aggregation, independent of its proteinase activity, to promote infection and virulence in oral candidiasis.
INTRODUCTION
Candida albicans is a commensal fungus that is often part of the oral microflora of healthy people. Loss of host immunity, HIV infection, corticosteroid use, or alteration of the oral microflora following antibiotic therapies permits a pathogenic transition of C. albicans to cause oropharyngeal candidiasis (OPC) (1, 2). Acute pseudomembranous candidiasis is one of the most common forms of OPC, in which C. albicans forms white patches on the surface of the buccal mucosa, tongue, or soft palate. These superficial fungal plaques can be lifted from underlying tissues for purposes of clinical diagnosis and analysis (3).
C. albicans expresses specific sets of virulence factors that promote hypha formation and adhesion and invasion of host tissues (4). Secreted aspartyl proteinases (Saps) are recognized virulence factors because they degrade host proteins to provide nitrogen for fungal cell metabolism, contribute to adherence, facilitate fungal epithelial and endothelial penetration, and are immunogenic during infection (5–7). Microbial proteinases are classified as serine, cysteine, metallo-, or aspartyl proteinases according to the site of catalytic hydrolysis of substrate peptide bonds; however, C. albicans produces only aspartyl proteinases (5, 6).
C. albicans expresses a family of 10 SAP genes that are clustered into groups SAP1 to SAP3, SAP4 to SAP6, SAP7, SAP8, and SAP9 and SAP10 based upon their sequence homologies and pH activities (8, 9). Sap1 through Sap8 are processed and transported via the secretory pathway to produce released extracellular enzymes, whereas Sap9 and Sap10 are glycosylphosatidylinositol (GPI)-anchored cell proteins. Thus, C. albicans Sap1 to -8 account for all secreted (extracellular) proteinase activity, and they are exclusively aspartyl proteinases (5, 6, 9). Each C. albicans Sap protein has a distinct substrate cleavage site and pH optimum. Sap1 to Sap3 and Sap8 have activity at lower pH values (2.5 to 5.0), whereas Sap4 to Sap6 have better activity at higher pH values (8, 10). C. albicans Sap expression levels and substrate activities are regulated by cell morphotype and environmental cues, so that SAP1 to SAP3 are expressed predominantly in yeast cells, whereas hyphal cells express mainly SAP4 to SAP6. However, hypha-specific secreted Sap4 to -6 were found to have substrate ranges and enzymatic activities similar to those of yeast cell-secreted Sap2, despite having different in vitro activities (5, 11, 12).
The plasticity of Sap secretion profiles and enzymatic activities has created a challenge to understanding the in vivo functions of C. albicans Sap proteins. C. albicans SAP4, SAP5, and SAP6 expression levels were found to be elevated in both mucosal and systemic infections (12, 13). However, cross-sectional studies of C. albicans SAP gene expression in human OPC showed that SAP2, as well as SAP4 to SAP6, were predominantly expressed in OPC patients, as well as Candida carriers (5, 13–16). C. albicans recovered from murine OPC showed that Sap4 to -6 were highly expressed during infection; however, other studies found a role for Sap1 to -6 in fungal invasion and damage to oral and vaginal epithelial mucosal surfaces (5, 14, 16–21). Thus, functional analyses of the abilities of individual Saps to promote virulence in mucosal infection has been inconclusive, due to different expression levels during the course of infection.
In addition to their classical role as proteinases, some studies have pointed to a role of C. albicans Saps in mediating fungal adhesion to and colonization of host tissues. High proteolytic activity of C. albicans was correlated with increased adhesion to human buccal epithelial cells (17, 22) and increased organ (spleen and kidney) colonization in mice (23, 24). However, these studies compared fungal adhesion of C. albicans cells pretreated with pepstatin A (a proteinase inhibitor that specifically inhibits most aspartyl proteinases) rather than using gene deletion mutants. Thus, it is not clear which of the C. albicans Sap family members might have a role in adherence, nor is the mechanism by which they contribute to adhesion to mucosal tissues known. Two hypotheses for how Saps promote fungal adherence to host cells have been proposed. In the first, secreted Saps modify the surfaces of host cells by their proteinase activity to expose proteins that are more favorable ligands for C. albicans binding. Alternatively, fungal cell surface Saps themselves serve as ligands that are able to bind host cells independently of their proteolytic activity (5).
We examined these alternative hypotheses by using a highly virulent C. albicans SAP8 deletion mutant that overexpresses SAP6 to understand the role of Saps in OPC. We determined for the first time that Sap6 functions as a hyphal-morphotype-specific cell-cell adhesion molecule independently of its proteinase activity and that this adhesion is mediated through its RGD motif. These results suggest a new role for hypha-specific fungal aggregation as a virulence factor mediated by Sap6.
MATERIALS AND METHODS
Strains.
C. albicans CAI4 (with URA3 replaced at the RPS1 locus using a Clp10 plasmid) was used as the wild-type (WT) control and in animal experiments. C. albicans Δsap6 was constructed using the URA blaster method (23). Both alleles were disrupted by two cycles of URA blasting using a hisG-URA3-hisG cassette. Each integration and disruption step was verified using PCR. For overexpression of SAP5 and SAP6, full-length gene fragments were cloned into a CIp10 plasmid to obtain pCIp10-SAP5 and pCIp10-SAP6. These plasmids were linearized using the NcoI restriction enzyme and transformed into the CAI4 (URA−) strain. The correct integration and orientation of transformed SAP5 and SAP6 genes within the RPS1 locus were confirmed by PCR. Strain auxotrophy after URA removal using fluoroorotic acid (FOA) was verified. C. albicans Δsap8 (URA+) and Δsap4/5/6 (URA+) were kindly provided by B. Hube (Jena, Germany). All the strains used in this study had similar growth rates in vitro. C. albicans ALS mutant strains (URA+) were provided by S. Filler (University of California—Los Angeles), and C. albicans Δrbt1 (URA+) was provided by A. Johnson (University of California—San Francisco). The strains used are listed in Table 1.
TABLE 1.
C. albicans strains used in this study
| Strain | Description | URA statusa | Reference |
|---|---|---|---|
| CAI4 | Δura3::imm434 Δura3::imm434 RPS1 Δrps1::CIp10-URA3 | + | This study |
| Δsap8 | Δsap8::hisG Δsap8::hisG-URA3-hisG | + | 32 |
| Δsap8/SAP8 | Δsap8::hisG Δsap8::hisG RPS1 Δrps1::CIp10-SAP8-URA3 | + | 32 |
| Δsap4/5/6 | Δsap6::hisG Δsap6::hisGΔsap4::hisG Δsap4::hisG Δsap5::hisG Δsap5::hisG RPS1 Δrps1::CIp10-URA3 | + | 23 |
| Δsap6 | Δsap6::hisG Δsap6::hisG-URA3-hisG | + | This study |
| SAP5 OE | Δura3::imm434 Δura3::imm434 RPS1 Δrps1::CIp10-SAP5-URA3 | + | This study |
| SAP6 OE | Δura3::imm434 Δura3::imm434 RPS1 Δrps1::CIp10-SAP6-URA3 | + | This study |
| Δals1/Δals3 | als1::hisG als1::hisG als3::dpl::200 als3::dpl200 | + | 48 |
| Δrbt1 | Δrbt1::hisG Δrbt1::hisG-URA3-hisG | + | 49 |
+, positive.
Murine model of oral candidiasis.
To determine the virulence of C. albicans strains in oral candidiasis, immunosuppressed mice (C57BL/6J; 6- to 8-week-old females; Jackson Laboratory) were infected sublingually as described previously (25). The animal protocols were approved by the University at Buffalo Institutional Animal Care and Use Committee (ORB06042Y). Ten mice were used in each experimental group, and experiments were repeated at least twice. The mice were immunosuppressed using 225 mg/kg of body weight cortisone 21-acetate (Sigma-Aldrich; C3130-5G) subcutaneously on days −1, +1, and +3 of infection. Anesthetized mice were infected sublingually with cotton balls carrying 1 × 107 fungal cells for 2 h. The mice were monitored daily for weight loss. On the fifth day postinfection, the mice were weighed and sacrificed by cervical dislocation under anesthesia, and the tongues were collected immediately. To quantify levels of Candida infection, one-half of the tongue tissue was weighed, homogenized, and plated on yeast extract-peptone-dextrose (YPD) plates to obtain the number of CFU per gram of tongue tissue. The other half of the tongue was fixed immediately in 10% formalin and embedded in paraffin, and thin sections were stained with periodic acid-Schiff (PAS) stain for histological analysis. Some tongue tissues were stained by terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) at the histology facility at Roswell Park Cancer Institute to detect apoptotic cells. To quantify the fungal load in kidneys, kidneys were removed aseptically, weighed, homogenized, and plated on YPD agar to enumerate the CFU per gram of kidney tissue. For isolation of RNA from fungal tongue plaques, Candida plaques were lifted whole from the underlying tongue tissue and stored immediately in RNAlater (Life Technologies) for further use. For microscopic analysis, fungal plaques removed from the tongue surface were fixed immediately in 10% buffered formalin and stored at 4°C. Statistical differences between groups (n = 10) were determined by Student's t test.
Total RNA isolation and real-time PCR.
To reduce the amount of murine cell RNA in the tongue plaque samples, tongue plaques were resuspended in 1 ml of TRIzol reagent (Life Technologies) for 5 min to dissolve the mammalian cells and centrifuged at 2,000 × g for 5 min. The cell pellet was resuspended in 1 ml of TRIzol reagent and vortexed (4 cycles; 6 m/s) with 0.45-μm glass beads using a FastPrep-24 instrument (MP Biomedicals). Lysed cells (1 ml) were collected, and chloroform (200 μl) was added and then mixed vigorously for 15 s and maintained for 2 to 3 min at room temperature. The cell lysate was centrifuged at 11,500 × g for 10 min at 4°C to separate the RNA-containing upper aqueous layer, which was collected and mixed with 0.5 volume of 100% ethanol to precipitate total RNA from the Candida cells. The total precipitated RNA was further purified using an RNeasy kit from Qiagen according to the manufacturer's instructions. Following isolation, RNA purity and concentrations were determined using an Agilent Bioanalyzer 2100 (Agilent Technologies). cDNA was synthesized for each sample using an iScript cDNA synthesis kit (Bio-Rad) following the manufacturer's instructions, with equal amounts of RNA (2 μg in a 20-μl reaction mixture).
To quantify SAP gene expression, synthesized cDNA (1 μl) was used to amplify transcripts of selected genes using an iCycler iQ real-time PCR detection system (Bio-Rad) as described previously (26). The standard curve generated from cDNA dilutions from each sample was used to quantitate mRNAs for the genes of interest using ACT1 as a normalized control for each condition. The results were expressed as an average of triplicate samples, and differences between experimental groups were evaluated for significance using an unpaired Student t test and analyzed with Prism 5.0 software.
Protein isolation and Western blotting.
Candida plaques collected from mouse tongues were suspended in 1% Triton X-100 for 10 min at room temperature to remove murine cell contamination. The Candida cells were pelleted by centrifugation at 3,000 × g for 5 min and stored at −80°C. For protein extraction, the cell pellets were placed on ice and resuspended in 300 µl 10% trichloroacetic acid (TCA) buffer (10 mM Tris-HCl, pH 8.0, 10% trichloroacetic acid, 25 mM NH4OAc, 1 mM sodium EDTA). Total cellular lysates were isolated by disrupting cells using acid-washed glass beads (as described above). Samples were placed on ice for 5 min between cycles. The beads were removed, and the samples were centrifuged at 4°C for 10 min at 15,000 × g. The supernatant was removed and resuspended in 150 μl of buffer (0.1 M Tris-HCl, pH 11.0, 3% SDS). Samples were boiled for 5 min and then centrifuged at 15,000 × g for 30 s. The normalized protein content (20 μg) was separated by SDS-PAGE on 12% gels and transferred to nitrocellulose membranes. After transfer, the membranes were incubated with primary antibodies at 4°C for 16 h in 5% bovine serum albumin (BSA) buffer (0.5 g BSA, 10 ml Tris-buffered saline with Tween 20 [TBST]), followed by washing with TBST. For Cek1 phosphorylation, anti-phospho-p42/44 MAP kinase (MAPK) (ERK1/2 Thr202/Tyr204) rabbit monoclonal antibody (P-Cek1) (Signaling Technology) was used as the primary antibody. Cek1 protein was used as a loading control and detected by a polyclonal Cek1 antibody (raised against two fragments of Cek1 protein, from amino acids 86 to 101 and 111 to 125, by Genemed Synthesis, Inc.). This Cek1 antibody recognizes Cek1p, as well as its close homologue Cek2p. Goat anti-rabbit IgG-horseradish peroxidase (HRP) (Jackson ImmunoResearch Laboratories, Inc.) was used as the secondary antibody. The membranes were then incubated with secondary antibodies at 25°C for 1 h in blocking buffer, washed, and used for detection using a SuperSignal West Pico detection kit (Thermo Scientific).
Epithelial cell damage assay.
SCC-15 epithelial cells (ATCC CRL-1623) were routinely cultured in Dulbecco's modified Eagle's medium (DMEM)-F12 (Lonza AG) with 10% fetal bovine serum (FBS) (Gibco) at 37°C in 5% CO2 in a CO2 incubator. Epithelial cell damage caused by C. albicans WT and SAP deletion strains was determined by release of lactate dehydrogenase (LDH) from epithelial cells following 24 h of incubation with C. albicans strains using the Cytotoxicity Detection kit (Cayman Chemical). For these assays, 200 μl (1 × 105 cells/ml) of SCC-15 epithelial cells was seeded per well in 96-well tissue culture plates (Corning Inc., USA) and allowed to grow for 24 h to 95% confluence. Confluent epithelial cells were washed twice with Dulbecco's phosphate-buffered saline (PBS) (Gibco), and 100 μl DMEM-F12 with 2% FBS per well was added before infection with C. albicans strains. Cultures of C. albicans WT and SAP deletion strains grown overnight at 30°C in YPD (BD Biosciences) were washed in PBS and diluted to 5 × 105 cells/ml in DMEM-F12 without FBS, and cells (100 μl) were added to each well at a multiplicity of infection (MOI) of 1. The Candida and epithelial cells were coincubated for 24 h at 37°C in 5% CO2, and then the plate was centrifuged at 500 × g for 5 min, and 100 μl of the culture supernatant from each well was collected and measured at A492 using a Bio-Tek plate reader. Uninfected SCC-15 cells in DMEM-F12 and C. albicans alone were used as controls. Uninfected SCC-15 cells in DMEM-F12 supplemented with 1% Triton X-100 for 1 h were used as a positive control. Each experiment was performed at least twice in triplicate, and differences between experimental groups were evaluated for significance using an unpaired Student t test.
Proteinase activity assay.
Culture supernatants from C. albicans planktonic growth cells (yeast nitrogen base [YNB] medium supplemented with 2% glucose at 37°C for 12 h at 220 rpm) were obtained after centrifugation at 4,000 × g for 10 min and filter sterilized by passing them through a 0.22-μm syringe filter. The supernatants from C. albicans 12-h biofilms were collected as described previously (27). Culture supernatants of C. albicans WT and SAP deletion and overexpression strains were tested for their proteolytic activities using a fluorescent-proteinase assay kit (Thermo Scientific) with slight modifications. C. albicans culture supernatants (100 μl) were incubated with 100 μl of fluorescein-labeled casein (FTC) solution (100 μg/ml, prepared in 0.1 M sodium citrate buffer, pH 5.0) at 37°C for 1 h. Digestion of FTC into smaller fragments resulted in loss of fluorescence that was measured at 485/538 nm using a microplate reader. Proteolytic activity in culture supernatants was determined using a standard curve plotted against the concentration of the reference proteinase trypsin and relative fluorescence units (RFU) of the FTC substrate. Each experiment was performed at least twice, and differences between groups were evaluated for significance using an unpaired Student t test.
Recombinant Sap purification and labeling.
Pichia pastoris clones expressing C. albicans Sap5, Sap6, and Sap5 and Sap6 in which both RGD sequences were deleted (Sap5ΔRGD and Sap6ΔRGD) were kindly provided by Michel Monod (Centre Hospitalier Universitaire Vaudois, Lausanne, Switzerland) and Jordan Tang (Oklahoma Medical Research Foundation, Oklahoma City, OK). Recombinant proteins (rSaps) were expressed and purified from P. pastoris as described previously (28, 29). Briefly, culture supernatants from P. pastoris were concentrated 100-fold using a Vivaspin20 (Sartorious) and dialyzed overnight at 4°C against 10 mM sodium citrate buffer (pH 5.0). The dialyzed culture supernatants were loaded on a DEAE Sephadex A-25 column (GE Healthcare), washed with 10 mM sodium citrate buffer, and eluted with 100 mM sodium citrate buffer (pH 5.0). The eluted proteins were then loaded onto a hydroxyapatite (Sigma-Aldrich) column washed with 50 mM sodium phosphate buffer (pH 7.0) and eluted with 150 mM sodium phosphate buffer (pH 7.0). The recombinant proteins (rSap5, rSap6, rSap5ΔRGD, and rSap6ΔRGD) were further concentrated using Amicon centrifugal filter units (MiIlipore) and quantified using a bicinchoninic acid (BCA) protein assay (Thermo Scientific), and their purity was verified by SDS-PAGE. The proteinase activity of each purified recombinant Sap was determined using a fluorescent-proteinase assay as described above. The recombinant Sap proteins were heat inactivated by autoclaving at 120°C for 20 min, and the loss of proteinase activity was confirmed using a fluorescent-proteinase assay. rSap6 was labeled with fluorescein isothiocyanate (FITC) (Sigma-Aldrich), using 50 μl of FITC (1 mg/ml in dimethyl sulfoxide [DMSO]) added to 1 ml of rSap6 (2 mg/ml) buffered in 0.1 M sodium carbonate buffer (pH 9) and incubated overnight at 4°C in the dark. FITC-labeled rSap6 (F-rSap6) was purified using a Sephadex G25 column and stored at 4°C until further use.
Adherence assay.
For adherence, SCC-15 oral epithelial cells were grown to confluence in 24-well tissue culture plates at a cell density of 1 × 105 cells/ml/well. Confluent cells were serum starved overnight prior to experiments. Epithelial monolayers were infected with C. albicans WT and SAP strains prepared as for the cell damage assay described above. In experiments with rSaps, C. albicans cells were preincubated with 10 μM rSap6 or rSap5 for 30 min at 37°C and washed twice with PBS. C. albicans cells were allowed to adhere to epithelial cells for 90 min at 37°C in 5% CO2. Following incubation, nonadherent cells were removed by washing the wells twice with PBS. Adherent Candida cells were collected by lysis of epithelial cells using 1 ml of 0.1% Triton X-100 per well and plated on YPD agar plates in triplicate for quantification of average numbers of CFU. The initial inoculum was also plated in triplicate on YPD agar to obtain the total number of CFU. Percent adhesion was calculated as follows: (average number of adherent CFU/average total number of CFU) × 100. The experiments were performed on two separate occasions. Statistical differences were assessed using Student's t test.
Cell culture and microscopy.
C. albicans cells were cultured overnight in YPD broth, diluted to an optical density at 600 nm (OD600) of 0.3 in prewarmed YNB medium supplemented with 1.25% GlcNAc, and incubated for 3 h at 37°C to induce germination (30). Cells were collected by centrifugation (100 × g), and germination was observed using a Zeiss AxioImager fluorescence microscope. In some experiments, YNB supplemented with 10% fetal bovine serum (Gibco) was used for induction of germination.
Fungal tongue plaques were resuspended in 1 ml 1% Triton X-100 (Sigma-Aldrich) for 10 min in order to dissolve murine epithelial cells and then recovered by centrifugation at 3,000 × g for 5 min. Candida cells were stained with calcofluor white M2R (Sigma Aldrich) and visualized under a microscope at ×20 magnification.
For Sap6 binding experiments, C. albicans cells (yeast and hyphal forms) were fixed with 4% paraformaldehyde, incubated with F-rSap6 (10 μM) for 1 h at 37°C, centrifuged at 3,000 × g for 5 min, and washed twice to separate cells from unbound F-rSap6. Control cells were incubated with FITC alone. The cells were visualized with a Zeiss AxioImager fluorescence microscope at ×63 magnification using the FITC channel.
Microcolony formation was examined as described previously (31). Briefly, for induction of microcolonies, single colonies of C. albicans CAI4 and SAP deletion strains were inoculated and grown overnight in YPD broth at 30°C at 220 rpm, washed twice with PBS, and diluted to 1 × 103 cells/ml in PBS. A total of 100 μl of cells for each strain was inoculated into 500 μl RPMI medium (Gibco) per well of a 24-well cell culture plate (Corning Inc., USA) and incubated at 37°C for 24 h in the presence of 5% CO2. Microcolonies were observed under a Zeiss AxioImager fluorescence inverted microscope at ×20 magnification.
Cell aggregation assays.
To study the effects of Saps on cell-cell aggregation, C. albicans cells were grown overnight in YPD broth, diluted to an OD600 of 0.3, and grown under hypha-inducing conditions (prewarmed YNB medium containing 1.25% GlcNAc or 10% FBS at 37°C) or nongerminating conditions (YNB medium containing 2% glucose at 30°C) for 1 h or 3 h. The C. albicans cultures were divided, and one group was incubated with purified rSaps (rSap5, rSap6, rSap5ΔRGD, and rSap6ΔRGD) at concentrations of 2.5 and 10 μM for 15 min, while the other served as a control. After incubation, the cells were fixed using 4% paraformaldehyde, and images were captured with a Zeiss AxioImager fluorescence microscope. Each experiment was performed in duplicate and repeated on three different days. Cell-cell aggregation was quantified by measuring the average aggregate diameter of 50 aggregates per strain using Axiovision4 software.
For inhibition experiments, 10 μM RGD peptide (Sigma-Aldrich) or an anti-integrin antibody (anti-integrin αM CBR1/5; Santa Cruz Biotechnology) at 1:100 dilution was added to C. albicans cells previously grown for 1 h under hypha-inducing conditions (as described above) and incubated with RGD peptide or anti-integrin αM antibody (IgG1) for another 30 min. C. albicans cells were collected by centrifugation at 4,000 × g for 5 min and washed twice with PBS. Cells pretreated with RGD peptide or anti-integrin αM antibody were then incubated with 10 μM rSap6 for 15 min, and cell aggregation was measured microscopically, or they were incubated with F-rSap6 (10 μM) and examined microscopically for cell binding. A control isotype antibody (IgG1) was also used at 1:100 dilution for inhibition experiments.
RESULTS
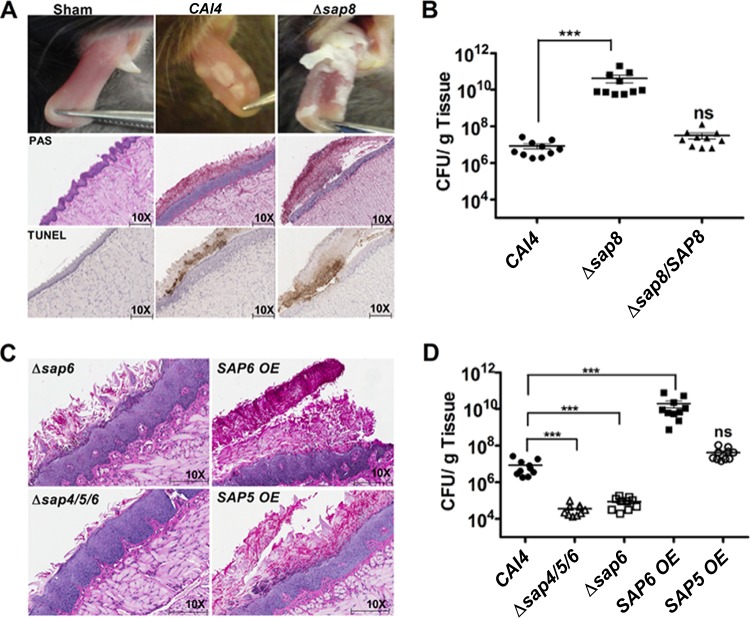
C. albicans Sap8 mutants are hypervirulent in OPC.
Our previous work suggested a possible role for Saps, and in particular Sap8, in the processing of signaling mucin Msb2 in C. albicans in vitro (32). To directly assess the role of SAP8 in vivo, we examined the virulence of a Δsap8 strain in murine oral candidiasis. Mice infected sublingually with the Δsap8 strain developed thick fungal patches that covered large areas of the dorsal surface of the tongue by day 5 compared to the smaller and thinner fungal plaques produced by WT CAI4 cells (Fig. 1A, top row) and the complemented Δsap8/SAP8 cells (data not shown). Mice infected with the Δsap8 strain lost significantly more body weight by day 5 and appeared sicker throughout the duration of infection than those infected with the wild-type CAI4 strain, while mice infected with the Δsap8/SAP8 complemented strain had weight loss similar to that of mice infected with the WT (see Fig. S1A in the supplemental material). Histological analysis of tongues in Δsap8 strain-infected mice confirmed our visual observations, in that these plaques consisted of a thicker fungal mass with greater invasion of the superficial epithelial layer accompanied by loss of its overall architecture compared with WT-infected mice (Fig. 1A, middle row). TUNEL staining showed higher numbers (a 38% increase) of apoptotic cells in tongue epithelial layers of Δsap8 strain-infected mice than in WT-infected mice, confirming more extensive cell damage within the epithelial layers (Fig. 1A, bottom row). These apoptotic epithelial cells were often free of the underlying epithelium and embedded in candidal plaques. C. albicans WT infection resulted in 107 CFU/g tongue, similar to previous studies (25), while infection with C. albicans Δsap8 caused 3-log-fold higher numbers, which were restored to WT levels in a complemented Δsap8/SAP8 strain (Fig. 1B). Immunosuppression is required for sustained infection by C. albicans cells in this murine model; however, the Δsap8 strain was able to infect 104 CFU/g tongue tissue in nonimmunosuppressed mice (data not shown), further illustrating the increased virulence of the strain. We further investigated whether dissemination occurred by day 5 of oral infection with the Δsap8 strain. We found that kidney tissues had 103 CFU/g of C. albicans Δsap8 cells while kidneys from mice orally infected with the Δsap8/SAP8 strain had no detectable fungal cells (see Fig. S1B in the supplemental material). Thus, the weight loss of mice infected with the Δsap8 strain is likely due to disseminated disease, as well as oral infection.
FIG 1.
C. albicans SAP6 expression elevates the severity of infection in oral candidiasis. (A) Mice infected with the C. albicans Δsap8 strain had more extensive and thicker fungal tongue lesions than mice infected with the CAI4 strain. PAS and TUNEL staining of the tongue epithelium showed denser fungal plaques and more extensive apoptosis in tongues infected with C. albicans Δsap8. (B) Infection with C. albicans Δsap8 resulted in 3- to 4-log-fold higher numbers of CFU/g of tongue tissue (***, P < 0.001; ns, not significant), which was restored to WT levels in a Δsap8/SAP8 complemented strain. (C and D) Infection with the C. albicans SAP6 OE strain resulted in dense fungal tongue plaques and 4-fold higher numbers of CFU/g of tongue tissue, similar to infection with the Δsap8 strain, while infection with the Δsap6 and Δsap4/5/6 strains showed thinner fungal plagues and 2-fold lower numbers of CFU/g of tongue tissue than infection with the CAI4 strain. In contrast, infection with the SAP5 OE strain was similar in all respects to infection with the CAI4 strain. Horizontal lines in panels B and D represent statistical comparison between strains.
Since SAP8 is one of 10 members of the SAP gene family with overlapping functions, we anticipated that the higher virulence of the Δsap8 strain might be the result of altered expression levels of other SAP genes. Therefore, SAP1 to SAP10 expression levels were measured by quantitative PCR (qPCR) from candidal tongue plaques harvested from Δsap8 strain-infected animals at day 5 and compared with SAP gene expression from CAI4 tongue plaques also collected at day 5 (Table 2). Only SAP5 and SAP6 genes were more highly expressed (2-fold and 4-fold, respectively) in the Δsap8 strain than in WT cells from tongue plaques (Table 2). To determine if this SAP gene expression profile was unique to cells collected from infected tongues, we examined the Δsap8 strain with WT cells grown on agar surfaces with 10% serum. Similar to the in vivo data, SAP5 and SAP6 genes were increased by 2-fold and 3-fold in Δsap8 cells compared to WT cells grown on agar, recapitulating the selective increase in expression of these genes found in vivo. However, Δsap8 cells grown on agar had a significant reduction in expression levels of SAP1 to SAP3, SAP7, and SAP9 compared to WT cells grown on agar. Differences in ACT1 expression between in vitro agar surfaces and in vivo tongue samples did not permit direct comparison of SAP genes between conditions (agar versus tongues). Thus, we hypothesized that the selective increase in either SAP6 or SAP5 expression could be responsible for the increased virulence observed in the Δsap8 strain.
TABLE 2.
Expression profile of secreted aspartyl proteinase genes from tongue plaques or agar surfaces
| Origin | Strain | Expression levela |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SAP1 | SAP2 | SAP3 | SAP4 | SAP5 | SAP6 | SAP7 | SAP8 | SAP9 | SAP10 | ||
| Tongue plaque | WT | 0.05 ± 0.01 | 1.25 ± 0.05 | 0.12 ± 0.02 | 2.07 ± 0.51 | 5.32 ± 2.03 | 9.14 ± 2.83 | 0.33 ± 0.02 | 0.43 ± 0.02 | 0.16 ± 0.08 | 0.19 ± 0.07 |
| Δsap8 | 0.11 ± 0.03 | 1.43 ± 0.70 | 0.12 ± 0.01b | 3.74 ± 1.08 | 12.29 ± 1.95c | 38.21 ± 5.08d | 0.18 ± 0.07 | ND | 0.05 ± 0.02 | 0.18 ± 0.04 | |
| Agar surface | WT | 3.64 ± 1.08 | 4.44 ± 1.05 | 2.63 ± 0.96 | 2.24 ± 0.63 | 12.72 ± 2.36 | 21.34 ± 4.49 | 4.31 ± 1.34 | 0.14 ± 0.09 | 4.30 ± 0.94 | 1.64 ± 0.03 |
| Δsap8 | 1.16 ± 0.16b | 2.38 ± 0.32 | 0.21 ± 0.04b | 2.04 ± 0.21 | 22.90 ± 5.59c | 63.79 ± 6.44d | 0.43 ± 0.10c | ND | 1.24 ± 0.58c | 2.38 ± 0.35 | |
Gene expression levels were calculated as a ratio to targeted gene/ACT1 expression calculated from a cDNA standard curve. Differences in expression values between the WT and Δsap8 strains for each gene and each growth condition were measured using Student's t test. ND, not determined.
P < 0.05.
P < 0.01.
P < 0.001.
To test this, we compared C. albicans strains overexpressing Sap5 (SAP5 OE) and Sap6 (SAP6 OE) with Δsap6, Δsap4/5/6, and Δsap8 mutants for virulence during OPC (Fig. 1C and D). Mice infected sublingually with the SAP6 OE strain had 3-log-fold higher numbers at day 5 (1010 CFU/g tongue), which was the same as mice infected with the Δsap8 strain. In contrast, infection levels using the SAP5 OE strain were equal to WT levels, whereas mice infected with the Δsap6 or Δsap4/5/6 strain had significantly (P < 0.01) reduced levels of infection (∼104 CFU/g) (Fig. 1D). Thus, higher expression levels of Sap6 in the SAP6 OE strain were positively correlated with higher infection levels in OPC, as well as increased weight loss (see Fig. S1A in the supplemental material) and kidney dissemination (data not shown). The histology of the tongue tissues from mice infected with the SAP6 OE strain was similar to that produced by infection with the Δsap8 strain in that SAP6 OE candidal plaques were very thick and matted and underlying tissues had substantial disruption of the normal epithelial architecture (Fig. 1C). In contrast, fungal plaques overlying tissues infected with the Δsap6 or Δsap4/5/6 strain were only half as thick as WT plaques and caused less destruction of the underlying tongue epithelium, and animals had less weight loss from oral infection (see Fig. S1A in the supplemental material).
Fungal plaques of the Δsap8 and SAP6 OE strains have dense colony architectures.
Since the fungal tongue plaques produced by infection were very thick and cohesive, we removed them from underlying diseased tissues in large sections in order to examine their architecture microscopically by staining with calcofluor white (Fig. 2, top). Both Δsap8 and SAP6 OE plaques showed striking similarities in that they contained densely aggregated colonies with interwoven mats of elongated hyphae whereas WT CAI4, Δsap6, and Δsap8/SAP8 (data not shown) plaques were thinner and contained shorter hyphae with fewer interlocking hyphal regions (Fig. 2). We also compared the colony morphologies of strains grown on plastic surfaces using RPMI media under 5% CO2 and found that the Δsap6 mutant had substantially reduced microcolony formation compared to the WT, whereas the Δsap8 and SAP6 OE strains formed larger and denser microcolonies with more extensive hyphal projections (Fig. 2). These results suggested that SAP6 has a role in the formation of a more cohesive colony architecture and perhaps hyphal extension both in vitro and in vivo.
FIG 2.
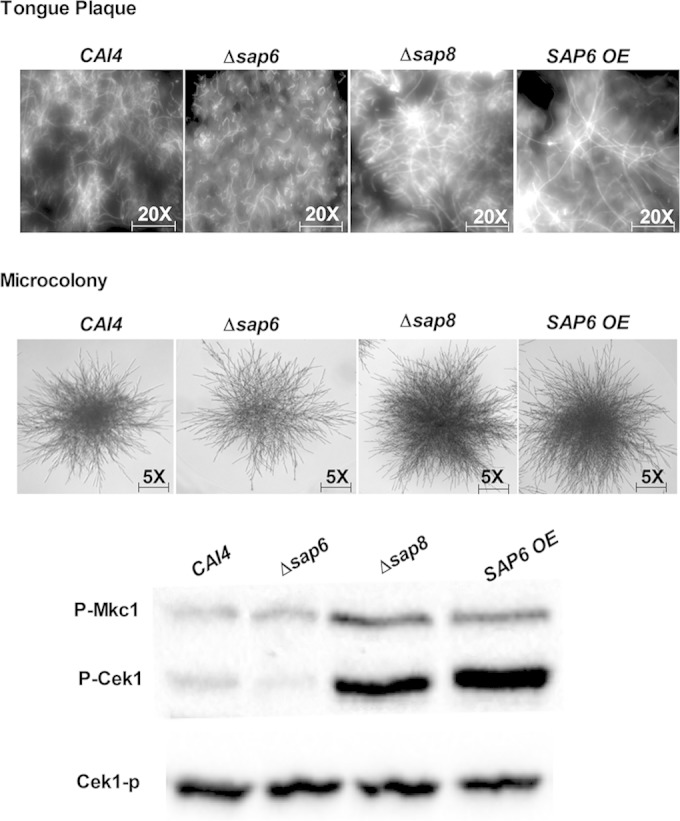
Architectures of in vivo fungal plaques and microcolony formation of the Δsap8 and SAP6OE strains show similar higher density and elongated filaments. (Top) Fungal plaques were removed from tongues, stained with calcofluor white, and observed under a DAPI (4′,6-diamidino-2-phenylindole) filter at ×20 magnification. Plaques from tongues infected with the Δsap8 and SAP6 OE strains contained fungal cells with longer and more compact hyphae than plaques from tongues infected with the Δsap6 and WT CAI4 strains. (Middle) Microcolonies of the Δsap8 and SAP6 OE strains grown under 5% CO2 were denser and more highly filamentous than those of the CAI4 strain, while the Δsap6 strain had the thinnest microcolony formation, although filament length was similar to that of the WT. (Bottom) Higher levels of Cek1 phosphorylation were found in fungal cells from tongue plaques infected with the Δsap8 and SAP6 OE strains than in tongue plaques infected with the CAI4 and Δsap6 strains.
Since Saps are involved in the Cek1 MAPK filamentation pathway (32) and Δsap8 tongue plaques were highly filamentous, we compared levels of Cek1 phosphorylation from Δsap8, Δsap6, and WT cells harvested from tongue plaques. After normalization by protein content, Δsap8 cells from tongue plaques had higher levels of Cek1 and Mkc1 phosphorylation than the Δsap6 and WT cells (Fig. 2, bottom), showing that increased Cek1 and Mkc1 signaling accompanied higher filamentation of Δsap8 cells in vivo.
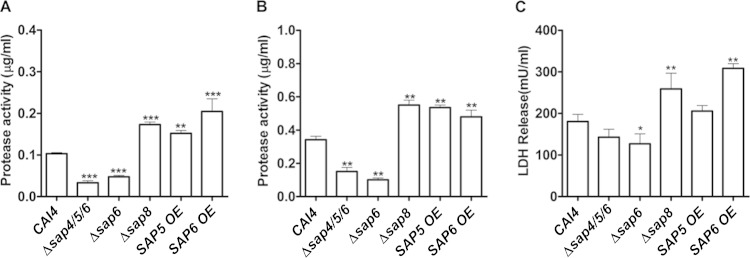
Secretion of Sap6 is not correlated with cell damage in oral epithelial cells.
We next examined whether differences in virulence among the strains were correlated with the amount of Sap proteinase secreted. The total Sap proteinase secreted into the media was measured both in planktonic growth (Fig. 3A) and in 12-h biofilms (Fig. 3B). Both Δsap6 and Δsap4/5/6 strains had more than a 50% reduction in total proteinase secretion compared with WT cells under both planktonic and biofilm growth conditions, while the Δsap8, SAP5 OE, and SAP6 OE strains had an average 50% increase in proteinase secretion under both growth conditions. To assess whether proteinase secretion levels affected fungus-induced epithelial cytotoxicity, an oral epithelial cell line (SCC-15 monolayers) was infected with the respective strains at an MOI of 1:10, and subsequent cell damage was measured by LDH release (Fig. 3C). Infection with the SAP6 OE and Δsap8 strains showed significantly (P < 0.001) greater cell damage (increased by 30%) that positively correlated with increased levels of secreted Sap6 in these strains. However, this relationship did not extend to the Δsap6 strain, as only a 20% decrease in cell killing was observed despite more than 50% reduction in proteinase secretion. Furthermore, the SAP5 OE strain showed no significant difference from the WT in LDH release, although it showed 2-fold more total proteinase secretion. Also, there was no significant difference in epithelial cell damage after infection with the Δsap4/5/6 strain, despite 2-fold less proteinase secretion. Overall, we did not find a correlation between Sap6 proteinase levels and epithelial cell death as measured by LDH release, and thus, cell death as a result of high levels of secreted Sap6 in the SAP6 OE and Δsap8 strains is likely not the only reason for their high virulence.
FIG 3.
Protease secretion and epithelial cell damage were both increased only in the Δsap8 and SAP6 OE strains. (A and B) Total proteinase activity after 12 h in either planktonic (A) or biofilm (B) growth was quantified using FTC solution as the substrate. Under both culture conditions, the total proteinase activity was 2-fold higher in the Δsap8, SAP5 OE, and SAP6 OE strains (**, P < 0.01; ***, P < 0.001) than in the CAI4 strain. (C) An epithelial cell line, SCC-15, was infected with the CAI4, Δsap4/5/6, Δsap6, Δsap8, SAP5 OE, and SAP6 OE strains for 12 h, and the cell damage was measured by LDH release. There was no significant change in LDH release in SCC-15 cells infected with the Δsap4/5/6 and SAP5 OE strains compared to cells infected with the CAI4 strain. Both the Δsap8 and SAP6 OE strains induced significant cell damage (*, P < 0.05; **, P < 0.01). The results represent the averages from triplicate samples from two independent experiments. The error bars indicate standard deviations.
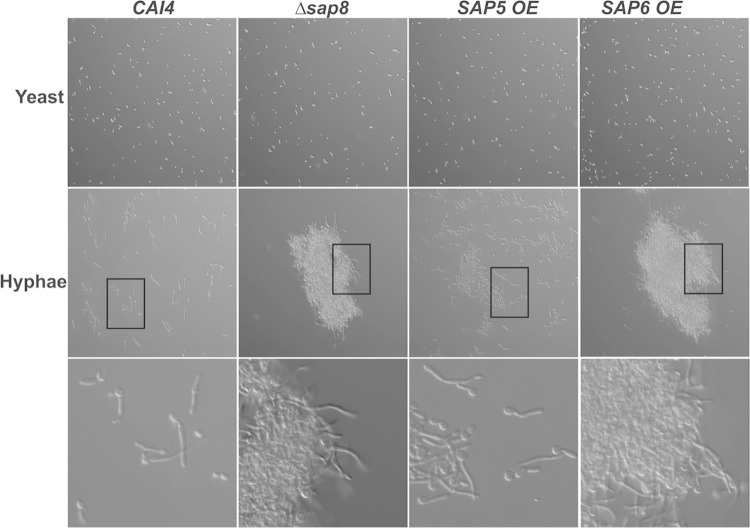
Sap6 causes concentration-dependent aggregation in germinated cells independent of its proteolytic activity.
Since Δsap8 and SAP6 OE plaques had extensive hyphal formation, we next compared the relative ability of each strain to form hyphae after incubation with YNB medium containing 1.25% GlcNAc at 37°C for 3 h (Fig. 4). While the percentages of germinated cells and hyphal lengths were similar among all the strains upon exposure to GlcNAc, we unexpectedly observed very substantial aggregation of germinated C. albicans Δsap8 cells that did not occur in germinated WT cells exposed to the same conditions. This aggregation was similar upon hyphal induction with FBS but did not occur in yeast phase Δsap8 cells. Germinated SAP6 OE cells formed even larger cellular aggregates, while germination of SAP5 OE cells produced only small cell aggregates (Fig. 4). Neither overexpression strain formed cell aggregates as blastospores. Both Δsap6 and Δsap4/5/6 strains formed typical hyphae; however, neither strain formed aggregates upon germination (data not shown). To determine whether Sap proteinase activity was required for this aggregation phenotype, cells were treated with pepstatin A (10 μM) under germination conditions; however, cells did not undergo yeast-to-hyphal transition in the presence of pepstatin A, likely a result of impaired MAPK signaling (32), so we could not test for aggregation.
FIG 4.
Germination of Δsap8 and SAP6 OE cells resulted in formation of large cell-cell aggregates. Cells were grown at 37°C for 3 h to form hyphae and observed under ×20 magnification. (Top row) No aggregation was found in yeast form cells of the CAI4, Δsap8, SAP5 OE, and SAP6 OE strains. (Middle and bottom rows) Hypha formation was accompanied by the formation of large aggregates in the Δsap8 and SAP6 OE strains, but not in the wild-type CAI4 strain, while the SAP5 OE strain formed smaller aggregates. The boxed areas in the middle row are shown at higher magnification in the bottom row.
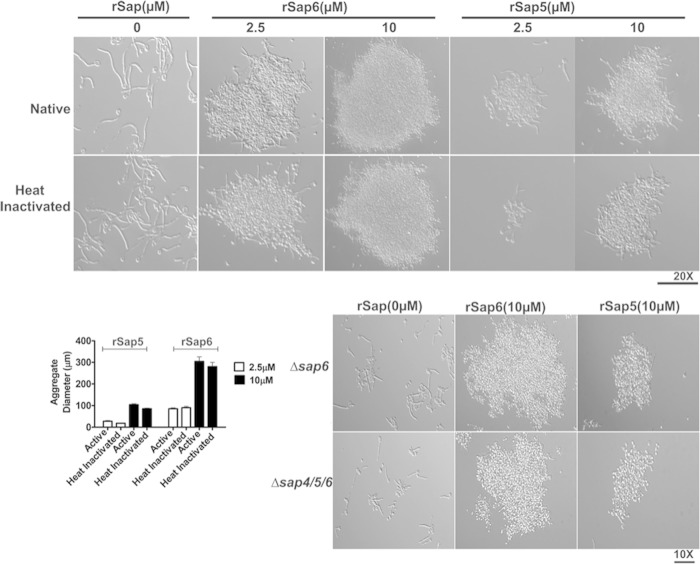
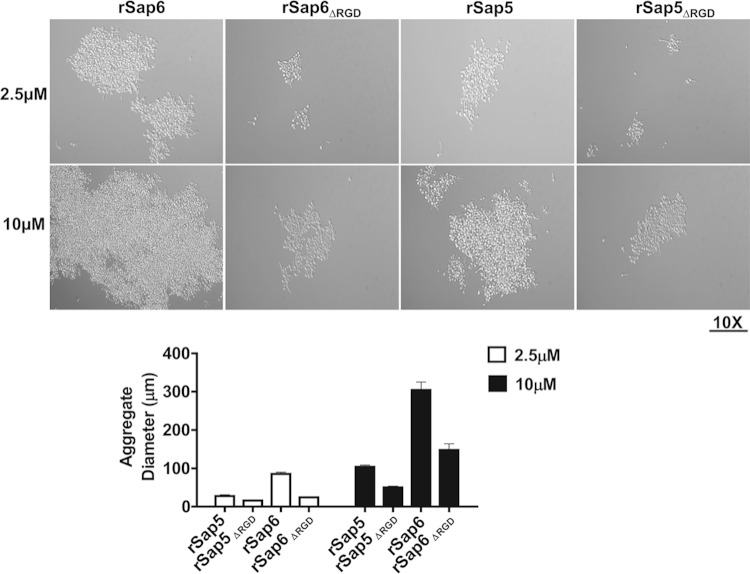
Since aggregation was found only in germinated cells with high expression levels of Sap6 and to a lesser extent in cells overexpressing Sap5, we hypothesized that secreted Sap5 or Sap6 might be responsible for the observed aggregation phenotype. WT cells were germinated in YNB medium containing 1.25% GlcNAc for 3 h, and then, rSap5 and rSap6 (2.5 μM and 10 μM) were added to the cells and incubated for 15 min (Fig. 5). Germinated WT cells formed aggregates after the addition of 2.5 μM rSap6, and even larger aggregates were formed after the addition of 10 μM rSap6 (Fig. 5, top). Addition of rSap5 resulted in cell aggregates that were only one-third the size of rSap6 aggregates, and many germinated cells remained unaggregated in the presence of rSap5. Since Saps have enzymatic activity as proteinases, we asked whether cell aggregation might be a result of increased exposure of underlying cell wall ligands as a result of proteolytic digestion by rSap6 or rSap5. To test this, rSap5 and rSap6 proteinase activities were abolished by heat treatment (120°C for 15 min), and the resulting enzyme inactivation was confirmed by proteinase assay. Addition of heat-inactivated rSap6 or rSap5 to germinated WT cells did not change the levels of aggregation as measured by aggregate diameter compared to native Saps (Fig. 5), showing that Sap5 or Sap6 enzyme activity is not required for aggregation. To confirm this, a proteinase inhibitor (pepstatin A; 10 μM) was incubated with rSaps, and then, the mixture was added to already germinated cells. Addition of the pepstatin A proteinase inhibitor did not change the levels of cell aggregation by either rSap6 or rSap5 (data not shown), confirming that the cellular aggregation induced by Saps is independent of its proteolytic function.
FIG 5.
Addition of Sap6 to CAI4 cells causes hypha-specific cell-cell adhesion independent of its protease activity. (Top) CAI4 cells were germinated at 37°C for 3 h and then incubated with either native or heat-inactivated (120°C for 20 min) rSap6 or rSap5 (2.5 or 10 μM) for 15 min and observed microscopically. At least 50 different fields were measured for aggregate diameter using AxioImager software and averaged. Addition of native rSap6 (2.5 μM) to germinated CAI4 cells resulted in the formation of large and compact aggregates whose size and density were tripled when incubated with 10 μM rSap6. Addition of rSap5 resulted in the formation of smaller (one-quarter the size) aggregates than rSap6. (Bottom left) There was no significant difference in the aggregation of germinated CAI4 cells incubated with heat-inactivated rSap6 or rSap5 compared to native protein. (Bottom right) Germinated cells that did not produce Sap6 (Δsap6 or Δsap4/5/6) showed cellular aggregation similar to that of CAI4 cells after addition of 10 μM rSap6 and rSap5. The error bars indicate standard deviations.
We next examined whether cell-cell adhesion could be induced by addition of Sap6 or Sap5 to cells that do not express Sap4, -5, or -6. C. albicans Δsap6 and Δsap4/5/6 strains were allowed to germinate for 3 h and then incubated with rSap6 or rSap5 (Fig. 5, bottom). Both Δsap6 and Δsap4/5/6 cells had levels of aggregation similar to those of WT cells following addition of Sap5 or Sap6, showing that extracellular Sap6 or Sap5 alone is sufficient to induce aggregation in filamented cells and that this aggregation does not require the cell wall modification induced by expression and release of Sap4, -5, or -6 at the cell surface. However, in all cases, Sap6 had a much greater ability to induce cell-cell adhesion than its closest homologue, Sap5.
Sap6 binds only to germinated C. albicans cells.
As Sap6 is a secreted protein that is highly expressed during filamentation (11, 12), we examined whether binding of Sap6 was specific to hyphae. F-rSap6 was incubated for 1 h with WT CAI4 cells that either were yeast form or were germinated by incubation with YNB medium supplemented with 1.25% GlcNAc at 37°C. As expected from the lack of cell-cell adhesion of yeast cells incubated with rSap6, there was no detectable F-rSap6 associated with the surfaces of yeast cells, and no subsequent binding of F-rSap6 to yeast cells was observed even after 12 h of incubation (Fig. 6). In contrast, germinated cells had abundant surface-associated F-rSap6 along hyphae (that was more intense at the hyphal tips), although binding was also observed with the mother cell. Thus, Sap6 binding is not specific to hyphae, although only germinated cells bind Sap6. These data suggested that cell wall changes accompanying germination, rather than hypha-specific proteins, are permissive for Sap6 binding. To explore this, we examined the roles of three major hypha-specific adhesins, Als1, Als3, and Rbt1, in Sap6-mediated cell-cell adhesion. C. albicans Δals1/Δals3 and Δrbt1 cells were incubated under hypha-inducing conditions for 3 h to induce robust hyphae, and then, rSap6 was added to the cells (see Fig. S2 in the supplemental material). Both Δals1/Δals3 and Δrbt1 mutants displayed cell aggregation similar to that of WT cells upon addition of Sap6, showing that Als1, Als3, and Rbt1 adhesins are not required for Sap6-mediated cell-cell aggregation.
FIG 6.
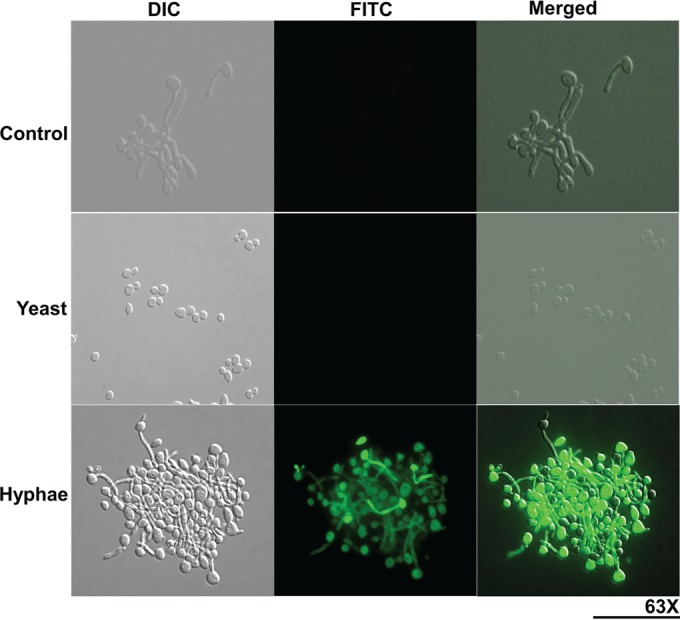
Sap6 binds to the surfaces of germinated C. albicans cells. Both yeast and germinated cells of the CAI4 strain were incubated with FITC-labeled rSap6 (10 μM) for 1 h and observed under ×63 magnification using the FITC channel. Germinated cells were incubated with 10 μM FITC alone as a control. There was no detectable binding of FITC-rSap6 to yeast phase cells, while germinated cells had abundant surface binding of FITC-rSap6 to hyphae and with the mother cell. DIC, differential interference contrast.
Sap6 increased adherence of C. albicans to oral epithelial cells.
To determine whether Saps might also promote C. albicans adhesion to oral mucosa, we examined the adherence of C. albicans Δsap8, Δsap6, and SAP6 OE strains to SCC-15 oral epithelial cells. Adhesion of the SAP6 OE and Δsap8 strains to oral epithelial cells was significantly (P < 0.05) higher than that of WT cells, while that of the Δsap6 strain was significantly (P < 0.05) reduced (Fig. 7A). There was no difference between the adhesion of C. albicans Δsap4/5/6 and SAP5 OE strains and that of WT cells. To further examine whether this increase in adhesion to oral epithelial cells might be due to Sap6 itself, rSap6 or rSap5 was first added to C. albicans. Preincubation with rSap6 increased adhesion of WT cells by 20% (P < 0.01) and also restored adhesion of both the ∆sap4/5/6 and Δsap6 strains to WT levels with rSap6 (Fig. 7B). Interestingly, addition of rSap6 to Δsap8 cells did not further increase its already elevated adhesion to oral epithelial cells. Preincubation of C. albicans cells with rSap5 did not alter the adhesion of any of these strains (Δsap8, Δsap6, or Δsap4/5/6) (data not shown).
FIG 7.
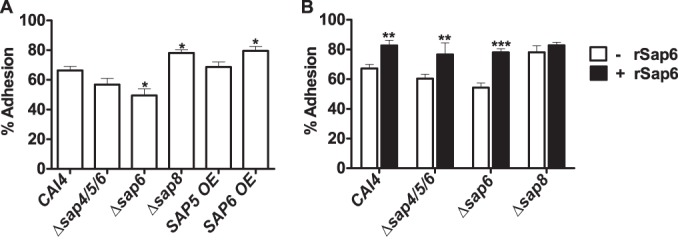
Sap6 increases Candida adhesion to host epithelial cells. (A) SCC-15 epithelial cell monolayers were infected with the CAI4 and SAP deletion strains for 90 min. After extensive washing, the adherent cells were harvested using 0.1% Triton X-100 and plated on YPD agar plates, and the plates were incubated at 30°C for 24 h to determine the numbers of CFU of adherent fungal cells. Both the Δsap8 and SAP6 OE strains had significantly increased adhesion to epithelial cells, whereas the Δsap6 strain showed reduction in adhesion compared to the CAI4 strain (*, P < 0.05). (B) Candida cells were preincubated with rSap6 (10 μM) for 30 min at 37°C, washed twice with PBS, and then added to epithelial cells. The CAI4, Δsap4/5/6, and Δsap6 strains showed significantly increased adhesion when preincubated with rSap6 (**, P < 0.01; ***, P < 0.001). The results represent the averages of triplicate samples from at least two independent experiments. The error bars indicate standard deviations.
The Sap 6 integrin-binding motif (RGD) mediates cell aggregation.
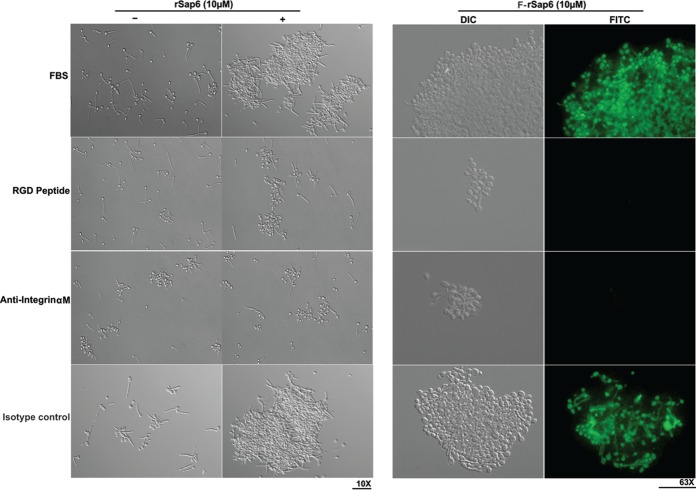
Since Sap4 to -6 all contain RGD molecular motifs known to bind to epithelial cell surface integrins to induce cell adhesion (29), we asked whether this RGD motif might mediate hyphal aggregation induced by Sap6 and Sap5. Germinated WT cells were incubated with rSap6ΔRGD or rSap5ΔRGD (2.5 μM and 10 μM) and assayed for cell-cell aggregation. We found a significant reduction (more than 60% at either 2.5 μM or 10 μM) in cell-cell adhesion using rSap6ΔRGD compared to native rSap6 protein, and a similar reduction (about 50%) was observed for cells preincubated with rSap5ΔRGD (Fig. 8). Alternatively, WT cells were germinated for 30 min in the presence of RGD peptide (10 μM) or anti-integrin αM antibody (1:100) and washed to remove unbound peptide or antibody, and then the cells were incubated with rSap6. Pretreatment of germinated C. albicans cells with RGD peptide resulted in a reduction in cell aggregation (about 50%) similar to that found for rSap6ΔRGD (Fig. 9, left). Pretreatment with anti-integrin αM antibody resulted in some cell aggregation of control Candida cells; however, no further aggregation was observed following addition of rSap6. The incubation of WT cells with the control isotype IgG1 did not affect aggregation or cell surface binding of Sap6. These data show that a C. albicans cell surface RGD binding receptor, recognized by this anti-integrin antibody, is involved in hyphal aggregation mediated by Sap6. As expected, there was no observable binding of F-rSap6 to germinated cells preincubated with either RGD peptide or anti-integrin αM antibody (Fig. 9, right). Thus, an RGD motif in Sap6 mediates hyphal cell-cell adhesion.
FIG 8.
An RGD motif in Sap6 and Sap5 is required for cell aggregation. (Top) Germinated CAI4 cells were incubated with rSap6, rSap5, or rSap5 and rSap6 in which both RGD sequences were deleted (rSap6ΔRGD and rSap5ΔRGD) at 2.5 μM or 10 μM for 15 min. Cell aggregates from at least 50 different fields were observed by microscope (×10 magnification) and analyzed using AxioImager software to measure the average diameter. (Bottom) Germinated cells incubated with rSap6ΔRGD or rSap5ΔRGD had more than 60% reduction in aggregate diameter compared to rSap6 and rSap5 at 2.5 μM and 10 μM. The error bars indicate standard deviations.
FIG 9.
Sap6-mediated aggregation and cell binding were inhibited by RGD peptide or integrin antibodies. (Left) Germinated CAI4 cells were first incubated with RGD peptide (10 μM) or with anti-integrin αM antibody (1:100 dilution), washed, and then incubated with rSap6 (10 μM) to induce cell aggregation. Germinated cells preincubated with RGD peptide had significant reduction in rSap6-induced cell adhesion. −, absence; +, presence. (Right) Binding of Sap6 to the hyphal cell surface was inhibited in CAI4 cells preincubated with RGD peptide or anti-integrin αM antibody. An isotype antibody was used as a control. The cells were observed microscopically at ×10 magnification for cellular aggregation and at ×63 magnification for binding.
DISCUSSION
Although it is known that Sap protein production is closely associated with yeast-to-hyphal transition and that Saps contribute to adherence and virulence, our data show for the first time a nonenzymatic role for Sap6 in linking these functions through cell-cell aggregation. It has been shown that SAP2 and SAP4 to SAP6 were highly expressed in both carriers and oral candidiasis patients, suggesting a state of “permanent interaction” between these Saps and oral host tissues (5). Others found elevated SAP8, SAP5, and SAP6 expression in human denture stomatitis clinical isolates (33) and that SAP6, followed by SAP5, was among the most highly expressed genes in mouse tongue plaques during oral candidiasis (34). We unexpectedly found a hypervirulent phenotype of C. albicans cells lacking SAP8 that was phenocopied by C. albicans overexpressing SAP6, but not SAP5, in mouse oral infections. Furthermore, deletion of SAP6 alone resulted in attenuated virulence, similar to cells lacking SAP4 to SAP6 genes, showing a major role for Sap6, but not Sap5, in virulence in oral candidiasis. The striking phenotype in oral infection with cells overexpressing Sap6 was the density, thickness, and cohesiveness of fungal plaques compared with WT plaques. Although the total proteinase activity was significantly higher in mutants overexpressing Sap6 (Δsap8 and SAP6 OE strains), we found only a small increase in oral epithelial cell damage in vivo, as well as in vitro. Thus, our hypervirulence model pointed to a minimal role for proteinase activity in pathogenesis of oral candidiasis.
In line with these in vivo results, both the Δsap8 and SAP6 OE strains exhibited strong cell-cell aggregation upon yeast-to-hypha transition that was not observed in the same cells in yeast phase. This aggregation could be reproduced by addition of rSap6 or heat-inactivated rSap6 to germinated WT cells, thus showing that Sap6 functions as an adhesin independently of its proteinase activity. However, only germinated cells exhibited this aggregation, suggesting that cell surface molecules that permit Sap6-mediated cell-cell aggregation are exposed upon germination. It is unclear from our experiments whether Sap proteinases have an enzymatic role in cell wall remodeling to expose these adhesins, either directly or through MAPK signaling (32, 35), since experiments to block Sap enzymatic activity with pepstatin A also inhibited germination. It is known that Sap9, a GPI-anchored cell wall proteinase, activates several cell surface proteins, including adhesins Ywp1, Hwp1, and Rbt1, by enzymatic cleavage (35). Thus, it is likely that Saps have dual roles, one enzymatic and involved in germination and a second, nonenzymatic, function for cell-cell aggregation that is mainly carried out by Sap6.
Our experiments found that addition of Sap6 to C. albicans increased its adhesion to oral epithelial cells. However, these results could be due to adherence of larger cell aggregates rather than to Sap6 increasing fungal adhesion to epithelial cells. Interestingly, our histological examination of in vivo tongue plaques in Δsap8 infection showed significantly thicker plaques, similar to elevated adhesion as a result of cell aggregates. Nevertheless, greater cell-cell aggregation would result in higher numbers of CFU per unit of tongue tissue and higher morbidity in animals, suggesting that increased aggregation is also a virulence mechanism.
We observed that high colonization of tongue mucosal surfaces by C. albicans Δsap8 was also accompanied by profuse inflammation of epithelial tissues (as well as apoptotic cells) typical of a proinflammatory cytokine response. Saps are involved in activating proinflammatory responses in reconstituted human vaginal epithelium (RHVE) cells, as well as monocytes (27, 36). However, little is known about oral epithelial cell proinflammatory responses during OPC, and particularly whether secreted Sap6 is able to modulate the level of proinflammatory responses in oral epithelial cells, similarly to monocytes.
One surprising finding from our data was the functional differences between the related proteinases Sap5 and Sap6. Although Sap5 and Sap6 are both 419 amino acid residues in length, there is approximately 19% difference in the primary sequence, so that they share 80.9% sequence homology. The crystal structure of Sap5 has been solved (37) and extrapolated to the protein structure for the Sap4 to -6 subgroup. There is a highly conserved secondary structure of the middle and back regions of Sap5 that contains the aspartic proteinase active-site cleft required for substrate and inhibitor (pepstatin A) binding (37). There are also three extended loop structures (arms 1, 2, and 3) surrounding this cleft, among which the arm 1 loops of Sap4 to -6 contain at least one integrin-binding motif (RGD) at the surface-exposed tip of the loop (29). Sap4 has a single RGD motif, while Sap5 contains an RGDKGD motif and Sap6 has two sequential RGDRGD integrin-binding motifs. No other Saps contain this motif, suggesting that integrin binding is a biological function only of Sap4 to -6. Indeed, these motifs were shown to be involved in Sap4 to -6 binding to integrin molecules on A549 epithelial cells and could be inhibited by RGD-containing peptides or by substituting Sap RGD motifs (29). RGD motifs in fungal hyphae of basidiomycetes and other plant fungal pathogens are also able to facilitate cell-cell aggregation to form thick biofilms and mediate adherence to host cells (38, 39). Since variation in the RGD motif affects ligand binding affinity to a particular integrin molecule (40), divergence of this motif between Sap5 and Sap6 could be one of the possible reasons for the differences we found in aggregation and virulence.
The identities of C. albicans cell surface molecules exposed during germination that are bound by Sap6 are not yet known. Our evidence shows that major hypha-specific adhesins, including Als1, Als3, and Rbt1, are not involved. Our experiments using recombinant Sap6 lacking an RGD motif showed that binding to the C. albicans cell wall was greatly reduced, suggesting that a cell surface ligand for RGD is involved. Fungal cells bind with host ligands, including fibronectin, laminin, or iC3b (all of which contain RGD motifs), through integrin-like and fibronectin receptors (38, 41). C. albicans itself expresses a 185-kDa cell surface integrin-like protein (Int1) with limited structural similarity to leukocyte integrin αM that affects cell adhesion and aggregation (42–45). A C. albicans cell wall fibronectin-binding protein also binds the RGD region of fibronectin, as well as other extracellular matrix proteins (38, 46, 47). Our data found that addition of anti-integrin antibodies to germinated cells reduced Sap6 binding, further supporting a role for integrin-like and perhaps other as-yet-uncharacterized integrin-like cell wall proteins in Sap6 binding.
We report here a novel functional role of the secreted aspartyl proteinase Sap6 as a C. albicans adhesin that is mediated by its RGD motif. Our results show that Sap6 is an important virulence factor in oral candidiasis and also establish Sap6 as a multifunctional protein that acts as both an adhesin and a proteinase during infection. These new findings may provide an alternative therapeutic modality for candidiasis through modulation of Sap6 structure or function.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by awards R01DE010641 and R01DE022720 (M.E.) from the National Institute of Dental and Craniofacial Research, National Institutes of Health.
We thank Wade J. Sigurdson, Confocal Microscopy Facility, University at Buffalo, for assistance with microscopy. We also gratefully acknowledge Bernhard Hube, Scott Filler, A. D. Johnson, Michel Monod, and Jordan Tang for providing strains and plasmids.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00282-15.
REFERENCES
- 1.Cassone A, Cauda R. 2012. Candida and candidiasis in HIV-infected patients: where commensalism, opportunistic behavior and frank pathogenicity lose their borders. AIDS 26:1457–1472. doi: 10.1097/QAD.0b013e3283536ba8. [DOI] [PubMed] [Google Scholar]
- 2.Calderone RA, Clancy CJ. 2012. Candida and candidiasis, 2nd ed, p 137–154. ASM Press, Washington, DC. [Google Scholar]
- 3.Pappas PG, Kauffman CA, Andes D, Benjamin DK Jr, Calandra TF, Edwards JE Jr, Filler SG, Fisher JF, Kullberg BJ, Ostrosky-Zeichner L, Reboli AC, Rex JH, Walsh TJ, Sobel JD. 2009. Clinical practice guidelines for the management of candidiasis: 2009. Clin Infect Dis 48:503–535. doi: 10.1086/596757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaffin WL. 2008. Candida albicans cell wall proteins. Microbiol Mol Biol Rev 72:495–544. doi: 10.1128/MMBR.00032-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naglik JR, Challacombe SJ, Hube B. 2003. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev 67:400–428. doi: 10.1128/MMBR.67.3.400-428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naglik J, Albrecht A, Bader O, Hube B. 2004. Candida albicans proteinases and host/pathogen interactions. Cell Microbiol 6:915–926. doi: 10.1111/j.1462-5822.2004.00439.x. [DOI] [PubMed] [Google Scholar]
- 7.Schaller M, Borelli C, Korting HC, Hube B. 2005. Hydrolytic enzymes as virulence factors of Candida albicans. Mycoses 48:365–377. doi: 10.1111/j.1439-0507.2005.01165.x. [DOI] [PubMed] [Google Scholar]
- 8.Aoki W, Kitahara N, Miura N, Morisaka H, Yamamoto Y, Kuroda K, Ueda M. 2011. Comprehensive characterization of secreted aspartic proteases encoded by a virulence gene family in Candida albicans. J Biochem 150:431–438. doi: 10.1093/jb/mvr073. [DOI] [PubMed] [Google Scholar]
- 9.Hube B, Naglik J. 2001. Candida albicans proteinases: resolving the mystery of a gene family. Microbiology 147:1997–2005. [DOI] [PubMed] [Google Scholar]
- 10.Koelsch G, Tang J, Loy JA, Monod M, Jackson K, Foundling SI, Lin X. 2000. Enzymic characteristics of secreted aspartic proteases of Candida albicans. Biochim Biophys Acta 1480:117–131. doi: 10.1016/S0167-4838(00)00068-6. [DOI] [PubMed] [Google Scholar]
- 11.Chen YC, Wu CC, Chung WL, Lee FJ. 2002. Differential secretion of Sap4-6 proteins in Candida albicans during hyphae formation. Microbiology 148:3743–3754. [DOI] [PubMed] [Google Scholar]
- 12.Felk A, Kretschmar M, Albrecht A, Schaller M, Beinhauer S, Nichterlein T, Sanglard D, Korting HC, Schafer W, Hube B. 2002. Candida albicans hyphal formation and the expression of the Efg1-regulated proteinases Sap4 to Sap6 are required for the invasion of parenchymal organs. Infect Immun 70:3689–3700. doi: 10.1128/IAI.70.7.3689-3700.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naglik JR, Moyes D, Makwana J, Kanzaria P, Tsichlaki E, Weindl G, Tappuni AR, Rodgers CA, Woodman AJ, Challacombe SJ, Schaller M, Hube B. 2008. Quantitative expression of the Candida albicans secreted aspartyl proteinase gene family in human oral and vaginal candidiasis. Microbiology 154:3266–3280. doi: 10.1099/mic.0.2008/022293-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naglik JR, Newport G, White TC, Fernandes-Naglik LL, Greenspan JS, Greenspan D, Sweet SP, Challacombe SJ, Agabian N. 1999. In vivo analysis of secreted aspartyl proteinase expression in human oral candidiasis. Infect Immun 67:2482–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Staib P, Kretschmar M, Nichterlein T, Hof H, Morschhauser J. 2000. Differential activation of a Candida albicans virulence gene family during infection. Proc Natl Acad Sci U S A 97:6102–6107. doi: 10.1073/pnas.110031497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tavanti A, Pardini G, Campa D, Davini P, Lupetti A, Senesi S. 2004. Differential expression of secretory aspartyl proteinase genes (SAP1-10) in oral Candida albicans isolates with distinct karyotypes. J Clin Microbiol 42:4726–4734. doi: 10.1128/JCM.42.10.4726-4734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaller M, Korting HC, Schafer W, Bastert J, Chen W, Hube B. 1999. Secreted aspartic proteinase (Sap) activity contributes to tissue damage in a model of human oral candidosis. Mol Microbiol 34:169–180. doi: 10.1046/j.1365-2958.1999.01590.x. [DOI] [PubMed] [Google Scholar]
- 18.Schaller M, Schackert C, Korting HC, Januschke E, Hube B. 2000. Invasion of Candida albicans correlates with expression of secreted aspartic proteinases during experimental infection of human epidermis. J Investig Dermatol 114:712–717. doi: 10.1046/j.1523-1747.2000.00935.x. [DOI] [PubMed] [Google Scholar]
- 19.Korting HC, Hube B, Oberbauer S, Januschke E, Hamm G, Albrecht A, Borelli C, Schaller M. 2003. Reduced expression of the hyphal-independent Candida albicans proteinase genes SAP1 and SAP3 in the efg1 mutant is associated with attenuated virulence during infection of oral epithelium. J Med Microbiol 52:623–632. doi: 10.1099/jmm.0.05125-0. [DOI] [PubMed] [Google Scholar]
- 20.Lermann U, Morschhauser J. 2008. Secreted aspartic proteases are not required for invasion of reconstituted human epithelia by Candida albicans. Microbiology 154:3281–3295. doi: 10.1099/mic.0.2008/022525-0. [DOI] [PubMed] [Google Scholar]
- 21.Correia A, Lermann U, Teixeira L, Cerca F, Botelho S, da Costa RM, Sampaio P, Gartner F, Morschhauser J, Vilanova M, Pais C. 2010. Limited role of secreted aspartyl proteinases Sap1 to Sap6 in Candida albicans virulence and host immune response in murine hematogenously disseminated candidiasis. Infect Immun 78:4839–4849. doi: 10.1128/IAI.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kretschmar M, Hube B, Bertsch T, Sanglard D, Merker R, Schroder M, Hof H, Nichterlein T. 1999. Germ tubes and proteinase activity contribute to virulence of Candida albicans in murine peritonitis. Infect Immun 67:6637–6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanglard D, Hube B, Monod M, Odds FC, Gow NA. 1997. A triple deletion of the secreted aspartyl proteinase genes SAP4, SAP5, and SAP6 of Candida albicans causes attenuated virulence. Infect Immun 65:3539–3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ibrahim AS, Filler SG, Sanglard D, Edwards JE Jr, Hube B. 1998. Secreted aspartyl proteinases and interactions of Candida albicans with human endothelial cells. Infect Immun 66:3003–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conti HR, Shen F, Nayyar N, Stocum E, Sun JN, Lindemann MJ, Ho AW, Hai JH, Yu JJ, Jung JW, Filler SG, Masso-Welch P, Edgerton M, Gaffen SL. 2009. Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J Exp Med 206:299–311. doi: 10.1084/jem.20081463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Puri S, Lai WK, Rizzo JM, Buck MJ, Edgerton M. 2014. Iron-responsive chromatin remodelling and MAPK signalling enhance adhesion in Candida albicans. Mol Microbiol 93:291–305. doi: 10.1111/mmi.12659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pietrella D, Rachini A, Pandey N, Schild L, Netea M, Bistoni F, Hube B, Vecchiarelli A. 2010. The inflammatory response induced by aspartic proteases of Candida albicans is independent of proteolytic activity. Infect Immun 78:4754–4762. doi: 10.1128/IAI.00789-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borg-von Zepelin M, Beggah S, Boggian K, Sanglard D, Monod M. 1998. The expression of the secreted aspartyl proteinases Sap4 to Sap6 from Candida albicans in murine macrophages. Mol Microbiol 28:543–554. doi: 10.1046/j.1365-2958.1998.00815.x. [DOI] [PubMed] [Google Scholar]
- 29.Wu H, Downs D, Ghosh K, Ghosh AK, Staib P, Monod M, Tang J. 2013. Candida albicans secreted aspartic proteases 4-6 induce apoptosis of epithelial cells by a novel Trojan horse mechanism. FASEB J 27:2132–2144. doi: 10.1096/fj.12-214353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simonetti N, Strippoli V, Cassone A. 1974. Yeast-mycelial conversion induced by N-acetyl-D-glucosamine in Candida albicans. Nature 250:344–346. doi: 10.1038/250344a0. [DOI] [PubMed] [Google Scholar]
- 31.Mayer FL, Wilson D, Jacobsen ID, Miramon P, Grosse K, Hube B. 2012. The novel Candida albicans transporter Dur31 is a multi-stage pathogenicity factor. PLoS Pathog 8:e1002592. doi: 10.1371/journal.ppat.1002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puri S, Kumar R, Chadha S, Tati S, Conti HR, Hube B, Cullen PJ, Edgerton M. 2012. Secreted aspartic protease cleavage of Candida albicans Msb2 activates Cek1 MAPK signaling affecting biofilm formation and oropharyngeal candidiasis. PLoS One 7:e46020. doi: 10.1371/journal.pone.0046020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramage G, Coco B, Sherry L, Bagg J, Lappin DF. 2012. In vitro Candida albicans biofilm induced proteinase activity and SAP8 expression correlates with in vivo denture stomatitis severity. Mycopathologia 174:11–19. doi: 10.1007/s11046-012-9522-2. [DOI] [PubMed] [Google Scholar]
- 34.Fanning S, Xu W, Solis N, Woolford CA, Filler SG, Mitchell AP. 2012. Divergent targets of Candida albicans biofilm regulator Bcr1 in vitro and in vivo. Eukaryot Cell 11:896–904. doi: 10.1128/EC.00103-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schild L, Heyken A, de Groot PW, Hiller E, Mock M, de Koster C, Horn U, Rupp S, Hube B. 2011. Proteolytic cleavage of covalently linked cell wall proteins by Candida albicans Sap9 and Sap10. Eukaryot Cell 10:98–109. doi: 10.1128/EC.00210-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaller M, Korting HC, Borelli C, Hamm G, Hube B. 2005. Candida albicans-secreted aspartic proteinases modify the epithelial cytokine response in an in vitro model of vaginal candidiasis. Infect Immun 73:2758–2765. doi: 10.1128/IAI.73.5.2758-2765.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borelli C, Ruge E, Lee JH, Schaller M, Vogelsang A, Monod M, Korting HC, Huber R, Maskos K. 2008. X-ray structures of Sap1 and Sap5: structural comparison of the secreted aspartic proteinases from Candida albicans. Proteins 72:1308–1319. doi: 10.1002/prot.22021. [DOI] [PubMed] [Google Scholar]
- 38.Hostetter MK. 2000. RGD-mediated adhesion in fungal pathogens of humans, plants and insects. Curr Opin Microbiol 3:344–348. doi: 10.1016/S1369-5274(00)00101-6. [DOI] [PubMed] [Google Scholar]
- 39.Yasuda T, Shishido K. 1997. Aggregation of yeast cells induced by the Arg-Gly-Asp motif-containing fragment of high-molecular-mass cell-adhesion protein MFBA, derived from the basidiomycetous mushroom Lentinus edodes. FEMS Microbiol Lett 154:195–200. doi: 10.1111/j.1574-6968.1997.tb12643.x. [DOI] [PubMed] [Google Scholar]
- 40.Scarborough RM, Naughton MA, Teng W, Rose JW, Phillips DR, Nannizzi L, Arfsten A, Campbell AM, Charo IF. 1993. Design of potent and specific integrin antagonists. Peptide antagonists with high specificity for glycoprotein IIb-IIIa. J Biol Chem 268:1066–1073. [PubMed] [Google Scholar]
- 41.Luo S, Poltermann S, Kunert A, Rupp S, Zipfel PF. 2009. Immune evasion of the human pathogenic yeast Candida albicans: Pra1 is a Factor H, FHL-1 and plasminogen binding surface protein. Mol Immunol 47:541–550. doi: 10.1016/j.molimm.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 42.Calderone R. 1998. The INT1 of Candida albicans. Trends Microbiol 6:300–303. doi: 10.1016/S0966-842X(98)01321-3. [DOI] [PubMed] [Google Scholar]
- 43.Gale CA, Bendel CM, McClellan M, Hauser M, Becker JM, Berman J, Hostetter MK. 1998. Linkage of adhesion, filamentous growth, and virulence in Candida albicans to a single gene, INT1. Science 279:1355–1358. doi: 10.1126/science.279.5355.1355. [DOI] [PubMed] [Google Scholar]
- 44.Kinneberg KM, Bendel CM, Jechorek RP, Cebelinski EA, Gale CA, Berman JG, Erlandsen SL, Hostetter MK, Wells CL. 1999. Effect of INT1 gene on Candida albicans murine intestinal colonization. J Surg Res 87:245–251. doi: 10.1006/jsre.1999.5755. [DOI] [PubMed] [Google Scholar]
- 45.Bendel CM, Kinneberg KM, Jechorek RP, Erlandsen SL, Sahar DE, Wells CL. 2000. The Candida albicans INT1 gene facilitates cecal colonization in endotoxin-treated mice. Shock 13:453–458. doi: 10.1097/00024382-200006000-00006. [DOI] [PubMed] [Google Scholar]
- 46.Penn C, Klotz SA. 1994. Binding of plasma fibronectin to Candida albicans occurs through the cell binding domain. Microb Pathog 17:387–393. doi: 10.1006/mpat.1994.1084. [DOI] [PubMed] [Google Scholar]
- 47.Chaffin WL, Lopez-Ribot JL, Casanova M, Gozalbo D, Martinez JP. 1998. Cell wall and secreted proteins of Candida albicans: identification, function, and expression. Microbiol Mol Biol Rev 62:130–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nobile CJ, Schneider HA, Nett JE, Sheppard DC, Filler SG, Andes DR, Mitchell AP. 2008. Complementary adhesin function in C. albicans biofilm formation. Curr Biol 18:1017–1024. doi: 10.1016/j.cub.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Braun BR, Head WS, Wang MX, Johnson AD. 2000. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 156:31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.