Abstract

Thermophilic proteins have found extensive use in research and industrial applications because of their high stability and functionality at elevated temperatures while simultaneously providing valuable insight into our understanding of protein folding, stability, dynamics, and function. Cyclophilins, constituting a ubiquitously expressed family of peptidyl–prolyl isomerases with a range of biological functions and disease associations, have been utilized both for conferring stress tolerances and in exploring the link between conformational dynamics and enzymatic function. To date, however, no active thermophilic cyclophilin has been fully biophysically characterized. Here, we determine the structure of a thermophilic cyclophilin (GeoCyp) from Geobacillus kaustophilus, characterize its dynamic motions over several time scales using an array of methodologies that include chemical shift-based methods and relaxation experiments over a range of temperatures, and measure catalytic activity over a range of temperatures to compare its structure, dynamics, and function to those of a mesophilic counterpart, human cyclophilin A (CypA). Unlike those of most thermophile/mesophile pairs, GeoCyp catalysis is not substantially impaired at low temperatures as compared to that of CypA, retaining ~70% of the activity of its mesophilic counterpart. Examination of substrate-bound ensembles reveals a mechanism by which the two cyclophilins may have adapted to their environments through altering dynamic loop motions and a critical residue that acts as a clamp to regulate substrate binding differentially in CypA and GeoCyp. Fast time scale (pico- to nanosecond) dynamics are largely conserved between the two proteins, in accordance with the high degree of structural similarity, although differences do exist in their temperature dependencies. Slower (microsecond) time scale motions are likewise localized to similar regions in the two proteins with some variability in their magnitudes yet do not exhibit significant temperature dependencies in either enzyme.
The cyclophilins make up a widely distributed family of proteins, found across all kingdoms of life and known to be absent in only a small number of bacteria and archaea.1,2 Among organisms with cyclophilins, they are found in every cell and are often present as multiple isoforms (e.g., 17 isoforms exist in humans, eight in Saccharomyces cerevisiae, and two in Escherichia coli).3,4 Most, although not all, cyclophilins catalyze the cis–trans isomerization of peptidyl–prolyl bonds. Cyclophilins play a role in a range of biological functions, including as chaperones in protein folding and trafficking, in multiple signal transduction pathways, in pre-mRNA splicing, and as extracellular signaling molecules.1,5–9 Multiple viruses, including HIV-1 and hepatitis C, have been shown to utilize human cyclophilins in promoting viral replication and infectivity.10,11 Increasingly, cyclophilins are also being recognized for their dual roles in driving a number of cancers and other inflammatory diseases, acting both intracellularly to protect tumor cells against stresses, including hypoxia and high levels of reactive oxygen species, and extracellularly as cytokines driving disease progression.8,12–16 Among the many biological roles identified for cyclophilins, their ability to provide tolerance to a range of stresses, including high salinity, oxidative stress, osmotic stress, infection, cold, and heat, has been identified in many species.17–20 The specific mechanism by which cyclophilins provide these various stress appears to be multifaceted; however, the breadth of protection provided has led most studies to hypothesize that, in general, tolerance is mediated through protein chaperone activity as cyclophilins act to maintain protein homeostasis and promote proper protein folding.21
Aside from their broad biological relevance, cyclophilins, and namely the prototypical cyclophilin, human cylophilin A (CypA), have been used extensively to study the relationship among enzyme structure, dynamics, and function, yielding important insights into the role of inherent protein motions in regulating and/or directing catalysis. Specifically, early work on CypA indicated that the inherent dynamic motions of the protein correlate strongly with rates of catalytic turnover, suggesting that dynamics have been evolutionarily tuned for function.22 More recently, however, it has become clear that the dynamic landscape of CypA is significantly more complex than originally thought, with significant cross-talk between distinct dynamic segments.23 A powerful tool for revealing this dynamic landscape in CypA, as well as in studying dynamic motions in other systems, has been measuring motions over a range of temperatures.22 As fruitful as this approach has been, CypA’s reduced long-term stability above ~30 °C has limited the range over which these studies can be conducted. While two thermophilic proteins with cyclophilin-like folds have been previously characterized, they are catalytically inactive as peptidyl–prolyl isomerases and have likely evolved to fulfill some other function.24 Therefore, probing the structure, dynamics, and function of a catalytically active thermophilic cyclophilin counterpart would allow us to determine the degree of evolutionary conservation among cyclophilins.
In this study, we have characterized the structure, dynamics, and enzymatic function of the sole cyclophilin (GeoCyp) encoded in the genome of the thermophilic bacterium Geobacillus kaustophilus and compared them to those of the prototypical human homologue, CypA. Initially discovered by Takami et al. in deep sea sediment of the Mariana Trench, G. kaustophilus live at an optimal temperature of 60 °C, with a maximal temperature of 74 °C.25,26 GeoCyp is 49% similar and 37% identical to CypA and, on the basis of secondary structural predictions, adopts many, if not all, of the same secondary structural elements as CypA.27 As we have shown previously, GeoCyp is much more thermostable than CypA; while CypA denatures entirely with a Tm of 51 °C, GeoCyp exhibits a single secondary structural transition at 68 °C yet maintains some structure with significant secondary structural elements as high as 95 °C.27 Additionally, while CypA precipitates from solution when maintained above ~30 °C for an extended period of time, GeoCyp is structurally stable at 40 °C for at least several weeks. Therefore, we have determined the NMR solution structure of GeoCyp, revealing a typical cyclophilin fold but with several shortened loops relative to CypA. By generating ensembles of substrate-bound GeoCyp, we have identified specific conformational changes with a key residue involved in substrate binding and revealed a conserved electrostatic mechanism of isomerization compared to that described in our recent study of human CypA.28 We have also quantitatively characterized GeoCyp’s ability to bind and catalyze isomerization of a peptide substrate. Finally, combining experimental and computational approaches, we compared dynamic motions between GeoCyp and CypA over fast (pico- to nanosecond) and slower (microsecond) time scales. These studies reveal that, despite the vast evolutionary time separating the two cyclophilins and the drastically different environments for which the two proteins are adapted, their in vitro binding and catalytic functions are remarkably similar, and that they likewise exhibit similar dynamic profiles over a broad range of time scales and temperatures. These findings contrast with most previous findings for other thermophile/mesophile enzyme pairs, hinting that the specific roles of cyclophilins within the cell may have subjected the proteins to evolutionary pressures different from those of other protein families previously studied.
MATERIALS AND METHODS
Protein Expression and Purification
CypA and GeoCyp were expressed and purified as previously published, finishing with the purified protein in 50 mM Na2HPO4, 1 mM DTT, and 1 mM EDTA (pH 6.5).22,27 To allow for amide proton exchange, for deuterated proteins, pellets were lysed in 5 M guanidine hydrochloride, 100 mM Tris, and 100 mM EDTA (pH 7.5) and then dialyzed for 24 h into 1 M arginine, 100 mM Tris, and 100 mM NaCl (pH 7.5) and then into the nickel equilibration buffer (GeoCyp) or SP equilibration buffer (CypA). From this point, all purifications proceeded as described above. The peptide substrate was purified as previously published, finishing with a suspension in 50 mM Na2HPO4, 1 mM DTT, and 1 mM EDTA (pH 6.5).29
NMR Assignments
The model peptide was assigned (Table 1 of the Supporting Information) by collecting 15N HSQC, 13C HSQC, and HH TOCSY experiments on a 13C-and 15N-labeled peptide at 25 °C using a Varian 600 MHz spectrometer, with assignments at other temperatures determined by following amide peak positions. All spectra were processed using NMRPipe, and all data were analyzed using the CCPNmr analysis software package.30,31
Measuring Catalytic Efficiency
Catalytic efficiency was measured via the ZZ-exchange NMR experiment. Isotopically 15N-labeled peptide substrate (1 mM) was mixed with 1 or 20 μM protein, depending on the temperature used (20 μM for 0–20 °C and 1 μM for 30–45 °C). Data were collected with mixing times of 0, 0.036, 0.072, 0.144, 0.24, 0.3, 0.36, 0.54, 0.72, 0.9, 1.08, and 1.2 s on a Varian 600 MHz spectrometer and fit, via linear least-squares fitting, to the equations described by Farrow et al.32 The trans state of the peptide was found to comprise 89% of the total peptide by measuring peak intensities in the absence of enzyme. Cis, trans, and both exchange peaks of Leu 7 and cis, trans, and one exchange peak (the other is overlapped with another residue) of Asp 6 were simultaneously fit to determine kexeff values. Longitudinal relaxation was found to be nearly identical for both cis and trans conformations of both Asp 6 and Leu 7, so a single longitudinal relaxation parameter was used. For CypAR55A, CypAR55A/A103R, Geo-CypR47A, and GeoCypR47A/R92A, 1 mM 15N-labeled peptide was mixed with 100 μM protein, and data were collected at 30 °C using mixing times of 0, 0.192, 0.384, 0.576, 0.768, 1.014, 1.152, and 1.344 s.
Measuring Binding Affinity
Binding affinity was measured by NMR titration. 15N HSQC spectra were collected on 500 μM 15N-labeled protein in the presence of 0, 0.1, 0.2, 0.5, 1, and 2 mM peptide substrate on a Varian 900 MHz spectrometer. For peaks with significant chemical shift changes upon titration, chemical shift changes were least-squares fit individually to the steady state equilibrium binding equation below.
where F([L]) is the ligand-dependent chemical shift change, Fmax is the chemical shift change upon full saturation, [P] is the total protein concentration, [L] is the total ligand concentration, and KD is the dissociation constant. All chemical shifts that could be fit well individually (r2 > 0.99; indicating they are in the fast exchange regime needed to accurately calculate binding affinity) were then fit simultaneously, yielding a single dissociation constant determined for each protein.
NMR Relaxation Experiments
15N TROSY CPMG-RD experiments were conducted with 1 mM deuterated, isotopically 15N-labeled CypA and GeoCyp on a Varian 900 MHz spectrometer with a cryogenically cooled probe. Data were collected at 0, 10, 20, and 30 °C for CypA, as indicated, using constant time relaxation periods of 50, 60, 80, and 90 ms. Data were collected at 0, 10, 20, and 30 °C for GeoCyp, using constant time relaxation periods of 60, 70, 90, and 100 ms. R1 relaxation was measured on 0.5 mM [15N]CypA or GeoCyp, using mixing times of 10, 30, 50, 70, 90, and 110 ms.
NMR Solution Structure Determination
15N-edited and 13C-edited NOESY experiments were conducted with isotopically 15N- and 13C-labeled GeoCyp and used with previously determined chemical shift assignments27 to identify long-range interactions. Chemical shifts were analyzed using TALOS+33 and used to guide Rosetta34 fragment analysis. NOESY peak assignments were analyzed using CYANA version 2.135 and converted to Rosetta constraint format using the CS-Rosetta Toolkit (www.csrosetta.org). The RASREC-Rosetta algorithm36 was then used to calculate an ensemble of 20 lowest-scoring structures. This was run using the Janus supercomputing cluster at the University of Colorado, employing Message Passing Interface (MPI) over 528 CPUs. Violation analysis of the resulting ensemble of structures was performed using PDBStat37 and PSVS analysis (psvsnesg.org). Electrostatic potentials were determined using the APBS web server38 (www.poissonboltzmann.org).
Molecular Dynamics Ensembles
Ensembles of GeoCyp were generated as previously described for CypA.28 Briefly, the bound cis and bound trans states of GeoCyp were modeled starting from the free state of GeoCyp determined in this work.28 The two bound states where then simulated using the Amber99SB*-ILDN force field in explicit TIP3P water for 100 ns each at 300 K; the two final structures were then used as the starting structures for a chemical shift and NOE replica-averaged restrained simulation.28,39–42 CamShift was used to back-calculate the chemical shifts from both replicas at each time step. The force constant for the chemical shift restraints was set to 5.2 kJ/mol, and the force constant for the NOEs was set to 250 kJ mol−1 nm−2 with a bottom flat potential that is zero between 0.3 and 0.5 nm.43 Each replica has evolved through a series of annealing cycles between 300 and 450 K (100 ps at 300 K, 100 ps during which the temperature increased linearly to 450 K, 100 ps of constant-temperature molecular dynamics at 450 K, and 300 ps during which the temperature decreased linearly to 300 K). Each replica has evolved for a total nominal time of 150 ns. The final ensembles comprise all the 300 K structures sampled by both replicas after the first 50 ns. The averaged NOE and chemical shift restraints were added to GROMACS by using PLUMED2 and ALMOST.44–46
Electric Field Calculations
The electric field in the active site of GeoCyp has been calculated on the center of mass of the Gly–Pro peptide bond, using the partial charges of the force field for the whole GeoCyp. The z, x, and y components of the field were defined, following our previous work,28 as the normal to the ring plane defined by the N, Cα, and Cγ atoms of the proline residue; the Gly-C′–N-Pro peptide bond; and the normal to the such defined xz plane, respectively. The electric field calculated for CypA with the partial charges is in remarkable agreement with that calculated ab initio. Indeed, the average difference between the two methods is ~1 MV/cm.
Sequence Alignment
All Bacillaceae cyclophilins in the NCBI RefSeq database (www.ncbi.nlm.nih.gov/refseq) for which a full species name was indicated were included in the analysis. Alignment was perform using the Clustal Omega sequence alignment program.47
RESULTS
GeoCyp Structure Determination and Comparison of It to That of Human CypA
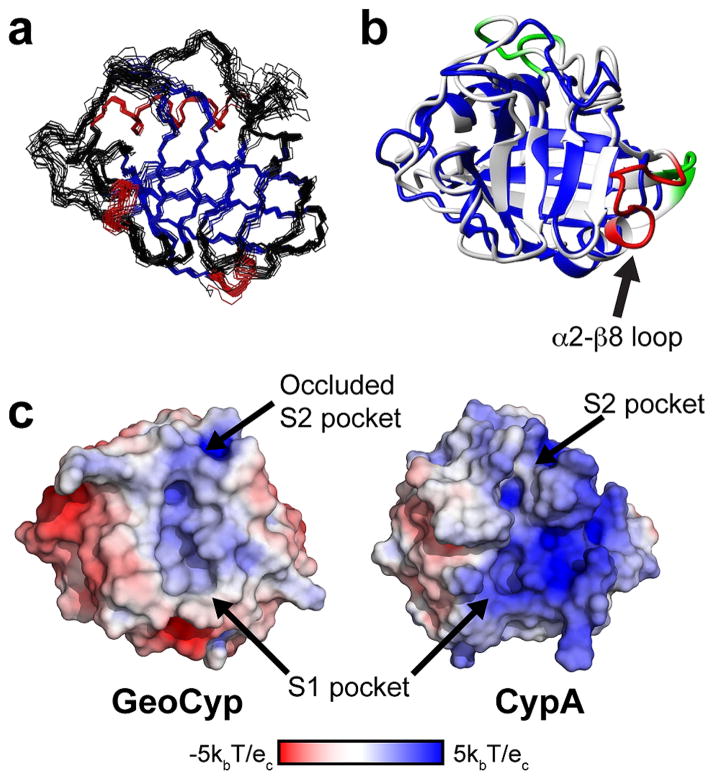
As we have previously shown GeoCyp to be a catalytically active, thermophilic cyclophilin,27 here we sought to structurally compare it to a mesophilic counterpart. Using the CS-ROSETTA36 implementation of the ROSETTA34 structure prediction platform, we determined the NMR solution structure of GeoCyp with a backbone rmsd among the 20 lowest-energy structures of 0.72 Å [Protein Data Bank entry 2MVZ (see Figure 1a and Table 1 for full statistics)]. GeoCyp adopts a typical cyclophilin fold, consisting of eight antiparallel β-strands arranged in a β-barrel, with two or three turn α-helices capping either end of the barrel, and an additional short 3–10 helix aligned parallel to the β-strands within the catalytic active site. Despite being only 41% identical, the β-barrel structures of GeoCyp and CypA are nearly superimposable, as are the 3–10 helix and the first of the α-helices (backbone rmsd of α-helices and β-sheets of 0.95 Å vs a global backbone rmsd of 1.55 Å). Figure 1b shows an alignment of CypA with the lowest-energy GeoCyp structure determined. GeoCyp contains several significantly shortened loops relative to CypA. Residues 12–15, 43–45, and 77–79 in CypA are absent from GeoCyp (Figure 1b, green); additionally, a 12-residue span (Figure 1b, red) comprising the final turn of the second α-helix (α2) and a loop between the helix and final β-sheet (β8) is absent from GeoCyp, which instead contains a two-residue stretch directly linking the truncated α-helix to the β-sheet. This α2–β8 loop comprises CypA residues 143–154 that are replaced by GeoCyp residues 132 and 133.
Figure 1.
Structural comparison of GeoCypA and CypA. (a) Solution structure of GeoCyp. The 20 lowest-energy structures are shown, with β-sheets colored blue and helices colored red. (b) Superimposed structures of CypA (from the previously generated molecular dynamics ensemble,28 white) and GeoCyp (lowest-energy structure, blue). Three short loop deletions from CypA are colored green, while the large α2–β8 loop deletion is colored red. (c) Surface representations of free CypA and GeoCyp structures. Structures are colored according to surface charge, with blue indicating basic charge and red indicating acidic charge. S1 and S2 binding pockets are indicated, including the occluded S2 pocket of GeoCyp.
Table 1.
Structural Statistics for the GeoCyp RASREC Rosetta Structuresa
| no. of residues | 146 |
| no. of NOE-based distance restraints | |
| NOE distance restraints (violations of ≥0.5 Å) | 1687 (146 ± 10) |
| intraresidue | 387 |
| inter-residue | 1300 |
| sequential (|i − j| = 1) | 548 |
| medium-range (|i − j| < 4) | 265 |
| long-range (|i − j| > 5) | 487 |
| no. of other restraints | |
| φ + ψ dihedral angle restraints (violations of ≥5°)b | 224 (29 ± 3.6) |
| average rmsd from the average structurec | |
| backbone (Å) | 0.72 ± 0.14 |
| heavy atom (Å) | 1.2 ± 0.14 |
| Ramachandran plot summaryd (%) | |
| most favored regions | 84.5 |
| allowed regions | 14.8 |
| generally allowed regions | 0.6 |
| disallowed regions | 0.1 |
| deviations from idealized geometry | |
| bond lengths (Å) | 0.011 |
| bond angles (deg) | 0.4 |
Statistics are given for the 20 best scored structures.
Torsion angle restraints derived from TALOS+.
The rmsds calculated using the iCing server.
Analysis performed using PROCHECK.
Previous studies have shown that thermophilic proteins tend to be shorter in sequence length and contain deletions in solvent-exposed loops relative to their mesophilic homologues, suggesting that the overall shorter length of GeoCyp, including the significantly shortened α2–β8 loop, and perhaps other loop deletions, may play roles in enhancing the stability of GeoCyp.48,49 While the α2–β8 loop is a region of structural diversity among cyclophilins, the extent of the deletion in GeoCyp is present in only two other cyclophilin-like domains whose structures have been determined, both from other thermophilic bacteria, Archaeoglobus fulgidus and Thermotoga maritima.24 While these other two thermophilic proteins do adopt typical cyclophilin folds, they retain a very low degree of homology to catalytically active cyclophilins, including lacking the absolutely conserved catalytic arginine (Arg 55 in CypA and Arg 47 in GeoCyp), indicating that they are not functional isomerases. Alternatively, the deletions of residues 12–15 and 43–45 have been previously identified in other cyclophilins, including other human homologues.4
While GeoCyp retains most of the canonical cyclophilin active site residues, there are two exceptions: His 110 in GeoCyp, which is typically a tryptophan but is present as a histidine in several catalytically active cyclophilins, and Val 53, which is typically a methionine but occasionally is replaced by other hydrophobic residues.4 Notably, in the unbound state for which we have determined the structure, the GeoCyp “gatekeeper” residues adopt an occluded conformation, with Arg 92 and Thr 64 blocking the hydrophobic groove into which substrate residues N-terminal of the isomerized proline generally fit (Figure 1c). While some cyclophilins with occluded active sites are still competent to bind substrates, bulky and occluding gatekeeper residues have been associated with a reduction or ablation of substrate binding.4 GeoCyp retains the hydrophobic active site characteristic of cyclophilins, and a charge distribution similar to that of CypA, with a number of basic residues flanking the active site pockets (Figure 1c).
Differential Conformational Sampling between Geo-Cyp and CypA When They Are Bound to a Peptide Substrate
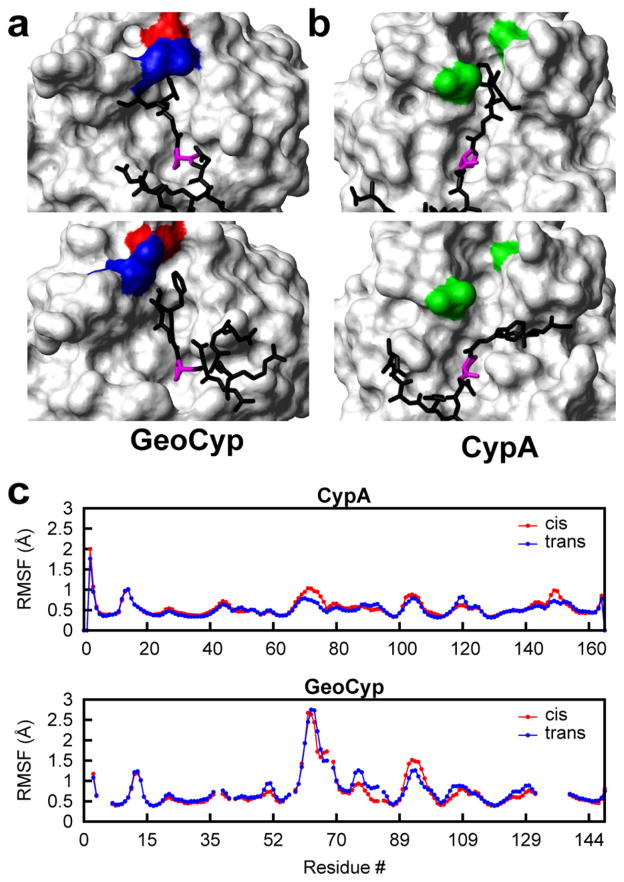
We have previously reported a peptide substrate for cyclophilins, GSFGPDLRAGD, a slightly modified version of one identified by Piotukh et al. in a phage display screen, in which the G-P peptide bond is readily isomerized by both CypA and GeoCyp (see Table 1 of the Supporting Information for peptide NMR assignments).27,29,50 The peptide is representative of a large class of putative biological cyclophilin targets, and amino acid substitutions have shown that residues Phe 3 and Gly 4 are critical in mediating the cyclophilin-substrate binding interaction.50 To investigate the conformational landscape of GeoCyp during turnover of this peptide, we docked the peptide to the solution structure using internuclear Overhauser effect (NOE) distance restraints and generated an ensemble of structures in the bound cis and bound trans forms, following a previously described method applied to CypA.28,42 The peptide binds in a mode similar to that of CypA, consistent with chemical shift perturbations observed upon addition of the substrate, which map to the canonical active site in both proteins (Figure 1 of the Supporting Information). As shown in panels a and b of Figure 2, in both CypA and GeoCyp, the substrate proline localized to the hydrophobic S1 pocket, while Phe 3 of the peptide samples into and out of the S2 pocket in the peptide-bound ensembles. While the structure of the CypA S2 pocket remains relatively static throughout the ensemble, the loops creating the S2 pocket are highly dynamic in GeoCyp, permitting access to the pocket despite the occluded nature of the pocket in the unbound form. This mobility is reflected in the significantly higher root-mean-square fluctuation (rmsf) values of these loops in GeoCyp compared to CypA (Figure 2c). In particular, Arg 92 in GeoCyp is highly mobile, acting both to clamp Phe 3 in place when in the pocket and forming π–cation interactions with the Phe 3 aromatic ring when out of the pocket (Figure 2a).
Figure 2.
(a) Representative structures, with the peptide in the trans conformation, from the molecular dynamics ensemble of GeoCyp bound to the model peptide. Within the ensemble, Phe 3 of the peptide samples the S2 pocket (top structure) and also exits the S2 pocket (bottom structure). Residues Arg 92 (blue) and Asp 66 (red) serve to clamp Phe 3 of the peptide into the pocket or partially occlude the pocket when Phe 3 is not in it. (b) Representative structures, with the peptide in the trans conformation, from the previously generated molecular dynamics ensemble of CypA bound to the model peptide.28 Phe 3 of the peptide also samples in (top structure) and out (bottom structure) of the S2 pocket within the CypA ensemble; however, the S2 pocket structure remains fully open, and no clamping occurs. Residues Ala 103 and Gly 75, homologous to Arg 92 and Asp 66 in GeoCyp, respectively, are colored green. (c) Root-mean-square fluctuation (rmsf) values for the backbone residues of CypA and GeoCyp in chemical shift-guided molecular dynamics simulations of the proteins bound to the peptide substrate in the cis (red) and trans (blue) conformations. GeoCyp residue numbers are shifted to match CypA, such that homologous residues are in line with one another.
A Gatekeeper Residue That Regulates Substrate Binding
Given the apparent role of S2 pocket-adjacent residues in regulating substrate binding in the substrate-bound GeoCyp ensembles, we sought to experimentally examine the role of Arg 92 (Ala 103 in CypA), as the bulkiest “gatekeeper” residue present and because of the apparent π–cation interactions between the side-chain guanidinium group and the substrate phenol ring when Phe 3 shifts out of the S2 pocket. Homologous swap mutations were thus generated in each protein (CypAA103R and GeoCypR92A). NMR titrations with the peptide substrate revealed that the mutations strengthened and weakened binding in CypA and GeoCyp, respectively, such that, in each case, the presence of arginine led to tighter binding (Table 2), consistent with clamping of the substrate and/or π–cation interactions as identified in ensembles of GeoCyp. In addition to binding, catalytic efficiency was measured for both wild-type and mutant proteins. Because of the reversible nature of the proline isomerization process, catalysis is measured via a ZZ-exchange NMR experiment in which the substrate is 15N-labeled and a low, catalytic concentration (20 μM) of protein is added. The time-dependent appearance of cross-peaks indicates turnover and can be used to determine an effective exchange rate (kexeff), which is a measurement of catalytic efficiency as previously described.29,32 This assay is not a measure of the catalytic rate on the enzyme that is not readily measurable but, instead, is a measure of the catalytic efficiency at the particular catalytic enzyme concentration used. Nonetheless, ZZ-exchange is an effective way to compare the catalytic rates of cyclophilin-mediated isomerization. As shown in Table 2, CypAA103R and GeoCypR92A catalyzed less and more efficiently than the wild-type proteins, respectively, indicating that the increase in substrate affinity caused by the presence of arginine at this position corresponds to a reduction in the rate of catalytic turnover. Analysis of cyclophilin protein sequence within the Bacillaceae family, of which G. kaustophilus is a member, reveals that only thermophilic bacteria contain an arginine at this site (see Discussion). Thus, evolution has apparently fine-tuned the S2 pocket to mediate a trade-off between a reduced binding affinity and a higher rate of turnover.
Table 2.
Binding Affinities and Catalytic Efficiencies for CypA, GeoCyp, and Their Mutants
| protein | KD (μM) | kexeff (s−1) |
|---|---|---|
| CypAWT | 76 ± 3a | 10.0 ± 0.5 |
| CypAA103R | 42 ± 3 | 7.6 ± 0.3 |
| GeoCypWT | 39 ± 2 | 4.7 ± 0.2 |
| GeoCypR92A | 118 ± 2 | 7.1 ± 0.3 |
Errors are fit errors from a single experiment.
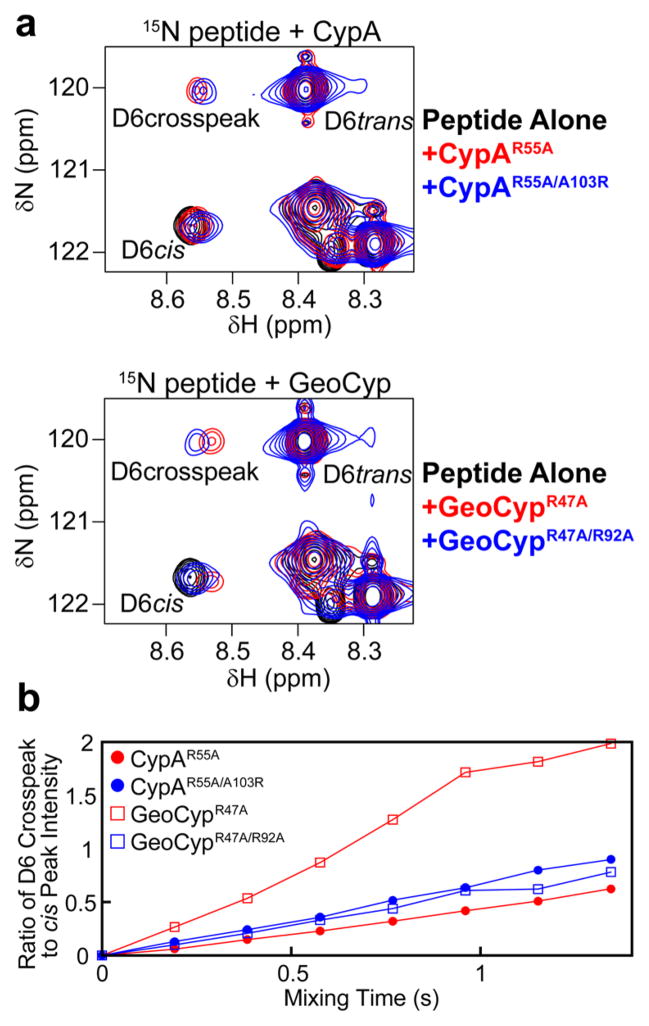
To further probe this trade-off between substrate binding and catalytic turnover in cyclophilins, we sought to examine the impact of altering affinity in the context of a second mutation that is known to significantly reduce the background binding affinity. We therefore generated the R92A and A103R swap mutations in the context of mutation to the “catalytic” arginine (Arg 55 in CypA and Arg 47 in GeoCyp). In addition to significantly reducing binding affinity, mutation of the catalytic arginine alone also nearly, but not entirely, ablates catalysis (see cross-peaks in Figure 3a, only in the presence of enzyme). For these double mutants (CypAR55A/A103R and GeoCypR47A/R92A), both proteins still bind more tightly with an arginine than alanine at the GeoCyp-92/CypA-103 site, as evidenced by a larger change in the chemical shift of 15N-labeled peptide peaks upon addition of protein (Figure 3a). As shown in Figure 3b, however, catalytic turnover follows an opposite trend when the catalytic arginine mutation is present, with CypAR55A/A103R catalyzing more efficiently than CypAR55A and GeoCypR47A catalyzing more efficiently than GeoCypR47A/R92A. Because significant line broadening occurs due to protein binding at the high concentrations needed to observe turnover in the context of mutation to the catalytic arginine, kexeff cannot be accurately quantitated. However, the ratio of cis peak intensity to cross-peak intensity is used as a proxy for the rate of turnover, where a larger slope corresponds to faster turnover (Figure 3b). Collectively, these data provide a rationale for the trade-off between binding affinity and substrate turnover in cyclophilins, wherein binding must be sufficiently tight to engage the substrate yet sufficiently weak to allow for substrate release after turnover.
Figure 3.
(a) ZZ-exchange spectra for the peptide substrate alone (black), with 100 μM protein with mutation to the “catalytic” residue CypAR55A or GeoCypR47A (red), and with 100 μM protein with mutation to both the “catalytic” residue and swap mutant CypAR55A/A103R or GeoCypR47A/R92A (blue). All spectra were collected at 30 °C, with a mixing time of 0.768 s. For both CypA and GeoCyp, arginine in the swap mutation position leads to tighter binding, as evidenced by larger chemical shift changes of the Leu 6 cis peak and cross-peak. (b) In the context of mutation of the “catalytic” arginine to an alanine, tighter binding corresponds to faster turnover. Because of significant line broadening due to protein binding at the high concentrations needed to observe turnover in the context of mutation to the catalytic arginine, kexeff cannot be accurately determined, so the ratio of cis peak intensity to cross-peak intensity is used as a proxy for rate of turnover, where a larger slope corresponds to faster turnover.
An Active Site Electric Field Is Conserved between CypA and GeoCyp
As described above, we utilized chemical shift-guided molecular dynamics methods to assess the mechanism by which cyclophilins catalyze proline isomerization. Recently, we demonstrated the existence of an electric field within the CypA active site in the –Z direction (defined as normal to the pyrrolidine ring of the isomerized proline) that acts, in a so-called “electrostatic handle” mechanism, to facilitate isomerization. Specifically, this electric field exists in both the cis bound and trans bound states and functions to reduce the energy of the ω = 90° transition state barrier by ~30 kJ/mol.28 To determine whether this catalytic mechanism is also conserved, we analyzed the ensembles of peptide-bound GeoCyp described above. As shown in Figure 4, GeoCyp exhibits a similar –Z electric field within the active site in both the cis and trans conformations, with a mean value of ~30 MV/ cm, compared to a mean value of ~45 MV/cm previously identified in CypA. Relative to CypA, this weaker field in GeoCyp, along with the tighter binding affinity identified above, explains the reduction in catalytic efficiency (Table 2). These results suggest that GeoCyp and CypA utilize an evolutionarily conserved mechanism of isomerization that is fine-tuned with respect to binding via the S2 binding pocket.
Figure 4.
GeoCyp exhibits a –Z electric field within the active site. Population distribution of the Z electrostatic fields in GeoCyp in the cis (black) and trans (gray) peptide conformations, where Z is defined as normal to the pyrrolidine ring of Pro 5 in the peptide substrate.
GeoCyp and CypA Exhibit Comparable Temperature-Dependent Activities
Thermophilic enzymes are generally optimally functional at the source organism’s optimal temperature, exhibiting a significant reduction in catalytic turnover at lower temperatures.51–58 As such, we measured the catalytic efficiency (kexeff) of both CypA and GeoCyp, using ZZ-exchange with 15N-labeled peptide substrate, over the range of temperatures that can be accessed by NMR spectroscopy. For the ZZ-exchange-based catalytic experiment, kexeff can be accurately measured only between ~0.5 and 30 s−1. To remain within this range, two different enzyme concentrations were used, 20 μM for 0–20 °C and 1 μM for 30–45 °C, with 1 mM 15N-labeled peptide used in all cases. As shown in Figure 5, over the range of 0–37 °C, GeoCyp consistently catalyzes isomerization at ~70% of the rate of CypA. Only at 45 °C, above the physiologically optimal temperature for CypA and approaching its denaturation temperature of 51 °C,27 does the increase in turnover rate with temperature slow for CypA and continue for GeoCyp such that they catalyze turnover at a comparable rate. Unlike many previously studied thermophiles, therefore, GeoCyp does not exhibit a substantial impairment of function at lower temperatures relative to that of its mesophilic counterpart, CypA.
Figure 5.
Catalytic efficiency (kexeff) for CypA and GeoCyp from 0 to 45°C. 15N-labeled peptide (1 mM) and 20 μM (0–20 °C) or 1 μM (30–45 °C) unlabeled protein were used. Error bars represent fit errors within a single experiment.
Dynamics Are Similar over Multiple Time Scales between CypA and GeoCyp, with Variability in Temperature Dependence and Magnitude
Given the surprisingly high activity of GeoCyp at low temperatures, we decided to examine whether, like many other thermophilic proteins, GeoCyp is hyperstabilized at low and moderate temperatures or, as might be predicted from our functional data, GeoCyp exhibits dynamics similar to those of CypA over these temperatures. NMR relaxation experiments were thus collected over a range of temperatures for both CypA and GeoCyp. As described in more detail below, by utilizing the Carr–Purcell–Meiboom–Gill relaxation dispersion (CPMG-RD) experiment, we found that GeoCyp exhibits weak self-association on the millisecond time scale, precluding direct analysis of these motions. To determine whether self-association would significantly impact measurement of faster time scale dynamics, we examined both fast time scale (pico- to nanosecond) dynamics via longitudinal (R1) relaxation experiments and slower (microsecond) motions via R20, the Rex-independent component of transverse relaxation that was estimated via the CPMG-RD experiment collected with a νcpmg of 1000 s−1.59,60 As shown in Figure 2 of the Supporting Information, we collected data at multiple concentrations and found minimal concentration-dependent changes to either R20 or R1 values, indicating that the weak self-association predominantly impacts motions on the slower, millisecond time scale and allowing comparison of CypA and GeoCyp dynamics over faster time scales such that we are able to determine the degree to which dynamics are stabilized in GeoCyp. We attempted to determine order parameters (S2) for both CypA and GeoCyp by measuring R1, R2, and 1H–15N heteronuclear NOE relaxation values and applying Modelfree 4.15 via the FAST-Modelfree implementation.61–63 However, using either an isotropic or axially symmetric diffusion model, a large number of residues were unable to be fit by any set of model-free parameters for either CypA (34 of 144 residues unassigned) or GeoCyp (24 of 120 residues unassigned), indicating that the model-free formalism is unable to accurately describe the dynamics of these cyclophilins, perhaps because of the large number of residues exhibiting micro- to millisecond internal motions (nearly 50% in CypA23). We have therefore directly compared R1 and R20 values between CypA and GeoCyp.
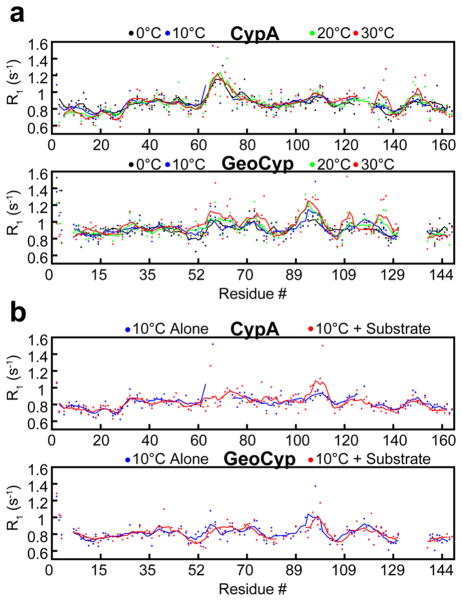
For CypA and GeoCyp, regions with elevated R1 values localize well to homologous regions within the proteins (Figure 6), corresponding to the high degree of structural similarity (Figure 1).64 Namely, these regions include the two large active site loops (residues 60–80 and 100–110 in CypA and residues 57–76 and 89–99 in GeoCyp), as well as the β7–α2 loop (residues 135–137 in CypA and residues 124–126 in GeoCyp). For each protein, R1 data were collected over a range of temperatures from 0 to 30 °C. Fast time scale dynamics in CypA are largely unaffected by temperature over this range, aside from slight elevations in R1 in the β7–α2 and α2–β8 loops. GeoCyp, however, exhibits significant temperature-dependent changes in active site R1 values over the same temperature range, including both large active site loops, suggesting that on the pico- to nanosecond time scale, GeoCyp may exhibit some of the low-temperature dynamic dampening that has been identified in other thermophilic proteins relative to their mesophilic counterparts.51,52
Figure 6.
(a) R1 relaxation rates for CypA and GeoCyp at 0 °C (black), 10 °C (blue), 20 °C (green), and 30 °C (red). Baseline values were subtracted off of each data set to normalize values between temperatures, facilitating comparison of dynamic variations. (b) R1 relaxation rates for CypA and GeoCyp at 10 °C alone (blue) and in the presence of saturating concentrations (4 mM) of the peptide substrate (red). For all plots, dots represent individual measurements and lines depict a five-residue moving average. GeoCyp residue numbers are shifted to match those of CypA, such that homologous residues are in line with one another.
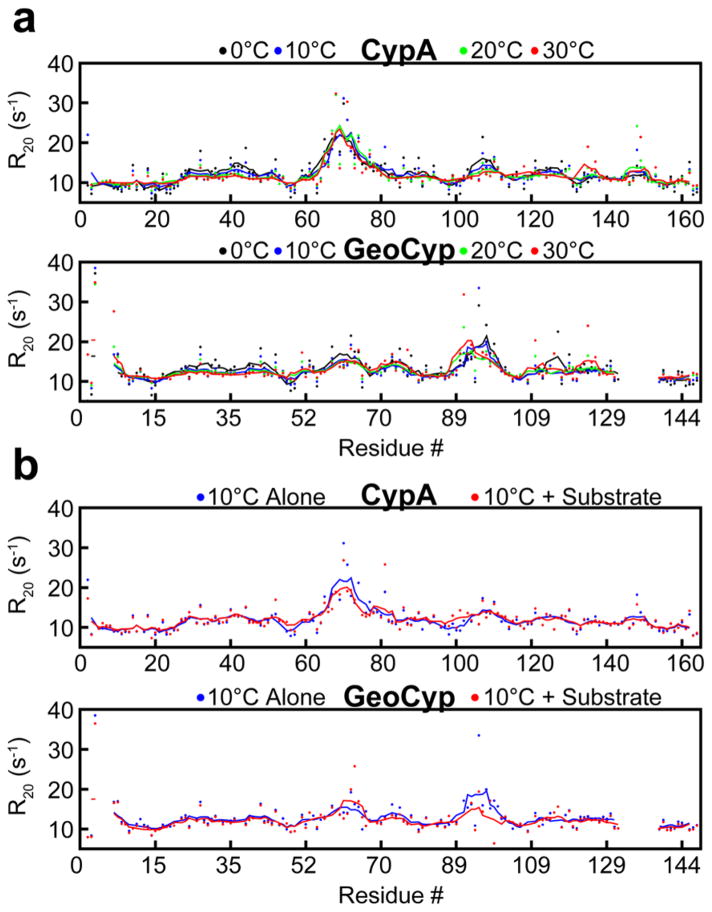
R20 relaxation rates were determined for both GeoCyp and CypA over the same temperature ranges as for R1 relaxation. As shown in Figure 7, localized variability in R20 relaxation is largely independent of temperature for both GeoCyp and CypA, indicating a lack of low-temperature dynamic dampening of GeoCyp on the microsecond time scale. R20 relaxation exhibits similar patterns throughout the two proteins, including the largest elevations mapping predominantly to loops within the active site, although with variability in the magnitudes. Notably, as with R1 values, loop 89–99 in GeoCyp exhibits much higher R20 values relative to those of the homologous loop in CypA, residues 100–110, consistent with the elevated flexibility identified in the molecular dynamics ensembles and required to access the occluded S2 binding pocket (Figures 1c and 2c).
Figure 7.
(a) R20 relaxation rates for CypA and GeoCyp at 0 °C (black), 10 °C (blue), 20 °C (green), and 30 °C (red). Baseline values were subtracted off of each data set to normalize values between temperatures, facilitating comparison of dynamic variations. (b) R20 relaxation rates for CypA and GeoCyp at 10 °C alone (blue) and in the presence of saturating concentrations (4 mM) of the peptide substrate (red). For all plots, dots represent individual measurements and the lines depict a five-residue moving average. GeoCyp residue numbers are shifted to match those of CypA, such that homologous residues are in line with one another.
Interestingly, neither R1 nor R20 values were dramatically impacted upon addition of saturating concentrations of the peptide substrate in CypA or GeoCyp (Figures 6b and 7b). The only significant changes seen are reductions in R20 values in the active site loops of each protein, likely corresponding to a reduced mobility upon substrate binding.
GeoCyp Weakly Self-Associates on the Millisecond Time Scale
The CPMG-RD experiment allows quantitative measurement of rates of motion in the slow micro- to millisecond range (~100–5000 s−1), which have previously been linked to enzymatic function in CypA.22,65 As deuteration has been previously shown to drastically improve the quality of CPMG-RD experiments applied to CypA, we generated uniformly 2H- and 15N-labeled GeoCyp.23 Previous studies have demonstrated that there is no dependence of CPMG-RD on protein concentration for CypA, indicating that no weak self-association is contributing to measured chemical exchange (Rex). To test for self-association of GeoCyp, we measured 15N CPMG-RD on GeoCyp over a concentration range of 0.5–2 mM and found that, unlike CypA, GeoCyp does exhibit concentration-dependent chemical exchange, indicating weak self-association (see Figure 3 of the Supporting Information). 15N CPMG-RD is a particularly sensitive means for monitoring weak self-association that is not readily observed in 15N HSQC spectra because of relatively small chemical shift perturbations that are induced. However, the high solubility of GeoCyp allowed us to collect a 15N HSQC spectrum at 9 mM and pinpoint the chemical shift perturbations (see Figure 3 of the Supporting Information). The majority of chemical shift changes between 0.5 and 9 mM mapped to the active site, suggesting that interactions between hydrophobic active site residues underlie this weak association. Thus, in the case of GeoCyp, this weak self-association precludes quantitative determination of self-association-independent rates on the millisecond time scale.
DISCUSSION
In this study, we have combined both recently developed chemical shift-based methods and standard NMR relaxation experiments to perform a comprehensive comparison of the structure, dynamics, and catalytic mechanism of a thermophilic/mesophilic pair of cyclophilins. GeoCyp adopts a typical cyclophilin fold, albeit with shortened loops commonly found in thermophilic proteins. Strikingly, despite the >20 °C difference in optimal growth temperature of humans and G. kaustophilus and the 17 °C increase in thermal melt transition of GeoCyp over CypA, catalytic activity is minimally reduced (~30%) for GeoCyp compared to that of CypA (Figure 5).27 This reduction in the level of catalysis appears to be mediated predominantly through higher binding affinity, with an additional, independent contribution from a somewhat reduced active site electric field. These findings contrast with multiple other studies of thermophile/mesophile pairs, for which thermophilic protein activities are significantly impaired at lower temperatures and become efficient only as the optimal organismal temperature is approached. Given cyclophilins’ roles in responding to cellular stress, including to heat and cold stress, perhaps maintenance of significant catalytic activity across a range of temperatures allows GeoCyp to respond efficiently to these stresses, especially given the temperature extremes that may be experienced in and around a deep-sea vent. Further investigation of other thermophilic cyclophilins may reveal whether this feature is unique to GeoCyp or common across thermophilic cyclophilins.
Studies here illustrate how cyclophilins have dynamically evolved to fine-tune mechanism. Specifically, structural analysis of GeoCyp in the free and substrate-bound forms has revealed a loop in the “gatekeeper” region of the protein that occludes the S2 binding pocket in the free enzyme but is nonetheless sufficiently mobile to permit binding to the substrate. The homologous mutational analysis of Arg 92 in GeoCyp (Ala 103 in CypA) highlights the evolutionary trade-off that exists between substrate affinity and rates of turnover. A previous study66 examining a different cyclophilin substrate demonstrated that rates of substrate release are comparable to rates of enzyme-bound isomerization; therefore, off-rates and isomerization rates both significantly impact kexeff, such that a reduced off-rate, caused by an arginine at residue GeoCyp-92/CypA-103, likewise reduces kexeff. However, under conditions of significantly weakened binding caused by mutation of Arg 47 in GeoCyp (Arg 55 in CypA), the rapid rate of substrate release becomes a limiting factor in isomerization, such that the increased binding affinity now increases kexeff. These data appear to be consistent with the optimal temperatures at which the mesophilic/thermophilic cyclophilins function; CypA binding affinity for the peptide substrate is highly temperature-dependent, indicating a significant entropic cost to binding. Thus, at the lower temperatures under which CypA operates, Ala 103 maintains sufficiently weak binding to allow efficient catalysis. In contrast, as the entropic cost of binding increases with the higher temperatures under which GeoCyp exists, the tighter binding that is mediated by loop clamping by Arg 92 functions to increase catalytic efficiency.
Our hypothesis of evolutionary tuning of the S2 loop is further bolstered by comparison of cyclophilin protein sequences across the Bacillaceae family. As shown in Table 2 of the Supporting Information, among Bacillaceae cyclophilins annotated in the RefSeq database, only in the closely related thermophilic genera Geobacillus and Anoxybacillus67 is the homologous site occupied by an arginine. The site contains alanine, or occasionally threonine or serine, throughout the other 71 members of the family, as in human CypA. Combined with our functional mutagenesis data, these results indicate an evolutionarily tuned functional role for this S2 site in regulating substrate interactions. The one other thermophilic Bacillaceae cyclophilin annotated, from Caldalkalibacillus thermarum,68 does contain an alanine at this homologous site, indicating that acquisition of this arginine adaptation is not universal among all thermophiles.
Numerous studies have focused on the interplay among protein dynamics, stability, and enzymatic function among thermophiles. In addition to maintaining stability at high temperatures, thermophilic proteins face the related challenge of retaining sufficient dynamic mobility to function efficiently. Given this balance, many studies have promoted the “corresponding state hypothesis” wherein evolution has tuned the dynamics of proteins to render all members of a given family equally dynamic at their source organisms’ optimal temperatures, rendering them hyperstabilized at lower temperatures and unstable at higher temperatures.53,54,56,57,69,70 Other studies, however, have refuted this notion, demonstrating cases for which thermophilic proteins exhibit dynamics comparable or elevated relative to that of a mesophile.58,71–73 The contradictions in these studies may reflect variability in the mechanisms of stabilization between different protein families but are also likely influenced by the time scale of dynamics that are observed, which can vary substantially depending on the technique utilized.71 Given the unusually high activity of GeoCyp at low temperatures relative to that of CypA (Figure 5) when compared to those of other thermophile/mesophile pairs, analyses of GeoCyp dynamics are unable to contribute directly to the debate over the corresponding state hypothesis; without substantial impairment of function at lower temperatures, the hypothesis makes no prediction about how motions should be affected. However, the temperature-dependent increases in deviations of R1 relaxation observed in GeoCyp but not CypA (Figure 6) suggest that comparable fast (pico- to nanosecond) time scale mobility exists in both proteins at lower temperatures, and that these motions increase more dramatically in GeoCyp with temperature. These findings are in line with several computational and experiment studies of other thermophiles, which found more fast time scale mobility at multiple temperatures when compared to that of mesophiles.71–74 Among these proteins, it appears that a more dynamic folded form of the protein contributes to a reduced entropic folding penalty, especially as temperature increases, such that more flexibility may actually stabilize the folded form.
In conclusion, while some dynamic variability exists between CypA and GeoCyp that may contribute to GeoCyp stability (elevated fast time scale dynamics) and function (magnitudes of loop motions around the S2 pocket), the analyses presented here of CypA and GeoCyp largely highlight the high degree of structural, dynamic, and mechanistic conservation in the cyclophilin family.
Supplementary Material
Acknowledgments
Funding
M.J.H. is supported by the Earleen and Victor Bolie Scholarship Fund and National Institutes of Health (NIH) Applications 5T32GM008730-13 and 1F31CA183206-01A1. E.Z.E. is supported by NIH Application 1RO1GM107262-01A1.
NMR experiments were conducted at several facilities described herein. The High Magnetic Field Laboratory (NHMFL) is supported by Cooperative Agreement DMR 0654118 between the National Science Foundation and the State of Florida. The Environmental Molecular Sciences Laboratory is a national scientific user facility sponsored by the Department of Energy’s Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory. The Rocky Mountain 900 Facility support by NIH Grant GM68928. This work utilized the Janus supercomputer, which is supported by the National Science Foundation (Grant CNS-0821794) and the University of Colorado. The Janus supercomputer is a joint effort of the University of Colorado, the University of Colorado Denver, and the National Center for Atmospheric Research.
ABBREVIATIONS
- GeoCyp
cyclophilin from G. kaustophilus
- CypA
human cyclophilin A
- rmsd
root-mean-square deviation
- rmsf
root-mean-square fluctuation
- NMR
nuclear magnetic resonance
- NOE
nuclear Overhauser effect
- kexeff
effective exchange rate
- CPMG-RD
Carr–Purcell–Meiboom–Gill relaxation dispersion
- HSQC
heteronuclear single-quantum coherence
Footnotes
The authors declare no competing financial interest.
Chemical shift changes for CypA and GeoCyp upon peptide binding, concentration dependence of millisecond dynamics in GeoCyp, Rex but not R1 or R20 values that are impacted by self-association in GeoCyp, NMR chemical shift assignments for the peptide substrate, and a sequence alignment of cyclophilins from Bacillaceae. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/ acs.biochem.5b00263.
References
- 1.Gothel SF, Marahiel MA. Peptidyl-prolyl cis-trans isomerases, a superfamily of ubiquitous folding catalysts. Cell Mol Life Sci. 1999;55:423–436. doi: 10.1007/s000180050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bang H, Pecht A, Raddatz G, Scior T, Solbach W, Brune K, Pahl A. Prolyl isomerases in a minimal cell. Catalysis of protein folding by trigger factor from Mycoplasma genitalium. Eur J Biochem. 2000;267:3270–3280. doi: 10.1046/j.1432-1327.2000.01355.x. [DOI] [PubMed] [Google Scholar]
- 3.Arevalo-Rodriguez M, Wu X, Hanes SD, Heitman J. Prolyl isomerases in yeast. Front Biosci. 2004;9:2420–2446. doi: 10.2741/1405. [DOI] [PubMed] [Google Scholar]
- 4.Davis TL, Walker JR, Campagna-Slater V, Finerty PJ, Paramanathan R, Bernstein G, MacKenzie F, Tempel W, Ouyang H, Lee WH, Eisenmesser EZ, Dhe-Paganon S. Structural and biochemical characterization of the human cyclophilin family of peptidyl-prolyl isomerases. PLoS Biol. 2010;8:e1000439. doi: 10.1371/journal.pbio.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferreira PA, Orry A. From Drosophila to humans: Reflections on the roles of the prolyl isomerases and chaperones, cyclophilins, in cell function and disease. J Neurogenet. 2012;26:132–143. doi: 10.3109/01677063.2011.647143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brazin KN, Mallis RJ, Fulton DB, Andreotti AH. Regulation of the tyrosine kinase Itk by the peptidyl-prolyl isomerase cyclophilin A. Proc Natl Acad Sci USA. 2002;99:1899–1904. doi: 10.1073/pnas.042529199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horowitz DS, Lee EJ, Mabon SA, Misteli T. A cyclophilin functions in pre-mRNA splicing. EMBO J. 2002;21:470–480. doi: 10.1093/emboj/21.3.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sherry B, Yarlett N, Strupp A, Cerami A. Identification of cyclophilin as a proinflammatory secretory product of lipopolysaccharide-activated macrophages. Proc Natl Acad Sci USA. 1992;89:3511–3515. doi: 10.1073/pnas.89.8.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki J, Jin ZG, Meoli DF, Matoba T, Berk BC. Cyclophilin A is secreted by a vesicular pathway in vascular smooth muscle cells. Circ Res. 2006;98:811–817. doi: 10.1161/01.RES.0000216405.85080.a6. [DOI] [PubMed] [Google Scholar]
- 10.Thali M, Bukovsky A, Kondo E, Rosenwirth B, Walsh CT, Sodroski J, Gottlinger HG. Functional association of cyclophilin A with HIV-1 virions. Nature. 1994;372:363–365. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- 11.Watashi K, Ishii N, Hijikata M, Inoue D, Murata T, Miyanari Y, Shimotohno K. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol Cell. 2005;19:111–122. doi: 10.1016/j.molcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 12.Hong F, Lee J, Song JW, Lee SJ, Ahn H, Cho JJ, Ha J, Kim SS. Cyclosporin A blocks muscle differentiation by inducing oxidative stress and inhibiting the peptidyl-prolyl-cis-trans isomerase activity of cyclophilin A: Cyclophilin A protects myoblasts from cyclosporin A-induced cytotoxicity. FASEB J. 2002;16:1633–1635. doi: 10.1096/fj.02-0060fje. [DOI] [PubMed] [Google Scholar]
- 13.Choi KJ, Piao YJ, Lim MJ, Kim JH, Ha J, Choe W, Kim SS. Overexpressed cyclophilin A in cancer cells renders resistance to hypoxia- and cisplatin-induced cell death. Cancer Res. 2007;67:3654–3662. doi: 10.1158/0008-5472.CAN-06-1759. [DOI] [PubMed] [Google Scholar]
- 14.Jin ZG, Lungu AO, Xie L, Wang M, Wong C, Berk BC. Cyclophilin A is a proinflammatory cytokine that activates endothelial cells. Arterioscler, Thromb, Vasc Biol. 2004;24:1186–1191. doi: 10.1161/01.ATV.0000130664.51010.28. [DOI] [PubMed] [Google Scholar]
- 15.Yurchenko V, Constant S, Eisenmesser E, Bukrinsky M. Cyclophilin-CD147 interactions: A new target for anti-inflammatory therapeutics. Clin Exp Immunol. 2010;160:305–317. doi: 10.1111/j.1365-2249.2010.04115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li M, Zhai Q, Bharadwaj U, Wang H, Li F, Fisher WE, Chen C, Yao Q. Cyclophilin A is overexpressed in human pancreatic cancer cells and stimulates cell proliferation through CD147. Cancer. 2006;106:2284–2294. doi: 10.1002/cncr.21862. [DOI] [PubMed] [Google Scholar]
- 17.Sykes K, Gething MJ, Sambrook J. Proline isomerases function during heat shock. Proc Natl Acad Sci USA. 1993;90:5853–5857. doi: 10.1073/pnas.90.12.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang P, Cardenas ME, Cox GM, Perfect JR, Heitman J. Two cyclophilin A homologs with shared and distinct functions important for growth and virulence of Cryptococcus neoformans. EMBO Rep. 2001;2:511–518. doi: 10.1093/embo-reports/kve109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruan SL, Ma HS, Wang SH, Fu YP, Xin Y, Liu WZ, Wang F, Tong JX, Wang SZ, Chen HZ. Proteomic identification of OsCYP2, a rice cyclophilin that confers salt tolerance in rice (Oryza sativa L.) seedlings when overexpressed. BMC Plant Biol. 2011;11:34. doi: 10.1186/1471-2229-11-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trivedi DK, Bhatt H, Pal RK, Tuteja R, Garg B, Johri AK, Bhavesh NS, Tuteja N. Structure of RNA-interacting cyclophilin A-like protein from Piriformospora indica that provides salinity-stress tolerance in plants. Sci Rep. 2013;3:3001. doi: 10.1038/srep03001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumari S, Roy S, Singh P, Singla-Pareek SL, Pareek A. Cyclophilins: Proteins in search of function. Plant Signaling Behav. 2013;8:e22734. doi: 10.4161/psb.22734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eisenmesser EZ, Bosco DA, Akke M, Kern D. Enzyme dynamics during catalysis. Science. 2002;295:1520–1523. doi: 10.1126/science.1066176. [DOI] [PubMed] [Google Scholar]
- 23.Schlegel J, Armstrong GS, Redzic JS, Zhang F, Eisenmesser EZ. Characterizing and controlling the inherent dynamics of cyclophilin-A. Protein Sci. 2009;18:811–824. doi: 10.1002/pro.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ai X, Li L, Semesi A, Yee A, Arrowsmith CH, Li SS, Choy WY. Hypothetical protein AF2241 from Archaeoglobus fulgidus adopts a cyclophilin-like fold. J Biomol NMR. 2007;38:353–358. doi: 10.1007/s10858-007-9172-8. [DOI] [PubMed] [Google Scholar]
- 25.Takami H, Kobata K, Nagahama T, Kobayashi H, Inoue A, Horikoshi K. Biodiversity in deep-sea sites located near the south part of Japan. Extremophiles. 1999;3:97–102. doi: 10.1007/s007920050104. [DOI] [PubMed] [Google Scholar]
- 26.Takami H, Takaki Y, Chee GJ, Nishi S, Shimamura S, Suzuki H, Matsui S, Uchiyama I. Thermoadaptation trait revealed by the genome sequence of thermophilic Geobacillus kaustophilus. Nucleic Acids Res. 2004;32:6292–6303. doi: 10.1093/nar/gkh970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holliday MJ, Zhang F, Isern NG, Armstrong GS, Eisenmesser EZ. 1H, 13C, and 15N backbone and side chain resonance assignments of thermophilic Geobacillus kaustophilus cyclophilin-A. Biomol NMR Assignments. 2014;8:23–27. doi: 10.1007/s12104-012-9445-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Camilloni C, Sahakyan AB, Holliday MJ, Isern NG, Zhang F, Eisenmesser EZ, Vendruscolo M. Cyclophilin A catalyzes proline isomerization by an electrostatic handle mechanism. Proc Natl Acad Sci USA. 2014;111:10203–10208. doi: 10.1073/pnas.1404220111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bahmed K, Henry C, Holliday M, Redzic JS, Ciobanu M, Zhang F, Weekes C, Sclafani RA, Degregori J, Eisenmesser EZ. Extracellular cyclophilin-A stimulates ERK1/2 phosphorylation in a cell-dependent manner but broadly stimulates nuclear factor κB. Cancer Cell Int. 2012;12:19. doi: 10.1186/1475-2867-12-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 31.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 32.Farrow NA, Zhang O, Forman-Kay JD, Kay LE. A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. J Biomol NMR. 1994;4:727–734. doi: 10.1007/BF00404280. [DOI] [PubMed] [Google Scholar]
- 33.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lange OF, Rossi P, Sgourakis NG, Song Y, Lee HW, Aramini JM, Ertekin A, Xiao R, Acton TB, Montelione GT, Baker D. Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples. Proc Natl Acad Sci USA. 2012;109:10873–10878. doi: 10.1073/pnas.1203013109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guntert P, Mumenthaler C, Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 36.Lange OF, Baker D. Resolution-adapted recombination of structural features significantly improves sampling in restraint-guided structure calculation. Proteins. 2012;80:884–895. doi: 10.1002/prot.23245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhattacharya A, Tejero R, Montelione GT. Evaluating protein structures determined by structural genomics consortia. Proteins. 2007;66:778–795. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- 38.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Best RB, Hummer G. Optimized molecular dynamics force fields applied to the helix-coil transition of polypeptides. J Phys Chem B. 2009;113:9004–9015. doi: 10.1021/jp901540t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Camilloni C, Robustelli P, De Simone A, Cavalli A, Vendruscolo M. Characterization of the conformational equilibrium between the two major substates of RNase A using NMR chemical shifts. J Am Chem Soc. 2012;134:3968–3971. doi: 10.1021/ja210951z. [DOI] [PubMed] [Google Scholar]
- 43.Kohlhoff KJ, Robustelli P, Cavalli A, Salvatella X, Vendruscolo M. Fast and accurate predictions of protein NMR chemical shifts from interatomic distances. J Am Chem Soc. 2009;131:13894–13895. doi: 10.1021/ja903772t. [DOI] [PubMed] [Google Scholar]
- 44.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 45.Tribello GA, Bonomi M, Branduardi D, Camilloni C, Bussi G. Plumed 2: New Feathers for an Old Bird. Comput Phys Commun. 2014;185:604–613. [Google Scholar]
- 46.Fu B, Sahakyan AB, Camilloni C, Tartaglia GG, Paci E, Caflisch A, Vendruscolo M, Cavalli A. ALMOST: An All Atom Molecular Simulation Toolkit for Protein Structure Determination. J Comput Chem. 2014;35:1101–1105. doi: 10.1002/jcc.23588. [DOI] [PubMed] [Google Scholar]
- 47.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38:W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Russell RJ, Ferguson JM, Hough DW, Danson MJ, Taylor GL. The crystal structure of citrate synthase from the hyperthermophilic archaeon pyrococcus furiosus at 1.9 Å resolution. Biochemistry. 1997;36:9983–9994. doi: 10.1021/bi9705321. [DOI] [PubMed] [Google Scholar]
- 49.Thompson MJ, Eisenberg D. Transproteomic evidence of a loop-deletion mechanism for enhancing protein thermostability. J Mol Biol. 1999;290:595–604. doi: 10.1006/jmbi.1999.2889. [DOI] [PubMed] [Google Scholar]
- 50.Piotukh K, Gu W, Kofler M, Labudde D, Helms V, Freund C. Cyclophilin A binds to linear peptide motifs containing a consensus that is present in many human proteins. J Biol Chem. 2005;280:23668–23674. doi: 10.1074/jbc.M503405200. [DOI] [PubMed] [Google Scholar]
- 51.Lam SY, Yeung RC, Yu TH, Sze KH, Wong KB. A rigidifying salt-bridge favors the activity of thermophilic enzyme at high temperatures at the expense of low-temperature activity. PLoS Biol. 2011;9:e1001027. doi: 10.1371/journal.pbio.1001027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolf-Watz M, Thai V, Henzler-Wildman K, Hadjipavlou G, Eisenmesser EZ, Kern D. Linkage between dynamics and catalysis in a thermophilic-mesophilic enzyme pair. Nat Struct Mol Biol. 2004;11:945–949. doi: 10.1038/nsmb821. [DOI] [PubMed] [Google Scholar]
- 53.D’Amico S, Marx JC, Gerday C, Feller G. Activity-stability relationships in extremophilic enzymes. J Biol Chem. 2003;278:7891–7896. doi: 10.1074/jbc.M212508200. [DOI] [PubMed] [Google Scholar]
- 54.Collins T, Meuwis MA, Gerday C, Feller G. Activity, stability and flexibility in glycosidases adapted to extreme thermal environments. J Mol Biol. 2003;328:419–428. doi: 10.1016/s0022-2836(03)00287-0. [DOI] [PubMed] [Google Scholar]
- 55.Georlette D, Damien B, Blaise V, Depiereux E, Uversky VN, Gerday C, Feller G. Structural and functional adaptations to extreme temperatures in psychrophilic, mesophilic, and thermophilic DNA ligases. J Biol Chem. 2003;278:37015–37023. doi: 10.1074/jbc.M305142200. [DOI] [PubMed] [Google Scholar]
- 56.Zavodszky P, Kardos J, Svingor A, Petsko GA. Adjustment of conformational flexibility is a key event in the thermal adaptation of proteins. Proc Natl Acad Sci USA. 1998;95:7406–7411. doi: 10.1073/pnas.95.13.7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bae E, Phillips GN., Jr Structures and analysis of highly homologous psychrophilic, mesophilic, and thermophilic adenylate kinases. J Biol Chem. 2004;279:28202–28208. doi: 10.1074/jbc.M401865200. [DOI] [PubMed] [Google Scholar]
- 58.Butterwick JA, Loria JP, Astrof NS, Kroenke CD, Cole R, Rance M, Palmer AG., III Multiple time scale backbone dynamics of homologous thermophilic and mesophilic ribonuclease HI enzymes. J Mol Biol. 2004;339:855–871. doi: 10.1016/j.jmb.2004.03.055. [DOI] [PubMed] [Google Scholar]
- 59.Palmer AG, III, Kroenke CD, Loria JP. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- 60.Loria JP, Berlow RB, Watt ED. Characterization of enzyme motions by solution NMR relaxation dispersion. Acc Chem Res. 2008;41:214–221. doi: 10.1021/ar700132n. [DOI] [PubMed] [Google Scholar]
- 61.Lipari G, Szabo A. Model-Free Approach to the Interpretation of Nuclear Magnetic-Resonance Relaxation in Macromolecules. 2 Analysis of Experimental Results. J Am Chem Soc. 1982;104:4559–4570. [Google Scholar]
- 62.Cole R, Loria JP. FAST-Modelfree: A program for rapid automated analysis of solution NMR spin-relaxation data. J Biomol NMR. 2003;26:203–213. doi: 10.1023/a:1023808801134. [DOI] [PubMed] [Google Scholar]
- 63.Mandel AM, Akke M, Palmer AG., III Backbone dynamics of Escherichia coli ribonuclease HI: Correlations with structure and function in an active enzyme. J Mol Biol. 1995;246:144–163. doi: 10.1006/jmbi.1994.0073. [DOI] [PubMed] [Google Scholar]
- 64.Knoll AH, Javaux EJ, Hewitt D, Cohen P. Eukaryotic organisms in Proterozoic oceans. Philos Trans R Soc, B. 2006;361:1023–1038. doi: 10.1098/rstb.2006.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kern D, Eisenmesser EZ, Wolf-Watz M. Enzyme dynamics during catalysis measured by NMR spectroscopy. Methods Enzymol. 2005;394:507–524. doi: 10.1016/S0076-6879(05)94021-4. [DOI] [PubMed] [Google Scholar]
- 66.Kern D, Kern G, Scherer G, Fischer G, Drakenberg T. Kinetic analysis of cyclophilin-catalyzed prolyl cis/trans isomerization by dynamic NMR spectroscopy. Biochemistry. 1995;34:13594–13602. doi: 10.1021/bi00041a039. [DOI] [PubMed] [Google Scholar]
- 67.Goh KM, Kahar UM, Chai YY, Chong CS, Chai KP, Ranjani V, Illias R, Chan KG. Recent discoveries and applications of Anoxybacillus. Appl Microbiol Biotechnol. 2013;97:1475–1488. doi: 10.1007/s00253-012-4663-2. [DOI] [PubMed] [Google Scholar]
- 68.Xue Y, Zhang X, Zhou C, Zhao Y, Cowan DA, Heaphy S, Grant WD, Jones BE, Ventosa A, Ma Y. Caldalkalibacillus thermarum gen. nov., sp nov., a novel alkalithermophilic bacterium from a hot spring in China. Int J Syst Evol Microbiol. 2006;56:1217–1221. doi: 10.1099/ijs.0.64105-0. [DOI] [PubMed] [Google Scholar]
- 69.Merz A, Yee MC, Szadkowski H, Pappenberger G, Crameri A, Stemmer WP, Yanofsky C, Kirschner K. Improving the catalytic activity of a thermophilic enzyme at low temperatures. Biochemistry. 2000;39:880–889. doi: 10.1021/bi992333i. [DOI] [PubMed] [Google Scholar]
- 70.Varley PG, Pain RH. Relation between stability, dynamics and enzyme activity in 3-phosphoglycerate kinases from yeast and Thermus thermophilus. J Mol Biol. 1991;220:531–538. doi: 10.1016/0022-2836(91)90028-5. [DOI] [PubMed] [Google Scholar]
- 71.Fitter J. Structural and dynamical features contributing to thermostability in α-amylases. Cell Mol Life Sci. 2005;62:1925–1937. doi: 10.1007/s00018-005-5079-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grottesi A, Ceruso MA, Colosimo A, Di Nola A. Molecular dynamics study of a hyperthermophilic and a mesophilic rubredoxin. Proteins. 2002;46:287–294. doi: 10.1002/prot.10045. [DOI] [PubMed] [Google Scholar]
- 73.Wintrode PL, Zhang D, Vaidehi N, Arnold FH, Goddard WA., III Protein dynamics in a family of laboratory evolved thermophilic enzymes. J Mol Biol. 2003;327:745–757. doi: 10.1016/s0022-2836(03)00147-5. [DOI] [PubMed] [Google Scholar]
- 74.Hernandez G, Jenney FE, Jr, Adams MW, LeMaster DM. Millisecond time scale conformational flexibility in a hyperthermophile protein at ambient temperature. Proc Natl Acad Sci USA. 2000;97:3166–3170. doi: 10.1073/pnas.040569697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.