Abstract
There is a consensus in the medical profession of the pressing need for novel antimicrobial agents due to issues related to drug resistance. In practice, solutions to this problem to a large degree lie with the identification of new and vital targets in bacteria and subsequently designing their inhibitors. We consider SecA a very promising antimicrobial target. In this review, we compile and analyze information available on SecA to show that inhibition of SecA has a multitude of consequences. Furthermore, we discuss issues critical to the design and evaluation of SecA inhibitors.
In the last few decades, the global emergence of drug-resistant, especially extensively drug-resistant and pan-drug resistant bacterial strains has raised severe healthcare concerns [1,2]. Bacteria including methicillin-resistant Streptococcus aureus (MRSA), Clostridium difficile, Streptococcus pneumonia, Mycobacterium tuberculosis (multidrug-resistant (MDR), extensively-drug resistant (XDR) Pseudomonas aeruginosa, Klebsiella species, Acinetobacter, Neisseria gonorrhoeae, Campylobacter and Salmonella have been classified as ‘High Priority Antibiotic Resistant Bacteria’ by the US Center for Disease Control and Prevention. Recently in the USA, the dissemination of carbapenem-resistant Enterobacteriaceae and its subtypes such as New Delhi metallo-β-lactamase resistant strains have raised added concerns due to the high mortality rates of these new strains [3–5].
The need for new antimicrobials
As one can imagine, the widespread emergence of drug-resistant bacteria has become a major public health concern in recent years. The urgent need for new antimicrobial agents cannot be overstated and developing drugs with novel mechanisms of action or against new targets is more imperative than ever [6,7]. However, any new antimicrobials effective against drug-resistant strains will not be used as the first line of treatment options (for good reasons). This means that there is not much money to be made. Therefore, the pharmaceutical industry is essentially staying away or at least not focusing on new antimicrobials [7]. Other than improved analogs of existing antibiotics, last three decades have seen only two new antibiotics (linezolid [8] and daptomycin [9]), whereas platensimycin [10] has emerged as a promising clinical candidate.
The focus in the field is on the search for antimicrobials with new mechanisms of action and/or against new targets instead of analog design along the lines of existing drugs. To help put this review in a broad perspective, we will start by pressing upon the need for novel targets with the focus being on SecA. This will be followed by discussions of the challenges involved in targeting SecA and screening strategies adopted to circumvent those issues. We will also put forth a comparison of known SecA inhibitors and the assay techniques employed therein. Our idea is to bring together the scattered pieces of the literature dedicated to developing SecA inhibitors and push forward the idea of SecA being an indispensable target, discuss the unique advantages of targeting SecA, and address technical issues that one has to consider in developing new SecA inhibitors. We hope this review will kindle the interests of the scientific community and stimulate more research towards designing drugs targeting SecA.
What makes for a good antimicrobial target?
For the discovery of new antimicrobials with the ability to combat drug resistance, novel targets are desired. Desirable features of an ideal target should at least include the following. First, the target should play an indispensable function in bacterial survival without any existing alternative pathways for its mitigation and compensation. Second, a genus-wide distribution of the target offers the possibility of developing broad-spectrum antimicrobials. Third, the pathogenic target must not have closely related human homologs, so as to minimize the potential cytotoxicity issues in humans. Fourth, the target should contribute vitally to bacterial virulence and pathogenicity [11,12].
The Sec-dependent protein translocase consists of oligomer complex of SecYEG and SecDF•YajC as membrane proteins [13,14] and SecA functions as an ATPase that provides the energy for the Sec-dependent protein translocation. When SecA is bound to the SecYEG complex, acidic phospholipids and a precursor protein such as proOmpA (the precursor of outer membrane protein A), it becomes fully active as an ATPase and a protein translocase [14,15]. In all bacteria, SecA plays an essential role as an ATPase in the protein translocation machinery. SecA is known to be critical for bacterial survival, and is responsible for the secretion of many vital proteins as well as some toxins and additional virulence factors [16–19]. Due to the fact that SecA plays an indispensable role in the secretion of bacterial toxins, is essential for survival of a broad-spectrum of bacteria, and unlike SecYEG there are no SecA counterpart in mammalian cells, SecA makes for an ideal target for antimicrobial development. In addition, because SecA is a membrane protein in its translocation functional form, there is an added advantage, in other words SecA inhibitors can directly access SecA without the need to enter the cytoplasmic space. Thus drug permeation and intracellular concentration are less of an issue with these inhibitors. Moreover, most efflux pumps consist of membrane proteins with signal peptides, especially in Gram-negative bacteria; thus inhibition of SecA can also be expected to affect the assembly of functional efflux. Efflux pumps are an important issue to address in overcoming the effect of multidrug resistance (MDR). The majority of bacteria only have one SecA homologue, however in some Gram-positive bacteria pathogen, there are two SecA homologues [20–28]. SecA1 is the conventional SecA, critical for the secretion of many proteins with a Sec-dependent signal peptide, and essential for the viability of bacteria [20,29–30]. SecA2 is less conservative than SecA1, involved in secretion of specific proteins related with virulence, and is not essential for the majority of bacteria containing two SecA homologues [23–25,27,31–32] except in Corynebacterium glutamicum [33] and Clostridium difficile [34]. Thus, SecA2 is not a good primary antimicrobial target as the conventional SecA (SecA1). SecA2 shares some homology as the conventional SecA and has ATPase activity [28,35] but it does not have exactly the same biochemistry property as the conventional SecA in terms of SecYEG interaction [28], ATPase activity [36], ADP releasing [37] and substrate(s) [28,38]. In the current review, we are focused on how the conventional SecA could be a potential drug target.
Key issues in the field include: whether SecA inhibition is sufficient to achieve antimicrobial effect; the unique consequence of attenuated virulence factor secretion through SecA inhibition; and, whether the functional form of SecA is accessible from the periplasm (in Gram-negative) or the extracellular matrix (in Gram-positive), which has implications on the ability for SecA inhibitors to bypass the effect of efflux pumps. As is detailed later in the text, available information seems to suggest that the answer is yes to all three questions. Additionally, because SecA is functional both in solution and in cellular membrane and intact SecA has an inhibitory C-terminal domain, the truncated enzyme assay system used has direct relevance to the likelihood of success in identifying SecA inhibitors which would have antimicrobial effect. Below we discuss in detail about this target, the current state of research, and future directions.
SecA is an ATPase, an integral membrane protein & a protein-conducting channel
Structure of SecA
In order to achieve a good level of understanding for inhibitor design and the various issues to be discussed, it is important that we come to an adequate appreciation of the structural features of this protein existing in various forms, its conformational changes and related subunit complexation. SecA as the central component of the Sec machinery is conserved across bacteria. It functions as an ATPase, assisting in the transport of proteins [39] and interacting with other components of the Sec machinery. In E. coli cells, there is relatively high concentration of SecA (8 μM) [40], which corresponds to around 13,000 copies per cell if assuming a cellular volume of 2.75 μm3 [3,41]. SecA is a soluble protein and localizes both to the cytosol and the cytoplasmic membrane [42]. It is believed that most soluble SecA forms homodimers in the cytosol [21,24–25,40,43–44]. This dimerization process is dynamic, and the dissociation constant was estimated to be in the low μM range (Kd ~ 0.1 μM) [43]. A more recent study estimated the Kd to be 0.76 nM using an equilibrium technique and dual-color fluorescence-burst analysis (DCFBA) [45]. However, the sensitive nature of the dimerization process, which is influenced by many factors such as salt, detergents or temperature, has provided debatable results amongst various studies [46]. No matter what the specific number for Kd is under a given condition, one thing is clear: these numbers are all much lower than the estimated concentration of SecA. Thus one would expect that SecA predominantly exists in the dimer form in the cytosol. However, the cytosolic form is not directly related to protein translocation and thus would not be the target of drug design. Furthermore, the exact implication of inhibiting the cytosolic soluble SecA is not well studied.
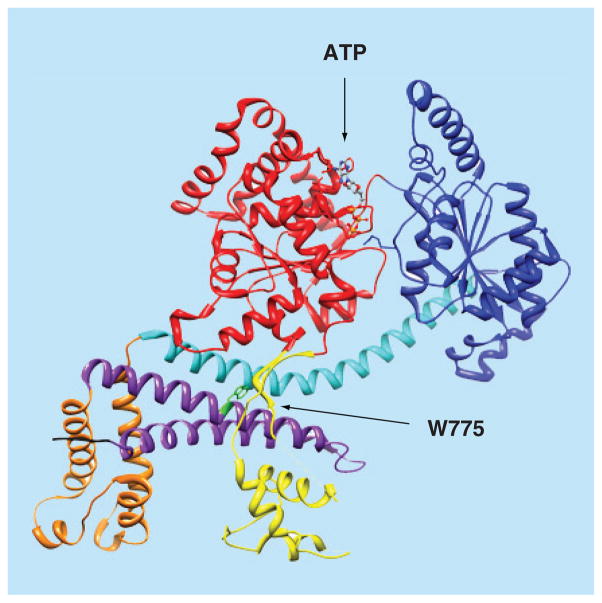
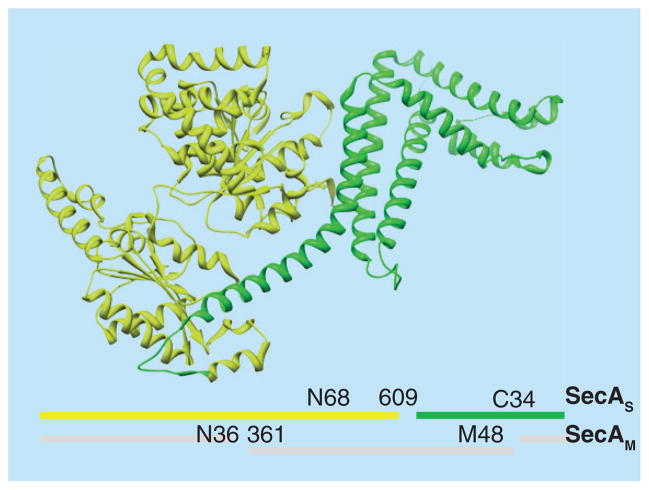
It is the consensus that each SecA protomer contains several substructural domains (Figure 1). The nucleotide binding domain (NBD) and the intramolecular regulator of ATPase activity 2 domain (IRA2) forms the ‘DEAD’ motor, the main catalytic moiety of SecA. The ATP binding site of SecA is located at the interface of the NBD and the IRA2 domains. SecA is a member of the superfamily II (SF2) DExH/D proteins, the majority of which are helicases [47,48]. Other than helicases, to achieve the unique functions of SecA, the preprotein binding domain (PBD) and the C-terminal domain are two key domains that contribute to SecA substrate specificity. The PBD is composed of two subdomains: an antiparallel β strand (stem) connecting PBD and NBD, and a bilobate globular domain (bulb) [49,50]. The C-terminal domain is composed of four substructures: the wing domain (WD), the α-helical scaffold domain (SD), the intramolecular regulator of ATP activity 1 domain (IRA1), and a C-terminal linker (CTL).
Figure 1. 3D structure of the Escherichia coli SecA monomer.
Color coding of SecA domains: NBD, red; IRA2, dark blue; SD, cyan; HWD, orange; PBD, yellow; IRA1, purple; and CTL (first 4 residues), black. Also shown are the bound ATP molecule (in ball and stick, colored by elements) and C34 regulating W775 residue (green). The figure was created using the UCSF Chimera package, using the coordinates of the EcSecA (PDB code 2FSG) [51].
For color images please see online www.future-science.com/doi/full/10.4155/FMC.15.42
The structures of SecA (Table 1) have been studied using x-ray crystallography, tNMR spectroscopy [52], cryo-EM [53–55], atomic force microscopy and small angle neutron scattering (SANS) [33] and small angle x-ray scattering [56]. Most of these studies revealed an equilibrium between monomeric and dimeric forms of SecA with one exception, which revealed a monomeric structure [57]. Interestingly, in each of the SecA structures, the protomer structures are very similar and yet the orientation of each protomer relative to one another can be quite different. In most dimers, their C-domains face opposite directions creating antiparallel structures (Figure 2A) [51,58–60]; while one has a parallel orientation (Figure 2B) [61]. These results further demonstrate the sensitive nature of the dimerization process. All these variance further demonstrate the complexity and difficulty of designing SecA inhibitors. On the other hand, design of form-specific SecA inhibitors can turn out to be an effective strategy for finding potent inhibitors. So far the SecA structures are all from soluble form, not membrane form interacting with lipids.
Table 1.
X-ray crystallography structures of SecA.
| Organism | PDB entry | Ligand | Additional structures | Ref. |
|---|---|---|---|---|
| Mycobacterium tuberculosis | 1NL3 | 1NKT (ADP, Mg2+) | [59] | |
| Thermus thermophiles | 2IPC | [61] | ||
| Escherichia coli | 3BXZ | ADP, Mg2+, spermidine | [62] | |
| E. coli | 2FSF | 2FSG (ATP), 2FSH (AMP-PNP), 2FSI (ADP) | [51] | |
| Bacillus subtilis | 1M6N | 1M74 (ADP, Mg2+,SO42−) | [63] | |
| B. subtilis | 3JV2 | ADP, Mg2+, peptide | [64] | |
| B. subtilis | 3DL8 | [65] | ||
| B. subtilis | 1TF5 | 1TF2 (ADP, Mg2+) | [66] | |
| B. subtilis | 2IBM | ADP | [60] | |
| Thermotoga maritime | 3JUX | ADP, Mg2+ | [64] | |
| T. maritime | 3DIN | ADP, Mg2+, BEF | [65] |
Figure 2. Space filling models of SecA dimers.
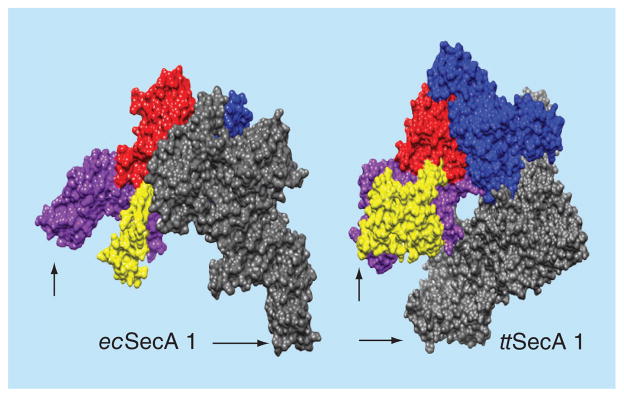
Dimeric SecA proteins were structurally aligned on one of their protomers (the nucleotide binding domain NBD, red; the intramolecular regulator of ATPase 2 IRA2, dark blue; the protein binding domain PBD, yellow and the C-domain, purple) so as to demonstrate the variable position that the second (grey) protomer occupies; arrows indicate C-terminus. The figure was created using the UCSF Chimera package [67,68]. The structures used are: Escherichia coli (ecSecA1; 2FSF) [51]. Thermus thermophilus (ttSecA; 2IPC) [61].
For color images please see online www.future-science.com/doi/full/10.4155/FMC.15.42
The transport process
As a major component of the bacterial Sec-system, domain organization of SecA was revealed by crystallographic structures from different species [51,57–61] and biochemical analyses [50,69–72]. Despite the fact that several structural and biochemical studies have been done over the last two decades, the exact mechanism of SecA-mediated protein translocation is still not fully understood. Some studies have concluded that SecA functions as a dimer [44,73–75] while other studies propose that monomeric SecA is the key component in protein translocation [65,76–77]. Meanwhile, both monomeric [65,76,78] and dimeric [73,75,78–79] SecA were found to bind SecYEG. Details of the SecA-SecYEG-mediated secretion process have been discussed elsewhere [63,80–82]. In one prevailing view of the transport process, the following events occur (Figure 3) [83–85]: the preprotein SecA either directly or via a chaperone protein such as SecB [52,86]; the preprotein bound SecA binds to the lateral gate SecY [66] of t he S ecYEG heterotrimeric complex; electrostatic gate opens [87] due to allosteric regulation; ADP is released from the SecA motor domains [86–88] and SecA C-terminal domain mediated suppression relieves; SecA acquires the translocation ATPase activity [15,89]; and SecYEG channel loosens [90] and gets ready to push the preproteins across the cytoplasmic membranes. As the final step of the secretion, the signal peptide is cleaved by signal peptidase in the membrane leading to the release of the mature protein, which gets folded later. In terms of inhibitor design, especially when considering using computational approaches to guide the design, it will be important to keep in mind of the conformational changes associated with the various steps of the transport process.
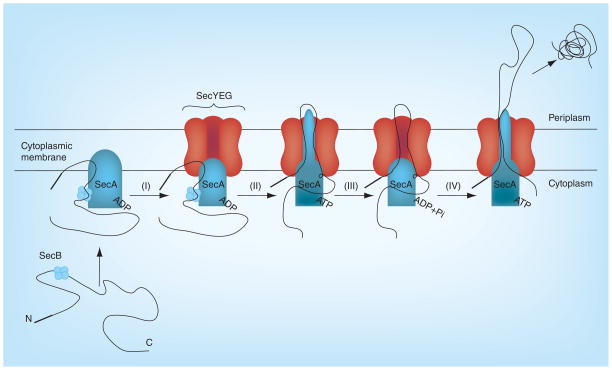
Figure 3. Model of the Sec-dependent preprotein secretion pathway in bacteria.
(I) Membrane SecA complexed with the SecB and preprotein binds to SecYEG; (II) SecA acquires the translocation ATPase activity and inserts itself into SecYEG channel; (III) SecYEG channel loosens and gets ready to push the pre-proteins; and (IV) the preprotein is pushed into the periplasm across the cytoplasmic membrane.
Adapted with permission from [91] © Elsevier (2011).
In 2011, a reciprocating piston model (Figure 4) was proposed by Driessen and co-workers [46] as another SecA/SecYEG-dependent translocation pathway model. It divides the whole translocation process into two parts. The initiation of protein translocation includes Steps 1–4 and the translocation cycle is illustrated in Steps 5–9. First, a SecA homodimer binds to the SecYEG channel (Step 1) [73,75] initiating the ATPase activity by changing the conformation of SecA (Step 2) [15,87]. Within the dimer, one SecA is anchored to SecYEG and the PBD of this protomer shows a clamp-like structure caused by a dramatic conformational change [65], and the other protomer, in the meantime, only interacts with SecYEG [45]. In the cytosol, homotetrameric chaperone protein SecB binds to the mature region of newly synthesized preproteins keeping them partially unfolded and transfers them to SecA. SecB interacts with both C-termini of the SecA dimer separately (Step 3) [92–95]. Subsequently, ATP binds with SecA and triggers SecB release, and the signal sequence and the adjacent mature region of the preprotein are pushed into the SecYEG channel forming a hairpin-like structure (insertion step, Step 4) [96–98]. Now the initiation part of protein translocation is complete.
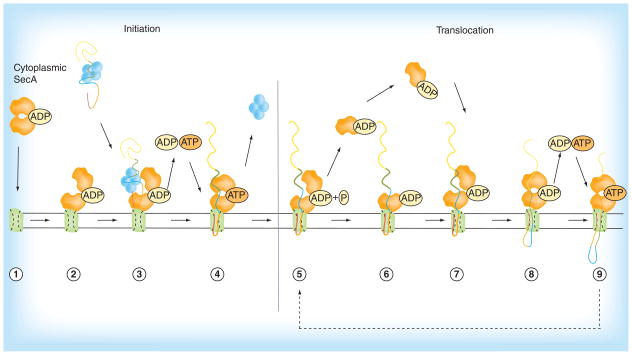
Figure 4. A proposed reciprocating piston model of the preprotein secretion pathway in bacteria.
Steps 1–4: Initiation of protein translocation is commenced by SecA cytoplasmic homodimer (1) and ends with the release of SecB chaperone (4). Steps 5–9: The translocation cycle is initiated with the release of SecA protomer (5) which is recycled back into the translocation cycle (6–7), finally culminating in the release of preprotein into the periplasm (9).
Reproduced with permission from [46] © Springer (2011).
The second part of the SecA mediated protein translocation starts from Step 5. During this step, one of the dimeric SecA protomer powered by ATP hydrolysis dissociates from the preprotein and the remaining SecA prevents backsliding of the polypeptide chain in the SecYEG channel. In addition, an assumed deinsertion step (Step 6), which is accelerated by the proton motive force (PMF) [99–103], is led by the conformational change of SecYEG bound SecA during ATP hydrolysis. SecA from the soluble SecA pool in cytosol rebinds to the preprotein (Step 7) and to the remaining SecA protomer. This rebinding of preprotein segment and subsequent SecYEG bound SecA causes compression of the polypeptide chain and forces it into the channel [85,104]. This ATP-independent translocation is responsible for the insertion of a 2–2.5 kDa peptide segment (Step 8) [105,106]. Then, ATP binding, insertion of SecA into SecYEG and translocation of another 2–2.5 kDa peptide segment occurs (Step 9) [65,83–84]. Repeating Steps 5–9 leads to translocation of 4–5 kDa or 25–30 amino acids in each cycle [100, 105–107] until the preprotein is fully transported.
The ATPase activity of SecA is regulated by the NBD & the IRA2 domains
Further complicating the picture is the fact that the SecA ATPase activity is regulated by various domains. This aspect is important in deciding whether to use the intact SecA or some truncated form for activity screening. This aspect will also be discussed in a later section. The opening and closing of the nucleotide-binding cleft, which is located in the gap between the NBD and IRA2 domains, play a key role in ATPase activity regulation [15,71,108]. The complex conformational changes upon binding the DEAD motor and C-terminal domain of SecA to SecYEG complex control the interactions between NBD and IRA2. Binding with lipids or SecYEG in the membrane weakens the interactions between the NBD and IRA2 domains and stimulates the ATPase activity. The SecA form with elevated ATPase activity with lipids/membranes is named ‘membrane ATPase’ [15,72].
Besides, binding of the DEAD motor and C-terminal domain of SecA to the SecYEG complex triggers conformational changes at the interface among the IRA1, PBD and CTL domains. These allosteric changes also increase the SecA affinity for signal peptides. Consequently, binding of the signal peptide near the stem region of the PBD-NBD interface is suggested to cause a large rotation of the bulb domain, which drives trapping of the first amino-terminal segment of mature preprotein domains [64,69,86]. This binding further increases the ATPase activity of SecA and converts it into a translocation ATPase. At this stage, the C-terminal domain inhibition effect is totally relieved and the gate 1 salt bridge is opened at the base of the DEAD motor [87]. Next, the DEAD motor ADP affinity is lost while the IRA2 detaches from the NBD and becomes disordered [87,109]. ADP released from the DEAD motor induces the PBD (with the trapped preprotein) conformational changes [50,109–110] and the dissociation of SecB [92]. Later, ATP binds to the empty nucleotide-binding cleft, and brings subsequent conformational changes that cause insertion of SecA along with bound pre-protein into the SecYEG channel. Finally, ATP hydrolysis drives a small segment of the preprotein release from SecA into the SecYEG channel [66]. The SecA returns to the NBD-IRA2 tight interaction state that forces SecA to exit the SecYEG channel while the PBD moves further along the pre-protein chain [83,84] and reattaches onto the following pre-protein segment, which causes ADP release and allows a new round of ATP binding and hydrolysis [111].
SecA functions as a protein-conducting channel in the membrane
SecA exists in two forms: a soluble and a membrane form. The membrane SecA interacts with phospholipids and integrates into membranes. The previously mentioned models for protein translocation view SecYEG as the core of the protein conducting channel in the membrane with SecA as a motor to transport proteins across the membrane (Figure 3, Wickner’s Model) [83]. However, in a major breakthrough that suggests an additional alternative model, it was found that SecA alone in liposomes can function as a protein-conducting channel [112] to promote protein translocation and ion-channel activity (Figure 5) [113]. This model [114] is strongly supported by earlier findings that SecA can integrate into the membrane in two forms, SecAM and SecAS [115–117], and can form ring-like pores structures in lipids [53,55,118] that provide the physical basis for the protein-conducting channel. In this model, the asymmetric SecA dimer is essential for functions; the membrane SecAM protomer function as the channel, while the SecAS as ATPase powering precursor peptides through the SecAM. The SecA-alone channels can be reconstituted with SecYEG to function as the prevailing SecA-SecYEG channels [119]. The finding that SecA functions as a protein-conducting channel as well as ATPase in the membrane strongly supports the notion that SecA is accessible from the periplasmic space (Gram-negative) extracellular matrix (Gram-positive) by inhibitors, to be discussed below. Although the Tai model is not considered the prevailing model, increasing amount of evidence seems to agree with this model of a second SecA channel. It is entirely possible that SecA has more than one functional form and does not have to exist in one uniform model in bacteria. Therefore, all the proposed models are not mutually exclusive.
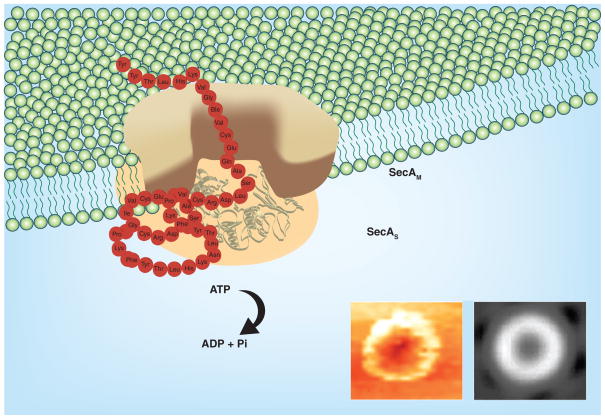
Figure 5. A working model of SecA in the membrane.
The Tai model [114]: SecA dimer [54,118] alone acts as the protein conducting-channel to promote protein translocation and ion-channel activity [113 62,120–122]. There exist two forms of integral SecA in the membranes [115,116]. SecAS has the same conformation as in soluble form, and SecAM is specific for lipids, acting as channels [53–55].
Reproduced from [114].
Established methods to test inhibitory effects on the functions of SecA
Because SecA is a membrane protein, exists in multiple forms interacting with many components, and is also soluble, developing appropriate assays that are robust and reliable, and accurately reflect the physiological state of the target is not an easy task. However, such assays are crucial for the inhibitor development effort.
Presently, there are several ways to test SecA inhibition (Table 2). The ATPase activity can be examined using different forms of SecA in solution, such as intrinsic SecA (regulated ATPase), truncated SecA without the C-terminal inhibitory sequence in solution (e.g., EcN68, unregulated ATPase), SecA in membrane (membrane ATPase), and SecA in complexation with SecYEG and precursor in membrane (translocation ATPase) [15]. There is also an electrophysiology/channel activity assay developed, which closely mimics the physiological conditions of opening channels [123,124]. The most important assays reflecting the whole translocation process are of course the inhibition of precursor translocation across the membrane vesicles [125] and liposomes [119]. Assaying inhibition of each form of SecA provides a partial understanding of the targeted SecA form; however, these assays when combined together can be used to obtain a reliable and more accurate understanding of the actual form/stage of protein translocation inhibited. Finally, the antimicrobial growth assay is used to correlate the enzyme inhibitory activity to bacterial protein secretion and growth in cells. It is important to emphasize that SecA inhibition studies do need to involve all different methods in order to achieve a thorough understanding of the ability for these inhibitors to inhibit SecA ATPase. The following section provides a detailed discussion of these assays.
Table 2.
Assays for testing SecA inhibition.
| No. | Assay | Information | Ref. |
|---|---|---|---|
| 1 | ATPase assay | Investigates the activity of SecA on ATP hydrolysis | [126–128] |
| 2 | SecA-LacZ reporter assay | Detects SecA inhibitors by investigating expression of SecA inhibition due to autogenous regulation | [129] |
| 3 | Antisense-mRNA assay | Identifies SecA inhibitors by using antisense-mRNA to block SecA translation and causing bacterial sensitization | [124] |
| 4 | Protein translocation assay | Examines the protein transport activity of SecA through vesicles and liposomes | [130–132] |
| 5 | Protein ion-channel activity assay | Measures the ion-channel activity of SecA | [113,133] |
ATPase assay
The assay involves determination of free inorganic phosphate formed as a result of ATP hydrolysis. In doing so, the malachite green colorimetric method is most commonly used [126–128]. In screening for potential inhibitors, the most obvious one is the use of the whole SecA in determining how an inhibitor modifies the ATPase activity. However, whole SecA is in a regulated and closed-state largely controlled by its regulatory/inhibitory C-terminal (C34) domain. Therefore this is a minimally active state of SecA. Thus, results generated using the whole SecA do not truly reflect the ability for the inhibitor to inhibit the fully active ATPase, which is the case when SecA is in membrane and in live bacteria. Another assay method uses truncated SecA with only the catalytic domain (N68) in solution (e.g., EcSecAN68, unregulated ATPase Figure 6). Because of the lack of the inhibitory C-terminal domain, this assay is very sensitive. It has also been demonstrated that results from assays using the truncated form parallels that of membrane SecA assays [125]. Another way of avoiding the intrinsic inhibition effect of the C-terminal domain is by using a mutant with elevated intrinsic activities. For example, in EcSecA residue W775 is important for the C34 regulation [134], which is located at the interface of the SD and the IRA1 domains (Figure 3). Replacing the bulky tryptophan with a small hydrophobic alanine weakens the interaction of the SD and the IRA1 domains and elevates the intrinsic ATPase activity by fivefold compared with the wild-type EcSecA [109,134]. Therefore, using this mutated W775A EcSecA allows the benefit of the full-length enzyme without the intrinsic inhibition effect of the C-terminal domain [63].
Figure 6. Two separable soluble domains and lipid-specific domain of SecA.
Two forms of SecA exist in the membrane: SecAS, which is similar to the soluble form with two separable domains: N68 and C34, and the other SecAM with the N36 and M48 domains spanning the lipid membrane [114]. X-ray ribbon structure of EcSecA with N68 (yellow) and C34 (green) is shown.
For color images please see online www.future-science.com/doi/full/10.4155/FMC.15.42
SecA-LacZ reporter assay
This assay makes use of autogenously regulated SecA expression [129]. The design was based on the belief that under normal protein secretion in E. coli, SecA controls its own expression, by binding to its own mRNA, blocking the translation active site and inhibiting its expression. On the other hand, inhibition of protein secretion dissociates the SecA-mRNA complex and causes an up-regulation of SecA. This screening strategy involves identification of compounds that enhance SecA expression through inhibition of protein secretion, presumably due to SecA inhibition or inhibition of any member of Sec machinery. Therefore, this screening strategy can be used to identify inhibitors of various components associated with Sec machinery functioning such as SecB, SecDF and SecYEG as well as SecA. However, it should be noted that there are other factors that could also regulate SecA expression as discussed in one review [135].
Antisense-mRNA based assay
The strategy involves using an antisense mRNA to bind selectively to the mRNA regions encoding SecA, thereby blocking its translation [124]. This decreased translation causes subsequent sensitization of bacteria towards SecA inhibitors. A two-plate agar based differential sensitivity assay including the antisense sequence RNA (AS-RNA) sensitized strain and the wild type controls are used to compare the differences in growth inhibition.
Protein translocation assay
Various in vitro functional assays have been developed to examine the protein transport activity of SecA. One method involved monitoring SecA-dependent translocation of AlkProPhoA(Cys-)3 [130] through SecYEG bearing membrane vesicles. Translocation of the preprotein is assayed measuring accessibility to protease K. A similar assay used proOmpA [131,127] translocation through membrane vesicles expressing SecYEG. Malachite green is used to quantify the ATP consumption by energy-motor SecA during the translocation process. Instead of membranes, proteoliposome reconstitution system of purified SecYEG and SecA has also been established to demonstrate the involvement of these proteins in translocation. A SecA-liposomes translocation assay without SecYEG as protein-conducting channel has also been developed [123]. It was found that SecA forms ring-like structures upon binding to anionic phospholipids, inserts into the membrane and thus can alone promote the translocation of proOmpA or OmpA into E. coli phospholiposomes, at an optimal concentration of 1–2 μM, 10 times higher than SecYEG-containing membranes. The in vitro translocation efficiency of proteins was similarly determined by their inaccessibility to proteinase K, and amount of translocated protein was analyzed by Western blotting followed by quantification.
Protein ion-channel activity assay
The protein/ion channel activity assay developed by our lab [113,133] is a semiphysiological assay for electrophysiological measurements of protein-channel. The sample mixture is injected into the dark pole site of oocytes using Nanoject II injector and the ion current is recorded for 1 min, 3 h after the injection. It truly mimics the cell’s physiological conditions, is easy to use with multiple oocytes (n = 20–30) for each variable, requires nanogram amounts of materials and studies the translocation by exogenous SecA through membrane vesicles/liposomes embedded in individual oocytes. Due to their large size, oocytes can easily accommodate various manipulations and electrode penetration and the recording noise is very low due to the large number of channels measured in such experiments (calculated to be 200,000–1,000,000 channels) [133]. It is important to emphasize that the results from the truncated SecA assay and the electrophysiology oocyte assays seem to parallel that of antimicrobial results, which adds further support to its near physiological nature.
Reported SecA inhibitors
The suitability of SecA as a target for the development of antibacterial agents has been increasingly recognized [118, 125, 129–131] and thus there have been efforts in developing SecA inhibitors. For a detailed discussion, readers are referred to an extensive review written by Segers et al. [91], which discusses in-depth information available on small-molecule SecA inhibitors. One special point that we would like to make is the possibility of SecA to be accessible by inhibitors from the periplasm (Gram-negative) or the extracellular matrix (Gram-positive). This means the SecA inhibitors may have the ability to bypass the effect of efflux pumps and hence can potentially diminish drug resistance caused due to efflux pumps, that is, the MDR issues.
Tables 3 & 4 provide a comparative summary of the various inhibitors that have been reported and the screening strategies employed therein. Prior to the recent efforts, inorganic azide at mM range [136–139] (1, Table 3) was the only known SecA inhibitor and the SecA binding mechanism of sodium azide has been extensively probed [84,106,140]. However, sodium azide is unlikely to be developed for therapeutic applications given its human toxicity and inhibitory activity against a diverse array of other enzymes [141–145].
Table 3.
Comparative account of various SecA inhibitors and the assays used.
| Inhibitor ID | Compound name and structure | Source (Year) | Inhibition constants (IC50) and assay used | Ref. |
|---|---|---|---|---|
| 1 | NaN3 Inorganic azide |
Synthetic (1990) | Translocation EcSecA: 5 mM Intrinsic and membrane-bound SecA ATPase: no effect |
[137] |
| 2 |
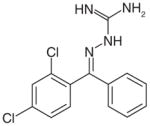 Imino-containing molecule |
Synthetic (2000) | Antibacterial: 6.2 μM (S. aureusRN 8081) Assay: SecA-lacZ reporter fusion to identify compounds that enhance SecA expression through inhibition of protein secretion |
[129] |
| 3 |
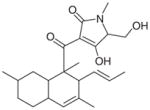 CJ-21058 |
CL47745, an unidentified fungus (2002) | SecA Translocation ATPase: 38.4 μM Antibacterial: 12.0 μM (MRSA, Enterococcus faecalis) Assay: Monitoring ATP hydrolysis using the SecA mediated translocation of proOmpA through the inner membrane vesicles bearing the heterotrimeric SecYEG complex |
[132] |
| 4 |
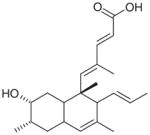 Pannomycin |
Geomyces pannorum (Fungus) (2009) | Antibacterial: B. subtilis (0.4 mM) S. aureus (1.4 mM) E. faecalis (1.4 mM) Assay: antisense-based screening strategy |
[124] |
| 5 |
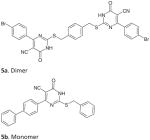 5-cyano-6-aryl-2-thiouracils derivatives |
Synthetic (2010) | Intrinsic SecATPase (EcN68): 2.0 μM (5b) Antibacterial (EcNR698): 20 μM (5a) Assay: ATPase malachite green assay |
[146] |
| 6 |
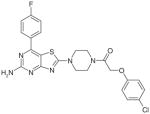 Thiazolo [4,5-d] pyrimidine derivatives |
Synthetic (2011) | Intrinsic SecA ATPase: 135 μM Translocation SecA ATPase:200 μM E. coli or S. aureus SecA Assay: ATPase malachite green assay Protein translocation using membrane vesicles containing overexpressed SecYEG and the E. coli preprotein AlkProPhoA(Cys-)3 |
[130] |
| 7 |
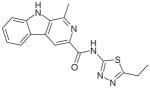 Indole derivative |
Synthetic (2012) | Intrinsic SecA ATPase inhibition: 0.25 μM (Candidatus Liberibacter asiaticus) Antibacterial: 0.76 mM (A. tumefaciens) Assay: ATPase Malachite green assay |
[147] |
Table 4.
A comparative account of Rose Bengal and Erythrosine B analogs’ SecA inhibitors and the assays used.
| Inhibitor ID | Compound name and structure | Source (year) | Inhibition constants (IC50s) and assay used
|
Ref. | |||
|---|---|---|---|---|---|---|---|
| ATPase | 8a | 8b | 8c | ||||
| 8 |
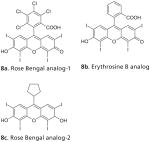
|
Synthetic (2012) | In vitro (IC50/μM) | [125,150] | |||
| Unregulated/truncatedN68 | 0.5 | 2 | ND | ||||
| Intrinsic ATPase | 25 | 21 | ND | ||||
| Membrane bound ATPase | 5 | 12 | ND | ||||
| Translocation ATPase | 0.9 | 10 | 2.5 | ||||
| In vivo (MIC/μM) | |||||||
| E. coli NR698 (leaky mutant) | 3.1 | 250–500 | 1.0 | ||||
| B. subtilis 168 | 3.1 | 250–500 | 6.0 | ||||
| E. coli (wild type) | >1000 | >1000 | ND | ||||
Later on, through virtual screening, random screening and natural product isolation, inhibitors of various structural classes have been identified. An imino-containing synthetic lipophilic compound [129] (2, Table 3) and CJ-21058 (3, Table 3), a natural product isolated from CL47745 (unidentified fungus), were found to inhibit SecA.
Another report of a natural product SecA inhibitor employed the antisense-based screening method, as discussed above. Pannomycin (4, Table 3) (from the extract of a leaf-litter fungus Geomyces Pannorum) was found using antisense-based screening method [124]. Pannomycin was shown to possess antibacterial activity, although; no direct in vitro SecA inhibition was reported.
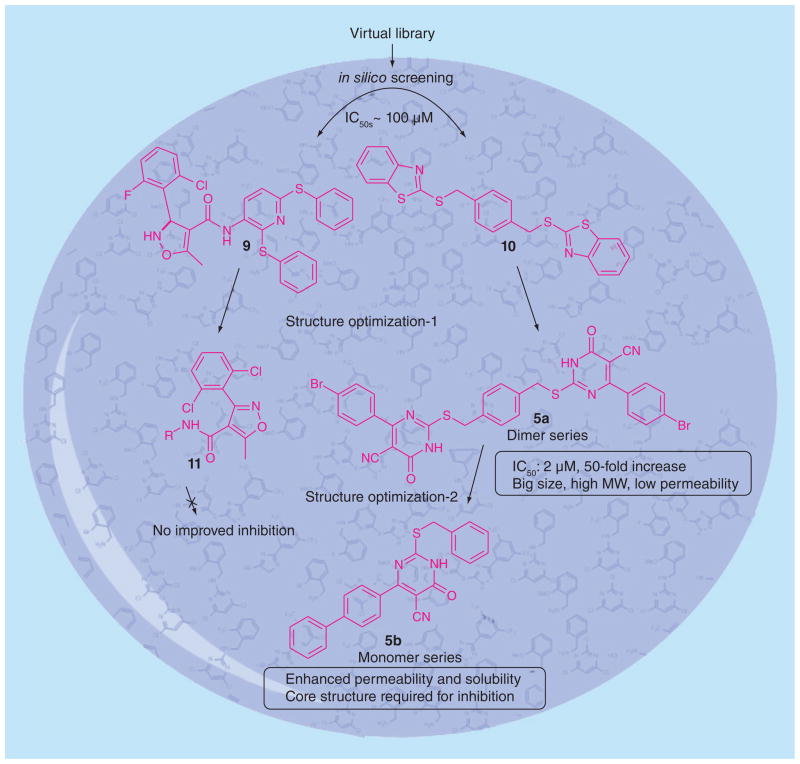
Efforts in our lab led to the discovery of the first ever synthetic SecA inhibitors, as shown in Figure 7. An in silico screening of small molecule ligand libraries (~115,000 compounds) was carried out using structure-based virtual screening methods against the E. coli SecATPase. The top ranked compounds were further tested for in vitro inhibition of ATPase activity against E. coli N68. Two modest inhibitors (Figure 7, 9 and 10) showed IC50 values in the range of 100 μM [148]. For a rationally guided design, the two hits obtained were docked into the enzyme active site and binding interactions were examined to aid in further structural optimization, which led to the isoxazolecarboxamide series (9). In the second series (10), different aryl structures flanking the central ring led to 5-cyano-6-aryl-2-thiouracils derivatives D and E (5a and 5b, Table 3). A simplified ‘monomer’ series of compounds was also prepared to understand the core structural need for inhibition. The compounds were tested in vitro using EcN68 SecA and the whole EcSecA [146]. Bacterial growth inhibition studies were done with an outer membrane leaky mutant NR698 having increased drug permeability [149] and wild-type E. coli strain MC4100. The result showed that the dimer series compound 5a possess low micromolar inhibition (IC50 = 2 μM), which is 50-fold more potent than the hit compound 9 (IC50 = 100 μM). For growth inhibition, the ‘monomer’ compound 5a exhibited the most potent activity against NR698 (IC50 = 20 μM), whereas the ‘dimer’ compound 5b did not exhibit significant antimicrobial activities. However, neither 5a nor 5b showed inhibition effects against wild type E. coli strain MC4100. Such results suggested that the permeability of 5a against NR698 and 5b against MC4100 might be a key factor [139].
Figure 7.
Structure optimization of 5-cyano-6-aryl-2-thiouracils derivatives.
After the publication of 5-cyano-6-aryl-2-thiouracil inhibitors, of a series of thiazolo [4, 5-d] pyrimidine derivatives (6, Table 3) was reported as SecA inhibitors [130]. Their screening strategy involved using an in vitro malachite green method and recombinant E. coli or S. aureus SecA. The compounds were also tested for inhibition of protein translocation using E. coli preprotein AlkProPhoA(Cys-)3 translocation through SecYEG containing membrane vesicles. Compound 6 was reported to have an IC50 value of 135 μM against EcSecA intrinsic ATPase and 200 μM for SecA translocation ATPase.
Due to the existence of SecA in different forms, it is truly important to screen inhibitors against various forms of SecA to seek correlation with antimicrobial results. Such correlation would indicate that inhibition of SecA was conducted within a state similar to that of physiological form in bacteria. Therefore, in one study, we used assays to individually assess inhibition by Rose Bengal (RB) and Erythrosine B (EB) on different SecA forms and found that the SecA inhibition results from channel activity assay and truncated SecA inhibition correlate with antimicrobial results the best. Specifically, inhibition assays involved truncated/unregulated SecA, regulated SecA (intrinsic, membrane-bound and translocase), SecA-mediated protein translocation and the antimicrobial growth inhibition using an outer membrane-leaky mutant NR698 and wild type strains [125]. As seen in Table 4 (8a and 8b), there are large differences in the sensitivity shown by different forms of SecA ATPases towards inhibitors. Such differences could largely be due to the effect of conformational changes affecting binding. The intrinsic SecA ATPase, which is the native and regulated form, shows higher IC50 values for RB (25 μM) and EB (21 μM) other forms of SecA. The membrane-bound form shows IC50 of 5 μM for RB and 12 μM for EB. The translocation ATPase shows IC50 values of 0.9 μM and 10 μM for RB and EB inhibition, respectively. The effect of inhibitors on SecA-dependent protein translocation of precursor proOmpA into the membrane vesicles was also studied. Both RB and EB were found to inhibit its translocation with IC50 values of 0.25 μM and 4 μM, respectively. We know that Sec dependent protein translocation maintains normal physiology and is crucial for viability; therefore the effect of inhibitors on bacterial growth was studied. The MIC values observed for Gram-negative bacteria (E. coli > 1 mM) are much higher than for Gram-positive (B. subtilis, 3.1 μM) and the leaky mutant (E. coli NR698, 3.1 μM), suggesting that outer membrane acts as a barrier to inhibitor permeability. More recently, we reported improved RB analogs showing inhibition in SecA ATPase, ion-channel and bacterial growth assays [150]. Compound 8c (Table 4) shows ion-channel inhibition with IC50 of 2.5 μM and several fold enhanced antimicrobial potency with MIC ~ 1 μM against (leaky mutant) E. coli NR698, as compared with RB. SAR studies on 20 analogs revealed that the xanthene ring on RB is essential for inhibition; the chlorinated benzoate position (8a, Table 4) can tolerate fairly substantial modifications; and the carboxylate group and the aryl ring are not required for activity [151,152].
In a recent study, SecA inhibitors were reported for a citrus plant bacterium Candidatus Liberibacter asiaticus (Las) responsible for causing Huanglongbing citrus disease [147]. A homology model of Las SecA was derived from E. coli SecA and used for virtual screening studies. The 20 hits obtained were tested for SecA ATPase activity inhibition using malachite green spectrophotometric method and antimicrobial growth inhibition was studied for A. tumefaciens. The most potent SecA inhibitor obtained was 8a with IC50 of 0.25 μM; and the MIC of 0.76 mM (A. tumefaciens) as shown in Table 4. This is the very first example of inhibitors targeting SecA of plant-infecting bacteria. Since, SecA is highly conserved in nature and the fact that homology model of Las SecA was based upon E. coli SecA, it may be worthwhile to test the inhibition of these compounds in different bacterial SecA.
Crowther et al. [153] have developed a whole-cell-based inhibitor screening strategy for the SecA machinery and demonstrated its high-throughput utility using various inhibitors. It involves using an engineered E. coli strain, which produces beta-galactosidase protein (β-gal) having the LamB sequence, causing it to be transported via Sec machinery. Transport of β-gal across SecYEG causes its inactivation; hence, the activity readout of β-gal is affected. This approach holds great advantage in screening for inhibitors of entire SecA machinery without having a specific target and works like antimicrobial (growth-based) assays in allowing only drug-like molecules to come up as hits. However, the screening results do not point to a specific molecular target as the reason for the observed effect.
In summary, seven structurally distinct classes of SecA inhibitors have been identified so far, along with the development of reliable assays, allowing screening of inhibitors against SecA ATPase under different conditions. These specific screening assays have helped in achieving a good understanding of the effects of inhibitors on various stages of protein translocation, and can lead to efficient and selective drug designing strategies. It needs to be emphasized that the electrophysiological assay using oocytes provides the best correlation with antimicrobial results. This is not surprising, since this is the only one having SecA in a physiological membrane structure, which mimics the original physiological state for SecA. A comparison of the enzymatic and microbial growth inhibition activities (Table 3) of various SecA inhibitors presses upon the need for improving antimicrobial growth inhibition potency. For example, RB/EB analogs and indole derivatives (Tables 3 & 4, 7 and 8) show the most potent enzyme inhibition, but antimicrobial properties of these compounds need further improvement. RB (8a) and EB (8b) show no inhibition (up to 1 mM) against the wild-type E. coli and indole derivatives (7) show minimal bactericidal concentration of 0.76 mM against A. tumefaciens. However, effective inhibition is seen (Table 4) against the leaky OM mutant with increased drug permeability (EcNR698: 8a, 3.1 μM, 8c, 1.0 μM) and Gram-positive (B. subtilis: 8a, 3.1 μM, 8c, 6.0 μM) bacteria. Clearly, these are due to the well-known outer membrane permeability issues in Gram-negative bacteria, which further suggest the need for overcoming outer membrane permeability problems. To an extent, this can be addressed by enhancing the drug-likeness of such compounds through modification of the structural features in order to improve physicochemical properties such as MW, logP and logS. Till date, membrane permeability poses one of the most challenging trials for medicinal chemists in the process of drug discovery.
Conclusions & future perspective
Thus far, ample results have demonstrated that indeed inhibition of SecA leads to antimicrobial effects and SecA is a valid target for the development of new antimicrobials. A critical issue in this area is the selection of the appropriate assays for SecA inhibition, which allows both throughput and accuracy. In this regard, the results from a combination of assays using truncated SecA, channel activity, and liposomal protein translocation seem to correlate well with antimicrobial effects. There are initial indications that SecA inhibition leads to attenuated secretion of virulence factors. Many results suggest that effect of SecA inhibitors can be more than just bactericidal/bacteriostatic and SecA inhibition can greatly attenuate pathogenicity. Furthermore, there are also indications that the potency of SecA inhibitors is not affected by the expression level of efflux pumps. Such results suggest that SecA may have the ability to overcome the effect of efflux by (1) directly accessing the target (SecA) and/or (2) impeding the integration of efflux pumps into bacterial membrane, especially in Gram-negative bacteria due to the presence of signal peptide sequence on efflux pumps recognizable for SecA Given the wide-spread nature of efflux pumps in bacteria and its importance in drug-resistance [154–160], such a finding by itself is of extraordinary novelty and significance. Thus SecA represent a very unique and promising target for antimicrobial development. The labs of Wang and Tai have collected ample evidence [125,127,150–152 (and some unpublished data, J Cui, AS Chaudhary, J Jin, PC Tai and B Wang, 2011-present)] to support the various points described. We hope our earlier review [161] will help stimulate significant research in this area, leading to the development of SecA-targeting novel antimicrobials.
Executive summary.
Given the substantial increase in the prevalence of drug resistance bacteria, there is an urgent need for developing new antimicrobials with novel mechanism of action.
We discuss the desired qualities of a good and ideal antimicrobial target in reference to SecA. The significance of SecA as an ATPase and an integral membrane protein along with its role in protein translocation in conjunction with SecYEG system is described.
Furthermore, SecA as an ATPase, an integral membrane protein, and a protein-conducting channel is discussed in great details. This includes a discussion of structure and mechanisms of protein translocation in
SecYEG/SecA. We also discuss the regulatory mechanisms for SecA activity such as SecA’s NBD and IRA2 domains.
Also, the established methods to test inhibitory effects on the functions of SecA, including assays such as SecA-lacZ reporter assay, antisense-mRNA based assay, ion-channel activity assay and others are described.
We provide our review and analysis of reported SecA inhibitors and highlight a future perspective by discussing the need for effective and accurate assays for assessing SecA activity.
Key terms
- Broad-spectrum antimicrobials
Antimicrobials capable of killing a wide range of bacteria by modulating a broad spectrum antimicrobial target
- SecYEG
Heterotrimeric complex comprising of SecY, SecE and SecG proteins, forming a channel in bacterial membrane for preprotein translocation
- Multidrug resistance
Nonsusceptibility of microbes to at least one antimicrobial agent in three or more categories
- SecA ATPase
Bacterial enzyme that possesses intrinsic ATPase activity, which is stimulated by lipids, and that catalyzes the translocation of preproteins through SecYEG machinery by hydrolyzing ATP
Footnotes
For reprint orders, please contact reprints@future-science.com
Financial & competing interests disclosure
Partial support for this work came from the National Institutes of Health (AI104168). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
- 1.Falagas ME, Karageorgopoulos DE. Pandrug resistance (PDR), extensive drug resistance (XDR), and multidrug resistance (MDR) among Gram-negative bacilli: need for international harmonization in terminology. Clin Infect Dis. 2008;46(7):1121–1122. doi: 10.1086/528867. [DOI] [PubMed] [Google Scholar]
- 2.Falagas ME, Koletsi PK, Bliziotis IA. The diversity of definitions of multidrug-resistant (MDR) and pandrugresistant (PDR) Acinetobacter baumannii and Pseudomonas aeruginosa. J Med Microbiol. 2006;55(12):1619–1629. doi: 10.1099/jmm.0.46747-0. [DOI] [PubMed] [Google Scholar]
- 3.Queenan AM, Bush K. Carbapenemases: the Versatile β-Lactamases. Clin Microbiol Rev. 2007;20(3):440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naas T, Nordmann P, Vedel G, Poyart C. Plasmid-mediated carbapenem-hydrolyzing β-Lactamase KPC in a Klebsiella pneumoniae isolate from France. Antimicrob Agents Chemother. 2005;49(10):4423–4424. doi: 10.1128/AAC.49.10.4423-4424.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez F, Endimiani A, Ray AJ, et al. Carbapenem-resistant Acinetobacter baumannii and Klebsiella pneumoniae across a hospital system: impact of post-acute care facilities on dissemination. J Antimicrob Chemother. 2010;65(8):1807–1818. doi: 10.1093/jac/dkq191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levy SB, Marshall B. Antibacterial resistance worldwide: causes, challenges and responses. Nat Med. 2004;10(12):S122–S129. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- 7.Taubes G. The bacteria fight back. Science. 2008;321(5887):356–361. doi: 10.1126/science.321.5887.356. [DOI] [PubMed] [Google Scholar]
- 8.Birmingham MC, Rayner CR, Meagher AK, et al. Linezolid for the treatment of multidrug-resistant, Gram-positive infections: experience from a compassionate-use program. Clin Infect Dis. 2003;36(2):159–168. doi: 10.1086/345744. [DOI] [PubMed] [Google Scholar]
- 9.Robbel L, Marahiel MA. Daptomycin, a bacterial lipopeptide synthesized by a nonribosomal machinery. J Biol Chem. 2010;285(36):27501–27508. doi: 10.1074/jbc.R110.128181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Soisson SM, Young K, et al. Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature. 2006;441(7091):358–361. doi: 10.1038/nature04784. [DOI] [PubMed] [Google Scholar]
- 11.Saier MH. Protein secretion and membrane insertion systems in Gram-negative bacteria. J Membr Biol. 2006;214(2):75–90. doi: 10.1007/s00232-006-0049-7. [DOI] [PubMed] [Google Scholar]
- 12.Vrontou E, Economou A. Structure and function of SecA, the preprotein translocase nanomotor. Biochim Biophys Acta. 2004;1694:67–80. doi: 10.1016/j.bbamcr.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 13.Mori H, Ito K. The Sec protein-translocation pathway. Trends Microbiol. 2001;9:494–500. doi: 10.1016/s0966-842x(01)02174-6. [DOI] [PubMed] [Google Scholar]
- 14.Van Klompenburg W, Ridder AN, van Raalte AL, et al. In vitro membrane integration of leader peptidase depends on the Sec machinery and anionic phospholipids and can occur posttranslationally. FEBS Lett. 1997;413(1):109–114. doi: 10.1016/s0014-5793(97)00888-0. [DOI] [PubMed] [Google Scholar]
- 15.Lill R, Dowhan W, Wickner W. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell. 2006;60(2):271–280. doi: 10.1016/0092-8674(90)90742-w. [DOI] [PubMed] [Google Scholar]
- 16.Chitlaru T, Gat O, Gozlan Y, et al. Differential proteomic analysis of the Bacillus anthracis secretome: distinct plasmid and chromosome CO2-dependent cross talk mechanisms modulate extracellular proteolytic activities. J Bacteriol. 2006;188(10):3551–3571. doi: 10.1128/JB.188.10.3551-3571.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rigel NW, Braunstein M. A new twist on an old pathway – accessory Sec systems. Mol Microbiol. 2008;69(2):291–302. doi: 10.1111/j.1365-2958.2008.06294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt MG, Rollo EE, Grodberg J, et al. Nucleotide-sequence of the SecA gene and Seca(Ts) mutations preventing protein export in Escherichia Coli. J Bacteriol. 1988;170(8):3404–3414. doi: 10.1128/jb.170.8.3404-3414.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siboo IR, Chaffin DO, Rubens CE, et al. Characterization of the accessory Sec system of Staphylococcus aureus. J Bacteriol. 2008;190(18):6188–6196. doi: 10.1128/JB.00300-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braunstein M1, Brown AM, Kurtz S, Jacobs WR., Jr Two nonredundant SecA homologues function in mycobacteria. J Bacteriol. 2001;183(24):6979–6990. doi: 10.1128/JB.183.24.6979-6990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bensing BA, Sullam PM. An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Mol Microbiol. 2002;44(4):1081–1094. doi: 10.1046/j.1365-2958.2002.02949.x. [DOI] [PubMed] [Google Scholar]
- 22.Lenz LL, Portnoy DA. Identification of a second Listeria secA gene associated with protein secretion and the rough phenotype. Mol Microbiol. 2002;45(4):1043–1056. doi: 10.1046/j.1365-2958.2002.03072.x. [DOI] [PubMed] [Google Scholar]
- 23.Lenz LL1, Mohammadi S, Geissler A, Portnoy DA. SecA2-dependent secretion of autolytic enzymes promotes Listeria monocytogenes pathogenesis. Proc Natl Acad Sci USA. 2003;100(21):12432–12437. doi: 10.1073/pnas.2133653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen QH, Wu Fives-Taylor PM. Investigating the role of secA2 in secretion and glycosylation of a fimbrial adhesin in Streptococcus parasanguisFW213. Mol Microbiol. 2004;53(3):843–856. doi: 10.1111/j.1365-2958.2004.04116.x. [DOI] [PubMed] [Google Scholar]
- 25.Takamatsu D, Bensing BA, Sullam PM. Genes in the accessory sec locus of Streptococcus gordonii have three functionally distinct effects on the expression of the platelet-binding protein GspB. Mol Microbiol. 2004;52(1):189–203. doi: 10.1111/j.1365-2958.2004.03978.x. [DOI] [PubMed] [Google Scholar]
- 26.Zelazny AM1, Calhoun LB, Li L, Shea YR, Fischer SH. Identification of Mycobacterium species by secA1 sequences. J Clin Microbiol. 2005;43(3):1051–1058. doi: 10.1128/JCM.43.3.1051-1058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siboo IR1, Chaffin DO, Rubens CE, Sullam PM. Characterization of the accessory Sec system of Staphylococcus aureus. J Bacteriol. 2008;190(18):6188–6196. doi: 10.1128/JB.00300-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bensing BA, Seepersaud R, Yen YT, Sullam PM. Selective transport by SecA2: an expanding family of customized motor proteins. Biochim Biophys Acta. 2014;1843(8):1674–1686. doi: 10.1016/j.bbamcr.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo XV, Monteleone M, Klotzsche M, et al. Silencing Mycobacterium smegmatis by using tetracycline repressors. J Bacteriol. 2007;189(13):4614–4623. doi: 10.1128/JB.00216-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48(1):77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 31.Monk IR, et al. Morphotypic conversion in Listeria monocytogenes biofilm formation: biological significance of rough colony isolates. Appl Environ Microbiol. 2004;70(11):6686–94. doi: 10.1128/AEM.70.11.6686-6694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Braunstein M, Espinosa BJ, Chan J, Belisle JT, Jacobs WR., Jr SecA2 functions in the secretion of superoxide dismutase A and in the virulence of Mycobacterium tuberculosis. Mol Microbiol. 2003;48(2):453–464. doi: 10.1046/j.1365-2958.2003.03438.x. [DOI] [PubMed] [Google Scholar]
- 33.Caspers M, Freudl R. Corynebacterium glutamicum possesses two secA homologous genes that are essential for viability. Arch Microbiol. 2008;189(6):605–610. doi: 10.1007/s00203-008-0351-0. [DOI] [PubMed] [Google Scholar]
- 34.Fagan RP, Fairweather NF. Clostridium difficile has two parallel and essential Sec secretion systems. J Biol Chem. 2011;286(31):27483–27493. doi: 10.1074/jbc.M111.263889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hou JM, D’Lima NG, Rigel NW, et al. ATPase activity of Mycobacterium tuberculosis SecA1 and SecA2 proteins and its importance for SecA2 function in macrophages. J Bacteriol. 2008;190(14):4880–4887. doi: 10.1128/JB.00412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bensing BA, Sullam PM. Characterization of Streptococcus gordonii SecA2 as a paralogue of SecA. J Bacteriol. 2009;191(11):3482–3491. doi: 10.1128/JB.00365-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D’Lima NG, Teschke CM. ADP-dependent conformational changes distinguish Mycobacterium tuberculosis SecA2 from SecA1. J Biol Chem. 2014;289(4):2307–2317. doi: 10.1074/jbc.M113.533323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Q1, Wu H, Kumar R, Peng Z, Fives-Taylor PM. SecA2 is distinct from SecA in immunogenic specificity, subcellular distribution and requirement for membrane anchoring in Streptococcus parasanguis. FEMS Microbiol Lett. 2006;264(2):174–181. doi: 10.1111/j.1574-6968.2006.00455.x. [DOI] [PubMed] [Google Scholar]
- 39.Eser M, Ehrmann M. SecA-dependent quality control of intracellular protein localization. Proc Natl Acad Sci USA. 2003;100(23):13231–13234. doi: 10.1073/pnas.2234410100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akita M, Shinkai A, Matsuyama S, Mizushima S. SecA, an essential component of the secretory machinery of Escherichia coli, exists as homodimer. Biochem Biophys Res Commun. 1991;74(1):211–216. doi: 10.1016/0006-291x(91)90507-4. [DOI] [PubMed] [Google Scholar]
- 41.Moran U, Phillips R, Milo R. SnapShot: key numbers in biology. Cell. 2010;141(7):1262–1262. doi: 10.1016/j.cell.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 42.Cabelli RJ, Dolan KM, Qian LP, et al. Characterization of membrane-associated and soluble states of SecA protein from wild-type and SecA51(TS) mutant strains of Escherichia coli. J Biol Chem. 1991;266(36):24420–24427. [PubMed] [Google Scholar]
- 43.Woodbury RL, Hardy SJS, Randall LL. Complex behavior in solution of homodimeric SecA. Protein Sci. 2002;11(4):875–882. doi: 10.1110/ps.4090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Driessen AJM. SecA, the peripheral subunit of the Escherichia coli precursor protein translocase, is functional as a dimer. Biochemistry. 1993;32(48):13190–13197. doi: 10.1021/bi00211a030. [DOI] [PubMed] [Google Scholar]
- 45.Kusters I, van den Bogaart G, Kedrov A, et al. Quaternary structure of SecA in solution and bound to SecYEG probed at the single molecule level. Structure. 2011;19(3):430–439. doi: 10.1016/j.str.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 46.Kusters I, Driessen AM. SecA, a remarkable nanomachine. Cell Mol Life Sci. 2011;68(12):2053–2066. doi: 10.1007/s00018-011-0681-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cordin O, Banroques J, Tanner NK, et al. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 48.Tanner NK, Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol Cell. 2001;8(2):251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- 49.Cooper DB, Smith VF, Crane JM, et al. SecA, the motor of the secretion machine, binds diverse partners on one interactive surface. J Mol Biol. 2008;382(1):74–87. doi: 10.1016/j.jmb.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Papanikou E, Karamanou S, Baud C, et al. Identification of the preprotein binding domain of SecA. J Biol Chem. 2005;280(52):43209–43217. doi: 10.1074/jbc.M509990200. [DOI] [PubMed] [Google Scholar]
- 51.Papanikolau Y, Papadovasilaki M, Ravelli RBG, et al. Structure of dimeric SecA, the Escherichia coli preprotein translocase motor. J Mol Biol. 2007;366(5):1545–1557. doi: 10.1016/j.jmb.2006.12.049. [DOI] [PubMed] [Google Scholar]
- 52.Gelis I, Bonvin AMJJ, Keramisanou D, et al. Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR. Cell. 2007;131(4):756–769. doi: 10.1016/j.cell.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bu Z, Wang L, Kendall DA. Nucleotide binding induces changes in the oligomeric state and conformation of Sec A in a lipid environment: a small-angle neutron-scattering study. J Mol Biol. 2003;332(1):23–30. doi: 10.1016/s0022-2836(03)00840-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen Y, Pan X, Tang Y, et al. Full-length Escherichia coli SecA dimerizes in a closed conformation in solution as determined by cryo-electron microscopy. J Biol Chem. 2008;283(43):28783–28787. doi: 10.1074/jbc.C800160200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen Y, Tai PC, Sui SF. The active ring-like structure of SecA revealed by electron crystallography: conformational change upon interaction with SecB. J Struct Biol. 2007;159(1):149–153. doi: 10.1016/j.jsb.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shilton B, Svergun DI, Volkov VV, et al. Escherichia coli SecA shape and dimensions. FEBS Lett. 1998;436(2):277–282. doi: 10.1016/s0014-5793(98)01141-7. [DOI] [PubMed] [Google Scholar]
- 57.Osborne AR, Clemons WM, Rapoport TA. A large conformational change of the translocation ATPase SecA. Proc Natl Acad Sci USA. 2004;101(30):10937–10942. doi: 10.1073/pnas.0401742101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hunt JF, Weinkauf S, Henry L, et al. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science. 2002;297(5589):2018–2026. doi: 10.1126/science.1074424. [DOI] [PubMed] [Google Scholar]
- 59.Sharma V, Arockiasamy A, Ronning DR, et al. Crystal structure of Mycobacterium tuberculosis SecA, a preprotein translocating ATPase. Proc Natl Acad Sci USA. 2003;100(5):2243–2248. doi: 10.1073/pnas.0538077100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zimmer J, Li WK, Rapoport TA. A novel dimer interface and conformational changes revealed by an X-ray structure of B. subtilis SecA. J Mol Biol. 2006;364(3):259–265. doi: 10.1016/j.jmb.2006.08.044. [DOI] [PubMed] [Google Scholar]
- 61.Vassylyev DG, Mori H, Vassylyeva MN, et al. Crystal structure of the translocation ATPase SecA from Thermus thermophilus reveals a parallel, head-to-head dimer. J Mol Biol. 2006;364(3):248–258. doi: 10.1016/j.jmb.2006.09.061. [DOI] [PubMed] [Google Scholar]
- 62.Nikaido H, Zgurskaya HI. Antibiotic efflux mechanisms. Curr Opin Infect Dis. 1999;12:529–536. doi: 10.1097/00001432-199912000-00001. [DOI] [PubMed] [Google Scholar]
- 63.Segers K, Klaassen H, Economou A, et al. Development of a high-throughput screening assay for the discovery of small-molecule SecA inhibitors. Anal Biochem. 2011;413(2):90–96. doi: 10.1016/j.ab.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 64.Zimmer J, Rapoport TA. Conformational flexibility and peptide interaction of the translocation ATPase SecA. J Mol Biol. 2009;394(4):606–612. doi: 10.1016/j.jmb.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 2008;455(7215):936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Osborne AR, Rapoport TA. Protein translocation is mediated by oligomers of the SecY complex with one SecY copy forming the channel. Cell. 2007;129(1):97–110. doi: 10.1016/j.cell.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 67•.Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera – a visualization system for exploratory research and analysis. J Comp Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from the NIH (National Center for Research Resources grant 2P41RR001081, National Institute of General Medical Sciences grant 9P41GM103311) [DOI] [PubMed] [Google Scholar]
- 68.Tang Y, Pan X, Chen Y, et al. Dimeric SecA couples the preprotein translocation in an asymmetric manner. PloS ONE. 2011;6(1):e16498. doi: 10.1371/journal.pone.0016498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baud C, Karamanou S, Sianidis G, et al. Allosteric communication between signal peptides and the SecA protein DEAD motor ATPase domain. J Biol Chem. 2002;277(16):13724–13731. doi: 10.1074/jbc.M200047200. [DOI] [PubMed] [Google Scholar]
- 70.Karamanou S, Vrontou E, Sianidis G, et al. A molecular switch in SecA protein couples ATP hydrolysis to protein translocation. Mol Microbiol. 1999;34(5):1133–1145. doi: 10.1046/j.1365-2958.1999.01686.x. [DOI] [PubMed] [Google Scholar]
- 71.Sianidis G, Karamanou S, Vrontou E, et al. Cross-talk between catalytic and regulatory elements in a DEAD motor domain is essential for SecA function. EMBO J. 2001;20(5):961–970. doi: 10.1093/emboj/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Price A, Economou A, Duong F, et al. Separable ATPase and membrane insertion domains of the SecA subunit of preprotein translocase. J Biol Chem. 1996;271(49):31580–31584. doi: 10.1074/jbc.271.49.31580. [DOI] [PubMed] [Google Scholar]
- 73.De Keyzer J, Van der Sluis EO, Spelbrink REJ, et al. Covalently dimerized SecA is functional in protein translocation. J Biol Chem. 2005;280(42):35255–35260. doi: 10.1074/jbc.M506157200. [DOI] [PubMed] [Google Scholar]
- 74.Jilaveanu LB, Oliver D. SecA Dimer Cross-Linked at Its Subunit Interface Is Functional for Protein Translocation. J Bacteriol. 2006;188(1):335–338. doi: 10.1128/JB.188.1.335-338.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jilaveanu LB, Zito CR, Oliver D. Dimeric SecA is essential for protein translocation. Proc Natl Acad Sci USA. 2005;102(21):7511–7516. doi: 10.1073/pnas.0502774102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Or E, Navon A, Rapoport T. Dissociation of the dimeric SecA ATPase during protein translocation across the bacterial membrane. EMBO J. 2002;21(17):4470–4479. doi: 10.1093/emboj/cdf471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Or E, Boyd D, Gon S, et al. The Bacterial ATPase SecA Functions as a Monomer in Protein Translocation. J Biol Chem. 2005;280(10):9097–9105. doi: 10.1074/jbc.M413947200. [DOI] [PubMed] [Google Scholar]
- 78.Duong F. Binding, activation and dissociation of the dimeric SecA ATPase at the dimeric SecYEG translocase. EMBO J. 2003;22(17):4375–4384. doi: 10.1093/emboj/cdg418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tziatzios C, Schubert D, Lotz M, et al. The Bacterial Protein–Translocation Complex: SecYEG Dimers Associate with One or Two SecA Molecules. J Mol Biol. 2004;340(3):513–524. doi: 10.1016/j.jmb.2004.04.076. [DOI] [PubMed] [Google Scholar]
- 80.Papanikou E, Karamanou S, Economou A. Bacterial protein secretion through the translocase nanomachine. Nat Rev Microbiol. 2007;5(11):839–851. doi: 10.1038/nrmicro1771. [DOI] [PubMed] [Google Scholar]
- 81.Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450(7170):663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 82.Lycklama a Nijeholt JA, Driessen AJM. The bacterial Sectranslocase: structure and mechanism. Philos Trans R Soc Lond B Biol Sci. 2012;2367(1592):1016–1028. doi: 10.1098/rstb.2011.0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Economou A, Wickner W. SecA promotes preprotein translocation by undergoing Atp-driven cycles of membrane insertion and deinsertion. Cell. 1994;78(5):835–843. doi: 10.1016/s0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 84.Economou A, Pogliano JA, Beckwith J, et al. SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell. 1995;83(7):1171–1181. doi: 10.1016/0092-8674(95)90143-4. [DOI] [PubMed] [Google Scholar]
- 85.Jilaveanu LB, Oliver DB. In vivo membrane topology of Escherichia coli SecA ATPase reveals extensive periplasmic exposure of multiple functionally important domains clustering on one face of SecA. J Biol Chem. 2007;282(7):4661–4668. doi: 10.1074/jbc.M610828200. [DOI] [PubMed] [Google Scholar]
- 86.Gouridis G, Karamanou S, Gelis I, et al. Signal peptides are allosteric activators of the protein translocase. Nature. 2009;462(7271):363–368. doi: 10.1038/nature08559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Karamanou S, Gouridis G, Papanikou E, et al. Preprotein-controlled catalysis in the helicase motor of SecA. EMBO J. 2007;26(12):2904–2914. doi: 10.1038/sj.emboj.7601721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fak JJ, Itkin A, Ciobanu DD, et al. Nucleotide exchange from the high-affinity ATP-binding site in SecA is the rate-limiting step in the ATPase cycle of the soluble enzyme and occurs through a specialized conformational state. Biochemistry. 2004;43(23):7307–7327. doi: 10.1021/bi0357208. [DOI] [PubMed] [Google Scholar]
- 89.Smith PA, Powers ME, Roberts TC, et al. In vitro activities of arylomycin natural-product antibiotics against Staphylococcus epidermidis and other coagulase-negative Staphylococci. Antimicrob Agents Chemother. 2011;55(3):1130–1134. doi: 10.1128/AAC.01459-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li W, Schulman S, Boyd D, et al. The plug domain of the SecY protein stabilizes the closed state of the translocation channel and maintains a membrane seal. Mol Cell. 2007;26(4):511–521. doi: 10.1016/j.molcel.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 91.Segers K, Anné J. Traffic jam at the bacterial Sec Translocase: targeting the SecA nanomotor by small-molecule inhibitors. Chem Biol. 2011;18(6):685–698. doi: 10.1016/j.chembiol.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 92.Fekkes P, vanderDoes C, Driessen AJM. The molecular chaperone SecB is released from the carboxy-terminus of SecA during initiation of precursor protein translocation. EMBO J. 1997;16(20):6105–6113. doi: 10.1093/emboj/16.20.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Randall LL, Crane JM, Liu G, et al. Sites of interaction between SecA and the chaperone SecB, two proteins involved in export. Protein Sci. 2004;13(4):1124–1133. doi: 10.1110/ps.03410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mao C, Hardy SJS, Randall LL. Maximal efficiency of coupling between ATP hydrolysis and translocation of polypeptides mediated by SecB requires two protomers of SecA. J Bacteriol. 2009;191(3):978–984. doi: 10.1128/JB.01321-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Suo Y, Hardy SJS, Randall LL. Orientation of SecA and SecB in complex, derived from disulfide cross-linking. J Bacteriol. 2011;193(1):190–196. doi: 10.1128/JB.00975-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Engelman DM, Steitz TA. The spontaneous insertion of proteins into and across membranes: the helical hairpin hypothesis. Cell. 1981;23(2):411–422. doi: 10.1016/0092-8674(81)90136-7. [DOI] [PubMed] [Google Scholar]
- 97.Joly JC, Wickner W. The SecA and SecY subunits of translocase are the nearest neighbors of a translocating preprotein, shielding it from phospholipids. EMBO J. 1993;12(1):255–263. doi: 10.1002/j.1460-2075.1993.tb05651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cannon KS, Or E, Clemons WM, et al. Disulfide bridge formation between SecY and a translocating polypeptide localizes the translocation pore to the center of SecY. J Cell Biol. 2005;169(2):219–225. doi: 10.1083/jcb.200412019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tani K, Tokuda H, Mizushima S. Translocation of ProOmpA possessing an intramolecular disulfide bridge into membrane vesicles of Escherichia coli Effect of membrane energization. J Biol Chem. 1990;265(28):17341–17347. [PubMed] [Google Scholar]
- 100.Shiozuka K, Tani K, Mizushima S, et al. The proton motive force lowers the level of ATP required for the in vitro translocation of a secretory protein in Escherichia coli. J Biol Chem. 1990;265(31):18843–18847. [PubMed] [Google Scholar]
- 101.Nouwen N, de Kruijff B, Tommassen J. prlA suppressors in Escherichia coli relieve the proton electrochemical gradient dependency of translocation of wild-type precursors. Proc Natl Acad Sci USA. 1996;93(12):5953–5957. doi: 10.1073/pnas.93.12.5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nishiyama K-i, Fukuda A, Morita K, et al. Membrane deinsertion of SecA underlying proton motive force-dependent stimulation of protein translocation. EMBO J. 1999;18(4):1049–1058. doi: 10.1093/emboj/18.4.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bessonneau P, Besson V, Collinson I, et al. The SecYEG preprotein translocation channel is a conformationally dynamic and dimeric structure. EMBO J. 2002;21(5):995–1003. doi: 10.1093/emboj/21.5.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Driessen AJ, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem. 2008;77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- 105.Schiebel E, Driessen AJ, Hartl FU, et al. Delta mu H+ and ATP function at different steps of the catalytic cycle of preprotein translocase. Cell. 1991;64(5):927–939. doi: 10.1016/0092-8674(91)90317-r. [DOI] [PubMed] [Google Scholar]
- 106.Van der Wolk JPW, de Wit JG, Driessen AJM. The catalytic cycle of the Escherichia coli SecA ATPase comprises two distinct preprotein translocation events. EMBO J. 1997;16(24):7297–7304. doi: 10.1093/emboj/16.24.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Benach J, Chou Y-T, Fak JJ, et al. Phospholipid-induced monomerization and signal-peptide-induced oligomerization of SecA. J Biol Chem. 2003;278(6):3628–3638. doi: 10.1074/jbc.M205992200. [DOI] [PubMed] [Google Scholar]
- 108.Robson A, Gold VAM, Hodson S, et al. Energy transduction in protein transport and the ATP hydrolytic cycle of SecA. Proc Natl Acad Sci USA. 2009;106(13):5111–5116. doi: 10.1073/pnas.0809592106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Keramisanou D, Biris N, Gelis I, et al. Disorder-order folding transitions underlie catalysis in the helicase motor of SecA. Nat Struct Mol Biol. 2006;13(7):594–602. doi: 10.1038/nsmb1108. [DOI] [PubMed] [Google Scholar]
- 110.Ding HY, Mukerji I, Oliver D. Nucleotide and phospholipid-dependent control of PPXD and C-domain association for SecA ATPase. Biochemistry. 2003;42(46):13468–13475. doi: 10.1021/bi035099b. [DOI] [PubMed] [Google Scholar]
- 111.De Keyzer J, van der Does C, Kloosterman TG, et al. Direct demonstration of ATP-dependent release of SecA from a translocating preprotein by surface plasmon resonance. J Biol Chem. 2003;278(32):29581–29586. doi: 10.1074/jbc.M303490200. [DOI] [PubMed] [Google Scholar]
- 112.Wang HW, Chen Y, Yang H, et al. Ring-like pore structures of SecA: implication for bacterial protein-conducting channels. Proc Natl Acad Sci USA. 2003;100(7):4221–4226. doi: 10.1073/pnas.0737415100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hsieh YH, Zhang H, Lin BR, et al. SecA alone can promote protein translocation and Ion-channel activity:SecYEG increases efficiency and signal peptide specificity. J Biol Chem. 2005;286:44702–44709. doi: 10.1074/jbc.M111.300111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.You Z, Liao M, Zhang H, et al. Phospholipids induce conformational changes of SecA to form membrane-specific domains: AFM structures and implication on protein-conducting channels. PloS ONE. 2013;8(8):e72560. doi: 10.1371/journal.pone.0072560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chen X, Brown T, Tai PC. Identification and characterization of protease-resistant SecA fragments: secA has two membrane-integral forms. J Biol Chem. 1998;180(3):527–537. doi: 10.1128/jb.180.3.527-537.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen X, Xu H, Tai PC. A significant fraction of functional SecA is permanently embedded in the membrane. SecA cycling on and off the membrane is not essential during protein translocation. J Biol Chem. 1996;271(47):29698–29706. doi: 10.1074/jbc.271.47.29698. [DOI] [PubMed] [Google Scholar]
- 117.Hu HJ, Holley J, He J, et al. To be or not to be: predicting soluble SecAs as membrane proteins. IEEE Trans NanoBioscience. 2007;6(2):168–179. doi: 10.1109/tnb.2007.897486. [DOI] [PubMed] [Google Scholar]
- 118.Wang H, Na B, Yang H, et al. Additional in vitro and in vivo evidence for SecA functioning as dimers in the membrane: dissociation into monomers is not essential for protein translocation in Escherichia coli. J Biol Chem. 2008;190(4):1413–1418. doi: 10.1128/JB.01633-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hsieh YH, Zhang H, Wang H, et al. Reconstitution of functionally efficient SecA-dependent protein-conducting channels: transformation of low-affinity SecA-liposome channels to high-affinity SecA-SecYEG-SecDF. YajC channels. Biochem Biophys Res Commun. 2013;431(3):388–392. doi: 10.1016/j.bbrc.2013.01.042. [DOI] [PubMed] [Google Scholar]
- 120.Tang Y, Pan X, Tai PC, et al. Electron microscopic visualization of asymmetric precursor translocation intermediates: SecA functions as a dimer. Sci China Life Sci. 2010;53(9):1049–1056. doi: 10.1007/s11427-010-4061-x. [DOI] [PubMed] [Google Scholar]
- 121.Yang YB, Lian J, Tai PC. Differential translocation of protein precursors across SecY-deficient membranes of Escherichia coli: SecY is not obligatorily required for translocation of certain secretory proteins in vitro. J Bacteriol. 1997;179(23):7386–7393. doi: 10.1128/jb.179.23.7386-7393.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nithianantham S, Shilton BH. Analysis of the isolated SecA dead motor suggests a mechanism for chemical–mechanical coupling. J Mol Biol. 2008;383(2):380–389. doi: 10.1016/j.jmb.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 123.Hsieh YH, Zhang H, Lin BR, et al. SecA alone can promote protein translocation and Ion-channel activity:SecYEG increases efficiency and signal peptide specificity. J Biol Chem. 2005;286:44702–44709. doi: 10.1074/jbc.M111.300111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Parish CA, Cruz Mdl, Smith SK, et al. Antisense-guided isolation and structure elucidation of pannomycin, a substituted cis-decalin from Geomyces pannorum. J Nat Prod. 2008;72(1):59–62. doi: 10.1021/np800528a. [DOI] [PubMed] [Google Scholar]
- 125.Huang Y-J, Wang H, Gao F-B, et al. Fluorescein analogues inhibit SecA ATPase: the first sub-micromolar inhibitor of bacterial protein translocation. ChemMedChem. 2012;7(4):571–577. doi: 10.1002/cmdc.201100594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gouridis G, Karamanou S, Koukaki M, et al. In vitro assays to analyze translocation of the model secretory preprotein alkaline phosphatase. Methods Mol Biol. 2010;619:157–172. doi: 10.1007/978-1-60327-412-8_10. [DOI] [PubMed] [Google Scholar]
- 127.Lanzetta PA, Alvarez LJ, Reinach PS, et al. An improved assay for nanomole amounts of inorganic phosphate. Anal Biochem. 1979;100(1):95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- 128.Mitchell C, Oliver D. Two distinct ATP-binding domains are needed to promote protein export by Escherichia coli SecA ATPase. Mol Microbiol. 1993;10(3):483–497. doi: 10.1111/j.1365-2958.1993.tb00921.x. [DOI] [PubMed] [Google Scholar]
- 129.Alksne LE, Burgio P, Hu W, et al. Identification and analysis of bacterial protein secretion inhibitors utilizing a seca-lacz reporter fusion system. Antimicrob Agents Chemother. 2000;44(6):1418–1427. doi: 10.1128/aac.44.6.1418-1427.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jang M-Y, Jonghe SD, Segers K, et al. Synthesis of novel 5-amino-thiazolo[4,5-d]pyrimidines as E. coli and S. aureus SecA inhibitors. Biorg Med Chem. 2011;19(1):702–714. doi: 10.1016/j.bmc.2010.10.027. [DOI] [PubMed] [Google Scholar]
- 131.Chen LL, Tai PC. ATP Is Essential for protein translocation into Escherichia-Coli membrane-vesicles. Proc Natl Acad Sci USA. 1985;82(13):4384–4388. doi: 10.1073/pnas.82.13.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sugie Y, Inagaki S, Kato Y, et al. CJ-21,058, a New SecA inhibitor isolated from a fungus. J Antibiot. 2002;55(1):25–29. doi: 10.7164/antibiotics.55.25. [DOI] [PubMed] [Google Scholar]
- 133.Lin BR, Gierasch LM, Jiang C, et al. Electrophysiological studies in Xenopus oocytes for the opening of Escherichia coli SecA-dependent protein-conducting channels. J Membr Biol. 2006;214(2):103–113. doi: 10.1007/s00232-006-0079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Vrontou E, Karamanou S, Baud C, et al. Global coordination of protein translocation by the SecA IRA1 switch. J Biol Chem. 2004;279(21):22490–22497. doi: 10.1074/jbc.M401008200. [DOI] [PubMed] [Google Scholar]
- 135.Ito YK, Chiba S, Pogliano K. Divergent stalling sequences sense and control cellular physiology. Biochem Biophys Res Commun. 2010;393(1):1–5. doi: 10.1016/j.bbrc.2010.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Loew O. Ueber das Verhalten des Azoimids zu lebenden Organismen. Ber deut chem Ges. 1891;24:2947–2953. [Google Scholar]
- 137.Oliver DB, Cabelli RJ, Dolan KM, et al. Azide-resistant mutants of Escherichia coli alter the SecA protein, an azide-sensitive component of the protein export machinery. Proc Natl Acad Sci USA. 1990;87(21):8227–8231. doi: 10.1073/pnas.87.21.8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Klein M, Hofmann B, Klose M, et al. Isolation and characterization of a Bacillus subtilis secA mutant allele conferring resistance to sodium azide. FEMS Microbiol Lett. 1994;124(3):393–397. doi: 10.1111/j.1574-6968.1994.tb07314.x. [DOI] [PubMed] [Google Scholar]
- 139.Nakane A, Takamatsu H, Oguro A, et al. Acquisition of azide-resistance by elevated SecA ATPase activity confers azide-resistance upon cell growth and protein translocation in Bacillus subtilis. Microbiology. 1995;141(Pt 1):113–121. doi: 10.1099/00221287-141-1-113. [DOI] [PubMed] [Google Scholar]
- 140.Eichler J, Wickner W. The SecA subunit of Escherichia coliPreprotein Translocase Is Exposed to the Periplasm. J Bacteriol. 1998;180(21):5776–5779. doi: 10.1128/jb.180.21.5776-5779.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bowler MW, Montgomery MG, Leslie AG, et al. How azide inhibits ATP hydrolysis by the F-ATPases. Proc Natl Acad Sci USA. 2006;103(23):8646–8649. doi: 10.1073/pnas.0602915103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yoshikawa S, Shinzawa-Itoh K, Nakashima R, et al. Redox-coupled crystal structural changes in bovine heart cytochrome c oxidase. Science. 1998;280(5370):1723–1729. doi: 10.1126/science.280.5370.1723. [DOI] [PubMed] [Google Scholar]
- 143.Stoddard BL, Ringe D, Petsko GA. The structure of iron superoxide dismutase from Pseudomonas ovalis complexed with the inhibitor azide. Protein Eng. 1990;4(2):113–119. doi: 10.1093/protein/4.2.113. [DOI] [PubMed] [Google Scholar]
- 144.Yound JM, Wang JH. The nature of binding of competitive inhibitors to alcohol dehydrogenases. J Biol Chem. 1971;246(9):2815–2821. [PubMed] [Google Scholar]
- 145.Zaitsev VN, Zaitseva I, Papiz M, et al. An X-ray crystallographic study of the binding sites of the azide inhibitor and organic substrates to ceruloplasmin, a multicopper oxidase in the plasma. J Biol Inorg Chem. 1999;4(5):579–587. doi: 10.1007/s007750050380. [DOI] [PubMed] [Google Scholar]
- 146.Chen W, Huang Y-j, Gundala SR, et al. The first low μM SecA inhibitors. Bioorg Med Chem. 2010;18(4):1617–1625. doi: 10.1016/j.bmc.2009.12.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Akula N, Zheng H, Han FQ, et al. Discovery of novel SecA inhibitors of Candidatus Liberibacter asiaticus by structure based design. Bioorg Med Chem. 2011;21(14):4183–4188. doi: 10.1016/j.bmcl.2011.05.086. [DOI] [PubMed] [Google Scholar]
- 148.Li M, Huang Y-J, Tai PC, et al. Discovery of the first SecA inhibitors using structure-based virtual screening. Biochem Biophys Res Commun. 2008;368(4):839–845. doi: 10.1016/j.bbrc.2008.01.135. [DOI] [PubMed] [Google Scholar]
- 149.Ruiz N, Falcone B, Kahne D, et al. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell. 2005;121:307–317. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 150.Cui J, Jin J, Hsieh Y-H, Yang H, et al. Design, Synthesis and biological evaluation of rose bengal analogues as seca inhibitors. ChemMedChem. 2013;8(8):1384–1393. doi: 10.1002/cmdc.201300216. [DOI] [PubMed] [Google Scholar]
- 151.Hsieh YH, Huang J-S, Jin Y-J, et al. Mechanisms of Rose Bengal inhibition on SecA ATPase and ion channel activities. Biochem Biophys Res Commun. 2014;454:308–312. doi: 10.1016/j.bbrc.2014.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chaudhary AS, Jin J, Chen PC, Tai W, Wang B. Design, syntheses and evaluation of 4-oxo-5-cyano thiouracils as SecA inhibitors. Bioorg Med Chem. 2015;23:105–117. doi: 10.1016/j.bmc.2014.11.017. [DOI] [PubMed] [Google Scholar]
- 153.Crowther GJ, Quadri SA, Shannon-Alferes BJ, et al. A Mechanism-based whole-cell screening assay to identify inhibitors of protein export in Escherichia coli by the sec pathway. J Biomol Screen. 2012;17(4):535–541. doi: 10.1177/1087057111431606. [DOI] [PubMed] [Google Scholar]
- 154.Nikaido H, Zgurskaya HI. Antibiotic efflux mechanisms. Curr Opin Infect Dis. 1999;12(6):529–536. doi: 10.1097/00001432-199912000-00001. [DOI] [PubMed] [Google Scholar]
- 155.Masaoka Y, Ueno Y, Morita Y, et al. A two-component multidrug efflux pump, EbrAB, in Bacillus subtilis. J Biol Chem. 2000;182(8):2307–2310. doi: 10.1128/jb.182.8.2307-2310.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Van Bambeke F, Balzi E, Tulkens PM. Antibiotic efflux pumps. Biochem Pharmacol. 2000;60(4):457–470. doi: 10.1016/s0006-2952(00)00291-4. [DOI] [PubMed] [Google Scholar]
- 157.Lomovskaya O, Watkins W. Inhibition of efflux pumps as a novel approach to combat drug resistance in bacteria. J Mol Microbiol Biotech. 2001;3(2):225–236. [PubMed] [Google Scholar]
- 158.Markham PN, Neyfakh AA. Efflux-mediated drug resistance in Gram-positive bacteria. Curr Opin Microbiol. 2001;4(5):509–514. doi: 10.1016/s1369-5274(00)00243-5. [DOI] [PubMed] [Google Scholar]
- 159.Levy SB. Active efflux, a common mechanism for biocide and antibiotic resistance. J App Microbiol. 2002;92:65S–71S. [PubMed] [Google Scholar]
- 160.Zhang Y, Eric Ballard C, Zheng SL, et al. Design, synthesis, and evaluation of efflux substrate-metal chelator conjugates as potential antimicrobial agents. Bioorg Med Chem lett. 2007;17(3):707–711. doi: 10.1016/j.bmcl.2006.10.094. [DOI] [PubMed] [Google Scholar]
- 161.Chen W, Chaudhary A, Cui J, et al. SecA inhibitors: next generation antimicrobials. J Chin Pharm Sci. 2012;21:526–530. [Google Scholar]