Abstract
We previously reported an upregulation of the clock transcript BMAL1, correlating with TIMP1 expression in fresh-frozen samples from papillary thyroid carcinoma (PTC). Since frozen postoperative biopsy samples are difficult to obtain, we aimed to validate the application of high-precision NanoString analysis for formalin-fixed paraffin-embedded (FFPE) thyroid nodule samples and to screen for potential biomarkers associated with PTC. No significant differences were detected between fresh-frozen and FFPE samples. NanoString analysis of 51 transcripts in 17 PTC and 17 benign nodule samples obtained from different donors and in 24 pairs of benign and PTC nodules, obtained from the same donor (multinodular goiters), confirmed significant alterations in the levels of BMAL1, c-MET, c-KIT, TIMP1, and other transcripts. Moreover, we identified for the first time alterations in CHEK1 and BCL2 levels in PTC. A predictive score was established for each sample, based on the combined expression levels of BMAL1, CHEK1, c-MET, c-KIT and TIMP1. In combination with BRAF mutation analysis, this predictive score closely correlated with the clinicopathological characteristics of the analyzed thyroid nodules. Our study identified new thyroid transcripts with altered levels in PTC using the NanoString approach. A predictive score correlation coefficient might contribute to improve the preoperative diagnosis of thyroid nodules.
Keywords: papillary thyroid carcinoma, NanoString analysis, FFPE, biomarkers, circadian clock
INTRODUCTION
Thyroid cancer is the most common endocrine malignancy with increasing incidence over the last decade [1]. Well-differentiated thyroid carcinomas, comprising papillary thyroid carcinoma (PTC, 85%) and follicular thyroid carcinoma (FTC, 15%), represent the vast majority of thyroid malignancies. Poorly differentiated and undifferentiated (anaplastic) thyroid carcinomas (PDTC and ATC) are less common but more aggressive [2]. Fine-needle aspiration (FNA) biopsy represents the gold standard for the preoperative diagnosis of PTC with an accuracy of 90% and is recommended for the clinical evaluation of all suspicious non-secreting thyroid nodules ≥ 1cm [3, 4]. Thyroid FNA allows the accurate preoperative recognition of thyroid malignancies, except for 30% of indeterminate cases, where the distinction between benign and malignant nodules is impossible based on cytological features. At present, genetic tests including the combination of BRAF V600E and RAS point mutations and RET-PTC and PAX8-PPARγ rearrangements are considered to provide high specificity and positive predictive value for PTC and FTC diagnosis (“rule in” tests). However, the sensitivity of these mutation analyses is low. Moreover, RAS or PAX8/PPARγ mutations are also found in a subset of follicular adenomas, therefore limiting their predictive value. In contrast, gene expression classifier tests such as Afirma have an excellent negative predictive value, with a limited specificity (“rule out” tests). The presence of the BRAFV600E mutation in PTC is one of the best-defined prognostic markers today, demonstrating also a strong association with disease aggressiveness ([5] and references therein). Moreover, molecular biomarkers with strongly altered expression levels in PTC, including TIMP1 [6], c-KIT [7], c-MET [8], or TPO (thyroperoxidase) [9] have been proposed to be predictive of PTC. Of note, the BRAFV600E mutation exhibited correlation with changes in TIMP1 and TPO expression levels in PTC [6].
Our recent study has revealed that circadian clocks are operative in human thyroid tissue with altered properties in PTC and PDTC nodules. Moreover, the expression levels of core clock genes BMAL1 and CRY2 were altered in PTC tissue samples, as compared to normal thyroid and benign nodules [3]. Interestingly, alterations in BMAL1 levels strongly correlated with those of TIMP1 in the same PTC nodules. This study further underscores a tight connection between circadian clock alterations and cell malignant transformation in general, and revealed a possible role of the circadian clock, or of core clock components, in human thyroid malignancies. Moreover, it suggested an opening for employing circadian core clock components as potential diagnosis markers for thyroid malignancies [10]. In line with our findings, a recent report suggested that levels of the nuclear receptors REV-ERBα and RORα are significantly altered in PTC and that these alterations correlate with the presence of the BRAFV600E mutation [11].
In spite of substantial efforts [12–15], at present there are still no reliable preoperative tests/diagnosis markers available for patients with indeterminate thyroid FNA cytology. Therefore, the search for reliable preoperative markers of malignancy for thyroid nodules with indeterminate cytology stays of utmost clinical importance [16]. In an attempt to further characterize the involvement of the core clock machinery in thyroid malignancies and to search for additional reliable markers for PTC diagnosis, we performed comparative large-scale transcript expression analysis by probe-based NanoString, evaluated as one of the most accurate approaches for gene expression assessment [17]. Obtaining a large number of fresh-frozen postoperative biopsy samples represents a significant challenge for large-scale transcript analysis. We therefore explored an alternative approach by employing archival formalin-fixed paraffin-embedded (FFPE) samples, which are more readily available. Although formalin fixation affects nucleic acid quality, it has been recently suggested that NanoString nCounter technology allows for reliable gene expression measurements, using RNA extracted from oral squamous cell carcinoma and malignant breast tissue FFPE samples [18, 19]. We thus aimed at validating NanoString as a method of choice for FFPE thyroid sample gene expression analysis, and to apply this approach for a rationally designed search for potential biomarker candidates by comparison of transcript expression in PTC versus benign thyroid nodules.
RESULTS
FFPE thyroid nodule samples allow for a reliable estimation of transcript expression levels in comparison to fresh-frozen samples assessed by NanoString nCounter™ analysis
To validate the application of the NanoString nCounter™ approach for thyroid nodule sample analysis, FFPE and fresh-frozen tissue samples were obtained from 8 donors (4 benign nodules and 4 PTC in each group of samples; see Table 1 for donor characteristics). The expression levels of 34 transcripts (See Methods and Supplemental Table 1, code-set 1) were assessed in RNA samples extracted from FFPE and fresh-frozen tissues by NanoString nCounter™ technology and compared between frozen and FFPE sample groups (Supplemental Table 2). The statistical analysis revealed significant differences in 2 out of 34 transcripts in the benign and PTC groups, respectively, with the fold difference fixed at ≥ 2 and a raw P-value < 0.05. No transcripts showed significant difference in their false discovery rate (FDR) 5% (see Supplemental Table 2 for details). Moreover, high Pearson correlation coefficients were observed for each pair of FFPE and frozen sample (average of r2 = 0.84; 0.63 < r2 < 0.99). The results of these proof-of-principle experimental series were in good agreement with previous reports [18, 19] and strongly suggested that RNA extracted from FFPE samples represents a reliable material for NanoString analysis despite its high level of degradation.
Table 1. Donor characteristics and diagnosis.
| Code set | Case | Sex | Age, years | Operation time | Size (cm), malignant nodule | Histologic diagnosis |
|---|---|---|---|---|---|---|
| PTC | ||||||
| 1 | 1* | M | 56 | 13h 40min | 1.8 | IClassical PTC, pT1b |
| 2 | F | 54 | 12h 50min | 3.0 | IIClassical PTC, pT3pN0 | |
| 3 | F | 72 | 8h 30min | 5.0 | IIClassical PTC, pT3pN1a | |
| 4 | F | 63 | 8h 45min | 4.1 | IIClassical PTC, pT3 (m) | |
| 5* | M | 25 | 8h 30min | 3.2 | IIClassical PTC, pT2pN1a | |
| 6* | F | 51 | 12h 50min | 2.5 | IIClassical PTC, pT3 (m) pN1a | |
| 7* | F | 30 | 8h 35min | 1.5 | IIClassical PTC, pT1bpN0 | |
| 2 | 8 | M | 39 | 13h 15min | 0.9 | IFollicular variant PTC, pT1a pN1b |
| 9 | F | 31 | 13h 30min | 3.0 | IIClassical PTC, pT2 | |
| 10 | M | 26 | 14h 00min | 3.2 | IIMixed PTC, pT2 | |
| 11 | F | 64 | 13h 40min | 2.0 | IITall cell PTC, pT3 pN1a | |
| 12 | F | 42 | 10h 30min | 5.0 | IIClassical PTC, pT3pN1b | |
| 13 | F | 21 | 12h 45min | 2.5 | IIClassical PTC, pT2 | |
| 14 | M | 79 | 9h 45min | 2.1 | IIClassical PTC, pT3 (m) | |
| 15 | F | 41 | 10h 55min | 2.3 | IIClassical PTC, pT3 (m) pN1b | |
| 16 | F | 48 | 9h 20min | 2.0 | IIClassical PTC, pT3 pN1b | |
| 17 | F | 71 | 9h 20min | 4.0 | IIClassical PTC, pT3 pN0 | |
| Benign | ||||||
| 1 | 18 | F | 46 | 8h 25min | ||
| 19 | F | 40 | 8h 40min | |||
| 20 | F | 57 | 10h 40min | |||
| 21* | F | 68 | 9h 30min | |||
| 22* | M | 49 | 10h 30min | |||
| 23* | F | 47 | 10h 30min | |||
| 24* | F | 52 | 8h 20min | |||
| 2 | 25 | F | 50 | 11h 10min | ||
| 26 | F | 41 | 9h 00min | |||
| 27 | F | 45 | 9h 30min | |||
| 28 | F | 69 | 9h 50min | |||
| 29 | F | 62 | 10h 00min | |||
| 30 | F | 61 | 12h 00min | |||
| 31 | F | 66 | 11h 45min | |||
| 32 | F | 46 | 10h 05min | |||
| 33 | F | 45 | 8h 20min | |||
| 34 | F | 50 | 11h 35min | |||
According to the histologic type and invasiveness, PTC samples were divided into two groups corresponding to the clinical aggressiveness:
less aggressive PTC
more aggressive PTC
Cases with fresh-frozen and FFPE samples analyzed for methodology validation (see Suppl. Table 2).
NanoString analysis of transcript expression patterns in benign and PTC thyroid nodules obtained from distinct donors
Our recent study had revealed alterations in the expression levels of the core clock transcripts BMAL1 and CRY2 in PTC in comparison to benign nodules and healthy tissue, as measured by qRT-PCR [3]. In order to further explore the connection between core clock gene expression levels and PTC progression and to search for additional reliable PTC preoperative diagnostics markers, we conducted the NanoString analysis of 17 PTC and 17 benign FFPE samples, obtained during thyroid surgery from distinct donors (see Table 1 for donor characteristics and postoperative diagnosis). 51 genes, comprising those related to thyroid function, core clock, cell cycle and apoptosis, were selected for analysis in benign and PTC FFPE samples (Supplemental Table 1). Several transcripts, previously demonstrated to exhibit strong expression level changes in PTC, such as TIMP1, c-MET and c-KIT [6, 7, 20, 21], were included for additional validation of the methodology and for the correlation analysis. Our study revealed that the levels of TIMP1 and c-MET were 6-fold upregulated, while those of c-KIT were 11-fold downregulated in PTC (Table 3, upper part), which is in good agreement with the works of others [6, 7, 20, 21], and our own previous study [3]. Furthermore, the levels of VEGFR1, PPARγ, TG (thyroglobulin), DIO2, and ALDH1 transcripts were moderately (2 - 5-fold) downregulated. The levels of SLC26A4 (pendrin) and TPO were strongly downregulated on average, however the expression levels of both transcripts varied vastly among the samples, and therefore the changes were not statistically significant (P-value > 0.05). Our results are in good agreement with previous studies that have assessed these genes individually in PTC samples using immunohistochemistry and quantitative RT-PCR [22–26]. In addition to confirming previous findings, a large-scale screening by NanoString analysis allows for the combined high-precision evaluation of changes in the expression levels of a large number of transcripts in the same PTC sample.
Table 3. Altered transcript expression in PTC samples as compared to benign counterparts from different donors.
| Gene | P-value (PTC vs benign) | P-value with FDR (PTC vs benign) | Fold change | Total number of samples | Number of samples with expression value > 50 (linear scale) |
|---|---|---|---|---|---|
| Without consideration of clinical aggressiveness | |||||
| BCL2 | 5.86 × 10−11 | 3.51 × 10−10 | −3.17 | 34 | 34 |
| BMAL1 | 5.56 × 10−10 | 2.50 × 10−9 | 4.44 | 34 | 34 |
| CHEK1 | 8.88 × 10−4 | 1.45 × 10−3 | 2.97 | 34 | 32 |
| c-KIT | 4.60 × 10−7 | 1.38 × 10−6 | –10.80 | 34 | 32 |
| c-MET | 5.15 × 10−13 | 9.27 × 10−12 | 5.96 | 34 | 34 |
| PPARγ | 6.33 × 10−6 | 1.42 × 10−5 | −3.96 | 34 | 31 |
| TG | 2.71 × 10−6 | 6.96 × 10−6 | −3.85 | 34 | 34 |
| TIMP1 | 1.02 × 10−11 | 9.18 × 10−11 | 6.27 | 34 | 34 |
| VEGFR1 | 2.42 × 10−4 | 4.36 × 10−4 | –2.18 | 34 | 34 |
| ALDH1 | 3.97 × 10−8 | 6.75 × 10−7 | –5.69 | 20 | 20 |
| DIO2 | 2.51 × 10−3 | 5.34 × 10−3 | –3.61 | 20 | 20 |
| With consideration of clinical aggressiveness: more aggressive PTC (group II in Table 1) | |||||
| AKT2 | 6.75 × 10−5 | 1.35 × 10−4 | −2.01 | 32 | 32 |
| BCL2 | 1.28 × 10−10 | 7.70 × 10−10 | −3.24 | 32 | 32 |
| BMAL1 | 1.74 × 10−9 | 7.82 × 10−9 | 4.05 | 32 | 32 |
| CHEK1 | 1.30 × 10−3 | 1.79 × 10−3 | 3.01 | 32 | 30 |
| CRY2 | 1.40 × 10−8 | 5.02 × 10−8 | −2.02 | 32 | 32 |
| c-KIT | 3.22 × 10−7 | 9.65 × 10−7 | −12.21 | 32 | 30 |
| c-MET | 2.48 × 10−12 | 4.47 × 10−11 | 5.99 | 32 | 32 |
| PPARγ | 1.61 × 10−6 | 3.61 × 10−6 | −4.46 | 32 | 29 |
| TG | 1.14 × 10−6 | 2.92 × 10−6 | −4.23 | 32 | 32 |
| TIMP1 | 5.38 × 10−11 | 4.85 × 10−10 | 5.91 | 32 | 32 |
| VEGFR1 | 7.51 × 10−5 | 1.35 × 10−4 | −2.37 | 32 | 32 |
| ALDH1 | 1.49 × 10−7 | 2.53 × 10−6 | −5.64 | 19 | 19 |
| DIO2 | 3.81 × 10−3 | 8.10 × 10−3 | −3.65 | 19 | 19 |
The fold changes between the transcript expression levels in PTC and benign (B) samples were calculated as follows: Fc (fold change) PTC/B = 2(average log(PTC)-average log(B)) where PTC and B are normalized expression values obtained by NanoString analysis for the respective samples.
Importantly, levels of the core clock transcript BMAL1 exhibited a 4-fold upregulation in the studied PTC samples, which is in agreement with our recent findings [3]. Moreover, our analysis revealed for the first time that the cell-cycle related transcript CHEK1 and the apoptosis-related transcript BCL2 were 3 fold up- and downregulated in PTC, respectively (Table 3, upper part).
To address possible correlations between disease progression and gene expression changes, we next analyzed transcript changes solely in samples with a more aggressive form of the disease according to histologic type and invasiveness, which represented 88.2% (15 out of 17) of PTC cases in this group (see Table 3, lower part). In these PTC samples, CRY2 and AKT2 transcripts exhibited a 2-fold downregulation, in addition to the transcripts altered for the entire group of samples (compare upper and lower parts of Table 3). Of note, a 2-fold downregulation of CRY2 expression was previously observed by us employing qRT-PCR in a smaller subset of samples [3]. Changes for AKT2 in PTC samples have not been previously reported.
NanoString analysis of transcript expression pattern in benign thyroid nodules and PTC pairs obtained from the same donors (multinodular goiter surgeries)
We next analyzed a second group of samples comprising 24 pairs of benign and PTC samples obtained from multinodular goiter surgeries. As for the first group of patients, surgeries were performed during the same time window (see Table 2 for patient characteristics and diagnosis). In addition to increasing the number of donors, comparing PTC and benign nodule samples, derived from the same donors, aimed to ensure that the observed differences in transcript expression are resulting from tumor progression solely and not from genetic background differences among the subjects. NanoString analysis of 51 transcript levels was performed in these 48 FFPE samples (24 pairs; Table 2). As demonstrated in Table 4, upper part, only CHEK1, c-KIT, c-MET and TIMP1 exhibited 2 - 3-fold differences in PTC samples compared to their benign counterparts. These differences are clearly less pronounced if compared to the respective differences that we obtained in the first part of the study (compare fold changes in the upper parts of Tables 3 and 4). Of note, in contrast to the first group of analyzed PTC samples, with the majority being in the category of clinically more aggressive subtypes (88.2%), only about half (54.2 %) of the multinodular goiter PTCs were diagnosed postoperatively as aggressive (compare Table 2 to Table 1). Separate analysis of a subset of the multinodular goiter samples with more aggressive PTC subtypes revealed 2 - 4-fold upregulation in the levels of CHEK1, c-MET and TIMP1, and a 2-4-fold downregulation of BCL2, c-KIT, TG and PPARγ (Table 4, lower part). BMAL1 was 1.9-fold upregulated in this subgroup of PTC samples (Table 4, lower part). These results are generally in agreement with those obtained in the first part of the study, although observed fold changes are less pronounced. This difference might be attributed to the more homogeneous genetic background between benign and PTC nodules in this group, ensuring that the differences are solely associated with PTC malignancy progression and also to the overall less aggressive form of PTC in this group.
Table 2. Donor characteristics and diagnosis: multinodular goiter cases.
| Code set | Case | Sex | Age, years | Operation time | Size (cm), malignant tumor | Histologic diagnosis |
|---|---|---|---|---|---|---|
| PTC/Benign | ||||||
| 1 | 35/36 | F | 38 | 11h 35min | 3.8 | IFollicular variant PTC, pT2 (m) |
| 37/38 | F | 79 | 9h 35 min | 3.5 | IFollicular variant PTC, pT2 (m) | |
| 39/40 | F | 42 | 9h 45min | 2.5 | IFollicular variant PTC, pT2 | |
| 41/42 | F | 24 | 10h 40min | 3.9 | IFollicular variant PTC, pT2 | |
| 43/44 | F | 59 | 12h 55min | 1.1 | IClassical PTC, pT1b (m) | |
| 45/46 | M | 29 | 8h 00min | 5.5 | IIClassical PTC, pT3 pN0 | |
| 47/48 | M | 65 | 9h 05min | 5.0 | IIClassical PTC, pT3 | |
| 49/50 | F | 31 | 12h 40min | 1.7 | IIClassical PTC, pT3 | |
| 51/52 | F | 51 | 12h 50min | 2.5 | IIClassical PTC, pT3 (m) pN1a | |
| 53/54 | F | 74 | 10h 10min | 0.9 | IIClassical PTC, pT3 | |
| 55/56 | M | 47 | 8h 00min | 3.5 | IIClassical PTC, pT2 (m) pN0 | |
| 57/58 | F | 49 | 9h 30min | 2.2 | IIClassical PTC, pT3 (m) pN1a | |
| 2 | 59/60 | F | 63 | 15h 00min | 2.2 | IFollicular variant PTC, pT2 |
| 61/62 | F | 58 | 9h 10min | 1.5 | IFollicular variant PTC, pT1b | |
| 63/64 | F | 49 | 8h 10min | 1.3 | IFollicular variant PTC, pT1b | |
| 65/66 | F | 55 | 14h 00min | 1.1 | IFollicular variant PTC, pT1b | |
| 67/68 | M | 63 | 17h 00min | 3.1 | IFollicular variant PTC, pT2 | |
| 69/70 | F | 66 | 8h 20min | 3.5 | IClassical PTC, pT2 | |
| 71/72 | F | 69 | 13h 10min | 2.8 | IIOncocytic PTC, pT2 | |
| 73/74 | F | 36 | 14h 00min | 1.5 | IIClassical PTC, pT1b | |
| 75/76 | F | 35 | 8h 20min | 1.5 | IIFollicular variant PTC, pT3(m) pN1a | |
| 77/78 | F | 29 | 8h 25min | 1.3 | IIClassical PTC, pT1b | |
| 79/80 | F | 76 | 12h 50min | 1.2 | IIClassical PTC, pT1b | |
| 81/82 | F | 41 | 13h 45min | 1.1 | IISolid PTC, pT1b | |
less aggressive PTC
more aggressive PTC
Table 4. Altered transcript expression in PTC samples as compared to benign counterparts from the same donors (multinodular goiter cases).
| Gene | P-value (PTC vs benign) | P-value with FDR (PTC vs benign) | Fold change | Total number of samples | Number of samples with expression value > 50 (linear scale) |
|---|---|---|---|---|---|
| Without consideration of clinical aggressiveness | |||||
| CHEK1 | 2.63 × 10−4 | 1.15 × 10−3 | 2.05 | 48 | 47 |
| c-KIT | 2.38 × 10−6 | 2.39 × 10−5 | −3.23 | 48 | 48 |
| c-MET | 2.66 × 10−6 | 2.39 × 10−5 | 2.73 | 48 | 48 |
| TIMP1 | 2.00 × 10−3 | 6.00 × 10−3 | 2.01 | 48 | 48 |
| With consideration of clinical aggressiveness: more aggressive PTC (group II in Table 2) | |||||
| BCL2 | 3.08 × 10−6 | 1.38 × 10−5 | −2.11 | 26 | 26 |
| BMAL1 | 7.30 × 10−4 | 1.64 × 10−3 | 1.93 | 26 | 26 |
| CHEK1 | 1.32 × 10−3 | 2.08 × 10−3 | 2.16 | 26 | 26 |
| c-KIT | 1.59 × 10−6 | 9.63 × 10−6 | −4.25 | 26 | 26 |
| c-MET | 5.64 × 10−11 | 1.02 × 10−9 | 4.55 | 26 | 26 |
| PPARγ | 1.59 × 10−3 | 2.21 × 10−3 | −2.81 | 26 | 25 |
| TG | 1.60 × 10−6 | 9.63 × 10−6 | −3.03 | 26 | 26 |
| TIMP1 | 1.22 × 10−5 | 4.38 × 10−5 | 3.38 | 26 | 26 |
Lastly, we performed a combined analysis of all 41 PTC samples enrolled in this study (17 in the first part and 24 in the second part), and compared those to the 17 benign samples from the first part of the study (Table 5), resulting in a total of 58 samples. The 24 benign nodules from the second group of samples were not included in this analysis, to satisfy independent samples criteria for statistical analysis and to ensure homogenous comparison among the 58 samples derived from different donors. Most of the transcript changes, identified in the first part of the study, were also confirmed by this combined analysis of 41 PTC samples (compare Table 3 and 5). Interestingly, VDR appeared 3-fold upregulated in the combined PTC group, in good agreement with recent work [27]. Of note, even though the fold-change of the core clock transcripts CRY2, PER2 and PER3 was below the cut-off value (2-fold change) in this group of PTC samples, a tendency towards a statistically significant 1.5 – 2-fold downregulation was observed, consistent with the qRT-PCR results obtained by us previously [3]. Taking into account the clinical characteristics of the PTC cases, an analysis performed on the more aggressive cases only (Table 5, lower part) confirmed significant alterations in the levels of all transcripts appearing in Table 3, except for AKT2 and CRY2, which were now downregulated slightly below 2-fold and thus qualified as non-significant. Importantly, the 3-fold upregulation of BMAL1 and CHEK1, and the 3-fold downregulation of BCL2 in the overall PTC group provided further validation of these genes as potential new biomarkers for PTC.
Table 5. Altered transcript expression in PTC versus benign samples (combined PTC samples).
| Gene | P-value (PTC vs benign) | P-value with FDR (PTC vs benign) | Fold change | Total number of samples | Number of samples with expression value > 50 |
|---|---|---|---|---|---|
| Without consideration of clinical aggressiveness | |||||
| BCL2 | 1.86 × 10−6 | 1.42 × 10−5 | −2.21 | 58 | 58 |
| BMAL1 | 3.15 × 10−6 | 1.42 × 10−5 | 2.87 | 58 | 58 |
| CHEK1 | 5.48 × 10−6 | 1.97 × 10−5 | 2.97 | 58 | 56 |
| c-KIT | 2.43 × 10−6 | 1.42 × 10−5 | −5.64 | 58 | 56 |
| c-MET | 5.41 × 10−8 | 9.74 × 10−7 | 3.48 | 58 | 58 |
| PPARγ | 1.64 × 10−3 | 2.95 × 10−3 | −2.55 | 58 | 54 |
| TG | 6.38 × 10−4 | 1.28 × 10−3 | −2.39 | 58 | 58 |
| TIMP1 | 1.34 × 10−5 | 3.45 × 10−5 | 3.39 | 58 | 58 |
| ALDH1 | 2.66 × 10−3 | 1.97 × 10−2 | −2.55 | 32 | 32 |
| VDR | 2.38 × 10−3 | 2.11 × 10−2 | 3.15 | 26 | 25 |
| With consideration of clinical aggressiveness: more aggressive PTC (group II in Tables 1,2) | |||||
| BCL2 | 2.33 × 10−9 | 2.10 × 10−8 | −2.70 | 45 | 45 |
| BMAL1 | 1.15 × 10−7 | 3.46 × 10−7 | 3.49 | 45 | 45 |
| CHEK1 | 9.81 × 10−6 | 2.21 × 10−5 | 3.10 | 45 | 43 |
| c-KIT | 7.07 × 10−8 | 2.55 × 10−7 | −7.82 | 45 | 43 |
| c-MET | 1.30 × 10−12 | 2.35 × 10−11 | 4.81 | 45 | 45 |
| PPARγ | 2.30 × 10−5 | 4.60 × 10−5 | −3.56 | 45 | 41 |
| TG | 2.03 × 10−7 | 5.22 × 10−7 | −3.52 | 45 | 45 |
| TIMP1 | 2.05 × 10−8 | 9.24 × 10−8 | 4.79 | 45 | 45 |
| ALDH1 | 2.63 × 10−4 | 2.30 × 10−3 | −3.26 | 25 | 25 |
| DIO2 | 2.24 × 10−2 | 4.23 × 10−2 | −2.41 | 25 | 25 |
| VDR | 2.85 × 10−3 | 2.28 × 10−2 | 3.36 | 20 | 19 |
Alterations of BMAL1, CHEK1, c-MET, c-KIT and TIMP1 expression levels in PTC exhibit pair-wise correlations
Given that the NanoString approach allows for the assessment of numerous transcript levels within the same samples, we next addressed possible correlations among transcript changes upon PTC. Pair-wise correlation analysis was performed between transcripts, which exhibited the most marked alterations in both studied cohorts. Analysis of the combined set of 58 PTC samples enrolled in this study (Tables 1, 2) revealed that c-KIT expression levels inversely correlated with all the other transcript changes, exhibiting strong correlation with c-MET and moderate correlation with BMAL1, CHEK1 and TIMP1 (Figure 1). BMAL1 level changes were strongly positively correlated with those of c-MET and TIMP1 and weakly with CHEK1 (Figure 1). In line with these findings, our recent study suggested a positive correlation between BMAL1 and TIMP1 transcript levels, assessed by qRT-PCR in the same PTC samples [3]. CHEK1 exhibited a weak to moderate positive correlation with c-MET and TIMP1, while c-MET and TIMP1 exhibited a strong positive correlation among each other in the same PTC samples (Figure 1). Our joint NanoString analysis of BMAL1, CHEK1, c-MET, c-KIT and TIMP1 levels in the same PTC samples suggest that expression level changes in these transcripts might be correlated. Therefore, this group of transcripts represents a plausible cluster of markers whose collective changes are associated with PTC development and which might be potentially predictive for PTC diagnosis.
Figure 1. Pair-wise correlations between BMAL1, CHEK1, TIMP1, c-KIT and c-MET transcript changes in PTC.
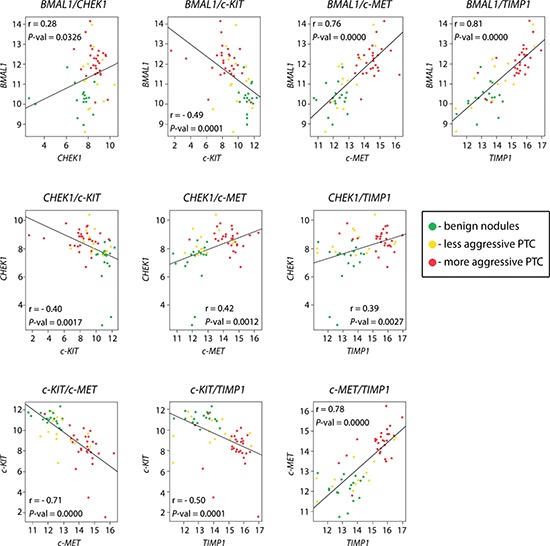
Pearson correlation analysis revealed pair-wise correlations of gene expression in PTC (combined samples from 1st and 2nd cohorts, Tables 1 and 2) and benign (1st cohort, Table 1) samples. The correlation strength was based on Evans classification (see Methods for details), with a coefficient r value < 0.20 reflecting very weak correlation; 0.20 – 0.39 – weak; 0.40 – 0.59 – moderate; 0.60 – 0.79 – strong; and > 0.80 - very strong. The dots at each graph are representing normalized respective gene expression values.
BRAFV600E mutation analysis in PTC samples
The BRAFV600E mutation is associated with PTC in 40–70% of cases [9]. Although the benefit of assessing the BRAFV600E mutational status preoperatively, as a method to determine the extent of thyroidectomy and lymph node dissection, remains uncertain, it might be considered useful in order to support the preoperative diagnosis, especially in cases that are potentially malignant (Bethesda category V). In an attempt to acquire additional valuable characteristics of the PTC nodules, analyzed in this study by NanoString, we conducted a BRAFV600E mutation analysis in the same samples. As listed in Table 6, 81% (13/16) of PTC samples from the first group, diagnosed as classical PTC, exhibited the BRAFV600E mutation, which is in agreement with a previous report [6]. In the multinodular goiter PTC cases only 12 PTC samples were of the classical type, with 67% (8/12) exhibiting the BRAFV600E mutation (Table 6, lower part).
Table 6. BRAF exon 15 mutation analysis.
| Group | Case | BRAF exon 15 | Histologic diagnosis | |
|---|---|---|---|---|
| PROT | DNA | |||
| PTC samples (Table 1) | 1 | p.V600E | c.1799T>A | IClassical PTC, pT1b |
| 2 | WT | WT | IIClassical PTC, pT3pN0 | |
| 3 | p.V600E | c.1799T>A | IIClassical PTC, pT3pN1a | |
| 4 | p.V600E | c.1799T>A | IIClassical PTC, pT3 (m) | |
| 5 | p.V600E | c.1799T>A | IIClassical PTC, pT2pN1a | |
| 6 | p.V600E | c.1799T>A | IIClassical PTC, pT3 (m) pN1 | |
| 7 | p.V600E | c.1799T>A | IIClassical PTC, pT1bpN0 | |
| 8 | p.V600E | c.1799T>A | IFollicular variant PTC, pT1a pN1b | |
| 9 | p.V600E | c.1799T>A | IIClassical PTC, pT2 | |
| 10 | p.V600E | c.1799T>A | IIMixed PTC, pT2 | |
| 11 | p.V600E | c.1799T>A | IITall cell PTC, pT3 pN1a | |
| 12 | WT | WT | IITall cell PTC, pT3 pN1a | |
| 13 | WT | WT | IIClassical PTC, pT2 | |
| 14 | not accessible | IIClassical PTC, pT3 (m) | ||
| 15 | p.V600E | c.1799T>A | IIClassical PTC, pT3 (m) pN1b | |
| 16 | p.V600E | c.1799T>A | IIClassical PTC, pT3 pN1b | |
| 17 | p.V600E | c.1799T>A | IIClassical PTC, pT3 pN0 | |
| Multinodular goiter PTC samples (Table 2) | 35 | WT | WT | IFollicular variant PTC, pT2 (m) |
| 37 | WT | WT | IFollicular variant PTC, pT2 (m) | |
| 39 | WT | WT | IFollicular variant PTC, pT2 | |
| 41 | WT | WT | IFollicular variant PTC, pT2 | |
| 43 | WT | WT | IClassical PTC, pT1b (m) | |
| 45 | p.V600E | c.1799T>A | IIClassical PTC, pT3 pN0 | |
| 47 | WT | WT | IIClassical PTC, pT3 | |
| 49 | p.V600E | c.1799T>A | IIClassical PTC, pT3 | |
| 51 | p.V600E | c.1799T>A | IIClassical PTC, pT3 (m) pN1a | |
| 53 | p.V600E | c.1799T>A | IIClassical PTC, pT3 | |
| 55 | p.V600_K601>Ep.R603Q | c.1799_1801delTGAWT | IIClassical PTC, pT2 (m) pN0 | |
| 57 | WT | WT | IIClassical PTC, pT3 (m) pN1a | |
| 59 | WT | WT | IFollicular variant PTC, pT2 | |
| 61 | WT | WT | IFollicular variant PTC, pT1b | |
| 63 | WT | WT | IFollicular variant PTC, pT1b | |
| 65 | WT | WT | IFollicular variant PTC, pT1b | |
| 67 | WT | WT | IFollicular variant PTC, pT2 | |
| 69 | WT | WT | IClassical PTC, pT2 | |
| 71 | WT | WT | IIOncocytic PTC, pT2 | |
| 73 | p.V600E | c.1799T>A | IIClassical PTC, pT1b | |
| 75 | p.V600E | c.1799T>A | IIFollicular variant PTC, pT3(m) pN1a | |
| 77 | WT | WT | IIClassical PTC, pT1b | |
| 79 | p.V600E | c.1799T>A | IIClassical PTC, pT1b | |
| 81 | WT | WT | IISolid PTC, pT1b | |
less aggressive PTC
more aggressive PTC
Mutation nomenclature is based on the COSMIC data base (Catalogue of Somatic Mutations in Cancer) Sanger Institute: WT– BRAF wild type; pV600E, pV600_K601>E, p. R603Q – mutation at BRAF protein level; c.1799T>A, c.1799_1801delTGA, c1808G>A – mutation at BRAF nucleotide level.
Predictive score for PTC diagnosis based on BMAL1, CHEK1, TIMP1, c-MET, and c-KIT combined expression level changes and on BRAFV600E mutation analysis
In an attempt to correlate the degree of expression level changes of BMAL1, CHEK1, TIMP1, c-MET and c-KIT with the histological diagnosis, we aimed at establishing a gene expression-based predictive score, taking into account collective biomarker changes [28, 29]. Along with such a gene expression-based score, the BRAFV600E mutation status was employed as an additional parameter for establishing a correlation with the postoperative clinical evaluation.
A final predictive score was established for each biological sample, based on the expression levels of five distinctive genes BMAL1, CHEK1, c-KIT, c-MET and TIMP1, which exhibited stable changes in PTC and correlated among each other using the Linear Prediction Score (LPS; for details see Supplemental Methods; [29]). To test the performance of the score, a receiver operating characteristic (ROC) analysis was performed, and ROC curves were established (see Supplemental Figure 1 and Supplemental Methods).
Our results indicate that at the threshold = 0.27, based on empirical curve analysis (Supplemental Figure 1), our predictive score discriminates PTC from benign cases with 90% sensitivity and 100% specificity in the validation set. The thus obtained scores were plotted for each sample of the three sample groups and divided according to their clinical characteristics into: benign, less aggressive PTC and more aggressive PTC (Figure 2). Among the total of 58 samples, 4 samples only (7%) were misclassified according to the expression-based score analysis. In addition, striking correlations were observed between higher scores and PTC aggressiveness (Figure 2). Turning to BRAFV600E mutation mapping in the same samples, 19 out of 27 PTC samples with more aggressive histological type bore the BRAFV600E mutation, while in the less aggressive histological group only 2 out of 13 samples had the mutation (samples marked as triangles at Figure 2; Supplemental Table 3). Thus established gene expression-based score, combined with BRAFV600E mutation analysis, might help to distinguish between benign and PTC samples, especially for more aggressive (classical PTC) cases. Although these results strongly suggest a predictive value for PTC diagnosis, follow-up studies with a higher number of samples will be required to estimate the here proposed coefficient validity.
Figure 2. Scatter plot of gene expression-based predictive scores correlated to PTC biological aggressiveness and BRAF mutation status.
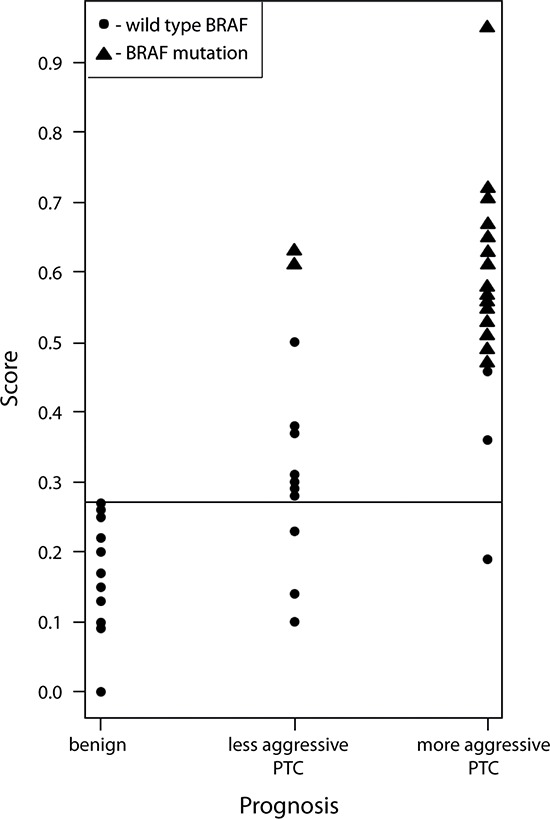
The gene expression-based score for benign and PTC samples was calculated based on joint expression levels of BMAL1, CHEK1, c-KIT, c-MET and TIMP1 transcripts. Thus obtained score values were plotted for three clusters: benign, less aggressive PTC and more aggressive PTC, and allowed a clear distinction between these groups. BRAFV600E mutation was strongly associated with samples characterized by the highest predictive score values and with more aggressive PTC diagnosis.
DISCUSSION
RNA quantification in archival FFPE human thyroid nodule samples using NanoString analysis
NanoString nCounterTM, a color-coded probe-based method, has been reported to attain superior gene expression quantification results in comparison to qRT-PCR [19, 30]. A major limitation of RNA quantification studies performed in fresh post-operative thyroid nodules, including our own recent study [3], is the low number of available samples, in particular for the malignant nodules. To overcome this, we turned to much more readily available archival FFPE material. Our analysis suggests that FFPE samples provide a reliable alternative for NanoString analysis as compared to fresh-frozen tissue biopsy samples (Supplemental Table 2). In summary, our study validates the NanoString method for assessing transcript expression in FFPE human thyroid nodule samples and is in good agreement with recent work performed with FFPE samples from different tissues [18, 19]. Strong advantages of the NanoString approach are high precision due to direct probe hybridization and collective assessment of a large number of transcripts in the same sample. The use of FFPE samples greatly increases the available material and thus statistical relevance of the study. However, the significant costs of NanoString screening might represent a serious drawback if employed for routine diagnosis. Therefore, alternative cost-efficient methods for the screening of a large number of samples, for example by multiplex qPCR analysis of the newly identified subset of biomarkers, will be more appropriate for clinical applications.
Altered transcript expression in human PTC: identifying new and confirming previously reported potential biomarkers by NanoString analysis
The essential cell cycle component CHEK1 and the apoptosis related gene BCL2, previously reported to be associated with a number of malignancies [31–34], were identified for the first time by our screening to exhibit significant changes in their expression levels in PTC (Tables 3–5). CHEK1 was significantly upregulated in all PTC samples, however it did not exhibit a clear correlation with PTC aggressiveness and displayed only a weak to moderate correlation with other analyzed genes (Tables 3–5, Figure 1). Providing further insights into the causality and molecular mechanisms of these alterations in cell cycle, apoptosis, as well as core clock key components with thyroid malignancy development might be of great interest (see below).
In good agreement with a number of previous studies [6–8], NanoString analysis of two groups of samples (total number of 41 benign and 41 PTC samples) revealed that TIMP1, c-KIT and c-MET exhibited highly significant alterations in PTC with pronounced fold changes and low variability among the samples (Tables 3–5). The role of these transcripts in oncogenic transformation in general and their association with human PTC has been well established [35–37]. Of note, we observed pair-wise correlations between these genes, namely TIMP1 and c-MET exhibited strong positive correlation, while c-KIT was strongly and moderately inversely correlated with c-MET and TIMP1, respectively (Figure 1). Consistently with previous studies, these transcript level changes correlated with the presence of the BRAFV600E mutation (Figure 2; [6]).
We consider the multinodular goiter samples the most reliable ones for studying gene expression changes upon thyroid malignancies, as differences between benign and PTC nodules in this group are attributed only to tumor progression. Of note, the newly identified PTC-associated markers CHEK1 and BCL2 were significantly altered in a subgroup with more severe histological prognosis (Table 4, lower part). C-KIT, c-MET and TIMP1 exhibited the strongest alterations in these samples. Since PPARγ and TG exhibited at least a 3-fold downregulation and little variability in multinodular goiter PTC samples, compared to their benign counterparts from the same subjects (Table 4), these genes might represent additional valuable candidates for the here presented combined predictive score. TPO, previously proposed to be predictive of PTC and associated with the BRAFV600E mutation status [9], indeed showed a strong downregulation in the first group of samples albeit its high variability. Strikingly, its mRNA levels did not change significantly in the multinodular goiter sample group. The same was true for SLC26A4, making the predictive value of these two genes for PTC rather problematic.
Correlation between circadian clock alterations and human PTC development
Our study has validated the significant upregulation of the core clock gene BMAL1 in PTC samples, previously shown by us in a smaller subset of samples as strongly upregulated in PTC and FTC [3]. Based on the comparison of 41 pairs of PTC and benign nodules in this study, BMAL1 expression reliably increased about 3-fold (1.93 – 4.4-fold in different groups of samples; Tables 3–5). The fold increase of the BMAL1 transcript was lower than previously observed, which might be due to the less aggressive character and the higher number of PTC cases enrolled in this study. However, BMAL1 upregulation was observed in all of the analyzed samples, with low variability among the samples (P-value < 0.05 for all the groups; Tables 3–5), making this alteration highly significant. Additionally, the core clock component REV-ERBα exhibited a 2 - 3-fold upregulation in PTC in both cohorts of analyzed samples; however, the variability was extremely high for this transcript (P-value > 0.05) and therefore it was not considered significant. This result is highly consistent with our previous analysis [3], where REV-ERBα exhibited a tendency of being 2-fold upregulated, which was defined as non-significant due to high variability among the samples. Of note, REV-ERBα levels were recently reported to be significantly upregulated in PTC [11]. This discrepancy regarding REV-ERBα between our results and the work by others might be attributed to the differences in PTC sampling and/or data analysis. Finally, we observed a non-significant but stable tendency in PTC for the downregulation of CRY2 (average of 1.9-fold; P-value < 0.05; Table 3), PER2 and PER3 (1.4-fold downregulation; P-value < 0.05), again being consistent with our own recent observations [3]. By contrast, DBP, RORα, CRY1, CLOCK and PER1 transcripts, assessed in the same samples, did not exhibit any alterations in PTC (not shown). In summary, by NanoString analysis of 41 pairs of benign and PTC samples we further validated our recent findings on expression level changes of certain core clock components in human PTC, including the significant upregulation of BMAL1.
Of note, BMAL1 transcript levels showed a strong positive correlation with those of TIMP1 and c-MET, and were moderately inversely correlated with c-KIT within the same PTC samples (Figure 1; [3]). These correlations further underscore a potential link between BMAL1 expression and possibly function, as well as thyroid tumor progression, and strongly argue that the observed correlations might have functional significance. Of note, CHEK1, identified for the first time by this study as significantly upregulated in PTC, has been previously found to be regulated by BMAL1 [38]. We are therefore not only suggesting CHEK1 as a potential new biomarker for PTC but are also further confirming the connection between the circadian clock, cell cycle regulation, and malignant transformation.
There is accumulating evidence on the important role of core clock components for cell cycle progression and timing of cell division [39]. The circadian oscillator gates cytokinesis to defined time windows in in vitro cultured fibroblasts and regulates key components of the cell cycle, such as WEE1, CCNB1 and CDC2 in mouse liver cells in vivo (reviewed in [10]). Of note, PER2 has been suggested to play a key role as tumor suppressor by regulating DNA damage response pathways [40]. This finding is however in controversy with more recent study outcomes, demonstrating that deficiency in either Per1 or Per2 genes does not make mice more tumor-prone [52], suggesting that the role of the PER proteins as bona fide tumor suppressors needs to be reevaluated. BMAL1 has been recently demonstrated to play an essential role in ROS homeostasis, oxidative stress response [41, 42], cell cycle control (reviewed in [43]), senescence [44] and mTOR signaling [45]. All these pathways are disrupted in various malignancies and are implicated in cancer development; the observed changes in BMAL1 expression in thyroid tumors may result in disturbances in these pathways. BMAL1 knockout mice exhibited reduced life span and premature aging [41]. Taking together, these findings and data presented here support the important role of BMAL1 in the development of thyroid malignancies. At present, we cannot discriminate whether these changes in core clock components are important for the process of malignant transformation, or rather represent an adaptation to the transformation itself. It will be of interest to test this hypothesis by manipulating the expression levels of BMAL1 in human primary thyrocytes, derived from benign or malignant nodules, with subsequent analysis of their oncogenic properties. Exploring the potential role of circadian clock components in the malignant transformation of human thyroid nodules represents an important step forward in our understanding of the molecular link between clock function, thyroid tissue physiology and pathophysiology of malignant nodules. Moreover, providing new insights into the role of the circadian clock in human thyroid malignancies might have important clinical perspectives, as discussed below.
Towards a reliable correlation coefficient for PTC diagnosis: combined gene expression-based score and BRAF mutation analysis
Importantly, we established in this study a correlation coefficient between changes in the collective expression levels of BMAL1, CHEK1, TIMP1, c-KIT and c-MET, and the BRAFV600E mutation status and combined this with clinical evaluation obtained after surgery. Such generated predictive score exhibited 90% sensitivity and 100% specificity (Supplemental Figure 1; Figure 2; Supplemental Table 3). A previous study [29], which used a similar approach for establishing a linear predictive score, obtained 88% specificity based on 27 gene expression levels in a cohort of 109 subjects for the diagnosis of diffuse large B-cell lymphoma. The specificity value obtained in this study is well comparable to the here established score characteristics. Even though our predictive score is only indicative at this point and demands rigorous confirmation in follow-up studies, it will help if confirmed to increase the reliability of preoperative PTC diagnosis.
The next important challenge will be to apply a similar screening approach for FTC nodules, representing the group with the highest number of suspicious nodules, without clear preoperative diagnosis. We recently provided evidence [3] that human thyrocyte oscillator changes, observed upon PTC, were even more remarkable in PDTC in in vitro cultured primary cells, suggesting that in synchronized primary thyrocytes clock alterations might become more pronounced during malignancy progression. Thus, further analysis of core clock components in FTC and PDTC biopsy samples by NanoString analysis, as was done in this study for PTC, will shed additional light on the connection between thyroid malignancy progression and changes in core clock components. Moreover, exploring the implication of thus identified markers for PTC, and possibly FTC, for preoperative analysis using FNA material would be an additional important step forward towards improving the preoperative diagnosis of suspicious thyroid nodules. Indeed, given that the preoperative diagnosis of malignant thyroid nodules is still far from providing reliable results in numerous cases of suspicious or indeterminate nodules [46], it is of highest importance to launch a prospective study that validates the application of changes in core clock, cell cycle and thyroid function gene expression levels. In combination with the BRAFV600E mutation status and clinical examination, such a predictive score will allow for more reliable preoperative diagnosis of thyroid carcinomas.
METHODS
Study participants and thyroid tissue sampling
FFPE samples from benign and PTC human thyroid nodules were obtained from the archive of the Pathology Department, Geneva University Hospital. Donor characteristics are summarized in Table 1 (benign and PTC nodule samples from different donors) and Table 2 (benign and PTC nodules obtained from the same donors, multinodular goiter surgery cases). Samples from surgeries performed in the time window between 8 AM and 2 PM were chosen, with two exceptions (59/60 and 67/68, Table 2). Malignant tumors were classified by histopathological analysis according to the World Health Organization Classification of Thyroid Tumors [50] and staged according to the AJCC Cancer Staging Manual 7th ed. In cases where fresh-frozen and FFPE samples were compared, one part of the obtained thyroid tissue was deep-frozen and kept for tissue transcript analysis, the other part was processed to establish FFPE samples. Fresh thyroid tissue samples (1 cm3) were obtained from patients undergoing thyroidectomy for thyroid cancer or a suspicious nodule. Written informed consent was obtained from each patient and the study protocol was approved by the local Ethics Committee (CER 11–014).
RNA extraction from fresh-frozen and FFPE samples
Frozen tissue biopsies were homogenized using a Polytron homogenizer (Kinematica AG), and total RNA was extracted using the RNA spin II kit (Macherey-Nagel) according to manufacturer's instructions. For RNA extraction from FFPE samples 3 μm thick tissue sections were deparaffinized in xylol, proteins were digested overnight, and RNA was subsequently extracted using the High Pure miRNA isolation kit (Roche) according to the manufacturer's instructions.
Gene expression quantification using multiplexed, color-coded probe pairs (NanoString nCounterTM)
51 candidate genes were selected for analysis, including genes involved in thyroid function, circadian clock, cell cycle, and apoptosis, based on our own previous study and on literature search. Probes were designed and synthesized by NanoString nCounter™ technologies (Supplemental Table 1). 200 – 400 ng of total RNA, extracted from fresh-frozen or FFPE samples, were hybridized with multiplexed NanoString probes, as described in [17]. Background correction was made by subtracting the mean + 2 SD from the raw counts obtained with negative controls. Values <1 were fixed to 1 to avoid negative values after log transformation. Counts for target genes were then normalized with the geometric mean of three house-keeping genes (HPRT1, RLP13A and ACTB) selected as the most stable ones using the geNorm algorithm [47]. Normalized data were log2 transformed for further analyses.
DNA extraction and BRAF mutation PCR analysis
For analysis of the BRAFV600E mutation 3 μm thick FFPE tissue sections were deparaffinized in xylol, proteins were digested and DNA was subsequently extracted in the QIAcube using the QIAamp DNA FFPE Tissue Kit (QIAGEN). PCR was performed as previously described [48], using the following primers: forward 5′aaactcttcataatgcttgctctg3′ and reverse 5′ggccaaaaatttaatcagtgga3′PCR products were separated on an agarose gel, purified using the peqGold gel extraction kit (Peqlab) and sequenced in a capillary automatic sequencer (Applied Biosystems 96-cappilary 3730xl).
Statistical analysis
To assess differences in gene-expression values between the different groups, ANOVA tests with contrasts (significant P-values < 0.05) were performed using Partek Genomics Suite (http://www.partek.com). P-values were corrected for multiple testing by use of the false-discovery rate (FDR) method of Benjamini and Hochberg [49]. A conservative significance threshold of 5% FDR associated with a fold change value of 2 or more was applied (see Supplementary Methods for more details on the statistical analysis). A merge of the results obtained in two independent NanoString runs was performed as described in Supplemental methods. Pair-wise Pearson correlation analysis was applied to test correlation between gene expression data obtained by NanoString. Correlation strength was interpreted as proposed by Evans [51].
SUPPLEMENTARY METHODS
Acknowledgments
We are grateful to Didier Chollet and Mylene Docquier (iGE3 Genomics Platform) for help in performing the NanoString experiments, and to Ursula Loizides-Mangold for critical reading of the manuscript. This work was funded by the Fondation pour la lutte contre le cancer et pour des recherché medico-biologiques.
Footnotes
DISCLOSURE SUMMARY
The authors declare no conflict of interest.
REFERENCES
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Elisei R, Molinaro E, Agate L, Bottici V, Masserini L, Ceccarelli C, Lippi F, Grasso L, Basolo F, Bevilacqua G, Miccoli P, Di Coscio G, Vitti P, Pacini F, Pinchera A. Are the clinical and pathological features of differentiated thyroid carcinoma really changed over the last 35 years? Study on 4187 patients from a single Italian institution to answer this question. J Clin Endocrinol Metab. 2010;95:1516–1527. doi: 10.1210/jc.2009-1536. [DOI] [PubMed] [Google Scholar]
- 3.Mannic T, Meyer P, Triponez F, Pusztaszeri M, Le Martelot G, Mariani O, Schmitter D, Sage D, Philippe J, Dibner C. Circadian clock characteristics are altered in human thyroid malignant nodules. J Clin Endocrinol Metab. 2013;98:4446–4456. doi: 10.1210/jc.2013-2568. [DOI] [PubMed] [Google Scholar]
- 4.Auger M. Hurthle cells in fine-needle aspirates of the thyroid: A review of their diagnostic criteria and significance. Cancer cytopathology. 2014;122:241–249. doi: 10.1002/cncy.21391. [DOI] [PubMed] [Google Scholar]
- 5.Xing M, Haugen BR, Schlumberger M. Progress in molecular-based management of differentiated thyroid cancer. Lancet. 2013;381:1058–1069. doi: 10.1016/S0140-6736(13)60109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ilie MI, Lassalle S, Long-Mira E, Hofman V, Zangari J, Benaim G, Bozec A, Guevara N, Haudebourg J, Birtwisle-Peyrottes I, Santini J, Brest P, Hofman P. In papillary thyroid carcinoma, TIMP-1 expression correlates with BRAF (V600E) mutation status and together with hypoxia-related proteins predicts aggressive behavior. Virchows Archiv: an international journal of pathology. 2013;463:437–444. doi: 10.1007/s00428-013-1453-x. [DOI] [PubMed] [Google Scholar]
- 7.Tomei S, Mazzanti C, Marchetti I, Rossi L, Zavaglia K, Lessi F, Apollo A, Aretini P, Di Coscio G, Bevilacqua G. c-KIT receptor expression is strictly associated with the biological behaviour of thyroid nodules. Journal of translational medicine. 2012;10:7. doi: 10.1186/1479-5876-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sierra JR, Tsao MS. c-MET as a potential therapeutic target and biomarker in cancer. Therapeutic advances in medical oncology. 2011;3:S21–35. doi: 10.1177/1758834011422557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romei C, Ciampi R, Faviana P, Agate L, Molinaro E, Bottici V, Basolo F, Miccoli P, Pacini F, Pinchera A, Elisei R. BRAFV600E mutation, but not RET/PTC rearrangements, is correlated with a lower expression of both thyroperoxidase and sodium iodide symporter genes in papillary thyroid cancer. Endocrine-related cancer. 2008;15:511–520. doi: 10.1677/ERC-07-0130. [DOI] [PubMed] [Google Scholar]
- 10.Philippe J, Dibner C. Thyroid Circadian Timing: Roles in Physiology and Thyroid Malignancies. Journal of biological rhythms. 2014 doi: 10.1177/0748730414557634. [DOI] [PubMed] [Google Scholar]
- 11.Mond M, Alexiadis M, Eriksson N, Davis MJ, Muscat GE, Fuller PJ, Gilfillan C. Nuclear receptor expression in human differentiated thyroid tumors. Thyroid : official journal of the American Thyroid Association. 2014;24:1000–1011. doi: 10.1089/thy.2013.0509. [DOI] [PubMed] [Google Scholar]
- 12.Makki FM, Taylor SM, Shahnavaz A, Leslie A, Gallant J, Douglas S, Teh E, Trites J, Bullock M, Inglis K, Pinto DM, Hart RD. Serum biomarkers of papillary thyroid cancer. Journal of otolaryngology - head & neck surgery = Le Journal d'oto-rhino-laryngologie et de chirurgie cervico-faciale. 2013;42:16. doi: 10.1186/1916-0216-42-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salajegheh A, Dolan-Evans E, Sullivan E, Irani S, Rahman MA, Vosgha H, Gopalan V, Smith RA, Lam AK. The expression profiles of the galectin gene family in primary and metastatic papillary thyroid carcinoma with particular emphasis on galectin-1 and galectin-3 expression. Exp Mol Pathol. 2014;96:212–218. doi: 10.1016/j.yexmp.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Tuttle RM, Leboeuf R, Martorella AJ. Papillary thyroid cancer: monitoring and therapy. Endocrinology and metabolism clinics of North America. 2007;36:753–778. doi: 10.1016/j.ecl.2007.04.004. vii. [DOI] [PubMed] [Google Scholar]
- 15.Fallahi P, Giannini R, Miccoli P, Antonelli A, Basolo F. Molecular diagnostics of fine needle aspiration for the presurgical screening of thyroid nodules. Current genomics. 2014;15:171–177. doi: 10.2174/1389202915999140404100347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexander EK, Kennedy GC, Baloch ZW, Cibas ES, Chudova D, Diggans J, Friedman L, Kloos RT, LiVolsi VA, Mandel SJ, Raab SS, Rosai J, Steward DL, Walsh PS, Wilde JI, Zeiger MA, et al. Preoperative diagnosis of benign thyroid nodules with indeterminate cytology. N Engl J Med. 2012;367:705–715. doi: 10.1056/NEJMoa1203208. [DOI] [PubMed] [Google Scholar]
- 17.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, James JJ, Maysuria M, Mitton JD, Oliveri P, Osborn JL, Peng T, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nature biotechnology. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 18.Norton N, Sun Z, Asmann YW, Serie DJ, Necela BM, Bhagwate A, Jen J, Eckloff BW, Kalari KR, Thompson KJ, Carr JM, Kachergus JM, Geiger XJ, Perez EA, Thompson EA. Gene expression, single nucleotide variant and fusion transcript discovery in archival material from breast tumors. PLoS One. 2013;8:e81925. doi: 10.1371/journal.pone.0081925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reis PP, Waldron L, Goswami RS, Xu W, Xuan Y, Perez-Ordonez B, Gullane P, Irish J, Jurisica I, Kamel-Reid S. mRNA transcript quantification in archival samples using multiplexed, color-coded probes. BMC Biotechnol. 2011;11:46. doi: 10.1186/1472-6750-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.da Silveira Mitteldorf CA, de Sousa-Canavez JM, Leite KR, Massumoto C, Camara-Lopes LH. FN1, GALE, MET, and QPCT overexpression in papillary thyroid carcinoma: molecular analysis using frozen tissue and routine fine-needle aspiration biopsy samples. Diagnostic cytopathology. 2011;39:556–561. doi: 10.1002/dc.21423. [DOI] [PubMed] [Google Scholar]
- 21.Pusztaszeri MP, Sadow PM, Faquin WC. CD117: a novel ancillary marker for papillary thyroid carcinoma in fine-needle aspiration biopsies. Cancer cytopathology. 2014;122:596–603. doi: 10.1002/cncy.21437. [DOI] [PubMed] [Google Scholar]
- 22.Fenton C, Patel A, Dinauer C, Robie DK, Tuttle RM, Francis GL. The expression of vascular endothelial growth factor and the type 1 vascular endothelial growth factor receptor correlate with the size of papillary thyroid carcinoma in children and young adults. Thyroid : official journal of the American Thyroid Association. 2000;10:349–357. doi: 10.1089/thy.2000.10.349. [DOI] [PubMed] [Google Scholar]
- 23.Karger S, Berger K, Eszlinger M, Tannapfel A, Dralle H, Paschke R, Fuhrer D. Evaluation of peroxisome proliferator-activated receptor-gamma expression in benign and malignant thyroid pathologies. Thyroid : official journal of the American Thyroid Association. 2005;15:997–1003. doi: 10.1089/thy.2005.15.997. [DOI] [PubMed] [Google Scholar]
- 24.Ringel MD, Anderson J, Souza SL, Burch HB, Tambascia M, Shriver CD, Tuttle RM. Expression of the sodium iodide symporter and thyroglobulin genes are reduced in papillary thyroid cancer. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2001;14:289–296. doi: 10.1038/modpathol.3880305. [DOI] [PubMed] [Google Scholar]
- 25.Arnaldi LA, Borra RC, Maciel RM, Cerutti JM. Gene expression profiles reveal that DCN, DIO1, and DIO2 are underexpressed in benign and malignant thyroid tumors. Thyroid : official journal of the American Thyroid Association. 2005;15:210–221. doi: 10.1089/thy.2005.15.210. [DOI] [PubMed] [Google Scholar]
- 26.Belguith-Maalej S, Rebuffat SA, Charfeddine I, Mnif M, Nadir RF, Abid M, Ghorbel A, Peraldi-Roux S, Ayadi H, Hadj-Kacem H. SLC26A4 expression among autoimmune thyroid tissues. Immunobiology. 2011;216:571–578. doi: 10.1016/j.imbio.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 27.Clinckspoor I, Hauben E, Verlinden L, Van den Bruel A, Vanwalleghem L, Vander Poorten V, Delaere P, Mathieu C, Verstuyf A, Decallonne B. Altered expression of key players in vitamin D metabolism and signaling in malignant and benign thyroid tumors. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 2012;60:502–511. doi: 10.1369/0022155412447296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brutsche MH, Joos L, Carlen Brutsche IE, Bissinger R, Tamm M, Custovic A, Woodcock A. Array-based diagnostic gene-expression score for atopy and asthma. The Journal of allergy and clinical immunology. 2002;109:271–273. doi: 10.1067/mai.2002.121530. [DOI] [PubMed] [Google Scholar]
- 29.Wright G, Tan B, Rosenwald A, Hurt EH, Wiestner A, Staudt LM. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci U S A. 2003;100:9991–9996. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulkarni MM. Digital multiplexed gene expression analysis using the NanoString nCounter system. Curr Protoc Mol Biol. 2011 doi: 10.1002/0471142727.mb25b10s94. Chapter 25:Unit25B 10. [DOI] [PubMed] [Google Scholar]
- 31.Eun YG, Hong IK, Kim SK, Park HK, Kwon S, Chung DH, Kwon KH. A Polymorphism (rs1801018, Thr7Thr) of BCL2 is Associated with Papillary Thyroid Cancer in Korean Population. Clinical and experimental otorhinolaryngology. 2011;4:149–154. doi: 10.3342/ceo.2011.4.3.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sochalska M, Tuzlak S, Egle A, Villunger A. Lessons from gain- and loss-of-function models of pro-survival Bcl2 family proteins: implications for targeted therapy. The FEBS journal. 2015 doi: 10.1111/febs.13188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim MK, Min DJ, Wright G, Goldlust I, Annunziata CM. Loss of compensatory pro-survival and anti-apoptotic modulator, IKKepsilon, sensitizes ovarian cancer cells to CHEK1 loss through an increased level of p21. Oncotarget. 2014;5:12788–12802. doi: 10.18632/oncotarget.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Best A, James K, Dalgliesh C, Hong E, Kheirolahi-Kouhestani M, Curk T, Xu Y, Danilenko M, Hussain R, Keavney B, Wipat A, Klinck R, Cowell IG, Cheong Lee K, Austin CA, Venables JP, et al. Human Tra2 proteins jointly control a CHEK1 splicing switch among alternative and constitutive target exons. Nature communications. 2014;5:4760. doi: 10.1038/ncomms5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maroun CR, Rowlands T. The Met receptor tyrosine kinase: a key player in oncogenesis and drug resistance. Pharmacology & therapeutics. 2014;142:316–338. doi: 10.1016/j.pharmthera.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 36.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang J, Wu YL, Chen BJ, Zhang W, Tanaka Y, Sugiyama H. The C-kit receptor-mediated signal transduction and tumor-related diseases. International journal of biological sciences. 2013;9:435–443. doi: 10.7150/ijbs.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kelleher FC, Rao A, Maguire A. Circadian molecular clocks and cancer. Cancer letters. 2014;342:9–18. doi: 10.1016/j.canlet.2013.09.040. [DOI] [PubMed] [Google Scholar]
- 39.Dibner C, Schibler U. Circadian timing of metabolism in animal models and humans. Journal of internal medicine. 2015 doi: 10.1111/joim.12347. [DOI] [PubMed] [Google Scholar]
- 40.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 41.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Musiek ES, Lim MM, Yang G, Bauer AQ, Qi L, Lee Y, Roh JH, Ortiz-Gonzalez X, Dearborn JT, Culver JP, Herzog ED, Hogenesch JB, Wozniak DF, Dikranian K, Giasson BI, Weaver DR, et al. Circadian clock proteins regulate neuronal redox homeostasis and neurodegeneration. J Clin Invest. 2013;123:5389–5400. doi: 10.1172/JCI70317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khapre RV, Samsa WE, Kondratov RV. Circadian regulation of cell cycle: Molecular connections between aging and the circadian clock. Ann Med. 2010;42:404–415. doi: 10.3109/07853890.2010.499134. [DOI] [PubMed] [Google Scholar]
- 44.Khapre RV, Kondratova AA, Susova O, Kondratov RV. Circadian clock protein BMAL1 regulates cellular senescence in vivo. Cell Cycle. 2011;10:4162–4169. doi: 10.4161/cc.10.23.18381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khapre RV, Kondratova AA, Patel S, Dubrovsky Y, Wrobel M, Antoch MP, Kondratov RV. BMAL1-dependent regulation of the mTOR signaling pathway delays aging. Aging (Albany NY) 2014;6:48–57. doi: 10.18632/aging.100633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sigstad E, Paus E, Bjoro T, Berner A, Groholt KK, Jorgensen LH, Sobrinho-Simoes M, Holm R, Warren DJ. The new molecular markers DDIT3, STT3A, ARG2 and FAM129A are not useful in diagnosing thyroid follicular tumors. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2012;25:537–547. doi: 10.1038/modpathol.2011.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kimura ET, Nikiforova MN, Zhu Z, Knauf JA, Nikiforov YE, Fagin JA. High prevalence of BRAF mutations in thyroid cancer: genetic evidence for constitutive activation of the RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma. Cancer research. 2003;63:1454–1457. [PubMed] [Google Scholar]
- 49.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Statistics in medicine. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 50.DeLellis R, Lloyd R, Heitz P, Eng C. The International Agency for Research on Cancer. In: Lloyd R, Heitz P, Eng C, editors. Pathology and genetics of tumors of endocrine organs (International Agency for Research on Cancer-World Health Organization Classification of Tumors) International Agency for Research on Cancer; 2004. [Google Scholar]
- 51.Evans JD. Straightforward statistics for the bahavioral sciences. Brooks/Cole Publishing Company. 1996 [Google Scholar]
- 52.Antoch M, Toshkov I, Kuropatwinski KK, Jackson M. Deficiency in PER proteins has no effect on the rate of spontaneous and radiation-induced carcinogenesis. Cell Cycle. 2013;12:3673–80. doi: 10.4161/cc.26614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


