Abstract
The Hox complex contains 39 genes clustered into four groups involved in cell differentiation and development. We cloned full-length sequence of Hoxc11 gene from water buffalo Bubalus bubalis, assessed its copy number, localized the same onto the chromosome 5, and studied its evolutionary conservation across the species. Northern hybridization of Hoxc11 showed a 2.2 kb band in the tissues analyzed. Real-Time PCR showed highest expression of Hoxc11 gene in lung followed by spleen, spermatozoa, and testis. Six interacting partners of this gene showed higher expression in spleen, lung, testis, and spermatozoa. During the early stages of development, Hoxc11 and its interacting partners both showed lower expression, which then became prominent during the age of 1–3 years, regressed drastically thereafter, and remained so until the animal's life time (∼20 years). The high expression of Hoxc11 and its interacting partners in spermatozoa and testis during the onset of puberty suggests its likely role in the differentiation of gonads and subsequent reproductive activities. Additional work on Hoxc11 especially, in the context of respiratory, immunological, and in/fertility in other species, including humans would be useful for establishing its broader biological significance towards the enrichment of functional and comparative genomics.
Introduction
Molecular events involved in the developmental processes and the resultant phenotypes have attracted a great deal of attention. Several genes are involved in tissue and skeletal development, organogenesis, and cell repair system. Of these, homeobox genes seem to be the predominant ones participating in these events, although the mechanism still remains unclear (Lewis, 2000).
A typical eukaryotic genome contains clustered Homeobox genes, termed Hox in nonhuman vertebrates and HOX in humans belonging to class Antennapedia (ANTP). The Hox genes have been found in all the animals examined thus far, and known to be involved in pattern formation (Daftary and Taylor, 2006). There are 39 Hox genes ordered in four clusters (Hox A, B, C, and D) organized into 13 homologues. These genes have evolved by tandem quadruplication from the ancestral cluster and have diverged in vertebrates and invertebrates during the course of genome evolution (Di-Poï et al., 2010). At the molecular level, Hox genes encode a highly conserved DNA-binding domain, known as homeodomain of approximately 60 amino acids. In mammals, axis positioning, tissue determination, organogenesis, and skeletal ontogeny during development involve coordinated expression, tight regulatory role, and interplay of the homeobox genes with their interacting partners (Daftary and Taylor, 2006; Limura and Pourquié, 2007).
Most of the Hox genes have largely been studied during the embryonic stages of development in the context of pattern formation across the species, including humans (Sordino et al., 1995; Goodman, 2002; Wang et al., 2009; Di Bonito et al., 2013). However, information on the expression of these genes in the adult animals is not available. A perusal of literature shows that 5′ Hox C genes particularly, Hoxc11 has been partially characterized. In mouse, Hoxc11 is shown to express in the posterior region in developing limbs and gut (Ahn and Ho, 2008; Freitas et al., 2012). Hox is reported to act as selector genes; accordingly, its expression within a certain body segment determines one particular pathway of development over the others (Hoegg and Meyer, 2005). Understanding the mechanisms by which Hoxc11 gene regulates the morphological features requires the identification of downstream targets within its genetic and developmental pathways. We studied the expression of different interacting partners of Hoxc11 gene, which includes Homeobox A9 (Hoxa9), POU class 3 Homeobox 3 (POU3F3), Nucleoporin 98 (NUP98), Meis Homeobox 1 (MEIS1), DEAD/H (Asp-Glu-Ala-Asp) box polypeptide 10 (DDX10), and T-box transcription factor T-box 4 (TBX4) reported earlier (Miller et al., 2003; Pineault et al., 2004; Iwasaki et al., 2005; Bai et al., 2006). Most of these genes encode DNA-binding transcription factors involved in regulating gene expression thus controlling morphogenesis, embryogenesis, differentiation, and other developmental processes (Sumiyama et al., 1996; Shen et al., 1999; Simon, 1999; Nakao et al., 2000; Wang et al., 2010; Franks and Hetzer, 2013;Yuan and Braun, 2013), whereas others have been implicated in carcinogenesis (Jia et al., 2013; Thol et al., 2013). Studies have shown that Hoxc11 and its interacting partners have been implicated in acute myeloid leukemia development (Jankovic et al., 2008), hematopoietic stem cell regulation (Palmqvist et al., 2007), nephrogenic mesoderm specification during early kidney and hind limb development (Naiche and Papaioannou, 2003; Gong et al., 2007). Aberrant expression of Hoxc11 has been reported in several cancer cell lines encompassing breast (Raman et al., 2000; Makiyama et al., 2005), renal (Cillo et al., 1992), bladder (Cantile et al., 2003), cervical (Hung et al., 2003), prostate (Huang et al., 2007), ovarian (Naora et al., 2001), and thyroid (Takahashi et al., 2004). These cancer cell lines may prove to be a rich resource to uncover mutational landscape of this gene and modulated expression, if any, compared with that in normal cells. Similar analysis of the interacting partners of Hoxc11 gene in cancerous cases would provide much clearer picture on the overall organizational and expressional changes of these genes in the context of normal and affected genomes narrowing the search for possible cancer-specific biomarker(s).
Water buffalo (Bubalus bubalis) contributes immensely to the agricultural economy in the Indian subcontinent and South Asian countries through milk, meat, hide, and fuel. In addition, the animal is used for draught purposes. However, less information is available on the genomics of this species. So far, neither the expression of Hoxc11 gene in the adult animals has been explored nor its involvement in reproduction is established. Also, its role beyond the pattern formation has not yet been scrutinized. We undertook characterization of the Hoxc11 gene in water buffalo, isolated its full-length sequence, assessed its copy number status, localized it onto the chromosomes and studied the tissue, spermatozoa, and age-specific expression. We also conducted in silico analysis to deduce Hoxc11 interacting partners and ascertained their expression across the tissues, spermatozoa, and in the blood samples of different age groups of buffaloes. Detailed understanding on the genomics of Hoxc11 gene in buffalo is envisaged to be useful in augmenting our knowledge on its role in context of genome analysis in general and animal biotechnology in particular.
Materials and Methods
Sample collection
Blood, brain, heart, kidney, liver, lung, spleen, testis, and ovary from water buffalo (seven animals) were collected from the Gazipur slaughter house, New Delhi, India, with the help of an on-site veterinary officer. Buffalo semen samples were procured from an in vitro fertilization (IVF) center (Frozen Semen Production Center, Chak Gajaria), in Lucknow (U.P), India. Goat, cattle, and sheep blood samples were obtained from the owner of the animals with the help of veterinarian only for the purpose of research work (Srivastava et al., 2008; Pathak et al., 2010; Kumar et al., 2011). Blood samples from different age groups (45 days to 20 years) of buffalo were collected from the local dairy and Gazipur slaughter house (Srivastava et al., 2007). DNA samples of pig, human, chimpanzee, rat, rhinoceros, tiger, cat, and fish were available in the lab from the other projects (Srivastava et al., 2008; Pathak et al., 2010). Blood samples from the endangered species were procured earlier with due permissions from the competent authorities of the State and Union Government of India. All these samples were procured strictly in accordance with the guidelines of the Institute's Ethics and Biosafety committees and due approvals were taken from these committees.
Isolation of total RNA and cDNA synthesis
Tissue samples from both the sexes of buffalo were collected as mentioned above. Total RNA was isolated from the blood and tissues of buffalo using TRIzol (Molecular Research Center, Inc.) following the standard protocol (Srivastava et al., 2008). RNA from the semen samples was isolated following the standard protocol (Srivastava et al., 2009). The presence of DNA was ruled out by PCR using β-actin primers [GenBank: DQ661647]. Following this, 3–5 μg of mRNA was reverse transcribed into cDNA using the commercially available high-capacity cDNA RT kit (Applied Biosystems). The success of cDNA synthesis was confirmed by PCR amplification using a set of bubaline-derived β-actin (forward: 5′CAGATCATGTTCGAGACCTTCAA3′ and reverse: 5′GATGATCTTGATCTTCATTGTGCTG3′) primers. Genomic DNA from the blood was extracted according to the standard phenol–chloroform procedure (John and Ali, 1997).
Cloning and isolation of Hoxc11 gene
Using cDNA from buffalo testis and three pairs of primers based on Bos taurus Hoxc11 gene [GenBank: AC_000162.1], full-length sequence from B. bubalis was isolated. Details of the primer sequences, Tm and corresponding size of the amplicons are given in Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/dna). The PCR-generated amplicons were processed independently for cloning into pGEMT-Easy cloning vector (Promega). The screening for positive clones was done using colony PCR and further confirmed by restriction digestion with EcoR1 (Fermentas). The recombinant clones were then sequenced on the Applied Biosystems 3130xl genetic analyzer using the BigDye® Terminator v3.1 cycle sequencing kits (Applied Biosystems) following standard protocols (Premi et al., 2007). We sequenced a total of ten recombinant clones (pLH1-pLH10) to ascertain interclonal variation. The sequences were found to be identical across the clones. The full-length sequence of B. bubalis Hoxc11 gene was submitted to the NCBI database.
Multiple sequence alignment of Hoxc11 gene across the species and construction of phylogenetic tree
To assess the homology of Hoxc11 gene across the species, database search was performed using BLAST (www.ncbi.nlm.nih.gov/blast/blast.cgi). On further analysis, sequences from 16 other species were retrieved from the NCBI database (www.ncbi.nlm.nih.gov/gorf/gorf.html) and subjected to sequence alignment employing ClustalW (www.ebi.ac.uk/clustalw) and ClustalX 2.0.10 (Larkin et al., 2007). The species showing homology of >89% were taken into consideration for the construction of phylogenetic tree employing the MEGA 5.2 software (Tamura et al., 2011).
Cross hybridization of buffalo Hoxc11 gene with DNA from other species
Approximately, 1–2 μg of heat-denatured genomic DNA from 12 different species namely goat Capra hircus, cattle Bos indicus, sheep Ovis aries, pig Sus scrofa, human Homo sapiens, chimpanzee Pan troglodytes, rat Rattus rattus, rhinoceros Rhinoceros unicornis, tiger Tigris tigri, cat Felis catus, and fish Labeo rohita, including water buffalo B. bubalis was slot blotted onto the nylon membrane (Amersham) and UV crosslinked. Hoxc11 recombinant plasmid (pLH1) was labeled with [32P] α-dCTP using the RediPrime™ II kit (Amersham Pharmacia Biotech) and used as probe for hybridization with the genomic DNA of these species following standard procedures (Rawal et al., 2012). 2×SSC and β-actin were used as negative and internal controls, respectively.
Copy number estimation of Hoxc11 gene
The copy number of Hoxc11 per haploid genome was calculated using absolute quantitation using the SYBR green assay and Sequence Detection System-7500 (Applied Biosystems). Real-time qPCR assays were performed in a 15 μL reaction volume containing 7.5 μL 2×SYBR Green® PCR Master Mix (Applied Biosystems), genomic DNA (0.5, 1.0 and 2.0 ng), forward and reverse primers at final concentration of 1 μM. Primers and assay conditions were similar to those used for relative expression studies (Supplementary Table S1). Copy number estimation for Hoxc11 was done using buffalo blood genomic DNA as template and 10-fold serial dilutions of the recombinant plasmid (pLH1) specific to the gene in the range of 30×102–30×107 copies (assuming haploid genome of bovine animals=3.3 pg per cell). Reaction specificity was confirmed with melting curves analysis. The copies of Hoxc11 in buffalo genome were then extrapolated as per established protocols (Kumar et al., 2011).
Chromosomal localization of Hoxc11 gene by fluorescence in situ hybridization
Approximately, 400 μL of buffalo blood was cultured for chromosome preparation following standard protocols (Rawal et al., 2012). Fluorescence in situ hybridization (FISH) was carried out using commercially available Pan troglodytes Hoxc11 (CH251-635D2) [GenBank: AC184055] bacterial artificial chromosome (BAC) clone as the probe. The BAC clone was procured from BACPAC Resources Centre Oakland. DNA from cosmid was isolated and sequences were confirmed with gene-specific primers used for endpoint PCR. The BAC clone was labeled with Texas Red tagged dCTP (Invitrogen) using the nick translation kit from Abott Molecular, Inc. Probe so prepared was then used for FISH on the metaphase chromosomes following standard procedures (Rawal et al., 2012). Slides were counterstained with DAPI, screened under the Olympus Fluorescence Microscope (BX51) and images were captured with the Olympus U-CMAD-2 CCD camera. Chromosomal mapping of Hoxc11 was ascertained following the International System for Chromosome Nomenclature (ISCND 2000) established for Bovids (Cribiu et al., 2001).
Northern blot hybridization of buffalo Hoxc11
For northern blot analysis, 10 μg of total RNAs from different somatic tissues and gonads of buffalo were resolved on 1.2%/2.2 M formaldehyde gel by electrophoresis. The RNAs were transferred onto Hybond N+ membrane (Amersham Biosciences). The blots were prehybridized in a solution for 4 h and later hybridized overnight at 42°C in 1x hybridization solution [5×SSC, 50% Formamide, 5×Denhardt's solution, 1% SDS and 100 μg/mL heat-denatured sheared nonhomologous DNA (single-stranded salmon sperm DNA)] (Srivastava et al., 2007). A recombinant plasmid (pLH1) containing Hoxc11 gene was used as the probe following the labeling protocols mentioned above. Bubaline-derived β-actin gene probe was used as a positive control. After hybridization, the blots were washed at low stringency in 2×SSC in 0.1% SDS at 42°C for 10–15 min and at high stringency with 0.1% SSC in 0.1% SDS at 65°C once for 5 min. The blots were then exposed to Kodak XAR-5 film at −80°C for 4–16 h.
Expression of Hoxc11 and its interacting partners by RT-PCR
In silico analysis employing Search Tool for the Retrieval of Interacting Genes/Proteins (STRING 9.05) (http://string-db.org/) uncovered six interacting partners of Hoxc11 gene namely Hoxa9, POU3F3, NUP98, MEIS1, DDX10, and TBX4 (Supplementary Fig. S1). The mRNA sequences for all the six interacting partners were retrieved from the NCBI database ([www.ncbi.nlm.nih.gov/gorf/gorf.html] Table 1). Following this, RT-PCR was conducted for studying the expression of Hoxc11 and its interacting partners. Approximately, 50 ng cDNA from different somatic tissues, gonads, and spermatozoa of buffalo and gene-specific internal primers designed by the Primer Express Software V3.0 (ABI) (Supplementary Table S1) were used. These RT-PCR reactions were conducted to ensure the success of the subsequent Real-Time PCR. Thus, the resultant amplicons obtained from these reactions were in the range 58–83 bp.
Table 1.
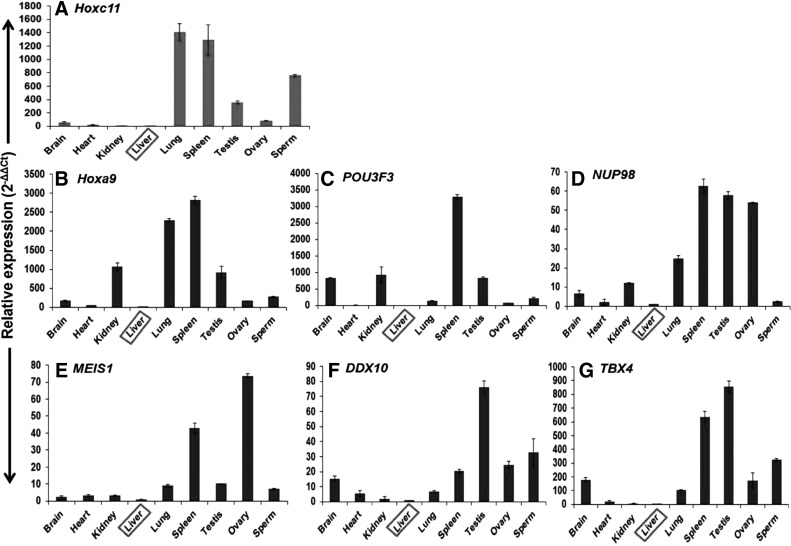
Relative Expression of the Hoxc11 Gene and Its Interacting Partners in Different Somatic Tissues, Gonads, and Spermatozoa of Bubalus bubalis
| Relative expression (in folds 2−ΔΔCt) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| S. No. | Gene | Accession number | Brain | Heart | Kidney | Liver | Lung | Spleen | Testis | Ovary | Spermatozoa |
| 1 | Hoxc11 | KJ959631 | 58.98601 | 16.36073 | 3.080226 | Cb | 1409.592 | 1288.554 | 353.0729 | 80.08948 | 759.6635 |
| 2 | Hoxa9 | NM_001105617.2 | 182.0342 | 53.76215 | 1073.461 | Cb | 2291.497 | 2819.249 | 920.8925 | 171.8848 | 274.7837 |
| 3 | POU3F3 | XM_001787846.3 | 844.695 | 8.909229 | 944.0955 | Cb | 153.316 | 3299.845 | 851.0235 | 84.14354 | 226.1481 |
| 4 | NUP98 | XM_002693428 | 6.5639 | 2.1382 | 12.0252 | Cb | 24.7879 | 62.6586 | 57.8229 | 53.9436 | 2.4123 |
| 5 | MEIS1 | NM_001083507.1 | 2.305303 | 3.36703 | 3.140457 | Cb | 9.062812 | 42.78931 | 10.17238 | 73.67969 | 7.0320 |
| 6 | DDX10 | NM_001076881.2 | 15.2676 | 5.4252 | 2.0142 | Cb | 6.7594 | 20.5858 | 76.2619 | 24.6456 | 32.8440 |
| 7 | TBX4 | NM_001192193.1 | 180.0867 | 22.5936 | 4.5966 | Cb | 106.1851 | 636.2647 | 853.7116 | 171.7094 | 324.6576 |
Cb denotes calibrator tissue (expression value 1). The bold numerical values indicate the maximum expression of a gene in the given sample.
Quantitative expression by real-time PCR of Hoxc11 and its interacting partners
To ascertain the quantitative expression of Hoxc11 and its interacting partners, we conducted Real-Time PCR using cDNA from across the somatic tissues, gonads, and spermatozoa as template. Thereon, assays were performed using Power SYBR® green (Part no. 4367659; ABI) on Sequence Detection System 7500 (Applied Biosystems). Similarly, age-related expression of Hoxc11 and its six interacting partners was carried out using cDNA isolated from blood lymphocytes of different age groups of buffaloes and the same set of Real-Time primers (Supplementary Table S1). GAPDH [GenBank: XR_083674.1] primers (forward: GCAAGTTCCACGGCACAGT and reverse: GATGGTGATGGCCTTTCCAT) were used to normalize the values of each cDNA sample. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was taken as an internal control for each reaction. Primers giving single peak in the dissociation curve and a standard curve having a slope value of −3.3 to −3.5 with a regression coefficient (R)2 value >0.99 were used. The PCR cyclic conditions comprise an initial denaturing step at 95°C, followed by 40 cycles each of 95°C for 15 s and 60°C for 1 min. For each experimental set, nontemplate reaction was included as a negative control. To adjust for variations in the input sample, the average Ct values for the individual target genes were normalized against the average Ct values for the housekeeping gene (GAPDH) [ΔCt(Target)=[Ct(Target)–Ct(GAPDH)]. To achieve maximum efficiency of the primers, the amplicon size was kept small (57–83 bp) so that the expression levels of the target genes remain 2−ΔΔCt (Pathak et al., 2010). Based on this, expression levels of the desired genes mentioned earlier in different tissues, spermatozoa, and blood samples of different age group of animals were ascertained.
Results
Hoxc11 gene in water buffalo B. bubalis
The amplified PCR products corresponding to Hoxc11 gene are shown in Supplementary Figure S2. Clone I (pHoxc11_c1-1380 bp) covered nucleotides from 1 to 1380 bp, clones II (pHoxc11_c2-1400 bp), and III (pHoxc11_c3-930 bp) covered nucleotides from 1200 to 2599 bp and 2501 to 3430 bp, respectively. The B. bubalis Hoxc11 gene of 3430 bp was deduced from the different overlapping fragments. In silico analysis showed the presence of two exons. The first exon covered 109–892 base pairs and had a 5′UTR of 60 base pairs (109–168 bp). The second exon covering nucleotides from 2167 to 2581 bp consists of 182 bp 3′UTR from 2400 to 2581 bp. The two exons are separated by a 1273 bp intron. The exon–intron boundaries are in agreement with the potential exon–intron donor site (5′ end of the intron) at 893 nucleotide position and an acceptor site (3′end of the intron) at 2166 nucleotide position. The full-length sequence of B. bubalis Hoxc11 mRNA [GenBank: KJ959631] of 1199 bp, had a coding region corresponding to 304 amino acids with 60.39% GC content (Supplementary Fig. S3).
Phylogenetic status of Hoxc11 gene
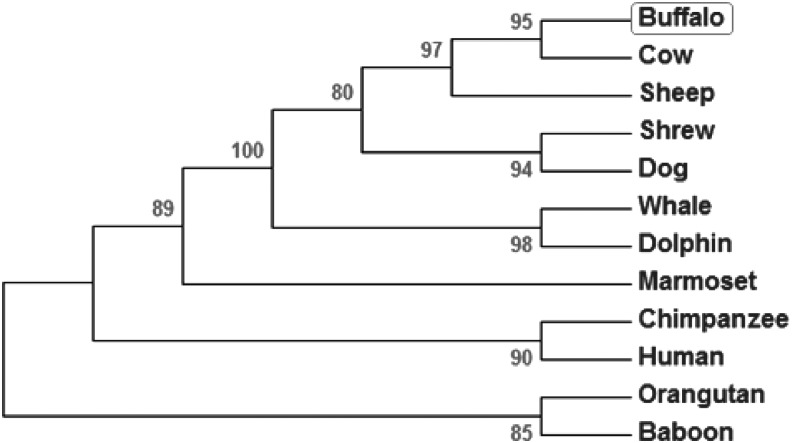
B. bubalis Hoxc11, mRNA, and predicted amino acid sequences aligned with those of 16 other species using ClustalW and ClustalX showed more than 89% homology both at the nucleotide and protein levels suggesting high degree of conservation (Table 2 and Supplementary Fig. S4). Phylogenetic tree constructed by maximum parsimony showed the evolutionary status of Hoxc11 gene in different species placing buffalo closer to cattle (Fig. 1).
Table 2.
Homology Status of Hoxc11 Gene Across the Species with That of Buffalo Both at the Transcriptional and Translational Levels
| Homology with buffalo Hoxc11 (%) | |||||||
|---|---|---|---|---|---|---|---|
| S. No. | Species (Scientific name) | Species (common name) | Accession number | mRNA length (bp) | Amino acid residues (aa) | mRNA | Amino acids |
| 1 | Bubalus bubalis | Buffalo | KJ959631 | 1199 | 304 | 100 | 100 |
| 2 | Ovis aries | Sheep | XM_004007385 | 822 | 273 | 95.26 | 89.38 |
| 3 | Cavia porcellus | Guinea Pig | XM_003476246 | 915 | 304 | 95.74 | 96.38 |
| 4 | Ailuropoda melanoleuca | Giant Panda | XM_002923428 | 915 | 304 | 95.96 | 96.71 |
| 5 | Canis lupus familiaris | Dog | XM_003433496.3 | 969 | 305 | 95 | 95.39 |
| 6 | Sorex araneus | Shrew | XM_004601549 | 1182 | 304 | 95.41 | 97.04 |
| 7 | Bos taurus | Cow | NM_001192873 | 1199 | 304 | 98.58 | 98.03 |
| 8 | Tursiops truncatus | Bottlenose Dolphin | XM_004310785 | 2041 | 305 | 96.83 | 97.04 |
| 9 | Callithrix jacchus | Marmoset | XM_002752542 | 2046 | 304 | 94.86 | 95.39 |
| 10 | Orcinus orca | Killer Whale | XM_004274234 | 2047 | 305 | 97.16 | 97.04 |
| 11 | Papio anubis | Olive Baboon | XM_003906472 | 2047 | 304 | 94.54 | 94.74 |
| 12 | Pan troglodytes | Chimpanzee | XM_509104 | 2050 | 304 | 95.3 | 95.72 |
| 13 | Odobenus rosmarus | Walrus | XM_004406198 | 2053 | 304 | 96.17 | 96.38 |
| 14 | Pongo abelii | Sumatran Orangutan | XM_003778046 | 2053 | 304 | 94.97 | 95.72 |
| 15 | Equus caballus | Horse | XM_003365248 | 2054 | 306 | 96.28 | 97.04 |
| 16 | Felis catus | Cat | XM_003988781 | 2072 | 304 | 96.5 | 97.04 |
| 17 | Homo sapiens | Human | NM_014212 | 2100 | 304 | 95.41 | 96.05 |
The accession number, mRNA length, and amino acid residues, and their maximum homology status both at the nucleotide and amino acid levels are mentioned.
FIG. 1.
Phylogenetic status of Hoxc11 gene across different species. The tree was constructed based on the nucleotide sequences using the MEGA 5.2 software employing maximum parsimony with Subtree Prunning–Regrafting (SPR) algorithm. Bootstrap values are indicated at the branch points. The bootstrap consensus tree was inferred from 1000 replicates to represent the evolutionary history of the taxa analyzed.
Cross hybridization of Hoxc11 with other species
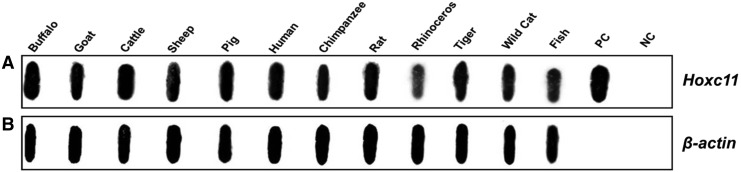
To ascertain the evolutionary conservation, we cross hybridized Hoxc11 clone with the genomic DNA of 12 different species and detected positive signals in all the samples. As expected, uniform signal was detected in these samples upon hybridization with β-actin gene used as control (Fig. 2A, B).
FIG. 2.
Cross hybridization of Hoxc11 gene with genomic DNA from different species. (A) Note the presence of signal in the species analyzed given on top of the panel. PC denotes the positive control (recombinant plasmid) and NC, the negative control (2×SSC). (B) β-actin was used as an internal control.
Copy number and chromosomal localization of Hoxc11 gene in buffalo
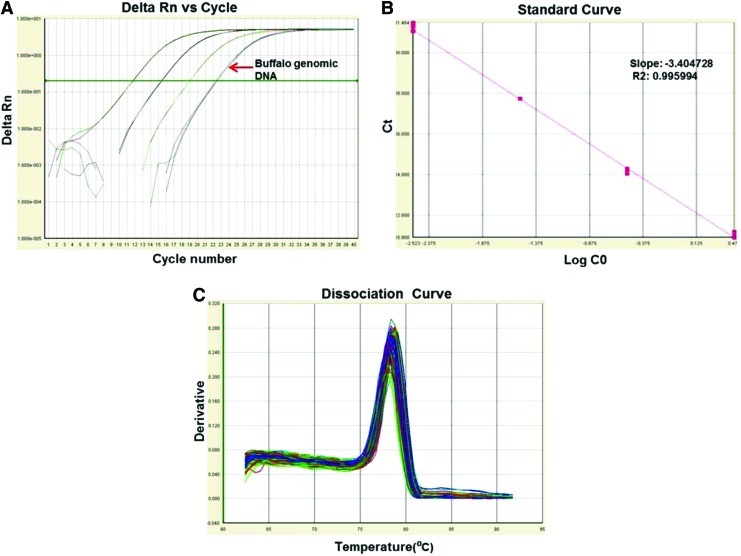
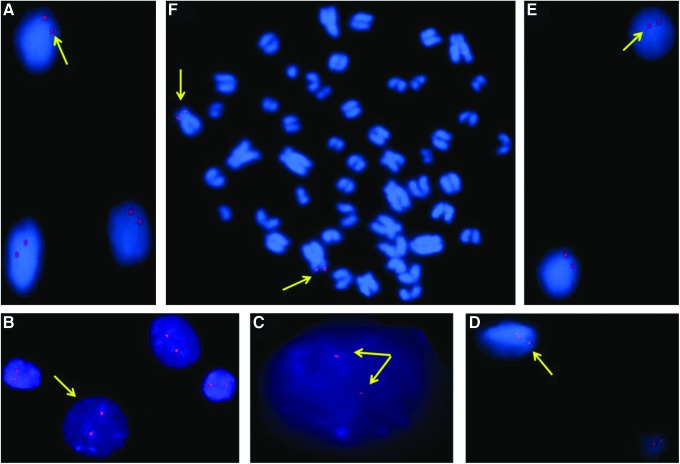
We studied copy number status of Hoxc11 gene using the SYBR green and Real-Time PCR. Representative amplification plot along with its corresponding standard and dissociation curves are given in Figure 3A–C. The B. bubalis Hoxc11 gene was found to have one copy per haploid genome. Chromosomal mapping using FISH (fluorescence in situ hybridization) revealed the presence of Hoxc11 on buffalo metacentric chromosome 5 with corresponding signals on the interphase nuclei (Fig. 4).
FIG. 3.
Copy number status of Hoxc11 gene assessed by Real-Time PCR. Amplification plot was based on 10-fold dilution series of the plasmid containing the Hoxc11 insert. (A) Delta Rn vs. Cycle showing amplification plot of the standard plasmid and genomic DNA of water buffalo. (B) Standard curve with a slope value of −3.4, arrow indicates the genomic DNA. (C) Dissociation curve showing single peak, substantiates primer specificity with the target DNA. Color images available online at www.liebertpub.com/dna
FIG. 4.
Localization of Hoxc11 gene on buffalo metacentric chromosome using Fluorescence in situ hybridization. Note the signals shown by arrows on the interphase nuclei (A–E) and metacentric chromosome 5 (F). All the pictures were captured at the scale of 20 μm. Color images available online at www.liebertpub.com/dna
Expression of buffalo Hoxc11 based on northern hybridization
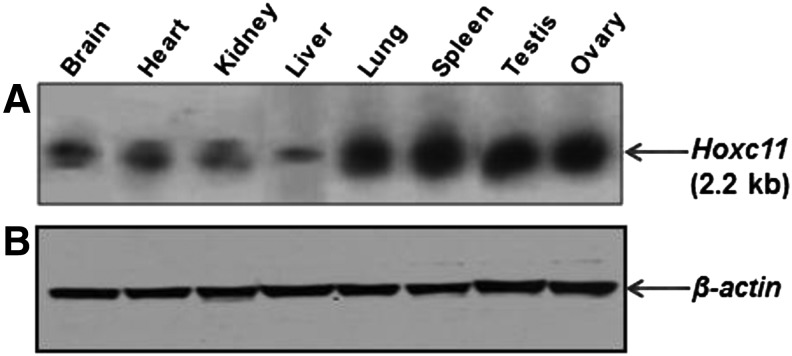
Total RNA from the different tissues of buffalo resolved on the agarose gel used for northern blot hybridization detected a single band (2.2 kb) with varying intensity. Higher expression was observed in the lung, spleen, testis, and ovary. On prolonged exposure of the blot to the X- ray film, faint bands were also observed in the brain, heart, kidney, and liver (Fig. 5). The altered signal intensities uncovered in northern blot corroborated with subsequent RT-PCR-based expression data.
FIG. 5.
Northern blot hybridization of Hoxc11 gene using total RNA from different tissues of buffalo. (A) The tissues used are given on top of the panel. Note varying signals across the tissues. (B) β-actin was used as control.
Relative expression of Hoxc11 and its interacting partners
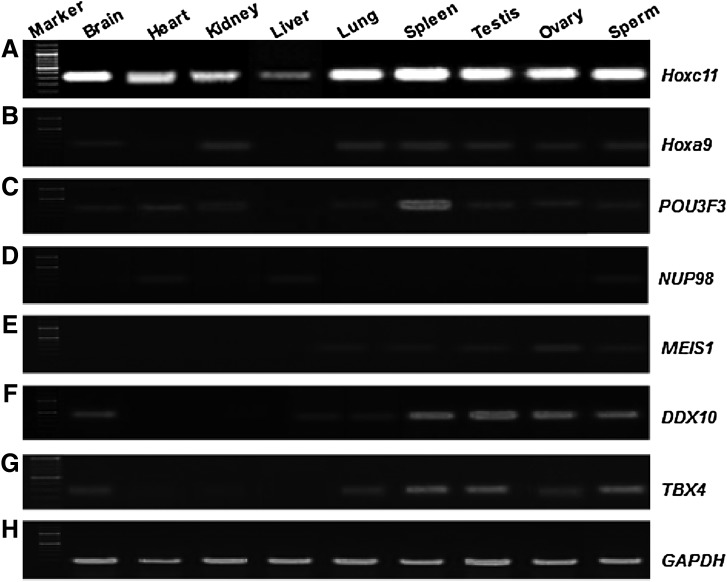
RT-PCR analysis using gene-specific internal primers of Hoxc11 and cDNA from across the tissues and spermatozoa detected a band of 58 bp with higher expression in the lung, spleen, spermatozoa, testis, and ovary and relatively lower expression in other tissues (Fig. 6A). These results were in accordance with that of the northern blot analysis. The RT-PCR expression studies conducted on six interacting partners Hoxa9, POU3F3, NUP98, MEIS1, DDX10, and TBX4 of Hoxc11 gene across different tissues and spermatozoa detected 60–83 bp amplicons (Fig. 6B–G). The RT-PCR results were substantiated by quantitative expression of Hoxc11 and its interacting partners across the tissues and spermatozoa employing Real-Time PCR. Quantitative expression analysis showed the highest expression of Hoxc11 in lung followed by that in spleen, spermatozoa, and testis. Similarly, Hoxc11 interacting partners showed higher expression in spleen, lung, spermatozoa, and testis compared with that in liver taken as calibrator (Fig. 7A–G and Table 1). These results suggest the regulatory role of Hoxc11 gene in maintaining the tissues and spermatozoa-specific expression of other genes.
FIG. 6.
RT-PCR-based expression of Hoxc11 gene and its interacting partners in different tissues and spermatozoa of buffalo. RT-PCR results were obtained using gene-specific internal primers and cDNA from different somatic tissues, gonads, and spermatozoa. The gene IDs are given on the right of the panels (A–G) and the sample IDs on top of the panels. The quality and quantity of the cDNA samples were normalized using GAPDH primers shown at the bottom (H). For size marker, 50 bp ladder was used. GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
FIG. 7.
Quantitative expression of Hoxc11 and its interacting partners in different tissues and spermatozoa of buffalo. Representative expression of Hoxc11 gene and its six interacting partners, Hoxa9, POU3F3, NUP98, MEIS1, DDX10, and TBX4 across the tissues given in panels (A–G). The bar represents the expression levels of the genes in folds. The gene IDs are mentioned on the top left corner and sample IDs, below the panels. Liver was taken as the calibrator for all the genes (boxed). The error bars indicate the reproducibility of the experiments. Note the maximum expression of Hoxc11 in lung followed by spleen, testis, and spermatozoa. For details, see Table 1.
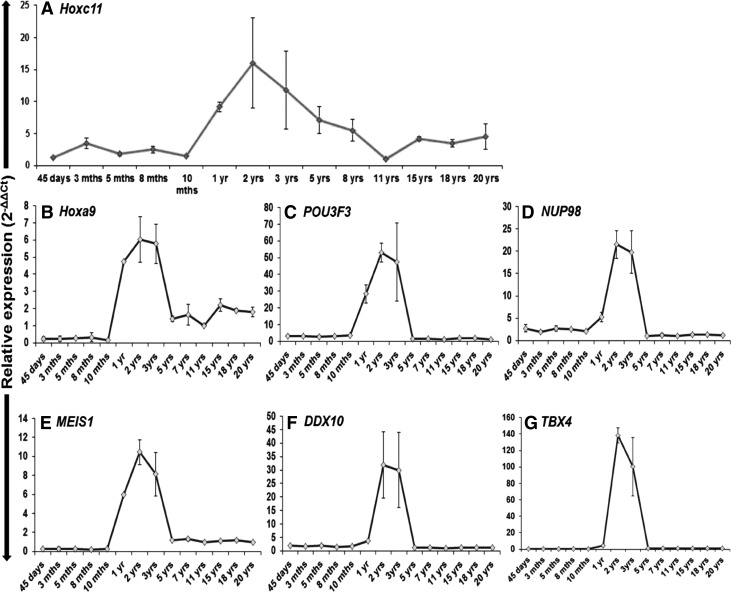
Age-specific expression of Hoxc11 and its interacting partners
To gain an insight into the expression of Hoxc11 and its interacting partners during the course of development, we assessed their expression using Real-Time PCR in water buffalo among different age groups of animals ranging from 45 days to 20 years. The minimum expression of Hoxc11 and its interacting partners was detected in the blood lymphocytes of 45 days old animals. Following this, a gradual but consistent increase in the expression ranging from 0.17 to 3.4-folds during 3–10 months of age was noticed. A sharp increase (approximately four-folds) in the expression was detected in 1-year-old animal as compared with that of 10 months. The higher expression of Hoxc11 and its interacting partners was sustained in 2–3 years aged animals. However, the expression of these genes was drastically regressed after 3 years of age and remained so till the age of 20 years (Fig. 8 and Table 3). From the present study and information available in the literature (Di Rocco et al., 1997; Mann and Affolter, 1998; Liang et al., 2013), we construe that Hoxc11 and its interacting partners work in cohort not only to regulate the developmental events of the organism but also play an important role in its sustenance (Fig. 8).
FIG. 8.
Relative expression of Hoxc11 and its interacting partners in different age groups of buffalo. The quantitative expression carried out using the cDNA isolated from the blood lymphocytes of different age groups of animals. Note the enhanced expression of Hoxc11 (A) and its interacting partners (B–G) in the animals of one year of age and sustenance of the same during 2–3 years followed by their gradual decrease till 20 years of age. The error bars indicate the reproducibility of the experiments.
Table 3.
Relative Expression of Hoxc11 Gene and Its Interacting Partners Across Different Age Groups of Bubalus bubalis
| Relative expression (in folds 2−ΔΔCt) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S. No. | Gene | 45 Days | 3 months | 5 months | 8 months | 10 months | 1 year | 2 years | 3 years | 5 years | 7 years | 11 years | 15 years | 18 years | 20 years |
| 1 | Hoxc11 | 1.25 | 3.48 | 1.80 | 2.51 | 1.52 | 9.18 | 16.02 | 11.8 | 7.1 | 5.49 | cb | 4.15 | 3.48 | 4.48 |
| 2 | Hoxa9 | 0.22 | 0.24 | 0.25 | 0.32 | 0.17 | 4.73 | 6.03 | 5.77 | 1.40 | 1.65 | cb | 2.2 | 1.9 | 1.80 |
| 3 | POU3F3 | 3.32 | 3.26 | 2.88 | 2.98 | 3.70 | 28.33 | 52.9 | 47.44 | 1.70 | 1.59 | cb | 1.87 | 1.75 | 1.00 |
| 4 | NUP98 | 2.74 | 1.95 | 2.64 | 2.54 | 2.12 | 5.31 | 21.51 | 19.81 | 1.07 | 1.23 | cb | 1.30 | 1.29 | 1.15 |
| 5 | MEIS1 | 0.23 | 0.29 | 0.27 | 0.20 | 0.26 | 5.94 | 10.46 | 8.14 | 1.17 | 1.29 | cb | 1.10 | 1.19 | 0.97 |
| 6 | DDX10 | 1.86 | 1.58 | 2.04 | 1.43 | 1.65 | 3.70 | 31.93 | 29.95 | 1.17 | 1.29 | cb | 1.12 | 1.27 | 1.08 |
| 7 | TBX4 | 0.42 | 0.50 | 0.47 | 0.55 | 0.26 | 4.48 | 138.37 | 100.24 | 1.26 | 1.13 | cb | 1.25 | 1.14 | 0.98 |
Cb denotes calibrator tissue (expression value 1). The age groups having maximum expression are in bold.
Discussion
Hoxc11 gene has been studied in different organisms during the embryonic stages (Hostikka and Capecchi, 1998; Liang et al., 2013), but its significance and that of its interacting partners with respect to adult organisms remained unknown. While the collective action of HOX family genes has been well documented, information on an individual gene belonging to this family and its expression in a large mammalian species has remained elusive. In mouse, Hoxc11 amino acid sequence of the homeobox is ∼93% identical to its published paralogs (Hoxa11 and Hoxd11) from other vertebrates, whereas the same is only 65% identical with homeoboxes of neighboring Hox genes (Hostikka and Capecchi, 1998). In the present study, in silico analysis showed a high degree of conservation (>89%) of B. bubalis Hoxc11 gene across the species not only with respect to CDS, but also to 5′and 3′ UTRs. Despite varying length of mRNA, the coding region of Hoxc11 corresponding to 304 amino acids remained faithfully conserved across the species. This high level of conservation is the testimony of its biological requirement in different cell and tissue systems. However, 5′ and 3′ UTRs seem to be equally critical in maintaining the regulatory roles of Hoxc11 gene across the species (Table 2 and Supplementary Fig. S4).
In vertebrates, Hoxc11 conforms to the concept of spatial and temporal colinearity along the primary body axis and is expected to show a high expression in the posterior region of the organism (Zákány et al., 2004; Pearson et al., 2005; Schorderet and Duboule, 2011). Previous studies have shown that the Hoxc11 gene is expressed during the early stages of mouse embryonic development in different tissues like pelvis, kidney, posterior urethra, and gut (Hostikka and Capecchi, 1998). In a recent study, from our laboratory, we localized the HOXC11 protein in the nuclei of gonads and different somatic tissues of buffalo employing immunohistochemistry. Subsequently, immunoblotting using specific antibodies showed differential expression of buffalo HOXC11 protein reflecting its possible tissue-specific function (Sharma et al., 2014). In the present study, we have demonstrated varying levels of Hoxc11 gene expression in an adult buffalo. In accordance with the abovementioned studies, Real-Time analysis showed highest expression of buffalo Hoxc11 in lung followed by that in spleen, spermatozoa, and testis suggesting its role in respiration, immunotransaction and gonadal functions. Thus, besides its involvement in the morphogenesis of the posterior region, Hoxc11 seems to play equally significant role in the anterior region as evident from its enhanced lung expression. The higher expression of Hoxc11 interacting genes in spleen, lung, testis, and spermatozoa indicates close coordination of this gene with other associated genes. Significantly, we demonstrated high expression of DDX10 gene in the testis and spermatozoa, which is in accordance with the earlier report on its potential role in germ cell development and spermatogenesis (Abdelhaleem et al., 2003). Moreover, a relatively lower expression of NUP98, MEIS1, and DDX10 may be taken as a reflection of their diverse functions in somatic tissues and gonads. Thus, Hoxc11 at the transcriptional level acts as a decoding system in regulating the expression of its interacting partners in different somatic tissues, gonads, and spermatozoa in buffalo, thereby fulfilling its broader biological mandates. Taken together, Hoxc11 seems to have attributes similar to that of a pleiotropic gene.
Buffalo is a complex species as its reproductive biology is riddled with unknown and unfathomed facts. Buffalo heifers usually attain puberty when they reach about 55–60% of their adult body weight. Although the age of puberty may vary ranging from 18 to 46 months under different genotype, nutrition, management, and climatic conditions, the females exhibit first oestrus during 15–18 months (Jainudeen and Hafez, 1993; Barile, 2005). Till date, neither the involvement of Hoxc11 expression in the reproduction of buffalo is demonstrated nor is there any report on its sustained expression throughout the life of the animal. Based on the upregulation of Hoxc11 and its interacting partners noticed during 1–3 years of age along with high expression in testis and spermatozoa that coincides with the onset of puberty, we infer their involvement in reproduction and spermatogenesis. Further, gradual decrease in Hoxc11 expression and that of its interacting partners in the animals after 3 years and beyond, until the age of 20 years, suggests its sustained requirement even in the adult animals. This is evident from the hormonal changes (LH, FSH, progesterone, and estradiol-170) and differentiation of the sexual/reproductive organs in the animals (Singh et al., 2001; Perera et al., 2005; Singh et al., 2006). Thus, Hoxc11 gene expression correlates with the specific stages of gonadal differentiation. Taken together, Hoxc11 gene warrants attention in the overall realm of genome analysis focusing on animal research to augment deeper understanding of several key phenomena such as physiology, immunology, endocrine systems, and reproduction. Also, establishing a correlation based on the genetic architecture of Hoxc11 gene and its resultant phenotype may help in breed delineation corresponding to genetic basis of eliteness or other physical and physiological attributes of the animal.
Conclusions
The higher expression of Hoxc11 and its interacting partners in testis, spermatozoa, and during 1–3 years of age of the animal reflects its likely role during and after fertilization, in gonadal differentiation, and reproduction. This gene, functions beyond pattern formation during the development as evident from our studies on adult animals. Pursuance of Hoxc11 especially in the context of respiratory, immunological, and in/fertility would enrich our understanding on the animal research in general and animal biotechnology in particular.
Supplementary Material
Acknowledgments
This work was supported by a core grant from the Department of Biotechnology (DBT) of the National Institute of Immunology, New Delhi and research grants nos. BT/PR11805/MED/12/424/2009, BT/PR14102/AAQ/01/438/2010 from DBT, New Delhi and no. SR/SO/AS-115/2012 from the Department of Science and Technology (DST), New Delhi to SA. SA acknowledges the award of the J.C. Bose National Fellowship by DST, New Delhi and equipment donation from the Alexander Von Humboldt Foundation, Bonn, Germany. Technical assistance from Khem Singh Negi is acknowledged.
Author's Contributions
S.A. and L.R. conceived and designed the research work. L.R. performed the experiments and in silico analysis and drafted the article. L.R., D.P., and N.S. analyzed and interpreted the data. S.A. scrutinized the data analysis, revised the article critically, and provided overall supervision. All the authors read and approved the final article.
Disclosure Statement
The author(s) declare they have no competing interests.
References
- Abdelhaleem M., Maltais L., and Wain H. (2003). The human DDX and DHX gene families of putative RNA helicases. Genomics 81, 618–622 [DOI] [PubMed] [Google Scholar]
- Ahn D., and Ho R.K. (2008). Tri-phasic expression of posterior Hox genes during development of pectoral fins in zebrafish: implications for the evolution of vertebrate paired appendages. Dev Biol 322, 220–233 [DOI] [PubMed] [Google Scholar]
- Bai X.T., Gu B.W., Yin T., Niu C., Xi X.D., Zhang J., Chen Z., and Chen S.J. (2006). Trans-repressive effect of NUP98-PMX1 on PMX1-regulated c-FOS gene through recruitment of histone deacetylase 1 by FG repeats. Cancer Res 66, 4584–4590 [DOI] [PubMed] [Google Scholar]
- Barile V.L. (2005). Reproductive Efficiency in Female Buffaloes. In Buffalo Production and Research. Regional Office for Europe Technical Series 67 Borghese A., ed. (Inter-regional Cooperative Research Network on Buffalo, FAO Regional Office for Europe, Rome: ). pp. 77–108 [Google Scholar]
- Cantile M., Cindolo L., Napodano G., Altieri V., and Cillo C. (2003). Hyperexpression of locus C genes in the HOX network is strongly associated in vivo with human bladder transitional cell carcinomas. Oncogene 22, 6462–6468 [DOI] [PubMed] [Google Scholar]
- Cillo C., Barba P., Freschi G., Bucciarelli G., Magli M.C., and Boncinelli E. (1992). HOX gene expression in normal and neoplastic human kidney. Int J Cancer 51, 892–897 [DOI] [PubMed] [Google Scholar]
- Cribiu E.P., Di Berardino D., Di Meo G.P., Eggen A., Gallagher D.S., Gustavsson I., Hayes H., Iannuzzi L., Popescu C.P., and Rubes J., et al. (2001). International system for chromosome nomenclature of domestic bovids (ISCNDB 2000). Cytogenet Cell Genet 92, 283–299 [DOI] [PubMed] [Google Scholar]
- Daftary G.S., and Taylor H.S. (2006). Endocrine Regulation of HOX Genes. Endocr Rev 27, 331–355 [DOI] [PubMed] [Google Scholar]
- Di Bonito M., Narita Y., Avallone B., Sequino L., Mancuso M., Andolfi G., Franzè A.M., Puelles L., Rijli F.M., and Studer M., et al. (2013). Assembly of the Auditory Circuitry by a Hox Genetic Network in the Mouse Brainstem. PLoS Genet 9, e1003249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rocco G., Mavilio F., and Zappavigna V. (1997). Functional dissection of a transcriptionally active, target-specific Hox-Pbx complex. EMBO J 16, 3644–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di-Poï N., Montoya-Burgos J.I., Miller H., Pourquié O., Milinkovitch M.C., and Duboule D. (2010). Changes in Hox genes structure and function during the evolution of the squamate body plan. Nature 464, 99–103 [DOI] [PubMed] [Google Scholar]
- Franks T.M., and Hetzer M.W. (2013). The role of Nup98 in transcription regulation in healthy and diseased cells. Trends Cell Biol 23, 112–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas R., Gómez-Marín C., Wilson J.M., Casares F., and Gómez-Skarmeta J.L. (2012). Hoxd13 contribution to the evolution of vertebrate appendages. Dev Cell 23, 1219–1229 [DOI] [PubMed] [Google Scholar]
- Gong K.Q., Yallowitz A.R., Sun H., Dressler G.R., and Wellik D.M. (2007). A Hox-Eya-Pax complex regulates early kidney developmental gene expression. Mol Cell Biol 27, 7661–7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman F.R. (2002). Limb malformations and the human Hox genes. Am J Med Genet 112, 256–265 [DOI] [PubMed] [Google Scholar]
- Hoegg S., and Meyer A. (2005). Hox clusters as models for vertebrate genome evolution. Trends Genet 21, 421–424 [DOI] [PubMed] [Google Scholar]
- Hostikka S.L., and Capecchi M.R. (1998). The mouse Hoxc11 gene: genomic structure and expression pattern. Mech Dev 70, 133–145 [DOI] [PubMed] [Google Scholar]
- Huang L., Pu Y., Hepps D., Danielpour D., and Prins G.S. (2007). Posterior Hox gene expression and differential androgen regulation in the developing and adult rat prostate lobes. Endocrinology 148, 1235–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung Y.C., Ueda M., Terai Y., Kumagai K., Ueki K., Kanda K., Yamaguchi H., Akise D., and Ueki M. (2003). Homeobox gene expression and mutation in cervical carcinoma cells. Cancer Sci 94, 437–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki M., Kuwata T., Yamazaki Y., Jenkins N.A., Copeland N.G., Osato M., Ito Y., Kroon E., Sauvageau G., and Nakamura T. (2005). Identification of cooperative genes for NUP98-HOXA9 in myeloid leukemogenesis using a mouse model. Blood 105, 784–793 [DOI] [PubMed] [Google Scholar]
- Jainudeen M.R., and Hafez E.S.E. (1993). Cattle and buffalo. In Reproduction in Farm Animals. 6th ed. Hafez E.S.E., ed. (Lea and Febiger, Philadelphia, USA: ), pp. 315–329 [Google Scholar]
- Jankovic D., Gorello P., Liu T., Ehret S., La Starza R., Desjobert C., Baty F., Brutsche M., Jayaraman P.S., and Santoro A., et al. (2008). Leukemogenic mechanisms and targets of a NUP98/HHEX fusion in acute myeloid leukemia. Blood 111, 5672–5682 [DOI] [PubMed] [Google Scholar]
- Jia X.H., Zhu L.P., Li J.C., and Wang C.C. (2013). Expression of homeobox gene HOXA9 in childhood acute leukemia, and its clinical significance. Zhongguo Dang Dai Er Ke Za Zhi 15, 268–272 [PubMed] [Google Scholar]
- John M.V., and Ali S. (1997). Synthetic DNA-based genetic markers reveal intra-interspecies DNA sequence variability in the Bubalus bubalis related genomes. DNA Cell Biol 16, 369–378 [DOI] [PubMed] [Google Scholar]
- Kumar S., Gupta R., and Ali S. (2011). Molecular mining of alleles in water buffalo Bubalus bubalis and characterization of the TSPY1 and COL6A1 genes. PLoS One 6, e24958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., and Lopez R., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lewis M.T. (2000). Homeobox genes in mammary gland development and neoplasia. Breast Cancer Res 2, 158–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Franks T.M., Marchetto M.C., Gage F.H., and Hetzer M.W. (2013). Dynamic association of NUP98 with the human genome. PLoS Genet 9, e1003308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limura T., and Pourquié O. (2007). Hox genes in time and space during vertebrate body formation. Dev Growth Differ 49, 265–275 [DOI] [PubMed] [Google Scholar]
- Makiyama K., Hamada J., Takada M., Murakawa K., Takahashi Y., Tada M., Tamoto E., Shindo G., Matsunaga A., and Teramoto K., et al. (2005). Aberrant expression of HOX genes in human invasive breast carcinoma. Oncol Rep 13, 673–679 [PubMed] [Google Scholar]
- Mann R.S., and Affolter M. (1998). Hox proteins meet more partners. Curr Opin Genet Dev 8, 423–429 [DOI] [PubMed] [Google Scholar]
- Miller G.J., Miller H.L., van Bokhoven A., Lambert J.R., Werahera P.N., Schirripa O., Lucia M.S., and Nordeen S.K. (2003). Aberrant HOXC expression accompanies the malignant phenotype in human prostate. Cancer Res 63, 5879–5888 [PubMed] [Google Scholar]
- Naiche L.A., and Papaioannou V.E. (2003). Loss of Tbx4 blocks hindlimb development and affects vascularization and fusion of the allantois. Development 130, 2681–2693 [DOI] [PubMed] [Google Scholar]
- Nakao K., Nishino M., Takeuchi K., Iwata M., Kawano A., Arai Y., and Ohki M. (2000). Fusion of the nucleoporin gene, NUP98, and the putative RNA helicase gene, DDX10, by inversion 11 (p15q22) chromosome translocation in a patient with etoposide-related myelodysplastic syndrome. Intern Med 39, 412–415 [DOI] [PubMed] [Google Scholar]
- Naora H., Yang Y.Q., Montz F.J., Seidman J.D., Kurman R.J., and Roden R.B. (2001). A serologically identified tumor antigen encoded by a Homeobox gene promotes growth of ovarian epithelial cells. Proc Natl Acad Sci USA 98, 4060–4065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmqvist L., Pineault N., Wasslavik C., and Humphries R.K. (2007). Candidate genes for expansion and transformation of hematopoietic stem cells by NUP98-HOX fusion genes. PLoS One 2, e768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak D., Srivastava J., Samad R., Parwez I., Kumar S., and Ali S. (2010). Genome-wide search of the genes tagged with the consensus of 33.6 repeat loci in buffalo Bubalus bubalis employing minisatellite-associated sequence amplification. Chromosome Res 18, 441–458 [DOI] [PubMed] [Google Scholar]
- Pearson J.C., Lemons D., and McGinnis W. (2005). Modulating Hox gene functions during animal body patterning. Nat Rev Genet 6, 893–904 [DOI] [PubMed] [Google Scholar]
- Perera B.M.A.O., Abeygunawardena H., Vale W.G., and Chantalakhana C. (2005). Buffalo. In Livestock and Wealth Creation–Improving the Husbandry of Animals Kept by Poor People in Developing Countries. Livestock Production Programme, Owen E., Kitalyi A., Jayasuriya N., and Smith T., eds. (Natural Resources International Limited, UK: ), pp. 451–471 [Google Scholar]
- Pineault N., Abramovich C., Ohta H., and Humphries R.K. (2004). Differential and common leukemogenic potentials of multiple NUP98-Hox fusion proteins alone or with Meis1. Mol Cell Biol 24, 1907–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premi S., Srivastava J., Chandy S.P., and Ali S. (2007). AZFc somatic microdeletions and copy number polymorphism of the DAZ genes in human males exposed to natural background radiation. Hum Genet 121, 337–346 [DOI] [PubMed] [Google Scholar]
- Raman V., Martensen S.A., Reisman D., Evron E., Odenwald W.F., Jaffee E., Marks J., and Sukumar S. (2000). Compromised HOXA5 further can limit P53 expression in human breast tumours. Nature 405, 974–978 [DOI] [PubMed] [Google Scholar]
- Rawal L., Ali S., and Ali S. (2012). Molecular mining of GGAA tagged transcripts and their expression in water buffalo Bubalus bubalis. Gene 492, 290–295 [DOI] [PubMed] [Google Scholar]
- Schorderet P., and Duboule D. (2011). Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS Genet 7, e1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M., Rawal L., Panwar D., Sehgal N., and Ali S. (2014). Differential expression of Homeobox C11 protein in water buffalo Bubalus bubalis and its putative 3D structure. BMC Genomics 15, 638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W.F., Rozenfeld S., Kwong A., Köm ves L.G., Lawrence H.J., and Largman C. (1999). HOXA9 forms triple complexes with PBX2 and MEIS1 in myeloid cells. Mol Cell Biol 19, 3051–3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon H. (1999). T-box genes and the formation of vertebrate forelimb- and hindlimb specific pattern. Cell Tissue Res 296, 57–66 [DOI] [PubMed] [Google Scholar]
- Singh A.K., Brar P.S., Nanda A.S., and Prakash B.S. (2006). Effect of suckling on basal and GnRH-induced LH release in post-partum dairy buffaloes. Anim Reprod Sci 95, 244–250 [DOI] [PubMed] [Google Scholar]
- Singh B., Dixit V.D., Singh P., Georgie G.C., and Dixit V.P. (2001). Plasma inhibin levels in relation to steroids and gonadotrophins during oestrous cycle in buffalo. Reprod Domest Anim 36, 163–167 [PubMed] [Google Scholar]
- Sordino P., van der Hoeven F., and Duboule D. (1995). Hox gene expression in teleost fins and the origin of vertebrate digits. Nature 375, 678–681 [DOI] [PubMed] [Google Scholar]
- Srivastava J., Premi S., Kumar S., and Ali S. (2009). Expressional dynamics of minisatellite 33.15 tagged spermatozaol transcriptome in Bubalus bubalis. BMC Genomics 10, 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava J., Premi S., Kumar S., and Ali S. (2008). Organization differential expression of the GACA/GATA tagged somatic spermatozoal transcriptomes in Buffalo Bubalus bubalis. BMC Genomics 9, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava J., Premi S., Kumar S., Parwez I., and Ali S. (2007). Characterization of Smoc-1 uncovers two transcript variants showing differential tissue and age specific expression in Bubalus bubalis. BMC Genomics 8, 436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumiyama K., Washio-Watanabe K., Saitou N., Hayakawa T., and Ueda S. (1996). Class III POU genes: generation of homopolymeric amino acid repeats under GC pressure in mammals. J Mol Evol 43, 170–178 [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Hamada J., Murakawa K., Takada M., Tada M., Nogami I., Hayashi N., Nakamori S., Monden M., and Miyamoto M., et al. (2004). Expression profiles of 39 HOX genes in normal human adult organs and anaplastic thyroid cancer cell lines by quantitative real-time RT-PCR system. Exp Cell Res 293, 144–153 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., and Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thol F., Kölking B., Hollink I.H., Damm F., van den Heuvel-Eibrink M.M., Michel Zwaan C., Bug G., Ottmann O., Wagner K., and Morgan M., et al. (2013). Analysis of NUP98/NSD1 translocations in adult AML and MDS patients. Leukemia 27, 750–754 [DOI] [PubMed] [Google Scholar]
- Wang K., Shi D., Zhu P., Dai J., Zhu L., Zhu H., Lv Y., Zhao B., and Jiang Q. (2010). Association of a single nucleotide polymorphism in Tbx4 with developmental dysplasia of the hip: a case-control study. Osteoarthritis Cartilage 18, 1592–1595 [DOI] [PubMed] [Google Scholar]
- Wang Z., Yuan L., Rossiter S.J., Zuo X., Ru B., Zhong H., Han N., Jones G., Jepson P.D., and Zhang S. (2009). Adaptive evolution of 59 HoxD genes in the origin and diversification of the cetacean flipper. Mol Biol Evol 26, 613–622 [DOI] [PubMed] [Google Scholar]
- Yuan X., and Braun T. (2013). An unexpected switch: regulation of cardiomyocyte proliferation by the homeobox gene meis1. Circ Res 113, 245–248 [DOI] [PubMed] [Google Scholar]
- Zákány J., Kmita M., and Duboule D. (2004). A dual role for Hox genes in limb anterior-posterior asymmetry. Science 304, 1669–1672 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.