Background: The inbred mouse strain DBA/2J shows impaired hippocampal memory formation, which has been attributed to postsynaptic changes.
Results: DBA/2J shows reduced expression of exocytosis proteins, paired-pulse facilitation, and number of synaptic vesicles.
Conclusion: Proteomic, ultrastructural, and physiological investigations suggest a deficit in presynaptic function in DBA/2J.
Significance: This is the first study describing the DBA/2J hippocampal proteome and ultrastructure.
Keywords: electrophysiology, exocytosis, neuroscience, neurotransmitter release, proteomics, synaptic plasticity, RASAL1, iTRAQ, presynaptic
Abstract
The inbred strains C57BL/6J and DBA/2J (DBA) display striking differences in a number of behavioral tasks depending on hippocampal function, such as contextual memory. Historically, this has been explained through differences in postsynaptic protein expression underlying synaptic transmission and plasticity. We measured the synaptic hippocampal protein content (iTRAQ (Isobaric Tags for Relative and Absolute Quantitation) and mass spectrometry), CA1 synapse ultrastructural morphology, and synaptic functioning in adult C57BL/6J and DBA mice. DBA mice showed a prominent decrease in the Ras-GAP calcium-sensing protein RASAL1. Furthermore, expression of several presynaptic markers involved in exocytosis, such as syntaxin (Stx1b), Ras-related proteins (Rab3a/c), and rabphilin (Rph3a), was reduced. Ultrastructural analysis of CA1 hippocampal synapses showed a significantly lower number of synaptic vesicles and presynaptic cluster size in DBA mice, without changes in postsynaptic density or active zone. In line with this compromised presynaptic morphological and molecular phenotype in DBA mice, we found significantly lower paired-pulse facilitation and enhanced short term depression of glutamatergic synapses, indicating a difference in transmitter release and/or refilling mechanisms. Taken together, our data suggest that in addition to strain-specific postsynaptic differences, the change in dynamic properties of presynaptic transmitter release may underlie compromised synaptic processing related to cognitive functioning in DBA mice.
Introduction
Inbred and gene-specific mutant mouse strains have provided a basis for exploration of the impact of genetic variation on structural, physiological, and molecular factors that modulate learning and memory. A number of strains display prominent differences in behavioral tasks depending on hippocampal function (1). In particular, DBA/2J (DBA)3 is outperformed by the C57Bl/6J (C57) strain in tasks for aversive memory formation and maintenance (2). Various deficits in tasks for spatial memory, such as Barnes and Morris water maze (3–5), and nonspatial memory, such as cued and contextual fear conditioning (5, 6), have been reported for DBA mice. These memory impairments may be linked to a reduced capacity for the maintenance of θ-burst LTP in the CA1 area of the hippocampus (Schaffer collateral pathway) in DBA compared with C57 (5, 7). θ-burst activity is assumed important in novel context exploration (8), an important factor in the formation of spatial and fear conditioning memories. Induction of LTP appeared unaffected unless interburst intervals were extremely compressed to 3 s (8, 9).
In search of the principal molecular mechanisms underlying the physiological and behavioral phenotype of DBA mice, studies have mainly focused on postsynaptic proteins. For instance, changes in the level and activity of signaling proteins such as protein kinase C (PKC), the protein kinase A pathway, and mitogen-activated protein kinase specifically affect behavioral and physiological performance (10, 11). Differences in the level and activity of some of the above proteins have also been shown to affect memory formation in DBA mice (11–13). For example, treatment with oxiracetam increased membrane-bound PKC, which was associated with improved memory performance of DBA mice in the Morris water maze (14).
However, the diverse physiological responses to the temporal pattern of stimulation may be insufficiently explained by postsynaptic mechanisms only. A reduced capacity for paired-pulse facilitation (PPF) has been observed (5), which suggests the presence of changes in presynaptic dynamics and short term plasticity that may further contribute to the physiological and behavioral phenotype in DBA mice. Because a comprehensive view of the synaptic composition, morphology, and short term plasticity in these strains is lacking, we performed an integrative analysis of the synaptic differences between DBA and C57 in adult (10–13 weeks) mice.
We observed decreased expression levels of presynaptic markers involved in exocytosis, together with a lower number of synaptic vesicles and presynaptic cluster size in DBA mice, and no changes in postsynaptic density or active zone. Moreover, DBA mice displayed decreased paired-pulse facilitation and enhanced short term depression of glutamatergic synapses. These results suggest an alteration in transmitter release or refilling mechanisms that, in addition to known postsynaptic mechanisms, may contribute to learning and memory differences in these strains.
Experimental Procedures
Animals
Male C57Bl6/J and DBA/2J were obtained from Charles River (Lyon, France) and were individually housed with cage enrichment and water and food ad libitum on a 12/12-h rhythm lights on/off with lights on at 7 a.m. All experiments were approved by the Animal Users Care Committee of the VU University and complied with the European Council Directive (86/609/EEC).
Synaptic Membrane Preparation, iTRAQ Labeling, Two-dimensional Liquid Chromatography, and Mass Spectrometry (MSMS)
Synaptic membranes were isolated from hippocampi of C57 and DBA animals and iTRAQ-labeled for quantitation as described previously (15–17). In brief, we used eight animals per strain and pooled hippocampi from two mice as a single sample. After trypsin protein digestion, samples were labeled simultaneously in a 4-plex iTRAQ experiment (iTRAQ reagents 113–116) and processed for two-dimensional liquid chromatography followed by tandem mass spectrometry (4700 Proteomics Analyzer; Applied Biosystems) as described previously (18). This experiment was performed twice with independent replicates, yielding an n = 4 (C57) versus n = 4 (DBA) comparison of biological independent samples.
Protein Identification
MSMS spectra were annotated against a concatenated target-decoy database of the Uniprot Mouse reference sequences database (version 07/2013) using Mascot Server software (MatrixScience version 2.3.01). Database searches were performed with trypsin/Pro specificity, in which trypsin cleaves at the C terminus of Lys-Arg, unless followed by Pro, specificity allowing no missed cleavages. Modifications on lysine residues and N termini and methylthio modifications on cysteine residues were set as fixed modifications, whereas iTRAQ modifications on tyrosine residues and oxidation of methionine residues were allowed as variable modifications. Mass tolerance was 200 ppm for precursor ions and 0.4 Da for fragment ions. For each spectrum, the best scoring peptide sequence was used. Protein inference was performed using in- modified IsoformResolver software aiming for consistent protein assignment of peptides across experiments (19). False discovery rates (FDR) for peptide and protein identifications (threshold at 5%) were established using Mayu software (20). FDR of protein identification was based on “unique” (assigned to a single globally inferred protein) spectra exclusively.
SNP Peptide Removal
Based on information in dbSNP (version 5/28/2013) on nonsynonymous SNP sites differing between the two strains, an exclusion list of tryptic protein fragments that have a different amino acid sequence in C57Bl6/J and DBA/2J was deduced. Peptides present in the SNP exclusion list were not used during iTRAQ protein quantification as these peptides may cause a bias in the iTRAQ signal and mapping to the proteome (for excluded peptides please refer to supplemental Fig. 1). Protein quantification was carried out with the remaining non-SNP-containing peptides.
Protein Quantification
For protein quantification, only unique spectra within 5% peptide identification FDR were used. If multiple spectra were assigned to the same peptide sequence in an experiment, only the spectrum with the highest ion score was used for quantification. Spectra with very low iTRAQ reporter signals (maximum intensity of any of the reporter ions less than 100) were removed. Only proteins with a minimum of two unique peptides in each experiment passing the above criteria and within the 5% protein level FDR were considered for quantification. After correction for isotope impurities, iTRAQ reporter ions were log2-transformed, error-corrected, and normalized by variance-stabilizing normalization implemented in the VSN r-package (21). Normalized log-transformed iTRAQ reporter intensities of each spectrum were centered to the average intensity of all iTRAQ reporter ions in the respective spectrum to obtain standardized values. Sample level of protein abundance was determined by taking the average centered iTRAQ reporter intensities of the respective iTRAQ reporters of all spectra assigned to that protein. Prior to further analysis, hierarchical clustering of samples was performed as a quality control (supplemental Fig. 2), showing normal biological variation between genotypes without any batch effects. Statistical evaluation of protein abundance differences was performed with the SAM r-package (22) calculating the permutation-derived false discovery rate (q value) as described previously (17).
Immunoblotting
Proteins were separated on a SDS gel and electroblotted onto PVDF membrane (Bio-Rad). After blocking and incubation of the first antibody in PBS/Tween, 5% milk powder overnight at 4 °C, β-tubulin 1:2000 (Sigma), Stxbp1 (Munc18) 1:5000, and Rab3c 1:1000 (kind gift from M. Verhage, Dept. of Functional Genomics, VU University, Amsterdam, Netherlands), raphillin3A 1:10,000 (Transduction Laboratories, Lexington, KY), RASAL1 1:100 (GenScript, Piscataway), GluA1 1:1000 (Genscript, Piscataway, NJ), and GluA2/GluN1/GluN2a/GluN2b 1:1000 (University of California at Davis/National Institutes of Health), the blot was washed and incubated for 1 h at room temperature with HRP or alkaline phosphatase-conjugated secondary antibody (GE Healthcare, Diegem, Belgium, 1:10,000). Before being used for immunoblotting, all antibodies were checked for specificity, i.e. whether they showed a band that migrated at the size of the predicted molecular weight on immunoblot. Immunodetection was performed using the enhanced chemi-fluorescence immunoblotting detection system for alkaline phosphatase-conjugated secondary antibody (GE Healthcare) or SuperSignal West Femto for HRP-conjugated secondary antibody (Thermo Scientific, Rockford, IL), and blots were scanned with the FLA-5000 (Fuji Photo Film Corp.) or with the LI-COR Odyssey system, respectively. Relative amounts of immunoreactivity were quantified using ImageJ (National Institutes of Health, Bethesda). To correct for input differences, either Coomassie staining (the upper or lower half of the same gel for enhanced chemi-fluorescence) or the stain-free (TCE containing gel for Femto) activated signal was used.
Electron Microscopy
Mice (n = 4 per strain) were subjected to transcardiac perfusion with a mixture of glutaraldehyde (2.5%) and paraformaldehyde (4%) in 0.1 m cacodylate (caco) buffer, pH 7.4. From the fixed brain, 50-μm-thick coronal slices containing the CA1 (including stratum radiatum) were postfixed (1% OsO4) and stained with 1% uranyl acetate. After embedding in Epon, ultrathin sections (∼90 nm) were collected on 400 mesh copper grids and stained with uranyl acetate and lead citrate. For each condition, docked vesicles, total vesicle number, postsynaptic density (PSD), active zone (AZ) length, and vesicle cluster surface (nm2) were measured on digital images taken at ×100,000 magnification using software in individual electron micrographs in which a single synapse was counted from each of the four perfusion-fixed mice per strain using a Jeol (Peabody, MA) 1010 electron microscope. The observer was not informed about the strain.
The active zone membrane was recognized as a specialized part of the pre-synaptic plasma membrane that contained a clear density that is always opposed to the PSD, and this structure also contains nearby docked synaptic vesicles. The cluster size indicates the size (perimeter or surface) occupied by the total pool of synaptic vesicles (docked + undocked) present per synapse.
Electrophysiological Analysis of Short Term Plasticity
Horizontal hippocampal brain slices (300 μm thick) were prepared in ice-cold artificial cerebrospinal fluid containing the following: 125 mm NaCl, 3 mm KCl, 1.25 mm NaH2PO4, 3 mm MgSO4, 1 mm CaCl2, 26 mm NaHCO3, 10 mm glucose (∼300 mosm). Slices were cut on a vibrating microtome and placed in artificial cerebrospinal fluid in a submerged-style holding chamber and bubbled with carbogen (95% O2, 5% CO2) containing the following: 125 mm NaCl, 3 mm KCl, 1.25 mm NaH2PO4, 1 mm MgSO4, 2 mm CaCl2, 26 mm NaHCO3, 10 mm glucose. Slices were left for 1 h to recover before recording began. Recordings were done in artificial cerebrospinal fluid (same as holding chamber), and slices were perfused in a submerged recording chamber at 28–32 °C. To block GABAA receptor-mediated synaptic currents, we added SR-95531 (gabazine, 10 μm, Tocris) to the recording solution. Patch pipettes (3–5 megohms) were pulled from standard-wall borosilicate tubing and were filled with intracellular solution containing the following (in mm): 140 potassium gluconate, 9 KCl; 10 HEPES; 4 K2-phosphocreatine; 4 ATP (magnesium salt); 0.4 GTP (pH 7.2–7.3, pH adjusted with KOH; 290–300 mosm). After whole-cell configuration, the internal solution was allowed to diffuse for 5 min into the cell prior to the onset of recording. The membrane potential was held at −70 mV.
Schaffer collateral fibers were stimulated using an extracellular electrode positioned in the stratum radiatum, and responses were recorded in CA1 pyramidal cells, selected on the basis of their morphology, and visualized using differential interference contrast microscopy. Moderate stimulation was used to measure synaptic facilitation or depression in response to presynaptic trains (of 10 pulses) over a range of frequencies (5–100 Hz). For each frequency, the stimulus train was repeated 20 times, with a 15-s delay between each sweep. Sweeps at each frequency were equally divided into two groups, one at the start and one at the end of the experiment, allowing time-dependent changes in the responses to be identified.
For the AMPA/NMDA receptor current ratios, recordings were made using pipette medium containing 120 mm cesium gluconate, 10 mm CsCl, 8 mm NaCl, 2 mm MgATP, 10 mm phosphocreatine, 0.2 mm EGTA, 10 mm HEPES, 0.3 mm Tris-GTP, and 1 mm QX-314. Single excitatory postsynaptic currents (EPSCs) were evoked by extracellular stimulation of the Schaffer collaterals. Cells were initially held at −70 mV to obtain AMPA receptor-mediated currents followed by depolarization to +40 mV to obtain AMPA and NMDA receptor-mediated currents. For each membrane potential, the evoked postsynaptic responses were repeated 20 times. The peak AMPA receptor current was measured from average traces obtained at −70 mV, and the NMDA receptor current was measured from traces obtained at +40 mV at a time point where AMPA receptor currents decayed to less than 20% from the peak amplitude.
Statistical Analyses
Proteomics data were corrected for multiple testing, using the permutation-derived false discovery rate (q value) of the tool at a q value <10% (two class unpaired, log2-scaled, T-statistic, 1000 permutations, automatic estimation of s0 factor, k-nearest neighbors (k = 10)) together with a p < 0.05 (unpaired Student's t test) to increase the stringency of analysis. Data for immunoblotting and ultrastructural analysis were analyzed by unpaired Student's t test at p < 0.05. Over-representation was analyzed by assigning cellular localization and functional class using a manually curated list (23) and a hypergeometric distribution test. For electrophysiological data, analysis of variance with repeated measures was used (within subjects effect Huynh-Feldt after Mauchly's test for sphericity), and post hoc unpaired Newman-Keuls test at p < 0.05. All data are given as mean ± S.E.
Results
Proteomics Profiling of the Synaptic Membrane Fraction
To detect changes in expression in the proteomes of hippocampal synaptic membrane fractions of C57 and DBA mice, we used the well established iTRAQ quantitative proteomics analysis (16–18). Genetic differences (single nucleotide polymorphisms, SNPs) between the strains resulted in a number peptides containing nonsynonymous SNPs not included in the reference (C57) proteome. As these peptides may cause a bias in the iTRAQ signal and mapping to the proteome, they were excluded from normalization and analysis (supplemental material).
In total, 403 proteins were identified with the presence of at least two unique peptides (confidence interval of >95%). Using manually curated ontologies for subcellular localization and function of synaptic proteins (23), we classified these proteins to create an impression of the hippocampal synaptic proteome (Fig. 1B). With respect to cellular localization, we detected cytosolic and mitochondrial proteins (49.1%) and proteins related to the synaptic part (membrane, membrane-associated, vesicle; 25.1%). Based on the functional annotation, we found a high percentage of nonmitochondrial proteins involved in intracellular signal transduction, structural plasticity, and exocytosis (30%).
FIGURE 1.
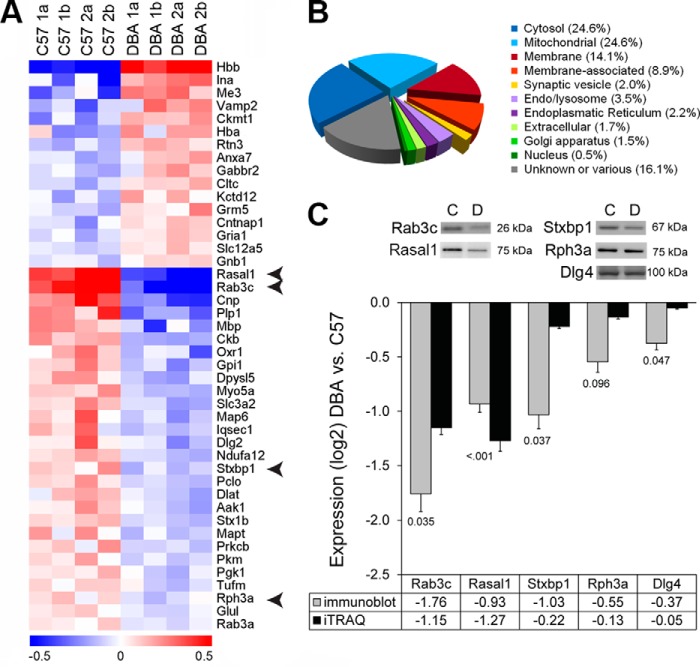
Strain expression differences in the hippocampal synaptic proteome. A, 41 differentially expressed proteins across strains showed a clear distribution for strain for proteins that were expressed higher (red) as well as lower (blue) in DBA mice. The color indicates the relative abundance on a log2 scale. Arrows indicate proteins used for validation with immunoblot. B, in the full dataset, a large fraction (25.1%) of hippocampal proteins detected by iTRAQ was either in or associated with the synaptic membrane and synaptic vesicles. C, immunoblotting confirmed the differential expression (labels below the bars show p values) of several presynaptic proteins as found by iTRAQ analysis. Examples are shown for each blot that was reprobed for the different antibodies.
As glial cells in the hippocampus may also contribute to synaptic plasticity as part of tripartite synapses (24, 25), we compared our data to curated lists of specific glial genes (26) to get an impression of cell type contribution. The majority of proteins was of neuronal origin (30%) or not expressed in a single cell type (49.4%). Glial contribution originated mostly from astrocytes (14.6%), but proteins of oligodendrocyte (5.2%) and microglial (0.8%) origin were also found.
Subsequently, we analyzed differences in protein abundance between C57 and DBA. From the quantitative proteomics dataset, 41 proteins showed statistically significant differences both at p < 0.05 and FDR q <0.1 (Fig. 1A and Table 1). We were able to confirm the differential expression of several regulated proteins (Fig. 1). Highly down-regulated proteins in the DBA strain were Rasal1 and Rab3c. Associated ontology (GO) terms indicate that these proteins are both involved in small GTPase-mediated signal transduction (accessed via Biomart, April 2014).
TABLE 1.
Regulated proteins identified in the hippocampal synaptic membrane fraction
For each protein, the UniProt protein accession and official gene symbol are listed, together with the number of unique peptides quantified in two sets of iTRAQ experiments (a and b). Strain differences were analyzed by two statistical methods (SAM using permutations to address the multiple testing problem and Newman-Keuls test). Proteins with a q value (SAM) of <10% and a p value (Newman-Keuls) of <0.05 were considered differentially expressed. The differential expression of DBA compared with C57 is indicated as a change on a log2 scale. The proteins are grouped according to molecular function as used for the overrepresentation analysis (cf. Fig. 2).
Strain-specific Differences Are More Pronounced in Presynaptic than in Postsynaptic Proteins
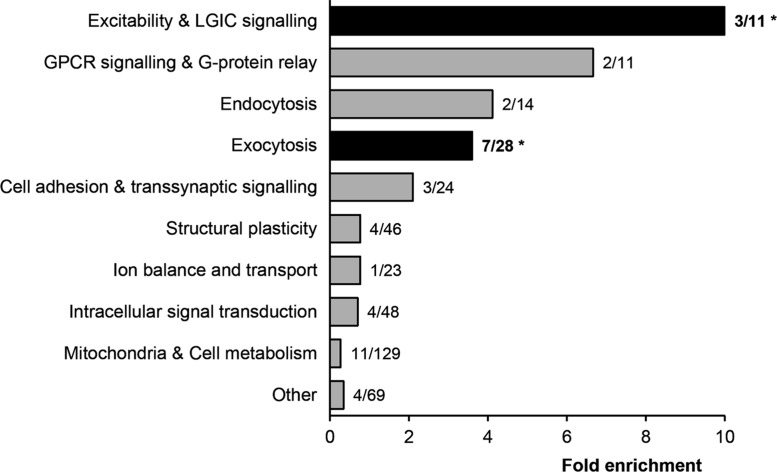
To gain insight into the type of proteins differentially expressed between strains, we performed over-representation analysis on the 41 regulated proteins. With respect to cellular localization, synaptic vesicle proteins were over-represented (p = 0.005). Although we detected a high proportion of cytosolic and mitochondrial proteins, these categories were not over-represented. For the functional annotation, proteins involved in exocytosis (p = 0.004) and excitability and ligand-gated ion channel (LGIC) signaling (p = 0.018) were overrepresented (Fig. 2).
FIGURE 2.
Over-representation analysis of strain-specific hippocampal protein expression. The 403 proteins identified by iTRAQ were grouped according to molecular function, and fold enrichment (bars) was determined. Labels show the number of differentially expressed proteins over the total number of proteins detected in that category (e.g. for exocytosis: 7 out of 28 proteins were significantly regulated). Both exocytosis (p = 0.004) and excitability and LGIC signaling (p = 0.018) were significantly over-represented in this analysis (determined with a hypergeometric test).
In the category “exocytosis,” we found an increased level of the vesicle-associated protein Vamp2 (1.26-fold change). However, most proteins were decreased in expression (Fig. 1C), including members of the SNARE complex and associated proteins (syntaxin 1B, 0.87-fold; Stxbp1/Munc18, 0.86-fold change) and Rab/Ras GTPases (Rab3A, 0.93-fold; Rab3C 0.45-fold change). Remarkably, we found an additional Rab/Ras GTPase that was prominently lowered in DBA mice (0.41-fold change), RasGAP-activating-like protein 1 (RASAL1).
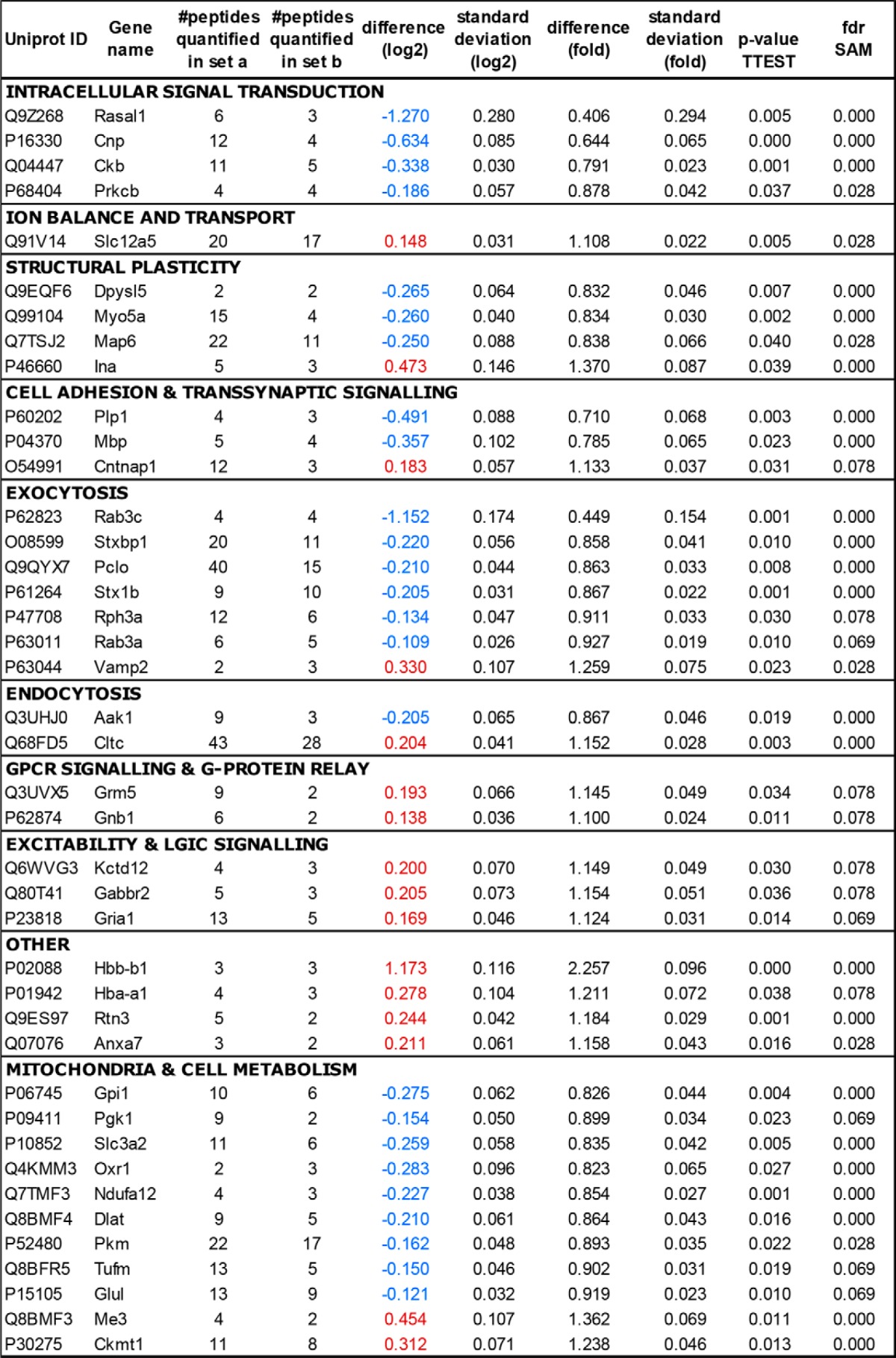
In the category excitability, the AMPA glutamate receptor subunit 1 (GluA1, 1.12-fold change), GABA-B2 receptor, and its auxiliary subunit Kctd12 (both 1.15-fold change) were elevated. However, when we examined additional glutamate receptor subunits by immunoblotting, only the NMDA receptor subunit GluN2A was significantly decreased (Fig. 3, A and B). We obtained AMPA/NMDA receptor current ratios to test whether these small changes in postsynaptic protein levels would translate into functional changes at postsynaptic sites. However, DBA mice do not display a significantly lower AMPA/NMDA receptor current ratio (Fig. 3C).
FIGURE 3.
Glutamate receptor signaling is relatively unaffected. A, strain differences for glutamate (AMPA and NMDA) receptor (R) subunits were analyzed by immunoblot. B and C, lower expression of GluN2A (p = 0.013; n = 7 each strain) did not significantly affect AMPA/NMDA receptor current ratios (p = 0.205; C57, n = 12 slices from eight mice; DBA, n = 13 slices from six mice). Arrowheads indicate the correct molecular mass of GluA1 and GluA2 (∼100 kDa), and GluN1 (∼105 kDa), and GluN2A and GluN2B (∼165 kDa). The blot of GluA2 shows additional bands as it was reprobed.
In summary, DBA mice primarily showed regulation (average 1.3-fold up- or down-regulation) of proteins involved in the vesicle cycle (exocytosis n = 7 and endocytosis n = 2), with a major change in Rab3C and RASAL1. In contrast, the up-regulation (average 1.18-fold) of postsynaptic proteins involved in excitability and LGIC signaling (n = 3) appeared to be more ambiguous.
Reduced Vesicle Pool with Normal Synapse Morphology
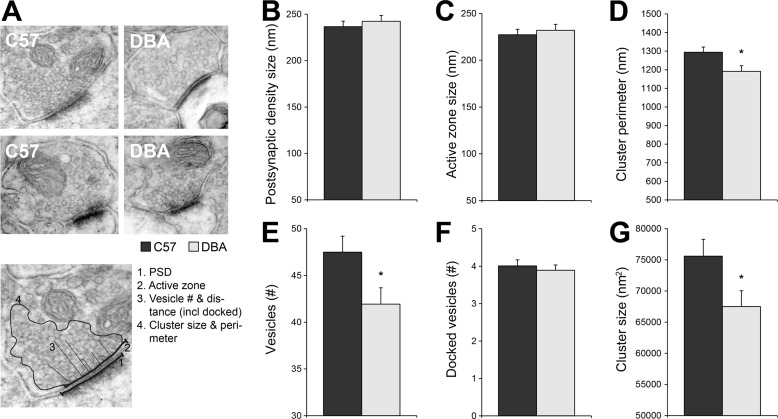
To analyze whether changes in levels of presynaptic proteins are related to morphological alterations, we performed an ultrastructural analysis on synapses of the CA1 stratum radiatum (Fig. 4A). The synaptic contact, determined by the length of the PSD and the AZ, was unchanged (Fig. 4, B and C). However, DBA mice appeared to have a reduced recycling/reserve vesicle pool, as both the number of vesicles (Fig. 4E) and the presynaptic cluster, i.e. the size of the set of vesicles, was significantly decreased in DBA mice (Fig. 4, D and G). The number of docked vesicles per AZ length did not differ, suggesting that the readily releasable pool was not affected (Fig. 4F).
FIGURE 4.
Ultrastructural analysis of CA3 to CA1 synapses. A, when C57 was compared with DBA, no significant differences were observed for the length of the PSD (B; p = 0.598) or the AZ (C; p = 0.713) in the CA1 stratum radiatum and receiving CA3 synapses. However, DBA mice showed less synaptic vesicles (E; p = 0.025) and a decrease in cluster size (G; p = 0.036) and cluster perimeter (p = 0.014; D). F, although the total number of vesicles was lower in DBA mice, the number of docked vesicles was not affected. The analysis consisted of n = 125 C57 and n = 120 DBA synapses from four animals per strain.
Weakened Presynaptic Short Term Plasticity in DBA Mice
The observed decrease in the number of vesicles and cluster size could affect short term presynaptic plasticity. We therefore made whole-cell recordings from CA1 pyramidal neurons and stimulated Schaffer collateral inputs in acute hippocampal slices (Fig. 5A). Using different pulse intervals, we found that C57 and DBA mice differ in enhanced and facilitated postsynaptic depolarization with repetitive stimulation upon a single afferent volley.
FIGURE 5.
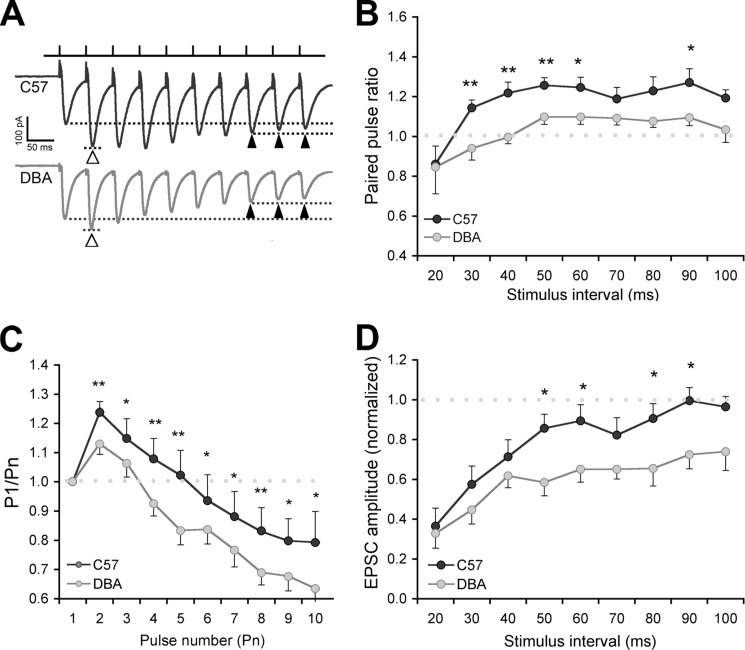
Paired-pulse ratio is affected in DBA mice. A, schematic showing the short term plasticity protocol. Schaffer collateral fibers were stimulated using a 10-pulse, variable frequency stimulation train, and EPSCs were recorded in CA1 pyramidal cells. The first two stimuli in a train were used for paired-pulse analysis. B, paired-pulse facilitation in C57 and DBA pyramidal cells (interpulse interval; 50 ms). Each pair of points (connected by gray lines) shows the average 1st and 2nd EPSCs from individual experiments (mean of 20 stimulus trains). Points connected by a thick black line show group averages. Note the increase in current (more negative amplitude) in the 2nd pulse in C57 mice. EPSCs in the 1st pulse were not different between the two strains.
PPF was significantly impaired (F(1.18) = 10.02, p = 0.005, Figs. 5B and 6A) in DBA compared with C57 mice. In response to low frequency synaptic input (<20 Hz; 50–60- and 90-ms interpulse interval), PPF was reduced, and in the high frequency range (25–50 Hz; 20–40-ms interpulse interval), no PPF was observed in DBA mice, whereas this only occurred in C57 at the highest input frequency (Fig. 6B).
FIGURE 6.
Paired-pulse ratio is affected in DBA mice. A, example trace of single experiments (50-ms interpulse interval) showing paired-pulse facilitation in response to the first two stimuli in the train (open arrowheads, dotted lines) in C57 (12 slices from eight animals) but hardly in DBA (13 slices in six animals). The last three pulses from each train were averaged to study short term plasticity (closed arrowheads, dotted lines), which shows depression in DBA. EPSCs are averaged responses from 20 sweeps (stimulus artifacts truncated). B, effect of interstimulus interval on paired-pulse facilitation. Paired-pulse ratio is plotted against the interpulse interval. Facilitation is reduced in DBA mice, mainly with high frequency synaptic input. C, at the 50-ms interval (20 Hz), the 4th pulse leads to depression in DBA mice compared with the 7th to 10th pulse in C57 mice. D, effect of interstimulus interval on short term plasticity. Whereas high frequency (20–50 Hz; 20–40 ms) input leads to depression in both lines, DBA mice show a clear depression also at low frequency input (10–20 Hz; 50–100 ms). *, p < 0.05; **, p < 0.01.
Furthermore, short term depression occurred more readily in DBA (F(1.18) = 6.47, p = 0.020; Fig. 6, C and D) when we measured amplitudes of the last three EPSC responses in a stimulus train (Fig. 6A). This effect was most obvious at low synaptic input frequencies and also present when all EPSC responses were analyzed (P1/Pn; Fig. 6C). DBA mice show synaptic depression at a faster rate, which, in the absence of clear postsynaptic glutamate receptor effects, could relate to a faster depletion of vesicles. This would functionally corroborate the smaller cluster size observed in DBA mice (cf. Fig. 4).
Discussion
Differences in learning and memory in aversive tasks between the common inbred strains C57Bl/6J and DBA/2J have, at the molecular level, mostly been attributed to changes in postsynaptic proteins. We show that additionally the significant proteome changes (reduced expression of proteins involved in exocytosis) and presynaptic changes (reduced vesicle pool and decreased facilitation, short term plasticity) reducing the capacity for vesicle release may underlie the learning phenotype in DBA mice.
Proteomics analysis of the hippocampal synaptic membrane fraction revealed a marked decrease in expression levels of proteins involved in exocytosis in DBA mice. Significantly regulated proteins were found in the three main groups regulating neurotransmitter release as follows: members of the SNARE complex, regulators thereof from the Sec1/Munc18 homologue protein family (Syntaxin 1B, Stxbp1), and Rab/Ras family of small GTPases (Rab3a and Rab3c). The SNARE and Sec1/Munc proteins are required for vesicle fusion with the plasma membrane and thus play a key role in synaptic plasticity (27, 28). Interestingly, Munc18 levels and recruitment to the synapse are controlled in a PKC-dependent manner (27). Additionally, the Rab/Ras GTPases Rab3A and Rab3C were differentially expressed. Studies in quadruple Rab3(A to D) knock-out mice (29) have found that these proteins modulate calcium-induced fusion of vesicles. However, with Rab3A expression (triple knock-out) there was no apparent decrease in evoked responses. We compared our dataset with observed regulation of hippocampal synaptic membrane proteins 4 h after contextual fear conditioning (plasticity related to learning) (30). It is of interest to note that we found overlap between datasets in six (out of seven) exocytosis proteins that were differentially expressed in DBA mice. Given that this strain shows impaired performance in contextual fear conditioning (6), one might speculate that the baseline difference (for DBA) in these synaptic plasticity proteins may contribute to the disrupted learning of this task. However, this would need further validation within the framework of a learning paradigm.
With regard to alterations in postsynaptic content, the elevation of proteins involved in LGIC signaling, such as the AMPA glutamate receptor subunit 1 (GluA1), could not be validated by immunoblot. The relatively small change, if any, appeared to be in agreement with other studies (31, 32) that found the basal receptor density in hippocampal slices to be unaffected. Alterations are probably more related to AMPA receptor function, such as calcium-induced ligand binding (31). We did, however, detect a reduction in NMDA receptor subunit 2A, although this did not significantly alter the AMPA/NMDA receptor current ratio. Furthermore, level and activity of hippocampal PKC, especially of the γ isoform, have previously been found to influence the performance of DBA mice in spatial memory tasks (11, 12). Although we did not determine activity of the three detected isoforms in our dataset (α, β, and γ), only the PKC-β level was slightly reduced in the hippocampal synaptic membrane fraction of DBA mice.
In addition, we found differential expression of the myelin-associated proteins myelin basic protein and CNP (2′-3′-cyclic-nucleotide 3′-phosphodiesterase), suggesting strain-dependent differences in white matter content. This would, however, need further confirmation by using brain regions with high white matter content and a less selective isolation method (e.g. total tissue homogenate).
The presynaptic phenotype at the proteome level was further supported by ultrastructural analysis revealing a reduced availability of synaptic vesicles. Compared with C57, DBA mice had significantly reduced recycling/reserve vesicle pools (lower number of synaptic vesicles and smaller presynaptic cluster size) in hippocampal CA1 area synapses. The readily releasable pool (number of docked vesicles) was not affected, suggesting that the proteomic changes mainly affect later steps in the vesicle pathway (priming, fusion, or vesicle return). We detected several differentially expressed proteins associated with these processes. We found reduced myosin Va (Myo5a), which is an important transport motor for vesicles and interacts with the microtubule-stabilizing protein MAP6/STOP (33, 34). The latter protein (also decreased in our dataset) is likely important for regulating vesicle cluster size, as knock-out mice for MAP6 were found to have depleted synaptic vesicle pools and impaired synaptic plasticity (35).
In line with the proteomics and EM results, we observed changed short term plasticity in DBA mice compared with C57 mice. PPF at both 40- and 90-ms intervals (25 and 11 Hz) was significantly impaired in DBA compared with C57 mice, in concurrence with the study by Nguyen et al. The degree of impairment in paired-pulse facilitation was most prominent at high stimulus frequencies (interval of less than 40 ms). Furthermore, we found increased short term depression that occurred at lower stimulus frequencies in DBA mice. Impairment of fast vesicle recycling may result in a frequency-dependent increase in short term depression in hippocampal CA1 synapses, which may emerge in early phases (300 ms at 20 Hz) of stimulation (36). Taken together, these observations suggest differences in replenishment of the readily releasable pool in DBA mice. The physiological phenotype suggests that DBA synapses are more easily depleted due to the reduced number of vesicles located to the synapse, as observed in our ultrastructural analysis. In response to this depletion, we may find increased synaptobrevin (Vamp2) as it appears to provide positive feedback for vesicle refill upon high frequency stimulation (37). Indeed, differences in the induction and maintenance of hippocampal LTP have mainly been found when high frequency or θ burst stimulation is used in DBA mice (7, 9, 38). This suggests that the impaired performance of DBA mice in aversive tasks with normal learning in appetitive spatial tasks (2) might be explained through a higher demand on the (compromised) hippocampal neurotransmission.
Remarkably, we find prominently lowered RASAL1 in DBA mice. This protein is a member of the GAP1 family of Ras GTPase-activating proteins (RasGAP), which includes GAP1m, GAP1IP4BP, and CAPRI (39, 40). Members of this family show dual specificity; they have both Ras and Rap GTPase-activating protein activity, a property that has also been found for SynGAP (41, 42). Although the gene is highly expressed in hippocampal CA1 (43), information on its function in neurons is lacking. However, CAPRI and RASAL1 have been found to act as a Ca2+ sensor by in vitro characterization; upon an increase of free intracellular calcium, they become associated with the plasma membrane, which increases their catalytic activity to locally deactivate Ras (39, 44). The translocation of RASAL1 to the membrane is reversible and occurs in synchrony with the frequency of Ca2+ oscillations, a behavior known from other “decoders” of calcium signals involved in memory formation as follows: PKC, calmodulin, and calmodulin-dependent protein kinase II (45–49). More importantly, RASAL1 contains two N-terminal C2 domains (C2A and C2B) that have high sequence homology with the synaptotagmin protein family. These proteins are known regulators of activity-dependent release of neurotransmitters and able to promote the membrane fusion step of exocytosis (50). Using experiments previously applied to synaptotagmin 1 function, Sot et al. (51) found that RASAL1 was also able to sense membrane curvature in vesicles. Considering its calcium and membrane-related sensing properties, we speculate that RASAL1 might be involved in the reduced functional plasticity in DBA mice by affecting vesicle exocytosis or endocytosis.
In conclusion, we found significant differences in the presynaptic proteome, ultrastructure, and plasticity in DBA mice compared with C57 that could reduce the capacity for vesicle release. The underlying mechanism may be the combined effect of reduced expression of vesicle (exocytosis) proteins and changed calcium signaling by RASAL1. The observed presynaptic changes may, in addition to described postsynaptic differences, further explain the learning phenotype that has been described for DBA mice.
Supplementary Material
Acknowledgments
We thank Rolinka van der Loo and Ruud Wijnands for technical assistance. Electron microscopy was performed at the VU/VUmc EM facility.
This work was supported by a NeuroBsik grant to the Dutch Mouse Phenomics consortium (to A. B. S., K. W. L., and S. S.), the Center for Medical Systems Biology (to A. B. S., K. W. L., and S. S.), Netherlands Consortium for Systems Biology Grant 050-060-409 (to M. L. and S. S.), Dutch Medical Research Council Grant ZonMW 912-04-022, and HEALTH-2009-2.1.2-1 EU-FP7 “SynSys” Grant 242167 (to A. B. S., K. W. L., and S. S.).

This article contains supplemental Table S1 and Figs. S1 and S2.
- DBA
- DBA/2J
- PPF
- paired-pulse facilitation
- C57
- C57BL/6J
- SNP
- single nucleotide polymorphism
- FDR
- false discovery rate
- PSD
- postsynaptic density
- AZ
- active zone
- EPSC
- excitatory postsynaptic current
- LGIC
- ligand-gated ion channel
- SAM
- statistical analysis for microarray
- iTRAQ
- Isobaric Tags for Relative and Absolute Quantitation
- LTP
- long-term potentiation.
References
- 1. Schimanski L. A., Nguyen P. V. (2004) Multidisciplinary approaches for investigating the mechanisms of hippocampus-dependent memory: a focus on inbred mouse strains. Neurosci. Biobehav. Rev. 28, 463–483 [DOI] [PubMed] [Google Scholar]
- 2. Youn J., Ellenbroek B. A., van Eck I., Roubos S., Verhage M., Stiedl O. (2012) Finding the right motivation: genotype-dependent differences in effective reinforcements for spatial learning. Behav. Brain Res. 226, 397–403 [DOI] [PubMed] [Google Scholar]
- 3. Upchurch M., Wehner J. M. (1988) Differences between inbred strains of mice in Morris water maze performance. Behav. Genet. 18, 55–68 [DOI] [PubMed] [Google Scholar]
- 4. Holmes A., Wrenn C. C., Harris A. P., Thayer K. E., Crawley J. N. (2002) Behavioral profiles of inbred strains on novel olfactory, spatial and emotional tests for reference memory in mice. Genes Brain Behav. 1, 55–69 [DOI] [PubMed] [Google Scholar]
- 5. Nguyen P. V., Abel T., Kandel E. R., Bourtchouladze R. (2000) Strain-dependent differences in LTP and hippocampus-dependent memory in inbred mice. Learn. Mem. 7, 170–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. André J. M., Cordero K. A., Gould T. J. (2012) Comparison of the performance of DBA/2 and C57BL/6 mice in transitive inference and foreground and background contextual fear conditioning. Behav. Neurosci. 126, 249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schimanski L. A., Nguyen P. V. (2005) Impaired fear memories are correlated with subregion-specific deficits in hippocampal and amygdalar LTP. Behav. Neurosci. 119, 38–54 [DOI] [PubMed] [Google Scholar]
- 8. Otto T., Eichenbaum H., Wiener S. I., Wible C. G. (1991) Learning-related patterns of CA1 spike trains parallel stimulation parameters optimal for inducing hippocampal long term potentiation. Hippocampus 1, 181–192 [DOI] [PubMed] [Google Scholar]
- 9. Nguyen P. V., Duffy S. N., Young J. Z. (2000) Differential maintenance and frequency-dependent tuning of LTP at hippocampal synapses of specific strains of inbred mice. J. Neurophysiol. 84, 2484–2493 [DOI] [PubMed] [Google Scholar]
- 10. Izquierdo I., McGaugh J. L. (2000) Behavioural pharmacology and its contribution to the molecular basis of memory consolidation. Behav. Pharmacol. 11, 517–534 [DOI] [PubMed] [Google Scholar]
- 11. Wehner J. M., Sleight S., Upchurch M. (1990) Hippocampal protein kinase C activity is reduced in poor spatial learners. Brain Res. 523, 181–187 [DOI] [PubMed] [Google Scholar]
- 12. Bowers B. J., Christensen S. C., Pauley J. R., Paylor R., Yuva L., Dunbar S. E., Wehner J. M. (1995) Protein and molecular characterization of hippocampal protein kinase C in C57BL/6 and DBA/2 mice. J. Neurochem. 64, 2737–2746 [DOI] [PubMed] [Google Scholar]
- 13. Paylor R., Baskall-Baldini L., Yuva L., Wehner J. M. (1996) Developmental differences in place-learning performance between C57BL/6 and DBA/2 mice parallel the ontogeny of hippocampal protein kinase C. Behav. Neurosci. 110, 1415–1425 [DOI] [PubMed] [Google Scholar]
- 14. Fordyce D. E., Clark V. J., Paylor R., Wehner J. M. (1995) Enhancement of hippocampally-mediated learning and protein kinase C activity by oxiracetam in learning-impaired DBA/2 mice. Brain Res. 672, 170–176 [DOI] [PubMed] [Google Scholar]
- 15. Li K. W., Hornshaw M. P., Van Der Schors R. C., Watson R., Tate S., Casetta B., Jimenez C. R., Gouwenberg Y., Gundelfinger E. D., Smalla K.-H., Smit A. B. (2004) Proteomics analysis of rat brain postsynaptic density. Implications of the diverse protein functional groups for the integration of synaptic physiology. J. Biol. Chem. 279, 987–1002 [DOI] [PubMed] [Google Scholar]
- 16. Counotte D. S., Li K. W., Wortel J., Gouwenberg Y., Van Der Schors R. C., Smit A. B., Spijker S. (2010) Changes in molecular composition of rat medial prefrontal cortex synapses during adolescent development. Eur. J. Neurosci. 32, 1452–1460 [DOI] [PubMed] [Google Scholar]
- 17. Klemmer P., Meredith R. M., Holmgren C. D., Klychnikov O. I., Stahl-Zeng J., Loos M., van der Schors R. C., Wortel J., de Wit H., Spijker S., Rotaru D. C., Mansvelder H. D., Smit A. B., Li K. W. (2011) Proteomics, ultrastructure, and physiology of hippocampal synapses in a fragile X syndrome mouse model reveal presynaptic phenotype. J. Biol. Chem. 286, 25495–25504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li K. W., Miller S., Klychnikov O., Loos M., Stahl-Zeng J., Spijker S., Mayford M., Smit A. B. (2007) Quantitative proteomics and protein network analysis of hippocampal synapses of CaMKIIα mutant mice. J. Proteome Res. 6, 3127–3133 [DOI] [PubMed] [Google Scholar]
- 19. Meyer-Arendt K., Old W. M., Houel S., Renganathan K., Eichelberger B., Resing K. A., Ahn N. G. (2011) IsoformResolver: A peptide-centric algorithm for protein inference. J. Proteome Res. 10, 3060–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reiter L., Claassen M., Schrimpf S. P., Jovanovic M., Schmidt A., Buhmann J. M., Hengartner M. O., Aebersold R. (2009) Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry. Mol. Cell. Proteomics 8, 2405–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Karp N. A., Huber W., Sadowski P. G., Charles P. D., Hester S. V., Lilley K. S. (2010) Addressing accuracy and precision issues in iTRAQ quantitation. Mol. Cell. Proteomics 9, 1885–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tusher V. G., Tibshirani R., Chu G. (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U.S.A. 98, 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ruano D., Abecasis G. R., Glaser B., Lips E. S., Cornelisse L. N., de Jong A. P., Evans D. M., Davey Smith G., Timpson N. J., Smit A. B., Heutink P., Verhage M., Posthuma D. (2010) Functional gene group analysis reveals a role of synaptic heterotrimeric G proteins in cognitive ability. Am. J. Hum. Genet. 86, 113–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bernardinelli Y., Muller D., Nikonenko I. (2014) Astrocyte-synapse structural plasticity. Neural Plast. 2014, 232105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Verbich D., Prenosil G. A., Chang P. K., Murai K. K., McKinney R. A. (2012) Glial glutamate transport modulates dendritic spine head protrusions in the hippocampus. Glia 60, 1067–1077 [DOI] [PubMed] [Google Scholar]
- 26. Goudriaan A., de Leeuw C., Ripke S., Hultman C. M., Sklar P., Sullivan P. F., Smit A. B., Posthuma D., Verheijen M. H. (2014) Specific glial functions contribute to schizophrenia susceptibility. Schizophr. Bull. 40, 925–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cijsouw T., Weber J. P., Broeke J. H., Broek J. A., Schut D., Kroon T., Saarloos I., Verhage M., Toonen R. F. (2014) Munc18–1 redistributes in nerve terminals in an activity- and PKC-dependent manner. J. Cell Biol. 204, 759–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jahn R., Fasshauer D. (2012) Molecular machines governing exocytosis of synaptic vesicles. Nature 490, 201–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schlüter O. M., Schmitz F., Jahn R., Rosenmund C., Südhof T. C. (2004) A complete genetic analysis of neuronal Rab3 function. J. Neurosci. 24, 6629–6637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rao-Ruiz P., Carney K. E., Pandya N., van der Loo R. J., Verheijen M. H., van Nierop P., Smit A. B., Spijker S. (2015) Time-dependent changes in the mouse hippocampal synaptic membrane proteome after contextual fear conditioning. Hippocampus 10.1002/hipo.22432 [DOI] [PubMed] [Google Scholar]
- 31. Ménard C., Valastro B., Martel M.-A., Martinoli M.-G., Massicotte G. (2004) Strain-related variations of AMPA receptor modulation by calcium-dependent mechanisms in the hippocampus: contribution of lipoxygenase metabolites of arachidonic acid. Brain Res. 1010, 134–143 [DOI] [PubMed] [Google Scholar]
- 32. Zilles K., Wu J., Crusio W. E., Schwegler H. (2000) Water maze and radial maze learning and the density of binding sites of glutamate, GABA, and serotonin receptors in the hippocampus of inbred mouse strains. Hippocampus 10, 213–225 [DOI] [PubMed] [Google Scholar]
- 33. Wöllert T., Patel A., Lee Y.-L., Provance D. W., Jr., Vought V. E., Cosgrove M. S., Mercer J. A., Langford G. M. (2011) Myosin5a tail associates directly with Rab3A-containing compartments in neurons. J. Biol. Chem. 286, 14352–14361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rudolf R., Bittins C. M., Gerdes H.-H. (2011) The role of myosin V in exocytosis and synaptic plasticity. J. Neurochem. 116, 177–191 [DOI] [PubMed] [Google Scholar]
- 35. Andrieux A., Salin P. A., Vernet M., Kujala P., Baratier J., Gory-Fauré S., Bosc C., Pointu H., Proietto D., Schweitzer A., Denarier E., Klumperman J., Job D. (2002) The suppression of brain cold-stable microtubules in mice induces synaptic defects associated with neuroleptic-sensitive behavioral disorders. Genes Dev. 16, 2350–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ertunc M., Sara Y., Chung C., Atasoy D., Virmani T., Kavalali E. T. (2007) Fast synaptic vesicle reuse slows the rate of synaptic depression in the CA1 region of hippocampus. J. Neurosci. 27, 341–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lvov A., Chikvashvili D., Michaelevski I., Lotan I. (2008) VAMP2 interacts directly with the N terminus of Kv2.1 to enhance channel inactivation. Pflugers Arch. 456, 1121–1136 [DOI] [PubMed] [Google Scholar]
- 38. Jones M. W., Peckham H. M., Errington M. L., Bliss T. V., Routtenberg A. (2001) Synaptic plasticity in the hippocampus of awake C57BL/6 and DBA/2 mice: interstrain differences and parallels with behavior. Hippocampus 11, 391–396 [DOI] [PubMed] [Google Scholar]
- 39. Walker S. A., Kupzig S., Bouyoucef D., Davies L. C., Tsuboi T., Bivona T. G., Cozier G. E., Lockyer P. J., Buckler A., Rutter G. A., Allen M. J., Philips M. R., Cullen P. J. (2004) Identification of a Ras GTPase-activating protein regulated by receptor-mediated Ca2+ oscillations. EMBO J. 23, 1749–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yarwood S., Bouyoucef-Cherchalli D., Cullen P. J., Kupzig S. (2006) The GAP1 family of GTPase-activating proteins: spatial and temporal regulators of small GTPase signalling. Biochem. Soc. Trans. 34, 846–850 [DOI] [PubMed] [Google Scholar]
- 41. Kupzig S., Deaconescu D., Bouyoucef D., Walker S. A., Liu Q., Polte C. L., Daumke O., Ishizaki T., Lockyer P. J., Wittinghofer A., Cullen P. J. (2006) GAP1 family members constitute bifunctional Ras and Rap GTPase-activating proteins. J. Biol. Chem. 281, 9891–9900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Krapivinsky G., Medina I., Krapivinsky L., Gapon S., Clapham D. E. (2004) SynGAP-MUPP1-CaMKII synaptic complexes regulate p38 MAP kinase activity and NMDA receptor-dependent synaptic AMPA receptor potentiation. Neuron 43, 563–574 [DOI] [PubMed] [Google Scholar]
- 43. Lein E. S., Hawrylycz M. J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A. F., Boguski M. S., Brockway K. S., Byrnes E. J., Chen L., Chen L., Chen T.-M., Chin M. C., Chong J., Crook B. E., Czaplinska A., Dang C. N., Datta S., Dee N. R., Desaki A. L., Desta T., Diep E., Dolbeare T. A., Donelan M. J., Dong H.-W., Dougherty J. G., Duncan B. J., Ebbert A. J., Eichele G., Estin L. K., Faber C., Facer B. A., Fields R., Fischer S. R., Fliss T. P., Frensley C., Gates S. N., Glattfelder K. J., Halverson K. R., Hart M. R., Hohmann J. G., Howell M. P., Jeung D. P., Johnson R. A., Karr P. T., Kawal R., Kidney J. M., Knapik R. H., Kuan C. L., Lake J. H., Laramee A. R., Larsen K. D., Lau C., Lemon T. A., Liang A. J., Liu Y., Luong L. T., Michaels J., Morgan J. J., Morgan R. J., Mortrud M. T., Mosqueda N. F., Ng L. L., Ng R., Orta G. J., Overly C. C., Pak T. H., Parry S. E., Pathak S. D., Pearson O. C., Puchalski R. B., Riley Z. L., Rockett H. R., Rowland S. A., Royall J. J., Ruiz M. J., Sarno N. R., Schaffnit K., Shapovalova N. V., Sivisay T., Slaughterbeck C. R., Smith S. C., Smith K. A., Smith B. I., Sodt A. J., Stewart N. N., Stumpf K.-R., Sunkin S. M., Sutram M., Tam A., Teemer C. D., Thaller C., Thompson C. L., Varnam L. R., Visel A., Whitlock R. M., Wohnoutka P. E., Wolkey C. K., Wong V. Y., Wood M., Yaylaoglu M. B., Young R. C., Youngstrom B. L., Yuan X. F., Zhang B., Zwingman T. A., Jones A. R. (2007) Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 [DOI] [PubMed] [Google Scholar]
- 44. Liu Q., Walker S. A., Gao D., Taylor J. A., Dai Y.-F., Arkell R. S., Bootman M. D., Roderick H. L., Cullen P. J., Lockyer P. J. (2005) CAPRI and RASAL impose different modes of information processing on Ras due to contrasting temporal filtering of Ca2+. J. Cell Biol. 170, 183–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Oancea E., Meyer T. (1998) Protein kinase C as a molecular machine for decoding calcium and diacylglycerol signals. Cell 95, 307–318 [DOI] [PubMed] [Google Scholar]
- 46. Violin J. D., Zhang J., Tsien R. Y., Newton A. C. (2003) A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. J. Cell Biol. 161, 899–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Craske M., Takeo T., Gerasimenko O., Vaillant C., Török K., Petersen O. H., Tepikin A. V. (1999) Hormone-induced secretory and nuclear translocation of calmodulin: oscillations of calmodulin concentration with the nucleus as an integrator. Proc. Natl. Acad. Sci. U.S.A. 96, 4426–4431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. De Koninck P., Schulman H. (1998) Sensitivity of CaM kinase II to the frequency of Ca2+ oscillations. Science 279, 227–230 [DOI] [PubMed] [Google Scholar]
- 49. Giese K. P., Mizuno K. (2013) The roles of protein kinases in learning and memory. Learn. Mem. 20, 540–552 [DOI] [PubMed] [Google Scholar]
- 50. Südhof T. C., Rizo J. (2011) Synaptic vesicle exocytosis. Cold Spring Harb. Perspect. Biol. 3, a005637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sot B., Behrmann E., Raunser S., Wittinghofer A. (2013) Ras GTPase activating (RasGAP) activity of the dual specificity GAP protein Rasal requires colocalization and C2 domain binding to lipid membranes. Proc. Natl. Acad. Sci. U.S.A. 110, 111–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.