Abstract
The frequent lack of a positive and timely microbiological diagnosis in patients with lower respiratory tract infection (LRTI) is an important obstacle to antimicrobial stewardship. Patients are typically prescribed broad-spectrum empirical antibiotics while microbiology results are awaited, but, because these are often slow, negative, or inconclusive, de-escalation to narrow-spectrum agents rarely occurs in clinical practice. The aim of this study was to develop and evaluate two multiplex real-time PCR assays for the sensitive detection and accurate quantification of Streptococcus pneumoniae, Haemophilus influenzae, Staphylococcus aureus, Moraxella catarrhalis, Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii. We found that all eight bacterial targets could be reliably quantified from sputum specimens down to a concentration of 100 CFUs/reaction (8333 CFUs/mL). Furthermore, all 249 positive control isolates were correctly detected with our assay, demonstrating effectiveness on both reference strains and local clinical isolates. The specificity was 98% on a panel of nearly 100 negative control isolates. Bacterial load was quantified accurately when three bacterial targets were present in mixtures of varying concentrations, mimicking likely clinical scenarios in LRTI. Concordance with culture was 100% for culture-positive sputum specimens, and 90% for bronchoalveolar lavage fluid specimens, and additional culture-negative bacterial infections were detected and quantified. In conclusion, a quantitative molecular test for eight key bacterial causes of LRTI has the potential to provide a more sensitive decision-making tool, closer to the time-point of patient admission than current standard methods. This should facilitate de-escalation from broad-spectrum to narrow-spectrum antibiotics, substantially improving patient management and supporting efforts to curtail inappropriate antibiotic use.
Keywords: Bacteria, molecular diagnostics, PCR, quantitative, respiratory infection
Introduction
The frequent lack of a positive and timely microbiological diagnosis in patients with lower respiratory tract (LRT) infection (LRTI) is an important obstacle to antimicrobial stewardship. Patients are typically prescribed broad-spectrum empirical antibiotics while microbiology results are awaited, but, because these are often slow, negative, or inconclusive, de-escalation to narrow-spectrum agents rarely occurs in clinical practice.
Current standard diagnostic methods for respiratory bacteria are culture-based and typically take 24–72 h [1]. Culture also has low sensitivity; a positive microbiological diagnosis may only be made in approximately 30% of patients with community-acquired pneumonia [2]. As a wide range of pathogens can cause LRTI, it is common to request a mixture of different diagnostic tests, including culture, antigen testing, and serology, on a number of different specimen types [3]. In contrast, the routine use of viral multiplex real-time PCR (mRT-PCR) assays allows a single respiratory specimen to be screened for a large number of respiratory viruses easily within the working day [4].
We sought to improve the sensitivity and turn-around time of microbiological diagnosis of LRTI in our centre by developing a quantitative bacterial mRT-PCR approach. At the same time, we wanted to simplify the process by testing the same LRT specimen extract used for viral mRT-PCR and using the same PCR platforms, thus fitting in with our current workflow and minimizing costs. A key requirement was the ability to determine the bacterial load in order to provide information that may be clinically useful in excluding LRT specimen contamination with oral commensal bacteria.
Real-time PCR assays for some common respiratory bacteria have been described, but no single assay covers the wide range of Gram-positive and Gram-negative pathogens required for LRTI diagnosis. Furthermore, some assays have a lack of sensitivity and/or specificity for organisms such as Streptococcus pneumoniae and Haemophilus influenzae, owing to the use of suboptimal gene targets [3,5–10]. Several molecular respiratory pathogen panels are currently commercially available or in development. Typically, these are more expensive than PCRs developed in-house, lack a full range of viral and bacterial targets, are non-quantitative, or require specific testing platforms [11–13].
The aim of this study was to develop and evaluate two mRT-PCR assays for the sensitive detection and accurate quantification of the following eight respiratory bacterial pathogens: S. pneumoniae, H. influenzae, Staphylococcus aureus, and Moraxella catarrhalis (mRT-PCR 1), and Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii (mRT-PCR 2).
Materials and methods
Positive and negative control isolates
The positive control bacterial strains used in assay validation were as follows: S. pneumoniae American Type Culture Collection (ATCC) 49619, H. influenzae ATCC 9007, Staphylococcus aureus ATCC 29213, M. catarrhalis local laboratory reference strain, E. coli ATCC 35218, K. pneumoniae National Collection of Type Cultures (NCTC) 13442, P. aeruginosa ATCC 27853, and A. baumannii NCTC 13424. Phocine herpes virus (PhHV) was obtained as a viral cell culture stock from the Department of Virology, University Hospital Rotterdam, for use as an internal control. Plasmids containing assay target genes were generated by cloning PCR products with the pGEM-T Easy vector system (Promega, Southampton, UK). Plasmid extracts were diluted in carrier polyA RNA (Qiagen, Manchester, UK) at 0.05 mg/L in ten-fold dilution series for use in PCR optimization and as quantification standards. A large panel of control isolates were selected to include organisms targeted by the mRT-PCR 1 and mRT-PCR 2 assays, and 88 isolates closely related to the target organisms and/or commonly found in the respiratory tract as pathogens or commensals (Table 1). Isolates were obtained from the Royal Infirmary of Edinburgh Clinical Microbiology Laboratory, Scottish Haemophilus, Legionella, Meningococcus and Pneumococcus Reference Laboratory, Scottish MRSA Reference Laboratory, Scottish Bacterial Sexually Transmitted Infection Reference Laboratory, ATCC and NCTC Public Health England. Chlamydophila pneumoniae and Chlamydophila psittaci were commercially supplied as DNA extracts (Vircell, Granada, Spain). Well-characterized clinical isolates were from respiratory specimens wherever possible, and were identified by colonial morphology, standard biochemical methods, VITEK-2 (bioMérieux, Basingstoke, UK), Microflex matrix-assisted laser desorption ionization time-of-flight mass spectrometry (Bruker, Coventry, UK), and sequencing, as appropriate.
Table 1.
Specificity panel
| Organism | No. | Source/strain |
|---|---|---|
| Positive control isolates | ||
| Streptococcus pneumoniae | 36 | Clinical isolates (25); SHLMPRL isolates (10); ATCC 49619 |
| Haemophilus influenzae | 47 | Clinical isolates (25); SHLMPRL isolates (18) (serotypes A, B, D, E, F, NTHi); ATCC 9007 (serotype C); ATCC 49766 (NTHi); ATCC 49247 (NTHi); NCTC 8468 |
| Staphyloccoccus aureus | 41 | Clinical isolates (25); SMRSARL isolates (10); MRSA S113; ATCC 25923; ATCC 1026; ATCC BAA-976; ATCC BAA-977; ATCC 29213 |
| Moraxella catarrhalis | 26 | Clinical isolates (25); NCTC 11020 |
| Escherichia coli | 28 | Clinical isolates (25); NCTC 13476; ATCC 25922; ATCC 35218 |
| Klebsiella pneumoniae | 28 | Clinical isolates (25); NCTC 13442; NCTC 13443; NCTC 13439 |
| Pseudomonas aeruginosa | 26 | Clinical isolates (25); ATCC 27853 |
| Acinetobacter baumannii | 17 | Clinical isolates (16); NCTC 13424 |
| Negative control isolates (n = 88) | ||
| Abiotrophia adiacens | 1 | NCTC 13000 |
| Acinetobacter calcoaceticus | 1 | Clinical isolate |
| Acinetobacter haemolyticus | 1 | NCTC 12155 |
| Acinetobacter. johnsonii | 1 | NCTC 12154 |
| Acinetobacter junii | 1 | NCTC 12153 |
| Acinetobacter lwoffii | 1 | Clinical isolate |
| Acinetobacter nosocomialis | 1 | Clinical isolate |
| Actinomyces israelii | 1 | Clinical isolate |
| Aggregatibacter actinomycetemcomitans | 1 | NCTC 9710 |
| Aggregatibacter segnis | 1 | SHLMPRL isolate |
| Arcanobacterium haemolyticum | 1 | Clinical isolate |
| Bordetella parapertussis | 1 | NCTC 5952 |
| Bordetella pertussis | 1 | NEQAS 1505 |
| Chlamydophila pneumoniae (DNA) | 1 | CM-1 (VR1360) |
| Chlamydophila psittaci (DNA) | 1 | 6BC |
| Corynebacterium striatum | 1 | NCTC 764 |
| Eikenella corrodens | 1 | ATCC BAA-1152 |
| Enterobacter hormacecei | 1 | ATCC 700323 |
| Enterococcus casseliflavus | 1 | ATCC 700327 |
| Enterococcus faecalis | 1 | ATCC 51299 |
| Fusobacerium necrophorum | 1 | Clinical isolate |
| Gemella morbillorum | 1 | Clinical isolate |
| Haemophilus aphrophilus | 1 | Clinical isolate |
| Haemophilus haemolyticus | 1 | NCTC 10659 |
| Haemophilus parahaemolyticus | 1 | Clinical isolate |
| Haemophilus parainfluenzae | 10 | Clinical isolate |
| Haemophilus paraphrohaemolyticus | 1 | NCTC 10670 |
| Kingella kingae | 1 | NCTC 10529 |
| Klebsiella oxytoca | 1 | Clinical isolate |
| Legionella pneumophila | 1 | Clinical isolate |
| Micrococcus luteus | 1 | Clinical isolate |
| Moraxella atlantae | 1 | NCTC 11091 |
| Moraxella lacunata | 1 | NCTC 11011 |
| Moraxella nonliquefaciens | 1 | NCTC 10464 |
| Moraxella osloensis | 1 | Clinical isolate |
| Mycoplasma pneumoniae | 1 | NCTC 10119 |
| Mycoplasma salivarium | 1 | NCTC 10113 |
| Neisseria cinerea | 1 | SBSTIRL isolate |
| Neisseria gonorrhoeae | 1 | SBSTIRL isolate |
| Neisseria lactamica | 1 | SBSTIRL isolate |
| Neisseria meningitidis | 1 | SBSTIRL isolate |
| Neisseria mucosa | 1 | SBSTIRL isolate |
| Neisseria perflava | 1 | SBSTIRL isolate |
| Neisseria sicca | 1 | SBSTIRL isolate |
| Neisseria subflava | 1 | SBSTIRL isolate |
| Peptostreptococcus magnus | 1 | Clinical isolate |
| Porphyromonas gingivalis | 1 | NCTC 11834 |
| Prevotella intermedia | 1 | NCTC 13070 |
| Pseudomonas fluorescens | 1 | NCTC 10038 |
| Pseudomonas putida | 1 | NCTC 10936 |
| Pseudomonas stutzeri | 1 | NCTC 10475 |
| Staphylococcus capitis | 1 | SMRSARL isolate |
| Staphylococcus epidermidis | 3 | ATCC 12228, SMRSARL isolate (2) |
| Staphylococcus haemolyticus | 2 | SMRSARL isolate |
| Staphylococcus hominis | 1 | SMRSARL isolate |
| Staphylococcus intermedius | 1 | SMRSARL isolate |
| Staphylococcus lugdunensis | 1 | SMRSARL isolate |
| Staphylococcus pseudintermedius | 1 | SMRSARL isolate |
| Staphylococcus saphrophyticus | 1 | Clinical isolate |
| Staphylococcus warneri | 1 | SMRSARL isolate |
| Stenotrophomonas maltophila | 1 | ATCC 17666 |
| Streptococcus agalactiae | 1 | Clinical isolate |
| Streptococcus anginosus | 1 | Clinical isolate |
| Streptococcus bovis | 1 | NCTC 8177 |
| Streptococcus constellatus | 1 | Clinical isolate |
| Streptococcus gordonii | 1 | Clinical isolate |
| Streptococcus intermedius | 1 | Clinical isolate |
| Streptococcus mitis | 1 | SHLMPRL isolate |
| Streptococcus mutans | 1 | Clinical isolate |
| Streptococcus oralis | 1 | Clinical isolate |
| Streptococcus parasanguinis | 2 | SHLMPRL isolate, clinical isolate |
| Streptococcus pyogenes | 1 | Clinical isolate |
| Streptococcus salivarius | 1 | ATCC 19258 |
| Streptococcus sanguinis | 1 | Clinical isolate |
| Ureaplasma urealyticum | 1 | NCTC 10177 |
ATCC, American Type Culture Collection; MRSA, methicillin-resistant Staphylococcus aureus; NCTC, National Collection of Type Cultures, Public Health England; NTHi, non-typeable Haemophilus influenzae; SBSTIRL, Scottish Bacterial Sexually Transmitted Infection Reference Laboratory; SHLMPRL, Scottish Haemophilus, Legionella, Meningococcus and Pneumococcus Reference Laboratory; SMRSARL, Scottish MRSA Reference Laboratory.
Nucleic acid extraction
Pure cultures of control bacterial isolates were suspended in saline to 0.5 McFarland standard concentration, and total nucleic acid was extracted with the DNeasy Blood and Tissue kit (Qiagen), following the protocol for Gram-positive bacteria according to the manufacturer's instructions. Crude cell lysates were also made by boiling 150 μL of cell suspension for 10 min, centrifuging for 1 min at 11 000 g, and removing the supernatant for testing. Sputum specimens were initially centrifuged to pellet purulent material, and then physically homogenized in sterile viral transport medium with sterile glass beads. Total nucleic acid was extracted from 200 μL of clinical specimen with the following protocol: 60 min of incubation with 4 μL of enzymatic lysis buffer (20 mM Tris, pH 8, 2 mM EDTA, 1.2% Triton X-100) + 2 μL of lysozyme (100 mg/mL) at 37°C, 60 min of incubation with 25 μL of proteinase K (Qiagen) at 56°C, nucliSENS easyMAG (bioMérieux) automated extraction with PhHV internal control added to the lysis buffer, and elution in 100 μL.
Assay design
Candidate assay targets for the eight bacteria of interest were chosen on the basis of published validation data. The National Centre for Biotechnology Information standard nucleotide Basic Local Alignment Search Tool was used to check candidate oligonucleotide sequence matches to targets in the GenBank database. Assays were then redesigned or modified with Beacon Designer (Premier Biosoft, Palo Alto, CA, USA) and Autodimer [14] in order to optimize for multiplex performance. Optimized oligonucleotide sequences were checked by the Basic Local Alignment Search Tool against the GenBank database for in silico specificity. Sequences were also checked against alignments of all target gene sequences deposited in GenBank for the species of interest to check in silico sensitivity. On the basis of these assessments, eight targets were selected for pathogen detection, with four targets per mRT-PCR assay. The composition of each mRT-PCR assay is detailed in Table 2. Discrimination of each target in the reaction was achieved with the use of oligonucleotide probes labelled with one of four fluorophores: 6-FAM, Texas Red, Yakima Yellow, and Cy5. In order to assess the quality of LRT specimens, a real-time quantitative PCR assay was also designed with Beacon Designer (Premier Biosoft) and the RefSeq gene NG_007073.2 for the detection of the human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene. An already validated in-house real-time PCR assay for the PhHV gB gene was used to detect PCR inhibition [15].
Table 2.
Oligonucleotide sequences
| Assay | Organism | Gene target | Oligonucleotide | Final reaction concentration (μM) | Reference |
|---|---|---|---|---|---|
| mRT-PCR 1 | Streptococcus pneumoniae | Autolysin (lytA) | Forward: ACGCAATCTAGCAGATGAAGCA | 0.10 | [17] |
| Reverse: TCGTGCGTTTTAATTCCAGCT | 0.10 | ||||
| Probe: YY-TGCCGAAAACGCTTGATACAGGGAG-BHQ1 | 0.05 | ||||
| Haemophilus influenzae | l-Fuculokinase (fucK) | Forward: ATGGCGGGAACATCAATGA | 0.15 | [20] | |
| Reverse: ACGCATAGGAGGGAAATGGTT | 0.15 | ||||
| Probe: FAM-CGGTAATTGGGATCCAT-MGB | 0.10 | ||||
| Staphylococcus aureus | Thermostable nuclease (nuc) | Forward: AGCATCCTAAAAAAGGTGTAGAGA | 0.15 | [22] | |
| Reverse: CTTCAATTTTMTTTGCATTTTCTACCA | 0.15 | ||||
| Probe: TEX-TTTTCGTAAATGCACTTGCTTCAGGACCA-BHQ2 | 0.10 | ||||
| Moraxella catarrhalisa | Outer membrane protein (copB) | Forward: CGTGTTGACCGTTTTGACTTT | 0.15 | Modified from [23] | |
| Reverse: CATAGATTAGGTTACCGCTGACG | 0.15 | ||||
| Probe: Cy5-ACCGACATCAACCCAAGCTTTGG–BHQ3a | 0.10 | ||||
| mRT-PCR 2 | Escherichia colib | Conserved protein, function unknown (yccT) | Forward: ATCGTGACCACCTTGATT | 0.25 | Modified from [24] |
| Reverse: TACCAGAAGATCGACATC | 0.25 | ||||
| Probe: TEX-CATTATGTTTGCCGGTATCCGTTT-BHQ2 | 0.10 | ||||
| Klebsiella pneumoniae | Citrate synthase (gltA) | Forward: AGGCCGAATATGACGAAT | 0.25 | Modified from [24] Probe: this publication |
|
| Reverse: GGTGATCTGCTCATGAA | 0.25 | ||||
| Probe: YY-ACTACCGTCACCCGCCACA-BHQ1 | 0.10 | ||||
| Pseudomonas aeruginosa | DNA gyrase subunit B (gyrB) | Forward: CCTGACCATCCGTCGCCACAAC | 0.25 | [25] | |
| Reverse: CGCAGCAGGATGCCGACGCC | 0.25 | ||||
| Probe: FAM-CCGTGGTGGTAGACCTGTTCCCAGACC-BHQ1 | 0.10 | ||||
| Acinetobacter baumannii | OXA-51-like β-lactamase (blaOXA-51-like) | Forward: TTTAGCTCGTCGTATTGGACT | 0.125 | Modified from [28] | |
| Reverse: CCTCTTGCTGAGGAGTAATTTT | 0.125 | ||||
| Probe: Cy5-TGGCAATGCAGATATCGGTACCCA-BHQ3a | 0.05 | ||||
| Specimen quality control | Human | Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | Forward: TTGTCTCACTTGTTCTCT | 0.30 | This publication |
| Reverse: ATGGGAGTTGTTTTCTTG | 0.30 | ||||
| Probe: FAM-CTCGTCTTCTGTCATCTCTGCTG-BHQ1 | 0.20 | ||||
| Internal control for inhibition | PhHV | Glycoprotein B (gB) | Forward: GGGCGAATCACAGATTGAATC | 0.30 | [15] |
| Reverse: GCGGTTCCAAACGTACCAA | 0.30 | ||||
| Probe:Cy5-TTTTTATGTGTCCGCCACCATCTGGATC–BHQ3a | 0.10 |
Fluorophores: BHQ, Black Hole Quencher; TEX, Texas Red; YY, Yakima Yellow.
mRT-PCR, multiplex real-time PCR; PhHV, phocine herpes virus.
Moraxella catarrhalis assay detects Moraxella lacunata and Moraxella nonliquefaciens.
Escherichia coli assay detects Shigella species.
Real-time PCR
Reactions were carried out on the ABI 7500 Fast instrument (Applied Biosystems, Paisley, UK). mRT-PCR assays were carried out in a total volume of 20 μL, comprising 10 μL of QuantiFast Multiplex PCR mastermix (Qiagen), 2 μL of nuclease-free water (Promega), 2 μL of oligonucleotide mixture (Table 2), and 6 μL of nucleic acid extract. Cycle parameters were 95°C for 5 min, followed by 45 cycles of 95°C for 45 s and 60°C for 45 s. GAPDH real-time PCR was carried out with the same reaction components, but with different cycle parameters: 95°C for 5 min, followed by 45 cycles of 95°C for 30 s and 60°C for 30 s. PhHV PCR was carried out with 10 μL of Express qPCR Universal SuperMix (Invitrogen, Paisley, UK), 1 μL of oligonucleotide mixture (Table 2), and 9 μL of nucleic acid extract. Cycle parameters were 95°C for 20 s, followed by 45 cycles of 95°C for 3 s and 60°C for 30 s. Runs were accepted if negative (no template) controls were negative and positive controls for each amplification target were positive. For quantification, mixed plasmid dilution series ranging from 6 × 101 to 6 × 106 gene copies/reaction were included in each run. ABI 7500 Fast System SDS software v. 1.4 (Applied Biosystems) was used to construct a six-point standard curve and extrapolate a quantitative result. Runs were accepted if standard curves were linear (reaction efficiency of 90–119%, R2 > 0.98). Clinical specimens were classified as positive and quantifiable for a bacterial target if the bacterial load was ≥100 CFUs/reaction. Quantitative results were accepted if the internal control was positive with a quantification cycle (Cq) value within the range ±1 log (Cq ± 3.33) difference from negative extraction controls. CFUs/reaction were assumed to be equivalent to gene copies/reaction, because all of the bacterial target genes are single copy. Conversion of gene copies/reaction to CFUs/mL was based on a 6-μL input per PCR reaction drawn from 100 μL of nucleic acid extract concentrated from 200 μL of specimen.
Analytical performance
PCR efficiency (E = 10(−1/slope) – 1) was estimated by testing at least ten replicates of ten-fold mixed plasmid dilution series in different runs, and was acceptable between the values of 0.90 and 1.10. The quantitative range was determined from the range of ten-fold mixed plasmid dilutions that gave optimal linearity, efficiency, standard deviation and coefficient of variation between replicates. Analytical specificity was measured by testing DNA extracted from the large panel of positive and negative control isolates detailed above. Analytical sensitivity was estimated with surplus anonymized sputum specimens spiked before extraction with target bacterial species (mock specimens). These were tested at five different bacterial spike concentrations in triplicate per run, for four runs. A probit analysis was carried out to estimate the limit of detection of each assay (Minitab v.17). Precision by repeatability was used as a proxy for analytical accuracy, as reference standards for quantification of the eight bacterial pathogens are not available. To encompass between-run and within-run variation in both the extraction process and the PCR process, mock specimens at three different bacterial spike concentrations were tested in triplicate per run, for four runs on different days, with the same operator and equipment. To assess performance in quantification of mixed infections, plasmids were mixed to give unequal mixtures for three targets at 200 000, 20 000 and 200 gene copies/reaction. This was carried out for every combination of the four targets for both the mRT-PCR 1 assay and the mRT-PCR 2 assay (Table 3).
Table 3.
Matrix for triple mixture composition
| Mixture | 200 gene Copies/reaction | 20 000 gene copies/reaction | 200 000 gene copies/reaction |
|---|---|---|---|
| 1 | A | B | C |
| 2 | A | B | D |
| 3 | A | C | D |
| 4 | A | D | C |
| 5 | A | C | B |
| 6 | A | D | B |
| 7 | B | A | C |
| 8 | B | A | D |
| 9 | B | C | D |
| 10 | B | D | C |
| 11 | B | C | A |
| 12 | B | D | A |
| 13 | C | A | B |
| 14 | C | A | D |
| 15 | C | B | D |
| 16 | C | D | B |
| 17 | C | B | A |
| 18 | C | D | A |
| 19 | D | A | B |
| 20 | D | A | C |
| 21 | D | B | C |
| 22 | D | C | B |
| 23 | D | B | A |
| 24 | D | C | A |
For multiplex real-time PCR (mRT-PCR) 1: A = Streptococcus pneumoniae; B = Haemophilus influenzae; C = Staphylococcus aureus; D = Moraxella catarrhalis.
For mRT-PCR 2: A = Escherichia coli; B = Klebsiella pneumoniae; C = Pseudomonas aeruginosa; D = Acinetobacter baumannii.
Performance on clinical LRT specimens
Anonymized residual sputum specimens from patients with radiologically confirmed pneumonia and bronchalveolar lavage (BAL) fluid specimens from critical-care patients were collected between September 2012 and March 2015 following routine microbiological culture. For sputum specimens, a fleck of pus was inoculated directly onto Columbia horse blood agar and chocolate blood bacitracin agar (Oxoid, Basingstoke, UK), spread for the production of discrete colonies, and incubated for 48 h at 37°C in 5% CO2; microscopy was not carried out. Isolates were identified with standard biochemical methods and/or matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Results for sputum specimens were reported semiquantitatively in the following format: small numbers—colonies present in the well of the plate only; moderate numbers—colonies present in the first streak out from the well; and large numbers—colonies present in the second or third streak out from the well. Microbiological culture of BAL fluid specimens was carried out as for sputum specimens, with the exception that cultures were quantitative; plates were inoculated evenly with 10 μL of fluid, and results were reported as CFUs/mL. Residual specimens were stored at –80°C before being tested retrospectively for this study. Testing was carried out in accordance with local ethical approval (South East Scotland SAHSC Human Annotated BioResource reference No. 10/S1402/33). Datasets were subjected to a normality test, and differences between non-parametric data were tested for significance with the Mann–Whitney test (Minitab v.17).
Results
Assay design
The eight bacterial pathogens chosen for the mRT-PCR 1 and mRT-PCR 2 assays were the species most commonly detected by conventional culture-based methods in patients with pneumonia in a recent local study [2]. Assay development and validation were carried out according to recent guidance [16]. The lytA target was chosen for S. pneumoniae because it is well characterized, sensitive and specific for the identification of S. pneumoniae, and superior to other targets, such as ply [17,18]. The commonly used protein D-encoding gene hpd#3 assay target for H. influenzae was not employed in this study, owing to the known cross-reaction with Haemophilus aphrophilus [19]. The fucK target was chosen instead, because it had similar sensitivity for the detection of H. influenzae and discrimination from Haemophilus haemolyticus, and also performed slightly better than hpd#3 in a recent reference laboratory-based study [20,21]. The nuc target was chosen for Staphylococcus aureus detection because it has also been well evaluated in a reference laboratory setting [22]. Use of the copB gene target is widespread for the detection of M. catarrhalis, but some assays have primer mismatches with published M. catarrhalis copB sequences, so an assay in a more conserved region of the gene was chosen [23]. For the detection of E. coli and K. pneumoniae, assays targeting yccT and gltA, respectively, were used [24]. Owing to the high level of homology between E. coli and Shigella species, it was not possible to find an E. coli-specific assay target. However, as Shigella species are very unlikely to be found in the respiratory tract, this was not deemed to be clinically relevant in the context of LRTI diagnosis. The gyrB target was chosen for the detection of P. aeruginosa rather than the commonly used ecfX target, because of its slightly higher detection rate and specificity on clinical isolates [25–27]. The blaOXA-51-like gene chosen for A. baumannii detection has been found in A. baumannii isolates, with a few exceptions, and has not been detected in other Acinetobacter species [28–30].
Estimation of PCR efficiency, linearity, and quantitative range
Amplification was linear and efficient for all targets within the quantitative range 60–6 000 000 gene copies/reaction, with the exception of the gyrB target, which had a quantitative range of 600–6 000 000 gene copies/reaction (Table 4). Within this range, all replicates tested were positive, with Cq value standard deviations of 0.34–2.29 and coefficients of variation of 1.68–8.85% for all targets. Outside the quantitative range, assays were non-linear and less efficient, and low-concentration replicates were not consistently detected.
Table 4.
Analytical sensitivity and limit of detection; assay slope, linearity and efficiency were calculated for a quantitative range of 60–6 000 000 gene copies/reaction for all targets except for Pseudomonas aeruginosa, which had a quantitative range of 600–6 000 000 gene copies/reaction
| Number of replicates (%) detected for input bacterial spike, CFUs/reaction |
Limit of detection (95% level), CFUs/reaction | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Slope | Linearity (R2) | Efficiency | 100 000 CFUs/PCR (8 333 333 CFUs/mL) | 1000 CFUs/PCR (83 333 CFUs/mL) | 100 CFUs/PCR (8333 CFUs/mL) | 10 CFUs/PCR (833 CFUs/mL) | 1 CFU/PCR (83 CFUs/mL) | |||
| mRT-PCR 1 | Streptococcus pneumoniae | –3.18 | 0.92 | 1.06 | 12/12 (100) | 12/12 (100) | 12/12 (100) | 12/12 (100) | 2/9 (22) | 2.1 |
| Haemophilus influenzae | –3.11 | 0.97 | 1.10 | 12/12 (100) | 12/12 (100) | 12/12 (100) | 12/12 (100) | 12/12 (100) | 0.8 | |
| Staphylococcus aureus | –3.33 | 0.98 | 1.00 | 12/12 (100) | 12/12 (100) | 12/12 (100) | 7/9 (78) | 7/9 (78) | 15.5 | |
| Moraxella catarrhalis | –3.18 | 0.92 | 1.06 | 12/12 (100) | 12/12 (100) | 12/12 (100) | 6/12 (50) | 3/12 (25) | 23.1 | |
| mRT-PCR 2 | Escherichia coli | –3.27 | 0.95 | 1.05 | 12/12 (100) | 12/12 (100) | 11/11 (100) | 10/12 (83) | 8/12 (67) | 18.5 |
| Klebsiella pneumoniae | –3.43 | 0.93 | 1.00 | 12/12 (100) | 12/12 (100) | 11/11 (100) | 12/12 (100) | 8/12 (67) | 1.5 | |
| Pseudomonas aeruginosa | –3.46 | 0.96 | 0.96 | 12/12 (100) | 12/12 (100) | 10/11 (91) | 0/12 (0) | 0/12 (0) | 104.6 | |
| Acinetobacter baumannii | –3.20 | 0.96 | 1.08 | 12/12 (100) | 12/12 (100) | 11/11 (100) | 12/12 (100) | 5/12 (42) | 2.1 | |
| Control | Human | –3.37 | 0.99 | 0.98 | NA | NA | NA | NA | NA | NA |
mRT-PCR, multiplex real-time PCR; NA, not assessed.
Analytical sensitivity
Analytical sensitivity and limit of detection were estimated with nine to 12 replicates of sputum specimens spiked with bacteria at five different concentrations, from 1 to 100 000 CFUs/reaction (Table 4). Both the mRT-PCR 1 assay and the mRT-PCR 2 assay were highly sensitive for the detection of all bacteria in mock specimens down to the level of 100 CFUs/reaction, except for P. aeruginosa, for which the assays were approximately 1 log less sensitive. Limits of detection were approximately 1–100 CFUs/reaction (corresponding to 83–8333 CFUs/mL), depending on the target.
Analytical specificity
Coverage of a large panel of control isolates was 100% for all assay targets, and there were no cross-reactions between targets within the mRT-PCR 1 assay and the mRT-PCR 2 assay. Specificity against a panel of 88 isolates closely related to the target organisms and/or commonly found in the respiratory tract as pathogens or commensals was 100% for all targets, with the exception of copB, which was 98% specific, owing to cross-reactivity with Moraxella lacunata and Moraxella nonliquefaciens (Tables 5 and 6). However, the copB assay did not detect Moraxella osloensis or Moraxella atlantae.
Table 5.
Analytical specificity summary for multiplex real-time PCR 1 detection (no. (%))
| Isolates | Streptococcus pneumoniae | Haemophilus influenzae | Staphylococcus aureus | Moraxella catarrhalis |
|---|---|---|---|---|
| Streptococcus pneumoniae | 36/36 (100) | 0/36 (0) | 0/36 (0) | 0/36 (0) |
| Haemophilus influenzae | 0/47 (0) | 47/47 (100) | 0/47 (0) | 0/47 (0) |
| Staphylococcus aureus | 0/41 (0) | 0/41 (0) | 41/41 (100) | 0/41 (0) |
| Moraxella catarrhalis | 0/26 (0) | 0/26 (0) | 0/26 (0) | 26/26 (100) |
| Panel of 88 respiratory and related organisms + Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii | 0/92 (0) | 0/92 (0) | 0/92 (0) | 2/92a (2) |
Moraxella lacunata and Moraxella nonliquefaciens were copB positive.
Table 6.
Analytical specificity summary for multiplex real-time PCR 2 detection (no. (%))
| Isolates | Escherichia coli | Klebsiella pneumoniae | Pseudomonas aeruginosa | Acinetobacter baumannii |
|---|---|---|---|---|
| Escherichia coli | 28/28 (100) | 0/28 (0) | 0/28 (0) | 0/28 (0) |
| Klebsiella pneumoniae | 0/28 (0) | 28/28 (100) | 0/28 (0) | 0/28 (0) |
| Pseudomonas aeruginosa | 0/26 (0) | 0/26 (0) | 26/26 (100) | 0/26 (0) |
| Acinetobacter baumannii | 0/17 (0) | 0/17 (0) | 0/17 (0) | 17/17 (100) |
| Panel of 88 respiratory and related organisms + Streptococcus pneumoniae, Haemophilus influenzae, Staphylococcus aureus and Moraxella catarrhalis | 0/92 (0) | 0/92 (0) | 0/92 (0) | 0/92 (0) |
Precision (repeatability)
Precision was measured by comparing bacterial load quantification by the mRT-PCR 1 assay and the mRT-PCR 2 assay for 11–12 replicates of PCR-negative sputum specimens spiked with each of the eight target bacteria at high (100 000 CFUs/reaction), medium (1000 CFUs/reaction) and low (100 CFUs/reaction) concentrations. The numbers of replicates with bacterial load quantified within a 1 log CFUs/reaction range for high, medium and low target concentrations were 96 of 96 (100%), 89 of 96 (92.7%), and 86 of 92 (93.5%), respectively. The most precise assays were for S. pneumoniae and H. influenzae, because all replicates were within a 1 log CFUs/reaction range. The least precise assays were for Staphylococcus aureus, M. catarrhalis, and P. aeruginosa; these quantified 33 of 36 (91.7%), 32 of 36 (88.9%) and 32 of 35 (91.4%) replicates, respectively, within the 1 log range. Overall, the results indicated that reliable bacterial load quantification from sputum specimens was achievable with both assays down to a concentration of 100 CFUs/reaction, which is equivalent to 8333 CFUs/mL.
Detection of targets in unequal triple mixtures
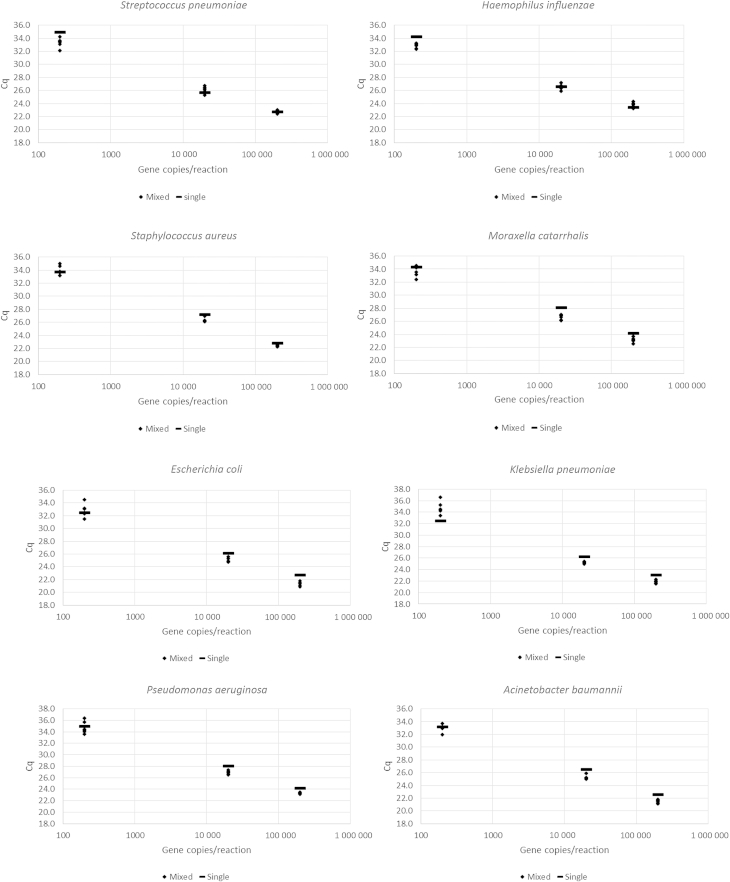
The Cq value for each target in a mixture was similar to the Cq value for that target alone, and all values were tightly clustered, with standard deviations within the range 0.14–1.10 (Fig. 1). Therefore, all components within a triple mixture, including the minority component, could be accurately quantified despite the presence of other components at different concentrations.
Fig. 1.
Distribution of Cq values in triple unequal mixture (n = 6) and single (n = 1) plasmid positive control material for each assay target at three concentrations (200, 20 000 and 200 000 gene copies/reaction).
Testing of clinical specimens
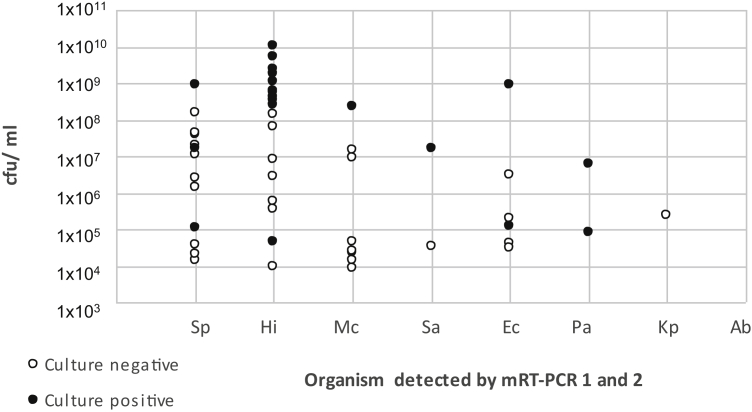
Twenty consecutive culture-negative sputum specimens obtained from individual patients with pneumonia were retrospectively tested with the novel assays. Specimens had a median GAPDH concentration of 139 659 gene copies/reaction (range: 10 853–1 030 000 gene copies/reaction). Twelve of 20 (65%) specimens were positive with the mRT-PCR 1 and mRT-PCR 2 assays, with seven (58.3%) single infections and five (41.7%) mixed infections (Table 7). In addition, a further 20 consecutive sputum specimens, culture positive for any of the eight bacterial pathogens targeted by the mRT-PCR 1 and mRT-PCR 2 assays, were retrospectively tested with the novel assays. These specimens had a median GAPDH concentration of 134 000 gene copies/reaction (range: 4876–729 000 gene copies/reaction), which was not significantly different from that in the culture-negative group (p 0.968). In 20 of 20 (100%) sputum specimens, the mRT-PCR 1 and mRT-PCR 2 assays detected the same bacterial species grown in culture (Table 7). Furthermore, the novel PCR assays detected an additional 15 bacterial infections, giving a total of 11 (55%) single infections and nine (45%) mixed infections. By semiquantitative sputum culture, the majority of isolates present were reported as large numbers of organisms, corresponding to between 4.68 × 104 CFUs/mL and 1.10 × 1010 CFUs/mL as measured by the mRT-PCR assay (Table 7). However, bacterial loads were significantly lower in the culture-negative group (median: 3.50 × 105 CFUs/mL) than in the culture-positive group (median: 3.33 × 108 CFUs/mL) (p 0.0001) (Fig. 2). Finally, 20 consecutive BAL fluid specimens obtained from individual patients admitted to critical care were retrospectively tested with the novel assays (Table 7). GAPDH was quantified in 18 of 20 BAL fluid specimens, and this gave a median GAPDH concentration of 24 720 gene copies/reaction (range: 892–176 356 gene copies/reaction), which was significantly lower than in sputum specimens (p 0.0006). Concordant results for bacterial targets in the mRT-PCR assays were achieved for 18 of 20 specimens, with seven additional bacterial infections being identified by PCR. Two specimens gave discordant results for bacterial targets present in the mRT-PCR assays: one specimen grew 3.0 × 102 CFUs/mL of S. aureus and was PCR negative; a second specimen grew 1.0 × 104 CFUs/mL of K. pneumoniae and was K. pneumoniae PCR negative at just below the 100 CFUs/reaction cut-off for the assay. When culture-positive specimens were detected by the mRT-PCR assays, quantitative PCR bacterial loads were in the same order of magnitude as or higher than those of quantitative culture (Table 7). Candida species and Morganella morganii were identified in five specimens, but targets for these organisms were not present in the PCR assays. In three specimens, overgrowth of respiratory flora is likely to have masked the existence of respiratory pathogens; culture results were reported as ‘no significant growth’ or ‘upper respiratory tract flora’, but the mRT-PCR assays identified >104 CFUs/mL of at least one pathogen.
Table 7.
Bacterial detection by multiplex real-time PCR (mRT-PCR) 1 and mRT-PCR 2 assays in lower respiratory tract specimens (sputa, n = 40; bronchoalveolar lavage (BAL), n = 20)
| Specimen type (no. of specimens) | Standard culture | mRT-PCR 1 and mRT-PCR 2 result |
|---|---|---|
| Sputum | No growth | Haemophilus influenzae 2.81 × 106 CFUs/mL |
| Sputum | No growth | H. influenzae 5.56 × 105 CFUs/mL |
| Sputum | No growth | H. influenzae 6.31 × 107 CFUs/mL |
| Sputum | No growth | H. influenzae 3.43 × 105 CFUs/mL |
| Sputum | No growth | Streptococcus pneumoniae 3.87 × 107 CFUs/mL |
| Sputum | No growth | S. pneumoniae 4.26 × 107 CFUs/mL |
| Sputum | No growth | Escherichia coli 1.91 × 105 CFUs/mL |
| Sputum | No growth |
H. influenzae 1.47 × 108 CFUs/mL S. pneumoniae 1.99 × 104 CFUs/mL |
| Sputum | No growth |
H. influenzae 8.42 × 106 CFUs/mL Moraxella catarrhalis 1.46 × 104 CFUs/mL |
| Sputum | No growth |
H. influenzae 3.50 × 105 CFUs/mL E. coli 4.18 × 104 CFUs/mL |
| Sputum | No growth |
H. influenzae 9.76 × 103 CFUs/mL S. pneumoniae 1.43 × 106 CFUs/mL M. catarrhalis 8.74 × 103 CFUs/mL |
| Sputum | No growth |
S. pneumoniae 1.12 × 107 CFUs/mL E. coli 3.11 × 104 CFUs/mL |
| Sputum (eight specimens) | No growth | Negative |
| Sputum | H. influenzae (LN) | H. influenzae 2.52 × 109 CFUs/mL |
| Sputum | H. influenzae (LN) | H. influenzae 1.14 × 109 CFUs/mL |
| Sputum | H. influenzae (LN) | H. influenzae 4.06 × 108 CFUs/mL |
| Sputum | H. influenzae (LN) | H. influenzae 5.48 × 108 CFUs/mL |
| Sputum | H. influenzae (LN) | H. influenzae 5.10 × 109 CFUs/mL |
| Sputum | H. influenzae (LN) | H. influenzae 3.24 × 108 CFUs/mL |
| Sputum | H. influenzae (SN) |
H. influenzae 2.49 × 108 CFUs/mL S. pneumoniae 2.47 × 106 CFUs/mL |
| Sputum | H. influenzae (LN) |
H. influenzae 3.41 × 108 CFUs/mL S. pneumoniae 1.58 × 108 CFUs/mL |
| Sputum | H. influenzae (LN) |
H. influenzae 1.10 × 1010 CFUs/mL M. catarrhalis 2.22 × 104 CFUs/mL |
| Sputum | H. influenzae (LN) |
H. influenzae 4.68 × 104 CFUs/mL M. catarrhalis 9.42 × 106 CFUs/mL |
| Sputum | H. influenzae (LN) |
H. influenzae 1.77 × 109 CFUs/mL S. pneumoniae 3.79 × 104 CFUs/mL M. catarrhalis 2.45 × 104 CFUs/mL Staphylococcus aureus 3.49 × 104 CFUs/mL |
| Sputum | S. pneumoniae (LN) | S. pneumoniae 8.67 × 108 CFUs/mL |
| Sputum | S. pneumoniae (LN) |
S. pneumoniae 1.65 × 107 CFUs/mL H. influenzae 9.42 × 103 CFUs/mL M. catarrhalis 4.43 × 104 CFUs/mL |
| Sputum | S. pneumoniae (LN) |
S. pneumoniae 1.12 × 105 CFUs/mL H. influenzae 6.28 × 108 CFUs/mL M. catarrhalis 1.42 × 107 CFUs/mL E. coli 2.95 × 106 CFUs/mL Klebsiella pneumoniae 2.36 × 105 CFUs/mL |
| Sputum | E. coli (LN) | E. coli 8.92 × 108 CFUs/mL |
| Sputum | E. coli (LN) |
E. coli 1.17 × 105 CFUs/mL S. pneumoniae 1.42 × 104 CFUs/mL |
| Sputum | Pseudomonas aeruginosa (MN) | P. aeruginosa 8.27 × 104 CFUs/mL |
| Sputum | P. aeruginosa (LN) |
P. aeruginosa 6.02 × 106 CFUs/mL S. pneumoniae 1.97 × 107 CFUs/mL |
| Sputum | M. catarrhalis (LN) | M. catarrhalis 2.40 × 108 CFUs/mL |
| Sputum | Staphylococcus aureus (MN) | Staphylococcus aureus 1.59 × 107 CFUs/mL |
| BAL (seven specimens) | No growth | Negative |
| BAL | No growth | H. influenzae 9.71 × 103 CFUs/mL |
| BAL | No significant growth | H. influenzae 5.07 × 106 CFUs/mL |
| BAL | No significant growth |
E. coli 1.29 × 105 CFUs/mL Staphylococcus aureus 1.85 × 104 CFUs/mL |
| BAL | Upper respiratory tract flora Candida albicans 2 × 102 CFUs/mL |
S. pneumoniae 4.30 × 105 CFUs/mL |
| BAL | Candida albicans 7 × 103 CFUs/mL | Negative |
| BAL | Candida glabrata | Negative |
| BAL | Morganella morganii 1 × 104 CFUs/mL | Negative |
| BAL |
E. coli 2 × 103 CFUs/mL S. pneumoniae 1 × 104 CFUs/mL H. influenzae 1 × 104 CFUs/mL |
E. coli 3.31 × 105 CFUs/mL S. pneumoniae 2.90 × 106 CFUs/mL H. influenzae 6.21 × 107 CFUs/mL |
| BAL | P. aeruginosa >1 × 104 CFUs/mL | P. aeruginosa 2.21 × 105 CFUs/mL |
| BAL | E. coli >1 × 104 CFUs/mL |
E. coli 1.48 × 108 CFUs/mL (M. catarrhalis target just below cut-off) |
| BAL |
E. coli 3 × 102 CFUs/mL Candida tropicalis >1 × 104 CFUs/mL |
E. coli 8.38 × 105 CFUs/mL H. influenzae 3.26 × 105 CFUs/mL |
| BAL | Staphylococcus aureus 3 × 102 CFUs/mL | Negative |
| BAL | K. pneumoniae 1 × 104 CFUs/mL |
Staphylococcus aureus 5.71 × 104 CFUs/mL (K. pneumoniae target just below cut-off) |
LN, large numbers; MN, moderate numbers; SN, small numbers.
Fig. 2.
Bacterial loads calculated by multiplex real-time PCR (mRT-PCR) in culture-positive (n = 20) and culture-negative (n = 20) sputum specimens from patients with pneumonia. Ab, Acinetobacter baumannii; Ec, Escherichia coli; Hi, Haemophilus influenzae; Kp, Klebsiella pneumoniae; Mc, Moraxella catarrhalis; Pa, Pseudomonas aeruginosa; Sa, Staphylococcus aureus; Sp, Streptococcus pneumoniae.
Discussion
We have described the development and validation of two quantitative mRT-PCR assays for eight important respiratory bacterial pathogens, and demonstrated their potential as an improved diagnostic tool in LRTI. Through in silico analysis and in vitro experimentation, we have designed assays that sensitively and specifically detect the targeted bacterial species. Bacterial targets could be reliably quantified from sputum specimens down to a concentration of 100 CFUs/reaction or approximately 8000 CFUs/mL.
A key strength of our study was that all 249 positive control isolates were correctly detected with our assay, demonstrating its effectiveness on both reference strains and local clinical isolates. Concordance with culture for 20 culture-positive sputum specimens was also 100%. In addition, specificity on a panel of nearly 100 negative control isolates was 98%. Furthermore, the two assays quantified bacterial load accurately when three bacterial targets were present in mixtures of varying concentrations, mimicking likely clinical scenarios in LRTI. These assays are combined with controls for specimen quality and reaction inhibition, and run on a standard fast real-time PCR platform, enabling results to be produced within 6 h of specimen receipt. To the best of our knowledge, there are no other quantitative molecular assays for LRTI diagnosis that have been as extensively validated and meet these criteria.
One of the weaknesses of our study was that an extended extraction protocol was required before processing on an automated platform, increasing manual complexity and turn-around time. This was necessary for efficient extraction of Staphylococcus aureus DNA from sputum specimens, as we had previously found that mock specimens containing Staphylococcus aureus spikes were underquantified with standard automated extraction only. Enzymatic lysis of Staphylococcus aureus with achromopeptidase has been described as both rapid and efficient, and therefore warrants further investigation as an alternative method in this setting [31]. However, when particularly mucopurulent specimens, such as sputa, are used, an additional proteinase K incubation step may still be required, in order to fully break down the material before addition to an automated extraction system. Despite the limitations of the current protocol, results are achievable in 6 h, owing to the use of automated systems and fast mRT-PCR, thus enabling same-day reporting for specimens received in the morning. This is still quicker than culturing even the faster-growing Gram-negative pathogens in our panel, and means that a comprehensive multi-pathogen result is available at a single time-point.
We set criteria for acceptance of quantification results based on the Cq of the internal control falling within a 1 log range; outside of this range, quantitative results were not accepted, owing to the potential for partial PCR inhibition leading to inaccuracy of measurement. However, as our quantitative mRT-PCR and internal control assays utilize different reagents, they may not be equally affected by the presence of PCR inhibitors, and therefore a bacterial load output from an assay meeting the quality control criteria could still potentially be an underestimate. This was not an issue during the sputum bacterial spiking experiments carried out here, although these were limited in number. Similarly, testing of BAL fluid specimens showed that quantitative PCR bacterial loads were generally higher than those obtained with quantitative culture, but BAL fluid is significantly less cellular than sputum, and would therefore be expected to be less inhibitory.
Furthermore, we tested only a small number of sputum and BAL specimens for comparison with culture in a proof-of-principle experiment. However, this demonstrated the potential clinical utility of our molecular assays and the suitability of these two contrasting LRT specimen types. It was clear from our small collection of clinical specimens that there is an added benefit with mRT-PCR; it can detect more bacterial infections than culture alone, and provide a simultaneous quantitative output. Specimens that were culture positive for any of the eight bacterial pathogens targeted by the mRT-PCR assays had higher bacterial loads than those that were culture negative, indicating the increased sensitivity of PCR as compared with culture. This may prove to be particularly useful in the hospital setting, where patients are more likely to have prior antibiotic exposure before specimens are taken, thus reducing bacterial burden and viability.
A significant issue that we discovered during development was that the copB assay for M. catarrhalis cross-reacted with M. nonliquefaciens and M. lacunata isolates. Although several other authors have used assays based on the copB gene to target M. catarrhalis [23,32–34], none have reported its specificity against M. nonliquefaciens, and only one study tested M. lacunata, with no cross-reactivity being detected [32]. In silico analysis did not predict copB cross-reactions, because of a lack of genomic sequence data and copB gene sequences for Moraxella species in GenBank. M. nonliquefaciens and M. lacunata are members of the normal respiratory microbiota, but rarely cause respiratory tract infection themselves [35]. The paucity of sequence data makes design of a more specific M. catarrhalis assay problematic, and therefore the clinical relevance of this cross-reaction is uncertain at present. However, the flexibility of an in-house mRT-PCR format means that, once further sequence information is available, a modified M. catarrhalis assay could be readily incorporated and revalidated. In the meantime, we recommend that other groups who currently use the copB gene target for M. catarrhalis be aware of the potential for cross-reactivity with some Moraxella species when reporting results from direct detection on respiratory specimens. Furthermore, as the copB assay did not cross-react with M. osloensis or M. atlantae, this assay cannot be regarded as a genus-specific Moraxella species assay. Cross-reactivity is also an issue for E. coli PCR assays, owing to the high level of homology between E. coli and Shigella species. However, as Shigella species are very unlikely to be found in the respiratory tract, this is unlikely to be diagnostically relevant in a respiratory context.
In order to enable accurate molecular diagnosis of LRTI, a specimen should ideally be: (a) obtained from the LRT rather than the upper respiratory tract (URT); and (b) of good quality, i.e. purulent material rather than salivary in the case of sputum. This is because of the potential for detection of colonization rather than infection when URT specimens are used, and the potential for contamination of LRT specimens with oral flora during sampling. For this reason, our assays have been developed for use on sputum and BAL fluid specimens rather than throat swabs or nasopharyngeal aspirates, and have an in-built measure of cellular content for quality control (GAPDH). Microscopy can be carried out to determine the quality of a sputum specimen based on the numbers of polymorphonuclear leukocytes and squamous epithelial cells [1], but GAPDH is present in all cells, and therefore does not allow such discrimination. Sputum microscopy is not routinely carried out in our centre, so we do not know to what extent our cellular load outputs were likely to be influenced by contaminating epithelial cells from the URT, but we deliberately used only macroscopically purulent material from sputum specimens for PCR in this study for a direct comparison with culture.
Rates of URT asymptomatic carriage of S. pneumoniae and H. influenzae in adults have been studied most frequently by PCR, and range from 2% and 39%, depending on the setting [36,37]. Furthermore, molecular studies using paired specimens have shown that the bacteria and viruses detected in the URT may not accurately reflect the organisms detected in the LRT [38–40]. However, quantification of bacterial DNA load may be important in distinguishing infection from asymptomatic colonization; most work described to date has focused on S. pneumoniae, and a cut-off of 104 to 105 gene copies/mL has typically been described [3,41–43]. Therefore, our assay could be used to investigate whether similar clinical cut-offs can be determined for other respiratory bacteria and for conditions other than pneumonia. In comparison with semiquantitative sputum culture in patients with pneumonia, we found that, although the majority of isolates were reported as ‘large numbers’ of organisms in monomicrobial infections, a wide range of CFUs/mL was measured by quantitative PCR and many organisms were detected in mixed infections in different proportions. In some cases, the organism grown in culture was not the organism with the highest bacterial load in a polymicrobial infection as determined by quantitative PCR. Overall, these observations may indicate the tendency of one organism to overgrow others in culture, leading to under-reporting of mixed infections. Our assay could also be used in settings where quantitative culture of respiratory specimens is the norm and rapid diagnosis is of key importance. For example, the use of quantitative culture for BAL fluid is standard for the microbiological diagnosis of ventilator-acquired pneumonia, with a cut off of 104 CFUs/mL [44]. As proof-of-principle, we were able to quantify bacterial loads in BAL fluid specimens from critical-care patients with comparable results to those obtained with culture, and many of the major ventilator-acquired pneumonia pathogens are included in our assay. Therefore, the use of mRT-PCR assays could provide a faster quantitative result in this setting. Although not validated for use here, there is also scope to use our mRT-PCR assays on whole blood specimens, as higher DNA loads have been found to correlate with disease severity in some studies [45–47].
In conclusion, a quantitative molecular test for the key bacterial causes of LRTI has the potential to provide a more sensitive decision-making tool, closer to the time-point of patient admission, than current standard methods. A faster and more accurate microbiological diagnosis should facilitate de-escalation from broad-spectrum to narrow-spectrum antibiotics, substantially improving patient management and supporting efforts to curtail inappropriate antibiotic use.
Transparency declaration
The authors have no conflicts of interest to declare for this work.
Acknowledgements
This work was supported by grant ETM-250 from the Chief Scientist Office Scotland. The authors would like to thank L. Mackenzie and I. Serrano for assistance with specimen collection, and the staff of the Royal Infirmary of Edinburgh Clinical Microbiology Laboratory, Scottish Haemophilus, Legionella, Meningococcus and Pneumococcus Reference Laboratory, Scottish MRSA Reference Laboratory and Scottish Bacterial Sexually Transmitted Infection Reference Laboratory for kindly providing clinical isolates.
Editor: G. Greub
References
- 1.Public Health England. Investigation of bronchoalveolar lavage, sputum and associated specimens. UK Standards for Microbiology Investigations 2014; B 57: Issue 2.5. http://www.hpa.org.uk/SMI/pdf.
- 2.Chalmers J.D., Taylor J.K., Singanayagam A., Fleming G.B., Akram A.R., Mandal P. Epidemiology, antibiotic therapy, and clinical outcomes in health care-associated pneumonia: QA UK cohort study. Clin Infect Dis. 2011;53:107–113. doi: 10.1093/cid/cir274. [DOI] [PubMed] [Google Scholar]
- 3.Johansson N., Kalin M., Tiveljung-Lindell A., Giske C.G., Hedlund J. Etiology of community-acquired pneumonia: Increased microbiological yield with new diagnostic methods. Clin Infect Dis. 2010;50:202–209. doi: 10.1086/648678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Templeton K.E., Scheltinga S.A., Beersma M.F., Kroes A.C., Claas E.C. Rapid and sensitive method using multiplex real-time PCR for diagnosis of infections by influenza A and influenza B viruses, respiratory syncytial virus, and parainfluenza viruses 1, 2, 3, and 4. J Clin Microbiol. 2004;42:1564–1569. doi: 10.1128/JCM.42.4.1564-1569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdeldaim G.M., Stralin K., Korsgaard J., Blomberg J., Welinder-Olsson C., Herrmann B. Multiplex quantitative PCR for detection of lower respiratory tract infection and meningitis caused by Streptococcus pneumoniae, Haemophilus influenzae and Neisseria meningitidis. BMC Microbiol. 2010;10:310. doi: 10.1186/1471-2180-10-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curran T., Coyle P.V., McManus T.E., Kidney J., Coulter W.A. Evaluation of real-time PCR for the detection and quantification of bacteria in chronic obstructive pulmonary disease. FEMS Immunol Med Microbiol. 2007;50:112–118. doi: 10.1111/j.1574-695X.2007.00241.x. [DOI] [PubMed] [Google Scholar]
- 7.Kais M., Spindler C., Kalin M., Ortqvist A., Giske C.G. Quantitative detection of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis in lower respiratory tract samples by real-time PCR. Diagn Microbiol Infect Dis. 2006;55:169–178. doi: 10.1016/j.diagmicrobio.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Rice L.M., Reis A.H., Jr., Mistry R., Khan H., Khosla P., Bharya S. Design and construction of a single tube, quantitative endpoint, LATE-PCR multiplex assay for ventilator-associated pneumonia. J Appl Microbiol. 2013;115:818–827. doi: 10.1111/jam.12281. [DOI] [PubMed] [Google Scholar]
- 9.Rios-Licea M.M., Bosques F.J., Arroliga A.C., Galindo-Galindo J.O., Garza-Gonzalez E. Quadruplex real-time quantitative PCR assay for the detection of pathogens related to late-onset ventilator-associated pneumonia: a preliminary report. J Microbiol Methods. 2010;81:232–234. doi: 10.1016/j.mimet.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 10.Stralin K., Backman A., Holmberg H., Fredlund H., Olcen P. Design of a multiplex PCR for Streptococcus pneumoniae, Haemophilus influenzae, Mycoplasma pneumoniae and Chlamydophila pneumoniae to be used on sputum samples. APMIS. 2005;113:99–111. doi: 10.1111/j.1600-0463.2005.apm1130203.x. [DOI] [PubMed] [Google Scholar]
- 11.Benson R., Tondella M.L., Bhatnagar J., Carvalho Mda G., Sampson J.S., Talkington D.F. Development and evaluation of a novel multiplex PCR technology for molecular differential detection of bacterial respiratory disease pathogens. J Clin Microbiol. 2008;46:2074–2077. doi: 10.1128/JCM.01858-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bogaerts P., Hamels S., de Mendonca R., Huang F.D., Roisin S., Remacle J. Analytical validation of a novel high multiplexing real-time PCR array for the identification of key pathogens causative of bacterial ventilator-associated pneumonia and their associated resistance genes. J Antimicrob Chemother. 2013;68:340–347. doi: 10.1093/jac/dks392. [DOI] [PubMed] [Google Scholar]
- 13.Jamal W., Al Roomi E., AbdulAziz L.R., Rotimi V.O. Evaluation of Curetis Unyvero, a multiplex PCR-based testing system, for rapid detection of bacteria and antibiotic resistance and impact of the assay on management of severe nosocomial pneumonia. J Clin Microbiol. 2014;52:2487–2492. doi: 10.1128/JCM.00325-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vallone P.M., Butler J.M. AutoDimer: a screening tool for primer-dimer and hairpin structures. Biotechniques. 2004;37:226–231. doi: 10.2144/04372ST03. [DOI] [PubMed] [Google Scholar]
- 15.Niesters H.G.M. Quantification of viral load using real-time amplification techniques. Methods. 2001;25:419–429. doi: 10.1006/meth.2001.1264. [DOI] [PubMed] [Google Scholar]
- 16.Saunders N., Zambon M., Sharp I., Siddiqui R., Bermingham A., Ellis J. Guidance on the development and validation of diagnostic tests that depend on nucleic acid amplification and detection. J Clin Virol. 2013;56:260–270. doi: 10.1016/j.jcv.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 17.Carvalho Mda G., Tondella M.L., McCaustland K., Weidlich L., McGee L., Mayer L.W. Evaluation and improvement of real-time PCR assays targeting lytA, ply, and psaA genes for detection of pneumococcal DNA. J Clin Microbiol. 2007;45:2460–2466. doi: 10.1128/JCM.02498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wessels E., Schelfaut J.J., Bernards A.T., Claas E.C. Evaluation of several biochemical and molecular techniques for identification of Streptococcus pneumoniae and Streptococcus pseudopneumoniae and their detection in respiratory samples. J Clin Microbiol. 2012;50:1171–1177. doi: 10.1128/JCM.06609-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X., Mair R., Hatcher C., Theodore M.J., Edmond K., Wu H.M. Detection of bacterial pathogens in Mongolia meningitis surveillance with a new real-time PCR assay to detect Haemophilus influenzae. Int J Med Microbiol. 2011;301:303–309. doi: 10.1016/j.ijmm.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Meyler K.L., Meehan M., Bennett D., Cunney R., Cafferkey M. Development of a diagnostic real-time polymerase chain reaction assay for the detection of invasive Haemophilus influenzae in clinical samples. Diagn Microbiol Infect Dis. 2012;74:356–362. doi: 10.1016/j.diagmicrobio.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 21.Zhang B., Kunde D., Tristram S. Haemophilus haemolyticus is infrequently misidentified as Haemophilus influenzae in diagnostic specimens in Australia. Diagn Microbiol Infect Dis. 2014;80:272–273. doi: 10.1016/j.diagmicrobio.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 22.Pichon B., Hill R., Laurent F., Larsen A.R., Skov R.L., Holmes M. Development of a real-time quadruplex PCR assay for simultaneous detection of nuc, Panton–Valentine leucocidin (PVL), mecA and homologue mecALGA251. J Antimicrob Chemother. 2012;67:2338–2341. doi: 10.1093/jac/dks221. [DOI] [PubMed] [Google Scholar]
- 23.Dunne E.M., Manning J., Russell F.M., Robins-Browne R.M., Mulholland E.K., Satzke C. Effect of pneumococcal vaccination on nasopharyngeal carriage of Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis, and Staphylococcus aureus in Fijian children. J Clin Microbiol. 2012;50:1034–1038. doi: 10.1128/JCM.06589-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clifford R.J., Milillo M., Prestwood J., Quintero R., Zurawski D.V., Kwak Y.I. Detection of bacterial 16S rRNA and identification of four clinically important bacteria by real-time PCR. PLoS One. 2012;7:e48558. doi: 10.1371/journal.pone.0048558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anuj S.N., Whiley D.M., Kidd T.J., Bell S.C., Wainwright C.E., Nissen M.D. Identification of Pseudomonas aeruginosa by a duplex real-time polymerase chain reaction assay targeting the ecfX and the gyrB genes. Diagn Microbiol Infect Dis. 2009;63:127–131. doi: 10.1016/j.diagmicrobio.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 26.Kidd T.J., Ramsay K.A., Hu H., Bye P.T.P., Elkins M.R., Grimwood K. Low rates of Pseudomonas aeruginosa misidentification in isolates from cystic fibrosis patients. J Clin Microbiol. 2009;47:1503–1509. doi: 10.1128/JCM.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Gall F., Le Berre R., Rosec S., Hardy J., Gouriou S., Boisame-Gastrin S. Proposal of a quantitative PCR-based protocol for an optimal Pseudomonas aeruginosa detection in patients with cystic fibrosis. BMC Microbiol. 2013;13:143. doi: 10.1186/1471-2180-13-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chuang Y.C., Chang S.C., Wang W.K. High and increasing Oxa-51 DNA load predict mortality in Acinetobacter baumannii bacteremia: implication for pathogenesis and evaluation of therapy. PLoS One. 2010;5:e14133. doi: 10.1371/journal.pone.0014133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turton J.F., Woodford N., Glover J., Yarde S., Kaufmann M.E., Pitt T.L. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J Clin Microbiol. 2006;44:2974–2976. doi: 10.1128/JCM.01021-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zander E., Higgins P.G., Fernandez-Gonzalez A., Seifert H. Detection of intrinsic blaOXA-51-like by multiplex PCR on its own is not reliable for the identification of Acinetobacter baumannii. Int J Med Microbiol. 2013;303:88–89. doi: 10.1016/j.ijmm.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 31.Paule S.M., Pasquariello A.C., Hacek D.M., Fisher A.G., Thomson R.B., Jr., Kaul K.L. Direct detection of Staphylococcus aureus from adult and neonate nasal swab specimens using real-time polymerase chain reaction. J Mol Diagn. 2004;6:191–196. doi: 10.1016/S1525-1578(10)60509-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greiner O., Day P.J., Altwegg M., Nadal D. Quantitative detection of Moraxella catarrhalis in nasopharyngeal secretions by real-time PCR. J Clin Microbiol. 2003;41:1386–1390. doi: 10.1128/JCM.41.4.1386-1390.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirama T., Yamaguchi T., Miyazawa H., Tanaka T., Hashikita G., Kishi E. Prediction of the pathogens that are the cause of pneumonia by the battlefield hypothesis. PLoS One. 2011;6:e24474. doi: 10.1371/journal.pone.0024474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith-Vaughan H., Byun R., Nadkarni M., Jacques N.A., Hunter N., Halpin S. Measuring nasal bacterial load and its association with otitis media. BMC Ear Nose Throat Disord. 2006;6:10. doi: 10.1186/1472-6815-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakwinska O., Bastic Schmid V., Berger B., Bruttin A., Keitel K., Lepage M. Nasopharyngeal microbiota in healthy children and pneumonia patients. J Clin Microbiol. 2014;52:1590–1594. doi: 10.1128/JCM.03280-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Obasi C.N., Barrett B., Brown R., Vrtis R., Barlow S., Muller D. Detection of viral and bacterial pathogens in acute respiratory infections. J Infect. 2014;68:125–130. doi: 10.1016/j.jinf.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stralin K., Korsgaard J., Olcen P. Evaluation of a multiplex PCR for bacterial pathogens applied to bronchoalveolar lavage. Eur Respir J. 2006;28:568–575. doi: 10.1183/09031936.06.00006106. [DOI] [PubMed] [Google Scholar]
- 38.Goddard A.F., Staudinger B.J., Dowd S.E., Joshi-Datar A., Wolcott R.D., Aitken M.L. Direct sampling of cystic fibrosis lungs indicates that DNA-based analyses of upper-airway specimens can misrepresent lung microbiota. Proc Natl Acad Sci USA. 2012;109:13769–13774. doi: 10.1073/pnas.1107435109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wurzel D.F., Marchant J.M., Clark J.E., Mackay I.M., Wang C.Y.T., Sloots T.P. Respiratory virus detection in nasopharyngeal aspirate versus bronchoalveolar lavage is dependent on virus type in children with chronic respiratory symptoms. J Clin Virol. 2013;58:683–688. doi: 10.1016/j.jcv.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huijskens E.G., Rossen J.W., Kluytmans J.A., van der Zanden A.G., Koopmans M. Evaluation of yield of currently available diagnostics by sample type to optimize detection of respiratory pathogens in patients with a community-acquired pneumonia. Influenza Other Respir Viruses. 2014;8:243–249. doi: 10.1111/irv.12153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abdeldaim G.M., Stralin K., Olcen P., Blomberg J., Herrmann B. Toward a quantitative DNA-based definition of pneumococcal pneumonia: a comparison of Streptococcus pneumoniae target genes, with special reference to the Spn9802 fragment. Diagn Microbiol Infect Dis. 2008;60:143–150. doi: 10.1016/j.diagmicrobio.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 42.Albrich W.C., Madhi S.A., Adrian P.V., van Nierkerk N., Mareletsi T., Cutland C. Use of a rapid test of pneumococcal colonization density to diagnose pneumococcal pneumonia. Clin Infect Dis. 2012;54:601–609. doi: 10.1093/cid/cir859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stralin K., Herrmann B., Abdeldaim G., Olcen P., Holmberg H., Molling P. Comparison of sputum and nasopharyngeal aspirate samples and of the PCR gene targets lytA and Spn9802 for quantitative PCR for rapid detection of pneumococcal pneumonia. J Clin Microbiol. 2014;52:83–89. doi: 10.1128/JCM.01742-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chastre J., Fagon J.Y. Ventilator-associated pneumonia. Am J Respir Crit Care Med. 2002;165:867–903. doi: 10.1164/ajrccm.165.7.2105078. [DOI] [PubMed] [Google Scholar]
- 45.Peters R.P., de Boer R.F., Schuurman T., Gierveld S., Kooistra-Smid M., van Agtmael M.A. Streptococcus pneumoniae DNA load in blood as a marker of infection in patients with community-acquired pneumonia. J Clin Microbiol. 2009;47:3308–3312. doi: 10.1128/JCM.01071-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rello J., Lisboa T., Lujan M., Gallego M., Kee C., Kay I. Severity of pneumococcal pneumonia associated with genomic bacterial load. Chest. 2009;136:832–840. doi: 10.1378/chest.09-0258. [DOI] [PubMed] [Google Scholar]
- 47.Werno A.M., Anderson T.P., Murdoch D.R. Association between pneumococcal load and disease severity in adults with pneumonia. J Med Microbiol. 2012;61:1129–1135. doi: 10.1099/jmm.0.044107-0. [DOI] [PubMed] [Google Scholar]