Abstract
Choline is an essential nutrient, and the amount needed in the diet is modulated by several factors. Given geographical differences in dietary choline intake and disparate frequencies of single-nucleotide polymorphisms (SNPs) in choline metabolism genes between ethnic groups, we tested the hypothesis that 3 SNPs that increase dependence on dietary choline would be under negative selection pressure in settings where choline intake is low: choline dehydrogenase (CHDH) rs12676, methylenetetrahydrofolate reductase 1 (MTHFD1) rs2236225, and phosphatidylethanolamine-N-methyltransferase (PEMT) rs12325817. Evidence of negative selection was assessed in 2 populations: one in The Gambia, West Africa, where there is historic evidence of a choline-poor diet, and the other in the United States, with a comparatively choline-rich diet. We used 2 independent methods, and confirmation of our hypothesis was sought via a comparison with SNP data from the Maasai, an East African population with a genetic background similar to that of Gambians but with a traditional diet that is higher in choline. Our results show that frequencies of SNPs known to increase dependence on dietary choline are significantly reduced in the low-choline setting of The Gambia. Our findings suggest that adequate intake levels of choline may have to be reevaluated in different ethnic groups and highlight a possible approach for identifying novel functional SNPs under the influence of dietary selective pressure.—Silver, M. J., Corbin, K. D., Hellenthal, G., da Costa, K.-A., Dominguez-Salas, P., Moore, S. E., Owen, J., Prentice, A. M., Hennig, B. J., Zeisel, S. H. Evidence for negative selection of gene variants that increase dependence on dietary choline in a Gambian cohort.
Keywords: diet and selection, adequate intake levels, phosphatidylethanolamine-N-methyltransferase, choline dehydrogenase, methylenetetrahydrofolate dehydrogenase
Choline is an essential nutrient (1) with functional relevance in a wide array of biologic pathways, including epigenetic modulation of gene expression, brain development, hepatic lipid homeostasis, and energy metabolism. Choline is positioned at the intersection of 1-carbon metabolism pathways, which generate methyl groups from choline, methionine, and folate that are essential for biologic methylation reactions (2). Two key phenotypes emerge when dietary choline is limited in humans. The most prominent is in the liver, where accumulation of lipids is concurrent with increased markers of damage, such as elevated serum liver enzymes and hepatocyte apoptosis. A smaller subset of individuals exhibit a muscle phenotype characterized by elevated serum creatine phosphokinase from muscle. These symptoms resolve when choline is reintroduced into the diet (3–6). Furthermore, there is an extensive body of literature demonstrating the metabolic and health consequences of inadequate choline intake, ranging from neural tube defects to cancer, in various ethnic groups (3, 6–11).
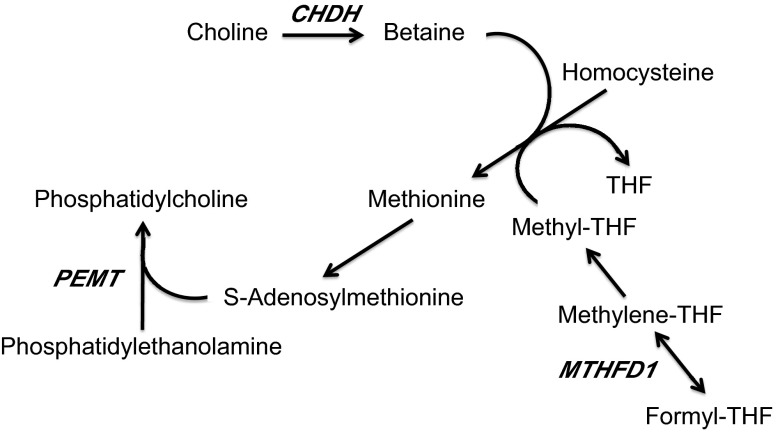
Adequate intake (AI) for choline, established from observations of choline intake in healthy U.S. adults, is 425–550 mg/d (1, 12). However, the requirement for choline is modulated by several factors, including sex, menopausal status (5), and the gut microbiome (13). Genetic variation also plays a role, and 3 functional single-nucleotide polymorphisms (SNPs) in particular are known to increase dependence on dietary choline. These are hereafter referred to as choline-dependent (CD) SNPs: choline dehydrogenase (CHDH), rs12676; methylenetetrahydrofolate reductase 1 (MTHFD1), rs2236225; and phosphatidylethanolamine-N-methyltransferase (PEMT), rs12325817 (3, 6) (Fig. 1).
Figure 1.
Metabolic pathways modulated by CHDH, PEMT, and MTHFD1. Choline is oxidized to form betaine by CHDH. Betaine is used as a methyl donor in the formation of methionine. MTHFD catalyzes the formation of methyltetrahydrofolate, which is an alternative methyl donor in the formation of methionine. Methionine is used to form S-adenosylmethionine, which is necessary in the methylation of phosphatidylethanolamine to form phosphatidylcholine. Genetic polymorphisms in CHDH, PEMT, and MTHFD1 increase dependence on dietary choline by modulating the formation of choline and its utilization as a methyl donor.
Several lines of evidence demonstrate a role for CD SNPs in affecting metabolism and dependence on dietary choline. CHDH encodes a mitochondrial protein that catalyzes the first irreversible step in the oxidation of choline to betaine. Premenopausal female carriers of the T allele of CHDH rs12676 (a nonsynonymous coding SNP) have greater dependence on dietary choline (3). In men, this allele is also associated with lower sperm CHDH protein levels (14). Individuals with this SNP need more choline precursor to drive production of this reaction’s product, betaine, which is necessary for methylation reactions.
MTHFD1 encodes a folate-metabolizing enzyme that catalyzes 3 reactions that direct the flow of 1-carbon folates (15); the formation of 5-methyl-terahydrofolate (5-methyl-THF) is practically irreversible in vivo, but the interconversion of 5,10-methylene-THF and 10-formyl-THF is closer to equilibrium (6, 16). Thus, 5,10-methylene-THF may be directed by MTHFD1, either toward homocysteine methylation or away from it. The MTHFD1 rs2236225 polymorphism (a nonsynonymous coding SNP) increases the flux between 5,10-methylene-THF and 10-formyl-THF and thereby reduces the flux between 5,10-methylene-THF and 5-methyl-THF, making less 5-methyl-THF available for homocysteine remethylation. When 5-methyl-THF is not available, more betaine from choline is needed for homocysteine remethylation (6, 17). Carriers of the A allele of MTHFD1 rs2236225 thus have an increased dependence on dietary choline (6).
PEMT encodes an enzyme that sequentially methylates phosphatidylethanolamine to generate phosphatidylcholine, a source of choline (18). PEMT expression is induced by estrogen, and PEMT rs12325817 is a promoter SNP that abrogates estrogen-mediated induction of the gene (19). Female carriers of the C allele of this SNP (on the coding strand) are more susceptible to development of organ dysfunction when eating a low-choline diet (3–5), because they are less able to induce the gene with estrogen and thereby make less of their own choline (in the form of phosphatidylcholine). It is reasonable to suggest that women with CD SNPs who are eating low-choline diets deliver less choline to the fetus (via the placenta) and that this could negatively affect fetal outcome (20). There may also be effects on the establishment of methylation patterns in the epigenome of the very early embryo, in that these are known to be sensitive to nutrients in the 1-carbon pathway (21).
The distribution of multiple SNPs in genes within the 1-carbon metabolism pathway varies across different ethnic groups, and these genetic patterns are associated with different health outcomes (22, 23). Differences in the distribution of CD SNPs are particularly evident between populations of Caucasian and African descent (22, 23). The diversity in access to choline in various regions of the world led us to hypothesize that the disparate frequency of functional variants in choline metabolism is influenced by dietary selective pressures. Using 2 independent statistical methods, we tested this hypothesis of choline-mediated selective pressure by comparing 2 populations: one in The Gambia (GAM) with a choline-poor diet (24–28), and the other composed of individuals of Caucasian/European descent (EUR) from North Carolina in the United States, with a relatively choline-rich diet (29–32). Furthermore, we compared allele frequencies of CD SNPs in GAM and EUR cohorts with those observed in another African population [HapMap (International HapMap Project, National Center for Biotechnology Information, Bethesda, MD, USA)]: the Maasai in Kinyawa, Kenya; MKK), an ethnic population that is genetically more similar to Gambians, but with a traditional diet that is relatively high in choline (33).
MATERIALS AND METHODS
North Carolina clinical cohort
The individuals included in this study were men and women from 3 previously reported studies (4, 5, 34). Briefly, these studies examined the amount of dietary choline needed for optimal health and the role played by genetic variation. In one study, dietary choline restriction produced liver and muscle phenotypes in subjects who were inpatients at the Clinical and Translational Research Center, University of North Carolina (UNC) Chapel Hill School of Medicine. There were 3 phases to the study. The baseline phase provided a diet with adequate choline (550 mg/70 kg per day). The choline depletion phase provided 50 mg choline per day. The final repletion phase reintroduced adequate choline into the diets (5). The second study was similar to the first, but the focus was on women and the importance of estrogen for endogenous choline synthesis (4). In the third study, pregnant women were examined to determine whether total choline intake, SNPs, or both influence the amount of choline and its metabolites found in breast milk and plasma (34). Written informed consent was obtained from all participants, and the Institutional Review Board at UNC Chapel Hill approved all protocols. The samples used in the study included 162 Caucasian individuals from whom sufficient DNA was available for genotyping. Three first-degree relatives were excluded, leaving 159 subjects (17 males and 142 females) for analysis.
Gambian study cohort
We selected women who participated in 1 of 3 studies in The Gambia (24, 35, 36), for whom a DNA sample was available for genotyping and excluded all first-degree relatives, so that 241 subjects were available for the study. Briefly, all women were recruited between 2009 and 2010 in the Kiang West district of rural Gambia, from the 36 villages in the catchment area of the Medical Research Center (MRC) International Nutrition Group’s field station at MRC Keneba (http://www.ing.mrc.ac.uk). Written informed consent was obtained from all participants, and the joint Gambian Government/MRC Ethics Committee approved all procedures.
Gene and variant selection and genotyping
Gene variants used in this study were those selected for a previous investigation that targeted SNP mapping to genes in the choline pathway and the intersecting folate and methionine pathways (±5 kb from gene boundaries, to assess the role of distal regulatory elements); in peripherally related genes that metabolize choline containing lipids; or in genes with a direct relationship to fatty liver, a choline-mediated phenotype (22). The set of genotyped SNPs included the 3 CD SNPs that were the focus of this study (CHDH rs12676, MTHFD1 rs2236225, and PEMT rs12325817), because they have been associated with an increased dependence on dietary choline (3–6, 34, 37) and have known functional effects on choline metabolism (14, 15, 19, 37, 38). For this study, we included 226 SNPs genotyped in both the GAM and EUR cohorts, but removed 12 SNPs for which there is limited evidence of an influence on dietary choline requirements (23), but no functional data, as these may otherwise have biased our analysis. Thus, of the remaining 214 SNPs, 3 are the CD SNPs and the remainder lack any published evidence of a role in modulating choline requirements, as is necessary for our statistical tests to be valid. Details of further SNP filtering procedures are given below.
Samples were genotyped as described in several publications (6, 19, 22, 23). Briefly, 98% of SNPs were genotyped with an oligo-specific extension-ligation assay on a custom Golden Gate array (Illumina, Inc., San Diego, CA, USA) (39). We used an in-house real-time PCR assay for the PEMT rs12325817 SNP (22, 23), because it cannot be genotyped on the Illumina platform. Four SNPs in the EUR cohort were genotyped by alternative methods. Two CD SNPs, rs12676 and rs2236225, had a subset of samples that failed on the Illumina platform, so they were genotyped via matrix-assisted laser desorption/ionization–time-of-flight (MALDI-TOF) primer-extension assay (Sequenom, Inc., San Diego, CA, USA) (22). Two other SNPs, rs3733890 and rs4244599, were part of targeted investigations before implementation of the custom Illumina array. They were genotyped via MALDI-TOF mass spectrometry and real-time PCR, respectively, as described elsewhere (6, 19).
MKK genotypes
We downloaded MKK genotypes for 95 unrelated individuals (42 males, 43 females), genotyped at 1,457,897 SNPs as part of HapMap3 (40). A majority of the 214 study SNPs genotyped in GAM and EUR were not present in the HapMap data, including 2 of the 3 CD SNPs. All missing SNPs were therefore imputed using IMPUTE2 (41), with phase 1 data from the 1000 Genomes project (EMBL-EBI, Hinxton, United Kingdom) as a reference panel. HapMap MKK genotypes were converted from the hg18 to hg19 genome build using liftOver (http://hgdownload.cse.ucsc.edu/admin/exe/linux.x86_64/liftover) before imputation. Metrics for imputation quality indicated that the 2 CD SNPs were imputed with high confidence (IMPUTE2 info = 0.98 and certainty = 0.99 for rs12676; info = 0.97, certainty = 0.99 for rs12325817). IMPUTE2 metrics for internal cross-validation of existing sample genotypes against imputed values indicated that imputation was successful (>95% overall concordance; Supplemental Table 1). Thirty-four SNPs could not be confidently aligned with GAM and EUR allele calls because they had complementary alleles that made strand direction difficult to assign. Thus, 180 SNPs remained for the MKK cohort before SNP filtering.
SNP filtering
Our statistical tests for selection treat missing and monomorphic SNPs differently and perform different cross-cohort comparisons. For this reason, SNP filtering strategies vary, and we consider these for each test separately.
Method 1: pairwise cross-cohort comparisons
For each cross-cohort comparison, only SNPs with genotype data across both cohorts were considered (GAM vs. EUR: 214 SNPs considered; GAM vs. MKK and MKK vs. EUR: 180). All SNPs with a genotype call rate <90% in either cohort were removed (GAM vs. EUR: 3 SNPs removed; GAM vs. MKK 2; MKK vs. EUR: 1). Because nonzero minor allele frequencies (MAFs) are necessary to calculate variance-adjusted statistics, we further removed all SNPs that were monomorphic in either cohort (GAM vs. EUR: 16 SNPs removed; GAM vs. MKK: 6 SNPs; MKK vs. EUR: 12 SNPs). Finally, because the statistical test assumes that SNPs are independent, for each cross-cohort comparison, we measured pairwise correlations between all SNPs in each cohort and retained only 1 of each pair of SNPs with an r2 ≥ 0.8 in either cohort (GAM vs. EUR: 21 SNPs removed; GAM vs. MKK 23 SNPs; MKK vs. EUR: 26 SNPs). This process left 174 SNPs for the GAM vs. EUR analysis, 149 SNPs for GAM vs. MKK and 141 SNPs for MKK vs. EUR.
Method 2: population genetic model
This method can accommodate SNPs that are missing in only 1 cohort or are monomorphic in 1–2 of the 3 cohorts. We therefore considered all 214 SNPs for this analysis, but recorded SNPs with a genotype call rate <90% in any cohort as missing for that cohort (1 EUR SNP and 2 GAM SNPs). We further removed 2 SNPs that were monomorphic across all 3 cohorts and performed linkage disequilibrium (LD) filtering across all 3 cohorts using the same r2 threshold as described for method 1, which resulted in the removal of another 38 SNPs, leaving 174 SNPs for the method 2 analysis. To generate empirical probabilities to test against the null hypothesis of no negative selection in the GAM cohort at the CD SNPs, we used 144 of these 174 SNPs that were nonmissing in all 3 cohorts. However, we note that results were very similar when we used all 210 SNPs that were nonmissing in the Gambia cohort for this analysis.
Statistical tests for selection
Variation in SNP allele frequencies, both within and between populations, may be driven by selection or by random processes of genetic drift. Genetic drift can lead to SNPs being driven to fixation or lost entirely from a population simply by random chance (42). It is also possible for variants to arise de novo in a population through mutation. It is therefore important to allow for the possibility that any or all of these factors may be the cause of variation in allele frequencies when looking for evidence of selection at any particular SNP. We used 2 statistical tests for assessing evidence of negative selection at CD SNPs in the GAM sample.
Method 1: pairwise cross-cohort comparisons
Methods for assessing evidence of selection generally rely on dense genotyping around SNPs or genes of interest (43). Because we did not have access to such data, we instead tested each SNP independently, using a statistical test that compares allele frequency changes of CD SNPs to an empirical null distribution of the same test statistic calculated for other genotyped SNPs not known to increase dependence on dietary choline. We performed 3 separate cross-cohort comparisons: GAM vs. EUR; MKK vs. EUR; and MKK vs. EUR. Here, we describe our method for assessing evidence of negative selection in the GAM vs. EUR cohorts. The corresponding tests for the other 2 cross-cohort comparisons proceed in a similar manner.
For each SNP and in each cohort, we recorded the SNP MAF, where the minor allele is defined as the less frequent allele in the EUR population. Note that by applying this parameter, the functional variant known to increase dependence on dietary choline is the minor allele for all 3 CD SNPs in all cohorts. We next calculated the change in MAF for SNP j as
where mGAM and mEUR are the minor allele frequencies in the GAM and EUR populations, respectively. The mean change in MAF for a set S of 3 SNPs is then given by
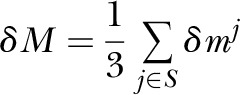 |
The distribution of this test statistic under the null, where all SNPs are subject to the same random fluctuations, is obtained by calculating the mean change in MAF for all 862,924 possible combinations of 3 SNPs drawn from the complete set of 174 markers. A significance measure for the alternative hypothesis that the 3 CD SNPs are under negative selection may then be computed as the proportion of all possible values for the test statistic that show a mean decrease in MAF at least as small as δMCD, where δMCD is the value of δM, when S is the set of CD SNPs. An implicit assumption is that all SNPs are independent, and, for this reason, in a preprocessing step, we filtered SNPs by LD, ensuring maximum pairwise LD r2 = 0.8. The accuracy of our method is particularly sensitive to violations of nonindependence at CD SNPs, and we therefore present the pairwise r2 coefficients for these in Table 1.
TABLE 1.
Pairwise r2 coefficients for 3 CD SNPs in each cohort
| SNPs | r2(GAM) | r2 (EUR) | r2 (MKK) |
|---|---|---|---|
| rs12325817a, rs2236225 | 0.020 | 0.024 | 0.003 |
| rs2236225, rs12676a | 0.008 | 0.006 | 0.028 |
| rs12325817a, rs12676a | 0.003 | 0.000 | 0.005 |
MKK imputed allele.
We calculated a variance-adjusted probability to account for differences in the distribution of minor allele dosage at each SNP, by computing a Welch-type t statistic for the mean change in MAF at SNP j as
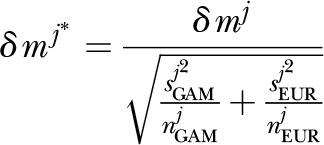 |
where sjGAM is the sample variance in minor allele dosage in the Gambian cohort, njGAM is the number of recorded genotypes for SNP j, and so on. This calculation allows us, for example, to down-weight large changes in MAF between cohorts where variance in minor allele dosage within one or both cohorts is large, or the number of genotyped SNPs is relatively small. Variance-adjusted significances are then calculated by permutation as outlined above, with
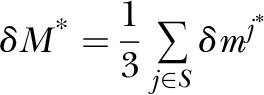 |
Summary statistics for all SNPs are presented in Supplemental Table 2.
Method 2: population genetic model
This test calculates the probability of observing the sampled data based on a standard population genetics model that assumes no selection (44). The setup and model are very similar to that described in Beaumont and Balding (45), differing only in the mechanistic details of inference. In particular, the model assumes that the 3 populations originate from a common ancestral population, equivalent to a tree merging the 3 groups via 2 internal nodes, with SNP allele frequencies changing from generation to generation, as they are subject to processes of random drift. In addition to allowing a joint comparison of the allele frequencies across all 3 cohorts at once, this test is expected to be more powerful than the method 1 test if the underlying model is an accurate summary of the real historical processes affecting the populations’ allele frequencies.
As in method 1, we define the minor allele to be the less frequent allele in the
EUR population. At each SNP, we assume the minor allele count X
in a given cohort (i.e., where X
 {G, C, M}, where G = GAM, C = EUR, M = MKK) follows a binomial
(nX,pX)
distribution, with nX the number of nonmissing sampled
haplotypes and pX the (unknown) frequency of the minor
allele for the given population at this SNP. As in Balding and Nichols (44), we assumed that
pX follows a β distribution with mean
(pX) = pA and
Var(pX) = dX
pA (1 − pA). Here,
pA is the (unknown) ancestral allele frequency for
this minor allele, equivalent in this 3-population case to the allele frequency at
the junction in the tree where all 3 populations merge, and
dX measures the relative drift in population X from
this ancestral frequency. In this scenario, we can integrate out
pX analytically, giving Pr(X |
pA, dX) which follows a
β-binomial distribution. At the given SNP, the joint probability of the
minor allele counts for all 3 cohorts, conditional on the
dX of each, is:
{G, C, M}, where G = GAM, C = EUR, M = MKK) follows a binomial
(nX,pX)
distribution, with nX the number of nonmissing sampled
haplotypes and pX the (unknown) frequency of the minor
allele for the given population at this SNP. As in Balding and Nichols (44), we assumed that
pX follows a β distribution with mean
(pX) = pA and
Var(pX) = dX
pA (1 − pA). Here,
pA is the (unknown) ancestral allele frequency for
this minor allele, equivalent in this 3-population case to the allele frequency at
the junction in the tree where all 3 populations merge, and
dX measures the relative drift in population X from
this ancestral frequency. In this scenario, we can integrate out
pX analytically, giving Pr(X |
pA, dX) which follows a
β-binomial distribution. At the given SNP, the joint probability of the
minor allele counts for all 3 cohorts, conditional on the
dX of each, is:
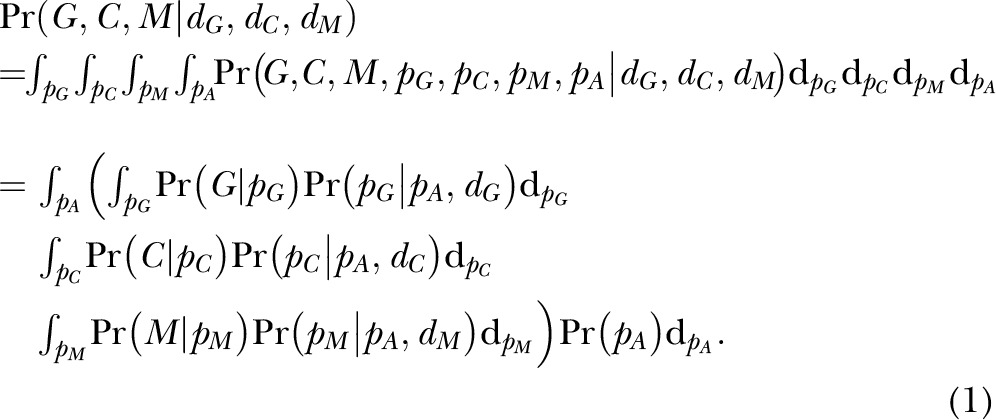 |
We assume
Pr(pA) follows a uniform distribution and integrate
out pA numerically to calculate Eq. 1. Assuming independence across
the 174 SNPs remaining after our LD-pruning procedure (see SNP filtering, above),
we find the maximum likelihood estimates (MLEs) of {dG, dC,
dM} by maximizing the joint likelihood of Eq. 1 across all 174 SNPs over a
3-dimensional grid. Assuming that the frequencies at most of these 174 SNPs are
not affected by selection, this method provides estimates of the genome-wide
expected drift value for each population's allele frequency relative to the
ancestral frequency value, under a neutral model with no selection. Letting
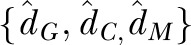 be our maximum likelihood values of
be our maximum likelihood values of
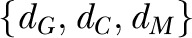 we next use Eq.
1 to calculate:
we next use Eq.
1 to calculate:
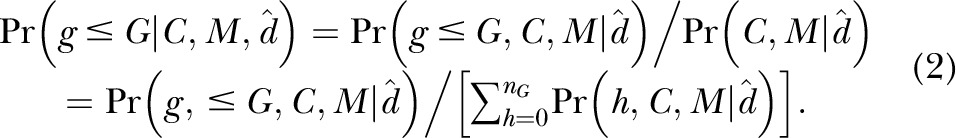 |
for each of 144 LD-pruned SNPs
with nonmissing data in all 3 cohorts. For each SNP, this calculation gives the
probability of observing a minor allele count less than or equal to that sampled
in the GAM cohort, given the minor allele counts sampled in the EUR and MKK
cohorts and our inferred drift values for each population. We next take the
average of Eq. 2 across the 3 CD
SNPs. Finally, analogous to the permutation procedure in method 1, we found the
average across all 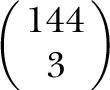 = 487,344 subsets of 3-SNP combinations and calculated the
proportion of such 3-SNP averages that were smaller than those of the 3 CD SNPs.
This proportion provided an empirical probability that tested the null hypothesis
that the allele frequencies for GAM at the 3 CD SNPs follow the above neutral
beta-binomial model vs. the 1-sided alternative model in which
the CD SNP GAM frequencies are smaller than that expected under the neutral model.
Full details are given in Supplemental Methods 1.
= 487,344 subsets of 3-SNP combinations and calculated the
proportion of such 3-SNP averages that were smaller than those of the 3 CD SNPs.
This proportion provided an empirical probability that tested the null hypothesis
that the allele frequencies for GAM at the 3 CD SNPs follow the above neutral
beta-binomial model vs. the 1-sided alternative model in which
the CD SNP GAM frequencies are smaller than that expected under the neutral model.
Full details are given in Supplemental Methods 1.
RESULTS
Principal component analyses (PCAs) of 144 SNPs in common across the 3 cohorts (GAM, EUR, and MKK) revealed the extent to which these vary in their genetic background (Fig. 2). The results support our hypothesis that EUR and MKK represent interesting choline-rich comparator populations, one (MKK) with a genetic background similar to that of GAM and the other (EUR) with a genetic background that is more distinct.
Figure 2.
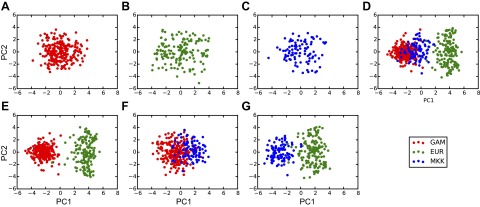
Cross-cohort comparisons confirm that GAM and MKK individuals are more closely related genetically than are EUR individuals. Plots illustrate the first 2 principal components from (A–C) 1-, (E–G) 2-, or (D) 3-cohort PCAs. PCAs illustrate interindividual differences at 144 SNPs across the 3 cohorts.
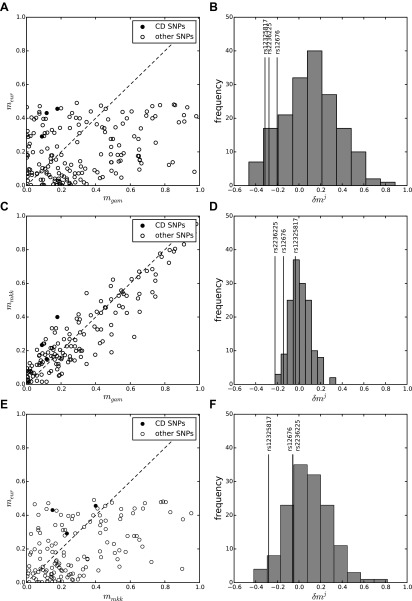
We first assessed evidence of negative selection of CD SNPs by using a method based on pairwise cross-cohort comparisons (method 1). We compared MAFs in GAM vs. EUR, GAM vs. MKK, and MKK vs. EUR. Cross-cohort MAF distributions are presented in Fig. 3. This figure shows a wide distribution in MAF differences across all tested SNPs in each cross-cohort comparison, although these differences are markedly reduced when the more genetically similar African populations are compared (middle plots). The 3 CD SNPs (black filled circles) show a lower MAF (negative δmj) in GAM compared to EUR (top right plot). MAFs for CD SNPs in each cohort are presented in Table 2. Results of statistical tests for evidence of negative selection at CD SNPs are presented in Table 3. These provide strong evidence of negative selection of CD SNPs in GAM compared with both EUR and MKK (adjusted P = 0.007, 0.002), and weaker evidence of negative selection in MKK compared with EUR (adjusted P = 0.04). The evidence of negative selection was strongest in GAM vs. MKK, because the observed reductions in CD SNP MAFs took place against a genetic background where there was relatively little overall difference in MAFs between the 2 cohorts (Fig. 2 and middle right-hand plot in Fig. 3). Right-hand plots in Fig. 3 reveal that the 3 CD SNPs showed a relatively large reduction in MAF compared to background in GAM vs. EUR, whereas only rs2236225 (MTHFD1) and rs12676 (CHDH) showed such a reduction in GAM vs. MKK and only rs12325817 (PEMT) in MKK vs. EUR.
Figure 3.
Cross-cohort MAF distributions illustrate MAF differences at CD SNPs compared to genetic background. MAF comparisons are shown for (A) GAM vs. EUR, (C) GAM vs. MKK, and (E) MKK vs. EUR. Note that the minor allele is defined for the EUR cohort, so that, in the top and bottom plots, the EUR MAF, mEUR ≤ 0.5 for all SNPs, and the possible change in MAF for SNP j in the non-EUR cohort ranges from −0.5 to 1. SNPs with reduced MAF in (A, C) GAM and (E) MKK are located to the left of the dashed black line of parity. These include the 3 CD SNPs (filled circles). B, D, F) distribution of MAF differences, δmj for each cross-cohort comparison. In each case, δmj is defined as the SNP MAF in the cohort on the y-axis subtracted from the SNP MAF for the cohort on the x-axis. Solid black vertical lines illustrate δmj for the 3 CD SNPs.
TABLE 2.
Minor allele frequencies at 3 CD SNPs in the GAM, MKK, and EUR cohorts
| SNPs | Minor (major) allele from EUR data | MAF_GAM | MAF_MKK | MAF_EUR |
|---|---|---|---|---|
| rs12676a,b | T(G) | 0.09 | 0.23 | 0.29 |
| rs2236225 | A(G) | 0.18 | 0.40 | 0.46 |
| rs12325817a,b | C(G) | 0.12 | 0.15 | 0.43 |
MKK-imputed allele.
Reported based on the reverse genome strand, because these genes are transcribed from that strand (dbSNP build 141).
TABLE 3.
Statistical tests for evidence of negative selection at 3 CD SNPs, according to cross-cohort comparison method 1
| Comparison | Null hypothesis tested | SNPs tested (n) | Unadjusted P | Variance-adjusted P |
|---|---|---|---|---|
| GAM vs. EUR | CD SNP MAFs are not significantly reduced in GAM compared with EUR | 174 | 0.004 | 0.007 |
| GAM vs. MKK | CD SNP MAFs are not significantly reduced in GAM compared with MKK | 149 | 0.002 | 0.002 |
| MKK vs. EUR | CD SNP MAFs are not significantly reduced in MKK compared with EUR | 141 | 0.03 | 0.04 |
We performed a further, independent test to identify negative selection of CD SNPs by using an alternative population genetic model that compares SNP frequencies across all 3 cohorts simultaneously (method 2). Results are presented in Table 4. Although this method tests a slightly different null hypothesis—namely, whether the GAM data at the CD SNPs follow expectations under a neutral model, given the EUR and MKK data—the results strongly support the findings of method 1. In particular, we found strong evidence of negative selection of CD SNPs in GAM than in EUR and MKK (P = 0.008 by permutation) and no evidence of negative selection in MKK vs. EUR and GAM (P = 0.7).
TABLE 4.
Statistical tests for evidence of negative selection at 3 CD SNPs, according to population genetic model-based method 2
| Comparison | Null hypothesis tested | SNPs tested (n) | Permutation P |
|---|---|---|---|
| GAM vs. (EUR+MKK) | CD SNP MAFs are not significantly reduced in GAM compared with EUR and MKK | 144 | 0.008 |
| MKK vs. (EUR+GAM) | CD SNP MAFs are not significantly reduced in MKK compared with EUR and GAM | 144 | 0.7 |
DISCUSSION
Choline deficiency has known deleterious effects on health (3, 6–11) and reproduction (20). Although the essentiality of choline in the diet has been tested directly only in U.S. populations where the 2 most prominent races are Caucasian and African American (3–6), the biologic consequences of inadequate choline intake have been demonstrated in a wide range of human (3, 6–11) and rodent studies (46–48). It is therefore biologically plausible that where dietary choline is restricted, a genetically optimized choline metabolism would be likely to confer a survival advantage, irrespective of ethnicity or geographic location.
Numerous statistical methods have been developed for identifying genomic regions undergoing selection (see Ref. 49 for a review). Several rely on capturing multiple variants within each locus (50–52) or require densely genotyped data (53, 54). In this study, we instead used 2 independent statistical methods that enable analysis of sparsely genotyped unlinked SNPs, to assess for evidence of negative selection of SNPs that increase dependence on dietary choline in populations with divergent access to choline-containing foods. The first method used a standard statistical test based on cross-cohort comparisons of observed differences in allele frequencies in the GAM, EUR, and MKK cohorts, and compared MAF changes in known CD SNPs against other genotyped SNPs not known to affect dependence on dietary choline. In the second method, we modeled observed MAF differences in a population genetic model closely related to work described by Beaumont and Balding (45) that describes processes of drift that lead to genetic divergence between populations over time. As in method 1, we assumed that the non-CD SNPs are neutral, both when inferring the relative levels of drift separating populations’ allele frequencies and when generating an empirical null distribution to calculate probabilities. It remains a possibility that 1 or more of these “background” SNPs could influence dependence on dietary choline, potentially biasing our test statistics in one or the other direction, depending on whether minor alleles increase or decrease this dependence. Indeed, an interesting finding that warrants further investigation is the presence of other SNPs that have very high or very low MAFs in GAM vs. EUR (Fig. 3A, top left and bottom right quadrants). These represent promising candidates for future functional studies. Both statistical approaches assume that SNPs are independent after LD pruning, although we note that results changed little when no SNPs were excluded based on LD.
The population in GAM is a good model for low choline availability. Our study cohort is from the Kiang West district in rural Gambia, where mean choline intake in women was recently estimated to be 155 mg/d, with only 2.8% of the women consuming intakes above 425 mg/d (24). This level of intake is in line with historic evidence and documentation describing the traditional Gambian diet, which is rice-based and low in choline-rich foods, such as meats, milk, and eggs (25–28). In contrast, in the U.S. choline-rich foods are abundant in the current food supply, and the mean choline intake is ∼2 times higher than in The Gambia (32, 55). Investigations of traditional foods in the United States suggest an abundance of foods of animal origin (30, 31), which supports the likelihood of higher choline availability in Caucasian immigrant populations in the United States than in GAM during evolutionarily relevant time frames. It is notable that current intakes of choline in Europe are similar to those in the United States (56) and are in agreement with traditional foods consumed in Europe (57). Therefore, although there is a lack of direct evidence on historic diets, current intakes in GAM, the United States, and Europe align with traditional diets and support our characterization of low choline intake in GAM relative to that in the United States and Europe. Despite the inherent difficulty in characterizing historic diets in evolutionary studies, there is evidence supporting recent and continuous diet-driven selection in humans (58). Although we focused on dietary choline because of the known effects of choline deficiency in humans and the modulation of these effects by specific genetic variants, we acknowledge the possibility that other 1-carbon nutrients could influence the negative selective pressure that we addressed in this study.
Our evidence that negative selection occurs at 3 functional CD SNPs in different genes that independently modulate choline metabolism supports our hypothesis that the observed MAF changes are unlikely to have occurred by chance. These findings were strengthened by observed shifts in MAF in MKK, a population that is genetically similar to GAM (59, 60), but with a traditionally much higher intake of choline from foods such as milk, meat, and blood (33). It is therefore striking that a cross-cohort comparison of GAM vs. MKK provided equally strong evidence of negative selection at CD SNPs, supporting the argument that MAF differences are due to differences in choline intake, rather than chance or some other factor. Our use of MKK HapMap genotypes required that we impute multiple missing SNPs to enable a comparison with existing EUR and GAM data. Genotype imputation is an established method for inferring missing genotypes, although imputation accuracy can vary between populations and genomic regions (61). Internal cross-validation checks confirmed that imputation of missing genotypes for MKK data was successful. We note that neither of our statistical methods is able to distinguish between the equivalent scenarios of negative selection of CD SNPs in GAM and positive selection of CD SNPs in MKK and EUR. However, given the known deleterious effects of these SNPs in conditions of low dietary choline, we consider the former scenario to be the most probable.
The results presented here are consistent with those in other studies showing the influence of diet on gene selection. A prominent example is the genotype-mediated persistence of lactase functionality, and thus the ability to digest lactose in milk, in populations with high dairy intake such as the Maasai (62). It is interesting that this persistence occurs in parallel with positive selection of lipid metabolism gene variants that are cardioprotective (63). In this population, the high cholesterol and fat intake from the traditional diet is not accompanied by the high blood cholesterol levels and increased incidence of cardiovascular disease that is seen in European populations where lactase function persists in the absence of the positive selection of lipid metabolism variants and in an environment where high fat, high cholesterol foods are common (63). This suggests that the mismatch between diet and the genes involved in the metabolic pathways of these dietary components in Europeans contribute to adverse health outcomes. The selection for lactase persistence is estimated to have occurred 7500 years ago, suggesting that relatively recent dietary influences can modify the persistence of genetic variants (64). Additional support for the influence of diet on genetic variation is the positive selection in populations with high starch intake of additional copies of the salivary amylase gene which encodes the enzyme responsible for starch hydrolysis (65). The switch to high-starch diets occurred approximately 10,000 years ago after the transition from hunter-gathering to farming, providing additional support for the influence of relatively recent dietary exposures on the genome (66). These diet-genome interactions are believed to optimize metabolic requirements in humans (67), which fits with our hypothesis that in The Gambia, choline metabolism was genetically optimized to adjust for a diet low in sources of choline.
In this study, low dietary choline correlated with a reduced frequency of alleles that increase dependence on dietary choline. This finding could have health implications if there is a mismatch between choline intake and a population’s endogenous capacity to produce choline and its metabolites. For example, a recent report on food patterns in MKK shows a shift from a traditional high-choline diet composed primarily of meat, milk, and blood [which averages approximately 58 mg of choline per 100 g food (68)] to one composed primarily of milk, maize, and beans (69) [which averages about 15 mg choline per 100 g food (68)]. This shift could have health consequences for future generations of Maasai, whose genotypes are adapted to a high-choline diet. Our finding that SNPs that influence choline requirements occur at different frequencies across populations raises the possibility that current recommended intake levels for choline are not optimal across all populations and that they may need to be reevaluated to account for genetic differences. Finally, current methods for identifying functional genetic variants are labor and cost intensive, involving computationally intensive genome-wide screens combined with large epidemiologic studies or in-depth phenotyping in clinical studies. In this study, we offer a relatively simple alternative approach, whereby differences in the frequency of genetic variants within nutrient-relevant metabolic pathways across populations with divergent levels of nutrient intake can highlight putative functional SNPs that warrant further investigation.
Supplementary Material
Acknowledgments
The authors thank the women from Kiang West, The Gambia, for their participation; their laboratory technicians, field assistants, nurses, data entry clerks, and other staff at MRC Keneba, The Gambia, and A. J. Fulford, Ph.D., for support in data management; Joseph Galanko, Ph.D., for assistance with data organization; the research subjects in the North Carolina clinical cohorts for participating; Leslie Fischer for clinical study management; and Catherine Walker for a critical reading of the manuscript. The Gambian studies that provided data for the analyses were supported by Wellcome Trust Grant WT086369MA (to B.J.H.), U.K. Medical Research Council core funding Grant MC-A760-5QX00 to the International Nutrition Group, and the United Kingdom Department for the International Development (DFID) under the MRC/DFID Concordat agreement. Work completed by Dr. Zeisel’s research team was supported by grants from the Bill and Melinda Gates Foundation and Grant DK56350 from the U.S. National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases. G.H. is jointly funded by Grant 098386/Z/12/Z from the Wellcome Trust and by a Sir Henry Dale Fellowship from the Royal Society. B.J.H. and S.H.Z. share senior authorship of this article.
Glossary
- AI
adequate intake
- CD
choline dependence/dependent
- CHDH
choline dehydrogenase
- EUR
European
- GAM
The Gambia
- LD
linkage disequilibrium
- MKK
Maasai in Kinyawa, Kenya
- MALDI-TOF
matrix-assisted laser desorption/ionization–time-of-flight
- MAF
minor allele frequency
- MTHFD1
methylenetetrahydrofolate reductase 1
- PCA
principal component analysis
- PEMT
phosphatidylethanolamine-N-methyltransferase
- SNP
single-nucleotide polymorphism
- THF
tetrahydrofolate
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- 1.Institute of Medicine (US) Standing Committee on the Scientific Evaluation of Dietary Reference Intakes and Its Panel on Folate, Other B Vitamins, and Choline. (1998) Choline. In Dietary Reference Intakes for Folate, Thiamin, Riboflavin, Niacin, Vitamin B12, Pantothenic Acid, Biotin, and Choline, Vol. 1, pp. 390–422, National Academies Press, Washington, D.C. [PubMed] [Google Scholar]
- 2.Corbin K. D., Zeisel S. H. (2012) The nutrigenetics and nutrigenomics of the dietary requirement for choline. Prog. Mol. Biol. Transl. Sci. 108, 159–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Da Costa K. A., Kozyreva O. G., Song J., Galanko J. A., Fischer L. M., Zeisel S. H. (2006) Common genetic polymorphisms affect the human requirement for the nutrient choline. FASEB J. 20, 1336–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fischer L. M., da Costa K. A., Kwock L., Galanko J., Zeisel S. H. (2010) Dietary choline requirements of women: effects of estrogen and genetic variation. Am. J. Clin. Nutr. 92, 1113–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischer L. M., daCosta K. A., Kwock L., Stewart P. W., Lu T. S., Stabler S. P., Allen R. H., Zeisel S. H. (2007) Sex and menopausal status influence human dietary requirements for the nutrient choline. Am. J. Clin. Nutr. 85, 1275–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohlmeier M., da Costa K. A., Fischer L. M., Zeisel S. H. (2005) Genetic variation of folate-mediated one-carbon transfer pathway predicts susceptibility to choline deficiency in humans. Proc. Natl. Acad. Sci. USA 102, 16025–16030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchman A. L. (2009) The addition of choline to parenteral nutrition. Gastroenterology 137(5, Suppl)S119–S128 [DOI] [PubMed] [Google Scholar]
- 8.Detopoulou P., Panagiotakos D. B., Antonopoulou S., Pitsavos C., Stefanadis C. (2008) Dietary choline and betaine intakes in relation to concentrations of inflammatory markers in healthy adults: the ATTICA study. Am. J. Clin. Nutr. 87, 424–430 [DOI] [PubMed] [Google Scholar]
- 9.Shaw G. M., Carmichael S. L., Yang W., Selvin S., Schaffer D. M. (2004) Periconceptional dietary intake of choline and betaine and neural tube defects in offspring. Am. J. Epidemiol. 160, 102–109 [DOI] [PubMed] [Google Scholar]
- 10.Zeng F. F., Xu C. H., Liu Y. T., Fan Y. Y., Lin X. L., Lu Y. K., Zhang C. X., Chen Y. M. (2014) Choline and betaine intakes are associated with reduced risk of nasopharyngeal carcinoma in adults: a case-control study. Br. J. Cancer 110, 808–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang C. X., Pan M. X., Li B., Wang L., Mo X. F., Chen Y. M., Lin F. Y., Ho S. C. (2013) Choline and betaine intake is inversely associated with breast cancer risk: a two-stage case-control study in China. Cancer Sci. 104, 250–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeisel S. H., da Costa K. A. (2009) Choline: an essential nutrient for public health. Nutr. Rev. 67, 615–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spencer M. D., Hamp T. J., Reid R. W., Fischer L. M., Zeisel S. H., Fodor A. A. (2011) Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology 140, 976–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson A. R., Lao S., Wang T., Galanko J. A., Zeisel S. H. (2012) Choline dehydrogenase polymorphism rs12676 is a functional variation and is associated with changes in human sperm cell function. PLoS ONE 7, e36047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Field M. S., Shields K. S., Abarinov E. V., Malysheva O. V., Allen R. H., Stabler S. P., Ash J. A., Strupp B. J., Stover P. J., Caudill M. A. (2013) Reduced MTHFD1 activity in male mice perturbs folate- and choline-dependent one-carbon metabolism as well as transsulfuration. J. Nutr. 143, 41–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horne D. W. (2003) Neither methionine nor nitrous oxide inactivation of methionine synthase affect the concentration of 5,10-methylenetetrahydrofolate in rat liver. J. Nutr. 133, 476–478 [DOI] [PubMed] [Google Scholar]
- 17.Pomfret E. A., daCosta K. A., Zeisel S. H. (1990) Effects of choline deficiency and methotrexate treatment upon rat liver. J. Nutr. Biochem. 1, 533–541 [DOI] [PubMed] [Google Scholar]
- 18.Ridgway N. D., Vance D. E. (1992) Phosphatidylethanolamine N-methyltransferase from rat liver. Methods Enzymol. 209, 366–374 [DOI] [PubMed] [Google Scholar]
- 19.Resseguie M. E., da Costa K. A., Galanko J. A., Patel M., Davis I. J., Zeisel S. H. (2011) Aberrant estrogen regulation of PEMT results in choline deficiency-associated liver dysfunction. J. Biol. Chem. 286, 1649–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeisel S. H. (2013) Nutrition in pregnancy: the argument for including a source of choline. Int. J. Womens Health 5, 193-199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waterland R. A., Jirtle R. L. (2003) Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol. Cell. Biol. 23, 5293–5300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corbin K. D., Abdelmalek M. F., Spencer M. D., da Costa K. A., Galanko J. A., Sha W., Suzuki A., Guy C. D., Cardona D. M., Torquati A., Diehl A. M., Zeisel S. H. (2013) Genetic signatures in choline and 1-carbon metabolism are associated with the severity of hepatic steatosis. FASEB J. 27, 1674–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Da Costa K. A., Corbin K. D., Niculescu M. D., Galanko J. A., Zeisel S. H. (2014) Identification of new genetic polymorphisms that alter the dietary requirement for choline and vary in their distribution across ethnic and racial groups. FASEB J. 28, 2970–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dominguez-Salas P., Moore S. E., Cole D., da Costa K. A., Cox S. E., Dyer R. A., Fulford A. J., Innis S. M., Waterland R. A., Zeisel S. H., Prentice A. M., Hennig B. J. (2013) DNA methylation potential: dietary intake and blood concentrations of one-carbon metabolites and cofactors in rural African women. Am. J. Clin. Nutr. 97, 1217–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nutrition Country Profile Republic of The Gambia. (2010) Nutrition and Consumer Protection Division, Food and Agriculture Organization of the United Nations, Rome
- 26.Murphy, P. K. (1981) The Foods and Cooking of Rural Gambia: National and Regional Styles of Cookery. Oxford Symposium on Food and Cookery pp. 290-298 [Google Scholar]
- 27.Park, M. (2000) Travels in the Interior Districts of Africa. Duke University Press, Durham, NC.
- 28.Hudson G. J. (1995) Food intake in a west African village: estimation of food intake from a shared bowl. Br. J. Nutr. 73, 551–569 [DOI] [PubMed] [Google Scholar]
- 29.Cho E., Zeisel S. H., Jacques P., Selhub J., Dougherty L., Colditz G. A., Willett W. C. (2006) Dietary choline and betaine assessed by food-frequency questionnaire in relation to plasma total homocysteine concentration in the Framingham Offspring Study. Am. J. Clin. Nutr. 83, 905–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cordain L., Eaton S. B., Sebastian A., Mann N., Lindeberg S., Watkins B. A., O’Keefe J. H., Brand-Miller J. (2005) Origins and evolution of the Western diet: health implications for the 21st century. Am. J. Clin. Nutr. 81, 341–354 [DOI] [PubMed] [Google Scholar]
- 31.Dyson L. (2000) American cuisine in the 20th century. Food Reviews 23, 2-7 [Google Scholar]
- 32.Wallace T. C., McBurney M., Fulgoni V. L. III (2014) Multivitamin/mineral supplement contribution to micronutrient intakes in the United States, 2007-2010. J. Am. Coll. Nutr. 33, 94–102 [DOI] [PubMed] [Google Scholar]
- 33.Kuhnlein, H. V., Erasmis, B., and Spigelski, D., eds. (2009) The Maasai food system and food and nutrition security. In Indigenous Peoples’ Food Systems: The Many Dimensions of Culture, Diversity and Environment for Nutrition and Health. pp. 231–249, Food and Agriculture Organization of the United Nations, Centre for Indigenous Peoples’ Nutrition and Environment, Rome [Google Scholar]
- 34.Fischer L. M., da Costa K. A., Galanko J., Sha W., Stephenson B., Vick J., Zeisel S. H. (2010) Choline intake and genetic polymorphisms influence choline metabolite concentrations in human breast milk and plasma. Am. J. Clin. Nutr. 92, 336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dominguez-Salas P., Moore S. E., Baker M. S., Bergen A. W., Cox S. E., Dyer R. A., Fulford A. J., Guan Y., Laritsky E., Silver M. J., Swan G. E., Zeisel S. H., Innis S. M., Waterland R. A., Prentice A. M., Hennig B. J. (2014) Maternal nutrition at conception modulates DNA methylation of human metastable epialleles. Nat. Commun. 5, 3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore S. E., Fulford A. J., Darboe M. K., Jobarteh M. L., Jarjou L. M., Prentice A. M. (2012) A randomized trial to investigate the effects of pre-natal and infant nutritional supplementation on infant immune development in rural Gambia: the ENID trial: Early Nutrition and Immune Development. BMC Pregnancy Childbirth 12, 107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ivanov A., Nash-Barboza S., Hinkis S., Caudill M. A. (2009) Genetic variants in phosphatidylethanolamine N-methyltransferase and methylenetetrahydrofolate dehydrogenase influence biomarkers of choline metabolism when folate intake is restricted. J. Am. Diet. Assoc. 109, 313–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carroll N., Pangilinan F., Molloy A. M., Troendle J., Mills J. L., Kirke P. N., Brody L. C., Scott J. M., Parle-McDermott A. (2009) Analysis of the MTHFD1 promoter and risk of neural tube defects. Hum. Genet. 125, 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan J. B., Oliphant A., Shen R., Kermani B. G., Garcia F., Gunderson K. L., Hansen M., Steemers F., Butler S. L., Deloukas P., Galver L., Hunt S., McBride C., Bibikova M., Rubano T., Chen J., Wickham E., Doucet D., Chang W., Campbell D., Zhang B., Kruglyak S., Bentley D., Haas J., Rigault P., Zhou L., Stuelpnagel J., Chee M. S. (2003) Highly parallel SNP genotyping. Cold Spring Harb. Symp. Quant. Biol. 68, 69–78 [DOI] [PubMed] [Google Scholar]
- 40.Altshuler D. M., Gibbs R. A., Peltonen L., Altshuler D. M., Gibbs R. A., Peltonen L., Dermitzakis E., Schaffner S. F., Yu F., Peltonen L., Dermitzakis E., Bonnen P. E., Altshuler D. M., Gibbs R. A., de Bakker P. I., Deloukas P., Gabriel S. B., Gwilliam R., Hunt S., Inouye M., Jia X., Palotie A., Parkin M., Whittaker P., Yu F., Chang K., Hawes A., Lewis L. R., Ren Y., Wheeler D., Gibbs R. A., Muzny D. M., Barnes C., Darvishi K., Hurles M., Korn J. M., Kristiansson K., Lee C., McCarrol S. A., Nemesh J., Dermitzakis E., Keinan A., Montgomery S. B., Pollack S., Price A. L., Soranzo N., Bonnen P. E., Gibbs R. A., Gonzaga-Jauregui C., Keinan A., Price A. L., Yu F., Anttila V., Brodeur W., Daly M. J., Leslie S., McVean G., Moutsianas L., Nguyen H., Schaffner S. F., Zhang Q., Ghori M. J., McGinnis R., McLaren W., Pollack S., Price A. L., Schaffner S. F., Takeuchi F., Grossman S. R., Shlyakhter I., Hostetter E. B., Sabeti P. C., Adebamowo C. A., Foster M. W., Gordon D. R., Licinio J., Manca M. C., Marshall P. A., Matsuda I., Ngare D., Wang V. O., Reddy D., Rotimi C. N., Royal C. D., Sharp R. R., Zeng C., Brooks L. D., McEwen J. E.; International HapMap 3 Consortium (2010) Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Howie B. N., Donnelly P., Marchini J. (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gillespie, J. H. (2004) Population Genetics: A Concise Guide, 2nd ed., John Hopkins University Press, Baltimore, MD [Google Scholar]
- 43.Akey J. M., Eberle M. A., Rieder M. J., Carlson C. S., Shriver M. D., Nickerson D. A., Kruglyak L. (2004) Population history and natural selection shape patterns of genetic variation in 132 genes. PLoS Biol. 2, e286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balding D. J., Nichols R. A. (1995) A method for quantifying differentiation between populations at multi-allelic loci and its implications for investigating identity and paternity. Genetica 96, 3–12 [DOI] [PubMed] [Google Scholar]
- 45.Beaumont M. A., Balding D. J. (2004) Identifying adaptive genetic divergence among populations from genome scans. Mol. Ecol. 13, 969–980 [DOI] [PubMed] [Google Scholar]
- 46.Mehedint M. G., Craciunescu C. N., Zeisel S. H. (2010) Maternal dietary choline deficiency alters angiogenesis in fetal mouse hippocampus. Proc. Natl. Acad. Sci. USA 107, 12834–12839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schattenberg J. M., Galle P. R. (2010) Animal models of non-alcoholic steatohepatitis: of mice and man. Dig. Dis. 28, 247–254 [DOI] [PubMed] [Google Scholar]
- 48.Wang B., Majumder S., Nuovo G., Kutay H., Volinia S., Patel T., Schmittgen T. D., Croce C., Ghoshal K., Jacob S. T. (2009) Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology 50, 1152–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lachance J., Tishkoff S. A. (2013) Population genomics of human adaptation. Annu. Rev. Ecol. Evol. Syst. 44, 123–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McDonald J. H., Kreitman M. (1991) Adaptive protein evolution at the Adh locus in Drosophila. Nature 351, 652–654 [DOI] [PubMed] [Google Scholar]
- 51.Tajima F. (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fay J. C., Wu C. I. (2000) Hitchhiking under positive Darwinian selection. Genetics 155, 1405–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sabeti P. C., Reich D. E., Higgins J. M., Levine H. Z., Richter D. J., Schaffner S. F., Gabriel S. B., Platko J. V., Patterson N. J., McDonald G. J., Ackerman H. C., Campbell S. J., Altshuler D., Cooper R., Kwiatkowski D., Ward R., Lander E. S. (2002) Detecting recent positive selection in the human genome from haplotype structure. Nature 419, 832–837 [DOI] [PubMed] [Google Scholar]
- 54.Voight B. F., Kudaravalli S., Wen X., Pritchard J. K. (2006) A map of recent positive selection in the human genome. PLoS Biol. 4, e72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yonemori K. M., Lim U., Koga K. R., Wilkens L. R., Au D., Boushey C. J., Le Marchand L., Kolonel L. N., Murphy S. P. (2013) Dietary choline and betaine intakes vary in an adult multiethnic population. J. Nutr. 143, 894–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dalmeijer G. W., Olthof M. R., Verhoef P., Bots M. L., van der Schouw Y. T. (2008) Prospective study on dietary intakes of folate, betaine, and choline and cardiovascular disease risk in women. Eur. J. Clin. Nutr. 62, 386–394 [DOI] [PubMed] [Google Scholar]
- 57.Weichselbaum, E., Benelam, B., and Costa, H. S. (2009) Synthesis report No 6: Traditional Foods in Europe. Institute of Food Research, Norwich, UK. Available at: http://www.eurosfaire.prd.fr/7pc/documents/1263815283_traditional_foods_can_sustain_european_cultures.pdf. Accessed December 2, 2014
- 58.Nielsen R., Hellmann I., Hubisz M., Bustamante C., Clark A. G. (2007) Recent and ongoing selection in the human genome. Nat. Rev. Genet. 8, 857–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abraham G., Inouye M. (2014) Fast principal component analysis of large-scale genome-wide data. PLoS ONE 9, e93766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jallow M., Teo Y. Y., Small K. S., Rockett K. A., Deloukas P., Clark T. G., Kivinen K., Bojang K. A., Conway D. J., Pinder M., Sirugo G., Sisay-Joof F., Usen S., Auburn S., Bumpstead S. J., Campino S., Coffey A., Dunham A., Fry A. E., Green A., Gwilliam R., Hunt S. E., Inouye M., Jeffreys A. E., Mendy A., Palotie A., Potter S., Ragoussis J., Rogers J., Rowlands K., Somaskantharajah E., Whittaker P., Widden C., Donnelly P., Howie B., Marchini J., Morris A., SanJoaquin M., Achidi E. A., Agbenyega T., Allen A., Amodu O., Corran P., Djimde A., Dolo A., Doumbo O. K., Drakeley C., Dunstan S., Evans J., Farrar J., Fernando D., Hien T. T., Horstmann R. D., Ibrahim M., Karunaweera N., Kokwaro G., Koram K. A., Lemnge M., Makani J., Marsh K., Michon P., Modiano D., Molyneux M. E., Mueller I., Parker M., Peshu N., Plowe C. V., Puijalon O., Reeder J., Reyburn H., Riley E. M., Sakuntabhai A., Singhasivanon P., Sirima S., Tall A., Taylor T. E., Thera M., Troye-Blomberg M., Williams T. N., Wilson M., Kwiatkowski D. P.; Wellcome Trust Case Control Consortium; Malaria Genomic Epidemiology Network (2009) Genome-wide and fine-resolution association analysis of malaria in West Africa. Nat. Genet. 41, 657–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang L., Li Y., Singleton A. B., Hardy J. A., Abecasis G., Rosenberg N. A., Scheet P. (2009) Genotype-imputation accuracy across worldwide human populations. Am. J. Hum. Genet. 84, 235–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schlebusch C. M., Sjödin P., Skoglund P., Jakobsson M. (2013) Stronger signal of recent selection for lactase persistence in Maasai than in Europeans. Eur. J. Hum. Genet. 21, 550–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wagh K., Bhatia A., Alexe G., Reddy A., Ravikumar V., Seiler M., Boemo M., Yao M., Cronk L., Naqvi A., Ganesan S., Levine A. J., Bhanot G. (2012) Lactase persistence and lipid pathway selection in the Maasai. PLoS ONE 7, e44751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Itan Y., Powell A., Beaumont M. A., Burger J., Thomas M. G. (2009) The origins of lactase persistence in Europe. PLOS Comput. Biol. 5, e1000491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perry G. H., Dominy N. J., Claw K. G., Lee A. S., Fiegler H., Redon R., Werner J., Villanea F. A., Mountain J. L., Misra R., Carter N. P., Lee C., Stone A. C. (2007) Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 39, 1256–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Patin E., Quintana-Murci L. (2008) Demeter’s legacy: rapid changes to our genome imposed by diet. Trends Ecol. Evol. (Amst.) 23, 56–59 [DOI] [PubMed] [Google Scholar]
- 67.Babbitt C. C., Warner L. R., Fedrigo O., Wall C. E., Wray G. A. (2011) Genomic signatures of diet-related shifts during human origins. Proc. Biol. Sci. 278, 961–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.USDA National Nutrient Database for Standard Reference, Release 26. (2013) U.S. Department of Agriculture, Agricultural Research Service. Nutrient Data Laboratory Home Page. Available at: http://www.ars.usda.gov/ba/bhnrc/ndl/. Accessed June 27, 2014
- 69.Knoll N., Kuhnt K., Kyallo F. M., Kiage-Mokua B. N., Jahreis G. (2011) High content of long-chain n-3 polyunsaturated fatty acids in red blood cells of Kenyan Maasai despite low dietary intake. Lipids Health Dis. 10, 141 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.