Abstract
Trinucleotide repeat (TNR) expansions are the underlying cause of more than 40 neurodegenerative and neuromuscular diseases, including myotonic dystrophy and Huntington’s disease, yet the pathway to expansion remains poorly understood. An important step in expansion is the shift from a stable TNR sequence to an unstable, expanding tract, which is thought to occur once a TNR attains a threshold length. Modeling of human data has indicated that TNR tracts are increasingly likely to expand as they increase in size and to do so in increments that are smaller than the repeat itself, but this has not been tested experimentally. Genetic work has implicated the mismatch repair factor MSH3 in promoting expansions. Using Saccharomyces cerevisiae as a model for CAG and CTG tract dynamics, we examined individual threshold-length TNR tracts in vivo over time in MSH3 and msh3Δ backgrounds. We demonstrate, for the first time, that these TNR tracts are highly dynamic. Furthermore, we establish that once such a tract has expanded by even a few repeat units, it is significantly more likely to expand again. Finally, we show that threshold- length TNR sequences readily accumulate net incremental expansions over time through a series of small expansion and contraction events. Importantly, the tracts were substantially stabilized in the msh3Δ background, with a bias toward contractions, indicating that Msh2-Msh3 plays an important role in shifting the expansion-contraction equilibrium toward expansion in the early stages of TNR tract expansion.
Keywords: Saccharomyces cerevisiae, Msh2-Msh3, mismatch repair, trinucleotide repeat tract
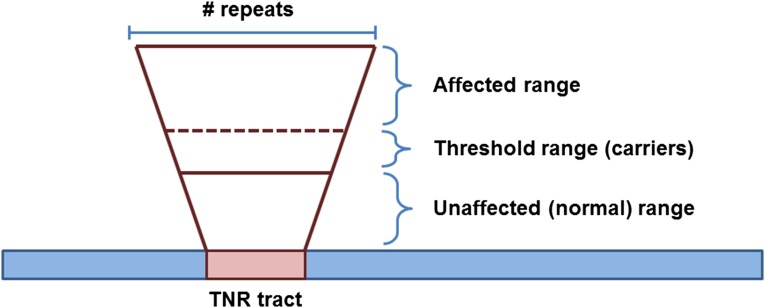
THE EXPANSION of trinucleotide repeat (TNR) sequences is the underlying cause of over 40 neurodegenerative and neuromuscular diseases (Castel et al. 2010; McMurray 2010). TNR sequences made of (CNG)n repeats are of particular interest because of their role in causing Huntington’s disease (HD) and myotonic dystrophy type 1 (DM1), as well as a number of other diseases (McMurray 2010). TNR tracts within the normal range (which is tract dependent) are stably maintained within that range (Figure 1). However, through a mechanism(s) that remains unclear, a TNR tract can expand, increasing the number of repeats within the tract. Initially, this brings the tract into a threshold-length range (Gannon et al. 2012; Concannon and Lahue 2014) (Figure 1 and Figure 2), in which these somewhat longer tracts are not pathogenic but are increasingly susceptible to expansion; individuals with this phenomenon are carriers for disease. Once a tract has expanded sufficiently, it crosses a threshold; tracts above this threshold (which is disease specific) are pathogenic and cause disease (Figure 1). As the size of the tract increases, it becomes increasingly unstable and prone to changes in length, particularly expansions.
Figure 1.
Size of trinucleotide repeat (TNR) tract determines disease phenotype. The normal range of a TNR tract is present in unaffected individuals. This range varies depending on the TNR sequence and position with respect to the relevant gene. As the TNR tract gets larger, it enters the threshold or premutation range. These individuals are typically unaffected by disease but are carriers because these somewhat larger tracts are prone to further expansion. Once the TNR tract expands into the affected range, individuals are symptomatic and are affected by disease.
Figure 2.
Distribution of tract size following initial expansion. Whisker plot of the median tract size following selection on 5-FOA plates in MSH3 and msh3Δ strains. The (CAG)25 and (CTG)25 tracts were examined. The plot was created using GraphPad Prism. Both the median and mean sizes of the CAG tracts are statistically different from the CTG tracts (Wilcox rank, t-test, P < 0.0001). The median and mean CAG tract lengths in the MSH3 background are statistically different from the CAG tract in the msh3Δ background, whereas the CTG tract size is similar in both strain backgrounds (Wilcox rank, t-test, P < 0.0001).
The dynamic behavior of TNR tracts that are within the threshold range (i.e., more susceptible to expansions) and the manner in which they occur in vivo remain unclear. Studies of TNR tract length changes have largely relied on end point experiments and therefore do not address the dynamic behavior of the tracts, i.e., the rate at which tracts continue to expand. Modeling of human data has predicted that threshold-length TNR tracts will continue to increase in length in increments smaller than the repeat itself (Higham et al. 2012; Morales et al. 2012; Higham and Monckton 2013), although this has never been demonstrated directly. Single-sperm typing studies in humans demonstrated small expansion and contraction events (one–two repeats) in TNR sequences in Kennedy’s disease, HD, and DM1 patients (Zhang et al. 1994; Leeflang et al. 1995, 1999; Martorell et al. 2004), particularly in alleles near the pathogenic threshold (Zhang et al. 1994; Leeflang et al. 1995; Castel et al. 2010). These observations are consistent with the existence of an equilibrium between expansion and contraction events. As the tract length increased in sperm cells, there was significant bias toward expansions vs. contractions. The sizes of the observed expansion and contraction events were similar to our in vitro observations (one–two repeats) (Kantartzis et al. 2012), in contrast to larger expansion events observed in nondividing cells or postmitotic neurons (McMurray 2010).
One factor known to contribute to (CNG)n tract expansions is the mismatch repair (MMR) complex Msh2-Msh3 (MutSβ in mammals). Typically, Msh2-Msh3 recognizes and binds insertion-deletion loops (IDLs) that result from DNA polymerase slippage events, often within repetitive sequences (Lovett 2004; Li 2008). These are then targeted for excision and resynthesis. Strikingly, rather than correcting them, Msh2-Msh3 promotes TNR expansions in both mammalian somatic and germ cells (Castel et al. 2010; McMurray 2010). This difference is likely related to the propensity of (CNG)n sequences to form secondary structures once they have slipped and are single stranded owing to the inherent complementarity of the C’s and G’s within the repeat sequence (Castel et al. 2010; McMurray 2010) and to the manner in which Msh2-Msh3 interacts with these unique structures (Lang et al. 2011). As the tract length increases, the potential complexity of the secondary structure increases (Gacy and McMurray 1998; Pearson and Sinden 1998; Slean et al. 2013). Nonetheless, loss of MSH2 or MSH3 leads to a significant decrease in expansion events in mouse models of HD (Manley et al. 1999; Owen et al. 2005) and MD1 (van den Broek et al. 2002; Foiry et al. 2006). Similarly, Msh3 promotes expansions in human cells (Gannon et al. 2012; Halabi et al. 2012). We recently demonstrated a significant decrease in expansion of both CAG and CTG repeat tracts in Saccharomyces cerevisiae in an msh3Δ background (Kantartzis et al. 2012).
The role that Msh2-Msh3 plays in promoting TNR expansions remains unclear. Our in vitro results indicate that yeast Msh2-Msh3 interferes with proper Okazaki fragment processing by Rad27 (Fen1) and Cdc9 (Lig1) in the presence of a dynamic CTG or CAG repeat tract, leading to small incremental expansions and providing mechanistic insight into the role of Msh2-Msh3 in replication-dependent TNR expansions (Kantartzis et al. 2012). This model predicts that Msh2-Msh3 modulates TNR dynamics in vivo. Subsequently, Stevens et al. (2013) demonstrated that Msh2-Msh3 can promote expansions in vitro using human cell culture extracts in a replication-independent manner, consistent with a role for Msh2-Msh3 in promoting expansions in somatic as well as germline cells (Castel et al. 2010; McMurray 2010).
To address the question of threshold-length tract behavior and the role that MSH3 plays in that behavior, we have established S. cerevisiae as a model to visualize and characterize individual tract dynamics in dividing cells as a function of time through multiple generations. We observed highly dynamic DNA sequences with progressive, incremental expansion in an MSH3 background. The TNR tract dynamics were substantially altered in the absence of MSH3, resulting in more stable repeat sequences. These observations indicate that MSH3 is an important factor in promoting the transition of a TNR tract into the pathogenic range.
Methods and Materials
Strains and media
All yeast transformations were performed using the lithium acetate method (Gietz et al. 1992). Yeast strains were derived from the S288c background and are detailed in Supporting Information, Table S1. In addition to the msh3Δ::KANMX strain, constructed by amplifying a chromosomal msh3Δ::KANMX fragment from the yeast deletion collection that was integrated into the MSH3 chromosomal location in FY86 (Kantartzis et al. 2012), we constructed a second msh3Δ strain in FY86. The msh3Δ::hisG strain was constructed by integration of a hisG-URA3-hisG cassette into the MSH3 locus using pEAI88 (Lyndaker et al. 2008). Integration was selected on synthetic medium lacking uracil. The URA3 gene was selected against by growth on 5-FOA, which selects for recombination between the two hisG moieties (Alani et al. 1987), resulting in a single hisG interrupting the MSH3 ORF. The tract dynamics in the two msh3Δ backgrounds were indistinguishable.
TNR substrates were integrated into MSH3 and msh3Δ strains, as described previously (Miret et al. 1998; Dixon et al. 2004; Kantartzis et al. 2012). Briefly, each plasmid was digested with Bsu36I and then transformed to allow integration by homologous recombination at the LYS2 chromosomal locus. Integrants were selected on synthetic medium lacking histidine. Single integration of the TNR substrate at the correct location was confirmed by Southern blot, as described previously (Dixon et al. 2004). Three independent single integrants for each substrate were selected for further analysis. This was repeated in all three strain backgrounds. Prior to performing the expansion assay, each strain was struck out on synthetic medium lacking histidine and uracil to obtain single colonies. Individual colonies were analyzed by colony PCR to ensure that they contained the proper number of repeats (Dixon et al. 2004). A subset also was sequenced to confirm the tract using SO299 as a primer (5′-ACTTGGGGAGAGGTGCG) (Dixon et al. 2004; Kantartzis et al. 2012).
Growth curves
Saturated cultures were diluted 1:100 in synthetic medium lacking histidine. Absorbance (A600) of the cultures was measured every 45 min for 10 hr. The cultures were allowed to saturate overnight and were then diluted again and the growth curve repeated. There was no difference in the growth curves of the different strains tested or between the first and second days of growth, with a doubling time of approximately 140 min once the cells reached exponential phase (Figure S1 and data not shown). In 24 hr, the cells underwent approximately six cell divisions. For microcolony growth curves, individual cells were tracked using a dissection microscope with a micromanipulator, with examination every 30 min for 10 hr. The daughter cells were removed from the mother following each cell division and placed elsewhere on the plate. There was no observable difference in the timing of the cell cycle between the MSH3 and the msh3Δ strains, which both exhibited a doubling time of approximately 140 min. Generation times of growth in liquid and on solid medium were confirmed by counting the number of cells in culture and in colonies of different sizes using a hemacytometer.
Liquid time course protocol
Expanded CAG or CTG tracts in either the wild-type or msh3Δ background were selected by growth on SC medium lacking histidine (to maintain the tract construct) and containing 5-FOA (to select for expansions that abrogate URA3 expression; United States Biological), as described previously (Miret et al. 1998; Kantartzis et al. 2012). The size of the tract was analyzed by colony PCR using SO295 and SO296 (Kantartzis et al. 2012). The PCR cycle consisted of one round at 95° for 5 min and then 35 cycles at 95° for 2 min, 53° for 1 min, and 72° for 1 min, followed by a final extension at 72° for 10 min. The PCR products were digested with SphI and AflII and then electrophoresed through a 12% polyacrylamide gel in 1× TBE buffer, and the gel was stained with ethidium bromide (EtBr, 0.5 μg/ml) to determine tract size. The gels were imaged on a GelDoc system (Bio-Rad) and quantified by ImageQuant (Molecular Dynamics).
Three independent colonies were selected for expansion events in each genetic background, i.e., (CTG)25 and (CAG)25 tracts in MSH3 and msh3Δ . Each colony was resuspended in 106 mL of SC medium lacking histidine (SC-his) and grown to saturation at 30° with shaking. Then 5 ml of each saturated culture was used to prepare genomic DNA (gDNA). From each saturated culture, a logarithmically growing (log-phase) culture and a stationary-phase culture were established. For the logarithmic culture, 10 ml of SC-his was inoculated with 200 μl of the saturated culture and grown at 30° in a shaker for 24 hr. This dilution was repeated every 24 hr for the duration of the time course. The remainder of each saturated culture was collected by centrifugation, and the pellet was washed twice with sterile water and then resuspended in 100 mL of sterile distilled water. These stationary-phase cultures were maintained at 30° in the shaker for the duration of the time course. The stationary-phase cell pellet was washed and resuspended in fresh distilled water every 48 hr.
Every 24 hr, 5 ml from each log- and stationary-phase culture was used to prepare gDNA. On days 1, 7, and 14, dilutions from each culture were plated on SC-his to get individual colonies. PCR was performed, as described earlier, to amplify the tract using the gDNA from each time point as a template. Colony PCR was performed on individual colonies from days 7 and 14.
PCR controls
PCR has been used extensively to examine TNR tract sizes (Zhang et al. 1994; Leeflang et al. 1995, 1999; Miret et al. 1998; Dixon et al. 2004). Nonetheless, we performed several controls to ensure that these observed expansions were not PCR artifacts. First, we performed PCR reactions using the CAG or CTG tract plasmid (used to integrate the tract into the yeast chromosome) (Miret et al. 1998; Kantartzis et al. 2012) as a template alongside every set of PCR reactions. A subset of these reactions is shown in Figure S2 and clearly demonstrates the ability of Taq polymerase to accurately and reproducibly amplify a tract of 25 repeats. In over 100 PCR reactions using the plasmid as a template, the major product was a 75-base-pair fragment in every case. Minor products were observed occasionally at larger sizes and likely represent background due to polymerase error. Based on Southern blots of 20 independent plasmid amplification reactions, ≥85% (±1.5% SEM) of the total product is the 75-base-pair unexpanded tract (Figure S3, right, and data not shown). As an additional control, we performed five independent colony PCR reactions simultaneously from the same colony to assess reproducibility of Taq in amplifying both expanded and unexpanded tracts by colony PCR (Figure S4). The resulting pattern of PCR products was very consistent and reproducible.
Mixing experiment
To determine whether the ratio of PCR products was representative of the input DNA template, we performed PCR reactions that contained different ratios of genomic DNA from cells with either an unexpanded and an expanded TNR tract. The DNA was amplified and then quantified (Figure S5). gDNA was prepared from cells carrying an unexpanded tract and from cells carrying an expanded tract using standard techniques. The DNA concentration of each preparation was measured by NanoDrop (Thermo Scientific). The gDNA from both backgrounds was used as a template for PCR at different concentration ratios. Ratios of 1:1, 1:5, 1:10, and 1:20 unexpanded-expanded and expanded-unexpanded were tested as templates in the standard PCR reaction (described earlier).
The ratio of unexpanded product to expanded product correlated with the input DNA, although at a 1:1 ratio there was a notable bias toward amplification of the unexpanded tract. This reassured us that we were not preferentially amplifying the expanded products and thereby overestimating the expansion rate. Furthermore, the PCR output was a reasonable representation of the input template, and we are more likely to err on the side of underestimating expansion events when performing PCR with a nonhomogeneous template population. We do note that this may lead us to underestimate expansions in the msh3Δ background as well.
Colony PCR time course protocol
Expanded TNR tracts were selected, and the tract size was analyzed and measured, as described earlier. Tracts of different length were selected for this experiment. Colonies with expanded tracts were incubated continuously at 30° for 10–14 days . Colony PCR was performed every 24 hr and the tract analyzed by PCR and digestion. Colony PCR was performed either from the same side of the colony or from around the perimeter of the colony with similar results. When taken from around the colony, the samples were taken from four locations in rotations corresponding to 12, 3, 6, and 9 o’clock. Because the cells were always sampled from the perimeter of the colony, we assumed a generation time of ∼140 min, as described earlier.
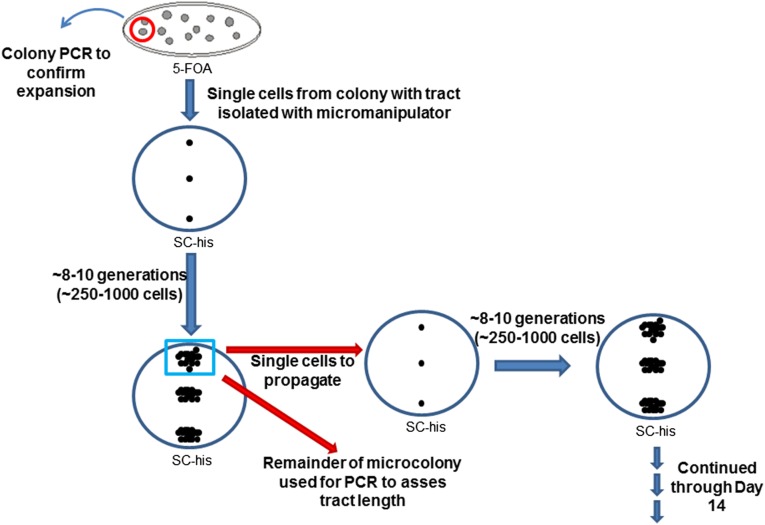
Microcolony PCR time course protocol
Expanded TNR tracts were selected, and the tract size was analyzed and measured, as described earlier. Colonies with expanded tracts were resuspended in sterile water. Each suspension was run in a line on an SC-his plate. Using a micromanipulator on a dissection microscope, three single cells from each suspension were selected for the time course. Each plate was incubated at 30° for 15–20 hr, until the microcolony contained between 250 and 1000 cells; this was the size necessary for reproducible PCR results. Three individual cells were removed from the microcolony to continue the time course. (Three cells were selected to ensure that at least one microcolony arose in the next round.) The remaining cells were used for colony PCR to assess the size of the TNR tract. The plate then was incubated at 30° for another 15–20 hr to allow growth of the four individual cells to a microcolony. One of the four cells was selected to propagate and continue the time course. The time course was continued through 14 cycles. At each step, tract sizes were analyzed by PCR, as described earlier. Time points in which no PCR amplification was observed were excluded from our analysis.
Southern blotting
To verify that the PCR products contained the TNR tract, Southern blots were performed (Sambrook and Russell 2001). Briefly, PCR reactions were electrophoresed through a 12% acrylamide gel (1× TBE). The gel was transferred to a nylon membrane, cross-linked and hybridized with a radiolabeled probe, as described previously (Dixon et al. 2004; Kantartzis et al. 2012). The probe was a PCR product amplified from pBL69 (Miret et al. 1998) with SO295 and SO296 and then digested with SphI. The resulting fragment was radiolabeled using the Takara Random Priming Kit. The membrane was washed, dried, and exposed to a PhosphorImager screen and imaged and quantified, where applicable, in ImageQuant.
Mutation-rate calculations
To calculate the mutation rates, the probability of a change in tract size was treated as a binomial distribution with P = proportion of tracts with a change in length and q = proportion of tracts with no length change. The rate of tract change was defined as the number of changes observed per number of cells examined per generation. For the liquid time courses, the mutation rate was derived from the number of tract changes observed per colonies tested divided by 126 generations per culture. For the colony and microcolony time courses, the proportion of tract changes was calculated as the number of tract changes divided by the number time points examined divided by the number of generations from one time point to the next. Growth curve experiments in both liquid medium and on plates were used to determine the number of generations for these calculations (Figure S1 and data not shown). To calculate 95% confidence intervals (95% C.I.) on the mutation rates, the F-statistic was used (Zar 1999; Foster et al. 2006).
Plasmid retention assays
Three independent MSH3 isolates with large tracts, ranging from 33 to 48 repeats, were transformed with a low-copy plasmid (ARS CEN) carrying the KANMX cassette, which confers resistance to G418 (Wach et al. 1994). Three independent MSH3 isolates with small tracts, ranging from 25 to 30 repeats, were transformed with a low-copy plasmid (ARS CEN) carrying the NATMX cassette, which confers resistance to nourseothricin (Goldstein and McCusker 1999). The two plasmids had indistinguishable retention rates in the absence of selection (data not shown). In each of three cultures, one isolate with a large tract and one isolate with a small tract were mixed 1:1 and were taken through a time course in SC-his for 6 days (see the section Liquid time course protocol earlier) in the absence of selection for either plasmid. In the first set, a 40-repeat tract and a 25-repeat (unexpanded) tract were mixed. In the second set, a 38-repeat tract and a 31-repeat tract were mixed. In the third set, a 33-repeat tract and a 25-repeat tract were mixed. Every 24 hr, a sample of the culture was plated on SC-his medium. Each individual colony was tested for growth on YPD medium containing either G418 or nourseothricin. The percentages of cells retaining resistance for one drug or the other are plotted in Figure S6.
Results
The major goal of this study was to determine how a TNR tract behaves as it approaches a pathogenic length. To do this, we adapted the in vivo URA3-based reporter system in S. cerevisiae that we used previously (Miret et al. 1998; Kantartzis et al. 2012) and examined larger threshold-length (CNG)n tract dynamics over time in the presence and absence of MSH3. Previous work indicated that 25-repeat tracts approach the threshold length in this system (Miret et al. 1998; Concannon and Lahue 2014). Therefore, we started with (CTG)25 and (CAG)25 TNR tracts and selected single colonies with expanded tracts, as described previously (see Materials and Methods) (Miret et al. 1998; Kantartzis et al. 2012). We took this analysis further by determining (1) whether expanded tracts continue to expand, (2) at what rate tracts expand, and (3) in what increments expansions occur. Throughout this work we will follow the following convention: a CTG or CAG tract refers to the sequence of the nascent DNA on the lagging strand. Thus, a CTG tract refers to CAG on the template strand, and the newly replicated DNA will contain the CTG tract.
We took three different approaches to answering these questions using PCR techniques to monitor tract length. First, we performed liquid time course experiments to determine whether additional expansions accumulated in replicating cells within a reasonable time frame (Figure 3, Figure 4, and Figure 5). Once a tract has expanded, it is not possible to select for further expansions; this tested the feasibility of our approach. Second, we sought to observe dynamic tract size changes in a single colony over time (Figure 6, Figure 7, Figure 8, and Figure 9). The expanded tract of an individual growing colony was monitored through multiple generations in the absence of selection. Third, we isolated individual cells from colonies with expanded tracts and monitored tract dynamics over a well-defined number of generations (Figure 10 and Figure 11). In each case, approximate rates of tract changes were determined (Table 1). Combined, these three approaches allowed us to monitor tract length dynamics through multiple generations to document the types of tract changes that occurred in vivo and the rates at which they occurred. Each set of experiments was done in the presence and absence of MSH3 to determine its role in tract dynamic behavior.
Figure 3.
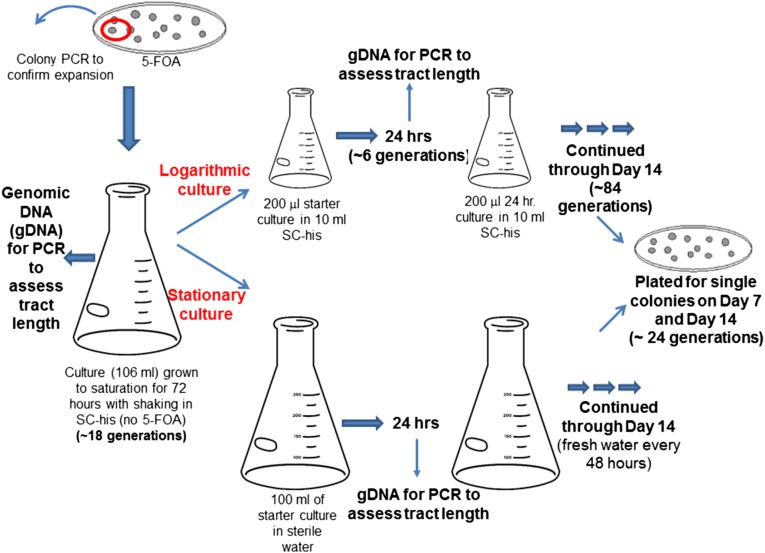
Schematic of the liquid time course experiment protocols. Individual colonies with TNR tract expansions were selected on plates containing 5-FOA; the tract increase was confirmed by colony PCR in each case. A single colony with an expanded tract was used to inoculate a large culture that was grown to saturation over 72 hr (∼18 generations). From this population, genomic DNA (gDNA) was isolated. Then parallel log- and stationary-phase cultures were established (see Materials and Methods) and propagated for 14 days. gDNA was prepared from each culture every 24 hr (∼6 generations) and then subjected to PCR to evaluate tract size. A sample from each log- and stationary-phase cultures was diluted and plated to isolate individual colonies (∼26 generations), which then were subjected to PCR to assess tract length. This experiment is akin to a mutation-accumulation experiment, although we limited the number of cell divisions (∼6 per time point) to mitigate any fitness effects of tract changes. This is an end point experiment that indicated that additional expansions are observable in a 2-week time span. Further, it allowed us to observe general trends in the population as well as looking at individual tract lengths from single colonies. See Materials and Methods for additional details.
Figure 4.
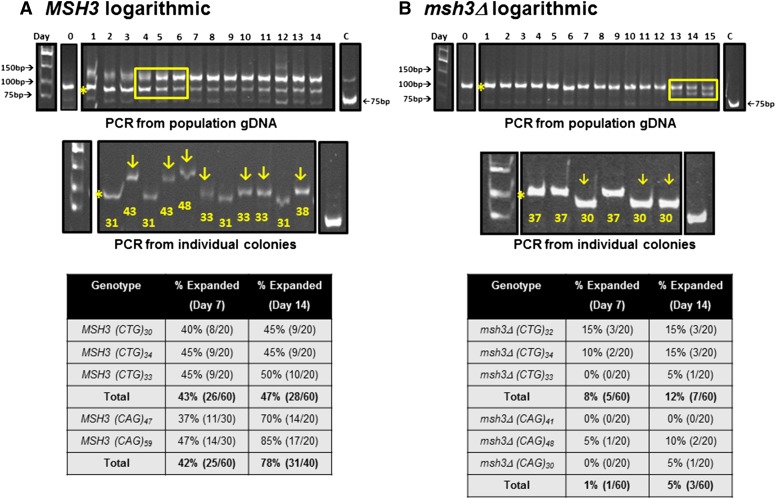
Liquid time course experiments in MSH3 and msh3Δ logarithmic cultures to determine population tract dynamics. TNR expansion events in the MSH3 or msh3Δ background were selected and confirmed by PCR (see Materials and Methods) and log-phase cultures were established from individual isolates (see Figure 3 for cartoon). (A) MSH3(CTG). (B) msh3Δ(CTG). Top: The TNR tract from gDNA isolated from the MSH3 log-phase cultures was amplified, digested, and analyzed by electrophoresis. In these examples, the initial expansion (indicated by the asterisk) contained 31 repeats for MSH3 and 37 repeats for msh3Δ. The numbers across the top of the gel indicate the day of the time course. The boxed region in A (MSH3) indicates the progressive accumulation of a larger tract. The boxed region in B (msh3Δ) indicates contraction events that result in a smaller tract. The cultures went through approximately six generations in a 24-hr period (see Materials and Methods). Middle: Samples from day 14 cultures were plated on minimal medium lacking histidine to obtain individual colonies. Colony PCR was performed to amplify the TNR tract from colonies to determine individual tract lengths within the population. The arrows indicate tracts that have incurred an additional expansion. The number below each tract indicates the number of repeats within each tract. Control PCR reactions using the TNR plasmid (C) were performed alongside each set as a marker for the 75-base-pair tract (25 repeats). We occasionally observed higher-molecular-weight products with the control plasmid, but these represent a minor population of ≤15% of the total PCR product (Figure S2 and Figure S3). The asterisk indicates the predominant expansion product in the starting culture. Bottom: Summary of expansion frequency on days 7 and 14 in the different genetic backgrounds tested based on PCR amplification of tracts from individual colonies.
Figure 5.
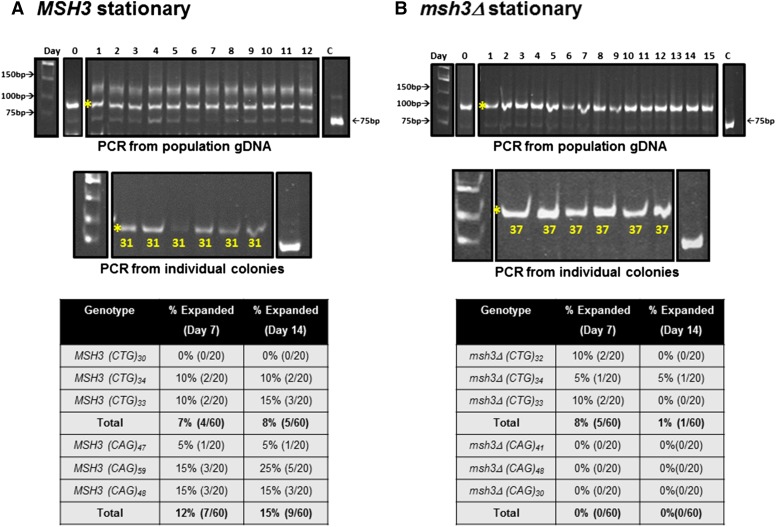
Liquid time course experiments in MSH3 and msh3Δ stationary-phase cultures to determine population tract dynamics. Stationary-phase cultures were maintained in parallel with log-phase cultures (see Figure 3 for cartoon). The examples shown here for (A) MSH3 and (B) msh3Δ match those shown in Figure 3. Top: The TNR tract was amplified from gDNA isolated from the stationary-phase culture, digested, and analyzed by electrophoresis. In both MSH3 and msh3Δ, the population remained unchanged over time. The numbers across the top of the gel indicate the day of the time course. Middle: Samples from day 14 cultures were plated on minimal medium lacking histidine to obtain individual colonies. Colony PCR was performed to amplify individual TNR tracts from the stationary-phase population to determine tract lengths. These tracts were quite stable. Control PCR reactions using the TNR plasmid (C) were performed alongside each set. The asterisk indicates the predominant expansion product in the starting culture. Bottom: Summary of expansion frequency on days 7 and 14 in the different genetic backgrounds tested based on PCR amplification of tracts from individual colonies. For more detail, see Table S4.
Figure 6.
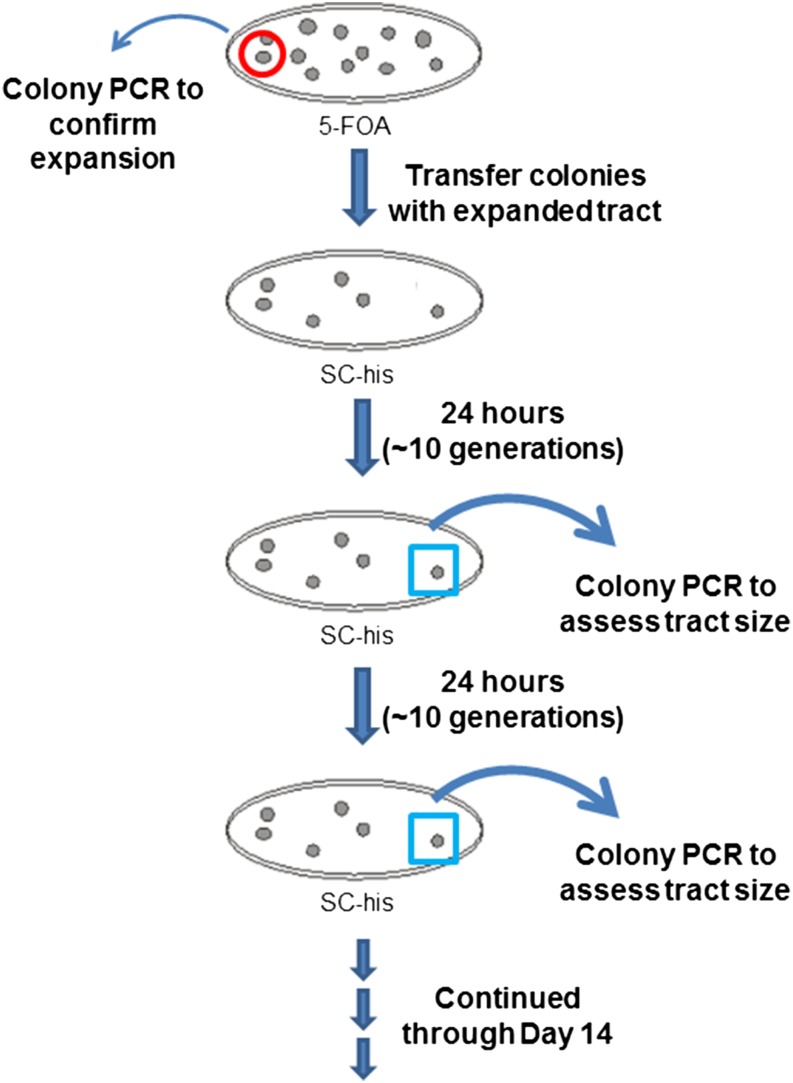
Schematic of the colony time course experiment protocol. Individual colonies with TNR tract expansions were selected on plates containing 5-FOA; the tract increase was confirmed by colony PCR in each case. A single colony with an expanded tract was transferred to nonselective medium and allowed to continue to grow. For each time course, the same colony was subjected to colony PCR every 24 hr (∼10 generations), sampling from the perimeter of the colony where the cells continued to grow. This approach allowed us to examine the dynamics of a single TNR tract as a function of time. Because the PCR is derived from a subsample of an actively dividing colony, we are necessarily examining a mixed population, albeit derived from the same original cell. See Materials and Methods for additional details.
Figure 7.
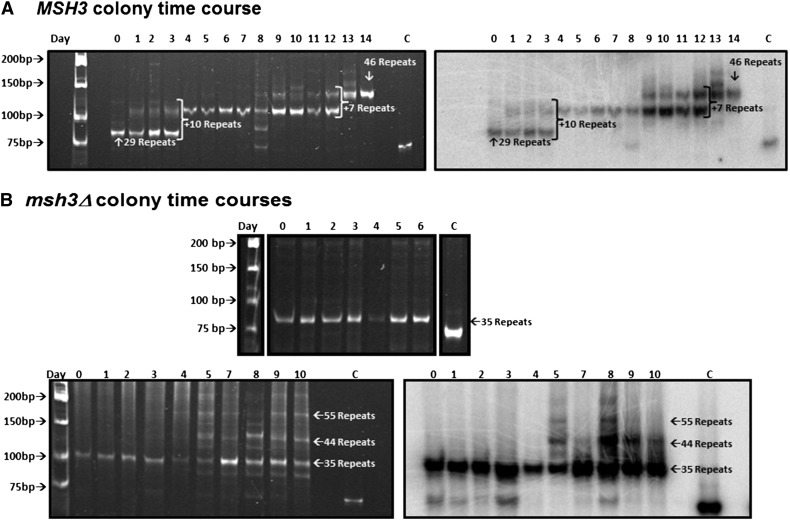
Progressive expansion events in MSH3 but not in msh3Δ. A. MSH3. Expansion events were selected in the MSH3(CTG)25 background and confirmed by PCR. Individual colonies were followed over several days; colony PCR was performed on each colony every 24 hr for 14 days to amplify the TNR tract (see Figure 6 for cartoon). Left: The tracts from time course E4 were resolved on a 12% polyacrylamide gel and stained with EtBr. The numbers across the top of the gel indicate the day of the time course. Right: Southern blot of the gel in left panel to demonstrate that the expansion products contain TNR sequence. The lanes marked C in each panel indicate the 75-base-pair tract amplified from the TNR plasmid control. (B) msh3Δ. Expansion events were selected in the msh3Δ(CTG)25 background and confirmed by PCR. Individual colonies were followed over several days; colony PCR was performed on each colony every 24 hr for 10 days to amplify the TNR tract. Top: The tracts from time course V1 were resolved on a 12% polyacrylamide gel and strained with EtBr. The numbers across the top of the gel indicate the day of the time course. Bottom left: The tracts from time course C8 were resolved on a 12% polyacrylamide gel and stained with EtBr. The numbers across the top of the gel indicate the day of the time course. Bottom right: Southern blot of the gel in lower left panel to demonstrate that the expansion products indicated by the arrows contain TNR sequence. The lanes marked C in each panel indicate the 75-base-pair tract amplified from the TNR plasmid control.
Figure 8.
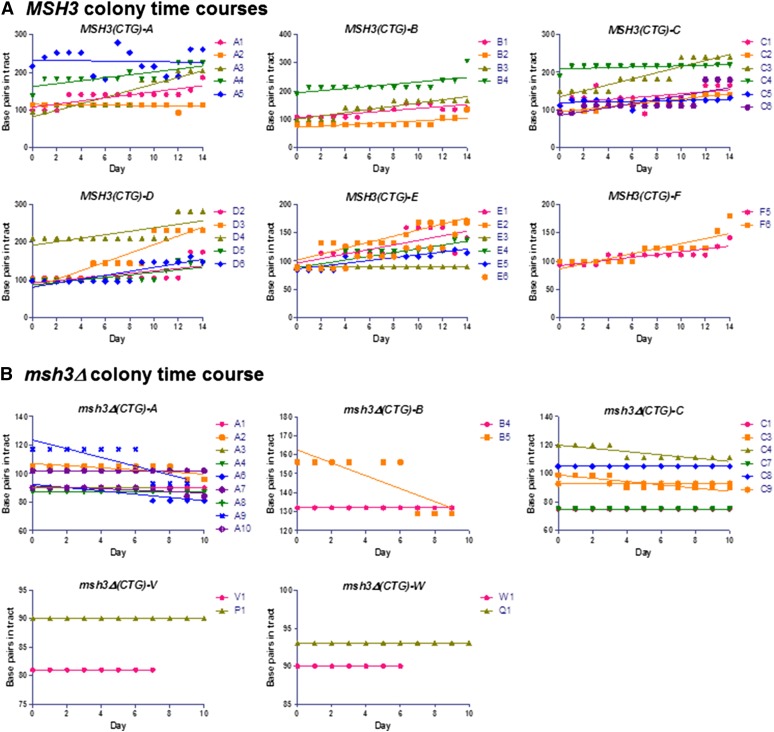
Colony time courses in MSH3 and msh3Δ backgrounds. The TNR tract size was plotted as a function of time to demonstrate trends in the data. Each panel represents an independent (CTG)25 integrant in the different strain backgrounds. Each curve in a single panel represents the time course of an independent expansion event in that background. (A) MSH3. All MSH3 colony time courses were plotted. Each plot shows the tract size changes in independent colonies from a single MSH3(CTG)25 isolate. A total of 64 time courses were performed: 28 showed progressive expansion, 3 showed tract stability, 11 showed no amplification (loss of tract), and 22 showed high background with multiple PCR products. This last category was difficult to interpret but exhibited trends similar to those analyzed in Figure 8A; 9 tracts exhibited increasing trends, and 4 tracts appeared stable. (B) msh3Δ. All msh3Δ time courses were plotted, and linear regressions were calculated. A total of 64 time courses were performed: 0 showed progressive expansion, 23 showed tract stability, 7 tracts showed contractions, 26 showed no amplification (loss of tract), and 6 showed high background with multiple PCR products. Linear regressions were plotted for each time course to illustrate the general trends in tract length.
Figure 9.
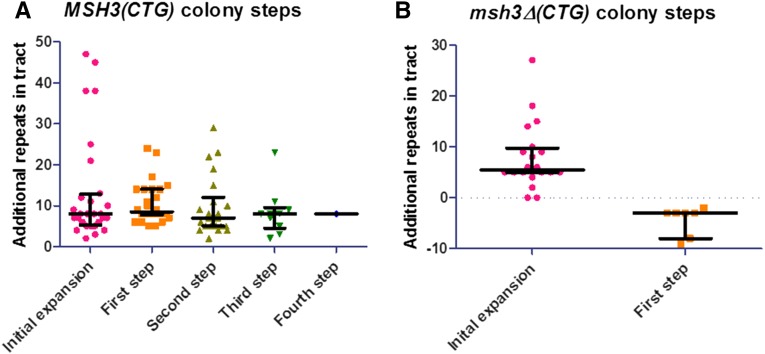
Sizes of each step (tract size change) in colony time course experiments. Each progressive tract length change, defined as the loss of one tract length and the concomitant appearance of a different tract length, was plotted as a function of size for MSH3 (A) and msh3Δ (B) using GraphPad Prism. The median and interquartile ranges for each step are indicated in black. (A) MSH3. The initial expansion size selected on 5-FOA is indicated (pink circles). The orange squares indicate the size of each individual tract upon the first tract length change or step. Similarly, the second, third, and fourth steps are shown (olive triangles, green inverted triangles, and blue diamonds, respectively). The median size of each step is essentially unchanged. (B) msh3Δ. The sizes of the initial expansion events are shown (pick circles); the median is comparable to that of the MSH3 time courses. Only one progressive step was observed (orange squares), and this was a step down to a short tract length.
Figure 10.
Schematic of the microcolony time course experiment protocol. Individual colonies with TNR tract expansions were selected on plates containing 5-FOA; the tract increase was confirmed by colony PCR in each case. Instead of initiating the time course with a potentially mixed population, the microcolony time course was started with single cells from a colony with an expanded tract. These cells were restricted to between 8 and 10 cell divisions, with an eye to minimizing the heterogeneity within the population. See Materials and Methods for additional details.
Figure 11.
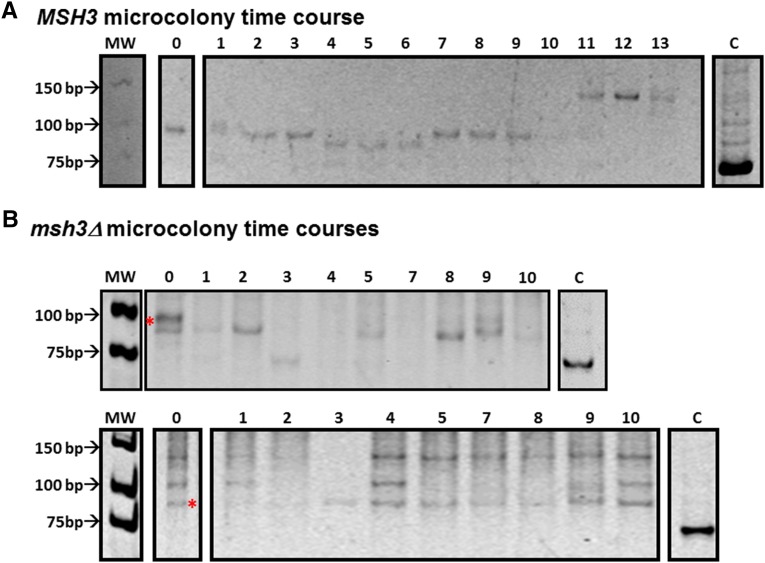
Dynamic changes in TNR tracts starting from a single cell. Expansion events were selected in either the MSH3(CTG)25 (A) or msh3Δ(CTG)25 (B) background and confirmed by PCR. Individual cells from these colonies were isolated and allowed to undergo 8–10 rounds of replication, resulting in a microcolony approximately 250–1000 cells in size. A single cell was then taken from this microcolony to propagate another microcolony. The remainder was used to amplify the TNR tract by PCR to determine tract length (see Figure 10 for cartoon). (A) An example of TNR tract dynamics in an MSH3 microcolony (F from Table 2). There are multiple changes in the TNR tract over time. (B) Two examples of TNR tract dynamics in an msh3Δ microcolony (N and Q, respectively, from Table 3). The red asterisk indicates the position of the initial expansion tract size, as determined by Southern blot. The upper bands in the bottom panel do not contain TNR tract sequences, as determined by Southern blot (data not shown). The tract appears more stable than in the MSH3 background. Both gels are 12% polyacrylamide gels stained with EtBr; the images of been inverted for ease of viewing. The lanes marked C in each panel indicate the 75-base-pair tract amplified from the TNR plasmid control. The numbers across the top of the gels indicate the time point.
Table 1. Summary of rates of tract length change calculated from liquid, colony, and microcolony time courses.
| Genotype | Rates of tract length changes | ||
|---|---|---|---|
| Liquida (95% C.I.)b | Colony (95% C.I.) | Microcolony (95% C.I) | |
| MSH3(CTG) | 1.2 × 10−3 (9.0 × 10−4 to 1.6 × 10−3) | 1.5 × 10−2 (1.2 × 10−2 to 1.9 × 10−2) | 3.5 × 10−2 (2.8 × 10−2 to 4.2 × 10−2)e |
| MSH3(CAG) | 3.1 × 10−3 (2.4 × 10−3 to 3.4 × 10−3) | NDc | ND |
| msh3Δ(CTG) | 3.1 × 10−4 (1.3 × 10−4 to 6.0 × 10−4) | 3.6 × 10−3 (1.5 × 10−3 to 7.2 × 10−3)d | 1.6 × 10−2 (1.1 × 10−2 to 2.2 × 10−2)f |
| msh3Δ(CAG) | 1.3 × 10−4 (2.9 × 10−4 to 3.7 × 10−4) | ND | ND |
| MSH3(CTG)25 | <5 × 10−5g,h | ND | <1.2 × 10−3g,h |
| MSH3(CAG)25 | 5.1 × 10−5h | ND | ND |
Expansions only.
95% C.I.s determined by F-statistic.
Not determined.
Predominantly contractions.
Slight bias toward expansions.
Bias toward contractions.
No tract changes observed.
Low numbers precluded accurate calculation of 95% C.I.
Prior to selecting for expansion events, colonies were screened for unexpanded tracts by colony PCR (Miret et al. 1998; Dixon et al. 2004; Kantartzis et al. 2012). In the process of screening the tracts, we noted differences between the MSH3 and msh3Δ strain backgrounds. The ratio of unexpanded, expanded, and contracted tracts was significantly different in the MSH3 vs. the msh3Δ strains (χ2: P = 0.0038). In particular, the relative ratios of expanded to unexpanded tracts and expanded to contracted tracts were significantly higher in MSH3 vs. msh3Δ strains (Fisher’s exact test: P = 0.0073 and 0.0061, respectively), consistent with a role for MSH3 in promoting TNR expansions. These data also indicated that msh3Δ strains exhibit an increase in contraction events, as suggested previously (Schweitzer and Livingston 1997; Castel et al. 2010).
Distinct expansion tracts in CAG and CTG repeat tracts
To measure the size of each initial expansion event in CTG and CAG tracts, the TNR tract from a 5-FOA-resistant colony was amplified by PCR (Miret et al. 1998; Dixon et al. 2004; Kantartzis et al. 2012), and the product was digested with SphI and AflII (see Materials and Methods), generating a DNA fragment that contains only the TNR tract. Although PCR has been used extensively to examine TNR tract sizes (Zhang et al. 1994; Leeflang et al. 1995, 1999; Miret et al. 1998; Dixon et al. 2004), we performed several controls to demonstrate that the observed expansions were not PCR artifacts (see Materials and Methods) (Figure S2, Figure S3, Figure S4, and Figure S5). With an unexpanded tract, the PCR product is 75 base pairs (25 CNG repeats); an expanded tract is larger, and a contracted tract is smaller (Figure S3). The size of each fragment was interpolated from a standard curve generated by the migration of low-molecular-weight DNA standards (New England Biolabs) electrophoresed alongside the PCR products.
In MSH3 cells, the median size of the initial expansion event, i.e., the expansion observed following selection on 5-FOA medium, was 42 repeats for the CAG tract (range 31–67 repeats) and 33 repeats for the CTG tract (range 26–43 repeats) (Figure 2 and Table S2), an increase of 17 and 8 repeats, respectively. The larger expansion size in the presence of the CAG tract is consistent with previous work (Miret et al. 1998) and is likely due to the relative stability of CTG and CAG secondary structures. CAG repeats form less stable structures than CTG repeats (Marquis Gacy et al. 1995; Pearson and Sinden 1996; Bacolla et al. 2008), and therefore, a larger expansion is predicted to be required to stabilize the secondary structure sufficiently to promote an expansion (Miret et al. 1998). Importantly, these size ranges put the tracts near the pathogenic threshold for CAG and CTG repeats in coding regions in humans (Figure 1) (Leeflang et al. 1999; Castel et al. 2010; McMurray 2010).
In two different msh3Δ backgrounds (see Materials and Methods) (Table S1), expansion events were less frequent, as reported previously (Kantartzis et al. 2012). The median initial tract length was 31 repeats in the CAG tract (range 31–48 repeats) and 33.5 in the CTG tract (range 28–39 repeats), an increase of 6 and 8 repeats, respectively (Figure 2 and Table S2). It is unclear why the size of the CAG tract initial expansion is significantly smaller in the msh3Δ vs. the MSH3 background (Figure 2), while the CTG tract expansions were comparable in both backgrounds.
Expanded TNR tracts continue to expand in vivo
Using the same yeast reporter system, Rolfsmeier et al. (2001) demonstrated increased rates of CTG tract expansions and contractions as tract length increased. Therefore, we predicted that the initial expansion events that we observed, all ≥29 repeats, similarly would be less stable than the 25-repeat tract, increasing the probability of observing additional expansion events over time. However, although we can select for the initial expansion event, it is not possible to select for additional expansion events in this system. Therefore, to test the feasibility of screening expanded CAG and CTG tracts for additional expansions, we performed 14-day time course experiments in parallel liquid cultures (Figure 3). Three independent isolates (single colonies) with expanded CAG or CTG repeat tracts in MSH3 or msh3Δ backgrounds, representing a single initial expansion event (day 0 in Figure 4 and Figure 5), were selected and used to inoculate starting cultures for a total of 12 independent time course experiments. Isolates with different initial tract sizes were selected for these experiments (Table S2). The growth curves of MSH3 and msh3Δ strains carrying either CAG or CTG repeat tracts were indistinguishable (Figure S1 and data not shown); the cells went through approximately six generations every 24 hr. From each starter culture, we established parallel log- and stationary-phase cultures in liquid medium (Figure 3).
gDNA was isolated from each log- and stationary-phase culture every 24 hr and used as a template for PCR to assess TNR tract sizes within the population of the culture. A representative time course is shown in Figure 4A (top). Cells from each culture also were plated at days 7 and 14; individual colonies were used as a template for colony PCR to amplify individual TNR tracts at different time points (Figure 4B, middle). There was no selection for expansion events during the time course, i.e., no 5-FOA. Therefore, the tracts were free to remain stable, to expand, or to contract.
In the MSH3 background, the cells readily accumulated additional expansions in actively dividing cells (Figure 4); tracts in stationary cultures were substantially more stable (see next subsection). In both CAG and CTG tract time courses, analysis of the gDNA revealed that the tract size exhibited a general shift in the population to a larger size (Figure 4A, top). The band representing the major initial expansion event of 31 repeats (asterisk in Figure 4A) became less prominent over the time course. At the same time, a higher-molecular-weight product accumulated and became more prominent as the time course progressed. We note that the larger fragment was present at the beginning of the time course, presumably due to additional expansion events that occurred during the 3 days required to bring the cultures to saturation prior to setting up parallel cultures (see Materials and Methods) (Figure 3). Two lines of evidence suggest that this pattern is not a result of a selective advantage of cells with larger repeat tracts. First, there were only about six generations from one time point to the next (Figure S1). Second, we performed plasmid retention assays in which cells with short tracts were transformed with a plasmid conferring resistance to ClonNAT and cells containing a long tract were transformed with a plasmid conferring resistance to G418. Equal numbers of each cell type were mixed and allowed to grow in the absence of selection for 6 days. Both plasmids were present at a 1:1 ratio throughout the time course (Figure S6), indicating that neither strain has a selective advantage.
To determine whether the higher-molecular-weight tract represented (1) a single-larger expansion event that accumulates within the population over time or (2) an average of multiple different-sized expansion events within the population, we performed colony PCR on individual colonies plated on days 7 and 14. Colony PCR clearly demonstrated that a variety of tract lengths existed within the population (Figure 4A, middle) indicating multiple independent expansion events. CTG tracts from individual colonies revealed that 43 and 47% of the CTG tracts had sustained an additional expansion event at days 7 and 14 of the time course, respectively. Similarly, the CAG tracts exhibited a high frequency of additional expansion: 42 and 78% expansion at days 7 and 14, respectively (Figure 4A, bottom). These frequencies and the growth rate, allowed us to calculate expansion rates of 1.2 × 10−3 per cell generation (95% C.I.: 9.0 × 10−4 to 1.6 × 10−3) for MSH3(CTG) and 3.1 × 10−3 (95% C.I.: 2.4 × 10−3 to 3.4 × 10−3) per cell generation in the MSH3(CAG) background (Table 1). These rates are approximately two orders of magnitude higher than we observed when we performed time courses starting with unexpanded (CTG)25 or (CAG)25 tracts (Table 1 and Figure S7). Notably, the rates of initial expansion (i.e., starting with an unexpanded tract) in the time course assays performed here were consistent with our previous measurements for the initial expansions using the selective assay (Kantartzis et al. 2012). These data indicate that small increases in tract length significantly increase the rate of expansion. Furthermore, these rates may be underestimates; the further expanded tracts that we observed could have arisen through more than one expansion event. It is noteworthy that the CAG tract, which has a lower expansion rate when starting at 25 repeats (Miret et al. 1998; Kantartzis et al. 2012), was more likely to continue expanding than the CTG tract, perhaps a result of the longer starting tract sizes, consistent with the idea that stability of the secondary structure that forms within the tract affects the probability of expansion (Miret et al. 1998; Rolfsmeier et al. 2001). Once the CAG tract has increased sufficiently to generate a more stable secondary structure, it expands at least as rapidly as the CTG repeat tract.
Expanded tracts are more stable in msh3Δ background
In contrast to the MSH3 strains, both CAG and CTG expanded tracts in the msh3Δ strains were much more stable when propagated in liquid culture (Figure 4B). We observed no additional expansions in the msh3Δ background when the gDNA was analyzed (Figure 4B, top, and data not shown). Intriguingly, analysis of the gDNA from one msh3Δ time course revealed contraction events starting on day 12 (Figure 4B, top). Colony PCR of tracts derived from individual cells similarly revealed very few additional expansion events (12% for CTG tracts, 5% for CAG tracts) (Figure 4B, middle and bottom); the CTG tracts were as likely to contract as to expand (12% expansions vs. 8% contractions). The expansion rate for msh3Δ(CTG) was 3.1 × 10−4 per cell generation (95% C.I.: 1.3 × 10−4 to 6.0 × 10−4), fourfold lower that in the MSH3 background. Similarly, the expansion rate in the msh3Δ(CAG) background was ∼20-fold lower than in MSH3 strains at 1.3 × 10−4 (95% C.I.: 2.9 × 10−5 to 3.7 × 10−4) per cell generation (Table 1).
These data demonstrate that CNG repeat tracts become increasingly unstable and much more susceptible to expansion once they have crossed the “stability” threshold, i.e., ≥29 repeats in this system, particularly in the presence of MSH3. These data also indicate that the presence of MSH3 promotes not only the initial expansion event (or events) that allow for selection in this system but also subsequent expansion events, at least within this threshold range. Therefore, we can use expanded tracts as a starting point to examine tract dynamics; there are likely to be changes within a time frame of several days. Furthermore, the initial expansions are relatively small, increasing to a total of ≤50 repeats and representing early events in the progression of an expanded tract.
Expansions were enhanced in dividing cells
In contrast to CNG repeat tracts in replicating cells, the tracts from the stationary-phase cultures in both the MSH3 and msh3Δ backgrounds were more stable (Figure 5), indicating that a significant proportion of the expansion events depend on DNA replication. Based on analysis of the gDNA from MSH3 cultures, the major initial expansion event (asterisk) remains the major PCR product throughout the time course; there is little or no accumulation of additional expansion events (Figure 5A, top). Colony PCR from individual colonies similarly revealed that the tracts were very stable in stationary phase (Figure 5A, middle and bottom). We observed that 7 and 8% of the CTG tracts were further expanded on days 7 and 14, respectively, while 12 and 15% of the CAG tracts had additional expansions on days 7 and 14, respectively.
The frequency of expansions in the msh3Δ background also was lower in the stationary-phase cultures (Figure 5B). There were neither expansions nor contractions when the gDNA was analyzed. And there were very few additional expansions observed from the colony PCR analysis, again confirming that DNA replication is important in promoting these additional expansion events. We do note, however, that even in the stationary phase, there were more additional expansion events in the MSH3 background than in the msh3Δ background. With the CTG tract, we observed 8 and 1% expansions on days 7 and 14, respectively; we observed no expansions in the CAG stationary phase cultures (Figure 5B, bottom). This is consistent with the observation that Msh2-Msh3 promotes CNG expansions in postmitotic (nondividing) tissues in mammalian systems (McMurray 2010; Kovalenko et al. 2012).
Expanded TNR tracts increase in discrete steps
To follow tract dynamics as a function of time, we selected independent expanded tracts on 5-FOA medium and confirmed their sizes by PCR, as described earlier. Colonies with a range of initial tract sizes (i.e., initial expansion events) were selected for further analysis . We chose to focus on CTG tracts because the range of initial expansion events was more similar in the MSH3 and msh3Δ backgrounds than with CAG tracts (Figure 2 and Table S2). Each colony was then transferred to nonselective medium (no 5-FOA) so that the tract was free to expand or contract (Figure 6). Colony PCR was performed on the same colony every 24 hr for 10–14 days to assess tract length. Additional expansions were observed in most of the time courses, and the increase in the size of the tract was progressive (Figures 7A and Figure 8A). While multiple tract sizes often were observed at the same time point, the overall size of the predominant tract increased over time, with a concomitant loss of the smaller-sized tracts. We refer to this as a progressive tract length change. Typically, two–three discrete progressive changes, or increments, were observed over 14 days, indicating that the expansions occur in steps. We confirmed that these PCR products contained the TNR tract by Southern blot using a tract-specific (CTG) probe (Figure 7A, right). We also performed PCR controls to rule out the possibility that larger tracts were preferentially amplified (see Materials and Methods) (Figure S5).
In Figure 8A, we plotted the progressive changes in tract length as a function of time for all the MSH3(CTG) time courses that we evaluated. There is a clear upward trend in tract length in most of the time courses, although contractions and stable tracts are also evident. Importantly, these changes were discrete increases in size, which we refer to as steps. Overall, the tracts appeared to expand in relatively small increments, or steps, over time (Figure 7A and Figure 8A). Even cells with very large initial expansions exhibited incremental increases in this range. For example, two colonies (Figure 8A, B4 and C4) with initial expansions of 38 repeats (for total lengths of 63 repeats) had subsequent increases of only 8 and 9 repeat lengths, respectively. Conversely, one 70-repeat tract (Figure 8A, D4) sustained a 24-repeat increase. Therefore, in our data set, there does not seem to be a strict correlation between tract length and the size of additional expansion events in the tract length range that we investigated. All the increases were smaller than the initial tract length. These data therefore suggest that the longer tract expansions are the result of multiple expansion events.
To assess the size of each step and determine whether there is a correlation in step size with respect to starting tract length, we plotted the number of additional repeat units we observed at each tract length change for all the time courses plotted in Figure 8A (Figure 9A). Thus, the pink circles demonstrate the number of repeat units we observed in each initial expansion event, i.e., after selection on 5-FOA. The median size of the initial expansion was 24 base pairs (8 repeats). An additional increase in tract length, i.e., the first step beyond the initial expansion, was plotted using the orange squares, and the subsequent increases, or second, third, and fourth steps, were similarly plotted. Thus, only the time courses that exhibited progressive tract length changes are represented in this figure. We note that there was not a strict correlation in the timing of the tract length changes (Figure 8A), and therefore, the size of the tract changes was plotted independent of time point. When all expansions were taken into account, the median size of the tract length increase, or step, was 24 base pairs (8 repeats); the average increase was 11 repeats (±18 repeats, 2 SD), but the range was large: 2–47 repeats. The median size of each step was 8 repeats and did not increase with increasing tract size within the range of tract lengths that we observed (Figure 9A). In fact, the increase in tract size decreased after the initial expansion, as did the range in expansion events.
When individual msh3Δ colonies with an expanded TNR tract were followed over time, the TNR tracts appeared more stable (Figures 7B, Figure 8B, and Figure 9B), consistent with our observations in the population-based time course (Figure 4). In 64 time courses, we observed no evidence of progressive expansion (Figures 7B and Figure 8B), as defined earlier with the concomitant loss of the smaller tract length. We did observe additional PCR products in some cases, but this resulted in a mixed population, with ≥75% of the tract remaining at the original expansion size (Figure 7B). The results of the mixing experiments (Figure S5B) suggest that this ratio indicates an approximately fivefold excess of unexpanded tract template in the reaction. In these events, we observed a total of one–two tract species in addition to the original tract size, which continued to dominate the population. We also observed contractions of the predominant PCR product in seven independent time courses (Figure 8B). In contrast, no progressive contractions were observed in the MSH3 background, although some individual contractions were observed (Figure 8A). Southern blotting confirmed the presence of the CTG tract in each of these DNA fragments (Figure 7B and data not shown). Overall, taking only progressive tract length changes into account, the rate of change in tract length in msh3Δ cells was 3.6 × 10−3 per generation (95% C.I.: 1.5 × 10−3 to 7.2 × 10−3); the changes observed were all contractions. In contrast, the rate of tract length change in MSH3 cells, again including only progressive tract length changes, was 1.5 × 10−2 per generation (95% C.I.: 1.2 × 10−2 to 1.9 × 10−2), fourfold higher than in the msh3Δ cells, and the observed changes were almost exclusively expansions (Table 1). We note that these are likely underestimates because minor expansion and contraction products were observed in both MSH3 and msh3Δ time courses. Furthermore, the extent of expanded products observed may depend on the section of the colony sampled. Nonetheless, the presence of Msh2-Msh3 appears to shift the dynamic equilibrium toward expansion over time, ultimately resulting in longer and longer TNR tracts. The absence of the complex stabilizes the tract and permits an increase in contraction events, possibly as a result of the MMR deficiency (see Discussion).
TNR tracts are highly dynamic as they expand
The incremental expansions of the TNR tract observed within individual colonies were smaller than the tract length but were typically larger than the increments we observed in vitro (Kantartzis et al. 2012), although they were on the same order of expansions previously observed by single-sperm typing in humans (Leeflang et al. 1995, 1999). To determine whether each expansion event represented a single larger event or the sum of multiple small events, indicating a highly dynamic TNR tract, we initiated time courses with a single cell rather than a potentially mixed population within a colony. First, we selected a colony that contained an expanded TNR tract. From this colony we isolated individual cells using a micromanipulator (Figure 10). These single cells were allowed to go through approximately 8–10 cell divisions (and therefore 8–10 rounds of replication), resulting in a microcolony of ∼250–1000 cells. Individual cells from each microcolony were selected to propagate a new microcolony to continue the time course. The remainder of the microcolony was used as a template for PCR.
In the MSH3 background, starting with tracts between 30 and 33 repeats, we observed very dynamic behavior of the tract, with several small changes in tract size occurring over the course of 2 weeks (Figure 11A and Table 2). Multiple products were frequently observed at each time point, indicating a mixed tract population within the microcolony. We quantified the predominant tract at each time point and observed both expansions and contractions in similar quantities (slight bias toward expansions) and of similar sizes, many of which were one, two, or three TNRs in length (i.e., three, six, or nine nucleotides) (Table 4). Although the total number of events is low, we calculated an approximate rate of tract size change per generation (expansions and contractions) of 3.5 × 10−2 per generation (95% C.I.: 2.8 × 10−2 to 4.2 × 10−2), assuming 10 generations per time point (Table 1). The changes in tract size observed on this time scale were smaller than those observed in the colony time course and more closely resembled the size of expansions observed in vitro in the presence of Msh2-Msh3, Rad27 (Fen1), and Cdc9 (Lig1) (Kantartzis et al. 2012). Therefore, in a wild-type (MSH3) background, threshold-length TNR tracts demonstrated a tendency to gradually increase in size. Notably, even in tracts where there was no net change in tract length at the end of the time course, the tract length did change during the time course (i.e., C, D, E, and G in Table 2).
Table 2. MSH3 microcolony tract dynamics.
| TNR tract dynamics of individual MSH3 microcolonies | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Day | A | B | C | D | E | Fa | G | H | I | J | M | R | P | S | 9 |
| Initial tractb | 32 | 30 | 32 | 33 | 31 | 30 | 31 | 32 | 34 | 30 | 30 | 31 | 33 | 32 | 31 |
| 1 | 0c | 0 | 0 | 5 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 3 | 0 | 0 | −5 | 0 | −2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | −3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | −7 |
| 4 | 2 | −2 | 0 | −2 | −3 | −2 | −2 | 0 | −3 | 5 | 0 | −4 | −3 | 0 | 7 |
| 5 | 1 | 0 | 1 | 3 | 3 | 0 | 2 | −3 | 3 | 0 | 0 | 4 | 3 | 0 | 0 |
| 6 | −1 | 0 | 0 | 0 | 2 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 7 | 1 | 0 | −3 | 0 | 0 | 2 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | −4 |
| 8 | 0 | 0 | −2 | 0 | −7 | 0 | 8 | −5 | 0 | 2 | 0 | 0 | 0 | 0 | 3 |
| 9 | −1 | 0 | 4 | 0 | 0 | 0 | 0 | −3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 10 | 1 | 0 | 0 | 7 | 7 | 4 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 11 | 0 | 0 | 0 | −7 | −2 | 6 | −8 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| 12 | 11 | 0 | 0 | −4 | −2 | 0 | −2 | 0 | 0 | −2 | 0 | 0 | 0 | 0 | 0 |
| 13 | —d | −1 | 0 | — | 4 | 0 | 9 | 14 | 3 | 5 | 0 | 4 | −3 | 0 | 0 |
| Final tracte | 46 | 27 | 32 | 30 | 33 | 40 | 38 | 46 | 37 | 42 | 30 | 35 | 30 | 32 | 31 |
| Net changef | 14 | −3 | 0 | −3 | 2 | 10 | 7 | 14 | 3 | 12 | 0 | 4 | −3 | 0 | 0 |
Corresponds to gel in Figure 11A.
Initial expansion size following selection in the presence of 5-FOA in number of repeats.
Change in number of repeats in tract.
No amplification of tract.
Number of repeats.
Net change in number of repeats within tract.
Table 4. Trinucleotide repeat tract dynamics in microcolony time courses.
| Genotype | Type of event | Number of events | Median size of event (range in number of repeats) |
|---|---|---|---|
| MSH3(CTG)a | Expansion | 38 | 3 (1–14) |
| Contraction | 30 | 3 (1–8) | |
| Final TNR tract length | 33 (27–46) | ||
| msh3Δ(CTG)b | Expansion | 10 | 3 (1–6) |
| Contraction | 18 | 2 (1–7) | |
| Final TNR tract length | 30 (29–32) |
Ninety-six total time courses were initiated: 32 time courses were completed to at least day 12; of these,13 exhibited TNR tract dynamics, 2 exhibited a stable tract length, 15 exhibited no tract amplification, and 2 were uninterpretable due to high background.
One-hundred and five total time courses were initiated: 37 time course were completed to at least day 10; of these, 14 exhibited TNR tract dynamics, 9 exhibited a stable tract length, and 14 exhibited no tract amplification.
In the absence of MSH3, the tracts appeared more stable overall (Figure 11B and Table 3), with an estimated mutation rate per generation of 1.6 × 10−2 per generation (95% C.I.: 1.1 × 10−2 to 2.2 × 10−2) (Table 1), half the rate observed in the presence of MSH3. We do note that additional PCR products were observed in some of these time courses. However these additional bands did not change in size over the course of the experiment, indicating stability. Furthermore, in several cases, including the lower panel of Figure 11B, Southern blots indicated that the upper bands did not contain TNR sequences (data not shown). When changes did occur, the median size of the increments was similar to that observed in the MSH3 background. However, the tracts in the msh3Δ strains were more likely to contract than to expand (10 expansions, 18 contractions) (Table 4), in contrast to roughly equal proportions of expansions and contractions in the MSH3 background, with a bias toward expansions (38 expansions, 30 contractions) (Table 4).
Table 3. msh3Δ microcolony tract dynamics.
| TNR tract dynamics of individual msh3Δ microcolonies | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Day | A | B | C | D | E | F | G | H | I | J | K | L | M | Na | O | P | Qb | R | S | T | U | V | W | |
| Initial tractc | 30 | 31 | 30 | 30 | 31 | 32 | 32 | 32 | 32 | 29 | 30 | 30 | 32 | 30 | 30 | 36 | 29 | 29 | 30 | 30 | 29 | 29 | 30 | |
| 1 | 0d | 0 | 0 | 0 | 0 | −3 | −2 | −2 | 0 | 0 | 0 | 0 | −2 | −1 | 0 | — | 0 | 0 | 0 | 0 | 0 | −2 | 0 | |
| 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | −7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 3 | —e | — | — | — | — | — | — | — | — | — | −5 | 0 | −5 | −5 | — | 0 | 0 | 0 | — | — | 0 | — | — | |
| 4 | — | — | — | — | 0 | 0 | 0 | −1 | — | — | 0 | 0 | 0 | 5 | — | — | 0 | — | — | — | 0 | — | — | |
| 5 | — | — | — | — | — | — | — | — | — | — | — | — | — | 0 | — | — | 0 | — | — | 0 | 0 | — | — | |
| 6 | 0 | — | 0 | 0 | 0 | 0 | — | 0 | 0 | — | 5 | 0 | 5 | 0 | 0 | — | 0 | — | — | 0 | — | — | — | |
| 7 | 0 | 0 | 0 | 0 | −1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | — | 0 | 0 | 0 | 0 | 0 | 0 | — | 0 | 0 | |
| 8 | −1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | — | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 6 | |
| 9 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | −3 | 0 | 0 | 0 | 0 | 1 | 0 | — | 0 | 0 | — | 0 | 0 | 0 | 0 | |
| 10 | 0 | 0 | 0 | −5 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | −1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | — | −1 | |
| 11 | 0 | 0 | 0 | 1 | — | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | — | — | 0 | — | 0 | 0 | — | — | — | −4 | |
| Final tractf | 29 | 31 | 30 | 30 | 31 | 29 | 30 | 31 | 32 | 29 | 30 | 30 | 30 | 29 | 30 | 29 | 29 | 29 | 30 | 30 | 29 | 31 | 28 | |
| Net changeg | −1 | 0 | 0 | 0 | 0 | −3 | −2 | −1 | 0 | 0 | 0 | 0 | −2 | −1 | 0 | −7 | 0 | 0 | 0 | 0 | 0 | 0 | −2 | |
Corresponds to gel in upper panel of Figure 11B.
Corresponds to gel in lower panel of Figure 11B.
Initial expansion size following selection in the presence of 5-FOA in number of repeats.
Change in number of repeats in tract.
No amplification of tract.
Number of repeats.
Net change in number of repeats within tract.
Discussion
We have, for the first time in any system, documented the dynamic behavior of threshold-length TNR tracts and tracked them over time in dividing cells. The rate of change in the size of threshold-length TNR tracts is remarkable, at two–three orders of magnitude higher than the stable (CNG)25 repeats in yeast (Miret et al. 1998; Rolfsmeier et al. 2001; Kantartzis et al. 2012). Thus, small increases in tract length led to substantially higher expansion rates. The rapid rate of change in tract length, both expansions and contractions, was particularly striking when threshold-length tracts were examined in microcolonies derived from a single cell. In contrast, we did not observe similar changes in an unexpanded (CTG)25 tract (Figure S8).
Previous studies have proposed a model for TNR expansions in dividing cells that involved incremental expansions (Leeflang et al. 1995, 1999; Martorell et al. 2004; Kantartzis et al. 2012). Here we have shown directly that incremental expansions do in fact occur in vivo within CTG repeat tracts. The size of the increments varies but can be as small as a single repeat unit. This is consistent with the behavior of CAG repeats at both the HD and androgen receptor loci in humans (Zhang et al. 1994; Leeflang et al. 1995). Recent work modeling human DM1 expansion data also has demonstrated that these expansions are likely to occur through small incremental steps (Higham et al. 2012; Morales et al. 2012; Higham and Monckton 2013). In induced pluripotent stem cells (iPSCs) from HD and DM1 patients, Du et al. (2013) observed that (CAG)∼46 and (CTG)∼57 tracts were stable after 12 and 16 passages, respectively. For comparison, yeast cells went through approximately 120 cell divisions during the 2-week time course experiments in this study. However, a larger CTG tract (∼126 repeats) expanded by approximately one repeat increment in each passage; a (CTG)∼773 tract expanded in larger increments (Du et al. 2013). These data are consistent with a stepwise increase in the early stages of TNR tract expansion.
Importantly, the absence of MSH3 alters the dynamics of the tract in this model system. Overall, in actively replicating cells, both the CAG and CTG tracts are more stable in the msh3Δ background, consistent with previous observations (Owen et al. 2005; Kantartzis et al. 2012). Expansions did occur, but at a significantly lower rate than in the MSH3 background, as described previously (Kantartzis et al. 2012). Furthermore, msh3Δ cells were much less likely to incur an additional expansion, and when additional expansions did occur, they were less likely to become the predominant population (Figures 3–11), despite the fact that the cell division time for the two strains was indistinguishable. Therefore, loss of MSH3 reduces the probability of incurring both an initiating and a secondary expansion event. However, contraction events were observed more frequently in msh3Δ strains (e.g., Figure 8 and Table 4). This is consistent with the results of Schweitzer and Livingston (1997), who demonstrated an increase in TNR contractions in msh2Δ cells using a different yeast reporter system. The increased rate of contraction may be related to the loss of IDL MMR in the absence of MSH3; Msh2-Msh3 preferentially corrects deletion events, i.e., loops on the template strand, during DNA replication (Schweitzer and Livingston 1997; Sia et al. 1997; Romanova and Crouse 2013). These observations support the suggestion that Msh3 is a potential therapeutic target (Castel et al. 2010) and underscore the need to develop a better understanding of tract dynamics in different genetic backgrounds. It will be of interest to determine whether the size of the contractions varies with MSH3 status.
It is striking that in the absence of selection for tracts larger than 30 repeats, we nonetheless observed a strong bias toward expansions in the MSH3 background with very few contractions. This is distinct from previous observations in yeast, where there was a strong bias toward contractions (Maurer et al. 1996; Miret et al. 1997; Rolfsmeier et al. 2001), although when expansions were observed, they were relatively small, ranging from 2 to ∼20 repeat lengths (Maurer et al. 1996; Miret et al. 1998), consistent with our observations. We performed a preliminary contraction assay and observed rates of contraction similar to those observed previously (Miret et al. 1997), indicating that there is nothing inherent in our strain background that predisposes toward expansions (data not shown) but rather that the sizes of the tracts themselves led to the shift toward expansions. As such, the threshold-length tract behavior that we observed in yeast mimics that which has been observed in human sperm cells and iPSCs and therefore is a good model for events in dividing cells. In this model we can look at large numbers of cells over many generations, making this a very flexible and informative approach to examining TNR tract dynamics. It is important to note that Msh2-Msh3-mediated expansions are just one mechanism by which TNR tracts increase in length. This yeast system is amenable to examining the contribution of additional genetic factors on tract dynamics in both deletion and mutation backgrounds (e.g., Debacker et al. 2012; Concannon and Lahue 2014)).
Although this study focused on actively replicating cells, we note that the frequency of additional expansions also was elevated in stationary-phase cultures relative to the unexpanded tract (Figure 5A); ∼10–15% of the tracts had an additional expansion event in the MSH3 background. Notably, this decreased in the absence of Msh3 (Figure 5B). Msh2-Msh3 has been shown to be important in promoting somatic tract expansions in postmitotic (nonreplicating) neurons, specifically in medium-spiny striatal neurons (MSNs) in a mouse model for HD (Kovalenko et al. 2012). Deletion of Msh2 eliminated most of the CAG expansions in the HD gene (HTT) in the MSNs. The decrease in expansions correlated with a reduction in HTT CAG-dependent phenotypes in the mice. In another study, Msh3 polymorphisms that altered Msh3 protein levels in mouse striatal tissues also altered the stability of the HD transgene CAG repeat tract: elevated Msh3 levels correlated with increased tract size (Tomé et al. 2013). Recent work has suggested that CAG expansions in somatic tissue also may occur in an incremental manner (Lee et al. 2011). A cohort of HD CAG knock-in mice was sampled at 2, 5, 9, 12, and 16 months of age, and CAG tract length was examined in the liver and striatum. In these tissues, there was an average increase in tract size of one repeat per month, although there was a much broader distribution in the striatum than in the liver. In vitro, human Msh2-Msh3 (MutSβ) stimulated expansions in a replication-independent system (Stevens et al. 2013). Therefore, monitoring TNR tracts in stationary-phase yeast cultures may be a powerful way to model tract dynamics in nonproliferating cells and elucidate the mechanism(s) of expansions in somatic cells.
Although we have not tested the effect of msh2Δ on dynamic TNR tract behavior, our in vivo and in vitro data (Kantartzis et al. 2012) examining TNR tracts are consistent with a model in which TNR secondary structures are bound and stabilized by Msh2-Msh3. Increased tract length increases the probability that alternative DNA structures will form, perhaps during Okazaki fragment processing or base excision repair, leading to the recruitment of Msh2-Msh3. So why are the structures not repaired? In vitro work with human MutSβ (Msh2-Msh3) has indicated that the Msh2-Msh3/TNR complex is distinct from the Msh2-Msh3/loop complex (Owen et al. 2009; Lang et al. 2011). Similarly, Msh2-Msh3 exhibits distinct nucleotide turnover behavior when bound to MMR vs. double-strand DNA break repair (DSBR) substrates (Kumar et al. 2013, 2014). It is not yet clear how these proposed kinetic differences could translate into increased expansions, but we favor the possibility that Msh2-Msh3-mediated ATP-binding and hydrolysis activities are altered in the presence of the TNR substrate, disrupting error-free MMR. Altered Msh2-Msh3 activity could block repair altogether or recruit an error-prone repair process that ultimately retains the insertion. Recent work has demonstrated that mammalian MutLγ (Mlh1-Mlh3) is required for somatic CAG expansions in a HD mouse; MutLα (Mlh1-Pms2) was dispensable (Pinto et al. 2013). Intriguingly, Rogacheva et al. (2014) demonstrated that yeast Mlh1-Mlh3 enhances Msh2-Msh3 DNA-binding activity, while Msh2-Msh3 stimulates Mlh1-Mlh3 endonuclease activity. Therefore, enhanced Msh2-Msh3 binding to TNR structures could inappropriately recruit Mlh1-Mlh3 during replication or outside of S phase and stimulate its endonuclease activity, leading to gap formation and DNA repair synthesis that incorporates incremental expansions (Gomes-Pereira et al. 2004; Pluciennik et al. 2013).
Disruption of Msh2-Msh3 has been proposed as an attractive therapeutic target for TNR expansion disease such as HD or DM1 (Panigrahi et al. 2005, 2006; Castel et al. 2010). Loss of MSH3 leads to a decrease in expansion events and promotes contractions. However, disruption of Msh2-Msh3 will have a negative impact on overall genome stability; in addition to MMR, Msh2-Msh3 has been implicated in DSBR (Surtees et al. 2004; Park et al. 2013; van Oers et al. 2013) and interstrand DNA cross-link repair (Takahashi et al. 2011; Barber et al. 2005; Zhao et al. 2009). Notably, many neuronally expressed genes contain repetitive sequences in their regulatory regions (Bacolla et al. 2008), the stability of which is likely at least partially dependent on Msh2-Msh3. Therefore, in considering Msh2-Msh3 as a molecular target, a clear mechanistic understanding of Msh2-Msh3 function in promoting repeats is vital to ensure that a balance is struck between promoting genome stability and genome instability in developing rational therapeutic strategies.
Supplementary Material
Acknowledgments
We are grateful to Mark Sutton, Robert Lahue, Catherine Freudenreich, and Eric Alani for comments on this manuscript and to members of the Surtees Laboratory for discussions of this work. Work in the Surtees Laboratory is supported by National Institutes of Health grant GM-087459. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Communicating editor: J. A. Nickoloff
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.177303/-/DC1
Literature Cited
- Alani E., Cao L., Kleckner N., 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116: 541–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacolla A., Larson J. E., Collins J. R., Li J., Milosavljevic A., et al. , 2008. Abundance and length of simple repeats in vertebrate genomes are determined by their structural properties. Genome Res. 18: 1545–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber L. J., Ward T. A., Hartley J. A., McHugh P. J., 2005. DNA interstrand cross-link repair in the Saccharomyces cerevisiae cell cycle: overlapping roles for PSO2 (SNM1) with MutS factors and EXO1 during S phase. Mol. Cell. Biol. 25: 2297–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel A. L., Cleary J. D., Pearson C. E., 2010. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat. Rev. Mol. Cell Biol. 11: 165–170. [DOI] [PubMed] [Google Scholar]
- Concannon C., Lahue R. S., 2014. Nucleotide excision repair and the 26S proteasome function together to promote trinucleotide repeat expansions. DNA Repair 13: 42–49. [DOI] [PubMed] [Google Scholar]
- Debacker K., Frizzell A., Gleeson O., Kirkham-McCarthy L., Mertz T., et al. , 2012. Histone deacetylase complexes promote trinucleotide repeat expansions. PLoS Biol. 10: e1001257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon M., Bhattacharyya S., Lahue R., 2004. Genetic assays for triplet repeat instability in yeast, pp. 29–45 in Trinucleotide Repeat Protocols, edited by Kohwi. Y. Humana Press, New York. [DOI] [PubMed] [Google Scholar]
- Du J., Campau E., Soragni E., Jespersen C., Gottesfeld J. M., 2013. Length-dependent CTG·CAG triplet-repeat expansion in myotonic dystrophy patient-derived induced pluripotent stem cells. Hum. Mol. Genet. 22: 5276–5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiry L., Dong L., Savouet C., Hubert L., Te Riele H., et al. , 2006. Msh3 is a limiting factor in the formation of intergenerational CTG expansions in DM1 transgenic mice. Hum. Genet. 119: 520–526. [DOI] [PubMed] [Google Scholar]
- Foster, P. L., L. C. Judith, and M. Paul, 2006 Methods for determining spontaneous mutation rates, pp. 195–213 in Measuring Biological Responses with Automated Microscopy (Methods in Enzymology Series, Vol. 414). Academic Press, New York. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, P. L., 2006 Methods for determining spontaneous mutation rates, pp.195–213 in DNA Repair, Part B (Methods in Enzymology Series, Vol. 409). Editors, J.L. Campbell and P. Modrich. Academic Press, New York. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gacy A. M., McMurray C. T., 1998. Influence of hairpins on template reannealing at trinucleotide repeat duplexes: a model for slipped DNA. Biochemistry 37: 9426–9434. [DOI] [PubMed] [Google Scholar]
- Gannon A.-M. M., Frizzell A., Healy E., Lahue R. S., 2012. MutSβ and histone deacetylase complexes promote expansions of trinucleotide repeats in human cells. Nucleic Acids Res. 40: 10324–10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D., Jean A. S., Woods R. A., Schiestl R. H., 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A. L., McCusker J. H., 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553. [DOI] [PubMed] [Google Scholar]
- Gomes-Pereira M., Fortune M. T., Ingram L., McAbney J. P., Monckton D. G., 2004. Pms2 is a genetic enhancer of trinucleotide CAG·CTG repeat somatic mosaicism: implications for the mechanism of triplet repeat expansion. Hum. Mol. Genet. 13: 1815–1825. [DOI] [PubMed] [Google Scholar]
- Halabi A., Ditch S., Wang J., Grabczyk E., 2012. DNA mismatch repair complex MutSβ promotes GAA·TTC repeat expansion in human cells. J. Biol. Chem. 287: 29958–29967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higham C. F., Monckton D. G., 2013. Modelling and inference reveal nonlinear length-dependent suppression of somatic instability for small disease associated alleles in myotonic dystrophy type 1 and Huntington disease. J. R. Soc. Interface 10: 20130605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higham C. F., Morales F., Cobbold C. A., Haydon D. T., Monckton D. G., 2012. High levels of somatic DNA diversity at the myotonic dystrophy type 1 locus are driven by ultra-frequent expansion and contraction mutations. Hum. Mol. Genet. 21: 2450–2463. [DOI] [PubMed] [Google Scholar]
- Kantartzis A., Williams G. M., Balakrishnan L., Roberts R. L., Surtees J. A., et al. , 2012. Msh2-Msh3 interferes with Okazaki fragment processing to promote trinucleotide repeat expansions. Cell Rep. 2: 216–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalenko M., Dragileva E., St. Claire J., Gillis T., Guide J. R., et al. , 2012. Msh2 acts in medium-spiny striatal neurons as an enhancer of CAG instability and mutant huntingtin phenotypes in Huntington’s disease knock-in mice. PLoS ONE 7: e44273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar C., Eichmiller R., Wang B., Williams G. M., Bianco P. R., et al. , 2014. ATP binding and hydrolysis by Saccharomyces cerevisiae Msh2-Msh3 are differentially modulated by mismatch and double-strand break repair DNA substrates. DNA Repair 18: 18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar C., Williams G. M., Havens B., Dinicola M. K., Surtees J. A., 2013. Distinct requirements within the Msh3 nucleotide binding pocket for mismatch and double-strand break repair. J. Mol. Biol. 425: 1881–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang W. H., Coats J. E., Majka J., Hura G. L., Lin Y., et al. , 2011. Conformational trapping of mismatch recognition complex MSH2/MSH3 on repair-resistant DNA loops. Proc. Natl. Acad. Sci. USA 108: E837–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-M., Pinto R. M., Gillis T., St. Claire J. C., Wheeler V. C., 2011. Quantification of age-dependent somatic CAG repeat instability in Hdh CAG knock-in mice reveals different expansion dynamics in striatum and liver. PLoS ONE 6: e23647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeflang E. P., Tavaré S., Marjoram P., Neal C. O. S., Srinidhi J., et al. , 1999. Analysis of germline mutation spectra at the Huntington’s disease locus supports a mitotic mutation mechanism. Hum. Mol. Genet. 8: 173–183. [DOI] [PubMed] [Google Scholar]
- Leeflang E. P., Zhang L., Tavare S., Hubert R., Srinidhi J., et al. , 1995. Single sperm analysis of the trinucleotide repeats in the Huntington’s disease gene: quantification of the mutation frequency spectrum. Hum. Mol. Genet. 4: 1519–1526. [DOI] [PubMed] [Google Scholar]
- Li G.-M., 2008. Mechanisms and functions of DNA mismatch repair. Cell Res. 18: 85–98. [DOI] [PubMed] [Google Scholar]
- Lovett S. T., 2004. Encoded errors: mutations and rearrangements mediated by misalignment at repetitive DNA sequences. Mol. Microbiol. 52: 1243–1253. [DOI] [PubMed] [Google Scholar]
- Lyndaker A. M., Goldfarb T., Alani E., 2008. Mutants defective in Rad1-Rad10-Slx4 exhibit a unique pattern of viability during mating-type switching in Saccharomyces cerevisiae. Genetics 179: 1807–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley K., Pugh J., Messer A., 1999. Instability of the CAG repeat in immortalized fibroblast cell cultures from Huntington’s disease transgenic mice. Brain Res. 835: 74–79. [DOI] [PubMed] [Google Scholar]
- Marquis Gacy A., Goellner G., Juranic N., Macura S., McMurray C. T., 1995. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell 81: 533–540. [DOI] [PubMed] [Google Scholar]
- Martorell L., Gámez J., Cayuela M. L., Gould F. K., McAbney J. P., et al. , 2004. Germline mutational dynamics in myotonic dystrophy type 1 males: allele length and age effects. Neurology 62: 269–274. [DOI] [PubMed] [Google Scholar]
- Maurer D. J., O’Callaghan B. L., Livingston D. M., 1996. Orientation dependence of trinucleotide CAG repeat instability in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 6617–6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray C. T., 2010. Mechanisms of trinucleotide repeat instability during human development. Nat. Rev. Genet. 11: 786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miret J. J., Pessoa-Brandão L., Lahue R. S., 1997. Instability of CAG and CTG trinucleotide repeats in Saccharomyces cerevisiae. Mol. Cell. Biol. 17: 3382–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miret J. J., Pessoa-Brandão L., Lahue R. S., 1998. Orientation-dependent and sequence-specific expansions of CTG/CAG trinucleotide repeats in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 95: 12438–12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales F., Couto J. M., Higham C. F., Hogg G., Cuenca P., et al. , 2012. Somatic instability of the expanded CTG triplet repeat in myotonic dystrophy type 1 is a heritable quantitative trait and modifier of disease severity. Hum. Mol. Genet. 21: 3558–3567. [DOI] [PubMed] [Google Scholar]
- Owen B. A., Lang W. H., McMurray C. T., 2009. The nucleotide binding dynamics of human MSH2-MSH3 are lesion dependent. Nat. Struct. Mol. Biol. 16: 550–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen B. A., Yang Z., Lai M., Gajec M., Badger J. d., et al. , 2005. (CAG)n-hairpin DNA binds to Msh2-Msh3 and changes properties of mismatch recognition. Nat. Struct. Mol. Biol. 12: 663–670. [DOI] [PubMed] [Google Scholar]
- Panigrahi G. B., Lau R., Montgomery S. E., Leonard M. R., Pearson C. E., 2005. Slipped (CTG)·(CAG) repeats can be correctly repaired, escape repair or undergo error-prone repair. Nat. Struct. Mol. Biol. 12: 654–662. [DOI] [PubMed] [Google Scholar]
- Panigrahi G. B., Slean M. M., Simard J. P., Gileadi O., Pearson C. E., 2006. Isolated short CTG/CAG DNA slip-outs are repaired efficiently by hMutSβ, but clustered slip-outs are poorly repaired. Proc. Natl. Acad. Sci. USA 107: 12593–12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. M., Huang S., Tougeron D., Sinicrope F. A., 2013. MSH3 mismatch repair protein regulates sensitivity to cytotoxic drugs and a histone deacetylase inhibitor in human colon carcinoma cells. PLoS ONE 8: e65369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C. E., Sinden R. R., 1996. Alternative structures in duplex DNA formed within the trinucleotide repeats of the myotonic dystrophy and fragile X loci. Biochemistry 35: 5041–5053. [DOI] [PubMed] [Google Scholar]
- Pearson C. E., Sinden R. R., 1998. Trinucleotide repeat DNA structures: dynamic mutations from dynamic DNA. Curr. Opin. Struct. Biol. 8: 321–330. [DOI] [PubMed] [Google Scholar]
- Pinto R. M., Dragileva E., Kirby A., Lloret A., Lopez E., et al. , 2013. Mismatch repair genes Mlh1 and Mlh3 modify CAG instability in Huntington’s disease mice: genome-wide and candidate approaches. PLoS Genet. 9: e1003930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluciennik A., Burdett V., Baitinger C., Iyer R. R., Shi K., et al. , 2013. Extrahelical (CAG)/(CTG) triplet repeat elements support proliferating cell nuclear antigen loading and MutLα endonuclease activation. Proc. Natl. Acad. Sci. USA 110: 12277–12282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogacheva M. V., Manhart C. M., Chen C., Guarne A., Surtees J., et al. , 2014. Mlh1-Mlh3, a meiotic crossover and DNA mismatch repair factor, is a Msh2-Msh3-stimulated endonuclease. J. Biol. Chem. 289: 5664–5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfsmeier M. L., Dixon M. J., Pessoa-Brandão L., Pelletier R., Miret J. J., et al. , 2001. Cis-elements governing trinucleotide repeat instability in Saccharomyces cerevisiae. Genetics 157: 1569–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanova N. V., Crouse G. F., 2013. Different roles of eukaryotic MutS and MutL complexes in repair of small insertion and deletion loops in yeast. PLoS Genet. 9: e1003920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W., 2001. Molecular Cloning: A Laboratory Manual, Ed. 3rd Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Schweitzer J. K., Livingston D. M., 1997. Destabilization of CAG trinucleotide repeat tracts by mismatch repair mutations in yeast. Hum. Mol. Genet. 6: 349–355. [DOI] [PubMed] [Google Scholar]
- Sia E., Kokoska R., Dominska M., Greenwell P., Petes T., 1997. Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol. Cell. Biol. 17: 2851–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slean M. M., Reddy K., Wu B., Nichol Edamura K., Kekis M., et al. , 2013. Interconverting conformations of slipped-DNA junctions formed by trinucleotide repeats affect repair outcome. Biochemistry 52: 773–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J. R., Lahue E. E., Li G.-M., Lahue R. S., 2013. Trinucleotide repeat expansions catalyzed by human cell-free extracts. Cell Res. 23: 565–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surtees J. A., Argueso J. L., Alani E., 2004. Mismatch repair proteins: key regulators of genetic recombination. Cytogenet. Genome Res. 107: 146–159. [DOI] [PubMed] [Google Scholar]
- Takahashi M., Koi M., Balaguer F., Boland C. R., Goel A., 2011 MSH3 mediates sensitization of colorectal cancer cells to cisplatin, oxaliplatin, and a poly(ADP-ribose) polymerase inhibitor. J. Biol. Chem. 286: 12157–12165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomé S., Manley K., Simard J. P., Clark G. W., Slean M. M., et al. , 2013. MSH3 polymorphisms and protein levels affect CAG repeat instability in Huntington’s disease mice. PLoS Genet. 9: e1003280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Broek W. J. A. A., Nelen M. R., Wansink D. G., Coerwinkel M. M., te Riele H., et al. , 2002. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum. Mol. Genet. 11: 191–198. [DOI] [PubMed] [Google Scholar]
- van Oers J. M. M., Edwards Y., Chahwan R., Zhang W., Smith C., et al. , 2013. The MutSβ complex is a modulator of p53-driven tumorigenesis through its functions in both DNA double-strand break repair and mismatch repair. Oncogene 33: 3939–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A., Brachat A., Pohlmann R., Philippsen P. O., 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793–1808. [DOI] [PubMed] [Google Scholar]
- Zar J. H., 1999. Biostatistical Analysis, Ed. 4th Prentice Hall, Upper Saddle River, NJ. [Google Scholar]
- Zhang L., Leeflang E. P., Yu J., Arnheim N., 1994. Studying human mutations by sperm typing: instability of CAG trinucleotide repeats in the human androgen receptor gene. Nat. Genet. 7: 531–535. [DOI] [PubMed] [Google Scholar]
- Zhao J., Jain A., Iyer R. R., Modrich P. L., Vasquez K. M., 2009. Mismatch repair and nucleotide excision repair proteins cooperate in the recognition of DNA interstrand cross-links. Nucleic Acids Res. 37: 4420–4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.