Abstract
Background:
Acinetobacter baumannii is one of the most important pathogens in hospital-acquired infections especially in intensive care units (ICUs). This opportunistic pathogen can be easily isolated from water, soil, and hospital facilities. A. baumannii as a nosocomial opportunistic pathogen is resistant to a wide range of antibiotics and responsible for multiple infections, including bacteremia, pneumonia, meningitis, urinary tract infections, and surgical wounds. The aim of this study was to determine frequency and resistance patterns of A. baumannii isolated in ICUs of Isfahan Hospitals.
Materials and Methods:
During 1 year period (2012-2013), 350 specimens were collected from ICUs of Isfahan hospitals. The isolates were characterized as A. baumannii by conventional phenotypic, biochemical tests and confirmed by PCR for OXA-51-like gene. Susceptibility of isolates was determined by standard disk diffusion method according to CLSI.
Results:
From total of 350 specimens, 43 isolates were A. baumannii. The antimicrobial patterns of isolates showed that 53.5% of isolates were resistant to amikacin, 83.7% to tetracyclin, 86% to ceftazidime, 90.7% to Trimethoprim sulfametoxazol, 93% to imipenem, cefepime, meropenem, ampicillin–sulbactam. All isolates were resistant to ciprofloxacin.
Conclusion:
This study showed a high resistance of A. baumannii to a wide range of antimicrobial agent. It is necessary to adopt appropriate strategies to control the spread of the bacteria in care unit centers and wards.
Keywords: A. baumannii, antibiotic resistance pattern, blaOXA-51-like gene, intensive care units, molecular method
INTRODUCTION
Acinetobacter baumannii, a gram-negative, non-fermenting cocobacilli, is mostly found in soil, different water sources, and many healthcare environments.[1,2] This bacteria is abundant in normal flora of skin and mucous membranes in humans and able to cause opportunistic infections such as upper respiratory tract infections, pneumonia, meningitis, septicemia, and urinary tract infection, particularly in intensive care units (ICUs).[3] A. baumannii is resistant to dehydration, UV, common chemical disinfectants and detergents.[4] This bacteria can survive in dry environmental conditions and is isolated from various parts of hospital such as curtain, bed, furniture and clinical equipment.[5] Therefore, it can disseminate from different sources but hands of hospital staff is the most common mode of spread.[6] Recently, A. baumannii appeared significantly in the form of endemic and epidemic infections in hospitals.[7,8] In the clinical setting A. baumannii is extremely dangerous, due to it is ability to colonize and infect severely immunocompromised patients in ICUs.[9,10] Few methods are available to eliminate A. baumannii from widely used hospital equipments, especially catheter-related tools.[11]
The resistance of A. baumannii to antimicrobial agents is mediated by all of the known major resistance mechanisms, including modification of target sites, enzymatic inactivation, active efflux and decreased influx of drugs.[12]
Recently, it has been shown that NaCl, a monovalent cation largely found in our skin, is associated with enhanced multidrug resistance (MDR) of A. baumannii.[12] This bacteria produces metallo-beta-lactamase (IMP-VIM and types of SIM) causing resistance to most beta-lactam.[13,14] A. baumannii harbored enzymes such as Oxacillinase OXA-51 which hydrolyze carbapenems. Also, three groups of irrelevant enzymes of OXA-23, OXA-24, OXA-58 have been defined.[15] Of 100 isolates of imipenem-resistant Acinetobacter spp. collected from Tehran hospitals in 2009 and 2010, 6 isolates produced metallo-beta-lactamases and 94 isolates produced OXA-type carbapenemase.[10] The mortality rates for A. baumannii infections are about 50% for bacteremia and approximately 23-75% for pneumonia.[16] In the ICU, the mortality rate is 54%.[17]
MDR phenotype in A. baumannii results when integron-borne resistance determinants against different classes of antibiotics coexist, giving rise to MDR gene cassettes.[18,19]
Most publications defined MDR A. baumannii strains as resistant to at least three of the following five drug classes: Cephalosporins, carbapenems, ampicillin–sulbactam, fluoroquinolones, and aminoglycosides.[20]
Treatment of MDR A. baumannii infections is difficult. Currently, carbapenems, as the drug of choice in treating MDR A. baumannii infections, are used. However, carbapenem-resistant strains are increasing.[21]
Due to the high prevalence rate of this infection as well as various patterns of antibiotic resistance in different geographical areas, an investigation on prevalence rate and antibiotic sensitivity pattern in different part of the world is essential. These data would provide useful information on distribution of resistance patterns and the possibility to choose the proper treatment strategy.
The aim of this study was to determine the prevalence and antimicrobial resistance pattern of A. baumannii isolated from patients hospitalized in ICUs of Isfahan hospitals.
MATERIALS AND METHODS
During the 1 year period (2012-2013), 350 specimens from different sources including catheter, blood, urine, wound, CSF, sputum, and eye were collected. The specimens were inoculated initially on blood agar (Merck) and MacConkey agar (Merck) medium and incubated for 24 h at 37°C. Conventional biochemical methods such as oxidase, citrate, urea urease, malonate consumption, oxidation and fermentation of sugars, motility and indole production were used to identify A. baumannii. The isolates were stored in BHI medium containing 15% glycerol at −20°C.
In order to confirm the strain, isolates were subjected to PCR against the OXA-51–like gene. DNA of the isolates was extracted by DNA extraction kit (Fermentas) according to the manufacturer's protocol. DNA concentration and purity was measured using a spectrophotometer.
PCR mixture was done in a total volume of 25 μl including 1 μl MgCl2 (1.5 mM), 0.3 μl Taq DNA polymerase (500 U), 2.5 μl 10x PCR buffer, 0.5 μl dNTP (200 μM), 1 μl of each primer) 10 pmol/ml) and 2 μl of DNA template (5 ng genomic DNA). The primer sets F: 5’-TAA TGC TTT GAT CGG CCT TG and R: 5’-CTT CG TGG ATT CGA CTT CAT was used for amplification of the OXA-51-like gene.[22] Pseudomonas aeruginosa ATCC 27853 and A. baumannii ATCC 19606 strains have been used as negative and positive control, respectively.
Amplification reactions were carried out using a DNA thermal cycler (Master Cycle Gradiant, Eppendrof, Germany) with the following program: One cycle of 5 min at 94°C, 30 cycles as follows: Denaturation at 94°C for 25 s, annealing at 53°C for 40 s, initial extension at 72°C for 50 s, followed by final extension of 6 min at 72°C. PCR products were separated by electrophoresis on a 1.5% agarose gel and visualized by staining with green viewer under UV light.[23,24]
The antibiotic sensitivity pattern was done according to disk diffusion or Kirby-Bauer method. For this purpose antibiotics including: Amikacin, tetracyclin, ceftazidime, and carbapenem (imipenem and meropenem), trimethoprim sulfametoxazol, ampicillin–sulbactam, ciprofloxacin (Rosco, Denmark) have been used. A fresh 24 h bacterial lawn of isolates has been used to prepare 0.5 McFarland (1.5 × 108 cfu/ml) bacterial concentration and the diameter of the inhibition of bacterial growth were measured and compared to the reference tables provided by CLSI (Clinical and Laboratory Standard Institute).[25] Statistical analysis was performed by SPSS software version 14, using Chi-square test (Chi-Square) and kappa coefficient. A P < 0.05 was considered as an indicator of significance.
RESULTS
Data presented in Figure 1 demonstrate the frequency of A. baumannii from different samples. As it is shown in Figure 1, most of the specimen belongs to catheters (30.27%). 32.6%[14] of isolates were of women and 67.4% samples[26] were of men. There was no significant relationship between age, sex and the infection [Table 1 and Figure 2] (P < 0.05).
Figure 1.
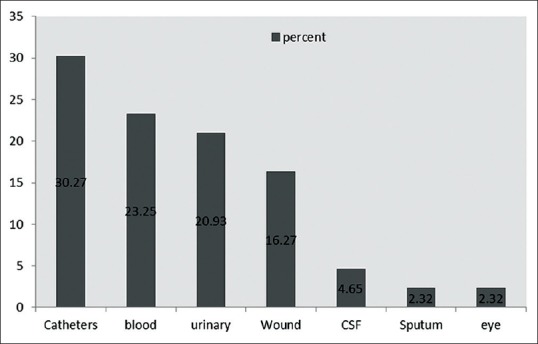
Distribution of Acinetobacter baumannii according to type of specimens in ICU
Table 1.
Frequency of patients according to gender

Figure 2.
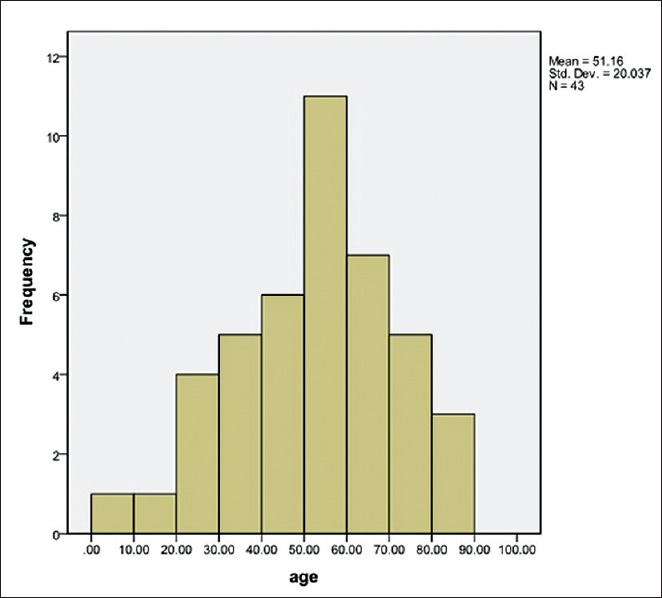
Frequency of patients according to age
From 350 specimen, 43 isolates were characterized as A. baumannii by phenotypic method. All of the isolates were confirmed by PCR. Figure 3 showed the result of PCR for the blaOXA-51-like gene. All of the strains harbored the blaOXA-51.
Figure 3.
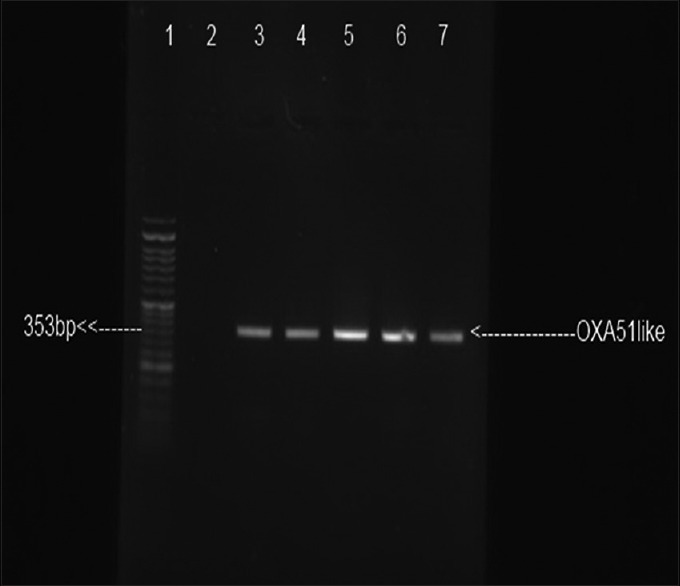
Electrophoresis result of blaOXA-51 amplification: lane 1: Size marker 50 bp; lane 2: Negative control P. aeruginosa ATCC 27853; lane 3: Positive control A. baumannii ATCC 19606; lanes 4–7: Clinical isolates
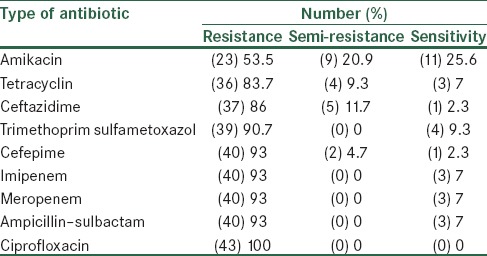
Our results showed that 53.5% of the isolates were resistant to amikacin, 83.7% to tetracyclin, 86% to ceftazidime, 90.7% to trimethoprim sulfametoxazol, 93% to cefepime, imipenem, meropenem, and ampicillin–sulbactam. All of isolates were resistant to ciprofloxacin [Table 2].
Table 2.
Antibiotics resistant patterns in Acinetobacter baumannii isolates
DISCUSSION
In recent years, nosocomial infections of A. baumannii, as an opportunistic pathogen, are increasing. Treatment of this bacteria especially MDR and broad-spectrum beta-lactamases strains is of major concern.[2] In our study from 350 samples collected in ICU, 43 (12.3%) isolates were of A. baumannii.
In a study in Tehran, from 100 collected samples in ICU of Rasoul Akram hospital, 21 samples (21%) were A. baumannii.[27] In another study of Rit and his colleagues in 2012 among 4180 clinical isolates 74.02% A.baumannii and 25.98% other types of Acinetobacter have been diagnosed.[28]
The present study showed a high percentage (100%) of MDR. The resistance rate to beta-lactam antibiotics, trimethoprim sulfametoxazol and tetracyclin was more than 80%. Resistance against imipenem and meropenem was more than 90%. The antibiotic resistance observed in this study was relatively similar to reports in other parts of Iran.
Sadeghifard and colleagues in 2010 showed that all isolates of A. baumannii had 100% resistance to aztreonam, cefotaxime, ceftazidime, ceftriaxone, meropenem and ticarcyline-clavulanate.[29] In another study these researcher showed that the isolated bacteria had 100% resistance to tetracyclin, 95/2% to gentamicin, amikacin, and 90/5% to ceftazidime.[27]
Ayan and colleagues reported that from 52 isolates, all of strains were resistant to piperacillin, cefepime, cefotaxime, ceftazidime, ceftriaxone, gentamicin, and aztreonam.[26]
Wang and colleagues (2003) described in an epidemic of drug-resistant strains of A. baumannii in the ICU, all the strains were resistant to aztreonam, amikacin, ampicillin–sulbactam, ceftazidime, cefepime, ciprofloxacin, gentamicin, imipenem, meropenem, piperacillin, tazobactam, and ticarcillin-clavlonic and sensitive to polymixine B.[30]
Other studies in Asia and the Middle East indicate prevalence of MDR A. baumannii in these regions.[31,32]
The difference in resistance pattern of A. baumannii is probably due to diversity in clinical samples, time of doing study and approaches of treatment in each geographical area. Comparing our results with other studies reveals that resistance of the bacteria to current antibiotics is increasing. In our study 32.6% of samples (14 persons) were of women and 67.4%[26] samples were of men and the highest age percentage was 50-60 years old [Figure 3] with the mean age of 51 years. Hujer, et al. in 2006 reported that from 75 A. baumannii isolates recovered from patients; 67 (89%) were of men and 8 (11%) were of women (mean age = 35 years).[33] Hello, et al. (2010) stated that the mean patient age was 57.8 years.[34] Similarly, Kwon, et al. (2007) indicated that the mean age of 80 A. baumannii-infected patients was 47 years, 57 (71.25%) of them were males and 23 (28.75%) were females.[35] In another study, Tsakris, et al., (2008) showed that the age of patients ranged from 22 to 83 years (mean age 68 years) and found the males outnumbered the females.[36] In our study from 43 clinically isolated A. baumannii the highest percentage (30.27%) was catheters followed by blood (23.25%), urine (20.93%), wound (16.27) and other clinical specimens (9.29%).
Amudhan, et al. (2011) found the highest isolation percentage (53.49%) was from respiratory secretions followed by 21.55%, 15.17%, 3.45%, 3.45% and 2.59%, respectively, from blood, wounds, cerebrospinal fluid, body fluids and urine.[37] Furthermore, Martins, et al. (2009) found from 53 clinically isolated A. baumannii the highest percentage (73.58%) was from respiratory tract followed by blood (16.98%), urine (3.77%) and other clinical specimens (5.66%).[38] Moreover, Mammina, et al. (2012) found that from 36 A. baumannii isolates, 26 (72.2%) isolates were from the respiratory tract infections.[39]
In a survey conducted in 2010 in Saudi Arabia from 1210 isolates collected from different samples such as breathing samples (469), blood (400), injury/tissues (235), urine (56), nasal swab (35), and CSF (15 from patients in section ICU composed, 40.9% A. baumannii, 19.4% Klebsiella pneumoniae and 16.3 Pseudomonas aeruginosa.[40]
ICU is of major concern of infection due to duration time of hospitalization and intensity of illness. Previous studies have shown A. baumannii as the most frequent nosocomial infection in the ICU.[41,42] Geographical data about resistance pattern of Acinetobacter would provide suitable information for treatment strategies. This study showed a high percentage of resistance to antimicrobial agent in A. baumannii. Therefore, it is necessary to adopt appropriate strategies to control the spread of these strains in ICU.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
REFERENCES
- 1.Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51:3471–84. doi: 10.1128/AAC.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: Emergence of a successful pathogen. Clin Microbiol Rev. 2008;21:538–82. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fagon JY, Chastere J, Domart Y, Trouillet JL, Gibert C. Mortality due to ventilator-associated pneumonia or colonization with Pseudomonas or Acinetobacter species: Assessment by quantitative culture of samples obtained by a protected specimen brush. Clin Infect Dis. 1996;23:538–42. doi: 10.1093/clinids/23.3.538. [DOI] [PubMed] [Google Scholar]
- 4.Murray PR, Baron EJ, Pfaller MA, Tenover FC, Yolken RH. 7th ed. Washington, DC: ASM Press; 1999. Manual of Clinical Microbiology; pp. 517–25. [Google Scholar]
- 5.Jawad A, Seifert H, Snelling AM, Heritage J, Hawkey PM. Survival of Acinetobacter baumannii on dry surfaces: Comparison of outbreak and sporadic isolates. J Clin Microbiol. 1998;36:1938–41. doi: 10.1128/jcm.36.7.1938-1941.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diomedi A. Acinetobacter baumannii pandrug-resistant: Update in epidemiological and antimicrobial managing issues. Rev Chilena Infectol. 2005;2:298–320. doi: 10.4067/s0716-10182005000600003. [DOI] [PubMed] [Google Scholar]
- 7.Dijkshoorn L, Van Vianen W, Degener JE, Michel MF. Typing of Acinetobacter calcoaceticus strains isolated from hospital patients by cell envelope protein profiles. Epidemiol Infect. 1987;99:659–67. doi: 10.1017/s0950268800066516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joly-Guillou ML. Clinical impact and pathogenicity of Acinetobacter. Clin Microbiol Infect. 2005;11:868–73. doi: 10.1111/j.1469-0691.2005.01227.x. [DOI] [PubMed] [Google Scholar]
- 9.Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. The population structure of Acinetobacter baumannii: Expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One. 2010;5:e10034. doi: 10.1371/journal.pone.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shahcheraghi F, Abbasalipour M, Feizabadi M, Ebrahimipour G, Akbari N. Isolation and genetic characterization of metallo-β-lactamase and carbapenamase producing strains of Acinetobacter baumannii from patients at Tehran hospitals. Iran J Microbiol. 2011;3:68–74. [PMC free article] [PubMed] [Google Scholar]
- 11.Barbolla RE, Centrón D, Maimone S, Rospide F, Salgueila C, Altclas J, et al. Molecular epidemiology of Acinetobacter baumannii spread in an adult intensive care unit under an endemic setting. Am J Infect Control. 2008;36:444–52. doi: 10.1016/j.ajic.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 12.Mihu MR, Martinez LR. Novel therapies for treatment of multi-drug resistant Acinetobacter baumannii skin infections. Virulence. 2011;2:97–102. doi: 10.4161/viru.2.2.15061. [DOI] [PubMed] [Google Scholar]
- 13.Lee K, Lee WG, Uh Y, Ha GY, Cho J, Chong Y, Korean Nationwide Surveillance of Antimicrobial Resistance Group VIM- and IMP-type metallo-beta-lactamase-producing Pseudomonas spp. and Acinetobacter spp. in Korean hospitals. Emerg Infect Dis. 2003;9:868–71. doi: 10.3201/eid0907.030012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee K, Yum JH, Yong D, Lee HM, Kim HD, Docquier JD, et al. Novel acquired metallo-beta-lactamase gene, bla (SIM-1), in a class 1 integron from Acinetobacter baumannii clinical isolates from Korea. Antimicrob Agents Chemother. 2005;49:4485–91. doi: 10.1128/AAC.49.11.4485-4491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bou G, Cerveró G, Domínguez MA, Quereda C, Martínez-Beltrán J. Characterization of a nosocomial outbreak caused by a multiresistant Acinetobacter baumannii strain with a carbapenem-hydrolyzing enzyme: High-level carbapenem resistance in A. baumannii is not due solely to the presence of beta-lactamases. J Clin Microbiol. 2000;38:3299–305. doi: 10.1128/jcm.38.9.3299-3305.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luna CM, Aruj PK. Nosocomial Acinetobacter pneumonia. Respirology. 2007;12:787–91. doi: 10.1111/j.1440-1843.2007.01147.x. [DOI] [PubMed] [Google Scholar]
- 17.Bassetti M, Righi E, Esposito S, Petrosillo N, Nicolini L. Drug treatment for multidrug-resistant Acinetobacter baumannii infections. Future Microbiol. 2008;3:649–60. doi: 10.2217/17460913.3.6.649. [DOI] [PubMed] [Google Scholar]
- 18.Seward RJ. Detection of integrons in worldwide nosocomial isolates of Acinetobacter spp. Clin Microbiol Infect. 1999;5:308–18. doi: 10.1111/j.1469-0691.1999.tb00149.x. [DOI] [PubMed] [Google Scholar]
- 19.Yum JH, Yi K, Lee H, Yong D, Lee K, Kim JM, et al. Molecular characterization of metallo-beta-lactamase-producing Acinetobacte rbaumannii andAcinetobacter genomospecies 3 from Korea: Identification of two new integrons carrying the bla (VIM-2) gene cassettes. J Antimicrob Chemother. 2002;49:837–40. doi: 10.1093/jac/dkf043. [DOI] [PubMed] [Google Scholar]
- 20.Peleg AY, Franklin C, Bell JM, Spelman DW. Dissemination of the metallo-beta-lactamase gene blaIMP-4 among gram-negative pathogens in a clinical setting in Australia. Clin Infect Dis. 2005;41:1549–56. doi: 10.1086/497831. [DOI] [PubMed] [Google Scholar]
- 21.Zarrilli R, Crispino M, Bagattini M, Barretta E, Popolo AD, Triassi M, et al. Molecular epidemiology of sequential outbreaks of Acinetobacter baumannii in an intensive care unit shows the emergence of carbapenem resistance. J Clin Microbiol. 2004;42:946–53. doi: 10.1128/JCM.42.3.946-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown S, Young HK, Amyes SG. Characterisation of OXA-51, a novel class D carbapenemase found in genetically unrelated clinical strains of Acinetobacter baumannii from Argentina. Clin Microbiol Infect. 2005;11:15–23. doi: 10.1111/j.1469-0691.2004.01016.x. [DOI] [PubMed] [Google Scholar]
- 23.Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006;27:351–3. doi: 10.1016/j.ijantimicag.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 24.Taherikalani M, Fatolahzadeh B, Emaneini M, Soroush S, Feizabadi MM. Distribution of different carbapenem resistant clones of Acinetobacter baumannii in Tehran Hospitals. New Microbiol. 2009;32:265–71. [PubMed] [Google Scholar]
- 25.Wayne, PA, USA: Clinical Laboratory Standards Institute; 2012. Clinical Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. 20th Informational Supplement: M100-S20. [Google Scholar]
- 26.Ayan M, Durmaz R, Aktas E, Durmaz B. Bacteriological, clinical and epidemiological characteristics of hospital-acquired Acinetobacter baumannii infection in a teaching hospital. J Hosp Infect. 2003;54:39–45. doi: 10.1016/s0195-6701(03)00076-8. [DOI] [PubMed] [Google Scholar]
- 27.Farivar AS, Nowroozi J, Emami M. The prevalence of Acinetobacter in sergical ICU in Rasoul Akram hospital in 2004-2005. Journal of Rafsanjan university of medical Sciences. 2005;4:342–7. [Google Scholar]
- 28.Rit K, Saha R. Niger Med J. Multidrug-resistant Acinetobacter infection and their susceptibility patterns in a tertiary care hospital. Niger Med J. 2012;53:126–8. doi: 10.4103/0300-1652.104379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sadeghifard N, Ranjbar R, Zaeimi J. Antimicrobial susceptibility, plasmid profiles, and RAPD-PCR typing of Acinetobacter bacteria. Asian Biomed (Res Rev News) 2010;4:901–11. [Google Scholar]
- 30.Wang SH, Sheng WH, Chang YY, Wang LH, Lin HC, Chen ML, et al. Healthcare-associated outbreak due to pan-drug resistant Acinetobacter baumanii in a surgical intensive care unit. J Hosp Infect. 2003;53:97–102. doi: 10.1053/jhin.2002.1348. [DOI] [PubMed] [Google Scholar]
- 31.Gopinath P, Geethapriya S, Jayakeerthana KH, Srivani R. Detection of certain virulence attributes and antimicrobial resistance pattern among clinical isolates of Acinetobacter baumannii. IJPBS. 2011;2:501–7. [Google Scholar]
- 32.Koh TH, Sng LH, Wang GC, Hsu LY, Zhao Y. IMP-4 and OXA beta-lactamases in Acinetobacter baumannii from Singapore. J Antimicrob Chemother. 2007;59:627–32. doi: 10.1093/jac/dkl544. [DOI] [PubMed] [Google Scholar]
- 33.Hujer KM, Hujer AM, Hulten EA, Bajaksouzian S, Adams JM, Donskey CJ, et al. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob Agents Chemother. 2006;50:4114–23. doi: 10.1128/AAC.00778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Hello S, Falcot V, Lacassin F, Mikulski M, Baumann F. Risk factors for carbapenem-resistant Acinetobacter baumannii infections at a tertiary care hospital in New Caledonia, South Pacific. Scand J Infect Dis. 2010;42:821–6. doi: 10.3109/00365548.2010.496087. [DOI] [PubMed] [Google Scholar]
- 35.Kwon KT, Oh WS, Song JH, Chang HH, Jung SI, Kim SW, et al. Impact of imipenem resistance on mortality in patients with Acinetobacter bacteraemia. J Antimicrob Chemother. 2007;59:525–30. doi: 10.1093/jac/dkl499. [DOI] [PubMed] [Google Scholar]
- 36.Tsakris A, Ikonomidis A, Poulou A, Spanakis N, Vrizas D, Diomidous M, et al. Clusters of imipenem-resistant Acinetobacter baumannii clones producing different carbapenemases in an intensive care unit. Clin Microbiol Infect. 2008;14:588–94. doi: 10.1111/j.1469-0691.2008.01996.x. [DOI] [PubMed] [Google Scholar]
- 37.Amudhan SM, Sekar U, Arunagiri K, Sekar B. OXA beta-lactamase-mediated carbapenem resistance in Acinetobacter baumannii. Indian J Med Microbiol. 2011;29:269–74. doi: 10.4103/0255-0857.83911. [DOI] [PubMed] [Google Scholar]
- 38.Martins AF, Kuchenbecker R, Sukiennik T, Boff R, Reiter KC, Lutz L, et al. Carbapenem-Resistant Acinetobacter baumannii producing the OXA-23 enzyme: Dissemination in southern Brazil. Infection. 2009;37:474–6. doi: 10.1007/s15010-009-9003-9. [DOI] [PubMed] [Google Scholar]
- 39.Mammina C, Palma DM, Bonura C, Aleo A, Fasciana T, Sodano C, et al. Epidemiology and clonality of carbapenem-resistant Acinetobacter baumannii from an intensive care unit in Palermo, Italy. BMC Res Notes. 2012;5:365. doi: 10.1186/1756-0500-5-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saeed NK, Kambal AM, El-Khizzi NA. Antimicrobial-resistant bacteria in a general intensive care unit in Saudi Arabia. Saudi Med J. 2010;31:1341–9. [PubMed] [Google Scholar]
- 41.Zolldann D, Haefner H, Poetter C, Buzello S, Sohr D, Luetticken R, et al. Assessment of a selective surveillance method for detecting nosocomial infections in patients in the intensive care department. Am J Infect Control. 2003;31:261–5. doi: 10.1067/mic.2003.72. [DOI] [PubMed] [Google Scholar]
- 42.Jeong SH, Bae IK, Kwon SB, Lee K, Yong D, Woo GJ, et al. Investigation of a nosocomial outbreak of Acinetobacter baumanii producing PER-1 extended-spectrum beta-lactamase in an intensive care unit. J Hosp Infect. 2005;59:242–8. doi: 10.1016/j.jhin.2004.09.025. [DOI] [PubMed] [Google Scholar]


