Summary
Chemical signals sensed on the periplasmic side of bacterial cells by transmembrane chemoreceptors are transmitted to the flagellar motors via the histidine kinase CheA, which controls the phosphorylation level of the effector protein CheY. Chemoreceptor arrays comprise remarkably stable supramolecular structures in which thousands of chemoreceptors are networked through interactions between their cytoplasmic tips, CheA, and the small coupling protein CheW. To explore the conformational changes that occur within this protein assembly during signalling, we used in vivo crosslinking methods to detect close interactions between the coupling protein CheW and the serine receptor Tsr in intact E. coli cells. We identified two signal-sensitive contacts between CheW and the cytoplasmic tip of Tsr. Our results suggest that ligand binding triggers changes in the receptor that alter its signalling contacts with CheW (and/or CheA).
Keywords: chemotaxis, chemoreceptor arrays, CheA
Introduction
The chemotactic behaviour of E. coli relies on exquisite activity control of the signalling histidine kinase CheA in response to chemical stimuli (see Hazelbauer and Lai, 2010 for a recent review on chemotaxis). CheA autophosphorylates and then donates its phosphoryl group to the response regulator CheY. Phospho-CheY interacts with the flagellar motors and causes a change in their direction of rotation from counterclockwise (CCW), the default swimming direction, to clockwise (CW), which initiates a tumble that reorients the cell’s heading. CheA control occurs within an ordered array of thousands of transmembrane chemoreceptor molecules, known as methyl-accepting chemotaxis proteins (MCPs), that are networked through interactions with CheA and the coupling protein CheW. Attractant binding to the receptor inhibits CheA, thus prolonging cell travel in the preferred direction. CheA deactivation is followed by a sensory adaptation process that restores pre-stimulus activity, mediated through reversible covalent modification of the chemoreceptors by the opposing activities of CheR, a methyltransferase, and CheB, a methylesterase.
The chemoreceptor cluster amplifies and integrates sensory information from chemoreceptors of different chemo-sensing specificities. Recent studies combining crystallographic and NMR structures of chemotaxis proteins and complexes with cryoelectron tomography of receptor arrays have suggested an architectural basis for signal amplification. The current picture of this remarkable supramolecular structure involves trimers of chemoreceptor dimers (Kim et al., 1999; Studdert and Parkinson, 2004) organized in a hexagonal lattice through interactions between the cytoplasmic tips of receptor molecules and two structurally homologous domains: the small protein CheW; and P5, the C-terminal domain of CheA (see models for array organization in Briegel et al., 2012; Liu et al., 2012; Li et al., 2013).
The interlocking network of receptor, CheW, and CheA molecules gives rise to an ultrastable structure (Erbse and Falke, 2009) that precludes component exchanges between signalling teams within the array (Studdert and Parkinson, 2005). Yet, binding of chemoeffectors at the periplasmic side of receptors controls CheA activity within the cluster with great sensitivity over several orders of magnitude. The conformational changes that mediate receptor signalling are not yet clear.
CheW shares with the P5 domain of CheA a structure consisting of two β-barrels sandwiching a hydrophobic core (Bilwes et al., 1999; Griswold et al., 2002; Fig. 1A and 1B). The central groove between the two subdomains has been implicated in the interaction with chemoreceptors by NMR shift studies (Vu et al., 2011; Wang et al., 2012), X-ray analyses of crystallized complexes (Park et al., 2006), and by genetic studies (Liu and Parkinson, 1991; Boukhvalova et al., 2002).
Fig. 1. Cysteine replacements in CheW and Tsr.
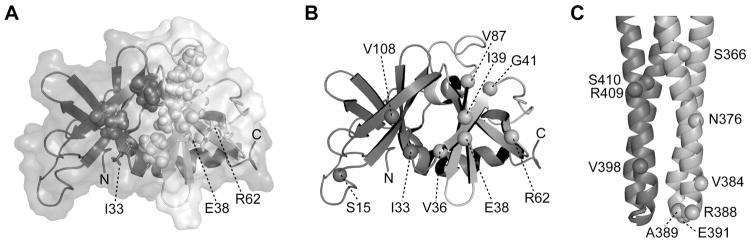
(A) CheW structure (PDB 2HO9, Li et al., 2007). Subdomains 1 and 2 are coloured in dark- and light-grey respectively. Residues in the hydrophobic groove that are postulated to interact with receptors are shown in space-fill representation. Residues I33, E38 and R62 are shown as sticks.
(B) Backbone structure of CheW. Labelled spheres indicate the residue positions chosen for cysteine reporters.
(C) The cytoplasmic hairpin tip of a Tsr dimer (PDB 1QU7, Kim et al., 1999). The spheres in the darker subunit show C-helix residues chosen for cysteine reporters; spheres in the lighter subunit show N-helix residues chosen for cysteine reporters.
The cytoplasmic domain of chemoreceptors is a four-helix, coiled-coil bundle with a hairpin turn at the membrane-distal tip (Fig. 1C). Residues in the N-helix near the hairpin tip contact both CheW and P5 (Vu et al., 2011; Wang et al., 2012; Li et al., 2013; Piasta et al., 2013) as well as their counterparts in other receptors to form trimers of dimers (Kim et al., 1999). In the context of the receptor array, the internal subunits of the receptor trimers contact one another, whereas their external subunits are free to engage in contacts with CheW or P5. The receptor helix C-terminal to the hairpin does not seem to play a major role in these interactions.
In the present work, we used in vivo crosslinking methods to investigate the interaction between CheW and MCPs. Single cysteine residues in the receptor and CheW formed disulphides in a signal-dependent manner. One of the CheW-receptor signal-sensitive disulphides involves a cysteine replacement at an arginine residue that is highly conserved both in CheW and in the structurally homologous P5 domain of CheA, together with a cysteine replacement at a position in the receptor involved in both receptor-receptor contacts and receptor-CheA contacts. Cells expressing the two cysteine-containing proteins displayed full chemotactic function, indicating that the changes in crosslinking efficiency reflect protein movements that occur during normal signalling.
Results
Formation of disulphides between Tsr/Tar and CheW in whole cells
NMR chemical shift studies performed with proteins from T. maritima have identified residues in CheW and in the chemoreceptor hairpin that may promote interaction between the two proteins (Vu et al., 2011). In CheW, the residues that contact the chemoreceptor are mainly situated in the groove between its two β-barrel subdomains (Fig. 1A). In the chemoreceptor, the residues that contact CheW lie in the N-helix of the tip-proximal region of the hairpin (Fig. 1C). Structural studies of protein complexes by pulsed dipolar ESR spectroscopy (Bhatnagar et al., 2010) and X-ray crystallography (Briegel et al., 2012; Li et al., 2013) also implicate the same regions of the two proteins in their interaction, although some differences exist in the proposed interfaces.
To explore the CheW-chemoreceptor interaction in intact E. coli cells, we introduced single cysteine replacements at different CheW residues of the proposed interaction surface (Fig. 1B), and co-expressed those CheW proteins with serine (Tsr) or aspartate (Tar) receptors carrying single-cysteine substitutions in the hairpin tip region (Fig. 1C), in the absence of other chemotaxis proteins. We then looked for disulphide bond formation between the two proteins by immunoblotting analysis of the resulting protein extracts. Co-expression of CheW-R62C and Tsr-V398C, for example, produced a crosslinked product (Fig. 2A, lane 5), that was dependent on the presence of the cysteine substitution in both proteins (Fig 2A, compare with lanes 4 and 6) and was eliminated by treatment of the samples with a reducing agent (dithiothreitol, not shown). A detectable level of crosslinked product required treatment of the cells with the oxidant diamide, consistent with the strongly reducing character of the bacterial cytoplasm (Fig. 2A, compare lanes +/−diamide). The crosslinking observed between CheW-R62C and Tsr-V398C was specific, as it did not happen when those reporter proteins were paired with other cysteine-containing partners (Fig. 2B). The occurrence of Tsr-Tsr crosslinking products was variable and their intensity was dependent on the presence of specific CheW cysteine-carrying mutants, suggesting that interactions with the CheW variants might affect the dynamics or geometry of Tsr molecules with respect to one another.
Fig. 2. Disulphide crosslinking between CheW and Tsr.
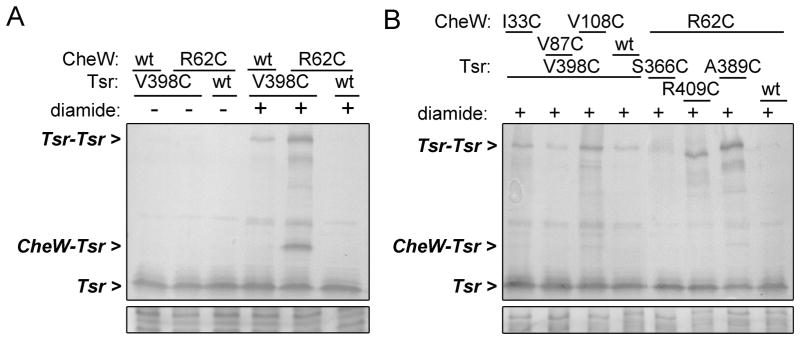
(A) Representative crosslinking result. UU1581 cells expressing wild-type or Cys-reporter versions of Tsr (pCS12 plasmid derivatives) and CheW (pPA770 plasmid derivatives) were grown to mid-log phase and treated or not with diamide. Tsr expression was induced at 0.45 μM sodium salicylate and CheW expression at 25 μM IPTG (physiological levels). Samples were analysed by SDS-PAGE in 10% acrylamide gels and visualized by immunoblotting with an antibody against the conserved cytoplasmic region of Tsr. Protein load of the samples was analysed by Coomassie staining; a representative region is shown (bottom panel).
(B) An experiment similar to that in (A) with various Tsr and CheW Cys-reporters.
In all, we tested nine Cys-substituted Tsr or Tar receptors and nine Cys-substituted CheW proteins in pairwise combinations (Table 1). The Tsr and Tar reporter proteins were all stably expressed at wild-type levels. Their ability to mediate chemotaxis ranged from fully functional to nonfunctional (Suppl. Fig. 1A). The CheW reporter proteins were more variable, both with respect to expression level and to function (Suppl. Fig. 1B).
Table 1.
Disulphide cross-linking between CheW and Tsr.
| CheW
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| S15 | I33 | V36 | E38 | I39 | G41 | R62 | V87 | V108 | ||
| Tsr | S366 | − | − | − | − | − | − | − | − | − |
| N376* | − | − | − | − | − | − | − | − | − | |
| V384 | + | − | − | − | − | − | − | − | − | |
| R388 | − | + | − | − | − | − | − | − | − | |
| A389 | − | − | − | − | − | − | − | − | − | |
| E391 | ++ | − | − | − | − | + | − | − | − | |
| V398** | + | − | − | − | − | − | ++ | − | − | |
| R409 | − | − | − | − | − | − | − | − | − | |
| S410* | − | − | − | − | − | − | − | − | − | |
Assays were done with the equivalent Tar replacements.
Assays were done with either Tsr or Tar replacement.
UU1581 cells expressing cysteine-substituted Tsr or Tar at physiological levels (0.45 μM sodium salicylate, pCS12 plasmid derivatives) and cysteine-substituted CheW at over-expression levels (50–100 μM IPTG, pPA770 plasmid derivatives) were treated with diamide, and the crosslinking products were analysed by immunoblotting with an anti-CheW polyclonal antibody. Band intensity was determined with the software ImageQuant (Amersham). The occurrence of covalent binding between Tsr and CheW under the assayed conditions was calculated as the percentage of crosslinking product relative to the total amount of Tsr in each sample. Categories correspond to − (<20%), + (20–40%) or ++ (>40%).
We found two conspicuous crosslinking products and several less prominent ones (Table 1). Both of the major reporter pairs were detected at physiological expression levels of CheW, indicating that they were not due to over-expression artefacts (not shown and Fig. 2A). Crosslinking between CheW-S15C and Tsr-E391C had been described between the equivalent residues in T. maritima proteins in an in vitro study (Bhatnagar et al., 2010). Thus, the occurrence of a strong in vivo crosslinking signal between S15, located near the N-terminus of CheW, and E391 at the hairpin turn, is consistent with the predicted proximity between subdomain 1 of CheW and the hairpin tip. The other strong crosslinking signal involved R62, a highly conserved residue in subdomain 2 of CheW, and V398, a residue in the C-helix of the receptor hairpin that had previously been implicated in dimer-dimer interactions within the trimer of dimers (Studdert and Parkinson, 2004). The minor crosslinking products were more variable in occurrence and intensity in different experiments. Several of them involved the S15C CheW reporter, suggesting that the N-terminus of CheW might be flexible and able to adopt different positions with respect to the receptor.
Influence of ternary complex formation on crosslinking efficiency
To assess whether the extent of crosslinking was different in the context of receptor-CheA-CheW ternary signalling complexes, the reporter pairs were tested at physiological expression levels in strains with or without CheA expressed from the chromosome. Initial tests were done with reporters expressed from compatible plasmids in strains UU1610 (CheA+) and UU1613 (CheA−). Both strains lacked the modification enzymes CheR and CheB to simplify the pattern of receptor bands. Both strains also encode a cysteine-tagged aspartate receptor (Tar-S364C), but this receptor site does not crosslink to any of the CheW reporters (Table 1, Tsr-S366C) and is not relevant to the experiment.
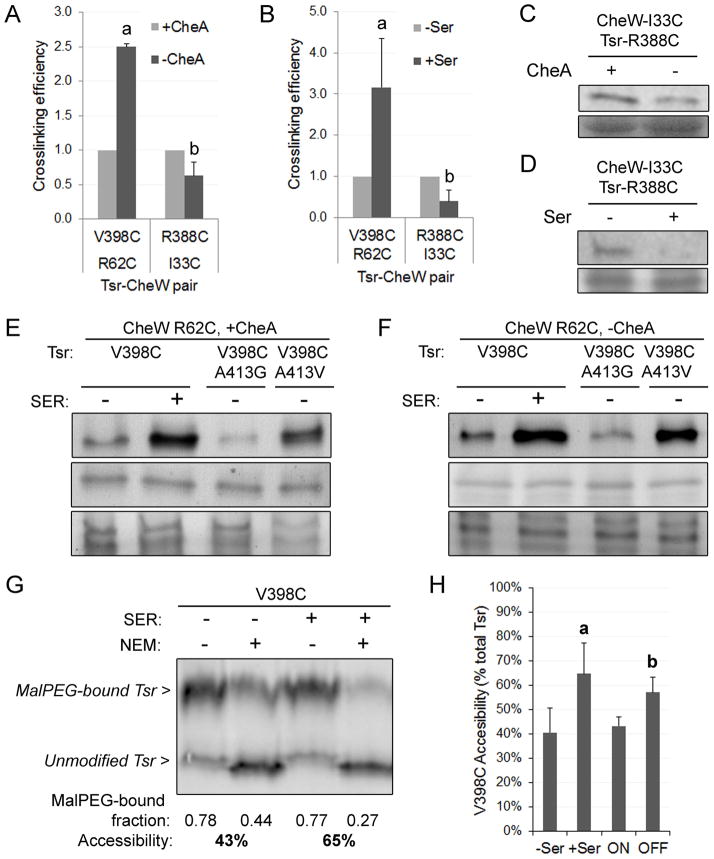
We observed a strong signal for the pair CheW-R62C // Tsr-V398C whose intensity was reproducibly higher in the absence of CheA (Fig. 3A). For the remaining pairs, we did not observe a significant change in crosslinking efficiency in the absence or presence of CheA, with the exception of the weak pair CheW-I33C // Tsr-R388C, which showed a small, but reproducible, decrease in crosslinking efficiency in the absence of CheA (Fig. 3A, example gel in Fig. 3C). The opposing effect of CheA on these two pairs suggests that the reduced crosslinking efficiency of the CheW-R62C // Tsr-V398C pair in the presence of CheA is not simply due to competition between CheA and CheW for binding to the receptors, as suggested for ternary complexes in vitro (Asinas and Weis, 2006). Rather, the CheA effects might indicate a slightly different interaction between CheW and receptors when the proteins are in the context of ternary complexes and assembled chemoreceptor clusters. The increase in crosslinking efficiency for the pair Tsr-V398C//CheW-R62C and the decrease for the pair Tsr-R388C//CheW-I33C in cells lacking CheA was also observed, with similar changes in magnitude, when UU1581 (no MCPs, no CheA) and MDP15 (no MCPs, CheA+) were used as host strains (Fig. 3E and 3F for the pair Tsr-V398C//CheW-R62C and data not shown).
Fig. 3. Efficiency of disulphide formation between CheW and Tsr under different conditions.
(A–D) Cells expressing CheW-R62C and Tsr-V398C, or CheW-I33C and Tsr-R388C (pPA770 or pCS12 plasmid derivatives respectively), were treated with diamide. Protein samples were analysed by Coomassie staining and by immunoblotting. Band intensity was determined with the software ImageQuant (Amersham). The intensity of the crosslinking band was corrected for any variations in the amount of protein loaded in each lane and then normalized to the control conditions (light-grey bars). Crosslinking efficiency is defined as the fold-change in the intensity of the crosslinking band product relative to control condition. Error bars indicate the standard error of each mean. a, b letters indicate significant changes relative to control samples (light-grey bars, p < 0.05).
(A) Strains UU1610 (CheA+) and UU1613 (CheA−) were used. Protein samples were analysed by Coomassie staining and by immunoblotting with an antibody against the conserved cytoplasmic region of Tsr. The mean of three independent experiments is shown for each reporter pair.
(B) MDP15 (CheA+) cells carrying the corresponding plasmids were treated with diamide immediately after the addition of serine. Protein samples were analysed by Coomassie staining and by immunoblotting with an anti-CheW polyclonal antibody. Values are the mean of four (R62C-V398C) or three (I33C-R388C) independent experiments.
(C) Representative result of (A) for CheW-I33C // Tsr-R388C crosslinking pair.
(D) Representative result of (B) for CheW-I33C // Tsr-R388C crosslinking pair.
(E, F) Cells expressing Tsr-V398C variants and CheW-R62C were incubated in the absence or presence of 10 mM serine and treated with diamide. Protein samples were analysed by SDS-PAGE and immunoblotting with an anti-CheW polyclonal antibody. For simplicity, only the crosslinking product is shown (top panel). The same samples were analysed by immunoblotting with an anti-Tsr antibody (middle panel) and by Coomassie staining (bottom panel) after treatment with DTT to reduce disulphide crosslinks. The A413G alteration locks Tsr in the “ON” state; A413V locks Tsr output in the “OFF” state.
(E) Experiment carried out in strain MDP15 (CheA+).
(F) Experiment carried out in strain UU1581 (CheA−).
(G) Cell samples from the experiment of panel (E) were treated or not with 50 μM NEM to block solvent-exposed thiol groups. Denatured protein samples were then treated with Mal-PEG to mark thiol groups that were not blocked by NEM. Samples were analysed by SDS-PAGE and immunoblotting with anti-Tsr antibody. Band intensity was determined with the software ImageQuant (Amersham). The fraction of Tsr that was MalPEG-modified and the resulting accessibility values were calculated as explained in Experimental Procedures.
(H) Accessibility is expressed as the percentage of NEM-blocked Tsr (i.e. solvent-accessible) relative to total Tsr amount. The mean of three independent experiments is shown. Error bars indicate the standard error of each mean. a, b letters indicate significant changes relative to “-Ser” and “ON” conditions respectively (p < 0.05).
Signalling-dependent changes in crosslinking efficiency
Based on the CheA-dependent effects described above, we chose two reporter pairs (CheW-I33C // Tsr-R388C and CheW-R62C // Tsr-V398C) to assess the effects of attractant stimuli on crosslinking efficiency in ternary signalling complexes. The crosslinking tests were performed in the presence or absence of a saturating level of the attractant serine in strain MDP15 (CheA+), with expression of the plasmid-encoded reporters at physiological levels. We found that the attractant stimulus substantially enhanced crosslinking between CheW-R62C and Tsr-V398C (Fig. 3B and Fig. 4E, lanes 1 and 2), whereas it reduced crosslinking between CheW-I33C and Tsr-R388C (Fig. 3B and 3D). The divergent serine responses displayed by the two crosslinking pairs could reflect a signal-state difference in the interaction between CheW and the receptor. To test this possibility, we measured crosslinking of the CheW-R62C // Tsr-V398C pair using receptor alterations that lock output in the kinase-active, ON state (A413G; P. Ames and J.S. Parkinson, unpublished results) or the kinase-inactive, OFF state (A413V; Ames and Parkinson, 1994). Crosslinking efficiency was significantly higher for the receptor locked in the OFF state (Tsr-V398C/A413V, Fig. 3E, lane 4) than it was for the locked-ON receptor (Tsr-V398C/A413G, Fig. 3E, lane 3).
Fig. 4. Suppression of CheW-R62* and CheA-R555* functional defects by Tsr-V398C.
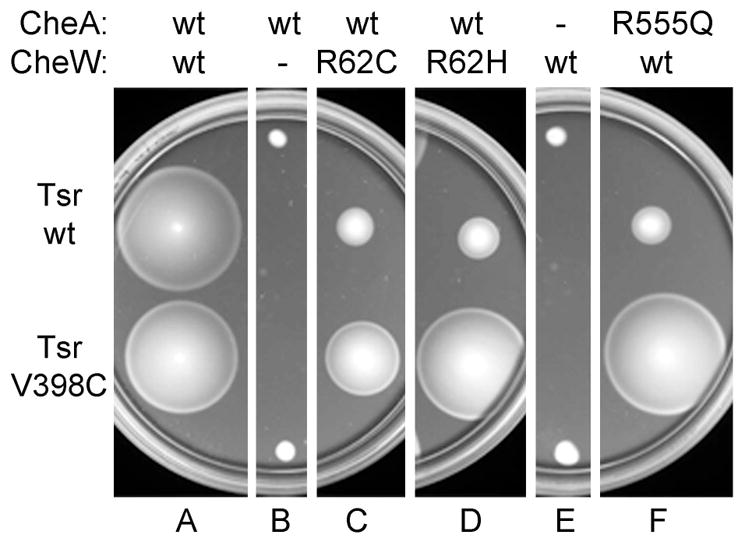
Tryptone soft-agar plates with chloramphenicol 12.5 μg/ml, ampicillin 50 μg/ml, sodium salicylate 0.45 μM and IPTG 25 μM, were inoculated with strains carrying different versions of CheW or CheA in the chromosome, and expressing compatible plasmids pRZ33 (CheY and CheZ) and pCS12 (wild-type Tsr) or pCS12 V398C (Tsr-V398C). Plates were incubated at 32.5 °C for 9 hours. Used strains were UU2700 (wild-type CheA, wild-type CheW), MDP21 (wild-type CheA, no CheW), MDP23 (wild-type CheA, CheW-R62C), MDP22 (wild-type CheA, CheW-R62H), MDP26 (no CheA, wild-type CheW) and MDP28 (CheA-R555Q, wild-type CheW).
The signal state difference in crosslinking efficiency for the CheW-R62C // Tsr-V398C pair might reflect a direct signal-induced conformational change in the receptor. Alternatively, the conformational change might depend on the ternary signalling complex, in which case it should only occur in the presence of CheA. However, the increase in CheW-R62C // Tsr-V398C crosslinking efficiency upon treatment with serine and with the locked-OFF version of the receptor also occurred in the absence of CheA (strain UU1581; Fig. 3F). This result indicates that different signalling states influence the CheW-receptor crosslinking interaction through direct conformational effects on the receptor.
To look for more direct evidence of that conformational change, we measured the in vivo accessibility of Tsr residue V398C in different signalling states. Briefly, our assay involved treatment of intact cells with N-ethyl maleimide (NEM), which reacts with solvent-exposed thiol groups, followed by denaturation and labelling of unblocked groups by maleimide-polyethylene glycol (MalPEG) treatment. The more exposed a cysteine residue is, the more readily it becomes blocked by NEM and, thus, remains unmodified by MalPEG. In this experiment, we used a low NEM concentration to detect slight differences in the accessibility of the reporter thiol group. To control the signalling state of the receptor complexes, the experiments were done in MDP15 cells expressing CheW-R62C and Tsr-V398C (or the ON or OFF derivatives) at physiological levels. An example gel is shown in Fig. 3G, and quantitative data in Fig. 3H. We found that residue V398C was significantly more solvent-exposed, i.e., more easily blocked by NEM, in the presence of serine than in its absence (Fig. 3G, compare lanes 2 and 4, and Fig. 3H). Similarly, V398C was more accessible in the OFF-state mutant receptor than in the ON-state mutant receptor (Fig. 3H). We conclude that solvent exposure of Tsr residue V398C is higher in the kinase-off signalling state and that this accessibility increase reflects the conformational change that also enhances the efficiency of crosslinking to the CheW-R62C reporter.
Chemotactic properties of Tsr-V398C and CheW-R62C mutants
When assessed individually, Tsr-V398C mediates normal serine chemotaxis in soft agar assays (Studdert and Parkinson, 2004), whereas CheW-R62C does not (approximately 45% of wild-type colony size; Liu and Parkinson, 1991). Although amino acid replacements at this highly conserved arginine residue cause CheW functional defects, they do not seem to affect chemoreceptor interaction or assembly of ternary complexes. For example, CheW-R62H cannot support chemotaxis, but in vitro exhibits a normal binding affinity for the Tar receptor and a normal ability to assemble kinase-activity ternary complexes (Boukhvalova et al., 2002). The presence of the V398C replacement in Tsr suppressed the chemotactic defect in CheW-R62C (Fig. 4, panel C), indicating that the signal-dependent changes in crosslinking efficiency for this pair of mutants indeed reflect changes in the relative orientation of the two reporter proteins that take place in the context of cells that show chemotaxis to serine.
The equivalent position to CheW-R62 in the homologous P5 domain of CheA (residue 555) is also a highly conserved arginine. Amino acid replacements at this CheA position also impair chemotactic ability, but do not affect binding to CheW or other signalling parameters when the mutant proteins are assayed in vitro (Zhao and Parkinson, 2006a, 2006b). Tsr-V398C also alleviates the functional defects of CheA-R555Q (Fig. 4, panel F), and those of CheW-R62H (Fig. 4, panel D). The suppression of analogous defects in the two proteins by Tsr-V398C suggests that the conserved arginine residues in CheW and the P5 domain of CheA may play similar signalling roles.
Discussion
CheW-chemoreceptor contacts in intact cells
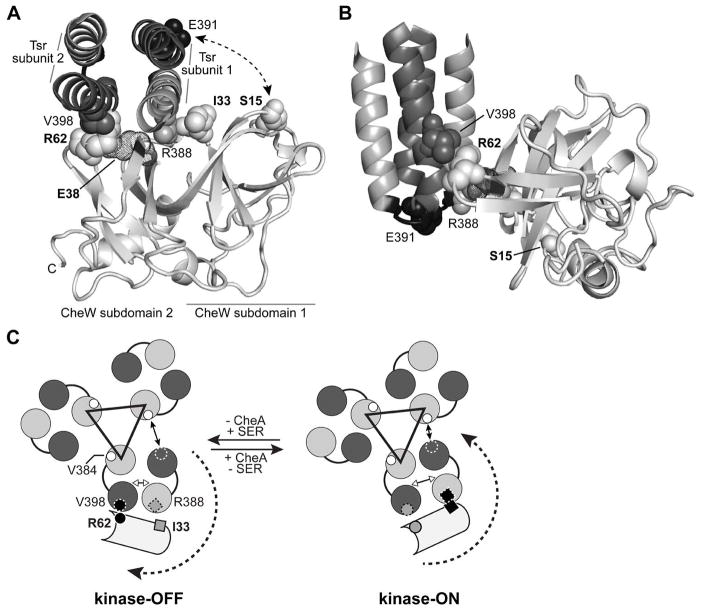
The CheW and receptor residues that formed disulphide bridges in vivo in the present study lie relatively close one to another in the models for the chemoreceptor array built from crystallographic and tomographic data (Briegel et al., 2012; Liu et al., 2012; Li et al., 2013; see Suppl. Fig. 2). Considering the available atomic structures for CheW and for the dimeric receptor hairpin, the crosslinking signals we identified are consistent with a CheW molecule interacting with both subunits of the same receptor dimer (Fig. 5A). In this view, the N-terminus of CheW (subdomain 1) is close to the hairpin tip of one receptor subunit (subunit 1, CheW-S15C // Tsr-E391C crosslinking) and the groove between the two CheW subdomains interacts with the N-helix of the same receptor subunit (CheW-I33C // Tsr-R388C crosslinking). Subdomain 2 of CheW contacts the C-helix of receptor subunit 2 (CheW-R62 // Tsr-V398C crosslinking).
Fig. 5. CheW-Tsr contacts during signalling.
(A) Interaction of the E. coli CheW protein (light grey) with the Tsr N-helix (grey) and C-helix (dark grey) at the hairpin tip. The docked structure was modelled from the atomic coordinates for the corresponding Thermotoga protein complex (PDB 3UR1, Briegel et al., 2012). Only residues 373–409 of Tsr are shown, viewed in the membrane-proximal to -distal direction. Residues that formed disulphide crosslinks in the present work are shown space-filled. CheW residue E38, which is thought to form a salt bridge with CheW-R62 (Ortega et al., 2013b), is shown in dot representation.
(B) Side view of the modelled CheW-Tsr complex. The structure shown in (A) was rotated 90° about the X and Y axes to show the juxtaposition of the Tsr-V398 (dark grey) and CheW-R62 (light grey) side chain atoms. The loop that joins the N- and C-helices of Tsr is shaded black; residue E391 (space-filled) defines its mid-point.
(C) Model of the signalling-related motions of CheW interacting with the Tsr trimer of dimers. The Tsr N- and C-helices are depicted as grey and dark grey circles. The black triangle connects the N-helices of the inner subunits at the trimer axis that make most of the trimer-stabilizing contacts between receptor dimers. A CheW molecule (light grey) is shown interacting with the outer helices of one Tsr dimer. CheW-R62C (small circle) crosslinks most efficiently with Tsr-V398C (small circle with dashed circumference) in the kinase-OFF signalling state (black circles, left). CheW-I33C (small square) crosslinks most efficiently with Tsr-R388C (small square with dashed border) in the kinase-ON signalling state (black squares, right). The receptor molecules are thought to undergo conformational shifts during signalling that rotate the C-helix of the outer subunit of one dimer toward or away from the N-helix of the inner subunit of the neighbouring dimer. In consequence, the interaction between V398 in the C-helix of the outer subunit (small dashed circle outline) changes its distance relative to V384 in the N-helix of the inner subunit (small white circles). The distances between helices in the trimer of dimers are based on the molecular dynamics study done by Ortega et al 2013a (Fig. 5 in cited reference).
This interaction model agrees with the long-postulated interaction between the CheW subdomain cleft and the N-helix of the receptor hairpin (Liu and Parkinson, 1991; Boukhvalova et al., 2002; Griswold et al., 2002). Moreover, in a recent NMR study, the C-helix residue equivalent to Tsr-V398 in a T. maritima soluble receptor (residue 156 of Tm14) was one of the receptor residues that showed a large chemical shift upon interaction with CheW (Vu et al., 2011). In that study, nine other significantly perturbed receptor residues were located in the N-helix, immediately preceding the hairpin tip.
CheW-chemoreceptor contacts in the context of ternary signalling complexes
The presence of CheA caused a notable decrease in crosslinking efficiency for the pair Tsr-V398C // CheW-R62C (Figs. 3A, 3E and 3F) and a reproducible increase in the weaker crosslinking signal for the pair Tsr-R388C // CheW-I33C (Figs. 3A and 3B). These behaviours suggest that the observed changes in crosslinking efficiency reflect a change in the relative position of CheW with respect to the receptor within the ternary complex rather than a simple receptor-binding competition between CheA and CheW. Interestingly, an attractant stimulus also elicited opposite changes in crosslinking efficiency for the two reporter pairs: The crosslinked product for Tsr-V398C // CheW-R62C increased, whereas that for Tsr-R388C // CheW-I33C decreased (Figs. 3B, 3C, 3E and 3F). These results suggest that the presence of CheA promotes the interaction between CheW and the receptor that takes place in the kinase-ON conformation. However, even in the absence of CheA, attractant had a significant effect on crosslinking efficiency (Fig. 3F), indicating that ligand binding induces a direct conformational change in the receptor molecule that affects its interaction with CheW. Whereas the crosslinking pair CheW-I33C/Tsr-R388C was unable to mediate serine taxis (not shown) and both proteins were partially or totally non-functional when tested individually (Mowery et al, 2008; Fig. S1 A and B), the more conspicuous signalling-dependent crosslinking pair (CheW-R62C/Tsr-V398C) mediated clear responses to serine when both proteins were co-expressed, indicating their functional proficiency.
A recent in vitro study has described the interactions between the chemoreceptor tip and the CheW-like P5 domain of CheA (Piasta et al., 2013). That study used disulphide crosslinking between the cytoplasmic domain of E. coli Tsr and the P5 domain of S. typhimurium CheA to assess the proximity between reporter sites in a functionally active complex. Although attractant stimuli did not alter disulphide formation rates for reporter pairs at the interface between the N-helix of the receptor hairpin tip and the groove between β-barrels in P5, there was a signal-dependent change in the rate of disulphide formation between a residue located in subdomain 2 of P5 and residue 398 from the C-helix of the receptor tip. Those results suggested that, in response to attractant binding, the P5 subunit bound to the N-helix of the receptor (from the external subunit in the trimer of dimers), shifts its subdomain 2 closer to the C-helix of the other subunit (the internal one) of the same chemoreceptor dimer. Our crosslinking results suggest the existence of a similar movement for a receptor-bound CheW in response to an attractant stimulus (see scheme in Fig. 5C). It is worth noticing that the study of Piasta et al. describes the interaction between residue 398 in Tsr and the residue equivalent to E533 in E. coli CheA, which presumably forms a salt bridge with R555 (Ortega et al., 2013b), the counterpart to the CheW R62 residue that interacts with Tsr-V398 (this study).
We also observed that the presence of attractant caused a significant increase in the solvent exposure of residue 398 (Figs. 3G and 3H). This increase in accessibility might occur at the residue engaged in interaction with CheW (or CheA P5 domain), and/or by the corresponding residue in the other subunit of the dimer, which might loosen its interaction with the neighbouring dimer in the trimer upon receiving the attractant stimulus (see scheme in Fig. 5C).
The attractant-elicited conformational changes that favour the contact between CheW, bound to the N-helix of a Tsr subunit, and the C-helix of the other subunit of the dimer are also consistent with a recent molecular dynamics simulation of the receptor hairpin tip biased toward ON or OFF outputs by different modification states (Ortega et al., 2013a). The authors proposed that the OFF conformation of the chemoreceptor dimer is correlated with a shortened distance between the C-helix of the internal subunit in the trimer of dimers and the N′-helix belonging to the external subunit of the same dimer (see scheme in Fig. 5C). This scenario is consistent both with the increased contact between the pair Tsr-V398C and CheW-R62C and also with the increased accessibility of residue 398 upon an attractant stimulus.
Chemoreceptor contacts with CheW and P5: Are they perfectly equivalent?
The structural homology between CheW and the P5 subdomain of CheA suggests that these two proteins might interact with the receptor tip in a comparable manner. Crystallographic and NMR chemical shift studies support this view, albeit leaving open the possibility of slight differences between the two interactions. NMR studies using T. maritima proteins showed that interaction with CheW, P5, or a CheW/P5 complex perturbed residues in the same region of the receptor tip, however the directions and magnitudes of the observed changes differed (Vu et al., 2011; Wang et al., 2012). Two crystallographic studies of T. maritima ternary complexes have identified two different interaction interfaces on the receptor tip for CheW and P5. The crystal structure reported in Briegel et al. (2012) seems to describe very well the interface between the receptor and CheW. In that study the P5 domain was postulated to be in a non-native orientation. In contrast, the structure reported in Li et al. (2013) shows a P5 domain interacting with the region of the receptor previously implicated in the CheW interaction, with CheW bound further from the receptor tip, in a seemingly non-native fashion. The comprehensive crosslinking study of Piasta et al. (2013) clearly supports an interaction between P5 and the receptor matching that described by Li et al. (2013).
The distances between beta carbons of the two signalling-dependent crosslinking pairs found in this work best correlate with the structure reported in Briegel et al., 2012 (Fig. 5A, Suppl. Fig. 2). Whereas the study of Piasta et al. (2013) identified the interaction between Tsr-A387 and St CheA-L545 (equivalent to I33 in Ec CheW) as insensitive to attractants, we found in this work that the interaction between the contiguous residue in Tsr (R388C) and CheW-I33C showed small, but reproducible, changes in response to attractants.
Taken together, these observations suggest that CheW and the CheA-P5 domain interact with chemoreceptors in a similar but possibly slightly different way.
A suggested signalling role for CheW-R62/CheR-R555
Replacements at the conserved arginine residues CheW-R62 or CheA-R555 disrupt chemotaxis in soft agar plates but otherwise do not cause any significant defect in ternary complex formation or kinase activation or control when tested in vitro (Boukhvalova et al., 2002; Zhao and Parkinson, 2006a and b). The observed interaction between the C-helix residue 398 in Tsr and CheW-R62C (this work) or a nearby residue in CheA-P5 (Piasta et al., 2013), together with the observation that a cysteine replacement at V398 suppresses the chemotaxis defect of arginine replacement mutants of the two proteins, suggests that the nature of the interaction is similar in both the crosslinking and suppression pairs. We speculate that the interaction between the hydrophobic portion of the conserved arginine side chain (R62 in CheW or R555 in CheA) and Tsr-V398 at the periphery of the trimer of dimers stabilizes the OFF conformation of the ternary complex. The crosslinking observed between Tsr V398C, situated in the C-helix of the inner subunit, and residues in subdomain 2 of CheW (this work) or CheA-P5 (Piasta et al., 2013) suggests that in order to reach the OFF state the outer subunit of the same dimer moves so that its N-helix (to which CheW or P5 are bound) gets closer to the C-helix of the inner subunit (see double-headed white arrow in Fig. 5C, left). Such a conformational change might be opposed by the strong hydrophobic interaction between Tsr-V398 from the outer subunit and Tsr-V384 from the neighbouring dimer (double-headed black arrow in Fig. 5C). In the absence of an arginine residue at CheW-62/CheA-555 positions, the attractant-induced movement might not be sufficiently stabilized, resulting in a non-functional array. However, a replacement that weakens the dimer-to-dimer interaction might compensate for this defect and restore the ability of the conformational change to occur.
Experimental procedures
Bacterial strains
Strains were derivatives of the E. coli K12 strain RP437 (Parkinson and Houts, 1982) and are listed in Table 2.
Table 2.
Bacterial strains.
| Strain | Relevant Genotype | Source or reference |
|---|---|---|
| RP1078 | (cheW-tap)DE2217 | Liu and Parkinson, 1989 |
| RP8607 | (tsr)DE7028 (cheW-tap)DE2217 (trg)DE100 | Mowery et al., 2008 |
| UU1581 | (flhD-flhB)DEtr4 (tsr)DE7028 (trg)DE100 | Studdert and Parkinson, 2005 |
| UU1610 | tar-S364C (tsr)DE7028 (trgDE100) (tap-cheB)DE2241 cheW(Am)113 | Studdert and Parkinson, 2005 |
| UU1613 | tar-S364C (tsr)DE7028 (trgDE100) (tap-cheB)DE2241 (cheA-cheW)DE2167 | Studdert and Parkinson, 2005 |
| UU2612 | (tar-tap)DE4530 (tsr)DE5547 (aer)DE1 (trg)DE4543 | Zhou et al., 2011 |
| UU2700 | (cheY-cheZ)DE1215 (tar-tap)DE4530 (tsr)DE5547 (aer)DE1 (trg)DE4543 | Han and Parkinson, 2014 |
| MDP15 | cheW::kan-ccdB (tar-cheZ)DE4211 (tsr)DE5547 (aer)DE1 (trg)DE4543 | This work |
| MDP21 | cheW::kan-ccdB (cheY-cheZ)DE1215 (tar-tap)DE4530 (tsr)DE5547 (aer)DE1 (trg)DE4543 | This work |
| MDP22 | cheW-R62H (cheY-cheZ)DE1215 (tar-tap)DE4530 (tsr)DE5547 (aer)DE1 (trg)DE4543 | This work |
| MDP23 | cheW-R62C (cheY-cheZ)DE1215 (tar-tap)DE4530 (tsr)DE5547 (aer)DE1 (trg)DE4543 | This work |
| MDP26 | cheA::ccdB-kan (cheY-cheZ)DE1215 (tar-tap)DE4530 (tsr)DE5547 (aer)DE1 (trg)DE4543 | This work |
| MDP28 | cheA-R555Q (cheY-cheZ)DE1215 (tar-tap)DE4530 (tsr)DE5547 (aer)DE1 (trg)DE4543 | This work |
Plasmids
Plasmids derived from pACYC184 (Chang and Cohen, 1978), which confers chloramphenicol resistance, were: pKG116 [salicylate-inducible expression vector] (Buron-Barral et al., 2006); pCS12 [salicylate-inducible wild-type tsr] (Studdert and Parkinson, 2005); and pCS66 [salicylate-inducible wild-type 6XHis-tagged tar] (Studdert and Parkinsion, 2005). Plasmids derived from pBR322 (Bolivar et al., 1977), which confers ampicillin resistance, were: pCJ30 [isopropyl β-D-thiogalactopyranoside (IPTG)-inducible expression vector] (Bibikov et al., 1997), pRZ33 [IPTG-inducible cheYZ] (Lai and Parkinson, unpublished results) and pPA770 [IPTG-inducible cheWΔ8] (Studdert and Parkinson, 2005). pPA770 encodes a fully functional CheW protein that lacks eight N-terminal residues of wild-type CheW.
Site-directed mutagenesis
Mutations were introduced into plasmids with the QuikChange Site-Directed Mutagenesis Kit (Stratagene). For tsr or tar mutations, pCS12 or pCS66 were used as template plasmids. For cheW mutations, pPA770 was used as the template plasmid. Candidate mutants were verified by sequencing the entire protein-coding region.
Chemotaxis assay in semi-solid agar plates
Cells carrying wild-type or mutant versions of CheW or CheA in the chromosome and carrying deletions in all the chemoreceptors, CheY and CheZ genes, were transformed with plasmids carrying IPTG-inducible CheYZ genes and sodium salicylate wild-type or cysteine-substituted Tsr. Plasmid-carrying cells were inoculated on tryptone semi-solid agar plates with 0.25% agar (Parkinson, 1976) containing 12.5 μg ml-1 chloramphenicol, 50 μg ml-1 ampicillin, 0.45 μM sodium salicylate and 50 μM IPTG. Plates were incubated for 9 h at 32.5 °C.
Immunoblotting
Cells were pelleted by centrifugation (6000 x g) and resuspended at an OD600 of 2 in 10 mM potassium phosphate (pH 7.0) and 0.1 mM EDTA. Cells from 0.5 ml of the suspension were pelleted and lysed by boiling in 50 μl of sample buffer (Laemmli, 1970). Proteins released from the lysed cells were analysed by electrophoresis in sodium dodecyl sulphate-containing polyacrylamide gels (SDS-PAGE) and visualized by immunoblotting with an antiserum directed against the highly conserved portion of the Tsr signalling domain (Ames and Parkinson, 1994) in 10% acrylamide, 0.05% bisacrylamide gels (Studdert and Parkinson, 2004), or an antiserum against CheW in 17.5% acrylamide, 0.47% bisacrylamide gels (Cardozo et al., 2010). Either Cy5-labeled (Amersham) or alkaline phosphatase-conjugated (Sigma) goat anti-rabbit immunoglobulin were used as secondary antibodies. Cy5-labeled antibodies were detected with a Storm 840 fluorimager (Amersham); alkaline phosphatase-conjugated antibodies were developed with nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (both from Promega) and converted to grey scale images with a digital scanner. All gel images were analysed with ImageQuant (Amersham).
Disulphide crosslinking
Cells were grown at 30°C to mid-exponential phase in tryptone broth containing 25 μg ml−1 of chloramphenicol, 100 μg ml−1 of ampicillin, and the appropriate concentrations of sodium salicylate and IPTG, harvested by centrifugation, and resuspended at OD600 = 2 in 10 mM potassium phosphate (pH 7) and 0.1 mM EDTA. Cell suspensions (0.5 ml) were incubated for 45 min at 30°C with 0.5 mM diamide. Reactions were quenched by the addition of 10 mM N-ethyl maleimide (NEM). Cells were pelleted and then lysed by boiling in 50 μl of sample buffer. Proteins released from the lysed cells were analysed by SDS/PAGE in 10% acrylamide, 0.05% bisacrylamide gels and visualized by immunoblotting with Tsr or CheW antiserum.
For analysis of the stimulus effect on cross-linking efficiency, cells were washed with phosphate buffer, resuspended at an OD600 = 2 in the same buffer and treated with 10 mM L-serine before diamide treatment.
In Vivo Accessibility of Thiol Groups
Accessibility assays were done essentially as described by Massazza et al. (2011). Cells expressing Tsr-V398C at physiological concentrations were grown to mid-log phase, harvested by centrifugation, and resuspended at an OD600 = 2 in 10 mM potassium phosphate (pH 7.0) and 0.1 mM EDTA. Cell suspensions (0.5 ml) were treated or not with 50 μM NEM for 15 min at room temperature. Reactions were quenched by the addition of 20 mM dithiothreitol for 15 min at room temperature. Cells were pelleted, washed twice with potassium phosphate buffer to remove excess dithiothreitol, and then lysed by boiling in 50 μl of sample buffer (Laemmli, 1970). Unblocked thiol groups were then labelled by treatment with 5 mM MalPEG (5 kDa) (Boehringer Mannheim) for 15 min at room temperature. Total protein extracts were analysed by immunoblotting with Tsr antiserum as described before. Gel images were analysed with ImageQuant (Amersham), and the accessibility of thiol groups was calculated as follows: the percentage of total MCP with shifted mobility in the NEM-treated samples was expressed as a fraction of that percentage in the untreated cells. This fraction represented the residues that were not accessible for blockage by the NEM treatment. Accessibility could then be expressed as 1 - (fraction of inaccessible residues).
Statistics
Error bars are reported as standard errors for the indicated means. Tests of statistical significance employed the unidirectional Student’s t test for paired variables.
Supplementary Material
Acknowledgments
We thank Peter Ames and Run-Zhi Lai (University of Utah) for providing strains and plasmids and for their help and assistance with strain construction. This work was supported by research grant PIP 154 (to C.A.S.) from the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Argentina, and by research grant GM19559 (to J.S.P.) from the National Institute of General Medical Sciences. The Protein-DNA Core Facility at the University of Utah receives support from National Cancer Institute grant CA42014 to the Huntsman Cancer Institute. A.P. is a fellow and C.A.S. a Career Investigator from CONICET, Argentina.
References
- Ames P, Parkinson JS. Constitutively signaling fragments of Tsr, the Escherichia coli serine chemoreceptor. J Bacteriol. 1994;176:6340–6348. doi: 10.1128/jb.176.20.6340-6348.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asinas AE, Weis RM. Competitive and cooperative interactions in receptor signaling complexes. J Biol Chem. 2006;281:30512–30523. doi: 10.1074/jbc.M606267200. [DOI] [PubMed] [Google Scholar]
- Bhatnagar J, Borbat PP, Pollard AM, Bilwes AM, Freed JH, Crane BR. Structure of the ternary complex formed by a chemotaxis receptor signaling domain, the CheA histidine kinase, and the coupling protein CheW as determined by pulsed dipolar ESR spectroscopy. Biochemistry. 2010;49:3824–3841. doi: 10.1021/bi100055m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikov SI, Biran R, Rudd KE, Parkinson JS. A signal transducer for aerotaxis in Escherichia coli. J Bacteriol. 1997;179:4075–4079. doi: 10.1128/jb.179.12.4075-4079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilwes AM, Alex LA, Crane BR, Simon MI. Structure of CheA, a signal-transducing histidine kinase. Cell. 1999;96:131–141. doi: 10.1016/s0092-8674(00)80966-6. [DOI] [PubMed] [Google Scholar]
- Bolivar F, Rodriguez RL, Greene PJ, Betlach MC, Heyneker HL, Boyer HW. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- Boukhvalova MS, Dahlquist FW, Stewart RC. CheW binding interactions with CheA and Tar. Importance for chemotaxis signaling in Escherichia coli. J Biol Chem. 2002;277:22251–22259. doi: 10.1074/jbc.M110908200. [DOI] [PubMed] [Google Scholar]
- Briegel A, Li X, Bilwes AM, Hughes KT, Jensen GJ, Crane BR. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc Natl Acad Sci U S A. 2012;109:3766–3771. doi: 10.1073/pnas.1115719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buron-Barral MC, Gosink KK, Parkinson JS. Loss- and gain-of-function mutations in the F1-HAMP region of the Escherichia coli aerotaxis transducer Aer. J Bacteriol. 2006;188:3477–3486. doi: 10.1128/JB.188.10.3477-3486.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo MJ, Massazza DA, Parkinson JS, Studdert CA. Disruption of chemoreceptor signalling arrays by high levels of CheW, the receptor-kinase coupling protein. Mol Microbiol. 2010;75:1171–1181. doi: 10.1111/j.1365-2958.2009.07032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AC, Cohen SN. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbse AH, Falke JJ. The core signaling proteins of bacterial chemotaxis assemble to form an ultrastable complex. Biochemistry. 2009;48:6975–6987. doi: 10.1021/bi900641c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold IJ, Zhou H, Matison M, Swanson RV, McIntosh LP, Simon MI, Dahlquist FW. The solution structure and interactions of CheW from Thermotoga maritima. Nat Struct Biol. 2002;9:121–125. doi: 10.1038/nsb753. [DOI] [PubMed] [Google Scholar]
- Han XS, Parkinson JS. Unorthodox sensory adaptation site in the Escherichia coli serine chemoreceptor. J Bacteriol. 2014;196:641–649. doi: 10.1128/JB.01164-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer GL, Lai WC. Bacterial chemoreceptors: providing enhanced features to two-component signaling. Curr Opin Microbiol. 2010;13:124–132. doi: 10.1016/j.mib.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KK, Yokota H, Kim SH. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Li Y, Hu Y, Fu W, Xia B, Jin C. Solution structure of the bacterial chemotaxis adaptor protein CheW from Escherichia coli. Biochem Biophys Res Commun. 2007;360:863–867. doi: 10.1016/j.bbrc.2007.06.146. [DOI] [PubMed] [Google Scholar]
- Li X, Fleetwood AD, Bayas C, Bilwes AM, Ortega DR, Falke JJ, Zhulin IB, Crane BR. The 3.2 A resolution structure of a receptor: CheA:CheW signaling complex defines overlapping binding sites and key residue interactions within bacterial chemosensory arrays. Biochemistry. 2013;52:3852–3865. doi: 10.1021/bi400383e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Hu B, Morado DR, Jani S, Manson MD, Margolin W. Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells. Proc Natl Acad Sci U S A. 2012;109:E1481–1488. doi: 10.1073/pnas.1200781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JD, Parkinson JS. Role of CheW protein in coupling membrane receptors to the intracellular signaling system of bacterial chemotaxis. Proc Natl Acad Sci U S A. 1989;86:8703–8707. doi: 10.1073/pnas.86.22.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JD, Parkinson JS. Genetic evidence for interaction between the CheW and Tsr proteins during chemoreceptor signaling by Escherichia coli. J Bacteriol. 1991;173:4941–4951. doi: 10.1128/jb.173.16.4941-4951.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massazza DA, Parkinson JS, Studdert CA. Cross-linking evidence for motional constraints within chemoreceptor trimers of dimers. Biochemistry. 2011;50:820–827. doi: 10.1021/bi101483r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowery P, Ostler JB, Parkinson JS. Different signaling roles of two conserved residues in the cytoplasmic hairpin tip of Tsr, the Escherichia coli serine chemoreceptor. J Bacteriol. 2008;190:8065–8074. doi: 10.1128/JB.01121-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega DR, Yang C, Ames P, Baudry J, Parkinson JS, Zhulin IB. A phenylalanine rotameric switch for signal-state control in bacterial chemoreceptors. Nat Commun. 2013a;4:2881. doi: 10.1038/ncomms3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega DR, Mo G, Lee K, Zhou H, Baudry J, Dahlquist FW, Zhulin IB. Conformational coupling between receptor and kinase binding sites through a conserved salt bridge in a signaling complex scaffold protein. PLoS Comput Biol. 2013b;9:e1003337. doi: 10.1371/journal.pcbi.1003337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Borbat PP, Gonzalez-Bonet G, Bhatnagar J, Pollard AM, Freed JH, Bilwes AM, Crane BR. Reconstruction of the chemotaxis receptor-kinase assembly. Nat Struct Mol Biol. 2006;13:400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- Parkinson JS. cheA, cheB, and cheC genes of Escherichia coli and their role in chemotaxis. J Bacteriol. 1976;126:758–770. doi: 10.1128/jb.126.2.758-770.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson JS, Houts SE. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J Bacteriol. 1982;151:106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piasta KN, Ulliman CJ, Slivka PF, Crane BR, Falke JJ. Defining a key receptor-CheA kinase contact and elucidating its function in the membrane-bound bacterial chemosensory array: a disulfide mapping and TAM-IDS Study. Biochemistry. 2013;52:3866–3880. doi: 10.1021/bi400385c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studdert CA, Parkinson JS. Crosslinking snapshots of bacterial chemoreceptor squads. Proc Natl Acad Sci U S A. 2004;101:2117–2122. doi: 10.1073/pnas.0308622100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studdert CA, Parkinson JS. Insights into the organization and dynamics of bacterial chemoreceptor clusters through in vivo crosslinking studies. Proc Natl Acad Sci U S A. 2005;102:15623–15628. doi: 10.1073/pnas.0506040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu A, Wang X, Zhou H, Dahlquist FW. The Receptor-CheW binding interface in bacterial chemotaxis. J Mol Biol. 2011 doi: 10.1016/j.jmb.2011.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Vu A, Lee K, Dahlquist FW. CheA-receptor interaction sites in bacterial chemotaxis. J Mol Biol. 2012;422:282–290. doi: 10.1016/j.jmb.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Parkinson JS. Mutational analysis of the chemoreceptor-coupling domain of the Escherichia coli chemotaxis signaling kinase CheA. J Bacteriol. 2006a;188:3299–3307. doi: 10.1128/JB.188.9.3299-3307.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Parkinson JS. Cysteine-scanning analysis of the chemoreceptor-coupling domain of the Escherichia coli chemotaxis signaling kinase CheA. J Bacteriol. 2006b;188:4321–4330. doi: 10.1128/JB.00274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Ames P, Parkinson JS. Biphasic control logic of HAMP domain signalling in the Escherichia coli serine chemoreceptor. Mol Microbiol. 2011;80:596–611. doi: 10.1111/j.1365-2958.2011.07577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.