Evolution by natural selection includes two main steps: the generation of heritable variations (e.g., mutations) and the differential proliferation of the variants in the environment. When the neo-Darwinists synthesized a modern view of natural selection and genetics in the early 20th century, they specified a simplifying assumption that Darwin (12) had not: that the rates of formation of mutations would be independent of exposure to selective environments (e.g., see reference 47). Thus, evolution, and the mutations driving it, should be constant and gradual. That some spontaneous mutations form independently of interaction with the environment is certainly true (42, 46, 52; see also many subsequent papers). These form before an organism encounters a selective environment, with a definable relationship to cell divisions (“growth-dependent mutations”), probably because many result from DNA replication errors. However, work with several microbial assay systems indicates the existence of additional mutation pathways that appear to be induced in response to the environment (reviewed in references 16, 58, and 60). These mutation mechanisms, called stationary-phase or stress-induced mutation, operate specifically under growth-limiting stress and may sometimes produce mutations that confer a growth advantage in the growth-limiting environment, called adaptive mutations. The problem is, are they really something different from growth-dependent mutations?
WHICH MODEL?
Three general models for the origin of apparent adaptive mutations have garnered intense interest because of their evolutionary implications (reviewed in references 10, 16, 58, and 60). Directed mutation (DM) models suggested the provocative possibility that mutations might be targeted specifically to those that relieve the stress (e.g., see reference 9), an idea tinged with Lamarckism. In hypermutation (HM) models, mutation rates increase genome wide in response to stress, stimulating both nonadaptive and adaptive mutations (e.g., see references 25, 53, 58, and 70 and see also references 13 and 55), in harmony with Darwinism but appearing incompatible with the neo-Darwinist constraint of constant, gradual evolutionary change (implying constant mutation rates) (e.g., see reference 47). Cryptic-growth (CG) models specify constant mutation rates (in accordance with the neo-Darwinist constraint) but that extra DNA replications (not observed by the investigator) in rare growing cells give the appearance of enhanced mutation under stress and even of mutagenesis targeted to selected genes (e.g., see references 2, 23, 34, 43, 44, and 54). These general models, and the evolutionary consequences, can be distinguished by elucidating the molecular mechanism(s) of the mutagenesis. In their simplest form, CG models predict mutation mechanisms identical to those of growth-dependent mutation, whereas DM and HM models predict different mechanisms of mutation, one directed preferentially to selected genes (DM models) and the other affecting many genes (HM models).
MANY MECHANISMS AND HYPERMUTATION AS A GENERAL STRATEGY
No single mutational mechanism underlies the many cases of stationary-phase mutations reported for different bacteria and for yeasts. A variety of molecular mechanisms that appear different from growth-dependent mutation are implicated (though some similarities in the stationary-phase mechanisms are becoming apparent) (reviewed in references 16, 58, and 60; see also references 32 and 33). This discourages CG models for these systems. Implying that HM is a general, multimechanism strategy of bacteria, the vast majority of 787 natural isolates of Escherichia coli from diverse habitats worldwide display variable levels of increased general mutability under starvation (3). (CG models are unlikely to be responsible for the appearance of increased mutation frequencies in the starved, aging colonies in this assay because, for the small number of strains in which the genetic requirements of stress-induced mutation was examined, these differed from the genetic requirements for growth-dependent mutation in those strains [3, 67], arguing against standard growth-dependent mutation mechanisms having occurred.) Additional data suggest that HM mechanisms speed evolution in the real world, regardless of whether HM mechanisms were selected for that group benefit or as an incidental by-product of error-prone DNA metabolism processes selected for other reasons (3). Thus, HM appears to be a general bacterial strategy.
THE E. COLI Lac SYSTEM
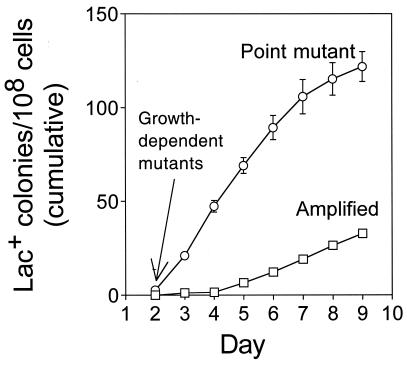
The most mechanistic information is known about the E. coli Lac frameshift-reversion system (8), for which HM models (e.g., see references 41, 45, 58, and 69) and CG models (e.g., see references 2 and 34) are currently under debate. In this assay, cells in which the chromosomal lac operon is deleted and which carry the lacIZ33 +1 frameshift allele on an F′ conjugative plasmid are plated on minimal lactose medium, selecting Lac+ frameshift reversion mutants (8). Growth-dependent Lac+ revertants formed before plating are visible as colonies by about 2 days of incubation (Fig. 1). Additional Lac+ colonies arise over the next several days (Fig. 1) from a population of cells showing no net growth (8). These are adaptive mutants formed after exposure to the lactose starvation medium (8, 30, 50).
FIG. 1.
Adaptive point mutation and adaptive gene amplification are separate outcomes that arise with different kinetics. Point mutants have compensatory frameshift reversions in the lac gene (19, 61) and also carry high frequencies of other mutations genome wide (24, 57, 70). lac-amplified clones carry 20 to 50 tandem repeats of 7- to 40-kb DNA segments spanning the leaky lac frameshift allele, which allow growth on lactose medium without a compensatory frameshift mutation (30). lac-amplified clones do not carry high levels of extra mutations genome wide (30). Shown is a cumulative plot. The lac-amplified clones comprise typically ≥40% of new colonies from day 8 onward (30). Data are from reference 30.
POINT MUTANTS AND AMPLIFIED ADAPTIVE Lac+ COLONIES
Two distinct classes of adaptive revertants are seen (Fig. 1): compensatory frameshift revertants (Lac+ point mutants) (19, 61) and strains carrying gene amplification of the leaky lacIZ33 allele as 20 to 50 tandem copies of 7- to 40-kb, F′-carried repeats (lac-amplified clones) (30). These allow growth on lactose medium without a compensatory frameshift mutation (30; see references 2, 18, 36, and 68 for previous descriptions of lac amplification). Although rare initially, lac-amplified clones constitute ≥40% of new colonies from day 8 onward, a major adaptive outcome in this system (30).
DIFFERENT MECHANISMS EXCLUDE SIMPLE CRYPTIC-GROWTH MODELS
Whereas simple CG models predict the same mutation mechanism(s) for growth-dependent and adaptive reversions, DM and HM models predict different mechanisms. Both the point mutations (19, 61) and amplifications (30) are different from growth-dependent Lac+ reversions (19, 61) and form via different mechanisms (see below) arguing against simple CG models. For point mutation, DM models are untenable because mutations accumulate in genes other than lac (5, 17, 24, 57, 70; discussed below), whereas HM models are supported by many aspects of the mutation mechanism as follows.
POINT MUTATION MECHANISM: ERROR-PRONE DNA DOUBLE-STRAND-BREAK REPAIR IMPLICATED
The proteins for double-strand-break repair by homologous recombination are required for adaptive and not growth-dependent Lac frameshift reversions (22, 28, 29). We suggested that error-prone repair of DNA double-strand breaks or ends could generate adaptive point mutants (28).
F-transfer proteins, but not conjugative transfer of the F′ plasmid, are also required for point mutation (20, 21, 23). Single-strand nicks made at the transfer origin could lead to high levels of double-strand ends on the F′ plasmid, promoting Lac+ reversion by error-prone repair there (22, 40, 56, 59). Supporting this idea, double-strand-break-repair-protein-dependent mutations also accumulate in the E. coli chromosome (5) but at a roughly 20-fold lower frequency than in the F′ plasmid (17). This suggests that the same mechanism operates in both places but is more active in F′, probably because frequent single-strand nicks yield more double-strand breaks and ends there (5, 59). (Repair by homologous recombination should be available even to stationary-phase cells, roughly 40% of which contain more than one chromosome [and the fraction with more than one F′ plasmid is likely to be higher] [1]. Duplicated genome segments are also reasonable candidates for partner DNA for homologous double-strand-break repair.) But double-strand ends also arise in the chromosome and must be repaired there (e.g., see reference 11). We suggest that during starvation stress, a mutagenic stress response leads to their repair being error prone, promoting mutation (5) as follows.
SOS RESPONSE, ERROR-PRONE DNA POLYMERASE, AND DECREASED MISMATCH REPAIR PROMOTE POINT MUTATION
A salient feature of HM models is that they propose stress responses that increase the general mutation rate specifically during the stress. The classical mutagenic stress response of E. coli is the SOS DNA damage response (66). SOS induction is required specifically for adaptive (and not growth-dependent) point mutation (8, 48) as is the SOS-regulated, error-prone DNA polymerase DinB (Pol IV) (14, 49, 51). DinB is responsible for ∼85% of the point mutations (49). Neither SOS nor DinB is required for adaptive amplification (49). Also contributing to general HM in the point mutation mechanism, the postreplicative mismatch repair system becomes limiting transiently during adaptive mutation (27; but also see references 15 and 26). This could be caused by mismatch repair being overwhelmed by excess DNA polymerase errors (27) made by DinB (14, 49). However limiting mismatch repair activity occurs, the combination of an error-prone DNA polymerase and limiting mismatch repair is expected to be mutagenic genome wide, supporting HM models.
POINT MUTATION IS NOT DIRECTED MUTATION
Two kinds of studies show that point mutation is not directed to the lac gene. First, while Lac+ revertants accumulate in the population of starving cells, unselected mutations (reversions of a tet frameshift allele) also accumulate both in a tet gene near lac in the F′ plasmid (17) and in a chromosomal tet gene (5). These form via the same double-strand-break-repair-protein- and DinB-dependent mechanism as Lac+ point mutations in F′, although the frequency of the chromosomal reversions is ∼20-fold lower (perhaps because Tra-promoted double-strand ends in F′ are more frequent than chromosomal double-strand breaks, as discussed above). Second, Lac+ point mutants carry high frequencies of unselected mutations, as follows.
GENOME-WIDE HYPERMUTATION IN A CELL SUBPOPULATION (POINT MUTANTS ONLY)
The Lac+ point mutants carry high frequencies of unselected (secondary) mutations throughout their genomes, ∼50-fold higher than their Lac− neighbors starved on the same selective medium, which are similar to never-starved cells (24, 57, 70). Thus, only a subpopulation of the starved cells is hypermutated. This, plus the evidence in the previous section, is considered by most authors to have ruled out DM models and provided strong support for HM models for point mutation. The adaptive amplified clones are not similarly hypermutated and so do not descend from the same subpopulation (30).
The hypermutation that these subpopulation cells experience is transient (24, 57, 62, 70). What makes the subpopulation different from the main population is not known. Proposals include the suggestion that subpopulation cells are those that acquire DNA double-strand breaks (70) or induce an SOS response (48), leading to increased dinB expression (14, 49), or amplify the dinB gene, leading to its overexpression (39, 64). Any of these mechanisms, if they generated most Lac+ point mutants, would indicate an HM mechanism.
DOES THE HYPERMUTABLE SUBPOPULATION GENERATE MOST Lac+ POINT MUTANTS?
The question of whether the hypermutable subpopulation generates most Lac+ point mutants is critical for distinguishing HM models from current CG models (e.g., see references 34 and 64), which demand that most Lac+ not come from cells with an elevated mutation rate. It has been suggested that only 10% of the point mutants descend from the hypermutable cell subpopulation, the other 90% arising from cells not engaging in transient hypermutation (57). The 90% could then generate point mutants with no change in mutation rate via a CG model (63, 65). The idea is that some point mutants appear to contain more detectable secondary mutations than others (though the data on this are very few [57, 70]), so perhaps they have come from the hypermutable subpopulation whereas the others did not. This is possible, but not proven. However, suggesting that 90% of point mutations occur in cells with “normal” mutation rates is not compatible with ∼85% of point mutants requiring an SOS response (49), DinB (49), and limiting mismatch repair (27). Thus, a simpler hypothesis is that the vast majority of Lac+ point mutants arose by a single HM mechanism but that those with more secondary mutations remained in the transient mutable state longer before becoming Lac+ and exiting that state (Fig. 2) and so acquired more detectable secondary mutations (5, 6). Either different mutation rates, or different lengths of time spent being hypermutable, can account for the data. We suggest that the second possibility is simpler because it offers a single point mutation mechanism (HM) for most point mutants (5, 6, 58), which is harmonious with the requirements for DinB, SOS, and limiting mismatch repair for most point mutants (but see Fig. 2 for an alternative two-population model).
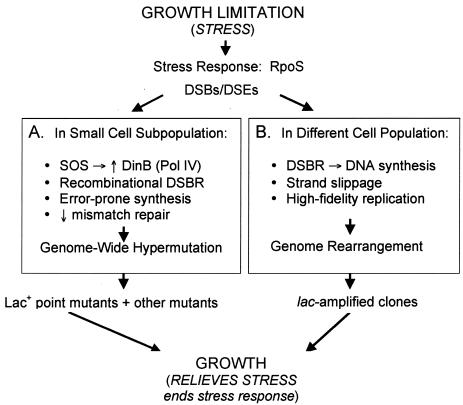
FIG. 2.
A stress-response (HM) model for adaptive point mutation and amplification (modified from references 30, 45, and 58). Starvation is proposed to promote RpoS-dependent stress responses that make the normally high-fidelity process of DNA double-strand-break repair (DSBR) error prone in two different ways. (A) Version I: in a small subpopulation of cells, genome-wide hypermutation is caused by high-level expression of the error-prone DNA polymerase DinB (Pol IV) (66, 71), which is upregulated both by the SOS response (37, 38) and also by RpoS (41), the stationary-phase and general stress-response transcription (σ) factor (35) required both for adaptive point mutation and amplification (45). Also in this population, mismatch repair becomes limiting transiently, perhaps via saturation by excess DNA polymerase errors (27) made by DinB (14, 49). The outcome is genome-wide hypermutation wherever DNA double-strand breaks (DSBs) or ends (DSEs) are formed and repaired. (Because more DSEs form in the F′ plasmid than in the chromosome, due to single-strand nicks made at the F transfer origin, there is more mutation on F′ [17] than on the chromosome [5], but the chromosome still experiences DSBR-protein- and DinB-dependent stationary-phase point mutations [5] because it still sometimes has DSEs to repair.) Version II: an alternative version (not drawn) of this idea, compatible with the hypothesis that two cell populations generate point mutations (57) (discussed in the text), is that both populations have a mutagenic stress response leading to increased DinB. (This is necessary because most point mutations [85%] are DinB dependent). However, one population is less mutable than the other only because it has less DinB, because, e.g., it experiences only RpoS, but not SOS, induction of DinB, whereas the other experiences both and makes more DinB (and then also saturates mismatch repair), making it more mutable. This alternative is more complicated than version I, but possible. (B) We suggest that amplification is provoked by error-prone DSBR of a different sort: that DNA synthesis primed during DSBR lacks the controls of replication from an origin and in starving cells is slower and more likely to stall due to limiting nucleotides. We suggest that upon stalling, template switching occurs (similar to recombination models discussed in reference 7; see also reference 72). This can produce a novel junction sequence in the DNA that leads to amplification either via promoting rolling circle replication or generating a duplication that amplifies to many copies by recombination (illustrated in reference 31). Because adaptive amplification is SOS and DinB independent (49), the role played by RpoS in adaptive amplification cannot be to up-regulate DinB (45). RpoS may play different roles in point mutation and amplification (45).
POINT MUTATION AND AMPLIFICATION ARE RpoS-DEPENDENT STRESS RESPONSES— A STRESS-RESPONSE (HM) MODEL
Finally, both adaptive point mutation and amplification are stress responses requiring the stationary-phase and general-stress-response transcription (σ) factor, RpoS (45). (This conclusion was also reached in reference 41; however, that paper did not show that the decreased yield of Lac+ colonies for rpoS mutants is not caused by any of several possible artifactual explanations discussed in references 4 and 45 and ruled out in reference 45.) This is incompatible with CG models, in which mutations occur because some cells are growing (not in stationary phase) and not undergoing a stress response (e.g., see references 2 and 34). This strongly supports HM models such as that shown in Fig. 2, in which both point mutation and genome rearrangement (amplification) are stress responses that are induced by starvation and turned off if cells happen to generate a mutation or amplification that allows them to grow in the growth-limiting environment. We have drawn amplification and point mutation as a branched pathway with RpoS acting early (Fig. 2) because of the known role of the RpoS regulon early during entry into stationary phase (35), but whether they are a branched pathway or two separate pathways is not yet determined. They are clearly two separate outcomes (Fig. 1) (30, 49), in contrast with the following model.
AMPLIFICATION MUTAGENESIS— A CRYPTIC-GROWTH MODEL
Amplification mutagenesis (AM) is a CG model that seeks to generate adaptive mutants without an increase in mutation rate (2, 34), as per the neo-Darwinist constraint. In AM, amplification of lac is a precursor to mutation, not a separate outcome. Mutations are proposed to occur in cells that are able to grow, forming small (micro)colonies, as a result of a preexisting duplication of the leaky lac allele (2, 34). During growth, more copies of lac accumulate (by amplification and growth of the colony), which increases the likelihood of a point mutation occurring in one of the copies. Subsequent replication, deamplification, and segregation would yield a cell that has acquired a Lac+ point mutation and lost the amplified DNA. No special features other than growth and replication are supposed to contribute to most Lac+ adaptive point mutations. To explain genome-wide hypermutation (6, 24, 57, 70), one version of AM proposes that amplified DNA per se induces the SOS response and DinB, which would also enhance Lac+ mutation (34). In another version, genome-wide hypermutation is caused by rare coamplification of dinB with lac (64). Both versions specify the important constraint that only hypermutation of unselected genes, and not most Lac+ adaptive point mutation, is caused by the increase in DinB error-prone polymerase. One way that this could be achieved would be if only 10% of point mutants originate in the hypermutable subpopulation whereas 90% originate from other cells with standard mutation rates.
THE AMPLIFICATION MUTAGENESIS MODEL DOES NOT FIT THE DATA
Most adaptive mutations require an error-prone DNA polymerase.
Eighty-five percent of point mutations require the DinB/Pol IV error-prone polymerase (49), whereas standard spontaneous growth-dependent Lac+ reversions do not (49, 51). This is incompatible with the idea that normal mutation rates (from normal generation-dependent replication errors) produce most adaptive mutations. If the AM model were to depart from this and invoke excess DinB for most point mutations, it would then be another HM model (similar to that shown in Fig. 2).
Selective growth with lac amplification is not sufficient to produce point mutants.
In the AM model, a key feature is that growth with lac amplification is sufficient to produce point mutants; no special conditions or stress responses are allowed (2, 34, 65). Several tests of this idea have failed to support it. First, when cells containing lac amplification were replated on lactose, only 1 of 680 resulting Lac+ colonies contained a point mutation, indicating that amplification does not promote point mutation (30) to the extent that the model demands. More than 23,000 cells from the 680 colonies were assayed (30), precluding sampling error as a likely reason for the failure to detect point mutants.
lac-amplified clones not “channeled” into point mutation by DNA polymerase errors.
If amplifications were the major intermediate converted by DNA polymerase errors into point mutations, as the AM model specifies (34), then decreasing polymerase errors (by blocking SOS induction or knocking out dinB) should not only decrease point mutations but should increase the amplification component of the curve (Fig. 1) proportionately. The point mutants lost should remain amplified and so contribute to the numbers of amplified colonies. This is not observed: both SOS and dinB defects decrease numbers of point mutants dramatically (∼85%) without increasing lac-amplified clones (49). These data support models such as that shown in Fig. 2, in which the channeling of DNA intermediates into either the amplification or point mutation pathway occurs at a step before generation of amplified DNA and in which lac amplification allows growth, deflecting cells from a mutagenic stress response.
Amplification is neither mutagenic nor associated with general hypermutation (30).
Lac adaptive mutation is a stress response, not a consequence of CG.
As discussed above, Lac+ point mutation and amplification require the stationary-phase and general stress response transcription factor, RpoS (45). This indicates that both are stress responses and also that they occur in stationary-phase (not actively growing) cells. The AM model specifies that Lac+ adaptive mutation is not a stress response and is a consequence of growth (2, 34, 65).
By contrast, all data reported so far are compatible with a branched-pathway, stress-responsive HM model (Fig. 2) in which either point mutation or amplification can lead to adaptive Lac+ colony formation, rapid growth, and cessation of the genome-altering stress responses. The process looks Darwinian but without the constraint of more conservative neo-Darwinists that mutation rates stay constant.
Acknowledgments
We gratefully acknowledge stimulating discussions with Ichizo Kobayashi, Hisaji Maki, Ivan Matic, Suzanne Rutherford, and John Cairns and with Rebecca Ponder and Andrew Slack, whose unpublished data contributed to the ideas in this paper. We thank Rob Dorit and Mary-Jane Lombardo for improving the manuscript.
This work was supported by Public Health Service grants R01-GM53158 (to S.M.R.) and R01-GM64022 (to P.J.H.).
REFERENCES
- 1.Akerlund, T., K. Nordstrom, and R. Bernander. 1995. Analysis of cell size and DNA content in exponentially growing and stationary-phase batch cultures of Escherichia coli. J. Bacteriol. 177:6791-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson, D. I., E. S. Slechta, and J. R. Roth. 1998. Evidence that gene amplification underlies adaptive mutability of the bacterial lac operon. Science 282:1133-1135. [DOI] [PubMed] [Google Scholar]
- 3.Bjedov, I., O. Tenaillon, B. Gérard, V. Souza, E. Denamur, M. Radman, F. Taddei, and I. Matic. 2003. Stress-induced mutagenesis in bacteria. Science 300:1404-1409. [DOI] [PubMed] [Google Scholar]
- 4.Bridges, B. A., P. L. Foster, and A. R. Timms. 2001. Effect of endogenous carotenoids on “adaptive” mutation in Escherichia coli FC40. Mutat. Res. 473:109-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bull, H. J., M.-J. Lombardo, and S. M. Rosenberg. 2001. Stationary-phase mutation in the bacterial chromosome: recombination protein and DNA polymerase IV dependence. Proc. Natl. Acad. Sci. USA 98:8334-8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bull, H. J., G. J. McKenzie, P. J. Hastings, and S. M. Rosenberg. 2000. Evidence that stationary-phase hypermutation in the E. coli chromosome is promoted by recombination. Genetics 154:1427-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bzymek, M., and S. T. Lovett. 2001. Instability of repetitive DNA sequences: the role of replication in multiple mechanisms. Proc. Natl. Acad. Sci. USA 98:8319-8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cairns, J., and P. L. Foster. 1991. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics 128:695-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cairns, J., J. Overbaugh, and S. Miller. 1988. The origin of mutants. Nature 335:142-145. [DOI] [PubMed] [Google Scholar]
- 10.Chicurel, M. 2001. Can organisms speed their own evolution? Science 292:1824-1827. [DOI] [PubMed] [Google Scholar]
- 11.Cox, M. M., M. F. Goodman, K. N. Kreuzer, D. J. Sherratt, S. J. Sandler, and K. J. Marians. 2000. The importance of repairing stalled replication forks. Nature 404:37-41. [DOI] [PubMed] [Google Scholar]
- 12.Darwin, C. 1859. The origin of species, 6th ed. Penguin Books, New York, N.Y.
- 13.Echols, H. 1981. SOS functions, cancer and inducible evolution. Cell 25:1-2. [DOI] [PubMed] [Google Scholar]
- 14.Foster, P. L. 2000. Adaptive mutation in Escherichia coli. Cold Spring Harbor Symp. Quant. Biol. 65:21-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster, P. L. 1999. Are adaptive mutations due to a decline in mismatch repair? The evidence is lacking. Mutat. Res. 436:179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foster, P. L. 1999. Mechanisms of stationary phase mutation: a decade of adaptive mutation. Annu. Rev. Genet. 33:57-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foster, P. L. 1997. Nonadaptive mutations occur in the F′ episome during adaptive mutation conditions in Escherichia coli. J. Bacteriol. 179:1550-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foster, P. L. 1994. Population dynamics of a Lac− strain of Escherichia coli during selection for lactose utilization. Genetics 138:253-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foster, P. L., and J. M. Trimarchi. 1994. Adaptive reversion of a frameshift mutation in Escherichia coli by simple base deletions in homopolymeric runs. Science 265:407-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foster, P. L., and J. M. Trimarchi. 1995. Adaptive reversion of an episomal frameshift mutation in Escherichia coli requires conjugal functions but not actual conjugation. Proc. Natl. Acad. Sci. USA 92:5487-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foster, P. L., and J. M. Trimarchi. 1995. Conjugation is not required for adaptive reversion of an episomal frameshift mutation in Escherichia coli. J. Bacteriol. 177:6670-6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foster, P. L., J. M. Trimarchi, and R. A. Maurer. 1996. Two enzymes, both of which process recombination intermediates, have opposite effects on adaptive mutation in Escherichia coli. Genetics 142:25-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galitski, T., and J. R. Roth. 1995. Evidence that F′ transfer replication underlies apparent adaptive mutation. Science 268:421-423. [DOI] [PubMed] [Google Scholar]
- 24.Godoy, V. G., F. S. Gizatullin, and M. S. Fox. 2000. Some features of the mutability of bacteria during nonlethal selection. Genetics 154:49-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall, B. G. 1990. Spontaneous point mutations that occur more often when advantageous than when neutral. Genetics 126:5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris, R. S., G. Feng, K. J. Ross, R. Sidhu, C. Thulin, S. Longerich, S. K. Szigety, P. J. Hastings, M. E. Winkler, and S. M. Rosenberg. 1999. Mismatch repair is diminished during stationary-phase mutation. Mutat. Res. 437:51-60. [PubMed] [Google Scholar]
- 27.Harris, R. S., G. Feng, K. J. Ross, R. Sidhu, C. Thulin, S. Longerich, S. K. Szigety, M. E. Winkler, and S. M. Rosenberg. 1997. Mismatch repair protein MutL becomes limiting during stationary-phase mutation. Genes Dev. 11:2426-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris, R. S., S. Longerich, and S. M. Rosenberg. 1994. Recombination in adaptive mutation. Science 264:258-260. [DOI] [PubMed] [Google Scholar]
- 29.Harris, R. S., K. J. Ross, and S. M. Rosenberg. 1996. Opposing roles of the Holliday junction processing systems of Escherichia coli in recombination-dependent adaptive mutation. Genetics 142:681-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hastings, P. J., H. J. Bull, J. R. Klump, and S. M. Rosenberg. 2000. Adaptive amplification: an inducible chromosomal instability mechanism. Cell 103:723-731. [DOI] [PubMed] [Google Scholar]
- 31.Hastings, P. J., and S. M. Rosenberg. 2002. In pursuit of a molecular mechanism for adaptive gene amplification. DNA Repair 1:111-123. [DOI] [PubMed] [Google Scholar]
- 32.Heidenreich, E., R. Novotny, B. Kneidinger, V. Holzmann, and U. Wintersberger. 2003. Non-homologous end joining as an important mutagenic process in cell cycle-arrested cells. EMBO J. 22:2274-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heidenreich, E., and U. Wintersberger. 2001. Adaptive reversion of a frameshift mutation in arrested Saccharomyces cerevisiae cells by simple deletions in mononucleotide repeats. Mutat. Res. 473:101-107. [DOI] [PubMed] [Google Scholar]
- 34.Hendrickson, H., E. S. Slechta, U. Bergthorsson, D. I. Andersson, and J. R. Roth. 2002. Amplification-mutagenesis: evidence that “directed” adaptive mutation and general hypermutability result from growth with a selected gene amplification. Proc. Natl. Acad. Sci. USA 99:2164-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horiuchi, T., S. Horiuchi, and A. Novick. 1963. The genetic basis of hypersynthesis of β-galactosidase. Genetics 48:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kenyon, C. J., and G. C. Walker. 1980. DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc. Natl. Acad. Sci. USA 77:2819-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim, S.-R., G. Maenhaut-Michel, M. Yamada, Y. Yamamoto, K. Matsui, T. Sofuni, T. Nohmi, and H. Ohmori. 1997. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an SOS gene product (DinB/P) enhances frameshift mutations in the absence of any exogenous agents that damage DNA. Proc. Natl. Acad. Sci. USA 94:13792-13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kofoid, E., U. Bergthorsson, E. S. Slechta, and J. R. Roth. 2003. Formation of an F′ plasmid by recombination between imperfectly repeated chromosomal Rep sequences: a closer look at an old friend (F′(128) pro lac). J. Bacteriol. 185:660-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuzminov, A. 1995. Collapse and repair of replication forks in Escherichia coli. Mol. Microbiol. 16:373-384. [DOI] [PubMed] [Google Scholar]
- 41.Layton, J. C., and P. L. Foster. 2003. Error-prone DNA polymerase IV is controlled by the stress-response sigma factor, RpoS, in Escherichia coli. Mol. Microbiol. 50:549-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lederberg, J., and E. M. Lederberg. 1952. Replica plating and indirect selection of bacterial mutants. J. Bacteriol. 63:399-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lenski, R. E., and J. E. Mittler. 1993. The directed mutation controversy and neo-Darwinism. Science 259:188-259. [DOI] [PubMed] [Google Scholar]
- 44.Lenski, R. E., M. Slatkin, and F. J. Ayala. 1989. Another alternative to directed mutation. Nature 337:123-124. [DOI] [PubMed] [Google Scholar]
- 45.Lombardo, M.-J., I. Aponyi, and S. M. Rosenberg. 2004. General stress response regulator RpoS in adaptive mutation and amplification in Escherichia coli. Genetics 166:669-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luria, S. E., and M. Delbrück. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mayr, E. 1982. The growth of biological thought, diversity, evolution, and inheritance. Belknap, Cambridge, Mass..
- 48.McKenzie, G. J., R. S. Harris, P. L. Lee, and S. M. Rosenberg. 2000. The SOS response regulates adaptive mutation. Proc. Natl. Acad. Sci. USA 97:6646-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKenzie, G. J., P. L. Lee, M.-J. Lombardo, P. J. Hastings, and S. M. Rosenberg. 2001. SOS mutator DNA polymerase IV functions in adaptive mutation and not adaptive amplification. Mol. Cell 7:571-579, 119. [DOI] [PubMed] [Google Scholar]
- 50.McKenzie, G. J., M.-J. Lombardo, and S. M. Rosenberg. 1998. Recombination-dependent mutation in Escherichia coli occurs in stationary phase. Genetics 149:1163-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McKenzie, G. J., D. B. Magner, P. L. Lee, and S. M. Rosenberg. 2003. The dinB operon and spontaneous mutation in Escherichia coli. J. Bacteriol. 185:3972-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Newcomb, H. B. 1949. Origin of bacterial variants. Nature 164:150. [DOI] [PubMed] [Google Scholar]
- 53.Ninio, J. 1991. Transient mutators: a semiquantitative analysis of the influence of translation and transcription errors on mutation rates. Genetics 129:957-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Partridge, L., and M. J. Morgan. 1988. Is bacterial evolution random or selective? Nature 336:22. [DOI] [PubMed] [Google Scholar]
- 55.Radman, M. 1975. SOS repair hypothesis: phenomenology of an inducible DNA repair which is accompanied by mutagenesis. Basic Life Sci. 5A:355-367. [DOI] [PubMed] [Google Scholar]
- 56.Rodriguez, C., J. Tompkin, J. Hazel, and P. L. Foster. 2002. Induction of a DNA nickase in the presence of its target site stimulates adaptive mutation in Escherichia coli. J. Bacteriol. 184:5599-5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rosche, W. A., and P. L. Foster. 1999. The role of transient hypermutators in adaptive mutation in Escherichia coli. Proc. Natl. Acad. Sci. USA 96:6862-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosenberg, S. M. 2001. Evolving responsively: adaptive mutation. Nat. Rev. Genet. 2:504-515. [DOI] [PubMed] [Google Scholar]
- 59.Rosenberg, S. M., R. S. Harris, and J. Torkelson. 1995. Molecular handles on adaptive mutation. Mol. Microbiol. 18:185-189. [DOI] [PubMed] [Google Scholar]
- 60.Rosenberg, S. M., and P. J. Hastings. 2003. Modulating mutation rates in the wild. Science 300:1382-1383. [DOI] [PubMed] [Google Scholar]
- 61.Rosenberg, S. M., S. Longerich, P. Gee, and R. S. Harris. 1994. Adaptive mutation by deletions in small mononucleotide repeats. Science 265:405-407. [DOI] [PubMed] [Google Scholar]
- 62.Rosenberg, S. M., C. Thulin, and R. S. Harris. 1998. Transient and heritable mutators in adaptive evolution in the lab and in nature. Genetics 148:1559-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roth, J. R., E. Kofoid, F. P. Roth, O. G. Berg, J. Seger, and D. I. Andersson. 2003.. Adaptive mutation requires no mutagenesis—only growth under selection—a response. Genetics 165:2319-2321.
- 64.Slechta, E. S., K. L. Bunny, E. Kugelberg, E. Kofoid, D. I. Andersson, and J. R. Roth. 2003. Adaptive mutation: general mutagenesis is not a programmed response to stress but results from rare coamplification of dinB with lac. Proc. Natl. Acad. Sci. USA 100:12847-12852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Slechta, E. S., J. Liu, D. I. Andersson, and J. R. Roth. 2002. Evidence that selected amplification of a bacterial lac frameshift allele stimulates Lac(+) reversion (adaptive mutation) with or without general hypermutability. Genetics 161:945-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sutton, M. D., B. T. Smith, V. G. Godoy, and G. C. Walker. 2000. The SOS response: recent insights into umuDC-dependent mutagenesis and DNA damage tolerance. Annu. Rev. Genet. 34:479-497. [DOI] [PubMed] [Google Scholar]
- 67.Taddei, F., J. A. Halliday, I. Matic, and M. Radman. 1997. Genetic analysis of mutagenesis in aging Escherichia coli colonies. Mol. Gen. Genet. 256:277-281. [DOI] [PubMed] [Google Scholar]
- 68.Tlsty, T. D., A. M. Albertini, and J. H. Miller. 1984. Gene amplification in the lac region of E. coli. Cell 37:217-224. [DOI] [PubMed] [Google Scholar]
- 69.Tompkins, J. D., J. L. Nelson, J. C. Hazel, S. L. Leugers, J. D. Stumpf, and P. L. Foster. 2003. Error-prone polymerase, DNA polymerase IV, is responsible for transient hypermutation during adaptive mutation in Escherichia coli. J. Bacteriol. 185:3469-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Torkelson, J., R. S. Harris, M.-J. Lombardo, J. Nagendran, C. Thulin, and S. M. Rosenberg. 1997. Genome-wide hypermutation in a subpopulation of stationary-phase cells underlies recombination-dependent adaptive mutation. EMBO J. 16:3303-3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wagner, J., P. Gruz, S. R. Kim, M. Yamada, K. Matsui, R. P. P. Fuchs, and T. Nohmi. 1999. The dinB gene encodes a novel E. coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol. Cell 4:281-286. [DOI] [PubMed] [Google Scholar]
- 72.Yoshiyama, K., K. Higuchi, H. Matsumura, and H. Maki. 2001. Directionality of DNA replication fork movement strongly affects the generation of spontaneous mutations in Escherichia coli. J. Mol. Biol. 307:1195-1206. [DOI] [PubMed] [Google Scholar]