Abstract
In humans, there is a strong correlation between sensitivity to substances of abuse and addiction risk. This differential tolerance to drugs has a strong genetic component. The identification of human genetic factors that alter drug tolerance has been a difficult task. For this reason and taking advantage of the fact that Drosophila responds similarly to humans to many drugs, and that genetically it has a high degree of homology (sharing at least 70% of genes known to be involved in human genetic diseases), we looked for genes in Drosophila that altered their nicotine sensitivity. We developed an instantaneous nicotine vaporization technique that exposed flies in a reproducible way. The amount of nicotine sufficient to “knock out” half of control flies for 30 minutes was determined and this parameter was defined as Half Recovery Time (HRT). Two fly lines, L4 and L70, whose HRT was significantly longer than control´s were identified. The L4 insertion is a loss of function allele of the transcriptional factor escargot (esg), whereas L70 insertion causes miss-expression of the microRNA cluster miR-310-311-312-313 (miR-310c). In this work, we demonstrate that esg loss of function induces nicotine sensitivity possibly by altering development of sensory organs and neurons in the medial section of the thoracoabdominal ganglion. The ectopic expression of the miR-310c also induces nicotine sensitivity by lowering Esg levels thus disrupting sensory organs and possibly to the modulation of other miR-310c targets.
Introduction
Nicotine addiction is a serious public health problem. Several studies in humans have provided evidence that there is a strong genetic component underlying nicotine and other substances addiction.
Assessing the genetic contribution to addiction in humans is, to say the least, complicated. It is very difficult to separate the environmental contribution from the genetic effects; therefore, the use of animal models to study the genetic basis of addiction is invaluable. Drosophila melanogaster and humans respond to pharmacological treatments in very similar ways. Flies become hyperactive after the exposure to cocaine, fall unconscious after ingesting a sufficient dose of benzodiazepines and have a very similar intoxication with alcohol as humans do, thus they have consistently been used to study the genetic basis of drug differential sensitivity and addiction [1,2]. Considering that, in humans, hypersensitivity to drugs correlates with an increased addiction risk [3,4], we established a protocol for the reproducible exposure of flies to nicotine and characterized the normal response of wild type (wt) flies to this exposure. Using this approach, we identified two P-element insertion lines (L4 and L70) that confer hypersensitivity to nicotine. L4 insertion causes a loss-of-function mutation of the escargot (esg) gene, whereas L70 insertion leads to the miss-expression of the miR-310 c. The miR-310 c codes for four microRNAs: miR-310, miR-311, miR-312 and miR-313 that share an identical seed sequence. Furthermore, we show that ectopic expression of the miR-310 c enhances esg phenotypes in the same way as the knock down of Esg by an RNAi, suggesting that these miRNAs downregulate esg expression. We also demonstrate that other esg loss-of-function alleles have a similar nicotine hypersensitive phenotype. It is important to note that this is the first time a behavioral phenotype is associated with the miR-310 c.
Results
L4 and L70 P{GawB} insertions confer an abnormal sensitivity to nicotine
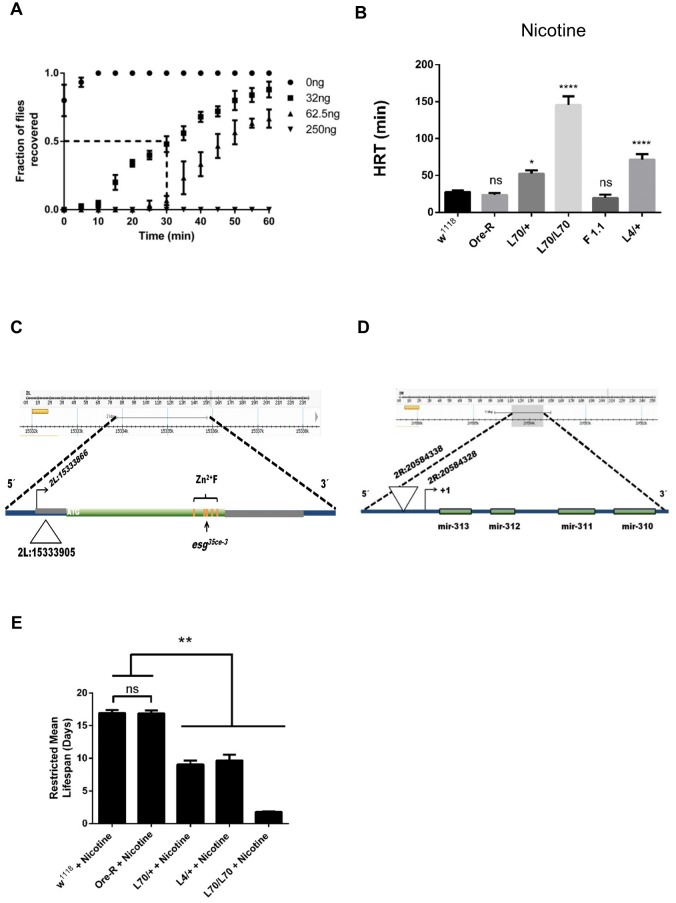
Based on McClung and Hirsh [5], we isolated mutants that have a differential response to nicotine compared to control lines, white 1118 (w 1118) and Oregon-R (Ore-R) flies. When flies are exposed to nicotine, they lose their negative geotaxis reflex for a period of time that is proportional to the amount of vaporized nicotine. In order to determine the standard amount of nicotine to be used, we treated w 1118 flies with different quantities of nicotine diluted in water to identify the sufficient amount of volatilized nicotine that allows half of the exposed flies to recover in 30 min. This parameter is defined as the Half Recovery Time (HRT). In our conditions w 1118 has a HRT of 30 min when exposed to 32 ng of nicotine. All further experiments were performed using this amount of nicotine. Line w 1118 was used as control and reference line, because it is the genetic background of the P{GawB} insertion collection and of all the other lines used in this work, Oregon-R was also used as an independent wt line and behaved identically to w 1118 thus strongly suggesting that this is the wt sensitivity to nicotine (Fig 1A). A collection of 200 lines with random P{GawB} insertions in their genome was screened, and from this process we isolated two lines, L4 and L70, which are hypersensitive to nicotine exposure. None of the other insertion lines tested showed any statistically significant differential sensitivity to nicotine when compared to the control lines. Heterozygous individuals of both lines, L4/+ and L70/+, have an augmented HRT of 71 and 52 min, respectively, compared to w 1118 (30 min). Homozygous L70 is even more sensitive having a HRT of 145 min, suggesting a robust dosage effect (Fig 1B), while homozygous L4 is lethal. The mock exposure of L4 and L70 to volatilized water (the nicotine vehicle) had no effect on their climbing capacity when compared with controls, demonstrating that the longer HRT is due to higher nicotine sensitivity and not to motility impairment (S1 Fig).
Fig 1. Genetic background determines Drosophila response to nicotine.
(A) Dose response of w 1118 flies to different concentrations of nicotine. The time needed for half of the flies to recover from a given concentration of nicotine exposure is defined as Half Recovery Time (HRT), note that mock exposure of flies (0 ng, closed circles) allows flies to recover nearly immediately. (B) HRTs are shown for different fly genotypes: w 1118 and Ore-R are controls, L70 has an insertion located at the 5’ of the miR-310 c, F1.1 is a revertant line, where the P-element insertion of L70 line has been precisely excised. L4 line has an insertion in esg. L70 and L4 showed nicotine hypersensitivity. (C) Schematic representation of the L4 P{GawB} insertion. The triangle represents the insertion site according to fly-base standard genome coordinates (release version 6). Zinc fingers are denoted by orange bands, the site of the esg 35ce-3 mutation is shown by an arrow. (D) Schematic representation of the L70 P{GawB} insertion. The inverted triangle represents the insertion site according to fly-base standard genome coordinates (release version 6). The arrow shows the miR-310 c cistron transcription initiation site. (E) L70 and L4 insertion lines are sensitive to chronic nicotine exposure. Restricted mean lifespan of mutant genotypes are significantly reduced compared to w 1118 when maintained in standard cornmeal food, supplemented with 0.5 mg/ml nicotine. All experiments were repeated at least 3 times with an n ≥ 100 flies. * = P≤ 0.01, ** = P≤ 0.001, **** = P<< 0.001, ns = not significant.
The insertion sites of P{GawB} in these mutants were identified using inverse PCR (L70) and plasmid rescue (L4) and confirmed by PCR amplification and sequencing of the insertion site. L4 insertion site is 39 nucleotides downstream of the transcription initiation site of the escargot gene (esg) (Fig 1C). L70 is homozygous viable and fertile and its insertion site is 10 nucleotides upstream of the transcription initiation site of the miR-310 c (Fig 1D). To demonstrate that the P{GawB} insertion causes hypersensitivity in L70, five independent precise excision lines were isolated by mobilizing the P{GawB} element. All these reverted lines showed normal sensitivity to nicotine (Fig 1B and S2 Fig).
In order to measure if the exposure route would have an effect on the sensitivity, and if the phenotype is different depending on an acute or chronic exposure, we exposed the flies to food containing 0.5 mg/ml of nicotine and measured their mean restricted lifespan. As shown in Fig 1E, L70/+ and L4/+ are significantly more sensitive to nicotine-containing food than control flies (w 1118 16.98 ± 0.41 days, Ore-R 16.87 ± 0.45 days, L70/+ 9.07 ± 0.61 days, L4/+ 9.68 ± 0.86 days, L70/L70 1.77 ± 0.09 days). Therefore, these lines are hypersensitive to nicotine regardless of the way they are exposed to it (volatilized or ingested).
L4 is an escargot loss-of-function mutation and causes nicotine hypersensitivity
To evaluate how the P-element insertion was affecting Esg levels in the L4/+ line, we performed western blot analysis using embryos because esg expression levels are reported to be highest at this developmental stage [6]. We found that in embryos, L4 has approximately half of Esg protein compared to the control line w 1118 (Fig 2A and 2B). We attempted to perform western blots of adult extracts but Esg could not be detected in these conditions. This result is consistent with reports showing a very low esg expression at this stage [7,8]. Additionally, we found that in approximately 8% of the population, L4 flies have the characteristic cuticle defects reported in other esg loss of function allelles, such as loss of bristles in the tergites and sternites, and loss of pigmentation in the dorsal abdomen [7]. These data strongly suggest that L4 leads to a loss of function of esg.
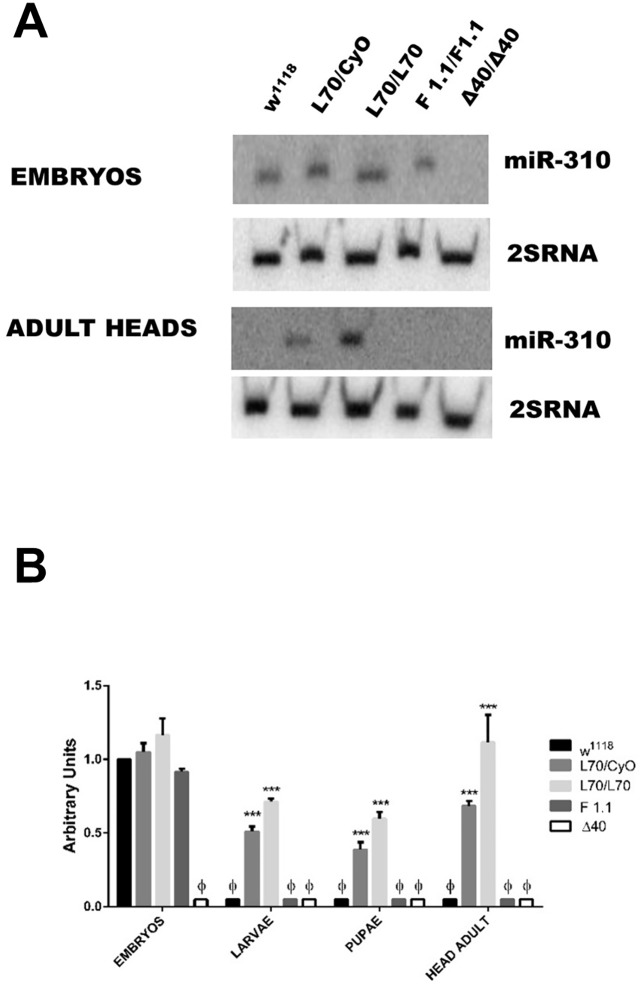
Fig 2. L4 P-element insertion causes Esg loss of function.
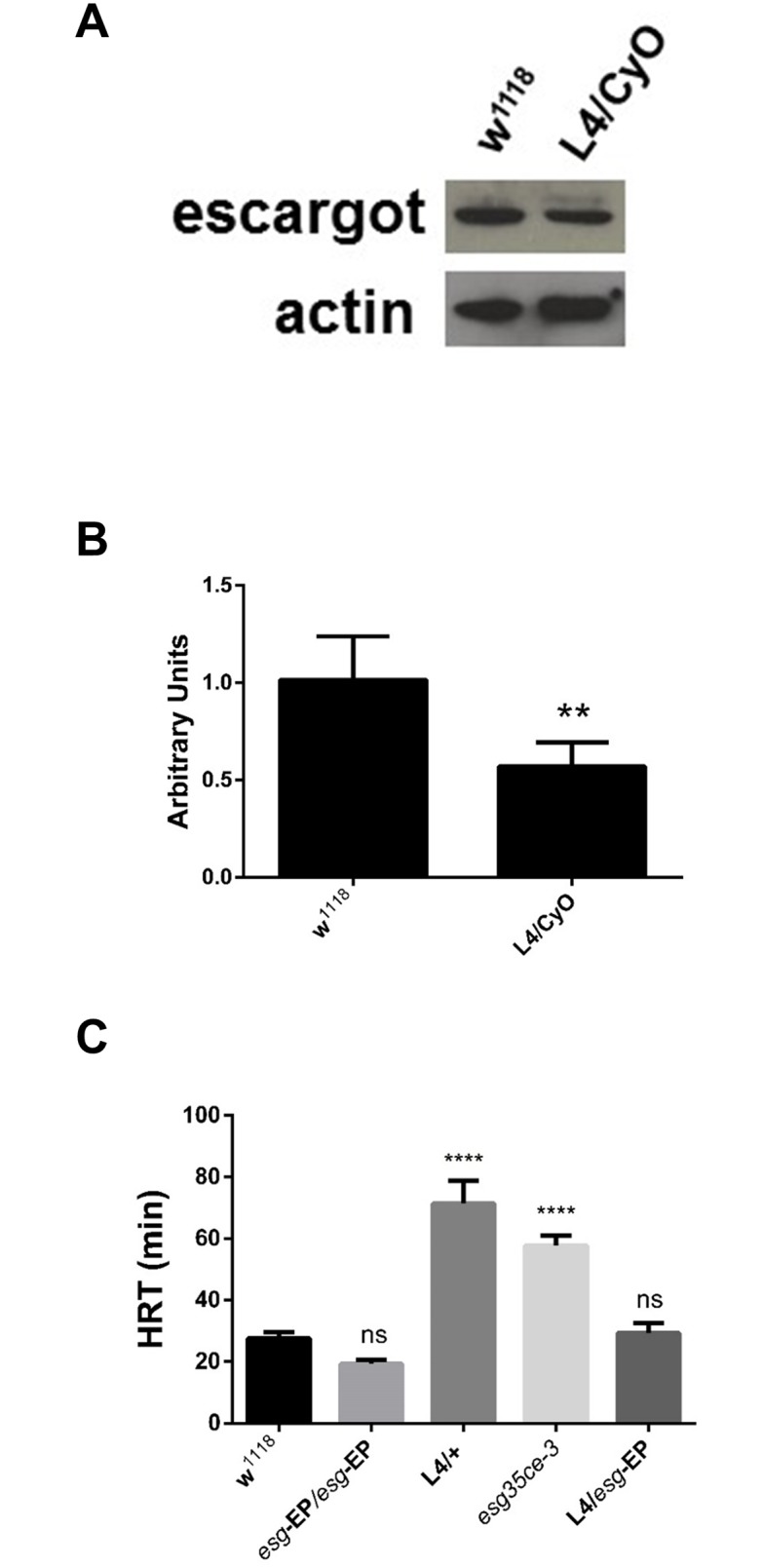
(A) Western blot of L4 embryos showing a reduced expression of the Esg protein compared to w 1118. (B) Quantitative densitometry of western blots. n = 3 (C) Half recuperation time (HRT) of the different esg alleles used. EP = P{EP}esg EU143, esg 35ce-3 (loss of function allele) ** = P≤ 0.001, **** = P<< 0.001.
In order to verify if the loss of function of esg is causing hypersensitivity to nicotine, we tested another esg mutant allele (esg 35Ce-3), that produces an inactive transcription factor due to a point mutation in the third Esg zinc-finger domain [8]. This allele also has a homozygous lethal phenotype and the sensitivity of heterozygous individuals to nicotine is almost identical to L4 heterozygous individuals (Fig 2C). In addition, the transheterozygous genotype L4/esg 35Ce-3 is lethal, therefore confirming that L4 is an esg loss of function allele. L4 is an enhancer trap insertion as it carries GAL4 in the P{GawB} element, driving the expression of any gene under the control of UAS sequences in the endogenous esg expression pattern. The EP insertion P{EP}esg EU143 can be used to express esg under the control of any GAL4 driver. Recovering the esg expression levels in the L4/ P{EP}esg EU143 (L4> P{EP}esg EU143) genotype led to normal sensitivity to nicotine (Fig 2C).
These results clearly indicate that a reduction of one half of the esg gene dosage is sufficient to reproduce the nicotine hyper-sensitive phenotype.
The L70 insertion causes the mirR-310-311-312-313 cluster (miR-310 c) to be miss-expressed
To evaluate how the P-element insertion was affecting the expression levels of the miR-310 c in L70 line, we performed northern blot analyses using embryos, larvae, pupas and adult flies. We found no difference between L70 and w 1118 embryos. However, the miR-310 c was expressed both in L70 adults and pupae whereas no expression, or extremely low expression, was detected in in the control line, w 1118, at these stages (Fig 3A and 3B and S3 Fig). It has been reported that the miR-310 c is strongly expressed during early embryo development (0–12 hrs), with its expression decreasing in 12 to 24 hrs embryos and being extremely low in larvae, pupae and adults [9]. These data indicate that the L70 insertion caused somehow a deregulation of the miR-310 c expression, provoking its ectopic expression at late developmental stages. As the insertion site of P{GawB} element was close to the transcription initiation site (10 bp), we presumed that the transcription initiation of this gene could be affected; however, by 5'RACE, we corroborated that the miR-310 c transcription initiation site in L70 adults is identical to the transcription initiation site of wt embryos (S4 Fig).
Fig 3. L70 P-element insertion causes ectopic expression of the miR-310 c in adult flies.
(A) Northern blots of fly embryos and adult heads show that there is no adult expression of the miR-310 transcript in adult tissues in w 1118 and revertant line F1.1 while there is expression of the miR-310 transcript in L70. Δ40 strain has a deletion of the complete miR-310 c and is used as a negative control. Similar results were found with the other miRs transcripts (S2 Fig). (B) Densitometric analysis of (A). Ф = not detectable, *** = P≤ 0.0001.
Nicotine sensitivity of L4 and L70 is not due to a general lack of fitness
To evaluate the possibility that L4 and L70 nicotine hypersensitivity is due a general lack of fitness, we performed tests with different environmental factors that may uncover some other type of fitness defects. Heat resistance and ether sensibility can be used as proxy of general fitness. Heat resistance can be evaluated using the floating Petri dish assay (see Materials and methods). Our data showed that L4 and L70 contend normally with heat (Fig 4A). The ether sensibility of L4 and L70 was also normal (Fig 4B). These data together with the fact that L4 and L70 flies have the same climbing abilities than the controls (S1 Fig) suggest that these mutant lines do not have a general lack of fitness.
Fig 4. L70 and L4 mutant lines are not hypersensitive to heat or ether.
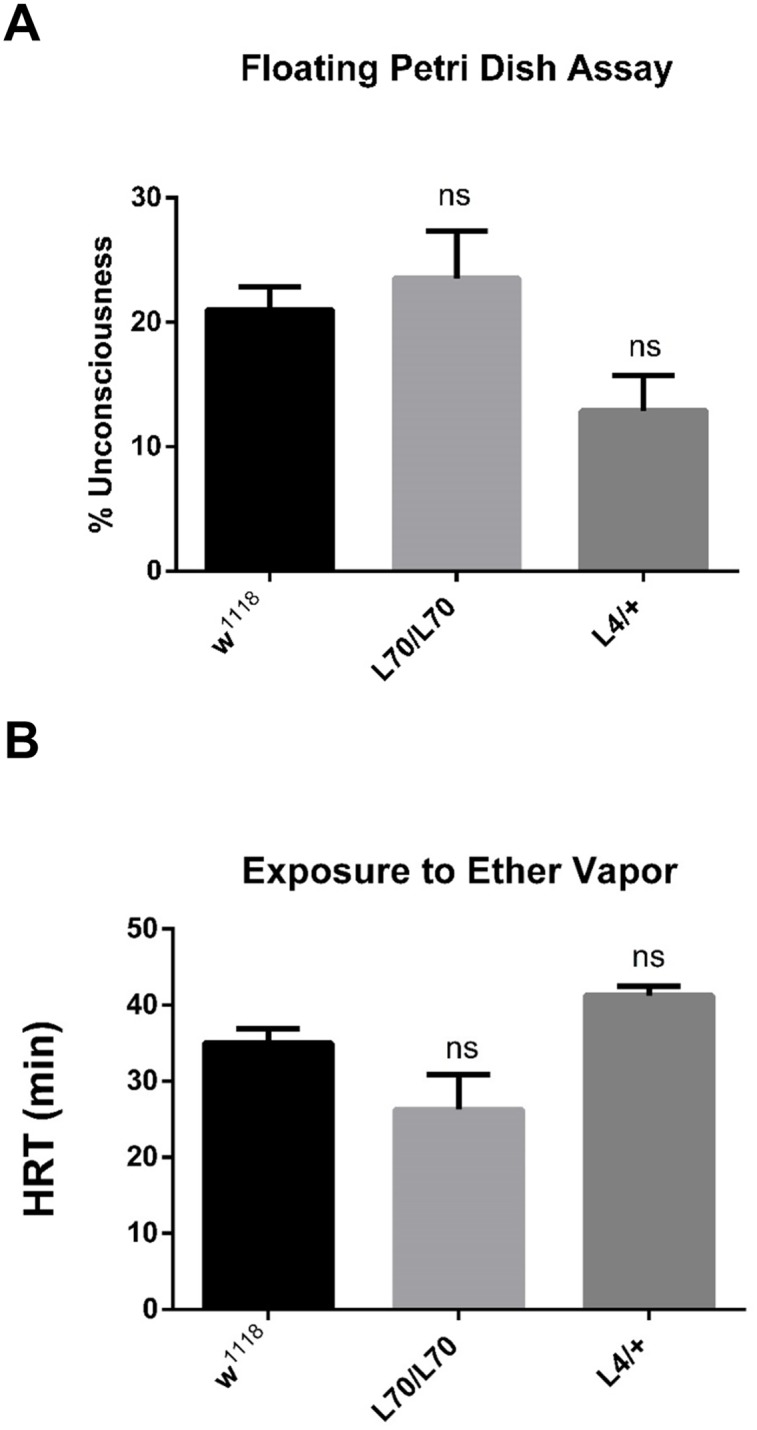
(A) Percentage of unconscious flies exposed to 45°C in the floating plate assay: L70 and L4 are as resistant to heat as w 1118. (B) HRT of L4 and L70 flies exposed to ether. No significant (ns) difference was detected in either condition.
Expressing small interference RNAs against esg or expressing the miR-310 c under the control of L4 enhances the mutant esg cuticle phenotype and nicotine sensitivity
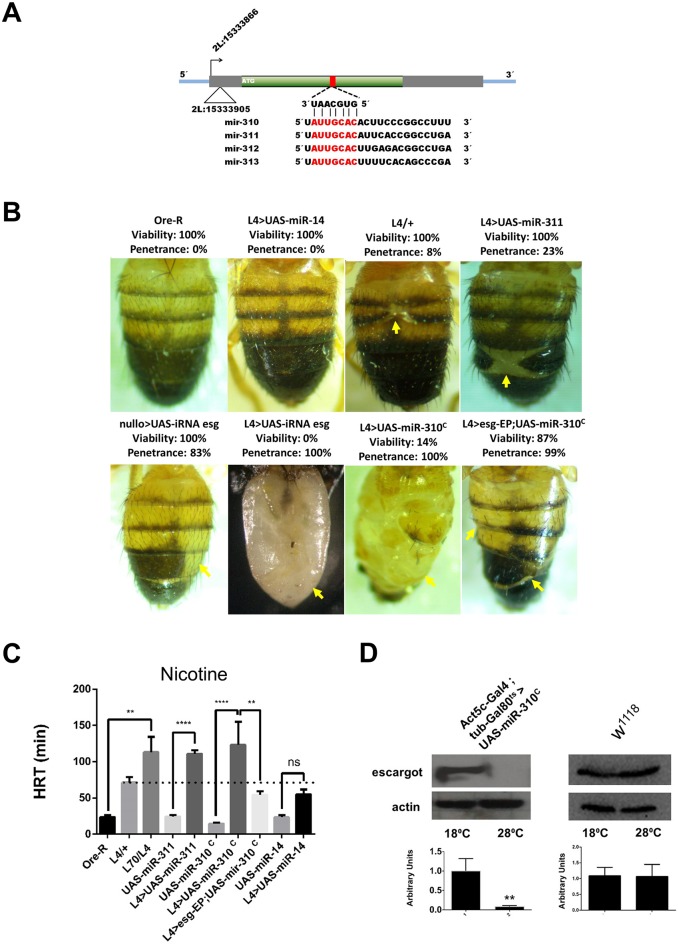
Analysis of the esg transcript using TargetScanFly 6.2 [10] showed that it contains one complementary target site for the seven nucleotide “seed” sequence shared by the four microRNAs in the miR-310 c. The target sequence is located in the 3’ end of esg’s open reading frame, from nucleotides 1113 to 1119 (Fig 5A). Interestingly, in the Drosophila esg homolog family members: snail, worniu and scratch this seed sequence is absent. We tested snail 18, a loss of function mutant, and it did not present any abnormal nicotine sensitivity thus suggesting that esg has a direct involvement in nicotine sensitivity and that the expression of the miR-310 c down-regulates esg. Although animal canonical regulation by microRNAs involves targeting of the transcript 3´UTR, there are well-documented cases showing that microRNA regulation is also achieved by targeting the coding region, especially in Drosophila[11,12]. Noteworthy, it has been shown that the miR-310 c regulates Khc-73 via a target sequence located in its ORF [13].
Fig 5. Genetic interaction of the miR-310 c with esg.
(A) Schematic representation of esg gene. The red rectangle represents the target sequences in esg’s ORF of miR-310, miR-311, miR-312 and miR-313. An alignment of the four mRNAs encoded in the miR-310 c is also shown, red characters show the seed sequence. (B) Phenotype penetrance of the L4 flies expressing small RNAs: Ore-R = wt abdomen, L4> UAS-miR-14 = miR-14 driven by L4 (note that there is no apparent phenotype) L4/+ = basal L4, L4>UAS-miR-311 = miR311 driven by L4, nullo>UAS-iRNA esg = short interference RNA against esg driven by nullo, L4>UAS-iRNA esg = short interference RNA against esg driven by L4, L4>UAS- miR-310 c = miR-310 c driven by L4,L4>esg-EP;UAS-miR-310 c = miR-310 c and esg from the EP driven by L4. Phenotype of the small interference RNA against esg is similar to the miR-311 and miR-310 c phenotype (cuticle phenotype is indicated with yellow arrows). Note that the presence of the esg-EP suppresses the effect of the miR-310 c expression (L4>esg-EP;UAS-miR-310 c). (C) HRTs become significantly longer when L70 and L40 genetically interact (L70/L4). This interaction can be phenocopied by the L4 induced expression of the miR-310 c (L4> UAS- miR-310 c) or miR-311 alone, suggesting that the miR-310 c down-regulates esg expression. Dashed line indicates the basal sensitivity of L4/+. n≥100, * = P ≤ 0.01, ** = P ≤ 0.001, **** = P<< 0.001, ns = not significant. (D) Western blots show that Esg protein levels can be conditionally ablated when the miR-310 c is expressed. The miR-310 c can only be expressed at 28°C in the Act5c-GAL4;Tub-Gal80ts>UAS-miR-310 c genotype, in these conditions esg becomes undetectable, while in the control (w 1118) remains unaffected, lower panels correspond to the densitometric analysis of the western blots, ** = P≤ 0.001.
MicroRNA target regulation is usually demonstrated using artificial constructs that have the microRNA target sequence fused to luciferase and measuring in vitro the reduction in luciferase activity. We are fortunate to have an in vivo and biologically relevant phenotypic reporter that is the classic loss of function esg phenotype. As mentioned above, loss of function of esg in the ectoderm during development causes characteristic cuticle defects whose extent is proportional to the amount of esg loss of function [7]. In order to demonstrate that cuticle defects can be used as a reporter of esg loss of function, we used RNA interference to down-regulate esg using the nullo promoter that is a low level constitutive driver which is expressed in all somatic tissues [14]. In this genotype (nullo>UAS-iRNA-esg) the cuticle phenotype is induced in 89% of the flies with 100% viability. Furthermore, when L4 is used to drive the UAS-iRNA-esg construct (L4>UAS-iRNA-esg), the cuticle phenotype penetrance is enhanced from 8%, in L4 by itself, to 100%. In this genotype, the esg phenotype is so severe that flies are unable to eclose and have to be dissected from the pupal case to evaluate cuticle damage (Fig 5B).
When the miR-310 c is expressed from the UAS-DsRed53-mir-310-313 line [13] using L4 as a driver we found that the cuticle phenotype penetrance increased to 100%, although it was less severe than the one induced by the UAS-iRNA-esg, as they have 14% of viability. The eclosed flies behaved normally but were as sensitive to nicotine as the L4/L70 flies (Fig 5C). These data suggest that miR-310 c is lowering Esg levels in a similar manner as the esg iRNA. This is also supported by the fact that individually expressed miR-311 has an enhanced cuticle phenotype with a penetrance of 23% with 100% viability (Fig 5B). The expression of the non-related microRNA miR-14 driven by L4 had no effect on nicotine HRT or cuticle phenotype (Fig 5B).
To confirm that the cuticle phenotype induced by the expression of the miR-310 c is specifically caused by a reduction of esg function, this phenotype was reverted by augmenting the esg gene dosage using the following genotype: L4>esg-EP;UAS-miR-310 c. In these flies, cuticle defects were partially rescued and viability increased from 14% to 87% (Fig 5B). Nicotine sensitivity was rescued as the HRT of this line decreased from 123 minutes, shown by the L4>UAS-miR-310 c, to 54 minutes in L4>esg-EP;UAS-miR-310 c (Fig 5C).
To further demonstrate that the miR-310 c overexpression reduces the amount of Esg, we performed a western blot analysis of embryos having the following genotype: Act5c-GAL4;tub-GAL80ts>miR-310 c. Embryos were used because in adult flies Esg levels are extremely low. The expression of this cluster during embryo development reduces esg protein to undetectable levels thus explaining the lethality observed when the miR-310 c is expressed early in development with a strong promoter (Fig 5D). Overall, these experiments clearly demonstrate that the miR-310 c is downregulating esg.
The phenotype of the ectopic expression of the miR-310 c can be phenocopied by a constitutive GAL4 driver but only when it is expressed late in development
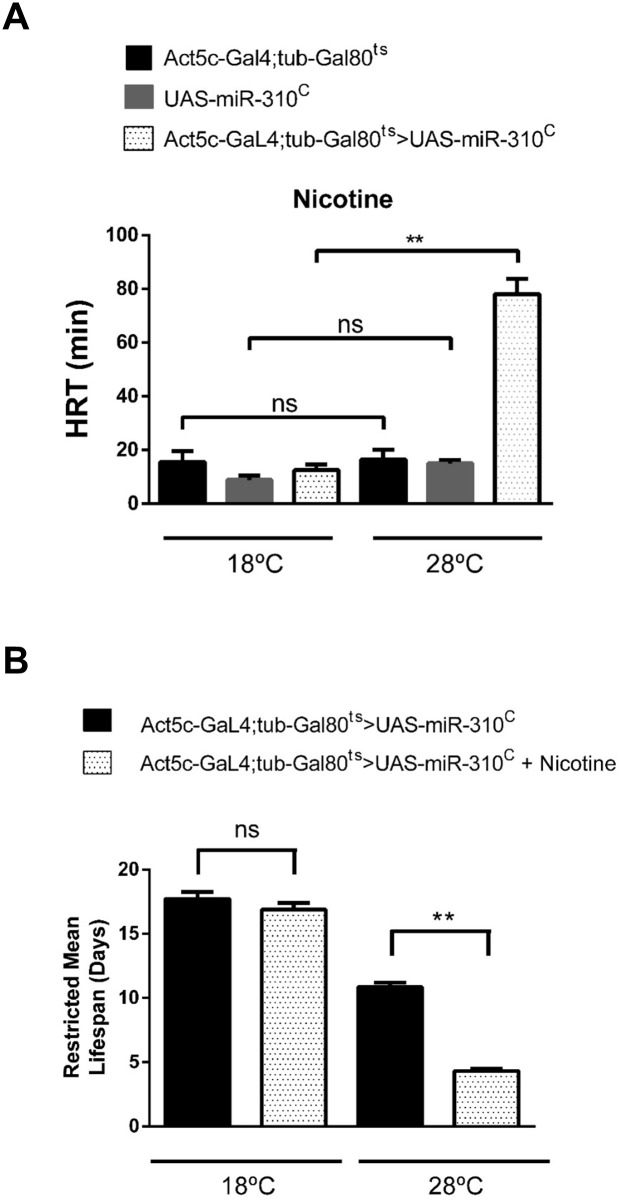
To demonstrate that the ectopic expression of the miR-310 c is enough to enhance the nicotine-sensitivity, we used the transgenic UAS-DsRed53-mir-310-313 line driven by a battery of GAL4 drivers of different strengths and directed to different target organs; however, no driver was able to direct the expression of the miR-310 c in such a way that it phenocopies L70 (S1 Table). A possible explanation for this result could be that the miR-310 c expression was not directed to the correct tissues, time and/or levels of expression. We attempted to drive the expression of the miR-310 c by strong constitutive GAL4 drivers such as the actin promoter but we only confirmed that constitutive strong expression of the microRNAs is lethal as previously reported (S1 Table) [15]. To overcome this issue, we decided to use the GAL4/GAL80ts system in order to control the time at which the miR-310 c was expressed. For this, we induced the miR-310 c expression by incubating flies at 28°C, the permissive temperature, at different times during development using the Act5c-GAL4;tub-Gal80ts driver. Under these conditions, flies were viable (60%) only when the expression of the miR-310 c was induced after sixteen days of development at 18°C when they are just at the end of pupation or after eclosion. Flies expressing miR-310 c became as sensitive to nicotine as the L70 line after 72 hrs of induction (Fig 6A). Also, these flies are more sensitive to nicotine-containing food thus fully phenocoping L70 (Fig 6B).
Fig 6. Temporally controlled expression of the miR-310 c in adult flies phenocopies L70 mutant line.
(A) The HRT of the line Act5c-GAL4;Tub-Gal80ts>UAS-miR-310 c is significantly longer only when it is at the GAL80 permissive temperature (28°C). Embryos were reared at 18°C for sixteen days, just before eclosion pupae were transferred to 28°C and maintained at this temperature for 3 days before assaying, controls were kept at 18°C, n ≥ 100. (B) Survival of the line Act5c-GAL4;Tub-Gal80ts>UAS-miR-310 c is significantly reduced. Restricted mean life of Act5c-GAL4;Tub-Gal80ts>UAS-miR-310 c genotype is significantly reduced when maintained in standard cornmeal food supplemented with 0.5 mg/ml nicotine at the GAL80 permissive temperature n ≥ 100 flies, n = 3 in all experiments, ** = P ≤ 0.001, ns = not significant.
The gene esg is expressed in labial imaginal discs
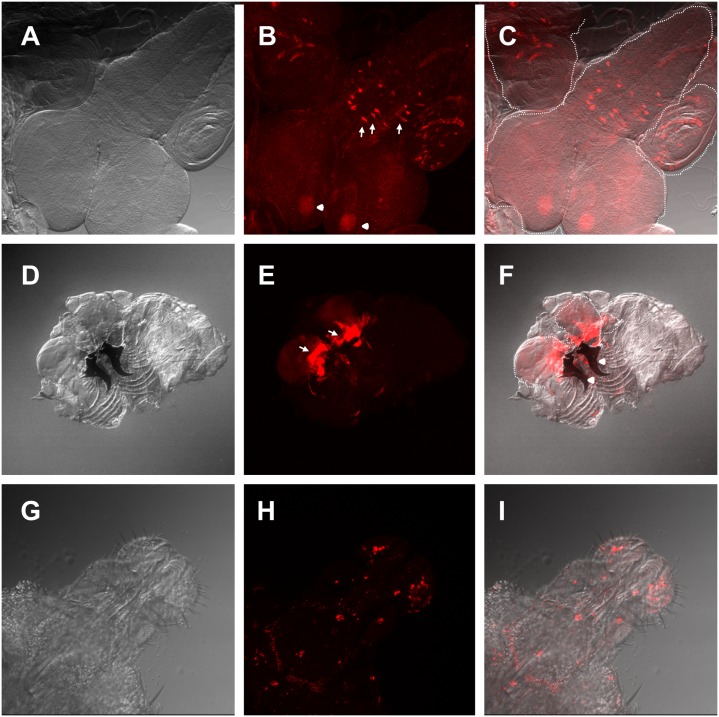
Trying to understand the cellular basis of these phenotypes, we characterized the L4 expression pattern by crossing it with UAS-GFP and found that it has the same expression pattern as the one reported for esg: embryonic ectoderm and histoblasts; larval leg and wing imaginal discs, brain, spiracles and tracheae; adult intestinal stem cells, malpighian tubules, testis and salivary glands [7,16–19]. Our data shows that esg is also expressed in the third instar larval mushroom bodies and the medial section of the thoracoabdominal ganglion, where motor neurons that innervate wings and legs are located. (Fig 7A–7C). Importantly, we found that esg is expressed in the proximal region of the third instar larvae labial imaginal discs (Fig 7D–7F) and in the labial palp of the late pupae (Fig 7G–7I) where the taste organs are developing. The lack of esg may be altering the normal differentiation of these organs causing hypersensitivity to nicotine.
Fig 7. Expression pattern of the esg L4 enhancer trap.
(A-C) L4>UAS-GFP third instar larval central nervous system (CNS) and adjacent imaginal discs A) Differential interference contrast (DIC) microscopy image. (B) Confocal maximal projection of an immunohistochemistry against GFP. Arrows show the medial section of the thoracoabdominal ganglion esg expressing neurons and arrowheads show mushroom bodies. (C) Overlay of DIC and confocal images. (D-F) L4>UAS-GFP third instar larval inverted maxillary region that includes labial imaginal discs. (D) DIC microscopy image. (E) Confocal maximal projection of GFP fluorescence. Arrows show the expression domain in the proximal labial discs. Arrowheads show mandibles. (F) Overlay of (D) and (E). (G-I) L4>UAS-Stinger late pupal proboscis. (G) DIC microscopy image. (H) Confocal maximal projection of stringer fluorescence. (I) Overlay of (H) and (I). White dash-lines show the border of the CNS and labial imaginal discs, respectively.
Transcriptome analysis shows that esg loss of function affects genes involved in chemical perception and cuticle development
We sequenced the transcriptome of L4 and L70 heads and compared each of the mutant lines with both control lines (w 1118 and Ore-R). To identify the biological processes altered in our mutants, we used the resources of the Gene Ontology Consortium [20,21] to do an enrichment of biological process analysis, using the database of genes whose ontology is supported by experimental data. For this analysis, we only considered genes that were consistently up or downregulated in the mutant lines when compared with both control lines. We used as cutoff a “double control log2 ratio” (see Materials and methods) of 1 or greater, that is equivalent to a minimum of a 2 fold increase when compared to the controls’ levels of expression; or -1 or lower, that is equivalent to a minimum reduction of half of the controls’ levels of expression. Computed genes with no mutant phenotype, molecular feature or determined function (CGs followed by an integer) were not included. Using these criteria, we found that in L4, 507 genes were upregulated and 233 were downregulated. The biological processes in which the affected genes in L4 participate are related to male gamete generation, cuticle development and detection of chemical stimulus: odorant, pheromones, taste and chemical receptors (S2 Table). For L70, 164 genes were upregulated while 233 were downregulated. The affected genes in L70 are involved in the following biological processes: detection of chemical stimulus involved in sensory perception (specifically smell), neurological system process, detection of pheromones, sexual reproduction and nervous system development (S3 Table). These data are in agreement with our observed phenotypes and also with previous literature as both esg and the miR-310 c had been reported to be required for male gametogenesis and neural development [22,23].
Discussion
In this work, we identified two mutant lines that are statistically significant more sensitive to nicotine when compared to control flies. One of them is L4, a loss of function allele that reduces the levels of the transcription factor Esg by half, as demonstrated by western blot analysis. The hypersensitivity of this P-element insertion mutant is clearly reproduced by esg 35Ce-3, a loss-of-function mutant that expresses a function-less full length Esg protein with one of its zinc fingers inactivated by a missense point mutation. This loss of function is further confirmed by the fact that the L4/esg 35Ce-3 genotype is lethal, demonstrating that they belong to the same complementation group. In addition, the induced expression of esg by L4 using an EP insertion in the esg gene completely rescues the normal nicotine phenotype; thus, confirming that the loss of half the dosage of esg is enough to confer hypersensitivity to nicotine.
L4 shows the classical esg cuticle phenotype previously reported for other esg alleles. The esg knock-down obtained by RNAi severely enhances cuticle phenotype, in the same way as the miss-expression of the miR-310 c does, either by its own promoter or by using the nullo driver, consequently demonstrating that this cluster is down-regulating the esg gene. The esg downregulation was also shown in embryos, where the miR-310 c over expression completely abates Esg production.
We found esg expression in the proximal labial imaginal disc, the precursor of the proboscis, where the main gustatory organs reside. In order to explain the nicotine hypersensitivity we propose that the loss of esg function in the labial discs during development is promoting premature differentiation of these sensory organ precursors affecting their cellular fate and causing ectopic expression of chemical receptors. Esg, and the two other members of the family, Worniu and Snail, participate in neuroblast development and differentiation by mediating asymmetric cell divisions that has been shown to be involved in the correct development of sensory bristles and other taste specialized sensilla [24,25]. Esg affects aspects of the differentiation process in intestinal stem cells and male germ cells by maintaining stemness and repressing differentiation [26]. This Esg function might be conserved during gustatory organ development and differentiation and the development of neurons of the medial section of the thoracoabdominal ganglion. This hypothesis is supported by the fact that other Snail family members have a similar role in the nervous system [27,28], and by our sequencing data that showed that many genes involved in sensory and chemical perception are affected when there is esg loss of function. The change in cell fate hypothesis that we propose is further supported by the fact that esg is involved in olfactory neuron fate determination as the over-expression of Esg together with Nerfin induces the appearance of CO2 sensitive neurons that do not exist in the wild type [29]. The fact that that esg is expressed in the labial discs, proboscis and labella and that, in our transcriptome analysis, the expression of several bitter taste and odorant receptors were affected is consistent with an aberrant chemical sensilla differentiation caused by the lowered expression of esg, either through its downregulation via miRs or by mutation. The lack of esg could also be affecting other sections of the nervous system such as the medial section of the thoracoabdominal ganglion, where many of the motor neurons that control limb movement are located, thus contributing to the loss of negative geotaxis reflex of esg mutants when exposed to nicotine.
The other nicotine hypersensitive line, L70, showed ectopic expression of the miR-310 c during pupation and in adult flies when it is not normally expressed. This altered miR-310 c expression is sufficient to induce nicotine hypersensitivity, as demonstrated by ectopically expressing the miR-310 c late in pupal development under the control of a strong constitutive promoter. The hypersensitivity shown by L70 can be explained if the ectopic expression of these micro-RNAs downregulates esg, thus phenocopying esg loss of function. The stronger phenotype observed in homozygous L70 compared to L4 can be explained by the fact that the miRs in this cluster have additional target genes that affect neural physiology. The known targets of the miR-310 c are β-catenin, dTCF [22] and Khc-73 [13]. Khc-73 is important for the modulation of synaptic strength, so its downregulation could be altering the intensity of the response to nicotine; βcatenin and TCF could be involved in neuron determination and synapse physiology via the Wg/Wnt pathway.
It is worth to mention that our work does not exclude metabolic pathways involved in detoxification, a line of inquiry that must be pursued in the future. We need to further investigate the precise levels of Esg during metamorphosis in these lines and to analyze in greater detail both the architecture and physiology of the adult nervous system in these lines.
In conclusion, we show that esg is important for the development of organs involved in chemical perception and that the ectopic expression of the miR-310 c modulates esg expression.
Materials and Methods
Volatilized nicotine sensitivity test
Groups of 10 male flies 0–3 days post-eclosion were put into an empty vial (VWR vial O.D. x H = 25 x 95 mm), where they were exposed for 15 seconds to volatilized nicotine (N3976 SIGMA-ALDRICH). Nicotine solution was volatilized by placing 2 μl on a 3.0 mm Nichrome filament (NI30PF World Precision Instruments, Inc.), which was introduced into the vial and heated to 250°C to achieve instantaneous volatilization. To titrate the amount of nicotine to be used 0 to 1000 ng of nicotine were tested. The flies were then transferred into a clean vial labeled with a perimetral line 2.5 cm from the bottom. The negative geotaxis reflex was measured every 5 minutes. Flies that crossed the line in a time no longer than 10 seconds after being tapped to the bottom were considered recovered and removed from the vial. The observation time was performed until 100% of the flies recovered. Half Recovery Time (HRT) was defined as the time at which 50% of the flies recovered their geotaxis reflex. The amount of 32 ng of nicotine in 2 μl caused an HRT of approximately 30 minutes for the line w 1118; therefore, we used this amount in the rest of the tests. For statistical analysis of the data one-way ANOVA was used and a P ≤ 0.01 was considered significant. All graphs and statistical analyses were done in GraphPad Prism 6.01.
Chronic exposure to nicotine
Newly eclosed male flies were placed in standard corn meal agar food (control) or with nicotine (experimental) at a concentration of 0.5 mg/ml in groups of 20–30 flies. Flies were changed every 24 hours to a new vial and the number of dead flies was registered until there were no more flies alive. Assays were performed at 18°C, 25°C and 28°C. For survival analysis, we used the Kaplan-Meier estimator using the OASIS: online application for the survival analysis of lifespan assays performed in aging research (http://sbi.postech.ac.kr/oasis/) [30].
Floating Petri dish assay
The avoidance of noxious heat assay or hot water floating Petri dish assay was performed by introducing 30 adult male flies in a 14 cm plastic Petri dish sealed with vinyl electric tape. Petri dishes were floated in a 45°C water bath for 30 min. in darkness. Fit flies migrate to the roof of the petri dish and avoid becoming unconscious because of the heat shock, flies found unconscious were considered unfit [31]. For statistical analysis of the data one-way ANOVA was used and P ≤ 0.01 was considered significant. Graphs and statistical analyses were done in GraphPad Prism 6.01.
Exposure to ether vapors
Groups of ten flies were placed for one minute in an empty standard Drosophila vial covered with a cotton plug that had 1 ml of ether. Flies were transferred to a fresh vial and the HRT was determined. The experiment was repeated at least ten times. For statistical analysis of the data one-way ANOVA was used and P ≤ 0.01 was considered significant. Graphs and statistical analyses were done in GraphPad Prism 6.01.
Mapping of L70 and L4 mutations
L70 mutant line was mapped using inverse PCR [32]. PstI and XbaI enzymes were used (NEB, Inc.) to digest genomic DNA. DNA was ligated with T4 ligase and fragments were amplified with the following oligonucleotides: F5'GCACGTTTGCTTGTTGAGAGGAAAGG3’ and R5'CGGTAAGCTAGCTTCGGCTATCG3'. The products obtained were sequenced and subsequently mapped to the Drosophila genome using BLAST analysis in http://flybase.org/blast/. The insertion site was mapped to 2R:20584338. L4 mutant line was mapped by plasmid rescue technique [32,33]. PstI enzyme (NEB, Inc.) was used to digest genomic DNA and obtain pBluescript fused to adjacent genomic DNA. Three clones were obtained and sequenced using the T7 primer 5´TAATACGACTCACTATAGGG3’. Sequences obtained were identical and subsequently mapped to the Drosophila genome using BLAST analysis in http://flybase.org/blast/. The insertion mapped to site 2L: 15333905. Insertion sites were confirmed by PCR and the resulting products were sequenced. The oligonucleotides used were: 5'CACAACCTTTCCTCTCAACAA3' and 5'CTGACGTTGATCAGTTCCTCG3’ for the 5´ end of the P{GawB} insertion and oligos 5'TAGCCGAGTGCCGCGATTG3' and 5'CTGACGTTGATCAGTTCCTCG3’ for the 3' end.
Northern-blot
Total RNA was prepared using Trizol reagent (Invitrogen, Carlsbad, CA), according to manufacturer’s directions. Total RNA (10–20 μg) was separated in a 15% poyacrylamide/7 M urea/0.5X TBE gel and transferred to Hybond-N+ membrane (Amersham, Piscataway, NJ). Hybridizations were carried out at 42°C with the Ultra-Hyb Oligo solution (Ambion, Austin, TX) using as probes DNA oligonucleotides labeled with P32-γATP (PerkinElmer) at their 5′ end using T4 polynucleotide kinase (Fermentas). Oligonucleotides used: miR-310-5’AAAGGCCGGGAAGTGTGCAATA3’, miR-311-5’tcaggccggtgaatgtgcaata3’, miR-312-5’tcaggccgtctcaagtgcaata3’, miR-313-5’tcgggctgtgaaaagtgcaata3’ and as a loading control 2SRNA-5´TACAACCCTCAACCATATGTAGTCCAAGCA. Two washes using 2x SSC/0.1% SDS were carried for 30 min at 42°C. Autoradiographs were analyzed using a Typhoon PhosphorImager (Amersham). All membranes were exposed for 24 hours and control hybridizations were exposed for 5 minutes.
5’ RACE
Total RNA was extracted from 3 day adult heads as above. 5´RACE was performed using the FirstChoice RLM-RACE Kit according to manufacturer´s instructions including a third round of nested PCR. Primers used:
First PCR: Del1Fwd 5´-GATTCGTTTTAAAGTAATCAGG-3´ and 5' RACE Outer Primer 5'-GCTGATGGCGATGAATGAACACTG-3'.
Second nested PCR: Del2Fwd 5´-GCTAATTGTTAGTATAAGGCA-3´ and 5' RACE Inner Primer 5'-CGCGGATCCGAACACTGCGTTTGCTGGCTTTGATG-3'.
Third nested PCR: miR-313 5´-TCGGGCTGTGAAAAGTGCAATA-3´ and 5' RACE Inner Primer 5'-CGCGGATCCGAACACTGCGTTTGCTGGCTTTGATG-3'.
PCR product was purified using Amicon Ultra-0.5 MILLIPORE columns and directly sequenced using primer miR-313.
Western-blot
Total proteins from fly embryos aged 0–14 hrs were extracted in lysis buffer (250 mM sucrose, 50 mM Tris-HCl (pH 7.5), 5 mM EDTA, 25 mM KCl, 5 mM MgCl2 and 1% SDS) containing protease inhibitors. Protein extracts (50–100 μg) were subjected to 12% SDS-polyacrylamide gel electrophoresis, and transferred to Immobilon polyvinylidene difluoride membranes (MILLIPORE) for 2 hrs at 200 mA on a Mighty Small Transphor (Amersham Pharmacia Biotech). Blots were blocked overnight with 5% nonfat dry milk in PBS 1X containing 0.1% Tween-20. Custom made anti-Escargot antibodies (rabbit polyclonal, EZBiolab) were added at a dilution of 1:1000 for two hours and developed using the SuperSignal West Pico Chemiluminiscent Substrate (Thermo Scientific). After developing, blots were stripped by incubation at 55°C in a buffer containing: 62 mM TRIS-HCL pH = 6.8, 2% SDS and 0.1 M β-Mercaptoethanol. After stripping blots were assayed with anti-Actin antibody (Developmental Hybridoma Bank) using a dilution 1:3500 and developed as above. All blots were detected using Kodak radiographic film. Quantitation was performed using ImageJ [34] using the gel quantitation utility.
Transcriptome analysis
Total RNA from fly male heads was extracted using Trizol reagent (Invitrogen, Carlsbad, CA), according to manufacturer’s directions. RNA was stabilized using RNA stable tubes from Biomatrica and processed according to manufacturer’s instructions. Fifty μg of total RNA per line were sent for deep sequencing to the Beijing Genome Institute (BGI Americas corporation). BGI provided the following services: 1. Data filtering that includes removing adaptors contamination and low-quality reads from raw reads. 2. Assessment of sequencing (Alignment Statistics, randomness assessment of sequencing, distribution of reads on the reference genome). 3. Gene expression and annotation (Gene coverage and coverage depth). 4. Gene expression difference analysis (this tables are available as provided by BGI in S4–S7 Tables) which were obtained with the NOIseq differential expression method [35]. Sample (L4 or L70) and control lines (w 1118 and Ore-R) were sequenced as independent duplicates. FPKMs for each duplicate were averaged and the mean log2 ratio was calculated using the following formula: (mean sample FPKM)/(mean control FPKM). Since we were using two different control strains, in order to avoid possible genetic background effects we specifically selected genes that were consistently up or down regulated in the sample when compared to both control lines. We obtained a “double control log2 ratio” by averaging the mean log2 ratios from both comparisons of each sample with both controls. Only genes that had a “double control log2ratio” bigger than 1 or smaller than -1 were selected for the gene ontology analysis. The genes that satisfied these requirements were analyzed with the tools from the Gene Ontology Consortium [20,21] to do an enrichment of biological process analysis, using the database of genes whose ontology is supported by experimental data. Raw data available at: http://www.ncbi.nlm.nih.gov/bioproject/288398. Accession numbers: SRR2080216, SRR2080214, SRR2080213, SRR2080215, SRR2080211, SRR2080210, SRR2080212.
Immunohistochemistry
Tissue was dissected in PBS and fixed in PBS 4% formaldehyde. Tissue was then permeabilized with 0.2% TritonX-100 and blocked for 30 min with 5% goat preimmune serum at room temperature. The primary antibody (mouse anti-GFP, 1:10, developmental studies hybridoma bank) was added, incubated overnight at 4°C and washed 3 times in PBS 0.2% TritonX-100 at room temperature. The secondary antibody (Cy3conjugated, 1:300, goat anti-mouse, Rockland Immunochemicals, Inc., Gilbertsville,PA) was added and incubated for 90 min and washed 3 times in PBS 0.2% TritonX-100 at room temperature. Samples were mounted in Citifluor (Ted Pella Inc).
Confocal Imaging
Images were collected at the “Laboratorio Nacional de Microscopía Avanzada (LNMA)” in an inverted Confocal multiphotonic Olympus FV1000, using optimal confocal pinhole. Images were analyzed and reconstructed using the open source ImageJ program [34].
Stocks and crosses
Flies were raised on standard yeast medium at 18°C, 25°C and 28°C. Oregon-R and w 1118 strains were obtained from the Drosophila stock center in Bloomington, Indiana. L4 (w 1118 ; P{GawB-GAL4. esg} L4) and L70 (w 1118 ; P{GawB-GAL4. miR310/313} L70), transgenic lines were obtained in our laboratory by mobilization of P-element P{GawB}. The P-element in L70 was mobilized by crossing to the Δ2–3 transposase source. Revertant lines (F 1.1, F 1.2, F 2, F 5 and F 7.2) were recovered by screening for loss of the white marker, the insertion region was sequenced and revertant lines were tested for nicotine sensitivity. UAS-iRNA esg (Bloomington Stock # 34063), snail 18 (Bloomington Stock # 2311), Δ40 line (deletion of cluster miR-310 c), UAS-DsRed-53-miR-310-313 were kindly provided by Pejmun Haghighi Lab (McGill University), individual UAS-LUC-miR-311, UAS-LUC-miR-14 and all other lines used in this work are available at the Bloomington Drosophila Stock Center (NIH P40OD018537).
Supporting Information
All mutant genotypes recovered immediately after exposure to vaporized water (nicotine 0 ng).
(TIF)
P{GawB} was mobilized using the transposase source Δ2–3. Five independent lines: F1.1, F1.2, F2, F5, F7.2 (data for F1.1 is in Fig 1) were recovered, all of them having a precise excision that was confirmed by DNA sequence. HRT from all revertant lines did not show significant differences from the control lines. ns = not significant.
(TIF)
(A) Northern blot and their quantitative analysis using RNA from embryos. (B) Northern blot and their quantitative analysis using RNA from adult male thorax. (C) Northern blot and their quantitative analysis using RNA from adult male abdomen. Probes used are indicated at the right of each hybridization result. There is practically no adult expression of the miR-310, miR-311 and miR-312 transcripts from wt, Δ40 and F1.1 revertant line in adult tissues tested, while there is high expression of the transcripts in L70 mutant line. miR-313 was not detectable. *** = P << 0.001, Ф = non detectable.
(TIF)
FirstChoice RLM-RACE Kit was used to determine the transcription initiation site of the miR-310c. A) Arrow shows third nested PCR product of w 1118 and sequence histogram. 1) MWM 2) Primers miR-313/Inner5´RACE, w 1118 cDNA. 3) Primers miR-313/Inner5´RACE, no cDNA B) Arrow shows third nested PCR product of L70 and sequence histogram. 1) MWM 2) Primers miR-313/Inner5´RACE, L70 cDNA. 3) Primers miR-313/Inner5´RACE, no cDNA C) Sequence alignment of w1118 and L70 PCR products.
(TIF)
(PDF)
(DOCX)
(DOCX)
(DOCX)
(XLS)
(XLS)
(XLS)
(XLS)
Acknowledgments
We thank René Hernández Vargas for technical support, Santiago Becerra for oligonucleotide synthesis, Jorge Arturo Yanez Ponce de Leon for DNA sequencing, Andrés Saralegui Amaro and Jaime Arturo Pimentel Cabrera from the Laboratorio Nacional de Microscopía Avanzada (LNMA) support in confocal imaging.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Consejo Nacional de Ciencia y Tecnología (CONACyT, Mexico), (http://www.conacyt.mx/), (grant no. 131685 to ER), scholarship no. 481914 to Fernando Rosales, and scholarship no. 255052 to Iván Sánchez. Universidad Nacional Autonoma de Mexico; Direccion General de asuntos del Personal Académico. Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica (http://dgapa.unam.mx/html/papiit/papit.html), grant no. IN203611 and IN204214. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Heberlein U, Tsai LT-Y, Kapfhamer D, Lasek AW. Drosophila, a genetic model system to study cocaine-related behaviors: a review with focus on LIM-only proteins. Neuropharmacology. 2009;56 Suppl 1: 97–106. 10.1016/j.neuropharm.2008.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Devineni AV, Heberlein U. Preferential ethanol consumption in Drosophila models features of addiction. Curr Biol. 2009;19: 2126–32. 10.1016/j.cub.2009.10.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Velàzquez-Marrero C, Seale GE, Treistman SN, Martin GE. BK Channel β4 Subunit Influences Sensitivity and Tolerance to Alcohol by Altering its Response to Kinases. J Biol Chem. 2014289(42):29261–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Morel C, Fattore L, Pons S, Hay YA, Marti F, Lambolez B, et al. Nicotine consumption is regulated by a human polymorphism in dopamine neurons. Mol Psychiatry. Nature Publishing Group; 2013;19: 930–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McClung C, Hirsh J. Stereotypic behavioral responses to free-base cocaine and the development of behavioral sensitization in Drosophila. Curr Biol. 1998;8: 109–12. [DOI] [PubMed] [Google Scholar]
- 6. Marygold SJ, Leyland PC, Seal RL, Goodman JL, Thurmond J, Strelets VB, et al. FlyBase: improvements to the bibliography. Nucleic Acids Res. 2013;41: D751–7. 10.1093/nar/gks1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hayashi S, Hirose S, Metcalfe T, Shirras A. Control of imaginal cell development by the escargot gene of Drosophila. Development. 1993;118: 105–115. [DOI] [PubMed] [Google Scholar]
- 8. Fuse N, Hirose S, Hayashi S. Diploidy of Drosophila imaginal cells is maintained by a transcriptional repressor encoded by escargot. Genes Dev. 1994;8: 2270–2281. [DOI] [PubMed] [Google Scholar]
- 9. Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, et al. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5: 337–50. [DOI] [PubMed] [Google Scholar]
- 10. Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17: 1850–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mandke P, Wyatt N, Fraser J, Bates B, Berberich SJ, Markey MP. MicroRNA-34a modulates MDM4 expression via a target site in the open reading frame. PLoS One. 2012;7: e42034 10.1371/journal.pone.0042034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schnall-Levin M, Zhao Y, Perrimon N, Berger B. Conserved microRNA targeting in Drosophila is as widespread in coding regions as in 3’UTRs. Proc Natl Acad Sci U S A. 2010;107: 15751–6. 10.1073/pnas.1006172107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tsurudome K, Tsang K, Liao EH, Ball R, Penney J, Yang J-S, et al. The Drosophila miR-310 cluster negatively regulates synaptic strength at the neuromuscular junction. Neuron. 2010;68: 879–93. 10.1016/j.neuron.2010.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kunwar PS, Starz-Gaiano M, Bainton RJ, Heberlein U, Lehmann R. Tre1, a G protein-coupled receptor, directs transepithelial migration of Drosophila germ cells. PLoS Biol. Public Library of Science; 2003;1: E80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tang T, Kumar S, Shen Y, Lu J, Wu M-L, Shi S, et al. Adverse interactions between micro-RNAs and target genes from different species. Proc Natl Acad Sci U S A. 2010;107: 12935–40. 10.1073/pnas.1007591107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tanaka-Matakatsu M, Uemura T, Oda H, Takeichi M, Hayashi S. Cadherin-mediated cell adhesion and cell motility in Drosophila trachea regulated by the transcription factor Escargot. Development. 1996;122: 3697–705. [DOI] [PubMed] [Google Scholar]
- 17. Samakovlis C, Manning G, Steneberg P, Hacohen N, Cantera R, Krasnow MA. Genetic control of epithelial tube fusion during Drosophila tracheal development. Development. 1996;122: 3531–6. [DOI] [PubMed] [Google Scholar]
- 18. Whiteley M, Noguchi PD, Sensabaugh SM, Odenwald WF, Kassis JA. The Drosophila gene escargot encodes a zinc finger motif found in snail-related genes. Mech Dev. 1992;36: 117–27. [DOI] [PubMed] [Google Scholar]
- 19. Biteau B, Karpac J, Supoyo S, Degennaro M, Lehmann R, Jasper H. Lifespan extension by preserving proliferative homeostasis in Drosophila Kim SK, editor. PLoS Genet. Public Library of Science; 2010;6: e1001159 10.1371/journal.pgen.1001159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. The Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2014;43: D1049–1056. 10.1093/nar/gku1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8: 1551–66. 10.1038/nprot.2013.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pancratov R, Peng F, Smibert P, Yang S, Olson ER, Guha-Gilford C, et al. The miR-310/13 cluster antagonizes β-catenin function in the regulation of germ and somatic cell differentiation in the Drosophila testis. Development. 2013;140: 2904–16. 10.1242/dev.092817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Streit A, Bernasconi L, Sergeev P, Cruz A, Steinmann-Zwicky M. mgm 1, the earliest sex-specific germline marker in Drosophila, reflects expression of the gene esg in male stem cells. Int J Dev Biol. 2002;46: 159–66. [PubMed] [Google Scholar]
- 24. Cai Y, Chia W, Yang X. A family of snail-related zinc finger proteins regulates two distinct and parallel mechanisms that mediate Drosophila neuroblast asymmetric divisions. EMBO J. 2001;20: 1704–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Le Borgne R, Bellaïche Y, Schweisguth F. Drosophila E-Cadherin Regulates the Orientation of Asymmetric Cell Division in the Sensory Organ Lineage. Curr Biol. Elsevier; 2002;12: 95–104. [DOI] [PubMed] [Google Scholar]
- 26. Korzelius J, Naumann SK, Loza-Coll MA, Chan JS, Dutta D, Oberheim J, et al. Escargot maintains stemness and suppresses differentiation in Drosophila intestinal stem cells. EMBO J. 2014; 289(42):29261–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lai S-L, Miller MR, Robinson KJ, Doe CQ. The Snail family member Worniu is continuously required in neuroblasts to prevent Elav-induced premature differentiation. Dev Cell. 2012;23: 849–57. 10.1016/j.devcel.2012.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Voog J, D’Alterio C, Jones DL. Multipotent somatic stem cells contribute to the stem cell niche in the Drosophila testis. Nature. 2008;454: 1132–6. 10.1038/nature07173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hartl M, Loschek LF, Stephan D, Siju KP, Knappmeyer C, Kadow ICG. A new Prospero and microRNA-279 pathway restricts CO2 receptor neuron formation. J Neurosci. 2011;31: 15660–73. 10.1523/JNEUROSCI.2592-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang J-S, Nam H-J, Seo M, Han SK, Choi Y, Nam HG, et al. OASIS: online application for the survival analysis of lifespan assays performed in aging research. PLoS One. Public Library of Science; 2011;6: e23525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Neely GG, Hess A, Costigan M, Keene AC, Goulas S, Langeslag M, et al. A genome-wide Drosophila screen for heat nociception identifies α2δ3 as an evolutionarily conserved pain gene. Cell. 2010;143: 628–38. 10.1016/j.cell.2010.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang AM, Rehm EJ, Rubin GM. Recovery of DNA sequences flanking P-element insertions in Drosophila: inverse PCR and plasmid rescue. Cold Spring Harb Protoc. 2009;2009: pdb.prot5199. [DOI] [PubMed] [Google Scholar]
- 33. Hersberger M, Kirby K, Phillips JP, Würgler FE, Koller T, Widmer RM. A plasmid rescue to investigate mutagenesis in transgenic D. melanogaster. Mutat Res Mutagen Relat Subj. 1996;361: 165–172. [DOI] [PubMed] [Google Scholar]
- 34. Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tarazona S, García F, Ferrer A, Dopazo J, Conesa A. NOIseq: a RNA-seq differential expression method robust for sequencing depth biases. EMBnet.journal. 2012;17: 18. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All mutant genotypes recovered immediately after exposure to vaporized water (nicotine 0 ng).
(TIF)
P{GawB} was mobilized using the transposase source Δ2–3. Five independent lines: F1.1, F1.2, F2, F5, F7.2 (data for F1.1 is in Fig 1) were recovered, all of them having a precise excision that was confirmed by DNA sequence. HRT from all revertant lines did not show significant differences from the control lines. ns = not significant.
(TIF)
(A) Northern blot and their quantitative analysis using RNA from embryos. (B) Northern blot and their quantitative analysis using RNA from adult male thorax. (C) Northern blot and their quantitative analysis using RNA from adult male abdomen. Probes used are indicated at the right of each hybridization result. There is practically no adult expression of the miR-310, miR-311 and miR-312 transcripts from wt, Δ40 and F1.1 revertant line in adult tissues tested, while there is high expression of the transcripts in L70 mutant line. miR-313 was not detectable. *** = P << 0.001, Ф = non detectable.
(TIF)
FirstChoice RLM-RACE Kit was used to determine the transcription initiation site of the miR-310c. A) Arrow shows third nested PCR product of w 1118 and sequence histogram. 1) MWM 2) Primers miR-313/Inner5´RACE, w 1118 cDNA. 3) Primers miR-313/Inner5´RACE, no cDNA B) Arrow shows third nested PCR product of L70 and sequence histogram. 1) MWM 2) Primers miR-313/Inner5´RACE, L70 cDNA. 3) Primers miR-313/Inner5´RACE, no cDNA C) Sequence alignment of w1118 and L70 PCR products.
(TIF)
(PDF)
(DOCX)
(DOCX)
(DOCX)
(XLS)
(XLS)
(XLS)
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.