Abstract
Molecular diversity surveys have demonstrated that aquatic fungi are highly diverse, and that they play fundamental ecological roles in aquatic systems. Unfortunately, comparative studies of aquatic fungal communities are few and far between, due to the scarcity of adequate datasets. We combined all publicly available fungal 18S ribosomal RNA (rRNA) gene sequences with new sequence data from a marine fungi culture collection. We further enriched this dataset by adding validated contextual data. Specifically, we included data on the habitat type of the samples assigning fungal taxa to ten different habitat categories. This dataset has been created with the intention to serve as a valuable reference dataset for aquatic fungi including a phylogenetic reference tree. The combined data enabled us to infer fungal community patterns in aquatic systems. Pairwise habitat comparisons showed significant phylogenetic differences, indicating that habitat strongly affects fungal community structure. Fungal taxonomic composition differed considerably even on phylum and class level. Freshwater fungal assemblage was most different from all other habitat types and was dominated by basal fungal lineages. For most communities, phylogenetic signals indicated clustering of sequences suggesting that environmental factors were the main drivers of fungal community structure, rather than species competition. Thus, the diversification process of aquatic fungi must be highly clade specific in some cases.The combined data enabled us to infer fungal community patterns in aquatic systems. Pairwise habitat comparisons showed significant phylogenetic differences, indicating that habitat strongly affects fungal community structure. Fungal taxonomic composition differed considerably even on phylum and class level. Freshwater fungal assemblage was most different from all other habitat types and was dominated by basal fungal lineages. For most communities, phylogenetic signals indicated clustering of sequences suggesting that environmental factors were the main drivers of fungal community structure, rather than species competition. Thus, the diversification process of aquatic fungi must be highly clade specific in some cases.
Introduction
For a long time, fungi were thought to have negligible ecological functions in aquatic systems [1]. Only recently, it has been shown that aquatic fungal communities contribute to elemental cycles and mineralization processes [2]. These roles have been demonstrated for planktonic fungal communities [3–5], and for fungal communities of the marine subseafloor [6]. Differences in community structure result in different functional activities of the community that in return affect the functioning of the ecosystem. It is crucial to understand the community structure as a multi-player assemblage and to gain insights into the community composition, structure, and diversity [7,8].
Studies of aquatic fungal communities have benefited immensely from the development of molecular techniques. Thus, nowadays, information on fungal communities from a wide range of habitats exists, for example from deep-sea environments [9], hydrothermal vent ecosystems [10], coastal regions [3], anoxic regions [11], lakes [12,13], rivers [14] or associated to aquatic animals [15,16], plants [17,18] or algae [19]. In contrast to studies of terrestrial fungal communities, most of these studies targeted the 18S rRNA gene as marker region instead of the commonly used Internal Transcribed Spacers (ITS)–the “announced fungal barcode” [20].
The existing molecular information has rarely been used to get a more profound view on aquatic fungi. Currently, the sparseness of sequence data with contextual (meta) information poses a major obstacle for comparative studies and the construction of reference data sets. Although contextual data guidelines have been developed addressing this issue, most published data do not yet follow them [21]. Thus, essential contextual data has to be recovered by tedious manual screening of databases and publications. Here, fungal culture collections become important. They have the potential to provide high-quality sequences, which are combined with relevant and detailed contextual data [22].
In a first step, we generated a contextualized 18S rRNA gene sequence reference dataset for aquatic fungi. This dataset included all publicly available nearly full-length 18S rRNA gene sequences as well as sequences from the marine fungal culture collection of the GEOMAR Helmholtz Centre for Ocean Research Kiel (Germany). We tested our hypothesis that habitat-specific biomes exist for aquatic fungi using the created dataset. We included in our study phylogenetic-based pairwise community comparisons, analyses of the taxonomic composition and of phylogenetic signals.
Materials and Methods
Collection of aquatic fungal community studies
We used the “WEB OF SCIENCE” (http://apps.webofknowledge.com), “Google scholar” (http://scholar.google.com) and textbooks references to retrieve all published community studies containing fungal sequence data. The resources were screened using key words such as “community”, “marine”, “freshwater”, “fungi”, “eukaryotic” or a combination of these. Papers were screened for fungal sequence data. A study was deemed suitable for inclusion in our dataset if the following criteria were met: (i) published study and (ii) study detected at least 15 different fungal species or OTUs. All publications were additionally screened for the marker gene used, and this information was recorded. If several marker genes were used in the study, each marker gene was counted separately.
Sequence collection and alignment
In June 2013, high-quality nearly full-length fungal 18S rRNA gene sequences were obtained from release 111 of the SILVA SSU Ref dataset [23] (released July 2012). Briefly, sequences in the SILVA Ref dataset were retrieved from the European Nucleotide Archive (ENA) release 111 (http://www.ebi.ac.uk/ena/) and aligned with the SILVA Incremental Aligner (SINA) [24]. The sequences were then subjected to the following quality criteria: (i) sequence length ≥ 1200 bp, (ii) alignment identity value above or equal to 70, and (iii) alignment quality above 50. Based on the manually curated SILVA taxonomy, this dataset contained 13,501 fungal sequences. All further steps sequence alignment refinement and phylogenetic analyses were carried out with the ARB software [25].
We identified a subset of 1,537 unique sequences with aquatic origins [26]. We verified that all sequences from publications older than mid 2013 were present in the SILVA dataset, if they fulfilled the quality criteria. We added 247 fungal 18S rRNA gene sequences from the KSMP Kiel (Kultur Sammlung Mariner Pilze Kiel) culture collection (GEOMAR Helmholtz Centre for Ocean Research, Kiel, Germany) to the aquatic subset, and also aligned them using the SINA aligner following manual refinement. All of these sequences originated from marine-derived fungal cultures, were ≥ 1,200 bp in length and contained less than 5 ambiguous bases. Cultivation of fungi and DNA extraction were carried out as described in Wiese et al. [27]. Specifically, PCR for amplification of fungal 18S rRNA gene fragments was performed using puReTaq Ready-To-Go PCR Beads (GE Healthcare, Solingen, Germany) with the primers NS1 (5'-GTA GTC ATA TGC TTG TCT C-3´) and FR1 (5'-AIC CAT TCA ATC GGT AIT-3') according to Gomes et al. [28]. PCR parameters were set to the following values: initial denaturation (8 min at 94°C), 35 cycles of primer denaturation (30 sec at 94°C), annealing (45 sec at 48°C), and elongation (3 min at 72°C) followed by a final elongation step (10 min at 72°C). Closest relatives of the sequences were identified by sequence comparisons against the National Center for Biotechnology Information (NCBI) GenBank database (http://blast.ncbi.nlm.nih.gov/Blast.cgi) using BLASTN [29]. Sequences from fungal strains obtained during this study were submitted to the GenBank database and were assigned accession numbers KJ939309 –KJ939313 and KM096133 –KM096374, respectively (S1 Table).
Contextual data and trait analyses
We manually added structured contextual data to all sequences in this study [26]. This contextual data defines the environment where each fungal sequence originated. The new “environment” field allows EnvO-Lite controlled vocabulary with 20 different terms (http://obo.cvs.sourceforge.net/viewvc/obo/obo/ontology/environmental/EnvO-Lite-GSC.obo?revision=1.4&view=markup) [30]. These annotations are based on the contextual data fields like isolation source, title, and host that were already available in the SILVA SSU Ref dataset. Finally, sequences were assigned to seven EnvO-Lite defined habitat categories, namely animal-associated, aquatic, freshwater, hot spring, hydrothermal vent, marine, and sediment. The EnvO-Lite category “plant-associated” was exchanged for “plant/algae-associated”, as several sequences were derived from seaweed samples. Furthermore, two additional categories were created, namely seawater and detritus, as 17% of all sequences in our dataset originated from samples of these two habitat types [26]. Detailed information on habitat definition can be found in Table 1.
Table 1. Definition of the ten habitat categories used in this study.
All categories are based on EnvO-Lite controlled vocabulary with the exception of “detritus” and “seawater”. The EnvO-Lite defined habitat “plant-associated” was extended to include algal samples as well.
| Habitat category | Description |
|---|---|
| Animal-associated | A habitat that is in or on a living animal. Here "animal" denotes an individual of a species that is a sub-taxon of NCBITaxon:33208. The sequences of our dataset were derived exclusively from marine animals, with the exception of one. |
| Aquatic | A habitat that is in or on water. This category contains all sequences of our dataset that do not fall into freshwater or marine categories, such as peatlands with high amount of dissolved solids and low salt concentration. |
| Detritus | A habitat that is in or on death organic material. The sequences of our dataset were exclusively of marine origin, with the exception of one. |
| Freshwater | A habitat that is in or on a body of water containing low concentrations of dissolved salts and other total dissolved solids. |
| Hot spring | A spring that is produced by the emergence of geothermally-heated groundwater from the Earth's crust. |
| Hydrothermal vent | A fissure in the Earth's surface from which geothermally-heated water issues. |
| Marine | A habitat that is in a sea or ocean containing high concentrations of dissolved salts and other total dissolved solids (typically >35 grams dissolved salts per liter). This category contains all marine-origin sequences of our dataset, but could not be further assigned to a more specific category. |
| Plant/algae-associated | A habitat that is in or on a living plant or algae. This category contains all sequences of our dataset that were exclusively derived from marine plants and algae. |
| Seawater | Any water in a sea or ocean that is neither close to the bottom nor near the shore, and originating from the pelagic zone. |
| Sediment | Sediment is an environmental substance comprised of any particulate matter that can be transported by fluid flow and which eventually is deposited as a layer of solid particles\on the bed bottom of a body of water or other liquid. Sequences of our dataset originated from both marine and freshwater samples. |
It was unfortunately not possible to further subcategorize e.g. seawater into horizontal zonation of seawater column, or sediment into marine vs. freshwater sediment due to poor contextual data. Sequences assigned to the categories detritus, animal- and plant/algae-associated were all derived from marine samples (except two sequences from detritus and animal-associated categories).
Phylogenetic analysis
The original alignment was filtered using a 50% base conservation filter in the ARB software environment [25]. This filter left 1,562 informative alignment columns out of the complete alignment for phylogenetic analyses. Maximum likelihood tree reconstruction was performed with RAxML version 7.0.4 [31], applying the previously created base conservation filter, along with a termini filter to filter out any non-rRNA sequence overhangs. The GTRGAMMA model of DNA substitution has been chosen since the pinvar and the alpha parameter cannot be estimated independently [32]. RAxML was run using 100 rapid bootstrap inferences, using the bootstrap trees as starting trees. Eight choanoflagellate sequences were used as outgroup taxa, as choanoflagellates are one of the most closely related non-Fungi. The tree was edited with Inkscape, version 0.48 (http://inkscape.org).
Phylogenetic affiliation of basal fungal lineages
As the majority of sequences from basal fungal lineages were environmental sequences and had erroneous taxonomic assignments, further analyses were carried out to resolve their taxonomy. Sequences that grouped with basal fungal lineages in the phylogenetic tree reconstruction step were subjected to three additionally analyses. Firstly, BLASTN was performed at the NCBI BLAST website with default parameters [29]. Secondly, the phylogenetic placement of these sequences was checked in the eukaryotic guide-tree of the SILVA SSURef 111 dataset. Finally, current literature was consulted for more information on taxonomic position. The reported taxonomic affiliations for basal fungal lineages reflect the combined results of these three approaches (S2 Table).
Detection of differences in aquatic fungal communities
Fungal assemblages of the ten defined habitats (see 2.3) were analyzed in different ways. Firstly, the habitat-specific taxonomic compositions were inspected by subsampling sequences to the sequence number of the smallest community using the QIIME pipeline [33] (hot spring samples were excluded due to low sequence number). Taxonomic charts were constructed by clustering the subsampled sequences into operational taxonomic units based on 99% sequence identity (OTUs0.01), a commonly used threshold for clustering fungal 18S rDNA sequences [34]. Secondly, fungal assemblages were tested for significant differences between habitats. Thus, sequences were pooled according to their assignment to different habitat categories (see 2.3). These habitat–pooled sequences were subjected to a Significance test based on weighted UniFrac values [35]. The weighted UniFrac metric is the phylogenetic Kantorovich-Rubinstein distance between two sequence distributions in a phylogenetic tree [36]. It is, therefore, a mathematically well-defined metric for significant differences between communities. Permutation tests were carried out with the FastUniFrac suite [35] implemented in the Galaxy platform [37,38]. Testing involved 1,000 Monte Carlo steps (the maximum number permitted by the Galaxy server). To check for convergence we repeated each test forty times. We assumed significant difference if the Bonferroni corrected p-value was <0.05 each time. Finally, we tested for significant phylogenetic differences between sub-communities in each habitat. To this end we calculated weighted Unifrac distances for all pairs of sub-communities in each habitat that contained at least four sub-communities with 25 sequences or more. Proof of general significance (P<0.05) of phylogenetic beta-diversity between habitats was deduced by running an one-way ANOVA implemented in R version 3.1.0 [39]. We subsequently identified significantly different pairs of habitats using the Scheffé post hoc test (P<0.05).
Phylogenetic signals
Phylogenetic signals of different habitats were tested with the software PHYLOCOM v 4.2 [40], using the COMSTRUCT function. Phylogenetic relatedness was estimated via the net relatedness index (NRI; the average phylogenetic distance within a community) and the nearest taxa index (NTI; the average phylogenetic distance to the closest relative in a community). Phylogenetic signals can indicate either overdispersion or phylogenetic clustering of taxa in the communities. Thus, NRI shows how many taxa in a community are dispersed/clustered over the whole phylogenetic tree, while the NTI indicates dispersal/clustering on lower taxonomic levels. As a null model the “–m 2” function was chosen, to account for species in the phylogenetic tree lacking contextual data. Here, samples of the null communities were created with species randomly drawn from all taxa present in the phylogenetic tree. The number of randomizations was set to 9999. A p-value of P<0.05 against the random distributions was considered to indicate a significant phylogenetic signal. In a last step, NODESIG analyses were run to identify the clades in the phylogenetic tree that led to the detection of phylogenetic signals.
Results and Discussion
What 18S rRNA gene sequences can tell us about aquatic fungal communities
Fungal community surveys in aquatic and terrestrial realms have proceeded at different rates and depths, with surveys of aquatic fungi often lagging behind. This trend has recently changed and there is now more interest in understanding aquatic fungal communities. Several studies have linked aquatic fungi with ecosystem functioning [41]. The kindled interest is further fueled by the biotechnology industry. Enzymes from aquatic fungi show distinct physiological characteristics, such as high salt tolerance or barophilicity, which are highly prized in industrial applications [42].
In the past, studies of aquatic fungi relied on microscopic observations [43]. With the advent of the molecular era, these studies also started employing Sanger-sequencing like culture-trapped fungal isolates [19,44,45] or environmental clone libraries [46]. Aquatic fungal-specific surveys applying next generation high-throughput sequencing approaches just started in 2012 with only a handful of data currently available [13–15,47–50].
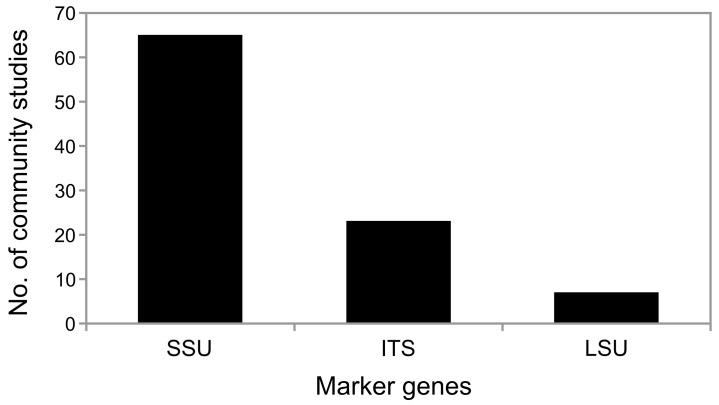
Most often, aquatic fungi are studied as part of eukaryotic or protistan communities [51–54], while molecular studies using fungal-specific primers are less common [48,55]. Therefore, the majority of sequence data on aquatic fungi is 18S rRNA gene sequences (Fig 1). The ITS regions perform poorly in inferring phylogenies [56] and thus, only 352 of the high-quality fungal ITS sequences derived from aquatic systems in the UNITE database [57] can be assigned to any taxonomic level (https://unite.ut.ee/, accessed January 2015). However, as the aquatic realm is a rich source of novel fungal species and groups [58,59], proper phylogenetic inference is needed to resolve their placement within the fungal tree of life. Thus, current aquatic fungal-specific studies use a double marker gene approach. They target the 18S rRNA gene and the ITS regions [48,60], which results in a deeper taxonomic resolution of the community [47].
Fig 1. Number of aquatic fungal community studies and the different fungal marker genes targeted.
For the summary all community studies containing fungal sequence information were counted, which targeted one of the following marker genes: ITS, LSU or SSU.
In addition to improve taxonomic classification, phylogeny-based community analyses give community surveys another flavor. They enable the inspection of the community from an evolutionary perspective [35]. It is possible to test the relevance of phylogenetic clades for the functioning of a community [61], or the lineage specific evolution as a response to specific factors [40]. Hierarchical clustering methods forming OTUs on an arbitrary threshold fail in this regard.
In summary, the accumulated “full-length”18S rRNA sequence data on aquatic fungi should not be disregarded. Instead, it should be viewed as a valuable resource leading to a better understanding of aquatic fungal communities.
A full-length 18S rRNA gene dataset for aquatic fungi
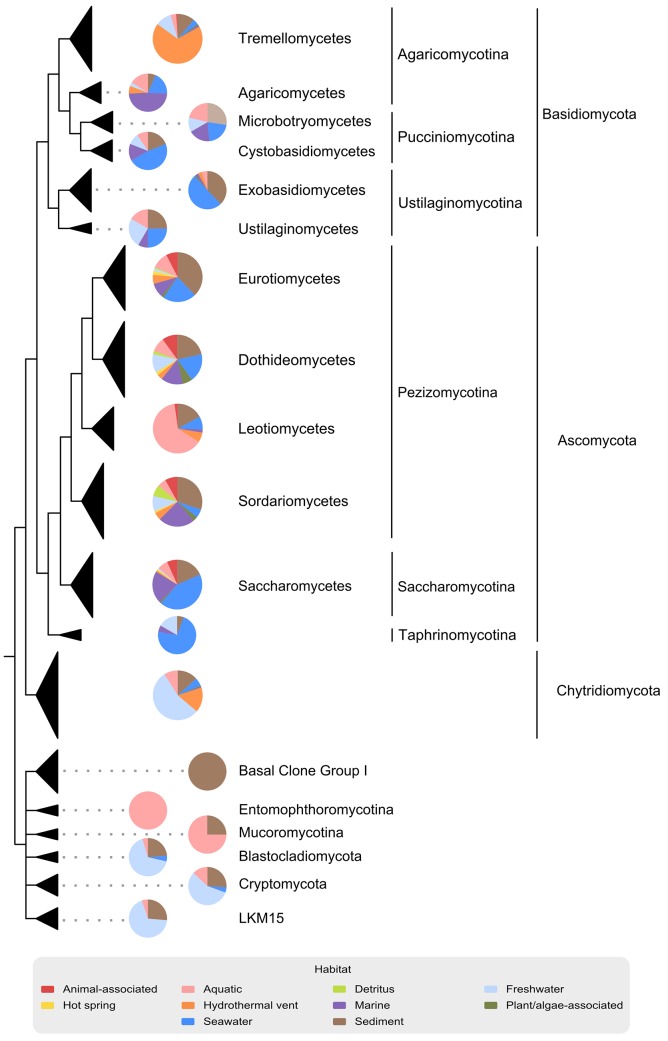
Our dataset is the first comprehensive in-depth survey of published nearly full-length 18S rRNA gene sequence data on aquatic-derived fungi (S1 Fig) and contains 1,784 fungal sequences [26]. Consistent with literature [58,62], Ascomycota was the largest group (Fig 2). The three main classes of Ascomycota—Eurotiomycetes, Dothideomycetes and Sordariomycetes (Fig 2)—are well known and dominant marine fungal lineages [63]. Interestingly, a high proportion of Basidiomycota sequences were also found in our dataset, with the majority being yeast fungi (Fig 2, S1 Fig). Recent studies on isolated Basidiomycota reported less than 100 species isolated from aquatic sources [62,64], but molecular data suggest that Basidiomycota diversity must be much higher in aquatic environments [9,17]. Chytridiomycota formed the third largest sequence group in our dataset. Furthermore, seven additional phyla/subphyla of basal fungal lineages were discovered. Of those, the recently described Cryptomycota [65] formed the largest sequence group (Fig 2; S2 Table). However, a reliable statement about the proportion of these seven groups cannot be made. Partly due to PCR primer bias, sequences associated with basal fungal lineages are often not detected in molecular surveys [11]. For instance, Cryptomycota normally accounts for only 0.02–4.5% of total sequence reads [66], and additionally, it is generally difficult to map these sequences to reference clades.
Fig 2. Schematic diagram of the phylogenetic tree using 18S rRNA gene sequences from aquatic fungi.
Sequence data were collected from the SILVA dataset and the KSMP-Kiel culture collection. Thickness of taxonomic triangles is proportional to the sequence amount of each taxonomic groups. Piecharts beside triangles show group-specific sequence assignments to the habitat types.
In order to investigate how the habitat affects fungal community assemblages, we had to link the sequence data with contextual data on sampling location (S1 Fig) and habitat type (Fig 2, Table 1). Most sequences were only sparsely annotated and were not following the “Minimum Information about a Marker Gene Sequence” requirements [21]. We added all relevant contextual data by manually screening databases and publications. Thus, our dataset is a valuable resource for further ecological studies, as well as a high quality reference dataset [26].
Using the modified EnvO-Lite controlled vocabulary, sequences could be categorized into ten different habitat categories (Table 1). The sequences were assigned to both marine and freshwater categories, as well as to organism-associated categories (Fig 2). Further, sampling locations showed global distribution over six continents and eight oceans (S1 Fig) [26].
Fungal community structure across aquatic habitats
1.1.1 Fungal communities in freshwater environments
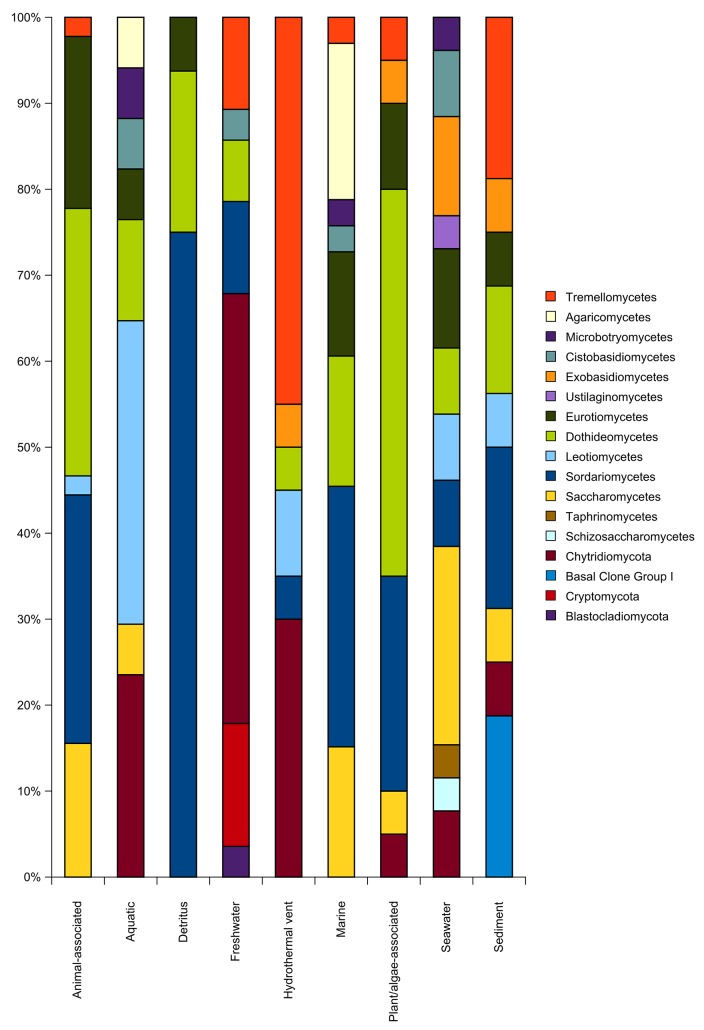
Freshwater fungal community structure differed significantly from all other habitat communities (Fig 3). This is similar to what has been observed for marine and freshwater microorganisms, which usually group in distinct marine and freshwater phylogenetic clusters [67]. Nearly 70% of all OTUs0.01 clustered in basal fungal lineages (Fig 4) and most of them belonged to the Chytridiomycota, which is known to be dominant in most freshwater samples [68,69]. Chytridiomycota play various roles as parasites [41] or in decomposing processes [5]. The other 30% of OTUs0.01 clustered equally in Asco- and Basidiomycota. We observed a tendency of taxonomic overdispersion in the freshwater habitat, mainly caused by these two phyla (Figs 5 and 6). Compared to other habitats, fungal assemblages of freshwater showed the highest diversity on the phylum level (Fig 4, subsampled data). The total amount of fungal sequences assigned to freshwater habitats clustered in several different fungal classes and subclasses (Fig 2). In contrast to other systems, freshwater systems are rich in carbon sources and structures, due to high terrestrial carbon input. Furthermore, freshwater systems are dynamic systems due to their physical nature, which degrade, sequester or translocate carbon [70]. In the pelagic zone of freshwater ecosystems, decreased amounts of soil organic carbon (C) associated with nutrient release from soil-microorganisms greatly influences the C dynamics, as well as the nutrient balance [71,72]. Fungi play essential roles in these freshwater nutrient processes [73]. As Chytridiomycota lack the ability to degrade cellulose [74], pre-processing of substrates by Ascomycota and Basidiomycota is essential to allow the colonization and use as a feeding source by other organisms [73,75]. Although several freshwater Ascomycota species have been intensively studied, about 70% of them were only reported once [62], explaining the tendency of overdispersion of higher fungi in our dataset.
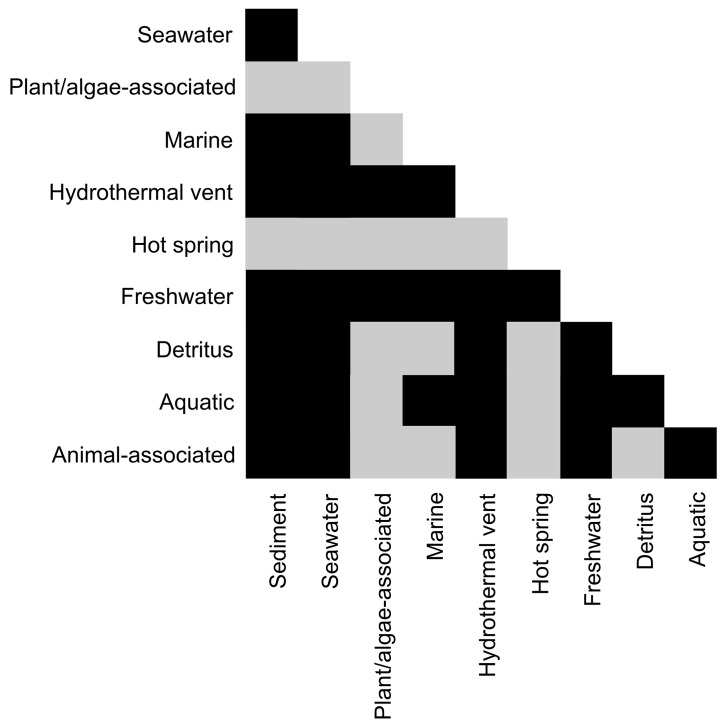
Fig 3. Impact of habitat on fungal assemblages.
Pairwise overlap of fungal assemblages according to habitat types; significant differences (P < 0.05) shown in black.
Fig 4. Taxonomic assemblages of different habitat types.
Subsampled assemblages inferred from clustering sequences at a 99% similarity level. OTUs0.01 counts were calculated for all habitat categories at order level and for basal fungal lineages at phylum/subphylum level. (Hot spring excluded due to low sequence number).
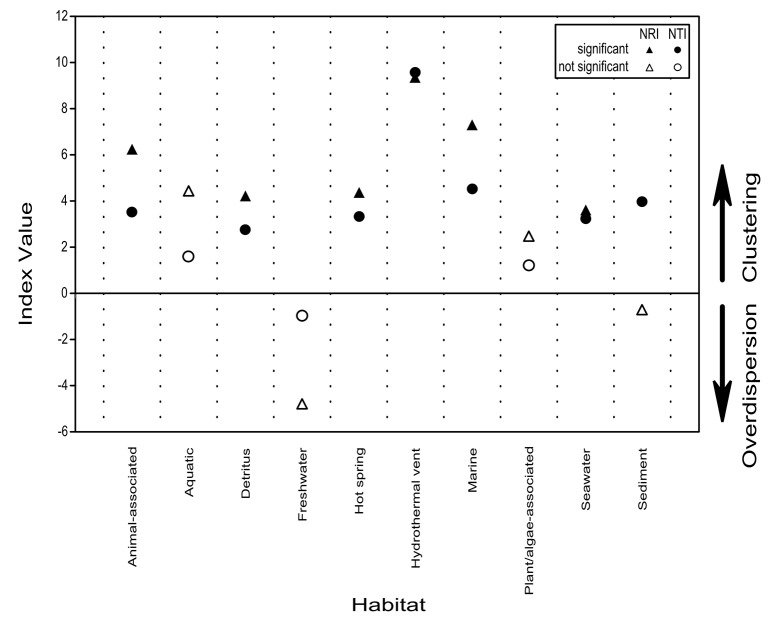
Fig 5. Phylogenetic signal analyses of aquatic fungal taxa present in different habitats.
Net Relatedness Index and Nearest Taxa Index demonstrate clustering or overdispersion of fungal taxa in a habitat over the whole pool of phylogeny or within particular terminal clades, respectively. Comparison of observed data against randomly generated samples, number of generations = 9999.
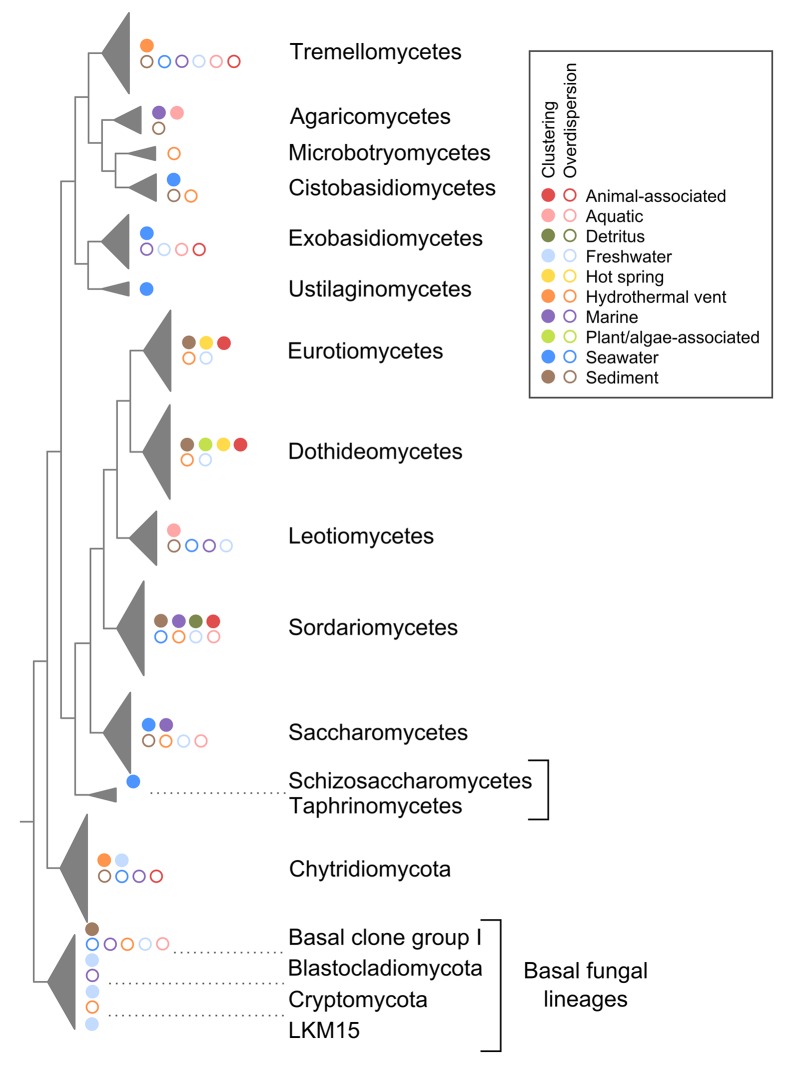
Fig 6. Positions of phylogenetic clustering or overdispersion within the phylogenetic tree tested by NODESIG analyses.
Only significant results are shown. Full circle, clustering signal; open circle, overdispersion.
Taxonomic diversity (Fig 4) and the observed phylogenetic diversity of the subsamples in our dataset (S2 Fig) could also be explained by the diverse ecology of freshwater systems. They can be divided into different groups, which belong either to the lentic systems such as lakes or ponds, or to lotic systems with a continuous water flow. However, even among lakes, environmental conditions such as physicochemical parameters are diverse [76]. Diverse environmental conditions within individual systems lead to distinct demands on fungal adaption and dispersal strategies [77].
1.1.2 Fungal communities in sediment environments
The NRI value of fungal assemblages from sediments was negative (Fig 5). This indicates a tendency (though not significant) of overdispersion at higher taxonomic levels (Fig 6). Thus, several groups of Ascomycota, Basidiomycota, Chytridiomycota, and other basal fungal lineages were reported. None of these formed a dominant group on higher taxonomic rank levels (Figs 2 and 4). Sediments show a large surface area, which is susceptible to abrasion. A key step for microorganisms to survive in this environment is their ability to attach to protected areas of grains or particles [78]. Additionally, fungi have to adapt to sediment innate conditions [6], such as a sharp oxygen gradient, particular nutrient and organic matter absorption capacity, or selective entrainment transport [79,80]. Hence, fungal assemblages of sediments were significantly different than assemblages of nearly all other habitat categories (Fig 3). The environmental pressure especially affected the phylogenetic diversity of Eurotiomycetes, Dothideomycetes, Sordariomycetes and the Basal Clone Group I, as these groups showed significant phylogenetic clustering (Fig 6). Phylotypes of the Basal Clone Group I have so far only exclusively been detected with molecular tools and mainly been found in sediment samples [81–83], with few exceptions [84,85]. Thus, it is unclear if the phylogenetic signal of the Basal Clone Group I is due to environmental pressure, or due to incomplete species data on members of this group. In contrast, Sordariomycetes and Dothideomycetes are among the largest groups of marine Ascomycota (Fig 2) and have a broad spectrum of morphology and functionality [63], where only specific subclades seem to be adapted to life in sediments (S1 Fig).
1.1.3 Fungal communities in seawater environments
The fungal assemblages of seawater (pelagic zone) showed significant phylogenetic clustering (Fig 5). As the larger groups within OTUs0.01 counts (Fig 4), yeast fungi of Basidiomycota and Ascomycota were mainly responsible for this signal (Fig 6). These results suggest that unicellular fungal forms dominate not only deep oceans [9], but also seawater. On the other hand, filamentous fungal forms have been reported to dominate locally [3] or seasonally [4]. They are also positively correlated with phytoplankton abundance, or associated with swimming food sources from detritus particles to nematodes [86]. In comparison to other habitat types, the fungal assemblage of seawater was the most diverse one with respect to the represented fungal richness on class level. One possibility could be intra-habitat specific differences as found for some sub-communities in our dataset (S2 Fig). Geographical and/or environmental barriers might exist. For example, carbon source fluxes are known to have a priming effect in freshwater ecosystems [87], and they could act in a similar manner in seawater as well. Local scale differences in fungal communities, e.g. in pelagic depth zones [88], have also been reported. However, more intensive sampling is needed to support such hypotheses and to determine the factors that impact the geographical dispersal of fungi in seawater at different scales.
1.1.4 Fungal communities in marine hydrothermal vent systems
The hydrothermal vent category was the only habitat with a dominant Basidiomycota fraction. About 50% of OTUs0.01 were assigned to this phylum (Fig 4). Basidiomycota taxa clustered mainly in clades known to contain yeast fungi (S1 Fig), which is the dominant fungal form in the deep sea [9]. They were also responsible, besides Chytridiomycota taxa, for the high NRI and NTI values. High NRI and NTI values indicate whole clade, as well as species exclusion (Figs 5 and 6) [89]. In hydrothermal vent regions, adaptation to high temperature is necessary. However, only a handful of described fungal species are recognized to be thermophilic or are able to adapt to temperatures above 45°C [90]. This could be the reason why we did not find a significant difference between fungal assemblages of hydrothermal vents and hot spring habitats (Fig 3). However, habitats with high temperatures are heterogeneous in terms of their physical and chemical characteristics [90]. The extremely high phylogenetic signal observed for hydrothermal vents, in contrast to hot spring habitats (Fig 5), suggests that environmental conditions other than temperature generate an extreme selection pressure in hydrothermal vents, where only highly adapted fungi can diversify. Our analyses suggest that such highly adapted fungi are found particularly within Tremellomycetes and Chytridiomycota (Fig 6).
1.1.5 Fungal communities associated with marine plant/algae
The fungal community composition found in marine plants/algae did not differ significantly from other habitats, except for freshwater and hydrothermal vent (Fig 3) and was dominated by Eurotiomycetes, Sordariomycetes and Dothideomycetes (Fig 4). These three Ascomycota groups could be detected in nearly all other habitat types (Fig 4). Dothideomycetes form the largest group within the phylum Ascomycota and exhibits a high ecological diversity [91]. Species belonging to this class have dramatically different genome sizes, and functional capabilities of individual species are highly variable [92], probably reflecting the wide spectrum of ecological adaptation in this fungal group.
The life cycles of plant-colonizing fungi often include stages independent of the host [43]. Furthermore, transmission of plant/algae-associated fungi may include spore dispersal, release of hyphal fragments, and passive distribution of infected or dead plant tissue [93]. These may be the reasons why marine plant/algae-associated fungal taxa were also detected in other marine habitats (S1 Fig) resulting into low number of significant differences in fungal assemblages (Fig 3). Our results suggest similarities between aquatic plant/algae-associated fungi and terrestrial root-colonizing fungi, which are detectable for a long time in soil prior to root infection [94]. In contrast to our findings, bacterial communities associated with the green algae Ulva australis are significantly different from communities sampled in the surrounding seawater [95]. Whether only one or both of these scenarios are plausible for aquatic fungal communities associated with plant/algae has to be verified in future studies.
1.1.6 Fungal communities of marine detritus
All fungal sequences of our dataset assigned to detritus habitat clustered exclusively in Eurotiomycetes, Sordariomycetes, and Dothideomycetes (Fig 4). These three fungal classes are the sources of diverse industrially-important enzymes or bioactive compounds [96]. The process of decomposition is very complex and demands specific enzymatic activities. These activities were detected within various functional groups of fungi, but not for all species of those groups [97]. Additionally, fungal elemental composition with respect to the available nutrients also seems to be an important benefit for fungi in this habitat type. Fungi with no elemental homeostasis have a competitive advantage over bacteria on nutrient-depleted detrital resources [98], explaining the limited taxonomic diversity and the phylogenetic clustering in detritus samples (Figs 4 and 5, S1 Fig).
Concluding Remarks
The most widely used marker gene of aquatic fungi in terms of available sequence amount is the 18S rRNA gene (Fig 1). Thus, the evaluation of existing 18S rRNA gene sequence data promises a more profound understanding on aquatic fungal communities. As the 18S rRNA gene sequence can be used for a phylogenetic evaluation apart from hierarchical clustering analysis of the fungal community, it can serve as a highly valuable complementary resource to ITS, providing deeper insights into fungal life in aquatic systems.
Here, we provide the first comprehensive dataset of nearly full-length 18S rRNA gene sequences of aquatic fungi with a rich set of manually curated contextual information, which can be used as a reference dataset including a phylogenetic tree [26]. Ascomycota are prevalent in the dataset (Fig 2). However, the observed Basidiomycota sequence richness and their distribution among diverse habitats (Fig 2) suggest a hitherto underestimation of this phylum in aquatic environments.
This dataset allowed us to show that habitat-specific biomes exist for aquatic fungal communities, as fungal assemblages of 28 out of 45 pairwise habitat comparisons were significantly different (P<0.05) from each other (Fig 3). Freshwater fungal assemblages differed to all other fungal assemblages and were dominated by the lineages of basal fungi. In contrast, the assemblage of the plant/algae habitat only differed significantly from two other assemblages showing that fungi associated to plants/algae are present in the surrounding habitats prior to host colonization.
Phylogenetic signals revealed several cluster effects suggesting that selection pressure within habitats was driven mainly by environmental factors and not by species competition (Fig 5). Thus, although no true environmental marine clade has yet been found [58], diversification processes of fungi in marine systems must have been highly clade specific in some cases.
As aquatic fungal community structures vary to a great extent with habitats, fungal species in aquatic environments seem to be very specific to their habitat, rather than being generalists. A large number of unknown fungal species are regularly detected in other studies [58], which further supports this idea.
Our combined dataset gives the first insights into general patterns of aquatic fungal communities. However, more in-depth aquatic fungal community surveys are urgently needed in order to achieve a more exhaustive perspective on fungal life in aquatic systems.
Supporting Information
The tree was rooted with sequences of eight species of the Choanoflagellates. Branch support was estimated with 100 bootstrap calculations. Only bootstrap values ≥ 50% are indicated. Sequences derived from the KSMP-Kiel culture collection are printed in bold. Color code, contextual data: colored background of fungal taxa, habitat occurrence of fungal taxa; colored lines beside sequence full name, geographical origin of fungal taxa. Subcategories of the geographical origin displayed as letter to the right of the colored stripe. *, wrong full name annotation of sequence.
(PDF)
Length of boxes shows variation of phylogenetic distances inferred from pairwise sample comparisons. Black lines, median value. Different letters indicate significant differences (P<0.05) as determined by one-way ANOVA test followed by a Scheffé post hoc test. Number of sub-communities/habitat: four (except sediment: seven).
(PDF)
After inspecting all BLASTN hits, sequences were assigned to a submission name.
(PDF)
Fungal sequences were subjected to BLASTN analysis and checked for their taxonomic placement in the eukaryotic guide-tree of the SILVA release 111. Sequences were classified depending on combined results from the methods mentioned above, as well as literature searches.
(PDF)
Acknowledgments
We thank two anonymous reviewers for helpful comments on our manuscript. This study was supported by the University of Bremen, Germany. We thank Cecilia Duprè and Martin Diekmann for fruitful discussions about biodiversity indices. The authors are grateful to Andrea Schneider for processing the molecular work with the strains from the KSMP-Kiel. We also thanks the the Institute of Clinical Molecular Biology in Kiel (Germany) for providing Sanger sequencing, which was in part supported by the DFG Cluster of Excellence “Inflammation at Interfaces” and “Future Ocean”.
Data Availability
Sequences from fungal strains obtained during this study were submitted to the GenBank database and were assigned to accession numbers KJ939309 – KJ939313 and KM096133 – KM096374, respectively. All data supporting this article can be accessed via the Dryad Digital Repository (http://dx.doi.org/10.5061/dryad.7fv64). The following files can be found: (i) a newick-formatted file of the phylogenetic tree, (ii) a file including all information on sequences used in this study and assignment of sequences to the diverse habitat categories, (iii) a list indicating which sequences cluster together at a 99% similarity level, and (iv) OTU count per habitat category.
Funding Statement
The study was supported by the University of Bremen, Germany (http://www.uni-bremen.de/). Sanger sequencing partly supported by the German Research Foundation (DFG) (http://www.dfg.de/en/) Cluster of Excellence “Inflammation at Interfaces” and “Future Ocean”.
References
- 1. Kirchman DL (2008) Microbial ecology of the oceans Hoboken, USA: John Wiley & Sons. [Google Scholar]
- 2. Raghukumar C (2012) Biology of marine fungi Heidelberg: Springer. [Google Scholar]
- 3. Gao Z, Johnson ZI, Wang GY (2010) Molecular characterization of the spatial diversity and novel lineages of mycoplankton in Hawaiian coastal waters. Isme Journal 4: 111–120. 10.1038/ismej.2009.87 [DOI] [PubMed] [Google Scholar]
- 4. Gutierrez MH, Pantoja S, Tejos E, Quinones RA (2011) The role of fungi in processing marine organic matter in the upwelling ecosystem off Chile. Marine Biology 158: 205–219. [Google Scholar]
- 5. Kagami M, Amano Y, Ishii N (2012) Community structure of planktonic fungi and the impact of parasitic chytrids on phytoplankton in Lake Inba, Japan. Microbial Ecology 63: 358–368. 10.1007/s00248-011-9913-9 [DOI] [PubMed] [Google Scholar]
- 6. Orsi W, Biddle JF, Edgcomb V (2013) Deep sequencing of subseafloor eukaryotic rRNA reveals active fungi across marine subsurface provinces. PLoS ONE 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Prosser JI (2012) Ecosystem processes and interactions in a morass of diversity. FEMS Microbiology Ecology 81: 507–519. 10.1111/j.1574-6941.2012.01435.x [DOI] [PubMed] [Google Scholar]
- 8. Konopka A (2009) What is microbial community ecology? ISME Journal 3: 1223–1230. 10.1038/ismej.2009.88 [DOI] [PubMed] [Google Scholar]
- 9. Bass D, Howe A, Brown N, Barton H, Demidova M, et al. (2007) Yeast forms dominate fungal diversity in the deep oceans. Proceedings of the Royal Society B-Biological Sciences 274: 3069–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Le Calvez T, Burgaud G, Mahe S, Barbier G, Vandenkoornhuyse P (2009) Fungal diversity in Deep-Sea hydrothermal ecosystems. Applied and Environmental Microbiology 75: 6415–6421. 10.1128/AEM.00653-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jebaraj CS, Raghukumar C, Behnke A, Stoeck T (2010) Fungal diversity in oxygen-depleted regions of the Arabian Sea revealed by targeted environmental sequencing combined with cultivation. FEMS Microbiology Ecology 71: 399–412. 10.1111/j.1574-6941.2009.00804.x [DOI] [PubMed] [Google Scholar]
- 12. Lefevre E, Letcher PM, Powell MJ (2012) Temporal variation of the small eukaryotic community in two freshwater lakes: emphasis on zoosporic fungi. Aquatic Microbial Ecology 67: 91–105. [Google Scholar]
- 13. Monchy S, Sanciu G, Jobard M, Rasconi S, Gerphagnon M, et al. (2011) Exploring and quantifying fungal diversity in freshwater lake ecosystems using rDNA cloning/sequencing and SSU tag pyrosequencing. Environmental Microbiology 13: 1433–1453. 10.1111/j.1462-2920.2011.02444.x [DOI] [PubMed] [Google Scholar]
- 14. Duarte S, Barlocher F, Trabulo J, Cassio F, Pascoal C (2015) Stream-dwelling fungal decomposer communities along a gradient of eutrophication unraveled by 454 pyrosequencing. Fungal Diversity 70: 127–148. [Google Scholar]
- 15. Amend AS, Barshis DJ, Oliver TA (2012) Coral-associated marine fungi form novel lineages and heterogeneous assemblages. ISME Journal 6: 1291–1301. 10.1038/ismej.2011.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chukanhom K, Hatai K (2004) Freshwater fungi isolated from eggs of the common carp (Cyprinus carpio) in Thailand. Mycoscience 45: 42–48. [Google Scholar]
- 17. Sakayaroj J, Preedanon S, Supaphon O, Jones EBG, Phongpaichit S (2010) Phylogenetic diversity of endophyte assemblages associated with the tropical seagrass Enhalus acoroides in Thailand. Fungal Diversity 42: 27–45. [Google Scholar]
- 18. Magnes M, Hafellner J (1991) Ascomyceten auf Gefäßpflanzen an Ufern von Gebirgsseen in den Ostalpen. Biblioth Mycol 139: 1–182. [Google Scholar]
- 19. Zuccaro A, Schulz B, Mitchell JI (2003) Molecular detection of ascomycetes associated with Fucus serratus . Mycological Research 107: 1451–1466. [DOI] [PubMed] [Google Scholar]
- 20. Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, et al. (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences of the United States of America 109: 6241–6246. 10.1073/pnas.1117018109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yilmaz P, Kottmann R, Field D, Knight R, Cole JR, et al. (2011) Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nature Biotechnology 29: 415–420. 10.1038/nbt.1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nakagiri A (2012) Culture collections and maintenance of marine fungi In: Jones EBG, Pang KL, editors. Marine fungi. Berlin Boston: Walter de Gruyter; pp. 501–508. [Google Scholar]
- 23. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, et al. (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Research 41: D590–D596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pruesse E, Peplies J, Glockner FO (2012) SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28: 1823–1829. 10.1093/bioinformatics/bts252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ludwig W, Strunk O, Westram R, Richter L, Meier H, et al. (2004) ARB: a software environment for sequence data. Nucleic Acids Research 32: 1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Panzer K, Yilmaz P, Weiß M, Reich L, Richter M, et al. (2015) Data from: Identification of habitat-specific biomes of aquatic fungal communities using a comprehensive nearly full-length 18S rRNA dataset enriched with contextual data. Dryad Digital Repository. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wiese J, Ohlendorf B, Blumel M, Schmaljohann R, Imhoff JF (2011) Phylogenetic identification of fungi isolated from the marine sponge Tethya aurantium and identification of their secondary metabolites. Marine Drugs 9: 561–585. 10.3390/md9040561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gomes NCM, Fagbola O, Costa R, Rumjanek NG, Buchner A, et al. (2003) Dynamics of fungal communities in bulk and maize rhizosphere soil in the tropics (vol 69, pg 3758, 2003). Applied and Environmental Microbiology 69: 5737–5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. Journal of Molecular Biology 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 30.Morshed A, Aryal J, Dutta R (2013) Environmental spatio-temporal ontology for the Linked open data cloud. 2013 12th Ieee International Conference on Trust, Security and Privacy in Computing and Communications (Trustcom 2013): 1907–1912.
- 31. Stamatakis A, Ludwig T, Meier H (2005) RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21: 456–463. [DOI] [PubMed] [Google Scholar]
- 32. Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, et al. (2010) QIIME allows analysis of high-throughput community sequencing data. Nature Methods 7: 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Anderson IC, Campbell CD, Prosser JI (2003) Potential bias of fungal 18S rDNA and internal transcribed spacer polymerase chain reaction primers for estimating fungal biodiversity in soil. Environmental Microbiology 5: 36–47. [DOI] [PubMed] [Google Scholar]
- 35. Hamady M, Lozupone C, Knight R (2010) Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. Isme Journal 4: 17–27. 10.1038/ismej.2009.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Evans SN, Matsen FA (2012) The phylogenetic Kantorovich-Rubinstein metric for environmental sequence samples. Journal of the Royal Statistical Society Series B-Statistical Methodology 74: 569–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goecks J, Nekrutenko A, Taylor J, Team G (2010) Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biology 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, et al. (2005) Galaxy: A platform for interactive large-scale genome analysis. Genome Research 15: 1451–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Team RDC (2008) R: A language and environment for statistical computing Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 40. Webb CO, Ackerly DD, Kembel SW (2008) Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics 24: 2098–2100. 10.1093/bioinformatics/btn358 [DOI] [PubMed] [Google Scholar]
- 41. Jobard M, Rasconi S, Sime-Ngando T (2010) Diversity and functions of microscopic fungi: a missing component in pelagic food webs. Aquatic Sciences 72: 255–268. [Google Scholar]
- 42. Panno L, Bruno M, Voyron S, Anastasi A, Gnavi G, et al. (2013) Diversity, ecological role and potential biotechnological applications of marine fungi associated to the seagrass Posidonia oceanica . New Biotechnology 30: 685–694. 10.1016/j.nbt.2013.01.010 [DOI] [PubMed] [Google Scholar]
- 43. Kohlmeyer J, Kohlmeyer E (1979) Marine Mycology: The higher fungi New York: Academic Press. [Google Scholar]
- 44. Yuen TK, Hyde KD, Hodgkiss IJ (1999) Interspecific interactions among tropical and subtropical freshwater fungi. Microbial Ecology 37: 257–262. [DOI] [PubMed] [Google Scholar]
- 45. Burgaud G, Le Calvez T, Arzur D, Vandenkoornhuyse P, Barbier G (2009) Diversity of culturable marine filamentous fungi from deep-sea hydrothermal vents. Environmental Microbiology 11: 1588–1600. 10.1111/j.1462-2920.2009.01886.x [DOI] [PubMed] [Google Scholar]
- 46. Lai XT, Cao LX, Tan HM, Fang S, Huang YL, et al. (2007) Fungal communities from methane hydrate-bearing deep-sea marine sediments in South China Sea. ISME Journal 1: 756–762. [DOI] [PubMed] [Google Scholar]
- 47. Arfi Y, Buee M, Marchand C, Levasseur A, Record E (2012) Multiple markers pyrosequencing reveals highly diverse and host-specific fungal communities on the mangrove trees Avicennia marina and Rhizophora stylosa . FEMS Microbiology Ecology 79: 433–444. 10.1111/j.1574-6941.2011.01236.x [DOI] [PubMed] [Google Scholar]
- 48. Wang X, Singh P, Gao Z, Zhang XB, Johnson ZI, et al. (2014) Distribution and diversity of planktonic fungi in the West Pacific Warm Pool. PLoS ONE 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bellemain E, Davey ML, Kauserud H, Epp LS, Boessenkool S, et al. (2013) Fungal palaeodiversity revealed using high-throughput metabarcoding of ancient DNA from arctic permafrost. Environmental Microbiology 15: 1176–1189. 10.1111/1462-2920.12020 [DOI] [PubMed] [Google Scholar]
- 50. Arfi Y, Marchand C, Wartel M, Record E (2012) Fungal diversity in anoxic-sulfidic sediments in a mangrove soil. Fungal Ecology 5: 282–285. [Google Scholar]
- 51. Lefevre E, Bardot C, Noel C, Carrias JF, Viscogliosi E, et al. (2007) Unveiling fungal zooflagellates as members of freshwater picoeukaryotes: evidence from a molecular diversity study in a deep meromictic lake. Environmental Microbiology 9: 61–71. [DOI] [PubMed] [Google Scholar]
- 52. Bik HM, Sung W, De Ley P, Baldwin JG, Sharma J, et al. (2012) Metagenetic community analysis of microbial eukaryotes illuminates biogeographic patterns in deep-sea and shallow water sediments. Molecular Ecology 21: 1048–1059. 10.1111/j.1365-294X.2011.05297.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stoeck T, Bass D, Nebel M, Christen R, Jones MDM, et al. (2010) Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Molecular Ecology 19: 21–31. 10.1111/j.1365-294X.2009.04480.x [DOI] [PubMed] [Google Scholar]
- 54. Taib N, Mangot JF, Domaizon I, Bronner G, Debroas D (2013) Phylogenetic affiliation of SSU rRNA genes generated by massively parallel sequencing: New insights into the freshwater protist diversity. PLoS ONE 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nikolcheva LG, Cockshutt AM, Barlocher F (2003) Determining diversity of freshwater fungi on decaying leaves: Comparison of traditional and molecular approaches. Applied and Environmental Microbiology 69: 2548–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Richards TA, Jones MDM, Leonard G, Bass D (2012) Marine fungi: Their ecology and molecular diversity. Annual Review of Marine Science, Vol 4 4: 495–522. [DOI] [PubMed] [Google Scholar]
- 57. Kõljalg U, Larsson KH, Abarenkov K, Nilsson RH, Alexander IJ, et al. (2005) UNITE: a database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytologist 166: 1063–1068. [DOI] [PubMed] [Google Scholar]
- 58. Manohar CS, Raghukumar C (2013) Fungal diversity from various marine habitats deduced through culture-independent studies. FEMS Microbiology Letters 341: 69–78. 10.1111/1574-6968.12087 [DOI] [PubMed] [Google Scholar]
- 59. Jones EBG (2011) Are there more marine fungi to be described? Botanica Marina 54: 343–354. [Google Scholar]
- 60. Singh P, Raghukumar C, Verma P, Shouche Y (2011) Fungal community analysis in the Deep-Sea sediments of the Central Indian Basin by culture-independent approach. Microbial Ecology 61: 507–517. 10.1007/s00248-010-9765-8 [DOI] [PubMed] [Google Scholar]
- 61. Martiny AC, Treseder K, Pusch G (2013) Phylogenetic conservatism of functional traits in microorganisms. ISME Journal 7: 830–838. 10.1038/ismej.2012.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shearer CA, Descals E, Kohlmeyer B, Kohlmeyer J, Marvanova L, et al. (2007) Fungal biodiversity in aquatic habitats. Biodiversity and Conservation 16: 49–67. [Google Scholar]
- 63. Jones EB, Pang KL (2012) Marine fungi and fungal-like organisms Berlin/Boston: De Gruyter. [Google Scholar]
- 64. Jones EBG, Fell JW (2012) Basidiomycota In: Jones EBG, Pang KL, editors. Marine Fungiand fungal-like organisms. Berlin Boston: Walter de Gruyter; pp. 49–64. [Google Scholar]
- 65. Jones MDM, Forn I, Gadelha C, Egan MJ, Bass D, et al. (2011) Discovery of novel intermediate forms redefines the fungal tree of life. Nature 474: 200–U234. 10.1038/nature09984 [DOI] [PubMed] [Google Scholar]
- 66. Livermore JA, Mattes TE (2013) Phylogenetic detection of novel Cryptomycota in an Iowa (United States) aquifer and from previously collected marine and freshwater targeted high-throughput sequencing sets. Environmental Microbiology 15: 2333–2341. 10.1111/1462-2920.12106 [DOI] [PubMed] [Google Scholar]
- 67. Logares R, Brate J, Bertilsson S, Clasen JL, Shalchian-Tabrizi K, et al. (2009) Infrequent marine-freshwater transitions in the microbial world. Trends in Microbiology 17: 414–422. 10.1016/j.tim.2009.05.010 [DOI] [PubMed] [Google Scholar]
- 68. Sparrow FK (1960) Aquatic Phycomycetes. Ann Arbor, USA: University of Michigan Press. [Google Scholar]
- 69. Barr DJS (2001) Chytridiomycota In: McLaughlin DJ, McLaughlin EG, Lemke P, editors. The Mycota. New York, NY: Springer-Verlag; pp. 93–112. [Google Scholar]
- 70. Fisher SG, Likens GE (1973) Energy flow in Bear Brook, New Hampshire—Integrative approach to stream ecosystem metabolism. Ecological Monographs 43: 421–439. [Google Scholar]
- 71. Otto S, Balzer W (1998) Release of dissolved organic carbon (DOC) from sediments of the NW European Continental Margin (Goban Spur) and its significance for benthic carbon cycling. Progress in Oceanography 42: 127–144. [Google Scholar]
- 72. Sowerby A, Emmett BA, Williams D, Beier C, Evans CD (2010) The response of dissolved organic carbon (DOC) and the ecosystem carbon balance to experimental drought in a temperate shrubland. European Journal of Soil Science 61: 697–709. [Google Scholar]
- 73. Baldy V, Gessner MO, Chauvet E (1995) Bacteria, fungi and the breakdown of leaf-litter in a large river. Oikos 74: 93–102. [Google Scholar]
- 74. Gleason FH, Kagami M, Lefèvre E, Sime-Ngando T (2008) The ecology of chytrids in aquatic ecosystems: roles in food web dynamics. Fungal Biology Reviews 22: 17–25. [Google Scholar]
- 75. Gessner MO, Chauvet E, Dobson M (1999) A perspective on leaf litter breakdown in streams. Oikos 85: 377–384. [Google Scholar]
- 76. Goncalves VN, Vaz ABM, Rosa CA, Rosa LH (2012) Diversity and distribution of fungal communities in lakes of Antarctica. FEMS Microbiology Ecology 82: 459–471. 10.1111/j.1574-6941.2012.01424.x [DOI] [PubMed] [Google Scholar]
- 77. Goh TK, Hyde KD (1996) Biodiversity of freshwater fungi. Journal of Industrial Microbiology & Biotechnology 17: 328–345. [Google Scholar]
- 78. Findlay RH, Trexler MB, Guckert JB, White DC (1990) Laboratory study of disturbance in marine-sediments—Response of a microbial community. Marine Ecology Progress Series 62: 121–133. [Google Scholar]
- 79. Avramidis P, Bekiari V, Kontopoulos N, Kokidis N (2013) Shallow coastal lagoon sediment characteristics and water physicochemical parameters—Myrtari Lagoon, Mediterranean Sea, Western Greece. Fresenius Environmental Bulletin 22: 1628–1635. [Google Scholar]
- 80. Hedges JI, Hu FS, Devol AH, Hartnett HE, Tsamakis E, et al. (1999) Sedimentary organic matter preservation: A test for selective degradation under oxic conditions. American Journal of Science 299: 529–555. [Google Scholar]
- 81. Nagahama T, Takahashi E, Nagano Y, Abdel-Wahab MA, Miyazaki M (2011) Molecular evidence that deep-branching fungi are major fungal components in deep-sea methane cold-seep sediments. Environmental Microbiology 13: 2359–2370. 10.1111/j.1462-2920.2011.02507.x [DOI] [PubMed] [Google Scholar]
- 82. Takishita K, Yubuki N, Kakizoe N, Inagaki Y, Maruyama T (2007) Diversity of microbial eukaryotes in sediment at a deep-sea methane cold seep: surveys of ribosomal DNA libraries from raw sediment samples and two enrichment cultures. Extremophiles 11: 563–576. [DOI] [PubMed] [Google Scholar]
- 83. Tian F, Yu Y, Chen B, Li HR, Yao YF, et al. (2009) Bacterial, archaeal and eukaryotic diversity in Arctic sediment as revealed by 16S rRNA and 18S rRNA gene clone libraries analysis. Polar Biology 32: 93–103. [Google Scholar]
- 84. Stoeck T, Taylor GT, Epstein SS (2003) Novel eukaryotes from the permanently anoxic Cariaco Basin (Caribbean sea). Applied and Environmental Microbiology 69: 5656–5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Stoeck T, Hayward B, Taylor GT, Varela R, Epstein SS (2006) A multiple PCR-primer approach to access the microeukaryotic diversity in environmental samples. Protist 157: 31–43. [DOI] [PubMed] [Google Scholar]
- 86. Jones EBG (1971) Aquatic fungi In: Booth C, editor. Methods in microbiology. New York: Academic Press; pp. 731–742. [Google Scholar]
- 87. Bianchi TS (2012) The role of terrestrially derived organic carbon in the coastal ocean: A changing paradigm and the priming effect (vol 108, pg 19473, 2011). Proceedings of the National Academy of Sciences of the United States of America 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Bachy C, Lopez-Garcia P, Vereshchaka A, Moreira D (2011) Diversity and vertical distribution of microbial eukaryotes in the snow, sea ice and seawater near the North Pole at the end of the polar night. Frontiers in Microbiology 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Lozupone CA, Knight R (2008) Species divergence and the measurement of microbial diversity. FEMS Microbiology Reviews 32: 557–578. 10.1111/j.1574-6976.2008.00111.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Maheshwari R, Bharadwaj G, Bhat MK (2000) Thermophilic fungi: Their physiology and enzymes. Microbiology and Molecular Biology Reviews 64: 461–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hyde KD, Jones EBG, Liu JK, Ariyawansa H, Boehm E, et al. (2013) Families of Dothideomycetes . Fungal Diversity 63: 1–313. [Google Scholar]
- 92. Ohm RA, Feau N, Henrissat B, Schoch CL, Horwitz BA, et al. (2012) Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi. Plos Pathogens 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Monk KA, Samuels GJ (1990) Mycophagy in grasshoppers (Orthoptera, Acrididae) in Indo-Malayan rain forests. Biotropica 22: 16–21. [Google Scholar]
- 94. Danielsen L, Thurmer A, Meinicke P, Buee M, Morin E, et al. (2012) Fungal soil communities in a young transgenic poplar plantation form a rich reservoir for fungal root communities. Ecology and Evolution 2: 1935–1948. 10.1002/ece3.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Burke C, Thomas T, Lewis M, Steinberg P, Kjelleberg S (2011) Composition, uniqueness and variability of the epiphytic bacterial community of the green alga Ulva australis . ISME Journal 5: 590–600. 10.1038/ismej.2010.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Velmurugan N, Lee YS (2012) Enzymes from marine fungi: current research and future prospects In: Jones EBG, Pang KL, editors. Marine fungi and fungal-like organisms. Berlin Boston: Walter de Gruyter; pp. 441–474. [Google Scholar]
- 97.Sridhar KR (2012) Decomposition of materials in the sea. In: Jones EBG, Pang KL, editors. Marine fungi and fungal-like organisms. pp. 475–500.
- 98. Danger M, Chauvet E (2013) Elemental composition and degree of homeostasis of fungi: are aquatic hyphomycetes more like metazoans, bacteria or plants? Fungal Ecology 6: 453–457. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The tree was rooted with sequences of eight species of the Choanoflagellates. Branch support was estimated with 100 bootstrap calculations. Only bootstrap values ≥ 50% are indicated. Sequences derived from the KSMP-Kiel culture collection are printed in bold. Color code, contextual data: colored background of fungal taxa, habitat occurrence of fungal taxa; colored lines beside sequence full name, geographical origin of fungal taxa. Subcategories of the geographical origin displayed as letter to the right of the colored stripe. *, wrong full name annotation of sequence.
(PDF)
Length of boxes shows variation of phylogenetic distances inferred from pairwise sample comparisons. Black lines, median value. Different letters indicate significant differences (P<0.05) as determined by one-way ANOVA test followed by a Scheffé post hoc test. Number of sub-communities/habitat: four (except sediment: seven).
(PDF)
After inspecting all BLASTN hits, sequences were assigned to a submission name.
(PDF)
Fungal sequences were subjected to BLASTN analysis and checked for their taxonomic placement in the eukaryotic guide-tree of the SILVA release 111. Sequences were classified depending on combined results from the methods mentioned above, as well as literature searches.
(PDF)
Data Availability Statement
Sequences from fungal strains obtained during this study were submitted to the GenBank database and were assigned to accession numbers KJ939309 – KJ939313 and KM096133 – KM096374, respectively. All data supporting this article can be accessed via the Dryad Digital Repository (http://dx.doi.org/10.5061/dryad.7fv64). The following files can be found: (i) a newick-formatted file of the phylogenetic tree, (ii) a file including all information on sequences used in this study and assignment of sequences to the diverse habitat categories, (iii) a list indicating which sequences cluster together at a 99% similarity level, and (iv) OTU count per habitat category.