Our aim is to review and explain the capabilities and performance of currently available approaches for segmentation of lungs with pathologic conditions on chest CT images, with illustrations to give radiologists a better understanding of potential choices for decision support in everyday practice.
Abstract
The computer-based process of identifying the boundaries of lung from surrounding thoracic tissue on computed tomographic (CT) images, which is called segmentation, is a vital first step in radiologic pulmonary image analysis. Many algorithms and software platforms provide image segmentation routines for quantification of lung abnormalities; however, nearly all of the current image segmentation approaches apply well only if the lungs exhibit minimal or no pathologic conditions. When moderate to high amounts of disease or abnormalities with a challenging shape or appearance exist in the lungs, computer-aided detection systems may be highly likely to fail to depict those abnormal regions because of inaccurate segmentation methods. In particular, abnormalities such as pleural effusions, consolidations, and masses often cause inaccurate lung segmentation, which greatly limits the use of image processing methods in clinical and research contexts. In this review, a critical summary of the current methods for lung segmentation on CT images is provided, with special emphasis on the accuracy and performance of the methods in cases with abnormalities and cases with exemplary pathologic findings. The currently available segmentation methods can be divided into five major classes: (a) thresholding-based, (b) region-based, (c) shape-based, (d) neighboring anatomy–guided, and (e) machine learning–based methods. The feasibility of each class and its shortcomings are explained and illustrated with the most common lung abnormalities observed on CT images. In an overview, practical applications and evolving technologies combining the presented approaches for the practicing radiologist are detailed.
©RSNA, 2015
Introduction
Computed tomography (CT) is a vital diagnostic modality widely used across a broad spectrum of clinical indications for diagnosis and image-guided procedures. Nearly all CT images are now digital, thus allowing increasingly sophisticated image reconstruction techniques as well as image analysis methods within or as a supplement to picture archiving and communication systems (1). The first and fundamental step for pulmonary image analysis is the segmentation of the organ of interest (lungs); in this step, the organ is detected, and its anatomic boundaries are delineated, either automatically or manually (2). Errors in organ segmentation would generate false information with regard to subsequent identification of diseased areas and various other clinical quantifications, so accurate segmentation is a necessity.
The purpose of this article is to review and explain the capabilities and performance of currently available approaches for segmenting lungs with pathologic conditions on chest CT images, with illustrations to provide radiologists with a better understanding of potential choices for decision support in everyday practice. First, object segmentation is defined and explained, followed by summaries of the five major classes of lung segmentation: (a) thresholding-based, (b) region-based, (c) shape-based, (d) neighboring anatomy–guided, and (e) machine learning–based methods. Then hybrid approaches for generic lung segmentation in clinical practice are covered, as well as methods for evaluating the efficacy of segmentation. Finally, the current and future use of segmentation software for clinical diagnosis is discussed.
What Is Object Segmentation?
The aim of medical image segmentation is to extract quantitative information (eg, volumetric data, morphometric data, textural patterns–related information) with regard to an organ of interest or a lesion within the organ. In general, a segmentation problem can be considered as consisting of two related tasks: object recognition and object delineation. Object recognition is the determination of the target object’s whereabouts on the image or its location, whereas object delineation draws the object’s spatial extent and composition. Although object recognition is known as a high-level process, object delineation refers to a low-level process; and it is well known that humans are superior to computers at performing high-level vision tasks (3–6) such as object recognition. On the other hand, computational methods are better for low-level tasks such as object delineation and finding the exact spatial extent of the object (3,4,7). Image segmentation in this high- to low-level hierarchy is a combination of recognition and delineation steps (8). This hierarchical relation between the object recognition and object delineation steps is illustrated with an example of a pulmonary CT image and its segmentation (Fig 1). Note that in the object recognition step (Fig 1a), the left and right lung fields are identified through user interaction (ie, a high-level task); and in the object delineation step (Fig 1b), user-provided information is processed to find the exact boundary of the lung fields (ie, a low-level task).
Figure 1a.
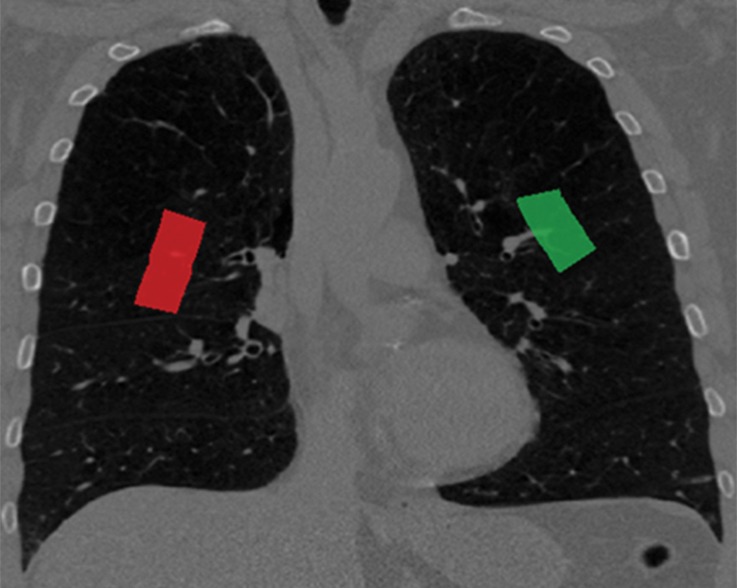
Example of the tasks of object recognition (a) and object delineation (b) for the left lung (green) and right lung (red) on a coronal CT image.
Figure 1b.
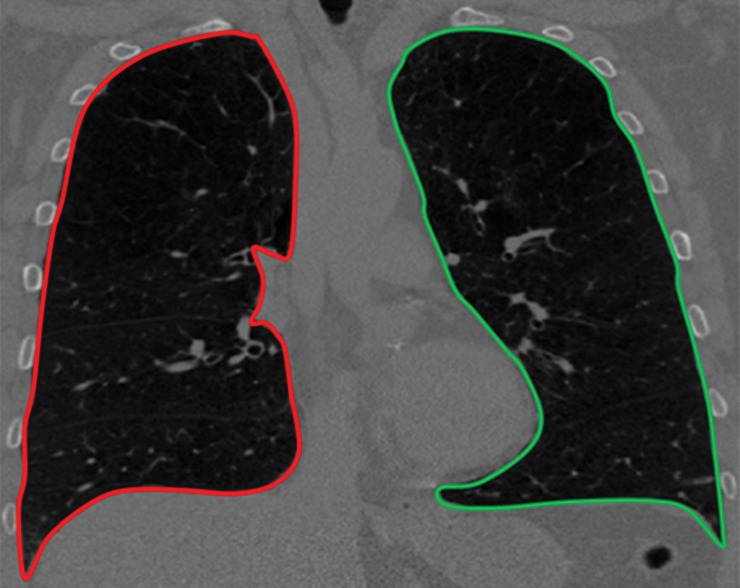
Example of the tasks of object recognition (a) and object delineation (b) for the left lung (green) and right lung (red) on a coronal CT image.
Because in vivo image analysis of lung diseases has become a necessity for clinical and research applications, it is important for radiologists to become familiar with the opportunities and challenges involved in automated segmentation of lungs on CT images. Because of recent technical advances in radiology and informatics, it may even be possible in the near future for radiologists to quantitatively assess disease severity as a percentage of total lung volume, which may influence how radiologists characterize the extent, severity, and morphologic evolution of the disease with longitudinal CT examinations (9).
Segmentation of lung fields is particularly challenging because differences in pulmonary inflation with an elastic chest wall can create large variability in volumes and margins when attempting to automate the segmentation of lungs. Moreover, the presence of disease in the lungs can interfere with software attempting to locate lung margins. For example, a consolidation along the pleural margin of the lungs may generate an erroneous delineation in which the consolidation is treated as outside the lungs because its attenuation characteristics are similar to other aspects of the soft tissue of nearby anatomic structures.
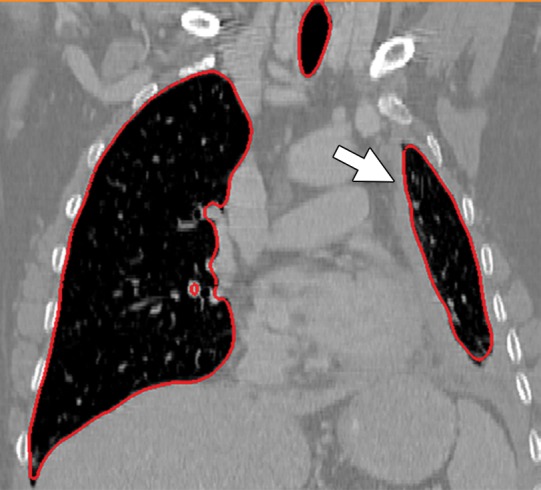
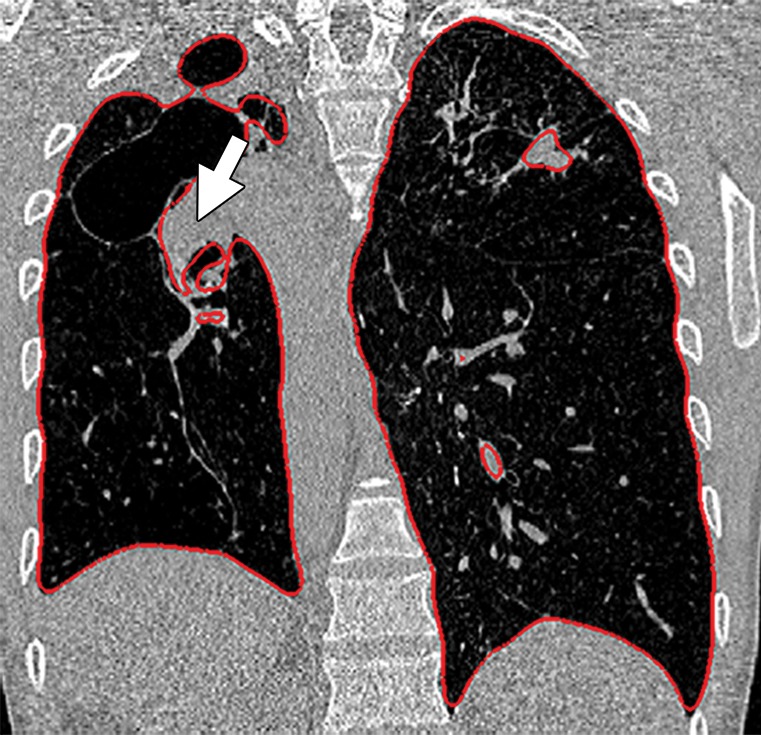
Historically, nearly all image segmentation approaches for lungs functioned well only with absent or minimal lung pathologic conditions. Those segmentation methods have been shown to be effective in the calculation of lung volume and the initiation of computer-aided detection systems (10), which is considered in a wide range of clinical applications (10–21). However, those segmentation methods fail to perform efficiently when a pathologic condition or abnormality is present in moderate to marked lung volumes or demonstrates complex patterns of attenuation (11–13,16–18). For example, cavities and consolidation can lead to inaccurate boundary identification (Fig 2). Similarly, the presence of pneumothorax or pleural effusion on a CT image can greatly distort the results of automated segmentation, hence leading to incorrect quantification (Fig 3).
Figure 2a.
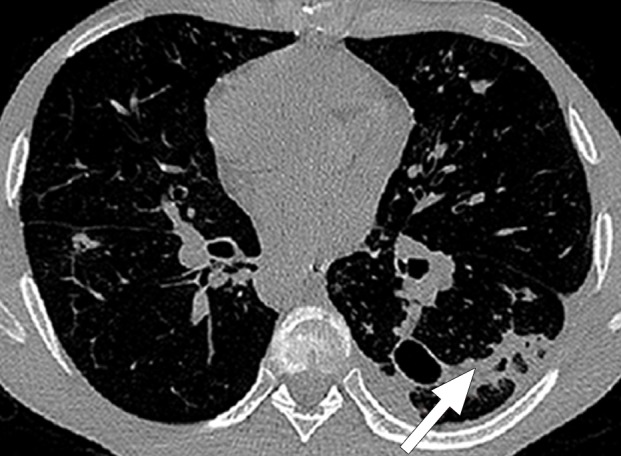
Inaccurate boundary identification. Axial (a, b) and coronal (c, d) CT images show that cavities and consolidation (arrow in a, c) can lead to inaccurate segmentation (red contours in b, d).
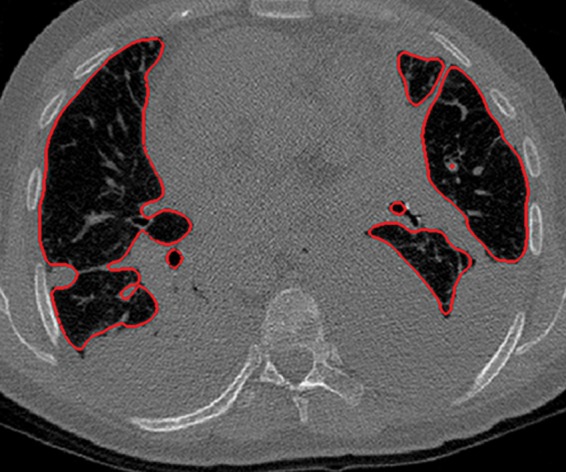
Figure 3a.
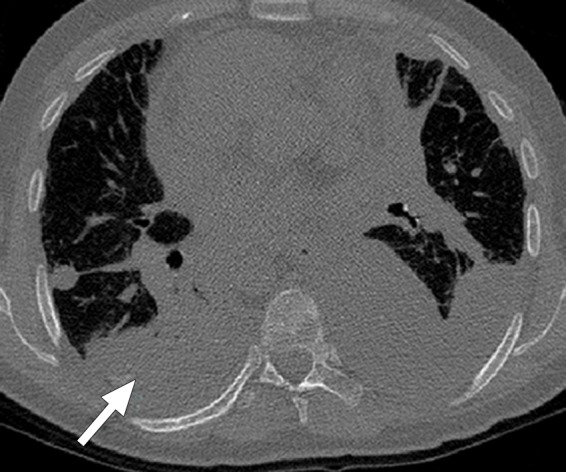
Distorted automated segmentation. Axial (a, b) and coronal (c, d) CT images show that pleural effusions (arrow in a, c) can lead to inaccurate segmentation (red contours in b, d).
Figure 2b.
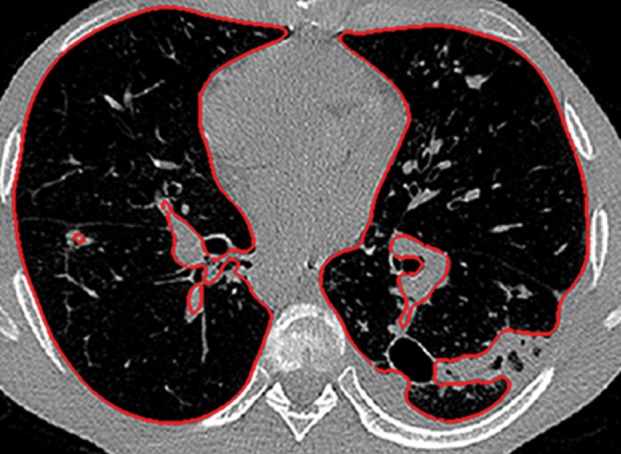
Inaccurate boundary identification. Axial (a, b) and coronal (c, d) CT images show that cavities and consolidation (arrow in a, c) can lead to inaccurate segmentation (red contours in b, d).
Figure 2c.
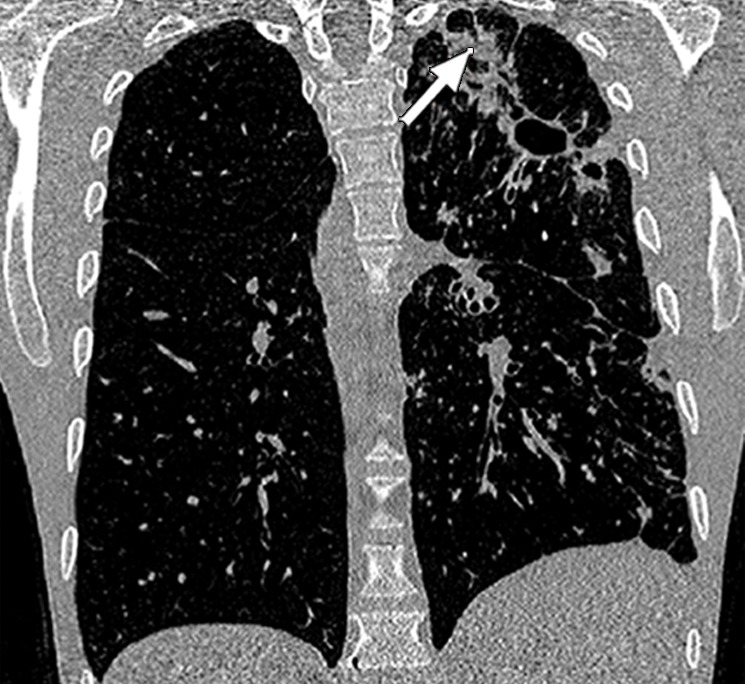
Inaccurate boundary identification. Axial (a, b) and coronal (c, d) CT images show that cavities and consolidation (arrow in a, c) can lead to inaccurate segmentation (red contours in b, d).
Figure 2d.
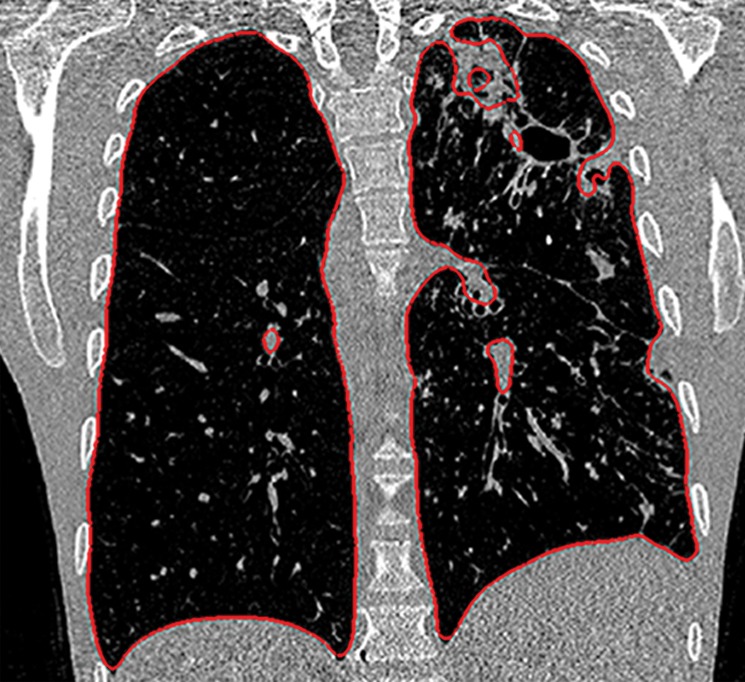
Inaccurate boundary identification. Axial (a, b) and coronal (c, d) CT images show that cavities and consolidation (arrow in a, c) can lead to inaccurate segmentation (red contours in b, d).
Figure 3b.
Distorted automated segmentation. Axial (a, b) and coronal (c, d) CT images show that pleural effusions (arrow in a, c) can lead to inaccurate segmentation (red contours in b, d).
Figure 3c.
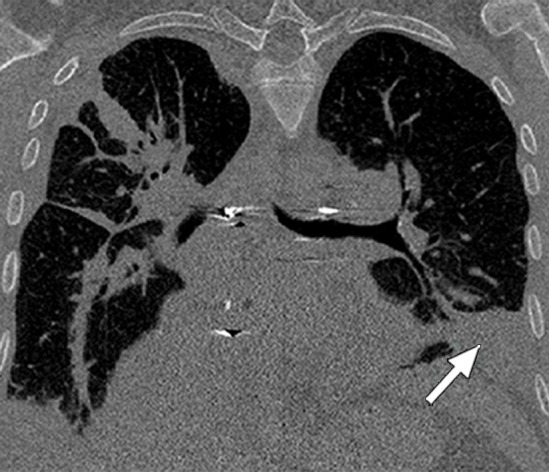
Distorted automated segmentation. Axial (a, b) and coronal (c, d) CT images show that pleural effusions (arrow in a, c) can lead to inaccurate segmentation (red contours in b, d).
Figure 3d.
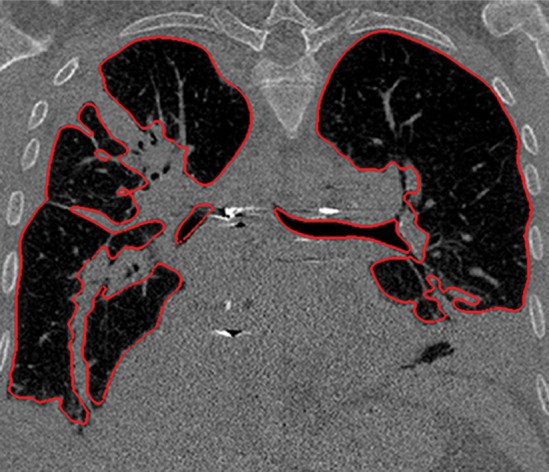
Distorted automated segmentation. Axial (a, b) and coronal (c, d) CT images show that pleural effusions (arrow in a, c) can lead to inaccurate segmentation (red contours in b, d).
Currently, no single segmentation method achieves a globally optimal performance for all cases. Although specialized methods (22–24) that are designed for a particular subset of abnormalities have been shown to be successful, only a few attempts at generic segmentation methods have been made so far (20,25,26). This fragmentation of available solutions contributes to the gap between clinical practitioners, who are the end users of radiologic image analysis techniques, and the informatics experts.
Herein, we intend to bridge this gap between practicing radiologists and informatics experts by first briefly providing an overview of the current lung segmentation methods on CT images. Then, after providing a description and the background of object segmentation and the Fleischner terminology for lung pathologic conditions, we present five major classes of lung segmentation methods: (a) thresholding-based, (b) region-based, (c) shape-based, (d) neighboring anatomy–guided, and (e) machine learning–based methods. Our focus in this review is to succinctly present the advantages and disadvantages of these approaches in terms of segmentation accuracy, ease of use, and computational cost (ie, memory or processor requirements, time needed for producing outputs). Therefore, a full-length description of each method is beyond the scope of this article, and interested readers are referred to relevant literature with the reference citations. Furthermore, we limit our review to the lung segmentation methods, not the quantification and detection of lung abnormalities, which can be a topic of review of computer-aided detection systems in lung diseases. However, the relationship between these two tasks of segmentation and quantification and also the necessary background on quantification and detection of lung abnormalities are provided in the article.
Drawings and diagrams are used throughout the article to illustrate a wide range of pulmonary abnormalities. It is important to mention that to achieve the best segmentation results, most techniques are used in combination with one another and may also include some primitive pre- or postprocessing steps to remove noise and other artifacts; the discussion of such combinations, however, is outside the scope of this article. The intent here is to help clinicians make the right choice when selecting image segmentation methods for pulmonary image analysis, without delving into algorithmic details about the methods’ functionality. Manual delineation techniques and assistive-manual methods are not discussed; instead, we focus only on fully automated methods, with a particular emphasis on the segmentation of lungs with pathologic conditions. However, it should be kept in mind that automated segmentation techniques should complement the radiologist’s work flow by saving time in measuring (27), selecting, and classifying various findings. Automated segmentation techniques are not a substitute for the radiologist’s clinical interpretations.
Image Segmentation Methods for Abnormal Lungs
Before describing the lung segmentation methods and evaluating their performance in cases with different pathologic conditions, it is pertinent to introduce common pathologic imaging patterns encountered on pulmonary CT images, to fully appreciate the performance of segmentation methods, as well as the difficulty of the problem with respect to the particular type and location of the abnormal imaging patterns. We refer our readers to the Fleischner Society glossary of commonly observed abnormal imaging patterns on lung CT images by Hansell et al (28).
We describe the most widely used techniques for lung segmentation on CT images and classify these techniques into five major classes. For each class, we summarize the advantages and disadvantages of the methods and provide example applications for which the methods have been shown to be successful (Table 1). In the subsequent subsections, the details of each segmentation class are explained.
Table 1:
Five Major Classes of CT Lung Segmentation Methods
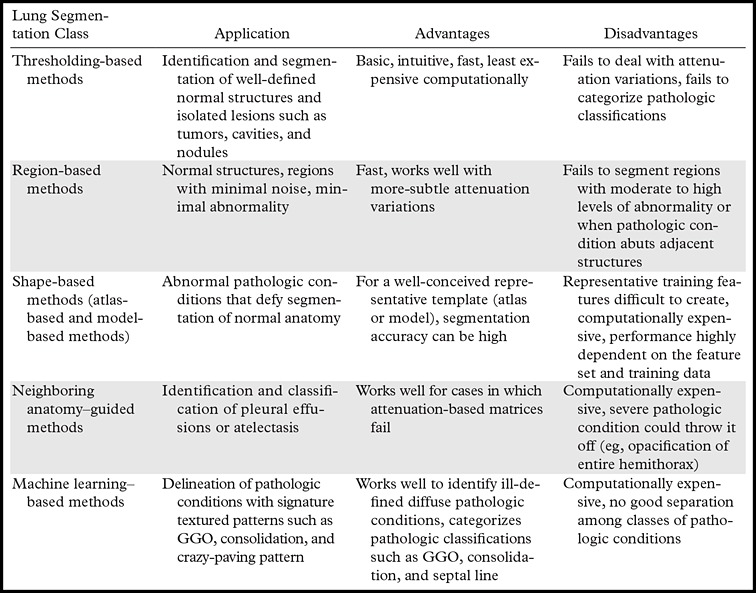
Note.—GGO = ground-glass opacity.
Thresholding-based Methods
Thresholding-based methods are the most basic and well-understood class of the segmentation techniques and are commonly used in most picture archiving and communication systems and third-party viewing applications because of their simplicity (29). Thresholding-based methods (2,30,31) segment the image by creating binary partitions that are based on image attenuation values, as determined by the relative attenuation of structures on CT images. A thresholding procedure attempts to determine attenuation values, termed threshold(s), that create the partitions by grouping together all image elements with attenuation values that satisfy the thresholding interval. The thresholding-based process is shown in a flowchart (Fig 4).
Figure 4.
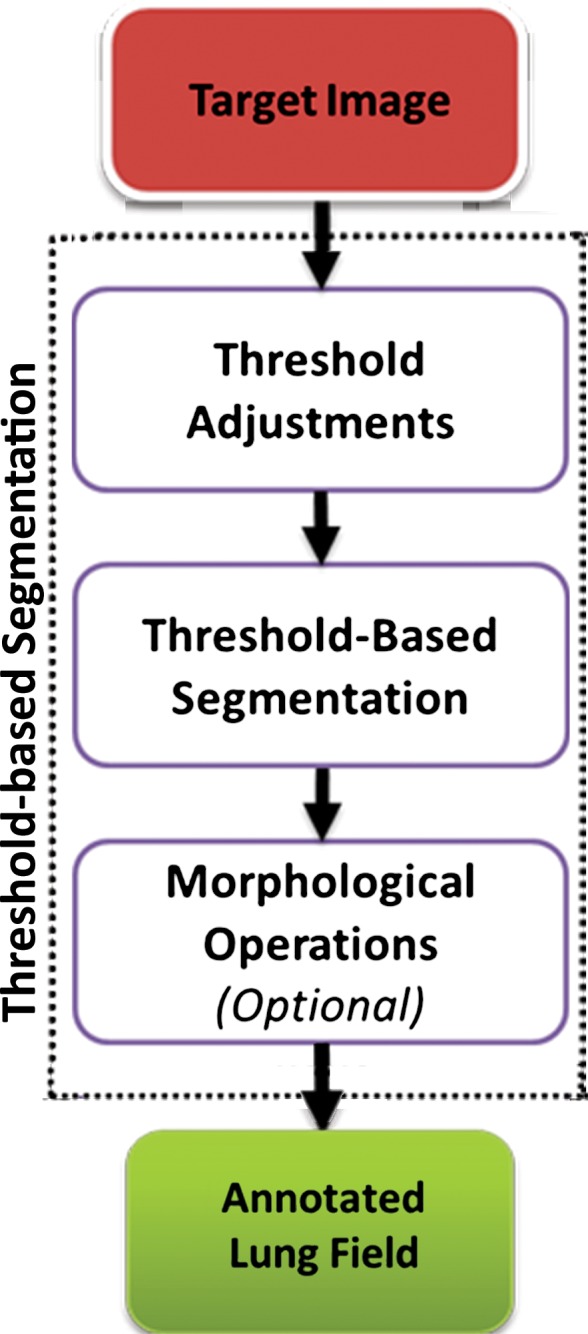
Flowchart of a thresholding-based method of lung segmentation. The attenuation numbers (in Hounsfield units) of the pixels are used to segment the lungs. False-positive findings and artifacts may still occur with this approach; therefore, morphologic operations can be conducted afterward.
Thresholding-based methods are simple and effective for obtaining segmentations from images with a well-defined contrast difference among the regions. Indeed, these methods usually perform better on CT images (10,11,26,32–39), compared with images obtained with other imaging modalities, because of the fact that the attenuation values, measured in Hounsfield units, have well-defined ranges for different tissue components on CT images. However, thresholding-based techniques do not typically take into account the spatial characteristics of the target objects (lungs). Moreover, these techniques are generally sensitive to noise and imaging artifacts, compared with the other classes of lung segmentation methods. The presence of abnormal imaging patterns affects this class of thresholding-based segmentation methods more than other methods because no spatial information and variability are considered during the segmentation process.
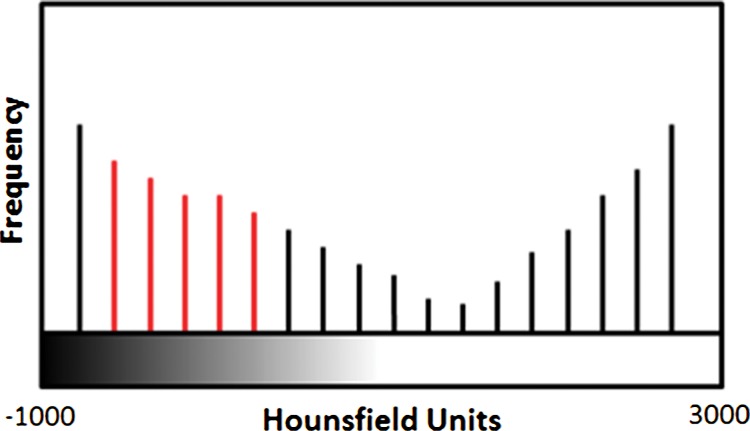
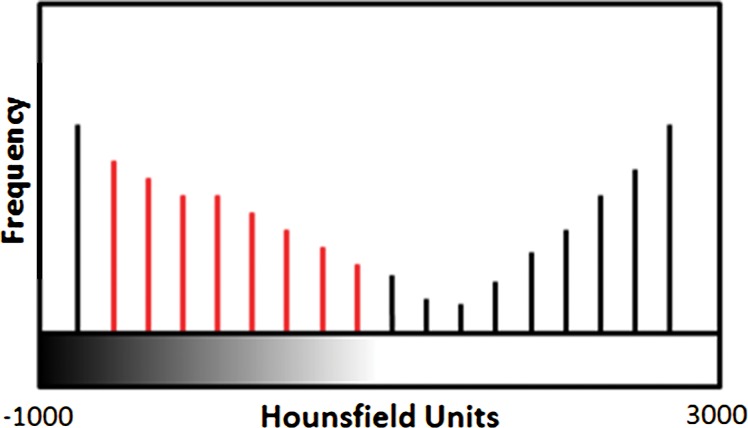
An overview of the thresholding-based segmentation method is shown in Figure 5, in which the upper and lower limits of the thresholding interval allow the selection of lung regions. Note that appropriate selection of the threshold parameters may be enough for segmenting lungs with minimal or no pathologic conditions because of the stable attenuation values of the air and lung fields. On the other hand, it may be difficult to include pathologic areas within the lung regions with thresholding-based approaches because the thresholding interval is often set to exclude adjacent tissues from lung fields, but pathologic regions (ie, consolidation) may share similar attenuation values to those of soft tissues. Figure 6 shows two examples in which pleural effusions and consolidations exist in the lung, and thresholding-based segmentation failed to delineate the lung boundaries correctly because of these abnormal imaging markers. Often, various morphologic operations or a manual false-positive removal process may be needed to correct the resulting segmentation. In terms of efficiency, thresholding-based methods are the fastest image segmentation methods, often taking only a few seconds, and yield completely reproducible segmentation.
Figure 5a.
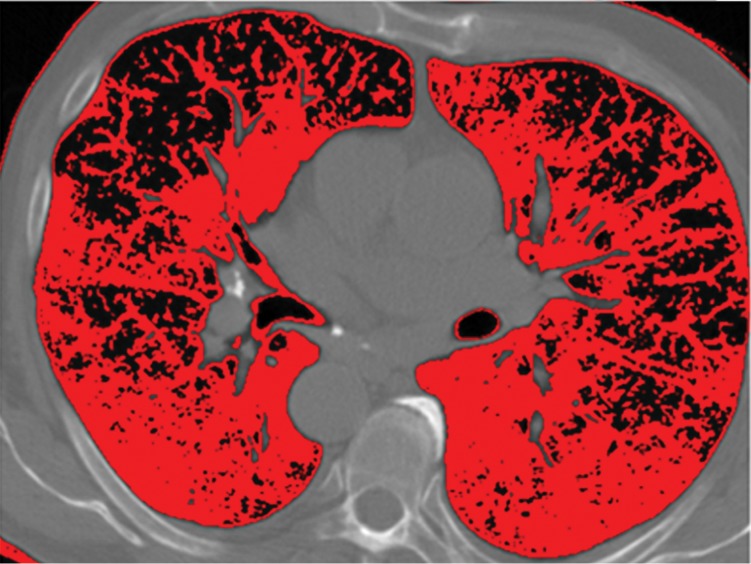
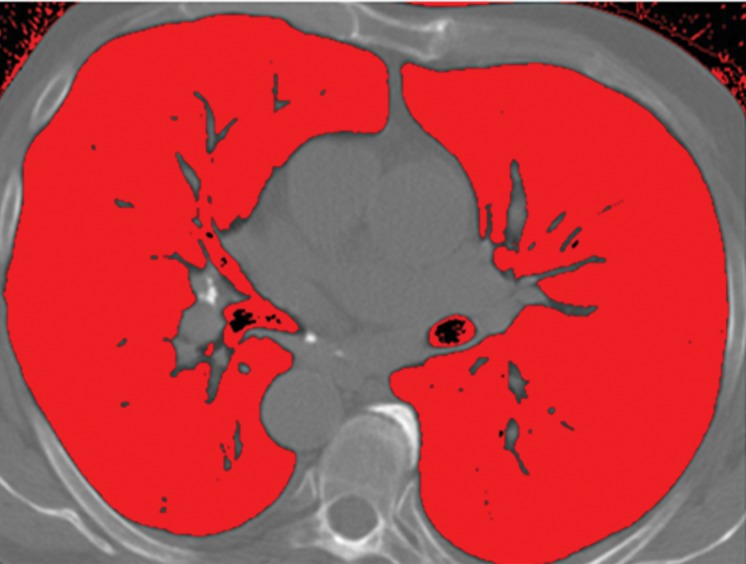
Schematic diagram providing an overview of the thresholding-based approach to lung segmentation. Graphs (a, b) show how the upper and lower threshold values (shown with red vertical lines in a, b) in Hounsfield units are adjusted to annotate the lungs on CT images (c, d). The suboptimal interval of attenuation in a results in excluded lung parenchyma (black regions in c) from the segmented lung regions (red), in comparison with the better attenuation interval in b, which results in better lung segmentation in d.
Figure 6a.
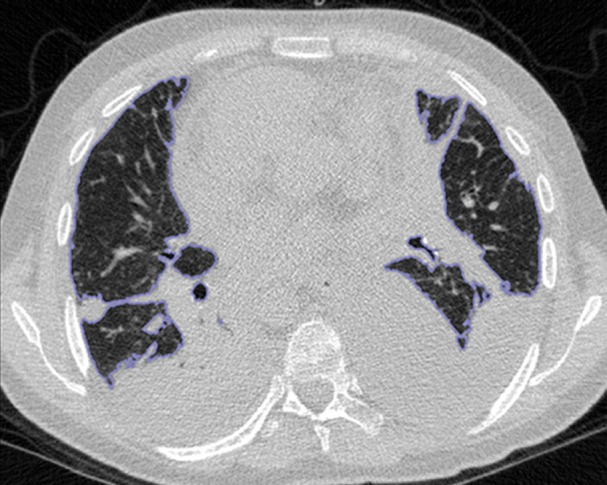
Inaccurate boundary identification. Blue contours are segmentation results for estimated lung boundaries. CT images show two examples of suboptimal results of thresholding-based delineation that are due to pleural effusions (a) and consolidations (b).
Figure 5b.
Schematic diagram providing an overview of the thresholding-based approach to lung segmentation. Graphs (a, b) show how the upper and lower threshold values (shown with red vertical lines in a, b) in Hounsfield units are adjusted to annotate the lungs on CT images (c, d). The suboptimal interval of attenuation in a results in excluded lung parenchyma (black regions in c) from the segmented lung regions (red), in comparison with the better attenuation interval in b, which results in better lung segmentation in d.
Figure 5c.
Schematic diagram providing an overview of the thresholding-based approach to lung segmentation. Graphs (a, b) show how the upper and lower threshold values (shown with red vertical lines in a, b) in Hounsfield units are adjusted to annotate the lungs on CT images (c, d). The suboptimal interval of attenuation in a results in excluded lung parenchyma (black regions in c) from the segmented lung regions (red), in comparison with the better attenuation interval in b, which results in better lung segmentation in d.
Figure 5d.
Schematic diagram providing an overview of the thresholding-based approach to lung segmentation. Graphs (a, b) show how the upper and lower threshold values (shown with red vertical lines in a, b) in Hounsfield units are adjusted to annotate the lungs on CT images (c, d). The suboptimal interval of attenuation in a results in excluded lung parenchyma (black regions in c) from the segmented lung regions (red), in comparison with the better attenuation interval in b, which results in better lung segmentation in d.
Figure 6b.
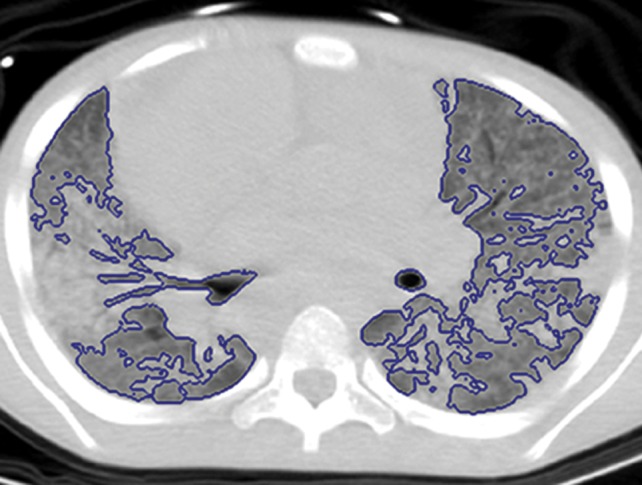
Inaccurate boundary identification. Blue contours are segmentation results for estimated lung boundaries. CT images show two examples of suboptimal results of thresholding-based delineation that are due to pleural effusions (a) and consolidations (b).
Region-based Methods
The main postulate of the region-based segmentation methods relies on the fact that neighboring pixels within one region have similar values (40). The best-known method of this class of segmentation methods is probably the region-growing method, in which the general procedure is to compare one pixel to its neighboring pixels, and if a predefined region criterion (ie, homogeneity) is met, then the pixel is said to belong to the same class as one or more of its neighbors (30,40–44). Although a predefined region criterion is critical in the region-growing method, the region-growing methods are more accurate and efficient, compared with the thresholding-based segmentation methods, because they include “region” criteria as well as spatial information (20,44). For applications in lung segmentation on CT images, region-based segmentation methods (particularly region growing) have been found to be useful for their efficiency and robustness in dealing with attenuation variations (caused by mild pathologic conditions and imaging artifacts) by reinforcing spatial neighborhood information and a regional term (20). Diagrams of the general approach used for region-based segmentation are shown in Figure 7.
Figure 7a.
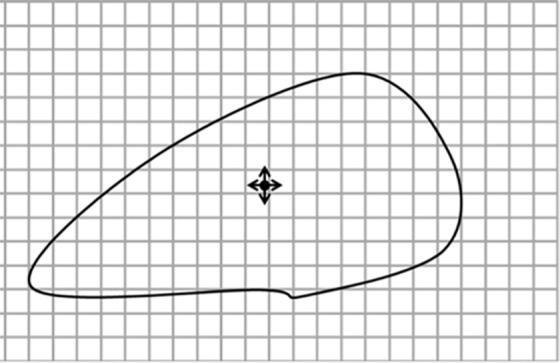
Diagrams of the general idea of region-based segmentation: Region-based segmentation approaches start with a seed point and then grow as they add neighboring pixels or voxels to the evolving annotation as long as the neighborhood criterion is satisfied. (a) Start of growing a region shows initial seed point (black circle) and directions of growth (arrows). (b) Growing process after a few iterations shows area grown so far (black area), current voxels being tested (gray circles), and potential directions of further growth (arrows). (c) Final segmentation (black area).
Figure 7b.
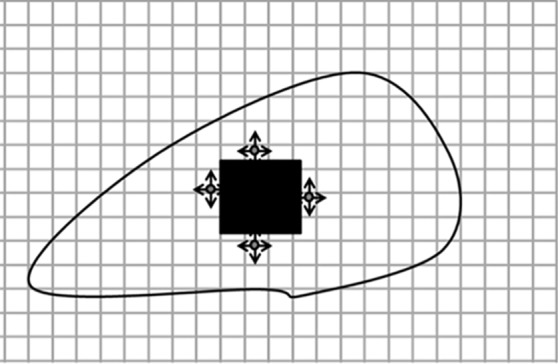
Diagrams of the general idea of region-based segmentation: Region-based segmentation approaches start with a seed point and then grow as they add neighboring pixels or voxels to the evolving annotation as long as the neighborhood criterion is satisfied. (a) Start of growing a region shows initial seed point (black circle) and directions of growth (arrows). (b) Growing process after a few iterations shows area grown so far (black area), current voxels being tested (gray circles), and potential directions of further growth (arrows). (c) Final segmentation (black area).
Figure 7c.
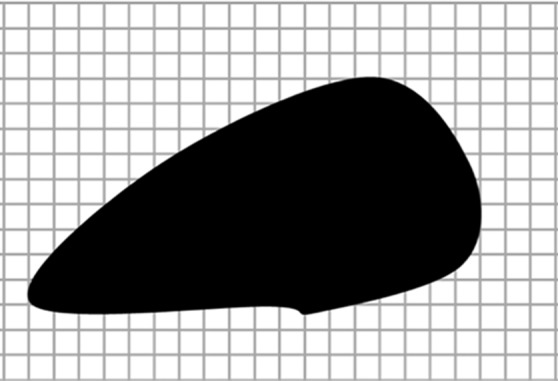
Diagrams of the general idea of region-based segmentation: Region-based segmentation approaches start with a seed point and then grow as they add neighboring pixels or voxels to the evolving annotation as long as the neighborhood criterion is satisfied. (a) Start of growing a region shows initial seed point (black circle) and directions of growth (arrows). (b) Growing process after a few iterations shows area grown so far (black area), current voxels being tested (gray circles), and potential directions of further growth (arrows). (c) Final segmentation (black area).
In addition to region growing, a number of other region-based segmentation methods have been introduced in the literature, including the watershed transform (45), graph cuts (46), random walks (47), and fuzzy connectedness (48). In watershed segmentation, the main idea underlying the method comes from geography; the idea is that of a landscape or topographic relief that is flooded by water, with watersheds being the division lines of the domains of attraction of rain falling over the region (45). Although watershed transformation is computationally feasible and therefore can be considered efficient, it has the drawback of an oversegmentation problem and hence is a less likely choice for the lung segmentation problem.
In contrast to watersheds, the graph-cut and the random walk methods of region-based segmentation are graph-based segmentation methods, and these two are considered globally optimal segmentations because of higher accuracy. Although in the graph-cut method, edges and attenuation (or texture) information are used to build an energy function to be minimized for the segmentation purpose (46), the probability of each pixel is computed according to the random walker concept, in which every pixel’s random walker first arrives at background or foreground cues provided by the users (47). Although random walk and graph-cut segmentation algorithms can be considered as efficient and accurate with regard to lung segmentation on CT images, these algorithms have not yet been shown to be successful when moderate to high amounts of pathologic findings exist in the lungs. Five well-established region-based segmentation methods are briefly summarized in Table 2, along with their most commonly used criteria for lung segmentation.
Table 2:
Common Region-based Methods and Their Main Criteria for Lung Segmentation on CT Images
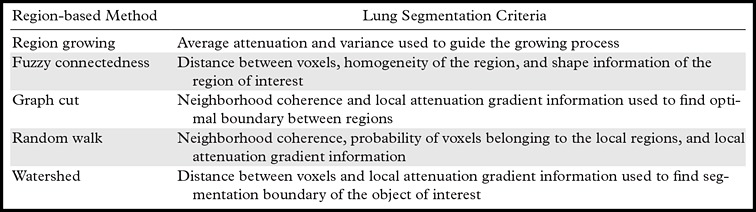
For region-based lung segmentation, the “seeded” scheme is commonly applied (Fig 8). In such cases, a small patch (seed) that is considered to be most representative of the target region (lung) is first identified. Seed points are the coordinates of a representative set of voxels belonging to the target organ to be segmented, and they can be selected either manually or automatically. Once the seed points are identified, a predefined neighborhood criterion is used to extract the desired region. Different methods feature different criteria for determining the lung boundaries. For instance, one possible criterion could be to grow the region until the lung edge is detected. As another example, region homogeneity can be used for convergence of the segmentation (40–44).
Figure 8.
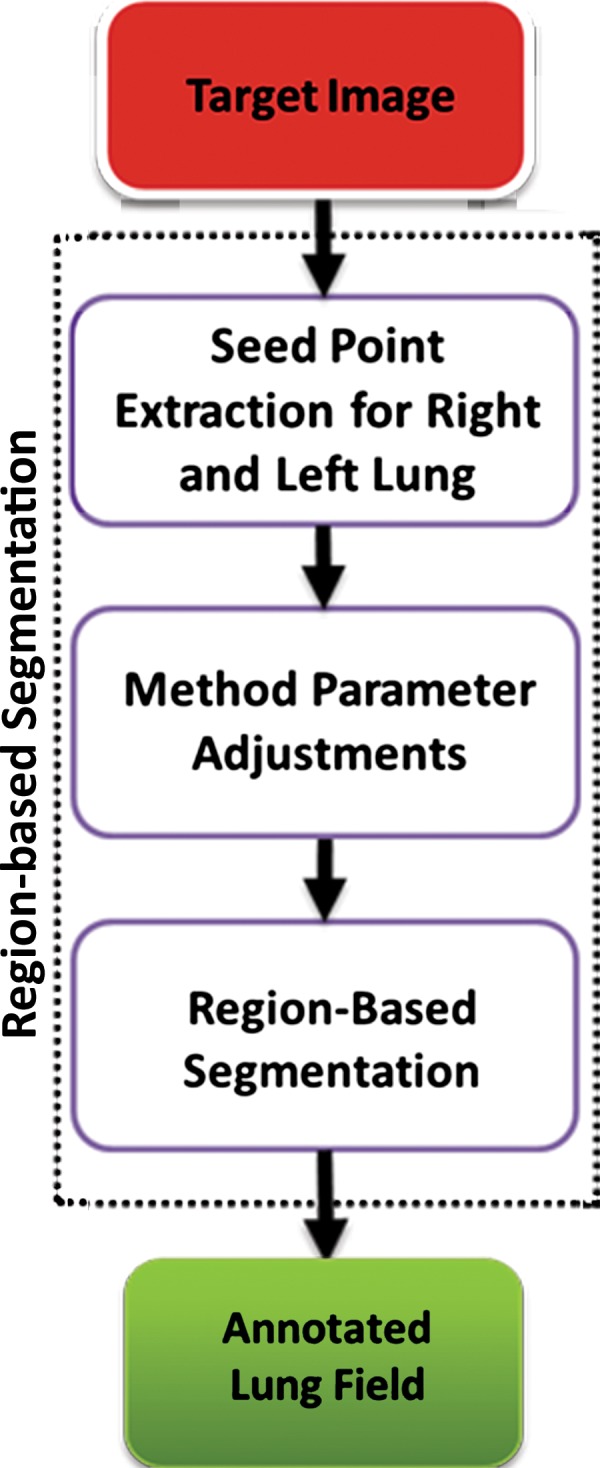
Flowchart of the region-based method of lung segmentation.
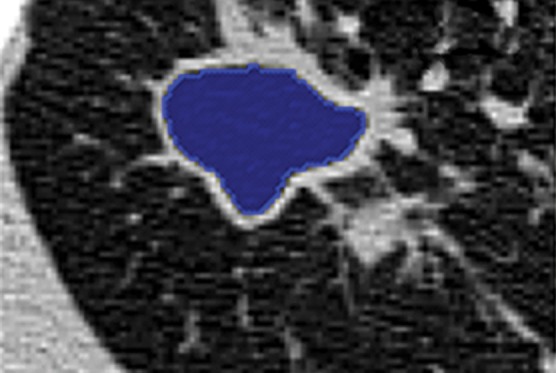
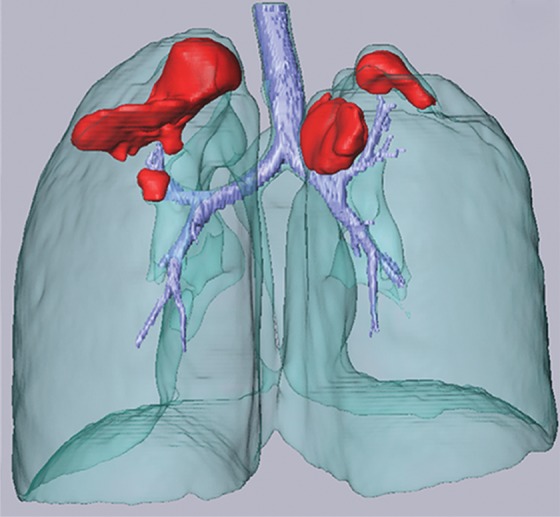
Region-based methods can be used for delineation of airways and pathologic conditions with homogeneous content such as cavities. With this ability, a single segmentation algorithm can be used to depict and quantify multiple organ and suborgan structures. An example is a single region-based segmentation approach that is applied to multiple structures in pulmonary image analysis: a cavity in the right upper lobe, the airways, and the lung fields (Fig 9).
Figure 9a.
Example in which a single region-based segmentation approach was used to delineate multiple pulmonary structures. On a given CT image (a), the lung fields (green in d), airways (light blue in d), and cavity regions (blue in b, c) were all segmented by using the same region-based segmentation approach. On the final segmentation image (d), all structures were depicted together, along with multiple cavities (red).
Figure 9b.
Example in which a single region-based segmentation approach was used to delineate multiple pulmonary structures. On a given CT image (a), the lung fields (green in d), airways (light blue in d), and cavity regions (blue in b, c) were all segmented by using the same region-based segmentation approach. On the final segmentation image (d), all structures were depicted together, along with multiple cavities (red).
Figure 9c.

Example in which a single region-based segmentation approach was used to delineate multiple pulmonary structures. On a given CT image (a), the lung fields (green in d), airways (light blue in d), and cavity regions (blue in b, c) were all segmented by using the same region-based segmentation approach. On the final segmentation image (d), all structures were depicted together, along with multiple cavities (red).
Figure 9d.
Example in which a single region-based segmentation approach was used to delineate multiple pulmonary structures. On a given CT image (a), the lung fields (green in d), airways (light blue in d), and cavity regions (blue in b, c) were all segmented by using the same region-based segmentation approach. On the final segmentation image (d), all structures were depicted together, along with multiple cavities (red).
Region-based segmentation methods serve as an efficient tool for extracting homogeneous regions such as lungs with no to mild pathologic conditions. In comparison with the thresholding-based methods, region-based methods generate more-precise lung segmentation results (13,20,30,49) without causing false positives in out-of-body regions with similar attenuation values. However, depending on the magnitude of noise and the precision of the neighborhood criteria, region-based methods can suffer from false negatives within the lung region and thus may demand further postprocessing, as shown in several cases in which region-based methods failed to perform well (Fig 10).
Figure 10a.
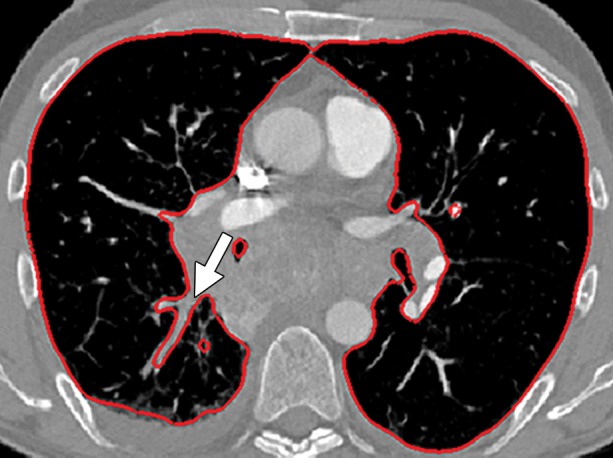
Potential failures of region-based segmentation methods. Six examples of potential failures of region-based segmentation methods show lung boundaries (red contours) and areas in which the algorithms fail (arrows). In particular, the structures that are excluded from lung segmentation are vascular structures (a, d), consolidations (b, c, f), and a pleural effusion (e). Compare with Figure 16, which shows optimal segmentation in similar cases with the use of the neighboring anatomy–guided segmentation method.
Figure 10b.
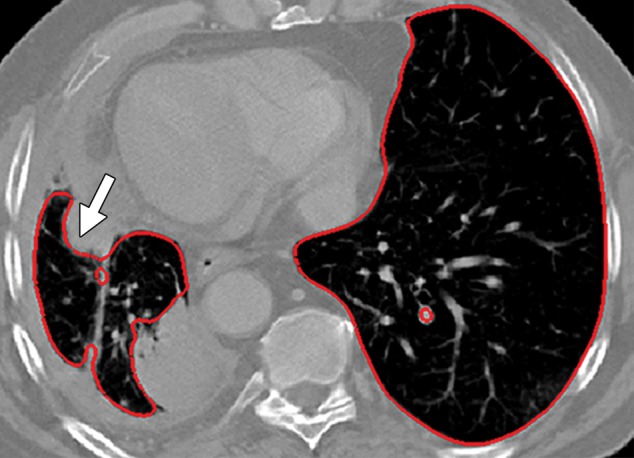
Potential failures of region-based segmentation methods. Six examples of potential failures of region-based segmentation methods show lung boundaries (red contours) and areas in which the algorithms fail (arrows). In particular, the structures that are excluded from lung segmentation are vascular structures (a, d), consolidations (b, c, f), and a pleural effusion (e). Compare with Figure 16, which shows optimal segmentation in similar cases with the use of the neighboring anatomy–guided segmentation method.
Figure 10c.
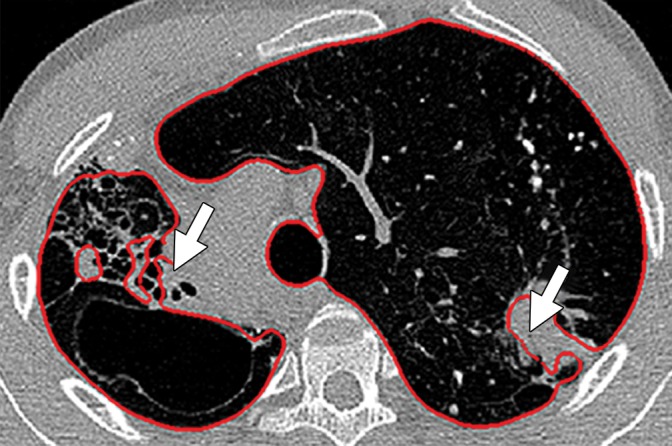
Potential failures of region-based segmentation methods. Six examples of potential failures of region-based segmentation methods show lung boundaries (red contours) and areas in which the algorithms fail (arrows). In particular, the structures that are excluded from lung segmentation are vascular structures (a, d), consolidations (b, c, f), and a pleural effusion (e). Compare with Figure 16, which shows optimal segmentation in similar cases with the use of the neighboring anatomy–guided segmentation method.
Figure 10d.
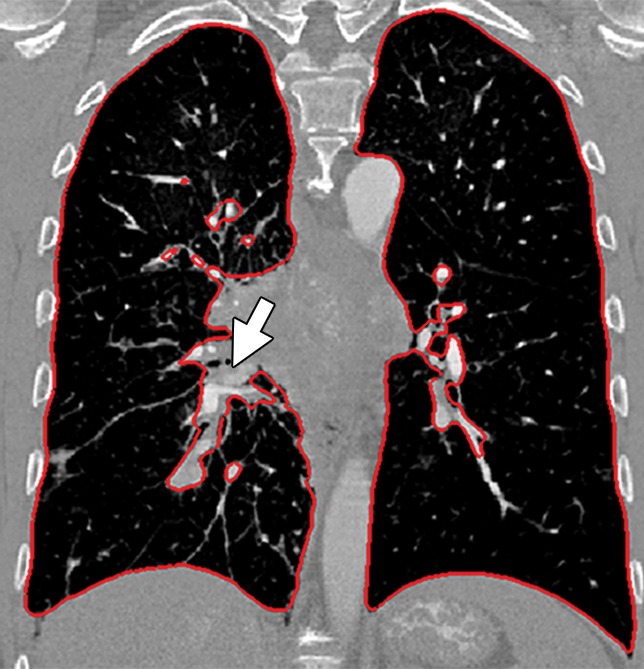
Potential failures of region-based segmentation methods. Six examples of potential failures of region-based segmentation methods show lung boundaries (red contours) and areas in which the algorithms fail (arrows). In particular, the structures that are excluded from lung segmentation are vascular structures (a, d), consolidations (b, c, f), and a pleural effusion (e). Compare with Figure 16, which shows optimal segmentation in similar cases with the use of the neighboring anatomy–guided segmentation method.
Figure 10e.
Potential failures of region-based segmentation methods. Six examples of potential failures of region-based segmentation methods show lung boundaries (red contours) and areas in which the algorithms fail (arrows). In particular, the structures that are excluded from lung segmentation are vascular structures (a, d), consolidations (b, c, f), and a pleural effusion (e). Compare with Figure 16, which shows optimal segmentation in similar cases with the use of the neighboring anatomy–guided segmentation method.
Figure 10f.
Potential failures of region-based segmentation methods. Six examples of potential failures of region-based segmentation methods show lung boundaries (red contours) and areas in which the algorithms fail (arrows). In particular, the structures that are excluded from lung segmentation are vascular structures (a, d), consolidations (b, c, f), and a pleural effusion (e). Compare with Figure 16, which shows optimal segmentation in similar cases with the use of the neighboring anatomy–guided segmentation method.
Furthermore, seed selection may require manual interactions or other intelligent algorithms to be run before the segmentation process, to find an appropriate voxel or region to start the delineation process (25). Some of the postprocessing operations can be summarized as follows. For instance, to control the parameters of the region-based segmentation and remove the inherent noise, images may be smoothed before the segmentation process. In addition, artifact removal can be done manually before starting the delineation algorithm. Alternatively, to remove artifacts or pathologic regions near the lungs, it may be more feasible to crop out the lung regions from the CT image and process the delineation algorithm on the newly defined region of interest in which artifacts do not exist anymore. Last, but not least, for challenging cases such as when nodules or pathologic conditions are near the lung boundary, attenuation remapping to the lung region as well as enhancing the lung boundary with edge detection can be useful for accurate segmentation without having failures (10–12,26,30,46).
In terms of efficiency, region-based segmentation methods can be considered efficient because the timings (a few seconds to a few minutes) and the computational cost reported in the literature are within the bounds of clinical utility (10–12,26,30,46). The repeatability of the region-based segmentation methods depends on the location of the seed points (if seeding-based segmentation); hence, different region-based methods have different robustness for repeatability. For instance, the fuzzy connectedness method (48) of image segmentation has been shown to be more robust in comparison with the graph-cut, random walk, and region-growing segmentation methods (48).
Shape-based Methods
Recently, the use of prior shape information about anatomic organs such as lungs has gained popularity in medical image segmentation, especially to segment organs with abnormalities that cannot be annotated by using the standard thresholding-based techniques. These shape-based techniques take either an atlas-based approach or a model-based approach to find the lung boundary. Figure 11 shows a generic overview of the shape-based lung segmentation methods. In this review, we combined model-based and atlas-based methods into a broader shape-based class because they both share similar algorithmic and semantic details. Furthermore, the advantages and disadvantages of using both methods are similar in terms of segmentation performance and time efficiency.
Figure 11.
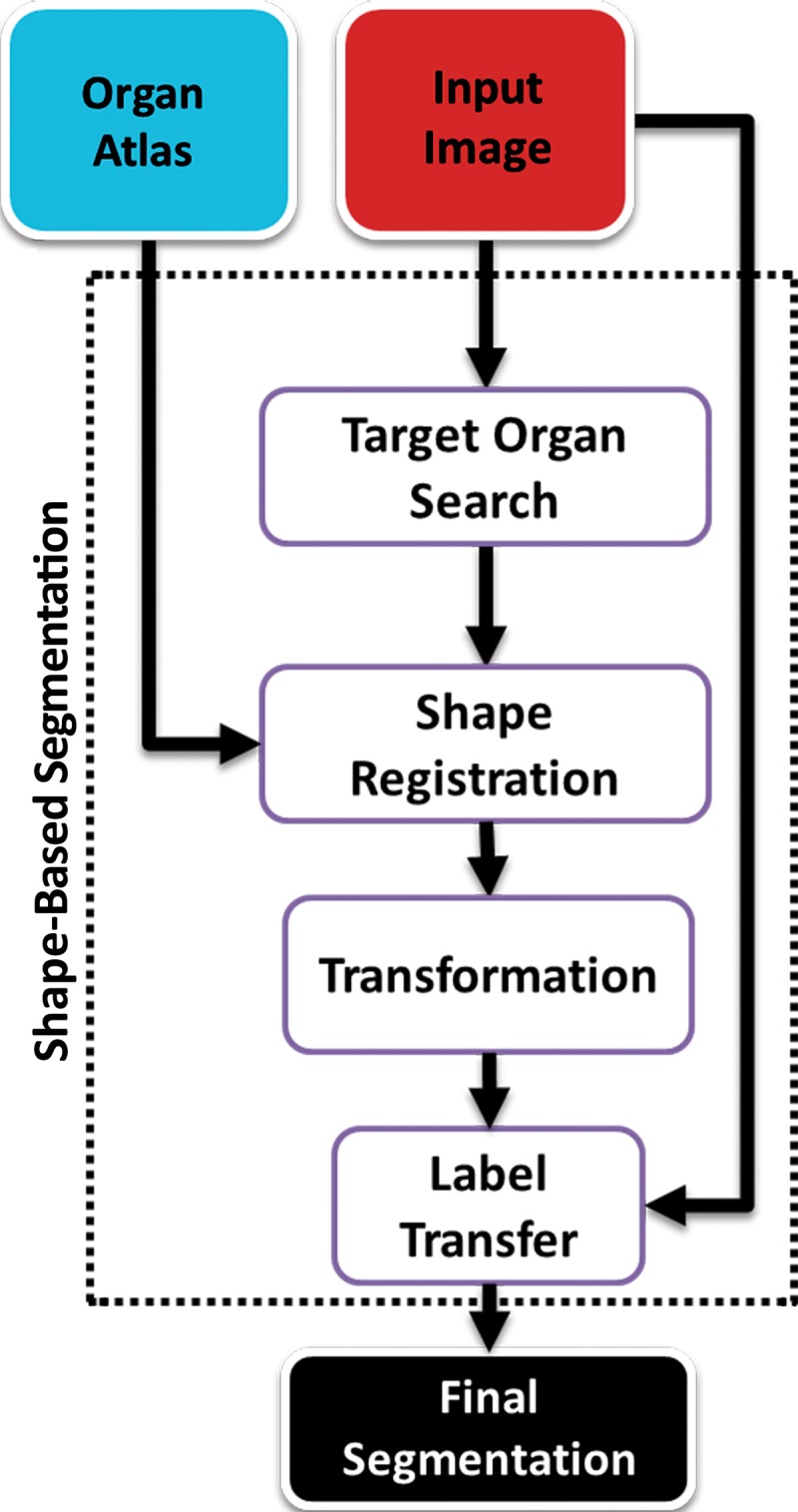
Generic overview flowchart of shape-based approaches to lung segmentation.
Atlas-based Methods.—Atlas-based methods use prior shape information about the target organ for recognition and delineation. An atlas consists of a template CT image and the corresponding labels of the thoracic regions. To perform segmentation, the template image is registered to the target image; once alignment is completed, labels of the atlas are propagated onto the target image (Fig 12). It should be noted that registration (alignment) is a difficult and ill-posed problem, although many registration methods are available with submillimeter accuracy.
Figure 12a.
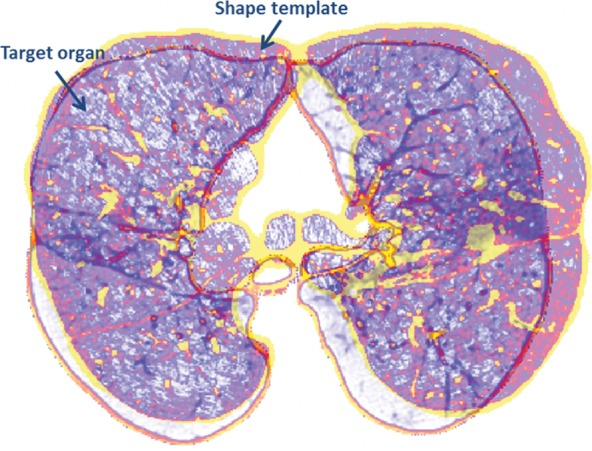
Atlas-based approach to lung segmentation. Atlas-based approaches often start with a template of the target organ (a). An image registration algorithm is then used to align the template to the target image such that the template can be transformed geometrically into the target image to identify lung tissues (b).
Figure 12b.
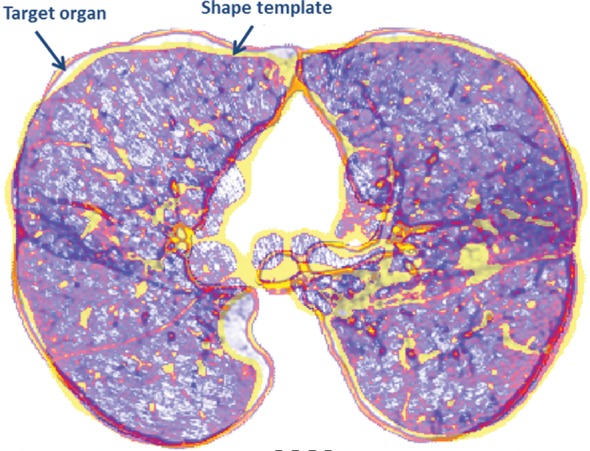
Atlas-based approach to lung segmentation. Atlas-based approaches often start with a template of the target organ (a). An image registration algorithm is then used to align the template to the target image such that the template can be transformed geometrically into the target image to identify lung tissues (b).
Atlas-based methods have been found to be useful in the segmentation of lungs with mild to moderate abnormalities (44,50,51); however, a robust representative anatomic atlas is often difficult to create because of large intersubject variabilities as well as differences related to the pathologic condition. For example, cases of scoliosis may be difficult to analyze if the atlas was created by using a population of normal spines (Fig 13).
Figure 13a.
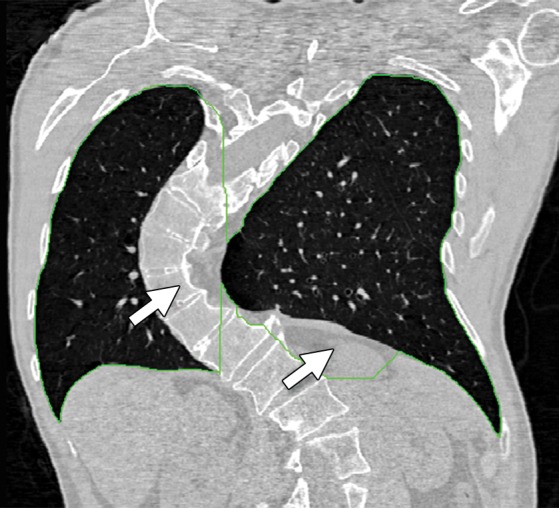
Example of a limitation of shape-based methods of lung segmentation. Because shape-based segmentation approaches assume a certain anatomic structure for the lungs, pathologic lungs with certain shape changes can be mis-segmented. In a severe case of scoliosis, although region- and thresholding-based methods performed well (b), a failure is observed with the shape-based method (a), with the boundary of the right lung (green contour) extending over the spine (arrow at left) and with the left lung boundary (green contour) spanning the medial left upper portion of the abdomen (arrow at right).
Figure 13b.
Example of a limitation of shape-based methods of lung segmentation. Because shape-based segmentation approaches assume a certain anatomic structure for the lungs, pathologic lungs with certain shape changes can be mis-segmented. In a severe case of scoliosis, although region- and thresholding-based methods performed well (b), a failure is observed with the shape-based method (a), with the boundary of the right lung (green contour) extending over the spine (arrow at left) and with the left lung boundary (green contour) spanning the medial left upper portion of the abdomen (arrow at right).
Model-based Methods.—Model-based methods use prior shape information, similar to atlas-based approaches; however, to better accommodate the shape variabilities, the model-based approaches fit either statistical shape or appearance models of the lungs to the image by using an optimization procedure. The aim in these models is to cope with the variability of the target organs that are being considered. Basically, the expected shape and local gray-level structure of a target object in the image are used to derive the segmentation process in such methods. The delineation is finalized when the model finds its best match for the CT data to be segmented.
The model-based approach naturally belongs to the top-down strategy in which recognition is followed by delineation. Unlike other low-level approaches such as thresholding- and region-based approaches, model-based methods consider both the global and local variation of the shape and texture; therefore, these methods are considered effective in handling the abnormal lung segmentation problem. In particular, because of the probabilistic nature of measuring variation in the training step in which expert knowledge is captured in the system, model-based methods work well in handling mild to moderate abnormalities and anatomic variability. On the other hand, similar to atlas-based approaches, a representative prior model covering diverse demographics is usually difficult to create. Finally, as a well-known deficiency of the model-based approaches, segmentation failure may be inevitable if the model is not initiated close enough to the actual boundary of the lungs.
Snakes, Active Contours, and Level Sets.—Boundary-based image segmentation methods such as snakes (52), or active contours (44), and level sets (53) are considered in the category of shape-based segmentation methods in this review. These algorithms are extensively used to locate object boundaries when boundary curves are defined within an image domain that can move under the influence of internal forces coming from within the curve itself and external forces computed from the image data. The internal and external forces are defined so that the boundary curves will conform to an object boundary or other desired features within an image (44,52,53). With regard to lung segmentation of CT images in the literature, only a few groups of investigators have used snakes and level sets (44,53). Although these methods are desirable and efficient when their points of initialization are located near the correct boundary, the method often fails when initialization of the algorithm is not so close to the actual boundaries. Moreover, when a pathologic condition exists inside the lung fields, it is easy for those methods to converge into an incorrect lung boundary, or boundary curve evolution may stop in the pathologic areas without converging into the lung boundaries (44,52,53).
The repeatability of the shape-based segmentation methods is the least robust among the five major classes of lung segmentation because most of the shape-based segmentation methods require a registration framework or localization of the model into the target image, and the initial position of the model or registration parameters can significantly affect the delineation results (3). The efficiency of the shape-based segmentation methods relies on the efficiency of the registration or localization algorithms, which often take more time than what is desired routinely in clinics (13).
Neighboring Anatomy–guided Methods
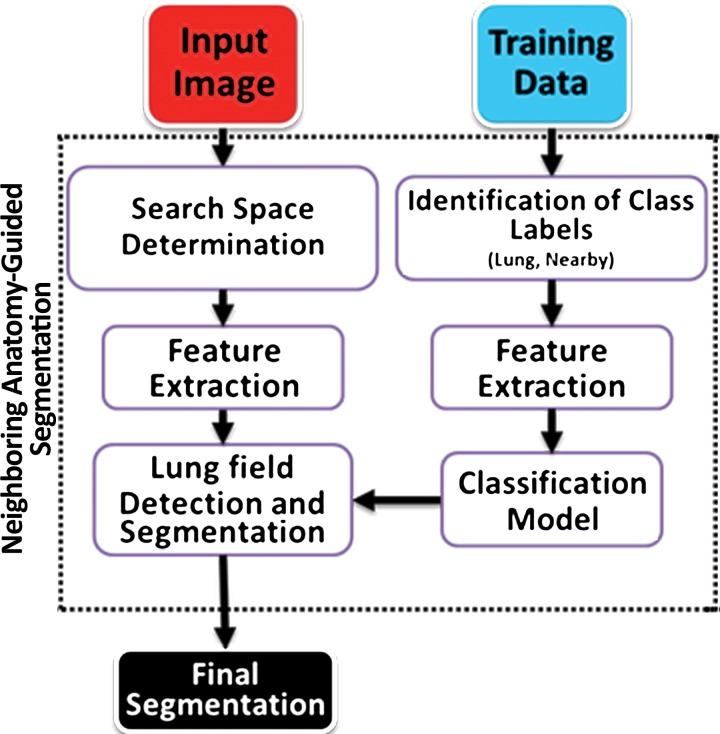
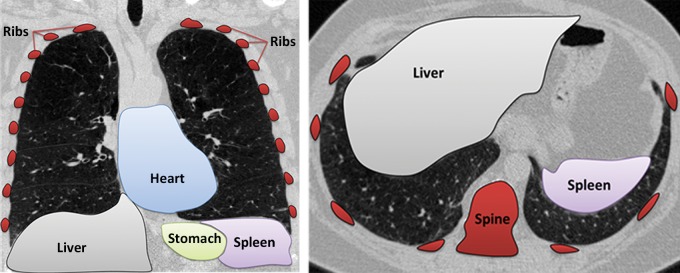
Neighboring anatomy–guided methods use the spatial context of neighboring anatomic objects of the lung (eg, rib cage, heart, spine) for delineating lung regions with optimal or near-optimal accuracy. The basic idea behind the use of neighboring organs for lung segmentation is to restrict the search space of the optimal boundary search and remove some of the false-positive findings automatically from the suboptimal segmentations. Once it is known, for instance, where the heart and rib cage are, then it is easier for a segmentation algorithm not to leak into those territories (Fig 14).
Figure 14.
Schematic diagrams provide an overview of the neighboring anatomy–guided method of segmentation. With this approach, individual organs can be identified on the basis of their expected locations.
These neighboring anatomy–guided methods are designed to handle cases in which, because of the presence of extreme abnormality or an imaging artifact, the lung regions cannot be delineated readily. Information on the neighboring anatomic structures is foreseen to have a great potential in lung image segmentation because the neighboring object interactions in the lung region are much stronger and are predictable. The flowchart shown in Figure 15 is an overview of the core concept in neighboring anatomy–guided lung segmentation. Note that, similar to model-based approaches, a prior model is necessary for the lungs and their neighboring structures.
Figure 15.
Flowchart of the neighboring anatomy–guided method of lung segmentation.
Neighboring anatomy–guided methods can be extremely helpful in the segmentation of lung regions in which discriminative attenuation and texture information is found to be either not available or not useful for annotation purposes (3,25). For example, a large amount of pleural fluid or extensive atelectasis can cause incorrect segmentation and measurements; however, the neighboring anatomy–guided segmentation approach can deliver optimal or near-optimal segmentation results by considering the rib cage, heart, liver, and other soft tissues adjacent to the lung parenchyma (Fig 16).
Figure 16a.
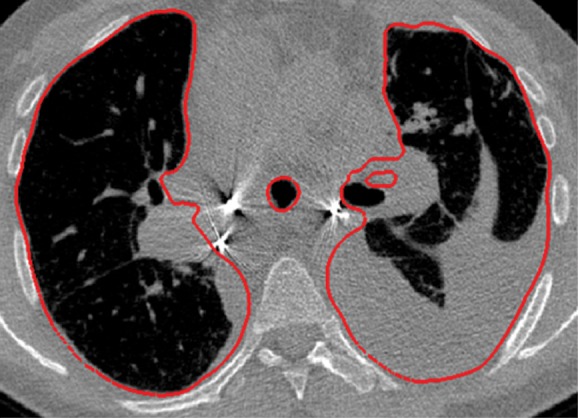
Examples of cases (large amounts of pleural fluid and extensive atelectasis) in which neighboring anatomy–guided segmentation methods produced successful lung delineations (red contours) on axial (a–c) and coronal (d–f) CT images.
Figure 16b.
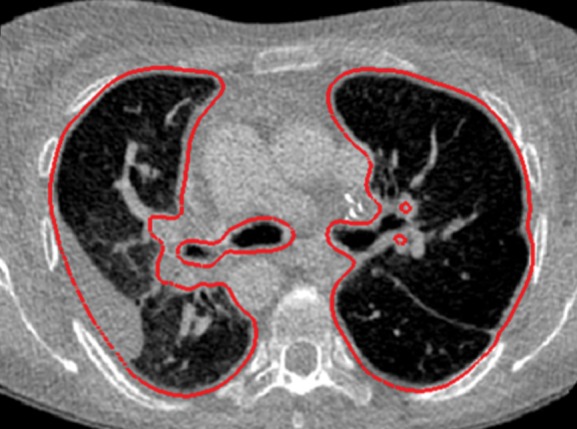
Examples of cases (large amounts of pleural fluid and extensive atelectasis) in which neighboring anatomy–guided segmentation methods produced successful lung delineations (red contours) on axial (a–c) and coronal (d–f) CT images.
Figure 16c.
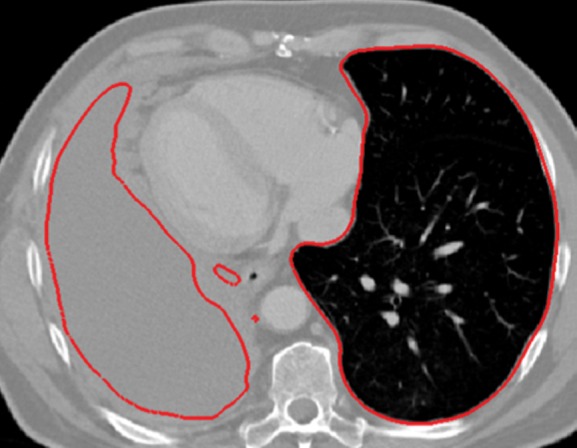
Examples of cases (large amounts of pleural fluid and extensive atelectasis) in which neighboring anatomy–guided segmentation methods produced successful lung delineations (red contours) on axial (a–c) and coronal (d–f) CT images.
Figure 16d.
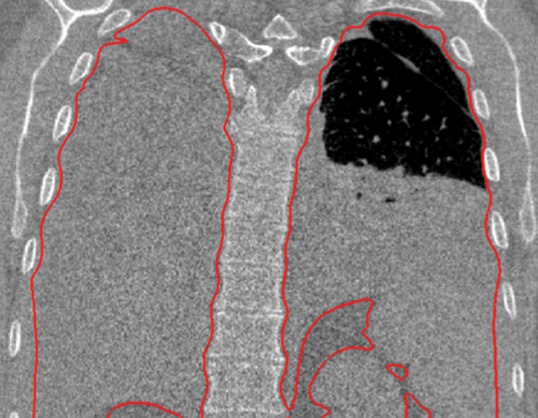
Examples of cases (large amounts of pleural fluid and extensive atelectasis) in which neighboring anatomy–guided segmentation methods produced successful lung delineations (red contours) on axial (a–c) and coronal (d–f) CT images.
Figure 16e.
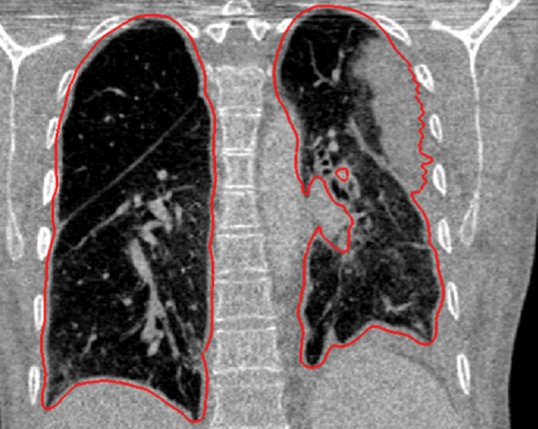
Examples of cases (large amounts of pleural fluid and extensive atelectasis) in which neighboring anatomy–guided segmentation methods produced successful lung delineations (red contours) on axial (a–c) and coronal (d–f) CT images.
Figure 16f.
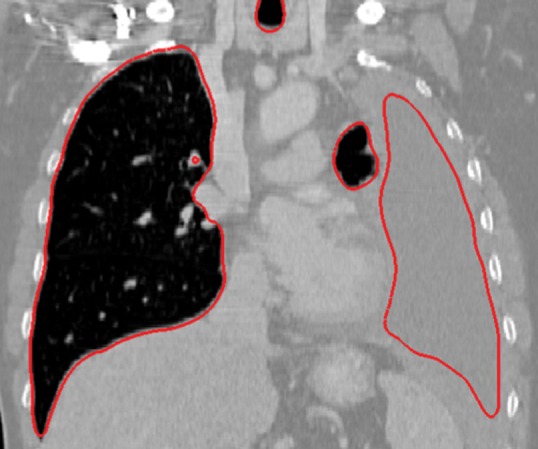
Examples of cases (large amounts of pleural fluid and extensive atelectasis) in which neighboring anatomy–guided segmentation methods produced successful lung delineations (red contours) on axial (a–c) and coronal (d–f) CT images.
Because of the success of neighboring anatomy–guided segmentation approaches in segmenting challenging cases pertaining to lung abnormalities, much work is currently in progress in this area, and more developed updates of neighboring anatomy–guided segmentation approaches are currently evolving (25,54). Despite the fact that these neighboring anatomy–guided methods are accurate, their performance greatly depends on the assumption of not having any abnormality in the neighboring structures of the lung (eg, rib cage, heart, spine), which can be difficult to guarantee if there are multifocal areas of disease in organs adjacent to pathologic lung regions. Furthermore, the efficiency of the methods greatly depends on the amount of pathologic findings existing in the lung (the larger the pathologic area, the slower the algorithm).
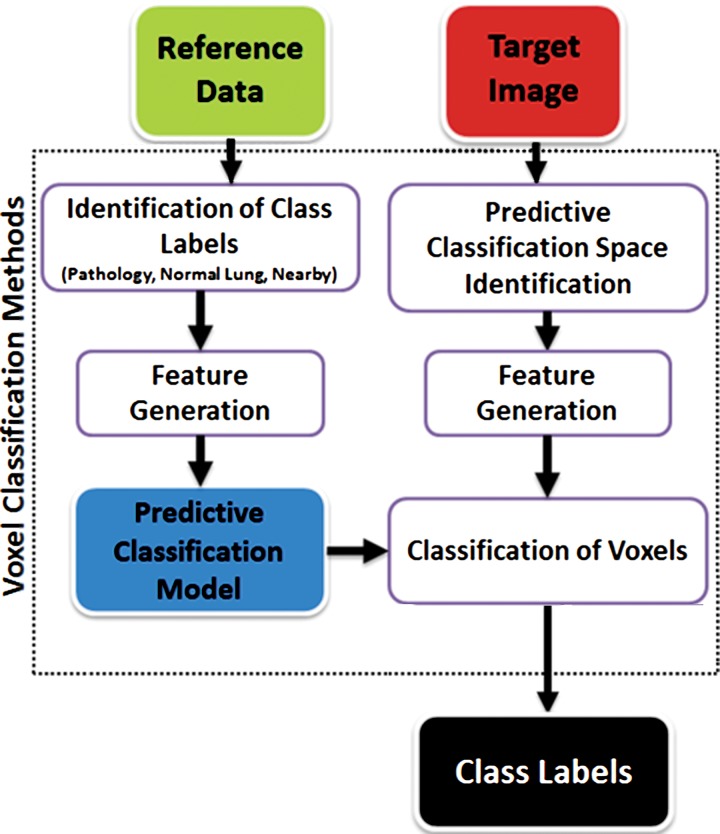
Machine Learning–based Methods
Machine learning–based methods, as the name implies, are concerned with the construction of systems that can learn from the data. Through exposure to data, the algorithm adjusts its parameters (termed features) for identifying structures and disease patterns. In machine learning–based methods, one aims to predict the lung abnormalities on the basis of the features extracted from the data and to include these features in the segmentation process so that the system discriminates the correct lung boundaries. Pattern recognition and machine learning techniques provide decision support for a vast spectrum of lung abnormalities. In practice, these methods use a set of training data containing observations, called image patches (ie, small image blocks), and their anatomic labels (eg, normal lung tissue, pathology, neighboring soft tissue). Figure 17 shows some of these patches pertaining to different abnormal classes; each patch is used along with its class label. The training data are then used to determine to which anatomic class a new, never-seen-before observation belongs. Each individual observation is analyzed with a set of quantifiable properties that are termed features. The selection of the most appropriate set of features depends on the labeling task at hand and is an area of active research.
Figure 17.

Image patches. The five most commonly observed and used normal and abnormal imaging patterns are shown as image patches. Because machine learning–based classification algorithms often require supervised training for abnormalities, image patches (ie, small image blocks) will be extracted and used in the determination of normal and abnormal classes for the classification process during the lung segmentation. GGO = ground-glass opacity.
The simplest feature is the image attenuation itself. However, more sophisticated methods for complex image processing tasks have been developed (2,25,26). For instance, Hu et al (2) combined a thresholding-based approach with a dynamic programming approach to separate right and left lung fields, and this process was followed by a sequence of morphologic operations to smooth the irregular boundaries along the mediastinum. As another example, Hua et al (54) used a graph-search algorithm by combining attenuation, gradient, boundary smoothness, and rib information for segmentation of diseased lungs. More recently, Mansoor et al (25) developed an approach in which machine learning–based algorithms were used to detect a large spectrum of pathologic conditions, and this approach was combined with region-based segmentation of lung fields and neighboring anatomy–guided segmentation correction processes.
The basic steps of the machine learning–based methods are summarized in Figure 18. The classification algorithm examines each pixel or voxel to determine its class label; therefore, machine learning–based methods are often called pixel- or voxel-based classification methods. Although it may be computationally expensive to assess all of the pixels for identification of pathologic conditions, the high accuracy of the classification rates and the existence of parallel computing and powerful workstations make machine learning–based segmentation methods attractive (12).
Figure 18.
Flowchart of machine learning–based lung segmentation. First, a model is built by using features extracted from reference image data (see Fig 17). Then, for any given test image, newly extracted features are used to define the pixel classes: pathologic condition or normal.
Paradoxically, although machine learning–based strategies are often developed for pulmonary computer-aided detection systems for the identification of particular lung abnormalities, all of those methods require the lungs to be segmented before the identification task. However, the segmentation of lungs is erroneous if machine learning–based methods are not used for separating the abnormal tissue from normal tissues. On CT images, abnormal imaging patterns can be successfully detected with machine learning–based methods and be included in the final delineation of the lungs (Fig 19).
Figure 19a.
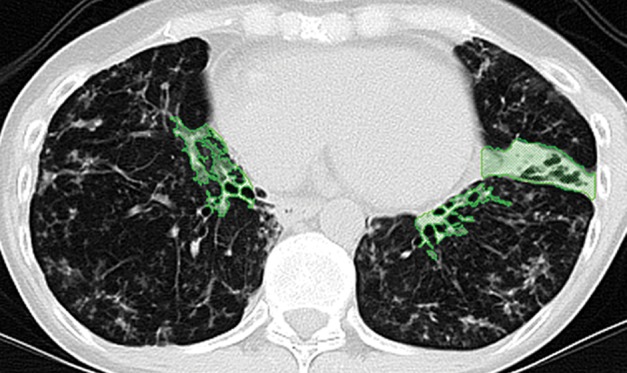
Examples of successful machine learning–based segmentation. Machine learning–based methods can identify various abnormal imaging patterns (green areas), such as consolidation and an atelectatic segment of the lingula (a), areas of ground-glass opacity (b), brochiectatic air bronchograms (c), patchy consolidation and a cavity (d), and consolidation and crazy-paving pattern (e). Because of these successful depictions, the final lung delineations were not erroneous.
Figure 19b.
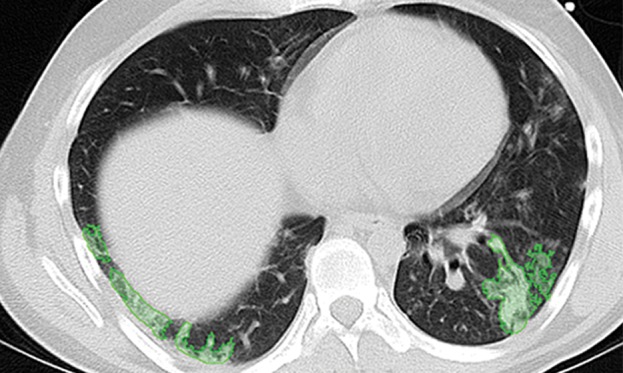
Examples of successful machine learning–based segmentation. Machine learning–based methods can identify various abnormal imaging patterns (green areas), such as consolidation and an atelectatic segment of the lingula (a), areas of ground-glass opacity (b), brochiectatic air bronchograms (c), patchy consolidation and a cavity (d), and consolidation and crazy-paving pattern (e). Because of these successful depictions, the final lung delineations were not erroneous.
Figure 19c.
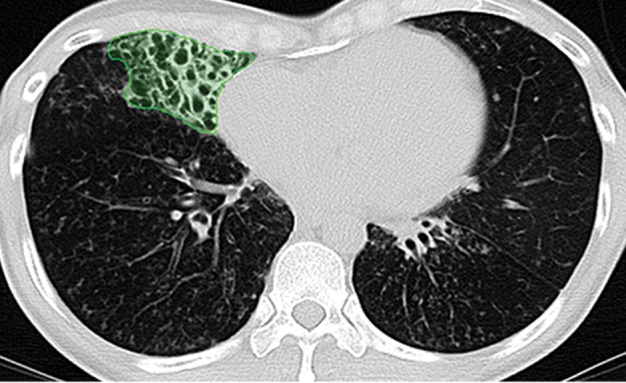
Examples of successful machine learning–based segmentation. Machine learning–based methods can identify various abnormal imaging patterns (green areas), such as consolidation and an atelectatic segment of the lingula (a), areas of ground-glass opacity (b), brochiectatic air bronchograms (c), patchy consolidation and a cavity (d), and consolidation and crazy-paving pattern (e). Because of these successful depictions, the final lung delineations were not erroneous.
Figure 19d.
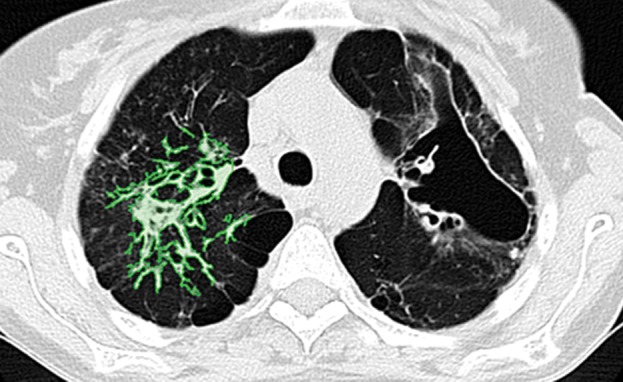
Examples of successful machine learning–based segmentation. Machine learning–based methods can identify various abnormal imaging patterns (green areas), such as consolidation and an atelectatic segment of the lingula (a), areas of ground-glass opacity (b), brochiectatic air bronchograms (c), patchy consolidation and a cavity (d), and consolidation and crazy-paving pattern (e). Because of these successful depictions, the final lung delineations were not erroneous.
Figure 19e.
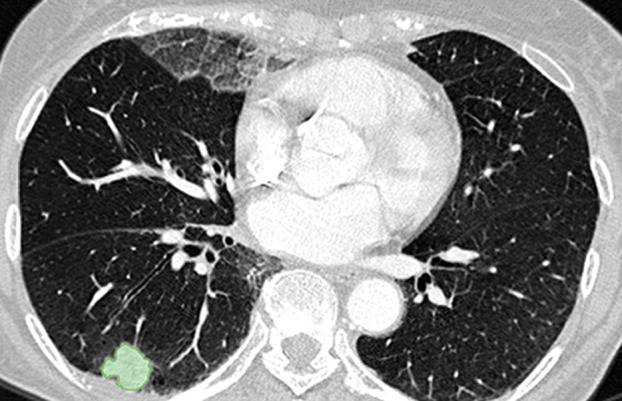
Examples of successful machine learning–based segmentation. Machine learning–based methods can identify various abnormal imaging patterns (green areas), such as consolidation and an atelectatic segment of the lingula (a), areas of ground-glass opacity (b), brochiectatic air bronchograms (c), patchy consolidation and a cavity (d), and consolidation and crazy-paving pattern (e). Because of these successful depictions, the final lung delineations were not erroneous.
Machine learning–based methods are useful for detecting and quantifying pathologic conditions; hence, these methods are considered to be the core part of the lung segmentation process as it examines every single voxel on the CT image and results in both lung and pathologic areas in the same framework (55–59). However, the main disadvantage of machine learning–based approaches is that depending on the complexity of the feature set, these approaches are computationally expensive and usually cannot model structural information (such as global shape or appearance information of the lungs) because only small patches are considered as features to be submitted into the classifiers. Another disadvantage is the difficulty in extracting a representative training set that spans anatomic and physiologic variabilities among different subjects while not overfitting the classification model. Machine learning–based methods are reproducible and therefore highly desirable. However, machine learning–based methods have the least efficiency among the five major classes of lung segmentation because of the pixel-by-pixel assessment of class labels (pathologic conditions vs lungs) (60–64).
Hybrid Approaches to Generic Lung Segmentation in Clinical Practice
In consideration of the anatomic variabilities in the clinical data, no single segmentation method can provide a generic solution to be used in clinical practice. Therefore, recently developed practical applications have concentrated on intelligently concatenating multiple segmentation strategies to provide a global generic solution. For instance, Mansoor et al (25) proposed a novel pathologic lung segmentation method that couples region-based approaches with neighboring anatomy constraints and a machine learning–based pathology recognition system for the delineation of lung fields. The proposed framework works in multiple stages; during stage 1, a modified fuzzy connectedness segmentation algorithm (a region-based segmentation approach) is used to perform the initial lung parenchyma extraction. During the second stage, texture-based local features are used to segment abnormal imaging patterns (consolidations, ground glass, interstitial thickening, tree-in-bud pattern, honeycombing, nodules, and micronodules) that are missed during the first stage of the algorithm. This refinement stage is further complemented by a neighboring anatomy–guided segmentation approach to include abnormalities that are texturally similar to neighboring organs or pleura regions. Therein, texture means spatial arrangement of an image or repeated patterns in a region of image. Although hybrid methods are feasible when varying numbers or types of abnormalities exist in the lungs, creating the class labels in the machine learning part of the hybrid methods, as well as their parameter training, is not trivial. In another example, Hua et al (54) used a graph-search algorithm to find anatomic constraints such as ribs and then constrained the graph-cut algorithm for finding the lung boundary. There are also initial preprocessing steps in which possible pathologic regions are detected and included in the segmentation. However, this approach may require accurately defined seed sets for identifying background and foreground objects.
Methods to Evaluate the Efficacy of Segmentation
For quantitative evaluation of image segmentation, three factors should be considered: precision (reliability), accuracy (validity), and efficiency (viability). Assessing precision requires segmentation tasks to be repeated, and variation is reported through a statistical test. Accuracy, on the other hand, denotes the “degree” to which segmentation agrees with the ground truth (reference standard or surrogate truth). To assess efficiency, both the computational time and user time for algorithm training and execution should be measured (65). Although the individual significance of these three methods may change depending on the application, segmentation methods must be compared by using all three factors. For accuracy assessment, a need exists for a ground truth or reference standard that is often obtained through manual drawings of the boundary of target organs by expert observers. Because automated segmentation methods are aiming to at least approximate human manual segmentation, the use of expert delineation of an object as a reference standard is widely accepted in the image processing literature.
The two most commonly used ideas for determining the accuracy of segmentation are (a) region-based measurement (spatial overlap) and (b) boundary-based measurement. Several metrics exist for spatial overlap, but the Dice similarity coefficient is the most widely used in the literature (66). The Dice similarity coefficient (DSC) is given as
where VGT is the reference standard segmentation (ground truth), Vtest is the segmentation performed by using any of the methods, |•| is the size operator, and ∩ is the intersection operator, which determines the overlapping area between the reference standard and the test segmentation.
In addition to region-based measurements, boundary-based measurements such as the Haussdorf distance (66) can be used for a complementary evaluation metric to the Dice similarity coefficient for measuring boundary mismatches. The Haussdorf distance determines how far apart two boundaries are from each other and analyzes the shape similarities of the two methods. It should be noted that although many authors report only the Dice similarity coefficient or the total segmentation volume values, the use of the Dice similarity coefficient alone is not sufficient for determining the accuracy of a segmentation algorithm. For instance, it is highly possible for a segmentation method to produce the same volume as the volume of the reference standard, but the method may have sensitivity less than 50% (ie, segmentation leaks into nonobject territory with some amount of volume, but still the same volume of reference standard can be obtained). Figure 20 exemplifies this problem, such that Vtest1 is closer to the reference standard than Vtest2, given that the ground truth is VGT, although Vtest1, Vtest2, and VGT have identical volumes.
Figure 20a.
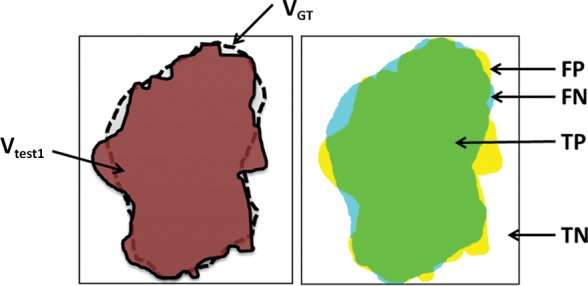
Schematic diagrams of sensitivity and specificity metrics with color-coded condition–test outcome pairs: true positive (TP) (green area), true negative (TN) (white area), false positive (FP) (yellow area), and false negative (FN) (blue area). (a) Sensitivity = 94.69%; specificity = 94.19%. (b) Sensitivity = 72.99%; specificity = 78.16%. VGT = reference standard segmentation (ground truth), Vtest = lung segmentation (brown area) obtained by using any of the segmentation methods.
Figure 20b.
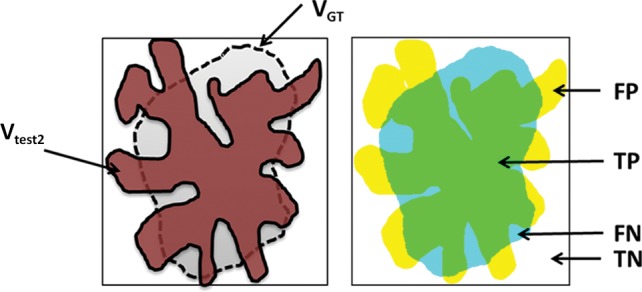
Schematic diagrams of sensitivity and specificity metrics with color-coded condition–test outcome pairs: true positive (TP) (green area), true negative (TN) (white area), false positive (FP) (yellow area), and false negative (FN) (blue area). (a) Sensitivity = 94.69%; specificity = 94.19%. (b) Sensitivity = 72.99%; specificity = 78.16%. VGT = reference standard segmentation (ground truth), Vtest = lung segmentation (brown area) obtained by using any of the segmentation methods.
Sensitivity and specificity are yet other statistical quantifiers for the binary classification test of evaluating the segmentation accuracy. Sensitivity, or the true-positive rate, measures the ratio of actual positives that are correctly identified (also known as true positives, or TP) to all positives, P (ie, the sum of the correctly identified positives [TP] and the incorrectly identified negatives); the incorrectly identified negatives are also known as false negatives (FN). Sensitivity is calculated as follows: Sensitivity = TP/P = TP/(TP + FN).
Specificity (sometimes known as the true-negative rate) measures the ratio of the negatives that are correctly identified (also known as true negatives, or TN) to all negatives (N). Specificity is calculated as follows: Specificity = TN/N = TN/(TN + FP), where FP denotes false positives, or incorrectly identified positives.
A perfect segmentation would be described as 100% sensitive (ie, labeling all pixels belonging to the target object [eg, the lung] correctly) and 100% specific (ie, not labeling any voxel from the background as belonging to target object). Figure 20 uses the same mock example to explain sensitivity and specificity.
Efficiency (viability) and precision (reliability) are two other metrics that should be reported along with accuracy measurements (ie, sensitivity and specificity). To assess precision, one needs to choose a figure of merit, repeat the segmentation considering all sources of variation, and determine variations in the figure of merit with statistical analysis. To assess efficiency, both the computational time and user time required for algorithm training and for algorithm execution should be measured and analyzed. Because precision, accuracy, and efficiency factors have an influence on one another, it is suggested that all of these measurements should be reported together (65).
Discussion
The increasing role of software and image processing in clinical radiology underpins the need for greater awareness among radiologists of how software can identify structures and lesions and yield quantitative characteristics about these objects on the image. Some areas of radiology are utilizing computer-aided detection methods for lesion identification, such as in the diagnosis of lung and breast nodules (67,68). However, the future potential of computer-aided detection in radiology is substantial because segmentation algorithms continue to improve in regard to the quality of output and the efficiency of these methods in radiologists’ work flow. In particular, the contribution to the accurate longitudinal assessment of disease progression and response to treatment will enable optimal individualized patient management.
This review presents a general overview of segmentation methods, which software uses to identify a structure or lesion, draw contours around the object’s boundaries, and then extract that structure for three-dimensional assessment apart from surrounding structures. In Table 3, all classes of lung segmentation methods and their advantages, disadvantages, and possible failures with respect to different lung abnormality types observed on CT images are summarized (25,60). These classifications are based on the evidence of published work in the literature (13,17,25,33,35,37,60–64,68) and the available computer-aided detection systems for pulmonary pathologic identifications. With this detailed comparison, we hope that our readers can compare and contrast the available lung segmentation methods for their specific lung segmentation tasks and determine the most suitable one for the task at hand. Additionally, we hope that radiologists, by understanding the limitations and strengths of these approaches, can become more familiar with the technical trends affecting the development of radiologic software and computer-assisted methods for lung disease. In fact, computer-aided detection systems are not intended to replace radiologists but rather to be complementary to their diagnostic tasks. Indeed, the radiologist can override descriptions output by automated systems, thus having the last say in the diagnostic process.
Table 3:
Effectiveness of Each Major Class of Methods for Segmentation of the Lung Fields and Various Abnormal Imaging Patterns of Lung Diseases
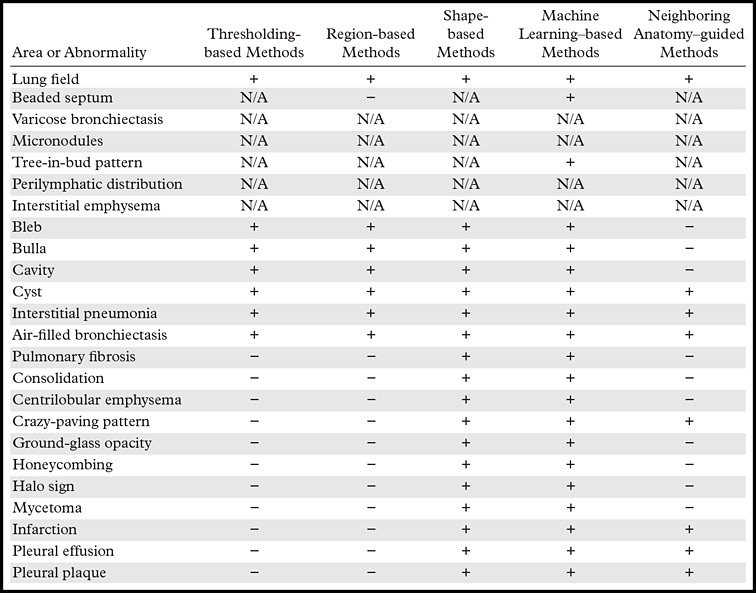
Note.—N/A = not available or not implemented for that particular pattern, + = method can be used successfully, − = method may fail.
Segmentation has been far more successful for nonmoving fixed structures such as the brain; segmentation of the lungs must address the challenges of dynamic chest volumes in the respiratory cycle. Moreover, some pathologic conditions in the lungs have limited the utility of lung segmentation because algorithms have often failed to accurately compute lung volumes when consolidations, masses, effusions, or pneumothorax is present. Recent advances (13,17,33,35,37,61–64,68), however, are starting to overcome these limitations by producing segmentation results that include the lesion(s) in a three-dimensional rendering, so that lung pathologic conditions can be calculated as a percentage of the total lung volume for assessing severity and yielding rates of change in disease with serial CT examinations.
Until recently, segmentation of lung fields was performed manually by using input from radiologists having expertise in diagnostic criteria and anatomic landmarks. However, progress in software quality and computational efficiency has made automated lung segmentation methods available to replace some manual measurements. At the current time, lung segmentation methods have not been amalgamated into single approaches or unified platforms using a single user interface. This fragmentation of these software approaches into incompatible packages has limited the effectiveness of implementing lung segmentation as a standard method in the clinical context. Most methods are designed for a particular subset of imaging abnormality. These approaches work well within their realm of particular imaging and anatomic characteristics but fail to address other subsets of lesions that may be on the image(s). Future efforts will seek to improve the quality of segmentation (of lung volumes and disease), increase the efficiency of these software platforms (for facile use in the clinical work flow), and unify the algorithms so that the user interface can seamlessly integrate with picture archiving and communication systems.
Conclusion
We provide a critical appraisal of the current approaches to lung segmentation on CT images to assist clinicians in making better decisions when selecting the tools for lung field segmentation. We divided the lung field segmentation methods into five broad categories, with an overview of relative advantages and disadvantages of the methods belonging to each group. We believe that this synopsis and the subsequent recommendations will supplement the role of radiologists in the diagnostic approach while guiding the selection and application of automated segmentation tools for pulmonary analysis.
Footnotes
Funding: The research was supported by the Intramural Programs of the Center for Infectious Disease Imaging, the National Institute of Allergy and Infectious Diseases, and the National Institute of Biomedical Imaging and Bioengineering, at the National Institutes of Health. All authors are or were employees of the National Institutes of Health.
Supported in part by the Center for Research in Computer Vision of the University of Central Florida.
Presented as an education exhibit at the 2013 RSNA Annual Meeting.
L.R.F. has provided disclosures (see “Disclosures of Conflicts of Interest”); all other authors have disclosed no relevant relationships.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Disclosures of Conflicts of Interest.—: L.R.F. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: holds a patent for a multi–-gray-scale overlay window (U.S. patent no. 8,406,493 B2); CRADA with Carestream Health. Other activities: disclosed no relevant relationships.
References
- 1.Twair AA, Torreggiani WC, Mahmud SM, Ramesh N, Hogan B. Significant savings in radiologic report turnaround time after implementation of a complete picture archiving and communication system (PACS). J Digit Imaging 2000;13(4):175–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu S, Hoffman EA, Reinhardt JM. Automatic lung segmentation for accurate quantitation of volumetric X-ray CT images. IEEE Trans Med Imaging 2001;20(6):490–498. [DOI] [PubMed] [Google Scholar]
- 3.Bagci U, Chen X, Udupa JK. Hierarchical scale-based multiobject recognition of 3-D anatomical structures. IEEE Trans Med Imaging 2012;31(3):777–789. [DOI] [PubMed] [Google Scholar]
- 4.Marr D, Poggio T. A computational theory of human stereo vision. Proc R Soc Lond B Biol Sci 1979;204(1156):301–328. [DOI] [PubMed] [Google Scholar]
- 5.Poggio T, Ullman S. Vision: are models of object recognition catching up with the brain? Ann N Y Acad Sci 2013;1305: 72–82. [DOI] [PubMed] [Google Scholar]
- 6.Serre T, Wolf L, Bileschi S, Riesenhuber M, Poggio T. Robust object recognition with cortex-like mechanisms. IEEE Trans Pattern Anal Mach Intell 2007;29(3):411–426. [DOI] [PubMed] [Google Scholar]
- 7.Marr D, Ullman S, Poggio T. Bandpass channels, zero-crossings, and early visual information processing. J Opt Soc Am 1979;69(6):914–916. [DOI] [PubMed] [Google Scholar]
- 8.Chen X, Udupa JK, Bagci U, Zhuge Y, Yao J. Medical image segmentation by combining graph cuts and oriented active appearance models. IEEE Trans Image Process 2012;21(4):2035–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bagci U, Kramer-Marek G, Mollura DJ. Automated computer quantification of breast cancer in small-animal models using PET-guided MR image co-segmentation. EJNMMI Res 2013;3(1):49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sluimer I, Schilham A, Prokop M, van Ginneken B. Computer analysis of computed tomography scans of the lung: a survey. IEEE Trans Med Imaging 2006;25(4): 385–405. [DOI] [PubMed] [Google Scholar]
- 11.Armato SG, 3rd, Sensakovic WF. Automated lung segmentation for thoracic CT impact on computer-aided diagnosis. Acad Radiol 2004;11(9):1011–1021. [DOI] [PubMed] [Google Scholar]
- 12.Bağcı U, Bray M, Caban J, Yao J, Mollura DJ. Computer-assisted detection of infectious lung diseases: a review. Comput Med Imaging Graph 2012;36(1):72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bagci U, Foster B, Miller-Jaster K, et al. A computational pipeline for quantification of pulmonary infections in small animal models using serial PET-CT imaging. EJNMMI Res 2013;3(1):55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christe A, Brönnimann A, Vock P. Volumetric analysis of lung nodules in computed tomography (CT): comparison of two different segmentation algorithm softwares and two different reconstruction filters on automated volume calculation. Acta Radiol 2014;55(1):54–61. [DOI] [PubMed] [Google Scholar]
- 15.Cui H, Wang X, Feng D. Automated localization and segmentation of lung tumor from PET-CT thorax volumes based on image feature analysis. Conf Proc IEEE Eng Med Biol Soc 2012;2012:5384–5387. [DOI] [PubMed] [Google Scholar]
- 16.Diciotti S, Lombardo S, Falchini M, Picozzi G, Mascalchi M. Automated segmentation refinement of small lung nodules in CT scans by local shape analysis. IEEE Trans Biomed Eng 2011;58(12):3418–3428. [DOI] [PubMed] [Google Scholar]
- 17.Elinoff JM, Bagci U, Moriyama B, et al. Recombinant human factor VIIa for alveolar hemorrhage following allogeneic stem cell transplantation. Biol Blood Marrow Transplant 2014;20(7):969–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo Y, Zhou C, Chan HP, et al. Automated iterative neutrosophic lung segmentation for image analysis in thoracic computed tomography. Med Phys 2013;40(8):081912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun S, Bauer C, Beichel R. Automated 3-D segmentation of lungs with lung cancer in CT data using a novel robust active shape model approach. IEEE Trans Med Imaging 2012;31(2):449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Li F, Li Q. Automated segmentation of lungs with severe interstitial lung disease in CT. Med Phys 2009;36(10):4592–4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Z, Bagci U, Kubler A, et al. Computer-aided detection and quantification of cavitary tuberculosis from CT scans. Med Phys 2013;40(11):113701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pu J, Roos J, Yi CA, Napel S, Rubin GD, Paik DS. Adaptive border marching algorithm: automatic lung segmentation on chest CT images. Comput Med Imaging Graph 2008;32(6):452–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sluimer IC, Niemeijer M, van Ginneken B. Lung field segmentation from thin-slice CT scans in presence of severe pathology. In: Fitzpatrick JM, Sonka M, eds. Proceedings of SPIE: medical imaging 2004—image processing. Vol 5370. Bellingham, Wash: International Society for Optical Engineering, 2004; 1447. [Google Scholar]
- 24.Yao J, Bliton J, Summers RM. Automatic segmentation and measurement of pleural effusions on CT. IEEE Trans Biomed Eng 2013;60(7):1834–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mansoor A, Bagci U, Xu Z, et al. A generic approach to pathological lung segmentation. IEEE Trans Med Imaging 2014;33(12):2293–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sluimer I, Prokop M, van Ginneken B. Toward automated segmentation of the pathological lung in CT. IEEE Trans Med Imaging 2005;24(8):1025–1038. [DOI] [PubMed] [Google Scholar]
- 27.Folio LR, Sandouk A, Huang J, Solomon JM, Apolo AB. Consistency and efficiency of CT analysis of metastatic disease: semiautomated lesion management application within a PACS. AJR Am J Roentgenol 2013;201(3):618–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hansell DM, Bankier AA, MacMahon H, McLoud TC, Müller NL, Remy J. Fleischner Society: glossary of terms for thoracic imaging. Radiology 2008;246(3):697–722. [DOI] [PubMed] [Google Scholar]
- 29.Cooke RE, Jr, Gaeta MG, Kaufman DM, Henrici JG, inventors; Agfa Corporation, assignee . Picture archiving and communication system. U.S. patent 6,574,629. June 3, 2003.
- 30.Brown MS, McNitt-Gray MF, Mankovich NJ, et al. Method for segmenting chest CT image data using an anatomical model: preliminary results. IEEE Trans Med Imaging 1997;16(6):828–839. [DOI] [PubMed] [Google Scholar]
- 31.Kemerink GJ, Lamers RJ, Pellis BJ, Kruize HH, van Engelshoven JM. On segmentation of lung parenchyma in quantitative computed tomography of the lung. Med Phys 1998;25(12):2432–2439. [DOI] [PubMed] [Google Scholar]
- 32.Ko JP, Betke M.. Chest CT: automated nodule detection and assessment of change over time—preliminary experience. Radiology 2001;218(1):267–273. [DOI] [PubMed] [Google Scholar]
- 33.Armato SG, 3rd. Image annotation for conveying automated lung nodule detection results to radiologists. Acad Radiol 2003;10(9):1000–1007. [DOI] [PubMed] [Google Scholar]
- 34.Armato SG, 3rd. Computerized analysis of mesothelioma on CT scans. Lung Cancer 2005;49(suppl 1):S41–S44. [DOI] [PubMed] [Google Scholar]
- 35.Armato SG, 3rd, Altman MB, Wilkie J, et al. Automated lung nodule classification following automated nodule detection on CT: a serial approach. Med Phys 2003;30(6): 1188–1197. [DOI] [PubMed] [Google Scholar]
- 36.Armato SG, 3rd, Oxnard GR, Kocherginsky M, Vogelzang NJ, Kindler HL, MacMahon H. Evaluation of semiautomated measurements of mesothelioma tumor thickness on CT scans. Acad Radiol 2005;12(10):1301–1309. [DOI] [PubMed] [Google Scholar]
- 37.Giger ML, Karssemeijer N, Armato SG, 3rd. Computer-aided diagnosis in medical imaging. IEEE Trans Med Imaging 2001;20(12):1205–1208. [DOI] [PubMed] [Google Scholar]
- 38.Reeves AP, Biancardi AM, Apanasovich TV, et al. The Lung Image Database Consortium (LIDC): a comparison of different size metrics for pulmonary nodule measurements. Acad Radiol 2007;14(12):1475–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roy AS, Armato SG, 3rd, Wilson A, Drukker K. Automated detection of lung nodules in CT scans: false-positive reduction with the radial-gradient index. Med Phys 2006;33(4):1133–1140. [DOI] [PubMed] [Google Scholar]
- 40.Adams R, Bischof L. Seeded region growing. IEEE Trans Pattern Anal Mach Intell 1994;16(6):641–647. [Google Scholar]
- 41.Hojjatoleslami SA, Kittler J. Region growing: a new approach. IEEE Trans Image Process 1998;7(7):1079–1084. [DOI] [PubMed] [Google Scholar]
- 42.Pavlidis T, Liow YT. Integrating region growing and edge detection. IEEE Trans Pattern Anal Mach Intell 1990;12(3):225–233. [Google Scholar]
- 43.Tremeau A, Borel N. A region growing and merging algorithm to color segmentation. Pattern Recognit 1997;30(7):1191–1203. [Google Scholar]
- 44.Zhu SC, Yuille A. Region competition: unifying snakes, region growing, and Bayes/MDL for multiband image segmentation. IEEE Trans Pattern Anal Mach Intell 1996;18(9):884–900. [Google Scholar]
- 45.Mangan AP, Whitaker RT. Partitioning 3D surface meshes using watershed segmentation. IEEE Trans Vis Comput Graph 1999;5(4):308–321. [Google Scholar]
- 46.Boykov Y, Jolly MP. Interactive organ segmentation using graph cuts. In: Delp SL, DiGoia AM, Jaramaz B, eds. Medical Image Computing and Computer-assisted Intervention: MICCAI 2000. Berlin, Germany: Springer-Verlag, 2000; 276–286. [Google Scholar]
- 47.Grady L. Random walks for image segmentation. IEEE Trans Pattern Anal Mach Intell 2006;28(11):1768–1783. [DOI] [PubMed] [Google Scholar]
- 48.Udupa JK, Samarasekera S. Fuzzy connectedness and object definition: theory, algorithms, and applications in image segmentation. Graph Models Image Proc 1996;58(3): 246–261. [Google Scholar]
- 49.Xu Z, Bagci U, Foster B, Mansoor A, Mollura DJ. Spatially constrained random walk approach for accurate estimation of airway wall surfaces. Med Image Comput Comput Assist Interv 2013;16(pt 2):559–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li B, Christensen GE, Hoffman EA, McLennan G, Reinhardt JM. Establishing a normative atlas of the human lung: intersubject warping and registration of volumetric CT images. Acad Radiol 2003;10(3):255–265. [DOI] [PubMed] [Google Scholar]
- 51.Zhang L, Reinhardt JM. 3D pulmonary CT image registration with a standard lung atlas. In: Chen CT, Clough AV, eds. Proceedings of SPIE: medical imaging 2000—physiology and function from multidimensional images. Vol 3978. Bellingham, Wash: International Society for Optical Engineering, 2000; 67. [Google Scholar]
- 52.Xu C, Prince JL. Snakes, shapes, and gradient vector flow. IEEE Trans Image Process 1998;7(3):359–369. [DOI] [PubMed] [Google Scholar]
- 53.Vese LA, Chan TF. A multiphase level set framework for image segmentation using the Mumford and Shah model. Int J Comput Vis 2002;50(3):271–293. [Google Scholar]
- 54.Hua P, Song Q, Sonka M, et al. Segmentation of pathological and diseased lung tissue in CT images using a graph-search algorithm. In: IEEE International Symposium on Biomedical Imaging: from nano to macro. New York, NY: Institute of Electrical and Electronics Engineers, 2011; 2072–2075. [Google Scholar]
- 55.Song Y, Cai W, Zhou Y, Feng DD. Feature-based image patch approximation for lung tissue classification. IEEE Trans Med Imaging 2013;32(4):797–808. [DOI] [PubMed] [Google Scholar]
- 56.Xu Y, Sonka M, McLennan G, Guo J, Hoffman EA. MDCT-based 3-D texture classification of emphysema and early smoking related lung pathologies. IEEE Trans Med Imaging 2006;25(4):464–475. [DOI] [PubMed] [Google Scholar]
- 57.Yao J, Dwyer A, Summers RM, Mollura DJ. Computer-aided diagnosis of pulmonary infections using texture analysis and support vector machine classification. Acad Radiol 2011;18(3):306–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Korfiatis PD, Karahaliou AN, Kazantzi AD, Kalogeropoulou C, Costaridou LI. Texture-based identification and characterization of interstitial pneumonia patterns in lung multidetector CT. IEEE Trans Inf Technol Biomed 2010;14(3):675–680. [DOI] [PubMed] [Google Scholar]
- 59.Bagci U, Yao J, Wu A, et al. Automatic detection and quantification of tree-in-bud (TIB) opacities from CT scans. IEEE Trans Biomed Eng 2012;59(6):1620–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Rikxoort EM, van Ginneken B. Automated segmentation of pulmonary structures in thoracic computed tomography scans: a review. Phys Med Biol 2013;58(17): R187–R220. [DOI] [PubMed] [Google Scholar]
- 61.Bagci U, Miller-Jaster K, Yao J, et al. Automatic quantification of tree-in-bud patterns from CT scans. Proc IEEE Int Symp Biomed Imaging 2012;2012:1459–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bağcı U, Yao J, Caban J, Palmore TN, Suffredini AF, Mollura DJ. Automatic detection of tree-in-bud patterns for computer assisted diagnosis of respiratory tract infections. Conf Proc IEEE Eng Med Biol Soc 2011;2011:5096–5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bagci U, Yao J, Caban J, Suffredini AF, Palmore TN, Mollura DJ. Learning shape and texture characteristics of CT tree-in-bud opacities for CAD systems. Med Image Comput Comput Assist Interv 2011;14(pt 3):215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Caban JJ, Yao J, Bagci U, Mollura DJ. Monitoring pulmonary fibrosis by fusing clinical, physiological, and computed tomography features. Conf Proc IEEE Eng Med Biol Soc 2011;2011:6216–6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Udupa JK, Leblanc VR, Zhuge Y, et al. A framework for evaluating image segmentation algorithms. Comput Med Imaging Graph 2006;30(2):75–87. [DOI] [PubMed] [Google Scholar]
- 66.Sonka M, Hlavac V, Boyle R. Image processing, analysis, and machine vision. 3rd ed. Toronto, Ontario: Thomson, 2008. [Google Scholar]
- 67.Awai K, Murao K, Ozawa A, et al. Pulmonary nodules at chest CT: effect of computer-aided diagnosis on radiologists’ detection performance. Radiology 2004;230(2):347–352. [DOI] [PubMed] [Google Scholar]
- 68.Joo S, Yang YS, Moon WK, Kim HC. Computer-aided diagnosis of solid breast nodules: use of an artificial neural network based on multiple sonographic features. IEEE Trans Med Imaging 2004;23(10):1292–1300. [DOI] [PubMed] [Google Scholar]