Significance
Glioblastoma is the most common type of adult brain cancer, with near-uniform fatality within 2 y of diagnosis. Therapeutic failure is thought to be related to small subpopulations of cells that exhibit tumorigenicity, the cellular capacity to reconstitute the entire tumor mass. One fundamental issue is whether tumorigenicity exists within a static subpopulation of cells or whether the capacity is stochastically acquired. We provide evidence that tumorigenicity is a cellular property that is durable yet undergoes low-frequency stochastic changes. We showed that these changes are driven by lysine-specific demethylase 1 (LSD1)-mediated epigenetic (heritable non-DNA sequence-altering) modifications that impact expression of key transcription factors, which in turn govern transitions between tumorigenic states. These findings harbor implications for glioblastoma therapeutic development.
Keywords: epigenomics, glioblastoma, neoplastic stem cells
Abstract
The available evidence suggests that the lethality of glioblastoma is driven by small subpopulations of cells that self-renew and exhibit tumorigenicity. It remains unclear whether tumorigenicity exists as a static property of a few cells or as a dynamically acquired property. We used tumor-sphere and xenograft formation as assays for tumorigenicity and examined subclones isolated from established and primary glioblastoma lines. Our results indicate that glioblastoma tumorigenicity is largely deterministic, yet the property can be acquired spontaneously at low frequencies. Further, these dynamic transitions are governed by epigenetic reprogramming through the lysine-specific demethylase 1 (LSD1). LSD depletion increases trimethylation of histone 3 lysine 4 at the avian myelocytomatosis viral oncogene homolog (MYC) locus, which elevates MYC expression. MYC, in turn, regulates oligodendrocyte lineage transcription factor 2 (OLIG2), SRY (sex determining region Y)-box 2 (SOX2), and POU class 3 homeobox 2 (POU3F2), a core set of transcription factors required for reprogramming glioblastoma cells into stem-like states. Our model suggests epigenetic regulation of key transcription factors governs transitions between tumorigenic states and provides a framework for glioblastoma therapeutic development.
Glioblastoma is the most common primary brain cancer and remains one of the deadliest of malignancies despite contemporary treatment strategies (1), with near-uniform fatality within 2 y of diagnosis (2, 3). There is growing evidence that the lethality of this tumor is driven by subpopulations of cells with properties of self-renewal and tumorigenicity (4)—that is, the capacity to generate phenocopies of the original tumor when transplanted (5, 6). How glioblastoma cells retain or gain tumorigenicity while the bulk of the tumor does not remains a fundamental question. Conceptualization of this phenomenon includes the elite and stochastic models (7). The elite model states that restricted cell subpopulations harbor intrinsic tumorigenic properties that cannot be acquired once lost. The stochastic model, on the other hand, presupposes that all cells within a population are intrinsically comparable in their ability to spontaneously acquire or lose tumorigenicity.
Epigenetic alterations are stable, long-term changes in cellular phenotype that are not due to variations in DNA sequence (8). One means by which epigenetic alterations impact cell phenotype involves modulation of transcriptional activity via histone modification (9). Here we demonstrate that glioblastoma tumorigenicity is best conceptualized by a hybrid elite-stochastic model governed by histone modification through the lysine-specific demethylase 1 (LSD1; aka KDM1A) (10). This modification, in turn, influences the expression of key transcription factors required to reprogram glioblastoma cells into a stem-like state, including avian myelocytomatosis viral oncogene homolog (MYC) (11), oligodendrocyte lineage transcription factor 2 (OLIG2), SRY (sex determining region Y)-box 2 (SOX2), and POU class 3 homeobox 2 (POU3F2) (12). Our results suggest that the framework governing glioblastoma tumorigenicity parallels that described in mammalian development, where cell fate is dictated by epigenetic and hierarchical regulation of a core set of transcription factors (13).
Results
Examination of Models of Glioblastoma Tumorigenicity.
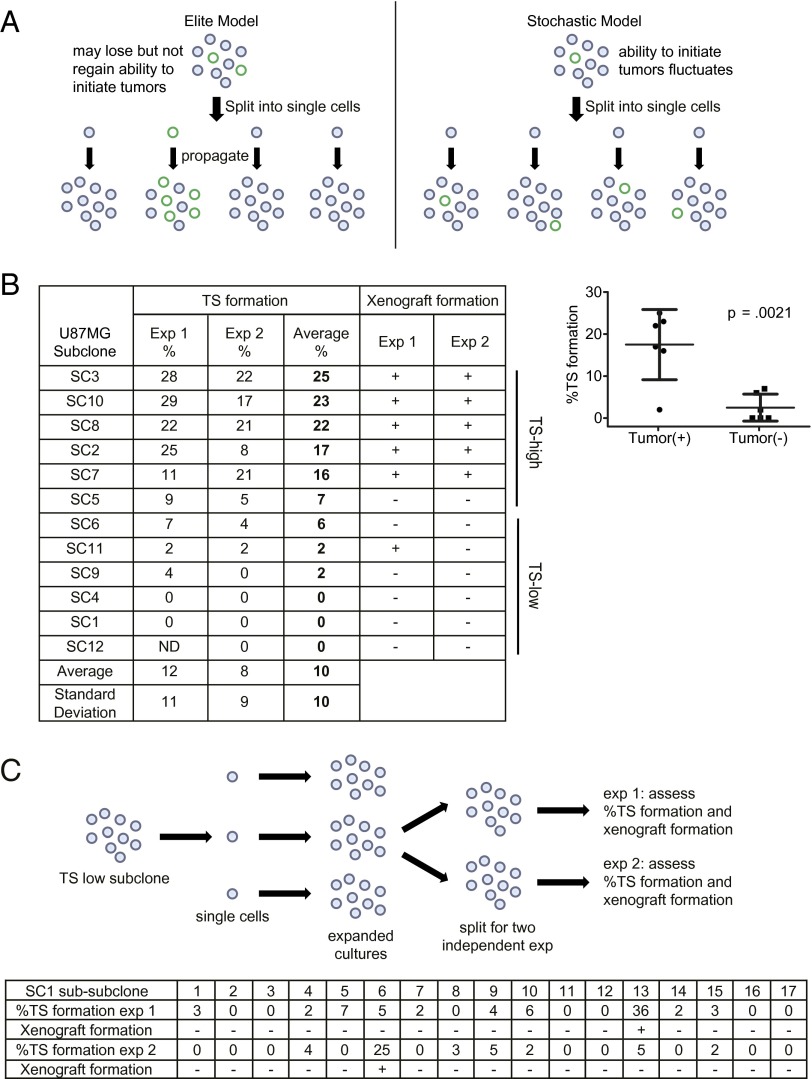
Two accepted assays of tumorigenicity include tumor-sphere (TS) formation—that is, cellular capacity to propagate in serum-free media as aggregates in suspension (14)—and xenograft growth—that is, cellular capacity to initiate tumor formation upon transplant into immunocompromised mice (5). A purely elite model predicts two distinct types of subclones: (i) clones derived from elite cells that are capable of both TS and xenograft formation and (ii) clones derived from nonelite cells with neither capability. On the other hand, a purely stochastic model predicts that cells derived from all subclones will show equal capacities for both TS and xenograft formation (Fig. 1A).
Fig. 1.
Glioblastoma tumorigenicity shows hybrid features of elite and stochastic models. (A) Schematic representation of each model and its predictions. Colored circles represent cells with (green) and without (gray) capacity for tumor formation. (B) Subclones of U87MG were passaged under serum/adherent conditions and tested for ability of single-cell suspensions to propagate as TSs. In parallel, the subclones were tested for ability to form s.c. xenografts. A plus sign denotes formation of at least one tumor in the five mice injected with the various subclones. Results of two independent experiments are shown (Left). Vertical scatter plot depicts frequencies of TS formation grouped by xenograft formation ability per Exp 1 (Right). Error bars, SD. (C) Subclones of a low TS-forming subclone, U87MG-SC1, were tested for TS and xenograft formation capacity. (Top) Schematic depicting the experiment. For each subclone, cells were split into two independent cultures when the cell number reached 1 × 105 and passaged for 20 additional passages before single-cell TS formation and xenograft assays. (Bottom) Percentages of single-cell suspensions from each U87MG-SC1 subclone that formed TS as well as the xenograft formation capacity of each subclone. A plus sign denotes formation of at least one tumor in the five mice injected (Bottom).
To test these models, we isolated subclones derived from single cells of three well-characterized, patient-derived glioblastoma lines with tumorigenic properties: U87MG, a line established for over 40 y (15); 83, a line that underwent ∼30 passages (16); and CMK3, a line that underwent <5 passages (17). To assess the extent of their genetic homogeneity, the subclones were profiled by Affymetrix Human Mapping 250K Nsp Arrays and whole exome sequencing (Fig. S1 and Datasets S1–S3). We observed ∼1% of genetic variation between the various subclones of U87MG and 83. We did not uncover any recurrent genetic events associated with tumorigenicity. Moreover, the level of genetic variation detected was within the range of systematic or genotyping errors. We therefore considered these subclones to be genomically isogenic within statistical error of SNP arrays and exome sequencing.
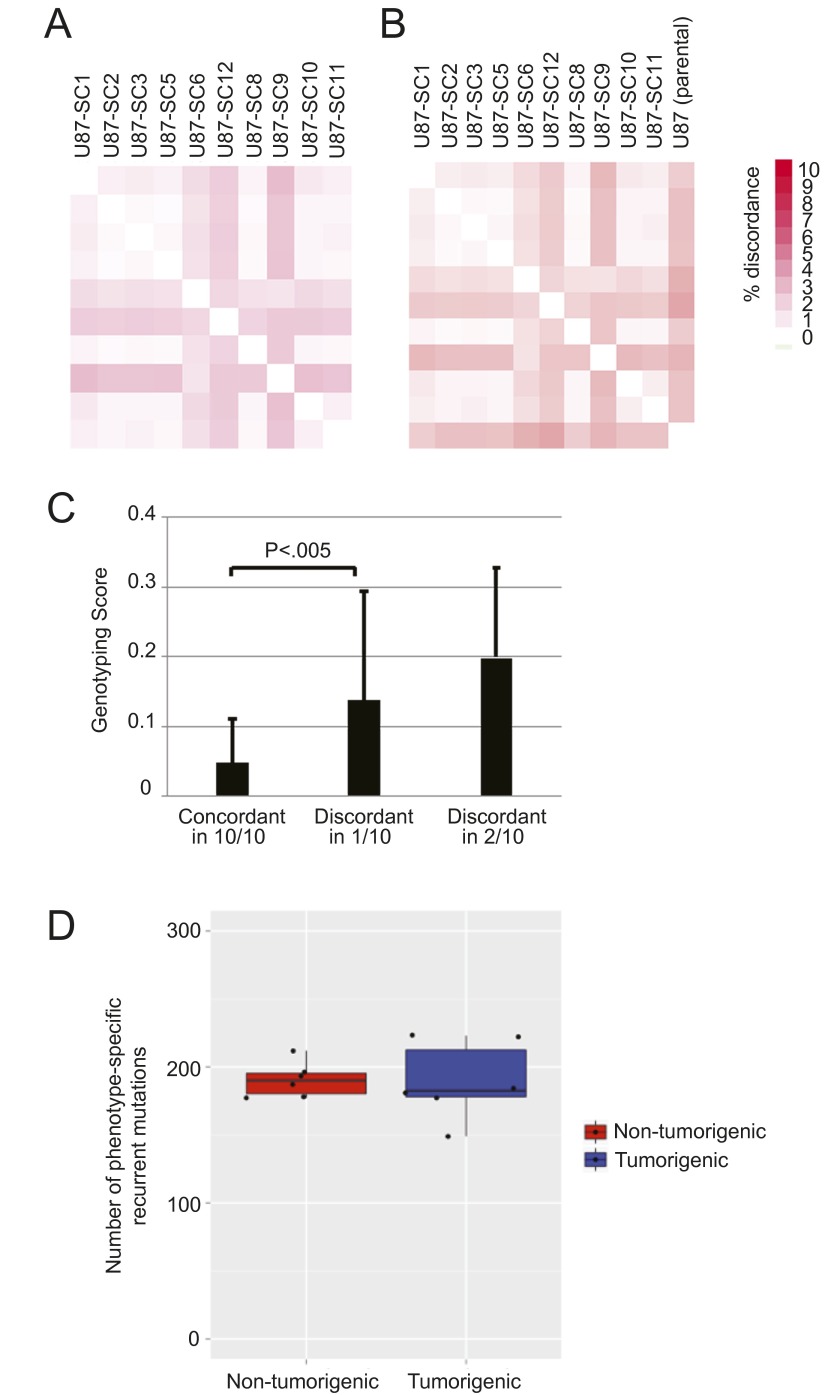
Fig. S1.
Differences in glioblastoma tumorigenicity among U87MG subclones are unlikely due to genetic changes. (A) Genotyping of U87MG subclones using Affymetrix Human Mapping 250K NSP arrays. Heat map representing the fraction of discordant genotypes between each pair of clones out of ∼251,650 loci called in both samples. (B) Heat map representing the fraction of discordant genotypes between each clone and the parental U87MG cell line at the ∼80,398 loci called in both Illumina and Affymetrix arrays. (C) Genotype quality score (decreasing scale) between scores at 228,774 × 10 genotypes concordant in all 10 clones and scores at 2,435 and 920 × 2 genotypes discordant in one or two clones, respectively. Statistical differences between groups were assessed using the paired Student’s t test; error bars represent SD. Of all the loci, 98% were concordant in all tested U87MG subclones; the low level of discordance is attributable to systematic errors in the genotyping, as the quality scores of the concordant genotypes were significantly different from those of discordant genotypes. (D) Number of recurrent mutations specific to nontumorigenic versus tumorigenic subclones of the 83 and U87MG glioblastoma cell lines, determined by whole exome sequencing of six subclones of each phenotype. P = 0.55 by Student’s t test, showing no significant difference that would otherwise suggest tumorigenesis is linked to a gross difference in mutational load.
Approximately 50% of subclones generated from the established glioblastoma line U87MG were capable of both TS and xenograft formation (Fig. 1B). Moreover, these capacities were mostly stable between independent experiments, suggesting that tumorigenicity is substantially an intrinsic property of the subclones. Similar clone-to-clone variations in tumorigenicity were observed for 83 and CMK3 (Fig. S2 A and B). These clone-to-clone variations support the elite model of tumorigenicity. Further, we observed that the degree of variance (e.g., subclone-to-subclone variation) in TS formation among the U87MG subclones exceeded that expected from experimental handling (Fig. S2C), suggesting that the intrinsic properties of these subclones contributed to the overall variance of TS formation.
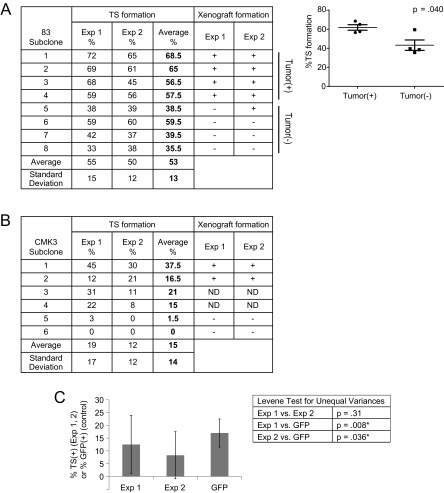
Fig. S2.
TS and xenograft formation assays in short-term–passaged glioblastoma lines recapitulate findings in the established glioblastoma line U87MG. (A) Subclones of the glioblastoma line 83 were passaged under serum/adherent conditions and tested for ability of single-cell suspensions to propagate as TSs. In parallel, the subclones were tested for ability to form s.c. xenografts. A plus sign denotes formation of at least one tumor in the five mice injected with the various subclones. Results of two independent experiments are shown (Left). Vertical scatter plot depicts frequencies of TS formation grouped by xenograft formation ability per Exp 1 (Right). (B) As above, but with the primary glioblastoma line CMK3. (C) Comparison of variances in frequencies of TS formation of U87MG subclones, relative to that expected from variation in experimental handling. For the latter, experiments were repeated 10 times using a mixture of 20% GFP-expressing U87MG and 80% non–GFP-expressing U87MG, which approximates the deterministic model (Left). The Levene test for equality of variances was performed, with asterisks denoting statistically different variances (Right).
On the other hand, we noted that a few subclones exhibited changes in tumorigenicity, such as U87MG-SC11 (Fig. 1B). This observation suggested a stochastic component to tumorigenicity. To further characterize this stochastic component, we isolated 17 single cell-derived subclones from U87MG-SC1, a line incapable of TS or xenograft formation, and tested whether these subclones can spontaneously acquire these properties of tumorigenicity. Importantly, for each subclone, cells were split into two independent cultures when the cell number reached 1 × 105. The cultures were then assessed in terms of TS- and xenograft-forming capacities after 20 additional passages (Fig. 1C). In the first set of cultures, one U87MG-SC1 subclone acquired the capacities for TS and xenograft formation, whereas in the second set a distinct subclone acquired these capacities. These observations suggest that a stochastic event occurring after the culture split induced tumorigenicity. Overall, our findings suggest that tumorigenicity in glioblastoma cells is best described by a hybrid model that is largely deterministic (elite) but with opportunities for dynamic (stochastic) interchange between nontumorigenic and tumorigenic states.
Requirement of MYC Expression in Glioblastoma Tumorigenicity.
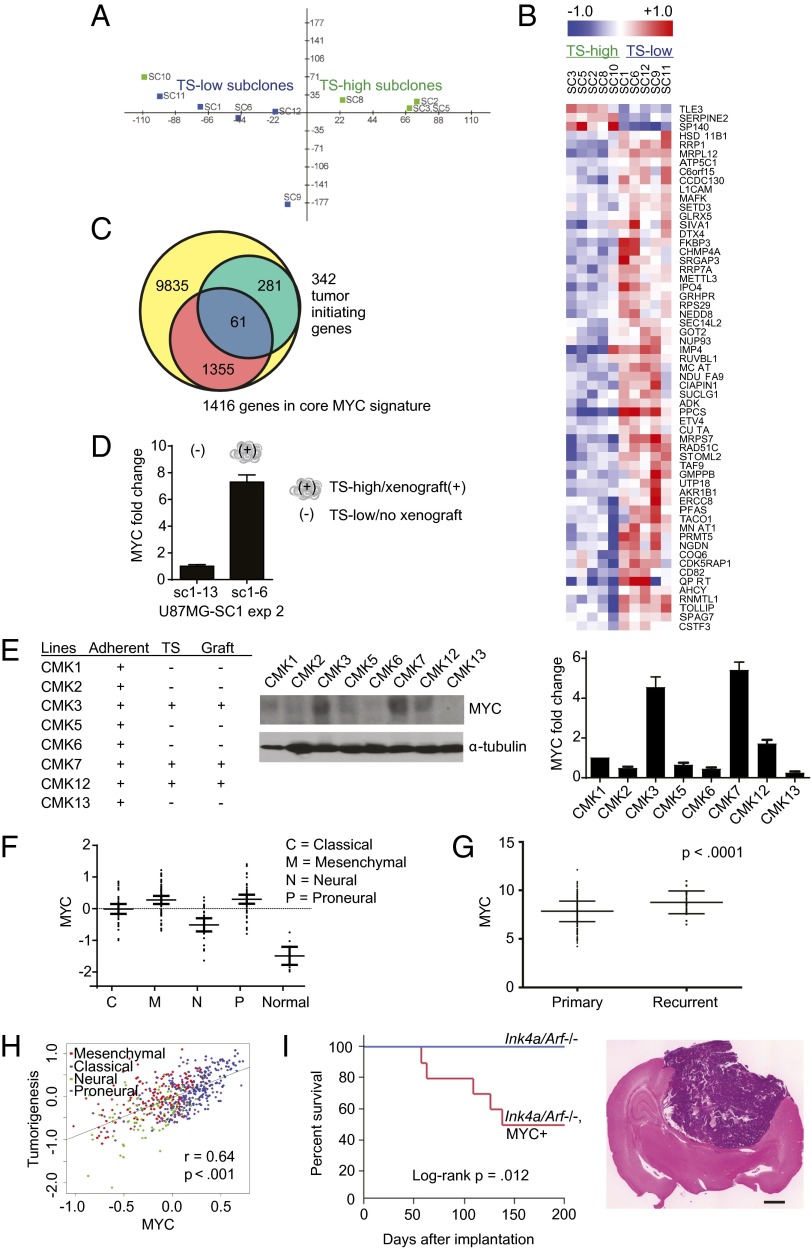
To elucidate the molecular determinants that mediate tumorigenicity, we performed expression profiling of the U87MG subclones. Realizing the hazards of signature creation from a limited dataset (18), we tested whether the subclones of differing tumorigenicity can be distinguished based on a previously published gene signature that correlated with tumorigenicity in a series of short-term–passaged, patient-derived glioblastoma lines (19). Principal component analysis and hierarchical clustering using this signature partitioned our subclones by their intrinsic tumorigenicity (Fig. 2 A and B). This observation suggested that the physiology underlying tumorigenicity in U87MG is analogous to that previously observed in short-term–passaged glioblastoma lines. Supporting this hypothesis, tumorigenic subclones of U87MG exhibited increased expression of CD133, a cell surface marker associated with glioblastoma tumorigenicity (5, 6) (Fig. S3A).
Fig. 2.
MYC expression associates with tumorigenicity of glioblastoma cells. (A) Principal component analysis of U87MG subclones showing high versus low propensities for propagation as TSs using a tumorigenicity signature (17) that partitioned the subclones. (B) Heat map showing genes that differed significantly in expression between TS high versus low subclones. (C) Venn diagram showing enrichment of core MYC-driven genes in the tumorigenicity signature. Blue, tumorigenicity genes overlapping with the core MYC signature; green, tumorigenicity genes not in the core MYC signature; red, core MYC signature genes not in the tumorigenicity signature; yellow, genes in neither set. Overlap of the core MYC signature and the tumorigenicity signature was significantly higher than would be expected by chance (17.8% vs. 12.3% of total genes; χ2 two-tailed P < 0.0001). Genes are listed in Dataset S4. (D) qRT-PCR showed increased MYC expression in a U87MG-SC1 subclone with high tumorigenicity (sc1–6) relative to a subclone with low tumorigenicity (sc1–13) (Fig. 1C, exp 2). Error bars, SD. (E) Elevated MYC expression in tumorigenic, clinical glioblastoma specimens. Eight freshly resected glioblastoma specimens were dissociated and tested for growth as adherent lines, as TS, and as s.c. flank xenografts in nude mice (Left). (Middle) Western blot of MYC expression. (Right) Densitometric quantitation of MYC expression normalized to α-tubulin. Error bars, SD. (F) Elevated MYC expression in TCGA glioblastoma samples (grouped by molecular subtype) relative to 10 normal brain samples profiled by the same microarrays. Error bars, 95% confidence interval. (G) Elevated MYC expression in TCGA recurrent glioblastomas relative to newly diagnosed tumors (profiled by the same microarray platform). Error bars, SD. (H) Correlation of a core MYC signature and a glioblastoma xenograft tumorigenicity signature (24) (Materials and Methods). The color of each dot denotes the TCGA molecular subtype for each tumor. Correlation was analyzed by linear regression (P < 0.01, r = 0.64). (I) Tumorigenicity of non–tumor-forming Cdkn2a (Ink4a/Arf)-null NSCs/NPCs following exogenous MYC expression. (Left) Survival of mice bearing orthotopic transplants of 105 NSCs/NPCs transduced without or with human MYC cDNA retroviral constructs. [H&E-stained whole-brain section scale bar, 1,000 µm (Right).]
Fig. S3.
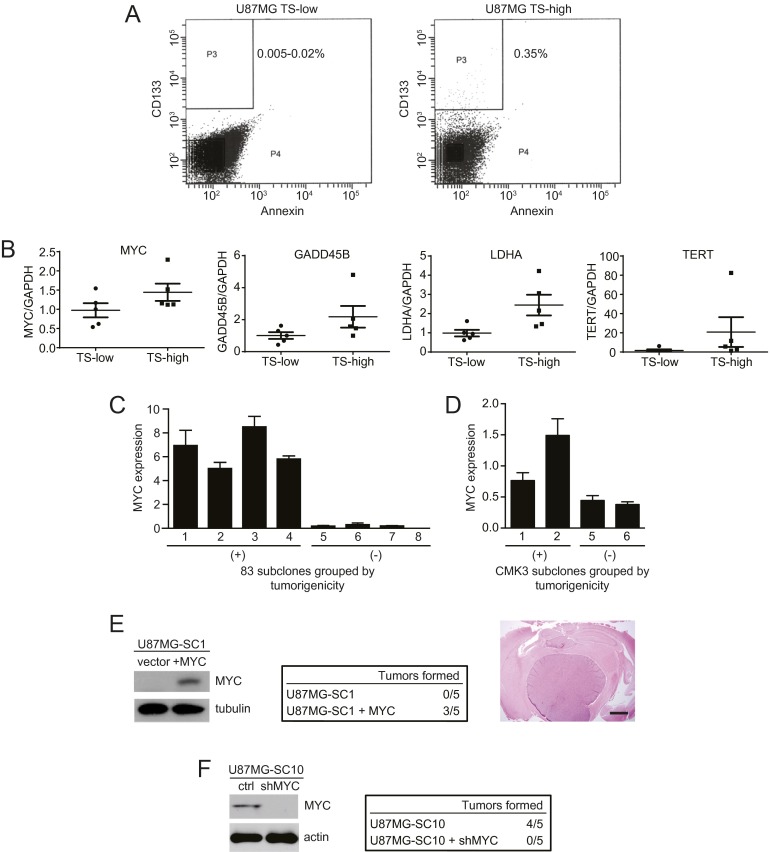
Modulation of MYC expression alters glioblastoma tumorigenicity. (A) Flow cytometry to quantify the CD133+ population in a U87MG subclone showing low versus high tumorigenicity. The y axis, CD133 staining; x axis, annexin staining. (B) Expression of MYC and MYC target genes quantified by RT-PCR in U87MG subclones grouped by low (TS-low) versus high (TS-high) propensity for TS formation. Error bars, SD. (C) Assessment by RT-PCR of relative MYC expression in subclones of the glioblastoma line 83 grouped by tumorigenicity (Fig. S2A). (D) Assessment by RT-PCR of relative MYC expression in subclones of the glioblastoma line CMK3 grouped by tumorigenicity (Fig. S2B). (E) Western blot of MYC expression in U87MG-SC1, a U87MG subclone that showed no tumorigenicity, without versus with exogenous MYC expression (Left). Exogenous MYC expression restored tumorigenicity of U87MG-SC1 when injected intracranially in nude mice; n = 5 per group (Center). [H&E-stained whole-brain section scale bar, 1,000 µm (Right).] (F) Western blot of MYC expression in U87MG-SC10, a tumorigenic U87MG subclone, without versus with MYC knockdown by doxycycline-inducible MYC shRNA (ctrl, no doxycycline; shMYC, doxycycline added at day 0) (Left). MYC knockdown abolished tumorigenicity of U87MG-SC10 when injected intracranially in nude mice; n = 5 per group (Right).
Pathway analysis of our tumorigenicity signature did not reveal enrichment for any particular biologic processes. We next explored whether these genes were commonly regulated by a master transcription factor. To this end, we identified gene signatures associated with transcription factors implicated in glioblastoma pathogenesis (20, 21) and determined their overlap with the tumorigenicity signature (19). Using this approach, we identified MYC (21). Of the 342 genes in the tumorigenicity signature that were evaluable across microarray platforms, 61 (17.8%) overlapped with the 1,416 evaluable genes in the MYC signature. This overlap was significantly higher than would be expected by chance (1,416/11,532 total genes, or 12.3%; χ2 two-tailed P < 0.0001; Fig. 2C and Dataset S4). Supporting the importance of MYC as a tumorigenicity determinant, tumorigenic U87MG subclones consistently exhibited increased expression of MYC as well as established MYC target genes, including growth arrest and DNA-damage–inducible beta (GADD45B) (22), lactate dehydrogenase A (LDHA) (23), and telomerase reverse transcriptase (TERT) (24) (Fig. S3B). Notably, spontaneous acquisition of tumorigenicity by U87MG-SC1 subclones (Fig. 1C) was accompanied by increased MYC expression (Fig. 2D). MYC expression also tracked closely with tumorigenicity in the glioblastoma lines 83 and CMK3 (Fig. S3 C and D).
As further evidence of the critical role of MYC as a determinant of tumorigenicity, its exogenous expression in U87MG-SC1 restored cellular capacity for xenograft formation (Fig. S3E). Further, MYC silencing in a tumorigenic U87MG subclone, U87MG-SC10, abolished its tumorigenicity (Fig. S3F).
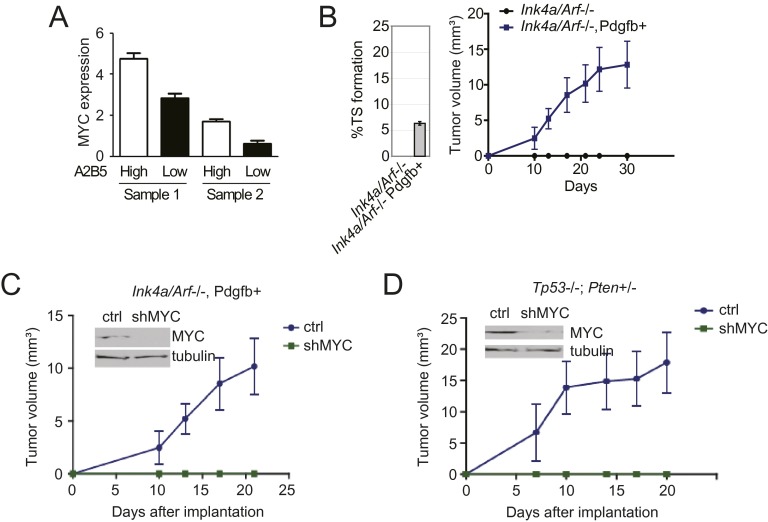
To gain assurance that the findings are not specific to established glioblastoma lines, we performed experiments using freshly isolated human glioblastoma specimens. Each specimen was immediately processed for MYC immunoblotting, cell culture, and xenograft growth in nude mice. The three specimens out of eight that could be cultured as TSs and formed xenografts also harbored the highest expression levels of MYC (Fig. 2E). Of note, lines derived from the two specimens with the highest MYC expression (CMK3 and CMK7) propagated as xenografts after 10 serial passages, whereas the line derived from the specimen with the lower MYC expression (CMK12) failed to propagate beyond 10 passages. Second, we tested whether subpopulations from a single glioblastoma specimen differing in tumorigenicity differed in MYC expression. A2B5 is a cell surface ganglioside epitope expressed on glial progenitors and marks glioblastoma cells with enhanced tumorigenicity (25). When cells from fresh glioblastoma specimens from two independent patients were FACS sorted into high and low A2B5-expressing subpopulations, those showing high expression showed 2–3-fold higher MYC expression relative to those with low expression (Fig. S4A).
Fig. S4.
Direct and indirect modulation of MYC expression impacts glioblastoma tumorigenicity. (A) Correlation of MYC expression and the cell surface tumorigenicity marker A2B5. Cells from two freshly resected glioblastomas were sorted by high versus low A2B5 expression. MYC expression in the sorted cells was assessed by RT-PCR. Error bars, SD. (B) Single-cell suspensions were prepared from a glioblastoma line derived from an Ink4a/Arf−/− GEMM upon exogenous Pdgfb expression (27) versus an Ink4a/Arf−/− NSC/NPC line and tested for capacity to propagate as TSs (Left). These lines were also tested for capacity for nude mouse s.c. flank xenograft growth; n = 5 per group (Right). (C) Tumorigenicity of GEMM glioblastoma lines following MYC silencing. Ink4a/Arf−/−, Pdgfb+ cells were transduced with doxycycline-inducible MYC shRNA, injected into the flanks of nude mice, and followed for tumor growth. Ctrl, no doxycycline; shMYC, doxycycline given starting 7 d after implantation; n = 5 per group. Error bars, SD. (D) As C, with Tp53−/−; Pten+/− cells (29).
To analyze the association of MYC expression with tumorigenicity in a larger number of clinical glioblastoma specimens, we took a bioinformatics approach. Using The Cancer Genome Atlas (TCGA) glioblastoma database, we found that MYC expression is elevated in all glioblastoma subtypes relative to normal human cerebrum (Fig. 2F). Moreover, MYC expression was elevated in recurrent glioblastomas relative to newly diagnosed glioblastomas (Fig. 2G). Expression of a MYC core signature (21) further correlated with a gene signature associated with tumorigenicity in glioblastomas (26) (r = 0.64; Fig. 2H). These results support an association between MYC expression and tumorigenicity.
We additionally confirmed the association between MYC expression and tumorigenicity in three genetically engineered murine models (GEMMs) of glioblastoma. Cyclin-dependent kinase inhibitor 2A (Cdkn2a, also known as Ink4a/Arf)-null neural stem/progenitor cells (NSCs/NPCs) (27), which are not tumorigenic, formed lethal intracranial tumors upon exogenous MYC expression (Fig. 2I). Exogenous expression of platelet-derived growth factor beta (PDGFB) in the NSC/NPC line increased its tumorigenicity (27, 28) (Fig. S4B). This tumorigenicity was abolished upon doxycycline-inducible MYC shRNA silencing in this line (Fig. S4C). Similar loss of tumorigenicity following MYC shRNA silencing was also observed in a line derived from an hGFPA-Cre+: Tp53lox/lox; Ptenlox/+ GEMM (29) (Fig. S4D). Overall, these observations demonstrated consistent associations between MYC expression and tumorigenicity.
MYC Modulates Transcription Factors That Mediate Tumorigenicity.
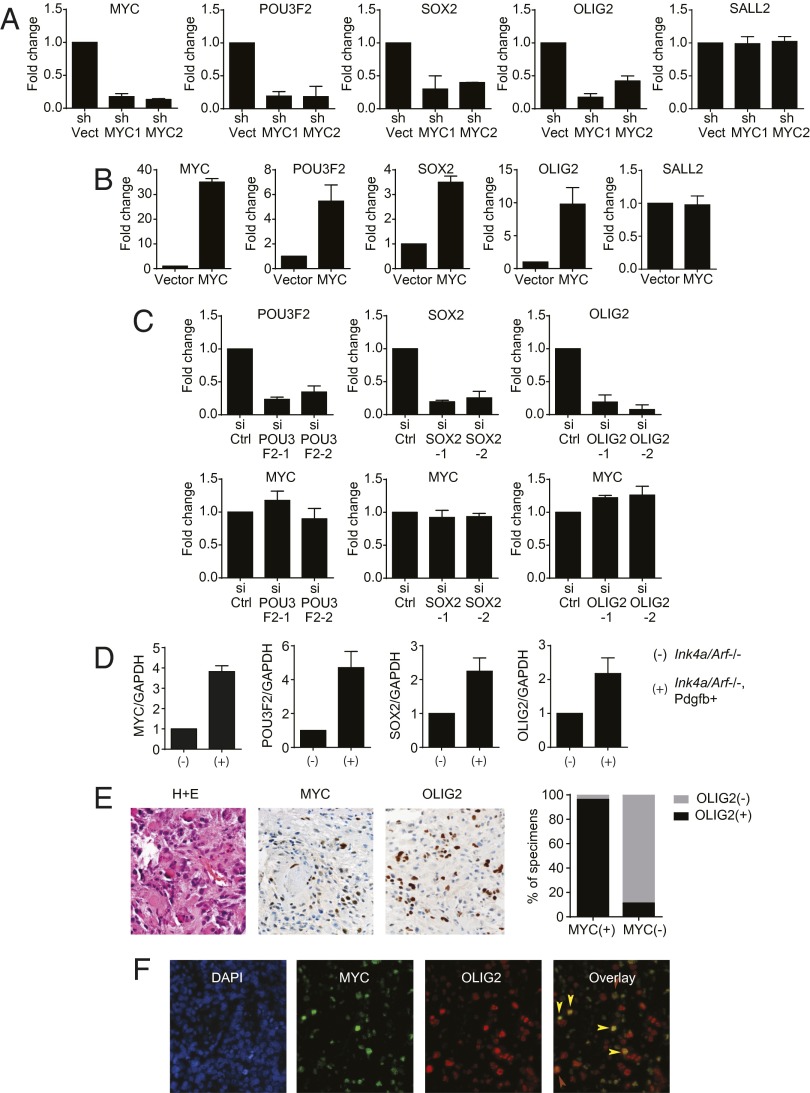
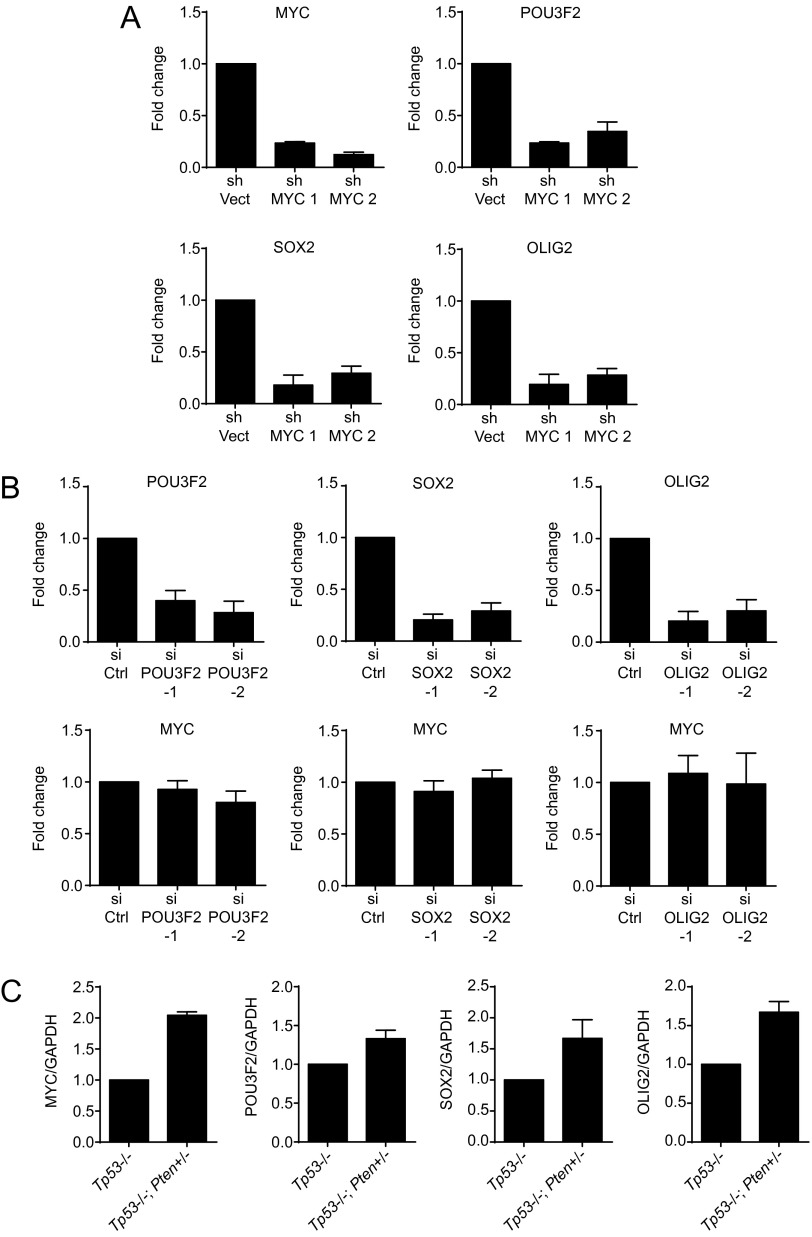
Having established the association between MYC and tumorigenicity, we next investigated the molecular mechanism underlying this association. Core transcription factors that mediate reprogramming of glioblastoma into more stem-like states were recently identified (12). Because tumorigenicity is a property associated with a stem-like state (5), we examined the genetic interaction between MYC and these core transcription factors. MYC shRNA knockdown in U87MG decreased the expression levels of three of the four transcription factors sufficient for glioblastoma reprogramming, including OLIG2, SOX2, and POU3F2 (Fig. 3A). This result was recapitulated using the glioblastoma line 83 (16) (Fig. S5A). Additionally, exogenous MYC expression in U87MG induced the expression of these reprogramming factors (Fig. 3B). In contrast, siRNA knockdown of POU3F2, SOX2, or OLIG2 in U87MG did not affect MYC expression, suggesting that MYC is not downstream of these three transcription factors (Fig. 3C). These results were also recapitulated in the 83 glioblastoma line (Fig. S5B). Moreover, the expression levels of MYC, OLIG2, SOX2, and POU3F2 were elevated in a tumorigenic murine Cdkn2a (Ink4a/Arf), Pdgfb+ glioblastoma line (27, 28) relative to its nontumorigenic precursor Cdkn2a (Ink4a/Arf) astrocyte line (Fig. 3D). Similar results were observed in a tumorigenic murine hGFPA-Cre+: Tp53lox/lox; Ptenlox/+ glioblastoma line and its nontumorigenic precursor hGFPA-Cre+: Tp53lox/lox astrocyte line (29) (Fig. S5C).
Fig. 3.
MYC expression regulates expression of downstream transcription factors (POU3F2, SOX2, and OLIG2) that mediate tumorigenicity. (A) Silencing of MYC by independent shRNAs suppressed expression of POU3F2, SOX2, and OLIG2 (qRT-PCR). Error bars, SD. (B) Exogenous MYC expression in U87MG induced expression of POU3F2, SOX2, and OLIG2 (qRT-PCR). Error bars, SD. (C) Silencing of POU3F2, SOX2, and OLIG2 by independent siRNAs (Top) did not affect MYC expression (Bottom, qRT-PCR). Error bars, SD. (D) Increased expression of MYC, POU3F2, SOX2, and OLIG2 in a tumorigenic Cdkn2a (Ink4a/Arf), Pdgfb+ glioblastoma line relative to its nontumorigenic precursor Cdkn2a (Ink4a/Arf) astrocyte line (qRT-PCR). Error bars, SD. (E) IHC staining for MYC and OLIG2 in a panel of 65 human glioblastoma specimens. (Left) Representative staining in one specimen. (Right) Percentages of specimens staining positive versus negative for OLIG2 grouped by MYC staining. (F) Colocalization by IF of MYC and OLIG2 in a representative human glioblastoma specimen, with MYC in green, OLIG2 in red, and colocalization in yellow (marked by arrowheads).
Fig. S5.
MYC expression drives expression of downstream transcription factors that promote glioblastoma stem-like states. (A) Assessment by RT-PCR of relative expression of MYC and transcription factors that mediate reprogramming of glioblastoma into more stem-like states (12), following MYC knockdown by two independent shRNAs in the glioblastoma line 83. (B) Assessment by RT-PCR of relative expression of the reprogramming transcription factors (Top) and MYC (Bottom), following knockdown of each transcription factor by two independent siRNAs in the glioblastoma line 83. Error bars, SD. (C). Assessment by RT-PCR of relative expression of the reprogramming transcription factors and MYC in nontumorigenic Tp53−/− astrocytes compared with tumorigenic Tp53−/−; Pten+/− glioblastoma cells. The tumorigenic glioblastoma line exhibited increased expression of MYC, POU3F2, SOX2, and OLIG2 relative to the nontumorigenic precursor astrocyte line. Error bars, SD.
Consistent with our hypothesis of a MYC-OLIG2 regulatory axis, immunohistochemical (IHC) analysis of a panel of 65 human glioblastoma specimens showed a tight association between MYC and OLIG2 (30) staining. Of the specimens with high MYC staining, 96% showed high OLIG2 staining (Materials and Methods). In contrast, only 11% of specimens with undetectable MYC staining showed high OLIG2 staining (P < 0.05; Fig. 3E). Colabeling immunofluorescence (IF) experiments further demonstrated that between 7% and 15% of glioblastoma cells stained positive for both MYC and OLIG2 (Fig. 3F).
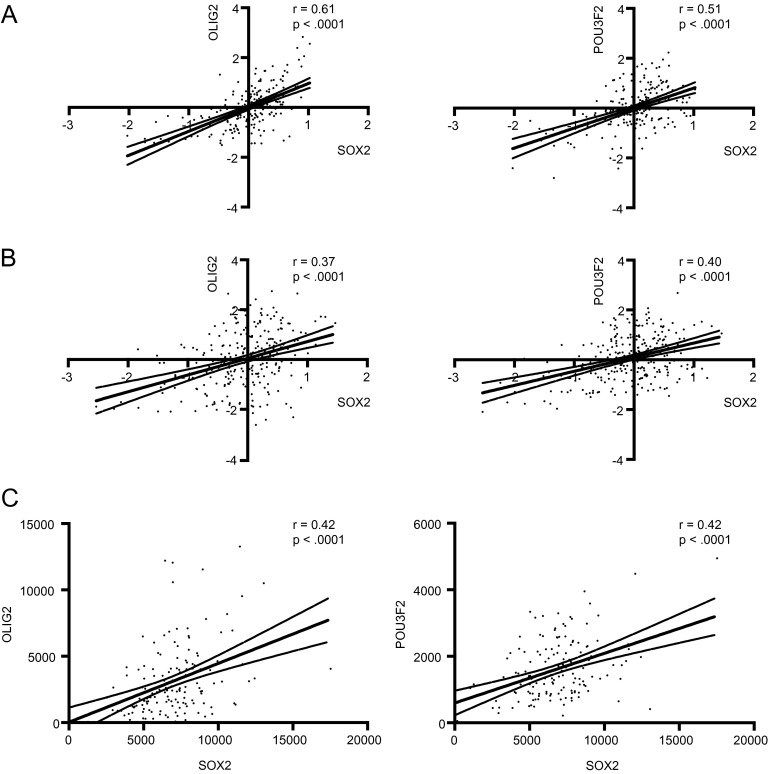
Analysis of TCGA glioblastoma trancriptome datasets revealed that the mRNA expression of OLIG2, SOX2, and POU3F2 closely correlated with one another (Fig. S6), supporting their regulation by a common regulator. Further supporting this hypothesis, immunostaining of SOX2, like OLIG2, closely tracked with MYC (Fig. S7).
Fig. S6.
Correlation of SOX2, OLIG2, and POU3F2 expression in human glioblastoma specimens, using published microarray and RNAseq gene expression data. (A) TCGA microarray dataset (70). (B) Validation dataset comprised of four publically available datasets used in Verhaak et al. (70). (C) TCGA RNAseq data of mostly nonoverlapping samples compared with A. All correlations were analyzed by linear regression. P values indicate significance of nonzero slope.
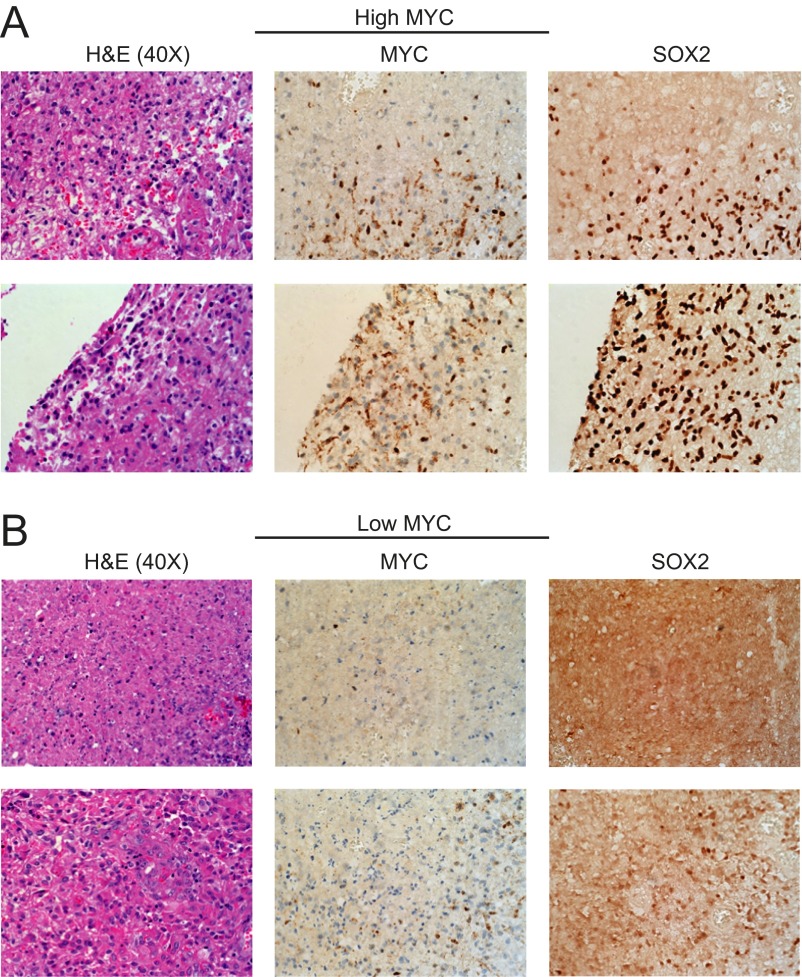
Fig. S7.
IHC staining for MYC and SOX2 in human glioblastoma specimens grouped by (A) high versus (B) low MYC expression. H&E staining at 40× magnification is also shown for each specimen.
Epigenetic Regulation of MYC Expression by LSD1.
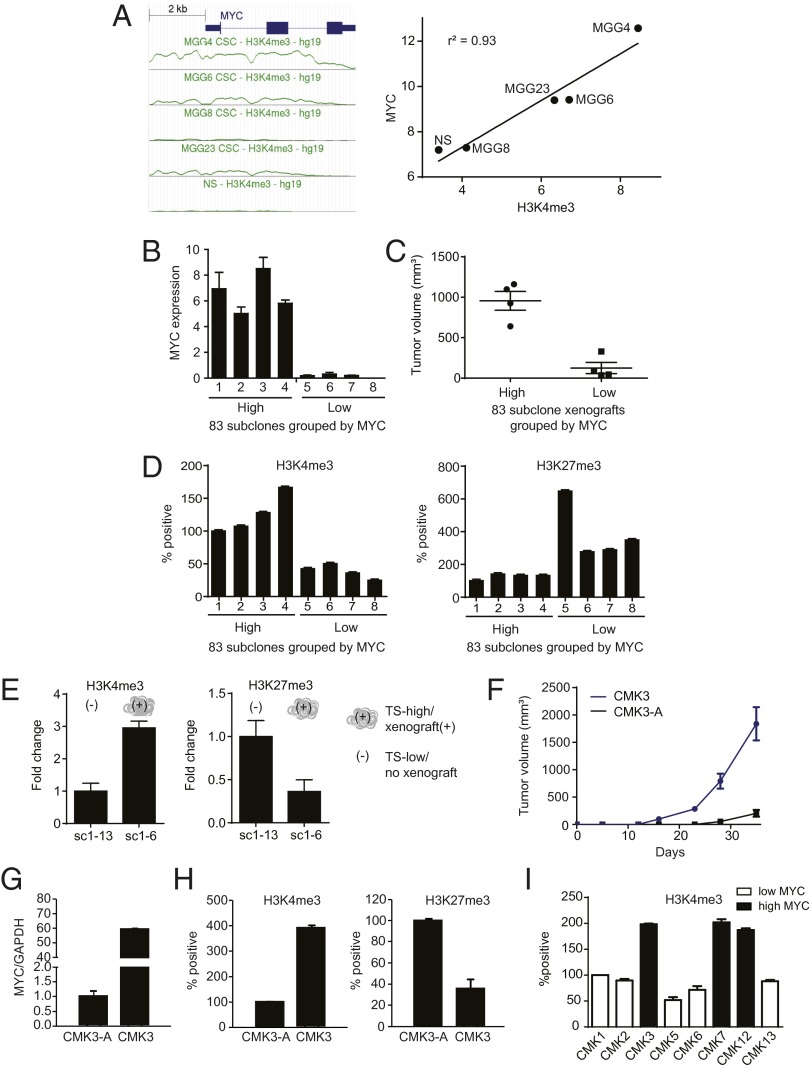
Despite the near isogenicity of the U87MG and 83 subclones (Fig. S1), the subclones exhibit differing tumorigenicity (Fig. 1B). We thus hypothesized that tumorigenicity and MYC expression are epigenetically modulated. Trimethylation of histone 3 lysine 4 (H3K4me3) and lysine 27 (H3K27me3) are histone marks found in chromatin regions with modulated transcriptional activity (31–33). H3K4me3 favors relaxed chromatin and increased transcription, whereas H3K27me3 typically represses transcription. We first tested the association between MYC expression and the abundance of H3K4me3 in a panel of short-term–passaged glioblastoma lines that differed in tumorigenicity (34). We observed that the abundance of H3K4me3 at the MYC locus correlated closely with MYC expression (r2 = 0.93; Fig. 4A).
Fig. 4.
Tumorigenicity is associated with histone 3 trimethylation favoring high MYC expression. (A) ChIP-seq analysis of MYC locus (MYC gene + 2 kb upstream) H3K4me3 abundance in four glioblastoma (CSC) lines and a NSC line (31) (Left) revealed an association between H3K4me3 abundance and MYC expression. (Right) Mean abundance of MYC locus H3K4me3 plotted against MYC expression and analyzed by linear regression. (B) Assessment by RT-PCR of relative MYC expression in eight subclones of the glioblastoma line 83 (14). (C) Association of MYC expression and s.c. flank xenograft tumor growth among the eight 83 subclones. (D) Association of MYC locus H3K4me3/H3K27me3 (by ChIP), MYC expression, and tumorigenicity in the eight 83 subclones. (E) Association of MYC locus H3K4me3/H3K27me3 (by ChIP) and tumorigenicity in U87MG-SC1 subclones differing in tumorigenicity (Fig. 1C, exp 2). (F) Comparison of volumetric growth of nude mouse s.c. flank xenografts of a primary glioblastoma line propagated under TS (CMK3) versus adherent/serum (CMK3-A) conditions; n = 5 per group. (G) Culturing conditions that suppressed CMK3 tumorigenicity (CMK3-A) decreased MYC expression (qRT-PCR). (H) Culturing conditions that suppressed CMK3 tumorigenicity (CMK3-A) decreased MYC locus H3K4me3 while inducing MYC locus H3K27me3 (ChIP). (I) Association between MYC expression and MYC locus H3K4me3 in a panel of eight freshly resected glioblastoma specimens, including three that are tumorigenic and show high MYC expression and five that are nontumorigenic and show low MYC expression (Fig. 2E). All error bars, SD.
We next performed chromatin immunoprecipitation (ChIP) to assess the relative abundance of H3K4me3 and H3K27me3 at the MYC locus in a panel of subclones of the glioblastoma line 83 (16) that differed in MYC expression (Fig. 4B) and tumorigenicity (Fig. 4C). We found that high H3K4me3 tracked closely with tumorigenicity and high MYC expression, whereas high H3K27me3 tracked closely with lack of tumorigenicity and low MYC expression (Fig. 4D). Moreover, a U87MG-SC1 subclone that spontaneously acquired tumorigenicity and high MYC expression exhibited higher H3K4me3 and lower H3K27me3 levels at the MYC locus relative to a nontumorigenic U87MG-SC1 subclone (Fig. 4E).
To better establish chromatin remodeling as a mechanism for regulation of MYC expression, we next determined whether perturbations that alter tumorigenicity simultaneously altered H3K4me3 to H3K27me3 distribution at the MYC locus. Culturing CMK3 (17), normally cultured as TSs in serum-free media, in the presence of serum significantly diminished its tumorigenicity relative to cells cultured under TS conditions (Fig. 4F). This loss of tumorigenicity was associated with a decrease in MYC expression (Fig. 4G) and an increase in the ratio of H3K27me3 to H3K4me3 at the MYC locus (Fig. 4H). Finally, in the eight freshly resected glioblastoma specimens (Fig. 2E), the three tumorigenic, high-MYC specimens harbored significantly higher levels of H3K4me3 at the MYC locus relative to the remaining nontumorigenic specimens (Fig. 4I). These results support our hypothesis that epigenetic regulation of MYC serves as a determinant of glioblastoma tumorigenicity.
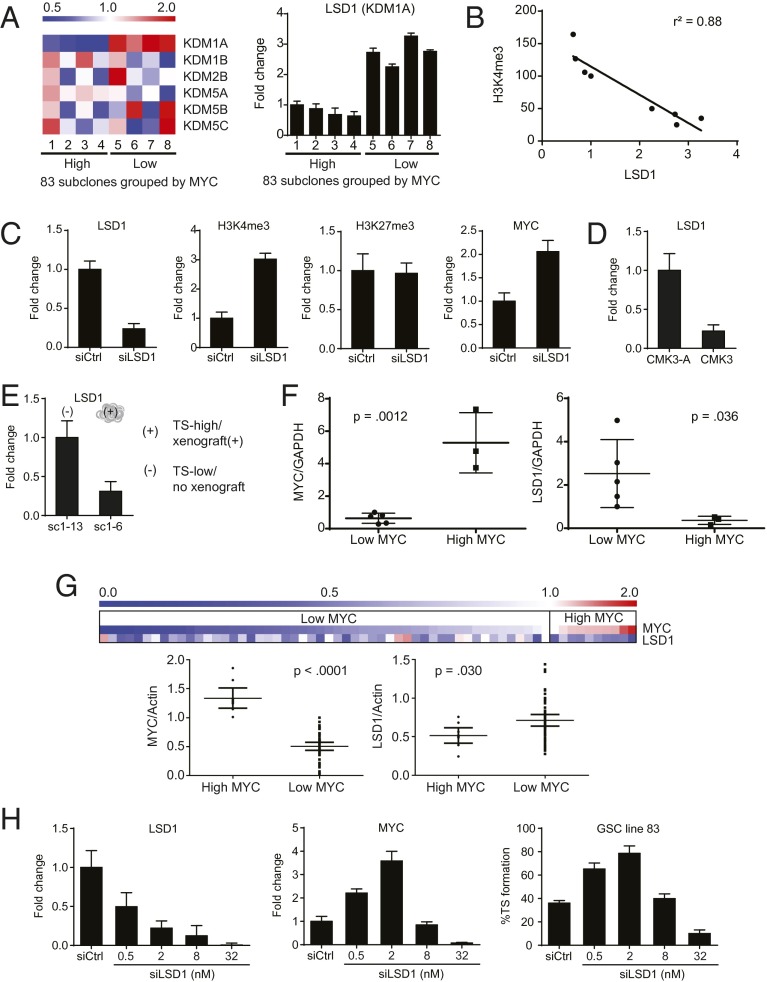
The KDM family of histone lysine demethylases is primarily responsible for regulating histone marks including H3K4 and H3K27 trimethylation (35). We hypothesized that members of this family may modulate the relative abundance of H3K4 trimethylation at the MYC locus. To test this hypothesis, we conducted a targeted RT-PCR screen to identify members of the KDM family previously shown to modulate H3K4 methylation and determine if any members exhibit expression patterns that correlated with MYC expression. Of those tested, a correlation was found only for LSD1 (KDM1A) (Fig. 5A). LSD1 catalyzes the demethylation of H3K4me2 and H3K4me1 and promotes loss of H3K4me3 to repress gene expression (10, 36). As such, we hypothesized that LSD1 expression regulates H3K4me3 homeostasis at the MYC locus. Supporting this hypothesis, LSD1 mRNA levels inversely correlated with H3K4me3 at the MYC locus for all subclones of the glioblastoma line 83 (Fig. 5B). LSD1 silencing in the glioblastoma line 83 induced increased H3K4me3 abundance at the MYC locus and increased MYC expression (Fig. 5C). Moreover, suppression of CMK3 tumorigenicity by serum addition led to increased LSD1 expression (Fig. 5D). Additionally, a U87MG-SC1 subclone that spontaneously acquired tumorigenicity (and increased H3K4me3 at the MYC locus) exhibited decreased expression of LSD1 relative to a nontumorigenic subclone (Fig. 5E). The tumorigenic U87MG subclones consistently exhibited higher MYC and lower LSD1 expression relative to the nontumorigenic subclones (Fig. S8A). Finally, decreased LSD1 expression tracked closely with increased MYC H3K4me3 and MYC expression in both the murine Cdkn2a (Ink4a/Arf), Pdgfb+ and the hGFPA-Cre+: p53lox/lox; Ptenlox/+ glioblastoma models (Fig. S8B).
Fig. 5.
MYC locus H3K4me3 and MYC expression are associated with LSD1 expression. (A) Assessment by RT-PCR of relative expression of six H3K4-specific histone lysine demethylases in subclones of the glioblastoma line 83 grouped by MYC expression, shown as a heat map with blue designating below average expression and red designating above average expression. Only LSD1 (KDM1A) showed statistically significant association with MYC expression (Right). Error bars, SD. (B) Correlation of MYC locus H3K4me3 abundance and LSD1 expression in subclones of the 83 glioblastoma cell line. Correlation was analyzed by linear regression. (C) LSD1 siRNA silencing in U87MG induced accumulation of H3K4me3 at the MYC locus and MYC expression (RT-PCR) without affecting H3K27me3 abundance (ChIP). (D) Culturing conditions that suppressed CMK3 tumorigenicity (CMK3-A) induced LSD1 expression (qRT-PCR). (E) A tumorigenic U87MG-SC1 subclone (sc1–6) showed decreased LSD1 expression relative to a nontumorigenic subclone (sc1–13; Fig. 1C, exp 2). (F) Association between MYC and LSD1 expression (qRT-PCR) in eight freshly resected glioblastoma specimens. Error bars, SD. (G) Assessment by RT-PCR of MYC and LSD1 expression in single cells isolated from one freshly dissected glioblastoma specimen, shown as a heat map with blue designating below average expression and red designating above average expression (Top). LSD1 expression levels are grouped by high versus low MYC expression (Bottom), demonstrating that high MYC-expressing cells harbored lower LSD1 expression (P = 0.03). Error bars, 95% confidence interval. (H) Titration of LSD1 knockdown in 83 by increasing amounts of LSD1 siRNA, assessed by RT-PCR (Left). (Center) Corresponding MYC expression assessed by RT-PCR. (Right) Percentages of single-cell suspensions of 83 cells that propagated as TSs following graded LSD1 knockdown. All error bars, SD.
Fig. S8.
(A) Expression of MYC and LSD1 quantified by RT-PCR in U87MG subclones grouped by low (TS-low) versus high (TS-high) propensity for TS formation. Error bars, SD. (B) Expression of LSD1 quantified by RT-PCR in nontumorigenic Ink4a/Arf−/− astrocytes compared with Ink4a/Arf−/−, Pdgfb+ glioblastoma cells, and Tp53−/− astrocytes compared with tumorigenic Tp53−/−; Pten+/− glioblastoma cells.
To demonstrate the clinical pertinence of our tissue culture findings, we characterized the LSD1 expression levels in our panel of eight freshly resected glioblastoma specimens. We found that the three tumorigenic, high-MYC specimens harbored significantly lower expression of LSD1 relative to the remaining nontumorigenic specimens (Fig. 5F). Moreover, single glioblastoma cells were isolated from one freshly dissected clinical specimen without culture and subjected to quantitative PCR (qPCR) analysis of MYC and LSD1 expression. Consistent with our MYC IHC stain (Fig. 3E), we found that ∼10% of the analyzed cells exhibited significantly elevated MYC mRNA expression. The LSD1 expression in these high MYC-expressing cells was significantly lower relative to the low MYC-expressing cells (Fig. 5G). In aggregate, our results suggest that LSD1 regulates transitions in tumorigenicity through epigenetic modulation of MYC.
Interestingly, previous reports suggest that LSD1 silencing or pharmacologic inhibition exerts antineoplastic effects (12, 37). In contrast, our results suggest that LSD1 knockdown induces MYC expression and tumorigenicity. We hypothesized that the discrepant results may be the consequence of differing levels of LSD1 knockdown. Efficient silencing of LSD1 may induce cell death. However, transient, partial silencing of the LSD1 level may facilitate transitions between tumorigenic states, as we observed. Supporting this hypothesis, titration of LSD1 siRNA transfection revealed that the lower range of siRNA transfection was conducive to tumorigenicity, as evidenced by increased TS formation, whereas higher ranges were associated with cell death and lowered MYC expression (Fig. 5H).
Discussion
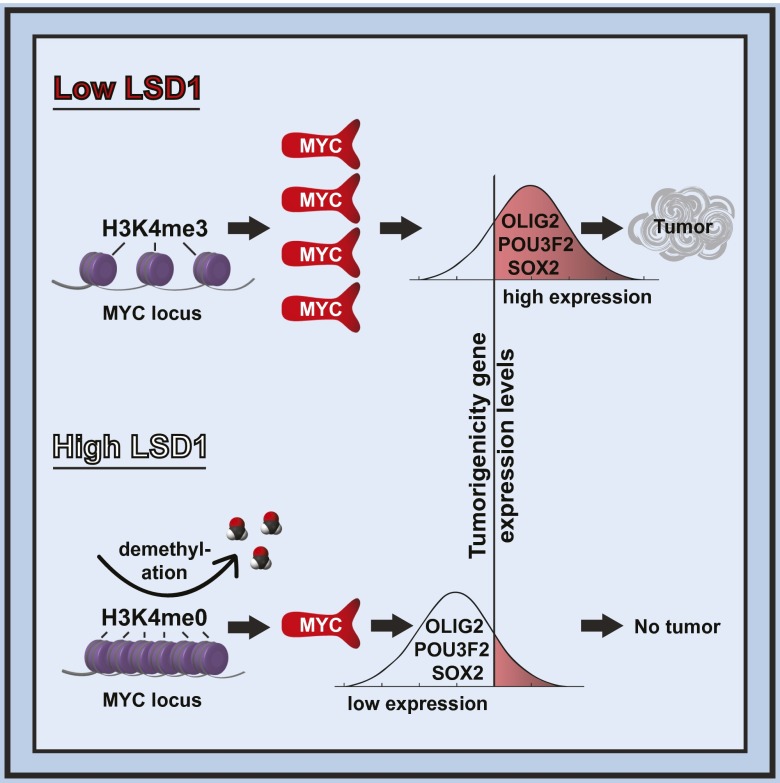
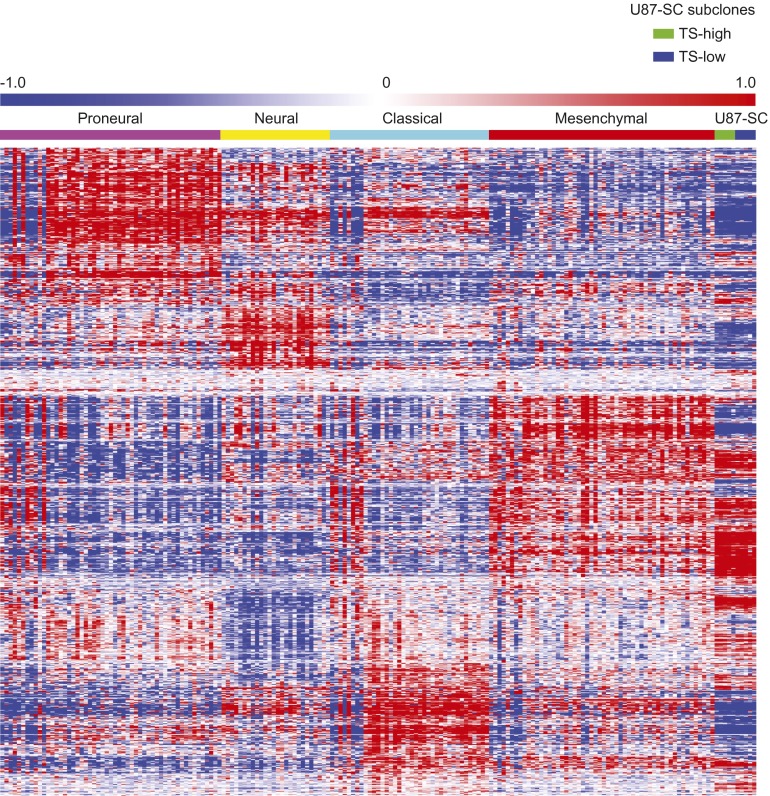
Although elegantly engineered murine models have been created for in vivo studies of glioblastoma tumorigenicity (38, 39), their pertinence to the human disease warrants careful interpretation (40). Ultimately, thoughtful interpretation of these results in the context of clinically derived specimens and cell lines is warranted. In this context, we studied how cell subpopulations retain or gain tumorigenicity using glioblastoma cell lines that underwent variable numbers of passages and validated our observations using clinical specimens. Our results are largely consistent with studies in murine models indicating that tumorigenicity is largely an intrinsic property of the cell (38, 39). However, tumorigenicity can also be gained or lost as a result of what appear to be stochastic fluctuations (41) in the expression of epigenetic regulators such as LSD1 (10). These changes initiate altered regulation of a cascade of transcription factors capable of cellular reprogramming (42), including MYC (43), ultimately transforming the cellular capacity for tumorigenicity (Fig. 6). Our results indicate that this reprogramming did not fundamentally alter the expression of genes that determine the glioblastoma subtype (Fig. S9). The capacity for spontaneous transition in cell states provides a potential mechanism for acquired therapeutic resistance and warrants consideration in rational design of combinatorial therapy. Importantly, it places an emphasis on targeting the transition between nontumorigenic and tumorigenic states, rather than a static population of elite cells.
Fig. 6.
Model figure. Tumorigenicity is largely an intrinsic property of the cell. However, tumorigenicity can also be gained or lost as a result of stochastic fluctuations. This transition is governed by epigenetic regulation through LSD1. Low LSD1 expression is associated with high MYC locus H3K4me3, which promotes high expression of MYC and downstream MYC target genes including OLIG2, POU3F2, and SOX2. These factors, in turn, promote a tumorigenic cell state.
Fig. S9.
Heat maps showing hierarchical clustering of genes that comprise the TCGA molecular subtype classifier (70) for TCGA samples grouped by molecular subtype (70) compared with U87MG subclones grouped by high versus low TS-forming capacity, the latter to assess whether there are global differences in gene expression based on tumorigenic potential.
There is emerging or renewed interest in targeting the genes implicated in our study [i.e., LSD1 (37, 44) and MYC (45, 46)] as an anticancer strategy. Notably, these genes, and downstream factors including OLIG2 and SOX2, modulate cellular phenotypes in a dose-dependent manner (47–49). Consequently, their expression is tightly regulated during normal development (50, 51). Our observation that LSD1 silencing can promote tumorigenicity or cell death depending on the level of knockdown represents an extreme version of this dose dependency. This may partly explain the variability in responses to LSD1 inhibition observed in differing contexts. For instance, LSD1 inhibitors have been shown to inhibit the growth of teratocarcinoma, embryonic carcinoma, and seminoma cells (44). On the other hand, LSD1 depletion increased migration and invasion of androgen-independent prostate cancer cells (52). The dichotomy of this “Janus” effect (53) bears relevance given the nonuniform pharmacologic distribution of therapeutic agents within tumors (54, 55). The mechanistic basis for the dichotomy is thus an important area of active exploration.
Molecular and phenotypic cell-to-cell variability within a clonal population, shown here for LSD1, has been previously described for other genes (56). The observed heterogeneity in LSD1 and MYC expression within glioblastoma cell populations is somewhat anti-intuitive, particularly in the context of long-term–passaged cell lines, where one may expect the more robust tumorigenic cells to eventually dominate the population. It remains unclear whether heterogeneity is maintained because nontumorigenic cells contribute to the overall in vitro or in vivo fitness of the cell population (57), from stochasticity inherent to molecular interactions (58, 59) or from interaction with the tumor microenvironment (60). Understanding the basis of cell individuality within cancer may be of value in shaping therapeutic paradigms.
Materials and Methods
Cell Lines, Culture, and TS Formation Assays.
U87MG and derived subclones were propagated in DMEM + 15% (vol/vol) FBS + 100 U/mL penicillin/100 ng/mL streptomycin in tissue culture-treated plates at 37 °C in humidified 5% (vol/vol) CO2. The glioblastoma lines CMK3 and 83 were passaged as TSs in complete NeuroCult NS-A (Stemcell Technologies), including 20 ng/mL recombinant human (rh) EGF + 10 ng/mL rh bFGF (FGF2) + 2 µg/mL heparin + 100 U/mL penicillin/100 ng/mL streptomycin (TS media). Primary glioblastoma lines CMK1–13 were derived from fresh surgical specimens after written informed consent. Subclones of U87MG, CMK3, and 83 were derived by plating of single cells. Approximately 10% of the plated cells gave rise to propagating subclones. Specimens were dissociated and propagated under serum/adherent and TS conditions.
Single-cell TS formation assays were performed by placing single cells by limiting dilution into individual wells of ultra-low attachment 96-well plates (Corning Incorporated) containing 100 µL TS media. After 2 wk, wells were scored as positive if by visual inspection they contained an aggregate of ≥16 cells. Approximately 100 single cells were scored in each experiment.
For MYC knockdown, MYC shRNA sequence TRCN0000416981 (The RNAi Consortium, Broad Institute of MIT and Harvard) was cloned into the Tet-pLKO.1-puro doxycycline-inducible vector. For exogenous expression, cells were transduced with MYC via the pMIG vector as previously described (61).
Xenograft Studies.
The s.c. xenografts were generated by injecting in the flanks of homozygous NCr nude mice (Charles River Laboratories) either trypsinized cells propagated under serum/adherent conditions and suspended in PBS or cells propagated under TS growth conditions. Tumor xenograft volume was assessed by a pair of calipers according to the formula π(length)(width)2/6. Intracranial xenografts were generated by anesthetizing mice with ketamine 150 mg/kg and xylazine 12 mg/kg i.p. (Phoenix Pharmaceuticals) before head fixation in a stereotactic frame (Stoelting), injection of cells through a 27-gauge needle for 2 min at 2 mm lateral and posterior to the bregma and 3 mm below the dura, and incision closure with Vetbond (3M Co.). Tumor specimens were formalin fixed and paraffin embedded before sectioning and hematoxylin/eosin (H&E) staining, IF, or immunohistochemistry studies.
Details on murine tumor model studies are provided in SI Materials and Methods.
Immunostaining and Molecular Techniques.
ChIP and qPCR were performed as previously described (62, 63). Normalized Ct (∆Ct) values were calculated by subtracting the Ct obtained with input DNA from that obtained with immunoprecipitated DNA [∆Ct = Ct (IP) – Ct (input)].
For single-cell qPCR, ∼2,000 cells isolated from a freshly resected clinical specimen were loaded onto a 17–25 µm C1 single-cell Auto Prep IFC chamber (Fluidigm), and cell capture was performed according to the manufacturer’s instructions. Capture efficiency was 77% (74/96 wells occupied with a single cell) as determined by fluorescent microscopy. Both the empty wells (7) and doublet-occupied wells (15) were noted and excluded from further analysis. Upon capture, reverse transcription and cDNA preamplification were performed according to the manufacturer’s instructions (Fluidigm). cDNA was then harvested and assessed using Flex Six ChIP to examine the expression levels of MYC, LSD1, GAPDH, and ACTIN.
Primers, antibodies, and detailed immunostaining methods are provided in SI Materials and Methods.
Statistics.
Murine survival following xenograft implantation was plotted using the Kaplan–Meier method and compared using the log-rank test. The Levene test for equality of variances (64) was used to assess for statistically significant differences in the variability of tumor-sphere (TS) formation rates among subclones compared with control. To demonstrate enrichment of genes driven by MYC in the tumorigenicity signature, χ2 analysis was performed as a two-tailed test with P < 0.05. All analyses were performed using SAS JMP 9.0.
Bioinformatics.
SNP analyses.
Whole-genome DNA was extracted from 10 clones using a DNA extraction kit (Qiagen) and profiled using Human Mapping 250K NSP Arrays (Affymetrix). The 10 sample .CEL files were analyzed using Genotyping Console Version 4.1.3.840 (Affymetrix) following the manufacturer’s instructions. The dataset is available under Gene Expression Omnibus (GEO) accession no. GSE56316.
Gene expression analyses.
RNA samples from glioblastoma cell lines including U87MG subclones in the midlogarithmic growth phase were analyzed using Affymetrix HT HG-U133A arrays. Robust Multichip Average (RMA) gene-centric expression summary values were computed using RMAExpress (65), using a custom chip definition file (CDF) generated using AffyProbeMiner (66). The dataset is available under GEO accession no. GSE54967.
Principal component analysis was performed using MultiExperiment Viewer v4.6 (67) with the subset of genes in a published tumorigenicity signature (19). Significance Analysis of Microarrays (68) was performed to identify genes whose expression levels differed significantly between high versus low TS-forming U87MG subclones.
TCGA analyses.
Preprocessed level 3 data were obtained from TCGA (69) for 582 glioblastoma specimens. Specimens were categorized according to TCGA subtype—proneural, neural, mesenchymal, or classical—as previously defined (70). A normalized expression value for each gene was calculated by subtracting the gene’s mean expression value across the dataset and then dividing by its SD. Signature scores were calculated by summing the normalized expression values of all genes up-regulated in the signature and subtracting the normalized expression values of all genes down-regulated in the signature. Signatures used include the core MYC regulated genes (21) and the xenograft tumorigenicity genes (26). The resultant signature scores were plotted, and Pearson’s correlation coefficients were determined.
Epigenetic analyses of histone modifications.
Published ChIP-seq data (34) were downloaded to assess H3K4me3 abundance at the MYC locus (MYC gene + 2 kb upstream) of four glioblastoma (CSC) and an NSC line. Mean H3K4me3 abundance was plotted against MYC expression determined by Affymetrix HG-U133 Plus 2 arrays using probeset 202431_s_at. Correlation was analyzed by linear regression.
Ethics Statement.
Research involving human participants was approved by the Dana-Farber/Harvard Cancer Center institutional review board under protocol 07–231. Animal work was conducted in accordance with the National Research Council (NRC) Guide for the Care and Use of Laboratory Animals and was approved by the Dana-Farber Cancer Institute Institutional Animal Care and Use Committee under protocol 05–056 and the University of California San Diego Institutional Animal Care and Use Committee under protocol S13070.
SI Materials and Methods
Murine Tumor Model Studies.
Cdkn2a (Ink4a/Arf)-null primary NSC/NPCs were isolated by microdissection of the subventricular zones of 6-wk-old male Cdkn2a-null C57BL/6 mice (B6.129-Cdkn2atm1Rdp; NCI Frederick). These were maintained as TSs in DMEM/F12 with 20 ng/mL EGF, 20 ng/mL basic FGF, B27 supplement without vitamin A (Invitrogen), 200 ng/mL heparan sulfate, and 100 U/mL penicillin/100 ng/mL streptomycin at 37 °C in 5% humidified CO2. The NSC/NPCs were infected with retrovirus prepared from the mammalian expression vector pMXs-IG with IRES-GFP engineered to express MYC as previously described (71), and the resulting mixture of GFP-positive and -negative cells was cultured as TSs and used for implantation after one passage. The 105 transduced cells were implanted into the brains of female C57BL/6 mice 6–8 wk old.
Tumors were generated in the brains of Cdkn2a (Ink4a/Arf)-null mice via overexpression of Pdgfb as previously described (27, 72). Brain tumors were also generated in an hGFAP-Cre+: p53lox/lox; Ptenlox/+ GEMM, and glioblastoma cell lines were established from these tumors, as previously described (29, 73).
IF, IHC, and Western Blotting.
The following antibodies were used: MYC (manual IF and IHC) (cat. no. 631206; Cell Signaling Technology), MYC (automated IHC) (cat. no. Ab32072; Abcam), OLIG2 (cat. no. AB9610; Millipore), α-tubulin (cat. no. T9026; Sigma), and Actin (cat. no. A5316; Sigma).
For manual IF and IHC studies, slides were incubated in a hybridization oven at 60 °C for 60 min to remove secondary paraffin layers. Antigen retrieval was accomplished by incubation in citrate buffer at subboiling temperature for 10 min. Primary antibodies were applied and incubated at 4 °C overnight. For IHC, the reaction was visualized by diaminobenzidine (DAB) (Vector Laboratories).
For automated IHC, a Ventana Discovery Ultra (Ventana Medical Systems) was used. Tissue sections were cut from blocks of formalin-fixed paraffin tumor tissue and mounted on Superfrost plus slides (Fisher Scientific) and dried at room temperature overnight. Slides were depariffined, and antigen retrieval was performed using CC1 for 24 min at 95 °C. Primary antibodies were incubated at 37 °C for 32 min. Due to the low-level expression of MYC, we used a tyramide-based amplification to enhance the signal-to-noise ratio. For single IHC, DAB was used as the chromagen followed by a hematoxylin counterstain. For dual IHC, DAB was used for the first chromagen, followed by Discovery Purple (Ventana Medical Systems) with Fast Green used for the counterstain. For dual automated IF, MYC detection was done by an amplification process followed by a biotinylated anti-HRP (Jackson Immunoresearch). The secondaries were SA-Alexa 488 (1:600) and Alexa 594 (1:400×), both from Invitrogen.
For MYC IHC, the samples were scored as low (<5% staining) or high (>5% staining). For MIB IHCs, the samples were scored as low (<20% staining) or high (>20% staining). To determine the percentage of dual-labeled cells for MYC and MIB-1, we used sections labeled with DAB and Discovery Purple as the chromagens. Sections were analyzed under bright field observation on a Zeiss Axioimager M2 microscope using Nuance software (PerkinElmer). An image cube was captured using the Nuance spectral imaging camera, which takes a bright field image at every 10-nm wavelength allowing for discrete identification of multiple overlapping chromagens in the same cellular compartment. For each chromagen, a threshold was determined for specific staining. After the threshold was set, the software determined the percentage of single- and dual-labeled cells. Total number of nuclei within the field was counted using Fast green.
For xenograft protein extract preparation for Western blotting, s.c. tumors were surgically removed and stored in liquid nitrogen. A piece of frozen tumor was ground with a mortar and pestle under liquid nitrogen, and the ground powder was then suspended in cell lysis buffer (9803; Cell Signaling Technology).
RNA Interference Reagents.
The following constructs were used: MYC, TRCN0000416981 (shMYC-1) (The RNAi Consortium) and TRCN0000312580 (shMYC-2) (Sigma); POU3F2, SR303643 (2 nM) (OriGene Technologies); SOX2, SR304528 (2 nM) (OriGene Technologies); OLIG2, SR306903 (2 nM) (OriGene Technologies); and LSD1, SR307947 (2 nM) (OriGene Technologies).
Quantitative RT-PCR and ChIP.
The following ChIP primers and antibodies were used: H3K4me3, 5′-CTCCCGTCTAGCACCTT-3′ and 5′-GAGATGTGTCTGCCTGTTC-3′ (no. 39915; Active Motif); and H3K27me3, 5′-CTGCACTCCCAGCTACT-3′ and 5′-CCCACGTCTTAGAAACTCATC-3′ (no. ab6002; Abcam).
The following quantitative RT-PCR primers were used: MYC, 5′-ACTCTGAGGAGGAACAAGAA-3′ and 5′-TGGAGACGTGGCACCTCTT-3′; GADD45B, 5′-TACGAGTCGGCCAAGTTGATG-3′ and 5′-GGATGAGCGTGAAGTGGATTT-3′; LDHA, 5′-ATGGCAACTCTAAAGGATCAGC-3′ and 5′-CCAACCCCAACAACTGTAATCT-3′; TERT, 5′-AAATGCGGCCCCTGTTTCT-3′ and 5′-CAGTGCGTCTTGAGGAGCA-3′; OLIG2, 5′-CCAGAGCCCGATGACCTTTTT-3′ and 5′-CACTGCCTCCTAGCTTGTCC-3′; SOX2, 5′-GCCGAGTGGAAACTTTTGTCG-3′ and 5′-GGCAGCGTGTACTTATCCTTCT-3′; POU3F2, 5′-CGGCGGATCAAACTGGGATTT-3′ and 5′-TTGCGCTGCGATCTTGTCTAT-3′; SALL2, 5′-CAGCGGAAACCCCAACAGTTA-3′ and 5′-GAGGGTCAGTAGAACATGCGT-3′; LSD1 (KDM1A), 5′-TGACCGGATGACTTCTCAAGA-3′ and 5′-GTTGGAGAGTAGCCTCAAATGTC-3′; KDM1B, 5′-CTCTCCTGTGGGGAACATTTC-3′ and 5′-GACTAGGTTCGGTTTTGCCATT-3′; KDM2B, 5′-GGGTTCCCCTGATATTTCGAGA-3′ and 5′-GCTCCCCACTAGGAGTTTGAC-3′; KDM5A, 5′-AGCCGAGTTGGGAGGAGTT-3′ and 5′-TGGACTCTTGGAGTGAAACGA-3′; KDM5B, 5′-AGTGGGCTCACATATCAGAGG-3′ and 5′-CAAACACCTTAGGCTGTCTCC-3′; KDM5C, 5′-GGGTCCGACGATTTCCTACC-3′ and 5′-ATGCCCGATTTCTCTGCGATG-3′; EZH2, 5′-AATCAGAGTACATGCGACTGAGA-3′ and 5′-GCTGTATCCTTCGCTGTTTCC-3′; GAPDH, 5′-ATCATCCCTGCCTCTACTGG-3′ and 5′-GTCAGGTCCACCACTGACAC-3′; and 18S rRNA, 5′-GGGAGGTAGTGACGAAAAATAACAAT-3′ and 5′-TTGCCCTCCAATGGATCCT-3′.
The following quantitative RT-PCR primers for single cells were used: LSD1, 5′-TGACCGGATGACTTCTCAAGA-3′ and 5′-GTTGGAGAGTAGCCTCAAATGTC-3′; MYC, 5′-CTTCTCTCC GTCCTCGGATTCT-3′ and 5′-GAAGGTGATCC AGACTCTGACCTT-3′; GAPDH, 5′-ACCCAGAAGACTGTGGATGG-3′ and 5′-TTCTAGACGGCAGGTCAGGT-3′; and Actin, 5′-TGAAGTGTGACGTGGACATC-3′ and 5′-GGAGGAAGCAATGATCT-3′.
Whole Exome Subclone Sequencing.
The exome sequencing library was prepared using Agilent SureSelect Version 4 (Agilent Technologies). After quantification, the indexed libraries were pooled in equimolar amounts and sequenced on Illumina HiSeq instruments. The resulting reads were aligned to the hg19 reference genome using Burrows--Wheeler aligner (BWA) MEM (BWA v0.7.10); duplicate reads were removed using Picard MarkDuplicates v1.115. The reads were further realigned around known indels from the single nucleotide polymorphism database (dbSNP) using the GATK v3.3.0 (IndelRealignment). Finally the variants were simultaneously called across all 12 clones using the Varscan v2.3 mpileup2cns command. Variants properly genotyped in all 12 clones and with an average coverage greater than 10 reads were analyzed. Functional annotation of the variants was performed using Annovar (version 2014-07-14).
Supplementary Material
Acknowledgments
We thank Dr. Bing Ren for his expertise and helpful discussions. C.C.C. is supported by the Doris Duke Charitable Foundation Clinical Scientist Development Award (www.ddcf.org), the Sontag Foundation Distinguished Scientist Award (www.sontagfoundation.org), the Burroughs Wellcome Fund Career Awards for Medical Scientists (www.bwfund.org), the Kimmel Scholar award (www.kimmel.org), and a Discovery Grant from the American Brain Tumor Association (www.abta.org). Y.M. and W.H. are supported by the China National Natural Science Foundation (81001115; www.nsfc.gov.cn) and China National Funds for Distinguished Young Scientists (81025013; www.nsfc.gov.cn).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. W.A.W. is a Guest Editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE56316 and GSE54967).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1501967112/-/DCSupplemental.
References
- 1.Ng K, Kim R, Kesari S, Carter B, Chen CC. Genomic profiling of glioblastoma: Convergence of fundamental biologic tenets and novel insights. J Neurooncol. 2012;107(1):1–12. doi: 10.1007/s11060-011-0714-2. [DOI] [PubMed] [Google Scholar]
- 2.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R, et al. European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups National Cancer Institute of Canada Clinical Trials Group Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 4.Bartek J, Jr, et al. Key concepts in glioblastoma therapy. J Neurol Neurosurg Psychiatry. 2012;83(7):753–760. doi: 10.1136/jnnp-2011-300709. [DOI] [PubMed] [Google Scholar]
- 5.Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 6.Zhou BB, et al. Tumour-initiating cells: Challenges and opportunities for anticancer drug discovery. Nat Rev Drug Discov. 2009;8(10):806–823. doi: 10.1038/nrd2137. [DOI] [PubMed] [Google Scholar]
- 7.Yamanaka S. Elite and stochastic models for induced pluripotent stem cell generation. Nature. 2009;460(7251):49–52. doi: 10.1038/nature08180. [DOI] [PubMed] [Google Scholar]
- 8.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358(11):1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 9.Waldmann T, Schneider R. Targeting histone modifications—Epigenetics in cancer. Curr Opin Cell Biol. 2013;25(2):184–189. doi: 10.1016/j.ceb.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 11.Kim J, et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143(2):313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suvà ML, et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157(3):580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447(7143):425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 14.Steuer AF, Rhim JS, Hentosh PM, Ting RC. Survival of human cells in the aggregate form: Potential index of in vitro cell transformation. J Natl Cancer Inst. 1977;58(4):917–921. doi: 10.1093/jnci/58.4.917. [DOI] [PubMed] [Google Scholar]
- 15.Pontén J, Macintyre EH. Long term culture of normal and neoplastic human glia. Acta Pathol Microbiol Scand. 1968;74(4):465–486. doi: 10.1111/j.1699-0463.1968.tb03502.x. [DOI] [PubMed] [Google Scholar]
- 16.Mao P, et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci USA. 2013;110(21):8644–8649. doi: 10.1073/pnas.1221478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akers JC, et al. MiR-21 in the extracellular vesicles (EVs) of cerebrospinal fluid (CSF): A platform for glioblastoma biomarker development. PLoS ONE. 2013;8(10):e78115. doi: 10.1371/journal.pone.0078115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Venet D, Dumont JE, Detours V. Most random gene expression signatures are significantly associated with breast cancer outcome. PLOS Comput Biol. 2011;7(10):e1002240. doi: 10.1371/journal.pcbi.1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen R, et al. A hierarchy of self-renewing tumor-initiating cell types in glioblastoma. Cancer Cell. 2010;17(4):362–375. doi: 10.1016/j.ccr.2009.12.049. [DOI] [PubMed] [Google Scholar]
- 20.Croce CM. Oncogenes and cancer. N Engl J Med. 2008;358(5):502–511. doi: 10.1056/NEJMra072367. [DOI] [PubMed] [Google Scholar]
- 21.Kim J, Lee JH, Iyer VR. Global identification of Myc target genes reveals its direct role in mitochondrial biogenesis and its E-box usage in vivo. PLoS One. 2008;3(3):e1798. doi: 10.1371/journal.pone.0001798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engelmann A, Speidel D, Bornkamm GW, Deppert W, Stocking C. Gadd45 beta is a pro-survival factor associated with stress-resistant tumors. Oncogene. 2008;27(10):1429–1438. doi: 10.1038/sj.onc.1210772. [DOI] [PubMed] [Google Scholar]
- 23.Shim H, et al. c-Myc transactivation of LDH-A: Implications for tumor metabolism and growth. Proc Natl Acad Sci USA. 1997;94(13):6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu KJ, et al. Direct activation of TERT transcription by c-MYC. Nat Genet. 1999;21(2):220–224. doi: 10.1038/6010. [DOI] [PubMed] [Google Scholar]
- 25.Tchoghandjian A, et al. A2B5 cells from human glioblastoma have cancer stem cell properties. Brain Pathol. 2010;20(1):211–221. doi: 10.1111/j.1750-3639.2009.00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joo KM, et al. Patient-specific orthotopic glioblastoma xenograft models recapitulate the histopathology and biology of human glioblastomas in situ. Cell Reports. 2013;3(1):260–273. doi: 10.1016/j.celrep.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 27.Hambardzumyan D, Amankulor NM, Helmy KY, Becher OJ, Holland EC. Modeling adult gliomas using RCAS/t-va technology. Transl Oncol. 2009;2(2):89–95. doi: 10.1593/tlo.09100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, et al. Genome-wide shRNA screen revealed integrated mitogenic signaling between dopamine receptor D2 (DRD2) and epidermal growth factor receptor (EGFR) in glioblastoma. Oncotarget. 2014;5(4):882–893. doi: 10.18632/oncotarget.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng H, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455(7216):1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ligon KL, et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53(4):503–517. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008;27(2):406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 33.Koch CM, et al. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007;17(6):691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rheinbay E, et al. An aberrant transcription factor network essential for Wnt signaling and stem cell maintenance in glioblastoma. Cell Reports. 2013;3(5):1567–1579. doi: 10.1016/j.celrep.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Højfeldt JW, Agger K, Helin K. Histone lysine demethylases as targets for anticancer therapy. Nat Rev Drug Discov. 2013;12(12):917–930. doi: 10.1038/nrd4154. [DOI] [PubMed] [Google Scholar]
- 36.Adamo A, et al. LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat Cell Biol. 2011;13(6):652–659. doi: 10.1038/ncb2246. [DOI] [PubMed] [Google Scholar]
- 37.Singh MM, et al. Inhibition of LSD1 sensitizes glioblastoma cells to histone deacetylase inhibitors. Neuro-oncol. 2011;13(8):894–903. doi: 10.1093/neuonc/nor049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen J, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488(7412):522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wee B, Charles N, Holland EC. Animal models to study cancer-initiating cells from glioblastoma. Front Biosci (Landmark Ed) 2011;16:2243–2258. doi: 10.2741/3851. [DOI] [PubMed] [Google Scholar]
- 40.Diede SJ, et al. Fundamental differences in promoter CpG island DNA hypermethylation between human cancer and genetically engineered mouse models of cancer. Epigenetics. 2013;8(12):1254–1260. doi: 10.4161/epi.26486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raj A, van Oudenaarden A. Nature, nurture, or chance: Stochastic gene expression and its consequences. Cell. 2008;135(2):216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 43.Gabay M, Li Y, Felsher DW. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb Perspect Med. 2014;4(6) doi: 10.1101/cshperspect.a014241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J, et al. Novel histone demethylase LSD1 inhibitors selectively target cancer cells with pluripotent stem cell properties. Cancer Res. 2011;71(23):7238–7249. doi: 10.1158/0008-5472.CAN-11-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Annibali D, et al. Myc inhibition is effective against glioma and reveals a role for Myc in proficient mitosis. Nat Commun. 2014;5:4632. doi: 10.1038/ncomms5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stellas D, et al. Therapeutic effects of an anti-Myc drug on mouse pancreatic cancer. J Natl Cancer Inst. 2014;106(12) doi: 10.1093/jnci/dju320. [DOI] [PubMed] [Google Scholar]
- 47.Liu Z, et al. Induction of oligodendrocyte differentiation by Olig2 and Sox10: Evidence for reciprocal interactions and dosage-dependent mechanisms. Dev Biol. 2007;302(2):683–693. doi: 10.1016/j.ydbio.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 48.Taranova OV, et al. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006;20(9):1187–1202. doi: 10.1101/gad.1407906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schuhmacher M, Eick D. Dose-dependent regulation of target gene expression and cell proliferation by c-Myc levels. Transcription. 2013;4(4):192–197. doi: 10.4161/trns.25907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cai J, et al. A crucial role for Olig2 in white matter astrocyte development. Development. 2007;134(10):1887–1899. doi: 10.1242/dev.02847. [DOI] [PubMed] [Google Scholar]
- 51.Hutton SR, Pevny LH. SOX2 expression levels distinguish between neural progenitor populations of the developing dorsal telencephalon. Dev Biol. 2011;352(1):40–47. doi: 10.1016/j.ydbio.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 52.Ketscher A, et al. LSD1 controls metastasis of androgen-independent prostate cancer cells through PXN and LPAR6. Oncogenesis. 2014;3:e120. doi: 10.1038/oncsis.2014.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song NY, Surh YJ. Janus-faced role of SIRT1 in tumorigenesis. Ann N Y Acad Sci. 2012;1271:10–19. doi: 10.1111/j.1749-6632.2012.06762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Minchinton AI, Tannock IF. Drug penetration in solid tumours. Nat Rev Cancer. 2006;6(8):583–592. doi: 10.1038/nrc1893. [DOI] [PubMed] [Google Scholar]
- 55.Simpson-Herren L, Noker PE. Diversity of penetration of anti-cancer agents into solid tumours. Cell Prolif. 1991;24(4):355–365. doi: 10.1111/j.1365-2184.1991.tb01164.x. [DOI] [PubMed] [Google Scholar]
- 56.Huang S. Non-genetic heterogeneity of cells in development: More than just noise. Development. 2009;136(23):3853–3862. doi: 10.1242/dev.035139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Inda MM, et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010;24(16):1731–1745. doi: 10.1101/gad.1890510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297(5584):1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 59.Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: From theories to phenotypes. Nat Rev Genet. 2005;6(6):451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 60.Sneddon JB, Werb Z. Location, location, location: The cancer stem cell niche. Cell Stem Cell. 2007;1(6):607–611. doi: 10.1016/j.stem.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451(7175):141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 62.Chaffer CL, et al. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell. 2013;154(1):61–74. doi: 10.1016/j.cell.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li J, et al. Epigenetic suppression of EGFR signaling in G-CIMP+ glioblastomas. Oncotarget. 2014;5(17):7342–7356. doi: 10.18632/oncotarget.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Levene H. Robust tests for equality of variances. In: Olkin I, editor. Contributions to Probability and Statistics; Essays in Honor of Harold Hotelling. Stanford Univ Press; Stanford, CA: 1960. pp. 278–292. [Google Scholar]
- 65.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 66.Liu H, et al. AffyProbeMiner: A web resource for computing or retrieving accurately redefined Affymetrix probe sets. Bioinformatics. 2007;23(18):2385–2390. doi: 10.1093/bioinformatics/btm360. [DOI] [PubMed] [Google Scholar]
- 67.Saeed AI, et al. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 68.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Verhaak RG, et al. Cancer Genome Atlas Research Network Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shimizu T, et al. c-MYC overexpression with loss of Ink4a/Arf transforms bone marrow stromal cells into osteosarcoma accompanied by loss of adipogenesis. Oncogene. 2010;29(42):5687–5699. doi: 10.1038/onc.2010.312. [DOI] [PubMed] [Google Scholar]
- 72.Shih AH, et al. Dose-dependent effects of platelet-derived growth factor-B on glial tumorigenesis. Cancer Res. 2004;64(14):4783–4789. doi: 10.1158/0008-5472.CAN-03-3831. [DOI] [PubMed] [Google Scholar]
- 73.Alcantara Llaguno S, et al. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell. 2009;15(1):45–56. doi: 10.1016/j.ccr.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.