Abstract
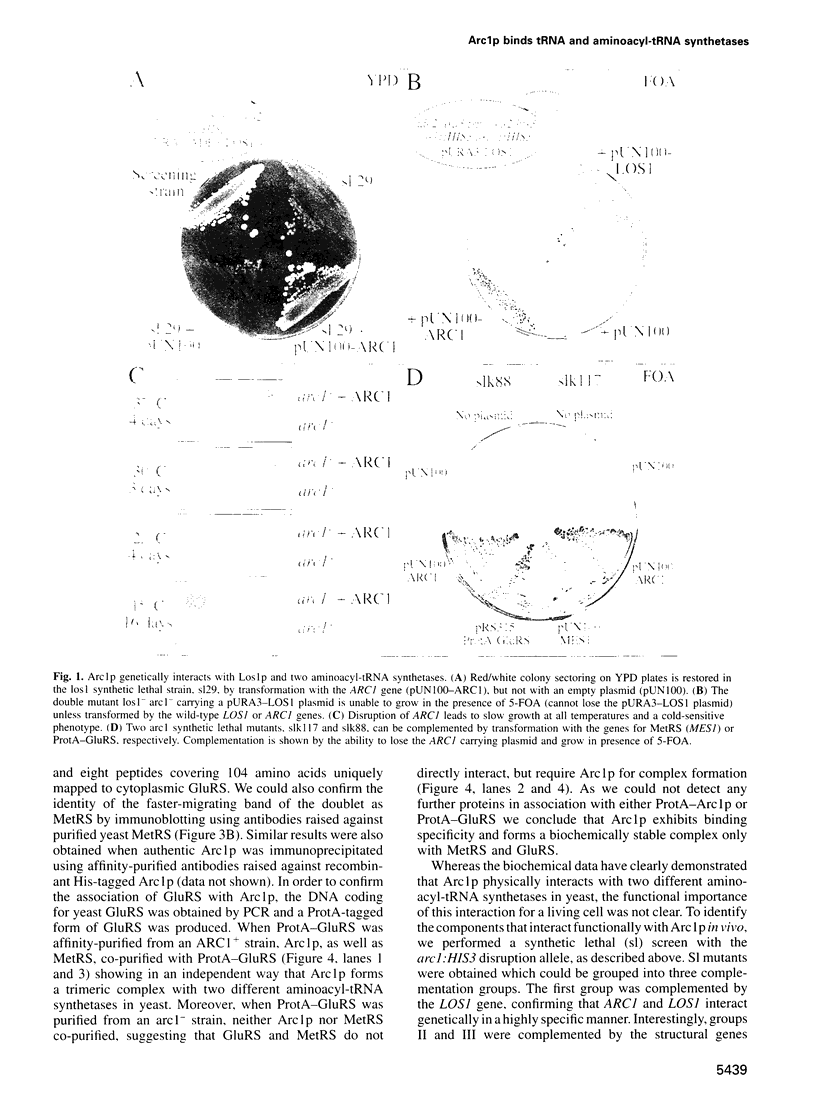
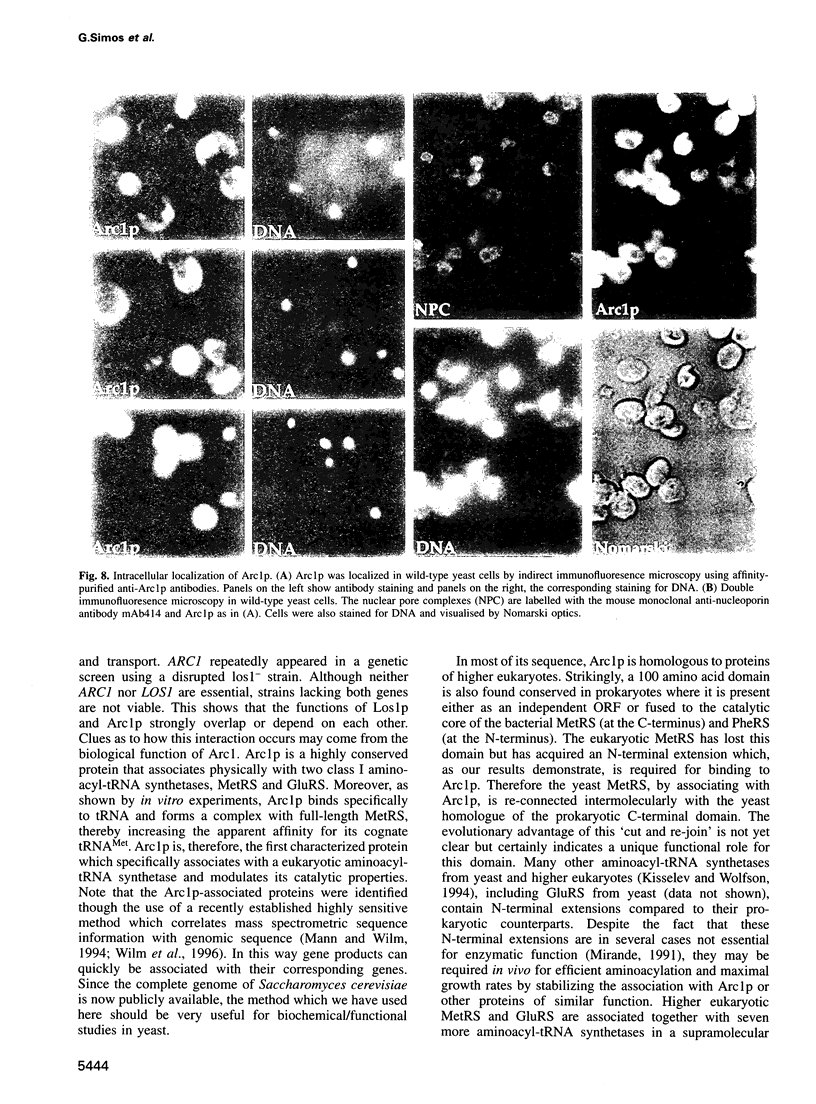
Arc1p was found in a screen for components that interact genetically with Los1p, a nuclear pore-associated yeast protein involved in tRNA biogenesis. Arc1p is associated with two proteins which were identified as methionyl-tRNA and glutamyl-tRNA synthetase (MetRS and GluRS) by a new mass spectrometry method. ARC1 gene disruption leads to slow growth and reduced MetRS activity, and synthetically lethal arc1- mutants are complemented by the genes for MetRS and GluRS. Recombinant Arc1p binds in vitro to purified monomeric yeast MetRS, but not to an N-terminal truncated form, and strongly increases its apparent affinity for tRNAMet. Furthermore, Arc1p, which is allelic to the quadruplex nucleic acid binding protein G4p1, exhibits specific binding to tRNA as determined by gel retardation and UV-cross-linking. Arc1p is, therefore, a yeast protein with dual specificity: it associates with tRNA and aminoacyl-tRNA synthetases. This functional interaction may be required for efficient aminoacylation in vivo.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Despons L., Senger B., Fasiolo F., Walter P. Binding of the yeast tRNA(Met) anticodon by the cognate methionyl-tRNA synthetase involves at least two independent peptide regions. J Mol Biol. 1992 Jun 5;225(3):897–907. doi: 10.1016/0022-2836(92)90409-d. [DOI] [PubMed] [Google Scholar]
- Elledge S. J., Davis R. W. A family of versatile centromeric vectors designed for use in the sectoring-shuffle mutagenesis assay in Saccharomyces cerevisiae. Gene. 1988 Oct 30;70(2):303–312. doi: 10.1016/0378-1119(88)90202-8. [DOI] [PubMed] [Google Scholar]
- Fasiolo F., Gibson B. W., Walter P., Chatton B., Biemann K., Boulanger Y. Cytoplasmic methionyl-tRNA synthetase from Bakers' yeast. A monomer with a post-translationally modified N terminus. J Biol Chem. 1985 Dec 15;260(29):15571–15576. [PubMed] [Google Scholar]
- Frantz J. D., Gilbert W. A novel yeast gene product, G4p1, with a specific affinity for quadruplex nucleic acids. J Biol Chem. 1995 Sep 1;270(35):20692–20697. doi: 10.1074/jbc.270.35.20692. [DOI] [PubMed] [Google Scholar]
- Grandi P., Doye V., Hurt E. C. Purification of NSP1 reveals complex formation with 'GLFG' nucleoporins and a novel nuclear pore protein NIC96. EMBO J. 1993 Aug;12(8):3061–3071. doi: 10.1002/j.1460-2075.1993.tb05975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris C. L., Kolanko C. J. Aminoacyl-tRNA synthetase complex in Saccharomyces cerevisiae. Biochem J. 1995 Jul 1;309(Pt 1):321–324. doi: 10.1042/bj3090321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haselbeck R. C., Greer C. L. Minimum intron requirements for tRNA splicing and nuclear transport in Xenopus oocytes. Biochemistry. 1993 Aug 24;32(33):8575–8581. doi: 10.1021/bi00084a026. [DOI] [PubMed] [Google Scholar]
- Hurt D. J., Wang S. S., Lin Y. H., Hopper A. K. Cloning and characterization of LOS1, a Saccharomyces cerevisiae gene that affects tRNA splicing. Mol Cell Biol. 1987 Mar;7(3):1208–1216. doi: 10.1128/mcb.7.3.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmolowski A., Boelens W. C., Izaurralde E., Mattaj I. W. Nuclear export of different classes of RNA is mediated by specific factors. J Cell Biol. 1994 Mar;124(5):627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao J., Houck K., Fan Y., Haehnel I., Libutti S. K., Kayton M. L., Grikscheit T., Chabot J., Nowygrod R., Greenberg S. Characterization of a novel tumor-derived cytokine. Endothelial-monocyte activating polypeptide II. J Biol Chem. 1994 Oct 7;269(40):25106–25119. [PubMed] [Google Scholar]
- Kerjan P., Cerini C., Sémériva M., Mirande M. The multienzyme complex containing nine aminoacyl-tRNA synthetases is ubiquitous from Drosophila to mammals. Biochim Biophys Acta. 1994 Apr 21;1199(3):293–297. doi: 10.1016/0304-4165(94)90009-4. [DOI] [PubMed] [Google Scholar]
- Kisselev L. L., Wolfson A. D. Aminoacyl-tRNA synthetases from higher eukaryotes. Prog Nucleic Acid Res Mol Biol. 1994;48:83–142. doi: 10.1016/s0079-6603(08)60854-5. [DOI] [PubMed] [Google Scholar]
- Kranz J. E., Holm C. Cloning by function: an alternative approach for identifying yeast homologs of genes from other organisms. Proc Natl Acad Sci U S A. 1990 Sep;87(17):6629–6633. doi: 10.1073/pnas.87.17.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M., Wilm M. Electrospray mass spectrometry for protein characterization. Trends Biochem Sci. 1995 Jun;20(6):219–224. doi: 10.1016/s0968-0004(00)89019-2. [DOI] [PubMed] [Google Scholar]
- Mann M., Wilm M. Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal Chem. 1994 Dec 15;66(24):4390–4399. doi: 10.1021/ac00096a002. [DOI] [PubMed] [Google Scholar]
- Mattaj I. W. RNA recognition: a family matter? Cell. 1993 Jun 4;73(5):837–840. doi: 10.1016/0092-8674(93)90265-r. [DOI] [PubMed] [Google Scholar]
- Meinnel T., Mechulam Y., Dardel F., Schmitter J. M., Hountondji C., Brunie S., Dessen P., Fayat G., Blanquet S. Methionyl-tRNA synthetase from E. coli--a review. Biochimie. 1990 Aug;72(8):625–632. doi: 10.1016/0300-9084(90)90126-2. [DOI] [PubMed] [Google Scholar]
- Mirande M. Aminoacyl-tRNA synthetase family from prokaryotes and eukaryotes: structural domains and their implications. Prog Nucleic Acid Res Mol Biol. 1991;40:95–142. doi: 10.1016/s0079-6603(08)60840-5. [DOI] [PubMed] [Google Scholar]
- Nagy E., Rigby W. F. Glyceraldehyde-3-phosphate dehydrogenase selectively binds AU-rich RNA in the NAD(+)-binding region (Rossmann fold). J Biol Chem. 1995 Feb 10;270(6):2755–2763. doi: 10.1074/jbc.270.6.2755. [DOI] [PubMed] [Google Scholar]
- Negrutskii B. S., Stapulionis R., Deutscher M. P. Supramolecular organization of the mammalian translation system. Proc Natl Acad Sci U S A. 1994 Feb 1;91(3):964–968. doi: 10.1073/pnas.91.3.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Schimmel P. Aminoacyl tRNA synthetases: general scheme of structure-function relationships in the polypeptides and recognition of transfer RNAs. Annu Rev Biochem. 1987;56:125–158. doi: 10.1146/annurev.bi.56.070187.001013. [DOI] [PubMed] [Google Scholar]
- Schimmel P., Ribas de Pouplana L. Transfer RNA: from minihelix to genetic code. Cell. 1995 Jun 30;81(7):983–986. doi: 10.1016/s0092-8674(05)80002-9. [DOI] [PubMed] [Google Scholar]
- Schlaich N. L., Hurt E. C. Analysis of nucleocytoplasmic transport and nuclear envelope structure in yeast disrupted for the gene encoding the nuclear pore protein Nup1p. Eur J Cell Biol. 1995 May;67(1):8–14. [PubMed] [Google Scholar]
- Senger B., Aphasizhev R., Walter P., Fasiolo F. The presence of a D-stem but not a T-stem is essential for triggering aminoacylation upon anticodon binding in yeast methionine tRNA. J Mol Biol. 1995 May 26;249(1):45–58. doi: 10.1006/jmbi.1995.0279. [DOI] [PubMed] [Google Scholar]
- Senger B., Despons L., Walter P., Fasiolo F. The anticodon triplet is not sufficient to confer methionine acceptance to a transfer RNA. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10768–10771. doi: 10.1073/pnas.89.22.10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K., Fabre E., Tekotte H., Hurt E. C., Tollervey D. Yeast nucleoporin mutants are defective in pre-tRNA splicing. Mol Cell Biol. 1996 Jan;16(1):294–301. doi: 10.1128/mcb.16.1.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W. C., Selvakumar D., Stanford D. R., Hopper A. K. The Saccharomyces cerevisiae LOS1 gene involved in pre-tRNA splicing encodes a nuclear protein that behaves as a component of the nuclear matrix. J Biol Chem. 1993 Sep 15;268(26):19436–19444. [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996 Mar 1;68(5):850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simos G., Tekotte H., Grosjean H., Segref A., Sharma K., Tollervey D., Hurt E. C. Nuclear pore proteins are involved in the biogenesis of functional tRNA. EMBO J. 1996 May 1;15(9):2270–2284. [PMC free article] [PubMed] [Google Scholar]
- Singh R., Green M. R. Sequence-specific binding of transfer RNA by glyceraldehyde-3-phosphate dehydrogenase. Science. 1993 Jan 15;259(5093):365–368. doi: 10.1126/science.8420004. [DOI] [PubMed] [Google Scholar]
- Stapulionis R., Deutscher M. P. A channeled tRNA cycle during mammalian protein synthesis. Proc Natl Acad Sci U S A. 1995 Aug 1;92(16):7158–7161. doi: 10.1073/pnas.92.16.7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szweykowska-Kulinska Z., Senger B., Keith G., Fasiolo F., Grosjean H. Intron-dependent formation of pseudouridines in the anticodon of Saccharomyces cerevisiae minor tRNA(Ile). EMBO J. 1994 Oct 3;13(19):4636–4644. doi: 10.1002/j.1460-2075.1994.tb06786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobian J. A., Drinkard L., Zasloff M. tRNA nuclear transport: defining the critical regions of human tRNAimet by point mutagenesis. Cell. 1985 Dec;43(2 Pt 1):415–422. doi: 10.1016/0092-8674(85)90171-0. [DOI] [PubMed] [Google Scholar]
- Walter P., Weygand-Durasevic I., Sanni A., Ebel J. P., Fasiolo F. Deletion analysis in the amino-terminal extension of methionyl-tRNA synthetase from Saccharomyces cerevisiae shows that a small region is important for the activity and stability of the enzyme. J Biol Chem. 1989 Oct 15;264(29):17126–17130. [PubMed] [Google Scholar]
- Wilhelm M. L., Reinbolt J., Gangloff J., Dirheimer G., Wilhelm F. X. Transfer RNA binding protein in the nucleus of Saccharomyces cerevisiae. FEBS Lett. 1994 Aug 1;349(2):260–264. doi: 10.1016/0014-5793(94)00683-0. [DOI] [PubMed] [Google Scholar]
- Wilm M., Mann M. Analytical properties of the nanoelectrospray ion source. Anal Chem. 1996 Jan 1;68(1):1–8. doi: 10.1021/ac9509519. [DOI] [PubMed] [Google Scholar]
- Wilm M., Shevchenko A., Houthaeve T., Breit S., Schweigerer L., Fotsis T., Mann M. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature. 1996 Feb 1;379(6564):466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- Wimmer C., Doye V., Grandi P., Nehrbass U., Hurt E. C. A new subclass of nucleoporins that functionally interact with nuclear pore protein NSP1. EMBO J. 1992 Dec;11(13):5051–5061. doi: 10.1002/j.1460-2075.1992.tb05612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Lockshin C., Herbert A., Winter E., Rich A. Zuotin, a putative Z-DNA binding protein in Saccharomyces cerevisiae. EMBO J. 1992 Oct;11(10):3787–3796. doi: 10.1002/j.1460-2075.1992.tb05464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]