Abstract
Vestibular schwannomas (VSs), the most common tumors of the cerebellopontine angle, arise from Schwann cells lining the vestibular nerve. Pharmacotherapies against VS are almost non‐existent. Although the therapeutic inhibition of inflammatory modulators has been established for other neoplasms, it has not been explored in VS. A bioinformatic network analysis of all genes reported to be differentially expressed in human VS revealed a pro‐inflammatory transcription factor nuclear factor‐kappa B (NF‐κB) as a central molecule in VS pathobiology. Assessed at the transcriptional and translational level, canonical NF‐κB complex was aberrantly activated in human VS and derived VS cultures in comparison to control nerves and Schwann cells, respectively. Cultured primary VS cells and VS‐derived human cell line HEI‐193 were treated with specific NF‐κB siRNAs, experimental NF‐κB inhibitor BAY11‐7082 (BAY11) and clinically relevant NF‐κB inhibitor curcumin. Healthy human control Schwann cells from the great auricular nerve were also treated with BAY11 and curcumin to assess toxicity. All three treatments significantly reduced proliferation in primary VS cultures and HEI‐193 cells, with siRNA, 5 μM BAY11 and 50 μM curcumin reducing average proliferation (±standard error of mean) to 62.33% ± 10.59%, 14.3 ± 9.7%, and 23.0 ± 20.9% of control primary VS cells, respectively. These treatments also induced substantial cell death. Curcumin, unlike BAY11, also affected primary Schwann cells. This work highlights NF‐κB as a key modulator in VS cell proliferation and survival and demonstrates therapeutic efficacy of directly targeting NF‐κB in VS.
Keywords: Vestibular schwannoma, Network analysis, NF-κB, TNF, BAY 11-7082, Curcumin
Highlights
Bioinformatic network analysis revealed NF‐κB as a central node in VS pathobiology.
Canonical NF‐κB complex is aberrantly active in human VS and derived VS cultures.
NF‐κB siRNA & inhibitor BAY 11‐7082 reduce proliferation and survival in VS cultures.
Clinically relevant drug curcumin reduces proliferation and survival in VS cultures.
Our work demonstrates therapeutic efficacy of directly targeting NF‐κB in VS.
Abbreviations
- BAY11
BAY 11-7082
- GAN
great auricular nerve
- IκBα
inhibitor of kappa B alpha
- Iκκ
inhibitor of kappa B alpha kinase
- NF-κB
nuclear factor-kappa B
- SC
Schwann cell
- TNF
tumor necrosis factor
- VS
vestibular schwannoma
1. Introduction
Vestibular schwannomas (VSs) are the fourth most common intracranial tumors (Mahaley et al., 1990). Although histologically non‐malignant, they can cause multiple cranial neuropathies and even death due to their location in the cerebellopontine angle and potential for brainstem compression. Currently, main treatment modalities for growing VSs are surgical resection and stereotactic radiotherapy. Although interest in pharmacotherapies against VS is increasing (Plotkin et al., 2012), none are FDA approved. This is partially because drugs such as bevacizumab, which shrink some VSs, have substantial side effects, including renal failure, which may outweigh potential benefits (Plotkin et al., 2012). Therefore, there is an unmet medical need to establish well‐tolerated pharmacotherapies to prevent VS growth. Although much is known about the different pathways implicated in VS pathobiology, the interconnectedness among these pathways has not been studied extensively.
To identify the major orchestrators of VS growth, we conducted the first comprehensive network analysis of the published genes aberrantly expressed in sporadic VS. Nuclear factor‐kappa B (NF‐κB), a transcription factor known for mediating the physiological inflammatory response and pathologic inflammation in several diseases, including neoplastic growth (Hoesel and Schmid, 2013), was identified as a central factor in a top‐ranking network. Although NF‐κB has been connected to other molecules in VS, NF‐κB activation and the accompanying inflammation have not been directly explored as therapeutic targets against sporadic VS. However, level of infiltration of CD163+ tumor‐associated macrophages, known to pathologically promote tumor growth and survival, correlates with human VS growth rate, motivating research to investigate the inflammatory pathways that may promote VS growth (de Vries et al., 2013).
NF‐κB can regulate the transcription of over 300 downstream genes, resulting in differential influences on cell growth, proliferation and survival depending on the stimulus (Gilmore, 2014). NF‐κB's therapeutic inhibition has been investigated in several cancers because of its role in pathological inflammation accompanying neoplastic growth (Hoesel and Schmid, 2013). NF‐κB is especially relevant for VS since merlin, the protein encoded by the NF2 gene, acts as a negative regulator of the NF‐κB pathway (Kim et al., 2002) and merlin is dysfunctional in majority of VSs (Lee et al., 2012, 2012). Additionally, Axl, a member of the TAM family of receptor tyrosine kinases, regulates overexpression of survivin and cyclin D1 through NF‐κB, leading to enhanced survival, cell‐matrix adhesion and proliferation of cultured VS cells (Ammoun et al., 2013). NF‐κB also regulates p75‐associated VS proliferation and apoptosis (Ahmad et al., 2014).
We investigated NF‐κB's aberrance in human VS and the therapeutic potential of NF‐κB inhibition. Our results suggest that the NF‐κB pathway is aberrantly activated in VS and VS‐derived cultures compared to healthy nerves and SCs, respectively. NF‐κB inhibition in primary VS cells and a VS‐derived human cell line using NF‐κB siRNA, an experimental NF‐κB inhibitor BAY 11‐7082 (BAY11) and a clinically relevant inhibitor curcumin decreased proliferation and survival of the tumor cells. Our work provides novel insight into NF‐κB's expression and role in VS pathobiology and demonstrates therapeutic efficacy of directly targeting NF‐κB in VS.
2. Materials and methods
2.1. Ingenuity Pathway Analysis
A literature search was performed with PubMed using MeSH terms neuroma, acoustic, proteins, genes, gene expression, gene expression regulation, gene expression profiling, micorarray analysis, DNA mutational analysis, immunohistochemistry, enzyme‐linked immunosorbent assay, tumor suppressor proteins, DNA and RNA. Only human studies with relevant controls and explicit description of statistical criteria were selected. Differentially expressed molecules were analyzed on April 14th 2011 using Ingenuity Pathway Analysis (IPA, Ingenuity Systems, Inc.) version 9.0, while setting a cutoff value to 2. Molecules reported to be up‐ or down‐regulated qualitatively were assigned a value 2 or −2, respectively. To avoid a bias toward molecules with extreme differential expression, the absolute maximal value for fold change was set to 100 for molecules with a greater change. The maximal number of molecules per network was 35. The most interconnected molecule in each network is known as the hub.
2.2. Specimen collection
Freshly harvested human specimens of sporadic VS and control great auricular nerve (GAN) were collected from indicated surgeries, placed in saline and transported to the laboratory on ice. The study protocols were approved by Human Studies Committee of Massachusetts General Hospital and Massachusetts Eye and Ear Infirmary, and conducted in accordance with the Helsinki Declaration.
2.3. Real time quantitative polymerase chain reaction
Expression of genes in the NF‐κB pathway was measured using real time quantitative PCR (qPCR). Human VS or GAN tissue was placed in RNA Later (Qiagen) temporarily. RNA was extracted using RNeasy Mini‐Kit (Qiagen) and reverse‐transcribed to cDNA with Taqman Reverse Transcription Reagent kit (Applied Biosystems), as previously described (Stankovic et al., 2009). qPCR was performed using Applied Biosystems 7700 Sequence Detection System with TaqMan Primers (Applied Biosystems) for NFΚB1 (encoding p50 subunit of the NF‐κB heterodimer, Hs01042010_m1), RELA (encoding p65 subunit of the NF‐κB heterodimer required for activation, Hs01042010_m1), TNF (encoding tumor necrosis factor, an inducer for NF‐κB, Hs01042010_m1), RANK (encoding receptor activator of nuclear factor‐kappa B, Hs00187192_m1), NFΚB2 (Hs01028901_g1), REL (Hs00968440_m1), and RELB (Hs00232399_m1) and for downstream genes with κB sites, namely CCND1 (encoding cyclin D1, Hs00765553_m1), BCL2 (encoding B‐cell lymphoma 2, Hs00608023_m1), CSF2 (encoding colony stimulating factor 2, Hs00929873_m1), and XIAP (encoding X‐linked inhibitor of apoptosis, Hs00745222_s1). The reference gene was ribosomal RNA 18S (Hs9999901_s1).
2.4. Protein extraction and quantification
Translation and activation of the NF‐κB pathway components were investigated through western blot analysis. Total protein was extracted on ice from freshly harvested specimens of VS and GAN in RIPA buffer supplemented with protease and phosphatase inhibitors (Roche Applied Sciences). The lysate was isolated by centrifugation and stored at −80 °C. Equal protein was loaded per lane, separated on a 4–20% Tris‐glycine gel (Invitrogen) and transferred onto a Polyvinylidene fluoride membrane (Millipore). The membrane was blocked and probed with Cell Signaling Technology antibodies against NF‐κB phosphorylated (P‐) p65 (#3033), NF‐κB p65 (#8242), inhibitor of kappa B alpha (IκBα, #11930) or NF‐κB p50 (Abcam, #ab7971), followed by secondary antibodies (Jackson‐Immuno Research). Membranes were visualized with ChemiDoc XRS+ (Bio‐Rad Laboratories). Band densities were quantified using ImageJ and normalized to GAPDH expression (Cell Signaling Technology, #5174).
2.5. Immunohistochemistry
Human VS and GAN specimens were fixed in 4% PFA, transferred to PBS, embedded in paraffin, sectioned, deparaffinized with xylene, washed in PBS, permeabilized with Triton‐X 100 (Integra) for 5 min, blocked in normal horse serum and incubated with primary antibodies against S100 (Dako, #Z0311) or p50 (Abcam, #ab7971) and corresponding fluorescent secondary antibodies (Jackson‐Immuno Research). Nuclei were labeled with Hoechst stain (Invitrogen). The tissue was visualized and imaged using Carl Zeiss 2000 upright microscope.
2.6. Primary human Schwann cell and vestibular schwannoma cell culture
Using sterile technique, freshly harvested VS or GAN tissue was rinsed in PBS, dissected in culture medium consisting of Dulbecco's Modified Eagle's medium with Ham's F12 mixture (DMEM/F12), 10% fetal bovine serum, 1% Penicillin/Streptomycin (Pen/Strep) and 1% GlutaMAX, dissociated in Hyaluronidase and Collagenase (all from Life Technologies) overnight and cultured for 2–4 weeks, as previously described (Dilwali et al., 2014). Human VS cell line HEI‐193, derived from a patient with neurofibromatosis type 2 (NF2), was obtained from Dr. Giovanni at the House Ear Institute (Hung et al., 2002, 2002).
2.7. Pharmacologic treatment of VS cultures with BAY 11‐7082, curcumin and siRNA
For siRNA treatment, cultured primary VS cells or HEI‐193 cells were placed in antibiotic‐ and serum‐free media overnight as instructed by manufacturer. The next day, the cells were incubated with siRNAs targeting NF‐κB genes RELA (#s11915) and NFΚB1 (#s9504), with control cells being treated with scrambled siRNA (#TR30015), for 5 days (all purchased from Life Technologies). The vehicle used for siRNA delivery was RNAiMax (#13778030, Life Technologies). Some cultures were incubated with a fluorescent red oligo (Life Technologies) with vehicle to assess transfection efficiency.
For pharmacologic treatment, cultured primary VS cells, primary SCs and HEI‐193 cells were treated for 48 h with NF‐κB inhibitors BAY 11‐7082 (BAY11, #sc‐200615) or curcumin (#sc‐200509A) (both purchased from Santa Cruz Biotechnology) in media fortified with antibiotics and serum. BAY11 or curcumin, diluted in 100% DMSO, were mixed to the accurate concentrations in media and applied to the cultures (with DMSO concentration in media being 1% maximum), alongside no treatment (NT) with DMSO alone.
2.8. Quantification of proliferation and apoptosis
After treatment, proliferation or apoptosis was assessed as previously detailed (Dilwali et al., 2014). Briefly, cell proliferation was quantified by adding 5‐Bromo‐2′‐Deoxyuridine (BrdU, Invitrogen) to the cultured cells 20 h prior to fixation. Primary antibodies against BrdU (AbD Serotec, #OBT0030G), S100 (Dako, #Z031129), or p50 (Abcam, #ab7971) were used. For assessing apoptosis, terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL, Roche Applied Sciences) was applied for 1 h at 37 °C and 0.5 h at RT on the shaker after fixation and permeabilization. Cells were counted by an investigator (S.D.) blinded to the treatment conditions. Cells were counted in ≥3 fields. Cell proliferation and apoptosis were reported as percent BrdU positive and TUNEL positive nuclei, respectively. As a validation for TUNEL staining, apoptosis was also assessed using immunocytochemistry by the expression of cleaved caspase‐3 in cells treated with siRNA or curcumin. Antibody against cleaved caspase‐3 (Cell Signaling Technology, #9661) was utilized. The inhibitors were compared to the control group by normalizing the percent change in proliferation or percent of apoptosis in comparison to the non‐treated cells.
2.9. Electrophoretic mobility shift assay (gel shift assay)
The gel shift assay was performed using the LightShift Chemiluminescent EMSA Kit (Thermo Scientific, #20148) according to the manufacturer's manual. The 6% DNA retardation gels and biotin labeled NF‐κB binding site oligos (sense: 5′‐AGTTGAGGGGACTTTCCCAGGC‐biotin‐3′ and anti‐sense: 5′‐TCAACTCCCCTGAAAGGGTCCG‐biotin‐3′), and non‐labeled oligos were purchased from Invitrogen. The nuclear extract of VS tumors and control GAN tissues were purified using the Nuclear Extraction Kit (Abcam, #ab113474).
2.10. Statistical analyses
Networks from IPA were statistically analyzed with the right‐tailed Fisher's exact test; p < 0.05 was considered significant. For qPCR, western blot and treatment of cultured cells, two‐tailed t‐test was used to assess significance with a p < 0.05 considered significant after a Benjamini‐Hochberg correction for multiple hypotheses.
3. Results
3.1. Network analysis reveals nuclear factor‐kappa B (NF‐κB) as a central modulator of VS growth
Of the 622 articles identified, 19 met our inclusion criteria (Aarhus et al., 2010; Archibald et al., 2010; Bian et al., 2005; Cayé‐Thomasen et al., 2010; Cioffi et al., 2010; Dayalan et al., 2006; Doherty et al., 2008; Kramer et al., 2010; Lassaletta et al., 2009; O'Reilly et al., 2004; Patel et al., 2008; Plotkin et al., 2009; Sawaya and Highsmith, 1988; Saydam et al., 2011; Seol et al., 2005; Stankovic et al., 2009; Szeremeta et al., 1995; Thomas et al., 2005; Welling et al., 2002), generating 221 molecules eligible for network analysis: 162 overexpressed and 59 underexpressed molecules in sporadic VS. IPA generated a total of 19 networks. Supplementary Table S1 shows the hubs of the top 14 networks. Here we focus on validation of the hub of the second most significant network (p = 10−33, Figure 1). We focus on NF‐κB because it is a key pro‐inflammatory transcription factor that could be an important therapeutic target in VS (Ammoun et al., 2013), and TNFα, an inducer of NF‐κB, was the hub of another top‐ranking network (Supplementary Table S1).
Figure 1.
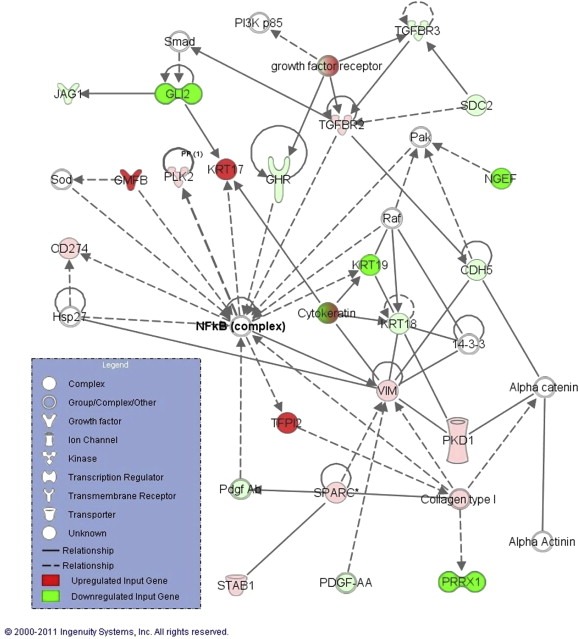
A highly significant network (p = 10−33) that connects molecules reported to be upregulated (green) or downregulated (red) in VS with other molecules from IPA (white). The hub of this network is nuclear factor‐kappa B (NF‐κB) complex. Solid lines represent direct and dashed lines represent indirect interactions.
3.2. Vestibular schwannomas have aberrant expression and activation of the canonical NF‐κB pathway
The canonical and non‐canonical NF‐κB pathways were investigated in VS compared to GAN using qPCR, western blot and immunohistochemistry. The qPCR data are expressed as the average with range of expression in parentheses and “n” indicating the number of different tumors or control nerves. In the canonical pathway, expression of genes NFΚB1 (encoding the p50 subunit) and RELA (encoding the p65 subunit) tended to be higher in VSs (n = 10) compared to GANs (n = 10), albeit not significantly, with p = 0.18 and 0.17, respectively (Figure 2A). Non‐canonical components REL, RELB and NFΚB2 exhibited different patterns of expression. REL was 3.1 (1.0–9.4) fold higher in VSs (n = 13) than in GANs (n = 10) (p = 0.01, Figure 2A). NFΚB2 had the same average expression in VSs as GANs (p = 0.22, Figure 2A). Interestingly, RELB was 0.4 (0.2–1.0) fold downregulated in VSs (n = 13) compared to GANs (range 0.5–2.1, n = 10) (p = 0.02, Figure 2A).
Figure 2.
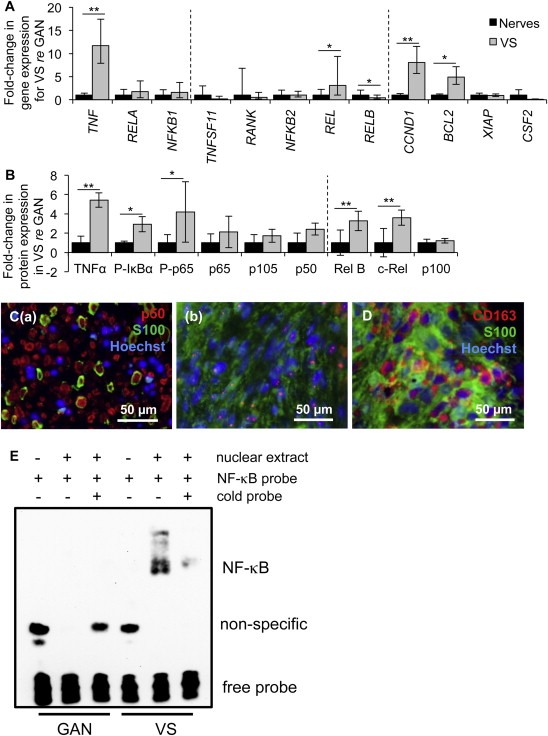
NF‐κB is aberrantly activated in VS. A. NF‐κB pathway expression in human VSs (n ≥ 9 tumors) versus GANs (n ≥ 8 nerves) as measured through qPCR. Dashed lines separate genes by groups, being genes associated with canonical NF‐κB pathway, non‐canonical NF‐κB pathway and downstream targets of NF‐κB. Error bars represent range. B. NF‐κB pathway expression in human VSs (n ≥ 4) versus GANs (n ≥ 4) as quantified through western blot analysis. P‐means phosphorylated protein. Dashed line separates canonical and non‐canonical proteins. Error bars represent SD. C. Representative images of p50 expression (red), as visualized through immunohistochemistry, in (a) VS and (b) GAN specimens. Schwann or schwannoma cells are labeled with S100 (green). D. Representative image of CD163 expression (red), schwannoma cells (S100, green). GAN and VS expression is shown in black and gray bars, respectively (A, B). *p < 0.05, **p < 0.01. Nuclei are labeled with Hoechst (blue) in (C, D). E. Gel shift results show interaction between nuclear extracts of GAN tissue (pooled from 6 different patients, lanes 2–3) or VS tissue (pooled from 4 different patients, lanes 5–6) with the NF‐κB binding site. The interaction could be disrupted in VS nuclear extracts by adding excessive unlabeled NF‐κB binding site (lane 6).
Exploring the downstream genes with κB binding sites by qPCR, two genes under canonical NF‐κB control were significantly upregulated in VSs (n = 15) relative to GANs (n = 15): pro‐proliferative CCND1 at 8.1 (5.7–11.5) fold (p = 0.0007) and anti‐apoptotic BCL2 at 4.9 (3.3–7.1) fold (p = 0.02, Figure 2A). The ranges in GAN were 0.7–1.4 and 0.8–1.3 for CCND1 and BCL2, respectively. Anti‐apoptotic XIAP was equally expressed in VSs (n = 12) and GANs (n = 7) (p = 0.18, Figure 2A). Pro‐proliferative CSF2 tended to be downregulated, albeit not significantly (p = 0.11, Figure 2A), in VSs (n = 9) compared to GANs (n = 7).
Upstream regulator of the canonical NF‐κB pathway, gene TNF encoding TNFα, was expressed at 11.7 (7.9–17.4) fold higher levels in VSs (n = 10) than in GANs (range 0.7–1.5, n = 10) (p = 0.003, Figure 2A). Upstream regulator of the non‐canonical NF‐κB pathway, RANKL gene TNFS11 tended to be downregulated in VSs (n = 10) than in GANs (n = 10), although not significantly (p = 0.20, Figure 2A).
Following qPCR, NF‐κB translation and activation were assessed. Western blot analysis revealed that NF‐κB canonical pathway was substantially activated in VSs compared to GANs. TNFα activates inhibitor of kappa B kinase (Iκκ), which phosphorylates inhibitor of kappa B alpha (IκBα), enabling the heterodimer of NF‐κB p65 and p50 to phosphorylate and relocate to the nucleus to promote transcription of genes important for survival and proliferation (Karin, 1999). Western blot data are summarized as average fold change ± standard deviation, with “n” indicating the number of different tumors or nerves. The representative western blot results are shown in Supplementary Figure S1 and the statistical results are shown in Figure 2B. The internal control protein, GAPDH, was not significantly different between VSs and GANs (p = 0.36). NF‐κB p65 (encoded by the RELA gene) and p105 (encoded by the NFΚB1 gene) had an insignificant trend of being more abundant in VSs (n = 7–10) than in GANs (n = 7–9), with p = 0.09 and = 0.14, respectively (Figure 2B). The phosphorylated form of p65 was 4.2 ± 3.1 fold more expressed in VSs (n = 9) compared to GANs (n = 8, p = 0.03, Figure 2B). p105's derived subunit p50 likewise showed tendency for higher expression, albeit non‐significant, in VSs (n = 15) compared to GANs (n = 11, p = 0.10, Figure 2B). NF‐κB's canonical inducer, TNFα, was 5.4 ± 0.7 fold more abundant in VS (n = 4) than in GAN (n = 4, p = 0.001, Figure 2B), demonstrating the same trend as seen through qPCR. The phosphorylated form of IκBα was also 2.8 ± 0.8 fold higher in VSs (n = 4) than in GANs (n = 4, p = 0.01, Figure 2B).
The expression of NF‐κB non‐canonical proteins c‐Rel (encoded by REL gene) and p100 (encoded by NFΚB2 gene) mirrored the corresponding mRNA expression: c‐Rel was 3.6 ± 0.8 fold more expressed in VSs (n = 7) than in GANs (n = 7, p = 0.003, Figure 2B). p100 was not different in VSs (n = 4) compared to GANs (n = 4, p = 0.42, Figure 2B). Interestingly, expression of Rel B protein (encoded by RELB) was significantly, 3.3 ± 1fold higher in VSs (n = 7) than in GANs (n = 7, p = 0.006), demonstrating the opposite trend from the corresponding mRNA. Taken together, these results demonstrate presence and basal activation of NF‐κB in GANs and VSs, and consistently higher activation of the canonical NF‐κB pathway in VSs.
Immunohistochemistry in 5 different VS and 4 different GAN samples verified that NF‐κB was active in VS as the p50 subunit localized to the nuclei in VS specimens (Figure 2C (b)), thus corroborating the western blot results demonstrating a higher level of phosphorylation, and hence activation of NF‐κB in VSs. GAN specimens showed minimal p50 nuclear localization although p50 was present in the cytoplasm (Figure 2C (a)). S100, a marker for SCs, highlights schwannoma cells in VS specimens and SCs encasing the nerve fibrils in GANs (Figure 2C (a)). Additionally, CD163‐positive tumor‐associated macrophages were present in the same VSs at substantially higher levels than in GANs (Figure 2D), indicating an aberrant inflammatory presence in VS as described previously (de Vries et al., 2013). Taken together, our results demonstrating activation of p50 and several other proteins associated with NF‐κB in VS expand on the previous finding of p65 activation in VS cells (Ammoun et al., 2013).
To further confirm the activation of the NF‐κB pathway in VS tissues, the gel shift assay was performed and the DNA binding activities in VS tissues and GAN tissues were compared. To provide enough nuclear extracts for the assay, VS tissues were pooled from 4 patients and GAN tissues were pooled from 6 patients. The binding of NF‐κB p65‐p50 heterodimer to its binding site was avid in the VS nuclear extract and not detectable in the GAN nuclear extract (Figure 2E). The interaction in the VS nuclear extract could be blocked by adding the excess non‐labeled oligos containing the NF‐κB binding site (Figure 2E, lane 6). These results indicate that the NF‐κB activity was specifically elevated in VS tissues.
3.3. Specific NF‐κB knockdown decreases proliferation and survival of VS cultured cells
The NF‐κB canonical pathway was also expressed and activated at significantly higher levels in primary VS cultures (n = 6 different tumors) compared to SC cultures (n = 6 different nerves) (Figure 3A). NF‐κB p65 and its phosphorylated form had 1.9 ± 0.4 fold (p = 0.01) and 2.8 ± 0.4 fold (p = 0.02) higher expression in VS cells compared to SCs, respectively. NF‐κB p105 and its derived subunit p50 were present in cultures, although not at significantly higher levels than in SCs (p = 1.0 and p = 0.06, respectively, Figure 3A).
Figure 3.
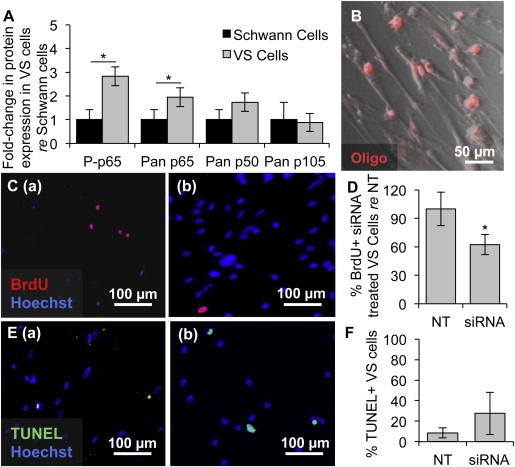
NF‐κB is aberrantly activated in primary VS cultures and its siRNA‐mediated knockdown decreases proliferation. A. NF‐κB expression in cultured human VSs (n ≥ 6 tumors) normalized to expression in SC cultures (n ≥ 6 nerves) as quantified through western blot analysis. P‐means phosphorylated protein. Error bars represent SD. B. Representative image of effective transfection of a fluorescently labeled oligonucleotide (oligo, red) in primary VS cells. C. Representative proliferation images are shown for (a) scrambled siRNA or (b) siRNA treated primary VS cells. BrdU in nuclei (red) marks proliferating cells. D. Quantification of proliferation changes after siRNA treatment in primary VS cells normalized to proliferation in control scrambled siRNA treated (NT) cells (n = 4 results were from 4 independent results from cultures of two different patients). E. Representative cell death images are shown for (a) scrambled siRNA and (b) siRNA treated primary VS cells. TUNEL (green) in nuclei marks dying cells. F. Quantification of cell death rate after siRNA treatment of primary VS cells as measured by TUNEL staining (n = 3 different cultures). Error bars represent SD for panels D and F. *p = 0.025, re = compared to. Nuclei are labeled with Hoechst (blue) in (C, E).
Applying siRNAs targeting RELA and NFKB1 concurrently decreased proliferation, as measured by nuclear BrdU staining, and cell survival, as measured by the TUNEL assay. Results are summarized as average ± standard error of mean (SEM), with “n” referring to the number of cultures from different specimens. Proliferation changes are normalized to each culture's proliferation rate. Transfection efficiency of approximately 94 ± 3% was achieved in primary VS cells (n = 3), as assessed by transfection of a fluorescent red‐labeled oligo (Figure 3B). The siRNA‐mediated knockdown of NFKB1 and RELA in VS cells was detected using western blot and the results are shown in Supplementary Figures S2A and S2B, respectively. Basal proliferation in VS cultures treated with scrambled siRNA was 6.5% ± 2.6% (n = 4, Figure 3C (a), D). Proliferation significantly decreased to 62.33% ± 10.59% after siRNA treatment (n = 4, p = 0.025, Figure 3C (b), D). Percentage of VS cells treated with scrambled siRNA only exhibiting TUNEL staining was 8.59% ± 4.92% (n = 3, Figure 3E (a), F). Apoptosis tended to increase, although insignificantly, to 27.54% ± 20.53% in VS cultures treated with NF‐κB siRNA (n = 3, p = 0.53, Figure 3E (b), F). Similar results were also observed using anti‐cleaved caspase‐3 immunocytochemistry. NF‐κB siRNA transfection in VS increased the percentage of cells that expressed cleaved caspase‐3 from 2.01% ± 1.24% to 7.44% ± 7.15%; however, the difference was not statistically significant (n = 3, p = 0.13, Fig S3A).
3.4. NF‐κB small‐molecule inhibitor BAY 11‐7082 decreases proliferation and survival selectively in primary VS and HEI‐193 cells
Primary VS cells, control SC cultures and the HEI‐193 cell line were treated with BAY11. BAY11 treatment significantly decreased the activity of the NF‐κB pathway as shown by the decrease of phosphorylated p65 in western blot (Fig. S2C, left). Results are reported using the same format and meaning of “n” and “p” as for siRNA application. Treatment with 1 and 5 μM BAY11 changed proliferation in VS cells to 54.7 ± 22.8% (n = 5, p = 0.15, Figure 4A (b), B) and 14.3 ± 9.7% (n = 4, p = 0.002, Figure 4A (c), B) of the non‐treated cells (NT, Figure 4A (a)), respectively. The apoptotic rate changed from 1.1 ± 0.27% (Figure 4C (a), D) in the NT VS cells to 36 ± 13% (n = 7, p = 0.06. Figure 4C (b), D) and 47 ± 12% (n = 8, p = 0.02, Figure 4C(c), D) in cells treated with 1 μM and 5 μM BAY11, respectively.
Figure 4.
NF‐κB inhibitor BAY11‐7082 leads to selective decrease in proliferation and survival of VS cells. A. Representative proliferation images for primary VS cultures treated with (a) no treatment (NT), (b) 1 μM and (c) 5 μM BAY11‐7082 (BAY11). BrdU in nuclei (red) marks proliferating cells, S100 (green) marks schwannoma cells. B. Quantification of proliferation changes after treatment with BAY11 at different concentrations (given in μM) in primary VS cells, primary SCs and HEI‐193 NF2 VS cell line, all normalized to proliferation in control NT cells (n ≥ 3). C. Representative cell death images are shown for primary VS cultures treated with (a) NT, (b) 1 μM and (c) 5 μM BAY11‐7082 (BAY11). TUNEL (green) in nuclei marks dying cells. D. Quantification of cell death rate after treatment with BAY11 at different concentrations (given in μM) in primary VS cells, primary SCs and HEI‐193 NF2 VS cell line (n ≥ 3). *p < 0.05, **p < 0.01, re = compared to. Error bars represent SEM. Nuclei are labeled with Hoechst (blue) in (A, C).
In the control SC cultures, normalized proliferation rates did not change significantly, being 100.0 ± 34.7%, 165.2 ± 125.1% (p = 0.70), 133.2 ± 101.1% (p = 0.69), 130.2 ± 65.6% (p = 0.78) for NT cells, 1 μM, 5 μM and 1 mM BAY11, respectively (n = 3, Figure 4B). SCs demonstrated higher apoptosis only at the highest, 1 mM BAY11 treatment. NT, 1 μM, 5 μM or 1 mM treated GAN cells exhibited apoptosis rates of 2.0 ± 0.9%, 1.0 ± 0.7% (p = 0.53), 0.7 ± 0.7% (p = 0.47) and 36.5 ± 26.5% (p = 0.43), respectively (n = 3, Figure 4D). These control experiments suggest that 5 μM BAY11 has the greatest therapeutic promise against VS without being toxic to SCs.
BAY11 treatment also decreased HEI‐193 cell survival in a dose‐dependent manner. HEI‐193 cells had very high basal proliferation rates of 84.9 ± 11.7% (n = 3). NT, 10 μM, 100 μM and 1 mM BAY11 treated HEI‐193 cells exhibited normalized proliferation rates of 100.0 ± 13.8%, 65.6 ± 24.6% (n = 5, p = 0.25), 9.1 ± 4.9% (n = 5, p = 0.006) and 4.3 ± 3.3% (n = 5, p = 0.003), respectively (Figure 4B). NT, 1, 5, 10, 100 μM and 1 mM BAY11 treated HEI‐193 cells exhibited apoptotic rates of 1.3 ± 0.8%, 1.3 ± 0.5% (n = 6, p = 0.22), 1.9 ± 1.2% (n = 6, p = 0.26), 1.1 ± 1.1% (n = 5, p = 0.63), 58.8 ± 21.5% (n = 5, p = 0.04) and 55.3 ± 16.9% (n = 5, p = 0.02), respectively (Figure 4D).
3.5. Clinically‐relevant NF‐κB inhibitor curcumin decreases proliferation and survival in cultured primary VS cells, NF2 VS cell line and primary SCs
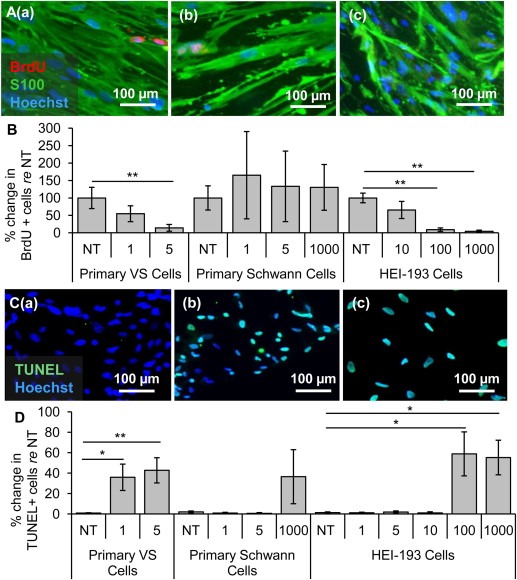
Curcumin, a natural, well‐tolerated NF‐κB inhibitor that is currently used in many clinical trials for various neurological, inflammatory and neoplastic diseases, ranging from Alzheimer's disease to colon cancer (Hatcher et al., 2008), was tested in VS cells. Treatment of curcumin significantly decreased the activity of the NF‐κB pathway as shown by the decrease of phosphorylated p65 in western blot (Fig. S2, right). Results are reported using the same format and meaning of “n” and “p” as for siRNA application. Proliferation decreased in a dose‐dependent manner in VS cultures, with VS cells receiving NT, 5, 20 and 50 μM curcumin (Figure 5A (a–d, respectively)) exhibiting normalized proliferation rates of 100.0% ± 30.5%, 141.8% ± 95.2% (p = 0.57), 23.0 ± 20.9% (p = 0.03) and 9.8 ± 5.3 (p = 0.0005) (n = 3, Figure 5B). Apoptosis also increased in a dose‐dependent manner, with VS cells receiving NT, 5, 20 or 50 μM curcumin (Figure 5C (a–d, respectively)) exhibiting apoptotic rates of 0.3 ± 0.1, 11.6 ± 6.3% (n = 8, p = 0.37), 1.8 ± 1.0% (n = 3, p = 0.37) and 73.3 ± 6.3% (n = 7, p = 0.0005) (Figure 5D). The effect of curcumin treatment on apoptosis in VS cells was also investigated by cleaved caspase‐3 immunocytochemistry. The results demonstrate a statistically significant apoptosis‐inducing effect of 50 μM curcumin in VS cells (n = 3, p = 0.02, Fig. S3B). Taken together, these results suggest that the decrease of NF‐κB by curcumin may be the mechanism leading to apoptosis in VS cells.
Figure 5.
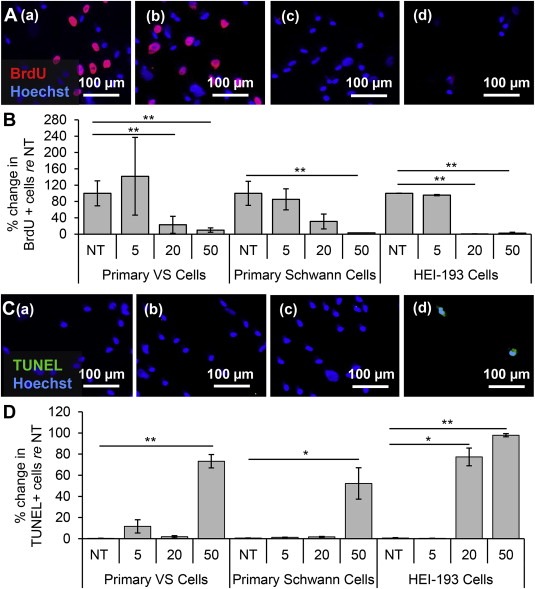
Clinically‐relevant NF‐κB inhibitor curcumin leads to selective decrease in proliferation and survival of VS cells. A. Representative proliferation images for primary VS cultures treated with (a) no treatment (NT), (b) 5, (c) 20, and (d) 50 μM curcumin. BrdU in nuclei (red) marks proliferating cells. B. Quantification of proliferation changes after treatment with curcumin at 5, 20, and 50 μM in primary VS cells, primary Schwann cells and HEI‐193 NF22 VS cell line, all normalized to proliferation in control NT cells (n ≥ 3); C. Representative cell death images are shown for primary VS cultures treated with (a) NT, (b) 5, (c) 20, and (d) 50 μM curcumin. TUNEL (green) marks dying cells. D. Quantification of cell death rate after treatment with curcumin at 5, 20, and 50 μM in primary VS cells, primary Schwann cells and HEI‐193 NF2 VS cell line (n ≥ 3). *p < 0.05, **p < 0.01, re = compared to. Error bars represent SEM. Nuclei are labeled with Hoechst (blue) in (A, C).
Surprisingly, in contrast to the seemingly well‐tolerated profile for curcumin in humans, curcumin decreased proliferation and increased apoptosis in control SC cultures at concentrations comparable to those efficacious in VS cultures. Proliferation tended to decrease in a dose‐dependent manner, with SCs receiving NT, 5, 20, 50 μM curcumin exhibiting normalized proliferation rates of 100.0% ± 29.6%, 85.3 ± 25.7% (n = 4, p = 0.33), 31.0 ± 18.3% (n = 4, p = 0.13) and 3.14% (n = 1, p = 0.04) (Figure 5B); the trend became significant only at the highest tested dose. Apoptosis had the same trend with the highest dose leading to a significant increase in cell death. NT, 5, 20 or 50 μM treated GAN cells exhibited apoptotic rates of 0.6 ± 0.2, 1.3 ± 0.3% (n = 4, p = 0.16), 1.7 ± 0.4% (n = 4, p = 0.31) and 52.2 ± 14.9% (n = 5, p = 0.03) (Figure 5D). Nonetheless, the doses up to 20 μM seemed selectively cytostatic against primary VS cells.
Intriguingly, the HEI‐193 cells were more susceptible to curcumin than primary VS cells or healthy SCs. Proliferation decreased drastically with dose increases: HEI‐193 cells receiving NT, 5, 20 or 50 μM curcumin exhibiting normalized proliferation rates of 100.0% ± 0.2%, 95.0 ± 1.6% (p = 0.12), 0.4 ± 0.4% (p = 0.0001) and 2.3 ± 2.3% (p = 0.001) (n = 3, Figure 5B). Apoptosis increased drastically at 20 μM, in contrast to the primary VS cells exhibiting apoptosis at 50 μM. HEI‐193 cells receiving NT, 5, 20 or 50 μM curcumin exhibited apoptotic rates of 0.5 ± 0.4, 0.3 ± 0.1% (n = 5, p = 0.32), 77.3 ± 8.4% (n = 3, p = 0.02) and 97.8 ± 1.5% (n = 4, p = 0.00003) (Figure 5D).
4. Discussion
Conducting the first comprehensive network analysis of molecules implicated in VS pathobiology, we identified and validated NF‐κB as a central regulator. We also found direct interactors of NF‐κB such as PDGF (Olson et al., 2007), which have been implicated in VS progression, to be the hubs of other significant networks (Supplementary Table S1). Although others have suggested that NF‐κB is activated in VS cells via upstream stimulation such as with p75 signaling (Ahmad et al., 2014), we find that NF‐κB is inherently highly active in human VS tissue and derived primary VS cells. The apparent disparity may be due to differences in detection methods or sample processing. As all prior experiments had been conducted on cultured cells and cell lines, we are the first to show that the aberrant NF‐κB activation occurs also in the freshly resected VS tissue and cannot be deemed an artifact of culturing.
Our analysis of expression signatures of the downstream NF‐κB genes in VS suggests a unique NF‐κB target gene program in VS, as may be expected in pathologic inflammation (Hoesel and Schmid, 2013). Since NF‐κB is highly expressed by immature SCs during development, progressively declining from pre‐myelinating SCs to near absence in mature myelinating SCs (Nickols et al., 2003), our findings are also consistent with VSs exhibiting a gene expression profile akin to immature SCs (Hung et al., 2002, 2002). Pre‐existing upregulation of NF‐κB in VSs, along with a few defining mutations in other genes, could enable neoplastic proliferation of non‐myelinating SCs.
Using a pre‐clinical model of primary human VS cells, we demonstrate potential therapeutic efficacy of directly targeting NF‐κB via experimental and clinical inhibitors. Our work with freshly harvested VS samples from different patients captures the variability of NF‐κB aberrance in different VSs. Our results suggest that therapeutic targeting of NF‐κB may be generally effective against VSs, not only against a small subset of VSs. By utilizing three different modalities to inhibit NF‐κB: (1) highly‐specific siRNAs against the NF‐κB p50 and p65, (2) a pharmacologic inhibitor BAY11 and (3) a clinically‐relevant, natural inhibitor curcumin, we affirm NF‐κB's role in VS proliferation and survival. We reinforce previous findings that siRNA mediated NF‐κB knockdown in primary VS cells reduces proliferation and survival (Ammoun et al., 2013), and expand on them by using more clinically relevant inhibitors.
A small molecule NF‐κB inhibitor BAY11 showed a high level of efficacy and specificity against VS cells. Although BAY11 has been characterized as an effective inhibitor of NF‐κB by inhibiting IκK activation, recently BAY11 has been recognized to target many other pro‐inflammatory molecules, including TNFα (Lee et al., 2012, 2012). Future work is needed to determine whether the therapeutic efficacy of BAY11 against VS cells is solely due to NF‐κB inhibition. As BAY11 was not cytotoxic in primary SCs and has been well tolerated in vivo in murine tumor xenograft studies (Dewan et al., 2003), future exploration of BAY11 against VS in animal models in vivo is warranted.
Curcumin, a clinically relevant NF‐κB inhibitor that has been tested in many clinical trials (Hatcher et al., 2008), inhibited proliferation and promoted apoptosis of both primary VS cells and HEI‐193 VS cells. Curcumin's greater effectiveness against HEI‐193 VS cells at a lower dose than required for primary VS cells suggests a higher therapeutic efficacy against NF2‐derived than sporadic VSs. The dosage curve of curcumin resembles a previously established dosage curve for HEI‐193 cells in a study that focused on another molecule through which curcumin may be acting: Hsp70 (Angelo et al., 2011). A follow‐up study by the same authors investigating curcumin's direct binding partners did not reveal the NF‐κB complex being a target in HEI‐193 cells (Angelo et al., 2013), although the authors had previously reported inhibition of phosphorylation of Protein Kinase B (AKT), an upstream regulator of NF‐κB activation (Angelo et al., 2011; Bai et al., 2009). This is in contrast to the large body of literature that shows curcumin's role as an NF‐κB inhibitor, via inhibition of TNFα‐induced IκB degradation (Hatcher et al., 2008), and as a general inhibitor of inflammation (Hatcher et al., 2008; Marin et al., 2007). It is important to acknowledge that although curcumin was found to be efficacious against colon cancer and Alzheimer's disease in animal and human studies, therapeutic and toxicity profiles of curcumin have not been comprehensively elucidated (Burgos‐Morón et al., 2010). Some clinical trials have noted nausea and diarrhea in patients taking curcumin (Burgos‐Morón et al., 2010). Since the levels of curcumin that led to primary VS and SC death were comparable, more research is required on curcumin's toxicity profile, best formulation and administration methods and its efficacy in brain diseases. Importantly, however, curcumin has recently been shown to have otoprotective effect against aminoglycoside toxicity and the associated hearing loss (HL) (Salehi et al., 2014). Since most VS patients present with HL, future studies are needed to explore whether curcumin could attenuate both VS growth and the associated HL simultaneously.
By establishing aberrance of several molecules involved in the NF‐κB pathway and efficacy of NF‐κB inhibition selectively in VS cells via several inhibitors, we demonstrate NF‐κB as a potential pharmacologic target against VS. However, possible future clinical targeting of NF‐κB has to be considered carefully given that NF‐κB is an important signaling node that most cells rely on.
Conflicts of interest
All authors have no conflicts of interest.
Funding
The funding sources were National Institute of Deafness and Other Communication Disorders (KO8DC010419‐D1 to K.M.S.; T32DC00038 to K.M.S., S.D.), Department of Defense (W81XWH‐14‐1‐0091 to K.M.S.) and the Bertarelli Foundation (K.M.S.). The funding sources had no direct involvement in this work.
Supporting information
The following are the supplementary data related to this article:
Figure S1 Representative western blot images showing the expression of NF‐κB pathway proteins in GAN and VS cells. GAPDH is used as housekeeping protein for all western blots. A. Expression of NF‐κB p105 and its derived subunit p50, Rel B, c‐Rel and p100 in control GAN tissue (lanes 1–4) versus VS tissue (lanes 5–8). Black arrow marks true band for p50. B. Expression of TNFα and P‐IκBα in control GAN tissue (lanes 1–4) versus VS tissue (lanes 5–8) (top panel); expression of NF‐κB p65 in control GAN tissue (lanes 1–3) versus VS tissue (lanes 4–6) (middle panel); phosphorylation of NF‐κB p65 in control GAN tissue (lanes 1–3) versus VS tissue (lanes 4–6) (bottom panel). C. Expression of NF‐κB p50, phosphorylated (P‐) p65 and total p65 in VS tumor cultures (lanes 1–3) versus GAN‐derived Schwann cell (SC) cultures (lanes 4–6). The statistical results are shown in Figure 2B.
Figure S2 Expression of NF‐κB pathway proteins after treatment with siRNA, BAY, and Curcumin. A. Expression of NF‐κB p50 in VS cells after transfection with siRNA targeting NFKB1. B. Expression of NF‐κB p65 in VS cells after transfected with siRNA targeting RELA. C. Phosphorylation of NF‐κB p65 in VS cells after BAY11 (lanes 1–2) and Curcumin treatment (lanes 3–4).
Figure S3 Cleaved caspase‐3 activation after treatment of primary VS cells with NFκB siRNA and Curcumin. A. Quantification of cleaved caspase‐3 positive cells after treatment with control siRNA (vehicle only or scrambled siRNA) or siRNA targeting NFKB1 and RELA (n = 3, p = 0.13). B. Quantification of cleaved caspase‐3 positive cells for NT cells and after treatment with 5, 20, and 50 μM Curcumin (n = 3). *p < 0.05.
Table S1 Characteristics of IPA networks connecting molecule implicated in VS pathobiology. The networks are ranked according to statistical significance, with network score being –log (p‐value).
Acknowledgments
We are grateful to Drs. Emerick, McKenna, Barker and Martuza for assisting in specimen collection.
Supplementary data 1.
1.1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2015.03.009.
Dilwali Sonam, Briët Martijn C., Kao Shyan-Yuan, Fujita Takeshi, Landegger Lukas D., Platt Michael P., Stankovic Konstantina M., (2015), Preclinical validation of anti-nuclear factor-kappa B therapy to inhibit human vestibular schwannoma growth, Molecular Oncology, 9, doi: 10.1016/j.molonc.2015.03.009.
Contributor Information
Sonam Dilwali, Email: sdilwaliutsw@gmail.com.
Martijn C. Briët, Email: martijnbriet@gmail.com
Shyan-Yuan Kao, Email: Shyan-Yuan_Kao@meei.harvard.edu.
Takeshi Fujita, Email: takeshi_fujita@meei.harvard.edu.
Lukas D. Landegger, Email: Lukas_landegger@meei.harvard.edu
Michael P. Platt, Email: Michael.Platt@bmc.org
Konstantina M. Stankovic, Email: konstantina_stankovic@meei.harvard.edu
References
- Aarhus, M. , Bruland, O. , Sætran, H.A. , Mork, S.J. , Lund-Johansen, M. , Knappskog, P.M. , 2010. Global gene expression profiling and tissue microarray reveal novel candidate genes and down-regulation of the tumor suppressor gene CAV1 in sporadic vestibular schwannomas. Neurosurgery 67, 998–1019. [DOI] [PubMed] [Google Scholar]
- Ahmad, I. , Yue, W.Y. , Fernando, A. , Clark, J.J. , Woodson, E.A. , Hansen, M.R. , 2014. p75NTR is highly expressed in vestibular schwannomas and promotes cell survival by activating nuclear transcription factor kappaB. Glia 62, 1699–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammoun, S. , Provenzano, L. , Zhou, L. , Barczyk, M. , Evans, K. , Hilton, D.A. , Hafizi, S. , Hanemann, C.O. , 2013. Axl/Gas6/NFκB signaling in schwannoma pathological proliferation, adhesion and survival. Oncogene 33, 336–346. [DOI] [PubMed] [Google Scholar]
- Angelo, L.S. , Maxwell, D.S. , Wu, J.Y. , Sun, D. , Hawke, D.H. , McCutcheon, I.E. , Slopis, J.M. , Peng, Z. , Bornmann, W.G. , Kurzrock, R. , 2013. Binding partners for curcumin in human schwannoma cells: biologic implications. Bioorg. Med. Chem. 21, 932–939. [DOI] [PubMed] [Google Scholar]
- Angelo, L.S. , Wu, J.Y. , Meng, F. , Sun, M. , Kopetz, S. , McCutcheon, I.E. , Slopis, J.M. , Kurzrock, R. , 2011. Combining curcumin (diferuloylmethane) and heat shock protein inhibition for neurofibromatosis 2 treatment: analysis of response and resistance pathways. Mol. Cancer Ther. 10, 2094–2103. [DOI] [PubMed] [Google Scholar]
- Archibald, D.J. , Neff, B.A. , Voss, S.G. , Splinter, P.L. , Driscoll, C.L.W. , Link, M.J. , Dong, H. , Kwon, E.D. , 2010. B7–H1 expression in vestibular schwannomas. Otol. Neurotol. 31, 991–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, D. , Ueno, L. , Vogt, P.K. , 2009. Akt-mediated regulation of NF-κB and the essentialness of NF-κB for the oncogenicity of PI3K and Akt. Int. J. Cancer 125, 2863–2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian, L. , Tirakotai, W. , Sun, Q. , Zhao, W. , Shen, J. , Luo, Q. , 2005. Molecular genetics alterations and tumor behavior of sporadic vestibular schwannoma from the People's Republic of China. J. Neurooncol. 73, 253–260. [DOI] [PubMed] [Google Scholar]
- Burgos-Morón, E. , Calderón-Montaño, J.M. , Salvador, J. , Robles, A. , López-Lázaro, M. , 2010. The dark side of curcumin. Int. J. Cancer 126, 1771–1775. [DOI] [PubMed] [Google Scholar]
- Cayé-Thomasen, P. , Borup, R. , Stangerup, S. , Thomsen, J. , Nielsen, F.C. , 2010. Deregulated genes in sporadic vestibular schwannomas. Otol. Neurotol. 31, (2) 256–266. [DOI] [PubMed] [Google Scholar]
- Cioffi, J.A. , Yue, W.Y. , Mendolia-Loffredo, S. , Hansen, K.R. , Wackym, P.A. , Hansen, M.R. , 2010. MicroRNA-21 overexpression contributes to vestibular schwannoma cell proliferation and survival. Otol. Neurotol. 31, 1455–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries, M. , Briaire-de Bruijn, I. , Malessy, M.J.A. , de Bruïne, Sica, F.T. , van, d.M. , Hogendoorn, P.C.W. , 2013. Tumor-associated macrophages are related to volumetric growth of vestibular schwannomas. Otol. Neurotol. 34, 347–352. [DOI] [PubMed] [Google Scholar]
- Dayalan, A.H.P.P. , Jothi, M. , Keshava, R. , Thomas, R. , Gope, M.L. , Doddaballapur, S.K. , Gope, R. , 2006. Age dependent phosphorylation and deregulation of p53 in human vestibular schwannomas. Mol. Carcinog. 45, (1) 38–46. [DOI] [PubMed] [Google Scholar]
- Dewan, M.Z. , Terashima, K. , Taruishi, M. , 2003. Rapid tumor formation of human T-cell leukemia virus type 1-infected cell lines in novel NOD-SCID/γcnull mice: suppression by an inhibitor against NF-kB. J. Virol. 77, 5286–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilwali, S. , Patel, P.B. , Roberts, D.S. , Basinsky, G.M. , Harris, G.J. , Emerick, K. , Stankovic, K.M. , 2014. Primary culture of human Schwann and schwannoma cells: improved and simplified protocol. Hear. Res. 315, 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty, J.K. , Ongkeko, W. , Crawley, B. , Andalibi, A. , Ryan, A.F. , 2008. ErbB and Nrg: potential molecular targets for vestibular schwannoma pharmacotherapy. Otol. Neurotol. 29, 50–57. [DOI] [PubMed] [Google Scholar]
- Gilmore, T. , 2014. NF-kB Transcription Factors http://www.bu.edu/nf-kb/ [Google Scholar]
- Hatcher, H. , Planalp, R. , Cho, J. , Torti, F.M. , Torti, S.V. , 2008. Curcumin: from ancient medicine to current clinical trials. Cell Mol. Life Sci. 65, 1631–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoesel, B. , Schmid, J. , 2013. The complexity of NF-kappaB signaling in inflammation and cancer. Mol. Cancer 12, 86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung, G. , Li, X. , Faudoa, R. , Xeu, Z. , Kluwe, L. , Rhim, J.S. , Slattery, W. , Lim, D. , 2002. Establishment and characterization of a schwannoma cell line from a patient with neurofibromatosis 2. Int. J. Oncol. 20, 475–482. [PubMed] [Google Scholar]
- Hung, G. , Colton, J. , Fisher, L. , Oppenheimer, M. , Faudoa, R. , Slattery, W. , Linthicum, F. , 2002. Immunohistochemistry study of human vestibular nerve schwannoma differentiation. Glia 38, 363–370. [DOI] [PubMed] [Google Scholar]
- Karin, M. , 1999. How NF-B is activated: the role of the IB kinase (IKK) complex. Oncogene 18, 6867–6874. [DOI] [PubMed] [Google Scholar]
- Kim, J.Y. , Kim, H. , Jeun, S. , Rha, S.J. , Kim, Y.H. , Ko, Y.J. , Won, J. , Lee, K. , Rha, H.K. , Wang, Y. , 2002. Inhibition of NF-kappaB activation by merlin. Biochem. Biophys. Res. Commun. 296, 1295–1302. [DOI] [PubMed] [Google Scholar]
- Kramer, F. , Stöver, T. , Warnecke, A. , Diensthuber, M. , Lenarz, T. , Wissel, K. , 2010. BDNF mRNA expression is significantly upregulated in vestibular schwannomas and correlates with proliferative activity. J. Neurooncol. 98, (1) 31–39. [DOI] [PubMed] [Google Scholar]
- Lassaletta, L. , Martinez-Glez, V. , Torres-Martín, M. , Rey, J.A. , Gavilán, J. , 2009. cDNA microarray expression profile in vestibular schwannoma: correlation with clinical and radiological features. Cancer Genet. Cytogenet. 194, 125–127. [DOI] [PubMed] [Google Scholar]
- Lee, J. , Rhee, M.H. , Kim, E. , Cho, J.Y. , 2012. BAY 11-7082 is a broad-spectrum inhibitor with anti-inflammatory activity against multiple targets. Mediators Inflamm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.D. , Kwon, T.J. , Kim, U. , Lee, W. , 2012. Genetic and epigenetic alterations of the NF2 gene in sporadic vestibular schwannomas. PLoS One 7, e30418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahaley, M.S.J. , Mettlin, C. , Natarajan, N. , Laws, E.R.J. , Peace, B.B. , 1990. Analysis of patterns of care of brain tumor patients in the United States: a study of the Brain Tumor Section of the AANS and the CNS and the Commission on Cancer of the ACS. Clin. Neurosurg. 36, 347–355. [PubMed] [Google Scholar]
- Marin, Y.E. , Wall, B.A. , Wang, S. , 2007. Curcumin downregulates the constitutive activity of NF-κB and induces apoptosis in novel mouse melanoma cells. Melanoma Res. 17, 274–283. [DOI] [PubMed] [Google Scholar]
- Nickols, J.C. , Valentine, W. , Kanwal, S. , Carter, B.D. , 2003. Activation of the transcription factor NF-kappaB in Schwann cells is required for peripheral myelin formation. Nat. Neurosci. 6, 161–167. [DOI] [PubMed] [Google Scholar]
- Olson, C.M. , Hedrick, M.N. , Izadi, H. , Bates, T.C. , Olivera, E.R. , Anguita, J. , 2007. p38 mitogen-activated protein kinase controls NF-κB transcriptional activation and tumor necrosis factor alpha production through RelA phosphorylation mediated by mitogen- and stress-activated protein kinase 1 in response to Borrelia burgdorferi Antigens. Infect. Immun. 75, 270–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly, B.F. , Kishore, A. , Crowther, J.A. , Smith, C. , 2004. Correlation of growth factor receptor expression with clinical growth in vestibular schwannomas. Otol. Neurotol. 25, 791–796. [DOI] [PubMed] [Google Scholar]
- Patel, A.K. , Alexander, T.H. , Andalibi, A. , Ryan, A.F. , Doherty, J.K. , 2008. Vestibular schwannoma quantitative polymerase chain reaction expression of estrogen and progesterone receptors. Laryngoscope 118, 1458–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin, S.R. , Merker, V.L. , Halpin, C. , Jennings, D. , McKenna, M.J. , Harris, G.J. , Barker, F.G. , 2012. Bevacizumab for progressive vestibular schwannoma in neurofibromatosis type 2: a retrospective review of 31 patients. Otol. Neurotol. 33, 1046–1052. [DOI] [PubMed] [Google Scholar]
- Plotkin, S.R. , Stemmer-Rachamimov, A. , Barker, F.G. , Halpin, C. , Padera, T.P. , Tyrrell, A. , Sorensen, A.G. , Jain, R.K. , di Tomaso, E. , 2009. Hearing improvement after bevacizumab in patients with neurofibromatosis type 2. N. Engl. J. Med. 361, 358–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi, P. , Akinpelu, O.V. , Waissbluth, S. , Peleva, E. , Meehan, B. , Rak, J. , Daniel, S.J. , 2014. Attenuation of cisplatin ototoxicity by otoprotective effects of nanoencapsulated curcumin and dexamethasone in a guinea pig model. Otol. Neurotol. 35, 1131–1139. [DOI] [PubMed] [Google Scholar]
- Sawaya, R. , Highsmith, R. , 1988. Plasminogen activator activity and molecular weight patterns in human brain tumors. J. Neurosurg. 68, 73–79. [DOI] [PubMed] [Google Scholar]
- Saydam, O. , Senol, O. , Würdinger, T , Mizrak, A. , Ozdener, G.B. , Stemmer-Rachamimov, A.O. , Yi, M. , Stephens, R.M. , Krichevsky, A.M. , Saydam, N. , Brenner, G.J. , Breakefield, X.O. , 2011. miRNA-7 attenuation in Schwannoma tumors stimulates growth by upregulating three oncogenic signaling pathways. Cancer Res. 71, 852–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol, H.J. , Jung, H.W. , Park, S.H. , Hwang, S.K. , Kim, D.G. , Paek, S.H. , Chung, Y.S. , Sub Lee, C. , 2005. Aggressive vestibular schwannomas showing postoperative rapid growth – their association with decreased p27 expression. J. Neurooncol. 75, 203–207. [DOI] [PubMed] [Google Scholar]
- Stankovic, K.M. , Mrugala, M.M. , Martuza, R.L. , Silver, M. , Betensky, R.A. , Nadol, J.B. , Stemmer-Rachamimov, A. , 2009. Genetic determinants of hearing loss associated with vestibular schwannomas. Otol. Neurotol. 30, 661–667. [DOI] [PubMed] [Google Scholar]
- Szeremeta, W. , Monsell, E.M. , Rock, J.P. , Caccamo, D.V. , 1995. Proliferation indicies of vestibular schwannomas by Ki-67 and proliferating cell nuclear antigen. Otol. Neurotol. 16, 616–619. [PubMed] [Google Scholar]
- Thomas, R. , Prabhu, P.D.A. , Mathivanan, J. , Rohini, Sivakumar, D. , Jayakumar, P.N. , Devi, B.I. , Satish, S. , Sastry, K.V.R. , Gope, R. , 2005. Altered structure and expression of RB1 gene and increased phosphorylation of pRb in human vestibular schwannomas. Mol. Cell. Biochem. 271, 113–121. [DOI] [PubMed] [Google Scholar]
- Welling, D.B. , Lasak, J.M. , Akhmametyeva, E. , Ghaheri, B. , Chang, L. , 2002. cDNA microarray analysis of vestibular schwannomas. Otol. Neurotol. 23, 736–748. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the supplementary data related to this article:
Figure S1 Representative western blot images showing the expression of NF‐κB pathway proteins in GAN and VS cells. GAPDH is used as housekeeping protein for all western blots. A. Expression of NF‐κB p105 and its derived subunit p50, Rel B, c‐Rel and p100 in control GAN tissue (lanes 1–4) versus VS tissue (lanes 5–8). Black arrow marks true band for p50. B. Expression of TNFα and P‐IκBα in control GAN tissue (lanes 1–4) versus VS tissue (lanes 5–8) (top panel); expression of NF‐κB p65 in control GAN tissue (lanes 1–3) versus VS tissue (lanes 4–6) (middle panel); phosphorylation of NF‐κB p65 in control GAN tissue (lanes 1–3) versus VS tissue (lanes 4–6) (bottom panel). C. Expression of NF‐κB p50, phosphorylated (P‐) p65 and total p65 in VS tumor cultures (lanes 1–3) versus GAN‐derived Schwann cell (SC) cultures (lanes 4–6). The statistical results are shown in Figure 2B.
Figure S2 Expression of NF‐κB pathway proteins after treatment with siRNA, BAY, and Curcumin. A. Expression of NF‐κB p50 in VS cells after transfection with siRNA targeting NFKB1. B. Expression of NF‐κB p65 in VS cells after transfected with siRNA targeting RELA. C. Phosphorylation of NF‐κB p65 in VS cells after BAY11 (lanes 1–2) and Curcumin treatment (lanes 3–4).
Figure S3 Cleaved caspase‐3 activation after treatment of primary VS cells with NFκB siRNA and Curcumin. A. Quantification of cleaved caspase‐3 positive cells after treatment with control siRNA (vehicle only or scrambled siRNA) or siRNA targeting NFKB1 and RELA (n = 3, p = 0.13). B. Quantification of cleaved caspase‐3 positive cells for NT cells and after treatment with 5, 20, and 50 μM Curcumin (n = 3). *p < 0.05.
Table S1 Characteristics of IPA networks connecting molecule implicated in VS pathobiology. The networks are ranked according to statistical significance, with network score being –log (p‐value).


